Abstract
Infection with Zika virus (ZIKV), a mosquito-borne flavivirus has been casually linked with increased congenital microcephaly in Brazil from 2015 through 2016. Sensitive and specific diagnosis of patients with Zika fever (ZIKF) remains critical for patient management. We developed a ZIKV NS5 qRT-PCR assay by combining primers described by Balm et al. and a new Taqman probe. The assay was evaluated and compared with another assay described by Lanciotti et al. (ZIKV 1107) using 51 blood and 42 urine samples from 54 suspected ZIKV patients. ZIKV NS5 performed better in terms of sensitivity with more samples detected as ZIKV-positive (n = 37) than ZIKV 1107 (n = 34) for urine, and ZIKV-positive (n = 29) than ZIKV 1107 (n = 26) for blood. Both assays displayed good overall agreement for urine (κappa = 0.770) and blood (κappa = 0.825) samples. Improved availability of validated diagnostic tests, such ZIKV NS5 qRT-PCR, will be critical to ensure adequate and accurate ZIKV diagnosis.
Introduction
Zika virus (ZIKV) is an enveloped, positive-sense, single-stranded RNA virus belonging to the Flaviviridae family1. First isolated in 1947 from a sentinel rhesus macaque in Uganda2, ZIKV remained an obscure pathogen until the 2007 outbreak on the Yap Islands in the Federated States of Micronesia3,4, followed by a larger epidemic in French Polynesia in 2013 and 20145,6. ZIKV outbreaks subsequently occurred throughout 2014 to 2016 on other Pacific islands6,7. In early 2015, ZIKV was identified for the first time in Brazil8. Within a year, ZIKV had spread throughout continental South America and into Central America, the Caribbean, and Mexico8,9. ZIKV has also re-emerged in Asia10, with imported or autochthonous ZIKV infections being reported in Asian countries, including Cambodia, Indonesia, Thailand and Singapore11.
Zika infection is an undifferentiated systemic febrile illness with a short-lived fever, non-specific rash, conjunctivitis and arthralgia3,12. It is rarely life threatening and maybe sometimes completely asymptomatic3. With the spread of ZIKV to the Americas and its association with a marked increase in the incidence of Guillain-Barré syndrome (GBS) in French Polynesia5,13,14 and congenital Zika syndrome in Brazil15–17, sensitive and accurate diagnosis of patients with ZIKV infection is critical to ongoing epidemiologic surveillance, patient management and eventually treatment of the disease.
In endemic countries such as Brazil, where different arboviruses have been circulating for several years18, the detection of ZIKV is more complicated and very challenging. Specifically, the similarity and non-specific nature of the clinical symptoms provoked by these viruses impede correct differential diagnosis during the febrile phase. Moreover, there is a high level of cross-reactivity among different flaviviruses19. A study done in Rio de Janerio estimated that more than 60% of the Brazilian population are seropositive for dengue virus (DENV)20. Furthermore, recent outbreaks of Yellow Fever Virus (YFV)21 and the YFV vaccination program in Brazil22 have made ZIKV detection by serological tests extremely difficult.
Therefore, the recommended gold standard for ZIKV diagnosis in flaviruses endemic areas is molecular-based detection of ZIKV RNA in patients’ specimens during the acute phase of virus infection23,24. Several ZIKV reverse transcription PCR (RT-PCR) assays have been reported23, and the most well-reported assay is that from the 2007 Yap Islands epidemic4. It comprises of 2 one-step real-time RT-PCR (qRT-PCR) reactions, targeting the ZIKV pre-membrane (prM) and envelope (E) genes (referred to as the ZIKV 860 and ZIKV 1107 respectively). In this study, we compared a laboratory-developed qRT-PCR (ZIKV NS5) with ZIKV 1107 using clinical samples collected from 54 suspected ZIKV cases between day 1 and 6 of post illness onset. We analyzed 51 blood samples and 42 urine samples from these suspected ZIKV patients. All of them had symptoms compatible with acute ZIKV infection, including fever, myalgia, rash and conjuctivitis. In addition, urine and blood samples from 20 heathy individuals negative for ZIKV were also include as controls.
Methods
Patients
In this study, samples from patients presenting acute febrile disease for less than 7 days from February to June of 2016 were assessed. All patients were admitted to hospitals through their respective emergency departments (ED) at Campinas city, Southeast of Brazil. We recruited a total of 54 ZIKV-suspected patients (42 females, 12 males, mean age 37 years ± 16 years) based on clinical signs, such as the presence of low fever, rash, myalgia and conjunctivitis.
