Abstract
Drosophila melanogaster plasmatocytes, the phagocytic cells among hemocytes, are essential for immune responses, but also play key roles from early development to death through their interactions with other cell types. They regulate homeostasis and signaling during development, stem cell proliferation, metabolism, cancer, wound responses, and aging, displaying intriguing molecular and functional conservation with vertebrate macrophages. Given the relative ease of genetics in Drosophila compared to vertebrates, tools permitting visualization and genetic manipulation of plasmatocytes and surrounding tissues independently at all stages would greatly aid a fuller understanding of these processes, but are lacking. Here, we describe a comprehensive set of transgenic lines that allow this. These include extremely brightly fluorescing mCherry-based lines that allow GAL4-independent visualization of plasmatocyte nuclei, the cytoplasm, or the actin cytoskeleton from embryonic stage 8 through adulthood in both live and fixed samples even as heterozygotes, greatly facilitating screening. These lines allow live visualization and tracking of embryonic plasmatocytes, as well as larval plasmatocytes residing at the body wall or flowing with the surrounding hemolymph. With confocal imaging, interactions of plasmatocytes and inner tissues can be seen in live or fixed embryos, larvae, and adults. They permit efficient GAL4-independent Fluorescence-Activated Cell Sorting (FACS) analysis/sorting of plasmatocytes throughout life. To facilitate genetic studies of reciprocal signaling, we have also made a plasmatocyte-expressing QF2 line that, in combination with extant GAL4 drivers, allows independent genetic manipulation of both plasmatocytes and surrounding tissues, and GAL80 lines that block GAL4 drivers from affecting plasmatocytes, all of which function from the early embryo to the adult.
Keywords: macrophage, plasmatocyte, hemocyte, FACS, imaging, Genetics of Immunity
Drosophila plasmatocytes are well known for their immune functions in combatting bacteria, fungi, and viruses through phagocytosis and siRNA production (Braun et al. 1998; Elrod-Erickson et al. 2000; Lemaitre and Hoffmann 2007; Tassetto et al. 2017). Yet recent years have revealed the many ways in which they also play crucial roles in development and homeostasis, contacting and exchanging signals with surrounding cells. This has expanded the repertoire of functions that plasmatocytes are known to carry out to protect the organism; their patrolling serves not only to detect and destroy foreign invaders, but also to assess defects in endogenous cell states and stimulate corrective cellular responses. Many of the processes they affect and the molecular pathways they use to do so are conserved with vertebrate macrophages, making Drosophila plasmatocytes an excellent model system (Wynn et al. 2013; Ratheesh et al. 2015).
Plasmatocytes influence development in several different ways. They migrate widely in the embryo to phagocytose and thus clear cells that have undergone programmed cell death (Tepass et al. 1994; Zhou et al. 1995; Franc et al. 1996). As they move, plasmatocytes secrete extracellular matrix (ECM) components, which assemble into a stable basal lamina whose presence affects later steps in development (Fessler and Fessler 1989; Olofsson and Page 2005; Martinek et al. 2008; Matsubayashi et al. 2017). This effect can occur by the ECM providing a substrate for cell movement or by binding Dpp, a BMP family member, and influencing its signaling (Olofsson and Page 2005; Bunt et al. 2010; Van De Bor et al. 2015). These developmental functions are conserved in vertebrates. Vertebrate macrophages also engulf apoptotic cells during development (Gouon-Evans et al. 2000; Leers et al. 2002), and show molecular conservation with Drosophila in some of the receptors they use to recognize dying cells (Franc et al. 1996; Fadok et al. 1998; Manaka et al. 2004; Greenberg et al. 2006; Kurucz et al. 2007; Wu et al. 2009). Vertebrate macrophages secrete the ECM component collagen (Schnoor et al. 2008), which can bind BMP family members (Vukicevic et al. 1994; Sieron et al. 2002).
Plasmatocytes are also crucial for maintaining the organism after it has formed. They alter responses to damage in the gut, regulating stem cell proliferation by secreting stimulatory factors (Ayyaz et al. 2015; Chakrabarti et al. 2016). Plasmatocytes kill tumor cells by expressing TNFα (Parisi et al. 2014), or stimulate their invasion if tumors also express activated Ras, through MMP1 induction by TNFα-induced JNK signaling (Cordero et al. 2010; Pérez et al. 2017). Plasmatocytes can even alter metabolism and aging; upon engulfing lipids, they induce JAK-STAT signaling in surrounding tissues, which modulates insulin sensitivity, hyperglycemia, fat storage, and lifespan (Woodcock et al. 2015). Conservation with vertebrates is seen for these processes as well. Vertebrate macrophages alter gut stem cell proliferation to promote regeneration; they may also use BMP to do so as BMP2 inducible kinase is upregulated in responding gut tissues (Pull et al. 2005). Vertebrate macrophages can promote tumor induction of MMPs and invasion by secreting TNFα (Hagemann et al. 2005). Finally, vertebrate macrophages also participate in an inflammatory response to obesity that leads to insulin insensitivity (Weisberg et al. 2003; Xu et al. 2003; Patsouris et al. 2008; Pollard 2009), as is seen in the Drosophila response to lipid ingestion (Woodcock et al. 2015). Thus, conservation is seen between vertebrates and Drosophila in the ways in which immune cells and surrounding tissues affect one another and the molecular pathways they use to do so.
The genetic power of Drosophila melanogaster can help elucidate how plasmatocytes regulate organismal development and homeostasis, and how tissues signal their state to plasmatocytes to induce responses. Yet such studies require tools that are lacking, ones that permit the live imaging or manipulation of plasmatocyte behavior along with the modulation and visualization of other cells. Here, we describe a set of tools designed to facilitate these approaches and demonstrate that they function at all stages of the Drosophila life cycle. These lines will thus greatly aid investigations of the manifold interactions of Drosophila plasmatocytes with other tissues from birth to death, enabling insights that can be relevant for vertebrate systems.
Materials and Methods
Cloning
Standard molecular biology methods were used, and all constructs were first tested for functionality by transfection into the plasmatocyte-like S2 cell line (Schneider 1972; Woodcock et al. 2015) (a gift from Frederico Mauri of the Knoblich laboratory at IMBA, Vienna) before injection into flies. Restriction enzymes BSiWI, PstI, and AscI were obtained from New England Biolabs (Frankfurt, Germany); XbaI and EcoRI were from Promega (Mannheim, Germany). PCR amplifications were performed with CloneAmp HiFi PCR Premix from Clontech’s European distributor Takara Bio Europe (Saint-Germaine en Laye, France) using a peqSTAR 2× PCR machine from PEQLAB (Erlangen, Germany). All Infusion cloning was conducted using an Infusion HD Cloning kit obtained from Clontech’s European distributor (see above); relevant oligos were chosen using the Infusion primer tool at the Clontech website http://bioinfo.clontech.com/infusion/convertPcrPrimersInit.do.
Construction of srpHemo-3XmCherry
A 2.5 kb XbaI-EcoRI fragment, which contains three repeats of mCherry, was cloned from pJJH1295 (Bakota et al. 2012; Evans et al. 2014) (a gift from Jürgen Heinisch, Addgene plasmid #36914), into the multiple cloning site of pCaSpeR4 (a gift from Leonie Ringrose, IMBA, Vienna). Subsequently, a 4.3 kb fragment of the srp promoter was amplified from plasmid srpHemoA (Brückner et al. 2004) (a gift from K. Brückner) by PCR with the following primers:
5′-CGAGGTCGACTCTAGAAAATTTTGATGTTTTTAAATAGTCTTATCAGCAATGGCAA-3′.
5′-ACGAAGCTTCTCTAGATATGGGATCCGTGCTGGGGTAGTGC-3′.
This fragment was cloned upstream of the 3xmCherry fragment at the XbaI site by Infusion cloning to create DSPL172.
Construction of srpHemo-H2A::3XmCherry
A 458 bp fragment containing the first 124 amino acids from histone H2A was amplified from pKS23b, a gift from Kristen Senti and Julius Brennecke at IMBA, using the following primers:
5′-AGAGAAGCTTCGTACGCGTACGATGTCTGGACGTGGAAAAG-3′.
5′-CGACCTGCAGCGTACGCGTACGGCCGCCGCCTCTAGACACTT-3′.
This fragment was placed by Infusion cloning at the BSiWI site of DSPL172, downstream of the srp promoter and upstream of the 3XmCherry fragment, with the linker sequence SRGGGRTRTLQV to create DSPL216.
Construction of srpHemo-moe::3XmCherry
An 869 bp fragment from the Moesin cDNA SD10366 (DGRC) (Rubin et al. 2000) containing amino acids 370–646, and thus the ERM domain of the protein, was amplified by PCR using the following primers:
5′-AGAGAAGCTTCGTACGATGGACACCATCGATGTGCA-3′.
5′-CGACCTGCAGCGTACGCATGTTCTCAAACTGATCG-3′.
This fragment was cloned as above at the BSiwI site in DSPL172, downstream of the srp promoter and upstream of the 3xmCherry fragment, with the linker MRTLQVD.
Construction of srpHemo-QF2
A 4.3 kb fragment containing the srpHemo promoter was amplified from the srpHemoA plasmid (Brückner et al. 2004) (a gift from K. Brückner) using the following primers:
5′-TTATGCTAGCGGATCCAAATTTTGATGTTTTTAAATAGTCTTATCAGCAAT GGCAA-3′.
5′-TGGCATGTTGGAATTCTATGGGATCCGTGCTGGGGTAGTGC-3′.
This fragment was used to replace the synaptobrevin promoter in the nsyb-QF2 plasmid (Riabinina et al. 2015) (a gift from C. Potter). The synaptobrevin promoter was released by a digest with BamHI and EcoRI and replaced by srpHemo using Infusion cloning.
Construction of srpHemo-GAL80
A 4.3 kb fragment of the srp promoter was amplified from plasmid srpHemoA (Brückner et al. 2004) (a gift from K. Brückner) by PCR with the following primers:
5′-GCATGTCGACCTCGAGAAATTTTGATGTTTTTAAATAGTCTTATCAGCAATGGCAA-3′.
5′-CTCCCGGGTACTCGAGTATGGGATCCGTGCTGGGGTAGTGC-3′.
This fragment was cloned by infusion into the (w+) attB plasmid (a gift from Jeff Sekelsky, Addgene plasmid #30326) at the XhoI site to create DSPL237.
