ERF17 transcriptional regulation activity in apple fruit peel degreening increases with serine repeat number.
Abstract
Through natural or human selection, many fleshy fruits have evolved vivid external or internal coloration, which often develops during ripening. Such developmental changes in color are associated with the biosynthesis of pigments as well as with degreening through chlorophyll degradation. Here, we demonstrated that natural variation in the coding region of the gene ETHYLENE RESPONSE FACTOR17 (ERF17) contributes to apple (Malus domestica) fruit peel degreening. Specifically, ERF17 mutant alleles with different serine (Ser) repeat insertions in the coding region exhibited enhanced transcriptional regulation activity in a dual-luciferase reporter assay when more Ser repeats were present. Notably, surface plasmon resonance analysis showed that the number of Ser repeats affected the binding activity of ERF17 to the promoter sequences of chlorophyll degradation-related genes. In addition, overexpression of ERF17 in evergreen apples altered the accumulation of chlorophyll. Furthermore, we demonstrated that ERF17 has been under selection since the origin of apple tree cultivation. Taken together, these results reveal allelic variation underlying an important fruit quality trait and a molecular genetic mechanism associated with apple domestication.
Genetic variants of cultivated plant species derived from wild progenitors undergo both natural and human selection for a wide variety of traits. In this regard, variation in color attributes is an important consideration in the breeding programs of many fleshy fruit species (Sun-Waterhouse et al., 2013; Faramarzi et al., 2014; Wang et al., 2015), with colorful-skinned fruits often being favored by consumers. A range of compounds can confer color, but the red, purple, and blue coloration of leaves, flowers, fruit peels, and flesh is usually a consequence of the accumulation of the anthocyanin class of flavonoid pigments. In addition to anthocyanin biosynthesis, color change in ripening fruit often is associated with a concurrent degreening as a result of chlorophyll (Chl) degradation (Lai et al., 2015).
Much has been learned about the anthocyanin biosynthetic pathway and the associated regulatory genes (Espley et al., 2007; Gonzalez et al., 2008; An et al., 2012, 2015; Albert et al., 2014; Yoshida et al., 2015). In apple (Malus domestica), the Myeloblastosis (MYB) transcription factors MdMYB1, MdMYBA, MYB10, and MYB110a have been described as involved in anthocyanin production (Takos et al., 2006; Ban et al., 2007; Espley et al., 2007; Chagné et al., 2013). However, the molecular basis for the natural variation of Chl degradation before coloring during apple fruit ripening remains largely undefined.
Chl degradation-related genes and enzymes have been studied in a range of plant species, including nonyellow coloring1 (NYC1) and NYC1-like (NOL) in rice (Oryza sativa) and Arabidopsis (Arabidopsis thaliana; Kusaba et al., 2007; Horie et al., 2009). Interestingly, of these two genes, loss-of-function mutations in either NYC1 or NOL in rice, but only NYC1 in Arabidopsis, lead to a stay-green phenotype (Kusaba et al., 2007; Horie et al., 2009; Jibran et al., 2015). Another of the Chl degradation-related enzymes, chlorophyllase (CLH), is known to persist, expressing during Chl breakdown in senescent leaves (Matile et al., 1996). Previous studies also have characterized pheophytinase (PPH) as a plastid-localized enzyme that is central to Chl breakdown in Arabidopsis (Eckardt, 2009; Schelbert et al., 2009). The tomato (Solanum lycopersicum) sipph mutant is defective in Chl breakdown in leaves, resulting in elevated levels of pheophytin (Guyer et al., 2014). Similarly, in Arabidopsis, the absence of red chlorophyll catabolite reductase (RCCR) results in leaf cell death as a result of the accumulation of photodynamic red Chl catabolite, while pheophorbide a oxygenase (PAO) also is required for the detoxification of Chl catabolites (Pruzinská et al., 2007).
The biochemical mechanisms of Chl degradation have been studied extensively, but less is known about the regulation of the associated genes. The function of the regulator, nonyellowing1/stay-green1 (NYE1/SGR1), which is responsible for the green cotyledon trait of Mendel’s pea (Pisum sativum; Hörtensteiner, 2009), has been demonstrated in diverse species. In addition, a transcription factor (NYE2) that promotes Chl degradation has been identified in Arabidopsis (Wu et al., 2016), and two MYB transcription factors from tomato, Golden2-like1 (GLK1) and GLK2, regulate tomato fruit Chl development (Powell et al., 2012). Moreover, Chl degradation is triggered by environmental stimuli and phytohormones, such as ethylene (Oeller et al., 1991; Hörtensteiner, 2006; Pino et al., 2008). In rice, expression of the ethylene-responsive genes Sub1A and Sub1C is up-regulated during Chl degradation in leaves (Fukao et al., 2006). This is consistent with the observation that four ethylene response factors, CitERF5, CitERF6, CitERF7, and CitERF13, play roles in citrus fruit skin degreening (Xie et al., 2014). Subsequently, dual-luciferase and yeast one-hybrid assays show that CitERF13 specifically recognizes and binds to the CitPPH promoter, resulting in an increase in CitPPH transcript levels (Yin et al., 2016). However, despite such insights, the relationship between ERF function and variation in fruit coloration (degreening) is not well understood.
In this study, we provide evidence that variation in the pattern of Ser residues in the ERF17 coding region in the Malus genus is responsible for differences in the transcriptional regulatory activity of ERF17 and that this accounts for the varied Chl degradation rate in apple (Malus spp.) peel. Our findings suggest that the Ser repeat number influences the ability of ERF17 to bind Chl degradation-related genes.
RESULTS
An ERF17 Mutant Allele Is Associated with Apple Fruit Peel Degreening
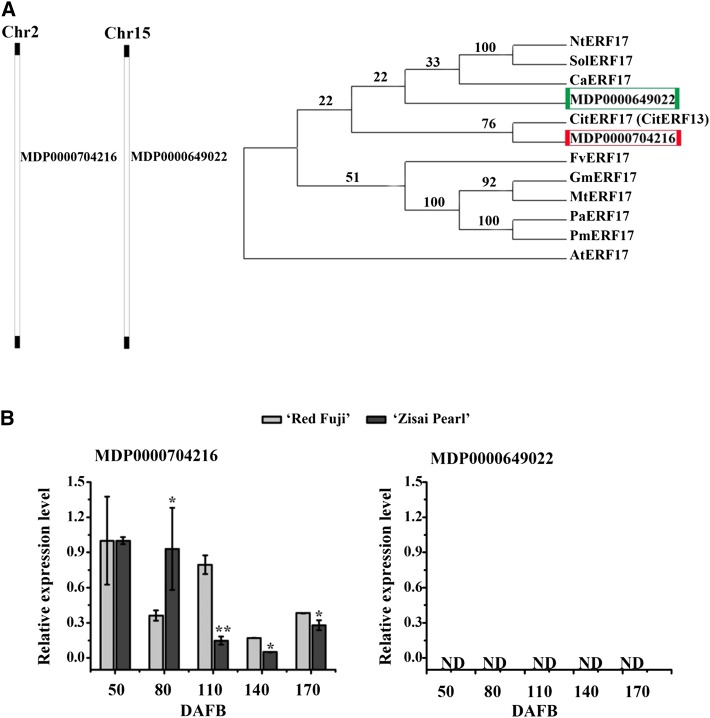
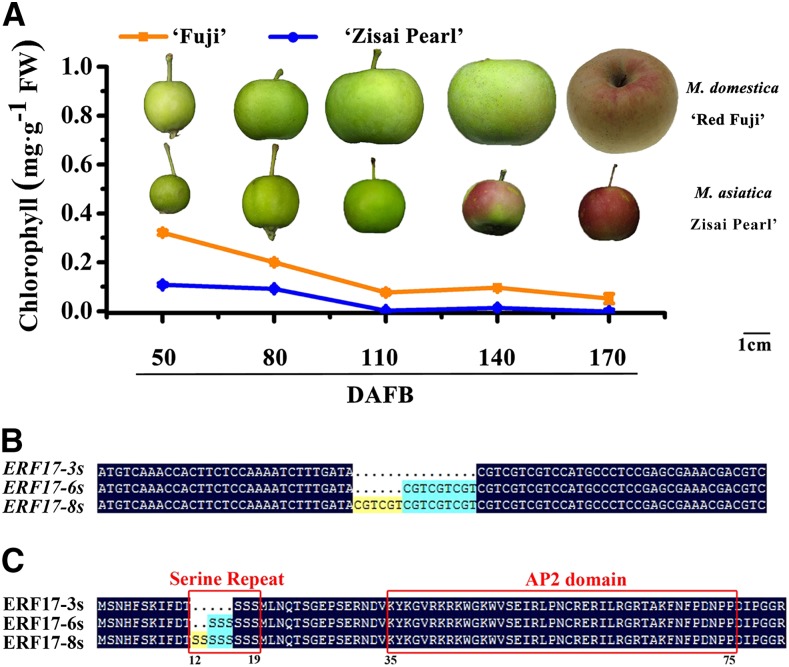
Since CitERF13 has been identified as a key transcription factor in citrus fruit skin degreening (Yin et al., 2016), we targeted apple ERF genes for analysis. Phylogenetic analysis identified two genes in the apple genome (MDP0000649022 and MDP0000704216) that are closely related to CitERF17 (Fig. 1A). We determined that MDP0000704216 was expressed in the apple fruit peel, while MDP0000649022 was not (Fig. 1B). We observed that the Chl content of the fruit peel of Malus asiatica ‘Zisai Pearl’ was lower than that in the peel of Malus domestica ‘Red Fuji’ during fruit development (Fig. 2A). To further investigate the molecular basis of the fruit peel degreening phenotype, we analyzed the ERF17 (MDP0000704216) gene sequences from cv Zisai Pearl, Red Fuji, and their progeny. We amplified DNA fragments with different numbers of Ser repeat units for the equivalent regions in cv Zisai Pearl and Red Fuji: the former is homozygous with three Ser repeats, while cv Red Fuji is heterozygous with six or eight Ser repeats in the equivalent region, +36 to +57 bp downstream of the ATG start site. Alleles containing three, six, and eight Ser repeats were named ERF17-3S, ERF17-6S, and ERF17-8S, respectively (accession nos. MF498883, MF498882, and MF498881; Fig. 2, B and C).