Both whole blood and urine samples were obtained from 39 patients, while only urine was collected from 3 other patients, and only blood from another 12 patients. The negative control group comprised of 20 age-matched healthy individuals without signs of infection within 30 days prior to sample collection. Whole blood and urine samples were obtained from these healthy individuals. Hence in total, 93 clinical specimens from ZIKV-suspected patients together with 40 specimens from negative control individuals were assessed. All samples were tested negative by RT-PCR for DENV.
Ethics statements
This study was approved by the Research Ethics Committee of the University of Campinas (CEP 053407/2016; CEEA: 56793516.0.0000.54), in accordance with the tenets of the Declaration of Helsinki for human research. Written informed consent was obtained from all patients.
RNA extraction
Viral RNA from whole blood and urine samples was extracted using the easyMAG® automated extractor (BioMerieux, Quebec, Canada), according to manufacturer’s instructions.
ZIKV real-time RT-PCR assays
ZIKV 1107 was the reference assay used25–27, performed with the following modifications: Cycling conditions were the following: 45 °C for 1 min (RT step); 95 °C for 5 min; 45 cycles of 95 °C for 15 s, and 60 °C for 1 min. ZIKV primers and probe primers were used at final concentrations of 400 nM and 200 nM. In parallel, NS5-targeted qRT-PCR was performed as previously described28 with an additional novel lab-designed FAM-labeled molecular beacon probe (TACCAGGAGGAAGGATGTATG) (ZIKV NS5). Cycling conditions and primer/probe concentrations used were similar to that of ZIKV 1107 with the exception of the annealing and extension temperature at the 56 °C for 1 min. Assay exclusivity of ZIKV NS5 was confirmed by testing viral RNA extracted from the following viruses (Supplementary Table 1), and no cross-reactions were identified: Dengue viruses (DENV serotype 1–4), West Nile virus (WNV), yellow fever virus (YFV), chikungunya virus (CHIKV) and o’nyong-nyong virus (ONNV). Analytical sensitivity was also determined using quantitated ZIKV RNA transcripts and the lower limit of detection was estimated as 10 copies for the NS5 gene target. Copy numbers of ZIKV RNA were determined by using the Ribogreen RNA specific Quantitiation Kit (Invitrogen, Carlsbad, USA). RNA transcripts ranging from 2 × 106 to 0.2 copies were performed in pentaplicates to construct standard curves for both qRT-PCR assays to estimate the copy number of ZIKV in patient samples. All qRT-PCR assays were performed on the CFX96 Touch™ Real-Time PCR Detection System (BioRad, Hercules, USA) using QuantiNova Probe RT-PCR Kit (Qiagen, Hilden, Germany) in 25 µL reactions with 3 µL RNA template. qRT-PCRs with cycle threshold (Ct) values higher than 40 cycles were considered negative.
Statistical analysis
For statistical analysis, two-tailed Fisher’s exact tests, unpaired t tests, and kappa statistics were performed using Prism 7 software (GraphPad Software Inc, San Diego, USA).
Results
In the present study, we validated primers described by Balm et al. with the addition of a novel probe specific for ZIKV detection using a one-step Taqman qRT-PCR assay (ZIKV NS5). Clinical samples were collated from 54 patients admitted to hospitals facilities in Campinas, São Paulo state, Brazil. These patients had symptoms frequently associated with ZIKV infection, including fever, myalgia, rash and conjunctivitis. The average age of the patients was 37 years ± 16 years (6–65 years) and 77.8% (n = 42) of them were females. The mean period between the onset of symptoms and sampling was 2.92 days (±1.31, ranging from 1 to 6 days). All clinical samples were collected in the first half of 2016, during the rainy season, probably during the first ZIKV outbreak in state of São Paulo, Brazil.
The sensitivities of ZIKV 1107 and ZIKV NS5 were evaluated and compared using respective standard curves (Fig. 1A) for estimating ZIKV RNA copies in patient samples (Supplementary Tables 2 and 3). ZIKV RNA transcripts with known concentrations were analyzed by both qRT-PCR assays in replicas of 5. Results indicated that these 2 qRT-PCR assays did not differ in their sensitivities (P > 0.05, t tests) (Fig. 1A, Supplementary Figure 1). Both PCR efficiencies, as reflected by the R2 values of the respective standard curves (0.9945 for ZIKV NS5 and 0.9944 for ZIKV 1107), were high (R2 > 0.90). Of the 54 patients suspected for ZIKV infection, 30 had ZIKV RNA detected in blood and 39 had ZIKV RNA detected in urine using one of the qRT-PCR assays evaluated in this study.
