Four Leu-rich repeat extensin chimeras, LRX8–11, are redundantly involved in pollen germination and pollen tube growth in Arabidopsis.
Abstract
During pollen tube growth, the walls of the tube provide the mechanical strength resisting turgor pressure to protect two sperm cells. Cell wall proteins may play an important role in this process. Pollen tube cell wall proteins known as leucine-rich repeat extensins (LRXs) harbor a leucine-rich repeat domain and an extensin domain. In this study, the functions of four pollen-expressed LRXs, LRX8, LRX9, LRX10, and LRX11 (LRX8–11), were characterized in Arabidopsis (Arabidopsis thaliana). LRX8–11 displayed a consistent expression pattern in mature pollen grains and pollen tubes. In a phenotypic analysis of four single mutants, six double mutants, four triple mutants, and a quadruple mutant, the triple and quadruple mutant plants displayed markedly reduced seed set and decreased male transmission efficiency accompanied by compromised pollen germination and pollen tube growth. GFP-fused LRX8, LRX10, and LRX11 were found to be localized to pollen tube cell walls. An immunohistochemical analysis of pollen tube cell wall polysaccharides showed an increase in the amount of rhamnogalacturonan I in the subapical walls of pollen tubes of the lrx9 lrx10 lrx11 and lrx8 lrx9 lrx11 mutants and a decrease in the content of fucosylated xyloglucans in lrx8 lrx9 lrx11 compared with wild-type plants. Moreover, the callose content in the apical walls of pollen tubes increased in the lrx8 lrx9 lrx11 mutant. In conclusion, we propose that LRX8–11 function synergistically to maintain pollen tube cell wall integrity; thus, they play critical roles in pollen germination and pollen tube growth.
Double fertilization is a crucial and complex process in angiosperms, which have nonmotile sperm cells and rely on siphonogamy for reproduction (Feijó, 2010). When pollen grains germinate on the stigma of a flower, they develop polarized pollen tubes that move through transmitting tracts to deliver sperm cells to an embryo sac; in this way, double fertilization is achieved (Dresselhaus et al., 2016). Thus, rapid pollen tube growth is essential for successful double fertilization in flowering plants. The structure and physiological processes of pollen tubes have been studied extensively. A pollen tube is a self-organized system with a highly polarized growth pattern that requires coordinated vesicular transport, a highly dynamic cytoskeleton, cell wall deposition and remodeling, ROP GTPase-based signaling, reactive oxygen species, and calcium gradients (Krichevsky et al., 2007; Qin and Yang, 2011; Kroeger and Geitmann, 2012; Guan et al., 2013). Importantly, the pollen tube wall provides sufficient mechanical strength to resist high turgor pressure, maintain cell shape, and protect sperm cells. It also plays important roles in cell expansion and signaling (Gu and Nielsen, 2013; Hepler et al., 2013).
The plant cell wall is a dynamic and complicated structure that consists mainly of cellulose, hemicellulose, pectin, and a little protein (Lerouxel et al., 2006; Sorek and Turner, 2016). Despite their low abundance, plant cell wall proteins have structural and signaling functions and participate in many crucial processes, including cell-cell communication, wound responses, pollen tube guidance, and embryogenesis (Showalter, 1993; Cassab, 1998). Most plant cell wall proteins are glycosylated proteins with highly repetitive sequences, similar to collagen in the extracellular matrix of animals (Showalter et al., 2010; Mohnen and Tierney, 2011). However, our understanding of the functional mechanisms of plant cell wall proteins in somatic cells is limited, and even less is known about the cell wall proteins in growing pollen tubes.
Extensins (EXTs), which bear a Ser-Hyp3–5 repeat motif, were the first structural cell wall proteins to be characterized (Lamport, 1967). Some EXTs contain a conserved hydrophobic Tyr-X-Tyr motif (where X indicates any amino acid) that is responsible for intermolecular and intramolecular cross-linking with other extensins (Fry, 1982; Brady and Fry, 1997; Ringli, 2010). EXTs are induced by wounding and tensile stress, and they are thought to mediate cell wall rigidity in cells that are no longer expanding (Merkouropoulos et al., 1999). In Arabidopsis (Arabidopsis thaliana), there are 20 classical EXT genes. Overexpression of AtEXT1 has been shown to increase the thickness of inflorescence stems (Roberts and Shirsat, 2006), while the knockout of AtEXT3 resulted in the production of rsh (for root-, shoot-, hypocotyl-defective), a lethal embryogenesis mutant that displays incomplete wall development. EXT3 is a self-assembling molecule that may form scaffolds that serve as the template for pectin assembly (Cannon et al., 2008). In comparison, the ext18 mutant exhibits multiple defects in vegetative growth, pollen germination, and pollen tube growth (Choudhary et al., 2015).
Leucine-rich repeat (LRR) EXTs (LRXs) are chimeric proteins that contain an LRR domain and an EXT domain (Baumberger et al., 2003a). The LRR domain mediates protein-protein interactions; many proteins containing this domain are involved in plant development, pathogen defense, and signal transduction (Osakabe et al., 2013). There are two classes of LRX genes, which are distinguished based on their expression patterns: the first class is expressed in vegetative tissue, such as TOML-4 in tomato (Solanum lycopersicum) and LRX1 to LRX7 in Arabidopsis (Zhou et al., 1992); while the second class is expressed specifically in pollen, such as pollen extensin-like1 (Pex1) in maize (Zea mays; Rubinstein et al., 1995b) and LRX8, LRX9, LRX10, and LRX11 (LRX8–11) in Arabidopsis (Baumberger et al., 2001).
Several vegetative tissue-expressed LRXs have been reported to be involved in wound responses, plant development, and root hair elongation. In tomato, TOML-4 expression is induced by wounding (Zhou et al., 1992). In Arabidopsis, LRX1 and LRX2 are highly expressed in root hair cells; lrx1 and lrx1 lrx2 mutant plants display severe defects in root hair elongation (Baumberger et al., 2003b). However, the mutants repressor of lrx1 (rol1) and rol5 were shown to suppress the mutant phenotype of lrx1 by modifying the content and structure of its cell walls (Diet et al., 2006; Leiber et al., 2010). Double and triple mutants of LRX3, LRX4, and LRX5, which are expressed in leaves and stems, exhibit retarded plant development and changes in several cell wall polysaccharides (Draeger et al., 2015). In maize, the LRX protein Pex1 is localized to the intine of pollen grains and callose-rich pollen tube cell walls (Rubinstein et al., 1995b). However, the biological functions of pollen-expressed LRX8–11 in Arabidopsis are elusive.
In this study, the functions of Arabidopsis LRX8–11 were explored. LRX8–11 were found to be highly expressed in mature pollen and pollen tubes. Notably, triple mutants of these genes and a quadruple mutant exhibited significantly reduced fertility. Male transmission efficiency in these mutants also was markedly decreased. Furthermore, the triple and quadruple mutant plants displayed impaired pollen germination and pollen tube growth in vitro and in vivo. Finally, an immunolabeling assay for cell wall polymers in the pollen tube wall revealed an increased amount of rhamnogalacturonan I (RGI) in the subapical walls of pollen tubes of the lrx9 lrx10 lrx11 and lrx8 lrx9 lrx11 mutants and a decreased amount of fucosylated xyloglucans in lrx8 lrx9 lrx11. Additionally, the callose content in the apical walls of pollen tubes increased in the lrx8 lrx9 lrx11 mutant. Together, these findings reveal the essential role of LRX8–11 in pollen germination and tube growth in Arabidopsis.
RESULTS
LRX8–11 Are Expressed Mainly in Pollen Grains and Growing Pollen Tubes
LRX8 was identified from a differential proteomic analysis of apoplastic proteins during pollen germination (Ge et al., 2011). The functions of LRX8 and its homologs LRX9, LRX10, and LRX11 were investigated in this study. The phylogenetic analysis revealed that LRX8 has the highest homology with LRX9 and that LRX10 is highly homologous with LRX11 in Arabidopsis (Supplemental Fig. S1). In addition, homologs of LRX8 were found in tomato, maize, rice (Oryza sativa, indica group), and Brachypodium distachyon but not in Escherichia coli, Saccharomyces cerevisiae, Chlamydomonas reinhardtii, or Catharanthus roseus. By aligning the complete amino acid sequences of LRX8–11, we found that the LRR domain is highly conserved, whereas the EXT domains of the proteins differ greatly in arrangement and number of Pro-rich repeats (Supplemental Fig. S2).
The expression profiles of LRX8–11 were initially investigated by reverse transcription (RT)-PCR. As shown in Supplemental Figure S3A, LRX8–11 consistently showed high-level expression in open flowers, inflorescences, and siliques. LRX8 showed low-level expression in roots, stems, and rosette leaves, while LRX10 showed low-level expression in roots and cauline leaves (Supplemental Fig. S3A). To examine the expression patterns of LRX8–11 further, the putative promoters of these genes (containing 2,040, 2,070, 2,048, and 2,075 bp of sequence located upstream of the start codon [ATG], respectively) were fused individually with the GUS reporter gene. The resulting constructs were introduced into wild-type Arabidopsis. Consistent with our RT-PCR results, GUS signals corresponding to ProLRX8:GUS, ProLRX9:GUS, ProLRX10:GUS, and ProLRX11:GUS were detected mainly in open flowers (Supplemental Fig. S3, B, F, J, and N). LRX9 also was expressed in siliques and rosette leaves (Supplemental Fig. S3, F and G). In dissected open flowers, GUS signals corresponding to ProLRX8:GUS, ProLRX9:GUS, ProLRX10:GUS, and ProLRX11:GUS were found predominantly in pollen at anther stage 13, while LRX8 and LRX9 also were detected in filaments and sepals, respectively (Supplemental Fig. S3, D, H, L, and P and E, I, M, and Q, respectively).
