Summary
Transcriptional pausing by RNA polymerases (RNAPs) is a key mechanism to regulate gene expression in all kingdoms of life and is a prerequisite for transcription termination. The essential bacterial transcription factor NusA stimulates both pausing and termination of transcription, thus playing a central role. Here, we report single-particle electron cryo-microscopy reconstructions of NusA bound to paused E. coli RNAP elongation complexes with and without a pause-enhancing hairpin in the RNA exit channel. The structures reveal four interactions between NusA and RNAP that suggest how NusA stimulates RNA folding, pausing, and termination. An asymmetric translocation intermediate of RNA and DNA converts the active site of the enzyme into an inactive state, providing a structural explanation for the inhibition of catalysis. Comparing RNAP at different stages of pausing provides insights on the dynamic nature of the process and the role of NusA as a regulatory factor.
Keywords: transcription, RNA polymerase structure, transcriptional pausing, NusA, his-pause, cryo-EM
Graphical Abstract
Highlights
-
•
Two cryo-EM reconstructions of paused RNAP elongation complexes bound by NusA
-
•
NusA provides positively charged cavity for RNA structures and stabilizes pause
-
•
Asymmetric translocation intermediate explains transcriptional pausing
-
•
Dynamic process of pausing reflected by RNAP global conformational changes
Guo et al. present cryo-EM structures of paused RNAP bound to NusA. NusA interacts through four points with RNAP and forms a positively charged cavity above the pause-stabilizing RNA hairpin. An asymmetric, half-translocated RNA-DNA hybrid (RNA post-translocated, template DNA pre-translocated) explains transcriptional pausing. NusA further stabilizes the paused RNAP.
Introduction
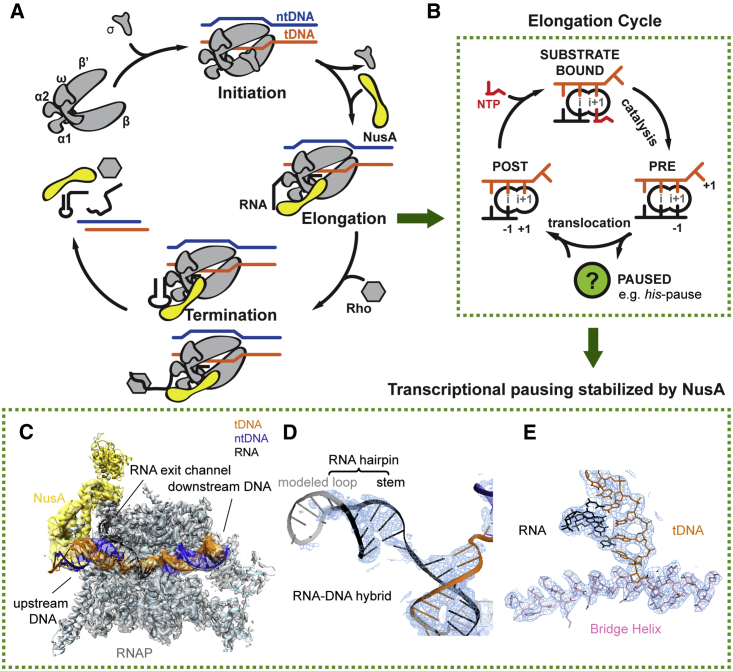
DNA is transcribed into RNA by a protein enzyme called RNA polymerase (RNAP) in all three kingdoms of life. In bacteria, five protein subunits (α2ββ′ω) form a universally conserved core architecture and harbor the functionally relevant sites. Transcription is divided into initiation, elongation, and termination phases (Figures 1A and S1A).
Figure 1.
Schematics of Transcription and Cryo-EM Reconstruction of hisPEC-NusA
(A) Transcription occurs in three distinct phases. During initiation bacterial RNAP binds Sigma factor to bind promoter DNA (top left). After initiation, RNAP enters the elongation phase (right). During elongation transcription factors like NusA (yellow) join. In bacteria, two pathways lead to termination (bottom). At intrinsic terminators, a hairpin in the nascent RNA destabilizes the EC. At Rho terminators, Rho ATPase causes transcript release. NusA stimulates both pathways.
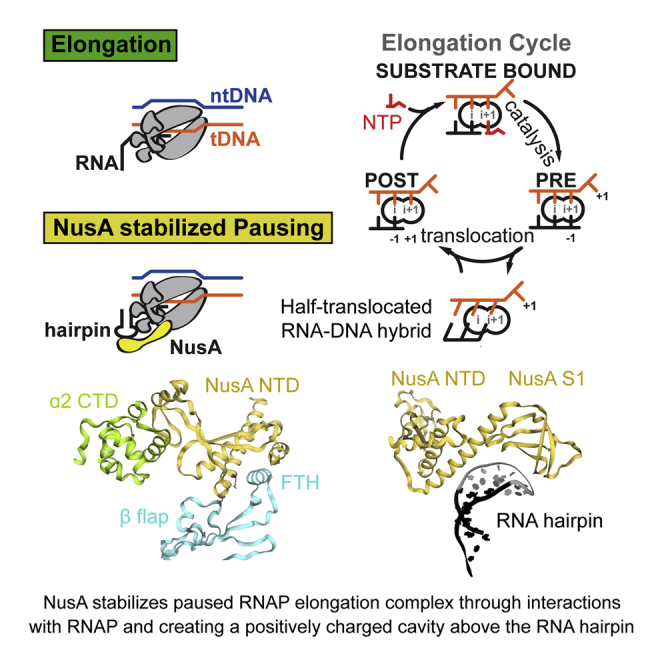
(B) In a post-translocated EC, the RNA 3′ end (position −1) occupies the i-site, while the i+1-site holds the +1 tDNA base and is open for the next NTP substrate to bind (left). Once the correct NTP substrate (red) is bound (top) catalysis can take place leading to a pre-translocated EC, where the RNA 3′ end now occupies the i+1-site (right). Translocation moves RNAP along the DNA by one base pair. When RNAP pauses, it enters an offline state that competes with elongation (green circle).
(C) Overview of hisPEC-NusA cryo-EM structure with RNAP in gray, tDNA in orange, ntDNA in blue, RNA in black, and NusA in yellow envelope.
(D) Representative cryo-EM density (blue mesh) for RNA-DNA hybrid and RNA hairpin is shown with cartoon model superimposed (tDNA orange, ntDNA blue, RNA black).
(E) Representative cryo-EM density (blue mesh) for the active site revealed side chains of the bridge helix (pink) and nucleotides of tDNA (orange) and RNA (black).
See also Table 1 and Figures S1, S2, and S4.
During elongation, RNAP translocates along DNA, progressing through several states. However, elongation is frequently interrupted by offline states, which compete with nucleotide addition. These are called transcriptional pauses (Figure 1B). Pausing regulates gene expression on many levels: (1) it affects RNA synthesis rates and synchronizes transcription and translation (Landick et al., 1985); (2) it facilitates RNA folding (Pan et al., 1999); (3) it enables transcription factor binding to elongation complexes (ECs) (Artsimovitch and Landick, 2002); (4) it plays a role in the regulation by riboswitches (Wickiser et al., 2005); (5) it is a prerequisite for termination (Gusarov and Nudler, 1999, Kassavetis and Chamberlin, 1981); and (6) it is an important, rate limiting step in early elongation of many genes in metazoans (Core and Lis, 2008). Pausing is triggered by the underlying DNA sequence and genome-wide studies identified a consensus pause sequence in E. coli (Larson et al., 2014, Vvedenskaya et al., 2014). Kinetic studies suggested pausing occurs through at least two distinct mechanisms that share a common intermediate called the elemental pause. Class I pauses are stabilized by nascent RNA hairpin structures within the RNAP exit channel. Class II pauses cause RNAP to backtrack along DNA (Artsimovitch and Landick, 2000). One of the best-characterized class I pauses is found in the leader region of the E. coli his operon (his-pause). The his-pause synchronizes transcription and translation, and an active-site rearrangement was proposed to inhibit nucleotide addition (Toulokhonov et al., 2007). Transcription elongation factors further modulate pauses. One of these essential factors, which is conserved in Bacteria and Archaea, is called NusA (Ingham et al., 1999, Shibata et al., 2007). NusA has been studied for over 40 years, and its gene was identified as a requirement for phage lambda protein N-mediated antitermination (Friedman and Baron, 1974). Likewise, NusA is a component in antitermination complexes required for rRNA transcription (Vogel and Jensen, 1997). NusA has also been implicated in facilitating RNA folding (Pan et al., 1999). However, the most apparent role of NusA is to stimulate class I (hairpin stabilized) pauses and intrinsic as well as Rho-dependent termination (Artsimovitch and Landick, 2000). Although structural approaches have been used to shed light on the diverse roles of NusA in complex with RNAP (Said et al., 2017, Yang et al., 2009), we lack high-resolution information to gain mechanistic insights.
NusA is a flexible, multi-domain protein (Figure S1B; Worbs et al., 2001). The N-terminal domain (NusA-NTD) is necessary and sufficient to enhance pausing (Ha et al., 2010). It was proposed to interact with the β subunit flap-tip helix (FTH) and with the pause enhancing RNA hairpin loop (Ha et al., 2010, Mah et al., 1999, Yang et al., 2009). Three RNA binding domains (S1, and two K-homology, KH1 and KH2) follow and some bacterial species, including E. coli, have two C-terminal acidic repeats (AR1, AR2). AR2 can bind the C-terminal domain of one of the RNAP α subunits (α-CTD). This binding releases an auto-inhibitory interaction between AR2 and KH1 and promotes RNA binding to NusA (Mah et al., 2000, Schweimer et al., 2011).
X-ray crystallography provided insights into pausing intermediates. This includes backtracked eukaryotic RNA polymerase II and bacterial RNAPs proposed to be in the elemental pause (Cheung and Cramer, 2011, Wang et al., 2009, Weixlbaumer et al., 2013). However, we lack an understanding of hairpin-mediated pause stabilization and its modulation by transcription factors. Active site extrusion of the RNA 3′ end interrupts transcription in a class II pause, but we do not know what halts catalysis in a class I pause. Here, we report single-particle electron cryo-microscopy (cryo-EM) reconstructions of functional, paused E. coli RNAP ECs at the his-pause bound by NusA with and without a hairpin in the RNA exit channel at 3.6 and 4.1 Å resolution, respectively. The structures explain the inhibition of catalysis and how RNAP accommodates a hairpin in the exit channel and allow us to propose how NusA stimulates RNA folding and stabilizes the paused state. The structures also define new interactions between NusA and RNAP, which we biochemically verified. In addition, the results allow us to speculate about translocation and how NusA may stimulate intrinsic, hairpin-mediated transcription termination, and its role in the process of transcriptional pausing.
Results
Assembly of a Functional, Paused E. coli RNAP EC In Vitro
E. coli RNAP ECs were formed using nucleic acid scaffolds (short DNA and RNA oligonucleotides mimicking a transcription bubble) upstream of the well-characterized his-pause and extended to the pause site to adopt a paused state (hisPEC) (Figure S1C; Chan and Landick, 1993, Kyzer et al., 2007). Pause escape can be measured when GTP is added to extend the RNA further (Figures S1C and S1D). In presence of NusA (hisPEC-NusA), pause dwell times increased 3- to 4-fold, consistent with previous reports (Figure S1E; Artsimovitch and Landick, 2000, Kyzer et al., 2007, Toulokhonov et al., 2001).