Figure 1.
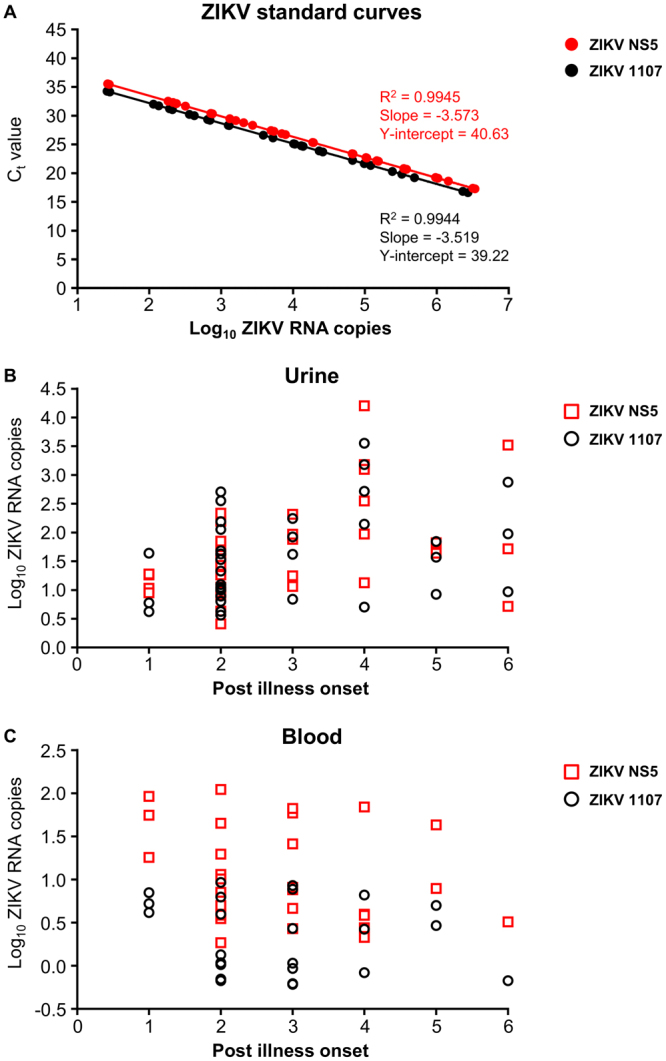
Results of qRT-PCR from patients, Campinas, Brazil, 2016. (A) Standard curves of ZIKV NS5 and ZIKV 1107 generated from ZIKV RNA transcripts and extracted ZIKV viral RNA. Comparison between extrapolated ZIKV RNA copies from qRT-PCR of (B) urine and (C) blood samples with post illness onset.
Results for 69 positive samples from ZIKV-suspected patients were stratified by day of post illness onset to examine the pattern of viral load in both urine (Fig. 1B) and blood (Fig. 1C) samples. A relatively high frequency of positive ZIKV detection across one to six days post illness onset was observed for urine (Fig. 1B) when compared to blood (Fig. 1C) samples. ZIKV RNA copies were observed to be highest in urine samples at four days post illness onset. Interestingly, a decreasing trend was seen in blood samples with only one sample showing positive detection at 6 days post illness onset (Fig. 1C). ZIKV RNA copies extrapolated by the respective standard curves (ZIKV 1107 and ZIKV NS5) showed an overlapping range of between 10 to 104 copies for urine samples (Supplementary Table 2, Fig. 1B). However, in blood samples, ZIKV copies extrapolated using the ZIKV NS5 standard curve reflected higher viral copy numbers compared than that from using the ZIKV 1107 curve, with a difference of almost 10 copies (Supplementary Table 3, Fig. 1C). Interestingly, we observed that results from ZIKV NS5 were able to present the trend of viral kinetics in blood samples where the mean of viral load at 1 to 6 days post illness onset were estimated at 33.12, 11.12, 15.86, 10.24, 12.72 and 1.07 copies as compared to 3.28, 1.22, 2.01, 1.36, 3.23 and 0.22 when using ZIKV 1107.
