Kara et al. show that ACKR4 regulates early activated B cell differentiation fate. ACKR4 limits the early proliferation of activated B cells by restricting their initial access to the splenic interfollicular zone.
Abstract
Activated B cells can initially differentiate into three functionally distinct fates—early plasmablasts (PBs), germinal center (GC) B cells, or early memory B cells—by mechanisms that remain poorly understood. Here, we identify atypical chemokine receptor 4 (ACKR4), a decoy receptor that binds and degrades CCR7 ligands CCL19/CCL21, as a regulator of early activated B cell differentiation. By restricting initial access to splenic interfollicular zones (IFZs), ACKR4 limits the early proliferation of activated B cells, reducing the numbers available for subsequent differentiation. Consequently, ACKR4 deficiency enhanced early PB and GC B cell responses in a CCL19/CCL21-dependent and B cell–intrinsic manner. Conversely, aberrant localization of ACKR4-deficient activated B cells to the IFZ was associated with their preferential commitment to the early PB linage. Our results reveal a regulatory mechanism of B cell trafficking via an atypical chemokine receptor that shapes activated B cell fate.
Introduction
Differentiation of activated B cells during the initial stages of T cell–dependent antibody responses proceeds simultaneously along pathways leading to early (extrafollicular) plasmablasts (PBs), germinal center (GC) B cells, and GC-independent, early memory B cells. These pathways differ in their spatiotemporal emergence, the longevity of their end products, their affinity for antigens, and their functional capacity (Taylor et al., 2012) and are considered important for establishing robust and diverse antibody responses. Adoption of these fates is controlled in part by B cell–trafficking receptors, which are dynamically regulated after antigen engagement to enable B cell access to antigens, interactions with T cells, and positioning in distinct lymphoid niches that foster the formation of immediate or long-lasting, antigen-specific antibody responses (Pereira et al., 2010). How antigen-activated B cells regulate their response to the several chemoattractants to which they may be simultaneously or sequentially exposed is uncertain. It is, however, potentially crucial as a mechanism in determining stoichiometry in the distribution of B cells along the differentiation pathways that generate the effector B cells of the immune response.
A key event in the initiation of T cell–dependent humoral immune responses is the CCR7-directed migration of antigen-engaged B cells toward, and subsequent EBI2/CXCR5/CCR7-dependent distribution along, the border between the T cell and B cell zones (Reif et al., 2002; Okada et al., 2005; Chan et al., 2009; Gatto et al., 2009, 2011; Pereira et al., 2009; Hannedouche et al., 2011; Kelly et al., 2011). Cognate T and B cell interactions at this interface drive EBI2-mediated relocalization to the interfollicular and outer follicular regions in which activated B cells initially proliferate (Chan et al., 2009; Gatto et al., 2009; Kelly et al., 2011; Kerfoot et al., 2011). Proliferating B cells subsequently trifurcate their differentiation trajectories, adopting a chemoattractant receptor profile that drives their positioning to lymphoid microenvironments that promote their effector function. Early PB differentiation is coupled with the induction of CXCR4 and down-regulation of CXCR5 and CCR7, which repositions these cells in extrafollicular niches and the splenic red pulp (Hargreaves et al., 2001). These PBs are short lived and elicit the first line of antigen-specific antibody defense (Smith et al., 1996). GC-committed B cells down-regulate EBI2 (Gatto et al., 2009; Pereira et al., 2009) but maintain CXCR4 and CXCR5 expression (Allen et al., 2004), drawing them into the follicular dendritic cell–rich follicle center where GCs form. Another subset of B cells ultimately adopts a trafficking receptor profile that allows its continuous recirculation through the blood and secondary lymphoid organ follicles as early memory B cells, which retain their germline-encoded antibody. Whether the spatiotemporal control of B cell chemoattractant responsiveness, which is a crucial component of activated B cell differentiation, is stochastic or is intrinsic to the identified receptors and ligands and whether other receptors are involved remain unknown.
Recent studies have shown that a subfamily of atypical chemokine receptors regulates cellular migration (Nibbs and Graham, 2013). These receptors are uncoupled from the classic chemokine receptor-signal transduction machinery, do not induce cell migration, are mainly expressed outside the hematopoietic compartment, and mediate chemokine removal or redistribution in vivo (Nibbs and Graham, 2013). Atypical chemokine receptor 4 (ACKR4) binds CCR7 ligands CCL19 and CCL21 and the CCR9 ligand CCL25 and, thus, regulates their bioavailability in vivo without initiating cellular migration (Gosling et al., 2000; Comerford et al., 2006, 2010; Heinzel et al., 2007; Bunting et al., 2013; Ulvmar et al., 2014; Lucas et al., 2015; Bryce et al., 2016). However, despite the important role of CCR7 in the development of T cell–dependent antibody responses, the function of ACKR4 in this context is unknown. We now report an important, B cell–intrinsic role for ACKR4 in regulating B cell differentiation during the initial stages of the T cell–dependent humoral immune response.
Results and discussion
Although a previous study (Heinzel et al., 2007) concluded that ACKR4 is expressed exclusively by cells of nonhematopoietic origin in unimmunized mice, we detected ACKR4 transcripts and protein expression by GC B cells (Fig. 1, A and B). To investigate the possible functions for hematopoietic ACKR4 in T cell–dependent humoral immunity, we used bone marrow (BM) chimerism to generate mice in which ACKR4 deficiency was restricted to the hematopoietic compartment (H-Ackr4−/−). We immunized these H-Ackr4−/− mice with sheep red blood cells (SRBCs) and observed an increased frequency of GC B cells at all time points assessed after immunization but most prominently on day 5 relative to hematopoietic WT (H-WT) mice (Fig. 1 C). The number of T follicular helper (TFH) cells, mediators of GC B cell selection and proliferation, and T follicular regulatory (TFR) cells, implicated in regulating the magnitude of the GC reaction, were also increased in immunized H-Ackr4−/− mice relative to controls (Fig. S1 A). The formation of early PBs, despite a lack of detectable ACKR4 expression, in H-Ackr4−/− mice was also enhanced on day 5 of the response (Fig. 1 D). These data reveal a negative regulatory role for ACKR4 expression in the hematopoietic compartment on early PB and GC B cell development. To determine whether this effect was intrinsic to B cell expression of ACKR4, we reconstituted lethally irradiated mice with a 4:1 mixture of BM recovered from B cell–deficient (μMT) mice that lack endogenous B cells (Kitamura et al., 1991) and either WT (B-WT) or Ackr4−/− (B-Ackr4−/−) mice. Immunizing B-Ackr4−/− mice with SRBC reproduced the increased numbers of splenic GC B cells, TFH cells, and early PBs seen in immunized H-Ackr4−/− mice (Fig. 1, E and F; and Fig. S1 B). Collectively, these data reveal a B cell–intrinsic regulatory role for ACKR4 in the early stages of the B cell response to a T cell–dependent antigen.
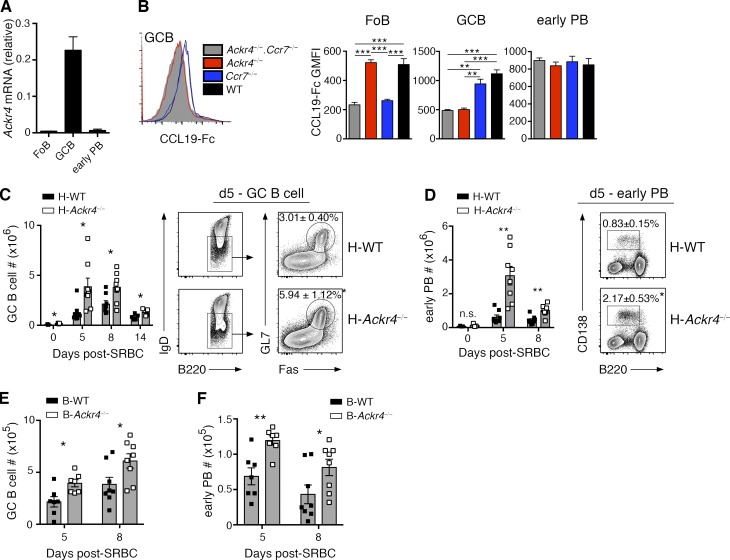
Figure 1.
