Abstract
AIM:
To evaluate the diagnostic performance of MDI and temocillin disk (30 μg) for detection of carbapenem-resistant Enterobacteriaceae in comparison to real-time PCR.
MATERIAL AND METHODS:
Fifty specimens submitted to the Microbiology Laboratory of Ain Shams University Hospitals and showed resistance to carbapenem drugs through routine culture and susceptibility testing, were assessed by both temocillin disk (30 μg) and MDI set to detect carbapenem-resistant Enterobacteriaceae. Results were compared to real-time PCR for detection of carbapenemase genes blaKPC, blaNDM, blaOXA–48-like, blaVIM, and blaIMP.
RESULTS:
Our work revealed that most of the CPE isolates were Klebsiella species (62%) followed by E. coli (24%), Serratia (10%) and Citrobacter (4%). Phenotypic detection of carbapenem-resistant classes revealed OXA - 48 in 96% of isolates, followed by MBLs (82%), and KPC (34%). All isolates were negative for AmpC. Detection of the genes by real-time PCR showed that the predominance was for the blaOXA-48 gene (96%) then blaVIM (94%) followed by blaNDM (54%), blaKPC (46%) and finally blaIMP (40%). Evaluation of the MDI set against PCR showed sensitivity (82.1%) and specificity (70%). The temocillin disk had 97.9% sensitivity and 50% specificity. The evaluation of Temocillin disk and MDI in combination for detection of carbapenem-resistant Enterobacteriaceae showed 99.7% sensitivity and 35% specificity.
CONCLUSION:
Adding Temocillin disk to Mastdisks ID inhibitor combination set provides a simple, easy, rapid and highly sensitive test that can be used for screening and classification of carbapenem-resistant Enterobacteriaceae. However, it still needs confirmation by molecular techniques.
Keywords: Enterobacteriaceae, Carbapenemases, Mastdisks inhibitor, Temocillin
Introduction
Multidrug-resistant organisms are markedly increasing among bacterial species, substantially, Carbapenem-resistant Enterobacteriaceae which have been reported worldwide [1].
Carbapenems are considered the drugs of the last choice for treating multidrug-resistant pathogens due to their high clinical efficacy and safety [2]. There is a major problem concerning resistance to carbapenem groups of drugs as it limits treatment options and thereby obligates for the use of antibiotics with high toxicity like tigecycline and colistin [3]. The most common mechanism of carbapenem resistance is the production of carbapenemase enzymes [2].
At first, carbapenemases were chromosomal - mediated in a few specific species, but they are now plasmid - mediated, or both chromosomally - and plasmid-mediated. This leads to the much more aggressive spread of resistance due to horizontal transmission among various bacterial species and genera [4].
Carbapenemases are classified according to their functional and molecular properties. Molecular classes A and D are the β - lactamases having serine at their active site, whereas molecular class B β - lactamases are all metalloenzymes with zinc at their active site [5].
The most clinically significant enzymes among the class A carbapenemases are K. pneumoniae carbapenemase (KPC) enzymes. KPC-producing K. pneumoniae are widely disseminated among several countries [6]. The class B beta-lactamases or Metallo – beta-lactamases (MBLs) have also been identified in various enterobacterial species, including K. pneumonia [7]. They are mainly NewDelhi Metallo – beta-lactamase (NDM - 1), Verona integrin - encoded Metallo – beta-lactamase (VIM), and Imipenemase (IMP) type enzymes. NDM - 1 is the most commonly identified worldwide. Carbapenem - hydrolysing oxacillinase - 48 (OXA - 48) is the most frequently reported class D beta-lactamase [8].
OXA - 48-producing isolates are frequently multidrug - resistant, as they combine multiple resistance mechanisms. This enzyme shows different hydrolysing activities against β - lactam antibiotics, with high activity against penicillins but only low activity against carbapenems. OXA - 48 carbapenemase has very weak activity against third -generation and fourth - generation cephalosporins; however, these are seldom a therapeutic option because other β - lactamases, such as extended -spectrum β - lactamases (ESBL), are frequently associated [9]. The problem of OXA - 48 is that it may go undetected and classified as susceptible with imipenem and meropenem MIC of (≤ 1 μg/ml) according to Clinical and Laboratory Standards Institute (CLSI, 2015) and (≤ 2 μg/ml) according to European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines respectively [11]. MIC determination alone is not accurate for the screening of OXA - 48.
