P-glycoprotein contributes to multidrug resistance by exporting a broad range of substrates across the cell membrane. Using molecular dynamics simulations, Barreto-Ojeda et al. identify key lipid-binding sites and reveal lipid access pathways toward the cavity of the transporter.
Abstract
P-glycoprotein (P-gp) exports a broad range of dissimilar compounds, including drugs, lipids, and lipid-like molecules. Because of its substrate promiscuity, P-gp is a key player in the development of cancer multidrug resistance. Although P-gp is one of the most studied ABC transporters, the mechanism by which its substrates access the cavity remains unclear. In this study, we perform coarse-grained molecular dynamics simulations to explore possible lipid access pathways in the inward-facing conformation of P-gp embedded in bilayers of different lipid compositions. In the inward-facing orientation, only lipids from the lower leaflet access the cavity of the transporter. We identify positively charged residues at the portals of P-gp that favor lipid entrance to the cavity, as well as lipid-binding sites at the portals and within the cavity, which is in good agreement with previous experimental studies. This work includes several examples of lipid pathways for phosphatidylcholine and phosphatidylethanolamine lipids that help elucidate the molecular mechanism of lipid binding in P-gp.
Introduction
P-glycoprotein (P-gp), a member of the ATP-binding cassette (ABC) transporter superfamily, is responsible for the translocation of a wide range of compounds across the plasma membrane (Chen et al., 1986; Loo and Clarke, 1994). P-gp is found in several tissues facing an excretory compartment (e.g., in the apical surface of epithelial cells of the liver, pancreas, and small intestine; Thiebaut et al., 1987). It is also found in the brush border of kidneys and the endothelial cells of the blood–brain barrier (Eckford and Sharom, 2006; Sharom, 2014). Natural substrates of P-gp are toxins and xenobiotics, against which P-gp provides a mechanism of defense by exporting them outside the cell. Substrates binding to P-gp are structurally dissimilar compounds (Aller et al., 2009; Leong et al., 2012; Zhang et al., 2014), which include metabolites, drug molecules like analgesics and antibiotics, drug-like molecules such as diagnostic dyes, and lipids (van Helvoort et al., 1996; Bosch et al., 1997; Ambudkar et al., 2003; Marcoux et al., 2013; Ferreira et al., 2015). In addition, several studies have linked the overexpression of ABC transporters, most notably P-gp, with mechanisms of resistance in various cancer cells (Fletcher et al., 2010). Because of its broad substrate specificity, P-gp confers multidrug resistance (MDR) by pumping several pharmacological agents to the extracellular medium, preventing their accumulation inside the cell (Gros and Buschman, 1993; Sharom, 2014). The development of MDR is considered one of the major challenges in cancer treatment (Ambudkar et al., 2003; Szakács et al., 2006; Chen et al., 2016), and because P-gp acts as a key player, the study of its underlying mechanism is considerably important to improve current chemotherapeutic treatments (Gottesman, 1993; Sharom, 2014).
In addition to having a drug transporter function, P-gp has also been proposed to act as a lipid flippase (Higgins and Gottesman, 1992), flipping lipids from the inner to the outer leaflet (Sharom, 2014). Direct interaction between lipid-based molecules and P-gp has been identified in previous studies (van Helvoort et al., 1996; Bosch et al., 1997; Marcoux et al., 2013; Sharom, 2014). Competition between transport and inhibition of ATPase activity suggests that lipids are not only active modulators of the transport mechanism of P-gp, but also of substrates (Sharom, 2011, 2014). The high affinity of the transporter for lipids and lipid-based molecules such as miltefosine and edelfosine, which are both phospholipid-based anticancer agents, has been linked with the flippase model (Sharom, 2014). Given the crucial role in cancer MDR, further understanding of the factors that determine lipid binding is essential for targeting P-gp inhibition as a means to increased drug delivery in chemotherapy treatments.
The P-gp structure consists of two units linked by a flexible linker. Each unit includes an N-terminal transmembrane domain (TMD) followed by a C-terminal nucleotide-binding domain (NBD). The TMDs, with six transmembrane helices each (TMD1, H1–H6; TMD2, H7–H12), form the translocation pathway for substrates. TMDs and NBDs are coupled by intracellular loops. As for any other ABC transporter, the substrate translocation mechanism of P-gp is ATP driven (Loo and Clarke, 1995). ATP binding and hydrolysis at the NBDs induce NBD dimerization and dissociation, which in turn drive conformational changes in the TMDs (Loo and Clarke, 1994; Aller et al., 2009). To allow transport, P-gp switches between inward-facing and outward-facing states, which is relevant for substrate binding and release, respectively (Sharom, 2014; Chen et al., 2016). In the inward-facing conformation (Li et al., 2014), the two TMDs (TMD1 and TMD2) form a cavity open toward the cytoplasm that is enclosed by helices H1, H3, H5, H7, H11, and H12. In addition, two portals (referred to as P1 and P2) allow access of molecules directly from the bilayer. P1 is constituted by helices H4 and H6, and P2 is constituted by H10 and H12. Although several theoretical and experimental studies have targeted substrate binding (Leong et al., 2012; Marcoux et al., 2013; Ferreira et al., 2015), the mechanism by which P-gp can recognize substrates remains elusive.
In this study, as a step toward a better understanding of how substrates access the translocation cavity, we use molecular dynamics (MD) simulations at a coarse-grained (CG) level of detail to investigate lipid pathways and lipid–protein interaction in P-gp using the Martini force field (Marrink et al., 2007). CG models are a powerful tool to investigate biomolecular processes of larger systems, or longer timescales, because of their simplified level of detail while maintaining chemical specificity (Marrink et al., 2007). CG models are particularly suitable to reach system size and timescales that are relevant to comprehensively study substrate pathways (Ingólfsson et al., 2014a). The Martini force field has been widely used to resolve lipid–protein interactions in diverse biomolecular systems (Marrink and Tieleman, 2013; Chavent et al., 2016). For example, Karo et al. (2012) explored the binding of mitochondrial creatine kinase to the membrane. Sengupta and Chattopadhyay (2012) analyzed the cholesterol-binding sites in the serotonin1A receptor. Schmidt et al. (2013) investigated phosphatidylinositol phosphate–binding events in the potassium channel Kir2.2.
Given the importance of the role of lipids as possible substrates in P-gp, we investigated lipid–protein interactions using the crystallographic structure of mouse P-gp in the inward-facing conformation (Li et al., 2014), which was embedded in membranes of different ratios of POPC and POPE lipids. To investigate the P-gp lipid pathways, we first analyzed lipid access to the cavity. In the Martini force field (Marrink et al., 2007), even though the headgroups of POPC and POPE are zwitterionic, the interaction parameters differ based on hydrogen-bonding capability and solvation-free energy, among other properties (Marrink et al., 2004; de Jong et al., 2013). Because only the headgroup of POPE is a donor for hydrogen bonds, a different lipid–protein interaction might be possible, and given the membrane composition, it may berelevant for mechanistic details. For this reason, we first evaluated a possible selectivity between phosphatidylcholine (PC) and phosphatidylethanolamine (PE) lipids. Second, we studied the interactions between lipids and protein residues at the portals and inside the cavity. Finally, we analyzed the dynamics of the lipids inside the cavity of P-gp and identified key interactions between the lipid headgroups and the lipid tails with the residues in the cavity. In agreement with previous experimental studies (Aller et al., 2009; Marcoux et al., 2013), our results highlight the importance of positively charged residues at the portals, such as Arg355, which may facilitate lipid access to the cavity via electrostatic interactions.
Materials and methods
We used the refined structure of mouse P-gp as the input structure for MD simulations (Protein Data Bank accession no. 4M1M; Li et al., 2014). We conducted CG MD simulations of P-gp embedded in lipid bilayers of a different composition (Table 1). Because the most abundant phospholipids in the upper and lower leaflet for the mammalian plasma membrane (Ingólfsson et al., 2014b) are PC and PE, our simulations include pure POPC and POPE bilayers, a 1:1 symmetric POPC/POPE bilayer, and two asymmetric bilayers: one with the same PC/PE ratio as reported for the plasma membrane (Ingólfsson et al., 2014b), and, for comparison, one with a PC/PE ratio inverted between the two leaflets. All simulations were performed using the GROMACS 4.6 package (Hess et al., 2008) and the standard Martini force field 2.2 for protein and 2.0 for lipids (Marrink et al., 2007; Hess et al., 2008; Monticelli et al., 2008; Chavent et al., 2016).
