Abstract
Animal development depends on not only the linear genome sequence that embeds millions of cis-regulatory elements, but also the three-dimensional (3D) chromatin architecture that orchestrates the interplay between cis-regulatory elements and their target genes. Compared to our knowledge of the cis-regulatory sequences, the understanding of the 3D genome organization in human and other eukaryotes is still limited. Recent advances in technologies to map the 3D genome architecture have greatly accelerated the pace of discovery. Here, we review emerging concepts of chromatin organization in mammalian cells, discuss the dynamics of chromatin conformation during development, and highlight important roles for chromatin organization in cancer and other human diseases.
Keywords: 3D chromatin organization, chromosome conformation capture, topologically associating domain, TAD, gene regulation
INTRODUCTION
Lineage-specific gene expression during development is driven by both gene promoters and distal enhancers (Kim & Shiekhattar 2015, Lee & Young 2013, Levine 2010, Ong & Corces 2011). Promoters initiate RNA synthesis by directing the loading of RNA polymerases and the accessory factors to transcription start sites. Enhancers confer tissue- and temporal-specific expression of target genes by recruiting sequence-specific transcription factors and chromatin remodeling complexes (Kim & Shiekhattar 2015, Lee & Young 2013, Levine 2010). In addition to enhancers and promoters, insulator elements play a key role in gene expression by limiting the action of enhancers and preventing the spreading of heterochromatin (Gaszner & Felsenfeld 2006). Thanks to efforts from several large-scale consortia, millions of candidate cis-regulatory elements have been cataloged in many model organisms, including human, mouse, and Drosophila (ENCODE Project Consortium 2012, modENCODE Consortium et al. 2010, Roadmap Epigenomics Consortium et al. 2015, Yue et al. 2014), paving the way for in-depth understanding of the gene-regulatory programs in these species.
To elucidate the gene-regulatory programs in humans and other species, delineating the interplay between enhancers and target promoters is necessary. This task is surprisingly nontrivial because enhancers can modulate transcription of target genes from a large genomic distance and regardless of relative orientations. Additionally, enhancers can regulate target genes that are not their immediate neighbors (Sanyal et al. 2012). Therefore, predicting enhancer-target relationships based simply on the linear genome sequence is not a solution (Levine 2010, Levine et al. 2014).
Early studies on the β-globin gene cluster and several other genetic loci demonstrated that spatial proximity between promoters and distal enhancers due to chromatin looping is critical for transcriptional regulation of developmentally regulated gene expression (Deng et al. 2012, 2014; Gaszner & Felsenfeld 2006; Tolhuis et al. 2002). Enhancer-target gene interactions appear to be restricted by insulator elements, which set the boundaries for their contacts (Gaszner & Felsenfeld 2006). Therefore, to decipher the target genes of enhancers and to understand the mechanisms of their action, it is necessary to consider how chromatin fibers are folded in 3D in different cell types and developmental stages.
Recent years have seen rapid progress in technologies for genome-wide analysis of 3D genome organization in eukaryotes. Additionally, chromatin organization in a growing number of cell types and species has been studied in greater and greater detail and scales. Here we provide an overview of our current understanding of 3D genome organization in mammalian genomes. We start with an introduction of the major tools used to map 3D genome structure, followed by a discussion of the general hierarchical structures in different cell types. We then focus on dynamics of 3D genome organization during various biological processes and their relationships with gene regulation. Finally, we illustrate recent examples showing how 3D genome conformation can inform us about the underlying molecular mechanisms of oncogene activation and congenital diseases.
TOOLS TO EXPLORE 3D GENOME ORGANIZATION
Two general approaches have been extensively used to study 3D genome organization: microscopy-based imaging tools and chromosome conformation capture (3C)-coupled sequencing methods (hereafter referred to as C-technologies). The imaging tools enable direct measurement of spatial distances between genomic loci and their movement in single cells but are limited in throughput, resolution, and genome coverage. Alternatively, C-technologies in combination with high-throughput sequencing can provide a genome-wide view of chromatin organization from populations of cells but might overlook cell-to-cell variations. The imaging-based technologies and C-technologies therefore provide complementary views of 3D genome organization, despite some discrepancies at specific regions (Dekker 2016, Williamson et al. 2014).
Imaging-Based Tools
Before the advent of C-technologies, the predominant method to study 3D genome organization was fluorescence in situ hybridization (FISH) (Langer-Safer et al. 1982). In FISH, the spatial distances between two or more loci are measured through hybridization of fluorescently labeled probes to DNA after fixation and mild denaturation and are visualized under light microscopy (Cremer et al. 2008). Because of the high levels of cell-to-cell variations, a large number of cells (often between 100 and 1,000) are generally needed to confidently and fully ascertain the spatial distances between a pair of loci.
Improvements have been made recently to address two major limitations of FISH: resolution and genome coverage. The spatial resolution of FISH is constrained by the diffraction limit of light sources, which is ~200 nm laterally and >500 nm axially (Walter et al. 2006). The genomic resolution depends on the sizes of the fluorescent probes used. The most frequently used fosmid probes are ~40 kb on average, making it difficult to resolve two DNA fragments spaced less than <100 kb apart along the genome. Improvement in the FISH resolution has been obtained by implementing super-resolution microscopy (reviewed in Lakadamyali & Cosma 2015) and short oligonucleotide-based probes (Beliveau et al. 2014). Conventional FISH assays characterize only a few specific loci simultaneously because of limited fluorescent labels. This limitation in genome coverage has been overcome by a multiplexed FISH method that employs multiple rounds of hybridizations and sequential imaging, enabling tracking of spatial positions of more than 30 genomic loci simultaneously (Wang et al. 2016).
Live-cell imaging can reveal the dynamic behaviors of interacting loci by tracing their 3D trajectories. For instance, the encounter frequency between a promoter and its distal regulatory element in pro-B cells may be determined via modeling using physical parameters extracted from live-cell imaging data (Lucas et al. 2014). However, live-cell imaging often requires laborious and time-consuming genetic engineering, which could potentially impede its general application. During recent years, increasing efforts have been made to directly label endogenous genomic loci, especially with the aid of dCas9 fusion proteins. Although technical challenges remain with these new technologies, such technologies promise to greatly expand our toolbox for analysis of dynamic 3D genome configuration in different species (reviewed in Chen et al. 2016).
C-Technologies
Besides direct visualization using image-based tools, 3D genome organization has also been successfully inferred from pairwise contact frequencies in the genome. In C-technologies, the nucleus is chemically fixed to preserve the 3D chromosome conformation, followed by DNA fragmentation and religation. If the two DNA fragments are spatially close at the time of cross-linking, they can be ligated, a process known as proximity ligation (Dekker et al. 2002). The frequency of the ligation products can be assayed to assess the spatial proximity between pairs of loci using PCR, DNA microarrays, or direct DNA sequencing (Dekker et al. 2002, Dostie et al. 2006, Lieberman-Aiden et al. 2009).
In recent years, next-generation DNA sequencing technologies have been combined with C-technologies, resulting in a proliferation of tools that map genomic organization with various coverage and resolution as outlined below (for a more detailed discussion of these methods, please see Schmitt et al. 2016b) (Figure 1).
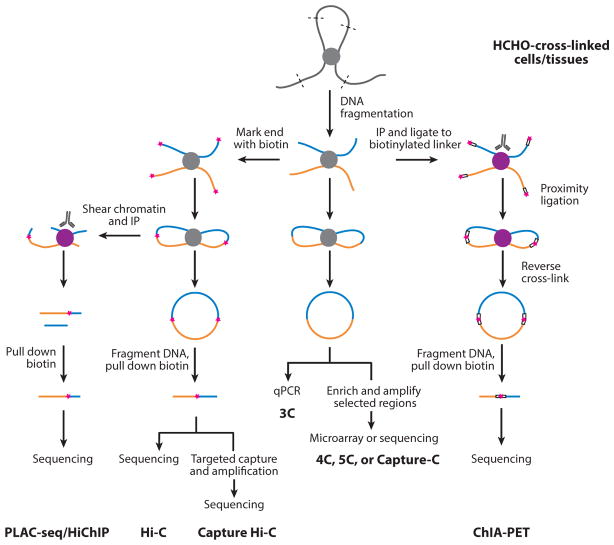
Figure 1.
Brief overview of the C-technologies. All C-technologies share the steps of formaldehyde cross-linking, DNA fragmentation, and proximity ligation but may differ in whether to label the fragment end with biotin, the strategy to enrich the regions of interest, and the quantification methods. The biotin label is represented by the pink star. Abbreviation: IP, immunoprecipitation.
3C: one to one
3C, the earliest version of C-technologies, measures contact frequency between two preselected loci (Dekker et al. 2002). In 3C, the proximity ligation products are amplified and quantified by PCR using a pair of primers matching the ends of two DNA fragments of interest. This technology’s resolution is determined by the restriction enzyme used for fragmentation: A four-cutter yields a resolution of ~256 bp and a six-cutter a resolution of ~4,096 bp. Despite its low throughput, 3C is extensively used to verify long-range interactions between genomic loci due to its high resolution and ease of use.
Hi-C: all to all
Hi-C offers unbiased measurement of all possible interactions across the genome via a combination of 3C and next-generation DNA sequencing (Lieberman-Aiden et al. 2009). Compared with 3C, the major modification of Hi-C is labeling DNA ends with biotin for subsequent enrichment of proximity ligation products. Different variants of Hi-C, including tethered conformation capture, DNase Hi-C, micro-C, and in situ Hi-C, have been invented to further improve the resolution or efficiency of proximity ligation and to reduce the background noise (Hsieh et al. 2015, Kalhor et al. 2011, Ma et al. 2015, Rao et al. 2014).
