Abstract
Regulatory Factor X (RFX) transcription factors (TFs) are best known for activating genes required for ciliogenesis in both vertebrates and invertebrates. In humans, eight RFX TFs have a variety of tissue-specific functions, while in the worm Caenorhabditis elegans, the sole RFX gene, daf-19, encodes a set of nested isoforms. Null alleles of daf-19 confer pleiotropic effects including altered development with a dauer constitutive phenotype, complete absence of cilia and ciliary proteins, and defects in synaptic protein maintenance. We sought to identify RFX/daf-19 target genes associated with neuronal functions other than ciliogenesis using comparative transcriptome analyses at different life stages of the worm. Subsequent characterization of gene expression patterns revealed one set of genes activated in the presence of DAF-19 in ciliated sensory neurons, whose activation requires the daf-19c isoform, also required for ciliogenesis. A second set of genes is downregulated in the presence of DAF-19, primarily in nonsensory neurons. The human orthologs of some of these neuronal genes are associated with human diseases. We report the novel finding that daf-19a is directly or indirectly responsible for downregulation of these neuronal genes in C. elegans by characterizing a new mutation affecting the daf-19a isoform (tm5562) and not associated with ciliogenesis, but which confers synaptic and behavioral defects. Thus, we have identified a new regulatory role for RFX TFs in the nervous system. The new daf-19 candidate target genes we have identified by transcriptomics will serve to uncover the molecular underpinnings of the pleiotropic effects that daf-19 exerts on nervous system function.
Keywords: RFX transcription factor, neuronal gene expression, dauer formation, roaming behavior, aldicarb resistance
CHARACTERIZING Regulatory Factor X (RFX) transcription factor (TF) function and identifying RFX target genes are key to understanding a wide range of diseases. Problems with RFX-controlled genes are linked to impaired immune function, cancer, and ciliopathies including developmental disorders, kidney disease, and deafness. RFX TFs regulate processes including mitosis in yeasts (Garg et al. 2015), ciliogenesis and cilia maintenance (Choksi et al. 2014), adaptive immune responses (Reith and Mach 2001), innate immunity (Xie et al. 2013), and maintenance of terminally differentiated hair cells in the mammalian ear (Elkon et al. 2015). RFX target genes encode ciliary components (e.g., Schafer et al. 2003; Jensen et al. 2016), phosphatases involved in carcinogenesis (Su et al. 2014), major histocompatibility complex genes (Meissner et al. 2012), and genes associated with dyslexia (Tammimies et al. 2016), to name a few.
RFX TFs share and are defined by a highly conserved winged helix DNA-binding domain (DBD) (Emery et al. 1996; Gajiwala et al. 2000) by which gene networks are regulated in both invertebrates and vertebrates. RFX TFs evolved early in the unikont lineage, and, though well known for controlling ciliogenesis, they apparently evolved after the origin of cilia (Chu et al. 2010; Piasecki et al. 2010). Thus, they likely have an ancient function. Humans have eight RFX genes (Aftab et al. 2008; https://www.ncbi.nlm.nih.gov/gene/731220). RFX TFs regulate target genes via a highly conserved cis-acting regulatory sequence called the X-box motif (Efimenko et al. 2005; Laurencon et al. 2007; Chu et al. 2012). The X-box sequence has been used effectively to identify many RFX target genes (e.g., Blacque et al. 2005; Chen et al. 2006; Henriksson et al. 2013).
Here, we describe novel functions for daf-19, the only RFX gene in the worm Caenorhabditis elegans. DAF-19 was the first RFX TF linked to the process of ciliogenesis (Swoboda et al. 2000; Senti and Swoboda 2008) and it was found also to regulate innate immunity (Xie et al. 2013). The C. elegans daf-19 locus encodes at least four related gene products with three transcriptional start sites (Craig et al. 2013). Two smaller isoforms, DAF-19C and DAF-19M (Figure 1A), have a role in ciliated sensory neurons (CSNs) only. The majority of CSNs, the only ciliated cell type in the worm, are generated during embryogenesis (Jensen et al. 2016). daf-19c, expressed in all CSNs via an internal promoter (Senti and Swoboda 2008; Craig et al. 2013), is sufficient to drive ciliogenesis in CSNs (Senti et al. 2009). DAF-19C alone is also sufficient to drive continued larval development in a food-rich environment as opposed to entering the dispersive and stress-resistant dauer larval stage (Senti and Swoboda 2008). DAF-19C regulates transcription of ciliary genes via X-box motifs supported by a nearby C-box enhancer element (Burghoorn et al. 2012). In contrast, the daf-19m isoform is expressed only in IL2 and male-specific CSNs, where it regulates the transcription of genes required for male mating behaviors, including those with orthologs implicated in polycystic kidney disease (Wang et al. 2010). Gene regulation by DAF-19M results in cilia specialization via a DAF-19C-independent cascade of gene expression (Wang et al. 2010).
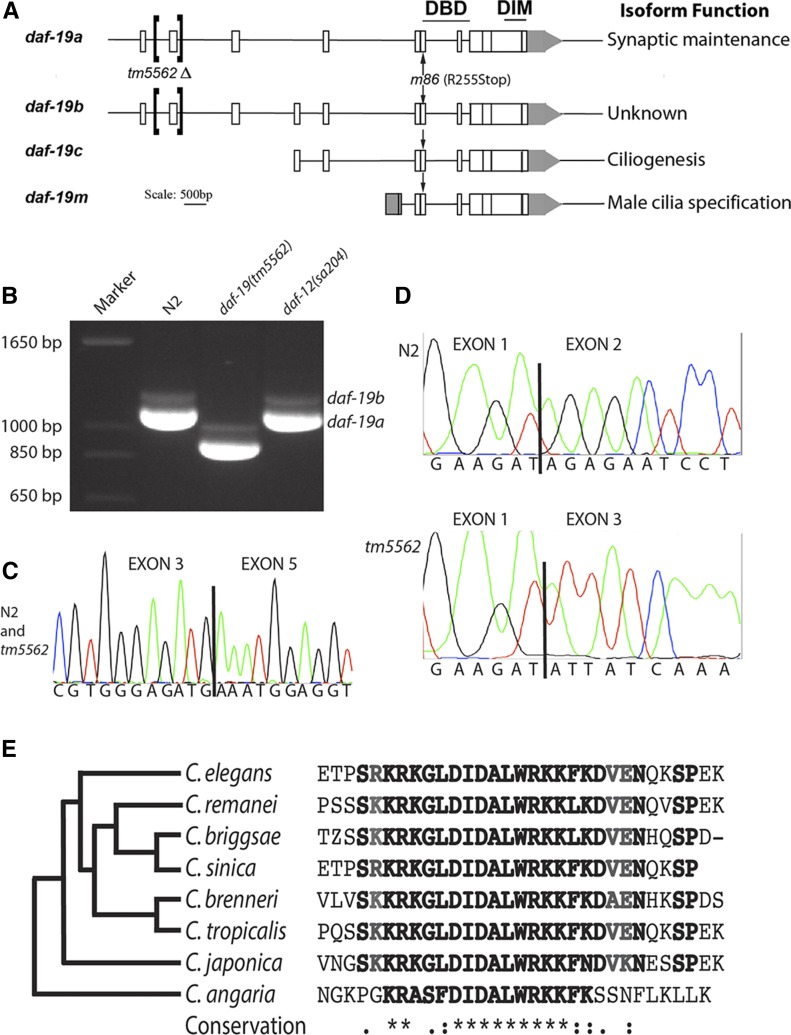
Figure 1.
daf-19 structure and characterization of daf-19(tm5562) RNA products. (A) Three promoters produce four related daf-19 RNA products, as shown. Conserved DNA-binding (DBD) and dimerization (DIM) domains are indicated above encoding exons; locations of a new deletion allele tm5562 and the null allele m86 are noted with brackets and arrows, respectively. (B) Reverse transcription (RT)-PCR using primers to exons 1 and 10 reveal a highly abundant 1123-bp daf-19a transcript from worms wild-type for daf-19, a 934-bp transcript from tm5562 worms, and a much less abundant daf-19b product that is 75-bp larger. (C) Sequencing trace from the smaller RT-PCR product shows splicing of exon 3–5 as in daf-19a. (D) Sequencing traces of exon 1 splicing in daf-19a cDNA products from wild-type N2 (top) and daf-19(tm5562) worms (below). (E) Translated sequences from exon 2 of daf-19 orthologs aligned using T-Coffee (Di Tommaso et al. 2011) and clustered (left) according to the phylogeny of Slos et al. (2017). Sequence identifiers: C. elegans, gi|71988112; C. remanei, gb|EFP05251.1; C. briggsae, emb|CAP22350.4; C. sinica, scaffold 899; C. brenneri, gb|EGT33833.1; C. tropicalis, scaffold 563.1; C. japonica, contig 15741.5; and C. angaria, contig 57337_2.
Roles for the larger DAF-19A/B isoforms extend into adulthood, including regulation of foraging behavior and synaptic protein maintenance (Senti and Swoboda 2008). daf-19a and b differ by one small, alternately spliced exon (Figure 1, A–C), are highly expressed in nonciliated neurons (Saito et al. 2013), and are also less highly expressed in the hypodermis and body wall muscle at all life stages (Senti and Swoboda 2008; Craig et al. 2013). Null daf-19(m86) mutant adults expressing only transgenic DAF-19C have apparently normal cilia and sensory functions, but display abnormal dwelling and roaming behaviors (Senti and Swoboda 2008), consistent with a synaptic defect in nonsensory neurons (Flavell et al. 2013). This defect was confirmed by pharmacological studies and a noted reduction in certain synaptic protein levels in adults. However, RNA levels of these proteins were not similarly decreased (Senti and Swoboda 2008), suggesting an indirect regulatory effect of DAF-19A/B on synaptic protein turnover. Thus, daf-19 promotes certain neuronal functions beyond ciliogenesis at the embryonic life stage, and these pleiotropic effects can be attributed to downstream effects of different daf-19-regulated genes. We sought to identify new DAF-19 target genes, particularly DAF-19A target genes that function in neurons, by characterizing postembryonic transcriptomes of daf-19(m86) null mutants and isogenic age-matched controls at the L1 larval and adult stages of development. Transcriptome approaches have previously successfully identified new RFX target genes expressed early in worm development (Chen et al. 2006; Phirke et al. 2011; Jensen et al. 2016) and in other organisms, including humans (e.g., Nelander et al. 2005; Wu et al. 2016).
Here, for the first time, we describe target genes of DAF-19A, in addition to new target genes of DAF-19C. Further, we characterize a new daf-19a/b isoform-specific mutation (tm5562) and demonstrate that DAF-19A/B expression is directly or indirectly responsible for the downregulation of target genes in certain nonsensory neurons, while DAF-19C primarily activates genes in CSNs. We propose that the intricate interplay of DAF-19 isoforms controls the expression of different gene batteries in defined sets of neurons, and that DAF-19 impacts neuronal functions ranging from basic synaptic maintenance to neuronal outputs in development (dauer formation) and behavior (foraging and locomotion).
