SUMMARY
Mesoderm development (MESD) is a 224 amino acid mouse protein that acts as a molecular chaperone for the low-density lipoprotein receptor (LDLR) family. Here, we provide evidence that the region 45–184 of MESD is essential and sufficient for this function and suggest a model for its mode of action. NMR studies reveal a β–α–β–β–α–β core domain with an α-helical N-terminal extension that interacts with the β sheet in a dynamic manner. As a result, the structural ensemble contains open (active) and closed (inactive) forms, allowing for regulation of chaperone activity through substrate binding. The mutant W61R, which is lethal in Drosophila, adopts only the open state. The receptor motif recognized by MESD was identified by in vitro-binding studies. Furthermore, in vivo functional evidence for the relevance of the identified contact sites in MESD is provided.
INTRODUCTION
The low-density lipoprotein receptor (LDLR) family (also known as LRPs) covers a wide range of physiologically important cellular functions. The founding member, LDLR, has mainly been characterized by its function in systemic cholesterol homeostasis. It rapidly binds cholesterol-rich LDL particles and triggers their internalization. Indeed, most family members are endocytic receptors, involved in the uptake of a wide variety of ligands. Besides this, several LDLR family members (including sorLA) are involved in the processing of the amyloid precursor protein (APP) (Andersen et al., 2005, 2006; Cam et al., 2005). In addition, members of the LDLR family are implicated in cellular-signaling events that control neuronal migration (Reelin binding to ApoER2 and VLDLR) (Sinagra et al., 2005), embryonic development (WNT proteins binding to LRP5 and LRP6) (He et al., 2004), vascular branching in the eye (Norrie binding to LRP5) (Xu et al., 2004), and endothelial cell proliferation (PDGF binding to LRP1) (Takayama et al., 2005). These diverse functions depend upon the presence of correctly folded and post-translationally modified receptors on the cell surface. The folding of proteins from the LDLR family is a difficult task, partly owed to the very high number of intramolecular disulfide bonds present in the complement-type repeat (CR) and epidermal growth factor (EGF) domains, as well as due to the very complex packing of six contiguous YWTD repeats into a six-bladed β-propeller structure functionally interacting with the C-terminal adjacent EGF domain (Jeon et al., 2001; Springer, 1998).
In mammalian cells the folding of this β-propeller/EGF motif in the endoplasmic reticulum (ER) is assisted by mesoderm development (MESD), the homolog of the Drosophila protein Boca (Culi and Mann, 2003; Culi et al., 2004; Hsieh et al., 2003). The physiological importance of MESD was indicated by the studies of Mesd-deficient mouse embryos. In this mouse model, lack of MESD results in disruption of embryonic polarity and lack of mesoderm differentiation (Culi and Mann, 2003; Hsieh et al., 2003). The embryonic defects were attributed to a lack of functional expression of LRP5 and LRP6, two members of the LDLR gene family that are indispensable for the canonical WNT-signaling pathway (Pinson et al., 2000; Tamai et al., 2000; Wehrli et al., 2000). In addition, MESD is essential in the visceral endoderm of the early postimplantation embryo for endocytosis and membrane localization of LRP2/Megalin (Lighthouse et al., 2010). Expression of these receptors on the cell surface is not only essential for proper embryonic development but also for adult functions. Thus, several additional roles have recently been described for LRP5/6 in adult mice and humans, e.g., in bone metabolism. ”Gain-of-function” mutations in LRP5 and/or LRP6 result in an increased bone density (Van Wesenbeeck et al., 2003; Zhang et al., 2004), and “Loss-of-function” mutations cause osteoporosis as well as hereditary eye disorders (Gong et al., 2001). Some of these mutations interfere with the interaction of LRP5 with WNT inhibitors rather than the binding of WNT proteins (Boyden et al., 2002; Mao et al., 2001, 2002). Some mutations also affect the binding of MESD to the receptor (Zhang et al., 2004; Carter et al., 2005), highlighting the importance of understanding receptor-ligand interactions at the molecular level.
The MESD core region corresponding to residues 104–177 (numbering according to the mouse homolog) forms a domain with a fold commonly found in nature (Köhler et al., 2006). However, the conserved area of MESD is not limited to this core domain and extends to the region between residue 43 and 103, which is essential for function (Köhler et al., 2006; Koduri and Blacklow, 2007). Despite the importance of MESD in, for example, LRP5 and LRP6 maturation, little is known about how this protein works as a molecular chaperone. Key to understanding MESD’s function is the identification of the interaction interface between MESD and the LDLR family members and development of a model for the MESD-assisted folding of the receptor motif consisting of a β-propeller and a C-terminal adjacent EGF domain.
Here, we present a structural model for MESD residues 45–184. According to this model, the central domain of MESD (residues 104–177) is extended N terminally by a large flexible loop region culminating in an N-terminal α helix. By investigating the internal dynamics by NMR, we demonstrated that the N-terminal helix loosely interacts with the β sheet of the core domain. By screening a peptide library containing small, unfolded receptor fragments, we identified a partly hydrophobic and partly electrostatic interaction interface between the unfolded receptors and MESD. Finally, in vivo studies of several MESD mutants provided evidence for the functional relevance of these hydrophobic and polar contacts. Based on these observations, we suggest a mechanism for the MESD-assisted folding of the β-propeller/EGF-domain motif.
RESULTS
An N-Terminal α Helix Extends from the MESD Core Domain
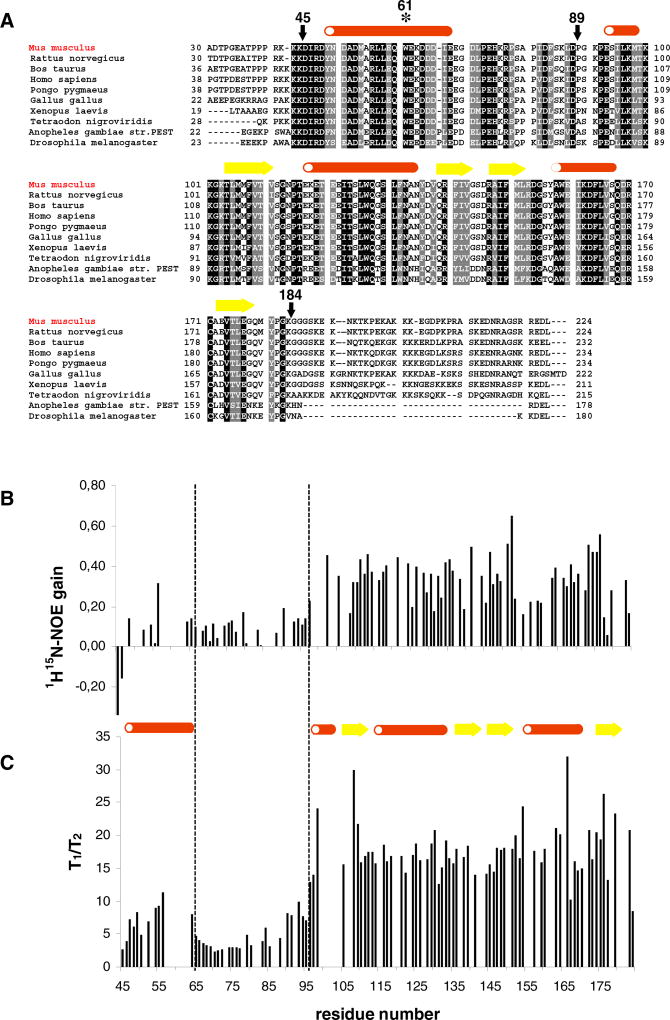
As described previously, MESD consists of a highly conserved core region corresponding to residues 43–184 (Figure 1A), with residues 104–177 forming a domain with a common type of fold but low sequence identity (Köhler et al., 2006). It is still unknown how the conserved residues K43–K103 interact with the core domain. Using 15N-HSQC-NMR spectroscopy of MESD truncations, we show that this region is comprised of residues with chemical shifts characteristic for α-helical secondary structures. In order to characterize the internal dynamics of MESD, we carried out measurements of the heteronuclear Nuclear Overhauser Effect (NOE) (Figure 1B) and amide nitrogen relaxation parameters (Figure 1C). These data were interpreted as evidence that residues 47–64 are relatively inflexible, corroborating a predicted α-helical structure. In contrast, residues 65–95 are highly flexible and are followed by the rigid core domain. The NOE contacts observed between residues 45 and 103 and the core domain could only be ambiguously assigned. Structure calculations based on these ambiguous distance restraints using CYANA (Guentert, 2004) did not converge with respect to an involvement of residues 45–103 in a globular structure (Figure 2A). However, a characteristic pattern of short-range contacts did support at least a temporary existence of an α-helical secondary structure among residues 49–68.
