Abstract
During antifungal drug treatment and hypoxia, genetic and epigenetic changes occur to maintain sterol homeostasis and cellular function. In this study, we show that SET domain-containing epigenetic factors govern drug efficacy to the medically relevant azole class of antifungal drugs. Upon this discovery, we determined that Set4 is induced when Saccharomyces cerevisiae are treated with azole drugs or grown under hypoxic conditions; two conditions that deplete cellular ergosterol and increase sterol precursors. Interestingly, Set4 induction is controlled by the sterol-sensing transcription factors, Upc2 and Ecm22. To determine the role of Set4 on gene expression under hypoxic conditions, we performed RNA-sequencing analysis and showed that Set4 is required for global changes in gene expression. Specifically, loss of Set4 led to an upregulation of nearly all ergosterol genes, including ERG11 and ERG3, suggesting that Set4 functions in gene repression. Furthermore, mass spectrometry analysis revealed that Set4 interacts with the hypoxic-specific transcriptional repressor, Hap1, where this interaction is necessary for Set4 recruitment to ergosterol gene promoters under hypoxia. Finally, an erg3Δ strain, which produces precursor sterols but lacks ergosterol, expresses Set4 under untreated aerobic conditions. Together, our data suggest that sterol precursors are needed for Set4 induction through an Upc2-mediated mechanism. Overall, this new sterol-signaling pathway governs azole antifungal drug resistance and mediates repression of sterol genes under hypoxic conditions.
Keywords: chromatin, SET4, hypoxia, antifungal drugs, epigenetics, sterol, gene expression, Saccharomyces cerevisiae
STEROLS are a major class of neutral lipids that affect important physical properties of membranes in eukaryotic cells, such as membrane fluidity, transport, and permeability (Parks and Casey 1995; Espenshade and Hughes 2007). The primary sterol found in the cell membranes of all fungi and protozoa is ergosterol (Espenshade and Hughes 2007; Weete et al. 2010; Dupont et al. 2012). Ergosterol is the yeast equivalent to cholesterol in humans and it has important structural and signaling functions necessary for the survival of the organism (Parks and Casey 1995; Mouritsen and Zuckermann 2004; Espenshade and Hughes 2007). In addition, ergosterol and cholesterol biosynthesis is of fundamental and medical interest because this pathway contains the targets of several antifungal and statin drugs (Parks and Casey 1995; Odds et al. 2003; Kathiravan et al. 2012).
In Saccharomyces cerevisiae and other yeast species, ergosterol biosynthesis is an oxygen-dependent pathway (Parks and Casey 1995; Kwast et al. 1998; Rosenfeld and Beauvoit 2003; Espenshade and Hughes 2007; Ishtar Snoek and Yde Steensma 2007). Under hypoxic or anaerobic conditions, ergosterol production is blocked, leading to its depletion (Rosenfeld and Beauvoit 2003; Joshua and Höfken 2017). Yeast respond to ergosterol depletion by signaling structural changes in the cell wall to facilitate uptake of exogenous ergosterol (Abramova et al. 2001; Alimardani et al. 2004; Ishtar Snoek and Yde Steensma 2007; Zavrel et al. 2013). Therefore, ergosterol homeostasis is a balance between ergosterol synthesis and sterol uptake from the exogenous environment.
To maintain ergosterol homeostasis, ergosterol (ERG) genes, which encode enzymes needed for ergosterol biosynthesis, are tightly controlled by transcription factors (Kwast et al. 2002; Davies et al. 2005; Davies and Rine 2006; Hickman and Winston 2007; Ishtar Snoek and Yde Steensma 2007; Joshua and Höfken 2017). Three transcription factors that are known to regulate ERG genes include Hap1, Ecm22, and Upc2 (Fytlovich et al. 1993; Vik and Rine 2001; Becerra et al. 2002; Ter Linde and Steensma 2002; Tamura et al. 2004; Davies et al. 2005; Davies and Rine 2006). Upc2 and Ecm22 are paralogs and activate genes in response to sterol levels. During sterol depletion using statin or antifungal azole drugs, ERG genes are induced by Upc2 and Ecm22 (Vik and Rine 2001; Wilcox et al. 2002; Davies et al. 2005; Davies and Rine 2006; Joshua and Höfken 2017).
Under aerobic conditions, Hap1 is a heme-dependent transcription factor that is required for the proper expression of mitochondrial respiratory and oxidative stress genes (Zhang and Hach 1999; Becerra et al. 2002). Hap1 is also needed for steady state ERG transcript levels and full induction of ERG genes when cells are treated with lovastatin (Fytlovich et al. 1993; Becerra et al. 2002; Tamura et al. 2004; Davies and Rine 2006). Under hypoxic conditions, Hap1 is a heme-independent transcription factor (Zhang and Hach 1999); however, depending on the strain, Hap1 acts as a transcriptional repressor or activator. In a S288C FY strain that expresses an integrated wild-type (WT) copy of HAP1, Hap1 functions together with the corepressor complex, Tup1, and Cyc8 to repress ERG genes (Hickman and Winston 2007). In addition, deletion of HAP1 resulted in an increase in ERG genes and other genes involved in ergosterol metabolism (Hickman and Winston 2007). In contrast, in a W303 strain that expresses an endogenous HAP1, ERG genes increase in expression when switched to hypoxic conditions (Davies and Rine 2006); although it was not determined if Hap1 mediated this increase. Nevertheless, an aGH1 strain lacking HAP1 showed a subset of genes were downregulated under hypoxia, suggesting Hap1 can act as a transcriptional activator (Lombardia et al. 2000). Currently, it is unclear what mediates these strain-specific differences.
Although the aforementioned transcription factors are necessary for regulating ergosterol biosynthesis, additional studies are needed to determine how this medically relevant pathway is regulated and how sterol-sensing transcription factors function with epigenetic regulators. Epigenetic regulators have been shown to mediate yeast growth under antifungal drug treatment. For example, deletions of BRE1, BRE2, and HOS2 results in a hypersensitive growth defect to the antifungal metabolite Brefeldin A (BFA) (Muren et al. 2001; South et al. 2013). Work from our laboratory demonstrated that loss of the H3K4 methyltransferase, Set1, or loss of the Set1 complex members that affect H3K4 methylation, has a hypersensitive growth defect in the presence of BFA (South et al. 2013). Interestingly, the BFA hypersensitivity observed in a set1∆ strain is due to decreased expression of conserved ERG genes resulting in decreased ergosterol production (South et al. 2013). Nevertheless, set1∆ strains become resistant to BFA when grown in the presence of ergosterol, which is due to the induction of sterol transporters and uptake of exogenously provided ergosterol (South et al. 2013). Although loss of SET3 resulted in BFA hypersensitivity and decreased expression of ERG genes, Set4, the Set3 paralog, was not investigated (South et al. 2013). Until this study, little was known about the biological and biochemical function of Set4.
In this study, we investigated the role of SET domain-containing epigenetic factors, in particular Set4, under conditions that alter ergosterol levels such as azole drug treatment and hypoxia. Initially, we determined that deletion of SET1 and SET3 results in hypersensitivity to the medically relevant antifungal drugs ketoconazole and fluconazole. In contrast, a deletion of SET4 results in azole drug resistance, indicating that SET domain proteins govern antifungal drug efficacy. However, we demonstrate that the azole-resistant phenotype is independent of changes in ERG11 or ABC transporter gene expression, mechanisms known to play a role in azole resistance. Interestingly, under ergosterol-limiting conditions, we show that Set4 expression is induced by the sterol-sensing transcriptional activators Upc2 and Ecm22. RNA-sequencing analysis determined that Set4 is required for ERG gene repression. Importantly, we show that Set4 directly targets the ergosterol gene promoters ERG11 and ERG3 under hypoxia and that Set4 recruitment is dependent on the transcriptional repressor, Hap1. Finally, we demonstrate that Set4 expression is upregulated in an erg3∆ strain, suggesting a precursor sterol, but not ergosterol, regulates Set4 levels. Overall, we have discovered a new sterol–Upc2 signaling pathway mediated by Set4 that governs azole drug efficacy and sterol homeostasis under hypoxic conditions.
Materials and Methods
Plasmids and yeast strains
All plasmids and yeast strains are described in Supplemental Material, Table S1 and Table S2 in File S7. Note that, depending on the yeast strain, there are genomic differences in HAP1. Most S. cerevisiae strains including W303 strains contain a WT copy of HAP1 (Davies and Rine 2006). Many, if not all, S288C strains including BY4741 and FY strains contain an in-frame Ty1 insertion near the 3′ end of the HAP1 gene (Gaisne et al. 1999; Hickman and Winston 2007). The hap1–Ty1 gene fusion is expressed and replaces 13 amino acids of the Hap1 C terminus as well as adds 32 amino acids from the Ty1 element (Gaisne et al. 1999). Because the hap1–Ty1 gene fusion has been shown to partially compromise the function of Hap1, the FY2609 strain (the “HAP1-corrected” strain) was also used (Gaisne et al. 1999; Hickman and Winston 2007). FY2609 was modified by replacing the hap1–Ty1 gene fusion with a WT copy of HAP1 (Hickman and Winston 2007).
The yeast plasmids used in this study were constructed and PCR verified as previously described (South et al. 2010, 2013; Mersman et al. 2012; Zhang et al. 2016). FLAG-tagged strains were generated using the N-ICE plasmid tagging system and PCR verified (Zhang et al. 2016).
Yeast growth conditions
Yeast growth and drug treatment were performed as previously described (Agarwal et al. 2003). The indicated strains were inoculated in SC media and grown to saturation overnight. Cells were diluted to an OD600 of 0.1 and recovered to log phase for 3 hr shaking at 30°. Prior to treatment, cells were collected for the untreated sample and zero time point. Cultures were treated at an OD600 of 0.2 with 56 µg/ml ketoconazole (Sigma-Aldrich, St. Louis, MO) dissolved in DMSO as previously described (Agarwal et al. 2003). Cells were collected every 3 hr. The indicated yeast strains were grown in YPD media to log phase under aerobic shaking conditions, or for 8 hr under hypoxia in a Bio-Bag Type A System (14-910-5; Fisher Scientific, Pittsburgh, PA). Cells were immediately pelleted and flash frozen.
Azole dilution assays and growth assays
For dilution assays, yeast strains were inoculated in SC media and grown to saturation overnight. Yeast strains were diluted to an OD600 of 0.1 and grown in SC media to log phase shaking at 30°. The indicated strains were spotted in fivefold dilutions starting at an OD600 of 0.01 on untreated SC plates or plates containing 1 µg/ml ketoconazole (Sigma-Aldrich) or 12 µg/ml fluconazole (Sigma-Aldrich). Plates were grown at 30° for 2–5 days. For growth assays, the indicated yeast strains were inoculated in SC media and grown to saturation overnight. Yeast strains were diluted to an OD600 of 0.1 and grown in SC media to log phase shaking at 30°. The indicated strains were diluted to an OD600 of 0.0001 in 100 μl SC media. Cells were left untreated or treated with 20 μg/ml ketoconazole and grown for 50 hr shaking at 30°. The cell density OD600 was determined every 2 hr using the Bio-Tek Synergy 4 multimode plate reader (http://www.biotek.com).
Yeast extraction and Western blot analysis
Whole cell extraction and Western blot analysis to detect 3×FLAG-Set4, 3×FLAG-Set3, histone modifications, and glucose-6-phosphate dehydrogenase (G6PDH) were performed as previously described (Briggs et al. 2001; Fingerman et al. 2005; Mersman et al. 2012). The polyclonal anti-FLAG rabbit antibody was used to detect 3×FLAG-Set4 and 3×FLAG-Set3 expression at 1:5000 (catalog F7425; Sigma-Aldrich). The H3K4 methylation-specific and anti-G6PDH antibodies were used as previously described (South et al. 2013; Harmeyer et al. 2015).
Gene expression analysis
RNA was isolated from cells by standard acid phenol purification. Reverse transcription was performed using the QuantiTect Reverse Transcription Kit (QIAGEN, Valencia, CA) per the manufacturer’s instructions. Primer Express 3.0 software was used for designing primers (see Table S3 in File S7) and quantitative real-time polymerase chain reaction (qRT-PCR) was performed as previously described (South et al. 2013; Zhang et al. 2016). Three biological replicates, including three technical replicates, were performed for all samples. Data were analyzed using the ΔΔCt method where ACT1 or RDN18-1 [18S ribosomal RNA (rRNA)] was used as an internal control. All samples were normalized to an untreated, untagged WT strain. The unpaired t-test was used to determine the statistical significance between two strains at the genes of interest. All statistical values were reported as the raw P-values. The mean, SD, SEM, and statistical significance for appropriate data can be found in Table S5 in File S7.
