Summary
N-linked glycosylation is a predominant post-translational modification of protein in eukaryotes, and its dysregulation is the etiology of several human disorders. The enzyme UDP-N-acetylglucosamine:dolichyl-phosphate N-acetylglucosaminephosphotransferase (GlcNAc-1-P-transferase, GPT) catalyzes the first and committed step of N-linked glycosylation in the endoplasmic reticulum membrane, and it is the target of the natural product tunicamycin. Tunicamycin has potent antibacterial activity by inhibiting the bacterial cell wall synthesis enzyme MraY, but its usefulness as an antibiotic is limited by off-target inhibition of human GPT. Our understanding of how tunicamycin inhibits N-linked glycosylation and efforts to selectively target MraY are hampered by a lack of structural information. Here we present crystal structures of human GPT in complex with tunicamycin. Our structural and functional analyses reveal the difference between GPT and MraY in their mechanisms of inhibition by tunicamycin. We demonstrate that this difference could be exploited for the design of MraY-specific inhibitors as potential antibiotics.
Main text
N-linked glycosylation of proteins is one of the most common post-translational modifications in mammals and is involved in many important physiological processes including synaptic transmission, cell adhesion, organellar trafficking, protein folding, and neurodevelopment. The integral membrane enzyme UDP-N-acetylglucosamine:dolichyl-phosphate N-acetylglucosaminephosphotransferase (GlcNAc-1-P-transferase or GPT; DPAGT1 gene in human or ALG7 in yeast) catalyzes the first and committed step of N-linked glycosylation on the cytosolic face of endoplasmic reticulum (ER) membrane 1. This reaction involves the transfer of N-acetylglucosamine-1-phosphate (GlcNAc-1-P) from UDP-N-acetylglucosamine (UDP-GlcNAc) to the carrier lipid dolichyl-phosphate (Dol-P). The dolichyl-diphosphate-N-acetylglucosamine (Dol-PP-GlcNAc) product is further processed by downstream enzymes and flipped into the ER lumen where processing is completed, before the sugars are transferred to the target protein by the oligosaccharyltransferase complex. Recent studies demonstrate that loss-of-function mutations in DPAGT1 are the etiologies of two types of human neurological diseases, congenital myasthenic syndrome (CMS) 13 and congenital disorder of glycosylation type lj (CDG-Ij) 2–4, and that overexpression of DPAGT1 is a critical contributor to the early development and progression of oral cancer 5,6. GPT is a member of the polyprenyl-phosphate-N-acetylhexosamine-1-phosphate-transferase (PNPT) superfamily, which also contains the bacterial paralogues MraY (peptidoglycan synthesis), WecA (lipopolysaccharide and enterobacterial common antigen synthesis), and TarO (teichoic acid synthesis) 7.
Tunicamycin is a nucleoside analog inhibitor of many PNPT superfamily enzymes 8,9. Tunicamycin was thought of as a transition state analog that mimics the substrates UDP-GlcNAc and dolichyl phosphate 10–14 (Supplementary Fig. 1), but it acts as a high-affinity competitive inhibitor for UDP-GlcNAc in mammalian GPT 15–18. Tunicamycin is widely used as a research tool to block N-linked glycosylation or as an ER stress inducer to activate the unfolded protein response 19–22. Tunicamycin also has potent antibacterial activity due to its competitive inhibition of the essential translocase MraY for its natural substrate UDP-MurNAc-pentapeptide 23,24. However, tunicamycin has not been used as an antibiotic in the clinic due to its cytotoxicity in mammalian cells arising from its off-target inhibition of GPT 19. Efforts to develop tunicamycin into an antibiotic are hampered by a dearth of structural information 25. Of the PNPT superfamily, only crystal structures of MraY have been published, either from Aquifex aeolicus (MraYAA) in the Mg2+-bound state 26 and bound to the competitive inhibitor muraymycin D2 27, or from Clostridium bolteae (MraYCB) in complex with tunicamycin 28. MraY contains a highly conserved active site, including ~34 invariant residues, that overlaps with the inhibitor binding sites; thus MraY orthologues are known to catalyze the same enzymatic reaction and share natural product inhibitors 23,26,27,29,30. The uridine moiety of both muraymycin D2 and tunicamycin was bound to the same location in the conserved active site of MraY, which is consistent with the idea that both tunicamycin and muraymycin D2 act as competitive inhibitors to the substrate UDP-MurNAc-pentapeptide (Supplementary Fig. 1) 23,27,28. Despite progress in our understanding of MraY, a crystal structure of GPT has remained elusive and many questions remain regarding the mechanism of GPT inhibition by tunicamycin. Does tunicamycin inhibit GPT and MraY by the same mechanism? Comparative studies of GPT and MraY are not only key to obtaining mechanistic insight into the inhibition of N-linked glycosylation by tunicamycin, but are also important as a platform for the design of antibiotics targeting MraY.
Toward these goals, we solved the crystal structure of human GPT in complex with tunicamycin. Structural analyses performed in parallel with biochemical and enzymatic studies demonstrate that GPT and MraY utilize different quaternary structure organization and mechanisms of inhibition by tunicamycin. Finally, we applied this knowledge to the design of an MraY-specific tunicamycin analog, which would serve as a blueprint for the development of MraY-specific antibiotics.
Structure of hGPT bound to tunicamycin
We determined the crystal structures of human GPT (hGPT) in complex with tunicamycin for both the canonical Pro129 variant (UniProt ID Q9H3H5) and a His129 variant (European Nucleotide Archive ID: AAG43168) to 3.10 Å and 2.95 Å resolution respectively. Both hGPT variants are functionally competent and they have comparable enzymatic activities (Supplementary Fig. 2). The final models were refined to good geometry (Table 1). Both models are structurally similar (Cα r.m.s.d ~0.8 Å). The canonical Pro129 model is used for all the figures except where noted. The electron density map for the hGPT structure was of excellent quality (Supplementary Fig. 3), allowing unambiguous placement of the entire tunicamycin core as well as the aliphatic tail (Supplementary Fig. 4), which was previously unresolved in the MraY-tunicamycin complex structure 28.
Table 1.
Data collection and refinement statistics.
| hGPT (canonical)* | hGPT (His129 variant)† | |
|---|---|---|
| Data collection | ||
| Space group | C2 | C2 |
| Cell dimensions | ||
| a, b, c (Å) | 212.27, 105.51, 149.39 | 210.62, 105.36, 148.08 |
| α, β, γ (°) | 90, 103.53, 90 | 90, 103.49, 90 |
| Wavelength (Å) | 0.9791 | 0.9791 |
| Resolution (Å) | 75.31 - 3.10 (3.21 - 3.10)a | 93.69 - 2.95 (3.06 - 2.95)a |
| CC1/2 | 0.96 (0.59) | 0.84 (0.76) |
| I/σ(I) | 13.65 (0.66) | 17.51 (0.56) |
| Rpim | 0.068 (>1.0) | 0.079 (>1.0) |
| Redundancy | 79.9 (69.1) | 112.4 (112.8) |
| Completeness (%) | 97.5 (96.3) | 92.6 (54.7) |
| Completeness‡ (%) | 60.1 (18.4) | 55.3 (15.7) |
| Ellipsoidal completeness‡ (%) | 94.9 (99.8) | 94.8 (99.5) |
| Refinement | ||
| Resolution (Å) | 75.31 - 3.10 (3.21 - 3.10)a | 93.69 - 2.95 (3.06 - 2.95)a |
| No. reflections | 35100 (1071) | 36769 (1030) |
| Rwork/Rfree (%) | 25.6/28.9 | 26.0/29.1 |
| No. atoms | ||
| Protein | 11170 | 11280 |
| Tunicamycin/POPG | 360 | 364 |
| B factors | ||
| Protein | 44.71 | 41.80 |
| Tunicamycin/POPG | 51.08 | 46.50 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.003 | 0.003 |
| Bond angles (°) | 0.68 | 0.58 |
| Ramachandran (%) | ||
| Favored | 96.48 | 97.23 |
| Allowed | 3.52 | 2.77 |
| Outliers | 0.0 | 0.0 |
| Rotamer outliers | 0.0 | 0.0 |
| Clashscore | 4.66 | 1.57 |
Merged from 23 crystals.
