Arabidopsis CEL6 and MAN7 proteins affect cell morphology and silique dehiscence, which can be manipulated to different degrees by altering their activities.
Abstract
Cellulases, hemicellulases, and pectinases play important roles in fruit development and maturation. Although mutants with defects in these processes have not been reported for cellulase or hemicellulase genes, the pectinases ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1) and ADPG2 were previously shown to be essential for silique dehiscence in Arabidopsis (Arabidopsis thaliana). Here, we demonstrate that the cellulase gene CELLULASE6 (CEL6) and the hemicellulase gene MANNANASE7 (MAN7) function in the development and dehiscence of Arabidopsis siliques. We found that these genes were expressed in both vegetative and reproductive organs and that their expression in the silique partially depended on the INDEHISCENT and ALCATRAZ transcription factors. Cell differentiation was delayed in the dehiscence zone of cel6 and man7 mutant siliques at early flower development stage 17, and a comparison of the spatio-temporal patterns of CEL6 and MAN7 expression with the locations of delayed cell differentiation in the cel6 and man7 mutants revealed that CEL6 and MAN7 likely indirectly affect the timing of cell differentiation in the silique valve at this stage. CEL6 and MAN7 were also found to promote cell degeneration in the separation layer in nearly mature siliques, as cells in this layer remained intact in the cel6 and man7 mutants and the cel6-1 man7-3 double mutant, whereas they degenerated in the wild-type control. Phenotypic studies of single, double, triple, and quadruple mutants revealed that higher-order mutant combinations of cel6-1, man7-3, and adpg1-1 and adpg2-1 produced more severe silique indehiscent phenotypes than the corresponding lower-order mutant combinations, except for some combinations involving cel6-1, man7-3, and adpg2-1. Our results demonstrate that the ability of the silique to dehisce can be manipulated to different degrees by altering the activities of various cell wall-modifying enzymes.
Cellulose, hemicellulose, and pectin are common components of plant cell walls (Keegstra, 2010) that need to be modified or degraded during cell differentiation and organ abscission and dehiscence in plants. Cellulases, hemicellulases, and pectinases in plants are responsible for the modification or degradation of the three cell wall components, respectively. Although the biochemical reactions catalyzed by these enzymes are generally understood, knowledge of their biological functions in developmental processes is limited. Most notably, genetic studies of a combined effect of loss of function in more than one type of these enzymes on a plant developmental process have not been conducted. Such studies can yield insight into how the three types of enzymes together affect the same plant developmental process, which may be relevant to improving agriculturally important traits of crops. For example, the kinetics of ripening, abscission, and dehiscence of crop organs or seeds can conceivably be manipulated by altering the activities of one or more of these enzymes.
Plant pectinases are responsible for degrading pectin that is mostly located in the middle lamella where cell-to-cell adhesion occurs. Pectinases are expected to play a major role in cell separation during abscission and dehiscence. Indeed, multiple investigations have provided experimental evidence for the role of pectinases in abscission and dehiscence. The endo-polygalacturonase (PG; a pectinase) gene RDPG1 in Brassica napus has been found to be expressed in the fruit dehiscence zone (Petersen et al., 1996). A promoter region of this gene also drives expression of the GUS gene in abscission and dehiscence zones and the style during pollen tube growth in Arabidopsis (Arabidopsis thaliana; Sander et al., 2001). Conversely, the promoter of ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), an Arabidopsis homolog of RDPG1, drives expression in the anther and fruit dehiscence zones and the seed abscission zone in B. napus (Jenkins et al., 1999). positional sterility2, a pectinase in tomato (Solanum lycopersicum), is a homolog of ADPG1 in Arabidopsis and is required for anther dehiscence (Gorguet et al., 2009). These findings suggest functional conservation of pectinases in abscission and dehiscence in plants. In Arabidopsis, five PG genes have been found to be expressed at locations where cell separation occurs, which include the abscission zones of floral organs and the dehiscence zones in anthers and siliques (González-Carranza et al., 2007). Loss of function in ADPG1 caused delayed shedding of Arabidopsis floral organs (González-Carranza et al., 2007). Ogawa et al. (2009) reported that the three closely related Arabidopsis PGs, ADPG1, ADPG2, and QRT2, are involved in multiple cell separation events in reproductive organs, with ADPG1 and ADPG2 being essential for silique dehiscence. Ogawa et al. (2009) also showed that the expression of ADPG1 and ADPG2 in the silique dehiscence zone and the seed abscission zone, and the expression of ADPG1 in these two cell separation zones, depends on the transcription factors INDEHISCENT (IND) and HECATE3, respectively. IND is known as a regulator of cell division and differentiation in the dehiscence zone in the silique and is required for silique dehiscence (Liljegren et al., 2004; Wu et al., 2006; van Gelderen et al., 2016). In tomato, expression of genes encoding pectinases in the pedicel abscission zone depends on the MADS-box transcription factors J, MC, and SIMBP21, and a similar mechanism is predicted to occur in apple (Malus domestica; Nakano et al., 2015). Parallel to their roles in abscission and dehiscence, pectinases also participate in fleshy fruit ripening (García-Gago et al., 2009; Roongsattham et al., 2012; Fabi et al., 2014).
Plant cellulases degrade cellulose, which is a major component of the cell wall. Cellulases are also known to be associated with abscission and dehiscence processes in many plant species (Abeles, 1969; Lashbrook et al., 1994; del Campillo and Bennett, 1996; Gonzalez-Bosch et al., 1997; Trainotti et al., 1997; Lane et al., 2001; Du et al., 2014). Along with the pectinase genes, expression of a cellulase gene in the pedicel abscission zone is also dependent on the J, MC, and SIMBP21 transcription factors in tomato (Nakano et al., 2015). Genetic evidence shows that the cellulase encoded by the RSW2 gene functions in anther dehiscence in Arabidopsis (Lane et al., 2001). Like pectinases, cellulases also act in fleshy fruit ripening (Christoffersen et al., 1984; Lashbrook et al., 1994; Harpster et al., 1997).
In addition to the above-mentioned functions in late developmental processes, pectinases and cellulases also act in cell differentiation in early developmental processes. The pectinase ZePG1 is localized on the secondary wall thickenings of differentiating tracheary elements and phloem regions in Zinnia elegans, suggesting a role for ZePG1 in the differentiation of tracheary elements and other cells (Nakashima et al., 2004). A tomato pectinase is also predicted to function in vascular tissue differentiation in addition to its role in seed germination (Sitrit et al., 1999). Expression of the tomato cellulase, Cel4, is correlated with rapid cell expansion in pistils, hypocotyls, and leaves (Brummell, et al., 1997). Similarly, the Arabidopsis cellulase KORRIGAN is also correlated with rapid cell elongation, and the korrigan mutant is an extreme dwarf with pronounced alterations in the primary cell wall, revealing that cellulose modifications by KORRIGAN is coupled to cellulose synthesis in the cell wall (Nicol et al., 1998; Lane et al., 2001). Interestingly, the pectin composition in the cell wall in korrigan is affected even though KORRIGAN is not expected to directly affect pectin metabolism (His et al., 2001). Homologs of KORRIGAN also participate in cellulose formation in the secondary wall in Populus (Yu et al., 2014). Moreover, another xylem-specific cellulase in Populus is required for normal secondary cell wall formation in the xylem (Yu et al., 2013).
Because hemicellulose is another common component of plant cell walls (Heredia et al., 1995), plant hemicellulases are expected to play roles similar to those of cellulases and pectinases in plant development. However, available experimental evidence showing the functions of plant hemicellulases in plant development is very limited. To our knowledge, to date, the biological functions of plant hemicellulases have been established only for the seed germination process (Iglesias-Fernández et al., 2011, 2013; Martínez-Andújar et al., 2012).
Twenty-five cellulase genes (Urbanowicz et al., 2007) and eight mannanase (enzyme catalyzing the degradation of mannan, a type of hemicellulose molecule) genes (Yuan et al., 2007) have been predicted to exist in the Arabidopsis genome. None of these genes has been shown to act in a dehiscence process in Arabidopsis. Here, we show that CELLULASE6 (CEL6) and MANNANASE7 (MAN7) likely indirectly affect the timing of cell differentiation in the silique valve and promote silique dehiscence by facilitating cell disintegration in the separation layer. We also show that loss-of function mutations of CEL6, MAN7, ADPG1, and ADPG2 reduce the ability of the silique to dehisce in an additive fashion. Our results may be useful for engineering crops with desired dehiscence kinetics.
RESULTS
CEL6 and MAN7 Are Expressed in the Silique and Their Expression Is Partially Dependent on IND and ALC
To identify genes that encode uncharacterized cell wall-degrading enzymes that function in silique dehiscence in Arabidopsis, we first conducted in silico searches (see “Methods”). This effort resulted in the identification of 39 cellulase and other cell wall-degrading enzyme genes (Supplemental Table S1). To determine which of these genes are expressed at a relatively high level in late silique development, we searched their mean-normalized expression levels in siliques with seeds containing late heart to midtorpedo embryos using the AtGenExpress Visualization Tool (http://jsp.weigelworld.org/expviz/expviz.jsp?experiment=development&normalization=normalized&probesetcsv). The chosen stage is the latest available for microarray experiments with silique tissue, which should fall within flower development stage 17 (Smyth et al., 1990; Le et al., 2010). We decided to select the genes that have a mean-normalized expression level of >1 at this stage, which yielded 12 genes (Supplemental Table S2). We next determined which of the 12 genes are coexpressed with the transcription factor gene ALCATRAZ (ALC) in ATTED-II (http://atted.jp/), because ALC is a basic helix-loop-helix (bHLH) transcription factor that positively regulates silique dehiscence (Rajani and Sundaresan, 2001). Only 2 of the 12 genes, AT4G39010 and MAN7 (AT5G66460), were found to be within the first 1,000 genes coexpressed with ALC. AT4G39010 encodes a cellulase (Urbanowicz et al., 2007) and MAN7 encodes a hemicellulase. For convenience and clarity, hereafter, AT4G39010 is named CEL6, following the previously designated CEL1-5 (Shani et al., 1997; Yung et al., 1999; del Campillo et al., 2004; Urbanowicz et al., 2007).
