Abstract
Distal enhancers are thought to play important roles in the spatiotemporal regulation of gene expression during embryonic development, but few predicted enhancer elements have been shown to affect transcription of their endogenous genes or to alter phenotypes when disrupted. Here, we demonstrate that a 123.6-kb deletion within the mouse Slc25a13 gene is associated with reduced transcription of Dlx5, a gene located 660 kb away. Mice homozygous for the Slc25a13 deletion mutation [named hyperspin (hspn)] have malformed inner ears and are deaf with balance defects, whereas previously reported Slc25a13 knockout mice showed no phenotypic abnormalities. Inner ears of Slc25a13hspn/hspn mice have malformations similar to those of Dlx5−/− embryos, and Dlx5 expression is severely reduced in the otocyst but not the branchial arches of Slc25a13hspn/hspn embryos, indicating that the Slc25a13hspn deletion affects otic-specific enhancers of Dlx5. In addition, transheterozygous Slc25a13+/hspn Dlx5+/− mice exhibit noncomplementation with inner ear dysmorphologies similar to those of Slc25a13hspn/hspn and Dlx5−/−embryos, verifying a cis-acting effect of the Slc25a13hspn deletion on Dlx5 expression. CRISPR/Cas9-mediated deletions of putative enhancer elements located within the Slc25a13hspn deleted region failed to phenocopy the defects of Slc25a13hspn/hspn mice, suggesting the possibility of multiple enhancers with redundant functions. Our findings in mice suggest that analogous enhancer elements in the human SLC25A13 gene may regulate DLX5 expression and underlie the hearing loss that is associated with split-hand/-foot malformation 1 syndrome. Slc25a13hspn/hspn mice provide a new animal model for studying long-range enhancer effects on Dlx5 expression in the developing inner ear.
Keywords: Dlx5, Slc25a13, SHFM1, inner ear, distal enhancer
RECENT genome-wide studies of chromatin and transcription factor signatures have revealed the prevalence of cis-regulatory elements (enhancers) in the genome and their potential importance in determining spatial, temporal, and quantitative patterns of gene expression (Visel et al. 2009; Bulger and Groudine 2011; Daniel et al. 2014; Shlyueva et al. 2014; Coppola et al. 2016). Classical gene-centric studies, which rely on phenotypic consequences to identify enhancer function, have identified only a few human diseases that are caused by enhancer disruptions, and these usually involve dysregulation of key developmental control genes (Kleinjan and van Heyningen 2005). The distal-less homeobox proteins DLX5 and DLX6 are developmentally important transcription factors thought to be regulated by multiple tissue-specific enhancers (Birnbaum et al. 2012), and deletions or disruptions of these enhancers are thought to contribute to the various anomalies associated with split-hand/-foot malformation 1 (SHFM1) (Rasmussen et al. 2016), the SHFM syndrome mapped to chromosome 7q21 [Online Mendelian Inheritance in Man (OMIM) 220600].
Clinical features of the SHFM1 locus sometimes include craniofacial anomalies and hearing loss as well as the characteristic limb malformation (Ignatius et al. 1996; Haberlandt et al. 2001; Tackels-Horne et al. 2001; Wieland et al. 2004; Bernardini et al. 2008; Saitsu et al. 2009). Linkage mapping and analysis of cytogenetic abnormalities have defined the SHFM1 chromosomal region to a ∼1.5-Mb interval that includes five known genes: DYNC1I1, SLC25A13, SEM1, DLX5, and DLX6 (Scherer et al. 1994; Marinoni et al. 1995; Haberlandt et al. 2001; Tackels-Horne et al. 2001; Wieland et al. 2004; van Silfhout et al. 2009; Brown et al. 2010; Birnbaum et al. 2012). DLX5 and DLX6 are considered the primary candidate genes for SHFM1 because of their role in appendicular skeletal development (Robledo et al. 2002), and because mice with double mutations of these genes exhibit bilateral limb defects typical of SHFM1 (Merlo et al. 2002a; Robledo et al. 2002; Robledo and Lufkin 2006). DLX5 intragenic mutations have been found in a few SHFM1 patients, supporting an underlying role for this gene in the SHFM1 limb malformation (Shamseldin et al. 2012; Sowinska-Seidler et al. 2014; Wang et al. 2014) and in the hearing loss associated with SHFM1D (Shamseldin et al. 2012). Most cases of SHFM1, however, are associated with chromosomal aberrations—including deletions, inversions, and translocations—that do not directly include DLX5 or DLX6 (Birnbaum et al. 2012; Rasmussen et al. 2016). Disruptions of tissue-specific, long-range enhancers that regulate DLX5/DLX6 expression are thought to underlie most cases of SHFM1 (Birnbaum et al. 2012). Although predicted enhancer elements for SHFM1-associated phenotypes were shown to enhance transcription of reporter genes in transgenic zebrafish and mice, their effects on endogenous Dlx5 or Dlx6 expression and the phenotypic consequences of their loss have not been determined.
Though most studies have focused on DLX5 and DLX6 dysfunction or dysregulation as the primary causes for SHFM1, indirect evidence from tissue expression patterns suggests that other genes in the region may also contribute to the phenotype. For example, the solute carrier family 25 member 13 gene (SLC25A13) lies within the 1.2-Mb SHFM1 minimal critical region, and the orthologous gene in mice (Slc25a13) is expressed in limb buds, branchial arches, and otic vesicle (Sinasac et al. 1999; del Arco et al. 2002; Birnbaum et al. 2012); suggesting a possible role for this gene in development of the limb, craniofacial, and inner ear anomalies of the SHFM1 phenotype. Mice with targeted and gene trap mutations of Slc25a13 (Sinasac et al. 2004; Contreras et al. 2007), however, have normal limb morphology. Likewise, human mutations of SLC25A13, although they underlie cases of citrullinemia (CTLN2; OMIM 603471) and neonatal intrahepatic cholestasis (NICCD; OMIM 605814), do not result in SHFM1-like abnormalities or hearing loss (Saheki and Kobayashi 2002).
Here, we report on the discovery and characterization of a new, naturally occurring, recessive mouse mutation named hyperspin (hspn) and show that it is a 123.6-kb deletion within the Slc25a13 gene. Most homozygous mutant mice (Slc25a13hspn/hspn) are viable and fertile; however, in contrast to the normal phenotype of previously reported Slc25a13 mutations (Sinasac et al. 2004; Contreras et al. 2007), they exhibit rapid circling and head-shaking behaviors typical of vestibular dysfunction and they are deaf with malformed inner ears. Because of the similarities in the inner ear dysmorphologies of Slc25a13hspn/hspn and Dlx5 knockout mice (Acampora et al. 1999; Depew et al. 1999; Merlo et al. 2002b), we hypothesized and provide evidence that the loss of long-range cis-regulatory elements located within the Slc25a13hspn deletion causes a reduction in Dlx5 expression during inner ear development. The mammalian inner ear is a very complex and intricate structure that requires a highly orchestrated pattern of cell-type and stage-specific transcriptional regulation for its development. Our discovery of distal-acting, cis-regulatory elements that specifically enhance Dlx5 expression in the otocyst of mouse embryos adds to our understanding of how such precise patterning of gene expression is achieved.
Materials and Methods
Mouse husbandry
All mice in this study were obtained from research or production colonies and housed in the Research Animal Facility at The Jackson Laboratory (JAX) in Bar Harbor, ME. JAX is accredited by the American Association for the Accreditation of Laboratory Animal Care. All procedures involving the use of mice were approved by the JAX Institutional Animal Care and Use Committee and were performed in accordance with the guidelines and regulations of the US National Institutes of Health Office of Laboratory Animal Welfare and the Public Health Service Policy on the Humane Care and Use of Laboratory Animals.
Embryonic stages of mice were determined from timed matings by observing females for vaginal plugs. The day of the plug was considered embryonic day 0.5 (E0.5).
The hspn mutation of Slc25a13
The recessive hspn mutation arose spontaneously in the CXJ1/SalkJ recombinant inbred strain, and was shown to have occurred on the SJL/J-derived strand of DNA, as determined by markers that are polymorphic between BALB/c and SJL/J. Mutant mice (hspn/hspn) are identified by their hyperactive circling behavior. To alleviate the severe skin lesions that occurred in mutant mice of the CXJ1/SalkJ-hspn strain, a congenic inbred strain was developed by repeated backcrossing of mutant mice to the C57BL/6J inbred strain until achieving the equivalent of 10 backcross generations, followed by inbreeding to generate the B6.SJL(C)-Slc25a13 hspn/Kjn congenic strain which is available from JAX as stock #5679. All studies described here were done with mice of the B6.SJL(C)-Slc25a13 hspn/Kjn congenic strain.
Development of Slc25a13 knockout mice
A knockout allele of the Slc25a13 gene was produced using a combination of recombineering (Liu et al. 2003; Warming et al. 2005) and standard molecular cloning techniques. As diagramed in Supplemental Material, Figure S1, a 10,696-bp fragment of the Slc25a13 gene containing the targeted exon 5 was retrieved from a bacterial artificial chromosome and cloned into a targeting vector. Exon 5 (140 bp) was chosen for targeting because it is present in all known protein-coding transcripts of the gene and its deletion would cause an early disruption of the reading frame. A PGK-Neo selection cassette was then integrated into the targeting vector by homologous recombination. After electroporation of the targeting vector into C57BL/6N JM8A3 embryonic stem (ES) cells, ES cells with the targeted mutant allele were identified by G418 selection and confirmed by Southern blot and loss-of-allele assays. Verified ES cells were then microinjected into C57BL/6NJ host blastocysts. Progeny from host females were selected for the presence of PGK-Neo and the mutant Slc25a13 allele and used to establish a homozygous C57BL/6J-Slc25a13tm2Kjn/Kjn knockout line. The floxed PGK-Neo cassette was removed from this line by mating with mice of the B6.Cg-Tg(Sox2-cre)1Amc/J strain (stock #8454; JAX). The resulting progeny from this cross, which were heterozygous for the Slc25a13 knockout allele but lacking the PGK-Neo cassette and the Cre transgene, were intercrossed to establish the homozygous knockout line STOCK-Slc25a13tm2.1Kjn/Kjn (stock #29351; JAX), which was used for the studies described here.
