Abstract
Microbial cells have developed sophisticated multicomponent structures and machineries to govern basic cellular processes, such as chromosome segregation, gene expression, cell division, mechanosensing, cell adhesion and biofilm formation. Because of the small cell sizes, subcellular structures have long been difficult to visualize using diffraction-limited light microscopy. During the last three decades, optical and force nanoscopy techniques have been developed to probe intracellular and extracellular structures with unprecedented resolutions, enabling researchers to study their organization, dynamics and interactions in individual cells, at the single-molecule level, from the inside out, and all the way up to cell–cell interactions in microbial communities. In this Review, we discuss the principles, advantages and limitations of the main optical and force nanoscopy techniques available in microbiology, and we highlight some outstanding questions that these new tools may help to answer.
The functioning of a microbial cell relies on a complex set of multicomponent cellular structures. These structures often adopt specific shapes, reside in specific subcellular spaces, and carry out functions specific to these properties. Examples can be found both inside and outside the cell. Inside the cell, large assemblies formed by bacterial cytoskeletal proteins play important roles in cell division, chromosome segregation and cell shape maintenance1. On the outside of the cell, the macromolecular cell wall structure forms a barrier to separate the inner cell from its environment and fulfils essential functions such as defining cell shape, cell motility and cell adhesion processes2,3.
A current challenge in cellular and molecular microbiology is to understand how these cellular structures and machineries are assembled at the correct time and space to achieve their functions. Clearly, microscopy is one powerful method to investigate these cellular structures. However, the small sizes of bacterial cells, often in the range of ~1 μm, are not much larger than that allowed by diffraction in conventional light microscopy (~200–300 nm), making it impossible to visualize these structures, or to assess their assembly dynamics and interactions with other cellular partners, in nanoscopic details. Using much shorter wavelengths as in electron microscopy (EM) methods, such as electron cryo-tomography (ECT), provides views of microbial structures with much higher resolution, but requires substantial instrumentation and cannot be carried out in live cells.
Since the late 1980s, scientists have attempted to break the diffraction barrier in light microscopy. One branch of efforts concentrated on the ‘near-field’, in which a tiny probe is placed near the sample and scanned, thus providing images of surfaces with a resolution that is no longer limited by diffraction. Among near-field microscopy methods, atomic force microscopy (AFM), also referred to as force nanoscopy, is particularly well suited for microbiology as it can typically reach resolutions of a few nanometres for surface components of living cells. Another branch of efforts focused on the diffraction-limited ‘far-field’, in which super-resolution microscopy, also called optical nanoscopy, can probe the localization and motion of single molecules in live cells with a resolution in the range of a few tens of nanometres. These nanoscopy techniques, when further combined with biochemical and genetic interrogations, have profoundly impacted our perception of microbial structures, enabling us for the first time to analyse the organization, dynamics and interactions of cellular structures and machineries at the single molecule level, throughout the cell, and further up to intercellular interactions in microbial communities. In this Review, we describe the main principles of force and optical nanoscopy modalities currently available and discuss distinct advantages and limitations of each. We then survey recent breakthroughs that have been made possible using these new tools, going from the interior of the cell to the cell surface.
Principles of nanoscopy methods
Currently there is a large number of different nanoscopy methods available for imaging and probing small microbial cells. While each comes with different acronyms and varied detection schemes, there are common operational principles as detailed below.
Force nanoscopy
Invented in 1986, AFM was initially designed for imaging surfaces. The key breakthrough was the realization that samples can be imaged at atomic or molecular resolution, without using an incident beam of photons or electrons, but by measuring the near-field interaction between a sharp tip and the surface4. The tip is brought in contact with the specimen and raster scans the surface while sensing the minute forces (in the piconewton range) acting on the tip. A piezoelectric scanner moves the tip (or sample) in 3D with high accuracy. The tip is mounted on a soft cantilever made of silicon or silicon nitride. The deformation of the cantilever is measured with a laser beam, enabling the determination of the force on the tip. AFM can work in various environments, including aqueous buffers, thus making it appropriate to analyse biological systems under physiological conditions without the need for labelling or fixation.
AFM is commonly used for imaging the topography of biological structures, with a resolution hundreds of times better than that allowed by the optical diffraction limit. As the technique shows exquisite sensitivity for surfaces, it is ideally suited for imaging membranes and cell walls5,6 (Fig. 1, top). Several imaging modes may be used, which essentially differ in the way the tip is scanned on the surface. In the contact mode, the tip always touches the sample surface and is scanned at a constant applied force. In the dynamic (or intermittent) mode, the tip is oscillated to lower the forces during imaging, thereby minimizing deformation and alteration of the biological specimen. An important issue for reliable imaging of microbiological structures is sample preparation. While isolated structures such as membranes and cell walls can be readily imaged following adsorption on mica, whole cells need to be firmly immobilized on a solid substrate, for instance by attachment onto glass slides coated with positively charged macromolecules like polylysine or by mechanical trapping into porous polymer membranes5. Advances in instrumentation, data recording and interpretation, and sample preparation, have enabled researchers to routinely capture molecular details of microbial structures5,7. As images are recorded in buffer and in real-time, it is possible to track dynamic changes during processes like cell growth or interaction with antibiotics5. While conventional AFMs have a rather slow time resolution (one image per min), high-speed AFMs (HS-AFMs) have recently been developed, in which the use of small cantilevers and improved electronics enables imaging biosystems with millisecond resolution8. This has opened new vistas for studying physiologically relevant molecular and cellular dynamics8.
Figure 1. Probing the microbial cell from the inside out using nanoscopy.
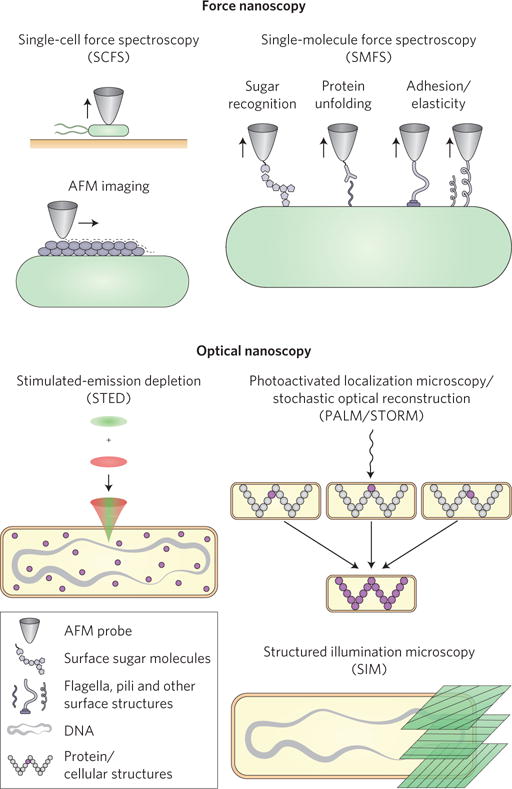
Top, force nanoscopy probes the outside of the cell. AFM imaging unravels the architecture of membranes and cell walls with nanoscale resolution. Whereas SMFS measures the localization, adhesion, elasticity and interactions of individual surface molecules and appendages, SCFS quantifies the forces driving the adhesion of whole cells. Bottom, optical nanoscopy probes the inside of the cell. The three commonly used imaging modes, STED, PALM/STORM, and SIM are shown. STED combines an excitation beam (green) and a depletion beam (red) to generate an excitation volume smaller than a diffraction-limited spot, which then raster scans the sample to generate a super-resolved image. PALM/STORM stochastically activates single molecules (purple) to acquire their positions subsequently, and reconstructs a super-resolution imaging by adding all the molecules’ positions together. SIM uses wide-field illumination of different excitation patterns and takes advantage of the generated moiré fringes (convolutions between green illumination patterns and cellular structures) to extract the information about the underlying structure.
Besides imaging, AFM may also be used as an ultrasensitive force probe, yielding direct information on the localization, adhesion, elasticity and interactions of individual molecules9,10 (Fig. 1, top). In this mode, called force spectroscopy, interaction forces are measured by recording the deflection of the cantilever while the tip is moved up and down, thus yielding a force–distance curve. Furthermore, acquiring spatially resolved force–distance curves makes it possible to map interactions and properties of surfaces9. Mapping and functionally analysing single molecules using so-called single-molecule force spectroscopy (SMFS) requires labelling the tip with specific ligands, like antibodies or lectins, and then measuring the characteristic adhesion force between the ligand and its receptor9,11. Biofunctionalization of the tip is achieved using crosslinker molecules that anchor ligands firmly at low density, while maintaining their mobility and functionality. In microbiology, SMFS can help us understand how cell surface proteins such as adhesins assemble into nanodomains on the surface of living cells12,13. SMFS may also be exploited to pull on single molecules in order to learn about their elasticity, a property that plays an important role in cell behaviour13,14. These single-molecule manipulations have shown that microbial molecules and structures feature unanticipated mechanical responses when subjected to force (for example, protein unfolding and unzipping, pilus extension and spring properties), which contribute significantly to cellular functions such as mechanosensing and adhesion.
Single-cell force spectroscopy (SCFS), a variation of SMFS, measures forces between a single cell and a target surface (Fig. 1, top)15. This modality is highly useful to understand the extent to which single-molecule properties and interactions contribute to the behaviour of whole cells. Here, a key issue is to attach a single cell to the cantilever while at the same time ensuring that cell viability and functionality are maintained. An assay was recently proposed for preparing single microbial probes by using colloidal cantilevers and a bio-inspired polydopamine wet adhesive16,17. The method is versatile, nondestructive and provides precise control of cell positioning, therefore enabling reliable single-cell analysis. Today, the combination of SMFS and SCFS is increasingly used in microbiology to decipher forces driving microorganism–microorganism, microorganism–host, and microorganism–substrate interactions. Importantly, dissecting the molecular nature of measured forces generally requires integrating AFM with modern tools of molecular genetics (mutant strains deficient for cell wall constituents, expression of specific proteins and their mutants in cell display models).
There is much interest in correlating the nanostructure of microbial systems with their chemical and biophysical properties. Multiparametric imaging is a new force-spectroscopy-based AFM modality that can simultaneously image the structure of a sample and quantify its properties and interactions, at the speed of conventional imaging (~30 sec per image)18. This technology has been applied to the native purple membrane from Halobacterium salinarum, which is composed of the light-driven proton-pump bacteriorhodopsin19,20. The adhesion and elasticity of native bacteriorhodopsins were observed at a resolution of a few nanometres19, and the flexibility of individual proteins was mapped and correlated with the protein crystal structure20. Applied to living microbial cells, multiparametric imaging has enabled observation of single filamentous bacteriophages extruding from living bacteria21, mapping adhesive nanodomains on fungal pathogens22, and unravelling structural and physical dynamics of the Staphylococcus aureus cell wall23 and extracellular matrix substances24. As such, this novel modality provides a means of relating surface structure with biophysical properties, which is essential to understand cell surface functions. Despite the numerous advantages of force nanoscopy highlighted above, there are also a number of limitations that are important to keep in mind (Box 1).
