Abstract
Secretory phospholipases A2 (sPLA2s) are potent components of mammalian innate-immunity antibacterial mechanisms. sPLA2 enzymes attack bacteria by hydrolyzing bacterial membrane phospholipids, causing membrane disorganization and cell lysis. However, most Gram-negative bacteria are naturally resistant to sPLA2. Here we report a novel resistance mechanism to mammalian sPLA2 in Escherichia coli, mediated by a phospholipid repair system consisting of the lysophospholipid transporter LplT and the acyltransferase Aas in the cytoplasmic membrane. Mutation of the lplT or aas gene abolished bacterial lysophospholipid acylation activity and drastically increased bacterial susceptibility to the combined actions of inflammatory fluid components and sPLA2, resulting in bulk phospholipid degradation and loss of colony-forming ability. sPLA2-mediated hydrolysis of the three major bacterial phospholipids exhibited distinctive kinetics and deacylation of cardiolipin to its monoacyl-derivative closely paralleled bacterial death. Characterization of the membrane envelope in lplT- or aas-knockout mutant bacteria revealed reduced membrane packing and disruption of lipid asymmetry with more phosphatidylethanolamine present in the outer leaflet of the outer membrane. Moreover, modest accumulation of lysophospholipids in these mutant bacteria destabilized the inner membrane and rendered outer membrane–depleted spheroplasts much more sensitive to sPLA2. These findings indicated that LplT/Aas inactivation perturbs both the outer and inner membranes by bypassing bacterial membrane maintenance mechanisms to trigger specific interfacial activation of sPLA2. We conclude that the LplT/Aas system is important for maintaining the integrity of the membrane envelope in Gram-negative bacteria. Our insights may help inform new therapeutic strategies to enhance host sPLA2 antimicrobial activity.
Keywords: membrane lipid, phospholipase A, bacteria, phospholipid turnover, membrane structure, membrane biogenesis, bacterial resistance, lysophospholipid, membrane envelope, outer membrane asymmetry, phospholipase A2
Introduction
The ability of humans and other mammalian species to combat a wide array of potentially invasive bacterial species depends, in part, on diverse cellular and humoral antibacterial innate immune systems (1, 2). Among the latter, secretory phospholipases A2 (sPLA2s)2 can act directly against many Gram-positive bacteria and against both Gram-positive and Gram-negative bacteria when present in concert with other cellular and humoral host defense systems (3–8, 10). sPLA2 catalyze the breakdown of membrane phospholipid (PL) by hydrolyzing the acyl ester bond at the sn-2 position, generating free fatty acid and detergent-like lysophospholipid (LPL) (11). sPLA2 are present in various tissues, tears, and inflammatory fluids, including at high levels in blood plasma of patients with acute bacterial infections. Among the family of 10 different sPLA2 in humans, the group IIA isoform (sPLA2-IIA) is considered the most potent antibacterial sPLA2 (3, 10, 12–15). However, under at least certain circumstances (e.g. combined action with membrane attack complex (MAC)), other sPLA2 including the “pancreatic” sPLA2-IB can also act on Gram-negative bacteria and contribute to host antibacterial action (6, 13, 16, 17).
sPLA2s act most efficiently by binding to PL-rich interfaces from which individual PL molecules can diffuse into the active site pocket of the bound enzyme and be degraded, and the products can be replaced by another PL substrate molecule from this interface (namely interfacial activation mechanism) (14, 18). sPLA2s, especially group IIA, can penetrate through the cell wall of Gram-positive bacteria to directly bind to the membrane, resulting in the high susceptibility of many Gram-positive species (3). In contrast, Gram-negative bacteria exhibit much greater intrinsic resistance to sPLA2, which has been attributed to the unique structure of their additional outer envelope layer, the outer membrane (OM) (3, 19). The OM of Gram-negative bacteria is arranged with an asymmetric lipid distribution: phospholipids (mainly phosphatidylethanolamine (PE)) comprise the inner leaflet of the membrane, whereas the outer leaflet contains the complex glycolipid lipopolysaccharide (LPS). The restriction of OM PLs to the inner leaflet of the OM precludes, under normal conditions, access of an extracellular sPLA2 to the bacterial PL. The neutrophil-derived bactericidal/permeability-increasing protein (BPI) and MAC facilitate sPLA2 attack by disrupting the LPS-rich outer leaflet of the OM (1, 6). However, even under these conditions, the bactericidal potency of sPLA2 toward many Gram-negative bacteria (e.g. Escherichia coli, Salmonella typhimurium, Pseudomonas aeruginosa, and Neisseria meningitidis) is still limited, requiring doses of sPLA2 often beyond their physiological concentrations (1, 6, 13, 20, 21). Thus, this intrinsic resistance may help Gram-negative pathogens survive host antibacterial mechanisms and also limits potential therapeutic application of sPLA2 toward Gram-negative bacteria-mediated infections.
Of note, Gram-negative bacteria uniquely contain a PL repair system comprised of two inner membrane (IM)-associated proteins: LPL transporter (LplT) and acyltransferase-acyl carrier protein synthetase (Aas) (22) (see Fig. 1a). LplT promotes energy-independent transbilayer migration of each of the major LPL metabolites in Gram-negative bacteria (i.e. lyso-phosphatidylethanolamine (LPE), lyso-phosphatidylglycerol (LPG), and lyso-cardiolipin) from the periplasmic to the cytoplasmic leaflet of the IM (23). Aas is a peripheral membrane protein on the cytoplasmic side of the IM (24), positioned to convert transported LPLs to their respective parent diacyl-PL species (PE or PG) or a triacyl form of cardiolipin (CL) (23). The potentially membrane-disruptive properties of LPLs raised the possibility that the LplT/Aas protein system plays a housekeeping role to minimize accumulation of LPLs generated de novo as by-products of normal bacterial envelope modification processes, as in the biogenesis of OM lipoproteins (25, 26). Deletion of lplT or aas gene, however, resulted in only a modest elevation of LPL in E. coli and no change in bacterial growth or viability (22, 23, 26). In contrast, OM-depleted spheroplasts generated from E. coli cells that lacked LplT or Aas function exhibited increased sensitivity to venom-derived sPLA2 (23), suggesting that LplT/Aas provide a “self-defense” mechanism to counteract the membrane-disruptive effects of LPLs produced under host-stressed conditions that include activation of sPLA2 action.
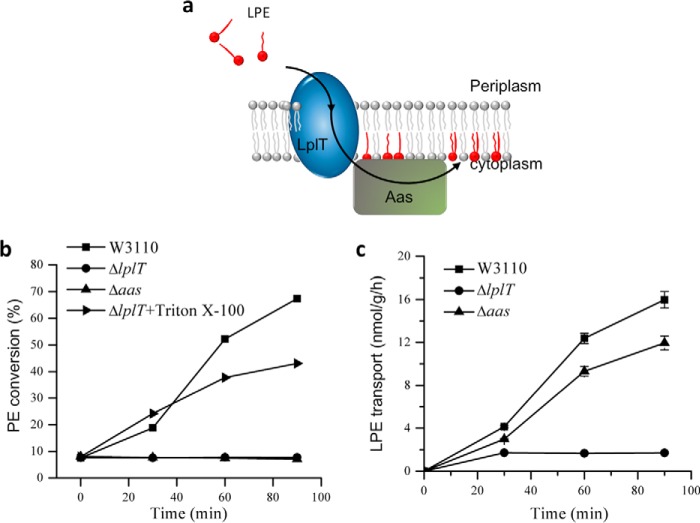
Figure 1.
LplT/Aas-mediated LPL acylation in E. coli. a, cartoon model showing lysophospholipid transporter LplT importing LPE from the periplasm across the cytoplasmic membrane for acylation by acyltransferase Aas to generate PE. b, LPE acylation assay of E. coli W3110 WT, ΔlplT, and Δaas strains. The activities were calculated based on conversion (%) of [32P]LPE to [32P]PE shown on TLC images in Fig. S2. c, LPE transport assays using spheroplasts generated from E. coli W3110 WT, ΔlplT, and Δaas cells. The transport activity of LplT in each sample was calculated based on the acquired radioactivity in the spheroplasts.
To test this hypothesis, we used E. coli as a model to explore the role of LplT/Aas in the resistance of intact Gram-negative bacteria to mammalian sPLA2. Our findings demonstrate a remarkable role for cytoplasmic membrane LplT/Aas in protecting E. coli intact bacteria from the attack of sPLA2. Unexpectedly, our data indicated that the resistance mediated by LplT/Aas is not attributed to any direct repairing of sPLA2-generated LPLs. Instead, LplT and Aas cooperatively function to maintain the ability of both the OM and IM to act as structural barriers to initial interfacial activation of sPLA2. The conservation of the lplT/aas loci in many Gram-negative bacteria, including those pathogenic species listed above, raises the possibility of a conserved mechanism of resistance in Gram-negative microorganisms. This study provides the first evidence of a physiological role of LplT/Aas in bacterial defense and suggests new strategies for rendering Gram-negative bacterial pathogens more sensitive to host antimicrobial activities.