A 1307 bp fragment containing GAL80 was amplified with the following primers from pAC-GAL80 (Potter et al. 2010; a gift from Liqun Luo, Addgene plasmid #24346):
5′-CTTCTGCAAGGCGCGCCCAATCAAAATGGATTACAACAAAAGGAG-3′.
5′-CGGTGCCTAGGCGCGCCTACCGGTAGACATGATAAGATACATTGATG-3′.
This fragment was introduced into DSPL237, downstream of the srp fragment at the AscI site, using Infusion cloning to create DSPL322.
Drosophila melanogaster stocks
Flies were raised on standard agar, cornmeal, molasses, and yeast food containing 1.5% Nipagin bought from IMBA (Vienna, Austria). Adults were placed in cages in a Percival DR 36VL incubator maintained at 29° and 65% humidity, and embryos were collected on standard plates prepared in house from apple juice, sugar, agar, and Nipagin, and treated with yeast from Lesaffre (Marcq, France). This applies to all experiments except the QF2 movie, whose fly husbandry conditions are described below. repo-GAL4 and QUAS-CD8::GFP were obtained from the Bloomington Drosophila Stock Center, UAS-moe::mCherry from P. Martin (Millard and Martin 2008), hml-dsRed from K. Brückner, and srp-GAL4 UAS-2xeGFP from R. Reuter.
Embryo immunohistochemistry
Embryos were fixed with a standard 18.5% formaldehyde/heptane fix for 20 min followed by methanol devitellinization. mCherry embryos were visualized directly after fixation, rehydration, and mounting. srp-moe::GFP embryos were rehydrated and underwent antibody staining, using standard protocols and overnight incubation with a 1:500 dilution of GFP antibody (Aves Labs, Tigard, OR), followed (after washing) by incubation for 2 hr with a 1:500 dilution of Goat anti-Chicken Alexa Fluor 488 secondary (Invitrogen, Carlsbad, CA). Embryos to be stained with Lz Ab (DSHB, Iowa City, IA) were heat-fixed using standard protocols and incubated overnight with a 1:20 dilution of the antibody, followed after washing by incubation for 2 hr with a 1:500 dilution of Goat anti-Mouse Alexa Fluor 488 secondary antibody (Invitrogen). After washing, they were mounted in Vectashield Mounting Medium (Vector Labs, Burlingame) on 76 × 26 mm slides from Glasfabrik Karl Hecht (Sondheim, Germany) with 22 × 40 mm coverslips, No. 1 thickness (VWR International, Radnor, PA).
Microscopy
Embryo images were taken with an Inverted LSM700 Confocal Microscope from Zeiss (Jena, Germany), using a Plain-Apochromat 20×/NA 0.8 Air Objective. Larvae and adult flies were imaged with a Leica M205 FA Stereo Microscope, a Leica Planapo 2.0× objective, and a Leica DFC3000G camera (Wetzlar, Germany). Larvae were anesthetized for 10–15 sec with a FlyNap Anesthetic Kit (ArtNr 173010, Carolina Biological Supply Company, Burlington, NJ), rinsed 2× in water, then examined under the stereomicroscope. Adult flies were anesthetized for 3 min in FlyNap, and then immediately examined under the stereomicroscope. For imaging on the confocal, larvae and adults were prepared as described, and then mounted in Halocarbon 200 oil (CatNr: 25073-100, Polysciences Inc., Warrington, PA) in a sandwich of a plastic frame, a YSI 5685 Membrane Kit 002 (ArtNr: 1518-9862, Yellow Springs Instrument Co., Yellow Springs, OH) and a cover glass (CatNr: 631-014724X50 mm, thickness 1.5, VWR) immediately prior to visualization.
Macrophage cell counts
Embryos were analyzed at stage 15–16 for total plasmatocyte number using Imaris (Bitplane) by detecting all the plasmatocyte nuclei as spots.
Time-lapse imaging
For the srpHemo-H2A::3xmCherry time-lapse movies, embryos were dechorionated in 50% bleach for 4 min, washed with water, and mounted in halocarbon oil 27 (Sigma) between a coverslip and an oxygen-permeable membrane (YSI), as described above. The anterior dorsolateral region of the embryo was imaged on an inverted multiphoton microscope (TrimScope II, LaVision) equipped with a W Plan-Apochromat 40×/1.4 oil immersion objective (Olympus). mCherry was imaged at 1100 nm excitation wavelengths, using a Ti-Sapphire femtosecond laser system (Coherent Chameleon Ultra) combined with optical parametric oscillator technology (Coherent Chameleon Compact OPO). Excitation intensity profiles were adjusted to tissue penetration depth and Z-sectioning for imaging was set at 1 µm for tracking. For long-term imaging, movies were acquired for 180–200 min with a frame rate of 40 sec. All embryos were imaged with a temperature control unit set to 28.5°.
For the srp-QF2 QUAS-mCD8::GFP repo-GAL4 UAS-moe::mCherry time-lapse movies, flies were left to lay eggs on grape juice/agar plates overnight at 25°. Embryos were dechorionated in bleach. Stage 15 embryos of the appropriate genotype were identified based on the absence of balancer chromosomes expressing fluorescent markers, and mounted in 10S Voltatef oil (VWR) between a glass coverslip and a gas-permeable Lumox culture dish (Greiner), as described previously (Milchanowski et al. 2004; Evans et al. 2010). Movie images were taken at room temperature every 15 min on an Ultraview spinning disk microscope (PerkinElmer) equipped with a 20× NA 0.5 Plan-Neofluar air objective. Maximum projection images were made from ∼40 µm of Z stacks taken every 3 µm. Image processing was done by using ImageJ.
For the srpHemo-moe::3xmCherry time-lapse movies, embryos were dechlorinated in bleach for 1:15 min, and stage 15 embryos were identified and mounted in a slide covered with a double-sided sticky tape, oriented ventrally, and covered with 10S Voltatef oil (VWR) and a glass coverslip, as described in Stramer et al. (2010). Movie images were taken at room temperature every 5 sec on a Zeiss LSM 880 microscope, using Airyscan and a 63×/1.40 Oil DIC objective. Maximum projection images were made from ∼17 µm of Z stacks taken every 1 µm. Image processing was done by ImageJ.
Transgenic line production
The srpHemo-GAL80 construct was injected into lines y1 M{vas-int.Dm}ZH-2A w*; M{3xP3-RFP.attP}ZH-51D (BL 24483) and y1 M{vas-int.Dm}ZH-2A w*; M{3xP3-RFP.attP}ZH-86Fb (BL 24749), obtained from Peter Duchek of IMBA, to produce inserts on the second and third chromosomes through C31-mediated integration (Bischof et al. 2007). Our srpHemo-QF2 driver was injected into yw; p[w3′, y+, attP16a (Okulski et al. 2011) to produce an insert on the second chromosome. After injection, all male survivors were crossed to w; Sp/CyO; PrDr/TM3Ser virgins. After hatching, we screened for transformants based on eye color and crossed them again to w; Sp/CyO; PrDr/TM3Ser virgins to get rid of the integrase inserted on the X chromosome. We kept three transformants/landing site.
All other vectors were co-injected into w1118 (BL-3605) using standard injection methods, along with a helper plasmid Δ2-3 (Robertson et al. 1988) that permits P element transposase expression. w+ transformants were selected and double balanced.
qPCR
RNA was isolated from ∼50,000 mCherry-positive or mCherry-negative cells using an RNeasy Plus Micro Kit (QIAGEN), following the manufacturer’s protocol. Of the resulting RNA, 50 ng was used for cDNA synthesis using the Sensiscript RT Kit (QIAGEN) and oligo dT primers. A Takyon qPCR Kit (Eurogentec) was used to mix qPCR reactions based on the provided protocol, using the following primers:
mCherry: 5′-ACATCCCCGACTACTTGAAGC-3′ and 5′ ACCTTGTAGATGAACTCGCCG-3′
which were designed using Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastDescAd).
Pvr: 5′-GTGACTTTGGTCTGGCTCG-3′ and 5′-GATTCCAGCGCCAGC-3′.
RhoL: 5′-CCTGAGCTATCCCAGTACCAA-3′ and 5′-ACCACTTGCTTTTCACGTTTTC-3′.
Drpr: 5′-TCCACCTATCGCATTAAACACC-3′ and 5′-ACAGTCCCTCACAATACGGTT-3′.
RpL32: 5′-AGCATACAGGCCCAAGATCG-3′ and 5′-TGTTGTCGATACCCTTGGGC-3′.
These four sets of primers were obtained from FlyPrimerBank (http://www.flyrnai.org/FlyPrimerBank). qPCR was run on a LightCycler 480 (Roche) and data were analyzed using LightCycler 480 Software and Prism.
FACS analysis
Embryos were collected for 1 hr from adult w−; srpHemo-3xmCherry flies and aged for an additional 4 hr, all at 29°. Embryos collected from w− flies were processed in parallel and served as a negative control. Embryos were dissociated using a procedure adapted from Estrada and Michelson (2008). Embryos were dechorionated with fresh 50% bleach for 5 min, thoroughly rinsed with water, and blotted on a dry towel. Next, 30 mg of embryos were transferred with a paintbrush into a dounce homogenizer. Subsequent procedures were carried out at 4° or on ice, and all the solutions were cooled. Homogenizers were filled with 10 ml of Seecof saline (6 mM Na2HPO4, 3.67 mM KH2PO4, 106 mM NaCl, 26.8 mM KCl, 6.4 mM MgCl2, and 2.25 mM CaCl2 at a pH of 6.8) and embryos were homogenized with 10 vertical strokes. The resulting suspensions from three homogenizers were collected into a 50 ml Falcon tube (Corning, NY) and centrifuged at 500 rpm for 6 min 30 sec to pellet tissue debris. The supernatant was collected into a separate 50 ml Falcon tube and centrifuged at 1250 rpm for 10 min to precipitate cells. The supernatant was discarded, and the cell pellet was resuspended in 20 ml RPMI media with 10% FBS, which was then split into two 10 ml Falcon tubes. Next, 1 ml of heat-inactivated FBS was slowly pipetted down to the bottom of each of the tubes, which were subsequently centrifuged at 1250 rpm for 10 min to separate out the dead cells that remained in the upper phase after centrifugation. The resulting cell pellet was resuspended in 500 ml of Schneider’s media with 25% (0.2 μM filtered) heat-inactivated FBS and 2 mM EDTA (to reduce calcium dependent adhesion and thus the formation of clumps). The cell suspension was filtered to remove cell clumps using a Falcon 12 × 75 mm Polystyrene tube with a cell strainer cap containing a 35 μm nylon mesh. The cells were analyzed or sorted using a FACS Aria III (BD) flow cytometer. Emission signals for mCherry (600LP, 610/20), dsRed (583/15), and near infrared (755LP, 780/60) were detected. Data were analyzed with FloJo (Tree Star) software. The cells from the dissociated negative control w− embryos were sorted to set a baseline plot. A sample of the cells from the dissociated srpHemo-3xmCherry embryos was stained with 2 μg/μl Propidium Iodide and almost no dead cells were detected upon sorting. Macrophages from these same srpHemo-3xmCherry embryos were sorted based on mCherry fluorescence into 2 ml Eppendorf tubes with 50 μl of Schneider’s Drosophila media.
