The kinetics of changes in the ethylene transcriptome in light-grown roots combined with mutant analyses reveal a central role of the ETR1 receptor and a limited role of EIN3 in root development.
Abstract
Transcriptomic analyses with high temporal resolution provide substantial new insight into hormonal response networks. This study identified the kinetics of genome-wide transcript abundance changes in response to elevated levels of the plant hormone ethylene in roots from light-grown Arabidopsis (Arabidopsis thaliana) seedlings, which were overlaid on time-matched developmental changes. Functional annotation of clusters of transcripts with similar temporal patterns revealed rapidly induced clusters with known ethylene function and more slowly regulated clusters with novel predicted functions linked to root development. In contrast to studies with dark-grown seedlings, where the canonical ethylene response transcription factor, EIN3, is central to ethylene-mediated development, the roots of ein3 and eil1 single and double mutants still respond to ethylene in light-grown seedlings. Additionally, a subset of these clusters of ethylene-responsive transcripts were enriched in targets of EIN3 and ERFs. These results are consistent with EIN3-independent developmental and transcriptional changes in light-grown roots. Examination of single and multiple gain-of-function and loss-of-function receptor mutants revealed that, of the five ethylene receptors, ETR1 controls lateral root and root hair initiation and elongation and the synthesis of other receptors. These results provide new insight into the transcriptional and developmental responses to ethylene in light-grown seedlings.
Transcriptomic data sets have changed the way we understand molecular events that mediate important biological responses, including hormone responses (Fortes et al., 2011; Eremina et al., 2016) and developmental mechanisms (Hale et al., 2016; Gupta et al., 2017). The addition of another dimension, time, magnifies the insights we can obtain from transcriptomic data. The transcriptome is highly dynamic; the same comparison of transcriptomes between treatments or conditions might yield very different results depending on the time of sampling. A time-course approach uses this complexity as an advantage. By examining changes in a transcriptome at multiple time points, we can see the progression of transcriptional changes, revealing patterns and pathways (Yosef and Regev, 2011). This temporal resolution also provides layers of information beyond apparent up- or down-regulation. It also allows greater confidence in the reproducibility of those data as changes are replicated across time. Even more significantly, we can observe when changes in a transcript’s levels peak, and group transcripts based on kinetics, as well as the direction of change. These kinetic patterns can then be correlated with other biological information, such as gene function, protein-protein interactions, targets for transcription factor binding, and the timing of events on a cell, tissue, or organism level (Nagano et al., 2012). In particular, this approach can be useful for connecting early signaling events, later transcriptional changes, and their ultimate developmental responses.
Plant responses to the gaseous hormone ethylene are an excellent model for studying these relationships between signaling, transcriptional changes, and development. The ethylene signaling pathway was uncovered by molecular genetic approaches, taking advantage of the profound developmental changes observed in dark-grown seedlings in response to elevated ethylene (Bleecker et al., 1988; Guzmán and Ecker, 1990); yet, the transcriptional changes that connect signaling and development are incompletely characterized, especially in light-grown tissues. In the model plant Arabidopsis (Arabidopsis thaliana), there are five ethylene receptors that act as negative regulators of the pathway (Chang et al., 1993; Schaller and Bleecker, 1995; Hua and Meyerowitz, 1998; Sakai et al., 1998). These five ethylene receptors are not equal, with subfunctionalization observed for different responses in different tissues and developmental stages (Wang et al., 2003; Binder et al., 2004b, 2006; Qu et al., 2007; Liu et al., 2010; McDaniel and Binder, 2012; Wilson et al., 2014a; Bakshi et al., 2015). This subfunctionalization is likely due to diversity in receptor structure and signaling capabilities (O’Malley et al., 2005; Wang et al., 2006; Shakeel et al., 2013; Bakshi et al., 2015). In air, these receptors act through a RAF-like kinase, CTR1 (Kieber et al., 1993), to inhibit signaling through the EIN2 protein, whose catalytic activity is not yet known (Alonso et al., 1999; Qiao et al., 2009, 2012; Ju et al., 2012). The application of ethylene inhibits the receptors, leading to lower CTR1 activity, which, in turn, causes a reduction in the phosphorylation of EIN2; this leads to a decrease in the ubiquitination of EIN2 and a rise in EIN2 protein levels, allowing for the proteolytic release of the C-terminal portion of the protein via an unidentified protease (Kieber et al., 1993; Qiao et al., 2009, 2012; Chen et al., 2011; Ju et al., 2012; Wen et al., 2012). The C-terminal portion of EIN2 modulates two transcription factors, EIN3 and EIN3-LIKE1 (EIL1), leading to the majority of ethylene responses (Chao et al., 1997; Solano et al., 1998; Alonso et al., 1999; Guo and Ecker, 2003; Yanagisawa et al., 2003; Binder et al., 2004b; Gagne et al., 2004; Qiao et al., 2012). In dark-grown shoot tissues treated with ethylene, it has been observed that EIN2 stabilizes the EIN3 transcription factor (An et al., 2010; Wen et al., 2012), which has a central role in mediating the ethylene triple response (short and wide hypocotyl and exaggerated apical hook; Alonso et al., 2003; Guo and Ecker, 2003; Yanagisawa et al., 2003). Some EIN3 targets are known, and in dark-grown seedlings, EIN3-regulated transcriptional changes have been observed to occur in waves (Chang et al., 2013). What is less clear is the role of EIN3 in ethylene responses in all contexts, such as roots or other tissues in light-grown seedlings, and which transcriptional responses to ethylene drive these tissue growth condition-specific developmental responses.
The kinetics of transcriptional responses to ethylene are likely to be of importance in Arabidopsis roots, where developmental responses occur on multiple time scales. In Arabidopsis, root architecture is defined by the elongation of the primary root and the development and elongation of lateral (branching) roots, which are covered in root hairs. Lateral root development is a complex process that involves the reprogramming of differentiated cells in the primary root to dedifferentiate and form a new root that recapitulates the developmental program of primary roots (Péret et al., 2009). The development of root hairs from epidermal cells also provides additional surface area to the root system (Cutter, 1978). This complex branching architecture of roots is critical for plant health, as roots are essential for the uptake of water and nutrients into the plant. Differences in root structure have been correlated with traits such as enhanced drought resistance (Yu et al., 2008; Zhan et al., 2015), which will be increasingly important for improved agriculture in a changing climate.
Ethylene mediates several different developmental changes in roots, namely, it inhibits primary root growth and lateral root development but stimulates root hair development and elongation (Guzmán and Ecker, 1990; Kieber et al., 1993; Tanimoto et al., 1995; Rahman et al., 2002; Seifert et al., 2004; Ivanchenko et al., 2008; Negi et al., 2008, 2010). Ethylene-mediated inhibition of root elongation can take effect in as few as 5 min (Le et al., 2001), while changes in lateral root development and root hair formation may take hours to become statistically significant (Lewis et al., 2013). Together with previous observations of wave-like transcriptional responses (Chang et al., 2013), this suggests that the kinetics of transcriptional changes may be an important layer of information for understanding ethylene-mediated transcriptional changes and how they control development.
To provide new insight into the ethylene gene regulatory networks that control the development of roots in light-grown seedlings, we performed a time-course transcriptomic study of Arabidopsis roots. We treated roots with the direct ethylene precursor, 1-aminocyclopropane-1-carboxylic acid (ACC), and compared the transcript abundance with that of a time-matched control. We observed many transcripts that responded to ACC in a time-dependent manner and grouped these transcripts according to the kinetics of changes. By comparing transcripts in this data set with two others from dark-grown seedlings and roots, we identified tissue- and environment-specific transcriptional responses. We used these clusters to demonstrate that EIN3 and Ethylene Response Factors (ERFs) bind to a subset of root ethylene-responsive transcripts, targeting the most rapidly and positively regulated response clusters in these light-grown samples. Examining these changes with high temporal resolution also led to insights into gene function and how they might connect to developmental changes. We also examined the ethylene receptors, finding different roles for receptors in distinct root developmental responses. Together, these experiments provide insight into the networks of signaling and transcriptional responses that drive root development.
RESULTS
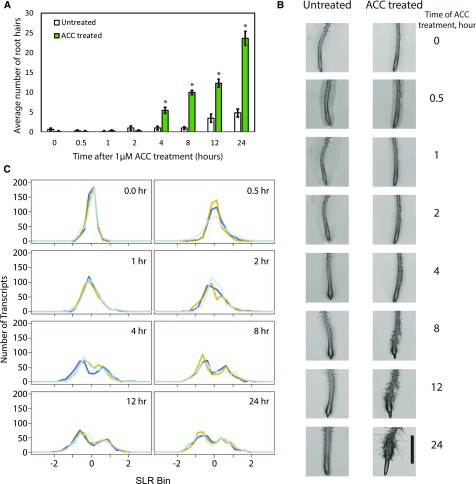
ACC-Treated Roots Show Changes in Root Hair Numbers and Transcript Abundance across a 24-h Time Course
To identify a relevant time course of ACC treatment for our microarray analyses that spanned the time line of developmental effects, we examined the kinetics of stimulation of root hair initiation. We transferred 4-d-old seedlings to new medium with or without the ethylene precursor ACC at 1 µm and quantified root hairs at eight time points: 0, 0.5, 1, 2, 4, 8, 12, and 24 h after transfer (Fig. 1, A and B). A two-way ANOVA found significant differences between control and ACC treatments, and a Tukey’s posthoc test was performed to determine the time points at which ACC had a significant effect (Supplemental Table S1). ACC-treated roots showed significantly more root hairs than untreated controls at 4 h and at all later time points (P < 0.0001). This demonstrates that changes in root hair initiation and elongation in response to ACC occur during the 24 h after treatment, illustrating that these eight time points provide a developmentally pertinent time line for microarray analysis.
Figure 1.
Kinetics of ACC-induced root hair formation and transcript abundance changes. A, Four-day-old seedlings were transferred to new medium containing 1 µm ACC for the indicated times. Root hairs were imaged at the indicated times, and the average number and se of root hairs for 12 to 18 seedlings are reported. Asterisks indicate significant differences (P < 0.0001) between ACC-treated roots and the time-matched controls. B, Differential interference contrast images of root hair growth over time in untreated and ACC-treated Col-0 roots. Size bar = 1 mm. C, A series of histograms representing the SLR distribution of the 449 DE transcripts at each time point demonstrates that most transcripts decrease in abundance upon ACC treatment, with the most profound changes occurring beginning at 4 h when developmental responses are detected. Three biological replicates are represented individually.
To detect genes whose expression is regulated by ethylene, we performed a transcriptome analysis of roots treated with ACC, a solid precursor of ethylene gas that could be added to growth substrates. Previous analyses have indicated that the root transcriptional and developmental responses to ACC mirror those of ethylene under our treatment conditions (Negi et al., 2008; Lewis et al., 2011b). As one goal of these experiments was to compare responses to two plant hormones, ethylene and auxin, it was optimal to use a solid that could be added to the growth medium to elevate both hormones. We isolated RNA from roots at the same eight time points used for root hair quantification. For each time point, plants were transferred to control medium or medium containing 1 µm ACC for the indicated times before root tissue was harvested for RNA extraction and microarray analysis. This analysis was performed simultaneously with a previously published auxin (indole-3-acetic acid [IAA]) transcriptome time course, sharing common control samples (Lewis et al., 2013).
The microarray data for each treatment replicate were analyzed separately. A strict differential expression filtering approach was used to identify genes that were differentially expressed (DE) in the treatment compared with controls (Lewis et al., 2013). This process, as outlined in Supplemental Figure S1, first identified transcripts with a signal intensity above the Affymetrix background signal at every time point. The signal log ratio (SLR; log base 2 of fold change) was calculated for each transcript relative to an averaged time-matched control. Transcripts with an SLR > 0.5 (corresponding to a 1.4-fold change, chosen to identify robust abundance changes) for at least one time point, and with a consistent magnitude and pattern of change, were identified. This filtering resulted in a group of 449 transcripts with robust and consistent changes. The identity of each transcript and its abundance relative to time-matched controls are reported in Supplemental File S1.
The global distribution of SLRs for DE genes across the time course indicates that the majority of ACC transcripts have SLRs close to 0 at the 0-h time point (Fig. 1C), which is consistent with the control and treatment groups beginning with similar transcriptional profiles. For the 0.5-, 1-, and 2-h time points, the peak flattens and spreads, showing that few transcripts have changes in this early time window. However, at the 4-h time point, the data set yields a discernible bimodal distribution, indicating that many transcripts have either reduced or elevated abundance relative to the time-matched controls at this time point. This timing is consistent with the timing of ACC-induced root hair formation (Fig. 1A). It is interesting that most transcripts have lower abundance than the control samples (as seen by the higher peak on the negative side of the SLR bin). This distribution pattern remains through the 24-h time point, suggesting that some genes may maintain differential expression even beyond the scope of our time course. This pattern across the time course implies an ethylene response whereby a small number of genes respond rapidly, many others respond more slowly, and most of those genes are repressed rather than induced.