Results
Coupling of LplT and Aas in phospholipid repair
In prokaryotes, functionally related genes are often co-transcribed in one bicistronic operon. The genes aas and lplT are adjacently located in their own operon in the genomes of E. coli and many other Gram-negative bacterial species (22). Our previous spheroplast study was carried out using the E. coli BL21(DE3) strain (23). To avoid any genetic variation, our current study was performed in the background of a standard E. coli W3110 strain. Two single gene knockout strains, ΔlplT and Δaas, were generated using the λ-Red recombination approach (Fig. S1a and Table S1). Deletion of these genes did not affect bacterial growth in LB broth (Fig. S1b). Their loss of LPL acylation (ΔlplT and Δaas) activity or LPL transport (ΔlplT) was confirmed using the spheroplast-based [32P] LPE acylation and transport assays (Fig. 1, b and c, and Fig. S2a). The functional coupling of LplT and Aas was further demonstrated by treating ΔlplT spheroplasts with 1% Triton X-100 detergent (Fig. 1b and Fig. S2b). These assays are consistent with our previous observation (23) that LplT and Aas operate as a coupled functional unit that provides a unique cytoplasmic membrane PL repairing mechanism in E. coli.
Deletion of the lplT or aas genes increases sensitivity of E. coli to sPLA2-dependent killing
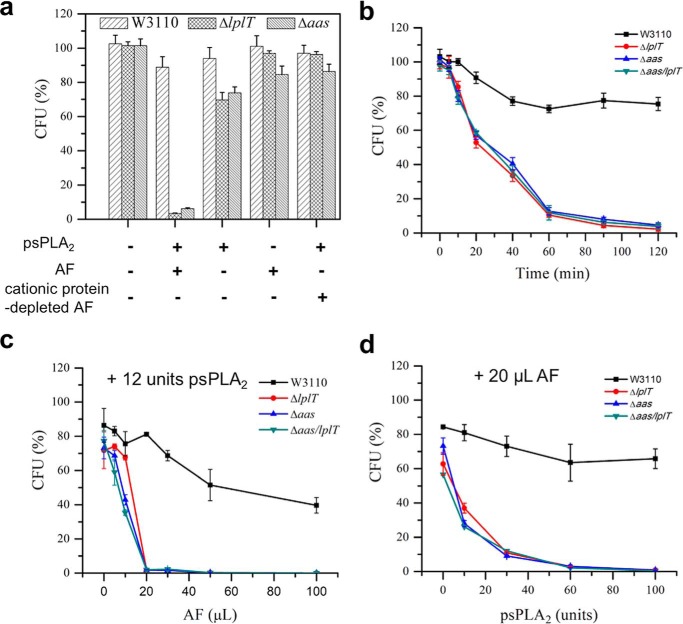
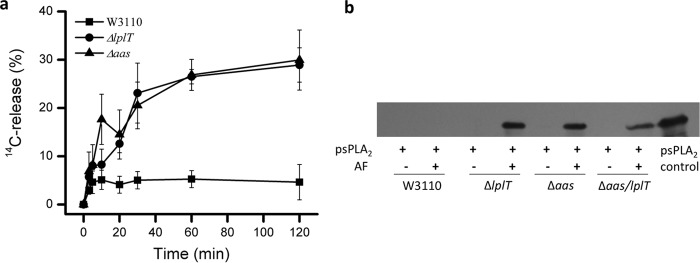
E. coli, including rough strains (producing truncated LPS), are intrinsically resistant to sPLA2, including all 10 human isoforms tested at μm concentration (6, 13, 15). Exposure of E. coli to host non-enzymatic antibacterial proteins producing alterations of the OM (e.g. BPI, MAC) increases bacterial sensitivity to mammalian sPLA2, including sPLA2-IB (“pancreatic”). To test the possible role of LplT and/or Aas on the sensitivity of E. coli to sPLA2, we incubated E. coli WT, ΔlplT, and Δaas strains with purified porcine pancreatic sPLA2 (referred to as psPLA2) and/or a previously characterized rabbit inflammatory (ascitic) fluid (AF) that contains BPI, several other rabbit granulocyte-derived cationic antimicrobial peptides and proteins, and each of the components of the complement system needed to form MAC (6, 21, 27, 28). Under the conditions tested, WT E. coli was resistant to AF and/or psPLA2, as judged by colony-forming units (CFUs) (Fig. 2a). In sharp contrast, both ΔlplT and Δaas strains exhibited much greater sensitivity to combined treatment with AF + psPLA2 (>95% of the mutant cells killed within 1 h) (Fig. 2a). The greater sensitivity of the mutant strains was also manifest kinetically, with nearly all of the mutant bacteria killed within 2 h, with a half-time of ∼30 min (Fig. 2b). Maximal killing of the mutant strains required the combined and dose-dependent presence of both AF and psPLA2 (Fig. 2, a, c, and d) and the presence in AF of cationic antimicrobial proteins (e.g. BPI) (Fig. 2a). Virtually identical results were obtained using three different AF (Fig. S3), including one collected from a complement (C)6-deficient rabbit that is unable to form the membrane attack complex of complement (C5b-8)9n (6). We found that both ΔlplT and Δaas cells in lag, logarithmic growth or stationary phase were highly prone to treatment with AF + psPLA2 and their sensitivities were indistinguishable, whereas the resistance of WT was apparent at each phase of growth (Fig. S4). We also tested a E. coli double knockout strain Δaas/lplT in which the entire aas/lplT locus was deleted in the genome (Fig. S1). The increased sensitivity of the ΔlplT, Δaas, and Δaas/lplT strains was nearly identical in all tested conditions (Fig. 2, b–d). These data strongly suggest that the susceptibility of E. coli to psPLA2 in the presence of AF can be significantly enhanced by disrupting either the lplT or aas gene and thus imply an essential role of LplT/Aas in protecting the bacterium from sPLA2 attack.
Figure 2.
Deletion of lplT or aas gene increases susceptibility of E. coli to psPLA2. a, viability tests of E. coli W3110 WT, ΔlplT, and Δaas strains treated with 12 units of psPLA2, 20 μl of AF, or 20 μl of cationic protein-depleted AF for 90 min. b, time-course viability assays of E. coli W3110 WT, ΔlplT, Δaas, and Δaas/lplT strains (with 12 units psPLA2, with 20 μl of AF). c and d, titration experiment of E. coli viability in the presence of 12 units of psPLA2 and indicated volume of AF (c) and in the presence of 20 μl of AF and psPLA2 (d) as indicated. The reaction time was 90 min. All data are expressed as CFUs (%) compared with untreated WT control at zero time.
LPL acylation confers bacterial resistance to sPLA2
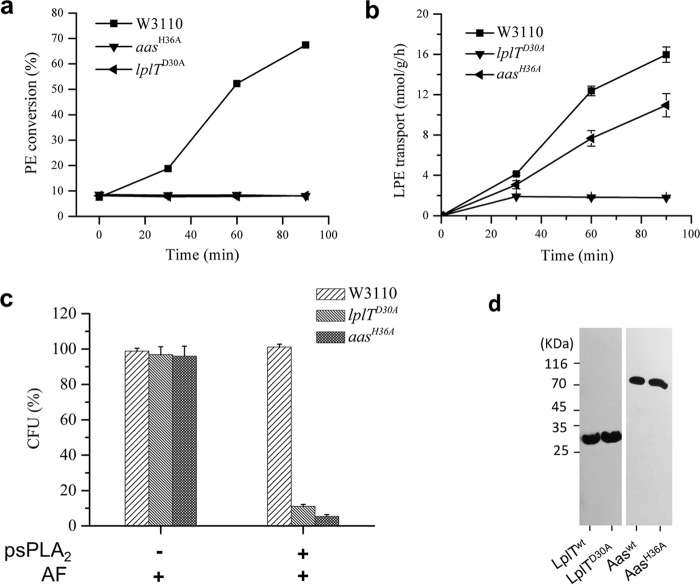
We next asked whether the increased susceptibility of ΔlplT and Δaas strains is caused by their loss of LPL acylation activity or by the gene deletions per se, given the fact that the membrane proteins LplT and Aas are also structural components of E. coli membrane envelope. To address this question, we generated two chromosomal single residue knockin mutant strains: lplTD30A and aasH36A. Asp30 is a conserved residue localized on the first transmembrane domain of LplT and was predicted to be important for substrate binding (25). By substituting Asp30 with an alanine, the lplTD30A strain completely abrogated its LPE transport activity (Fig. 3a). The bifunctional Aas protein harbors two catalytic domains: an N-terminal acyltransferase PlsC domain and a C-terminal acyl-ACP synthetase ACS domain. His36 is an essential catalytic residue in the PlsC domain (29). Spheroplasts generated from lplTD30A or aasH36A cells lack the ability to acylate LPE (Fig. 3b and Fig. S2). We next examined these two knockin strains for their sensitivity to psPLA2. The results showed that both lplTD30A and aasH36A strains exhibited high sensitivity to psPLA2, and the extent of their susceptibility was nearly identical to that observed in the knockout strains (Fig. 3c). Note that expression of both LplTD30A and AasH36A proteins was similar to that of the respective WT proteins (Fig. 3d). Taken together, these results strongly support the notion that the LPL acylation function of LplT/Aas confers increased bacterial resistance to psPLA2-dependent killing.
