During cytokinesis, centrosomal asters inhibit cortical contractility at the cell poles. Mangal et al. provide molecular insight into this phenomenon, showing that TPXL-1, which localizes to astral microtubules, activates Aurora A kinase to clear contractile ring proteins from the polar cortex.
Abstract
During cytokinesis, a signal from the central spindle that forms between the separating anaphase chromosomes promotes the accumulation of contractile ring components at the cell equator, while a signal from the centrosomal microtubule asters inhibits accumulation of contractile ring components at the cell poles. However, the molecular identity of the inhibitory signal has remained unknown. To identify molecular components of the aster-based inhibitory signal, we developed a means to monitor the removal of contractile ring proteins from the polar cortex after anaphase onset. Using this assay, we show that polar clearing is an active process that requires activation of Aurora A kinase by TPXL-1. TPXL-1 concentrates on astral microtubules coincident with polar clearing in anaphase, and its ability to recruit Aurora A and activate its kinase activity are essential for clearing. In summary, our data identify Aurora A kinase as an aster-based inhibitory signal that restricts contractile ring components to the cell equator during cytokinesis.
Introduction
Cytokinesis partitions the contents of the mother cell to the two daughter cells to complete cell division. In animal cells, cytokinesis is accomplished by constriction of a cortical contractile ring that assembles in response to activation of the small GTPase RhoA (Piekny et al., 2005; Jordan and Canman, 2012). After anaphase onset, removal of an inhibitory phosphorylation recruits the RhoA guanine nucleotide exchange factor (GEF) ECT2 to the cell cortex (Su et al., 2011), where it activates RhoA. RhoA, in turn, activates the formins, which assemble long actin filaments that make up the ring (Otomo et al., 2005), and Rho-kinase, which promotes the assembly and recruitment of myosin II (Matsumura et al., 2011). Contractile rings also include septin filaments and the filament cross-linker anillin (D’Avino, 2009; Piekny and Maddox, 2010).
To ensure that each daughter cell inherits an equivalent genomic complement, contractile ring assembly is directed by the anaphase spindle. Contractile ring proteins accumulate on the equatorial cortex in part because of a signal arising from the central spindle that forms between the separating chromosomes (von Dassow, 2009; Green et al., 2012; D’Avino et al., 2015; Mishima, 2016). At the same time, an inhibitory signal that prevents the accumulation of contractile ring proteins on the polar cortex has been proposed to arise from the centrosomal microtubule asters (von Dassow, 2009; Green et al., 2012; D’Avino et al., 2015; Mishima, 2016) or from kinetochores (Rodrigues et al., 2015).
Recent work in Drosophila melanogaster and cultured human cells suggests that kinetochore-localized protein phosphatase 1 (PP1) promotes cortical actin clearing by dephosphorylating ezrin/radixin/moesin proteins as the chromosomes approach the polar cortex during anaphase (Rodrigues et al., 2015). However, the centrosomal microtubule asters can also inhibit contractile ring protein accumulation at the poles. Selective disassembly of dynamic astral microtubules broadens the equatorial RhoA zone/contractile ring, and laser ablation of single asters in sea urchin embryos shifts the active RhoA zone toward the ablated aster (von Dassow et al., 2009), suggesting that astral microtubules limit the zone where contractile ring proteins can accumulate (Bement et al., 2005; Foe and von Dassow, 2008; Murthy and Wadsworth, 2008; von Dassow et al., 2009; Zanin et al., 2013). Placing microtubule asters in proximity to the cortex also suppresses cortical contractility in Caenorhabditis elegans embryos and grasshopper spermatocytes (Werner et al., 2007; Chen et al., 2008). Despite evidence for an aster-based signal that inhibits cortical contractility, its molecular identity has remained unknown.
Results and discussion
Contractile ring proteins are actively cleared from the polar cortex after anaphase onset
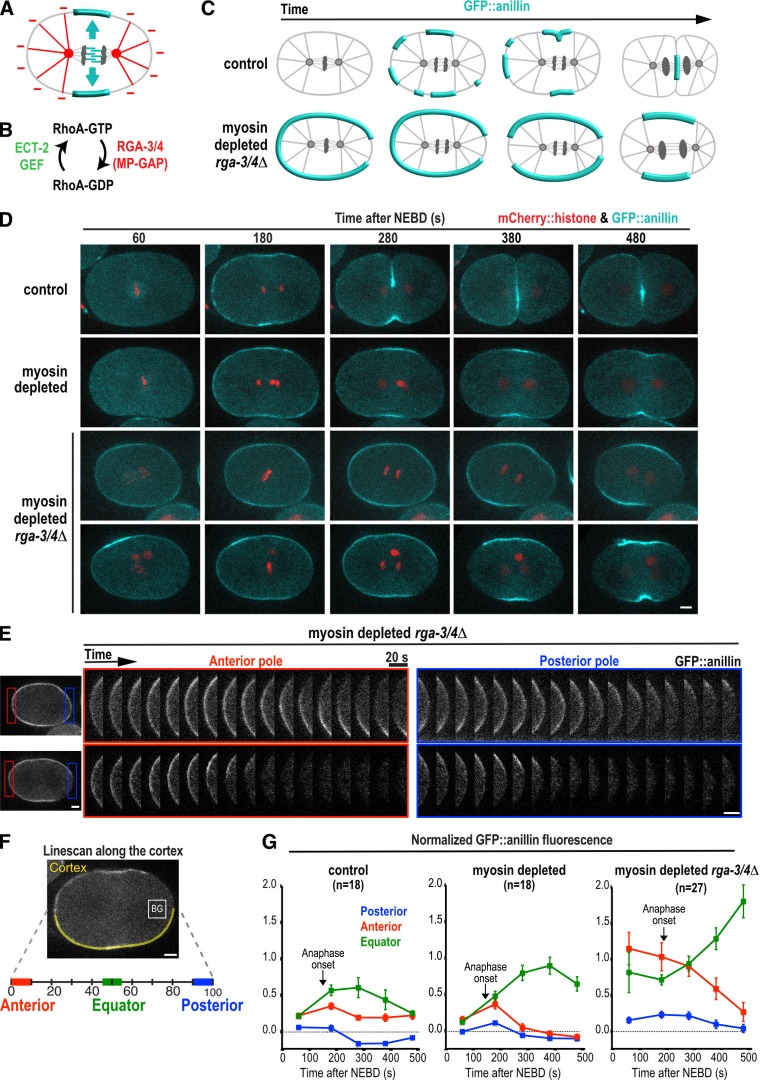
To explore the mechanism that prevents contractile ring proteins from accumulating at the cell poles (Fig. 1 A), we established an assay monitoring cortical levels of a GFP fusion with the contractile ring protein anillin (GFP::anillin) during the first division of the C. elegans embryo (Fig. 1, C–G). We chose anillin because it mirrors the localization of active RhoA (Piekny and Glotzer, 2008) and is recruited independently of myosin II (Fig. 1 D; Maddox et al., 2005), which allows monitoring of cortical patterning when contractility is inhibited.
Figure 1.
Contractile ring proteins are actively cleared from the polar cortex after anaphase onset. (A) The spindle provides a stimulatory signal that promotes the equatorial accumulation of contractile ring proteins (cyan) and an inhibitory signal that prevents their accumulation on the polar cortex (red). (B) During mitosis, RhoA is activated by the ECT-2 GEF and inactivated by MP-GAP (RGA-3/4 in C. elegans). (C) Schematics illustrating the localization of the contractile ring marker GFP::anillin during the first division of control (top) and myosin-depleted rga-3/4Δ (bottom) embryos. (D) Representative time-lapse series of the first division of control (n = 9, top row) and myosin-depleted (n = 9, second row) embryos expressing GFP::anillin (cyan) and mCherry::histone (red). Two representative myosin-depleted rga-3/4Δ embryos are shown (bottom two rows): one in which GFP::anillin is evenly distributed around the cortex before anaphase onset (n = 6/14; third row), and one in which GFP::anillin is absent from the posterior cortex before anaphase onset (n = 8/14; bottom row). Time is seconds after NEBD. (E) Kymographs of the anterior and posterior poles in the myosin-depleted rga-3/4Δ embryos shown in D beginning 180 s after NEBD. (F) GFP::anillin fluorescence on the anterior, posterior, and equatorial cortex was quantified as shown after subtraction of mean cytoplasmic background (BG, measured in box). The mean GFP::anillin fluorescence in each region was normalized by dividing by the mean intensity at the anterior pole in myosin-depleted rga-3/4Δ embryos 180 s after NEBD. (G) Normalized cortical GFP::anillin fluorescence is plotted for the anterior, posterior, and equatorial cortex in control, myosin-depleted, and myosin-depleted rga-3/4Δ embryos. Arrows mark the mean point of anaphase onset for each condition. Error bars are SEM; n = number of linescans. Bars, 5 µm.
