Abstract
Oncogene-induced DNA replication stress contributes critically to the genomic instability present in cancer1–4. However, elucidating how oncogenes deregulate DNA replication has been impeded by the difficulty in mapping replication initiation sites on the human genome. In this study, using a sensitive assay to monitor nascent DNA synthesis in early S phase, we identified thousands of replication initiation sites in cells before and after induction of the oncogenes CCNE1 or MYC. Remarkably, both oncogenes induced firing of a novel set of DNA replication origins that mapped within highly transcribed genes. These ectopic origins were normally suppressed by transcription during G1, but precocious entry into S phase, prior to all genic regions having been transcribed, allowed firing of origins within genes in cells with activated oncogenes. Forks from oncogene-induced origins were prone to collapse, as a result of conflicts between replication and transcription, and were associated with DNA double-strand break formation and chromosomal rearrangement breakpoints both in our experimental system and in a large cohort of human cancers. Thus, firing of intragenic origins caused by premature S phase entry represents a mechanism of oncogene-induced DNA replication stress that is relevant for genomic instability in human cancer.
Studies addressing the mechanisms underlying oncogene-induced DNA replication stress have implicated as possible causes: reduced or increased origin firing, depletion of nucleotides, shortage of replication factors, reduced fork elongation rates, increased transcription and replication-transcription conflicts 5–13. However, no study has yet mapped, in a genome-wide manner, DNA replication and transcription in cells, before and after oncogene activation, which could provide important insights towards understanding oncogene-induced DNA replication stress.
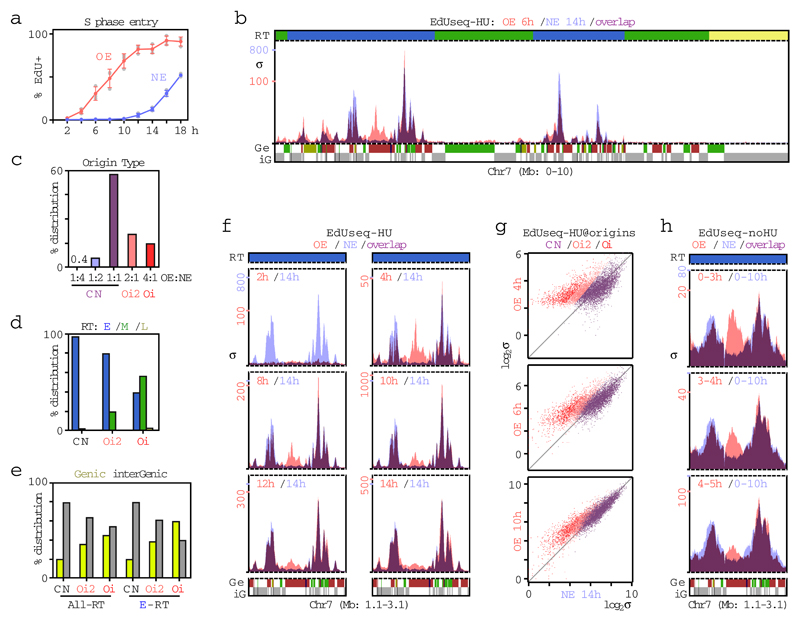
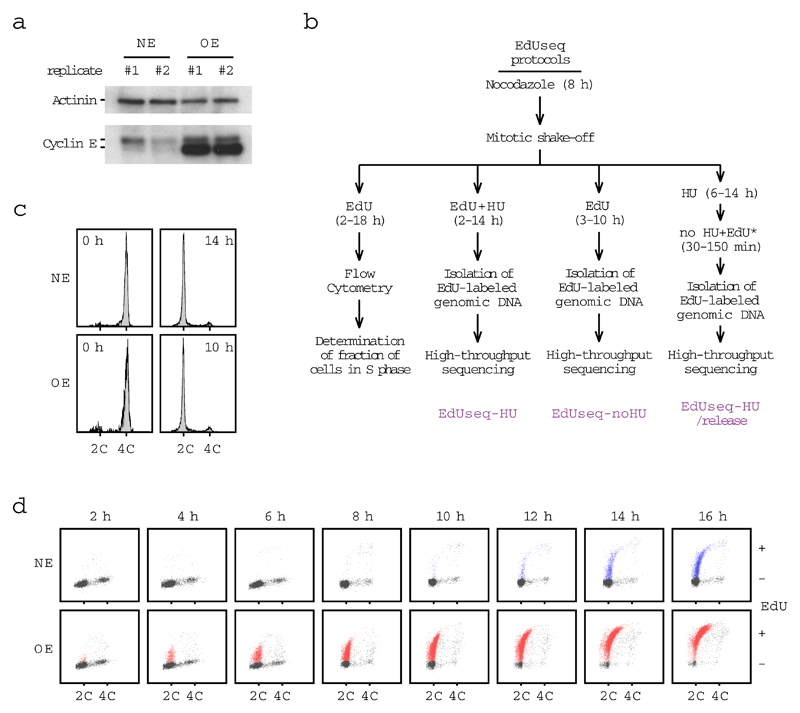
We studied U2OS human cell lines, in which inducible activation of the proto-oncogenes CCNE1 (cyclin E) or MYC leads to DNA replication stress6,13–15. Amplification of the CCNE1 and MYC genes are among the most frequent genetic changes in human cancers2–4. We first focused on the cyclin E system. As previously shown5,14,16, overexpression of cyclin E shortened the length of G1, from about 10-12 h for cells with normal cyclin E activity, to as little as 2-4 h (Fig. 1a and Extended Data Fig. 1). To examine DNA replication initiation (origin firing), cells with normal levels of cyclin E and cells overexpressing cyclin E were harvested by mitotic shake-off and allowed to proceed through the cell cycle in the presence of 5-ethynyl-2’-deoxyuridine (EdU) and hydroxyurea (HU) (refs 17–19). EdU-labeled DNA was isolated and subjected to high-throughput sequencing (EdUseq-HU; Extended Data Fig. 1b, c). The data, analyzed at a genomic bin resolution of 10 Kb, yielded well-resolved peaks, corresponding to the regions where DNA replication initiated (Fig. 1b and Supplementary Table 1). Of 6,164 identified peaks, 927 were induced strongly, at least four-fold, in the cyclin E overexpressing cells and will be referred to as oncogene-induced (Oi) origins; 1,281 were induced modestly by cyclin E overexpression and will be referred to as Oi2 origins; and 3,956 were of similar magnitude in the normal and cyclin E overexpressing cells and will be referred to as constitutive origins (Fig. 1b, c).
Figure 1. Firing of novel origins upon cyclin E overexpression.
a, Percentage of EdU positive cells (mean and SD; n=3 independent experiments; gray symbols, individual data points) at the indicated times after mitotic shake-off. OE, overexpression of cyclin E; NE, normal levels of cyclin E.
b, Replication initiation profiles (EdUseq-HU) at a representative genomic region in OE and NE cells harvested 6 and 14 h after mitotic shake-off, respectively. RT, replication timing (blue, early; green, mid; yellow, late S phase); Ge, genes (green, forward direction of transcription; red, reverse; yellow, unspecified; blue, multiple genes within bin); iG, intergenic (gray). Bin resolution, 10 Kb; ruler scale, 100 Kb; σ, sigma.
c, Classification of origins based on adjusted σ value ratios in OE over NE cells: CN, constitutive < 2-fold; Oi2, oncogene-induced 2 > 2-fold; Oi, oncogene-induced > 4-fold.
d, e, Distribution of CN, Oi2 and Oi origins according to RT (d) (E, early; M, mid; L, late S phase) and gene annotation (e) (All-RT, all RT domains; E-RT, early S RT genomic domains).
f, g, Replication initiation profiles (EdUseq-HU) at a representative genomic region (f) and scatter plots of EdUseq-HU values at all origins (g) in NE and OE cells at the indicated times after mitotic shake-off.
h, Replication initiation profiles (EdUseq-noHU) at a representative genomic region in OE and NE cells. EdU was present during the indicated times following mitotic shake-off.
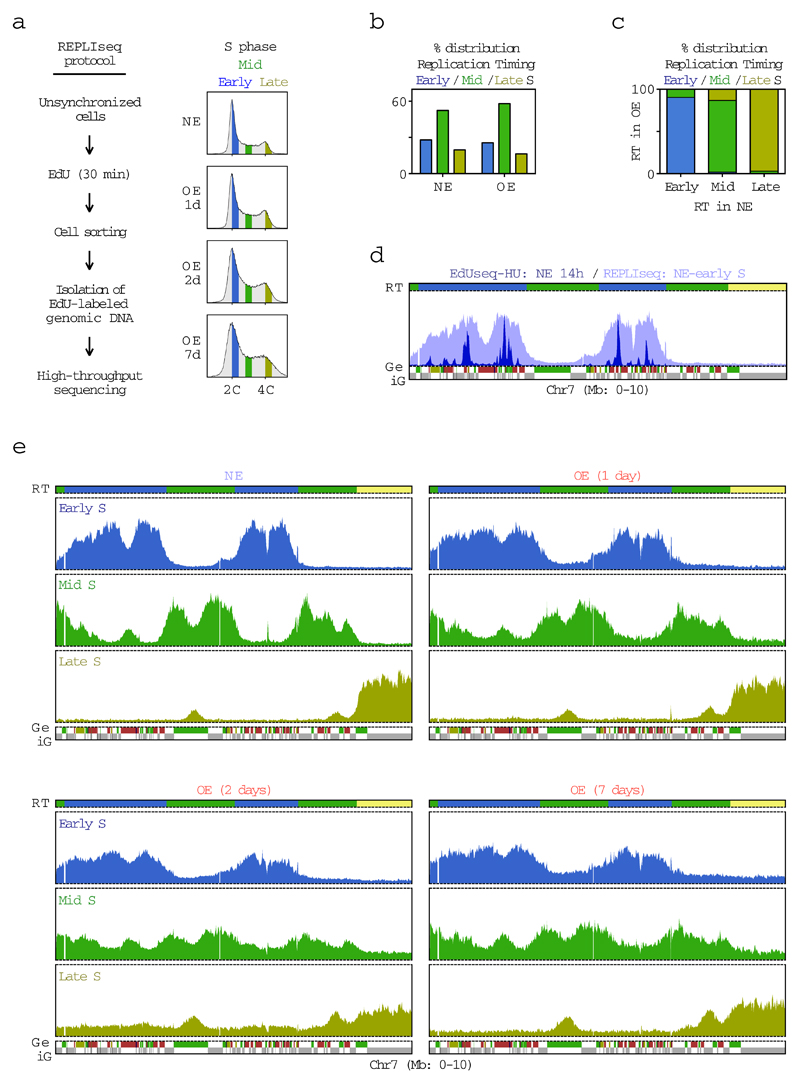
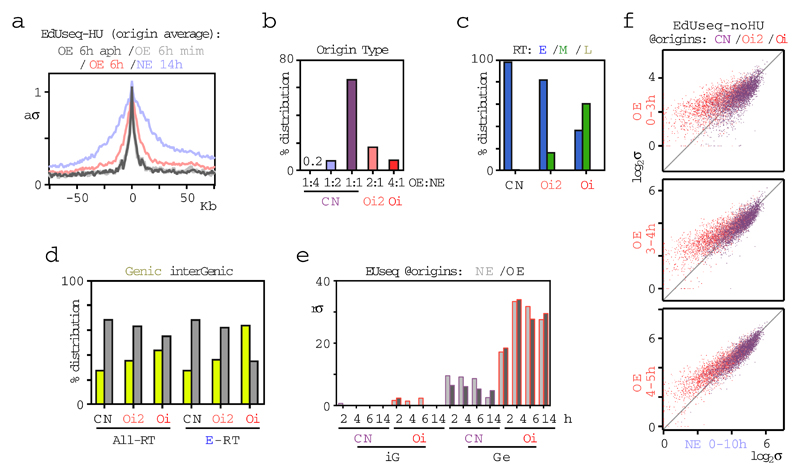
To determine whether constitutive and Oi origins were qualitatively different, we examined their genomic distribution in relation to replication timing and gene annotation. Early, mid and late S phase replicating domains were mapped by REPLIseq (ref. 20) (Extended Data Fig. 2). The constitutive origins were present exclusively in early S domains, as expected of cells that had just entered S phase, whereas, the Oi origins exhibited a broader distribution encompassing early and mid S domains (Fig. 1d). In regard to gene annotation, while the constitutive origins mapped predominantly to intergenic regions, a substantial fraction of the Oi origins, particularly those in early S domains, mapped within protein-coding genes (Fig. 1e). Similar results were obtained when the resolution of origin mapping was increased from 10 Kb to 1 Kb by treating the cells with mimosine or aphidicolin, in addition to HU, to more robustly arrest fork progression after origin firing (Extended Data Fig. 3 and Supplementary Table 2).
