Abstract
N-glycosylation is one of the most common protein post-translational modifications in eukaryotes and has a relatively conserved core structure between fungi, animals and plants. In plants, the biosynthesis of N-glycans has been extensively studied with all the major biosynthetic enzymes characterized. However, few studies have applied advanced mass spectrometry to profile intact plant N-glycopeptides. In this study, we use hydrophilic enrichment, high-resolution tandem mass spectrometry with complementary and triggered fragmentation to profile Arabidopsis N-glycopeptides from microsomal membranes of aerial tissues. A total of 492 N-glycosites were identified from 324 Arabidopsis proteins with extensive N-glycan structural heterogeneity revealed through 1110 N-glycopeptides. To demonstrate the precision of the approach, we also profiled N-glycopeptides from the mutant (xylt) of β-1,2-xylosyltransferase, an enzyme in the N-glycan biosynthetic pathway. This analysis represents the most comprehensive and unbiased collection of Arabidopsis N-glycopeptides revealing an unsurpassed level of detail on the micro-heterogeneity present in N-glycoproteins of Arabidopsis. Data are available via ProteomeXchange with identifier PXD006270.
The N-glycosylation of proteins is a prevalent post-translational modification (PTM) found in eukaryotes including microbes, animals and plants. The modification is important for proper protein function and can affect folding, enzyme activity and protein-protein interactions (1). The core structure of N-glycans is comprised of N-acetylglucosamine (GlcNAc)1 and mannose (Man) (Man3GlcNAc2) and is conserved across kingdoms although some unusual N-glycan core structures can be found in proteins derived from Archaea (2). In eukaryotes, most N-linked glycosylation occurs on asparagine residues at the canonical consensus sequence N-X-S/T, where “X” can be any amino acid except proline, although non-consensus sequences have been reported (3).
The sequential biosynthesis of N-glycans within the endomembrane system is a highly-conserved process between eukaryotes. However, differences in the maturation processes result in the glycan structural diversity observed between kingdoms. In higher plants, the initial steps occur at the cytosolic side of the endoplasmic reticulum (ER) with dolichol phosphate (DolP) acting as the acceptor for the initial glycosylation steps to form a Man5GlcNAc2-DolP structure. In the ER, additional Man and glucose (Glc) molecules are added from Dol-linked donors to form Glc3Man9GlcNAc2-DolP (4). The oligosaccharide is then transferred to the nascent polypeptide via the oligosaccharyltransferase (OST) complex. Before entering the Golgi apparatus, the three Glc molecules and a single Man are removed in processes involving the calnexin (CNX) and calreticulin (CRT) cycle and ER quality-control (ERQC) processes resulting in a correctly folded glycoprotein with Man8GlcNAc2 glycan structures (supplemental Fig. S1). Once in the Golgi apparatus, the Man residues are trimmed by mannosidases to form a Man5GlcNAc2 structure. This is followed by the addition of a GlcNAc residue by β-1,2-N-acetylglucosaminyltransferase I (GnT1) (5). This step is crucial for the downstream maturation processes involving the removal of Man residues by Golgi mannosidases and the addition of xylose (Xyl) by β1–2-xylosyltransferase (XYLT) and fucose (Fuc) by α1–3-fucosyltransferases (FUT11/12) resulting in the archetypal complex N-glycan structure GlcNAc2Man3XylFucGlcNAc2. Further maturation process and post-Golgi processing of this structure results in extensive heterogeneity of the N-glycan structure (supplemental Fig. S1). Several recent reviews provide extensive detail on the biosynthesis of N-glycans in plants (4, 6).
The characterization N-linked glycopeptides by mass spectrometry (MS) remains challenging because of physio-chemical properties of the glycopeptide, incomplete fragmentation during CID and micro-heterogeneity of the N-glycan structure (7). Consequently, initial approaches sought to remove the N-glycan structures and profile the resultant peptides by MS or even the released carbohydrate structures themselves. More recently high-resolution MS coupled with complementary fragmentation techniques have enabled the direct characterization of N-glycopeptides. Over the past two decades numerous studies have defined the N-glycan structures in plants by MS (5, 8–10). However, these profiles are not associated with a polypeptide sequence. More recently, several reports have employed endoglycosidases e.g. PNGase A/F, to remove N-glycans from enriched glycopeptide and glycoprotein preparations before their identification by MS (3, 11). Collectively these studies have defined over 2000 N-glycosites from the reference plant Arabidopsis (12). However, because an N-glycosidase was employed to increase peptide identification by MS, these results lack any N-glycan structural information.
In the past year, two studies have characterized intact N-glycopeptides from plants using high resolution MS. A quantitative survey targeting glycoproteins associated with chilling stress in Arabidopsis seedlings identified 105 proteins containing 174 glycosites enriched using hydrophilic interaction chromatography (HILIC) (13). However, the study employed low resolution ion trap-based collision-induced dissociation (CID) with some higher-energy collisional dissociation (HCD) which resulted in a number of unusual N-glycan structures reported (13). Current approaches for N-glycopeptide identifications from complex samples now employ complementary fragmentation techniques, such as electron-transfer dissociation (ETD) and HCD to reveal information about N-glycopeptides for unambiguous assignments (14). Such a strategy was recently applied to Arabidopsis inflorescence samples enriched using wheat germ agglutinin (15). The study characterized 348 glycosites from 270 proteins and highlighted the importance of ETD in the unambiguous assignment of the peptide sequence and the presence of the GlcNAc oxonium ion in HCD spectra. Although the MS analysis was untargeted, the authors reported that over 30% of the unique glycoforms characterized (110 sites) contained a single N-GlcNAc structure and that the high-Man N-glycan structures were the dominant N-glycans in Arabidopsis (15). The study was unable to identify the archetypal and most abundant complex N-glycan in plants, namely the GlcNAc2Man3XylFucGlcNAc2 complex-type, or any structure containing both a Fuc and Xyl moiety (5, 16, 17). The use of lectin weak affinity chromatography likely biased the resultant N-glycopeptide population and although the data set comprising sites and structural heterogeneity is the largest yet reported, it may have inadvertently excluded populations of N-glycans that remained “cryptic” to the lectin, an observation also reported by the authors (15).
In this study, we report the analysis of tryptic N-glycopeptides derived from a microsomal membrane preparation of aerial tissues using HILIC enrichment followed by high resolution tandem MS employing complementary fragmentation techniques (HCD and ETD) to produce a robust and unbiased profile of N-glycopeptides from Arabidopsis. In total, we have reproducibly identified 1110 distinct glycopeptides from over 324 N-glycoproteins from Arabidopsis revealing extensive structural heterogeneity at these sites.
