Abstract
Accumulating evidence implicates monopolar spindle-one-binder protein (MOB)2 as an inhibitor of nuclear-Dbf2-related kinase (NDR) by competing with MOB1 for interaction with NDR1/2. NDR/large tumor suppressor (LATS) kinases may function similarly to yes-associated protein (YAP) kinases and be considered as members of the Hippo core cassette. MOB2 appears to serve roles in cell survival, cell cycle progression, responses to DNA damage and cell motility. However, the underlying mechanisms involved remain unclarified. In the present study, it was demonstrated that the knockout of MOB2 by clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein 9 promoted migration and invasion, induced phosphorylation of NDR1/2 and decreased phosphorylation of YAP in SMMC-7721 cells when compared with the blank vector-transduced cells. By contrast, the overexpression of MOB2 resulted in the opposite results. Mechanistically, MOB2 regulated the alternative interaction of MOB1 with NDR1/2 and LATS1, which resulted in increased phosphorylation of LATS1 and MOB1 and thereby led to the inactivation of YAP and consequently inhibition of cell motility. The results of the present study provide evidence of MOB2 serving a positive role in LATS/YAP activation by activating the Hippo signaling pathway.
Keywords: monopolar spindle-one-binder protein 2, hippo pathway, yes-associated protein, nuclear-Dbf2-related kinase, large tumor suppressor, monopolar spindle-one-binder protein 2
Introduction
Monopolar spindle-one-binder proteins (MOBs) are highly conserved from yeast to mammals. MOBs function as signal transducers in signaling pathways via their interactions with the nuclear Dbf2-related (NDR)/large tumor suppressor (LATS) family of kinases (1–3). To date, at least six different human MOB genes (MOB1A, MOB1B, MOB2, MOB3A, MOB3B and MOB3C) have been identified (1). Among them, MOB1A/B may interact directly with NDR1/2 and LATS1/2 and enhance their activity via the Hippo signaling pathway (1,2). By contrast, MOB2 interacts specifically with NDR1/2 kinases, but not with LATS1/2 kinases in mammalian cells (4–6). Specifically, MOB2 and MOB1 may compete for binding with the same NDR1/2 N-terminal regulatory domain, where MOB1 binds to NDR1/2 to promote the kinase activity of NDR1/2 and MOB2 interacts with NDR1/2 to interfere with the activity of NDR1/2 (4–6). Although MOB2 has been potentially linked to cell cycle progression and the DNA damage response in the context of NDR kinase signaling (1,4,7), the biological role of MOB2 has not yet been fully clarified.
An inhibitory effect of MOB2 on the migration and invasion of human hepatocellular carcinoma (HCC) cell lines SMMC-7721 and HepG2 has been previously described (8). However, the underlying molecular mechanism remains unclarified. In the present study, the effects of MOB2 on the activation of NDR/LATS kinases and the molecular mechanism through which MOB2 regulates LATS/yes-associated protein (YAP) activation were investigated.
Materials and methods
Cell lines and culture conditions
Human HCC cell line SMMC-7721 and human 293T cells, purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China), were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 µg/ml streptomycin and 100 U/ml penicillin, and maintained in a humidified incubator with 5% CO2 at 37°C.
Construction and lentiviral infection
The lentiviral vectors were prepared, and the lentiviruses encoding MOB2 (LV-MOB2) and control lentiviruses (LV-C) were generated and purified. Viral titers were determined by the Shanghai GeneChem Co., Ltd. (Shanghai, China). Following lentiviral infection, 1.0 µg/ml puromycin (cat. no. sc-205821; Santa Cruz Biotechnology, Inc., Dalla, TX, USA) was subsequently used to select stably transduced cell lines for two weeks. The cell lines that express a stable expression of control or MOB2 were established and screened by western blotting as previously described (8).
For clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein 9 (Cas9)-mediated MOB2 gene knockout, the single-guide RNA (sgRNA) targeting MOB2 was generated using the online CRISPR Design Tool (http://crispr.mit.edu/), and the sgRNA-MOB2 sequence is 5′-AGAAGCCCGCTGCGGAGGAG-3′. The lentiCRISPRv2 vector (Addgene, Inc., Cambridge, MA, USA) harboring a puromycin resistance cassette was digested using BsmBI and ligated using annealing oligonucleotides (forward, 5′-CACCGAGAAGCCCGCTGCGGAGGAG-3′ and reverse, 5′-AAACCTCCTCCGCAGCGGGCTTCTC-3′). Once the sequence was verified by sequencing, the constructs were transfected into 293T cells, which were grown to 70–80% confluence in a 10 cm dish, using EndoFectin Lenti reagent (GeneCopoeia, Inc., Rockville, MD, USA) together with the lentiviral packaging vectors pSPAX2 and pCMV-VSV-G (all from Addgene, Inc.). After transfection for 48 h, the viral particles were harvested and purified, and 1.5×106 SMMC-7721 cells were seeded in a 10 cm dish were infected with the indicated lentiviruses in the presence of polybrene (5 µg/ml; Shanghai GeneChem Co., Ltd.) for 14 h at 37°C. The infected SMMC-7721 cells were selected using puromycin 6 days following successful lentiviral transduction, followed by monoclonalization. The knockout of MOB2 expression was screened using western blotting. To construct the vector that express short hairpin RNA (shRNA) against human yes-associated protein (YAP) (shYAP), the primers: Forward, 5′-GATCCGCTGGTCAGAGATACTTCTTAATTCAAGAGATTAAGAAGTATCTCTGACCAGCTTTTTTA-3′ and reverse, 5′-CGCGTAAAAAAGCTGGTCAGAGATACTTCTTAATCTCTTGAATTAAGAAGTATCTCTGACCAGCG-3′ were synthesized and annealed, and followed by cloning into the BamH I and MluI sites of the pLent-U6-GFP-Puro vector (ViGene Biosciences Inc., Rockville, MD, USA), and the non-silencing control shRNA vector (shNC) was also generated. The MOB2 knockout SMMC-7721 cells were transfected with shYAP or with shNC for 72 h using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The knockdown of YAP expression was screened by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blotting.