Figure 3.
E. coli resistance is determined by the function of LplT and Aas. a, LPE acylation assay of E. coli W3110, lplTD30A, and aasH36A strains. The activities were calculated based on conversion (%) of LPE to PE shown on TLC images in Fig. S2. b, LPE transport assays using spheroplasts generated from E. coli W3110 WT, lplTD30A, and aasH36A cells. The transport activity of LplT in each sample was calculated based on the acquired radioactivity in the spheroplasts. c, viability tests of E. coli W3110 WT, lplTD30A, and aasH36A strains (+ 12 units of psPLA2, ± 20 μl AF) for 90 min. The data are expressed as CFU (%) compared with untreated WT control. d, Western blotting analysis using anti-His antibody to examine LplTWT, LplTD30A, AasWT, or AasH36A protein expressed in E. coli. Cell lysates containing the same amount of total protein were loaded for each sample.
PL degradation is facilitated by inactivation of LplT/Aas
The increased susceptibility of ΔlplT and Δaas strains may reflect increased PL hydrolysis mediated by psPLA2. Under normal growth conditions, three major E. coli PLs (PE, PG, and CL) are present in the IM, whereas PE is relatively enriched in the OM (30). To better characterize the action of psPLA2, WT and mutant E. coli were metabolically prelabeled with [32Pi] and then incubated for 2 h with AF ± psPLA2. At the times indicated, the lipids were extracted and resolved by TLC. The identity of each PL/LPL species was unambiguously determined by comparing its migration rate with that of PL/LPL standards on TLC (Fig. S5). Metabolic radiolabeling of the WT and mutant bacteria were generally similar, with PE representing 60–65 mol %, PG representing 20 mol %, CL representing 7–8 mol %, and other components representing in toto <10 mol % of the total labeled PL pools (zero time in Fig. 4a and Table S2).
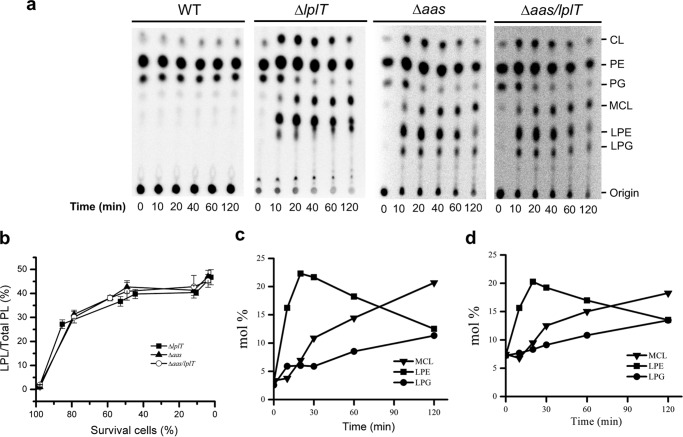
Figure 4.
Inactivation of LplT/Aas facilitates psPLA2-mediated PL degradation in E. coli. a, TLC images of PL extracted from 32P-labeled E. coli cells of W3110 E. coli WT, ΔlplT, Δaas, and Δaas/lplT strains. The cells were treated with 12 units of psPLA2 and 20 μl of AF for indicated times. b, kinetics of PL degradation in ΔlplT, Δaas, and Δaas/lplT cells. PL degradation is expressed as mol % of net LPL (LPE + LPG + MCL) compared with total PL in each sample. Degradation (mol %) of each individual PL of ΔlplT (c) and Δaas (d) was obtained from panel a and Table S2.
In the presence of AF + psPLA2, no breakdown of PL nor accumulation of LPL was found in the WT parent strain (Fig. 4a). In contrast, massive PL degradation occurred in both Δaas and ΔlplT strains under the same treatment conditions (not in the presence of AF only; Fig. S6), reflected by accumulation of multiple LPLs (Fig. 4a). Overall, the kinetics of degradation of the various PL species were very similar in the Δaas, Δlpl, and Δaas/lplT strains. The levels of LPL in the two mutant strains rose to nearly 45% of the total radiolabeled PL pool when nearly all bacteria were killed (Fig. 4b and Table S2). However, the catabolic kinetics of individual PL was remarkably different (Fig. 4, c and d). LPE was initially the most prominent LPL breakdown product, representing 17 mol % of the total PL pool after just 10 min incubation. Most (>80%) of the treated mutant bacteria were viable at this time (Fig. 2b), consistent with psPLA2 action initially limited to the OM. In contrast, accumulation of lyso-derivatives of PG and CL progressed more slowly and continuously throughout the 2-h incubation period, equaling (LPG) or exceeding (mono-acyl CL (MCL)) net accumulation of LPE at 2 h (Fig. 4, c and d). Indeed, an acute induction of CL was apparent in both mutant strains when the mutant bacteria were treated with both AF + psPLA2 (not AF alone; Fig. S6), resulting in a nearly 2-fold increase in CL levels within the first 10 min of incubation (Fig. 4a). After 2 h of incubation, the level of MCL exceeded by nearly three times the amount of CL initially present in the bacteria before treatment with AF + psPLA2. sPLA2-catalyzed deacylation of CL is stepwise in vitro (Fig. S5) (23). However, only the mono-acyl form was detected in AF + psPLA2-treated E. coli (ΔlplT and Δaas) (Fig. 4a). In contrast to the kinetics of LPE accumulation, the rate and extent of CL hydrolysis much more closely paralleled loss of bacterial viability (Fig. S7, a and b), consistent with psPLA2 action in the IM. These results demonstrate that inactivation of LplT/Aas facilitates the PL hydrolytic activity of sPLA2 on the bacterial envelope.
LplT/Aas prevent the binding of psPLA2 to the E. coli outer membrane
The lack of LPL accumulation in WT E. coli treated with AF + psPLA2 (Fig. 4a) could reflect either extremely efficient reacylation of psPLA2-triggered LPL or an unexpected effect of LplT/Aas on the resistance of E. coli to psPLA2 attack. To distinguish these two possibilities, the same reaction conditions were examined with [14C]-oleic acid–labeled cells and 1.5% albumin to trap the released free fatty acids (and LPL) in the extracellular medium and thus preclude LPL reacylation. Consistent with the TLC assays, both ΔlplT and Δaas strains showed progressive PL degradation, with nearly 30% of radiolabeled bacterial fatty acids recovered in the extracellular medium after 2 h of incubation, whereas there was little detectable fatty acid released from the WT bacteria (Fig. 5a). These findings thus indicate an unexpected effect of LplT/Aas on the resistance of E. coli to psPLA2 attack.
Figure 5.
LplT/Aas prevent AF-dependent binding of psPLA2 to E. coli. a, free fatty acid release assay of E. coli W3110 WT, ΔlplT, and Δaas cells. ∼106 E. coli cells labeled with [14C]oleic acid were incubated with 20 μl of AF and/or 12 units of psPLA2. The radioactivity in the supernatant fraction was determined by scintillation counting. PL degradation is expressed as the percentage of total 14C radioactivity in the reaction that is recovered as the products of hydrolysis. b, Western blotting analysis of psPLA2 pulldown assay of W3110 WT, ΔlplT, Δaas, and Δaas/lplT cells in the presence of psPLA2 and ± AF. psPLA2 bound with the cells was detected using anti-psPLA2 antibody.
To further test this possibility, we also examined psPLA2 binding to the WT and mutant bacteria, because sPLA2 binding to target E. coli is an important determinant of its action (20, 32). As shown in Fig. 5b, psPLA2 binding was detected to the mutant but not the WT bacteria expressing functional LplT and Aas and only when added together with AF. Thus, these results suggest that, surprisingly, the function of LplT and Aas maintains the resistance of E. coli to psPLA2 by occluding psPLA2 from the bacterial outer envelope.