EXPERIMENTAL PROCEDURES
Plant Material and Growth
Arabidopsis thaliana (L.) Heynh. Columbia-0 (Col-0) and xylt seeds were obtained from the Arabidopsis Biological Resource Center (ABRC, http://abrc.osu.edu/). The T-DNA insertion line Salk_04226 (xylt) was used for the β-1,2-xylosyltransferase (XylT) mutant (At5g55500). Plants were grown in growth chamber under long day growth conditions (16 h light and 8 h dark) at 22 °C.
Enrichment of N-glycopeptides from Arabidopsis Seedlings
The aerial part of 3-week-old seedlings (rosette, n = 6) or florets (n = 1) and stems (n = 1) from 6-week-old plants were harvested and microsomal membranes prepared according to previous methods (18). Briefly, around 1 g of Arabidopsis material was harvested and homogenized with a mortar and pestle in 8 ml extraction buffer (50 mm HEPES-KOH (pH 6.8), 0.4 m sucrose, 1 mm dithiothreitol (DTT), 5 mm MnCl2 and 5 mm MgCl2). The homogenate was filtered through two layers of Miracloth and centrifuged at 3000 × g for 10 min. The supernatant was then centrifuged at 100,000 × g for 30 min and the pellet containing endomembrane proteins (around 500 μg of total protein) was resuspended in 100 μl of 7 m urea in 100 mm ammonium bicarbonate. DTT was added to a final concentration of 10 mm and the sample incubated at 60 °C for 1 h. After cooling, iodoacetamide (IAA) was added to a final concentration of 100 mm and incubated at room temperature for 45 min. The sample was diluted to 1 m urea with 100 mm ammonium bicarbonate. Trypsin was added (1:25 w/w) and proteins digested overnight at 37 °C. Acetic acid was added to a final concentration of 1% (v/v) and the digest centrifuged at 13,000 g for 5 min at room temperature. Peptides were purified by Sep-Pak plus C18 cartridges (Waters Corporation). Glycopeptides were batch enriched using a HILIC SPE (50 to 450 μl, The Nest Group). Spin columns were conditioned per supplier instructions, washed with 500 μl of water, twice with 500 μl of 80% acetonitrile and 1% TFA, then the sample added. HILIC spin columns were washed twice with 500 μl of 80% acetonitrile and 1% TFA, and the sample eluted in a stepwise fashion with 200 μl of 70% acetonitrile and 1% TFA, followed by 200 μl 60% acetonitrile and 1% TFA and finally 200 μl 50% acetonitrile and 1% TFA. The enriched N-glycopeptides were dried using a vacuum concentrator and then desalted using ZipTipC18 Pipette Tips (Merck, KGaA) following manufacturer instructions and eluting to a final volume of 25 μl in 0.1% formic acid.
Identification of N-glycopeptides by Tandem Mass Spectrometry
The enriched glycopeptides were analyzed using an Orbitrap Fusion™ Lumos™ Tribrid™ Mass Spectrometer (Thermo Fischer Scientific) fitted with a nano-flow HPLC (Ultimate 3000 RSLC, Thermo Fisher Scientific). The nano-LC system was equipped with an Acclaim Pepmap nano-trap column (Thermo Fisher Scientific - C18, 100 Å, 75 μm × 2 cm) and an Acclaim Pepmap RSLC analytical column (Thermo Fisher Scientific - C18, 100 Å, 75 μm × 50 cm). For each LC-MS/MS experiment, 5 μl of the purified glycopeptide mix was loaded onto the enrichment (trap) column at an isocratic flow of 5 μl min−1 containing 3% acetonitrile and 0.1% formic acid for 6 min before the enrichment column was switched in-line with the analytical column. The eluents used for the LC were 0.1% (v/v) formic acid (solvent A) and 100% acetonitrile/0.1% formic acid (v/v). The gradient applied was 3% B to 20% B for 95 min, 20% B to 40% B in 10 min, 40% B to 80% B in 5 min and maintained at 80% B for the final 5 min before equilibration for 10 min at 3% B. The MS system was operated in positive ion mode at a resolution of 120,000 in full scan mode using data-dependent acquisition (DDA). Two types of MS/MS analysis were performed on samples (supplemental Fig. S2), HCD triggered ETD or ETD only (supplemental Table S1). For ETD only, MS2 ETD was triggered for ions greater than 50,000 with a charge state between 3 and 8, at a resolution of 15,000 and an AGC target of 50,000 and Activation Q of 0.25 using charge dependent reaction times of 11.59 ms (+6), 16.69 ms (+5), 26.08 ms (+4), and 46.37 ms (3+). For HCD triggered ETD, the MS2 was operated in HCD mode with a resolution of 30,000, AGC target of 50,000, Activation Q of 0.25, EThcD (False) and Collision Energy of 30% for ions above 50,000 with a charge state between 3 and 8. ETD fragmentation was undertaken at a resolution of 30,000 using charge dependent reaction times of 11.59 ms (+6), 16.69 ms (+5), 26.08 ms (+4) and 46.37 ms (3+). An AGC target of 300,000 for the precursor ion was triggered when one of the following ions was detected in the top 20 ions in the HCD fragment spectra: 138.0545 (GlcNAc, fragment 1), 163.06 (Hex), 186.076 (GlcNAc, fragment 2), 204.0967 (GlcNAc) or 366.1396 (ManGlcNAc). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (19) partner repository with the data set identifier PXD006270 (http://www.ebi.ac.uk/pride/archive/projects/PXD006270). A key outlining the raw data filenames at the ProteomeXchange to samples described in this study is available in supplemental Table S1.
Spectral Data Interrogation
The spectral data were interrogated using Byonic (Protein Metrics) version 2.6 through Proteome Discoverer™Software (Thermo Fisher Scientific) version 2.1 against The Arabidopsis TAIR10 protein database (20) including standard contaminants (27,416 sequences). A threshold for fragment spectra was applied, where fragment ions less than 1% signal-to-noise were discarded. The Byonic parameters were: cleavage site(s): RK, cleavage side (trypsin): C-terminal, digestion specificity: fully specific, missed cleavages: 2, precursor mass tolerance: 5 ppm, fragment type: both HCD and ETD, fragment mass tolerance for HCD and ETD: 10 ppm and 20 ppm respectively, fixed modifications: Carbamidomethyl/+57.021464 @C and variable modifications using an in-house plant N-glycan database (supplemental Table S2) @ N. Advanced settings included, the charge states (3, 4, 5) apply to unassigned spectra and skip bad spectra; precursor isotope off by X is Too high (wide); maximum precursor mass is 10,000; precursor and charge assignments is compute from MS1; maximum # of precursors per scan is 2; smooth width (m/z) is 0.01; the peptide output options are automatic score cut; the protein output options are protein 1% FDR (or 20 reverse count) calculated using the target/decoy approach. Peptide spectrum matches (PSMs) were exported from the Proteome Discoverer™ Software and imported into KNIME (21). PSMs were filtered to only include peptides with a glycan modification and log probability ( Log Prob ) of > 4 for HCD (p < 0.0001) spectra or > 2 for ETD spectra (p < 0.01). The Log Prob is the absolute value of the log10 of the posterior error probability (PEP), which considers the Byonic score, delta, precursor mass error, digestion specificity, and so forth (10 features in all). This resulted in an FDR < 1% for all PSMs (FDR 2D) (22) and are outlined in supplemental Table S3. Reported glycan structures (composition and linkage) are inferred based on the mass for reported N-glycan structures found in plants as outlined in supplemental Table S2. Annotated spectra for all matches are available at ProteomeXchange (.byrslt file sets) and can be viewed using the Byonic Viewer (https://www.proteinmetrics.com). To compile the final collection of reproducible N-glycopeptides (supplemental Table S4), each PSM had to satisfy the following criteria: have both HCD and ETD matches, unless experimentally validated by a previous Arabidopsis N-glycan study (3, 11, 13, 15). Finally, N-glycopeptides were then only accepted if observed in at least two of the eight biological replicates.