Wound-healing assay
In total, 5.0×105 of the SMMC-7721 cells that overexpressed MOB2 (LV-MOB2), MOB2-knocked out cells (LV-sgMOB2) and the corresponding vector controls (LV-C and LV-sgC) were seeded onto 6-well culture plates and serum-starved overnight at 37°C. The cell monolayers were wounded by scratching with a sterile 200 µl plastic pipette tip and gently washed three times with phosphate-buffered saline (PBS) and captured under a phase-contrast microscope at ×100 magnification and marked as 0 h. The cells were further cultured with DMEM supplemented with 1% FBS at 37°C for 48 h, and wound closure was observed and captured under an inverted microscope at a magnification of ×100. The relative migration of cells was calculated. All experiments were performed in triplicate and repeated three times.
Transwell assay
Transwell migration and invasion assays were performed using Boyden chambers (diameter, 6.5 mm; pore size, 8.0 µm; Corning Incorporated, Corning, NY, USA) as described previously (8). The migrated or invaded cells on the lower surface of the inserts were fixed with methanol for 15 min at room temperature, stained with 0.1% crystal violet for 20 min at room temperature and counted from six random fields using a phase-contrast microscope at a magnification of ×100 per insert from triplicate wells. A total of three separate experiments were performed.
RNA extraction and RT-qPCR
Total RNA was isolated and purified from the SMMC-7721 cells that overexpressed MOB2 (LV-MOB2), MOB2-knocked out cells (LV-sgMOB2) and the corresponding vector controls (LV-C and LV-sgC) using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was obtained using the HiScript First Strand cDNA Synthesis kit (Vazyme, Piscataway, NJ, USA) according to the manufacturer's protocol. qPCR was performed using the SYBR Green qPCR system (Takara Biotechnology, Co., Ltd., Dalian, China) according to the manufacturer's protocol, and the qPCR thermocycling conditions were 95°C for 5 min followed by 40 cycles of 95°C for 10 sec and 60°C for 34 sec. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as an internal control. The primer sequences were as follows: Connective tissue growth factor (CTGF) forward, 5′-AGGAGTGGGTGTGTGACGA-3′ and reverse, 5′-CCAGGCAGTTGGCTCTAATC-3′; cysteine-rich angiogenic inducer 61 (CYR61) forward, 5′-AGCCTCGCATCCTATACAACC-3′ and reverse, 5′-TTCTTTCACAAGGCGGCACTC-3′; YAP forward, 5′-CTCGAACCCCAGATGACTTC-3′ and reverse, 5′-CCAGGAATGGCTTCAAGGTA-3′; and GAPDH forward, 5′-GCACCGTCAAGGCTGAGAAC-3′ and reverse, 5′-TGGTGAAGACGCCAGTGGA-3′. The relative expression of the indicated mRNAs (normalized to GAPDH expression) was calculated using the 2−ΔΔCq method as described (9). All experiments were performed in triplicate and repeated three times.
Western blot analysis
Protein extraction and subsequent western blotting were performed following standard methods as described previously (8). In brief, the cells were lysed using RIPA buffer (Beyotime Institute of Biotechnology, Haimen, China) supplemented with protease inhibitors for 30 min at 4°C, and proteins were quantified using the Bradford method, and approximately 40 µg cellular proteins were separated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA). The membranes were blocked with 5% non-fat dried milk in tris-buffered saline with Tween-20 for 2 h at room temperature, and incubated with primary antibodies at 4°C overnight followed by incubation for 2 h at room temperature with horseradish peroxidase (HRP)-conjugated secondary antibodies. The bands were detected using the Pierce ECL Plus western blotting substrate (Thermo Fisher Scientific, Inc.). GAPDH served as the loading control, and quantification of the western blotting results was performed using ImageJ 2× software (National Institutes of Health, Bethesda, MD, USA). A total of three independent experiments were performed. The antibodies used were as follows: Rabbit polyclonal anti-MOB2 (cat. no. SAB1301138; 1:500; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany); rabbit polyclonal anti-NDR1/2 (cat. no. sc-271703; 1:200; Santa Cruz Biotechnology, Inc., Dalla, TX, USA); rabbit monoclonal anti-YAP (cat. no. 14074; 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA), rabbit monoclonal anti-pS127YAP (cat. no. 13008; 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA), rabbit monoclonal anti-pS397YAP (cat. no. 13619; 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA), rabbit monoclonal anti-LATS1 (cat. no. 3477; 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA), rabbit monoclonal anti-pY1079LATS1 (cat. no. 8654; 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA), rabbit monoclonal anti-pS909 LATS1 (cat. no. 9157; 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA), rabbit monoclonal anti-MOB1 (cat. no. 13730; 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA), rabbit monoclonal anti-pY35 MOB1 (cat. no. 8699; 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA), HRP-conjugated anti-mouse IgG and HRP-linked anti-rabbit IgG (cat. no. 7076 and 7074, respectively; 1:2,000; Cell Signaling Technology, Inc., Danvers, MA, USA); mouse monoclonal anti-GAPDH (cat. no. KC-5G4; 1:1,000; KangChen Bio-tech Co., Ltd., Shanghai, China). The rabbit polyclonal antibody against NDR1/2 (pThr444/442) was generated as described previously (10,11). The peptides (KDWVFINYT(PO4)YKRFEG) were synthesized and conjugated to keyhole limpet hemocyanin in the laboratory. The rabbit injections and bleed collection were performed by DGpeptidesCo., Ltd (Hangzhou, China), and the antisera were purified and extensively characterized in the laboratory.