Effects of LplT/Aas inactivation on the physical and compositional properties of the outer membrane
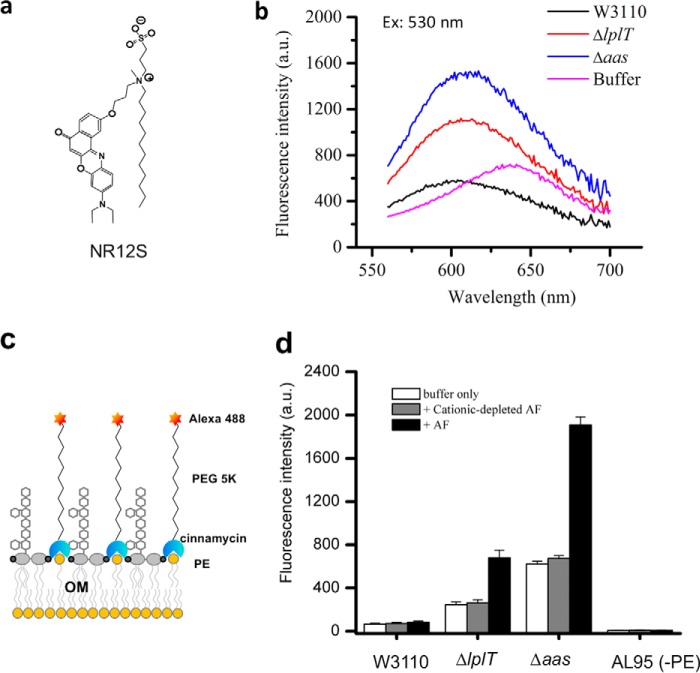
Enhanced psPLA2 binding and catalysis toward the mutant strains suggested that the natural resistance of the OM to sPLA2 depended on the functional integrity of LplT/Aas. If so, we predicted that the properties of the outer leaflet of the OM would be modified in the absence of functional LplT or Aas in a way conducive to sPLA2 action. We tested this hypothesis by comparing the interaction of the WT and mutant strains with two different probes of membrane outer leaflets: (i) Nile Red-based NR12S whose amphiphilic anchor (Fig. 6a) precludes transbilayer migration of the dye (33) and (ii) PE-specific probe Ro 09-0198 (cinnamycin)–PEG5000–Alexa 488 (34) that is also restricted to the surface of the outer leaflet by virtue of the linkage of PEG5000 (Fig. 6c). Incubation of NR12S with WT, ΔlplT, and Δaas E. coli resulted in both an increase in the maximum fluorescence of the dye in comparison to its properties in aqueous buffer and a blue shift of its emission spectrum (Fig. 6b), indicative of insertion of the dye into the (outer) membrane in each of these strains. However, in both of the mutant strains, the fluorescence intensity of the dye was substantially higher than in WT bacteria, and the wavelength of their fluorescence maximum exhibited a slight red shift (Fig. 6b). These findings are consistent with looser (less ordered) intermolecular packing within the outer leaflet of the two mutant strains.
Figure 6.
Alterations of the outer membrane in ΔlplT and Δaas strains. a, chemical structure of amphiphilic fluorescent probe NR12S. b, emission spectra of fluorescent probe NR12S in W3110 WT, ΔlplT, and Δaas cells after excitation at 530 nm. c, assay of bacterial OM asymmetry by determining PE at the bacterial surface. d, changes in fluorescent intensity following the addition of the Ro 09-0198–PEG5000–Alexa 488 conjugate into W3110 WT and mutant cells are shown.
One possible explanation for the different interactions of NR12S with the mutant bacteria is a role for LplT/Aas in maintaining the extreme lipid asymmetry of the OM of WT E. coli, restricting phospholipids (e.g. PE) to the inner leaflet and enriching the more ordered LPS in the outer leaflet. Binding of fluorescent Ro 09-0198–PEG5000–Alexa 488 was significantly greater in the mutant versus the WT strains (Fig. 6d), consistent with an increased amount of accessible PE on the surface of E. coli when the bacteria lack functional LplT or Aas. In contrast, no bacterial-bound fluorescence was detected when the probe was incubated with a PE-deficient E. coli strain AL95 (35), confirming that bacterial binding of Ro 09-0198–PEG5000–Alexa 488 was PE-dependent. Preincubation of the mutant, but not WT bacteria with a low dose of AF further increased binding of the fluorescent Ro 09-0198 probe (Fig. 6d), whereas the cationic protein-depleted AF had no effect. These results strongly suggest that inactivation of LplT/Aas resulted in a less extreme lipid asymmetry of the OM, which was further amplified by superimposed OM-perturbing effects of the cationic components in AF (e.g. BPI).
Absence of LplT/Aas function does not promote activation of bacterial OM PagP or PldA
Two mechanisms known to contribute to the extreme lipid asymmetry of the OM are mediated by phospholipase A (PldA) and lipid A palmitoyltransferase (PagP), which are also potentially linked to the function of LplT/Aas. Both enzymes normally remain inactive in bacteria but can be activated to generate LPL when PL is present in the outer leaflet of the OM (36, 37). There was no significant increase in LPL or free fatty acid in the ΔlplT/Δaas strains, even in the presence of AF (without psPLA2) (Table S2 and Fig. S8), indicating no activation of PldA under these conditions. PagP catalyzes palmitoylation of lipid A by using externalized PL as acyl donor to convert PE + hexa-acylated lipid A (LPS) to hepta-acylated LPS and LPL. However, lipid A released from the extracted LPS of both WT and mutant strains and analyzed by mass spectrometry revealed only hexa-acylated lipid A (Fig. S9), indicating that OM PagP is not activated in the absence of LplT/Aas.
LplT/Aas protect bacterial IM from sPLA2 attack
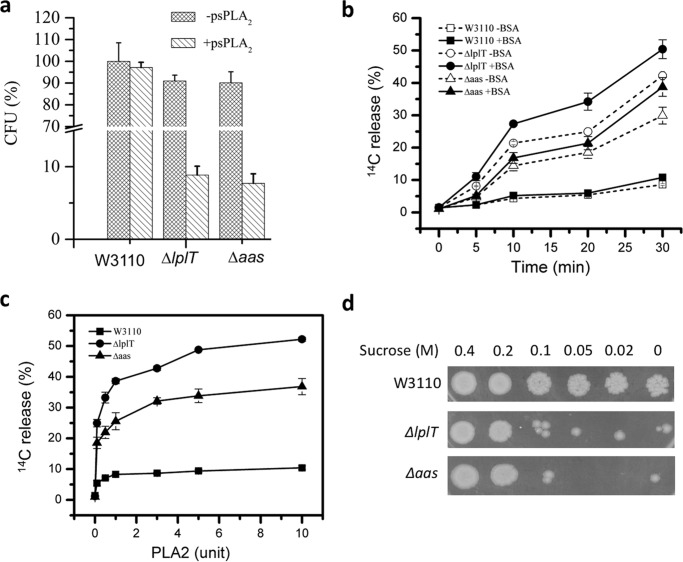
The extent of PL degradation by the combined action of AF + psPLA2 in the mutant strains suggests strongly that PL breakdown and accumulation of LPL under bactericidal conditions extend to the IM where essential energy-dependent and -generating machinery reside. We recently showed that OM- and cell wall-depleted spheroplasts generated from WT E. coli BL21(DE3) cells were resistant to added venom sPLA2, whereas the corresponding spheroplasts from mutant (ΔlplT or Δaas) bacteria were sensitive (23). These observations were confirmed and extended using spheroplasts generated from the WT and mutant W3110 strains used in this study. In contrast to the requirements for action on intact bacteria, psPLA2 added alone (i.e. without AF), even at relatively low enzyme concentrations, produced extensive PL degradation in ΔlplT or Δaas spheroplasts (Figs. 7, a–c). By contrast, WT spheroplasts were highly resistant to the added psPLA2, even with albumin present to preclude recycling of the PL breakdown products. These findings indicate that the presence of LplT/Aas during bacterial growth and spheroplast generation markedly increases the resistance of the IM to psPLA2.
Figure 7.
LplT/Aas protect the IM from sPLA2 attack. a, susceptibility of spheroplasts generated from E. coli W3110 WT, ΔlplT, or Δaas cells after treated with 12 units of psPLA2. The data are expressed as percentages of recovered CFU compared with untreated spheroplasts from the WT control. b, free fatty acids release assay of W3110 WT, ΔlplT, or Δaas spheroplasts (± 1.5% BSA). c, psPLA2 titration experiment for free fatty acid release assay. Spheroplasts made from [14C]oleic acid-labeled E. coli cells were incubated with indicated concentrations of psPLA2 at 37 °C for 20 min. d, stability test of WT, ΔlplT, or Δaas spheroplasts (for details see “Experimental procedures”).
In the absence of added psPLA2, the spheroplasts from both the WT and mutant strains were stable in 0.5 m sucrose and equally able to regenerate CFU (Fig. 7a). Incubation with psPLA2 under these conditions caused >90% reduction in regeneration of CFU from the mutant spheroplasts but had no effect on the WT spheroplasts (Fig. 7a). In the absence of added psPLA2, the mutant spheroplasts were much more susceptible to hypo-osmotic shock (Fig. 7d). Taken together, these findings suggest that the function of LplT and Aas contributes to the stability of the IM, with vulnerability to psPLA2 a particularly sensitive marker of IM instability (see “Discussion”).
