Abstract
The RNA-binding protein, UPF1, is best known as the central factor in the nonsense-mediated RNA decay (NMD) pathway. Feng et al. now report a new function for UPF1 – it is an E3 ubiquitin ligase that specifically promotes the decay of a key pro-muscle transcription factor: MYOD. UPF1 achieves this through its RING-like domain, which confers ubiquitin E3 ligase activity. Physiological relevance for UPF1’s E3 ligase activity is supported by two findings: (i) UPF1 suppresses myogenesis and (ii) disruption of UPF1’s E3 ligase activity alleviates this repression. In the future, it will be important to define other protein substrates of UPF1-driven ubiquitination and to determine whether this biochemical activity is responsible for some of UPF1’s many biological functions, including in development and stress responses. The exciting findings presented by Feng et al. open up the possibility that protein turnover and RNA turnover are coupled processes.
Keywords: Nonsense Mediated Decay, development, ubiquitination, proteasome, myogenesis, E3 ligase
Introduction
NMD is a RNA degradation pathway originally discovered by virtue of its ability to degrade aberrant RNAs harboring premature termination codons (PTCs). In this role, NMD serves as a quality control pathway that protects cells from the potential deleterious effects of the truncated protein products arising from PTC-containing mRNAs [1]–[3]. More recently, NMD has been shown to selectively degrade a subset of normal mRNAs in species spanning the phylogenetic scale [1], [4]. In addition, it has been shown that NMD activity varies between cell types, tissues, and developmental stages and a number of post-transcriptional regulatory mechanisms modulating NMD activity have been identified [5]. These new findings have led to the view that modulations in NMD magnitude lead to alterations in the transcriptome to achieve specific biological outcomes. For example, NMD is regarded as being important for a wide variety of developmental events, based on the developmental defects that occur as a result of loss or knockdown of NMD factors in a variety of species [6]. While most studies have focused on how loss of RNA decay might contribute to these defects, it is also possible that auxiliary roles of NMD factors could instead be responsible, a theme we touch on later in this commentary.
Arguably the most critical protein in the NMD pathway is UPF1, a RNA helicase involved in nearly every step of the NMD process, from target discrimination [7], to remodeling the NMD-mRNA complex and enhancing nuclease access to the mRNA [8]. UPF1 is thought to mediate the degradation of NMD substrates through a phosphorylation/dephosphorylation cycle. While the precise role of this cycle remains unclear, evidence suggests that phosphorylation of UPF1 is critical for NMD, while dephosphorylation allows for UPF1 recycling [9].
UPF1 has been ascribed many biological roles. For example, UPF1 is thought to be critical for early embryonic development in mammals, based on the finding that Upf1-null mice suffer from early embryonic lethality [10]. Loss of UPF1 also causes embryonic lethality in zebrafish and flies [11][12], but has only minimal impact on simpler organisms such as C. elegans and S. cerevisiae [6]. While UPF1’s underlying mechanism of action in development is largely unknown, some progress has been made. For example, UPF1 is downregulated during the process of myogenesis [13], suggesting that this downregulatory response accelerates and possibly amplifies the generation of muscle cells during myogenesis. By analogy, UPF1 downregulation also occurs during neural differentiation; gain- and loss-of-function experiments have shown this is both necessary and sufficient to drive this differentiation event [14][15].
While it is often assumed that these biological activities of UPF1 derive from its well-established role in NMD, UPF1 also functions in other pathways. For example, it participates in histone mRNA decay as well as the staufen (STAU)1-mediated RNA decay (SMD) pathway [9]. In histone RNA decay, stem-loop-binding protein (SLBP) binds to a conserved stem-loop structure found in the 3′ end of histone mRNAs and recruits UPF1 to trigger mRNA decay [9]. In SMD, mRNAs containing a STAU1-binding site downstream of a termination codon in the 3′UTR are bound by STAU1 and UPF1, resulting in UPF1 phosphorylation and RNA decay [9]. There is also evidence that UPF1 functions in genome maintenance, including in DNA replication, DNA damage responses [16], and telomere replication [17][18]. A common thread in all these roles is nucleic acid, which is consistent with UPF1’s biochemical function as an RNA/DNA helicase. It is also consistent with the fact that UPF1’s various functions are perturbed by mutations in the region of UPF1 encodings its helicase domain (Fig. 1 and references therein).
