Abstract
Centromeres are the chromosomal sites of assembly for kinetochores, the protein complexes that attach to spindle fibers and mediate separation of chromosomes to daughter cells in mitosis and meiosis. In most multicellular organisms, centromeres comprise a single specific family of tandem repeats—often 100–400 bp in length—found on every chromosome, typically in one location within heterochromatin. Drosophila melanogaster is unusual in that the heterochromatin contains many families of mostly short (5–12 bp) tandem repeats, none of which appear to be present at all centromeres, and none of which are found only at centromeres. Although centromere sequences from a minichromosome have been identified and candidate centromere sequences have been proposed, the DNA sequences at native Drosophila centromeres remain unknown. Here we use native chromatin immunoprecipitation to identify the centromeric sequences bound by the foundational kinetochore protein cenH3, known in vertebrates as CENP-A. In D. melanogaster, these sequences include a few families of 5- and 10-bp repeats; but in closely related D. simulans, the centromeres comprise more complex repeats. The results suggest that a recent expansion of short repeats has replaced more complex centromeric repeats in D. melanogaster.
Keywords: ChIP, Cid, rotational phasing, prod
THE separation of chromosomes in mitosis and meiosis is orchestrated by the kinetochore, a protein complex usually found at one location on each chromosome, termed the centromere. The kinetochore attaches chromosomes to spindle microtubules and mediates alignment on the metaphase plate, senses tension, and controls entry into anaphase (Musacchio and Desai 2017; Salmon and Bloom 2017). A key protein of the kinetochore is a centromeric variant of histone H3 (cenH3), which forms specialized nucleosomes that wrap centromeric DNA (Steiner and Henikoff 2015). Despite their conserved function, both centromeres and cenH3s evolve rapidly (Henikoff et al. 2001; Rosin and Mellone 2017), with little conservation of centromere sequences between closely related species (Lee et al. 2005; Gong et al. 2012). Despite sequence divergence, in most plants and animals centromeres have a common organization: they are embedded in heterochromatin and typically comprise megabase-scale arrays of tandem repeats (satellite DNAs) that are recalcitrant to genome assembly methods, with repeat monomers often of lengths of ∼100–400 bp (Melters et al. 2013). A single repeat family typically dominates the centromeres of all chromosomes in a species, and is partially occupied by cenH3 nucleosomes (Schueler et al. 2001; Cheng et al. 2002; Zhong et al. 2002; Nagaki et al. 2003; Henikoff et al., 2015).
This pattern does not apply to Drosophila melanogaster. Instead, major satellite repeats are mostly tandem arrays of very short 5- to 12-bp sequences, often following the pattern RRNRN or RRNRNRN, where R is a purine and N is any nucleotide (Lohe and Brutlag 1986). The distribution of these short satellites, which have been mapped by in situ hybridization to specific bands in heterochromatin (Lohe et al. 1993), appears to preclude the possibility of a single repeat family found at the centromere of every chromosome. Nor does it appear that any satellite sequence is restricted to centromeres. A summary of localization of selected satellites in prior studies is given in Table 1.
Table 1. Selected candidate centromeric satellite repeats in D. melanogaster.
| Repeat | Prior localizationa | Localization confirmed by FISH in this report | Anti-Cid ChIP enrichment in this report |
|---|---|---|---|
| AATAT | X,Y, 4; centromeric (X)b | X, 4; centromeric (X) | Yes |
| AATAG | X, Y, 2, 4; noncentromeric | 2; noncentromeric | Yes |
| AAGAG | X, Y, 2, 3, 4; centromeric (X)c | Noncentromeric (X) | No |
| Prodsat: AATAACATAG | 2, 3; pericentromericd | — | Yes |
| Dodecasatellite: CGGTCCCGTACT or GGTCCCGTACT | 3; centromerice | — | No |
| 359-bp repeat | X; noncentromericb | — | No |
The centromere of the D. melanogaster minichromosome Dp 1187 which is derived from In(1)sc8, an inversion of the X chromosome, was one of the first metazoan centromeres investigated in molecular detail (Le et al. 1995; Murphy and Karpen 1995; Sun et al. 1997, 2003). Deletion derivatives defined a 440-kb region that was necessary for full centromere function, and which encompassed complex DNA from transposons embedded in uniform arrays of the satellites AATAT and TTCTC (AAGAG) from left to right. Fluorescent in situ hybridization (FISH) mapping of AAGAG on the pericentric inversion In(1LR)pn2b demonstrated that this pentamer is present on the short right arm of the X chromosome (Koryakov et al. 1999). Heterochromatic banding patterns on a series of secondary rearrangements of the paracentric inversion In(1)pn2a mapped the centromere to the heterochromatic band corresponding to AATAT (Tolchkov et al. 2000). This cast doubt on whether the AAGAG satellite is required for normal X centromere function, and raised the question of whether AATAT is found at centromeres on other chromosomes. AATAT is distributed throughout chromosome 4, but is generally undetectable by in situ hybridization on chromosome 2, and does not appear to be centromeric on chromosome 3 (Lohe et al. 1993; Jagannathan et al. 2017); indicating that it is most likely not at the centromeres of these metacentric autosomes.
The centromere of chromosome 3 was mapped by using a SuUR mutant or the double mutant SuUR Su(var)3-906, both of which suppress underreplication of heterochromatic sequences in salivary gland polytene chromosomes, and also by using otu mutants, which polytenize chromosomes of pseudonurse cells. The centromere was found to be the constriction between blocks of the 10-mer AATAACATAG and the 11- or 12-mer GGTCCCGTACT or CGGTCCCGTACT, known as dodecasatellite (Koryakov et al. 2002; Andreyeva et al. 2007; Garavís et al. 2015). The AATAACATAG 10-mer is also known as Prodsat because, during mitosis, it is specifically bound by the protein encoded by proliferation disruptor (prod) (Platero et al. 1998; Torok et al. 2000). prod mutants have defects in chromosome condensation near centromeres 2 and 3 and in anaphase chromatid separation (Torok et al. 1997). Simultaneous immunolocalization of the Prod protein with the Drosophila CENP-A protein found that they are immediately adjacent on chromosomes 2 and 3 (Blower and Karpen 2001), indicating that both proteins may occupy Prodsat at the centromeres of chromosomes 2 and 3. Alternatively, colocalization of CENP-A with dodecasatellite on chromatin fibers suggested that dodecasatellite may be all or part of the centromere on chromosome 3 (Garavís et al. 2015).
In the sibling species D. simulans, which has been separated from D. melanogaster by ∼5 MY (Tamura et al. 2004), dodecasatellite is similarly near the centromeres of chromosomes 2 and 3 (Carmena et al. 1993; Jagannathan et al. 2017). This is in contrast to many other satellites, whose locations change dramatically in D. simulans.
A common method for identifying centromere sequences in other organisms has been chromatin immunoprecipitation (ChIP) of cenH3. Because immunoprecipitates (IPs) of ChIP experiments always have background DNA from the whole genome, centromere sequences are identified as those that are quantitatively enriched over the same sequences in the input DNA (Takahashi et al. 2000; Zhong et al. 2002; Nagaki et al. 2003, 2004; Marques et al. 2015). Here we use a native ChIP protocol to identify centromere sequences in D. melanogaster and D. simulans. We find that a few families of tandem repeats, including AATAT, AATAG, and Prodsat, are enriched at D. melanogaster centromeres. In D. simulans, we find larger complex tandem repeats at the centromeres. We show that centromere repeats have been expanding in both species. Our results indicate rapid divergence of centromeres in these species and suggest that small repeats are replacing older, complex repeats, especially in D. melanogaster.
Materials and Methods
Nomenclature
The Drosophila cenH3 variant is encoded by the centromere identifier (cid) locus. Early phylogenetic analyses failed to establish orthology of the Cid protein with the vertebrate cenH3 protein CENP-A (Malik and Henikoff 2003; Baker and Rogers 2006; Dawson et al. 2007; Postberg et al. 2010); however, recent sequencing of cenH3 genes from a broad range of insects supported their orthology with CENP-A (Drinnenberg et al. 2014), and here we refer to the Cid protein as CENP-A. We continue to refer to the antibodies used to bind Drosophila CENP-A as anti-Cid antibodies because they cross-react with CENP-A proteins only in D. melanogaster and close relatives. We use the traditional designations (Carmena et al. 1993; Lohe et al. 1993; Torok et al. 2000) for common short satellites (Table 1), and the terms “simcent1” and “simcent2” for newly identified centromere sequences in D. simulans.