Figure 1.
Identification of two ERF17 genes in the apple genome. A, Phylogenetic analysis of homologous ERF17 transcription factors. MDP0000704216 and MDP0000649022 locate on apple chromosome 2 (Chr2) and chromosome 15 (Chr15), respectively. The numbers on the tree represent nucleotide substitutions. B, Relative expression of MDP0000704216 (left) and MDP0000649022 (right) in peels of cv Red Fuji (gray) and cv Zisai Pearl (black) various days after flower blossom (DAFB). An actin fragment was amplified as an internal control. Values are means ± sd (n = 3). Asterisks indicate statistically significant differences by Student’s t test (*, P < 0.05 and **, P < 0.01) compared with cv Red Fuji peel. ND, Not detected.
Figure 2.
Chl degradation of fruit peel in cv Red Fuji and Zisai Pearl during development and variation in ERF17 gene and ERF17 protein sequences. A, Appearance of fruits and Chl content during development. Error bars indicate sd of three biological replicates. DAFB, Days after flower blossom; FW, fresh weight. Bar = 1 cm. B, Variations in the ERF17 cDNA fragment between the ERF17-3S, ERF17-6S, and ERF17-8S alleles (for full sequences, see Supplemental Fig. S1). C, Variations in the ERF17 amino acid sequences between the ERF17-3S, ERF17-6S, and ERF17-8S alleles.
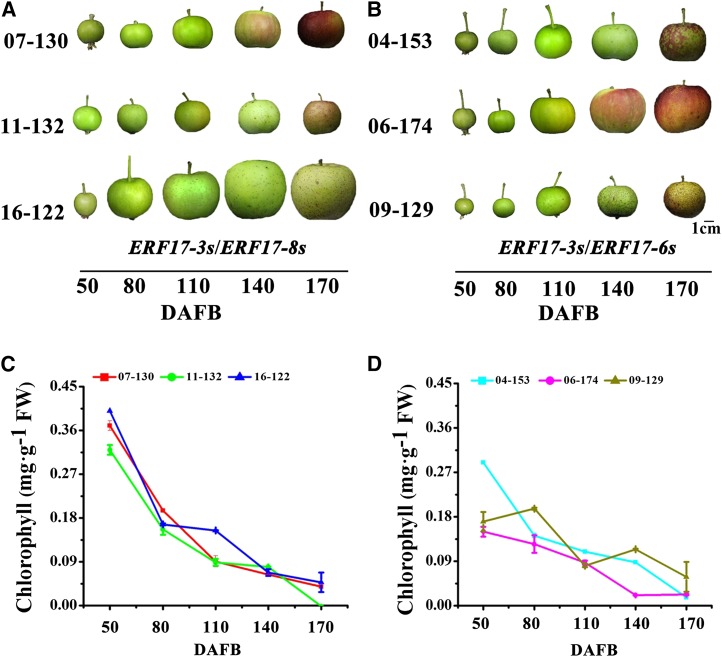
The association between the ERF17 allele and the fruit peel degreening phenotype was confirmed by analyzing the progeny from a cross between cv Zisai Pearl (ERF17-3S) and cv Red Fuji (ERF17-6S /ERF17-8S). Using the segregating population, a quantitative trait locus (QTL) for Chl content of the fruit peel was located on linkage group 2, and ERF17 (MDP0000704216) is located in the interval spanned by MdSNPui03272 and CTG1061980 of that QTL (Supplemental Fig. S1). The genotype of the progeny was ERF17-3S/ERF17-6S or ERF17-3S/ERF17-8S. The ERF17-3S/ERF17-8S genotype showed a higher Chl degradation rate than the ERF17-3S/ERF17-6S genotype, consistent with the Ser repeat number contributing to the phenotype and revealing a positive association between the number of Ser repeats and the peel Chl degradation rate (Fig. 3). We also observed a strong negative correlation (R2 =1, P < 0.05) between the peel Chl degradation rate, a notable phenotypic symptom of peel degreening, and ERF17 expression in cv Zisai Pearl and Red Fuji (Supplemental Table S1). Moreover, the lower Chl content in the peel was associated with lower fruit firmness and an increased fruit expansion phenotype (Supplemental Table S2). These results, combined with the genetic analysis, suggest that the ERF17 mutant allele confers fruit peel degreening.
Figure 3.
Analyzing progeny from a cross between cv Red Fuji and Zisai Pearl. A, Appearance of ERF17-3S/ERF17-8S genotype fruits during development. Bar = 1 cm. B, Appearance of ERF17-3S/ERF17-6S genotype fruits during development. C, Chl degradation during development of the ERF17-3S/ERF17-8S genotype. D, Chl degradation during development of the ERF17-3S/ERF17-6S genotype. DAFB, Days after flower blossom; FW, fresh weight. Error bars in C and D indicate sd of three biological replicates.
The Ser Repeat Number in the Coding Region of ERF17 Alters Its Transcriptional Regulatory Activity
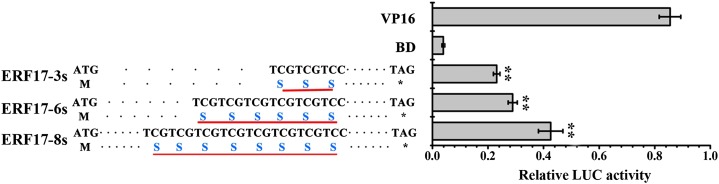
The Ser repeat units (amino acids 12–19) locate before the APETALA2 (AP2) domain (amino acids 35–75; Fig. 2C). The Ser repeats also exist in Citrus spp., Arabidopsis, tomato, and tobacco (Nicotiana tabacum), and the numbers of Ser repeats vary in different species. We hypothesized that the different numbers of Ser repeats within the ERF17 coding region might affect the basal transcriptional regulatory activity of ERF17 (Supplemental Fig. S2C). To test this, we examined the transcriptional regulatory activity of different ERF17 alleles using a dual-luciferase reporter assay in Arabidopsis protoplasts. The coding sequences of the ERF17 proteins were fused to the DNA sequence encoding the GAL4 reporter DNA-binding domain (which is a transcriptional activator protein and controls the structural genes of Gal/melibiose utilization in yeast) to generate pRT-BD-ERF17 effector plasmids. The GAL4-dual-luciferase (GAL4-LUC) reporter plasmid was generated from pUC19 (Ohta et al., 2001), which contains the firefly LUC reporter gene driven by the minimal TATA box of the 35S promoter and five copies of GAL4-binding elements. The pPTRL (Renilla reniformis luciferase) driven by the 35S promoter was used as a control. Compared with the pRT-BD negative control, all the ERF17 alleles showed transcriptional regulatory activity, and we observed a strong positive association between activity and the number of Ser repeats (Fig. 4).
Figure 4.
Transcriptional regulation activity of ERF17 allele proteins. A dual-luciferase reporter assay of ERF17 allele proteins using an Arabidopsis protoplast system is shown. The plasmid ratio used (pTRL:reporter:effector) was 1:6:6. Values are means ± sd (n = 3). Asterisks indicate statistically significant differences by Student’s t test (*, P < 0.05 and **, P < 0.01) compared with the binding domain (BD).
