During development, cell proliferation is regulated, ensuring that tissues reach their correct size and shape. Forest et al. show that the Drosophila melanogaster scaffold protein big bang (Bbg) controls epithelial tissue growth without affecting epithelial polarity and architecture. Bbg interacts with spectrins at the apical cortex and promotes Yki signaling and actomyosin contractility.
Abstract
During development, cell numbers are tightly regulated, ensuring that tissues and organs reach their correct size and shape. Recent evidence has highlighted the intricate connections between the cytoskeleton and the regulation of the key growth control Hippo pathway. Looking for apical scaffolds regulating tissue growth, we describe that Drosophila melanogaster big bang (Bbg), a poorly characterized multi-PDZ scaffold, controls epithelial tissue growth without affecting epithelial polarity and architecture. bbg-mutant tissues are smaller, with fewer cells that are less apically constricted than normal. We show that Bbg binds to and colocalizes tightly with the β-heavy–Spectrin/Kst subunit at the apical cortex and promotes Yki activity, F-actin enrichment, and the phosphorylation of the myosin II regulatory light chain Spaghetti squash. We propose a model in which the spectrin cytoskeleton recruits Bbg to the cortex, where Bbg promotes actomyosin contractility to regulate epithelial tissue growth.
Introduction
In animals, the fine regulation of cell numbers and cell size ensures that tissues and organs reach their correct size and shape to fulfill their function. Intercellular signaling pathways, and in particular their effects on the cell cycle and cell death programs, are key mediators to coordinate this harmonious growth. Hippo signaling represents one key pathway regulating tissue size and cell proliferation, in particular for epithelial tissues. First identified using genetic screens in Drosophila melanogaster, the core Hippo pathway is composed of two cytoplasmic kinases, Hippo (MST1/2 in humans; Harvey et al., 2003; Wu et al., 2003) and Warts (Wts; LATS1/2), which act to phosphorylate and exclude out of the nucleus the transcription coactivator Yorkie (Yki; YAP and TAZ). Mutations in hippo or warts lead to a stabilization of unphosphorylated Yki in the nucleus and overproliferation. Recent evidence points to a major role of the Hippo pathway in the biology of stem cells and as a tumor suppressor in many cancers (Halder and Johnson, 2011; Yu and Guan, 2013; Meng et al., 2016).
This core Hippo pathway integrates many upstream inputs, in particular from the apical scaffolds Expanded (Ex; FRMD6), Merlin (Mer; NF2; Hamaratoglu et al., 2006; Badouel et al., 2009; Yin et al., 2013), and Kibra (Kib; KIBRA; Baumgartner et al., 2010; Genevet et al., 2010; Yu et al., 2010) acting redundantly to recruit and activate the core kinases and to trap Yki in an inactive cortical compartment. Among the many other upstream activators of the Hippo pathway, the cell architecture and cytoskeletal tension have recently emerged as a key, but still poorly understood, regulator. Indeed, in epithelial cells, different members of the apical–basal polarity machinery such as Crumbs (Crb; Chen et al., 2010; Grzeschik et al., 2010; Ling et al., 2010; Robinson et al., 2010) modulate the activity and localization of the upstream scaffolds Ex, Mer, and Kib or directly impinge on Wts/LATS activity on Yki/YAP/TAZ (Schroeder and Halder, 2012; Yu and Guan, 2013). Different studies have highlighted the role of the actin cytoskeleton. Capped or destabilized actin filaments promote the sequestration of Yki/YAP/TAZ out of the nucleus, whereas filamentous actin polymerization and contractile actin networks favor Yki/YAP/TAZ activity (Dupont et al., 2011; Fernández et al., 2011; Sansores-Garcia et al., 2011; Gaspar and Tapon, 2014). Although the level of activated phosphorylated nonmuscle myosin II light chain (MLC; p-MLC) is critical for this effect of the actin cytoskeleton on Yki/YAP/TAZ activity, it is, however, not clear to what extent the core Hippo kinases cassette mediate this effect (Dupont et al., 2011; Codelia et al., 2014; Gaspar and Tapon, 2014).
Recent studies have described the negative role on Yki/YAP/TAZ activity of the spectrin-based cytoskeleton both in Drosophila and in human cell lines. Spectrin mutant cells accumulate Yki/YAP/TAZ in the nucleus and slightly overproliferate (Deng et al., 2015; Fletcher et al., 2015; Wong et al., 2015). Even though conserved across evolution, this role of spectrins must be tissue specific because not all spectrin mutant tissues in Drosophila show enhanced proliferation (Ng et al., 2016). Spectrins form an elastic submembranous network of heterotetramers of α and β subunits, which cross-links actin fibers (Bennett and Baines, 2001; Baines, 2009; Stabach et al., 2009). In Drosophila, there is one α and two β isoforms: β and β-heavy (βH; also known as karst [Kst]). In epithelial cells, α/β dimers are found basolaterally, whereas α/βH dimers are localized apically (Thomas and Kiehart, 1994; Lee et al., 1997; Thomas et al., 1998; Thomas and Williams, 1999; Zarnescu and Thomas, 1999). Spectrins prevent Yki activity in the eye and wing epithelial tissues, but the exact mechanism is still unclear and has been proposed to involve either the clustering and activation of the upstream Hippo activators as well as spectrin-binding partners Crb and Ex (Fletcher et al., 2015) or alternatively, the relaxation of the contractile actomyosin network and down-regulation of active p-MLC (Deng et al., 2015).
To better understand the regulation of epithelial cell numbers and proliferation, we have been interested in the role of apical scaffolds and, in particular, of PDZ domains containing scaffolds, as they represent attractive candidates to link apical–basal polarity cues, adhesion, and the cytoskeleton to integrate numerous stimuli. In this study, we report that Drosophila big bang (Bbg), a poorly characterized multi-PDZ scaffold (Kim et al., 2006) that had previously been linked to midgut homeostasis (Bonnay et al., 2013) and border cell migration in the adult Drosophila female egg chamber (Aranjuez et al., 2012), controls epithelial tissue growth without affecting epithelial polarity and architecture and that bbg mutant tissues are smaller with fewer cells. We show that Bbg binds to and colocalizes tightly with βH-Spectrin/Kst at the apical cortex, but contrary to spectrins, Bbg promotes Yki activity. Unlike spectrins, Bbg promotes the accumulation of a dense apical F-actin network and of activated p-MLC. We propose a model in which, through its stabilization of a contractile-prone actin cytoskeleton, Bbg positively regulates Yki activity and epithelial tissue growth.
Results
Bbg is an apical scaffold regulating epithelial cell numbers
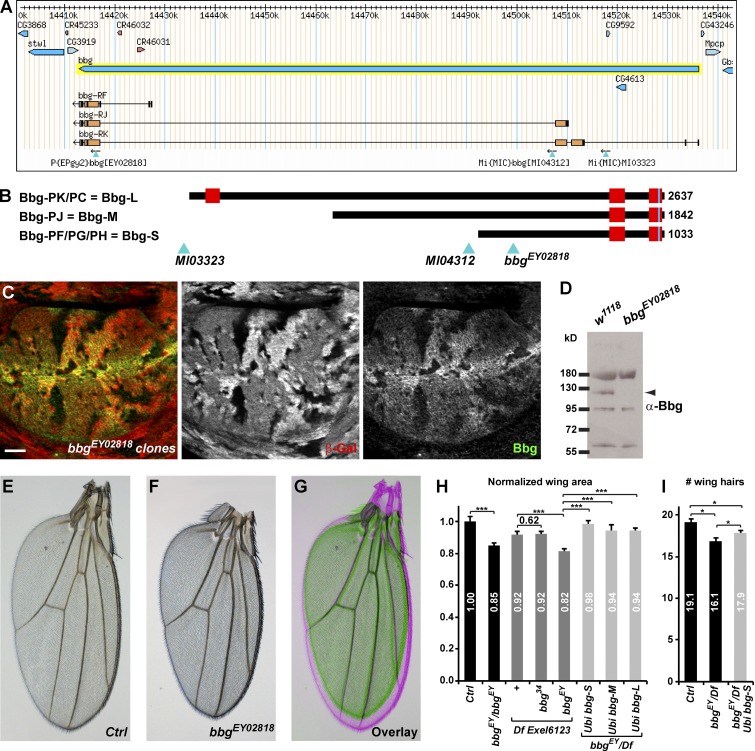
Bbg encodes for a large protein with two or three PDZ domains depending on isoform: a long isoform encoding a 2,637-aa-long protein with three PDZs, referred to as Bbg-L (Fig. 1 B) or short and intermediate isoforms coding for 1,033-aa-long and 1,842-aa-long proteins with only two PDZs, which are referred to as Bbg-S and Bbg-M, respectively (Fig. 1 B). PDZ domains are protein–protein interaction domains, suggesting that Bbg could act as a large protein scaffold. To study the function of bbg, we first raised an antibody directed at the last two PDZ domains shared by all Bbg isoforms (anti-Bbg). Bbg was expressed ubiquitously at the level of the apical cortex of epithelial cells. In third instar larval wing imaginal discs in particular, Bbg was expressed in all cells with a strong enrichment at the level of the dorsoventral boundary in the central wing pouch region. This staining was completely lost in clones of cells mutant for the transposable P element P{EPgy2}EY02818, inserted in a common exon to all bbg isoforms (Fig. 1 A), validating both the specificity of the anti-Bbg antibody and suggesting that the bbgEY02818 mutation is a protein null (Fig. 1 C). Western blot analysis on whole larval head extracts highlighted a specific band at ∼120 kD corresponding with the predicted size of the Bbg-S isoform (Fig. 1 D; note that this antibody also recognized a 60-, a 95-, and a 180-kD band, all of which were not specific).
Figure 1.
bbgcontrols adult Drosophila wing size. (A) A bbg locus (adapted from Flybase GBrowse) showing the main bbg transcript isoforms and the transposable elements used in this study. (B) Diagram representation of the main three Bbg protein isoforms. Red boxes represent PDZ domains. Light blue triangles indicate the localization of the transposable elements used with respect to the translated Bbg proteins. (C) bbgEY02818 mutant clones in the larval wing disc marked by the absence of β-galactosidase (red, middle). In mutant cells, Bbg protein was totally absent (green, right). Bar, 25 µm. (D) Western blot of cleared lysates from third instar larval heads of control (w1118; left) and bbgEY02818 (right) reveals a specific band at ∼120 kD for Bbg. (E–G) Effect of the bbg mutations on adult wing size. bbgEY02818 homozygous wings (F and green in G) were misshapen and smaller than controls (E and pink in G). (H) Quantifications of the bbg small wing phenotype expressed as ratios compared with the mean sizes (area) of the control (Ctrl) wings. The bbg34 allele had no effect. Providing ubiquitously expressed bbg transgenes (bbg-S, bbg-M, and bbg-L) in the bbgEY02818 mutant background significantly rescued the adult small wing phenotype. SD is shown; n = 18–20 independent female wings. (I) Quantification of wing cell numbers. Cells were estimated by counting the number of wing hairs along a vertical above the posterior cross vein between the L3 and L4 veins. SEM is shown; unpaired two-tailed Student’s t test; *, P < 0.05; ***, P < 0.001; n = 19 independent female wings.