MS Data Processing and Analysis
The areas (XICs) used for occupancy graphs were only obtained for the monoisotopic peak from [M+3H]3+ ions and only from the precursors for identified MS2 spectra (PSMs), using the Precursor Ions Quantifier node in Proteome Discoverer™ Software (Thermo Fisher Scientific) version 2.1. The peak areas were normalized for each separate MS run using the total peak area, then the average normalized peak area was used when N-glycopeptides were observed across multiple runs (minimum of 2). The list of all identified N-glycans meeting these criteria are outlined in supplemental Table S3 in the “Area” column. The subcellular location of proteins was obtained from the SUBcellular Arabidopsis (SUBA) database using the SUBAcon consensus score (23).
Experimental Design and Statistical Rationale
To define the Arabidopsis N-glycoproteome, N-glycopeptides from 8 independent replicates were enriched and analyzed by MS and only N-glycopeptides identified in 2 independent analyses were accepted to define the resultant data set of 1110 glycopeptides (supplemental Fig. S2). A minimum of n = 3 independent biological replicates was employed for STDERR, the (n) employed is detailed in figure legends.
RESULTS
Characterization of N-glycopeptides from Arabidopsis
We employed hydrophilic interaction chromatography (HILIC) to enrich N-glycopeptides from complex lysates. Because N-linked glycosylation occurs in the endomembrane, we isolated Arabidopsis microsomes (3000 to 100,000 × g) from 1 g FW of aerial tissues and digested proteins overnight with trypsin before enrichment of N-glycopeptides using HILIC SPE (supplemental Fig. S2). All samples were analyzed by tandem MS employing HCD product ion triggered ETD on an Orbitrap Fusion Lumos. To obtain further identifications, some samples were analyzed in duplicate or triplicate. Some samples were analyzed using an ETD only method to obtain further complementary fragmentation data (supplemental Fig. S2). A total of six rosette samples were initially analyzed to establish N-glycan diversity. Because a recent profile of N-glycans from floral tissue had indicated that it was dominated by N-GlcNAc and Man-rich structures (15), we also analyzed a sample from flowers. Furthermore, previous reports have indicated that N-glycans structures containing Lea epitopes are only found in specific tissues of Arabidopsis, such as stems (24); therefore, we also enriched N-glycans from this tissue. Analysis of the 8 independent samples (six rosette, one flower and one stem) using a stringent score cut-off yielded over 2159 distinct N-glycopeptides from 556 Arabidopsis proteins with various combinations of HCD and/or ETD fragmentation spectra (supplemental Table S3). Less than 10% of total PSMs in supplemental Table S3 (9168) were matched using ETD spectra. A final validated collection of Arabidopsis N-glycopeptides was obtained by applying the following criteria: a glycopeptide was identified in two independent samples and a glycosite required independent HCD and ETD spectra. The following exception was applied—glycosites with only either HCD or ETD spectra were included if sites had previously been characterized by tandem MS (3, 11, 13, 15). This process yielded 1110 distinct N-glycopeptides comprising 492 N-glycosites and 56 distinct N-glycan structures from 324 Arabidopsis proteins (supplemental Table S4). Of the 1110 N-glycopeptides in this filtered set, over 40% were matched with both HCD and ETD spectra. From the 492 N-glycosites reported here, a total of 476 (97%) had been reported by previous Arabidopsis N-glycoproteomic studies (3, 11, 13, 15).
Types of N-glycans in Arabidopsis
To examine the distribution of N-glycan heterogeneity in Arabidopsis we used the XIC for the filtered wild-type N-glycopeptides (supplemental Table S5) and analyzed the abundance of N-glycan structures found in Arabidopsis (Fig. 1). The high-mannose type of N-glycan comprised around 30% of the structures and are found in the ER and early Golgi (supplemental Fig. S1). This structural class was mainly comprised of Man5GlcNAc2, but also featured other forms including Man6–9GlcNAc2. The most abundant N-glycan structures in Arabidopsis are the complex-types (45%) and are defined as structures produced in the cis-Golgi after GlcNAcylation (25). This structural type is exemplified by GlcNAc2Man3XylFucGlcNAc2, an N-glycan structure that dominated this class along with GlcNAcMan3XylFucGlcNAc2. The hybrid-type structures are intermediate N-glycan structures and are relatively minor components of the glycopeptide population (< 5%), with GlcNAcMan4XylFucGlcNAc2 being the most prominent of this type. The paucimannose structural type represent β-N-acetylhexosaminidase processed N-glycans (25) and are found in around 20% of identified N-glycopeptides. The most abundant example we identified was the Man3XylFucGlcNAc2 structure. The distribution of structures highlighted (Fig. 1) are comparable to that found when N-glycan structures have been hydrolyzed from total protein extracts of Arabidopsis and profiled by MS (Table I). In contrast, a recent study reported that high-mannose structural class dominated the N-glycan population and that the GlcNAc-only class was a prominent structural form in Arabidopsis (15). Although we identified a handful of GlcNAc only N-glycan structures in our survey, their proportion compared with other structural classes is minor (Fig. 1).
Fig. 1.
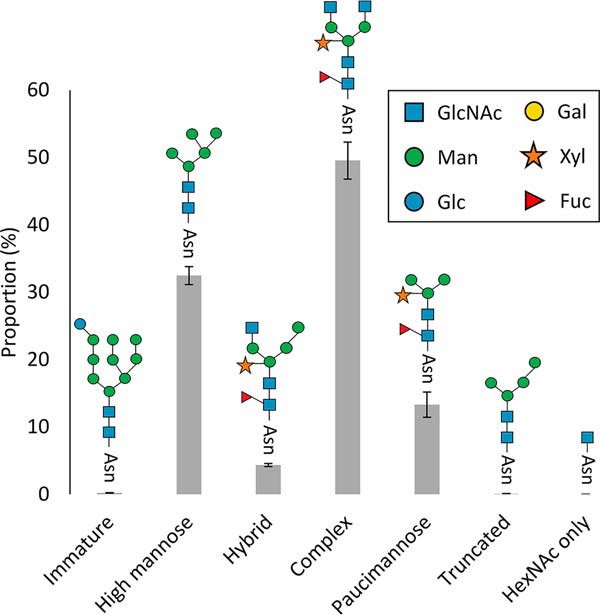
Proportion of N-glycan structures found in Arabidopsis wild-type N-glycopeptides. The 1110 N-glycopeptides identified from rosette, stem, and flower samples were divided into six structural classes (immature, high-mannose, hybrid, complex, paucimannose, truncated, and GlcNAc only) based on previous analyses (supplemental Table S2). The areas (XICs) for each N-glycan structure from a given experiment (n = 8) were extracted and used to calculate the proportion for each structural class. The numbers are the percentage of total normalized monoisotopic peak area for specific glycan types with S.E. The most common representative structure for each N-glycan class is shown.