ERF17 Regulation of Chl Degradation-Related Gene Expression
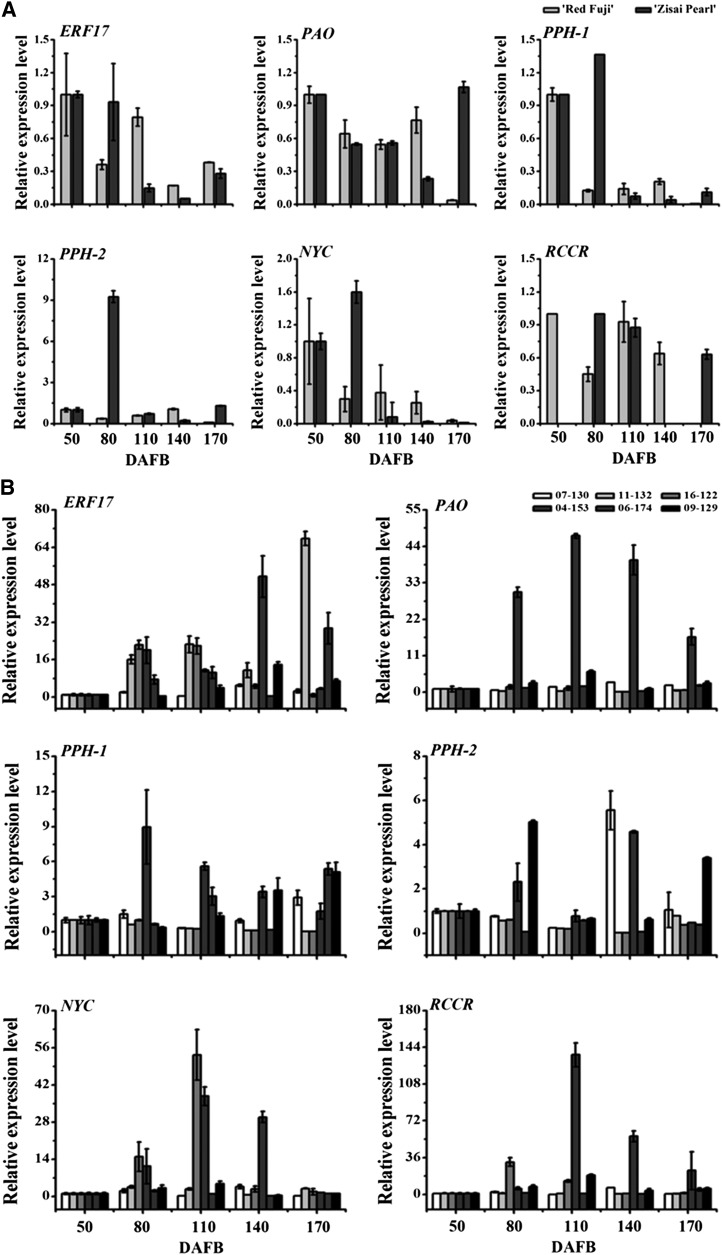
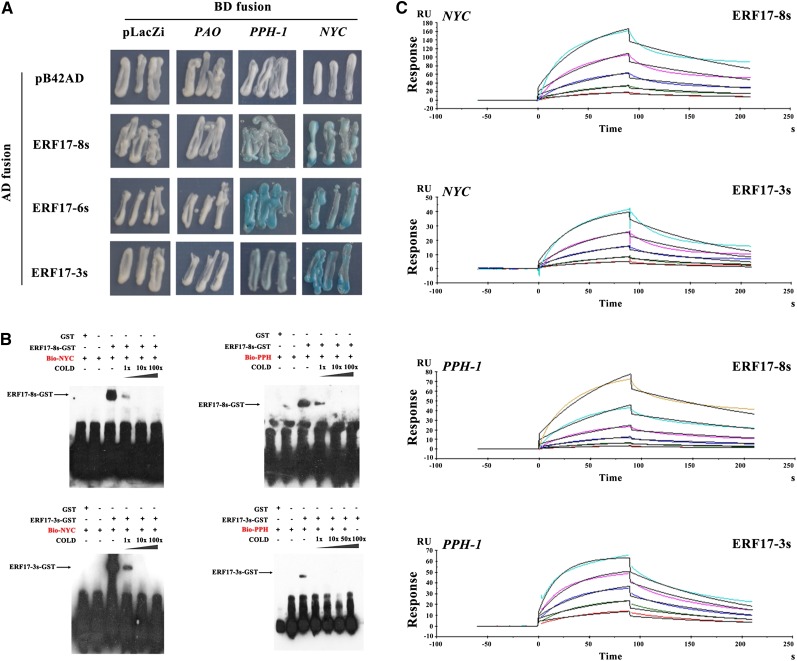
To determine whether ERF17 regulates the expression of Chl degradation-related genes, we investigated the binding of ERF17 to the promoters of degreening-related genes identified in the apple genome database (https://www.rosaceae.org/tools/ncbi; Supplemental Fig. S3). Two known target motifs for AP2/ERF transcription factors, DRE and GCC-box motifs (Stockinger et al., 1997), were found in the PAO (MDP0000315206), PPH (MDP0000312878), and NYC (MDP0000124013) promoter regions (Supplemental Fig. S4). We also measured the expression of these genes in the fruit peels of cv Zisai Pearl, Red Fuji, and their progeny during fruit development and found that ERF17 expression was positively correlated with that of PPH and NYC (R2 = 0.96, P < 0.05 and R2 = 0.97, P < 0.05, respectively; Fig. 5; Table I). We next investigated the ability of ERF17 to bind to the promoter of the Chl degradation-related genes using yeast one-hybrid analysis and found that ERF17-3S, ERF17-6S, and ERF17-8S bound to both the PPH and NYC promoters (Fig. 6A).
Figure 5.
ERF17 and Chl degradation-related gene expression in fruit peels during development. A, Gene expression in fruit peels of cv Red Fuji and Zisai Pearl during development. B, Gene expression in fruit peels from a cross between cv Red Fuji and Zisai Pearl: 07-130, 11-132, and 16-122 represent the ERF17-3S/ERF17-8S genotype, and 04-153, 06-174, and 09-129 represent the ERF17-3S/ERF17-6S genotype. Error bars indicate sd of three biological replicates. DAFB, Days after flower blossom.
Table I. Correlation analysis between the expression of ERF17 and Chl degradation-related genes.
| ERF17 | CLH | NYC | PAO | PPH-1 | PPH-2 | RCCR | |
|---|---|---|---|---|---|---|---|
| ERF17 | 1.00 | 0.76 | 0.97a | 0.42 | 0.96b | 0.76 | 0.65 |
| CLH | 1.00 | 0.67 | 0.46 | 0.58 | 0.21 | 0.82 | |
| NYC | 1.00 | 0.19 | 0.99** | 0.73 | 0.76 | ||
| PAO | 1.00 | 0.17 | 0.45 | −0.95 | |||
| PPH-1 | 1.00 | 0.79 | 0.74 | ||||
| PPH-2 | 1.00 | 0.32 | |||||
| RCCR | 1.00 |
P < 0.05. bP < 0.01.
Figure 6.
ERF17 regulates Chl degradation-related genes. A, Yeast one-hybrid assay showing the activity of LacZ reporters driven by the PPH and NYC promoters and activated by activation domain (AD) fusion with ERF17-8S and ERF17-3S (blue color). The empty vectors pB42AD and pLacZi were used as negative controls (white color). BD, Binding domain. B, Gel-shift analysis of ERF17-8S and ERF17-3S binding to the promoters of PPH and NYC. EMSAs of a GST-labeled probe with ERF17-8S and ERF17-3S proteins are shown. DNA probes represent the promoters of PPH and NYC (probe sequences are shown in Supplemental Table S3). Protein/DNA complexes are indicated by arrows. C, SPR binding profiles of ERF17-8S and ERF17-3S proteins to the promoters of PPH and NYC. Protein concentrations were 275 nm in all lines but the bottom line, for which it was 17.2 nm. RU, Response units.
To further investigate the effect of the number of Ser repeats on ERF17-binding activity using electrophoretic mobility shift assays (EMSAs), we expressed ERF17-3S and ERF17-8S in Escherichia coli as recombinant GST-tagged proteins, which were then purified and incubated with DNA probes corresponding to the PPH or NYC promoter (Supplemental Table S3). ERF17-3S and ERF17-8S both bound to biotin-labeled PPH and NYC probes (Fig. 6B), although we observed differences in the intensity of the signal. To quantify a potential difference in binding affinity, we used surface plasmon resonance (SPR) with immobilized oligonucleotides. ERF17-8S had lower dissociation rate constant and equilibrium dissociation constant values compared with those of ERF17-3S, suggesting that ERF17-8S has higher affinity for the PPH and NYC promoters and that the binding stability of ERF17-8S is stronger than that of ERF17-3S (Fig. 6C; Table II). Taken together, these results suggest that the Ser repeat number is a key factor in the binding of ERF17 to the promoters of Chl degradation-related genes.