Figure 1. The Conserved Core Region of MESD Is Characterized by Strongly Changing Flexibilities.
(A) Sequence alignment of ten MESD homologs from mouse (NCBI-ID: Q9ERE7), rat (Q5U2R7), cow (Q3T0U1), human (Q14696), orangutan (Q5R6F1), chicken (Q5ZKK4), African clawed frog (Q4V7K5), puffer fish (Q4STS2), mosquito (Q7Q9R4), and Drosophila fly (Q9V4N7): black, identity; gray, similarity. The secondary structural elements are mapped to the mouse sequence; after every ten residues of the mouse MESD sequence, a space is included. Arrows mark the MESD truncations used for NMR structural studies. The asterisk shows the location of the W61R mutation. Interestingly, the homolog from chicken does not contain the characteristic C-terminal ER retention signal (REDL).
(B) The signal gain due to the heteronuclear 1H-15N-NOE, given as the ratio of the signal intensities of a 1H-15N-NOE spectrum and a reference spectrum without NOE transfer, is associated with the residue number. A low signal gain reflects high flexibility. (C) The ratio of T1 and T2 relaxation rate is associated with the residue number. A low T1/T2 ratio illustrates high flexibility. The dashed lines mark the domain boundaries.
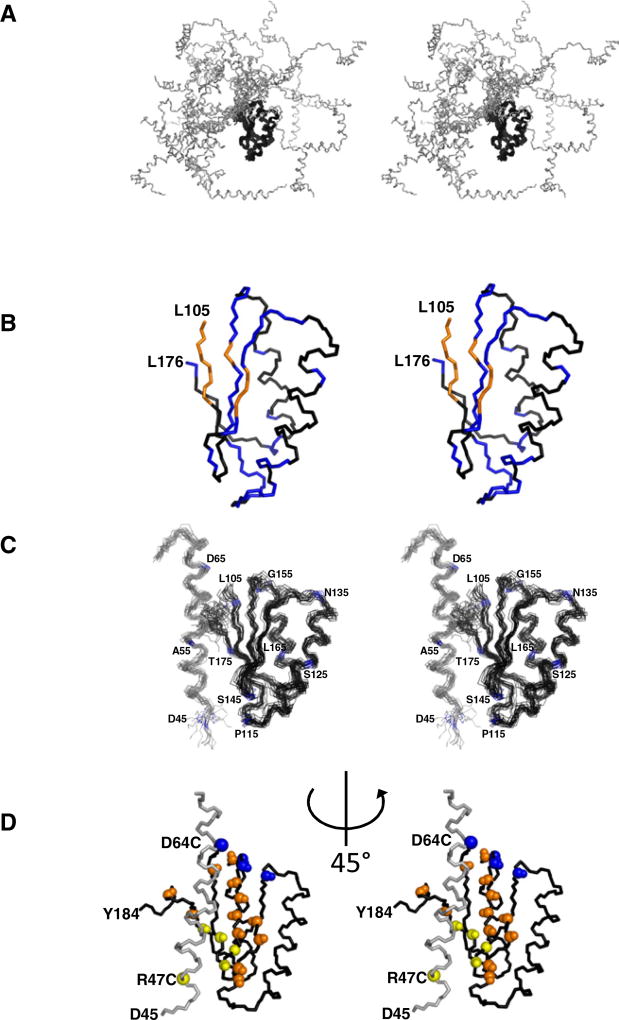
Figure 2. The Model for the Structure of the Conserved Core Region of MESD Corresponding to Residues 45–184.
(A) The “open form” backbone view of the 20 energetically lowest conformers of MESD45–184 achieved by a structure calculation using the CYANA protocol (Guentert, 2004) for the assignment of ambiguous NOE distance restraints. Only the “core domain” (residues 104–177, black) converges to a globular structure.
(B) NMR solution structure of MESD89–184. The amide signals of all colored residues are shifted in the 1H-15N-HSQC-NMR spectrum of MESD45–184. Δδ1H > 0.02 ppm or Δδ15N > 0.2 ppm. The amide signals of blue-marked residues are back shifted to the core domain chemical shifts in the 45–184 fragment carrying the W61R mutation. W61R mutant amide resonances of the orange-colored residues are not in superposition with any of the wild-type truncation.
(C) The “closed form” model structure of MESD45–184 based on the chemical shift perturbations shown in (B). The model has been enforced by 656 possible assignments of NOE distance restraints, 226 dihedral angle restraints empirically obtained from Talos (Cornilescu et al., 1999), and 36 RDCs. Residues 71–103 of the flexible loop region are omitted.
(D) Relaxation enhancement by introducing paramagnetic labels. Backbone amide groups affected (>50% reduction of peak height) by attachment of the paramagnetic MTSL label to C47 or C64 are colored in yellow and blue, respectively. Orange-colored backbone NH groups are affected from both spin labels. All views are in stereo. N and C termini (D45 and Y184) are marked. Orientation of (D) is rotated anticlockwise by 45° with respect to (A)–(C). See also Figure S1.
By comparing the 15N-HSQC-spectra of MESD45–184 and MESD89–184, we identified the residues of the core domain that show shifts of NMR signals as a result of their intramolecular interaction with residues 45–103 (Figure 2B). Based on these chemical shift perturbations, we developed a model structure of the MESD core region corresponding to residues 45–184. The model is enforced by a set of 656 possible assignments of ambiguous NOE distances, supplemented by orientational information of amide bonds based on 36 residual dipolar couplings (RDCs), of which four RDCs are located within the N-terminal helix as well as 226 empirically determined dihedral angles (Cornilescu et al., 1999). Table 1 provides a summary of the structural statistics. This model predicts that the N-terminal α helix interacts with the β sheet of the core domain via hydrophobic contacts (Figure 2C). The core domain maintains the structural integrity within MESD45–184, as evidenced by a high number of distance constraints. The structures of the core domain in both MESD fragments are superimposing very well (see Figure S1 available online).
Table 1.
NMR and Refinement Statistics for the Model Structure of MESD45–184
| Protein | |
|---|---|
| NMR Distance and Dihedral Constraints | |
| Distance constraints | |
| Total NOE | 656 |
| Intraresidue | 0 |
| Inter-residue | 656 |
| Sequential (|i − j| = 1) | 296 |
| Medium range (|i − j| ≤ 4) | 108 |
| Long range (|i − j| > 4) | 252 |
| Hydrogen bonds | 106 |
| Total dihedral angle restraints (Talos; Cornilescu et al. [1999]) | 226 |
| ϕ | 113 |
| Ψ | 113 |
| Total RDCs | 36 |
| Within α1 | 4 |
| Structure statistics | |
| Violations (average of 20 conformers) | |
| Distance constraints (>0.3 Å) | 0.1 |
| Dihedral angle constraints (>5°) | 0.05 |
| Maximum dihedral angle violation (°) | 5.5 |
| Maximum distance constraint violation (Å) | 0.36 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 0.7 |
| Average pairwise rmsd among 20 refined structures (Å) | |
| Heavy | 1.2 |
| Backbone | 0.7 |
| MolProbity Clashscore (raw score/Z score)220 | 26.35/−3.00 |
Circular dichroism (CD) spectra of MESD89–184 account for an α-helical content of 38 residues (data not shown), well in agreement with the structure of the core domain comprising 39 residues within α helices (Köhler et al., 2006). However, the CD spectra of MESD45–184 indicate an α-helical content of 47 residues, whereas characteristic short-range NOEs support an α-helical secondary structure for 58 residues. This is taken as evidence that both the “open” and “closed” form (Figures 2A and 2C) are present in the ensemble average observed by CD spectroscopy.
The presented NMR-based “closed” structural form of MESD45–184 (Figure 2C) is further supported by relaxation enhancement observed after introducing paramagnetic labels on both ends of helix α1 (Figure 2D). Amide hydrogens in the vicinity of these labels show remarkably increased T1 relaxation and lead to strongly reduced signal intensities. Figure 2D illustrates the affected areas with respect to the position of the label. After introducing the paramagnetic label at the beginning of helix α1 (R47C-MTSL), the signals of the yellow amide groups vanish. The paramagnetic label at the end of this helix (D64C-MTSL) exclusively affects the blue amides; whereas the orange amide groups are affected by both labels. These data strongly support the orientation and position of helix α1, as shown in our model.