For the cells overexpressing cyclin E, origin firing was examined at multiple time points after mitotic exit, revealing that the Oi origins fired predominantly in the cells with the shortest G1 phases (less than 6 h) (Fig. 1f, g and Supplementary Table 3). As expected, origin firing was not observed prior to S phase entry, i.e. within the first 2 h after mitotic exit. Furthermore, the Oi origins initiated DNA replication also in the absence of HU, arguing that they were not dormant origins (Fig. 1h and Extended Data Fig. 3f). Thus, oncogene activation led to the firing of novel replication origins within genomic domains normally devoid of replication initiation in cells that entered S phase prematurely.
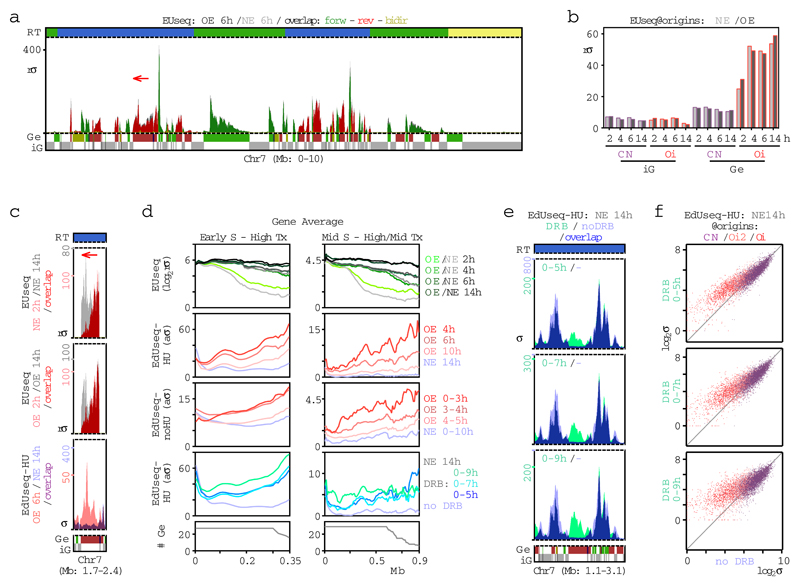
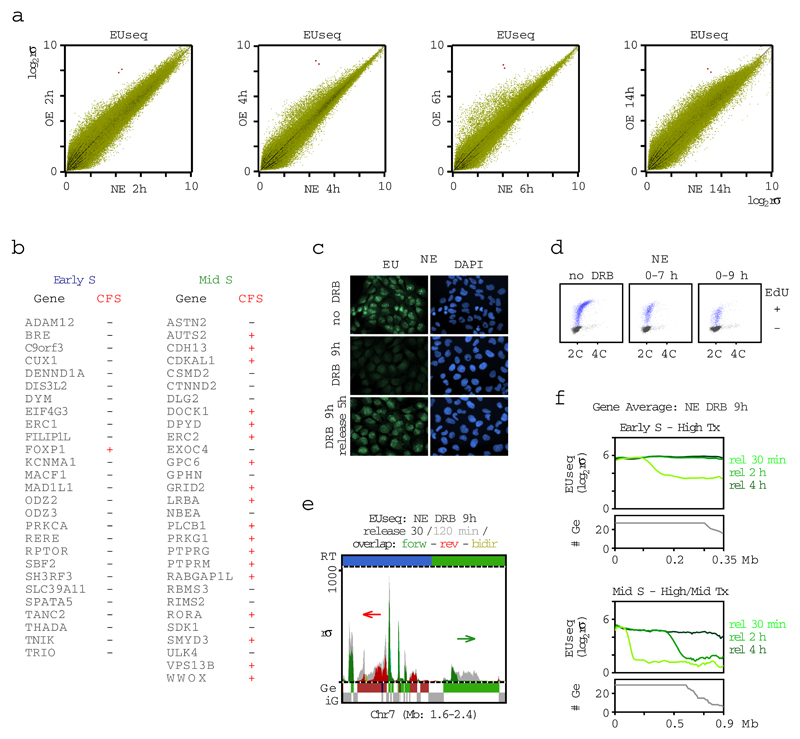
The aberrant firing of intragenic Oi origins could be related to deregulation of transcription, but the newly-synthesized transcript profiles (EUseq) of cells examined at multiple time points after mitotic exit, showed that cyclin E overexpression did not impact transcription genome-wide (Fig. 2a and Extended Data Fig. 4a) or at the genomic sites where the Oi origins fired (Fig. 2b and Extended Data Fig. 3e). Interestingly, whereas the constitutive origins, including the genic ones, mapped to non-transcribed or weakly transcribed regions, the genic Oi origins mapped to sites that were highly transcribed at all time points examined, except for the early time point of 2 h after mitotic exit (Fig. 2b and Extended Data Fig. 3e). Thus, replication at the sites of Oi origins may initiate before these sites are transcribed. Indeed, specific examples (Fig. 2c) and averages of transcription and replication along large genes (Fig. 2d and Extended Data Fig. 4b) revealed that 2 h after mitotic exit, the transcription wave front had not yet reached the 3’ end of genes, where most of the Oi origins were located.
Figure 2. Suppression of intragenic origin firing by transcription.
a, Newly-synthesized transcript profiles (EUseq) at a representative genomic region in cells with normal cyclin E (NE) levels or overexpressing cyclin E (OE), 6 h after mitotic shake-off (OE: dark gray; NE: light gray; overlap: color; direction of transcripts: green, forward; red, reverse; yellow, bidirectional). Replication timing (RT) and gene (Ge/iG) annotations are as in Fig. 1b. rσ, relative sigma.
b, Median transcript levels (EUseq) at the genomic bins corresponding to genic (Ge) and intergenic (iG), constitutive (CN) and oncogene-induced (Oi) origins in NE (light) and OE (dark gray) cells at the indicated times after mitotic shake-off.
c, Transcription (EUseq) and replication initiation (EdUseq-HU) profiles at the indicated times after mitotic shake-off for the gene marked by the arrow in (a).
d, Average transcription (EUseq) and replication initiation (EdUseq-HU or EdUseq-noHU) along the length of large (>0.35 Mb for early S; >0.65 Mb for mid S), transcribed (Tx) genes in OE, NE and DRB-treated NE cells at various time points after mitotic shake-off. High Tx, upper tercile; mid Tx, middle tercile; rσ, relative σ; aσ, adjusted σ; #Ge, number of averaged genes.
e, f, Replication initiation (EdUseq-HU) profiles at a representative genomic region (e) and scatter plots of EdUseq-HU values at all origins (f) in control (no DRB) and DRB-treated (0-5, 0-7 or 0-9 h) NE cells harvested 14 h after mitotic shake-off.
Transcription has been proposed to inactivate intragenic origins prior to S phase entry21,22. Thus, one interpretation of our results is that intragenic origins fired upon oncogene-induced premature S phase entry, because transcription did not have the time needed to reach the end of the transcription units. To test this hypothesis, we used 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) to inhibit transcription elongation for the first 5, 7 or 9 h of G1 in cells with normal levels of cyclin E. The DRB treatment did not decrease the length of G1, but it reduced the time during which transcription was active in G1 (Extended Data Fig. 4c-f), and, consistent with our hypothesis, it led to firing of intragenic origins at the same genomic positions as the Oi origins (Fig. 2d-f).
Inducible activation of the proto-oncogene MYC in U2OS cells led to a similar shortening of the G1 phase and firing of intragenic Oi origins, many of which overlapped with the cyclin E-induced Oi origins (Extended Data Fig. 5). Myc activation affected transcription more broadly than cyclin E overexpression, but, as observed in the cyclin E system, constitutive and Oi origins were associated with sites of low and high transcriptional activity, respectively, highlighting the mechanistic similarities between cyclin E and Myc-induced intragenic origin firing (Extended Data Fig. 5i-k).
HeLa cells, a well-characterized cancer cell line, entered S phase with similar kinetics as U2OS cells overexpressing cyclin E. Analysis of DNA replication initiation at 6 and 14 h after mitotic shake-off, revealed firing of the same Oi origins, as characterized in the U2OS cell systems, for the cells with the shortest G1 phases (Extended Data Figs 6 and 7). In contrast, Oi origins did not fire in RPE1 cells, which are not transformed and which have long G1 phases (Extended Data Figs 6 and 7). Notwithstanding the differences in Oi origin firing, the majority of constitutive origins were shared in all cell lines examined (Extended Data Fig. 5h, 6 and 7).