Table II. Affinity parameters of SPR assays.
| Protein | DNA | Association Rate Constant | Dissociation Rate Constant | Equilibrium Dissociation Constant |
|---|---|---|---|---|
| m−1 s−1 | s−1 | m | ||
| ERF17-8S | PPH | 1.480 × 104 | 4.521 × 10−3 | 3.055 × 10−7 |
| ERF17-3S | PPH | 0.969 × 104 | 8.352 × 10−3 | 8.623 × 10−7 |
| ERF17-8S | NYC | 5.686 × 104 | 5.208 × 10−3 | 9.159 × 10−8 |
| ERF17-3S | NYC | 6.638 × 104 | 8.648 × 10−3 | 1.303 × 10−7 |
Overexpression of an ERF17 Allele in Evergreen Apple Alters the Accumulation of Chl
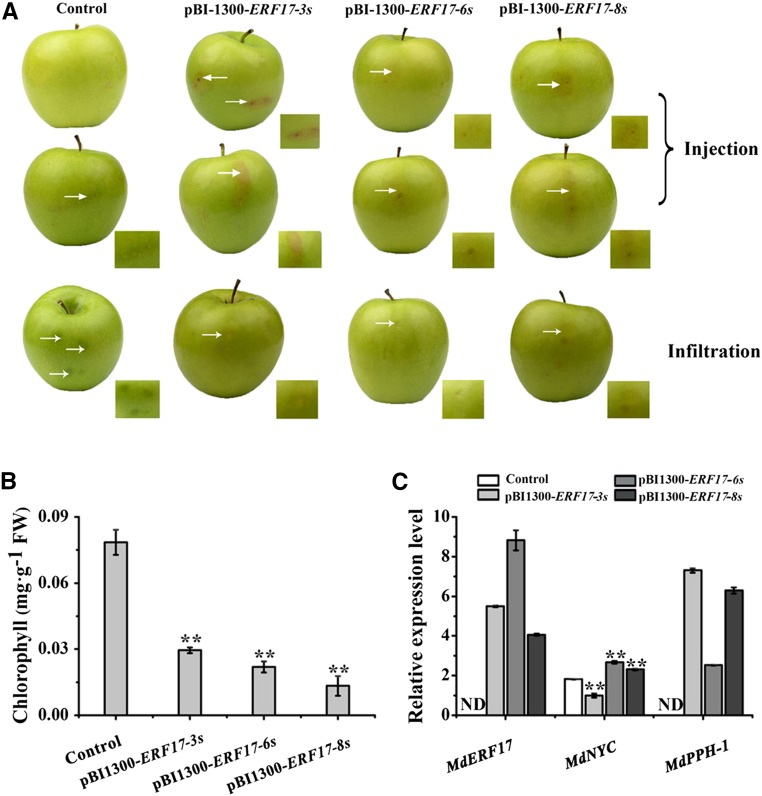
Due to the long juvenile period involved in obtaining stably transformed transgenic apple fruits, we chose to use a transient expression assay involving Agrobacterium tumefaciens infiltration to overexpress ERF17 in apple fruit. The fruit of M. domestica ‘Granny Smith’ is green, and ERF17 expression is undetectable in this cultivar (Fig. 7C). To investigate whether overexpressing ERF17 in cv Granny Smith would cause the degradation of peel Chl and to study the effects of the Ser repeat number on ERF17 gene function, A. tumefaciens-harboring vectors to constitutively express different ERF17 alleles (35S::ERF17-3S, 35S::ERF17-6S, or 35S::ERF17-8S) were separately injected or infiltrated into cv Granny Smith fruits. We observed the induction in ERF17 transcript levels (Fig. 7C), and apple fruits overexpressing ERF17 showed a rapid loss of green color in the peel at the injection sites, resulting in significant bleaching, coincident with the appearance of a reddish color (Fig. 7A). Chl content analyses revealed lower levels of Chl accumulation in transgenic 35S::ERF17-8S fruit than in 35S::ERF17-3S and 35S::ERF17-6S fruit (Fig. 7B). Moreover, compared with the control, the expression of PPH and NYC was significantly higher when ERF17 was overexpressed, with the highest levels in 35S::ERF17-8S fruit, followed by 35S::ERF17-6S and 35S::ERF17-3S (Fig. 7C). These findings indicate that ERF17 contributes to Chl degradation in apple fruit peel and that the Ser repeat number in the coding region of ERF17 influences the fruit peel degreening phenotype.
Figure 7.
ERF17 is essential for Chl degradation in apple fruit. A, Apple fruit peel coloration. Three ERF17 genotypes were overexpressed in apple fruits by two A. tumefaciens-mediated transient transformation methods (injection and infiltration). Apples injected and infiltrated with the empty pBI1300 vector were used as controls. B, Chl content around the infiltration sites of apple peels in mg g−1 fresh weight (FW). C, Relative expression levels of MdERF17, MdNYC, and MdPPH in apple fruit peels around the infiltration sites determined using reverse transcription real-time quantitative PCR (RT-qPCR). An actin fragment was amplified as an internal control. Values are means ± sd (n = 3) in B and C. Asterisks indicate statistically significant differences by Student’s t test (**, P < 0.01) compared with the control. ND, Not detected.
Since we observed a reddish color in the injection sites of cv Granny Smith, we hypothesized that ERF17 might alter anthocyanin accumulation during fruit ripening. To test this, we used the fruits of a local apple, cv TaiShanZaoXia, harvested at the green peel stage before ripening for another set of infiltrations with the same A. tumefaciens lines as those used above. We observed that the apples transiently overexpressing ERF17 showed darker red coloration than the controls, and higher levels of anthocyanins were detected in the transgenic apples expressing 35S::ERF17-8S than fruit expressing 35S::ERF17-3S or 35S::ERF17-6S (Supplemental Fig. S5). Moreover, transcripts of the anthocyanin biosynthesis genes chalcone synthase and dihydroflavonol 4-reductase and the anthocyanin biosynthesis regulator MYB1 were present at higher levels in the transiently ERF17-overexpressing fruit peels compared with control fruits (Supplemental Fig. S6).
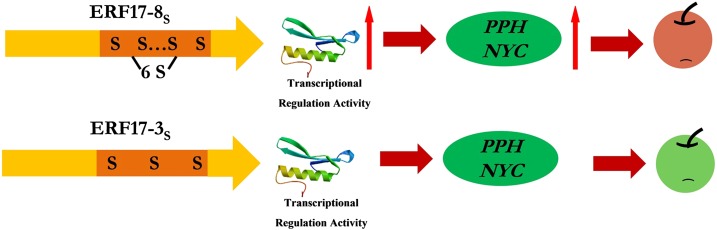
Based on the data presented in this study, we propose a model for the apple peel degreening regulatory module where the increased Ser repeat number in the coding region of ERF17 enhances its transcriptional regulatory activity and its binding ability to the promoter of Chl degradation-related genes. This leads to apple peel color changes associated with an increased fruit peel degreening rate and the biosynthesis of pigments (Fig. 8).
Figure 8.
Schematic model of the regulation of the apple peel degreening regulatory module. The increased Ser repeat number in the coding region of ERF17 enhances its transcriptional regulatory activity and its binding ability to the promoter of Chl degradation-related genes. NYC and PPH are required for Chl degradation, which can contribute to apple fruit pigment. An elevated expression level of NYC and PPH improves apple fruit peel degradation and promotes pigment accumulation.
Evolutionary Analysis of the ERF17 Allele
We hypothesized that the ERF17 allele has undergone selection during the process of apple fruit cultivation and the selection for color variants. To test this, we allelotyped the ERF17 loci of 72 accessions belonging to five different Malus species (Phipps et al., 1990; Yao et al., 2015), which represent the diversity of the five Malus species. Tajima’s D tests (Tajima, 1989) detected departure from neutrality for the ERF17 genes of M. domestica, while there was no significant departure for the ERF17 locus in the other four species, indicating that the ERF17 allele has been under selection during apple cultivation (Table III).