Genes with Related Functions Cluster into Groups with Similar Ethylene Responses
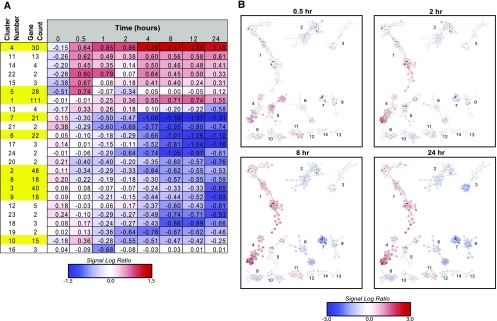
We might expect that transcripts with common regulatory controls and/or downstream functions might show similar patterns of abundance change. To identify these patterns, the 449 DE genes were clustered using the consensus clustering option in the SC2ATmd program (which can be downloaded from https://github.com/AmyOlex/SC2Atmd; Olex and Fetrow, 2011) and an empirically derived threshold for transcripts with common behavior 83% of the time. This threshold was empirically determined to maximize the information that could be obtained from the resulting clusters: higher thresholds resulted in many singletons and small clusters that were not useful for statistical analyses, whereas lower thresholds gave a few large clusters that did not adequately represent the diversity of temporal patterns. The 83% threshold grouped the transcripts into 24 clusters, with 49 single transcripts (singletons) remaining with unique temporal expression patterns that were dissimilar to transcripts in these clusters (Fig. 2). The patterns of the three replicates for each transcript also can be examined, as shown for four clusters in Supplemental Figure S2. Within a given cluster, the behavior of the transcripts is relatively consistent, although there is slight variation from the shared pattern between replicates and transcripts. Averaging the SLR values of every transcript in a cluster at each time point gives a representative profile of that cluster (Fig. 2A). These clusters are ordered by direction of change, rate of change, and magnitude of change. This view shows that there are more clusters that are down-regulated than up-regulated, consistent with the overall SLR profile in Figure 1. The kinetics of these clusters is highly varied, with rapid or more slowly executed responses and with response durations that are transient or sustained. For example, cluster 5 shows early and transient changes, while cluster 1 shows late and sustained change. Clusters 1 and 7 are particularly interesting, as they show increased and decreased transcripts, respectively, that change with the kinetics that parallel the kinetics of ACC-induced root hair initiation.
Figure 2.
Clusters of transcripts with similar profiles of temporal changes with early, middle, and late responses to ACC treatment can be visualized by heat maps and cluster networks. A, Consensus clusters were labeled (Cluster Number) from largest (DE cluster 1) to smallest (DE clusters 20–24). Gene Count represents the number of transcripts in a given cluster; the top 10 largest clusters are highlighted in yellow. The time-course profile for a cluster represents the average SLR at each time point. B, The cluster network of the 13 largest DE clusters is shown at four representative time points, revealing the degree of similarity of temporal responses within clusters. Nodes represent individual genes, and edges connect genes that clustered together in at least 83% of clustering iterations. For full time-course cluster networks, see Supplemental Figure S3. For both A and B, the red-blue scale represents positive and negative SLRs, respectively; white represents an SLR of zero (see color scale).
We also represented the clusters as networks to provide additional insight into the similarity of responses within each cluster. In the network diagram, transcripts are shown as nodes and a connecting edge represents two transcripts that are clustered together in 83% or more of the clustering iterations. The length of the edges in these networks is based on the number of node connections, where many connections result in shorter edges (as determined by Cytoscape’s layout algorithm), but does not indicate physical protein interactions or any other molecular interaction. The networks at 0.5, 2, 8, and 24 h are shown in Figure 2B, with the complete time course shown in Supplemental Figure S3. The layout of nodes in a cluster is determined by the degree (the number of attached vertices) of each node, so groups of transcripts that frequently cluster with one another will form tight subclusters. For example, cluster 6 has two groups of highly connected transcripts that form tight subclusters, while several other transcripts clustered with fewer additional transcripts at the 83% threshold. In contrast, cluster 7 is tightly connected, with every transcript connected to most other transcripts, suggesting a high degree of similarity between the SLR patterns of these transcripts. To understand the functional significance of these genes with coordinated expression patterns, we asked whether these clusters also contained conserved biological functions.
To determine if genes that cluster together may be functionally related, a Gene Ontology (GO) enrichment analysis was performed. This analysis determined which, if any, GO annotations appear in a cluster more often than would be expected by chance. Significantly enriched (P < 0.05) annotations were found in most of the 10 largest clusters (Table I). Some annotations were expected, such as ethylene response and two-component signal transduction (cluster 4). Others, such as the annotations for cell wall processes in cluster 9, which are reduced in abundance at late time points, suggest a mechanism that may modulate root growth and development.
Table I. Some clusters are significantly enriched in gene annotations, suggesting that genes with related processes respond similarly to ACC, with clusters in order of response rate.
| Cluster | Gene Count | Cluster Regulation | Overrepresented Cluster Annotations | Transcript Changes with SLR ≥ 0.05 |
|---|---|---|---|---|
| 4 | 30 | Up | Ethylene/hormone response, two-component signal transduction | 30 min to 24 h |
| 5 | 28 | Complex | Oxidoreductase activity | 30 min to 24 h |
| 7 | 21 | Down | None | 1 to 24 h |
| 10 | 15 | Down | None | 2 to 4 h |
| 1 | 111 | Up | None | 4 to 24 h |
| 2 | 48 | Down | Transferase activity | 4 to 24 h |
| 6 | 22 | Down | Metal/ion binding | 4 to 24 h |
| 8 | 18 | Down | None | 8 to 24 h |
| 9 | 18 | Down | Cell wall biogenesis, biosynthesis, and organization | 12 to 24 h |
| 3 | 40 | Down | Oxidoreductase activity | 24 h |
Differentially Expressed Genes Are Predominantly Unchanged across the Control Time Course
There are two possible scenarios whereby ACC reduced transcript abundance, resulting in negative SLR values. Either the abundance of transcripts in the treated samples decreases while the control values are held constant, or the transcripts in the treated samples are constant and the controls increase over time. The question of which of these is happening is an important one, because they represent different biological responses. In the first scenario, ethylene regulation decreases transcript levels from a steady state; in the second, ethylene regulation prevents a preprogrammed increase in transcript levels.
To resolve these two possibilities, we ran a separate filtering analysis on both the control samples alone and the ACC-treated samples alone. For each set of samples, a new SLR was calculated, this time using the 0-h time point as the control value, which we call the control-only SLRs and the ACC-only SLRs. We then ran the same filters used in the DE analysis to determine if transcripts exhibited significant changes in abundance relative to time zero. The results of these analyses for a subset of genes in several of the original clusters described above can be seen in Supplemental Figure S4, where the SLR ratios calculated with time-matched controls are compared with SLR calculated for the ACC-treated or control samples normalized to the time zero samples. What is immediately apparent is that, in control samples, the abundance of these transcripts remains essentially unchanged across the time course of these experiments while changing substantially in ACC-treated samples. Indeed, for most transcripts, the ACC-only SLR profile parallels the original DE SLR profile very closely. The results of the filtering steps indicate that most transcripts passed all filters in the ACC-treated samples, with only four failing the SLR filter and five failing the consistency filters, as noted in Supplemental File S2. Many of the control samples failed the SLR filter, consistent with limited change in these samples. This suggests that, for most of these transcripts, ACC treatment results in a change from a baseline expression level, with transcripts at relatively constant levels in untreated time-matched controls. This pattern holds when examining the entire data set of 449 DE genes; the results of this analysis for the 449 transcripts in our filtered data set are summarized in Supplemental File S2.
Present-Absent Analysis Identified Additional Genes of Interest
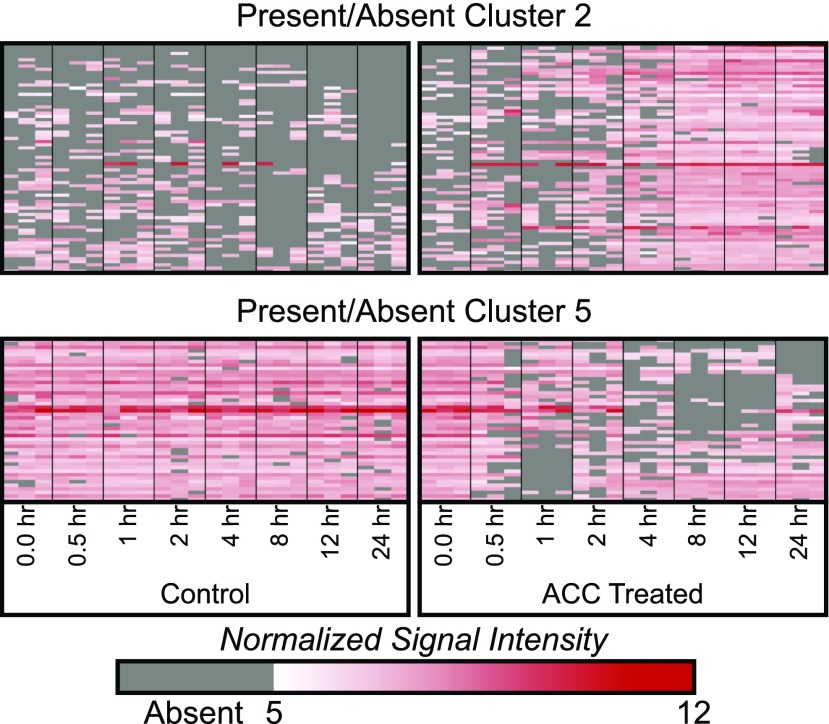
Transcripts that accumulate at low levels may not be detected over the chip background and, therefore, will have a detection P value that does not pass the Affymetrix-defined present threshold. Any gene with at least one absent measurement was filtered out by the DE analysis criteria, and a fold change was not calculated because it is inappropriate to calculate an SLR when a transcript abundance has a zero value (which is any value not above the Affymetrix-defined background) in either the control or treatment. However, transcripts that accumulate at background levels in either the control or ACC treatment conditions could still be of interest. To identify these transcripts, a present-absent (PA) analysis was performed using a previously outlined strategy (Lewis et al., 2013). Samples where a transcript did not pass the detection P value threshold were defined as absent and those that passed were defined as present. For this analysis, we make the assumption that, if a gene changes from present in the control to absent in the treatment or vise versa, then the experimental treatment has modified the abundance of transcripts for this gene in a meaningful way. Thus, we can search for consistent patterns of present-absent or absent-present in the control-ACC comparisons at each time point to identify genes that are consistently modified by these criteria. An additional 375 genes were identified and clustered in a similar manner to the 449 DE genes. These transcript identities and transcript abundance are found in Supplemental File S3, and heat maps of two clusters are shown in Figure 3. Some clusters show apparent up-regulation by ACC (absent-present pattern), including cluster 3 at the 12-h time point and cluster 2 across the 8- to 24-h time points, while others show apparent down-regulation by ACC (present-absent pattern), such as clusters 4 and 5 (Fig. 3; Supplemental Fig. S5). We say apparent because there is no way to know precisely if the gene is up- or down-regulated, since half the data are missing; however, if a gene has a signal strong enough to be detected on the control chip, and then the same gene is listed as absent on the experimental chip, we assume that the transcript abundance was reduced to below background levels in response to ethylene.
Figure 3.
PA analysis reveals additional genes with consistent, time-dependent changes in transcript abundance. Heat maps represent patterns of transcript abundance for two representative PA clusters. Columns represent three replicates for each time point; each row is an individual transcript. The white-red color scale represents the normalized signal intensity, and gray designates absent values that did not pass the Affymetrix detection cutoff.
Like the DE clusters, PA gene clusters were analyzed for GO enrichment. Cluster 5 has a present-absent pattern (or down-regulation) and is enriched in cell wall-related annotations, similar to the DE cluster 9. Other annotations found in various clusters (1, 2, 4, and 6) include oxidation/reduction, cytochrome P450, hormone signaling, and heat response, respectively (Table II). Interestingly, three of the six clusters were found to have significant (P < 0.05) enrichment in genes encoding transcription factor and/or transcriptional regulatory proteins. As transcription factors that had altered expression levels would, in turn, affect the levels of downstream transcripts, transcription factors provide a possible mechanism for long-term developmental effects of elevated ethylene.
Table II. GO analysis results for absent-present (AP) clusters.
| Cluster | Gene Count | Cluster Patterns | Overrepresented Cluster Annotations |
|---|---|---|---|
| 1 | 126 | Various, including AP at 8 or 24 h | Iron/oxygen/ion binding |
| 2 | 73 | AP at 4 to 24 h | Signal transduction, immune response, transcriptional regulation/transcription factors |
| 3 | 65 | AP at 8 to 24 h (most strongly at 12 h) | Photosynthetic machinery |
| 4 | 47 | PA at 1 to 24 h | Membrane |
| 5 | 46 | Various PA | Membrane |
| 6 | 18 | AP at 2 to 24 h | None |
Cell- and Tissue Type-Specific Expression Patterns of DE Transcripts Identify Groups of Transcripts with Unique Localization Patterns
This transcriptomic data set reveals transcript abundance changes in response to elevated ethylene with high temporal resolution but contains transcripts isolated from a number of tissue and cell types. The presence of high-spatial-resolution genome-wide transcript abundance maps for roots (Brady et al., 2007) allowed the identification of groups of transcripts from our DE data set to be examined for cell type (Supplemental Fig. S6) and tissue type (Supplemental Fig. S7) expression patterns. We overlaid this pattern on a color-coded network of our top 10 clusters. The similarity in pattern of related cell types (lateral roots and pericycle cells from which lateral roots emerge) or the clusters of genes linked to the elongation zone compared with the maturation zone suggest groups of ACC-regulated transcripts with coordinated spatial expression patterns. In a limited number of cases, we see that these transcripts come from common ACC time-course clusters, such as a group of genes with a high level of expression in the developing and maturing xylem that is enriched in genes from DE clusters 3 and 9 (Supplemental Fig. S6).
Time-Independent Analysis of Function Reveals Pathways Enriched in ACC-Responsive Transcripts
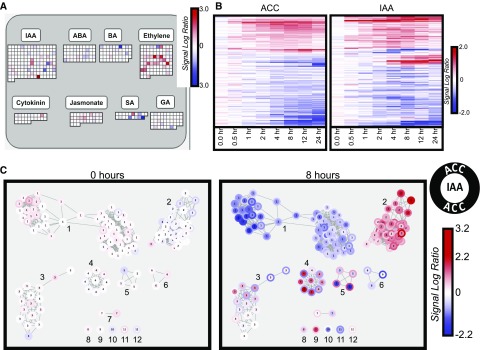
The above functional analyses of ACC-responsive genes rely on the similarity of their kinetic responses. However, it also may be possible that genes within a given pathway or functional group may respond at different points in the time course. In order to discover functionally related genes in a time-independent manner, the microarray data were collapsed across the time course. For each transcript, the largest magnitude average SLR at any time point was identified, yielding the most extreme change for any given gene. All transcripts were visualized using MapMan (Thimm et al., 2004) to identify relative enrichment of pathways and processes. Both the MapMan analysis and the functional annotation analysis described above revealed groups of transcripts with annotations linking them to ethylene responses or signaling pathways (Fig. 4; Table I). The metabolism overview (Supplemental Fig. S8) revealed an enrichment of genes involved in cell wall processes. This is consistent with the results of the GO analysis described earlier. It also revealed a number of transcripts annotated as controlling secondary metabolism linked to the glucosinolate, terpenoid, phenylpropanoid, and anthocyanin pathways that had altered abundance in response to ACC treatment.