Although Hi-C has a similar theoretical resolution with 3C, its practical resolution is restricted by sequencing depth. For instance, to achieve ~1-Mb, 10-kb, and 1-kb resolution would require approximately 8.4 million, 2 billion, and 6.5 billion total reads, respectively (Jin et al. 2013, Lieberman-Aiden et al. 2009, Rao et al. 2014). Given the high sequencing cost of high-resolution Hi-C, a variety of C-technologies with sacrificed genome coverage have been developed to cost-efficiently examine the 3D chromatin structure at selected loci/regions with high resolution (see below).
Whereas the conventional Hi-C protocol measures the average contact frequencies in millions of cells, single-cell Hi-C techniques have been developed to analyze chromosome conformation in individual cells and to uncover cell-to-cell variability. The first single-cell Hi-C study relied on manual isolation of single nuclei after in situ proximity ligation, followed by proximity ligation product enrichment and sequencing library construction. Due to the low efficiency, the procedure gives rise to approximately 10,000 unique contacts per cell on average for tens of cells at a time (Nagano et al. 2013). Recently, this protocol was substantially enhanced in both genome coverage and throughput, leading to the acquisition of Hi-C maps from ~3,000 single cells with a median of 182,000 unique contacts per cell (Nagano et al. 2017). Independently, high-throughput single-cell Hi-C has also been carried out by combining combinatorial indexing and Hi-C, achieving impressive throughput and coverage (Ramani et al. 2017). Whereas the above single-cell Hi-C methods start from populations of cells for restriction enzyme digestion and proximity ligation, performing such steps on single cells was recently achieved (Flyamer et al. 2017, Stevens et al. 2017). Stevens et al. (2017) combined imaging with an in-nucleus Hi-C process, yielding 37,000–122,000 contacts for eight individual cells. By contrast, single-nucleus Hi-C employing whole-genome amplification that skips biotin enrichment revealed up to 1.9 million unique contacts per oocyte (Flyamer et al. 2017).
4C: one to all
4C allows for genome-wide identification of all possible interacting partners for one specific locus of interest, or viewpoint (Simonis et al. 2009). By using primers designed to target the viewpoint, all ligation products containing this viewpoint sequence can be amplified and then quantified by high-throughput sequencing. 4C can be used to easily achieve a resolution of a few kilobases with several million sequencing reads.
5C: many to many
5C was developed to measure contact frequencies among DNA fragments within a finite number of target regions (Dostie et al. 2006). To detect specific proximity ligation products in these regions, a pool of 5C primers are used for multiplexed capture, followed by ligation-mediated amplification. Typically, 5C is used to assess chromatin interactions among regions that are several hundred kilobases to a few megabases in size. Compared to Hi-C, 5C greatly reduces the number of sequencing reads required to reach the same resolution. A study employing 5C to examine ~7-Mb regions can achieve ~4-kb resolution with ~20 million reads, whereas similar analysis with Hi-C requires billions of reads (Phillips-Cremins et al. 2013, Rao et al. 2014).
ChIA-PET: many to many
ChIA-PET (chromatin interaction analysis by paired-end tag), which combines ChIP and proximity ligation and sequencing, detects contacts from a subset of genomic loci associated with a particular protein (Fullwood et al. 2009). In ChIA-PET, cross-linked chromatins are sonicated and then subjected to immunoprecipitation by using antibodies against the protein of interest, followed by proximity ligation, amplification of proximity ligation products, and sequencing. Similar to 5C, ChIA-PET is a cost-effective method to identify long-range interactions at high resolution. In a recent study, CTCF ChIA-PET achieved ~100-bp resolution (Tang et al. 2015). A robust ChIA-PET experiment often requires a few hundred million cells as starting material, due to its inefficiency of proximity ligation after chromatin sonication.
Capture-C/Capture Hi-C: many to all
When genome-wide identification of contacting partners for tens of selected loci is desired, such information can be acquired with multiple 4C experiments. However, if the research focus is on hundreds of loci or more, such as all promoters across the genome, further scaling up of 4C is impractical. To generate such many-to-all contact maps, alternative strategies are needed to enrich all ligation junctions with at least one end falling into a preselected pool.
One way to this end is combining 3C or Hi-C with targeted capture and sequencing (Capture-C/Capture Hi-C) (Dryden et al. 2014, Hughes et al. 2014). In Capture-C/Capture Hi-C, a 3C or Hi-C library is subject to targeted capture by hybridization to pools of DNA or RNA oligos to enrich for the proximity ligation products corresponding to specific regions. The resulting DNA is sequenced and analyzed to reveal spatial contacts centered at these regions. This technique has been used successfully to identify chromatin interactions centered on human and mouse promoters in many cell types (Hughes et al. 2014, Javierre et al. 2016, Mifsud et al. 2015, Schoenfelder et al. 2015a).
PLAC-seq/HiChIP: many to all
Very recently, PLAC-seq and HiChIP were introduced as another many-to-all approach that enables identification of long-range interactions associated with specific proteins of interest (Fang et al. 2016, Mumbach et al. 2016). In these methods, first DNA fragmentation and in situ proximity ligation are performed in the cross-linked cells just as in in situ Hi-C, and then chromatin immunoprecipitation using specific antibodies is carried out, followed by enrichment of biotinylated ligation junctions. In contrast to Capture-C/Capture Hi-C, PLAC-seq/HiChIP does not require synthesis of capture probes and is a versatile method to map long-range interactions associated with different categories of genomic regions with relatively low sequencing cost. This method uses substantially fewer cells and simpler procedures than does ChIA-PET.
GENERAL FEATURES OF 3D GENOME ORGANIZATION
Overwhelming data suggest that the genome is not randomly folded in the interphase nucleus (Figure 2). Several common features of genome organization have been reported in diverse species and cell types. At the largest scale, each chromosome occupies a defined volume in the nucleus, which is referred to as a chromosome territory (CT). At the subchromosome scale, two compartments can be observed, with each composed of multiple noncontinuous regions of several to tens of megabases in size. At a finer scale, topologically associating domains (TADs) have been delineated that show remarkable conservation across cell types and species. Compartments and TADs can be further substratified into subcompartments and sub-TADs (or contact domains), respectively. Finally, tens of thousands of long-range chromatin loops, spanning hundreds of kilobases, have also been defined in mammalian cells (Rao et al. 2014, Tang et al. 2015).
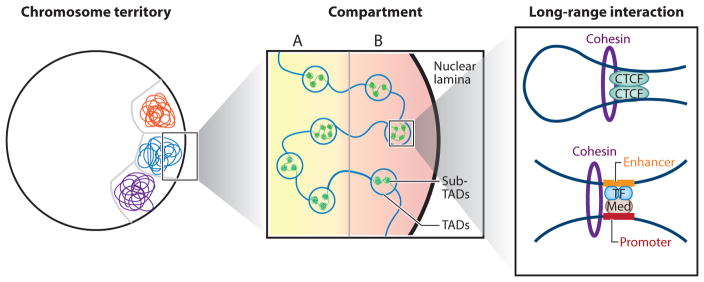
Figure 2.
Hierarchical genome organization in mammals. From a large to a fine scale, chromosome territory, compartment A/B, topologically associating domains (TADs), sub-TADs, and long-range interaction can be observed (left to right). (Left) Each chromosome territory is denoted by different colors. Only three chromosomes are shown. (Middle) Compartments A and B are indicated by a yellow and pink background, respectively. Compartment B is correlated with nuclear lamina. TADs are represented by large indigo circles, whereas the inner sub-TADs are shown as smaller green circles. (Right) Two types of long-range interactions are shown: One interaction is a CTCF-CTCF loop mediated by the Cohesin complex, whereas the other is between the promoter and the enhancer that often involves the Mediator (Med) complex, the Cohesin complex, and lineage-specific transcription factors.
Chromosome Territories
In the interphase, each chromosome occupies a specific region of a nucleus known as a CT (reviewed in Cremer & Cremer 2010). Overlaps between different chromosomes are restricted to the borders of CTs (Branco & Pombo 2006). CTs can be visualized using FISH, from which their positions in the nucleus and the neighborhood CTs can be determined (Bolzer et al. 2005). Segregation of chromosomes into nonoverlapping territories is also supported by Hi-C data sets, as evidenced by much greater intrachromosomal contact probability than for interchromosomal contacts (Lieberman-Aiden et al. 2009). This principle led to the successful use of Hi-C data for haplotype phasing (Selvaraj et al. 2013) and to de novo scaffolding of contigs (Burton et al. 2013, Kaplan & Dekker 2013).
CTs in the nucleus are not randomly distributed, and several features of CT positioning have been observed. First, nuclear CT positions are partially conserved after mitosis, yielding nonrandomly altered patterns in daughter cells compared to mother cells (Parada et al. 2003). Second, despite this cell-to-cell variation, individual CTs have their preferences for radial positioning, which largely depend on their genomic properties. In general, small, gene-rich chromosomes are located close to the center of the nucleus, whereas larger, gene-poor ones are located near the nuclear periphery (Croft et al. 1999). Third, CT positioning also correlates with cell type–specific factors such as replication timing and transcriptional activity. Early-replicating loci and active genes tend to localize in the nuclear interior, whereas late-replicating loci and repressed genes have a preference for nuclear periphery (Grasser et al. 2008, Takizawa et al. 2008).
Compartments
The Hi-C data, after normalization and converting to an observed/expected matrix, display a plaid pattern, suggesting the existence of two compartments or nuclear neighborhoods (Figure 3). Principal-component analysis on the observed/expected matrix partitions each chromosome into compartments A and B based on the first principal component (Lieberman-Aiden et al. 2009). Compartment A regions are early replicating, contain a high density of genes, and exhibit strong mRNA expression activities. They are enriched for H3K36me3 and DNaseI hypersensitive sites (Lieberman-Aiden et al. 2009). In contrast, compartment B sequences are late replicating and overlap strongly with lamina-associated domains (Ryba et al. 2010). Compartment A and B partition is cell type specific; 36% of the human genome shows compartment A/B switching during stem cell differentiation (Dixon et al. 2015), and this number increases to ~60% when 14 additional primary tissues are analyzed (Schmitt et al. 2016a).