Box 1. Limitations of nanoscopy approaches.
Force nanoscopy
Surface-sensitive technique, so cannot probe intracellular structures.
AFM imaging and force spectroscopy (SMFS, SCFS) are not routine and require several months of advanced training to obtain reliable results.
Success will depend on the quality of sample and tip preparation protocols, the accuracy of recording conditions, data collection and interpretation, the type of specimen being analysed (live cells versus purified structures), and the availability of proper controls (blocking experiments, mutant strains).
Not all microbiological specimens will be well suited for AFM. Softer cells, like Gram-negative bacteria, will generally feature poor data because the cell is deformed or damaged due to the local pressure applied by the tip, or because the tip is contaminated by loosely bound molecules.
Imaging speed has always been a limiting factor and recent HS-AFM instruments may not be appropriate for imaging live cells.
Conventional SMFS and SCFS assays are low-throughput, preparation of single-molecule and single-cell probes is time-consuming, meaning performing statistically significant numbers of measurements is difficult.
Optical nanoscopy
High spatial resolution in optical nanoscopy comes at the expense of temporal resolution, and requires the optimal combination of both imaging and labelling conditions.
Fluorescent labelling of molecules of interest may alter the biological function/localization of labelled molecules, thus requiring careful validation of the functionality of the labelled molecules prior to imaging.
Phototoxicity of cells caused by long time illumination of activation and excitation light prevents time-lapse live cell studies.
Reconstructed images can be easily affected by sample drifting during imaging, mis-registration of multi-colour channels, and/or image reconstruction algorithms. Calibrations and control samples using fiducial markers should be used to avoid artefacts.
Optical nanoscopy
To probe the inside of a cell without being invasive, fluorescence light microscopy is ideal. The ultimate goal of resolving a fluorescent cellular structure with nanoscopic details is to obtain the spatial coordinates of all molecules making up the structure with nanometre precision.
Stimulated emission depletion (STED) microscopy overcomes the diffraction barrier by optically confining the excitation beam in a confocal microscope to a spot smaller than a diffraction-limited area25. The confinement can be achieved by overlaying two beams: an excitation beam to induce fluorescence; and a donut-shaped STED beam to inhibit fluorescence at the outer rim of the excitation beam (Fig. 1, bottom). As such, only molecules at the centre of the excitation beam, that is, where the inhibition STED beam intensity is zero, are allowed to fluoresce, hence effectively shrinking the point spread function (PSF) of the fluorescent spot. The two overlaid beams then scan point-to-point across a sample the same way as that in a confocal laser scanning microscope to generate a super-resolution image26. Thus, the spatial resolution is determined by the size of the intensity-zero centre of the STED beam, and the temporal resolution is determined by the scanning speed. Currently, STED can routinely reach a spatial resolution of ~40–80 nm with a frame rate of ~1 s−1, but a recent development using an ultrafast electro-optical scanning technique achieved an impressive 5–10 ms per frame with ~70 nm resolution27. STED is also naturally capable of 3D imaging with an axial resolution of up to 40 nm because of its confocal-like setup28.
In wide-field microscopy, one commonly used optical nanoscopy is single-molecule localization-based microscopy (SMLM) such as PALM (photoactivated localization microscopy), STORM (stochastic optical reconstruction microscopy) and other derivatives29–32. The basic concept is that if a particular feature of a molecule can be isolated from surrounding molecules in the same focal volume, and used to determine the molecule’s position with greater spatial precision than the focal volume, each individual molecule’s coordinates can then be sequentially determined and finally superimposed to reconstruct a super-resolved image (Fig. 1, bottom). To realize this concept, two key components, single-molecule detection and photoswitchable fluorophores, are required. Single-molecule detection allows the localization of the centroid position of a single molecule’s PSF with a few nanometre’s precision, effectively ‘breaking’ the diffraction barrier33,34. Photoswitchable fluorophores allow the stochastic activation of individual fluorophores one at a time within a diffraction-limited area by a low dosage of the activation light, effectively isolating single fluorophores from the others in the same area35. Along the axial direction, sub-diffraction-limit resolution can also be achieved by a variety of different methods36–38. For example, a simple setup using cylindrical-lens-induced astigmatism can achieve an axial resolution of ~80 nm (ref.39), and the two-objective, interference-based iPALM (interferometric PALM) can reach ~15 nm axial resolution40.
The spatial resolution of SMLM is limited by the following three factors: (1) the precision in localizing single fluorophores, which is mainly influenced by the number of photons a fluorophore can emit34; (2) labelling density (ρ) of the cellular structure, or sampling frequency, dictated by the Nyquist criterion, d = 2/√ρ for 2D imaging41; and (3) the spread of repeat localizations of same molecules42,43. When the first two factors are not limiting, the spread of repeat localizations of same molecules determines the actual spatial resolution in SMLM imaging. Currently SMLM can routinely reach a spatial resolution of ~30 nm in 2D, but the temporal resolution is low because often tens of thousands of images are required to reconstruct one super-resolution image. However, in a recent study, by using a fast CMOS (complementary metal-oxide-semiconductor) camera and a combination of noise-minimizing strategies, an impressive frame rate of up to 32 super-resolution images per second has been achieved in both fixed and live mammalian cells44, raising the promise of SMLM’s application to microbiology.
A different concept of wide-field super-resolution imaging, structured illumination microscopy45,46 (SIM, Fig. 1, bottom), utilizes a predetermined, sinusoidal illumination pattern with alternating maxima and minima to excite a fluorescently labelled cellular structure. The emitted fluorescence from the structure produces the so-called moiré fringes, which are the product of both the structure pattern and the illumination pattern. Moiré fringes contain fine details of the underlying cellular structure, and are coarse enough to be easily visualized by conventional fluorescence microscopy. Thus, by applying multiple (>3, often 10–15) illumination patterns with different orientation and phases to the same sample, information of the underlying cellular structure can be mathematically extracted45. The spatial resolution of SIM is determined by the spatial frequency of the spacing of the illumination pattern. In the linear excitation regime the spatial resolution in 2D can only be improved by a factor of two to ~120 nm. In 3D, SIM can reach an axial resolution of ~300 nm by applying a spatial illumination pattern where out-of-focus light is not detected due to interference46. However, because only ~10–15 illumination patterns are needed to reconstruct a super-resolution image, SIM can achieve a fast frame rate of less than 1 s−1. Thus, it is possible to probe the dynamics of cellular structures in super-resolution even in fast growing bacterial cells. In addition, SIM does not require special photoswitchable fluorophores and is fully compatible with regular fluorescent proteins. Because of these properties, when temporal resolution is the primary concern, SIM is often the best choice. Recent employment of nonlinearity to generate high frequency harmonics such as that in saturated SIM (SSIM)47,48 can further push the spatial resolution to ~50 nm, hence holding promise for achieving both high temporal and spatial resolution.
While different in formats, all optical nanoscopy methods such as STED, SMLM, and SIM rely on one central principle to achieve sub-diffraction-limit resolution. That is, by switching on and off the fluorescence of a subpopulation of molecules within a diffraction-limited area, molecules that are spatially indistinguishable from each other can be separated in time. This key principle of temporal separation by switching on and off fluorescence unifies all optical nanoscopy methods49. Variations of the key concept led to the development of a diverse set of optical nanoscopy methods with overly abundant acronyms. For example, turning molecules off by ground state depletion (GSD) instead of stimulated emission led to GSD microscopy50; using reversibly photoswitchable fluorophores to reduce the depletion beam intensity substantially for less photo-damage of the sample led to RESOLFT (ref.51). Using fluorescence change of fluorophores upon binding and unbinding to DNA or membrane led to PAINT (points accumulation for imaging in nanoscale topography52,53) or BALM (binding activated localization microscopy)54. Readers are referred to excellent articles for in-depth discussions of operational principles of these methods55, selection of fluorophores56,57, practical concerns and applications58,59, and quantitative analyses60,61. In Box 1 we summarize the major limitations of optical nanoscopy.
Applications in microbiology
By employing new physical principles and imaging technologies, force and optical nanoscopy techniques have provided novel insights into the organization, dynamics and functions of the microbial cell. Here, we survey some of the new biological insights learned from various nanoscopy methods in microbiology and discuss potential new directions, going through cellular structures from the inside to the outside of the cell.
Spatial organization of the nucleoid
Genetic and biochemical studies have shown that the bacterial chromosome is spatially organized into segregated, activity-insulated domains62. Optical nanoscopy has complemented population-based studies by directly visualizing the overall morphology and cellular positions of the nucleoid at the single-cell level (Fig. 2).
Figure 2. Spatial organization of bacterial nucleoid.
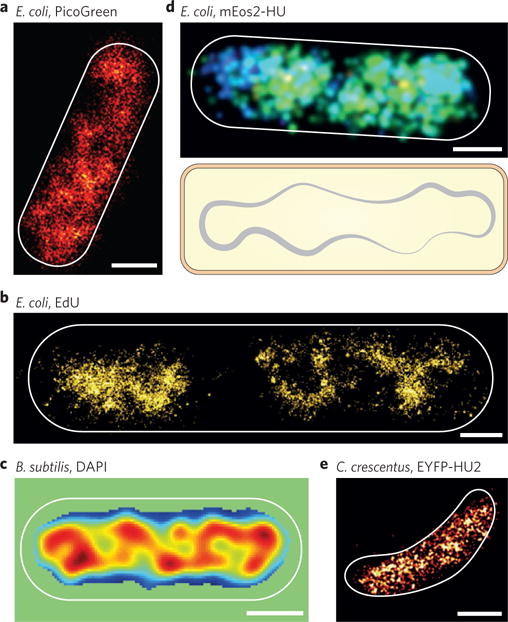
a, BALM image of a fixed E. coli cell showing voids and inhomogeneous nucleoid organization when labelled with PicoGreen54. b, STORM image of a fixed E. coli cell showing asymmetric and heterogeneous nucleoid structures63. The cell was incubated with EdU in LB medium, subsequently fixed and labelled with Alexa647 by click chemistry. c, 3D-SIM image of a DAPI-stained live B. subtilis cell showing high-density chromosomal regions (red peaks)67. d, Top, a live E. coli cell grown in minimal M9 medium showed dispersed, rather uniform distribution of mEos2-labelled HU protein in 3D super-resolution imaging70. Bottom, schematic of a bacterial nucleoid. e, Super-resolution image of EYFP-HU2 in a fixed C. crescentus cell grown in minimal M2G medium69. All cell contours are indicated by white outlines. Scale bars, 500 nm. Figure reproduced from: ref.54, ACS (a); ref.63, Elsevier (b); ref.67, Elsevier (c); ref.70, AAAS (d); ref.69, Biophys. Soc. (e).
