MCTPs exhibit diverse and overlapping patterns of gene expression and protein localization, and might be involved in Arabidopsis development with potential functional divergence or redundancy.
Abstract
Multiple C2 domain and transmembrane region proteins (MCTPs) are evolutionarily conserved in eukaryotic organisms and may function as signaling molecules that mediate trafficking of other regulators. Although there are a large number of MCTPs found in the plant lineage, biological information on most plant MCTPs remains unknown. Here, we report systematic characterization of 16 members in the entire Arabidopsis (Arabidopsis thaliana) MCTP family. Using GUS and GFP reporter assays, we reveal their distinct or overlapping patterns of gene expression and protein localization in developing Arabidopsis plants and Nicotiana benthamiana leaf epidermal cells. We further analyze in vivo effects of three C2 domains on the regulatory role of MCTP1 (FTIP1) in flowering time control in Arabidopsis, demonstrating that these C2 domains may be cooperative to mediate FTIP1 function during the floral transition. Through examining all available T-DNA insertional mutants of Arabidopsis MCTPs, we further reveal that mctp6-1 significantly enhances the late-flowering phenotype of ftip1-1 possibly through affecting FLOWERING LOCUS T in different manners, exemplifying that different MCTPs additively regulate a specific plant developmental process. Taken together, our results suggest functional divergence or redundancy of MCTP members in Arabidopsis and provide a community resource for further understanding various MCTP functions in plant development.
Division or differentiation of plant cells is highly determined by the positional and developmental information perceived by these cells. Cell-to-cell communication plays an important role in this process and is crucial to mediate signals from diverse pathways. Transduction of extracellular signals by receptor-like kinases or direct transfer of transcription factors between cells is part of this sort of communication required for modulating plant growth and development in response to environmental and internal signals (Van Norman et al., 2011; Wu and Gallagher, 2012), which is usually regulated by both intercellular and intracellular trafficking pathways.
Plants have evolved an elaborate endomembrane system for proper compartmentation of signaling molecules that facilitate trafficking of proteins or other macromolecules among endomembrane compartments (Contento and Bassham, 2012). Recent findings have suggested that a group of highly conserved multiple C2 domain and transmembrane region proteins (MCTPs), each of which contains three to four C2 domains at the N terminus and one to four transmembrane regions at the C terminus, could function as important signaling molecules that mediate trafficking of other regulators in plant cells (Shin et al., 2005; Fulton et al., 2009; Liu et al., 2012, 2013; Song et al., 2017). The C2 domain is one of the most prevalent eukaryotic lipid binding domains and could serve as a docking module that targets proteins to specific intracellular membrane by forming phospholipid complexes (Nalefski and Falke, 1996). In membrane trafficking proteins, different C2 domains always bear different conserved sequences, implying that multiple C2 domains in these proteins may function cooperatively rather than additively for a specific cellular function (Cho and Stahelin, 2006).
Sequence analyses have revealed that while MCTPs are evolutionarily conserved (Nalefski and Falke, 1996; Shin et al., 2005; Lek et al., 2012; Liu et al., 2013), there is a significantly increased number of MCTP repertoire in the plant lineage compared to the number of counterparts in animals. This implies more diverse and specific functions of MCTPs in regulating cellular processes in plants. There are a total of 16 MCTPs in the Arabidopsis (Arabidopsis thaliana) genome, among which QUIRKY (QKY) and FT-INTERACTING PROTEIN1 (FTIP1) have been suggested to affect macromolecular trafficking (Fulton et al., 2009; Liu et al., 2012). QKY interacts with a Leu-rich repeat receptor-like kinase STRUBBELIG (SUB), thus affecting intercellular communication that contributes to the relevant plant organogenesis mediated by SUB (Fulton et al., 2009; Trehin et al., 2013; Vaddepalli et al., 2014). FTIP1 is associated with endoplasmic reticulum (ER) membrane and mediates intercellular trafficking of the florigen protein FLOWERING LOCUS T (FT) specifically from companion cells to sieve elements, thus controlling flowering time in Arabidopsis (Liu et al., 2012). The closest ortholog of FTIP1 in rice (Oryza sativa), OsFTIP1, plays a similar role in mediating rice flowering time under long days through affecting trafficking of a rice counterpart of FT, RICE FLOWERING LOCUS T1, from companion cells to sieve elements (Song et al., 2017). Interestingly, both QKY and FTIP1 have been found to be specifically localized in plasmodesmata that are important conduits for exchange of molecules among plant cells (Liu et al., 2012; Vaddepalli et al., 2014), suggesting that these MCTPs might serve as essential regulators for mediating key intercellular signaling through plasmodesmata.
To systematically investigate MCTPs in Arabidopsis, in this study we used both GUS and GFP reporter assays to explore gene and protein expression of 16 members in the entire Arabidopsis MCTP family, respectively. We examined dynamic tissue- and developmental-specific GUS expression driven by 5′ upstream sequences of MCTP members and revealed their distinct or overlapping expression patterns in developing Arabidopsis plants. We also investigated the subcellular localization of MCTP members by transiently expressing GFP-MCTPs in Nicotiana benthamiana leaf epidermal cells. To understand the function of various C2 domains in MCTPs, we further analyzed in vivo effects of three C2 domains on the regulatory role of FTIP1 in flowering time control in Arabidopsis. In addition, through examining all available T-DNA insertional mutants of Arabidopsis MCTPs, we showed that mctp6-1 significantly enhanced the late-flowering phenotype of ftip1-1 possibly through affecting FT, exemplifying that MCTPs additively regulate a specific plant developmental process. Taken together, our results demonstrate functional divergence or redundancy of MCTP members in Arabidopsis and establish a community resource for further understanding MCTP function in macromolecular trafficking in plants.
RESULTS
Identification of MCTP Family Proteins in Arabidopsis
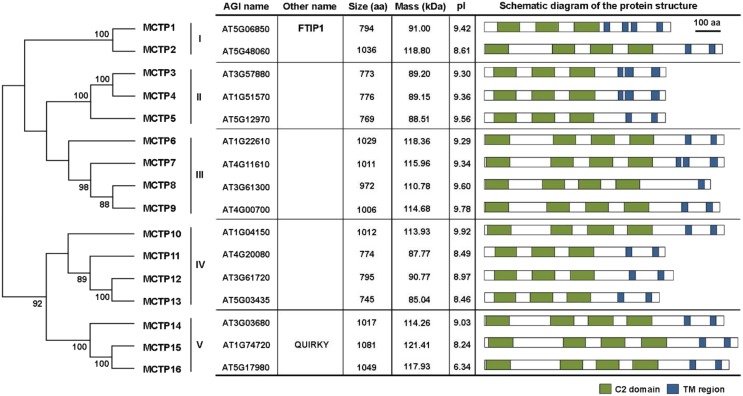
As QKY and FTIP1 were the first two MCTP proteins identified in Arabidopsis (Fulton et al., 2009; Liu et al., 2012), we used the FTIP1 protein sequence as a query to search for putative MCTPs using BLAST program in The Arabidopsis Information Resource and identified a total of 16 MCTP members, designated MCTP1-16, each of which contains three to four N-terminal C2 domains and one to four C-terminal transmembrane regions (Fig. 1; Supplemental Fig. S1).
Figure 1.
Classification of MCTP family proteins in Arabidopsis. Five groups of MCTP proteins are defined based on phylogenetic analysis of Arabidopsis MCTPs shown on the left. The phylogenetic tree was generated with MEGA6 using the neighbor-joining algorithm. Numbers on the major branches indicate bootstrap values (>50%) in 1,000 replicates. For each MCTP, the Arabidopsis Genome Initiative (AGI) gene number, other name, amino acid number, molecular weight, pI of the predicted protein, and schematic diagram of major motifs are indicated in the table on the right. The prediction of protein motifs is based on SMART (http://smart.embl-heidelberg.de). C2 domains and transmembrane (TM) regions are labeled as green and blue boxes, respectively. aa, Amino acids.