Immunoprecipitation for mass spectrometry analysis
For analysis of Set4 protein interaction partners, 200 ml of the 3×FLAG-Set4 and the untagged WT strains were grown to log phase under hypoxia for 8 hr. Whole cell lysate was prepared from harvested cells by lysing cells with glass beads and bead beating. Cells were lysed with 2 ml of lysis buffer (20 mM HEPES, pH 7.5, 350 mM NaCl, 10% glycerol, 0.1% Tween 20) containing protease and phosphatase inhibitors as previously described (Mersman et al. 2012). For immunoprecipitation, 60 μl of Protein G Magnetic Dynabeads were conjugated with 20 μl M2 FLAG antibody and immunoprecipitated with 2 ml of whole cell lysate for 2 hr rotating at 4°. Proteins were resolved on a 10% SDS-PAGE gel and whole lanes were cut out for in-gel trypsin digestion. For details on mass spectrometry (MS) analysis see File S4 and the supplemental methods in File S6.
Chromatin immunoprecipitation
ZipChIP was performed as previously described (Harmeyer et al. 2015). Briefly, 50 ml cultures were grown to log phase (OD600 of 0.6) in YPD media at 30° under aerobic shaking conditions or 8 hr of hypoxia. Cells were formaldehyde cross-linked and harvested as previously described (Harmeyer et al. 2015). Cell lysates were precleared with 5 µl of unbound Protein G magnetic beads for 30 min rotating at 4°. A total of 200 µl of precleared lysate was immunoprecipitated with 10 µl of Protein G magnetic beads (10004D; Life Technologies) conjugated to 1 µl of M2 FLAG antibody (F1804; Sigma-Aldrich) or MYC antibody (9E10). Probe sets used in qRT-PCR are described in Table S4 in File S7.
RNA sequencing
The BY4741 WT and set4Δ strains were grown to log phase in aerobic and hypoxic conditions for 8 hr in YPD media. Total RNA of three biological replicates for each condition and sample were isolated by standard acid phenol purification, treated with DNase (Ambion), and total RNA was purified using standard acid phenol purification. The quality of the RNA was tested using an Agilent Bioanalyzer 2100 using the High Sensitivity DNA Chip. The complementary DNA library was prepared by the Purdue Genomics Facility using the TruSeq Stranded Kit with poly(A) selection (Illumina) according to the manufacturer’s instructions. For more details on RNA-sequencing analysis see the supplemental methods in File S6.
Data availability
Strains are available upon request. File S5 and File S7 contain additional tables and figures including the strain list and genotypes, gene expression primers, and chromatin immunoprecipitation (ChIP) probe sets. File S1, File S2, File S3, and File S4 include gene expression data from the RNA-sequencing analysis and peptide hits from the MS analysis. Genome-wide RNA-sequencing data are available at Gene Expression Omnibus under accession number GSE107492.
Results
Set1 and Set3 govern azole drug sensitivity
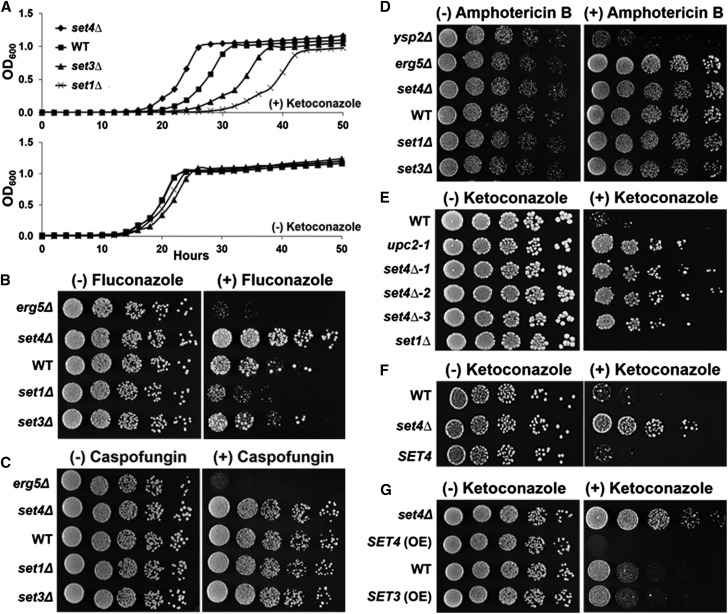
Our previous work determined that loss of Set1 or Set3 resulted in hypersensitivity to BFA due to the decreased expression of the ergosterol gene, ERG11, which encodes the enzyme inhibited by azole antifungal drugs such as ketoconazole and fluconazole (Odds et al. 2003; South et al. 2013). To test the role of SET domain proteins in drug sensitivity, particularly Set4, we performed liquid growth assays in the presence and absence of the medically relevant azole drug, ketoconazole, using WT, set1Δ, set3Δ, and set4Δ strains. All strains grew similarly without the drug (Figure 1A), while set1Δ and set3Δ strains showed hypersensitivity toward 20 µg/ml ketoconazole when compared to WT (Figure 1A) and similarly to what we observed for BFA (South et al. 2013). Interestingly, the set4Δ strain grew better than WT in the presence of ketoconazole (Figure 1A), indicating that Set3 and Set4 have different biological functions.
Figure 1.
SET domain proteins govern azole antifungal drug sensitivity and resistance. (A) Growth curve of indicated BY4741 strains over a 50-hr time course in SC media with 20 µg/ml ketoconazole or SC media. (B–E) Dilution assays of the indicated BY4741 strains spotted on SC plates containing 12 µg/ml fluconazole, 0.5 µg/ml caspofungin, 0.5 µg/ml amphotericin B, or 1 µg/ml ketoconazole. (F) Dilution assay of BY4741 WT or set4Δ strains transformed with plasmids containing SET4 from its endogenous promoter or empty vector spotted on SC-Ura plates with 1 µg/ml ketoconazole. (G) Dilution assay of BY4741 WT, set4Δ, and overexpressed SET4 and SET3 strains. Strains were spotted on SC plates with 1 µg/ml ketoconazole.
To determine if the two subclasses of azole drugs, imidazole and triazole, would behave similarly, we performed dilution assays on plates containing the triazole drug, fluconazole, using WT, set1Δ, set3Δ, and set4Δ strains. Similar to what was observed for liquid growth assays with ketoconazole, the set1Δ strain showed hypersensitivity when grown on plates containing 12 µg/ml fluconazole (Figure 1, A and B), while the set3∆ strain did not have an apparent growth difference (Figure 1B). Intriguingly, the set4Δ strain grew better than WT on fluconazole plates, suggesting that the loss of SET4 leads to azole drug resistance (Figure 1B). The DMSO-treated WT, set1Δ, set3∆, and set4Δ strains grew similarly, demonstrating that the changes in observed growth are due to azole drug treatment (Figure 1, A and B). Additionally, the erg5Δ strain was used as positive control for fluconazole sensitivity. Altogether, these results demonstrate that SET domain proteins govern the efficacy of both the imidazole and triazole subclasses of azole antifungal drugs.
To characterize the extent of SET domain proteins in drug sensitivity and resistance, we tested whether set1Δ, set3Δ, and set4Δ strains were sensitive to other classes of antifungal drugs including the echinocandin drug, caspofungin, and the polyene drug, amphotericin B (Odds et al. 2003; Campoy and Adrio 2017). Dilution assays were performed on plates containing 0.25 µg/ml caspofungin or 0.5 µg/ml amphotericin B. In contrast to the azole drug assays, the set1Δ, set3Δ, and set4Δ strains grew like WT on plates containing either caspofungin or amphotericin B; indicating that the phenotypes observed in the set1Δ, set3Δ, and set4Δ strains were specific to the azole class of antifungal drugs (Figure 1, C and D). The erg5Δ and ysp2Δ strains were used as positive controls for drug sensitivity (Markovich et al. 2004; Gatta et al. 2015). To further analyze the set4Δ strain, we tested whether the resistant-like growth observed in a set4Δ strain treated with azole drugs grows similarly to upc2-1, a well-known, azole-resistant mutant (Flowers et al. 2012). Dilution assays were performed on ketoconazole plates spotted with three independent knockouts of SET4 and the upc2-1 mutant strain. All three set4Δ strains grew like the upc2-1 mutant and grew better than WT, providing strong evidence that yeast strains lacking Set4 will consequently lead to azole drug resistance (Figure 1E). Again, all strains grew similarly on plates without the drug, showing that all strains were equally spotted, and the set1Δ strain was used as a control for azole sensitivity (Figure 1E).
To confirm that loss of Set4 promotes azole drug resistance, we performed a rescue experiment by adding back SET4 to the set4∆ strain and analyzing yeast growth in the presence and absence of ketoconazole using a dilution plate assay. The WT and set4Δ strains transformed with an empty yeast plasmid vector or SET4 driven from the endogenous promoter were spotted on SC-Ura plates with or without 1 µg/ml ketoconazole. The WT strain transformed with empty vector and the set4Δ strain transformed with endogenous SET4 grew similarly on ketoconazole plates, whereas the set4Δ strain transformed with empty vector showed resistant growth on ketoconazole plates (Figure 1F). All three strains grew similarly on SC-Ura plates without the drug (Figure 1F).
Because a set4Δ strain is resistant to ketoconazole and fluconazole, we hypothesized that overexpression of Set4 would lead to azole hypersensitivity. To test this hypothesis, we integrated a constitutive PYK1 promoter upstream of SET4 and SET3 using our N-ICE plasmid system and performed a dilution assay on ketoconazole and fluconazole plates (Zhang et al. 2016). Overexpression of Set4 resulted in hypersensitive growth compared to WT under ketoconazole or fluconazole treatment, suggesting that too much Set4 negatively affects cell viability (Figure 1G). Overexpression of Set3 grew similarly to WT, further demonstrating that Set4 and Set3 play distinct roles in governing azole efficacy (Figure 1G). The set4∆ strain was used as a control to show drug resistance (Figure 1G). All strains grew similarly on SC plates, indicating that overexpression of Set4 or Set3 did not alter growth conditions. To verify that Set4 and Set3 were overexpressed in the strains generated using the N-ICE plasmid tagging system, qRT-PCR and Western blot analysis were used to detect Set4 and Set3 expression levels (Figure S1, A and B, in File S5). As expected, Set4 and Set3 expression increased significantly in the overexpression strains compared to the endogenous tagged strains (Figure S1, A and B, in File S5). Overall, these results establish that the loss of SET4 is responsible for azole resistance and that cellular levels of Set4 are important for governing this phenotype.
Set4 expression is induced under azole antifungal drug treatment
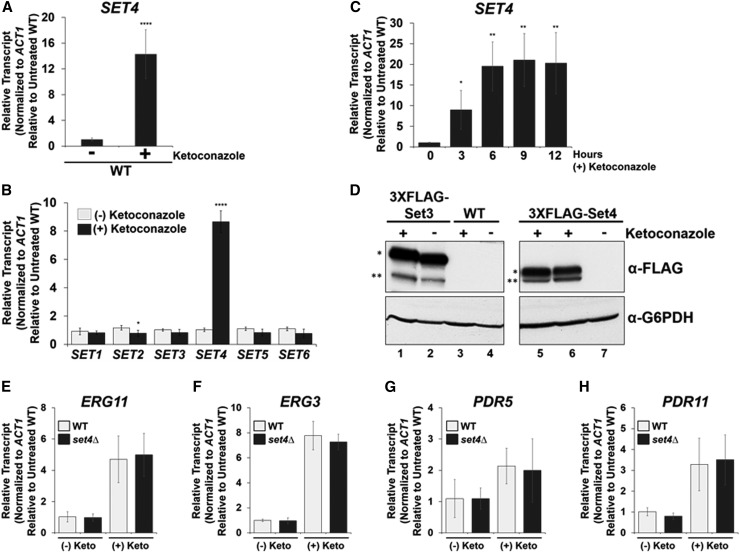
Because Set4 levels are critical for altering azole resistance, endogenous SET4 messenger RNA (mRNA) levels were quantified from WT cells that were treated with ketoconazole or DMSO by qRT-PCR analysis. Interestingly, SET4 transcript levels increase significantly (16-fold) following 3 hr of treatment with 56 µg/ml ketoconazole (Figure 2A). Actin mRNA levels were used to normalize SET4 transcript levels and the ketoconazole-treated WT sample was set relative to DMSO-treated WT cells. These results are consistent with a microarray study that showed increased SET4 expression upon treatment with ketoconazole (Agarwal et al. 2003).
Figure 2.