In fixed cells, using the reversible binding and unbinding of PicoGreen, a dye that increases its fluorescence ~1000-fold upon its binding to the major groove of double-stranded DNA (dsDNA), fine fibres and small voids inside the nucleoid were observed that were not discernible in conventional light microscopy54 (Fig. 2a). In other studies (Fig. 2b), chromosomal DNA was directly labelled by incorporating a thymidine analogue EdU (5-ethynyl-2′deoxyuridine) during DNA replication, which is then conjugated with a photoswitchable dye63–65 by performing the ‘click’ reaction66. The use of a bright organic dye such as Alexa647 led to much-improved resolution, revealing a cycle of decondensation and recondensation of the nucleoid upon replication initiation and completion, and previously unseen hetero-structures of the nucleoid with regions connected by thin fibres63,65 (Fig. 2b). In live cells, DAPI (4′,6-diamidino-2-phenylindole), another dsDNA intercalating dye that can diffuse through bacterial membrane, has been used in SIM to visualize the nucleoid structure in both Bacillus subtilis67 (Fig. 2c) and Escherichia coli cells68. The ~2-fold increase of resolution in SIM is enough to visualize high-density chromosomal regions (HDRs) heterogeneously distributed along the long axis of B. subtilis cells (Fig. 2c) and also E. coli cells grown in rich medium67,68.
Alternatively, researchers have fused non-specific DNA-binding proteins (commonly nucleoid-associated proteins, NAPs) to photoactivatable fluorescent proteins (PA-FPs) and visualized their spatial distributions by SMLM as a proxy for the underlying nucleoid organization. However, in both E. coli and Caulobacter crescentus, all NAPs investigated so far showed rather dispersed, uniform distribution with no distinct organization patterns (Fig. 2d,e)69,70. This discrepancy is likely due to differences of the nucleoid organization and dynamics between live and fixed cells, growth conditions, or bacterial species. Further investigations, combined with mutational and functional analyses, will likely provide new insight into the roles of these NAPs in organizing the nucleoid.
Labelling of the nucleoid as described permits the imaging of the global nucleoid structure in super-resolution, but is not sequence-specific. FROS (Fluorescent repressor operator system71) uses the binding of a repressor protein such as LacI or TetR fused with a FP to tandem arrays of operator sites to label chromosomal DNA segment specifically. A recent study modified the FROS method and achieved a precision of ~15 nm in determining the cellular position of labelled DNA sites72, allowing looped and unlooped conformations of a short (~2 Kbps) chromosomal DNA segment to be distinguished in live E. coli cells72. If further scaled up to allow systematic, multiplexed sequence-specific imaging, this method may make it possible to investigate how the spatial organization of the nucleoid is coupled to specific gene expression activities.
Distribution and dynamics of DNA-associated machineries
Many DNA-associated molecular machineries exhibit complex spatial distributions and dynamics on the nucleoid. Here optical nanoscopy, in particular SMLM imaging, is often complemented by single-molecule tracking (SMT) to probe where these proteins localize to, how fast they are moving, and how they change in spatial distribution and mobility in response to different cellular signals or genetic mutations (Fig. 3). From these measurements it is possible to deduce reaction kinetics and dissect molecular mechanisms of these machineries at the single-molecule level in live cells.
Figure 3. Spatial distribution and dynamics of DNA-associated machineries.
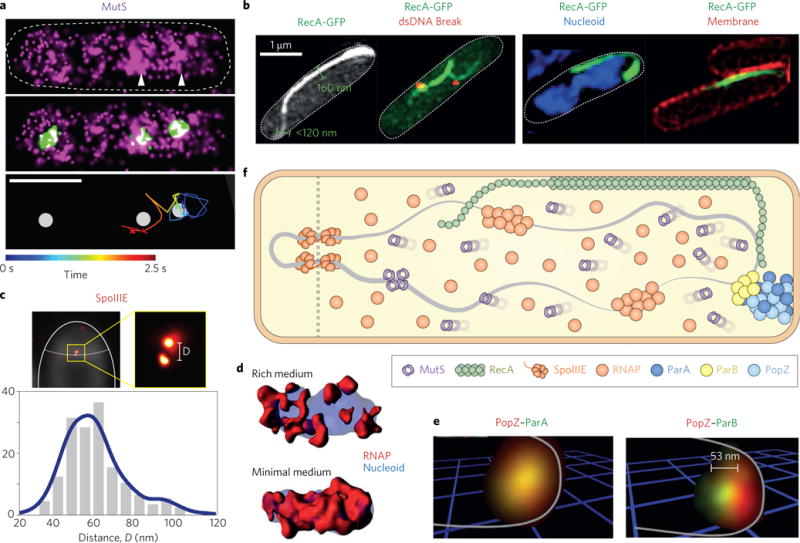
a, Spatial distribution of MutS-PAmCherry in a live E. coli cell (top), superimposed with DnaX-Citrinie-labelled replisome (middle, green spots), and a representative time-coloured SMT trajectory of MutS-PAmCherry showing its slowed diffusion in the replisome regions (bottom, white spots)73. Scale bar, 1 μm. b, 3D-SIM images of E. coli cells showing RecA-GFP alone, with ParB-mCherry labelled dsDNA break ends (red spots), with DAPI-stained DNA (blue), or with FM4-64 stained membrane (red)77. The thickness of RecA bundles was measured at 160 nm on average, and the thinner filaments at the ends of the bundle were measured <120 nm, the resolution limit of SIM. c, PALM imaging to identify the spatial arrangement of SpoIIIE motor complexes at the septum between the forespore and mother cell in B. subtilis85. Top left, a diffraction-limited, FM5-95 stained membrane image of a ΔσE B. subtilis cell overlaid with the corresponding PALM image of SpoIIIE-tdEos. Cell and septum are outlined in white. Top right, zoomed PALM image (red foci) of SpoIIIE-tdEos showed clear separation between two complexes. Bottom, the histogram of the separation distance D measured from individual dual foci showed a mean separation value at 55 nm. d, Surface rendering of 3D-SIM images of the nucleoid (DAPI, blue) and RNAP-GFP (red) in live E. coli cells showed highly clustered RNAP distribution in rich medium, but more uniform distribution in minimal medium68. e, Two-colour PALM imaging of PAmCherry-PopZ (red) with ParAG16V-EYFP (green, left) or EYFP-ParB (green, right) showing that inactive ParAG16V colocalizes with PopZ, but ParB is displaced ~53 nm away from PopZ (ref.87). f, A cartoon summarizing cellular localizations and spatial organizations of DNA-associated machineries as depicted in panels a–e. Figure reproduced from: ref.73, PNAS (a); ref.77, NPG (b); ref.85, eLife (c); ref.68, PNAS (d); ref.87, PNAS (e).
Using two-colour PALM imaging and SMT, B. subtilis MutS, an enzyme involved in DNA mismatch repair73, was observed to sample rapidly the entire nucleoid, but dwell at the replisome region (Fig. 3a,f). Importantly, MutS was found only to identify errors in newly replicated DNA through its transient association with the replisome. Using a similar strategy, the spatial search and repair pattern of DNA polymerase I and ligase upon DNA methylation damage have been mapped with high resolution, providing quantitative measurement of the search time, binding time, and enzymatic rates of these two enzymes in live cells74. Additionally, PALM imaging helped discover that in E. coli, Pol V, the translesion DNA synthesis enzyme75, was initially sequestered on the inner membrane upon the induction of SOS response; only at a late stage was it released in a RecA-dependent manner into the cytosol to perform translesion DNA synthesis76. Consistent with this observation, it was recently shown by a combination of SIM imaging and biochemical assays that in response to dsDNA break and genetic mutations, RecA forms bundled filaments along the inner membrane through its interaction with anionic phospholipids77,78 (Fig. 3b). With much enhanced resolution, these studies revealed that RecA bundles are likely composed of multiple laterally associated RecA filaments, with thinner RecA-ssDNA filaments extending from both ends (Fig. 3b,f), which dynamically sample the inner cell compartment likely in search for homology77,78.
Similar strategies also revealed interesting spatial distribution patterns and diffusive behaviours of RNA polymerase (RNAP)64,68,79–81. RNAP forms dense clusters of ~150 nm in size preferably at the edge of nucleoid in fast growing cells, but distributes more homogenously and becomes more mobile in slow-growing or transcription-inhibited cells64,68,81 (Fig. 3d). Furthermore, two-colour SIM imaging found that dense RNAP clusters colocalize with transcription anti-termination factors NusA and NusB (ref.82), likely suggesting that multiple RNAP molecules cluster on ribosomal RNA (rrn) operons83, although it remains unknown whether these RNAP clusters are actively engaged in transcription.
Finally, quantitative PALM imaging and two-colour super-resolution colocalization studies have helped elucidate molecular mechanisms of DNA-associated machineries by providing precise cellular positioning and molecular counting measurements. SpoIIE, a DNA translocase responsible for exporting the replicated chromosome from the mother cell to the forespore compartment during sporulation in B. subtilis84, was shown to form paired DNA channels on each side of the sporulation septum, the separation of which is too small (<50 nm) to be visible by conventional fluorescence microscopy85,86 (Fig. 3c,f). Another example is that ParB and ParA, two proteins involved in chromosome segregation in C. crescentus, were found to localize with ~50 nm difference with respect to the pole organizing protein PopZ (Fig. 3e,f), revealing the differential roles of PopZ in modulating these two proteins’ spatial organization and dynamics87.
Cellular localization and morphology of cytoskeletal structures
It is now well documented that a large number of bacterial proteins, in particular polymeric proteins such as actin and tubublin homologues, form subcellular structures that have specific shapes and cellular locations. Optical nanoscopy can probe spatial features and organization of these structures at unprecedented resolution, providing new information regarding their possible molecular compositions, assembly mechanisms and functional roles (Fig. 4).
Figure 4. Intracellular location and morphology of cytoskeletal structures.