For structural studies we assembled hisPECs directly at the pause site in buffer conditions used for grid freezing and purified them by size-exclusion chromatography. Pause escape kinetics of directly reconstituted hisPECs recapitulate the features observed when walking the EC to the pause site, including the effects of NusA (Figures S1F–S1H and S2A).
Structure Determination of the E. coli hisPEC-NusA Complex
An EM reconstruction of the hisPEC-NusA (see STAR Methods for details) revealed electron density attributable to upstream and downstream DNA, the RNA-DNA hybrid, the RNA hairpin in the RNA exit channel, NusA on top of the RNA exit channel, and details of the active site (Figures 1C–1E and S2). The density for NusA was resolved to lower resolution compared to the rest due to its flexibility. 3D classification led to five reconstructions, of which four show different conformations of NusA relative to RNAP. Reconstruction 5 (PEC-NusA) refined to 4.1 Å resolution, lacked hairpin density, and showed weak density for NusA (Figure S3). Reconstruction 1 (hisPEC-NusA) could be refined to 3.6 Å overall with higher resolution at the center, resolving ordered amino acid side chains and base pairs, and lower resolution at the periphery (Figures 1E and S4A–S4C; Table 1). It represents a conformational average of NusA and is the reconstruction we used for most of our analysis (Figure S3B).
Table 1.
Refinement and Model Statistics for the Two Reconstructions of hisPEC-NusA and PEC-NusA, Related to Figure 1
| Data Collection | ||
|---|---|---|
| Particles | 1,038,883 | |
| Pixel size (Å) | 1.1 | |
| Defocus range (μm) | 0.8–3.5 | |
| Voltage (kV) | 300 | |
| Electron dose (e- Å-2) | 53 | |
| hisPEC-NusA | PEC-NusA | |
| Model composition | ||
| Non-hydrogen atoms | 30,201 | 26,512 |
| Protein residues | 3,868 | 3,184 |
| RNA bases | 21 | 10 |
| DNA bases | 70 | 70 |
| Ligands (Zn2+/Mg2+) | 2/1 | 2/1 |
| Refinement | ||
| Resolution (Å) | 3.6 | 4.1 |
| Map sharpening B-factor (Å2) | 142.4 | 130.4 |
| Average B factor (Å2) | 119 | 182 |
| Root-mean-square deviations (RMSDs) | ||
| Bond lengths (Å) | 0.007 | 0.002 |
| Bond angles (°) | 0.927 | 0.506 |
| Ramachandran plot | ||
| Favored (%) | 90.38 | 93.65 |
| Allowed (%) | 9.54 | 6.29 |
| Outliers (%) | 0.08 | 0.06 |
| Molprobity | ||
| Clash score | 3.65 | 8.81 |
| Rotamer outliers (%) | 0.40 | 0.04 |
| Overall score | 1.69 | 1.89 |
Global Conformational Changes in the hisPEC-NusA Complex
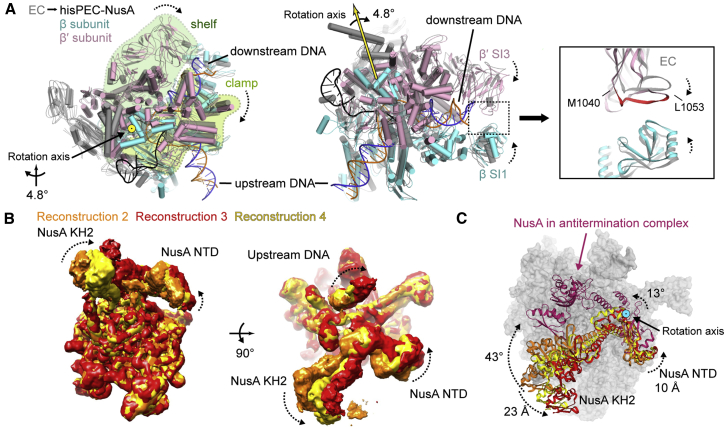
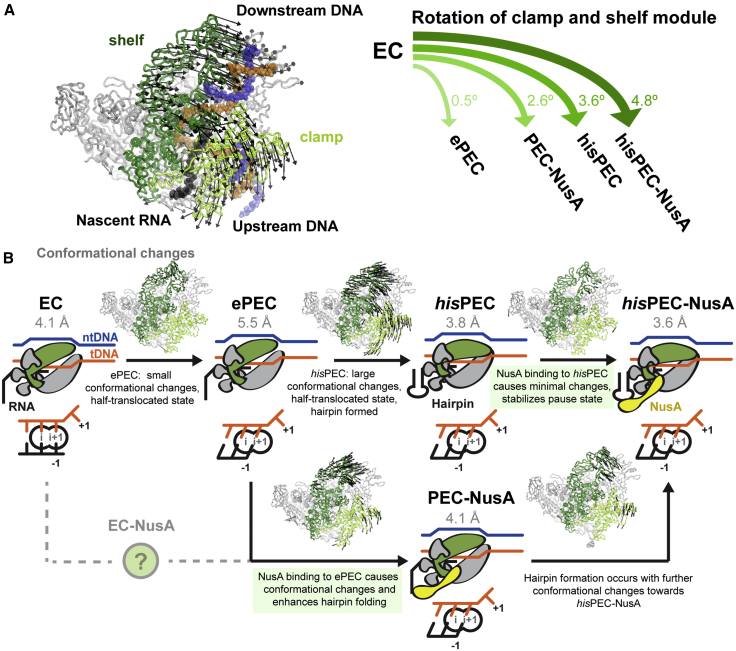
Global conformational changes occurred in all five subunits of RNAP comparing the hisPEC-NusA with a canonical EC structure using the RNAP core module as a reference (Table S2; Kang et al., 2017). Most prominently the RNAP clamp and shelf rotate relative to the core module by ∼4.8° and tilt the downstream DNA relative to the upstream DNA accordingly. The central rotation axis crosses shelf and core and is approximately parallel to the bridge helix (Figure 2A; Movie S1). While the RNAP β1 domain moves away from the main nucleic acid binding channel, β2 moves closer bringing the tips of the β and β′ subunit pincers closer together (Figure 2A; Movie S1). Although not resolved in our maps, this could lead to interactions between residues in the β2 domain sequence insertion I (SI1) and residues in the trigger loop insertion SI3 (Figure 2A; Table S2). Deletions in SI3 (β′ΔT1045-L1053, β′ΔM1040-1048), which might disrupt these interactions, reduce his-pause half-lives 10-fold (Conrad et al., 2010). The global conformational changes appear to be critical for the his-pause and lead to changes on the local level, which we will discuss in the following sections.
Figure 2.
Conformational Changes in the hisPEC-NusA and Structural Heterogeneity of NusA
(A) Superposition of hisPEC-NusA and EC (PDB ID 6ALH) (Kang et al., 2017). Difference in the RNAP clamp and shelf, β, β′ subunit pincers are shown (hisPEC-NusA clamp, light green; shelf, dark green; β, cyan; β′, pink; EC, gray). The RNAP clamp and shelf rotate by 4.8° relative to the core (left). The tips of the pincers are closer, possibly allowing interactions between the β and β′ subunits (right, β′ M1040 to L1053 identified in mutational study are highlighted).
(B) Superposition of three reconstructions shows that RNAP is stable, while NusA and upstream DNA are flexible.
(C) Overlay of three models of NusA orientations (from reconstruction 2, 3, 4) and NusA in antitermination complex (PDB ID 5MS0) (Said et al., 2017). NusA needs to rotate more than 40 degrees from hisPEC to antitermination complex. The rotation axis is highlighted.
See also Figure S3.
Conformational Heterogeneity of NusA in the hisPEC-NusA Complex
Classification led to 4 reconstructions with clear density for NusA. Superposition showed conformational heterogeneity was restricted to NusA and upstream DNA (Figure 2B). However, no distinct states were identified where NusA would refine to high resolution after 3D classification (Figure S3). Compared to the rest, the local resolution in regions corresponding to NusA is lower. In particular, the C-terminal portion can only be seen at low contour level and in low-resolution maps (Figure S3). However, crystal and nuclear magnetic resonance (NMR) structures of all E. coli NusA domains readily fit into the density and allowed us to assess the conformational freedom for the factor (Said et al., 2017, Schweimer et al., 2011). NusA rotates around a pivot point close to the N terminus and close to one of the primary interaction points with RNAP (see next section). In addition, the NTD, S1, KH1, and KH2 domains are flexibly linked with each other and undergo bending motions. The combination of these movements results in displacements of up to 10 Å in the head domain of NusA-NTD and of up to 23 Å at the C-terminal end of KH2 (Figure 2C; Movie S1). The extent of displacement must be even larger for AR1 and AR2, but those regions are disordered in some classes. In summary, NusA has a large conformational freedom in the hisPEC-NusA due to its intrinsic flexibility consistent with previous structural studies on the isolated protein (Drögemüller et al., 2015, Ma et al., 2015, Worbs et al., 2001).
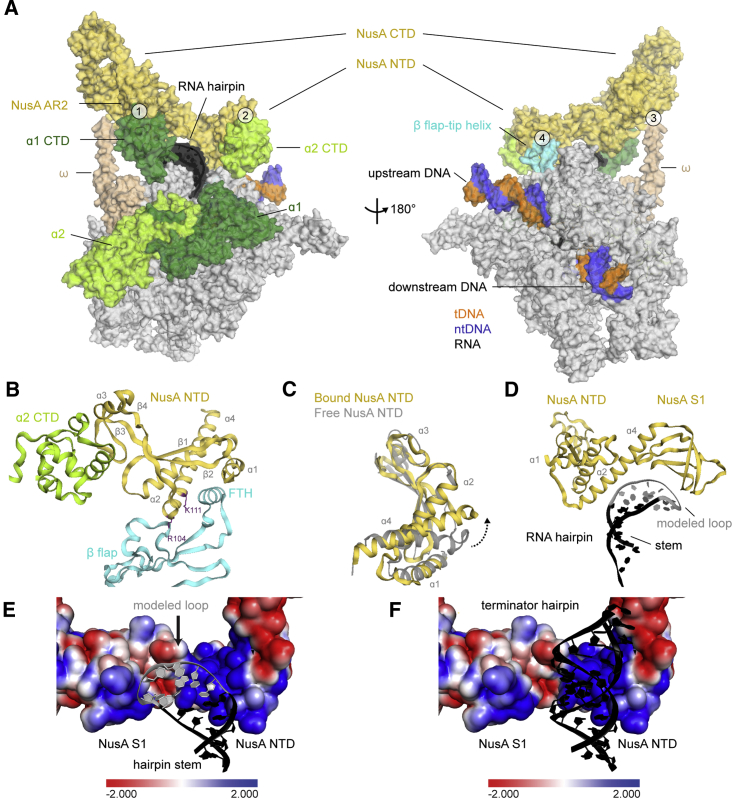
NusA Interacts with the hisPEC through at Least Four Contact Points
Low-resolution maps revealed extra density on the N and C terminus of NusA. On the C terminus, the NMR structure of a complex between NusA-AR2 and the CTD of an RNAP α subunit (α1-CTD) could be fit (PDB ID: 2JZB; Schweimer et al., 2011; Figures 3A and S5A; Movie S1). Additional density extended from the second RNAP α subunit (α2-CTD) toward NusA-NTD. We were able to fit α2-CTD (Figures 3A, S5A, and S5B), revealing an interaction with NusA-NTD not seen before (Figure 3B; Movie S1). At low contour levels, the linker between the N- and CTDs of α2 becomes visible. The α-CTDs are flexibly linked and support NusA’s large conformational freedom (Figures 2B and 2C).