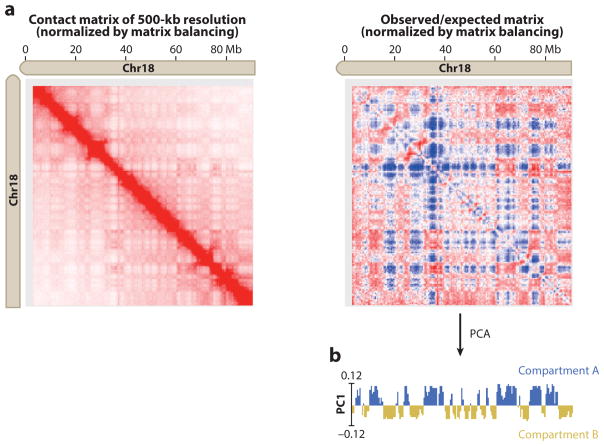
Figure 3.
Hi-C data reveal two types of compartments in each chromosome. (a) A plaid pattern emerges after transforming the Hi-C contact matrix (left) to the observed/expected matrix (right), suggesting the presence of two compartments. Contacts within the same compartment are enriched, whereas contacts between different compartments are depleted. The Hi-C data are from mouse embryonic stem cells (Fang et al. 2016) and are visualized with Juicebox (Durand et al. 2016). (b) Principal-component analysis (PCA) on the observed/expected matrix partitions each chromosome into two compartments, A and B, based on the first principal component (PC1).
Although the presence of compartment A/B is illustrated by Hi-C data in a population of cells, whether such organization is conserved in each individual cell is still unclear. Wang et al. (2016) answer this question by tracing the spatial positions of multiple genomic loci along the individual chromosomes using multiplexed FISH and confirm the existence of compartment A/B at the single-cell level. In line with the implication of depleted contacts between different compartments, compartments A and B are separated in space and are organized in a spatially polarized manner (Wang et al. 2016).
With a higher-resolution Hi-C map, compartments A and B can be further divided into five subcompartments, A1–A2 and B1–B3, and each compartment associates with a specific pattern of histone modifications (Rao et al. 2014). Interestingly, five principal chromatin types (two active and three repressed) are also characterized in Drosophila cells (Filion et al. 2010), suggesting that similar compartment organization may be conserved in metazoa.
Topologically Associating Domains
In the exploration of contact maps from Hi-C or 5C with 40-kb resolution or higher, self-associating chromosomal domains emerge and are referred to as topological domains or TADs (Figure 4a) (Dixon et al. 2012, Nora et al. 2012). In mouse embryonic stem cells (ESCs), more than 90% of the genomes are organized into ~2,200 TADs with a median size of 880 kb (Dixon et al. 2012). Similar domain organization has also been observed (albeit in a smaller size) in nonmammalian genomes such as Drosophila (Sexton et al. 2012), zebrafish (Gómez-Marín et al. 2015), Caenorhabditis elegans (Crane et al. 2015), and yeast (Hsieh et al. 2015, Mizuguchi et al. 2015). Contact frequency between genomic loci in the same TADs is severalfold higher than the contact frequency of loci belonging to different TADs. In agreement with the observation from C-technologies, one pair of loci from the same TAD is also spatially closer than the other pair of similar genomic distance but from different TADs (Dixon et al. 2012). Interestingly, TAD-like structures were not detected in Hi-C maps generated from Arabidopsis thaliana (Wang et al. 2015), suggesting that plants may employ distinct mechanisms for chromatin folding.
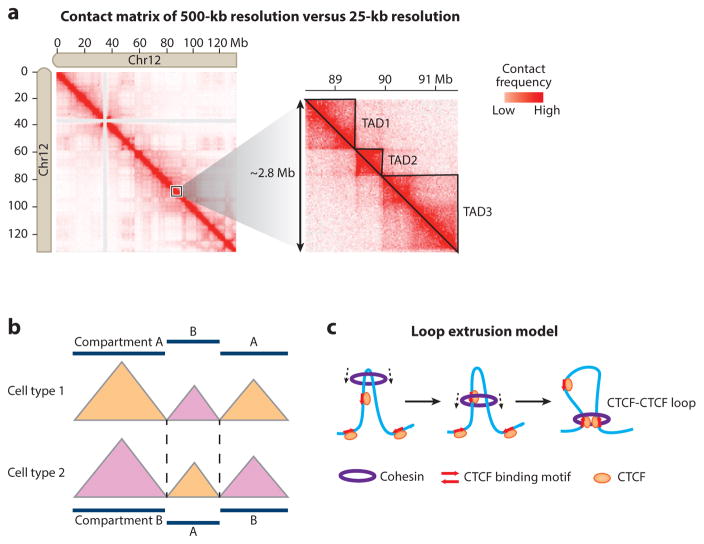
Figure 4.
Identification and characterization of topologically associating domains (TADs). (a) (Left) Cis-contact matrix for chromosome 12 from human embryonic stem cells at 500-kb resolution. (Right) A detailed contact matrix of an ~2.8-Mb region at 25-kb resolution reveals TAD organization as triangles. TAD1–3 are manually annotated and indicated by solid lines. The Hi-C data are taken from Dixon et al. (2012) and are visualized with Juicebox (Durand et al. 2016). (b) The position of TAD boundaries is largely conserved between different cell types, but TADs may transit between different compartments. (c) TAD formation by loop extrusion. The Cohesin complex forms a ring structure and travels along the DNA fiber, extruding a progressively larger loop until it is stalled by bound CTCF with convergent orientation (red arrows).
Multiple lines of evidence support TAD as a fundamental unit of genome organization in different species (Dekker & Heard 2015, Dixon et al. 2016). First, the positioning of TAD is relatively stable across cell types and appears to be independent of tissue-specific gene expression or histone modifications. During ESC differentiation, genome-wide switching of compartments A and B occurs, whereas TAD positioning remains stable (Figure 4b) (Dixon et al. 2015). In the sperm nucleus, where the majority of histones are replaced by protamines, the identified TADs also display a distribution similar to those from fibroblast cells (Battulin et al. 2015). Second, TAD positioning is evolutionarily conserved; 50–70% of TAD boundaries are shared between human and mouse ESCs (Dixon et al. 2015). Third, the activity of promoters and enhancers within the same TAD appears to be weakly coordinated (Dixon et al. 2015, Nora et al. 2012, Zhan et al. 2017). Furthermore, a reporter gene randomly inserted in different places in the mouse genome mimics expression patterns of genes within the same TAD (Symmons et al. 2014). Fourth, TAD is a stable unit of replication-time regulation. Replication domains of ~400–800 kb whose positions are invariant across different cell types and species had been previously identified with microscopy (Hiratani et al. 2008, Jackson & Pombo 1998, Ryba et al. 2010). A recent study showed that the boundaries of replication domains exhibit a nearly one-to-one correspondence with TAD boundaries, suggesting that the TAD is equivalent to the replication domain (Pope et al. 2014).
The relatively stable nature of the TAD raises the questions of what mechanisms demarcate TAD boundaries and establish TADs. Hints come from observations that TAD boundaries are enriched for multiple factors, including H3K4me3, H3K36me3, transcription start sites, housekeeping genes, tRNA genes, short interspersed nuclear elements, and CTCF binding sites. Among them, CTCF binding sites and housekeeping genes are particularly strongly enriched; 75% and 33% of all the TAD boundaries in mouse ESCs associate with CTCF and housekeeping genes within 20 kb, respectively (Dixon et al. 2012). These results suggest that both CTCF binding and high levels of transcription activity may contribute to TAD formation.
Earlier work has implicated CTCF as a key player in genome organization (reviewed by Phillips & Corces 2009). Recent studies have further supported the role of this DNA binding protein in the formation of TADs. Using RNAi to deplete 80% of CTCF, Zuin et al. (2014) observed reduced intra-TAD interactions and increased inter-TAD interactions, suggesting weakening, but not the disappearance of, TAD boundaries. To explain how CTCF-mediated long-range interactions can drive the formation of TADs, a loop extrusion model is proposed in which a chromatin motor complex (such as the Cohesin complex) loads onto DNA and then extrudes a progressively larger loop until it is stalled by CTCF binding at convergent orientation, thus setting up TADs (Figure 4c) (Sanborn et al. 2015). In polymer simulations, this model not only gives rise to interaction patterns that agree well with the observed TAD structures in Hi-C data but also makes accurate predictions for new chromatin loop formation after deletion of anchoring CTCF binding sites (Fudenberg et al. 2016, Sanborn et al. 2015). Nevertheless, a substantial fraction of TAD boundaries lack CTCF occupancy (Dixon et al. 2012), suggesting alternative, CTCF-independent mechanisms responsible for TAD formation.
Given the enrichment of housekeeping genes and active marks at TAD boundaries, some boundaries can be simply defined by the distribution of highly transcribed genes. Previous studies show that long strings of nucleosomes fold into secondary structures and further fold into compact tertiary structures, which may result in the formation of TADs. By contrast, at TAD boundaries such higher-order structures are destabilized and are disrupted by a high level of histone acetylation at active genes (reviewed in Pepenella et al. 2014). Using polymer simulations, Ulianov et al. (2016) demonstrated that the TAD partitioning in Drosophila is based primarily on the different packing abilities of active and inactive genes. Similar analysis is yet to be performed on mammalian genomes, and further studies are required to determine how much histone acetylation may contribute to the TAD formation in mammals.
