CAG/CTG trinucleotide repeat expansions cause several degenerative neurological and muscular diseases. Koch et al. show that the chromatin remodeling...
Keywords: ISWI chromatin remodeler, CAG repeat expansion, transcription-coupled repair, base excision repair
Abstract
CAG/CTG trinucleotide repeats are unstable sequences that are difficult to replicate, repair, and transcribe due to their structure-forming nature. CAG repeats strongly position nucleosomes; however, little is known about the chromatin remodeling needed to prevent repeat instability. In a Saccharomyces cerevisiae model system with CAG repeats carried on a YAC, we discovered that the chromatin remodeler Isw1 is required to prevent CAG repeat expansions during transcription. CAG repeat expansions in the absence of Isw1 were dependent on both transcription-coupled repair (TCR) and base-excision repair (BER). Furthermore, isw1∆ mutants are sensitive to methyl methanesulfonate (MMS) and exhibit synergistic MMS sensitivity when combined with BER or TCR pathway mutants. We conclude that CAG expansions in the isw1∆ mutant occur during a transcription-coupled excision repair process that involves both TCR and BER pathways. We observed increased RNA polymerase II (RNAPII) occupancy at the CAG repeat when transcription of the repeat was induced, but RNAPII binding did not change in isw1∆ mutants, ruling out a role for Isw1 remodeling in RNAPII progression. However, nucleosome occupancy over a transcribed CAG tract was altered in isw1∆ mutants. Based on the known role of Isw1 in the reestablishment of nucleosomal spacing after transcription, we suggest that a defect in this function allows DNA structures to form within repetitive DNA tracts, resulting in inappropriate excision repair and repeat-length changes. These results establish a new function for Isw1 in directly maintaining the chromatin structure at the CAG repeat, thereby limiting expansions that can occur during transcription-coupled excision repair.
EXPANSION of trinucleotide repeats (TNRs) beyond a stable threshold number is the cause of multiple heritable neurodegenerative diseases, including Huntington’s disease, myotonic dystrophy, and multiple spinocerebellar ataxias (SCAs) (McMurray 2010; Usdin et al. 2015). Changes in TNR length (contractions and expansions) occur due to the formation of stable secondary structures when the DNA is transiently single-stranded. Molecular processes including DNA replication, DNA repair, and transcription involve transiently single-stranded DNA and have been implicated in repeat instability (López Castel et al. 2010; Usdin et al. 2015; Polleys et al. 2017). The known disease-causing repeat expansions occur in transcribed regions, and several previous studies have shown that transcription through trinucleotide repeats contributes to instability (Bowater et al. 1997; Parniewski et al. 1999; Schumacher et al. 2001; Lin et al. 2006; Jung and Bonini 2007; Lin and Wilson 2007). Factors involved in transcription-coupled repair (TCR) of DNA and base-excision repair (BER) play a role in transcription-associated instability, providing a mechanism for the insertion of extra repeat units (Parniewski et al. 1999; Lin et al. 2006; Jung and Bonini 2007; Kovtun et al. 2007). A study using an SCA1 disease mouse model found that TCR plays an important role in creating expansions in neuronal cells that lead to disease progression (Hubert et al. 2011). Additionally, the presence of R-loops, stable DNA:RNA hybrids that form during transcription, can promote CAG repeat instability (Lin et al. 2010; Reddy et al. 2011, 2014; Lin and Wilson 2012; Su and Freudenreich 2017).
The CAG repeat is a strong nucleosome-positioning element (Wang and Griffith 1995; Godde and Wolffe 1996; Volle and Delaney 2012). Several chromatin factors have been shown to impact repeat stability, including the CTCF protein, which organizes chromatin loop domains, a DNA methyltransferase (Dnmt1), which maintains CpG methylation, and histone deacetylase complexes (HDACs), which condense chromatin structure through histone deacetylation (Gorbunova et al. 2004; Jung and Bonini 2007; Libby et al. 2008; Debacker et al. 2012; Gannon et al. 2012). It was also recently shown that both histone H4 tail acetylation and the RSC chromatin remodeler are important in preventing CAG expansions from occurring during sister chromatid recombination (House et al. 2014). These discoveries have led to the proposal that the unusual chromatin structure associated with expanded repeats may play a role in repeat expansions (Dion and Wilson 2009). Chromatin-remodeling proteins are necessary to slide nucleosomes to allow for access to the DNA during transcription, replication, and repair. However, a role for chromatin remodeling in protecting against repeat instability during transcription has not been investigated.
The ISWI chromatin-remodeling complexes have been shown to remodel chromatin during transcription in both yeast and mammalian cells (Petty and Pillus 2013). ISWI proteins contain an ATPase domain that is necessary for its remodeling activity, using energy from ATP hydrolysis to slide or reposition nucleosomes (Tsukiyama et al. 1999). ISWI proteins contain a HAND-SANT-SLIDE domain that makes multiple contacts with linker DNA, and increases the affinity and specificity for nucleosomes (Mueller-Planitz et al. 2013). In yeast, the SANT domain is necessary for Isw1-remodeling activity but is not needed for its repressive function (Pinskaya et al. 2009). Crystal structures have revealed that a dinucleosome substrate is used for chromatin remodeling (Grüne et al. 2003; Yamada et al. 2011). ISWI binding to linker DNA relieves inhibition by the NegC domain of ISWI that controls its ATPase activity in the presence of appropriate nucleosomal substrates (Clapier and Cairns 2012).
In yeast, there are two additional known nucleosome-spacing enzymes, Isw2 and Chd1. While Isw1 can either repress or promote transcription depending on the proteins it interacts with (Morillon et al. 2003; Vary et al. 2003), Isw2 mainly has a repressive effect on transcription by positioning nucleosomes that obstruct transcription (Goldmark et al. 2000; Whitehouse et al. 2007). Chd1 acts with Isw1 to maintain genome-wide nucleosome organization (Gkikopoulos et al. 2011). By chromatin immunoprecipitation (ChIP) sequencing, Isw1 and Chd1 were found to be enriched at nucleosome-depleted regions, suggesting that the remodelers act on DNA that has been transiently unwrapped during transcription (Zentner et al. 2013). At coding regions, Isw1 and Chd1 prevent histone exchange during transcription elongation (Smolle et al. 2012). In general, Chd1 mostly acts in genes that have shorter nucleosome spacing and Isw1 acts in genes with longer spacing (Ocampo et al. 2016). In the absence of both Isw1 and Chd1, nucleosome organization is grossly perturbed, especially within coding regions, although some nucleosome locations such as favored binding sequences are unaffected (Gkikopoulos et al. 2011).
Here, we report the discovery that Isw1, a conserved chromatin remodeler that functions during transcription, is required to maintain CAG repeat stability in a yeast model system. CAG repeat expansion frequency is significantly increased when ISW1 is deleted, and this increase is dependent on transcription through the repeat. Through genetic dissection of several DNA repair pathways, we found this increase in expansions to be dependent on the TCR pathway as well as the BER pathway. Multiple hypotheses were investigated to better understand the reason for the increase in excision repair-induced CAG expansions in the absence of Isw1. We conclude that Isw1 functions downstream of RNA Polymerase II (RNAPII) to inhibit the formation of CAG repeat expansions. We propose that a defect in nucleosome reassembly and spacing after passage of RNAPII allows the formation of DNA structures, which provoke excision repair and subsequent repeat expansions.
Materials and Methods
Yeast strains and YACs
Gene deletions were created using one-step gene replacement and gene disruptions were confirmed by PCR. The pMET25-RNH1 strain was created using the pYM-N35 plasmid (Janke et al. 2004). The construction of the CAG-85 URA3-YAC (URA3-YAC) was described previously (Callahan et al. 2003). The CAG-85 ADE2-URA3-YAC was made by first subcloning ADE2 from pRS402 into pHZ1 to construct pEP1. Then, 85 CAG repeats were cloned into pEP1, and the desired YAC was obtained by linearizing the plasmid and transforming into yeast containing the URA3-YAC. The CAG-100 pGAL1URA3-YAC was made by subcloning the CAG repeat and CYC1 terminator sequence into pYES2. The terminator sequence, CAG repeats, and pGAL1 were subcloned into pVS20, and the desired YAC was obtained by linearizing the final plasmid and transforming into yeast containing a YAC truncated at the G4T4 sequence. The construction of the Ttef1-CAG-70-Tcyc1 URA3-YAC (2T-YAC) has been described previously (Su and Freudenreich 2017).
CAG repeat stability assay
Stability assays were performed on the YACs depicted in Figure 1A, Figure 2A, and Figure 4A, with primers newCAGfor/newCAGrev for the CAG-85 URA3-YAC (URA3-YAC) and pGAL1 CAG-100 URA3-YAC, primers CAGfor60bp/CAGrev35bp for the CAG-85 ADE2-URA3-YAC, and primers T720B/CTGrev2 for the Ttef1-CAG-70-Tcyc1 URA3-YAC (2T-YAC), as previously described (Sundararajan et al. 2010), with the following modifications for the CAG-100 pGAL1URA3-YAC: the colony was resuspended in YC-Leu + 2% raffinose and grown for one doubling. The culture was then split into two tubes, washed twice with sterile water, and resuspended in YC-Leu + 2% glucose (not induced) or YC-Leu + 2% galactose (induced), then grown for six to seven doublings and plated for single colonies on YC-Leu-Ura plates. CAG repeat changes were assessed by analyzing PCR reactions on a high-resolution 2% Metaphor agarose (Lonza) gel. All stability (expansion and contraction) data are in Supplemental Material, Table S2 in File S1. Primer sequences are available upon request.
