Abstract
Pancreatic cancer is characterized by chemoresistance after several cycles of chemotherapy, which is a major issue responsible for treatment failure of pancreatic cancer. Therefore, it is necessary to explore the specific mechanism underlying chemotherapeutic resistance to overcome this issue. Here we report that miR-1266 is dramatically elevated and correlates with poor survival and chemotherapy response in pancreatic cancer patients. Upregulation of miR-1266 enhanced the chemoresistance of pancreatic cancer cells to gemcitabine (GEM) in vitro and in vivo; conversely, inhibition of miR-1266 yielded the opposite effect. Importantly, silencing of miR-1266 restored the sensitivity of pancreatic cancer cells to GEM in a dose-dependent manner in vivo. Furthermore, our results demonstrate that miR-1266 promotes resistance of pancreatic cancer cells to GEM by targeting multiple negative regulators of the STAT3 and NF-κB pathways, including SOCS3, PTPN11, ITCH, and TNIP1, leading to constitutive activation of STAT3 and NF-κB signaling. Thus, our findings clarify a novel mechanism by which miR-1266 induces chemotherapeutic resistance in pancreatic cancer, indicating that miR-1266 may be used as chemotherapeutic response indicator. Antagomir-1266 as a chemotherapeutic sensitizer, in combination with GEM, may serve as a rational regimen in the treatment of chemotherapy-resistant pancreatic cancer.
Keywords: miR-1266, chemotherapeutic resistance, pancreatic cancer, NF-kB signaling, STAT3 signaling
Introduction
Pancreatic cancer is a devastating malignancy with a dismal prognosis because of the advanced stage at the time of diagnosis.1, 2 Surgical resection is currently the only therapeutic method with potentially curative possibility. However, the 5-year survival rate is still no more than 20%, even after resection.3 Traditionally, gemcitabine (GEM) is generally used to treat inoperable pancreatic cancer. Although novel therapeutic approaches, such as combination chemotherapy of nab-paclitaxel and GEM, have favorable prospects, they only prolonged survival by a few months.4, 5 The failure of chemotherapy in pancreatic cancer patients is principally due to therapeutic resistance following a long period of treatment, which is called acquired resistance. Therefore, we urgently need to explore the underlying molecular mechanism of chemotherapy resistance in pancreatic cancer and develop effective therapeutic avenues to overcome these obstacles.
MicroRNAs (miRNAs) are a diverse family of small non-coding RNAs (19–25 nt) involved in many biological processes, including proliferation, differentiation, the cell cycle, and apoptosis.6, 7, 8 Mechanistically, miRNAs work by binding to the 3′ UTR of downstream mRNAs, resulting in mRNA degradation or repression of translation, and function as either tumor suppressors or inducers in different kinds of cancer.9, 10, 11, 12, 13, 14 Emerging evidence indicates that miRNAs are considered important regulators of drug resistance in pancreatic cancer.15 In one study, miR-200a, miR-200b, and miR-200c were all downregulated in GEM-resistant pancreatic cancer cells.16 Evidence also indicates that oncogenic miR-181c is dramatically increased in pancreatic cancer tissues and cells, which contributes to chemoresistance by inactivating the Hippo signaling pathway in pancreatic cancer.17 The above results imply that miRNAs may contribute to drug resistance in pancreatic cancer.
It is worth noting that a large number of signaling pathways, including signal transducer and activator of transcription 3 (STAT3) and the nuclear factor κB (NF-κB) are involved in chemotherapy resistance and represented an area of intense investigation. STAT3 is a member of the STAT protein family and is triggered by a broad spectrum of cytokines and growth factors, including interleukin 6 (IL-6), IL-10, leptin, and c-src, via phosphorylation of tyrosine 705.18, 19 Constitutive activation of STAT3 has a prominent effect on resistance to some anti-cancer drugs.20 NF-κB is involved in many biological processes, including apoptosis, tumor metastasis, and cell survival.21, 22 Recent evidence has demonstrated that the NF-κB signaling pathway represents an important basis for the occurrence of drug resistance.22, 23
Several studies revealed that miRNAs play a crucial role in the resistance of various tumors cells to a broad range of anticancer drugs by regulating these signaling pathways.16, 24, 25, 26, 27 Consequently, it is possible that the tweaking of chemotherapy drugs with miRNAs would facilitate the development of novel therapeutic approaches to improve the efficacy of treatment. In this study, we report that miR-1266 is robustly elevated in pancreatic cancer tissues and significantly associated with poor survival and chemotherapy response in patients with pancreatic cancer. Furthermore, our results show that miR-1266 protects pancreatic cancer cells from apoptosis induced by GEM by targeting multiple negative regulators, including SOCS3, PTPN11, ITCH, and TNIP1, resulting in constitutive activation of the STAT3 and NF-κB signaling pathways. Importantly, we find that inhibition of miR-1266 sensitizes pancreatic cancer cells to GEM in a dose-dependent manner in vivo. In summary, the combination of traditional chemotherapeutic drugs with miR1266 may be a new strategy to cure pancreatic cancer, which provides the preclinical theoretical proof for pancreatic cancer therapy.
Results
miR-1266 Is Upregulated in Pancreatic Cancer and Correlated with Poor Prognosis
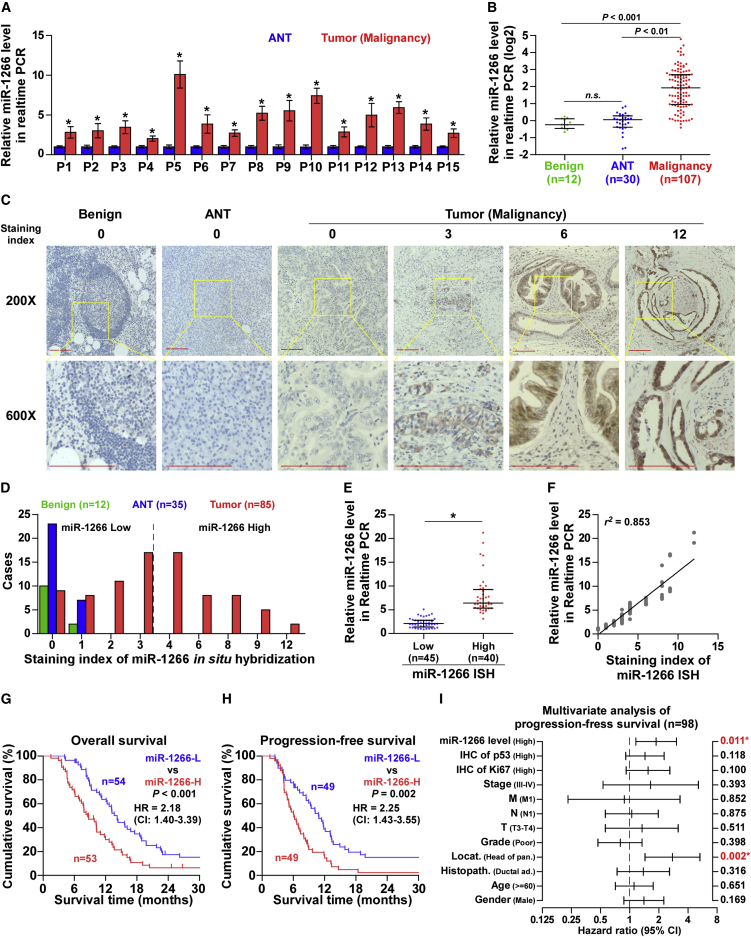
To discern potential targetable miRNAs in pancreatic cancer, we analyzed pancreatic cancer miRNA sequencing datasets from The Cancer Genome Atlas (TCGA) and E-GEOD-32678 and found that miR-1266 expression was markedly upregulated in pancreatic cancer tissues compared with normal pancreatic tissues (Figures S1A and S2B). Consistently, we observed that level of miR-1266 expression was upregulated in primary pancreatic cancer tissues compared with that in the matched adjacent normal tissue (ANT) as well as in benign pancreatic lesions (Figures 1A and 1B). To determine the major sources of miR-1266 in primary pancreatic cancer samples, in situ hybridization (ISH) was performed. As shown in Figure 1C, miR-1266 expression was mainly detected in pancreatic cancer cells, and its expression was lower or absent in stromal cells. The staining index of miR-1266 ISH in pancreatic cancer tissues was dramatically higher than that in ANT or benign pancreatic lesions (Figure 1D). Furthermore, pancreatic cancer tissues with low staining scores of miR-1266 ISH displayed lower miR-1266 expression compared with that with high staining scores of miR-1266 ISH by real-time PCR (Figure 1E), indicating that there was a positive linear correlation between staining scores of miR-1266 ISH and miR-1266 mRNA expression (Figure 1F). Statistical analysis of pancreatic cancer samples revealed that increased miR-1266 expression strongly correlated with M classification, clinical stage, chemotherapeutic response, and overall and progression-free survival (Figures 1G and 1H; Tables 1 and 2). Univariate Cox regression analysis indicated that pancreatic cancer patients with high miR-1266 expression had shorter overall survival and progression-free survival compared with patients with low miR-1266 expression (Tables 3 and 4). Multivariate Cox regression analysis revealed that miR-1266 may be an independent factor for predicting poor overall and progression-free survival (Figure 1I; Tables 3 and 4).Similarly, high miR-1266 expression predicted a poor prognosis in the TCGA pancreatic cancer miRNA datasets (Figures S1C and S1D). Thus, these results suggest that upregulation of miR-1266 might be involved in human pancreatic cancer progression.
Figure 1.
miR-1266 Is Upregulated in Pancreatic Cancer and Correlated with a Poor Prognosis
(A) Real-time PCR analysis of miR-1266 expression in 15 primary pancreatic cancer tissues compared with matched adjacent normal tissues. Transcript levels were normalized to U6 expression. *p < 0.05. (B) Real-time PCR analysis of miR-1266 expression in 107 freshly collected pancreatic cancer tissue samples, 30 adjacent normal pancreatic tissues, and 12 benign pancreatic lesions, including 3 pancreatic cyst samples, 4 pancreatitis samples, 1 pancreatic inflammation and cyst sample, 3 microcystic adenoma samples, and 1 dystrophic calcification sample. Transcript levels were normalized to U6 expression. Lines represent median and lower/upper quartiles. (C) Representative images of low-power (×200, top) or high-power (×600, bottom) fields of miR-21 expression by in situ hybridization (ISH) in pancreatic cancer tissues with different ISH scores (0, 3, 6, and 12), adjacent normal pancreatic tissue (ANT), and benign pancreatic lesions (Benign). Scale bars, 100 μm. (D) The number of pancreatic cancer tissues, ANT, and benign pancreatic lesions in different staining index groups of ISH. (E) Real-time PCR analysis of miR-1266 in 40 pancreatic cancer tissues with high ISH scores compared with 45 pancreatic cancer tissues with low ISH scores. The median ISH score in pancreatic cancer tissues was used to stratify low and high ISH scores. *p < 0.05. (F) Correlation of miR-1266 expression level with ISH scores in pancreatic cancer tissues. (G and H) Kaplan-Meier analysis of overall (G) and progression-free (H) survival curves of patients with pancreatic cancer with high miR-1266 expression (more than median, n = 51) versus low miR-1266 expression (less than median, n = 51). The median of miR-1266 expression levels in 102 pancreatic cancer tissues was used to stratify the high and low expression levels of miR-1266. The data shown in the scatterplot and bar graph were determined by the median with interquartile range and median with SD. p < 0.001, log rank test. (I) Multivariate Cox regression analysis to evaluate the significance of the association between miR-1266 expression and progression-free survival. Hazard ratio (HR) values were obtained by log2 transformation.