After genetic background cleaning, the bbgEY02818 mutation was viable and fertile, but homozygous individuals exhibited smaller adult external structures such as wings or legs; bbgEY02818 mutant wings were ∼15% smaller than WT controls (Fig. 1, E–H). This effect was almost as strong as that of bbgEY02818 put over Df(3L)Exel6123, a genetically mapped deficiency covering the bbg locus (18% reduction), suggesting that bbgEY02818 behaved genetically as a strong hypomorph. Importantly, this was efficiently rescued by transgenes driving ubiquitously WT forms of Bbg (bbg-S–rescued wings were only 2% smaller than controls, and bbg-M– and bbg-L–rescued wings only 6% smaller; Fig. 1 H), showing that the small wing phenotype was indeed caused by mutations in the bbg gene. It is noteworthy that Df(3L)Exel6123 gave a dominant small wing phenotype of ∼8%, opening the possibility that the stronger phenotype of bbgEY02818 when put over the deficiency could be caused by additional haploinsufficient mutations.
Despite not being detected by Western blots, we tested whether the Bbg-M and Bbg-L isoforms could be implicated in bbg function. We therefore generated isoform-specific mutations using recombinase-mediated cassette exchange (Venken et al., 2011). We replaced the two MiMIC insertions flanking two large exons found only in the bbg-M and bbg-L isoforms (Fig. 1 A) with cassettes carrying a mini-white gene (for easier selection) and a loxP site. When put in trans together with a source of Cre recombinase, the two loxP sites recombined, generating a precise deletion of these two exons at high frequency (Fig. S1). However, the bbg mutant generated, called bbg34, appeared completely normal and fertile with no discernable external phenotype, suggesting that neither the bbg-L nor the bbg-M isoform had any noticeable role, at least on the viability or external morphology of Drosophila. Furthermore, when put over the Df(3L)Exel6123 deficiency, bbg34 did not produce smaller wings (Fig. 1 H), further validating that the bbg-L and bbg-M isoforms did not affect wing size. However, even though bbg-L and bbg-M are not necessary for the control of wing size, they appear sufficient because the Bbg-L and Bbg-M isoforms were still able to rescue the bbgEY02818 mutant small wing phenotype. Indeed, Bbg-M and Bbg-L contain all of the domains of Bbg-S and therefore likely could compensate for its absence.
Collectively, these results show that Bbg-S (referred simply as Bbg hereafter) is ubiquitously expressed in imaginal discs and controls the growth of wing tissues. To investigate whether these smaller adult wings could be reflecting fewer cells in the tissue, we estimated the number of cells along a line between the L3 and L4 veins above the posterior cross-vein. As each adult wing cell normally produces a single distally pointing wing hair, wing hair numbers can be used as a proxy to wing cell numbers. Along this fixed landmark, bbg mutant wings had fewer cells (16.1 on average) than controls (19.1). This was rescued by a WT bbg-S transgene (17.9; Fig. 1 I), suggesting that the bbg mutant small adult wings were at least in part a result of fewer cells being produced or retained.
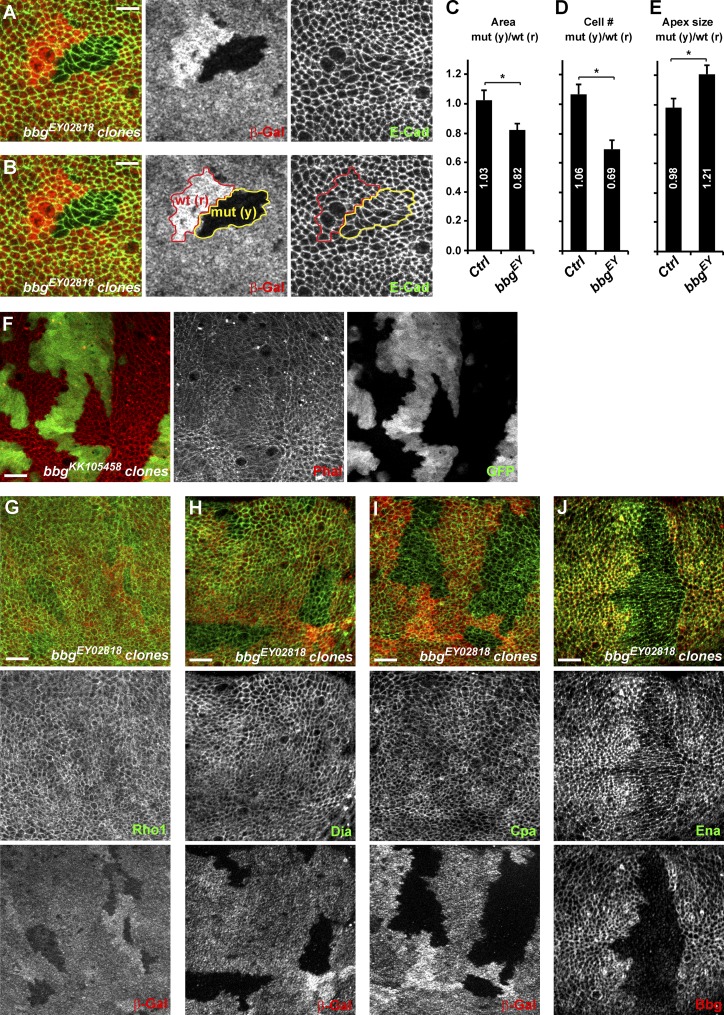
Using mitotic clonal analysis, we then investigated in the central pouch region of third instar wing discs whether at this early stage, bbg mutant clones had fewer cells than control (Fig. 2, A and B). Bbg mutant clones were on average 18% smaller than their WT twin spots when total area was considered (control empty clones revealed no difference with their twin spots; Fig. 2 C) and had ∼30% fewer cells (Fig. 2 D). The small size effect of bbg mutants appeared therefore more pronounced at earlier stages than in adult wings, suggesting that compensatory mechanisms could be playing after larval stages. Alternatively, this difference could also reflect the difference in the experimental systems used: whole mutant wing in the adult measurement, compared with small mutant clones competing with WT cells in the larval setup. Bbg mutant clones had fewer cells, but bbg mutant cells were wider and more apically relaxed than controls as their mean apical size (total clone area divided by number of cells) was ∼21% bigger (Fig. 2 E). Given that the apical size of cells varies widely in the larval wing disc depending on their position (more packed at the center and more stretched at the periphery), we always compared the size of equivalently located clones (bbg mutant with their twin spots) to eliminate this position bias.
Figure 2.
bbg controls cell number and cell size. (A and B) Mitotic clones for bbgEY02818 in the central region of third instar wing imaginal discs. Homozygous mutant clones are marked by the absence of β-galactosidase (β-Gal; red; middle panels) and highlighted by the yellow outline (mut (y)) in the right two panels in B. Twin spots, which were fully WT for bbg and arose at the same time as the bbg mutant clones, are marked by the bright β-Gal staining (two copies), and are highlighted by the red outline (wt (r)) in the middle and right panels in B. Apical cell outlines are revealed by E-Cad staining (green; right). Bars, 5 µm. (C–E) Quantification of the mean FRT80B bbgEY02818 mutant clone total area (C), cell number (D), and apical cell size (E) and shown as the mean ratio between individual mutant tissue clones (yellow) and their corresponding WT twin spots (red). Controls were “empty” clones performed with FRT80B. SEM is shown; unpaired two-tailed student t test; *, P < 0.05; n = 10 clone pairs from independent discs. (F) Overexpression clones marked by GFP (green, right) leading to the depletion of bbg and showing the organization of the cortical apical F-actin (red, middle). A strong decrease of apical F-actin was seen in bbg-depleted cells. (G–J) bbgEY02818 mutant clones in the larval wing disc marked by the absence of β-galactosidase (red, bottom) and marked for the actin regulators Rho1 (G; green, middle), Dia (H; green, middle), Cpa (I; green, middle), and Ena (J; green, middle). Cpa and Ena cortical levels were reduced in bbg mutant cells. Bars, 10 µm.
Bbg regulates cortical actin
The size of the apical surface is controlled by the network of actin cables that belt the apical cortex. Strikingly, in bbg-depleted cells (using the strong bbgKK105458 RNAi line), less filamentous cortical actin was detected, as shown by phalloidin staining (Fig. 2 F), suggesting that the defects in apical constriction could be a consequence of an impaired apical cortical actin cytoskeleton. We therefore investigated whether Bbg could be regulating the localization and levels of different actin regulators, focusing on the key actin cytoskeleton regulator and small GTPase Rho1 (Settleman, 2001), on the actin filament nucleators responsible for actin cables growth such as the formin diaphanous (Dia; Afshar et al., 2000), on the destabilizing factors such as capping protein α (Cpa; Fernández et al., 2011), which through their occupancy of the barbed end of actin cables prevent their further growth and ultimately lead to actin cable destruction, and on the capping protein inhibitor enabled (Ena; VASP in mammals; Bear and Gertler, 2009). All were localized at the apical cortex of wing imaginal disc cells. In bbg mutant cells, the level and localization of Rho1 were unaffected, suggesting that the actin filament defects are not a consequence of weakened or altered Rho1 recruitment at the cortex (Fig. 2 G). Similarly, in bbg mutant cells, the level and localization of Dia remained unaffected (Fig. 2 H), suggesting that the effects of bbg mutants on cortical F-actin accumulation are unlikely to be a consequence of a reduced Dia-mediated F-actin nucleation. A weak reduction in Cpa cortical levels could be observed in bbg mutant cells (Fig. 2 I). Because cpa loss leads to F-actin accumulation (Fernández et al., 2011; Sansores-Garcia et al., 2011), the F-actin reduction observed in bbg mutant cells cannot be explained by this weaker Cpa recruitment. However, even though not grossly affected by bbg mutations, more subtle interactions between cpa and bbg could be taking place. We thus tested whether the effects of cpa loss could be modified by bbg mutants. In the wing pouch of third instar larvae, cpa-depleted cells (through the use of the strong cpaKK108554 or cpaHMS02249 RNAi lines) transiently accumulated actin (increased phalloidin staining; Fig. S2 A) and were extruded basally out of the epithelia (Fernández et al., 2011). The further depletion of bbg imposed its phenotype on cortical actin, and instead of accumulating actin, cpa bbg double mutant cells showed less cortical actin (Fig. S2 B). However, similarly to cpa mutant cells, cpa bbg double mutant cells were also extruded out of the epithelia, and very few mutant cells were recovered in the wing pouch. Among the actin regulators tested, only Ena was affected, and bbg mutant cells had lower levels of cortical Ena (Fig. 2 J). Ena has been shown to regulate F-actin elongation in several Drosophila epithelia (Grevengoed et al., 2003; Gates et al., 2007), and it is tantalizing to propose that the effects of Bbg on cortical actin are mediated at least in part through Ena. However, we did not observe any obvious cortical F-actin alterations similar to those seen in bbg mutants by modulating Ena levels either through overexpression or RNAi-mediated knockdown (Fig. S2, C and D). These results, even though obtained with RNAi-hypomorphic combinations, suggest that the cortical F-actin fibers in bbg mutant cells are insensitive to Cpa or to the inhibitory role Ena plays on acting capping proteins (Bear and Gertler, 2009), suggesting that the F-actin stabilization by Bbg is likely mediated by other mechanisms than the modulation of the Cpa–Ena complex.