We classified these 16 Arabidopsis MCTPs into five clades (Fig. 1) through phylogenetic analysis based on multiple sequence alignment. These 16 MCTPs share relatively conserved amino acids in the three C2 domains and the transmembrane regions (Supplemental Fig. S2), while the overall amino acid identities among these family members ranged from 33% to 93% (Supplemental Fig. S3). Notably, the N-terminal regions of these MCTPs contain highly variable amino acid sequences with variable lengths (Supplemental Fig. S2), indicating that the N-terminal region may contribute to the functional divergence among MCTPs.
In silico chromosome mapping showed that the 16 MCTP genes were dispersed throughout the Arabidopsis genome and located in all the chromosomes except chromosome II (Supplemental Fig. S4). Interestingly, the closest homologs, such as MCTP3 and MCTP4, are located separately in different chromosomes. This scattered distribution of closest homologs is possibly due to the extensive reshuffling and divergent evolution following massive duplications.
Phylogenetic Analysis of MCTP Homologs in Plants
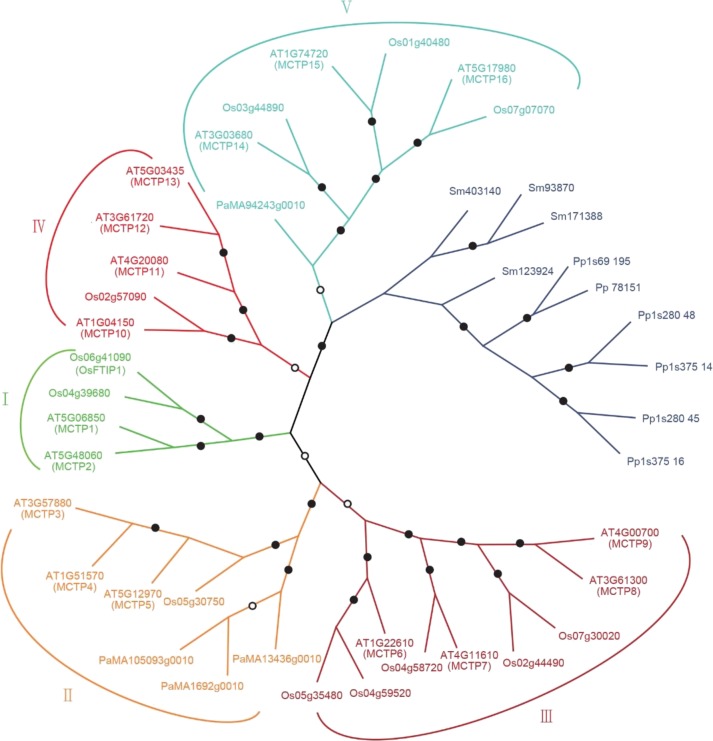
To identify MCTP homologs in plant genomes, we performed BLASTP or TBLASTN search in protein and genome databases using FTIP1 as a query sequence and found MCTP homologs in all land plant genomes that have been sequenced. The evolutionary relationship among MCTPs in different species was analyzed in detail using neighbor-joining and maximum likelihood methods. We included the angiosperm eudicot Arabidopsis, the monocot rice, the gymnosperm Norway spruce (Picea abies), lycophyte (Selaginella moellendorffii), and moss (Physcomitrella patens) in phylogenetic analysis (Fig. 2). It is noteworthy that MCTPs exist in both lycophytes, which are the oldest living lineage of vascular plants, and mosses, which are the early nonvascular land plants. These MCTPs are present in a separate clade representing its early formation in seedless plants. The number of MCTPs in seed plants is greatly expanded and classified into five groups (I–V) based on strong bootstrap values (Fig. 2). In contrast, there are a limited number of MCTPs in animals, such as human and mouse (Supplemental Fig. S5). A large number of MCTP repertoires in the plant lineage imply that as immovable organisms, plants may evolve to generate more functionally specialized MCTPs to mediate cellular activities in response to changing developmental or environmental stimuli.
Figure 2.
Phylogenetic relationships of MCTP homologs in different plant species. Proteins from different species are indicated by a two-letter prefix (AT, Arabidopsis thaliana; Os, Oryza sativa; Pa, Picea abies; Sm, Selaginella moellendorffii; Pp, Physcomitrella patens). All available gene names are also indicated. For each node, the level of statistical support by the neighbor-joining method using the Jones-Thornton-Taylor with discrete Gamma distribution (+G) model and the maximum likelihood method inferred using the LG+G model is marked by a filled circle if both values are above 80% bootstrap support or an open circle if both values are higher than 50% bootstrap support.
Analysis of pMCTP:GUS Activity in Arabidopsis Vegetative Tissues
Among all MCTP genes in Arabidopsis, only the expression patterns of MCTP1 (FTIP1) and MCTP15 (QKY) have already been reported (Liu et al., 2012; Trehin et al., 2013; Vaddepalli et al., 2014). To understand temporal and spatial expression patterns of all Arabidopsis MCTP genes, we generated pMCTP:GUS reporter lines, in which the GUS gene was driven by 1.5 to 3 kb of the 5′ genomic region upstream of each MCTP coding sequence. At least 15 independent transgenic lines were created for each construct. Since most of the transgenic lines for each construct showed consistent GUS staining patterns, we selected one representative line each for further monitoring the detailed expression patterns in various tissues at different developmental stages. In general, none of the pMCTP:GUS reporter lines exhibited constitutive expression patterns, while the 5′ upstream sequences of MCTPs in the same clade often conferred distinct GUS expression patterns in various tissues, suggesting diverse functions of MCTPs in various developmental processes in the life cycle of Arabidopsis.
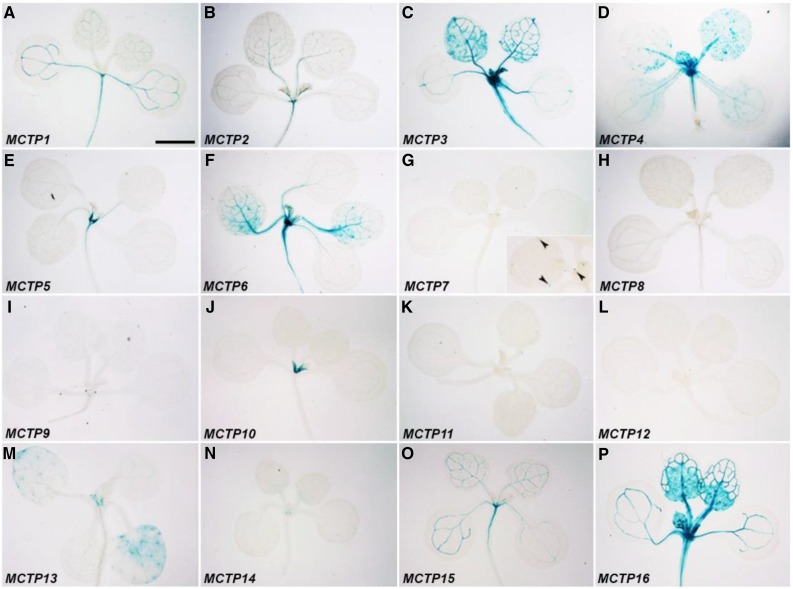
We first examined pMCTP:GUS reporter lines at the vegetative phase by staining of 11-d-old whole seedlings (Fig. 3; Supplemental Fig. S6A) and roots of 5-d-old seedlings (Fig. 4; Supplemental Fig. S6, B and C). All lines except for MCTP11 displayed GUS staining to various extents in either shoots and/or roots. In the aerial part, MCTP1 (FTIP1), MCTP2, MCTP3, MCTP6, MCTP15 (QKY), and MCTP16 lines all displayed GUS activity in vascular tissues (Fig. 3, A–C, F, O, and P). Their staining patterns were grouped into three different types. GUS expression of MCTP3, MCTP6, and MCTP16 lines was early detected in whole protruding young leaves, but mainly found in vascular tissues of adult leaves at later stages (Fig. 3, C, F, and P). GUS staining of MCTP1 (FTIP1) and MCTP15 (QKY) lines was relatively uniform in vascular tissues throughout cotyledons and rosette leaves (Fig. 3, A and O), whereas staining of the MCTP2 line was relatively weak in secondary and tertiary veins compared to primary veins (Fig. 3B). MCTP3, MCTP4, MCTP5, MCTP6, and MCTP16 lines exhibited GUS staining in the shoot apex region (Fig. 3, C–F and P), while MCTP9, MCTP10, MCTP13, and MCTP14 lines displayed GUS activity in incipient leaf primordia (Fig. 3, I, J, M, and N). The MCTP7 line exhibited GUS staining specifically in the hydathode region (Fig. 3G).
