Abstract
Triple-negative breast cancer (TNBC) has high rates of local recurrence and distant metastasis, partially due to its high invasiveness. The Forkhead box C1 (FOXC1) transcription factor has been shown to be specifically overexpressed in TNBC and associated with poor clinical outcome. How TNBC’s high invasiveness is driven by FOXC1 and its downstream targets remains poorly understood. In the present study, pathway-specific PCR array assays revealed that WNT5A and matrix metalloproteinase-7 (MMP7) were upregulated by FOXC1 in TNBC cells. Interestingly, WNT5A mediates the upregulation of MMP7 by FOXC1 and the WNT5A-MMP7 axis is essential for FOXC1-induced invasiveness of TNBC cells in vitro. Xenograft models showed that the lung extravasation and metastasis of FOXC1-overexpressing TNBC cells were attenuated by knocking out WNT5A, but could be restored by MMP7 overexpression. Mechanistically, FOXC1 can bind directly to the WNT5A promoter region to activate its expression. Engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP), coupled with mass spectrometry, identified FOXC1-interacting proteins including a group of heterogeneous nuclear ribonucleoproteins involved in WNT5A transcription induction. Finally, we found that WNT5A activates NF-κB signaling to induce MMP7 expression. Collectively, these data demonstrate a FOXC1-elicited non-canonical WNT5A signaling mechanism comprising NF-κB and MMP7 that is essential for TNBC cell invasiveness, thereby providing implications toward developing an effective therapy for TNBC.
Introduction
Triple-negative breast cancer (TNBC) is a heterogenous group of tumors that lack immunohistochemical staining or overexpression of the estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2).1 Compared with ER-positive or HER2-positive breast cancer, TNBC is associated with a high histologic grade, frequent distant metastasis, and high mortality rate.1,2 Although it has been intensively studied, the mechanism underlying the poor prognosis of TNBC remains to be elucidated.
The WNT signaling pathway has been demonstrated to be involved in tumorigenesis.3 WNT signaling is frequently activated in TNBC as opposed to other subtypes of breast cancer.4,5 Recent studies have established the transcription factor forkhead box C1 (FOXC1) as a pivotal diagnostic and prognostic biomarker for basal-like breast cancer (BLBC), a subtype of breast cancer which is characterized by basal cytokeratin expression and shares many similar clinicopathologic traits with TNBC.6,7 Elevated FOXC1 expression is associated with a worse overall survival of breast cancer patients.6,8 In this study, we explored the FOXC1-associated mechanism underlying the invasiveness of TNBC cells. Our results show that WNT5A, a ligand that mediates non-canonical β-catenin-independent WNT signaling, was markedly induced by FOXC1 in TNBC cells. Upregulated WNT5A activated NF-κB signaling, which in turn induced matrix metalloproteinase-7 (MMP7) expression. This WNT5A-NF-κB-MMP7 axis elicited by FOXC1 is essential for the invasiveness of TNBC cells, and may be further exploited to develop new treatment for TNBC.