In addition to CTCF, the Cohesin complex, and active genes, a recent simulation study also suggests that TAD organization may be regulated by DNA supercoiling (Benedetti et al. 2014). Indeed, the boundaries of supercoiling domains and TADs partially overlap (Naughton et al. 2013). Additionally, topoisomerase II beta (TOP2B) also colocalizes with CTCF and Cohesin at TAD boundaries (Uusküla-Reimand et al. 2016). Further experiments are needed to demonstrate the functional role of TOP2B and supercoiling in the formation of TADs and chromatin loops.
Sub–Topologically Associating Domains and Insulation Neighborhoods
TADs can be further divided into smaller sub-TADs observed from high-resolution 5C of mouse ESCs (Phillips-Cremins et al. 2013). The later genome-wide, 1-kb-resolution in situ Hi-C data reveal a similar size of structure in human GM12878, termed as contact domains (medium size 185 kb) (Rao et al. 2014). Like TADs, sub-TADs display the self-association feature with a decrease in contact frequency across sub-TAD boundaries, and some sub-TAD boundaries are associated with CTCF/Cohesin-mediated interactions (Rao et al. 2014). However, compared to TADs, sub-TADs are less conserved across cell/tissue types and appear to be related to cell type–specific gene expression (Berlivet et al. 2013, Phillips-Cremins et al. 2013). The variant sub-TAD partitioning may be caused by cell type–specific enhancer-promoter interactions mediated by Mediator/Cohesin and lineage-specific proteins (Phillips-Cremins et al. 2013).
The presence of smaller structural units within TADs has also been reported by analyzing CTCF and Cohesin ChIA-PET data. Young and colleagues observed that the chromatin loops anchored by CTCF and Cohesin served as the units of gene regulation (Dowen et al. 2014, Hnisz et al. 2016a). The regions defined by such chromatin loops, which were termed as insulated neighborhoods, appear to constrain the enhancer-promoter interactions within the same neighborhood. Perturbation of the anchors by genetic engineering and/or naturally occurred mutations led to gene dysregulation (Dowen et al. 2014, Hnisz et al. 2016b, Ji et al. 2016).
Chromatin Loops
According to the polymeric nature of chromatin fibers, two distant genomic loci would contact each other at very low frequency due to random collision (Bornfleth et al. 1999, Lieberman-Aiden et al. 2009, Marshall et al. 1997). However, certain loci have a significantly higher contact frequency than expected by chance, and such long-range interactions may provide insights into transcription regulation at distal regulatory elements. The link between long-range chromatin interactions and transcriptional activation has been demonstrated by the study of β-globin and γ-globin genes. In these cases, creation of ectopic interactions between the promoters and locus control regions is sufficient to trigger transcriptional activation (Deng et al. 2012, 2014).
Given that the sizes of individual regulatory elements range from several hundred bases to a few kilobases, it is imperative that chromatin looping interactions be identified at near-kilobase resolution. To achieve this, high-resolution C-data covering a portion of or even the entire genome have been generated for various cell types (Javierre et al. 2016, Jin et al. 2013, Krijger et al. 2016, Li et al. 2012, Phillips-Cremins et al. 2013, Rao et al. 2014, Sanyal et al. 2012, Schoenfelder et al. 2015a, Tang et al. 2015, Y. Zhang et al. 2013). Despite the differences in the cell type studied and the algorithms used for long-range interaction identification, universal features of long-range interactions have been revealed. First, such interactions often occur in the same TADs or sub-TADs (Jin et al. 2013), and extremely long range interactions are very rare except for specific genomic regions, such as Hox genes (Joshi et al. 2015). Second, active promoters, enhancers, and CTCF binding sites are frequently involved in the long-range interactions (Jin et al. 2013, Sanyal et al. 2012). In addition to promoter-enhancer interactions, promoter-promoter and enhancer-enhancer interactions are widespread to form complex networks (Li et al. 2012, Sanyal et al. 2012, Y. Zhang et al. 2013).
These studies have revealed at least two different types of long-range interactions. Interactions of the first type are constitutive, are invariant across cell types, and demarcate TAD or sub-TAD boundaries (Dowen et al. 2014, Ji et al. 2016, Phillips-Cremins et al. 2013). Such chromatin interactions are often found in the form of DNA loops mediated by CTCF and the Cohesin complex (Rao et al. 2014). A majority of these CTCF-CTCF loops are found in a convergent orientation (Rao et al. 2014, Tang et al. 2015), which helps to explain why only a small portion of all CTCF binding sites are involved in domain boundary demarcation. This orientation preference is also one of the major assumptions in the loop extrusion model to effectively simulate TAD structures (see above). The importance of the orientation of CTCF binding motifs in loop formation is further supported by genetic engineering (de Wit et al. 2015, Guo et al. 2015, Sanborn et al. 2015). The other type of long-range interaction is more cell type specific and is often associated with enhancers and enhancer-associated factors as well as with the Cohesin complex and the Mediator complex (Ji et al. 2016, Phillips-Cremins et al. 2013).
DYNAMIC 3D GENOME ORGANIZATION AND GENE REGULATION
Although some aspects of the 3D genome architecture are conserved in different cell types, notable reorganization can happen in distinct biological processes. For example, dramatic changes in chromatin conformation occur during mitosis and X chromosome inactivation. In contrast, during differentiation and reprogramming, more subtle changes that may contribute to cell type–specific gene expression take place.
3D Genome Reorganization During the Cell Cycle
Upon entering mitosis, the chromosomes begin to condense and become visible under the light microscope (Paweletz 2001). The molecular details of the internal organization of mitotic chromosomes have been investigated with C-technologies (Naumova et al. 2013). At M phase, previously described features of interphase chromosome organization, such as TADs and compartments, appear to be completely erased. Instead, mitotic chromosomes acquire a homogeneous folding state regardless of local sequence and chromosome identity. Such mitotic chromosome conformation is shared between different cell types, suggesting a common organizational principle of mitotic chromosomes. Combining the experimental data and polymer simulation results, Naumova et al. (2013) propose that mitotic chromosomes are folded in two steps: First, linear compaction is achieved by the formation of stochastic arrays of consecutive loops (80–120 kb in size), probably through loop extrusion mediated by SMC complexes (Goloborodko et al. 2016). Second, axial compression occurs to form the cylindrical shape of a mitotic chromosome.
After mitosis, cell type–specific chromosome conformation must be rapidly reestablished in daughter cells to ensure that the same set of genes can be expressed properly. This process is proposed to start from TAD formation, which likely relies on specific bookmarks of TAD boundaries on mitotic chromosomes. Afterward, the long-range interactions within TADs are established, and compartment A/B is formed by stochastic self-assembly of TADs in a cell type–specific manner (Dekker 2014).
At different stages of interphase, chromosome conformations have also been tracked, and they are largely invariant. The A/B compartment and the TAD borders are stable across early G1, mid-G1, and S phases (Naumova et al. 2013). However, the strength of A/B compartmentalization and TAD insulation is subject to change quantitatively through interphase correlated with DNA replication, as demonstrated by a recent single-cell Hi-C study (Nagano et al. 2017).
3D Genome Organization of the Inactive X Chromosome
Homologous chromosomes other than the X chromosome have highly similar high-order chromatin structure (Darrow et al. 2016, Deng et al. 2015, Dixon et al. 2015, Giorgetti et al. 2016, Rao et al. 2014): The active X chromosome (Xa) has TADs of regular size, whereas the inactive X chromosome (Xi) is partitioned into two large, contiguous domains, with a general absence of TADs. Similar Xi organization is conserved across the human, rhesus macaque, and mouse. The boundary of two large domains on Xi lies near the DXZ4 macrosatellite and its orthologs, which are bound by CTCF on Xi but not on Xa (Darrow et al. 2016, Horakova et al. 2012, Rao et al. 2014).
Both Xist repression and the DXZ4 element contribute to the formation of Xi-specific conformation (Darrow et al. 2016, Giorgetti et al. 2016, Minajigi et al. 2015). Loss of Xist partially restores the TADs on the Xi, converting it to a more Xa-like conformation (Minajigi et al. 2015). In contrast, the recruitment of Xist to Xa promotes the formation of a domain boundary at DXZ4, a signature of Xi conformation (Giorgetti et al. 2016). The mechanism of Xist regulating Xi organization may be repelling of Cohesin binding at specific loci (Minajigi et al. 2015). In contrast to Xist ablation, deletion of DXZ4 causes fusion of the two large domains as well as a further reduction of TAD signals on Xi (Darrow et al. 2016, Giorgetti et al. 2016). Additionally, loss of DXZ4 also influences the histone modification patterns on Xi and has a minor effect on Xi transcription (Darrow et al. 2016).
3D Genome Reorganization During Embryonic Stem Cell Differentiation
Genome-wide 3D genome reorganization during stem cell differentiation has been recently examined in human embryonic stem cells (hESCs) (Dixon et al. 2015). Consistent with the notion that TADs serve as stable organizational units, TAD boundary positioning is largely unchanged when hESCs differentiate into four distinct early embryonic cell lineages. However, both intra-TAD and inter-TAD interactions change in a way corresponding to the alterations in transcription levels and epigenetic states. Upon differentiation, hundreds of TADs display a significant overall increase or decrease in intradomain interaction frequencies. TADs that gain more interactions show increased levels of active epigenetic marks, upregulation of gene expression, and a shift from compartment B to A. By contrast, TADs with decreased chromatin interactions tend to display downregulation of gene expression and a shift from compartment A to B.
Whereas the above study focuses mainly on ESC differentiation, a more comprehensive analysis including primary human tissues uncovers an additional tissue type–specific feature of genome organization termed FIREs (frequently interacting regions) (Schmitt et al. 2016a). FIREs are regions that exhibit unusually high levels of local contact frequency (typically within 200 kb). Unlike long-range interactions that connect two genomic regions, FIREs involve multiple partners in the neighborhood and represent local chromatin interaction hot spots. FIREs are highly tissue specific, are located near cell identity genes, and are enriched for active enhancers and superenhancers, suggesting that a more complex hub-like spatial structure is present around tissue-specific genes to regulate their expression. Establishment of FIREs relies on the Cohesin complex because depletion of Cohesin causes a dramatic reduction of local interaction frequencies at FIREs in both human and mouse cells.