Rapid detection of carbapenemases is crucial to ensure early detection and implementation of control measures in hospitals. Preliminary screening by Disk Diffusion should be confirmed by Modified Hodge test (MHT) according to the recommendation of CLSI [12]. However, MHT requires at least 24 - 48 h and thereby it is time-consuming. Also, it lacks specificity and may give false positive results or fail to detect MBLs [13].
Molecular techniques remain the reference standard for the identification and differentiation of carbapenemases. The most common of which is PCR. Sequencing is sometimes done following PCR for precise identification of a carbapenemase, rather than just its group (e.g. VIM - type, KPC - type, NDM -type, and OXA - 48 - type) [13].
Materials and Methods
Patient population
This study was conducted on 50 specimens randomly selected from different clinical specimens submitted to the Microbiology Laboratory of Ain Shams University Hospitals for routine culture and susceptibility testing that showed resistance to carbapenem drugs by both Kirby Bauer disk diffusion method (They were nonsusceptible, i.e., intermediate or resistant to the carbapenems if zone diameter was < 23 for meropenem (10 μg) and imipenem (10 μg) and Broth microdilution method if MICs were > 1 μg/ml for meropenem and imipenem (10). The isolates included 31 (62%) Klebsiella species, 12 (24%) E.coli, 5 (10%) Serratia and 2 (4%) Citrobacter.
Most disks ID inhibitor combination disks (MDI)
Most disks ID inhibitor combination disks (MDI) (Mast Diagnostics) method was performed according to the manufacturer’s instructions. Four disks were included: disk A, containing a carbapenem (meropenem, 10 μg); disk B, consisting of meropenem (10 μg) and an MBL inhibitor; disk C, consisting of meropenem (10 μg) with a KPC inhibitor; and disk D, containing meropenem (10 μg) with an AmpC inhibitor. The interpretation of the test is as follows. The zone of inhibition of disk A is compared to the zones of inhibition of each of disks B, C, and D. If disk B shows a zone difference of ≥ 5 mm from disk A, the organism is considered as producing MBL activity as in (Figure 1). If disk C shows a zone difference of ≥ 4 mm from disk A, the organism considered as producing KPC activity. If disk C and disk D both show a zone difference of ≥ 5 mm from disk A, the organism is considered as producing AmpC activity coupled with porin loss (impermeability).
Figure 1.
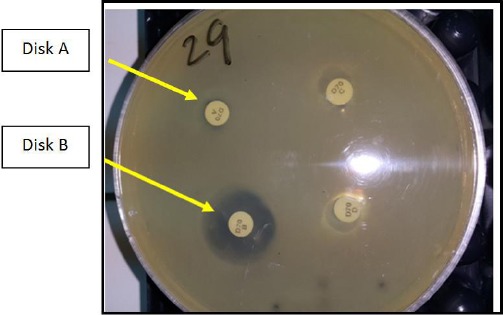
Mastdisks inhibitor combination set showing MBL producing strain (Disk B > disk A by 5 mm)
Detection of OXA 48
Temocillin testing using a modified zone diameter cut - off of < 12 mm (Figure 2) was used as a tool for discriminating Ambler class D carbapenemase producers [14].
Figure 2.
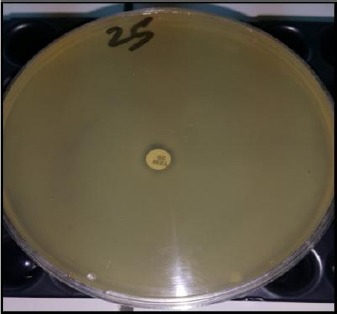
Disc diffusion susceptibility test showing Temocillin resistant strain (OXA48 producer)