Bbg associates with the spectrin cytoskeleton
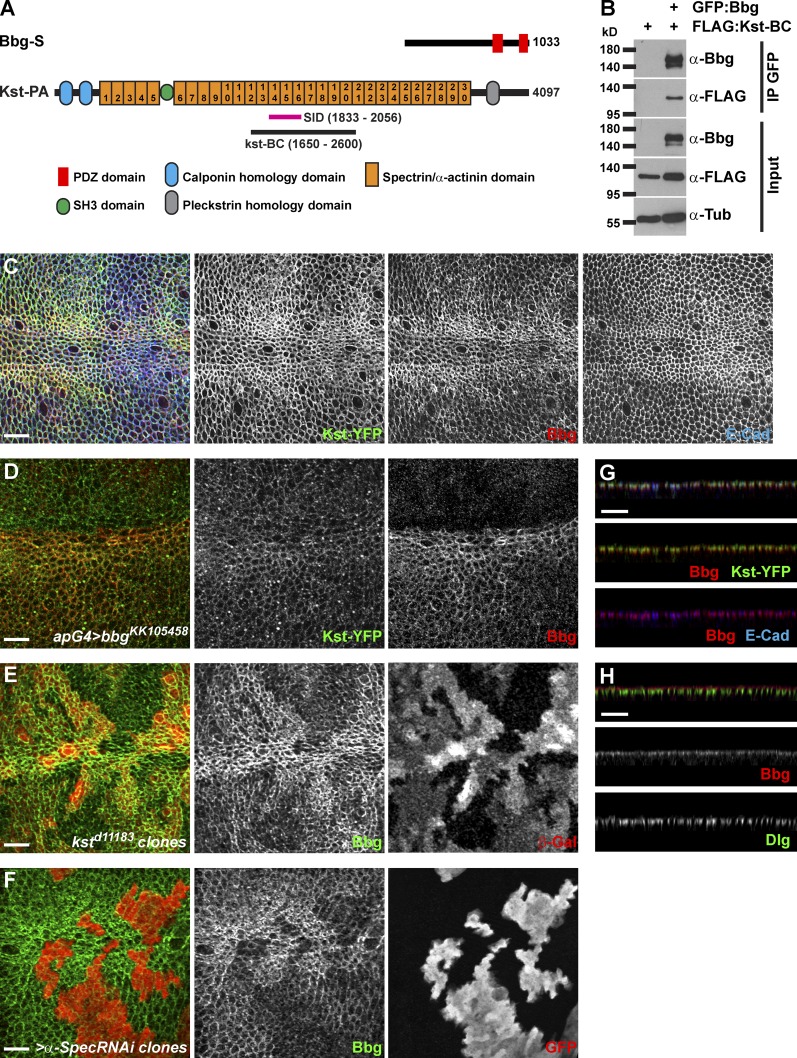
To identify the mechanisms by which bbg could be acting, we sought to identify the Bbg protein partners by performing whole-genome yeast two-hybrid (Y2H) screens using the Bbg-S isoform as a bait. Among the 177 Bbg-interacting clones recovered, 58 corresponded with βH-Spectrin, also known as Kst in Drosophila, and three clones corresponded with β-Spectrin. The spectrin cytoskeleton is made of tetramers of two α and two β spectrin chains that arrange in a lattice all around the cell cortex (Bennett and Baines, 2001; Baines, 2009; Stabach et al., 2009). In imaginal disc epithelial cells, there are two types of β chains: the basolateral β-Spectrin and the apical βH-Spectrin Kst (Thomas and Kiehart, 1994; Thomas and Williams, 1999; Zarnescu and Thomas, 1999). Kst was the top Bbg interactor in terms of number of prey clones recovered.
To validate these Y2H results, we performed coimmunoprecipitation (coIP) experiments in cultured Drosophila S2 cells. Kst is a huge 4,097-aa-long protein which is too big to work with in coIPs. The sequence shared by all the isolated Kst prey clones defined a potential minimal necessary interacting fragment or selected interaction domain (SID) spanning aa 1,833–2,056, centered around the spectrin/α-actinin repeat 15 (Fig. 3 A). We therefore generated a Flag-tagged Kst fragment encompassing the SID Kst-BC (aa 1,650–2,600; Fig. 3 A). In coIPs, Flag–Kst-BC was detected in the GFP immunoprecipitate only in the presence of GFP-Bbg, showing that Kst and Bbg interacted physically (Fig. 3 B).
Figure 3.
Bbg binds to the spectrin cytoskeleton. (A) Schematic diagrams of Bbg and the βH-Spectrin Kst structures. In pink is shown the minimal overlap of the Kst Y2H prey clones interacting with Bbg (SID). In black is shown the newly generated Kst-BC fragment used in coIP experiments in B. (B) The Kst-BC fragment coimmunoprecipitates with Bbg. Bottom: Western blot of cleared lysates from cells expressing either FLAG alone (negative control) or FLAG-tagged Kst-BC with GFP-tagged Bbg. Top: Western blot after immunoprecipitation (IP) with anti-GFP. FLAG-tagged Kst-BC was detected only when GFP-Bbg was present. (C) In larval wing imaginal disc epithelial cells, Bbg (red) colocalized with Kst (green). Apical cell junctions were revealed by E-Cad (blue). Note that both Bbg and Kst were found in a cortical ring just inside the membrane-bound belt of E-Cad. (D) The RNAi-mediated depletion of bbg in dorsal wing disc cells using the apterous-Gal4 driver (apG4; top part of the image) did not affect the localization of Kst (Kst-YFP; green, middle), whereas Bbg (red, right) was completely absent. (E and F) In kst mutant clones marked by the absence of β-Gal (E; red, right) or in clones expressing an α-Spec RNAi transgene marked by the presence of GFP (F; red, right), Bbg (green, middle) was only weakly recruited to the cortex. (G) Z section corresponding with the image in C and showing the overlap between Bbg (red, middle and bottom) and Kst-YFP (green, middle) or E-Cad (blue, bottom). (H) Z section showing that Bbg (red, middle) was localized apically to the septate junctions marker Dlg (green, bottom). Bars, 10 µm.
One possible role of this binding is that Bbg and Kst could regulate each other’s localization. In wing disc epithelial cells, Bbg accumulated as an apical cortical ring. In particular, along the dorsoventral midline, Bbg was found in a very thick submembranous compartment (Fig. 3 C) colocalizing very intimately with Kst, as indicated by the Kst-YFP fusion protein at the level of the adherens junctions (AJs; marked by E-cadherin [E-Cad]; Fig. 3 G), and apically of the septate junctions (marked by discs-large [Dlg]; Fig. 3 H). Given this colocalization, we then tested whether Kst and Bbg control each other’s localization. In bbg-depleted cells (using the strong bbgKK105458 RNAi line driven in all dorsal cells by apterous-Gal4), Kst localization and levels were normal or very subtly lowered (Fig. 3 D). Similarly, in bbg mutant cells, the levels and localization of apicobasal markers and scaffolds such as aPKC (subapical compartment), E-Cad, Arm, Pyd (AJs), or Dlg (septate junctions) were not affected (Fig. S3), indicating that the overall polarity of bbg mutant cells was not affected. In kst mutant cells, however, Bbg cortical localization was strongly reduced, even though it was not completely abolished, and low levels of Bbg were retained at the cortex (Fig. 3 E). α-Spec–depleted cells showed similar effects (Fig. 3 F), showing that not only Kst but the spectrin cytoskeleton was required for Bbg apical cortical localization. The residual cortical Bbg, observed even when the core α-Spectrin subunit was absent (Fig. 3 F), suggests that two pools of cortical Bbg exist: a pool associated with the spectrin cytoskeleton and a smaller pool of Bbg more tightly associated with the membrane and which might be recruited through alternative mechanisms such as interaction with other binding partners or with lipids (Bbg presents a putative palmitoylation site at its C terminus).
However, we note that despite its binding and its action to stabilize Bbg in a broad subcortical domain, Kst is unlikely to mediate the effect of Bbg on F-actin. Indeed, in contrast to bbg-mutant cells, kst mutant tissues had no changes in cortical F-actin (Fig. S2 E) as reported previously (Deng et al., 2015). Strikingly, in tissues mutant for kst and bbg, less cortical actin was detected (Fig. S2 F), further showing that even though Bbg and βH-Spectrin form a complex, the effect of Bbg on cortical F-actin is independent of Kst. Our observations do not rule out, however, that possible redundant mechanisms might account for the lack of effect of Kst on F-actin, but if they do, they are unlikely to be mediated by an effect on Rho1 or Cpa, because similarly to what we observed for bbg, in kst mutant cells, the levels and localization of Rho1 and Cpa were unaffected (Fig. S2, G and H).
Collectively, these results show that Kst and Bbg interact physically and that this interaction is responsible for the cortical apical ring accumulation of Bbg in Drosophila wing disc epithelial cells, but they suggest that the cortical Kst-independent pool of Bbg is the active pool controlling cortical actin accumulation.
Bbg, spectrins, and the Hippo pathway
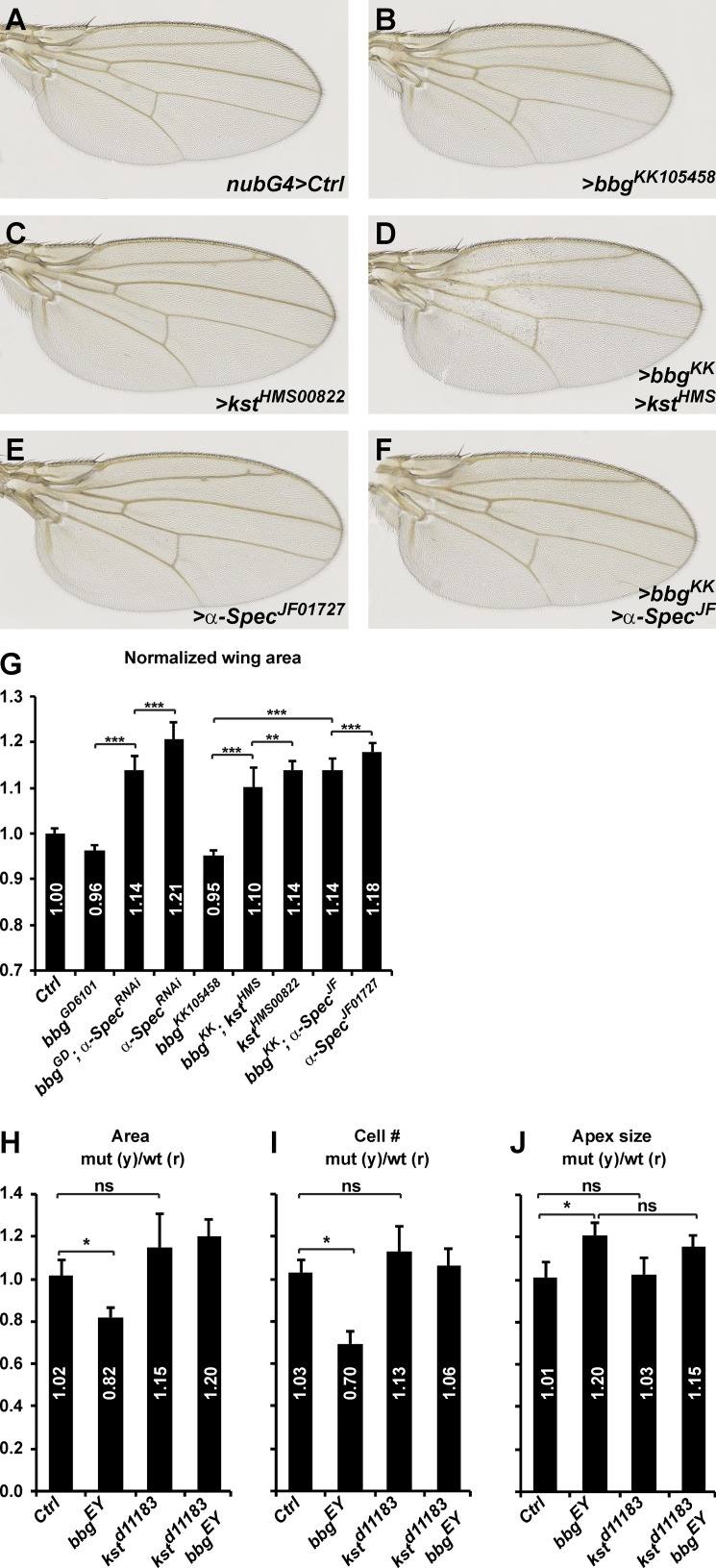
Apart from stabilizing apical cortical F-actin, Bbg also regulates wing tissue growth. We first sought to study the relationship between Bbg and spectrins in the control of wing growth. We thus recombined the two strong kstd11183 and bbgEY02818 mutations, which are both located on chromosome 3L. The strong hypomorph kstd11183 is pupal lethal, reflecting the pleiotropic role of the spectrin cytoskeleton. Unfortunately, the double mutant kstd11183 bbgEY02818 did not produce viable adults, and the relationship between Bbg and Kst on adult wing growth could only be studied using inducible localized RNAi-mediated knockdown, in this case in the whole wing pouch territory, with the nubbin-Gal4 driver. Similarly to bbgEY02818 mutations, bbgRNAi wings were smaller than controls (∼5% smaller; Fig. 4, A, B, and G). In contrast and as previously reported (Deng et al., 2015; Fletcher et al., 2015), kst or α-Spec RNAi-mediated knockdown generated bigger wings than controls (14–18% bigger; Fig. 4, A, C, E, and G). The roles of bbg and spectrins are therefore opposite. These 5–15% effects on wing growth by RNAi of bbg and spectrins are within the range of weak mutations in the Hippo pathway such as kibra (Fig. S4 A). We then investigated whether any of the effects would impose on the others and generated double RNAi lines. Lowering the dose of bbg in the context of impaired spectrin levels led to wing tissues that were smaller than spectrinRNAi alone but still bigger than bbgRNAi alone (Fig. 4, D, F, and G). Even though RNAi-based interactions could be difficult to interpret as they usually reflect partial knockdowns, these results suggest that bbg and spectrins act either at the same level or independently in the control of wing tissue growth.