Results and Discussion
WNT5A is up-regulated by FOXC1 in TNBC cells
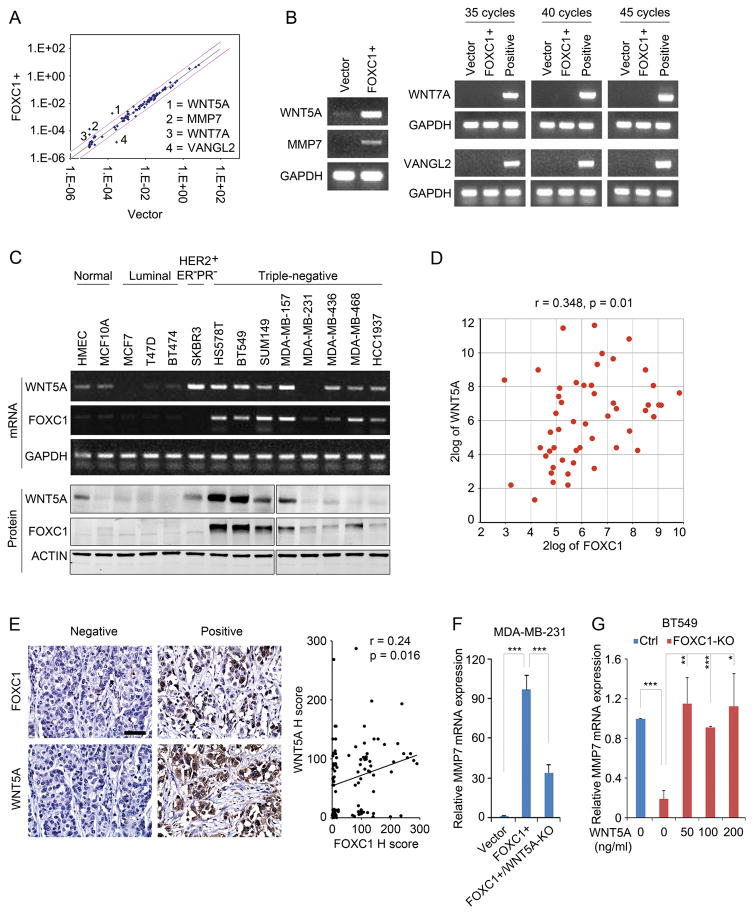
Because FOXC1 expression and WNT signaling are associated with TNBC, we tested whether FOXC1 regulates WNT pathways. Given that FOXC1 does not activate β-catenin-dependent WNT signaling in TNBC cells,9 we performed the WNT signaling pathway PCR array assay using control and FOXC1-overexpressing MDA-MB-231 human TNBC cells to explore the potential effect of FOXC1 on WNT signaling in an unbiased manner. WNT ligands, WNT5A and WNT7A, and the WNT target gene MMP7 were significantly up-regulated in FOXC1-overexpressing cells, whereas VANGL2, a receptor of WNT ligands, was down-regulated (Fig. 1A). The up-regulation of WNT5A and MMP7 mRNA by FOXC1 was subsequently confirmed by PCR assays (Fig. 1B), but the expression of WNT7A and VANGL2 mRNA was undetectable in the same assays when testing different amplification cycles (Fig. 1B). This discrepancy might be caused by the extremely low endogenous levels of WNT7A and VANGL2 mRNA in MDA-MB-231 cells, which resulted in the very large variation of the Ct (cycle threshold) values in the PCR array assay (data not shown).
Figure 1.
WNT5A is up-regulated by FOXC1 in TNBC cells. A, WNT signaling PCR array assays in control and FOXC1-overexpressing (FOXC1+) MDA-MB-231 cells (All the cell lines used in this study were acquired from American Type Culture Collection (ATCC) and maintained according to ATCC instructions). FOXC1-overexpressing stable cells were established as described previously.9 pCMV6 empty vector and pCMV6-FOXC1 were from Origene. WNT signaling PCR array was performed by using RT2 Profiler PCR Array Human WNT Signaling Pathway Plus kit (Qiagen) according to the manufacturer’s instructions. B, PCR detection of the genes identified in PCR array assays. Positive control mRNAs for WNT7A and VANGL2 were from HCC1937 and SUM149 cells, respectively. GAPDH was used as an internal control. Primers used were: PCR: WNT5A-forward: 5′-AAGCCCTAATTACCGCCGTC-3′, WNT5A-reverse: 5′-TCCAATGGA CTTCTTCATGGC-3′; MMP7-forward: 5′-GCTACAGTGGGAACAGGCTC-3′, MMP7-reverse: 5′-GGGATCTCTTTGCCCCACAT-3′; WNT7A-forward: 5′-GATCAAGCAGAA TGCCCGGA-3′, WNT7A-reverse: 5′-CTGCACGTGTTGCACTTGAC-3′; VANGL2-forward: 5′-GGGGGTGACCAGACTCAAGA-3′, VANGL2-reverse: 5′-CTCTTAGAGCGGTGTCGG TC-3′. GAPDH-forward: 5′-ATGGGTGTGAACCATGAGAA-3′; GAPDH-reverse: 5′-GT GCTAAGCAGTTGGTGGT G-3′. C, PCR and western blotting assays of FOXC1 and WNT5A expression in various normal mammary epithelial cells and breast cancer cell lines. GAPDH and ACTIN were used as internal controls for mRNA and protein, respectively. The primary antibodies used were WNT5A (1:300, MAB645, R&D systems), FOXC1 (1:500, sc-21394, Santa Cruz), and ACTIN (1:1000, sc-1616, Santa Cruz). D, correlation analysis between FOXC1 and WNT5A mRNAs in 51 breast cancer cell lines. Analysis was performed using the online tool (http://hgserver1.amc.nl/cgi-bin/r2/main.cgi). E, correlation analysis of FOXC1 and WNT5A proteins based on IHC staining in TNBC samples (n = 100). Scale