Table 1.
The Clinicopathological Characteristics of 107 Patients with Pancreatic Cancer
| Parameters | Number of Cases |
|---|---|
| Gender | |
| Female | 45 |
| Male | 62 |
| Histopathology | |
| Ductal ad. | 84 |
| Other | 23 |
| Grade | |
| High/moderate | 70 |
| Poor | 37 |
| N Stage | |
| N0 | 37 |
| N1 | 70 |
| Clinical Stage | |
| I | 19 |
| II–IV | 88 |
| Treatment Response | |
| CR/PR | 43 |
| SD/PD | 64 |
| IHC Status of Ki67 | |
| Low | 54 |
| High | 53 |
| Age (years) | |
| <60 | 46 |
| ≥60 | 61 |
| Location | |
| Head of pancreas | 76 |
| Other | 31 |
| T Stage | |
| T1–T2 | 27 |
| T3–T4 | 80 |
| M Stage | |
| M0 | 103 |
| M1 | 4 |
| Survival Status | |
| Alive | 16 |
| Dead | 91 |
| Progression Status | |
| Positive | 87 |
| Negative | 11 |
| NA | 9 |
| IHC Status of p53 | |
| Low | 50 |
| High | 57 |
Other histopathology includes adenocarcinoma not otherwise specified, mucinous carcinoma, and neuroendocrine carcinoma. Other locations include body of pancreas, tail of pancreas, and unidentified location. NA, not available; IHC, immunological histological chemistry; CR, complete response; PR, partial response; SD, stable disease; PD, clinical progressive disease; ductal ad., ductal adenocarcinoma.
Table 2.
The Relationship between miR-1266 and Clinicopathological Characteristics in 107 Patients with Pancreatic Cancer
| Parameters | Number of Cases | miR-1266 Expression |
p Values | |
|---|---|---|---|---|
| Low | High | |||
| Gender | ||||
| Female | 45 | 23 | 22 | 0.908 |
| Male | 62 | 31 | 31 | |
| Age (years) | ||||
| <60 | 46 | 26 | 20 | 0.277 |
| ≥60 | 61 | 28 | 33 | |
| Histopathology | ||||
| Ductal ad. | 84 | 54 | 40 | 0.228 |
| Other | 23 | 10 | 13 | |
| Location | ||||
| Head of pancreas | 76 | 37 | 39 | 0.564 |
| Other | 31 | 17 | 14 | |
| Grade | ||||
| High/moderate | 70 | 38 | 32 | 0.277 |
| Poor | 37 | 16 | 21 | |
| T Stage | ||||
| T1–T2 | 27 | 16 | 11 | 0.291 |
| T3–T4 | 80 | 38 | 42 | |
| N Stage | ||||
| N0 | 37 | 22 | 15 | 0.176 |
| N1 | 70 | 32 | 38 | |
| M Stage | ||||
| M0 | 103 | 54 | 49 | 0.040* |
| M1 | 4 | 0 | 4 | |
| Clinical Stage | ||||
| I | 19 | 14 | 5 | 0.026* |
| II–IV | 88 | 40 | 48 | |
| Treatment Response | ||||
| CR/PR | 43 | 14 | 29 | 0.004* |
| SD/PD | 64 | 39 | 25 | |
| IHC Status of Ki67 | ||||
| Low | 54 | 30 | 24 | 0.228 |
| High | 53 | 24 | 29 | |
| IHC Status of p53 | ||||
| Low | 50 | 32 | 18 | 0.009* |
| High | 57 | 22 | 35 | |
Other histopathology includes adenocarcinoma not otherwise specified, mucinous carcinoma, and neuroendocrine carcinoma. *p <0.05. NA, not available; IHC, immunological histological chemistry; CR, complete response; PR, partial response; SD, stable disease; PD, clinical progressive disease; ductal ad., ductal adenocarcinoma.
Table 3.
Univariate and Multivariate Analysis of Factors Associated with Overall Survival in 107 Pancreatic Cancer Patients
| Characteristics | Univariate Analysis |
Multivariate Analysis |
||
|---|---|---|---|---|
| HR (95% CI) | p Values | HR (95% CI) | p Values | |
| Gender (male) | 1.08 (0.72–1.62) | 0.719 | 1.02 (0.65–1.60) | 0.930 |
| Age (≥ 60 years) | 1.06 (0.70–1.61) | 0.782 | 1.38 (0.87–2.19) | 0.176 |
| Histopathology (ductal ad.) | 1.25 (0.75–2.10) | 0.394 | 1.26 (0.70–2.26) | 0.435 |
| Location (head of pancreas) | 2.28 (1.38–3.77) | 0.001∗ | 1.62 (0.88–2.98) | 0.121 |
| Grade (poor) | 1.05 (0.68–1.64) | 0.815 | 0.83 (0.50–1.38) | 0.482 |
| T stage (T3–T4) | 2.48 (1.46–4.24) | 0.001∗ | 1.24 (0.61–2.53) | 0.549 |
| N stage (N1) | 1.85 (1.17–2.92) | 0.008∗ | 0.93 (0.50–1.71) | 0.810 |
| M stage (M1) | 0.98 (0.36–2.68) | 0.972 | 0.35 (0.11–1.10) | 0.072 |
| Clinical stage (III–IV) | 4.04 (2.07–7.89) | 4.04 | 2.55 (0.95–6.82) | 0.062 |
| IHC status of Ki67 (high) | 2.40 (1.57–3.69) | <0.001∗ | 1.82 (1.09–3.03) | 0.022∗ |
| IHC status of p53 (high) | 1.38 (0.91–2.09) | 0.127 | 1.27 (0.81–1.98) | 0.299 |
| miR-1266 (high) | 2.07 (1.36–3.14) | 0.001∗ | 1.83 (1.14–2.95) | 0.013∗ |
p <0.05. HR, hazard ratio; CI, confidence interval; IHC, immunological histological chemistry.
Table 4.
Univariate and Multivariate Analysis of Factors Associated with Progression-free Survival in 98 Pancreatic Cancer Patients
| Characteristics | Univariate Analysis |
Multivariate Analysis |
||
|---|---|---|---|---|
| HR (95% CI) | p Values | HR (95% CI) | p Values | |
| Gender (male) | 1.19 (0.79–1.79) | 0.399 | 1.40 (0.87–2.25) | 0.169 |
| Age (≥ 60 years) | 1.02 (0.66–1.55) | 0.945 | 1.11 (0.71–1.75) | 0.651 |
| Histopathology (ductal ad.) | 1.25 (0.74–2.13) | 0.408 | 1.38 (0.74–2.58) | 0.316 |
| Location (head of pancreas) | 3.04 (1.83–5.08) | <0.001∗ | 2.76 (1.44–5.32) | 0.002∗ |
| Grade (poor) | 1.05 (0.68–1.63) | 0.822 | 0.80 (0.47–1.34) | 0.398 |
| T stage (T3–T4) | 3.05 (1.73–5.37) | <0.001∗ | 1.34 (0.56–3.17) | 0.511 |
| N stage (N1) | 1.99 (1.25–3.17) | 0.004∗ | 1.05 (0.56–1.97) | 0.875 |
| M stage (M1) | 0.98 (0.31–3.13) | 0.978 | 0.88 (0.23–3.31) | 0.852 |
| Clinical stage (III–IV) | 3.90 (2.03–7.48) | <0.001∗ | 1.64 (0.53–5.13) | 0.393 |
| IHC status of Ki67 (high) | 2.33 (1.50–3.62) | <0.001∗ | 1.55 (0.92–2.60) | 0.100 |
| IHC status of p53 (high) | 1.36 (0.89–2.08) | 0.151 | 1.44 (0.91–2.27) | 0.118 |
| miR-1266 (high) | 2.16 (1.39–3.35) | 0.001∗ | 1.87 (1.16–3.04) | 0.011∗ |
p <0.05. HR, hazard ratio; CI, confidence interval; IHC, immunological histological chemistry.
Upregulation of miR-1266 Is Associated with Poor Chemotherapy Response in Pancreatic Cancer
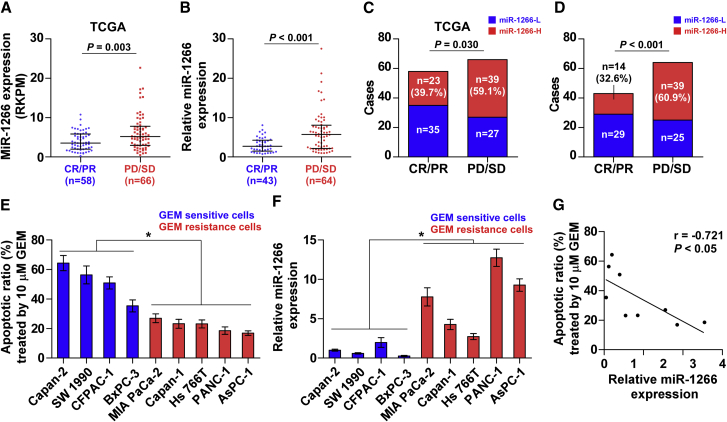
Because cytostatic treatment is the only therapeutic option for the majority of pancreatic cancer patients, we investigated whether miR-1266 plays a role in the chemotherapeutic response in publicly available datasets from TCGA as well as our samples. Notably, we found that miR-1266 expression and the percentage of high expression of miR-1266 was elevated in pancreatic cancer patients with a poor chemotherapy response (progressive disease [PD] and stable disease [SD]) compared with those with a good response (complete response [CR] and partial response [PR]) (Figures 2A–2D), indicating that miR-1266 significantly correlated with a poor chemotherapy response. We further analyzed the correlation of miR-1266 expression with chemotherapeutic response in 9 pancreatic cancer cell lines. The apoptotic ratio was calculated after all cells were treated with 10 μM GEM. As shown in Figure 2E, five pancreatic cancer cell lines, including MIA PaCa-2, Capan-1, HS 766T, PANC-1, and AsPC-1, displayed more resistance to GEM compared with the other 4 pancreatic cancer cell lines. We further examined the expression of miR-1266 in these pancreatic cancer cells, and it was interesting to find that the expression levels of miR-1266 in GEM-resistant pancreatic cancer cells were generally higher than in GEM-sensitive cells (Figure 2F), indicating that miR-1266 expression negatively correlates with chemotherapeutic response in pancreatic cancer cells (Figure 2G). Furthermore, Kaplan-Meier survival analysis revealed that patients with PD/SD exhibited shorter overall survival (p < 0.001; hazard ratio = 2.98, 95% confidence interval [CI] = 1.94 to 4.57; Figure S2A) and progression-free survival (p < 0.001; hazard ratio = 3.47, 95% CI = 2.19 to 5.47; Figure S2B), which was consistent with TCGA profiles (Figures S2C and S2D). These observations reveal that miR-1266 promotes chemoresistance in pancreatic cancer.