Figure 4.
bbgcan modify the overgrowth of spectrin mutants. (A–F) Representative adult female Drosophila wings after nubbin-Gal4 (nubG4)-driven RNAi knockdown of the indicated genotypes. Lowering bbg dosage can modify (reduce) the overgrowth induced by kst or α-Spec RNAi. (G) Quantification of the effects shown in A–F. The wing area for each genotype is expressed as a ratio compared with the mean wing area of the nubG4>UAS GFP controls (Ctrl). SD is shown; unpaired two-tailed Student’s t test on raw data; **, P < 0.01; ***, P < 0.001; n = 17–22 independent female wings. (H–J) Quantification of the mean FRT80B mutant clone total areas (H), cell numbers (I), and apical cell sizes (J) of the indicated genotypes shown as the mean ratio between individual mutant tissue clones and their corresponding WT twin spots as shown in Fig. 2. Controls are “empty” clones performed with FRT80B. SEM is shown; unpaired two-tailed Student’s t test; *, P < 0.05; n = 10 clone pairs from independent discs.
But although the kstd11183 bbgEY02818 double mutant chromosome could not be used to monitor adult wing size and morphology, we could recover double mutant clones in third instar wing discs. Although bbg mutant clones were smaller and possessed fewer cells than their twin spots, kst mutant clones were slightly larger and possessed slightly more cells, even though this failed to reach statistical significance (Fig. 4, H and I). Interestingly, double mutant clones were also slightly larger, with slightly more cells. Finally, we did not detect any difference in the apical size of kst mutant cells (Fig. 4 J), suggesting that Kst does not control apical constriction. This is reminiscent of the normal apical cortical F-actin belt seen in kst mutant cells (Fig. S2 E). Strikingly, in kst bbg double mutant cells, the apex was enlarged, reminiscent of the bbg phenotype and of the impaired cortical actin of kst bbg double mutant cells (Fig. S2 F).
Collectively, these results suggest that even though spectrins (Kst and α-Spec) are required for the apical cortical enrichment of Bbg, Kst and Bbg have separable functions regarding cortical actin enrichment and apical constriction. Furthermore, Kst and Bbg have opposite effects on wing tissue growth and appear to act either independently or at the same level, converging on a common growth regulator. Recent studies demonstrated that in Drosophila and in human cell lines, the spectrin cytoskeleton controls epithelial growth by antagonizing the activity of the proproliferation and antiapoptosis transcription coactivators Yki and YAP (Deng et al., 2015; Fletcher et al., 2015; Wong et al., 2015). In Drosophila, mutant wing tissues for α-Spec or for βH-Spectrin exhibit an elevated Yki activity and grow larger than controls. Similarly, human cell lines with impaired SPECTRINS exhibit reduced inactive phosphorylated YAP levels.
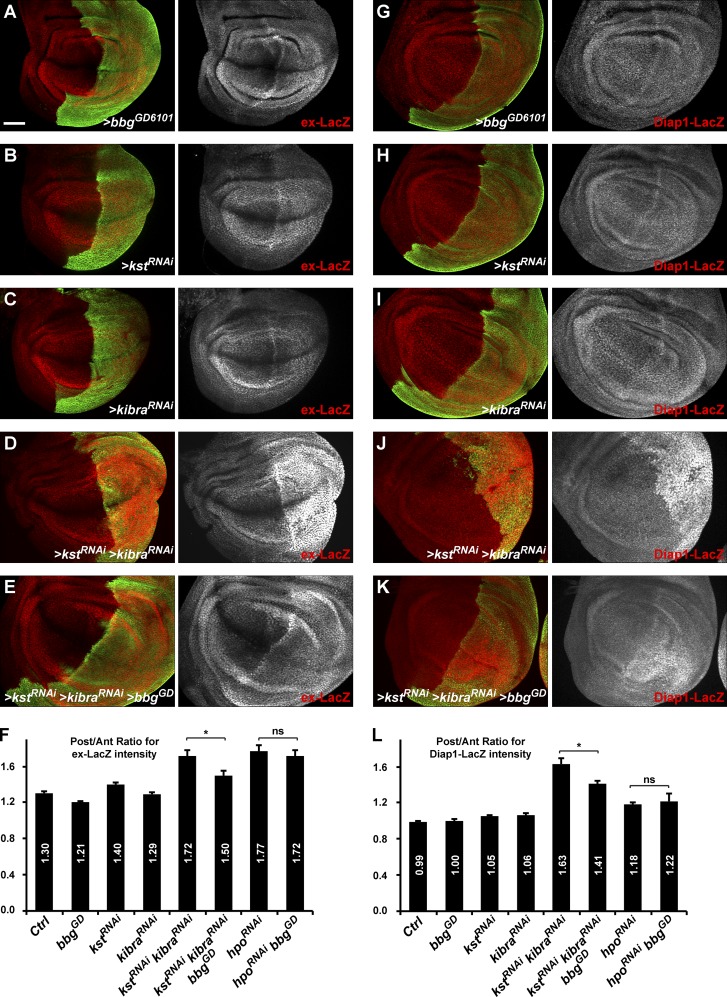
We thus investigated whether the spectrin-binding Bbg could also be controlling Yki activity and whether bbg and spectrins have opposite effects as suggested by their adult wing phenotypes (smaller and bigger than WT, respectively; Fig. 1, E and F; Deng et al., 2015; Fletcher et al., 2015). When driven in the posterior part of the wing disc (under the control of the hh-Gal4 driver), the RNAi-mediated knockdown of bbg (Fig. 5, A and F) led to a subtle decreased expression of the Yki activity reporter ex-LacZ (Harvey and Tapon, 2007). Conversely, a slight increase in ex-LacZ reporter activity was observed when kst was impaired (Fig. 5, B and F). These effects were subtle and could not be observed for Diap1-LacZ, another Yki activity reporter (Wu et al., 2008). Although the effects observed were very modest, these results suggest that bbg acts positively on Yki activity. To better monitor the effects of bbg on these Yki-activity reporters, we used two sensitized backgrounds in which Yki-activity was enhanced, thus allowing for a better detection of a putative decreased activity as predicted by the bbg adult wing phenotype. Such sensitized backgrounds have been helpful in revealing the role of other Hippo pathway regulators such as crumbs or spectrins on ex-LacZ levels (in combination with kibra; Ling et al., 2010; Fletcher et al., 2015).
Figure 5.
bbg controls Yki activity. (A–E and G–K) Pouch region of third instar larva wing imaginal discs. Effects of hh-Gal4–driven RNAi knockdown in the posterior compartment marked by GFP (green) on the expression of the Yki activity reporters ex-LacZ (A–E, red) and Diap1-LacZ (G–K, red). The expression of ex-LacZ or Diap1-LacZ was almost not affected when bbg (A and G), kst (B and H), or kibra (C and I) are knocked down individually. Although a strong Yki activity was detected after double RNAi for kst and kibra (D and J), this was partially suppressed by bbg RNAi (E and K). Bar, 50 µm. (F and L) Quantifications of the effects presented in A–E for ex-LacZ (F) and in G–K for Diap1-LacZ (L). Results are presented as means of ratios calculated for each experimental disc between the mean gray intensity (reflecting reporter expression) of the posterior compartment (GFP positive) and the mean gray intensity of the corresponding anterior compartment. Note that in control discs, the expression of ex-LacZ was stronger in the posterior compared with the anterior. SEM is shown; unpaired two-tailed Student’s t test; *, P < 0.05; n = 10–13 independent discs.
Indeed and as reported previously (Fletcher et al., 2015), in a kibraRNAi-sensitized background, the RNAi-mediated knockdown of kst promoted a robust increase in the expression of both ex-LacZ and Diap1-LacZ reporters (note that in the conditions used, kibraRNAi alone did not have any significant effect; Fig. 5, C, D, I, and J). Interestingly, this effect of the double kstRNAi kibraRNAi was suppressed by coexpressing an RNAi construct for bbg (Fig. 5, E, F, K, and L), further showing that bbg acts positively on Yki activity either downstream or in parallel with kst. Importantly, even if the spectrin cytoskeleton stabilized a large pool of Bbg in an apical cortical ring, a small pool of Bbg remained tightly associated with the plasma membrane in spectrins mutants, which was further affected by bbgRNAi in these experiments. However, bbg knockdown did not have any effect on the up-regulation of both ex-LacZ and Diap1-LacZ reporters induced by the knockdown of the core kinase hippo (hpoRNAi; Fig. 5, F and L; and Fig. S4, B–E). These results therefore suggest that Bbg promotes Yki activity during the growth of larval wing discs but that its action lies at the level of the upstream regulators of the Hippo pathway such as Kib or spectrins and not downstream of the core kinases.