Table 1. Lipid composition of the bilayers.
| System | Composition |
|---|---|
| S1 | POPC |
| S2 | POPE |
| S3 | POPC/POPE 1:1 |
| S4 | Upper leaflet POPC/POPE 16:3, lower 11:16 |
| S5 | Upper leaflet POPC/POPE 11:16, lower 16:3 |
Simulation setup
The atomistic structure of P-gp was converted to the corresponding CG model using the Martinize (de Jong et al., 2013) building tool. An elastic network was applied for atom pairs within a 1.0-nm cutoff distance (Periole et al., 2009). All protein bilayer systems were built using the INSANE (INSert membrANE) CG tool (Wassenaar et al., 2015). Membranes were solvated with standard Martini water. Ions were also added to neutralize the protein and to reach a final concentration of 150 mM NaCl. Each simulation box of 140 × 140 × 180 Å contained one copy of P-gp in CG representation, ∼600 lipids, ∼23,000 CG water, ∼250 Na+, and ∼260 Cl−.
Energy minimization for all systems was performed using the steepest-descent algorithm with a force tolerance of 100 kJ mol−1nm−1 for convergence. For electrostatic interactions, we used a shift function to turn off the interactions between 0 and 1.2 nm. The relative dielectric constant was set to εr = 15, as is standard in nonpolarizable Martini. All subsequent equilibration simulations were performed for 8 ns in the NPT ensemble with a 10-fs time step. The Berendsen barostat (Berendsen et al., 1984) kept the systems at one bar with a relaxation time constant of τp = 1 ps, with semiisotropic pressure coupling. The reference temperature was set at 310K and controlled by a velocity-rescaling thermostat (Bussi et al., 2007) with a relaxation time constant of τT = 0.5 ps. Position restraints on the protein, with a force constant of 1,000 kJ mol−1nm−2, were applied gradually, first on all of the proteins, and then on the backbone beads only. Production runs were performed with a 20-fs integration time step. In the production simulations, the Berendsen barostat (Berendsen et al., 1984) was used to maintain the systems at one bar with a relaxation time constant of τp = 5 ps. The reference temperature of 310K was controlled by the velocity rescaling thermostat (Bussi et al., 2007) with a relaxation time constant of τT = 2 ps. The electrostatic interactions were treated as described above for energy minimization. For production run, each system was simulated for 20 µs.
Lipid access and lipid–protein interaction analysis
Lipid access
The main cavity is defined here as formed by the residues on the helices lining the inner portion of the P-gp cavity (i.e., the side chain of the residues located along the helices H1, H3, and H5 and H7, H11, and H12). Residues on H2, H8, and H9 (external helices) and H4, H6, H10, and residues Tyr994 to Ser1002 of H12 (portals) were excluded. A lipid was considered as accessing the cavity if the corresponding PO4 bead was found for >200 consecutive nanoseconds within 8 Å of any of the side chain residues of the main cavity. Among different values, the 8 Å cutoff was the optimum distance to evaluate access events with no overestimation. The window of 200 ns guarantees that the access events are not caused by thermal fluctuations. Numerical analyses were performed using open-source Python NumPy and MDTraj libraries (van der Walt et al., 2011; McGibbon et al., 2015).
Lipid occupancy inside the P-gp cavity
A contact between a lipid molecule and the cavity was counted if the corresponding PO4 bead was located within 5 Å from any of the residues in the cavity of P-gp. We will refer to this as the contact condition. Initially, our analysis considered all possible residue–phosphate pairs interacting within 5, 8, and 10 Å. We found ∼450 residues of P-gp interacting with lipid headgroups within a cutoff of 10 Å. When the cutoff distance was reduced to 7 Å, the number of residues involved in lipid–protein interactions decreased by 60% compared with the 10-Å cutoff. Because our aim was to find the most representative lipid-binding sites, we reduced the cutoff to 5 Å, which registered ∼160 residues corresponding to the top 30% set of residues interacting with lipid headgroups. For each system, the number of lipids that satisfied the contact condition at the same time was calculated, representing the number of lipids that simultaneously remained inside the cavity of P-gp. For this, the restriction that a contact must be at least 200 ns was not applied. We also calculated the time during which a given number of lipid molecules occupied the cavity simultaneously.
Lipid–protein interactions
Portals
To elucidate the lipid pathways in P-gp, we performed lipid–protein interaction analysis to identify the residues at the portals of P-gp following the contact condition. The portals of the protein are delimited by helices H4 and H6 (P1) and H10 and H12 (P2). We counted a binding event when the PO4 bead of a lipid molecule was located within 5 Å of the residues at the portals of P-gp. At the portals, the residues considered were Lys230 to His241 of H4, Ser345 to Gly356 of H6, Leu875 to Lys883 of H10, and Tyr994 to Ser1002 of H12. The analysis of binding events was performed on the lipid molecules selected from the previously described lipid access. Lipid-binding sites were identified as the top five residues for which the contact condition was satisfied during the last 15 µs of simulation time. The maximum contact time registered was normalized to 1.0. A color map distinguishes three levels of interaction: very high interactions are between 0.8 and 1(red); high interactions are between 0.4 and 0.7 (magenta); and moderate interactions are <0.4 (gray). The level of interaction was calculated for residues at the portals (Fig. 3) and in the cavity (Fig. 4).
Cavity of P-gp
Similar to the analysis of the lipid–protein interactions at the portals, a lipid molecule was considered bound if the corresponding PO4 bead was located within 5 Å of the residues at the cavity of P-gp for at least 200 ns. The lipid-binding sites and the levels of interaction follow the same definition as explained above for the top 10 residues, in addition to those interacting exclusively with PC (blue) and PE (cyan), as illustrated in Fig. 4.
Sequence alignment
The sequences were retrieved from the UniProt Knowledgebase (accession nos. P21447, P08183, P21440, and P21439; The UniProt Consortium, 2017) for mouse and human P-gp and mouse and human ABCB4, respectively. The sequence alignment was performed using MAFFT (Katoh et al., 2002; Katoh and Standley, 2013).
Lipids inside the cavity
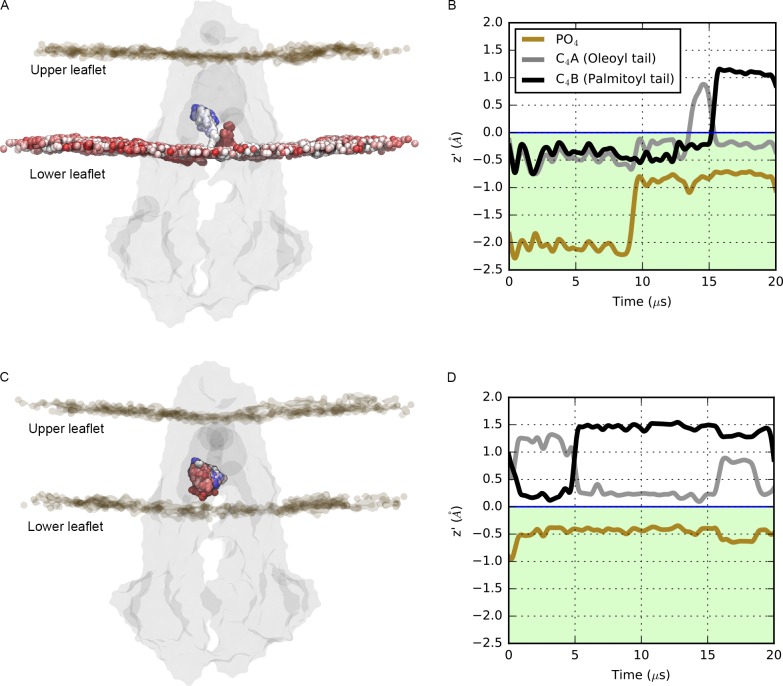
To analyze the dynamics of the lipid headgroups and tails inside the P-gp cavity, we considered the PO4 bead of each lipid and the last bead of each tail (i.e., C4A and C4B beads). The lipid-binding sites for each tail were identified by the contact condition. For each frame, we defined the middle plane of the membrane z’ = 0 as the mean z coordinates between the upper and lower leaflet. Region I is defined as the highest region in the P-gp cavity, z’ > 0; region II corresponds to the region between the lower leaflet and the middle plane of the bilayer, where z’ < 0. Measurements of the z coordinate for the phosphate (PO4) bead and the last bead of each tail (oleoyl tail, C4A bead; palmitoyl tail, C4B bead) were performed.