E. coli ATCC 25922 and Klebsiella ATCC 700603 were included as negative control.
Genotypic Identification
All test isolates were subcultured on blood agar overnight at 36°C, and five selected colonies were suspended in 1.5 ml microtubes containing 180 µL sterile distilled water, then whole genomic DNA was extracted by QIAamp DNA Mini kit (QIAGEN Sample and Assay Technologies, Germany) according to manufacturer’s instruction. Suitable primers each targeting selected regions of the blaKPC, blaNDM, blaOXA – 48 - like blaVIM and blaIMP genes were used (Table 1). Amplification reactions were performed in a final volume of 25 μL containing 12.5 μL 2 x HRM PCR Master Mix (Thermo Scientific, Lithuania, EU), and two primer mix, 9.5 μL RNase - free H2O, and one μL of the template DNA. The optimized cycling protocol for High Resolution Melting (HRM) Analysis was as follow: initial denaturation at 95°C for 5 minutes followed by 40 cycles at 95°C for 10 seconds, 55°C for 30 seconds, and 72°C for 10 seconds, followed by HRM analysis of 65 - 95°C for 2 seconds (Rotor-Gene, QIAGEN) [15].
Table 1.
Primers targeting selected regions of the blaKPC, blaNDM, blaOXA-48-like, blaVIM, and blaIMP genes (Monteiro et al. 2012)
| Template | Primer | Sequence (5’-3’) |
|---|---|---|
| blaOXA-48 | Forward primer | TGTTTTTGGTGGCATCGAT |
| Reverse primer | GTAAMRATGCTTGGTTCGC | |
| blaNDM-1 | Forward primer | TTGGCCTTGCTGTCCTTG |
| Reverse primer | ACACCAGTGACAATATCACCG | |
| blaKPC | Forward primer | TCGCTAAACTCGAACAGG |
| Reverse primer | TTACTGCCCGTTGACGCCCAATCC | |
| blaIMP | Forward primer | GAGTGGCTTAATTCTCRATC |
| Reverse primer | AACTAYCCAATAYRTAAC | |
| blaVIM | Forward primer | GTTTGGTCGCATATCGCAAC |
| Reverse primer | AATGCGCAGCACCAGGATAG |
Results
Phenotypic detection of carbapenem-resistant classes using carbapenemase detection set by (MastDiagnostics) and temocillin disk revealed that the highest positive results were for OXA48 (detected in 96% of samples), followed by MBLs (82%), then for KPC (34%), while all samples were negative for AmpC.
Some specimens showed two or more positive carbapenemase producing enzymes, 46% of specimens were positive for both OXA 48 and MBL, whereas 32% specimens were positive for OXA 48, MBL and KPC in combination. Only 2% showed positive results for both OXA 48 and KPC.
By applying real-time PCR (singleplex) to detect blaKPC, blaNDM, blaOXA – 48 - like, blaVIM and blaIMP genes in our samples, blaOXA – 48 - like gene showed the highest percentage (96%) followed by blaVIM, blaNDM, blaKPC, and blaIMP with (94%) (54%), (46%), and (40%), respectively (Figure 3).
Figure 3.
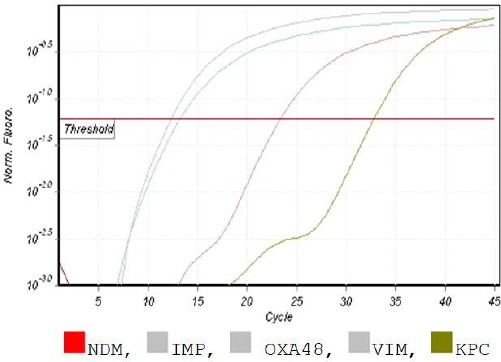
Melt data for HRM; The isolate was positive for OXA48, IMP, KPC, VIM and negative for NDM
Statistical evaluation of the diagnostic performance of the carbapenemase detection set and temocillin disk against PCR showed, a significant difference between both methods regarding KPC and MBLs (P-value < 0.001). However, no significant difference was detected between temocillin disk and PCR regarding OXA - 48 (P value was 2.239).
By comparing the results of temocillin disk specifically used for detection of OXA 48 and PCR, 47 out of 50 specimens were true positive (97.9%) and one specimen was a false negative (2.1%). One out of two specimens was true negative (50%), and one specimen was false positive (50%). Temocillin disk had 97.9% sensitivity, 50% specificity, 97.95% PPV, 50% NPV and efficacy was 73.95%.
Evaluation of class A (KPC) detection by using disk C compared to PCR detection of the blaKPC gene showed that 15 out of 23 specimens were true positive (65.2%) and eight were a false negative (34.8%). Whereas 25 out of 27 were true negative (92.6%) and two were false positive (7.4%). Sensitivity, specificity, PPV, NPV was 65.2%, 92.6%, 88.24%, 75.78% respectively and efficacy was 78.9%.