Auditory-evoked brain stem response
Hearing in mice was assessed by auditory-evoked brain stem response (ABR) threshold analysis. Mice were anesthetized with an intraperitoneal injection of tribromoethanol (0.2 ml of 20 mg/ml stock per 10 g of body weight), and then placed on a 37° temperature-controlled pad in a sound-attenuating chamber. Needle electrodes were placed just under the skin, with the active electrode placed between the ears just above the vertex of the skull, the ground electrode between the eyes, and the reference electrode underneath the left ear. High-frequency transducers were placed just inside the ear canal and computer-generated sound stimuli were presented at defined intervals. Thresholds were determined for broadband clicks and 8-, 16-, and 32-kHz pure-tone stimuli by increasing the sound pressure level (SPL) in 10-dB increments followed by 5-dB increases and decreases to determine the lowest level at which a distinct ABR wave pattern could be recognized. Stimulus presentation and data acquisition were performed using the Smart EP Evoked Potential System (Intelligent Hearing Systems, Miami, FL).
Scanning electron microscopy
For scanning electron microscopy (SEM) analysis, inner ears were dissected and fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer for 4 hr at 4°, and then subjected to two 10 min washes in 0.1 M phosphate buffer. The cochlear shell and the stria vascularis were dissected away to expose the organ of Corti, and the sample was then postfixed with 1% osmium tetroxide in 0.1 M phosphate buffer, treated with thiocarbohydrazide, dehydrated, and critical-point dried (Self et al. 1998). After mounting, samples were sputter coated to produce a 15-nm gold/palladium coating and examined at 20 kV (Z = 15) on a Hitachi (Tokyo, Japan) S-3000 Scanning Electron Microscope. Images were processed and preserved using Quartz.PCI (version 5.1) software (Quartz, Vancouver, Canada).
Adult inner ear whole-mount preparations
Whole-mount preparations of adult inner ears were used to examine otoconia and gross morphology. Inner ears were dissected and flushed with neutral-buffered formalin (NBF) through a hole at the cochlear apex. Inner ears were then immersed in NBF, rinsed with distilled deionized water, and then dehydrated by 2-hr immersions in 70% ethanol, 95% ethanol, and two dehydration rounds in 100% ethanol, and then dried well. Inner ears were then cleared and stored in methyl salicylate. Inner ear whole mounts were viewed using polarized light on a Leica MZ 125 dissecting microscope, and images were captured and archived using the Leica DFC 425 camera and Leica Application Suite version 3.6.0 (Leica Microsystems, Wetzlar, Germany).
Inner ear paintfills
Paintfill analysis of embryonic inner ears was adapted and performed as previously described (Kiernan 2006). Briefly, embryonic mice (E15.5) were decapitated and the heads bisected and fixed overnight in Bodian’s fixative. Heads were then dehydrated in ethanol serial washes, and cleared overnight in methyl salicylate. Several microliters of paint suspension (1% white gloss latex paint or white-out correction fluid in methyl salicylate) were injected into the middle turn of the cochlea. The paintfilled inner ear was then dissected away from the rest of the head.
Whole-mount RNA in situ hybridization
Dlx5 expression in whole-mount embryos was examined by in situ hybridization. RNA from newborn mouse inner ears was reversely transcribed with the iScript cDNA Synthesis Kit (1708890; Bio-Rad, Hercules, CA), and a template complementary DNA for SP6 transcription was amplified using PCR primers DLX5F (5′-CAGGTGAAAATCTGGTTTCAGAAC-3′) and Sp6-DLX5R (5′-GCGATTTAGGTGACACTATAGGCACCATTGATAGTGTCCACAGTTG-3′) and used to produce a digoxigenin (DIG)-labeled RNA probe (11175025910; Roche). Embryos from timed matings were collected at E9.5 and E10.5 and genotyped by PCR of tail-tip DNA. RNA in situ hybridization was performed according to previously described methods (Piette et al. 2008). Homozygous hspn and wild-type embryos were transferred to clean vials containing 4% paraformaldehyde (PFA) in PBS, the solution was refreshed, and fixation continued at 4° overnight while gently rocking. The following day, embryos were dehydrated through a series of methanol washes and stored in 100% methanol. Embryos were rehydrated using methanol/PBS with Tween 20 (PBST) washes and incubated for 9.5 min in 10 μg/ml proteinase K, rinsed, and refixed in 4% PFA/0.2% glutaraldehyde in PBST. Embryos were washed in 50% hybridization solution/50% PBST for 3 min, in 100% hybridization solution for 3 min, and then prehybridized for 3 hr in fresh 100% hybridization solution at 65°. Denatured DIG-labeled riboprobe was then added to the embryos and hybridized overnight at 70° while rocking. The next day, the embryos were washed and incubated with 1 ml of antibody buffer for 2 hr while rocking. After the antibody buffer was removed, preblocked anti-DIG antibody was added and incubated at 4° overnight. The next day, embryos were washed and incubated in 1 ml of BM Purple (11442074001; Sigma-Aldrich, St. Louis, MO) covered in foil while rocking overnight. When sufficient staining was achieved, embryos were washed with stop solution followed by methanol dehydration. Whole embryos were transferred to a small petri dish and imaged under an inverted light microscope.
Genetic mapping and genotyping of the hspn mutation
Genomic DNA for mapping and genotyping was prepared from tail tips by the HotSHOT procedure (Truett et al. 2000). PCR reactions were performed using the 5 PRIME MasterTaq Kit (catalog number 2200210) in accordance with the manufacturer’s instructions, and run in a Bio-Rad Peltier Thermal Cycler. Amplification consisted of one cycle of denaturation at 97° for 30 sec followed by 40 cycles, each consisting of 94° for 30 sec, 58° for 30 sec, and 72° for 30 plus 1 sec per cycle. After the 40 cycles, the final product was extended for 10 min at 72°. PCR products were visualized on 3% NuSieve (Lonza Bioscience, Rockland, ME) agarose gels.
To genetically map the hspn mutation, female mice from the B6.SJL(c)-Slc25a13hspn/Kjn congenic strain were mated to CAST/EiJ males. The resulting F1 hybrid progeny (+/hspn) were intercrossed and F2 mice were analyzed for linkage analysis with genetic markers on all chromosomes. Only phenotypically mutant mice recognized by their rapid circling behavior (presumed hspn/hspn) were used for the initial hspn map location, and remaining F2 progeny with nonmutant phenotypes were subsequently used for fine mapping. Linkage associations were identified by the concordance between hspn genotypes deduced from mouse phenotypes and genotypes of microsatellite DNA markers typed by PCR amplification. Data analysis was performed using the Map Manager QTX (http://www.complextrait.org/archive/2002/html/manly.html) and Microsoft Office Excel (http://office.microsoft.com) software programs.
A three-primer PCR assay was developed to distinguish hspn and wild-type alleles. A wild-type-specific forward primer (WT-F, 5′-GGTCGTAGCTCGTGTCTTCC-3′), a hspn-specific forward primer (mut-F, 5′-CTCTCTGCAGGTTTCCCTTG-3′), and a common reverse primer (common-R, 5′-CAATGAGAACTGAGTATTAGAGGCTG-3′) were combined in a PCR reaction under conditions as described above. The PCR product size for the PCR product with the hspn deletion mutation is 205 bp, and the PCR product size for the wild-type allele is 160 bp.
Candidate gene DNA sequence analysis
PCR reactions for DNA sequencing were performed in the same manner as described above for microsatellite markers in genetic mapping. The PCR primers used to amplify specific regions of each gene evaluated were selected by flanking each evaluated exon using the Primer3 computer program (http://bioinfo.ut.ee/primer3/). PCR products were purified with the QIAquick PCR Purification Kit (QIAGEN, Valencia, CA), sequenced using the Applied Biosystems (Foster City, CA) BigDye Terminator ready reaction kit (version 3.1), and data analysis was performed using Applied Biosystems Sequencing Analysis Software (version 5.2).
Mouse strains with CRISPR/Cas9-mediated deletions
The formal names and JAX stock numbers for the inbred mouse strains with CRISPR/Cas9-generated deletions corresponding to specific regions within the Slc25a13 gene are shown in Table S1. These strains were used for in vivo tests of putative Dlx5 enhancer effects. To generate the Slc25A13 deletions, zygotes were collected from naturally mated superovulated female C57BL/6J mice and pronuclear microinjected using ∼2–5 pl of 10 mm Tris, 0.5 mM EDTA, pH 7.5 (IDT); a 0.2 unit/µl RNase inhibitor (RNAsin; Promega, Madison, WI) with Cas9 messenger RNA (mRNA) at 100 ng/µl (Trilink Biotechnologies); plus two single guide RNAs (sgRNAs) at 50 ng/µl each, flanking the targeted region, as described previously (Low et al. 2016). The 25-kb region deletion also used an oligonucleotide (200-mer, 3 ng/µl IDT) bridging the two cut sites. When possible, truncated sgRNAs were used to reduce the probability of off-target effects (Fu et al. 2014). sgRNA targeting regions were designed using the Web-based software packages ZiFiT (http://zifit.partners.org/ZiFiT/), CRISPR Design (http://crispr.mit.edu), and Breaking-Cas (http://bioinfogp.cnb.csic.es/tools/breakingcas/). Microinjected zygotes were transferred to pseudopregnant females and carried to term. At weaning age, genomic DNA from tail tip or ear punches was used to identify founders of interest by PCR amplification with primers spanning the deleted region, followed by Sanger sequencing. Putative founder mice showing deletions of the appropriate size and location were crossed to wild-type C57BL/6J mice to produce N1 generation mice. After final characterization of the transmitted alleles in N1 mice, selected individuals were intercrossed to establish homozygous lines. The individual sequences for each target and the PCR primers used to identify the alleles are shown in Table S1.