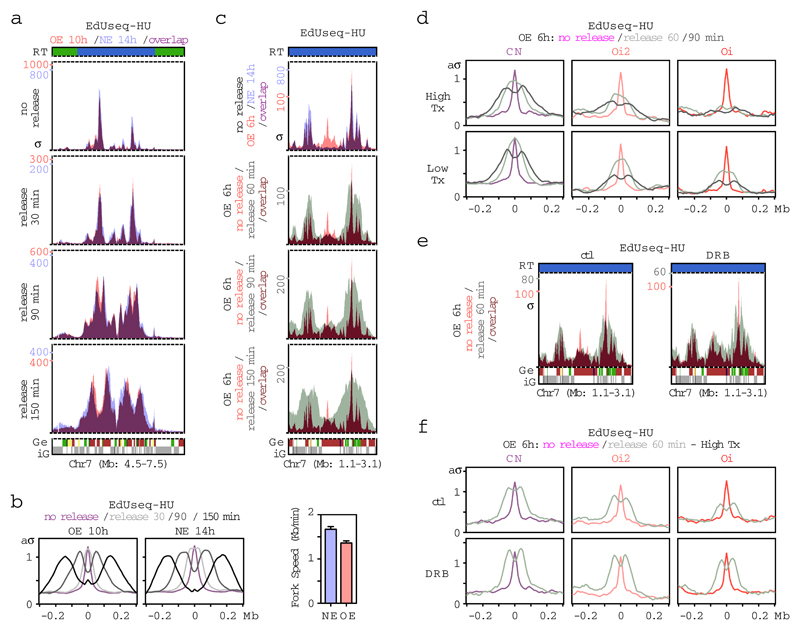
To determine whether Oi origin firing could lead to DNA replication stress, we compared fork progression from constitutive and Oi origins. To monitor constitutive origins, cells expressing normal or high levels of cyclin E were arrested with HU for 14 and 10 h after mitotic exit, respectively, and then released from the HU block for various time periods (30 min-2.5 h) with EdU being added for the last 30 min before harvesting the cells (EdUseq-HU/release) (Fig. 3a). There was robust incorporation of EdU label around the constitutive origins at all release time points examined. Although fork progression from constitutive origins was slower in cyclin E overexpressing cells, previously attributed to the increased number of origins6,7, there was no evidence of fork collapse (Fig. 3a, b). To monitor Oi origins, we repeated the experiment with cyclin E overexpressing cells that were arrested with HU for only 6 h after mitotic exit, a time point at which Oi origins have fired with high efficiency. Interestingly, the Oi origins failed to recover from the HU block, indicating fork collapse, whereas, in the same cells, forks from the constitutive origins did not collapse (Fig. 3c, d and Supplementary Table 4). The degree of fork collapse correlated with the level of transcription at the sites of Oi origins (Fig. 3d) and, treating the cells with the transcription inhibitor DRB as the cells were entering S phase, rescued fork collapse (Fig. 3e, f and Supplementary Table 4), suggesting that replication-transcription conflicts were the underlying cause. This conclusion was further supported by analysis of cells with normal levels of cyclin E, in which Oi origins were induced to fire by inhibiting transcription in early G1 (Extended Data Fig. 8).
Figure 3. Collapse of Oi forks due to conflicts with transcription.
a, Fork progression profiles (EdUseq-HU/release) at a representative genomic region in cells overexpressing cyclin E (OE) or with normal cyclin E (NE) levels that were arrested with HU for 10 or 14 h, respectively, after mitotic shake-off and then released for the indicated time periods. EdU was added 30 min before harvesting the cells. The no release (EdUseq-HU) profile is shown as reference. Replication timing (RT) and gene (Ge/iG) annotations are as in Fig. 1b. σ, sigma.
b, Genome-wide average fork progression at constitutive (CN) origins in OE and NE cells from the experiment shown in (a). Fork speeds were determined from the distances traveled by the forks between the 90 and 150 min release time points. aσ, adjusted σ.
c, Replication initiation (EdUseq-HU) profiles at a representative genomic region in OE and NE cells (upper panel, reference) and fork progression profiles of OE cells released from a 6 h HU block for the indicated times and labeled with EdU for 30 min before harvesting (lower panels, EdUseq-HU/release).
d, Genome-wide average fork progression at constitutive (CN) and oncogene-induced (Oi2 and Oi) origins located in highly or lowly transcribed (Tx) regions (upper and lower terciles, respectively) from the experiment shown in (c).
e, Rescue of fork progression (EdUseq-HU/release) at Oi origins within a representative genomic region in DRB-treated OE cells. DRB was added 4 h after mitotic shake-off and kept until harvesting. ctl, control (no DRB).
f, Genome-wide average fork progression at CN, Oi2 and Oi origins located in highly transcribed (Tx) regions from the experiment shown in (e).
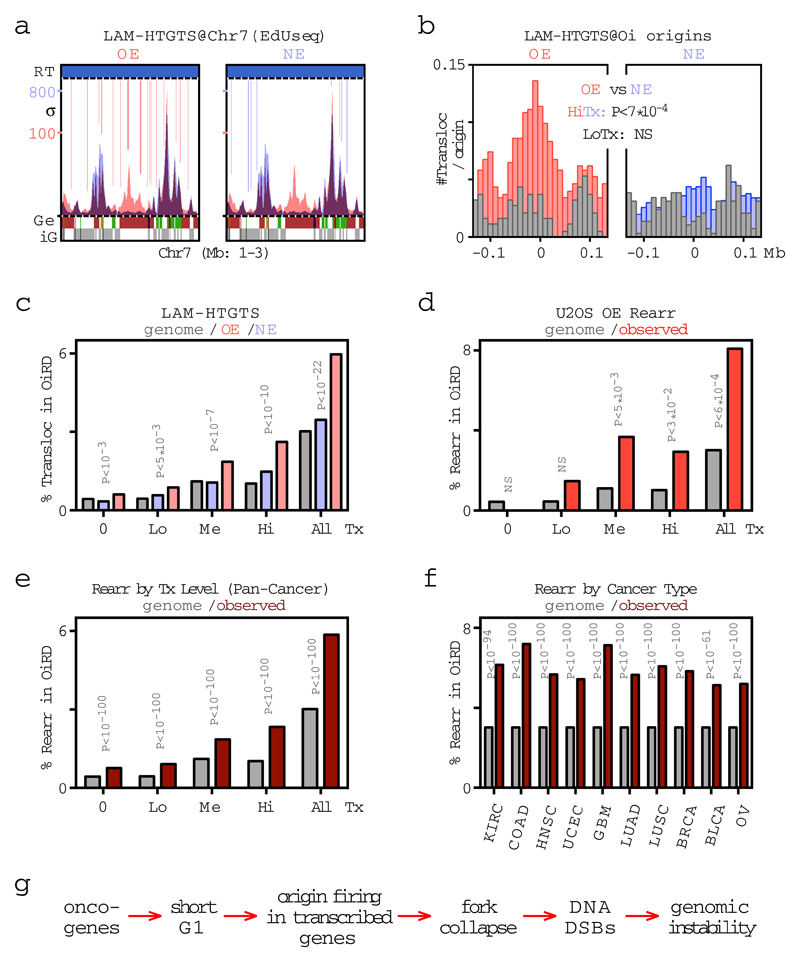
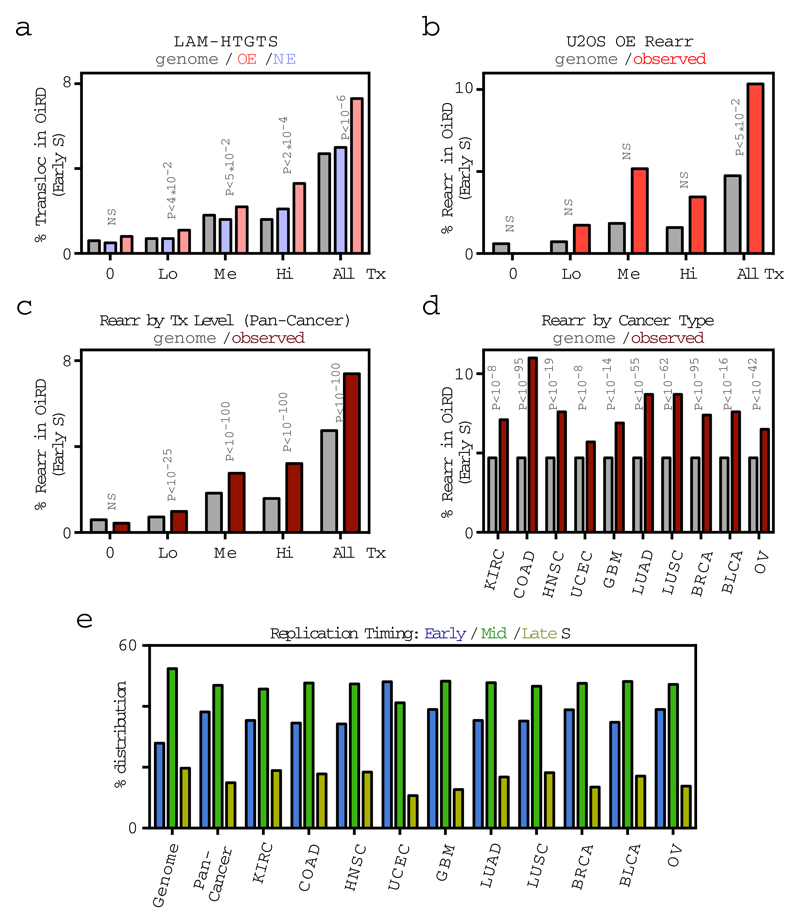
We next examined whether fork collapse at Oi origins led to the formation of DNA DSBs. We studied unsynchronized cells expressing normal or high levels of cyclin E in the absence of exogenous, replication stress-inducing agents, such as HU. DNA DSBs leading to translocations with a CRISPR/Cas9-induced site-specific DNA DSB were identified using the linear amplification-mediated high-throughput genome-wide translocation sequencing (LAM-HTGTS) assay23. Translocations were mapped to Oi origins and also, more broadly, to the genomic domains replicated from these origins (Oi replication initiation domains), which we identified from the replication initiation profiles of cells that were not treated with HU (Fig. 1h). Translocation breakpoints were enriched at Oi origins specifically in cyclin E overexpressing cells (Fig. 4a, b). The effect was dependent on the level of transcription, since only Oi origins at highly transcribed loci were significantly associated with translocations (Fig. 4b). Similarly, the percentage of translocations mapping to Oi replication initiation domains significantly increased upon cyclin E overexpression, and the identified translocations mapped preferentially to those domains having mid or high levels of transcription (Fig. 4c and Extended Data Fig. 9a). Similar findings were obtained by examining breakpoints of previously identified gross chromosomal rearrangements (amplifications and deletions) in the same cyclin E-inducible cells14 and in a large cohort of human cancers4 (Fig. 4d-f, Extended Data Fig. 9b-d and Supplementary Table 5).