For each genotype, 15 third-instar larvae were collected directly from bottles with a brush. Prior to homogenization, they were rinsed in water to dislodge any fly food residue and kept on ice to immobilize them. For each genotype, eight pairs of male and female adult flies were collected after CO2 anesthesia in an eppendorf tube and kept on ice till homogenization. Homogenization of larvae and adults, and FACS analysis, proceeded as for the embryos above, except that the LIVE/DEAD Fixable Near-IR Dead Cell Stain kit (Thermo Fisher Scientific) was utilized on a sample according to the manufacturers’ instructions. Data shown is representative of FACS from three independent experiments.
Data availability
All plasmids and Drosophila strains created in this study are available from the authors upon request. All strains are also available through the Bloomington Drosophila Stock Center.
Results
Direct fusion lines visualize plasmatocyte nuclei, the cytoplasm, and the cytoskeleton in the embryo
Visualizing plasmatocytes in fixed and live specimens is essential for understanding how these cells interact with surrounding tissues. Previous studies have labeled plasmatocytes by using various GAL4 drivers to activate UAS-reporters (Table 1; Evans et al. 2014). However, this approach prevents the simultaneous use of other GAL4 drivers to independently affect or image separate tissues. Direct fusions have been made of plasmatocyte-specific promoters to fluorescent proteins (Table 2; Evans et al. 2014), but none of these expressed at all stages of the life cycle. Additionally, the expression that many displayed was weak and, in some lines, was also present in large extraneous tissues, making live plasmatocyte detection and FACS analysis challenging. Therefore, we fused the srpHemo promoter that guides specific plasmatocyte expression in the embryo (Brückner et al. 2004) to three copies of mCherry (Shaner et al. 2004; Bakota et al. 2012), a red fluorescent monomer with a rapid maturation time, low photobleaching, and the ability to survive fixation with fluorescence intact. We also fused the first 124 amino acids of Histone H2A to mCherry, concentrating the signal in the nucleus to facilitate cell counting and tracking. There is little autofluorescence in the embryo in the red spectrum, and thus these srpHemo-H2A::3xmCherry lines displayed extremely brightly fluorescing plasmatocytes with little background starting at embryonic stage 8 and continuing through stage 17; the signal was still strongly visible after fixation with heat, formaldehyde, and paraformaldehyde—utilizing methanol, ethanol, or a hand-held needle to devitellinize—without any antibody staining required (formaldehyde/methanol is shown in Figure 1A). In contrast, the plasmatocyte fluorescence in the previously constructed srp-moe::GFP (Moreira et al. 2010) does not survive fixation (data not shown) and is weak when viewed live [Supplemental Material, Figure S1, A and B; asterisks in A show autofluorescent yolk granules as plasmatocytes only become evident live at stage 10 (data not shown)]. Upon staining with an antibody against GFP, plasmatocytes can be observed starting at stage 8 but are accompanied by strong extraneous expression in the amnioserosa (arrow in Figure S1, C and D), which is also seen live (arrow in Figure S1B) but not observed in live or fixed negative controls (data not shown). Thus, utilizing three copies of mCherry fused directly to the srpHemo promoter produces a plasmatocyte marker that is brightly visible in live or fixed embryos without antibody staining from early embryonic stages onwards.
Table 1. GAL4 Driver lines previously utilized for Plasmatocyte expression.
| Promoter Source | Tissue Expression of Reporter | Time of UAS-Reporter Expression in Plasmatocytes | References for Creation and Expression | ||
|---|---|---|---|---|---|
| Embryo | Larva | Adult | |||
| serpent (srpHemo) | P, LG in larva and adult PC | St 10–17 | L1+, L2+/−, L3+/− | + | Brückner et al. (2004), Zaidman-Rémy et al. (2012) |
| serpent (srp) | P, LG, FB, embryonic midgut, amnioserosa, larval and adult PC, larval SG | St 10–17 | L1+, L2+/−, L3+/− | + | Crozatier et al. (2004), Milchanowski et al. (2004), Avet-Rochex et al. (2010) |
| croquemort | P in adult, internal tissues | St 12–17 | — | + | Olofsson and Page (2005) (embryo) Clark et al. (2011) (adult) |
| peroxidasin | P, LG from L2 on, in larva and adult PC, weak FB in L3 | St 12–17 | L1-L3 | ++ | Stramer et al. (2005) (embryo) Stofanko et al. (2008) (larva) Ghosh et al. (2015) (adult) |
| glial cells missing | P, lateral glia | St 10–17 | — | — | Bernardoni et al. (1997), Olofsson and Page (2005), Avet-Rochex et al. (2010) |
| hemese | 80% of circulating P, sessile P, sections of midgut, SG | — | L3 | — | Zettervall et al. (2004) |
| hemolectin | P, LG | — | L2, L3 | + | Avet-Rochex et al. (2010), Sinenko and Mathey-Prevot (2004), Woodcock et al. (2015) (adult) |
| collagen | P, LG cortical zone, and FB at all stages | St 13–17 | L1–L3 | + | Asha et al. (2003), Avet-Rochex et al. (2010) |
| singed | P | St 11–17 | — | Zanet et al. (2012) | |
| eater | P, LG | — | L3 | — | Tokusumi et al. (2009) |
UAS, upstream activating sequence; P, Plasmatocytes; LG, lymph gland; PC, Pericardial cells; St, stage; FB, Fat Body; SG, salivary gland.
Table 2. Direct fusion lines for plasmatocyte visualization: previously published and described in this paper.
| Promoter Source-Reporter Utilized | Reporter Utilized: Tissue Expression | Time and Level of Reporter Expression in Plasmatocytes | References for Creation and Expression | ||
|---|---|---|---|---|---|
| Embryo | Larva | Adult | |||
| Previously published | |||||
| hemolectin-DsRed | P, CC, LG | — | +++L2–L3 | +/− | Makhijani et al. (2011) |
| hemolectin-DsRed::nls | P, CC, LG | — | +++L2–L3 | +/− | Clark et al. (2011), Makhijani et al. (2011) |
| eater-DsRed | P | — | +/−L3 | — | Tokusumi et al. (2009), Ayyaz et al. (2015) |
| eater-GFP | P | — | +/−L3 | — | Sorrentino et al. (2007) |
| serpent-moe::GFP | P, LG, in embryo amnioserosa | + | +/−L1–EL3 | — | Moreira et al. (2010), Razzell et al. (2013) |
| This paper | |||||
| srpHemo-3xmCherry and derivatives | P, CC until embryonic stage 15, cortical zone of LG, | +++ | +++ | +++ | This paper |
| PC in second and third-instar larvae and adult, SGS from embryonic stage 16 till LL3 | St 8–17 | L1–L3 | |||
P, Plasmatocytes; CC, Crystal Cells; LG, lymph gland; SGS, Stomatogastric nervous system; PC, Pericardial cells; St, stage.
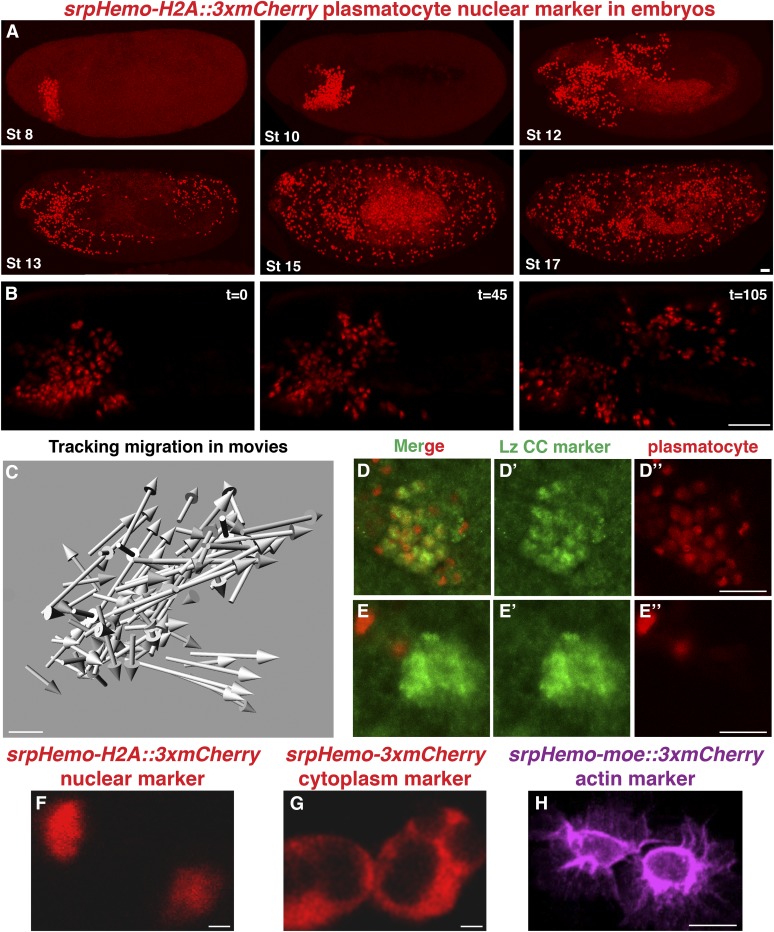
Figure 1.