Figure 3.
Interactions between NusA and hisPEC
(A) Surface representation of hisPEC-NusA. RNAP α1 subunit (forest), α2 subunit (lime), ω subunit (wheat), β flap tip helix (cyan), NusA (yellow), RNA hairpin (black), and upstream and downstream DNA (tDNA orange, ntDNA blue) are indicated. The four interaction points are (1) RNAP α1-CTD and NusA-AR2; (2) RNAP α2-CTD and NusA-NTD; (3) RNAP ω and NusA-KH1/KH2; (4) RNAP FTH and NusA-NTD.
(B) Cartoon representation of the interaction between α2-CTD (lime), β-flap region (cyan), and NusA-NTD (yellow). Secondary structure elements and residues identified in mutational studies are labeled.
(C) Comparison of NusA-NTD bound to the hisPEC (yellow) with the solution structure of NusA-NTD (gray) (PDB ID 2KWP). Dashed arrow indicates conformational changes of helix α4 as a result of binding to RNAP.
(D) Cartoon representation of NusA-NTD and S1 domain (yellow) above the RNA stem (black) and modeled hairpin loop (gray).
(E) Electrostatic surface potential of NusA above RNA hairpin (black) shows positively charged regions (blue). The modeled loop is shown in gray.
(F) Like (E) but with the model of a terminator hairpin superimposed on the pause hairpin. A much longer terminator hairpin could be easily accommodated by NusA and would align with a positively charged surface of NusA.
See also Figure S5.
A further interaction occurs between the C-terminal end of the RNAP ω subunit and the interface between the two KH domains (Figures 3A and S5C). The C-terminal residues of ω are often disordered in RNAP structures (Kang et al., 2017, Murakami, 2013) but are stabilized through their interaction with NusA.
Near the RNA exit channel, we observed density for the highly mobile FTH, which is often disordered in ECs (Kang et al., 2017, Vassylyev et al., 2007). The FTH is stabilized in a position distal to the RNA exit channel. It inserts its hydrophobic face (residues L901, L902, I905, and F906) into a hydrophobic cavity created by the α1, α2, and α4 helices of NusA-NTD. This requires a conformational change in the NTD, consistent with data from solution studies (Figures 3B, 3C, S5D, and S5E; Movie S1; Ma et al., 2015).
In the RNA exit channel, density for the 5-bp stem of the RNA hairpin was evident, but the loop region is disordered (Figure 1D). NusA-NTD and S1 extend the exit channel and form a concave, positively charged cradle above the hairpin (Figures 3D and 3E). Interestingly, modeling suggests that NusA could accommodate longer, folded RNA structures, like terminator hairpins, in the present conformation (Figure 3F). Together with NusA-S1, the β′-dock and β′-zinc finger (Table S2) form a positively charged pore providing a path for the nascent transcript leading to the KH domains (Figure S5F).
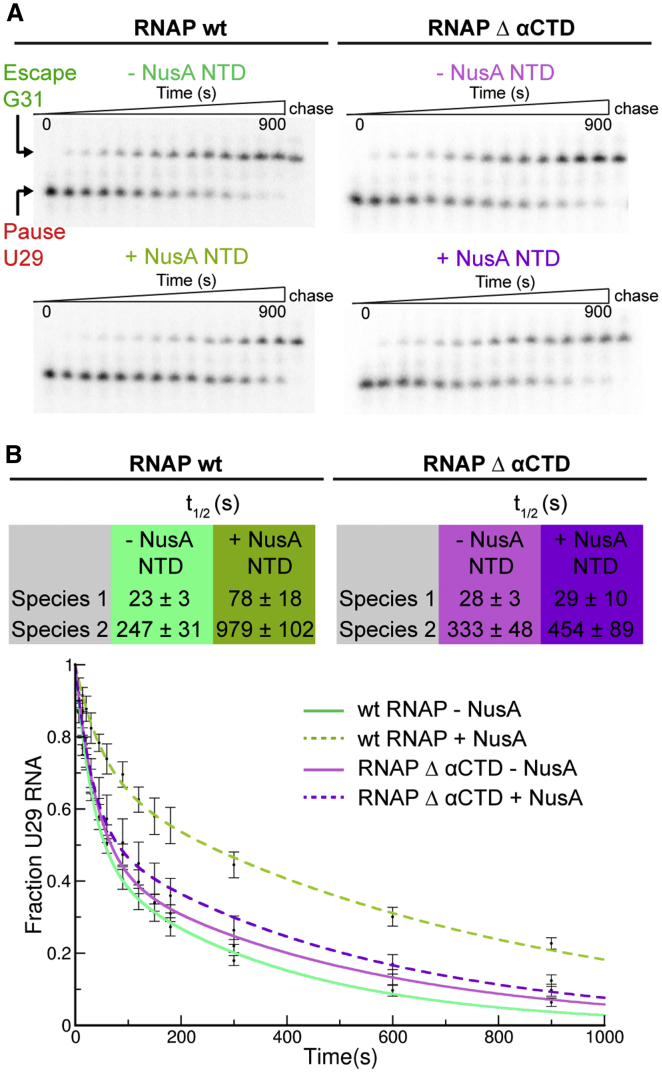
RNAP α-CTD Is Essential for NusA-NTD-Mediated Enhancement of Hairpin Pauses
We wanted to test whether the interaction between NusA-NTD and α2-CTD plays a role in pause enhancement. NusA-NTD is sufficient to enhance pauses, lacks the auto inhibitory AR2 domain, and thus should not require the RNAP α-CTDs unless the interaction with the NusA-NTD is important (Ha et al., 2010). We purified a mutant RNAP without α-CTDs (RNAP-Δα-CTD) (Twist et al., 2011). While NusA-NTD enhanced the his-pause of wild-type RNAP, it failed to do so for RNAP-Δα-CTD (Figures 4A and 4B). Thus, at least for NusA-NTD, the interaction with α2-CTD is important for the enhancement of the hairpin pause.
Figure 4.
Wild-Type RNAP and RNAP-Δα-CTD Respond Differently to NusA-NTD
(A) Wild-type RNAP (RNAPwt) hisPECs were elongated with GTP in the absence (green) or presence (olive) of NusA-NTD. RNAP-Δα-CTD hisPECs were elongated with GTP in absence (pink) or presence (purple) of NusA-NTD. Assays were carried out as described in STAR Methods. Representative gels are shown.
(B) The fraction of RNA29 remaining from at least three independent experiments was plotted as a function of reaction time. The rate of pause escape was determined by nonlinear regression of [U29] versus time using a double exponential decay. Double exponential decay suggested two kinetic species of RNAP with different pause half-lives (t1/2) as seen in the table. For RNAPwt, NusA-NTD enhanced pausing for both species 3- to 4-fold, but this is not true for RNAP-Δα-CTD. Data are represented as mean ± SEM.
The RNA Exit Channel Accommodates the Pause Hairpin
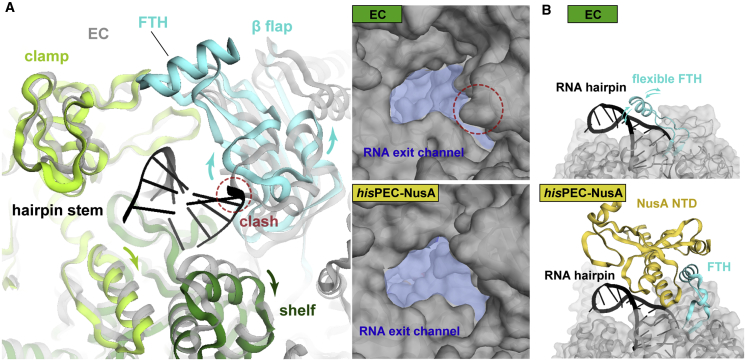
The RNA exit channel, formed by the RNAP shelf, clamp, and β-flap modules, is wider in the hisPEC-NusA complex compared to an EC to accommodate the RNA hairpin. Structural superposition shows the hairpin stem would clash with protein elements in the channel of ECs (Figure 5A; Movie S1). The FTH, which is usually highly flexible in ECs, has previously been shown to delay RNA duplex formation in the exit channel but the effect was relieved by NusA (Hein et al., 2014). Our structure shows NusA stabilizes the FTH in a position distal to the RNA hairpin, explaining how NusA relieves steric interference of duplex formation by the FTH (Figure 5B).
Figure 5.
Conformational Changes in the RNA Exit Channel
(A) Conformational changes of RNA exit channel from EC (gray) (PDB ID 6ALH) (Kang et al., 2017) to hisPEC-NusA (β flap, cyan; shelf, forest; clamp, lime). Dashed circle indicates clash between EC and hairpin stem (left). RNA exit channel (blue area) expands from EC to hisPEC-NusA (right).
(B) Comparison of the FTH (cyan) in the hisPEC-NusA and EC structures. In ECs, the FTH is usually flexible (one possible orientation close to the RNA hairpin is shown here). Binding of NusA-NTD (yellow) to the FTH stabilizes it in a distal position to the RNA hairpin.