We compared results of class B (MBL) detection by using disk B with PCR to detect blaVIM, blaNDM, and blaIMP genes. It was found that 40 out of 46 (87%) specimens were true positive and six (13%) was a false negative. Whereas three out of 4 (75%) specimens were true negative and one (25%) was a false positive. Sensitivity, specificity for PPV, NPV was 87%, 75%, 97.5%, 33% respectively and efficacy was 90.2%.
Finally, the whole detection set was evaluated according to PCR. It was shown that 33 out of 40 specimens were true positive (82.5%) and seven were a false negative (17.5%). Whereas seven out of ten specimens were true negative (70%) and three were false positive (30%). Overall sensitivity, specificity, PPV, NPV were 82.1%, 70%, 91.6%, 50% respectively and efficacy was 85.7%.
Evaluation of the combined use of Temocillin disk and Mastdisks ID inhibitor combination set (MDI) in parallel for detection of carbapenem-resistant Enterobacteriaceae showed 99.7% sensitivity and 35% specificity.
A high percentage of positive isolates for OXA 48 (96%) and the low percentage of negative OXA 48 isolates (4%) resulted in Low specificity for Temocillin disk as well as specificity of Temocillin disk and MDI in combination. One isolate was a false negative (2.1%), one out of two isolates (that were negative by PCR) was true negative (50%), and one was false positive (50%).
Discussion
Antibiotic resistance is a major cause for concern in Enterobacteriaceae family. The rate of resistance is increasing especially carbapenem-resistant Enterobacteriaceae. Therefore, the need for a simple and accurate method is crucial for detection of these bacteria.
Some phenotypic confirmation tests have been performed for the detection of carbapenemase-producing Enterobacteriaceae. These include; modified Hodge test (MHT), inhibitor - based methods using metal chelators for MBLs (e.g., MBL Etest) and boronic acid for KPCs [13].
In this study, Mastdisks ID inhibitor combination disks kit (MDI) (Mast Diagnostics) was evaluated for the detection of Enterobacteriaceae producing carbapenemases. Sanjeev and Mehra (2015), stated that simultaneous using of both inhibitors (PBA for KPC detection and DPA for MBL detection) as in our study, seems to restrict the activity of both carbapenemases against meropenem, allowing the detection of isolates that co-produce these enzymes in almost all cases [16]. Their study has shown the combined use of two inhibitors can detect and differentiate carbapenemase production. However, Mastdisks combination inhibitor set was not designed to detect OXA – 48 - like B -lactamase in Enterobacteriaceae. That is why we performed disk diffusion assay with temocillin disk to detect OXA – 48 - like producing Enterobacteriaceae. The high - level resistance to the B - lactam temocillin might be presumptive evidence for the presence of OXA - 48 [17].
We found that 96% of our isolates were sensitive to temocillin disk used for OXA - 48 detection. In areas where OXA - 48 producers are predominant, temocillin testing using a modified zone diameter cut - off of < 12 mm [18], could be an easy and useful tool for discriminating Ambler class D carbapenemase producers from extended spectrum b - lactamases and/or AmpC - producing isolates among carbapenem – non - susceptible Enterobacteriaceae isolates. About temocillin, 98.1% (317/323) of the OXA – 48 - producing Enterobacteriaceae isolates displayed disk inhibition zones of < 12 mm compared with only 10.0%(92/919) of the carbapenemase - negative isolates [14].
Also, 82% and 34% were positive for MBLs using disk B and KPC using disk C respectively. These results were concordant with a study conducted by Doyal and co-workers (2012) [19], who stated that 100%, 93% and 49% of their isolates were positivity for OXA - 48, MBLs and KPC detection respectively. Another study conducted by Andrea et al. (2014) [20], they found that 92% of their CPE isolates were OXA 48 positive, 49.5% were harbouring double positive KPC and MBL when used the Mastdisks ID inhibitor combination. Although their study was in Italy, their results were similar to our results, so we can conclude that class D is prevalent there like in Egypt while other classes are not.