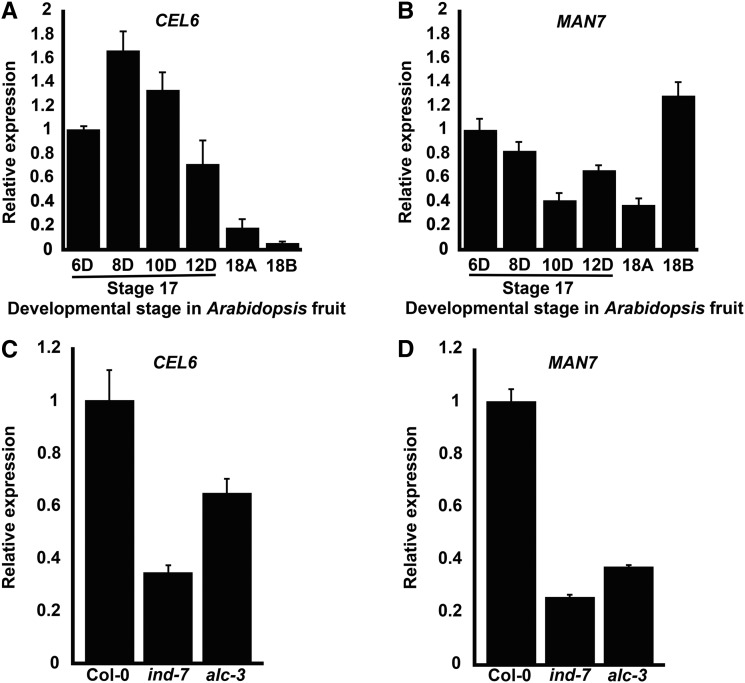
To confirm and further characterize the expression of the above identified genes, we conducted RT-qPCR experiments for the transcripts of CEL6 and MAN7 during stages 17 and 18 of floral development in the wild type (Col-0). Stage 17 starts when all the floral organs (not including the carpels) shed and ends with the silique turning yellow, and stage 18 is marked by the yellow and yet not dehisced silique (Smyth et al., 1990). In this investigation, we further divide stage 17 into 17A (∼4–6 d after anthesis) and 17B (∼6–12 d after anthesis), stage 18 into 18A (light yellow) and 18B (yellow), and stage 19 into 19A (initial dehiscence) and 19B (dehiscence extended from an initial dehiscence area; Supplemental Fig. S1). CEL6 and MAN7 were expressed at relatively high levels throughout stage 17, and MAN7 even attained its highest expression level at stage 18B (Fig. 1, A and B). We therefore focused on CEL6 and MAN7 in subsequent investigation for their likely relevance to silique dehiscence based on the expression results.
Figure 1.
RT-qPCR analysis of CEL6 and MAN7 expression in siliques of Col-0 and the ind-7 and alc-3 mutants. A, CEL6 expression at different developmental stages of Col-0. B, MAN7 expression at different developmental stages of Col-0. C, CEL6 expression at stage 17B in Col-0 and the ind-7 and alc-3 mutant. D, MAN7 expression at stage 17B in Col-0 and the ind-7 and alc-3 mutant. 6D-12D in A and B and similar designations in subsequent figures denote the number of days after anthesis. Shown in each plot are average relative expression levels ± se with the value of the first bar on the left being 1.
We next determined the expression levels of CEL6 and MAN7 in late stage-17 (10 d after anthesis) siliques of Col-0, the ind-7 mutant, and the alc-3 mutant to test if their expression is regulated by IND and ALC. IND is also a bHLH transcription factor and a major regulator of silique dehiscence (Liljegren et al., 2004; Wu et al., 2006; van Gelderen et al., 2016), but it was not included in the earlier coexpression screening for silique dehiscence-related wall-degrading enzyme genes because it is absent in the microarray data due to its highly specific expression in a small number of cells in the dehiscence zone. The mutants ind-7 (SALK_058083) and alc-3 (SALK_103763) are newly identified alleles that each exhibited an indehiscent phenotype similar to those of previously reported ind and alc alleles (Liljegren et al., 2004; Wu et al., 2006; Rajani and Sundaresan, 2001). RT-qPCR studies indicated that ind-7 and alc-3 were null or nearly null alleles (Supplemental Fig. S2). We chose ind-7 and alc-3 because they and other plant lines used in this investigation were in the Col-0 background, which provided a condition for consistent comparisons of phenotypes and gene expression levels. The transcript levels of CEL6 in ind-7 and alc-3 were ∼30% and 60% of the wild-type level, respectively (Fig. 1C). The transcript levels of MAN7 in ind-7 and alc-3 were ∼25% and 35% of the wild-type level, respectively (Fig. 1D). These results indicate that IND and ALC positively regulate the expression of CEL6 and MAN7 in late stage-17 siliques, validating and extending the in silico findings that CEL6 and MAN7 are expressed in stage-17 siliques and coexpressed with ALC.
Expression Domains of CEL6 and MAN7 Largely Overlap in Vegetative and Reproductive Organs and Shift in Location during Silique Development
To investigate whether CEL6 and MAN7 are expressed in tissues that are relevant to silique dehiscence, we generated transgenic lines harboring either the pCEL6:GUS transgene or pMAN7:GUS transgene. In the seedlings of the pCEL6:GUS lines, the GUS signal was detected throughout the cotyledons and first leaf, but the signal was increasingly restricted to the margins and vascular tissue in later formed leaves (Supplemental Fig. S3, A–E). The GUS signal was in the root and root primordia except the mature root tip, and the junction region between the root and the hypocotyl is highly stained for the GUS signal (Supplemental Fig. S3, F–H). The GUS signal was also in parenchyma cells of seed coat origin (Supplemental Fig. S3I). In the inflorescence, the GUS signal was in all floral organs and pollen, and in young siliques (stages 13–16) the GUS signal was mostly in the basal region and the stigma-style region (Supplemental Figs. S3, J–L, and S4, A and B). In stage-17 siliques, the GUS signal appeared to diminish (Supplemental Fig. S4, C–F). However, in stage-18 siliques, new GUS signal appeared mostly in the region encompassing the replum and the valve margins and in the funiculi (Fig. 2, A and B). The patterns of GUS signal in the pMAN7:GUS plants were almost identical to those in the pCEL6:GUS plants (Supplemental Fig. S5; Fig. 2, I and J), except that the signal was also observed in the root cap (Supplemental Fig. S5F).
Figure 2.
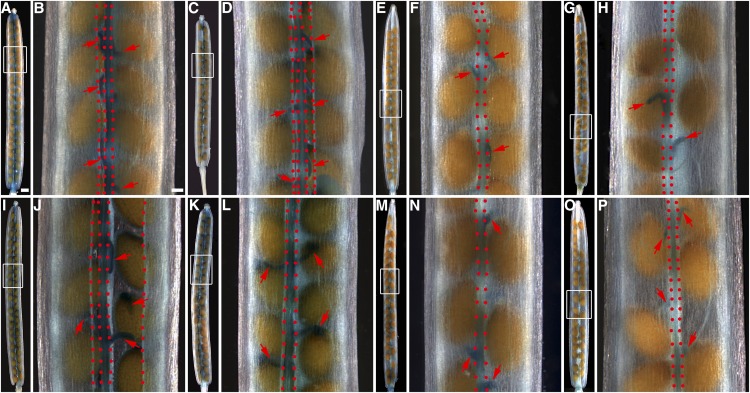
GUS staining patterns in siliques of the GUS-promoter and GUS-protein fusion lines of the CEL6 and MAN7 genes in the wild-type, ind-7, and alc-3 backgrounds. A and B, Siliques of a pCEL6:GUS line. C and D, Silique of a pCEL6:CEL6-GUS line. E and F, Silique of a pCEL6:GUS line in the ind-7 mutant background. G and H, Silique of a pCEL6:GUS line in the alc-3 mutant background. I and J, Silique of a pMAN7:GUS line. K and L, Silique of a pMAN7:MAN7-GUS line. M and N, Silique of a pMAN7:GUS line in the ind-7 mutant background. O and P, Silique of a pMAN7:GUS line in the alc-3 mutant background. B, D, F, H, J, L, N, and P, Higher magnification images of the rectangular areas in the images immediately to the left, respectively. Red arrows indicate funiculi. In dehisced siliques (B, D, and J), vertically aligned red dots delineate the replum (right two dotted lines in B or two central dotted lines in D and J) and mark the freed valve margins. In nondehisced siliques (F, H, L, N, and P), the dotted lines mark the junction areas between the valves and the replum. Bar in A = 500 μm for A, C, E, G, I, K, M, and O and bar in B = 100 μm for B, D, F, H, J, L, N, and P.
To further validate the expression patterns of CEL6 and MAN7, we created Arabidopsis lines that expressed the CEL6-GUS fusion protein by the pCEL6:CEL6-GUS transgene or the MAN7-GUS fusion protein by the pMAN7:MAN7-GUS transgene. GUS staining of these lines showed that these fusion proteins were expressed in patterns overall similar to those observed with their promoter-GUS lines (Supplemental Figs. S3–S7; Fig. 2, A–D and I–L). The GUS signal was weaker and more restricted to certain regions in some of the vegetative and reproductive organs but stronger in the style before stage 17 in the pCEL6:CEL6-GUS lines than in the pCEL6:GUS lines (Supplemental Figs. S3, S4, A–L, and S6). A similar trend was observed in the vegetative organs for the pMAN7:MAN7-GUS and pMAN7:GUS lines, with the GUS signal being weaker in the former lines, except that the GUS signal was stronger in the vasculature in the leaves of pMAN7:MAN7-GUS plants than in those of pMAN7:GUS plants (Supplemental Figs. S5, A–I, and S7, A–I). The GUS signals in the reproductive organs appeared to be at similar or higher levels in the pMAN7:MAN7-GUS lines compared with those in the pMAN7:GUS lines (Supplemental Figs. S4, M–X, S5, J–L, and S7, J–L; Fig. 2, I–L). Compared with the pMAN7:GUS lines, at stages 15 and 16, the GUS signal in the pMAN7:MAN7-GUS lines extended from the base and the style of the silique into the valves, but the valves still appeared to be free of the signal along most of their lengths. In stage-18 siliques, for both CEL6 and MAN7, the GUS signals were overall similar in intensity and location between the promoter-fusion and protein-fusion lines (Fig. 2, A–D and I–L). Semithin sections of GUS-stained stage-17B siliques of the four types of transgenic lines further showed that the GUS signals were mostly in the valve margins and the replum (Supplemental Fig. S8). These results indicate that the expression domains of CEL6 and MAN7 largely overlap in vegetative and reproductive organs and developmentally shift in location in the silique.