Data availability
All mouse strains examined in this study are commercially available from JAX. All data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
The inner ear phenotype of hspn mutant mice
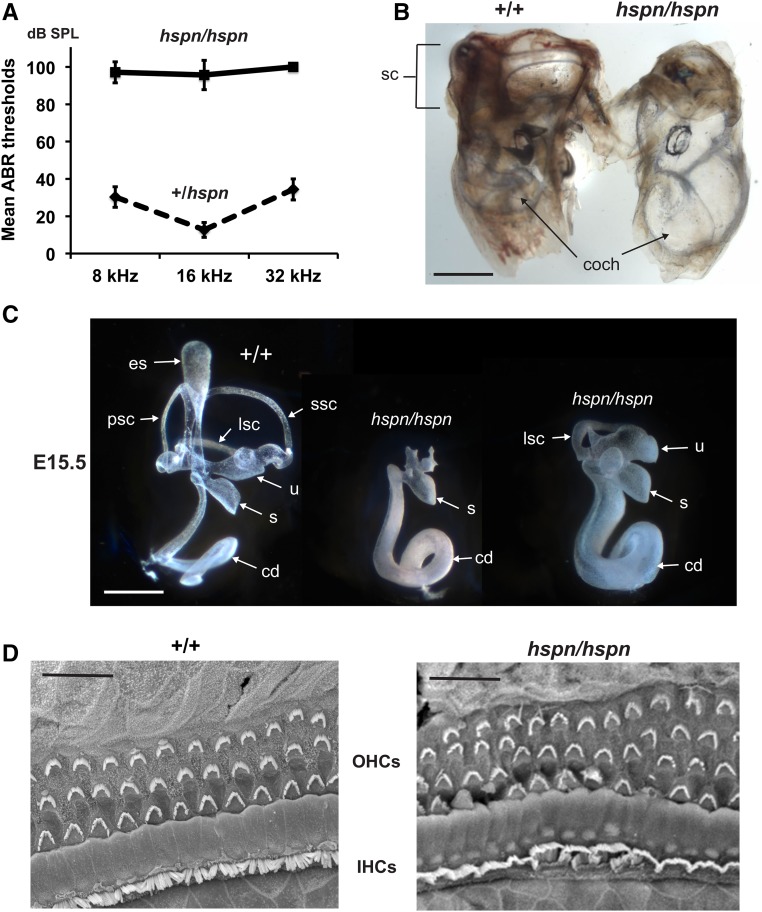
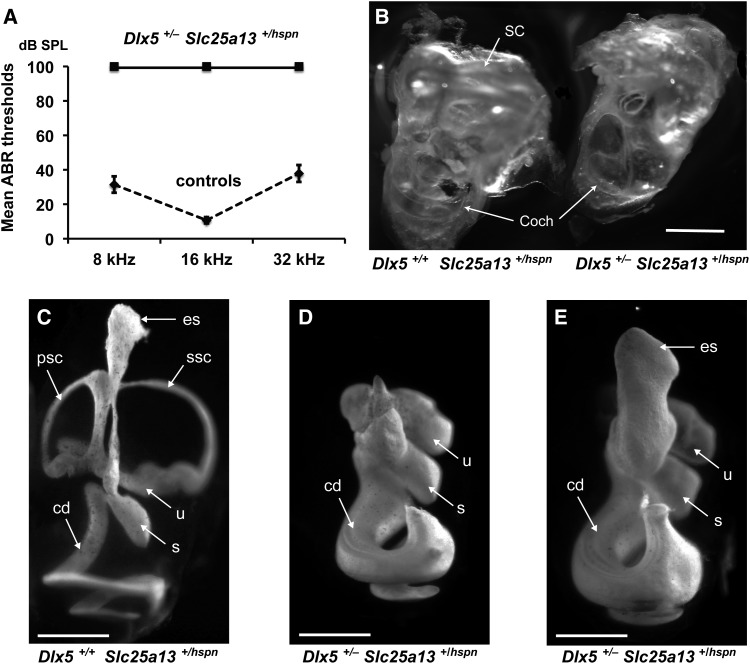
hspn mutant mice (hspn/hspn) exhibit rapid circling and head-shaking behavior and are unable to swim. These behaviors indicate inner ear dysfunction, which often includes hearing impairment. We therefore used ABR thresholds to assess the hearing sensitivity of individual hspn mutant and control mice (Figure 1A). At 4–5 weeks of age, ABR wave patterns could not be detected for pure-tone 8-, 16-, and 32-kHz stimuli in hspn/hspn mutant mice even at the highest sound intensity presented (100-dB SPL), thus demonstrating that these mice are profoundly deaf. Heterozygous mice (+/hspn) exhibited normal behavior and swimming ability, and their ABR thresholds at 5–12 weeks of age did not differ from those of littermate wild-type (+/+) mice, confirming the recessive nature of the hspn mutation.
Figure 1.
Inner ear phenotype of hspn/hspn mutant mice. (A) Hearing assessment by ABR. Average ABR thresholds of +/hspn heterozygous control mice (N = 32) tested at 5–12 weeks of age compared with those of hspn/hspn mutants (N = 7) tested at 4–5 weeks of age. The +/hspn control mice exhibited normal thresholds at all test frequencies (8, 16, and 32 kHz), whereas the highly elevated ABR thresholds of hspn/hspn mutant mice indicated profound hearing impairment. Error bars represent SD. (B) Cleared whole mounts of inner ears from 4-week-old adult mice. Inner ears of the adult hspn/hspn mutant shows reduced development of the semicircular canals (sc) and a much wider and shorter cochlea (coch) than that of the +/+ control. Bar, 1 mm. (C) Paintfills of the membranous labyrinths of inner ears from E15.5 embryos. Compared with the normal morphology of +/+ controls, inner ears of hspn/hspn mutants lack an endolymphatic sac (es) and duct, have a severe reduction in semicircular canal development, and exhibit a swollen and shortened cochlear duct (cd). The saccule (s) and cochlear duct were present in all hspn/hspn mutant inner ears examined; however, the extent of dorsal structure development varied. Posterior semicircular canals (psc) and superior semicircular canals (ssc) never developed in mutant inner ears, but there was variable development of the lateral semicircular canal (lsc) and utricle (u). Bar, 0.5 mm. (D) SEM surface images of the organ of Corti in the midapical region of the cochlea from a 4-week-old hspn/hspn mutant mouse and an age-matched +/+ control. Although a single row of inner hair cells (IHCs) and three rows of outer hair cells (OHCs) are present in both mutant and control mice, the hair cells of hspn/hspn mutant mice are disorganized, with an extra row of OHCs near the cochlear apex. Bar, 20 μm.
Examination of cleared whole-mount preparations of inner ears from adult mice (5 weeks of age) revealed that the inner ears of hspn mutant mice are small and malformed compared with those of control mice (Figure 1B). In hspn mutants, the cochlea appears abnormally swollen with fewer turns than controls, and all three semicircular canals appear absent, reduced, or malformed. To investigate the observed inner ear malformations in more detail and at an earlier time point, E15.5 embryos were collected for inner ear paintfill analysis (Kiernan 2006). Inner ears of hspn mutant embryos displayed variable dysmorphologies, with the most severe defects occurring in the dorsal, vestibular part of the inner ear (Figure 1C). In inner ears of all mutant embryos examined (N = 7), the endolymphatic duct and sac and the superior and posterior semicircular canals were missing, and—in all but one—the lateral semicircular canal and utricle were also missing. Although the cochlea is present in all mutant inner ears, the cochlear duct appears to be undercoiled and wider than that of controls.
Because the gross structure of the cochlea of hspn/hspn mutants appeared only moderately affected, we investigated potential cellular defects within the cochlear duct, which contains the sensory hair cells. We used SEM to examine organ of Corti surface preparations from six wild-type and four hspn mutant mice at 4 weeks of age (Figure 1D). Throughout the basal and middle turns of the hspn mutant cochlea, we observed the normal pattern of three rows of outer hair cells and single row of inner hair cells with properly aligned hair bundles, but with occasional missing hair bundles, indicating the onset of hair cell degeneration. However, the apical turn of the cochlea contained an additional fourth and occasional fifth row of outer hair cells and abnormal orientations of hair bundles. The increase in hair-cell rows in the apical region is likely related to the shortened cochlea, possibly due to incomplete convergent extension (Ma et al. 2000; Pauley et al. 2006; Kopecky et al. 2011). Overall, however, hair-cell formation and the general patterning of the cochlear duct do not appear significantly altered in the hspn mutants. The profound deafness of hspn mutant mice (Figure 1A) is most likely a consequence of the failure to develop a functional endolymphatic sac (Figure 1C). The endolymphatic sac regulates ion transport and fluid homeostasis in the inner ear. A failure of fluid absorption by the endolymphatic sac would cause dilatation of the membranous labyrinth, as seen in hspn mutant embryos (Figure 1C), and result in the increased cochlear diameter observed in adult mutants (Figure 1B). A similar expansion of endolymph volume and ionic imbalance during inner ear development is thought to underlie the deafness exhibited by mice with Slc26a4 (Kim and Wangemann 2010), Foxi1 (Hulander et al. 2003), Atp6v0a4 (Lorente-Canovas et al. 2013), and Atp6v1b1 (Tian et al. 2017) mutations.
Genetic mapping and molecular characterization of the hspn mutation
hspn mutant mice from the B6.SJL-Slc25a13 hspn/Kjn congenic strain were used for all phenotypic and genetic analyses. To genetically map the hspn mutation, we analyzed mutant (hspn/hspn) F2 progeny from an intercross of B6.SJL-Slc25a13 hspn/Kjn × CAST/EiJ F1 hybrids. Cosegregation analysis of 211 F2 mutant (hspn/hspn) mice with genetic markers on all chromosomes localized the hspn mutation to a 1.2-Mb region on chromosome 6 (Chr 6), between D6Mit296 (Chr 6: 5.7-Mb position; GRCm38) and D6Mit139 (Chr 6: 6.9 Mb). The Dlx5 and Dlx6 genes are located in this region and were considered good candidates for hspn because mice with targeted mutant alleles of Dlx5, though they exhibit perinatal lethality, have dysmorphic inner ears similar to what we observed for hspn/hspn mice (Depew et al. 1999). However, exon sequence analysis of Dlx5 and its neighbor, Dlx6, including splice recognition sequences, revealed no DNA abnormalities and RT-PCR of inner ear mRNA for each of these genes produced amplicons of appropriate size and sequence.