Figure 3.
GUS staining of 11-d-old pMCTP:GUS lines grown under long days. Representative GUS staining of pMCTP1:GUS (A), pMCTP2:GUS (B), pMCTP3:GUS (C), pMCTP4:GUS (D), pMCTP5:GUS (E), pMCTP6:GUS (F), pMCTP7:GUS (G), pMCTP8:GUS (H), pMCTP9:GUS (I), pMCTP10:GUS (J), pMCTP11:GUS (K), pMCTP12:GUS (L), pMCTP13:GUS (M), pMCTP14:GUS (N), pMCTP15:GUS (O), and pMCTP16:GUS (P). The inset in (G) shows a higher magnification of leaves with GUS staining in hydathodes (arrowheads). Bar = 2 mm.
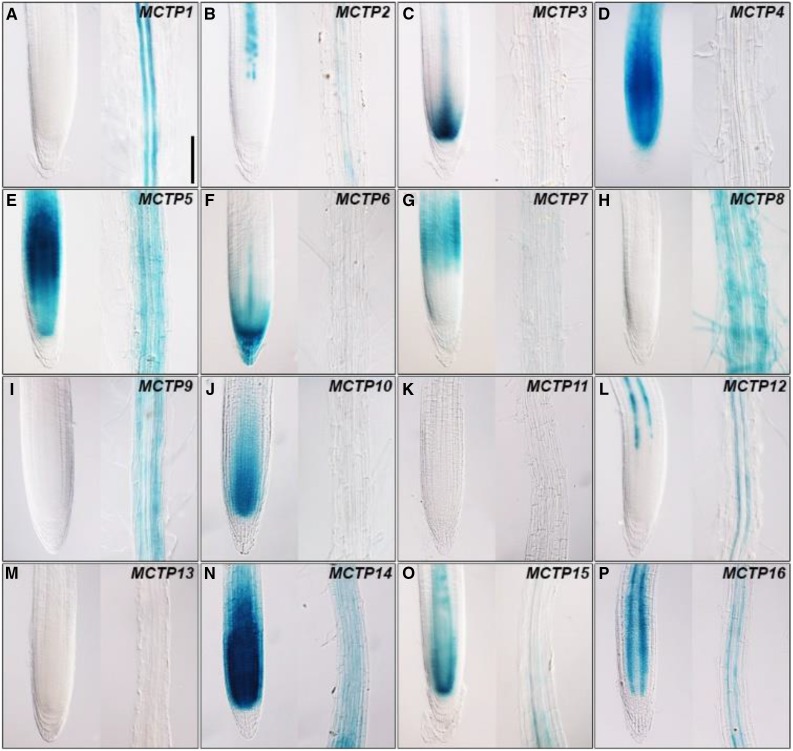
Figure 4.
GUS staining of the meristematic (left panel) and elongation zones (right panel) of pMCTP:GUS roots 5 d after germination grown on Murashige and Skoog plates under long days. Representative GUS staining of pMCTP1:GUS (A), pMCTP2:GUS (B), pMCTP3:GUS (C), pMCTP4:GUS (D), pMCTP5:GUS (E), pMCTP6:GUS (F), pMCTP7:GUS (G), pMCTP8:GUS (H), pMCTP9:GUS (I), pMCTP10:GUS (J), pMCTP11:GUS (K), pMCTP12:GUS (L), pMCTP13:GUS (M), pMCTP14:GUS (N), pMCTP15:GUS (O), and pMCTP16:GUS (P). Bar = 100 µm.
All pMCTP:GUS reporter lines except for those containing MCTP11 and MCTP13 upstream sequences showed various GUS staining patterns in roots (Fig. 4). Compared to other lines, there was relatively specific GUS activity detected in root vascular tissues of MCTP1 (FTIP1), MCTP2, MCTP12, MCTP15 (QKY), and MCTP16 lines, among which only GUS staining of the MCTP1 (FTIP1) line did not extend to the meristematic zone (Fig. 4, A, B, L, O, and P). MCTP4, MCTP5, MCTP10, and MCTP14 lines exhibited strong GUS activity throughout the meristematic zone (Fig. 4, D, E, J, and N), whereas GUS activity of the MCTP7 line was only restricted in the basal meristem (Fig. 4G). MCTP6 and MCTP8 lines were the only ones that displayed GUS activity in the root cap (Fig. 4F) and root hair (Fig. 4H), respectively.
Analysis of pMCTP:GUS Activity in Arabidopsis Reproductive Tissues
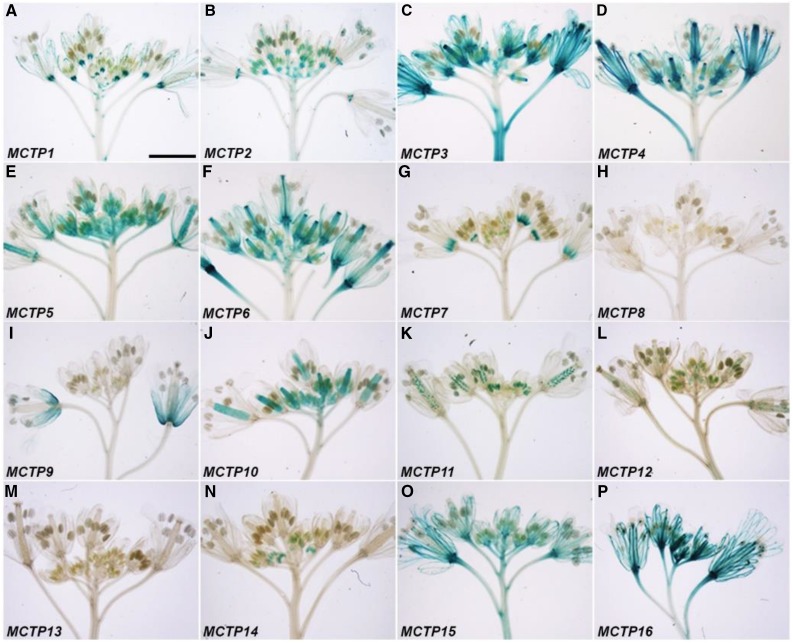
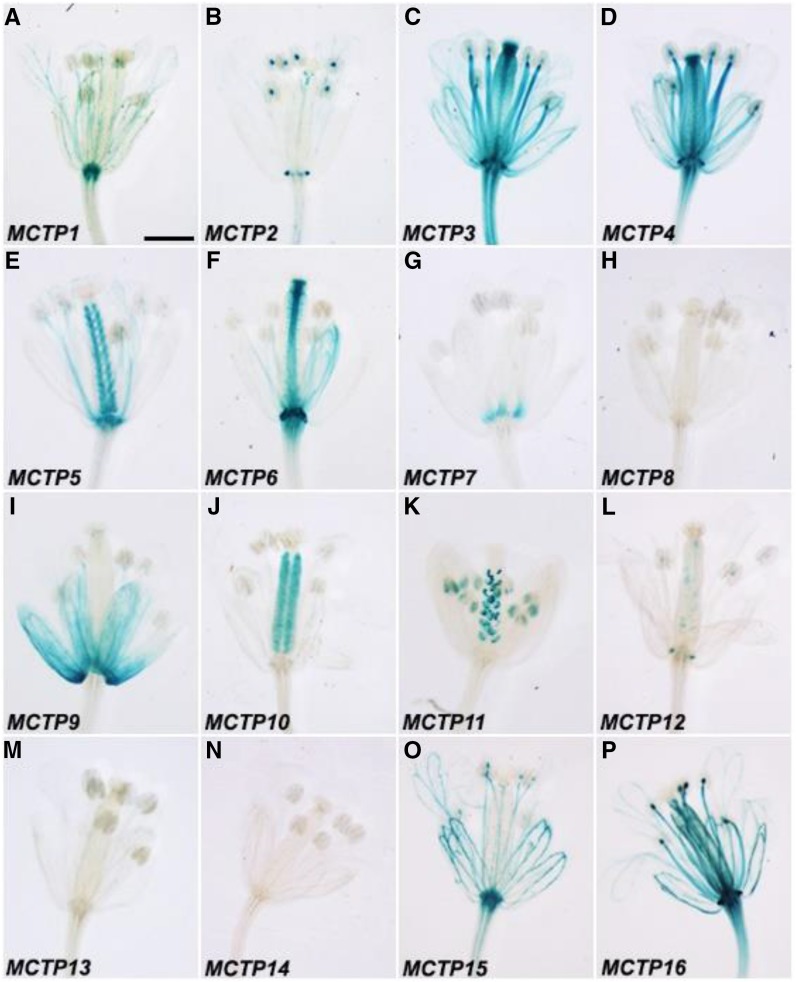
Except for MCTP8 and MCTP13, the upstream sequences of the other MCTPs were able to drive GUS expression with diverse patterns in inflorescences bearing flowers at various stages (Figs. 5 and 6; Supplemental Fig. S6, D and E). There were consistent GUS staining patterns detected in flowers at different stages in many MCTP lines, such as MCTP1 (FTIP1), MCTP3, MCTP6, and MCTP16 (Fig. 5, A, C, F, and P), whereas other lines showed changes in GUS staining in developing flowers. For example, GUS staining of the MCTP9 line was only detected in flowers at late stages (Figs. 5I and 6I). In contrast, GUS activity of the MCTP14 line was only observed in stamens of young flowers, but disappeared in old flowers (Figs. 5N and 6N). Interestingly, both MCTP7 and MCTP11 lines displayed dynamic changes in GUS expression in various floral organs at different stages. GUS staining of the MCTP7 line was detected in gynoecia of young flowers, but later confined to the distal end of filaments of old flowers (Figs. 5G and 6G). GUS activity of the MCTP11 line gradually decreased in pollen grains, but increased in ovules of developing flowers (Figs. 5K and 6K).
Figure 5.