B cell ACKR4 limits early PB and GC B cell responses. (A) Quantitative PCR analysis of Ackr4 transcript abundance in sorted Fo B cells (B220+IgD+Fas−GL7−), GC B cells (B220+IgD−Fas+GL7+), and early PB (B220lo/-CD138+) relative to the housekeeping gene Rplp0 (means ± SD). (B) Representative histogram of CCL19–Fc staining on GC B cells from Ackr4−/−.Ccr7−/− (cannot bind CCL19–Fc), Ackr4−/− (CCR7 staining), Ccr7−/− (ACKR4 staining), and WT (ACKR4/CCR7 staining) mice. Graphs plot geometric mean fluorescence intensity (GMFI) of CCL19-Fc staining on Fo B cells, GC B cells, and early PBs, gated as in A (n = 5 mice/genotype; means ± SEM). (C and D) H-Ackr4−/− and H-WT mice were generated by reconstituting lethally irradiated B6.Ly5.1 mice with BM from Ackr4−/− or WT mice, respectively. Representative plots of splenic GC B cell (C) and early PB (D) populations day 5 after SRBC immunization. Numbers in plots present the means ± SEM frequency of GC B cells per total B220+ cells (C) or early PBs per spleen (D). Graphs present the total number of GC B cells (C) and early PB (D) per spleen on the days indicated after immunization. n = 3 (day 0); n = 8 (day 5); n = 8 (day 8); and n = 5 (day 14) mice/genotype; means ± SEM). (E and F) B cell–Ackr4−/− (B-Ackr4−/−) and B-WT mice were generated by reconstituting lethally irradiated B6.Ly5.1 mice with a mixture of BM from μMT (80%) and Ackr4−/− mice or WT mice (20%), respectively. Graphs plot the total number of GC B cells (E) and early PB (F) per spleen on the days indicated after SRBC. (A–F) n = 7 (day 5) and n = 8 (day 8) mice/genotype; means ± SEM. Data are representative of two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (B) One-way analysis of variance with the Bonferroni multiple-comparisons test. (C–F) Two-tailed, unpaired Student’s t test or two-tailed, nonparametric Mann-Whitney test, as appropriate.
ACKR4 expression by GC B cells (Fig. 1, A and B), and their disproportionate expansion in immunized H-Ackr4−/− and B-Ackr4−/− mice (Fig. 1, C and F), prompted us to investigate whether ACKR4 functioned in GC biology. ACKR4 internalizes and degrades CCR7 ligands without initiating cellular migration (Gosling et al., 2000; Townson and Nibbs, 2002; Comerford et al., 2006; Heinzel et al., 2007). However, CCL21 staining appeared to be normal around established Ackr4-deficient GCs in histologic sections of mesenteric LNs from H-Ackr4−/− mice (Fig. 2 A). This suggests that GC B cell expression of ACKR4 does not determine the abundance or localization of CCL21 outside GCs. GC B cells cycle between light zones (LZs) and dark zones (DZs) of the GCs, reflecting the iterative process of affinity-based selection, proliferation, and Ig somatic hypermutation that drives antibody affinity maturation (Victora and Nussenzweig, 2012). Flow cytometric assessment of DZ (CXCR4hiCD86lo) and LZ (CXCR4loCD86hi) GC B cell phenotypes (Victora et al., 2010; Bannard et al., 2013) revealed no differences between H-WT and H-Ackr4−/− GCs (Fig. 2 B). Further, GC B cell proliferation and apoptosis were equivalent between H-WT and H-Ackr4−/− mice as measured by BrdU incorporation, DNA content, and Annexin V analyses (Fig. 2, C and D). Assessment of affinity maturation using Ig heavy chain–variable (V) region sequencing of individual (4-hydroxy-3-nitrophenyl)acetyl (NP)–specific IgG1+ GC B cells revealed that overall somatic mutations and the affinity-enhancing W to L replacement at position VH33 accumulated at similar rates in Ackr4-deficient and -sufficient GCs (Fig. 2 E). Serum antibody titers of NP-specific IgG were enhanced in H-Ackr4−/− mice at day 21 after immunization; however, the ratio of NP5- (high affinity) to NP32- (global affinity) reactive IgG was equivalent between H-WT and H-Ackr4−/− mice (Fig. 2 F). In addition, we addressed whether ACKR4 influenced GC B cell–affinity maturation in a cell-intrinsic manner. To do this, we crossed Ackr4−/− mice to SWHEL Ig transgenic mice (SWHEL.Ackr4−/−) and measured GC B cell–affinity maturation in competition with WT SWHEL B cells after transfer. SWHEL B cells express a B cell receptor (BCR) derived from the hen egg lysozyme (HEL)–specific HyHEL10 mAb (Brink et al., 2015). Immunization with the reduced-affinity HEL mutant HEL3X (acid dissociation constant [Ka] = 1 × 107 M−1) elicits a SWHEL GC B cell response characterized by a Y53D substitution in the Ig heavy chain V region, which increases HyHEL10 affinity to HEL3X by >100-fold (Paus et al., 2006; Chan et al., 2012; Brink et al., 2015). The additionally reduced affinity HEL mutant HEL4X (Ka = 2.3 × 105 M−1) does not bind the SWHEL BCR in a physiologically relevant manner but can bind the Y53D mutant form of HyHEL10 selected during the SWHEL GC response to HEL3X (Chan et al., 2012), thereby providing a metric for affinity maturation in the anti–HEL3X response. WT (CD45.1/2) and Ackr4−/− (CD45.2) SWHEL B cells were cotransferred into CD45.1 congenic recipient mice and immunized with HEL3X-SRBC (Fig. 2 G). On day 9, DZ/LZ phenotypes (Fig. 2 H) and the frequency of HEL4X-binding GC B cells (Fig. 2 I) were equivalent between WT and Ackr4−/− SWHEL GC B cells, indicating that, in this system, the absence of ACKR4 on GC B cells does not compromise the processes of affinity-based selection or establishment of DZ and LZ phenotypes in the presence of competing ACKR4-sufficient GC B cells. Collectively, these data show GC B cell proliferation, apoptosis, DZ/LZ phenotype ratio, somatic hypermutation, and affinity maturation to be unaltered in Ackr4-deficient GCs and suggest, instead, that B cell expression of ACKR4 negatively regulates, specifically, the pre-GC stages of development of the B cell response.
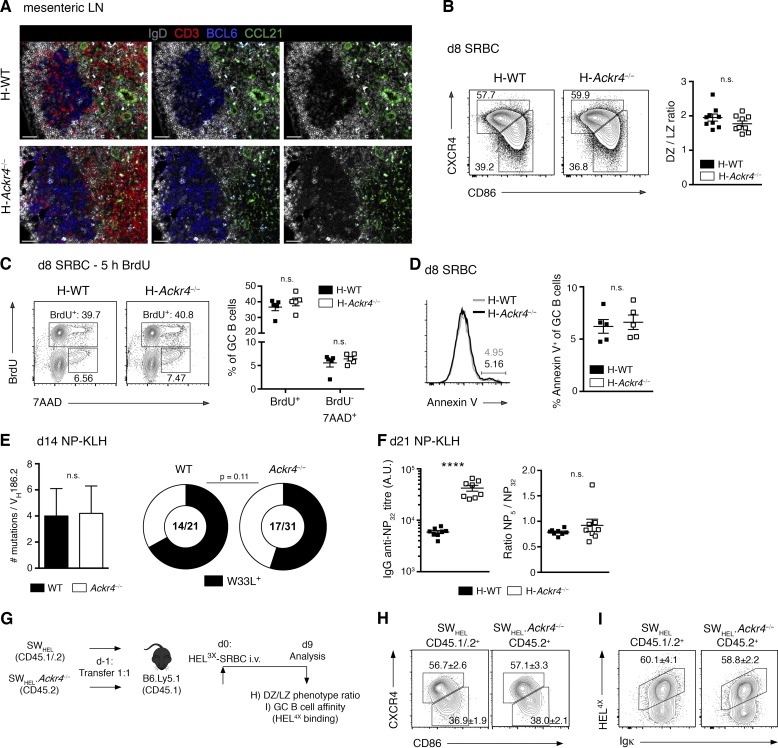
Figure 2.
ACKR4 in GC biology. (A) Representative histology of CCL21 (green) abundance surrounding mesenteric LN GCs in H-WT and H-Ackr4−/− mice. BCL-6, blue; IgD, white; and CD3, red. Right: IgD and CCL21 staining. Middle: IgD, BCL6, and CCL21 staining. Left: All stains merged. Bars, 50 µm. n = 2–3 mice/genotype. (B) Representative DZ (CXCR4hiCD86lo) and LZ (CXCR4loCD86hi) phenotype staining and ratios among splenic GC B cells (B220+IgD−Fas+GL7+) from H-WT and H-Ackr4−/− mice day 8 after SRBC. n = 8 mice/genotype (means ± SEM). (C) BrdU (2 mg) was administered i.p. 5 h before euthanasia. Representative BrdU and 7-aminoactinomycin D (DNA content) staining and quantitation of splenic GC B cells from H-WT and H-Ackr4−/− mice day 8 after SRBC. n = 5 mice/genotype (means ± SEM). (D) Representative staining and quantitation of Annexin V+ splenic GC B cells day 8 after SRBC. n = 5 mice/genotype. (E) Total mutation (left; means ± SD) and high-affinity W33L mutation (right) frequency per VH186.2 gene segment from single WT and Ackr4−/− NP+IgG1+ splenic GC B cells day 14 after NP-KLH immunization. Numbers in doughnut plots indicate the number of W33L+ sequences of the total VH186.2+ sequences analyzed. (F) Total (NP32) and the NP5/NP32 titer of serum NP-specific IgG antibodies in H-WT and H-Ackr4−/− mice day 21 after NP-KLH immunization. n = 8 mice/genotype (means ± SEM). (G) Experimental schematic. WT (CD45.1/2) and Ackr4−/− (CD45.2) SWHEL B cells were cotransferred into B6.Ly5.1 (CD45.1) mice and immunized the next day with HEL3X-SRBC. (H and I) Representative flow cytometric DZ/LZ ratio (H) and HEL4X (I; high-affinity) staining on concurrent WT and Ackr4−/− SWHEL GC B cells (B220+HEL+CD138−GL7+) on day 9. n = 5 mice. Numbers in plots present means ± SEM (A–I). Data are representative of one (A and E) and two (B–D and F–I) independent experiments. ****, P < 0.0001. (B–D and F) Two-tailed, unpaired Student’s t test. (E) Two-tailed, unpaired Student’s t test (left), Fisher’s exact test (right, doughnut plots). (H and I) Two-tailed, unpaired Student’s t test.