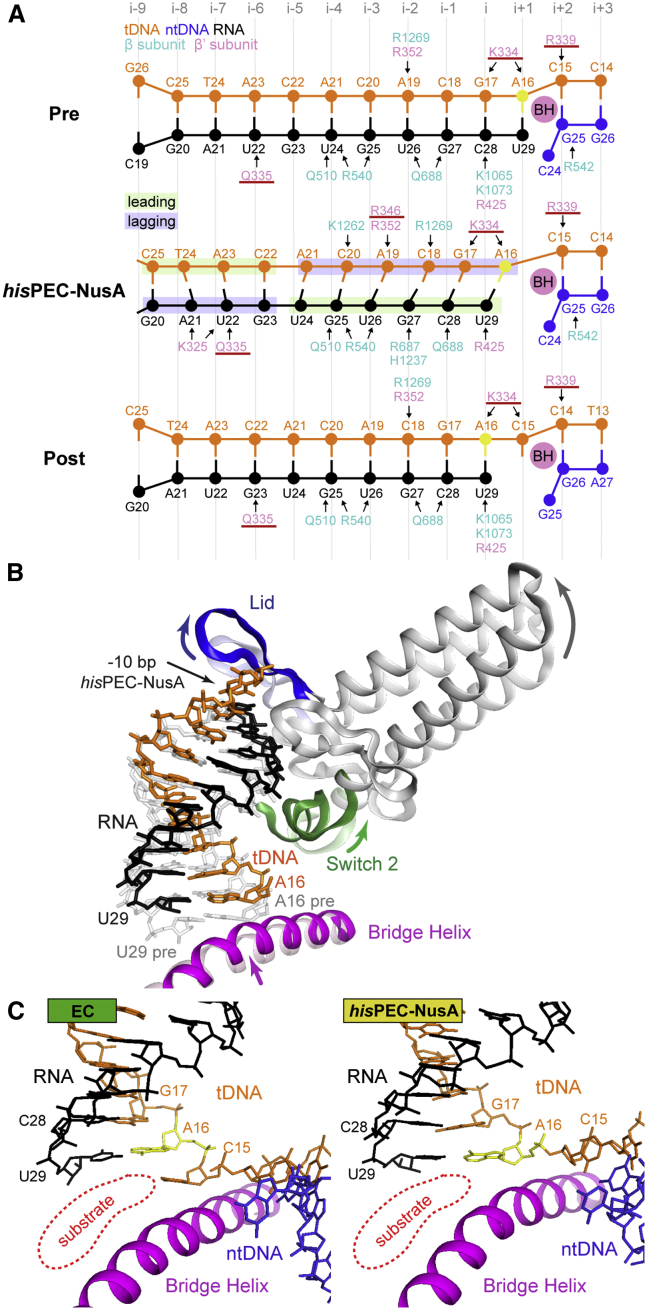
The RNA-DNA Hybrid Adopts a Conformation Different from ECs
We used the structurally rigid core module to compare the hisPEC-NusA to EC structures (Kang et al., 2017, Vassylyev et al., 2007). We noticed two critical differences for the RNA-DNA hybrid in the hisPEC-NusA reconstruction: (1) The RNA-DNA hybrid contains 10 bp. The terminal base pair upstream of the RNA-DNA hybrid (position −10; RNA G20, template DNA (tDNA) C25) is shifted upstream relative to its position in pre- or post-translocated states (Figures 6A and S6A). The lid loop, which is part of the RNAP clamp module, moved upstream as a result of global conformational changes and provides space for the −10 bp (Figures 6B and S6B). (2) The overall conformation of the RNA-DNA hybrid is different compared to ECs. In the hisPEC-NusA, the hybrid base pairs are tilted to various degrees. The RNA strand adopts a post-translocated state, yet the base in position −10 (G20) still pairs with the tDNA. The tDNA on the other hand has not fully translocated and appears in an intermediate, half-translocated position (Figures 6A and 6B). A16 of the tDNA (position −1) moved between the nucleoside triphosphate (NTP) binding site (i+1-site) and the i-site but pairs to the 3′ end of the RNA, which moved to the post-translocated position (i-site) (Figures 6A–6C, and S6A; Movie S1). The next tDNA base downstream (C15, position +1) is still paired to the non-template DNA (ntDNA) strand (G25). It cannot enter the active site, is separated from it by the bridge helix, and thus cannot bind any incoming NTP substrate. The bridge helix connects the RNAP core and shelf modules and is more kinked in the hisPEC-NusA compared to ECs as a result of the global conformational changes. This active site conformation halts the nucleotide addition cycle and provides a structural explanation for the inhibition of catalysis (Figure 6C). Importantly, the same half-translocated hybrid conformation is observed in the hisPEC without NusA by Kang et al., 2018 (this issue of Molecular Cell). Thus, it is not an effect of NusA binding but is likely characteristic for any hairpin stabilized paused state. Kang et al. also propose that SI3 (Table S2) cannot adopt the position required for trigger loop folding in the hisPEC conformation. This provides an additional obstacle for catalysis (Kang et al., 2018).
Figure 6.
RNA-DNA Hybrid Comparison between hisPEC-NusA and EC Structures
(A) Schematic illustration of polar interactions between RNAP and the RNA-DNA hybrid in pre- (top), and post-translocated states (bottom), and for the hisPEC-NusA (middle). Hybrid movement of the hisPEC-NusA was estimated using the ribose moieties of the pre- and post-translocation complex as references. Ribose sugars are shown as circles, bases, and phosphates are shown as lines. Arrows indicate polar interactions. Residues of the RNAP Switch 2 are underlined.
(B) Comparison of Switch 2 (green), clamp helices (gray), lid loop (blue), and bridge helix (pink) between EC (transparent) and hisPEC-NusA (solid). A superposition of a modeled pre-translocated hybrid (gray transparent) and the hybrid of the hisPEC-NusA is also shown (color). In the hisPEC-NusA, the lid loop moved upstream providing space for the −10 base pair. Switch 2, connected to the lid loop through the clamp helices, also moved upstream but maintained contacts to the downstream tDNA and upstream RNA bases it would contact in the pre-translocated state. The bridge helix is slightly kinked.
(C) Active site comparison between the post-translocated EC structure (hisPEC sequence modeled based on PDB ID 6ALH; (Kang et al., 2017) and the hisPEC-NusA. The substrate-binding site is highlighted (red). In contrast to a post-translocated state (EC, left), the next incoming tDNA base (C15) has not yet accommodated in the active site in the hisPEC-NusA because of the half-translocated hybrid.
See also Figures S6 and S7.
For 14% of our particles density for NusA was weak (Figure S3A, reconstruction 5, PEC-NusA, 4.1 Å). At low contour levels density corresponding to NusA-NTD can be seen, and we assume NusA adopts a wider range of conformations in this subset (Figure S3B). No hairpin is visible and the conformation of RNAP is an intermediate between hisPEC-NusA and EC (shelf and clamp rotated by ∼2.6°; Table S1). The RNA-DNA hybrid is still half-translocated and thus RNAP is paused (Figure S6C).
The Half-Translocated State Appears to be a Translocation Intermediate
We modeled pre- and post-translocated ECs using available crystal and EM structures (Kang et al., 2017, Vassylyev et al., 2007). Structural superposition of the models with the hisPEC-NusA suggests an asymmetric translocation progress of the RNA-DNA hybrid relative to RNAP. The upstream tDNA, and downstream RNA are leading, while the downstream tDNA, and upstream RNA are lagging (Figures 6A and S6A).
We compared the RNAP contacts with the RNA-DNA hybrid in different states. RNAP forms equivalent contacts to the hybrid between a pre- and a post-translocated complex, except they are shifted downstream by one nucleotide respectively (Figure 6A, compare top to bottom; Kang et al., 2017).
However, RNAP conformational changes in the hisPEC-NusA altered the contacts with the hybrid: (1) Some contacts are the same as for a post-translocated EC, consistent with the notion that these regions have finished translocation. (2) Others made by residues K334, Q335, and R339 from Switch 2 (Table S2) to the downstream tDNA and upstream RNA are the same as in a pre-translocated EC, consistent with the notion that parts of the hybrid have not finished translocation (Figure 6A). Interestingly, regions contacted by Switch 2 have not completed translocation and, as a result, the RNA-DNA hybrid moved in an asymmetric fashion (Figure 6B).
Discussion
Here, we report a 3.6-Å cryo-EM structure of a hairpin stabilized paused RNAP EC bound to the essential transcription elongation factor NusA (hisPEC-NusA, Figures 1C–1E). A subset of particles allowed us to obtain a 4.1 Å reconstruction of a paused complex without hairpin and more loosely bound NusA (PEC-NusA, Figure S3). The hisPEC-NusA structure explains why catalysis halts in the paused state, provides insights into its dynamic nature, and allows us to propose how NusA prolongs the pause. The structure also provides an opportunity to speculate about translocation of RNAP and about the role of NusA in transcription termination.
Implications of NusA as a Regulatory Factor of Transcription
Our structure revealed four protein-protein interaction points between the hisPEC and NusA (Figure 3A). First, NusA-CTD interacts with the RNAP α1-CTD in agreement with previous findings (Figures 3A and S5A; Mah et al., 2000, Schweimer et al., 2011).
Second, NusA-NTD binds α2-CTD (Figures 3B and S5B). This interaction is consistent with results from NMR titration studies (Drögemüller et al., 2015), except it was interpreted as an interaction between the β′ subunit and the NusA-NTD head (helix α3, sheets β3, β4, and the N-terminal part of β2). We confirmed the functional relevance of this interaction to enhance the hairpin pause (Figure 4). Presumably, it increases the overall affinity and restricts the conformational freedom of NusA-NTD.
Third, the C-terminal end of the RNAP ω subunit binds between the two NusA KH domains (Figures 3A and S5C). Consistent with this observation, truncation of both KH domains affect NusA affinity for RNAP but not function in E. coli (Ha et al., 2010). Importantly, RNA binding to the KH domains, as seen in previous crystal structures, likely requires to break this interaction (Beuth et al., 2005, Said et al., 2017).
Fourth, the NusA-NTD binds the RNAP FTH, which has also been established by cross-linking, low-resolution negative stain EM, NMR, and mutagenesis (Ha et al., 2010, Ma et al., 2015, Toulokhonov et al., 2001, Yang et al., 2009). Furthermore, deletion of the FTH abolished the pause enhancing effect of NusA-NTD (Ha et al., 2010, Toulokhonov et al., 2001). Our structure is consistent and provides a more detailed picture of the FTH interaction with a hydrophobic pocket in NusA-NTD (Figures 3B, S5D, and S5E). Mutations R104A and K111A in NusA helix α4 led to total loss of NusA activity to enhance pausing (Ma et al., 2015). Although not resolved in our reconstruction, K111 could interact with the FTH, while R104 could interact with one of the linkers connecting the FTH to the flap domain (Figure 3B).
The elongated structure and flexibly linked domains of NusA result in a large degree of conformational freedom (Figures 2B, 2C, and S3). In addition, NusA rotates relative to RNAP around a pivot point close to the FTH interaction, consistent with the flexible nature of the FTH. Likewise, to reach the NusA orientation observed in a λN-dependent antitermination complex, it needs to rotate by ∼43° around the same rotation axis (Figure 2C; Said et al., 2017). We suggest the interaction with the FTH serves as an important anchor point to provide NusA with enough flexibility to interact with a plethora of factors involved in pausing, termination, anti-termination, and DNA repair (Cohen et al., 2009, Said et al., 2017).
Apart from these interactions, NusA provides a surface of positively charged residues known to bind RNA (Beuth et al., 2005, Said et al., 2017). The RNA exit channel widens into a positively charged funnel at the surface of RNAP formed by the β′-zinc finger, the β′-dock, and the β-flap. NusA-NTD extends this positively charged channel using conserved residues (Figure S5G). The cavities formed by NusA can accommodate structured RNA and may facilitate RNA folding (Figure 3E). This would explain NusA effects on termination and cotranscriptional RNA folding (Gusarov and Nudler, 2001, Pan et al., 1999). The hairpin loop is disordered, but modeling suggests the positively charged cavities formed by NusA-NTD and S1 would surround it (Figures 3E and S5G). This explains how NusA is able to protect the loops of pause hairpins from RNase cleavage (Ha et al., 2010, Toulokhonov and Landick, 2003). Furthermore, the β′-zinc finger, the β′-dock, Switch3, and the C-terminal residues of the β subunit form a positively charged groove through which the RNA may be guided along the positively charged surface of NusA (Figures S5F and S5G).
Pause hairpin modifications that shift the stem upstream and increase the available surface for NusA to interact with (longer stem, longer spacer between hairpin and hybrid, or formation of long RNA duplex using antisense oligonucleotides) all increase the effect of NusA (Kolb et al., 2014, Toulokhonov et al., 2001). Opposing changes (shorter loop or no hairpin) reduce the effect of NusA (Ha et al., 2010, Toulokhonov et al., 2001). Thus, any change that may increase NusA-RNA interactions enhances the effect of NusA on pause duration. Interactions with structured RNA could restrict and/or stabilize NusA in a conformation required for pause enhancement. This is supported by our PEC-NusA reconstruction, where NusA is even more flexible without a hairpin.
Strikingly, in the PEC-NusA, RNAP adopts an intermediate conformation between EC and hisPEC-NusA presumably caused by NusA binding (relative to an EC, the clamp and shelf rotated ∼4.8° for hisPEC-NusA but only ∼2.6° for PEC-NusA, Figure 7A; Table S1).
Figure 7.
Comparison with Other Paused Complexes and Model for the his-Pause
(A) The shelf and clamp module rotate relative to their position in a EC (left). The extent of rotation is different for various intermediates determined in this work (PEC-NusA, hisPEC-NusA) and by Kang et al. (2018) (ePEC, hisPEC).