Table III. Nucleotide polymorphisms in ERF17 for Malus spp.
m = number of sequences, S = number of segregating sites, ps = S/m, Θ = ps/a1, π = nucleotide diversity, and D is the Tajima test statistic.
| Species | m | ps | Θ | π | D | Neutral Mutation Range (Tajima’s D) |
|---|---|---|---|---|---|---|
| M. asiatica | 7 | 0.025054 | 0.010226 | 0.010271 | 0.024563 | −1.498 to 1.728 |
| M. baccata | 10 | 0.021361 | 0.007551 | 0.005696 | −1.17371 | −1.559 to 1.719 |
| M. prunifolia | 13 | 0.047918 | 0.015442 | 0.017332 | 0.551679 | −1.580 to 1.708 |
| M. domestica | 29 | 0.062706 | 0.015967 | 0.008435 | −1.77898 | −1.581 to 1.714 |
| M. sieversii | 13 | 0.030438 | 0.010079 | 0.008785 | −0.58541 | −1.580 to 1.708 |
The number of Ser repeats in the ERF17 coding region explains most of the apple peel coloring variation among the 22 genus Malus germplasm accessions (Supplemental Fig. S7; Supplemental Table S4). The Ser repeat number of ERF17 positively correlated with fruit peel color phenotypes, revealing a close association of ERF17 with the green fruit peel trait. Most M. domestica cultivars and their ancestral species, Malus sieversii, are ERF17-8S genotypes, and most wild species are ERF17-3S or ERF17-3S/ERF17-8S genotypes (Supplemental Table S4). Moreover, this result revealed a possible selection for ERF17.
DISCUSSION
Recently, Volk et al. (2015) reported that M. domestica has two distinct chloroplast haplotypes that are shared with four other species: Malus sylvestris, Malus orientalis, M. sieversii, and Malus prunifolia. It is likely that apple has been domesticated many times throughout history, with selection of individual wild genotypes made in different geographic regions on different continents (e.g. Asia and Europe). Different selection criteria may have been based on ethnic or cultural origin. Fruit color is a characteristic for which there has likely been selection, associated with the attraction of seed-dispersing animals. The typical stay-green apple cultivar, cv Granny Smith, is popular in some regions specifically for its green color. Consistent with this idea, in our evolutionary analysis of ERF17, the lower DNA sequence diversity, together with the fixation of the ERF17 allele in M. domestica species, suggests that the allele has been under selection.
Genetic variants of cultivated crops derived from wild progenitors of plants under both natural and human selection provide vital sources of this type of genetic variation. Variation in the expression of genes associated with agriculturally important traits due to transposon insertion or differential expansion of microsatellite repeat sequences also has been observed (Sureshkumar et al., 2009). Previously, we reported that a core-promoter element insertion can enhance iron-regulated transporter1 gene expression (Zhang et al., 2017). In addition to promoter variation, single-nucleotide polymorphisms or insertions/deletions in coding regions also may affect gene function. Therefore, the identification of functional single-nucleotide polymorphisms or insertions/deletions in genes coupled with analyses of their effects on phenotypes is a potentially useful strategy to elucidate their impact on gene function for variety improvement. In addition to identifying the ERF17 allele, our study reveals a regulatory mechanism associated with its natural variation. The use of highly heterozygous Malus genotypes allowed us to determine that different numbers of Ser repeat insertions in the coding region of ERF17 provide the basis of the regulation of apple peel degreening during ripening. Notably, there is allelic variation in tomato, with four or five Ser repeats in the ERF17 homolog SolERF017 (Solyc12g009240.1). Solanum pimpinellifolium fruit are red, and SolERF017 in this species contains five Ser repeats. However, Solanum pennellii fruit are green, and SolERF017 from this species has four Ser repeats (https://solgenomics.net/; Lin et al., 2014). These data suggest that relationships between Ser repeat numbers and fruit color may exist in multiple species; however, coloration mechanisms in different species are varied and complex and need further investigation. Our results establish that repeat Ser insertions can enhance transcriptional regulatory activity, which may represent a general tool for variety improvement.
There is strong collinearity between large segments of apple chromosomes 3 and 11, 5 and 10, 9 and 17, and 13 and 16 as well as between shorter segments of chromosomes 1 and 7, 2 and 7, 2 and 15, 4 and 12, 12 and 14, 6 and 14, and 8 and 15 (Velasco et al., 2010). Our phylogenetic analysis showed that two ERF17 genes, MDP0000649022 and MDP0000704216, are located in duplicated regions on chromosomes 2 and 15. Preservation by relative dosage occurs when deletion of one copy of a duplicate pair relative to its interacting partner has negative fitness consequences (Freeling, 2009; Birchler and Veitia, 2012). There will then be selection to maintain both copies in a stoichiometric balance, at least initially, following a whole-genome duplication (Conant et al., 2014). Our results show the two collinear ERF17 alleles varied in their genomic sequence (Supplemental Fig. S1) and that MDP0000649022, which is located on chromosome 15, was not expressed in apple fruit. Based on RNA sequencing analyses of cv Golden Delicious apple fruit (flesh) taken at 14 developmental stages (Yang et al., 2015) and cv Greensleeves (apple leaves; Wu et al., 2015), MDP0000704216 is expressed in the fruit flesh but MDP0000649022 is not, and neither ERF17 gene is expressed in apple leaves. We suggest that, following whole-genome duplication, the duplicated ERF17 genes exhibited nonfunctionalization, where one of the gene copies was physically lost or pseudogenized.
Descriptions of stay-green mutants with a defect in a catalytic or regulatory gene involved in Chl breakdown have been reported (Vicentini et al., 1995; Thomas et al., 1996; Moser and Matile, 1997; Akhtar et al., 1999; Roca et al., 2004; Hörtensteiner, 2006; Barry et al., 2008; Borovsky and Paran, 2008). These stay-green mutants are impaired in particular steps in the Chl degradation pathway, rather than showing a general down-regulation. The fruit of M. domestica cv Granny Smith has a stay-green peel with an ERF17 genotype of ERF17-3S/ERF17-8S, and the ERF17 transcript is undetectable in the peel. Complementing ERF17 in cv Granny Smith caused a degradation of peel Chl, which, in turn, suggests that ERF17 deficiency causes the stay-green phenotype of apple fruit peel. The varied ERF17 expression in the Malus genus led to our hypothesis that variation in ERF17 regulatory pathways for apple peel degreening may exist.
Fleshy fruit ripening involves major biochemical and physiological changes in texture, flavor, and color (Seymour et al., 2013), which are controlled by ethylene in climacteric fruit. Accordingly, we observed a correlation between Chl content and fruit ripening index (Supplemental Table S3). ERF is an important transcription factor in the ethylene signaling pathway (Klee and Giovannoni, 2011; Rodrigues et al., 2014) and is involved in the regulation of ethylene biosynthesis. In tomato, LeERF2 activates the expression of ethylene biosynthetic genes (Zhang and Huang, 2010), while in banana (Musa acuminata), MaERF9 positively regulates 1-aminocyclopropane-1-carboxylic acid synthase (MaACS1) and 1-aminocyclopropane-1-carboxylic acid oxidase activity by binding to their promoters (Xiao et al., 2013). In apple, MdERF3 enhances the transcription of MdACS1 by binding to its promoter (Li et al., 2016), and jasmonic acid promotes ethylene biosynthesis through the regulation of MdERF genes and ethylene biosynthetic genes by MdMYC2 (Li et al., 2017). We observed that the apple progeny with lower Chl content in the peel were associated with lower fruit firmness and a reduced fruit expansion phenotype (Supplemental Table S3). Furthermore, we found that overexpression of ERF17 promoted fruit red coloration (Supplemental Figs. S4 and S5). In pear (Pyrus spp.), the ERF/AP2 transcription factor PyERF3 was found to interact with PyMYB114 and its partner PybHLH3 to coregulate anthocyanin biosynthesis (Yao et al., 2017). Taken together, these results suggest that variations in the ERF17 allele may differentially affect apple peel reddening by regulating ethylene biosynthesis or may regulate anthocyanin biosynthesis directly.
Natural variation provided an important mechanism for evolutionary change and phenotypic variation. Here, we show that a difference in Ser insertion number in a coding region of a regulatory gene influences color, which is under selection. This discovery has implications for the development of novel varieties.
MATERIALS AND METHODS
Plant Material
An F1 population derived from a cross between Malus asiatica ‘Zisai Pearl’ and Malus domestica ‘Red Fuji’ was used in this study. The parents were crossed in 2007, and the hybrids (young seedlings from seeds) were planted in the experimental orchard of the China Agricultural University (Changping, Beijing, China) and maintained using standard management techniques. Fruit were harvested at 50, 80, 110, 140, and 170 d after flower blossom. Fruit peels were sampled and frozen in liquid nitrogen for later analysis, with each sample comprising three biological replicates and each replicate including three fruits on one plant.