Table I. Comparison of major classes of N-glycans detected in plants.
| N-glycan Class | This Study (2018) Arabidopsisa | Strasser et al., (2005/2006) (5, 16) Arabidopsisa | Pedersen et al., (2017) (10) Lotus japonicusa | Elbers et al., (2001) (9) Tobaccoa | Ma et al., (2016) (13) Arabidopsisb | Henquet et al., (2008) (8) Arabidopsisa | Yoo et al., (2015) (17) Arabidopsisa | Xu et al., (2016) (15) Arabidopsisb |
|---|---|---|---|---|---|---|---|---|
| Immature | 0.2 | – | – | – | 3.2 | – | – | 2.7 |
| High-mannose | 32.5 | 26.1 | ∼ 49 | 10.7 | 37.7 | 48.0 | ∼ 38 | 43.1 |
| Hybrid | 4.4 | – | – | – | 11.5 | – | – | 2.9 |
| Complex | 49.6 | 45.9 | ∼ 30 | 34.3 | 29.8 | 24.0 | ∼ 39 | - |
| Paucimannose | 13.3 | 28.0 | ∼ 21 | 52.7 | 14.7 | 28.1 | ∼ 23 | 1.5 |
| Truncated | 0.1 | – | – | – | 3.2 | – | – | 25.0 |
| GlcNAc only | – | – | – | – | – | – | – | 24.9 |
aabundances determined by reported XIC.
babundances determined using reported occurrence (number).
The Micro-heterogeneity of N-glycan Structures in Arabidopsis Proteins
An examination of individual proteins revealed that many of the identified sites exhibited varying levels of N-glycan structural micro-heterogeneity. Using the relative abundance (XIC) of each N-glycopeptide within a replicate (supplemental Table S3) and then normalizing these distributions across the six Arabidopsis rosette replicates, we generated a heatmap highlighting the structural heterogeneity found at a given N-glycosite (Fig. 2). N-glycosites exhibited differing patterns ranging from high-mannose structures (e.g. AT2G01720.1, DEIGnISTSHLR) to paucimannose structural types (e.g. AT3G18080.1, nATAEITVDQYHR). These distributions reflected the overall abundances observed in Fig. 1, with minimal proportions of hybrid structures observed for any of these glycosites. Interestingly, we could find examples of different glycosites from the same protein (e.g. AT4G08850.1) harboring different proportions of N-glycan structural types, namely LEnLTLDDNHFEGPVPK which was mainly found with Man9GlcNAc2 (high-mannose) and LnGSIPSEIGR which was observed with GlcNAc2Man3XylFucGlcNAc2, a complex structure (Fig. 2). The heatmap highlights the structural variations found at a given N-glycosite and could reflect a proteins functional state or localization within the endomembrane. For example, in the case of AT4G08850.1 (MIK2) which is a receptor kinase involved in pollen guidance, it appears to exist with two distinct N-glycan structures (immature/high-mannose and complex) at two different N-glycosites. It is conceivable that a significant proportion of this protein remains within the ER under quality control before its release to the plasma membrane.
Fig. 2.
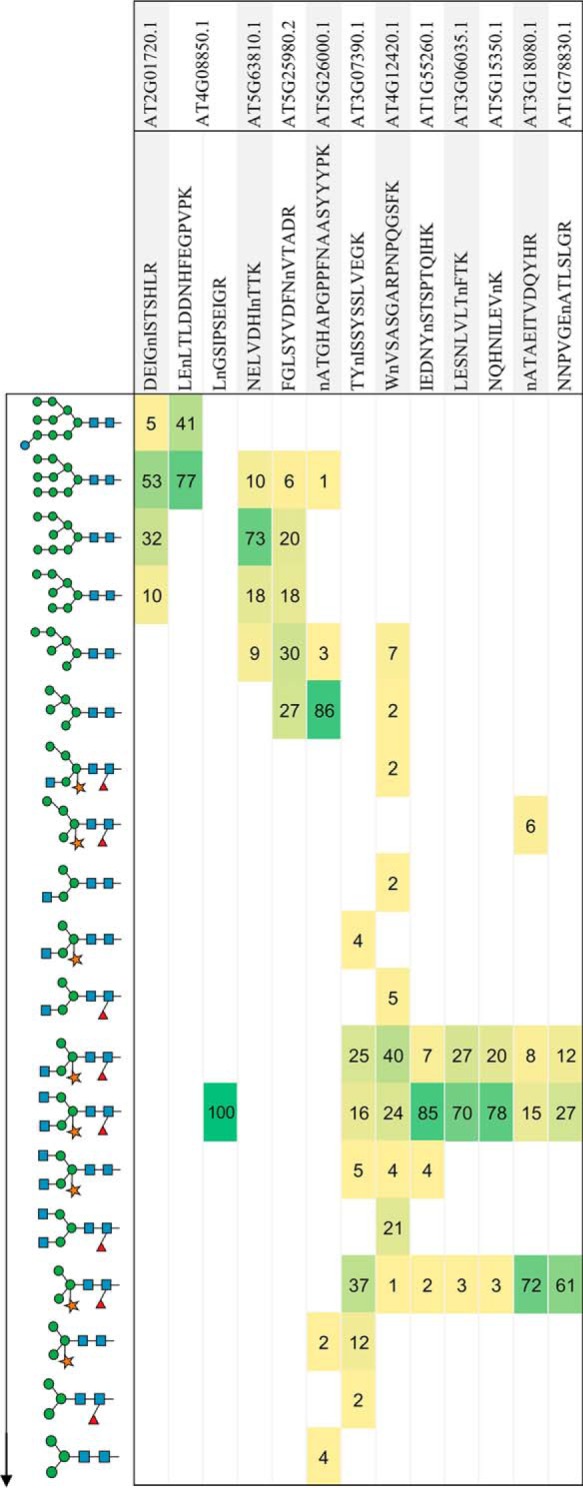
Heatmap highlighting N-glycan heterogeneity of N-glycopeptides. A range of N-glycopeptides were manually selected to highlight the structural heterogeneity patterns observed in glycoproteins from wild-type rosette (n = 6). A proportion was calculated from the area of the XIC for each N-glycan structure for a given glycopeptide for each experiment and then averaged. These patterns highlight a prominent structural class for a given glycopeptide, for example, immature, high-mannose, complex and paucimannose. Structures are shown in a generalized linear sequence representing the maturation of the N-glycan structure in plants (top to bottom).