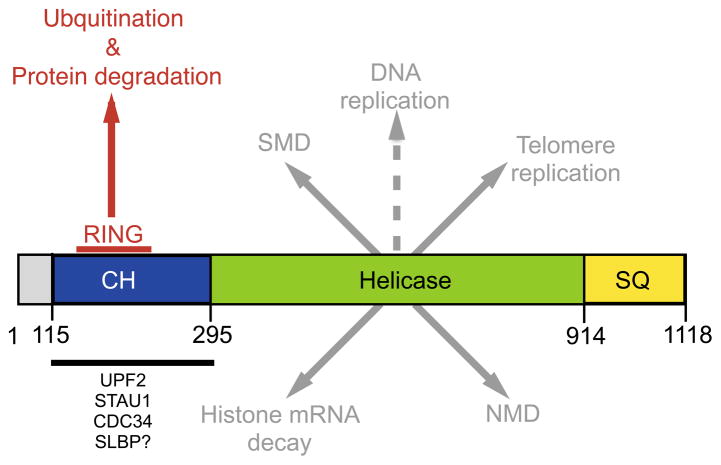
Figure 1.
Domain structure of UPF1 with its various molecular functions highlighted. Domain structure adapted from Dehghani-Tafti et al. [37]. The CH domain is involved in binding several factors (highlighted below the bold line). It is not currently known whether this domain binds SLBP, but seems likely given the similarities between SMD and histone mRNA decay [9]. The RING domain reported by Feng et al. [19] is contained within the CH domain (highlighted by the red line). Mutations within the RING domain disrupt UPF1’s ubiquitination and protein degradation activity [19]. Mutations that disrupt the RNA helicase activity of UPF1 disrupt UPF1’s function in NMD, [38] SMD [39], telomere replication [18] and histone mRNA decay [36]. It has not been directly tested whether the helicase domain is required for UPF1’s putative function in DNA replication, but seems likely given that it is a nucleic acid-dependent process. The SQ domain contains numerous serine residues that are phosphorylated by SMG1 to drive UPF1 function.
Given all these functions for UPF1, one might surmise that the surprises are over. Writing recently in Molecular Cell, Feng et al. report strong evidence for an exciting new function for UPF1 – it is enzymatically involved in ubiquitination and thereby promotes protein decay [19]. They provide a biological link for this new function by showing that human UPF1 represses myogenesis through its activity as an E3 ubiquitin ligase and that it promotes the decay of the myogenesis-promoting transcription factor MYOD.
Discovery that UPF1 is an E3 ubiquitin ligase
UPF1 represses myogenesis
To determine whether UPF1 has a causal role in myogenesis, Feng et al. performed UPF1 knockdown and overexpression experiments. They found that UPF1 knockdown accelerated the differentiation of the immortalized human myoblast cell lines 54-1 and MB135. They observed a similar effect of UPF1 knockdown in two primary human skeletal myoblast cell cultures. Conversely, UPF1 overexpression slowed myogenesis. Together, this data provided strong evidence that UPF1 represses human myogenesis, at least in vitro.
What physiological role might this myogenic-inhibitory activity have? Previous work showed that UPF1 levels and NMD efficiency are decreased when mouse myoblasts undergo differentiation in vitro [13]. These findings lead to a model in which reduced UPF1 levels allow for myogenesis to proceed (Fig. 2).