Figure 4. Oi origins are associated with DNA DSB formation and genomic rearrangements.
a, Translocations identified by LAM-HTGTS within a representative genomic region in cells overexpressing cyclin E (OE) and cells with normal cyclin E (NE) activity shown as vertical lines (red and blue, respectively, with color intensity reflecting the number of translocations per genomic bin) in the context of the replication initiation profiles of these cells (EdUseq-HU).
b, Number of translocations (#Transloc) per origin in OE and NE cells at oncogene-induced (Oi) origins and surrounding genomic bins. Data are plotted separately for origins mapping to highly (red, blue) and lowly (gray) transcribed (Tx) sites (upper and lower terciles, respectively) and statistical comparisons between OE and NE samples were performed by random permutation tests.
c-f, Mapping of translocations (Transloc; n=27,364) identified by LAM-HTGTS (c), genomic rearrangement (Rearr; n=136) breakpoints identified previously14 in the same cells overexpressing cyclin E (OE) (d) and genomic rearrangement (Rearr; n=490,711) breakpoints from a TCGA pan-cancer dataset4 (e, f) to genomic regions replicated from Oi origins (oncogene-induced replication initiation domains, OiRDs). The fraction of translocations/breakpoints mapping to OiRDs is shown for non-transcribed (0), low (Lo), medium (Me) and highly (Hi) transcribed genomic bins, and for all genomic bins (c-e) or for all genomic bins in common cancer types (f). The distribution of OiRDs in the genome is shown in gray. Statistical comparisons are between NE (blue) and OE (pink) samples (c) or between observed (red) and genomic (gray) frequencies (d-f) and were performed either by random permutation tests (c, d) or by calculating z-scores (e, f). NS, not significant. KIRC, kidney renal cell; COAD, colon adenocarcinoma; HNSC, head and neck squamous cell; UCEC, uterine cervix; GBM, glioblastoma multiformae; LUAD, lung adenocarcinoma; LUSC, lung squamous cell; BRCA, breast; BLCA, bladder; OV, ovary.
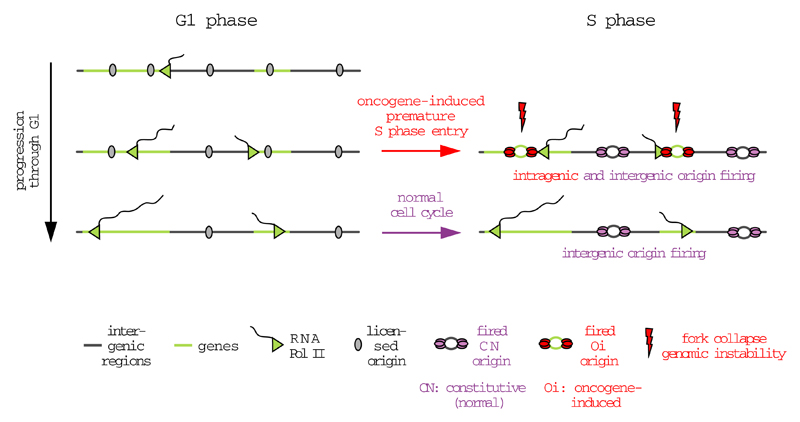
g, Proposed mechanism for oncogene-induced DNA replication stress.
The genome-wide replication initiation and transcription profiles, described here, provide new insights regarding how oncogenes induce DNA replication stress. We observed that oncogenes induce the firing of novel replication origins, which, unlike the constitutive origins, are intragenic and give rise to replication forks that are prone to collapse. Therefore, oncogene-induced DNA replication stress does not involve all replisomes, but, rather the subset derived from the novel, intragenic origins. This latter subset was also associated with a higher frequency of genomic rearrangements in cancer. Our study did not interrogate the late replicating part of the genome, which is where most common fragile sites map24–26. Nevertheless, more chromosomal rearrangements in human cancers map to early, than late, replicating genomic domains (Extended Data Fig. 9e) and early replicating fragile sites have also been identified27.
The collapse of forks initiating from intragenic, oncogene-induced origins could be attributed to replication-transcription conflicts, whereas forks from intergenic, constitutive origins, did not collapse, even when replicating highly transcribed genes. The different behavior may relate to the fact that head-on replication-transcription collisions, the most damaging type28, cannot be prevented when origins fire within genes. However, for constitutive origins, which are intergenic, head-on collisions can be avoided by a genomic organization, including replication fork barriers, that favors co-directionality of replication with transcription29–30. Alternatively, forks might be particularly sensitive to replication-transcription conflicts shortly after origin firing, before, for example, lagging strand synthesis has converted the initial origin bubble to double-stranded DNA daughter molecules, which would explain why forks from intragenic origins are prone to collapse.
Our study also helps explain how oncogenes induce firing of intragenic origins. We observed that transcription suppresses origin firing from within genes. In normal cell cycles, the length of G1 is sufficient for transcription to inactivate origins across the entire length of genes. However, oncogenes, which shorten drastically the length of G1, leave insufficient time for transcription to inactivate all intragenic origins (Fig. 4g and Extended Data Fig. 10). This concept of transcription erasing intragenic origins fits with the well-known observation that the largest genes in the human genome are late replicating, which would, in principle, provide more time for transcription to reach the 3’ end of these genes before they initiate replication. This mechanism also helps reconcile how shortening of the G1 phase, a typical outcome of oncogene activation, leads to aberrant origin firing and DNA replication stress.
Methods
Cell culture
U2OS cells inducibly overexpressing cyclin E (U2OS-CE) were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen, Cat. No. 11960), supplemented with 10% fetal bovine serum (FBS; Invitrogen, Cat. No. 10500), penicillin-streptomycin-glutamine (Invitrogen, Cat. No. 10378-016), G418 400 µg/ml (Invitrogen, Cat. No. 10131-027), puromycin 1 µg/ml (Sigma, Cat. No. P8833) and tetracycline 2 µg/ml (Sigma, Cat. No. T7660). At the indicated number of days before the experiment, the cells were split into two aliquots. One aliquot was cultured in media without tetracycline to induce cyclin E overexpression (OE cells) and the other in media with 1µg/ml doxycycline (Sigma, Cat. No. D3447) to maintain low levels of ectopic cyclin E expression (NE cells). U2OS cells inducibly activating Myc (U2OS-MycER) were maintained in Dulbecco’s modified Eagle’s medium without phenol red (Invitrogen, Cat. No. 31053), supplemented with 10% fetal bovine serum and penicillin-streptomycin-glutamine. At the indicated number of days before the experiment, MycER activity was induced by 100 nM of 4-hydroxytamoxifen (4-OHT; Sigma, Cat. No. H7904) dissolved in methanol. HeLa cells were maintained in Dulbecco’s modified Eagle’s medium, supplemented with 10% fetal bovine serum and penicillin-streptomycin-glutamine. hTERT-RPE1 retinal pigment epithelial cells were purchased from the American Type Culture Collection (ATCC) and were cultured in Dulbecco's Modified Eagle Medium/Ham's F-12 (Invitrogen, Cat. No. 12634-010), supplemented with 10% fetal bovine serum, penicillin-streptomycin-glutamine and Hygromycin B (Sigma, Cat. No. H3274).
Antibodies, fluorescence and immunoblotting
Antibodies specific for Cyclin E (Novocastra, Cat. No. NCL- CYCLINE), α-Actinin (Millipore, Cat. No. 05-384) and c-Myc (Cell Signalling, Cat. No. 5605) were obtained from the indicated vendors. Immunofluorescence and immunoblotting were performed, as previously described14. EU staining was performed using the Click-iT RNA Alexa Fluor 488 Imaging Kit (ThermoFisher, Cat. No. C10329) according to the manufacturer’s instructions. Cell nuclei were counterstained with DAPI.
REPLIseq
U2OS-CE cells were cultured for 1, 2 or 7 days with or without doxycycline. The day before the experiment, the cells were re-seeded in order to obtain a 70% confluency the following day. EdU (Invitrogen, Cat. No. A10044) was added at a concentration of 25 µM for 30 minutes to the asynchronously growing cells. The cells were then harvested, fixed in 90% methanol overnight and permeabilized with 0.2% triton-X in PBS. EdU was coupled to a cleavable biotin linker (Azide-PEG(3+3)-S-S-biotin) (Jena Biosciences, Cat. No. CLK-A2112-10) using the Click-it Kit (Invitrogen Cat. No. C-10420). Genomic DNA was stained with propidium iodide (Sigma, Cat. No. 81845) in combination with RNAse (Roche, Cat. No. 11119915001) and the cells were then sorted into 3 cell cycle populations according to DNA content using a MoFlo Astrios flow sorter (Beckman Coulter) present at the Flow Cytometry platform of the Medical Faculty of the University of Geneva. DNA isolated from the sorted cells was purified by phenol-chloroform extraction and ethanol precipitation and subjected to EdU-labeled DNA isolation (see EdU-labeled DNA isolation and Sequencing below). The REPLIseq samples are listed in Supplementary Table 6.
Flow-cytometry assessment of S-phase entry
U2OS-CE cells were cultured with or without doxycycline for two days; whereas, U2OS-MycER were cultured with or without 4OHT for three days. Cells were treated with 100 ng/ml nocodazole (Tocris, Cat. No. 1228) for 8 h to induce mitotic arrest, except RPE1 cells that were treated with 200ng/ml nocodazole. Mitotic cells were isolated by shake-off, washed with PBS, released in warm media containing 25 µM EdU (Invitrogen, Cat. No. A10044) and then collected every two hours and fixed with 90% methanol overnight. The cells were prepared for flow cytometry using the Click-it Kit (Invitrogen Cat. No. C-10420) according to the manufacturer’s instructions. The genomic DNA was stained with propidium iodide (Sigma, Cat. No. 81845) in combination with RNAse (Roche, Cat. No. 11119915001). EdU-DNA content profiles were then acquired by flow cytometry (Gallios, Beckman Coulter) to assess the percentage of cells that entered S phase in each condition at each time point.
EdUseq
U2OS-CE cells were cultured with or without doxycycline for two days and U2OS-MycER cells were cultured with or without 4OHT for three days before being exposed to 100 ng/ml nocodazole (Tocris, Cat. No. 1228) for 8 h to induce mitotic arrest. HeLa and RPE1 cells were treated with 100ng/ml or 200ng/ml of nocodazole, respectively, for 8 h. Cells in mitosis were isolated by shake-off, washed with PBS, released in warm media containing 2 mM hydroxyurea (HU, Sigma, Cat. No. H8627) and 25 µM EdU (Invitrogen, Cat. No. A10044). When indicated DRB (5,6-Dichloro-1-β-D-ribofuranosylbenzimidazole, Sigma, Cat. No. D1916) (75 µM), mimosine (Sigma, Cat. No. M0253) (1mM) or aphidicolin (Sigma, Cat. No. A0781) (1µM) were also added to the tissue culture media. The cells were then collected at the indicated time points and fixed with 90% methanol overnight. The EdUseq-HU samples are listed in Supplementary Table 7.
In a second series of experiments (EdUseq-noHU), after mitotic shake-off, the cells were released in media without HU. 25 µM EdU was added directly to the media or one hour before the cells were collected as indicated. The EdUseq-noHU samples are listed in Supplementary Table 8.
In a third series of experiments (EdUseq-HU/release), after mitotic shake-off, the cells were released in media containing HU, but not EdU. After the indicated time of incubation, HU was removed and the cells were released in warm media. 25 µM EdU was added 30 min before collecting the cells, which were then fixed with 90% methanol overnight. To inhibit transcription elongation, DRB (5,6-Dichloro-1-β-D-ribofuranosylbenzimidazole, Sigma, Cat. No. D1916) was added at 75 µM to the cells for the indicated time points. The EdUseq-HU/release samples are listed in Supplementary Table 9.