GUS staining of inflorescence apices of pMCTP:GUS lines grown under long days. Representative GUS staining of pMCTP1:GUS (A), pMCTP2:GUS (B), pMCTP3:GUS (C), pMCTP4:GUS (D), pMCTP5:GUS (E), pMCTP6:GUS (F), pMCTP7:GUS (G), pMCTP8:GUS (H), pMCTP9:GUS (I), pMCTP10:GUS (J), pMCTP11:GUS (K), pMCTP12:GUS (L), pMCTP13:GUS (M), pMCTP14:GUS (N), pMCTP15:GUS (O), and pMCTP16:GUS (P). Bar = 3 mm.
Figure 6.
GUS staining of open flowers of pMCTP:GUS lines grown under long days. Representative GUS staining of pMCTP1:GUS (A), pMCTP2:GUS (B), pMCTP3:GUS (C), pMCTP4:GUS (D), pMCTP5:GUS (E), pMCTP6:GUS (F), pMCTP7:GUS (G), pMCTP8:GUS (H), pMCTP9:GUS (I), pMCTP10:GUS (J), pMCTP11:GUS (K), pMCTP12:GUS (L), pMCTP13:GUS (M), pMCTP14:GUS (N), pMCTP15:GUS (O), and pMCTP16:GUS (P). Bar = 2 mm.
While most of the pMCTP:GUS reporter lines exhibited GUS staining patterns that were consistent with the gene expression data available in Arabidopsis eFP browser (Winter et al., 2007), inconsistent patterns were found for MCTP14. MCTP14 was ubiquitously expressed in Arabidopsis based on the microarray data in eFP browser, while the pMCTP14:GUS line displayed specific staining mainly in incipient leaf primordia at the vegetative stage (Fig. 3N) and in stamens of young flowers (Fig. 5N). It is possible that the 5′ upstream sequence included in the pMCTP14:GUS construct may not contain all essential cis-elements that are required for conferring endogenous MCTP14 expression in Arabidopsis.
Subcellular Localization of the MCTPs in Arabidopsis
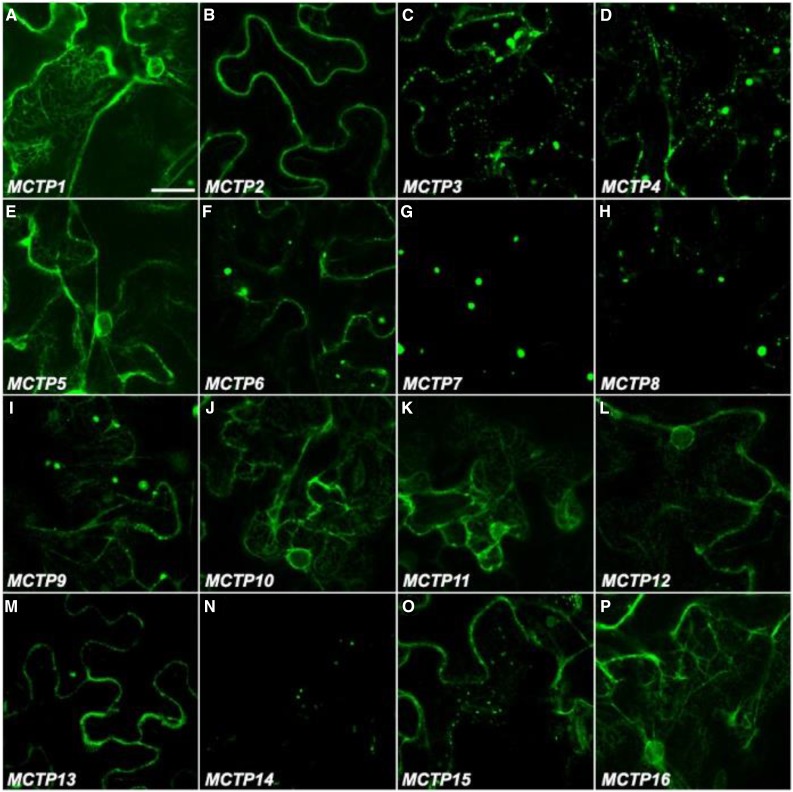
To determine the subcellular localization of 16 MCTPs, we transiently expressed their full-length open reading frames fused with the GFP reporter (GFP-MCTPs) in N. benthamiana leaf epidermal cells (Fig. 7) and observed generally four different types of subcellular localization patterns in tobacco cells. The first group of MCTPs, including MCTP1 (FTIP1), MCTP5, MCTP10, MCTP11, MCTP12, and MCTP16, were associated with the ER network (Fig. 7, A, E, J–L, and P), which is substantiated by colocalization of representative MCTPs in this group with an ER marker, RFP-HDEL (Supplemental Fig. S7A; Nelson et al., 2007). The second group of MCTPs comprising MCTP3, MCTP4, MCTP6, MCTP9, MCTP13, and MCTP15 (QKY) was detected in both the cytosol and plasma membrane (Fig. 7, C, D, F, I, M, and O). MCTPs in the third group, including MCTP7, MCTP8, and MCTP14, were only localized in small structures within cells, possibly intracellular vesicles (Fig. 7, G, H, and N), while MCTP2 in the last group was only localized in the plasma membrane (Fig. 7B). To further understand the concrete subcellular localization of MCTPs in the second and third groups, we coexpressed 35S:GFP-MCTPs with the fluorescence-tagged organelle markers for Golgi (35S:Man49-CFP; Nelson et al., 2007) and endosomes (35S:RFP-RabF2b; Jaillais et al., 2006). We observed that MCTP3, MCTP4, MCTP6, and MCTP7 partially resided in the endosomal compartments (Supplemental Fig. S7B), while MCTP14 and MCTP15 were partially localized in the Golgi (Supplemental Fig. S7C). Distinct subcellular localization patterns of the MCTPs in tobacco cells indicate that different MCTPs may exert distinct functions in plant cells.