Discussion
Here, we demonstrate a novel role of LplT and Aas in the resistance of E. coli to host sPLA2. In the experiments shown, we purposely used low doses of AF to dilute out sPLA2-IIA present in AF (5) and to eliminate any psPLA2-independent killing of either WT or mutant strains. Under identical conditions in which no detectable PL degradation and LPL accumulation and no diminution in bacterial viability were seen in the WT strain, >90% of the mutant bacteria were killed (Figs. 2a and 4a). This degree of killing paralleled degradation of >40% of the prelabeled PL and their accumulation as LPL, suggesting accumulation of LPL in the IM where they could exert toxic effects on the lamellar structure of bilayer and essential energy-generating and -dependent metabolic systems.
We began these studies anticipating that the absence of LplT/Aas function would increase bacterial susceptibility to added sPLA2 by allowing increased net accumulation of LPL produced during sPLA2 treatment. However, the absence of measurable PL degradation (Fig. 4a) in the WT strain expressing both LplT and Aas precluded testing a role of LplT/Aas-mediated PL repair during sPLA2 treatment on restraining the consequences of sPLA2 action. Ironically, this limitation of our experimental design made possible detection of an unexpected role of LplT/Aas function during bacterial growth on the physical properties and composition of the outer leaflet of the OM. The relevance of these effects to psPLA2 action is strongly supported by the correlation of these OM changes with the ability of psPLA2 to act on the mutant but not WT bacteria. This was most evident in the studies of the PE-specific/OM outer leaflet-restricted PE-specific probe Ro 09-0198–PEG5000–Alexa 488 that showed maximal interactions with AF-treated mutant strains (Fig. 6d), matching the conditions most favorable for psPLA2 surface binding and action on the intact bacteria (Fig. 5). Although other surface modifications yielding access to phosphorylethanolamine are possible, we believe that the mobilization of increased outer leaflet phosphatidylethanolamine (Fig. 6d) is most consistent with changes promoting psPLA2 binding and interfacial activation, as well as the looser lipid packing revealed by the studies with NR12S (Fig. 6b).
The absence of detectable PldA or PagP activation under identical experimental conditions (Figs. S7 and S8) suggests that smaller perturbations in the lipid organization of the OM are sufficient for psPLA2 activation in comparison to requirements for activation of these endogenous OM lipid-modifying enzymes. The absence of PldA and PagP activation may in fact be important, leaving accessible substrate PL available to the added sPLA2. The ability of initially limited alterations of the OM leads to the massive net PL degradation observed (Figs. 4a and 5a) and may be explained by the ability of products of sPLA2 action (e.g. LPL), unlike those of PagP (e.g. hyperacylated lipid A and LPL), to exacerbate membrane instability and thereby promote conditions favorable for further sPLA2 action, especially in the setting of no LPL reacylation (i.e. no LplT/Aas function). Detergent-like LPL can disturb lamellar membrane structure, because of its bending elasticity and local curvature-modulating features, promoting substrate binding and catalysis by sPLA2 (18, 38). Further perturbation of the OM would be expected to lead to greater mobilization of PE in the outer leaflet and activation of PagP, resulting in conversion of hexa- to hepta-acylated LPS, increased lipid packing in the outer leaflet, and resistance to membrane-active cationic antibacterial peptides and proteins including those present in AF (39). However, the rapid breakdown of PE by psPLA2 could preclude this bacterial stress response. The apparent absence of PagP activity also suggests that inactivation of LplT/Aas does not trigger the bacterial virulence regulatory system PhoP/PhoQ (40). The absence of induction of these protective bacterial responses may help further explain the remarkable sensitivity of the LplT/Aas mutants to the combined action of host co-factors for sPLA2 in AF and psPLA2 that we observed.
The remarkable differences in psPLA2 susceptibility of IM spheroplasts recovered from WT versus ΔlplT and Δaas mutant strains (Fig. 7a) parallel relatively small differences in LPL accumulation in these IM-rich cell-free fractions (4 mol % from the WT strain and ∼8 mol % in the mutant strains) (22, 23). These findings thus also suggest that initially small lipid differences can be sufficient for much greater differences in sPLA2 action, especially in the setting of absent LplT/Aas function. The resistance of the WT spheroplasts to psPLA2 was somewhat surprising given the sensitivity of cell-free membrane protoplasts from the Gram-positive bacteria Staphylococcus aureus and Bacillus subtilis (3). These findings suggest important differences in the sensitivity of phospholipids within the corresponding cytoplasmic (inner) membranes of Gram-negative and Gram-positive bacteria to host sPLA2 that may contribute to the greater bactericidal potency of mammalian sPLA2 toward Gram-positive bacteria.
It is more challenging to understand how LplT and Aas residing in or tightly associated with the IM can affect the changes in the OM. OM lipid asymmetry as an essential protective strategy of Gram-negative bacteria is believed to be facilitated by multiple mechanisms including the cytoplasmic Mla system, which promotes retrograde transport of diacyl PLs from OM to IM (41). The OM conformation is also mediated by the processes of membrane biogenesis including those for LPS, which also occur in the IM (19, 30). Whether accumulation of LPL as a result of LplT/Aas inactivation alters the conformation and function of these biogenetic systems involved in the envelope homeostasis is unknown. It has been shown that LPL directly modifies the activity of bacterial membrane proteins such as mechanosensitive channel MscL (42). Indeed, the acute accumulation of CL at the early stage of the psPLA2 action (Fig. 4a and Table S2) suggested that bacterial de novo PL biosynthetic enzymes were stimulated by the accumulation of LPL. Characterization of the function and lipid requirement of membrane biosynthetic proteins in the ΔlplT and Δaas strains may help to understand the role of LplT/Aas in maintaining bacterial OM lipid asymmetry.
In summary, we have demonstrated a new Gram-negative bacterial resistance mechanism against host sPLA2 using E. coli and psPLA2 that depends on the conserved LPL transport and acylation system (LplT/Aas) that is unique to Gram-negative bacteria. Given the conservation of this system in many other Gram-negative bacteria and the similar interfacial activation requirements of the various sPLA2, we predict that the principles revealed in this study should apply to other strains and species of Gram-negative bacteria and other sPLA2 including sPLA2-IIA, a hallmark of mammalian inflammatory responses to infection (3, 4, 7, 12). The continuing development and spread of antibiotic resistance genes among Gram-negative bacteria, normal members of the microbiome and pathogens alike, have increased interest in the design and development of compounds that, rather than targeting bacterial functions crucial for viability per se, instead target sites that would render host defense systems mobilized during infection and inflammation more effective (43). The fact that inactivation of LplT and Aas does not affect bacterial normal growth (Fig. S1b) but specifically disarms their resistance to sPLA2-mediated host antibacterial attack fits this description well. Therefore, the experimental model described herein provides a template for testing LplT/Aas as new drug targets dealing with Gram-negative bacterial resistance in the future.
Experimental procedures
Materials and strains
The rabbit AFs used in this study were previously isolated from sterile inflammatory peritoneal exudates in New Zealand White rabbits following intraperitoneal injection with oyster glycogen-saturated saline (6, 27) and stored at −80 °C prior to use. To deplete rabbit AF of cationic proteins, AF was incubated with CM-Sephadex resin equilibrated in 0.9% (w/v) saline buffered with 2.5 mm Tris-HCl, pH 7.5, for 1 h followed by recovery of the unbound fraction that contained greater than 95% of the total AF protein content but was fully (>98%) depleted of cationic proteins (28). sPLA2 purified from porcine pancreas and Crotalus adamanteus venom were purchased from Sigma or Worthington. [32P]PO4 and [14C]oleic acid were purchased from MP Biochemicals or PerkinElmer Life Sciences. [32P]LPL was generated by digestion of [32P]PL using venom PLA2 as previously described (23). 1-Oleoyl-2-hydroxy-sn-glycero-3-phosphoethanolamine (18:1 LPE) was a product of Avanti Polar Lipids. Anti-psPLA2 antibody was purchased from Lifespan Biosciences. E. coli strains W3110 and AL95 and plasmids used for bacterial genetics were gifts from Drs. William Dowhan or Jiqiang Ling (University of Texas, McGovern Medical School). NR12S (N-[3-[[9-(diethylamino)-5-oxo-5H-benzo[a]phenoxazin-2-yl]oxy]propyl]-N-methyl-N-(3-sulfopropyl)-1-dodecanaminium) was kindly provided by Dr. Andrey S. Klymchenko (University of Strasbourg). Ro 09-0198 and Alexa 488–PEG5000–NHS were purchased from Cayman or Nanocs.