For all three series of EdUseq experiments, after fixing, the cells were permeabilized with 0.2% triton-X in PBS. EdU was coupled to a cleavable biotin-azide linker (Azide-PEG(3+3)-S-S-biotin) (Jena Biosciences, Cat. No. CLK-A2112-10) using the reagents of the Click-it Kit (Invitrogen, Cat. No. C-10424). The DNA was then purified by phenol-chloroform extraction and ethanol precipitation and subjected to EdU-labeled DNA isolation (see EdU-labeled DNA isolation and Sequencing below).
EdU-labeled DNA isolation and sequencing
Genomic DNA was sonicated to a size range of 100 to 500 bp with a bioruptor sonicator (Diagenode). EdU-labeled DNA fragments were then isolated using Dynabeads MyOne streptavidin C1 (Invitrogen, Cat. No. 65001) according to the manufacturer’s instructions with minor modifications. Briefly, for each sample, the beads were washed three times with Binding and Washing Buffer 1x (5 mM Tris-HCl pH 7.5, 0.5 mM EDTA, 1 M NaCL, 0.5% Tween-20) using a magnet. After washing, the beads were resuspended to twice the original volume with Binding and Washing Buffer 2x, mixed with an equal volume of sonicated EdU-labeled DNA and incubated for 15 min on a rotating wheel at room temperature. The beads were then washed three times with Binding and Washing Buffer 1x and once with TE using the magnet. The EdU-labeled DNA was eluted by incubating the streptavidin beads with 2% ß-mercaptoethanol (Sigma, Cat. No. M6250) for 1 h at room temperature. The eluted DNA for REPLIseq was purified by phenol-chloroform extraction followed by ethanol precipitation before being prepared for Illumina single-end sequencing. The eluted DNA for EdUseq was directly used for library preparation. The libraries were made by the Genomics Platform of the University of Geneva using the TruSeq ChIP Sample Prep Kit (Illumina, Cat. No. IP-202-1012). 100 bp single-end read sequencing reactions were then performed on an Illumina Hi-Seq 2500 or Illumina Hi-Seq 4000 sequencer.
In order to compare the levels of EdU incorporation among the various REPLIseq samples, the EdU-labeled genomic DNA isolated from NE and OE 2d U2OS-CE cells was spiked with a constant amount of EdU-labeled DNA prepared from mouse embryo fibroblasts (MEFs) before isolating the EdU-labeled DNA. This permitted calibration of the amount of EdU incorporation per cell among the various samples, by dividing the number of sequencing reads of EdU-labeled human DNA by the number of reads of EdU-labeled mouse DNA and by the fraction of EdU-positive cells in that sample.
EUseq
For newly-synthesized transcripts sequencing (EUseq), cells were synchronized in mitosis and released in 2 mM HU, as in EdUseq. For the series of experiments for which transcription elongation was inhibited, DRB (5,6-Dichloro-1-β-D-ribofuranosylbenzimidazole, Sigma, Cat. No. D1916) was added at 75 µM to the cells for the indicated time periods. EU (5-Ethynyl-uridine, Jena Biosciences, Cat. No. CLK-N002-10) was added to the cells at a concentration of 0.5 mM 30 minutes before the cells were collected at the indicated time points. RNA was then extracted and purified using TRIzol (Invitrogen, Cat. No. 15596) and isopropanol precipitation. Nascent RNA was biotinylated and purified using the reagents of the Click-iT Nascent RNA Capture Kit (Invitrogen, Cat. No. C-10365) according to the manufacturer instructions, but replacing the biotin-azide from the kit by the cleavable biotin-azide (Azide-PEG(3+3)-S-S-biotin) (Jena Biosciences, Cat. No. CLK-A2112-10). The EU-labeled RNA was then isolated using Dynabeads MyOne streptavidin C1 (Invitrogen, Cat. No. 65001) according to the manufacturer’s instructions with minor modifications. Briefly, the beads (50 µl of beads per µg of RNA) were washed three times with Binding and Washing Buffer 1x (5 mM Tris-HCl pH 7.5, 0.5 mM EDTA, 1 M NaCL, 0.5% Tween-20) followed by two 2 min washes in Solution A (0.1 M NaOH, 0.05 M NaCl) and two washes in Solution B (0.1 M NaCl) using a magnet. After washing, the beads were resuspended to twice the original volume with Binding and Washing Buffer 2x, and mixed with an equal volume of EU-labeled RNA. The RNA had been previously heated at 70°C and placed back on ice to remove secondary structures. The mix was incubated for 30 min on a rotating wheel at room temperature. The beads were then washed three times with Binding and Washing Buffer 1x and once with RNAse free-water using the magnet. The EU-labeled RNA was finally eluted by incubating the streptavidin beads with 2% ß-mercaptoethanol (Sigma, Cat. No. M6250) for 1 h at room temperature. Sequencing libraries were prepared by the Genomics Platform of the University of Geneva using the TruSeq Stranded Total RNA with Ribo-Zero Gold (Illumina, Cat. No. RS-122-2301) omitting the ribo-depletion step. 100 bp single-end read sequencing reactions were performed on an Illumina Hi-Seq 2500 or Illumina Hi-Seq 4000 sequencer. The EUseq samples are listed in Supplementary Table 10.
LAM-HTGTS
Translocations were detected in NE and OE U2OS-CE cells using LAM-HTGTS as previously described in Hu, J. et al.23 omitting the optional enzyme blocking step. DNA double-strand breaks (DSBs) were induced at a bait site located in an early replicating intergenic region of chromosome 9, using a guide RNA (gRNA-chr9: CACCGAGGAAACTGAGTCACAGGCT, chr9: 21685208-21685227) in combination with the Cas9 nuclease (Addgene, Cat. No. 48138, pX458, pSpCas9(BB)-2A-GFP). Cells with normal expression of cyclin E and cells in which cyclin E overexpression had been induced for three days were transfected with the Cas9:gRNA-chr9 plasmid and collected two days after transfection for the LAM-HTGTS procedure. Non-transfected cells were also collected as negative control sample. The primers were designed according to23:
Bio-primer-chr9: /5-biotin/AAGTCTCTCCAGCCAAGAACAG
-
I5-Nested-chr9: ACACTCTTTCCCTACACGACGCTCTTCCGATCT-BARCODE-
-GGAAAGGGTAGTGGGAGGTAGAAAGC
Paired-end read sequencing reactions (100 bp) were performed on an Illumina Hi-Seq 2500 sequencer. The LAM-HTGTS samples are listed in Supplementary Table 11.
Sequence alignment and calculation of sigma values
Sequence reads were aligned to the masked human genome assembly (GRCh37/hg19) using the Burrows-Wheeler Aligner algorithm31, retaining only the reads with the highest quality score. Custom Perl scripts were then developed to process the data for analysis and visualization. First, the chromosomes were split into 10 Kb bins and the sequence reads were assigned to their respective bin. Then, to correct for sequencing bias across the genome (reflecting experimental biases and differences in the number of masked base pairs per bin), the number of sequence reads per bin (SeqRpB) was normalized using the number of reads previously obtained by sequencing genomic DNA from the same cells (referred to as adjust sample and representing a total of more than 342 million reads)14 using the formula:
where, NormSeqRpB stands for normalized sequence reads per bin and AdjRpB for adjust reads per bin. Genomic bins were retained for further analysis only if the number of AdjRpB per bin was within the range of 25-10,000 (average 1,241 AdjRpB), resulting in a human genome assembly of 275,491 genomic bins corresponding to chromosomes 1-22 and chromosome X (since U2OS cells were derived from a female patient). After normalization of the sequence reads, standard deviation (SD) values were calculated for each genomic bin. This permitted the calculation of sigma (σ) values for each genomic bin according to the formula:
For all samples, the mean background NormSeqRpB value was much smaller than the peak NormSeqRpB values, but was, nevertheless, subtracted to lead to more accurate sigma value estimates. The sigma values were then used to plot the data and perform all subsequent analyses, thus, allowing comparison of samples with different background levels and providing a quick way to ascertain whether the observed peaks were statistically significant.
To calculate the genome-wide, mean background NormSeqRpB value, we used a differential function to determine the fraction of the genome with background signal (i.e. the genomic bins lacking true signal); the mean NormSeqRpB value of these genomic bins corresponded to the genome-wide, background NormSeqRpB mean value. NormSeqRpB SDs were calculated for each genomic bin. First, the fraction of the genome with background signal was sorted into 200 equal subfractions, according to the number of AdjRpB, with each subfraction corresponding to a range of AdjRpB, spanning the entire possible range of 25-10,000. The NormSeqRpB values of these subfractions were then used to calculate background NormSeqRpB SD values for each subfraction. The SD values of all the subfractions were plotted against the mean AdjRpB of their corresponding subfractions, resulting in a power regression curve of the type:
where a and b are constants. For all samples, the power regression equations fit the data with coefficients of determination (R2) greater than 0.9. The determined values of the a and b constants were then used to calculate a SD for each genomic bin (including the bins with true signal) from its AdjRpB. For the EUseq data, most background genomic bins had NormSeqRpB values equal to zero; thus, for these samples, we calculated relative, rather than absolute SD and sigma values.
The EdUseq-HU datasets were graphed after subtracting the mean background NormSeqRpB value; all other datasets (including the EdUseq-HU/release and EdUseq-noHU datasets) were graphed without background subtraction.
The computer codes to perform the analyses described above are included in the Supplementary Information section.
Identification and classification of replication origins from EdUseq-HU data
A peak-finding algorithm searched for local maxima. Each local maximum was then evaluated on the basis of its sigma value and shape and retained as a peak, only if its values exceeded predefined sigma and shape thresholds. One peak list comprised the peaks identified in the EdUseq-HU 4 h OE dataset, while a second list comprised the peaks identified in the 14 h NE dataset. The peaks that had been identified in both datasets at exactly the same genomic bin were then used to calculate an adjustment factor (AdjFactor) using the formula:
The sigma values of all genomic bins of the 4 h OE sample were then multiplied by this adjustment factor. A new peak list was then generated by including all peaks from both datasets, irrespective of whether the peak was present in both samples or only in one sample. Peaks that mapped to adjacent genomic bins (i.e. within 10 Kb) in the two datasets were considered to correspond to a single origin and assigned to the genomic bin with the highest sigma value (original sigma for the 14 h NE dataset and adjusted sigma for the 4 h OE dataset).
For every peak in the merged peak list (irrespective of whether the peak had been identified in the NE or OE or both samples), the sigma values at its genomic position in the NE and OE samples (adjusted sigma for the OE sample) were obtained and compared. If the ratio of the OE:NE sigma values was greater than 4, then the origin was considered as oncogene-induced (Oi); otherwise, if the ratio was greater than 2, but lower than 4, the origin was considered as Oi2. All other origins were considered to be constitutive (CN). The assignment of origins into the constitutive (CN), Oi2 and Oi classes listed above, facilitated comparisons among the various samples using power regression curves of the type:
where a and b are constants.