Cortical GFP::anillin fluorescence was quantified by performing linescans along the cortex from the anterior to the posterior pole in central plane confocal images (Fig. 1 F). In control embryos, GFP::anillin appeared on the cortex at the equator and anterior pole concurrent with anaphase onset (Fig. 1, D and G, control 180-s panel). The signal on the polar cortex then dropped over the ensuing 100 s (200–300 s after nuclear envelope breakdown [NEBD]; Fig. 1, D and G). The signal at the equator also dropped after the 300-s time point, because the constricting contractile ring moved away from the cell surface. To isolate the effects of the spindle-based signaling that patterns the cortex from component rearrangement caused by myosin-based cortical compression and expansion, we analyzed GFP::anillin after depleting the heavy chain of myosin II by RNAi. In myosin-depleted embryos, the contractile ring does not constrict, and GFP::anillin levels on the equatorial cortex rose continuously over the 200-s interval after anaphase onset. Levels of anillin on the polar cortex declined during this interval in a fashion similar to controls, suggesting that the reduction in anillin levels is not a downstream consequence of ring constriction (Fig. 1, D and G, 200–400 s after NEBD; and Video 1).
In control and myosin-depleted embryos, low levels of anillin are transiently observed on the polar cortex at anaphase onset (Fig. 1 G). The subsequent reduction in the amount of anillin on the polar cortex could be caused by a reduction of a signal that promotes anillin accumulation or by activation of an active clearing mechanism. To distinguish between these possibilities, we increased the amount of anillin on the cortex by eliminating the major GTPase-activating protein (GAP) activity that opposes RhoA activation (Fig. 1 B). In C. elegans, this activity is encoded by the paralogous RGA-3 and RGA-4 proteins, which are homologues of human MP-GAP (Schmutz et al., 2007; Schonegg et al., 2007; Zanin et al., 2013). To monitor cortical patterning in the absence of component rearrangement resulting from myosin-based cortical compression and expansion, we also depleted myosin. In myosin-depleted rga-3/4Δ embryos, GFP::anillin was present at high levels on the equatorial and polar cortex before anaphase onset, allowing us to monitor how signaling by the anaphase spindle alters this pattern (Fig. 1, C–G). As in control embryos, the initial accumulation of GFP::anillin on the cortex at the posterior pole was more variable than at the anterior pole (Fig. 1, D and E; and Video 2). Depleting PAR-2 or PAR-1 did not significantly alter this distribution, suggesting that it is not caused by the posterior PAR proteins (unpublished data). Because of the variable initial accumulation on the posterior pole, we compared cortical GFP::anillin dynamics at the anterior pole to those at the cell equator. In myosin-depleted rga-3/4Δ embryos, levels of GFP::anillin on the equatorial cortex were high at metaphase and then increased over the 300 s after anaphase onset (Fig. 1, D–G, 180–500 s after NEBD). In contrast, at the anterior pole, levels of cortical GFP::anillin were high at metaphase and then decreased over the 300 s after anaphase onset (Fig. 1, D–G). We conclude that an active mechanism clears contractile ring proteins from the polar cortex after anaphase onset.
Kinetochore-localized PP1 is not required for polar clearing
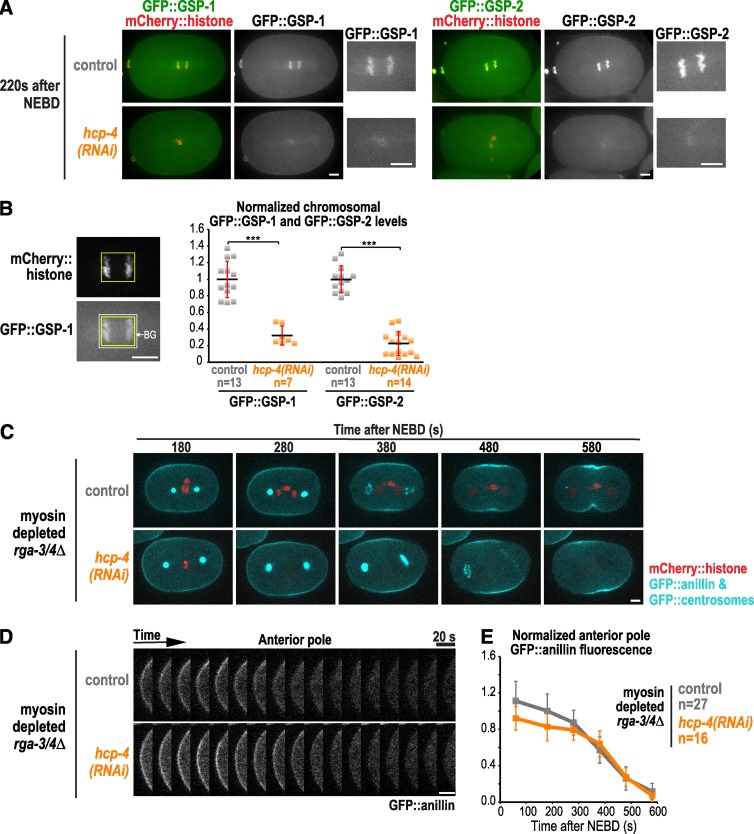
Work in Drosophila sensory organ precursor cells suggested that clearing of cortical actin from the cell poles is mediated by PP1 on kinetochores that approach the cortex as the chromosomes separate (Rodrigues et al., 2015). C. elegans has four catalytic PP1 subunits: GSP-1, -2, -3, and -4. Because GSP-3 and -4 are sperm specific (Wu et al., 2012), we focused on GSP-1 and GSP-2, which both localize to kinetochores and anaphase chromosomes (Hattersley et al., 2016; Kim et al., 2017). To test whether this pool of PP1 is involved in polar clearing, we depleted the C. elegans CENP-C homologue (called HCP-4). CENP-C depletion blocks the recruitment of all outer kinetochore components (Oegema et al., 2001; Desai et al., 2003), including the two known to provide conserved PP1 docking sites, KNL-1 and MEL-28 (Espeut et al., 2012; Hattersley et al., 2016). Imaging GFP::GSP-1 and GFP::GSP-2 confirmed that levels of both PP1 subunits on chromosomes were greatly reduced in hcp-4(RNAi) embryos (Fig. 2, A and B). HCP-4 depletion also blocks chromosome segregation, which would prevent residual chromatin-associated PP1 from moving toward the poles during anaphase. Imaging myosin-depleted rga-3/4Δ embryos expressing a GFP-tagged centrosome marker in addition to GFP::anillin revealed essentially identical clearing of GFP::anillin from the polar cortex in control and hcp-4(RNAi) embryos (Fig. 2, C–E; and Video 3). These data indicate that polar clearing of anillin is not mediated by kinetochore-localized PP1 in the C. elegans embryo. The difference from the prior analysis could be related to the fact that kinetochores are much further from the cortex in embryonic systems with large asters than in sensory organ precursor or cultured cells.
Figure 2.
Kinetochore-localized PP1 is not required for polar clearing. (A) Representative images of control (n = 13) and hcp-4(RNAi) embryos expressing mCherry::histone and in situ fusions of GFP with GSP-1 (left; n = 7) or GSP-2 (right; n = 14). (B) Mean chromosomal GFP::GSP-1 and GFP::GSP-2 fluorescence was measured 200 s after NEBD. n = number of embryos; error bars are the SD; p-values are two-tailed Student’s t test (***, P < 0.001). (C) Time-lapse series of myosin-depleted rga-3/4Δ embryos expressing GFP::anillin, a GFP::centrosome marker (GFP::SPD-5) and mCherry::histone. Representative control (top; n = 14) and hcp-4(RNAi) embryos (bottom; n = 9) are shown. (D) Kymographs of the anterior pole in the embryos in C beginning 180 s after NEBD. (E) Normalized cortical GFP::anillin fluorescence is plotted for the anterior pole. The control data are reproduced from Fig. 1 G for comparison. n = number of linescans; error bars are SEM. Bars, 5 µm.