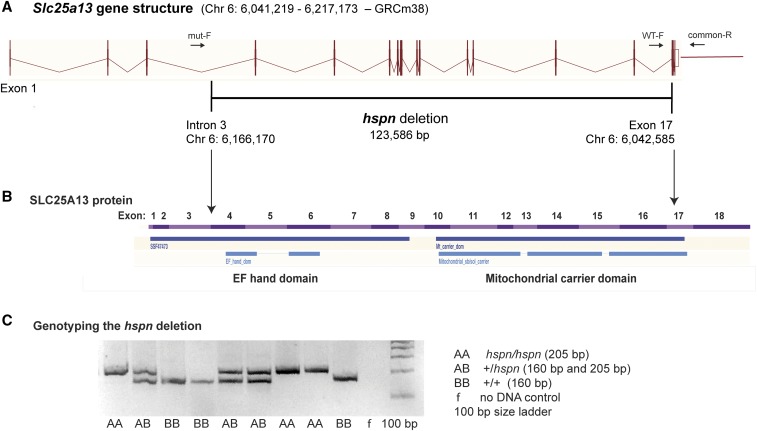
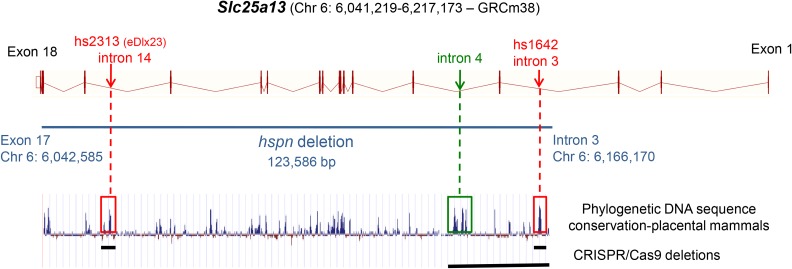
Linkage analysis of 81 additional mutant F2 mice from the same linkage cross and typing of additional polymorphic CA-repeat markers spanning the 1.2-Mb candidate interval further refined it to a 0.8-Mb region (Chr 6: 5.86–6.66 Mb; GRCm38), which definitively eliminated Dlx5 and Dlx6 (Chr 6: 6.87–6.88 Mb) as candidate genes. The refined hspn candidate region included part of Dync1i1 (Chr 6: 5.73–6.22 Mb) and all of Slc25a13 (Chr 6: 6.04–6.22 Mb) and Sem1 (Chr 6: 6.56–6.58 Mb). Upon exon sequence analysis of these genes, a large deletion was detected in the Slc25a13 gene, but no DNA alterations were found in exonic regions of Dync1i1 or Sem1. The presence or absence of small PCR products from multiple locations spanning the genomic region around Slc25a13 further refined the extent of the deletion. Comparisons of PCR-amplified DNA sequences between wild-type and hspn/hspn mice using primers corresponding to sites on either side of the deletion precisely defined the deletion as extending from within intron 3 to exon 17 of Slc25a13 (Chr 6: 6,042,586- to 6,166,170-bp position; GRCm38), deleting 123.6 kb of genomic DNA (Figure 2A). No additional chromosomal rearrangements were detected in the 0.8-Mb hspn candidate region. Most of the 123.6-kb deletion is comprised of noncoding introns but also contains exons 4–17, which encode most of the SLC25A13 protein, including the EF-hand and mitochondrial carrier domains (Figure 2B). A three-primer PCR assay was developed to distinguish hspn and wild-type alleles (Figure 2C; Materials and Methods).
Figure 2.
Molecular characterization of the Slc25a13hspn mutation. (A) Diagramatic representation of the exon–intron genomic structure of Slc25a13. The position of the 123,586-bp hspn deletion, extending from intron 3 to exon 17, is shown as a horizontal black line. Positions of PCR primers (mut-F, WT-F, and common-R) used to genotype mice for presence or absence of the hspn deletion are shown as ↔. The wild-type allele is amplified with the WT-F forward primer and the common-R reverse primer, and the hspn deletion allele is amplified with mut-F and common-R primers. (B) Structural diagram of the SLC25A13 protein. Regions of the protein encoded by the 18 exons of the Slc25a13 gene are shown as alternating pink and purple bands. Most of the protein is deleted by the hspn mutation (indicated by the area between the ↓’s), including both the EF-hand and mitochondrial domains. The structural diagrams for the Slc25a13 gene and SLC25A13 protein were derived from the Ensembl genome browser. (C) Results from the three-primer PCR assay for the presence or absence of the hspn deletion using the mut-F, WT-F, and common-R primers shown in (A). The PCR product size for the Slc25a13 allele with the hspn deletion is 205 bp, and the PCR product size for the wild-type (+) allele is 160 bp; both product sizes are detected in +/hspn heterozygotes.
Generation and phenotype of a targeted Slc25a13 knockout mutation
The protein encoded by Slc25a13 has a strongly bipartite structure (Figure 2B): the N-terminal domain includes two sets of calcium-binding EF hands, and the C-terminal domain functions as a mitochondrial membrane-bound transporter that participates in the malate aspartate shuttle which supplies aspartate from the mitochondria to the cytosol (Palmieri et al. 2001). Human SLC25A13 mutations perturb the mitochondrial transporter domain of SLC25A13, but do not alter the N-terminal EF-hand domain (Saheki and Kobayashi 2002). The previously reported mouse mutation Slc25a13tm1Lct deletes exon 10 and mimics the human mutation 851del4 (Sinasac et al. 2004). DNA sequences downstream of exon 10, which encode the C-terminal mitochondrial transporter domain, are eliminated in this mutation; but the upstream region encoding the N-terminal EF-hand domain may still be transcribed. It is possible, therefore, that the Slc25a13tm1Lct mutation produces a truncated protein isoform that retains some function during development, which may disrupt inner ear development and be at least partially responsible for the hspn mutant phenotype.
To address this possibility and to assess the hearing sensitivity of Slc25a13 knockout mice (which has not been previously reported), we generated a new knockout mutation of Slc25a13 with a deletion of exon 5 (Figure S1) which disrupts the reading frame and eliminates both domains of the protein (Figure 2). Mice homozygous for the new knockout mutation (STOCK-Slc25a13tm2.1Kjn/Kjn)—like previously reported genetically engineered Slc25a13 mutants—are viable, fertile, and do not exhibit any overt abnormalities. In contrast to Slc25a13hspn/hspn mutant mice, the Slc25a13tm2.1Kjn knockout mice (Slc25a13−/−) do not exhibit the circling or head-tossing behaviors characteristic of vestibular dysfunction and they have normal ABR thresholds (Figure S2), indicating normal inner ear function. The severe phenotype of hspn mutant mice, therefore, is not caused by loss of protein function, but must rather be due to the loss of regulatory sequences within the noncoding regions of the Slc25a13 gene.
The mouse Slc25a13hspn deletion in relation to the SHFM1 locus on human chromosome 7
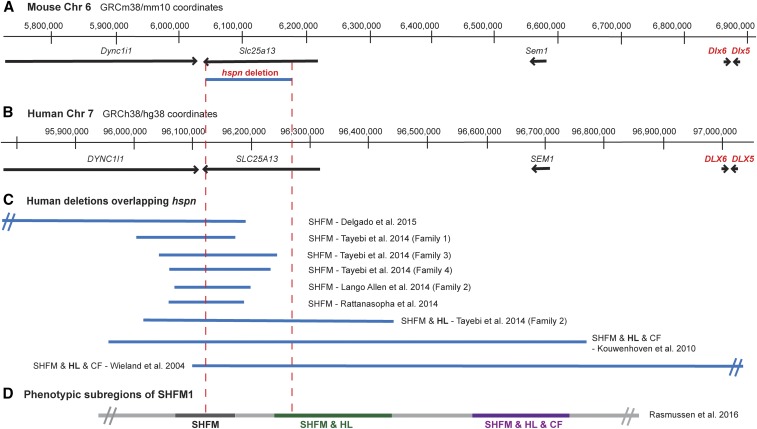
The arrangement of genes within the SHFM1 region of human chromosome 7 (DYNC1L1, SLC25A13, SEM1, DLX5, and DLX6) corresponds closely with that of the orthologous genes on proximal mouse Chr 6 (Figure 3, A and B). Chromosomal deletions and breakpoints in SHFM1 patients have been associated with isolated SHFM, SHFM with hearing loss, and SHFM with hearing loss and craniofacial abnormalities (Rasmussen et al. 2016). Three phenotypic subregions of the SHFM1 locus were identified by the correspondence of these different phenotypes with the sites and extents of the chromosomal aberrations, some of which are shown in Figure 3C. The Slc25a13hspn deletion on mouse Chr 6 overlaps with part of the SHFM with hearing loss subregion of the SHFM1 locus (Figure 3D). The different SHFM1 phenotypes resulting from the chromosomal abnormalities are thought to be the result of disruptions of different cis-acting elements that regulate tissue-specific expression of DLX5 and, to a lesser extent, DLX6.
Figure 3.
Chr 6 map position of the mouse Slc25a13hspn deletion relative to Dlx5/6 and the corresponding SHFM1 region of human chromosome 7. (A) The region of mouse Chr 6 that corresponds to the human chromosome 7q21.3 region of SHFM1. Positions of the Dync1i1, Slc25a13, Shfm1, Dlx6, and Dlx5 genes are shown as horizontal black lines with arrowheads indicating the direction of transcription. The position of the Slc25a13hspn intragenic deletion is shown as a horizontal blue line. (B) The corresponding SHFM1 locus of human chromosome 7. Positions of the DYNCLI1, SLC25A13, SEM1, DLX6, and DLX5 genes are shown as horizontal black lines with arrowheads indicating the direction of transcription. (C) Deletions in SHFM1 patients that overlap with the position of the mouse Slc25a13hspn deletion (demarcated by the area between the vertical red dotted lines). The SHFM1 deletions (Wieland et al. 2004; Kouwenhoven et al. 2010; Lango Allen et al. 2014; Rattanasopha et al. 2014; Tayebi et al. 2014; Delgado and Velinov 2015) were associated with isolated SHFM, SHFM with hearing loss (SHFM & HL), or SHFM with hearing loss and craniofacial abnormalities (SHFM1 & HL & CF). The diverse SHFM1 phenotypes are thought to be the result of disrupted long-range enhancer effects on DLX5/DLX6 expression. (D) The three phenotypic subregions of the SHFM1 locus inferred from their correlations with chromosomal deletions and inversion break points (Rasmussen et al. 2016). The Slc25a13hspn deletion overlaps with part of the SHFM & HL subregion.