Azole antifungal drug treatment induces Set4 expression. (A and B) Expression of indicated genes was determined in WT cells treated with DMSO or 56 µg/ml ketoconazole for 3 hr by qRT-PCR analysis. Statistical analysis identified significant changes for SET2 and SET4 expression (* P < 0.05 and **** P < 0.0001, respectively). (C) SET4 expression was determined in WT cells treated with DMSO or 56 µg/ml ketoconazole by qRT-PCR analysis. Gene expression analysis was set relative to the DMSO-treated WT and expression was normalized to actin mRNA levels (ACT1). Data were analyzed from three biological replicates with three technical replicates. Error bars represent SD. * P < 0.05, ** P < 0.01. (D) Western blot analysis of Set3 and Set4 protein levels under DMSO and 56 µg/ml ketoconazole treatment. Lane 6 represents 3 hr of ketoconazole treatment. Lanes 1, 3, and 5 represent 6 hr of ketoconazole treatment. Lanes 2, 4, and 7 show untreated Set3, untagged WT, and Set4, respectively. G6PDH was used as a loading control. * indicates 3×FLAG-Set3 and 3×FLAG-Set4 protein levels, respectively. ** denotes protein degradation bands. (E–H) Gene expression analysis (qRT-PCR) of the indicated genes in WT and set4∆ strains treated with DMSO or 56 μg/ml ketoconazole for 6 hr. The indicated mRNA transcript levels were normalized to ACT1 and set relative to the DMSO-treated WT [indicated as (-) Keto]. Error bars represent the SD of three biological replicates each with three technical repeats. Gene expression and Western blot analyses were performed using the BY4741 strain.
Because deletions of SET1 and SET3 result in sensitivity to azole treatment, we tested whether transcript levels of genes encoding SET domain proteins are affected by ketoconazole treatment. WT cells were treated with ketoconazole and qRT-PCR was used to analyze SET1–SET6 expression. SET1, SET3, SET5, and SET6 transcript levels were not significantly altered in WT cells treated with ketoconazole; however, SET2 expression showed a 38% decrease in transcript levels under ketoconazole (Figure 2B). The SET1–SET6 mRNA levels from the ketoconazole-treated samples were set relative to their respective gene in DMSO-treated conditions and normalized to ACT1.
To characterize the induction of SET4 in response to ketoconazole, we treated WT cells with 56 µg/ml ketoconazole and cells were collected over time. As determined by qRT-PCR analysis, SET4 transcript levels peaked at 6 hr (20-fold increase) postketoconazole treatment (Figure 2C). ACT1 was used to normalize SET4 mRNA levels and the ketoconazole-treated samples were set relative to the DMSO WT cells, indicated by the zero time point. To determine if the increase in SET4 expression following ketoconazole treatment correlated with an increase in protein levels, Set4 and Set3 were N-terminally 3×FLAG tagged, using our N-ICE plasmid tagging system, at their respective loci and expressed from their endogenous promoter (Zhang et al. 2016). Western blot analysis of the 3×FLAG-Set4 strain showed an increase in Set4 protein following 3 and 6 hr of ketoconazole treatment (Figure 2D, lanes 6 and 5, respectively). Surprisingly, Set4 levels were undetectable in the DMSO-treated sample, indicating that Set4 expression is extremely low or not expressed under standard yeast growth conditions (Figure 2D, lane 7). Western blot analysis of 3×FLAG-Set3 protein levels showed minimal differences in Set3 protein levels in the DMSO- and ketoconazole-treated samples (Figure 2D, lanes 1 and 2, respectively). G6PDH was used as a loading control. The untagged WT was used as a negative control for ketoconazole- and DMSO-treated strains (Figure 2D, lanes 3 and 4, respectively). Overall, these results demonstrate that Set4 expression is induced under azole treatment and provides the first evidence of an inducible SET domain-containing protein in S. cerevisiae and, to our knowledge, in other eukaryotes.
Set4 does not affect the expression of genes known to be involved in azole resistance
In recent years, fungal pathogens such as Candida albicans and C. glabrata have developed resistance to azole antifungal drugs by upregulating the gene encoding the azole drug target, ERG11, or upregulating genes encoding the ABC transporters PDR5 and PDR11 (Cowen et al. 2014; Whaley et al. 2016; Campoy and Adrio 2017). To determine if the azole resistance observed in a set4∆ strain is correlated with an upregulation of ERG11, PDR5, or PDR11 compared to WT; qRT-PCR analysis was performed at the indicated genes in WT and set4∆ strain treated with DMSO or 56 μg/ml ketoconazole for 6 hr. However, no statistical difference in the expression of ERG11, ERG3, PDR5, and PDR11 between WT and the set4∆ strain was observed (Figure 2, E–H). Therefore, Set4 does not mediate drug resistance through known resistant genes but may regulate other genes that are unknown to be involved in azole drug resistance.
Set4 expression is highly induced under hypoxic conditions
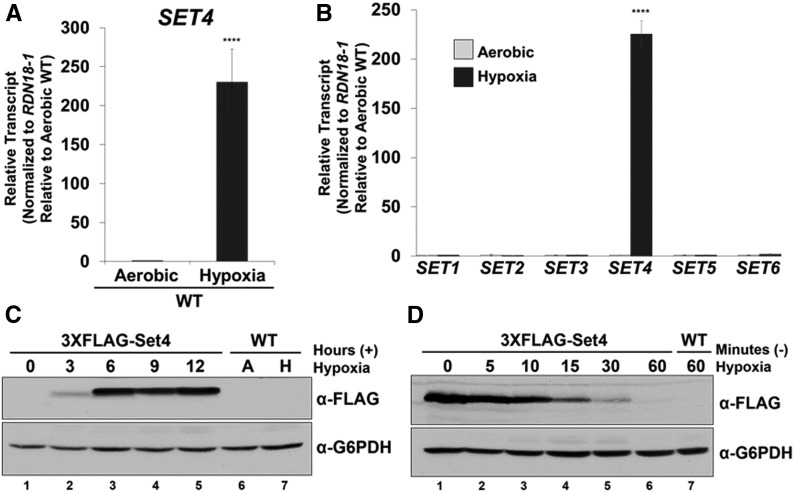
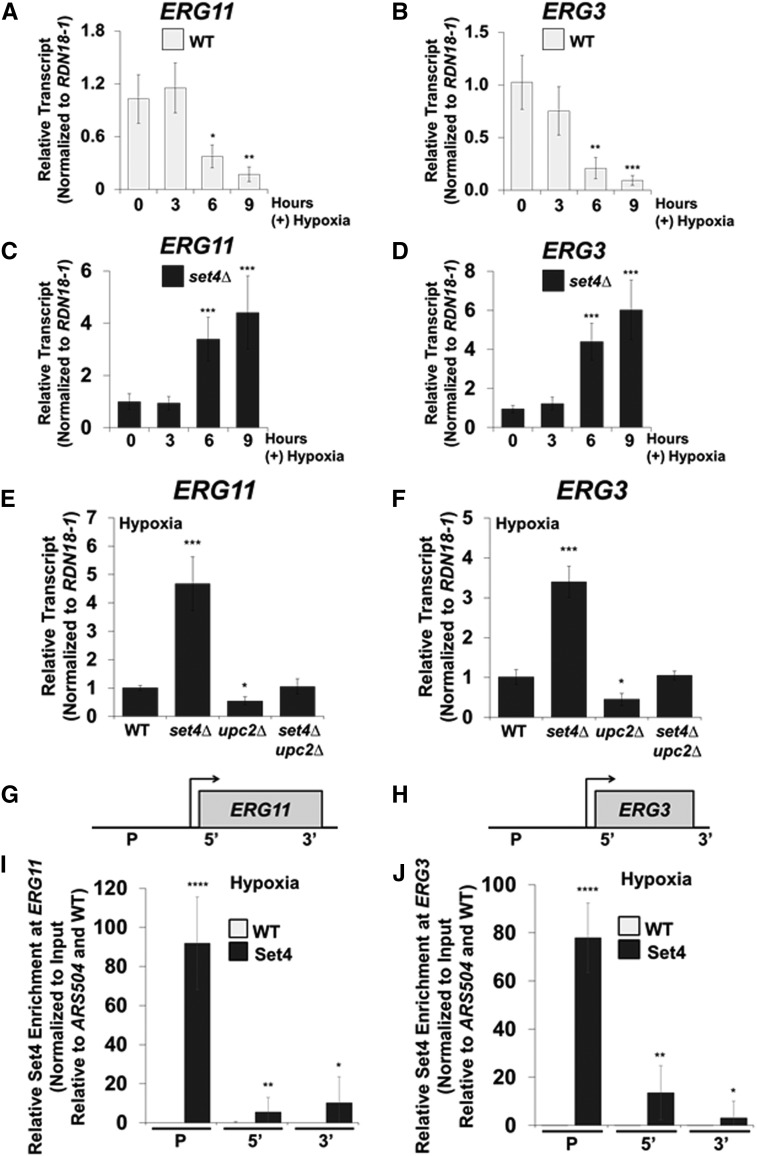
Because yeast are not typically exposed to azole drugs, we wanted to determine a physiological condition where Set4 is induced. Interestingly, azole drugs decrease cellular ergosterol levels through inhibition of Erg11 (Odds et al. 2003; Campoy and Adrio 2017). A similar condition occurs when yeast are grown under low oxygen such as hypoxia or anaerobic conditions (Rosenfeld and Beauvoit 2003; Espenshade and Hughes 2007; Ishtar Snoek and Yde Steensma 2007). Under hypoxia, ERG genes are repressed in strains that have full-length HAP1, and many of the enzymes in the ergosterol biosynthesis pathway have limited function since they are oxygen dependent (Rosenfeld and Beauvoit 2003; Hon et al. 2005; Hickman and Winston 2007). To determine if Set4 expression increased under hypoxia, we performed qRT-PCR analysis on WT cells grown under aerobic conditions or 8 hr of hypoxia. Strikingly, SET4 transcript levels were highly upregulated (∼200-fold) under hypoxia (Figure 3A). SET4 transcript levels were normalized to RDN18-1 and set relative to the aerobic WT strain. These results demonstrate that hypoxia is a physiological condition that significantly induces SET4 expression in S. cerevisiae.
Figure 3.
Hypoxia induces Set4 expression. (A and B) SET4 transcript level was determined in WT cells grown under aerobic conditions or 8 hr of hypoxia by qRT-PCR analysis. **** P < 0.0001. (B) Expression of SET1–SET6 was determined in WT cells grown under aerobic conditions or 8 hr of hypoxia by qRT-PCR analysis. Gene expression analyses were set relative to the aerobic WT using the 18S rRNA (RDN18-1) as the internal control to normalize transcript levels. Data were analyzed from three biological replicates with three technical replicates. Error bars represent SD. SET4 was the only gene that significantly changed in expression (**** P < 0.0001). (C) Western blot analysis of Set4 protein induction over time under hypoxia. Aerobic (A) and hypoxia (H). The untagged WT was used as a negative control. (D) Western blot analysis of Set4 protein levels following release from hypoxic conditions. Lane 1 represents Set4 protein levels following 8 hr of hypoxia. Lanes 2–7 indicate Set4 protein levels following release from hypoxic conditions. G6PDH was used as a loading control. Gene expression and Western blot analyses were performed in BY4741 strains.
To determine if genes encoding other SET domain proteins modulate their expression in response to hypoxia, we analyzed SET1–SET6 transcript levels by qRT-PCR under aerobic and hypoxic conditions (Figure 3B). SET4 transcript levels increased significantly (P < 0.0001) under hypoxic conditions (∼200-fold), whereas no significant gene expression changes were observed for the remaining genes encoding SET domain proteins (Figure 3B). RDN18-1 levels were used to normalize transcript levels and each gene was set to the appropriate aerobic sample. These results are consistent with the SET4 induction under ketoconazole, indicating that conditions that decrease cellular ergosterol levels lead to induction of Set4 expression.
To determine if the Set4 protein was also induced under hypoxic conditions, we grew the 3×FLAG-Set4 strain driven from the endogenous SET4 promoter over time. Western blot analysis of the 3×FLAG-Set4 strain showed an increase in Set4 protein levels after 3 hr of hypoxia (Figure 3C, lane 2). Set4 proteins levels peaked at 6 hr and remained detectable through 12 hr of hypoxic treatment (Figure 3C, lanes 3–5). To further characterize Set4 protein expression levels, we examined how rapidly Set4 expression decreased when switching from hypoxic to aerobic conditions. The 3×FLAG-Set4 strain was grown under hypoxic conditions for 8 hr and released into aerobic shaking conditions (Figure 3D). Cells were collected 5, 10, 15, 30, and 60 min following the switch from hypoxia to aerobic conditions (Figure 3D). Western blot analysis of 3×FLAG-Set4 protein levels demonstrated that Set4 expression decreased rapidly and is undetectable following 60 min of aerobic growth (Figure 3D, lanes 2–6). G6PDH was used as a loading control and the untagged WT strains grown under aerobic or hypoxic conditions were used as negative controls (Figure 3C, lanes 6 and 7, and Figure 3D, lane 7). These results show that Set4 expression is tightly regulated under aerobic conditions. Altogether, our results suggest that proper Set4 expression is needed under environmental conditions that alter sterol levels so that appropriate cellular adaptation occurs.