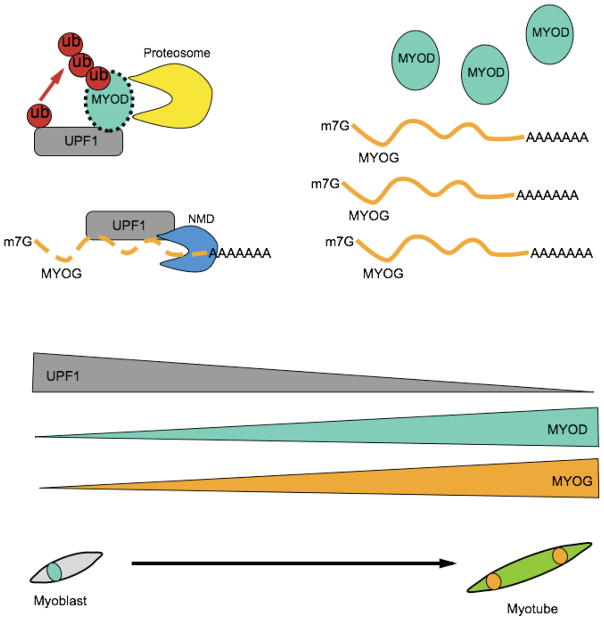
Figure 2.
Model for UPF1’s role in myogenesis. In myoblasts, UPF1 levels are high allowing UPF1 to degrade the MYOG mRNA [13] through its NMD activity, and to degrade MYOD protein through its E3 ubiquitin ligase activity [19]. UPF1’s negative effect on MYOD protein and MYOG mRNA levels correlates with the low levels of these factors in myoblasts and allows UPF1 to suppress myogenesis. When myoblasts differentiate into myotubes, UPF1 levels drop, which may allow MYOG mRNA and MYOD protein to escape degradation, correlating with the high levels of these factors seen in differentiated muscle cells (myotubes).
UPF1 targets the myogenic transcription factor, MYOD, for protein decay
One mechanism by which UPF1 could inhibit myogenesis is by degrading the mRNA encoding the pro-myogenic transcription factor, MYOGENIN (MYOG), as this RNA was previously shown to be a NMD target [13]. However, Feng et al. found that MYOG mRNA level did not increase in response to UPF1 knockdown until 2 days after induction of differentiation, which made it a poor candidate to dictate the modulatory effects of UPF1 on differentiation.
To obtain a perspective on the range of RNAs regulated by UPF1 in human myoblasts, the authors turned to RNAseq analysis. This analysis revealed that a relatively large number of genes dysregulated in response to UPF1 knockdown are targets of a master myogeneic transcription factor in the same subfamily as MYOG, called MYOD. This result suggested that UPF1 modulates the MYOD transcriptional program. It also raised the possibility that MYOD is a key target of NMD responsible for the ability of UPF1 to repress myogenesis. In support of MYOD mRNA being a direct NMD target transcript, the steady-state level of MYOD mRNA increased in response to UPF1 knockdown. However, MYOD mRNA stability was not increased by UPF1 knockdown, as would have been expected if it were a direct target. Pre-mRNA analysis revealed that the increase in MYOD steady state mRNA levels in response to UPF1 knockdown likely resulted from increased transcription, rather than inhibited RNA decay. This finding illustrates the challenges of defining NMD target mRNAs, as mere upregulation of an mRNA in response to NMD perturbation is clearly not sufficient to define a direct target of the NMD pathway.
These results implied that UPF1 somehow decreases MYOD protein level by a means other than destabilizing MYOD RNA. In further support, the authors found that UPF1 knockdown increased MYOD protein level before a measurable increase in MYOD steady-state mRNA level. These surprising results motivated Feng et al. to determine whether instead UPF1 promotes MYOD protein destabilization. To test this, they made use of the protease inhibitor MG132 and found that it perturbed UPF1’s ability to decrease MYOD protein level, consistent with the possibility that UPF1 promotes the destabilization of MYOD protein.