Our findings in H-Ackr4−/− and B-Ackr4−/− mice indicated that ACKR4 negatively regulates both early PB and GC B cell responses (Fig. 1, C–G). Recent evidence has indicated that a single B cell clone can enter all three possible differentiation fates, and that this positively correlates with the magnitude of their early proliferation and resistance to apoptosis (Taylor et al., 2015). Based on these observations and our data, we hypothesized that ACKR4 functions in undifferentiated, activated B cells to negatively regulate both early PB and GC B cell responses. This hypothesis predicts that Ackr4-deficient B cells will preferentially enter early PB and GC B compartments in an environment in which Ackr4−/− and WT B cells are in competition. To test this, we studied Ackr4−/− early PB and early GC B cell responses (day 5 after SRBC administration) in the context of mixed BM chimeras. In mice reconstituted with an equal mixture of Ackr4−/− and WT BM, ACKR4 deficiency enhanced early PB and early GC B cell responses relative to concurrently activated WT cells (Fig. 3 A). To extend these observations, we studied concurrent WT (CD45.1/2) and Ackr4−/− (CD45.2) SWHEL B cell responses to the reduced affinity HEL mutant HEL2X (Ka = 8 × 107 M−1). This response forms early PBs, GC B cells, and a population of cells (B220+Ig[HEL]hiGL7-) that likely encompass early memory B cells (Chan et al., 2009; Brink et al., 2015) by day 5 after immunization. At this time point, Ackr4−/− SWHEL B cells outcompeted WT SWHEL B cells in the early PB and GC B and B220+HELhiGL7− cell populations, most prominently among SWHEL early PBs (Fig. 3 B). We hypothesized further that enforced expression of ACKR4 prior to B cell activation by antigens may limit entry into early PB and GC B cell compartments. We tested this by generating Ackr4 knockin mice (Rosa26LSL-Ackr4; Fig. S2 A) and making expression conditional to follicular (Fo) B cells by introducing a Cre recombinase driven by the regulatory elements of Cd23 (Cd23Cre; Kwon et al., 2008). Fo B cells from Cd23Cre/+.Rosa26LSL-Ackr4/+ mice expressed functional ACKR4 (Fig. S2, B and C) and had frequencies of splenic Fo B cells and marginal-zone B cells that were equivalent to WT littermates (Fig. S2 D). Transgenic ACKR4 expression by Fo B cells did not alter CCR7 or CXCR5 expression at rest or after anti–IgM stimulation (Fig. S2 E), but, consistent with the known function of ACKR4 as a scavenger of CCR7 ligands (Comerford et al., 2006, 2010), inhibited anti-IgM–stimulated B cell migration toward CCL21, but not CXCL13, a non-ACKR4 ligand in mice (Townson and Nibbs, 2002; Fig. S2 F). Supporting the hypothesis that ACKR4 negatively regulates activated B cell differentiation, in mixed BM chimeric mice reconstituted with Cd23Cre/+.Rosa26LSL‑Ackr4/+ and WT BM, ACKR4 transgenic B cells were less represented among early PB and GC B cells that form on day 5 of the SRBC response (Fig. 3 C). These data suggest that enforced ACKR4 expression in Fo B cells limits their ability to form early PB and GC B cells when in competition with WT cells. Collectively, we conclude that ACKR4 negatively regulates early PB and GC B cell responses in a B cell–intrinsic manner. Furthermore, these experiments did not reveal a cell-intrinsic role for ACKR4 in TFH or TFR development (Fig. S1 C), suggesting that the enhanced T cell responses observed in H-Ackr4−/− and B-Ackr4−/− mice (Fig. S1, A and B) are secondary to enhanced B cell responses, an observation that is in keeping with recent evidence, indicating that the magnitude of the TFH cell response is proportional to the magnitude of the GC B cell response (Baumjohann et al., 2013).
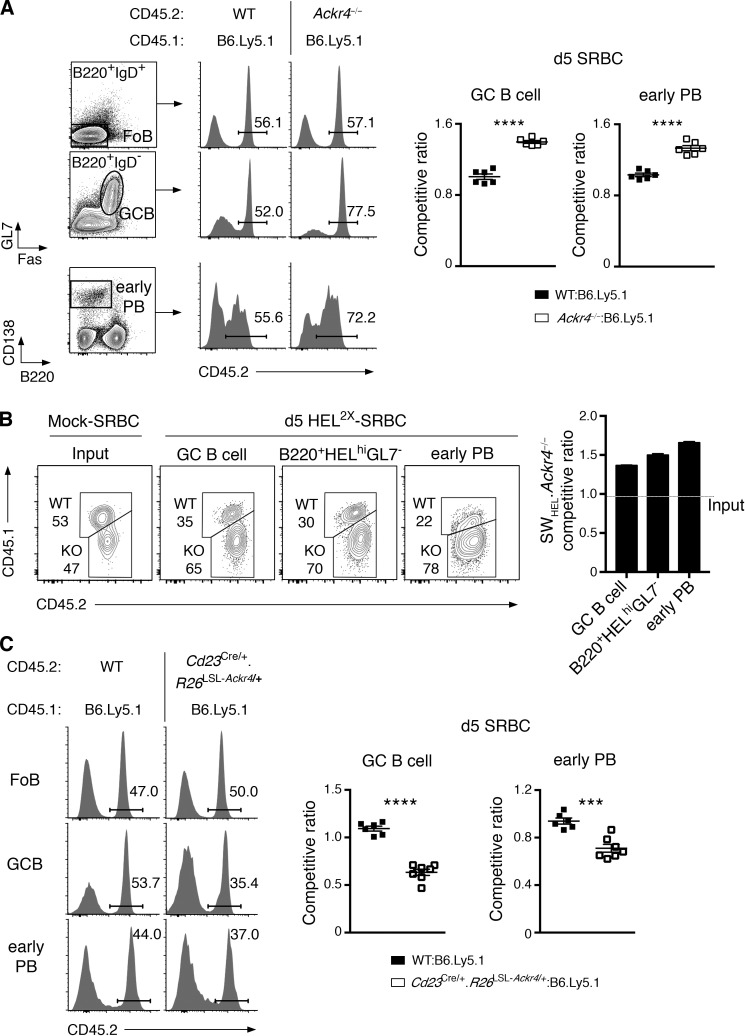
Figure 3.
ACKR4 limits early B cell responses in a B cell–intrinsic manner. (A) Mixed BM chimeras were generated with an equal mixture of B6.Ly5.1 (CD45.1) and WT or Ackr4−/− (both CD45.2) BM. Representative plots of CD45.2+ cells among Fo B cells (B220+IgD+Fas−GL7−), GC B cells (B220+IgD−Fas+GL7+), and early PBs (B220lo/-CD138+) 5 d after SRBC. Gating of these populations is shown. Competitive ratios of GC B cells (left graph) and early PBs (right graph) are plotted as the frequencies of CD45.2+ cells in each compartment, normalized to the frequencies of CD45.2+ cells in the concurrent Fo B cell compartment. n = 6 mice/genotype (means ± SEM). (B) WT (CD45.1/2) and Ackr4−/− (CD45.2) SWHEL B cells were cotransferred into B6.Ly5.1 (CD45.1) mice and immunized with HEL2X-SRBC (n = 4 mice) or mock-conjugated SRBCs (n = 3 mice). Contribution of WT and Ackr4−/− cells to SWHEL GC B cell (B220+Igκ+HELintGL7+CD138−), early PBs (B220lo/-Igκ+HEL+CD138+), and B220+HELhiGL7− (also Igκ+CD138−) cell populations 5 d after immunization. The SWHEL.Ackr4−/− competitive ratio is plotted as the ratio of Ackr4−/− to WT among SWHEL effector cell compartments from day 5 HEL2X-SRBC–immunized mice, normalized to the input ratio (as determined from mock-SRBC immunized mice analyzed on day 5 [means ± SEM]). (C) Cd23Cre/+.Rosa26LSL-Ackr4/+ mixed BM chimeras were generated and analyzed as in A (n = 6–7 mice/genotype; mean ± SEM). (A–C) Data are representative of two (B and C) and three (A) independent experiments. Two-tailed, unpaired Student’s t test. ***, P < 0.001; ****, P < 0.0001.