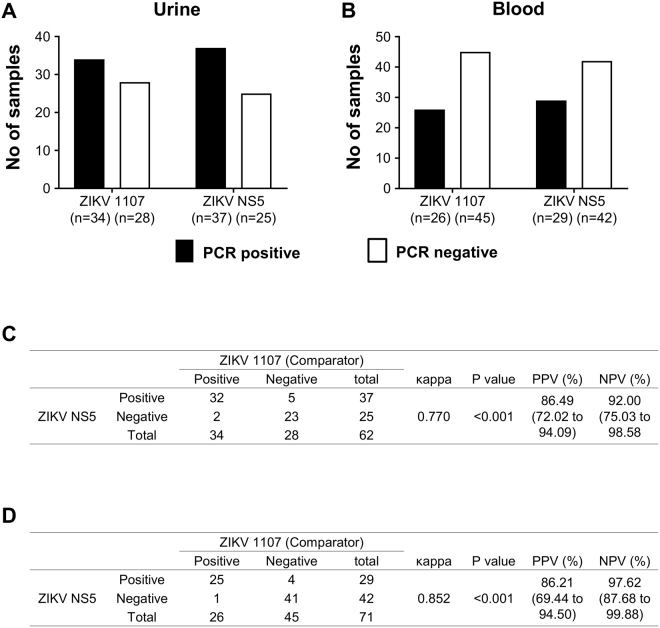
Samples that were tested ZIKV negative were indicated as ZIKV-PCR negative samples as reflected in Fig. 2C and D. ZIKV detection rates between the 2 qRT-PCR assays were comparable (P > 0.05, Fisher’s test). 34 urine samples were detected as positive with ZIKV 1107, and 37 positive urine samples using ZIKV NS5 (Fig. 2A). Frequency of positive detection by ZIKV NS5 was also higher for blood samples with 3 more samples detected as ZIKV positive (n = 29), compared to ZIKV 1107 (n = 26) (Fig. 2B). The 2 qRT-PCR assays displayed good overall agreement for both urine (κappa = 0.770) (Fig. 2C) and blood (κappa = 0.825) samples (Fig. 2D). The sensitivity and specificity of ZIKV NS5 in comparison to ZIKV 1107 were 94.11% and 82.14% for urine, 96.15% and 91.11% for blood respectively. The positive and negative predictive values were 86.49% and 92.00% for urine samples (Fig. 2C), while 86.21% and 97.62% for blood samples (Fig. 2D).
Figure 2.
Comparison between ZIKV 1107 and ZIKV NS5. (A) Urine and (B) blood samples were subjected to ZIKV qRT-PCR detection. Bar-charts show the number of samples which are PCR positive or negative. Using mean Ct value = 40 as cut-off, extrapolated ZIKV RNA copies are: ZIKV 1107–0.601, ZIKV NS5–1.497. (C) Overall agreement between ZIKV 1107 and ZIKV NS5 was assessed by κappa test for (C) urine and (D) blood samples. Statistical significance was measured using 2-sided Fisher exact test between the number of samples tested ZIKV positive by ZIKV 1107 or ZIKV NS5. PPV, positive predictive value; NPV, negative predictive value shown with 95% confidence interval.
Discussion
Low levels of viremia (≤9.26 copies) among ZIKV-infected patients were observed in this study. These estimates were also consistent with that observed in patients from the Yap outbreak (mean quantifiable viremia was 4.4 copies/mL of serum, standard deviation (SD) 0.94)4, and patients from the Nicaraguan outbreak (mean quantifiable viremia was 4.7 copies/mL of serum, SD 0.97)29. Such low viremia at disease presentation provides a likely explanation for the short window period of less than 5 days for ZIKV detection in blood4,25,26,30,31. Future studies with larger numbers of positive specimens will provide further characteristics to the dynamics of ZIKV levels in blood so as to determine if there are correlations of virus load with disease severity and immune responses.
Even though only acute samples of up to 6 days post illness onset were included in the study, ZIKV was still detected at higher titres in urine samples than blood samples of these patients (ZIKV 1107 range was 3.66–3568.61 copies). Moreover, the diagnostic utility of using urine samples was recently confirmed whereby ZIKV RNA was not only detectable in urine at a higher load but with a longer duration than in serum26,32. The clinical relevance of testing for ZIKV RNA in urine would allow diagnosis of acute infection after viremia has resolved, extending the window for ZIKV detection24. Hence, optimal diagnosis of acute ZIKV infection may require testing of multiple specimen types.