Construction of E. coli knockout strains
Chromosomal in-frame mutants were generated in the background of E. coli W3110 strain using λ-Red homologous recombination approach as described (44). The primers used for mutagenesis are listed in Table S1. For gene knockout mutants, linear gene displacement cassettes containing a chloramphenicol resistance region and homologous sequences were PCR-amplified from the plasmid template pKD3. The PCR products were transformed into competent cells of E. coli W3110 cells carrying the plasmid pKD46 by electroporation. λ-Red recombinase in the competent cells was induced by adding l-arabinose in the medium. Transformants were incubated in LB broth containing ampicillin and 10 mm l-arabinose for 2 h at 32 °C for homologous recombination and then selected on LB agar plates containing chloramphenicol at 30 °C overnight. Replacement of the target genes by the chloramphenicol resistance gene was verified by PCR. The temperature-sensitive plasmid pKD46 was cured by incubation at 37 °C in LB broth. Excision of resistance cassettes was carried out using the helper plasmid pCP20 at 30 °C. Temperature-sensitive plasmid pCP20 was further cured by overnight incubation at 42 °C in LB broth. Positive mutants were examined by the loss of chloramphenicol resistance and by PCR-based DNA sequencing.
Construction of E. coli knockin strain
Chromosomal knockin mutant strains were generated using the Kn-pBAD-ccdB selectable cassette in two steps. The heat-inducible plasmid pSIM6 was first transformed into the W3110 cells to prepare λ-Red–induced cells. In the first step, a Kn-pBAD-ccdB cassette flanked by homologous sequences was amplified using PCR. The PCR product was transformed into the cells by electroporation. Transformants were immediately suspended into 2 ml of LB broth supplemented with 1% glucose and grown at 32 °C for 2 h to facilitate homologous recombination. Recombinants were selected on LB agar plates supplemented with ampicillin, kanamycin, and 1% glucose at 30 °C overnight. Replacement of the target gene by the Kn-pBAD-ccdB cassette was verified by PCR and by comparing bacterial growth onto LB plates supplemented with 1% glucose or 1% arabinose. Prior to the second knockin step, mutations corresponding to H36A and D30A was introduced into plasmids harboring the aas or lplT gene using site-directed mutagenesis. The PCR fragment containing aasH36A or lplTD30A mutation was transformed into the cells containing the Kn-pBAD-ccdB cassette. After electroporation, the cells were incubated in 2 ml of LB broth supplemented with 1% glucose at 32 °C for 2 h. Knockin mutants were selected on LB agar plates containing ampicillin and 1% arabinose at 37 °C overnight. Replacement of the Kn-pBAD-ccdB cassette by the mutant DNA was verified by PCR and examined on LB plates supplemented with 1% arabinose. The pSIM6 plasmid was cured by growing positive candidates overnight at 37 °C in LB broth and then tested for sensitivity to ampicillin. Knockin mutations were further confirmed by PCR-based DNA sequencing.
Bacterial cells used in all assays were prepared using similar approaches. Briefly, E. coli cells were grown overnight in LB broth at 37 °C and then diluted (1:100) into fresh medium for continuous growth from lag phase (A600 = 0.1), to logarithmic phase (A600 = 0.5), and finally to stationary phase (A600 = 4.0). All experiments were carried out using E. coli cells harvested at the logarithmic phase if not indicated.
Preparation of E. coli spheroplasts
E. coli cells were washed three times and resuspended in 10 mm HEPES, pH 7.5, 0.75 m sucrose, 10 mm MgSO4, and 2.5% (w/v) LiCl. After addition of 1 mg/ml lysozyme, cell suspensions were ice-chilled and then warmed to room temperature, followed by incubation with gentle shaking at 30 °C for 30 min. Intact spheroplasts were collected by centrifugation (3,000 × g for 10 min) at room temperature and suspended at 10 mg/ml total protein in the above buffer without LiCl. Spheroplast formation and stability were thoroughly monitored nephelometrically by comparing the A600 of a 100-μl spheroplast solution to 2 ml of either plain water or a solution of 10 mm MgCl2/0.45 m sucrose, respectively.
LPL acylation assay
LPL acylation activity was measured by monitoring conversion of LPE to PE in spheroplasts as we previously described (23). [32P]LPE (∼100,000 cpm) mixed with synthetic 18:1 LPE of 200 μm in ethanol was used as substrate. 10 μm (final concentration) of substrates was added into spheroplast solutions. At the indicated time, the reactions were terminated by adding chloroform-methanol (1:2 v:v). The lipids were extracted and separated by TLC and analyzed using phosphorimaging. The acylation activity is expressed as a percentage of LPE to PE conversion.
LplT transport assay
LplT transport activity was measured by detecting import of [32P]LPE into spheroplasts using an approach we established previously (23). Briefly, 10 μm (final concentration) of substrates was added into 0.5 ml of spheroplast solutions. At the indicated time, the samples were layered onto 0.15 ml of 22% perchloric acid and 0.50 ml of silicone oil and centrifuged at 14,000 × g for 5 min at room temperature to separate spheroplasts from free LPE substrate. After centrifugation, the sample moves through silicone oil, and the radioactivity of collected spheroplasts was measured using a liquid scintillation counter to calculate transport activity.
Analysis of LplT and Aas mutant protein expression
LplT and Aas proteins were overproduced using a T7 expression vector in E. coli BL21(DE3) strains as previously described (23). The genes encoding LplT or Aas from E. coli were cloned into a pET28a plasmid to fuse with a N-terminal His6 tag. The mutations were introduced into the respective vector using a standard site-directed mutagenesis method. Overproduction of WT and mutant proteins was performed in antoinduction medium (9) at 37 °C overnight. The cells were harvested and ruptured by sonication. Cell lysates at similar protein concentrations were subjected to SDS-PAGE, transferred to immunoblots, and probed with anti-His antibody.
Bacterial viability assay
sPLA2-mediated bactericidal tests were performed using the protocol previously described with slight modifications (16, 28). Prior to assays, E. coli cells were washed three times and suspended in a buffer containing 40% HBSS and 1.5% BSA. 100 μl of cell suspension (∼106 cells) was mixed with rabbit AF and/or psPLA2 at 37 °C for the indicated times. The reactions were terminated by diluting into 1 ml of LB broth, and then 100 μl of a dilution was immediately spread on a LB agar plate. CFU was determined after 16 h of incubation at 37 °C.
TLC PL analysis
PL analysis was carried out using 32P-labeled E. coli cells by TLC. To prepare [32P] bacteria, E. coli were grown in LB broth supplemented with 5 μCi/ml [32P] PO4. The assay was performed using the same protocol for viability test as described above. At indicated time, the reactions were stopped by adding chloroform followed by lipid extraction using the acidic Bligh and Dyer method as described previously (23). The lipids were loaded onto a silica gel G thin-layer plate presoaked with boric acid and developed with chloroform/methanol/ammonia/H2O (60:37.5:1:3, v/v/v/v). The air-dried plate was exposed to a storage phosphor screen (Eastman Kodak) overnight. Individual PL was visualized and quantified using phosphorimaging (Bio-Rad) to calculate PL content expressed as mol % of the total PL pool.
Free fatty acid release assay
sPLA2 activity was determined by measuring 14C-labeled fatty acid hydrolyzed from bacterial cells as previously described (31). 14C-Labeled cells were prepared in LB broth containing 0.5 μCi/ml [1-14C]oleic acid. After labeling for 2 h at 37 °C, the cells were pelleted and then incubated in fresh LB broth for additional 30 min at 37 °C. The cells were washed with 1% BSA in sterile PBS to remove unincorporated radioactive precursors and then suspended in PBS. The assay was carried out using the same protocol of viability test as described above. The reactions were stopped by pelleting intact cells. The supernatant containing radiolabeled free fatty acid and lyso compounds was collected and quantified by a liquid scintillation counter.
sPLA2 binding assay
100 μl of E. coli cells (∼109) in a buffer containing 40% HBSS, and 1.5% BSA was incubated with 6 units of psPLA2 in the presence or absence of 20 μl of AF at 37 °C for 30 min. The cells were pelleted and washed three times with the same buffer and then analyzed by Western blotting analysis using anti-psPLA2 antibody.
NR12S fluorescence assay
1 ml of E. coli cells were washed three times with buffer containing 0.1 m HEPES, pH 7.5, and 100 mm KCl and suspended in 100 μl of same buffer. The cells were incubated with 20 nm NR12S (3 mm in DMSO stock) at room temperature for 30 min. The cells were washed three times with the same buffer. Fluorescence spectra were measured on a fluorescence spectrophotometer at the excitation wavelength of 530 nm, and the emission was recorded within the range between 560 and 700 nm wavelength.