Materials and Methods
C. elegans strains and culture conditions
C. elegans strains (Supplemental Material, Table S1) were cultured at 20° following standard procedures (Brenner 1974) except for LU455, daf-19(m86), grown at 15° to obtain adults for behavioral assays. Gene expression studies at multiple life stages required the daf-12(sa204) allele to fully suppress the highly penetrant dauer formation constitutive (Daf-c) phenotype conferred by daf-19(m86). him-5(e1490) was used to generate males. The daf-19(tm5562) mutant was obtained from the Mitani laboratory collection and was outcrossed to wild-type N2 six times. Three primers [upstream (U), downstream (D), and poison (P)] identified the tm5562 deletion allele during outcrossing: U5, 5′-AATGACCTTCACACGGTGTC-3′; D5, 5′-CACACCGGGTGCTTCACCAT-3′; and P5, 5′-GCGCACAATAGGCTCCAAGC-3′. The 865-bp deletion was confirmed by sequencing PCR amplicons after outcrossing. daf-19(tm5562/+) heterozygous and associated control animals were generated by mating to a strain carrying an integrated fluorescent marker.
L1- and adult-stage worms were collected for transcriptome analysis from JT204 daf-12(sa204) and JT6924 daf-19(m86); daf-12(sa204). To collect L1-stage worms, worms were grown on agarose-containing solid egg-NGM medium for 6–7 days until a sizeable gravid adult worm population was observed. Embryos were isolated by hypochlorite treatment and grown to the threefold stage in S-Basal medium with Escherichia coli OP50 with gentle agitation at 20°. A second hypochlorite treatment removed hatched worms and dead eggs. Threefold-stage embryos were further grown on OP50-seeded NGM agarose plates for 5 hr to obtain selectively enriched L1-stage worms. To obtain adult-stage worms, a synchronized hypochlorite-treated population was grown to the L4 stage on NGM agarose plates. Worms were gently rinsed from plates and gravity-settled for 5 min, washed with M9, and settled for 3 min, followed by a repetition with 1 min of settling. Early-stage larvae and embryos were left in each supernatant. This procedure was repeated after worms fed on agarose media for another 24 hr. Two-day-old well-fed adults were harvested 50 hr past the L4 stage.
RNA extraction, cDNA preparation and labeling, and microarray data generation
Harvested worms were passed through three freeze and thaw cycles using liquid nitrogen. For reverse transcriptase (RT)-PCR and microarray analysis, total RNA from the L1 stage was isolated using a standard phenol–chloroform extraction procedure following homogenization; total RNA from adults was prepared using TRIzol followed by a QIAGEN (Valencia, CA) RNAeasy kit following the manufacturer’s directions. The quantity and quality of the extracted RNA was determined using an Agilent Bioanalyzer 2100 (Agilent, Santa Clara, CA). A standard eukaryotic target preparation protocol using 5 μg of total RNA was conducted for each array, as described in the Affymetrix GeneChip Eukaryotic Sample labeling protocol (Affymetrix). Four independent RNA preparations from L1 larvae and three from adults of each strain were used for one GPL200 Affymetrix C. elegans genome array hybridization each; array data were analyzed as described in the GeneChip Expression Analysis technical manual (Affymetrix). In vitro transcription, fragmentation, hybridization, staining, and scanning were performed by the Bioinformatics and Expression Analysis core facility, Karolinska Institute, Stockholm-Huddinge, Sweden (www.bea.ki.se). Primers for RT-PCR analysis of daf-19 were 5′-GCCATCGACGAGCAGTGTG-3′ and 5′-CATGCAAGGAGAGACGCTG-3′. Additional primers for sequencing were 5′-CTTACGAGGTGTTCCAGACGA-3′, 5′-TCGTCTGGAACACCTCGTAAG-3′ and 5′-AGACGGATCGGATGAGCTTTC-3’. RT-PCR used the SuperScript III One-Step RT-PCR system (Invitrogen, Carlsbad, CA).
Transcriptome analysis
Subsequent to scanning, microarrays were subjected to a range of low-stringency analyses, including image analyses, signal summarization, and normalization. Expression reports were examined to confirm that all internal control values were within the acceptable range, as defined by the Affymetrix Data Analysis Fundamentals (Affymetrix). Data passing all quality control measures were utilized for further statistical analyses.
CEL files obtained from microarray hybridizations were imported into robust multichip average (RMA) Express (version 0.3; http://rmaexpress.bmbolstad.com), and expression signal values were calculated using an RMA expression summary and quantile normalization technique. BRB Array Tools (version 3.3; http://linus.nci.nih.gov/BRB-ArrayTools.html) were used to identify genes with statistically signiϕicant variation in expression. The probability threshold was set at a maximum of 0.05 (P-value ≤ 0.05) for genes to be considered statistically differentially expressed in wild-type and mutant populations. Genes with a signal variation of 1.5-fold or greater were selected for subsequent experiments. To reduce false discoveries, a class comparison test was conducted using a multivariate permutation test with a confιdence level of 97% (L1 analysis) and 90% (adults). This analysis revealed that 403 probes identified 370 unique differentially regulated genes (n = 235 downregulated and n = 135 upregulated) in daf-19 mutant L1 larvae. Such a permutation is very similar to that utilized by the Significance Analysis of Microarrays (SAM) analyses (Tusher et al. 2001). Additional lists of genes were generated using SAM (version 2.2) with a false discovery rate (FDR) of ≤ 5% (Q-value ≤ 5%) for L1 analysis and 10% for adult analysis. Thirty repeated runs of SAM were performed using variable random seed numbers for each run. During each run of SAM, 100 permutations were performed. Probes appearing with a bootstrap frequency of 24 (80% of total runs) were further considered for signal variation filtering, used to selectively identify genes with a 1.5-fold or greater variation between the two genetic conditions (L1 larvae: n = 154 downregulated and n = 58 upregulated). The list of genes differentially regulated in adults was generated similarly: RMA normalization followed by class comparison with a multivariate permutation, a FDR of 0.1 and 70% confidence level (n = 120 downregulated genes, n = 75 upregulated). SAM analysis yielded a list of 119 unique genes (n = 74 downregulated; n = 45 upregulated) that is completely contained within the larger list of differentially regulated genes. Comparisons between transcriptomes (threefold-stage embryos, Phirke et al. (2011); L1 larvae, this work; and adults, this work) were undertaken using the gene lists generated by class comparison (Table S2, Table S3, and Table S4).
Transcriptional GFP-fusion gene expression constructs
Gene expression was assessed exclusively using transcriptional GFP reporter constructs generated by inserting 1–3 kb of DNA upstream of genes of interest into the poly-cloning site of the GFP reporter plasmid pPD95.75 (Table S5). Germline transformations by microinjection (Mello et al. 1991) used one of two methods. In one set, GFP constructs (40 ng/μl) and the transformation marker elt-2p::mCherry [10 ng/μl; gift from Gert Jansen; Burghoorn et al. (2010)] were microinjected with carrier DNA at 50 ng/μl. Worms expressing mCherry in the intestine were analyzed for GFP expression. Alternatively, GFP constructs (70 ng/μl) were co-injected with 30 ng/μl unc-122p::DsRed with no carrier DNA. Worms with DsRed expression in coelomocytes were analyzed for GFP expression. Transgene GFP expression patterns were characterized in ≥ 30 L1 adult-stage worms per strain using a Leica TCS SP5 II confocal microscope; adult expression patterns are shown. For gakh-1 and del-4 age-dependent expression analysis, N = 130 and 40 per strain, respectively. Worms were immobilized using 10 mM NaN3 on 2.0% agarose pads. Two daf-19 translational constructs fused isoform-specific endogenous promoters to partial cDNAs expressing either daf-19a (pGG67_3) or daf-19c (pGG14) [described in Senti and Swoboda (2008)]; these two constructs were used for isoform-specific rescue experiments.
Dye-filling and behavioral assays
Fluorescent dye-filling assays:
Starich et al. (1995) using DiI (Molecular Probes, Eugene, Or) were used to confirm cilia defects in daf-19 mutant animals and to assist with neuronal identification in wild-type worms.
Dauer formation assays:
Twenty gravid hermaphrodites were placed on OP50 for 6 hr, and dauer larvae were enumerated after 5 days (15°), 4 days (20°), or 3 days (25°) of growth. Tukey’s pairwise comparisons of arcsine-transformed data were used to detect differences in average dauer production between strains for three replicate experiments. All behavioral and life history assays were done blind to strain identity.
Early life history:
Thirty gravid hermaphrodites were placed on OP50 for 2 hr to produce tightly synchronized populations. The life stage of each offspring was assessed every 8 hr. Tukey’s pairwise comparisons were used to detect significant differences in percent of worms at particular life stages in three replicate experiments.
Roaming assays:
L4 worms were grown on fresh OP50 lawns for 12 hr, after which 30 adult worms were individually picked to the center of a bacterial lawn and allowed to forage for 1 hr at 19° and constant humidity. The proportion of OP50-covered 5-mm squares crossed by tracks was enumerated (cf. Senti and Swoboda 2008). Replicate assays were repeated on seven different dates. Data were arcsine-transformed for statistical analysis. Two-way ANOVA analysis revealed assay date to be a significant variable; the behavior of each strain was compared using Tukey’s pairwise comparisons for each assay date.
Aldicarb assays:
Twenty hermaphrodites, 30 hr post-L4 stage, were placed on OP50-seeded NGM agar containing 500 μM aldicarb. Complete paralysis of the head was scored as in Mahoney et al. (2006). Data from each of six replicate assays were compared using a survivorship curve comparison test (Pyke and Thompson 1986).
Data availability
All microarray data sets were submitted to the Gene Expression Omnibus database (Edgar and Barrett 2006) under accession number GSE96068. C. elegans strains (Table S1) are available upon request. Gene lists generated from this study as well as from Phirke et al. (2011) are included in Table S2, Table S3, and Table S4. Table S5 lists all the primers used and Table S6 lists transgene constructs with apparent DAF-19-independent expression.
Results
Identification of DAF-19 target genes in L1 larvae and adult worms
To identify novel DAF-19 target genes, we employed whole-genome microarrays of daf-19(m86) null mutants and isogenic wild-type worms from synchronized L1 larvae and 2-day old adults. To add to the comparably prepared gene lists from embryos (Chen et al. 2006; Phirke et al. 2011 in Table S2), a comparison of mutant and wild-type populations (cf. Materials and Methods) uncovered large sets of differentially regulated genes in L1 larvae (Table S3) and adult worms (Table S4).