It was observed that quantification by the ZIKV NS5 qRT-PCR reflected lower Ct values and therefore higher copy numbers than that from ZIKV 1107 qRT-PCR. It has been reported that in flavivirus infection, NS5 expression is higher than that of the E protein33. Results also showed that 34 urine samples were detected positive with ZIKV 1107, and 37 positive samples using ZIKV NS5. On the other hand, 29 whole blood samples were detected positive with ZIKV NS5, compared to 26 positive samples by ZIKV 1107. This indicates that the ZIKV NS5 is more sensitive. Therefore, flavivirus NS5 could act as a good target for molecular diagnostics for improved sensitivity.
ZIKV diagnostic testing relies on serology and RT-PCR. Immunoglobin (Ig) M antibodies usually appear during the first week after symptom onset, and their appearance is rapidly followed by the appearance of IgG antibodies4,25. Although IgM testing may be able to identify recent ZIKV infections, a positive IgM serology still requires confirmation with a plaque reduction neutralization test (PRNT) due to cross-reactivity with other flaviviruses, such as DENV34. Particularly, serological testing in populations with high DENV exposure and YFV vaccination could be extremely difficult due to cross-reactivity issues. Hence, RT-PCR has an advantage over serology due to its specificity. Additionally, ZIKV RNA detection by RT-PCR is an indication of acute ZIKV infection, although persistence has been observed in urine, saliva, tears and semen25,31,35–37.
The worldwide spread of ZIKV and its association with fetal neurologic abnormalities has led to unprecedented interest in the virus23. In Brazil, a high percentage of people get concurrent infection with more than one DENV serotype periodically. Thus, people typically assume episodes of rash-febrile illness to be DENV, and only seek medical assistance when critical clinical manifestations appear38,39. Since complications generally manifest only after 5 days post illness onset, urine samples are rarely collected. Therefore, the use of this improved qRT-PCR assay will be useful for molecular diagnosis of ZIKV in blood samples. Sensitive and accurate diagnosis of patients with ZIKV infection is critical to ongoing epidemiologic surveillance, management of patients with an undifferentiated febrile illness and assessment of therapies. Importantly, improved availability of validated diagnostic tests, such as the ZIKV NS5 qRT-PCR, will be critical to understand and ensure an adequate, timely and accurate laboratory response.
Electronic supplementary material
Acknowledgements
We thank the study subjects for their participation and clinical teams within the network in patient recruitment, sample collection and data entry. This work is supported by core research grants provided to the Singapore Immunology Network by the Biomedical Research Council (BMRC), and also partially supported by the BMRC A*STAR-led Zika Virus Consortium Fund (project number: 15/1/82/27/001), Agency for Science, Technology and Research (A*STAR), Singapore. In addition, this work is supported by Sao Paulo Research Foundation (FAPESP –2016/00194-8). FTMC is a research fellow from CNPq (Brazilian National Council for Scientific and Technological Development).
Author Contributions
G.P.M., J.A.L., L.C.C., C.W.A., M.R.R., R.A., E.A., R.P.J., A.R.R.F. and Zika-Unicamp Network contributed the samples, virological and clinical data. C.C.J., J.J.L.T., Y.W.K., F.T.M.C., J.L.P.M. and L.F.P.N. conceived and designed the experiments. C.C.J. and P.L.P. performed the experiments. J.J.L.T., Y.W.K., F.T.M.C., J.L.P.M. and L.F.P.N. analyzed the data. J.J.L.T., Y.W.K., F.T.M.C., J.L.P.M. and L.F.P.N. drafted the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Carla Cristina Judice and Jeslin J. L. Tan contributed equally to this work.
A comprehensive list of consortium members appears at the end of the paper
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-22159-2.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jose Luiz Proenca-Modena, Email: jlmodena@unicamp.br.