The power curve was converted to a linear regression curve:
to facilitate plotting of the data as scatter plots and calculation of coefficients of determination (R2). A similar analysis was performed for the cells with inducible activation of Myc and for the EUseq data.
Assignment of replication timing domains
The early, mid and late S phase REPLIseq data were calibrated by spiking the samples with a known quantity of mouse genomic EdU-labeled DNA. After assignment of the REPLIseq reads to genomic bins and adjustment of the number of reads, as described above for the EdUseq data, the number of early, mid and late S phase reads were compared for each genomic bin. If one sample (early, mid or late S) accounted for more than half of the total reads for a specific genomic bin, then that bin was assigned to the corresponding replication timing domain. The assignment of replication timing domains used for further analysis was based on the NE samples, which showed sharp REPLIseq profiles.
Assignment of genic and intergenic domains
RefSeq gene annotations were used to compile a list of all human protein-coding genes and their position in the genome. Genomic bins were defined as being purely genic, if they mapped entirely within protein-coding genes; purely intergenic, if the bins mapped entirely within intergenic sequences; or mixed, if they encompassed both genic and intergenic sequences. The analysis of the distribution of origins in the genome considered the pure intergenic and mixed genic/intergenic bins as intergenic and the pure genic bins as genic.
Determination of average EUseq and EdUseq signals along large genes
EUseq relative sigma (rσ) values were converted to a log2 scale for all the subsequent analysis described in this section. Genes over 200 Kb in size were identified in the early and mid replicating parts of the genome using RefSeq gene annotations and the average EUseq log2(rσ) value across the lengths of these genes at 14 h after mitotic exit was used to classify the genes according to their level of transcription (high, upper tercile; medium, middle tercile; and low, lower tercile). EUseq datasets corresponding to different times after exit from mitosis were then adjusted relative to each other by comparing the sum of their EUseq log2(rσ) values corresponding to the first 5 genomic bins of each large gene. Then, average EUseq log2(rσ) values were plotted as a function of the distance from the 5-prime end of the gene. The five most 3-prime genomic bins of each gene were trimmed and not included in the analysis, as in some genes EdUseq signal from origins located in intergenic bins adjacent to the gene was spilling over into the 3-prime end of the gene. Average EdUseq values (linear σ) were also plotted along gene length. Sigma values of the OE samples were adjusted relative to the sigma values of the NE samples.
Calculation of fork speed
Fork speeds were calculated from EdUseq datasets of NE and OE cells treated with HU for 14 and 10 h, after mitotic exit, respectively, and EdUseq-HU/release datasets (90 or 150 min release). The 10% tallest constitutive peaks in the genome were initially selected. Then, the peak finding algorithm described above was used to identify peaks in the EdUseq-HU/release 90 and 150 min datasets on either side of the origins. The positions of peaks identified with high confidence in the release datasets, were then used to calculate the distance forks traveled between the 90 and 150 min time points. The same list of origins (N=325) was examined in the NE and OE samples.
Analysis of fork collapse
To study fork collapse, a set of origins was selected for which fork progression could be monitored without interference from neighboring origins. The criterion for selection was that within 20 genomic bins of the position of the origin being examined, there was no other origin that had a sigma value equal to or greater than the sigma value of the origin being selected. Selected origins were further classified according to their transcription level (upper and lower terciles, as defined above) at the time the cells were released from the HU arrest. The number of origins, thus selected, in each category were: constitutive CN-high tx, 67; constitutive CN-low tx, 233; Oi2-high tx, 39; Oi2-low tx, 55; Oi-high tx, 57; Oi-low tx, 47. Adjusted averages (relative to the no release data) of EdUseq and EdUseq-HU/release datasets were then calculated for each origin category. A similar analysis, using the same set of origins, was performed for the cells treated with the transcription elongation inhibitor DRB.
Identification of translocations by LAM-HTGTS and mapping to Oi origins
Paired-end sequencing reads were aligned independently to the masked human genome assembly (GRCh37/hg19) using the Burrows-Wheeler Aligner mem algorithm31 and duplicate reads were filtered out. Read pairs corresponding to junctions between the bait site and another genomic region and containing the junction sequence in one of the two reads were retained. Furthermore, because DNA double-strand ends induced by fork collapse in S phase will most likely be repaired by microhomology-mediated end joining, paired ends were further required to have a 2-5 base pair microhomology junction. Translocation breakpoints identified by LAM-HTGTS in OE and NE cells were then mapped to Oi origins. A subset of all Oi origins was used for this analysis, requiring that within 10 genomic bins of the position of the origin being examined, there was no other origin that had a sigma value equal to or greater than twice the sigma value of the origin being selected. Selected origins were further classified according to their transcription level (upper and lower terciles, as defined above) 14h after the cells were released from HU arrest. The number of selected origins were: Oi-high tx, 108; Oi-low tx, 62. The number of translocations mapping to each genomic bin was divided by the number of origins (Oi-high tx or Oi-low tx); for the translocations identified in the NE cells, the average number of translocations per bin was further adjusted by the ratio of the total number of LAM-HTGTS in the NE and OE samples to allow comparisons between the samples. A permutation analysis was performed to evaluate whether the observed differences between the number of identified translocations mapping to Oi origins in the OE and NE samples were statistically significant.
Identification of Oi replication initiation domains (OiRDs)
The sigma values of the EdUseq-noHU datasets from OE cells incubated for the first 3 h after mitotic exit with EdU and from the NE cells incubated for the first 10 h after mitotic exit with EdU (Fig. 1h) were converted to log2 values. A linear regression curve of the values corresponding to the genomic positions of the Oi2 origins in an OE:NE plot was then used to assign all genomic bins that had EdUseq signal above background to Oi replication initiation domain (OiRD) bins, using as criterion that the log2 OE:NE sigma values of the bin were more than 0.6 units to the left of the Oi2 curve.
Mapping of translocations and genomic rearrangement breakpoints to OiRDs
The translocation breakpoints identified by LAM-HTGTS (n=16,629 and n=10,735 for NE and OE cells, respectively), the breakpoints of genomic rearrangements (n=136, Extended Data Table 2; derived from 81 rearrangements - for rearrangements less than 100 Kb long, a single breakpoint was calculated corresponding to the center position of the rearrangement) identified by us in the same U2OS cells overexpressing cyclin E for three weeks14 and the breakpoints of rearrangements (deletions and amplifications, n=490,711) present in a cohort of ~5,000 human cancers4 were mapped to the Oi replication initiation domains (OiRDs). The frequency of OiRDs in the entire genome served as reference. The analysis was performed in the context of the entire genome (Fig. 4c-f) and, also, for only the early S replicating part of the genome (Extended Data Fig. 9a-d), where about half of the OiRDs mapped. Statistical comparisons for the LAM-HTGTS and U2OS breakpoint rearrangement data were performed using random permutations; for the TCGA data, the observed and genomic frequencies were used to calculate z-scores, from which P values were determined.
Code availability
Computer codes and data files used to process and plot the data are available as Supplementary Information. Other codes are available upon request to the corresponding author.
Data availability
The fastq sequencing data and associated information described in this study have been deposited in the Sequence Read Archive (SRA) as BioProject PRJNA397123.
Extended Data
Extended Data Figure 1. Experimental setup to study S phase entry and DNA replication initiation.
a, Cyclin E protein levels, as determined by immunoblotting, in cells with normal levels of cyclin E (NE) and cells overexpressing cyclin E (OE) (2.5 days after tetracycline withdrawal). Actinin serves as a loading control. This is a representative example of more than ten independent replicates.
b, Experimental outline of the protocol used to monitor S phase entry by flow cytometry and of the EdUseq protocols: EdUseq-HU, EdUseq-noHU and EdUseq-HU/release. *, indicates that EdU was added 30 min before harvesting the cells.
c, Flow cytometry profiles of cells with normal levels of cyclin E (NE) and cells overexpressing cyclin E (OE), after mitotic shake-off (0 h) and 14 and 10 h later, respectively, after the cells had been released in media containing HU and EdU. 2C and 4C, DNA content of G1 and G2 cells, respectively. This is a representative example of more than ten independent replicates.
d, DNA content versus EdU incorporation flow cytometry plots of NE and OE cells. EdU-positive NE and OE cells were gated blue and red, respectively. 2C and 4C, DNA content of G1 and G2 cells, respectively. The gating strategy for these data is shown in Supplementary Information Fig. S1.
Extended Data Figure 2. Identification of replication timing domains by REPLIseq.
a, Experimental outline of the REPLIseq protocol and FACS profiles of cells with normal levels of cyclin E (NE) and cells overexpressing cyclin E (OE) for 1, 2 or 7 days (1d, 2d and 7d, respectively). Cells were sorted according to DNA content into early (blue), mid (green) and late (yellow) S phase fractions. 2C and 4C, DNA content of G1 and G2 cells, respectively.
b, Assignment of replication timing (RT) domains. The fractions of the genome that were replicated in early, mid or late S phase were determined on the basis of the REPLIseq profiles of the NE and 2d OE cells.
c, Distribution of early, mid and late replication timing bins in 2d OE cells according to their replication timing in NE cells.
d, Comparison of the origin firing profile determined by EdUseq-HU (Fig. 1b) and the early S replication profile determined by REPLIseq in NE cells. Replication timing (RT) domains and genic/intergenic (Ge/iG) regions are as in Fig. 1b. Bin resolution, 10 Kb; ruler scale, 100 Kb.
e, REPLIseq profiles of the first 10 Mb of chr7 of cells expressing normal levels of cyclin E (NE) or overexpressing cyclin E (OE) for 1, 2 or 7 days (1d, 2d and 7d, respectively). Profiles are shown separately for the cells in early, mid and late S phase. Replication timing (RT) domains and genic/intergenic (Ge/iG) regions are as in Fig. 1b. Bin resolution, 10 Kb; ruler scale, 100 Kb.
Extended Data Figure 3. Further characterization of replication origins.
a, Adjusted average sigma values at 1 Kb resolution around 1,828 origins refined by performing the EdUseq-HU protocol in the additional presence of mimosine or aphidicolin (light and dark gray, respectively), compared to OE and NE cells treated only with HU (pink and blue, respectively). aσ, adjusted σ.
b, Distribution of the subset of origins refined at 1 Kb resolution (1,828 origins) relative to origin type, as determined by the original 10 Kb resolution assignment: CN, constitutive; Oi2, oncogene-induced 2; Oi, oncogene-induced.
c, Distribution of constitutive (CN) and oncogene-induced (Oi2 and Oi) origins, refined at 1 Kb resolution (1,828 origins), according to replication timing (RT) domains (E, early; M, mid; L, late S phase).
d, Distribution of constitutive (CN) and oncogene-induced (Oi2 and Oi) origins, refined at 1 Kb resolution (1,828 origins), according to gene annotation in all replication timing domains (all-RT) or only in the early S phase replicating domains (E-RT).
e, Transcription (EUseq) levels (median) in NE (light gray) and OE (dark gray) cells at sites of constitutive (CN) and oncogene-induced (Oi) origins, refined at 1 Kb resolution, at various time points after mitotic shake-off. Origins mapping to genic (Ge) or intergenic (iG) genomic bins were plotted separately. rσ, relative σ.
f, Scatter plots of EdUseq-noHU σ values (log2) at all origins (CN, purple; Oi2, pink; Oi, red) at 10 Kb resolution for NE vs OE cells not treated with HU (Fig. 1h). EdU was present during the indicated time points.