Merged from 31 crystals.
After applying anisotropy correction in STARANISO.
Values in parentheses are for highest-resolution shell.
Ellipsoidal completeness was calculated by STARANISO. All other statistics were calculated in PHENIX.
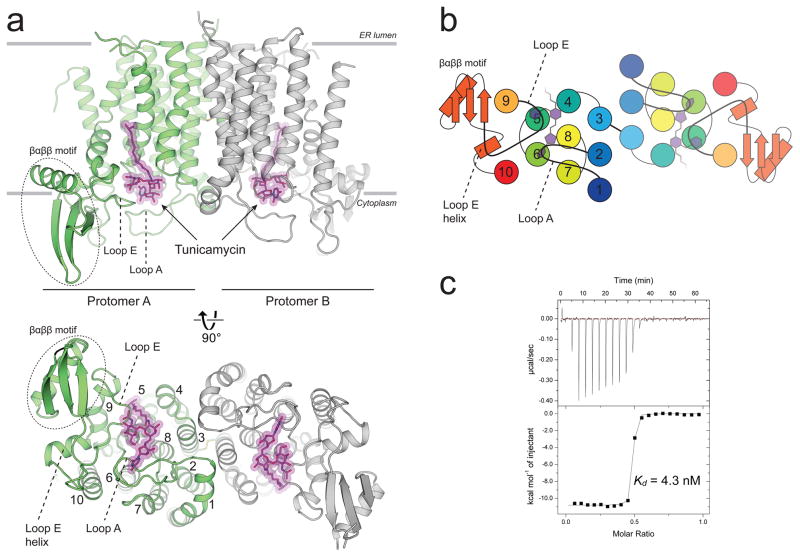
hGPT crystallized as a homodimer with one tunicamycin molecule bound to the active site of each protomer near the cytosolic side of the ER membrane (Fig. 1a). hGPT adopts the canonical PNPT superfamily fold of 10 transmembrane helices, with the active site framed on one side by the C-terminal segment of TM 9 (TM 9b) that is bent outward into the membrane. Notably, hGPT contains a large insertion in loop E between TMs 9b and 10, which folds into an extended βαββ motif not seen in MraY (Supplementary Fig. 5). This βαββ motif protrudes into the cytosol and connects to TM10 via the loop E helix (Fig. 1a). Tunicamycin is bound in an enclosed pocket formed mainly by TMs 4, 5, 6, 8 and almost completely covered by cytoplasmic loops A (between TMs 1 and 2) and E (between TMs 9 and 10) (Fig. 1a – 1b). The aliphatic tail of tunicamycin protrudes into a groove between TMs 4, 5, and 9, which has been suggested to be the binding site for the polyprenyl moiety of undecaprenyl phosphate (C55-P) in MraY 26. Tunicamycin is a high-affinity binder to hGPT, which we characterized by isothermal titration calorimetry (ITC) with a dissociation constant (Kd) of 5.6 nM in detergent micelles (Fig. 1c).
Figure 1. The crystal structure of human GPT in complex with tunicamycin reveals the structural basis for high affinity binding.
a, hGPT crystallized as a homodimer with one tunicamycin molecule (magenta sticks) bound to the cytosolic active site cavity of each protomer, tightly enclosed by loops A and E. The two protomers are related by pseudo-twofold rotational symmetry and are covalently linked by a disulfide bond between TM3 of each protomer. b, Cartoon representation of the cytoplasmic view, rainbow colored from N-terminus (blue) to C-terminus (red). c, Representative ITC raw data (top) and binding isotherm (bottom) for tunicamycin interacting with wild-type hGPT; Kd = 4.3 nM, ΔH° = −10.8 kcal/mol. This ITC experiment was performed in triplicate (technical replicates (5.6+/−1.3 nM, mean +/− S.E.M.)).
Tunicamycin binding to GPT versus MraY
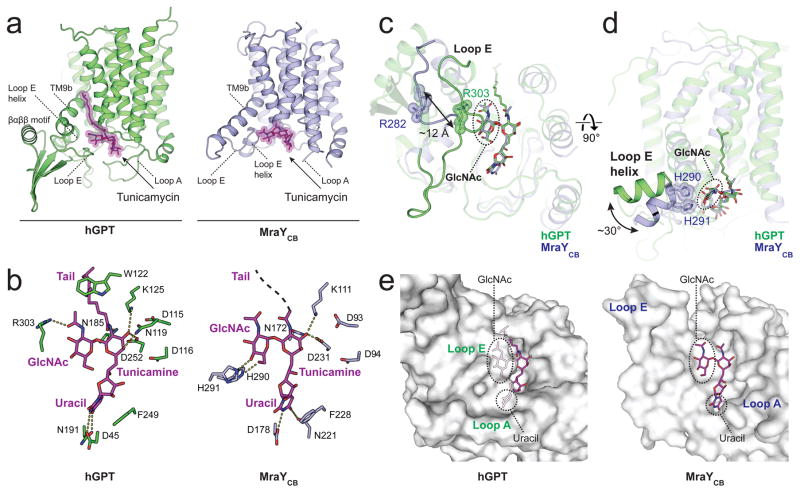
To gain further insight into the binding of tunicamycin to hGPT, we next compared the hGPT-tunicamycin complex to the MraYCB-tunicamycin complex 28 (Fig. 2a). Tunicamycin is composed of uracil, tunicamine, GlcNAc, and an aliphatic tail 14 (Supplementary Fig. 1). The binding of the uracil, tunicamine, and aliphatic tail moieties of tunicamycin to hGPT is analogous to their binding to MraYCB with noted differences (Fig. 2b). The uracil moiety of tunicamycin is stabilized by a conserved π-π stacking interaction with either Phe249 (hGPT) or Phe228 (MraYCB) 28, which was also observed in the crystal structure of MraYAA in complex with muraymycin D2 27. This presumably resembles the interaction formed between hGPT or MraY and the uracil moiety of their respective UDP-sugar substrates (Supplementary Fig. 1). The core tunicamine moiety of tunicamycin is stabilized by hydrogen bonds with Asp252 in hGPT (Asp231 in MraYCB) in both hGPT and MraYCB. However, the tunicamine forms additional hydrogen bond interactions with Asn119 in hGPT that are not seen in the MraYCB-tunicamycin complex. The amide linkage to the aliphatic tail of tunicamycin is stabilized by an interaction with the invariant Asn185 (Asn172 in MraYCB, but the amide linkage is flipped 180° in MraYCB possibly due to the aliphatic tail being unresolved). The tail is further locked into place by Trp122 in hGPT but not in MraYCB (Pro108).
Figure 2. Tunicamycin binding to GPT is analogous but distinct from that to MraY.
a, Tunicamycin (magenta sticks) binds to the active site of both hGPT (green) and bacterial MraYCB (blue, PDB 5JNQ) near the cytoplasmic side of the membrane. The aliphatic tail is not resolved in the MraY-tunicamycin structure. b, Interactions between tunicamycin and either hGPT or MraYCB. c, Loop E of hGPT is more extensive and closer to tunicamycin than it is the MraYCB structure, bringing the invariant residue Arg303 into contact with the GlcNAc moiety of tunicamycin. The corresponding residue in MraYCB (Arg282) does not interact with tunicamycin. d, The Loop E helix of hGPT is swiveled by 30 degrees towards the horizontal plane as compared to MraYCB. e, Tunicamycin is almost completely enclosed in the binding pocket of hGPT, as compared to the solvent-exposed groove of MraY. In hGPT, the uracil and GlcNAc moieties of tunicamycin are completely covered by loop A and loop E respectively, while are both solvent-exposed in MraY.