bar = 50 μm. Immunohistochemistry (IHC) was performed as described previously.9 Formalin-fixed paraffin-embedded TNBC tissue microarray slides were from US Biomax (BR10011a). Primary antibodies used were FOXC1 (1:50, Onconostic Technologies) and WNT5A (1:500, ab86720, Abcam). The IHC staining intensity was presented using the pathological H-score and the regression correlation was analyzed. F and G, real-time PCR analysis of MMP7 mRNA expression in MDA-MB-231 cells (F) and BT549 cells (G). MMP7 primers were: MMP7-forward: 5′-AAGTGGTCACCTACAGGATCG-3′, MMP7-reverse: 5′-TGGCCCATCAAATGGGTAGG -3′. FOXC1-KO BT549 cells were treated with different concentrations of recombinant WNT5A protein (R&D Systems) for 4 hours. The knockout of FOXC1 and WNT5A was performed by using CRISPR/Cas9 (#52961, Addgene).35 The guide RNA (gRNA) sequences were: FOXC1: 5′-GGGTGCGAGTACACGCTCAT-3′; WNT5A: 5′-TTCAATTACAACCTGGGCGA-3′. The bar graph indicates mean ± SD, n = 3. *, p < 0.05, **, p < 0.01, ***, p < 0.001.
Consistent with our results, earlier studies showed that MMP7 is up-regulated by FOXC1 and their expression is positively correlated with each other in breast cancer cells.10 WNT5A has also been shown to be expressed in TNBC samples.11 Here, we examined the association between the expression of FOXC1 and WNT5A in normal human mammary epithelial and breast cancer cells. As shown in Fig. 1C, the expression of FOXC1 positively correlated with WNT5A in both mRNA and protein levels in breast cancer cells. This positive correlation was also observed in a microarray dataset of 51 breast cancer cell lines12 (Fig. 1D). The regulation of WNT5A by FOXC1 was further confirmed in other TNBC cell lines, subgrouped by Lehmann et al.13 and Neve et al.,14 except SUM149 cells (Supplementary Fig. 1A–C). Notably, FOXC1 induced WNT5A mRNA, but not protein expression in MDA-MB-468 cells (Supplementary Fig. 1A and B). We speculate that the discrepancy between detected WNT5A protein and mRNA levels in MDA-MB-468 cells might be attributed to protein translation and stability, as well as secretion of WNT5A protein. The positive correlation between FOXC1 and WNT5A protein levels was further validated by immunohistochemistry assays performed in a TNBC tissue microarray (Fig. 1E). Taken together, these results demonstrate that WNT5A is up-regulated by FOXC1 in TNBC cells.
Because MMP7 is a downstream target of tumor-associated macrophage-derived WNT5A,15 we tested whether FOXC1 induction of MMP7 is in fact mediated by WNT5A in TNBC. To do so, WNT5A was knocked out by CRISPR/Cas9 in FOXC1-overexpressing MDA-MB-231 cells (Supplementary Fig. 1D). We found that the FOXC1-induced MMP7 gene expression was attenuated in the WNT5A-knockout cells (Fig. 1F). Moreover, the expression of MMP7 in BT549 and HCC1806 cells, which have high levels of endogenous FOXC1, was suppressed after knocking out FOXC1 and restored by recombinant WNT5A protein treatment (Fig. 1G; Supplementary Fig. 1A and E). These results suggest that MMP7 is regulated by a FOXC1-WNT5A signaling axis in TNBC cells.