Occupancy maps
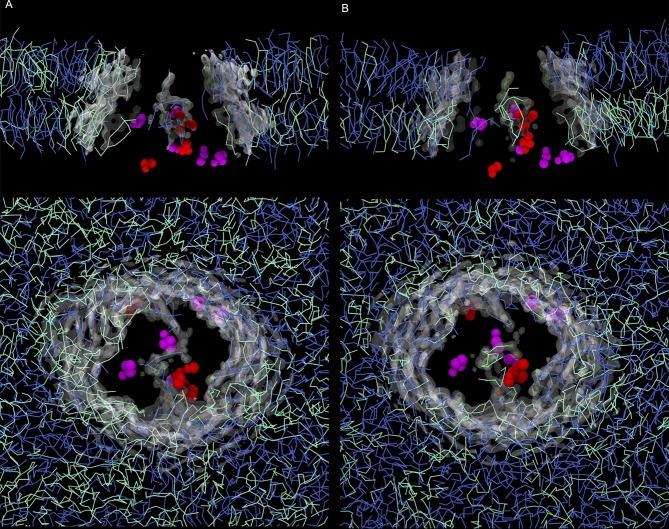
Fractional lipid occupancy was calculated for POPE and POPC lipid types over the last 15 µs of each simulation system using the VolMap plugin in VMD 1.9.1 with a grid resolution of 1 Å (Humphrey et al., 1996). For the systems with pure POPC or POPE bilayers, maximum occupancy was found at 0.78 and 0.72, respectively. The maximum occupancy was found at 0.73 for the system with a 1:1 POPC/POPE ratio and at 0.78 and 0.74 for the asymmetric bilayer with the plasma ratio and the asymmetric system with the inverted plasma ratio, respectively. For visualization purposes, the isosurfaces in Fig. 6 are displayed at 20%, 50%, and 70% of the maximum value, set at 0.72 for all the systems, and the maps are superimposed with the protein structure extracted from the last frame of each simulation.
Results
Lipid access events were identified by the interaction between the lipid headgroup and residues of helices H1, H3, and H5 and H7, H11, and H12 following the contact condition (see Materials and methods, Lipid access). The number of lipid access events was determined by the number of lipid headgroups accessing the main cavity for both PC and PE lipid types. From our simulations, we found that these events involve only lipid molecules from the lower leaflet.
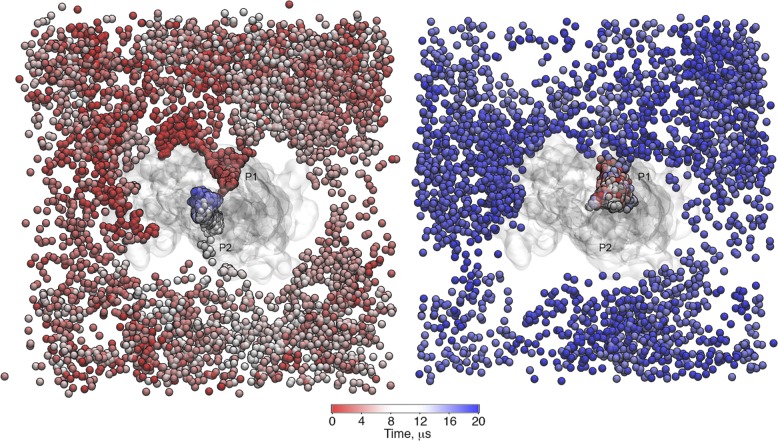
The level of detail from the CG approach, together with the extensive sampling for lipid diffusion in our simulations, allowed us to observe reversible lipid binding in the cavity of P-gp as well as lipid confinement inside the cavity. Two examples are illustrated in Fig. 1. On the left, at 0 µs (red), the selected lipid is outside the cavity; during the first microsecond of the simulation, it entered the cavity through P1, where it remained for <1 µs. After ∼9 µs (white), the lipid again entered the cavity through P2, where it stayed for the remaining simulation time (blue). The right panel shows a lipid molecule that remained for ∼16 µs inside the cavity before diffusing outside. Despite the differences between POPE and POPC regarding the headgroup subtype previously mentioned, the results show no indication of selectivity toward any of the lipid types (Fig. 2 A). The differences in the number of access events for POPC and POPE lipids in the different membranes are not substantial. In what follows, we generally combine the results for both lipids from all the simulation systems.
Figure 1.
Two examples of PO4 bead trajectories found in the pure POPC bilayer. (Left) At 0 µs, the lipid molecule resides outside of the cavity. It accesses the cavity twice during the simulation time, first during the first microsecond of the simulation through P1 and then after ∼9 µs through P2. (Right) The selected lipid molecule remains inside the cavity for ∼16 µs before exiting through P1. P-gp is shown as a transparent surface, and the PO4 bead of the selected lipids is shown as a sphere, colored as a function of time.
Figure 2.
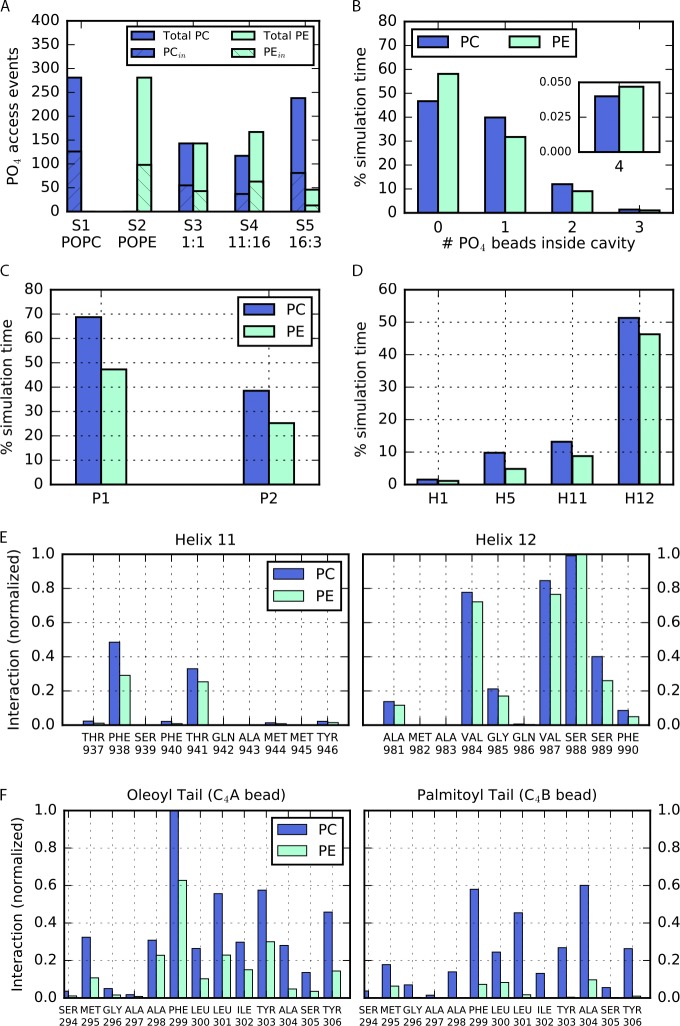
Number of lipid access events, lipid occupancy, and lipid-binding sites in the cavity and portals of P-gp. (A) Number of lipid access events. For each system (Table 1, S1–S5), the total number of lipid molecules PC (blue) and PE (cyan) in the lower leaflet is shown with the number of access events observed for each lipid type. (B) Lipid occupancy in the cavity. The number of PC or PE molecules occupying the cavity during the total simulation time is shown. Up to four lipid molecules can access the cavity simultaneously. (C) Lipid binding at the portals. Amount of simulation time for which binding events are detected at P1 and P2 for both PC and PE lipids. (D) Lipid binding in the cavity. Amount of simulation time for which binding events are detected for transmembrane helices H1, H5, H11, and H12. The interactions were detected along H1, H5, H11, and H12. Binding events at H3, H7, and H10 were not observed in our simulations. (E and F) Binding sites of lipid headgroups and tails in the cavity of P-gp normalized over the maximum number of interactions. (E) Mainly residues along H11 and H12 registered interactions with PO4 beads. (F) Binding sites for the oleoyl and palmitoyl tail were found mainly at H5.