Expression of CEL6 and MAN7 Is Reduced in Late Silique Development in ind-7 and alc-3
Because IND and ALC are specifically expressed in the valve margin in late silique development (Liljegren et al., 2004; Wu et al., 2006; Rajani and Sundaresan, 2001) and the RT-qPCR results showed that the expression of CEL6 and MAN7 are partially dependent on IND and ALC, we tested whether the reduced expression of CEL6 and MAN7 occurs in the valve margin in the ind-7 and alc-3 mutants. We introduced the pCEL6:GUS or pMAN7:GUS constructs into these mutants by crossing and GUS-stained stage-18 siliques from the mutant plants containing one of the two transgenes. The results show that the GUS signal from either pCEL6:GUS or pMAN7:GUS was absent or at a very low level in the valve margins but retained in the replum region and the funiculi in ind-7 (Fig. 2, A, B, E, F, I, J, M, and N). Similar results occurred with the GUS signals in alc-3 (Fig. 2, A, B, G, H, I, J, O, and P). These results are consistent with the results in Figure 1, C and D, suggesting that IND and ALC positively regulate the expression of CEL6 and MAN7 in the valve margin.
Identification of the cel6 and man7 Mutants
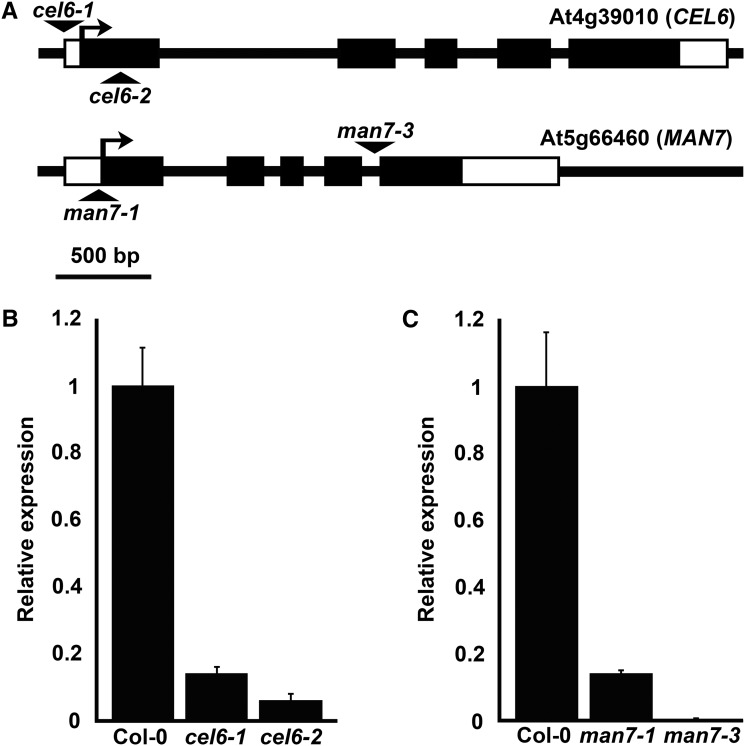
To investigate the functions of CEL6 and MAN7 in silique development, we obtained two T-DNA-insertion mutant alleles for each of the two genes (see also “Methods”). By conducting PCRs to detect the presence of a T-DNA insertion, we confirmed the locations of the T-DNA insertions in these mutants as described in TAIR (Supplemental Fig. S9). The cel6-1 mutant (SALK_060505C) had a T-DNA insertion at a position 86 bp upstream of the start codon of CEL6, and cel6-2 (WiscDsLox485-488K15) had a T-DNA insertion in the first exon of CEL6 (Fig. 3A). The man7-1 mutant (GABI_747H02), which was previously characterized as a mutant defective in seed germination (Iglesias-Fernández et al., 2011), has a T-DNA insertion in the 5′-UTR (12 bp from the start codon) of MAN7, and the man7-3 mutant (SAIL_424_H03) has a T-DNA insertion in the fourth intron of MAN7 (Fig. 3A). We then investigated the transcript levels of the two genes in stage-17 siliques of the mutants and the Col-0 control by RT-qPCR. Results from this investigation showed that the transcript levels of CEL6 and MAN7 in their respective mutants were ∼14% (in cel6-1), 6% (in cel6-2), 14% (in man7-1), and 0.2% (in man7-3) of the wild-type level (Fig. 3, B and C), suggesting that they were null or nearly null alleles.
Figure 3.
Identification of the cel6-1, cel6-2, man7-1, and man7-3 mutants. A, Positions of the T-DNA insertions in At4g39010 (CEL6) and At5g66460 (MAN7). Black boxes and the lines between them represent exons and introns, respectively. Open boxes represent the predicted 5′- and 3′-untranslated regions. Triangles indicate the positions of the T-DNA insertions. B, Average relative expression levels (±se) of CEL6 in stage-17 siliques of Col-0 and the cel6-1 and cel6-2 mutants. The Col-0 expression level is defined as 1. C, Average relative expression levels (±se) of MAN7 in stage-17 siliques of Col-0 and the man7-1 and man7-3 mutants. The Col-0 expression level is defined as 1.
Cell Differentiation in the Dehiscence Zone Is Delayed in Early Stage 17 Siliques of cel6 and man7 Mutants
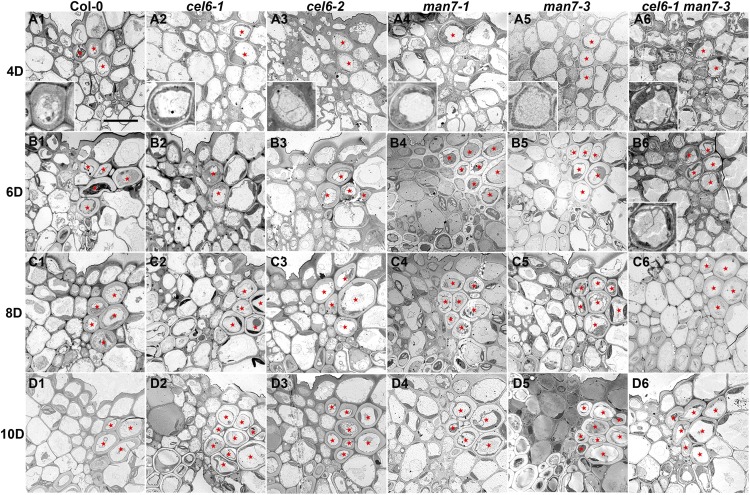
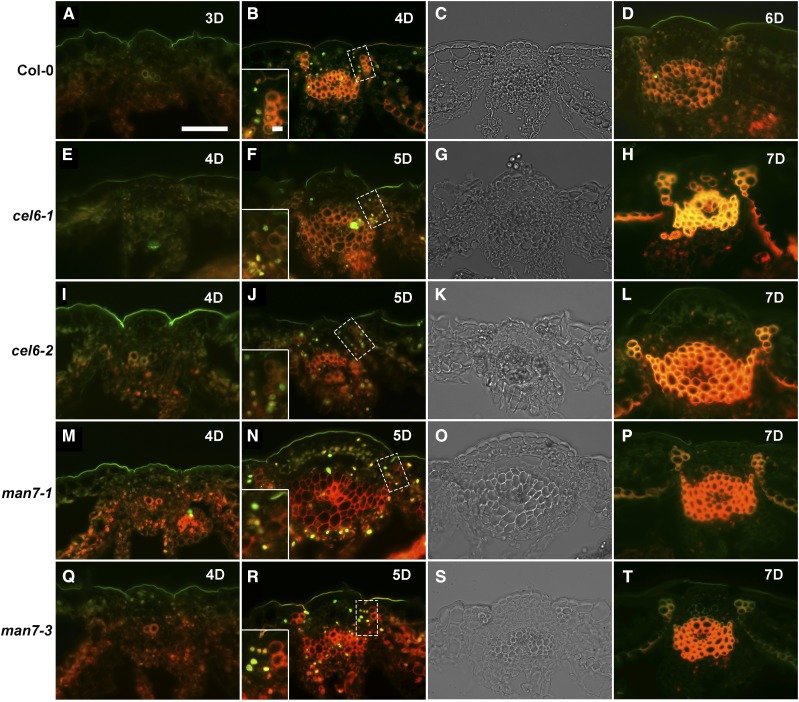
In the process of characterizing the phenotypes of the cel6 and man7 mutants, we observed that silique development in these mutants was delayed compared to the wild type. In particular, progression of silique development from stage 15 to stage 18A in Col-0, on average, took 14 d, while the same developmental process was significantly delayed by 2 to 3 d in the single and double mutants (t test, P < 10−36, n = 50). Because most of the silique elongation and lateral enlargement occurs in stage 17 and our initial goal of the research was to determine the roles of CEL6 and MAN7 in silique dehiscence, we first focused on the cellular features in the dehiscence zone in stage-17 siliques. The dehiscence zone in an Arabidopsis silique is a longitudinally narrow region where small centrally located parenchyma cells in the separation layer are flanked by a group of cells with thickened and lignified cell walls on the valve side and relatively large parenchyma cells on the replum side (Wu et al., 2006). In siliques of Col-0, 4, 6, 8, and 10 d after anthesis, the lignified cells in the dehiscence zone were readily observed (Fig. 4, A1, B1, C1, and D1). It was difficult to detect such cells in the cel6 and man7 single mutants and the cel6-1 man7-3 double mutant 4 d after anthesis as the cells in the corresponding region had thin cell walls (Fig. 4, A2–A6). Such cells in the valve margin were convincingly observed in the single mutants 6 d after anthesis (Fig. 4, B2–B5), but not in the cel6-1 man7-3 double mutant (Fig. 4B6). Only at the subsequent stages, that is 8 and 10 d after anthesis, did these cells in the double mutant appear to attain wall thicknesses similar to those in Col-0 and the single mutants at the corresponding stages (Fig. 4, C1–C6 and D1–D6). These observations indicate that mutations in these genes cause a delay in secondary wall thickening in this group of cells in the valve margin, and the double mutant had a more severe phenotype in this regard than the single mutants. CEL6 and MAN7 proteins, therefore, normally promote secondary wall thickening in this group of cells in the valve margin.
Figure 4.
Transmission electron microscopy images of transverse sections of stage-17 siliques of Col-0 and the cel6 and man7 mutants. A1 to D1, Col-0. A2 to D2, The cel6-1 mutant. A3 to D3, The cel6-2 mutant. A4 to D4, The man7-1 mutant. A5 to D5, The man7-3 mutant. A6 to D6, The cel6-1 man7-3 double mutant. A1 to A6, Siliques at stage 4D (4 d after anthesis). B1 to B6, Siliques at stage 6D. C1 to C6, Siliques at stage 8D. D1 to D6, Siliques at stage 10D. Red stars in Col-0 indicate lignified cells, and in the mutants either cells presumably destined to be lignified or lignified cells, in the dehiscence zone. Insets in A1 to A6 and B6 show enlargements of such cells in the corresponding images. Bar = 10 μm for all images (excluding the insets).