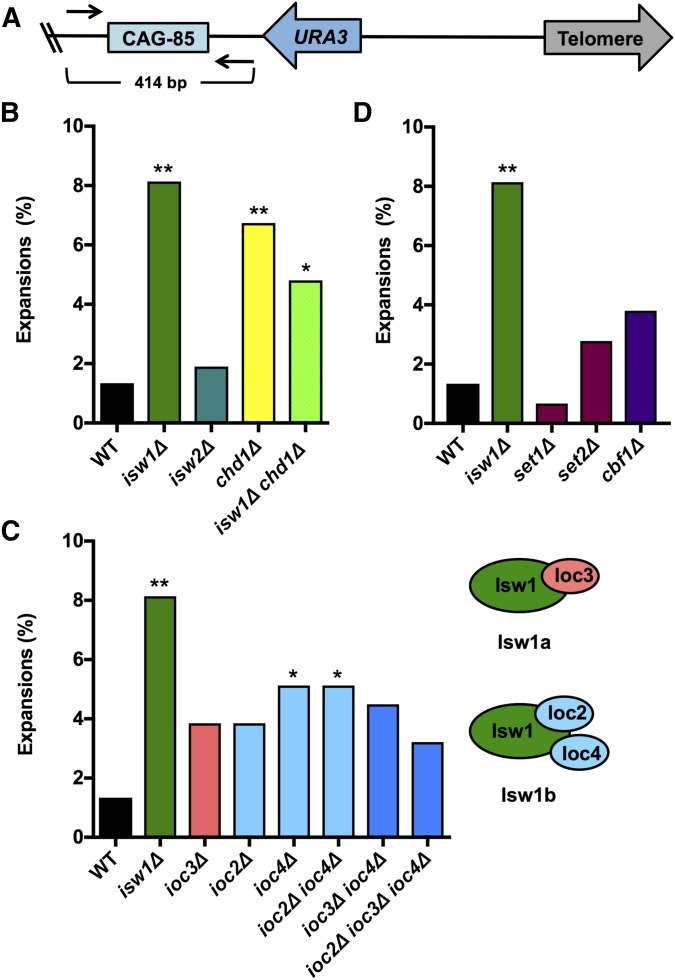
Figure 1.
Isw1 prevents CAG-85 repeat expansions. (A) A CAG-85 tract is on the right arm of the URA3-YAC (YAC CF1), where the URA3 gene is 198 bp from CAG-85 and 4.2 kb from the telomere. Repeat length was measured by PCR amplification using primers (black arrows) upstream and downstream from the repeat, with an intact CAG-85 repeat resulting in a 414-bp product. The frequency of CAG-85 expansions was measured in strains lacking (B) ISW1 or other genes encoding chromatin remodeling proteins (ISW2 and CHD1), (C) genes encoding subunits of Isw1 complexes Isw1a (IOC3) or Isw1b (IOC2 and IOC4) individually or in combination with deletion of another subunit, and (D) genes encoding proteins with known functions of recruiting Isw1 to chromatin (SET1, SET2, and CBF1). Expansion frequencies were tested for significant deviation from wild-type (WT) frequency using Fisher’s exact test, * P < 0.05 and ** P < 0.01.
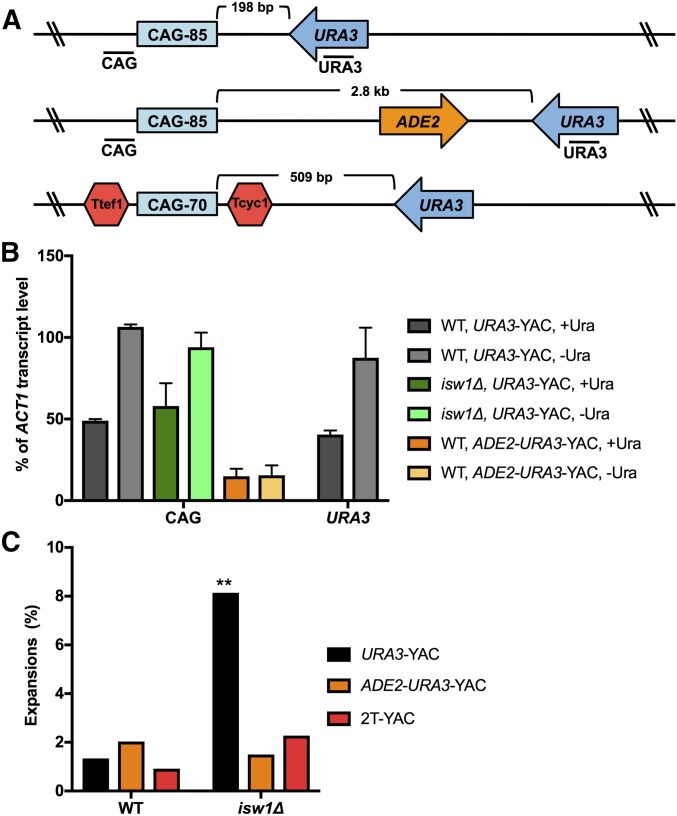
Figure 2.
Expansions occurring in the absence of ISW1 are dependent on transcription through the repeat tract. (A) Three YAC constructs were tested for repeat instability: the CAG-85 URA3-YAC (URA3-YAC) (top); the CAG-85 ADE2-URA3- YAC, where the URA3 gene has been moved far away (2.8 kb) from CAG-85 (middle); and the Ttef1-CAG-70-Tcyc1 URA3 YAC (2T-YAC), where transcription terminators (Ttef1, Tcyc1) flank the CAG repeat (bottom). Locations of PCR amplicons next to the CAG repeat and URA3 used in quantitative RT-PCR (qRT-PCR) are indicated. (B) qRT-PCR of wild-type (WT) and isw1∆ strains containing the URA3-YAC, and WT containing the ADE2-URA3-YAC in noninducing (+Ura) and inducing (−Ura) conditions. CAG transcript level is normalized to the level of ACT1 transcript and is presented as percent of ACT1 transcript level. qRT-PCR data are from two independent experiments; error bars indicate the SD. (C) The frequency of CAG repeat expansions was measured in WT and isw1∆ strains containing the CAG-85 URA3-YAC (URA3-YAC), CAG-85 ADE2-URA3-YAC, and Ttef1-CAG-70-Tcyc1 URA3-YAC (2T-YAC). Expansion frequencies were tested for significant deviation from WT using Fisher’s exact test, ** P < 0.01.
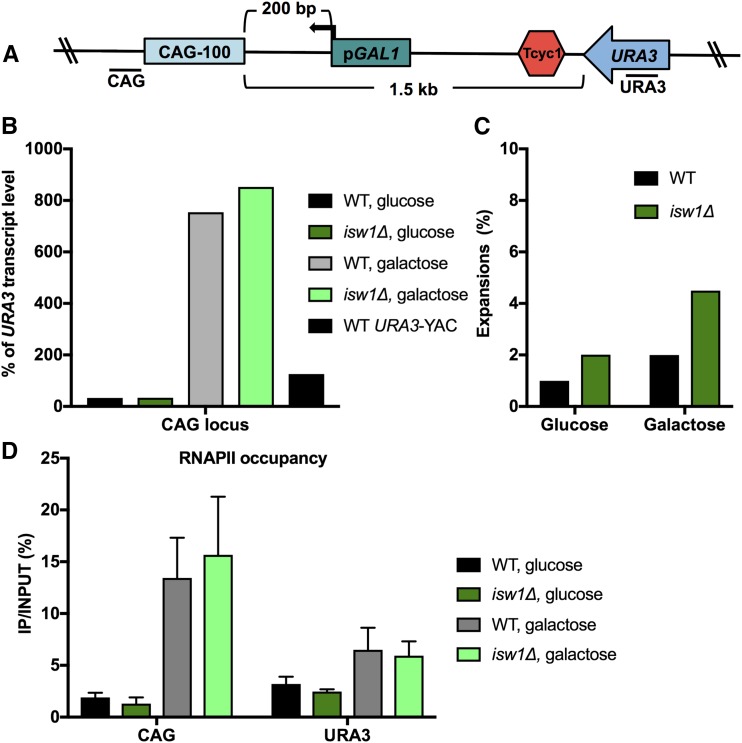
Figure 4.
High levels of transcription of the repeat reduce the requirement for Isw1 in repeat maintenance. (A) Inducible YAC construct CAG-100 pGAL1 URA3-YAC contains the pGAL1 promoter next to the CAG repeat and a transcription terminator (Tcyc1) after URA3 to terminate URA3 transcription. Locations of PCR amplicons next to the CAG repeat and URA3 used in quantitative RT-PCR (qRT-PCR) are indicated. (B) qRT-PCR analysis of CAG transcript level in wild-type (WT) and isw1∆ strains in noninducing (glucose) and inducing (galactose) conditions. CAG transcript level is normalized to the level of URA3 transcript and is presented as percent of URA3 transcript level. (C) The frequency of CAG-100 expansions was measured in WT and isw1∆ strains in noninducing (glucose) and inducing (galactose) conditions. (D) RNA polymerase II (RNAPII) occupancy next to the CAG repeat and within URA3 on the YAC was measured by RNAPII chromatin immunoprecipitation, followed by quantitative PCR in WT and isw1∆ strains in noninducing (glucose) and inducing (galactose) conditions. RNAPII immunoprecipitated is shown as % immunoprecipitated (IP)/INPUT; the average of three experiments with SEM is shown.
MMS sensitivity assay
Yeast strains were grown to saturation, normalized to OD600 of 1.0, then fivefold serial dilutions were made. Ten microliters of each dilution was spotted on plates for growth control (YC-Leu-Ura) and plates containing MMS (0.03–1%). Plates were incubated at 30° for 3–7 days before images were captured.
pGAL1 induction conditions
To induce pGAL1 for RT-PCR, ChIP, and micrococcal nuclease (MNase assays), yeast cultures were grown in YC-Leu + 2% raffinose to OD600 1.0–1.6. Then, the culture was split into two tubes, washed twice with sterile water, and resuspended in YC-Leu + 2% glucose (not induced) or YC-Leu + 2% galactose (induced). The cultures were then incubated at 30° for 1 hr and cells were collected for experimental analysis.
Quantitative RT-PCR (qRT-PCR)
Total RNA was isolated from yeast cell pellets using the Illustra RNAspin mini kit (GE Healthcare), following the manufacturer’s directions. RT-PCR was performed using Superscript First Strand Synthesis kit (Life Technologies) with 1 μg RNA, following the manufacturer’s protocol, using random hexamers for priming. RT-PCR samples were analyzed using quantitative PCR (qPCR) with SYBR green PCR mastermix (Roche) on a 7300 real-time PCR system (Applied Biosystems, Foster City, CA). qPCR reactions were run in duplicate. Transcript levels were normalized to ACT1 or URA3 levels. URA3 normalization was done in experiments with galactose induction because ACT1 RNA levels change significantly in these growth conditions, whereas URA3 levels do not. qPCR primer sequences are available upon request.