Subcellular Locations of N-glycan Structures in Arabidopsis
To ascertain whether the type of N-glycan structure revealed information about a protein's subcellular location, we examined the subcellular distributions of identified glycoproteins using their most abundant N-glycan structures as the representative structural type for a given glycoprotein (supplemental Table S5). Using protein subcellular locations as defined by the SUBcellular Arabidopsis database (SUBA), we found that broad N-glycan structural types were indeed associated with distinct subcompartments of the endomembrane system (Fig. 3). Glycoproteins that are localized in the ER mainly contain high-mannose structures, whereas Golgi and plasma membrane localized glycoproteins retain complex structures and glycoproteins destined for the vacuole and extracellular space are dominated by paucimannose structures. These observations are generally in agreement with what is known about the subcellular partitioning of N-glycan biosynthesis in plants (supplemental Fig. S1). The analysis confirms previous findings, such as that the activity of the plasma membrane residing β-N-acetylhexosaminidase (HEXO3) appears to be directed against secreted glycoproteins and not those residing at the plasma membrane (25).
Fig. 3.
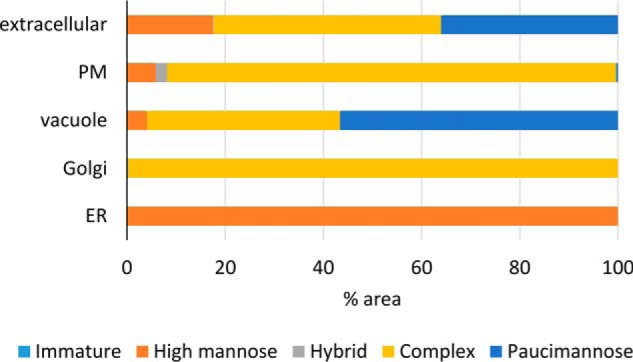
Subcellular distribution N-glycan structural classes. The most prominent N-glycan structural class for a glycoprotein was used as the representative structural form. The area of each glycopeptide (XIC) was used to estimate N-glycan structural proportions in each sample (n = 6, wild-type rosette). Representative N-glycan structures were only assigned if the proportion was greater than 60% averaged across the experiments. A total of 270 proteins were employed in this analysis. The subcellar locations (SUBAcon) were obtained from the SUBA database. Only proteins found in the endomembrane were included, namely extracellular, plasma membrane (PM), vacuole, Golgi, and endoplasmic reticulum (ER).
Structural Heterogeneity of N-glycans from the β-1,2-xylosyltransferase Mutant
To verify our glycopeptide enrichment and profiling approach and to highlight the analytical subtlety of the data, we profiled N-glycopeptides from a mutant in the N-glycan biosynthetic pathway. The Arabidopsis xylt mutant (26) harbors an insertion at the At5g55500 locus which encodes a β-1,2-XylT responsible for the addition of a β-1,2-Xyl to the maturing N-glycan structure within the Golgi apparatus (27, 28). Enriched N-glycopeptides from rosette material from xylt mutants comprising 3 biological replicates were analyzed by MS. Over the three replicates, a total of 363 unique glycosites from 236 proteins were identified (supplemental Table S3). Only those glycosites that had been unambiguously identified in the wild-type samples (supplemental Table S4) were considered for further analyses. To examine structural heterogeneity, areas (XIC) for N-glycopeptides from the xylt mutant and the wild-type rosette samples were extracted as previously described and major N-glycan structures compared (Fig. 4). The comparison indicates that there is little difference in proportions of high-mannose structures observed between the xylt mutant and wild type. However, as expected the production of complex glycan structures containing Xyl was virtually undetectable in the xylt mutants. The inability to add β-1,2-Xyl resulted in a significant increase in the proportion of complex and paucimannose N-glycan structures lacking Xyl e.g. increased abundance of GlcNAc2Man3FucGlcNAc2 and Man3FucGlcNAc2 with associated decrease of GlcNAc2Man3XylFucGlcNAc2 and Man3XylFucGlcNAc2 when compared with wild type (Fig. 4).
Fig. 4.
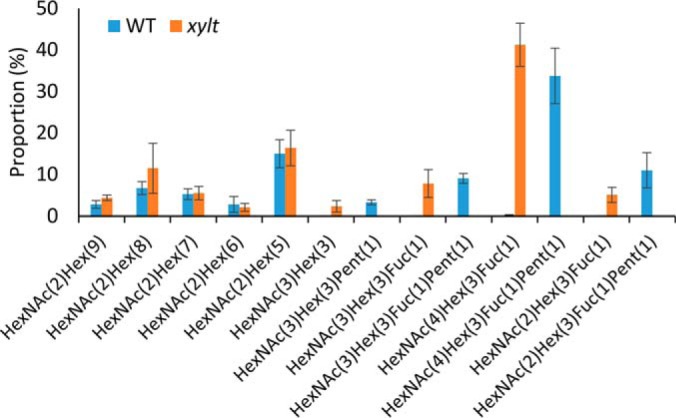
A comparison of N-glycan distributions between wild type and the xylt mutant. The areas (XICs) for N-glycopeptides from wild-type rosette (n = 6) and xylt rosette (n = 3) were extracted and used to calculate the proportion of major structures. The values are the percentage of total normalized monoisotopic peak area for specific glycan types with S.E.
DISCUSSION
The application of an N-glycopeptide enrichment method coupled to high-resolution tandem MS incorporating complementary fragmentation (HCD and ETD) has revealed the extent of N-glycan micro-heterogeneity for nearly 500 N-glycosites from 324 proteins from the reference plant Arabidopsis. Although 97% of these N-glycosites have been previously reported, the depth of data highlights the differential N-glycan maturation process between N-glycoproteins and at specific N-glycosites. The proportion of N-glycan structures reported in this study is very similar to previously reported profiles for N-glycan structures from N-glycoproteins of Arabidopsis (5, 16, 17). This includes the occurrence of Man5GlcNAc2, GlcNAcMan3XylFucGlcNAc2, GlcNAc2Man3XylFucGlcNAc2, and Man3XylFucGlcNAc2 which collectively comprise the majority (ca. 70%) of observed N-glycan structures in wild-type Arabidopsis (Fig. 4).