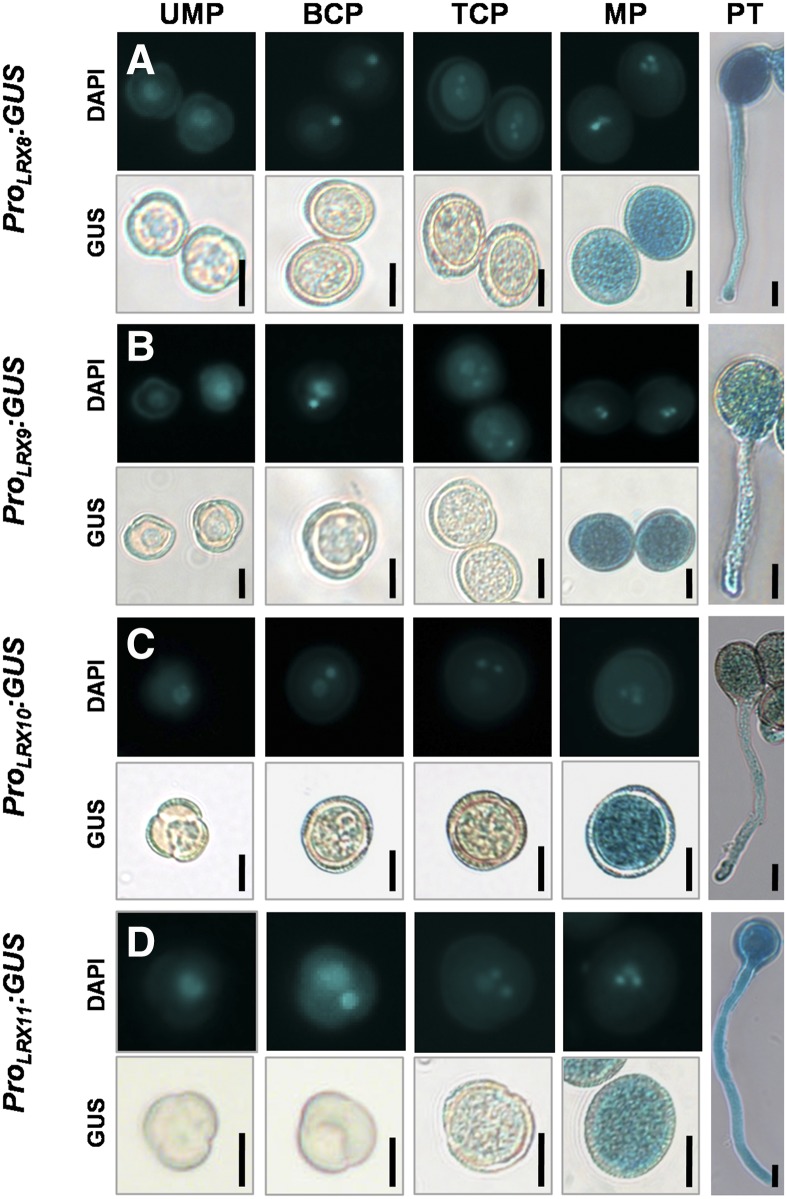
To explore LRX8–11 expression in developing microspores and pollen tubes, 4′,6-diamidino-2-phenylindole (DAPI) staining combined with GUS staining was performed. Notably, LRX8–11 exhibited high-level expression in mature pollen grains and growing pollen tubes, while a weak GUS signal was detected in uninuclear microspores, bicellular pollen, and early tricellular pollen (Fig. 1). Therefore, LRX8–11 are preferentially expressed in mature pollen grains and growing pollen tubes, suggesting a crucial role for LRX8–11 in pollen and pollen tube growth.
Figure 1.
LRX8–11 are highly expressed in mature pollen grains and growing pollen tubes. The detection of GUS activity in 5-week-old wild-type plants expressing ProLRX8:GUS, ProLRX9:GUS, ProLRX10:GUS, or ProLRX11:GUS is shown. Developing male gametophytes were stained with DAPI and GUS. UMP, Uninuclear microspore; BCP, bicellular pollen; TCP, early tricellular pollen; MP, mature pollen grain; PT, pollen tube. Bars = 10 μm.
A Disruption in LRX Expression Causes a Significant Reduction in Fertility
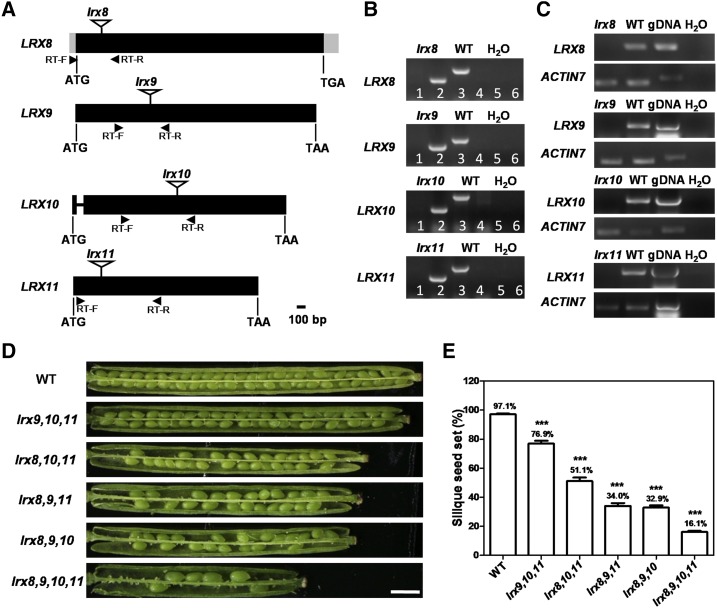
To explore the biological functions of LRX8–11, T-DNA insertion mutants were obtained from the Arabidopsis Biological Resource Center (ABRC; Fig. 2A). LRX8, LRX9, and LRX11 contain no introns, while LRX10 contains two exons and one intron. DNA-level identification revealed that the mutants of LRX8 (AT3G19020; SALK_132358), LRX9 (AT1G49490; SALK_137156), LRX10 (AT2G15880; SALK_087083C), and LRX11 (AT4G33970; SALK_076356) carried the T-DNA insertion in an exon (Fig. 2B). Transcriptional analyses showed that the transcription of LRX8–11 in homozygous lrx8, lrx9, lrx10, and lrx11 mutant plants was knocked out, respectively (Fig. 2C).
Figure 2.
LRX8–11 disruption caused a significant reduction in fertility. A, Schematic diagrams of the genomic loci and positions of the T-DNA insertions in LRX8–11. Exons are represented by black boxes, introns by a line, and untranslated regions by gray boxes. The location of each T-DNA insertion is indicated by an inverted triangle. B, DNA level identification of lrx8, lrx9, lrx10, and lrx11. The primers in the odd lanes are LP and RP, and those in the even lanes are LBb1.3 and RP. C, RT-PCR showed no LRX8–11 transcripts in open flowers from lrx8, lrx9, lrx10, and lrx11 plants, respectively. ACTIN7 served as a loading control. Amplification was performed for 25 cycles for ACTIN7 and 35 cycles for LRX8–11. gDNA, Genomic DNA. D, The mutants exhibited reduced seed set. Bar = 1 mm. E, Statistical analysis of silique seed set in wild-type (WT) and triple and quadruple mutant plants. All values are based on three biological replicates (n = 103, 43, 29, 45, 63, and 89). Error bars show se. Asterisks indicate values that differed significantly from the wild type (***, P < 0.001, calculated using Student’s t test).
Next, a detailed phenotypic analysis was performed using lrx8, lrx9, lrx10, and lrx11 single mutant and wild-type plants. During vegetative and reproductive growth, the lrx8, lrx9, lrx10, and lrx11 single mutants displayed no obvious differences from wild-type plants, including plant height, branching, flowering, and silique seed set (Supplemental Figs. S4A and S5). Subsequently, genetic crosses were performed to produce six double mutants (lrx8 lrx9, lrx8 lrx10, lrx8 lrx11, lrx9 lrx10, lrx9 lrx11, and lrx10 lrx11), four triple mutants (lrx8 lrx9 lrx10, lrx8 lrx9 lrx11, lrx8 lrx10 lrx11, and lrx9 lrx10 lrx11), and the quadruple mutant lrx8 lrx9 lrx10 lrx11 for further investigation.
During vegetative growth, the six double mutants, four triple mutants, and quadruple mutant displayed no obvious defects compared with wild-type plants (Supplemental Fig. S4, B and C). However, during reproductive growth, the lrx8 lrx9 and lrx8 lrx10 mutants showed slightly reduced fertility (Supplemental Fig. S5). The four triple mutants and the quadruple mutant exhibited significantly reduced fertility compared with wild-type plants (Fig. 2D; Supplemental Fig. S4C). Among the four triple mutants, the siliques of lrx8 lrx9 lrx10 had the lowest seed set (32.9%), followed by lrx8 lrx9 lrx11 (34%), lrx8 lrx10 lrx11 (51.1%), and lrx9 lrx10 lrx11 (76.9%); these results were significantly different from those in the wild type. The seed set in the quadruple mutant (lrx8 lrx9 lrx10 lrx11) was only 16.1% (Fig. 2E). Thus, all of the triple mutants and the quadruple mutant exhibited significantly reduced fertility.
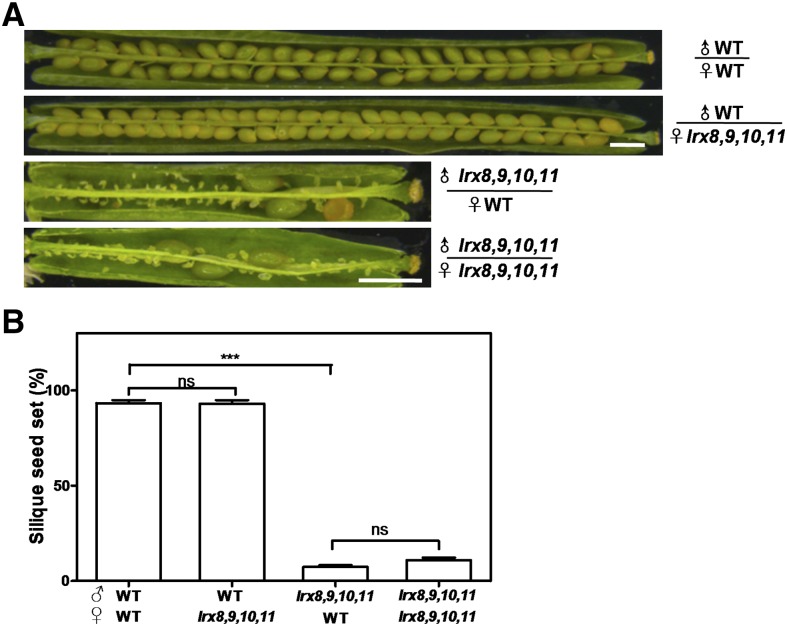
To determine the reason for this decrease in fertility, reciprocal crosses between wild-type plants and the quadruple mutant lrx8 lrx9 lrx10 lrx11 were performed, and the resulting siliques were dissected (Fig. 3A). When lrx8 lrx9 lrx10 lrx11 was used as the female parent and the wild type as the male parent, the silique seed set was 93%, compared with 93.2% for a wild-type self-cross, suggesting that the female gametophyte in lrx8 lrx9 lrx10 lrx11 functioned normally. However, when lrx8 lrx9 lrx10 lrx11 was used as the male parent and the wild type as the female parent, the seed set was 7.4%, which is not significantly different from the rate (10.9%) for self-crossed lrx8 lrx9 lrx10 lrx11 plants (Fig. 3B). Thus, the reduced fertility of lrx8 lrx9 lrx10 lrx11 was due to defects in the male gametophyte, implying that the function of the male gametophyte in lrx8 lrx9 lrx10 lrx11 was seriously impaired while that of the female gametophyte was unaffected.
Figure 3.
Reciprocal cross-pollination of lrx8 lrx9 lrx10 lrx11 and wild-type (WT) plants. A, Siliques were harvested at ∼15 d after hand pollination and dissected for statistical analysis. Bars = 1 mm. B, Statistical analysis of silique seed set. Values are based on three biological replicates (n = 52, 50, 51, and 41). Error bars represent se (***, P < 0.001; ns, not significant, calculated using Student’s t test).
Next, to determine whether a loss of function of LRX8–11 would affect embryo development, we observed embryo development in wild-type, lrx8 lrx9 lrx11, and lrx8 lrx9 lrx10 lrx11 plants from 1 to 8 d after pollination. Both mutants displayed normal embryogenesis compared with the wild type (Supplemental Fig. S6).