The cells of the separation layer are expected to undergo programmed cell death (PCD) during silique development. Morphological characteristics of plant cell PCD include ameboid-shaped nuclei, condensed chromatin, and cellular disorganization (Vanyushin et al., 2004; van Doorn et al., 2011; Bar-Dror et al., 2011). Ameboid-shaped nuclei and condensed chromatin near the nuclear periphery were observed in the separation layer 4 and/or 6 d after anthesis in the wild type (Supplemental Fig. S10, A–D) and the mutants (Supplemental Fig. S10, G–J and M–P). Degenerated cells without the typical cytoplasm-nuclear organization were observed in the separation layer at late stage 17 (Supplemental Fig. S10, E, F, K, L, Q, and R). To further characterize PCD in the separation layer, we conducted the terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay to detect nuclear DNA fragmentation, a hallmark of PCD (Bar-Dror et al., 2011), with sections of siliques of 3 to 13 d after anthesis. In Col-0, strong TUNEL signals were first detected in a region that overlapped with the separation layer (between the fluorescent lignified cells in the valve margin and the replum) 4 d after anthesis (Fig. 5B). Strong signals were also detected 5 d after anthesis but not at other stages in the same region in Col-0 (Fig. 5, A, C and D; Supplemental Fig. S11, A–D). In the cel6-1, cel6-2, man7-1, and man7-3 mutants, strong signals were detected in the same region only 5 d after anthesis (Fig. 5, F, J, N, and R) but not at other stages (Fig. 5, E, G–I, K–M, O–Q, S, and T; Supplemental Fig. S11, E–T). The same results were obtained in at least three siliques for each genotype at each stage. Thus, PCD in the separation layer starts at early stage 17 in the wild type and the mutants and is delayed in the mutants. In Figure 5, the fluorescence of the lignified cells in the valve margin apparently resulted from the thickened secondary cell walls. Such fluorescent cell walls first appeared in Col-0 siliques four days after anthesis (Fig. 5B), whereas in the siliques of the mutants it seemed to first weakly appear 5 d after anthesis (Fig. 5, F, J, N, and R). The delayed occurrence of the fluorescent cell walls in the valve margin is consistent with the delayed secondary wall thickening in these cells described earlier (Fig. 4). The effects of the mutations on the delay of cell differentiation in the dehiscence zone were observed in cross sections at different locations along the longitudinal axis of the siliques. As reported earlier, the expression of CEL6 and MAN7 in the silique is only in the apical (including the stigma, style, and adjacent fruit wall) and basal regions prior to and during stage 17 (Supplemental Fig. S4). The observed developmental delay in the dehiscence zone thus likely indirectly results from the loss of function in CEL6 or MAN7.
Figure 5.
TUNEL signals in silique cross sections of Col-0 and the cel6-1, cel6-2, man7-1, and man7-3 mutants. All images are merged TUNEL (green) and PI (red; nuclear stain) fluorescence images, except for C, G, K, O, and S, which are bright-field images corresponding to images immediately to the left, respectively. Insets show enlargements of the rectangular areas marked in the corresponding images. Bar in the inset in B = 5 µm for all the insets, and bar in A = 50 μm all the other images.
Cell Morphology and Integrity in the Separation Layer in Late Silique Development Are Altered in cel6 and man7 Mutants
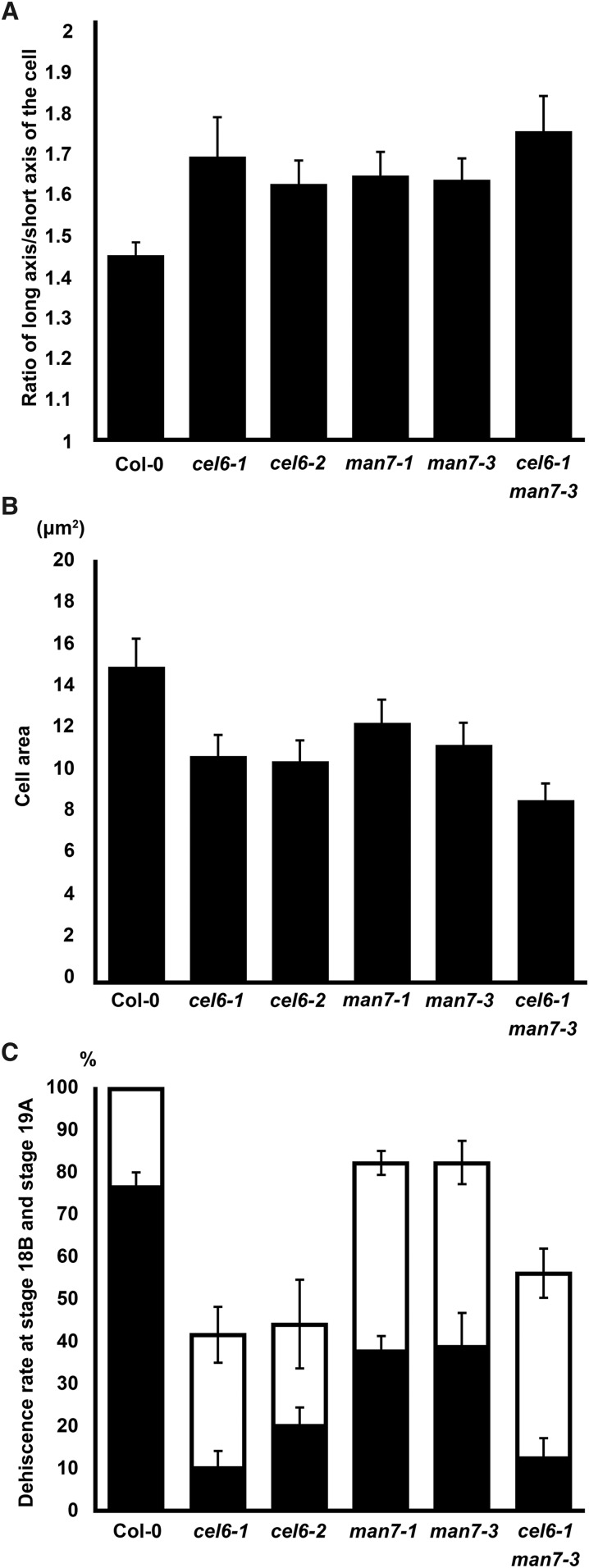
Because CEL6 and MAN7 are expressed in the dehiscence zone in late silique development (Fig. 2, A–D and I–L), we predicted that CEL6 and MAN7 participate in the degradation of cell walls in the separation layer. Indeed, cell walls in the separation layer in the wild type appeared to be weakened toward late stage 17 as the walls were wavy and the neighboring cells were increasingly interdigitated (Supplemental Fig. S10, C–E), in contrast with the lack of such changes in the mutants even at late stage 17 (Supplemental Fig. S10, I–K and O–Q). The cell size and shape in the separation layer also appeared to differ between the wild type and the mutants, presumably due to the difference in cell wall rigidity between them when the cells were under stress and/or strain. We thus measured the cell lengths and widths in the separation layer in TEM images of late stage-17 siliques in the wild type and the mutants, and calculated the length-to-width ratio and the area (product of the length and width) of each measured cell. The ratio and area were deemed to be indicators of the cell shape and size, respectively. The average length-to-width ratio of Col-0 siliques was 1.44 ± 0.04 (se), which was significantly smaller than the averages (between 1.62 and 1.75 with standard errors ranging from 0.05 to 0.1) of the cel-6 and man-7 single mutants and the cel6-1 man7-3 double mutant (Fig. 6A; t test, P ≤ 0.02). The average cell area of Col-0 was 15.0 ± 1.3 µm2, which was significantly larger than those of cel6-1 (10.7 ± 1.1 µm2), cel6-2 (10.5 ± 1.0 µm2), man7-3 (11.2 ± 1.1 µm2), and cel6-1 man7-3 (8.6 ± 0.8 µm2; Fig. 6B; t test, P ≤ 0.03), although it was not significantly different from that of man7-1 (12.3 ± 1.1 µm2; Fig. 6B; t test, P = 0.13). These results indicate that the mutations altered the cell shape and size in the separation layer in the single and double mutants. The greater effect of the mutations on the cell size in the double mutant compared to that in the single mutants suggests that the mutations in the two genes are additive in affecting cell morphology in the separation layer.
Figure 6.
Silique phenotypes in Col-0 and the cel6 and man7 mutants. A, Average ratios (±se) of long axis to short axis of cells in the separation layer. B, Average cell areas (±se) of cells in the separation layer. C, Average dehiscence rates (±se) of two consecutive siliques during the stage 18B to 19A transition. Black bars are of the younger siliques and open bars of the older siliques.
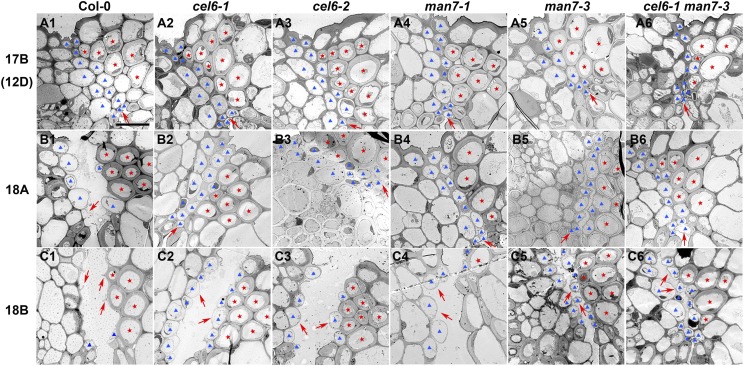
To obtain additional evidence that the mutations affect separation layer in late silique development, we further investigated cell morphology in the separation layer in late stage-17 and stage-18 siliques in the wild type and mutants. We first noticed that the cells in the separation layer were not as organized and distinct in the Col-0 accession as in the Ler accession (Fig. 7; Wu et al., 2006). The cells in the separation layer near the inner epidermis were disintegrating at late stage 17 in Col-0 (arrow, Fig. 7A1), but such cells remained intact in all the single mutants and the double mutant at the corresponding stage (Fig. 7A2–7A6). At stage 18A, the siliques of Col-0 were usually easily shattered along the separation layer during specimen preparation for the TEM study, although most of the cells in separation layer, except the small cells near the inner epidermis, were still intact (Fig. 7B1). The siliques of the single mutants and the double mutant usually remained indehiscent during specimen preparation for the TEM study, and all cells in the separation layer remained intact (Fig. 7B2–7B6). Cells in the separation layer in Col-0 all seemed broken at stage 18B in the sections (Fig. 7C1). By contrast, most, if not all, cells of the separation layer in the single mutants and the double mutant remained intact at stage 18B (Fig. 7C2–7C6). These observations suggest that cell walls in the separation layer in the mutants were stronger than those in the wild type, preventing the bursting of these cells during the sample preparation process. Therefore, CEL6 and MAN7 are involved in the degradation of cell walls in the separation layer during silique maturation.