Figure 2.
Upregulation of miR-1266 Is Associated with a Poor Chemotherapy Response in Pancreatic Cancer
(A and B) In the TCGA dataset (A) and our pancreatic cancer specimens (B), miR-1266 expression levels were much higher in pancreatic cancer patients with a poor chemotherapy response (progressive disease [PD] or stable disease [SD]) than in those with a good response (complete response [CR] or partial response [PR]). (C and D) Percentages and numbers of samples showed high or low miR-1266 expression in patients with different chemotherapy responses in the TCGA dataset (C) and our pancreatic cancer specimens (D). The 102 pancreatic cancer tissues were obtained from patients with gemcitabine-based treatment. (E) Apoptotic ratio of nine different pancreatic cancer cell line after treatment with 10 μm GEM. (F) Expression levels of miR-1266 in pancreatic cancer cells after treatment with 10 μm GEM. (G) Correlation of miR-1266 expression level with apoptotic ratio in pancreatic cancer cells after treatment with 10 μM GEM.
miR-1266 Promotes Chemoresistance in Pancreatic Cancer In Vitro
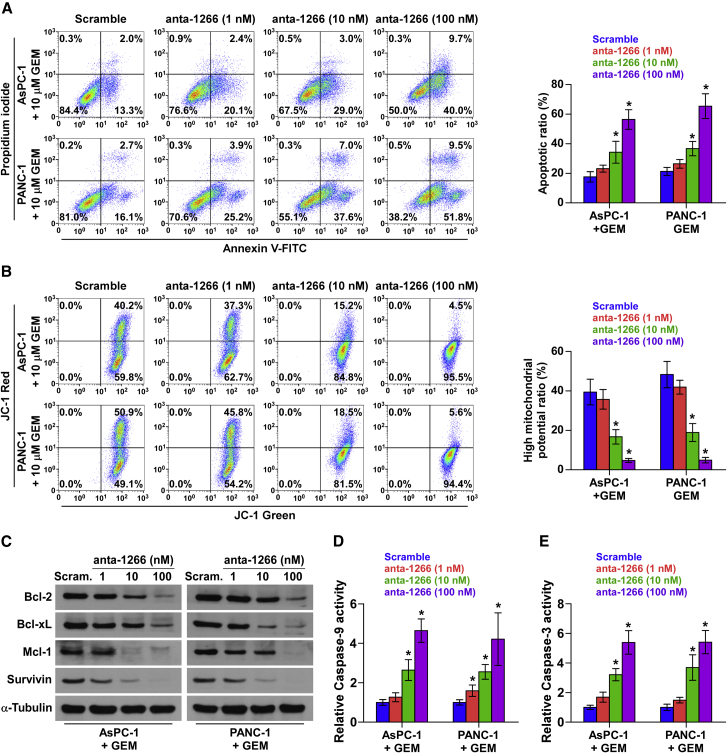
The role of miR-1266 in the chemoresistance of pancreatic cancer was further examined by treatment with the first-line anti-pancreatic cancer drug GEM in vitro. Based on the expression level of miR-1266 shown in Figure 2F, we first endogenously silenced miR-1266 by transfecting different concentrations of antagomir-1266 (anta-1266) in AsPC-1 and PANC-1 cells (Figure S3A). As shown in Figure 3A, we found that silencing of miR-1266 dramatically increased the apoptosis rate of AsPC-1 and PANC-1 cells treated with GEM in a dose-dependent manner. However, only a high concentration of anta-1266 (100 nM) could increase the apoptotic rates of AsPC-1 and PANC-1 cells in the absence of GEM treatment (Figure S3B). A mitochondrial membrane potential assay was performed, and the results showed that silencing miR-1266 decreased the mitochondrial potential of AsPC-1 and PANC-1 cells under treatment of GEM in a dose-dependent manner (Figure 3B). Moreover, the effect of miR-1266 on apoptotic protection was further confirmed by examining the expression levels of the anti-apoptotic proteins Bcl-2, Bcl-xL, Mcl-1, and Survivin and the activity of caspase-9 and caspase-3. As shown in Figures 3C–3E, downregulation of miR-1266 decreased Bcl-2, Bcl-xL, Mcl-1, and Survivin expression but enhanced the activity of caspase-9 and caspase-3 in a dose-dependent manner. Therefore, these results demonstrate that silencing miR-1266 attenuates the resistance of pancreatic cancer cells to GEM in vitro.
Figure 3.
miR-1266 Promotes Chemoresistance in Pancreatic Cancer In Vitro
(A) Annexin V-FITC/PI staining of the indicated cells treated with gemcitabine (10 μM) for 36 hr. Error bars represent the mean ± SD of three independent experiments. *p < 0.05. (B) JC-1 staining showed that silencing miR-1266 decreased the mitochondrial potential in a dose-dependent manner in pancreatic cancer cells. Error bars represent the mean ± SD of three independent experiments. *p < 0.05. (C) Western blotting analysis of Bcl-2, Bcl-xL, Mcl-1, and Survivin in the indicated cells. (D and E) Analysis of the activities of caspase-9 (D) and caspase-3 (E) as detected by the cleaved forms of these two proteins. Error bars represent the mean ± SD of three independent experiments. *p < 0.05.
We further exogenously overexpressed miR-1266 via virus transduction in BxPC-3 and SW1990 cells (Figure S3A) because the expression levels of miR-1266 were lower than in other cells (Figure 2F). As shown in Figure S3B, we found that miR-1266 overexpression reduced the apoptosis rates of BxPC-3 and SW1990 cells in the absence of GEM treatment, which was more obvious in BxPC-3 and SW1990 cells treated with GEM (Figure S3C). In addition, miR-1266 overexpression increased the mitochondrial potential of BxPC-3 and SW1990 cells under treatment with GEM (Figure S3D). Moreover, overexpression of miR-1266 increased Bcl-2, Bcl-xL, Mcl-1, and Survivin expression but reduced the activity of caspase-9 or caspase-3 (Figures S3E–S3G). The effects of miR-1266 were not cytotoxic, as assessed by MTT assay of proliferation (Figure S3H). Collectively, these results demonstrate that miR-1266 promotes pancreatic cancer cells resistance to GEM in vitro.
Inhibition of miR-1266 Sensitizes Pancreatic Cancer Cells to Gemcitabine In Vivo
We next assessed whether inhibition of miR-1266 might accelerate chemotherapy drug function in vivo. Mice were randomly divided into three groups (n = 8/group). For the first group, AsPC-1-Vector cells were inoculated subcutaneously (2 × 106 cells per injection) in the left dorsal flanks, and, for the other two groups, all mice were subcutaneously injected with 2 × 106 AsPC-1 cells and treated with 100 μL anta-1266 (0.1 mmol/L) or 100 μL anta-1266 (1 mmol/L) through the lateral tail vein every 4 days. After 10 days of inoculation, mice were intraperitoneally injected with 50 mg/kg GEM twice each week for 4 weeks (Figure S4A). The tumor volumes and weights were differentially decreased in the anta-1266 plus GEM group compared with the scramble control (Figures S4A–S4C). Importantly, the combined anta-1266 and GEM treatment markedly restricted tumor growth to low volumes and prolonged the overall survival of mice (Figures S4B–S4D). Moreover, we investigated the effect of anta-1266 on the chemotherapeutic response in another pancreatic cancer cell line, PANC-1, with high concentration of anta-1266 (1 mmol/L). Consistently, the tumor volumes and weights were differentially decreased in the anta-1266 plus GEM group compared with the scramble group (Figures S4E and S4F). In addition, tumors formed by miR-1266-downexpressing AsPC-1 and PANC-1 cells had increased caspase-3 activity (Figure S4G). We further examined the effect of overexpression of miR-1266 on chemoresistance to see whether upregulating miR-1266 induces GEM resistance in BxPC-3 cells in vivo. As shown in Figures S4H and S4I, the tumor volumes and weights were significantly increased in the miR-1266 overexpression plus GEM group compared with the control groups. Collectively, these findings indicate that silencing miR-1266 restores the chemotherapeutic sensitivity of pancreatic cancer cells to GEM in vivo.
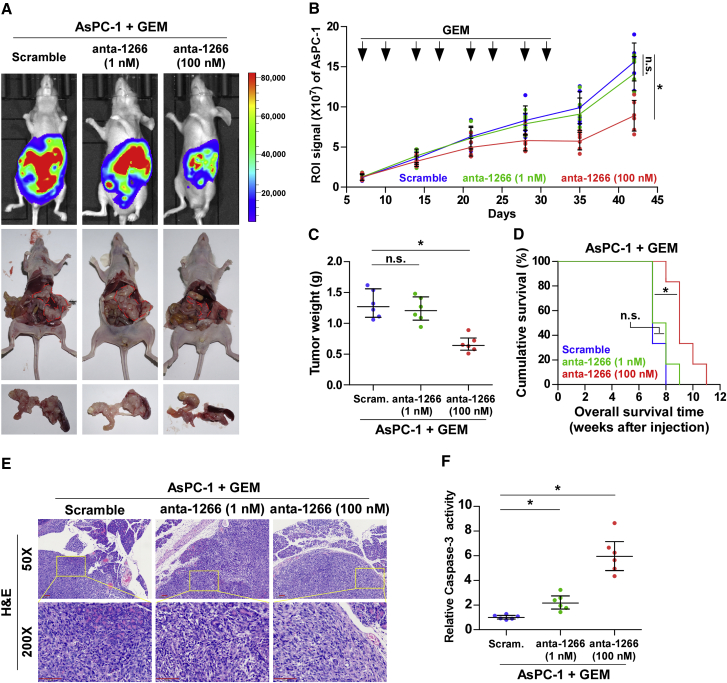
We next investigated the therapeutic efficacy of anta-1266 in the chemoresistance of pancreatic cancer cells to GEM in an orthotopic xenograft mouse model. As shown in Figures 4A–4D, only a high dose of anta-1266 (1 mmol/L) following lateral tail vein administration dramatically reduced the tumor volumes and weights and increased the overall survival of mice. However, a low dose of anta-1266 (0.1 mmol/L) had no significant effect on pancreatic tumor progression (Figures 4A–4D). H&E staining from the indicated tumors of mice showed that only a high dose of anta-1266 decreased the tumor burden in the pancreas of orthotopic xenograft mice (Figure 4E). Strikingly, the capsule of the pancreas was still complete after administration of a high dose of anta-1266 (Figure 4E). Furthermore, caspase-3 activity was robustly elevated in mice injected with high dose of anta-1266 (Figure 4F). Collectively, our results demonstrate that a high dose of anta-1266 effectively improves the chemotherapeutic sensitivity of pancreatic cancer cells to GEM in vivo.