Notably, the interactions formed with the GlcNAc moiety of tunicamycin are significantly different between hGPT and MraYCB (Fig. 2c–d). Due to the elongated loop E in hGPT, the invariant Arg303 is flipped inward and interacts with the N-acetyl group of the GlcNAc moiety; however, the corresponding residue Arg282 in MraYCB is moved completely away from tunicamycin. The loop E helix of hGPT does not interact with the GlcNAc moiety of tunicamycin, unlike the loop E helix of MraYCB (His290 and His291) which is oriented at a 30° angle relative to that in hGPT. The GlcNAc moiety of tunicamycin has been proposed to mimic the GlcNAc moiety of the endogenous GPT substrate UDP-GlcNAc, or the MurNAc moiety in the MraY substrate UDP-MurNAc-pentapeptide 11–13. These differences in tunicamycin binding could reflect the different substrate specificity of hGPT and MraY.
Another marked difference between the tunicamycin binding pockets of GPT and MraY is solvent accessibility. The GlcNAc and the uracil moieties of tunicamycin are completely enclosed by loop E and loop A in hGPT, respectively, which was not observed in MraYCB (Fig 2e). Consequently, tunicamycin is sequestered in the active site pocket of hGPT, but is exposed to the cytosol when bound to the shallow groove of MraYCB.
Distinct dimer interfaces of GPT and MraY
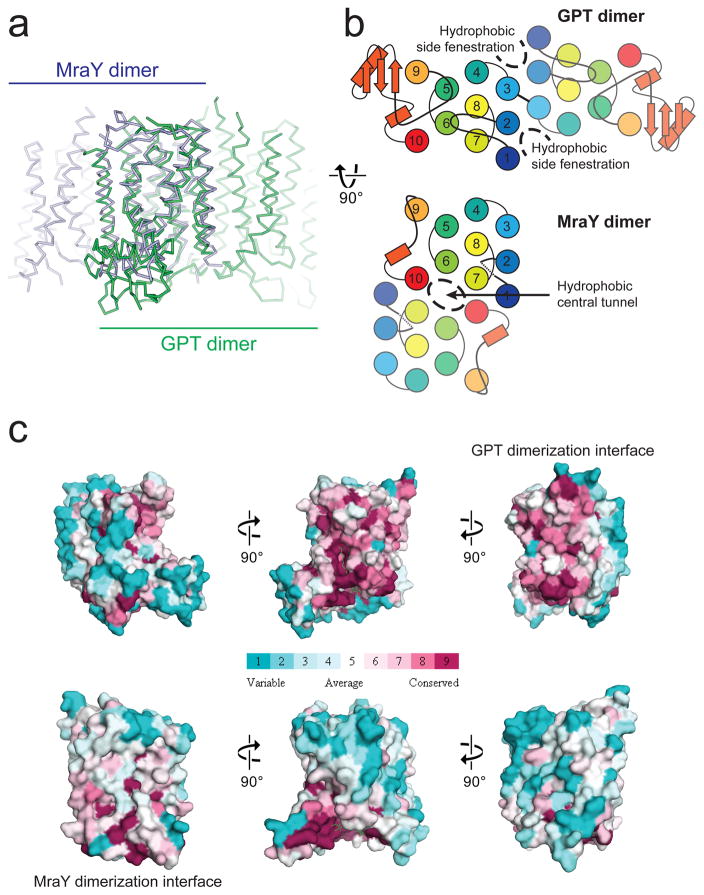
Both GPT and MraY were previously shown to form a dimer in the membrane 26,31. Cooperativity within the GPT dimer was proposed for its function, as a dominant-negative effect was observed by co-expression of the inactive Arg303Lys mutant with the wild-type GPT 31. Strikingly, comparison of the hGPT and MraY structures reveal dramatically different homo-dimerization interfaces, occurring at opposite sides of the protomer (Fig. 3a). The GPT dimer is formed by TMs 2, 3, 4 and part of TM 1, while the MraY dimer is formed by TM 10 and parts of TMs 1 and 7 (Fig. 3b). The different dimerization interfaces are supported by sequence conservation data: the GPT dimerization interface is conserved among GPT sequences but not MraY sequences, and the reverse is true for the MraY dimerization interface (Fig. 3c). An unusual membrane-buried disulfide bond is found between Cys106 of each protomer in the canonical hGPT structure, which is absent in the MraY dimer. The conserved nature of this disulfide bond among GPT orthologues provides further evidence that this is the physiological dimerization interface of GPT (Supplementary Fig. 5). We recognized that while the core of the GPT dimerization interface is buried, two hydrophobic fenestrations penetrate the dimer interface from the sides, both containing strong snake-like electron density which we assigned as a phospholipid tail from the 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG) added during protein purification (Fig 3b and Supplementary Fig. 6). The MraY dimer interface contains a central hydrophobic tunnel separating the TM 6 of each protomer, which has been suggested to be occupied by lipid molecules (Fig. 3b) 26. It is increasingly recognized that interfacial lipids are important for stabilizing the membrane protein oligomers 32. Consistent with this idea, the phosphatidylglycerol-dependence of GPT activity was reported in early studies 33,34, suggesting that the observed lipid-dependent regulation of GPT activity might be due to its stabilizing effects on the dimer interface. The opposite dimerization interfaces between GPT and MraY, as well as the resultant difference in lipid accessibility to the interface, suggest that GPT and MraY potentially may be regulated differently by phospholipids.
Figure 3. Divergence between GPT and MraY is underpinned by their different dimerization interfaces.
a, Superposition of hGPT (green) with MraYCB (blue) reveals strikingly different dimerization interfaces, despite the conserved fold of each protomer. b, The different dimerization interfaces of GPT and MraY result in altered membrane accessibility. Hydrophobic side fenestrations penetrate the GPT dimer, while a central hydrophobic tunnel occupies the MraY dimer. c, The interface for GPT dimerization is conserved only among GPT orthologue sequences but not among MraY sequences, and vice versa is true for MraY. Surfaces are colored in increasing conservation from cyan (least) to magenta (most). Sequence conservation was mapped onto the hGPT and MraYCB structures using 30 orthologue sequences for each alignment.
Distinct roles of magnesium
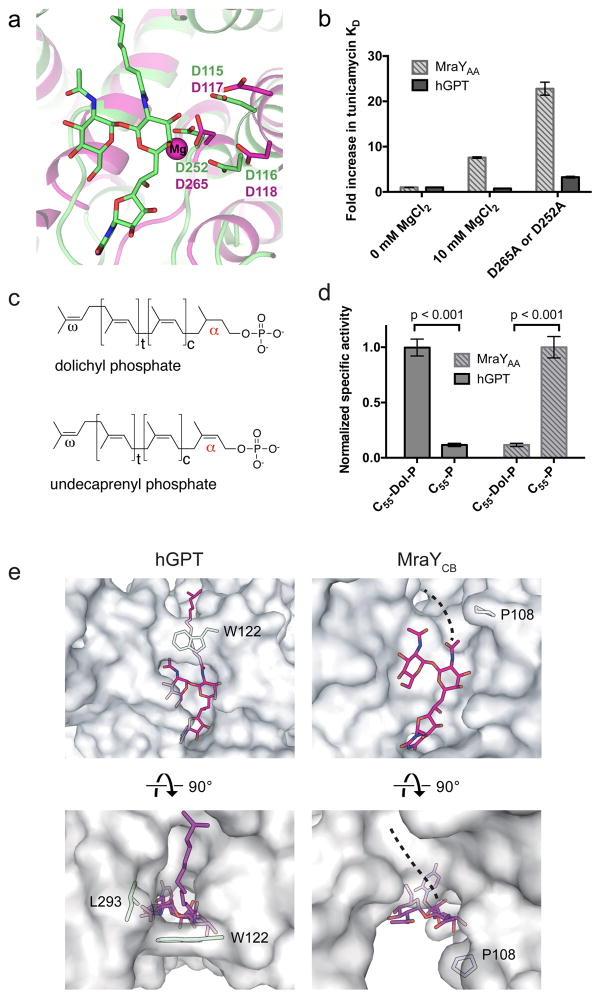
Magnesium is a required cofactor for GPT 35 and MraY 30 enzymatic activity. Extensive studies on MraY catalysis have revealed that three key aspartate residues (Asp116, Asp117, and Asp265 in MraYAA), conserved throughout the PNPT superfamily, are critical for enzymatic activity 26,29. Crystallographic studies unambiguously identified one of the aspartates, Asp265 in MraYAA (Asp231 and Asp252 in MraYCB and hGPT, respectively) as the coordinating residue of the magnesium cofactor (Fig. 4a).