A high prevalence of CPE can be found in southern Europe and Asia, Greece, Italy, Turkey, and Israel than in other parts of the world. Historical/cultural relationship and exchange of populations with other countries of high prevalence can influence the type of CPE. Cross-border transfer of patients, travel, medical tourism and refugees might also play an important role. This is particularly true for the spread of OXA - 48 in France and Belgium from North Africa, and in Germany, probably from Turkey, or the identification in UK of NDM - 1 producers of Indian origin. The increasing frequency of CPE could be related to the increasing prevalence of OXA - 48 producers in many European countries and the exportation of KPC producers from Greece and Italy to other European countries [21].
Some of our isolates showed double or triple positive for carbapenemase enzymes. Both OXA 48 and MBL were positive in 23 isolates (46%), whereas 32% showed positive results for OXA48, MBL and KPC altogether. Only (2%) were positive for OXA48 and KPC together (2%). This means that OXA - 48 was positive in most our isolates in combination with other classes. OXA - 48 enzyme in Enterobacteriaceae, can give weak level carbapenem resistance and with cross-resistance with KPC enzymes and MBLs tend to confer broader effects on the resistance profile of the host strain [22]. OXA - 48 enzyme was positive in most of our isolates combined with other resistant classes that made OXA - 48 carbapenemase production more evident and which otherwise couldn’t be detected if present alone.
In this study, we applied real-time PCR to detect blaKPC, blaNDM, blaOXA – 48 - like blaVIM, and blaIMP genes. OXA 48 gene showed the highest percentage (96%) then VIM (94%) followed by NDM (54%), KPC (46%) and finally IMP (40%). This could be explained by the fact that Amber class D is weak and needs the coexistence of other classes to be expressed and so had the highest percentage. Also, in a study conducted by Samar (2016); blaOXA – 48 -like gene’s percentage was the highest (28.6%), followed byblaKPC (19%), blaVIM (9.5%) and finally blaNDM (2.4%) [23].
Most disks ID inhibitor combination disks had a sensitivity and specificity of 87% and 75% respectively for detecting MBL producers, 65.2% and 92.6% respectively, for detecting KPC producers. In our study, evaluation of carbapenemase detection set for detection of all classes of CPE in comparison to PCR showed sensitivity 82.1%, specificity 70%, PPV 91.6% and NPV 50%. Samar [25]; in her study stated that the combined disk test showed sensitivity 100% and specificity 88.9%. Doyal and co-workers (2012), stated that overall, the sensitivity and specificity for MDI were 78% and 93% respectively, MDI performed well for the detection of KPCs and NDMs but poorly for VIMs, IMPs, and OXA – 48 - like enzymes [19].
On the other hand, temocillin disk had 97.9% sensitivity, 50% specificity, 97.95% PPV and 50% NPV and efficacy was 73.95%. Another study conducted by Woodford et al. (2014) in England, the sensitivity and specificity of temocillin were 90.7% and 88.2% respectively with PPV 27.6 % and NPV 99.5% [24].
Huang and co-workers (2014), support previous data and confirm that high - level resistance to temocillin is a highly sensitive and specific phenotypic surrogate marker of OXA - 48 productions in strains with decreased susceptibility to a carbapenem, although in itself it could not differentiate OXA - 48 from VIM - type carbapenemases [18], [25].
However, the overall sensitivity and specificity of both Temocillin disk and carbapenemase detection set tests in our study for detection of carbapenem-resistant Enterobacteriaceae was 99.7% and 35% respectively.
Low specificity for Temocillin disk as well as specificity of Temocillin disk and MDI in combination could be explained by the high percentage of positive isolates for OXA 48 (96%) and the low percentage of negative isolates (4%). Only two isolates were negative by PCR, one was true negative (50%), and the other was false positive (50%) thus reducing Temocillin disk test specificity.
So, adding Temocillin disk to carbapenemase detection set provides an easy, inexpensive, rapid and highly sensitive test for screening of a large number of isolates for CPE. However, it still needs confirmation by molecular techniques.
Acknowledgement
We would like to express our gratitude for prof. Ibrahim K. Ali for his great support and active guidance throughout this work.
Footnotes
Funding: This research was funded by the National Research Centre - Egypt
Competing Interests: The authors have declared that no competing interests exist
Financial support
This research was funded by the National Research Centre - Egypt.