Figure 4.
Ethylene regulates many hormone synthesis, signaling, and response genes. A, Image generated using the Regulation Overview in the MapMan Application Software. Squares under a given hormone represent genes/transcripts annotated to that hormone’s synthesis, signaling, or response. The color scale designates the greatest absolute SLR across the time course for each transcript. IAA, Indole-3-acetic acid; ABA, abscisic acid; BA, brassinosteroid; SA, salicylic acid; GA, gibberellic acid. B, The 139 ACC and IAA DE overlap transcripts, with average SLR across the time course shown for both hormone treatments. C, Network map showing clusters of 139 ACC and IAA DE overlap transcripts.
Some ACC-Responsive Transcripts Also Respond to Treatment with IAA
Ethylene and auxin control many of the same plant processes, sometimes synergistically, sometimes antagonistically (Muday et al., 2012). In roots, for example, ethylene and auxin both inhibit primary root elongation (Swarup et al., 2002) and stimulate root hair growth (Pitts et al., 1998), but ethylene inhibits while auxin stimulates lateral root development (Ivanchenko et al., 2008; Negi et al., 2010; Lewis et al., 2011a). Given these relationships, we were interested in the shared and divergent transcriptional responses to both ethylene and auxin in roots.
In a previously published study, our laboratory used the same experimental and statistical methods described here to examine the kinetics of the transcriptional responses of Arabidopsis roots to IAA, the most prevalent auxin. We identified 1,246 transcripts that were DE in response to IAA and identified 498 transcripts via a PA analysis.
To identify transcripts whose abundance changed in response to both ACC and IAA, we compared the lists of DE and PA transcripts from both ACC and IAA experiments, yielding 139 transcripts in common for the DE data sets, and 124 transcripts were found in both PA data sets. No transcripts from one DE list were found on the opposite PA list. We clustered the 139 DE overlap transcripts using the same methods as for the ACC DE 449 transcripts. The majority of these transcripts responded in the same direction and with comparable magnitudes in both treatments (Fig. 4B). A minority responded in opposing ways to the two treatments: nine (6.5%) showed increased abundance in response to ACC but decreased abundance in response to IAA, and 14 (10.1%) showed the opposite response.
We clustered the 139 DE overlap transcripts using the methods described previously and visualized these clusters in network diagrams. Figure 4C clearly shows two large clusters with transcripts that move in the same direction in both treatments (cluster 1 with decreasing abundance and cluster 2 with increasing abundance) and several smaller clusters where transcripts changed in opposite directions between the two treatments. These transcripts may be of particular interest in relation to lateral root development, which is oppositely regulated by ethylene and auxin. This subset of oppositely regulated transcripts was too small for GO analysis; however, we did observe that the DE overlap as a whole was highly enriched in cell wall-related genes. The patterns in the PA overlap are not as distinct, but it appears that, for many of the transcripts, their response to IAA was less different from the controls than their response to ACC. That is, where transcripts were present in the control, they tended to be absent in ACC and present in IAA, and vice versa.
To better understand the interplay between ethylene and auxin transcriptional changes, we also performed a comparison of the 449 DE transcripts with a previous transcriptome study of ethylene and auxin treatment in roots of dark-grown seedlings at a single time point of treatment (Stepanova et al., 2007). Those authors examined transcriptional changes in response to ethylene or auxin treatment in the wild type and in mutants with altered responses to these hormones (ein2-5 and aux1-7). Their analysis found 511 transcripts regulated by ethylene, 899 regulated by IAA, and 191 regulated by both hormones. Our data set of 449 included 80 transcripts that they found to be ethylene regulated, with 24 of these also auxin regulated, so 7% of their ethylene-regulated transcripts were in our data set and 26% of the transcripts from our set were in their data set. This comparison suggests that there are large differences in transcripts regulated by auxin and ethylene in dark-grown roots compared to roots of light-grown seedlings, consistent with insufficient comparisons between ethylene responses as a function of light conditions.
Comparison with Ethylene-Responsive Transcripts from Dark-Grown Seedlings
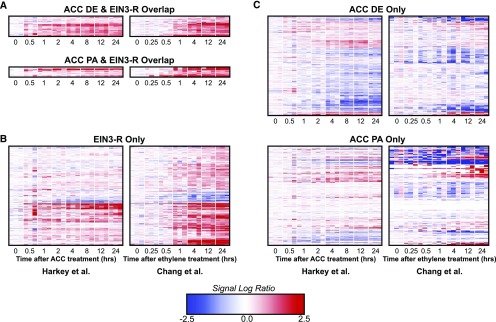
We were interested in further comparison of our data set with other ethylene response transcriptomics, to examine the effect of light on ethylene transcriptional response. In a previously published study, Chang et al. (2013) examined transcriptional responses to ethylene over a time course using RNA sequencing (RNA-seq). Their experimental methods differed from ours in that they used ethylene gas and whole, dark-grown seedlings. They also focused on transcripts for genes that were bound by EIN3 in a separate chromatin immunoprecipitation sequencing (ChIP-seq) experiment. They separated transcripts into two primary groups: EIN3-R transcripts, which are bound by EIN3 and respond to ethylene treatment, and EIN3-NR transcripts, which are bound by EIN3 but do not respond to ethylene treatment. They also had a third group, EIN3-ND transcripts, which are bound by EIN3 but were not detected in the RNA-seq experiment.
We compared our DE and PA transcripts with these three lists of genes and identified some overlap. Out of 375 EIN3-R transcripts, 37 also were found in our DE list (8.2% of DE transcripts and 5.5% of EIN3-R transcripts) and 12 were found in our PA list (3.1% of PA transcripts and 3.2% of EIN3-R transcripts); out of 886 EIN3-NR transcripts, 32 also were found in our DE list (7.1% of DE transcripts and 3.6% of EIN3-NR transcripts) and 16 were found in our PA list (4.2% of PA transcripts and 1.8% of EIN3-NR transcripts). No DE transcripts and four PA transcripts were found in the EIN3-ND list. A full list of all transcripts in these overlapping and not overlapping sets is available in Supplemental File S4.
Transcripts that were found in both the EIN3-R list and our DE list appear to have similar patterns of abundance change, suggesting that they are regulated in roots as in other tissues (Fig. 5A). We consider this group of genes to be tissue- and condition-independent ethylene transcriptional targets. Of transcripts from the EIN3-R list that were not DE in our data set (Fig. 5B), a subset appears to have similar patterns of increasing abundance in our microarray but were filtered out during our DE analysis due to a magnitude change below our threshold. However, the majority of transcripts did not have the same level of response in our microarray as in the Chang et al. (2013) set, suggesting that these may be genes that are regulated in other tissues but not in roots.
Figure 5.
Most ACC DE and PA transcripts are not EIN3 targets. The EIN3-R label represents transcripts that were found to be EIN3 targets and ethylene regulated by Chang et al. (2013) in dark-grown whole seedlings. A, Transcripts that were identified as DE or PA in our data set and as EIN3-R by Chang et al. (2013) B, Transcripts that were found to be EIN3-R, but not DE or PA, in our data set. C, Transcripts that were found to be DE or PA in our data set but not EIN3-R in the Chang et al. (2013) data set.
Surprisingly, nearly as many transcripts were found in the EIN3-NR list (which are defined by Chang et al. [2013] as not regulated by ethylene due to a fold change of less than 1.5) and genes that we identify as DE (with a fold change of 1.4; Supplemental Fig. S9) as in the EIN3-R and DE overlap. We hypothesize that many of the EIN3-NR transcripts were not DE because of the diversity of tissues or etiolated growth. In contrast, these transcripts had more substantial changes in our root-specific study. It is also possible that many of our transcripts were not in this data set because they were not regulated by EIN3.
To further explore whether the limited overlap was due to differences in tissue used (whole seedlings versus roots), transcriptome profiling approach (RNA-seq versus microarray), or could be accounted for by the different methods of filtering, we performed our own filtering methods on the Chang et al. (2013) data using the reads per kilobase per million (RPKM) data the authors provided. Due to differences in the type of data, a few adjustments were made to the filtering steps. In lieu of the detection P value cutoff, we required that a gene have an RPKM ≥ 1 in every sample. There were not time-matched controls, so we calculated the SLR for every time point using time zero as the control. Otherwise, all filtering was performed identically. From this, we identified 971 transcripts that met our criteria for DE (Supplemental File S5). Of those 971, only 71 overlapped with our ACC DE transcripts (Supplemental Fig. S10). Of transcripts that were Chang et al. (2013) DE but not ACC DE, some were not detected in the microarray because there was no probe for them present, some appeared to have a less distinct or consistent response in the ACC data set, but many have no observable response in the ACC data set, suggesting that these responses occur only in shoot tissues. A similar pattern holds in reverse for transcripts that were ACC DE but not Chang et al. (2013) DE. We looked at the 71 common transcripts identified in both data sets with our filtering methods. This method found some of the same transcripts as the Chang et al. (2013) filters, with 16 EIN3-R targets and four EIN3-NR transcripts, respectively. The limited overlap between these two sets of transcripts demonstrates the differences between our respective analyses. However, the 16 transcripts that were found to be ethylene regulated in our data set and by Chang et al. (2013) in both their original analysis and via our DE pipeline may be of particular interest as gold standard EIN3-dependent, ethylene-responsive transcripts. The proteins these transcripts encode include the ethylene receptors ERS1 and ERS2 and other members of the ethylene signaling pathway, CTR1 and RTE1, as well as two transcription factors.
Another important finding of these comparisons is that most of our DE transcripts (84.6%) and most of our PA transcripts (91.2%) do not show up in either the original EIN3-R or EIN3-NR lists (Fig. 5C), meaning that they were not detected by the EIN3 ChIP-seq experiment. This suggests that these transcripts may not be direct targets of EIN3 but may be targets of other EIL-family transcription factors, downstream targets of EIN3/EIL regulation, or regulated by a different set of transcription factors than those detected in dark-grown seedlings. As EIN3 mediates direct, primary responses to ethylene, we expect some of the more slowly regulated transcripts in our DE data set to be regulated by transcription factors that are downstream of EIN3. To ask if EIN3 is enriched in specific clusters, especially the rapidly induced clusters, we looked for EIN3-binding sites in our 10 largest DE clusters.
EIN3 Is Not Responsible for All Transcriptional or Developmental Responses to ACC in Roots
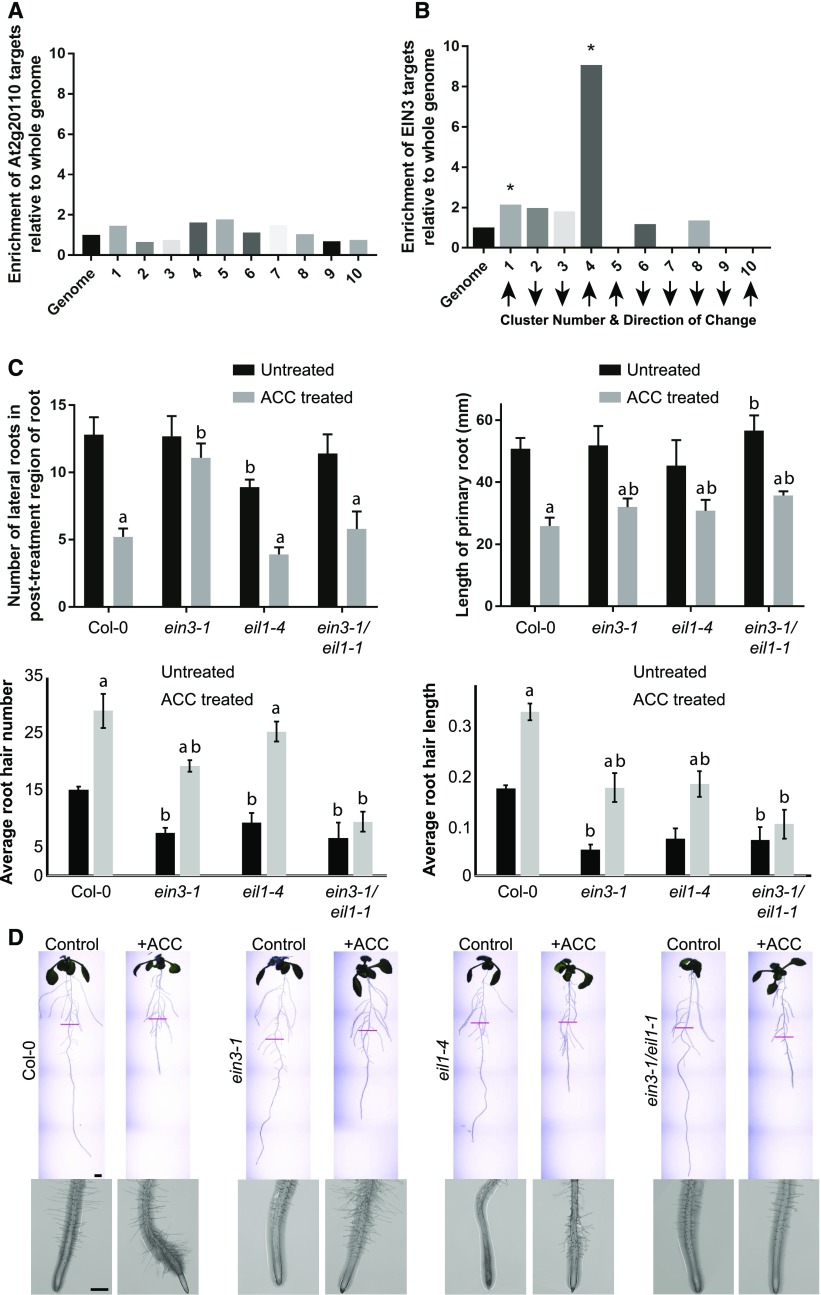
To identify EIN3 targets in the DE 449 transcripts, we used a publicly available data set from O’Malley et al. (2016). They identified transcription factor targets through DAP-seq, a technique similar to ChIP-seq, which uses affinity-tagged transcription factors to pull down their associated DNA, rather than transcription factor antibodies. Out of 1,118 genes identified as EIN3 targets by this method, 57 were DE in our data set. This represents a statistically significant (P < 10−4) enrichment of EIN3 targets in our data set (7% of DE transcripts versus 4% of the genome), which is logical given EIN3’s role in ethylene signaling. We asked whether these transcripts from genes bound by EIN3 and At2g20110, a transcription factor of unknown function, were equally distributed within the DE clusters. The binding sites for At2g20110 were not enriched in any clusters. In contrast, for EIN3, several clusters had no EIN3 targets or had a percentage of EIN3 targets not statistically higher than the genome (Fig. 6, A and B). Only two clusters, 1 and 4, were identified as significantly enriched in EIN3 targets. Both of these clusters are early up, meaning that they have transcripts that increase in abundance at early time points. These clusters also contain several ethylene signaling genes that are targeted by EIN3, including ERS1 and ERS2, ETR2, and EBF2. This suggests that EIN3 primarily plays a role in regulating early transcripts, such as those in clusters 1 and 4, whose transcript abundance reached an SLR > 0.5 at 4 h and 30 min, respectively, and that other transcription factors are responsible for later transcriptional changes.