Figure 4. MraY and GPT bind tunicamycin and magnesium differently and they are selective for distinct lipid substrates.
a, Structural overlay of hGPT-tunicamycin (green) and Mg2+-bound MraYAA (magenta, PDB ID: 4J72). Labeled residues in magenta are known to be critical for MraYAA activity and Asp265 coordinates the magnesium cofactor (magenta sphere). b, Fold increase in Kd of tunicamycin with either hGPT or MraY as measured by ITC. The Kd of tunicamycin for hGPT without added MgCl2 is 5.6 ± 1.3 nM (triplicate, technical replicates). The Kd of tunicamycin for MraYAA without added MgCl2 is 37 ± 1 nM (triplicate, technical replicates). Data are shown as the mean of three technical replicates ± s.e.m. c, Chemical structures of dolichyl phosphate and undecaprenyl phosphate. d, TLC-based specific activity assay for hGPT and MraYAA in the presence of either dolichyl phosphate (C55-dol-P) or undecaprenyl phosphate (C55-P). hGPT is selective for the saturated α-isoprenyl position in its substrate dolichyl-phosphate, while MraY is selective for the unsaturated α-isoprenyl position in its substrate undecaprenyl-phosphate. Measurements were performed in triplicate (technical replicates). Specific activity measurements were normalized relative to the C55-Dol-P condition for hGPT and to the C55-P condition for MraY. Data are shown as the mean of normalized specific activity ± s.e.m. Significance was determined with P < 0.001, two-tailed Student’s t-test. e, Cytoplasmic view of the tunicamycin lipid tail binding sites in hGPT and MraYCB. In GPT, the aliphatic tail of tunicamycin packs against the conserved Trp122, which forms a lipid-binding tunnel. By contrast, the corresponding residue in MraYCB (Pro108) is unable lock the lipid tail into place, leaving the hydrophobic groove exposed.
It has been suggested that Mg2+ is required for tunicamycin binding to MraY and GPT, because the tunicamine moiety of tunicamycin is thought to mimic the diphosphate moiety in the transition state of the enzymatic reaction between nucleotide sugar and lipid carrier 10,12,13. However, comparison of the tunicamycin-bound hGPT and the Mg2+-bound apo MraYAA structures shows that Asp265 (Asp252 in hGPT) coordinates Mg2+ in MraYAA, but interacts directly with tunicamycin in hGPT, suggesting Mg2+ and tunicamycin likely compete for binding (Fig. 4a)26. If our hypothesis is correct, we predict that Mg2+ is not required for tunicamycin binding, and we also expect reduced tunicamycin binding with increasing concentrations of Mg2+. We performed ITC by titrating tunicamycin into either MraYAA or hGPT in the presence or absence of MgCl2 (Fig. 4b). We used MraYAA for our assays because the structure, function, and inhibition of this orthologue have been well studied and MraY orthologues share common mechanisms of catalysis and inhibition 23,26,27,29,30. We found that tunicamycin binds to both MraYAA and hGPT in the absence of Mg2+, and the addition of 10 mM MgCl2 resulted in an eight-fold increase in Kd for tunicamycin with MraYAA. Mutating the Mg2+-coordinating residue in MraYAA (Asp265) to alanine increased the Kd for tunicamycin by over twenty-fold. On the contrary, the addition of 10 mM MgCl2 has no appreciable effect on tunicamycin binding to GPT. Furthermore, mutation of Asp252 to alanine increased Kd of tunicamycin by only about three-fold. In summary, our data suggest that the role of Mg2+ in tunicamycin binding is substantially different in GPT and MraY; while Mg2+ competes with tunicamycin binding to MraY, it has no detectable effect on tunicamycin binding to GPT. These observations are contrary to the previous notion that Mg2+ is essential for tunicamycin binding to these two enzymes 12,13.
Distinct lipid substrate specificity of GPT and MraY
GPT and MraY utilize different lipid substrates. GPT utilizes dolichyl phosphate, the lipid carrier for eukaryotic N-linked glycosylation, while MraY utilizes undecaprenyl phosphate, the lipid carrier for peptidoglycan biosynthesis. A major difference between these two polyprenyl phosphates is the α-isoprene unit closest to the phosphate group: dolichyl phosphate has a saturated bond at this position while undecaprenyl phosphate has an unsaturated bond (Fig. 4c). To test the selectivity of GPT and MraY for their respective lipid substrates, hGPT and MraYAA enzymatic reactions were carried out in the presence of either undecaprenyl phosphate (C55-P) or C55 dolichyl phosphate (C55-dol-P) (Fig. 4d). hGPT exhibits strong selectivity for C55-dol-P with nearly no observed activity in the presence of C55-P. Conversely, MraYAA shows clear preference for C55-P over C55-dol-P. This result is consistent with the structural architecture of the hGPT and MraYCB active sites (Fig. 4e). Tunicamycin contains a lipid tail that is thought to overlap with the lipid substrate binding sites in GPT and MraY. In hGPT, the lipid tail of tunicamycin fits snugly into a deep pocket, which is trapped by the conserved Trp122 (Fig. 4e left). This highly defined lipid tunnel is likely required to constrain dolichyl phosphate, which has higher rotational freedom due to the saturated α-isoprene unit. By contrast, the tunicamycin lipid tail binding site in MraY is a more shallow and wide groove than the analogous site in GPT (Fig. 4e right). The unsaturated α-isoprene unit of undecaprenyl phosphate has a limited degree of rotational freedom and its assumed geometry probably guides binding to the shallow MraY lipid tail binding groove. Structural differences between the lipid tail binding sites in GPT and MraY likely underpins the high selectivity each enzyme exhibits for its respective polyprenyl substrate.