Reduced Dlx5 expression in otocysts of Slc25a13hspn/hspn embryos
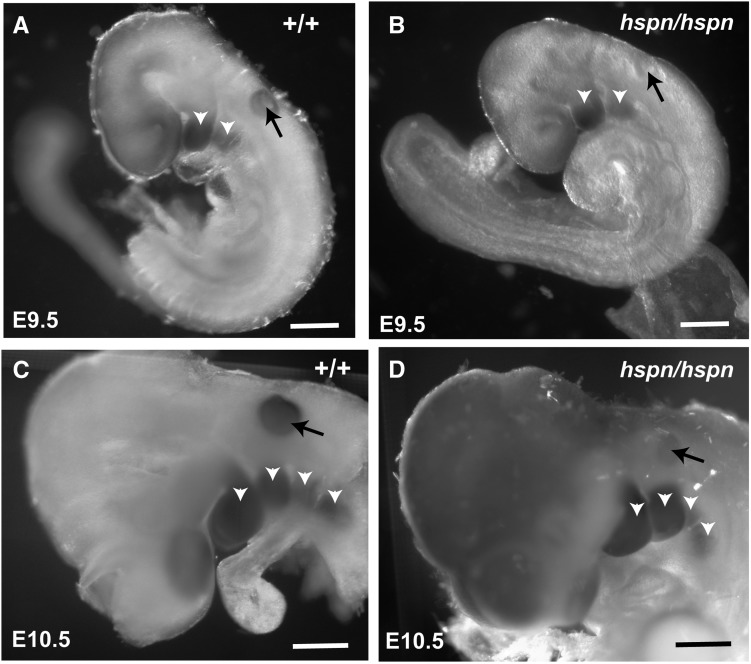
The inner ear phenotype of Slc25a13hspn/hspn mice (Figure 1) is very similar to that reported for Dlx5 knockout mice (Acampora et al. 1999; Depew et al. 1999; Merlo et al. 2002b), which show loss or underdevelopment of semicircular canals and other dorsal inner ear structures. Dlx6 is less important than Dlx5 for otic development; inner ears of Dlx6 knockout mice are like those of wild-type mice, with normal dorsally derived vestibular structures (Jeong et al. 2008). To test the hypothesis that regulatory elements located within the Slc25a13hspn deletion regulate otic expression of Dlx5, we examined Dlx5 expression in Slc25a13hspn/hspn embryos and controls by RNA in situ hybridization (Figure 4). Dlx5 was strongly expressed in the branchial arches and the otic vesicle of wild-type embryos at E9.5 (N = 5) and E10.5 (N = 2). Slc25a13hspn/hspn embryos at E9.5 (N =10) and E10.5 (N = 4) also showed strong expression of Dlx5 in the branchial arches, however, expression in the otic vesicle was much reduced or absent. This expression pattern confirms that the Slc25a13hspn deletion has an otic-specific negative effect on Dlx5 expression, which likely accounts for the similar inner ear phenotypes of Dlx5−/− and Slc25a13hspn/hspn mutant mice.
Figure 4.
Decreased Dlx5 expression in the otic vesicle of Slc25a13hspn/hspn embryos. Whole mounts of embryos were hybridized with a Dlx5 antisense RNA probe. Wild-type Slc25a13 (+/+) embryos at E9.5 (A) and E10.5 (C) showed expression of Dlx5 in the branchial arches (open arrowheads) and in the otic vesicle (solid arrows). Although Slc25a13hspn/hspn (hspn/hspn) mutant embryos (B and D) showed strong Dlx5 expression in the branchial arches, expression appeared absent in the otic vesicle at E9.5 (B) and severely reduced in the otic vesicle at E10.5 (D). Bars, 500 μm.
Partial lethality of Slc25a13hspn/hspn mutants and Dlx5+/− Slc25a13hspn/+ transheterozygotes
Dlx5−/− mice exhibit perinatal lethality. To estimate lethality of Slc25a13hspn/hspn homozygotes, we analyzed breeding records for the B6.SJL(c)-Slc25a13hspn/Kjn mouse colony. Slc25a13+/hspn × Slc25a13hspn/hspn (Table 1A) and Slc25a13+/hspn × Slc25a13+/hspn (Table 1B) matings both produced lower-than-expected numbers of Slc25a13hspn/hspn progeny, with weaning age (3–4 weeks) survival estimates of 28–38% for females and 51–55% for males (Table 1, A and B). For the combined progeny of both matings, the difference in the number of surviving female (57) and male (88) Slc25a13hspn/hspn mice was statistically significant (Chi square 6.63, 1 d.f.; P = 0.01).
Table 1. Viability estimates for Slc25a13hspn/hspn and Dlx5+/− Slc25a13+/hspn mice at weaning age.
| Observed number of progeny for each genotype | Expected genotype ratio | Chi square (probability) | Semilethal genotype | ||||
|---|---|---|---|---|---|---|---|
| Expected numbera | Estimated viabilityb (%) | ||||||
| A. Mating: Slc25a13+/hspn × Slc25a13hspn/hspn | |||||||
| Slc25a13hspn/hspn | Slc25a13+/hspn | 1:1 | Slc25a13hspn/hspn | ||||
| Female | 36 | 128 | 51.6 (P < 0.0001) | 128 | 28 | ||
| Male | 57 | 103 | 13.2 (P < 0.005) | 103 | 55 | ||
| B. Mating: Slc25a13+/hspn × Slc25a13+/hspn | |||||||
| Slc25a13hspn/hspn | Slc25a13+/+ or +/hspn | 1:3 | Slc25a13hspn/hspn | ||||
| Female | 21 | 168 | 19.4 (P < 0.0001) | 56 | 38 | ||
| Male | 31 | 182 | 12.4 (P < 0.005) | 60.7 | 51 | ||
| C. Mating: Dlx5+/− × Slc25a13hspn/hspn | |||||||
| Dlx5+/− Slc25a13+/hspn | Dlx5+/+ Slc25a13+/hspn | 1:1 | Dlx5+/KO Slc25a13+/hspn | ||||
| 5 (3 females, 2 males) | 54 (24 females, 30 males) | 40.7 (P < 0.0001) | 54 | 9 | |||
| D. Mating: Dlx5+/− × Dlx5+/+ | |||||||
| Dlx5+/− | Dlx5+/+ | 1:1 | No significant difference | ||||
| 76 | 69 | 0.34 (P > 0.5) | |||||
| E. Mating: Dlx5+/− × Slc25a13−/− | |||||||
| Dlx5+/− Slc25a13+/− | Dlx5+/+ Slc25a13+/− | 1:1 | No significant difference | ||||
| 6 | 6 | 0 (P > 0.9) | |||||
Expected number of progeny if no lethality.
Observed/expected number × 100.
To further test the hypothesis that cis-acting regulatory elements deleted by the hspn mutation of Slc25a13 reduce Dlx5 expression and decrease embryonic viability, we examined phenotypes of mice that were transheterozygous for Slc25a13hspn and Dlx5− alleles. The B6(Cg)-Dlx5tm1(cre/ERT2)Zjh/J strain (stock #29895; JAX) was used as our source for the Dlx5 knockout allele. Progeny were examined from matings of heterozygous Dlx5+/− (Slc25a13+/+) mice with homozygous Slc25a13hspn/hspn (Dlx5+/+) mutant mice (Table 1C). As expected, all 59 progeny produced from these matings were Slc25a13+/hspn; however, most (54) progeny at weaning age were Dlx5+/+ rather than Dlx5+/−, indicating a high rate of lethality for mice that had inherited the Dlx5− allele in trans configuration with the Slc25a13hspn mutation. Only 5 of the 59 progeny produced from this cross were Dlx5+/− Slc25a13+/hspn transheterozygotes, for an estimated viability at weaning age of only 9% (Table 1C). Although lethality was higher in the transheterozygotes (81%) than in Slc25a13hspn/hspn mice (45–72%), it was less severe than that exhibited by Dlx5−/− mice, which all die shortly after birth (Acampora et al. 1999; Depew et al. 1999). Dlx5+/− heterozygotes (Table 1D) and Dlx5+/− Slc25a13+/− transheterozygotes (Table 1E) showed no evidence of lethality at weaning age, demonstrating that it is noncomplementation of the Slc25a13hspn and Dlx5− mutant alleles that is responsible for Dlx5+/− Slc25a13+/hspn lethality. These results verify the cis-acting nature of the regulatory elements deleted by the Slc25a13hspn mutation.
Inner ear phenotype of Dlx5+/− Slc25a13+/hspn transheterozygotes
Only 5 out of a total of 59 progeny produced from Dlx5+/− × Slc25a13hspn/hspn matings were Dlx5+/− Slc25a13+/hspn transheterozygotes (Table 1C), and these were the only ones that exhibited a pronounced circling and head-bobbing phenotype indicating inner ear dysfunction. We tested these five mice at 5–10 weeks of age for ABR thresholds and examined the morphology of their inner ears. All of the transheterozygotes were deaf with no detectable ABR even at the maximum 100-dB SPL test stimulus (Figure 5A). Their inner ears were grossly malformed with undeveloped dorsal structures and enlarged cochlear ducts (Figure 5B), a phenotype similar to that of adult Slc25a13hsp/hspn mice (Figure 1B). In contrast, all 20 Dlx5+/− Slc25a13+/− transheterozygotes produced from Dlx5+/− × Slc25a13−/− matings had normal ABR thresholds at 5–8 weeks of age.
Figure 5.
The phenotype of Dlx5+/− Slc25a13+/hspn transheterozygotes is similar to that of Dlx5−/− knockout mice. (A) Mice between 5 and 10 weeks of age were tested for ABR thresholds at 8-, 16-, and 32-kHz frequencies. All three Dlx5+/− Slc25a13+/hspn transheterozygotes lacked ABRs even at the highest test stimulus (100-dB SPL), whereas all eight control mice (three Dlx5+/+ Slc25a13+/+, two Dlx5+/+ Slc25a13+/hspn, and three Dlx5+/− Slc25a13+/+) exhibited normal thresholds. Error bars represent SD. (B) Cleared inner ears from an adult (10 weeks old) Dlx5+/+ Slc25a13+/hspn control and a Dlx5+/− Slc25a13+/hspn transheterozygous littermate. Inner ears of Dlx5+/− Slc25a13+/hspn transheterozygotes lack fully developed semicircular canals (SC) and have a malformed cochlea (Coch) with enlarged duct. Bar, 1 mm. (C–E) Paintfills of the membranous labyrinths of inner ears from E15.5 embryos. Inner ears of Dlx5+/+ Slc25a13+/hspn controls (C) have normal morphology, whereas inner ears of Dlx5+/− Slc25a13+/hspn transheterozygotes (D and E) lack dorsal structures and have malformed and thickened cochlear ducts. An enlarged endolymphatic sac was seen in one (E) but not all (D) inner ears of transheterozygotes. cd, cochlear duct; es, endolymphatic sac; psc, posterior semicircular canal; s, saccule; ssc, superior semicircular canal; u, utricle. Bars, 0.5 mm.