Direct fusion lines allow fluorescent visualization of plasmatocyte nuclei, the cytoplasm, or the cytoskeleton in embryos from stage (St) 8 onwards. (A) Fixed srpHemo-H2A::3xmCherry embryos display strong fluorescence in the nuclei of plasmatocytes starting from St 8 and continuing throughout embryogenesis. Embryonic St is indicated in the lower left of each panel. (B) Stills from a two-photon movie of a srpHemo-H2A::3xmCherry St 10–12 embryo illustrating the low level of endogenous autofluorescence in the yolk. Three successive time points are shown, with the intervening time in minutes indicated in the upper right of each panel. (C) Arrows indicating relative displacement of macrophage nuclei in 90 min of live imaging starting at St 10. (D and E) Close-ups of merged confocal images of srpHemo-H2A::3xmCherry embryos stained with Lz antibody (D’ and E’) as a marker of crystal cells (CC). We see mCherry expression in CC until St 14 (D”), but no longer at St 15 (E”). Live image of individual plasmatocytes visualized with a two-photon microscope with (F) srpHemo-H2A::3xmCherry or (G) srpHemo-3xmCherry, or (H) with a Zeiss confocal microscope from an srpHemo-moe::3xmCherry embryo demonstrating nuclear, cytoplasmic, or actin labeling, respectively. All embryos are positioned with anterior to the left and dorsal at the top. Scale bars correspond to 20 μM in (A), 40 μM in (B), 20 μM in (C and D), 2 μM in (F and G), and 10 μM in (H).
We demonstrated the effectiveness of our srpHemo-H2A::3xmCherry nuclear line for tracking in live embryos by making two-photon movies of plasmatocyte migration from the head into the germband in stage 10–12 embryos (File S1) (Figure 1B). There was much less autofluorescence at the 1100 wavelength used for mCherry than at the 980 nm used for eGFP in the yolk and, particularly usefully, in the vitelline membrane, where absorption of laser energy through autofluorescence at 980 nm frequently leads to membrane rupture and death of the embryo during movie acquisition. The brightness of the mCherry signal also permitted the use of low laser power for effective imaging and thus less photobleaching. Analysis of plasmatocyte displacement based on tracking the nuclei with Imaris software revealed distinct paths of migration within the anterior, corresponding to the different directions ultimately chosen (Figure 1C). Efficient localization of srpHemo-H2A::3xmCherry to the nucleus (Figure 1F) also permitted easy determination of total plasmatocyte cell counts from confocal images using Imaris; we detected 592 ± 48 cells (n = 24) by analyzing wild-type stage 16 embryos, somewhat less than the 700 previously counted at stage 11 based on an antibody marker (Tepass et al. 1994). Thus, the nuclear-localized mCherry permits automated plasmatocyte tracking and counting, and eliminates many of the problems that occur with live two-photon imaging of GFP.
We assessed if our srpHemo-H2A::3xmCherry line also directs expression in crystal cells. These cells are born along with plasmatocytes from the mesoderm, migrate to a location around the proventriculus, and remain there during embryogenesis (Lebestky et al. 2000). In larvae and adults, they mobilize to enhance melanization in response to wounds or wasp egg infection (Galko and Krasnow 2004; Dudzic et al. 2015). We used an antibody recognizing Lozenge, a crystal cell marker, and observed colocalization with srpHemo-H2A::3xmCherry (Figure 1, D–D”) through much of embryogenesis, but by stage 15 (Figure 1, E–E”) no mCherry colocalization was detected. We also observed extraneous expression in the stomatogastric nervous system starting at stage 16/17 (data not shown). Given that there are 35 crystal cells (Milchanowski et al. 2004) and that we detect ∼600 total cells, we conclude that 94% of all embryonic cells labeled with srpHemo-H2A::3xmCherry before stage 15 are plasmatocytes.
We further created srpHemo-3xmCherry lines to produce plasmatocytes with a labeled cytoplasm (Figure 1G and Figure S1, E and E’), which are useful for studies examining direct contact of plasmatocytes with other tissues as well as their phagocytosis of pathogens and apoptotic cells. To visualize polymerized actin in plasmatocytes during studies of migration, we fused the mCherry with the C-terminal part of moesin that had been previously used to detect actin (Edwards et al. 1997). In these actin-binding srpHemo-moe::3xmCherry lines, we could detect filopodial and lamellipodial extensions within the plasmatocytes in live and fixed embryos (Figure 1H and Figure S1, F and F’), and could make time-lapse movies of plasmatocyte actin dynamics (File S2). Although the expression is weaker than in the cytoplasmic version, plasmatocytes can still be easily seen from all of these lines in fixed heterozygous embryos (Figure S1, E–G’), which allows analysis of the heterozygous progeny that arise, for example, during RNAi crosses. Heterozygotes can also be used in live imaging (nuclear line is shown in File S3, movie stills in Figure S1, H and H’). These lines were inserted at random positions on the second and third chromosomes, and are viable as homozygous embryos. Thus, our lines fusing the srpHemo promoter to 3xmCherry, either on its own or combined with other protein domains, permitted easy visualization of either the cytoplasm, nuclei, or actin cytoskeleton of plasmatocytes in the embryo in multiple contexts.
Direct fusion lines visualize plasmatocyte nuclei, the cytoplasm, and the cytoskeleton in larvae and adults
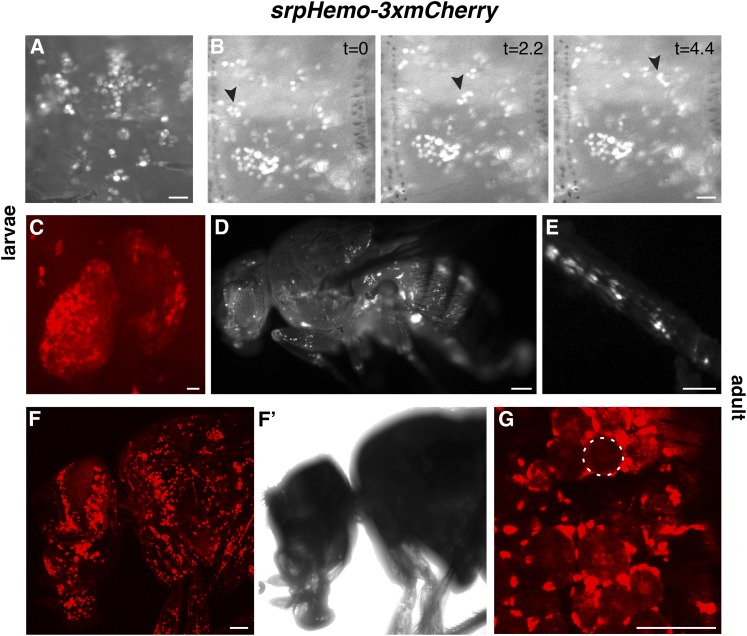
These lines also permitted clear visualization of individual plasmatocytes in larvae and adults. In larvae, the characteristic pattern of resident plasmatocytes sitting in the body wall pockets (Makhijani et al. 2011) was most easily evident live through a stereomicroscope for the cytoplasmic 3xmCherry (Figure 2A), although it was also visible in the nuclear- and actin-localized forms (Figure S2, A and B). Individual plasmatocytes from the srpHemo-3xmCherry lines were also visible in these conditions floating in the hemolymph (Figure 2B and File S4), thereby allowing detection of the fluid flow. We frequently observed clusters of floating plasmatocytes adhering to a darker nonfluorescing droplet (most visible in File S4 when examining the cells indicated with an arrow in Figure 2B). The cortical zone of the third-instar larval lymph gland was labeled by mCherry (Figure 2C) along with 40 pericardial cells, pairs of large (50 μM) oval cells in a repeating pattern along the dorsal vessel that allow the heartbeat to be easily visualized (Figure S2C and File S5) (Das et al. 2007). We also observed this pericardial fluorescence in two other lines that visualize plasmatocytes, srpHemo-GAL4 UAS-GFP (Brückner et al. 2004) and pxn-GAL4 UAS-GFP (Stramer et al. 2005), but not in hml-DsRed (Makhijani et al. 2011) (data not shown). Expression was also seen in the stomatogastric nervous system during larval stages (Figure S2D). In fixed or live larvae, plasmatocytes were also visible deep within the body, at depths of ≤130 μm with confocal imaging (Figure S2, E and E’ shows a fixed first-instar srpHemo-3xmCherry with a 3D projection of plasmatocytes). At adult stages, cytoplasmic- (Figure 2, D and E), nuclear-, and actin-targeted mCherry-labeled plasmatocytes (Figure S2, G–I) were visible in the head, thorax, and legs using a stereomicroscope. Confocal images of live srpHemo-3xmCherry adults detected plasmatocytes within the body, at depths up to 94 μm (Figure 2, F and F’). The discovery of plasmatocytes encircling cells in the fat body (Figure 2G) is particularly interesting given recent results demonstrating their role in regulating metabolism (Woodcock et al. 2015). While in larvae the srpHemo-3xmCherry signal is similar to that seen in third-instar hml-DsRed larvae (data not shown), in the adult the srpHemo-3xmCherry signal is much brighter (compare Figure S2, F’ and G’) and can be easily detected in heterozygotes of all constructs (Figure S2, G’–I’). This allows direct detection of the presence of the chromosome in adults, greatly facilitating crosses. srp-moe::GFP, the other direct fusion line expressed beyond a single stage (Table 1), is much weaker than srpHemo-3xmCherry in second- and early third-instar larvae (Figure S3, A–C), and not detectible in late third-instar larvae and adults even with a confocal microscope (Figure S3, D–K). Thus, the srpHemo-3xmCherry lines permit visualization of plasmatocytes in live and fixed samples without antibody staining from the embryo to the adult.
Figure 2.
Direct fusion lines allow live imaging of plasmatocytes in larvae and adults. (A) Live image of plasmatocytes sitting on the body wall of a srpHemo-3xmCherry larva, viewed through the cuticle with a stereomicroscope. (B) Time-lapse imaging of plasmatocytes in a srpHemo-3xmCherry larva filmed through a stereomicroscope. Three successive time points separated by 2.2 sec each are shown. Arrowhead indicates a group of cells that float in the hemolymph while most other cells remain attached to the body wall. (C) Confocal image of labeling by srpHemo-3xmCherry third-instar larval lymph gland. (D) Live image of plasmatocytes in a srpHemo-3xmCherry adult and in a close-up of (E) the leg viewed through a stereomicroscope. (F–G) Live image of a srpHemo-3xmCherry adult viewed with a confocal microscope. (F) 3D projection of plasmatocytes in the head, proboscis, and thorax. (F’) transmitted light view of the adult fly imaged in (F). (G) Single confocal slice showing plasmatocytes encircling adult fat body cells (one indicated with white circle). Scale bars correspond to 500 μM in (A–C), 250 μM in (D and E), and 100 μm in (F–G).