Immunoprecipitation
Immunoprecipitation was performed using the Pierce Classic IP kit (cat. no. 26146; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Briefly, the cells were washed with PBS three times and lysed at 4°C for 10 min in IP lysis/wash buffer (pH 7.4) containing 0.025M Tris, 0.15M NaCl, 0.001M EDTA, 1% NP-40 and 5% glycerol, and incubated for 20 min at 4°C. The samples were mixed periodically. Subsequent to centrifugation at 13,000 × g and 4°C for 15 min to pellet the cell debris, the protein concentration of the supernatant was determined using the Bradford method, and equivalent quantities of protein (800 µg) were pre-cleared using control agarose resin (component of the Pierce Classic IP kit). The pre-cleared cell lysates were incubated with the rabbit polyclonal anti-NDR1/2 (3 µg per sample) or rabbit monoclonal anti-LATS1 (dilution, 1:100) antibodies overnight at 4°C to form the immune complex followed by incubation at 4°C with 20 µl Protein A/G Plus Agarose beads (component of the Pierce Classic IP kit) for 1 h. Following four washes with the IP lysis/wash buffer, the beads were washed once with conditioning buffer and heated at 100°C for 10 min in Lane Marker Sample buffer (both buffers are components of the Pierce Classic IP kit) supplemented with dithiotheritol to a final concentration of 20 mM. The samples were then subjected to SDS-PAGE and western blotting as aforementioned.
Statistical analysis
All data were expressed as the mean ± standard deviation. Statistical analysis was performed using SPSS 19.0 software (SPSS, Inc., Chicago, IL, USA). The analyses were performed using unpaired Student's t-test or a one-way analysis of variance followed by Tukey's post-hoc test for multiple comparisons. P<0.05 was considered to indicate a statistically significant difference.
Results
Knockout of MOB2 positively regulates the migration and invasion of SMMC-7721 cells
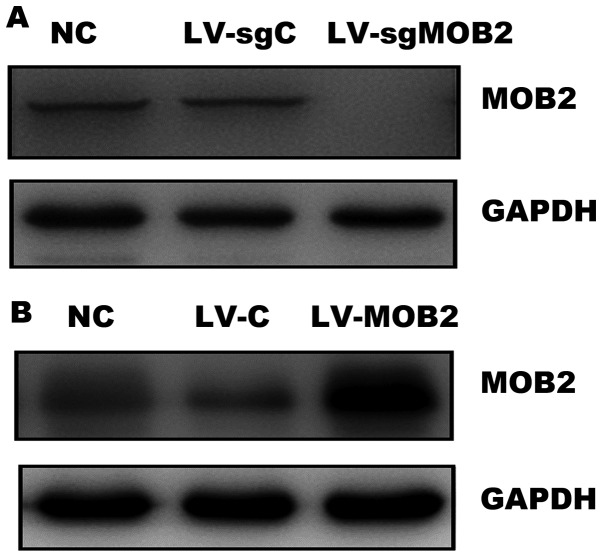
The inhibitory effects of MOB2 on the motility of the HCC cell lines SMMC-7721 and HepG2 were previously reported, where migration and invasion were markedly suppressed in MOB2-overexpressing cells and cell motility was decreased in MOB2-knocked down cells (8). In the present study, the aim was to further confirm this role of MOB2 in the regulation of migration and invasion of HCC cells. MOB2-knockout SMMC-7721 cells were generated by infecting the cells with lentiCRISPRv2 viruses that encode the Cas9 nuclease and single-guide RNA (sgRNA) that targets MOB2. Gene knockout was confirmed by western blotting. As presented in Fig. 1A, the expression of MOB2 was undetectable in cells that stably express MOB2-targeted sgRNA (LV-sgMOB2) when compared with the blank vector-transduced cells (LV-sgC) and the normal control cells (NC). In addition, the efficiency of MOB2 overexpression was also confirmed in LV-MOB2-transduced SMMC-772 cells by western blotting (Fig. 1B). Therefore, SMMC-7721 cells that stably overexpress MOB2, SMMC-7721 MOB2 knockout cells and their corresponding blank vector-transduced cells (LV-sgC and LV-C) were successfully established in order to perform subsequent experiments.
Figure 1.
Knockout and overexpression of MOB2 in lentiviral-transduced SMMC-7721 cells. SMMC-7721 cells that stably express MOB2, MOB2 sgRNA or the corresponding vector control were lysed. The (A) knockout and (B) overexpression of MOB2 were confirmed using western blotting (n=3). MOB2, monopolar spindle-one-binder protein 2; sgRNA, single-guide RNA; LV-C, control lentivirus; LV-sgMOB2, MOB2-knockout cells; LV-MOB2, MOB2-overexpressing cells; NC, normal control.
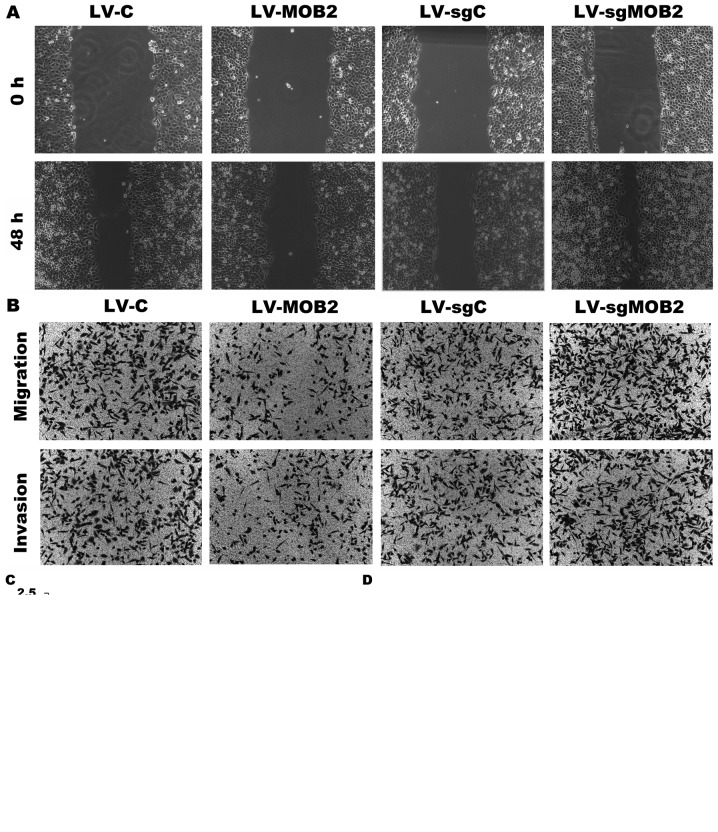
Subsequently, wound-healing and Transwell assays were performed in order to evaluate the effect of the knockout or overexpression of MOB2 on the migration and invasion of SMMC-7721 cells. The results of the wound-healing assay revealed that MOB2 knockout cells were able to repair the wound areas significantly faster compared with LV-sgC-transduced cells while MOB2-overexpressing SMMC-7721 cells exhibited a decreased wound-healing capacity compared with LV-C-transduced cells (P<0.05; Fig. 2). The effect of MOB2 on the migratory and invasive capacities of SMMC-7721 cells was also assessed using Transwell assays. Compared with LV-sgC, the knockout of MOB2 significantly increased the number of cells that migrated and invaded, whereas opposite results were observed in MOB2-overexpressing cells when compared with LV-C (Fig. 2). Therefore, these results clearly demonstrate that silencing MOB2 significantly promoted cell migration and invasion. By contrast, the overexpression of MOB2 significantly decreased the motility of SMMC-7721 cells, which is consistent with the results of a previous study (8), thus providing further confirmation that MOB2 regulates the migratory and invasive abilities of HCC cells.