WNT5A promoter is activated by a FOXC1 transactivator complex
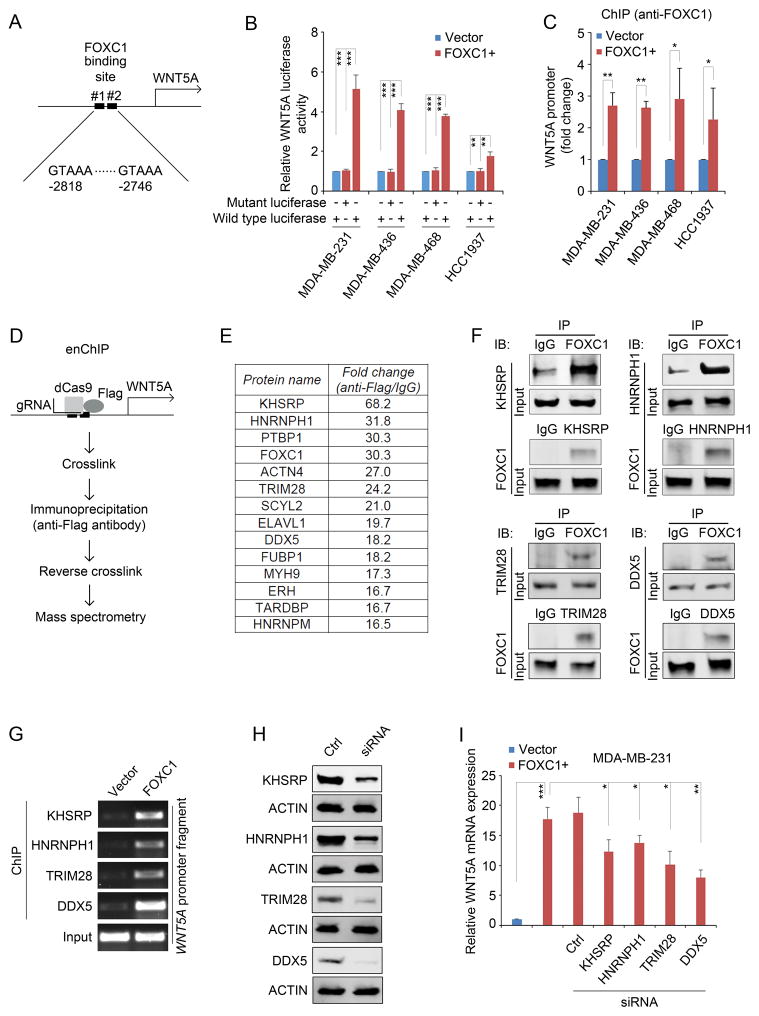
To test whether FOXC1 directly binds to the WNT5A promoter, we analyzed the sequence of the WNT5A promoter region and found two potential FOXC1-binding sites with the core consensus sequence of GTAAA16 (Fig. 2A). Accordingly, two 177bp fragments of the WNT5A promoter containing wild-type or mutant FOXC1-binding sites were cloned into a pGL4 reporter construct and luciferase reporter assays were performed. As shown in Fig. 2B, FOXC1 dramatically increased WNT5A luciferase activity in TNBC cells. This result was further confirmed by a ChIP assay in which the WNT5A promoter fragment was enriched by an immunoprecipitation assay using an anti-FOXC1 antibody (Fig. 2C). To gain insight into the molecular mechanism by which FOXC1 upregulates WNT5A transcription, we adopted the engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) assay17 (Fig. 2D). Subsequent mass spectrometry analysis identified proteins that bind to the WNT5A promoter together with FOXC1 (Fig. 2E and Supplementary Table 1). The interaction between FOXC1 and several representative proteins (i.e., KHSRP, HNRNPH1, TRIM28, and DDX5) from the proteomics analysis was further confirmed by reciprocal co-immunoprecipitation assays (Co-IP) (Fig. 2F). ChIP assays also demonstrated the binding of the FOXC1-interacting proteins to the WNT5A promoter region containing FOXC1-binding sites (Fig. 2G). In addition, these binding interactions were enhanced when FOXC1 was increased (Fig. 2G). The binding of KHSRP and HNRNPH1 to the WNT5A promoter was also confirmed by biotinylated oligonucleotide precipitation assays (Supplementary Fig. 2A). Individual knockdown of these proteins reduced FOXC1-induced WNT5A mRNA expression to a varying extent (Fig. 2H and I). Taken together, these data suggest that FOXC1 binds directly to the promoter of WNT5A and collaborates with other nuclear proteins to induce WNT5A expression.