Lipid occupancy inside the P-gp cavity
Using the contact definition described in Materials and methods, no PO4 beads of PC or PE lipids were found in the cavity for ∼45% and ∼60% of the simulation time, respectively (Fig. 2 B). Instead, water molecules were found inside. Overall, a single PC lipid remains in the cavity for ∼40% of the time, whereas a single PE remains during ∼30% of the simulation time. Transiently, up to a maximum of four lipid molecules can occupy the cavity simultaneously, although for short periods of time (<0.05%), as shown in Fig. 2 B.
Lipid–protein interactions at the portals
To identify the binding sites for POPC or POPE lipids, we looked at the residues located at the portals (helices H4 and H6 at P1; H10 and H12 at P2) that, during the simulation time, interacted with the headgroup of each lipid type. Every time a residue satisfied the contact condition, one binding event was counted (see Materials and methods). As shown in Fig. 2 C, overall, P1 registered more binding events than P2 for both lipid types.
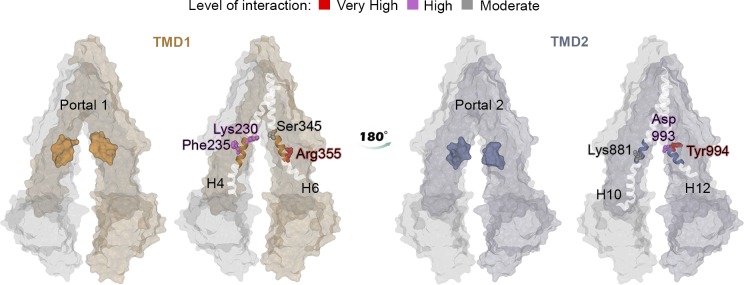
We then calculated the number of contacts between the PO4 beads to identify key binding residues at the portals involved in the lipid transit. Despite the chemical difference between POPC and POPE headgroups, the predominant binding sites found at the portals were the same for both lipid types: Arg355 located at H6 and Tyr994 at H12 (Fig. 3). Additional interactions were registered with the residues Lys230 and Lys235 at H4, Ser345 at H6, and Lys881 and Asp993 at H12.
Figure 3.
Binding residues for PC and PE lipids at the portals of P-gp. Binding residues are highlighted according to the color map. The maximum contact time registered was normalized to 1.0. A color map distinguishes three levels of interaction: very high interactions correspond to between 0.8 and 1 (red); high interactions correspond to between 0.4 and 0.7 (magenta); and moderate interactions correspond to <0.4 (gray). Lys230, Phe235, Ser345, and Arg355 at P1 and Asp993 and Tyr994 at P2 registered the top highest 10% of interactions.
Lipid–protein interactions inside the cavity of P-gp
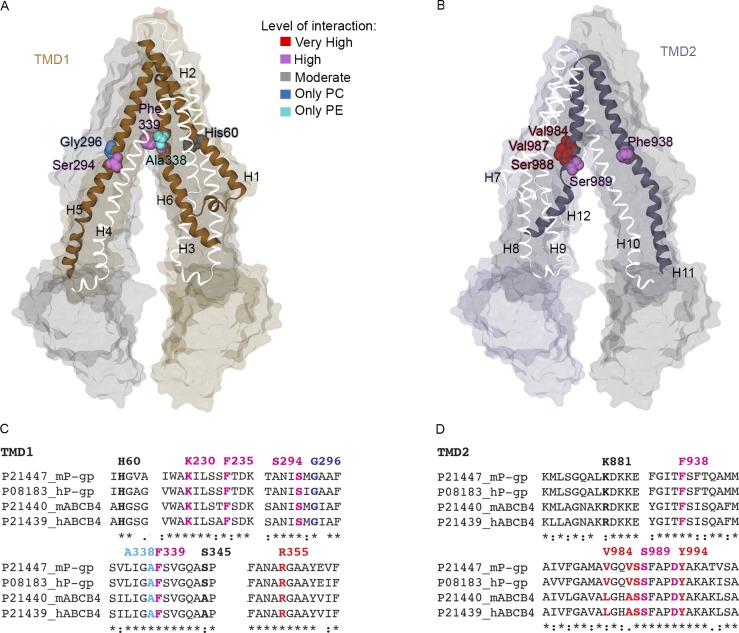
Similar to the previous analysis for the residues at the portals, we identified residues inside the cavity of P-gp involved in interactions with lipid molecules. PC and PE binding events mostly involved residues of H12 (Fig. 2 D). PC registered over ∼50% of the simulation time. PE lipid binding was observed over ∼45% of the simulation time. Binding residues of H1, H5, and H11 registered interactions during <15% of the time, whereas residues at H3, H7, and H10 did not show binding events over the course of the simulation. Inside the cavity, the lipid–protein interactions mainly involved the residues Val984, Val987, and Ser988 at H12 and residues Phe938 and Thr941 located along H11 (Fig. 2 E and Fig. 4 B), all of them in the upper half of the P-gp cavity. Additional binding events were found for residues His60 of H1, Ser294 of H5, and Phe339 of H6 in TMD1 (Fig. 4 A). We observed a very high similarity between the PC and PE binding residues. Only two binding residues showed selectivity for lipid types: Ala338 for PC and Gly266 for PE, both with moderate interaction. The set of identified binding residues is conserved in mouse and human P-gp and ABCB4, as shown in the sequence alignment in Fig. 4 (C and D). In addition, a hot spot for lipid binding was observed at Arg828 on H9. However, given the orientation of the residue pointing toward the external side of the protein, the observed lipid–protein interaction does not occur in the cavity. This result will be addressed in the Discussion.
Figure 4.
Binding residues in the cavity of P-gp. (A and B) Residues Ser294 and Phe339 of TMD1 (A) and Val984, Val897, Ser988, and Ser989 of TMD2 (B) are highlighted in the atomistic representation of P-gp. Gly296 and Ala338 registered interactions only with headgroups of PC (blue) and PE (cyan), respectively. (C and D) Sequence alignment of TMD1 (C) and TMD2 (D) for human (h) and mouse (m) P-gp and ABCB4 highlighting the significant interacting residues, colored according to the same color scheme as in A and B and Fig. 3.
Dynamics of the lipid molecules in the P-gp cavity
To illustrate the dynamics of the PO4 bead and lipid tails inside the cavity of P-gp, we calculated the mean z coordinate over windows of 1 µs and plotted over the course of the simulation time (Fig. 5). In all our simulations, we observed that the PO4 bead remained mainly in the lower region of the membrane (Fig. 5, A and C) for both lipid types PC and PE. The tails of the lipids, however, occupy distinct regions and extend to the upper region of the cavity, where oscillatory and flipping events were detected (Fig. 5, B and D). We also identified the lipid tail–binding sites in the uppermost region of the cavity, as established by the contact condition (see Materials and methods). For the tails, we observed binding events mostly at H5 and, to a lesser extent, at H7. No binding residues were found on H1, H3, H11, or H12. Residues Phe299 and Leu301 showed very high interaction with the tails for both lipid types. Frequent interactions were also identified between the oleoyl tail and Tyr303 and Tyr306 and between the palmitoyl tail and Phe299 and Ala304 (Fig. 2 F).
Figure 5.
Dynamics of headgroups and tails inside the cavity of P-gp. (A–D) The mean z coordinate profile for the headgroup beads (PO4) and tail beads (C4A and C4B) plotted over 20 µs of simulation for a lipid that accesses the cavity two times (A and B) and a lipid that remains in the cavity (C and D). In A and C, P-gp is shown in the gray surface, and the position of the selected PO4 bead is shown as a colored sphere, from red to white to blue, as a function of time following the color map in Fig. 1. The z profiles of the PO4 bead (brown line) and the C4A and C4B beads (gray and black lines) were calculated over windows of 1 µs.