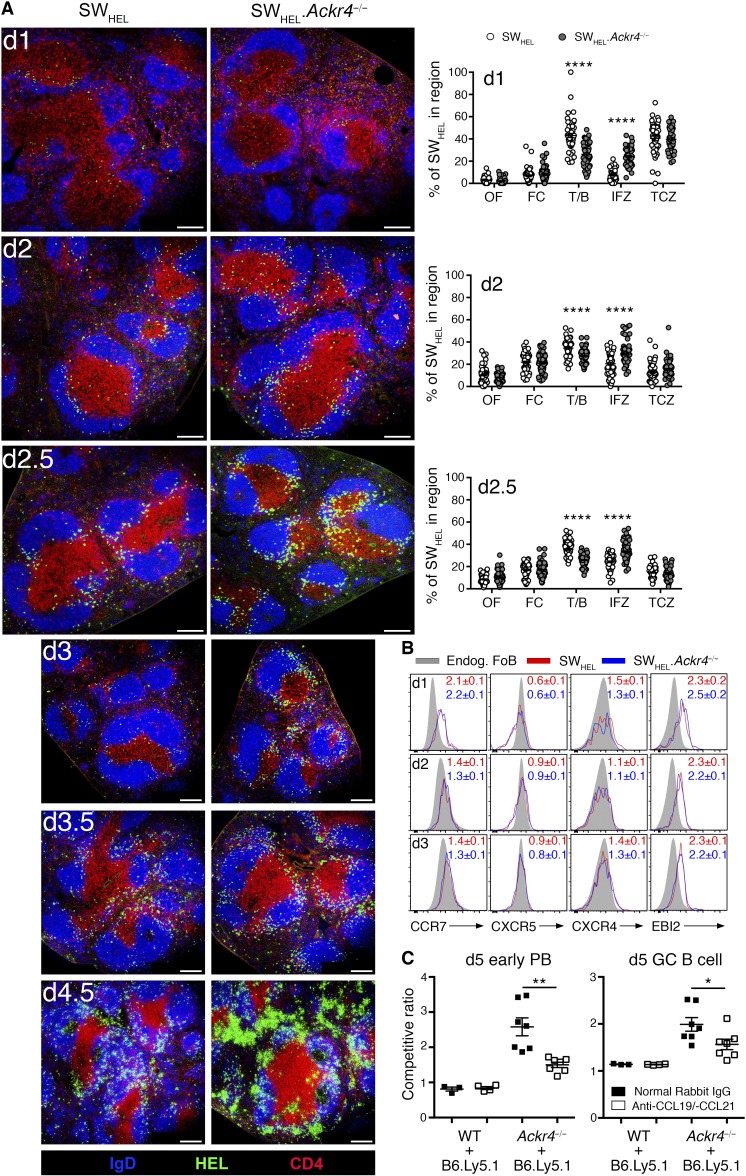
To examine ACKR4-dependent regulation of the initial antigen-engaged B cell differentiation in more detail, we tracked early WT and Ackr4−/− SWHEL B cell responses to HEL2X using mixed SWHEL B cell transfers. Profiling CFSE dilution revealed a cell-intrinsic proliferative advantage for Ackr4−/− over WT SWHEL B cells as early as day 2 (Fig. 4 A), which was increasingly apparent as the response progressed (Fig. 4 B). Notably, the magnitude of the advantage for Ackr4−/− SWHEL B cells on day 3 of the reaction (KO:WT = 1.26 ± 0.01) was similar to their advantage among the day 5 GC B cell (KO:WT = 1.36 ± 0.01; Fig. 3 B) and B220+HELhiGL7− cell (KO:WT = 1.49 ± 0.02; Fig. 3 B) populations. These findings suggest that enhanced early proliferation of responding Ackr4−/− SWHEL B cells may contribute to their accumulation among the effector cell compartments by day 5 of the reaction (Fig. 3 B). In experiments in which WT or Ackr4−/− SWHEL B cells were transferred into separate recipients, Ackr4−/− SWHEL B cells were detected at greater frequencies by day 2.5, and remained more abundant during the first 5.5 d of the response (Fig. 4 C). By day 5.5, mice receiving Ackr4−/− SWHEL B cells had an increased total number of early PBs, GC B cells, and B220+HELhiGL7− cells per spleen compared with controls (Fig. 4 D). Ackr4−/− SWHEL B cells showed a greater propensity to form early PBs (Fig. 4, E and F), which was apparent by flow cytometry on day 4.5 (Fig. 4 G) and translated to increased circulating anti–HEL IgM and IgG1 titers and kinetics (Fig. 4 H). Together, these findings suggest that ACKR4 limits early antigen-engaged B cell proliferation in a B cell–intrinsic manner, reducing the number of antigen-engaged B cell precursors available for early PB and GC B cell differentiation.
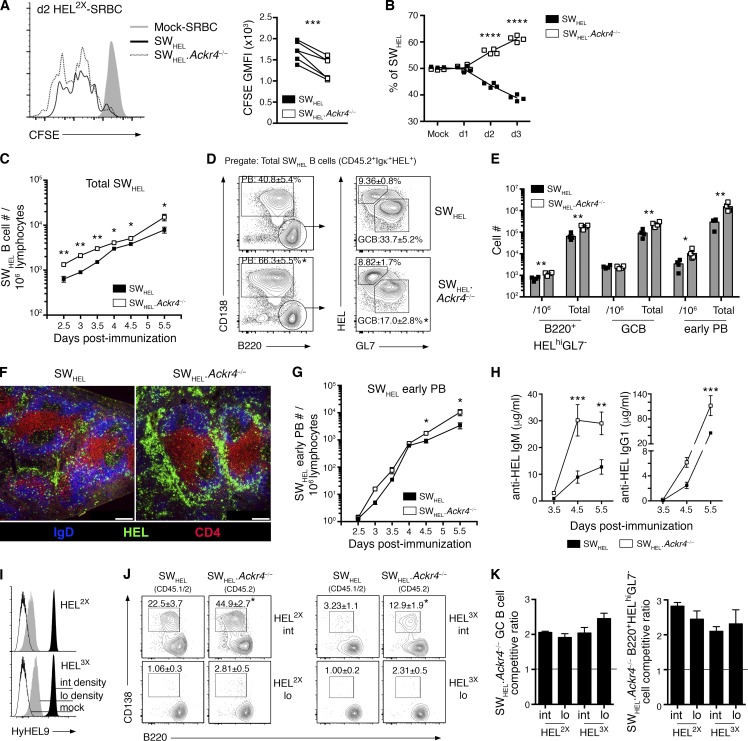
Figure 4.
ACKR4 limits early proliferation of antigen-engaged B cells and limits early PB responses. (A and B) WT (CD45.1/2) and Ackr4−/− (CD45.2) SWHEL B cells were cotransferred into B6.Ly5.1 (CD45.1) mice and immunized with HEL2X-SRBC. (A) Representative flow-cytometric CFSE profiles and analysis of CFSE GMFI among responding SWHEL B cells (B220+Igκ+HEL+CD138−) on day 2 (n = 6). Undivided SWHEL B cells were analyzed from mock-SRBC–immunized recipients (n = 2). (B) Contribution of WT and Ackr4−/− among responding SWHEL B cells (B220+Igκ+HEL+CD138−) over the first 3 d of the HEL2X-SRBC response (n = 4 mice/time point). (C–H) WT or Ackr4−/− (both CD45.2) SWHEL B cells were transferred into B6.Ly5.1 (CD45.1) mice and immunized with HEL2X-SRBC (n = 4 mice/genotype per time point). (C) Frequency of total responding SWHEL B cells (CD45.2+Igκ+HEL+) per 106 splenocytes (means ± SEM). (D) Representative plots of early PB and GC B cell and B220+HELhiGL7− cell responses as a frequency of total responding SWHEL B cells (plots are pregated CD45.2+Igκ+HEL+) on day 5.5. Numbers in plots represent the means ± SEM frequency of early PBs, GC B, or B220+HELhiGL7− cells as the frequency of the total responding SWHEL B cells. (E) Frequency of SWHEL B220+HELhiGL7− cells, GC B cells and early PBs (gated as shown in D) per 106 splenocytes and total number per spleen on day 5.5 (means ± SEM). (F) Representative histology of responding SWHEL B cells on day 5.5 (n = 4 mice/genotype). Bar, 200 µm. HEL, green; IgD, blue; and CD4, red. (G) Frequency of SWHEL early PBs (gated as in E) per 106 splenocytes (means ± SEM). (H) Anti-HEL IgM and IgG1 serum concentration as determined by ELISA (see Materials and methods; means ± SEM). (I–K) WT (CD45.1/2) and Ackr4−/− (CD45.2) were cotransferred into B6.Ly5.1 (CD45.1) recipients and immunized with HEL2X (intermediate affinity) or HEL3X (low affinity) conjugated to SRBCs at intermediate or low epitope densities (n = 4 mice/genotype per condition). (I) HEL2X or HEL3X were conjugated to SRBCs at intermediate (black histogram; 100 µg/ml; SRBC) and low (gray histogram; 5 µg/ml; SRBC) epitope densities and detected using HyHEL9 mAb. Mock-conjugated SRBCs (open histogram) are shown for comparison. (J) Representative plots of concurrent WT and Ackr4−/− SWHEL early PBs on day 5 (pregated Igκ+HEL+ and CD45.1+CD45.2+ [WT] or CD45.1−CD45.2+ [Ackr4−/−]). Numbers in plots represent the mean (± SEM) frequency of early PB as the frequency of total responding SWHEL B cells. (K) SWHEL.Ackr4−/− competitive ratio is plotted as the ratio of Ackr4−/− to WT among SWHEL GC B cell (left) or B220+HELhiGL7− cell (right) compartments (gated as in D), normalized to the input ratio, as determined from mock-SRBC–immunized mice. n = 4 mice (means ± SEM) on day 5. (A–K) *, P < 0.05; **, P < 0.01; ****, P < 0.0001; data are representative of two independent experiments. (A and J) Two-tailed, paired Student’s t test. (B and C–H) Two-tailed, unpaired Student’s t test. (C–K) Means ± SEM.