Fly stocks and cell lines
The embryonic cell lines S2-DRSC from D. melanogaster (hereafter S2) and ML82-19a from D. simulans were obtained from the Drosophila Genomics Resource Center, grown as recommended, and used for ChIP. S2 cells are approximately tetraploid for chromosomes 2 and 3, triploid for the X, and diploid for chromosome 4, with some variability (Lee et al. 2014). ML82-19a cells are diploid for autosomes with a single X chromosome (Supplemental Material, Figure S1A). Neither line has a Y chromosome, and our cell line experiments cannot address the nature of the Y centromere.
A Cid-GFP construct (Henikoff et al. 2000) was injected into w1118 flies and a stable line, P[Cid-GFP]8-10, was obtained in which CENP-A-GFP localizes to D. melanogaster centromeres (Figure S1B). CENP-A-GFP is undetectable by Western blots in protein extracts of these flies using either anti-Cid or anti-GFP antibodies; however endogenous CENP-A is detected with anti-CidM antibody, which we take as evidence that CENP-A-GFP has a very low expression level. T(1;3)eH2, ev/In(3R)C, e l(3)e was previously described (Henikoff 1980). Oregon R (D. melanogaster) was obtained from the Bloomington Stock Center, and w501 (D. simulans) was a gift from H. Malik.
Antibodies, immunocytology, and microscopy
Two independent rabbit anti-Cid antibodies were used to confirm results in D. melanogaster. The anti-CidH antibody, raised to the epitope acetyl-CAKRAPRPSANNSKSPNDD-amide, has been previously described (Henikoff et al. 2000). The anti-CidM antibody, raised to the epitope MPRHSRAKRAPRPSA, is a gift from H. Malik. Although the amino acid sequences of CENP-A in both D. melanogaster and D. simulans are identical in the region of the epitopes, only anti-CidM localizes to centromeres in D. simulans tissue culture cells and larval brains (Figures S1 and S2). Chicken anti-GFP antibody was from ThermoFisher Scientific (Waltham, MA). Anti-phospho-H3S10 antibody was Millipore 06-570 (Billerica, MA).
Dissection and fixation of larval brains for antibody detection followed (Larracuente and Ferree 2015), with the omission of acetic acid, which interferes with detection of CENP-A for both anti-Cid antibodies. Slides were collected in PBS with Tween 20 (PBST), blocked with Odyssey Block or PBST plus 10% goat serum for 30 min, and then incubated for 1 hr at room temperature or overnight at 4° with anti-Cid or anti-GFP antibodies diluted 1:1000 in blocking solution. Slides were then washed 3× 5 min with blocking solution, incubated for 1 hr at room temperature with a fluorescent secondary antibody (Jackson ImmunoResearch) diluted 1:100 in blocking solution, washed again 3× 5 min in blocking solution or PBS, stained with DAPI solution, mounted with a cover glass using Vectashield, and sealed with nail polish. Antibody detection in tissue culture cells was similar, except that cells were allowed to adhere to coverslips for at least 1 hr before swelling with sodium citrate and spinning coverslips in a Cytospin 4 at 1900 rpm for 1 min before fixing in 2% formaldehyde for 15 min. Subsequent steps were the same as for brains, except carried out on coverslips. Chromosomes were visualized using a DeltaVision microscope and software (Applied Precision, now GE Healthcare). Images were false colored using ImageJ (Schneider et al. 2012).
FISH
End-labeled fluorescent oligonucleotide probes and unlabeled probes were ordered from Eurofins Genomics (Louisville, KY). Unlabeled oligonucleotide probes were labeled with digoxigenin-11-dUTP (Roche) or biotin-16-dUTP using terminal deoxynucleotidyl transferase (Gibco/BRL) according to the manufacturer’s instructions. Probes for pentamers were 5′-(AATAT)10-3′, 5′-(AAGAG)10-3′, and 5′-[Alexa488]-(AATAG)10-3′. Probes for 10-mers were 5′-[Alexa594]-(AATAGAATTG)3-3′ and 5′-[Alexa594]- (AATAGAAGAG)3-3′. The simcent1 probe was 5′-[Alexa488]-AGTAAGTACTTATGTTGTTTTGATAATCGGCAATCAGACTC-3′.
Larval brain squashes were prepared and fixed as for immunolocalization, except that the fixative was 2% formaldehyde, 45% acetic acid. Hybridization was carried out according to Larracuente and Ferree (2015), or as follows: chromosomes were denatured in 0.07 M NaOH for 3 min, followed by a wash in 2× SSC for 5 min, dehydrated in 70% ethanol 2× 5 min and 95% ethanol 2× 5 min, and then allowed to air dry. Slides were prehybridized with hyb mix (50% formamide, 10% dextran sulfate, 3× SSC buffer) for 30 min at 25 or 37°. Probes (∼30 ng in 2 μl) were heated at 100° for 3 min, mixed with 12 μl of hyb mix, added to the slide, covered with a cover glass, sealed with rubber cement, and incubated in a moist chamber at 25° overnight (18° for AATAT). Slides were washed 2× 30 min in 50% formamide, 2× SSC, and then 3× 5 min in PBS or 0.1× SSC. For probes labeled with digoxigenin or biotin, slides were incubated for 1 hr with anti-digoxigenin fluorescent antibody or streptavidin-rhodamine and washed 3× 5 min in PBS. Slides were stained, mounted, and sealed as above. For combined CENP-A detection and FISH, immunolocalization and FISH protocols were carried out successively.
ChIP
Approximately 108 S2 cells or ML82-19a cells were harvested per sample and subjected to native ChIP in buffer containing 0.05% SDS to solubilize the kinetochore as described (Skene and Henikoff 2017). MNase digestion was for 5 or 15 min at 37°. Protein G Dynabeads were from Invitrogen (Carlsbad, CA). For embryo ChIP, 0- to 8-hr embryos were collected, washed and dechorionated as described (Steiner et al. 2012), and stored at −80°. Approximately 1 g of frozen embryos per sample was ground in liquid nitrogen, and nuclei were prepared as described (Steiner et al. 2012). Purified nuclei were then processed for ChIP as above (Skene and Henikoff 2017). Camelid GFP-nAb beads were obtained from Allele Biotechnology (San Diego, CA). Sequencing libraries were prepared as described (Neiman et al. 2012) using 14 PCR cycles with a 10 sec, 60° combined annealing/extension step.
Sequence analysis
The 250-bp, single-end reads from input and IP libraries of S2 cells were trimmed using Trim Galore! version 0.3.7 with the parameters: quality 20, adapter AGATCGGAAGAGC, stringency 3, phred 33, and length 25. For trimmed reads that occurred more than once, CD-HIT-EST version 4.6 (Fu et al. 2012) was used to cluster identical sequences, selecting the longest sequence, using the parameters -n 10 -r 1 -M 10000. The IP clusters with the most contributing reads were used to make reference sequences. Reads from input and IP libraries were mapped using Burrows–Wheeler Aligner (BWA) version 0.7.12-r1039 (Li and Durbin 2009) to the IP clusters, and the 100 clusters with the most mapped reads for input and IP were selected as reference sequences. For ML82-19a cells, reference sequences were generated from a 10-min MNase digest of chromatin. BWA was used to map 25- × 25-bp reads from input and IP libraries to the clusters. The 100 clusters with the most mapped reads for input and IP were selected as reference sequences. The reference sequences were analyzed for tandem repeats using the Tandem Repeats Finder server (https://tandem.bu.edu/trf/trf.html) with default parameters (Benson 1999).
The “grep” function of Unix was used to search for and count 9- to 16-mers and their reverse complements in raw Illumina sequencing reads. Counts of each candidate 9- to 16-mer were added to counts of its reverse complement, and then normalized to the total number of raw reads for either the input or IP libraries. Enrichment was calculated as the ratio of normalized IP counts to normalized input counts. Similar counts were made from sequencing data for Drosophila species downloaded from the Sequence Read Archive (SRA), normalizing to total reads to determine abundance. The National Center for Biotechnology Information Blast server was used to identify clones homologous to simcent1 and simcent2 sequences. Alignments were made using the Clustal Omega server of the European Molecular Biology Laboratory–European Bioinformatics Institute and adjusted by hand. Dot matrices were made with Dotter (Sonnhammer and Durbin 1995).
Data availability
Fly stocks and antibodies are available upon request. Figure S1 shows cell line karyotypes and centromere staining with anti-CidM and anti-GFP. Figure S2 shows hybridization of AATAG and 10-mer probes to D. simulans neuroblasts. File S1 and File S2 contain reference sequences for input and IP for S2 cells. File S3 and File S4 contain the reference sequences for ML82-19a cells. File S5 and File S6 display alignments of reference sequences containing simcent1 and simcent2, respectively. Table S1 lists the candidate 9- to 16-mers. Table S2 contains the SRA accessions counted for Drosophila species. Sequencing data are available at Gene Expression Omnibus with the accession number GSE105100.