Figure 4.
Anta-1266 Re-sensitizes Pancreatic Cancer Cells to Gemcitabine in Orthotopic Xenografts
(A) Xenograft model in nude mice. Shown are representative images of tumors in mice injected with scramble, a low dose of anta-1266 (0.1 mmol/L), and a high dose of anta-1266 (1 mmol/L). Mice were euthanized, and tumors from each experimental group were excised. (B) After 7 days of inoculating AsPC-1 cells, mice were intraperitoneally injected with 50 μg/g gemcitabine (GEM) twice each week for 4 weeks. Region of interest (ROI) signaling in the low-dose anta-1266, high-dose anta-1266, and scramble groups was measured from the fifth day at 5-day intervals. Data presented are the mean ± SD. (C) Tumor weights of each group. (D) The overall survival of mice in the indicated groups. (E) Representative H&E staining images from the indicated tumors of mice. Scale bars, 100 μm. (F) Caspase-3 activity in the tumor tissues formed by the low-dose anta-1266, high-dose anta-1266, and scramble groups.
miR-1266 Activates the STAT3 and NF-κB Signaling Pathways
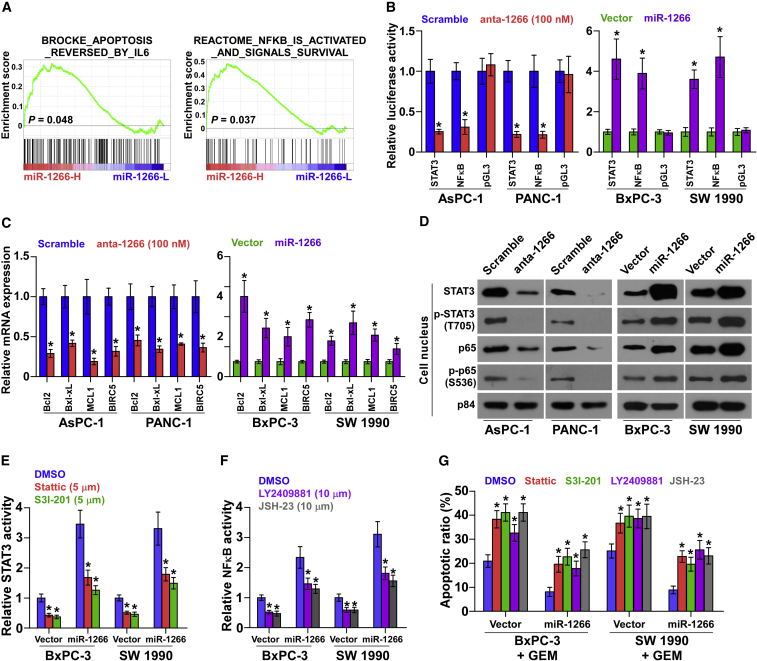
We next explored the mechanism underlying the stimulatory effect of miR-1266 on chemoresistance in pancreatic cancer. We performed a gene set enrichment analysis (GSEA) of miR-1266 expression against the oncogenic signature collection of the Molecular Signatures Database (MSigDB) was performed and indicated that miR-1266 overexpression significantly and positively correlated with the IL-6- and NF-κB-regulated anti-apoptosis signatures (“BROCKE_APOPTOSIS_REVERSED_BY_IL6” and “REACTOME_NFKB_IS_ACTIVATED_AND_SIGNALS_SURVIVAL”) (Figure 5A). These results suggest that miR-1266 may regulate the IL-6/STAT3 and NF-κB signaling pathways, which have been reported to promote chemoresistance.28, 29 As shown in Figures 5B and 5C, we found that miR-1266 overexpression significantly increased STAT3- and NF-κB-dependent luciferase activity as well as the expression levels of multiple downstream genes, including Bcl2, Bcl-xL, MCL1, and BIRC5 in BxPC-3 and SW1990 cells. Silencing miR-1266 repressed the STAT3- and NF-κB-dependent luciferase activity and expression levels of these downstream genes in AsPC-1 and PANC-1 cells. Moreover, cellular fractionation and western blotting analysis revealed that overexpression of miR-1266 increased, whereas silencing of miR-1266 reduced nuclear accumulation of STAT3 and NF-κB/p65 as well as the phosphorylated levels of STAT3 and NF-κB/p65 in the nucleus in pancreatic cancer cells (Figure 5D). Thus, these results demonstrate that miR-1266 activates the STAT3 and NF-κB signaling pathways.
Figure 5.
miR-1266 Activates the STAT3 and NF-κB Signaling Pathways
(A) Gene set enrichment analysis (GSEA) revealed that miR-1266 expression significantly and positively correlated with the IL-6- and NF-κB-regulated anti-apoptosis signatures. (B) STAT3 and NF-κB transcriptional activity was assessed by luciferase reporter constructs in the indicated cells. Error bars represent the mean ± SD of three independent experiments. *p < 0.05. (C) Real-time PCR analysis of Bcl2, Bcl-xL, MCL1, and BIRC5 in the indicated cells. Error bars represent the mean ± SD of three independent experiments. *p < 0.05. (D) Western blotting of nuclear STAT3 and NF-κB/p65 expression and the phosphorylated levels of STAT3 and NF-κB/p65 in the nucleus. The nuclear protein p84 was used as the nuclear protein marker. (E) STAT3 activity under treatment of the inhibitors Stattic (5 μM) and S3I-201 (100 μM) in the indicated cells. Error bars represent the mean ± SD of three independent experiments. *p < 0.05. (F) NF-κB activity under treatment with the inhibitors LY2409881 (10 μM) and JSH-23 (10 μM) in the indicated cells. Error bars represent the mean ± SD of three independent experiments. *p < 0.05. (G) Annexin V-FITC/PI staining of the indicated cells after treatment with Stattic, S3I-201, LY2409881, and JSH-23. Error bars represent the mean ± SD of three independent experiments. *p < 0.05.
STAT3 and NF-κB Activation Is Essential for miR-1266-Induced Chemoresistance
We then explored the functional significance of STAT3 and NF-κB signaling in the chemoresistance of pancreatic cancer cells by treatment with signaling inhibitors. As shown in Figure S5, the STAT3 inhibitors Stattic and S3I-201 and the NF-κB inhibitors LY2409881 and JSH-23 caused potent inhibition of STAT3 and NF-κB reporter activities in a dose-dependent manner. Notably, the stimulatory effects of miR-1266 on STAT3 and NF-κB activity were impaired by these inhibitors (Figures 5E and 5F). Moreover, inhibition of STAT3 and NF-κB signaling abrogated the protective effect on cell apoptosis by miR-1266 overexpression (Figure 5G). These results suggest that activation of STAT3 and NF-κB signaling is critical for miR-1266-induced chemoresistance in pancreatic cancer cells.
miR-1266 Targets Multiple Negative Regulators of STAT3 and NF-κB Signaling
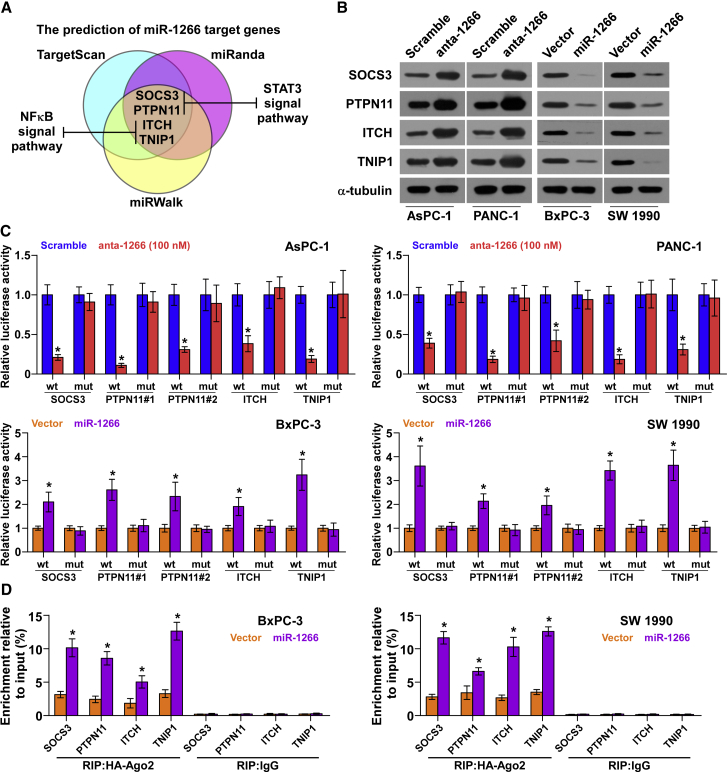
Using publicly available algorithms, including TargetScan (http://www.targetscan.org/vert_71/),30 miRanda (http://34.236.212.39/microrna/home.do),31 and miRWalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk/),32 we found that multiple negative regulators of STAT3 and NF-κB signaling, including SOCS3, PTPN11, ITCH, and TNIP1, may be potential targets of miR-1266 (Figures 6A and S6A). Western blot analysis revealed that miR-1266 overexpression reduced the expression levels of SOCS3, PTPN11, ITCH, and TNIP1. In contrast, silencing of miR-1266 increased the expression of SOCS3, PTPN11, ITCH, and TNIP1, suggesting that miR-1266 negatively regulates these proteins (Figure 6B). Furthermore, a luciferase assay showed that miR-1266 overexpression attenuated, whereas inhibition of miR-1266 elevated, the reporter activity driven by the 3′ UTRs of these transcripts but not by the mutant 3′ UTRs of these transcripts within miR-1266-binding seed regions in BxPC-3 and PANC-1 cells (Figure 6C). Moreover, a microribonucleoprotein (miRNP) immunoprecipitation (IP) assay revealed an association of miR-1266 with SOCS3, PTPN11, ITCH, and TNIP1 transcripts (Figure 6D), further indicating direct repressive effects of miR-1266 on these targets. Importantly, the tumor injected with a high dose of anta-1266 showed increased expression levels of SOCS3, PTPN11, ITCH, and TNIP1 compared with the scramble group in vivo (Figure S4J). In addition, individual silencing of these targets rescued the STAT3 and NF-κB activity repression in miR-1266-silencing cells (Figures S6B and S6C), demonstrating that SOCS3, PTPN11, ITCH, and TNIP1 were functional effectors of miR-1266 on regulating STAT3 and NF-κB signaling. Collectively, our results suggest that miR-1266 directly targets SOCS3, PTPN11, ITCH, and TNIP1, leading to constitutive activation of STAT3 and NF-κB signaling.
Figure 6.
miR-1266 Targets Multiple Negative Regulators of STAT3 and NF-κB Signaling
(A) Predicted miR-1266 target sequence in 3′ UTRs of SOCS3, PTPN11, ITCH, and TNIP1. (B) Western blotting of SOCS3, PTPN11, ITCH, and TNIP1 expression in the indicated cells. α-Tubulin served as the loading control. (C) Luciferase assay of cells transfected with pmirGLO-3UTR reporter in miR-1266-overexpressing and -silencing cells. (D) miRNP IP assay showing the association between miR-1266 and SOCS3, PTPN11, ITCH, and TNIP1 transcripts in pancreatic cancer cells. Pull-down of IgG antibody served as the negative control.