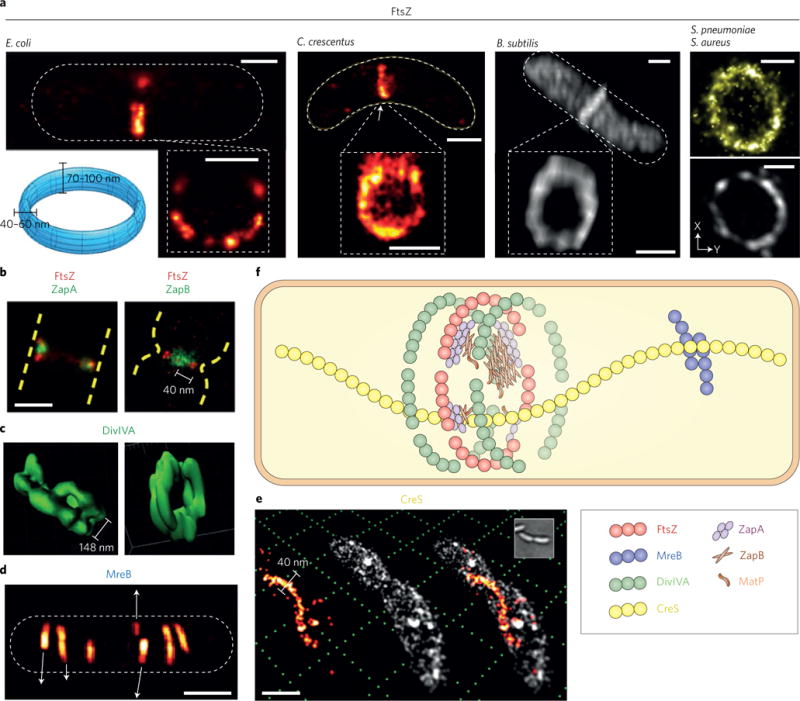
a, 3D-PALM or SIM imaging showed patchy, discontinuous FtsZ-ring structures in different bacterial species. From left to right: FtsZ-mEos3.2 in E. coli100, FtsZ-Dendra2 in C. crescentus92, FtsZ-GFP in B. subtilis91, FtsZ-spDendra2 in S. pneumoniae94 and in S. aureus91. A blue toroid at the bottom left shows the dimensions of the midcell zone the FtsZ ring occupies in most bacterial species measured. b, Two-colour PALM imaging in live E. coli cells shows that ZapA (Z-ring associated protein A) largely colocalizes with the Z-ring while ZapB is displaced inward ~40 nm (ref.103). c, SIM imaging shows that DivIVA, a peripheral membrane protein targeting FtsZ antagonizer MinC protein to membrane in B. subtilis, forms a double ring flanking the septum, and hence likely regulating the midcell positioning of the FtsZ-ring101. d, SIM imaging of GFP-MreB in B. subtilis showed MreB filaments moving independently along both directions of the membrane104. e, 3D, two-colour super-resolution imaging of Nile Red-stained membrane (white) and EYFP-labelled CreS, an intermediate-filament like protein that determines the crescent shape of C. crescentus110. The thickness of CreS filament was measured at ~40 nm. Scale bars, 500 nm except d (1 μm). f, Cartoon of cytoskeletal structures observed by optical nanoscopy as depicted in panels a–e. Figure reproduced from: ref.100, Wiley (a, E. coli); ref.92, PNAS (a, C. crescentus); ref.91, PLoS (a, B. subtilis, S. aureus); ref.94, ASM (a, S. pneumoniae); ref.103, PLoS (b); ref.101, ASM (c); ref.104, Biophys. Soc. (d); ref.110, PNAS (e).
The most studied subcellular structure in microbial optical nanoscopy so far is the bacterial cytokinesis ring formed by the essential division protein FtsZ and its partners. The FtsZ protein is a tubulin homologue and a GTPase88,89; it polymerizes at the midcell to form a ring-like structure, called the Z-ring, to recruit all other division proteins and initiate cytokinesis. While appearing as a smooth ring under conventional light microscopy, optical nanoscopy revealed that the Z-ring is a discontinuous, patchy structure in many bacterial species90–96, with FtsZ clusters loosely and heterogeneously organized in a toroid zone of ~70–100 nm by 40–60 nm (refs90,92,97–100; Fig. 4a). Most remarkably, due to much improved resolution, other proteins originally believed to form structures that are dependent on and colocalize with the Z-ring, were found to localize to places adjacent to, but distinct from, that of the Z-ring, providing new insight into their roles in positioning and stabilizing the Z-ring101–103 (Fig. 4b,c).
Beside FtsZ, actin homologues MreB and ParA, and intermediate-filament-like protein CreS also forms polymeric structures. Under super-resolution, the patchy, irregular foci formed by MreB, the protein involved in maintaining the rod-cell shape in many bacterial cells, resolved into thin, sometimes discontinuous filaments of variable lengths104–107. Importantly, live cell SIM–STED imaging observed length-dependent moving velocity of individual filaments (Fig. 4d), leading to a novel mechanistic model in which multiple cell wall synthesis motors are coupled on the same MreB filament and hence exert concerted peptidoglycan (PG) synthesis104. The other actin homologue ParA, a protein involved in chromosome segregation, was found in one study to form a narrow (~40 nm thickness) linear structure spanning the long axis of C. crescentus cells, possibly providing a track for the directional movement of the segregating chromosome guided by the ParB-parS complex108. Finally, CreS, or crescentin, a protein required for maintaining the crescent shape of C. crescentus109, has recently been imaged together with the surrounding cell membrane in live cells, both with 20–40 nm resolution in 3D (Fig. 4e)110.
Assembly and dynamics of membrane proteins
Microorganisms are surrounded by cell envelopes consisting of an inner membrane, a cell wall made of PG layers, and for Gram-negative bacteria, an outer membrane. Cell envelopes play essential roles, such as defining cell shape and division, resisting turgor pressure, providing receptor sites for viruses and antibiotics, and controlling cell adhesion and biofilm formation. At this level, optical nanoscopy continues to enable the mapping of unique membrane distribution patterns and dynamics of a variety of membrane proteins, including chemotaxis receptors111, sporulation complexes112, toxins113, respiratory systems114 and surface sugar metabolizing complexes115, and helped elucidate possible mechanisms of how a membrane protein’s localization and dynamics are coupled to its biological function. Yet here, force nanoscopy offers unique advantages over optical nanoscopy in probing structural and biophysical properties of cell surfaces with unmatched spatiotemporal resolution.
Force nanoscopy has enabled probing of cell surface structures directly in live cells116. AFM captured the surface ultrastructure of spores from the fungus Phanerochaete chrysosporium, showing that the surface was uniformly covered with crystalline protein layers (rodlets) with a periodicity of 10 nm (ref.116). However, the highest resolution was achieved on isolated membranes. Since the first molecular images of native E. coli OmpF porin surfaces were reported in 1995117, AFM has become an increasingly popular technique for observing protein assemblies and their structural changes in bacterial membranes, including surface layers, outer membranes from Gram-negative bacteria, and intracytoplasmic membranes of purple photosynthetic bacteria4–7. AFM may also be combined with cryo-EM to resolve the architecture of surface layers like the exosporium in B. cereus and B. thuringiensis spores118.
Currently, there is much effort in applying HS-AFM in microbiology. A prominent example is the fast imaging of the purple membrane, which contains 2D arrays of the light-driven proton pump bacteriorhodopsin. The structure of individual bacteriorhodopsin trimers could be observed at a speed of 100 ms per image119, and the motion of monomers and trimers was visualized, suggesting that the protein arrays were in dynamic association–dissociation equilibrium. A pioneering dynamic imaging study revealed structural changes of bacteriorhodopsin in response to light within 1 s (Fig. 5a)120. Under illumination with green light, cytoplasmic parts of monomers were brought into contact with neighbouring trimers and a cooperative interplay between excited proteins was observed. Additional observations brought novel insights into the trimer–trimer interaction energy and the amino acids involved in trimer interactions121.
Figure 5. Imaging the structure and dynamics of single proteins in bacterial membranes.
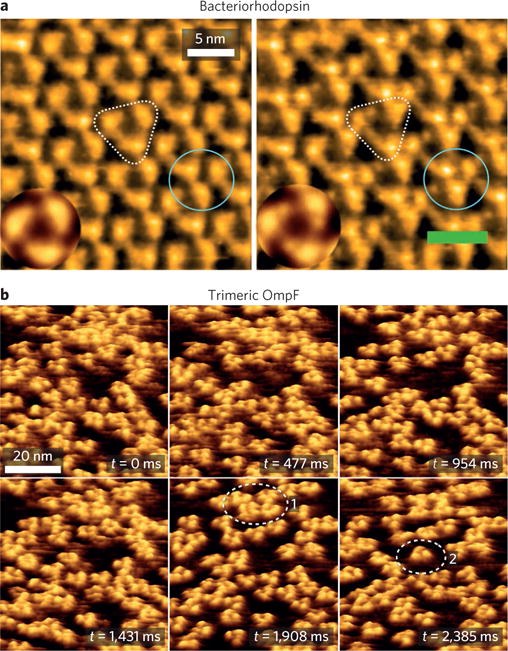
a, High-speed AFM images of bacteriorhodopsin documenting structural differences under dark (left) and green light illumination (right)8. The circles and triangles highlight trefoils and trimers (insets: averaged images of trimers). b, High-speed AFM images of the diffusion and interaction dynamics of trimeric OmpF (ref.8). Unlike protein trimers outlined in (1), the protein outlined in (2) is highly mobile. Figure reproduced from ref.8, ACS.
The high-speed technology has also provided key information about the diffusion of membrane proteins, which are often coupled to their functional states. An elegant example is the characterization of the diffusion and interaction of the OmpF porin122 (Fig. 5b). The motion of about 70 OmpF trimers was tracked and individual subunits were resolved, revealing wide distribution in the membrane due to diffusion-limited aggregation. Protein motion correlated with the local density of proteins, but single protein molecules were also found to diffuse freely.
Three-dimensional architecture and synthesis mode of cell wall
PG is a major component of bacterial cell walls, and has received considerable attention in view of its role in controlling cell shape, growth and division. Force and optical nanoscopy methods are often used in conjunction with each other to probe not only the overall architecture of PG with nanometre precision, but also its remodelling and synthesis dynamics with specificity. Together with EM, they have disclosed a diversity of PG supramolecular structures in different bacterial species123,124.
For example, while all Gram-positive microorganisms have thick cell walls, their PG architectures are drastically different from each other. Analysis of purified sacculi from B. subtilis revealed PG nanocables running parallel to the short cell axis and led to a model in which glycan strands polymerize to form PG ropes125. The coccal bacterium S. aureus showed a complex surface pattern with rings and knobble architectures associated with nascent and old PG synthesis respectively, and a large belt of PG forming a piecrust structure in the cell division plane126. Ovococcal bacteria such as Streptococcus pneumonia, Enterococcus facelis and Lactococcus lactis, however, showed relatively smooth surfaces, with thin PG bands parallel to the short axis of the cell and no visible twists associated with cable-like structures127. In the Gram-negative bacterium E. coli, high-resolution AFM revealed features running parallel to the plane of the purified sacculi and bands of porosity. Further combined with STORM imaging, it was suggested that only porous regions of PG are permissive for synthesis123. Of note, cell wall architectures have also been explored in living cells: AFM imaging revealed networks of PG fibres and nanocables on Bacillus atrophaeus spores128 and L. lactis cells129, respectively.
In optical nanoscopy, labelling of PG using dye-conjugated wheat germ agglutinin (WGA, binds to GlcNAc residues130), fluorescent d-amino acids (FDAAs, can be incorporated into PG when included in growth media131,132) and fluorescent vancomycin (binds to terminal D-Ala–D-Ala residues133) have allowed novel PG synthesis modes to be identified in a variety of different bacterial species93,134–136. Alternatively, labelling of proteins involved in PG synthesis and remodelling using PA-FP fusions96,137, immunofluorescence136,138, or fluorescent antibiotics that specifically bind to PBPs (penicillin binding proteins139) has revealed distinct cellular localization patterns of these enzymes with respect to other cell division and cell wall components, providing new insight into their roles at different stages of cell cycle.