Extended Data Figure 4. Further characterization of transcription profiles and effects of DRB.
a, Genome-wide comparison (all genomic bins) of newly-synthesized transcripts (EUseq) in cells expressing normal levels of cyclin E (NE) vs cells overexpressing cyclin E (OE) cells at the indicated time points after mitotic shake-off. The two genomic bins mapping to the CCNE1 (cyclin E) gene are colored red; rσ, relative sigma.
b, List of early and mid S large genes along which replication initiation and transcription profiles were plotted in Fig. 2d. For each gene, the association with a common fragile site (CFS) is indicated24.
c, Inhibition of transcription by DRB. EU incorporation was monitored by fluorescence microscopy in control, DRB-treated (9 h), and in DRB-treated (9 h) and released (5 h) cells. The nuclei of the cells were counterstained with DAPI.
d, DNA content versus EdU incorporation flow cytometry plots of NE cells 14 h after mitotic shake-off. The cells were treated with DRB for the indicated times. EdU-positive NE cells were gated in blue. 2C and 4C, DNA content of G1 and G2 cells, respectively.
e, Newly-synthesized transcript profiles (EUseq) at a representative genomic region in NE cells treated with DRB for 9 h after mitotic shake-off and then released for 30 or 120 min (release 30 min, dark gray; release 120 min, light gray; overlap: color; direction of transcription: green, forward; red, reverse; yellow, bidirectional). The red arrow indicates the transcription of the gene harboring oncogene-induced origins at our example locus on chromosome 7 and the green arrow indicates another large gene in this locus. Replication timing (RT) domain and gene (Ge/iG) annotations are as in Fig. 1b; rσ, relative sigma.
f, Average transcription (EUseq, log2rσ) in NE cells treated with DRB for 9 h after mitotic-shake-off and then released for 30, 120 or 240 min, along the length of large genes (>0.35 Mb for early S and >0.65 Mb for mid S genes). The genes are grouped according to replication timing (RT; early, mid S) and level of transcription (high Tx, upper tercile; mid Tx, middle tercile). rσ, relative σ; #Ge, number of genes averaged at each position.
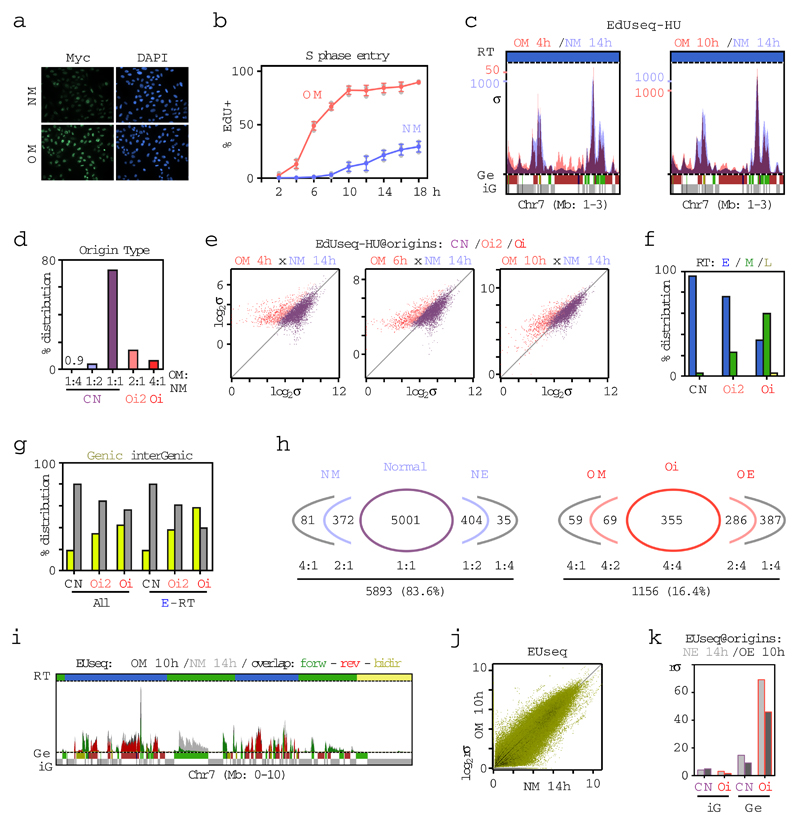
Extended Data Figure 5. Accelerated entry into S phase and firing of novel intragenic origins upon Myc activation.
a, Myc activation (3 days after adding 4-OHT), as determined by immunfluorescence, in cells with non-induced (NM) and induced (OM) Myc activity. Nuclei were counterstained with DAPI. Representative images from two independent experiments are shown.
b, Quantification of EdU positive cells at different time points after mitotic shake-off. Means and SDs were calculated from three independent experiments; gray symbols, individual data points. NM, normal Myc activity; OM, induced Myc activity.
c, Replication initiation (EdUseq-HU) profiles at a representative genomic region in OM and NM cells, harvested at the indicated times after mitotic shake-off. Peak heights are represented as sigma values (σ). Replication timing (RT) domains and gene annotations (Ge/iG) are as in Fig. 1b. Bin resolution, 10 Kb; ruler scale, 100 Kb.
d, Classification of constitutive (CN) and oncogene-induced (Oi2 and Oi) origins based on relative height ratios in OM versus NM cells (OM:NM).
e, Scatter plots of EdUseq-HU σ values at origins (CN, purple; Oi2, pink; Oi, red) in NM vs OM cells at the indicated time points after mitotic shake-off.
f, Distribution of CN, Oi2 and Oi origins in OM and NM cells according to RT domains (E, early; M, mid; L, late S phase).
g, Distribution of CN, Oi2 and Oi origins in OM and NM cells according to gene annotation in all replication timing domains (all-RT) or only in the early S phase replicating domains (E-RT).
h, Relative adjusted sigma ratios of replication origins identified in NE (normal cyclin E activity), NM, OE (cyclin E overexpression) or OM cells. Left, number of origins identified in NE or NM cells grouped according to their relative height ratios between these two cell lines. Right, number of Oi origins identified in OE or OM cells grouped according to their level of induction relative to the NE and NM cells, respectively.
i, Newly-synthesized transcript profiles (EUseq) at a representative genomic region in OM and NM cells 10 h and 14 h after mitotic shake-off, respectively (NM: light gray; OM: dark gray; overlap: green, forward; red, reverse; yellow, bidirectional direction of transcription). Replication timing (RT) domain and gene (Ge/iG) annotations are as in (c).
j, Genome-wide comparison (all genomic bins) of transcription in OM vs NM cells 10 h and 14 h after mitotic shake-off, respectively. rσ, relative σ.
k, Median transcription (EUseq) levels in NM (light gray) and OM (dark gray) cells at CN and Oi origins mapping to genic (Ge) or intergenic (iG) genomic bins at 14 and 10 h after mitotic shake-off, respectively.
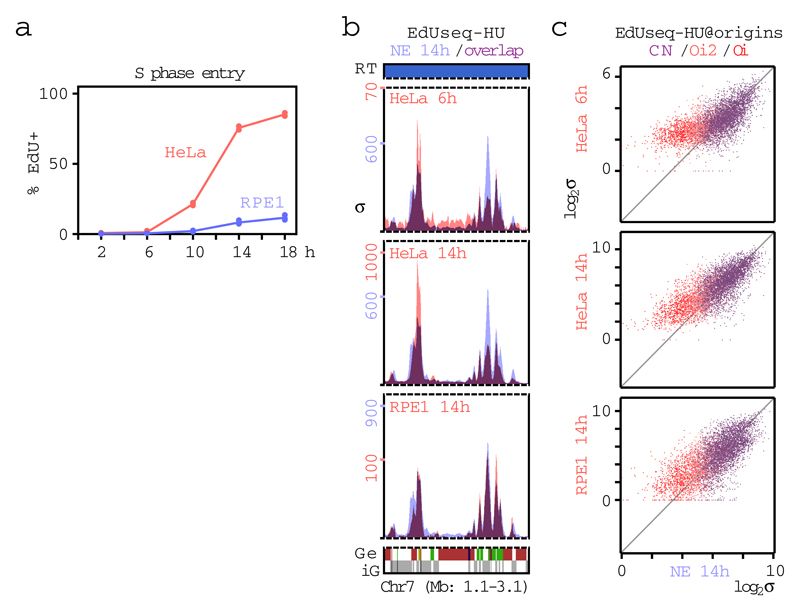
Extended Data Figure 6. S phase entry and replication initiation profiles of HeLa and RPE1 cells.
a, Percentage of EdU positive HeLa and RPE1 cells at different time points after mitotic shake-off (0 h). Means and individual data points are shown from two independent experiments.
b, Replication initiation (EdUseq-HU) profiles at a representative genomic region in HeLa and RPE1 cells at the indicated time points after mitotic shake-off. The profile of U2OS cells expressing normal levels of cyclin E (NE, blue) serves as reference.
c, Scatter plots of EdUseq-HU σ values (log2) at all origins (CN, purple; Oi2, pink; Oi, red) in HeLa and RPE1 cells vs U2OS cells with normal levels of cyclin E (NE) at the indicated time points after mitotic shake-off.