The C. elegans TPX2 homologue, TPXL-1, has a direct role in clearing anillin and F-actin from the polar cortex independent of its role in controlling spindle length
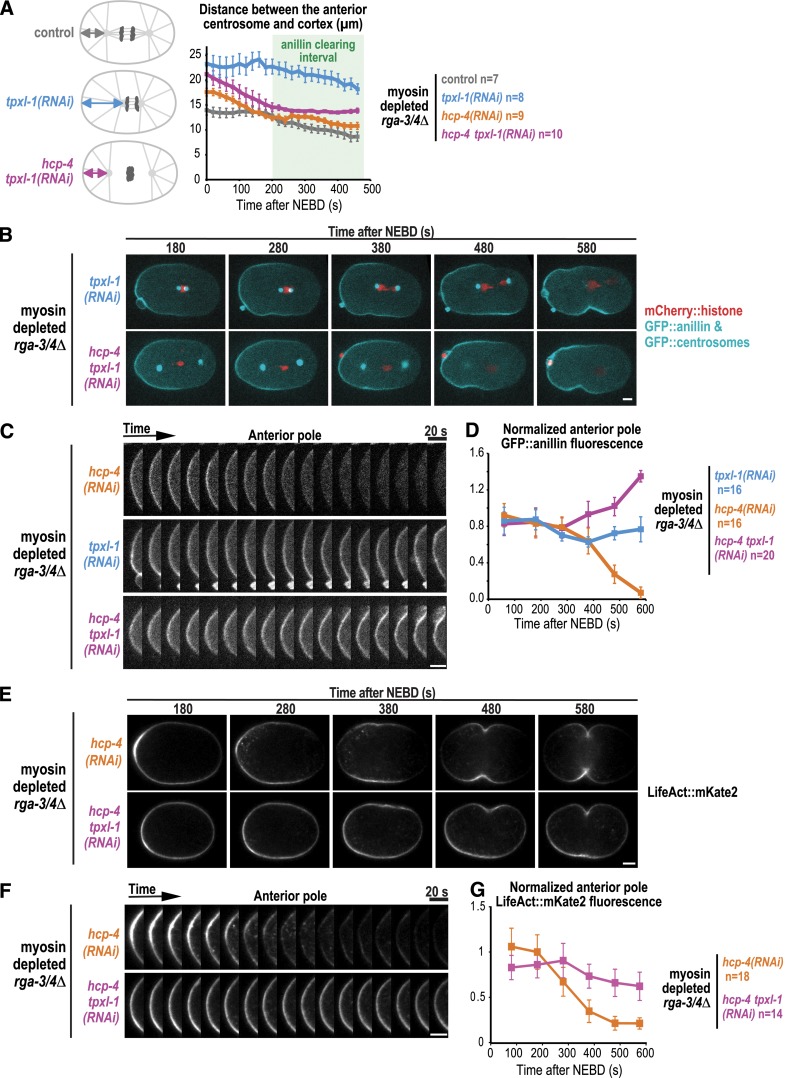
In an attempt to determine whether the proximity of the centrosomal asters is important for polar clearing, we analyzed embryos expressing the GFP centrosome marker and GFP::anillin that were depleted of the Aurora A activator TPXL-1 (Ozlü et al., 2005). In TPXL-1–depleted embryos, spindles are short because of the reduced length of kinetochore microtubules but elongate at a normal rate during anaphase (Ozlü et al., 2005). The shorter spindle length resulting from TPXL-1 depletion increased the distance between the anterior centrosome and cortex approximately twofold in myosin-depleted rga-3/4Δ embryos during the interval when contractile ring proteins are cleared (Fig. 3 A; 200–400 s after NEBD). Analysis in the same embryos revealed that depleting TPXL-1 also prevented clearing of GFP::anillin from the anterior cortex (Fig. 3, B–D; and Video 4).
Figure 3.
The C. elegans TPX2 homologue, TPXL-1, is required for polar clearing. (A) TPXL-1 depletion shortens kinetochore microtubules and increases the distance between the centrosomal aster and the anterior pole; codepletion of HCP-4, which disrupts kinetochore microtubules, rescues this distance. The distance between the anterior centrosome marked with GFP::SPD-5 and the anterior cell pole is plotted for the indicated conditions. n = number of embryos. (B) Time-lapse series of myosin-depleted rga-3/4Δ embryos expressing GFP::anillin, a GFP::centrosome marker (SPD-5, cyan) and mCherry::histone (red). Representative tpxl-1(RNAi) (top; n = 8) and hcp-4 tpxl-1(RNAi) (bottom; n = 10) embryos are shown. (C) Kymographs of the cortex at the anterior pole in the embryos in B beginning 180 s after NEBD. (D) Normalized cortical GFP::anillin fluorescence is plotted for the anterior pole; n = number of linescans. Images and quantification for hcp-4(RNAi) in C and D were reproduced from Fig. 2 (D and E) for comparison. (E) Time-lapse series of myosin-depleted rga-3/4Δ embryos expressing LifeAct::mKate2. Representative hcp-4(RNAi) (top; n = 9) and hcp-4 tpxl-1(RNAi) (bottom; n = 7) embryos are shown. (F) Kymographs of the cortex at the anterior pole in the embryos in E beginning 180 s after NEBD. (G) Normalized cortical LifeAct::mKate2 fluorescence is plotted for the anterior pole; n = number of linescans. All error bars are SEM. Bars, 5 µm.
Depleting TPXL-1 could prevent clearing by increasing the distance between the aster and the polar cortex. Alternatively, TPXL-1 could have a direct role in clearing. To distinguish between these possibilities, we took advantage of the fact that disrupting kinetochore assembly by codepleting HCP-4 along with TPXL-1 overcomes the short kinetochore-microtubule phenotype and allows separation of the centrosomal asters by cortical pulling forces, restoring them to a near-normal position (Fig. 3 A; Ozlü et al., 2005; Lewellyn et al., 2010). Despite restoration of the distance between the centrosome and polar cortex, GFP::anillin was not cleared in hcp-4 tpxl-1(RNAi) embryos (Fig. 3, B–D; and Video 4), suggesting that TPXL-1 is required for clearing even when the asters are in a normal position.
To investigate whether filamentous actin (F-actin) is also cleared from the polar cortex in a TPXL-1-dependent manner, we analyzed myosin-depleted rga-3/4Δ embryos expressing the F-actin binding probe LifeAct::mKate2. Similar to GFP::anillin, LifeAct::mKate2 was cleared from the anterior cell pole in hcp-4(RNAi) but not in hcp-4 tpxl-1(RNAi) embryos (Fig. 3, E–G; and Video 5). We conclude that TPXL-1 has a direct role in clearing anillin and F-actin from the polar cortex that is independent of its role in controlling spindle length.