We determined whether DAF-19-regulated transcriptomes were enriched for particular types of genes and whether that enrichment changed during development. We found that the breadth of gene categories represented by DAF-19-regulated transcriptomes narrowed as development proceeded from embryos to L1-stage larvae to adults (Table 1). Based on the timing of ciliogenesis, we expected that DAF-19-regulated transcriptomes from embryos and L1 larvae would be enriched for genes involved in ciliogenesis, while this would not be the case for the adult-stage gene list. Such a finding would be consistent with the results from Jensen et al. (2016), who used a gene expression profiling approach to track ciliary gene expression during development. ;Our DAF-19-regulated transcriptome from L1-stage larvae was grouped into nine gene ontology clusters with enrichment scores > 1.5 (Huang et al. 2009), each containing ≥ 10 genes, including clusters representing cilia, dauer formation, signaling, and aging. By comparison, the DAF-19-regulated transcriptome from adults was enriched for smaller sets of genes involved in signaling, aging, and proteolysis. Both of these transcriptomes displayed an even stronger reduction in clusters when compared to the threefold-stage embryo DAF-19-regulated transcriptome (Phirke et al. 2011) (Table 1 and Table S2).
Table 1. Annotation clusters of differentially regulated genes.
| Threefold embryo | Score | L1 | Score | Adult | Score |
|---|---|---|---|---|---|
| Cilium assembly/dauer | 10.67 | Cuticle/collagen | 12.04 | Signal peptide | 3.53 |
| Cilium biogenesis/protein transport | 4.85 | Cilium assembly/behavior | 5.21 | Aging | 2.99 |
| Mitosis/cell cycle | 4.04 | Cilium assembly/dauer | 4.17 | Proteolysis | 1.69 |
| Vulval location/dauer entry | 3.85 | Lipid modification | 3.31 | Lipase | 1.52 |
| Glutathione transferase | 3.66 | Disulfide bond/cuticle | 2.08 | 172 IDs, 17 clusters | |
| Protein processing/ubiquitin | 3.24 | Signal peptide | 1.85 | ||
| Cilium morphogenesis/behavior | 3.21 | Aging | 1.82 | ||
| Glucuronidation | 3.08 | Proteolysis | 1.52 | ||
| Cytoskeleton | 2.86 | Neuropeptide | 1.48 | ||
| Endoplasmic reticulum | 2.54 | 364 IDs, 37 clusters | |||
| Spindle/polar body extrusion | 2.48 | ||||
| Nucleosome | 2.45 | ||||
| ER lumen | 2.22 | ||||
| Histone | 2.03 | ||||
| Oxidoreductase | 2.03 | ||||
| Kinetochore/centromere | 1.97 | ||||
| Tetratricopeptide-like helical | 1.91 | ||||
| Cyclin | 1.90 | ||||
| Glucose/ribitol dehydrogenase | 1.77 | ||||
| EF-hand domain | 1.68 | ||||
| ATP-binding | 1.54 | ||||
| 909 IDs, 21 clusters > 1.5 |
Gene annotation clusters of differentially regulated genes. Genes identified as putative targets of daf-19 in threefold stage embryos (Phirke et al. 2011), L1 larvae, and 2-day-old adults were clustered using the DAVID Bioinformatics Resource 6.7 with a medium classification stringency (Huang et al. 2009). Clusters with scores of ≥ 1.5 are shown. IDs, identifiers; DAVID, database for annotation, visualization and integrated discovery; ER, endoplasmic reticulum.
Comparisons with the Cilia database CilDB (Arnaiz et al. 2014) also indicated an enrichment of known ciliary genes in the gene list from L1 larvae, but not in the gene list from adults. Overall, CilDB identifies 15% of C. elegans genes as ciliary based on two or more published studies. Using the same criteria, we identified > 21% of the gene list from L1 larvae as ciliary, while only 11% of the adult gene list were known ciliary genes. Core ciliary genes (e.g., bbs and nphp genes) important for the process of ciliogenesis (Choksi et al. 2014) were represented with statistical significance only on the gene lists from embyros and L1 larvae, indicating that the maintenance of cilia at the adult stage is very likely much less dependent on DAF-19 regulation. Lastly, there was significant enrichment (4–10-fold) in all three lists (threefold-stage embryo, L1 larvae, and adults) for genes expressed in neurons, with expression patterns becoming more restrictive as development proceeds (Figure S1). In light of the suggested function of DAF-19A/B in regulating the maintenance of protein levels at neuronal synapses, we note the presence in the adult gene list of 12 genes involved in proteolysis (Table 1), three of which we characterized in more detail: spg-20, anp-1, and skr-12.
For further analyses we chose a number of genes based on one of the following criteria: (1) a suggested neuronal expression pattern, (2) a WormBase annotation suggesting a connection to daf-19 mutant phenotypes such as protein stability or innate immunity, or (3) no known function or expression pattern (see Table S5). In total, we assessed expression patterns of 44 candidate daf-19 target genes using transcriptional GFP fusions, 32 of which have identified human orthologs (Shaye and Greenwald 2011). Out of the 44 genes, 33 showed strong GFP expression that was characterized further, whereby expression patterns were always compared between isogenic strains differing only by the daf-19 allele. Gene choices were not filtered for their presence in an operon; four such genes were analyzed: three that are the first gene of an operon and one, ddn-1, that was the second [based on Allen et al. (2011)]. Characteristics of daf-19 target genes and their expression patterns are summarized in Table 2 and Table S6.
Table 2. daf-19-dependent target genes.
| Mutant/WT fold-change | |||||||
|---|---|---|---|---|---|---|---|
| C. elegans Gene IDs | Gene function | 3Xa | L1 | Adult | Cilia database | Expression pattern in wild-type worms | Expression pattern in daf-19(null) worms |
| asic-2 T28F4.2 191981_at | Predicted sodium channel | 0.76 | 0.54 | 0.65 | Yes | IL2 neurons, two fainter OLs | Two faint OLs |
| spg-20 F57B10.9 185820_at | Orthologous to SPARTIN | 1.09 | 1.24 | 1.58 | Yes | IL2 neurons, most neurons posterior of nerve ring, body wall muscle | Most neurons posterior of nerve ring, body wall muscle |
| ddn-1 B0507.10 180569_at | Unknown | 1.17 | 0.29 | 1.02 | No | IL2, ASG, URX (rarer), AFD (faint), ASI (faint), ASE, I5, M5, intestine, muscle | ASE, I5, M5, intestine, some muscle |
| eppl-1 T01B11.2 190153_s_at | Transaminase activity | 0.89 | 0.62 | 1.16 | Yes | ASG, ASE, one tail neuron (PQR), pharynx, intestine, hypodermis | Tail neuron (PQR), pharynx, intestine, hypodermis |
| del-4 T28B8.5 190395_at | Predicted sodium channel | 1.14 | 0.40 | 1.33 | Yes | ASE, AIY, ASG (fainter), AIA, PHA, PQR (up to three neurons in tail) | AIA, ASG, up to three per head neurons in L1; 0–Two head neurons in adults, up to four tail neurons at all ages |
| mapk-15 C05D10.2 none | Mitogen-activated protein kinase | N/A | N/A | N/A | No | IL2, OLQ, CEPD/V, ASK, AFD, ADF, ADL, ASG, ASI, ASH, ASE, ADE, PHA, PHB, PQR | OLQ, CEPD/V, ASK, AFD, ADF, ADL, ASG, ASI, ASH, ASE, ADE, PQR |
| ddn-2 F35C5.11 184057_at | TGF-B activated receptor | 0.17 | 0.20 | 0.65 | Yes | ILs, two/three amphids, intestine | Two/three amphids, Intestine |
| xbx-9 C15C8.1 179348_at | unknown | 0.44 | 0.35 | 0.82 | Yesa | 3 CSNs, ADF | ADF |
| skr-12 C52D10.6 189189_s_at | Homolog of SKp1, ubiquitin-ligase complex | 2.11 | 0.96 | 0.55 | No | ASI, ASK (rarer), intestine and pharynx | IL1s, OLQs (rarer), AFD, ADF, ASG, ASE, ASI, ASK, intestine, parts of pharynx |
| gakh-1 F46G11.3 192321_s_at | Cyclin G-associated kinase | 0.47 | 0.67 | 0.45 | Yesa | AIY (v. rarely), RIF, two–four tail neurons in adults. L3 have up to six GFP+ head neurons. | URA, URB, IL2 (larvae only), BAG (<L4), AVA (rarer), AVB, SIAD/V, AIN (rarer), M4 (rarer), AIY, RIF, 2–4 tail neurons |
| rgs-8.1 F52D2.2 189970_s_at | Regulator of G protein signaling | 0.66 | 0.85 | 0.34 | Yes | PVT, faint intestine | RID, I2, 1 DV neuron, PVQ, PQRL/R, faint intestine |
| mapk-15 C05D10.2 none | Mitogen-activated protein kinase | N/A | N/A | N/A | No | Much less robust in IL1 | IL1 |
Summary of target gene expression. Characteristics and observed expression patterns of daf-19 target genes are summarized. Gene functions were obtained from WormBase WS258. Fold-change is mutant versus wild-type expression (Phirke et al. 2011). Identification as a ciliary gene in Cilia database v. 3 (Arnaiz et al. 2014) is indicated. Expression patterns are summarized from at least 30 worms per strain equally divided by L1/L2, L3, L4, and adult life stages, except gakh-1 and del-4, where N = 130 and 45, respectively. Genetic backgrounds were daf-19-12(sa204) with a daf-19 null background [daf-19(m86)], or daf-19(+/+), where some age-dependent differences in expression were noted. Sites of differential expression (fold-change differences ≥ 1.5-fold) are highlighted in bold.
Phirke et al. (2011) only.
The presence of DAF-19 activates gene expression
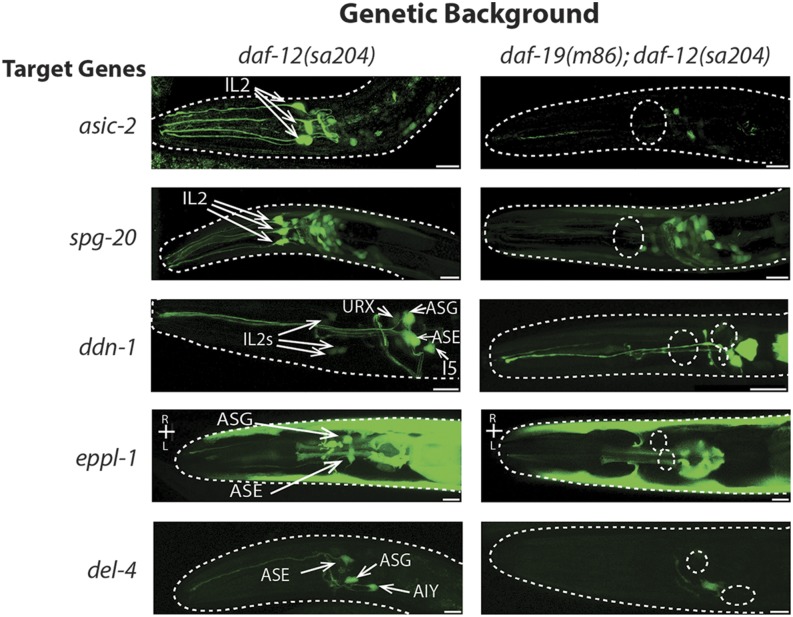
We compared the expression of putative target gene transcriptional fusions in daf-19 null and wild-type genetic backgrounds to determine where and in which way target gene expression was DAF-19-dependent. Our transcriptome analysis revealed new genes activated in the nervous system in the presence of DAF-19. Seven gene promoter fusions showed clear evidence of activation by the presence of wild-type DAF-19 in neurons, mostly limited to subsets of CSNs (Table 2). The inner labial (IL2) neurons were a common site of daf-19 target gene activation; expression of asic-2, spg-20, and ddn-1 (downstream of daf-19) (Figure 2), as well as ddn-2 [F35C5.11 in Phirke et al. (2011)] and mapk-15 (Piasecki et al. 2017), in IL2 neurons required daf-19. IL2 neuron identity was confirmed by colocalization with a cho-1::mCherry reporter (Pereira et al. 2015) or lack of colocalization with eat-4::mCherry (Serrano-Saiz et al. 2013) (Figure S2). ddn-1::gfp is also differentially expressed in other CSNs; we observed robust activation in the presence of DAF-19 in ASG, and less pronounced activation in URX (not a CSN), ASI, and AFD, the latter identified by eat-4 colocalization.