Figure 2.
WNT5A promoter is activated by a FOXC1 transactivator complex. A, diagrammatic illustration of putative FOXC1 binding sites in the WNT5A promoter. B, luciferase assays of cells transfected with the pGL4-WNT5A promoter luciferase reporter constructs containing wild-type or mutant FOXC1 binding sites. pGL4-WNT5A promoter plasmids was constructed by cloning wild-type (GTAAA) or mutant (ACCGC) 177 base pairs (−2868 to −2692, from the translation start site) of WNT5A promoter fragment into pGL4 luciferase reporter vector (Promega). The β-Galactosidase expression vector was from Promega. Cells seeded in 12-well plates were transfected with 500ng luciferase plasmids and 500ng expression plasmids using Lipofectamine 2000 (Invitrogen). Two hundred ng of β-galactosidase expression plasmid was used as an internal control. After 48h, cells were harvested and 20μl extracts were analyzed using the Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Luciferase activity was measured using a luminometer (Glomax multi detection system, Promega). The bar graph indicates mean ± SD, n = 3. **, p < 0.01, ***, p < 0.001. C, ChIP assays using an anti-FOXC1 antibody to pull down the WNT5A promoter-protein complex. ChIP assays were performed using the EZ-ChIP Chromatin Immunoprecipitation Kit (EMD Millipore) according to the manufacturer’s instructions. Anti-FOXC1 (sc-21394, Santa Cruz) antibody-immunoprecipitated DNA was analyzed by real-time PCR. The primers were: WNT5A-forward: 5′-AGACTGTAAAATGCCCACAGGT-3′, WNT5A-reverse: 5′-TCAAAGCTCCCCTTGGGAC-3′. The bar graph indicates mean ± SD, n = 3. *, p < 0.05, **, p < 0.01. D, diagrammatic illustration of the enChIP assay. E, the top enriched proteins that bind to the WNT5A promoter together with FOXC1. enChIP was performed as described previously.17,36 Briefly, cells were transfected with 3×FLAG-dCas9 (#51240, Addgene) and pSIR-neo-WNT5A (WNT5A gBlocks were sub-cloned into pSIR-neo (#51128, Addgene)). Guide RNA (gRNA) sequences for WNT5A: #1: 5′-CCTGGGGGCGATTTGCCGGG-3′; #2: 5′-GAAGTGGTCAAGGTTTACAG-3′; #3: 5′-TAAGGCCGCACACGCCCTGG-3′. Cells were crosslinked and chromatins were immunoprecipitated with an anti-FLAG antibody (F1804, Sigma-Aldrich). After reverse crosslinking, proteins were analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS) essentially as described.37 Briefly, following in-gel protein digestion, tryptic peptides were separated on a 50 cm EASY-Spray C18 column, and analyzed by an LTQ Orbitrap Elite mass spectrometer in the data-dependent acquisition mode. MS data were searched against the Uniprot Human database (released on 01/22/2016) with MaxQuant (v1.5.5.1).38 Protein quantification was performed using spectral counting.39 F, co-immunoprecipitation (Co-IP) assays to confirm the binding between FOXC1 and the factors identified in the enChIP assays. Co-IP was performed as described previously.9 Briefly, the whole cell lysates were extracted using IP buffer (50mM Tris-HCl (pH7.4), 150mM NaCl, 2mM EDTA-2Na, 1% NP40, 10% Glycerol) and immunoprecipitated with antibody-bound agarose beads (Thermo). Protein complexes were analyzed by western blotting analysis. The antibodies used in Co-IP assay were the same as the ones used in the western blotting. G, ChIP assays using different antibodies in control or FOXC1-overexpressing MDA-MB-231 cells. Immunoprecipitated WNT5A promoter fragments were subject to PCR assays. Primers used were: WNT5A-forward: 5′-AGACTGTAAAATGCCCACAG GT-3′, WNT5A-reverse: 5′-TCAAAGCTCCCCTTGGGAC-3′. H, western blotting analysis of the proteins in control siRNA- or protein-specific siRNA-transfected MDA-MB-231 cells. ACTIN was used as an internal control. KHSRP, HNRNPH1, TRIM28, and DDX5 siRNAs were from Santa Cruz. The primary antibodies used were: KHSRP (1:1000, #13398, Cell Signaling Technology), HNRNPH1 (1:500, sc-10042, Santa Cruz), TRIM28 (1:1000, #4123, Cell Signaling Technology), and DDX5 (1:1000, #9877, Cell Signaling Technology). I, real-time PCR analysis of WNT5A mRNA expression in MDA-MB-231 cells transfected with different siRNAs. Primers used were: WNT5A-forward: 5′-CCCTCGCCATGAAGAAGTCCA-3′, WNT5A-reverse: 5′-CATACCTAGCGACCACCAAGA -3′. The bar graph indicates mean ± SD, n = 3. *, p < 0.05, **, p < 0.01, ***, p < 0.001.