Chromatin immunoprecipitation (ChIP)
ChIP was performed as previously described (Pearson and Moore 2014) with the following adjustments. Samples were sonicated at 4° to shear the chromatin to within 250–500 bp on a Bioruptor sonicator (Diagenode). Immunoprecipitation was performed using 1 μg anti-RNAPII (CTD4H8; Santa Cruz). Protein G dynabeads (Life Technologies) were used for immunoprecipitation. The dynabeads were prewashed in FA lysis buffer containing 0.1% SDS, including blocking with 1 mg/ml BSA. Chromatin was incubated with antibody and beads for 4 hr at 4°. Input and immunoprecipitated DNA levels were quantified by qPCR using SYBR green PCR mastermix (Roche) on a 7300 real-time PCR system (Applied Biosystems). qPCR reactions were run in duplicate. qPCR primer sequences are available upon request.
MNase assay
The MNase assay procedure was adapted from a previous study (Wu and Winston 1997). Yeast cell pellets were resuspended in sorbitol buffer (1.1 M sorbitol, 20 mM KPO4, pH 7, and 0.5 mM CaCl2) with 10 mM DTT and incubated for 15 min while shaking at 30°. Cell pellets were resuspended in 1 ml sorbitol buffer with zymolyase 100T (1 mg/ml) and incubated at 30°. The cells were washed twice with 10 ml sorbitol buffer and pelleted (1200 rpm for 6 min at 4°). The pellet was then resuspended in buffer A (1 M sorbitol, 50 mM NaCl, 10 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 1 mM β-mercaptoethanol, and 0.5 mM spermidine) with 1× protease inhibitor cocktail to a total volume of 1 ml. Aliquots of 200 μl of cell slurry were added to 200 μl of 37° prewarmed buffer A with 0.15% Triton-X 100 and MNase (0, 0.25, 2.5, 7.5, and 15 units), incubated at 37° for 5 min, and stopped by incubating for 15 min at 37° with 40 μl of Stop Buffer (250 mM EDTA and 5% SDS). The DNA was isolated by adding 160 μl 5M potassium acetate to the samples, incubating on ice for 30 min, and pelleting debris (14,000 rpm for 15 min at 4°). The supernatant was added to an equal volume of isopropanol to precipitate the DNA and collected by centrifugation (14,000 rpm for 30 min at 4°). The DNA pellet was resuspended in 500 μl 1× TE with 100 μg/ml RNase A and incubated at 37° for 1 hr. The DNA was extracted twice using an equal volume of chloroform and precipitated overnight by adding 67 μl 7.5 M ammonium acetate and 500 μl isopropanol with 1 μl of glycogen. The resulting DNA was washed twice with 70% ethanol and resuspended in 30 μl 1× TE.
The Southern probe was created by amplifying a 313-bp region 14-bp upstream of the CAG repeat (primers #2045 and #348). The probe was radioactively labeled with 32P using the Random Primed DNA Labeling Kit (Roche), according to the manufacturer’s directions. The labeled probe was added to 20 ml 65° Church’s buffer.
MNase-digested DNA was run on a 1.5% agarose gel at 80 V for 6 hr, washed for 10 min in depurination solution (0.25 M HCl), 30 min in denaturation solution (0.5 M NaOH and 1.5 M NaCl), and 30 min in neutralization solution (1 M Tris pH 7.4 and 1.5 M NaCl), and then transferred to a nitrocellulose fiber nylon membrane (Sigma [Sigma Chemical], St. Louis, MO). The DNA was cross-linked and then placed in prehybridization buffer of 65° preheated Church’s Buffer for 15 min. Prehybridization buffer was removed, and the probe was hybridized to the membrane in hybridization solution (Church’s Buffer with a 32P-labeled DNA probe) overnight at 65° in a hybridization oven. The blots were washed in Blot Wash 1 (1× SSC and 0.1% SDS) at 65° twice for 15 min each. The blots were exposed on film for 3 days at −80°. Pairs of samples (wild-type or isw1∆, ± transcription induction) were digested with the same batch of MNase, and growth conditions, digestion times, and reagents were kept constant to facilitate direct comparison.
Data availability
Yeast strains used in this study are listed in Table S1 in File S1. Strains are available upon request. Raw instability data and associated P-values are listed in Table S2 in File S1.
Results
The Isw1 chromatin-remodeling protein prevents CAG repeat expansions
The CAG repeat is one of the strongest known nucleosome-positioning elements (Wang and Griffith 1995; Godde and Wolffe 1996; Volle and Delaney 2012), which potentially creates a barrier to transcription and DNA repair. Therefore, we sought to understand the contribution of chromatin remodeling to CAG repeat stability. Utilizing a CAG-85 repeat on a YAC, we identified the isw1∆ mutant in a screen for mutants with either increased CAG repeat fragility or instability (Gellon et al. 2011). Upon further testing, the most robust phenotype of strains lacking Isw1 was an increased frequency of CAG repeat expansions, determined using a PCR assay that can detect both expansions and contractions (Figure 1A). The CAG fragility rate and contraction frequency were not significantly different from the wild-type strain (data not shown and Table S1 in File S1). As a comparison, we tested the related chromatin remodeler genes ISW2 and CHD1. A deletion of CHD1 exhibited significantly increased CAG repeat expansions compared to wild-type, while isw2∆ CAG repeat expansion frequency was unchanged (Figure 1B). Because Isw1 and Chd1 act together to maintain genome-wide nucleosome organization (Gkikopoulos et al. 2011), we tested a double mutant. Although the deletion of both genes has additive effects on genome-wide nucleosome placement (Gkikopoulos et al. 2011), there was no further effect on repeat expansions when both genes were deleted (Figure 1B). Highly favored nucleosome-binding positions are less affected by the deletion of both remodelers (Gkikopoulos et al. 2011), which could explain this observation. Therefore, we focused on understanding the role of Isw1 in protecting against CAG repeat expansions.
The Isw1 ATPase is found in two complexes: Isw1a, along with subunit Ioc3, and Isw1b, along with subunits Ioc2 and Ioc4 (Tsukiyama et al. 1999; Vary et al. 2003). Isw1a positions dinucleosomes in promoter proximal regions and represses transcription initiation, whereas Isw1b positions nucleosomes in coding regions and promotes transcriptional elongation (Morillon et al. 2003, 2005). Isw1b also coordinates elongation with transcriptional termination and pre-mRNA processing (Morillon et al. 2003). We tested deletion of Isw1 subunit members individually and in combination to identify which Isw1 function is involved in suppressing CAG repeat expansions. This analysis supported a greater role for the Isw1b complex in the prevention of expansions, since deletion of IOC2 and IOC4 resulted in a significant increase in CAG repeat expansions. However, the effect on expansions upon deletion of the accessory complex members was not as profound as deletion of the gene encoding the Isw1 ATPase (Figure 1C), suggesting either that Isw1 may also function independently of Ioc2 and Ioc4 in preventing repeat expansions or that their deletion does not completely abolish ISWI activity. There is some previous evidence that Isw1 can function independently of the accessory complex members in its role in transcriptional silencing of rDNA (Mueller and Bryk 2007).
Next, we wanted to understand the contribution of known pathways that recruit Isw1 to chromatin in its role in preventing CAG repeat expansions. At some genes, Isw1 is recruited to chromatin by specific chromatin marks, such as methylation of histone H3 lysine 4 (H3K4) and histone H3 lysine 36 (H3K36) (Santos-Rosa et al. 2003; Maltby et al. 2012; Smolle et al. 2012). Set1 is a histone methyltransferase (HMT) that methylates H3K4, and Set2 is a HMT that trimethylates H3K36 and mediates Isw1b occupancy over certain coding regions, although not all Isw1 functions are Set1- or Set2-dependent (Smolle et al. 2012). Cbf1 is a DNA-binding protein that has been shown to recruit Isw1 to chromatin, an interaction required for chromatin remodeling at the promoter-proximal Cbf1-binding motif CACGTG (Kent et al. 2004). We tested these genes for their contribution to CAG repeat instability and found that none of the gene deletions resulted in significantly increased CAG repeat expansions (Figure 1D), suggesting that Isw1’s function in preventing CAG repeat expansions is independent of recruitment by Cbf1 or histone methylation by Set1 or Set2.
Isw1 prevents CAG repeat expansions from arising during transcription
The Isw1 chromatin-remodeling protein has a well-characterized role in transcription; therefore, we wanted to determine whether CAG repeat expansions in isw1∆ cells were dependent on transcription. The YAC used in our initial CAG repeat instability analysis contains a URA3 gene < 200 bp from the CAG repeat, with the direction of URA3 transcription toward the CAG repeat (Figure 2A). qRT-PCR analysis of the URA3-YAC showed high levels of transcription of a sequence proximal to the CAG repeat (Figure 2B). The level of CAG repeat transcription was similar to the level of URA3 transcript and was increased when URA3 expression was induced by eliminating uracil from the media cells were grown in, indicating that the CAG repeat transcript was likely due to readthrough transcription from URA3 (Figure 2B). Notably, transcript levels did not change in isw1∆ cells compared to wild-type in strains containing the URA3-YAC (Figure 2B). To test the role of transcription, two additional YACs were utilized: the CAG-85 ADE2-URA3-YAC, where the URA3 marker gene is located 2.8 kb from the CAG repeat with a convergently oriented ADE2 gene between URA3 and the CAG repeat, and a Ttef1-CAG-70-Tcyc1 URA3-YAC (2T-YAC), in which the CAG repeat is flanked by transcriptional terminator sequences (Figure 2A). There is minimal transcription of the CAG repeat on either the ADE2-URA3-YAC (60–80% reduction; Figure 2B) or the 2T-YAC (∼70% reduction; Su and Freudenreich 2017). Tellingly, no increase in CAG repeat expansions was observed in isw1∆ strains containing either of the constructs, with minimal transcription through the CAG repeat (Figure 2C). These results indicate that Isw1 plays an important role during transcription to prevent repeat instability.