Figure 6.
EIN3 is not responsible for all transcriptional or developmental responses to ACC. A and B, Enrichment of transcription factor target genes (O’Malley et al., 2016), with the bars representing fold change of the genome background in the top 10 ACC DE clusters. A, Relative abundance of binding sites of a transcription factor for which no significant enrichment was found. B, The relative abundance of EIN3 binding sites is enhanced in specific clusters, with asterisks indicating significant enrichments at P < 0.05 using a binomial test. C, The number of lateral roots was quantified in the presence and absence of ACC in Col-0, ein3, eil1, and the ein3 eil1 double mutant. D, Primary root elongation was measured in the presence and absence of ACC in Col-0, ein3-1, eil1-1, and the ein3-1 ein1-1 double mutant. E and F, The number and length of root hairs in the presence and absence of ACC in Col-0, ein3-1, eil1-1, and the ein3-1 ein1-1 double mutant. aSignificant difference between control versus ACC treatment within genotypes; bsignificant difference between the indicated genotype and Col-0 with the same treatments, at P < 0.05. G, Representative whole-root and root hair images of Col-0, ein3, eil1, and ein3 eil1. Magenta lines represent the length of the primary root at the time of transfer to control or ACC treatment plates. Bars = 2 mm in whole-root images and 0.5 mm in root hair images.
We asked whether EIN3’s role in early transcriptional changes was responsible for the phenotype seen in ACC-treated roots. The root developmental phenotype was examined in ein3 and eil1 single and double mutants. These single mutants retained some ethylene sensitivity, as evidenced by decreased lateral root numbers and primary root length after ACC treatment (Fig. 6, C, D, and G). In comparison, the ein3 eil1 double mutant had few root hairs and a nonsignificant ACC response (Fig. 6, E–G). These responses differ from those of the ein2 mutant, which shows no significant changes in root length, lateral root number, and root hair formation (see Figs. 9 and 10 below). These results differ from the total loss of long-term ethylene responses in dark-grown ein3 eil1 seedlings (Alonso et al., 2003; Binder et al., 2004a) and suggest that other transcriptional machinery is needed to explain the residual root elongation and lateral root responses in this double mutant.
Figure 9.
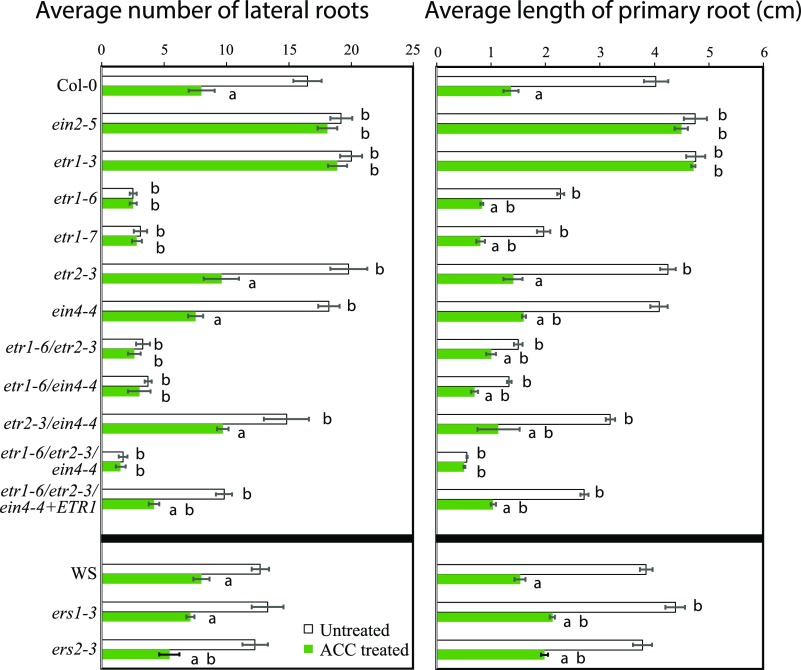
ACC inhibits lateral root initiation and primary root elongation through different ethylene receptors. Five-day-old seedlings were transferred to medium containing 1 µm ACC, and the lateral root number and primary root length were quantified after another 5 d. Averages and se of 30 seedlings are reported. The significance of ACC treatment within a genotype and between Col-0 and the indicated genotypes was assessed by ANOVA and Tukey’s posthoc test. aSignificant difference between control and treatment with ACC within a genotype, at P < 0.05; bsignificant difference between parental lines and mutant genotypes.
Figure 10.
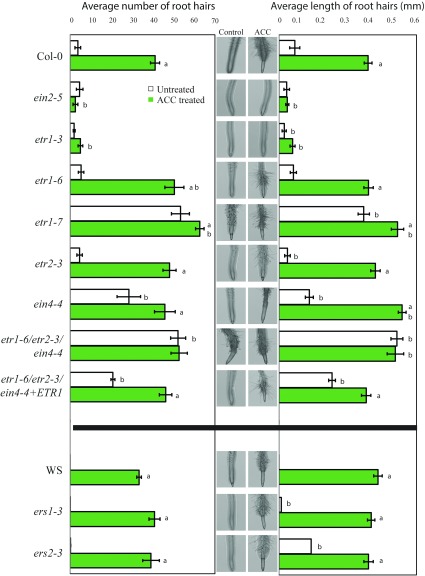
ETR1 and EIN4 both function in ACC-induced root hair initiation and elongation. ACC induces root hair formation and elongation through specific ethylene receptors. Four-day-old seedlings were treated with 1 µm ACC for 24 h. The number of root hairs is shown on the left and the length of root hairs shown on the right, with representative images for each genotype and treatment. Averages and se are reported for 12 to 18 seedlings. aSignificant difference between control and treatment with ACC within a genotype, at P < 0.05; bsignificant difference between parental lines and mutant genotypes.
ERFs Bind to Regulatory Regions of Genes in Some, But Not All, Clusters
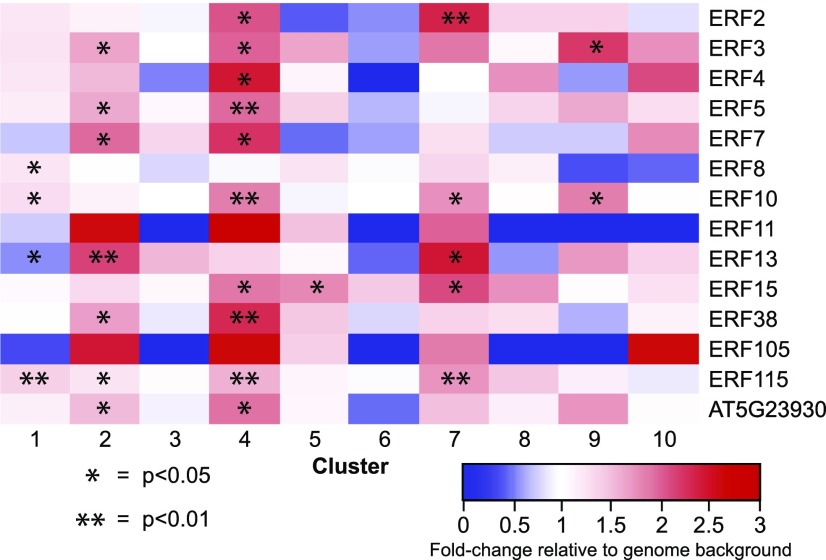
ERFs are transcription factors that are downstream of EIN3/EIL1 and are important for ethylene transcriptional responses (Licausi et al., 2013). We performed a DAP-seq enrichment analysis for the 14 ERFs for which DAP-seq data were available (Fig. 7). Like EIN3, most of these ERFs showed enrichment of targets in cluster 4 and, to a lesser extent, in cluster 1. Clusters 2, 5, 7, and 9 also show ERF enrichment to varying degrees. Some clusters showed no significant enrichment, and for some ERFs and clusters, there was an underrepresentation of ERF targets compared with the genome background. This is most striking in cluster 6. Like EIN3, this suggest that these known ERFs control a subset of the ACC transcriptional responses.
Figure 7.
Binding sites for ERFs are enriched in a subset of clusters. Enrichment of ERF target genes (O’Malley et al., 2016) in the top 10 ACC DE clusters reported relative to the whole genome. Significant differences were determined using a binomial test, with asterisks indicating P < 0.05 (*) and P < 0.01 (**).
Ethylene Receptors Show Distinct Transcript Response Kinetics
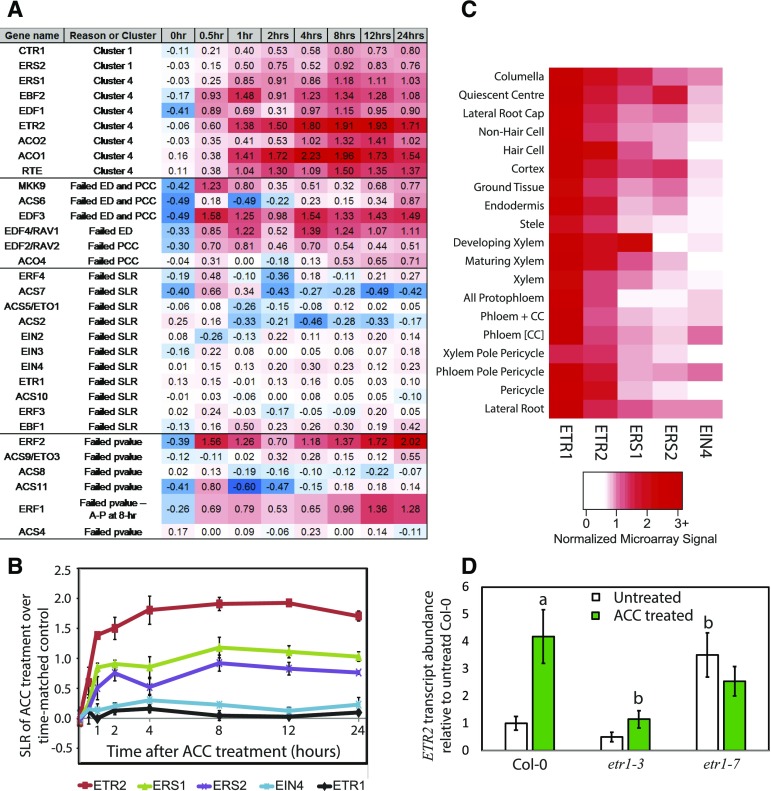
We also used a directed approach to follow the levels of transcripts of genes known to be ethylene regulated or that function in ethylene signaling or synthesis (Chen et al., 2007; Konishi and Yanagisawa, 2008). We queried the unfiltered data set of transcripts normalized to time-matched controls to examine the behavior of transcripts encoding ethylene synthesis and early ethylene signaling proteins, and the SLR of these genes and how they fared in our filtering analysis are indicated in Figure 8A. Ethylene synthesis genes, ACO1 and ACO2, were DE, while ACO4/EFE had an SLR > 0.5 but failed the Pearson’s correlation coefficient (PCC) filter for consistency between replicates. Several ACC synthase genes had apparent changes (SLR > 0.5 or < −0.5) but failed at various filtering steps due to inconsistent changes or low abundance at single time points.
Figure 8.
Ethylene controls many ethylene synthesis, signaling, and response genes, including expression of the ethylene receptor ETR2 in an ETR1-dependent manner. A, For each ethylene-related transcript queried from our data set, the results represent the DE cluster where it is found or where it failed in the filtering process. SLRs represent averages of three biological replicates. Red, white, and blue designate positive, zero-value, and negative SLRs, respectively (see color scale). B, The SLR patterns for five ethylene receptor transcripts show different kinetic responses to ACC treatment. SLRs are reported as averages with se as a function of time after ACC treatment from the microarray data set. C, The cell type-specific expression pattern of each of the receptors illustrated with relative levels of transcripts using a percentage scale for the normalized signal values from Brady et al. (2007). D, Transcript abundance in receptor mutants as determined by qRT-PCR. Nine-day-old seedlings were transferred to agar medium with and without ACC at 1 µm for 24 h before RNA extraction and quantification by qRT-PCR. Averages of three biological replicates are reported. The significance of ACC treatment within a genotype and between Col-0 and the indicated genotypes was assessed by ANOVA and Tukey’s posthoc test, and significant differences are indicated. aSignificant difference between treatments, at P < 0.0003; bsignificant difference between genotypes within treatments, at P < 0.003.