Survival and inner ear morphology was also examined in E15.5 embryos from a timed mating of Dlx5+/− and Slc25a13hspn/hspn mice. Of 13 embryos produced from two litters, 7 were genotyped as Dlx5+/+ Slc25a13+/hspn and 6 as Dlx5+/− Slc25a13+/hspn; demonstrating normal survival at this age. Paintfills of four inner ears from two E15.5 Dlx5+/+ Slc25a13+/hspn controls showed normal morphology (Figure 5C), whereas four inner ears from two littermate Dlx5+/− Slc25a13+/hspn transheterozygotes lacked dorsal structures and had malformed and thickened cochlear ducts (Figure 5, D and E), similar to the inner ears of Slc25a13hsp/hspn (Figure 1A) and Dlx5−/−(Depew et al. 1999; Merlo et al. 2002b) embryos. Paintfill analysis revealed some variability in the inner ear phenotype of the E15.5 Dlx5+/− Slc25a13+/hspn transheterozygotes, with inner ears from one of the two embryos examined showing development of the endolymphatic sac and duct (Figure 5E). We conclude that the inner ear malformations of Dlx5+/− Slc25a13+/hspn transheterozygotes are caused by a severe reduction in Dlx5 expression during otic development, which results from a loss of cis-regulatory Dlx5 enhancer elements on one chromosome (the Slc25a13hspn deletion) combined with a loss of a functional Dlx5 allele on the other chromosome (the Dlx5− knockout mutation).
In vivo deletion analysis of putative Dlx5 enhancer elements
We deleted two subregions of the hspn deletion that contain putative enhancer elements and tested the in vivo effects of these subdeletions on inner ear function. We used CRISPR/Cas9-mediated genome editing in C57BL/6J zygotes to produce separate deletions corresponding to two putative Dlx5 enhancer elements (hs1642 and hs2313) located within the genomic region deleted by the Slc25a13hspn mutation (Figure 6). The hs1642 and hs2313 elements are included in the VISTA Enhancer Browser (Visel et al. 2007), a resource for experimentally validated human and mouse noncoding fragments with gene enhancer activity assessed by reporter gene expression in transgenic mice. In assays of E11.5 mouse embryos, the hs1642 element showed enhancer activity in forebrain, hindbrain, and neural tube; and the hs2313 element, equivalent to the eDlx#23 enhancer (Birnbaum et al. 2012), showed enhancer activity in the otic vesicle, forebrain, branchial arch, and limb bud. Deletions of hs1642 (Δhs1642) and hs2313 (Δhs2313) were verified by DNA sequence analysis, and separate homozygous mouse strains were developed for each deletion. Details of the deletions and mouse strains are given in Table S1. All mice examined from the hs1642 and hs2313 deletion strains showed normal morphology and behavior, with no indications of vestibular dysfunction. Three mice from the hs1642 deletion strain and four from the hs2313 deletion strain were tested for ABR thresholds at 2–3 months of age, and all exhibited normal thresholds similar to those of age-matched controls. To test for cis-regulatory effects of the putative enhancer deletions, Slc25a13 Δhs1642/Δhs1642 and Slc25a13 Δhs2313/Δhs2313 mice were crossed with Dlx5+/− mice. All transheterozygous mice produced from these crosses (18 Slc25a13+/Δhs1642 Dlx5+/− and 11 Slc25a13+/Δhs3213 Dlx5+/−) showed normal behavior and normal ABR thresholds when tested at 4–13 weeks of age.
Figure 6.
Putative enhancer elements within the Slc25a13hspn deletion. Locations of two putative enhancer elements, hs2313 (Chr6: 6,058,554–6,059,028; GRCm38) and hs1642 (Chr 6: 6,163,229–6,163,674), that lie within the hspn-deleted region (shown as horizontal blue line) of Slc25a13 were obtained from the VISTA Enhancer Browser (http://enhancer-test.lbl.gov/frnt_page.shtml). The hs2313 element is equivalent to the eDlx#23 enhancer (Birnbaum et al. 2012). DNA sequences of both elements show a high degree of phylogenic conservation for noncoding DNA (peaks enclosed by red boxes). An additional noncoding region of high sequence conservation occurs in intron 4 (peak enclosed by green box). Specific deletions corresponding to these conserved regions were created using CRISPR/Cas9 genome editing, and the extent of these deletions are shown by horizontal black lines at the bottom of the figure. The gene structure diagram for Slc25a13 is derived from the Ensembl genome browser, and the phylogenetic conservation diagram is from the University of California, Santa Cruz Genome Browser comparative genomics track.
Because homozygous deletions of the hs1642 and hs2313 elements had no detectable negative phenotypic effects, we used CRISPR/Cas9 genome editing to produce larger (25 kb) deletions within the hspn-deleted region of Slc25a13. The deletions targeted the 6,142,000- 6,166,000-bp segment of mouse Chr 6 (GRCm38), chosen because it contains a phylogenetically conserved region in intron 4 of the Slc25a13 gene as well as the putative hs1642 enhancer element (Figure 6), and because the deletion covers the only region of the hspn deletion that overlaps with the corresponding region of the human SHFM1 locus predicted to associate with hearing loss (Figure 3). Three independent inbred strains of mice (line 6, line 12, and line 54) were produced that span the 25-kb targeted region of mouse Chr 6, and details of these strains and their deletions are given in Table S1. None of the mice from these homozygous deletion strains exhibited behaviors suggestive of vestibular dysfunction. A total of 10 mice from line 6, 3 from line 12, and 6 from line 54 were evaluated for hearing at 1–4 months of age, and all exhibited normal ABR thresholds.
The lack of any discernable vestibular or auditory dysfunction in mice from the strains listed in Table S1 indicates that the individual deletions of the putative enhancer regions are not by themselves responsible for the inner ear dysfunction of Slc25a13hspn/hspn mice.
Discussion
We present several lines of evidence demonstrating that the mutation underlying the congenital hearing loss, vestibular dysfunction, and dysmorphic inner ears of hspn mutant mice is a large intragenic deletion extending from within intron 3–4 to exon 17 of Slc25a13. First, the hspn mutation was mapped to a genomic interval that contains the Slc25a13 gene, and upon DNA sequencing of all exons in the genetically determined candidate region, only the Slc25a13 gene was mutated. Second, the chromosomal deletion identified in hspn genomic DNA is confined to a region within the Slc25a13 gene. Third, there is complete concordance between the mutant mouse phenotype and homozygosity for the Slc25a13 intragenic deletion in all mice examined. Nonetheless, there is a possibility that extraexonic sequence alterations outside of the Slc25a13 deletion, but still within the genetically defined critical region of the hspn mutation, may actually be responsible for the dysregulation of the downstream Dlx5 gene. However, because the spontaneous hspn mutation occurred recently in an inbred strain, only a single DNA sequence difference is expected between the coisogenic mutant strain and the control strain.
The inner ear abnormalities of Slc25a13hspn/hspn mutant mice (Figure 1) are similar to those reported for SHFM1 patients with hearing loss (Haberlandt et al. 2001; Wieland et al. 2004), with variable loss of vestibular structures and a thick, but shortened, cochlear duct. They are likewise similar to the inner ear defects of Dlx5 knockout mice (Acampora et al. 1999; Depew et al. 1999; Merlo et al. 2002b). Although Dlx5 and Dlx6 appear to be functionally redundant for jaw development, Dlx6 is less important than Dlx5 for otic development, and inner ears of Dlx6 knockout mice appear similar to those of wild-type mice, with normal vestibular structures (Jeong et al. 2008). The dorsal otocyst, where Dlx5 is expressed, gives rise to the anterior and posterior semicircular canals and the endolymphatic duct. Consistent with this developmental pattern, the saccule and cochlea of Dlx5 knockout and Slc25a13hspn/hspn mice appear less affected than the vestibular apparatus, which is very reduced in size with loss of or incorrectly formed semicircular canals and endolymphatic duct. In Dlx5 knockout mice (Acampora et al. 1999; Depew et al. 1999; Merlo et al. 2002b), as in Slc25a13hspn/hspn mutant mice (Figure 1), the severity of dysmorphology of the inner ears varies among individual mutants, even on inbred strain backgrounds.
Here we show by RNA in situ hybridization analysis that Dlx5 expression is greatly reduced or absent in the otic vesicle of Slc25a13hspn/hspn embryos (Figure 4), and that Dlx5+/− Slc2a13+/hspn transheterozygotes are deaf with malformed inner ears (Figure 5). Together, these results provide strong evidence that the loss of cis-acting regulatory elements located in the Slc25a13hspn gene reduce Dlx5 expression and are responsible for the dysmorphic inner ears and deafness of Slc25a13hspn/hspn mutant mice. Non-otic abnormalities that are commonly observed in mice homozygous for targeted Dlx5 knockout mutations, such as multiple defects of craniofacial structures (Acampora et al. 1999; Depew et al. 1999; Merlo et al. 2002b), are not observed in Slc25a13hspn/hspn mice. This is likely due to the fact that the regulatory elements deleted by the hspn mutation are specific to Dlx5 expression in the developing inner ear, as evidenced by loss of Dlx5 expression in the otocyst but not the branchial arches of Slc25a13hspn/hspn embryos (Figure 4). Although the Slc25a13hspn deletion is located ∼700 kb away from Dlx5, there is precedence for such long-range noncoding elements having regulatory effects on gene expression (Kleinjan and van Heyningen 2005; Noonan and McCallion 2010). For example, another developmentally important gene, Shh, has a long-range enhancer located 1 Mb away—in the intron of another gene—that regulates expression in the developing limb (Lettice et al. 2003).
Only ∼30% of female and 50% of male Slc25a13hspn/hspn mice survive to weaning age (Table 1). To account for this 50–70% lethality, Dlx5 expression in Slc25a13hspn/hspn mice must be detrimentally reduced in another organ during development that is more critical for viability than the inner ear. Dlx5 is expressed in many organ systems at various stages of embryonic development, but a definitive cause for neonatal death in Dlx5 knockout mice has not been determined (Depew et al. 1999). The Dlx5 expression levels related to lethality appear to be dosage sensitive because lethality at weaning age is 50–70% in Slc25a13hspn/hspn mice, 90% in Dlx5+/− Slc2a13+/hspn transheterozygotes (Table 1), and 100% in Dlx5−/− knockout mice shortly after birth (Acampora et al. 1999; Depew et al. 1999; Merlo et al. 2002b).