The N-Terminal Helix (47–69) Is Functionally Essential
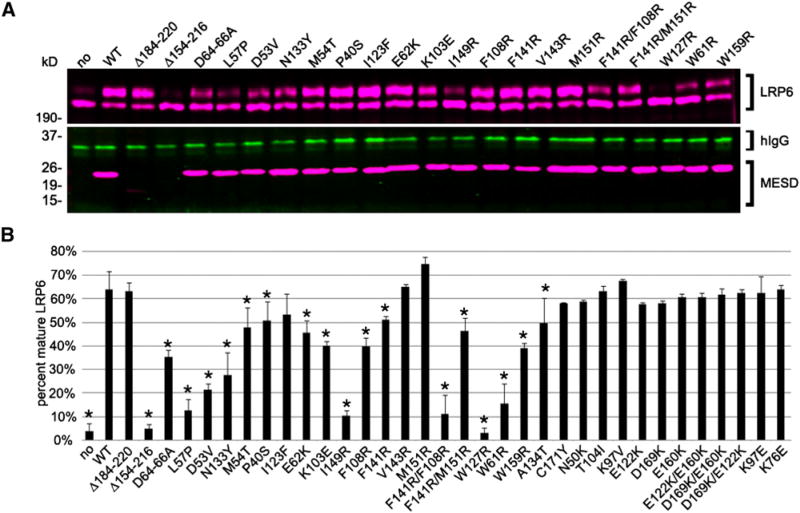
To identify residues important for MESD function, we screened a collection of Mesd truncations, and randomly PCR-generated or site-directed mutations using a cell culture-based LDLR-related protein-6 (LRP6) maturation assay (Hsieh et al., 2003) (Figure 3). In this assay we compared the efficiency of wild-type MESD and mutant MESD to promote maturation of LRP6 from a low-molecular weight ER-retained form to a mature higher molecular weight glycosylated form. In the absence of exogenously added MESD, approximately 94% of LRP6 is retained in the ER. In contrast, wild-type MESD promoted maturation of over 60% of total LRP6. Despite the apparent destabilizing effect of deleting the C-terminal domain (Δ184–220) (Figure 3A), mature LRP6 is detected at levels comparable to transfections with wildtype Mesd. These data suggest that the nonconserved and disordered C-terminal region of MESD is dispensable for function. In contrast, a slightly larger internal deletion (Δ154–216) eliminated MESD activity, suggesting that the disruption of the core domain eliminates MESD function or structural integrity.
Figure 3. Mutations in MESD Reduce Trafficking of Full-Length LRP6.
The ability of mutant MESD to traffic LRP6 was determined by assessing the maturation of LRP6 in cotransfection assays with MESD. (A) Representative western blot analyses of LRP6 maturation. The top 6% SDS-PAGE gel (>190 kDa) separates the glycosylated mature LRP6-Rho (upper band, magenta) from the lower molecular weight ER-retained LRP6-Rho (lower band, magenta). The lower 12% SDS-PAGE gel (<37 kDa) resolves hIgG (transfection control, green) and FLAG-tagged MESD (magenta). Proteins were visualized, and fluorescence was quantified using the Odyssey Infrared Imaging System (LI-COR Biosciences). (B) The efficiency of LRP6 maturation was determined by calculating the percentage of the mature membrane form (upper band) out of total LRP6 (upper and lower bands). Asterisks (*) highlight mutations that result in a significant (<0.05) reduction or enhancement of LRP6 maturation compared to wild-type (as determined by one-way ANOVA). Data and error bars indicate average ± standard deviation. Note that F141R/M151R is not significantly different compared to individual F141R or M151R mutations.
In contrast to MESD truncations, the MESD substitutions did not reduce mutant MESD in the cell compared to wild-type (Figure 3A). Because misfolded proteins are generally targeted for degradation in the ER, these data suggest that these mutations do not alter the stability or structural integrity of MESD. Several MESD substitutions (L57P, D(64–66)A, D53V, W61R, E62K) provide compelling evidence that the N-terminal α helix (49–68) is important for function (Figures 3 and 4A). MESD L57P and W61R each reduced LRP6 maturation to less than 25% of the level observed with cotransfection of wild-type Mesd. These substitutions likely impair interaction between the MESD N terminus and core domain, or interfere with MESD interactions with the LRP6 β-propeller/EGF. Notably, W61R is analogous to the Drosophila boca mutation (Culi and Mann, 2003). The ability of MESD W61R to promote maturation of 16% of LRP6 receptor suggests that the boca mutant may retain residual function. Likewise, D53V, D64-66A, and E62K reduced MESD activity to 34%, 55%, and 71% of wild-type, respectively. These mutations illustrate the functional importance of negatively charged residues within the α-helical domain. Other N-terminal mutations that modestly affected MESD function include M54T (75% of WT) and P40S (80% of WT).
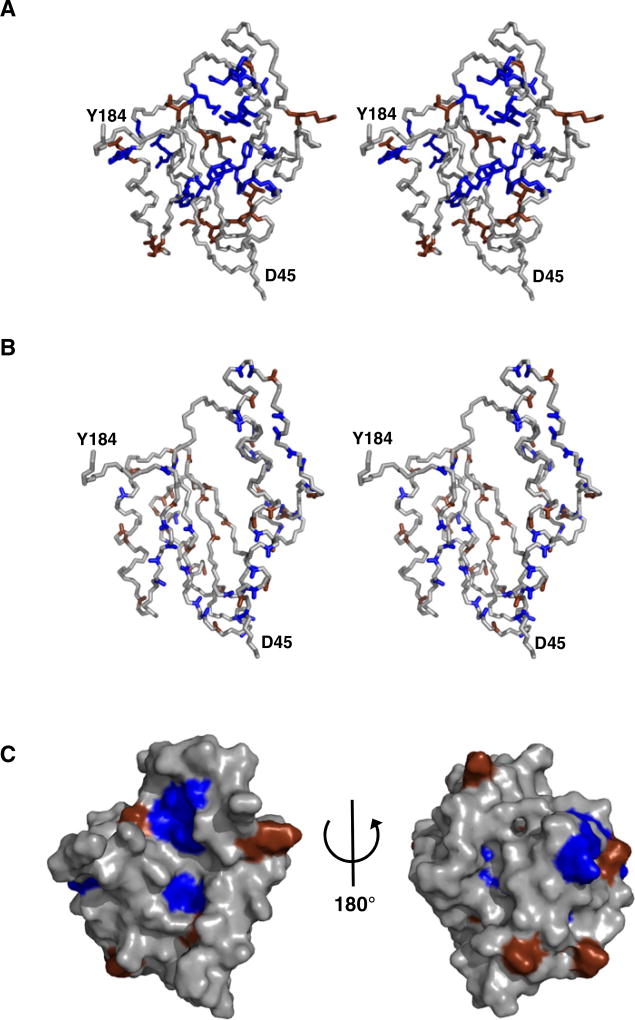
Figure 4. MESD Contains Surface-Located Hydrophobic and Acidic Residues Crucial for Function.
(A) Summary of LRP6 maturation assay. Single mutations of MESD reducing the level of mature LRP6 (blue) or not affecting the LRP6 maturation (brown) are mapped to the model structure of MESD45–184 as stereo view.
(B) The crucial hydrophobic and acidic areas are surface located, as shown by the increase of amide hydrogen T1 relaxation after addition of paramagnetic GdIII-DTPA to the sample buffer. The ratioT1Gd (III)/T1reference was lower than 0.9 for the amide hydrogens colored blue and higher than 0.9 for the brown-colored ones. Stereo view.
(C) Surface view of MESD45–184 with color coding as in (A).
Within the MESD core domain, I149R, F108R, and F141R highlight the importance of predicted exposed hydrophobic residues (Köhler et al., 2006) for MESD function. Although F141R and F108R individually reduced LRP6 maturation to 62% and 80% of the level observed with wild-type MESD, the combined mutations F141R/F108R reduced MESD function to 15% of wild-type. Similarly, I149R drastically reduced MESD activity to 17% of wild-type. In contrast, V143R and M151R did not significantly alter MESD activity, and F141R/M151R was not significantly different from F141R, suggesting that V143 and M151 are functionally distinct from F108, F141, and I149. Other mutations within the MESD core that significantly reduced MESD activity include W127R, N133Y, W159R, K103E, and A134T. W127R abolished MESD activity, whereas N133Y, W159R, K103E, and A134T modestly reduced LRP6 maturation to 43%–84% of wild-type activity.
In Figure 4A the location and functional relevance of the analyzed mutations were mapped to the model structure of MESD45–184. Residues making up the interface between helix α1 and the core domain are likely functionally indispensable. This includes residues K103 and D64, which likely form a salt bridge. The hydrophobic residues at the interface between helix α1 and the core domain, as well as the crucial acidic residues of helix α1, were further shown to be solvent exposed (Figures 4B and 4C), highlighting their likely involvement in receptor binding. Based on this model, the tryptophan residue substitutions may alter the MESD conformation by disrupting hydrophobic interactions without destabilizing MESD. Combined, these mutations provide functional evidence for the importance of the N-terminal α-helical extension as well as its transient interaction with exposed hydrophobic residues of the core domains β sheet.