The ethylene receptor isoforms have both redundant and nonredundant functions (Shakeel et al., 2013). Therefore, we were curious to know whether the application of ACC differentially affected the time course of receptor transcript accumulation in roots of light-grown plants. The levels of transcripts encoding the five ethylene receptors are plotted as a function of time after ACC treatment in Figure 8B. All five receptor transcripts are expressed in roots, with detection above the Affymetrix P value thresholds. ETR2, ERS1, and ERS2 transcripts, encoding three of the receptors, were differentially expressed, with ETR2 having the most profound change in abundance, while ETR1 and EIN4 both failed at the SLR cutoff (Fig. 8, A and B), consistent with prior reports in different tissues (Hua et al., 1998). The accumulation of ETR2 transcript is faster than has been observed previously for whole, dark-grown Arabidopsis seedlings (Binder et al., 2004b). These results indicate that the different receptor isoforms may have differing roles in the control of root growth and development. In support of this possibility, we examined the distribution of these receptor isoforms across the range of root tissue types using a previously published data set (Brady et al., 2007; Fig. 8C). The absolute levels of transcripts encoding ERS1 and ETR1 receptors are higher across these tissues but with variation between cell types evident, suggesting that these receptors may function to control distinct aspects of root development.
ETR1 Controls ACC-Modulated ETR2 Transcript Abundance
To examine the roles of ethylene receptors in controlling the expression of other ethylene receptors, we performed quantitative real-time (qRT)-PCR on wild-type and receptor mutant roots after treatment on control or ACC medium (Fig. 8D). The effect of ACC treatment on ETR2 transcript abundance was examined in Columbia-0 (Col-0), etr1-3 (a gain-of-function mutant), and etr1-7 (a loss-of-function mutant). A two-way ANOVA found significant differences between control and ACC treatment. The significance of specific comparisons was assessed using a Tukey’s posthoc test. In Col-0, ETR2 transcript abundance showed a 4-fold increase after ACC treatment, consistent with the microarray results. In the etr1-3 gain-of-function mutant, the ETR2 transcripts are not significantly different from Col-0 and do not change with ACC treatment. In the etr1-7 mutant, which is a loss-of-function mutant, ETR2 transcript abundance is elevated in control relative to Col-0 to levels not significantly different from ACC-treated Col-0 or etr1-7. These opposite responses in etr1-3 and etr1-7 are consistent with the absent and constitutive signaling in these two mutant alleles, respectively. These results suggest a pathway where ETR1 is responsible for ethylene-mediated increases in ETR2 transcripts. The role of ETR1 in controlling the synthesis of ERS1 and ERS2 transcripts also was examined in the etr1-3 mutant. No significant change in either transcript was detected in the presence or absence of ACC (Supplemental Fig. S11).
Specific Ethylene Receptors Modulate Lateral Root Initiation, Primary Root Elongation, and Root Hair Initiation and Elongation
The strategy of using ethylene receptor nulls to identify the receptors that function in distinct tissue or developmental processes has been productive, but it has not been used to examine root development (Shakeel et al., 2013; Gallie, 2015). To define the ethylene receptor isoforms that control root growth and development, we examined lateral root development, primary root elongation, and the initiation and elongation of root hairs in plants with gain-of-function and loss-of-function receptor mutations in the absence and presence of ACC. The number of lateral roots formed 5 d after treatment is shown in Figure 9. A two-way ANOVA found significant differences between control and ACC treatment. The dominant negative receptor mutant, etr1-3, as well as the ethylene-insensitive mutant, ein2-5, have increased lateral root numbers as compared with the wild type, with statistical significance examined with a Tukey’s post hoc test (Supplemental Table S2), which is consistent with previous reports (Ivanchenko et al., 2008; Negi et al., 2008; Muday et al., 2012). Lateral root formation is reduced ∼2-fold by ACC treatment in regions of roots formed after transfer to ACC in Col-0. In etr1-3 and ein2-5 mutants, the magnitude of the effect of ACC is reduced substantially, with only a 6% change.
We also examined lateral root development in receptor null or loss-of-function mutants (Fig. 9). The etr1-6 and etr1-7 null mutants formed statistically significantly fewer numbers of lateral roots relative to Col-0, forming only 15% and 19% of the number of lateral roots as Col-0, respectively. Both loss-of-function mutants had similar numbers of lateral roots in the presence and absence of ACC treatment, consistent with constitutive ethylene signaling due to loss of receptor function. This is in contrast with the phenotypes of the etr2-3, ein4-4, ers1-3, ers2-3, and etr2-3 ein4-4 mutants, which showed reduced lateral root abundance after ACC treatment. The ers1-3 and ers2-3 receptor null mutants in the Wassilewskija (Ws) background showed subtle differences in lateral root number relative to Ws in both the presence and absence of ACC.
We also used plants with mutations in multiple receptors and a complemented line to assess the role of these receptors. The etr1-6 etr2-3 mutant was not significantly different from etr1-6 in the absence or presence of ACC, while the slight difference between etr1-6 and etr1-6 ein4-4 was significantly different in the absence, but not in the presence, of ACC. The etr1-6 etr2-3 ein4-4 triple mutant exhibited a significant reduction in lateral root number relative to Col-0 in the control condition, with no change in number with ACC treatment. When etr1-6 etr2-3 ein4-4 was complemented with ETR1, there was a significant increase in lateral roots over the triple mutant, although the complementation was not complete, as the levels were still significantly different from those of Col-0. These data indicate that ETR1 has the major role, with a minor role for EIN4, in mediating lateral root formation upon treatment with ACC in Col-0, similar to the major role that ETR1 has in mediating ethylene-stimulated hypocotyl nutations (Binder et al., 2006) and the inhibition of ethylene responses by silver (McDaniel and Binder, 2012).
To determine the inhibitory effects of ACC on primary root elongation in the ethylene receptor mutants, we measured the length of the primary root before and after 5 d of 1 µm ACC treatment (Fig. 9). A two-way ANOVA found significant differences between control and ACC treatments (Supplemental Table S3). We observed greater root elongation in etr1-3 and ein2-5 than in Col-0 under control growth conditions or ACC treatment, which is consistent with previously published reports (Ivanchenko et al., 2008; Negi et al., 2008; Péret et al., 2009; Muday et al., 2012). Although untreated etr1-6 and etr1-7 had reduced root elongation to 66% and 47% of the primary root length of Col-0, respectively, root elongation was still sensitive to ACC, showing significant reduction upon ACC treatment relative to the untreated roots in both mutant genotypes, consistent with prior studies on roots of dark-grown seedlings (Hua and Meyerowitz, 1998; Cancel and Larsen, 2002). Mutants in other receptors had primary root lengths that were not significantly different from Col-0. The etr1-6 etr2-3 and etr1-6 ein4-4 double mutants show shorter primary roots than etr1-6 in the absence or presence of ACC, and the magnitude of response to ACC was reduced. The triple mutant has the shortest primary root, which was reduced compared with all the other genotypes and showed no effect of ACC treatment; however, the ETR1 complemented line is longer than the triple mutant and has a similar magnitude response to ACC as Col-0. Together, these data demonstrate a major role of ETR1 in modulating primary root elongation via ACC and suggest minor roles of ETR2 and EIN4 in this process.
Ethylene treatment also increases the number and length of root hairs (Tanimoto et al., 1995; Rahman et al., 2002), which are single-cell projections of the epidermis that increase the surface area of the root and aid in water and nutrient uptake (Cutter, 1978). We treated 4-d-old seedlings with 1 µm ACC for 24 h and quantified the number and length of root hairs (Fig. 10). A two-way ANOVA found significant differences in the number and length of root hairs between control and ACC treatments (Supplemental Tables S4 and S5). We observed a greater than 5-fold increase in the number and length of root hairs in Col-0 after ACC treatment (Fig. 10). Both ein2-5 and etr1-3 formed fewer, shorter root hairs in the presence of ACC than Col-0, and we observed no increase in the number or average length of root hairs in response to ACC in ein2-5, although etr1-3 showed a 2-fold increase in the number of root hairs after ACC treatment and these root hairs were shorter than those in Col-0. It has been observed that etr1-3 is not entirely ethylene insensitive with regard to the growth inhibition response in roots (Hall et al., 1999), and it appears that this incomplete insensitivity applies to root hair initiation as well. Interestingly, etr1-6 and etr1-7 have different responses to ACC treatment. In etr1-6, there is a higher induction of both root hair number and length by ACC, as compared with Col-0, while etr1-7 has a higher than wild-type number of root hairs under control conditions, with a smaller, additional induction by ACC than Col-0. This is consistent with descriptions of these two mutants, where etr1-7 generally has a more striking phenotype (Wilson et al., 2014b). The locations of the mutations are likely responsible for the different phenotypes of etr1-6 and etr1-7. The mutation occurs earlier in etr1-7, resulting in a shorter amino acid sequence in the translated protein compared with etr1-6 (Hua and Meyerowitz, 1998). This difference is important, as the N-terminal domain of ETR1 is known to have signaling properties through interactions with RTE1 (Gamble et al., 2002; Qu and Schaller, 2004; Xie et al., 2006; Qiu et al., 2012). Root hair numbers in etr2-3 in the presence of ACC are slightly and significantly greater than in Col-0, whereas ein4-4 has ∼6-fold more and ∼2-fold longer root hairs under control conditions, with significant differences from Col-0 detected in the absence and presence of ACC treatment. The etr1-6 etr2-3 ein4-4 triple mutant has the greatest number and length of root hairs under control conditions, with numbers greater than the single and double combinations of these mutations. When this line is transformed with ETR1, the number and length of lateral roots are decreased relative to the triple mutant, with a greater than 20-fold difference, but remain statistically different from Col-0. The magnitude of the responses suggests that ETR1 and EIN4 both play a role in modulating root hair initiation and elongation in response to ACC in Arabidopsis roots.
DISCUSSION
Examining transcriptional responses at a single time point during development or in response to enhanced hormone signaling limits our view of the progression of events. The challenge of large transcriptomic data sets is moving beyond long lists of genes to identify patterns and relationships that predict networks of genes that function together. Cascades of precisely ordered and sequentially activated transcription factors are the basis of many complex developmental processes (Davidson, 2010; Kurotaki et al., 2013). In recent years, transcriptomic data sets measured over a time course have provided substantial additional insight into understanding the gene regulatory networks that control development (Jaeger et al., 2012; Nagano et al., 2012). These data sets allow the observation of sequential series of transcriptional events that can be analyzed with sophisticated computational approaches to provide new insight into the networks of coordinated transcript changes. These approaches have uncovered the elaborately branched networks of regulons induced by a stimulus and coordinated by individual transcription factors (Yosef and Regev, 2011; Jaeger et al., 2012). We completed a time-course microarray of roots treated to elevate levels of the gaseous plant hormone ethylene that spanned the time window of ethylene-dependent changes in root growth and development, which revealed transcriptional regulators and signaling proteins not linked previously to ethylene signaling as well as conserved transcriptional responses that span tissue type and developmental context.
We identified changes in transcript abundance upon treatment of roots with ACC, an ethylene precursor, across eight time points that spanned 24 h. We quantified the induction of root hair formation across this same time course, demonstrating that the earliest transcript abundance changes precede this developmental response while the later time points are coincident with the enhanced formation and elongation of root hairs. The ability of ACC to inhibit elongation and block lateral root progression across this time course was demonstrated previously (Lewis et al., 2013). The root developmental changes in response to ACC treatment are the result of elevated ethylene (rather than direct effects of ACC), as the developmental responses are all blocked in the ethylene signaling mutants etr1-3 and ein2-5 (Figs. 9 and 10; Negi et al., 2008). Therefore, we can directly overlay root developmental changes on this root-specific ACC time course to look for temporal linkage between transcriptional and developmental responses.
The changes in transcript abundance across this data set were filtered in two independent ways (Lewis et al., 2013). The first approach applied a rigorous filtering to identify a DE data set. The 449 transcripts in this data set were above the Affymetrix detection threshold at every time point, were induced or repressed by more than 1.4-fold (SLR > 0.5), and had consistent patterns and magnitude of change in abundance in all three replicates, as judged by Pearson correlation and Euclidean distance (ED). For each gene, the SLR of transcript abundance in these data sets was clustered using k-means into 24 clusters with an empirically derived 83% consistency (Fig. 2). The distinct kinetics and magnitude of responses are best visualized by heat maps of whole clusters (Fig. 2A) or individual genes within select clusters (Supplemental Fig. S2). The consistency of behavior of transcript abundance changes within these clusters is evident in the cluster maps, in which the relative abundance of each transcript in each cluster is shown separately at each time point (Fig. 2B; Supplemental Fig. S3). A line connecting two nodes indicates that they met the 83% threshold; transcripts that are tightly connected in this way typically show a higher degree of correlation than those that are more spread out. For example, in cluster 7, all transcripts are highly correlated, while in cluster 3, one group of transcripts are tightly correlated while half the cluster shows less similarity. The clustering was initially performed with a range of consistency requirements, and when higher similarity of response for all transcripts was required, it resulted in clusters that looked like cluster 7, but there were many more singletons. A primary goal of clustering is to look for groups of genes with conserved function, and we chose to focus our analysis on clusters with slightly more variation in response (requiring 83% consistency) but of larger size to find statistically conserved functional annotations, as identified in Table I. We also can find clusters of transcripts that tightly overlay on the root hair kinetics in Figure 1, with transcripts increasing and decreasing in DE clusters 4 and 7, respectively, preceding root hair stimulation and with transcripts increasing and decreasing in clusters 1 and 6, respectively, at times coinciding with root hair developmental changes. These two groups of clusters may identify transcripts that drive development and execute the developmental changes, respectively.
We used a second filtering approach to identify some transcripts excluded by the DE filtering but that had robust responses to ACC treatment. A challenge with our initial filtering is that we eliminated transcripts that were not different from the background at any time points according to the Affymetrix detection P values, which eliminated transcripts that were not expressed in control but were induced by ACC treatment or whose abundance decreased to below background levels after treatment. This second filtering identified transcripts that were present (above background) at some time points while being absent (below background) at other time points, which generated a data set that we call PA. We could not calculate SLR for this data set, since it is mathematically inappropriate to use these zero values in fold change calculations, so we report the data directly for control and treatment samples. The PA data set also was filtered and clustered, heat maps were reported (Fig. 3; Supplemental Fig. S5), and functional annotations were obtained. The PA and DE data sets show similar patterns of transcript changes and have some overlapping functions, such as clusters with increased transcript abundance annotated with ethylene signaling or with decreased abundance annotated with cell wall function.