Set4 is necessary for global gene expression changes under hypoxic conditions
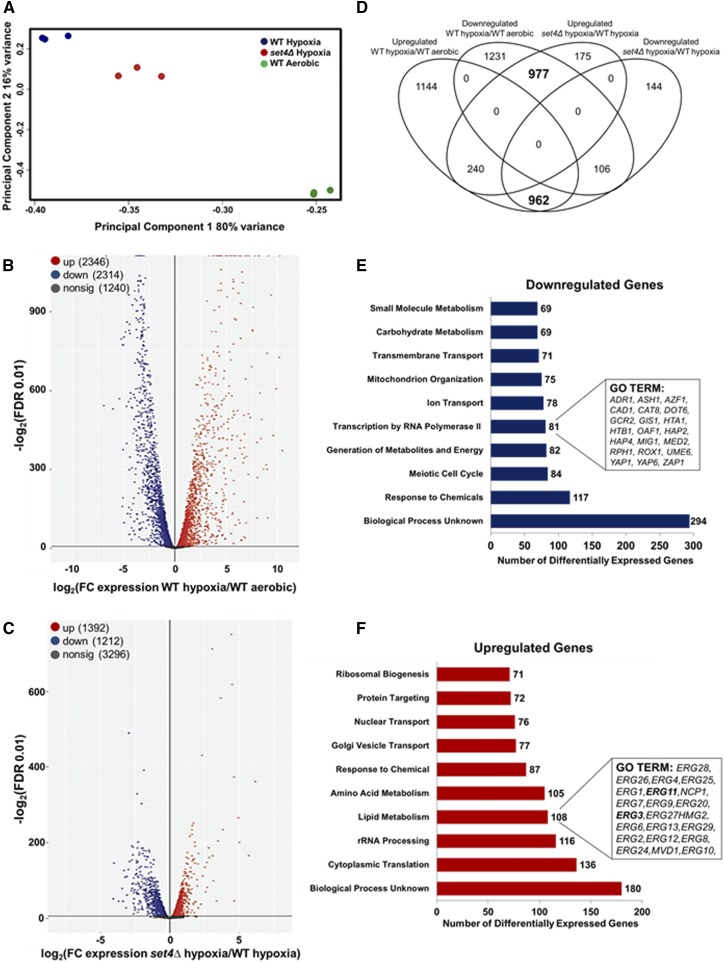
Transcriptional changes play a major role in yeast adaptation to various environmental conditions, and therefore vast epigenetic changes must occur for yeast to adapt and survive under hypoxic conditions (Ter Linde et al. 1999; Hickman et al. 2011; Bendjilali et al. 2017). To determine if induction of Set4 modulates hypoxic gene expression and adaptation in response to hypoxia, we performed RNA-sequencing analysis on the BY4741 isogenic WT and set4Δ strains grown under hypoxic conditions for 8 hr. After filtering the data, 5900 expressed genes were identified in the analysis covering >90% of the S. cerevisiae genome. Principal component analysis (PCA) was performed between aerobic and hypoxic samples. PCA showed that the biological replicates of each strain and condition cluster together (Figure 4A). Additionally, PCA demonstrated that the WT hypoxia, WT aerobic, and set4∆ hypoxia clusters are distinct from one another (Figure 4A). Importantly, the set4∆ and WT hypoxic sample sets are distinctly different but cluster closer together than the WT aerobic sample set (Figure 4A). This difference in sample clustering suggests that global changes in gene expression occur under hypoxia and when SET4 is deleted (Figure 4A).
Figure 4.
Set4 alters global levels of gene expression under hypoxic conditions. The genome-wide changes in gene expression under hypoxia were performed using BY4741 WT and set4Δ strains. (A) The PCA for WT and set4Δ hypoxic samples relative to WT aerobic samples based on the counts per million. (B) Volcano plot showing the significance [−log2 (FDR), y-axis] vs. the fold change (x-axis) of the DEGs identified in the WT hypoxic samples relative to WT aerobic samples. (C) Volcano plot showing the significance [−log2 (FDR), y-axis] vs. the fold change (x-axis) of the DEGs identified in the set4Δ hypoxic samples relative to WT hypoxic samples. Genes with significant differential expression (FDR < 0.01) in (B and C) are highlighted in red or blue for up- and downregulated genes, respectively. Gray highlighted genes are considered nonsignificant. (D) Venn diagram showing the number of genes identified as differentially expressed (FDR < 0.01). Bold numbers indicate a high overlap of genes predicted to be in common by chance based on Fisher’s exact test (P < 10−52). (E and F) GO terms of the Set4-dependent DEGs under hypoxic conditions. Downregulated genes refer to the DEGs that are dependent on Set4 for activation and the upregulated genes refer to the DEGs that are dependent on Set4 for repression. Significantly enriched groups of GO terms were identified for the DEGs from set4Δ and WT hypoxic samples. Bar plots show the number of DEGs in each GO group that are dependent on Set4 under hypoxia. The number of genes in each GO group is shown to the right of each bar. Genes identified are shown in the inset boxes.
EdgeR analysis was used to identify the differentially expressed genes (DEGs) under hypoxia vs. aerobic conditions in WT samples. A total of 2346 significantly upregulated genes (40% of the genes analyzed) and 2314 significantly downregulated genes (39% of the genes analyzed) were identified (Figure 4B and File S1). PCA and DEG analysis demonstrated by the volcano scatterplot {−log2 [false discovery rate (FDR)], y-axis} vs. the fold change (x-axis) of the DEGs indicate that the hypoxic WT strain is substantially and statistically different from the aerobic WT strain (Figure 4, A and B). Functional analysis indicated that a large number of genes induced under hypoxia are involved in cell stress, carbohydrate metabolism, cell wall maintenance, and lipid metabolism (File S1). Many genes identified in our genome-wide analysis have previously been found to respond to hypoxic conditions (Hickman et al. 2011; Bendjilali et al. 2017). For example, several studies—including this one—have found the following genes upregulated under hypoxia: ANB1, HEM13, CYC7, and COX5b; whereas CYC1 and COX5a were downregulated under hypoxia (File S1) (Ter Linde et al. 1999; Hickman and Winston 2007; Bendjilali et al. 2017). Furthermore, many cell wall genes are upregulated, including the DAN/TIR mannoprotein genes and the seripauperin (PAU) genes (File S1) (Ter Linde et al. 1999; Rachidi et al. 2000; Abramova et al. 2001; Cohen et al. 2001; Hickman et al. 2011; Bendjilali et al. 2017). In addition, the sterol transport genes PDR11 and AUS1 were upregulated (File S1) (Wilcox et al. 2002; Kohut et al. 2011).
The PCA and volcano plots also showed a statistically significant difference between the hypoxic set4∆ strain and the hypoxic WT (Figure 4, A and C). EdgeR analysis determined that 1392 genes were significantly upregulated (23% of genes analyzed) and 1212 genes were significantly downregulated (20% of genes analyzed) (FDR < 0.01) (Figure 4C and File S2). To determine the number of genes that overlap between the different strains and conditions tested, we plotted a Venn diagram representing the upregulated and downregulated genes for each comparison group (Figure 4D). According to the Venn diagram, a high degree of overlap was observed between genes that exhibited increased expression in WT cells under hypoxia and genes that were significantly downregulated in the set4∆ strain under hypoxia (962 genes, P < 10−52 by Fisher’s exact test) (Figure 4D). In contrast, the intersection between the DEGs exhibiting decreased expression in WT and a set4∆ strain was not larger than predicted by chance alone (106 genes, P > 0.99). These results reveal that Set4 is required directly or indirectly for the activation of a significant number of genes under hypoxia (Figure 4D).
In addition, 977 genes were downregulated in the comparison between WT under hypoxia and aerobic conditions but upregulated in the set4∆ strain under hypoxia, suggesting that Set4 is required to directly or indirectly repress these genes (Figure 4D). This high degree of overlap was also statistically significant and indicates that Set4 is required for the repression of a significant number of genes under hypoxia (977 genes, P < 10−52 by Fisher’s exact test). The intersection between the DEGs exhibiting increased expression in both WT and a set4∆ strain was not larger than predicted by chance alone (106 genes, P > 0.99) (Figure 4D). Together, these comparisons imply that Set4 is necessary to promote the transcriptional switch from aerobic to hypoxic conditions. Lastly, the GoSLIM database on the Saccharomyces Genome Database was used to categorize the biological processes, molecular functions, and pathways of the DEGs (Figure 4, E and F, and File S3). The top-10 gene ontology (GO) terms are shown in Figure 4, E and F. Additional GO terms are shown in File S3.
Many of the DEGs that were downregulated in the set4∆ strain included genes encoding meiotic cell cycle factors, factors that respond to chemical stress, genes involved in metabolite generation, and factors that mediate transcription by RNA polymerase II (Figure 4E and File S3). A majority of the DEGs that were upregulated in the set4∆ strain included genes involved in cytoplasmic translation, rRNA processing, amino acid metabolism, and lipid metabolism; suggesting that these genes are dependent on Set4 for repression under hypoxia (Figure 4F and File S3). Many genes upregulated in a set4∆ strain were not categorized in any biological process (File S3). Of these uncharacterized genes, 18 are PAU genes which have been hypothesized to play a role in cell wall remodeling (File S3) (Luo and van Vuuren 2009). Interestingly, 19 out of 20 genes involved in ergosterol metabolism were significantly upregulated in the set4∆ strain compared to WT (Figure 4F). These results indicate that Set4 is required for repressing genes involved in ergosterol metabolism. Because ERG genes are repressed under hypoxic conditions, it is likely that Set4 induction facilitates repression of the ERG genes and the entire ergosterol biosynthesis pathway for proper sterol homeostasis, cell wall remodeling, and uptake of exogenous sterols.
Set4 directly targets and enriches at ergosterol gene promoters under hypoxia
Cellular ergosterol levels are known to decrease in WT cells under hypoxia (Hickman et al. 2011; Bendjilali et al. 2017). To verify that ERG gene expression decreases in response to hypoxia, we performed qRT-PCR analysis in FY2609 WT cells grown under aerobic conditions and 3, 6, and 9 hr of hypoxia. ERG11 and ERG3 transcript levels decreased significantly in WT cells following 6 hr (65 and 81%, respectively) and 9 hr of hypoxic treatment (85 and 93%, respectively) (Figure 5, A and B). The significant decrease in ERG11 and ERG3 transcript levels correlates with the time at which Set4 expression peaked under hypoxic conditions (Figure 3C). In addition, our RNA-sequencing data clearly indicate that one role for Set4 is to repress genes involved in ergosterol biosynthesis (Figure 4F). To validate our RNA-sequencing result, we analyzed the ERG11 and ERG3 transcript levels by qRT-PCR in the FY2609 WT and set4Δ strains grown under hypoxia for 3, 6, and 9 hr of hypoxia. Consistent with our RNA-sequencing analysis, a set4Δ strain had approximately three- to sixfold more expression of ERG11 and ERG3 mRNA transcript levels compared to the WT strain at the 6 and 9 hr time points (Figure 5, C and D). To determine if this increase in ERG gene expression under hypoxia was specific to the set4Δ strain, we analyzed the ERG11 and ERG3 mRNA levels in WT, set4Δ, set3Δ, and the double deletion set4Δset3Δ strain under hypoxic conditions. In contrast to aerobic conditions, ERG11 and ERG3 transcript levels are not altered in a set3∆ strain under hypoxic conditions (Figure S2 in File S5). Furthermore, ERG11 and ERG3 transcript levels significantly increased in the set4Δ and set4Δset3Δ strains, demonstrating that Set4 specifically mediates ERG gene repression under hypoxic conditions (Figure S2 in File S5).
Figure 5.