The Transmission Efficiency of the Male Gametophyte Was Decreased Significantly in the Mutants
Next, a genetic analysis of several single, double, and quadruple mutants was conducted. The segregation ratios for selfed lrx8(+/−), lrx8(+/−) lrx9(−/−), and lrx8(−/−) lrx9(+/−) plants all deviated significantly from the expected Mendelian segregation ratio (1:2:1; Supplemental Table S1). Thus, a transmission efficiency experiment was performed. Using lrx8(+/−), lrx8(+/−) lrx9(−/−), and lrx8(−/−) lrx9(+/−) plants as the male parent and wild-type plants as the female parent, the male transmission efficiency decreased to 74.4%, 31.2%, and 62.3%, respectively, suggesting that the lrx8 single mutant and lrx8 lrx9 double mutant possessed defective male gametophytes.
Subsequently, a genetic analysis of quadruple mutant plants was performed. Four mutant combinations were used: lrx8(+/−) lrx9(−/−) lrx10(−/−) lrx11(−/−), lrx8(−/−) lrx9(+/−) lrx10(−/−) lrx11(−/−), lrx8(−/−) lrx9(−/−) lrx10(+/−) lrx11(−/−), and lrx8(−/−) lrx9(−/−) lrx10(−/−) lrx11(+/−). The segregation ratios for all combinations of selfed plants deviated remarkably from the classic Mendelian segregation ratio (Table I). Furthermore, the male transmission efficiency decreased remarkably to 0%, 13.7%, 46.6%, and 34.4%, respectively (Table I). However, the female transmission efficiency was not affected in any of the mutants, except lrx8 (+/−) lrx9 (−/−) lrx10 (−/−) lrx11 (−/−), at 74.5% (Table I). Therefore, a loss of function of LRX8–11 produced severe defects in the male gametophyte.
Table I. Segregation data from selfed progeny and a transmission efficiency analysis of the quadruple mutants.
Reciprocal crosses were performed between lrx heterozygous plants and wild-type (WT) plants. The transmission efficiency (TE) of gametes was calculated as follows: TE = number of progeny with the lrx allele/number of wild-type progeny × 100%. The observed TE was compared with the expected TE for normal gametes (100%). NA, Not applicable; TEF, female transmission efficiency; TEM, male transmission efficiency. For the χ2 test, degrees of freedom = 1, P < 0.05, χ2 > 3.84 or degrees of freedom = 2, P < 0.05, χ2 > 5.99.
| Parental Genotype (Female × Male) | Progeny Genotype |
TEF | TEM | χ2 | Expected |
|||
|---|---|---|---|---|---|---|---|---|
| +/+ | +/− | − /− | TE | χ2 | ||||
| lrx8(+/−) lrx9(−/−) lrx10(−/−) lrx11(−/−) ⊗ | 127 | 115 | 7 | NA | NA | 117 | NA | 5.99 |
| ♀WT × ♂ lrx8(+/−) lrx9(−/−) lrx10(−/−) lrx11(−/−) | 329 | 0 | NA | NA | 0% | 327 | 100% | 3.84 |
| ♀ lrx8(+/−) lrx9(−/−) lrx10(−/−) lrx11(−/−) × ♂WT | 231 | 172 | NA | 74.5% | NA | 8.34 | ||
| lrx8(−/−) lrx9(+/−) lrx10(−/−) lrx11(−/−) ⊗ | 120 | 128 | 32 | NA | NA | 57.4 | NA | 5.99 |
| ♀WT × ♂ lrx8(−/−) lrx9(+/−) lrx10(−/−) lrx11(−/−) | 227 | 38 | NA | NA | 13.7% | 179.8 | 100% | 3.84 |
| ♀ lrx8(−/−) lrx9(+/−) lrx10(−/−) lrx11(−/−) × ♂WT | 144 | 113 | NA | 78.5% | NA | 3.5 | ||
| lrx8(−/−) lrx9(−/−) lrx10(+/−) lrx11(−/−) ⊗ | 94 | 112 | 18 | NA | NA | 51.6 | NA | 5.99 |
| ♀WT × ♂ lrx8(−/−) lrx9(−/−) lrx10(+/−) lrx11(−/−) | 249 | 116 | NA | NA | 46.6% | 47.7 | 100% | 3.84 |
| ♀ lrx8(−/−) lrx9(−/−) lrx10(+/−) lrx11(−/−) × ♂WT | 170 | 173 | NA | 102% | NA | 0.012 | ||
| lrx8(−/−) lrx9(−/−) lrx10(−/−) lrx11(+/−) ⊗ | 161 | 223 | 44 | NA | NA | 64.72 | NA | 5.99 |
| ♀WT × ♂ lrx8(−/−) lrx9 (−/−) lrx10(−/−) lrx11 (+/−) | 224 | 77 | NA | NA | 34.4% | 70.8 | 100% | 3.84 |
| ♀ lrx8(−/−) lrx9(−/−) lrx10(−/−) lrx11(+/−) × ♂WT | 212 | 187 | NA | 88.2% | NA | 1.44 | ||
Pollen Germination and Pollen Tube Growth Were Impaired Significantly in the Triple Mutants and the Quadruple Mutant in Vitro and in Vivo
To investigate the reason for the decline in transmission efficiency of the male gametophyte, Alexander, fluorescein diacetate (FDA), and DAPI staining were used to examine pollen viability and cell nuclei development in single, double, triple, and quadruple mutant plants. However, no obvious difference was found compared with wild-type plants (Supplemental Fig. S7).
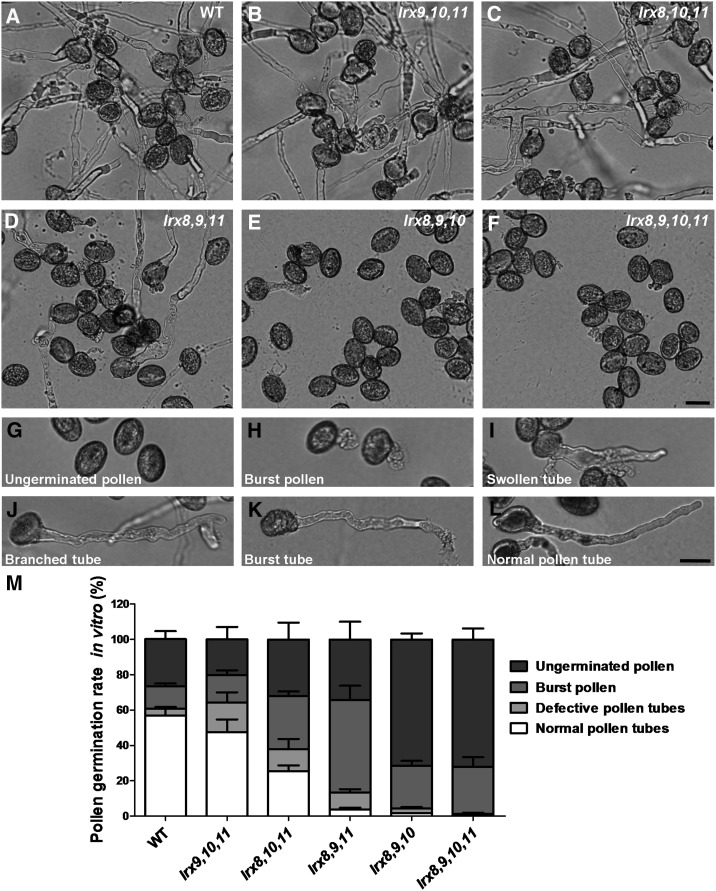
We next performed pollen germination and pollen tube growth assays in vitro using mutant and wild-type plants. The pollen germination rates in the single mutants lrx8, lrx9, lrx10, and lrx11 were comparable to those in the wild type (Supplemental Fig. S8). Among the six double mutants tested, only lrx8 lrx9 and lrx8 lrx10 exhibited significantly reduced pollen germination rates compared with the wild type (Supplemental Fig. S8).
In contrast, in the four triple mutants and the quadruple mutant, the pollen germination rates and pollen tube growth were remarkably impaired compared with the wild type (Fig. 4, A–F). According to the severity of the defect, the pollen grains and pollen tubes were divided into four types: (1) ungerminated pollen; (2) pollen grains that burst upon germination; (3) germinated pollen grains with defective pollen tubes, including swollen, branched, or burst tubes; and (4) successfully germinated pollen grains and normal pollen tubes (Fig. 4, G–L).
Figure 4.
The mutation of LRX8–11 caused defects in pollen germination and pollen tube growth in vitro. A to F, Pollen from wild-type (WT) and mutant plants was germinated in vitro for 5 h. Bar = 20 μm. G to L, Images of representative pollen grains and pollen tubes. G, Ungerminated pollen. H, Pollen that burst instantly upon germination. I, Swollen tube. J, Branched tube. K, Burst tube. L, Normal pollen tube. Bar = 20 μm. M, Statistical analysis of the pollen germination rates in wild-type and mutant plants. Data were collected from three independent experiments, and the number of pollen grains collected each time was greater than 300 (n > 1,000).
A statistical analysis of the quadruple mutant revealed that most of the pollen had either not germinated or burst, so the plants produced few pollen tubes (Fig. 4M). Among the four triple mutants, lrx8 lrx9 lrx10 displayed the most serious defects, with the highest proportions of ungerminated and burst pollen grains, while lrx9 lrx10 lrx11 exhibited the least serious defects, with the highest proportions of normally germinated pollen and defective pollen tubes; the results for the other two triple mutants (lrx8 lrx9 lrx11 and lrx8 lrx10 lrx11) were somewhere in the middle (Fig. 4M). The observed severity of the defects in pollen germination and pollen tube growth is consistent with the degree of impaired fertility detected among the mutants. Therefore, the triple mutants and the quadruple mutant exhibited dramatic defects in pollen germination and pollen tube growth in vitro.