A traditional model would be that UPF1 achieves this indirectly by degrading a mRNA encoding a protein involved in the stabilization of MYOD protein. However, several lines of previously published evidence suggested that UPF1 might directly act to destabilize MYOD. For example, proteomic studies had shown that UPF1 interacts with several components of the ubiquitination and proteasome machinery [20][21]. Furthermore, UPF1 itself contains an N-terminal cysteine- and histidine-rich domain that is structurally similar to the RING domain found in one of the two major types of eukaryotic E3 ubiquitin ligases [22]. Indeed, a previous study had found that Saccharomyces cerevisiae Upf1 uses this domain to undergo self-ubiquitination [23]. This study also showed that (i) this RING-like domain is conserved from yeast to human, (ii) the domain interacted with a yeast E2 conjugase, Ubc3, (iii) Upf1 underwent self-ubiquitination in vitro in a manner dependent on the RING domain. This study conducted in yeast indicated that there was precedent for the possibility that human UPF1 might have E3 ligase activity and thereby directly promote the decay of MYOD protein.
UPF1 degrades MYOD through UPF1’s E3 ligase activity
As a first step to investigate whether human UPF1 might have E3 ligase activity, Feng et al. performed molecular modeling and found that the putative RING domain in human UPF1 structurally mimics the RING domain of known RING E3 ubiquitin ligases. This analysis confirmed that human UPF1 likely has the E2-binding pocket essential for a functional E3 ligase. To empirically analyze whether human UPF1 is an E3 ligase, the authors made a mutant in the RING domain that was predicted to disrupt its E2-binding pocket. This RING domain mutant interacted poorly with the E2 conjugase, CDC34, the human ortholog of the yeast protein previously shown to be an activation partner of yeast Upf1’s E3 ligase activity [23]. Armed with this knowledge, the authors then tested whether human UPF1 might ubiquitinate MYOD. They made three observations supporting this notion. First, MYOD ubiquitination was reduced in cells expressing the UPF1 RING domain mutant. Second, MYOD and UPF1 physically interacted in a manner that was stabilized by disrupting UPF1’s RING domain. Third, ubiquitin associated with the UPF1 RING domain mutant, but not with WT-UPF1, suggesting that the mutant failed to transfer ubiquitin to its substrate. Importantly, this mutant did not impact the ability of UPF1 to function in NMD, suggesting that it could be used to distinguish between UPF1’s effects on NMD and ubiquitination.
To determine whether UPF1 destabilizes MYOD protein in muscle cells, the authors induced UPF1 expression from a tetracycline-regulated expression vector in stably transfected myoblast cells. They found that this led to rapid degradation of MYOD protein, as measured by half-life analysis. Degradation of the MYOD protein depended on UPF1’s RING domain, as expression of the UPF1 RING domain mutant did not trigger MYOD protein degradation. Together, these data provided compelling evidence that human UPF1 uses E3 ligase activity to degrade MYOD protein.
Evidence that UPF1 represses myogenesis through its RING domain
To assess the physiological significance of UPF1’s RING domain, Feng et al. compared the ability of myoblasts expressing inducible exogenous wild-type UPF1 and RING-mutant UPF1 to undergo differentiation. They found that in contrast to wild-type UPF1, which slowed myogenesis, mutant UPF1 actually led to a more rapid rate of myogenesis, as shown by a rapid (albeit modest) increase in myogeneic markers. This data suggested that the E3 ligase activity of UPF1 is required to inhibit myogenesis. Future studies will be needed to determine whether UPF1 inhibits myogenesis through destabilization of MYOD protein and/or other proteins, as discussed below.
Perspective
To what extent does UPF1 shape the proteome and how does it recognize its protein targets?