Results
FISH mapping the X centromere of D. melanogaster
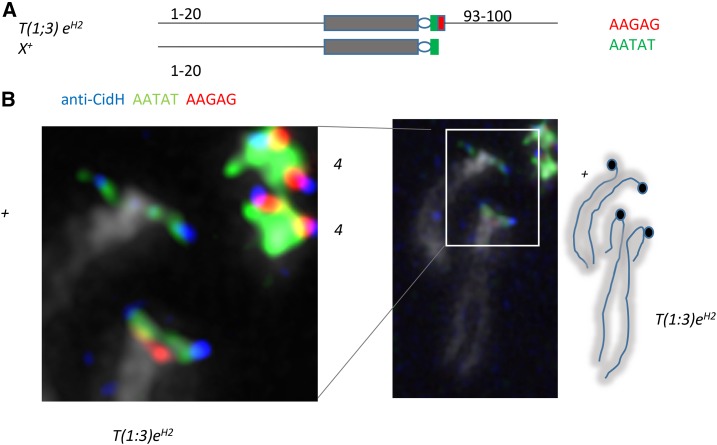
Previous studies had used chromosome rearrangements to map the X centromere in D. melanogaster to the heterochromatic band corresponding to a block of AATAT (Tolchkov et al. 2000), or used sequencing to map it to adjacent blocks of AATAT and CTCTT (= AAGAG) (Sun et al. 2003). To address this apparent discrepancy, we combined anti-CidH antibody staining with FISH to look at the translocation stock T(1;3)eH2/+, which has a rare break on the short right arm of the X chromosome (Figure 1A). Although the break does not separate AATAT and AAGAG repeats, fortuitously we found that the wild-type X chromosome in this stock lacks detectable AAGAG, which therefore appears to be unnecessary for the centromere. In chromosomes that appear to be under tension, the anti-CidH signal partially overlaps AATAT, apparently confirming that AATAT forms the X centromere (Figure 1B). In the same chromosome spread, partial overlap of AATAT, AAGAG, and anti-CidH signals on the fourth chromosome makes it unclear whether one or both of these pentamers are centromeric, underscoring the resolution limits of FISH for centromere mapping. We therefore turned to ChIP sequencing (ChIP-seq) to identify sequences directly bound by Drosophila CENP-A.
Figure 1.
AAGAG is not necessary for centromere function on the D. melanogaster X chromosome. (A) Translocation T(1;3) eH2 breaks on the short right arm of the X chromosome and at 93 on chromosome 3. (B) The wild-type X in the stock T(1;3) eH2/In(3R)C lacks detectable AAGAG and CENP-A signal overlaps with AATAT signal. Hybridization signal is not found under all of the anti-CidH signal, suggesting that the kinetochore interferes with hybridization.
D. melanogaster centromeres comprise 5- and 10-mers
ChIP for centromeres typically uses quantitative PCR to determine the enrichment of candidate centromeric repeats in the IP from an anti-cenH3 antibody, relative to the input DNA. This approach is not feasible when the candidate repeats are shorter than a typical PCR primer length. Instead, we used a counting approach to quantify the numbers of candidate sequence strings in raw reads in libraries made from the input and IP from ChIP experiments, using chromatin from the D. melanogaster cell line S2. To identify centromeric sequences in as unbiased a manner as possible, we selected as candidate sequences 9- to 16-mers (and their reverse complements) representing the 32 most abundant sequences identified in D. melanogaster by k-mers of 31 bp (Krassovsky and Henikoff 2014). The 9- to 16-mer candidate sequences consist of three or two tandem copies of 3- to 8-mer repeats or single copies of 10- to 15-mer repeats.
To identify additional candidate centromere sequences, we performed native ChIP-seq of CENP-A with 250-bp, single-end reads. Identical trimmed reads from the IP were clustered, selecting the longest read in a cluster. Both input and IP reads were mapped to the clusters (File S1 and File S2), ranking the clusters by the number of reads mapping to them, and selecting the top 100 clusters for both input and IP as reference sequences (Henikoff et al. 2015). Of the 100 reference sequences for the IP, 86 had at least two tandem copies of the Prodsat 10-mer AATAACATAG, whereas this sequence was in only 29 of the input reference sequences. A 20-mer of AATAG (four tandem copies) was found in five reference sequences of the IP compared with one input sequence. Together, these two repeats were found in 91 of the 100 IP reference sequences. We used Tandem Repeats Finder (Benson 1999) to identify additional repeats in the IP reference sequences. We found three tandem dimers or trimers that were unrepresented in the 32 most abundant short repeats and were apparently derived from and interspersed with Prodsat. We added these to our collection of candidate sequences, along with 10-mer negative controls selected from 5S DNA and from the complex 359-bp repeat family, which is not centromeric (Tolchkov et al. 2000; Sun et al. 2003). We also included 10-mer candidate centromere sequences identified in the D. simulans cell line ML82-19a (described further below). In total, we identified 74 9- to 16-mers and their reverse complements (Table S1) as candidate centromere sequences and controls.
We performed additional native ChIP-seq experiments with 25- × 25-bp paired-end reads, and counted the 9- to 16-mers in the raw reads of the inputs and IPs of all experiments. The 9- to 16-mers that were enriched at least twofold in the IPs of at least three experiments are shown in Table 2. The three most abundant sequences enriched in the IPs are Prodsat, AATAG, and AATAT; consistent with both our IP reference sequences and our FISH mapping of the X centromere. Other enriched candidates are also likely to be centromeric, but are more minor components, each comprising <5% of the number of counts for Prodsat, the most abundant centromeric sequence. The dodeca satellite, previously proposed to be part of the centromere of chromosome 3, and the AAGAG repeat, found near the centromere of chromosome 2 and in Dp1187 derived from the X chromosome (Sun et al. 2003; Garavís et al. 2015), are consistently depleted in our IPs. We therefore conclude that Prodsat, AATAG, and AATAT are the major components of D. melanogaster centromeres.
Table 2. Enrichment of sequences in CENP-A ChIP experiments in S2 cells.
| Repeat unita | IP countb | IP/IN 1c | IP/IN 2d | IP/IN 3e | IP/IN 4b | Median | Minimum | Maximum |
|---|---|---|---|---|---|---|---|---|
| AATAACATAG (Prodsat) | 15,020,471 | 3.06 | 14.78 | 6.76 | 8.96 | 7.9 | 3.1 | 14.8 |
| AATAGAATAG | 2,162,779 | 13.47 | 11.69 | 14.61 | 15.42 | 14 | 11.7 | 15.4 |
| AATATAATAT | 1,147,564 | 60.48 | 1.76 | 3.25 | 6.88 | 5.1 | 1.8 | 60.5 |
| ATTATATTTT | 527,674 | 2.04 | 0.98 | 2.03 | 3.03 | 2.0 | 0.98 | 3.0 |
| AAGATAAGAT | 90,458 | 4.74 | 0.91 | 9.1 | 2.67 | 3.7 | 0.9 | 9.1 |
| AGAATAACATATAAC | 68,335 | 11.24 | 36.35 | 61.09 | 89.7 | 48.7 | 11.2 | 89.7 |
| ATAACATATAACAT | 65,591 | 19.28 | 4.97 | 0.61 | 7.15 | 6.1 | 0.6 | 19.3 |
Candidate sequences enriched at least twofold in at least three experiments. Enrichment is the ratio of normalized IP counts to normalized input counts. IN, input.
Counts of sequences not in bold are <5% of those of Prodsat.
15 min MNase, anti-CidM, 250-bp single-end reads.
5 min MNase, anti-CidM, 25- × 25-bp reads.
5 min 2 × MNase, anti-CidH, 25- × 25-bp reads.
15 min MNase, anti-CidM, 25- × 25-bp reads.