(B) Model for RNAP entering the hairpin and NusA stabilized state at the his-pause. RNAP may convert to an ePEC when it encounters a pause sequence with a half-translocated RNA-DNA hybrid. Conformational changes in clamp and shelf module can be trapped by hairpin formation. Binding of NusA induces minimal additional changes and stabilizes paused conformation (top). Alternatively, NusA can bind an EC (resulting RNAP conformation unknown) or ePEC with half-translocated hybrid. RNAP adopts an intermediate conformation as a result of NusA binding. Hairpin formation (stimulated by NusA) leads to the final paused RNAP conformation (bottom). Active site schematics are shown (note that a pre-translocated EC was modeled based on 6ALH).
See also Table S1.
Comparing a reconstruction of a hairpin stabilized hisPEC without NusA (Kang et al., 2018) with our hisPEC-NusA shows only modest conformational changes in RNAP (clamp and shelf rotate an additional ∼1.2°). This suggests NusA binding to a paused complex with pre-formed hairpin mostly stabilizes the existing conformation and does not induce major further changes (Figure 7A).
Finally, using antisense RNAs, Hein et al. noticed that NusA stimulates the rate of RNA duplex formation in the exit channel. Deleting the FTH had the same effect (Hein et al., 2014). This suggests binding of NusA prevents the FTH from interfering with duplex formation.
In summary, we propose two roles for NusA in this context. (1) By guiding the nascent RNA along the positively charged surface, stabilizing the FTH and preventing it from interfering with duplex formation, NusA stimulates formation of hairpin structures in the RNA exit channel (Figure 5B). (2) NusA aids in forming and stabilizing the pause conformation of RNAP by protein-protein interactions with RNAP and electrostatic interactions with the RNA hairpin, thereby increasing pause lifetime.
Implications for the Role of NusA in Transcription Termination
At intrinsic terminators ECs pause at a U-tract and dissociate, and this depends on the formation of a terminator hairpin in the nascent transcript (Gusarov and Nudler, 1999). Unlike for pause hairpins, the terminator hairpin stem extends into the upstream region of the RNA-DNA hybrid. Consequently, it was proposed the terminator hairpin competes with and destabilizes the upstream end of the RNA-DNA hybrid (Gusarov and Nudler, 1999).
NusA stimulates intrinsic termination. Genome-wide studies in B. subtilis suggest that NusA has the most dramatic effect at terminators with weak hairpins and/or distal U-tract interruptions (Mondal et al., 2016). We anticipate that NusA stimulates intrinsic termination similarly to pausing. Thus, NusA could (1) increase the lifetime of the paused state; (2) stimulate terminator hairpin formation; and (3) stabilize terminator hairpins by providing a complementary, positively charged surface (Figures S5F and S5G).
Implications for Translocation
Different hypotheses have been proposed for the mechanism of RNAP translocation (Gelles and Landick, 1998, Komissarova and Kashlev, 1997), but transient intermediates are difficult to capture. To date, translocation intermediates have been proposed for a yeast RNA polymerase II α-amanitin complex (Brueckner and Cramer, 2008) and for single-subunit viral RNA-dependent RNAP where the two RNA strands where captured in an asymmetric translocation intermediate (Shu and Gong, 2016). Similar but different in details to the viral RNAP, the half-translocated RNA-DNA hybrid reported here also suggests to be a translocation intermediate resulting from asymmetric movement, where RNA leads, while tDNA lags behind (Figure 6A). Interestingly, the downstream DNA followed the shelf and clamp module and contacts are similar to a pre-translocated state, consistent with the observation that the tDNA strand has not fully translocated (Figures 6B and S7). Clamp and shelf rotation led to movement of Switch 2, which bridges the RNA-DNA hybrid by interactions with upstream RNA and downstream tDNA. Similarly Switch 2 may be involved in moving the hybrid during translocation. Switch 2 is, both spatially and in terms of primary sequence, in close proximity to the lid loop, which moved upstream to provide space for the −10 bp (Figures 6B and S6B). In the case of the his-pause, formation of an RNA hairpin and binding of NusA stabilized RNAP in a conformation where Switch 2 is in a position that corresponds to incomplete translocation. To finish translocation, Switch 2 needs to break its interactions and re-establish the equivalent ones to the corresponding bases downstream. In addition, the base pairs at position −10 and +1 need to melt so the tDNA base enters the active site. Interestingly, the consensus pause sequence predicts GC base pairs in both −10 and +1 positions and, as suggested by Vvedenskaya et al., may favor the observed state (Larson et al., 2014, Vvedenskaya et al., 2014).
Implications for Transcriptional Pausing
Temporary inhibition of catalysis is a key feature of a paused complex, but how does RNAP achieve this? The current model for transcriptional pausing proposes that RNAP first adopts an elemental paused state (ePEC) triggered by a pause inducing sequence and then isomerizes to more stable paused states like the hisPEC (Herbert et al., 2006, Neuman et al., 2003). Both reconstructions (hisPEC-NusA and PEC-NusA) show the same half-translocated RNA-DNA hybrid (Figure 6), which rendered RNAP catalytically inactive and provides the structural basis for the his-pause. It may likely be a feature of paused RNAP in general including the ePEC. In agreement, Kang et al. propose the RNA-DNA hybrid can adopt a half-translocated state with only minimal rotation of shelf and clamp module based on a 5.5 Å reconstruction (Figure 7; Kang et al., 2018).
Discrepancies to Previous Paused RNAP Structures
Three crystal structures of two bacterial RNAPs (T. thermophilus and T. aquaticus) were proposed to correspond to an ePEC (Weixlbaumer et al., 2013). A large rotation of the clamp module was observed, which is orthogonal to the one observed in the present reconstruction. Crystal packing interactions in the three crystal forms may have caused this difference. Other conformational changes in those structures are reminiscent of changes in the current one. This includes RNA exit channel expansion, bridge helix kinking and an intermediate translocation state. However, the details are different. For example, unlike in the present EM reconstruction, the bridge helix in the crystal structures was more strongly kinked and blocked the active site. As a result, the tDNA was post-translocated but the +1 tDNA base could not enter the active site.
Importantly, minimal scaffolds were used for the crystal structures (no upstream DNA duplex, no ntDNA bubble), which exhibited the expected biochemical behavior but lack the base in the tDNA to form a −10 bp. This likely affects the hybrid and active site conformation and could explain the differences. Finally, we cannot exclude species-specific differences.
A Model for Entering the his-Pause during Translocation
Immediately after catalysis RNAP is pre-translocated (Figure 1B). When it encounters the his-pause (or any sequence resembling the consensus pause), the −10 base pair in the hybrid and the +1 base pair in the downstream DNA duplex are GC base pairs. Melting and thus complete translocation is delayed and RNAP enters an otherwise transient, intermediate, half-translocated state, which halts nucleotide addition and corresponds to the ePEC (Figure 7B). One feature distinguishing the ePEC from an EC is the half-translocated hybrid, which halts nucleotide addition. High-resolution reconstructions of an ePEC in absence of transcription factors will be required to confirm that clamp and shelf rotation do not correlate with the half-translocated hybrid.
Global conformational changes, including clamp and shelf rotation, and exit channel expansion, as observed in the present reconstruction, could simply occur as a result of Brownian motion. Nucleation of a hairpin could block the exit channel and stabilize RNAP in the paused, half-translocated state. Additional binding of NusA would increase the lifetime of the paused state further (Figure 7B, top). Alternatively, NusA could bind an EC or ePEC and induce an intermediate state. The presence of NusA would stimulate hairpin formation and lead to the same end result (Figure 7B, bottom). Concomitant local changes like kinking of the bridge helix, movements of Switch 2 and the lid loop stabilize the asymmetric hybrid conformation.
To escape from the pause, the RNA-DNA hybrid needs to adopt a post-translocated state so a substrate can bind. Presumably, the equilibrium is strongly shifted to the paused state. However, in presence of NTPs, competition between NTP binding to a short-lived post-translocated state and the paused state may determine the rate of escape.
Conclusions
We have reconstituted a functional paused EC of E. coli RNAP at the well-characterized his-pause stabilized by the transcription elongation factor NusA. The complex shows how RNAP accommodates an RNA hairpin in the exit channel and explains why catalysis is inhibited, why a nascent hairpin stabilizes the paused state, how NusA prolongs the pause, and how it may aid RNA structure formation. We also speculate about translocation and the role of NusA at intrinsic terminators.
Follow-up studies will be of great interest. How does NusA interact with a EC or how can elongation factors like NusG reduce pausing? Mutational studies will allow us to test suggestions with respect to translocation and termination.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Escherichia coli LACR II rna-, rnb- (E. coli LOBSTR RNase I and II knock-out strain) | Andersen et al., 2013; and to be published | |
| Escherichia coli BL21(DE3)rpoA_HRV3C_CTD(His)10 (E. coli BL21(DE3) strain with HRV3C site in linker between α-NTD and α-CTD and a C-terminal decahistidine tag | Twist et al., 2011 | |
| Escherichia coli BL21(DE3) rna-, rnb- (E. coli BL21 RNase I and II knock-out strain) | Subbarayan and Deutscher, 2001 | |
| Chemicals, Peptides, and Recombinant Proteins | ||
| pVS11_rpoA_rpoB_rpoCHRV3C(His)10_rpoZ (E. coli RNAP co-expression plasmid for α-, β-, C-terminally His10-tagged β’-, and ω-subunits) | Twist et al., 2011 | |
| pACYC_Duet1_rpoZ (E. coli RNAP ω-subunit expression plasmid) | Twist et al., 2011 | |
| pVS10 (E. coli RNAP co-expression plasmid for α subunits with C-terminal (His)10-tag and an HRV3C site in linker between α-NTD and α-CTD, β-, β’-, and ω-subunits) | Twist et al., 2011 | |
| pET15b_10His_HRV3C_Ec_NusA_FL (E. coli NusA with cleavable N-terminal (His)10-tag) | This work | |
| pET15b_10His_HRV3C_Ec_NusA_NTD (E. coli NusA-NTD with N-terminal (His)10-tag) | This work | |
| Deposited Data | ||
| E. coli hisPEC-NusA | This work | PDB ID: 6FLQ |
| hisPEC-NusA density maps (hisPEC-NusA, 3 NusA orientations, low-res map used to place AR2-α1-CTD) | This work | EMD-4275 |
| E. coli PEC-NusA | This work | PDB ID: 6FLP |
| PEC-NusA density map | This work | EMD-4274 |
| Experimental Models: Organisms/Strains | ||
| Escherichia coli | ||
| Oligonucleotides | ||
| Template DNA (tDNA): CTCTGAATCTCTTCCAGCACAC ATCGGGACGTACTGACC |
This work | |
| Non-template DNA (ntDNA): GGTCAGTACGTCCCGTCG ATCTTCGGAAGAGATTCAGAG |
This work | |
| RNA27: CCUGACUAGUCUUUCAGGCGAUGUGUG | This work | |
| RNA29: CCUGACUAGUCUUUCAGGCGAUGUGUGCU | This work | |
| Software and Algorithms | ||
| Blocres | Cardone et al., 2013 | https://lsbr.niams.nih.gov/bsoft/programs/blocres.html |
| CCP4 suite | Winn et al., 2011 | http://www.ccp4.ac.uk/ |
| COOT v0.8.3 | Emsley and Cowtan, 2004 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| CryoSPARC | Punjani et al., 2017 | https://cryosparc.com |
| CTFFIND4 | Rohou and Grigorieff, 2015 | http://grigoriefflab.janelia.org/ctffind4 |
| EMAN v2.2 | Tang et al., 2007 | http://blake.bcm.edu/emanwiki/EMAN2 |
| Motioncor2 | Zheng et al., 2016 | http://msg.ucsf.edu/em/software/motioncor2.html |
| Phenix suite | Adams et al., 2010 | https://www.phenix-online.org/ |
| PyMOL | Schrodinger, 2015 | https://pymol.org/2/ |
| Relion v2.0 | Scheres, 2012 | https://www2.mrc-lmb.cam.ac.uk/relion/index.php/Main_Page |
| UCSF Chimera v1.11.2 | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/download.html |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Albert Weixlbaumer (albert.weixlbaumer@igbmc.fr).