Figure 7.
Subcellular localization of GFP-MCTPs in N. benthamiana leaf epidermal cells. Representative GFP fluorescence images of MCTP1 (FTIP1)-GFP (A), GFP-MCTP2 (B), GFP-MCTP3 (C), GFP-MCTP4 (D), GFP-MCTP5 (E), GFP-MCTP6 (F), GFP-MCTP7 (G), GFP-MCTP8 (H), GFP-MCTP9 (I), GFP-MCTP10 (J), GFP-MCTP11 (K), GFP-MCTP12 (L), GFP-MCTP13 (M), GFP-MCTP14 (N), GFP-MCTP15 (O), and GFP-MCTP16 (P). Bar = 30 µm.
Characterization of C2 Domains in MCTP1 (FTIP1)
Although multiple C2 domains have been suggested to either additively or individually act to mediate protein functions (Damer and Creutz, 1994; Cho and Stahelin, 2006; Marty et al., 2013), it is unclear how multiple C2 domains play roles in mediating MCTP functions in plants. To this end, we examined the role of each C2 domain in MCTP1 (FTIP1), which affects flowering time through mediating FT trafficking from companion cells to sieve elements (Liu et al., 2012).
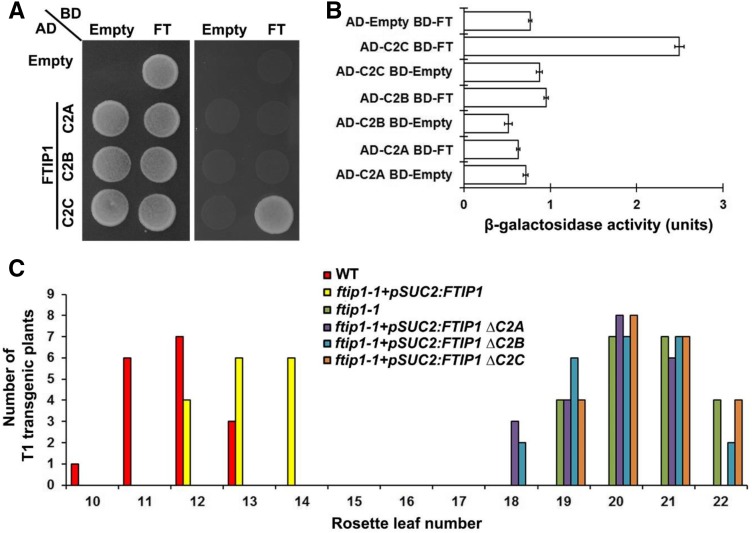
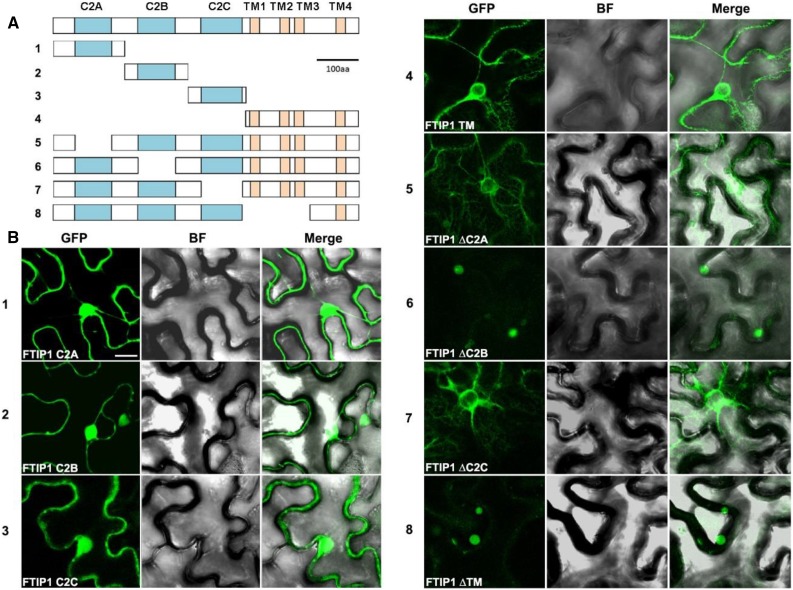
As the N-terminal region of FTIP1 containing three C2 domains interacts with FT (Liu et al., 2012), we tested the interaction of each C2 domain in the N-terminal region with FT by yeast two-hybrid assays and found that only the third C2 domain interacted with FT (Fig. 8, A and B), suggesting that this C2 domain is specifically required for FTIP1 interaction with FT. Since both FTIP1 and FT are colocalized to ER in plant cells (Liu et al., 2012), we further transiently expressed various truncated versions of FTIP1-GFP either containing individual C2 domains and the transmembrane region or with deletion of specific domain(s) in N. benthamiana leaf epidermal cells (Fig. 9A) to identify key domains required for FTIP1 subcellular localization (Fig. 9B). The GFP fusion proteins containing individual C2 domains were localized in both the plasma membrane and nucleus (Fig. 9B, panels 1–3), while the fusion protein bearing the C-terminal transmembrane region was obviously associated with the ER network (Fig. 9B, panel 4), demonstrating an important role of the transmembrane region in conferring FTIP1 localization into ER. Furthermore, among the FTIP1-GFP truncated proteins with deletion of individual domains, only deletion of the second C2 domain or part of the transmembrane region compromised FTIP1-GFP association with the ER network (Fig. 9B, panels 5–8). These results suggest that both the C-terminal transmembrane region and the second C2 domain of FTIP1 are essential for subcellular localization of FTIP1 into ER, although the second C2 domain itself is not sufficient for conferring this localization.
Figure 8.
Function of C2 domains of FTIP1 in the control of flowering time. A, Yeast two-hybrid assay of interaction between FT and each C2 domain of FTIP1. Transformed yeast cells were grown on SD-Trp/-Leu medium (left panel) and SD-His/-Trp/-Leu medium supplemented with 10 mm 3-amino-1,2,4-trizaole (right panel). B, Quantification of interaction between FT and each C2 domain of FTIP1 in yeast by β-galactosidase assays. C, Distribution of flowering time in T1 transgenic plants carrying the full-length or truncated versions (without each C2 domain) of the FTIP1 coding sequence in ftip1-1 background.
Figure 9.
Subcellular localization of truncated FTIP1 proteins in N. benthamiana leaf epidermal cells. A, Schematic diagrams depicting 8 different truncated versions of FTIP1 proteins fused with GFP as shown in B. C2 domains and transmembrane (TM) regions are labeled as cyan and orange boxes, respectively. B, Subcellular localization of 8 different truncated versions of FTIP1-GFP fusion proteins in N. benthamiana leaf epidermal cells. GFP, GFP fluorescence; BF, bright field; Merge, merge of GFP and bright field. Bar = 10 µm.
FTIP1 functions to mediate FT trafficking in the phloem so that the late-flowering phenotype of ftip1-1 can be rescued by the expression of FTIP1 coding sequence driven by the promoter of SUC TRANSPORTER2 (SUC2), which is active specifically in the phloem companion cells (Fig. 8C; Imlau et al., 1999; Liu et al., 2012). To examine the effect of each C2 domain on FTIP1 role in promoting flowering, we transformed ftip1-1 with various pSUC2:FTIP1 truncated constructs containing deletion of each C2 domain. Interestingly, all ftip1-1 lines transformed with the truncated versions of FTIP1 exhibited comparable flowering time to ftip1-1 (Fig. 8C), demonstrating that all the three C2 domains are essential for FTIP1 function in promoting flowering.