Comparisons of SWHEL B cell responses elicited by HEL antigens that differ in their affinity for HyHEL10 (Paus et al., 2006; Chan et al., 2009), or conjugated to SRBCs at varying epitope densities (Paus et al., 2006), have revealed that affinity and/or avidity of initial BCR-mediated activation positively regulates the initial expansion of antigen-engaged B cells. To investigate whether ACKR4-dependent changes to activated B cell responses were congruent across different initiating affinities and/or avidities of BCR-mediated activation, we studied WT (CD45.1/2) and Ackr4−/− (CD45.2) SWHEL B cell responses to intermediate (HEL2X)- or low (HEL3X)-affinity HEL proteins conjugated to SRBCs at intermediate or low epitope densities (Fig. 4 I; Paus et al., 2006; Chan et al., 2009). Ackr4−/− SWHEL B cells preferentially formed early PBs (Fig. 4 J) and outcompeted WT cells in GC B cell and B220+HELhiGL7- cell compartments 5 d after immunization when either HEL2X or HEL3X of equivalent intermediate epitope densities were used (Fig. 4 K). Conversely, when the epitope density was reduced (low density), both WT and Ackr4−/− SWHEL PB responses were barely detectable by flow cytometry on day 5 (Fig. 4 J), but the cell-intrinsic advantage for Ackr4−/− SWHEL GC B cell and B220+HELhiGL7- cell development at that time point remained apparent (Fig. 4 K). These data indicate that ACKR4-dependent negative regulation of B cell responses is independent of the initiating affinity or avidity of BCR-mediated activation.
To explore the relationship between ACKR4 function as a regulator of CCR7-dependent cellular migration and its negative regulation of activated B cell responses, we studied the distribution of WT and Ackr4−/− SWHEL B cells within the spleen after HEL2X-SRBC immunization using histology (Fig. 5 A). WT and Ackr4−/− SWHEL B cells were located throughout B cell follicles before immunization (unpublished data). Within 24 h, a proportion of WT SWHEL B cells had redistributed along the T/B border or had emigrated into the T cell zone. By days 2–2.5, most of the WT SWHEL B cells remained at the T/B border, although some redistribution to the interfollicular zone (IFZ; defined here as the lateral poles of the Fo B cell proximal to bridging regions, also referred to as the marginal-zone bridging channel) was apparent. In contrast, an increased proportion of Ackr4−/− SWHEL B cells were localized in the IFZ early during the response. On days 2–2.5, a larger proportion of Ackr4−/− SWHEL B cells were localized in the IFZ and were visibly more abundant than the WT SWHEL B cell response at that time, which became increasingly apparent during the next 60 h of the response. Whereas most WT SWHEL B cells were positioned in the IFZ and at the T/B border on days 3–3.5, a proportion of Ackr4−/− SWHEL B cells were distributed in the outer follicle, with the emergence of SWHEL B cell clusters in the outer follicular regions and the IFZ and the appearance of cells exhibiting a PB phenotype in the bridging channels on day 3, which became more obvious by days 3.5–4.5. These early ACKR4-dependent changes to the migratory patterns of SWHEL B cells were independent of detectable cell-intrinsic defects in CCR7, CXCR5, CXCR4, or EBI2 expression during the first 3 d of this response (Fig. 5 B). Thus, in the absence of ACKR4, a proportion of activated B cells preferentially home to the splenic IFZ during the early stages of the humoral immune response. Favorable positioning to interfollicular niches was accompanied with the enhanced expansion of Ackr4−/− SWHEL B cells in these zones and exaggerated early PB responses.
Figure 5.
ACKR4 limits early localization of antigen-engaged B cells to splenic IFZs. (A) WT or Ackr4−/− SWHEL B cells were transferred into WT recipients and immunized the next day with HEL2X-SRBC (days 1–2.5 analysis, transfer of 5 × 105 SWHEL B cells, immunization, 2 × 109 HEL2X-SRBC; and days 3–4.5 analysis, transfer of 2 × 105 SWHEL B cells, immunization, 109 HEL2X-SRBC). Representative histology of responding SWHEL B cells in the spleen on days 1, 2, 2.5, 3, 3.5, and 4.5 after immunization. Quantification of SWHEL B cell frequency per splenic region is depicted for days 1, 2, and 2.5 (see Materials and methods; means ± SEM). Bar, 200 µm. Blue, IgD; green, HEL; red, CD4; OF, outer follicle; FC, follicle center; T/B, T/B border; TCZ, T cell zone. Images are representative of five mice/genotype per time point; two-tailed, unpaired Student’s t test. (B) WT (CD45.1/2) and Ackr4−/− (CD45.2) SWHEL B cells were cotransferred into B6.Ly5.1 (CD45.1) mice and immunized with HEL2X-SRBC. Representative plots of CCR7, CXCR5, CXCR4, and EBI2 staining on responding SWHEL B cells (B220+Igκ+HEL+) relative to endogenous Fo B cells (CD45.1+CD45.2−B220+IgDhi). Numbers indicate the fold change (means ± SEM) in surface receptor GMFI on WT and Ackr4−/− SWHEL B cells relative to the concurrent endogenous Fo B cells (n = 5 mice/time point). (C) Mixed BM chimeras were generated with 80% BM from B6.Ly5.1 (CD45.1) and 20% from WT or Ackr4−/− (both CD45.2). Mice were administered polyclonal rabbit sera containing 500 µg anti–CCL19 and 500 µg anti–CCL21 or 1 mg normal rabbit IgG i.p. on days −1, 0, 2, and 4. Mice were immunized with SRBC on day 0 and analyzed on day 5. Competitive ratios of early PB (left; B220lo/-CD138+) and GC (right; B220+IgD−Fas+GL7+) B cells are plotted as the frequencies of CD45.2+ cells in each compartment normalized to the frequency of CD45.2+ cells in the concurrent Fo B cell (B220+IgD+Fas−GL7−) compartment (n = 4–7 mice/genotype; means ± SEM). Two-tailed unpaired Student’s t test. (A–C) Data are representative of two independent experiments. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
CCR7 guides activated B cell homing toward the T/B border but also contributes to the lateral spreading along that interface and positioning within the splenic IFZ (Reif et al., 2002; Okada et al., 2005; Gatto et al., 2011). To determine whether ACKR4-mediated regulation of early B cell responses was dependent on CCL19 and/or CCL21, these ligands were neutralized in mixed BM chimeras. This revealed that the advantage of ACKR4-deficient B cells to enter early PB and GC B cell compartments was dependent, at least in part, on physiological CCL19/CCL21 (Fig. 5 C).
Concluding remarks
Our findings establish ACKR4 as a B cell–intrinsic regulator of early PB and GC B cell responses. First, using ACKR4-deficient anti-HEL monoclonal B cells, we demonstrate that ACKR4 limits the early migration of antigen-engaged B cells to splenic interfollicular niches. Unrestricted access of activated B cells to these niches in the absence of ACKR4 was associated with their enhanced early expansion, which we propose increased the precursor pool of activated B cells available for subsequent differentiation into early PB and GC B cell fates. Further, we demonstrate that aberrant, splenic IFZ localization by antigen-engaged ACKR4-deficient anti-HEL B cells was accompanied by the preferential formation of early PB responses.
Existing evidence indicates that migration of activated B cells is predominantly shaped by their balanced responsiveness to CCR7, EBI2, CXCR5, and CXCR4 ligands (Pereira et al., 2010). Gatto et al. (2011) demonstrated that transfer of Cxcr5-deficient, activated B cells results in their exclusion from B cell follicles and accumulation in marginal-zone bridging channels; however, compound deletion of Cxcr5 with Ccr7 was shown to direct cells away from this niche and toward the outer regions of the follicle. These data indicate that, in addition to its established role as driving activated B cell migration toward the T cell zone (Reif et al., 2002), CCR7 also contributes to positioning activated B cells toward the IFZ and bridging zones of the spleen. Our experiments with SWHEL B cells indicate that deletion of ACKR4, a scavenger of CCR7 ligands, promotes responding B cell migration to splenic IFZ. We speculate that ACKR4 may function to “tune” early CCR7-dependent cues on a proportion of responding B cells, limiting their CCR7-driven homing to splenic IFZ. Our findings that physiological CCL19 and CCL21 were required, at least in part, for ACKR4-dependent changes to early PB and GC B cell responses supports a relationship between ACKR4 and CCR7 function in the regulation of activated B cell responses.