The discovery that UPF1 can act as an E3 ubiquitin ligase and thereby promote protein degradation, indicates that UPF1 can directly shape more than just the transcriptome. An important future research direction will be to identify the full gamut of proteins directly ubiquitinated by UPF1. A recent proteome-wide analysis of proteins with altered abundance upon UPF1 depletion could be used as a starting point for identifying proteins that are destabilized by UPF1 [24].
A major challenge in defining the extent of UPF1’s ability to shape the proteome will be dissociating UPF1’s RNA degradation activity from its ubiquitination/protein degradation activity, as both processes ultimately dictate protein level. In this regard, the UPF1 RING-domain mutant identified by Feng et al. that lacks E3 ligase activity and has normal NMD activity will be a powerful tool to distinguish whether UPF1 acts upon a given protein through its role in protein decay vs. RNA decay (or a combination of the two).
Another potential substrate of human UPF1’s E3 ligase activity is UPF1 itself, based on the finding that yeast Upf1 self-ubiquitinates in vitro [23]. This self-ubiquitination might serve as a homeostatic mechanism to buffer UPF1 levels, or it may act to regulate UPF1’s activity in response to external stimuli.
Regardless of whether UPF1’s protein targets are limited to MYOD and UPF1 itself, or extend to many more proteins, it will be important to determine how UPF1 identifies its substrates for ubiquitination. Are there specific features in the proteins that UPF1 recognizes, or does ubiquitination occur simply through proximity?
How, why, and to what extent does UPF1 modulate myogenesis?
How does UPF1 repress myogenesis? The authors favor the notion that UPF1 inhibits myogenesis through UPF1’s ability to destabilize MYOD protein. This notion is particularly compelling given that MYOD is a powerful and well-established pro-myogenic transcription factor [25]. However, it is also possible that UPF1 inhibits myogenesis through decay of some factor other than (or in addition to) MYOD. In the future, it will be important to perform rescue experiments with MYOD and other potential downstream targets to assess which possibility is correct.
The physiological relevance of UPF1’s modulatory role in myogenesis also remains to be determined. One possibility is that it is part of a feedback network to control muscle mass. Various stressors are known to inhibit NMD, probably as a means to regulate biological responses [26]. If exercise stress also inhibits NMD, this would be predicted to amplify myogenesis as a means to build muscle mass.
Currently the evidence in favor of UPF1 functioning in myogenesis is derived from in vitro cell models of myogenesis [13][19]. Is this also the case in vivo? To address this question in mice, it will be necessary to conditionally ablate Upf1 in myogenic cells, as global Upf1 knockout is embryonic lethal [10].
What is the relationship between mRNA and protein decay?
The discovery that UPF1 can act directly to initiate protein degradation raises the question as to whether this activity is coupled to UPF1’s RNA decay-promoting activity. Feng et al. showed that MYOD protein is destabilized by UPF1, while MYOD mRNA is not destabilized by UPF1, suggesting that—in human cells—the two processes are not coupled, at least in this instance. This hypothesis is further supported by the fact that disruption of the RING domain of human UPF1 does not compromise its NMD activity. However, studies in S. cerevisiae suggest that the two activities can be coupled. One line of evidence for this is the finding that mutations in the yeast UPF1 RING domain disrupt NMD [23]. Another line of evidence is studies demonstrating UPF1-dependent decay of peptides translated from PTC-containing transcripts [26][27]. In the future, it will be intriguing to determine whether the mechanism responsible for this UPF1 dependent peptide decay in yeast is UPF1’s E3 ligase activity.
If indeed it is found that a coupled RNA and protein decay system exists, such a system could be used to dramatically up- or down-regulate proteins to achieve physiological goals. For example, a coupled RNA and protein decay system would allow for efficient quality control; i.e., strong downregulation of mutant proteins from aberrant genes harboring PTCs. By both increasing the rate of RNA and protein decay, UPF1 could largely eliminate mutant proteins, thereby avoiding potential dominant-negative effects.