References
- 1.Singh-Moodley A. Perovic Antimicrobial susceptibility testing in predicting the presence of carbapenemase genes in Enterobacteriaceae in South Africa. BMC Infect Dis. 2016;16:536. doi: 10.1186/s12879-016-1858-7. https://doi.org/10.1186/s12879-016-1858-7 PMid:27716102 PMCid: PMC5050574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Queenan AM, Bush K. Carbapenemases the versatile beta-lactamases. Clin Microbiol Rev. 2007;20(3):440–458. doi: 10.1128/CMR.00001-07. https://doi.org/10.1128/CMR.00001-07 PMid:17630334 PMCid: PMC1932750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falagas M, Lourida P, Poulikakos P, et al. Antibiotic treatment of infections due to carbapenem-resistant Enterobacteriaceae: a systematic evaluation of the available evidence. Antimicrob Agents Chemother. 2014;58(2):654–663. doi: 10.1128/AAC.01222-13. https://doi.org/10.1128/AAC.01222-13 PMid:24080646 PMCid: PMC3910850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernández-Cuenca F, Rodríguez-Martínez JM, Gómez-Sánchez MA, et al. Production of a plasmid-encoded OXA-72 β-lactamase associated with resistance to carbapenems in a clinical isolate Acinetobacter junii. Int J Antimicrob Agents. 2012;39(1):93–94. doi: 10.1016/j.ijantimicag.2011.07.017. https://doi.org/10.1016/j.ijantimicag.2011.07.017 PMid:21982835. [DOI] [PubMed] [Google Scholar]
- 5.Agila K Pragasam, Sahni D Rani, Anandan S, et al. A Pilot Study on Carbapenemase Detection: Do We See the Same Level of Agreement as with the CLSI Observations. Journal of Clinical and Diagnostic Research. 2016;10(7):DC09–DC13. doi: 10.7860/JCDR/2016/16417.8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walther-Rasmussen J, Hoiby N. Class A carbapenemases. J Antimicrob Chemother. 2007;60:470–482. doi: 10.1093/jac/dkm226. https://doi.org/10.1093/jac/dkm226 PMid:17595289. [DOI] [PubMed] [Google Scholar]
- 7.Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17:1791–1798. doi: 10.3201/eid1710.110655. https://doi.org/10.3201/eid1710.110655 PMid:22000347 PMCid: PMC3310682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girmenia C, Serrao A, Canichella M. Epidemiology of Carbapenem Resistant Klebsiella pneumoniae Infections in Mediterranean Countries. Mediterr J Hematol Infect Dis. 2016;8(1):e2016032. doi: 10.4084/MJHID.2016.032. https://doi.org/10.4084/mjhid.2016.032 PMid:27441063 PMCid: PMC4943068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poirel L, Potron A, Nordmann P. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother. 2012;67:1597–1606. doi: 10.1093/jac/dks121. https://doi.org/10.1093/jac/dks121 PMid:22499996. [DOI] [PubMed] [Google Scholar]
- 10.Clinical Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, 25 thin formational supplements. CLSI, document M100-S25, Wayne PA. 2015 [Google Scholar]
- 11.Nordmann P, Gniadkowski M, Giske CG, et al. Identification and screening of carbapenemase-producing Enterobacteriaceae. Clinical Microbiology and Infection. 2012;18(5):432–438. doi: 10.1111/j.1469-0691.2012.03815.x. https://doi.org/10.1111/j.1469-0691.2012.03815.x PMid:22507110. [DOI] [PubMed] [Google Scholar]
- 12.Patel J, Cockerill F, Bradford P, et al. Clinical and Laboratory Standards Institute, Performance Standards for Antimicrobial Susceptibility Testing;Twenty first Informational Supplement. CLSI document M100-21. 3. Vol. 35. Wayne, PA: Clinical and Laboratory Standards Institute; 2011. [Google Scholar]
- 13.Nordmann P, Poirel L. Strategies for identification of carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother. 2013;68:487–489. doi: 10.1093/jac/dks426. https://doi.org/10.1093/jac/dks426 PMid:23104494. [DOI] [PubMed] [Google Scholar]