Figure 2.
Effect of MOB2 knockout or overexpression on the migration and invasion of SMMC-7721 cells. (A) Wound-healing assay was performed to assess the migration of SMMC7721 cells that overexpress MOB2 (LV-MOB2), MOB2-knocked out cells (LV-sgMOB2) and the corresponding vector controls (LV-C and LV-sgC, respectively) (magnification, ×100). (B) Representative Transwell cell migration and invasion assays in SMMC-7721 cells that overexpress MOB2, MOB2-knocked down cells (LV-sgMOB2) and the corresponding vector controls (LV-C and LV-sgC, respectively) (magnification, ×100) (C) Quantification of relative wound healing. (D) Quantification of relative numbers of migrated and invaded cells. All experiments were performed independently three times, and the data were presented as the mean ± standard deviation. *P<0.05 vs. the corresponding blank vector control (LV-C or LV-sgC). LV-C, control lentivirus; LV-sgMOB2, MOB2-knockout cells; LV-MOB2, MOB2-overexpressing cells; MOB2, monopolar spindle-one-binder protein 2; sgRNA, single-guide RNA.
MOB2-induced YAP phosphorylation is independent of NDR1/2 activation
Accumulating evidence has suggested that MOB2 may perform its functions by competing with MOB1 for interaction with NDR1/2, where the binding of MOB2 to NDR1/2 blocks the activity of NDR kinase, and the binding of MOB1 to NDR1/2 leads to increased activation of NDR1/2 kinases (1–4). NDR1/2 and LATS1/2 function as YAP kinases and may be considered as members of the Hippo core cassette (4,12,13). The phosphorylation of YAP leads to decreased nuclear YAP transcriptional activity, which serves critical roles in cell motility (14–17).
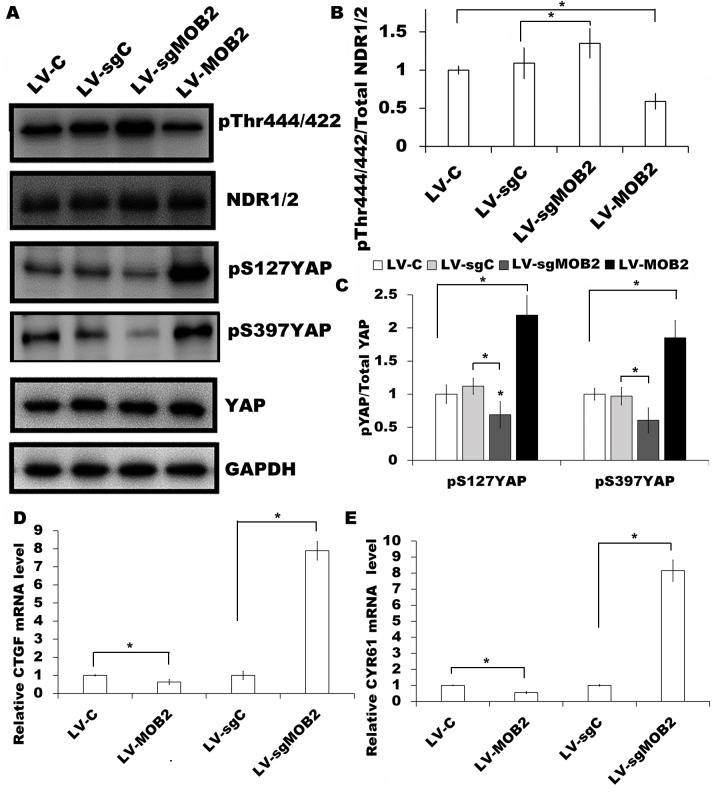
Therefore, in the present study, the expression of NDR1/2 and YAP in SMMC-7721 cells that stably overexpress MOB2, MOB2 knockout-SMMC-7721 cells and their corresponding vector control cells were examined. It was revealed that there was a significant decrease in the level of Thr444/442 phosphorylation (pT444/442) at the hydrophobic motif of NDR1/2 in MOB2-overexpressing cells compared with their corresponding vector control cells (P<0.05; Fig. 3A and B). There was also a significant increase in the level of YAP phosphorylation at S127 (pS127YAP) and S397 (pS397YAP) sites in MOB2-overexpressing cells compared with their corresponding control (P<0.05; Fig. 3C). By contrast, the knockout of MOB2 resulted in the significant upregulation of pT444/442 proteins and the downregulation of pS127YAP and pS397YAP compared with the corresponding vector controls (P<0.05; Fig. 3A-C). There were no significant differences observed between the empty vector-infected cells and the NC cells (data not shown). It is widely accepted that the phosphorylation of YAP results in its cytoplasmic retention and reduced nuclear YAP and transcriptional activity of TEA domain family (14–16). Therefore, RT-qPCR was performed in order to analyze the expression of CTGF and CYR61, two well-characterized YAP target genes (18,19), in SMMC-7721 cells MOB2 overexpressing-cells, MOB2 knockout cells and their corresponding vector control cells. It was revealed that the overexpression of MOB2 significantly decreased the expression of CTGF and CYR61, while the knockout of MOB2 significantly promoted the transcription of CTGF and CYR61 compared with the levels in their corresponding vector control cells (P<0.05; Fig. 3D and E).