One surprising finding in these data is that ACC treatment leads to more transcripts exhibiting reduced abundance than increased abundance. This is evident both in the histogram showing the number of transcripts as a function of SLR at each time point (Fig. 1) and in the DE and PA heat maps (Figs. 2 and 3; Supplemental Fig. S5). The negative SLR values in the DE data set could result from either decreased abundance in the treatment or increased abundance of transcripts in the untreated time-matched control data set. We find that most transcripts are relatively constant in the control data set, while the same transcripts decrease in abundance in response to ACC treatment (Supplemental Fig. S4). This differs from prior ethylene genome-wide data sets performed with dark-grown seedlings (Chang et al., 2013) or dark-grown roots (Stepanova et al., 2007), suggesting that ethylene may control root development in light-grown plants through very different machinery.
We also performed comparisons of this data set with a time-course data set (Chang et al., 2013). In that data set, transcripts whose abundance changed with ethylene treatment and that were regulated by the canonical ethylene transcriptional regulator EIN3 (Chao et al., 1997) were identified. We also performed a secondary DE analysis using the filtering approaches applied to our microarray data to examine the Chang et al. (2013) RNA data set using their RPKM values. This analysis identified 971 DE transcripts, which is approximately 40% of the total identified by the Chang et al. (2013) filtering. We identified a small number of transcripts in both lists that were also in our DE or PA data sets. There were a large number of their EIN3-regulated transcripts that showed no clear response in our data set (top half of Fig. 5B), that had more limited magnitude change, or that were inconsistent in pattern or magnitude change in our data set (Fig. 5B, middle and bottom). Similarly, we identified transcripts that changed robustly in our DE and PA data sets but whose abundance changes were weak or inconsistent in response to ethylene in their data set. Yet, in both of these two data sets, transcript changes span multiple time points, with sequential changes clearly indicating robust responses. This comparison leads us to conclude that there are very different sets of transcripts that change in these two data sets, consistent with the differences in growth conditions (light versus dark), tissue type (roots versus whole seedlings), and perhaps method for elevating ethylene (ethylene precursor versus ethylene gas). Developmental responses to ACC are lost in the ethylene-insensitive etr1-3 dominant negative and ein2-5 mutants (Figs. 9 and 10); therefore, it seems unlikely that a difference between ACC and ethylene treatment would explain most of these differences. When we filtered the Chang et al. (2013) data set using our statistical cutoffs, we again found limited overlap, suggesting that the difference is primarily in experimental methods (light versus dark grown and tissue specificity). To resolve how much variation is accounted for by tissue type, we compared with a second data set of dark-grown roots (Stepanova et al., 2005), which also yields limited overlap, consistent with light-dependent developmental differences. Within these three data sets, 25 transcripts were found in their respective ethylene-responsive groups, further supporting the idea of a small subset of genes that are important for ethylene response across multiple tissues, environmental conditions, and responses but with the majority of transcriptional changes happening in a context-dependent manner.Consistent with this conclusion, Figure 8A illustrates the abundance change of a variety of ethylene signaling and response transcripts in our data set, many of which are conserved across these three data sets (Table I).
To further explore the role of the canonical ethylene transcriptional machinery in this response, we asked whether EIN3, a critical upstream transcriptional regulator (Chao et al., 1997), played central roles in these root responses to ACC. We used a recently published DAP-seq data set in which the targets of EIN3 and a diversity of other transcription factors were identified across the Arabidopsis genome (O’Malley et al., 2016). We find that the EIN3 targets are found primarily in two DE clusters (1 and 4) that both show transcript abundance increases (Fig. 6). Rapidly induced cluster 4, annotated as ethylene responsive, is strongly enriched in EIN3-bound genes relative to the whole genome. These data are consistent with the model in which EIN3 binds to these genes (which include seven predicted transcription factors), which, in turn, control the transcription of other genes, leading to a network that can only be detected by high-resolution time-course transcriptomics. A similar pattern was found for ERFs, which also bind to targets in clusters 1 and 4 (Fig. 7). Additional enrichment was found in clusters 2 and 7, which contain seven and five ERF-binding sites, respectively, and in clusters 5 and 9, which are enriched in the binding of two ERFs. Together, these results reveal a subset of the ACC response clusters that are regulated by canonical ethylene transcriptional machinery, but a number of these other clusters may contain previously uncharacterized light-grown, root-specific transcripts whose regulation occurs via distinct mechanisms.
These data also suggest EIN3-independent pathways. We also tested the function of EIN3 in controlling root response as well as the EIL1 transcription factor using single and double mutants. In contrast to ein3 eil1 mutant seedlings grown in the dark, which show no response to prolonged ethylene treatment (Alonso et al., 2003), the ein3 eil1 double mutant shows inhibition of root growth and lateral root formation in response to treatment with ACC. In contrast, increased root hair initiation was not significant in the double mutant. This is consistent with a previous study that demonstrated a loss of the jasmonic acid induction of root hairs in the double mutant (Zhu et al., 2011). Those authors also showed no root hairs in the double mutant, but this may be explained by variations in growth conditions, such as sucrose supplementation in media and the age of seedlings. Together, these data indicate that both EIN3-dependent and -independent signaling controls specific aspects of root development.
Since the role of EIN3 is very different in roots as opposed to other tissue types, we also examined the regulation of and functional role of the five ethylene receptors to explore whether they too have different roles in roots of light-grown seedlings. Transcripts encoding the five ethylene receptors ETR1, ETR2, ERS1, ERS2, and EIN4 have different responses to ACC, with either no response or responses that differ in kinetics or magnitude of response (Fig. 8, A and B). Consistent with prior reports, ETR2, ERS1, and ERS2 are increased by treatments that elevated ethylene (Hua et al., 1998), but the more rapid and larger responses of ETR2 suggested the testable hypothesis that it had the most profound effect of ACC on root architecture. The dominant negative (or gain-of-function) ETR1 mutant (etr1-3) has been shown previously to be insensitive to the effects of ethylene on root elongation, lateral root development, and root hair initiation (Negi et al., 2008; Lewis et al., 2011b). We used null mutants in each of the five receptors and identified a strong role for ETR1 in controlling the root responses to ACC, but more subtle changes in development in null mutants in any of the other receptors, as summarized in Supplemental Table S6. We also examined multiple null mutants in two or three receptor genes, finding smaller and redundant roles for ETR2 and EIN4, especially in root hair formation. We used a triple mutant with profound developmental responses to find that these phenotypes are largely complemented with a genomic copy of ETR1. This is consistent with previously published data showing the importance of ETR1 in ethylene response in roots (Negi et al., 2008; Lewis et al., 2011b). Additionally, we examined ETR2 expression in etr1 mutant alleles and find that ETR2 levels are reduced in the gain-of-function allele and are elevated in the absence of ACC in the null allele, consistent with previous reports that ethylene induction of ETR2 protein levels is lost in etr1-1 (Chen et al., 2007). These results argue that the constitutively expressed ETR1 receptor has a predominant role in controlling root responses to ethylene, similar to its major role in controlling nutations and responses to silver ions (Shakeel et al., 2013).
To place this data set in the context of root development, we performed several other comparisons. First, we asked whether the ACC-regulated transcripts had cell type-specific expression patterns. We used previously published data sets that identify cell type-specific transcriptomes or transcriptomes that span the root tip developmental zone (Brady et al., 2007). We clustered these by common developmental response and asked whether any clusters were linked to specific cell or tissue expression patterns but did not find striking patterns, suggesting that ethylene response transcripts span developmental regions. This is consistent with limited tissue specificity due to the diffusion of the ethylene gas.
A second and more fruitful comparison was to look for overlaps with a root-specific IAA response data set, as the hormone IAA has different effects on root development than ethylene. Although auxin and ethylene both inhibit primary root elongation and stimulate root hair proliferation, these two hormones have opposite effects on lateral root initiation, with IAA stimulating and ACC and ethylene inhibiting this process. When we compare these two transcript abundance data sets, which were performed at the same time and with the same time-matched controls, we find 1,246 transcripts that respond to IAA, 449 that respond to ACC, and 139 in common. The majority of these show similar directional changes in response to both hormones with consistent increases (top of Fig. 4B) or decreases (bottom of Fig. 4B), while a small set shows opposite responses, consistent with transcripts that might differentially regulate lateral root inhibition. These differences are best seen in the cluster network maps shown in Figure 4C, which show opposite direction in the same subset of clusters. The transcripts in this overlap can be tested to ask about how they control root developmental responses to these two hormones.
The patterns we have revealed by comparison of our data set with other large-scale data sets, and by analysis of clusters with similar transcriptional changes for common gene functions or transcriptional regulators, demonstrate the effectiveness of a time-course approach to transcriptomics. When making comparisons with other data sets, it is especially important to have a high degree of confidence that the changes observed are real; our experimental design and filtering method mean that transcripts identified as ACC responsive were replicated across multiple time points and showed similar kinetics in three experimental replicates. Additionally, many of the transcripts we identified as ACC responsive in roots would have appeared unresponsive and uninteresting if we had chosen any single time point after treatment for analysis. Together, these two advantages of time-course transcriptomics helped us to identify responses to ethylene that were tissue and/or environment dependent along with responses shared across contexts. By looking for patterns in these transcript responses across time, we also were able to correlate changes with development and gene function, suggesting potential mechanisms for ethylene control of root development. Our comparison with the IAA data set led to a further narrowing of genes of interest specifically for ethylene and auxin cross talk in roots. We also were able to ask questions about receptor and transcription factor control of ethylene responses, discovering EIN3-independent as well as -dependent transcriptional changes in roots and specific roles for receptors in developmental changes. This data set provides an example of how time-course approaches can be useful within a hormone response and developmental context.
MATERIALS AND METHODS
Plant Growth and Arabidopsis Genotypes
Arabidopsis (Arabidopsis thaliana) Col-0 seeds were purchased from Lehle Seeds. The etr1-6, etr1-7, etr2-3, ein4-4, and ers2-3 mutants were obtained from Elliot Meyerowitz, and the ers1-3 mutant was obtained from Eric Schaller. All have been described previously (Hua and Meyerowitz, 1998; Qu et al., 2007). The etr1-6, etr1-7, etr2-3, and ein4-4 mutants are in the Col-0 background, and the ers1-3 and ers2-3 mutants are in the Ws background. The ein3-1 and eil1-4 single and ein3-1 eil1-1 double mutants are in the Col-0 background and were described previously (Binder et al., 2007). The combinatorial mutants and transformants have been described previously (Hua and Meyerowitz, 1998; Kim et al., 2011).
Plants were grown on 1× Murashige and Skoog medium (Caisson Laboratories), pH 5.6, Murashige and Skoog vitamins, and 0.8% agar, buffered with 0.05% MES (Sigma), and supplemented with 1% Suc. After stratification for 48 h at 4°C, plants were grown under 100 µmol m−2 s−1 continuous cool-white light. For phenotypic analyses, wild-type and mutant plants were grown on control medium for 5 d and then transferred to control and 1 µm ACC-containing media. Five days later, seedling images were captured with a scanner. Primary root growth was measured using Imaris 7.7.2 (Bitplane), and lateral root number was quantified manually.
ACC Treatments and RNA Isolation
RNA was isolated from seedlings grown on a nylon filter (03-100/32; Sefar Filtration) as described previously (Levesque et al., 2006). Plants were stratified and germinated subsequently on a filter pressed tightly against control medium with approximately 100 seedlings per plate. On day 5 after germination, the nylon was transferred to growth medium with and without 1 µm ACC for the given treatment time (0–24 h). After this time, roots were cut from seedlings and flash frozen in liquid nitrogen.
Frozen samples were ground in liquid nitrogen, and RNA isolation was performed according to the Qiagen plant RNeasy kit protocol, with the addition of the Qiagen RNase-free DNase treatment (Qiagen). After RNA isolation, samples were quantified by A260 using a Nanodrop spectrophotometer (Nanodrop Technologies). RNA concentrations were standardized to 150 ng µL−1 ± 10% by the addition of DEPC-treated 10 mm Tris-HCl, pH 8. Each sample yielded approximately 4.5 µg of RNA.
Microarray Analyses
RNA samples were sent to the Wake Forest University Comprehensive Cancer Center Microarray Shared Resource Center and were repurified on Qiagen RNeasy columns. The samples were analyzed on the Agilent Bioanalyzer and Eppendorf BioPhotometer for RNA integrity and concentration. Samples with RNA integrity values greater than 8 were carried forward for cDNA synthesis, labeling, and fragmentation. The samples were hybridized to the arrays and washed, and Affymetrix AGCC software was used to process the chips and perform image capturing. The resulting .CEL files were analyzed for quality assurance using internal Affymetrix parameters and custom signal distribution analyses developed in house. These CEL files have been posted to the Gene Expression Omnibus and can be found under accession number GSE84446.
Raw data were normalized by the microarray facility using systematic variation normalization as described previously (Chou et al., 2005), and the log2 of the signal intensity was reported along with the detection P value calculated by the Affymetrix software.