The lipid occupancy maps highlight the ordering of the lipids in the immediate proximity of P-gp (see Materials and methods). We observed high occupancy values inside the cavity, independent of the lipid composition (Fig. 6). The presence of the lipids spanned the entire available volume due to the lipid tails reaching the upper region, reinforcing the results showed in Fig. 5. The lipid organization inside the cavity did not significantly differ between the PC/PE ratios considered in this work (Fig. 6, A and B). In addition, hot spots of lipid occupancy were also detected at the portals, where lipids compete for access to the cavity.
Figure 6.
Lipid occupancy maps for the POPC/POPE (1:1) and plasma ratio PC/PE systems. (A) POPC/POPE. (B) PC/PE. (Top) Lateral view of the bilayer. The protein is omitted for clarity, and the occupancy maps are clipped to highlight the occupancy signal in the inner region of the cavity. (Bottom) Top view from the extracellular side. From the last frame of the corresponding simulation system, residues with a high degree of interaction are shown as magenta (Phe339 and Phe938) and red spheres (Val987, Ser988, Val984, Tyr994, and Arg355), following the same color scheme from Figs. 3 and 4. PE and PC lipids are shown in cyan and blue lines, respectively.
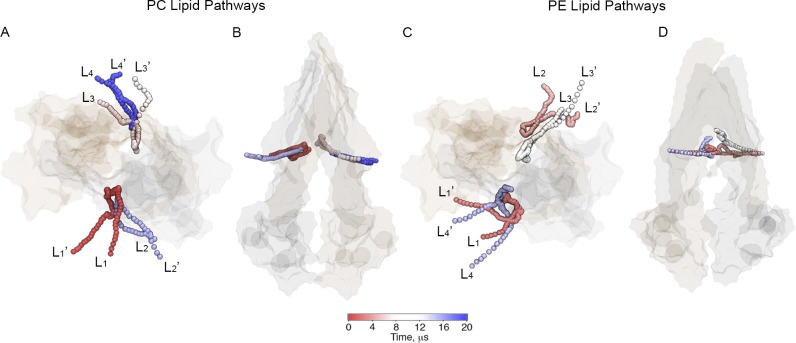
Lipid pathways
Our simulations enabled us to identify trajectories of lipids that access the cavity of P-gp. Examples of trajectories of PC and PE lipids along 20 µs of simulation time are illustrated in Fig. 7. Simultaneous lipid uptake through the same portal can take place, as shown in Fig. 7 (A and C), during the last 8 µs of simulation (light blue color) in S4 (Table 1). No unique lipid pathway was detected because the approach to the portals is different for different lipids. The residence time of the lipids inside the cavity is different for every lipid.
Figure 7.
Lipid pathways found for PC and PE lipid types in plasma membrane ratio PC/PE. Different lipid headgroups (PO4 beads) access the cavity of P-gp through both portals as a function of time during the 20-µs simulation. (A and B) Two different views of PC lipid pathways. (C and D) Two different views of PE lipid pathways. P-gp is shown in a transparent surface representation. Each lipid shown is labeled in the order in which it accesses the cavity along the simulation by subindex i = [1,…,4]. Before accessing the cavity, the lipids are labeled as Li; after leaving the cavity, the lipids are labeled as Li’.
Discussion
P-gp is one of the best characterized ABC transporters (Ambudkar et al., 2003; Picchianti-Diamanti et al., 2014). Its broad substrate specificity allows for the transport of many chemically diverse molecules (Sharom, 2014; Chufan et al., 2015; Subramanian et al., 2016), and to date, several studies have suggested poly-specific drug-binding sites in the cavity of P-gp (Loo and Clarke, 1994; Borst et al., 2000; Chufan et al., 2015; Neumann et al., 2017). MD simulations have been extensively used to provide molecular details on the interactions between substrates and specific regions of the protein (Borst et al., 2000), most of them assuming specific binding sites. For example, using umbrella sampling methods, Subramanian et al. (2015) investigated spontaneous binding for morphine and nicardipine. More recently, Syed et al. (2017) used the drug-binding site in the transmembrane region from docking studies to test interactions with P-gp.
For P-gp, it has been proposed that substrates access the cavity through the membrane (Eckford and Sharom, 2006; Subramanian et al., 2015), so lipid access pathways may also be relevant for elucidating drug transit and binding. The characterization of lipid-binding sites and lipid pathways in P-gp is relevant for a better understanding of drug binding to P-gp and other ABC transporters that are capable of translocating lipids (Tarling et al., 2013; Neumann et al., 2017). In addition, direct interaction between P-gp and lipid-based molecules such as miltefosine and edelfosine, which are lipid-based anticancer drug molecules, has been previously described (Eckford and Sharom, 2006; Syed et al., 2017). Toward this aim, we performed equilibrium CG MD simulations to explore how lipid molecules access the cavity of P-gp in the inward-facing conformation. With no previous assumption of specific binding sites, we identify key residues involved in the lipid binding at the portals and lipid occupancy in the cavity.
Lipid access to the cavity of P-gp
The structure of P-gp in the inward-facing conformation has two portals (P1 and P2) that provide access to the main cavity to molecules located in the lower leaflet of the membrane (Aller et al., 2009; Li et al., 2014). In all the analyzed simulations, we observed lipid diffusion toward the cavity of P-gp through the portals, but only for lipids of the inner leaflet, as previously suggested by Aller et al. (2009). No additional pathways for entrance to the cavity were detected. When comparing the two portals, a higher degree of lipid access was observed through P1 (Fig. 2 C). We link this result to the presence of the elbow helix of TMD2, which may screen the interactions at P2 between the lipid molecules and the residues located along H12, because it is partially covering H10. This feature also allows the direct exposure of Arg828 of H9, which faces the opening of P2, for lipid binding, although H9 is not included in the standard definition of portals followed in the literature (Aller et al., 2009; Li et al., 2014; Zhang et al., 2014).
A limitation of the currently available structures of P-gp (Jin et al., 2012; Li et al., 2014; Szewczyk et al., 2015; Nicklisch et al., 2016; Esser et al., 2017) is the missing linker that connects NBD1 with TMD2. The structure of the linker has been reported for the human cystic fibrosis transmembrane conductance regulator (Liu et al., 2017), another member of the ABC transporter superfamily that acts as an anion channel where channel opening can be hampered by the presence of the linker (Liu et al., 2017). In P-gp, this flexible region has been proposed to be important for substrate specificity; additionally, it affects the conformational changes driven by ATP binding and hydrolysis at the NBD (Hrycyna et al., 1998; Sato et al., 2009; Ferreira et al., 2012; Esser et al., 2017). Although the presence of this linker may modify the substrate diffusion toward the cavity of P-gp (Ferreira et al., 2015) because of its location in the cytosolic side, oriented toward the opening between the TMD1 and TMD2, we believe it is unlikely that the linker will affect the lipid-binding sites reported in this study.
Key lipid-binding residues in P-gp
At the portals
Our findings suggest that PC and PE lipids access the internal drug-binding pocket through the portals P1 and P2, which has also been reported for drug molecules in several studies (Loo and Clarke, 1994; Eckford and Sharom, 2006; Aller et al., 2009; Subramanian et al., 2015). It has also been suggested that a positively charged ring of Arg and Lys residues could drive substrates toward the cavity and promote lipid–protein interactions in ABC transporters in general via electrostatic interactions (Yu et al., 2015; Mehmood et al., 2016). From our simulations, two binding hot spots at P1 were identified: Lys230 of H4 and Arg355 of H6 (Fig. 3). This result is in perfect agreement with the experimental report from Marcoux et al. (2013), in which electrostatic interactions between the phosphate groups of lipid molecules and Lys230 were identified by mass spectrometry techniques. Lipid access events were also observed through P2, with key binding residues identified as Arg828 of H9, Lys881 of H10, and Asp993 and Tyr994 of H12 (Fig. 3). As mentioned above, Arg828 is not part of the canonical definition of portals (Li et al., 2014). However, because it is located at the bottom of H9 and its orientation is toward the exterior of the protein, facing H12, Arg828 is involved in the interactions for lipid access to the cavity through P2. Overall, the set of positively charged residues at the portals are key elements for lipid access because of the high affinity of Lys and Arg for phosphate groups via electrostatic interactions, promoting lipid entry into the cavity.