Molecular mechanisms of biofilm formation
A remarkable feature of microorganisms is their ability to switch between two lifestyles, namely free-living planktonic cells and biofilm-associated cells3. Two major players in biofilm formation are cell surface adhesins, which mediate adhesion to biomaterial and host surfaces, and extracellular matrix polymers that hold the cells together. AFM-based force spectroscopy has been used to study the binding mechanism of single molecules (SMFS), and to quantify the forces driving the adhesion of whole cells (SCFS). Much of our recent knowledge comes from the analysis of Staphylococcus epidermidis and S. aureus, which form biofilms on indwelling medical devices. Using SMFS, weak interactions (~100 pN) were measured between single fibronectin (Fn) molecules and fibronectin-binding proteins (FnBPs) from S. epidermidis140. FnBP binding forces were also measured on isolates from patients with an infected device, revealing a distinct binding force signature and specific single-amino-acid polymorphisms141. Notably, the S. epidermidis SdrG protein was found to bind to the blood plasma protein fibrinogen with a force of 2 nN, which is much stronger than the force measured so far for all other adhesins142. This strong binding originates from the high affinity ‘dock, lock, and latch’ (DLL) binding mechanism, which involves dynamic conformational changes leading to highly stable complexes. Unlike SdrG, the collagen-binding protein SdrF featured a dual-ligand-binding activity143. Adhesion to collagen-coated substrates was mediated by weak and strong bonds involving two distinct regions of the protein. The high dissociation rates of the bonds suggested they were less stable than DLL bonds.
The extracellular matrix mediates intercellular adhesion during the development of biofilms, but the underlying mechanisms are poorly understood. A widely investigated biofilm matrix component is the polycationic polysaccharide intercellular adhesin (PIA). Applying force nanoscopy to a clinically-relevant methicillin-resistant S. aureus (MRSA) strain, revealed the binding mechanism of PIA24 (Fig. 6a). Whereas multiparametric imaging showed that bacteria expressing PIA were surrounded by a soft and adhesive matrix, force measurements demonstrated that PIA mediates multivalent electrostatic interactions with polyanionic teichoic acids on the surface of neighbouring cells.
Figure 6. The matrix revolutions, dissecting the binding mechanisms of biofilm matrices.
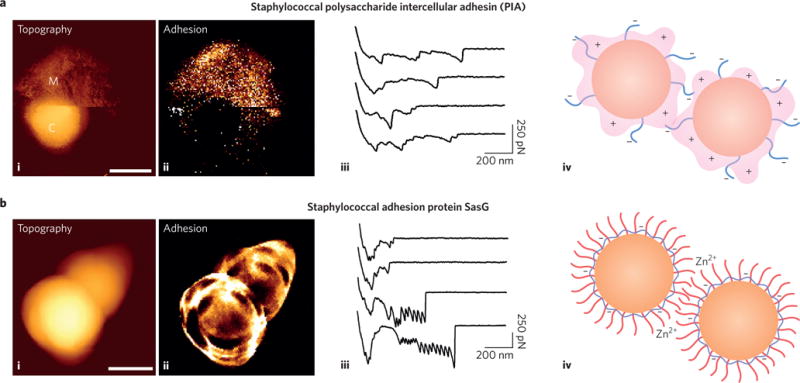
a, AFM unravels the molecular forces of the staphylococcal polysaccharide intercellular adhesin (PIA)24: (i) topographic and (ii) adhesion images of a living S. aureus cell expressing PIA (labels M and C highlight the PIA matrix and the cell surface, respectively); (iii) adhesion force signatures recorded between two cells; and (iv) the binding mechanism involving the multivalent electrostatic interaction of cationic PIA (pink) with polyanionic teichoic acids (blue) on the cell surface. b, AFM deciphers the zinc-dependent mechanical properties of the staphylococcal adhesion protein SasG (ref.23): (i) topographic and (ii) adhesion images of S. aureus cells expressing SasG recorded in the presence of zinc; (iii) adhesion force profiles recorded between two bacteria in the presence of zinc; and (iv) a proposed model for the zinc-dependent activation of intercellular adhesion (SasG proteins are in red, teichoic acids in blue). Scale bars, 1 μm. Figure reproduced from: ref.24, ACS (a); ref.23, PNAS (b).
There is evidence that the development of microbial biofilms also involves protein-based matrices. FnBPA proteins from S. aureus were shown to promote intercellular adhesion via low-affinity, zinc-dependent homophilic bonds between FnBPA domains on neighbouring cells144. Unlike the very strong and stable DLL bonds, homophilic bonds showed moderate strength and fast dissociation, which could be important for biofilm dissemination. For another S. aureus matrix protein SasG, AFM demonstrated zinc-dependent homophilic interactions between proteins on opposing bacteria, and unravelled the molecular elasticity of individual SasG molecules23 (Fig. 6b). While the protein mechanics measured on live cells was consistent with those measured on purified proteins145, absorption of zinc to the bacterial cell surface favoured SasG exposure and promoted homophilic binding of SasG proteins on opposing cells (Fig. 6b).
Cell–cell adhesion interactions have also been studied in fungal cells. For example, aggregation of the yeast Saccharomyces cerevisiae is mediated by lectin-like flocculin (Flo) proteins through molecular interactions that are not fully understood. Weak lectin–sugar interactions as well as strong unfolding forces associated with the force-induced extension of the hydrophobic tandem repeats of Flo proteins, have been shown to be involved in yeast flocculation146. The same behaviour was demonstrated for agglutinin-like sequence (Als) adhesins from C. albicans13, suggesting that lectin-like and Ig-like fungal adhesins have evolved similar domains with high mechanical strength to achieve adhesion. In C. albicans, single-molecule manipulations demonstrated that mechanical force triggers the formation of amyloid-like nanodomains of Als adhesins on the yeast cell surface, thus showing that microorganisms can use functional amyloids to activate adhesion, both through the lateral clustering of adhesins, and through amyloid bonds between cells.
Recently, SCFS has been implemented to study forces guiding microorganism–host interactions. The nanoscale adhesion forces between P. aeruginosa and host epithelial cells were shown to involve the extension of bacterial type IV pili and the formation of membrane tethers from host cells17. These mechanical responses may play a role in host colonization by increasing the adhesion lifetime of bacteria. In another study, the adhesion forces between C. albicans and macrophages were found to involve multiple specific molecular bonds between lectin receptors on the macrophage membrane and mannan carbohydrates on the fungal cell surface147. Furthermore, an innovative method combining multiparametric imaging with single bacterial probes was developed to map simultaneously the topography and adhesion properties of human skin at high spatiotemporal resolution148.
Optical nanoscopy has also provided insight into biofilm formation. Using an in vivo labelling strategy, the extracellular matrix of developing biofilms of Vibrio cholerae was imaged with single-molecule precision and three distinct levels of spatial organization of cells in the biofilm revealed, suggesting complementary architectural roles of the main matrix constituents149. In another study, biogenesis of bacterial membrane vesicles in Pseudomonas aeruginosa biofilms was investigated, and explosive cell lysis was found to play a crucial role in forming membrane vesicles that contribute to the structural integrity of the biofilm matrix150.
Combining the unique advantages of optical and force nanoscopy is particularly powerful in biofilm research. In recent work, AFM force measurement was combined with 3D-SIM imaging to show that cell surface localization of the LapA protein from Pseudomonas fluorescens correlated with adhesion forces, thus providing strong evidence that LapA function as a cell-surface biofilm adhesin151.
Towards novel applications in diagnosis and therapy
Force nanoscopy has also established itself as a valuable approach for diagnosis and therapy. An AFM cantilever-based technology152 was exploited to understand how cell wall PG interacts with the clinically important antibiotics vancomycin and oritavancin153, which suggested that mechanical stress alters the membrane and cell wall, thus leading to cell death. The device could also be used to better understand the influence of dosing and competing ligands on the functionality of the drugs. Another example involves monitoring the fluctuations of cantilevers in order to quickly assess the sensitivity and resistance of bacteria to antibiotics154. The technology could also determine the presence of viable microorganisms in complex, uncontrolled environments, such as soil and river samples155. These studies illustrate the power of AFM for studying the mechanism of action of antibiotics, and for helping select the most efficient treatments against pathogens.
Lastly, SCFS revealed the mechanism by which carbon nanoparticles (fullerenes) functionalized by multiple mannose residues are capable of blocking the adhesion of uropathogenic bacteria to their carbohydrate receptors via high-affinity multivalent bonds156. The authors suggested that this direct, label-free method could lead to novel applications in anti-adhesion therapy, that is, for the design of peptides or antibodies capable of treating microbial infections.
Conclusions
Characterization of multi-molecular structures and machineries of microbial cells is an essential step towards understanding cellular processes and functions, and could potentially lead to novel applications in medicine and biotechnology. Because of their small dimensions, microbial subcellular structures have long been difficult to study. Examples discussed here demonstrate that optical and force nanoscopy methods can tackle this problem with unprecedented resolution and sensitivity. Super-resolution and AFM techniques represent a powerful toolkit for probing the organization, dynamics, interactions and functionality of single molecules, from the inside out, and up to intercellular interactions, thereby allowing microbiologists to answer outstanding questions that were impossible to address before and to develop ultrasensitive assays for diagnosis and therapy.
Ultimately, the full potential of nanoscopy will be achieved when combining optical and force modalities. Establishing these correlated platforms in microbiology should allow the identification and tracking of specific cellular components, while probing their biophysical properties (adhesion, elasticity) simultaneously on the same single cell, thus contributing to the important connection between their structures and functions. Toward this goal, correlated AFM-fluorescence imaging has been exploited to track cell surface dynamics during cellular morphogenesis157,158. A recent study established a correlated SMLM and AFM imaging platform for localizing specific proteins within high-resolution AFM images159. Although correlative nanoscopy is still in its infancy, this approach offers promising prospects for the comprehensive analysis of the structure, dynamics and interactions of single molecules in microbial cells.
Acknowledgments
Work in the Dufrêne team was supported by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 693630), the National Fund for Scientific Research (FNRS), the FNRS-WELBIO (grant no. WELBIO-CR-2015A-05), the Federal Office for Scientific, Technical and Cultural Affairs (Interuniversity Poles of Attraction Programme), and the Research Department of the Communauté française de Belgique (Concerted research action). Y.F.D. is Research Director at the FNRS. Work in the Xiao lab is supported by National Institute of Health General Medicines 1R01GM086447-06, 1R01GM112008-01 (Multi-PI), National Science Foundation grant EAGER MCB1019000. Y.F.D. and J.X. thank Carla Coltharp and Xinxing Yang for their critical reading and suggestions of the manuscript.
Footnotes
Competing interests
The authors declare no competing financial interest.