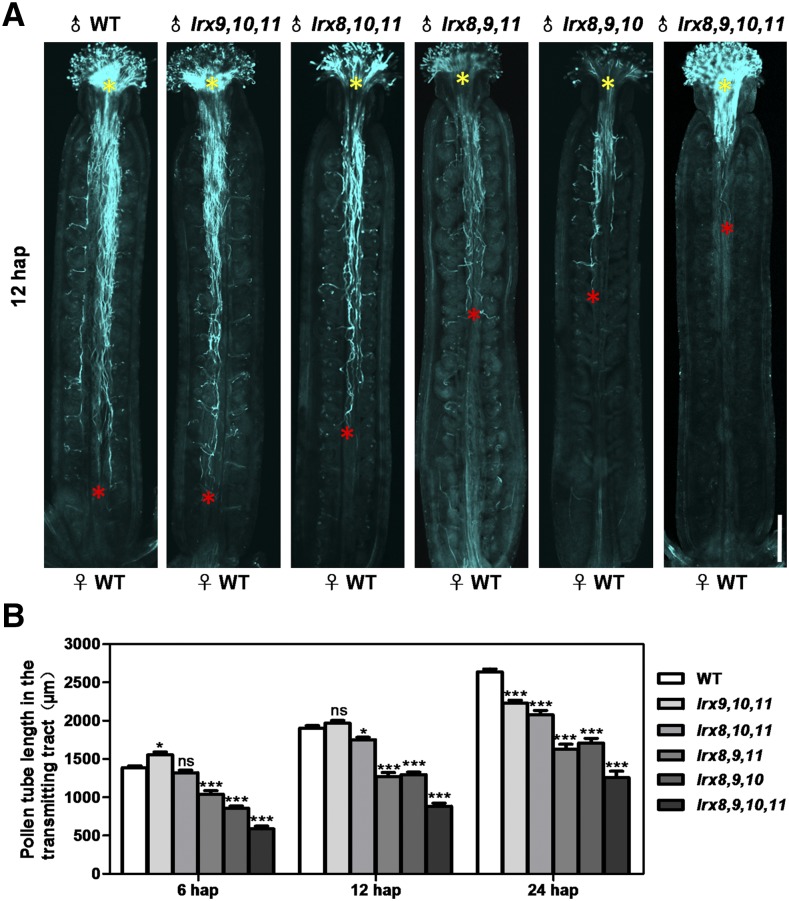
Subsequently, the pollen tube growth assay in vivo was conducted using four triple mutants and the quadruple mutant at 6, 12, and 24 h after pollination (hap). We also measured the length of the fastest pollen tubes of the wild type and mutants in the transmitting tract and performed statistical analysis (Fig. 5; Supplemental Fig. S9). At 6 hap, Aniline Blue staining revealed that the pollen tubes in the wild-type and all mutant plants were able to pass through the style and enter the transmitting tract. Notably, the pollen tube of the triple mutant lrx9 lrx10 lrx11 grew faster than that of the wild type. In contrast, two triple mutants (lrx8 lrx9 lrx11 and lrx8 lrx9 lrx10) and the quadruple mutant showed notably retarded growth compared with the wild type (Fig. 5B; Supplemental Fig. S9A). At 12 hap, the pollen tubes of the wild type and lrx9 lrx10 lrx11 almost reached the base of the transmitting tract, while the pollen tubes in the other three triple mutants and the quadruple mutant were still far away from the bottom of the transmitting tract (Fig. 5). At 24 hap, the length of the pollen tubes in all of the mutants tested was significantly shorter than that in the wild type (Fig. 5B; Supplemental Fig. S9B). Therefore, pollen tube growth in the triple mutants and the quadruple mutant was significantly compromised in vivo. These results indicate that LRX8–11 play critical roles in pollen germination and pollen tube growth.
Figure 5.
Pollen tube growth in the transmitting tract was compromised in the mutant plants. A, Growth of wild-type (WT) and mutant pollen tubes in the wild-type pistils at 12 hap as visualized by Aniline Blue staining. Yellow asterisks indicate the beginning of the pollen tubes, and red asterisks show the locations of the fastest pollen tubes. Bar = 200 μm. B, Statistical analysis of the absolute length of the pollen tube in the transmitting tract at 6, 12, and 24 hap. The data were collected from three independent experiments. n at 6 hap = 52, 43, 52, 52, 52, 52; n at 12 hap = 52, 48, 52, 45, 52, 52; and n at 24 hap = 52, 52, 52, 47, 52, 47. Error bars represent se (*, P < 0.05 and ***, P < 0.001; ns, not significant, calculated using two-way ANOVA).
Complementation of the Observed Mutant Phenotypes and an Analysis of the Subcellular Localization of LRX8, LRX10, and LRX11
To confirm that the observed mutant phenotypes resulted from the disruption of LRX8–11, we generated the following transgenic lines: ProLRX8:LRX8 and ProLRX8:LRX8-GFP(C) [C-terminal fusion] in lrx8 lrx9 lrx10, ProLRX10:LRX10 genome-GFP(C) in lrx8 lrx9 lrx10, and ProLRX11:LRX11-GFP(C) in lrx8 lrx9 lrx11. The transcription patterns and phenotypes of these transgenic lines were then examined (Supplemental Fig. S10). ProLRX8:LRX8/lrx8 lrx9 lrx10 T3-18-18, a single-copy homozygous line, restored the defects observed in lrx8 lrx9 lrx10 in terms of in vitro pollen germination and fertility (i.e. seed set). ProLRX8:LRX8-GFP(C), ProLRX10:LRX10-GFP(C), and ProLRX11:LRX11-GFP(C) also recovered the in vitro defects in pollen germination rate observed in lrx8 lrx9 lrx10 and lrx8 lrx9 lrx11 (Supplemental Fig. S11). Together, these results indicate that the loss of function of LRX8–11 caused the observed mutant phenotypes.
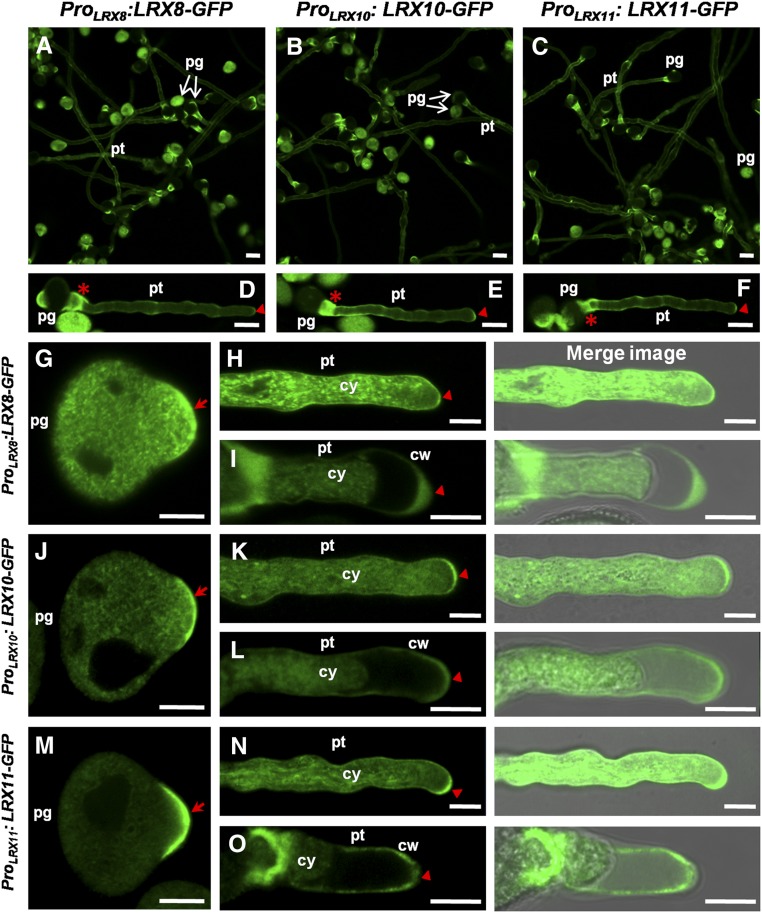
Next, the subcellular localization of LRX8, LRX10, and LRX11 was explored in pollen grains and pollen tubes. LRX8, LRX10, and LRX11 displayed similar subcellular localization patterns in germinated pollen grains and growing pollen tubes. GFP signals corresponding to LRX8-GFP, LRX10-GFP, and LRX11-GFP were detected in the collar region and tip of pollen tubes (Fig. 6, A–F). In pollen grains that had germinated for about 30 min, a strong fluorescence signal was detected in the tip of the germination site (Fig. 6, G, J, and M). In growing pollen tubes, strong fluorescence was detected upon further examination in the cytoplasm and the plasma membrane and/or cell walls of the pollen tubes (Fig. 6, H, K, and N; Supplemental Movie S1). To distinguish whether the signal was from the plasma membrane or the cell wall, a plasmolysis assay was performed. After plasmolysis, the majority of the signal was in the cytoplasm, and some was clearly detected in the cell walls of the pollen tubes (Fig. 6, I, L, and O). Thus, LRX8-GFP, LRX10-GFP, and LRX11-GFP are located in the cytoplasm and cell walls of growing pollen tubes.
Figure 6.
GFP-fused LRX8, LRX10, and LRX11 were located in the cytoplasm and walls of pollen tubes. Subcellular localization analysis of ProLRX8:LRX8-GFP(C), ProLRX10:LRX10-GFP(C), and ProLRX11:LRX11-GFP(C) in Arabidopsis is shown. A to C, Fluorescence images showing the GFP signals from LRX8-GFP, LRX10-GFP, and LRX11-GFP in pollen grains and pollen tubes cultured in vitro. pg, Pollen grains; pt, pollen tubes. Bars = 20 μm. D to F, Images of representative pollen tubes. Red asterisks mark the collar region, and red triangles indicate the pollen tube tip. Bars = 10 μm. G, J, and M, LRX8-GFP, LRX10-GFP, and LRX11-GFP localization in germinated pollen grains. Red arrows indicate the tip of the germination site. Bars = 10 μm. H, K, and N, LRX8-GFP, LRX10-GFP, and LRX11-GFP localization in growing pollen tubes. Red triangles indicate the tip of the pollen tube. cy, Cytoplasm. Bars = 5 μm. I, L, and O, A plasmolysis assay shows the locations of LRX8, LRX10, and LRX11 in the cytoplasm and walls of pollen tube cells. Red triangles indicate the tips of the pollen tubes. cw, Cell wall. Bars = 5 μm.
The Abundance of Pollen Tube Wall Polysaccharides Was Altered in the Mutants
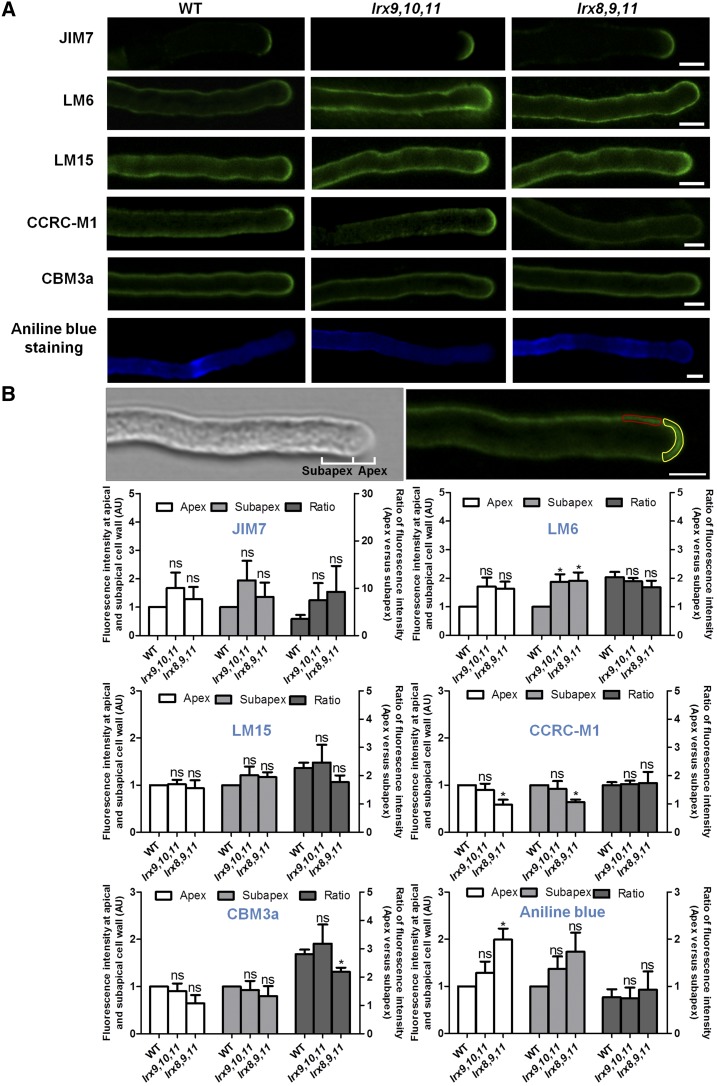
We next examined the distribution of pollen tube cell wall polysaccharides in two triple mutants (lrx9 lrx10 lrx11 and lrx8 lrx9 lrx11) and the wild type by immunolabeling. Several sugar-specific antibodies were used, including JIM7 (recognizes highly methylesterified homogalacturonan [HG]), LM6 (recognizes RGI), LM15 (recognizes nongalactosylated XyG), CCRC-M1 (recognizes fucosylated XyG), and cellulose-binding module 3a (CBM3a; recognizes crystalline cellulose). We detected and compared the abundance and distribution of cell wall polymers recognized by the antibodies (Fig. 7A). Moreover, the mean fluorescence intensity in the apex and subapex regions of pollen tubes, as well as the ratio between them, were measured and analyzed statistically (Fig. 7B).