TCR is the source of Isw1-dependent CAG repeat expansions
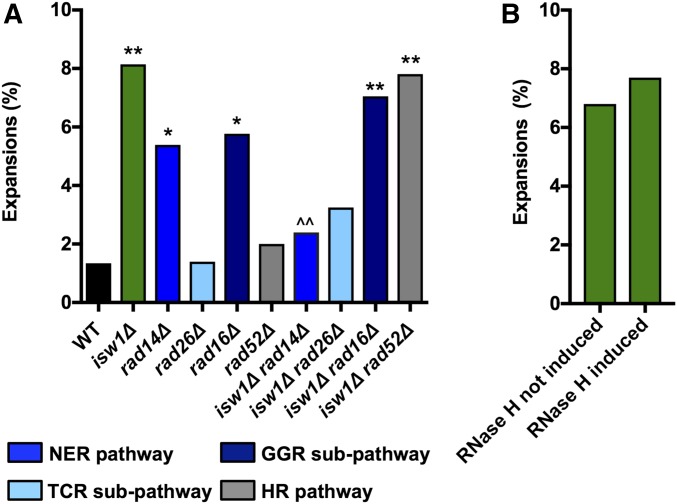
Since CAG repeat expansions occurring in the absence of Isw1 are transcription-dependent and CAG repeat expansions have previously been shown to arise during DNA TCR (Parniewski et al. 1999; Jung and Bonini 2007; Lin and Wilson 2007; Concannon and Lahue 2014), we tested whether these expansions in isw1∆ cells occur during TCR. We tested a general component of the nucleotide-excision repair (NER) pathway, RAD14 (human XPA), as well as proteins involved in the two subpathways of NER: RAD16 in the global genomic repair (GGR) subpathway, and RAD26 (hCSB) in the TCR subpathway. For comparison, we also tested a component of the homologous recombination (HR) pathway, RAD52, as HR has been shown to cause CAG repeat expansions during repair of DNA damage or stalled replication forks (Polleys et al. 2017). This analysis revealed that isw1∆-dependent expansions are suppressed by deletion of RAD14 and RAD26, indicating that expansions occurring in the absence of Isw1 are TCR-dependent (Figure 3A). In contrast, no reduction in isw1∆-dependent expansions was observed in the absence of RAD16 or RAD52; therefore, GGR and HR pathways are not involved.
Figure 3.
Expansions in isw1∆ are dependent on NER, but are independent of HR and R-loops. The frequency of CAG-85 expansions was measured in WT and isw1∆ strains (A) lacking genes involved in the NER pathway (RAD14), the TCR subpathway (RAD26), the GGR subpathway (RAD16), or the HR pathway (RAD52), and (B) isw1∆ strains containing pMET25-RNH1, to overexpress RNH1 and degrade R-loops, with pMET25 not induced or induced by growth in media containing or lacking methionine. Expansion frequencies were tested for significant deviation from WT using Fisher’s exact test; * P < 0.05 and ** P < 0.01. Expansion frequencies for double mutants were tested for significant deviation from isw1∆ using Fisher’s exact test; ^^ P < 0.01. GGR, global genomic repair; HR, homologous recombination; NER, nucleotide-excision repair; TCR, transcription-coupled repair; WT, wild-type.
R-loops have been previously implicated in CAG repeat instability (Lin et al. 2010; Reddy et al. 2011, 2014; Lin and Wilson 2012; Su and Freudenreich 2017) and, interestingly, recent research has revealed a novel function for Isw1 as an mRNA ribonucleoparticle export surveillance factor, which tethers transcripts that are not ready for export from the nucleus (Babour et al. 2016). R-loops form during transcription and therefore could explain why CAG repeat expansions are increased in isw1∆ strains if R-loops were increased in this background. We tested whether degrading R-loops by overexpressing RNH1, the gene that encodes RNase 1H, would affect CAG repeat stability. RNH1 overexpression was confirmed by RT-PCR in our strains (Figure S1 in File S1). RNase H induction did not suppress the expansion frequency in the isw1Δ strain (Figure 3B), implying that R-loops are not contributing to the expansions occurring in the absence of Isw1. Note that the isw1∆ phenotype, increased CAG repeat expansions, is different from the phenotypes we recently characterized in conditions of increased R-loops at an expanded CAG tract, which were repeat fragility and contractions that were dependent on cytosine deamination (Su and Freudenreich 2017).
RNAPII binding to the CAG repeat increases upon induced CAG repeat transcription, but is not the main cause of isw1∆-dependent expansions
One possible model to explain CAG repeat expansions in isw1∆ strains is that RNAPII stalls at the CAG repeat due to its movement being blocked by a nucleosome that is ordinarily moved by Isw1 chromatin remodeling. RNAPII stalling is hypothesized to trigger TCR (Brueckner et al. 2007). CAG repeat hairpins could form during TCR, leading to CAG repeat expansions (Usdin et al. 2015). To test whether CAG repeat transcription results in RNAPII stalling at the CAG repeat to cause expansions in isw1∆ strains, we designed a new YAC construct with inducible transcription of the CAG repeat. We inserted a transcriptional terminator sequence after the URA3 gene to decrease URA3 readthrough transcription and an inducible promoter, pGAL1, so we could turn transcription of the CAG repeat on and off (Figure 4A). RT-PCR analysis of this construct in noninducing (glucose) and inducing (galactose) conditions showed that this system works as expected, since high levels of transcript were detected at the CAG locus when the pGAL1 promoter was induced (22-fold over noninducing conditions; Figure 4B). In glucose conditions when transcription was suppressed, isw1∆-dependent expansions were low, consistent with the data shown in Figure 2. When CAG repeat transcription was induced, CAG repeat expansions were increased in isw1∆; however, the increase observed was not significant and not as high as observed for URA3 readthrough transcription levels (Figure 4C). These data suggest that very high levels of transcription may actually reduce the need for Isw1 to maintain stability at the CAG tract.
To further understand the impact of highly induced transcription, RNAPII occupancy was analyzed by ChIP. The ChIP results showed increased RNAPII stalling at the CAG repeat when transcription was induced in galactose; however, no difference in RNAPII binding was seen when wild-type and isw1∆ strains were compared (Figure 4D). These results show that Isw1 does not influence levels of RNAPII stalling within a CAG tract, and indicate that expansions in the absence of the Isw1 remodeler are unlikely to be due to excessive stalling of RNAPII.
BER also contributes to CAG expansions that arise in the absence of Isw1
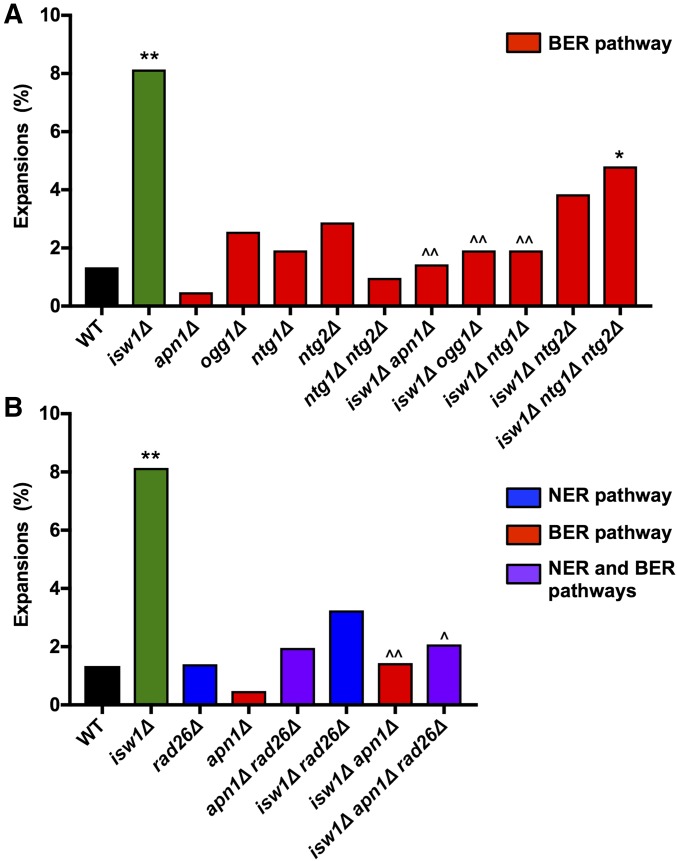
There is evidence that suggests cross talk between the TCR and BER pathways. For example, the TCR pathway repairs DNA damage typically repaired by the BER pathway in the absence of transcription (Kim and Jinks-Robertson 2010). Since CAG repeat expansions arising in the absence of Isw1 did not correlate with levels of RNAPII stalling, we decided to test whether they could also occur during other repair pathways, such as BER. Interestingly, expansions in isw1∆ were significantly decreased when the apurinic/apyrimidinic (AP) endonuclease APN1 or the DNA glycosylases OGG1, NTG1, and NTG2 were deleted (Figure 5A). Thus, the BER pathway contributes significantly to CAG expansions in the absence of Isw1, to the same or even a greater extent than TCR. To better understand whether these two pathways were acting independently or together, expansion frequency was tested in a triple mutant, isw1∆ apn1∆ rad26∆ (Figure 5B). CAG repeat expansions were suppressed to the same level in the triple mutant as in each double mutant, suggesting that the two pathways are working together to create the expansions observed in the absence of Isw1.
Figure 5.
CAG expansions in the absence of ISW1 are dependent on both base-excision repair (BER) and nucleotide-excision repair (NER). The frequency of CAG-85 expansions was measured in wild-type (WT) and isw1∆ strains (A) lacking genes involved in the BER pathway (APN1, OGG1, NTG1, and NTG2), and (B) double-mutant combinations of genes in the BER and NER pathways. Expansion frequencies were tested for significant deviation from WT using Fisher’s exact test; * P < 0.05 and ** P < 0.01. Expansion frequencies for double and triple mutants were tested for significant deviation from isw1∆ using Fisher’s exact test; ^ P < 0.05 and ^^ P < 0.01.