Figure 2.
Presence of DAF-19 activates expression of novel target genes in certain ciliated sensory neurons. Expression patterns of transcriptional fusions of daf-19 target gene promoters with the GFP gene in isogenic daf-12(sa204) and daf-19(m86); daf-12(sa204) adult hermaphrodites (N = 30 worms/strain analyzed). Views are lateral with dorsal at the top and anterior to the left, except strains expressing eppl-1::gfp. Neurons in which daf-19-dependent expression was found are identified. Cell bodies in which gene expression is absent in daf-19(m86) mutants are indicated by dotted circles. Bar, 10 μm.
Even though DAF-19C is expressed in many or all CSNs and DAF-19A/B in nearly all nonciliated neurons (Senti and Swoboda 2008), all daf-19 target genes examined appear to have RFX-independent expression in some neurons in addition to DAF-19 dependence in a subset of neurons (Table 2). For example, robust expression of spg-20::gfp was observed in the majority of neurons, but DAF-19-dependent activation was discernable only in IL2 neurons (Figure 2). Expression of eppl-1::gfp appeared DAF-19-dependent only in the CSNs ASE and ASG (Figure 2), but not in PQR or nonneuronal tissues, while DAF-19-dependent activation of xbx-9::gfp, found in our gene lists and previously described by Burghoorn et al. (2012), was restricted to three of four CSNs in which expression was observed. Finally, del-4::gfp expression was age-dependent in ASE, ASG, and several other unidentified neurons (Figure 2 and Table 2). ASE and ASG were identified by location, morphology, and by elimination: they do not dye-fill and are not cholinergic (Figure S2). In wild-type animals, del-4 expression decreased from eight amphid CSNs in L1–L2 larvae to half as many in adults. In daf-19(m86) null mutant worms, no more than three neuron pairs expressed del-4 in young larvae, and only one or two AIA neurons did so in adults. We conclude that DAF-19 activates del-4 expression in ASE and in AIY interneurons. In other studies, del-4 transcripts were found in ASE following serial analysis of gene expression (SAGE) analysis, and the gene contains an ASE motif (Etchberger et al. 2007) as well as an X-box motif (Efimenko et al. 2005).
DAF-19 presence downregulates some genes in the nervous system
Because two RFX orthologs in humans and yeast have been found to repress gene expression, we assessed expression patterns of several genes that appeared to be upregulated in the absence of DAF-19 in our gene lists. For the first time, we report that wild-type DAF-19 can downregulate target gene expression in both ciliated and nonciliated neurons. Three genes, all of which showed differential transcription in at least two life stages, were expressed in fewer neurons in wild-type worms than in daf-19(m86) null mutants. One additional gene, mapk-15, not identified by microarray analysis, was also downregulated by DAF-19 (Table 2).
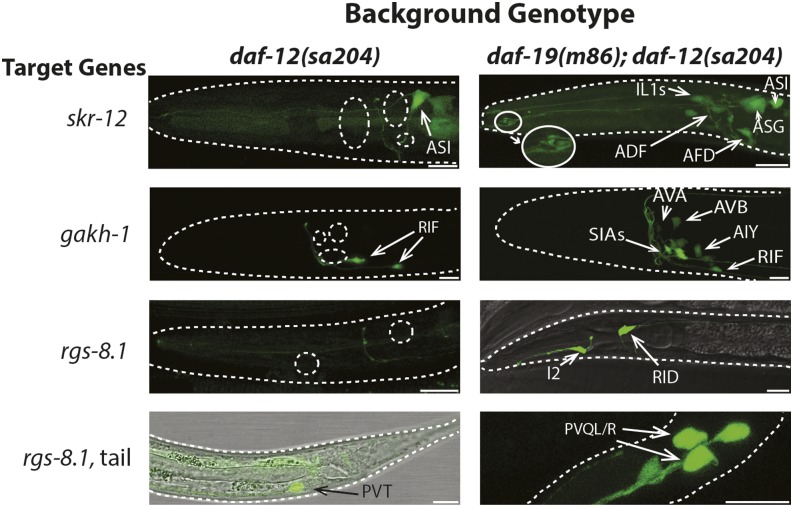
daf-19-dependent downregulation of skr-12 was seen in more than four sets of CSNs. Based on eat-4 colocalization, we identified skr-12 expression in the CSNs IL1, ASG, and AFD, and occasionally in the OLQ neurons in daf-19(m86) null mutants (Figure 3 and Figure S2). The characteristic microvilli at the tip of the AFD dendrites (Doroquez et al. 2014) were easily visible (Figure 3). Expression in two other CSNs, most likely ASE and ADF, was also downregulated by the presence of DAF-19. Two additional neurons in which skr-12 was downregulated could not be identified due to faint expression levels. Expression of skr-12 in the ASI and ASK amphid CSNs (identified through colocalization with DiI), as well as in the intestine and pharyngeal muscle, appeared to be daf-19-independent.
Figure 3.
DAF-19 presence downregulates expression of novel target genes in certain nonsensory neurons and ciliated sensory neurons. Expression patterns of transcriptional fusions of daf-19 target gene promoters with GFP in isogenic daf-12(sa204) and daf-19(m86); daf-12(sa204) adult hermaphrodites (N = 30 worms/strain analyzed, except for gakh-1, N = 130). Views are lateral with dorsal at the top and anterior to the left. Neurons in which daf-19-dependent expression was found are identified. Cell bodies in which gene expression is absent in worms wild-type for daf-19 are indicated by dotted circles. Microvilli at the dendritic tips of AFD sensory neurons (in the right skr-12 image) are magnified an additional 2×. Bar, 10 μm.
daf-19-dependent downregulation of gakh-1 was observed consistently in nonciliated neurons in an age-dependent manner. In wild-type adults, gakh-1::gfp was always expressed in the cholinergic RIF neurons and, rarely, in the AIY interneurons and the M4 pharyngeal motor neuron (Figure 3 and Figure S2). However, in daf-19(m86) mutants, the transgene was expressed in at least four additional sets of interneurons: AVB, AIY, AIN, and AVA (both less frequently), and either the SIAD and SIAV interneurons or possibly the SMBD motor neurons (Figure 3 and Figure S2). These latter neurons could not be unambiguously distinguished using cho-1::mCherry colocalization or by their morphology, but they never expressed gakh-1 in wild-type worms (N = 132). In L3 and younger larval daf-19 mutants, additional neuronal expression was seen in the glutamatergic BAG CSNs and, at low levels, in cho-1-expressing neurons anterior to the nerve ring (Figure S2). Sieburth et al. (2005) also reported gakh-1 expression in the intestine, pharynx, and head and tail neurons in wild-type worms.
Only in daf-19 mutant worms, rgs-8.1::gfp was consistently expressed in the I2 pharyngeal and nonciliated RID head neurons, as well as less frequently in four neurons of the tail (Figure 3 and Figure S2), including the PVQ interneuron pair, one of the DVA, B, or C neurons, and a single neuron directly posterior of PVQ. Expression in the PVT tail neuron was irregular in both wild-type and daf-19 mutant backgrounds. rgs-8.1 was also found to be differentially regulated in embryonic daf-19(m86) null mutant transcriptomes (Chen et al. 2006; Phirke et al. 2011).
In summary, expression of four target genes is downregulated only in the presence of DAF-19. This downregulation is specific to the nervous system.
Characterization of genes whose expression is (largely) independent of DAF-19
Fifteen gene promoter fusions yielded GFP expression in neurons that was (largely) independent of DAF-19 (Figure S3 and Table S6). This holds true also for genes expressed in both neurons and the intestine, or for genes (predominantly) expressed in the intestine, where they might be involved in innate immunity (Xie et al. 2013) (Figure S3 and Table S6). Three genes, anp-1, hex-1, and C06G3.6, showed a clear reduction in the frequency of neuronal expression in a daf-19(m86) null mutant background (Table S6). Seven genes were expressed in the intestine and not the nervous system, and another 14 were expressed in both tissues. In no case could we detect daf-19-dependent expression in the intestine (Table S6), even when neuronal expression was dependent on the presence of functional DAF-19 (N = 5 genes; Table 2). Similarly, though daf-19 has been reported to be expressed to some extent in body wall muscle and hypodermis (Senti and Swoboda 2008), expression of target genes in either tissue was indistinguishable between wild-type and daf-19(m86) null mutants (Table S6). Apparent DAF-19 independence of putative target gene expression could be due to incomplete control regions in our transcriptional fusion constructs, DAF-19 dependence only at embryonic stages of development that were not fully assessed, or false positives in the microarray data. Thus, our experiments revealed daf-19-dependent target gene expression in the nervous system only.
Characterization of tm5562, an isoform-specific allele of daf-19
To determine which of the abundant DAF-19 isoforms (A or C) is responsible for controlling target gene expression, we characterized a new daf-19 allele, tm5562 (Figure 1). tm5562 alters both daf-19a and b transcripts via deletion of exon 2, 582 bp of intron 1, and 93 bp of intron 2. RT-PCR revealed that tm5562 mutant worms produced RNAs shorter than wild-type by the predicted length of exon 2 (Figure 1B). Both daf-19a, in which exon 3 is spliced to exon 5, and the daf-19b isoform including exon 4 were affected (Figure 1, B–D). Only in RNA isolated from tm5562 mutants was exon 1 perfectly spliced to exon 3 in both isoforms (Figure 1D). Sequencing of RT-PCR amplicons as well as of genomic DNA revealed no other mutations through the DBD (data not shown). The tm5562 transcript is expected to produce in-frame proteins lacking the evolutionarily conserved exon 2 (Figure 1E).