To verify that these three sequences represent centromeres in flies as well as in S2 cells, and that they can be immunoprecipitated using an independent epitope, we performed ChIP using an anti-GFP antibody on P[Cid-GFP]8-10 embryos. These flies express a CENP-A-GFP fusion protein at a very low level, which localizes to centromeres (Figure S1B). Because anti-GFP is known to enrich for certain sequences in ChIP experiments (Teytelman et al. 2013), we also performed anti-GFP ChIP on wild-type Oregon R embryos as a control, and subtracted enrichment in Oregon R from enrichment in P[Cid-GFP]8-10. The sequences enriched at least twofold (Table 3) substantially match the results from S2 cells, including enrichment for AATAT, AATAG, AAGAT, and Prodsat. Since half of the embryos are male, it is possible that differences between the relative abundance of sequences in ChIP from embryos and from S2 cells reflect the inclusion of the Y centromere in embryos and aneuploidy in S2 cells, though it is also possible that these differences are simply experimental variation. By FISH, AATAG maps distally on the Y, while Prodsat was not found to be on the Y, and AATAT is known from multiple locations on the Y (Lohe et al. 1993); suggesting that either AATAT or AAGAT are likely candidates for the Y centromere. Enrichment of Prodsat, AATAT, AATAG, and AAGAT in both S2 cells and embryos using unrelated antibodies leads us to conclude that these sequences are major components of natural D. melanogaster centromeres.
Table 3. Enrichment of sequences in GFP ChIP from embryos.
| Repeat unit | IP counta | Cid-GFP IP/IN − OR IP/INb |
|---|---|---|
| AATAACATAG (Prodsat) | 371,367 | 6.75 |
| AATATAATAT | 215,558 | 4.44 |
| AAGATAAGAT | 53,939 | 13.05 |
| AATAGAATAG | 45,962 | 24.51 |
| AATAGAATTG | 5,061 | 3.18 |
Counts of AATAGAATTG (not in bold) are <5% of those of Prodsat. IN, input; OR, Oregon R.
Counts from Cid-GFP IP.
5 min MNase, anti-GFP, 25- × 25-bp reads. Enrichment of sequences in the IP was calculated separately for Cid-GFP and Oregon R embryos, and then subtracted.
Enrichment of AATAT in centromeres was expected both from our FISH mapping of the X centromere (Figure 1) and previous mapping (Tolchkov et al. 2000). Likewise, Prodsat has previously been mapped by FISH in or very near to the primary constrictions of chromosomes 2 and 3 (Platero et al. 1998; Koryakov et al. 2002), and Drosophila CENP-A abuts Prod protein (Blower and Karpen 2001), which binds to Prodsat (Torok et al. 2000). In contrast, AATAG was previously mapped by in situ hybridization using tritiated probes to the left of the centromere on chromosome 2, on the distal long arm of the Y chromosome (which is absent in S2 cells), and in small amounts to chromosome 4 (Lohe et al. 1993). We confirmed using an (AATAG)10 fluorescent probe that the main site of AATAG hybridization on chromosome 2 is separated from the anti-Cid signal (Figure 2A).
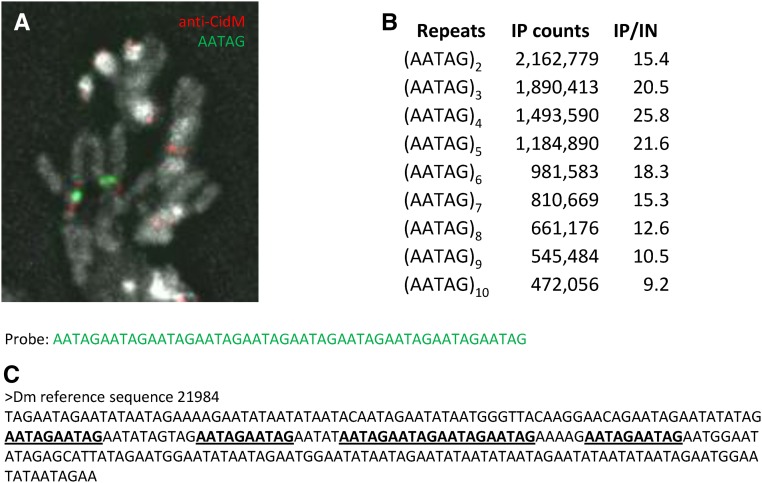
Figure 2.
AATAG repeats in D. melanogaster. (A) A fluorescent (AATAG)10 probe detects repeats left of the centromere on chromosome 2 in larval brain squashes. (B) Enrichment of varying numbers of (AATAG)n repeats in 250-bp reads from anti-CidM ChIP of S2 cells. (C) An example of a reference sequence containing (AATAG)2 repeats.
To address whether AATAG is present at centromeres in interrupted arrays that might not hybridize to our (AATAG)10 probe, we counted n-mers of (AATAG) in the 250-bp, single-end reads, with n ranging from 2 to 10 (Figure 2B). Although all n-mers were enriched in the IP relative to input, the greatest enrichment was for n = 4, indicating that tandem arrays of AATAG do seem to be predominantly interrupted with mismatched sequences in the IP. While 69% of the (AATAG)2 10-mers were also counted as (AATAG)4 20-mers, only 22% were counted as (AATAG)10 50-mers. Thus, we conclude that AATAG at centromeres is largely present as interrupted arrays rather than homogeneous arrays.
Four of the five (AATAG)4-containing reference sequences show interspersion of AATAG with other mismatched sequences, frequently AATAT, but not Prodsat (Figure 2C). The interrupting sequences generally maintain the 5-bp AANRN periodicity, as previously noted (Lohe and Brutlag 1987). In contrast to (AATAG)10 50-mers, which are present in only one of the five (AATAG)4-containing reference sequences, (AATAACATAG)5 50-mers occur in 68 of the 86 Prodsat-containing reference sequences. Only two of the Prodsat-containing reference sequences also have (AATAG)2, although the occurrence of a single AATAG among Prodsat repeats is fairly common. These patterns suggest that some or all of the (AATAG)2 counts in the ChIP may come from chromosome 4 (rich in AATAT), rather than chromosome 2 (rich in Prodsat).
D. simulans centromeres are enriched in complex sequences
Since centromeres evolve quickly (Henikoff et al. 2001), we also performed ChIP-seq on centromeres from the ML82-19a cell line from the sibling species D. simulans, which diverged from D. melanogaster ∼5 MYA (Tamura et al. 2004). Although D. simulans shares several repeats with D. melanogaster (Lohe and Roberts 1988; Jagannathan et al. 2017), others are not shared, and there has been no investigation of what sequences might be centromeric in D. simulans. We digested chromatin from ML82-19a cells with MNase for 10 min, made a library, and sequenced 250-bp, single-end reads, clustering them as before. We then performed ChIP-seq on ML82-19a chromatin and mapped 25- × 25-bp, paired-end reads from a 15 min MNase digest onto the clusters, selecting the 100 clusters with the most mapped reads for both input and IP as reference sequences from which to identify candidate centromeric sequences (File S3 and File S4).
Two complex sequence families that we termed simcent1 and simcent2 were present in 46 and 14 of the IP reference sequences, respectively, and one reference sequence contained sequence homologous to both. Neither of these families was found in the reference sequences for the input. Using Tandem Repeats Finder, we found that in both the simcent1 and simcent2 families, some but not all reference sequences have a local tandem duplication of a core sequence of ∼76 bp (simcent1) or 44 bp (simcent2) within the longer complex sequence. We also found that the 10-mer AATAGAATTG was in five of the IP reference sequences and none of the input sequences; whereas the 10-mer AATAGAAGAG was present in 27 of the IP sequences, and in 30 of the input sequences. An abundant 15-mer GAACAGAACATGTTC was present in 36 of the input sequences but in only one IP reference sequence, where it was a minor component at the end of AATAGAAGAG repeats. (AATAG)2 was present in 17 input and 13 IP sequences, predominantly interspersed within AATAGAAGAG or AATAGAATTG repeats. The 10-mer (AAGAG)2 was present in 31 of the input and 17 of the IP sequences, usually interspersed with AATAGAAGAG or in the sequence (AAGAG)2–3AACAA. Thus, while the D. simulans IP reference sequences contain both complex and simple repeats, only simcent1, simcent2, and the 10-mer AATAGAATTG are present in more IP sequences than input sequences.
We made alignments of the simcent1 and simcent2 reference sequences (File S5 and File S6) to select 10-mers from these sequences to count in the raw reads from the ChIP. From the simcent1 alignment, we selected 12 10-mers and their reverse complements, spanning a region of ∼200 bp that included the ∼76-bp duplicated sequence. From the simcent2 alignment, we selected five 10-mers and reverse complements, spanning 66 bp including both the 44-bp duplication and flanking sequence. We also selected the 10-mers AATAGAAGAG and AATAGAATTG, 10-mers from the abundant 15-mer repeat and one of its variants, and all the 9- to 16-mers from the most abundant short repeats of D. melanogaster plus 5S and 359-bp repeat controls, so that the same sequences were counted in both D. melanogaster and D. simulans ChIPs (Table S1). Sequences enriched at least twofold in the anti-CidM IP of two experiments are shown in Table 4. The complex simcent1 and simcent2 sequences were highly enriched and were the most abundant centromere sequences. The variability in the abundance of 10-mers nearly adjacent to one another in the simcent1 and simcent2 alignments probably reflects the relative conserva]tion of individual 10-mers across the repeat family, since we counted only exact matches. Among 5-mer and 10-mer tandem repeats, only AATAGAATTG was twofold enriched in two experiments; although (AATAT)2, (AATAG)2, (AAGAG)2, and AATAGAAGAG were all twofold enriched in one experiment, along with the 8-mers (ATATACAT)2 and (AATAATAT)2. Three additional 7-mers were enriched in all three experiments, but were <5% as abundant as most of the simcent1 and simcent2 10-mers (Table 4). We therefore conclude that simcent1 and simcent2 are the major centromere sequences in D. simulans, with some contribution from AATAGAATTG.