Figure 7.
Transmission electron microscopy images of transverse sections of stage 17B to stage 18 siliques of Col-0 and the cel6 and man7 mutants. A1 to C1, Col-0. A2 to C2, The cel6-1 mutant. A3 to C3, The cel6-2 mutant. A4 to C4, The man7-1 mutant. A5 to C5, The man7-3 mutant. A6 to C6, The cel6-1 man7-3 double mutant. A1 to A6, Siliques at stage 17B (12D). B1 to B6, Siliques at stage 18A. C1 to C6, Siliques at stage 18B. Red stars indicate lignified cells in the dehiscence zone. Blue triangles indicate presumed separation layer cells. Arrows in Col-0 indicate degenerating cells or wall stubs from degenerated cells in the separation layer, and arrows in the mutants indicate intact separation layer cells. Bar = 10 μm for all images.
Silique Dehiscence Is Impaired in cel6 and man7 Mutants
The siliques of the cel6 and man7 single and double mutants could dehisce, but their dehiscence appeared to occur at lower percentages (dehiscence rates) than the siliques of Col-0 during the transition from stage 18B to stage 19. To quantify these differences, we investigated the dehiscence rates of two consecutive siliques on the same inflorescence stem for the mutant and Col-0 samples. The younger one of these two siliques was at a stage between 18B and 19A (14 and 16 d after anthesis for Col-0 and the mutant lines, respectively), that is a still-hydrated yellow silique that might or might not have dehisced (Supplemental Fig. S12). By choosing these consecutive siliques in this developmental window, we likely captured siliques that were undergoing initial dehiscence, thus revealing how easily siliques dehisced on a plant. The chosen siliques were subject to careful examination under a dissecting scope to determine if they dehisced. Of the Col-0 plants, the average dehiscence rates of siliques at these two slightly different stages were 76% and 100%, respectively (Fig. 6C). The average dehiscence rates of the younger siliques of the single and double mutants ranged from 10% to 39%, and the average dehiscence rates of the older siliques 42% to 82%, significantly less than those of Col-0, respectively (t test, P < 10−5; Fig. 6C). The average dehiscence rates of the younger and older siliques of the cel6 mutants did not statistically differ from those of the double mutant, but they were significantly lower than those of the man7 mutants, respectively (t test, P < 0.02; Fig. 6C). These results indicate that the cel6 and man7 mutations impaired silique dehiscence, and the cel6-1 mutation was epistatic to the man7-3 mutation with respect to the dehiscence rate.
To further investigate the functions of CEL6 and MAN7, we overexpressed CEL6 or MAN7 in Col-0 using the cauliflower mosaic virus 35S promoter. We observed that overexpression of CEL6 and MAN7 either did not promote or only moderately promoted silique dehiscence in the transgenic lines (Supplemental Fig. S13), indicating that CEL6 and MAN7 are not major limiting factors for silique dehiscence in wild-type Arabidopsis.
The Silique Indehiscent Phenotypes of adpg1-1 and adpg1-1 adpg2-1 Are Enhanced by the cel6-1 and man7-3 Mutations
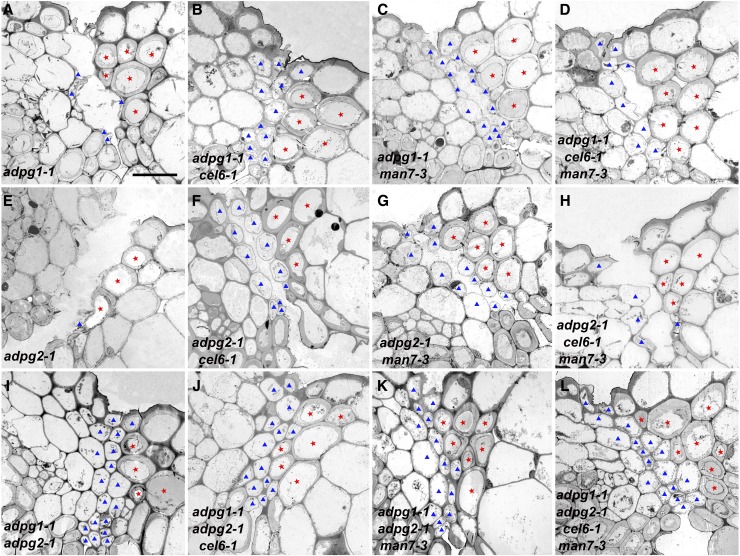
A previous investigation indicated that the pectinases ADPG1 and ADPG2 play a major and minor role, respectively, in silique dehiscence in Arabidopsis (Ogawa et al., 2009). Our investigation demonstrates that a cellulase (CEL6) and a hemicellulase (MAN7) also participate in silique dehiscence. To compare the effects of loss of function in one or more of these genes on silique dehiscence, all possible double, triple, and quadruple mutant lines of these four loci were generated. TEM characterization of cell morphology in the region encompassing the valve margin and the replum in stage-18B siliques was conducted. The adpg1-1 mutant siliques underwent incomplete dehiscence with collapsed or broken cells in the separation layer in the prepared samples (Fig. 8A). The adpg2-1 exhibited complete dehiscence with collapsed or broken cells in the separation layer (Fig. 8E). These observations are consistent with adpg1-1 and adpg2-1 having a severe indehiscent phenotype and a mild indehiscent phenotype, respectively. In both these mutants, cell degeneration occurred at the separation layer. The cel6-1 adpg1-1 double mutant was indehiscent with mostly intact cells at the separation layer (Fig. 8B). The man7-3 adpg1-1 double mutant exhibited cellular morphology similar to that in adpg1-1, but it appeared to have more intact cells in the separation layer than adpg1-1 did (Fig. 8, A and C). The cel6-1 man7-3 adpg1-1 triple mutant appeared similar in cell morphology at the separation layer to cel6-1 adpg1-1 (Fig. 8, B and D). These observations suggest that the effects of the adpg1-1, cel6-1, and man7-3 mutations on the cells in the separation layer were additive, with adpg-1 being primarily defective in cell separation at the middle lamella and cel6-1 and man7-3 being primarily defective in cell lysis. It is also likely that the CEL6 protein plays a greater role in cell lysis at the separation layer than MAN7 does in the normal silique dehiscence process, given the different severity levels of cell disintegration shown in Figure 8, B and C.
Figure 8.
Transmission electron microscopy images of transverse sections of stage 18B siliques in single, double, triple, and quadruple mutants. A, The adpg1-1 mutant. B, The cel6-1 adpg1-1 double mutant. C, The man7-3 adpg1-1 double mutant. D, The cel6-1 man7-3 adpg1-1 triple mutant. E, The adpg2-1 mutant. F, The cel6-1 adpg2-1 double mutant. G, The man7-3 adpg2-1 double mutant. H, The cel6-1 man7-3 adpg2-1 triple mutant. I, The adpg1-1 adpg2-1 double mutant. J, The cel6-1 adpg1-1 adpg2-1 triple mutant. K, The man7-3 adpg1-1 adpg2-1 triple mutant. L, The cel6-1 man7-3 adpg1-1 adpg2-1 quadruple mutant. Red stars indicate lignified cells in the dehiscence zone. Blue triangles indicate presumed broken or intact separation layer cells. Bar = 10 μm for all images.
Similar to the adpg2-1 single mutant, siliques of the double and triple mutants involving adpg2-1, cel6-1, and man7-3 all dehisced, but they showed intact cells in the separation layer in the processed samples (Fig. 8, F–H). Siliques of all the other triple and quadruple mutants in Figure 8 did not dehisce and contained intact cells in the separation layer in the processed samples (Fig. 8, I–L), further demonstrating that mutations in the pectinases and the cellulase and hemicellulase primarily affect cell separation at the middle lamella and cell lysis in this region, respectively.
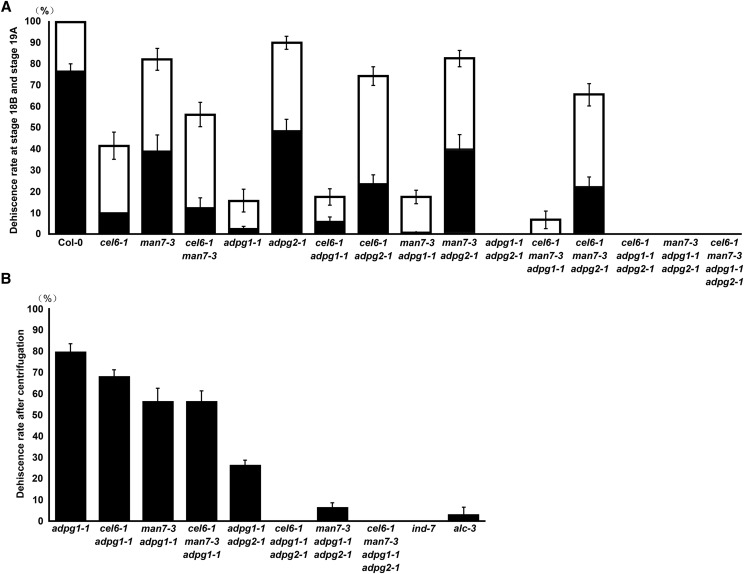
We also compared the silique dehiscence rates during the stage 18B-19A transition in the above lines. The adpg1-1 mutant had very low dehiscent rates for the two consecutive siliques on the same branch, which were not significantly different from those of the double mutants cel6-1 adpg1-1 and man7-3 adpg1-1, and the triple mutant cel6-1 man7-3 adpg1-1 (Fig. 9A; t test, P > 0.05). The double mutant cel6-1 adpg2-1 had lower and higher dehiscent rates than the adpg2-1 mutant and cel6-1 mutant, respectively (Fig. 9A; t test, P ≤ 0.01). The average dehiscence rates of the double mutant man7-3 adpg2-1 did not differ significantly from those of the man7-3 and adpg2-1 single mutants (Fig. 9A; t test, P > 0.14). These results did not clearly indicate whether cel6-1 or man7-3 further enhanced the silique indehiscent phenotype caused by the adpg1-1 mutation. On the other hand, cel6-1, but not man7-3, seemed to enhance the silique indehiscent phenotype caused by the adpg2-1 mutation.
Figure 9.