Set4 represses ergosterol genes and directly targets the promoters of ERG11 and ERG3 under hypoxia. (A and B) The mRNA transcript levels of ERG11 and ERG3 were determined in WT cells grown under aerobic or hypoxic conditions over time. qRT-PCR expression analysis in the WT strain was set relative to the aerobic or hypoxic WT using the 18S ribosome rRNA (RDN18-1) as the internal control to normalize transcript levels. (C and D) qRT-PCR analysis of ERG11 and ERG3 expression in set4Δ cells grown under aerobic or hypoxic conditions for 3, 6, or 9 hr. ERG11 and ERG3 expression in the set4∆ strain were normalized to RDN18-1 and set to the WT strain at the same relative time point. Data were analyzed from three biological replicates that had three technical replicates each. Error bars represent SD. * P < 0.05, ** P < 0.01, *** P < 0.005. (E and F) Gene expression analysis (qRT-PCR) of ERG11 and ERG3 under hypoxic conditions in WT, set4Δ, upc2Δ, and set4Δupc2Δ. The mRNA transcript levels were normalized to RDN18-1 and set relative to WT grown under hypoxia. * P < 0.05, *** P < 0.005. (G and H) Schematics of ERG11 and ERG3 loci with the specified positions of ChIP probes. (I and J) ChIP analysis of Set4 enriched at ERG11 and ERG3 was performed under hypoxic conditions using antibodies specific to a 3×FLAG tag for the detection of Set4. ChIP analyses were normalized to DNA input samples and set relative to the ARS504 and untagged WT. Error bars represent SD for three biological replicates with three technical replicates each. * P < 0.05, ** P < 0.01, **** P < 0.0001. Gene expression and ChIP analyses were performed using the FY2609 strain.
Both ERG11 and ERG3 mRNA transcript levels increase significantly in a set4∆ strain under hypoxia, suggesting that a transcriptional activator upregulated these ERG genes in the absence of Set4. Previously it has been shown that the transcriptional activator, Upc2, targets ERG genes under sterol limiting conditions (Vik and Rine 2001; Davies et al. 2005; Davies and Rine 2006). To determine whether ERG11 and ERG3 gene expression is upregulated by Upc2 when SET4 is deleted, qRT-PCR analysis was performed under hypoxic conditions in the WT, set4Δ, upc2Δ, and set4Δupc2Δ strains. Interestingly, both ERG11 and ERG3 mRNA transcript levels failed to increase in the upc2Δ and set4Δupc2Δ strains under hypoxic conditions, indicating that Upc2 is required to upregulate ERG11 and ERG3 expression in the absence of Set4 (Figure 5, E and F). Additionally, ERG11 and ERG3 transcript levels significantly decreased in a upc2Δ strain but not in the set4Δupc2Δ strain, suggesting that Set4 represses these genes further when UPC2 is deleted (Figure 5, E and F). The mRNA transcript levels were normalized to RDN18-1 and set relative to the hypoxic WT strain. Together, these data imply that Set4 and Upc2 oppose one another to regulate ERG gene expression under hypoxic conditions.
Because Set4 and Upc2 both regulate the expression of ERG11 and ERG3 (Figure 5, E and F), we hypothesized that Set4 directly targets ERG gene promoters to facilitate gene repression under hypoxia. To determine Set4 binding at ERG promoters, we performed ChIP analysis using our published ZipChIP method (Harmeyer et al. 2015). Probe sets were designed to the promoter and the 5′ and 3′ ORFs of ERG11 and ERG3 (Figure 5, G and H). Interestingly, ZipChIP analysis showed that 3×FLAG-Set4 was significantly enriched at the promoter regions of the ERG11 and ERG3 loci when compared with the untagged WT (Figure 5, I and J). In contrast, 3×FLAG-Set4 was minimally detected at the 5′ ORF and the 3′ ORF (Figure 5, I and J). The qRT-PCR analysis was normalized to DNA input levels and set relative to the autonomously replicating sequence, ARS504, and WT untagged strain. Altogether, our data demonstrate that Set4 represses ergosterol genes and directly targets the promoters of ERG11 and ERG3.
Although our data show that Set4 directly represses ERG genes, RNA sequencing suggests that Set4 may be needed for the direct activation of select downregulated DEGs, including oxidative stress (CTT1) and sporulation (SPS100) genes (Figure 4E). To test whether Set4 directly targets these genes, our ZipChIP analysis was performed. ChIP analysis showed minor Set4 enrichment at the promoters of CTT1 and SPS100 (5- and 17-fold, respectively) (Figure S3 in File S5). In addition, Set4 was slightly enriched (22-fold) at a constitutively expressed gene, PMA1, which was not differentially expressed between WT and set4Δ strains (Figure S3 in File S5). In contrast, Set4 promoter enrichment at ERG11 (92-fold) and ERG3 (79-fold) was significantly higher than at CTT1, SPS100, or PMA1 (compare Figure 5, I and J, with Figure S3 in File S5). These results demonstrate that Set4 has low promoter enrichment at DEGs found to be downregulated in a set4Δ strain. We expect that Set4 is either nonspecifically binding at these promoters or plays an indirect role in the activation of these genes. Additional studies are needed to better understand how Set4 functions at downregulated DEGs.
Set4 is targeted to ergosterol gene promoters by Hap1 under hypoxic conditions
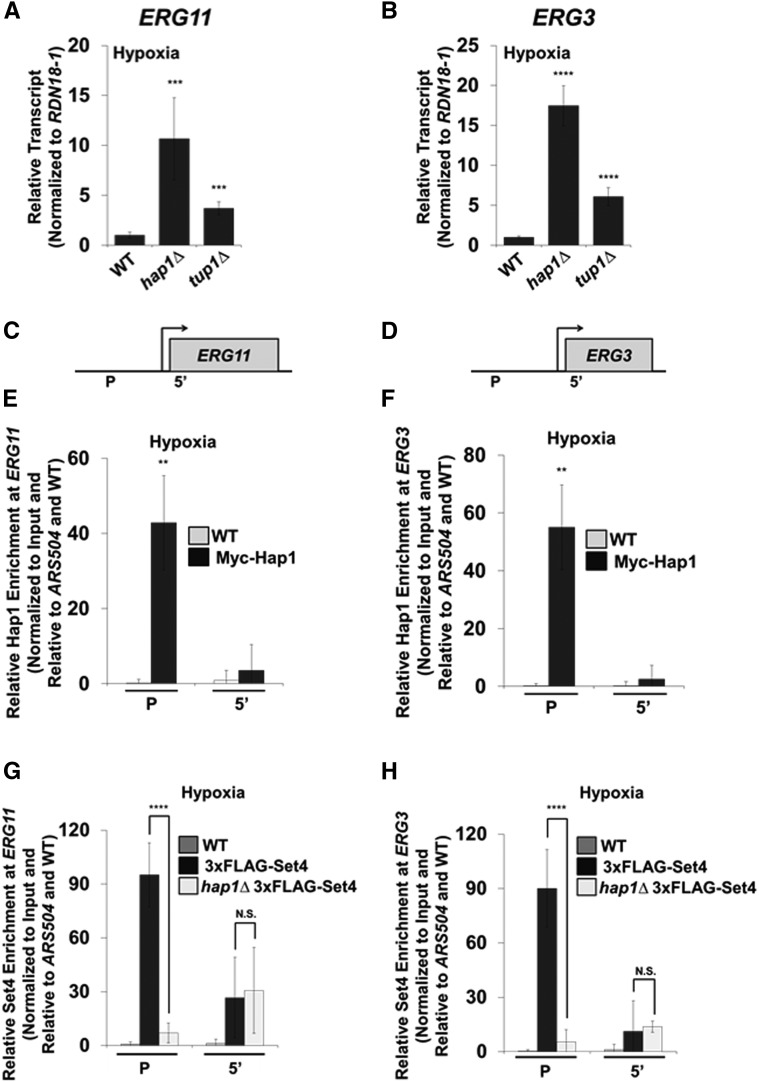
Our data show that Set4 functions as a direct repressor of ERG genes under hypoxic conditions (Figure 4 and Figure 5). Because Set4 enriches at the promoter of ERG genes, we hypothesized that Set4 may associate with a transcription factor to help facilitate gene repression under hypoxia. To determine the Set4-interacting partners under hypoxic conditions, we performed an immunoprecipitation assay with untagged WT and 3×FLAG-Set4 strains using an anti-FLAG-specific antibody from whole cell lysate harvested from hypoxic yeast cells. The immunoprecipitated samples were separated by SDS-PAGE and trypsin digested. The resulting peptides from the 3×FLAG-Set4 and untagged WT immunoprecipitation hypoxic samples were eluted and analyzed by MS. Peptide hits revealed that Set4 co-immunoprecipated with the transcription factor Hap1 and the transcriptional corepressor Tup1 under hypoxic conditions, suggesting that Set4 functions in a complex with Hap1 and Tup1 (File S4). Interestingly, Hap1 and Tup1 have been shown to repress ERG genes under hypoxic conditions (Hickman and Winston 2007).
To confirm that Hap1 and Tup1 repress ERG genes under our hypoxic conditions, we analyzed transcript levels by qRT-PCR in hap1∆ and tup1∆ strains, normalized to RDN18-1 and set relative to the WT strain. Consistent with published data, ERG11 and ERG3 expression levels increased significantly in the hap1∆ and tup1∆ strains (Figure 6, A and B) (Davies and Rine 2006; Hickman and Winston 2007). To determine if Hap1 directly targets the ERG11 and ERG3 loci under hypoxia, our ZipChIP was performed (Harmeyer et al. 2015). ChIP analysis showed that Myc-Hap1 is significantly enriched at the promoters of ERG11 and ERG3 (Figure 6, E and F), consistent with previous studies (Davies and Rine 2006; Hickman and Winston 2007). Importantly, the promoter probe sets were designed near (within one nucleosome distance) the Hap1 binding sites identified in the promoters of ERG11 (−788 and −641) and ERG3 (−358) by the YEASTRACT repository (Figure 6, C and D).
Figure 6.
The transcriptional repressor, Hap1, is required for Set4 binding to the promoters of ergosterol genes under hypoxic conditions. (A and B) Relative transcript levels of ERG11 and ERG3 were determined following 8 hr of hypoxia in WT, hap1Δ, and tup1Δ strains using qRT-PCR analysis. Transcript levels were set relative to the WT strain and expression levels were normalized to RDN18-1. *** P < 0.005, **** P < 0.0001. (C and D) Schematics of ERG11 and ERG3 loci with the specified positions of ChIP probes. (E and F) ChIP analysis of Hap1 under hypoxia at ERG11 and ERG3. ** P < 0.005. (G and H) ChIP analysis of Set4 under hypoxia in WT and hap1Δ strains at ERG11 and ERG3. ChIP analyses were normalized to DNA input samples and set relative to the untagged WT and the ARS504 loci. Error bars represent SD for three biological replicates with three technical replicates each. Gene expression and ChIP analyses were performed using the FY2609 strain. **** P < 0.0001. N.S., no significant difference.
To determine if Hap1 is necessary for Set4 recruitment to ERG gene promoters, we analyzed 3×FLAG-Set4 enrichment in WT and hap1∆ strains. Strikingly, 3×FLAG-Set4 promoter localization was significantly decreased in the hap1∆ strain, demonstrating that Hap1 is necessary for Set4 recruitment to ERG gene promoters under hypoxia (Figure 6, G and H). No significant difference in Set4 enrichment between the WT and hap1∆ strains was observed at the 5′ regions (Figure 6, G and H). Additionally, Western blot analysis demonstrated that 3×FLAG-Set4 protein expression levels were unaffected in a hap1Δ strain under hypoxia, verifying that the loss of Set4 enrichment at the promoter in a hap1Δ strain was not due to changes in global Set4 expression levels (Figure S4 in File S5). Altogether, these results demonstrate that Set4 associates and functions with Hap1 and the corepressor, Tup1, to maintain ergosterol homeostasis during hypoxic conditions.