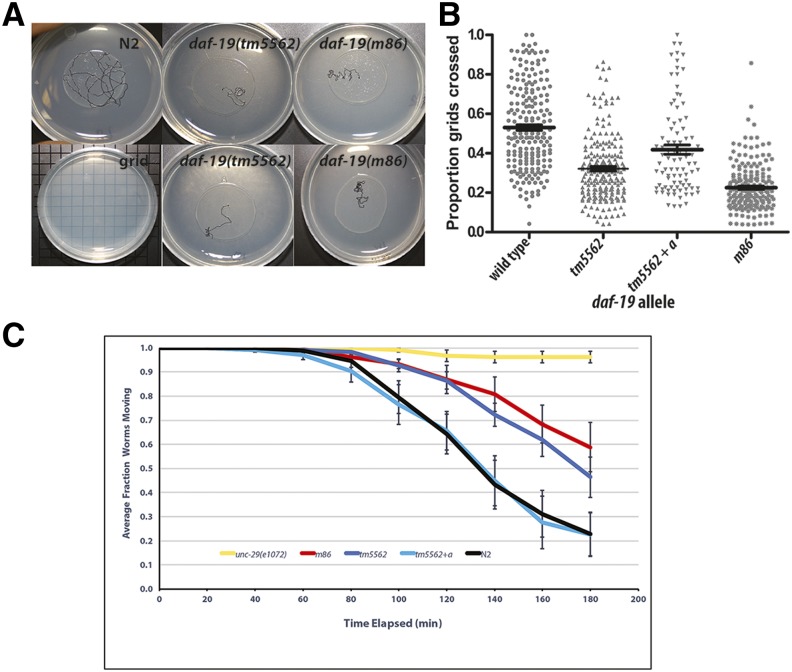
We determined that daf-19(tm5562) confers phenotypes expected of a daf-19a but not a daf-19c mutant, such as roaming and pharmacological defects previously reported (Senti and Swoboda 2008). Wild-type worms will both dwell in small areas of a bacterial lawn and will roam to (Ben Arous et al. 2009), or explore (Flavell et al. 2013), new areas. We quantified the roaming behavior of individual worms and detected a significant difference between strains (Figure 4, A and B). daf-19(tm5562) worms displayed less roaming behavior than did wild-type N2 worms on all seven assay dates (P < 0.003). In all assays, daf-19(tm5562) behavior was either indistinguishable from that of daf-19(m86) null mutants (P > 0.5) or displayed an intermediate phenotype, statistically distinguishable from both wild-type N2 (P < 0.002) and daf-19(m86) null mutants (P < 0.02). As was the case for daf-19(m86) (Senti and Swoboda 2008), the aberrant roaming behavior of daf-19(tm5562) worms was rescued by transgenic expression of a daf-19a-specific construct (P < 0.03) (Figure 4B).
Figure 4.
daf-19(tm5562) confers decreased roaming behavior and aldicarb resistance. (A) Each 1-day-old adult worm roamed freely from the center of an OP50 bacterial lawn for 1 hr at 19°: shown are representative worm tracks for worms of indicated genotypes. (B) Jitter plot of the proportion of grids crossed versus the total number of grids underneath the OP50 bacterial lawn; N = 30 worms per assay. tm5562 + a indicates transgene expression of daf-19a from its endogenous promoter. The median and 1 SEM are indicated. Tukey’s pairwise comparisons indicate that tm5562 worms roam less than wild-type N2 worms on all assay dates (P < 0.003), while tm5562 + a transgenic worms roam significantly more than tm5562 worms (P < 0.03). (C) Response to 500 μM aldicarb was assessed as in Mahoney et al. (2006). Error bars are 1 SEM. Pairwise comparisons of average paralysis curves (N = 6 assays) were compared (Pyke and Thompson 1986). tm5562 worms are not different from m86 worms (P = 0.56), whereas tm5562 + a transgenic worms differ from m86 worms (P = 0.02) and do not differ from wild-type N2 worms (P = 0.56).
We assessed synaptic function in daf-19(tm5562) mutants using resistance to aldicarb, a cholinesterase inhibitor. For comparison, daf-19(m86) 1-day-old adult worms were consistently resistant to aldicarb, though not to the level of age-matched unc-29(e1072) mutants lacking a subunit of the acetylcholine receptor (Figure 4C). Age-matched daf-19(tm5562) worms were resistant to aldicarb at levels statistically indistinguishable from those of daf-19(m86) worms in an average of six replicate assays (P = 0.56). This synaptic defect was rescued by transgenic expression of daf-19a (P = 0.02). Average aldicarb sensitivity of rescued worms was indistinguishable from that of wild-type N2 (P = 0.56) (Figure 4C).
We used the aldicarb assay to test whether daf-19(tm5562) is a hypermorphic or dominant negative allele. If so, we expect heterozygous tm5562/+ mutant animals to confer an aldicarb-resistant phenotype. Instead, we found that daf-19(tm5562/+) heterozygotes had a phenotype indistinguishable from that of wild-type N2 worms mated with the same fluorescently marked strain (Figure S4D) (cf. Materials and Methods). We conclude that daf-19(tm5562) is a hypomorphic or loss-of-function allele affecting both daf-19a and b isoforms.
As expected, the tm5562 allele did not appear to affect daf-19c function. While the daf-19(m86) null allele confers a highly penetrant Daf-c phenotype (Swoboda et al. 2000), daf-19(tm5562) did not confer a Daf-c phenotype in the presence of food at any temperature tested (Figure S4A). Normal progression from the first to second larval stages thus indicates that daf-19a/b is not responsible for the developmental decision to enter the dauer larval stage. Animals lacking DAF-19C do not develop cilia and therefore CSNs that normally take up the fluorescent, lipophilic DiI can no longer do so. The fact that daf-19(tm5562) worms dye-filled normally in both amphids (Figure S4B) and phasmids (data not shown), and males mated readily with hermaphrodites, suggests normal cilia development. These phenotypes are consistent with expression of functional daf-19c transcripts in daf-19(tm5562) worms.
Because daf-19 can affect development, we followed worm development in populations synchronized by a 2-hr time window (Figure S4C). We found that daf-19(tm5562) worms developed more slowly through the L3 stage, as indicated by the larger number of worms at this larval stage at 40 and 48 hr after egg laying (P = 0.00023 as compared to wild-type N2). We conclude that the time course of larval development of daf-19(tm5562) worms was slightly altered. Similar phenotypes, notably an L2 stage extension, have been found for Daf-c temperature-sensitive mutants of daf-2, when at the permissive temperature these worms do not enter the dauer stage (Ruaud et al. 2011; Olmedo et al. 2015). In summary, phenotypes conferred by daf-19(tm5562) are consistent with those of daf-19(m86) null mutant worms in which the daf-19c isoform is rescued by transgene expression (Senti and Swoboda 2008). These results indicate that the tm5562 allele strongly reduces or eliminates solely the function of the longer daf-19a and b isoforms (cf. Figure 1A). Rescue of tm5562 phenotypes by daf-19a expression suggests that DAF-19A plays an important and unique role in nervous system function.
daf-19 isoform-specific regulation of target gene expression
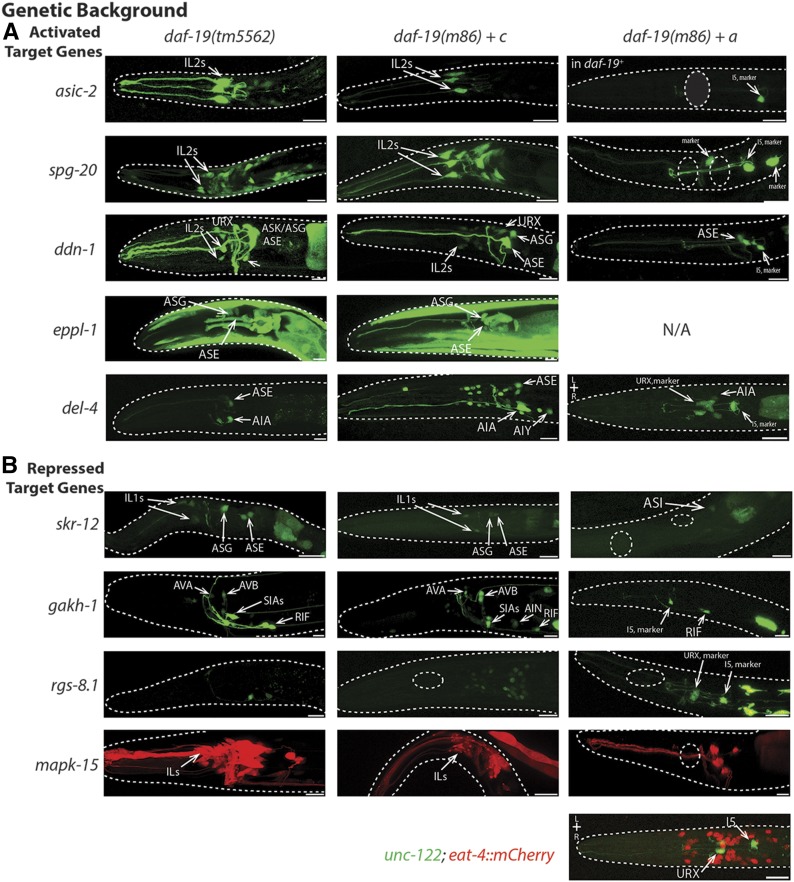
We used the daf-19(tm5562) genetic background to determine whether differential target gene expression required functional DAF-19A. For six target genes for which the presence of DAF-19 activated expression, we compared the expression patterns between daf-19(tm5562) mutants and daf-19(m86) null mutant worms that carried isoform-specific rescuing transgenes [notated as daf-19(0)+a or +c]. With no exceptions, these six target genes remained activated in a daf-19(tm5562) background (Figure 5 and Table 3). Consistent with this finding, the activated expression of these six target genes was rescued in daf-19(0)+c animals, whereas in daf-19(0)+a animals it was not rescued. Surprisingly, transgenic overexpression of daf-19a repressed the near-pan-neuronal expression of spg-20 as well as all expression of asic-2 (Figure 5), a phenotype we term ectopic repression (Table 3).
Figure 5.
Isoform-specific control of daf-19 target genes. (A) Target genes activated by the presence of DAF-19 are still activated in a daf-19(tm5562) background, where DAF-19C, but not DAF-19A, is functional. These genes are also activated when a transgene expressing DAF-19C, but not a DAF-19A transgene, is added to daf-19(m86) null mutant worms. Overexpression of DAF-19A represses expression of asic-2, spg-20, and mapk-15 even in cells where expression is typically independent of daf-19 (Figure 2). N/A = expression of target gene in muscle prevented identification of daf-19a co-injection marker. (B) Two target genes, skr-12 and gakh-1, are repressed in a daf-19 wild-type background, but are not repressed in a daf-19(tm5562) background when DAF-19A is nonfunctional. Expression of rgs-8.1 and mapk-15 is repressed in the presence of either isoform of DAF-19. Bar, 10 μm.