The phenotypic characteristics of fruit weight, fruit length, and fruit diameter were evaluated. Fruit firmness was determined and measured using a texture analyzer (TA.XT-21; Stable Micro Systems).
QTL Analysis
A total of 251 hybrid progeny from a cross between cv Zisai Pearl and Red Fuji were randomly selected for linkage map construction and QTL assays. A genetic map was available for QTL mapping (Tan et al., 2017). QTL detection was carried out using the MapQTL 6.0 software package (Van Ooijen, 2009). Interval mapping was performed for each trait, and the linkage maps and QTL positions were drawn using MapChart (Voorrips, 2002).
Extraction of DNA and Total RNA
Each 80- to 100-mg (fresh weight) sample was used to isolate DNA and total RNA according to the instructions of the N96 Plant Genomic DNA Kit (Tiangen Biotech) and the RNA prep Pure Plant Kit (Polysaccharides & Polyphenolics-rich; Tiangen Biotech), respectively. First-strand cDNA was synthesized using 1 μg of total RNA (ODA260/A280 > 1.8) from each sample using M-MLV reverse transcriptase (RNase H−; Takara), following the manufacturer’s instructions.
Chl Measurements
Chl concentration was determined after aqueous acetone extraction (Jung et al., 2016). Fresh fruit peels (0.8 or 1 g) were extracted with acetone.
HPLC Analysis of Anthocyanins
Anthocyanin concentrations in the peel were measured by HPLC as described by Zhang et al. (2014). Each 0.3-g sample was powdered in liquid nitrogen using a mortar and pestle before 1 mL of extraction buffer (70:27:2:1 [v/v] methanol:water:formic acid:trifluoroacetic acid) was added, and the mixture was stored overnight in the dark at 4°C. The mixture was then centrifuged at 4°C at 12,000g for 15 min, and each supernatant was filtered through a 0.22-μm Millipore filter. Trifluoroacetic acid:formic acid:water (0.1:2:97.9 [v/v]) was selected as mobile phase A and trifluoroacetic acid:formic acid:acetonitrile:water (0.1:2:48:49.9 [v/v]) was selected as mobile phase B. The gradients used were as follows: 0 min, 30% B; 10 min, 40% B; 50 min, 55% B; 70 min, 60% B; and 80 min, 30% B. Detection was performed at 520 nm for anthocyanin (Wu and Prior, 2005). All samples were analyzed in triplicate.
Amplification of Gene Sequences
Using 100 ng of first-strand cDNA, the full-length cDNAs of the ERF17 genes were amplified using gene-specific primers based on GenBank sequences (accession nos. MF498883, MF498882, and MF498881). PCR conditions were 5 min at 95°C followed by 35 cycles of 94°C for 30 s, 62°C ± 2°C for 30 s, and extension at 72°C, 700 bp min−1. The PCR products were purified and cloned directly into the pMD-19 (Takara) vector for sequencing. All primers used are listed in Supplemental Table S5.
RT-qPCR
RT-qPCR was performed with SYBR Premix Ex Taq II (Takara) in a CFX96 Real-Time PCR System (Bio-Rad). Relative levels of the gene transcripts were quantified by normalizing to the MdActin gene (nucleotide, CN938023) using the 2−ΔΔCT method (Livak and Schmittgen, 2001). PCR included preincubation at 95°C for 3 min followed by 40 cycles of denaturation at 94°C for 15 s, annealing at 58°C for 20 s, and dissociation at 65°C to 95°C for 0.1°C s−1. RT-qPCR analysis was repeated using three technical replicates. All primers are listed in Supplemental Table S5.
Transcriptional Activity Assays
The dual-luciferase method was used to analyze ERF17 regulatory activity in protoplasts according to a reported protocol (Wei et al., 2009). The GAL4-LUC reporter plasmid was generated from pUC19 (Ohta et al., 2001), which contains the firefly LUC reporter gene driven by the minimal TATA box of the 35S promoter of Cauliflower mosaic virus and five copies of GAL4-binding elements. The internal control pPTRL (Renilla reniformis luciferase) was driven by the 35S promotor (Wei et al., 2009). pRT-BD was constructed by insertion of the GAL4DBD coding region into the pRT107 vector by SacI/XbaI digestion (Wei et al., 2009).
The full-length coding sequences of ERF17-3S, ERF17-6S, and ERF17-8S were inserted into the pTRL-BD vector (Wei et al., 2015) using KpnI/XbaI digestion. The positive control (pRT-35S-BD-VP16) was constructed by insertion of VP16, a herpes simplex virus-encoded transcriptional activator protein, into pRT-BD. A pTRL:reporter:effector plasmid ratio of 1:6:6 was used in a cotransfection into Arabidopsis protoplasts using the PEG/CaCl2-mediated transfection method (Wei et al., 2015). After culturing for 16 h, luciferase assays were performed using the Promega dual-luciferase reporter assay system and a GloMax 20-20 luminometer (Promega). Relative LUC activity was defined as firefly LUC activity divided by R. reniformis LUC activity.
Yeast One-Hybrid Assay
The full-length ERF17-8s, ERF17-6s, and ERF17-3s cDNA PCR products were cloned into the pJG4-5 (pB42AD) vector for protein expression (Clontech Matchmaker One-Hybrid System user manual), while the NYC, PAO, and PPH promoters were ligated into the placZi vector. Constructs were cotransformed into yeast (Saccharomyces cerevisiae) strain EYG48 and selected on SD/Trp-Ura plates at 28°C. After 2 to 3 d, single yeast colonies were picked and cultured in liquid SD/Trp-Ura medium at 28°C with shaking at 200 rpm until they entered the logarithmic growth phase. Five microliters of bacteria solution was colored on an SD/Trp-Ura plate with X-gal added at 28°C. All transformations and screenings were performed three times. The primer sequences used are shown in Supplemental Table S5.
EMSA
The full-length ERF17-8s and ERF17-3s cDNA PCR products were subcloned into the pGEX-6p-1 expression vector (Liu et al., 2009). Bacterial growth and protein induction were performed as described by the manufacturer (Novagen). Following the induction of protein expression in the cells with 100 mL of 0.3 mm isopropyl-β-d-thiogalactoside and after washing and suspending in PBS (135 mm NaCl, 4.7 mm KCl, 10 mm Na2HPO4, and 2 mm NaH2PO4, pH 7.4), bacteria were stored at −80°C. The purity of the proteins was verified by SDS-PAGE analysis (Laemmli, 1970). The protein was quantified and then protein concentrations were adjusted by dilution in annealing buffer (100 mm Tris-HCl [pH 7.5], 10 mm EDTA, and 1 m NaCl), such that the probe was used at a concentration of 10 μm. The annealing conditions were 75°C for 30 min followed by slow annealing at room temperature for 2 h. Subsequently, 10 μL was diluted to a final concentration of 1 μm (in 100 μL of water). EMSA detection was performed as described by the manufacturer (Thermo; 89880). The probe sequences are listed in Supplemental Table S3.
Plant Transient Transformation
The coding sequences of the three ERF17 genes were amplified by PCR using cDNA as a template. The primers for the ERF17 coding sequence amplification were designed with XbaI and BamHI sites, which were used to ligate the fragment into the pBI1300 vector (Su et al., 2016). The Agrobacterium tumefaciens strain GV3103 containing pBI1300-ERF17s was transformed into cv Granny Smith and TaiShanZaoXia fruit via injection and vacuum infiltration (Tian et al., 2015). RT-qPCR was performed as described above. All primers are listed in Supplemental Table S5.
SPR
SPR measurements were performed using five dilutions (17.2, 34.4, 68.75, 137.5, and 275 nm) of the purified ERF proteins on a Biacore T200 platform using biotin-labeled oligonucleotides immobilized on SA chips (GE Healthcare). Data were analyzed with the Scrubber2-T200 software (BioLogic Software).
ERF17 Nucleotide Polymorphism Analyses in Malus Genotypes
To determine the DNA sequence diversity in the promoter of ERF17, DNA fragments (1,390 bp) were PCR amplified using the primers listed in Supplemental Table S5. DNA fragments were amplified from 72 Malus species. TransTaq DNA Polymerase High Fidelity (TransGen) was used in the PCRs to minimize DNA synthesis errors. The PCR products were purified and sequenced (Genewiz), and the sequences were analyzed using MEGA5.2 (Tamura et al., 2011).
Statistical Analysis
Significant differences were analyzed using the independent samples Student’s t test (SPSS Statistics 21; IBM). Mean values ± sd are presented for each set of three replicates using Data Processing System 7.05 (Wiley-Blackwell). Figures were made using Origin Pro 8 statistical software (OriginLab). Comparison and analysis of the ERF17 sequences were conducted with the advanced BLAST program at the National Center for Biotechnological Information (http//www.ncbi.nlm.nih.gov).