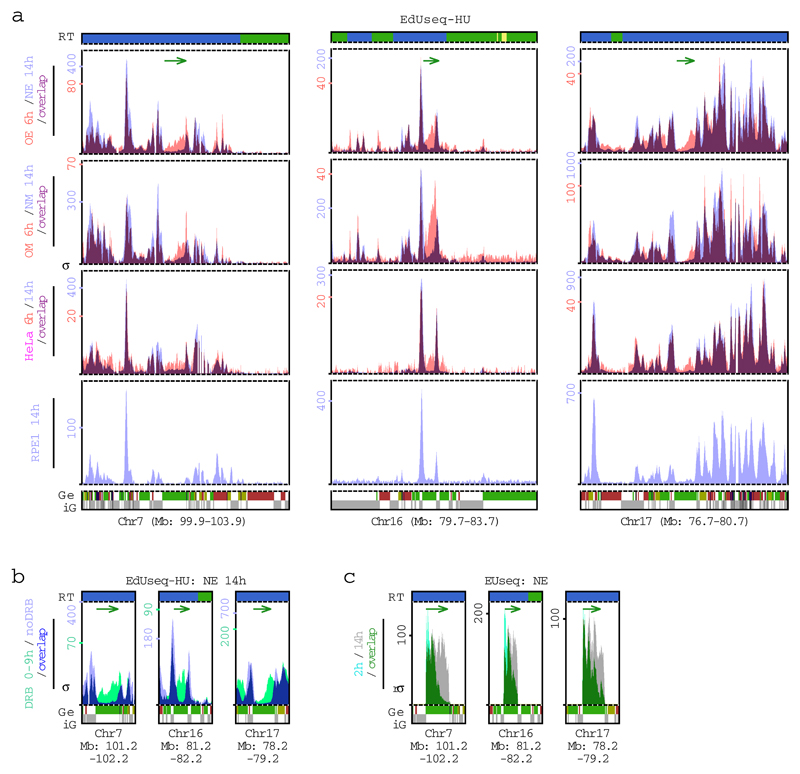
Extended Data Figure 7. Replication initiation and transcription profiles at selected genomic loci.
a, Replication initiation (EdUseq-HU) profiles at three genomic loci in different cells lines, from top to bottom: cells overexpressing cyclin E (OE) vs cells with normal cyclin E (NE) activity, harvested 6 and 14 h after mitotic shake-off, respectively; cells with induced Myc (OM) activity vs normal Myc (NM) activity, harvested 6 and 14 h after mitotic shake-off, respectively; HeLa cells harvested at 6 h vs 14 h after mitotic shake-off and RPE1 cells harvested 14 h after mitotic shake-off. Peak heights are represented as sigma values (σ). Replication timing (RT) domains and gene annotations (Ge/iG) are as in Fig. 1b. Bin resolution, 10 Kb; ruler scale, 100 Kb. The green arrows indicate the direction of transcription of the example gene of each locus harboring oncogene-induced origins.
b, Replication initiation (EdUseq-HU) profiles of control (noDRB) and DRB-treated (0-9 h) NE cells harvested 14 h after mitotic shake-off. The same genomic loci as in (a) are shown, focusing on the genes harboring the oncogene-induced origins. Replication timing (RT) domain and gene annotations (Ge/iG) are as in (a).
c, Newly-synthesized transcript profiles (EUseq) of NE cells, 2 and 14 h after mitotic shake-off (2h, light green; 14 h, gray; overlap, dark green) shown only for the example genes harboring the oncogene-induced origins shown in (a) (indicated by the green arrows). Replication timing (RT) domain and gene (Ge/iG) annotations are as in (a); rσ, relative sigma.
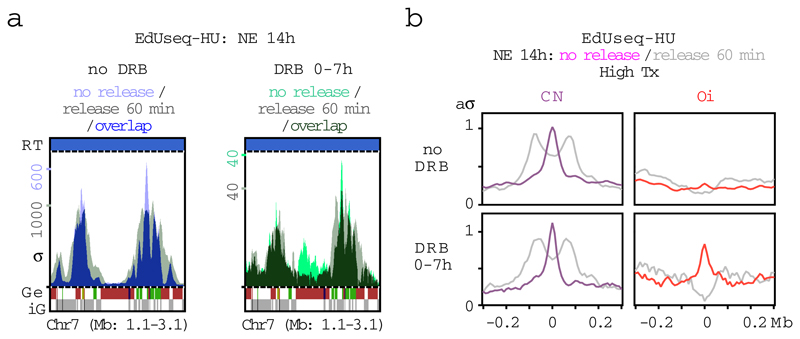
Extended Data Figure 8. Fork collapse at Oi origins induced in cells with normal levels of cyclin E by inhibiting transcription in early G1.
a, Replication initiation (EdUseq-HU, 14 h HU block) and fork progression (EdUseq-HU/release 60 min) profiles at a representative genomic region in U2OS cells with normal levels of cyclin E (NE) that were treated or not with DRB during the first 7 h of G1. Replication timing (RT) domains and gene annotations (Ge/iG) are as in Fig. 1b. Bin resolution, 10 Kb; ruler scale, 100 Kb.
b, Average fork progression (no release and 60 min release) at constitutive (CN) and oncogene-induced (Oi) origins located in highly transcribed regions in control and DRB treated (first 7 h of G1 phase) NE cells. aσ, adjusted average σ.
Extended Data Figure 9. Association of Oi origins with genomic rearrangements and replication timing profiles of cancer rearrangement breakpoints.
a, Mapping of translocations (Transloc; n=27,364) identified by LAM- HTGTS to genomic regions replicated from Oi origins (oncogene-induced replication initiation domains, OiRDs) with the analysis restricted to the early S replicating domains. The fraction of translocations mapping to OiRDs is shown for non-transcribed (0), low (Lo), medium (Me) and highly (Hi) transcribed genomic bins, as well as for all early S replicating bins. Statistical comparisons, using random permutation analysis, are between the NE (normal cyclin E activity, blue) and OE (cyclin E overexpression, pink) samples. The distribution of OiRDs in the genome (gray) is shown as reference.
b, Mapping of genomic rearrangement (Rearr; n=136) breakpoints, identified previously14 in the same cells overexpressing cyclin E (OE) to the oncogene-induced replication initiation domains (OiRDs), according to transcription levels, as in (a), with the analysis restricted to the early S replicating domains. Statistical comparisons, using random permutation analysis, are between observed (red) and genomic (gray) frequencies. NS, not significant.
c, Mapping of genomic rearrangement (Rearr; n=490,711) breakpoints from a TCGA pan-cancer dataset4 to the oncogene-induced replication initiation domains (OiRDs), according to transcription levels, as in (a), with the analysis restricted to the early S replicating domains. Statistical comparisons, using z-scores, are between observed and genomic frequencies.
d, Mapping of genomic rearrangement (Rearr; n=490,711) breakpoints in common cancer types from a TCGA pan-cancer dataset4 to the oncogene-induced replication initiation domains (OiRDs), with the analysis restricted to the early S replicating domains. KIRC, kidney renal cell; COAD, colon adenocarcinoma; HNSC, head and neck squamous cell; UCEC, uterine cervix; GBM, glioblastoma multiformae; LUAD, lung adenocarcinoma; LUSC, lung squamous cell; BRCA, breast; BLCA, bladder; OV, ovary. Statistical comparisons, using z-scores, are between observed (red) and genomic (gray) frequencies.
e, Distribution of cancer rearrangement breakpoints4 according to the replication timing data obtained from the REPLIseq experiment shown in Extended Data Fig. 2.
Extended Data Figure 10. Proposed mechanism for oncogene-induced DNA replication stress.
During the length of a normal G1 phase, transcription progressively inactivates intragenic origins, such that upon S phase entry origin firing is restricted to intergenic domains. Following oncogene activation, cells enter prematurely into S phase, prior to the inactivation of all intragenic origins. This results in bidirectional forks within highly transcribed genes, leading to conflicts between the replication and transcription machineries, fork collapse, DNA DSBs and genomic instability.
Supplementary Material
Supplementary Information is available in the online version of the paper.
Acknowledgements
We thank U. Schibler, R. Pillai, M. Docquier and present and past lab members for helpful discussions; J. Bartek for the U2OS cells inducibly overexpressing cyclin E; M. Eilers for the U2OS MycER cells; R. Beroukhim and S. Schumacher for access and help with TCGA cancer datasets; N. Roggli for help with the graphics scripts and the Flow Cytometry and Genomics platforms of the University of Geneva. This work was supported by grants from the European Commission (ONIDDAC) and the Swiss Science National Foundation.
Footnotes
Author Contributions T.D.H. and M.M. designed the experiments and wrote the paper; M.M. performed the experiments; T.D.H. wrote the computer scripts and performed the bioinformatic analyses with contributions by M.M.
Author Information The authors declare no competing financial interests.
References
- 1.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 2.Bignell GR, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ekholm-Reed S, et al. Deregulation of cyclin E in human cells interferes with prereplication complex assembly. J Cell Biol. 2004;165:789–800. doi: 10.1083/jcb.200404092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones RM, et al. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene. 2013;32:3744–3753. doi: 10.1038/onc.2012.387. [DOI] [PubMed] [Google Scholar]
- 7.Beck H, et al. Cyclin-dependent kinase suppression by WEE1 kinase protects the genome through control of replication initiation and nucleotide consumption. Mol Cell Biol. 2012;32:4226–4236. doi: 10.1128/MCB.00412-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Micco R, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 9.Bester AC, et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aird KM, et al. Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence. Cell Reports. 2013;3:1252–1265. doi: 10.1016/j.celrep.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toledo LI, et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155:1088–1103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- 12.Kotsantis P, et al. Increased global transcription activity as a mechanism of replication stress in cancer. Nat Commun. 2016;7 doi: 10.1038/ncomms13087. 13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartkova J, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 14.Costantino L, et al. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343:88–91. doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maya-Mendoza A, et al. Myc and Ras oncogenes engage different energy metabolism programs and evoke distinct patterns of oxidative and DNA replication stress. Mol Oncol. 2015;9:601–616. doi: 10.1016/j.molonc.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Resnitzky D, Gossen M, Bujard H, Reed SI. Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol Cell Biol. 1994;14:1669–1679. doi: 10.1128/mcb.14.3.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katou Y, et al. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- 18.MacAlpine DM. Coordination of replication and transcription along a Drosophila chromosome. Genes Dev. 2004;18:3094–3105. doi: 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karnani N, Dutta A. The effect of the intra-S-phase checkpoint on origins of replication in human cells. Genes Dev. 2011;25:621–633. doi: 10.1101/gad.2029711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen RS, et al. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad of Sci U S A. 2010;107:139–144. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sasaki T, et al. The Chinese hamster dihydrofolate reductase replication origin decision point follows activation of transcription and suppresses initiation of replication within transcription units. Mol Cell Biol. 2006;26:1051–1062. doi: 10.1128/MCB.26.3.1051-1062.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powell SK, et al. Dynamic loading and redistribution of the Mcm2-7 helicase complex through the cell cycle. EMBO J. 2015;34:531–543. doi: 10.15252/embj.201488307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu J, et al. Detecting DNA double-stranded breaks in mammalian genomes by linear amplification– mediated high-throughput genome-wide translocation sequencing. Nat Protoc. 2016;11:853–871. doi: 10.1038/nprot.2016.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson TE, et al. Large transcription units unify copy number variants and common fragile sites arising under replication stress. Genome Res. 2015;25:189–200. doi: 10.1101/gr.177121.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helmrich A, Ballarino M, Tora L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell. 2011;44:966–977. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 26.Letessier A, et al. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011;470:120–123. doi: 10.1038/nature09745. [DOI] [PubMed] [Google Scholar]
- 27.Barlow JH, et al. Identification of early replicating fragile sites that contribute to genome instability. Cell. 2013;152:620–632. doi: 10.1016/j.cell.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prado F, Aguilera A. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J. 2005;24:1267–1276. doi: 10.1038/sj.emboj.7600602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petryk N, et al. Replication landscape of the human genome. Nat Commun. 2016;7 doi: 10.1038/ncomms10208. 10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin MM, et al. Genome-wide depletion of replication initiation events in highly transcribed regions. Genome Res. 2011;21:1822–1832. doi: 10.1101/gr.124644.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The fastq sequencing data and associated information described in this study have been deposited in the Sequence Read Archive (SRA) as BioProject PRJNA397123.