Of note, some of the top enriched proteins have been shown to be involved in gene transcription. For example, ACTN4 can function as a transcriptional co-activator to promote the invasiveness of breast cancer cells.18 TRIM28 and ERH have been found to promote the accumulation of RNA polymerase II (PolII) to the transcriptional start site to initiate gene transcription.19,20 As a helicase, DDX5 can induce the recruitment of PolII to gene promoters to regulate breast cancer cell proliferation.21 Interestingly, KHSRP, HNRNPH1, PTBP1, and FUBP1 are heterogeneous nuclear ribonucleoproteins (HNRNPs), which typically bind to pre-mRNA and participate in gene splicing.22 Recent studies suggest that HNRNPs may also bind to DNA to regulate gene transcription. For example, FUBP1 can bind to an upstream element of the c-MYC promoter to regulate c-MYC mRNA expression.23
WNT5A activates NF-κB signaling in TNBC cells
WNT5A exerts its function mainly through activating β-catenin-independent signaling,24 which also has been observed in TNBC cells.25 However, it can simultaneously activate both β-catenin-independent and -dependent WNT signaling in TNBC cells to promote tumorigenicity.11 Since our previous study showed that FOXC1 does not activate β-catenin-dependent WNT signaling in TNBC cells,9 we next focused on the non-canonical pathways of WNT5A.
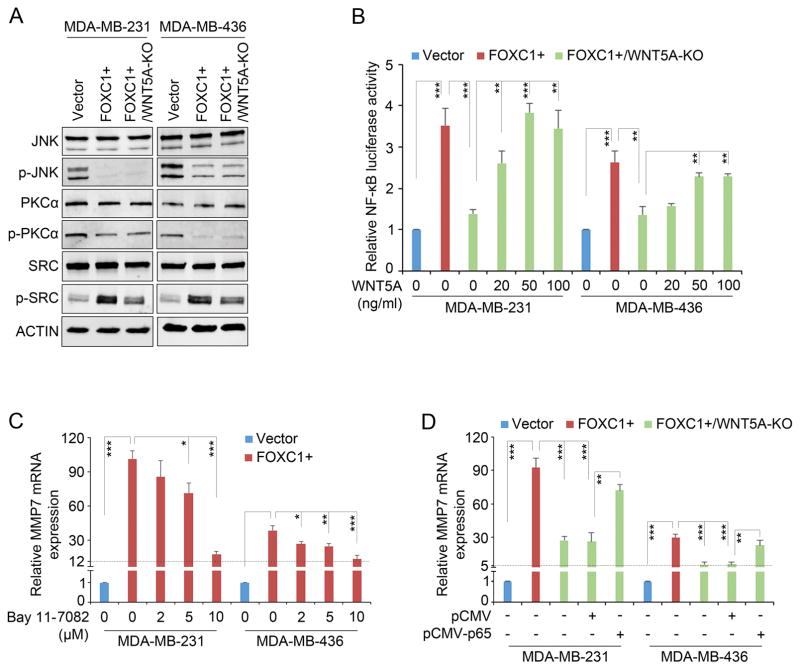
WNT5A can activate PKC and JNK pathways to promote the invasiveness of melanoma26 and breast cancer cells,15 respectively. However, we found that FOXC1 did not induce but, rather reduced the phosphorylation levels of PKC and JNK in TNBC cells (Fig. 3A). Surprisingly, FOXC1 increased SRC activation, which is partially dependent on WNT5A as WNT5A knockout reduced SRC activation (Fig. 3A and Supplementary Fig. 3A). SRC is a well-known proto-oncogene that is involved in multiple aspects of tumorigenesis. We found that the SRC inhibitor Dasatinib showed no effect on the FOXC1-induced MMP7 expression (Supplementary Fig. 3B) despite inhibiting SRC phosphorylation in FOXC1-overexpressing cells (Supplementary Fig. 3C), suggesting SRC signaling is not involved in MMP7 induction by the FOXC1-WNT5A axis. The functions of the FOXC1-induced activation of SRC signaling in TNBC cells need to be further characterized.