Bbg does not affect upstream Hippo pathway scaffolds
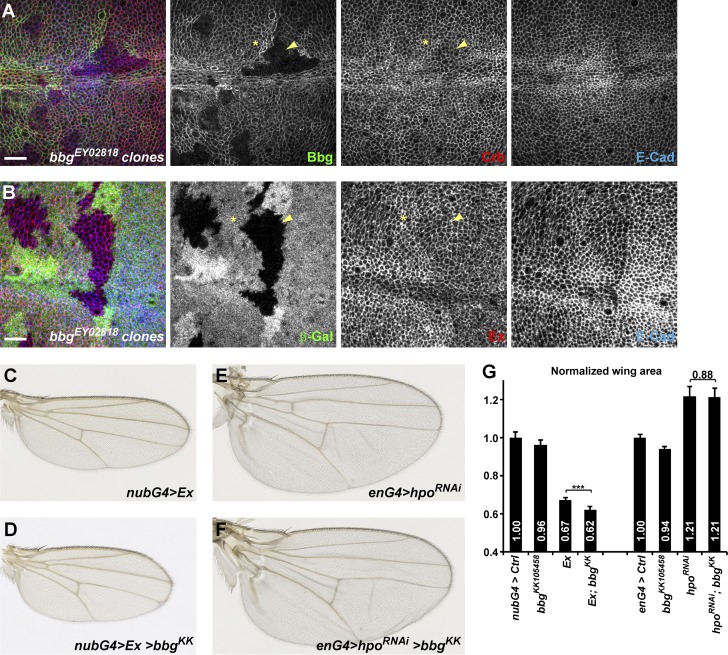
Even though the effects of bbg mutations or knockdown on total wing size were modest, they were nevertheless within the range of weak mutations in the Hippo pathway such as kibra (see Fig. 5 [C and I] on ex-LacZ and Diap1-LacZ expressions; see also overall wing size in Fig. S4 A). Collectively, these results suggest that Bbg, contrary to spectrins, promotes Yki activity and wing growth. We thus sought to identify by which mechanisms Bbg could be acting. Given that Bbg is an apical cortical scaffold affected in kst mutant tissues, we first investigated whether the localization of the two βH-Spectrin binding partners and Hippo pathway apical regulators Crb (Thomas et al., 1998; Zarnescu and Thomas, 1999; Médina et al., 2002; Pellikka et al., 2002) and Ex (Hamaratoglu et al., 2006; Maitra et al., 2006; Badouel et al., 2009; Fletcher et al., 2015) could be affected in bbg mutant tissues. In bbg mutant clones, the correct apical localization of neither Crb nor Ex was affected (Fig. 6, A and B). If anything, we could only detect in a few clones a very subtle down-regulation of Crb or Ex (compare stars and arrowheads in Fig. 6, A and B). However, this is not consistent with the positive role of Bbg on Yki because such a role predicts that bbg mutant tissues should exhibit an accumulation of upstream Hippo pathway activators. It appears therefore that Bbg does not control the localization of upstream Hippo pathway activators such as Crb or Ex. Furthermore, if the role of Bbg was to inhibit the activity rather than the localization of apical scaffolds, one would predict that a potent overexpression of these scaffolds would bypass this inhibition and that reducing bbg expression would not have any effect on wing size. Using genetic interactions, we could show that although nubbin-Gal4–driven overexpressed ex led to very reduced adult wings (33% reduction compared with controls; Fig. 6, C and G), the loss of bbg by RNAi produced only slightly reduced wings (4% reduction; Fig. 6 G). Strikingly, combining both led to even smaller wings in an additive fashion (Fig. 6, D and G), suggesting that ex and bbg act either independently or at the same epistatic level to control growth. We then investigated whether bbg could be branching on the Hippo pathway further downstream at the level of the core kinases. Although the loss of the hippo kinase by RNAi produced enlarged wings (21% increase compared with controls; Fig. 6, E and G), this effect could not be modified by combining it with bbgRNAi, reminiscent to what was seen with the Yki activity reporters ex-LacZ and Diap1-LacZ (Figs. 5 F and S4, B–E), further confirming that Bbg acts in parallel or downstream of the Hippo pathway upstream regulators (such as Kib, Ex, or Kst) but upstream of the core kinase Hippo to regulate wing tissue growth.
Figure 6.
The upstream Yki negative regulators Crb and Ex are not affected in bbg mutants. (A and B) In bbg mutant clones stained for E-Cad (blue) and marked by the absence of Bbg (A, red) or β-Gal (B, red), the localization and levels of Crb (A, green) or Ex (B, green) were not affected or weakly reduced (compare yellow arrowheads [mutant] with yellow asterisks [WT]). Bars, 10 µm. (C and D) Genetic interaction between bbg and ex. Although overexpressed ex under the control of the nubG4 driver led to strongly reduced wings (C), this effect was further enhanced when bbg was knocked down by RNAi (D). (E and F) Genetic interaction between bbg and hpo. Although the knockdown of hippo by RNAi under the control of the engrailed-Gal4 (enG4) driver led to strongly enlarged wings (E), this effect could not be modified by further impairing bbg (F). (G) Quantification of the effects shown in C–F. The wing area for each genotype was expressed as a ratio compared with the mean wing area of controls (nubG4>UAS GFP controls on the left, and enG4>UAS GFP controls on the right). SD is shown; unpaired two-tailed Student’s t test on raw data; ***, P < 0.001; n = 17–22 independent female wings.
Bbg controls MLC phosphorylation
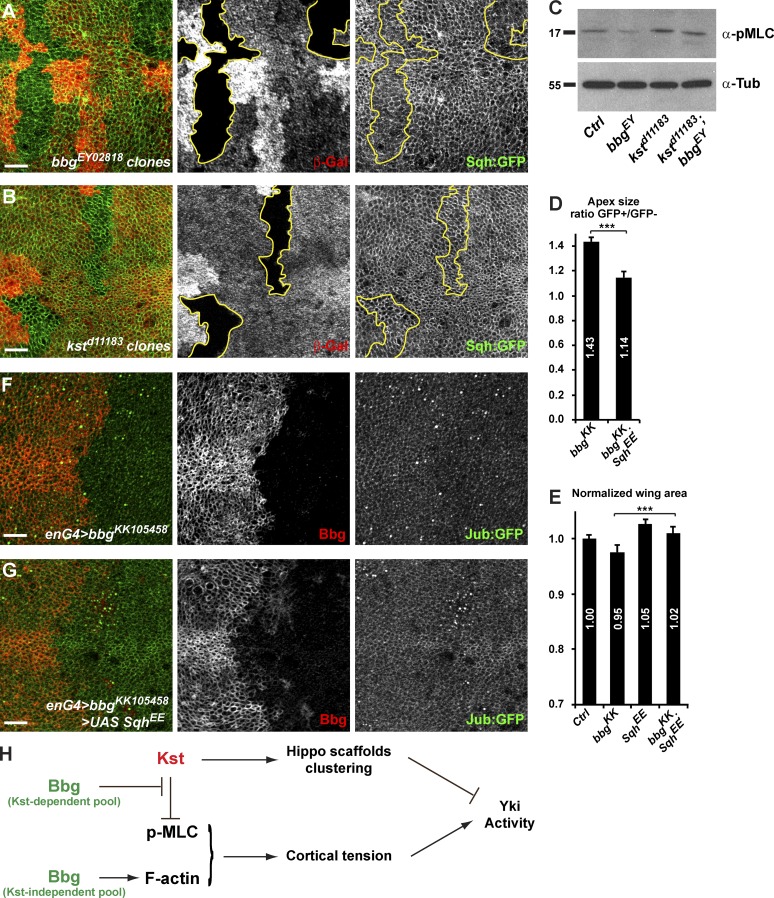
Besides apical scaffolds, the actin cytoskeleton, and in particular its tension, has recently been implicated in the control of Yki activity in growing epithelial tissues, and spectrins have been proposed to inhibit Yki activity by preventing the phosphorylation of the nonmuscle MLC (also known as Spaghetti squash; Sqh), the major regulator of actin cytoskeleton contractility (Rauskolb et al., 2014; Deng et al., 2015). In bbg mutant tissue, less filamentous cortical actin was detected, as shown by phalloidin staining (Fig. 2 F). We thus monitored the levels and activity of MLC in Drosophila wing discs using a Sqh-GFP fusion protein driven by the endogenous sqh promoter because there are no good antibodies to follow the endogenous Sqh protein (sqh>sqh-GFP; Martin et al., 2009). In bbg mutant cells, there was no gross effect on Sqh-GFP (Fig. 7 A; the weak reduction of apical Sqh-GFP that could be seen in bbg clones could reflect that bbg mutant cells were apically larger). As previously reported, we could not detect any changes in Sqh-GFP recruitment in kst mutant cells (Fig. 7 B; Deng et al., 2015). The activity of MLC is controlled by phosphorylation by a range of kinases including Rho-kinase (ROK), and p-MLC promotes contractility of the actin cytoskeleton (Rauskolb et al., 2014). We therefore also monitored the activity level of MLC using an anti–p-MLC antibody that cross-reacts in Drosophila on whole-disc protein extracts. Consistent with previously published results, p-MLC levels were increased in kst mutants (Fig. 7 C; Deng et al., 2015). Opposite to kst, in bbg mutant discs, p-MLC levels were decreased compared with WT controls. Strikingly, in kst bbg double mutant discs, p-MLC levels were also slightly increased, mimicking what was seen in single kst mutants (Fig. 7 C). These results show that Bbg promotes p-MLC accumulation and suggest that Kst might be acting downstream of Bbg, compatible with a model where Bbg prevents the destabilizing effect of Kst. These results suggest that the sparse actomyosin network in bbg mutants could be less amenable to contractile behavior. Indeed, when providing a constitutively activated version of Sqh (SqhEE, mimicking the activating phosphorylation of Sqh by ROK), bbg mutant cells were able to apically constrict again (Fig. 7 D), and the overall undergrowth effect of bbg knockdown on the adult wing size was suppressed (Fig. 7 E).
Figure 7.
Bbg controls p-Sqh levels and Jub cortical recruitment. (A and B) bbg (A) or kst (B) mutant clones (outlined in yellow) marked by the absence of the β-Gal marker (red) and the apical accumulation of the MLC regulatory subunit Sqh (Sqh-GFP; green). (C) Western blot of cleared lysates from third instar larval homozygous mutant wing discs reveals a decrease in p-Sqh (anti-pMLC) levels in bbg mutants (0.76 ± 0.20 times), whereas the levels were up-regulated in kst (1.44 ± 0.24) and kst bbg double mutants (1.51 ± 0.23). Tubulin (anti-Tub) was used as a loading control and for normalization. Molecular masses are given in kilodaltons. (D) Quantification of the mean apical cell size in overexpression clones for bbgRNAi (left) and for bbgRNAi with a constitutively active form of Sqh (SqhEE; right) and shown as the mean ratio between GFP-positive tissues (overexpressing) and GFP-negative tissues (nonexpressing controls) in individual experimental wing discs (see the Cell numbers and apical cell size section of Materials and methods). SEM is shown; unpaired two-tailed Student’s t test; n = 12 area pairs from independent discs. (E) Quantification of the wing area after SqhEE overexpression. Each genotype is expressed as a ratio compared with the mean wing area of the nubG4>UAS GFP controls (Ctrl). SD is shown; unpaired two-tailed Student’s t test based on raw data; ***, P < 0.001; n = 17–20 independent female wings. (F and G) Effect of bbg depletion marked by the absence of Bbg (red) on the apical recruitment of the Hippo-negative regulator Jub (Jub-GFP; green). Cells depleted for bbg (F) had slightly lower levels of Jub-GFP. This effect was suppressed when an activated form of Sqh (SqhEE) was coexpressed (G). Bars, 10 µm. (H) Model for the action of Bbg on the regulation of actin and growth in the Drosophila wing imaginal discs. Bbg bound to the spectrin cytoskeleton (Kst) accumulates at the cortex of epithelial cells, where it opposes the effect of Kst on MLC phosphorylation. Bbg, through its role on the actin cytoskeleton, promotes cortical tension and Jub recruitment, negatively impinging on the Hippo signaling pathway to promote proper growth of the tissue.