In addition to ESC differentiation, 3D genome reorganization has been investigated in the immune system. Upon antigen activation, naive B cells undergo dramatic changes in phenotype and transit to germinal center (GC) B cells. A recent study tracks 3D genome architecture during this transition and reveals that massive genome reorganization occurs, coordinating changes in expression and epigenetic regulation (Bunting et al. 2016). A GC B cell–specific interaction network is established around promoters of GC B phenotype–driving genes as well as active GC B enhancers bound by PU1 and SPID, which play critical roles in GC formation. As in ESC differentiation, the genomic regions that acquire long-range interactions during B cell maturation are associated with active epigenetic marks, including H3K27ac, H3K4me1, and H3K4me3.
3D Genome Organization in Pluripotent Stem Cells
Pluripotent stem cells have served as a model cell system to elucidate the mechanisms of 3D genome organization and to explore the relationships between dynamic chromatin organization and gene expression. Specific long-range interactions are established around the promoters of the key pluripotency genes Oct4, Nanog, and Sox2 (ONS) for their activation (Apostolou et al. 2013, Denholtz et al. 2013, Phillips-Cremins et al. 2013, Wei et al. 2013, H. Zhang et al. 2013). For instance, transcriptional activation of endogenous Oct4 during reprogramming relies on long-range intrachromosomal interactions between the Oct4 promoter and its distal enhancers; such interactions are mediated by Cohesin and Klf4. Depletion of either Cohesin or Klf4 in ESCs or induced pluripotent stem cells (iPSCs) disrupts those interactions and triggers cell differentiation, indicating a causal link between long-range interaction establishment and gene activation (Wei et al. 2013, H. Zhang et al. 2013). Similarly, Nanog-centered interactions also display a pluripotency-specific pattern, and knockdown of the mediating proteins Cohesin and/or Mediator leads to a significantly decreased reprogramming efficiency (Apostolou et al. 2013).
CTCF, the Cohesin complex, and the Mediator complex facilitate long-range interactions (Hadjur et al. 2009, Kagey et al. 2010, Splinter et al. 2006). Their specific roles in ESC-specific and constitutive interactions were revealed by comparing the 3D chromatin interactome between ESCs and neural progenitor cells (NPCs) around six key developmentally regulated genes (Oct4, Nanog, Nestin, Sox2, Klf4, and Olig1-Olig2) (Phillips-Cremins et al. 2013). CTCF and Cohesin cooccupancy is essential for the maintenance of constitutive interactions, whereas ESC-specific interactions are bridged by Cohesin and Mediator. This observation suggests that CTCF-dependent, Cohesin-mediated interactions tend to define invariant TADs and sub-TADs, whereas the CTCF-independent interactions are more likely to associate with lineage-specific enhancers. A recent study with genome-wide coverage confirms this classification (Ji et al. 2016). Application of Cohesin ChIA-PET to human ESCs also identified two categories of interactions: Cohesin/CTCF-mediated interactions and Cohesin/enhancer-mediated interactions. Cohesin-associated CTCF-CTCF loops connect boundaries of conserved TADs and are shared in both naive and primed human ESCs. By contrast, Cohesin-associated enhancer-promoter interactions are found within TADs, where differential regulation of genes occurs during the transition from the naive state to the primed state.
Besides CTCF, Cohesin, and Mediator, pluripotency factors are also integral to pluripotent cell–specific 3D genome architecture. First, ONS binding is enriched at ESC-specific interacting regions (de Wit et al. 2013, Denholtz et al. 2013, Phillips-Cremins et al. 2013). The contribution of ONS to ESC-specific interactions is further demonstrated by depletion of Oct4 or Nanog: Loss of either protein leads to a decrease in contact frequency specifically at its corresponding binding regions but does not affect the overall genome architecture. In addition, new contacts are established after the introduction of extra Nanog binding to the target genomic locus, underscoring the direct role of Nanog in mediating long-range interactions (de Wit et al. 2013).
Another notable feature of pluripotent genome organization is that Polycomb proteins shape repressive 3D spatial networks around developmentally important genes (Denholtz et al. 2013, Joshi et al. 2015, Schoenfelder et al. 2015b). In particular, extensive long-range promoter-promoter (both intra- and interchromosomal) interactions are observed at Hox gene clusters and are abolished upon depletion of subunits of Polycomb repressive complex 1 or 2 (Joshi et al. 2015, Schoenfelder et al. 2015b). However, such contacts are absent in naive ESCs cultured in 2i/LIF medium, suggesting that such contacts are not essential for maintenance of pluripotency (Joshi et al. 2015).
During reprogramming, massive 3D genome reorganization occurs, and somatic cells gradually acquire ESC-like genome structures (Beagan et al. 2016, Krijger et al. 2016). At the level of compartment A/B and TADs, the 3D genome organization of ESCs and iPSCs derived from different cell origins is largely identical (Krijger et al. 2016). Yet at a finer scale, detectable differences are observed between ESCs and NPC-derived iPSCs and are correlated with inaccurately reprogrammed gene expression. These incompletely rewired local chromatin interactions in NPC-derived iPSCs can be fixed by culturing in 2i/LIF media, probably through restoring CTCF occupancy after global demethylation (Beagan et al. 2016).
ROLES OF 3D GENOME CONFIGURATION IN DISEASE AND CANCER
Connecting Disease-Associated Variants to Target Genes
The majority of disease-associated variants uncovered by genome-wide association studies (GWAS) reside in noncoding sequences and are frequently found in or near cis-regulatory elements, raising the possibility that a substantial fraction of variants may contribute to disease pathogenesis by affecting transcription of specific genes (Hindorff et al. 2009, MacArthur et al. 2017, Maurano et al. 2012). Given that the target genes of distal cis-regulatory elements are not always the closest ones in linear distance, it is necessary to study disease-associated variants in the context of the 3D genome.
The power of 3D genome annotation in understanding the regulatory role of a noncoding variant by predicting its target gene was first demonstrated at specific loci through 3C or 4C-seq (Smemo et al. 2014, Visser et al. 2012). Now a genome-wide, 3D genome–based approach linking disease-associated genetic variants with their target genes is possible with high-resolution Capture Hi-C or Hi-C. The 3D promoter interactomes of 17 human primary hematopoietic cell types reveal more than 2,500 potential disease-associated genes linking to thousands of GWAS SNPs (Javierre et al. 2016). In a separate study, Hi-C contact maps from the human cerebral cortex are used to annotate the noncoding variants identified in schizophrenia GWAS (Won et al. 2016).
Gene Dysregulation by Alteration of Topologically Associating Domains
Because the TAD plays a critical role in securing promoter-enhancer interactions and limiting interdomain interactions (Symmons et al. 2016, Zhan et al. 2017), its alteration may also induce ectopic interactions between regulatory elements, leading to gene dysregulation. A number of studies have demonstrated that genetic manipulation of specific TAD boundaries can change the surrounding interaction patterns and can thus affect the expression of nearby genes. For example, when a 58-kb region encompassing a TAD boundary is deleted in mouse ESCs, interdomain interactions between adjacent TADs significantly increase, accompanied with upregulation of genes lying in the neighborhood TAD (Nora et al. 2012). Rather than by deleting a large genomic region, a more precise perturbation of a TAD boundary was achieved by targeting the CTCF-CTCF loop anchors that contribute to boundary formation. Similarly, loss of CTCF binding sites or inversion of the sequences altered local chromatin architecture and nearby gene expression (Dowen et al. 2014, Guo et al. 2015, Narendra et al. 2015).
Naturally occurring mutations may also disrupt TAD boundaries and in some cases lead to oncogene activation (Figure 5a). In IDH mutant gliomas, activities of TET proteins, which are involved in active DNA demethylation, are repressed, leading to hypermethylation of some CTCF binding sites and to reduced CTCF occupancy. As a result, the insulation function of the TAD boundary is impaired, allowing ectopic interactions between a constitutive enhancer and PDGFRA, an established glioma oncogene, to trigger the upregulation of PDGFRA (Flavahan et al. 2016). Oncogene activation through disruption of the TAD boundary is also observed in T cell acute lymphoblastic leukemia (T-ALL). In T-ALL, two pathogenesis genes, TAL1 and LMO2, are located near impaired TAD boundaries, and deletion of these two boundaries in HEK-293T cells resulted in the activation of both genes (Hnisz et al. 2016b). In addition, CTCF binding sites are frequently mutated in multiple cancer types, suggesting that TAD boundary disruption may be a common mechanism for oncogene activation (Katainen et al. 2015, Ji et al. 2016).
Figure 5.