Transcript Filtering and DE Calculation
A four-step filtering pipeline (Supplemental Fig. S1), described previously (Lewis et al., 2013), was used to process the normalized microarray data to identify genes with significant and consistent DE over time. The first step, the detection P value filter, identified transcripts that were reliably detected on the chip over background within each replicate data set independently. A transcript was retained if the detection P values were 0.06 or less (the Affymetrix recommended cutoff) for all time points (Supplemental Fig. S1, step 1). This is outlined at http://media.affymetrix.com/support/downloads/manuals/data_analysis_fundamentals_manual.pdf (May 18, 2015). For each replicate data set, the relative expression was reported as the SLR, which is equivalent to the log2 fold change. The SLR was calculated as the time-matched log2 ratio of the signal intensity at each ACC treatment time (for each replicate individually) relative to the average intensity of the control replicates at each time point. As data were already log2 transformed, this ratio calculation was simply a subtraction of the control value from the experimental value. Control replicates were averaged for each time point because control and experimental data sets were not paired; therefore, averaging the control provided a consistent baseline for replicate comparisons. These control values also were used for an IAA study that was performed simultaneously and was published previously (Lewis et al., 2013). After SLR values were calculated, the second step used them to identify transcripts that had a transcriptional response to ACC at some point during the time course (SLR filter; Supplemental Fig. S1, step 2). Any transcript with an SLR ≤ −0.5 or ≥ 0.5 (roughly a 1.4-fold change) for at least one time point was retained. The overlap filter (Supplemental Fig. S1, step 3) retained all transcripts that passed the detection P value and SLR filters in all replicates. The final step was the consistency filter (Supplemental Fig. S1, step 4), which identified transcripts that had consistent response to ACC over time. This filter calculated Pearson’s correlation coefficient (PCC) and the Euclidean distance (ED) for each transcript’s temporal profile to identify transcripts with a consistent pattern (PCC score) and magnitude (ED score) of expression between all replicates over time (Olex et al., 2010). All pairwise combinations of replicate temporal profiles were compared, resulting in three PCC and three ED scores for each transcript. Only two of the three PCC and ED scores were required to have PCC ≥ 0.7 and ED ≤ 1.09. The PCC cutoff was determined based on statistical reasoning, where any two data sets with a correlation greater than 0.7 were considered as highly correlated. The ED cutoff was chosen to be the median ED score over all three sets of scores for the entire filtered data set. Transcripts meeting or exceeding all filtering criteria were considered to be significantly and consistently expressed.
DE Calculation of Chang et al. (2013) Data
DE calculation using the data reported by Chang et al. (2013) was performed using calculated RPKM values provided by the authors. The calculation was performed as closely as possible to the methods described above, with the following differences. In place of the Affymetrix P value cutoff, transcripts were required to have an RPKM > 0 (i.e. to be detected) for all replicates. Since there was not a time-matched control data set, all SLRs were calculated using the average time-zero RPKM.
Consensus Clustering
Prior to clustering, a figure of merit (FOM) analysis was performed on the set of transcripts meeting the filtering criteria to identify the inherent number of clusters present in each replicate data set. The FOM also determined which clustering algorithm generated the most homogenous clusters with respect to the ED metric (k-means and hierarchical agglomerative clustering were compared; Yeung et al., 2001; Olex et al., 2007; Lewis et al., 2013). The DE filtered set of transcripts was clustered using the consensus clustering option provided by SC2ATmd (Olex and Fetrow, 2011), which was updated to allow users to specify a custom consensus threshold, where transcripts are included in a consensus cluster if they are grouped together less than 100% of the time. The calculated consensus matrix (Monti et al., 2003; Olex et al., 2010) is filtered based on the chosen threshold and converted to an adjacency matrix, where all values passing the threshold are changed to 1 and all other values are changed to 0. The adjacency matrix is then searched using MATLAB’s graphconncomp function, which uses a depth first search algorithm, to identify consensus clusters. Consensus clustering was run using K-means and ED with 10 starting clusters (parameters determined by the FOM analysis). A consensus threshold of 83%, where transcripts found in the same cluster 83% of the time would be included in the same consensus cluster, was chosen, as it returned clusters with consistent expression kinetics as well as a limited number of singletons compared with other thresholds (data not shown). Ten clustering iterations were performed per replicate.
PA Filtering and Clustering
PA filtering was performed as described previously (Klink et al., 2010) with some alterations. A pattern of PA was assigned if all three control replicates for a given time point had a detection P ≤ 0.04 (Affymetrix present call) and all ACC-treated replicates for the same time point had a detection P ≥ 0.06 (Affymetrix absent call). Similarly, a pattern of absent-present was assigned if the inverse filter was met (control P ≥ 0.06 and ACC P ≤ 0.04). Detection P values in the marginal range (0.06 > P > 0.04) were not considered, as those genes would have been included in the initial filtering analysis. A full list of genes passing the present-absent or the absent-present filter for at least one time point was constructed and is reported in Supplemental File S3.
The PA genes were clustered to identify groups with similar patterns over time. For each time point and condition (control or ACC) of a given gene, all three replicate signals were set to −1 if any one of the replicates was absent or marginal. Otherwise, if all three replicates were present (i.e. had detection P ≤ 0.04), then the signals were left intact for clustering. This modified data matrix was imported into the tool SC2ATmd for FOM analysis. The standard clustering tab in SC2ATmd (Olex and Fetrow, 2011) was then used to cluster these data into six clusters using k-means and ED, as determined by the FOM analysis.
Annotation Analysis of Microarray Data
The identification of significantly overrepresented annotations in each DE and PA cluster was performed using the analysis tool provided by AgriGO (Du et al., 2010; Tian et al., 2017). The genes for each cluster were imported into AgriGO’s Singular Enrichment Analysis tool using the Affymetrix probe identification number with the Arabidopsis gene model (TAIR9) chosen as background; all other options were left at their default setting. Annotation groups with P ≤ 0.05 were identified as significantly overrepresented.
Tissue and Cell Type Expression Patterns of the 449 DE Transcripts
Data were downloaded from http://www.plb.ucdavis.edu/labs/brady/software/BradySpatiotemporalData and queried for the 449 DE transcripts. The resulting data sets, both cell type and longitudinal slices, were analyzed using R (R Development Core Team, 2014). Distance was calculated using Pearson’s r2 (hyperSpec package; Beleites and Sergo, 2015), and transcripts were clustered using complete hierarchical clustering. Heat maps were generated using the heatmap.2 function from the gplots package (Warnes et al., 2016). Heat map colors correspond to the ventiles, or 20-quantiles, of the data.
Comparison of ACC-Regulated Transcripts with Other Data Sets
For the Chang et al. (2013) comparison, we used the lists of genes provided in their supplemental files and cross referenced these lists with our DE and PA lists to generate lists of overlapping and not overlapping genes. For each gene, we calculated SLRs for the Chang et al. (2013) data set as the log2 of the RPKM for a given sample divided by the average RPKM for time-zero samples using data shared by the Ecker laboratory. Heat maps were generated using these SLRs along with those from our data set, utilizing R (R Development Core Team, 2014) and the heatmap.2 function from the gplots package (Warnes et al. 2016). The order of the genes in the heat maps is based on complete hierarchical clustering using ED for our data set.
ACC and IAA Comparison
We cross referenced the lists of IAA DE and PA transcripts (Lewis et al., 2013) with the lists of ACC DE and PA transcripts. No transcripts were found to overlap across DE and PA data sets. This is likely because the same control samples were used, and transcripts that were identified as present-absent or absent-present tended to have more than one absent time point and, therefore, would show up in the PA list and not the DE list.
Both heat maps were generated using R (R Development Core Team, 2014) and the heatmap.2 function from the gplots package (Warnes et al.). The DE heat map was ordered with complete hierarchical clustering based on the ED on the 2- and 4-h time points. The PA overlap heat maps were ordered using complete hierarchical clustering based on the Canberra distance on the ACC data set. These parameters were chosen to most clearly bring out the patterns in each data set.
qRT-PCR
Samples containing 900 ng of RNA were used for cDNA synthesis with a 1:1 mixture of oligo(dT) and random hexamer primers and SuperScript III enzyme (Invitrogen). After digestion with RNase (Invitrogen), the A260 was measured using a Nanodrop spectrophotometer (Thermo Scientific) to ensure equal efficiency in the cDNA synthesis reactions between samples. qRT-PCR analysis using this cDNA was performed on a Roche Light Cycler 480 using SYBR Green detection chemistry. Primers specific to ETR2, ERS1, and ERS2 were used, and transcript abundance was quantified using a distinct standard curve for each primer.
Quantification of Primary Root and Root Hair Elongation and Lateral Root and Root Hair Initiation
For lateral root and primary root analysis, 5-d-old seedlings were transferred to control agar plates or agar plates containing 1 µm ACC for 5 d and imaged using an Olympus SZ61 stereoscope equipped with a DP27 color camera, utilizing cellSens software. Lateral root number was quantified manually, and primary root length was quantified using ImageJ or Imaris 7.7.2 (Bitplane). For root hair analysis, 4-d-old seedlings were transferred to control agar plates or agar plates containing 1 µm ACC for 1 d. Seedlings were imaged using a Leica MZ 16FA stereoscope with a Planapo 0.63× objective lens and an Infinity2-2C camera. Root hair numbers were quantified using ImageJ by counting all root hairs on one side of the root within a 1-mm length located 0.5 mm from the root tip. Average root hair length was quantified by measuring six representative root hairs per root.
Statistics
The number and length of root hairs, lateral root number, and primary length phenotypic data and qRT-PCR data were analyzed by two-way ANOVA. Tukey’s multiple comparison tests were then used to determine whether the differences between treatments within genotypes and the differences between genotypes within treatments were significant (GraphPad Prism 7).
For DAP-seq analyses, statistical significance was determined using a binomial test where the ratio of transcription factor targets in the cluster was compared with the ratio of transcription factor targets in the entire genome.
Accession Numbers
Arabidopsis Genome Initiative accession numbers for genes described in this article are as follows: ETR1, At1g66340; ERS1, At2g40940; EIN4, At3g04580; ETR2, At3g23150; ERS2, At1g04310; EIN2, At5g03280; EIN3, At3g20770; and EIL1, At2g27050. For all transcripts in Supplemental Files S1 to S5, the locus identifiers are included in the spreadsheets. The microarray data set has been deposited in the Gene Expression Omnibus under accession number GSE84446.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Flow chart of the filtering process.
Supplemental Figure S2. Clusters contain transcripts with similar patterns of abundance change.
Supplemental Figure S3. Cluster network maps for the whole time course expanded from Figure 2.
Supplemental Figure S4. Control-only and ACC-only analyses show that the majority of transcript abundance changes occur due to ACC treatment, while the control time course is relatively unchanged.
Supplemental Figure S5. PA analysis reveals additional genes with consistent, time-dependent changes in transcript abundance.
Supplemental Figure S6. The spatial pattern of the DE transcripts across root cell types reveals ACC-regulated genes with distinct accumulation patterns.
Supplemental Figure S7. The spatial pattern of the DE transcripts throughout root elongation and maturation reveals ACC-regulated genes with distinct accumulation patterns.
Supplemental Figure S8. MapMan metabolism overview shows that many genes related to cell wall processes and secondary metabolism respond to ACC treatment.
Supplemental Figure S9. Overlap comparison with Chang et al. (2013), expanded from Figure 5.
Supplemental Figure S10. DE analysis of Chang et al. (2013) data and comparison.
Supplemental Figure S11. ETR1 is not required for ACC-induced ERS1 or ERS2 transcript abundance.
Supplemental Table S1. P values from Student’s t tests comparing root hair numbers in untreated and ACC-treated Col-0 within time points.
Supplemental Table S2. P values from two-way ANOVA comparing lateral root numbers after 5 d of control or ACC treatment.
Supplemental Table S3. P values from two-way ANOVA comparing primary root length after 5 d of control or ACC treatment.
Supplemental Table S4. P values from two-way ANOVA comparing root hair numbers after 24 h of control or ACC treatment.
Supplemental Table S5. P values from two-way ANOVA comparing root hair lengths after 24 h of control or ACC treatment.
Supplemental Table S6. Table comparing root growth phenotypes among ethylene signaling mutants in untreated and ACC-treated roots.
Supplemental File S1. Details of the 449 transcripts that were DE after ACC treatment.
Supplemental File S2. Analysis of control and ACC-treated transcripts normalized to time zero.
Supplemental File S3. Details of the 375 transcripts in the present-absent data set.
Supplemental File S4. Detailed summary of transcripts identified by comparison with the Chang et al. data set using categories from Chang et al.
Supplemental File S5. Details on the transcripts identified by comparison to the Chang et al. data set using our DE analysis.
Acknowledgments
We appreciate the assistance of Lance Miller and the Cancer Biology Microarray facility in completion of the array analysis as well as the assistance of Lou Craddock in running the arrays and Jeff Chou in normalization. The Arabidopsis Biological Resource Center stock center (http://www.arabidopsis.org/abrc/) provided T-DNA insertion lines.
Footnotes
This work was supported by the National Science Foundation (NSF) MCB Systems and Synthetic Biology (MCB-1716279 to G.K.M., B.M.B., and A.L.O.), NSF Arabidopsis 2010 (IOS-0820717 to G.K.M. and J.S.F.), and NSF (MCB-1517032 to B.M.B.).
Articles can be viewed without a subscription.