Average dehiscence rates (±se) in the single, double, triple, and quadruple mutants. A, During the stage 18B to 19A transition. Black bars are of the younger siliques and open bars of the older siliques. B, Stage-19A siliques of naturally indehiscent or nearly indehiscent genotypes after the centrifugation impact. DR, average dehiscence rate.
Even though the siliques of the cel6-1 adpg1-1 and man7-3 adpg1-1 double mutants and the cel6-1 man7-3 adpg1-1 triple mutant all had low dehiscence rates, and the siliques of the adpg1-1 adpg2-1 double mutant, the cel6-1 adpg1-1 adpg2-1 and man7-3 adpg1-1 adpg2-1 triple mutants, and the quadruple mutants were all indehiscent (Fig. 9A), they might still differ in their ability to dehisce under a mechanical force. To test this prediction, we subjected yellow and dried siliques of these genotypes, adpg1-1, and the indehiscent mutants ind-7 and alc-3 to a centrifugation force (18,000 g) for 1 min in microcentrifuge tubes, and then investigated their dehiscence rates. The results showed that, on average, ∼80% of the siliques of adpg-1-1 dehisced after the centrifugation, which was significantly higher than those of cel6-1 adpg1-1, man7-3 adpg1-1, and cel6-1 man7-3 adpg1-1 (Fig. 9B; t test, P < 0.04). On average, ∼27% of the siliques of adpg1-1 adpg2-1 also dehisced after the centrifugation, which was significantly higher than those of man7-3 adpg1-1 adpg2-1 (Fig. 9B; t test, P < 10−4). The siliques of the cel6-1 adpg1-1 adpg2-1 triple mutant, the quadruple mutant, and the ind-7 mutant remained indehiscent after the centrifugation, while the siliques of the alc-3 mutant had an average dehiscence rate of ∼3% (Fig. 9B). These results revealed that the cel6-1 and man7-3 mutations enhanced the silique indehiscent phenotypes caused by the adpg1-1 mutation and the adpg1-1 adpg2-1 double mutations. There results also showed that the ind-7 and alc-3 mutants exhibited stronger silique indehiscent phenotypes than any of the single and double mutants and two of the triple mutants of the cell wall-degrading enzyme genes (Fig. 9B).
DISCUSSION
Multiple Classes of Cell Wall-Degrading Enzymes Participate in the Same Developmental Processes
Cell wall modifications are an integral part of plant development. Multiple components of the cell wall are likely modified during development, and each modification is catalyzed by a distinct class of cell wall-degrading enzyme. In this investigation, we found that cel6 and man7 mutants exhibited similar defects in silique development and dehiscence, and their expression patterns are also similar in vegetative and reproductive organs. These observations strongly suggest that cellulases and hemicellulases participate in the same developmental processes. Together with the findings of the roles of ADPG1 and ADPG2 in silique dehiscence (Ogawa et al., 2009), all three major classes of cell wall-degrading enzymes are now found to act in silique dehiscence. It will be interesting to explore whether the three classes of enzymes also modify cell walls in other developmental processes. It may be a paradigm that two or three classes of cell wall-degrading enzymes need to act on the same cell wall in the same time window to efficiently complete the cell wall changes required for normal growth and differentiation of a cell. After all, cellulase-hemicellulase complexes are widely used by bacteria and fungi to efficiently degrade plant materials (Dollhofer et al., 2015; Bensoussan et al., 2017). Regardless of whether the enzymes form a complex, the expression of cell wall-degrading enzymes such as CEL6, MAN7, and ADPG1 is likely regulated by the same transcription factors (Ogawa et al., 2009; this investigation).
Our GUS reporters indicate that both CEL6 and MAN7 are expressed in two homologous structures, the leaf margin and the silique valve margin. It is possible that cell differentiation in the leaf margin, as in the silique margin, needs the activities of CEL6, MAN7, and possibly other cell wall-degrading enzymes. CEL6, MAN7, ADPG1, and/or ADPG2 also share similar GUS staining patterns in the seed coat, funiculus/seed dehiscence zone, leaves, and floral organs (Ogawa et al., 2009; Iglesias-Fernández et al., 2013; this article), which suggests that their expression is under the control of a common mechanism and the biological processes require the coordinated actions of the three classes of cell wall-degrading enzymes.
Dehiscence and Abscission Likely Involve Both Cell Separation and Degeneration in Arabidopsis
Cell separation must occur in all dehiscence and abscission processes. Cell separation alone, without cell degeneration, can conceivably lead to dehiscence or abscission. However, in Arabidopsis silique dehiscence, as demonstrated in this report, both cell separation and degeneration occur in the separation layer. More importantly, both cell separation and degeneration influence the ability of the silique to dehisce, even though the former plays a bigger role than the latter. Based on the observation that CEL6 and MAN7 are coexpressed in multiple dehiscence and abscission zones in Arabidopsis, it may be predicted that cell degeneration occurs in multiple dehiscence and abscission processes. In fact, cell degeneration has been found to be associated with anther dehiscence in Arabidopsis (Sanders et al., 2000). Drawn upon from the Arabidopsis silique dehiscence case, it appears that the combination of cell separation and degeneration can make a dehiscence or an abscission process occur more easily than just cell separation can.
How Do CEL6 and MAN7 Indirectly Affect Silique Development during Stage 17?
Studies of various cell wall-degrading enzymes in plants have indicated that cell wall-degrading enzymes are directly involved in cell differentiation during plant development (Brummell, et al., 1997; Nicol et al., 1998; Sitrit et al., 1999; Lane et al., 2001; Nakashima et al., 2004; Yu et al., 2013, 2014). However, CEL6 and MAN7 seem to be indirectly involved in cell differentiation in the silique dehiscence zone during stage 17, based on their spatio-temporal expression patterns in stage-17 siliques in the promoter- and protein-GUS fusion lines, and the delayed secondary wall thickening and onset of the TUNEL signals in the dehiscence zone in their mutants. Then, how may CEL6 and MAN7 indirectly exert such effects? One possible answer to this question may be that the cel6 and man7 mutations slow down pollen tube growth, which delays fertilization and subsequent cell differentiation in the silique wall. It is known that the silique does not elongate if no fertilization occurs in Arabidopsis, underscoring the dependence of postanthesis silique development on the fertilization event. CEL6 and MAN7 are expressed in the pollen, the stigma, and the style prior to and during pollination (Supplemental Figs. S3–S7). Cells at any of these locations in the mutants may negatively impact pollen tube growth. To determine whether pollen tube growth is slowed at a particular step in the pollination-fertilization process and whether fertilization and the subsequent silique development are delayed in the mutants likely requires extensive studies of in vitro and in vivo pollen tube growth, seed development, and silique wall development outside the dehiscence zone in the mutants and the wild type.
The Ability of a Plant Part to Dehisce Can Be Manipulated to Both Large and Small Degrees Depending on the Type of Genes Altered
The dehiscence rates of the mutants in this investigation vary greatly, ranging from zero even under the centrifugation condition to close to the wild-type value when not centrifuged. A pattern also emerged in these plant lines: the mutants of the transcription factors ind-7 and alc-3 showed the strongest indehiscent phenotype, followed by, in the order of increasing dehiscence rates, the adpg1-1, cel6, man7, and adpg2-1 mutants. These results are consistent with the hypothesis that IND and ALC are at the top of the hierarchy of regulation and command the expression of numerous downstream genes involved in the dehiscence process. The major pectinase for silique dehiscence, ADPG1, ought to play a bigger role in the dehiscence processes than the cellulase CEL6 and hemicellulase MAN7, since its action on the pectin-rich outer cell wall layer is expected to impact dehiscence much more than the actions of CEL6 and MAN7 on the inner cell wall layer, which is rich in cellulose and hemicellulose. Even the more severe indehiscent phenotypes of the cel6 mutants compared to those of the man7 mutants may be explained by differential contributions of the cellulose and hemicellulose components of the cell wall to maintaining cell integrity in the separation layer. Our observations of the different dehiscence rates among the lines have potential implications to engineering desired dehiscent kinetics in crop species. Crop parts harvested for food often are concerned with the dehiscence of these parts. Different levels of an indehiscent trait may be desired for different crops and/or different storage and harvesting strategies. Our results demonstrate that the ability of a plant part to dehisce can be manipulated to different degrees by targeting the activities of different proteins involved in the dehiscence process.
CONCLUSION
By analyzing mutant phenotypes and gene expression patterns, we demonstrate that CEL6 and MAN7 affect cell differentiation in the silique and contribute to silique dehiscence. We also show that the ability of the silique to dehisce is differentially affected by the loss of function in the number and types of genes involved in the process.
MATERIALS AND METHODS
In Silico Screening
“Cellulase” was first used as a keyword to query www.arabidopsis.org to find cellulase genes. Protein sequences of these genes were used to perform BLAST searches in www.arabidopsis.org and www.ncbi.nlm.nih.gov/blast/Blast.cgi to identify additional cellulase and hemicellulase genes. Further identification of CEL6 and MAN7 as candidates for cell wall-degrading enzyme genes functioning in stage-17 siliques was based on their expression levels and coexpression with ALC according to publicly available microarray data (see also “Results”).
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) lines used in this study were in the Col-0 background. All mutant lines were obtained from the Arabidopsis Biological Resource Center at The Ohio State University, except for the man7-1 and adpg1-1 mutant lines that were from Nottingham Arabidopsis Stock Centre at The University of Nottingham. Plants were grown under long-day conditions (16 h light/8 h dark) at ∼22°C in a growth room or grow chamber on artificial soil or an agar medium at pH 5.7. The agar medium consisted of 4.3 g/L Murashige and Skoog salt mixture (Gibco, Thermo Fisher Scientific), 1% (w/v) Suc, and 0.8% (w/v) agar. For transgenic plant selection, antibiotics were added to the agar medium when the medium was cooled to ∼60°C.
RNA Extraction and Analysis
RNA samples were extracted using an RNeasy Plant Mini Kit (Qiagen) or E.Z.N.A. Plant RNA Kit (Omega Bio-tek) according to the manufacturers’ protocols. The plant materials used for RNA extraction were 10 to 20 siliques at each developmental stage for each genotype. Concentrations of the RNA samples were quantified by a NanoDrop 2000C spectrophotometer (Thermo Fisher Scientific). For cDNA synthesis, a total of 1 μg RNA from each sample was first treated with DNase I to eliminate genomic DNA contamination, and then used as a template for reverse transcription (QuantiTect Reverse Transcription Kit; Qiagen). The cDNA samples were used as templates for PCR or qPCR. The primers for the PCR or qPCR are listed in Supplemental Table S3. The qPCR was performed with the QuantiTect SYBR Green PCR Kit (Qiagen) using the CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories) or LightCycler 480 Real-Time PCR System (Roche), with three biological replicates and nine technical replicates for each sample type and ACTIN2 as the internal control. Primers for ACTIN2 are GTCGTACAACCGGTATTGTG-3 and GAGCTGGTCTTTGAGGTTTC.