Table 4. Enrichment of sequences in CENP-A ChIP experiments in ML82-19a cells.
| 10- to 14-mera | IP countsb | IP/IN 1c | IP/IN 2c | IP/IN 3b | Repeat family |
|---|---|---|---|---|---|
| ACAATCGTTT | 285,765 | 23.28 | 27.68 | 56.12 | simcent2 |
| TTGCTTTGAG | 165,104 | 26.41 | 22.00 | 50.96 | simcent2 |
| ACTGCAACGC | 149,863 | 20.52 | 20.66 | 39.06 | simcent2 |
| TAATGGTTTT | 114,881 | 3.12 | 2.13 | 3.52 | simcent1 |
| TTGTGTTTAC | 105,305 | 3.31 | 3.46 | 5.48 | simcent1 |
| AGTACTTATG | 87,881 | 15.39 | 16.23 | 19.70 | simcent1 |
| TGATAATCGG | 68,580 | 47.69 | 36.80 | 53.00 | simcent1 |
| TTTAATATTA | 51,479 | 3.66 | 2.50 | 3.34 | simcent1 |
| AAATAACACT | 50,478 | 3.74 | 4.07 | 4.88 | simcent1 |
| ATTATATTTT | 48,229 | 3.56 | 2.35 | 4.16 | simcent1 |
| CAATCAGACT | 38,353 | 35.92 | 30.76 | 38.84 | simcent1 |
| AAAACTACTT | 34,493 | 2.49 | 2.09 | 5.64 | simcent2 |
| AGCATGCAAC | 25,443 | 5.24 | 4.90 | 9.77 | simcent2 |
| AATAGAATTG | 22,660 | 2.01 | 0.92 | 2.19 | 10-mer |
| TCTGCGAGCC | 22,372 | 8.87 | 7.55 | 16.25 | simcent1 |
| ATTAGCGTTT | 4,247 | 5.40 | 4.11 | 5.72 | simcent1 |
| AACAAATAACAAAT | 1,278 | 5.56 | 2.75 | 6.36 | 7-mer |
| ATATAATATATAAT | 238 | 19.70 | 2.11 | 2.15 | 7-mer |
| AATAGACAATAGAC | 24 | d | 9.32 | 41.75 | 7-mer |
IN, input.
Candidate sequences enriched at least twofold in at least two experiments. Enrichment is the ratio of normalized IP counts to normalized input counts. Counts of sequences not in bold are <5% of those of ACAATCGTTT, the most abundant sequence counted in the IP.
15 min MNase, anti-CidM, 25- × 25-bp reads.
5 min MNase, anti-CidM, 25- × 25-bp reads.
Enrichment undefined due to absence in input.
It is evident from the simcent1 alignment that the simcent1 sequence family exists in multiple configurations, and is subject to numerous indels. To better understand the nature of the simcent1 family, we used blastn to search the D. simulans genome assembly with the core regions of the simcent1 sequences to identify homologous clones. As in the reference sequences, the simcent1 sequences in the clones exist in multiple complex arrangements, but a ∼500-bp periodicity can be discerned in clones JPYS01004644 and JPYS01000309 from D. simulans and in numerous clones from D. sechellia such as CH676566 and CH677222 (Figure 3). A 500-bp repeat present in D. simulans and the closely related D. mauritiana was previously described (Strachan et al. 1982), and we suggest that the simcent1 family corresponds at least in part to this repeat family. The simcent2 family is found in a 299-bp repeat in clone JPYS01006388, however the size is inconsistent in other clones such as JPYS01003943. This family appears to be absent in blast searches of the D. melanogaster and D. sechellia genomes.
Figure 3.
Periodicity of simcent1 repeats. Dot matrix plots of similarity for four clones with homology to simcent1 compared against themselves reveal repeats with a periodicity of ∼500 bp.
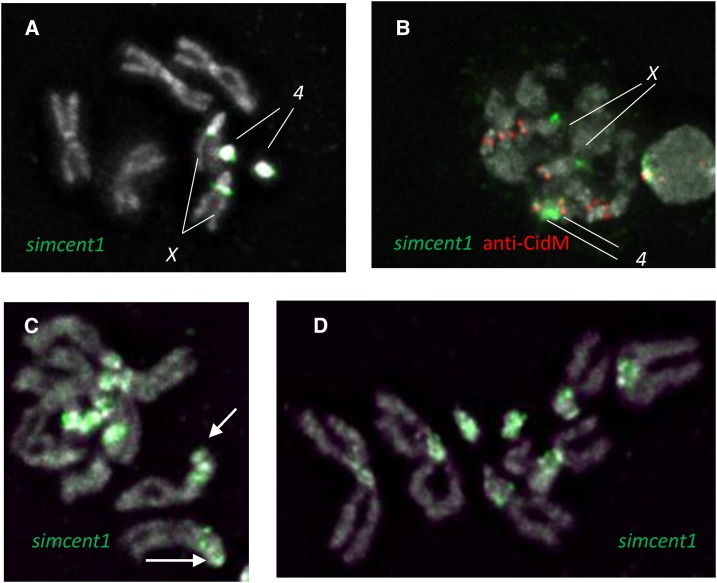
We used the core tandem duplication in the simcent1 sequence as a fluorescent probe for FISH to D. simulans chromosomes using formaldehyde fixation. This probe consistently hybridized to the X and 4 (Figure 4, A and B) in a pattern similar to that of a probe for (AATAG)10 (Figure S2, A and B). On the X chromosome, the simcent1 probe seems to hybridize as dots on the side of the X heterochromatin, remarkably reminiscent of centromere signals (Figure 4A). However, these dots are not at the centromeres, which are at the apparent end of the X chromosome, as in D. melanogaster (Figure 4B). In contrast, the simcent1 signal on chromosome 4 appears to touch the centromere. In hybridizations following fixation with formaldehyde and acetic acid, the simcent1 probe also hybridized more weakly to all D. simulans chromosomes (Figure 4, C and D); suggesting that it is detecting a diverged repeat family present in the centric heterochromatin of all chromosomes. Hybridization at the tip of the X (Figure 4C) may indicate the presence of diverged repeats at the X centromere. We are unable to use acetic acid with either of the anti-Cid antibodies as it eliminates anti-Cid signal, and we believe this variable hybridization of the simcent1 probe is the result of the absence or presence of acetic acid in the initial fixation, and that the weaker signal represents diverged repeats related to simcent1 present on all chromosomes.
Figure 4.
FISH of the simcent1 probe in D. simulans. (A) Hybridization of simcent1 to D. simulans larval neuroblasts using formaldehyde fixation. (B) simcent1 hybridization together with CENP-A detection. (C and D) Hybridization of simcent1 using formaldehyde fixation with 45% acetic acid. Arrow points to weak signal at the expected position of the X centromere.
We also made 30-bp probes for the 10-mers AATAGAATTG and AATAGAAGAG. Perhaps consistent with finding these 10-mers are interspersed with (AATAG)2 in the reference sequences, the (AATAGAATTG)3 probe has a hybridization pattern similar to (AATAG)10, with stronger hybridization to the fourth chromosome (where it may be centromeric) than to the noncentromeric site on the X (Figure S2C). The (AATAGAAGAG)3 probe hybridized strongly to the X and the tip of the Y, with no visible hybridization to any centromere (Figure S2D). Together these results are consistent with the conclusion that simcent1 is a major centromere component, while AATAGAATTG may be a centromere component on chromosome 4.