Recurrent Gains and Hypomethylation Contribute to miR-1266 Overexpression in Pancreatic Cancer Tissues
To clarify the underlying mechanism responsible for miR-1266 overexpression in pancreatic cancer tissues, we first analyzed the amplification status from a genetic perspective in the pancreatic cancer dataset from TCGA and found that recurrent gains (amplification) occurred in 6.7% of pancreatic cancer tissues (Figure S7A) and that miR-1266 expression levels in pancreatic cancer tissues with gains were higher than those without gains (Figure S7B). Consistently, our results indicated that gains were found in 6 of 85 pancreatic cancer tissues (approximately 7.1%) (Figure S7C) and that the expression level of miR-1266 in pancreatic cancer tissues with gains was robustly higher than in those without gains (Figure S7D). This finding indicates that recurrent gains are implicated in miR-1266 overexpression in a small portion of pancreatic cancer tissues and suggest that other unknown mechanisms may contribute to miR-1266 overexpression in pancreatic cancer tissues. With this question in mind, we further analyzed the methylation levels of miR-1266 in pancreatic cancer tissues from TCGA and found that the methylation level of miR-1266 in pancreatic cancer tissues was reduced compared with that in the ANT (Figure S7E), suggesting that hypomethylation may be an important factor contributing to overexpression of miR-1266 in pancreatic cancer tissues. To confirm this hypothesis, we further measured the methylation level of miR-1266 in our clinical samples, including benign pancreatic lesions tissues, ANT, and pancreatic cancer tissues by calculating the methylation ratio; namely, the methylation percentage in each tissue. As shown in Figure S7F, the case number of methylation ratio < 50% in pancreatic cancer tissues (n = 49, 57.7%) was obviously more than that in benign pancreatic lesions tissues and ANT. Furthermore, the lower the methylation ratio, the higher the expression levels of miR-1266 in pancreatic cancer tissues (Figure S7G). Taken together, our results indicate that recurrent gains and hypomethylation contribute to upstream regulation of miR-1266 overexpression in pancreatic cancer tissues.
Discussion
Although surgical resection has improved the quality of life, prolonged the survival time, and decreased the mortality of patients with advanced pancreatic cancer over the past decades, resistance to chemotherapy remains the principal aspect of treatment failure in patients with inoperable pancreatic cancer. The most common reason for acquired resistance to anticancer drugs is the formation of efflux pumps on the surface of tumor cells that eject anticancer drugs from inside the tumor cells to the outside.33 Other mechanisms of resistance, including insensitivity to drug-induced apoptosis, upregulation of DNA repair enzymes, induction of drug-detoxifying mechanisms, stromal proliferation, and reduced angiogenesis, play a crucial role in drug resistance.34, 35, 36, 37, 38, 39
Recent studies have also suggested that dysregulation of miRNAs contributes to chemoresistance in pancreatic cancer and may be potential targets in the treatment of pancreatic cancer.24, 27, 40, 41 Dhayat et al.25 found that miR-100 and miR-21, along with their respective targets PTEN and MDR-1, were identified as independent prognostic survival and chemotherapy response markers in stage II pancreatic adenocarcinoma (PDAC). Furthermore, accumulating literature indicates that therapy targeting miRNAs, such as miR-223 and miR-181c, re-sensitizes pancreatic cancer cells to chemotherapeutic drugs, further determining the roles of potential targets of miRNAs in the treatment of pancreatic cancer.17, 26 Here we found that miR-1266 was substantially overexpressed in pancreatic cancer and significantly correlated with short survival, high recurrence, and chemotherapy resistance. We demonstrated that miR-1266 promoted chemoresistance by activating the STAT3 and NF-κB pathways. More importantly, the delivery of anta-1266 dramatically sensitized pancreatic cancer cells to GEM treatment in vivo, determining the therapeutic efficacy of anta-1266 in pancreatic cancer chemotherapy. Taken together, our results propose that miR-1266 might be used as a chemotherapy response marker or potential therapeutic target for human pancreatic cancer.
Therapies targeting miRNAs are promising prospects in the treatment of cancers as versatile regulators of the human genome that orchestrate myriad cellular pathways to control cancer cell growth and apoptosis. Khalife et al.42 reported that high levels of miR-155 are associated with poor outcome in acute myeloid leukemia (AML). In vivo, MLN4924, an inhibitor of NEDD8-activating enzyme, reduced miR-155 expression and prolonged the survival of mice engrafted with leukemic cells, suggesting potential of miR-155 as a novel therapeutic target in AML.42 Another study from Jagannathan et al.43 indicated that miR-29b was significantly reduced in myeloma cells resistant to proteasome inhibitors, including bortezomib, carfilzomib, and ixazomib. Synthetically engineered miR-29b replacements impaired the growth of myeloma cells and xenotransplants, demonstrating potential application of miR-29b replacements as the first-in-class miR-based proteasome inhibitors to synergistically enhance the anti-tumor effect of bortezomib.43 These studies indicate that miRNA-targeting therapeutic approaches have potentially great therapeutic value in the treatment of cancer patients. In the current study, our results demonstrated that inhibition of miR-1266 by anta-1266 reversed the chemoresistance of AsPC-1 and PANC-1 cells to GEM in a dose-dependent manner in vitro. More importantly, the delivery of anta-1266 greatly resensitized pancreatic cancer cells to GEM in vivo, suggesting potentially applicable value of anta-1266 in the treatment of pancreatic cancer. Taken together, the results from our study and other studies indicate that the development of miRNA theragnostics will provide widely applicable avenues for the targeted delivery of personalized medicines to improve the chemotherapeutic resistance and outcome of cancer patients.
Dysregulation of apoptosis-associated genes induced by STAT3 and NF-κB signaling is one of the most common mechanisms of chemoresistance. For example, one study reported that mutant KRAS conferred drug resistance by upregulating Bcl-xL protein via the STAT3 signaling pathway in colorectal cancers.44 Another study found that manganese superoxide dismutase (MnSOD) induces the resistance of lung adenocarcinoma cells via the NF-κB/Bcl-2/Snail pathway.45 Consistently, the STAT3 and NF-κB signaling pathways have been identified to be constitutively activated in a variety of cancers; however, the molecular mechanism of constitutive activation of STAT3 and NF-κB signaling in pancreatic cancer remains unclear. In the present study, we found that overexpression of miR-1266 was able to simultaneously activate STAT3 and NF-κB signaling pathways by targeting SOCS3, PTPN11, ITCH, and TNIP1, resulting in resistance to chemotherapy drugs in pancreatic cancer. Thus, our study identified a novel mechanism for STAT3 and NF-κB signaling activation and pancreatic cancer chemoresistance.
Accumulating evidence indicates that another mechanism of miRNAs regulating chemoresistance is through modulating epithelial-to-mesenchymal transition (EMT).46 EMT is a pivotal step in tumor cell metastasis that includes consecutive processes of cells detaching, migrating, invading, dispersing, and finally residing in other organs.47 The role of EMT in tumor formation, dissemination, and metastasis is well established, but EMT-inducing chemoresistance provides a novel vision in the field of tumors. There is one study showing that inhibiting EMT enhanced the efficacy of chemotherapeutic drugs against pancreatic cancer cells.48 Furthermore, miR-223 rendered pancreatic cancer cells resistant to GEM by inducing EMT acquisition; importantly, inhibitors of miR-223 reversed the EMT phenotype and enhanced chemotherapy sensitivity.26 In the present study, we demonstrated that miR-1266 increased the resistance to GEM by targeting multiple negative regulators of the STAT3 and NF-κB signaling pathways. However, whether miR-1266 promotes chemoresistance of pancreatic cancer cells by modulating EMT still needs to be studied further.
In summary, our study reveals that miR-1266 upregulation plays an important role in chemotherapeutic resistance of pancreatic cancer as a critical activator of STAT3 and NF-κB signaling. Importantly, inhibition of miR-1266 sensitizes pancreatic cancer cells to chemotherapeutic drugs in a dose-dependent manner both in vitro and in vivo. Understanding the precise role of miR-1266 in pancreatic cancer pathogenesis and in the STAT3 and NF-κB signaling pathways promises to increase our knowledge of the biological basis of cancer development and may also facilitate the development of new therapeutic strategies against pancreatic cancer.
Materials and Methods
Cell Lines and Cell Culture
The human pancreatic cancer cell lines BxPC-3, PANC-1, AsPC-1, SW1990, Capan-1, Capan-2, CFPAC-1, MIA PaCa-2, and Hs 766T were obtained from the ATCC (USA), and all human pancreatic cancer cell lines were cultured as described in the ATCC protocol. The cell lines were maintained in RPMI 1640 (Invitrogen, USA) supplemented with 10% fetal bovine serum (FBS; HyClone, USA).
Patients and Tumor Tissues
A total of 15 adjacent normal pancreatic tissues from duodenal carcinoma, 12 benign pancreatic lesions, 15 paired pancreatic cancer tissues with matched ANTs, as well as other 92 separate pancreatic cancer tissues from patients treated with GEM-based chemotherapy were obtained during needle biopsy or surgery at the Sun Yat-sen University Cancer Center (Guangzhou, China) and the Affiliated Jiangmen Hospital of Sun Yat-sen University (Guangdong, China) between January 2008 and December 2012. The patients’ basic clinicopathological information is summarized in Tables 1, 5, and 6. The pancreatic cancer tissues, through radical resection, were microdissected to make pancreatic cancer tissues composed of at least 80% tumor tissues. The percentage of tumor tissues in pancreatic cancer tissues obtained from needle biopsy was at least more than 70%. The percentage of tumor tissues was evaluated by two independent professional pathologists. Patients were diagnosed based on clinical and pathological evidence, and the specimens were immediately snap-frozen and stored in liquid nitrogen tanks. For the use of these clinical materials for research purposes, patient consent and approval from the Institutional Research Ethics Committee were obtained. The median of miR-1266 expression was used to stratify high and low expression of miR-1266.
Table 5.
The Basic Information of 30 Cancer Patients for miR-1266 Expression Analysis in Pancreatic Normal Control Tissues
| Cases (n) | Percentage | |
|---|---|---|
| Disease | ||
| Pancreatic carcinoma | 15 | 50.0 |
| Duodenal carcinoma | 15 | 50.0 |
| Gender | ||
| Male | 16 | 53.3 |
| Female | 14 | 46.7 |
| Age | ||
| ≤60 | 14 | 46.7 |
| >60 | 16 | 53.3 |
Table 6.