Table 3. daf-19 isoform-specific target gene regulation.
| Expression in indicated worm genotype | |||||||
|---|---|---|---|---|---|---|---|
| Gene name | Gene ID | Wild-type daf-19 | daf-19 (tm5562) | daf-19(0) + daf-19c | daf-19(0) + daf-19a | Interpretation | |
| Target gene expression activated in the presence of daf-19 | |||||||
| asic-2 | T28F4.2 | Activated | Activated | Rescues activation | Ectopic repression | DAF-19C activates target gene; DAF-19A overexpression represses | |
| spg-20 | F57B10.9 | Activated | Activated | Rescues activation | Ectopic repression | DAF-19C activates target gene; DAF-19A overexpression represses | |
| ddn-1 | B0507.10 | Activated | Activated | Rescues activation | No rescue | DAF-19C activates target gene | |
| eppl-1 | T01B11.2 | Activated | Activated | Rescues activation | N/A | DAF-19C activates target gene | |
| del-4 | T28B8.5 | Activated | Activated | Rescues activation | Partial rescue | DAF-19C activates target gene | |
| mapk-15 | C05D10.2 | Activated | Activated | Rescues activation | Ectopic repression in head | DAF-19C activates target gene; DAF-19A overexpression represses | |
| Target gene expression repressed in the presence of daf-19 | |||||||
| skr-12 | C52D10.6 | Repressed | Activated | Activated | Ectopic repression | Repression requires DAF-19A; DAF-19A overexpression represses expression | |
| gakh-1 | F46G11.3 | Repressed | Activated | Activated | Rescues repression | Repression requires DAF-19A | |
| rgs-8.1 | F52D2.2 | Repressed | Repressed | Rescues repression | Rescues repression | Either isoform can repress | |
| mapk-15 | C05D10.2 | Repressed | Activated | Activated | Repressed in head | Repression requires DAF-19A | |
Isoform-specific control of target gene expression. The average expression phenotype of daf-19 target genes is shown in a daf-19(tm5562) mutant background (lacking functional DAF-19A) and in daf-19 null (m86) worms expressing either daf-19c or daf-19a from their endogenous promoters. ID, identifier; N/A, unc-122p::gfp co-injection marker not detectable due to target gene expression in muscle.
In contrast, downregulation of target genes appeared to depend primarily, but not exclusively, on the action of DAF-19A. In daf-19(tm5562) mutant worms, skr-12 and gakh-1 were highly expressed; that is, they were not repressed in a daf-19a mutant background, but they were repressed in an isogenic wild-type background (Figure 5 and Table 3). Overexpression of daf-19c did not change this lack of repression of gakh-1 and, usually, skr-12. In a few animals, skr-12 was repressed such that expression was limited to the ASE neurons (Figure 5). Overexpression of daf-19a fully rescued repression of gakh-1, yielding gene expression patterns exactly like that seen in wild-type worms, while repression of skr-12 expression by the presence of DAF-19A was more extensive than the wild-type pattern (Figure 3 and Figure 5). Thus, skr-12 and gakh-1 repression depended on functional DAF-19A.
To our surprise, rgs-8.1 expression in I2 and RID neurons could be repressed in the presence of either isoform of DAF-19. Expression in I2, RID, and in tail neurons was repressed in daf-19(tm5562) worms as well as in both daf-19(0)+a and +c worms. These data suggest that a particular stoichiometry of DAF-19 proteins is important for the correct expression of rgs-8.1. Note that in all cases, transgene presence was ascertained by the presence of the corresponding co-injection marker (cf. Materials and Methods and Figure 5). Lastly, a parallel project revealed that a kinase gene, mapk-15, appeared to be both activated and repressed by the presence of DAF-19. In the presence of DAF-19, mapk-15 is activated in IL2, PHA, and PHB, and many male tail CSNs, but is partially downregulated in IL1s and some of the neurons in the male tail (Piasecki et al. 2017). Expression in other CSNs, including many amphid head and the PQR tail CSNs, does not depend on DAF-19. We found mapk-15 activation in daf-19(tm5562) and in daf-19(0)+c worms, but not in daf-19(0)+a worms. Instead, overexpression of daf-19a repressed nearly all mapk-15 expression in the head, with variable repression in the hermaphrodite tail and no repression in the male tail, again suggesting that DAF-19 stoichiometry is important for correct gene expression in some neurons.
We conclude with the novel finding that expression of the DAF-19A isoform downregulates certain target genes in nonsensory neurons, while DAF-19C expression generally activates genes in CSNs and can, albeit rarely and restricted to small sets of neurons, also downregulate gene expression. These results are consistent with the described, mutually exclusive expression patterns of these two DAF-19 isoforms (Senti and Swoboda 2008).
Discussion
Using comparative transcriptomics during different life stages of the worm C. elegans, we have uncovered large sets of new target genes for DAF-19, the sole RFX TF in this organism. We can assign a number of these target genes to act in defined sets of neurons and, for some, at certain time points during development. DAF-19-dependent expression of these target genes extends beyond CSNs and the threefold stage of embryogenesis, by which time most cilia are formed (Sulston et al. 1983; Nechipurenko et al. 2016). Our work thereby greatly expands the suggested roles for DAF-19 and its target genes beyond a role in ciliogenesis (Choksi et al. 2014). Specifically, the presence of the larger DAF-19A isoform downregulates target genes in nonciliated neurons in larval through adult stages, while the smaller DAF-19C isoform primarily activates target genes in particular subsets of CSNs. Thus, daf-19 provides important contributions to the regulation and maintenance of neuron function throughout development and adulthood.
To determine which DAF-19 isoform regulates target gene expression, we characterized a novel daf-19 allele, tm5562, a deletion of exon 2. The homozygous mutant confers neuronal phenotypes expected of a daf-19a mutant, e.g., dwelling/roaming defects and aldicarb resistance, but does not affect ciliogenesis or confer abnormal dauer development as expected of a daf-19c mutation. Since daf-19(tm5562) lacks exon 2 and acts as a loss-of-function allele, we suggest that amino acids encoded by this exon play an important role in DAF-19A function. Sequence alignment of available daf-19 orthologs in different Caenorhabditis species reveals a highly conserved block of 22 amino acids (Figure 1E) in exon 2. Conservation of this sequence ranges from 80 to 90%. The conserved sequence is rich in positively charged amino acids but contains no currently recognized protein motif.
DAF-19-dependent downregulation of neuronal gene expression in C. elegans is a newly described phenomenon. We identified new target genes, skr-12, gakh-1, rgs-8.1, and the recently characterized mapk-15 (Piasecki et al. 2017), whose expression was downregulated by DAF-19A in a subset of neurons. In another study, downregulation by daf-19 was reported for the gene peli-1, but only when a canonical proximal X-box promoter motif was removed, and a less conserved, more distal X-box remained (Chu et al. 2012). None of the three downregulated genes identified in our transcriptomics approach harbor an identified, canonical X-box promoter motif, while the mapk-15 gene has a putative X-box motif (Blacque et al. 2005) located further upstream than is typical (Burghoorn et al. 2012; Piasecki et al. 2017). To what extent downregulation of target genes by daf-19 depends on X-box motifs, protein-binding partners, or possible indirect effects will be the subject of future studies.
RFX TFs have been found to repress target gene expression in other organisms. Human RFX1 is capable of auto-repression regulated by DNA damage and replication blocking (Lubelsky et al. 2005). Yeast RFX, Crt1, is also a transcriptional repressor that can repress its own promoter and that of other genes (Huang et al. 1998). Similar regulatory pathways control repression by these proteins in both humans and yeast (Lubelsky et al. 2005). C. elegans, DAF-19 is most similar to human RFX1-4 (Choksi et al. 2014). The finding that DAF-19A expression downregulates or represses target genes is thus conserved, even if the biological pathways in which the respective RFX TFs act may differ. It will be of interest to know whether target gene repression is linked to neuronal identity, as described by Hobert (2010).
Activation of target gene expression was shown to require the shorter DAF-19C isoform, known to be necessary for ciliogenesis. Target gene activation is known to be mediated through a canonical cis-acting X-box promoter motif (Blacque et al. 2005; Efimenko et al. 2005; Chen et al. 2006), as well as a nearby C-box enhancer element (Burghoorn et al. 2012). However, we found that four of the eight genes that were activated by the presence of DAF-19C do not contain a canonical X-box. Though this finding is novel for target genes activated by DAF-19C, Wang et al. (2010) found that DAF-19M target genes lack a canonical X-box, and Xie et al. (2013) found that an isoform of daf-19 activates the gene tph-1 in the ADF neurons and antimicrobial genes in the intestine through an X-box-independent mechanism. These authors suggest that either DAF-19 regulates these target genes through a different DNA sequence motif, an unrecognized degenerate X-box, or by partnering through its dimerization domain with another TF. Given that daf-19 lacks an apparent TF activation domain, the latter model appears likely. The fact that DAF-19 works with ATF-7 to regulate responses to pathogenic bacteria (Xie et al. 2013) lends further support to such a model. Alternatively, regulation of target genes that lack X-box motifs could be due to indirect effects of DAF-19C regulating other genes.
daf-19 alters gene expression in dauer-inhibiting and labial neurons
Mutations in TF genes often phenocopy neuronal ablation by causing misregulation of target genes, thereby affecting specific neuron function (Hobert 2016a). daf-19 null mutants are Daf-c, thus, one might expect to see differential gene regulation in neurons that inhibit dauer entry. Bargmann and Horvitz (1991) identified the ADF, ASI, and ASG neurons as repressing dauer development in noninducing conditions. Conversely the ASJ neurons are required for initiating dauer development (Schackwitz et al. 1996). Thus, we might expect to find-regulated gene expression in ADF, ASI, and ASG, but not in ASJ. We found that ddn-1, eppl-1, and del-4 were strongly activated by the presence of DAF-19C in ASG, ddn-1 was weakly activated by DAF-19C in ASI, and skr-12 expression was downregulated in ADF (Table 4), while none of the daf-19-regulated genes studied here were differentially expressed in ASJ neurons. It will be of interest to determine whether any genes regulated by daf-19 in this set of neurons are integral to the inhibition of dauer entry.
Table 4. Neurons in which daf-19 controls target gene expression.
| Neuron | Target genes | Selected neuron functions |
|---|---|---|
| IL2s | asic-2, spg-20, ddn-1 | Dauer behavior |
| URX | ddn-1 | Suppresses innate immunity, life span regulation |
| ASK | ddn-1 | Initiates local search behavior |
| AFD | ddn-1 | Ablation causes hypo-reversal |
| ASG | ddn-1, eppl-1, del-4 | Inhibits entry into dauer |
| ASI | ddn-1 | Suppresses turning, promotes dispersal, modulates innate immune response |
| ASE | del-4, eppl-1, ddn-1 | Food leaving behavior |
| AIY | del-4 | Suppresses turns and reversals, regulates life span, starvation response |
| I5 | ddn-1 | Regulates pharyngeal muscle relaxation |
| M5 | ddn-1 | Unknown |
| PHA, PHB | mapk-15 | Chemorepulsion, mate searching |
| IL1s | skr-12, mapk-15 | Spontaneous and faster foraging |
| OLQs | skr-12 | Spontaneous and faster foraging |
| BAG | gakh-1 | Controls life span, food leaving behavior |
| AFD | skr-12 | Ablation causes hypo-reversal |
| ADF | skr-12 | Inhibits entry into dauer, modulates NMJ transmission |
| ASG | skr-12 | Inhibits entry into dauer |
| I2 | rgs-8.1 | Unknown function |
| RID | rgs-8.1 | Unknown function |
| AVA | gakh-1 | Driver for backward locomotion |
| AVB | gakh-1 | Driver for forward locomotion |
| SIAD/V | gakh-1 | Nerve ring placement |
| AIN | gakh-1 | Unknown function |
| M4 | gakh-1 | Posterior isthmus peristalsis |
Neurons in which daf-19 presence activates or represses target gene expression, and associated neuron functions [from Altun et al. (2002–2017)] relevant to daf-19 mutant phenotypes. Neurons are listed top to bottom by their position in the worm, anterior to posterior. Ciliated sensory neurons are in bold. NMJ, neuromuscular junction.