Figure 7.
The abundance of cell wall polysaccharides in the pollen tube was changed in lrx9 lrx10 lrx11 (lrx9,10,11) and lrx8 lrx9 lrx11 (lrx8,9,11). A, Immunolabeling of pollen tube wall polysaccharides in wild-type (WT), lrx9,10,11, and lrx8,9,11 mutant plants using JIM7, LM6, LM15, CCRC-M1, and CBM3a. JIM7 labeled HG in the pollen tubes of wild-type, lrx9,10,11, and lrx8,9,11 plants. LM6 recognized RGI in the pollen tubes of wild-type, lrx9,10,11, and lrx8,9,11 plants. LM15 recognized nongalactosylated (XXXG motif) XyG in the pollen tubes of wild-type, lrx9,10,11, and lrx8,9,11 plants. CCRC-M1 recognized α-l-fucosylated XyG in the pollen tubes of wild-type, lrx9,10,11, and lrx8,9,11 plants. The labeling of crystalline cellulose was performed with CBM3a in the pollen tubes of wild-type, lrx9,10,11, and lrx8,9,11 plants. Callose was labeled using Aniline Blue in pollen tubes from wild-type, lrx9,10,11, and lrx8,9,11 plants. Bars = 5 μm. B, Quantitative analysis of the fluorescence intensity at the apical and subapical cell walls of the pollen tubes, and the ratio of the fluorescence intensity at the apex versus the subapex in wild-type, lrx9,10,11, and lrx8,9,11 plants. A single scan along the median of the pollen tube was used for quantification. The images in the top row show the apical and subapical regions of the pollen tube cell walls used for fluorescence intensity measurement. Apex, 0 to 3 μm away from the tip (marked in yellow); subapex, 3 to 8 μm away from the tip (marked in red). The fluorescence intensity of each region was quantified by measuring the mean gray value using ImageJ2x software. AU, Arbitrary fluorescence intensity units. Pollen grains were germinated in vitro for 3 h. Three biological repeats were performed, and 20 to 30 pollen tubes were used for each repeat. Error bars show se. Asterisks indicate values that differed significantly from the wild type (*, P < 0.05; ns, not significant, calculated using a two-way ANOVA and Student’s t test).
JIM7-labled HG deposition at the apex was a significant feature of wild-type pollen tubes. JIM7 labeling revealed that the lrx9 lrx10 lrx11 and lrx8 lrx9 lrx11 mutants showed no obvious difference compared with the wild type in the mean fluorescence intensity in the apex and subapex regions of the pollen tube walls as well as in the ratio between them (Fig. 7). RGI is a pectic polysaccharide that is highly branched with arabinan. LM6 recognizes RGI by binding to (1,5)-α-l-arabinan. In wild-type plants, LM6 label was distributed throughout the pollen tube walls but enriched at the tip. The mutants lrx9 lrx10 lrx11 and lrx8 lrx9 lrx11 displayed a much stronger signal in the subapical walls of pollen tubes when labeled by LM6, but the fluorescence intensity of the apical walls and the ratio of the apex to the subapex were not obviously affected in the mutants (Fig. 7).
LM15 and CCRC-M1 recognize nongalactosylated and fucosylated XyG, respectively. Our immunohistochemical results indicate no significant difference in LM15 fluorescence intensity between lrx9 lrx10 lrx11 or lrx8 lrx9 lrx11 and the wild type (Fig. 7). In comparison, CCRC-M1 fluorescence was reduced significantly at the apex and subapex of lrx8 lrx9 lrx11 pollen tubes, but the ratio between them was not changed (Fig. 7). The crystalline cellulose content, revealed by CBM3a labeling, was not obviously changed in the mutants lrx9 lrx10 lrx11 and lrx8 lrx9 lrx11, whereas the ratio of the fluorescence intensity at the apex to that at the subapex was reduced significantly in lrx8 lrx9 lrx11 (Fig. 7). Aniline Blue staining revealed a pronounced callose signal in the apical walls but not in the subapical walls of lrx8 lrx9 lrx11 pollen tubes compared with the wild type, and the ratio of the fluorescence intensity at the apex to that at the subapex was not changed significantly (Fig. 7). These data indicate that the loss of LRX8–11 function alters the abundance and distribution of pollen tube cell wall polysaccharides.
The Expression of Genes Involved in the Biosynthesis and/or Modification of Pollen Tube Cell Wall Polysaccharides
Immunohistochemical analyses showed that the polysaccharide composition of the pollen tube wall in two triple mutants was changed. Therefore, we selected several genes known to be involved in the biosynthesis and/or modification of cell wall polysaccharides in pollen grains and pollen tubes and performed a quantitative analysis of their transcript levels in lrx9 lrx10 lrx11 and lrx8 lrx9 lrx11. The selected genes included those encoding cellulose synthases (CESA1 and CESA3), cellulose synthase-like D proteins (CSLD1 and CSLD4), a pollen-specific pectin methylesterase1 (PPME1), pectin methylesterase48 (PME48), the PME-homologous protein VANGUARD1 (VGD1), a sialyltransferase-like protein (MALE GAMETOPHYTE DEFECTIVE2 [MGP2]), and an RGII xylosyltransferase (MGP4; Supplemental Fig. S12). However, quantitative real-time PCR revealed no significant change in the transcription of these genes in lrx9 lrx10 lrx11 and lrx8 lrx9 lrx11.
DISCUSSION
During pollen tube growth, the cell wall must synchronously maintain mechanical strength and plasticity (Geitmann, 2010; Mollet et al., 2013; Vogler et al., 2013). Despite their low abundance, structural proteins affect cell wall integrity; however, knowledge about the functions of these proteins in pollen tube cell walls is limited (Cassab, 1998; Geitmann, 2006). LRXs are cell wall proteins that typically harbor an LRR domain and an EXT domain (Baumberger et al., 2003a). In 1995, the first LRX protein, Pex1, was identified in maize. Pex1 is highly expressed in pollen and pollen tubes, particularly in pollen tube walls; thus, it has attracted the attention of scientists studying sexual reproduction (Rubinstein et al., 1995a, 1995b). However, until now, the function of LRXs in plants has been elusive. In this study, the biological functions of four pollen-expressed LRX proteins in Arabidopsis (LRX8–11) were characterized in detail. We found that LRX8–11 play redundant, crucial roles in pollen germination and pollen tube growth in Arabidopsis.
Genetic evidence, expression pattern analyses, and subcellular localization studies indicate the essential role of LRX8–11 in pollen germination and pollen tube growth in Arabidopsis. The transmission efficiency of the male gametophyte was reduced significantly in the lrx8 single mutant, lrx8 lrx9 double mutant, and lrx8 lrx9 lrx10 lrx11 quadruple mutant. The triple and quadruple mutants exhibited remarkably reduced fertility accompanied by poor pollen germination, deformed pollen tubes in vitro, and retarded pollen tube growth in vivo (Figs. 2 and 4). Similar to Pex1 in maize, LRX8–11 are expressed mainly in mature pollen and pollen tubes; GFP-fused LRX8, LRX10, and LRX11 were detected in pollen tube cell walls (Fig. 6), suggesting that the pollen-expressed LRXs in monocots and dicots have conserved functions (Ringli, 2005). We also found that, although LRX8–11 function redundantly, the importance of these four proteins was not consistent. LRX8 was the most important contributor, followed by LRX9, LRX10, and LRX11.
LRXs belong to the EXT family (Liu et al., 2016). Classic EXTs are induced by wounding and mechanical stress, and they are involved in maintaining cell wall rigidity (Merkouropoulos et al., 1999). A previous study of EXT3 suggested that EXTs participate in the formation of new cell walls (Cannon et al., 2008). Our data support this hypothesis. In the lrx8 lrx9 lrx10 and lrx8 lrx9 lrx10 lrx11 mutants, most of the pollen could not germinate or burst after germination. In triple mutant plants (lrx9 lrx10 lrx11, lrx8 lrx10 lrx11, and lrx8 lrx9 lrx11), some pollen could germinate, but the pollen tubes displayed morphological deformities or ruptured. Combined with the observed localization of LRX8, LRX10, and LRX11 to the apex and shank walls of pollen tubes (Fig. 6), LRX8–11 may be required for early cell wall deposition during pollen germination and cell wall assembly during pollen tube elongation. Notably, ext18 has a similar phenotype to double and triple mutants of LRX8–11, including a decreased pollen germination rate and burst pollen grains (Choudhary et al., 2015). However, the progeny of a self-crossed ext18(+/−) heterozygote segregated normally and the ext18 mutant exhibited multiple vegetative defects, which are different from double and triple mutants of LRX8–11. These data suggest a functional difference between LRX8–11 and EXT18 in plant growth. Other cell wall proteins also are reported to be involved in pollen tube growth. For example, arabinogalactan proteins (AGPs) are a class of highly glycosylated Hyp-rich proteins (Ellis et al., 2010; Pereira et al., 2015; Borassi et al., 2016). Mutations in Arabidopsis AGP6 and AGP11 and in Brassica campestris male fertility8 were shown to cause reduced pollen germination rates and impaired pollen tube growth (Levitin et al., 2008; Coimbra et al., 2009, 2010). Similarly, in tomato, the Gly-rich protein LeGRP92 has been implicated in pollen exine development, pollen viability, and poor germination (McNeil and Smith, 2010). Thus, cell wall proteins play important roles in maintaining the integrity of pollen tubes.
Unlike the situation in cells in vegetative tissue, the pollen tube cell wall is enriched with pectin and callose (Mollet et al., 2013). Biochemical and immunocytological analyses have shown that HG and RGI with arabinan are abundant pectic polysaccharides in pollen tube cell walls (Dardelle et al., 2010). In the lrx9 lrx10 lrx11 and lrx8 lrx9 lrx11 mutants, the amount of RGI in subapical walls of pollen tubes increased significantly compared with those in the wild type, while the content of highly methylated HG was not changed remarkably (Fig. 7). RGI is highly substituted by an arabinan side chain. LM6 labeling revealed that (1,5)-α-l-arabinan was distributed throughout the entire pollen tube, with a small increase at the apex. The pectic arabinan is reported to contribute to pollen development and pollen germination (Cankar et al., 2014; Stonebloom et al., 2016). The exact roles of arabinan in pollen tube growth are unclear. ARABINAN DEFICIENT1 and ARABINAN DEFICIENT2, which are putative arabinosyltransferases, may participate in the biosynthesis of RGI. However, the corresponding mutants exhibited no obvious growth defects (Harholt et al., 2006, 2012). It is thought that RGI regulates the porosity and generates the flexibility of cell walls (Baron-Epel et al., 1988; Verhertbruggen et al., 2009). Thus, an increase in branched RGI may influence the porosity of pollen tubes and render the wall unstable during pollen tube growth.