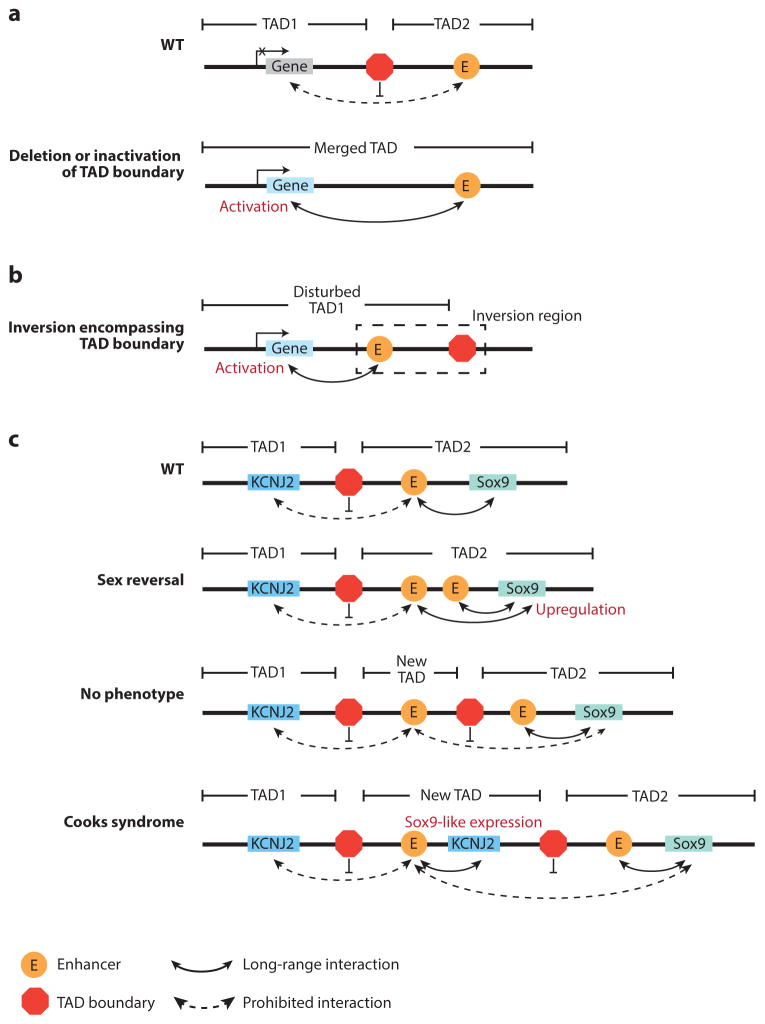
Dysregulation of gene expression by alteration of topologically associating domain (TAD) boundaries. (a) Deletion or inactivation of a TAD boundary can cause the merger of two neighborhood TADs, allowing the enhancer to activate new genes. (b) Similarly, inversion may also bring the enhancer and an irrelevant gene into the same TAD, leading to the gene’s activation. (c) The effect of duplication depends on its size and position, as exemplified around the Sox9 locus (Franke et al. 2016). WT denotes wild type.
Loss of TAD integrity has also been linked to congenital diseases, as illustrated by structural variations that occur in the EPHA4, Sox9, and LMNB1 loci (Franke et al. 2016, Giorgio et al. 2015, Lupiáñez et al. 2015). In these studies, the authors attempt to link the observed pheno- types to structural variations. This proved to be a difficult task if only changes in relevant gene dosages were considered. However, when the relative positions of the enhancer and promoter were examined in the context of TAD, the underlying mechanisms became apparent. One class of structural variations being investigated is deletion. When a TAD boundary is deleted, certain genes can be ectopically activated by enhancers in the neighboring TAD (Figure 5a). At the LMNB1 locus, a large deletion (~660 kb) upstream of the LMNB1 promoter eliminates a TAD boundary and allows for new chromatin interactions between at least three enhancers and the LMNB1 promoter, resulting in overexpression of LMNB1 and autosomal dominant adult-onset demyelinating leukodystrophy (Giorgio et al. 2015). Similarly, deletion of the centromeric or telomeric boundary of the EPHA4 TAD causes activation of IHH or PAX, leading to brachydactyly or polydactyly, respectively (Lupiáñez et al. 2015). By contrast, deletions that do not perturb the TAD boundary had no effect on the expression level of neighboring genes (Lupiáñez et al. 2015, Symmons et al. 2016). Besides deletions, inversions encompassing a TAD boundary can also bring enhancers and promoters from two different TADs into close proximity and trigger ectopic gene expression (Figure 5b). For example, F-syndrome can be caused by inversion of the centromeric boundary of the EPHA4 TAD, which activates WNT6 (Lupiáñez et al. 2015).
Duplications may also have functional consequences, depending on the sizes and positions of such duplications (Figure 5c). When the duplication involves no regulatory elements and is confined to one TAD (referred to as intra-TAD duplication), it generally has no major effect on the genes in the same TAD, because intra-TAD promoter-enhancer contact is robust and independent of genomic distance (Symmons et al. 2016). In contrast, if intra-TAD duplication covers an enhancer, the duplicated enhancer can introduce extra contacts with the targeted gene, leading to its overexpression. For instance, SOX9 can be upregulated by duplication of its regulatory region that contains a number of enhancers, causing sexual reversal (Franke et al. 2016). Also around the SOX9 locus, a larger duplication including both SOX9 enhancers and the adjacent TAD boundary results in no aberrant interactions or phenotype, which can be explained by the formation of a new TAD that prohibits the interaction between SOX9 and the duplicated enhancers. Further extended duplication encompassing the nearby KCNJ2 gene leads to a Sox9-like expression pattern of Kcnj2 and Cooks syndrome as a result of the extra copy of KCNJ2 being regulated by SOX9 enhancers (Franke et al. 2016).
Systematic computational analysis also confirms that considering TAD alterations is necessary when one is interpreting the consequence of structural variations. From this analysis, disruption of TAD boundary and subsequent enhancer misuse contribute to a substantial portion of copy-number variation–associated phenotypes (Ibn-Salem et al. 2014).
PERSPECTIVE
Our understanding of 3D genome organization has rapidly evolved in recent years, thanks to advances in C-technologies and imaging-based tools. We now appreciate that chromosome folding follows several common rules and that changes in 3D genome structure not only are a consequence of changes in gene expression but also could modulate gene expression. Importantly, disruption of higher-order chromatin structure (e.g., TADs) is recognized as a new mechanism of disease pathogenesis.
Future efforts should address several key questions. First, the molecular mechanisms that drive and maintain the hierarchical 3D genome structure have not been fully elucidated. Although CTCF-CTCF loops and the Cohesin complex have generally been accepted as a mechanism in TAD formation, this model cannot explain all experimental observations. Whether additional factors are involved in genome folding will require further investigation. Second, whether the principles of chromatin organization derived from populations of cells apply to single cells is still unclear. For example, the existence of compartment A/B in individual cells has been confirmed by FISH (Wang et al. 2016), and finer structures such as TADs were also observed in single-cell Hi-C data (Nagano et al. 2013). However, both imaging-based analysis and single-cell Hi-C studies have illustrated significant cell-to-cell variabilities in terms of 3D genome organization (Flyamer et al. 2017; Nagano et al. 2013, 2017; Ramani et al. 2017; Stevens et al. 2017), and it is unclear how such variabilities relate to gene regulation at the single-cell level. Furthermore, analysis of single-cell Hi-C data sets is challenging, given the sparsity of coverage (Flyamer et al. 2017; Nagano et al. 2013, 2017; Ramani et al. 2017; Stevens et al. 2017).
Because lineage-specific alteration of 3D structure often occurs within TADs or sub-TADs, acquiring maps of chromatin interactions at high resolution (ideally at a level capable of distinguishing individual regulatory elements) for a wide range of tissues or cells is necessary to delineate the influence of the 3D genome on lineage-specific gene regulation. Acquisition of such detailed maps for a wide spectrum of samples has been possible using relatively cost-effective targeted C-technologies such as Capture-C/Hi-C and PLAC-seq/HiChIP (Javierre et al. 2016; M. Yu, R. Fang, I. Juric, R. Hu, S. Preissl, M. Hu & B. Ren, unpublished results). We anticipate that high-resolution chromatin interaction maps from a wider range of tissues and cells will be generated in the near future.
Given the complexity of 3D genome organization as well as of gene regulation networks, there is no doubt that fully decoding their relationships will require multidisciplinary approaches. The 4D Nucleome project is one such effort with the goal of developing novel tools to map 3D genome organization and to generate reference maps of 3D genome architecture for different cell types and tissues (Dekker et al. 2017). One of the central goals of the consortium is to establish experimental guidelines and data standards for imaging and omics technologies. To achieve this goal, the consortium will study the chromatin organization of a panel of common cell lines by using a diverse array of omics- and imaging-based technologies. Systematic comparisons of different assays will be carried out to learn how different chromatin architectural features such as loops and TADs present themselves when examined using different methodologies. In particular, the integration of imaging- and omics-based data sets will lead to exciting new principles of chromatin organization. Results from this collaborative project are expected to greatly improve our understanding of nuclear organization and dynamics in mammalian cells and to provide important guidelines for expanding such studies to additional cell types and biological contexts.
Acknowledgments
We apologize to those authors whose work we fail to include in this review due to space constraints. We thank members of the Ren lab for comments on the manuscript. Work in the Ren lab is funded by the Ludwig Institute for Cancer Research, the California Institute for Regenerative Medicine (GC1R-06673), and the National Institutes of Health (U01 DK105541-01, R01 ES024984-02, U54 HG006997-04, R01 HG008135-01, U54 DK107977-01, 1UM1HL128773-01, and 2P50 GM085764-06).