N-glycan Structures Identified in Arabidopsis
In the past year, few studies have profiled N-glycopeptides and their corresponding structures using enrichment and tandem mass spectrometry. The recent quantitative analysis of N-glycans in response to chilling stress in Arabidopsis highlights the response of glycoproteins and specifically N-glycan structures, under this stress (13). The authors also employed HILIC enrichment of N-glycopeptides from Arabidopsis seedlings and report a collection of 504 N-glycopeptides comprising 174 N-glycosites with around 60% of these sites previously defined (3, 11). The diversity and profile of N-glycan structures reported by Ma et al., (13) is like that described in our report (Table I). There is only about a 12% overlap between N-glycosites outlined in our data set which could be caused by the source material, seedlings versus rosette, stem and florets. However, we did not observe major differences in N-glycan structural profiles or N-glycosites between rosette, flowers or stem material. The proportion and types of N-glycan structures outlined in our study and Ma et al., (13) matches the previously determined structural profiles identified in Arabidopsis (5, 16, 17), namely that the dominant structural class is a complex-type exemplified by GlcNAc2Man3XylFucGlcNAc2 (Table I). These observations contrast with the other recent report that employed high resolution MS with a glycopeptide enrichment strategy incorporating lectin weak affinity chromatography (15). These authors reported that the high-mannose structures dominated their glycopeptide profiles and that truncated N-glycan structures were nearly as abundant. Consequently, we divided our glycopeptides in similar classes for quantitation and comparison (Fig. 1) and specifically sought to profile N-glycans from Arabidopsis inflorescence material (florets) to match the tissue employed between the studies (supplemental Table S3). However, it is clear from our analysis and previous profiles that neither the truncated N-glycan structures nor glycopeptides harboring single GlcNAc residues are abundant in Arabidopsis. It is thus more likely that the lectin enrichment approach selectively enriches specific populations of N-glycopeptides. This preferential enrichment by lectin affinity chromatography for truncated N-glycans is supported by a recent study profiling N-glycopeptides containing single O-GlcNAc residues using the same affinity method (29). Although the N-glycan structural profiles outlined by Xu et al., (15) are enriched for a subpopulation of N-glycan structures, the majority of N-glycosites (76%) had previously been characterized (3), thus supporting the approach. The validity of the sites identified by Xu et al., (15) is exemplified by the confirmed characterization of an N-glycan at a non-consensus site- (Asn-X-Gly) on ATPERX34 (AT3G49120.1) in our study (supplemental Table S4). Thus, we provide independent confirmation of the existence of non-consensus N-glycan sites in plants, previously outlined in other species (3), although the reported N-glycan structure (paucimannose) in our analysis is likely more representative of the structural class at this N-glycosite.
The Lewisa Epitope and Plant N-glycans
This study represents the first report outlining the site-specific mapping and identification of N-glycans with the largest reported glycan structure in plants, those harboring two Lea epitopes and resulting in a Gal2Fuc2GlcNAc2Man3XylFucGlcNAc2 N-glycan structure. The rarity of the Lea structures in our data set is supported by prior reports indicating that the Lea epitope are specific to growth stages in Arabidopsis (root tips, seedlings and stems) and that they are relatively minor structures when profiled (24). Here we have identified three N-glycopeptides with both Lea antennae resulting in the most extensive N-glycan structure reported in plants. The three proteins AT1G52780.1 (DUF2921), AT2G38080.1 (laccase) and AT3G06035.1 (GPI-anchor protein) all appear to be functional members of either the plasma membrane or apoplast as would be expected for proteins harboring Lea epitopes (30). The annotated HCD spectra for the N-glycopeptide identified from AT3G06035.1 (TTQNLTILSK) containing two Lea epitopes is outlined in Fig. 5. The specificity of this structure in Arabidopsis may explain why two recent N-glycoproteomic studies had varying success in identifying N-glycopeptides with Lea epitopes. The study by Ma et al., (13) outlines several N-glycopeptides with Fuc3 but none of these candidates contains a Hex5 which would be required to form both Lea antennae. The report by Xu et al., (15) found no evidence of any peptides containing N-glycans structures with Lea epitopes, which could be caused by their enrichment procedures.
Fig. 5.
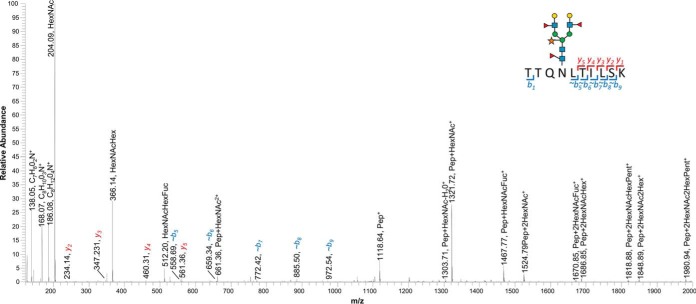
Annotated HCD spectra for an N-glycopeptide TTQNLTILSK containing two Lea epitopes. The precursor ion (1104.486 m/z) was identified in samples WT_Stem, WT_Ros-5 and WT_Ros-4 with resultant HCD spectra from all samples matching a well-characterized N-glycosite (3) from the protein AT3G06035 above the Log Prob threshold.
Subcellular Distribution of Glycoproteins
Extensive work defining subcellular proteomes in the reference plant Arabidopsis (31) allowed us to examine the subcellular distribution of glycoproteins in the endomembrane and relate to this the most prominent N-glycan structure(s). Our analysis demonstrated that the most prominent N-glycan structure is indicative of the functional location of a given glycoprotein within the endomembrane system. Thus, not surprisingly, glycoproteins with high-mannose structures were most likely to be associated with the ER, whereas proteins containing complex N-glycans were associated with the Golgi/plasma membrane and paucimannose structures were found associated with vacuolar and extracellular glycoproteins. Such conclusions are consistent with the current processes related to the partitioning of the N-glycan maturation process and protein secretion in plants (6, 25, 32). However, some protein trafficking can be achieved by unconventional protein secretion (UPS) pathways that bypass the Golgi (33, 34). Thus, a more detailed examination of these data could provide mechanisms to identify proteins that follow UPS pathways or sequestered proteins awaiting release or information about tertiary structures. For example, β-GALACTOSIDASE 10 (ATBGAL10, AT5G63810.1) has been identified in the extracellular proteome of Arabidopsis (31) and appears to be the main β-galactosidase acting on xyloglucan in the cell wall (35). ATBGAL10 contains three distinct N-glycosites, two with multiple high-mannose structures and the third (with few spectra) with an expected paucimannose structure (supplemental Table S4). The presence of these high-mannose structures could indicate that ATBGAL10 is either following a UPS pathway, is regulated in ER release or that these N-glycosites are folded in the protein before exit from the ER and unavailable for processing by Golgi resident mannosidases.