Chemical modification of tunicamycin selectively targets MraY over GPT
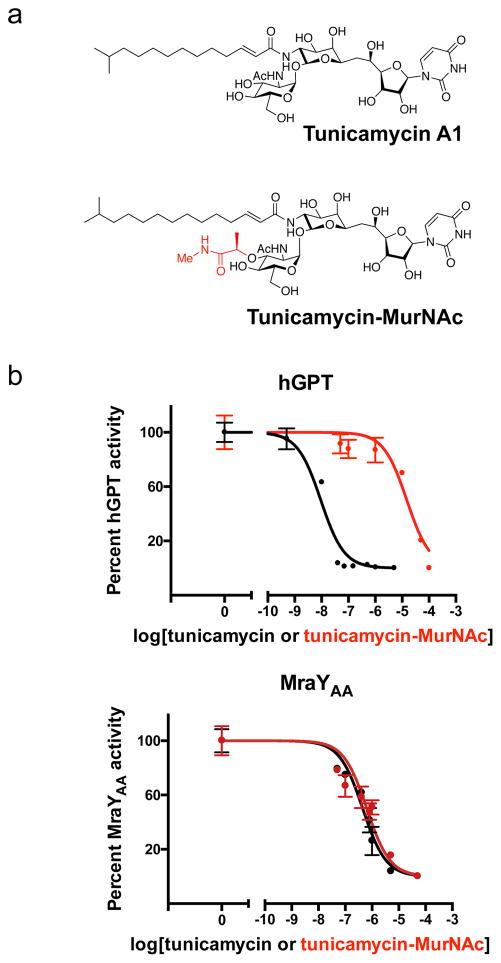
Efforts to develop tunicamycin as an antibiotic requires rational design to eliminate the cytotoxicity arising from its off-target effect on hGPT activity 25,36. Based on our structural analysis of the tunicamycin binding site, we designed a tunicamycin analog that replaces the GlcNAc sugar moiety in tunicamycin with a MurNAc sugar (Fig. 5a), which we synthesized as described in the recently published total synthesis of tunicamycin V (Supplementary Note 1)37. The rationale for this chemical modification is that (1) the binding pocket of hGPT formed by loop E tightly packs around the GlcNAc sugar (Fig. 2e) and likely cannot accommodate the bulkier MurNAc moiety, and (2) the natural substrate for MraY contains a MurNAc moiety, while the natural substrate of hGPT contains a GlcNAc moiety (Supplementary Fig. 1). We observed that the IC50 of tunicamycin for hGPT is about 9 nM and that of tunicamycin-MurNAc is about 15 μM, resulting in about a thousand-fold decrease in inhibition of hGPT by this simple modification of tunicamycin (Fig. 5b). By contrast, the activity of MraYAA is similarly inhibited by both tunicamycin and tunicamycin-MurNAc (Fig. 5b). This observation is consistent with the crystal structure of MraYCB bound to tunicamycin, which demonstrates that the binding site of the tunicamycin GlcNAc moiety is wider than that of hGPT and is therefore more likely to accommodate the larger MurNAc moiety (Fig. 2e). Therefore, we showed proof-of-principle to selectively target tunicamycin towards MraY over GPT at the protein level. Further investigation is required to evaluate the in vivo activity and toxicity of such analogs.
Figure 5. Chemically modifying tunicamycin can introduce ligand selectivity between hGPT and MraYAA.
a, Chemical structures of tunicamycin and its MurNAc derivative. The substructure highlighted in red differs from tunicamycin, being a MurNAc-like moiety rather than a GlcNAc moiety as in tunicamycin. b, IC50 measurements of tunicamycin and its MurNAc analog (tunicamycin-MurNac) with hGPT and MraYAA, respectively. hGPT is much less inhibited by tunicamycin-MurNAc than tunicamycin, while MraY appears to be similarly inhibited by both tunicamycin and tunicamycin-MurNAc. The IC50 for hGPT with tunicamycin is ~9 nM; for hGPT with tunicamycin-MurNAc is ~15 μM; for MraYAA with tunicamycin is ~450 nM; for MraYAA with tunicamycin-MurNAc is ~640 nM. Each IC50 value was determined using the TLC-based phosphoglycosyl transferase assay. Data are shown as the mean of triplicate measurements (technical replicates) ± s.e.m.
Discussion
Our structural and functional studies of hGPT provide the framework to elucidate the molecular basis of mutations that cause CDG-Ij and CMS13 (Supplementary Fig. 7). Intriguingly, distinct groups of mutations within DPAGT1 are responsible for either CDG-Ij or CMS13, and they are spread over hGPT rather than localized around the active site of hGPT (Supplementary Fig. 7). Systematic enzymatic, structural, and cellular studies are required to understand the effects of disease mutations on hGPT and the N-linked glycosylation.
GPT and MraY have drastically different dimer organization. It is unusual to have distinct dimerization organization among paralogues for enzymes. What is the possible reason for distinct dimerization of GPT compared to MraY? While the cooperativity of the GPT dimer and the importance of phosphatidyglycerol to GPT function were reported, no studies have been reported for the cooperativity of the MraY dimer. We observed that the two active sites are closer together in the GPT dimer than in the MraY dimer. In the GPT dimer, part of TM1 and loop A is closer to the symmetric axis (Fig. 1), potentially enabling communication between the two active sites within the dimer, which is not present in the MraY dimer. Therefore, we speculate that the distinct dimerization of GPT is to enable cooperativity, which adds another layer of complexity to the regulation of this eukaryotic system.
We showed that the mechanisms of tunicamycin binding to MraY and GPT are different in many respects. First, we found the role of Mg2+ is different in tunicamycin inhibition of GPT and MraY. Some potential reasons for this difference include, (1) GPT forms several additional interactions with tunicamycin, thereby making it less readily displaced by Mg2+, and (2) the Mg2+ binding site in GPT is different to that in MraY. Second, we found that the active site shape and solvent accessibility is significantly different between GPT and MraY. Difference in the active site shape could reflect the substantial difference in size between the GPT substrate UDP-GlcNAc (~600 Da) and the MraY substrate UDP-MurNAc-pentapeptide (~1200 Da) (Supplementary Fig. 1); the former could be easily captured in an enclosed pocket whereas the latter would require an exposed groove for binding. Third, the lipid substrate specificity is distinct in GPT and MraY, which is further supported by structural differences in the lipid binding site of each enzyme. We exploited one of these distinctions to design a tunicamycin analog with biased specificity toward MraY at the enzyme level. Our proof-of-principle studies with tunicamycin-MurNAc demonstrate that developing MraY-specific tunicamycin analogues is possible. In principle, we believe many of the features that distinguish MraY and GPT could be utilized for the development of MraY-specific tunicamycin analogs. For example, the lipid tail portion of tunicamycin can be also modified to reduce affinity for GPT.
Methods
Human GPT-tunicamycin complex expression, purification, and crystallization
The full-length DPAGT1 gene encoding human GPT (all residues 1-408, His129 variant, European Nucleotide Archive ID: AAG43168) was synthesized and subcloned into a modified pFastBac vector (Invitrogen) in frame with a C-terminal PreScission protease site, FLAG affinity tag and 10xHis affinity tag. hGPT (Pro129) variant construct was made by a mutagenesis with Pfu-Turbo DNA polymerase (Agilent) on hGPT (His129) construct. Baculovirus was produced according to the manufacturer’s protocol (Bac-to-Bac Baculovirus expression system, Invitrogen). For hGPT expression, Spodoptera frugiperda 9 (Sf9) insect cells were infected with a baculovirus at a density of 1.2 M cells mL−1 and grown at 27°C for 24 h in an orbital shaker. Tunicamycin (5 mgmL−1 in DMSO) was added into cultures at 1 mgL−1 and cells were grown for 48 h at 27°C. Cells were then harvested by centrifugation at 1710 rpm for 12 mins at 4°C. Cell pellets were resuspended in buffer A (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 μgmL−1 leupeptin, 1 μgmL−1 pepstatin, 1 μgmL−1 aprotinin, 1 mM PMSF and DNaseI) and lysed by sonication (5 × 30 pulses). All hGPT-tunicamycin complex purification steps were performed at 4°C. To solubilize the hGPT-tunicamycin complex, 40 mM n-dodecyl-β-D-maltopyranoside (DDM, Anatrace), 2 mgmL−1 iodoacetamide and 4 mM cholesteryl hemisuccinate tris salt (CHS, Anatrace) were added to the lysate and stirred at 4°C for 2 hrs. Insoluble material was removed by centrifugation (8000g, 30 min), and anti-FLAG resin was added to the supernatant and incubated for 1 h at 4°C. The resin was then washed with 10 column volumes of buffer B (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM DDM, 0.1 mM CHS). The hGPT-tunicamycin complex was eluted with 5 column volumes of elution buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1 mM CHS, 1 mM DDM, 0.1 mgmL−1 FLAG peptide, 0.1 mgmL−1 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG, Avanti Polar Lipids)). The FLAG and 10xHis affinity tags were removed by overnight incubation with PreScission protease at 4°C with 1 mM dithiothreitol (DTT) and 1 mM ethylenediaminetetraacetic acid (EDTA). The hGPT-tunicamycin complex was further purified by size-exclusion chromatography (Superdex 200 10/300 GL) with SEC buffer containing 20 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.3 mM decyl maltose neopentyl glycol (DMNG, Anatrace), 0.1 mM CHS, 0.1 mgmL−1 POPG, 2 mM DTT, 10 μM tunicamycin, 10 mM MgCl2).