The Basic Information of 12 Patients with Benign Pancreatic Lesions for miR-1266 Expression Analysis
| Cases (n) | Percentage | |
|---|---|---|
| Disease Type | ||
| Cyst | 3 | 25.0 |
| Inflammation | 4 | 33.3 |
| Inflammation & cyst | 1 | 8.3 |
| Microcystic adenoma | 3 | 25.0 |
| Dystrophic calcification | 1 | 8.3 |
| Gender | ||
| Male | 4 | 33.3 |
| Female | 8 | 66.6 |
| Age | ||
| ≤60 | 6 | 50.0 |
| >60 | 6 | 50.0 |
RNA Extraction, Reverse Transcription, and Real-Time PCR
Total RNA from tissues or cells was extracted using an RNA isolation kit (QIAGEN, USA) according to the manufacturer’s instructions. mRNA and miRNA were reverse-transcribed from total mRNA using the RevertAid First Strand cDNA synthesis kit (Thermo Scientific, USA) according to the manufacturer’s protocol. cDNA was amplified and quantified on a CFX96 system (Bio-Rad, USA) using iQ SYBR Green (Bio-Rad, USA). The primers are provided in Table 7. Real-time PCR was performed according to a standard method as described previously.49 Primers for U6 and miR-1266 were synthesized and purified by RiboBio (Guangzhou, China). U6 or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous control. The copy-number variation (CNV) of each sample of pancreatic cancer was examined using real-time PCR primer Hs05362117_cn through the TaqMan Copy Number Assay; TaqMan Copy Number Reference Assay RNase P and TaqMan Fast Advanced Master Mix were used as the loading control and amplification kit, respectively. Relative fold expressions were calculated with the comparative threshold cycle (2−ΔΔCt) method according to a previous study.50
Table 7.
A List of Primers Used in the Reactions for Real-Time RT-PCR
| Real-Time PCR Primer | |
|---|---|
| SOCS3-up | 5′-CAAGGACGGAGACTTCGATT-3′ |
| SOCS3-dn | 5′-AACTTGCTGTGGGTGACCAT-3′ |
| PTPN11-up | 5′-CTGCCTCCACACCAGTGATA-3′ |
| PTPN11-dn | 5′-GGAGCCTGAGCAAGGAGC-3′ |
| ITCH-up | 5′-GAGGCTACCCATTGAACCAA-3′ |
| ITCH-dn | 5′-AACGCCTTAACCCAAGGAAG-3′ |
| TNIP1-up | 5′-CAGCAGCAGGCTAGTGTGAC-3′ |
| TNIP1-dn | 5′-CTGCTTGTTCACTTCCAGCA-3′ |
| BCL2-up | 5′-GTGGATGACTGAGTACCTGAACC-3′ |
| BCL2-dn | 5′-AGACAGCCAGGAGAAATCAAAC-3′ |
| BCL2L1-up | 5′-GGTATTGGTGAGTCGGATCG-3′ |
| BCL2L1-dn | 5′-TGCTGCATTGTTCCCATAGA-3′ |
| MCL1-up | 5′-GAGGAGGACGAGTTGTACCG-3′ |
| MCL1 dn | 5′-ATGTCCAGTTTCCGAAGCAT-3′ |
| BIRC5-up | 5′-CCATTAACCGCCAGATTTGA-3′ |
| BIRC5-dn | 5′-TGTAGAGATGCGGTGGTCCT-3′ |
| GAPDH-up | 5′-ATTCCACCCATGGCAAATTC-3′ |
| GAPDH-dn | 5′-TGGGATTTCCATTGATGACAAG |
Plasmids, Small Interfering RNA, and Transfection
The human miR-1266 gene was PCR-amplified from genomic DNA and cloned into a pMSCV-puro retroviral vector (Clontech, Japan). The pSTAT3-luc, pNF-κB-luc, and control plasmids (Clontech, Japan) were used to examine transcription factor activity quantitatively. The 3′ UTR of human SOCS3, PTPN11, ITCH, and TNIP1 was PCR-amplified from genomic DNA and cloned into pmirGLO vectors (Promega, USA), and the list of primers used in clone reactions is presented in Table 8. anta-1266 is an RNA-like oligonucleotides that harbor various chemical modifications, including complete 2′-O-methylation of sugar, a phosphorothioate backbone, and a cholesterol moiety at the 3′ end. Anta-1266 and a scrambled control were synthesized and purified by RiboBio (Guangzhou, China), and the category number was miR30005920-1-10. Small interfering RNA (siRNA) for SOCS3, PTPN11, ITCH, and TNIP1 knockdown and the respective control RNA were synthesized and purified by RiboBio (Guangzhou, China). Transfection of miRNA, siRNAs, and plasmids was performed using Lipofectamine 3000 (Life Technologies, USA) according to the manufacturer’s instructions.
Table 8.
A List of Primers Used in the Reactions for Clone PCR
| Subcloning and Plasmid Construction | |
|---|---|
| miR-1266-clone-F | 5′-CAATTATTCAAAAGATGCTTGGG-3′ |
| miR-1266-clone-R | 5′-GGTATGGCCCTCCAGGTTAA-3′ |
| SOCS3-3UTR-44nt-clone-F | 5′-GCAAAGTTTGACTTGGATTGG-3′ |
| SOCS3-3UTR-1585nt-clone-R | 5′-CGTGGCACATGGCACAA-3′ |
| PTPN11-3UTR-198nt-clone-F | 5′-CTTCCCAATTACTCATTTCCTCA-3′ |
| PTPN11-3UTR-1993nt-clone-R | 5′-CCAAACTACCCCAAAGTCTCAA-3′ |
| ITCH-3UTR-1824nt-clone-F | 5′-ATTTCATGTACCCCAGGAAATAC-3′ |
| ITCH-3UTR-3549nt-clone-R | 5′-ACGCAAGTGAGGCAACCA-3′ |
| TNIP1-3UTR-30nt-clone-F | 5′-CCATGATTCGTGCAAATAGCT-3′ |
| TNIP1-3UTR-759nt-clone-R | 5′-GTCATTTGGCTCCACCTTCA-3′ |
Western Blot Analysis
The nuclear/cytoplasmic fraction was separated by using a cell fractionation kit (Cell Signaling Technology, USA) according to the manufacturer’s instructions, and the whole-cell lysates were extracted using RIPA buffer (Cell Signaling Technology). Western blot was performed according to a standard method as described previously.51 Briefly, the cell lysates were loaded with 10% loading buffer (Beyotime, China) and heated for 5 min at 100°C. Equal quantities of denatured protein samples were resolved on 10% SDS-polyacrylamide gels and then transferred onto polyvinylidene fluoride (PVDF) membranes (Roche, Switzerland). After blocking with 5% non-fat dry milk in Tris-buffered saline (TBS)/0.05% Tween 20, membranes were incubated with a specific primary antibody, followed by a horseradish peroxidase-conjugated secondary antibody. Proteins were visualized using enhanced chemiluminescence (ECL) reagents (Pierce, USA). Antibodies against Bcl-2, Bcl-xL, Mcl-1, Survivin, STAT3, p65, p84, SOCS3, PTPN11, ITCH, and TNIP1 were purchased from Abcam (Cambridge, MA, USA). The membranes were stripped and reprobed with an anti-α-tubulin antibody (Sigma-Aldrich, USA) as the loading control.
Flow Cytometric Analysis
Flow cytometric analysis of apoptosis was done using the fluorescein isothiocyanate (FITC) Annexin V Apoptosis Detection Kit I (BD, USA) and was performed as described by the protocol. Briefly, cells were dissociated with trypsin and resuspended at 1 × 106 cells/mL in binding buffer with 50 μL/mL FITC Annexin V and 50 μL/mL propidium iodide (PI). The cells were subsequently incubated for 15 min at room temperature and then analyzed with a Gallios flow cytometer (Beckman Coulter, USA). The cells’ inner mitochondrial membrane potential (Δψm) was detected by flow cytometry using the MitoScreen JC-1 staining kit (BD) and was performed as described in the protocol. Briefly, cells were dissociated with trypsin, resuspended at 1 × 106 cells/mL in assay buffer, and then incubated at 37°C for 15 min with 10 μl/mL JC-1. Before being analyzed by flow cytometer, cells were washed twice with assay buffer. Flow cytometry data were analyzed using FlowJo 7.6 software (Tree Star, USA).
Caspase-9 or Caspase-3 Activity Assays
The activity of caspase-9 or caspase-3 was analyzed by spectrophotometry using the Caspase-9 Colorimetric Assay Kit or Caspase-3 Colorimetric Assay Kit (Keygen, China) and was performed as described in the protocol. Briefly, 5 × 106 cells or 100 mg fresh tumor tissues were washed with cold PBS, resuspended in lysis buffer, and incubated on ice for 30 min. We mixed 50 μL cell suspension, 50 μL reaction buffer, and 5 μL caspase-3/9 substrate, and then incubated it at 37°C for 4 hr. The absorbance was measured at 405 nm, and bicinchoninic acid (BCA) protein quantitative analysis was used as the reference to normal each experiment groups.
Tumor Xenografts
All experimental procedures were approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University. 6-week-old BALB/c-nu mice were randomly divided into groups (n = 8/group), and the indicated cells (2 × 106) were inoculated subcutaneously into the inguinal folds of the nude mice. After 10 days of cell inoculation, the mice were injected intraperitoneally with 50 μg/g GEM twice per week for 4 weeks. In the experiment testing whether anta-1266 could enhance the sensitivity of pancreatic cancer cells to GEM, animals were injected with 100 μL anta-1266 (1 mmol/L), 100 μL anta-1266 (0.1 mmol/L), or antagomir scramble through the lateral tail vein every 4 days for 4 weeks. On day 45, tumors were detected by an IVIS imagining system (Caliper, USA), and then animals were euthanized and tumors were excised, weighed, and stored in liquid nitrogen tanks. For orthotopic xenografts, 2 × 106 AsPC-1 cells were inoculated subcutaneously into the inguinal folds of the nude mice. The tumors were resected aseptically when they measured ∼300 mm3 in volume. The tumor tissues were cut with scissors and minced into approximately 1 × 1 × 1 mm pieces in Hanks’ balanced salt solution containing 100 units/mL penicillin and 100 μg/mL streptomycin. Then, a tumor piece was transplanted into the pancreas as described previously.52 One week following orthotopic transplantation of tumor pieces, the mice were randomly divided into three groups (n = 6/group) and injected intraperitoneally with 50 μg/g GEM twice per week for 4 weeks. Meanwhile, the mice were injected with 100 μL anta-1266 (1 mmol/L), 100 μL anta-1266 (0.1 mmol/L), or antagomir scramble through the lateral tail vein every 4 days for 4 weeks. Tumor volume was determined using an external caliper and calculated using the equation (length [L] ×× width2 [W2])/2. On day 42, tumors were detected by an IVIS imagining system (Caliper, USA), and then the animals were euthanized and tumors were excised, weighed, and stored in liquid nitrogen tanks.