Our results indicate that the propensity of ACKR4-deficient SWHEL B cells to form early PB responses correlated with their favorable accumulation in the IFZ during the early stages of T cell–dependent humoral immunity. These data, together with published findings that (a) EBI2-deficient B cells, which are defective in their ability to access splenic IFZ and bridging channels, fail to form robust early PB responses (Gatto et al., 2009); (b) forced EBI2 expression on B cells promotes early PB responses (Gatto et al., 2009); and (c) targeted antigen delivery to splenic DCIR2+ dendritic cells, which localize to this niche, elicit robust early PB responses, and are implicated in promoting PB survival (García De Vinuesa et al., 1999; Chappell et al., 2012), support a model in which B cell migration at the early stages of activation has an important role in coordinating and balancing differentiation to early PB and GC B cell fates.
In summary, our results describe an in vivo, cell-intrinsic role for ACKR4 in shaping activated B cell differentiation and further our understanding of the cellular events that govern antibody production.
Materials and methods
Mice
All mice were on the C57BL/6J background and housed in specific pathogen-free conditions at Laboratory Animal Services, University of Adelaide (unless indicated otherwise). C57BL/6 (WT) and B6.Ly5.1 (B6.SJL Ptprca) were purchased from the Animal Resource Center and bred in house. Ackr4−/−, Ccr7−/−, μMT, Ackr4−/−.Ccr7−/− SWHEL (CD45.2), SWHEL (CD45.1/2), and SWHEL.Ackr4−/− (CD45.2) mice were bred and maintained in house. Cd23Cre mice were provided by M. Busslinger (Research Institute of Molecular Pathology, Vienna, Austria). Cd23Cre.Rosa26LSL-Ackr4/+ mice were generated by interbreeding and maintained in house. Mice used in experiments were gender- and age-matched animals and were between the ages of 6 and 12 wk. All animal experiments were approved by the Animal Ethics Committee of the University of Adelaide.
BM chimeras
Recipient mice were lethally irradiated with 1,000 rad (two doses of 500 rad) and reconstituted with 4–5 × 106 total BM cells i.v. of genotypes indicated in text. A minimum of 8 wk was allowed for reconstitution before experimentation.
Transfers, immunizations, and BrdU treatment
T cell–dependent humoral immune responses were induced with 2 × 109 SRBC (Applied Biological Products) or 50 µg NP-KLH (Biosearch Technologies) and precipitated in alum (Analar) i.p. HEL2X, HEL3X, and HEL4X, with varying affinities to HyHEL10, as described (Brink et al., 2015). Conjugation of HEL proteins to SRBCs was performed as described (Brink et al., 2015). Unless otherwise indicated, HEL conjugation to SRBCs was performed using 300 µg/ml HEL. Successful conjugation was confirmed using HyHEL9 mAb (Brink et al., 2015) before immunization. Unless otherwise indicated, splenocytes or Fo B cells purified by magnetic-activated cell sorting (CD43 Negative Isolation kit; Miltenyi Biotech) from SWHEL or SWHEL.Ackr4−/− mice containing 105 HEL-binding B cells were transferred i.v. into B6.Ly5.1-recipient mice. The next day, recipient mice were immunized i.v. with 109 HEL-conjugated SRBCs. For assessment of SWHEL B cell proliferation, splenocytes containing SWHEL B cells were CFSE loaded, as described (Quah et al., 2007), before transfer and immunization the next day. For BrdU experiments, 2 mg BrdU (Sigma-Aldrich) in 0.85% saline was injected i.p. 5 h before euthanasia.
Histology
For assessment of SWHEL B cell responses by histology (days 1–2.5), 5 × 105 HEL-binding B cells from SWHEL or SWHEL.Ackr4−/− mice were transferred i.v. into WT mice, which were immunized with 2 × 109 HEL2X-SRBC i.v. the next day. For assessment of SWHEL B cell responses by histology (days 3–5.5), 2 × 105 HEL-binding B cells from SWHEL or SWHEL.Ackr4−/− mice were transferred i.v. into WT mice, which were immunized with 109 HEL2X-SRBC i.v. the next day. Organs were frozen in Tissue-Tek optimal cutting temperature–embedding medium (Sakura Finetek). Cryostat sections (8 µm) were fixed in ice-cold acetone and stained, as previously described (Bunting et al., 2013). For detection of HEL-binding B cells, sections were first blocked with 30% normal horse serum and incubated with HELWT (100 ng/ml; Sigma-Aldrich), which was detected using unconjugated rabbit anti–HEL (polyclonal; Rockland) and goat anti–rabbit Ig-Alexa Fluor 488 (Life Technologies). For SWHEL histology, antibodies to IgD (11-26c; eBioscience) and CD4 (RM4-5; BD) were used. For the GC stain (Fig. 2 A), antibodies to IgD, CD3 (145-2C11; BD), BCL-6 (K112-91; BD), and CCL21 (goat polyclonal; R&D) were used. To enumerate transferred SWHEL B cell positioning in spleen sections, the outer follicle, follicle center, T/B interface, IFZs, and T cell zone per white pulp region were first defined (Gatto et al., 2011) in a blinded manner on images stained with IgD/CD4, with HEL-binding fluorescence removed. HEL-binding fluorescence was then merged with these images, and the total number of HEL-binding cells per white pulp area and HEL-binding cells in defined regions were enumerated by four independent researchers. Means ± SEM values across those four independent data sets are presented.
In vivo CCL19/CCL21 neutralization
Affinity-purified anti–mouse (m) CCL21 was generated and purified in house as described (Comerford et al., 2010). Anti–mCCL19 antibodies were raised in New Zealand white rabbits by immunization with full-length, synthetically manufactured CCL19, which was active in calcium mobilization and chemotaxis assays (Clark-Lewis et al., 1994). Serum IgG was purified from preimmunized bleeds (normal rabbit IgG [NRIgG]) and mCCL19- or mCCL21-immunized rabbits using Protein A columns (Millipore). The CCL19- or CCL21-neutralizing ability of these antibodies was confirmed in chemotaxis assays before their use in vivo. Recipient, mixed-BM chimeric mice were administered 500 µg affinity-purified rabbit anti–mCCL21 and 500 µg affinity-purified, rabbit anti–mCCL19 or 1 mg NRIgG i.p. on days −1, 0, 2, and 4. Mice were immunized with SRBCs i.p. on day 0 and analyzed 5 d later.
VH gene sequencing analysis of NP+IgG1+ GC B cells
Single NP+IgG1+ GC B cells were sorted from NP-KLH–immunized (day 14) mice using a BD FACSAria cell sorter. Two rounds of PCR were performed on cDNA using a single proximal 5′ primer for the J558 VH gene family (Ehlich et al., 1994; Smith et al., 2000), together with nested primers specific for Cγ1 (McHeyzer-Williams et al., 1991; Smith et al., 2000). Bands of expected size were purified, sequenced, and analyzed for VH186.2-containing sequences as described. For VH186.2+ clones, the region encoding amino acids 10–96 were compared in detail with the germline VH186.2 sequence as described (Smith et al., 2000).
Chemotaxis assay
Fo B cells purified by magnetic-activated cell sorting were activated with 5 µg/ml goat anti–mouse IgM (Jackson ImmunoResearch) for 24 h or rested overnight (unstimulated control). Various dilutions of recombinant mouse CCL21 (provided by the late I. Clark-Lewis) or CXCL13 (PeproTech) in 150 µl chemotaxis buffer (RPMI-1640 with 0.5% BSA and 20 mM Hepes) were added to the lower chambers of Transwell chemotaxis plates (96-well, 5-µm pore size; Corning). Cells were extensively washed in chemotaxis buffer and loaded into the upper chambers at 105 cells/well in 50 µl chemotaxis buffer and incubated for 3 h at 37°C. To enumerate B cell migration, cells were harvested from the bottom chambers, and B220+ cells were assessed by flow cytometry using a defined number of CaliBRITE beads (BD) as an internal reference. The migration index was calculated as described (Kara et al., 2013).
Chemokine-scavenging assay
FACS-sorted Fo B cells (2 × 105) were incubated in RPMI-1640 medium containing 5 or 10 ng/ml recombinant mouse CCL19 (R&D Systems) at 37°C for 3 h with inversion every 30 min. 100 µl of cell-free supernatant was then assessed for CCL19 concentration by ELISA as described previously (Comerford et al., 2010).
ELISA
Serum HEL-specific IgM and IgG1 concentrations were quantified using HyHEL10 IgM and IgG1 standards as described previously (Phan et al., 2003). NP-specific IgG was detected using NP5-BSA or NP32-BSA (10 µg/ml; Biosearch Technologies)–coating antigen. ELISAs were developed with 3,3′,5,5′-tetramethylbenzidine substrate.
Quantitative PCR
RNA was harvested using the Qiagen microRNeasy kit with on-column DNase treatment. cDNA synthesis was performed using Transcriptor First Strand cDNA synthesis kit (Roche) and used as the template in reactions with the LightCycler 480 SYBR Green Master Mix I (Roche). Relative Ackr4 transcript abundance was calculated with reference to the housekeeping gene Rplp0 using the formula: 2−ΔCT (ΔCT = CTAckr4 – CTRplp0); Rplp0, forward: 5′-AGATGCAGCAGATCCGCAT-3′, reverse: 5′-CAGTGAGCTTCCCGTTCAG-3′; Ackr4, forward: 5′-AGATGCAGCAGATCCGCAT-3′, reverse: 5′‑CAGTGAGCTTCCCGTTCAG-3′.