Structural Evidence for the Importance of Helix α1
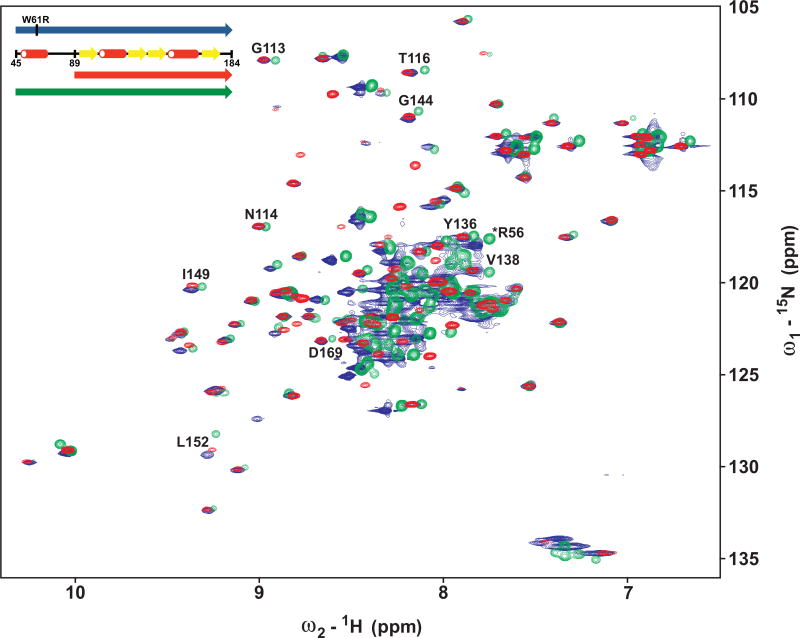
The essential role of helix α1 and its native interaction with the core domain’s β sheet could further be proven by the finding that the W61R mutation disrupts the positioning of the N-terminal helix (Figure 5). In effect, the chemical shift perturbations of the core domain signals that result from the interaction of helix α1 with the β sheet are back shifted in the W61R protein. In Figure 5, the HSQC spectra of the core domain MESD89–184 (red), wild-type MESD45–184 (green), and W61R mutant MESD45–184 (blue) are superimposed. In the case of the well-separated signals of G113, N114, T116, Y136, V138, G144, I149, L152, and D169, the spectra of the core domain (red) and the W61R mutant (blue) superpose very well, whereas the corresponding peaks are shifted in the spectra of the wild-type form (green). The affected residues are marked blue in Figure 2B. With the exception of the orange-marked residues, whose signals do not superimpose with any of the wild-type truncations, most β sheet residues in the mutant fragment are located in the same chemical environment as in the core domain. This suggests a disrupted interaction between β sheet and helix α1 in the W61R mutant. In addition, the remarkably increased line broadening of NMR signals in the chemical shift range between 7.5 and 8.5 ppm (Figure 5), as well as chemical shift perturbations between the mutant and wild-type MESD45–184 fragment of residues within helix α1 (e.g., R56), suggests a disruption of this N-terminal helix.
Figure 5. The MESD W61R Mutation Disrupts the Interaction between Helix α1 and Core Domain.
Overlay of the 1H-15N-HSQC-NMR spectra of MESD45–184 carrying the W61R mutation (blue), MESD45–184 of the wild-type (green), and MESD89–184 of the wild-type (red). The three fragments are shown schematically as an inset. Chemical shift perturbations between MESD89–184 and MESD45–184 due to the interaction of helix α1 with the core domain are back shifted in the mutated protein. The disruption of helix α1 in the mutated fragment leads to an increased signal line broadening in the 1H region 7.5–8.5 ppm, and to chemical shift perturbations of residues within α1 (marked by an asterisk).
MESD Binds to Hydrophobic Peptides that Are Interrupted by Positively Charged Residues
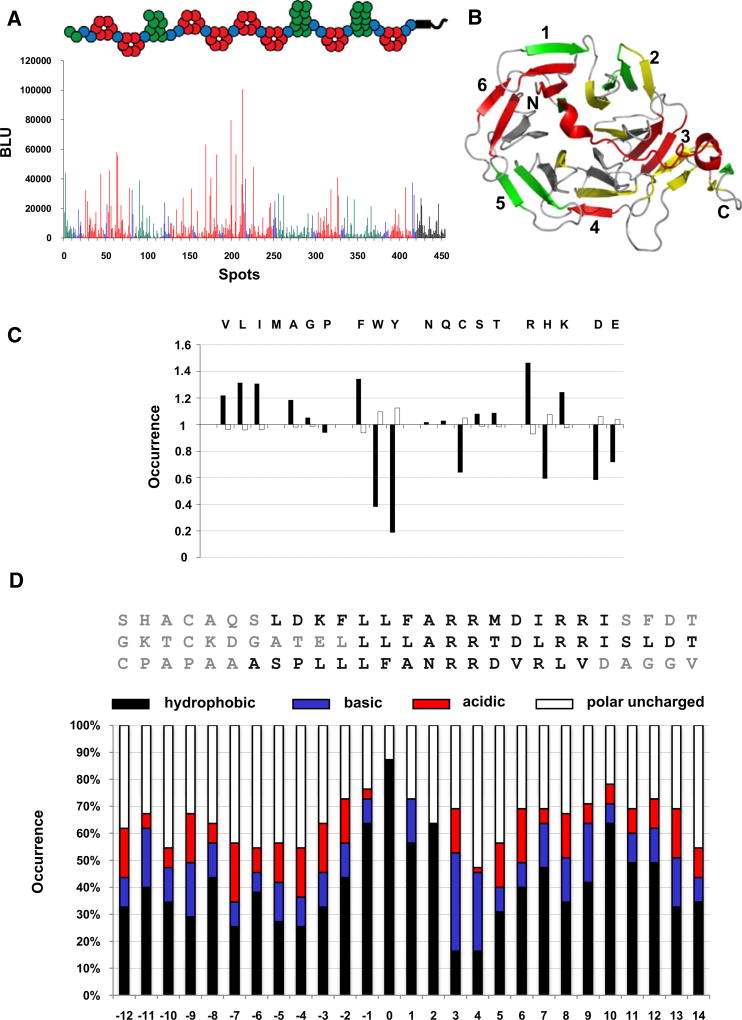
To obtain further insight into the MESD-assisted folding of the LDLR family, we characterized the interaction between MESD and the unfolded nascent receptors. To accomplish this we investigated the interaction between MESD and a cellulose-bound peptide library (Reineke et al., 1996) scanning the entire sequence of ten human LDLR family members. This peptide scan consisted of 16 residue-long fragments with six residue overlap from the human receptors LRP1, LRP1B, LRP2 (megalin), LRP4 (MEGF7), LRP5, LRP6, LDLR, VLDLR, LRP8 (ApoER2), and LR11 (SorLA). The large data set allowed a reliable statistical analysis of the receptor motifs recognized by MESD. The library was incubated with four different MESD constructs, namely the full-length MESD (30–224), the conserved part (45–184), the core domain (89–184), and the conserved part of the W61R mutant (45–184W61R). In the following the bound chaperone was detected by a chemiluminescent-based assay. The results for LRP1 are provided in Figure 6A and Figure S2 as a general example of the receptor chaperone-binding profile. The full-length MESD mainly bound receptor fragments from the β-propeller and the C-terminal adjacent EGF domain (Figure 6A), with preference for the first, third, and last propeller blade as well as the transition area to the adjacent EGF domain (Figure 6B). Only a few binding peptides in the complement-type region were identified. There are substantial differences in the distribution of amino acids within receptor fragments bound by MESD, compared to fragments without affinity to MESD (Figure 6C). MESD-bound receptor fragments are enriched in the hydrophobic residues valine, leucine, isoleucine, alanine, and phenylalanine, as well as in the positively charged residues arginine and lysine. In contrast, negatively charged residues, the aromatic residues tyrosine and tryptophan, as well as cysteines and histidines are unfavored. To determine the receptor motif recognized by MESD, we aligned sequences of the 55 best-binding fragments. These data provided evidence that MESD binds a core of several hydrophobic residues interrupted by some positively charged ones (Figure 6D), perfectly fitting the role as a molecular chaperone that assists folding of newly synthesized receptor peptide chains. The incubation of the membrane with the conserved part of MESD (45–184) results in the same pattern of bound receptor fragments (Figure S2B) as compared to the full-length protein. The incubation with the core domain (89–184), in contrast, results in a remarkably different binding pattern (Figure S2C). The interaction to the propeller domain is decreased with higher binding preference for the EGF domain itself and the complement-type region. Most intriguingly, the binding pattern of the lethal W61R mutant is very similar to MESD89–184 (Figure S2D) manifesting the structural similarity between these two MESD truncations and tearing them apart from the functional forms.
Figure 6. Interaction Studies between Native MESD and Short, Unfolded Receptor Fragments.