References
- Alonso JM, Hirayama T, Roman G, Nourizadeh SD, Ecker JR (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Solano R, Wisman E, Ferrari S, Ausubel FM, Ecker JR (2003) Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc Natl Acad Sci USA 100: 2992–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An F, Zhao Q, Ji Y, Li W, Jiang Z, Yu X, Zhang C, Han Y, He W, Liu Y, et al. (2010) Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22: 2384–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi A, Wilson RL, Lacey RF, Kim H, Wuppalapati SK, Binder BM (2015) Identification of regions in the receiver domain of the ETHYLENE RESPONSE1 ethylene receptor of Arabidopsis important for functional divergence. Plant Physiol 169: 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beleites C, Sergo V (2015) hyperSpec: a package to handle hyperspectral data sets in R. http://hyperspec.r-forge.r-project.org
- Binder BM, Mortimore LA, Stepanova AN, Ecker JR, Bleecker AB (2004a) Short-term growth responses to ethylene in Arabidopsis seedlings are EIN3/EIL1 independent. Plant Physiol 136: 2921–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder BM, O’Malley RC, Wang W, Moore JM, Parks BM, Spalding EP, Bleecker AB (2004b) Arabidopsis seedling growth response and recovery to ethylene: a kinetic analysis. Plant Physiol 136: 2913–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder BM, O’Malley RC, Wang W, Zutz TC, Bleecker AB (2006) Ethylene stimulates nutations that are dependent on the ETR1 receptor. Plant Physiol 142: 1690–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder BM, Walker JM, Gagne JM, Emborg TJ, Hemmann G, Bleecker AB, Vierstra RD (2007) The Arabidopsis EIN3 binding F-box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell 19: 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker AB, Estelle MA, Somerville C, Kende H (1988) Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241: 1086–1089 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Cancel JD, Larsen PB (2002) Loss-of-function mutations in the ethylene receptor ETR1 cause enhanced sensitivity and exaggerated response to ethylene in Arabidopsis. Plant Physiol 129: 1557–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Kwok S, Bleecker AB, Meyerowitz EM (1993) Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262: 539–544 [DOI] [PubMed] [Google Scholar]
- Chang KN, Zhong S, Weirauch MT, Hon G, Pelizzola M, Li H, Huang SSC, Schmitz RJ, Urich MA, Kuo D, et al. (2013) Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLife 2: e00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144 [DOI] [PubMed] [Google Scholar]
- Chen R, Binder BM, Garrett WM, Tucker ML, Chang C, Cooper B (2011) Proteomic responses in Arabidopsis thaliana seedlings treated with ethylene. Mol Biosyst 7: 2637–2650 [DOI] [PubMed] [Google Scholar]
- Chen YF, Shakeel SN, Bowers J, Zhao XC, Etheridge N, Schaller GE (2007) Ligand-induced degradation of the ethylene receptor ETR2 through a proteasome-dependent pathway in Arabidopsis. J Biol Chem 282: 24752–24758 [DOI] [PubMed] [Google Scholar]
- Chou JW, Paules RS, Bushel PR (2005) Systematic variation normalization in microarray data to get gene expression comparison unbiased. J Bioinform Comput Biol 3: 225–241 [DOI] [PubMed] [Google Scholar]
- Cutter E. (1978) The Epidermis: Plant Anatomy. Clowes & Sons, London [Google Scholar]
- Davidson EH. (2010) Emerging properties of animal gene regulatory networks. Nature 468: 911–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eremina M, Rozhon W, Poppenberger B (2016) Hormonal control of cold stress responses in plants. Cell Mol Life Sci 73: 797–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes AM, Agudelo-Romero P, Silva MS, Ali K, Sousa L, Maltese F, Choi YH, Grimplet J, Martinez-Zapater JM, Verpoorte R, et al. (2011) Transcript and metabolite analysis in Trincadeira cultivar reveals novel information regarding the dynamics of grape ripening. BMC Plant Biol 11: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JM, Smalle J, Gingerich DJ, Walker JM, Yoo SD, Yanagisawa S, Vierstra RD (2004) Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc Natl Acad Sci USA 101: 6803–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR. (2015) Ethylene receptors in plants: why so much complexity? F1000Prime Rep 7: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble RL, Qu X, Schaller GE (2002) Mutational analysis of the ethylene receptor ETR1: role of the histidine kinase domain in dominant ethylene insensitivity. Plant Physiol 128: 1428–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ecker JR (2003) Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677 [DOI] [PubMed] [Google Scholar]
- Gupta M, Bhaskar PB, Sriram S, Wang PH (2017) Integration of omics approaches to understand oil/protein content during seed development in oilseed crops. Plant Cell Rep 36: 637–652 [DOI] [PubMed] [Google Scholar]
- Guzmán P, Ecker JR (1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale MC, McKinney GJ, Thrower FP, Nichols KM (2016) RNA-seq reveals differential gene expression in the brains of juvenile resident and migratory smolt rainbow trout (Oncorhynchus mykiss). Comp Biochem Physiol Part D Genomics Proteomics 20: 136–150 [DOI] [PubMed] [Google Scholar]
- Hall AE, Chen QG, Findell JL, Schaller GE, Bleecker AB (1999) The relationship between ethylene binding and dominant insensitivity conferred by mutant forms of the ETR1 ethylene receptor. Plant Physiol 121: 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM (1998) Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94: 261–271 [DOI] [PubMed] [Google Scholar]
- Hua J, Sakai H, Nourizadeh S, Chen QG, Bleecker AB, Ecker JR, Meyerowitz EM (1998) EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell 10: 1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanchenko MG, Muday GK, Dubrovsky JG (2008) Ethylene-auxin interactions regulate lateral root initiation and emergence in Arabidopsis thaliana. Plant J 55: 335–347 [DOI] [PubMed] [Google Scholar]
- Jaeger PA, Doherty C, Ideker T (2012) Modeling transcriptome dynamics in a complex world. Cell 151: 1161–1162 [DOI] [PubMed] [Google Scholar]
- Ju C, Mee G, Marie J, Lin DY, Ying ZI, Chang J, Garrett WM, Kessenbrock M, Groth G, Tucker ML, et al. (2012) CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci USA 109: 19486–19491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Kim H, Helmbrecht EE, Stalans MB, Schmitt C, Patel N, Wen CK, Wang W, Binder BM (2011) Ethylene receptor ETHYLENE RECEPTOR1 domain requirements for ethylene responses in Arabidopsis seedlings. Plant Physiol 156: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink VP, Overall CC, Alkharouf NW, Macdonald MH, Matthews BF (2010) Microarray detection call methodology as a means to identify and compare transcripts expressed within syncytial cells from soybean (Glycine max) roots undergoing resistant and susceptible reactions to the soybean cyst nematode (Heterodera glycines). J Biomed Biotechnol 2010: 491217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi M, Yanagisawa S (2008) Ethylene signaling in Arabidopsis involves feedback regulation via the elaborate control of EBF2 expression by EIN3. Plant J 55: 821–831 [DOI] [PubMed] [Google Scholar]
- Kurotaki D, Osato N, Nishiyama A, Yamamoto M, Ban T, Sato H, Nakabayashi J, Umehara M, Miyake N, Matsumoto N, et al. (2013) Essential role of the IRF8-KLF4 transcription factor cascade in murine monocyte differentiation. Blood 121: 1839–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le J, Vandenbussche F, Van Der Straeten D, Verbelen JP (2001) In the early response of Arabidopsis roots to ethylene, cell elongation is up- and down-regulated and uncoupled from differentiation. Plant Physiol 125: 519–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, Blilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann JU, et al. (2006) Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol 4: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DR, Negi S, Sukumar P, Muday GK (2011a) Ethylene inhibits lateral root development, increases IAA transport and expression of PIN3 and PIN7 auxin efflux carriers. Development 138: 3485–3495 [DOI] [PubMed] [Google Scholar]
- Lewis DR, Olex AL, Lundy SR, Turkett WH, Fetrow JS, Muday GK (2013) A kinetic analysis of the auxin transcriptome reveals cell wall remodeling proteins that modulate lateral root development in Arabidopsis. Plant Cell 25: 3329–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DR, Ramirez MV, Miller ND, Vallabhaneni P, Ray WK, Helm RF, Winkel BSJ, Muday GK (2011b) Auxin and ethylene induce flavonol accumulation through distinct transcriptional networks. Plant Physiol 156: 144–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F, Ohme-Takagi M, Perata P (2013) APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol 199: 639–649 [DOI] [PubMed] [Google Scholar]
- Liu Q, Xu C, Wen CK (2010) Genetic and transformation studies reveal negative regulation of ERS1 ethylene receptor signaling in Arabidopsis. BMC Plant Biol 10: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel BK, Binder BM (2012) Ethylene receptor 1 (ETR1) is sufficient and has the predominant role in mediating inhibition of ethylene responses by silver in Arabidopsis thaliana. J Biol Chem 287: 26094–26103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti S, Tamayo P, Mesirov J, Golub T (2003) Consensus clustering: a resampling based method for class discovery and visualization of gene expression microarray data. Mach Learn 52: 91–118 [Google Scholar]
- Muday GK, Rahman A, Binder BM (2012) Auxin and ethylene: collaborators or competitors? Trends Plant Sci 17: 181–195 [DOI] [PubMed] [Google Scholar]
- Nagano AJ, Sato Y, Mihara M, Antonio BA, Motoyama R, Itoh H, Nagamura Y, Izawa T (2012) Deciphering and prediction of transcriptome dynamics under fluctuating field conditions. Cell 151: 1358–1369 [DOI] [PubMed] [Google Scholar]
- Negi S, Ivanchenko MG, Muday GK (2008) Ethylene regulates lateral root formation and auxin transport in Arabidopsis thaliana. Plant J 55: 175–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi S, Sukumar P, Liu X, Cohen JD, Muday GK (2010) Genetic dissection of the role of ethylene in regulating auxin-dependent lateral and adventitious root formation in tomato. Plant J 61: 3–15 [DOI] [PubMed] [Google Scholar]
- O’Malley RC, Huang SC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR (2016) Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165: 1280–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley RC, Rodriguez FI, Esch JJ, Binder BM, O’Donnell P, Klee HJ, Bleecker AB (2005) Ethylene-binding activity, gene expression levels, and receptor system output for ethylene receptor family members from Arabidopsis and tomato. Plant J 41: 651–659 [DOI] [PubMed] [Google Scholar]
- Olex AL, Fetrow JS (2011) SC2ATmd: a tool for integration of the figure of merit with cluster analysis for gene expression data. Bioinformatics 27: 1330–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olex AL, Hiltbold EM, Leng X, Fetrow JS (2010) Dynamics of dendritic cell maturation are identified through a novel filtering strategy applied to biological time-course microarray replicates. BMC Immunol 11: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olex AL, John DJ, Hiltbold EM, Fetrow JS (2007) Additional limitations of the clustering validation method figure of merit. In 45th ACM Southeast Annual Conference (Winston-Salem, NC). ACM, New York, pp 238–243 [Google Scholar]
- Péret B, De Rybel B, Casimiro I, Benková E, Swarup R, Laplaze L, Beeckman T, Bennett MJ (2009) Arabidopsis lateral root development: an emerging story. Trends Plant Sci 14: 399–408 [DOI] [PubMed] [Google Scholar]
- Pitts RJ, Cernac A, Estelle M (1998) Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J 16: 553–560 [DOI] [PubMed] [Google Scholar]
- Qiao H, Chang KN, Yazaki J, Ecker JR (2009) Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev 23: 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Shen Z, Huang SC, Schmitz RJ, Mark A, Briggs SP, Ecker JR (2012) Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338: 390–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L, Xie F, Yu J, Wen CK (2012) Arabidopsis RTE1 is essential to ethylene receptor ETR1 amino-terminal signaling independent of CTR1. Plant Physiol 159: 1263–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Hall BP, Gao Z, Schaller GE (2007) A strong constitutive ethylene-response phenotype conferred on Arabidopsis plants containing null mutations in the ethylene receptors ETR1 and ERS1. BMC Plant Biol 7: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Schaller GE (2004) Requirement of the histidine kinase domain for signal transduction by the ethylene receptor ETR1. Plant Physiol 136: 2961–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2014) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria
- Rahman A, Hosokawa S, Oono Y, Amakawa T, Goto N, Tsurumi S (2002) Auxin and ethylene response interactions during Arabidopsis root hair development dissected by auxin influx modulators. Plant Physiol 130: 1908–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Hua J, Chen QG, Chang C, Medrano LJ, Bleecker AB, Meyerowitz EM (1998) ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA 95: 5812–5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Bleecker AB (1995) Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science 270: 1809–1811 [DOI] [PubMed] [Google Scholar]
- Seifert GJ, Barber C, Wells B, Roberts K (2004) Growth regulators and the control of nucleotide sugar flux. Plant Cell 16: 723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakeel SN, Wang X, Binder BM, Schaller GE (2013) Mechanisms of signal transduction by ethylene: overlapping and non-overlapping signalling roles in a receptor family. AoB Plants 5: plt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Hoyt JM, Hamilton AA, Alonso JM (2005) A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17: 2230–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Likhacheva AV, Alonso JM (2007) Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19: 2169–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Parry G, Graham N, Allen T, Bennett M (2002) Auxin cross-talk: integration of signalling pathways to control plant development. Plant Mol Biol 49: 411–426 [DOI] [PubMed] [Google Scholar]
- Tanimoto M, Roberts K, Dolan L (1995) Ethylene is a positive regulator of root hair development in Arabidopsis thaliana. Plant J 8: 943–948 [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z (2017) agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res 15: 1833–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Esch JJ, Shiu SH, Agula H, Binder BM, Chang C, Patterson SE, Bleecker AB (2006) Identification of important regions for ethylene binding and signaling in the transmembrane domain of the ETR1 ethylene receptor of Arabidopsis. Plant Cell 18: 3429–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Hall AE, O’Malley R, Bleecker AB (2003) Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proc Natl Acad Sci USA 100: 352–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes GR, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, Lumley T, Maechler M, Magnusson A, Moeller S, et al. (2016) gplots: various R programming tools for plotting data. https://cran.r-project.org/web/packages/gplots/
- Wen X, Zhang C, Ji Y, Zhao Q, He W, An F, Jiang L, Guo H (2012) Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res 22: 1613–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RL, Bakshi A, Binder BM (2014a) Loss of the ETR1 ethylene receptor reduces the inhibitory effect of far-red light and darkness on seed germination of Arabidopsis thaliana. Front Plant Sci 5: 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RL, Kim H, Bakshi A, Binder BM (2014b) The ethylene receptors ETHYLENE RESPONSE1 and ETHYLENE RESPONSE2 have contrasting roles in seed germination of Arabidopsis during salt stress. Plant Physiol 165: 1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Liu Q, Wen CK (2006) Receptor signal output mediated by the ETR1 N terminus is primarily subfamily I receptor dependent. Plant Physiol 142: 492–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S, Yoo SD, Sheen J (2003) Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425: 521–525 [DOI] [PubMed] [Google Scholar]
- Yeung KY, Haynor DR, Ruzzo WL (2001) Validating clustering for gene expression data. Bioinformatics 17: 309–318 [DOI] [PubMed] [Google Scholar]
- Yosef N, Regev A (2011) Impulse control: temporal dynamics in gene transcription. Cell 144: 886–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Chen X, Hong YY, Wang Y, Xu P, Ke SD, Liu HY, Zhu JK, Oliver DJ, Xiang CB (2008) Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20: 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan A, Schneider H, Lynch JP (2015) Reduced lateral root branching density improves drought tolerance in maize. Plant Physiol 168: 1603–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, An F, Feng Y, Li P, Xue L, A M, Jiang Z, Kim JM, To TK, Li W, et al. (2011) Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci USA 108: 12539–12544 [DOI] [PMC free article] [PubMed] [Google Scholar]