Profiling the N-glycans of the β-1,2-xylosyltransferase Mutant
The Arabidopsis β-1,2- XylT belongs to glycosyltransferase (GT) family 61 and is responsible for the xylosylation of N-glycans (27). Arabidopsis XylT mutant plants do not exhibit altered phenotypes when grown under standard growth conditions, even with a complete absence of Xyl containing N-glycans (17, 26). A previous assessment of N-glycan profiles from xylt-1 plants had demonstrated that aside from the elimination of Xyl containing structures, the profiles were remarkably similar to wild-type samples (17). Our analysis has confirmed the absence of Xyl from N-glycopeptides in xylt plants providing a level of detail not previously outlined for any N-glycan pathway mutant in plants. The elimination of Xyl residues only had a minor impact on the resulting profile of N-glycan structures when compared with their wild-type counterparts. These minor differences could be attributed to the suboptimal activity of GnTII on structures that lack Xyl (17), resulting in a reduction in the rate of N-glycan maturation.
CONCLUSION
Protein glycosylation is a unique PTM because of extensive variations that can occur in the N-glycan structure. Consequently, mapping N-glycosites provides an incomplete picture about the composition of the PTM. In this report, we have outlined the benefits of uncovering structural heterogeneity of N-glycosites in the reference plant Arabidopsis using high-resolution tandem, MS coupled to complementary fragmentation techniques. The data significantly expands current knowledge in this area and clarifies recent observations concerning the types of N-glycan structures in plants.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD006270 (http://www.ebi.ac.uk/pride/archive/projects/PXD006270).
Supplementary Material
Acknowledgments
We thank Ching-Seng Ang at the Mass Spectrometry and Proteomics Facility, Bio21 Molecular Science & Biotechnology Institute (University of Melbourne) for assistance with mass spectrometry.
Footnotes
* This work was supported by Australian Research Council (ARC) Centre of Excellence in Plant Cell Walls grant [CE11091007]. JLH was supported by Australian Research Council Future Fellowships [FT130101165]. The work was also supported by the DOE Joint BioEnergy Institute (http://www.jbei.org) supported by the U. S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U. S. Department of Energy.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- GlcNAc
- N-acetylglucosamine
- ER
- endoplasmic reticulum
- DolP
- dolichol phosphate
- OST
- oligosaccharyltransferase
- CNX
- calnexin
- CRT
- calreticulin
- ERQC
- ER quality control
- HILIC
- hydrophilic interaction chromatography
- CID
- collision-induced dissociation
- HCD
- higher-energy collisional dissociation
- PSM
- peptide spectral matches
- ETD
- electron-transfer dissociation
- Man
- mannose
- Glc
- glucose
- Xyl
- xylose
- Fuc
- fucose
- Gal
- galactose
- N
- asparagine
- MS
- mass spectrometry
- MS/MS
- tandem mass spectrometry
- STDERR
- standard error
- XylT
- Beta-1,2-xylosyltransferase
- XIC
- extracted ion chromatogram
- T-DNA
- transfer deoxyribonucleic acid
- Col-0
- Columbio-0
- ABRC
- Arabidopsis Biological Resource Center
- DTT
- dithiothreitol
- HEPES
- 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- PNGase
- endoglycosidase
- TFA
- trifluoroacetic acid
- LC
- liquid chromatography
- PEP
- posterior error probability
- ppm
- parts per million
- Prob
- probability
- FDR
- false discovery rate
- SUBA
- subcellular Arabidopsis database
- ca
- circa
- ATBGAL10
- Arabidopsis thaliana beta-galactosidase 10
- Lea
- Lewis a epitope
- DUF
- domain of unknown function
- GPI
- glycophosphatidylinositol hexnac, N-acetylhexoseamine
- HEXO3
- beta-N-acetylhexosaminidase
- EThcD
- Electron-Transfer/Higher-Energy Collision Dissociation
- MS2
- second stage of mass spectrometry
- MS1
- first stage of mass spectrometry
- KNIME
- Konstanz information miner
- MIK2
- MDIS1-interacting receptor like kinase2
- SUBAcon
- SUBA consensus
- PTM
- post-translational modification
- AGC
- Automatic Gain Control
- PRIDE
- PRoteomics IDEntifications.
REFERENCES
- 1. Hebert D. N., Lamriben L., Powers E. T., and Kelly J. W. (2014) The intrinsic and extrinsic effects of N-linked glycans on glycoproteostasis. Nat. Chem. Biol. 10, 902–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jarrell K. F., Ding Y., Meyer B. H., Albers S. V., Kaminski L., and Eichler J. (2014) N-linked glycosylation in Archaea: a structural, functional, and genetic analysis. Microbiol. Mol. Biol. Rev. 78, 304–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zielinska D. F., Gnad F., Schropp K., Wisniewski J. R., and Mann M. (2012) Mapping N-glycosylation sites across seven evolutionarily distant species reveals a divergent substrate proteome despite a common core machinery. Mol. Cell 46, 542–548 [DOI] [PubMed] [Google Scholar]
- 4. Lannoo N., and Van Damme E. J. (2015) N-glycans: The making of a varied toolbox. Plant Sci. 239, 67–83 [DOI] [PubMed] [Google Scholar]
- 5. Strasser R., Stadlmann J., Svoboda B., Altmann F., Glossl J., and Mach L. (2005) Molecular basis of N-acetylglucosaminyltransferase I deficiency in Arabidopsis thaliana plants lacking complex N-glycans. Biochem. J. 387, 385–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Strasser R. (2016) Plant protein glycosylation. Glycobiology 26, 926–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ford K. L., Zeng W., Heazlewood J. L., and Bacic A. (2015) Characterization of protein N-glycosylation by tandem mass spectrometry using complementary fragmentation techniques. Front. Plant Sci. 6, 674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Henquet M., Lehle L., Schreuder M., Rouwendal G., Molthoff J., Helsper J., van der Krol S., and Bosch D. (2008) Identification of the gene encoding the alpha 1,3-mannosyltransferase (ALG3) in Arabidopsis and characterization of downstream N-glycan processing. Plant Cell 20, 1652–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Elbers I. J. W., Stoopen G. M., Bakker H., Stevens L. H., Bardor M., Molthoff J. W., Jordi W. J. R. M., Bosch D., and Lommen A. (2001) Influence of growth conditions and developmental stage on N-glycan heterogeneity of transgenic immunoglobulin G and endogenous proteins in tobacco leaves. Plant Physiol. 126, 1314–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pedersen C. T., Loke I., Lorentzen A., Wolf S., Kamble M., Kristensen S. K., Munch D., Radutoiu S., Spillner E., Roepstorff P., Thaysen-Andersen M., Stougaard J., and Dam S. (2017) N-glycan maturation mutants in Lotus japonicus for basic and applied glycoprotein research. Plant J. 91, 394–407 [DOI] [PubMed] [Google Scholar]