Luciferase Assay
Cells (4 × 104) were seeded in triplicate in 24-well plates and cultured for 24 h, and the luciferase assay was performed as described previously.53 Cells were transfected with 100 ng pSTAT3 or pNF-κB reporter luciferase plasmid or the pmirGLO-SOCS3-3′ UTR, -PTPN11-3′ UTR, -ITCH-3′ UTR, or -TNIP1-3′ UTR luciferase plasmid plus 5 ng pRL-TK Renilla plasmid (Promega) using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s recommendation. Luciferase and Renilla signals were measured 36 hr after transfection using the Dual Luciferase Reporter Assay Kit (Promega) according to the manufacturer’s protocol.
miRNA Immunoprecipitation
Cells were co-transfected with hemagglutinin (HA)-Ago2, followed by HA-Ago2 immunoprecipitation using HA antibody. Real-time PCR analysis of the IP material was used to test the association of the mRNA of SOCS3, PTPN11, ITHC and TNIP1 with the RNA-induced silencing complex (RISC). The specific processes were performed as described previously.54
High-Throughput Data Processing and Visualization
The clinical profile of pancreatic cancer dataset, copy number variation profile and methylation array profile were downloaded from TCGA (https://tcga-data.nci.nih.gov/tcga/). The analysis methods for the methylation array profile and copy number variation profile were different, and the specific procedures were as follows.
For the methylation array profile, we downloaded the DNA methylation dataset of pancreatic cancer in the 450K Methylation microarray from TCGA. We procured the β-value (representing the methylation level) of the related genes from the level 3 data of each sample. We exported the 4 miR-1266-related probe number from the methylation microarray data of 115 pancreatic cancer tissues and 10 ANTs using Excel 2010 and GraphPad 5 software, respectively, and calculated the differential β-value between the ANT and pancreatic cancer tissues and depicted the heatmap with MeV4.6 software.
For the CNV profile, we download the level 3 CNV dataset of pancreatic cancer in the SNP6.0 microarray from TCGA and analyzed the dataset with GISTIC2.0 software as described previously (all parameters as the default).55, 56 We procured the CNV number of each corresponding sample and defined the CNV number of the amplification and gain groups as “gain” and the sample without gains as “deletion” or “diploid.” We then analyzed the results using Excel 2010 and depicted each figure using GraphPad 5 software.
The analysis procedure for the GSEA was that we first downloaded the miRNA dataset of pancreatic cancer from TCGA and procured the expression value of the corresponding genes from the level 3 data of each sample (the unit was RNA sequencing [RNA-seq] by expectation maximization, reads per kilobase of exon model per million mapped reads [RKPM]). We analyzed the log2 value of each sample using Excel 2010 and GraphPad 5 and statistically analyzed the miRNA expression level of all pancreatic cancer tissues using a paired t test or unpaired t test. GSEA was performed with the RNaseqV2 dataset of pancreatic cancer from TCGA as the expression dataset. The high and low expression levels of miR-1266 were stratified by the medium expression level of miR-1266 in 178 pancreatic cancer tissues. GSEA was performed by Molecular Signatures Database v5.2 (http://software.broadinstitute.org/gsea/msigdb) (all processing parameters as the default).
ISH
In situ hybridization (ISH) was performed on PDAC tumors using locked nucleic acid (LNA) probes for miR-1266 (Exiqon, Vedbaek, Denmark). Paraffin-embedded tumors were deparaffinized, treated with proteinase K, and fixed in paraformaldehyde. The digoxigenin-labeled LNA probe was hybridized overnight. Slides were rinsed and incubated with anti-digoxigenin, a horseradish peroxidase (HRP)-linked antibody (catalog no. ZLI-9720, Zsbio, China), for 2 hr. The detection reaction was performed using the DAB Ready-to-Use Kit (catalog no. ZLI-9018, Zsbio, China). For each sample, the whole fields of each slide were analyzed by optical microscope. The ISH scores given by the two independent investigators were averaged for further comparative evaluation of the miR-1266 expression. Tumor cell proportion was scored as follows: 0 (no positive tumor cells), 1 (< 10% positive tumor cells), 2 (10%–35% positive tumor cells), 3 (35%–70% positive tumor cells), and 4 (> 70% positive tumor cells). The staining intensity was graded according to the following criteria: 0 (no staining), 1 (weak staining, light yellow), 2 (moderate staining, yellow brown), and 3 (strong staining, brown). The ISH score was calculated as the product of staining intensity score and the proportion of positive tumor cells. Using this method of assessment, we evaluated miR-1266 expression in pancreatic cancer samples by determining the staining intensity (SI), with scores of 0, 1, 2, 3, 4, 6, 8, 9, or 12. ISH score 6 was the median of all sample tissue SI scores. High and low expression of miR-1266 were stratified by the follow criteria. An ISH score of ≥ 4 was used to define tumors with high expression of miR-1266 and SI < 4 as tumors with low expression of miR-1266.
Methylation-Specific High-Resolution Melting Curve Analysis
DNA from pancreatic cancer samples was extracted using the TIANamp genomic DNA kit according to the manufacturer’s instructions (catalog no. DP304, Tiangen, China). Then DNA was subjected to bisulfite conversion with a bisulfite kit (catalog no. DP215, Tiangen, China). A methylation-specific high-resolution melting curve analysis was further performed using the HRM kit (EvaGreen, catalog no. FP210, Tiangen, China). The sequence of the primer used to detect the methylation levels of the cg06706204 location was as follows: forward, 5′-TGTCTAGCATCAAACAAGGCCA-3′; reverse, 5′-TGTAACTGCCATAACAGAACGGA-3′. A positive control of cg06706204(C) and negative control of cg06706204(T) were constructed using a T vector plasmid, and a standard curve was prepared according to 1:0, 3:1, 1:1, 1:3, and 0:1. The methylation ratio was calculated by methylation-specific high-resolution melting curve analysis.
Statistical Analysis
All values are presented as means ± SD. Significant differences were determined using GraphPad 5.0 software (USA). Student’s t test was used to determine statistical differences between two groups. The chi-square test was used to analyze the relationship between miR-1266 expression and clinicopathological characteristics. Survival curves were plotted using the Kaplan Meier method and compared by log rank test. Univariate and multivariate Cox regression analyses were performed with SPSS 21.0 (IBM, Chicago, IL, USA). p < 0.05 was considered significant. All experiments were repeated three times.
Author Contributions
L.S. developed ideas and drafted the manuscript. X.Z., D.R., and X.W. conducted the experiments and contributed to the analysis of data. X.L. and L.Y. contributed to the analysis of data. C.L. conducted the experiments. S.W. and J.Z. contributed to the analysis of data and revised the manuscript. X.P. edited the manuscript. All authors contributed to revise the manuscript and approved the final version for publication.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
This work was supported by the Ministry of Science and Technology of China (973 Program, 2014CB910604), the Natural Science Foundation of China (81325013, 81530082, 91529301, 91740118, 81773106), and the Science and Technology of Guangdong Province (2016A030308002).
Footnotes
Supplemental Information includes seven figures and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.01.004.
Supplemental Information
References
- 1.Kindler H.L. Front-line therapy of advanced pancreatic cancer. Semin. Oncol. 2005;32(6, Suppl 9):S33–S36. doi: 10.1053/j.seminoncol.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Kaur S., Kumar S., Momi N., Sasson A.R., Batra S.K. Mucins in pancreatic cancer and its microenvironment. Nat. Rev. Gastroenterol. Hepatol. 2013;10:607–620. doi: 10.1038/nrgastro.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oettle H., Post S., Neuhaus P., Gellert K., Langrehr J., Ridwelski K., Schramm H., Fahlke J., Zuelke C., Burkart C. Adjuvant chemotherapy with gemcitabine vs observation in patients undergoing curative-intent resection of pancreatic cancer: a randomized controlled trial. JAMA. 2007;297:267–277. doi: 10.1001/jama.297.3.267. [DOI] [PubMed] [Google Scholar]
- 4.Borazanci E., Von Hoff D.D. Nab-paclitaxel and gemcitabine for the treatment of patients with metastatic pancreatic cancer. Expert Rev. Gastroenterol. Hepatol. 2014;8:739–747. doi: 10.1586/17474124.2014.925799. [DOI] [PubMed] [Google Scholar]
- 5.Thota R., Pauff J.M., Berlin J.D. Treatment of metastatic pancreatic adenocarcinoma: a review. Oncology (Williston Park) 2014;28:70–74. [PubMed] [Google Scholar]
- 6.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 8.Shi X.B., Tepper C.G., deVere White R.W. Cancerous miRNAs and their regulation. Cell Cycle. 2008;7:1529–1538. doi: 10.4161/cc.7.11.5977. [DOI] [PubMed] [Google Scholar]
- 9.Ren D., Yang Q., Dai Y., Guo W., Du H., Song L., Peng X. Oncogenic miR-210-3p promotes prostate cancer cell EMT and bone metastasis via NF-κB signaling pathway. Mol. Cancer. 2017;16:117. doi: 10.1186/s12943-017-0688-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khew-Goodall Y., Goodall G.J. Myc-modulated miR-9 makes more metastases. Nat. Cell Biol. 2010;12:209–211. doi: 10.1038/ncb0310-209. [DOI] [PubMed] [Google Scholar]
- 11.Ren D., Wang M., Guo W., Huang S., Wang Z., Zhao X., Du H., Song L., Peng X. Double-negative feedback loop between ZEB2 and miR-145 regulates epithelial-mesenchymal transition and stem cell properties in prostate cancer cells. Cell Tissue Res. 2014;358:763–778. doi: 10.1007/s00441-014-2001-y. [DOI] [PubMed] [Google Scholar]
- 12.Ren D., Lin B., Zhang X., Peng Y., Ye Z., Ma Y., Liang Y., Cao L., Li X., Li R. Maintenance of cancer stemness by miR-196b-5p contributes to chemoresistance of colorectal cancer cells via activating STAT3 signaling pathway. Oncotarget. 2017;8:49807–49823. doi: 10.18632/oncotarget.17971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vetter G., Saumet A., Moes M., Vallar L., Le Béchec A., Laurini C., Sabbah M., Arar K., Theillet C., Lecellier C.H., Friederich E. miR-661 expression in SNAI1-induced epithelial to mesenchymal transition contributes to breast cancer cell invasion by targeting Nectin-1 and StarD10 messengers. Oncogene. 2010;29:4436–4448. doi: 10.1038/onc.2010.181. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Ren D., Wang M., Guo W., Zhao X., Tu X., Huang S., Zou X., Peng X. Wild-type p53 suppresses the epithelial-mesenchymal transition and stemness in PC-3 prostate cancer cells by modulating miR-145. Int. J. Oncol. 2013;42:1473–1481. doi: 10.3892/ijo.2013.1825. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z., Li Y., Ahmad A., Banerjee S., Azmi A.S., Kong D., Sarkar F.H. Pancreatic cancer: understanding and overcoming chemoresistance. Nat. Rev. Gastroenterol. Hepatol. 2011;8:27–33. doi: 10.1038/nrgastro.2010.188. [DOI] [PubMed] [Google Scholar]