Slc25a13 (Sinasac et al. 1999; del Arco et al. 2002; Birnbaum et al. 2012), Sem1 (Birnbaum et al. 2012), and Dlx5 (Acampora et al. 1999; Depew et al. 1999) are similarly expressed in branchial arches, limb bud, and otic vesicle of mouse embryos, yet expression in these tissues appears functionally important only for Dlx5. Expression of Slc25a13 in these tissues seems to be dispensable and may be an inadvertent consequence of its genomic location. The similar expression patterns of Slc25a13 and Dlx5, which have nothing in common except for their genomic location, suggests that they may be located in a regulatory landscape wherein tissue-specific coexpression of genes (which may or may not be functionally related) is regulated by a common control region or group of long-range enhancers, as has been described for the chromosomal regions around the mouse HoxD gene cluster (Spitz et al. 2003) and the Grem1 gene (Zuniga et al. 2004). Genomic clustering of coexpressed genes may be widespread in genomes, particularly in regions around developmental control genes (Spitz and Duboule 2008), and may correspond with topologically associating domains (Dekker and Heard 2015).
The SLC25A13 region homologous to the Slc25a13hspn deletion is included in the SHFM1 minimal region and overlaps with the subregion that is associated with hearing loss (Figure 3). Multiple long-range, tissue-specific enhancer elements that presumably regulate DLX5/6 expression have been identified at various sites in the SHFM1 locus that correspond with the limb, inner ear, and craniofacial abnormalities associated with this syndrome (Birnbaum et al. 2012). These putative enhancer elements were identified by their high degree of DNA sequence conservation across species (at least 77% identity for ≥100 bp between human and frog) followed by transgenic expression assays in zebrafish and mouse. The Slc25a13hspn deletion contains two of these putative enhancer elements, eDlx#23 (hs2313) and hs1642 (Figure 6). eDlx#23 enhanced reporter gene activity in the otic vesicle, forebrain, branchial arch, and limb bud, and was suggested to regulate DLX5 expression and underlie the hearing loss associated with some SHFM1 families (Birnbaum et al. 2012). Later studies, however, have described multiple SHFM1 families with deletions that include the eDlx#23 enhancer element but that do not exhibit hearing loss, indicating that deletion of eDlx#23 is not by itself responsible for SHFM1-associated hearing loss (Lango Allen et al. 2014; Rattanasopha et al. 2014; Tayebi et al. 2014; Delgado and Velinov 2015). Although the hs1642 enhancer element is present in all SHFM1 deletions associated with hearing loss, it does not show enhanced reporter gene activity in the otic vesicle of mouse embryos.
To directly test the functional importance of specific candidate enhancers in vivo, we used CRISPR/Cas9 genome editing to make deletions of the phylogenetically conserved eDlx#23 and hs1642 elements and also a deletion of a larger 25-kb DNA region containing additional conserved sequences (Figure 6 and Table S1). Mice homozygous for each of the putative enhancer deletions had no obvious phenotypic abnormalities and exhibited normal hearing. Deletions of other highly conserved elements, which also show in vivo enhancer activity in mouse transgenic assays, have likewise yielded mice without any noticeable abnormalities (Ahituv et al. 2007). The negative results from these studies demonstrate that transgenic enhancer assays do not reliably predict phenotypic relevance and may indicate that the conserved elements have a redundant regulatory function. Multiple and possibly redundant cis-enhancer elements appear to be pervasive in regulating developmental pathways (Cannavo et al. 2016). For example, multiple cis-regulatory elements, some of them partially redundant, have been located in a 1-Mb region upstream of POU3F4 and promote its expression during inner ear development (Naranjo et al. 2010). Additional CRISPR/Cas9-mediated subdeletions of the Slc25a13hspn deletion or multiple combinations of subdeletions may be needed to reduce Dlx5 expression enough to affect inner ear development. Across-species sequence conservation is not always a consistent feature of enhancers (Noonan and McCallion 2010), and less conserved sequences within the Slc25a13hspn deletion may also contribute to enhancer function. In addition to DNA sequence conservation, there are other means that could be employed to identify potential enhancer elements, including transcription factor binding sites, DNase hypersensitive sites, and chromatin immunoprecipitation sequencing for histone modifications and p300-bound regions (Shlyueva et al. 2014). The deletion strains listed in Table S1 provide valuable starting materials for future attempts to identify such enhancer elements.
In summary, we show here that loss of cis-regulatory elements within the Slc25a13hspn deletion decreases Dlx5 expression in the otic vesicle and is responsible for the inner ear dysmorphology and hearing loss of Slc25a13hspn/hspn mutant mice. Slc25a13hspn/hspn mice are viable as adults, in contrast to the perinatal lethality of Dlx5 knockout mice, and therefore can be used to study later-occurring effects of reduced Dlx5 expression in the inner ear. Our results in mice suggest that long-range DLX5 enhancer elements located in the human SLC25A13 gene may underlie the sensorineural hearing loss that is sometimes associated with SHFM1. The lack of limb or craniofacial abnormalities in Slc25a13hspn/hspn mice also suggests that disruptions of otic-specific DLX5 enhancers may underlie some cases of human nonsyndromic hearing impairment. The Slc25a13hspn mice provide a new model system for studying distal-acting enhancer elements that regulate Dlx5 expression during inner ear development.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300447/-/DC1.
Acknowledgments
We thank Sandra Gray for general mouse colony management and for the generation of linkage-cross mice and Cindy Avery of the Technology Evaluation and Development group for husbandry assistance with the mouse CRISPR deletion strains. We thank personnel of The Jackson Laboratory’s Genetic Engineering Technologies service for generation of the Slc25a13 knockout mouse and Pete Finger of the Electron Microscopy Service for tissue preparation and processing for scanning electron microscopy. This study was supported by US National Institutes of Health grant R01 DC-004301 (to K.R.J.). The Jackson Laboratory shared services are supported by National Institutes of Health grant P30 CA-034196.
Footnotes
Communicating editor: T. Magnuson
Literature Cited
- Acampora D., Merlo G. R., Paleari L., Zerega B., Postiglione M. P., et al. , 1999. Craniofacial, vestibular and bone defects in mice lacking the distal-less-related gene Dlx5. Development 126: 3795–3809. [DOI] [PubMed] [Google Scholar]
- Ahituv N., Zhu Y., Visel A., Holt A., Afzal V., et al. , 2007. Deletion of ultraconserved elements yields viable mice. PLoS Biol. 5: e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardini L., Palka C., Ceccarini C., Capalbo A., Bottillo I., et al. , 2008. Complex rearrangement of chromosomes 7q21.13-q22.1 confirms the ectrodactyly-deafness locus and suggests new candidate genes. Am. J. Med. Genet. A. 146A: 238–244. [DOI] [PubMed] [Google Scholar]
- Birnbaum R. Y., Everman D. B., Murphy K. K., Gurrieri F., Schwartz C. E., et al. , 2012. Functional characterization of tissue-specific enhancers in the DLX5/6 locus. Hum. Mol. Genet. 21: 4930–4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K. K., Reiss J. A., Crow K., Ferguson H. L., Kelly C., et al. , 2010. Deletion of an enhancer near DLX5 and DLX6 in a family with hearing loss, craniofacial defects, and an inv(7)(q21.3q35). Hum. Genet. 127: 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M., Groudine M., 2011. Functional and mechanistic diversity of distal transcription enhancers. Cell 144: 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E., Khoueiry P., Garfield D. A., Geeleher P., Zichner T., et al. , 2016. Shadow enhancers are pervasive features of developmental regulatory networks. Curr. Biol. 26: 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras L., Gomez-Puertas P., Iijima M., Kobayashi K., Saheki T., et al. , 2007. Ca2+ Activation kinetics of the two aspartate-glutamate mitochondrial carriers, aralar and citrin: role in the heart malate-aspartate NADH shuttle. J. Biol. Chem. 282: 7098–7106. [DOI] [PubMed] [Google Scholar]
- Coppola C. J., Ramaker R. C., Mendenhall E. M., 2016. Identification and function of enhancers in the human genome. Hum. Mol. Genet. 25: R190–R197. [DOI] [PubMed] [Google Scholar]
- Daniel B., Nagy G., Nagy L., 2014. The intriguing complexities of mammalian gene regulation: how to link enhancers to regulated genes. Are we there yet? FEBS Lett. 588: 2379–2391. [DOI] [PubMed] [Google Scholar]
- Dekker J., Heard E., 2015. Structural and functional diversity of topologically associating domains. FEBS Lett. 589: 2877–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Arco A., Morcillo J., Martinez-Morales J. R., Galian C., Martos V., et al. , 2002. Expression of the aspartate/glutamate mitochondrial carriers aralar1 and citrin during development and in adult rat tissues. Eur. J. Biochem. 269: 3313–3320. [DOI] [PubMed] [Google Scholar]
- Delgado S., Velinov M., 2015. 7q21.3 Deletion involving enhancer sequences within the gene DYNC1I1 presents with intellectual disability and split hand-split foot malformation with decreased penetrance. Mol. Cytogenet. 8: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew M. J., Liu J. K., Long J. E., Presley R., Meneses J. J., et al. , 1999. Dlx5 regulates regional development of the branchial arches and sensory capsules. Development 126: 3831–3846. [DOI] [PubMed] [Google Scholar]
- Fu Y., Sander J. D., Reyon D., Cascio V. M., Joung J. K., 2014. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32: 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberlandt E., Loffler J., Hirst-Stadlmann A., Stockl B., Judmaier W., et al. , 2001. Split hand/split foot malformation associated with sensorineural deafness, inner and middle ear malformation, hypodontia, congenital vertical talus, and deletion of eight microsatellite markers in 7q21.1-q21.3. J. Med. Genet. 38: 405–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulander M., Kiernan A. E., Blomqvist S. R., Carlsson P., Samuelsson E. J., et al. , 2003. Lack of pendrin expression leads to deafness and expansion of the endolymphatic compartment in inner ears of Foxi1 null mutant mice. Development 130: 2013–2025. [DOI] [PubMed] [Google Scholar]