TPXL-1 recruits Aurora AAIR-1 to astral microtubules but is not required for astral microtubule growth or nucleation
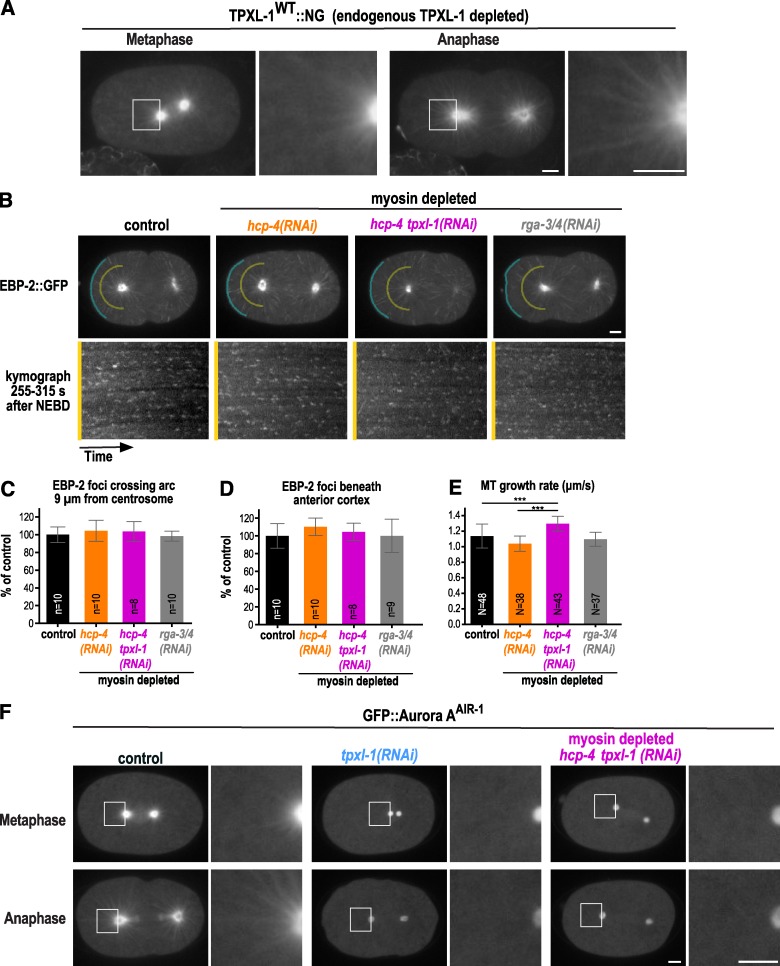
To follow TPXL-1 localization, we generated a functional RNAi-resistant transgene expressing TPXL-1 fused to mNeonGreen (TPXL-1WT::NG; Fig. S1). Consistent with prior work (Ozlü et al., 2005) and analysis by immunofluorescence (Fig. S2 A), TPXL-1WT::NG localized to the periphery of the pericentriolar material and along astral microtubules in metaphase and became more prominently localized to astral microtubules in anaphase (Fig. 4 A and Video 6). The vertebrate TPXL-1 homologue, TPX2, has been shown to promote microtubule-dependent microtubule nucleation in Xenopus laevis egg extracts (Petry et al., 2013). However, previous work in metaphase C. elegans embryos suggested that TPXL-1 depletion does not reduce microtubule nucleation and growth rates (Srayko et al., 2005) and that the microtubule density in the spindle is not reduced even though spindles are shorter (Greenan et al., 2010). To confirm these prior findings made in metaphase embryos, and ensure that TPXL-1 depletion also does not alter microtubule dynamics after anaphase onset when polar clearing occurs, we measured the nucleation and growth rates in embryos expressing the microtubule plus end–binding protein EBP-2::GFP.
Figure 4.
TPXL-1 recruits Aurora AAIR-1 to astral microtubules but is not required for astral microtubule growth or nucleation. (A) Confocal images of embryos expressing TPXL-1WT::NG (n = 10) after depletion of endogenous TPXL-1. (B) Projections of images acquired every 400 ms over a 4-s interval in embryos expressing EBP-2::GFP. Representative images are shown for a control embryo (n = 10) and for myosin-depleted hcp-4(RNAi) (n = 10), hcp-4 tpxl-1(RNAi) (n = 8), and rga-3/4(RNAi) (n = 10) embryos. To visualize EBP-2::GFP on microtubule tips without saturating the aster centers, a gamma of 1.2 was introduced in Photoshop. EBP-2 foci were counted in kymographs made along arcs 9 µm away from the anterior centrosome (yellow) and along the anterior cortex (blue) generated for the entire 1-min movie (255–315 s). (C and D) Graphs plot the mean number of EBP-2::GFP foci crossing the arcs 9 µm away from the anterior centrosome (C) and beneath the anterior cortex (D) as a percentage of the mean number in controls. Error bars are the SD; p-values are two-tailed Student’s t test (***, P < 0.001); n = number of embryos. (E) Microtubule growth rates measured 255–315 s after NEBD. Error bars are the SD; N = number of microtubules tracked in four or more embryos per condition. (F) Representative images of GFP::Aurora AAIR in control (n = 14), tpxl-1(RNAi) (n = 10), and myosin-depleted hcp-4 tpxl-1(RNAi) (n = 11) embryos. To visualize TPXL-1WT::NG and GFP::Aurora AAIR on astral microtubules without saturating the aster centers, gammas of 2.5 and 2.0 were introduced in Photoshop for the images in A and F, which were scaled equivalently across conditions. Bars, 5 µm.
Microtubule nucleation was monitored as described previously (Srayko et al., 2005); a circle with a radius of 9 µm centered on the anterior centrosome was drawn and kymographs were generated along the half of the circle facing the anterior cortex (Fig. 4 B). Consistent with prior work (Srayko et al., 2005), we found that microtubule nucleation decreases and the growth rate increases between metaphase and anaphase in control embryos (Fig. S2, C–E). Next, we analyzed TPXL-1–depleted embryos during the interval when polar clearing occurs (255–315 s after NEBD) and found that neither the number of nucleated microtubules crossing the 9-µm radius half circle (Fig. 4 B, yellow lines) nor the number of microtubules that reached the anterior cortex (Fig. 4 B, blue lines) were altered (Fig. 4, C and D; and Video 7). Rather than being decreased, the microtubule growth rate was slightly higher in hcp-4 tpxl-1(RNAi) embryos compared with controls (Fig. 4 E). Nucleation and growth rates were indistinguishable from controls in myosin-depleted rga-3/4(RNAi) embryos (Fig. 4, B–E), which exhibit comparable anillin dynamics to myosin-depleted rga-3/4Δ embryos (Fig. S2, F–H), excluding a role for RGA-3/4 in microtubule dynamics. RGA-3/4 was depleted by RNAi, because the ebp-2::gfp transgene was silenced in the rga-3/4Δ mutant. We conclude that neither TPXL-1 depletion nor the inhibition of rga-3/4 that we use to facilitate analysis of clearing leads to a change in microtubule dynamics that attenuates the asters.
Polar clearing requires the ability of TPXL-1 to activate Aurora A
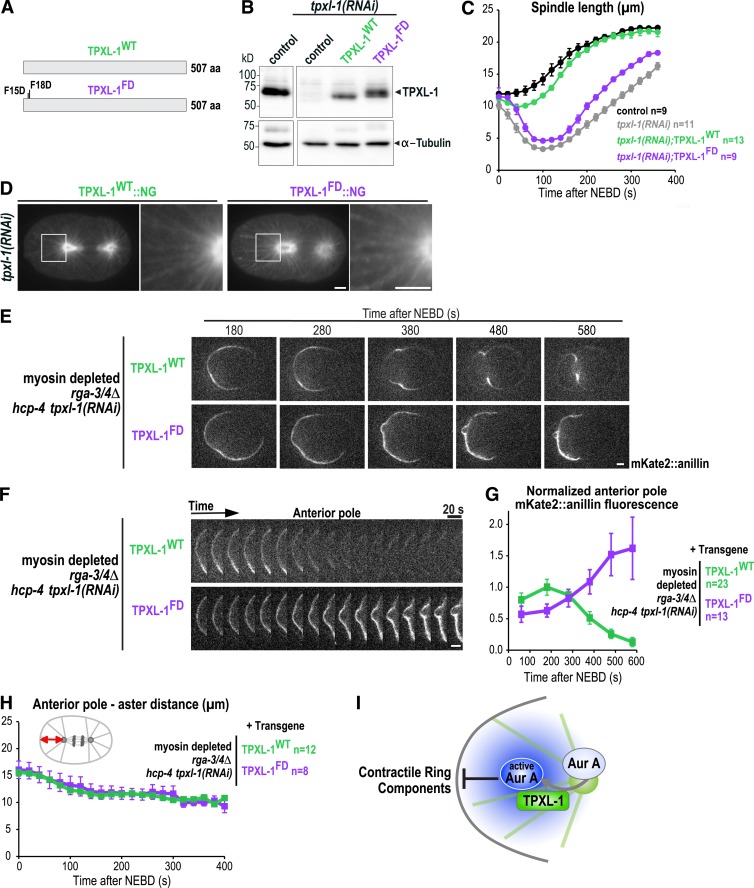
Like TPXL-1, Aurora A localizes to the centrosome and astral microtubules (Schumacher et al., 1998; Hannak et al., 2001; Motegi et al., 2006; Toya et al., 2011). We confirmed that the localization of Aurora A to astral microtubules depends on TPXL-1 by imaging a GFP fusion with Aurora A (Figs. 4 F and S2 B; and Video 8; Ozlü et al., 2005). To determine whether polar clearing requires the ability of TPXL-1 to recruit and activate Aurora A, we generated RNAi-resistant transgenes (untagged and tagged with NG) encoding WT TPXL-1 (TPXL-1WT) and a previously described mutant in which phenylalanine 15 and 18 are mutated to aspartic acid to prevent Aurora A binding and activation (TPXL-1FD; Ozlü et al., 2005; Bird and Hyman, 2008; Figs. 5 A and S1). The transgene-encoded proteins were expressed at levels comparable to endogenous TPXL-1 (Figs. 5 B and S1 B) and the WT, but not the FD mutant, protein rescued embryonic lethality and the short spindle phenotype resulting from endogenous TPXL-1 depletion (Figs. 5 C and S1, E and F). Both TPXL-1WT::NG and TPXL-1FD::NG localized to astral microtubules in anaphase after depletion of the endogenous protein (Fig. 5 D).