FTIP1 and MCTP6 Function Additively to Promote Flowering under Long Days
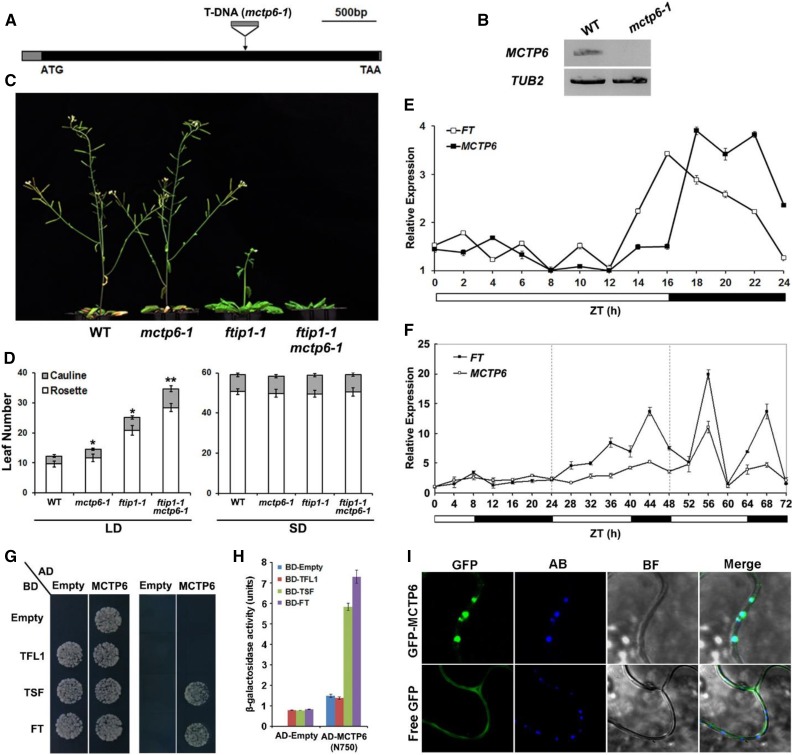
To understand the biological functions of Arabidopsis MCTPs during the floral transition, we isolated the available T-DNA insertional lines for 15 MCTPs from the Arabidopsis Biological Resource Center (Supplemental Fig. S8) and observed their growth phenotypes under long days. In addition to mctp1-1 (ftip1-1; SALK_013179) and mctp15 (qky-14; SALK_061045) that exhibited the late-flowering phenotype as previously reported (Liu et al., 2012; Trehin et al., 2013), mctp6-1 (SALK_145386) exhibited a similar flowering defect under long days (Fig. 10, A–D; Supplemental Fig. S8).
Figure 10.
The late-flowering phenotype of ftip1-1 is enhanced by mctp6-1. A, Schematic diagram shows the T-DNA insertion site in mctp6-1 (SALK_145386). The exon and 5′/3′ untranslated regions are indicated by black and gray boxes, respectively. The start codon (ATG) and stop codon (TAA) are labeled. B, MCTP6 expression is undetectable in mctp6-1 by semiquantitative PCR using the primers flanking the T-DNA insertion site. TUB2 expression is used as a control. C, ftip1-1 mctp6-1 flowers later than ftip1-1 or mctp6-1 under long days. D, Comparison of flowering time of mctp6-1, ftip1-1, and mctp6-1 ftip1-1 under long days (LDs) and short days (SDs). Error bars indicate sd. Single asterisks indicate a significant difference in flowering time between single mutants and wild-type plants, while a double asterisk indicates a significant difference in flowering time between double and single mutants (Student’s t test, P < 0.05). E, MCTP6 expression exhibits obvious diurnal oscillation in 9-d-old wild-type plants grown under LDs harvested at 2-h intervals over a 24-h period. FT expression was examined as a control. Sampling time was expressed in hours as Zeitgeber time (ZT), which is the number of hours after the onset of illumination. The lowest expression level of each gene is set as 1. Bars below the graph indicate the duration of day (white) and night (black). Error bars denote sd. F, MCTP6 expression is induced upon daylength extension. Wild-type seedlings were grown under SDs for 11 d before they were transferred to LDs. MCTP6 expression in seedlings were examined by quantitative real-time PCR at 4-h intervals for 3 d comprising one SD followed by two LDs. FT expression was examined as a control. The lowest expression level of each gene is set as 1. Error bars denote sd. G, Yeast two-hybrid assay of interaction between FT, TSF, or TFL1 and the N-terminal region of MCTP6 containing four C2 domains. Transformed yeast cells were grown on SD-Trp/Leu medium (left panel) and SD-His/-Trp/-Leu medium supplemented with 5 mm 3-amino-1,2,4-triazole (right panel). H, Quantification of interaction of FT, TSF, or TFL1 and the N-terminal region of MCTP6 containing four C2 domains in yeast by β-galactosidase assays. I, Subcellular localization of GFP-MCTP6 and free GFP in N. benthamiana leaf epidermal cells. Compared to free GFP, GFP-MCTP6 signal is colocalized with callose deposition staining with aniline blue, which indicates the position of plasmodesmata. GFP, GFP fluorescence; AB, aniline blue staining; BF, bright field image; Merge, merge of GFP, AB, and BF. Bar = 5 µm.
The full-length MCTP6 transcript was undetectable in mctp6-1 (Fig. 10B). Single mctp6-1 mutants flowered slightly later than wild-type plants, but significantly enhanced the late-flowering phenotype of ftip1-1 only under long days, whereas all of these single and double mutants did not exhibit a flowering defect under short days (Fig. 10, C and D), suggesting that FTIP1 and MCTP6 function additively to promote flowering specifically under long days. Consistently, we found that MCTP6 expression exhibited a diurnal oscillation under long days with peaks at night (Fig. 10E). Daylength shift experiments showed that like FT, expression of MCTP6 was substantially increased when shifted from short days to long days (Fig. 10F), demonstrating that MCTP6 expression is promoted by the photoperiod pathway. Furthermore, during the floral transition under long days, which occurred around 9 to 13 d after seed germination under our growth conditions, GUS staining of pMCTP6:GUS lines was consistently strong in vascular and mesophyll tissue of young leaves, but weak in older cotyledons and leaves (Fig. 3F; Supplemental Fig. S9).
To test whether MCTP6 and FTIP1 may share similar function to affect FT, we performed yeast two-hybrid assays and revealed the interaction of the truncated MCTP6 containing only the four C2 domains at the N terminus with FT and TWIN SISTER OF FT (TSF), but not with TERMINAL FLOWER1 (TFL1), all of which belong to the phosphatidylethanolamine-binding protein family (Fig. 10, G and H; Kobayashi et al., 1999; Wigge et al., 2005). In addition, we found that like FTIP1-GFP (Liu et al., 2012), GFP-MCTP6 was colocalized with callose deposition indicated by aniline blue staining, marking localization of GFP-MCTP6 in plasmodesmata (Fig. 10I). These observations imply that FTIP1 and MCTP6 could play additive roles in affecting FT during the floral transition.
DISCUSSION
Proteins containing multiple C2 domains and transmembrane region(s) are evolutionarily conserved and divided into four subgroups, including synaptotagmins, ferlins, tricalbins, and MCTPs, based on the number of C2 domains and the location of transmembrane region(s) (Lek et al., 2012). Although there are only a limited number of MCTPs, each containing three to four C2 domains at the N terminus and one to four transmembrane regions at the C terminus, in unicellular ancestors and animals, the number of MCTPs is significantly increased in higher plants (Fig. 2; Supplemental Fig. S5; Nalefski and Falke, 1996; Shin et al., 2005; Lek et al., 2012; Liu et al., 2013). This may cater to the requirement of more functionally specific MCTPs to mediate plant response to changing internal or environmental stimuli. Although a few plant MCTPs have been shown to be involved in membrane trafficking and fusion processes (Fulton et al., 2009; Liu et al., 2012, 2013; Vaddepalli et al., 2014; Song et al., 2017), biological information on most of plant MCTPs remains unknown.