Experimental Model and Subject Details
For plasmid construction, we used the Escherichia coli (E. coli) TOP10 strain (Invitrogen). For recombinant protein expression, we constructed an E. coli strain, called LACR II (Low Abundance of Cellular RNases). LACR II is derived from the E. coli LOBSTR strain (Andersen et al., 2013) with additional RNase deletions to lower the amount of RNase contamination in purified protein samples (details to be published elsewhere); E. coli strain BL21(DE3) rna- rnb- (Subbarayan and Deutscher, 2001) with RNase I and II knock out was a generous gift from the Deutscher lab; E. coli strain BL21(DE3)rpoA_HRV3C_CTD(His)10 (Twist et al., 2011) with a HRV3C site in the linker between the α subunit CTD and NTD was a generous gift from the Darst lab.
Method Details
Purification of RNAP, NusA, DNA and RNA
E. coli RNAP core enzyme with a C-terminally His10-tagged β’-subunit was overexpressed from pVS11_rpoA_rpoB_rpoCHRV3C(His)10_rpoZ in E. coli LACR II. To avoid substoichiometric amounts of ω, LACR II was co-transformed with pACYC_Duet1_rpoZ. For expression, 6 L of culture in LB (100 μg/ml Ampicillin, 34 μg/ml Chloramphenicol) were induced at an OD600 of 0.6-0.8 with 0.5 mM IPTG for 2 hours at 37°C. To purify E. coli RNAP-Δα-CTD, E. coli BL21(DE3)rpoA_HRV3C_CTD(His)10 was co-transformed with pVS10 and pACYC_Duet1_rpoZ. 6 L of culture in LB (100 μg/ml Ampicillin, 34 μg/ml Chloramphenicol) were induced at an OD600 of 0.6 with 0.5 mM IPTG for O/N at 18°C. E. coli RNAP lacking the α-CTDs was prepared as described previously with minor modifications (Twist et al., 2011). For both wild-type RNAP and RNAP-Δα-CTD purification, cells were harvested by centrifugation, resuspended in 5 volumes of lysis buffer (50 mM Tris-HCl, pH 8.0, 5% glycerol, 1 mM EDTA, 10 mM DTT, 0.1 mM PMSF, 1mM benzamidine, 10 μM ZnCl2, DNase I (0.5 μg/250 g cell), EDTA-free protease inhibitor cocktail (Sigma-Aldrich cOmplete, 1 tablet/50ml) and lysed using sonication. The lysate was cleared using centrifugation at 40,000 g. The RNAP components in the cell lysate were fractionated by polyethyleneimine precipitation followed by ammonium sulfate precipitation as described previously (Vassylyeva et al., 2002). The precipitate was resuspended in IMAC buffer (20 mM Tris-HCl, pH 8.0, 1 M NaCl, 5% glycerol, 5 mM β-mercaptoethanol, 0.1 mM PMSF, 1 mM Benzamidine, 10 μM ZnCl2), passed over a 20 mL Ni-IMAC Sepharose HP column (GE Healthcare) and eluted using a step-gradient into IMAC buffer plus 250 mM imidazole (0 mM Imidazole for 2CVs, 5 mM Imidazole wash for 2CVs, gradient from 5 to 40 mM Imidazole over 1 CV, 40 mM Imidazole for 5CV, and finally step to 250 mM Imidazole). Peak fractions were pooled, and dialyzed overnight in the presence of His-tagged HRV3C (PreScission) protease (1 mg HRV3C per 8 mg of protein) into dialysis buffer (20 mM Tris-HCl, pH 8.0, 1 M NaCl, 5% glycerol, 5 mM β-mercaptoethanol, 10 μM ZnCl2). Uncleaved protein, the cleaved His10-tag and HRV3C were selectively removed using the IMAC column and collecting the flow-through (containing cleaved RNAP). The sample was then dialyzed into Bio-Rex buffer (10 mM Tris-HCl, pH 8.0, 5% glycerol, 0.1 mM EDTA, 0.1 M NaCl, 1 mM DTT, 0.1 mM PMSF, 1 mM Benzamidine, 10 μM ZnCl2) until conductivity was ≤ 10 mS/cm. RNAP was then loaded on a 50 mL Bio-Rex 70 column (BIO-RAD) and eluted using a linear gradient into Bio-Rex buffer plus 1 M NaCl over 5 column volumes. The peak was concentrated and further purified by gel filtration using a HiLoad Superdex 200 PG 26/600 column (GE Healthcare) equilibrated with GF buffer (10 mM HEPES, pH 8.0, 0.5 M KCl, 1% Glycerol, 2 mM DTT, 0.1 mM PMSF, 1 mM benzamidine, 10 μM ZnCl2, 1 mM MgCl2). The final protein was dialyzed into EM buffer (10 mM HEPES, pH 8.0, 150 mM KOAc, 2 mM DTT, 10 μM ZnCl2, 5 mM MgOAc), concentrated to > 50 mg/ml and aliquots were flash frozen and stored at −80°C.
E. coli NusA or NusA-NTD with N-terminal His10-tags were overexpressed in E. coli BL21 (rna- rnb-) strain by inducing 6 L of culture in LB (50 μg/ml Ampicillin) at an OD600 of 0.7 with 1 mM IPTG for 3 hours at 37°C. Cells were harvested by centrifugation, resuspended in 5 volumes of lysis buffer (50 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 10 mM imidazole, 2 mM β-mercaptoethanol, 0.1 mM PMSF, 1mM benzamidine, DNase I (0.5mg/250 g cell), EDTA-free protease inhibitor cocktail (Sigma-Aldrich cOmplete, 1 tablet/50ml) and lysed using sonication. The lysates were cleared by centrifugation at 40,000 g for 30 minutes and loaded onto 2 × 5 mL HiTrap IMAC HP column (GE Healthcare) and eluted using a gradient into lysis buffer containing 250 mM imidazole over 10 CVs. Peak fractions were pooled, and dialyzed overnight in the presence of His-tagged HRV3C (PreScission) protease into lysis buffer plus 50 mM NaCl. Uncleaved protein, the His-tag and protease were selectively removed using the IMAC column and collecting the flow-through (containing cleaved NusA). The sample was then loaded on a 5ml HiTrap Q HP column (GE Healthcare). NusA was eluted using a gradient over 10 CVs into lysis buffer plus 1 M NaCl. The peak was concentrated and further purified by gel filtration using a Superdex 75 16/60 column equilibrated with GF buffer (10 mM HEPES, pH 8.0, 0.15 M NaCl, 0.1 mM EDTA, 1 mM DTT). The final protein was concentrated to > 50 mg/ml, aliquots were flash frozen, and stored at −80°C.
DNA (TriLink) and RNA (Dharmacon) oligonucleotides were chemically synthesized and gel purified by the manufacturer. RNA was deprotected following the protocols provided by the manufacturer. Both DNA and RNA were dissolved in RNase free water and aliquots were stored at −80°C.
Pause assays
E. coli RNAP or E. coli RNAP-Δα-CTD pause assays were carried out by a minimally modified limited step reconstitution assay (Kyzer et al., 2007). Nucleic acid scaffolds for pause assays were reconstituted using a 2-fold molar excess of ntDNA and tDNA over RNA27 (5 μM RNA final, Figure S1C), mixing them in reconstitution buffer (RB, 10 mM Tris-HCl, pH 8.0, 40 mM KCl, 5 mM MgCl2), incubating them for 2 min at 95°C, shifting to 75°C for 2 min followed by a shift to 45°C and slowly cooling them to 25°C in a PCR machine (1°C/min). ECs were formed 2 base pairs upstream of the E. coli his pause site (Figure S1C) in elongation buffer (EB, 10 mM HEPES-KOH pH 8.0, 100 mM KOAc, 5 mM MgOAc, 10 μM ZnCl2, 150 μM EDTA, 5% glycerol, 1 mM DTT) by mixing 0.5 μM scaffold with 1 μM RNAP in the presence of 20 μg/ml BSA. The RNA was labeled by first incubating with 32P-α-CTP (30 μCi) at 37°C, followed by addition of cold CTP (2 μM final) and UTP (100 μM final) resulting in a 29 nt long RNA (U29). To monitor the effect of NusA on the halted hisPEC (RNA29, position U29), NusA or NusA NTD (4 μM final) or elongation buffer EB was added and the complex was incubated for 5 min at 37°C. Transcription pause kinetics were determined by adding the next nucleotide (GTP, 10 μM final) at room temperature, taking samples at predetermined times and quenching with an equal volume of loading buffer (8 M urea, 20 mM EDTA pH 8, 5 mM Tris-HCl pH 7.5, 0.5% bromphenol blue, and 0.5% xylene cyanol). Samples were chased by adding 1 mM GTP and analyzed using denaturing polyacrylamide gels.
For cryo-EM studies, scaffolds were formed with tDNA, ntDNA and RNA29 directly at the his pause site (Figure S1F). To confirm functional complexes in cryo-EM studies, we used a direct reconstitution pause assay (Kyzer et al., 2007). RNA29 was 5′-labeled with 32P-γ-ATP using T4 polynucleotide kinase (NEB) and annealed with a 2-fold molar excess of tDNA, and ntDNA by incubating them in buffer RB for 2 min at 95°C and slowly cooling them to room temperature in a water bath. The hisPEC was formed directly at the his pause site (Figure S1F) in EM buffer plus 8 mM CHAPSO (EB2) by mixing 0.5 μM scaffold with 1 μM RNAP in the presence of 20 μg/ml BSA. To monitor the effect of NusA on the hisPEC (RNA29), NusA (4 μM final) or EM buffer was added and the complex was incubated for 5 min at 37°C. Transcription pause kinetics were determined by adding the next nucleotide (GTP, 10 μM final) at room temperature, taking samples at predetermined times and quenching with an equal volume of loading buffer (8 M urea, 20 mM EDTA pH 8, 5 mM Tris-HCl pH 7.5, 0.5% bromphenol blue, and 0.5% xylene cyanol). Samples were chased by adding 1 mM GTP and analyzed by separating on denaturing polyacrylamide gels.
For data analysis, gels were exposed to storage phosphor screens and quantified using a Typhoon PhosphorImager and ImageQuant software (GE Healthcare). The RNA species in each lane were quantified as a fraction of the total RNA in each lane and corrected for the non-reactive fraction remaining in the chase lane. To monitor the effect of NusA on hisPECs the rate of pause escape was determined by nonlinear regression of [U29] versus time (http://plasma-gate.weizmann.ac.il/Grace/) using a double exponential decay (Figures 4B, S1E, and S1H).