Figure 5.
Polar clearing requires the ability of TPXL-1 to activate Aurora A. (A) Schematics of the protein products of the WT and Aurora A–binding defective (FD) tpxl-1 transgenes. (B) Immunoblots of control (N2) worms and worms expressing TPXL-1WT or TPXL-1FD after depletion of endogenous TPXL-1 by RNAi were probed for TPXL-1 and α-tubulin as a loading control. (C) Spindle length calculated by measuring the distance between the centrosomes (Fig. S1 F) is plotted for control (black) and TPXL-1 depleted (tpxl-1(RNAi); gray) embryos and for embryos expressing TPXL-1WT (green) or TPXL-1FD (purple) after endogenous TPXL-1 depletion. n = number of embryos. (D) Confocal images of anaphase embryos expressing TPXL-1WT::NG (n = 10) or TPXL-1FD::NG (n = 11) after endogenous TPXL-1 depletion. To visualize TPXL-1::NG on astral microtubules without saturating the aster centers, a gamma of 2.5 was introduced in Photoshop. (E) Time-lapse series of myosin-depleted rga-3/4Δ embryos expressing mKate2::anillin and TPXL-1WT (n = 12) or TPXL-1FD (n = 8). Embryos were depleted of HCP-4 along with endogenous TPXL-1 to ensure comparable pole separation. (F) Kymographs of the anterior cortex of the embryos in E beginning 180 s after NEBD. (G) Normalized cortical mKate2::anillin fluorescence at the anterior pole; n = number of linescans. (H) Graph plotting the distance between the anterior aster and anterior pole. n = number of embryos. (I) Model illustrating how the activation of Aurora A by TPXL-1 on astral microtubules could generate a diffusible signal that inhibits the accumulation of contractile ring proteins on the polar cortex. All error bars are SEM. Bars, 5 µm.
The transgenes encoding untagged TPXL-1WT and TPXL-1FD were introduced into the rga-3/4Δ background with a transgene expressing mKate2::anillin, and polar clearing was monitored after depletion of endogenous TPXL-1. HCP-4 was also depleted to ensure that the distance between the anterior centrosome and the polar cortex was the same for embryos expressing the WT and FD mutant proteins (Fig. 5 H). Whereas TPXL-1WT supported clearing of mKate2::anillin from the anterior cortex, clearing failed in embryos expressing TPXL-1FD (Fig. 5, E–G; and Video 9). Thus, polar clearing requires the ability of TPXL-1 to bind and activate Aurora A.
Our work identifies TPXL-1 and Aurora A kinase as molecular components of an active aster-based mechanism that clears anillin and filamentous actin from the cell poles during cytokinesis. Although both proteins localize to astral microtubules during metaphase, this localization is much more prominent after anaphase onset. TPXL-1 is highly phosphorylated during mitosis (Heidebrecht et al., 1997; Wittmann et al., 2000; Shim et al., 2015), raising the possibility that cell cycle–based phosphoregulation controls this enrichment. Our data suggest that TPXL-1 binds astral microtubules during anaphase, where it recruits and activates Aurora A (Fig. 5 I). Because Aurora A has high turnover rates (Stenoien et al., 2003; Portier et al., 2007) and forms a gradient around monopolar spindles in Drosophila S2 cells (Ye et al., 2015) we favor a model in which Aurora A activated by TPXL-1 on astral microtubules diffuses to act on the adjacent cell cortex, where it clears contractile ring proteins by phosphorylating specific targets. Consistent with the idea that the local activation of Aurora A can suppress cortical contractility, prior work has shown that inhibiting PP6, the major T-loop phosphatase that negatively regulates Aurora A (Zeng et al., 2010), causes the ectopic accumulation of Aurora A on the cell cortex (Kotak et al., 2016) and globally suppresses cortical contractility (Afshar et al., 2010).
Our findings suggest that an aster-centered Aurora A gradient together with spindle midzone-centered Aurora B activity (Fuller et al., 2008) pattern the cortex during cytokinesis by acting as landmarks of low and high contractility within the cell. Future work will be needed to identify the relevant targets and determine whether Aurora A limits RhoA activation or acts on the RhoA-dependent contractile protein network.
Materials and methods
C. elegans strains
C. elegans strains are listed in Table S1. Strains were grown on NGM plates seeded with OP50 Escherichia coli at 20°C according to standard procedures (Stiernagle, 2006) except OD314 and ZAN43, which were maintained at 23°C or 25°C because of silencing of the GFP::anillin encoding transgene at lower temperatures. Single-copy transgenes integrated at specific chromosomal loci were generated using the MosSCI method (Frøkjaer-Jensen et al., 2008). To obtain single-copy insertions on chromosome II, young EG6699 adults were injected with TPXL-1WT, TPXL-1FD, TPXL-1WT::NG, or TPXL-1FD::NG containing pCFJ350 plasmids together with plasmids encoding coinjection markers (pMA122, pGH8, pCFJ190, and pCFJ104) and the transposase (pCFJ601). After 7–10 d, heat shock was performed for 1 h at 34°C, and surviving worms were screened for the absence of the mCherry-tagged array markers. The transgenes encoding mKate2::anillin (ANI-1) or LifeAct::mKate2 were generated by injecting appropriate constructs together with the plasmids encoding transposase and coinjection markers into EG8081 to obtain a single-copy insertion on chromosome IV. The transgene encoding GFP::Aurora AAIR-1 was generated by ballistic bombardment into DP38 (unc-119 (ed3); Praitis et al., 2001).
Transgene construction
Gibson cloning (E2611; NEB) was used to construct transgenes encoding WT and mNeonGreen tagged TPXL-1 (isoform A) in pCFJ350. Parts of exons 2 and 4 and the entire exon 3 of tpxl-1 were reencoded to render the transgenes RNAi-resistant. The intron between exon 2–3 was maintained and the introns between exons 3–4 and 4–5 were removed (Fig. S1 C). Because tpxl-1 is the second gene in an operon, the mex-5 promoter (488 nt) and the tbb-2 3′ UTR (330 nt; Zeiser et al., 2011) were used to drive expression (Fig. S1 D). Site-directed mutagenesis was used to mutate phenylalanine 15 and 18 of TPXL-1 to aspartic acid. mNeonGreen was codon-optimized for expression in C. elegans by using the codon adaption index (Redemann et al., 2011), and three introns were introduced. For the transgene encoding mKate2::anillin, Gibson cloning was used to insert a sequence encoding codon-optimized mKate2 provided by H. Bringmann (Max Planck Institute for Biophysical Chemistry, Göttingen, Germany; Turek et al., 2013) into the genomic ani-1 locus, including 541 nt of the ani-1 promoter and 329 nt of its 3′ UTR, and to clone this into pCFJ350. The transgene encoding LifeAct::mKate2 was generated by using Gibson cloning to insert the LifeAct::mKate2 sequence into the plasmid pCFJ151 containing mex-5 promoter and the tbb-2 3′ UTR. The transgene encoding GFP::Aurora AAIR-1 was generated by inserting the Aurora AAIR-1 cDNA into the pIC26 plasmid containing pie-1 regulatory sequences.