Different satellite repeats have expanded during divergence of Drosophila sibling species
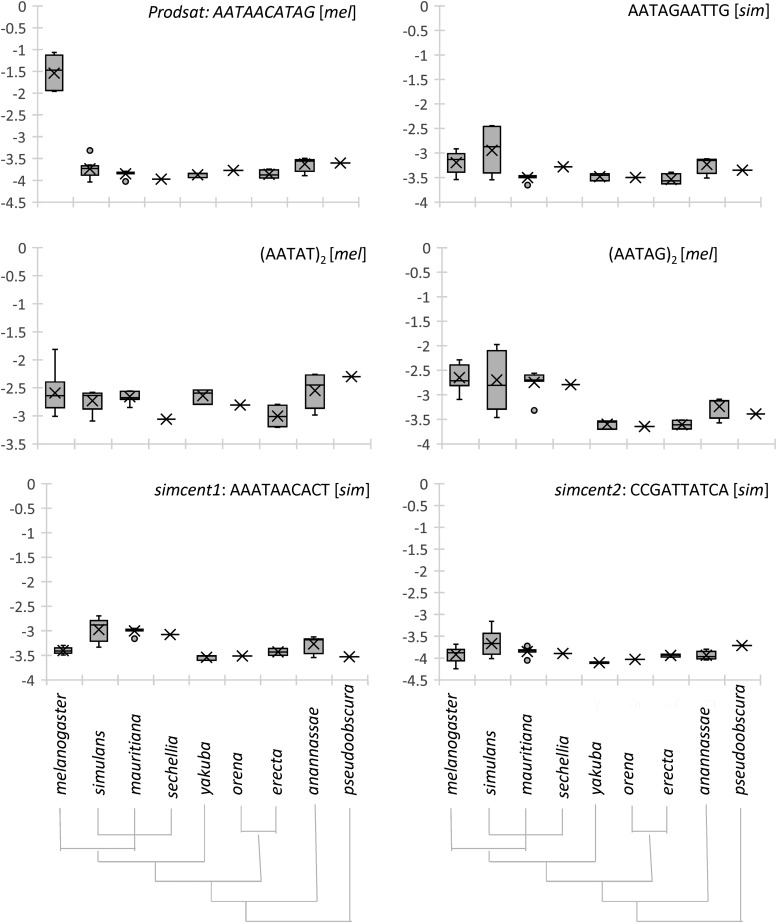
To better understand the evolution of the stark difference in centromeric repeats in D. melanogaster and D. simulans, we determined the abundance of these short and complex repeats in several Drosophila species with sequencing data available in the SRA (Figure 5). Although the amount of data differs for different species and there is substantial variation in data sets of the same species, several repeats appear to have expanded in the species in which they are enriched in centromeres. Most notably, Prodsat has expanded from its virtual absence in other species to comprise roughly 5 ± 2% of the genome in D. melanogaster. The simcent1 family, though far less abundant, appears to have expanded in the recently diverged simulans/sechellia/mauritiana clade in comparison with more distantly related species. Both simcent2 and AATAGAATTG have also expanded in D. simulans. (AATAG)2 and (AATAT)2 are slightly expanded in D. melanogaster compared with its closest relatives, although (AATAG)2 is comparable in D. simulans. Thus, most centromere sequences in these two species have undergone recent expansions.
Figure 5.
Abundance of simple and complex repeats in nine Drosophila species. All species are in the melanogaster subgroup, except D. ananassae, which is in the melanogaster group, and D. pseudoobscura, which serves as an outgroup. The cladograms at the bottom depict the relationships of the species. The y-axis represents the fraction of the reads that contain the repeat, presented on log10 scale. In the box and whisker plots, the box ends represent the second and third quartiles of the data separated by the median, while the whiskers represent the first and fourth quartiles. The mean is marked by X. For each centromere-enriched repeat, [mel] and [sim] indicate that in the corresponding species, the abundance of the repeat in anti-Cid IP was at least 5% of that of the most abundant centromere repeat in that species.
Discussion
We used ChIP with antibodies to the kinetochore protein CENP-A to identify sequences enriched in Drosophila centromeres. Our approach relied on counting short candidate centromere sequences to determine whether they are enriched at centromeres, and we therefore cannot exclude the possibility that there are other centromere sequences that we did not count. However, we think it is unlikely that there are other major centromere sequences because we would expect them to be captured in our 250-bp IP reference sequences.
In other organisms with known centromeric repeats, CENP-A typically occupies only a fraction of those repeats and, if this fraction is small, it will reduce the enrichment of repeats in ChIP experiments even if the ChIP is efficient. Thus the highly abundant Prodsat is less enriched than AATAG even though it is approximately seven times more abundant in the IP. We therefore consider both the abundance and enrichment of candidate sequences in the IP when judging their likely importance for centromere function.
A limitation of ChIP is that it cannot identify the chromosome(s) on which the enriched sequences occur. Because simple repeats are well mapped in D. melanogaster, our finding that AATAT and Prodsat are at centromeres is consistent with previous work (Torok et al. 1997; Tolchkov et al. 2000; Blower and Karpen 2001; Sun et al. 2003), suggesting that these repeats form the centromeres of chromosomes X (AATAT), 2, and 3 (Prodsat). Enrichment of AATAG was not anticipated by previous work, and it is uncertain which chromosome(s) uses this sequence as centromere, with chromosomes 2 and 4 being the most likely possibilities. Based on the greater enrichment of (AATAG)4, over longer arrays, much of the centromeric AATAG does not appear to be in extended homogeneous tandem arrays. Frequent heterogeneity in some AATAG repeats has been previously described (Lohe and Brutlag 1987).
Two satellites previously proposed as centromeric, AAGAG and dodecasatellite, were consistently depleted in anti-Cid ChIP from S2 cells and P[Cid-GFP]8-10 embryos. AAGAG was required for segregation of derivatives of the minichromosome Dp1187 (Sun et al. 1997) and comprises the bwD heterochromatic element (Platero et al. 1998), which behaves as a neocentromere (Platero et al. 1999). The selection process for Dp1187 or its derivatives could also have resulted in a neocentromere (Maggert and Karpen 2001). Fiber-FISH experiments found CENP-A on chromatin fibers containing dodecasatellite (Garavís et al. 2015). Although we do not dispute that such fibers occur, our data indicate that CENP-A bound to dodecasatellite is not a significant component of centromeres in S2 cells or P[Cid-GFP]8-10 embryos. Dodecasatellite forms secondary structures (Garavís et al. 2015) and might be more sensitive to MNase, leading to its overall depletion in ChIP, but it is unclear why this would result in preferential depletion of dodecasatellite in the IP if it were a significant component of centromeres.
In stark contrast to the 5-mer and 10-mer centromeric sequences in D. melanogaster, the complex simcent1 and simcent2 repeats were the most abundant and enriched centromere sequences in D. simulans. Larger complex repeats are typical of centromeric sequences (Melters et al. 2013) and are found in other Drosophila species in the buzzatii and Hawaiian picture-wing complexes (Miklos and Gill 1981; de Lima et al. 2017), as well as in other insects (Lorite et al. 2004; Mravinac et al. 2004). The short repeat AATAGAATTG was also enriched at D. simulans centromeres. AATAGAATTG is found on the X near the nucleolus in a peculiar hybridization pattern on the side of the chromosome, raising the possibility that it participates in some unusual sequence organization or structure there. Based on this noncentromeric location on the X, AATAGAATTG in the IP of anti-CidM more likely derives from chromosome 4. The weaker hybridization signal of simcent1 to centromeric regions of all chromosomes when using acetic acid fixation is typical for diverged repeat families. The variable hybridization pattern of simcent1 may reflect the combined effects of divergent sequences and reduced accessibility when omitting acetic acid in the initial formaldehyde fixation (Shapiro et al. 1978).
Our alignment of simcent1 family sequences (File S5) reveals divergence in the form of base pair substitutions, small indels, and rearrangements juxtaposing different sequences. Complex sequence arrangements of satellites have also been observed in the D. buzzatii complex (Kuhn et al. 2009). Indeed, our 41-bp simcent1 probe has seven to eight differences from the homologous sequences in the 500-bp repeat clone JPYS01004644. Although there is very limited cross-hybridization of the 500-bp repeats of D. simulans and D. erecta (Strachan et al. 1982), the D. erecta 500-bp repeats were also reported to hybridize to the centromeric regions of all chromosomes, including that of the acrocentric X where we see weak hybridization in D. simulans (Figure 4C); suggesting that this family may be ancestrally centromeric in the melanogaster subgroup.