As the primary polysaccharide in the pollen tube walls of Arabidopsis, callose is deposited mainly in the shank region, where it provides mechanical support (Parre and Geitmann, 2005; Nedukha, 2015). However, callose was detected in the apical walls in lrx8 lrx9 lrx11 plants, which exhibited slow-growing pollen tubes. Such ectopic deposition of callose may have reduced the plasticity of the pollen tube and hampered rapid pollen tube growth. We also found a decrease in the content of fucosylated XyG in lrx8 lrx9 lrx11 plants. In somatic Arabidopsis cells, xyloglucan is the most abundant hemicellulose and is thought to cross-link with cellulose (Scheller and Ulvskov, 2010). However, this link has not been proved in pollen tubes. Both fucosylated XyG and cellulose are localized to the inner cell walls in pollen tubes, implying a close relationship among them (Chebli et al., 2012). Therefore, the reduced content of fucosylated XyG may have affected the mechanical strength of the pollen tubes in lrx8 lrx9 lrx11. It is worth noting that the changes in wall components caused by the mutation may have altered the accessibility of the cell wall to antibodies or the overall thickness of the wall; either change could have influenced our immunolabeling data. A quantitative analysis of wall polysaccharides by gas chromatography-mass spectrometry could help resolve this issue. However, the expression levels of genes involved in the biosynthesis and/or modification of cell wall polysaccharides were not changed significantly in two triple mutants (lrx9 lrx10 lrx11 and lrx8 lrx9 lrx11) compared with the wild type.
How does the mutation of LRX8–11 cause changes in pollen cell wall polysaccharides? We cannot provide a complete explanation based on our results here. However, if we combine our results with those from previous studies of classic EXTs and LRX1 in Arabidopsis, clues can be found. First, EXTs are amphiphiles that self-assemble into scaffolds via covalent intramolecular and intermolecular cross-linking. Through acid-base interactions, EXTs and pectin may form EXT pectate, which may serve as the template for pectin deposition (Cannon et al., 2008; Lamport et al., 2011). Therefore, LRX8–11 also may play an important role in pectin deposition. Second, LRX1 is expressed specifically in root hairs, which, like pollen tubes, display tip growth. The disruption of LRX1 results in severe defects in root hair elongation (e.g. the swelling and rupture of root hairs; Baumberger et al., 2001). Interestingly, two suppressors, rol1 lrx1 and rol5 lrx1, which were shown to recover root hair growth in lrx1 plants, displayed significantly decreased RGI contents (Diet et al., 2006; Leiber et al., 2010). The authors of that study presumed that the decrease in RGI helped ameliorate the disordered cell wall in the root hairs of the lrx1 mutant. Our results show that the RGI content was elevated significantly in subapical walls of pollen tubes in lrx9 lrx10 lrx11 and lrx8 lrx9 lrx11 plants. Therefore, it is possibly that LRX8–11 may regulate RGI deposition in pollen tubes. However, if LRX8–11 serve as a scaffold for RGI deposition, the RGI content should have been reduced in the mutants. Another possibility is that LRX8–11 provide a template for other polysaccharides, which were not properly deposited in the triple mutants and, in turn, triggered a change in the RGI content. Beyond the EXT domain of LRXs, one should consider the LRR domain, which is relatively conserved throughout the LRX family (Baumberger et al., 2003a). Identifying the binding partner(s) of the LRR domain in LRXs will help uncover the function of LRXs in pollen tube cell walls.
In conclusion, LRX8–11, which are expressed preferentially in mature pollen and pollen tubes, play crucial roles in pollen germination and pollen tube growth, possibly by maintaining pollen tube wall integrity. Our work provides genetic evidence for the vital function of LRX8–11 in pollen tube growth and highlights the importance of cell wall proteins in plant growth.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) plants used in this research were of the Columbia (Col-0) ecotype. The T-DNA insertion mutants LRX8 (AT3G19020; SALK_132358), LRX9 (AT1G49490; SALK_137156), LRX10 (AT2G15880; SALK_087083C), and LRX11 (AT4G33970; SALK_076356) were obtained from the ABRC (http://www.arabidopsis.org). The seeds of wild-type and mutant plants were surface sterilized with 75% alcohol for 6 min, washed three times with sterile water, and plated on Murashige and Skoog (MS) medium. The transgenic plant seeds were sterilized in 2.5% sodium hypochlorite, washed three times with sterile water, and plated with 50 mg L−1 carbenicillin and 20 mg L−1 hygromycin. The MS plates were placed at 4°C for 3 d and then transferred to growth chambers under a 16-h-light/8-h-dark photoperiod at 22°C. After the seedlings had grown for ∼10 d on the MS medium, they were transplanted to soil under the above-mentioned conditions.
Mutant Genotyping
Mutant plants were identified by genotyping PCR, which was performed with the T-DNA insertion primer pairs shown in Supplemental Table S2. The primer on T-DNA is LBb1.3 (5′-ATTTTGCCGATTTCGGAAC-3′).
Genetic Analyses
All genetic analyses of LRX mutants were performed as described by Jiang et al. (2005) and Liu et al. (2011).
Expression Analyses
RT-PCR
Total RNA was extracted from various tissues in 5-week-old Arabidopsis plants using the Eastep Super Total RNA Extraction Kit (Promega; LS1040). Then, 1,000 ng of RNA was converted into single cDNAs using the PrimeScript RT Reagent Kit (Takara; RR037Q). The transcripts of all mutants were checked by RT-PCR using RT primers as described in Supplemental Table S4. ACTIN7 served as a loading control. Amplification was performed with 25 cycles for ACTIN7 and 35 cycles for LRX8 (633 bp), LRX9 (702 bp), LRX10 (1,138 bp), and LRX11 (436 bp).
GUS Staining
The putative promoters, which included 2,040, 2,070, 2,048, and 2,075 bp of sequence upstream of the start codon (ATG) of LRX8, LRX9, LRX10, and LRX11, respectively, were amplified from Col-0 genomic DNA by PCR using promoter primer pairs (Supplemental Table S3). These clones were fused with the GUS reporter gene in the vector pCAMBIA1300 (Cambia; http://www.cambia.org/daisy/cambia/home.html). All constructs were introduced to Col-0 plants of Arabidopsis using the inflorescence dipping method. Single-copy homozygous transgenic plants were obtained with the resistance screening used for GUS staining assays. The GUS staining solution contained 1 mg mL−1 X-GlcA, 0.5 mm potassium ferricyanide, 0.5 mm potassium ferrocyanide, 10 mm EDTA2Na, and 0.1% Triton X-100 in 50 mm sodium phosphate buffer, pH 7. The staining method was as follows. Samples including inflorescence and rosette leaves were fixed in precooled 90% acetone for 1 h, while pollen tubes cultured in vitro were not incubated in acetone. The samples were incubated at 37°C in the dark in the GUS staining solution. Next, inflorescence and rosette leaf chlorophyll was removed with several washes in 95% ethanol, and the tissues were treated with 75% ethanol (Yang et al., 1999).
Observation of Embryo Development
To track embryo development, open and flat flowers from wild-type and mutant plants were marked with colorful thread at 10 am for 8 d. Then, the marked siliques were picked up in a fixed solution (4% glutaraldehyde and 0.1% Triton X-100 in PBS, pH 7.4). The sample was transferred to a transparent solution (chloral hydrate, water, and glycerol at a ratio of 8:2:1) until the embryos became colorless (Deng et al., 2014). The embryos were examined and photographed using a microscope (Zeiss; AXIO Imager A1) under differential interference contrast optics.
Characterization of Pollen Grains
Pollen grain characterization was done by Alexander staining (Alexander, 1969), DAPI staining as described by McCormick (2004), and FDA staining as described by Pinney and Polito (1989).
Images of Alexander staining and DAPI staining were captured with a fluorescence microscope (Zeiss; AXIO Imager A1). The FDA staining images were captured with a laser confocal fluorescence microscope (Zeiss; LSM710).
Pollen Tube Growth in Vitro and in Vivo
For pollen germination in vitro, mature pollen grains were spread on solid medium containing 0.01% H3BO4, 5 mm CaCl2, 5 mm KCl, 1 mm MgSO4, 10% Suc, and 0.1% agarose as described (Boavida and McCormick, 2007). The samples were incubated for 5 or 6 h, and pollen germination results were examined with a microscope (Zeiss; AXIO Imager A1).
For pollen germination in vivo, emasculated flowers from wild-type plants were hand pollinated with wild-type and mutant pollen. The pollinated pistils were collected at 6, 12, and 24 hap and stained by Aniline Blue. The Aniline Blue staining method was as follows. Samples were fixed in an ethanol:acetic acid (3:1, v/v) solution and washed three times with 0.1 m PBS. The pollinated pistil was treated overnight in 8 m NaOH. After several washes with 0.1 m PBS, the pistil tissues were stained with decolorized Aniline Blue solution (0.1% Aniline Blue in 100 mm K3PO4 buffer, pH 11) overnight in the dark (Johnson-Brousseau and McCormick, 2004). Aniline Blue was purchased from Sangon Biotech (CAS no. 28631-66-5). All images were captured using an Olympus confocal fluorescence microscope (FV3000). A single scan along the median of the pollen tube was used for imaging. Images were captured using 405 nm for excitation (FV3000 in MATL mode; Olympus). The pollen tube length in the transmitting tract was measured using ImageJ2x.
Complementation and Subcellular Localization of the LRXs
We investigated whether LRX8 could complement the mutant phenotype. The LRX8 promoter and a genomic DNA fragment were cloned using specific primers for the LRX8 genome: F1/R1 and F2/R2. The two fragments were inserted into the pCambia1300-Pro35S:TNOS binary vector using PstI/SalI and SalI/XbaI. For subcellular localization, the LRX8 promoter and genomic DNA fragment were obtained using specific primers (LRX8 genome F1/R1 and F2/R2 No end) and cloned into the pCambia1300-Pro35S:GFP(C) binary vector, including a C-terminal-enhanced GFP fused with the target protein using PstI/SalI and SalI/XbaI. The fusion of LRX9 with GFP also was attempted, and the specific primers for the LRX9 genome were F1/R1 and F2/R2. The correct clone of LRX9 could not be isolated from Escherichia coli.
For the LRX10 promoter and genomic DNA fragment fused with GFP, the recombination process was similar to that described above. The specific primers used were LRX10 genome F1/R1 and F2/R2, and the restriction enzymes employed were PstI/NruI and NruI/XbaI. The LRX11 promoter and genomic DNA fragment were cloned with specific primers for the LRX11 genome, F1/R1 and F2/R2. Next, the two fragments were inserted into the pCambia1300-Pro35S:GFP(C) binary vector using PstI/HindIII and HindIII/XbaI.