FACS sorting from the embryo to the adult using the direct fusion cytoplasmic line
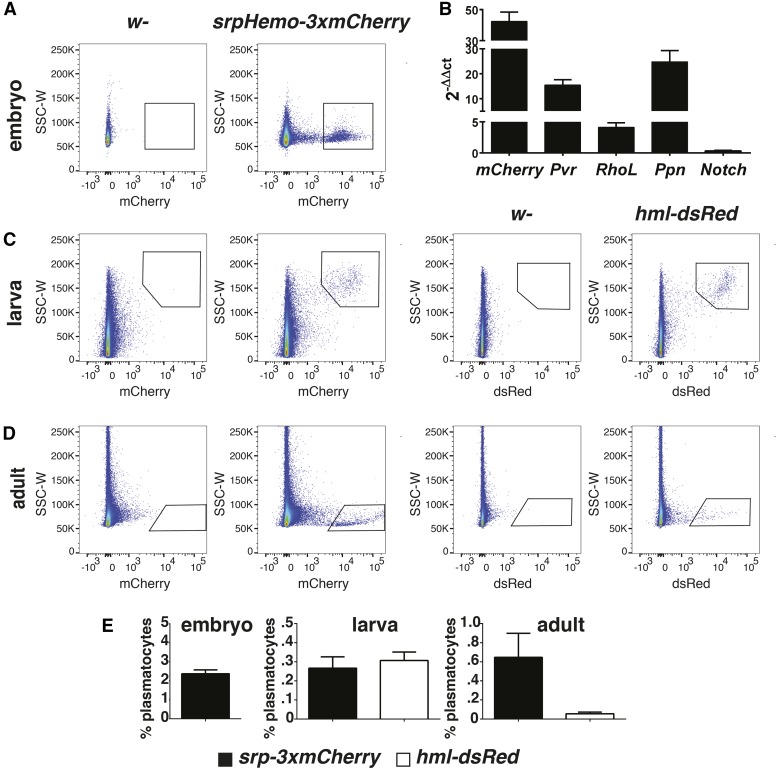
The srpHemo-3xmCherry line also facilitated purification of plasmatocytes by FACS. In stage 11 embryos, 2% of total cells from this line were mCherry-positive (Figure 3, A and E and Figure S4A). These cells were enriched for plasmatocyte markers such as Pvr, Papilin, and RhoL (Cho et al. 2002; Kramerova et al. 2003; Siekhaus et al. 2010), as assessed by qPCR (Figure 3B), but not for the broadly expressed gene Notch (Hartley et al. 1987), thus identifying the mCherry+ cells as plasmatocytes. We compared this line to the other extant direct fusion line in the red spectrum, hml-dsRed, which turns on in second-instar larvae. According to modENCODE data on FlyBase (http://flybase.org/reports/FBgn0029167.html), hemolectin is moderately expressed in LL3, almost absent in pupa, and shows low expression in adults, particularly females. We did not analyze srp-moe::GFP as it had strong extraneous expression in the embryo (Figure S1, B–D), was weak in second-early third-instar larvae, and showed no expression in late third-instar larvae and adults (Figure S3, A–K). The relative number of plasmatocytes was similar in srpHemo-3xmCherry and hml-DsRed in third-instar larvae (Figure 3, C and E), but we detected ≥10 times more fluorescent-positive plasmatocytes in srpHemo-3xmCherry than hml-DsRed adults (Figure 3, D and E), consistent with microscopic examination (Figure S2, F–G’), indicating very weak expression of hml-DsRed at this time. Using srpHemo-3xmCherry, we identified 0.25 and 0.6% of total cells as plasmatocytes in larvae and adults, respectively (Figure 3E and Figure S4, B and C). Thus, these srpHemo-derived constructs permit in vivo visualization and efficient FACS sorting, and analysis of plasmatocytes from the embryo to the adult independent of GAL4-based expression, unlike any other extant direct fusion line.
Figure 3.
The direct fusion srpHemo-3xmCherry line allows Fluorescence-Activated Cell Sorting (FACS) of plasmatocytes from embryos, larvae, and adults. (A) FACS plot of Side Scatter (SSC) vs. mCherry fluorescence signal in cells obtained from control w- and w-; srpHemo-3xmCherry and embryos, showing strong separation of mCherry+ signal from the remaining cells. (B) quantitative PCR conducted on cDNAs prepared from RNA isolated from mCherry+- and mCherry−-sorted cells using primers recognizing the plasmatocyte markers mCherry, Pvr, RhoL, Ppn, and Notch. The data are normalized to results for the housekeeping gene RpL32 and the graph shows the fold difference in signal observed between the mCherry+ (plasmatocytes) and mCherry− cells. (C and D) FACS plot of SSC vs. mCherry or DsRed fluorescence signal in cells obtained from srpHemo-3xmCherry, hml-DsRed, and control w-. In larvae (C), the two direct fusion lines show similar levels of fluorescent protein-positive plasmatocytes; however, in the adult (D), the number of DsRed+ plasmatocytes in hml-DsRed flies is strongly reduced when compared to the number of mCherry+ plasmatocytes in srpHemo-3xmCherry. (E) Quantification of plasmatocytes compared to total events detected during FACS analysis. Error bars in (B and E) represent SE of the mean. At least three independent experiments were conducted for each stage.
QF2 lines allowing genetic manipulation of plasmatocytes from the embryo to the adult
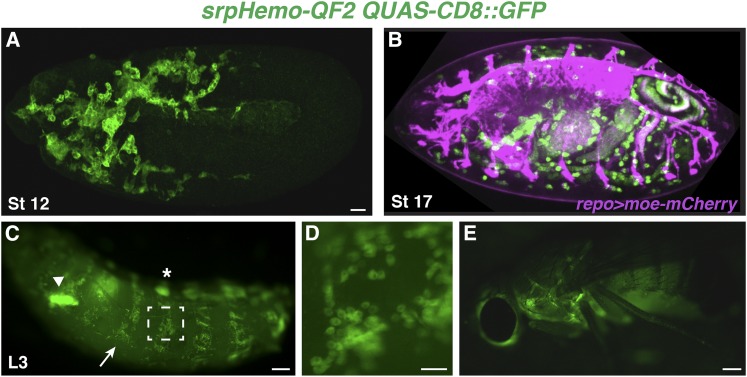
To permit genetic manipulation of plasmatocytes along with separate modulation of other tissues, we have taken advantage of the Q system (Potter et al. 2010; Potter and Luo 2011) and a nontoxic variant of the relevant transcription factor called QF2 (Riabinina et al. 2015). Our srpHemo-QF2 driver integrated at the attP16a landing site on the second chromosome can control the expression of QUAS constructs such as QUAS-CD8::GFP in plasmatocytes (Figure 4A), starting at embryonic stage 10. We additionally observed lower-level expression from srpHemo-QF2 either in the amnioserosa, mesoderm, and/or in punctate cells in the germband ectoderm in 11% of embryos (Figure S5, A and B). As QF2 does not bind to UAS sites, it can be combined with the known large repertoire of GAL4 drivers, which can then independently drive UAS constructs in other tissues. We illustrate this capability by combining srpHemo-QF2 QUAS-CD8::GFP with repo-GAL4 UAS-moe::mCherry to simultaneously label plasmatocytes and the embryonic nervous system (Figure 4B and File S6). In the larval stage, we see srpHemo-QF2-dependent expression detectable with a stereomicroscope again in the circulating and resident plasmatocyte population at the body wall during all larval stages (Figure 4, C and D), and in the third-instar larval lymph gland as well as pericardial cells (data not shown). Extraneous expression is seen in a subset of the fat body (arrowhead in Figure 4C). Plasmatocyte expression continues into the adult, which can be detected with a stereomicroscope (Figure 4E). Thus, the srpHemo-QF2 line permits the independent visualization or genetic modification of plasmatocytes and surrounding tissues.
Figure 4.
srpHemo-QF2 enables independent genetic manipulation of plasmatocytes and surrounding tissues in the embryo to the adult. (A) Confocal image of fixed stage (St) 12 srpHemo-QF2 QUAS-CD8::GFP embryo showing QF2 dependent expression in plasmatocytes. (B) Still from live imaging with a spinning disc microscope of a srpHemo-QF2 QUAS-mCD8-GFP/+; repo-GAL4 UAS-moe::mCherry/+ embryo demonstrating independent genetic control of plasmatocytes (in green) and the central nervous system (in purple). Anterior is to the left and the ventral side is up. (C–E) Stereomicroscope images of live samples. (C) srpHemo-QF2 QUAS-CD8::GFP third-instar larva showing expression in plasmatocytes (arrow), fat body (arrowhead), and cells along the dorsal vessel (asterisk). (D) Close up of plasmatocytes sitting on the body wall viewed through the cuticle from the region indicated in white box in (C). (E) srpHemo- QF2 QUAS-CD8::GFP adult. Anterior is to the left in all, dorsal is up in (A), (C–E), ventral is up in (B). Scale bars correspond to 20 μM in (A and B) and 500 μM in (C–E).