Ro 09-0198–based fluorescence assay
To prepare a PE-specific fluorescent dye, Ro 09-0198 was conjugated with Alexa 488–PEG5000–NHS via amine-NHS chemistry. Briefly, the two compounds were mixed in a molar ratio of 1:2 in a buffer of 50 mm NaHCO3 overnight at 4 °C. The reaction was stopped by adding 1 m lysine for 3 h. Prior to assays, E. coli cells were washed and then suspended in PBS saline. 20 μl AF, cationic protein-depleted AF or buffer was added into 0.5 ml of cells (A600 = 1) for 30 min and then washed twice with PBS supplemented with 2% LiCl. 1 μm of the Ro 09-0198 conjugate was mixed with bacteria for 30 min at 37 °C. The cells were washed three times with buffer to remove unbound conjugates. Fluorescence signals were measured at the following wavelengths: excitation at 490 nm and emission at 520 nm using a fluorescence spectrophotometer.
LC-ESI MS analysis of lipid A
To isolate lipid A, 50 ml E. coli cells were washed twice with 50 ml of PBS saline and then suspended in PBS (0.8 ml). Chloroform (1.0 ml) and methanol (2.0 ml) were added, and the mixtures were vigorously shaken at room temperature for 30 min to make the one-phase Bligh–Dyer mixture (chloroform:methanol:water = 1:2:0.8, v/v/v) and extract and remove PL. The mixtures were then centrifuged at 5,000 rpm for 30 min. The pellets were washed once with fresh one-phase Bligh–Dyer solution (3.5 ml) and centrifuged as above. The resulting pellets were suspended in 1.0 ml of sodium acetate (12.5 mm, pH 4.5) and heated at 100 °C for 30 min. After cooling to room temperature, the suspension was acidified to pH 1 with HCl and converted to a two-phase Bligh–Dyer mixture ((chloroform:methanol:PBS = 2:2:1.8, v/v/v) by adding 0.8 ml of PBS, chloroform, and methanol (2 ml each). The lower (organic) phase of each mixture was retrieved after phase partitioning via centrifugation at 5,000 rpm for 30 min and used for analysis.
Normal phase LC-ESI MS was performed using an Agilent 1200 Quaternary LC system coupled to a high resolution TripleTOF5600 mass spectrometer (Sciex, Framingham, MA). A Unison UK-Amino column (3 μm, 25 cm × 2 mm) (Imtakt USA, Portland, OR) was used. Mobile phase A consisted of chloroform/methanol/aqueous ammonium hydroxide (800:195:5, v/v/v). Mobile phase B consisted of chloroform/methanol/water/aqueous ammonium hydroxide (600:340:50:5, v/v/v/v). Mobile phase C consisted of chloroform/methanol/water/aqueous ammonium hydroxide (450:450:95:5, v/v/v/v). The elution program consisted of the following: 100% mobile phase A was held isocratically for 2 min and then linearly increased to 100% mobile phase B over 14 min and held at 100% B for 11 min. The LC gradient was then changed to 100% mobile phase C over 3 min, held at 100% C for 3 min, finally returned to 100% A over 0.5 min, and held at 100% A for 5 min. The total LC flow rate was 300 μl/min. The postcolumn splitter diverted ∼10% of the LC flow to the ESI source of the TF5600 mass spectrometer, with MS settings as follows: ion spray voltage = −4500 V, curtain gas = 20 p.s.i., ion source gas 1 = 20 p.s.i., declustering potential = −55 V, and focusing potential = −150 V. Nitrogen was used as the collision gas for MS/MS experiments. Data acquisition and analysis were performed using Analyst TF1.5 software (Sciex, Framingham, MA).
Spheroplasts stability test
Spheroplasts generated from 109 E. coli cells using the lysozyme-lithium method as above were suspended in 100 μl of solution containing different concentrations of sucrose and then kept at 25 °C for 10 min. 900 μl of LB broth supplemented with 0.45 m sucrose was added to spheroplasts. 3 μl of each samples was spotted on LB agar plate and incubated overnight at 37 °C.
Author contributions
Y. L., S. L., Z. G., W. M., J. W., and L. Z. data curation; Y. L., M. B., Z. G., J. W., and L. Z. formal analysis; Y. L., J. W., and L. Z. investigation; Y. L., M. B., S. L., Z. G., J. W., and L. Z. methodology; Y. L., J. W., and L. Z. writing-original draft; M. B., Z. G., J. W., and L. Z. resources; M. B., W. M., J. W., and L. Z. writing-review and editing; J. W. and L. Z. conceptualization; L. Z. supervision; L. Z. funding acquisition; L. Z. project administration.
Supplementary Material
Acknowledgments
We thank William Dowhan for providing E. coli strains and access to the phosphorimaging device, Jiqiang Ling for providing plasmids and helps for bacterial genetic mutations, and Andrey Klymchenko for providing the NR12S dye.
This work was supported by National Institutes of Health Grants R01GM097290 and R01GM098572 (to L. Z.) and R01GM61074 (to W. M.), and European Union Marie Skłodowska-Curie Grant H2020-MSCA-RISE-2015-690853 (to M. B.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1 and S2 and Figs. S1–S9.
- sPLA2
- secretory phospholipase A2
- OM
- outer membrane
- IM
- inner membrane
- LPS
- lipopolysaccharide
- MAC
- membrane attack complex
- BPI
- bactericidal/permeability-increasing protein
- PL
- phospholipids
- LPL
- lysophospholipids
- AF
- ascitic fluid
- psPLA2
- pancreatic phospholipase A2
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- CL
- cardiolipin
- LPE
- lysophosphatidylethanolamine
- LPG
- lysophosphatidylglycerol
- MCL
- monoacylated cardiolipin
- CFU
- colony-forming units
- ESI
- electrospray ionization.
References
- 1. Elsbach P., Weiss J., and Levy O. (1994) Integration of antimicrobial host defenses: role of the bactericidal/permeability-increasing protein. Trends Microbiol 2, 324–328 10.1016/0966-842X(94)90449-9 [DOI] [PubMed] [Google Scholar]
- 2. Nauseef W. M. (2007) How human neutrophils kill and degrade microbes: an integrated view. Immunol. Rev. 219, 88–102 10.1111/j.1600-065X.2007.00550.x [DOI] [PubMed] [Google Scholar]
- 3. Weiss J. P. (2015) Molecular determinants of bacterial sensitivity and resistance to mammalian group IIA phospholipase A2. Biochim. Biophys. Acta 1848, 3072–3077 10.1016/j.bbamem.2015.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nevalainen T. J., Graham G. G., and Scott K. F. (2008) Antibacterial actions of secreted phospholipases A2. Biochim. Biophys. Acta 1781, 1–9 10.1016/j.bbalip.2007.12.001 [DOI] [PubMed] [Google Scholar]
- 5. Weinrauch Y., Elsbach P., Madsen L. M., Foreman A., and Weiss J. (1996) The potent anti-Staphylococcus aureus activity of a sterile rabbit inflammatory fluid is due to a 14-kD phospholipase A2. J. Clin. Invest. 97, 250–257 10.1172/JCI118399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Madsen L. M., Inada M., and Weiss J. (1996) Determinants of activation by complement of group II phospholipase A2 acting against Escherichia coli. Infect. Immun. 64, 2425–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu Y., Raymond B., Goossens P. L., Njamkepo E., Guiso N., Paya M., and Touqui L. (2010) Type-IIA secreted phospholipase A2 is an endogenous antibiotic-like protein of the host. Biochimie 92, 583–587 10.1016/j.biochi.2010.01.024 [DOI] [PubMed] [Google Scholar]
- 8. Birts C. N., Barton C. H., and Wilton D. C. (2010) Catalytic and non-catalytic functions of human IIA phospholipase A2. Trends Biochem. Sci. 35, 28–35 10.1016/j.tibs.2009.08.003 [DOI] [PubMed] [Google Scholar]
- 9. Studier F. W. (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 10.1016/j.pep.2005.01.016 [DOI] [PubMed] [Google Scholar]
- 10. Femling J. K., Nauseef W. M., and Weiss J. P. (2005) Synergy between extracellular group IIA phospholipase A2 and phagocyte NADPH oxidase in digestion of phospholipids of Staphylococcus aureus ingested by human neutrophils. J. Immunol. 175, 4653–4661 10.4049/jimmunol.175.7.4653 [DOI] [PubMed] [Google Scholar]
- 11. Burke J. E., and Dennis E. A. (2009) Phospholipase A2 structure/function, mechanism, and signaling. J. Lipid Res. 50, S237–S242 10.1194/jlr.R800033-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grönroos J. O., Laine V. J., and Nevalainen T. J. (2002) Bactericidal group IIA phospholipase A2 in serum of patients with bacterial infections. J. Infect. Dis. 185, 1767–1772 10.1086/340821 [DOI] [PubMed] [Google Scholar]