All transgenic plants were screened for resistance to 50 mg L−1 carbenicillin and 20 mg L−1 hygromycin. In the T2 generation, resistance/sensitivity between 1.5 and 5.6, considered to be a single-copy insertion, were selected. In the T3 generation, homozygous transgenic plants were screened out using resistance markers.
For plasmolysis, the germinating pollen tubes on solid medium were soaked in a 40% (w/v) Suc solution for 30 min. The subcellular localization of LRX8, LRX10, and LRX11 in pollen tubes was visualized with a laser confocal fluorescence microscope (Zeiss; LSM710).
Phylogenetic Analysis
The amino acid sequence of LRX8 and 25 similar sequences identified by a BLAST search (Camacho et al., 2009) were aligned with the ClustalW tool applying default parameters. A phylogenetic tree was constructed by MEGA 6.06 using the neighbor-joining method (http://www.megasoftware.net/; Tamura et al., 2013).
The sequence alignment of LRX8, LRX9, LRX10, and LRX11 was achieved using DNAMAN version 6 (http://www.lynnon.com/).
Immunohistochemical Analysis of Pollen Tubes
Pollen germination in solid medium for 3 h in vitro was performed as described previously. Samples were fixed with 4% paraformaldehyde for 30 min at room temperature. Pollen tubes were washed with 0.1 m PBS three times for 5 min each. Samples were blocked with 1% BSA in 0.1 m PBS for 30 min and rinsed with 0.1% BSA (Solarbio) in 0.1 m PBS. Pollen tubes were incubated overnight at 4°C in the dark with the primary antibody (diluted with 0.1% BSA in 0.1 m PBS). After washing three times with 0.1% BSA in 0.1 m PBS, a secondary antibody was used, combined with fluorescein isothiocyanate (FITC). Samples were washed with 0.1% BSA in 0.1 m PBS three times and covered with a coverslip in antifade. Laser confocal fluorescence microscopy (Zeiss; LSM710) was performed to take photographs using 488-nm excitation light. Three independent biological repeats were performed; 20 to 30 pollen tubes were measured in each repeat (n > 60). Three batches of Arabidopsis plants grown for different lengths of time were used in every independent immunolabeling experiment. The relative value of the fluorescence intensity was presented in the mutants, with the wild-type signal defined as 1 in each biological repeat. The mean gray value was obtained through a statistical analysis performed using ImageJ2x. Data on the fluorescence intensity at the apical and subapical walls of pollen tubes were analyzed using a two-way ANOVA, while the fluorescence intensity ratio between the apex and subapex was examined using Student’s t test. All data were processed with GraphPad Prism 5 software.
The primary antibodies used in our study were LM6, LM15, CBM3a (University of Leeds; http://www.plantprobes.net/), JIM7, and CCRC-M1 (Complex Carbohydrate Research Center; https://www.ccrc.uga.edu/). LM6 recognizes (1,5)-α-l-arabinose oligomers (diluted 1:20). LM15 recognizes a linear tetrasaccharide in 1,4-β-d-galactans (diluted 1:20). Labeling for crystalline cellulose was performed with CBM3a (diluted 1:100). JIM7 label methyl esterified homogalacturonan (diluted 1:20). CCRC-M1 recognizes α-l-fucosylated xyloglucan (diluted 1:20).
For LM6, LM15, and JIM7 antibody detection, the secondary antibody anti-rat IgG-FITC (diluted 1:100; Abbkine) was used; for CCRC-M1 antibody detection, goat anti-mouse IgG-FITC (diluted 1:100; EarthOx) was used. For CBM3a antibody detection, monoclonal mouse anti-His monoclonal antibody (diluted 1:100; TransGen Biotech) was used, followed by subsequent incubation with goat anti-mouse IgG-FITC (diluted 1:100; EarthOx). Controls were performed by the incubation of pollen tubes with the secondary antibody alone.
Quantitative Real-Time PCR Assay
Total RNA was extracted from pollen germinated for 3 h in liquid germination medium in vitro using the Eastep Super Total RNA Extraction Kit (Promega; LS1040). Next, 1,000 ng of RNA was reverse transcribed into cDNA using the PrimeScript RT Reagent Kit (Takara; RR037Q). Quantitative real-time PCR was performed using UltraSYBR Mixture (CW0957M; CWBIO) and a Bio-Rad quantitative real-time PCR system. The reference genes were ACTIN8 and UBQ5. The relative expression levels were determined using the 2–ΔΔCT method. Three independent biological replicates and technical replicates of each sample were performed. The data were then analyzed using Bio-Rad CFX Manager and GraphPad Prism 5. The primers used for quantitative real-time PCR are described in Supplemental Table S5. The data were analyzed using a one-way ANOVA.
Primers Used in This Study
The primers used in this study are listed in Supplemental Tables S2 to S4.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: LRX8 (AT3G19020), LRX9 (AT1G49490), LRX10 (AT2G15880), LRX11 (AT4G33970), and ACTIN7 (AT5G098103). The genes involved in quantitative PCR were as follows: ACTIN8 (AT1G49240), UBQ5 (AT3G62250), CESA1 (AT4G32410), CESA3 (AT5G05170), CSLD1 (AT2G33100), CSLD4 (AT4G38190), PPME1 (AT1G69940), PME48 (AT5G07410), VGD1 (AT2G47040), MGP2 (AT1G08660), and MGP4 (AT4G01220).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Evolutionary relationships among the LRX amino acid sequences from Arabidopsis, tomato, maize, rice, and B. distachyon.
Supplemental Figure S2. Sequence alignment of LRX8–11.
Supplemental Figure S3. LRX8–11 are expressed mainly in open flowers and inflorescences.
Supplemental Figure S4. The triple mutants and the quadruple mutant showed normal vegetative growth but abnormal reproductive growth.
Supplemental Figure S5. Seed set of single and double mutant plants.
Supplemental Figure S6. Embryo development in wild type, lrx8 lrx9 lrx11, and lrx8 lrx9 lrx10 lrx11 from 1 to 8 d after pollination.
Supplemental Figure S7. The viability and nuclei of mutant pollen grains were normal in the mutants.
Supplemental Figure S8. In vitro pollen germination analyses of the single mutants and double mutants.
Supplemental Figure S9. Growth of wild-type and mutant pollen tubes in the wild-type pistils at 6 and 24 hap.
Supplemental Figure S10. RNA level identification of the transgenic rescue lines.
Supplemental Figure S11. LRX8, LRX10, and LRX11 expression almost rescued the in vitro pollen germination rates of the corresponding mutants.
Supplemental Figure S12. Expression of genes involved in the biosynthesis and/or modification of pollen tube cell wall polysaccharides.
Supplemental Table S1. Segregation data from selfed progeny and a transmission efficiency analysis of the single and double mutants.
Supplemental Table S2. Primers used for DNA and RNA level identification.
Supplemental Table S3. Primers used to prepare the constructs.
Supplemental Table S4. Primers used for quantitative real-time PCR.
Supplemental Table S5. Constructs used in this study.
Supplemental Movie S1. Subcellular localization of LRX8-GFP in growing pollen tubes.
Note Added in Proof
Recently, Ndinyanka Fabrice et al. (Ndinyanka Fabrice T, Vogler H, Draeger C, Munglani G, Gupta S, Herger AG, Knox JP, Grossniklaus U, Ringli C [2018] LRX proteins play a crucial role in pollen grain and pollen tube cell wall development. Plant Physiol 176: 1981–1992) and Sede et al. (Sede AR, Borassi C, Wengier DL, Mecchia MA, Estevez JM, Muschietti JP [2018] Arabidopsis pollen extensins LRX are required for cell wall integrity during pollen tube growth. FEBS Lett 592: 233–243) also published similar results about the functions of LRX8-11. Additionally, LRX8-11 may interact with pollen-expressed RALF4/19 (Mecchia MA, Santos-Fernandez G, Duss NN, Somoza SC, Boisson-Dernier A, Gagliardini V, Martínez-Bernardini A, Fabrice TN, Ringli C, Muschietti JP, Grossniklaus U [2017] RALF4/19 peptides interact with LRX proteins to control pollen tube growth in Arabidopsis. Science 358: 1600–1603).
Acknowledgments
We thank Christoph Ringli (Department of Plant and Microbial Biology, University of Zürich) for conducting a critical review of this study and for providing both helpful suggestions and seeds. We thank Yihua Zhou and Dr. Baocai Zhang (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for quantifying the cell wall sugars using gas chromatography-mass spectrometry. We also thank the ABRC for providing the seeds of the Arabidopsis T-DNA insertion mutants.
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31270357, 31300265, and 31770354), Program for Top Talents in University of Hebei Province (BJ2017048), Program for New Century Excellent Talents in University (NECT-12-0687), Key Project of Chinese Ministry of Education (grant no. 211019), Natural Science Foundation of Hebei Province (grant no. C2013205160), Foundation for High-Level Talents in Higher Education of Hebei, 100 Innovative Talents Program of Higher Education of Hebei (to Y.G.), and Science Foundation of Hebei Normal University.
Articles can be viewed without a subscription.