In the cavity
The funnel-shaped cavity of P-gp is lined by helices H1, H3, and H5 of TMD1 and H7, H11, and H12 of TMD2. Hydrophobic and aromatic residues in the inner cavity are important for substrate binding (Thiebaut et al., 1987; Aller et al., 2009; Martinez et al., 2014; Sharom, 2014; Mittra et al., 2017). Because of the amphipathic nature of lipids, we investigated the interactions between protein and lipid headgroups (PO4 beads) and lipid tails, respectively. The PO4 beads, once inside the cavity, established interactions mainly with residues of H11 and H12 (Fig. 2 D and Fig. 4 B). This result may reflect the structural architecture of the cavity of P-gp. The helical structure of H12 (residues 970–990) is interrupted by a second α helix (residues 991–994); therefore, the upper segment of H12 also belongs to the cavity. From our simulations, we observed interactions between the lipid headgroup and Phe938 on H11 (Fig. 2 E and Fig. 4 B), as reported by Aller et al. (2009) for the inhibitor verapamil. Additional interactions were also found for Ser989, Phe990, Pro992, Val984, Val987, and Ser988 on H12 and Thr941, Lys929, Lys 930, Met928, and Arg925 on H11 (Fig. 4). In the cavity of P-gp, our findings show the key role of Phe339 as a lipid-binding residue. This residue has also been shown to be involved in binding of the cyclic hexapeptide inhibitors cyclic-tris-(R)-valineselenazole (QZ59-RRR) and cyclic-tris-(S)-valineselenazole (QZ59-SSS; Li et al., 2014), while Zhang et al. (2015) corroborated the importance of Phe339 for doxorubicin access. If H12 drives many of the interactions between lipid headgroups and residues in the cavity, H5 appears to be the key player for the interactions between residues in the cavity of P-gp and the lipid tails (Fig. 2, E and F). Residues of H5 such as Leu300, Ile302, and Tyr303 were also reported as being binding sites for QZ59-RRR, QZ59-SSS, and verapamil (Aller et al., 2009). Overall, in the case of lipids, the tails extend toward the upper and more hydrophobic region of the cavity not explored by the PO4 beads (Fig. 5, B and D), with residues such as Phe299, Ile302, Tyr303, and Tyr306 mostly involved in the interactions with the oleoyl tail (Fig. 2 F). The location of substrates in the top of the cavity has been found in several experimental studies (Aller et al., 2009; Dolghih et al., 2011; Marcoux et al., 2013), in agreement with our simulations. Combined, these results reinforce the crucial role of H11, H12, and H5 in the interactions between protein and lipids as substrates.
Many ABC transporters are involved in lipid transport (Borst et al., 2000; Eckford and Sharom, 2009; Fletcher et al., 2010; Quazi and Molday, 2011; Neumann et al., 2017). In particular, several studies have revealed lipid transport capabilities not only for P-gp (Sharom, 1997, 2014; Eckford and Sharom, 2009), but also for other members of the ABCB subfamily, such as ABCB4, involved in the translocation of PC lipids into the bile (Linton, 2015; Zhao et al., 2015). The interplay between lipids and ABC transporters is rather complex, being dependent, for example, as shown for ABCB4, on specific lipid compositions and on the presence of other lipid transporters (Zhao et al., 2015). Although lipid transport is mainly assessed using modified fluorescent lipids for P-gp and ABCB4, mass spectrometry studies have provided further details on the binding of different lipids to P-gp and their presence inside its cavity together with drugs (Marcoux et al., 2013). In this context, our study describes the molecular details of lipids’ access to the main cavity of the transporter. Although we focused our simulations on mouse P-gp, our findings can be translated to related ABC transporters, as the residues we identified as important for lipid–protein interactions are conserved in human P-gp as well as in human and mouse ABCB4 (Fig. 4, C and D).
Conclusions
In this work, we identify lipid pathways for P-gp in the inward-facing conformation in bilayers with different PC/PE lipid ratios. By performing equilibrium MD simulations on a timescale of microseconds, we show that the lipid binding events in the cavity and at the portals of P-gp are accessible and measurable. This approach reveals that lipid access events involve lipids from the lower leaflet only and identifies key lipid-binding residues at the portals and inside the cavity of P-gp. We find no selectivity for PC versus PE lipid entry or lipid binding in P-gp. Future directions include lipid interactions with the outward-facing conformation, for which there is currently no experimental structure, interactions with less prevalent lipids, and ultimately, a molecular understanding of the full transport process of physiological substrates in a realistic lipid environment.
Acknowledgments
This work was supported by the Canadian Institutes of Health Research. Additional support came from Alberta Innovates - Health Solutions (AIHS) and Alberta Innovates - Technology Futures (AITF). R.-X. Gu is supported by fellowships from AIHS and the Canadian Institutes for Health Research (funding reference number MFE-140949). D.P. Tieleman is an AIHS Scientist and AITF Strategic Chair in (Bio)Molecular Simulation. Simulations were run on Compute Canada machines, supported by the Canada Foundation for Innovation and their partners. This work was undertaken, in part, thanks to funding from the Canada Research Chairs program.
The authors declare no competing financial interests.
Author contributions: E. Barreto-Ojeda carried out simulations and analyses and wrote an initial draft of the paper with contributions from V. Corradi and D.P. Tieleman. V. Corradi carried out analyses. V. Corradi and D.P. Tieleman conceived the study. All authors contributed to the interpretation of the results and the writing of the final manuscript.
José D. Faraldo-Gómez served as editor.
References
- Aller, S.G., Yu J., Ward A., Weng Y., Chittaboina S., Zhuo R., Harrell P.M., Trinh Y.T., Zhang Q., Urbatsch I.L., and Chang G.. 2009. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 323:1718–1722. 10.1126/science.1168750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambudkar, S.V., Kimchi-Sarfaty C., Sauna Z.E., and Gottesman M.M.. 2003. P-glycoprotein: from genomics to mechanism. Oncogene. 22:7468–7485. 10.1038/sj.onc.1206948 [DOI] [PubMed] [Google Scholar]
- Berendsen, H.J.C., Postma J.P.M., van Gunsteren W.F., DiNola A., and Haak J.R.. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690. 10.1063/1.448118 [DOI] [Google Scholar]
- Borst, P., Zelcer N., and van Helvoort A.. 2000. ABC transporters in lipid transport. Biochim. Biophys. Acta. 1486:128–144. 10.1016/S1388-1981(00)00053-6 [DOI] [PubMed] [Google Scholar]
- Bosch, I., Dunussi-Joannopoulos K., Wu R.L., Furlong S.T., and Croop J.. 1997. Phosphatidylcholine and phosphatidylethanolamine behave as substrates of the human MDR1 P-glycoprotein. Biochemistry. 36:5685–5694. 10.1021/bi962728r [DOI] [PubMed] [Google Scholar]
- Bussi, G., Donadio D., and Parrinello M.. 2007. Canonical sampling through velocity rescaling. J. Chem. Phys. 126:014101. 10.1063/1.2408420 [DOI] [PubMed] [Google Scholar]
- Chavent, M., Duncan A.L., and Sansom M.S.. 2016. Molecular dynamics simulations of membrane proteins and their interactions: from nanoscale to mesoscale. Curr. Opin. Struct. Biol. 40:8–16. 10.1016/j.sbi.2016.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.J., Chin J.E., Ueda K., Clark D.P., Pastan I., Gottesman M.M., and Roninson I.B.. 1986. Internal duplication and homology with bacterial transport proteins in the mdr1 (P-glycoprotein) gene from multidrug-resistant human cells. Cell. 47:381–389. 10.1016/0092-8674(86)90595-7 [DOI] [PubMed] [Google Scholar]