- 13. Degousee N., Ghomashchi F., Stefanski E., Singer A., Smart B. P., Borregaard N., Reithmeier R., Lindsay T. F., Lichtenberger C., Reinisch W., Lambeau G., Arm J., Tischfield J., Gelb M. H., and Rubin B. B. (2002) Groups IV, V, and X phospholipases A2s in human neutrophils: role in eicosanoid production and gram-negative bacterial phospholipid hydrolysis. J. Biol. Chem. 277, 5061–5073 10.1074/jbc.M109083200 [DOI] [PubMed] [Google Scholar]
- 14. Lambeau G., and Gelb M. H. (2008) Biochemistry and physiology of mammalian secreted phospholipases A2. Annu. Rev. Biochem. 77, 495–520 10.1146/annurev.biochem.76.062405.154007 [DOI] [PubMed] [Google Scholar]
- 15. Koduri R. S., Grönroos J. O., Laine V. J., Le Calvez C., Lambeau G., Nevalainen T. J., and Gelb M. H. (2002) Bactericidal properties of human and murine groups I, II, V, X, and XII secreted phospholipases A2. J. Biol. Chem. 277, 5849–5857 10.1074/jbc.M109699200 [DOI] [PubMed] [Google Scholar]
- 16. Weinrauch Y., Abad C., Liang N. S., Lowry S. F., and Weiss J. (1998) Mobilization of potent plasma bactericidal activity during systemic bacterial challenge: role of group IIA phospholipase A2. J. Clin. Invest. 102, 633–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Weiss J., Wright G., Bekkers A. C., van den Bergh C. J., and Verheij H. M. (1991) Conversion of pig pancreas phospholipase A2 by protein engineering into enzyme active against Escherichia coli treated with the bactericidal/permeability-increasing protein. J. Biol. Chem. 266, 4162–4167 [PubMed] [Google Scholar]
- 18. Berg O. G., Gelb M. H., Tsai M.-D., and Jain M. K. (2001) Interfacial enzymology: the secreted phospholipase A2-paradigm. Chem. Rev. 101, 2613–2654 10.1021/cr990139w [DOI] [PubMed] [Google Scholar]
- 19. Henderson J. C., Zimmerman S. M., Crofts A. A., Boll J. M., Kuhns L. G., Herrera C. M., and Trent M. S. (2016) The power of asymmetry: architecture and assembly of the Gram-negative outer membrane lipid bilayer. Annu. Rev. Microbiol. 70, 255–278 10.1146/annurev-micro-102215-095308 [DOI] [PubMed] [Google Scholar]
- 20. Weiss J., Inada M., Elsbach P., and Crowl R. M. (1994) Structural determinants of the action against Escherichia coli of a human inflammatory fluid phospholipase A2 in concert with polymorphonuclear leukocytes. J. Biol. Chem. 269, 26331–26337 [PubMed] [Google Scholar]
- 21. Qu X. D., and Lehrer R. I. (1998) Secretory phospholipase A2 is the principal bactericide for staphylococci and other gram-positive bacteria in human tears. Infect. Immun. 66, 2791–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harvat E. M., Zhang Y. M., Tran C. V., Zhang Z., Frank M. W., Rock C. O., and Saier M. H. Jr. (2005) Lysophospholipid flipping across the Escherichia coli inner membrane catalyzed by a transporter (LplT) belonging to the major facilitator superfamily. J. Biol. Chem. 280, 12028–12034 10.1074/jbc.M414368200 [DOI] [PubMed] [Google Scholar]
- 23. Lin Y., Bogdanov M., Tong S., Guan Z., and Zheng L. (2016) Substrate selectivity of lysophospholipid transporter LplT involved in membrane phospholipid remodeling In Escherichia coli. J. Biol. Chem. 291, 2136–2149 10.1074/jbc.M115.700419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jackowski S., Jackson P. D., and Rock C. O. (1994) Sequence and function of the aas gene in Escherichia coli. J. Biol. Chem. 269, 2921–2928 [PubMed] [Google Scholar]
- 25. Zheng L., Lin Y., Lu S., Zhang J., and Bogdanov M. (2017) Biogenesis, transport and remodeling of lysophospholipids in Gram-negative bacteria. Biochim. Biophys. Acta 1862, 1404–1413 10.1016/j.bbalip.2016.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hsu L., Jackowski S., and Rock C. O. (1991) Isolation and characterization of Escherichia coli K-12 mutants lacking both 2-acyl-glycerophosphoethanolamine acyltransferase and acyl-acyl carrier protein synthetase activity. J. Biol. Chem. 266, 13783–13788 [PubMed] [Google Scholar]
- 27. Zarember K., Elsbach P., Shin-Kim K., and Weiss J. (1997) p15s (15-kD antimicrobial proteins) are stored in the secondary granules of rabbit granulocytes: implications for antibacterial synergy with the bactericidal/permeability-increasing protein in inflammatory fluids. Blood 89, 672–679 [PubMed] [Google Scholar]
- 28. Weinrauch Y., Foreman A., Shu C., Zarember K., Levy O., Elsbach P., and Weiss J. (1995) Extracellular accumulation of potently microbicidal bactericidal/permeability-increasing protein and p15s in an evolving sterile rabbit peritoneal inflammatory exudate. J. Clin. Invest. 95, 1916–1924 10.1172/JCI117873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heath R. J., and Rock C. O. (1998) A conserved histidine is essential for glycerolipid acyltransferase catalysis. J. Bacteriol. 180, 1425–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Op den Kamp J. A. (1979) Lipid asymmetry in membranes. Annu. Rev. Biochem. 48, 47–71 10.1146/annurev.bi.48.070179.000403 [DOI] [PubMed] [Google Scholar]
- 31. Wright G. C., Weiss J., Kim K. S., Verheij H., and Elsbach P. (1990) Bacterial phospholipid hydrolysis enhances the destruction of Escherichia coli ingested by rabbit neutrophils. Role of cellular and extracellular phospholipases. J. Clin. Invest. 85, 1925–1935 10.1172/JCI114655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Forst S., Weiss J., Maraganore J. M., Heinrikson R. L., and Elsbach P. (1987) Relation between binding and the action of phospholipases A2 on Escherichia coli exposed to the bactericidal/permeability-increasing protein of neutrophils. Biochim. Biophys. Acta 920, 221–225 10.1016/0005-2760(87)90098-1 [DOI] [PubMed] [Google Scholar]
- 33. Kucherak O. A., Oncul S., Darwich Z., Yushchenko D. A., Arntz Y., Didier P., Mély Y., and Klymchenko A. S. (2010) Switchable nile red-based probe for cholesterol and lipid order at the outer leaflet of biomembranes. J. Am. Chem. Soc. 132, 4907–4916 10.1021/ja100351w [DOI] [PubMed] [Google Scholar]
- 34. Machaidze G., and Seelig J. (2003) Specific binding of cinnamycin (Ro 09-0198) to phosphatidylethanolamine: comparison between micellar and membrane environments. Biochemistry 42, 12570–12576 10.1021/bi035225b [DOI] [PubMed] [Google Scholar]
- 35. Bogdanov M., Heacock P. N., and Dowhan W. (2002) A polytopic membrane protein displays a reversible topology dependent on membrane lipid composition. EMBO J. 21, 2107–2116 10.1093/emboj/21.9.2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dekker N. (2000) Outer-membrane phospholipase A: known structure, unknown biological function. Mol. Microbiol. 35, 711–717 10.1046/j.1365-2958.2000.01775.x [DOI] [PubMed] [Google Scholar]
- 37. Jia W., El Zoeiby A., Petruzziello T. N., Jayabalasingham B., Seyedirashti S., and Bishop R. E. (2004) Lipid trafficking controls endotoxin acylation in outer membranes of Escherichia coli. J. Biol. Chem. 279, 44966–44975 10.1074/jbc.M404963200 [DOI] [PubMed] [Google Scholar]
- 38. Tatulian S. A. (2001) Toward understanding interfacial activation of secretory phospholipase A2 (PLA2): membrane surface properties and membrane-induced structural changes in the enzyme contribute synergistically to PLA2 activation. Biophys. J. 80, 789–800 10.1016/S0006-3495(01)76058-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bishop R. E. (2005) The lipid A palmitoyltransferase PagP: molecular mechanisms and role in bacterial pathogenesis. Mol. Microbiol. 57, 900–912 10.1111/j.1365-2958.2005.04711.x [DOI] [PubMed] [Google Scholar]
- 40. Groisman E. A. (2001) The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183, 1835–1842 10.1128/JB.183.6.1835-1842.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Malinverni J. C., and Silhavy T. J. (2009) An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc. Natl. Acad. Sci. U S A 106, 8009–8014 10.1073/pnas.0903229106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lundbaek J. A., and Andersen O. S. (1994) Lysophospholipids modulate channel function by altering the mechanical properties of lipid bilayers. J. Gen. Physiol. 104, 645–673 10.1085/jgp.104.4.645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sachdeva S., Palur R. V., Sudhakar K. U., and Rathinavelan T. (2017) E. coli group 1 capsular polysaccharide exportation nanomachinary as a plausible antivirulence target in the perspective of emerging antimicrobial resistance. Front. Microbiol. 8, 70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., and Mori H. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.