Figure 3.
Effect of MOB2 knockout and overexpression on the expression of NDR1/2 and YAP in SMMC-7721 cells. (A) Protein expression of NDR1/2 pT444/442, NDR1/2, YAP pS127, YAP pS397 YAP and YAP were analyzed in SMMC7721 cells that overexpress MOB2 (LV-MOB2), MOB2-knocked out cells (LV-sgMOB2) and the corresponding vector controls (LV-C and LV-sgC, respectively) by western blotting. GAPDH was used as a loading control (n=3). Densitometric analysis of the fold expression levels of (B) pT444/442 of NDR1/2 compared with total NDR1/2 and (C) pS127YAP and pS397YAP compared with total YAP. Data represents the mean ± SD of three independent experiments. *P<0.05 vs. the corresponding blank vector control (LV-C or LV-sgC). mRNA levels of (D) CTGF and (E) CYR61, two well-characterized YAP target genes, were analyzed from the indicated cells using reverse transcription-quantitative polymerase chain reaction and normalized to GAPDH expression. Data represents the mean ± SD (n=5). *P<0.05 vs. the corresponding blank vector control (LV-C or LV-sgC). MOB2, monopolar spindle-one-binder protein 2; NDR, nuclear-Dbf2-related kinase; YAP, yes-associated protein; p, phosphorylated; CTGF, connective tissue growth factor; CYR61, cysteine rich angiogenic inducer 61; LV-C, control lentivirus; LV-sgMOB2, MOB2-knockout cells; LV-MOB2, MOB2-overexpressing cells; LV-sgC, sgRNA control cells; sgRNA, single-guide RNA; SD, standard deviation.
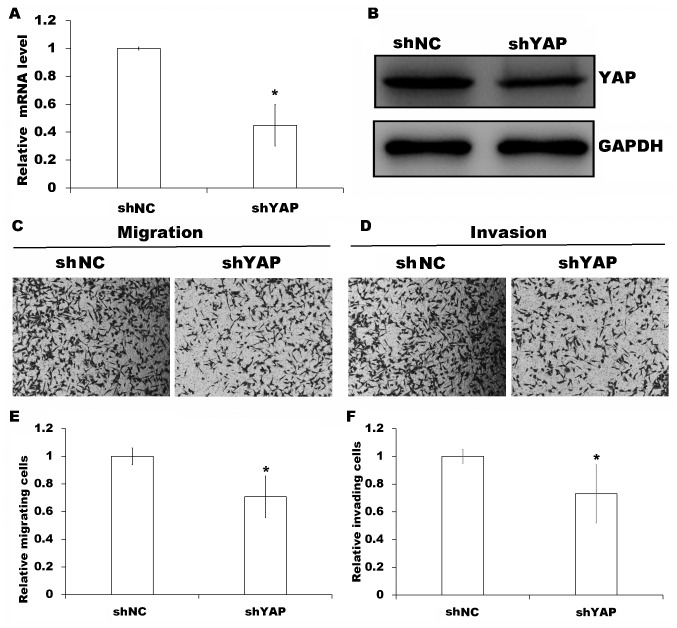
To evaluate the requirement of YAP for MOB2-regulated cell motility, YAP was knocked down in MOB2 knockout cells as verified by RT-qPCR and western blotting (P<0.05; Fig. 4A and B) followed by Transwell cell migration and invasion assays. Compared with the MOB2-silenced cells that were transfected with non-targeting shRNA vector (shNC), the silencing of YAP in MOB2 knockout cells significantly decreased cell migration and invasion (P<0.05; Fig. 4C-F), suggesting that YAP is involved in the modulation of the motility of SMMC-7721 cells via MOB2. Although NDR1/2 kinases have been reported to function as the upstream kinases of YAP and directly phosphorylate YAP (12), the results of the present study suggest that MOB2-mediated regulation of YAP phosphorylation appears to be independent of NDR1/2 activation.
Figure 4.
Effects of YAP knockdown on MOB2-regulated cell motility in MOB2-knockout SMMC-7721 cells. (A) Reverse transcription-quantitative polymerase chain reaction and (B) western blotting of YAP expression in MOB2 knockout SMMC-7721 cells following transfection with shYAP or shNC. Representative Transwell (C) migration (magnification, ×100) and (D) invasion assays (magnification, ×100). Quantification of results from Transwell (E) migration and (F) invasion assays. Data are presented as the mean ± standard deviation of three independent experiments. *P<0.05 vs. shNC. shRNA, small-hairpin RNA; MOB2, monopolar spindle-one-binder protein 2; shNC, non-targeting shRNA vector; shYAP, YAP-targeting shRNA; YAP, yes-associated protein; shRNA, short hairpin RNA.
MOB2-induced YAP activation may be mediated via activating MOB1-LATS signaling
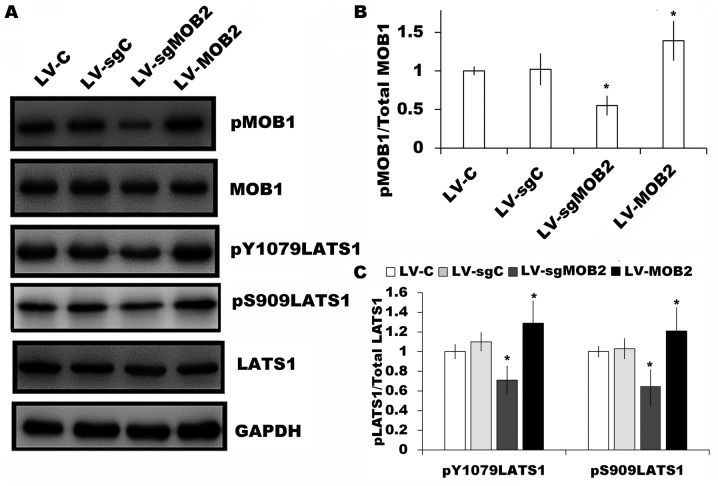
A key function of the Hippo tumor suppressor pathway is to inhibit the activity of YAP transcriptional co-activators by MOB1-LATS signaling through LATS-mediated direct phosphorylation of YAP (20,21). Therefore, the present study aimed to investigate the effect of MOB2 on the expression of MOB1 and LATS1 in SMMC-7721 cells. It was revealed that the overexpression of MOB2 significantly upregulated the levels of MOB1 phosphorylation (pMOB1) and LATS1 phosphorylation at Y1079 (pY1079LATS1) and S909 (pS909LATS1) sites compared with their corresponding controls (Fig. 5A-C). By contrast, the knockout of MOB2 significantly decreased the levels of pMOB1 and pLATS1 (pY1079LATS1 and pS909LATS1) compared with the corresponding vector control (P<0.05; Fig. 5A-C). Together, these results indicated that MOB2-induced YAP activation may be mediated through activating MOB1-LATS signaling.