In addition, a large number of genes are regulated by daf-19 in the labial neurons (Burghoorn et al. 2012; this study), known to generate dendritic arbors in dauer larvae and to direct dauer-specific behaviors (Schroeder et al. 2013). This suggests that further work could determine whether DAF-19 is one of the terminal selectors of labial neuron identity (cf. Hobert 2016b) as continued gene regulation in these neurons into adulthood suggests.
daf-19-dependent gene regulation in neurons involved in foraging and locomotion
Mutations in either major daf-19 isoform are associated with aberrant roaming behavior, specifically overdwelling and increased turning based on observed worm tracks (Figure 4; Senti and Swoboda 2008). Thus, one might predict that daf-19 target gene regulation would occur in neurons that inhibit dwelling and turning, or that stimulate roaming. That is, neurons, which upon ablation, lead to increased dwelling and reduced exploration would be expected to express DAF-19-dependent genes. These neurons include ASI and AIY, required to suppress reversals and turns associated with the local search state and dispersal (Gray et al. 2005; Cohen et al. 2009), and the ASE, ASI, and BAG neurons that function in adaptive food leaving behavior when food is depleted (Milward et al. 2011). In contrast, one would not expect to see daf-19-dependent gene expression in HSN or NSM in which tph-1 is responsible for suppressing exploration (Flavell et al. 2013).
We found differential expression of daf-19 target genes in neurons that both induce exploration and suppress dwelling behaviors. ddn-1 and del-4 were activated in the ASI and AIY neurons, respectively. Three genes were activated in the ASE neurons, while gakh-1 was repressed by daf-19a expression in the BAG neurons in larvae (Table 4). As predicted, we did not observe target gene regulation in HSN or NSM.
daf-19 regulates genes involved in protein homeostasis
We hypothesized that misregulation of genes involved in protein homeostasis contributes to neuronal phenotypes of adult daf-19(m86) mutants, based on detectable synaptic protein loss in these animals (Senti and Swoboda 2008). Vayndorf et al. (2016) also demonstrated that neuron remodeling during aging is modified by compromised protein homeostasis. Gene ontology analysis of the differential transcriptome of adult worms (Table 1 and Table S4) yielded an overrepresentation of putative daf-19 target genes with designations of ubiquitin and proteolysis. Of the 12 genes so identified, nine encode proteases or orthologs of the ubiquitin ligase system (http://www.wormbase.org). We analyzed expression patterns of three of these nine genes and found misregulation of all three in daf-19 mutant backgrounds. skr-12 is one of five SKP-1 ubiquitin ligase component orthologs (Edwards et al. 2009) in our adult differential transcriptome and it is downregulated by DAF-19A, while spg-20 encodes an ortholog of human SPG20, a ubiquitin ligase component implicated in spastic paraplegia (Karlsson et al. 2014), which is activated by the presence of DAF-19C. We also observed partial DAF-19-dependent activation of anp-1, which encodes an aminopeptidase (Table S6). In addition, Chu et al. (2012) reported that DAF-19 regulates peli-1, which encodes a protein with similarity to an E3 ubiquitin ligase. Modulation of peli-1 expression (Huang et al. 2002) is consistent with neuronal phenotypes associated with worms lacking only DAF-19A, while spg-20 mutants display lower crawling and thrashing speeds and are more sensitive to paraquat (Truong et al. 2015). In the future, it will be of interest to determine whether mutations of these or other ubiquitin components alter synaptic protein homeostasis.
The work described here, along with that reported by Chen et al. (2006), Phirke et al. (2011), and Jensen et al. (2016), provides an exhaustive list of novel daf-19 target genes at four points during worm development: from embryogenesis to the adult stage. We have characterized orthologs of human disease genes, including spg-20 (SPARTIN), mutations of which cause a spastic paraplegia called Troyer syndrome in humans (Patel et al. 2002), and gakh-1, encoding a possible regulator of clathrin-mediated membrane trafficking. Mutations of an orthologous human cyclin G-associated kinase (GAK) gene are implicated in susceptibility to Parkinson’s disease (Pankratz et al. 2009; Hamza et al. 2010). These two examples, but also others from our gene lists, will aid in studies of neuronal function and in understanding connections to human disease. Lastly, our discovery that DAF-19, the sole C. elegans RFX TF, modulates gene expression in nonciliated neurons opens up new avenues of research into the specification and function of nonsensory neurons.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300571/-/DC1.
Acknowledgments
E.A.D.S. thanks Bart De Stasio for statistical expertise and moral support, and the laboratory of Maureen Barr for a quiet place to write and wonderful colleagues, particularly Juan Wang for assistance with RT-PCR. Some worm strains were provided by the Caenorhabditis Genetics Center, funded by the National Institutes of Health (NIH) (P40 OD-010440). We acknowledge online resources, particularly WormBase and WormAtlas. Additional undergraduate students made contributions to this work: Kristen Bischel, Elisa Carloni, Christina Schaupp, and Lu Yu. E.A.D.S. was supported in this work by the NIH (R15 AG-16192-01) and by a Fulbright Research Fellowship with Sweden. A National Science Foundation (NSF)-Major Research Implementation grant (DBI 11-26711) to E.A.D.S., B.P.P., and colleagues provided a confocal microscope. Undergraduate summer stipends were provided by the NSF-Wisconsin Alliance for Minority Participation (402549) and Lawrence University. Work in the laboratory of P.S. was supported by the Swedish Research Council and by the Marcus Borgström, Torsten Söderberg, and Åhlén Foundations. P.S. also received support from the Swedish Foundation for Strategic Research and the Karolinska Institute (KI) Strategic Neurosciences Program. D.S.-T. received support from the KI in the form of a Ph.D. student (KID) scholarship.
Footnotes
Communicating editor: C. Kaplan
Literature Cited
- Aftab S., Semenec L., Chu J. S., Chen N., 2008. Identification and characterization of novel human tissue-specific RFX transcription factors. BMC Evol. Biol. 8: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M. A., Hillier L. W., Waterston R. H., Blumenthal T., 2011. A global analysis of C. elegans trans-splicing. Genome Res. 21: 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altun Z. F., Herndon L. A., Wolkow C. A., Crocker C., Lints R., et al. , 2002-2017. Wormatlas. Available at: http://www.wormatlas.org.
- Arnaiz O., Cohen J., Tassin A. M., Koll F., 2014. Remodeling Cildb, a popular database for cilia and links for ciliopathies. Cilia 3: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann C. I., Horvitz H. R., 1991. Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science 251: 1243–1246. [DOI] [PubMed] [Google Scholar]
- Ben Arous J., Laffont S., Chatenay D., 2009. Molecular and sensory basis of a food related two-state behavior in C. elegans. PLoS One 4: e7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque O. E., Perens E. A., Boroevich K. A., Inglis P. N., Li C. M., et al. , 2005. Functional genomics of the cilium, a sensory organelle. Curr. Biol. 15: 935–941. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghoorn J., Dekkers M. P. J., Rademakers S., de Jong T., Willemsen R., et al. , 2010. Dauer pheromone and G-protein signaling modulate the coordination of intraflagellar transport kinesin motor proteins in C. elegans. J. Cell Sci. 123: 2076–2083. [DOI] [PubMed] [Google Scholar]
- Burghoorn J., Piasecki B. P., Crona F., Phirke P., Jeppsson K. E., et al. , 2012. The in vivo dissection of direct RFX-target gene promoters in C. elegans reveals a novel cis-regulatory element, the C-box. Dev. Biol. 368: 415–426. [DOI] [PubMed] [Google Scholar]
- Chen N., Mah A., Blacque O. E., Chu J., Phgora K., et al. , 2006. Identification of ciliary and ciliopathy genes in Caenorhabditis elegans through comparative genomics. Genome Biol. 7: R126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choksi S. P., Lauter G., Swoboda P., Roy S., 2014. Switching on cilia: transcriptional networks regulating ciliogenesis. Development 141: 1427–1441. [DOI] [PubMed] [Google Scholar]
- Chu J. S. C., Baillie D. L., Chen N., 2010. Convergent evolution of RFX transcription factors and ciliary genes predated the origin of metazoans. BMC Evol. Biol. 10: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J. S. C., Tarailo-Graovac M., Zhang D., Wang J., Uyar B., et al. , 2012. Fine tuning of RFX/DAF-19-regulated target gene expression through binding to multiple sites in Caenorhabditis elegans. Nucleic Acids Res. 40: 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M., Reale V., Olofsson B., Knights A., Evans P., et al. , 2009. Coordinated regulation of foraging and metabolism in C. elegans by RFamide neuropeptide signaling. Cell Metab. 9: 375–385. [DOI] [PubMed] [Google Scholar]
- Craig H. L., Wirtz J., Bamps S., Dolphin C. T., Hope I. A., 2013. The significance of alternative transcripts for Caenorhabditis elegans transcription factor genes, based on expression pattern analysis. BMC Genomics 14: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Tommaso P., Moretti S., Xenarios I., Orobitg M., Montanyola A., et al. , 2011. T-coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39: W13–W17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroquez D. B., Berciu C., Anderson J. R., Sengupta P., Nicastro D., 2014. A high-resolution morphological and ultrastructural map of anterior sensory cilia and glia in Caenorhabditis elegans. Elife 3: e01948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Barrett T., 2006. NCBI GEO standards and services for microarray data. Nat. Biotechnol. 24: 1471–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards T. L., Clowes V. E., Tsang H. T. H., Connell J. W., Sanderson C. M., et al. , 2009. Endogenous spartin (SPG20) is recruited to endosomes and lipid droplets and interacts with the ubiquitin E3 ligases AIP4 and AIP5. Biochem. J. 423: 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimenko E., Bubb K., Mak H. Y., Holzman T., Leroux M. R., et al. , 2005. Analysis of xbx genes in C. elegans. Development 132: 1923–1934. [DOI] [PubMed] [Google Scholar]
- Elkon R., Milon B., Morrison L., Shah M., Vijayakumar S., et al. , 2015. RFX transcription factors are essential for hearing in mice. Nat. Commun. 6: 8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery P., Durand B., Mach B., Reith W., 1996. RFX proteins, a novel family of DNA binding proteins conserved in the eukaryotic kingdom. Nucleic Acids Res. 24: 803–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchberger J. F., Lorch A., Sleumer M. C., Zapf R., Jones S. J., et al. , 2007. The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev. 21: 1653–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell S. W., Pokala N., Macosko E. Z., Albrecht D. R., Larsch J., et al. , 2013. Serotonin and the neuropeptide PDF initiate and extend opposing behavioral states in C. elegans. Cell 154: 1023–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajiwala K. S., Chen H., Cornille F., Roques B. P., Reith W., et al. , 2000. Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature 403: 916–921. [DOI] [PubMed] [Google Scholar]