Fluorescence microscopy
For imaging C. elegans embryos, gravid hermaphrodites were dissected in a 4-µl drop of M9 buffer on an 18 × 18-mm coverslip, and the coverslip was inverted onto a 2% agarose pad. Because rga-3/4Δ embryos are partially osmosensitive, all rga-3/4Δ mutant strains were imaged without pressure (Zanin et al., 2013). In brief, worms were cut open in 4 µl L-15 blastomere culture medium (Edgar and Goldstein, 2012) and imaged on a 24 × 50-mm coverslip mounted on a metal slide (Monen et al., 2005). A Vaseline ring around the drop was used as a spacer, and a coverslip was added on top to prevent evaporation.
Images in Figs. 1, 2 C, 3 B, 5 E, and S1 F were acquired at 22°C on an UltraVIEW VoX spinning disk confocal microscope (PerkinElmer), which is controlled by Volocity 6.1.1. software (PerkinElmer) and attached to an Axio Observer D1 stand (Zeiss) that was equipped with a 63× 1.4-NA Plan-Apochromat oil immersion objective (Zeiss), EMCCD C9100-50 camera (Hamamatsu), and 488- and 561-nm lasers. Images in Figs. 3 E, 4, 5 D, and S2 (C and F) were acquired at 25°C on an eclipse Ti spinning disk confocal (Nikon), which was controlled by NIS Elements 4.51 and equipped with a 100× 1.45-NA Plan-Apochromat oil immersion objective, a 488-nm laser line, and a Andor DU-888 X-11056 camera (1,024 × 1,024 pixels). Images in Fig. 2 A were acquired at 20°C on an Andor Revolution XD Confocal System (Andor Technology) and a spinning disk confocal scanner unit (CSU-10; Yokogawa) mounted on an inverted microscope (TE2000-E; Nikon) equipped with 100× or 60× 1.4-NA Plan-Apochromat lenses, and outfitted with an electron multiplication back-thinned charged-coupled device camera (iXon; Andor Technology). All immunofluorescence images were taken on a laser scanning confocal TCS SP5 microscope (Leica) controlled by the Leica Application Suite Software 2.7.2. and equipped with a 63× 1.4-NA Plan-Apochromat oil immersion objective, and 405-, 488-, and 594-nm lasers.
Image quantifications
All quantification was done on raw images in Fiji (Schindelin et al., 2012). For the image analysis, NEBD was defined as the time point when mCherry::histone in the nucleus equilibrated with the cytoplasm. In strains without a mCherry::histone marker, NEBD was defined as the time point when the border of nucleus disappeared in transmitted light images. Linescans were drawn in Fiji along the cortex from the anterior to the posterior pole at defined time points after NEBD (Fig. 1 F). Sometimes the site of meiotic polar body extrusion resulted in a bright anterior signal at the cell cortex, and these cortices were excluded. The cytoplasmic background intensity was measured in a small box close to the posterior pole and subtracted from each cortex value. The mean anterior (0–10%), equatorial (45–55%), and posterior (90–100%) fluorescence intensity was calculated for each condition and plotted over time. All anillin fluorescence intensity values were normalized to the mean anterior fluorescence intensity 180 s after NEBD in myosin-depleted rga-3/4Δ embryos. The distance between the anterior pole and anterior aster was measured by drawing a line from the center of the centrosome (GFP::SDP-5) to the anterior pole (Figs. 3 A and 5 H). In one of seven ZAN248 and five of 12 ZAN249 embryos, GFP::SDP-5 was silenced, and in those the transmitted light images were used. To measure the chromosomal fluorescence intensity of GFP::GSP-1 and GFP::GSP-2 (Fig. 2 B), a rectangular box was drawn around the chromosome area on the mCherry::histone channel and the GFP fluorescence intensity was measured in the same box in GFP channel. Then the box was expanded by 5 pixels on each side, and the fluorescence intensity was measured. The signal intensity and the area difference between the original box and the expanded box were used to calculate the background intensity per pixel, which was subtracted from the intensity of original box. Fluorescent images were processed in Fiji and Adobe Photoshop Elements. If gamma scaling was applied in Photoshop, it is indicated in the figure legends. Graphs were plotted in Prism and Excel, and figures were assembled in Affinity Designer.
Quantification of microtubule dynamics
Microtubule nucleation rate was measured by acquiring EBP-2::GFP images at 400-ms intervals for a period of 1 min at 25°C (Srayko et al., 2005). In Fiji, an arc (29–30 µm long) was drawn 9 µm away from the anterior centrosome and underneath the anterior polar cortex. From these arcs, kymographs were generated for a 1-min time interval, and EBP-2::GFP dots were manually counted. To measure microtubule growth rate, individual EBP-2::GFP dots emerging from the anterior centrosome were manually tracked using Fiji Manual Tracking plugin. The position of EBP-2::GFP dot with respect to the centrosome was plotted at each time interval, and the growth rate of individual microtubules was calculated using the slope of each line (Excel). Microtubule growth rate was analyzed in at least three embryos, and 8–12 microtubules were tracked for each embryo.
Immunofluorescence
For immunofluorescence, gravid C. elegans hermaphrodites were cut open in 10 µl M9 on 0.1% poly-l-lysine–coated glass slides. Embryos were freeze-cracked in liquid N2 and fixed for 20 min in −20°C 100% methanol. Slides were washed in 1× PBS (0.2% Tween) for 5 min and incubated with rabbit anti-TPXL-1 (1:1,000; Ozlü et al., 2005; provided by A.A. Hyman, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany) or rabbit anti-Aurora AAir-1 (1:200; Hannak et al., 2001) combined with mouse anti–α-tubulin antibodies (1:250, DM1-α; T9026; Sigma-Aldrich) at 4°C overnight. Slides were washed 3× in 1× PBS (0.2% Tween) and incubated for 1 h at room temperature with anti-mouse Alexa Fluor 488 (1:500; A11001; Molecular Probes), anti-rabbit Alexa Fluor 594 (1:500; A21207; Molecular Probes), and 1 µg/ml Hoechst 33258 (861405; Sigma-Aldrich).
Immunoblotting
For immunoblotting, 30–40 adult worms were picked and washed multiple times in M9 buffer. Sample buffer was added, and worms were incubated at 95°C for 5 min and sonicated for 20 min (Zanin et al., 2011). Membranes were incubated with rabbit anti-TPXL-1 (1:1,000; Ozlü et al., 2005), mouse-anti α-tubulin (1:1,000) primary antibodies, and HRP-conjugated rabbit (170-6515; Bio-Rad) and mouse (170-6516; Bio-Rad) secondary antibodies.
C. elegans RNAi and lethality assays
For dsRNA production, the targeting region was amplified from C. elegans cDNA or genomic DNA by PCR with the T7-containing oligonucleotides listed in Table S2. The purified PCR products (Macherey-Nagel 740609) were used for T7 in vitro transcription (MEGAscript; AM1334; Ambion). L4 or young adults were injected with dsRNA, and the embryos of the injected worms were filmed after 24–28 h at 20°C or 20–24 h at 23°C. For codepletion experiments, dsRNAs were mixed at 1:1 ratio. Because the ebp-2::gfp transgene was silenced in the rga-3/4Δ mutant background, we depleted RGA-3/4 by RNAi to measure EBP-2::GFP dynamics. For tpxl-1(RNAi), similar depletion levels were observed after 24 h (Fig. 5 B) and 40 h (Fig. S1 B; both at 20°C). Embryonic lethality assays were performed at 20°C, and single injected worms were placed on NGM plates 24 h and killed 48 h after injection. The number of dead embryos and larvae was counted 24 h later.
Statistical analysis
Mean values are displayed with error bars representing either SEM or SD as indicated in the figure legends. Statistical significance was calculated using two-tailed Student’s t test after it was confirmed that the data displayed a normal distribution using the Kolmogorov–Smirnov test.
Online supplemental material
Fig. S1, related to Figs. 4 and 5, shows transgenes encoding untagged and mNG-tagged WT and FD mutant TPXL-1 and their ability to rescue embryonic lethality resulting from endogenous TPXL-1 depletion. Fig. S2, related to Fig. 4, shows the localization of endogenous TPXL-1 and Aurora AAIR-1, measurement of microtubule growth and nucleation rates in control embryos at metaphase and early and late anaphase, and GFP::anillin dynamics in myosin-depleted rga-3/4(RNAi) embryos. Table S1 lists C. elegans strains used in this study. Table S2 lists oligonucleotides used for dsRNA production. Video 1 is related to Fig. 1; Video 2 is related to Fig. 1; Video 3 is related to Fig. 2; Videos 4 and 5 are related to Fig. 3; Videos 6, 7, and 8 are related to Fig. 4; and Video 9 is related to Fig. 5.