Lisa F. P. Ng, Email: lisa_ng@immunol.a-star.edu.sg
The Zika-Unicamp Network:
Glaucia Maria Pastore, Helaine Maria Besteti Pires Mayer-Milanez, Carolina C. Ribeiro-do-Valle, Roseli Calil, Maria Laura Costa, João Renato Bennini Junior, Giuliane Jesus Lajos, Márcia Teixeira Garcia, Kleber Yotsumoto Fertrin, Maria Luiza Moretti, Marcos Tadeu Nolasco da Silva, Ana Carolina Coan, Maria Francisca Colella-Santos, Andrea Paula Bruno von Zuben, Marco Aurélio Ramirez Vinolo, Rodrigo Ramos Catharino, Gabriela Mansano do Nascimento, Matheus Martini, Ana Paula de Moraes, Ana Lucia Rodrigues Soledade, Daniel Augusto de Toledo Teixeira, Évellyn Ribeiro de Morais, Felipe Rebelo Santos, and Monique Fontana
References
- 1.Kuno G, Chang GJ. Full-length sequencing and genomic characterization of Bagaza, Kedougou, and Zika viruses. Arch Virol. 2007;152:687–696. doi: 10.1007/s00705-006-0903-z. [DOI] [PubMed] [Google Scholar]
- 2.Dick GW, Kitchen SF, Haddow AJ. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg. 1952;46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- 3.Duffy MR, et al. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med. 2009;360:2536–2543. doi: 10.1056/NEJMoa0805715. [DOI] [PubMed] [Google Scholar]
- 4.Lanciotti RS, et al. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis. 2008;14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao-Lormeau VM, et al. Zika virus, French polynesia, South pacific, 2013. Emerg Infect Dis. 2014;20:1085–1086. doi: 10.3201/eid2011.141380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derraik JG, Slaney D. Notes on Zika virus - an emerging pathogen now present in the South Pacific. Aust N Z J Public Health. 2015;39:5–7. doi: 10.1111/1753-6405.12302. [DOI] [PubMed] [Google Scholar]
- 7.Petersen LR, Jamieson DJ, Powers AM, Honein MA. Zika Virus. N Engl J Med. 2016;374:1552–1563. doi: 10.1056/NEJMra1602113. [DOI] [PubMed] [Google Scholar]
- 8.Campos GS, Bandeira AC, Sardi SI. Zika Virus Outbreak, Bahia, Brazil. Emerg Infect Dis. 2015;21:1885–1886. doi: 10.3201/eid2110.150847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fauci AS, Morens DM. Zika Virus in the Americas - Yet another Arbovirus threat. N Engl J Med. 2016;374:601–604. doi: 10.1056/NEJMp1600297. [DOI] [PubMed] [Google Scholar]
- 10.Duong V, Dussart P, Buchy P. Zika virus in Asia. Int J Infect Dis. 2016;54:121–128. doi: 10.1016/j.ijid.2016.11.420. [DOI] [PubMed] [Google Scholar]
- 11.Maurer-Stroh, S. et al. South-east Asian Zika virus strain linked to cluster of cases in Singapore, August 2016. Euro Surveill 21 (2016). [DOI] [PMC free article] [PubMed]
- 12.Musso D, Nilles EJ, Cao-Lormeau VM. Rapid spread of emerging Zika virus in the Pacific area. Clin Microbiol Infect. 2014;20:O595–596. doi: 10.1111/1469-0691.12707. [DOI] [PubMed] [Google Scholar]
- 13.Oehler, E. et al. Zika virus infection complicated by Guillain-Barre syndrome - case report, French Polynesia, December 2013. Euro Surveill 19 (2014). [DOI] [PubMed]
- 14.Ioos S, et al. Current Zika virus epidemiology and recent epidemics. Med Mal Infect. 2014;44:302–307. doi: 10.1016/j.medmal.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Mlakar J, et al. Zika virus associated with microcephaly. N Engl J Med. 2016;374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 16.Tetro JA. Zika and microcephaly: causation, correlation, or coincidence? Microbes Infect. 2016;18:167–168. doi: 10.1016/j.micinf.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 17.Driggers RW, et al. Zika virus Infection with prolonged maternal viremia and fetal brain abnormalities. N Engl J Med. 2016;374:2142–2151. doi: 10.1056/NEJMoa1601824. [DOI] [PubMed] [Google Scholar]
- 18.Figueiredo LT. The recent arbovirus disease epidemic in Brazil. Rev Soc Bras Med Trop. 2015;48:233–234. doi: 10.1590/0037-8682-0179-2015. [DOI] [PubMed] [Google Scholar]
- 19.Musso D, Gubler DJ. Zika Virus. Clin Microbiol Rev. 2016;29:487–524. doi: 10.1128/CMR.00072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honorio NA, et al. Spatial evaluation and modeling of Dengue seroprevalence and vector density in Rio de Janeiro, Brazil. PLoS Negl Trop Dis. 2009;3:e545. doi: 10.1371/journal.pntd.0000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dyer O. Yellow fever stalks Brazil in Zika’s wake. BMJ. 2017;356:j707. doi: 10.1136/bmj.j707. [DOI] [PubMed] [Google Scholar]