- Chen, Z., Shi T., Zhang L., Zhu P., Deng M., Huang C., Hu T., Jiang L., and Li J.. 2016. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family in multidrug resistance: A review of the past decade. Cancer Lett. 370:153–164. 10.1016/j.canlet.2015.10.010 [DOI] [PubMed] [Google Scholar]
- Chufan, E.E., Sim H.-M., and Ambudkar S.V.. 2015. Molecular basis of the polyspecificity of P-glycoprotein (ABCB1): recent biochemical and structural studies. Adv. Cancer Res. 125:71–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong, D.H., Singh G., Bennett W.F.D., Arnarez C., Wassenaar T.A., Schäfer L.V., Periole X., Tieleman D.P., and Marrink S.J.. 2013. Improved Parameters for the Martini Coarse-Grained Protein Force Field. J. Chem. Theory Comput. 9:687–697. 10.1021/ct300646g [DOI] [PubMed] [Google Scholar]
- Dolghih, E., Bryant C., Renslo A.R., and Jacobson M.P.. 2011. Predicting binding to p-glycoprotein by flexible receptor docking. PLOS Comput. Biol. 7:e1002083. 10.1371/journal.pcbi.1002083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckford, P.D.W., and Sharom F.J.. 2006. P-glycoprotein (ABCB1) interacts directly with lipid-based anti-cancer drugs and platelet-activating factors. Biochem. Cell Biol. 84:1022–1033. 10.1139/o06-196 [DOI] [PubMed] [Google Scholar]
- Eckford, P.D.W., and Sharom F.J.. 2009. ABC efflux pump-based resistance to chemotherapy drugs. Chem. Rev. 109:2989–3011. 10.1021/cr9000226 [DOI] [PubMed] [Google Scholar]
- Esser, L., Zhou F., Pluchino K.M., Shiloach J., Ma J., Tang W.K., Gutierrez C., Zhang A., Shukla S., Madigan J.P., et al. 2017. Structures of the Multidrug Transporter P-glycoprotein Reveal Asymmetric ATP Binding and the Mechanism of Polyspecificity. J. Biol. Chem. 292:446–461. 10.1074/jbc.M116.755884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, R.J., Ferreira M.-J.U., and Dos Santos D.J.V.A.. 2012. Insights on P-glycoprotein’s efflux mechanism obtained by molecular dynamics simulations. J. Chem. Theory Comput. 8:1853–1864. 10.1021/ct300083m [DOI] [PubMed] [Google Scholar]
- Ferreira, R.J., Ferreira M.-J.U., and Dos Santos D.J.V.A.. 2015. Do drugs have access to the P-glycoprotein drug-binding pocket through gates? J. Chem. Theory Comput. 11:4525–4529. 10.1021/acs.jctc.5b00652 [DOI] [PubMed] [Google Scholar]
- Fletcher, J.I., Haber M., Henderson M.J., and Norris M.D.. 2010. ABC transporters in cancer: more than just drug efflux pumps. Nat. Rev. Cancer. 10:147–156. 10.1038/nrc2789 [DOI] [PubMed] [Google Scholar]
- Gottesman, M.M. 1993. How cancer cells evade chemotherapy: sixteenth Richard and Hinda Rosenthal Foundation Award Lecture. Cancer Res. 53:747–754. [PubMed] [Google Scholar]
- Gros, P., and Buschman E.. 1993. The mouse multidrug resistance gene family: structural and functional analysis. Int. Rev. Cytol. 137C:169–197. [PubMed] [Google Scholar]
- Hess, B., Kutzner C., van der Spoel D., and Lindahl E.. 2008. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 4:435–447. 10.1021/ct700301q [DOI] [PubMed] [Google Scholar]
- Higgins, C.F., and Gottesman M.M.. 1992. Is the multidrug transporter a flippase? Trends Biochem. Sci. 17:18–21. 10.1016/0968-0004(92)90419-A [DOI] [PubMed] [Google Scholar]
- Hrycyna, C.A., Airan L.E., Germann U.A., Ambudkar S.V., Pastan I., and Gottesman M.M.. 1998. Structural flexibility of the linker region of human P-glycoprotein permits ATP hydrolysis and drug transport. Biochemistry. 37:13660–13673. 10.1021/bi9808823 [DOI] [PubMed] [Google Scholar]
- Humphrey, W., Dalke A., and Schulten K.. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38: 27–28. [DOI] [PubMed] [Google Scholar]
- Ingólfsson, H.I., Lopez C.A., Uusitalo J.J., de Jong D.H., Gopal S.M., Periole X., and Marrink S.J.. 2014a. The power of coarse graining in biomolecular simulations. Wiley Interdiscip. Rev. Comput. Mol. Sci. 4:225–248. 10.1002/wcms.1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingólfsson, H.I., Melo M.N., van Eerden F.J., Arnarez C., Lopez C.A., Wassenaar T.A., Periole X., de Vries A.H., Tieleman D.P., and Marrink S.J.. 2014b. Lipid organization of the plasma membrane. J. Am. Chem. Soc. 136:14554–14559. 10.1021/ja507832e [DOI] [PubMed] [Google Scholar]
- Jin, M.S., Oldham M.L., Zhang Q., and Chen J.. 2012. Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans. Nature. 490:566–569. 10.1038/nature11448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karo, J., Peterson P., and Vendelin M.. 2012. Molecular dynamics simulations of creatine kinase and adenine nucleotide translocase in mitochondrial membrane patch. J. Biol. Chem. 287:7467–7476. 10.1074/jbc.M111.332320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K., and Standley D.M.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30:772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K., Misawa K., Kuma K., and Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong, M.K., Chen H.-B., and Shih Y.-H.. 2012. Prediction of promiscuous p-glycoprotein inhibition using a novel machine learning scheme. PLoS One. 7:e33829. 10.1371/journal.pone.0033829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Jaimes K.F., and Aller S.G.. 2014. Refined structures of mouse P-glycoprotein. Protein Sci. 23:34–46. 10.1002/pro.2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton, K.J. 2015. Lipid flopping in the liver. Biochem. Soc. Trans. 43:1003–1010. 10.1042/BST20150132 [DOI] [PubMed] [Google Scholar]
- Liu, F., Zhang Z., Csanády L., Gadsby D.C., and Chen J.. 2017. Molecular Structure of the Human CFTR Ion Channel. Cell. 169:85–95.e8. 10.1016/j.cell.2017.02.024 [DOI] [PubMed] [Google Scholar]
- Loo, T.W., and Clarke D.M.. 1994. Reconstitution of drug-stimulated ATPase activity following co-expression of each half of human P-glycoprotein as separate polypeptides. J. Biol. Chem. 269:7750–7755. [PubMed] [Google Scholar]
- Loo, T.W., and Clarke D.M.. 1995. Covalent modification of human P-glycoprotein mutants containing a single cysteine in either nucleotide-binding fold abolishes drug-stimulated ATPase activity. J. Biol. Chem. 270:22957–22961. 10.1074/jbc.270.39.22957 [DOI] [PubMed] [Google Scholar]
- Marcoux, J., Wang S.C., Politis A., Reading E., Ma J., Biggin P.C., Zhou M., Tao H., Zhang Q., Chang G., et al. 2013. Mass spectrometry reveals synergistic effects of nucleotides, lipids, and drugs binding to a multidrug resistance efflux pump. Proc. Natl. Acad. Sci. USA. 110:9704–9709. 10.1073/pnas.1303888110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrink, S.J., and Tieleman D.P.. 2013. Perspective on the Martini model. Chem. Soc. Rev. 42:6801–6822. 10.1039/c3cs60093a [DOI] [PubMed] [Google Scholar]
- Marrink, S.J., de Vries A.H., and Mark A.E.. 2004. Coarse Grained Model for Semiquantitative Lipid Simulations. J. Phys. Chem. B. 108:750–760. 10.1021/jp036508g [DOI] [Google Scholar]
- Marrink, S.J., Risselada H.J., Yefimov S., Tieleman D.P., and de Vries A.H.. 2007. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 111:7812–7824. 10.1021/jp071097f [DOI] [PubMed] [Google Scholar]