References
- 1.Ghosal D, Lowe J. Collaborative protein filaments. Embo J. 2015;34:2312–2320. doi: 10.15252/embj.201591756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cloud-Hansen KA, et al. Breaching the great wall: peptidoglycan and microbial interactions. Nat Rev Microbiol. 2006;4:710–716. doi: 10.1038/nrmicro1486. [DOI] [PubMed] [Google Scholar]
- 3.Kolter R, Greenberg EP. Microbial sciences: The superficial life of microbes. Nature. 2006;441:300–302. doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- 4.Müller DJ, Dufrêne YF. Atomic force microscopy as a multifunctional molecular toolbox in nanobiotechnology. Nat Nanotech. 2008;3:261–269. doi: 10.1038/nnano.2008.100. [DOI] [PubMed] [Google Scholar]
- 5.Dufrêne YF. Towards nanomicrobiology using atomic force microscopy. Nat Rev Microbiol. 2008;6:674–680. doi: 10.1038/nrmicro1948. [DOI] [PubMed] [Google Scholar]
- 6.Engel A, Muller DJ. Observing single biomolecules at work with the atomic force microscope. Nat Struct Biol. 2000;7:715–718. doi: 10.1038/78929. [DOI] [PubMed] [Google Scholar]
- 7.Engel A, Gaub HE. Structure and Mechanics of Membrane Proteins. Annu Rev Biochem. 2008;77:127–148. doi: 10.1146/annurev.biochem.77.062706.154450. [DOI] [PubMed] [Google Scholar]
- 8.Ando T, Uchihashi T, Scheuring S. Filming Biomolecular Processes by High-Speed Atomic Force Microscopy. Chem Rev. 2014;114:3120–3188. doi: 10.1021/cr4003837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hinterdorfer P, Dufrêne YF. Detection and localization of single molecular recognition events using atomic force microscopy. Nat Methods. 2006;3:347–355. doi: 10.1038/nmeth871. [DOI] [PubMed] [Google Scholar]
- 10.Müller DJ, Helenius J, Alsteens D, Dufrêne YF. Force probing surfaces of living cells to molecular resolution. Nat Chem Biol. 2009;5:383–390. doi: 10.1038/nchembio.181. [DOI] [PubMed] [Google Scholar]
- 11.Strunz T, Oroszlan K, Schafer R, Guntherodt HJ. Dynamic force spectroscopy of single DNA molecules. Proc Natl Acad Sci USA. 1999;96:11277–11282. doi: 10.1073/pnas.96.20.11277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dupres V, et al. Nanoscale mapping and functional analysis of individual adhesins on living bacteria. Nat Methods. 2005;2:515–520. doi: 10.1038/nmeth769. [DOI] [PubMed] [Google Scholar]
- 13.Alsteens D, Garcia MC, Lipke PN, Dufrene YF. Force-induced formation and propagation of adhesion nanodomains in living fungal cells. Proc Natl Acad Sci. 2010;107:20744–20749. doi: 10.1073/pnas.1013893107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dupres V, et al. The yeast Wsc1 cell surface sensor behaves like a nanospring in vivo. Nat Chem Biol. 2009;5:857–862. doi: 10.1038/nchembio.220. [DOI] [PubMed] [Google Scholar]
- 15.Helenius J, Heisenberg CP, Gaub HE, Muller DJ. Single-cell force spectroscopy. J Cell Sci. 2008;121:1785–1791. doi: 10.1242/jcs.030999. [DOI] [PubMed] [Google Scholar]
- 16.Beaussart A, et al. Single-cell force spectroscopy of probiotic bacteria. Biophys J. 2013;104:1886–1892. doi: 10.1016/j.bpj.2013.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beaussart A, et al. Nanoscale adhesion forces of Pseudomonas aeruginosa type IV pili. ACS Nano. 2014;8:10723–10733. doi: 10.1021/nn5044383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dufrêne YF, Martínez-Martín D, Medalsy I, Alsteens D, Müller DJ. Multiparametric imaging of biological systems by force-distance curve–based AFM. Nat Methods. 2013;10:847–854. doi: 10.1038/nmeth.2602. [DOI] [PubMed] [Google Scholar]
- 19.Medalsy I, Hensen U, Muller DJ. Imaging and quantifying chemical and physical properties of native proteins at molecular resolution by force-volume AFM. Angew Chem Int Ed. 2011;50:12103–12108. doi: 10.1002/anie.201103991. [DOI] [PubMed] [Google Scholar]
- 20.Rico F, Su C, Scheuring S. Mechanical mapping of single membrane proteins at submolecular resolution. Nano Lett. 2011;11:3983–3986. doi: 10.1021/nl202351t. [DOI] [PubMed] [Google Scholar]
- 21.Alsteens D, Trabelsi H, Soumillion P, Dufrêne YF. Multiparametric atomic force microscopy imaging of single bacteriophages extruding from living bacteria. Nat Commun. 2013;4:2926. doi: 10.1038/ncomms3926. [DOI] [PubMed] [Google Scholar]
- 22.Formosa C, et al. Multiparametric imaging of adhesive nanodomains at the surface of Candida albicans by atomic force microscopy. Nanomed Nanotechnol Biol Med. 2015;11:57–65. doi: 10.1016/j.nano.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 23.Formosa-Dague C, Speziale P, Foster TJ, Geoghegan JA, Dufrêne YF. Zinc-dependent mechanical properties of Staphylococcus aureus biofilm-forming surface protein SasG. Proc Natl Acad Sci. 2015;113:410–415. doi: 10.1073/pnas.1519265113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Formosa-Dague C, et al. Sticky matrix: adhesion mechanism of the Staphylococcal polysaccharide intercellular adhesin. ACS Nano. 2016;10:3443–3452. doi: 10.1021/acsnano.5b07515. [DOI] [PubMed] [Google Scholar]
- 25.Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 26.Klar TA, Jakobs S, Dyba M, Egner A, Hell SW. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc Natl Acad Sci USA. 2000;97:8206–8210. doi: 10.1073/pnas.97.15.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider J, et al. Ultrafast, temporally stochastic STED nanoscopy of millisecond dynamics. Nat Methods. 2015;12:827–830. doi: 10.1038/nmeth.3481. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt R, et al. Mitochondrial cristae revealed with focused light. Nano Lett. 2009;9:2508–2510. doi: 10.1021/nl901398t. [DOI] [PubMed] [Google Scholar]
- 29.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 30.Hess ST, Girirajan TP, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heilemann M, et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew Chem Int Ed Engl. 2008;47:6172–6176. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- 33.van Oijen AM, Köhler J, Schmidt J, Müller M, Brakenhoff GJ. 3-Dimensional super-resolution by spectrally selective imaging. Chem Phys Lett. 1998;292:183–187. [Google Scholar]
- 34.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chozinski TJ, Gagnon LA, Vaughan JC. Twinkle, twinkle little star: photoswitchable fluorophores for super-resolution imaging. FEBS Lett. 2014;588:3603–3612. doi: 10.1016/j.febslet.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 36.Pavani SR, et al. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc Natl Acad Sci USA. 2009;106:2995–2999. doi: 10.1073/pnas.0900245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Juette MF, et al. Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nat Methods. 2008;5:527–529. doi: 10.1038/nmeth.1211. [DOI] [PubMed] [Google Scholar]
- 38.Tang J, Akerboom J, Vaziri A, Looger LL, Shank CV. Near-isotropic 3D optical nanoscopy with photon-limited chromophores. Proc Natl Acad Sci USA. 2010;107:10068–10073. doi: 10.1073/pnas.1004899107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang B, Wang W, Bates M, Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shtengel G, et al. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc Natl Acad Sci USA. 2009;106:3125–3130. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shroff H, Galbraith CG, Galbraith JA, Betzig E. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat Methods. 2008;5:417–423. doi: 10.1038/nmeth.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coltharp C, Kessler RP, Xiao J. Accurate construction of photoactivated localization microscopy (PALM) images for quantitative measurements. PLoS ONE. 2012;7:e51725. doi: 10.1371/journal.pone.0051725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Endesfelder U, Malkusch S, Fricke F, Heilemann M. A simple method to estimate the average localization precision of a single-molecule localization microscopy experiment. Histochem Cell Biol. 2014;141:629–638. doi: 10.1007/s00418-014-1192-3. [DOI] [PubMed] [Google Scholar]
- 44.Huang F, et al. Video-rate nanoscopy using sCMOS camera-specific single-molecule localization algorithms. Nat Methods. 2013;10:653–658. doi: 10.1038/nmeth.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gustafsson MG. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc. 2000;198:82–87. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- 46.Schermelleh L, et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gustafsson MG. Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. Proc Natl Acad Sci USA. 2005;102:13081–13086. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li D, et al. ADVANCED IMAGING. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science. 2015;349:aab3500. doi: 10.1126/science.aab3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hell SW. Microscopy and its focal switch. Nat Methods. 2009;6:24–32. doi: 10.1038/nmeth.1291. [DOI] [PubMed] [Google Scholar]
- 50.Hell SW, Kroug M. Ground-state-depletion fluorscence microscopy: A concept for breaking the diffraction resolution limit. Appl Phys B. 1995;60:495–497. [Google Scholar]
- 51.Hofmann M, Eggeling C, Jakobs S, Hell SW. Breaking the diffraction barrier in fluorescence microscopy at low light intensities by using reversibly photoswitchable proteins. Proc Natl Acad Sci USA. 2005;102:17565–17569. doi: 10.1073/pnas.0506010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharonov A, Hochstrasser RM. Wide-field subdiffraction imaging by accumulated binding of diffusing probes. Proc Natl Acad Sci USA. 2006;103:18911–18916. doi: 10.1073/pnas.0609643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jungmann R, et al. Single-molecule kinetics and super-resolution microscopy by fluorescence imaging of transient binding on DNA origami. Nano Lett. 2010;10:4756–4761. doi: 10.1021/nl103427w. [DOI] [PubMed] [Google Scholar]
- 54.Schoen I, Ries J, Klotzsch E, Ewers H, Vogel V. Binding-activated localization microscopy of DNA structures. Nano Lett. 2011;11:4008–4011. doi: 10.1021/nl2025954. [DOI] [PubMed] [Google Scholar]
- 55.Huang B, Babcock H, Zhuang X. Breaking the diffraction barrier: super-resolution imaging of cells. Cell. 2010;143:1047–1058. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dempsey GT, Vaughan JC, Chen KH, Bates M, Zhuang X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat Methods. 2011;8:1027–1036. doi: 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Durisic N, Laparra-Cuervo L, Sandoval-Alvarez A, Borbely JS, Lakadamyali M. Single-molecule evaluation of fluorescent protein photoactivation efficiency using an in vivo nanotemplate. Nat Methods. 2014;11:156–162. doi: 10.1038/nmeth.2784. [DOI] [PubMed] [Google Scholar]
- 58.Gahlmann A, Moerner WE. Exploring bacterial cell biology with single-molecule tracking and super-resolution imaging. Nat Rev Microbiol. 2014;12:9–22. doi: 10.1038/nrmicro3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Coltharp C, Xiao J. Superresolution microscopy for microbiology. Cell Microbiol. 2012;14:1808–1818. doi: 10.1111/cmi.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coltharp C, Yang X, Xiao J. Quantitative analysis of single-molecule superresolution images. Curr Opin Struct Biol. 2014;28:112–121. doi: 10.1016/j.sbi.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Small AR, Parthasarathy R. Superresolution localization methods. Annu Rev Phys Chem. 2014;65:107–125. doi: 10.1146/annurev-physchem-040513-103735. [DOI] [PubMed] [Google Scholar]