Figure 5.
Effect of MOB2 knockout and overexpression on the expression of MOB1 and LATS1 in SMMC-7721 cells. (A) The levels of pMOB1, MOB1, pY1079LATS1, pS909LATS1 and LATS1 protein expression were determined in SMMC-7721 cells that stably express MOB2, MOB2-knocked out cells and the corresponding vector controls by western blotting. GAPDH served as the loading control (n=3). The fold expression of (B) pMOB1 relative to total MOB1, and (C) pY1079LATS1 and pS909LATS1 relative to total LATS1 as determined by densitometric analysis. Data are presented as the mean ± standard deviation of three independent experiments. *P<0.05 vs. the corresponding vector control. MOB1, monopolar spindle-one-binder protein 1; MOB2, monopolar spindle-one-binder protein 2; LATS1, large tumor suppressor kinase 1; p-, phosphorylated; LV-C, control lentivirus; LV-sgMOB2, MOB2-knockout cells; LV-MOB2, MOB2-overexpressing cells; sgRNA, single-guide RNA.
MOB2 regulates the alternative interaction of MOB1 with LATS and NDR kinases
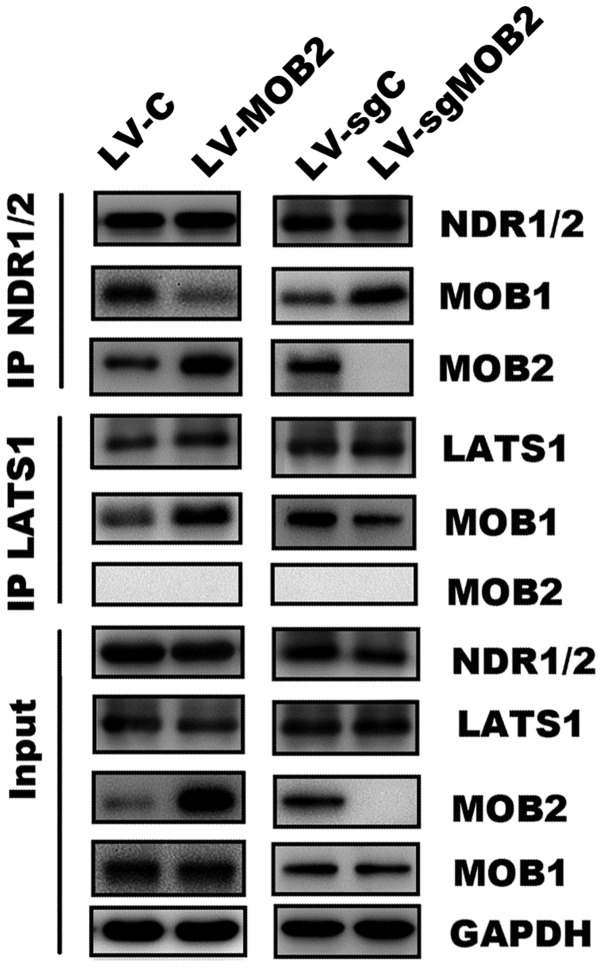
Mammalian NDR kinases are the only reported binding partners of MOB2, whereas MOB1 may associate with NDR and LATS kinases (1,5). To further dissect the molecular mechanism of MOB2 in LATS/YAP activation, immunoprecipitation assay was performed followed by western blotting. As hypothesized, it was confirmed that MOB2 may interact with NDR but not LATS, while MOB1 may bind to NDR1/2 and LATS1 (Fig. 6). It was revealed that MOB1 was enriched in LATS1 precipitates but reduced in NDR1/2 precipitates in MOB2-overexpressing cells. Furthermore, the knockout of MOB2 resulted in accumulation of MOB1 in NDR1/2 precipitates and reduction of MOB1 in LATS1 precipitates (Fig. 6). These observations demonstrate a role of MOB2 in regulating the alternative interaction of MOB1 with LATS1 and NDR1/2.
Figure 6.
MOB2 regulates the alternative interaction of MOB1 with LATS and NDR kinases. Protein expression of MOB1 and MOB2 following NDR1/2 or LATS1 immunoprecipitation in SMMC-7721 cells that stably express MOB2, MOB2 knockout cells and the corresponding vector control. A total of three independent experiments were performed. IP, immunoprecipitation; LV-C, control lentivirus; LV-sgMOB2, MOB2-knockout cells; LV-MOB2, MOB2-overexpressing cells; MOB1, monopolar spindle-one-binder protein 1; MOB2, monopolar spindle-one-binder protein 2; LATS1, large tumor suppressor kinase 1; NDR, nuclear-Dbf2-related kinase.
Discussion
In the present study, it was demonstrated that the knockout of MOB2 by CRISPR/Cas9 promoted the cell migration and invasion, induced NDR1/2 phosphorylation and decreased YAP phosphorylation in the SMMC-7721 HCC cell line. By contrast, the overexpression of MOB2 resulted in the opposite effect. It was additionally demonstrated that MOB2 exerts its regulation on cell motility at least in part via regulating the phosphorylation of YAP and the activity of YAP in the Hippo signaling pathway. The results of the present study further indicated a role of MOB2 in regulating the alternative interaction of MOB1 with LATS1 and NDR1/2, which result in increased phosphorylation of LATS1 and MOB1 and consequently the inactivation of YAP and regulation of cell motility. These results supported the hypothesis that MOB2 serves a positive role in the activation of LATS/YAP by activating the Hippo signaling pathway.