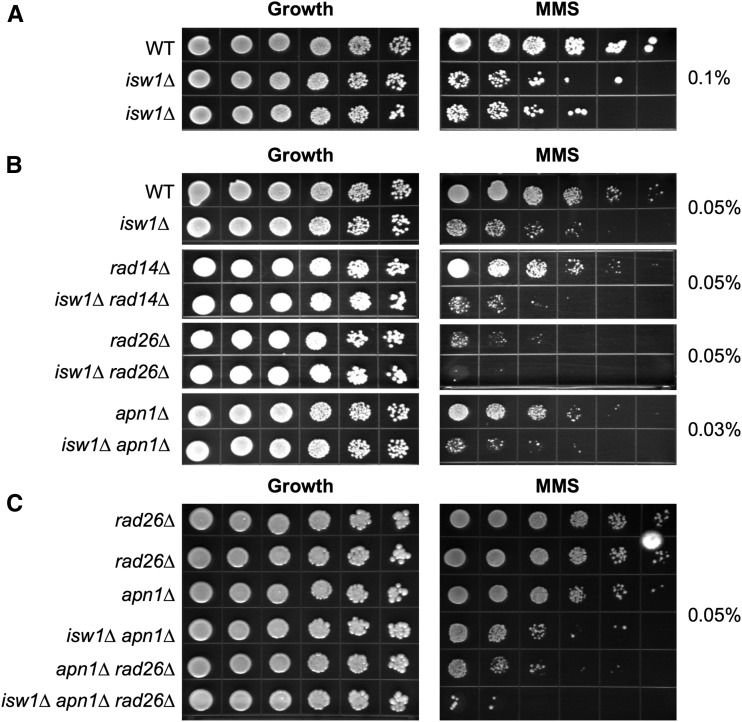
Deletion of ISW1 results in MMS sensitivity that is synergistic with NER and BER mutants
A previous study showed that deletion of ISW1 resulted in slight MMS sensitivity (Chen et al. 2010). MMS alkylates DNA bases, leading to damage that can be repaired by the BER or TCR pathways. We confirmed that deletion of the Isw1 ATPase results in MMS sensitivity in our background by comparing growth of wild-type and isw1∆ strains on plates containing MMS (Figure 6A and Figure S2 in File S1). Further phenotypic analysis of double mutants showed an increase in MMS sensitivity when both Isw1 and either the NER or BER pathway is disrupted (Figure 6B and Figure S2 in File S1). Lastly, MMS sensitivity was further increased in isw1∆ cells lacking both TCR and BER genes, to a greater extent than elimination of the TCR and BER pathways in the presence of Isw1 (e.g., compare isw1∆ apn1∆ rad26∆ to apn1∆ rad26∆ Figure 6C, or isw1∆ ogg1∆ rad26∆ to ogg1∆ rad26∆, Figure S2 in File S1). These findings show that the role of Isw1 in DNA repair is not limited to CAG/CTG repeat hairpins, but is important during repair of lesions caused by MMS. Furthermore, the additive or synergistic sensitivities indicate that Isw1 is not involved in BER or TCR per se, but that these pathways are acting in parallel. For example, the absence of Isw1 could create a chromatin environment that is more susceptible to MMS damage or CAG hairpin formation, which would then be acted upon by BER or TCR pathways.
Figure 6.
Deletion of ISW1 results in a synergistic increase in MMS sensitivity with nucleotide-excision repair (NER) and base-excision repair (BER) mutants. The indicated strains were plated on growth control plates (YC-Leu-Ura) or MMS plates to test the MMS sensitivity of (A) wild-type (WT) and isw1∆ strains on plates containing 0.1% MMS after 7 days of growth; (B) isw1∆ combined with deletion of NER, transcription-coupled repair (TCR), or BER genes on plates containing 0.03 or 0.05% MMS after 3 days of growth; and (C) isw1∆ combined with deletion of both NER and BER genes on plates containing 0.05% MMS after 3 days of growth.
Transcription reduces nucleosome occupancy over a CAG tract, and absence of Isw1 alters the nucleosome array
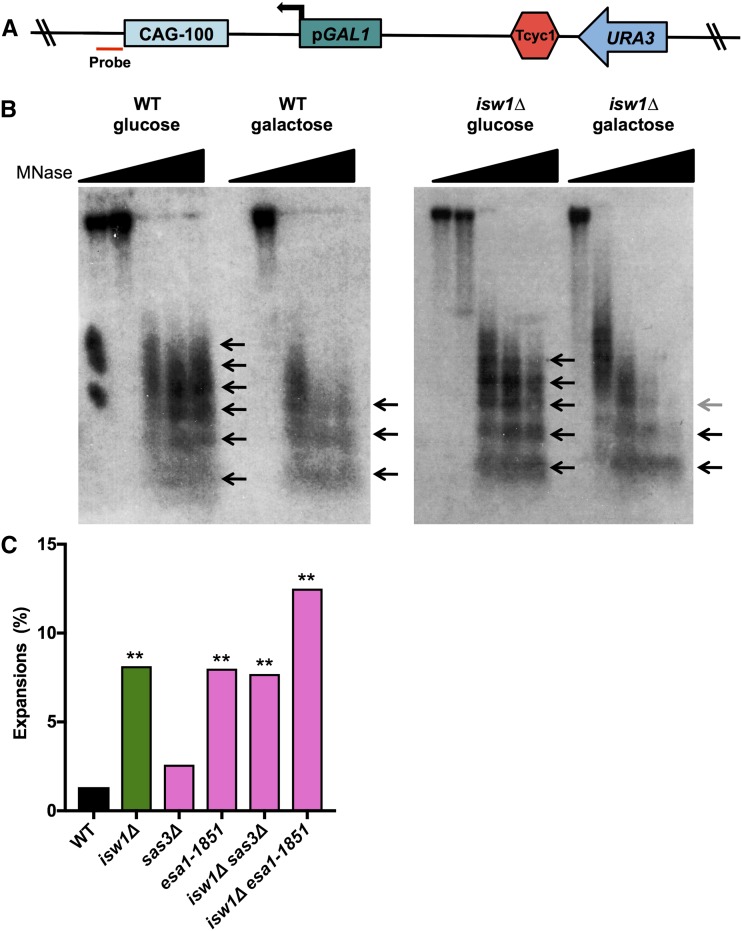
Since our data did not support a direct role for Isw1 in the TCR or BER pathways, we investigated whether Isw1’s role in preventing expansions is through its chromatin-remodeling activity during transcription. For example, Isw1 may be required to slide nucleosomes at the CAG repeat during transcription to prevent expansions. To test this model, we measured nucleosome positioning by indirect end-labeling of MNase-digested DNA at the CAG-100-pGAL1-URA3 YAC in wild-type and isw1∆ strains, in the presence or absence of induced transcription. A Southern blot probe was designed to detect the status of nucleosomes at the CAG repeat (Figure 7A). As predicted, the CAG repeat is a robust nucleosome-positioning element in wild-type cells, with an array of up to six spaced nucleosomes visible (Figure 7B, arrows). Transcription through the repeat reduced the association of nucleosomes at the CAG repeat, as there was a shift to shorter nucleosome arrays; only three are clearly present in galactose compared to five to six in glucose, indicating a more open chromatin structure (Figure 7B, left panel).
Figure 7.
Transcription reduces nucleosome occupancy over a CAG tract and isw1∆ alters the nucleosome array. (A) CAG-100 pGAL1 URA3-YAC construct showing location of the probe used in a Southern blot, 14-bp upstream of the CAG repeat (red). (B) Micrococcal nuclease (MNase) assay of wild-type (WT) and isw1∆ strains grown in noninducing (glucose) or inducing (galactose) conditions. The wedge indicates increasing MNase levels from 0 to 15 units; the 0-unit lane is missing in the WT galactose condition. The arrows indicate MNase-protected regions; an array of three to six distinguishable positioned nucleosomes is visible. Less-digested or less-positioned arrays appear as smears. (C) The frequency of CAG-85 expansions was measured in WT and isw1∆ strains with deletion of SAS3 or a catalytic mutation of ESA1 histone acetyltransferase (HAT) genes. Expansion frequencies were tested for significant deviation from WT using Fisher’s exact test; ** P < 0.01.
In addition to the notable change upon induction of transcription, there was a difference in the MNase sensitivity of chromatin at the CAG tract when comparing wild-type and isw1∆ mutants, with a shorter nucleosome array apparent in isw1∆ cells. For example, three positioned nucleosomes are clearly present at the highest MNase concentration in wild-type cells grown in galactose, while the di- and trinucleosome units are barely visible in the isw1∆ + galactose condition (Figure 7B). Wild-type cells also showed a longer nucleosome array in the absence of induced transcription compared to isw1∆, though this difference was subtler (six vs. five nucleosomes at the highest MNase concentration). The greater MNase sensitivity of the isw1∆ strain was confirmed in a second induction experiment (data not shown), as well as on the original CAG-85-URA3-YAC used for most of the instability experiments, where a much lower level of transcription through the repeat originated from the URA3 gene (Figure S3 in File S1). These results indicate that the chromatin structure of the transcribed CAG repeat is more open in the isw1∆ mutant compared to the wild-type strain.
Isw1 acts with histone H3/H4 acetylation to prevent CAG repeat expansions
Isw1 interacts physically with histone H3 and H4 (Pinskaya et al. 2009; Smolle et al. 2012), and a modification of histone H4, H4K16 acetylation, disrupts Isw1 chromatin remodeling (Corona et al. 2002; Shogren-Knaak et al. 2006). Isw1 exhibits genetic interaction with the NuA4 complex, containing the histone acetyltransferase (HAT) Esa1, to downregulate the expression of stress-induced genes (Lindstrom et al. 2006). Isw1 also interacts genetically with the HATs Sas3 and Gcn5, with Isw1 inactivation rescuing gcn5∆ sas3∆ synthetic lethality (Lafon et al. 2012). Therefore, we tested whether Isw1 might be working with either H3 or H4 histone acetylation to influence CAG instability. The CAG repeat expansions in isw1∆ remain increased in the absence of the H3 HAT, Sas3, and in the H4/H2A HAT mutant esa1-1851 (Figure 7C). However, the double mutant of isw1∆ with either sas3∆ or the esa1 mutation exhibited a less than additive expansion frequency. Therefore, Isw1 may be functioning together with histone acetylation to control nucleosome state in a manner that prevents CAG repeat instability.
Discussion
In this analysis, we have identified a function for budding yeast Isw1 in maintaining genomic stability, specifically in preventing the expansion of CAG repeats during transcription. CAG trinucleotide repeats are prone to both expansions and contractions, and the bias to expand in affected human cells is not well understood. However, the known expansion-prone CAG loci in humans are transcribed, and both BER and TCR pathways have been shown to cause repeat expansions in vitro and in mouse models (Kovtun et al. 2007; Hubert et al. 2011; Liu and Wilson 2012). Since chromatin remodeling must occur for DNA transactions, we hypothesized that this process would be an important component of facilitating transcription through CAG tracts. The fact that CAG/CTG repeats are one of the strongest known nucleosome-positioning elements may present an additional barrier to DNA transactions, and thus repeat stability may have a strong dependency on proper nucleosome remodeling and positioning. Since the ISWI complexes are highly conserved, we predict that they will also be important in preventing trinucleotide repeat expansions in human cells. Expansions are relatively rare in yeast compared to what is observed for repeats of equivalent length at human disease loci, yet we still observed a sixfold increase in CAG-85 expansion frequency in isw1Δ cells compared to wild-type. This suggests that altered chromatin structure during transcription could be a major factor in promoting disease-causing CAG expansions at human disease loci.