Size-exclusion chromatography fractions of GPT-tunicamycin complex were concentrated to 8–10 mgmL−1. Initial crystallization trials were performed with in-house crystal screens in 96-well sitting drop plates (Art-Robbins). Crystals were grown by sitting-drop vapor diffusion at 17°C in 0.2 M CaCl2, 0.05 M Tris-HCl, pH 8.5, 25–30% PEG 400. Crystals appeared within 3–4 days, at which point they were immediately harvested in liquid nitrogen without additional cryo-protectant.
Data collection and structure determination
We collected X-ray diffraction data at the NECAT 24-ID-C and 24-ID-E beamlines, as well as at the SERCAT 22-ID beamline (Advanced Photon Source, Argonne National Laboratory) with a wavelength of 0.9791 Å. Data were processed by XDS 38, and XDS-processed data from multiple isomorphous crystals were merged by POINTLESS and AIMLESS in BLEND 39–41. Due to severe diffraction anisotropy, merged data were subjected to ellipsoidal truncation and anisotropy correction in STARANISO 42. Anisotropy corrected data were used for molecular replacement using the publicly released structure of apo human GPT Val264Gly mutant (PDB ID: 5LEV) as search model in PHASER 43. Our hGPT-tunicamycin crystals were in C2 space group with two dimers in the asymmetric unit. The molecular replacement solution was first subjected to initial jelly-body refinement in the LORESTR pipeline 44, before manual model building in COOT 45 and refinement in REFMAC 46 and PHENIX.refine 47. Due to the higher resolution data of the hGPT(His129)-tunicamycin complex (2.95 Å resolution) than the canonical hGPT(Pro129)-tunicamycin complex (3.10 Å resolution), the initial round of molecular replacement and refinement was performed on the His129 data as described above. The model built from the His129 data was then transferred to the Pro129 data and a second round of model building and refinement was performed to produce the Pro129 structure. Structures were refined to a final Rwork/Rfree of 25.6/28.9% (Pro129 structure) and 26.0/29.1% (His129 structure). The final models were refined to excellent geometry (96–97% Ramachandran favored, 0% Ramachandran outliers, 0% rotamer outliers for both structures) and minimal clashes (clashscore of 4.66 for Pro129 and 1.57 for His129). Structural alignments and molecular graphics were created in PyMOL 48. MAFFT (“add new sequences to an existing alignment” mode) was used to expand the structure-guided sequence alignment to include more sequences 49. Sequence conservation was mapped onto the hGPT and MraYCB structures using the ConSurf server 50 using 30 orthologue seuqences for each alignment.
hGPT expression and purification for functional studies
For functional studies, the same DNA construct for hGPT expression was used, but tunicamycin was omitted from all the culture and protein purification steps. After adding baculovirus of hGPT wild-type or Asp252Ala mutant into 1.2 M Sf9 cells mL−1, cells were grown for 72 hrs at 27°C. Cells were then harvested by centrifugation at 1710 rpm for 12 mins at 4°C. The cell pellet was resuspended with buffer A (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 μgmL−1 leupeptin, 1 μgmL−1 pepstatin, 1 μgmL−1 aprotinin, 1 mM PMSF and DNaseI) and lysed by sonication (5 × 30 pulses). To solubilize hGPT, 40 mM DDM was added to the lysate and stirred at 4°C for 2 h. Insoluble material was removed by centrifugation (8000g, 30 min), and anti-FLAG resin was added to the supernatant and stirred for 1 h at 4°C. The resin was then washed with 10 column volumes of buffer B (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM DDM). hGPT was eluted with 5 column volumes of elution buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM DDM, 0.1 mgmL−1 FLAG peptide, 0.1 mgmL−1 POPG). FLAG and 10xHis affinity tag was removed by overnight incubation with PreScission protease at 4°C with 1 mM DTT and 1 mM EDTA. hGPT was further purified by size-exclusion chromatography (Superdex 200 10/300 GL) with SEC buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.3 mM DMNG, 0.1 mgmL−1 POPG, 2 mM DTT).
Isothermal titration calorimetry
MraYAA wild-type and Asp256Ala were purified as previously described 26 in a buffer containing 150mM NaCl, 20mM Tris-HCl pH 8.0, 5 mM DM and 2mM DTT. This same buffer was used to dilute the ligand, tunicamycin (Sigma-Aldrich). For one set of titrations, 10 mM MgCl2 was also included in the buffer. For MraYAA wild-type, 210 μM tunicamycin was titrated into 30 μM enzyme. hGPT wild-type and Asp252Ala were prepared as described above and tunicamycin was dissolved in the SEC buffer used to purify these enzymes. For MraYAA Asp265Ala, 300 μM tunicamycin was titrated into 30 μM enzyme. For hGPT WT and Asp252Ala, 240 μM tunicamycin was titrated into 40 μM enzyme. All titrations were performed in triplicate (technical replicates) at 30°C for hGPT and at 45°C for MraYAA using a MicroCal iTC200 (GE Healthcare). The total heat exchanged during each injection was fit to a single-site binding isotherm with Kd and ΔH° as independent parameters. Data were analyzed and figures were generated using Origin software (OriginLab Corp).
Thin-layer chromatography (TLC)-based enzyme assays
Enzymatic assays monitored either 1) the hGPT-mediated transfer of [14C]phospho-GlcNAc from [14C]UDP-GlcNAc to dolichyl phosphate (C55-Dol-P), forming [14C]Dol-PP-GlcNAc, or 2) the MraYAA-mediated transfer of [14C]phospho-MurNAc-pentapeptide from [14C]UDP-MurNAc-pentapeptide(DAP) ([14C]UM5A) to undecaprenyl phosphate (C55-P), forming [14C]Lipid I. A thin-layer chromatography (TLC)-based radiochemical assay 51 was used, which was optimized for hGPT and MraYAA-catalyzed reactions for the experiments described below. All reactions were quenched by spotting a 2-μL aliquot on a silica gel 60 thin layer chromatography (TLC) plate (EMD Millipore). The products and substrates were separated by TLC using isopropanol/ammonium hydroxide/water (6:3:1; v/v/v) as the mobile phase. The spots corresponding to product and substrate were visualized via Phosphorimager (Typhoon FLA 7000, GE Healthcare LifeSciences). The spot intensity was quantified using the ImageQuant TL (GE Healthcare LifeSciences). All experiments were performed in triplicate.
Specific activity measurements of hGPT wild-type His129 and Pro129 variants
The reaction buffer contained 70 mM Tris-HCl pH 8.0, 500 mM NaCl, 80 mM MgCl2, 5 mM DM, 1 mg/mL POPG, 10% glycerol, 500 μM C55-dol-P, 0.01 mM [14C]UDP-GlcNAc and 0.1 mM UDP-GlcNAc. The reactions were initiated with 200 nM hGPT His129 or Pro129, were incubated at 30°C for 20 min, and a 2-μL aliquot of each reaction was spotted on the TLC plate every 5 min. Reactions were performed in triplicate (technical replicates). The specific activity was calculated by dividing the rate of product formation (nmol/min) by the quantity of protein added in milligrams.
Lipid substrate dependence
The hGPT-catalyzed reactions contained the same buffer composition as described above. The MraYAA-catalyzed reaction buffer contained 100 mM Tris-HCl pH 8.0, 500 mM NaCl, 10 mM MgCl2, 20 mM CHAPS, and 150 μM [14C]UM5A. Each reaction included 500 μM of either C55-Dol-P or C55-P. Reactions were initiated by either 200 nM hGPT Pro129 or 50 nM MraYAA wild-type and were carried out for 12 min at 30°C (hGPT) or 5 min at 60°C (MraYAA). Reactions were performed in triplicate (technical replicates). Specific activity was calculated as described above and normalized to the maximally active reaction condition.