- Garg A., Futcher B., Leatherwood J., 2015. A new transcription factor for mitosis: in Schizosaccharomyces pombe, the RFX transcription factor Sak1 works with forkhead factors to regulate mitotic expression. Nucleic Acids Res. 43: 6874–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J. M., Hill J. J., Bargmann C. I., 2005. A circuit for navigation in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 102: 3184–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza T. H., Zabetian C. P., Tenesa A., Laederach A., Montimurro J., et al. , 2010. Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson’s disease. Nat. Genet. 42: 781–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson J., Piasecki B. P., Lend K., Burglin T. R., Swoboda P., 2013. Finding ciliary genes: a computational approach. Cilia. Pt B 525: 327–350. [DOI] [PubMed] [Google Scholar]
- Hobert O., 2010. Neurogenesis in the nematode Caenorhabditis elegans (October 4, 2010), WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.12.2, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O., 2016a A map of terminal regulators of neuronal identity in Caenorhabditis elegans. Wiley Interdiscip. Rev. Dev. Biol. 5: 474–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O., 2016b Terminal selectors of neuronal identity. Essays on developmental biology. P&T 116: 455. [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A., 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Huang M., Zhou Z., Elledge S. J., 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94: 595–605. [DOI] [PubMed] [Google Scholar]
- Huang N. N., Mootz D. E., Walhout A. J. M., Vidal M., Hunter C. P., 2002. MEX-3 interacting proteins link cell polarity to asymmetric gene expression in Caenorhabditis elegans. Development 129: 747–759. [DOI] [PubMed] [Google Scholar]
- Jensen V. L., Carter S., Sanders A. A. W. M., Li C., Kennedy J., et al. , 2016. Whole-organism developmental expression profiling identifies RAB-28 as a novel ciliary GTPase associated with the BBSome and intraflagellar transport. PLoS Genet. 12: e1006469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson A. B., Washington J., Dimitrova V., Hooper C., Shekhtman A., et al. , 2014. The role of spartin and its novel ubiquitin binding region in DALIS occurrence. Mol. Biol. Cell 25: 1355–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurencon A., Dubruille R., Efimenko E., Grenier G., Bissett R., et al. , 2007. Identification of novel regulatory factor X (RFX) target genes by comparative genomics in drosophila species. Genome Biol. 8: R195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubelsky Y., Reuven N., Shaul Y., 2005. Autorepression of rfx1 gene expression: functional conservation from yeast to humans in response to DNA replication arrest. Mol. Cell. Biol. 25: 10665–10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney T. R., Luo S., Nonet M. L., 2006. Analysis of synaptic transmission in Caenorhabditis elegans using an aldicarb-sensitivity assay. Nat. Protoc. 1: 1772–1777. [DOI] [PubMed] [Google Scholar]
- Meissner T. B., Liu Y., Lee K., Li A., Biswas A., et al. , 2012. NLRC5 cooperates with the RFX transcription factor complex to induce MHC class I gene expression. J. Immunol. 188: 4951–4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milward K., Busch K. E., Murphy R. J., de Bono M., Olofsson B., 2011. Neuronal and molecular substrates for optimal foraging in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 108: 20672–20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechipurenko I. V., Olivier-Mason A., Kazatskaya A., Kennedy J., McLachlan I. G., et al. , 2016. A conserved role for girdin in basal body positioning and ciliogenesis. Dev. Cell 38: 493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelander S., Larsson E., Kristiansson E., Mansson R., Nerman O., et al. , 2005. Predictive screening for regulators of conserved functional gene modules (gene batteries) in mammals. BMC Genomics 6: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo M., Geibel M., Artal-Sanz M., Merrow M., 2015. A high-throughput method for the analysis of larval developmental phenotypes in Caenorhabditis elegans. Genetics 201: 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratz N., Wilk J. B., Latourelle J. C., DeStefano A. L., Halter C., et al. , 2009. Genome-wide association study for susceptibility genes contributing to familial Parkinson disease. Hum. Genet. 124: 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel H., Cross H., Proukakis C., Hershberger R., Bork P., et al. , 2002. SPG20 is mutated in Troyer syndrome, an hereditary spastic paraplegia. Nat. Genet. 31: 347–348. [DOI] [PubMed] [Google Scholar]
- Pereira L., Kratsios P., Serrano-Saiz E., Sheftel H., Mayo A. E., et al. , 2015. A cellular and regulatory map of the cholinergic nervous system of C. elegans. Elife 4: e12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phirke P., Efimenko E., Mohan S., Burghoorn J., Crona F., et al. , 2011. Transcriptional profiling of C. elegans DAF-19 uncovers a ciliary base-associated protein and a CDK/CCRK/LF2p-related kinase required for intraflagellar transport. Dev. Biol. 357: 235–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piasecki B. P., Burghoorn J., Swoboda P., 2010. Regulatory factor X (RFX)-mediated transcriptional rewiring of ciliary genes in animals. Proc. Natl. Acad. Sci. USA 107: 12969–12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piasecki B. P., Sasani T. A., Lessenger A. T., Huth N., Farrell S., 2017. MAPK-15 is a ciliary protein required for PKD-2 localization and male mating behavior in Caenorhabditis elegans. Cytoskeleton (Hoboken) 74: 390–402. [DOI] [PubMed] [Google Scholar]
- Pyke D. A., Thompson J. N., 1986. Statistical-analysis of survival and removal rate experiments. Ecology 67: 240–245. [Google Scholar]
- Reith W., Mach B., 2001. The bare lymphocyte syndrome and the regulation of MHC expression. Annu. Rev. Immunol. 19: 331–373. [DOI] [PubMed] [Google Scholar]
- Ruaud A., Katic I., Bessereau J., 2011. Insulin/insulin-like growth factor signaling controls non-dauer developmental speed in the nematode Caenorhabditis elegans. Genetics 187: 337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T. L., Hashimoto S., Gu S. G., Morton J. J., Stadler M., et al. , 2013. The transcription start site landscape of C. elegans. Genome Res. 23: 1348–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schackwitz W. S., Inoue T., Thomas J. H., 1996. Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron 17: 719–728. [DOI] [PubMed] [Google Scholar]
- Schafer J. C., Haycraft C. J., Thomas J. H., Yoder B. K., Swoboda P., 2003. XBX-1 encodes a dynein light intermediate chain required for retrograde intraflagellar transport and cilia assembly in Caenorhabditis elegans. Mol. Biol. Cell 14: 2057–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder N. E., Androwski R. J., Rashid A., Lee H., Lee J., et al. , 2013. Dauer-specific dendrite arborization in C. elegans is regulated by KPC-1/Furin. Curr. Biol. 23: 1527–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti G., Swoboda P., 2008. Distinct isoforms of the RFX transcription factor DAF-19 regulate ciliogenesis and maintenance of synaptic activity. Mol. Biol. Cell 19: 5517–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti G., Ezcurra M., Löbner J., Schafer W. R., Swoboda P., 2009. Worms with a single functional sensory cilium generate proper neuron-specific behavioral output. Genetics 183: 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Saiz E., Poole R. J., Felton T., Zhang F., De La Cruz E. D., et al. , 2013. Modular control of glutamatergic neuronal identity in C. elegans by distinct homeodomain proteins. Cell 155: 659–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaye D. D., Greenwald I., 2011. OrthoList: a compendium of C. elegans genes with human orthologs. PLoS One 6: e20085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth D., Ch’ng Q., Dybbs M., Tavazoie M., Kennedy S., et al. , 2005. Systematic analysis of genes required for synapse structure and function. Nature 436: 510–517. [DOI] [PubMed] [Google Scholar]
- Slos D., Sudhaus W., Stevens L., Bert W., Blaxter M., 2017. Caenorhabditis monodelphis sp. n.: defining the stem morphology and genomics of the genus Caenorhabditis. BMC Zoology 2: 4. [Google Scholar]
- Starich T. A., Herman R. K., Kari C. K., Yeh W. H., Schackwitz W. S., et al. , 1995. Mutations affecting the chemosensory neurons of Caenorhabditis elegans. Genetics 139: 171–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J., Chiang H., Tseng P., Tai W., Hsu C., et al. , 2014. RFX-1-dependent activation of SHP-1 inhibits STAT3 signaling in hepatocellular carcinoma cells. Carcinogenesis 35: 2807–2814. [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G., Thomson J. N., 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100: 64–119. [DOI] [PubMed] [Google Scholar]
- Swoboda P., Adler H. T., Thomas J. H., 2000. The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol. Cell 5: 411–421. [DOI] [PubMed] [Google Scholar]
- Tammimies K., Bieder A., Lauter G., Sugiaman-Trapman D., Torchet R., et al. , 2016. Ciliary dyslexia candidate genes DYX1C1 and DCDC2 are regulated by regulatory factor X (RFX) transcription factors through X-box promoter motifs. FASEB J. 30: 3578–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong T., Karlinski Z. A., O’Hara C., Cabe M., Kim H., et al. , 2015. Oxidative stress in Caenorhabditis elegans: protective effects of spartin. PLoS One 10: e0130455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher V. G., Tibshirani R., Chu G., 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vayndorf E. M., Scerbak C., Hunter S., Neuswanger J. R., Toth M., et al. , 2016. Morphological remodeling of C. elegans neurons during aging is modified by compromised protein homeostasis. NPJ Aging Mech. Dis. 2: 16001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Schwartz H. T., Barr M. M., 2010. Functional specialization of sensory cilia by an RFX transcription factor isoform. Genetics 186: 1295–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Zhang J. F., Xu T., Xu L., Qiao J., et al. , 2016. Identification of therapeutic targets for childhood severe asthmatics with DNA microarray. Allergol. Immunopathol. (Madr.) 44: 76–82. [DOI] [PubMed] [Google Scholar]
- Xie Y., Moussaif M., Choi S., Xu L., Sze J. Y., 2013. RFX transcription factor DAF-19 regulates 5-HT and innate immune responses to pathogenic bacteria in Caenorhabditis elegans. PLoS Genet. 9: e1003324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All microarray data sets were submitted to the Gene Expression Omnibus database (Edgar and Barrett 2006) under accession number GSE96068. C. elegans strains (Table S1) are available upon request. Gene lists generated from this study as well as from Phirke et al. (2011) are included in Table S2, Table S3, and Table S4. Table S5 lists all the primers used and Table S6 lists transgene constructs with apparent DAF-19-independent expression.