Both TCR and BER excision repair pathways contribute to CAG expansions in the absence of ISWI
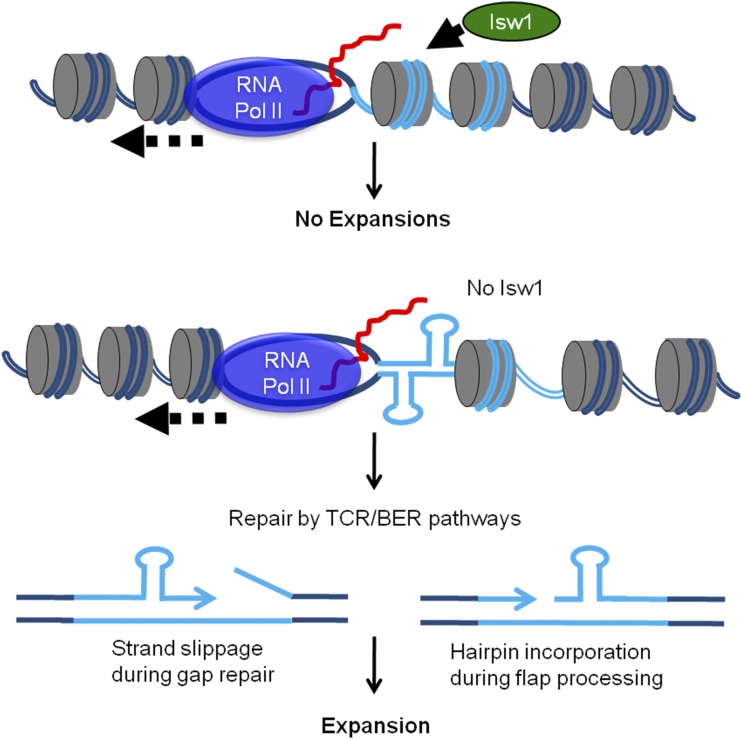
Our data show that the expansions that occur in the absence of Isw1 are dependent on proteins in both the TCR and BER pathways. One possibility we considered was that Isw1 acts during TCR or BER to facilitate proper repair. ISWI complexes have previously been implicated in DNA repair (Nakanishi et al. 2007; Chen et al. 2010; Erdel et al. 2010; Lans et al. 2010). In our system, the isw1∆ single mutant is sensitive to MMS, which could be consistent with a role for Isw1 in DNA repair. However, our MMS data indicate that Isw1 is acting in a separate pathway from both TCR and BER, as isw1∆ strains exhibit additive or synergistic MMS sensitivity when combined with mutants in the TCR or BER pathways. This is consistent with our CAG repeat expansion data that demonstrates differing roles for Isw1 and TCR or BER components; while expansions are increased in the absence of Isw1, stability is unaffected in TCR and BER single mutants. Thus, our results support the notion that TCR and BER are the cause of expansions in the absence of Isw1, which argues that Isw1 is not directly working in either pathway. Based on these observations, we favor the model that Isw1 is not directly functioning to facilitate the TCR or BER pathways, but rather in the absence of Isw1, DNA secondary structures form at the CAG repeat that are targets for either the TCR or BER pathways (Figure 8). There is extensive evidence from both in vitro and in vivo experiments in various model systems (bacteria, yeast, flies, and mice) that CAG expansions can occur during both TCR and BER pathways; see Liu and Wilson (2012), Usdin et al. (2015), Zhao and Usdin (2015), and Polyzos and McMurray (2017) for review. Expansions can occur during the gap-filling stage either by strand slippage or by incorporation of a hairpin that forms on the displaced flap, which renders the flap resistant to FEN1 processing (Figure 8). MutS complexes may have a role in stabilizing the looped-out structures to favor incorporation of a repeat expansion (Lai et al. 2016; Schmidt and Pearson 2016).
Figure 8.
Model for how Isw1 reestablishes proper chromatin structure after RNA polymerase II (RNA Pol II) passage to suppress repeat instability. In the absence of Isw1, nucleosome assembly, mobility, exchange, or modification behind RNA Pol II is altered, allowing for CAG or CTG hairpin formation, or increased DNA damage. This results in inappropriate excision repair by the transcription-coupled repair (TCR) or base-excision repair (BER) pathways, allowing CAG repeat expansion to occur.
An interesting observation was that deletion of TCR components (Rad26 or Rad14), BER glycosylases (Ogg1 and Ntg1/Ntg2), or the downstream BER AP endonuclease Apn1, all suppressed the CAG expansions in isw1∆ cells. One possibility is that all of the isw1∆-dependent CAG repeat expansions are generated through the BER pathway, since Rad26/hCSB has been shown to function in BER (Tuo et al. 2001; Stevnsner et al. 2008). The involvement of all steps of BER in the expansion pathway, from recognition (Ogg1 and Ntg1/2) to nick generation (Apn1), is a compelling argument for the importance of BER in generating the CAG expansions. Both OGG1 and NEIL1 glycosylases have been shown to be required for CAG expansions in mouse models (Kovtun et al. 2007; Mollersen et al. 2012; Budworth et al. 2015; Cilli et al. 2016). However, to our knowledge, Rad14/hXPA has not been characterized as having a role in BER; thus, suppression of CAG repeat expansions in the isw1∆ rad14∆ double mutant argues that the TCR pathway may be involved separately from the BER pathway. Also, mutants in both BER and TCR genes showed additive MMS sensitivity with each other and with isw1∆, consistent with separate pathways. Our results show that components of both pathways are important for preventing CAG expansions (Figure 5B), though we were unable to detect an additive effect in this assay since deletion of APN1 alone already reduced CAG expansions to the wild-type level. DNA lesions, such as abasic sites that are typically repaired by BER, are removed by TCR when they are located in the transcribed strand (Kim and Jinks-Robertson 2010), indicating that the two pathways could be working together. The exact connection between TCR and BER during transcription of repetitive DNA remains to be elucidated.
Isw1 may promote CAG repeat stability by reestablishing chromatin structure after transcription
Isw1 is acting to prevent CAG expansions at least partially in the context of the Isw1b complex (Figure 1C), suggesting that Isw1’s role during transcriptional elongation is relevant to CAG repeat maintenance. Since the ISWI complexes are important in the transcription of many genes, we considered whether the effect on CAG repeat expansions was direct or indirect. The suppression of expansions in the double mutants is not consistent with an indirect effect of isw1∆, for example by downregulation of TCR gene transcription, since deletion of the TCR gene should then have no, or minimal, effect. In addition, isw1∆ microarray data indicates no significant decrease in transcript levels of relevant NER or BER genes (Pinskaya et al. 2009). Also, the effect on nucleosome occupancy at the CAG repeat during transcription observed in isw1 mutants (Figure 7B), and detection of Isw1 binding to the CAG tract by ChIP (data not shown), support a direct role for Isw1 in preventing CAG repeat expansions.
One possible model for the increased CAG repeat expansions in isw1∆ strains was that there would be an increase in RNAPII stalling at the CAG tract; however, our data indicate that RNAPII does not require Isw1 chromatin remodeling to progress through the repeat. Isw1 promotes nucleosome reassembly and spacing after transit by RNA polymerase during transcriptional elongation, resulting in reestablishment of proper chromatin structure behind the elongation complex (Morillon et al. 2003; Gkikopoulos et al. 2011; Zentner et al. 2013). Thus, our result actually fits with the known role of ISWI in reestablishing the chromatin structure after transcription, and suggests that in its absence, a defect in nucleosome remodeling behind RNA polymerase, rather than ahead of the transcription bubble, is likely more relevant. Isw1 and Chd1 chromatin-remodeling activity also collaborate to remodel nucleosomes after replication, which allows for rapid chromatin organization after DNA synthesis (Yadav and Whitehouse 2016). These results indicate that ISWI is generally important for reestablishment of proper chromatin structure after disruption by polymerase passage; thus, in its absence, a greater opportunity for intrastrand DNA annealing could exist. The Clark laboratory showed that nucleosome spacing in isw1∆ cells is due to Chd1, with nucleosomes having a shorter spacing that cannot bind 1H, leading to decondensation of the chromatin fiber (Ocampo et al. 2016). Decondensation is not detectable by the assay we used, though the greater sensitivity to MNase digestion in isw1∆ cells is consistent with such an event. Additionally, transcription reduces nucleosome occupancy over the CAG repeat in both wild-type and isw1∆ cells, and transcription was also required for the increased CAG expansions in the isw1∆ background, consistent with a correlation between disruption of nucleosome structure and repeat instability. We did not detect a difference in nucleosome positioning, but since the CAG tract is a very strong nucleosome-positioning element, perhaps this overrides other effects on nucleosome position.
Somewhat surprisingly, while reducing transcription through the CAG tract virtually eliminated expansions occurring in isw1∆ strains, inducing a high level of transcription through the CAG tract did not significantly increase expansions. The level of transcription through the tract in our original YAC is similar to that through both the URA3 and ACT1 genes, which are housekeeping genes with medium–high transcription levels, whereas the GAL1 promoter induces very high levels of transcription, ∼7.5-fold greater than URA3 (Figure 2B). Thus, under very high levels of transcription, the need for Isw1 in maintaining repeat stability is lessened. It is possible that repeated passage of RNAPII disrupts CAG or CTG hairpin structures, or reduces the need for Isw1 to remodel nucleosomes after transcription. The level of transcription through the CAG tract on the URA3-YAC is likely more relevant to the situation found at the human genes containing expandable CAG repeats.
We present a model (Figure 8) in which lack of Isw1 creates a chromatin environment where nucleosome sliding or assembly does not occur normally after RNAPII passage, leading to a greater propensity for CAG or CTG hairpin formation, or increased DNA damage. During transcription, the DNA is rendered both transiently nucleosome-free and single-stranded. The single-stranded DNA could be more prone to DNA secondary structure formation if not efficiently repackaged into chromatin. The role of Isw1 in reestablishing the chromatin structure after transcription might be particularly important at repetitive regions because of their propensity to form deleterious DNA structures.