Accession Numbers
Sequence data from this article can be found in GenBank under the following accession numbers: ERF17-3S (MF498883), ERF17-6S (MF498882), and ERF17-8S (MF498881).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Genetic linkage map identified from the cv Zisai Pearl × Red Fuji cross population.
Supplemental Figure S2. Gene and protein sequence alignments.
Supplemental Figure S3. Phylogenetic analysis of CLH, NYC, PAO, PPH, and RCCR.
Supplemental Figure S4. Target motifs for AP2/ERF transcription factors in NYC, PAO, and PPH promoters.
Supplemental Figure S5. ERF17 overexpression in cv TaiShanZaoXia fruits by A. tumefaciens-mediated transient transformation.
Supplemental Figure S6. Anthocyanin concentrations and relative expression of anthocyanin biosynthesis genes in ERF17 transiently overexpressed cv TaiShanZaoXia fruits.
Supplemental Figure S7. Determination of the relationship between the ERF17 allele genotype and the Malus fruit coloring phenotype.
Supplemental Table S1. ERF17 expression rate and peel Chl degradation rate.
Supplemental Table S2. Correlation analysis between fruit maturity indices and relative Chl content.
Supplemental Table S3. Probe sequences used for the EMSA.
Supplemental Table S4. ERF17 genotypes in Malus germplasm accessions.
Supplemental Table S5. Primers used for gene cloning, RT-qPCR, LUC, yeast one-hybrid, and transient expression analyses.
Acknowledgments
We are indebted to technical engineer Jiawei Wu (GE Healthcare) for help with our SPR experiments. We thank Shouyi Chen and Wei Wei (State Key Laboratory of Plant Genomics, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) as well as Wenyuan Ruan (Institute of Agricultural Resources and Regional Planning, China Academy of Agricultural Sciences) for the gifts of transcriptional activity assay vectors and yeast one-hybrid assay vectors, respectively, and their advice on these assays.
Footnotes
This work was supported by the National Natural Science Foundation of China (no. 31401840), the Beijing Municipal Commission of Education (CEFF-PXM2017_014207_000043), the 111 Project (B17043), and Key Labs of both Nutrition and Physiology for Horticultural Crops and Physiology and Molecular Biology of Tree Fruit.
Articles can be viewed without a subscription.
References
- Akhtar MS, Goldschmidt EE, John I, Rodoni S, Matile P, Grierson D (1999) Altered patterns of senescence and ripening in gf, a staygreen mutant of tomato (Lycopersicon esculentum Mill.). J Exp Bot 50: 1115–1122 [Google Scholar]
- Albert NW, Davies KM, Lewis DH, Zhang H, Montefiori M, Brendolise C, Boase MR, Ngo H, Jameson PE, Schwinn KE (2014) A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26: 962–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An XH, Tian Y, Chen KQ, Liu XJ, Liu DD, Xie XB, Cheng CG, Cong PH, Hao YJ (2015) MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol 56: 650–662 [DOI] [PubMed] [Google Scholar]
- An XH, Tian Y, Chen KQ, Wang XF, Hao YJ (2012) The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J Plant Physiol 169: 710–717 [DOI] [PubMed] [Google Scholar]
- Ban Y, Honda C, Hatsuyama Y, Igarashi M, Bessho H, Moriguchi T (2007) Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol 48: 958–970 [DOI] [PubMed] [Google Scholar]
- Barry CS, McQuinn RP, Chung MY, Besuden A, Giovannoni JJ (2008) Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol 147: 179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA (2012) Gene balance hypothesis: connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci USA 109: 14746–14753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovsky Y, Paran I (2008) Chlorophyll breakdown during pepper fruit ripening in the chlorophyll retainer mutation is impaired at the homolog of the senescence-inducible stay-green gene. Theor Appl Genet 117: 235–240 [DOI] [PubMed] [Google Scholar]
- Chagné D, Lin-Wang K, Espley RV, Volz RK, How NM, Rouse S, Brendolise C, Carlisle CM, Kumar S, De Silva N, et al. (2013) An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol 161: 225–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Birchler JA, Pires JC (2014) Dosage, duplication, and diploidization: clarifying the interplay of multiple models for duplicate gene evolution over time. Curr Opin Plant Biol 19: 91–98 [DOI] [PubMed] [Google Scholar]
- Eckardt NA. (2009) A new chlorophyll degradation pathway. Plant Cell 21: 700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49: 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faramarzi S, Yadollahi A, Soltani BM (2014) Preliminary evaluation of genetic diversity among Iranian red fleshed apples using microsatellite markers. J Agric Sci Tech Iran 16: 373–384 [Google Scholar]
- Freeling M. (2009) Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol 60: 433–453 [DOI] [PubMed] [Google Scholar]
- Fukao T, Xu K, Ronald PC, Bailey-Serres J (2006) A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 18: 2021–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53: 814–827 [DOI] [PubMed] [Google Scholar]
- Guyer L, Hofstetter SS, Christ B, Lira BS, Rossi M, Hörtensteiner S (2014) Different mechanisms are responsible for chlorophyll dephytylation during fruit ripening and leaf senescence in tomato. Plant Physiol 166: 44–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie Y, Ito H, Kusaba M, Tanaka R, Tanaka A (2009) Participation of chlorophyll b reductase in the initial step of the degradation of light-harvesting chlorophyll a/b-protein complexes in Arabidopsis. J Biol Chem 284: 17449–17456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörtensteiner S. (2006) Chlorophyll degradation during senescence. Annu Rev Plant Biol 57: 55–77 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S. (2009) Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci 14: 155–162 [DOI] [PubMed] [Google Scholar]
- Jibran R, Sullivan KL, Crowhurst R, Erridge ZA, Chagné D, McLachlan ARG, Brummell DA, Dijkwel PP, Hunter DA (2015) Staying green postharvest: how three mutations in the Arabidopsis chlorophyll b reductase gene NYC1 delay degreening by distinct mechanisms. J Exp Bot 66: 6849–6862 [DOI] [PubMed] [Google Scholar]
- Jung S, Krippner J, Schubert S (2016) Is the photometric method using acetone extracts to determine chlorophyll concentrations applicable to Mg-deficient plants? J Plant Nutr Soil Sci 179: 439–442 [Google Scholar]
- Klee HJ, Giovannoni JJ (2011) Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet 45: 41–59 [DOI] [PubMed] [Google Scholar]
- Kusaba M, Ito H, Morita R, Iida S, Sato Y, Fujimoto M, Kawasaki S, Tanaka R, Hirochika H, Nishimura M, et al. (2007) Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 19: 1362–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lai B, Hu B, Qin YH, Zhao JT, Wang HC, Hu GB (2015) Transcriptomic analysis of Litchi chinensis pericarp during maturation with a focus on chlorophyll degradation and flavonoid biosynthesis. BMC Genomics 16: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Jiang Z, Zhang L, Tan D, Wei Y, Yuan H, Li T, Wang A (2016) Apple (Malus domestica) MdERF2 negatively affects ethylene biosynthesis during fruit ripening by suppressing MdACS1 transcription. Plant J 88: 735–748 [DOI] [PubMed] [Google Scholar]
- Li T, Xu Y, Zhang L, Ji Y, Tan D, Yuan H, Wang A (2017) The jasmonate-activated transcription factor MdMYC2 regulates ETHYLENE RESPONSE FACTOR and ethylene biosynthetic genes to promote ethylene biosynthesis during apple fruit ripening. Plant Cell 29: 1316–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Zhu G, Zhang J, Xu X, Yu Q, Zheng Z, Zhang Z, Lun Y, Li S, Wang X, et al. (2014) Genomic analyses provide insights into the history of tomato breeding. Nat Genet 46: 1220–1226 [DOI] [PubMed] [Google Scholar]
- Liu B, Li G, Sui X, Yin J, Wang H, Ren X (2009) Expression and functional analysis of porcine aminopeptidase N produced in prokaryotic expression system. J Biotechnol 141: 91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Matile P, Hortensteiner S, Thomas H, Krautler B (1996) Chlorophyll breakdown in senescent leaves. Plant Physiol 112: 1403–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser D, Matile P (1997) Chlorophyll breakdown in ripening fruits of Capsicum annuum. J Plant Physiol 150: 759–761 [Google Scholar]