- Ignatius J., Knuutila S., Scherer S. W., Trask B., Kere J., 1996. Split hand/split foot malformation, deafness, and mental retardation with a complex cytogenetic rearrangement involving 7q21.3. J. Med. Genet. 33: 507–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J., Li X., McEvilly R. J., Rosenfeld M. G., Lufkin T., et al. , 2008. Dlx genes pattern mammalian jaw primordium by regulating both lower jaw-specific and upper jaw-specific genetic programs. Development 135: 2905–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan A. E., 2006. The paintfill method as a tool for analyzing the three-dimensional structure of the inner ear. Brain Res. 1091: 270–276. [DOI] [PubMed] [Google Scholar]
- Kim H. M., Wangemann P., 2010. Failure of fluid absorption in the endolymphatic sac initiates cochlear enlargement that leads to deafness in mice lacking pendrin expression. PLoS One 5: e14041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinjan D. A., van Heyningen V., 2005. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 76: 8–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky B., Santi P., Johnson S., Schmitz H., Fritzsch B., 2011. Conditional deletion of N-Myc disrupts neurosensory and non-sensory development of the ear. Dev. Dyn. 240: 1373–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouwenhoven E. N., van Heeringen S. J., Tena J. J., Oti M., Dutilh B. E., et al. , 2010. Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet. 6: e1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lango Allen H., Caswell R., Xie W., Xu X., Wragg C., et al. , 2014. Next generation sequencing of chromosomal rearrangements in patients with split-hand/split-foot malformation provides evidence for DYNC1I1 exonic enhancers of DLX5/6 expression in humans. J. Med. Genet. 51: 264–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice L. A., Heaney S. J., Purdie L. A., Li L., de Beer P., et al. , 2003. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 12: 1725–1735. [DOI] [PubMed] [Google Scholar]
- Liu P., Jenkins N. A., Copeland N. G., 2003. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 13: 476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorente-Canovas B., Ingham N., Norgett E. E., Golder Z. J., Karet Frankl F. E., et al. , 2013. Mice deficient in H+-ATPase a4 subunit have severe hearing impairment associated with enlarged endolymphatic compartments within the inner ear. Dis. Model. Mech. 6: 434–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low B. E., Kutny P. M., Wiles M. V., 2016. Simple, efficient CRISPR-Cas9-mediated gene editing in mice: strategies and methods. Methods Mol. Biol. 1438: 19–53. [DOI] [PubMed] [Google Scholar]
- Ma Q., Anderson D. J., Fritzsch B., 2000. Neurogenin 1 null mutant ears develop fewer, morphologically normal hair cells in smaller sensory epithelia devoid of innervation. J. Assoc. Res. Otolaryngol. 1: 129–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinoni J. C., Stevenson R. E., Evans J. P., Geshuri D., Phelan M. C., et al. , 1995. Split foot and developmental retardation associated with a deletion of three microsatellite markers in 7q21.2-q22.1. Clin. Genet. 47: 90–95. [DOI] [PubMed] [Google Scholar]
- Merlo G. R., Paleari L., Mantero S., Genova F., Beverdam A., et al. , 2002a Mouse model of split hand/foot malformation type I. Genesis 33: 97–101. [DOI] [PubMed] [Google Scholar]
- Merlo G. R., Paleari L., Mantero S., Zerega B., Adamska M., et al. , 2002b The Dlx5 homeobox gene is essential for vestibular morphogenesis in the mouse embryo through a BMP4-mediated pathway. Dev. Biol. 248: 157–169. [DOI] [PubMed] [Google Scholar]
- Naranjo S., Voesenek K., de la Calle-Mustienes E., Robert-Moreno A., Kokotas H., et al. , 2010. Multiple enhancers located in a 1-Mb region upstream of POU3F4 promote expression during inner ear development and may be required for hearing. Hum. Genet. 128: 411–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan J. P., McCallion A. S., 2010. Genomics of long-range regulatory elements. Annu. Rev. Genomics Hum. Genet. 11: 1–23. [DOI] [PubMed] [Google Scholar]
- Palmieri L., Pardo B., Lasorsa F. M., del Arco A., Kobayashi K., et al. , 2001. Citrin and aralar1 are Ca(2+)-stimulated aspartate/glutamate transporters in mitochondria. EMBO J. 20: 5060–5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauley S., Lai E., Fritzsch B., 2006. Foxg1 is required for morphogenesis and histogenesis of the mammalian inner ear. Dev. Dyn. 235: 2470–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piette D., Hendrickx M., Willems E., Kemp C. R., Leyns L., 2008. An optimized procedure for whole-mount in situ hybridization on mouse embryos and embryoid bodies. Nat. Protoc. 3: 1194–1201. [DOI] [PubMed] [Google Scholar]
- Rasmussen M. B., Kreiborg S., Jensen P., Bak M., Mang Y., et al. , 2016. Phenotypic subregions within the split-hand/foot malformation 1 locus. Hum. Genet. 135: 345–357. [DOI] [PubMed] [Google Scholar]
- Rattanasopha S., Tongkobpetch S., Srichomthong C., Kitidumrongsook P., Suphapeetiporn K., et al. , 2014. Absent expression of the osteoblast-specific maternally imprinted genes, DLX5 and DLX6, causes split hand/split foot malformation type I. J. Med. Genet. 51: 817–823. [DOI] [PubMed] [Google Scholar]
- Robledo R. F., Lufkin T., 2006. Dlx5 and Dlx6 homeobox genes are required for specification of the mammalian vestibular apparatus. Genesis 44: 425–437. [DOI] [PubMed] [Google Scholar]
- Robledo R. F., Rajan L., Li X., Lufkin T., 2002. The Dlx5 and Dlx6 homeobox genes are essential for craniofacial, axial, and appendicular skeletal development. Genes Dev. 16: 1089–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saheki T., Kobayashi K., 2002. Mitochondrial aspartate glutamate carrier (citrin) deficiency as the cause of adult-onset type II citrullinemia (CTLN2) and idiopathic neonatal hepatitis (NICCD). J. Hum. Genet. 47: 333–341. [DOI] [PubMed] [Google Scholar]
- Saitsu H., Kurosawa K., Kawara H., Eguchi M., Mizuguchi T., et al. , 2009. Characterization of the complex 7q21.3 rearrangement in a patient with bilateral split-foot malformation and hearing loss. Am. J. Med. Genet. A. 149A: 1224–1230. [DOI] [PubMed] [Google Scholar]
- Scherer S. W., Poorkaj P., Allen T., Kim J., Geshuri D., et al. , 1994. Fine mapping of the autosomal dominant split hand/split foot locus on chromosome 7, band q21.3-q22.1. Am. J. Hum. Genet. 55: 12–20. [PMC free article] [PubMed] [Google Scholar]
- Self T., Mahony M., Fleming J., Walsh J., Brown S. D., et al. , 1998. Shaker-1 mutations reveal roles for myosin VIIA in both development and function of cochlear hair cells. Development 125: 557–566. [DOI] [PubMed] [Google Scholar]
- Shamseldin H. E., Faden M. A., Alashram W., Alkuraya F. S., 2012. Identification of a novel DLX5 mutation in a family with autosomal recessive split hand and foot malformation. J. Med. Genet. 49: 16–20. [DOI] [PubMed] [Google Scholar]
- Shlyueva D., Stampfel G., Stark A., 2014. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 15: 272–286. [DOI] [PubMed] [Google Scholar]
- Sinasac D. S., Crackower M. A., Lee J. R., Kobayashi K., Saheki T., et al. , 1999. Genomic structure of the adult-onset type II citrullinemia gene, SLC25A13, and cloning and expression of its mouse homologue. Genomics 62: 289–292. [DOI] [PubMed] [Google Scholar]
- Sinasac D. S., Moriyama M., Jalil M. A., Begum L., Li M. X., et al. , 2004. Slc25a13-knockout mice harbor metabolic deficits but fail to display hallmarks of adult-onset type II citrullinemia. Mol. Cell. Biol. 24: 527–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowinska-Seidler A., Badura-Stronka M., Latos-Bielenska A., Stronka M., Jamsheer A., 2014. Heterozygous DLX5 nonsense mutation associated with isolated split-hand/foot malformation with reduced penetrance and variable expressivity in two unrelated families. Birth Defects Res. A Clin. Mol. Teratol. 100: 764–771. [DOI] [PubMed] [Google Scholar]
- Spitz F., Duboule D., 2008. Global control regions and regulatory landscapes in vertebrate development and evolution. Adv. Genet. 61: 175–205. [DOI] [PubMed] [Google Scholar]
- Spitz F., Gonzalez F., Duboule D., 2003. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell 113: 405–417. [DOI] [PubMed] [Google Scholar]
- Tackels-Horne D., Toburen A., Sangiorgi E., Gurrieri F., de Mollerat X., et al. , 2001. Split hand/split foot malformation with hearing loss: first report of families linked to the SHFM1 locus in 7q21. Clin. Genet. 59: 28–36. [DOI] [PubMed] [Google Scholar]
- Tayebi N., Jamsheer A., Flottmann R., Sowinska-Seidler A., Doelken S. C., et al. , 2014. Deletions of exons with regulatory activity at the DYNC1I1 locus are associated with split-hand/split-foot malformation: array CGH screening of 134 unrelated families. Orphanet J. Rare Dis. 9: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C., Gagnon L. H., Longo-Guess C., Korstanje R., Sheehan S. M., et al. , 2017. Hearing loss without overt metabolic acidosis in ATP6V1B1 deficient MRL mice, a new genetic model for non-syndromic deafness with enlarged vestibular aqueducts. Hum. Mol. Genet. 26: 3722–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truett G. E., Heeger P., Mynatt R. L., Truett A. A., Walker J. A., et al. , 2000. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 29: 52, 54. [DOI] [PubMed] [Google Scholar]
- van Silfhout A. T., van den Akker P. C., Dijkhuizen T., Verheij J. B., Olderode-Berends M. J., et al. , 2009. Split hand/foot malformation due to chromosome 7q aberrations(SHFM1): additional support for functional haploinsufficiency as the causative mechanism. Eur. J. Hum. Genet. 17: 1432–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A., Minovitsky S., Dubchak I., Pennacchio L. A., 2007. VISTA Enhancer Browser–a database of tissue-specific human enhancers. Nucleic Acids Res. 35: D88–D92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A., Rubin E. M., Pennacchio L. A., 2009. Genomic views of distant-acting enhancers. Nature 461: 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Xin Q., Li L., Li J., Zhang C., et al. , 2014. Exome sequencing reveals a heterozygous DLX5 mutation in a Chinese family with autosomal-dominant split-hand/foot malformation. Eur. J. Hum. Genet. 22: 1105–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S., Costantino N., Court D. L., Jenkins N. A., Copeland N. G., 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland I., Muschke P., Jakubiczka S., Volleth M., Freigang B., et al. , 2004. Refinement of the deletion in 7q21.3 associated with split hand/foot malformation type 1 and Mondini dysplasia. J. Med. Genet. 41: e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga A., Michos O., Spitz F., Haramis A. P., Panman L., et al. , 2004. Mouse limb deformity mutations disrupt a global control region within the large regulatory landscape required for Gremlin expression. Genes Dev. 18: 1553–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All mouse strains examined in this study are commercially available from JAX. All data necessary for confirming the conclusions presented in the article are represented fully within the article.