Upc2 and Ecm22 are necessary for Set4 expression under hypoxic conditions and azole antifungal drug treatment
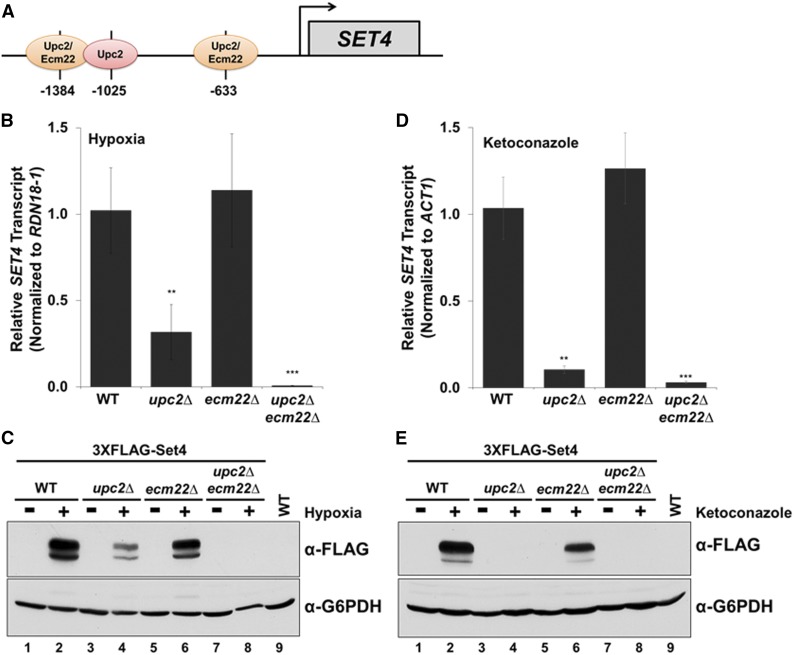
Because Set4 is induced under hypoxia and azole treatment, we wanted to determine the transcription factor(s) responsible for inducing Set4 expression. Previously, it has been shown that the transcriptional activators, Upc2 and Ecm22, upregulate gene targets in response to sterol depletion (Vik and Rine 2001; Davies et al. 2005; Davies and Rine 2006; Hickman et al. 2011). Under sterol depletion, Upc2 has been suggested to be the primary transcriptional regulator of ergosterol biosynthesis genes (Vik and Rine 2001; Davies et al. 2005; Davies and Rine 2006). Because sterol depletion occurs under hypoxia, we hypothesized that Upc2 and Ecm22, a paralog of Upc2, were necessary for Set4 induction under hypoxic conditions. First, the SET4 promoter was analyzed for Upc2 and Ecm22 binding sites (Figure 7A). Upc2 and Ecm22 are sterol regulatory element binding proteins that have been suggested to recognize the 7-bp DNA sequence TCGTATA known as the sterol regulatory element; Upc2 also recognizes the sequence TCGTTAAA (Vik and Rine 2001). The SET4 promoter contains three potential Upc2/Ecm22 binding sites predicted by the YEASTRACT repository database, including two binding sites for Upc2 and Ecm22 (−1384 and −633), and one site for Upc2 (−1025) (Figure 7A). To determine if SET4 expression is dependent on Upc2 and Ecm22 under hypoxia, we generated the following single and double deletion strains—upc2∆, ecm22∆, and upc2∆ecm22∆—and analyzed each deletion strain for SET4 mRNA levels. As predicted, the upc2∆ strain resulted in a significant decrease (72%) of SET4 transcript levels under hypoxia; however, the ecm22∆ strain did not affect SET4 expression (Figure 7B). Nevertheless, the upc2∆ecm22∆ double deletion strain abolished SET4 transcript levels under hypoxia (Figure 7B).
Figure 7.
Set4 expression under hypoxia or azole drug-treated conditions is primarily dependent on Upc2. (A) Schematic of the Upc2 and Ecm22 binding sites located in the SET4 promoter. (B and D) Using the indicated strains, SET4 expression was determined by qRT-PCR analysis. SET4 expression levels were normalized to RDN18-1 (hypoxia) or ACT1 (ketoconazole) and set relative to the aerobic or DMSO-treated WT strain. Data were analyzed from three biological replicates with three technical repeats. Each error bar represents SD. ** P < 0.01 and *** P < 0.005. (C and E) Western blot analysis of Set4 levels under the indicated conditions. G6PDH was used as a loading control. Gene expression and Western blot analyses were performed using the BY4741 strain.
To determine if the decrease in SET4 transcript levels in the upc2∆ and upc2∆ecm22∆ strains correlate with a decrease in protein levels, we generated the corresponding deletions—upc2∆, ecm22∆, and upc2∆ecm22∆—in the 3×FLAG-Set4 strain. Western blot analysis of the 3×FLAG-Set4 strains showed a moderate reduction in Set4 protein levels in the upc2∆ strain and a slight decrease in the ecm22∆ strain under hypoxia (Figure 7C, lanes 4 and 6). However, Set4 protein levels were completely abolished in the upc2∆ecm22∆ double deletion strain (Figure 7C, lane 8), confirming that Upc2 and Ecm22 are both necessary for the hypoxic-mediated induction of Set4 (Figure 7C). G6PDH was used as a loading control and an untagged WT grown under hypoxia was used as a negative control.
Studies have shown that Upc2 upregulates several gene targets including ERG genes, DAN/TIR genes, and PAU genes under antifungal drug treatment (Vik and Rine 2001; Wilcox et al. 2002; Davies et al. 2005; Davies and Rine 2006; Gallo-Ebert et al. 2013; Woods and Höfken 2016). To determine if Set4 expression is also dependent on Upc2 under azole drug treatment, we determined SET4 expression levels following 6 hr of treatment with 56 μg/ml ketoconazole in the WT, upc2∆, ecm22∆, and upc2∆ecm22∆ strains. Consistent with hypoxic conditions, SET4 transcript levels significantly decreased in the upc2∆ strain under ketoconazole treatment, albeit lower than observed under hypoxic conditions (Figure 7, B and D). Additionally, SET4 transcript levels were unchanged in the ecm22∆ strain (Figure 7D), while the upc2∆ecm22∆ strains show similar SET4 transcript levels as the upc2∆ (Figure 7D); suggesting that SET4 induction is completely dependent on Upc2. To determine if Set4 protein levels are solely dependent on Upc2 under azole drug treatment, we performed Western blot analysis in the 3×FLAG-Set4 strains containing upc2∆, ecm22∆, and upc2∆ecm22∆ double deletions. Consistent with SET4 transcript levels, Set4 protein expression is undetectable in the upc2∆ and upc2∆ecm22∆ double deletion strains treated with ketoconazole (Figure 7E, lanes 4 and 8). These results indicate that Upc2 is the primary transcription factor required to induce Set4 expression, while Ecm22 is partially responsible for Set4 expression in the absence of Upc2 under hypoxic conditions. In contrast, Upc2 is exclusively responsible for upregulating Set4 levels under azole treatment.
An erg3∆ strain induces the expression of Set4 under untreated aerobic conditions
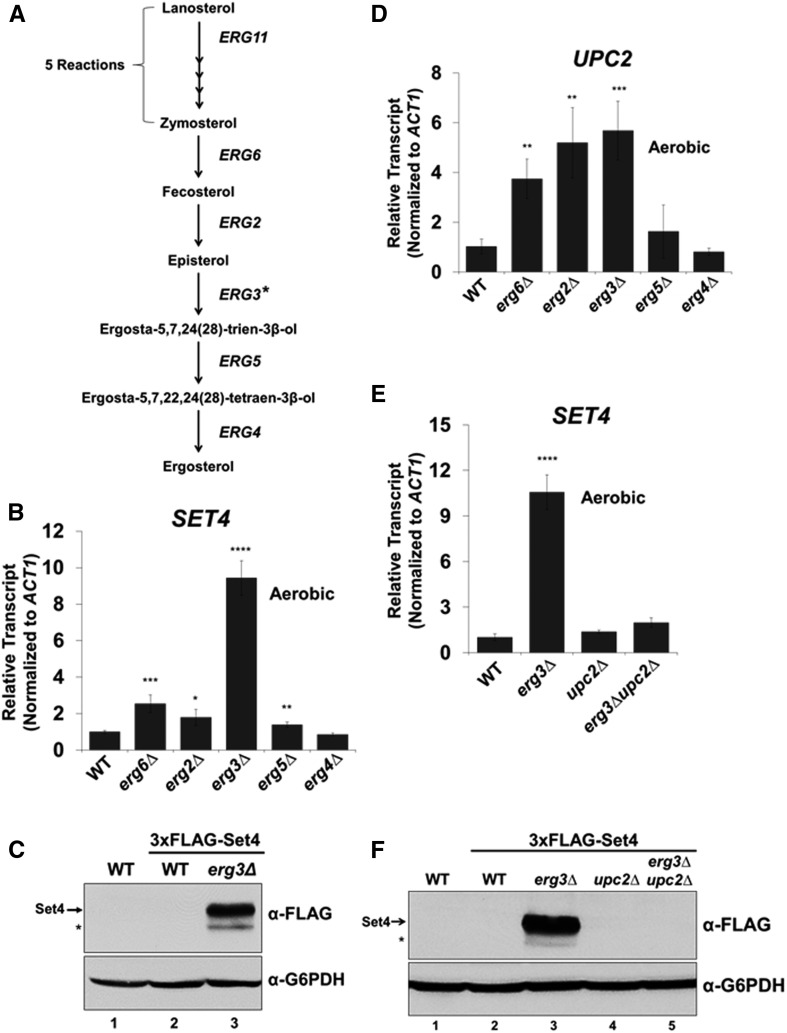
Because hypoxia and azole drug treatment are conditions that decrease ergosterol and increase sterol precursors, we hypothesized that Upc2 would upregulate Set4 expression under aerobic conditions in strains that are deficient in producing ergosterol but still produce precursor sterols. To test this hypothesis, we deleted the genes encoding the enzymes (ERG6, ERG2, ERG3, ERG5, and ERG4) needed for the final five steps of the ergosterol biosynthesis pathway (Figure 8A), and SET4 expression levels were examined by qRT-PCR. Intriguingly, the erg3∆ strain resulted in a significant increase (∼10-fold) in SET4 transcript levels, whereas SET4 expression increased threefold or less in the remaining ergosterol mutants (Figure 8B). To determine if Set4 protein levels induce in an erg3Δ strain, we deleted ERG3 in the 3×FLAG-Set4 strain. Western blot analysis revealed that 3×FLAG-Set4 protein was expressed in the erg3∆ strain, suggesting that increased levels of an ergosterol precursor(s) in the erg3∆ strain are involved in upregulating Set4 expression (Figure 8C and Figure S5 in File S5). Ergosterol precursors generated in an erg3∆ strain are episterol, ergosta-7-enol, ergosta-7,22,24(28)-trienol, and ergosta-7,22-dienol (Figure S5 in File S5) (Bard et al. 1993; Sanglard et al. 2003; Martel et al. 2010; Vale-Silva et al. 2012). Because Set4 expression is dependent on Upc2 under sterol depletion (Figure 7), UPC2 transcript levels were analyzed in the ergosterol mutants to determine whether UPC2 levels were upregulated in response to decreased ergosterol levels. The qRT-PCR analysis showed that UPC2 expression is significantly upregulated in the erg6∆, erg2∆, and erg3∆ strains and not significantly affected in the erg5∆ and erg4∆ strains; indicating that a decrease in ergosterol does not result in Upc2 induction (Figure 8D). To test if Upc2 is required for Set4 expression in the erg3∆ strain, we generated the erg3∆upc2∆ double deletion strain. Consistent with the requirement of Upc2 for the induction of Set4 under azole and hypoxic conditions, SET4 transcript level failed to increase in the erg3∆upc2∆ strain (Figure 8E). In addition, Western blot analysis revealed that 3×FLAG-Set4 expression was undetectable in the erg3∆upc2∆ double deletion strain, verifying that Set4 expression is solely dependent on Upc2 in the erg3∆ strain (Figure 8F, lane 5). Altogether, our data indicate that accumulation of a sterol precursor(s) in the erg3∆ strain works together with Upc2 to induce Set4 expression.
Figure 8.
Set4 is constitutively expressed in an erg3Δ strain under aerobic conditions by a precursor sterol and Upc2. (A) Modified pathway showing the final steps in ergosterol biosynthesis. The * on ERG3 represents potential pathways that modify episterol to generate additional sterol precursors (see Figure S5 in File S5). (B) qRT-PCR analysis of SET4 transcript levels in ergosterol mutant strains under aerobic conditions. Transcript levels were normalized to ACT1 and set relative to WT. (C) Western blot analysis of 3×FLAG-Set4 levels in WT and erg3∆ strain under aerobic conditions. The * indicates a likely protein degradation band. (D) Gene expression analysis by qRT-PCR of UPC2 in indicated strains under aerobic conditions. (E) SET4 mRNA levels determined by qRT-PCR in WT, erg3∆, upc2∆, and erg3∆upc2∆ under aerobic conditions. (B, D, E) * P < 0.05, ** P < 0.005, *** P < 0.0005, **** P < 0.0001. (F) Western blot analysis of Set4 protein levels in strains from E under aerobic conditions. G6PDH was used as a loading control and an untagged WT was used a negative control. The band under the Set4 band likely indicates protein degradation and is indicated by *. Gene expression data were normalized to ACT1 and set relative to WT. Error bars represents SD of three biological replicates. Gene expression and Western blot analyses were performed using the BY4741 strain.