- 11. Song W., Mentink R. A., Henquet M. G., Cordewener J. H., van Dijk A. D., Bosch D., America A. H., and van der Krol A. R. (2013) N-glycan occupancy of Arabidopsis N-glycoproteins. J. Proteomics 93, 343–355 [DOI] [PubMed] [Google Scholar]
- 12. Mann G. W., Calley P. C., Joshi H. J., and Heazlewood J. L. (2013) MASCP gator: an overview of the Arabidopsis proteomic aggregation portal. Front. Plant Sci. 4, 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ma J., Wang D., She J., Li J., Zhu J.-K., and She Y.-M. (2016) Endoplasmic reticulum-associated N-glycan degradation of cold-upregulated glycoproteins in response to chilling stress in Arabidopsis. New Phytol. 212, 282–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Desaire H. (2013) Glycopeptide analysis, recent developments and applications. Mol. Cell. Proteomics 12, 893–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu S. L., Medzihradszky K. F., Wang Z. Y., Burlingame A. L., and Chalkley R. J. (2016) N-glycopeptide profiling in Arabidopsis inflorescence. Mol. Cell. Proteomics 15, 2048–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Strasser R., Schoberer J., Jin C., Glossl J., Mach L., and Steinkellner H. (2006) Molecular cloning and characterization of Arabidopsis thaliana Golgi alpha-mannosidase II, a key enzyme in the formation of complex N-glycans in plants. Plant J. 45, 789–803 [DOI] [PubMed] [Google Scholar]
- 17. Yoo J. Y., Ko K. S., Seo H. K., Park S., Fanata W. I., Harmoko R., Ramasamy N. K., Thulasinathan T., Mengiste T., Lim J. M., Lee S. Y., and Lee K. O. (2015) Limited addition of the 6-Arm b1,2-linked N-acetylglucosamine (GlcNAc) residue facilitates the formation of the largest N-glycan in plants. J. Biol. Chem. 290, 16560–16572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zeng W., Ebert B., Parsons H. T., Rautengarten C., Bacic A., and Heazlewood J. L. (2017) Enrichment of Golgi membranes from Triticum aestivum (wheat) seedlings. In: Taylor N. L., and Millar A. H., eds. The Isolation of Plant Organelles and Structures: Methods and Protocols, pp. 131–150, Humana Press, New York: [DOI] [PubMed] [Google Scholar]
- 19. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lamesch P., Berardini T. Z., Li D., Swarbreck D., Wilks C., Sasidharan R., Muller R., Dreher K., Alexander D. L., Garcia-Hernandez M., Karthikeyan A. S., Lee C. H., Nelson W. D., Ploetz L., Singh S., Wensel A., and Huala E. (2012) The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Berthold M. R., Cebron N., Dill F., Gabriel T. R., Kotter T., Meinl T., Ohl P., Sieb C., Thiel K., and Wiswedel B. (2008) KNIME: The Konstanz Information Miner. In: Preisach C., Burkhardt H., Schmidt-Thieme L., and Decker R., eds. Data Analysis, Machine Learning and Applications. Studies in Classification, Data Analysis, and Knowledge Organization, pp. 319–326, Springer, Berlin, Heidelberg [Google Scholar]
- 22. Bern M. W., and Kil Y. J. (2011) Two-dimensional target decoy strategy for shotgun proteomics. J. Proteome Res. 10, 5296–5301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hooper C. M., Tanz S. K., Castleden I. R., Vacher M. A., Small I. D., and Millar A. H. (2014) SUBAcon: a consensus algorithm for unifying the subcellular localization data of the Arabidopsis proteome. Bioinformatics 30, 3356–3364 [DOI] [PubMed] [Google Scholar]
- 24. Strasser R., Bondili J. S., Vavra U., Schoberer J., Svoboda B., Glössl J., Léonard R., Stadlmann J., Altmann F., Steinkellner H., and Mach L. (2007) A unique β1,3-galactosyltransferase is indispensable for the biosynthesis of N-glycans containing Lewis a structures in Arabidopsis thaliana. Plant Cell 19, 2278–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liebminger E., Veit C., Pabst M., Batoux M., Zipfel C., Altmann F., Mach L., and Strasser R. (2011) β-N-acetylhexosaminidases HEXO1 and HEXO3 are responsible for the formation of paucimannosidic N-glycans in Arabidopsis thaliana. J. Biol. Chem. 286, 10793–10802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Strasser R., Altmann F., Mach L., Glossl J., and Steinkellner H. (2004) Generation of Arabidopsis thaliana plants with complex N-glycans lacking b1,2-linked xylose and core a1,3-linked fucose. FEBS Lett. 561, 132–136 [DOI] [PubMed] [Google Scholar]
- 27. Strasser R., Mucha J., Mach L., Altmann F., Wilson I. B., Glossl J., and Steinkellner H. (2000) Molecular cloning and functional expression of β1, 2-xylosyltransferase cDNA from Arabidopsis thaliana. FEBS Lett. 472, 105–108 [DOI] [PubMed] [Google Scholar]
- 28. Saint-Jore-Dupas C., Nebenfuhr A., Boulaflous A., Follet-Gueye M. L., Plasson C., Hawes C., Driouich A., Faye L., and Gomord V. (2006) Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway. Plant Cell 18, 3182–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu S. L., Chalkley R. J., Maynard J. C., Wang W., Ni W., Jiang X., Shin K., Cheng L., Savage D., Huhmer A. F., Burlingame A. L., and Wang Z. Y. (2017) Proteomic analysis reveals O-GlcNAc modification on proteins with key regulatory functions in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 114, E1536–E1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fitchette A. C., Cabanes-Macheteau M., Marvin L., Martin B., Satiat-Jeunemaitre B., Gomord V., Crooks K., Lerouge P., Faye L., and Hawes C. (1999) Biosynthesis and immunolocalization of Lewis a-containing N-glycans in the plant cell. Plant Physiol. 121, 333–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hooper C. M., Castleden I. R., Tanz S. K., Aryamanesh N., and Millar A. H. (2017) SUBA4: the interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res. 45, D1064–D1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shin Y. J., Castilho A., Dicker M., Sadio F., Vavra U., Grunwald-Gruber C., Kwon T. H., Altmann F., Steinkellner H., and Strasser R. (2017) Reduced paucimannosidic N-glycan formation by suppression of a specific beta-hexosaminidase from Nicotiana benthamiana. Plant Biotechnol. J. 15, 197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ding Y., Wang J., Wang J., Stierhof Y. D., Robinson D. G., and Jiang L. (2012) Unconventional protein secretion. Trends Plant Sci. 17, 606–615 [DOI] [PubMed] [Google Scholar]
- 34. van de Meene A. M., Doblin M. S., and Bacic A. (2017) The plant secretory pathway seen through the lens of the cell wall. Protoplasma 254, 75–94 [DOI] [PubMed] [Google Scholar]
- 35. Sampedro J., Gianzo C., Iglesias N., Guitian E., Revilla G., and Zarra I. (2012) AtBGAL10 is the main xyloglucan beta-galactosidase in Arabidopsis, and its absence results in unusual xyloglucan subunits and growth defects. Plant Physiol. 158, 1146–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD006270 (http://www.ebi.ac.uk/pride/archive/projects/PXD006270).