- 62.Dame RT, Tark-Dame M. Bacterial chromatin: converging views at different scales. Curr Opin Cell Biol. 2016;40:60–65. doi: 10.1016/j.ceb.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 63.Spahn C, Endesfelder U, Heilemann M. Super-resolution imaging of Escherichia coli nucleoids reveals highly structured and asymmetric segregation during fast growth. J Struct Biol. 2014;185:243–249. doi: 10.1016/j.jsb.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 64.Spahn C, Cella-Zannacchi F, Endesfelder U, Heilemann M. Correlative super-resolution imaging of RNA polymerase distribution and dynamics, bacterial membrane and chromosomal structure in Escherichia coli. Methods Appl Fluoresc. 2015;3:014005. doi: 10.1088/2050-6120/3/1/014005. [DOI] [PubMed] [Google Scholar]
- 65.Foo YH, Spahn C, Zhang H, Heilemann M, Kenney LJ. Single cell super-resolution imaging of E. coli OmpR during environmental stress. Integr Biol. 2015;7:1297–1308. doi: 10.1039/c5ib00077g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kolb HC, Finn MG, Sharpless KB. Click chemistry: diverse chemical function from a few good reactions. Angew Chem Int Ed Engl. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 67.Marbouty M, et al. Condensin- and replication-mediated bacterial chromosome folding and origin condensation revealed by Hi-C and super-resolution imaging. Mol Cell. 2015;59:588–602. doi: 10.1016/j.molcel.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 68.Stracy M, et al. Live-cell superresolution microscopy reveals the organization of RNA polymerase in the bacterial nucleoid. Proc Natl Acad Sci USA. 2015;112:E4390–E4399. doi: 10.1073/pnas.1507592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee SF, Thompson MA, Schwartz MA, Shapiro L, Moerner WE. Super-resolution imaging of the nucleoid-associated protein HU in Caulobacter crescentus. Biophys J. 2011;100:L31–33. doi: 10.1016/j.bpj.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang W, Li GW, Chen C, Xie XS, Zhuang X. Chromosome organization by a nucleoid-associated protein in live bacteria. Science. 2011;333:1445–1449. doi: 10.1126/science.1204697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- 72.Hensel Z, Weng X, Lagda AC, Xiao J. Transcription-factor-mediated DNA looping probed by high-resolution, single-molecule imaging in live E. coli cells. PLoS Biol. 2013;11:e1001591. doi: 10.1371/journal.pbio.1001591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liao Y, Schroeder JW, Gao B, Simmons LA, Biteen JS. Single-molecule motions and interactions in live cells reveal target search dynamics in mismatch repair. Proc Natl Acad Sci USA. 2015;112:E6898–E6906. doi: 10.1073/pnas.1507386112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Uphoff S, Reyes-Lamothe R, Garza de Leon F, Sherratt DJ, Kapanidis AN. Single-molecule DNA repair in live bacteria. Proc Natl Acad Sci USA. 2013;110:8063–8068. doi: 10.1073/pnas.1301804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tang M, et al. UmuD’(2)C is an error-prone DNA polymerase, Escherichia coli pol V. Proc Natl Acad Sci USA. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Robinson A, et al. Regulation of mutagenic DNA polymerase V activation in space and time. PLoS Genet. 2015;11:e1005482. doi: 10.1371/journal.pgen.1005482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lesterlin C, Ball G, Schermelleh L, Sherratt DJ. RecA bundles mediate homology pairing between distant sisters during DNA break repair. Nature. 2014;506:249–253. doi: 10.1038/nature12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rajendram M, et al. Anionic phospholipids stabilize RecA filament bundles in Escherichia coli. Mol Cell. 2015;60:374–384. doi: 10.1016/j.molcel.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bakshi S, Dalrymple RM, Li W, Choi H, Weisshaar JC. Partitioning of RNA polymerase activity in live Escherichia coli from analysis of single-molecule diffusive trajectories. Biophys J. 2013;105:2676–2686. doi: 10.1016/j.bpj.2013.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bakshi S, Siryaporn A, Goulian M, Weisshaar JC. Superresolution imaging of ribosomes and RNA polymerase in live Escherichia coli cells. Mol Microbiol. 2012;85:21–38. doi: 10.1111/j.1365-2958.2012.08081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Endesfelder U, et al. Multiscale spatial organization of RNA polymerase in Escherichia coli. Biophys J. 2013;105:172–181. doi: 10.1016/j.bpj.2013.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cagliero C, Zhou YN, Jin DJ. Spatial organization of transcription machinery and its segregation from the replisome in fast-growing bacterial cells. Nucleic Acids Res. 2014;42:13696–13705. doi: 10.1093/nar/gku1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jin DJ, Cagliero C, Martin CM, Izard J, Zhou YN. The dynamic nature and territory of transcriptional machinery in the bacterial chromosome. Front Microbiol. 2015;6:497. doi: 10.3389/fmicb.2015.00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bath J, Wu LJ, Errington J, Wang JC. Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell-prespore division septum. Science. 2000;290:995–997. doi: 10.1126/science.290.5493.995. [DOI] [PubMed] [Google Scholar]
- 85.Yen Shin J, et al. Visualization and functional dissection of coaxial paired SpoIIIE channels across the sporulation septum. eLife. 2015;4:e06474. doi: 10.7554/eLife.06474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fiche JB, et al. Recruitment, assembly, and molecular architecture of the SpoIIIE DNA pump revealed by superresolution microscopy. PLoS Biol. 2013;11:e1001557. doi: 10.1371/journal.pbio.1001557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ptacin JL, et al. Bacterial scaffold directs pole-specific centromere segregation. Proc Natl Acad Sci USA. 2014;111:E2046–E2055. doi: 10.1073/pnas.1405188111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.de Boer P, Crossley R, Rothfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature. 1992;359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- 89.RayChaudhuri D, Park JT. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature. 1992;359:251–254. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- 90.Fu G, et al. In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM) PLoS ONE. 2010;5:e12682. doi: 10.1371/journal.pone.0012680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Strauss MP, et al. 3D-SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: implications for triggering cytokinesis. PLoS Biol. 2012;10:e1001389. doi: 10.1371/journal.pbio.1001389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Holden SJ, et al. High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. Proc Natl Acad Sci USA. 2014;111:4566–4571. doi: 10.1073/pnas.1313368111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fleurie A, et al. MapZ marks the division sites and positions FtsZ rings in Streptococcus pneumoniae. Nature. 2014;516:259–262. doi: 10.1038/nature13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jacq M, et al. Remodeling of the Z-ring nanostructure during the Streptococcus pneumoniae cell cycle revealed by photoactivated localization microscopy. mBio. 2015;6:e01108–15. doi: 10.1128/mBio.01108-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Leisch N, et al. Growth in width and FtsZ ring longitudinal positioning in a gammaproteobacterial symbiont. Curr Biol. 2012;22:R831–R832. doi: 10.1016/j.cub.2012.08.033. [DOI] [PubMed] [Google Scholar]
- 96.Grangeon R, Zupan JR, Anderson-Furgeson J, Zambryski PC. PopZ identifies the new pole, and PodJ identifies the old pole during polar growth in Agrobacterium tumefaciens. Proc Natl Acad Sci USA. 2015;112:11666–11671. doi: 10.1073/pnas.1515544112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Coltharp C, Buss J, Plumer TM, Xiao J. Defining the rate-limiting processes of bacterial cytokinesis. Proc Natl Acad Sci USA. 2016;113:E1044–E1053. doi: 10.1073/pnas.1514296113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rowlett VW, Margolin W. 3D-SIM super-resolution of FtsZ and its membrane tethers in Escherichia coli cells. Biophys J. 2014;107:L17–L20. doi: 10.1016/j.bpj.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Biteen JS, Goley ED, Shapiro L, Moerner WE. Three-dimensional super-resolution imaging of the midplane protein FtsZ in live Caulobacter crescentus cells using astigmatism. ChemPhysChem. 2012;13:1007–1012. doi: 10.1002/cphc.201100686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lyu Z, Carla Coltharp C, Yang X, Xiao J. Influence of FtsZ GTPase activity and concentration on nanoscale Z-ring structure in vivo revealed by three-dimensional Superresolution imaging. Biopolymers. 2016;105:725–734. doi: 10.1002/bip.22895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Eswaramoorthy P, et al. Cellular architecture mediates DivIVA ultrastructure and regulates min activity in Bacillus subtilis. mBio. 2011;2:e00257–11. doi: 10.1128/mBio.00257-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Buss J, et al. In vivo organization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Mol Microbiol. 2013;89:1099–1120. doi: 10.1111/mmi.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Buss J, et al. A multi-layered protein network stabilizes the Escherichia coli FtsZ-ring and modulates constriction dynamics. PLoS Genet. 2015;11:e1005128. doi: 10.1371/journal.pgen.1005128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Olshausen PV, et al. Superresolution imaging of dynamic MreB filaments in B. subtilis—a multiple-motor-driven transport? Biophys J. 2013;105:1171–1181. doi: 10.1016/j.bpj.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Reimold C, Defeu Soufo HJ, Dempwolff F, Graumann PL. Motion of variable-length MreB filaments at the bacterial cell membrane influences cell morphology. Mol Biol Cell. 2013;24:2340–2349. doi: 10.1091/mbc.E12-10-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim SY, Gitai Z, Kinkhabwala A, Shapiro L, Moerner WE. Single molecules of the bacterial actin MreB undergo directed treadmilling motion in Caulobacter crescentus. Proc Natl Acad Sci USA. 2006;103:10929–10934. doi: 10.1073/pnas.0604503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Biteen JS, et al. Super-resolution imaging in live Caulobacter crescentus cells using photoswitchable EYFP. Nat Methods. 2008;5:947–949. doi: 10.1038/NMETH.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ptacin JL, et al. A spindle-like apparatus guides bacterial chromosome segregation. Nat Cell Biol. 2010;12:791–798. doi: 10.1038/ncb2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ausmees N, Kuhn JR, Jacobs-Wagner C. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell. 2003;115:705–713. doi: 10.1016/s0092-8674(03)00935-8. [DOI] [PubMed] [Google Scholar]