Discussion
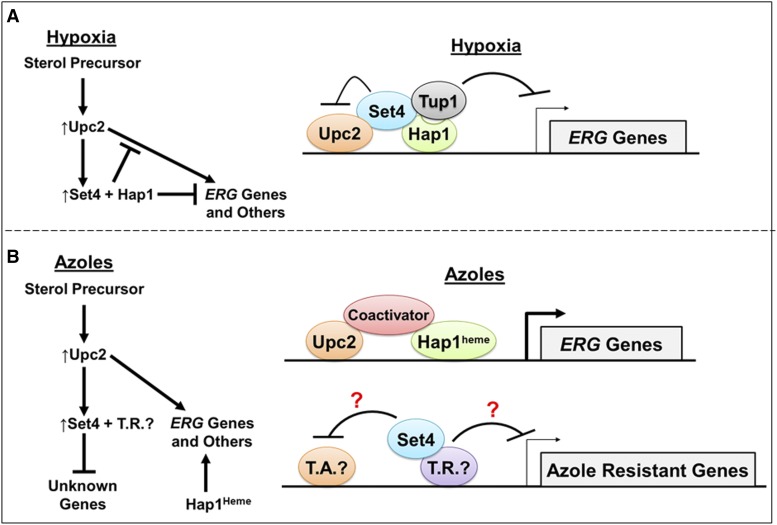
In this study, we established that Set4 is an inducible SET domain-containing protein that governs azole antifungal drug resistance, and directly represses ergosterol genes under hypoxic conditions by a Hap1-dependent mechanism. Initially, we identified that Set1, Set3, and Set4 govern drug efficacy to the medically relevant class of azole antifungal drugs. Upon further investigation, we surprisingly discovered that Set4 is not constitutively expressed under standard yeast growth conditions but is induced by the sterol-responsive transcriptional activators, Upc2 and Ecm22, under hypoxic conditions or azole drug treatment. Furthermore, we determined that Set4 modulates global gene expression under ergosterol-limiting conditions, and that under hypoxia Set4 is recruited to promoters by Hap1 to directly repress ERG3 and ERG11 expression. Therefore, Set4 plays a key role in cellular adaptation to environmental conditions that deplete ergosterol levels. Interestingly, Set4 expression is also upregulated in an untreated erg3∆ strain but not in an erg3∆upc2∆ double deletion strain, suggesting Upc2 and a precursor sterol(s)—but not ergosterol—control Set4 levels. Altogether, we have identified a new sterol-signaling pathway involving precursor sterols, sterol-sensing transcription factors, and Set4, which govern azole antifungal drug resistance and regulates the repression of sterol genes under hypoxic conditions.
Set4 has previously been defined as the Set3 paralog because SET4 was likely derived from SET3 during the whole-genome duplication of the Saccharomycotina species, and has been identified in S. cerevisiae, S. bayanus, S. castelli, and the opportunistic fungal pathogen C. glabrata (Pijnappel et al. 2001; Byrne and Wolfe 2005). Both Set4 and Set3 contain an N-terminal PHD finger and C-terminal SET domain; however, neither Set3 nor Set4 have been identified to have histone methyltransferase activity (Pijnappel et al. 2001). Additionally, the Set4 homologs UpSET (Drosophila melanogaster) and MLL5 (human, long isoform) have not been shown to have histone methyltransferase activity (Pijnappel et al. 2001; Fujiki et al. 2009; Sebastian et al. 2009; Mas et al. 2016; Zhang et al. 2017). On the other hand, MLL5 (human, short isoform) has been reported to have H3K4 methylation activity after MLL5 (short) has been post-translationally modified by O-GlcNac transferase (Fujiki et al. 2009). However, in S. cerevisiae, histone methylation by Set4 was undetectable both in vitro and in vivo when testing global levels of H3K4 methylation under aerobic and hypoxic conditions by Western blot analysis (Figure S6 in File S5). Currently, it is unknown whether Set4 has methyltransferase activity on histones or nonhistone substrates.
Like Set3, Set4 alters global gene expression; however, the mechanism by which Set4 modulates ergosterol gene expression is unknown but is likely different from Set3 (Kim et al. 2012). For example, loss of Set4 increases ERG11 and ERG3 transcript levels, whereas a deletion of Set3 does not affect these genes under hypoxia (Figure S2 in File S5). Under aerobic conditions, the Set3 complex has been shown to enrich at the 5′ regions of gene targets through associations with H3K4me2 and the Ser5-phosphorylated C-terminal domain of RNA polymerase II (Kim and Buratowski 2009; Govind et al. 2010). In addition, unlike the PHD finger of Set3, the PHD finger of Set4 does not bind to H3K4 methylated histones (Shi et al. 2007). Together, this suggests that Set3 and Set4 have evolved distinct biochemical functions.
Overall, our studies support a model where Upc2 induces SET4 to block Upc2-mediated ERG gene activation by a repressive complex consisting of Set4, Hap1, and Tup1 under hypoxia (Figure 5, E and F, and Figure 9A). Therefore, the Set4–Hap1–Tup1 complex functions in a feedback mechanism to regulate Upc2-mediated ERG gene activation (Figure 9A). In support of this hypothesis, deletion of SET4 results in increased expression of ERG11 and ERG3 compared to WT, whereas this increase is abolished in the upc2∆ and set4∆upc2∆ strains under hypoxia (Figure 5, E and F).
Figure 9.
Sterol precursor(s) lead to Upc2-facilitated induction of Set4 to repress genes required for antifungal drug resistance and sterol homeostasis under hypoxic conditions. (A) Model under hypoxia: Sterol-facilitated induction of Set4 by Upc2 mediates repression of ERG genes by association with the transcriptional repressors Hap1 and Tup1 to block the function of the transcriptional activator, Upc2. Hap1 under hypoxic conditions is heme independent and is considered a transcriptional repressor. Activation of ERG genes by Upc2 is blocked by the presence of Set4 and Hap1 as indicated by the inhibitory symbol. S288C strains expressing the hap1–Ty1 gene fusion partially repress ERG gene expression; whereas yeast strains expressing HAP1 will fully repress ERG genes. (B) Model under azole drug treatment: Sterol-facilitated induction of Set4 by Upc2 mediates repression of genes involved in azole resistance and likely associates with a transcriptional repressor (T.R.) to block a transcriptional activator (T.A.). Under azole treatment, ERG genes are Set4 independent but are activated by Upc2, Hap1heme, and coactivators. Hap1 under aerobic conditions is bound to heme and is considered a transcriptional activator.
Importantly, ERG11 and ERG3 gene expression increased similarly in the BY4741 set4∆ strain compared to the FY2609 set4∆ strain, indicating that neither the BY4741 strain expressing the hap1–Ty1 gene fusion nor the FY2609 strain expressing the WT HAP1 affects the function of Set4 (Figure S7 in File S5). Interestingly, ERG11 and ERG3 transcript levels increase by about twofold in the BY4741 WT strain; whereas ERG gene expression significantly decreases (Figure 5) in the FY2609 strain when switched to hypoxic conditions (Figure S8 in File S5). To our knowledge, this is the first observation indicating that the BY4741 strain expressing the hap1–Ty1 gene fusion is partially defective in repressing ERG genes (Figure S8 in File S5 and File S1). However, this is consistent with the findings that strains expressing the hap1–Ty1 gene fusion are partially defective in Hap1-mediated gene activation under aerobic conditions (Gaisne et al. 1999; Tamura et al. 2004).
In contrast, ERG11, ERG3, PDR5, and PDR11 expression increased similarly in WT and set4∆ strains treated with ketoconazole, indicating that these are Set4-independent genes (Figure 2, E–H). Therefore, under azole treatment, Set4 must regulate the expression of alternative gene targets unknown to play a role in azole drug resistance. We speculate that Set4 does not associate with Hap1 since Hap1 is bound to heme under aerobic conditions and acts as a transcriptional activator for ERG genes (Figure 9B). Genome-wide gene expression analysis and/or genetic suppressor screens will identify Set4 gene targets that contribute to azole drug resistance.
An important result revealed from our study was the observation that Set4 expression levels are upregulated in an erg3Δ strain (Figure 8). These data suggest that a precursor sterol, not ergosterol, is signaling induction of Set4 by an Upc2-mediated mechanism. In an erg3Δ strain, several ergosterol precursors have been indicated to accumulate under aerobic, untreated conditions in yeast such as episterol, ergosta-7-enol, ergosta-7,22,24(28)-trienol and ergosta-7,22-dienol (Figure S5 in File S5) (Sanglard et al. 2003; Martel et al. 2010; Vale-Silva et al. 2012). In a recent study, Upc2 was shown to bind ergosterol sequestering Upc2 outside of the nucleus; however, when ergosterol is depleted using treatment with fluconazole, Upc2 localizes to the nucleus (Yang et al. 2015). In an erg3Δ strain, ergosterol cannot be synthesized; therefore, the sterol precursors that accumulate in erg3Δ strain might bind to Upc2 or another factor, leading to Upc2 nuclear localization and Set4 expression. Understanding how Set4 functions in eukaryotic sterol homeostasis can provide insight into how SET domain proteins alter gene expression in hypoxic environments such as the human gut microbiota or tumor microenvironment. Furthermore, understanding the role of Set4 in drug resistance can provide insight into the development of new or improved antifungal drugs as well as develop novel strategies for treating fungal infections and antifungal drug resistance.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300554/-/DC1.
Acknowledgments
We thank Peter Cheung for reviewing this manuscript and Fred Winston for yeast strains. We thank the Purdue Bioinformatics Core for pipelines and bioinformatics software. This work was supported by the Purdue Department of Biochemistry, Bilsland Fellowship (to N.D.S.); the Purdue Biochemistry Bird Stair Fellowship (to N.D.S.); Ross Fellowship (to K.M.B.); Purdue Center for Cancer Research (grant P30CA023168: DNA Sequencing Shared Resource and Collaborative Core for Cancer Bioinformatics at Purdue); the Walther Cancer Foundation; and the Indiana University Simon Cancer Center (grant P30CA082709). Additional funding support was provided by the Purdue Executive Vice President for Research and Partnerships award (to S.D.B.), Purdue Center for Cancer Research Challenge Award (to S.D.B.), National Institute of Food and Agriculture 1007570 (S.D.B.), and the National Institutes of Health R01 EY-024905 (V.M.W.).