(A) The amount of bound MESD30–224 detected as chemiluminescence in Boehringer light units (BLU) for each fragment of the LRP1 receptor is illustrated. The fragments are colored with respect to the domain from which they are derived: red, β-propeller domain; blue, EGF domain; green, complement-type domain; black, others. The constitution of the LRP1 receptor is shown schematically as an inset.
(B) Preferred binding areas inside the β-propeller/EGF-domain motif: red-yellow-green-gray (much bound MESD→little bound MESD). LDLR β-propeller/EGF crystal structure from Jeon et. al. (2001).
(C) Amino acid occurrence in receptor fragments with binding affinity to MESD (black bars) versus fragments without detected binding to MESD (white bars). The values are the ratios to the relative occurrence of the respective residue within the whole peptide library.
(D) For determination of the receptor-binding motif, the sequences of receptor fragments with binding affinity to MESD (black letters) were aligned as shown exemplary for three fragments. After filling up the motif with flanking residues (gray letters), the occurrence of hydrophobic (black bars), positively charged (blue bars), negatively charged (red bars), as well as polar uncharged residues (white bars) at each position of the binding motif is shown. The position “0” refers to the start of the hydrophobic core motif.
See also Figure S2.
DISCUSSION
This work provides a structural basis for understanding diseases associated with misfolding of proteins from the LDLR family of type 1 receptors by investigating the conserved region of MESD corresponding to residues 45–184 using solution NMR, mutational analysis, and applying peptide libraries. As described recently, residues 104–177 fold into a conserved domain with a widespread β–α–β–β–α–β topology (Köhler et al., 2006). Here, we provide in vivo functional evidence and structural data that underscore the importance of MESD residues 45–103 for maturation of LRP6. Our results provide evidence for the formation of an α helix between residues 49 and 68, which temporarily interacts with the MESD “core domain” through hydrophobic contacts with the β sheet. In particular, residues M54, L57, L58, and W61 face the β sheet, where residues M106, F108, F141, V143, I149, and M151 form a well-suited contact area.
The interaction of the N-terminal helix with the β sheet is evident from chemical shift changes of the respective residues when comparing the spectra of the long (45–184) with those of the “core domain” (89–184) construct (Figure 5). However, the NMR data altogether suggest considerable structural dynamics with respect to the formation of a compact structure versus a partially disordered, “molten” population. The relaxation parameters provide strong evidence for a temporal release of the N-terminal helix from its position/interaction with the core domain. The T1/T2 ratio of residues 47–56 is half as large as the corresponding values of the core domain (107–177, Figure 1C), and the values for the linker region indicate very high flexibility. The absence of amide backbone HSQC signals for residues 57–63 indicates structural changes. Furthermore, extremely low intensities for long-range NOEs between α1 and the β sheet were observed. One example is the interaction between the aromatic NH of W61 and side-chain resonances of F141. Such an attenuation of distance-dependent cross peaks is also observed for protein-peptide complexes in case of an intermediate binding regime. As a result, CYANA calculations do not converge to a globular structure involving the N-terminal helix (Figure 2A). An interpretation of these tiny NOEs leads to the model shown in Figure 2C. Looking at all the data together, taking also into account the results from CD spectra, we conclude that the compact model (Figure 2C) is populated only to a certain extent. However, the experiments with paramagnetic tags, together with the values of four amide RDCs within helix 1 and further the chemical shift differences between MESD45–184 and the core domain, revealed the temporary location and orientation of the N-terminal helix. Most intriguingly, the N-terminal helix is not present in the mutant W61R, as evident from the overlay of HSQC spectra (Figure 5). Nearly all the “core domain” signals perturbed in the wild-type 45–184 fragment are back shifted in the protein carrying the W61R mutation (Figure 2B). The linker region (residues 69–103) is highly flexible, which leads to the exposure of a high number of acidic residues that may be exploited for receptor binding. Similar electrostatic contacts between a β-propeller structure and an intrinsically unstructured protein were demonstrated recently between the positively charged WD40 domain of Cdc4 and its acidic phosphorylated ligand, Sic1 (Mittag et al., 2008).
Chen et al. (2010) describe a very different structure for MESD41–184 (PDB-ID: 2KMI), including a differently folded core domain (104–177) with the α helices positioned on both sides of the central β sheet. The N-terminal helix, then showing an upside-down orientation as compared to our structure, interacts with the third α helix (117–135). Finally, the structure is described as a rigid, one-state conformer. In contrast, we have shown that MESD89–184 forms a domain with a very common fold (Köhler et al., 2006), and chemical shift changes between MESD89–184 and MESD45–184 are only notable for residues located in the core domain’s β sheet; no chemical shift changes are observed for residues located in helices. Therefore, our data do not agree with the structure described by Chen et al. (2010). Also, the positions of the core domain helices are well defined by a large number of constraints. Besides this, we could prove the orientation of the N-terminal helix in MESD45–184 by paramagnetic labeling as well as RDC measurements. In agreement with the confined chemical shift changes between the long and short forms of MESD, paramagnetic labels at both ends of α1 do not affect any helices of the core domain but exclusively increase the line width of β sheet signals.
The function of many chaperones depends on a hydrophobic surface patch whose availability is regulated by an ATP-dependent conformational change. In the case of MESD, we propose that a hydrophobic surface-located patch in the β sheet of the core domain is transiently covered by the N-terminal helix and subsequently released through exchange with a partially “molten” MESD structure. This conformational change may be supported by the presence of an appropriate binding partner for helix 1, namely the β-propeller/EGF motif. In line with this hypothesis, we observed hydrophobic and acidic residues in α1, which are both solvent exposed (Figure 4B) and crucial for the MESD function (Figure 4A). Consistent with this hypothesis, we observed that MESD interacts with unfolded receptor fragments isolated from the human LDLR family. Specifically, MESD binds to fragments from the β-propeller and the C-terminal adjacent EGF domain, where it binds to a receptor motif consisting of a core of several hydrophobic residues interrupted by basic amino acids. The interaction to the first and last blade of the β-propeller (Figure 6B), where the propeller ring is closed, as well as to the third blade, where the interaction interface to the adjacent EGF domain is built up, emphasizes the relevance of MESD for the folding of the β-propeller/EGF-domain motif.
According to investigations by Culi et al. (Culi and Mann, 2003; Culi et al., 2004), non-LDLR family receptors containing only a β-propeller without the C-terminal adjacent EGF domain, fold independently of MESD. For this reason it seems likely that the establishment of the native contacts between β-propeller and adjacent EGF domain is the primary MESD-dependent step during the folding process. However, to assemble these hydrophobic contacts, a folded propeller is most probably a prerequisite. Accordingly, we suggest a model for the mechanism by which MESD assists folding of the β-propeller/EGF motif that takes into consideration the structure-function studies above (Figure 7A). In this model a transient release of MESD’s N-terminal helix from the “core domain” exposes several hydrophobic residues, which facilitates the binding of helix 1 to receptor residues in the first propeller blade. Due to polar interactions, the positively charged propeller unit then wraps around the negatively charged N-terminal helix of MESD, mediating the closure of the propeller ring (Figure 7B). Finally, hydrophobic contacts between the β sheet of MESD and the EGF domain favor the critical positioning of the EGF within the third propeller blade (Figure 7B). This model suggests that the W61R boca mutation disrupts LDLR family folding because: (1) MESD is recruited to other interaction partners due to the permanent availability of the hydrophobic patch on the surface of the core domain; and (2) due to the corrupted N-terminal helix, a proper MESD interaction with the β-propeller unit does not take place. This proposed mechanism is further supported by the findings that MESD89–184 and MESD45–184W61R show a decreased interaction to β-propeller fragments, favoring the binding of the EGF domain and the complement-type region instead. Because these two nonfunctional MESD fragments omit a native N-terminal α helix, the hydrophobic β sheet patch is permanently exposed, which only allows the binding of the EGF domain and the complement-type region.