In addition to a direct role in remodeling nucleosomes at the repeat after transcription, Isw1 may also affect the nature of the histones at the repeat. The Workman laboratory showed increased trans-histone exchange in cells lacking Isw1 (Smolle et al. 2012), and such an event at the CAG repeat could result in a more open chromatin structure, allowing for DNA hairpin formation that initiates repair by TCR or BER, leading to expansions. Alternatively, H3 and H4 acetylation levels are increased in cells missing the ISW1 gene (Smolle et al. 2012), and this could impact repair. We previously showed that H3 and H4 acetylation and deacetylation is required to maintain CAG stability by promoting high-fidelity HR (Yang and Freudenreich 2010; House et al. 2014). We posited that histone acetyl marks are required for the recruitment of repair factors to the repeat, and that loss of acetylation (HAT mutants) or genome-wide overacetylation (HDAC mutants) leads to inefficient and low-fidelity repair due to a loss of a locus-specific signal (House et al. 2014). Though the expansions occurring in the isw1∆ mutant were due to BER and TCR rather than HR, overacetylation of H3 and H4 at the CAG tract could lead to inappropriate targeting of BER or TCR repair (Figure 8). Of note, CAG repeat expansion frequency in the sas3∆ or esa1 mutants was not additive with isw1∆, supporting a role for Isw1 within the same pathway as histone acetylation for repeat maintenance.
Conclusions
In summary, our data show that the Isw1 remodeler acts during transcription to control the stability of repetitive DNA, likely through reestablishment of proper chromatin structure after passage of RNAPII. Defects in this process lead to inappropriate excision repair and repeat instability. The function of ISWI remodelers is highly conserved in eukaryotic cells, and both the BER and TCR pathways have been shown to be causative for CAG expansions in mouse models (Kovtun et al. 2007, 2011; Goula et al. 2009; Liu et al. 2009; Hubert et al. 2011). The expansion mechanism operating in Isw1-defective cells was highly sensitive to transcription levels, with the greatest effect occurring at midrange transcription rates similar to those expected at many ORFs, including those containing expandable CAG repeats, and reduced effects for very low or very high transcription levels. Thus, subtle changes in transcription levels or chromatin structure could have magnified effects on the likelihood of repeat expansion. Why trinucleotide repeat expansions occur in particular cell types and developmental windows and not others is mysterious. There is evidence that expression levels of DNA repair factors (Goula et al. 2009; Mason et al. 2014) or chromatin states (Gorbunova et al. 2004; Libby et al. 2008; López Castel et al. 2011; Debacker et al. 2012; Gannon et al. 2012; House et al. 2014) are at play. Here, we show that the extent of Isw1 remodeling during transcription is an additional factor that determines repeat expansion frequencies.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300529/-/DC1.
Acknowledgments
We thank Jiahui Yang and Chen Li for help with generating strains, and Erica Polleys for experimental assistance with micrococcal nuclease assays. This research was supported by National Science Foundation grant MCB1330743 (to C.H.F.). M.R.K. was supported by the National Institutes of Health Institutional Research and Academic Career Development Awards Training in Education and Critical Research Skills postdoctoral program (K12 GM-074869).
Author contributions: M.R.K., N.C.M.H., and C.H.F. formulated the ideas, analyzed the data, and wrote the manuscript. The experiments were performed by M.R.K., N.C.M.H., C.M.C., R.M.J., C.G.S., C.E.J., E.A.P., and X.A.S. Funding was acquired by M.R.K. and C.H.F. C.H.F. was responsible for project administration.
Footnotes
Communicating editor: J. Surtees
Literature Cited
- Babour A., Shen Q., Dos-Santos J., Murray S., Gay A., et al. , 2016. The chromatin remodeler ISW1 is a quality control factor that surveys nuclear mRNP biogenesis. Cell 167: 1201–1214.e15. [DOI] [PubMed] [Google Scholar]
- Bowater R. P., Jaworski A., Larson J. E., Parniewski P., Wells R. D., 1997. Transcription increases the deletion frequency of long CTG.CAG triplet repeats from plasmids in Escherichia coli. Nucleic Acids Res. 25: 2861–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueckner F., Hennecke U., Carell T., Cramer P., 2007. CPD damage recognition by transcribing RNA polymerase II. Science 315: 859–862. [DOI] [PubMed] [Google Scholar]
- Budworth H., Harris F. R., Williams P., Lee D. Y., Holt A., et al. , 2015. Suppression of somatic expansion delays the onset of pathophysiology in a mouse model of Huntington’s disease. PLoS Genet. 11: e1005267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan J. L., Andrews K. J., Zakian V. A., Freudenreich C. H., 2003. Mutations in yeast replication proteins that increase CAG/CTG expansions also increase repeat fragility. Mol. Cell. Biol. 23: 7849–7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. H., Albuquerque C. P., Liang J., Suhandynata R. T., Zhou H., 2010. A proteome-wide analysis of kinase-substrate network in the DNA damage response. J. Biol. Chem. 285: 12803–12812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilli P., Ventura I., Minoprio A., Meccia E., Martire A., et al. , 2016. Oxidized dNTPs and the OGG1 and MUTYH DNA glycosylases combine to induce CAG/CTG repeat instability. Nucleic Acids Res. 44: 5190–5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier C. R., Cairns B. R., 2012. Regulation of ISWI involves inhibitory modules antagonized by nucleosomal epitopes. Nature 492: 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concannon C., Lahue R. S., 2014. Nucleotide excision repair and the 26S proteasome function together to promote trinucleotide repeat expansions. DNA Repair (Amst.) 13: 42–49. [DOI] [PubMed] [Google Scholar]
- Corona D. F., Clapier C. R., Becker P. B., Tamkun J. W., 2002. Modulation of ISWI function by site-specific histone acetylation. EMBO Rep. 3: 242–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debacker K., Frizzell A., Gleeson O., Kirkham-McCarthy L., Mertz T., et al. , 2012. Histone deacetylase complexes promote trinucleotide repeat expansions. PLoS Biol. 10: e1001257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion V., Wilson J. H., 2009. Instability and chromatin structure of expanded trinucleotide repeats. Trends Genet. 25: 288–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdel F., Schubert T., Marth C., Langst G., Rippe K., 2010. Human ISWI chromatin-remodeling complexes sample nucleosomes via transient binding reactions and become immobilized at active sites. Proc. Natl. Acad. Sci. USA 107: 19873–19878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon A. M., Frizzell A., Healy E., Lahue R. S., 2012. MutSβ and histone deacetylase complexes promote expansions of trinucleotide repeats in human cells. Nucleic Acids Res. 40: 10324–10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellon L., Razidlo D. F., Gleeson O., Verra L., Schulz D., et al. , 2011. New functions of Ctf18-RFC in preserving genome stability outside its role in sister chromatid cohesion. PLoS Genet. 7: e1001298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkikopoulos T., Schofield P., Singh V., Pinskaya M., Mellor J., et al. , 2011. A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science 333: 1758–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godde J. S., Wolffe A. P., 1996. Nucleosome assembly on CTG triplet repeats. J. Biol. Chem. 271: 15222–15229. [DOI] [PubMed] [Google Scholar]
- Goldmark J. P., Fazzio T. G., Estep P. W., Church G. M., Tsukiyama T., 2000. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103: 423–433. [DOI] [PubMed] [Google Scholar]
- Gorbunova V., Seluanov A., Mittelman D., Wilson J. H., 2004. Genome-wide demethylation destabilizes CTG.CAG trinucleotide repeats in mammalian cells. Hum. Mol. Genet. 13: 2979–2989. [DOI] [PubMed] [Google Scholar]
- Goula A. V., Berquist B. R., Wilson D. M., III, Wheeler V. C., Trottier Y., et al. , 2009. Stoichiometry of base excision repair proteins correlates with increased somatic CAG instability in striatum over cerebellum in Huntington’s disease transgenic mice. PLoS Genet. 5: e1000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüne T., Brzeski J., Eberharter A., Clapier C. R., Corona D. F., et al. , 2003. Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol. Cell 12: 449–460. [DOI] [PubMed] [Google Scholar]
- House N. C., Yang J. H., Walsh S. C., Moy J. M., Freudenreich C. H., 2014. NuA4 initiates dynamic histone H4 acetylation to promote high-fidelity sister chromatid recombination at postreplication gaps. Mol. Cell 55: 818–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert L., Jr., Lin Y., Dion V., Wilson J. H., 2011. Xpa deficiency reduces CAG trinucleotide repeat instability in neuronal tissues in a mouse model of SCA1. Hum. Mol. Genet. 20: 4822–4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., et al. , 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21: 947–962. [DOI] [PubMed] [Google Scholar]
- Jung J., Bonini N., 2007. CREB-binding protein modulates repeat instability in a Drosophila model for polyQ disease. Science 315: 1857–1859. [DOI] [PubMed] [Google Scholar]
- Kent N. A., Eibert S. M., Mellor J., 2004. Cbf1p is required for chromatin remodeling at promoter-proximal CACGTG motifs in yeast. J. Biol. Chem. 279: 27116–27123. [DOI] [PubMed] [Google Scholar]
- Kim N., Jinks-Robertson S., 2010. Abasic sites in the transcribed strand of yeast DNA are removed by transcription-coupled nucleotide excision repair. Mol. Cell. Biol. 30: 3206–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun I. V., Liu Y., Bjoras M., Klungland A., Wilson S. H., et al. , 2007. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature 447: 447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun I. V., Johnson K. O., McMurray C. T., 2011. Cockayne syndrome B protein antagonizes OGG1 in modulating CAG repeat length in vivo. Aging (Albany N.Y.) 3: 509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon A., Petty E., Pillus L., 2012. Functional antagonism between Sas3 and Gcn5 acetyltransferases and ISWI chromatin remodelers. PLoS Genet. 8: e1002994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y., Budworth H., Beaver J. M., Chan N. L., Zhang Z., et al. , 2016. Crosstalk between MSH2–MSH3 and polβ promotes trinucleotide repeat expansion during base excision repair. Nat. Commun. 7: 12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lans H., Marteijn J. A., Schumacher B., Hoeijmakers J. H., Jansen G., et al. , 2010. Involvement of global genome repair, transcription coupled repair, and chromatin remodeling in UV DNA damage response changes during development. PLoS Genet. 6: e1000941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby R. T., Hagerman K. A., Pineda V. V., Lau R., Cho D. H., et al. , 2008. CTCF cis-regulates trinucleotide repeat instability in an epigenetic manner: a novel basis for mutational hot spot determination. PLoS Genet. 4: e1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Wilson J. H., 2007. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol. Cell. Biol. 27: 6209–6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Wilson J. H., 2012. Nucleotide excision repair, mismatch repair, and R-loops modulate convergent transcription-induced cell death and repeat instability. PLoS One 7: e46807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Dion V., Wilson J. H., 2006. Transcription promotes contraction of CAG repeat tracts in human cells. Nat. Struct. Mol. Biol. 13: 179–180. [DOI] [PubMed] [Google Scholar]