GAL80 line blocking GAL4 action in plasmatocytes from the embryo to the adult
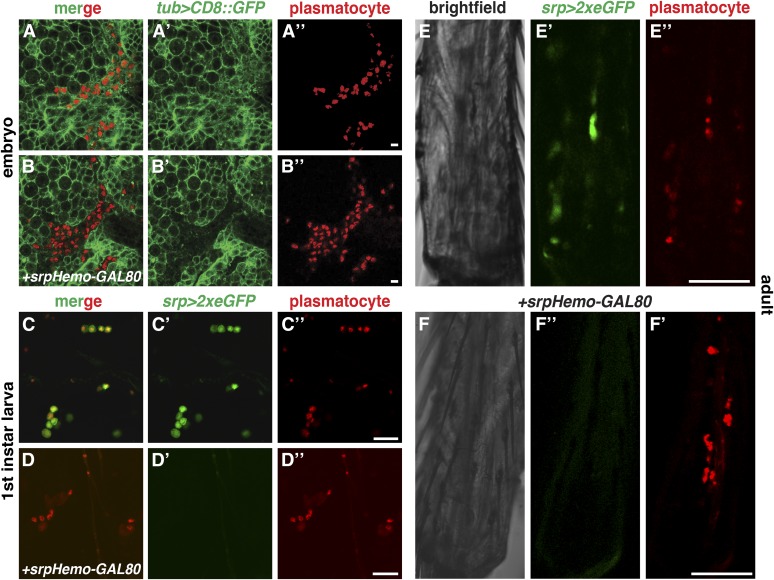
Finally, we wished to be able to genetically alter Drosophila using broadly expressed GAL4 drivers while not affecting plasmatocytes themselves. To this end, we utilized GAL80, which blocks the activity of GAL4 (Lee and Luo 1999), and created srpHemo-GAL80 lines. This construct was integrated on the second and third chromosome at the split white attP landing sites at ZH-51D and ZH-86Fb, which contain 3xP3-RFP and can be recognized in larvae by the remaining landing site red fluorescence in the brain (Figure S6A), hindgut (Figure S6B), and intersegmental nerves (asterisk in Figure S6E”), and in the top of the head (Figure S6, C and C’) (Bischof et al. 2007) in the adult, aiding detection of the chromosome in crosses. Should this extraneous RFP be deleterious for planned experiments, it can be eliminated from the line by expressing cre recombinase. To demonstrate the use of this construct, we visualized plasmatocytes using the above-described srpHemo-H2A::3xmCherry and utilized the ubiquitous driver tub-GAL4 to express UAS-CD8::GFP in the entire embryo (Figure 5, A–A”). The addition of srpHemo-GAL80 was able to block tub-GAL4-based expression of CD8::GFP in plasmatocytes (Figure 5, B and B”), but did not affect any of the surrounding cells (Figure 5, B and B’). To visualize this effect in larvae and adults, we shifted to GAL4-based expression of GFP just in plasmatocytes using srp-GAL4 UAS-2xeGFP. We observed the same capacity of the srpHemo-GAL80 to suppress the effect of GAL4 in plasmatocytes, resulting in no GFP expression in first- to third-instar larvae (compare Figure 5, C–C” to Figure 5, D–D” and Figure S6, D–D” to Figure S6, E–E”) and adults (compare Figure 5, E–E” to Figure 5, F–F”). We also noted that srp-GAL4 at these stages labeled only a subset of the plasmatocytes visualized with srpHemo-3xmCherry (Figure S6, D and E). Thus, srpHemo-GAL80 can insulate plasmatocytes from the effects of broadly expressed GAL4 drivers in the embryo, larva, and adult.
Figure 5.
srpHemo-GAL80 blocks the effect of GAL4 drivers on plasmatocytes. (A–F) Confocal images of fixed (A and B) and live (C–F) samples. (A–A”) Stage 11 srpHemo-H2A::3xmCherry/+, tub-GAL4 UAS-CD8::GFP/+, embryo showing the ubiquitous GAL4-dependent labeling of cell membranes by (A and A’) UAS-CD8::GFP, including in plasmatocytes labeled by the nuclear mCherry (A–A”). (B–B”) Stage 11 srpHemo-H2A::3xmCherry srpHemo-GAL80/+, tub-GAL4 UAS-CD8::GFP/+, embryo demonstrates that the expression of GAL80 in plasmatocytes labeled by nuclear mCherry (B”) leads to the suppression of CD8::GFP (B,B’). (C–C”) First-instar srpHemo-H2A::3xmCherry/+, srp-GAL4 UAS-2xeGFP/+, larva shows that plasmatocytes labeled by the nuclear mCherry also express cytoplasmic GFP. However, in a larva also carrying srpHemo-GAL80, the plasmatocytes (D) no longer express the GAL4-controlled GFP (D’ and D”). (E–E”) Legs of srpHemo-H2A::3xmCherry/+, srp-GAL4 UAS-2xeGFP/+, adults; GFP is expressed in plasmatocytes under GAL4 control (E’ and E”), but not in the presence of srpHemo-GAL80 (F’ and F”). Scale bars correspond to 20 μm in (A and B), 20 μm in (C and D), and 50 μm in (E and F).
Discussion
In recent years, plasmatocytes have been shown to be able to detect multiple physiological conditions, and produce adaptive and sometimes deleterious responses to them. Much of this work has focused on the signals sent from plasmatocytes to the surrounding tissues and the resulting effects (Ayyaz et al. 2015). To investigate the reverse aspect, namely how tissues signal to plasmatocytes and influence immune cell number or behavior, tools permitting the visualization or isolation of plasmatocytes in conditions where only surrounding tissues have been genetically altered are required. We have created three extremely bright lines that allow the easy detection of the plasmatocyte nucleus, cytoplasm, or actin cytoskeleton live or upon fixation from embryonic stage 8 until the adult in homozygotes and heterozygotes. The cytoplasmic line is particularly effective for FACS purification at all stages, facilitating quantitative assessment of the numbers of plasmatocytes and the levels of proteins expressed in them. This will also support next-generation sequencing analysis of the plasmatocyte transcriptome at many stages and eventually at the single-cell level. Our additional creation of srpHemo-QF2 and srpHemo-GAL80 facilitate targeted genetic manipulations in combination with other GAL4 drivers. Thus, we have produced a comprehensive set of tools permitting the analysis and genetic screening of plasmatocyte behaviors at all stages of the Drosophila life cycle.
Several of these new tools will permit experiments on plasmatocyte migration that were not feasible until now. Plasmatocytes are born from the mesoderm and start to migrate at embryonic stage 8, three stages and 3 hr before stage 11 when the previous visualization techniques using GAL4 and fluorescent reporters allowed their detection. Thus, the mechanisms that trigger the initiation of their movement, their coordination while they are in closer contact, or their choices to split into different paths (all of which occur prior to stage 10) have not been investigated. The extant direct fusion srp-moe::GFP line is weakly expressed in the embryo, absent in late larvae and adults, and utilizes a fluorophore whose activation and emission spectra is shared by many autofluorescent molecules in the fly. Thus, our srpHemo direct fusion lines that start expression at stage 8 will serve as the foundation for studies to address these migratory questions, with the nuclear line facilitating tracking and the actin labeling line aiding examination of the cytoskeletal underpinnings of this developmental movement. These lines will also aid investigations into the migration posited to underlie the final homing of plasmatocytes to their positions on the larval body wall, where they proliferate (Makhijani et al. 2011; Van De Bor et al. 2015), and to the dorsal clusters in the adult (Ghosh et al. 2015), which could shed light on resident macrophage homing in vertebrates.
The movement of plasmatocytes allows them to reach tissues where they are known to play important roles, responding to wounds (Stramer et al. 2005; Wood et al. 2006), engulfing dead cells (Tepass et al. 1994; Franc et al. 1996; Weavers et al. 2016a), promoting or killing tumors (Cordero et al. 2010; Parisi et al. 2014), regulating stem cell proliferation (Ayyaz et al. 2015; Van De Bor et al. 2015), and monitoring metabolism (Woodcock et al. 2015). The nature, though not the identity, of the cues that guide them to these tissues is somewhat understood for wounds (Razzell et al. 2013; Weavers et al. 2016b) and tumors (Pastor-Pareja et al. 2008), and remains completely unknown for the rest. Screens utilizing GAL4 expression of RNAi constructs in these tissues and monitoring plasmatocyte responses will be greatly aided by all three of our direct fusion lines, which are visible as heterozygotes. Such screens seeking to quantitatively examine effects on plasmatocyte proliferation throughout the organism should utilize FACS analysis and our srpHemo-3xmCherry line, which is effective from stage 8 to the adult. FACS analysis will detect changes in proliferation in both the lymph gland and the tissue-resident populations, as we see expression in plasmatocytes in both regions. If the chosen driver expresses broadly, our srpHemo-GAL80 can be used to block the activity of GAL4 in plasmatocytes from stage 9 in the embryo to the adult and allow the RNAi screen to only affect surrounding tissues. How tissues and plasmatocytes signal back and forth to one another can be investigated using our srpHemo-QF2 line in addition to extant GAL4 drivers to modulate the genetic behavior on both sides. If the process is only being investigated in L3 larvae and beyond, then the extant hml-QF2 (Lin and Potter 2016) can be used (Table 3). Thus, these lines should allow the identification of new mechanisms underlying plasmatocyte migration, and regulatory interactions between plasmatocytes and surrounding tissues at all stages of the Drosophila melanogaster life cycle.
Table 3. QF2 and GAL80 lines: previously published and described in this paper.
| Promoter Source-Txion Factor | Tissue Expression | Time and Level of Effect in Plasmatocytes | Reference for Creation and Expression | ||
|---|---|---|---|---|---|
| Embryo | Larva | Adult | |||
| hemolectin-QF2 | P, CC | +++L2–L3 | + | Expression based on hml-GAL4, same promoter (see Table 1) Lin and Potter (2016) | |
| srpHemo-QF2 | P, CC until embryonic stage 15, LG, PC in third-instar larvae and adult, small patch in larval FB | +++ | +++ | +++ | This paper |
| srpHemo-GAL80 | P, CC until embryonic stage 15, LG, PC in larvae and adult, SGS from embryonic stage 16 till LL3 | +++ | +++ | +++ | This paper |
P, Plasmatocytes; CC, Crystal Cells; LG, lymph gland; FB, Fat Body; SGS, Stomatogastric nervous system; PC, Pericardial Cells.
We hope that these reagents will also spur on new types of studies in the adult. The previously created hml-DsRed is visible in third-instar larvae, yet in adults hml-dsRed is hard to detect; our srpHemo-3xmCherry line (Figure 2, C–G and Figure 3D) thus enables experiments that were previously difficult. While plasmatocytes have been shown to regulate metabolism and affect aging (Woodcock et al. 2015), further investigations of how aging tissues signal to stimulate adaptive or deleterious plasmatocyte responses require direct visualization and FACS analysis of plasmatocytes in the adult. The role of other tissues in potentially influencing plasmatocyte responses to infection (Buchon et al. 2014) is another area that these lines could beneficially impact, by enabling screens as described above.