In this study, we have systematically characterized 16 MCTPs in Arabidopsis, which are grouped into five clades based on the phylogenetic analysis (Fig. 1). Examination of the GUS reporter lines driven by the 5′ upstream sequences of these MCTPs has revealed that none of the pMCTP:GUS reporter lines exhibit exactly the same GUS staining patterns at both vegetative and reproductive tissues. Notably, 5′ upstream sequences of MCTPs even in the same clades often drive GUS expression with distinct patterns in various tissues. For example, while MCTP1 (FTIP1) and MCTP2 lines generally exhibit GUS staining in vascular tissues, their distribution patterns in vegetative or reproductive tissues are different. In the aerial part of vegetative seedlings, GUS activity is uniformly detected in vascular tissues of cotyledons and rosette leaves of the MCTP1 (FTIP1) line (Fig. 3A), whereas GUS staining of the MCTP2 line is obvious in primary veins (Fig. 3B). In roots, although GUS activity is detectable in vascular tissues of both MCTP1 (FTIP1) and MCTP2 lines, the MCTP1 (FTIP1) line displays stronger GUS staining in the elongation zone than the MCTP2 line (Fig. 4, A and B). However, in the root meristematic zone, GUS staining completely disappears in the MCTP1 (FTIP1) line but is obvious in the MCTP2 line (Fig. 4, A and B). These different expression patterns indicate that even MCTPs sharing high sequence similarity might be differentially regulated to play distinct roles in various tissues. This is further substantiated by distinct subcellular localization patterns of GFP-MCTPs revealed in N. benthamiana leaf epidermal cells (Fig. 7).
Despite differential GUS expression patterns driven by the 5′ upstream sequences of different MCTPs, some pMCTP:GUS reporter lines display overlapping GUS activity in specific tissues. For example, MCTP1 (FTIP1), MCTP2, MCTP3, MCTP6, MCTP15 (QKY), and MCTP16 lines all exhibit GUS activity in vascular tissues of vegetative seedlings, albeit to different extents (Fig. 3, A–C, F, O, and P), implying that some MCTPs may act redundantly or additively in specific tissues during Arabidopsis development. Similarly, comparable subcellular localization exhibited by some MCTPs also indicates that some of them may participate redundantly or additively in similar cellular processes (Fig. 7). Thus, distinct or overlapping patterns of gene expression and subcellular localization revealed by our GUS and GFP reporter assays could be further investigated to explore functional divergence or redundancy of MCTP members in Arabidopsis.
The presence of multiple C2 domains at the N terminus is a signature feature of MCTPs, but their concrete functions in MCTPs remain unknown. Characterization of multiple C2 domains in MCTP1 (FTIP1) in this study has revealed specific roles of each C2 domain in FTIP1 (Figs. 8 and 9). Deletion of each C2 domain in FTIP1 almost completely compromises FTIP1 role in promoting flowering, suggesting that all the three C2 domains are required for FTIP1 function in the control of flowering time in Arabidopsis. Notably, the third C2 domain is specifically required for FTIP1 interaction with FT, while the second C2 domain, together with the C-terminal transmembrane region, is essential for MCTP1 (FTIP1) localization into the ER network. Since C2 domain has been suggested to act as a docking module targeting proteins to specific intracellular membrane (Nalefski and Falke, 1996), our findings suggest that multiple C2 domains in MCTPs may be cooperative to play an important role in mediating MCTP interaction with target protein(s), their subcellular localization, and subsequent biological effects.
Through examining the available T-DNA insertional lines for 15 Arabidopsis MCTPs, we have found that most of these lines do not display obvious defects under our growth conditions, implying possible functional redundancy among these MCTPs. Three T-DNA insertional lines, including mctp1-1 (ftip1-1) (Liu et al., 2012), mctp15 (qky-14) (Trehin et al., 2013), and mctp6-1, show the late-flowering phenotype (Fig. 10, A–D), suggesting that FTIP1, QKY, and MCTP6 are individually required for promoting flowering. Several pieces of evidence indicate that FTIP1 and MCTP6 affect flowering time in different manners. First, their gene expression patterns in tissues are fundamentally different. FTIP1 is specifically expressed in vascular tissues of cotyledons and rosette leaves, but not in the shoot apex (Liu et al., 2012; Fig. 3A), whereas MCTP6 is expressed in the shoot apex and early detected in whole protruding young leaves (Fig. 3F). Second, MCTP6 expression shows an obvious circadian rhythm under long days (Fig. 10E), which is in contrast to the nonrhythmic pattern of FTIP1 (Liu et al., 2012), indicating that they may respond differently to the photoperiod signal. Third, the subcellular localization patterns of FTIP1 and MCTP6 are also different. FTIP1 is associated with the ER network, whereas MCTP6 is mainly in intracellular compartments and the plasma membrane (Fig. 7, A and F). These observations suggest that FTIP1 and MCTP6 contribute to flowering time control in different ways at both cellular and tissue levels. Consistently, mctp6-1 significantly enhances the late-flowering phenotype of ftip1-1 under long days (Fig. 10, C and D). Despite these differences, FTIP1 and MCTP6 do share some common characteristics, such as interaction with FT, localization in plasmodesmata, and functioning in the photoperiod pathway. Thus, it will be interesting to further investigate their roles in the floral transition to understand whether these two MCTPs could play either simple additive or more complex cooperative roles in affecting FT.
CONCLUSION
In conclusion, our systematic analysis of 16 genes in the entire Arabidopsis MCTP family illustrates their complex gene expression and protein localization patterns in Arabidopsis. Through examining the roles of multiple C2 domains in FTIP1, we show that these signature domains in MCTPs may be cooperative to mediate MCTP function in plant development. In addition, various MCTPs, such as FTIP1 and MCTP6, could act in different manners to affect the same target to regulate a specific developmental process. Taken together, our findings reveal potential functional divergence or redundancy of MCTP members in Arabidopsis and establish a community resource for further mechanistic understanding MCTP function in plants.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants in the Columbia (Col) background were grown under long days (16 h light/8 h dark) or short days (8 h of light/16 h of dark) at 23°C. All T-DNA insertional mutants in the Col background were obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org).
Sequence Analysis
A primary BLASTP or TBLASTN search was performed using the Arabidopsis FTIP1 as a query sequence against protein and genome databases (Phytozome database, https://phytozome.jgi.doe.gov; Spruce genome database, http://congenie.org) to identify FTIP1 homologs. The sequences containing C2 domain and transmembrane regions were all included for further analysis. Each sequence was then searched against the protein Conserved Domain Database, SMART Database, and the Pfam Database to annotate domain modules. The program ProtTest was used for estimating the best model for each alignment by default, which identified LG+I+G+F as the best fit of the 112 examined evolutionary models according to Akaike Information Criterion statistics. The neighbor-joining phylogeny was performed by MEGA version 6.0 with 1,000 replicates. Since LG+I+G+F is not available in MEGA, we used the next best available JJT + G model and pairwise deletion. The maximum likelihood phylogeny was performed by PhyML software with 100 replicates and the best available LG + G model.
Plasmid Construction
pMCTP1(FTIP1):GUS and 35S:MCTP1(FTIP1)-GFP constructs were previously described (Liu et al., 2012). To generate other pMCTP:GUS reporter constructs, 1.5 to 3 kb of 5′ upstream sequence of each MCTP were amplified and cloned into pHY107 (Liu et al., 2007). To generate other 35S:GFP-MCTP reporter constructs, the cDNA encoding each full-length MCTP or each domain was amplified and cloned into pGreen-35S-GFP. To create various truncated versions of the constructs, pSUC2:FTIP1 and 35S:FTIP1-GFP (Liu et al., 2012) were mutagenized using a modified QuikChange site-directed mutagenesis approach. The primers used for plasmid construction are listed in Supplemental Table S1. Transgenic plants were generated in the Col background through Agrobacterium tumefaciens-mediated transformation and selected by Basta on soil.