- Oeller PW, Lu MW, Taylor LP, Pike DA, Theologis A (1991) Reversible inhibition of tomato fruit senescence by antisense RNA. Science 254: 437–439 [DOI] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipps JB, Robertson KR, Smith PG, Rohrer JR (1990) A checklist of the subfamily Maloideae (Rosaceae). Can J Bot 68: 2209–2269 [Google Scholar]
- Pino MT, Skinner JS, Jeknić Z, Hayes PM, Soeldner AH, Thomashow MF, Chen THH (2008) Ectopic AtCBF1 over-expression enhances freezing tolerance and induces cold acclimation-associated physiological modifications in potato. Plant Cell Environ 31: 393–406 [DOI] [PubMed] [Google Scholar]
- Powell ALT, Nguyen CV, Hill T, Cheng KL, Figueroa-Balderas R, Aktas H, Ashrafi H, Pons C, Fernández-Muñoz R, Vicente A, et al. (2012) Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 336: 1711–1715 [DOI] [PubMed] [Google Scholar]
- Pruzinská A, Anders I, Aubry S, Schenk N, Tapernoux-Lüthi E, Müller T, Kräutler B, Hörtensteiner S (2007) In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 19: 369–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca M, James C, Pruzinská A, Hörtensteiner S, Thomas H, Ougham H (2004) Analysis of the chlorophyll catabolism pathway in leaves of an introgression senescence mutant of Lolium temulentum. Phytochemistry 65: 1231–1238 [DOI] [PubMed] [Google Scholar]
- Rodrigues MA, Bianchetti RE, Freschi L (2014) Shedding light on ethylene metabolism in higher plants. Front Plant Sci 5: 665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelbert S, Aubry S, Burla B, Agne B, Kessler F, Krupinska K, Hörtensteiner S (2009) Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 21: 767–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour GB, Østergaard L, Chapman NH, Knapp S, Martin C (2013) Fruit development and ripening. Annu Rev Plant Biol 64: 219–241 [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M, Huang G, Zhang Q, Wang X, Li C, Tao Y, Zhang S, Lai J, Yang C, Wang Y (2016) The LEA protein, ABR, is regulated by ABI5 and involved in dark-induced leaf senescence in Arabidopsis thaliana. Plant Sci 247: 93–103 [DOI] [PubMed] [Google Scholar]
- Sun-Waterhouse D, Luberriaga C, Jin D, Wibisono R, Wadhwa SS, Waterhouse GIN (2013) Juices, fibres and skin waste extracts from white, pink or red-fleshed apple genotypes as potential food ingredients. Food Bioprocess Technol 6: 377–390 [Google Scholar]
- Sureshkumar S, Todesco M, Schneeberger K, Harilal R, Balasubramanian S, Weigel D (2009) A genetic defect caused by a triplet repeat expansion in Arabidopsis thaliana. Science 323: 1060–1063 [DOI] [PubMed] [Google Scholar]
- Tajima F. (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takos AM, Jaffé FW, Jacob SR, Bogs J, Robinson SP, Walker AR (2006) Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 142: 1216–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Lv S, Liu XY, Gao T, Li TH, Wang Y, Wu T, Zhang XZ, Han YP, Korban SS, et al. (2017) Development of high-density interspecific genetic maps for the identification of QTLs conferring resistance to Valsa ceratosperma in apple. Euphytica 213: 10 [Google Scholar]
- Thomas MB, Langewald J, Wood SN (1996) Evaluating the effects of a biopesticide on populations of the variegated grasshopper, Zonocerus variegates. J Appl Ecol 33: 1509–1516 [Google Scholar]
- Tian J, Han ZY, Zhang J, Hu Y, Song T, Yao Y (2015) The balance of expression of dihydroflavonol 4-reductase and flavonol synthase regulates flavonoid biosynthesis and red foliage coloration in crabapples. Sci Rep 5: 12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen JW. (2009) MapQTL 6.0. Kyazma, Wageningen, The Netherlands [Google Scholar]
- Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, et al. (2010) The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 42: 833–839 [DOI] [PubMed] [Google Scholar]
- Vicentini F, Hörtensteiner S, Schellenberg M, Thomas H (1995) Chlorophyll breakdown in senescent leaves: identification of the biochemical lesion in a stay-green genotype of Festuca pratensis Huds. New Phytol 129: 247–252 [DOI] [PubMed] [Google Scholar]
- Volk GM, Henk AD, Baldo A, Fazio G, Chao CT, Richards CM (2015) Chloroplast heterogeneity and historical admixture within the genus Malus. Am J Bot 102: 1198–1208 [DOI] [PubMed] [Google Scholar]
- Voorrips RE. (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93: 77–78 [DOI] [PubMed] [Google Scholar]
- Wang X, Wei Z, Ma F (2015) The effects of fruit bagging on levels of phenolic compounds and expression by anthocyanin biosynthetic and regulatory genes in red-fleshed apples. Process Biochem 50: 1774–1782 [Google Scholar]
- Wei W, Huang J, Hao YJ, Zou HF, Wang HW, Zhao JY, Liu XY, Zhang WK, Ma B, Zhang JS, et al. (2009) Soybean GmPHD-type transcription regulators improve stress tolerance in transgenic Arabidopsis plants. PLoS ONE 4: e7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Zhang YQ, Tao JJ, Chen HW, Li QT, Zhang WK, Ma B, Lin Q, Zhang JS, Chen SY (2015) The Alfin-like homeodomain finger protein AL5 suppresses multiple negative factors to confer abiotic stress tolerance in Arabidopsis. Plant J 81: 871–883 [DOI] [PubMed] [Google Scholar]
- Wu S, Li Z, Yang L, Xie Z, Chen J, Zhang W, Liu T, Gao S, Gao J, Zhu Y, et al. (2016) NON-YELLOWING2 (NYE2), a close paralog of NYE1, plays a positive role in chlorophyll degradation in Arabidopsis. Mol Plant 9: 624–627 [DOI] [PubMed] [Google Scholar]
- Wu T, Wang Y, Zheng Y, Fei Z, Dandekar AM, Xu K, Han Z, Cheng L (2015) Suppressing sorbitol synthesis substantially alters the global expression profile of stress response genes in apple (Malus domestica) leaves. Plant Cell Physiol 56: 1748–1761 [DOI] [PubMed] [Google Scholar]
- Wu XL, Prior RL (2005) Identification and characterization of anthocyanins by high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry in common foods in the United States: vegetables, nuts, and grains. J Agric Food Chem 53: 3101–3113 [DOI] [PubMed] [Google Scholar]
- Xiao YY, Chen JY, Kuang JF, Shan W, Xie H, Jiang YM, Lu WJ (2013) Banana ethylene response factors are involved in fruit ripening through their interactions with ethylene biosynthesis genes. J Exp Bot 64: 2499–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie XL, Shen SL, Yin XR, Xu Q, Sun CD, Grierson D, Ferguson I, Chen KS (2014) Isolation, classification and transcription profiles of the AP2/ERF transcription factor superfamily in citrus. Mol Biol Rep 41: 4261–4271 [DOI] [PubMed] [Google Scholar]
- Yang B, Dougherty L, Cheng L, Xu K (2015) A co-expression gene network associated with developmental regulation of apple fruit acidity. Mol Genet Genomics 290: 1247–1263 [DOI] [PubMed] [Google Scholar]
- Yao G, Ming M, Allan AC, Gu C, Li L, Wu X, Wang R, Chang Y, Qi K, Zhang S, et al. (2017) Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J 92: 437–451 [DOI] [PubMed] [Google Scholar]
- Yao JL, Xu J, Cornille A, Tomes S, Karunairetnam S, Luo Z, Bassett H, Whitworth C, Rees-George J, Ranatunga C, et al. (2015) A microRNA allele that emerged prior to apple domestication may underlie fruit size evolution. Plant J 84: 417–427 [DOI] [PubMed] [Google Scholar]
- Yin XR, Xie XL, Xia XJ, Yu JQ, Ferguson IB, Giovannoni JJ, Chen KS (2016) Involvement of an ethylene response factor in chlorophyll degradation during citrus fruit degreening. Plant J 86: 403–412 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Ma D, Constabel CP (2015) The MYB182 protein down-regulates proanthocyanidin and anthocyanin biosynthesis in poplar by repressing both structural and regulatory flavonoid genes. Plant Physiol 167: 693–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Lv Y, Wang Y, Rose JKC, Shen F, Han Z, Zhang X, Xu X, Wu T, Han Z (2017) TATA box insertion provides a selection mechanism underpinning adaptations to Fe deficiency. Plant Physiol 173: 715–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Song T, Li J, Tian J, Jin K, Yao Y (2014) Low medium pH value enhances anthocyanin accumulation in Malus crabapple leaves. PLoS ONE 9: e97904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Huang R (2010) Enhanced tolerance to freezing in tobacco and tomato overexpressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynthesis. Plant Mol Biol 73: 241–249 [DOI] [PubMed] [Google Scholar]