References
- Alexander MP. (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Baron-Epel O, Gharyal PK, Schindler M (1988) Pectins as mediators of wall porosity in soybean cells. Planta 175: 389–395 [DOI] [PubMed] [Google Scholar]
- Baumberger N, Doesseger B, Guyot R, Diet A, Parsons RL, Clark MA, Simmons MP, Bedinger P, Goff SA, Ringli C, et al. (2003a) Whole-genome comparison of leucine-rich repeat extensins in Arabidopsis and rice: a conserved family of cell wall proteins form a vegetative and a reproductive clade. Plant Physiol 131: 1313–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N, Ringli C, Keller B (2001) The chimeric leucine-rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana. Genes Dev 15: 1128–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N, Steiner M, Ryser U, Keller B, Ringli C (2003b) Synergistic interaction of the two paralogous Arabidopsis genes LRX1 and LRX2 in cell wall formation during root hair development. Plant J 35: 71–81 [DOI] [PubMed] [Google Scholar]
- Boavida LC, McCormick S (2007) Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J 52: 570–582 [DOI] [PubMed] [Google Scholar]
- Borassi C, Sede AR, Mecchia MA, Salgado Salter JD, Marzol E, Muschietti JP, Estevez JM (2016) An update on cell surface proteins containing extensin-motifs. J Exp Bot 67: 477–487 [DOI] [PubMed] [Google Scholar]
- Brady JD, Fry SC (1997) Formation of di-isodityrosine and loss of isodityrosine in the cell walls of tomato cell-suspension cultures treated with fungal elicitors or H2O2. Plant Physiol 115: 87–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cankar K, Kortstee A, Toonen MA, Wolters-Arts M, Houbein R, Mariani C, Ulvskov P, Jorgensen B, Schols HA, Visser RG, et al. (2014) Pectic arabinan side chains are essential for pollen cell wall integrity during pollen development. Plant Biotechnol J 12: 492–502 [DOI] [PubMed] [Google Scholar]
- Cannon MC, Terneus K, Hall Q, Tan L, Wang Y, Wegenhart BL, Chen L, Lamport DT, Chen Y, Kieliszewski MJ (2008) Self-assembly of the plant cell wall requires an extensin scaffold. Proc Natl Acad Sci USA 105: 2226–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassab GI. (1998) Plant cell wall proteins. Annu Rev Plant Physiol Plant Mol Biol 49: 281–309 [DOI] [PubMed] [Google Scholar]
- Chebli Y, Kaneda M, Zerzour R, Geitmann A (2012) The cell wall of the Arabidopsis pollen tube: spatial distribution, recycling, and network formation of polysaccharides. Plant Physiol 160: 1940–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary P, Saha P, Ray T, Tang Y, Yang D, Cannon MC (2015) EXTENSIN18 is required for full male fertility as well as normal vegetative growth in Arabidopsis. Front Plant Sci 6: 553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coimbra S, Costa M, Jones B, Mendes MA, Pereira LG (2009) Pollen grain development is compromised in Arabidopsis agp6 agp11 null mutants. J Exp Bot 60: 3133–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coimbra S, Costa M, Mendes MA, Pereira AM, Pinto J, Pereira LG (2010) Early germination of Arabidopsis pollen in a double null mutant for the arabinogalactan protein genes AGP6 and AGP11. Sex Plant Reprod 23: 199–205 [DOI] [PubMed] [Google Scholar]
- Dardelle F, Lehner A, Ramdani Y, Bardor M, Lerouge P, Driouich A, Mollet JC (2010) Biochemical and immunocytological characterizations of Arabidopsis pollen tube cell wall. Plant Physiol 153: 1563–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Zou W, Li G, Zhao J (2014) TRANSLOCASE OF THE INNER MEMBRANE9 and 10 are essential for maintaining mitochondrial function during early embryo cell and endosperm free nucleus divisions in Arabidopsis. Plant Physiol 166: 853–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diet A, Link B, Seifert GJ, Schellenberg B, Wagner U, Pauly M, Reiter WD, Ringli C (2006) The Arabidopsis root hair cell wall formation mutant lrx1 is suppressed by mutations in the RHM1 gene encoding a UDP-L-rhamnose synthase. Plant Cell 18: 1630–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draeger C, Fabrice TN, Gineau E, Mouille G, Kuhn BM, Moller I, Abdou MT, Frey B, Pauly M, Bacic A, et al. (2015) Arabidopsis leucine-rich repeat extensin (LRX) proteins modify cell wall composition and influence plant growth. BMC Plant Biol 15: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresselhaus T, Sprunck S, Wessel GM (2016) Fertilization mechanisms in flowering plants. Curr Biol 26: R125–R139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M, Egelund J, Schultz CJ, Bacic A (2010) Arabinogalactan-proteins: key regulators at the cell surface? Plant Physiol 153: 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijó JA. (2010) The mathematics of sexual attraction. J Biol 9: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC. (1982) Isodityrosine, a new cross-linking amino acid from plant cell-wall glycoprotein. Biochem J 204: 449–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge W, Song Y, Zhang C, Zhang Y, Burlingame AL, Guo Y (2011) Proteomic analyses of apoplastic proteins from germinating Arabidopsis thaliana pollen. Biochim Biophys Acta 1814: 1964–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geitmann A. (2006) The architecture and properties of the pollen tube cell wall. In Malhó R, ed, The Pollen Tube. Plant Cell Monographs, Vol 3 Springer, Berlin, pp 177–200 [Google Scholar]
- Geitmann A. (2010) How to shape a cylinder: pollen tube as a model system for the generation of complex cellular geometry. Sex Plant Reprod 23: 63–71 [DOI] [PubMed] [Google Scholar]
- Gu F, Nielsen E (2013) Targeting and regulation of cell wall synthesis during tip growth in plants. J Integr Plant Biol 55: 835–846 [DOI] [PubMed] [Google Scholar]
- Guan Y, Guo J, Li H, Yang Z (2013) Signaling in pollen tube growth: crosstalk, feedback, and missing links. Mol Plant 6: 1053–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harholt J, Jensen JK, Sørensen SO, Orfila C, Pauly M, Scheller HV (2006) ARABINAN DEFICIENT 1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis. Plant Physiol 140: 49–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harholt J, Jensen JK, Verhertbruggen Y, Søgaard C, Bernard S, Nafisi M, Poulsen CP, Geshi N, Sakuragi Y, Driouich A, et al. (2012) ARAD proteins associated with pectic arabinan biosynthesis form complexes when transiently overexpressed in planta. Planta 236: 115–128 [DOI] [PubMed] [Google Scholar]
- Hepler PK, Rounds CM, Winship LJ (2013) Control of cell wall extensibility during pollen tube growth. Mol Plant 6: 998–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Yang SL, Xie LF, Puah CS, Zhang XQ, Yang WC, Sundaresan V, Ye D (2005) VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell 17: 584–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Brousseau SA, McCormick S (2004) A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J 39: 761–775 [DOI] [PubMed] [Google Scholar]
- Krichevsky A, Kozlovsky SV, Tian GW, Chen MH, Zaltsman A, Citovsky V (2007) How pollen tubes grow. Dev Biol 303: 405–420 [DOI] [PubMed] [Google Scholar]
- Kroeger J, Geitmann A (2012) The pollen tube paradigm revisited. Curr Opin Plant Biol 15: 618–624 [DOI] [PubMed] [Google Scholar]
- Lamport DT. (1967) Hydroxyproline-O-glycosidic linkage of the plant cell wall glycoprotein extensin. Nature 216: 1322–1324 [Google Scholar]
- Lamport DT, Kieliszewski MJ, Chen Y, Cannon MC (2011) Role of the extensin superfamily in primary cell wall architecture. Plant Physiol 156: 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiber RM, John F, Verhertbruggen Y, Diet A, Knox JP, Ringli C (2010) The TOR pathway modulates the structure of cell walls in Arabidopsis. Plant Cell 22: 1898–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouxel O, Cavalier DM, Liepman AH, Keegstra K (2006) Biosynthesis of plant cell wall polysaccharides: a complex process. Curr Opin Plant Biol 9: 621–630 [DOI] [PubMed] [Google Scholar]
- Levitin B, Richter D, Markovich I, Zik M (2008) Arabinogalactan proteins 6 and 11 are required for stamen and pollen function in Arabidopsis. Plant J 56: 351–363 [DOI] [PubMed] [Google Scholar]
- Liu X, Wolfe R, Welch LR, Domozych DS, Popper ZA, Showalter AM (2016) Bioinformatic identification and analysis of extensins in the plant kingdom. PLoS ONE 11: e0150177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XL, Liu L, Niu QK, Xia C, Yang KZ, Li R, Chen LQ, Zhang XQ, Zhou Y, Ye D (2011) Male gametophyte defective 4 encodes a rhamnogalacturonan II xylosyltransferase and is important for growth of pollen tubes and roots in Arabidopsis. Plant J 65: 647–660 [DOI] [PubMed] [Google Scholar]
- McCormick S. (2004) Control of male gametophyte development. Plant Cell (Suppl) 16: S142–S153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil KJ, Smith AG (2010) A glycine-rich protein that facilitates exine formation during tomato pollen development. Planta 231: 793–808 [DOI] [PubMed] [Google Scholar]
- Merkouropoulos G, Barnett DC, Shirsat AH (1999) The Arabidopsis extensin gene is developmentally regulated, is induced by wounding, methyl jasmonate, abscisic and salicylic acid, and codes for a protein with unusual motifs. Planta 208: 212–219 [DOI] [PubMed] [Google Scholar]
- Mohnen D, Tierney ML (2011) Plants get Hyp to O-glycosylation. Science 332: 1393–1394 [DOI] [PubMed] [Google Scholar]
- Mollet JC, Leroux C, Dardelle F, Lehner A (2013) Cell wall composition, biosynthesis and remodeling during pollen tube growth. Plants (Basel) 2: 107–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedukha OM. (2015) Callose: localization, functions, and synthesis in plant cells. Cytol Genet 49: 49–57 [Google Scholar]
- Osakabe Y, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS (2013) Sensing the environment: key roles of membrane-localized kinases in plant perception and response to abiotic stress. J Exp Bot 64: 445–458 [DOI] [PubMed] [Google Scholar]
- Parre E, Geitmann A (2005) More than a leak sealant: the mechanical properties of callose in pollen tubes. Plant Physiol 137: 274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira AM, Pereira LG, Coimbra S (2015) Arabinogalactan proteins: rising attention from plant biologists. Plant Reprod 28: 1–15 [DOI] [PubMed] [Google Scholar]
- Pinney K, Polito V (1989) Olive pollen storage and in vitro germination. International Symposium on Olive Growing. 286: 207–210 [Google Scholar]
- Qin Y, Yang Z (2011) Rapid tip growth: insights from pollen tubes. Semin Cell Dev Biol 22: 816–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringli C. (2005) The role of extracellular LRR-extensin (LRX) proteins in cell wall formation. Plant Biosyst 139: 32–35 [Google Scholar]
- Ringli C. (2010) The hydroxyproline-rich glycoprotein domain of the Arabidopsis LRX1 requires Tyr for function but not for insolubilization in the cell wall. Plant J 63: 662–669 [DOI] [PubMed] [Google Scholar]
- Roberts K, Shirsat AH (2006) Increased extensin levels in Arabidopsis affect inflorescence stem thickening and height. J Exp Bot 57: 537–545 [DOI] [PubMed] [Google Scholar]
- Rubinstein AL, Broadwater AH, Lowrey KB, Bedinger PA (1995a) Pex1, a pollen-specific gene with an extensin-like domain. Proc Natl Acad Sci USA 92: 3086–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein AL, Marquez J, Suárez-Cervera M, Bedinger PA (1995b) Extensin-like glycoproteins in the maize pollen tube wall. Plant Cell 7: 2211–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P (2010) Hemicelluloses. Annu Rev Plant Biol 61: 263–289 [DOI] [PubMed] [Google Scholar]
- Showalter AM. (1993) Structure and function of plant cell wall proteins. Plant Cell 5: 9–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter AM, Keppler B, Lichtenberg J, Gu D, Welch LR (2010) A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol 153: 485–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek N, Turner S (2016) From the nucleus to the apoplast: building the plant’s cell wall. J Exp Bot 67: 445–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonebloom S, Ebert B, Xiong G, Pattathil S, Birdseye D, Lao J, Pauly M, Hahn MG, Heazlewood JL, Scheller HV (2016) A DUF-246 family glycosyltransferase-like gene affects male fertility and the biosynthesis of pectic arabinogalactans. BMC Plant Biol 16: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhertbruggen Y, Marcus SE, Haeger A, Verhoef R, Schols HA, McCleary BV, McKee L, Gilbert HJ, Knox JP (2009) Developmental complexity of arabinan polysaccharides and their processing in plant cell walls. Plant J 59: 413–425 [DOI] [PubMed] [Google Scholar]
- Vogler H, Draeger C, Weber A, Felekis D, Eichenberger C, Routier-Kierzkowska AL, Boisson-Dernier A, Ringli C, Nelson BJ, Smith RS, et al. (2013) The pollen tube: a soft shell with a hard core. Plant J 73: 617–627 [DOI] [PubMed] [Google Scholar]
- Yang WC, Ye D, Xu J, Sundaresan V (1999) The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev 13: 2108–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Rumeau D, Showalter AM (1992) Isolation and characterization of two wound-regulated tomato extensin genes. Plant Mol Biol 20: 5–17 [DOI] [PubMed] [Google Scholar]