These results support a model in which Bbg promotes a contractile actomyosin network at the cortex of wing epithelial cells through the stabilization of cortical F-actin and activated p-MLC. However, because kst bbg double mutant cells are still as apically relaxed as bbg mutant cells (Fig. 4 J) even though they appear to have higher p-MLC levels, the simple enrichment in activated Sqh is not the only factor controlling apical cell size. The architecture and levels of F-actin might also be critical. More studies are needed to better understand the respective contributions of F-actin, of p-MLC, and of their localization within epithelial cells on apical constriction.
Recently, a tensile actin–myosin network has been shown to promote the recruitment of the Hippo pathway–negative regulator and LIM domain–containing scaffold Ajuba (Jub). This actin-dependent Jub accumulation has been proposed to inhibit the Hippo pathway and increase Yki activity (Rauskolb et al., 2014) at least in part through the binding and inhibition of Wts (Das Thakur et al., 2010). We therefore investigated whether the positive effect of Bbg on Yki could be mediated through Jub relocalization. Indeed, the RNAi-mediated knockdown of bbg in the posterior wing compartment (using the engrailed-Gal4 driver) led to a reduction of Jub cortical recruitment in the posterior cells as evidenced using a Jub-GFP fusion protein (Fig. 7 F). This effect was suppressed by providing a constitutively active Sqh (SqhEE; Fig. 7 G), showing that experimentally promoting a more contractile actomyosin network in bbg-depleted cells is sufficient to restore Jub-GFP recruitment. Therefore, we propose that by stabilizing F-actin and phosphorylated active Sqh at the apical cortex, Bbg promotes Yki activity in wing discs at least in part through the local recruitment of Jub (Fig. 7 H) to sustain correct tissue growth. Furthermore, our results highlight a critical role for Bbg in controlling F-actin apical enrichment independently of Kst or of the Cpa–Ena capping complex and support a model where the pool of Bbg that is apically recruited by Kst acts there to inhibit the destabilizing role of Kst on p-MLC (Fig. 7 H).
Discussion
Investigating the role of PDZ-containing protein scaffolds, we describe in this study a new role for the previously reported bbg gene in regulating epithelial wing disc cell numbers. Even though our studies were primarily focused on wing imaginal discs, bbg must exert its role in other epithelial tissues such as imaginal leg discs because bbg mutant legs are ∼10% shorter than WT controls. Performing an unbiased Y2H screen, we show that the spectrin cytoskeleton, and in particular the βH-Spectrin subunit Kst, binds and stabilizes Bbg at the apical cortex. But even though spectrins recruit Bbg, Bbg plays an opposite role to that of spectrins on tissue growth and cell number control, and contrary to spectrins, Bbg promotes p-MLC enrichment and Yki activity. Based on detailed phenotypic analysis of bbg mutant cells, our work supports a role for Bbg independent of spectrins to stabilize a dense actomyosin network, promoting the recruitment of the Hippo pathway–negative regulator Jub, which was shown to then inhibit Wts activity (Rauskolb et al., 2014). Given the emerging link between actin dynamics and contractility and the regulation of the Hippo pathway (Dupont et al., 2011; Fernández et al., 2011; Sansores-Garcia et al., 2011; Yu and Guan, 2013; Gaspar and Tapon, 2014), we propose that through its role on the actomyosin cytoskeleton, bbg ultimately impinges on the activity of the terminal Hippo effector Yki to fine-tune epithelial cell numbers.
Spectrins and Bbg
The relationship between Kst and Bbg appears therefore complex. Kst binds and helps to recruit Bbg at the apical cortex, suggesting that Bbg could be acting downstream of the spectrin cytoskeleton. But even though spectrins stabilize Bbg at the cortex, Bbg and spectrins have opposite roles on overall wing growth and cell number. Furthermore, despite their physical association, Bbg acts independently of Kst to promote the apical enrichment of F-actin and more modestly of the myosin II regulatory subunit Sqh.
However, although Kst and Bbg have different roles on cortical actin, they converge to control the phosphorylation and activity of Sqh, albeit in opposite ways. Indeed, our study shows that Bbg promotes p-Sqh accumulation (Fig. 7 C), and previous research has shown that Kst prevents p-Sqh upstream of ROK (Deng et al., 2015). These opposite effects on p-Sqh are reminiscent to the opposite phenotypes of bbg and kst mutations on adult wing size (smaller and bigger, respectively). Given the one-way link between Kst and Bbg, namely that Kst controls Bbg accumulation, whereas Bbg has no effect on Kst (Fig. 3 E), one possible model is that through its binding and recruitment in a broad cortical ring, the Kst-trapped Bbg inhibits the negative role of Kst on p-Sqh. Indeed, we note that in kst or α-Spec mutant cells, some Bbg protein is still present at the cortex, suggesting that at least two pools of Bbg exist and that other mechanisms act in parallel with spectrins to promote Bbg cortical localization. Alternatively, the effect of Kst on Bbg localization could be completely independent from the action of Kst on tissue growth, and despite their physical interaction, Kst and Bbg could be acting through independent mechanisms converging on Sqh phosphorylation.
It appears therefore that the opposite effects of Bbg and Kst could be explained, at least in part, by their opposite effects on p-Sqh accumulation and contractility of the apical cortical actin network and their final effect on Yki activity: positive for Bbg and negative for Kst. We propose that the modification of Yki activity in kst mutants by the bbg mutation as reported by the ex-LacZ and Diap1-LacZ reporters reflects a modulation of the actin contractility (total F-actin levels and p-Sqh) by combining these two mutations.
Bbg stabilizes both apical F-actin and total p-Sqh levels, promoting a more contractile actomyosin cytoskeleton, a more constricted cellular apex, and the recruitment of the Wts inhibitor Jub. It is noteworthy that, even though Kst has been proposed to regulate Sqh activity, Kst does not appear to control Yki activity through Jub and Wts membrane recruitment (Deng et al., 2015) as would be predicted if Kst were indeed modifying actin cytoskeleton tension (Rauskolb et al., 2014). Alternatively, Kst has been proposed to prevent Yki function independently of actin regulation through the clustering of Hippo pathway upstream activators such as Crb (Fletcher et al., 2015). Future studies will help clarify some of these seemingly contradictory observations arising in part from the high degree of functional redundancy of the Hippo pathway upstream activators and the modular nature of the scaffolds involved. We note here that bbg cannot modify the hpo mutants, which could suggest that the actin-mediated regulation of Yki activity could be independent of the core Hippo pathway. Indeed, the role of the core Hippo and Wts kinases in response to mechanical stress or cytoskeletal changes is still unclear (Gaspar and Tapon, 2014). Alternatively, it could reflect the fact that core Hippo pathway members exhibit effects that are too strong to be modified. Future studies will help clarify whether the actin cytoskeleton influence on Yki is independent or not of the core Hippo pathway.
Bbg and cortical actin regulation
Even though regulating actin, Bbg does not have any obvious actin-binding domain. However, spectrins can bind to F-actin (Baines, 2009), but the role of Bbg on actin is unlikely mediated by the apical spectrin cytoskeleton because the organization or stability of F-actin fibers were unaffected in kst mutants even though Bbg levels and localization were strongly reduced (Thomas et al., 1998; Deng et al., 2015; this study). We observed, however, that a small fraction of Bbg was still present just under the membrane, suggesting that this small fraction of Bbg, which localizes independently of spectrins, is sufficient for Bbg-mediated cortical actin organization.
Among the different actin organizers that we could test, we could only detect a weak down-regulation of Ena in bbg mutant cells, suggesting that the capping protein inhibitor Ena/VASP could be implicated. During border cell migration, cortical Wts has been shown to phosphorylate and inhibit Ena activity, leading to a polarized and functional actin cytoskeleton (Lucas et al., 2013). It is therefore possible that through the recruitment of Jub and cortical inhibition of Wts (Rauskolb et al., 2014), Bbg promotes Ena enrichment. However, modulating Ena levels did not produce F-actin disruptions similar to those observed in bbg mutant cells, and the effect of Ena on cortical F-actin in wing disc cells must be more subtle. Furthermore, the enrichment in F-actin observed in cpa-deficient cells was completely suppressed by mutations in bbg. These striking observations suggest that the elongated actin filaments in cpa mutants require an intact bbg function to accumulate apically in epithelial cells. Further studies are therefore required to clarify the complex link between Bbg, spectrins, and cortical F-actin stabilization along with how this fine-tunes Yki activity.
A general role for Bbg
bbg mutants were originally described as more sensitive to knocking disturbances (“bang;” Kim et al., 2006), suggesting that they could have neuronal functions in sensing or responding to that stimulus. Bbg has also been implicated in gut immune tolerance, and bbg mutant flies die prematurely in cases of bacterial infection with signs of weakened gut epithelial barrier (Bonnay et al., 2013). Finally, a targeted RNAi screen identified bbg as a key regulator of border cell migration in the adult female egg chamber, with bbg-knockdown cells migrating slower than WT (Aranjuez et al., 2012). All these different phenotypes could be explained by subtle defects in the actin cytoskeleton, and it would be interesting to revisit these earlier studies in light of our results. In particular, both the spectrin cytoskeleton (Lee et al., 1997; Zarnescu and Thomas, 1999) and the Hippo pathway (Lucas et al., 2013) have been implicated in the regulation of border cell migration, which could represent a suitable system to study further the complex regulations between spectrins, actin dynamics, apical scaffolds, and the Hippo pathway.
Materials and methods
Drosophila genetics
The viable bbg34 allele was generated by exchanging the MiMIC cassettes to introduce loxP sites in the same orientation in the two MiMIC lines Mi{MIC}MI03323 and Mi{MIC}MI04312 that flank the two large 5′ exons coding for specific regions for the long (bbg-RC and bbg-RK) and medium (bbg-RJ) bbg isoforms. After providing a pulse of Cre recombinase, the precise deletion of these two exons was followed by PCR on genomic DNA (see Fig. S1).
The viable bbgEY02818 allele is a P-transposable element P{EPgy2}bbgEY02818 and was cleaned of an unrelated lethal mutation by six rounds of back crossing to a WT stock. Mapping of this P-element insertion was then confirmed by PCR on genomic DNA to disrupt the large 3′ exon common to all bbg isoforms.
All bbg alleles (bbg34 and bbgEY02818) were then recombined on FRT80B chromosomes to perform mitotic clonal analysis. The bbg loss-of-function phenotype was obtained either as bbgEY02818 homozygous or by crossing bbgEY02818 with the Df(3L)Exel6123 spanning the bbg locus.
The full-length bbg coding sequence corresponding with the long (bbg-RK), medium (bbg-RJ), or short (bbg-RF) isoforms were amplified by PCR as four different fragments and later reassembled by using conveniently located unique restriction sites using as template either the partial cDNA RE18302, newly generated random primed cDNAs from wing discs, or the BACPAC clones CH322-2L24 and CH322-132B5. These bbg isoforms were then cloned in the pKC26w-pUbiq rescue plasmid that allows expression of the cloned fragments under the ubiquitous ubiquitin promoter. All transgenes were inserted at the same chromosomal location (51C) using phiC31-mediated integration on the M{3xP3-RFP.attP}ZH-51C landing platform.
RNAi-mediated knockdown experiments were performed using the nubbin-Gal4 (all cells in the pouch central region of third instar wing discs), hedgehog-Gal4 or engrailed[e16E]-Gal4 (posterior compartment of the wing disc), apterous-Gal4 (dorsal wing disc), or randomly generated flip-out clones (using the y-, hsFLP122; Act FRT y+ FRT Gal4, UASt GFP line) to drive the expression of different RNAi-containing transgenes under upstream activation sequence (UAS) control. Crosses were usually cultivated at 30°C to allow maximum RNAi activity. The bbg RNAi lines used were the bbgGD6101 and bbgKK105458 from the Vienna Drosophila Research Centre, which were shown previously to promote very efficient bbg knockdown (Aranjuez et al., 2012). Genomic PCR following published guidelines (Vissers et al., 2016) showed that the bbgKK105458 contained only one insertion on platform 30B, and therefore was devoid of the reported “tiptop effect.”