Flow cytometry and sorting
Cells were stained as described previously (Kara et al., 2015) using antibodies specific for: CD45.2 (104), B220 (RA3-6B2), IgD (11-26c), IgDb (217–170), Fas (Jo2), GL7, CD86 (GL1), CD138 (281–2), Igκ (RMK-45), CD93 (AA4.1), CD21 (7G6), CD23 (B3B4), CD11b (M1/70), CD3 (145-2C11), CD4 (RM4-5), CD44 (IM7), PD-1 (J43), CXCR5 (2G8), and CCR7 (4B12; in house); and EBI2 (chicken polyclonal; Gatto et al., 2013), CXCR4 (2B11/CXCR4), Nrp-1 (3E12), and Foxp3 (FJK-16s). Unconjugated/biotinylated antibodies were detected using goat anti–rat IgG (Life Technologies), goat anti–human IgG-Fcγ fragment specific (Jackson ImmunoResearch), streptavidin-PE (Jackson ImmunoResearch), -Alexa Fluor 647 (Jackson ImmunoResearch), -BV510, -BV421, or -BV450. All antibodies and secondary reagents were purchased from BD, eBioscience, or BioLegend unless otherwise indicated. Dead cells were excluded using LIVE/DEAD fixable near-infrared dye (Molecular Probes). 7-aminoactinomycin D (eBioscience) was used to detect DNA content. BrdU staining was conducted using the BrdU Flow kit (BD). NP-binding was detected using NP-PE made in house (Smith et al., 2000). CCL19-Fc is a chimeric protein of mouse CCL19 fused to human IgG1 constant region (Reif et al., 2002). The Annexin V apoptosis detection kit (eBioscience) was used to detect Annexin V. Foxp3/TF staining buffer set (eBioscience) was used to detect FoxP3. For detection of HEL-binding cells, 2 × 106 splenocytes were incubated with soluble HEL (200 ng/ml), followed by detection of BCR-bound HEL using mAb HyHEL9 conjugated to Alexa Fluor 647 as described (Brink et al., 2015). Data were acquired on BD LSR II, BD FACSAria, or BD LSRFortessa flow cytometers. For sorting experiments, a BD FACSAria was used. Data were analyzed using FlowJo software (Tree Star).
Statistics
Data were analyzed with Prism (GraphPad Software) using either two-tailed (unpaired or paired, as appropriate) Student’s t tests (for normally distributed data sets comparing the mean of two samples), two-tailed nonparametric Mann-Whitney tests (for data sets that were determined by an F test not to have a normal distribution and where there was a comparison of the mean of two samples), or one-way analysis of variance with appropriate posttests as indicated in text (for comparisons of multiple samples). For all analyses, P < 0.05 was considered significant. Sample or experiment sizes were determined empirically for sufficient statistical power. No statistical tests were used to predetermine the size of experiments. No data points were excluded from statistical tests.
Online supplemental material
Fig. S1 shows that follicular T cell responses to SRBC immunization are enhanced in the absence of hematopoietic or B cell expression of ACKR4. Fig. S2 shows the generation and characterization of Rosa26LSL-Ackr4 knockin mice.
Supplementary Material
Acknowledgments
We thank Josef Nguyen (Royal Adelaide Hospital, Adelaide, Australia) for mouse irradiation; the staff of Laboratory Animal Services, University of Adelaide, for animal husbandry; Meinrad Busslinger for Cd23Cre mice; and Harald Hartweger for comments on the manuscript.
This work was supported in part by a grant from the Australian National Health and Medical Research Council (APP1105312) to S.R. McColl, J.G. Cyster, and I. Comerford, J.G. Cyster is an investigator of the Howard Hughes Medical Institute. E.E. Kara is supported by an Australian postgraduate award, a Norman and Patricia Polglase scholarship, and a National Health and Medical Research Council C.J. Martin Overseas Biomedical fellowship.
The authors declare no competing financial interests.
Author contributions: E.E. Kara designed, performed, and analyzed experiments and wrote the manuscript. C.R. Bastow, D.R. McKenzie, C.E. Gregor, K.A. Fenix, R. Babb, T.S. Norton, D. Zotos, L.B. Rodda, J.R. Hermes, and K. Bourne performed experiments; D.S. Gilchrist and R.J. Nibbs generated Rosa26LSL-Ackr4 mice; D.M. Tarlinton, C.G. Vinuesa, M. Alsharifi, R. Brink, G.R. Hill, and J.G. Cyster provided key reagents and critical discussions and edited the manuscript; I. Comerford and S.R. McColl conceptualized the project, designed the experiments, supervised the study, and wrote the manuscript.
References
- Allen C.D., Ansel K.M., Low C., Lesley R., Tamamura H., Fujii N., and Cyster J.G.. 2004. Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nat. Immunol. 5:943–952. 10.1038/ni1100 [DOI] [PubMed] [Google Scholar]
- Bannard O., Horton R.M., Allen C.D., An J., Nagasawa T., and Cyster J.G.. 2013. Germinal center centroblasts transition to a centrocyte phenotype according to a timed program and depend on the dark zone for effective selection. Immunity. 39:912–924. 10.1016/j.immuni.2013.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumjohann D., Preite S., Reboldi A., Ronchi F., Ansel K.M., Lanzavecchia A., and Sallusto F.. 2013. Persistent antigen and germinal center B cells sustain T follicular helper cell responses and phenotype. Immunity. 38:596–605. 10.1016/j.immuni.2012.11.020 [DOI] [PubMed] [Google Scholar]
- Brink R., Paus D., Bourne K., Hermes J.R., Gardam S., Phan T.G., and Chan T.D.. 2015. The SW(HEL) system for high-resolution analysis of in vivo antigen-specific T-dependent B cell responses. Methods Mol. Biol. 1291:103–123. 10.1007/978-1-4939-2498-1_9 [DOI] [PubMed] [Google Scholar]
- Bryce S.A., Wilson R.A., Tiplady E.M., Asquith D.L., Bromley S.K., Luster A.D., Graham G.J., and Nibbs R.J.. 2016. ACKR4 on Stromal Cells Scavenges CCL19 To Enable CCR7-Dependent Trafficking of APCs from Inflamed Skin to Lymph Nodes. J. Immunol. 196:3341–3353. 10.4049/jimmunol.1501542 [DOI] [PubMed] [Google Scholar]
- Bunting M.D., Comerford I., Seach N., Hammett M.V., Asquith D.L., Körner H., Boyd R.L., Nibbs R.J., and McColl S.R.. 2013. CCX-CKR deficiency alters thymic stroma impairing thymocyte development and promoting autoimmunity. Blood. 121:118–128. 10.1182/blood-2012-06-434886 [DOI] [PubMed] [Google Scholar]
- Chan T.D., Gatto D., Wood K., Camidge T., Basten A., and Brink R.. 2009. Antigen affinity controls rapid T-dependent antibody production by driving the expansion rather than the differentiation or extrafollicular migration of early plasmablasts. J. Immunol. 183:3139–3149. 10.4049/jimmunol.0901690 [DOI] [PubMed] [Google Scholar]
- Chan T.D., Wood K., Hermes J.R., Butt D., Jolly C.J., Basten A., and Brink R.. 2012. Elimination of germinal-center-derived self-reactive B cells is governed by the location and concentration of self-antigen. Immunity. 37:893–904. 10.1016/j.immuni.2012.07.017 [DOI] [PubMed] [Google Scholar]
- Chappell C.P., Draves K.E., Giltiay N.V., and Clark E.A.. 2012. Extrafollicular B cell activation by marginal zone dendritic cells drives T cell-dependent antibody responses. J. Exp. Med. 209:1825–1840. 10.1084/jem.20120774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Lewis I., Dewald B., Loetscher M., Moser B., and Baggiolini M.. 1994. Structural requirements for interleukin-8 function identified by design of analogs and CXC chemokine hybrids. J. Biol. Chem. 269:16075–16081. [PubMed] [Google Scholar]
- Comerford I., Milasta S., Morrow V., Milligan G., and Nibbs R.. 2006. The chemokine receptor CCX-CKR mediates effective scavenging of CCL19 in vitro. Eur. J. Immunol. 36:1904–1916. 10.1002/eji.200535716 [DOI] [PubMed] [Google Scholar]
- Comerford I., Nibbs R.J., Litchfield W., Bunting M., Harata-Lee Y., Haylock-Jacobs S., Forrow S., Korner H., and McColl S.R.. 2010. The atypical chemokine receptor CCX-CKR scavenges homeostatic chemokines in circulation and tissues and suppresses Th17 responses. Blood. 116:4130–4140. 10.1182/blood-2010-01-264390 [DOI] [PubMed] [Google Scholar]
- Ehlich A., Martin V., Müller W., and Rajewsky K.. 1994. Analysis of the B-cell progenitor compartment at the level of single cells. Curr. Biol. 4:573–583. 10.1016/S0960-9822(00)00129-9 [DOI] [PubMed] [Google Scholar]
- García De Vinuesa C., Gulbranson-Judge A., Khan M., O’Leary P., Cascalho M., Wabl M., Klaus G.G., Owen M.J., and MacLennan I.C.. 1999. Dendritic cells associated with plasmablast survival. Eur. J. Immunol. 29:3712–3721. [DOI] [PubMed] [Google Scholar]