Alternatively, no dedicated coupled RNA and protein decay system may exist. Instead, there may have been selection pressure over evolutionary time for UPF1 to take a multi-prong approach to achieve a given physiological goal. For example, UPF1 may dampen myogenesis by not only destabilizing MYOD protein but also by degrading mRNAs encoding other pro-myogenic components. This hypothesis is consistent with the finding that MYOG mRNA is a NMD target mRNA [13] (Fig. 2). It will be intriguing to test this “multi-prong” function hypothesis by performing rescue experiments with candidate UPF1 RNA and protein targets.
What is the biological importance of UPF1’s protein decay activity?
In the future, it will be critical to determine whether other biological functions ascribed to UPF1 come from its novel activity discovered by Feng et al. Examples of known roles of UPF1 include: (i) it promotes neural stemness [14][15], (ii) it acts as a tumor suppressor in cell lines [29] and (iii) it binds to members of the large long intersperse element-1 (LINE1) family and modulates both LINE1 expression and transposition [30]. Loss of UPF1’s ubiquitination activity could also contribute to a number of diseases associated with loss or reduction of UPF1, including human inflammatory myofibroblastic tumors [31], a rare form of human pancreatic cancer [32], and facio-scapulohumeral muscular dystrophy [33]. In addition, UPF1’s roles outside of NMD (Fig. 1) may involve its protein degradation function. For instance, it is tempting to speculate that UPF1’s function in histone metabolism might not only be through its ability to directly target histone mRNAs for degradation. For example, might UPF1 drive the decay of the SLBP protein, which is rapidly degraded by the proteosome at the end of S-phase [34] and is necessary for the shut off histone mRNA biosynthesis [35]. Given that UPF1 associates with SLBP to promote histone mRNA degradation [36], a reasonable hypothesis is that UPF1 may also be responsible for promoting the decay of SLBP.
In summary, UPF1 is a multifaceted molecule with diverse molecular and developmental roles. Given its many functions, it is not surprising that loss of UPF1 in early development results in embryonic lethality, and that loss-of-function mutations in UPF1 acquired later in development result in disease. In the future, it will be exciting to determine the extent to which UPF1’s E3 ligase activity shapes the proteome and drives biological processes. In the words of Benjamin Franklin, “Hide not your talents. They for use were made. What’s a sundial in the shade?”
Acknowledgments
This work was supported by National Institutes of Health grants to TP (T32 HD007203, F32GM113487) and MFW (RO1 GM111838).
References
- 1.Peccarelli M, Kebaara BW. Regulation of Natural mRNAs by the Nonsense-Mediated mRNA Decay Pathway. Eukaryot Cell. 2014 Sep;13(9):1126–1135. doi: 10.1128/EC.00090-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang Y-F, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007 Jan;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 3.Fatscher T, Boehm V, Gehring NH. Mechanism, factors, and physiological role of nonsense-mediated mRNA decay. Cell Mol Life Sci. 2015 Dec;72(23):4523–44. doi: 10.1007/s00018-015-2017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol. 2015 Nov;16(11):665–77. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- 5.Huang L, Wilkinson MF. Regulation of nonsense-mediated mRNA decay. Wiley Interdiscip Rev RNA. 2012;3(6):807–828. doi: 10.1002/wrna.1137. [DOI] [PubMed] [Google Scholar]