Previous studies have linked endogenous MOB2 to cell survival, cell cycle progression and DNA damage responses in the context of NDR kinase signaling (4,7). MOB2 directly phosphorylates YAP, and the inactivation of YAP by cytoplasmic retention is linked with the regulation of cell motility and proliferation (12,16–18,22–24), thereby establishing an association between NDR1/2 kinases with the Hippo signaling pathway on a cellular level. A previous study by the present authors has demonstrated that MOB2 serves an inhibitory role in the motility of the HCC cell lines SMMC-7721 and HepG2 (8). However, the details of the roles and underlying mechanisms involved in these processes remain unclarified. Hence, in the present study, the effect of MOB2 knockout by CRISPR/Cas9 and MOB2 overexpression on the migration and invasion of SMMC-7721 cells was investigated by wound-healing and Transwell assays. The data are consistent with the results of the previous study, where the overexpression of MOB2 decreased cell migration and invasion. However, the knockout of MOB2 decreased cell motility and markedly promoted the migration and invasion of SMMC-7721 cells. These results further demonstrated the role of MOB2 in regulating the migration and invasion of HCC cells.
NDR1/2 kinases and not LATS1/2 were reported to be the binding partners of MOB2. NDR1/2 kinases may function as the upstream kinases of YAP, which is the main downstream effector of the Hippo signaling pathway (1,4–7,12,18). The present study investigated whether MOB2 regulates the activation of NDR1/2 kinases and subsequently modulates the phosphorylation of YAP. It was revealed that phosphorylation at Thr444/442 of NDR1/2 was decreased in MOB2-overexpressing SMMC-7721 cells but increased in MOB2 knockout cells, suggesting that MOB2 may negatively regulate the activity of NDR1/2. Notably, it was additionally revealed that the overexpression of MOB2 resulted in the upregulation of YAP phosphorylation, while the knockout of MOB2 resulted in the downregulation of YAP phosphorylation, which is inconsistent with the function of NDR1/2 as YAP kinases (12). In addition, the functional significance of MOB2 expression on YAP activity was further evaluated. As hypothesized, there was a strong negative association between MOB2 expression and the expression of two YAP target genes, CYR61 and CTGF (P<0.05). Furthermore, in order to evaluate whether YAP is involved in MOB2-regulated cell motility, YAP was silenced in the MOB2 knockout cells. Compared with the MOB2-silenced cells that were transfected with non-targeting shRNA vector (shNC), the silencing of YAP in MOB2 knockout cells significantly decreased cell migration and invasion. Taken together, these results suggest that MOB2-mediated regulation of YAP phosphorylation appears to be independent of NDR1/2 activation.
Conventionally, the activated Hippo signaling pathway inhibits the transcriptional co-activator functions of YAP through the MOB1/LATS-mediated phosphorylation of YAP (1,25,26). Therefore, whether MOB2 expression exerts an effect on the expression of MOB1 and LATS in SMMC-7721 cells was examined. It was revealed that the overexpression of MOB2 increased the expression levels of pMOB1 and pLATS1. By contrast, the knockout of MOB2 expression decreased the expression levels of pMOB1 and pLATS1, thereby inhibiting YAP phosphorylation as aforementioned.
LATS kinases are activated by MOB1. MOB2 exerts its function upstream of NDR1/2 and functions as an inhibitor of MOB1-mediated NDR1/2 activation (27). An immunoprecipitation assay was performed in order to investigate whether the immunoprecipitation of MOB1 by NDR1/2 and LATS1 was affected by the overexpression or knockout of MOB2. It was revealed that the overexpression of MOB2 resulted in a notable reduction in the quantity of MOB1 that was immunoprecipitated with NDR1/2 but a substantial increase in the quantity of MOB1 that was immunoprecipitated with LATS1. By contrast, the knockout of MOB2 failed to accumulate the interactions of MOB1 with LATS1/2, and a more efficient interaction was displayed between MOB1 and NDR1/2, although further research is required. Taken together, these results demonstrate that MOB2 may compete with MOB1 for interaction with NDR1/2 and partially displace MOB1 from NDR1/2, which promotes the interaction of MOB1 with LATS1/2 and activates the Hippo signaling pathway.
In conclusion, the findings of the present study demonstrated that MOB2 regulates the alternative interaction of MOB1 with NDR1/2 and LATS1, which results in increased phosphorylation/activity of LATS1 and MOB1. This leads to the inactivation of YAP and consequently the inhibition of cell motility. Further study of the underlying mechanism as to how MOB2 knockout coordinates the Hippo signaling could be validated. However, these findings may reinforce MOB2 as a potential diagnostic or therapeutic target for HCC.
Acknowledgements
The present study was supported by the National Nature Science Foundation of China (grant no. 81672336), the Training Program of Innovation and Entrepreneurship for College Students in Jiangsu (grant nos. 201511117045Z and 201611117041Z) and the Open Research Fund Program of the Jiangsu Key Laboratory of Integrated Traditional Chinese and Western Medicine for Prevention and Treatment of Senile Diseases.