Mutant clones were generated at high frequency in the wing using abxUbxFLPase in combination with FRT80B bbg34, FRT80B bbgEY02818, or FRT80B kstd11183.
Bbg antibody generation
The sequences corresponding with aa 2,317–2,637 of the Bbg-PK protein (IVEEADPP…GPVKICFA) were cloned in frame with the glutathione S-transferase (GST) gene in the pGEX–6P-1 expression vector. The GST fusion protein was produced in Escherichia coli bacteria grown at 30°C after induction with 1 mM IPTG and purified on glutathione agarose beads (Thermo Fisher Scientific) before being eluted with an excess of free reduced glutathione (50 mM NaH2PO4, 300 mM NaCl, 1 mM DTT, 50 mM glutathione reduced, 1× cOmplete protease inhibitors [Roche], and 0.1% Triton X-100, pH 8). Rabbits were immunized with the purified protein (four rounds of injection with 200 µg each over a 3-mo period; Eurogentec). The best animal responder was then selected, and the serum was collected to produce the polyclonal rabbit anti-Bbg antibody.
Wing measurements
15–20 wings from independent females were mounted, imaged using NDP.view, and measured using ImageJ (National Institutes of Health). For quantification of RNAi effects, the ratio between the area of each experimental wing and the mean area of control wings (Gal4 driving a UASt GFP transgene) was calculated. Statistical significance was determined using unpaired, two-tailed Student’s t tests on raw wing measurements.
Cell numbers and apical cell size
In Figs. 2 and 4, mutant clones were generated by flippase-mediated FRT chromosome exchange. In this experimental setup, mutant clones and their twin spots originated from the same parental cell and should be of the same size and have the same number of cells. The apical size of cells was obtained by dividing clone area by the number of cells.
In Fig. 7, the mutant clones were generated by the overexpression of RNAi or UAS constructs. They were marked by GFP. In this setup, the size of the mutant GFP-positive tissue could not be compared with any WT counterpart, and we could not assess differences in clone size or cell numbers. Counting cells between equivalent area within each disc, the change in apical size of cells between control (GFP−) and mutant cells (GFP+) was evaluated.
Y2H screen
The Bbg-S Y2H screen was performed by Hybrigenics SA. Full-length Bbg-S (lacking its last 4 aa, which represent a potential palmitoylation site that could have interfered with the yeast nucleus targeting of the Bbg bait fusion) was cloned as an N-terminal LexA fusion in pB27 and used as a bait against a Drosophila whole embryo (0–24 h) cDNA library (RP2) in 0.0 mM 3-amino triazole. 7.39 × 107 clones were screened, from which 177 positive clones were recovered and retested under the same conditions as the screen. Putative positive hits such as Kst were confirmed by coIP in S2 cells.
Plasmids for cell culture
In Drosophila S2 cells, genes were expressed either by cotransfecting an Act5C-Gal4 plasmid (gift from B. Shilo, Weizmann Institute of Science, Rehovot, Israel) with UASt plasmids for the genes of interest or by using the Act5C promoter driven gateway destination vectors from the Drosophila Gateway Vector collection (http://emb.carnegiescience.edu/drosophila-gateway-vector-collection). The Kst fragments (Kst-A1–1,632, Kst-B1,633–1,903, Kst-C1,904–2,632, and Kst-D2,594–4,097) were described previously (Fletcher et al., 2015) and were used to generate the pAWF-Kst-BC1,650–2,600 construct by PCR using linker primers spanning over the codons corresponding with the Kst aa 1,898 and 1,909. Primers used were sense, 5′-GAAGCGACAGGAACAACTCAGAACTTCCTGGGAGAAC-3′, and anti-sense, 5′-GTTCTCCCAGGAAGTTCTGAGTTGTTCCTGTCGCTTC-3′.
Western blotting and immunoprecipitation
All immunoprecipitations were done in lysates from S2 cells grown in Schneider’s Drosophila medium (Gibco) supplemented with 10% FCS (Sigma-Aldrich), 100 U/ml penicillin, and 100 µg/ml streptomycin. 3.106 S2 cells were transfected using Effectene (QIAGEN). 48 h after transfection, S2 cells were lysed with Hepes lysis buffer (50 mM Hepes NaOH, pH 7.5, 150 mM NaCl, and 0.5% Triton X-100) supplemented with 1 mM DTT and cOmplete protease inhibitor cocktail. Half of the lysate was incubated with Sepharose beads with 3 µl of rabbit anti-GFP antibody (TP401; Torrey Pines Biolabs) for 1 h at 4°C. The beads were washed with Hepes lysis buffer four times.
After SDS-PAGE protein gel separation, proteins were visualized by immunoblotting using rabbit anti-GFP (1:2,000; TP401; Torrey Pines Biolabs), mouse anti-FLAG (1:5,000; F1804; Sigma-Aldrich), rabbit anti-pMLC2 (1:500; 3671; Cell Signaling Technology), and mouse antitubulin (1:10,000; DM1A; Sigma-Aldrich) antibodies.
Immunocytochemistry
Antibody staining of wing imaginal discs were performed using standard protocols. In short, larval heads containing the brain and imaginal discs were dissected in PBS, fixed for 20 min at RT in PBS with 4% formaldehyde, washed 3× for 10 min in PBS with 0.2% Triton X-100 (PBX), and blocked for 30 min in PBX with 0.1% BSA. Primary antibodies were then incubated overnight at 4°C under gentle agitation in PBX and 0.1% BSA at the indicated dilutions (see next paragraph). Samples were then washed 3× for 15 min in PBX, and fluorescent dye–coupled secondary antibodies were incubated for 90 min in PBX and 1% BSA at RT. After several washes in PBX, the stained tissues were then transferred to CitiFluor AF1 (Agar) mounting media for overnight equilibration. Individual discs were then dissected and mounted in CitiFluor. Images were acquired with an SP2-405 confocal microscope (Leica Microsystems) or an Apotome2 microscope (ZEISS) and processed using ImageJ.
Primary antibodies used were: mouse anti-Arm (1:25; N2 7A1; Developmental Studies Hybridoma Bank [DSHB]), mouse anti–β-galactosidase (1:25; 40-1a; DSHB), mouse anti-Crb (1:25; Cq4; DSHB), mouse anti-Dlg (1:25; 4F3; DSHB), mouse anti-Ena (1:200; 5G2; DSHB), mouse anti-Rho1 (1:25; p1D9; DSHB), rabbit anti-aPKC (1:500; anti-PKCz C-20; Santa Cruz Biotechnology, Inc.), rabbit anti-Bbg (1:5,000; polyclonal directed against Bbg aa 2,317–2,637 generated for this study), rabbit anti-Cpa (1:200; a gift from F. Janody; Amândio et al., 2014), rabbit anti-Dia (1:500; a gift from S. Wasserman; Afshar et al., 2000), rabbit anti-Ex (1:1,000; a gift from R. Fehon; Maitra et al., 2006), rabbit anti-GFP (1:200; TP401; Torrey Pines Biolabs), rabbit anti-Pyd (1:2,000; Djiane et al., 2011), and rat anti–E-Cad (1:25; DCAD2; DSHB). Actin fibers were visualized using TRITC-phalloidin (1:500; P1951; Sigma-Aldrich). Secondary antibodies used were conjugated to the Alexa Fluor 350, Alexa Fluor 488, Cy3, or Alexa Fluor 647 fluorochromes (1:500; Jackson ImmunoResearch Laboratories, Inc.).
Diap1-LacZ and ex-LacZ quantifications
Quantifications were performed as described previously (Djiane et al., 2014). In brief, in each wing disc, the mean level of gray was measured by ImageJ in an equivalent area in the anterior (non-GFP) and posterior (GFP) of the wing pouch, excluding the anterior/posterior and dorsal/ventral boundaries, which show discontinuities.
Online supplemental material
Four supplemental figures are provided, detailing the generation of the bbg34 mutant affecting specifically the bbg-M and bbg-L isoforms (Fig. S1), the effects of cpa, enabled, and karst on cortical F-actin in third instar wing discs (Fig. S2), the effects of bbg mutants on apicobasal markers (Fig. S3), and the relationship between bbg and the core Hippo pathway (Fig. S4).
Supplementary Material
Acknowledgments
We thank Amel Zouaz for help with Drosophila S2 cell cultures and François Schweisguth for discussions. We thank Richard Fehon, Georgina Fletcher, Florence Janody, Duojia Pan, Nic Tapon, Barry Thompson, Mirka Uhlirova, Koen Venken, Xiabao Wang, and Steven Wasserman for sharing flies, constructs, and antibodies. We acknowledge the Bloomington Drosophila Stock Center, the Vienna Drosophila Stock Center, the DGRC Kyoto Stock Center, and the Developmental Studies Hybridoma Bank for flies and antibodies.
This work was supported by grants from the Fondation ARC pour la Recherche sur le Cancer (PJA 20141201630), the Ligue Nationale Contre le Cancer (9FI12549ROSA), a European Commission Marie Curie Career Integration Grant (PCIG13-GA-2013-618371), and the ATIP/Avenir Programme.
The authors declare no competing financial interests.
Author contributions: conceived and analyzed experiments: E. Forest, L. Heron-Milhavet, and A. Djiane. Performed experiments: E. Forest, R. Logeay, C. Géminard, D. Kantar, L. Heron-Milhavet, F. Frayssinoux, and A. Djiane. Wrote the manuscript: A. Djiane.