GUS Staining
GUS staining was performed as previously reported with minor modifications (Yu et al., 2000). Tissues were infiltrated with staining solution (50 mm sodium phosphate buffer, pH 7.0, 0.5 mm potassium ferrocyanide, 0.5 mm potassium ferricyanide, and 0.5 mg/mL X-Gluc) in a vacuum chamber and subsequently incubated with staining solution at 37°C overnight.
Transient Expression of Proteins in Tobacco Leaf Epidermal Cells
The overnight A. tumefaciens cultures with expression vectors were harvested and resuspended in the infiltration buffer (10 mm MES, pH 5.6, 10 mm MgCl2, and 100 µM acetosyringone) with OD600 at 0.2. Infiltration solutions were infiltrated into the abaxial surface of 3-week-old tobacco (Nicotiana benthamiana) leaves with syringes. The leaves were examined 2 d after infiltration under confocal microscope.
Yeast Two-Hybrid Assay
To construct the vectors for yeast two-hybrid assays, the coding sequences of FT, each C2 domain of FTIP1, MCTP6 (N750), TSF, and TFL1 were amplified and cloned into pGADT7 or pGBKT7 (Clontech). The yeast two-hybrid assay was performed using the Yeastmaker Yeast Transformation System 2 (Clontech).
Expression Analysis
Total RNA was extracted using the FavorPrep Plant Total RNA Mini Kit (Favorgen) and reverse transcribed using the M-MLV Reverse Transcriptase (Promega). Quantitative real-time PCR was performed on three biological replicates using 7900HT Fast Real-Time PCR systems (Applied Biosystems) with Maxima SYBR Green/ROX qPCR Master Mix (Fermentas). The relative expression levels normalized to TUB2 were calculated as previously reported (Liu et al., 2007). Primers for real-time PCR are listed in Supplemental Table S1.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: MCTP1 (FTIP1), At5g06850; MCTP2, At5g48060; MCTP3, At3g57880; MCTP4, At1g51570; MCTP5, At5g12970; MCTP6, At1g22610; MCTP7, At4g11610; MCTP8, At3g61300; MCTP9, At4g00700; MCTP10, At1g04150; MCTP11, At4g20080; MCTP12, At3g61720; MCTP13, At5g03435; MCTP14, At3g03680; MCTP15 (QKY), At1g74720; MCTP16, At5g17980; TFL1, At5g03840; TSF, At4g20370; and FT, At1g65480.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Topology prediction of transmembrane helices of 16 MCTPs in Arabidopsis.
Supplemental Figure S2. Alignment of amino acid sequences of 16 MCTPs in Arabidopsis.
Supplemental Figure S3. Homology matrix tree of 16 MCTPs in Arabidopsis.
Supplemental Figure S4. Location of the MCTP Homologs on the Arabidopsis chromosomes.
Supplemental Figure S5. Phylogenetic analysis of MCTPs in animals.
Supplemental Figure S6. GUS staining in various tissues of wild-type plants grown under long days.
Supplemental Figure S7. Colocalization of representative GFP-MCTPs with various subcellular markers in N. benthamiana leaf epidermal cells.
Supplemental Figure S8. List of T-DNA insertion mutants of Arabidopsis MCTPs.
Supplemental Figure S9. GUS staining of developing pMCTP6:GUS seedlings grown under long days.
Supplemental Table S1. List of primers used in this study.
Acknowledgments
We thank the Arabidopsis Biological Resource Center for providing seeds and members of the Yu lab for discussion and comments on the manuscript.
Footnotes
Articles can be viewed without a subscription.
References
- Cho W, Stahelin RV (2006) Membrane binding and subcellular targeting of C2 domains. Biochim Biophys Acta 1761: 838–849 [DOI] [PubMed] [Google Scholar]
- Contento AL, Bassham DC (2012) Structure and function of endosomes in plant cells. J Cell Sci 125: 3511–3518 [DOI] [PubMed] [Google Scholar]
- Damer CK, Creutz CE (1994) Synergistic membrane interactions of the two C2 domains of synaptotagmin. J Biol Chem 269: 31115–31123 [PubMed] [Google Scholar]
- Fulton L, Batoux M, Vaddepalli P, Yadav RK, Busch W, Andersen SU, Jeong S, Lohmann JU, Schneitz K (2009) DETORQUEO, QUIRKY, and ZERZAUST represent novel components involved in organ development mediated by the receptor-like kinase STRUBBELIG in Arabidopsis thaliana. PLoS Genet 5: e1000355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlau A, Truernit E, Sauer N (1999) Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11: 309–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y, Fobis-Loisy I, Miège C, Rollin C, Gaude T (2006) AtSNX1 defines an endosome for auxin-carrier trafficking in Arabidopsis. Nature 443: 106–109 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Lek A, Evesson FJ, Sutton RB, North KN, Cooper ST (2012) Ferlins: regulators of vesicle fusion for auditory neurotransmission, receptor trafficking and membrane repair. Traffic 13: 185–194 [DOI] [PubMed] [Google Scholar]
- Liu C, Zhou J, Bracha-Drori K, Yalovsky S, Ito T, Yu H (2007) Specification of Arabidopsis floral meristem identity by repression of flowering time genes. Development 134: 1901–1910 [DOI] [PubMed] [Google Scholar]
- Liu L, Liu C, Hou X, Xi W, Shen L, Tao Z, Wang Y, Yu H (2012) FTIP1 is an essential regulator required for florigen transport. PLoS Biol 10: e1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhu Y, Shen L, Yu H (2013) Emerging insights into florigen transport. Curr Opin Plant Biol 16: 607–613 [DOI] [PubMed] [Google Scholar]
- Marty NJ, Holman CL, Abdullah N, Johnson CP (2013) The C2 domains of otoferlin, dysferlin, and myoferlin alter the packing of lipid bilayers. Biochemistry 52: 5585–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalefski EA, Falke JJ (1996) The C2 domain calcium-binding motif: structural and functional diversity. Protein Sci 5: 2375–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Shin OH, Han W, Wang Y, Südhof TC (2005) Evolutionarily conserved multiple C2 domain proteins with two transmembrane regions (MCTPs) and unusual Ca2+ binding properties. J Biol Chem 280: 1641–1651 [DOI] [PubMed] [Google Scholar]
- Song S, Chen Y, Liu L, Wang Y, Bao S, Zhou X, Teo ZW, Mao C, Gan Y, Yu H (2017) OsFTIP1-mediated regulation of florigen transport in rice is negatively regulated by the ubiquitin-like domain kinase OsUbDKγ4. Plant Cell 29: 491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trehin C, Schrempp S, Chauvet A, Berne-Dedieu A, Thierry AM, Faure JE, Negrutiu I, Morel P (2013) QUIRKY interacts with STRUBBELIG and PAL OF QUIRKY to regulate cell growth anisotropy during Arabidopsis gynoecium development. Development 140: 4807–4817 [DOI] [PubMed] [Google Scholar]
- Vaddepalli P, Herrmann A, Fulton L, Oelschner M, Hillmer S, Stratil TF, Fastner A, Hammes UZ, Ott T, Robinson DG, Schneitz K (2014) The C2-domain protein QUIRKY and the receptor-like kinase STRUBBELIG localize to plasmodesmata and mediate tissue morphogenesis in Arabidopsis thaliana. Development 141: 4139–4148 [DOI] [PubMed] [Google Scholar]
- Van Norman JM, Breakfield NW, Benfey PN (2011) Intercellular communication during plant development. Plant Cell 23: 855–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Gallagher KL (2012) Transcription factors on the move. Curr Opin Plant Biol 15: 645–651 [DOI] [PubMed] [Google Scholar]
- Yu H, Yang SH, Goh CJ (2000) DOH1, a class 1 knox gene, is required for maintenance of the basic plant architecture and floral transition in orchid. Plant Cell 12: 2143–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]