Figure 3.
WNT5A activates NF-κB signaling in TNBC cells. A, western blotting analysis of reported WNT5A pathways in TNBC cells. ACTIN was used as an internal control. The antibodies used were from Cell Signaling Technology at 1:1000 dilution: JNK (#9252), p-JNK (#9255), PKCα (#2056), p-PKCα (#9375), SRC (#2109), and p-SRC (#6943). B, luciferase assays in the cells transfected with NF-κB-responsive luciferase reporter construct and treated with recombinant WNT5A protein at different concentrations. NF-κB responsive luciferase reporter construct pGL4.32[luc2P/NF-κB-RE/Hygro] was from Promega. The bar graph indicates mean ± SD, n = 3. **, p < 0.01, ***, p < 0.001. C and D, real-time PCR analysis of MMP7 mRNA in cells treated with the NF-κB inhibitor Bay 11-7082 (Cayman Chemical) at different concentrations (C) or transfected with the empty vector pCMV or pCMV-p65 (D). The bar graph indicates mean ± SD, n = 3. *, p < 0.05, **, p < 0.01, ***, p < 0.001.
NF-κB signaling, which is highly active in TNBC cells and can be activated by FOXC1,27 mediates the functions of WNT5A in human dental pulp cells,28 so we tested the involvement of NF-κB signaling in the effect of FOXC1 on WNT5A in TNBC cells. We found that knocking out WNT5A impaired the FOXC1 induction of NF-κB-responsive luciferase activity, and WNT5A treatment abolished this reduction (Fig. 3B). In line with these results, FOXC1 knockout inhibited the NF-κB-responsive luciferase activity in HCC1806 cells, which was reversed by WNT5A treatment (Supplementary Fig. 3D). All these data suggest that FOXC1-induced WNT5A can activate NF-κB signaling in TNBC cells. Next, we treated FOXC1-overexpressing TNBC cells with different concentrations of the NF-κB inhibitor Bay 11-7082. As shown in Fig. 3C, inhibition of NF-κB substantially lowered the FOXC1-induced MMP7 expression. Similar results were also found in HCC1806 cells (Supplementary Fig. 3E). Of note, expression of KHSRP, the top factor identified in the enChIP-proteomics assay (Fig. 2E), was not affected by Bay 11-7082 treatment (Supplementary Fig. 3F), suggesting that the effect of Bay 11-7082 on MMP7 mRNA expression is selective. In agreement with the above results, p65 overexpression elevated MMP7 expression in WNT5A-KO TNBC cells (Fig. 3D and Supplementary Fig. 3G). These data suggest that NF-κB signaling mediates the effect of WNT5A on MMP7 expression in TNBC cells, which is supported by previous reports showing that NF-κB regulates MMP7 expression in cholangiocarcinoma and chondrosarcoma cells.29,30