IC50 measurements
The assay reaction mixtures contained the same buffer composition as described above for the hGPT- and MraYAA-catalyzed reactions, respectively. Reactions were initiated by either 200 nM hGPT Pro129 or 50 nM MraYAA and were carried out for 12 min at 30°C (hGPT) or 5 min at 60°C (MraYAA), which is within the enzymatic linear range for each enzyme. For IC50 measurements with tunicamycin and hGPT, the following concentrations of tunicamycin (μM) were used: 0, 0.0005, 0.01, 0.04, 0.07, 0.15, 0.5, 1, and 5. For IC50 measurements with tunicamycin-MurNAc and hGPT, the following concentrations of tunicamycin-MurNAc (μM) were used: 0, 0.05, 0.1, 1, 5, 10, 50, and 100. For IC50 measurements with tunicamycin and MraYAA, the following concentrations of tunicamycin (μM) were used: 0. 0.05, 0.1, 0.4, 0.8, 1, 5, and 50. The same concentration range was used for IC50 measurements with tunicamycin-MurNAc and MraYAA. All measurements were made in triplicate (technical replicates). Data were plotted in GraphPad Prism 6 and IC50 values were calculated using the log(inhibitor) vs. response – variable slope model.
Synthesis of tunicamycin-MurNAc
We synthesized tunicamycin-MurNAc as described in the recently published total synthesis of tunicamycin V (see Supplementary Note 1 for details) 37. In brief, the analog was synthesized by assemblage of a trichloroacetimidate of MurNAc N-methylamide derivative S5 as a glycosyl donor and a suitably protected tunicaminyluracil S6 as a glycosyl acceptor 37. Namely, treatment of S5 and S6 with TfOH in Et2O at 0 °C provided the desired 11′-β-1″-α-glycoside S7 in 69% yield in a highly stereoselective manner. Transformation of the azide group, deprotection of the phthaloyl group at the GalNAc moiety, and installation of the fatty acyl group gave S10. Finally, global deprotection of six acid labile protecting groups by BCl3 resulted in clean conversion to afford tunicamycin-MurNAc.
Data availability
Atomic coordinates and structure factors for the reported crystal structures are deposited in the Protein Data Bank under accession codes 6BW5 for the canonical hGPT (Pro129) and 6BW6 for the His129 variant. Any other data pertaining to this paper is available upon reasonable request.
Supplementary Material
Acknowledgments
Data for this study were collected at beamlines NECAT 24-ID-C and 24-ID-E, as well as at SERCAT 22-ID, both at the Advanced Photon Source. We thank Ben Chung for initial GPT biochemistry. We thank Ian Tickle and the STARANISO team for help with our anisotropic data analysis. This work was supported by the National Institutes of Health (R01GM120594 and R35NS097241 to S.-Y. L.), JSPS Grant-in-Aid for Scientific Research (B) (16H05097 to S.I.), Astellas Foundation for Research on Metabolic Disorders (to S.I.), Hokkaido University GFC, PSOU, funded by MEXT (to S.I), and BINDS from the Japan Agency for Medical Research and Development (to S.I.). Beamlines 24-ID-C and 24-ID-E are funded by P41 GM103403 and S10 RR029205.
Footnotes
Competing financial interests The authors declare no competing financial interests.
Author Contributions J.Y. performed GPT crystallization, data collection, and protein preparation for functional studies; E.H.M. performed all functional studies of GPT and MraY; B.K. assisted in sample purification for functional studies; A.K. and J.Y. performed X-ray data processing, model building and refinement. J.Y., A.K., B.K., and E.H.M. are under the guidance of S.-Y.L. S.I. designed tunicamycin-MurNAc. K.Y. synthesized the tunicamycin-MurNAc under the guidance of S.I. S.-Y.L., A.K., E.H.M., and J.Y. wrote the paper.
References
- 1.Lehrman MA. Biosynthesis of N-acetylglucosamine-P-P-dolichol, the committed step of asparagine-linked oligosaccharide assembly. Glycobiology. 1991;1:553–562. doi: 10.1093/glycob/1.6.553. [DOI] [PubMed] [Google Scholar]
- 2.Belaya K, et al. Mutations in DPAGT1 cause a limb-girdle congenital myasthenic syndrome with tubular aggregates. Am J Hum Genet. 2012;91:193–201. doi: 10.1016/j.ajhg.2012.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu X, et al. Deficiency of UDP-GlcNAc:Dolichol Phosphate N-Acetylglucosamine-1 Phosphate Transferase (DPAGT1) causes a novel congenital disorder of Glycosylation Type Ij. Hum Mutat. 2003;22:144–150. doi: 10.1002/humu.10239. [DOI] [PubMed] [Google Scholar]
- 4.Wurde AE, et al. Congenital disorder of glycosylation type Ij (CDG-Ij, DPAGT1-CDG): extending the clinical and molecular spectrum of a rare disease. Mol Genet Metab. 2012;105:634–641. doi: 10.1016/j.ymgme.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Liwosz A, Lei T, Kukuruzinska MA. N-glycosylation affects the molecular organization and stability of E-cadherin junctions. J Biol Chem. 2006;281:23138–23149. doi: 10.1074/jbc.M512621200. [DOI] [PubMed] [Google Scholar]
- 6.Nita-Lazar M, et al. Overexpression of DPAGT1 leads to aberrant N-glycosylation of E-cadherin and cellular discohesion in oral cancer. Cancer Res. 2009;69:5673–5680. doi: 10.1158/0008-5472.can-08-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lehrman MA. A family of UDP-GlcNAc/MurNAc: polyisoprenol-P GlcNAc/MurNAc-1-P transferases. Glycobiology. 1994;4:768–771. doi: 10.1093/glycob/4.6.768. [DOI] [PubMed] [Google Scholar]
- 8.Takatsuki A, Arima K, Tamura G. Tunicamycin, a new antibiotic. I. Isolation and characterization of tunicamycin. J Antibiot (Tokyo) 1971;24:215–223. doi: 10.7164/antibiotics.24.215. [DOI] [PubMed] [Google Scholar]
- 9.Takatsuki A, et al. The Structure of Tunicamycin. Agricultural and Biological Chemistry. 1977;41:2307–2309. doi: 10.1080/00021369.1977.10862856. [DOI] [Google Scholar]
- 10.Wang R, et al. A search for pyrophosphate mimics for the development of substrates and inhibitors of glycosyltransferases. Bioorg Med Chem. 1997;5:661–672. doi: 10.1016/s0968-0896(97)00005-9. [DOI] [PubMed] [Google Scholar]
- 11.Izumi M, Yuasa H, Hashimoto H. Bisubstrate analogues as glycosyltransferase inhibitors. Curr Top Med Chem. 2009;9:87–105. doi: 10.2174/156802609787354351. [DOI] [PubMed] [Google Scholar]
- 12.Price NP, Momany FA. Modeling bacterial UDP-HexNAc: polyprenol-P HexNAc-1-P transferases. Glycobiology. 2005;15:29R–42R. doi: 10.1093/glycob/cwi065. [DOI] [PubMed] [Google Scholar]
- 13.Xu L, Appell M, Kennedy S, Momany FA, Price NP. Conformational analysis of chirally deuterated tunicamycin as an active site probe of UDP-N-acetylhexosamine:polyprenol-P N-acetylhexosamine-1-P translocases. Biochemistry. 2004;43:13248–13255. doi: 10.1021/bi048327q. [DOI] [PubMed] [Google Scholar]
- 14.Elbein AD. Inhibitors of the biosynthesis and processing of N-linked oligosaccharide chains. Annu Rev Biochem. 1987;56:497–534. doi: 10.1146/annurev.bi.56.070187.002433. [DOI] [PubMed] [Google Scholar]
- 15.Keller RK, Boon DY, Crum FC. N-Acetylglucosamine- 1 -phosphate transferase from hen oviduct: solubilization, characterization, and inhibition by tunicamycin. Biochemistry. 1979;18:3946–3952. doi: 10.1021/bi00585a016. [DOI] [PubMed] [Google Scholar]