Supplementary Material
Acknowledgments
We thank A. Desai, B. Conradt, and T. Mikeladze-Dvali for discussions and comments on the manuscript and H. Harz, N. Lebedeva, and M. Schwarz for microscopy and technical support. We thank the Leonhardt and Conradt laboratories for sharing expertise, H. Bringmann for the mKate2 plasmid, and A.A. Hyman for the TPXL-1 antibody.
We thank the Caenorhabditis Genetic Center (funded by the National Institutes of Health Office of Research Infrastructure Programs P40 OD010440) for strains. This work was supported by grants to K. Oegema (National Institutes of Health; GM074207), E. Zanin (Deutsche Forschungsgemeinschaft, ZA619/3-1), and A.X. Carvalho (European Research Council; 640553–ACTOMYO). T. Kim was supported by a grant to Arshad Desai (National Institutes of Health; GM074215). K. Oegema receives salary and other support from the Ludwig Institute for Cancer Research. S. Mangal is a member of International Max Planck Research School for Molecular Life Sciences, and J. Sacher is a member of the Life Science Munich graduate program; both thank their programs for support.
The authors declare no competing financial interests.
Author contributions: E. Zanin and K. Oegema designed the experiments and wrote the paper. S. Mangal, J. Sacher, T. Kim, and E. Zanin performed all experiments and analyzed the data. D.S. Osório, A.X. Carvalho, and F. Motegi generated material used for the experiments.
References
- Afshar K., Werner M.E., Tse Y.C., Glotzer M., and Gönczy P.. 2010. Regulation of cortical contractility and spindle positioning by the protein phosphatase 6 PPH-6 in one-cell stage C. elegans embryos. Development. 137:237–247. 10.1242/dev.042754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bement W.M., Benink H.A., and von Dassow G.. 2005. A microtubule-dependent zone of active RhoA during cleavage plane specification. J. Cell Biol. 170:91–101. 10.1083/jcb.200501131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A.W., and Hyman A.A.. 2008. Building a spindle of the correct length in human cells requires the interaction between TPX2 and Aurora A. J. Cell Biol. 182:289–300. 10.1083/jcb.200802005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Foss M., Tseng K.-F., and Zhang D.. 2008. Redundant mechanisms recruit actin into the contractile ring in silkworm spermatocytes. PLoS Biol. 6:e209 10.1371/journal.pbio.0060209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Avino P.P. 2009. How to scaffold the contractile ring for a safe cytokinesis: Lessons from Anillin-related proteins. J. Cell Sci. 122:1071–1079. 10.1242/jcs.034785 [DOI] [PubMed] [Google Scholar]
- D’Avino P.P., Giansanti M.G., and Petronczki M.. 2015. Cytokinesis in animal cells. Cold Spring Harb. Perspect. Biol. 7:a015834–a18. 10.1101/cshperspect.a015834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Rybina S., Müller-Reichert T., Shevchenko A., Shevchenko A., Hyman A., and Oegema K.. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17:2421–2435. 10.1101/gad.1126303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar L.G., and Goldstein B.. 2012. Culture and Manipulation of Embryonic Cells. 107. Second edition New York: Elsevier Inc. 25 pp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeut J., Cheerambathur D.K., Krenning L., Oegema K., and Desai A.. 2012. Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore. J. Cell Biol. 196:469–482. 10.1083/jcb.201111107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe V.E., and von Dassow G.. 2008. Stable and dynamic microtubules coordinately shape the myosin activation zone during cytokinetic furrow formation. J. Cell Biol. 183:457–470. 10.1083/jcb.200807128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C., Davis M.W., Hopkins C.E., Newman B.J., Thummel J.M., Olesen S.-P., Grunnet M., and Jorgensen E.M.. 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40:1375–1383. 10.1038/ng.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller B.G., Lampson M.A., Foley E.A., Rosasco-Nitcher S., Le K.V., Tobelmann P., Brautigan D.L., Stukenberg P.T., and Kapoor T.M.. 2008. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 453:1132–1136. 10.1038/nature06923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R.A., Paluch E., and Oegema K.. 2012. Cytokinesis in animal cells. Annu. Rev. Cell Dev. Biol. 28:29–58. 10.1146/annurev-cellbio-101011-155718 [DOI] [PubMed] [Google Scholar]
- Greenan G., Brangwynne C.P., Jaensch S., Gharakhani J., Jülicher F., and Hyman A.A.. 2010. Centrosome size sets mitotic spindle length in Caenorhabditis elegans embryos. Curr. Biol. 20:353–358. 10.1016/j.cub.2009.12.050 [DOI] [PubMed] [Google Scholar]
- Hannak E., Kirkham M., Hyman A.A., and Oegema K.. 2001. Aurora-A kinase is required for centrosome maturation in Caenorhabditis elegans. J. Cell Biol. 155:1109–1116. 10.1083/jcb.200108051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattersley N., Cheerambathur D., Moyle M., Stefanutti M., Richardson A., Lee K.-Y., Dumont J., Oegema K., and Desai A.. 2016. A nucleoporin docks protein phosphatase 1 to direct meiotic chromosome segregation and nuclear assembly. Dev. Cell. 38:463–477. 10.1016/j.devcel.2016.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidebrecht H.J., Buck F., Steinmann J., Sprenger R., Wacker H.H., and Parwaresch R.. 1997. p100: A novel proliferation-associated nuclear protein specifically restricted to cell cycle phases S, G2, and M. Blood. 90:226–233. [PubMed] [Google Scholar]
- Jordan S.N., and Canman J.C.. 2012. Rho GTPases in animal cell cytokinesis: An occupation by the one percent. Cytoskeleton (Hoboken). 69:919–930. 10.1002/cm.21071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T., Lara-Gonzalez P., Prevo B., Meitinger F., Cheerambathur D.K., Oegema K., and Desai A.. 2017. Kinetochores accelerate or delay APC/C activation by directing Cdc20 to opposing fates. Genes Dev. 31:1089–1094. 10.1101/gad.302067.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S., Afshar K., Busso C., and Gönczy P.. 2016. Aurora A kinase regulates proper spindle positioning in C. elegans and in human cells. J. Cell Sci. 129:3015–3025. 10.1242/jcs.184416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewellyn L., Dumont J., Desai A., and Oegema K.. 2010. Analyzing the effects of delaying aster separation on furrow formation during cytokinesis in the Caenorhabditis elegans embryo. Mol. Biol. Cell. 21:50–62. 10.1091/mbc.E09-01-0089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox A.S., Habermann B., Desai A., and Oegema K.. 2005. Distinct roles for two C. elegans anillins in the gonad and early embryo. Development. 132:2837–2848. 10.1242/dev.01828 [DOI] [PubMed] [Google Scholar]
- Matsumura F., Yamakita Y., and Yamashiro S.. 2011. Myosin light chain kinases and phosphatase in mitosis and cytokinesis. Arch. Biochem. Biophys. 510:76–82. 10.1016/j.abb.2011.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima M. 2016. Centralspindlin in Rappaport’s cleavage signaling. Semin. Cell Dev. Biol. 53:45–56. 10.1016/j.semcdb.2016.03.006 [DOI] [PubMed] [Google Scholar]
- Monen J., Maddox P.S., Hyndman F., Oegema K., and Desai A.. 2005. Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nat. Cell Biol. 7:1248–1255. 10.1038/ncb1331 [DOI] [PubMed] [Google Scholar]
- Motegi F., Velarde N.V., Piano F., and Sugimoto A.. 2006. Two phases of astral microtubule activity during cytokinesis in C. elegans embryos. Dev. Cell. 10:509–520. 10.1016/j.devcel.2006.03.001 [DOI] [PubMed] [Google Scholar]