- 22.Verma R, Khanna P, Chawla S. Yellow fever vaccine: an effective vaccine for travelers. Hum Vaccin Immunother. 2013;10:126–128. doi: 10.4161/hv.26549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waggoner JJ, Pinsky BA. Zika virus: diagnostics for an emerging pandemic threat. J Clin Microbiol. 2016;54:860–867. doi: 10.1128/JCM.00279-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.St George K, et al. Zika virus testing considerations: lessons learned from the first 80 real-time reverse transcription-PCR-positive cases diagnosed in New York State. J Clin Microbiol. 2016;55:535–544. doi: 10.1128/JCM.01232-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bingham AM, et al. Comparison of Test Results for Zika Virus RNA in Urine, Serum, and Saliva Specimens from Persons with Travel-Associated Zika Virus Disease - Florida, 2016. MMWR Morb Mortal Wkly Rep. 2016;65:475–478. doi: 10.15585/mmwr.mm6518e2. [DOI] [PubMed] [Google Scholar]
- 26.Gourinat AC, O’Connor O, Calvez E, Goarant C, Dupont-Rouzeyrol M. Detection of Zika virus in urine. Emerg Infect Dis. 2014;21:84–86. doi: 10.3201/eid2101.140894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charrel RN, et al. Background review for diagnostic test development for Zika virus infection. Bull World Health Organ. 2016;94:574–584D. doi: 10.2471/BLT.16.171207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balm MN, et al. A diagnostic polymerase chain reaction assay for Zika virus. J Med Virol. 2012;84:1501–1505. doi: 10.1002/jmv.23241. [DOI] [PubMed] [Google Scholar]
- 29.Waggoner JJ, et al. Viremia and clinical presentation in Nicaraguan patients infected with Zika virus, Chikungunya virus, and Dengue virus. Clin Infect Dis. 2016;63:1584–1590. doi: 10.1093/cid/ciw589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Musso D, et al. Detection of Zika virus in saliva. J Clin Virol. 2015;68:53–55. doi: 10.1016/j.jcv.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 31.Jeong YE, et al. Viral and serological kinetics in Zika virus-infected patients in South Korea. Virol J. 2017;14:70. doi: 10.1186/s12985-017-0740-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kutsuna, S. et al. Two cases of Zika fever imported from French Polynesia to Japan, December 2013 to January 2014 [corrected]. Euro Surveill 19 (2014). [DOI] [PubMed]
- 33.Ong SP, Choo BG, Chu JJ, Ng ML. Expression of vector-based small interfering RNA against West Nile virus effectively inhibits virus replication. Antiviral Res. 2006;72:216–223. doi: 10.1016/j.antiviral.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 34.Rabe IB, et al. Interim Guidance for Interpretation of Zika Virus Antibody Test Results. MMWR Morb Mortal Wkly Rep. 2016;65:543–546. doi: 10.15585/mmwr.mm6521e1. [DOI] [PubMed] [Google Scholar]
- 35.Matheron S, et al. Long-Lasting Persistence of Zika Virus in Semen. Clin Infect Dis. 2016;63:1264. doi: 10.1093/cid/ciw509. [DOI] [PubMed] [Google Scholar]
- 36.Nicastri, E. et al. Persistent detection of Zika virus RNA in semen for six months after symptom onset in a traveller returning from Haiti to Italy, February 2016. Euro Surveill 21 (2016). [DOI] [PMC free article] [PubMed]
- 37.Tan JJL, et al. Persistence of Zika virus in conjunctival fluid of convalescence patients. Sci Rep. 2017;7:11194. doi: 10.1038/s41598-017-09479-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fares RC, Souza KP, Anez G, Rios M. Epidemiological Scenario of Dengue in Brazil. Biomed Res Int. 2015;2015:321873. doi: 10.1155/2015/321873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gibson G, et al. From primary care to hospitalization: clinical warning signs of severe dengue fever in children and adolescents during an outbreak in Rio de Janeiro, Brazil. Cad Saude Publica. 2013;29:82–90. doi: 10.1590/S0102-311X2013000500010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.