References
- 1.Hergovich A. MOB control: Reviewing a conserved family of kinase regulators. Cell Signal. 2011;23:1433–1440. doi: 10.1016/j.cellsig.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hergovich A. Regulation and functions of mammalian LATS/NDR kinases: Looking beyond canonical Hippo signalling. Cell Biosci. 2013;3:32. doi: 10.1186/2045-3701-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hergovich A. The roles of NDR protein kinases in Hippo signalling. Genes (Basel) 2016;7:E21. doi: 10.3390/genes7050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gundogdu R, Hergovich A. The possible crosstalk of MOB2 with NDR1/2 kinases in cell cycle and DNA damage signaling. J Cell Signal. 2016;1:125. doi: 10.4172/2576-1471.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohler RS, Schmitz D, Cornils H, Hemmings BA, Hergovich A. Differential NDR/LATS interactions with the human MOB family reveal a negative role for human MOB2 in the regulation of human NDR kinases. Mol Cell Biol. 2010;30:4507–4520. doi: 10.1128/MCB.00150-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bothos J, Tuttle RL, Ottey M, Luca FC, Halazonetis TD. Human LATS1 is a mitotic exit network kinase. Cancer Res. 2005;65:6568–6575. doi: 10.1158/0008-5472.CAN-05-0862. [DOI] [PubMed] [Google Scholar]
- 7.Gomez V, Gundogdu R, Gomez M, Hoa L, Panchal N, O'Driscoll M, Hergovich A. Regulation of DNA damage responses and cell cycle progression by hMOB2. Cell Signal. 2015;27:326–339. doi: 10.1016/j.cellsig.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu W, Zhang X, Qin H, Peng W, Xue Q, Lv H, Zhang H, Qiu Y, Cheng H, Zhang Y, et al. Modulation of tumor cell migration, invasion and cell-matrix adhesion by human monopolar spindle-one-binder 2. Oncol Rep. 2015;33:2495–2503. doi: 10.3892/or.2015.3855. [DOI] [PubMed] [Google Scholar]
- 9.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 10.Tamaskovic R, Bichsel SJ, Rogniaux H, Stegert MR, Hemmings BA. Mechanism of Ca2+-mediated regulation of NDR protein kinase through autophosphorylation and phosphorylation by an upstream kinase. J Biol Chem. 2003;278:6710–6718. doi: 10.1074/jbc.M210590200. [DOI] [PubMed] [Google Scholar]
- 11.Vichalkovski A, Gresko E, Cornils H, Hergovich A, Schmitz D, Hemmings BA. NDR kinase is activated by RASSF1A/MST1 in response to Fas receptor stimulation and promotes apoptosis. Curr Biol. 2008;18:1889–1895. doi: 10.1016/j.cub.2008.10.060. [DOI] [PubMed] [Google Scholar]
- 12.Zhang L, Tang F, Terracciano L, Hynx D, Kohler R, Bichet S, Hess D, Cron P, Hemmings BA, Hergovich A, Schmitz-Rohmer D. NDR functions as a physiological YAP1 kinase in the intestinal epithelium. Curr Biol. 2015;25:296–305. doi: 10.1016/j.cub.2014.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang L, Xue G, Grzmil M, Yang Z, Hergovich A, Hollaender GA, Stein JV, Hemmings BA, Matthias P. The kinases NDR1/2 act downstream of the Hippo homolog MST1 to mediate both egress ofthymocytes from the thymus and lymphocyte motility. Sci Signal. 2015;8:ra100. doi: 10.1126/scisignal.aab2425. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, George J, Deb S, Degoutin JL, Takano EA, Fox SB, AOCS Study group. Bowtell DD, Harvey KF. The Hippo pathway transcriptional co-activator, YAP, is an ovarian cancer oncogene. Oncogene. 2011;30:2810–2822. doi: 10.1038/onc.2011.8. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Li J, Li P, Wang Y, Liang Z, Jiang Y, Li J, Feng C, Wang R, Chen H, et al. Loss of DLG5 promotes breast cancer malignancy by inhibiting the Hippo signaling pathway. Sci Rep. 2017;7:42125. doi: 10.1038/srep42125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu FX, Zhang Y, Park HW, Jewell JL, Chen Q, Deng Y, Pan D, Taylor SS, Lai ZC, Guan KL. Protein kinase A activates the Hippo pathway to modulate cell proliferation and differentiation. Genes Dev. 2013;27:1223–1232. doi: 10.1101/gad.219402.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang S, Zhang L, Purohit V, Shukla SK, Chen X, Yu F, Fu K, Chen Y, Solheim J, Singh PK, et al. Active YAP promotes pancreatic cancer cell motility, invasion and tumorigenesis in a mitotic phosphorylation-dependent manner through LPAR3. Oncotarget. 2015;6:36019–36031. doi: 10.18632/oncotarget.5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moroishi T, Park HW, Qin B, Chen Q, Meng Z, Plouffe SW, Taniguchi K, Yu FX, Karin M, Pan D, Guan KL. A YAP/TAZ-induced feedback mechanism regulates Hippo pathway homeostasis. Genes Dev. 2015;29:1271–1284. doi: 10.1101/gad.262816.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayashi H, Higashi T, Yokoyama N, Kaida T, Sakamoto K, Fukushima Y, Ishimoto T, Kuroki H, Nitta H, Hashimoto D, et al. An imbalance in TAZ and YAP expression in hepatocellular carcinoma confers cancer stem cell-like behaviors contributing to disease progression. Cancer Res. 2015;75:4985–4997. doi: 10.1158/0008-5472.CAN-15-0291. [DOI] [PubMed] [Google Scholar]
- 20.Mo JS, Park HW, Guan KL. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 2014;15:642–656. doi: 10.15252/embr.201438638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mo JS, Yu FX, Gong R, Brown JH, Guan KL. Regulation of the Hippo-YAP pathway by protease-activated receptors (PARs) Genes Dev. 2012;26:2138–2143. doi: 10.1101/gad.197582.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 23.Haskins JW, Nguyen DX, Stern DF. Neuregulin 1-activated ERBB4 interacts with YAP to induce Hippo pathway target genes and promote cell migration. Sci Signal. 2014;7:ra116. doi: 10.1126/scisignal.2005770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huelter-Hassler D, Tomakidi P, Steinberg T, Jung BA. Orthodontic strain affects the Hippo-pathway effector YAP concomitant with proliferation in human periodontal ligament fibroblasts. Eur J Orthod. 2017;39:251–257. doi: 10.1093/ejo/cjx012. [DOI] [PubMed] [Google Scholar]
- 25.Oh H, Irvine KD. Yorkie: The final destination of Hippo signaling. Trends Cell Biol. 2010;20:410–417. doi: 10.1016/j.tcb.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vahid S, Thaper D, Gibson KF, Bishop JL, Zoubeidi A. Molecular chaperone Hsp27 regulates the Hippo tumor suppressor pathway in cancer. Sci Rep. 2016;6:31842. doi: 10.1038/srep31842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hergovich A, Schmitz D, Hemmings BA. The human tumour suppressor LATS1 is activated by human MOB1 at the membrane. Biochem Biophys Res Commun. 2006;345:50–58. doi: 10.1016/j.bbrc.2006.03.244. [DOI] [PubMed] [Google Scholar]