- Murthy K., and Wadsworth P.. 2008. Dual role for microtubules in regulating cortical contractility during cytokinesis. J. Cell Sci. 121:2350–2359. 10.1242/jcs.027052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K., Desai A., Rybina S., Kirkham M., and Hyman A.A.. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209–1226. 10.1083/jcb.153.6.1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otomo T., Otomo C., Tomchick D.R., Machius M., and Rosen M.K.. 2005. Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol. Cell. 18:273–281. 10.1016/j.molcel.2005.04.002 [DOI] [PubMed] [Google Scholar]
- Ozlü N., Srayko M., Kinoshita K., Habermann B., O’toole E.T., Müller-Reichert T., Schmalz N., Desai A., and Hyman A.A.. 2005. An essential function of the C. elegans ortholog of TPX2 is to localize activated aurora A kinase to mitotic spindles. Dev. Cell. 9:237–248. 10.1016/j.devcel.2005.07.002 [DOI] [PubMed] [Google Scholar]
- Petry S., Groen A.C., Ishihara K., Mitchison T.J., and Vale R.D.. 2013. Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell. 152:768–777. 10.1016/j.cell.2012.12.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekny A.J., and Glotzer M.. 2008. Anillin is a scaffold protein that links RhoA, actin, and myosin during cytokinesis. Curr. Biol. 18:30–36. 10.1016/j.cub.2007.11.068 [DOI] [PubMed] [Google Scholar]
- Piekny A.J., and Maddox A.S.. 2010. The myriad roles of Anillin during cytokinesis. Semin. Cell Dev. Biol. 21:881–891. 10.1016/j.semcdb.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Piekny A., Werner M., and Glotzer M.. 2005. Cytokinesis: Welcome to the Rho zone. Trends Cell Biol. 15:651–658. 10.1016/j.tcb.2005.10.006 [DOI] [PubMed] [Google Scholar]
- Portier N., Audhya A., Maddox P.S., Green R.A., Dammermann A., Desai A., and Oegema K.. 2007. A microtubule-independent role for centrosomes and aurora A in nuclear envelope breakdown. Dev. Cell. 12:515–529. 10.1016/j.devcel.2007.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., and Austin J.. 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 157:1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redemann S., Schloissnig S., Ernst S., Pozniakowsky A., Ayloo S., Hyman A.A., and Bringmann H.. 2011. Codon adaptation-based control of protein expression in C. elegans. Nat. Methods. 8:250–252. 10.1038/nmeth.1565 [DOI] [PubMed] [Google Scholar]
- Rodrigues N.T.L., Lekomtsev S., Jananji S., Kriston-Vizi J., Hickson G.R.X., and Baum B.. 2015. Kinetochore-localized PP1-Sds22 couples chromosome segregation to polar relaxation. Nature. 524:489–492. 10.1038/nature14496 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz C., Stevens J., and Spang A.. 2007. Functions of the novel RhoGAP proteins RGA-3 and RGA-4 in the germ line and in the early embryo of C. elegans. Development. 134:3495–3505. 10.1242/dev.000802 [DOI] [PubMed] [Google Scholar]
- Schonegg S., Constantinescu A.T., Hoege C., and Hyman A.A.. 2007. The Rho GTPase-activating proteins RGA-3 and RGA-4 are required to set the initial size of PAR domains in Caenorhabditis elegans one-cell embryos. Proc. Natl. Acad. Sci. USA. 104:14976–14981. 10.1073/pnas.0706941104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J.M., Ashcroft N., Donovan P.J., and Golden A.. 1998. A highly conserved centrosomal kinase, AIR-1, is required for accurate cell cycle progression and segregation of developmental factors in Caenorhabditis elegans embryos. Development. 125:4391–4402. [DOI] [PubMed] [Google Scholar]
- Shim S.Y., Perez de Castro I., Neumayer G., Wang J., Park S.K., Sanada K., and Nguyen M.D.. 2015. Phosphorylation of targeting protein for Xenopus kinesin-like protein 2 (TPX2) at threonine 72 in spindle assembly. J. Biol. Chem. 290:9122–9134. 10.1074/jbc.M114.591545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srayko M., Kaya A., Stamford J., and Hyman A.A.. 2005. Identification and characterization of factors required for microtubule growth and nucleation in the early C. elegans embryo. Dev. Cell. 9:223–236. 10.1016/j.devcel.2005.07.003 [DOI] [PubMed] [Google Scholar]
- Stenoien D.L., Sen S., Mancini M.A., and Brinkley B.R.. 2003. Dynamic association of a tumor amplified kinase, Aurora-A, with the centrosome and mitotic spindle. Cell Motil. Cytoskeleton. 55:134–146. 10.1002/cm.10120 [DOI] [PubMed] [Google Scholar]
- Stiernagle T. 2006. Maintenance of C. elegans (Feb. 11, 2006). WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su K.-C., Takaki T., and Petronczki M.. 2011. Targeting of the RhoGEF Ect2 to the equatorial membrane controls cleavage furrow formation during cytokinesis. Dev. Cell. 21:1104–1115. 10.1016/j.devcel.2011.11.003 [DOI] [PubMed] [Google Scholar]
- Toya M., Terasawa M., Nagata K., Iida Y., and Sugimoto A.. 2011. A kinase-independent role for Aurora A in the assembly of mitotic spindle microtubules in Caenorhabditis elegans embryos. Nat. Cell Biol. 13:708–714. 10.1038/ncb2242 [DOI] [PubMed] [Google Scholar]
- Turek M., Lewandrowski I., and Bringmann H.. 2013. An AP2 transcription factor is required for a sleep-active neuron to induce sleep-like quiescence in C. elegans. Curr. Biol. 23:2215–2223. 10.1016/j.cub.2013.09.028 [DOI] [PubMed] [Google Scholar]
- von Dassow G. 2009. Concurrent cues for cytokinetic furrow induction in animal cells. Trends Cell Biol. 19:165–173. [DOI] [PubMed] [Google Scholar]
- von Dassow G., Verbrugghe K.J.C., Miller A.L., Sider J.R., and Bement W.M.. 2009. Action at a distance during cytokinesis. J. Cell Biol. 187:831–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M., Munro E., and Glotzer M.. 2007. Astral signals spatially bias cortical myosin recruitment to break symmetry and promote cytokinesis. Curr. Biol. 17:1286–1297. 10.1016/j.cub.2007.06.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T., Wilm M., Karsenti E., and Vernos I.. 2000. TPX2, A novel Xenopus MAP involved in spindle pole organization. J. Cell Biol. 149:1405–1418. 10.1083/jcb.149.7.1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.-C., Go A.C., Samson M., Cintra T., Mirsoian S., Wu T.F., Jow M.M., Routman E.J., and Chu D.S.. 2012. Sperm development and motility are regulated by PP1 phosphatases in Caenorhabditis elegans. Genetics. 190:143–157. 10.1534/genetics.111.135376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye A.A., Deretic J., Hoel C.M., Hinman A.W., Cimini D., Welburn J.P., and Maresca T.J.. 2015. Aurora A kinase contributes to a pole-based error correction pathway. Curr. Biol. 25:1842–1851. 10.1016/j.cub.2015.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanin E., Dumont J., Gassmann R., Cheeseman I., Maddox P., Bahmanyar S., Carvalho A., Niessen S., Yates J.R. III, Oegema K., and Desai A.. 2011. Affinity Purification of Protein Complexes in C. elegans. 106. Second edition New York: Elsevier Inc. 34 pp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanin E., Desai A., Poser I., Toyoda Y., Andree C., Moebius C., Bickle M., Conradt B., Piekny A., and Oegema K.. 2013. A conserved RhoGAP limits M phase contractility and coordinates with microtubule asters to confine RhoA during cytokinesis. Dev. Cell. 26:496–510. 10.1016/j.devcel.2013.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiser E., Frøkjær-Jensen C., Jorgensen E., and Ahringer J.. 2011. MosSCI and gateway compatible plasmid toolkit for constitutive and inducible expression of transgenes in the C. elegans germline. PLoS One. 6:e20082–e20086. 10.1371/journal.pone.0020082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K., Bastos R.N., Barr F.A., and Gruneberg U.. 2010. Protein phosphatase 6 regulates mitotic spindle formation by controlling the T-loop phosphorylation state of Aurora A bound to its activator TPX2. J. Cell Biol. 191:1315–1332. 10.1083/jcb.201008106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.