Rapid change in animal and plant centromeres is thought to be a result of centromere drive, which was originally proposed to explain the rapid divergence of CENP-A between D. melanogaster and D. simulans (Malik and Henikoff 2001). Variant satellite arrays compete for recruitment of cenH3 and inclusion in the egg or megaspore during asymmetric female meiosis, in which only one meiotic product survives. It is therefore of interest that several centromere sequences– including Prodsat in D. melanogaster, and simcent1, simcent2, and AATAGAATTG in D. simulans– appear to be expanding over their relatively low levels in most sibling species in the melanogaster subgroup (Figure 5). In particular, Prodsat has expanded to become an order of magnitude more abundant than the other satellites counted here. It may have displaced the complex satellites present in D. simulans. What might be its advantage in centromere evolution? In rice, cenH3 nucleosomes exhibit rotational phasing of the DNA that wraps them (Zhang et al. 2013). A single turn of the DNA double helix is ∼10 bp, and sequences with 10-bp periodicity in WW dinucleotides (W = A or T) favor wrapping of nucleosomes by reducing the bending energy of wrapping (Struhl and Segal 2013). In particular, a 10-bp periodicity of AA dinucleotides, which is found in almost all 5-mer and 10-mer Drosophila satellites, is a “driving force” for nucleosome formation and minimizes bending energy (Prytkova et al. 2011), presumably stabilizing nucleosomes that may be under tension during anaphase. In addition to their 10-bp periodicity and sequences beginning with AA, an interesting feature of the centromere-enriched 5-mers and each half of the 10-mers is that almost all can be derived by a zero or one base change from AATAG (AAGAT requires two changes), probably reflecting sequence constraints on rotational phasing. Although short repeats are not common at centromeres, the 20-bp centromeric repeat of Astragulus sinicus (Tek et al. 2011) may serve the same proposed function as the 5- and 10-bp repeats of Drosophila.
If these repeats are advantageous for centromere formation, why are they not more common, particularly in other species of the melanogaster subgroup where they appear to have been present in low levels for millions of years? Rotational phasing at centromeres can be achieved without short repeats (Zhang et al. 2013), and a short repeat may need to reach a certain threshold expansion level in the right location before it can effectively recruit CENP-A nucleosomes to compete in female meiosis. In addition, if another protein binds to its sequence, its expansion may titrate the availability of the protein. The Prod protein binds specifically to Prodsat during mitosis in D. melanogaster, whereas it does not bind centric heterochromatin in D. simulans (Platero et al. 1998); suggesting that this binding is a new function that has become essential for condensation near the centromeres of chromosomes 2 and 3 (Torok et al. 1997) as Prodsat has expanded. Similarly, the complex 359-bp repeat, present in a large array on the X chromosome of D. melanogaster but in much smaller amounts on the autosomes of D. simulans, causes lethality in daughters of D. melanogaster males and D. simulans females because of mitotic defects associated with lagging 359-bp DNA during mitosis (Ferree and Barbash 2009); suggesting that a maternal protein in D.melanogaster is essential for proper condensation of the 359-bp repeats. Conversely, OdsH in D. mauritiana actively decondenses heterochromatin in D. simulans (Bayes and Malik 2009), leading to hybrid sterility. An imbalance between heterochromatic binding proteins and satellites may also be the cause of other hybrid incompatibilities and drive speciation (Satyaki et al. 2014). With this in mind, short repeats may be relatively uncommon because their amplification potentially provides tens of times more binding sites for matching DNA-binding proteins in the same length of DNA as more complex repeats, with potentially deleterious consequences.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300620/-/DC1.
Acknowledgments
We thank Harmit Malik for the anti-CidM antibody and for D. simulans w501 flies, Pete Skene and Kami Ahmad for technical advice, and Jorja Henikoff and Srinivas Ramachandran for help with data analysis and programming. We thank Christine Codomo for library preparation, the Fred Hutchinson Cancer Research Center Genomics Shared Resource for sequencing and data processing, Aaron Hernandez for cell line maintenance, and the Fred Hutchinson Cancer Research Center Imaging Shared Resource for use of their facility. We thank Daniel Barbash and anonymous reviewers for helpful suggestions on the manuscript.
Author contributions: P.T., S.K., and S.H. conceived the experiments; P.T. conducted the experiments; S.K. helped analyze data; and P.T., S.K., and S.H. wrote the manuscript.
Footnotes
Communicating editor: A. Houben
Literature Cited
- Andreyeva E. N., Kolesnikova T. D., Demakova O. V., Mendez-Lago M., Pokholkova G. V., et al. , 2007. High-resolution analysis of Drosophila heterochromatin organization using SuUR Su(var)3–9 double mutants. Proc. Natl. Acad. Sci. USA 104: 12819–12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker R. E., Rogers K., 2006. Phylogenetic analysis of fungal centromere H3 proteins. Genetics 174: 1481–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayes J. J., Malik H. S., 2009. Altered heterochromatin binding by a hybrid sterility protein in Drosophila sibling species. Science 326: 1538–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G., 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27: 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower M. D., Karpen G. H., 2001. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 3: 730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M., Abad J. P., Villasante A., Gonzalez C., 1993. The Drosophila melanogaster dodecasatellite sequence is closely linked to the centromere and can form connections between sister chromatids during mitosis. J. Cell Sci. 105: 41–50. [DOI] [PubMed] [Google Scholar]
- Cheng Z., Dong F., Langdon T., Ouyang S., Buell C. R., et al. , 2002. Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14: 1691–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson S. C., Sagolla M. S., Cande W. Z., 2007. The cenH3 histone variant defines centromeres in Giardia intestinalis. Chromosoma 116: 175–184. [DOI] [PubMed] [Google Scholar]
- de Lima L. G., Svartman M., Kuhn G. C. S., 2017. Dissecting the satellite DNA landscape in three Cactophilic Drosophila sequenced genomes. G3 (Bethesda) 7: 2831–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg I. A., deYoung D., Henikoff S., Malik H. S., 2014. Recurrent loss of CenH3 is associated with independent transitions to holocentricity in insects. eLife 3: e03676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferree P. M., Barbash D. A., 2009. Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol. 7: e1000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Niu B., Zhu Z., Wu S., Li W., 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28: 3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garavís M., Méndez-Lago M., Gabelica V., Whitehead S. L., González C., et al. , 2015. The structure of an endogenous Drosophila centromere reveals the prevalence of tandemly repeated sequences able to form i-motifs. Sci. Rep. 5: 13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Wu Y., Koblizkova A., Torres G. A., Wang K., et al. , 2012. Repeatless and repeat-based centromeres in potato: implications for centromere evolution. Plant Cell 24: 3559–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff J. G., Thakur J., Kasinathan S., Henikoff S., 2015. A unique chromatin complex occupies young alpha-satellite arrays of human centromeres. Sci. Adv. 1: e1400234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., 1980. A more conventional view of the “ebony” gene. Drosoph. Inf. Serv. 55: 61–62. [Google Scholar]
- Henikoff S., Ahmad K., Platero J. S., van Steensel B., 2000. Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. USA 97: 716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Ahmad K., Malik H. S., 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293: 1098–1102. [DOI] [PubMed] [Google Scholar]
- Jagannathan M., Warsinger-Pepe N., Watase G. J., Yamashita Y. M., 2017. Comparative analysis of satellite DNA in the Drosophila melanogaster species complex. G3 (Bethesda) 7: 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koryakov D. E., Alekseyenko A. A., Zhimulev I. F., 1999. Dynamic organization of the beta-heterochromatin in the Drosophila melanogaster polytene X chromosome. Mol. Gen. Genet. 260: 503–509. [DOI] [PubMed] [Google Scholar]
- Koryakov D. E., Zhimulev I. F., Dimitri P., 2002. Cytogenetic analysis of the third chromosome heterochromatin of Drosophila melanogaster. Genetics 160: 509–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krassovsky K., Henikoff S., 2014. Distinct chromatin features characterize different classes of repeat sequences in Drosophila melanogaster. BMC Genomics 15: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn G. C., Teo C. H., Schwarzacher T., Heslop-Harrison J. S., 2009. Evolutionary dynamics and sites of illegitimate recombination revealed in the interspersion and sequence junctions of two nonhomologous satellite DNAs in cactophilic Drosophila species. Heredity (Edinb) 102: 453–464. [DOI] [PubMed] [Google Scholar]