Footnotes
Communicating editor: A. Hinnebusch
Literature Cited
- Abramova N., Sertil O., Mehta S., Lowry C. V., 2001. Reciprocal regulation of anaerobic and aerobic cell wall mannoprotein gene expression in Saccharomyces cerevisiae. J. Bacteriol. 183: 2881–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal A. K., Rogers P. D., Baerson S. R., Jacob M. R., Barker K. S., et al. , 2003. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J. Biol. Chem. 278: 34998–35015. [DOI] [PubMed] [Google Scholar]
- Alimardani P., Regnacq M., Moreau-Vauzelle C., Ferreira T., Rossignol T., et al. , 2004. SUT1-promoted sterol uptake involves the ABC transporter Aus1 and the mannoprotein Dan1 whose synergistic action is sufficient for this process. Biochem. J. 381: 195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard M., Lees N. D., Turi T., Craft D., Cofrin L., et al. , 1993. Sterol synthesis and viability of erg11 (cytochrome P450 lanosterol demethylase) mutations in Saccharomyces cerevisiae and Candida albicans. Lipids 28: 963–967. [DOI] [PubMed] [Google Scholar]
- Becerra M., Lombardia-Ferreira L. J., Hauser N. C., Hoheisel J. D., Tizon B., et al. , 2002. The yeast transcriptome in aerobic and hypoxic conditions: effects of hap1, rox1, rox3 and srb10 deletions. Mol. Microbiol. 43: 545–555. [DOI] [PubMed] [Google Scholar]
- Bendjilali N., MacLeon S., Kalra G., Willis S. D., Hossian A. K., et al. , 2017. Time-course analysis of gene expression during the Saccharomyces cerevisiae hypoxic response. G3 (Bethesda) 7: 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs S. D., Bryk M., Strahl B. D., Cheung W. L., Davie J. K., et al. , 2001. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15: 3286–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne K. P., Wolfe K. H., 2005. The yeast gene order browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 15: 1456–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoy S., Adrio J. L., 2017. Antifungals. Biochem. Pharmacol. 133: 86–96. [DOI] [PubMed] [Google Scholar]
- Cohen B. D., Sertil O., Abramova N. E., Davies K. J., Lowry C. V., 2001. Induction and repression of DAN1 and the family of anaerobic mannoprotein genes in Saccharomyces cerevisiae occurs through a complex array of regulatory sites. Nucleic Acids Res. 29: 799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowen L. E., Sanglard D., Howard S. J., Rogers P. D., Perlin D. S., 2014. Mechanisms of antifungal drug resistance. Cold Spring Harb. Perspect. Med. 5: a019752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies B. S., Rine J., 2006. A role for sterol levels in oxygen sensing in Saccharomyces cerevisiae. Genetics 174: 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies B. S., Wang H. S., Rine J., 2005. Dual activators of the sterol biosynthetic pathway of Saccharomyces cerevisiae: similar activation/regulatory domains but different response mechanisms. Mol. Cell. Biol. 25: 7375–7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S., Lemetais G., Ferreira T., Cayot P., Gervais P., et al. , 2012. Ergosterol biosynthesis: a fungal pathway for life on land? Evolution 66: 2961–2968. [DOI] [PubMed] [Google Scholar]
- Espenshade P. J., Hughes A. L., 2007. Regulation of sterol synthesis in eukaryotes. Annu. Rev. Genet. 41: 401–427. [DOI] [PubMed] [Google Scholar]
- Fingerman I. M., Wu C. L., Wilson B. D., Briggs S. D., 2005. Global loss of Set1-mediated H3 Lys4 trimethylation is associated with silencing defects in Saccharomyces cerevisiae. J. Biol. Chem. 280: 28761–28765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers S. A., Barker K. S., Berkow E. L., Toner G., Chadwick S. G., et al. , 2012. Gain-of-function mutations in UPC2 are a frequent cause of ERG11 upregulation in azole-resistant clinical isolates of Candida albicans. Eukaryot. Cell 11: 1289–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki R., Chikanishi T., Hashiba W., Ito H., Takada I., et al. , 2009. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature 459: 455–459. [DOI] [PubMed] [Google Scholar]
- Fytlovich S., Gervais M., Agrimonti C., Guiard B., 1993. Evidence for an interaction between the CYP1(HAP1) activator and a cellular factor during heme-dependent transcriptional regulation in the yeast Saccharomyces cerevisiae. EMBO J. 12: 1209–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaisne M., Becam A. M., Verdiere J., Herbert C. J., 1999. A ‘natural’ mutation in Saccharomyces cerevisiae strains derived from S288c affects the complex regulatory gene HAP1 (CYP1). Curr. Genet. 36: 195–200. [DOI] [PubMed] [Google Scholar]
- Gallo-Ebert C., Donigan M., Liu H. Y., Pascual F., Manners M., et al. , 2013. The yeast anaerobic response element AR1b regulates aerobic antifungal drug-dependent sterol gene expression. J. Biol. Chem. 288: 35466–35477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatta A. T., Wong L. H., Sere Y. Y., Calderon-Norena D. M., Cockcroft S., et al. , 2015. A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. Elife 4: e07253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind C. K., Qiu H., Ginsburg D. S., Ruan C., Hofmeyer K., et al. , 2010. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol. Cell 39: 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmeyer K. M., South P. F., Bishop B., Ogas J., Briggs S. D., 2015. Immediate chromatin immunoprecipitation and on-bead quantitative PCR analysis: a versatile and rapid ChIP procedure. Nucleic Acids Res. 43: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman M. J., Winston F., 2007. Heme levels switch the function of Hap1 of Saccharomyces cerevisiae between transcriptional activator and transcriptional repressor. Mol. Cell. Biol. 27: 7414–7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman M. J., Spatt D., Winston F., 2011. The Hog1 mitogen-activated protein kinase mediates a hypoxic response in Saccharomyces cerevisiae. Genetics 188: 325–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon T., Lee H. C., Hu Z., Iyer V. R., Zhang L., 2005. The heme activator protein Hap1 represses transcription by a heme-independent mechanism in Saccharomyces cerevisiae. Genetics 169: 1343–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishtar Snoek I. S., Yde Steensma H., 2007. Factors involved in anaerobic growth of Saccharomyces cerevisiae. Yeast 24: 1–10. [DOI] [PubMed] [Google Scholar]
- Joshua I. M., Höfken T., 2017. From lipid homeostasis to differentiation: old and new functions of the zinc cluster proteins Ecm22, Upc2, Sut1 and Sut2. Int. J. Mol. Sci. 18: E772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiravan M. K., Salake A. B., Chothe A. S., Dudhe P. B., Watode R. P., et al. , 2012. The biology and chemistry of antifungal agents: a review. Bioorg. Med. Chem. 20: 5678–5698. [DOI] [PubMed] [Google Scholar]
- Kim T., Buratowski S., 2009. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell 137: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T., Xu Z., Clauder-Munster S., Steinmetz L. M., Buratowski S., 2012. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell 150: 1158–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohut P., Wustner D., Hronska L., Kuchler K., Hapala I., et al. , 2011. The role of ABC proteins Aus1p and Pdr11p in the uptake of external sterols in yeast: dehydroergosterol fluorescence study. Biochem. Biophys. Res. Commun. 404: 233–238. [DOI] [PubMed] [Google Scholar]
- Kwast K. E., Burke P. V., Poyton R. O., 1998. Oxygen sensing and the transcriptional regulation of oxygen-responsive genes in yeast. J. Exp. Biol. 201: 1177–1195. [DOI] [PubMed] [Google Scholar]
- Kwast K. E., Lai L. C., Menda N., James D. T., III, Aref S., et al. , 2002. Genomic analyses of anaerobically induced genes in Saccharomyces cerevisiae: functional roles of Rox1 and other factors in mediating the anoxic response. J. Bacteriol. 184: 250–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardia L. J., Cadahia-Rodriguez J. L., Freire-Picos M. A., Gonzalez-Siso M. I., Rodriguez-Torres A. M., et al. , 2000. Transcript analysis of 203 novel genes from Saccharomyces cerevisiae in hap1 and rox1 mutant backgrounds. Genome 43: 881–886. [DOI] [PubMed] [Google Scholar]
- Luo Z., van Vuuren H. J., 2009. Functional analyses of PAU genes in Saccharomyces cerevisiae. Microbiology 155: 4036–4049. [DOI] [PubMed] [Google Scholar]
- Markovich S., Yekutiel A., Shalit I., Shadkchan Y., Osherov N., 2004. Genomic approach to identification of mutations affecting caspofungin susceptibility in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 48: 3871–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel C. M., Parker J. E., Bader O., Weig M., Gross U., et al. , 2010. Identification and characterization of four azole-resistant erg3 mutants of Candida albicans. Antimicrob. Agents Chemother. 54: 4527–4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas Y. M. S., Barbon M., Teyssier C., Demene H., Carvalho J. E., et al. , 2016. The human mixed lineage leukemia 5 (MLL5), a sequentially and structurally divergent SET domain-containing protein with no intrinsic catalytic activity. PLoS One 11: e0165139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersman D. P., Du H. N., Fingerman I. M., South P. F., Briggs S. D., 2012. Charge-based interaction conserved within histone H3 lysine 4 (H3K4) methyltransferase complexes is needed for protein stability, histone methylation, and gene expression. J. Biol. Chem. 287: 2652–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouritsen O. G., Zuckermann M. J., 2004. What’s so special about cholesterol? Lipids 39: 1101–1113. [DOI] [PubMed] [Google Scholar]
- Muren E., Oyen M., Barmark G., Ronne H., 2001. Identification of yeast deletion strains that are hypersensitive to brefeldin A or monensin, two drugs that affect intracellular transport. Yeast 18: 163–172. [DOI] [PubMed] [Google Scholar]
- Odds F. C., Brown A. J., Gow N. A., 2003. Antifungal agents: mechanisms of action. Trends Microbiol. 11: 272–279. [DOI] [PubMed] [Google Scholar]
- Parks L. W., Casey W. M., 1995. Physiological implications of sterol biosynthesis in yeast. Annu. Rev. Microbiol. 49: 95–116. [DOI] [PubMed] [Google Scholar]
- Pijnappel W. W. M. P., Schaft D., Roguev A., Shevchenko A., Tekotte H., et al. , 2001. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 15: 2991–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachidi N., Martinez M. J., Barre P., Blondin B., 2000. Saccharomyces cerevisiae PAU genes are induced by anaerobiosis. Mol. Microbiol. 35: 1421–1430. [DOI] [PubMed] [Google Scholar]
- Rosenfeld E., Beauvoit B., 2003. Role of the non-respiratory pathways in the utilization of molecular oxygen by Saccharomyces cerevisiae. Yeast 20: 1115–1144. [DOI] [PubMed] [Google Scholar]
- Sanglard D., Ischer F., Parkinson T., Falconer D., Bille J., 2003. Candida albicans mutations in the ergosterol biosynthetic pathway and resistance to several antifungal agents. Antimicrob. Agents Chemother. 47: 2404–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian S., Sreenivas P., Sambasivan R., Cheedipudi S., Kandalla P., et al. , 2009. MLL5, a trithorax homolog, indirectly regulates H3K4 methylation, represses cyclin A2 expression, and promotes myogenic differentiation. Proc. Natl. Acad. Sci. USA 106: 4719–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Kachirskaia I., Walter K. L., Kuo J. H., Lake A., et al. , 2007. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J. Biol. Chem. 282: 2450–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South P. F., Fingerman I. M., Mersman D. P., Du H. N., Briggs S. D., 2010. A conserved interaction between the SDI domain of Bre2 and the Dpy-30 domain of Sdc1 is required for histone methylation and gene expression. J. Biol. Chem. 285: 595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South P. F., Harmeyer K. M., Serratore N. D., Briggs S. D., 2013. H3K4 methyltransferase Set1 is involved in maintenance of ergosterol homeostasis and resistance to Brefeldin A. Proc. Natl. Acad. Sci. USA 110: E1016–E1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Gu Y., Wang Q., Yamada T., Ito K., et al. , 2004. A hap1 mutation in a laboratory strain of Saccharomyces cerevisiae results in decreased expression of ergosterol-related genes and cellular ergosterol content compared to sake yeast. J. Biosci. Bioeng. 98: 159–166. [DOI] [PubMed] [Google Scholar]
- Ter Linde J. J., Steensma H. Y., 2002. A microarray-assisted screen for potential Hap1 and Rox1 target genes in Saccharomyces cerevisiae. Yeast 19: 825–840. [DOI] [PubMed] [Google Scholar]
- Ter Linde J. J., Liang H., Davis R. W., Steensma H. Y., van Dijken J. P., et al. , 1999. Genome-wide transcriptional analysis of aerobic and anaerobic chemostat cultures of Saccharomyces cerevisiae. J. Bacteriol. 181: 7409–7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale-Silva L. A., Coste A. T., Ischer F., Parker J. E., Kelly S. L., et al. , 2012. Azole resistance by loss of function of the sterol Δ5,6-desaturase gene (ERG3) in Candida albicans does not necessarily decrease virulence. Antimicrob. Agents Chemother. 56: 1960–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vik A., Rine J., 2001. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 6395–6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weete J. D., Abril M., Blackwell M., 2010. Phylogenetic distribution of fungal sterols. PLoS One 5: e10899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whaley S. G., Berkow E. L., Rybak J. M., Nishimoto A. T., Barker K. S., et al. , 2016. Azole antifungal resistance in Candida albicans and emerging non-albicans Candida species. Front. Microbiol. 7: 2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox L. J., Balderes D. A., Wharton B., Tinkelenberg A. H., Rao G., et al. , 2002. Transcriptional profiling identifies two members of the ATP-binding cassette transporter superfamily required for sterol uptake in yeast. J. Biol. Chem. 277: 32466–32472. [DOI] [PubMed] [Google Scholar]
- Woods K., Höfken T., 2016. The zinc cluster proteins Upc2 and Ecm22 promote filamentation in Saccharomyces cerevisiae by sterol biosynthesis-dependent and -independent pathways. Mol. Microbiol. 99: 512–527. [DOI] [PubMed] [Google Scholar]
- Yang H., Tong J., Lee C. W., Ha S., Eom S. H., et al. , 2015. Structural mechanism of ergosterol regulation by fungal sterol transcription factor Upc2. Nat. Commun. 6: 6129. [DOI] [PubMed] [Google Scholar]
- Zavrel M., Hoot S. J., White T. C., 2013. Comparison of sterol import under aerobic and anaerobic conditions in three fungal species, Candida albicans, Candida glabrata, and Saccharomyces cerevisiae. Eukaryot. Cell 12: 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Hach A., 1999. Molecular mechanism of heme signaling in yeast: the transcriptional activator Hap1 serves as the key mediator. Cell. Mol. Life Sci. 56: 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Novera W., Zhang Y., Deng L. W., 2017. MLL5 (KMT2E): structure, function, and clinical relevance. Cell. Mol. Life Sci. 74: 2333–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Serratore N. D., Briggs S. D., 2016. N-ICE plasmids for generating N-terminal 3 x FLAG tagged genes that allow inducible, constitutive, or endogenous expression in Saccharomyces cerevisiae. Yeast 34: 223–235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains are available upon request. File S5 and File S7 contain additional tables and figures including the strain list and genotypes, gene expression primers, and chromatin immunoprecipitation (ChIP) probe sets. File S1, File S2, File S3, and File S4 include gene expression data from the RNA-sequencing analysis and peptide hits from the MS analysis. Genome-wide RNA-sequencing data are available at Gene Expression Omnibus under accession number GSE107492.