Confirmation of T-DNA Insertion Mutants and Generation of Mutant Combinations
Homozygous T-DNA insertion mutants were confirmed using PCR with gene-specific and T-DNA primers (Supplemental Table S3) and the PCR Master Mix Kit (Qiagen). Genomic DNA samples used in the PCR were isolated according to Murray and Thompson (1980). PCR products were also sequenced to confirm the DNA insertion sites in each mutant. Antibiotic selections of transgenic lines were also employed to check the homozygosity of T-DNA insertion mutants, with kanamycin for the SALK lines, Basta for the SAIL line, hygromycin B for the WiscDsLox line, and sulfadiazine for the GABI-Kat line. The man-7-3 mutant was confirmed with GUS staining of its flowers.
Plasmid Construction and Transformation
The promoters of CEL6 (1,956 bp) and MAN7 (857 bp) were first cloned into the pMD19-T vector (Clontech) to create pMD19T-pCEL6 and pMD19T-pMAN7. These promoter fragments were used to replace the CMV 35S promoter in the pCAMBIA-1305.1 vector to create the pCEL6:GUS and pMAN7:GUS gene constructs. The same promoter fragments and their corresponding cDNA fragments were combined using the In-Fusion Cloning Kit (Clontech), and the newly created fragments were used to replace the 35S promoter in the pCAMBIA-1305.1 vector to create pCEL6:CEL6-GUS and pCAMBIA-pMAN7:MAN7-GUS, respectively. For overexpression of CEL6 and MAN7, each of their genomic coding regions was first cloned as two separate fragments into pMD19-T to circumvent the TA-cloning size limit. The two fragments were then combined using the In-fusion cloning system to form the full-length coding region of each gene in the pCAMBIA-1305.1 vector to create the pCAMBIA-OECEL6 and pCAMBIA-OEMAN7 gene constructs. The primers used in all the cloning steps are listed in Supplemental Table S4. Completed plasmids were sequenced to ensure that the plasmids were correct. Agrobacterium tumefaciens cells with a binary vector system including one of these plasmids were used to transform Col-0 plants (Clough and Bent, 1998). Transformants were selected on agar plates containing 15 mg/L hygromycin B (AG Scientific).
GUS Staining
Seedlings, inflorescences, and siliques were incubated overnight in GUS staining solution that contained 10 mm EDTA, 0.1% (v/v) Triton X-100, 100 µg/mL chloramphenicol, 2 mg/mL 5-bromo-4-chloro-3-indoxyl-β-d-glucuronide (X-Gluc), 0.5 mm ferric cyanide, and 0.5 mm ferrous cyanide in 50 mm sodium phosphate buffer (pH 7.0). The samples were cleared several times in 70% ethanol. Six or more independent lines for each transgene showed similar GUS staining patterns, and one of them is shown in Figure 2 and Supplemental Figures S3–S7.
TUNEL Assay
Small segments of siliques were fixed in 4% (w/v) paraformaldehyde in 0.1 m PBS overnight at 4°C, followed by three washes (15 min each) in the same buffer. Specimens were dehydrated in a graded ethanol series (15, 30, 45, 60, 70, 80, 90, and 100%), washed two times (20 min each) in dimethyl benzene, and infiltrated and embedded in Paraffin. Paraffin sections of 5 to 7 μm were cut using a rotary microtome (American Optical). The TUNEL assay was performed on these sections using the DeadEnd Fluorometric TUNEL System (Promega) according to the manufacturer’s instructions. Slides were then immediately stained with 10 μg/mL propidium iodide (Sigma-Aldrich) and mounted with SlowFade Gold antifade reagent (Invitrogen). Samples were observed and photographed under a Leica DM6 B microscope equipped with the Leica DFC550 imaging system (Deng et al., 2014).
Light Microscopy and Transmission Electron Microscopy for Other Morphological Studies
For sections for both light and transmission electron microscopy, fresh siliques were first embedded in 1% (w/v) agarose and quickly cut into small segments using a razor blade. Agarose embedding helped maintain the relative positions of the valves and the replum of a dehiscent or nearly dehiscent silique in subsequent tissue processing steps. The small segments of siliques were then fixed in 4% (w/v) paraformaldehyde and 3% (w/v) glutaraldehyde in 0.1 m PBS overnight at 4°C, followed by three washes (15 min each) in the same buffer. Samples were then postfixed in 1% (w/v) osmium tetroxide in PBS for 2 h at room temperature (25°C), followed by three washes (15 min each) in PBS. Specimens were dehydrated in a graded ethanol series (15, 30, 45, 60, 70, 80, 90, and 100%) and washed two times (10 min each time) in propylene oxide, and infiltrated and embedded in Epon 812 (SPI Supplies Division of Structure Probe). Polymerization proceeded for 24 h at 40°C, followed by 24 h at 60°C. For light microscopy, specimens were cut into 1- to 2-μm-thick sections using glass knives through Leica EM UC6 ultramicrotome (Leica), and stained with Toluidine Blue O. Similarly, 1- to 2-μm-thick sections of GUS-stained siliques were prepared after dehydration in a graded ethanol series starting from 70% ethanol without histological staining. For transmission electron microscopy, 70- to 90-nm-thick sections of the same embedded specimens were cut using a diamond knife (Diatome) on a Leica EM UC6 ultramicrotome, and collected by 150-mesh cuprum grids. Ultrathin sections were stained with uranyl acetate and lead citrate.
Light microscopes used for observation and photographing were a Leica S6D dissecting microscope equipped with the Leica EC3 imaging system , a Leica DMLB microscope equipped with the Leica DFC320 imaging system, and a Nikon Elipse 80i microscope equipped with the Nikon DS-Ri1 imaging system. The transmission electron microscope used was a Philips FEI-Tecnai 12 (FEI).
Characterization of Duration of Silique Development and Dehiscence
To investigate the duration between stage 15 and stage 18 siliques, at least 50 stage 15 flowers from four or more plants per genotype were labeled with colored strings and followed to stage 18. The long and short axes and areas of 50 cells in the separation layer in the transmission electron microscopy images were measured using SigmaScan Pro 5 (Systat Software) for each genotype. To determine the dehiscence rate, 15 pairs of consecutive siliques per plant and six or more plants per genotype were examined. The younger silique of the pair was yellow but still hydrated without an obviously dehiscent appearance to the naked eye. Silique dehiscence status was determined either by the naked eye if the dehiscence was apparent or under a dissecting microscope if otherwise. The dehiscence rate is expressed as a percentage of the dehiscent siliques of all the siliques examined. To further compare the dehiscence abilities of the genotypes that did not or largely did not naturally undergo dehiscence, a centrifugal force was first applied to dry siliques of these genotypes, and then the siliques were examined under a dissecting microscope. Individual dry siliques in 1.5-mL microcentrifuge tubes were spun at 18,000g for 1 min in a centrifuge (Eppendorf 5414D). For each genotype, 6 sets of 10 siliques per set were centrifuged. Statistical analysis (t test) was conducted in Microsoft Excel with 2 tails and unequal variance. The standard errors of the samples were calculated in SigmaScan Pro 5.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Examples of siliques of Col-0 at stages 17 to 19.
Supplemental Figure S2. Relative expression of IND and ALC in Col-0 and their respective mutants in stage-17 siliques (10D).
Supplemental Figure S3. GUS signals in vegetative and floral organs in pCEL6:GUS plants.
Supplemental Figure S4. GUS signals in stage 15 to 17 siliques.
Supplemental Figure S5. GUS signals in vegetative and floral organs in pMAN7:GUS plants.
Supplemental Figure S6. GUS signals in vegetative and floral organs in pCEL6:CEL6-GUS plants.
Supplemental Figure S7. GUS signals in vegetative and floral organs in pMAN7:MAN7-GUS plants.
Supplemental Figure S8. GUS signals in semithin sections of stage-17B (12D) siliques.
Supplemental Figure S9. Confirmation of the cel6 and man7 mutant alleles by PCR.
Supplemental Figure S10. Transmission electron microscopy images of transverse sections of early and late stage-17 siliques.
Supplemental Figure S11. TUNEL signals at stages not shown in Figure 5.
Supplemental Figure S12. Col-0 siliques as representatives of the younger siliques used in the dehiscence rate investigation.
Supplemental Figure S13. CEL6 or MAN7 expression levels and silique dehiscence rates in CEL6- and MAN7-overexpression lines.
Supplemental Table S1. Thirty-nine cellulase and other cell wall degrading enzyme genes identified by in silico searches.
Supplemental Table S2. Twelve cellulase and other cell wall degrading enzyme genes with a relative expression level >1 in stage 17 siliques according to searches with AtGenExpress Visualization Tool.
Supplemental Table S3. Primers for RT-PCR, qPCR, and T-DNA insertion identification.
Supplemental Table S4. Primers for gene cloning.
Acknowledgments
We thank Yixing Wang at Oklahoma State University and Lizhen Tao at South China Agricultural University for help on molecular experiments. We also thank Yaoguang Liu at South China Agricultural University for providing the vectors used in this investigation.
Footnotes
These authors contributed equally to the article.
This work was supported by grants from the National Natural Science Foundation of China (project nos. 31070159 and 31470293 to H.W.), by funds from South China Agricultural University to H.H., and by grants from Oklahoma Center for the Advancement of Science and Technology (PSB08-021 and PS13-006 to M.Y.).
Articles can be viewed without a subscription.