References
- Afshar K., Stuart B., and Wasserman S.A.. 2000. Functional analysis of the Drosophila diaphanous FH protein in early embryonic development. Development. 127:1887–1897. [DOI] [PubMed] [Google Scholar]
- Amândio A.R., Gaspar P., Whited J.L., and Janody F.. 2014. Subunits of the Drosophila actin-capping protein heterodimer regulate each other at multiple levels. PLoS One. 9:e96326 10.1371/journal.pone.0096326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranjuez G., Kudlaty E., Longworth M.S., and McDonald J.A.. 2012. On the role of PDZ domain-encoding genes in Drosophila border cell migration. G3 (Bethesda). 2:1379–1391. 10.1534/g3.112.004093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badouel C., Gardano L., Amin N., Garg A., Rosenfeld R., Le Bihan T., and McNeill H.. 2009. The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev. Cell. 16:411–420. 10.1016/j.devcel.2009.01.010 [DOI] [PubMed] [Google Scholar]
- Baines A.J. 2009. Evolution of spectrin function in cytoskeletal and membrane networks. Biochem. Soc. Trans. 37:796–803. 10.1042/BST0370796 [DOI] [PubMed] [Google Scholar]
- Baumgartner R., Poernbacher I., Buser N., Hafen E., and Stocker H.. 2010. The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev. Cell. 18:309–316. 10.1016/j.devcel.2009.12.013 [DOI] [PubMed] [Google Scholar]
- Bear J.E., and Gertler F.B.. 2009. Ena/VASP: towards resolving a pointed controversy at the barbed end. J. Cell Sci. 122:1947–1953. 10.1242/jcs.038125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett V., and Baines A.J.. 2001. Spectrin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Physiol. Rev. 81:1353–1392. 10.1152/physrev.2001.81.3.1353 [DOI] [PubMed] [Google Scholar]
- Bonnay F., Cohen-Berros E., Hoffmann M., Kim S.Y., Boulianne G.L., Hoffmann J.A., Matt N., and Reichhart J.-M.. 2013. big bang gene modulates gut immune tolerance in Drosophila. Proc. Natl. Acad. Sci. USA. 110:2957–2962. 10.1073/pnas.1221910110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-L., Gajewski K.M., Hamaratoglu F., Bossuyt W., Sansores-Garcia L., Tao C., and Halder G.. 2010. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc. Natl. Acad. Sci. USA. 107:15810–15815. 10.1073/pnas.1004060107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codelia V.A., Sun G., and Irvine K.D.. 2014. Regulation of YAP by mechanical strain through Jnk and Hippo signaling. Curr. Biol. 24:2012–2017. 10.1016/j.cub.2014.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Thakur M., Feng Y., Jagannathan R., Seppa M.J., Skeath J.B., and Longmore G.D.. 2010. Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr. Biol. 20:657–662. 10.1016/j.cub.2010.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H., Wang W., Yu J., Zheng Y., Qing Y., and Pan D.. 2015. Spectrin regulates Hippo signaling by modulating cortical actomyosin activity. eLife. 4:e06567 10.7554/eLife.06567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djiane A., Shimizu H., Wilkin M., Mazleyrat S., Jennings M.D., Avis J., Bray S., and Baron M.. 2011. Su(dx) E3 ubiquitin ligase-dependent and -independent functions of polychaetoid, the Drosophila ZO-1 homologue. J. Cell Biol. 192:189–200. 10.1083/jcb.201007023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djiane A., Zaessinger S., Babaoğlan A.B., and Bray S.J.. 2014. Notch inhibits Yorkie activity in Drosophila wing discs. PLoS One. 9:e106211 10.1371/journal.pone.0106211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S., et al. 2011. Role of YAP/TAZ in mechanotransduction. Nature. 474:179–183. 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- Fernández B.G., Gaspar P., Brás-Pereira C., Jezowska B., Rebelo S.R., and Janody F.. 2011. Actin-Capping Protein and the Hippo pathway regulate F-actin and tissue growth in Drosophila. Development. 138:2337–2346. 10.1242/dev.063545 [DOI] [PubMed] [Google Scholar]
- Fletcher G.C., Elbediwy A., Khanal I., Ribeiro P.S., Tapon N., and Thompson B.J.. 2015. The Spectrin cytoskeleton regulates the Hippo signalling pathway. EMBO J. 34:940–954. 10.15252/embj.201489642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar P., and Tapon N.. 2014. Sensing the local environment: actin architecture and Hippo signalling. Curr. Opin. Cell Biol. 31:74–83. 10.1016/j.ceb.2014.09.003 [DOI] [PubMed] [Google Scholar]
- Gates J., Mahaffey J.P., Rogers S.L., Emerson M., Rogers E.M., Sottile S.L., Van Vactor D., Gertler F.B., and Peifer M.. 2007. Enabled plays key roles in embryonic epithelial morphogenesis in Drosophila. Development. 134:2027–2039. 10.1242/dev.02849 [DOI] [PubMed] [Google Scholar]
- Genevet A., Wehr M.C., Brain R., Thompson B.J., and Tapon N.. 2010. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev. Cell. 18:300–308. 10.1016/j.devcel.2009.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grevengoed E.E., Fox D.T., Gates J., and Peifer M.. 2003. Balancing different types of actin polymerization at distinct sites. J. Cell Biol. 163:1267–1279. 10.1083/jcb.200307026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeschik N.A., Parsons L.M., Allott M.L., Harvey K.F., and Richardson H.E.. 2010. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr. Biol. 20:573–581. 10.1016/j.cub.2010.01.055 [DOI] [PubMed] [Google Scholar]
- Halder G., and Johnson R.L.. 2011. Hippo signaling: growth control and beyond. Development. 138:9–22. 10.1242/dev.045500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaratoglu F., Willecke M., Kango-Singh M., Nolo R., Hyun E., Tao C., Jafar-Nejad H., and Halder G.. 2006. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 8:27–36. 10.1038/ncb1339 [DOI] [PubMed] [Google Scholar]
- Harvey K., and Tapon N.. 2007. The Salvador-Warts-Hippo pathway - an emerging tumour-suppressor network. Nat. Rev. Cancer. 7:182–191. 10.1038/nrc2070 [DOI] [PubMed] [Google Scholar]
- Harvey K.F., Pfleger C.M., and Hariharan I.K.. 2003. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 114:457–467. 10.1016/S0092-8674(03)00557-9 [DOI] [PubMed] [Google Scholar]
- Kim S.Y., Renihan M.K., and Boulianne G.L.. 2006. Characterization of big bang, a novel gene encoding for PDZ domain-containing proteins that are dynamically expressed throughout Drosophila development. Gene Expr. Patterns. 6:504–518. 10.1016/j.modgep.2005.10.009 [DOI] [PubMed] [Google Scholar]
- Lee J.K., Brandin E., Branton D., and Goldstein L.S.. 1997. alpha-Spectrin is required for ovarian follicle monolayer integrity in Drosophila melanogaster. Development. 124:353–362. [DOI] [PubMed] [Google Scholar]
- Ling C., Zheng Y., Yin F., Yu J., Huang J., Hong Y., Wu S., and Pan D.. 2010. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc. Natl. Acad. Sci. USA. 107:10532–10537. 10.1073/pnas.1004279107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas E.P., Khanal I., Gaspar P., Fletcher G.C., Polesello C., Tapon N., and Thompson B.J.. 2013. The Hippo pathway polarizes the actin cytoskeleton during collective migration of Drosophila border cells. J. Cell Biol. 201:875–885. 10.1083/jcb.201210073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra S., Kulikauskas R.M., Gavilan H., and Fehon R.G.. 2006. The tumor suppressors Merlin and Expanded function cooperatively to modulate receptor endocytosis and signaling. Curr. Biol. 16:702–709. 10.1016/j.cub.2006.02.063 [DOI] [PubMed] [Google Scholar]
- Martin A.C., Kaschube M., and Wieschaus E.F.. 2009. Pulsed contractions of an actin-myosin network drive apical constriction. Nature. 457:495–499. 10.1038/nature07522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Médina E., Williams J., Klipfell E., Zarnescu D., Thomas G., and Le Bivic A.. 2002. Crumbs interacts with moesin and beta(Heavy)-spectrin in the apical membrane skeleton of Drosophila. J. Cell Biol. 158:941–951. 10.1083/jcb.200203080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z., Moroishi T., and Guan K.-L.. 2016. Mechanisms of Hippo pathway regulation. Genes Dev. 30:1–17. 10.1101/gad.274027.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng B.F., Selvaraj G.K., Santa-Cruz Mateos C., Grosheva I., Alvarez-Garcia I., Martín-Bermudo M.D., and Palacios I.M.. 2016. α-Spectrin and integrins act together to regulate actomyosin and columnarization, and to maintain a monolayered follicular epithelium. Development. 143:1388–1399. 10.1242/dev.130070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellikka M., Tanentzapf G., Pinto M., Smith C., McGlade C.J., Ready D.F., and Tepass U.. 2002. Crumbs, the Drosophila homologue of human CRB1/RP12, is essential for photoreceptor morphogenesis. Nature. 416:143–149. 10.1038/nature721 [DOI] [PubMed] [Google Scholar]
- Rauskolb C., Sun S., Sun G., Pan Y., and Irvine K.D.. 2014. Cytoskeletal tension inhibits Hippo signaling through an Ajuba-Warts complex. Cell. 158:143–156. 10.1016/j.cell.2014.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson B.S., Huang J., Hong Y., and Moberg K.H.. 2010. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Curr. Biol. 20:582–590. 10.1016/j.cub.2010.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansores-Garcia L., Bossuyt W., Wada K., Yonemura S., Tao C., Sasaki H., and Halder G.. 2011. Modulating F-actin organization induces organ growth by affecting the Hippo pathway. EMBO J. 30:2325–2335. 10.1038/emboj.2011.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder M.C., and Halder G.. 2012. Regulation of the Hippo pathway by cell architecture and mechanical signals. Semin. Cell Dev. Biol. 23:803–811. 10.1016/j.semcdb.2012.06.001 [DOI] [PubMed] [Google Scholar]
- Settleman J. 2001. Rac 'n Rho: the music that shapes a developing embryo. Dev. Cell. 1:321–331. 10.1016/S1534-5807(01)00053-3 [DOI] [PubMed] [Google Scholar]
- Stabach P.R., Simonović I., Ranieri M.A., Aboodi M.S., Steitz T.A., Simonović M., and Morrow J.S.. 2009. The structure of the ankyrin-binding site of beta-spectrin reveals how tandem spectrin-repeats generate unique ligand-binding properties. Blood. 113:5377–5384. 10.1182/blood-2008-10-184291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G.H., and Kiehart D.P.. 1994. Beta heavy-spectrin has a restricted tissue and subcellular distribution during Drosophila embryogenesis. Development. 120:2039–2050. [DOI] [PubMed] [Google Scholar]
- Thomas G.H., and Williams J.A.. 1999. Dynamic rearrangement of the spectrin membrane skeleton during the generation of epithelial polarity in Drosophila. J. Cell Sci. 112:2843–2852. [DOI] [PubMed] [Google Scholar]
- Thomas G.H., Zarnescu D.C., Juedes A.E., Bales M.A., Londergan A., Korte C.C., and Kiehart D.P.. 1998. Drosophila betaHeavy-spectrin is essential for development and contributes to specific cell fates in the eye. Development. 125:2125–2134. [DOI] [PubMed] [Google Scholar]
- Venken K.J.T., Schulze K.L., Haelterman N.A., Pan H., He Y., Evans-Holm M., Carlson J.W., Levis R.W., Spradling A.C., Hoskins R.A., and Bellen H.J.. 2011. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods. 8:737–743. 10.1038/nmeth.1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers J.H.A., Manning S.A., Kulkarni A., and Harvey K.F.. 2016. A Drosophila RNAi library modulates Hippo pathway-dependent tissue growth. Nat. Commun. 7:10368 10.1038/ncomms10368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K.K.L., Li W., An Y., Duan Y., Li Z., Kang Y., and Yan Y.. 2015. β-Spectrin regulates the hippo signaling pathway and modulates the basal actin network. J. Biol. Chem. 290:6397–6407. 10.1074/jbc.M114.629493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Huang J., Dong J., and Pan D.. 2003. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 114:445–456. 10.1016/S0092-8674(03)00549-X [DOI] [PubMed] [Google Scholar]
- Wu S., Liu Y., Zheng Y., Dong J., and Pan D.. 2008. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev. Cell. 14:388–398. 10.1016/j.devcel.2008.01.007 [DOI] [PubMed] [Google Scholar]
- Yin F., Yu J., Zheng Y., Chen Q., Zhang N., and Pan D.. 2013. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell. 154:1342–1355. 10.1016/j.cell.2013.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F.-X., and Guan K.-L.. 2013. The Hippo pathway: regulators and regulations. Genes Dev. 27:355–371. 10.1101/gad.210773.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Zheng Y., Dong J., Klusza S., Deng W.-M., and Pan D.. 2010. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev. Cell. 18:288–299. 10.1016/j.devcel.2009.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnescu D.C., and Thomas G.H.. 1999. Apical spectrin is essential for epithelial morphogenesis but not apicobasal polarity in Drosophila. J. Cell Biol. 146:1075–1086. 10.1083/jcb.146.5.1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.