- 16.Lehle L, Tanner W. The specific site of tunicamycin inhibition in the formation of dolichol-bound N-acetylglucosamine derivatives. FEBS Lett. 1976;72:167–170. doi: 10.1016/0014-5793(76)80922-2. [DOI] [PubMed] [Google Scholar]
- 17.Takatsuki A, Kohno K, Tamura G. Inhibition of Biosynthesis of Polyisoprenol Sugars in Chick Embryo Microsomes by Tunicamycin. Agricultural and Biological Chemistry. 1975;39:2089–2091. doi: 10.1080/00021369.1975.10861914. [DOI] [Google Scholar]
- 18.Tkacz JS, Lampen O. Tunicamycin inhibition of polyisoprenyl N-acetylglucosaminyl pyrophosphate formation in calf-liver microsomes. Biochem Biophys Res Commun. 1975;65:248–257. doi: 10.1016/s0006-291x(75)80086-6. [DOI] [PubMed] [Google Scholar]
- 19.Duksin D, Mahoney WC. Relationship of the structure and biological activity of the natural homologues of tunicamycin. J Biol Chem. 1982;257:3105–3109. [PubMed] [Google Scholar]
- 20.Faye L, Chrispeels MJ. Apparent Inhibition of beta-Fructosidase Secretion by Tunicamycin May Be Explained by Breakdown of the Unglycosylated Protein during Secretion. Plant Physiol. 1989;89:845–851. doi: 10.1104/pp.89.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koizumi N, Ujino T, Sano H, Chrispeels MJ. Overexpression of a gene that encodes the first enzyme in the biosynthesis of asparagine-linked glycans makes plants resistant to tunicamycin and obviates the tunicamycin-induced unfolded protein response. Plant Physiol. 1999;121:353–361. doi: 10.1104/pp.121.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oslowski CM, Urano F. Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol. 2011;490:71–92. doi: 10.1016/b978-0-12-385114-7.00004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brandish PE, et al. Modes of action of tunicamycin, liposidomycin B, and mureidomycin A: inhibition of phospho-N-acetylmuramyl-pentapeptide translocase from Escherichia coli. Antimicrob Agents Chemother. 1996;40:1640–1644. doi: 10.1128/aac.40.7.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura G, Sasaki T, Matsuhashi M, Takatsuki A, Yamasaki M. Tunicamycin Inhibits the Formation of Lipid Intermediate in Cell-free Peptidoglycan Synthesis of Bacteria. Agricultural and Biological Chemistry. 1976;40:447–449. doi: 10.1080/00021369.1976.10862071. [DOI] [Google Scholar]
- 25.Lukose V, Walvoort MTC, Imperiali B. Bacterial phosphoglycosyl transferases: initiators of glycan biosynthesis at the membrane interface. Glycobiology. 2017;27:820–833. doi: 10.1093/glycob/cwx064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung BC, et al. Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis. Science. 2013;341:1012–1016. doi: 10.1126/science.1236501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung BC, et al. Structural insights into inhibition of lipid I production in bacterial cell wall synthesis. Nature. 2016;533:557–560. doi: 10.1038/nature17636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hakulinen JK, et al. MraY-antibiotic complex reveals details of tunicamycin mode of action. Nat Chem Biol. 2017;13:265–267. doi: 10.1038/nchembio.2270. [DOI] [PubMed] [Google Scholar]
- 29.Al-Dabbagh B, et al. Active site mapping of MraY, a member of the polyprenyl-phosphate N-acetylhexosamine 1-phosphate transferase superfamily, catalyzing the first membrane step of peptidoglycan biosynthesis. Biochemistry. 2008;47:8919–8928. doi: 10.1021/bi8006274. [DOI] [PubMed] [Google Scholar]
- 30.Bouhss A, Trunkfield AE, Bugg TD, Mengin-Lecreulx D. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol Rev. 2008;32:208–233. doi: 10.1111/j.1574-6976.2007.00089.x. [DOI] [PubMed] [Google Scholar]
- 31.Dan N, Lehrman MA. Oligomerization of hamster UDP-GlcNAc:dolichol-P GlcNAc-1-P transferase, an enzyme with multiple transmembrane spans. J Biol Chem. 1997;272:14214–14219. doi: 10.1074/jbc.272.22.14214. [DOI] [PubMed] [Google Scholar]
- 32.Gupta K, et al. The role of interfacial lipids in stabilizing membrane protein oligomers. Nature. 2017;541:421–424. doi: 10.1038/nature20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plouhar PL, Bretthauer RK. A phospholipid requirement for dolichol pyrophosphate N-acetylglucosamine synthesis in phospholipase A2-treated rat lung microsomes. J Biol Chem. 1982;257:8907–8911. [PubMed] [Google Scholar]
- 34.Plouhar PL, Bretthauer RK. Restoration by phospholipids of dolichol pyrophosphate N-acetylglucosamine synthesis in delipidated rat lung microsomes. J Biol Chem. 1983;258:12988–12993. [PubMed] [Google Scholar]
- 35.Kaushal GP, Elbein AD. Purification and properties of UDP-GlcNAc:dolichyl-phosphate GlcNAc-1-phosphate transferase. Activation and inhibition of the enzyme. J Biol Chem. 1985;260:16303–16309. [PubMed] [Google Scholar]
- 36.Walvoort MT, Lukose V, Imperiali B. A Modular Approach to Phosphoglycosyltransferase Inhibitors Inspired by Nucleoside Antibiotics. Chemistry. 2016;22:3856–3864. doi: 10.1002/chem.201503986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto K, Yakushiji F, Matsumaru T, Ichikawa S. Total Synthesis of Tunicamycin V. Org Lett. 2018;20:256–259. doi: 10.1021/acs.orglett.7b03623. [DOI] [PubMed] [Google Scholar]
- 38.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/s0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foadi J, et al. Clustering procedures for the optimal selection of data sets from multiple crystals in macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2013;69:1617–1632. doi: 10.1107/s0907444913012274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evans PR. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr D Biol Crystallogr. 2011;67:282–292. doi: 10.1107/s090744491003982x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/s0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tickle IJ, Flensburg C, Keller P, Paciorek W, Sharff A, Vonrhein C, Bricogne G. STARANISO. Cambridge, United Kingdom: Global Phasing Ltd; 2017. [Google Scholar]
- 43.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kovalevskiy O, Nicholls RA, Murshudov GN. Automated refinement of macromolecular structures at low resolution using prior information. Acta Crystallogr D Struct Biol. 2016;72:1149–1161. doi: 10.1107/s2059798316014534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/s0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/s0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/s0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delano WL. The PyMol Molecular Graphics System. DeLano Scientific; 2002. [Google Scholar]
- 49.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Celniker G, et al. ConSurf: Using Evolutionary Data to Raise Testable Hypotheses about Protein Function. Isr J Chem. 2013;53:199–206. [Google Scholar]
- 51.Bouhss A, Crouvoisier M, Blanot D, Mengin-Lecreulx D. Purification and characterization of the bacterial MraY translocase catalyzing the first membrane step of peptidoglycan biosynthesis. J Biol Chem. 2004;279:29974–29980. doi: 10.1074/jbc.M314165200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structures are deposited in the Protein Data Bank under accession codes 6BW5 for the canonical hGPT (Pro129) and 6BW6 for the His129 variant. Any other data pertaining to this paper is available upon reasonable request.