- 6.Vicente-Crespo M, Palacios IM. Nonsense-mediated mRNA decay and development: shoot the messenger to survive? Biochem Soc Trans. 2010 Dec;38(6):1500–5. doi: 10.1042/BST0381500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee SR, Pratt GA, Martinez FJ, Yeo GW, Lykke-Andersen J. Target Discrimination in Nonsense-Mediated mRNA Decay Requires Upf1 ATPase Activity. Mol Cell. 2015 Aug;59(3):413–425. doi: 10.1016/j.molcel.2015.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franks TM, Singh G, Lykke-Andersen J. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense-mediated mRNA decay. Cell. 2010 Dec;143(6):938–50. doi: 10.1016/j.cell.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet. 2012 Apr;13(4):246–59. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet. 2001 Jan;10(2):99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- 11.Wittkopp N, Huntzinger E, Weiler C, Saulière J, Schmidt S, Sonawane M, Izaurralde E. Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol Cell Biol. 2009 Jul;29(13):3517–28. doi: 10.1128/MCB.00177-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metzstein MM, Krasnow MA. Functions of the Nonsense-Mediated mRNA Decay Pathway in Drosophila Development. PLoS Genet. 2006 Dec;2(12):e180. doi: 10.1371/journal.pgen.0020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev. 2009 Jan;23(1):54–66. doi: 10.1101/gad.1717309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruno IG, Karam R, Huang L, Bhardwaj A, Lou CH, Shum EY, Song HW, Corbett MA, Gifford WD, Gecz J, Pfaff SL, Wilkinson MF. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol Cell. 2011 May;42(4):500–10. doi: 10.1016/j.molcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lou CH, Shao A, Shum EY, Espinoza JL, Huang L, Karam R, Wilkinson MF. Posttranscriptional control of the stem cell and neurogenic programs by the nonsense-mediated RNA decay pathway. Cell Rep. 2014 Feb;6(4):748–64. doi: 10.1016/j.celrep.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azzalin CM, Lingner J. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr Biol. 2006 Feb;16(4):433–9. doi: 10.1016/j.cub.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 17.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric Repeat Containing RNA and RNA Surveillance Factors at Mammalian Chromosome Ends. Science (80- ) 2007 Nov;318(5851):798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 18.Chawla R, Redon S, Raftopoulou C, Wischnewski H, Gagos S, Azzalin CM. Human UPF1 interacts with TPP1 and telomerase and sustains telomere leading-strand replication. EMBO J. 2011 Aug;30(19):4047–58. doi: 10.1038/emboj.2011.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng Q, Jagannathan S, Bradley RK. The RNA Surveillance Factor UPF1 Represses Myogenesis via Its E3 Ubiquitin Ligase Activity. Mol Cell. 2017 Jul;67(2):239–251.e6. doi: 10.1016/j.molcel.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flury V, Restuccia U, Bachi A, Mühlemann O. Characterization of phosphorylation- and RNA-dependent UPF1 interactors by quantitative proteomics. J Proteome Res. 2014 Jun;13(6):3038–53. doi: 10.1021/pr5002143. [DOI] [PubMed] [Google Scholar]
- 21.Brannan KW, Jin W, Huelga SC, Banks CAS, Gilmore JM, Florens L, Washburn MP, Van Nostrand EL, Pratt GA, Schwinn MK, Daniels DL, Yeo GW. SONAR Discovers RNA-Binding Proteins from Analysis of Large-Scale Protein-Protein Interactomes. Mol Cell. 2016 Oct;64(2):282–293. doi: 10.1016/j.molcel.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadlec J, Guilligay D, Ravelli RB, Cusack S. Crystal structure of the UPF2-interacting domain of nonsense-mediated mRNA decay factor UPF1. RNA. 2006 Oct;12(10):1817–24. doi: 10.1261/rna.177606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi S, Araki Y, Ohya Y, Sakuno T, Hoshino SI, Kontani K, Nishina H, Katada T. Upf1 potentially serves as a RING-related E3 ubiquitin ligase via its association with Upf3 in yeast. RNA. 2008 Sep;14(9):1950–8. doi: 10.1261/rna.536308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sieber J, Hauer C, Bhuvanagiri M, Leicht S, Krijgsveld J, Neu-Yilik G, Hentze MW, Kulozik AE. Proteomic Analysis Reveals Branch-specific Regulation of the Unfolded Protein Response by Nonsense-mediated mRNA Decay. Mol Cell Proteomics. 