- 14.Huang T, Poirel L, Pierre Bogaerts P, et al. Temocillin and piperacillin/tazobactam resistance by disk diffusionas antimicrobial surrogate markers for the detection of carbapenemase-producing Enterobacteriaceae in geographical areas with a high prevalence of OXA-48 producers. J Antimicrob Chemother. 2014;69:445–450. doi: 10.1093/jac/dkt367. https://doi.org/10.1093/jac/dkt367 PMid:24055766. [DOI] [PubMed] [Google Scholar]
- 15.Monteiro J, Widen H R, Pignatari C A, et al. Rapid detection of carbapenemase genes by multiplex real-time PCR. Journal of Antimicrobial Chemotherapy. 2012;67(4):906–909. doi: 10.1093/jac/dkr563. https://doi.org/10.1093/jac/dkr563 PMid:22232516. [DOI] [PubMed] [Google Scholar]
- 16.Sanjeev K, Mehra SK. Performance of Modified Hodge Test and Combined Disk Test for Detection of Carbapenemases in Clinical Isolates of Enterobacteriaceae. Int J Curr Microbiol App Sci. 2015:772–783. [Google Scholar]
- 17.Glupczynski Y, Huang TD, Bouchahrouf W, et al. Rapid emergence and spread of OXA-48-producing carbapenem-resistant Enterobacteriaceae isolates in Belgian hospitals. Int J Antimicrob Agents. 2012;39:168–172. doi: 10.1016/j.ijantimicag.2011.10.005. https://doi.org/10.1016/j.ijantimicag.2011.10.005 PMid:22115539. [DOI] [PubMed] [Google Scholar]
- 18.Hartl R, Widhalm S, Kerschner H, et al. Temocillin and meropenem to diskriminate resistance mechanisms leading to decreased carbapenem susceptibility with focus on OXA-48 in Enterobacteriaceae. Clin Microbiol Infect. 2013;19:E230–232. doi: 10.1111/1469-0691.12146. https://doi.org/10.1111/1469-0691.12146 PMid:23397897. [DOI] [PubMed] [Google Scholar]
- 19.Doyal D, Peirano G, Lascols C, et al. Laboratory detection of Enterobacteriaceae that produce carbpenemases. J, Clin. Microbiol. 2012;50(12):3877–3880. doi: 10.1128/JCM.02117-12. https://doi.org/10.1128/JCM.02117-12 PMid:22993175 PMCid: PMC3503014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andrea B, Ilaria F, Antonietta C, et al. Comparison of phenotypic methods for the detection of carbapenem non-susceptible Enterobacteriaceae. J BMC Infectious Diseases. 2014;6:3–7. [Google Scholar]
- 21.Canton R, Akova M, Carmeli Y, et al. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clinical Microbiology and Infection. 2012;18(5):414–431. doi: 10.1111/j.1469-0691.2012.03821.x. https://doi.org/10.1111/j.1469-0691.2012.03821.x PMid:22507109. [DOI] [PubMed] [Google Scholar]
- 22.UK Stards for Microbiology Investigations (UK SMI) Detection of bacteria with carbapenem-hydrolysing β-lactamases (carbapenemases), PHE Bacteriology. 2015:60. [Google Scholar]
- 23.Samar S. Comparative evaluation of phenotypic and genotypic methods for detection of carbapenemases in clinically significant Klebsiella pneumonia isolates. Egyptian Journal of Medical Microbiology. 2016;25(1):109–116. https://doi.org/10.12816/0037099. [Google Scholar]
- 24.Woodford N, Pike R, Meunier D, et al. In vitro activity of temocillin against multidrug-resistant clinical isolates of Escherichia coli, Klebsiella spp. And Enterobacter spp., and evaluation of high-level temocillin resistance as a diagnostic marker for OXA-48 carbapenemase. J Antimicrob. Chemother. 2014;69:564–567. doi: 10.1093/jac/dkt383. https://doi.org/10.1093/jac/dkt383 PMid:24080500. [DOI] [PubMed] [Google Scholar]
- 25.Livermore DM, Warner M, Mushtaq S, et al. What remains against carbapenem-resistant Enterobacteriaceae?Evaluation of chloramphenicol, ciprofloxacin, colistin, fosfomycin, minocycline, nitrofurantoin, temocillin and tigecycline. Int J Antimicrob Agents. 2011;37:415–419. doi: 10.1016/j.ijantimicag.2011.01.012. https://doi.org/10.1016/j.ijantimicag.2011.01.012 PMid:21429716. [DOI] [PubMed] [Google Scholar]