- Lin Y., Dent S. Y., Wilson J. H., Wells R. D., Napierala M., 2010. R loops stimulate genetic instability of CTG.CAG repeats. Proc. Natl. Acad. Sci. USA 107: 692–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom K. C., Vary J. C., Jr., Parthun M. R., Delrow J., Tsukiyama T., 2006. Isw1 functions in parallel with the NuA4 and Swr1 complexes in stress-induced gene repression. Mol. Cell. Biol. 26: 6117–6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Wilson S. H., 2012. DNA base excision repair: a mechanism of trinucleotide repeat expansion. Trends Biochem. Sci. 37: 162–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Prasad R., Beard W. A., Hou E. W., Horton J. K., et al. , 2009. Coordination between polymerase beta and FEN1 can modulate CAG repeat expansion. J. Biol. Chem. 284: 28352–28366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López Castel A., Cleary J. D., Pearson C. E., 2010. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat. Rev. Mol. Cell Biol. 11: 165–170. [DOI] [PubMed] [Google Scholar]
- López Castel A., Nakamori M., Tomé S., Chitayat D., Gourdon G., et al. , 2011. Expanded CTG repeat demarcates a boundary for abnormal CpG methylation in myotonic dystrophy patient tissues. Hum. Mol. Genet. 20: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltby V. E., Martin B. J., Schulze J. M., Johnson I., Hentrich T., et al. , 2012. Histone H3 lysine 36 methylation targets the Isw1b remodeling complex to chromatin. Mol. Cell. Biol. 32: 3479–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason A. G., Tome S., Simard J. P., Libby R. T., Bammler T. K., et al. , 2014. Expression levels of DNA replication and repair genes predict regional somatic repeat instability in the brain but are not altered by polyglutamine disease protein expression or age. Hum. Mol. Genet. 23: 1606–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray C. T., 2010. Mechanisms of trinucleotide repeat instability during human development. Nat. Rev. Genet. 11: 786–799 (erratum: Nat. Rev. Genet. 11: 886). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollersen L., Rowe A. D., Illuzzi J. L., Hildrestrand G. A., Gerhold K. J., et al. , 2012. Neil1 is a genetic modifier of somatic and germline CAG trinucleotide repeat instability in R6/1 mice. Hum. Mol. Genet. 21: 4939–4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillon A., Karabetsou N., O’Sullivan J., Kent N., Proudfoot N., et al. , 2003. Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA polymerase II. Cell 115: 425–435. [DOI] [PubMed] [Google Scholar]
- Morillon A., Karabetsou N., Nair A., Mellor J., 2005. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol. Cell 18: 723–734. [DOI] [PubMed] [Google Scholar]
- Mueller J. E., Bryk M., 2007. Isw1 acts independently of the Isw1a and Isw1b complexes in regulating transcriptional silencing at the ribosomal DNA locus in Saccharomyces cerevisiae. J. Mol. Biol. 371: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Planitz F., Klinker H., Ludwigsen J., Becker P. B., 2013. The ATPase domain of ISWI is an autonomous nucleosome remodeling machine. Nat. Struct. Mol. Biol. 20: 82–89. [DOI] [PubMed] [Google Scholar]
- Nakanishi S., Prasad R., Wilson S. H., Smerdon M., 2007. Different structural states in oligonucleosomes are required for early vs. late steps of base excision repair. Nucleic Acids Res. 35: 4313–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocampo J., Chereji R. V., Eriksson P. R., Clark D. J., 2016. The ISW1 and CHD1 ATP-dependent chromatin remodelers compete to set nucleosome spacing in vivo. Nucleic Acids Res. 44: 4625–4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniewski P., Bacolla A., Jaworski A., Wells R. D., 1999. Nucleotide excision repair affects the stability of long transcribed (CTG*CAG) tracts in an orientation-dependent manner in Escherichia coli. Nucleic Acids Res. 27: 616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson E., Moore C., 2014. The evolutionarily conserved Pol II flap loop contributes to proper transcription termination on short yeast genes. Cell Rep. 9: 821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty E., Pillus L., 2013. Balancing chromatin remodeling and histone modifications in transcription. Trends Genet. 29: 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinskaya M., Nair A., Clynes D., Morillon A., Mellor J., 2009. Nucleosome remodeling and transcriptional repression are distinct functions of Isw1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 29: 2419–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polleys E. J., House N. C. M., Freudenreich C. H., 2017. Role of recombination and replication fork restart in repeat instability. DNA Repair (Amst.) 56: 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyzos A. A., McMurray C. T., 2017. Close encounters: moving along bumps, breaks, and bubbles on expanded trinucleotide tracts. DNA Repair (Amst.) 56: 144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy K., Tam M., Bowater R. P., Barber M., Tomlinson M., et al. , 2011. Determinants of R-loop formation at convergent bidirectionally transcribed trinucleotide repeats. Nucleic Acids Res. 39: 1749–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy K., Schmidt M. H., Geist J. M., Thakkar N. P., Panigrahi G. B., et al. , 2014. Processing of double-R-loops in (CAG).(CTG) and C9orf72 (GGGGCC).(GGCCCC) repeats causes instability. Nucleic Acids Res. 42: 10473–10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider R., Bernstein B. E., Karabetsou N., Morillon A., et al. , 2003. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol. Cell 12: 1325–1332. [DOI] [PubMed] [Google Scholar]
- Schmidt M. H., Pearson C. E., 2016. Disease-associated repeat instability and mismatch repair. DNA Repair (Amst.) 38: 117–126. [DOI] [PubMed] [Google Scholar]
- Schumacher S., Pinet I., Bichara M., 2001. Modulation of transcription reveals a new mechanism of triplet repeat instability in Escherichia coli. J. Mol. Biol. 307: 39–49. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M., Ishii H., Sun J. M., Pazin M. J., Davie J. R., et al. , 2006. Histone H4–K16 acetylation controls chromatin structure and protein interactions. Science 311: 844–847. [DOI] [PubMed] [Google Scholar]
- Smolle M., Venkatesh S., Gogol M. M., Li H., Zhang Y., et al. , 2012. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat. Struct. Mol. Biol. 19: 884–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevnsner T., Muftuoglu M., Aamann M. D., Bohr V. A., 2008. The role of Cockayne syndrome group B (CSB) protein in base excision repair and aging. Mech. Ageing Dev. 129: 441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X. A., Freudenreich C. H., 2017. Cytosine deamination and base excision repair cause R-loop-induced CAG repeat fragility and instability in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 114: E8392–E8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararajan R., Gellon L., Zunder R. M., Freudenreich C. H., 2010. Double-strand break repair pathways protect against CAG/CTG repeat expansions, contractions and repeat-mediated chromosomal fragility in Saccharomyces cerevisiae. Genetics 184: 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T., Palmer J., Landel C. C., Shiloach J., Wu C., 1999. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 13: 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuo J., Muftuoglu M., Chen C., Jaruga P., Selzer R. R., et al. , 2001. The Cockayne syndrome group B gene product is involved in general genome base excision repair of 8-hydroxyguanine in DNA. J. Biol. Chem. 276: 45772–45779. [DOI] [PubMed] [Google Scholar]
- Usdin K., House N. C., Freudenreich C. H., 2015. Repeat instability during DNA repair: insights from model systems. Crit. Rev. Biochem. Mol. Biol. 50: 142–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vary J. C., Jr., Gangaraju V. K., Qin J., Landel C. C., Kooperberg C., et al. , 2003. Yeast Isw1p forms two separable complexes in vivo. Mol. Cell. Biol. 23: 80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volle C. B., Delaney S., 2012. CAG/CTG repeats alter the affinity for the histone core and the positioning of DNA in the nucleosome. Biochemistry 51: 9814–9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. H., Griffith J., 1995. Expanded CTG triplet blocks from the myotonic dystrophy gene create the strongest known natural nucleosome positioning elements. Genomics 25: 570–573. [DOI] [PubMed] [Google Scholar]
- Whitehouse I., Rando O. J., Delrow J., Tsukiyama T., 2007. Chromatin remodelling at promoters suppresses antisense transcription. Nature 450: 1031–1035. [DOI] [PubMed] [Google Scholar]
- Wu L., Winston F., 1997. Evidence that Snf-Swi controls chromatin structure over both the TATA and UAS regions of the SUC2 promoter in Saccharomyces cerevisiae. Nucleic Acids Res. 25: 4230–4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav T., Whitehouse I., 2016. Replication-coupled nucleosome assembly and positioning by ATP-dependent chromatin-remodeling enzymes. Cell Rep. 15: 715–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Frouws T. D., Angst B., Fitzgerald D. J., DeLuca C., et al. , 2011. Structure and mechanism of the chromatin remodelling factor ISW1a. Nature 472: 448–453. [DOI] [PubMed] [Google Scholar]
- Yang J. H., Freudenreich C. H., 2010. The Rtt109 histone acetyltransferase facilitates error-free replication to prevent CAG/CTG repeat contractions. DNA Repair (Amst.) 9: 414–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner G. E., Tsukiyama T., Henikoff S., 2013. ISWI and CHD chromatin remodelers bind promoters but act in gene bodies. PLoS Genet. 9: e1003317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. N., Usdin K., 2015. The repeat expansion diseases: the dark side of DNA repair. DNA Repair (Amst.) 32: 96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Yeast strains used in this study are listed in Table S1 in File S1. Strains are available upon request. Raw instability data and associated P-values are listed in Table S2 in File S1.