Preparation and cryo-EM analysis of hisPEC-NusA
The hisPEC-NusA complex was assembled by mixing E. coli RNAP, nucleic acid scaffold (tDNA, ntDNA, RNA29, Figure S1F) and NusA with molar ratio of RNAP:scaffold:NusA = 1:3:5 in EM buffer (10 mM HEPES, pH 8.0, 150 mM KOAc, 2 mM DTT, 10 μM ZnCl2, 5 mM MgOAc) and incubated for 15 min at 37°C. Excess proteins and scaffold were removed by gel filtration using a Superdex 200 Increase 10/300 GL column (GE Healthcare) equilibrated in EM buffer. Before freezing, 8 mM CHAPSO was added to the samples. C-flat CF-1.2/1.3 400 mesh holey carbon grids were glow-discharged for 30 s prior to the application of 4 μL of the sample, and plunge-frozen in liquid ethane using a Vitrobot Mark IV (FEI) with 95% chamber humidity at 10°C. Images were recorded on a 300 keV Titan Krios (FEI) equipped with a K2 Summit camera (Gatan, Inc., Pleasanton, CA) operated in super-resolution counting mode with a super-resolution pixel size of 0.55 Å. The detector was placed at the end of a GIF Quantum energy filter (Gatan, Inc.), operated in zero-energy-loss mode with a slit width of 20 eV. To minimize the effects of coincidence loss, the dose rate used was ∼8 e-/pixel/s (equivalent to ∼6.6 e-/Å2 /s at the specimen level). The total exposure time was 8 s and intermediate frames were recorded every 0.2 s giving an accumulated dose of ∼53 e-/Å2 and a total of 40 frames per image. A total of 4,957 movies were recorded with a defocus range from −0.8 to −3.2 μm in super resolution mode. Nominal Magnification was 105,000x. Motion correction and dose weighting was performed using Motioncor2 (Zheng et al., 2016). Parameters of the contrast transfer function (CTF) of each micrograph were estimated with CTFFIND4 (Rohou and Grigorieff, 2015). In a first step, ∼20,000 particles were picked with the semi-automated swarm method using EMAN2 (Tang et al., 2007). Relion (Scheres, 2012) was used for the image processing workflow unless stated otherwise. Reference-free 2D classes were generated, five of which were used for template-based auto-picking after filtering them to 20 Å. After removing bad particles by 2D classification, 476,563 particles were used for 3D refinement. E. coli core RNAP derived from a crystal structure of E. coli holoenzyme (PDB ID 4YG2) (Murakami, 2013) was low pass filtered to 60 Å and used as a reference to yield an initial reconstruction. The reference did not contain any nucleic acids, or any additional ligands, and a ∼100-residue large domain (a part of sequence insertion 3, SI3; also known as β’i6; (Lane and Darst, 2010) was missing. Initial 3D classification using 4x4 binned particles without alignment resulted in several classes. Two contained weak NusA density and no hairpin density (PEC-NusA). Several other classes contained hisPEC-NusA with different orientations of NusA relative to the hisPEC (Figure S3). We merged 65966 particles without hairpin density, and used them in the program cryoSPARC (Punjani et al., 2017) for ab initio 3D classification and refinement. This yielded a structure (reconstruction 5, PEC-NusA) at 4.1 Å with weak density for NusA and no density for the RNA hairpin. During initial classification, one class (reconstruction 6) showed better density for NusA AR2 and RNAP α1 CTD, it was refined to 3.9 Å and low-pass filtered to 10 Å for model fitting. 379918 particles that showed density for both hisPEC and NusA were merged for subsequent refinement and 3D classification. Subsequent 3D classification using 2x2 binned particles without a mask yielded four classes with differences in densities corresponding to NusA and the upstream DNA duplex (Figure S3). Among these four classes, one major class (reconstruction 1) showed density that we consider being the average of NusA movements, while three other classes (reconstruction 2, 3, 4) represent extremes of the NusA orientations relative to the hisPEC (Figure S3B). The major class containing 157,100 particles was refined in cryoSPARC to a final resolution of 3.6 Å using the gold-standard Fourier shell correlation (FSC) 0.143 criterion. The three classes with extreme NusA orientations contained 10,381, 34,632 and 50,210 particles and were refined to 6.5 Å, 4.2 Å and 4.2 Å, respectively (Figure S3A). To avoid influences from the upstream DNA duplex on classification, we performed another 3D classification of the same particles with a mask excluding density from upstream DNA. This yielded four classes with differences only in the NusA orientations, without influence from upstream DNA duplex flexibility. These classes with different NusA orientations resembled those from unmasked 3D classification (data not shown) Local resolution calculation was performed using blocres (Cardone et al., 2013).
Structural modeling of hisPEC-NusA
To generate a model of hisPEC-NusA, we started by using models derived from the cryo-EM structure of E. coli RNAP core (PDB ID 6ALH), the crystal structure of E. coli NusA NTD, S1, KH1, KH2, and AR1 (PDB ID 5LM7), and an NMR structure of E. coli NusA AR2 domain in complex with Yersinia pseudotuberculosis RNAP α-subunit CTD (PDB ID 2JZB) (Kang et al., 2017, Said et al., 2017, Schweimer et al., 2011). The initial model was fit in COOT and refined using Phenix real-space-refine (Adams et al., 2010). Initially, RNAP and NusA domains were placed and refined as rigid bodies. Subsequently, after visual inspection and rebuilding, the model was further refined using secondary structure restraints. RNA-DNA hybrid and downstream DNA duplex were built de novo in COOT and real-space refined (Figures 1C–1E). Density for the RNA hairpin revealed only the 5 base pair stem but no defined density for the loop (Figure 1D). The 5 base pair hairpin stem was built de novo in COOT, while the 8-nucleotide long loop was modeled using existing loop structures with Rloom (Schudoma et al., 2010) and placed above the stem in COOT (Figure 1D). Due to conformational heterogeneity, the quality of the density for NusA was variable. Density for the NusA-NTD and RNAP α2-CTD allowed us to clearly identify secondary structure elements and place them followed by rigid-body refinement (Figure S5B). In contrast, the density for NusA S1, KH1, KH2 is poorly resolved and only allowed us to place these domains as rigid bodies. Density for NusA AR1 AR2 and RNAP α1 CTD are only visible in low-resolution maps where we could also fit crystal and NMR structures based on the shape and size of the density (Figure S5A). In the final five reconstructions with or without NusA, RNAP core adopted the same conformation. The map, which refined to 3.6 Å was used for model building. NusA was split into 6 rigid bodies based on its domain definition and were separately fitted and adjusted in COOT into the four reconstructions containing NusA. Model overfitting was evaluated by refining the models in one of the two independent half maps. The original atom positions were randomly displaced up to 0.5 Å and refined with restraints in Phenix against one of the two-independent half maps prior to FSC calculations. FSC curves were calculated between the resulting model and both the half maps, independently, for cross-validation (Figure S4F). Figures were prepared with UCSF Chimera (Pettersen et al., 2004) or PyMOL (Schrodinger, LLC, 2015).
Data and Software Availability
The accession number for the five cryo-EM reconstructions (hisPEC-NusA, three different NusA orientations, and the low-resolution reconstruction that allowed us to place the AR2-α1-CTD interaction using PDB: 2JZB) reported in this paper is EMD: 4275. The accession number for the PEC-NusA reconstruction reported in this paper is EMD: 4274. The accession numbers for the fitted models reported in this paper are PDB: 6FLQ (all atom model for hisPEC-NusA except for NusA and the two α-CTDs, which are modeled as Poly-Alanine) and 6FLP (all atom model for PEC-NusA except NusA was not modeled).
Acknowledgments
We thank Jin Young Kang, Seth A. Darst, and Robert Landick for insightful discussions and sharing their results. The authors were supported by the French Infrastructure for Integrated Structural Biology (FRISBI ANR-10-INBS-05, Instruct-ERIC, and grant ANR-10-LABX-0030-INRT, a French State fund managed by the Agence Nationale de la Recherche under the program Investissements d’Avenir ANR-10-IDEX-0002-02). The work was supported by a LabEx PhD fellowship to X.G. and the ERC starting grant TRANSREG (679734) to A.W.
Author Contributions
Conceptualization, A.W. and X.G.; Sample Preparation, X.G., J.C., and M.T.; Data Collection and Processing, X.G., A.G.M., C.C., G.P., and P.S.; Investigation and Analysis, X.G. and A.W.; Writing, X.G. and A.W.; Supervision, A.W.
Declaration of Interests
The authors declare no competing interests.
Published: March 1, 2018
Footnotes
Supplemental Information includes seven figures, two tables, and one movie and can be found with this article online at https://doi.org/10.1016/j.molcel.2018.02.008.
Supplemental Information
Movie S1 first shows an overview of RNAP (subunits labeled) and then illustrates the global conformational change as a results of entering the his-pause (RNAP modules labeled). The focus shifts to the NusA-NTD binding site (RNAP FTH cyan, RNA hairpin black). It shows the conformational change in NusA-NTD required to bind the RNAP FTH and stabilization of the FTH in a distal position relative to the RNA exit channel. Next we see the conformational flexibility of NusA and the four interaction points with RNAP (2 α-CTDs shades of green, FTH cyan, and C-terminal end of RNAP ω-subunit wheat). The movie then shows the conformational change in the RNA exit channel and how a hairpin would clash with the narrow channel seen in an EC. Finally the movie compares canonical translocation with the half-translocated state observed in the his-pause.