- 16.Li Y., VandenBoom T.G., 2nd, Kong D., Wang Z., Ali S., Philip P.A., Sarkar F.H. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69:6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen M., Wang M., Xu S., Guo X., Jiang J. Upregulation of miR-181c contributes to chemoresistance in pancreatic cancer by inactivating the Hippo signaling pathway. Oncotarget. 2015;6:44466–44479. doi: 10.18632/oncotarget.6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan Z.L., Guan Y.J., Wang L., Wei W., Kane A.B., Chin Y.E. Central role of the threonine residue within the p+1 loop of receptor tyrosine kinase in STAT3 constitutive phosphorylation in metastatic cancer cells. Mol. Cell. Biol. 2004;24:9390–9400. doi: 10.1128/MCB.24.21.9390-9400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silva C.M. Role of STATs as downstream signal transducers in Src family kinase-mediated tumorigenesis. Oncogene. 2004;23:8017–8023. doi: 10.1038/sj.onc.1208159. [DOI] [PubMed] [Google Scholar]
- 20.Zhao C., Li H., Lin H.J., Yang S., Lin J., Liang G. Feedback Activation of STAT3 as a Cancer Drug-Resistance Mechanism. Trends Pharmacol. Sci. 2016;37:47–61. doi: 10.1016/j.tips.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Karin M., Cao Y., Greten F.R., Li Z.W. NF-kappaB in cancer: from innocent bystander to major culprit. Nat. Rev. Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 22.Oya M., Takayanagi A., Horiguchi A., Mizuno R., Ohtsubo M., Marumo K., Shimizu N., Murai M. Increased nuclear factor-kappa B activation is related to the tumor development of renal cell carcinoma. Carcinogenesis. 2003;24:377–384. doi: 10.1093/carcin/24.3.377. [DOI] [PubMed] [Google Scholar]
- 23.Baldwin A.S. Control of oncogenesis and cancer therapy resistance by the transcription factor NF-kappaB. J. Clin. Invest. 2001;107:241–246. doi: 10.1172/JCI11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giovannetti E., Funel N., Peters G.J., Del Chiaro M., Erozenci L.A., Vasile E., Leon L.G., Pollina L.E., Groen A., Falcone A. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010;70:4528–4538. doi: 10.1158/0008-5472.CAN-09-4467. [DOI] [PubMed] [Google Scholar]
- 25.Dhayat S.A., Abdeen B., Köhler G., Senninger N., Haier J., Mardin W.A. MicroRNA-100 and microRNA-21 as markers of survival and chemotherapy response in pancreatic ductal adenocarcinoma UICC stage II. Clin. Epigenetics. 2015;7:132. doi: 10.1186/s13148-015-0166-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma J., Fang B., Zeng F., Ma C., Pang H., Cheng L., Shi Y., Wang H., Yin B., Xia J., Wang Z. Down-regulation of miR-223 reverses epithelial-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Oncotarget. 2015;6:1740–1749. doi: 10.18632/oncotarget.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang J.H., Voortman J., Giovannetti E., Steinberg S.M., Leon L.G., Kim Y.T., Funel N., Park J.K., Kim M.A., Kang G.H. Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS ONE. 2010;5:e10630. doi: 10.1371/journal.pone.0010630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhardwaj A., Sethi G., Vadhan-Raj S., Bueso-Ramos C., Takada Y., Gaur U., Nair A.S., Shishodia S., Aggarwal B.B. Resveratrol inhibits proliferation, induces apoptosis, and overcomes chemoresistance through down-regulation of STAT3 and nuclear factor-kappaB-regulated antiapoptotic and cell survival gene products in human multiple myeloma cells. Blood. 2007;109:2293–2302. doi: 10.1182/blood-2006-02-003988. [DOI] [PubMed] [Google Scholar]
- 29.Greten F.R., Weber C.K., Greten T.F., Schneider G., Wagner M., Adler G., Schmid R.M. Stat3 and NF-kappaB activation prevents apoptosis in pancreatic carcinogenesis. Gastroenterology. 2002;123:2052–2063. doi: 10.1053/gast.2002.37075. [DOI] [PubMed] [Google Scholar]
- 30.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dweep H., Sticht C., Pandey P., Gretz N. miRWalk--database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 33.Gottesman M.M., Pastan I.H. The Role of Multidrug Resistance Efflux Pumps in Cancer: Revisiting a JNCI Publication Exploring Expression of the MDR1 (P-glycoprotein) Gene. J. Natl. Cancer Inst. 2015;107:djv222. doi: 10.1093/jnci/djv222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Radin D., Lippa A., Patel P., Leonardi D. Lifeguard inhibition of Fas-mediated apoptosis: A possible mechanism for explaining the cisplatin resistance of triple-negative breast cancer cells. Biomed Pharmacother. 2016;77:161–166. doi: 10.1016/j.biopha.2015.12.022. [DOI] [PubMed] [Google Scholar]
- 35.Ojini I., Gammie A. Rapid Identification of Chemoresistance Mechanisms Using Yeast DNA Mismatch Repair Mutants. G3 (Bethesda) 2015;5:1925–1935. doi: 10.1534/g3.115.020560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Röckmann H., Schadendorf D. Drug resistance in human melanoma: mechanisms and therapeutic opportunities. Onkologie. 2003;26:581–587. doi: 10.1159/000074156. [DOI] [PubMed] [Google Scholar]
- 37.Aldinucci D., Celegato M., Casagrande N. Microenvironmental interactions in classical Hodgkin lymphoma and their role in promoting tumor growth, immune escape and drug resistance. Cancer Lett. 2016;380:243–252. doi: 10.1016/j.canlet.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 38.Mastri M., Rosario S., Tracz A., Frink R.E., Brekken R.A., Ebos J.M. The challenges of modeling drug resistance to antiangiogenic therapy. Curr. Drug Targets. 2016;17:1747–1754. doi: 10.2174/1389450117666151209123544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gottesman M.M. Mechanisms of cancer drug resistance. Annu. Rev. Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 40.Allen K.E., Weiss G.J. Resistance may not be futile: microRNA biomarkers for chemoresistance and potential therapeutics. Mol. Cancer Ther. 2010;9:3126–3136. doi: 10.1158/1535-7163.MCT-10-0397. [DOI] [PubMed] [Google Scholar]
- 41.Gisel A., Valvano M., El Idrissi I.G., Nardulli P., Azzariti A., Carrieri A., Contino M., Colabufo N.A. miRNAs for the detection of multidrug resistance: overview and perspectives. Molecules. 2014;19:5611–5623. doi: 10.3390/molecules19055611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khalife J., Radomska H.S., Santhanam R., Huang X., Neviani P., Saultz J., Wang H., Wu Y.Z., Alachkar H., Anghelina M. Pharmacological targeting of miR-155 via the NEDD8-activating enzyme inhibitor MLN4924 (Pevonedistat) in FLT3-ITD acute myeloid leukemia. Leukemia. 2015;29:1981–1992. doi: 10.1038/leu.2015.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jagannathan S., Vad N., Vallabhapurapu S., Vallabhapurapu S., Anderson K.C., Driscoll J.J. MiR-29b replacement inhibits proteasomes and disrupts aggresome+autophagosome formation to enhance the antimyeloma benefit of bortezomib. Leukemia. 2015;29:727–738. doi: 10.1038/leu.2014.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaanan A., Okamoto K., Kawakami H., Khazaie K., Huang S., Sinicrope F.A. The Mutant KRAS Gene Up-regulates BCL-XL Protein via STAT3 to Confer Apoptosis Resistance That Is Reversed by BIM Protein Induction and BCL-XL Antagonism. J. Biol. Chem. 2015;290:23838–23849. doi: 10.1074/jbc.M115.657833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen P.M., Cheng Y.W., Wu T.C., Chen C.Y., Lee H. MnSOD overexpression confers cisplatin resistance in lung adenocarcinoma via the NF-κB/Snail/Bcl-2 pathway. Free Radic. Biol. Med. 2015;79:127–137. doi: 10.1016/j.freeradbiomed.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 46.Fischer K.R., Durrans A., Lee S., Sheng J., Li F., Wong S.T., Choi H., El Rayes T., Ryu S., Troeger J. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voulgari A., Pintzas A. Epithelial-mesenchymal transition in cancer metastasis: mechanisms, markers and strategies to overcome drug resistance in the clinic. Biochim. Biophys. Acta. 2009;1796:75–90. doi: 10.1016/j.bbcan.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 48.Jiang J.H., Liu C., Cheng H., Lu Y., Qin Y., Xu Y.F., Xu J., Long J., Liu L., Ni Q.X., Yu X.J. Epithelial-mesenchymal transition in pancreatic cancer: Is it a clinically significant factor? Biochim. Biophys. Acta. 2015;1855:43–49. doi: 10.1016/j.bbcan.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 49.Guo W., Ren D., Chen X., Tu X., Huang S., Wang M., Song L., Zou X., Peng X. HEF1 promotes epithelial mesenchymal transition and bone invasion in prostate cancer under the regulation of microRNA-145. J. Cell. Biochem. 2013;114:1606–1615. doi: 10.1002/jcb.24502. [DOI] [PubMed] [Google Scholar]
- 50.Dai Y., Ren D., Yang Q., Cui Y., Guo W., Lai Y., Du H., Lin C., Li J., Song L., Peng X. The TGF-β signalling negative regulator PICK1 represses prostate cancer metastasis to bone. Br. J. Cancer. 2017;117:685–694. doi: 10.1038/bjc.2017.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang M., Ren D., Guo W., Huang S., Wang Z., Li Q., Du H., Song L., Peng X. N-cadherin promotes epithelial-mesenchymal transition and cancer stem cell-like traits via ErbB signaling in prostate cancer cells. Int. J. Oncol. 2016;48:595–606. doi: 10.3892/ijo.2015.3270. [DOI] [PubMed] [Google Scholar]
- 52.Furukawa T., Kubota T., Watanabe M., Kitajima M., Hoffman R.M. A novel “patient-like” treatment model of human pancreatic cancer constructed using orthotopic transplantation of histologically intact human tumor tissue in nude mice. Cancer Res. 1993;53:3070–3072. [PubMed] [Google Scholar]
- 53.Zhang X., Zhang L., Lin B., Chai X., Li R., Liao Y., Deng X., Liu Q., Yang W., Cai Y. Phospholipid Phosphatase 4 promotes proliferation and tumorigenesis, and activates Ca2+-permeable Cationic Channel in lung carcinoma cells. Mol. Cancer. 2017;16:147. doi: 10.1186/s12943-017-0717-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li X., Liu F., Lin B., Luo H., Liu M., Wu J., Li C., Li R., Zhang X., Zhou K., Ren D. miR-150 inhibits proliferation and tumorigenicity via retarding G1/S phase transition in nasopharyngeal carcinoma. Int. J. Oncol. 2017 doi: 10.3892/ijo.2017.3909. Published online March 10, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang X., Ren D., Guo L., Wang L., Wu S., Lin C., Ye L., Zhu J., Li J., Song L. Thymosin beta 10 is a key regulator of tumorigenesis and metastasis and a novel serum marker in breast cancer. Breast Cancer Res. 2017;19:15. doi: 10.1186/s13058-016-0785-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mermel C.H., Schumacher S.E., Hill B., Meyerson M.L., Beroukhim R., Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.