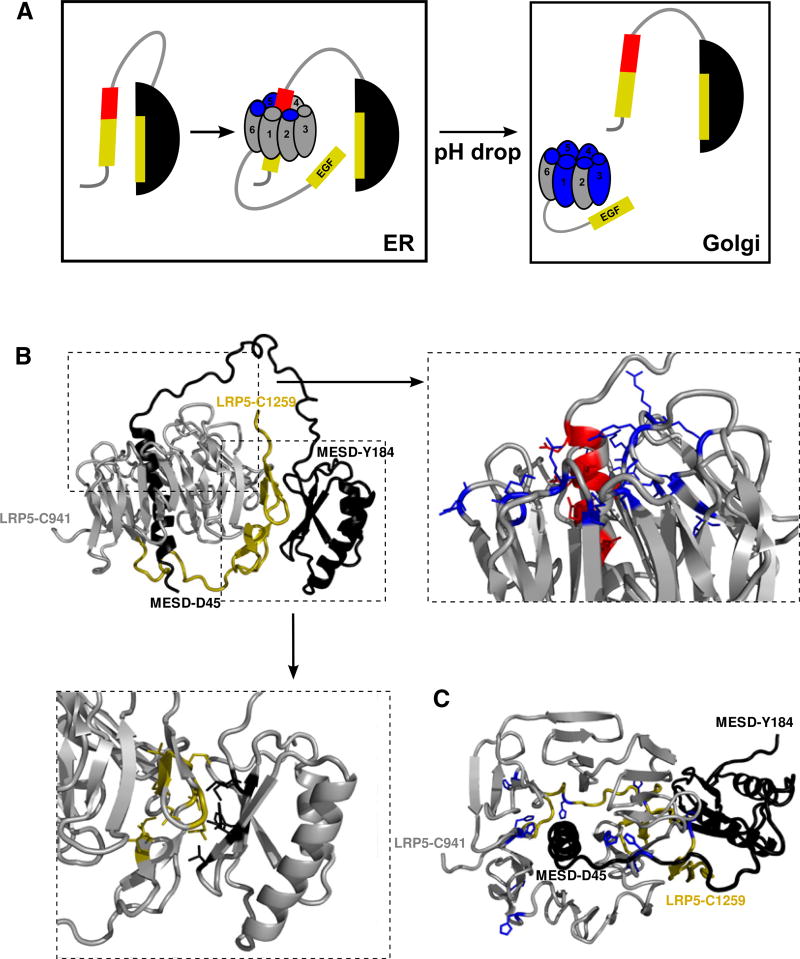
Figure 7. Proposed Model of the MESD-Assisted Folding of the β-Propeller/EGF Motif.
(A) Due to the weak hydrophobic contacts between helix α1 and core domain (black) within MESD, hydrophobic areas (yellow) are either transiently covered or surface exposed. Exposure of hydrophobic residues allows the interaction between MESD and hydrophobic β-propeller/EGF parts. Due to electrostatic interactions between the positively charged propeller center (blue) and the negatively charged part of helix α1 (red), the propeller wraps around helix α1. This mediates the closure of the propeller ring. Interaction between MESD and the EGF domain sets up the favorable interaction position between the EGF and the third β-propeller domain. Dissociation of MESD-receptor interaction could be triggered by a decrease in pH, as a result of the receptor transport to the Golgi, followed by a change in charge of histidine, leading to an increase in overall positive surface charge.
(B) Model structure of the C-terminal β-propeller/EGF motif of LRP5 bound to MESD45–184: MESD, black; β-propeller, gray; EGF domain, yellow. The N and C termini of MESD and β-propeller/EGF motive are marked. Detailed views illustrate the electrostatic contacts between acidic residues of MESD’s helix α1 (red) and positively charged propeller residues (blue) as well as the hydrophobic contacts between MESD’s β sheet and EGF domain (black and yellow, respectively). (C) Representation of histidine residues (blue) in the interface between MESD and β-propeller/EGF motif, which could act as pH switch leading to the complex dissociation.
The question remains how this MESD-receptor complex could dissociate to form the mature receptor. Because MESD shows neither binding nor hydrolysis of ATP (data not shown), its function must be facilitated through the transient nature of the contacts between helix α1 and the “core domain” and presumably be also regulated by competitive substrate binding. Other ATP-independent ER resident chaperones, e.g., HSP47 (Satoh et al., 1996), GRP94 (Wearsch and Nicchitta, 1997), or RAP (a highly specified chaperone crucial for folding of the CR-domains of the LDLR family) (Lee et al., 2006), use a pH change to regulate their substrate binding and release. Because receptor export from the ER into the Golgi is linked to a decrease in pH, the altered charge of several histidine residues in the MESD-receptor interface (e.g., LRP5 residues H965, H974, H1027, H1051, H1166, H1197, H1202, H1215, H1226, H1243) and, hence, an overall increase of the positive surface charge could lead to the dissociation of MESD from the mature receptor (Figures 7A and 7C). A very similar mechanism is described for the release of LDLR-bound ligands in the endosome (Beglova et al., 2004) as well as for the dissociation of the RAP chaperone from LDLR in the Golgi (Lee et al., 2006; Estrada et al., 2008). In all these cases, histidine residues are acting as a pH switch, triggering conformational changes and ligand release.
The tight binding between MESD and mature receptors described in the literature (Li et al., 2005; Lu et al., 2010) refers to interaction studies carried out with surface-located LRP5/6 receptors and exogenously added MESD. In detail, the unstructured highly positively charged C terminus of MESD could be shown to be important for this interaction. Furthermore, it could be shown that this exogenously added MESD competes with other ligands of LDLR family receptors at the cell surface making MESD to a promising drug target. Because the C-terminal ER retention signal REDL makes MESD to an ER resident chaperone, and free LRP receptors at the cell surface are crucial for the receptor function, the dissociation of the MESD-receptor complex within the cell is an essential process. The described tight interaction between MESD and folded receptors at the pH neutral cell surface supports our model of a pH-dependent dissociation within the Golgi. An increased overall charge of LDLR family receptors due to several histidines altering their charge within the acidic Golgi further explains the dissociation of the interaction between receptor and the positively charged MESD C terminus.
EXPERIMENTAL PROCEDURES
Preparation of MESD Proteins for NMR Measurements
Template DNA from Mesd was kindly provided by J. Herz (University of Texas Southwestern Medical Center, Dallas, TX, USA). MESD truncations were generated as described previously (Köhler et al., 2006).
For peptide library screens, unlabelled His-S-tagged MESD30–224, MESD45–184, MESD45–184W61R, and MESD89–184 were used.
NMR Measurements/Structure Calculation
NMR spectra were acquired at 300 K on Bruker DRX600, DMX750, and AV900 spectrometers equipped with cryoprobes. All experiments were performed with aqueous solutions of 1 mM protein in 20 mM Na2HPO4/NaH2PO4 buffer (pH 5.5) additionally containing 50 mM NaCl, 0.1 mM EDTA, 0.02% NaN3, and 10% D2O if not otherwise stated. All spectra were processed using Topspin 2.1 (Bruker Biospin GmbH). Assignments and structure calculations were performed as described previously (Köhler et al., 2006).
Twenty structures selected by overall energy from initial structure calculations were subjected to a refinement in water using CNS1.1 (Brünger et al., 1998) and the parallhdg5.3 force field (Linge et al., 2003). The final ensemble was analyzed by PSVS 1.3 (Bhattacharya et al., 2007). The Ramachandran plot analysis according to Morris et al. (1992) classifies 90.5% of the residues backbone torsion angles as most favored and 9.5% as additionally favored. Table 1 shows a summary of the structural statistics.
1H-15N RDCs were measured with a 1 mM uniformly 15N-labeled sample of MESD45–184 containing Pf1 phages (Profos). The phage concentration was increased until a HOD-signal splitting of 17 Hz was reached (Hansen et al., 1998). RDCs were extracted from un-decoupled 1H-15N-HSQC spectra as the difference of the 15N-J couplings between the sample with and without phages.
Relaxation rate measurements were performed as a series of 1H-15N-HSQC-type spectra (Farrow et al., 1994). Relaxation rates of the amide nitrogens were extracted from 11 spectra with delays of 12, 52, 102, 152, 202, 302, 402, 602, 902, 2002, and 5002 ms for T1, and 6, 10, 18, 26, 34, 42, 82, 122, 162, 202, and 242 ms for T2. To study the solvent exposure of MESD45–184, the T1 relaxation rates of the amide hydrogens in the absence and presence of GdIII-DTPA (Pintacuda and Otting, 2002) were determined. A GdIII-DTPA stock solution (50 mM diethylenetriaminepentaacetic acid [Sigma], 49 mM GdIIICl3 [Sigma] in 150 mM NaOH) was added to a 1 mM sample of MESD45–184 to a final concentration of 0.3 mM. The 1H-T1 relaxation rates were extracted from 11 spectra with delays of 10, 20, 30, 40, 50, 60, 70, 80, 90, 100, and 150 ms.
Site-Directed Paramagnetic Labeling of MESD45–184
To generate unique cysteine residue variants of MESD45–184, the native cysteine at position 171 was substituted by valine. Two single cysteine variants (C171V-R47C and C171V-D64C) were uniformly 15N labeled, followed by coupling of the nitroxide spin label methanethiosulfonate (MTSL; Toronto Research Chemicals) using a 10-fold molar excess dissolved in 50 µl d6-DMSO. After incubation for 4 hr at room temperature, the reaction was stopped by dialysis at 8°C against 20 mM Na2HPO4/NaH2PO4 buffer (pH 6.5) additionally containing 50 mM NaCl and 0.1 mM EDTA. The effect of the spin label was determined by comparing the peak intensities in 15N-1H-HSQC spectra taken on the protein with and without the paramagnetic label.