Given the wide range of processes Drosophila plasmatocytes have been shown to participate in, this set of tools will immediately prove useful to a broad number of scientists studying Drosophila development, aging, cancer, stem cells, wounds, immunity, and metabolism. Since plasmatocytes interact with tissues throughout the organism at all stages, these tools will also facilitate the discovery and investigation of many as yet unidentified regulatory processes. The genetic conservation observed between Drosophila and vertebrates strongly suggests that this future work will also prove beneficial for studies in higher organisms.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.300452/-/DC1.
Acknowledgments
We thank J. Brennecke, K. Brückner, P. Duchek, J. Heinisch, H. Gilbert, L. Luo, C. Potter, L. Ringrose, J. Sekelsky, K. Senti, and the Drosophila Genomics Resource Center, supported by National Institutes of Health (NIH) grant 2P40 OD-010949-10A1, for plasmids; F. Mauri and J. Knöblich for cell lines; and K. Brückner, P. Duchek, P. Martin, R. Reuter, the Bloomington Drosophila Stock Center (supported by NIH grant P40 OD-018537), and the Vienna Drosophila Resource Center for fly stocks. We utilized an antibody contributed by U. Banerjee, and produced by the Developmental Studies Hybridoma Bank, which was created by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the NIH, and is maintained at the University of Iowa. We thank the Life Scientific Service Units at the Institute of Science and Technology Austria for technical support, and assistance with microscopy and FACS analysis, and T. Hurd and P. Rangan for comments on the manuscript. A.G. and A.R. were supported by the Austrian Science Fund (FWF) grant DASI_FWF01_P29638S; A.R. by Marie Curie International Incoming Fellowship grant GA-2012-32950 BB: DICJI; M.R. by grant LSC16_021 from the NÖ Forschungs und Bildungsges.m.b.H. and the provincial government of Lower Austria; K.V. and S.W. by DOC Fellowships from the Austrian Academy of Sciences; and D.E.S. by Marie Curie Career Integration Grant 334077/IRTIM.
Footnotes
Communicating editor: D. Schneider
Literature Cited
- Asha H., Nagy I., Kovacs G., Stetson D., Andó I., et al. , 2003. Analysis of Ras-induced overproliferation in Drosophila hemocytes. Genetics 163: 203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avet-Rochex A., Boyer K., Polesello C., Gobert V., Osman D., et al. , 2010. An in vivo RNA interference screen identifies gene networks controlling Drosophila melanogaster blood cell homeostasis. BMC Dev. Biol. 10: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyaz A., Li H., Jasper H., 2015. Haemocytes control stem cell activity in the Drosophila intestine. Nat. Cell Biol. 17: 736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakota L., Brandt R., Heinisch J. J., 2012. Triple mammalian/yeast/bacterial shuttle vectors for single and combined Lentivirus- and Sindbis virus-mediated infections of neurons. Mol. Genet. Genomics 287: 313–324. [DOI] [PubMed] [Google Scholar]
- Bernardoni R., Vivancos V., Giangrande A., 1997. glide/gcm is expressed and required in the scavenger cell lineage. Dev. Biol. 191: 118–130. [DOI] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K., 2007. An optimized transgenesis system for Drosophila using germ-line-specific ϕC31 integrases. Proc. Natl. Acad. Sci. USA 104: 3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun A., Hoffmann J. A., Meister M., 1998. Analysis of the Drosophila host defense in domino mutant larvae, which are devoid of hemocytes. Proc. Natl. Acad. Sci. USA 95: 14337–14342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brückner K., Kockel L., Duchek P., Luque C. M., Rørth P., et al. , 2004. The PDGF/VEGF receptor controls blood cell survival in Drosophila. Dev. Cell 7: 73–84. [DOI] [PubMed] [Google Scholar]
- Buchon N., Silverman N., Cherry S., 2014. Immunity in Drosophila melanogaster — from microbial recognition to whole- organism physiology. Nature Publishing Group 14: 796–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunt S., Hooley C., Hu N., Scahill C., Weavers H., et al. , 2010. Hemocyte-secreted type IV collagen enhances BMP signaling to guide renal tubule morphogenesis in Drosophila. Dev. Cell 19: 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S., Dudzic J. P., Li X., Collas E. J., Boquete J.-P., et al. , 2016. Remote control of intestinal stem cell activity by haemocytes in Drosophila. PLoS Genet. 12: e1006089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho N. K., Keyes L., Johnson E., Heller J., Ryner L., et al. , 2002. Developmental control of blood cell migration by the Drosophila VEGF pathway. Cell 108: 865–876. [DOI] [PubMed] [Google Scholar]
- Clark R. I., Woodcock K. J., Geissmann F., Trouillet C., Dionne M. S., 2011. Multiple TGF-β superfamily signals modulate the adult Drosophila immune response. Curr. Biol. 21: 1672–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero J. B., Macagno J. P., Stefanatos R. K., Strathdee K. E., Cagan R. L., et al. , 2010. Oncogenic Ras diverts a host TNF tumor suppressor activity into tumor promoter. Dev. Cell 18: 999–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozatier M., Ubeda J.-M., Vincent A., Meister M., 2004. Cellular immune response to parasitization in Drosophila requires the EBF orthologue collier. PLoS Biol. 2: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D., Aradhya R., Ashoka D., Inamdar M., 2007. Post-embryonic pericardial cells of Drosophila are required for overcoming toxic stress but not for cardiac function or adult development. Cell Tissue Res. 331: 565–570. [DOI] [PubMed] [Google Scholar]
- Dudzic J. P., Kondo S., Ueda R., Bergman C. M., Lemaitre B., 2015. Drosophila innate immunity: regional and functional specialization of prophenoloxidases. BMC Biol. 13: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K. A., Demsky M., Montague R. A., Weymouth N., Kiehart D. P., 1997. GFP-moesin illuminates actin cytoskeleton dynamics in living tissue and demonstrates cell shape changes during morphogenesis in Drosophila. Dev. Biol. 191: 103–117. [DOI] [PubMed] [Google Scholar]
- Elrod-Erickson M., Mishra S., Schneider D., 2000. Interactions between the cellular and humoral immune responses in Drosophila. Curr. Biol. 10: 781–784. [DOI] [PubMed] [Google Scholar]
- Estrada B., Michelson A. M., 2008. A genomic approach to myoblast fusion in Drosophila, pp. 299–314 in Cell Fusion: Overviews and Methods, edited by Chen E. H. Humana Press, Totowa, NJ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans C. J., Liu T., Banerjee U., 2014. Drosophila hematopoiesis: markers and methods for molecular genetic analysis. Methods 68: 242–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans I. R., Zanet J., Wood W., Stramer B. M., 2010. Live imaging of Drosophila melanogaster embryonic hemocyte migrations. J. Vis. Exp. 36: 1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadok V. A., Warner M. L., Bratton D. L., Henson P. M., 1998. CD36 is required for phagocytosis of apoptotic cells by human macrophages that use either a phosphatidylserine receptor or the vitronectin receptor (alpha(v)beta(3)). J. Immunol. 161: 6250–6257. [PubMed] [Google Scholar]
- Fessler J. H., Fessler L. I., 1989. Drosophila extracellular matrix. Annu. Rev. Cell Biol. 5: 309–339. [DOI] [PubMed] [Google Scholar]
- Franc N. C., Dimarcq J. L., Lagueux M., Hoffmann J., Ezekowitz R. A., 1996. Croquemort, a novel Drosophila hemocyte/macrophage receptor that recognizes apoptotic cells. Immunity 4: 431–443. [DOI] [PubMed] [Google Scholar]
- Galko M. J., Krasnow M. A., 2004. Cellular and genetic analysis of wound healing in Drosophila larvae. PLoS Biol. 2: E239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Singh A., Mandal S., Mandal L., 2015. Active hematopoietic hubs in Drosophila adults generate hemocytes and contribute to immune response. Dev. Cell 33: 478–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouon-Evans V., Rothenberg M. E., Pollard J. W., 2000. Postnatal mammary gland development requires macrophages and eosinophils. Development 127: 2269–2282. [DOI] [PubMed] [Google Scholar]
- Greenberg M. E., Sun M., Zhang R., Febbraio M., Silverstein R., et al. , 2006. Oxidized phosphatidylserine–CD36 interactions play an essential role in macrophage-dependent phagocytosis of apoptotic cells. J. Exp. Med. 203: 2613–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann T., Wilson J., Kulbe H., Li N. F., Leinster D. A., et al. , 2005. Macrophages induce invasiveness of epithelial cancer cells via NF-kappa B and JNK. J. Immunol. 175: 1197–1205. [DOI] [PubMed] [Google Scholar]
- Hartley D. A., Xu T. A., Artavanis-Tsakonas S., 1987. The embryonic expression of the Notch locus of Drosophila melanogaster and the implications of point mutations in the extracellular EGF-like domain of the predicted protein. EMBO J. 6: 3407–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramerova I. A., Kramerov A. A., Fessler J. H., 2003. Alternative splicing of papilin and the diversity of Drosophila extracellular matrix during embryonic morphogenesis. Dev. Dyn. 226: 634–642. [DOI] [PubMed] [Google Scholar]
- Kurucz E., Márkus R., Zsámboki J., Folkl-Medzihradszky K., Darula Z., et al. , 2007. Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr. Biol. 17: 649–654. [DOI] [PubMed] [Google Scholar]
- Lebestky T., Chang T., Hartenstein V., Banerjee U., 2000. Specification of Drosophila hematopoietic lineage by conserved transcription factors. Science 288: 146–149. [DOI] [PubMed] [Google Scholar]
- Lee T., Luo L., 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22: 451–461. [DOI] [PubMed] [Google Scholar]
- Leers M. P. G., Björklund V., Björklund B., Jörnvall H., Nap M., 2002. An immunohistochemical study of the clearance of apoptotic cellular fragments. Cell. Mol. Life Sci. 59: 1358–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B., Hoffmann J., 2007. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25: 697–743. [DOI] [PubMed] [Google Scholar]
- Lin C.-C., Potter C. J., 2016. Editing transgenic DNA components by inducible gene replacement in Drosophila melanogaster. Genetics 203: 1613–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhijani K., Alexander B., Tanaka T., Rulifson E., Brückner K., 2011. The peripheral nervous system supports blood cell homing and survival in the Drosophila larva. Development 138: 5379–5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manaka J., Kuraishi T., Shiratsuchi A., Nakai Y., Higashida H., et al. , 2004. Draper-mediated and phosphatidylserine-independent phagocytosis of apoptotic cells by Drosophila hemocytes/macrophages. J. Biol. Chem. 279: 48466–48476. [DOI] [PubMed] [Google Scholar]