2016 May;15(5):1584–1597. doi: 10.1074/mcp.M115.054056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005 Aug;16(4–5):585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Goetz AE, Wilkinson M. Stress and the nonsense-mediated RNA decay pathway. Cell Mol Life Sci. 2017 Oct;74(19):3509–3531. doi: 10.1007/s00018-017-2537-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuroha K, Tatematsu T, Inada T. Upf1 stimulates degradation of the product derived from aberrant messenger RNA containing a specific nonsense mutation by the proteasome. EMBO Rep. 2009 Nov;10(11):1265–1271. doi: 10.1038/embor.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verma R, Oania RS, Kolawa NJ, Deshaies RJ. Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. Elife. 2013 Jan;2:e00308. doi: 10.7554/eLife.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang D, Zavadil J, Martin L, Parisi F, Friedman E, Levy D, Harding H, Ron D, Gardner LB. Inhibition of nonsense mediated RNA decay by the tumor microenvironment promotes tumorigenesis. Mol Cell Biol. 2011 Jul;31(17):3670–80. doi: 10.1128/MCB.05704-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor MS, LaCava J, Mita P, Molloy KR, Huang CRL, Li D, Adney EM, Jiang H, Burns KH, Chait BT, Rout MP, Boeke JD, Dai L. Affinity Proteomics Reveals Human Host Factors Implicated in Discrete Stages of LINE-1 Retrotransposition. Cell. 2013;155(5):1034–1048. doi: 10.1016/j.cell.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu J, Plank TD, Su F, Shi X, Liu C, Ji Y, Li S, Huynh A, Shi C, Zhu B, Yang G, Wu Y, Wilkinson MF, Lu Y. The nonsense-mediated RNA decay pathway is disrupted in inflammatory myofibroblastic tumors. J Clin Invest. 2016 Jun;126(8):3058–3062. doi: 10.1172/JCI86508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu C, Karam R, Zhou Y, Su F, Ji Y, Li G, Xu G, Lu L, Wang C, Song M, Zhu J, Wang Y, Zhao Y, Foo WC, Zuo M, Valasek MA, Javle M, Wilkinson MF, Lu Y. The UPF1 RNA surveillance gene is commonly mutated in pancreatic adenosquamous carcinoma. Nat Med. 2014 Jun;20(6):596–8. doi: 10.1038/nm.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng Q, Snider L, Jagannathan S, Tawil R, van der Maarel SM, Tapscott SJ, Bradley RK. A feedback loop between nonsense-mediated decay and the retrogene DUX4 in facioscapulohumeral muscular dystrophy. Elife. 2015 Jan;4 doi: 10.7554/eLife.04996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitfield ML, Zheng LX, Baldwin A, Ohta T, Hurt MM, Marzluff WF. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol Cell Biol. 2000 Jun;20(12):4188–98. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng L, Dominski Z, Yang XC, Elms P, Raska CS, Borchers CH, Marzluff WF. Phosphorylation of stem-loop binding protein (SLBP) on two threonines triggers degradation of SLBP, the sole cell cycle-regulated factor required for regulation of histone mRNA processing, at the end of S phase. Mol Cell Biol. 2003 Mar;23(5):1590–601. doi: 10.1128/MCB.23.5.1590-1601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaygun H, Marzluff WF. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat Struct Mol Biol. 2005 Sep;12(9):794–800. doi: 10.1038/nsmb972. [DOI] [PubMed] [Google Scholar]
- 37.Dehghani-Tafti S, Sanders CM. DNA substrate recognition and processing by the full-length human UPF1 helicase. Nucleic Acids Res. 2017 Jul;45(12):7354–7366. doi: 10.1093/nar/gkx478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lykke-Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000 Dec;103(7):1121–31. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 39.Park E, Gleghorn ML, Maquat LE. Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc Natl Acad Sci. 2013 Jan;110(2):405–412. doi: 10.1073/pnas.1213508110. [DOI] [PMC free article] [PubMed] [Google Scholar]