References
- Abeles FB. (1969) Abscission: role of cellulase. Plant Physiol 44: 447–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Dror T, Dermastia M, Kladnik A, Znidaric MT, Novak MP, Meir S, Burd S, Philosoph-Hadas S, Ori N, Sonego L, et al. (2011) Programmed cell death occurs asymmetrically during abscission in tomato. Plant Cell 23: 4146–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensoussan L, Moraïs S, Dassa B, Friedman N, Henrissat B, Lombard V, Bayer EA, Mizrahi I (2017) Broad phylogeny and functionality of cellulosomal components in the bovine rumen microbiome. Environ Microbiol 19: 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell DA, Bird CR, Schuch W, Bennett AB (1997) An endo-1,4-beta-glucanase expressed at high levels in rapidly expanding tissues. Plant Mol Biol 33: 87–95 [DOI] [PubMed] [Google Scholar]
- Christoffersen RE, Tucker ML, Laties GG (1984) Cellulase gene expression in ripening avocado fruit: the accumulation of cellulase mRNA and protein as demonstrated by cDNA hybridization and immunodetection. Plant Mol Biol 3: 385–391 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- del Campillo E, Bennett AB (1996) Pedicel breakstrength and cellulase gene expression during tomato flower abscission. Plant Physiol 111: 813–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Campillo E, Abdel-Aziz A, Crawford D, Patterson SE (2004) Root cap specific expression of an endo-beta-1,4-D-glucanase (cellulase): a new marker to study root development in Arabidopsis. Plant Mol Biol 56: 309–323 [DOI] [PubMed] [Google Scholar]
- Deng Y, Zou W, Li G, Zhao J (2014) TRANSLOCASE OF THE INNER MEMBRANE9 and 10 are essential for maintaining mitochondrial function during early embryo cell and endosperm free nucleus divisions in Arabidopsis. Plant Physiol 166: 853–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollhofer V, Podmirseg SM, Callaghan TM, Griffith GW, Fliegerová K (2015) Anaerobic fungi and their potential for biogas production. Adv Biochem Eng Biotechnol 151: 41–61 [DOI] [PubMed] [Google Scholar]
- Du M, Li Y, Tian X, Duan L, Zhang M, Tan W, Xu D, Li Z (2014) The phytotoxin coronatine induces abscission-related gene expression and boll ripening during defoliation of cotton. PLoS One 9: e97652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabi JP, Broetto SG, da Silva SL, Zhong S, Lajolo FM, do Nascimento JR (2014) Analysis of papaya cell wall-related genes during fruit ripening indicates a central role of polygalacturonases during pulp softening. PLoS One 9: e105685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Gago JA, Posé S, Muñoz-Blanco J, Quesada MA, Mercado JA (2009) The polygalacturonase FaPG1 gene plays a key role in strawberry fruit softening. Plant Signal Behav 4: 766–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Bosch C, del Campillo E, Bennett AB (1997) Immunodetection and characterization of tomato endo-beta-1,4-glucanase Cel1 protein in flower abscission zones. Plant Physiol 114: 1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Carranza ZH, Elliott KA, Roberts JA (2007) Expression of polygalacturonases and evidence to support their role during cell separation processes in Arabidopsis thaliana. J Exp Bot 58: 3719–3730 [DOI] [PubMed] [Google Scholar]
- Gorguet B, Schipper D, van Lammeren A, Visser RG, van Heusden AW (2009) ps-2, the gene responsible for functional sterility in tomato, due to non-dehiscent anthers, is the result of a mutation in a novel polygalacturonase gene. Theor Appl Genet 118: 1199–1209 [DOI] [PubMed] [Google Scholar]
- Harpster MH, Lee KY, Dunsmuir P (1997) Isolation and characterization of a gene encoding endo-beta-1,4-glucanase from pepper (Capsicum annuum L.). Plant Mol Biol 33: 47–59 [DOI] [PubMed] [Google Scholar]
- Heredia A, Jiménez A, Guillén R (1995) Composition of plant cell walls. Z Lebensm Unters Forsch 200: 24–31 [DOI] [PubMed] [Google Scholar]
- His I, Driouich A, Nicol F, Jauneau A, Höfte H (2001) Altered pectin composition in primary cell walls of korrigan, a dwarf mutant of Arabidopsis deficient in a membrane-bound endo-1,4-beta-glucanase. Planta 212: 348–358 [DOI] [PubMed] [Google Scholar]
- Iglesias-Fernández R, Barrero-Sicilia C, Carrillo-Barral N, Oñate-Sánchez L, Carbonero P (2013) Arabidopsis thaliana bZIP44: a transcription factor affecting seed germination and expression of the mannanase-encoding gene AtMAN7. Plant J 74: 767–780 [DOI] [PubMed] [Google Scholar]
- Iglesias-Fernández R, Rodríguez-Gacio MC, Barrero-Sicilia C, Carbonero P, Matilla A (2011) Three endo-β-mannanase genes expressed in the micropylar endosperm and in the radicle influence germination of Arabidopsis thaliana seeds. Planta 233: 25–36 [DOI] [PubMed] [Google Scholar]
- Jenkins ES, Paul W, Craze M, Whitelaw CA, Weigand A, Roberts JA (1999) Dehiscence-related expression of an Arabidopsis thaliana gene encoding a polygalacturonase in transgenic plants of Brassica napus. Plant Cell Environ 22: 159–167 [Google Scholar]
- Keegstra K. (2010) Plant cell walls. Plant Physiol 154: 483–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DR, Wiedemeier A, Peng L, Höfte H, Vernhettes S, Desprez T, Hocart CH, Birch RJ, Baskin TI, Burn JE, et al. (2001) Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-beta-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol 126: 278–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashbrook CC, Gonzalez-Bosch C, Bennett AB (1994) Two divergent endo-beta-1,4-glucanase genes exhibit overlapping expression in ripening fruit and abscising flowers. Plant Cell 6: 1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, et al. (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren SJ, Roeder AH, Kempin SA, Gremski K, Østergaard L, Guimil S, Reyes DK, Yanofsky MF (2004) Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 116: 843–853 [DOI] [PubMed] [Google Scholar]
- Martínez-Andújar C, Pluskota WE, Bassel GW, Asahina M, Pupel P, Nguyen TT, Takeda-Kamiya N, Toubiana D, Bai B, Górecki RJ, et al. (2012) Mechanisms of hormonal regulation of endosperm cap-specific gene expression in tomato seeds. Plant J 71: 575–586 [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Kato H, Shima Y, Ito Y (2015) Apple SVP family MADS-box proteins and the tomato pedicel abscission zone regulator JOINTLESS have similar molecular activities. Plant Cell Physiol 56: 1097–1106 [DOI] [PubMed] [Google Scholar]
- Nakashima J, Endo S, Fukuda H (2004) Immunocytochemical localization of polygalacturonase during tracheary element differentiation in Zinnia elegans. Planta 218: 729–739 [DOI] [PubMed] [Google Scholar]
- Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Höfte H (1998) A plasma membrane-bound putative endo-1,4-beta-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J 17: 5563–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Kay P, Wilson S, Swain SM (2009) ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are Polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 21: 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M, Sander L, Child R, van Onckelen H, Ulvskov P, Borkhardt B (1996) Isolation and characterisation of a pod dehiscence zone-specific polygalacturonase from Brassica napus. Plant Mol Biol 31: 517–527 [DOI] [PubMed] [Google Scholar]
- Rajani S, Sundaresan V (2001) The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr Biol 11: 1914–1922 [DOI] [PubMed] [Google Scholar]
- Roongsattham P, Morcillo F, Jantasuriyarat C, Pizot M, Moussu S, Jayaweera D, Collin M, Gonzalez-Carranza ZH, Amblard P, Tregear JW, et al. (2012) Temporal and spatial expression of polygalacturonase gene family members reveals divergent regulation during fleshy fruit ripening and abscission in the monocot species oil palm. BMC Plant Biol 12: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander L, Child R, Ulvskov P, Albrechtsen M, Borkhardt B (2001) Analysis of a dehiscence zone endo-polygalacturonase in oilseed rape (Brassica napus) and Arabidopsis thaliana: evidence for roles in cell separation in dehiscence and abscission zones, and in stylar tissues during pollen tube growth. Plant Mol Biol 46: 469–479 [DOI] [PubMed] [Google Scholar]
- Sanders PM, Lee PY, Biesgen C, Boone JD, Beals TP, Weiler EW, Goldberg RB (2000) The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12: 1041–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani Z, Dekel M, Tsabary G, Shoseyov O (1997) Cloning and characterization of elongation specific endo-1,4-beta-glucanase (cel1) from Arabidopsis thaliana. Plant Mol Biol 34: 837–842 [DOI] [PubMed] [Google Scholar]
- Sitrit Y, Hadfield KA, Bennett AB, Bradford KJ, Downie AB (1999) Expression of a polygalacturonase associated with tomato seed germination. Plant Physiol 121: 419–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainotti L, Spolaore S, Ferrarese L, Casadoro G (1997) Characterization of ppEG1, a member of a multigene family which encodes endo-beta-1,4-glucanase in peach. Plant Mol Biol 34: 791–802 [DOI] [PubMed] [Google Scholar]
- van Doorn WG, Beers EP, Dangl JL, Franklin-Tong VE, Gallois P, Hara-Nishimura I, Jones AM, Kawai-Yamada M, Lam E, Mundy J, et al. (2011) Morphological classification of plant cell deaths. Cell Death Differ 18: 1241–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gelderen K, van Rongen M, Liu A, Otten A, Offringa R (2016) An INDEHISCENT-controlled auxin response specifies the separation layer in early Arabidopsis fruit. Mol Plant 9: 857–869 [DOI] [PubMed] [Google Scholar]
- Vanyushin BF, Bakeeva LE, Zamyatnina VA, Aleksandrushkina NI (2004) Apoptosis in plants: specific features of plant apoptotic cells and effect of various factors and agents. Int Rev Cytol 233: 135–179 [DOI] [PubMed] [Google Scholar]
- Urbanowicz BR, Bennett AB, Del Campillo E, Catalá C, Hayashi T, Henrissat B, Höfte H, McQueen-Mason SJ, Patterson SE, Shoseyov O, et al. (2007) Structural organization and a standardized nomenclature for plant endo-1,4-beta-glucanases (cellulases) of glycosyl hydrolase family 9. Plant Physiol 144: 1693–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Mori A, Jiang X, Wang Y, Yang M (2006) The INDEHISCENT protein regulates unequal cell divisions in Arabidopsis fruit. Planta 224: 971–979 [DOI] [PubMed] [Google Scholar]
- Yu L, Chen H, Sun J, Li L (2014) PtrKOR1 is required for secondary cell wall cellulose biosynthesis in Populus. Tree Physiol 34: 1289–1300 [DOI] [PubMed] [Google Scholar]
- Yu L, Sun J, Li L (2013) PtrCel9A6, an endo-1,4-β-glucanase, is required for cell wall formation during xylem differentiation in populus. Mol Plant 6: 1904–1917 [DOI] [PubMed] [Google Scholar]
- Yuan JS, Yang X, Lai J, Lin H, Cheng ZM, Nonogaki H, Chen F (2007) The endo-beta-mannanase gene families in Arabidopsis, rice, and poplar. Funct Integr Genomics 7: 1–16 [DOI] [PubMed] [Google Scholar]
- Yung MH, Schaffer R, Putterill J (1999) Identification of genes expressed during early Arabidopsis carpel development by mRNA differential display: characterisation of ATCEL2, a novel endo-1,4-beta-D-glucanase gene. Plant J 17: 203–208 [DOI] [PubMed] [Google Scholar]