- Martinez, L., Arnaud O., Henin E., Tao H., Chaptal V., Doshi R., Andrieu T., Dussurgey S., Tod M., Di Pietro A., et al. 2014. Understanding polyspecificity within the substrate-binding cavity of the human multidrug resistance P-glycoprotein. FEBS J. 281:673–682. 10.1111/febs.12613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGibbon, R.T., Beauchamp K.A., Harrigan M.P., Klein C., Swails J.M., Hernández C.X., Schwantes C.R., Wang L.-P., Lane T.J., and Pande V.S.. 2015. MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophys. J. 109:1528–1532. 10.1016/j.bpj.2015.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehmood, S., Corradi V., Choudhury H.G., Hussain R., Becker P., Axford D., Zirah S., Rebuffat S., Tieleman D.P., Robinson C.V., and Beis K.. 2016. Structural and Functional Basis for Lipid Synergy on the Activity of the Antibacterial Peptide ABC Transporter McjD. J. Biol. Chem. 291:21656–21668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittra, R., Pavy M., Subramanian N., George A.M., O’Mara M.L., Kerr I.D., and Callaghan R.. 2017. Location of contact residues in pharmacologically distinct drug binding sites on P-glycoprotein. Biochem. Pharmacol. 123:19–28. 10.1016/j.bcp.2016.10.002 [DOI] [PubMed] [Google Scholar]
- Monticelli, L., Kandasamy S.K., Periole X., Larson R.G., Tieleman D.P., and Marrink S.-J.. 2008. The MARTINI Coarse-Grained Force Field: Extension to Proteins. J. Chem. Theory Comput. 4:819–834. 10.1021/ct700324x [DOI] [PubMed] [Google Scholar]
- Neumann, J., Rose-Sperling D., and Hellmich U.A.. 2017. Diverse relations between ABC transporters and lipids: An overview. Biochim. Biophys. Acta - Biomembr. 1859:605–618. [DOI] [PubMed] [Google Scholar]
- Nicklisch, S.C.T., Rees S.D., McGrath A.P., Gökirmak T., Bonito L.T., Vermeer L.M., Cregger C., Loewen G., Sandin S., Chang G., and Hamdoun A.. 2016. Global marine pollutants inhibit P-glycoprotein: Environmental levels, inhibitory effects, and cocrystal structure. Sci. Adv. 2:e1600001. 10.1126/sciadv.1600001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periole, X., Cavalli M., Marrink S.-J., and Ceruso M.A.. 2009. Combining an Elastic Network With a Coarse-Grained Molecular Force Field: Structure, Dynamics, and Intermolecular Recognition. J. Chem. Theory Comput. 5:2531–2543. 10.1021/ct9002114 [DOI] [PubMed] [Google Scholar]
- Picchianti-Diamanti, A., Rosado M.M., Scarsella M., Laganà B., and D’Amelio R.. 2014. P-glycoprotein and drug resistance in systemic autoimmune diseases. Int. J. Mol. Sci. 15:4965–4976. 10.3390/ijms15034965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quazi, F., and Molday R.S.. 2011. Lipid transport by mammalian ABC proteins. Essays Biochem. 50:265–290. 10.1042/bse0500265 [DOI] [PubMed] [Google Scholar]
- Sato, T., Kodan A., Kimura Y., Ueda K., Nakatsu T., and Kato H.. 2009. Functional role of the linker region in purified human P-glycoprotein. FEBS J. 276:3504–3516. 10.1111/j.1742-4658.2009.07072.x [DOI] [PubMed] [Google Scholar]
- Schmidt, M.R., Stansfeld P.J., Tucker S.J., and Sansom M.S.P.. 2013. Simulation-based prediction of phosphatidylinositol 4,5-bisphosphate binding to an ion channel. Biochemistry. 52:279–281. 10.1021/bi301350s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta, D., and Chattopadhyay A.. 2012. Identification of cholesterol binding sites in the serotonin1A receptor. J. Phys. Chem. B. 116:12991–12996. 10.1021/jp309888u [DOI] [PubMed] [Google Scholar]
- Sharom, F.J. 1997. The P-glycoprotein efflux pump: how does it transport drugs? J. Membr. Biol. 160:161–175. 10.1007/s002329900305 [DOI] [PubMed] [Google Scholar]
- Sharom, F.J. 2011. The P-glycoprotein multidrug transporter. Essays Biochem. 50:161–178. 10.1042/bse0500161 [DOI] [PubMed] [Google Scholar]
- Sharom, F.J. 2014. Complex Interplay between the P-Glycoprotein Multidrug Efflux Pump and the Membrane: Its Role in Modulating Protein Function. Front. Oncol. 4:41. 10.3389/fonc.2014.00041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, N., Condic-Jurkic K., Mark A.E., and O’Mara M.L.. 2015. Identification of Possible Binding Sites for Morphine and Nicardipine on the Multidrug Transporter P-Glycoprotein Using Umbrella Sampling Techniques. J. Chem. Inf. Model. 55:1202–1217. 10.1021/ci5007382 [DOI] [PubMed] [Google Scholar]
- Subramanian, N., Condic-Jurkic K., and O’Mara M.L.. 2016. Structural and dynamic perspectives on the promiscuous transport activity of P-glycoprotein. Neurochem. Int. 98:146–152. 10.1016/j.neuint.2016.05.005 [DOI] [PubMed] [Google Scholar]
- Syed, S.B., Arya H., Fu I.-H., Yeh T.-K., Periyasamy L., Hsieh H.-P., and Coumar M.S.. 2017. Targeting P-glycoprotein: Investigation of piperine analogs for overcoming drug resistance in cancer. Sci. Rep. 7:7972. 10.1038/s41598-017-08062-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakács, G., Paterson J.K., Ludwig J.A., Booth-Genthe C., and Gottesman M.M.. 2006. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 5:219–234. 10.1038/nrd1984 [DOI] [PubMed] [Google Scholar]
- Szewczyk, P., Tao H., McGrath A.P., Villaluz M., Rees S.D., Lee S.C., Doshi R., Urbatsch I.L., Zhang Q., and Chang G.. 2015. Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein. Acta Crystallogr. D Biol. Crystallogr. 71:732–741. 10.1107/S1399004715000978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarling, E.J., de Aguiar Vallim T.Q., and Edwards P.A.. 2013. Role of ABC transporters in lipid transport and human disease. Trends Endocrinol. Metab. 24:342–350. 10.1016/j.tem.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium . 2017. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45(D1):D158–D169. 10.1093/nar/gkw1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebaut, F., Tsuruo T., Hamada H., Gottesman M.M., Pastan I., and Willingham M.C.. 1987. Cellular localization of the multidrug-resistance gene product P-glycoprotein in normal human tissues. Proc. Natl. Acad. Sci. USA. 84:7735–7738. 10.1073/pnas.84.21.7735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Walt, S., Colbert S.C., and Varoquaux G.. 2011. The NumPy Array: A Structure for Efficient Numerical Computation. Comput. Sci. Eng. 13:22–30. 10.1109/MCSE.2011.37 [DOI] [Google Scholar]
- van Helvoort, A., Smith A.J., Sprong H., Fritzsche I., Schinkel A.H., Borst P., and van Meer G.. 1996. MDR1 P-glycoprotein is a lipid translocase of broad specificity, while MDR3 P-glycoprotein specifically translocates phosphatidylcholine. Cell. 87:507–517. 10.1016/S0092-8674(00)81370-7 [DOI] [PubMed] [Google Scholar]
- Wassenaar, T.A., Ingólfsson H.I., Böckmann R.A., Tieleman D.P., and Marrink S.J.. 2015. Computational Lipidomics with insane: A Versatile Tool for Generating Custom Membranes for Molecular Simulations. J. Chem. Theory Comput. 11:2144–2155. 10.1021/acs.jctc.5b00209 [DOI] [PubMed] [Google Scholar]
- Yu, J., Ge J., Heuveling J., Schneider E., and Yang M.. 2015. Structural basis for substrate specificity of an amino acid ABC transporter. Proc. Natl. Acad. Sci. USA. 112:5243–5248. 10.1073/pnas.1415037112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Sun T., Liang L., Wu T., and Wang Q.. 2014. Drug promiscuity of P-glycoprotein and its mechanism of interaction with paclitaxel and doxorubicin. Soft Matter. 10:438–445. 10.1039/C3SM52499J [DOI] [PubMed] [Google Scholar]
- Zhang, J., Li D., Sun T., Liang L., and Wang Q.. 2015. Interaction of P-glycoprotein with anti-tumor drugs: the site, gate and pathway. Soft Matter. 11:6633–6641. 10.1039/C5SM01028D [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Ishigami M., Nagao K., Hanada K., Kono N., Arai H., Matsuo M., Kioka N., and Ueda K.. 2015. ABCB4 exports phosphatidylcholine in a sphingomyelin-dependent manner. J. Lipid Res. 56:644–652. 10.1194/jlr.M056622 [DOI] [PMC free article] [PubMed] [Google Scholar]