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K.R., Leksa N.C., Schwartz T.U. Optimized E. coli expression strain LOBSTR eliminates common contaminants from His-tag purification. Proteins. 2013;81:1857–1861. doi: 10.1002/prot.24364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I., Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc. Natl. Acad. Sci. USA. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I., Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- Beuth B., Pennell S., Arnvig K.B., Martin S.R., Taylor I.A. Structure of a Mycobacterium tuberculosis NusA-RNA complex. EMBO J. 2005;24:3576–3587. doi: 10.1038/sj.emboj.7600829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueckner F., Cramer P. Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nat. Struct. Mol. Biol. 2008;15:811–818. doi: 10.1038/nsmb.1458. [DOI] [PubMed] [Google Scholar]
- Cardone G., Heymann J.B., Steven A.C. One number does not fit all: Mapping local variations in resolution in cryo-EM reconstructions. J. Struct. Biol. 2013;184:226–236. doi: 10.1016/j.jsb.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.L., Landick R. Dissection of the his leader pause site by base substitution reveals a multipartite signal that includes a pause RNA hairpin. J. Mol. Biol. 1993;233:25–42. doi: 10.1006/jmbi.1993.1482. [DOI] [PubMed] [Google Scholar]
- Cheung A.C.M., Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- Cohen S.E., Godoy V.G., Walker G.C. Transcriptional modulator NusA interacts with translesion DNA polymerases in Escherichia coli. J. Bacteriol. 2009;191:665–672. doi: 10.1128/JB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad T.M., Frazier M., Joyce A.R., Cho B.-K., Knight E.M., Lewis N.E., Landick R., Palsson B.Ø. RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc. Natl. Acad. Sci. USA. 2010;107:20500–20505. doi: 10.1073/pnas.0911253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L.J., Lis J.T. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drögemüller J., Strauß M., Schweimer K., Jurk M., Rösch P., Knauer S.H. Determination of RNA polymerase binding surfaces of transcription factors by NMR spectroscopy. Sci. Rep. 2015;5:16428. doi: 10.1038/srep16428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Friedman D.I., Baron L.S. Genetic characterization of a bacterial locus involved in the activity of the N function of phage lambda. Virology. 1974;58:141–148. doi: 10.1016/0042-6822(74)90149-4. [DOI] [PubMed] [Google Scholar]
- Gelles J., Landick R. RNA polymerase as a molecular motor. Cell. 1998;93:13–16. doi: 10.1016/s0092-8674(00)81140-x. [DOI] [PubMed] [Google Scholar]
- Gusarov I., Nudler E. The mechanism of intrinsic transcription termination. Mol. Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- Gusarov I., Nudler E. Control of intrinsic transcription termination by N and NusA: The basic mechanisms. Cell. 2001;107:437–449. doi: 10.1016/s0092-8674(01)00582-7. [DOI] [PubMed] [Google Scholar]
- Ha K.S., Toulokhonov I., Vassylyev D.G., Landick R. The NusA N-terminal domain is necessary and sufficient for enhancement of transcriptional pausing via interaction with the RNA exit channel of RNA polymerase. J. Mol. Biol. 2010;401:708–725. doi: 10.1016/j.jmb.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein P.P., Kolb K.E., Windgassen T., Bellecourt M.J., Darst S.A., Mooney R.A., Landick R. RNA polymerase pausing and nascent-RNA structure formation are linked through clamp-domain movement. Nat. Struct. Mol. Biol. 2014;21:794–802. doi: 10.1038/nsmb.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert K.M., La Porta A., Wong B.J., Mooney R.A., Neuman K.C., Landick R., Block S.M. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 2006;125:1083–1094. doi: 10.1016/j.cell.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham C.J., Dennis J., Furneaux P.A. Autogenous regulation of transcription termination factor Rho and the requirement for Nus factors in Bacillus subtilis. Mol. Microbiol. 1999;31:651–663. doi: 10.1046/j.1365-2958.1999.01205.x. [DOI] [PubMed] [Google Scholar]
- Kang J.Y., Olinares P.D.B., Chen J., Campbell E.A., Mustaev A., Chait B.T., Gottesman M.E., Darst S.A. Structural basis of transcription arrest by coliphage HK022 Nun in an Escherichia coli RNA polymerase elongation complex. eLife. 2017 doi: 10.7554/eLife.25478. Published online March 20, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.Y., Mishanina T.V., Bellecourt M.J., Mooney R.A., Darst S.A., Landick R. RNA polymerase accommodates a pause RNA hairpin by global conformational rearrangements that prolong pausing. Mol. Cell. 2018;69:802–815. doi: 10.1016/j.molcel.2018.01.018. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G.A., Chamberlin M.J. Pausing and termination of transcription within the early region of bacteriophage T7 DNA in vitro. J. Biol. Chem. 1981;256:2777–2786. [PubMed] [Google Scholar]
- Kolb K.E., Hein P.P., Landick R. Antisense oligonucleotide-stimulated transcriptional pausing reveals RNA exit channel specificity of RNA polymerase and mechanistic contributions of NusA and RfaH. J. Biol. Chem. 2014;289:1151–1163. doi: 10.1074/jbc.M113.521393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komissarova N., Kashlev M. RNA polymerase switches between inactivated and activated states By translocating back and forth along the DNA and the RNA. J. Biol. Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- Kyzer S., Ha K.S., Landick R., Palangat M. Direct versus limited-step reconstitution reveals key features of an RNA hairpin-stabilized paused transcription complex. J. Biol. Chem. 2007;282:19020–19028. doi: 10.1074/jbc.M701483200. [DOI] [PubMed] [Google Scholar]
- Landick R., Carey J., Yanofsky C. Translation activates the paused transcription complex and restores transcription of the trp operon leader region. Proc. Natl. Acad. Sci. USA. 1985;82:4663–4667. doi: 10.1073/pnas.82.14.4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane W.J., Darst S.A. Molecular evolution of multisubunit RNA polymerases: Structural analysis. J. Mol. Biol. 2010;395:686–704. doi: 10.1016/j.jmb.2009.10.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson M.H., Mooney R.A., Peters J.M., Windgassen T., Nayak D., Gross C.A., Block S.M., Greenleaf W.J., Landick R., Weissman J.S. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014;344:1042–1047. doi: 10.1126/science.1251871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Mobli M., Yang X., Keller A.N., King G.F., Lewis P.J. RNA polymerase-induced remodelling of NusA produces a pause enhancement complex. Nucleic Acids Res. 2015;43:2829–2840. doi: 10.1093/nar/gkv108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah T.F., Li J., Davidson A.R., Greenblatt J. Functional importance of regions in Escherichia coli elongation factor NusA that interact with RNA polymerase, the bacteriophage lambda N protein and RNA. Mol. Microbiol. 1999;34:523–537. doi: 10.1046/j.1365-2958.1999.01618.x. [DOI] [PubMed] [Google Scholar]
- Mah T.F., Kuznedelov K., Mushegian A., Severinov K., Greenblatt J. The alpha subunit of E. coli RNA polymerase activates RNA binding by NusA. Genes Dev. 2000;14:2664–2675. doi: 10.1101/gad.822900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal S., Yakhnin A.V., Sebastian A., Albert I., Babitzke P. NusA-dependent transcription termination prevents misregulation of global gene expression. Nat. Microbiol. 2016;1:15007. doi: 10.1038/nmicrobiol.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K.S. The X-ray crystal structure of Escherichia coli RNA polymerase Sigma70 holoenzyme. J. Biol. Chem. 2013;288:9126–9134. doi: 10.1074/jbc.M112.430900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman K.C., Abbondanzieri E.A., Landick R., Gelles J., Block S.M. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- Pan T., Artsimovitch I., Fang X.W., Landick R., Sosnick T.R. Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proc. Natl. Acad. Sci. USA. 1999;96:9545–9550. doi: 10.1073/pnas.96.17.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Rohou A., Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Said N., Krupp F., Anedchenko E., Santos K.F., Dybkov O., Huang Y.-H., Lee C.-T., Loll B., Behrmann E., Bürger J. Structural basis for λN-dependent processive transcription antitermination. Nat. Microbiol. 2017;2:17062. doi: 10.1038/nmicrobiol.2017.62. [DOI] [PubMed] [Google Scholar]
- Scheres S.H.W. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger, LLC, (2015). The PyMOL Molecular Graphics System, Version 1.8.
- Schudoma C., May P., Nikiforova V., Walther D. Sequence-structure relationships in RNA loops: Establishing the basis for loop homology modeling. Nucleic Acids Res. 2010;38:970–980. doi: 10.1093/nar/gkp1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweimer K., Prasch S., Sujatha P.S., Bubunenko M., Gottesman M.E., Rösch P. NusA interaction with the α subunit of E. coli RNA polymerase is via the UP element site and releases autoinhibition. Structure. 2011;19:945–954. doi: 10.1016/j.str.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata R., Bessho Y., Shinkai A., Nishimoto M., Fusatomi E., Terada T., Shirouzu M., Yokoyama S. Crystal structure and RNA-binding analysis of the archaeal transcription factor NusA. Biochem. Biophys. Res. Commun. 2007;355:122–128. doi: 10.1016/j.bbrc.2007.01.119. [DOI] [PubMed] [Google Scholar]
- Shu B., Gong P. Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation. Proc. Natl. Acad. Sci. USA. 2016;113:E4005–E4014. doi: 10.1073/pnas.1602591113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarayan P.R., Deutscher M.P. Escherichia coli RNase M is a multiply altered form of RNase I. RNA. 2001;7:1702–1707. [PMC free article] [PubMed] [Google Scholar]
- Tang G., Peng L., Baldwin P.R., Mann D.S., Jiang W., Rees I., Ludtke S.J. EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Toulokhonov I., Landick R. The flap domain is required for pause RNA hairpin inhibition of catalysis by RNA polymerase and can modulate intrinsic termination. Mol. Cell. 2003;12:1125–1136. doi: 10.1016/s1097-2765(03)00439-8. [DOI] [PubMed] [Google Scholar]
- Toulokhonov I., Artsimovitch I., Landick R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science. 2001;292:730–733. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- Toulokhonov I., Zhang J., Palangat M., Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol. Cell. 2007;27:406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Twist K.-A.F., Husnain S.I., Franke J.D., Jain D., Campbell E.A., Nickels B.E., Thomas M.S., Darst S.A., Westblade L.F. A novel method for the production of in vivo-assembled, recombinant Escherichia coli RNA polymerase lacking the α C-terminal domain. Protein Sci. 2011;20:986–995. doi: 10.1002/pro.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassylyev D.G., Vassylyeva M.N., Zhang J., Palangat M., Artsimovitch I., Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- Vassylyeva M.N., Lee J., Sekine S.I., Laptenko O., Kuramitsu S., Shibata T., Inoue Y., Borukhov S., Vassylyev D.G., Yokoyama S. Purification, crystallization and initial crystallographic analysis of RNA polymerase holoenzyme from Thermus thermophilus. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1497–1500. doi: 10.1107/S0907444902011770. [DOI] [PubMed] [Google Scholar]
- Vogel U., Jensen K.F. NusA is required for ribosomal antitermination and for modulation of the transcription elongation rate of both antiterminated RNA and mRNA. J. Biol. Chem. 1997;272:12265–12271. doi: 10.1074/jbc.272.19.12265. [DOI] [PubMed] [Google Scholar]
- Vvedenskaya I.O., Vahedian-Movahed H., Bird J.G., Knoblauch J.G., Goldman S.R., Zhang Y., Ebright R.H., Nickels B.E. Interactions between RNA polymerase and the “core recognition element” counteract pausing. Science. 2014;344:1285–1289. doi: 10.1126/science.1253458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Bushnell D.A., Huang X., Westover K.D., Levitt M., Kornberg R.D. Structural basis of transcription: Backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324:1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixlbaumer A., Leon K., Landick R., Darst S.A. Structural basis of transcriptional pausing in bacteria. Cell. 2013;152:431–441. doi: 10.1016/j.cell.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickiser J.K., Winkler W.C., Breaker R.R., Crothers D.M. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G.W., McCoy A. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worbs M., Bourenkov G.P., Bartunik H.D., Huber R., Wahl M.C. An extended RNA binding surface through arrayed S1 and KH domains in transcription factor NusA. Mol. Cell. 2001;7:1177–1189. doi: 10.1016/s1097-2765(01)00262-3. [DOI] [PubMed] [Google Scholar]
- Yang X., Molimau S., Doherty G.P., Johnston E.B., Marles-Wright J., Rothnagel R., Hankamer B., Lewis R.J., Lewis P.J. The structure of bacterial RNA polymerase in complex with the essential transcription elongation factor NusA. EMBO Rep. 2009;10:997–1002. doi: 10.1038/embor.2009.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Palovcak E., Armache J.-P., Cheng Y., Agard D. Anisotropic correction of beam-induced motion for improved single-particle electron cryo-microscopy. bioRxiv. 2016 doi: 10.1038/nmeth.4193. https://doi.org/10.1101/061960 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1 first shows an overview of RNAP (subunits labeled) and then illustrates the global conformational change as a results of entering the his-pause (RNAP modules labeled). The focus shifts to the NusA-NTD binding site (RNAP FTH cyan, RNA hairpin black). It shows the conformational change in NusA-NTD required to bind the RNAP FTH and stabilization of the FTH in a distal position relative to the RNA exit channel. Next we see the conformational flexibility of NusA and the four interaction points with RNAP (2 α-CTDs shades of green, FTH cyan, and C-terminal end of RNAP ω-subunit wheat). The movie then shows the conformational change in the RNA exit channel and how a hairpin would clash with the narrow channel seen in an EC. Finally the movie compares canonical translocation with the half-translocated state observed in the his-pause.