Footnotes
DISCLOSURE STATEMENT
B.R. is a cofounder of Arima Genomics, Inc. M.Y. is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Apostolou E, Ferrari F, Walsh RM, Bar-Nur O, Stadtfeld M, et al. Genome-wide chromatin interactions of the Nanog locus in pluripotency, differentiation, and reprogramming. Cell Stem Cell. 2013;12(6):699–712. doi: 10.1016/j.stem.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battulin N, Fishman VS, Mazur AM, Pomaznoy M, Khabarova AA, et al. Comparison of the three-dimensional organization of sperm and fibroblast genomes using the Hi-C approach. Genome Biol. 2015;16(1):77. doi: 10.1186/s13059-015-0642-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beagan JA, Gilgenast TG, Kim J, Plona Z, Norton HK, et al. Local genome topology can exhibit an incompletely rewired 3D-folding state during somatic cell reprogramming. Cell Stem Cell. 2016;18(5):611–24. doi: 10.1016/j.stem.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliveau BJ, Apostolopoulos N, Wu C-T. Visualizing genomes with Oligopaint FISH probes. Curr Protoc Mol Biol. 2014;105:14.23.1–20. doi: 10.1002/0471142727.mb1423s105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti F, Dorier J, Burnier Y, Stasiak A. Models that include supercoiling of topological domains reproduce several known features of interphase chromosomes. Nucleic Acids Res. 2014;42(5):2848–55. doi: 10.1093/nar/gkt1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlivet S, Paquette D, Dumouchel A, Langlais D, Dostie J, Kmita M. Clustering of tissue-specific sub-TADs accompanies the regulation of HoxA genes in developing limbs. PLOS Genet. 2013;9(12):e1004018. doi: 10.1371/journal.pgen.1004018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, et al. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLOS Biol. 2005;3(5):e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornfleth H, Edelmann P, Zink D, Cremer T, Cremer C. Quantitative motion analysis of subchromosomal foci in living cells using four-dimensional microscopy. Biophys J. 1999;77(5):2871–86. doi: 10.1016/S0006-3495(99)77119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLOS Biol. 2006;4(5):e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting KL, Soong TD, Singh R, Jiang Y, Béguelin W, et al. Multi-tiered reorganization of the genome during B cell affinity maturation anchored by a germinal center–specific locus control region. Immunity. 2016;45(3):497–512. doi: 10.1016/j.immuni.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JN, Adey A, Patwardhan RP, Qiu R, Kitzman JO, Shendure J. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat Biotechnol. 2013;31(12):1119–25. doi: 10.1038/nbt.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Guan J, Huang B. Imaging specific genomic DNA in living cells. Annu Rev Biophys. 2016;45(1):1–23. doi: 10.1146/annurev-biophys-062215-010830. http://dx.doi.org/10.1146/annurev-biophys-062215-010830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, et al. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015;523(7559):240–44. doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer M, Grasser F, Lanctôt C, Müller S, Neusser M, et al. Multicolor 3D fluorescence in situ hybridization for imaging interphase chromosomes. Methods Mol Biol. 2008;463(Chapter 15):205–39. doi: 10.1007/978-1-59745-406-3_15. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2(3):a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol. 1999;145(6):1119–31. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrow EM, Huntley MH, Dudchenko O, Stamenova EK, Durand NC, et al. Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. PNAS. 2016;113(31):E4504–12. doi: 10.1073/pnas.1609643113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Bouwman BAM, Zhu Y, Klous P, Splinter E, et al. The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature. 2013;501(7466):227–31. doi: 10.1038/nature12420. [DOI] [PubMed] [Google Scholar]
- de Wit E, Vos ESM, Holwerda SJB, Valdes-Quezada C, Verstegen MJAM, et al. CTCF binding polarity determines chromatin looping. Mol Cell. 2015;60(4):676–84. doi: 10.1016/j.molcel.2015.09.023. [DOI] [PubMed] [Google Scholar]
- Dekker J. Two ways to fold the genome during the cell cycle: insights obtained with chromosome conformation capture. Epigenet Chromatin. 2014;7(1):25. doi: 10.1186/1756-8935-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J. Mapping the 3D genome: aiming for consilience. Nat Rev Mol Cell Biol. 2016;17(12):741–42. doi: 10.1038/nrm.2016.151. [DOI] [PubMed] [Google Scholar]
- Dekker J, Belmont AS, Guttman M, Leshyk VO, Lis JT, et al. The 4D Nucleome Project. bioRxiv. 2017:103499. https://doi.org/10.1101/103499.
- Dekker J, Heard E. Structural and functional diversity of topologically associating domains. FEBS Lett. 2015;589(20A):2877–84. doi: 10.1016/j.febslet.2015.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295(5558):1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Denholtz M, Bonora G, Chronis C, Splinter E, de Laat W, et al. Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization. Cell Stem Cell. 2013;13(5):602–16. doi: 10.1016/j.stem.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lee J, Wang H, Miller J, Reik A, et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149(6):1233–44. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Rupon JW, Krivega I, Breda L, Motta I, et al. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell. 2014;158(4):849–60. doi: 10.1016/j.cell.2014.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Ma W, Ramani V, Hill A, Yang F, et al. Bipartite structure of the inactive mouse X chromosome. Genome Biol. 2015;16(1):152. doi: 10.1186/s13059-015-0728-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Gorkin DU, Ren B. Chromatin domains: the unit of chromosome organization. Mol Cell. 2016;62(5):668–80. doi: 10.1016/j.molcel.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518(7539):331–36. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16(10):1299–309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159(2):374–87. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden NH, Broome LR, Dudbridge F, Johnson N, Orr N, et al. Unbiased analysis of potential targets of breast cancer susceptibility loci by Capture Hi-C. Genome Res. 2014;24(11):1854–68. doi: 10.1101/gr.175034.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Robinson JT, Shamim MS, Machol I, Mesirov JP, et al. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 2016;3(1):99–101. doi: 10.1016/j.cels.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R, Yu M, Li G, Chee S, Liu T, et al. Mapping of long-range chromatin interactions by proximity ligation–assisted ChIP-seq. Cell Res. 2016;26(12):1345–48. doi: 10.1038/cr.2016.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143(2):212–24. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529(7584):110–14. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyamer IM, Gassler J, Imakaev M, Brandão HM, Ulianov SB, et al. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature. 2017;544(7648):110–14. doi: 10.1038/nature21711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke M, Ibrahim DM, Andrey G, Schwarzer W, Heinrich V, et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature. 2016;538(7624):265–69. doi: 10.1038/nature19800. [DOI] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. Formation of chromosomal domains by loop extrusion. Cell Rep. 2016;15(9):2038–49. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462(7269):58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7(9):703–13. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- Giorgetti L, Lajoie BR, Carter AC, Attia M, Zhan Y, et al. Structural organization of the inactive X chromosome in the mouse. Nature. 2016;535(7613):575–79. doi: 10.1038/nature18589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgio E, Robyr D, Spielmann M, Ferrero E, Di Gregorio E, et al. A large genomic deletion leads to enhancer adoption by the lamin B1 gene: a second path to autosomal dominant adult-onset demyelinating leukodystrophy (ADLD) Hum Mol Genet. 2015;24(11):3143–54. doi: 10.1093/hmg/ddv065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloborodko A, Imakaev MV, Marko JF, Mirny L. Compaction and segregation of sister chromatids via active loop extrusion. eLife. 2016;5:11202. doi: 10.7554/eLife.14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Marín C, Tena JJ, Acemel RD, López-Mayorga M, Naranjo S, et al. Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders. PNAS. 2015;112(24):7542–47. doi: 10.1073/pnas.1505463112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasser F, Neusser M, Fiegler H, Thormeyer T, Cremer M, et al. Replication-timing-correlated spatial chromatin arrangements in cancer and in primate interphase nuclei. J Cell Sci. 2008;121(11):1876–86. doi: 10.1242/jcs.026989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xu Q, Canzio D, Shou J, Li J, et al. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell. 2015;162(4):900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460(7253):410–13. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. PNAS. 2009;106(23):9362–67. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, et al. Global reorganization of replication domains during embryonic stem cell differentiation. PLOS Biol. 2008;6(10):e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Day DS, Young RA. Insulated neighborhoods: structural and functional units of mammalian gene control. Cell. 2016a;167(5):1188–200. doi: 10.1016/j.cell.2016.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016b;351(6280):1454–58. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horakova AH, Moseley SC, McLaughlin CR, Tremblay DC, Chadwick BP. The macrosatellite DXZ4 mediates CTCF-dependent long-range intrachromosomal interactions on the human inactive X chromosome. Hum Mol Genet. 2012;21(20):4367–77. doi: 10.1093/hmg/dds270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T-HS, Weiner A, Lajoie B, Dekker J, Friedman N, Rando OJ. Mapping nucleosome resolution chromosome folding in yeast by Micro-C. Cell. 2015;162(1):108–19. doi: 10.1016/j.cell.2015.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JR, Roberts N, McGowan S, Hay D, Giannoulatou E, et al. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat Genet. 2014;46(2):205–12. doi: 10.1038/ng.2871. [DOI] [PubMed] [Google Scholar]
- Ibn-Salem J, Köhler S, Love MI, Chung H-R, Huang N, et al. Deletions of chromosomal regulatory boundaries are associated with congenital disease. Genome Biol. 2014;15(9):423. doi: 10.1186/s13059-014-0423-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140(6):1285–95. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javierre BM, Burren OS, Wilder SP, Kreuzhuber R, Hill SM, et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell. 2016;167(5):1369–84. e19. doi: 10.1016/j.cell.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Dadon DB, Powell BE, Fan ZP, Borges-Rivera D, et al. 3D chromosome regulatory landscape of human pluripotent cells. Cell Stem Cell. 2016;18(2):262–75. doi: 10.1016/j.stem.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503(7475):290–94. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi O, Wang S-Y, Kuznetsova T, Atlasi Y, Peng T, et al. Dynamic reorganization of extremely long-range promoter-promoter interactions between two states of pluripotency. Cell Stem Cell. 2015;17(6):748–57. doi: 10.1016/j.stem.2015.11.010. [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467(7314):430–35. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol. 2011;30(1):90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Dekker J. High-throughput genome scaffolding from in vivo DNA interaction frequency. Nat Biotechnol. 2013;31(12):1143–47. doi: 10.1038/nbt.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katainen R, Dave K, Pitkänen E, Palin K, Kivioja T, et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet. 2015;47(7):818–21. doi: 10.1038/ng.3335. [DOI] [PubMed] [Google Scholar]