Interaction Studies with Receptor Fragments
The peptide library was prepared by automated spot synthesis on a N-modified cellulose-amino-hydroxypropyl ether (N-CAPE) membrane (Licha et al., 2000). The sequences of the following receptors were screened as 16 residue-long peptides that overlap by six residues: LRP1 (NCBI-ID: NP_002323); LRP1B (NP_061027); LRP2 (NP_004516); MEGF7 (BAA32468); LRP5 (NP_002326); LRP6 (XP_006874); VLDLR (XP_005453); ApoER2 (BAA09328); LDLR (XP_009082); and sorLA (AAC50891).
After treatment of the membrane with Blocking Buffer (10% 10× Blocking Buffer [Sigma], 146 mM saccharose in Tris-buffered saline [TBS]), the membrane was incubated with S-tag linked MESD (1 µM in Blocking Buffer) overnight at 4°C. For detection of bound MESD, S-protein-HRP-conjugate (Novagen; 1/5000 in Blocking Buffer) was used followed by incubation with Luminol (Uptilight HRP blot Reagent A and B; Interchim) and chemiluminescence detection. TBS was used for all washing steps.
LRP6 Maturation Assay
Expression Constructs
Construction of FLAG-MESD and LRP6-Rho was previously described (Hsieh et al., 2003). MESD-W61R, MESD-Δ184–220, and MESD-Δ154–216 were generated from FLAG-MESD by PCR amplification and subcloned into pcDNA3.1/V5-His-TOPO. EGFP-rho (EGFP/pRK5-SK) was a gift from Jen-Chih Hsieh (Stony Brook University). Human IgG heavy-chain plasmid (hIgG-pRK5) was previously described (Hsieh et al., 1999, 2003). Random amino acid substitutions were generated by PCR using QuikChange Site-Directed Mutagenesis Kit (Invitrogen).
Transfection of COS1 Cells
COS1 cells were seeded at 50% confluency in 12-well plates and transfected 24 hr later with a total of 1 µg plasmid DNA using Fugene 6 (Roche). Transfections contained 0.3 µg MESD, 0.4 µg LRP6-Rho (kindly provided by J. Hsieh), 0.1 µg hIgG plasmid, 0.1µg EGFP, and pCS2+ plasmid. The cells were detached from the plate in 1 ml of 5 mM EDTA/phosphate-buffered saline (PBS), collected by centrifugation in a microcentrifuge at 3000 rpm for 3 min, and lysed in 55 µl cold lysis buffer (1% Triton X-100/PBS containing final concentrations of 17 mg/ml aprotinin, 10 mg/ml benzamidine, 1 mg/ml leupeptin, 3 mg/ml antipain, 1 M PMSF). Ten microliters of lysates were used for SDS-PAGE in the presence of β-mercaptoethanol, followed by western blot analysis. All transfections were repeated at minimum three times. Representative blots are shown.
Western Blotting, Antibodies, Image Acquisition, and Quantitation
Proteins were transferred from SDS-PAGE to nitrocellulose membranes (0.45 µmProtran; VWR), blocked overnight in1% casein/TBS at 4°C, and incubated with primary antibodies and fluorescently labeled secondary antibodies each for 1 hr at room temperature. Membranes were washed with 1× TBST (0.05% Tween 20/TBS) 3 × 15 min before and after secondary antibody incubation. All antibodies were diluted in 1% casein/TBS. Rho-tagged LRP6 was detected using mouse monoclonal anti-rhodopsin (clone 1D4) at 1:1000. Flag-tagged MESD was detected using mouse monoclonal anti-FLAG (clone M2; Sigma) at 1:5000. Secondary antibody Alexa 680-labeled anti-mouse (Invitrogen) was used at 1:4,000, and human IgG heavy chain was detected using IRDye800-labeled anti-human IgG (Rockland) at 1:10,000. Membranes were scanned and bands of interest quantitated using the Odyssey Infrared Imaging System and software (LI-COR Biosciences).
Supplementary Material
Acknowledgments
We would like to thank J. Herz (Department of Molecular Genetics, University of Texas Southwestern Medical Center, Dallas, TX, USA), who provided us the DNA template for Mesd, and J. Hsieh for providing the Rho-tagged LRP6. We would also like to thank Prisca Boisguérin (Institute for Medical Immunology, Charité, Berlin, Germany) for the help in peptide spot synthesis, and X. Zhang for help in initial screening of MESD mutations. In addition, we wish to thank Dr. Robert S. Haltiwanger and Steven Smith for critical reading of the manuscript. This work was supported in part by a grant to B.C.H. from the NIH (GM 053964).
Footnotes
ACCESSION NUMBERS
The structure coordinates of the “open” and “closed” form of MESD (D45–K184) have been deposited in the Protein Data Bank under the accession codes 2RQM and 2RQK, respectively. Assigned resonances as well as restraints used within structure calculation have been deposited in the BMRB under the accession code 11076.
Supplemental Information includes two figures and can be found with this article online at doi:10.1016/j.str.2010.12.022.
Note Added in Proof
The fold of the core domain in our NMR structure is nicely corroborated by X-ray crystallographic investigations on the core domains of Boca, MESD and ceBMY-1 from D. melanogaster, M. musculus, and C. elegans, respectively, see the paper by Collins and Hendrickson in this issue of Structure (Collins and Hendrickson, 2011).
References
- Andersen OM, Reiche J, Schmidt V, Gotthardt M, Spoelgen R, Behlke J, von Arnim CA, Breiderhoff T, Jansen P, Wu X, et al. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc. Natl. Acad. Sci. USA. 2005;102:13461–13466. doi: 10.1073/pnas.0503689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen OM, Schmidt V, Spoelgen R, Gliemann J, Behlke J, Galatis D, McKinstry WJ, Parker MW, Masters CL, Hyman BT, et al. Molecular dissection of the interaction between amyloid precursor protein and its neuronal trafficking receptor SorLA/LR11. Biochemistry. 2006;45:2618–2628. doi: 10.1021/bi052120v. [DOI] [PubMed] [Google Scholar]
- Beglova N, Jeon H, Fisher C, Blacklow SC. Structural features of the low-density lipoprotein receptor facilitating ligand binding and release. Biochem. Soc. Trans. 2004;32:721–723. doi: 10.1042/BST0320721. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Tejero R, Montelione GT. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, Mitnick MA, Wu D, Insogna K, Lifton RP. High bone density due to a mutation in LDL-receptor-related protein 5. N. Engl. J. Med. 2002;346:1513–1521. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Cam JA, Zerbinatti CV, Li Y, Bu G. Rapid endocytosis of the low density lipoprotein receptor-related protein modulates cell surface distribution and processing of the beta-amyloid precursor protein. J. Biol. Chem. 2005;280:15464–15470. doi: 10.1074/jbc.M500613200. [DOI] [PubMed] [Google Scholar]
- Carter M, Chen X, Slowinska B, Minnerath S, Glickstein S, Shi L, Campagne F, Weinstein H, Ross M. Crooked tail (Cd) model of human folate-responsive neural tube defects is mutated in Wnt coreceptor lipoprotein receptor-related protein 6. Proc. Natl. Acad. Sci. USA. 2005;102:12843–12848. doi: 10.1073/pnas.0501963102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li Q, Liu C, Zhou B, Bu G, Wang J. NMR structure note: solution structure of the core domain of MESD that is essential for proper folding of LRP5/6. J. Biomol. NMR. 2010;47:283–288. doi: 10.1007/s10858-010-9426-8. [DOI] [PubMed] [Google Scholar]
- Collins MN, Hendrickson WA. Structural characterization of the Boca/Mesd maturation factors for LDL-receptor-type β-propeller domains. Structure. 2011;19(this issue):324–336. doi: 10.1016/j.str.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Culi J, Mann R. Boca, an endoplasmic reticulum protein required for wingless signaling and trafficking of LDL receptor family members in Drosophila. Cell. 2003;112:343–354. doi: 10.1016/s0092-8674(02)01279-5. [DOI] [PubMed] [Google Scholar]
- Culi J, Springer T, Mann R. Boca-dependent maturation of beta-propeller/EGF modules in low-density lipoprotein receptor proteins. EMBO J. 2004;23:1372–1380. doi: 10.1038/sj.emboj.7600132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada K, Fisher C, Blacklow SC. Unfolding of the Rap-D3 helical bundle facilitates dissociation of RAP-receptor complexes. Biochemistry. 2008;47:1532–1539. doi: 10.1021/bi702076y. [DOI] [PubMed] [Google Scholar]
- Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15 N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- Gong Y, Slee R, Fukai N, Rawadi G, Roman-Roman S, Reginato A, Wang H, Cundy T, Glorieux F, Lev D, et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- Guentert P. Automated NMR protein structure calculation with cyana. In: Downing A, editor. Methods in Molecular Biology. Clifton, NJ: Humana Press; 2004. pp. 353–378. [DOI] [PubMed] [Google Scholar]
- Hansen MR, Mueller L, Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat. Struct. Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- He X, Semenov M, Tamai K, Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: arrows point the way. Development. 2004;131:1663–1677. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- Hsieh JC, Kodjabachian L, Rebbert ML, Rattner A, Smallwood PM, Samos CH, Nusse R, Dawid IB, Nathans J. A new secreted protein that binds to Wnt proteins and inhibits their activities. Nature. 1999;398:431–436. doi: 10.1038/18899. [DOI] [PubMed] [Google Scholar]
- Hsieh JC, Lee L, Zhang L, Wefer S, Brown K, DeRossi C, Wines M, Rosenquist T, Holdener B. MESD encodes an LRP5/6 chaperone essential for specification of mouse embryonic polarity. Cell. 2003;112:355–367. doi: 10.1016/s0092-8674(03)00045-x. [DOI] [PubMed] [Google Scholar]
- Jeon H, Meng W, Takagi J, Eck M, Springer T, Blacklow S. Implications for familial hypercholesterolemia from the structure of the LDL receptor YWTD-EGF domain pair. Nat. Struct. Biol. 2001;8:499–504. doi: 10.1038/88556. [DOI] [PubMed] [Google Scholar]
- Koduri V, Blacklow S. Requirement for natively unstructured regions of mesoderm development candidate 2 in promoting low-density lipoprotein receptor-related protein 6 maturation. Biochemistry. 2007;46:6570–6577. doi: 10.1021/bi700049g. [DOI] [PubMed] [Google Scholar]
- Köhler C, Andersen OM, Diehl A, Krause G, Schmieder P, Oschkinat H. The solution structure of the core of mesoderm development (MESD), a chaperone for members of the LDLR-family. J. Struct. Funct. Genomics. 2006;7:131–138. doi: 10.1007/s10969-007-9016-5. [DOI] [PubMed] [Google Scholar]
- Lee D, Walsh J, Mikhailenko I, Yu P, Migliorini M, Wu Y, Krueger S, Curtis J, Harris B, Lockett S, et al. RAP uses a histidine switch to regulate its interaction with LRP in the ER and Golgi. Mol. Cell. 2006;22:423–430. doi: 10.1016/j.molcel.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Li Y, Chen J, Lu W, McCormick L, Wang J, Bu G. MESD binds to mature LDL-receptor-related protein-6 and antagonizes ligand binding. J. Cell Sci. 2005;118:5305–5314. doi: 10.1242/jcs.02651. [DOI] [PubMed] [Google Scholar]
- Licha K, Bhargava S, Rheinländer C, Becker A, Schneider-Mergener J, Volkmer-Engert R. Highly parallel nano-synthesis of cleavable peptide-dye conjugates on cellulose membranes. Tetrahedron Lett. 2000;41:1711–1715. [Google Scholar]
- Lighthouse JK, Zhang L, Hsieh JC, Rosenquist T, Holdener BC. MESD is essential for apical localization of megalin/LRP2 in the visceral endoderm. Dev. Dyn. 2010 doi: 10.1002/dvdy.22477. in press. Published online November 8, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- Lu W, Liu CC, Thottassery JV, Bu G, Li Y. Mesd is a universal inhibitor of Wnt coreceptors LRP5 and LRP6 and blocks Wnt/beta-catenin signaling in cancer cells. Biochemistry. 2010;49:4635–4643. doi: 10.1021/bi1001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao B, Wu W, Li Y, Hoppe D, Stannek P, Glinka A, Niehrs C. LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Nature. 2001;411:321–325. doi: 10.1038/35077108. [DOI] [PubMed] [Google Scholar]
- Mao B, Wu W, Davidson G, Marhold J, Li M, Mechler B, Delius H, Hoppe D, Stannek P, Walter C, et al. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature. 2002;417:664–667. doi: 10.1038/nature756. [DOI] [PubMed] [Google Scholar]
- Mittag T, Orlicky S, Choy WY, Tang X, Lin H, Sicheri F, Kay LE, Tyers M, Forman-Kay JD. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc. Natl. Acad. Sci. USA. 2008;105:17772–17777. doi: 10.1073/pnas.0809222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AL, MacArthur MW, Hutchinson EG, Thornton JM. Stereochemical quality of protein structure coordinates. Proteins. 1992;12:345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]
- Pinson KI, Brennan J, Monkley S, Avery BJ, Skarnes WC. An LDL-receptor-related protein mediates Wnt signalling in mice. Nature. 2000;407:535–538. doi: 10.1038/35035124. [DOI] [PubMed] [Google Scholar]
- Pintacuda G, Otting G. Identification of protein surfaces by NMR measurements with a pramagnetic Gd(III) chelate. J. Am. Chem. Soc. 2002;124:372–373. doi: 10.1021/ja016985h. [DOI] [PubMed] [Google Scholar]
- Reineke U, Sabat R, Kramer A, Stigler RD, Seifert M, Michel T, Volk HD, Schneider-Mergener J. Mapping protein-protein contact sites using cellulose-bound peptide scans. Mol. Divers. 1996;1:141–148. doi: 10.1007/BF01544952. [DOI] [PubMed] [Google Scholar]
- Satoh M, Hirayoshi K, Yokota S, Hosokawa N, Nagata K. Intracellular interaction of collagen-specific stress protein HSP47 with newly synthesized procollagen. J. Cell Biol. 1996;133:469–483. doi: 10.1083/jcb.133.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinagra M, Verrier D, Frankova D, Korwek K, Blahos J, Weeber E, Manzoni O, Chavis P. Reelin, very-low-density lipoprotein receptor, and apolipoprotein E receptor 2 control somatic NMDA receptor composition during hippocampal maturation in vitro. J. Neurosci. 2005;25:6127–6136. doi: 10.1523/JNEUROSCI.1757-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer TA. An extracellular beta-propeller module predicted in lipoprotein and scavenger receptors, tyrosine kinases, epidermal growth factor precursor, and extracellular matrix components. J. Mol. Biol. 1998;283:837–862. doi: 10.1006/jmbi.1998.2115. [DOI] [PubMed] [Google Scholar]
- Takayama Y, May P, Anderson RG, Herz J. Low density lipoprotein receptor-related protein 1 (LRP1) controls endocytosis and c-CBL-mediated ubiquitination of the platelet-derived growth factor receptor beta (PDGFR beta) J. Biol. Chem. 2005;280:18504–18510. doi: 10.1074/jbc.M410265200. [DOI] [PubMed] [Google Scholar]
- Tamai K, Semenov M, Kato Y, Spokony R, Liu C, Katsuyama Y, Hess F, Saint-Jeannet J, He X. LDL-receptor-related proteins in Wnt signal transduction. Nature. 2000;407:530–535. doi: 10.1038/35035117. [DOI] [PubMed] [Google Scholar]
- Van Wesenbeeck L, Cleiren E, Gram J, Beals R, Benichou O, Scopelliti D, Key L, Renton T, Bartels C, Gong Y, et al. Six novel missense mutations in the LDL receptor-related protein 5 (LRP5) gene in different conditions with an increased bone density. Am. J. Hum. Genet. 2003;72:763–771. doi: 10.1086/368277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wearsch PA, Nicchitta CV. Interaction of endoplasmic reticulum chaperone GRP94 with peptide substrates is adenine nucleotide-independent. J. Biol. Chem. 1997;272:5152–5156. doi: 10.1074/jbc.272.8.5152. [DOI] [PubMed] [Google Scholar]
- Wehrli M, Dougan S, Caldwell K, O’Keefe L, Schwartz S, Vaizel-Ohayon D, Schejter E, Tomlinson A, DiNardo S. Arrow encodes an LDL-receptor-related protein essential for wingless signalling. Nature. 2000;407:527–530. doi: 10.1038/35035110. [DOI] [PubMed] [Google Scholar]
- Xu Q, Wang Y, Dabdoub A, Smallwood PM, Williams J, Woods C, Kelley MW, Jiang L, Tasman W, Zhang K, Nathans J. Vascular development in the retina and inner ear: control by Norrin and Frizzled-4, a high-affinity ligand-receptor pair. Cell. 2004;116:883–895. doi: 10.1016/s0092-8674(04)00216-8. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang Y, Li X, Zhang J, Mao J, Li Z, Zheng J, Li L, Harris S, Wu D. The LRP5 high-bone-mass G171V mutation disrupts LRP5 interaction with Mesd. Mol. Cell. Biol. 2004;24:4677–4684. doi: 10.1128/MCB.24.11.4677-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.