- Gatto D., Paus D., Basten A., Mackay C.R., and Brink R.. 2009. Guidance of B cells by the orphan G protein-coupled receptor EBI2 shapes humoral immune responses. Immunity. 31:259–269. 10.1016/j.immuni.2009.06.016 [DOI] [PubMed] [Google Scholar]
- Gatto D., Wood K., and Brink R.. 2011. EBI2 operates independently of but in cooperation with CXCR5 and CCR7 to direct B cell migration and organization in follicles and the germinal center. J. Immunol. 187:4621–4628. 10.4049/jimmunol.1101542 [DOI] [PubMed] [Google Scholar]
- Gatto D., Wood K., Caminschi I., Murphy-Durland D., Schofield P., Christ D., Karupiah G., and Brink R.. 2013. The chemotactic receptor EBI2 regulates the homeostasis, localization and immunological function of splenic dendritic cells. Nat. Immunol. 14:446–453. 10.1038/ni.2555 [DOI] [PubMed] [Google Scholar]
- Gosling J., Dairaghi D.J., Wang Y., Hanley M., Talbot D., Miao Z., and Schall T.J.. 2000. Cutting edge: identification of a novel chemokine receptor that binds dendritic cell- and T cell-active chemokines including ELC, SLC, and TECK. J. Immunol. 164:2851–2856. 10.4049/jimmunol.164.6.2851 [DOI] [PubMed] [Google Scholar]
- Hannedouche S., Zhang J., Yi T., Shen W., Nguyen D., Pereira J.P., Guerini D., Baumgarten B.U., Roggo S., Wen B., et al. . 2011. Oxysterols direct immune cell migration via EBI2. Nature. 475:524–527. 10.1038/nature10280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves D.C., Hyman P.L., Lu T.T., Ngo V.N., Bidgol A., Suzuki G., Zou Y.R., Littman D.R., and Cyster J.G.. 2001. A coordinated change in chemokine responsiveness guides plasma cell movements. J. Exp. Med. 194:45–56. 10.1084/jem.194.1.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzel K., Benz C., and Bleul C.C.. 2007. A silent chemokine receptor regulates steady-state leukocyte homing in vivo. Proc. Natl. Acad. Sci. USA. 104:8421–8426. 10.1073/pnas.0608274104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kara E.E., Comerford I., Bastow C.R., Fenix K.A., Litchfield W., Handel T.M., and McColl S.R.. 2013. Distinct chemokine receptor axes regulate Th9 cell trafficking to allergic and autoimmune inflammatory sites. J. Immunol. 191:1110–1117. 10.4049/jimmunol.1203089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kara E.E., McKenzie D.R., Bastow C.R., Gregor C.E., Fenix K.A., Ogunniyi A.D., Paton J.C., Mack M., Pombal D.R., Seillet C., et al. . 2015. CCR2 defines in vivo development and homing of IL-23-driven GM-CSF-producing Th17 cells. Nat. Commun. 6:8644 10.1038/ncomms9644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly L.M., Pereira J.P., Yi T., Xu Y., and Cyster J.G.. 2011. EBI2 guides serial movements of activated B cells and ligand activity is detectable in lymphoid and nonlymphoid tissues. J. Immunol. 187:3026–3032. 10.4049/jimmunol.1101262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfoot S.M., Yaari G., Patel J.R., Johnson K.L., Gonzalez D.G., Kleinstein S.H., and Haberman A.M.. 2011. Germinal center B cell and T follicular helper cell development initiates in the interfollicular zone. Immunity. 34:947–960. 10.1016/j.immuni.2011.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura D., Roes J., Kühn R., and Rajewsky K.. 1991. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 350:423–426. 10.1038/350423a0 [DOI] [PubMed] [Google Scholar]
- Kwon K., Hutter C., Sun Q., Bilic I., Cobaleda C., Malin S., and Busslinger M.. 2008. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity. 28:751–762. 10.1016/j.immuni.2008.04.014 [DOI] [PubMed] [Google Scholar]
- Lucas B., White A.J., Ulvmar M.H., Nibbs R.J., Sitnik K.M., Agace W.W., Jenkinson W.E., Anderson G., and Rot A.. 2015. CCRL1/ACKR4 is expressed in key thymic microenvironments but is dispensable for T lymphopoiesis at steady state in adult mice. Eur. J. Immunol. 45:574–583. 10.1002/eji.201445015 [DOI] [PubMed] [Google Scholar]
- McHeyzer-Williams M.G., Nossal G.J., and Lalor P.A.. 1991. Molecular characterization of single memory B cells. Nature. 350:502–505. 10.1038/350502a0 [DOI] [PubMed] [Google Scholar]
- Nibbs R.J., and Graham G.J.. 2013. Immune regulation by atypical chemokine receptors. Nat. Rev. Immunol. 13:815–829. 10.1038/nri3544 [DOI] [PubMed] [Google Scholar]
- Okada T., Miller M.J., Parker I., Krummel M.F., Neighbors M., Hartley S.B., O’Garra A., Cahalan M.D., and Cyster J.G.. 2005. Antigen-engaged B cells undergo chemotaxis toward the T zone and form motile conjugates with helper T cells. PLoS Biol. 3:e150 10.1371/journal.pbio.0030150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus D., Phan T.G., Chan T.D., Gardam S., Basten A., and Brink R.. 2006. Antigen recognition strength regulates the choice between extrafollicular plasma cell and germinal center B cell differentiation. J. Exp. Med. 203:1081–1091. 10.1084/jem.20060087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira J.P., Kelly L.M., Xu Y., and Cyster J.G.. 2009. EBI2 mediates B cell segregation between the outer and centre follicle. Nature. 460:1122–1126. 10.1038/nature08226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira J.P., Kelly L.M., and Cyster J.G.. 2010. Finding the right niche: B-cell migration in the early phases of T-dependent antibody responses. Int. Immunol. 22:413–419. 10.1093/intimm/dxq047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T.G., Amesbury M., Gardam S., Crosbie J., Hasbold J., Hodgkin P.D., Basten A., and Brink R.. 2003. B cell receptor-independent stimuli trigger immunoglobulin (Ig) class switch recombination and production of IgG autoantibodies by anergic self-reactive B cells. J. Exp. Med. 197:845–860. 10.1084/jem.20022144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quah B.J., Warren H.S., and Parish C.R.. 2007. Monitoring lymphocyte proliferation in vitro and in vivo with the intracellular fluorescent dye carboxyfluorescein diacetate succinimidyl ester. Nat. Protoc. 2:2049–2056. 10.1038/nprot.2007.296 [DOI] [PubMed] [Google Scholar]
- Reif K., Ekland E.H., Ohl L., Nakano H., Lipp M., Förster R., and Cyster J.G.. 2002. Balanced responsiveness to chemoattractants from adjacent zones determines B-cell position. Nature. 416:94–99. 10.1038/416094a [DOI] [PubMed] [Google Scholar]
- Smith K.G., Hewitson T.D., Nossal G.J., and Tarlinton D.M.. 1996. The phenotype and fate of the antibody-forming cells of the splenic foci. Eur. J. Immunol. 26:444–448. 10.1002/eji.1830260226 [DOI] [PubMed] [Google Scholar]
- Smith K.G., Light A., O’Reilly L.A., Ang S.M., Strasser A., and Tarlinton D.. 2000. bcl-2 transgene expression inhibits apoptosis in the germinal center and reveals differences in the selection of memory B cells and bone marrow antibody-forming cells. J. Exp. Med. 191:475–484. 10.1084/jem.191.3.475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.J., Jenkins M.K., and Pape K.A.. 2012. Heterogeneity in the differentiation and function of memory B cells. Trends Immunol. 33:590–597. 10.1016/j.it.2012.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.J., Pape K.A., Steach H.R., and Jenkins M.K.. 2015. Humoral immunity. Apoptosis and antigen affinity limit effector cell differentiation of a single naïve B cell. Science. 347:784–787. 10.1126/science.aaa1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townson J.R., and Nibbs R.J.. 2002. Characterization of mouse CCX-CKR, a receptor for the lymphocyte-attracting chemokines TECK/mCCL25, SLC/mCCL21 and MIP-3beta/mCCL19: comparison to human CCX-CKR. Eur. J. Immunol. 32:1230–1241. [DOI] [PubMed] [Google Scholar]
- Ulvmar M.H., Werth K., Braun A., Kelay P., Hub E., Eller K., Chan L., Lucas B., Novitzky-Basso I., Nakamura K., et al. . 2014. The atypical chemokine receptor CCRL1 shapes functional CCL21 gradients in lymph nodes. Nat. Immunol. 15:623–630. 10.1038/ni.2889 [DOI] [PubMed] [Google Scholar]
- Victora G.D., and Nussenzweig M.C.. 2012. Germinal centers. Annu. Rev. Immunol. 30:429–457. 10.1146/annurev-immunol-020711-075032 [DOI] [PubMed] [Google Scholar]
- Victora G.D., Schwickert T.A., Fooksman D.R., Kamphorst A.O., Meyer-Hermann M., Dustin M.L., and Nussenzweig M.C.. 2010. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 143:592–605. 10.1016/j.cell.2010.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.