- Larracuente A. M., Ferree P. M., 2015. Simple method for fluorescence DNA In Situ hybridization to squashed chromosomes. J. Vis. Exp. 95: e52288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le M. H., Duricka D., Karpen G. H., 1995. Islands of complex DNA are widespread in Drosophila centric heterochromatin. Genetics 141: 283–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., McManus C. J., Cho D. Y., Eaton M., Renda F., et al. , 2014. DNA copy number evolution in Drosophila cell lines. Genome Biol. 15: R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. R., Zhang W., Langdon T., Jin W., Yan H., et al. , 2005. Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc. Natl. Acad. Sci. USA 102: 11793–11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe A., Roberts P., 1988. Evolution of satellite DNA sequenes in Drosophila, pp. 148–186 in Heterochromatin: Molecular and Structural Aspects, edited by Verma R. S. Cambridge University Press, New York. [Google Scholar]
- Lohe A. R., Brutlag D. L., 1986. Multiplicity of satellite DNA sequences in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 83: 696–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe A. R., Brutlag D. L., 1987. Adjacent satellite DNA segments in Drosophila structure of junctions. J. Mol. Biol. 194: 171–179. [DOI] [PubMed] [Google Scholar]
- Lohe A. R., Hilliker A. J., Roberts P. A., 1993. Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Genetics 134: 1149–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorite P., Carrillo J. A., Aguilar J. A., Palomeque T., 2004. Isolation and characterization of two families of satellite DNA with repetitive units of 135 bp and 2.5 kb in the ant Monomorium subopacum (Hymenoptera, Formicidae). Cytogenet. Genome Res. 105: 83–92. [DOI] [PubMed] [Google Scholar]
- Maggert K. A., Karpen G. H., 2001. The activation of a neocentromere in Drosophila requires proximity to an endogenous centromere. Genetics 158: 1615–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik H. S., Henikoff S., 2001. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157: 1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik H. S., Henikoff S., 2003. Phylogenomics of the nucleosome. Nat. Struct. Biol. 10: 882–891. [DOI] [PubMed] [Google Scholar]
- Marques A., Ribeiro T., Neumann P., Macas J., Novak P., et al. , 2015. Holocentromeres in Rhynchospora are associated with genome-wide centromere-specific repeat arrays interspersed among euchromatin. Proc. Natl. Acad. Sci. USA 112: 13633–13638 (erratum: Proc. Natl. Acad. Sci. USA 112: E6720). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melters D. P., Bradnam K. R., Young H. A., Telis N., May M. R., et al. , 2013. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 14: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklos G. L., Gill A. C., 1981. The DNA sequences of cloned complex satellite DNAs from Hawaiian Drosophila and their bearing on satellite DNA sequence conservation. Chromosoma 82: 409–427. [DOI] [PubMed] [Google Scholar]
- Mravinac B., Plohl M., Ugarkovic D., 2004. Conserved patterns in the evolution of Tribolium satellite DNAs. Gene 332: 169–177. [DOI] [PubMed] [Google Scholar]
- Murphy T. D., Karpen G. H., 1995. Localization of centromere function in a Drosophila minichromosome. Cell 82: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., Desai A., 2017. A molecular view of kinetochore assembly and function. Biology (Basel) 6: E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki K., Talbert P. B., Zhong C. X., Dawe R. K., Henikoff S., et al. , 2003. Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics 163: 1221–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki K., Cheng Z., Ouyang S., Talbert P. B., Kim M., et al. , 2004. Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36: 138–145. [DOI] [PubMed] [Google Scholar]
- Neiman M., Sundling S., Gronberg H., Hall P., Czene K., et al. , 2012. Library preparation and multiplex capture for massive parallel sequencing applications made efficient and easy. PLoS One 7: e48616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platero J. S., Csink A. K., Quintanilla A., Henikoff S., 1998. Changes in chromosomal localization of heterochromatin-binding proteins during the cell cycle in Drosophila. J. Cell Biol. 140: 1297–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platero J. S., Ahmad K., Henikoff S., 1999. A distal heterochromatic block displays centromeric activity when detached from a natural centromere. Mol. Cell 4: 995–1004. [DOI] [PubMed] [Google Scholar]
- Postberg J., Forcob S., Chang W. J., Lipps H. J., 2010. The evolutionary history of histone H3 suggests a deep eukaryotic root of chromatin modifying mechanisms. BMC Evol. Biol. 10: 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prytkova T. R., Zhu X., Widom J., Schatz G. C., 2011. Modeling DNA-bending in the nucleosome: role of AA periodicity. J. Phys. Chem. B 115: 8638–8644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin L. F., Mellone B. G., 2017. Centromeres drive a hard bargain. Trends Genet. 33: 101–117.10.1016/j.tig.2016.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon E. D., Bloom K., 2017. Tension sensors reveal how the kinetochore shares its load. BioEssays 39. DOI: 10.1002/bies.201600216.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyaki P. R., Cuykendall T. N., Wei K. H., Brideau N. J., Kwak H., et al. , 2014. The Hmr and Lhr hybrid incompatibility genes suppress a broad range of heterochromatic repeats. PLoS Genet. 10: e1004240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W., 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueler M. G., Higgins A. W., Rudd M. K., Gustashaw K., Willard H. F., 2001. Genomic and genetic definition of a functional human centromere. Science 294: 109–115. [DOI] [PubMed] [Google Scholar]
- Shapiro I. M., Moar M. H., Ohno S., Klein G., 1978. Acetic acid treatment denatures DNA while preserving chromosomal morphology during the in situ hybridization procedure. Exp. Cell Res. 115: 411–414. [DOI] [PubMed] [Google Scholar]
- Skene P. J., Henikoff S., 2017. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6: e21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer E. L., Durbin R., 1995. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167: GC1–GC10. [DOI] [PubMed] [Google Scholar]
- Steiner F. A., Henikoff S., 2015. Diversity in the organization of centromeric chromatin. Curr. Opin. Genet. Dev. 31: 28–35. [DOI] [PubMed] [Google Scholar]
- Steiner F. A., Talbert P. B., Kasinathan S., Deal R. B., Henikoff S., 2012. Cell-type-specific nuclei purification from whole animals for genome-wide expression and chromatin profiling. Genome Res. 22: 766–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachan T., Coen E., Webb D., Dover G., 1982. Modes and rates of change of complex DNA families of Drosophila. J. Mol. Biol. 158: 37–54. [DOI] [PubMed] [Google Scholar]
- Struhl K., Segal E., 2013. Determinants of nucleosome positioning. Nat. Struct. Mol. Biol. 20: 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Wahlstrom J., Karpen G., 1997. Molecular structure of a functional Drosophila centromere. Cell 91: 1007–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Le H. D., Wahlstrom J. M., Karpen G. H., 2003. Sequence analysis of a functional Drosophila centromere. Genome Res. 13: 182–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Chen E. S., Yanagida M., 2000. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288: 2215–2219. [DOI] [PubMed] [Google Scholar]
- Tamura K., Subramanian S., Kumar S., 2004. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol. Biol. Evol. 21: 36–44. [DOI] [PubMed] [Google Scholar]
- Tek A. L., Kashihara K., Murata M., Nagaki K., 2011. Functional centromeres in Astragalus sinicus include a compact centromere-specific histone H3 and a 20-bp tandem repeat. Chromosome Res. 19: 969–978. [DOI] [PubMed] [Google Scholar]
- Teytelman L., Thurtle D. M., Rine J., van Oudenaarden A., 2013. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc. Natl. Acad. Sci. USA 110: 18602–18607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolchkov E. V., Rasheva V. I., Bonaccorsi S., Westphal T., Gvozdev V. A., 2000. The size and internal structure of a heterochromatic block determine its ability to induce position effect variegation in Drosophila melanogaster. Genetics 154: 1611–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torok T., Harvie P. D., Buratovich M., Bryant P. J., 1997. The product of proliferation disrupter is concentrated at centromeres and required for mitotic chromosome condensation and cell proliferation in Drosophila. Genes Dev. 11: 213–225. [DOI] [PubMed] [Google Scholar]
- Torok T., Gorjanacz M., Bryant P. J., Kiss I., 2000. Prod is a novel DNA-binding protein that binds to the 1.686 g/cm(3) 10 bp satellite repeat of Drosophila melanogaster. Nucleic Acids Res. 28: 3551–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Talbert P. B., Zhang W., Wu Y., Yang Z., et al. , 2013. The CentO satellite confers translational and rotational phasing on cenH3 nucleosomes in rice centromeres. Proc. Natl. Acad. Sci. USA 110: E4875–E4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong C. X., Marshall J. B., Topp C., Mroczek R., Kato A., et al. , 2002. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14: 2825–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Fly stocks and antibodies are available upon request. Figure S1 shows cell line karyotypes and centromere staining with anti-CidM and anti-GFP. Figure S2 shows hybridization of AATAG and 10-mer probes to D. simulans neuroblasts. File S1 and File S2 contain reference sequences for input and IP for S2 cells. File S3 and File S4 contain the reference sequences for ML82-19a cells. File S5 and File S6 display alignments of reference sequences containing simcent1 and simcent2, respectively. Table S1 lists the candidate 9- to 16-mers. Table S2 contains the SRA accessions counted for Drosophila species. Sequencing data are available at Gene Expression Omnibus with the accession number GSE105100.