- Kim T-K, Shiekhattar R. Architectural and functional commonalities between enhancers and promoters. Cell. 2015;162(5):948–59. doi: 10.1016/j.cell.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijger PHL, Di Stefano B, de Wit E, Limone F, van Oevelen C, et al. Cell-of-origin-specific 3D genome structure acquired during somatic cell reprogramming. Cell Stem Cell. 2016;18(5):597–610. doi: 10.1016/j.stem.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakadamyali M, Cosma MP. Advanced microscopy methods for visualizing chromatin structure. FEBS Lett. 2015;589(20A):3023–30. doi: 10.1016/j.febslet.2015.04.012. [DOI] [PubMed] [Google Scholar]
- Langer-Safer PR, Levine M, Ward DC. Immunological method for mapping genes on Drosophila polytene chromosomes. PNAS. 1982;79(14):4381–85. doi: 10.1073/pnas.79.14.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152(6):1237–51. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. Transcriptional enhancers in animal development and evolution. Curr Biol. 2010;20(17):R754–63. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M, Cattoglio C, Tjian R. Looping back to leap forward: Transcription enters a new era. Cell. 2014;157(1):13–25. doi: 10.1016/j.cell.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148(1–2):84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas JS, Zhang Y, Dudko OK, Murre C. 3D trajectories adopted by coding and regulatory DNA elements: first-passage times for genomic interactions. Cell. 2014;158(2):339–52. doi: 10.1016/j.cell.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161(5):1012–25. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Ay F, Lee C, Gülsoy G, Deng X, et al. Fine-scale chromatin interaction maps reveal the cis-regulatory landscape of human lincRNA genes. Nat Methods. 2015;12(1):71–78. doi: 10.1038/nmeth.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45(D1):D896–901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, et al. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr Biol. 1997;7(12):930–39. doi: 10.1016/s0960-9822(06)00412-x. [DOI] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190–95. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mifsud B, Tavares-Cadete F, Young AN, Sugar R, Schoenfelder S, et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet. 2015;47(6):598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- Minajigi A, Froberg JE, Wei C, Sunwoo H, Kesner B, et al. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science. 2015;349(6245):aab2276. doi: 10.1126/science.aab2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi T, Barrowman J, Grewal SIS. Chromosome domain architecture and dynamic organization of the fission yeast genome. FEBS Lett. 2015;589(20A):2975–86. doi: 10.1016/j.febslet.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- modENCODE Consortium. Roy S, Ernst J, Kharchenko PV, Kheradpour P, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330(6012):1787–97. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumbach MR, Rubin AJ, Flynn RA, Dai C, Khavari PA, et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat Methods. 2016;13:919–22. doi: 10.1038/nmeth.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502(7469):59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Lubling Y, Várnai C, Dudley C, Leung W, et al. Cell cycle dynamics of chromosomal organisation at single-cell resolution. Nature. 2017;547(7661):61–67. doi: 10.1038/nature23001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra V, Rocha PP, An D, Raviram R, Skok JA, et al. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015;347(6225):1017–21. doi: 10.1126/science.1262088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton C, Avlonitis N, Corless S, Prendergast JG, Mati IK, et al. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat Struct Mol Biol. 2013;20(3):387–95. doi: 10.1038/nsmb.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, et al. Organization of the mitotic chromosome. Science. 2013;342(6161):948–53. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485(7398):381–85. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong C-T, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12(4):283–93. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada LA, Roix JJ, Misteli T. An uncertainty principle in chromosome positioning. Trends Cell Biol. 2003;13(8):393–96. doi: 10.1016/s0962-8924(03)00149-1. [DOI] [PubMed] [Google Scholar]
- Paweletz N. Walther Flemming: pioneer of mitosis research. Nat Rev Mol Cell Biol. 2001;2:72–75. doi: 10.1038/35048077. [DOI] [PubMed] [Google Scholar]
- Pepenella S, Murphy KJ, Hayes JJ. Intra- and inter-nucleosome interactions of the core histone tail domains in higher-order chromatin structure. Chromosoma. 2014;123(1–2):3–13. doi: 10.1007/s00412-013-0435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137(7):1194–211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria MEG, Sanyal A, Gerasimova TI, Lajoie BR, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153(6):1281–95. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope BD, Ryba T, Dileep V, Yue F, Wu W, et al. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515(7527):402–5. doi: 10.1038/nature13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani V, Deng X, Qiu R, Gunderson KL, Steemers FJ, et al. Massively multiplex single-cell Hi-C. Nat Methods. 2017;14:263–66. doi: 10.1038/nmeth.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159(7):1665–80. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium. Kundaje A, Meuleman W, Ernst J, Bilenky M, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, et al. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010;20(6):761–70. doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn AL, Rao SSP, Huang S-C, Durand NC, Huntley MH, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. PNAS. 2015;112(47):E6456–65. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489(7414):109–13. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AD, Hu M, Jung I, Xu Z, Qiu Y, et al. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep. 2016a;17(8):2042–59. doi: 10.1016/j.celrep.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AD, Hu M, Ren B. Genome-wide mapping and analysis of chromosome architecture. Nat Rev Mol Cell Biol. 2016b;17:743–55. doi: 10.1038/nrm.2016.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Furlan-Magaril M, Mifsud B, Tavares-Cadete F, Sugar R, et al. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res. 2015a;25(4):582–97. doi: 10.1101/gr.185272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Sugar R, Dimond A, Javierre B-M, Armstrong H, et al. Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome. Nat Genet. 2015b;47(10):1179–86. doi: 10.1038/ng.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj S, Dixon RJ, Bansal V, Ren B. Whole-genome haplotype reconstruction using proximity-ligation and shotgun sequencing. Nat Biotechnol. 2013;31(12):1111–18. doi: 10.1038/nbt.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148(3):458–72. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Simonis M, Klous P, Homminga I, Galjaard R-J, Rijkers E-J, et al. High-resolution identification of balanced and complex chromosomal rearrangements by 4C technology. Nat Methods. 2009;6(11):837–42. doi: 10.1038/nmeth.1391. [DOI] [PubMed] [Google Scholar]
- Smemo S, Tena JJ, Kim K-H, Gamazon ER, Sakabe NJ, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507(7492):371–75. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra R-J, Klous P, et al. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20(17):2349–54. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens TJ, Lando D, Basu S, Atkinson LP, Cao Y, et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature. 2017;544(7648):59–64. doi: 10.1038/nature21429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons O, Pan L, Remeseiro S, Aktas T, Klein F, et al. The Shh topological domain facilitates the action of remote enhancers by reducing the effects of genomic distances. Dev Cell. 2016;39(5):529–43. doi: 10.1016/j.devcel.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons O, Uslu VV, Tsujimura T, Ruf S, Nassari S, et al. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 2014;24(3):390–400. doi: 10.1101/gr.163519.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T, Meaburn KJ, Misteli T. The meaning of gene positioning. Cell. 2008;135(1):9–13. doi: 10.1016/j.cell.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, et al. CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell. 2015;163(7):1611–27. doi: 10.1016/j.cell.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra R-J, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10(6):1453–65. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Ulianov SV, Khrameeva EE, Gavrilov AA, Flyamer IM, Kos P, et al. Active chromatin and transcription play a key role in chromosome partitioning into topologically associating domains. Genome Res. 2016;26(1):70–84. doi: 10.1101/gr.196006.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uusküla-Reimand L, Hou H, Samavarchi-Tehrani P, Rudan MV, Liang M, et al. Topoisomerase II beta interacts with cohesin and CTCF at topological domain borders. Genome Biol. 2016;17(1):182. doi: 10.1186/s13059-016-1043-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser M, Kayser M, Palstra R-J. HERC2 rs12913832 modulates human pigmentation by attenuating chromatin-loop formation between a long-range enhancer and the OCA2 promoter. Genome Res. 2012;22(3):446–55. doi: 10.1101/gr.128652.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Joffe B, Bolzer A, Albiez H, Benedetti PA, et al. Towards many colors in FISH on 3D-preserved interphase nuclei. Cytogenet Genome Res. 2006;114(3–4):367–78. doi: 10.1159/000094227. [DOI] [PubMed] [Google Scholar]
- Wang C, Liu C, Roqueiro D, Grimm D, Schwab R, et al. Genome-wide analysis of local chromatin packing in Arabidopsis thaliana. Genome Res. 2015;25(2):246–56. doi: 10.1101/gr.170332.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Su J-H, Beliveau BJ, Bintu B, Moffitt JR, et al. Spatial organization of chromatin domains and compartments in single chromosomes. Science. 2016;353(6299):598–602. doi: 10.1126/science.aaf8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Gao F, Kim S, Yang H, Lyu J, et al. Klf4 organizes long-range chromosomal interactions with the Oct4 locus in reprogramming and pluripotency. Cell Stem Cell. 2013;13(1):36–47. doi: 10.1016/j.stem.2013.05.010. [DOI] [PubMed] [Google Scholar]
- Williamson I, Berlivet S, Eskeland R, Boyle S, Illingworth RS, et al. Spatial genome organization: contrasting views from chromosome conformation capture and fluorescence in situ hybridization. Genes Dev. 2014;28(24):2778–91. doi: 10.1101/gad.251694.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H, de la Torre–Ubieta L, Stein JL, Parikshak NN, Huang J, et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature. 2016;538(7626):523–27. doi: 10.1038/nature19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515(7527):355–64. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y, Mariani L, Barozzi I, Schulz EG, Blüthgen N, et al. Reciprocal insulation analysis of Hi-C data shows that TADs represent a functionally but not structurally privileged scale in the hierarchical folding of chromosomes. Genome Res. 2017;27(3):479–90. doi: 10.1101/gr.212803.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Jiao W, Sun L, Fan J, Chen M, et al. Intrachromosomal looping is required for activation of endogenous pluripotency genes during reprogramming. Cell Stem Cell. 2013;13(1):30–35. doi: 10.1016/j.stem.2013.05.012. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wong C-H, Birnbaum RY, Li G, Favaro R, et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature. 2013;504(7479):306–10. doi: 10.1038/nature12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin J, Dixon JR, van der Reijden MIJA, Ye Z, Kolovos P, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. PNAS. 2014;111(3):996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]