- 110.Lew MD, et al. Three-dimensional superresolution colocalization of intracellular protein superstructures and the cell surface in live Caulobacter crescentus. Proc Natl Acad Sci USA. 2011;108:E1102–E1110. doi: 10.1073/pnas.1114444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Greenfield D, et al. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 2009;7:e1000137. doi: 10.1371/journal.pbio.1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fredlund J, Broder D, Fleming T, Claussin C, Pogliano K. The SpoIIQ landmark protein has different requirements for septal localization and immobilization. Mol Microbiol. 2013;89:1053–1068. doi: 10.1111/mmi.12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Haas BL, Matson JS, DiRita VJ, Biteen JS. Single-molecule tracking in live Vibrio cholerae reveals that ToxR recruits the membrane-bound virulence regulator TcpP to the toxT promoter. Mol Microbiol. 2015;96:4–13. doi: 10.1111/mmi.12834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Llorente-Garcia I, et al. Single-molecule in vivo imaging of bacterial respiratory complexes indicates delocalized oxidative phosphorylation. Biochim Biophys Acta. 2014;1837:811–824. doi: 10.1016/j.bbabio.2014.01.020. [DOI] [PubMed] [Google Scholar]
- 115.Karunatilaka KS, Cameron EA, Martens EC, Koropatkin NM, Biteen JS. Superresolution imaging captures carbohydrate utilization dynamics in human gut symbionts. mBio. 2014;5:e02172. doi: 10.1128/mBio.02172-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dufrene YF, Boonaert CJ, Gerin PA, Asther M, Rouxhet PG. Direct probing of the surface ultrastructure and molecular interactions of dormant and germinating spores of Phanerochaete chrysosporium. J Bacteriol. 1999;181:5350–5354. doi: 10.1128/jb.181.17.5350-5354.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schabert F, Henn C, Engel A. Native Escherichia coli OmpF porin surfaces probed by atomic force microscopy. Science. 1995;268:92–94. doi: 10.1126/science.7701347. [DOI] [PubMed] [Google Scholar]
- 118.Kailas L, et al. Surface architecture of endospores of the Bacillus cereus/anthracis/thuringiensis family at the subnanometer scale. Proc Natl Acad Sci. 2011;108:16014–16019. doi: 10.1073/pnas.1109419108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Casuso I, Kodera N, Le Grimellec C, Ando T, Scheuring S. Contact-mode high-resolution high-speed atomic force microscopy movies of the purple membrane. Biophys J. 2009;97:1354–1361. doi: 10.1016/j.bpj.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shibata M, Yamashita H, Uchihashi T, Kandori H, Ando T. High-speed atomic force microscopy shows dynamic molecular processes in photoactivated bacteriorhodopsin. Nat Nanotech. 2010;5:208–212. doi: 10.1038/nnano.2010.7. [DOI] [PubMed] [Google Scholar]
- 121.Yamashita H, et al. Role of trimer–trimer interaction of bacteriorhodopsin studied by optical spectroscopy and high-speed atomic force microscopy. J Struct Biol. 2013;184:2–11. doi: 10.1016/j.jsb.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 122.Casuso I, et al. Characterization of the motion of membrane proteins using high-speed atomic force microscopy. Nat Nanotech. 2012;7:525–529. doi: 10.1038/nnano.2012.109. [DOI] [PubMed] [Google Scholar]
- 123.Turner RD, Hurd AF, Cadby A, Hobbs JK, Foster SJ. Cell wall elongation mode in Gram-negative bacteria is determined by peptidoglycan architecture. Nat Commun. 2013;4:1496. doi: 10.1038/ncomms2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Turner RD, Vollmer W, Foster SJ. Different walls for rods and balls: the diversity of peptidoglycan. Mol Microbiol. 2014;91:862–874. doi: 10.1111/mmi.12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hayhurst EJ, Kailas L, Hobbs JK, Foster SJ. Cell wall peptidoglycan architecture in Bacillus subtilis. Proc Natl Acad Sci. 2008;105:14603–14608. doi: 10.1073/pnas.0804138105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Turner RD, et al. Peptidoglycan architecture can specify division planes in Staphylococcus aureus. Nat Commun. 2010;1:26. doi: 10.1038/ncomms1025. [DOI] [PubMed] [Google Scholar]
- 127.Wheeler R, Mesnage S, Boneca IG, Hobbs JK, Foster SJ. Super-resolution microscopy reveals cell wall dynamics and peptidoglycan architecture in ovococcal bacteria. Mol Microbiol. 2011;82:1096–1109. doi: 10.1111/j.1365-2958.2011.07871.x. [DOI] [PubMed] [Google Scholar]
- 128.Plomp M, Leighton TJ, Wheeler KE, Hill HD, Malkin AJ. In vitro high-resolution structural dynamics of single germinating bacterial spores. Proc Natl Acad Sci. 2007;104:9644–9649. doi: 10.1073/pnas.0610626104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Andre G, et al. Imaging the nanoscale organization of peptidoglycan in living Lactococcus lactis cells. Nat Commun. 2010;1:27. doi: 10.1038/ncomms1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lotan R, Sharon N, Mirelman D. Interaction of wheat-germ agglutinin with bacterial cells and cell-wall polymers. Eur J Biochem. 1975;55:257–262. doi: 10.1111/j.1432-1033.1975.tb02158.x. [DOI] [PubMed] [Google Scholar]
- 131.Kuru E, et al. In Situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Ed Engl. 2012;51:12519–12523. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liechti GW, et al. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature. 2014;506:507–510. doi: 10.1038/nature12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sheldrick GM, Jones PG, Kennard O, Williams DH, Smith GA. Structure of vancomycin and its complex with acetyl-D-alanyl-D-alanine. Nature. 1978;271:223–225. doi: 10.1038/271223a0. [DOI] [PubMed] [Google Scholar]
- 134.Monteiro JM, et al. Cell shape dynamics during the staphylococcal cell cycle. Nat Commun. 2015;6:8055. doi: 10.1038/ncomms9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.van Teeseling MC, et al. Anammox Planctomycetes have a peptidoglycan cell wall. Nat Commun. 2015;6:6878. doi: 10.1038/ncomms7878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tsui HC, et al. Pbp2x localizes separately from Pbp2b and other peptidoglycan synthesis proteins during later stages of cell division of Streptococcus pneumoniae D39. Mol Microbiol. 2014;94:21–40. doi: 10.1111/mmi.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lee TK, et al. A dynamically assembled cell wall synthesis machinery buffers cell growth. Proc Natl Acad Sci USA. 2014;111:4554–4559. doi: 10.1073/pnas.1313826111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Land AD, et al. Requirement of essential Pbp2x and GpsB for septal ring closure in Streptococcus pneumoniae D39. Mol Microbiol. 2013;90:939–955. doi: 10.1111/mmi.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kocaoglu O, et al. Selective penicillin-binding protein imaging probes reveal substructure in bacterial cell division. ACS Chem Biol. 2012;7:1746–1753. doi: 10.1021/cb300329r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bustanji Y, et al. Dynamics of the interaction between a fibronectin molecule and a living bacterium under mechanical force. Proc Natl Acad Sci. 2003;100:13292–13297. doi: 10.1073/pnas.1735343100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lower SK, et al. Polymorphisms in fibronectin binding protein A of Staphylococcus aureus are associated with infection of cardiovascular devices. Proc Natl Acad Sci USA. 2011;108:18372–18377. doi: 10.1073/pnas.1109071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Herman P, et al. The binding force of the staphylococcal adhesin SdrG is remarkably strong. Mol Microbiol. 2014;93:356–368. doi: 10.1111/mmi.12663. [DOI] [PubMed] [Google Scholar]
- 143.Herman-Bausier P, Dufrêne YF. Atomic force microscopy reveals a dual collagen-binding activity for the staphylococcal surface protein SdrF. Mol Microbiol. 2015;99:611–621. doi: 10.1111/mmi.13254. [DOI] [PubMed] [Google Scholar]
- 144.Herman-Bausier P, El-Kirat-Chatel S, Foster TJ, Geoghegan JA, Dufrêne YF. Staphylococcus aureus fibronectin-binding protein A mediates cell-cell adhesion through low-affinity homophilic bonds. mBio. 2015;6:e00413–15. doi: 10.1128/mBio.00413-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gruszka DT, et al. Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein. Nat Commun. 2015;6:7271. doi: 10.1038/ncomms8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.El-Kirat-Chatel S, et al. Forces in yeast flocculation. Nanoscale. 2015;7:1760–1767. doi: 10.1039/c4nr06315e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.El-Kirat-Chatel S, Dufrêne YF. Nanoscale adhesion forces between the fungal pathogen Candida albicans and macrophages. Nanoscale Horiz. 2016;1:69–74. doi: 10.1039/c5nh00049a. [DOI] [PubMed] [Google Scholar]
- 148.Formosa-Dague C, et al. Forces between Staphylococcus aureus and human skin. Nanoscale Horiz. 2016;1:298–303. doi: 10.1039/c6nh00057f. [DOI] [PubMed] [Google Scholar]
- 149.Berk V, et al. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science. 2012;337:236–239. doi: 10.1126/science.1222981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Turnbull L, et al. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nature Commun. 2016;7:11220. doi: 10.1038/ncomms11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ivanov IE, et al. Atomic force and super-resolution microscopy support a role for LapA as a cell-surface biofilm adhesin of Pseudomonas fluorescens. Res Microbiol. 2012;163:685–691. doi: 10.1016/j.resmic.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Fritz J, et al. Translating biomolecular recognition into nanomechanics. Science. 2000;288:316–318. doi: 10.1126/science.288.5464.316. [DOI] [PubMed] [Google Scholar]
- 153.Ndieyira JW, et al. Nanomechanical detection of antibiotic–mucopeptide binding in a model for superbug drug resistance. Nat Nanotech. 2008;3:691–696. doi: 10.1038/nnano.2008.275. [DOI] [PubMed] [Google Scholar]
- 154.Longo G, et al. Rapid detection of bacterial resistance to antibiotics using AFM cantilevers as nanomechanical sensors. Nat Nanotech. 2013;8:522–526. doi: 10.1038/nnano.2013.120. [DOI] [PubMed] [Google Scholar]
- 155.Kasas S, et al. Detecting nanoscale vibrations as signature of life. Proc Natl Acad Sci. 2014;112:378–381. doi: 10.1073/pnas.1415348112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Beaussart A, Abellán-Flos M, El-Kirat-Chatel S, Vincent SP, Dufrêne YF. Force nanoscopy as a versatile platform for quantifying the activity of antiadhesion compounds targeting bacterial pathogens. Nano Lett. 2016;16:1299–1307. doi: 10.1021/acs.nanolett.5b04689. [DOI] [PubMed] [Google Scholar]
- 157.El-Kirat-Chatel S, Dufrêne YF. Nanoscale imaging of the Candida-macrophage interaction using correlated fluorescence-atomic force microscopy. ACS Nano. 2012;2012:10792–10799. doi: 10.1021/nn304116f. [DOI] [PubMed] [Google Scholar]
- 158.Andre G, et al. Fluorescence and atomic force microscopy imaging of wall teichoic acids in Lactobacillus plantarum. ACS Chem Biol. 2011;6:366–376. doi: 10.1021/cb1003509. [DOI] [PubMed] [Google Scholar]
- 159.Odermatt PD, et al. High-resolution correlative microscopy: bridging the gap between single molecule localization microscopy and atomic force microscopy. Nano Lett. 2015;15:4896–4904. doi: 10.1021/acs.nanolett.5b00572. [DOI] [PubMed] [Google Scholar]