- Martinek N., Shahab J., Saathoff M., Ringuette M., 2008. Haemocyte-derived SPARC is required for collagen-IV-dependent stability of basal laminae in Drosophila embryos. J. Cell Sci. 121: 1671–1680. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y., Louani A., Dragu A., Sánchez-Sánchez B. J., Serna-Morales E., et al. , 2017. A moving source of matrix components is essential for de novo basement membrane formation. Curr. Biol. 27: 3526–3534.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milchanowski A. B., Henkenius A. L., Narayanan M., Hartenstein V., Banerjee U., 2004. Identification and characterization of genes involved in embryonic crystal cell formation during Drosophila hematopoiesis. Genetics 168: 325–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard T. H., Martin P., 2008. Dynamic analysis of filopodial interactions during the zippering phase of Drosophila dorsal closure. Development 135: 621–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira S., Stramer B., Evans I., Wood W., Martin P., 2010. Prioritization of competing damage and developmental signals by migrating macrophages in the Drosophila embryo. Curr. Biol. 20: 464–470. [DOI] [PubMed] [Google Scholar]
- Okulski H., Druck B., Bhalerao S., Ringrose L., 2011. Quantitative analysis of polycomb response elements (PREs) at identical genomic locations distinguishes contributions of PRE sequence and genomic environment. Epigenetics Chromatin 4: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsson B., Page D. T., 2005. Condensation of the central nervous system in embryonic Drosophila is inhibited by blocking hemocyte migration or neural activity. Dev. Biol. 279: 233–243. [DOI] [PubMed] [Google Scholar]
- Parisi F., Stefanatos R. K., Strathdee K., Yu Y., Vidal M., 2014. Transformed epithelia trigger non-tissue-autonomous tumor suppressor response by adipocytes via activation of Toll and Eiger/TNF signaling. Cell Rep. 6: 855–867. [DOI] [PubMed] [Google Scholar]
- Pastor-Pareja J. C., Wu M., Xu T., 2008. An innate immune response of blood cells to tumors and tissue damage in Drosophila. Dis. Model. Mech. 1: 144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsouris D., Li P.-P., Thapar D., Chapman J., Olefsky J. M., et al. , 2008. Ablation of CD11c-positive cells normalizes insulin sensitivity in obese insulin resistant animals. Cell Metab. 8: 301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez E., Lindblad J. L., Bergmann A., 2017. Tumor-promoting function of apoptotic caspases by an amplification loop involving ROS, macrophages and JNK in Drosophila. Elife 6: e26747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard J. W., 2009. Trophic macrophages in development and disease. Nat. Rev. Immunol. 9: 259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter C. J., Luo L., 2011. Using the Q system in Drosophila melanogaster. Nat. Protoc. 6: 1105–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter C. J., Tasic B., Russler E. V., Liang L., Luo L., 2010. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell 141: 536–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pull S. L., Doherty J. M., Mills J. C., Gordon J. I., Stappenbeck T. S., 2005. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proc. Natl. Acad. Sci. USA 102: 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratheesh A., Belyaeva V., Siekhaus D. E., 2015. Drosophila immune cell migration and adhesion during embryonic development and larval immune responses. Curr. Opin. Cell Biol. 36: 71–79. [DOI] [PubMed] [Google Scholar]
- Razzell W., Evans I. R., Martin P., Wood W., 2013. Calcium flashes orchestrate the wound inflammatory response through DUOX activation and hydrogen peroxide release. Curr. Biol. 23: 424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riabinina O., Luginbuhl D., Marr E., Liu S., Wu M. N., et al. , 2015. Improved and expanded Q-system reagents for genetic manipulations. Nat. Methods 12: 219–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. M., Preston C. R., Phillis R. W., Johnsonschlitz D. M., Benz W. K., et al. , 1988. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics 118: 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G. M., Hong L., Brokstein P., Evans-Holm M., Frise E., et al. , 2000. A Drosophila complementary DNA resource. Science 287: 2222–2224. [DOI] [PubMed] [Google Scholar]
- Schneider I., 1972. Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol. 27: 353–365. [PubMed] [Google Scholar]
- Schnoor M., Cullen P., Lorkowski J., Stolle K., Robenek H., et al. , 2008. Production of type VI collagen by human macrophages: a new dimension in macrophage functional heterogeneity. J. Immunol. 180: 5707–5719. [DOI] [PubMed] [Google Scholar]
- Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N. G., Palmer A. E., et al. , 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22: 1567–1572. [DOI] [PubMed] [Google Scholar]
- Siekhaus D., Haesemeyer M., Moffitt O., Lehmann R., 2010. RhoL controls invasion and Rap1 localization during immune cell transmigration in Drosophila. Nat. Cell Biol. 12: 605–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieron A. L., Louneva N., Fertala A., 2002. Site-specific interaction of bone morphogenetic protein 2 with procollagen II. Cytokine 18: 214–221. [DOI] [PubMed] [Google Scholar]
- Sinenko S. A., Mathey-Prevot B., 2004. Increased expression of Drosophila tetraspanin, Tsp68C, suppresses the abnormal proliferation of ytr-deficient and Ras/Raf-activated hemocytes. Oncogene 23: 9120–9128. [DOI] [PubMed] [Google Scholar]
- Sorrentino R. P., Tokusumi T., Schulz R. A., 2007. The friend of GATA protein U-shaped functions as a hematopoietic tumor suppressor in Drosophila. Dev. Biol. 311: 311–323. [DOI] [PubMed] [Google Scholar]
- Stofanko M., Kwon S. Y., Badenhorst P., 2008. A misexpression screen to identify regulators of Drosophila larval hemocyte development. Genetics 180: 253–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stramer B., Wood W., Galko M. J., Redd M. J., Jacinto A., et al. , 2005. Live imaging of wound inflammation in Drosophila embryos reveals key roles for small GTPases during in vivo cell migration. J. Cell Biol. 168: 567–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stramer B., Moreira S., Millard T., Evans I., Huang C.-Y., et al. , 2010. Clasp-mediated microtubule bundling regulates persistent motility and contact repulsion in Drosophila macrophages in vivo. J. Cell Biol. 189: 681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassetto M., Kunitomi M., Andino R., 2017. Circulating immune cells mediate a systemic RNAi-based adaptive antiviral response in Drosophila. Cell 169: 314–325.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass U., Fessler L. I., Aziz A., Hartenstein V., 1994. Embryonic origin of hemocytes and their relationship to cell death in Drosophila. Development 120: 1829–1837. [DOI] [PubMed] [Google Scholar]
- Tokusumi T., Shoue D. A., Tokusumi Y., Stoller J. R., Schulz R. A., 2009. New hemocyte-specific enhancer-reporter transgenes for the analysis of hematopoiesis in Drosophila. Genesis 47: 771–774. [DOI] [PubMed] [Google Scholar]
- Van De Bor V., Zimniak G., Papone L., Cerezo D., Malbouyres M., et al. , 2015. Companion blood cells control ovarian stem cell niche microenvironment and homeostasis. Cell Rep. 13: 546–560. [DOI] [PubMed] [Google Scholar]
- Vukicevic S., Latin V., Chen P., Batorsky R., Reddi A. H., et al. , 1994. Localization of osteogenic protein-1 (bone morphogenetic protein-7) during human embryonic development: high affinity binding to basement membranes. Biochem. Biophys. Res. Commun. 198: 693–700. [DOI] [PubMed] [Google Scholar]
- Weavers H., Evans I. R., Martin P., Wood W., 2016a Corpse engulfment generates a molecular memory that primes the macrophage inflammatory response. Cell 165: 1658–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weavers H., Liepe J., Sim A., Wood W., Martin P., et al. , 2016b Systems analysis of the dynamic inflammatory response to tissue damage reveals spatiotemporal properties of the wound attractant gradient. Curr. Biol. 26: 1975–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg S. P., McCann D., Desai M., Rosenbaum M., Leibel R. L., et al. , 2003. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest. 112: 1796–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W., Faria C., Jacinto A., 2006. Distinct mechanisms regulate hemocyte chemotaxis during development and wound healing in Drosophila melanogaster. J. Cell Biol. 173: 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock K. J., Kierdorf K., Pouchelon C. A., Vivancos V., Dionne M. S., et al. , 2015. Macrophage-derived upd3 cytokine causes impaired glucose homeostasis and reduced lifespan in Drosophila fed a lipid-rich diet. Immunity 42: 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.-H., Bellmunt E., Scheib J. L., Venegas V., Burkert C., et al. , 2009. Glial precursors clear sensory neuron corpses during development via Jedi-1, an engulfment receptor. Nat. Neurosci. 12: 1534–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn T. A., Chawla A., Pollard J. W., 2013. Macrophage biology in development, homeostasis and disease. Nature 496: 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Barnes G. T., Yang Q., Tan G., Yang D., et al. , 2003. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J. Clin. Invest. 112: 1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidman-Rémy A., Regan J. C., Brandão A. S., Jacinto A., 2012. The Drosophila larva as a tool to study gut-associated macrophages: PI3K regulates a discrete hemocyte population at the proventriculus. Dev. Comp. Immunol. 36: 638–647. [DOI] [PubMed] [Google Scholar]
- Zanet J., Jayo A., Plaza S., Millard T., Parsons M., et al. , 2012. Fascin promotes filopodia formation independent of its role in actin bundling. J. Cell Biol. 197: 477–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zettervall C. J., Anderl I., Williams M. J., Palmer R., Kurucz E., et al. , 2004. A directed screen for genes involved in Drosophila blood cell activation. Proc. Natl. Acad. Sci. USA 101: 14192–14197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Hashimi H., Schwartz L. M., Nambu J. R., 1995. Programmed cell death in the Drosophila central nervous system midline. Curr. Biol. 5: 784–790. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All plasmids and Drosophila strains created in this study are available from the authors upon request. All strains are also available through the Bloomington Drosophila Stock Center.