WNT5A mediates the FOXC1-induced invasiveness of TNBC cells
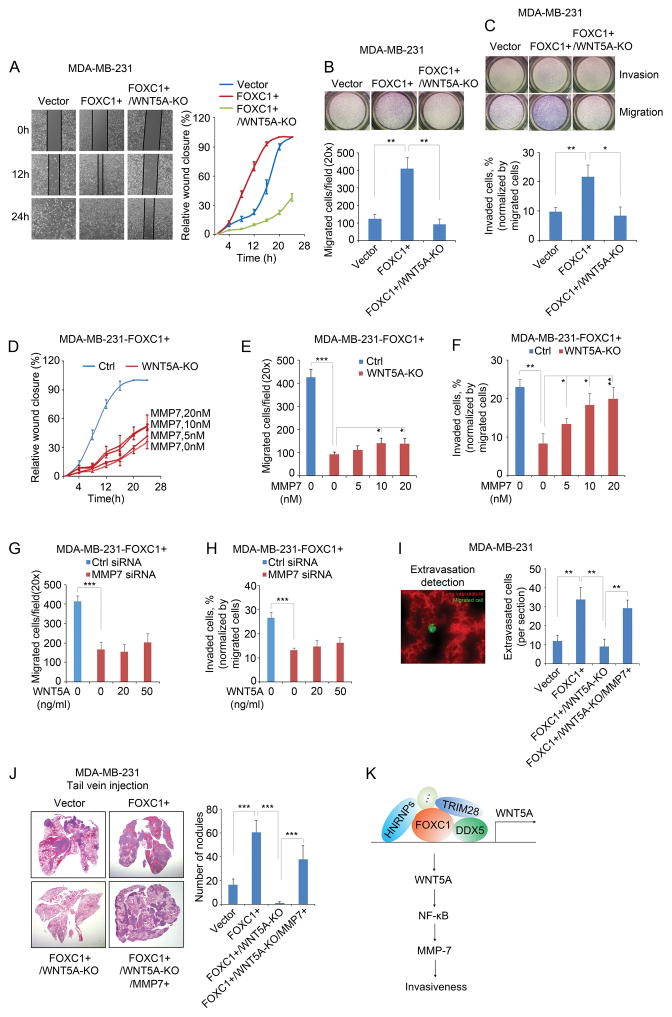
WNT5A has been shown to promote cancer cell migration and invasion and distant metastasis in many cancer subtypes.31 It can induce a mesenchymal phenotype and thus enhance the migration and invasion ability of human mammary epithelial cells.32 To test whether WNT5A mediates FOXC1-induced migration and invasion of TNBC cells, we first performed a wound healing assay. As shown in Fig. 4A, knocking out WNT5A significantly attenuated the migration of FOXC1-overexpressing MDA-MB-231 cells. This effect was further confirmed by the transwell migration assay (Fig. 4B). The Matrigel invasion assay also showed that WNT5A mediates FOXC1-induced invasion of TNBC cells (Fig. 4C). Of note, FOXC1-induced proliferation in TNBC cells was not affected by WNT5A knockout (Supplementary Fig. 4A).
Figure 4.
WNT5A mediates the FOXC1-induced invasiveness of TNBC cells. A, wound healing assays at different time points in different groups of MDA-MB-231 cells. Cells were seeded into a 12-well plate and allowed to grow to 90% confluence. The surface of each well was scratched by using a 1ml pipette tip. Cells were washed twice with culture medium to remove the detached cells. Pictures were taken at different time points and the gap distance was measured using ImageJ software. B and C, migration (B) and invasion (C) assays in different groups of MDA-MB-231 cells. 1×105 cells were re-suspended in 500μl serum-free medium and seeded into the upper compartment of Transwell chamber (Corning) or Matrigel invasion chamber (BD Biosciences) for migration or invasion assay, respectively. The lower compartment of the chamber was filled with 750 μl complete medium. After 4 hours (migration) or 8 hours (invasion) of incubation, cells left in the upper compartment were removed with a cotton swab, and the migrated or invaded cells were stained by using HEMA3 staining kit (Fisher Scientific). Pictures were taken and the cells were counted using ImageJ software. The bar graph indicates mean ± SD, n = 3. ***, p < 0.001. D, E, and F, FOXC1-overexpressing WNT5A-KO MDA-MB-231 cells were treated with different concentrations of recombinant MMP7 protein (Aviva Systems Biology) in wound healing assays (D), migration assay (E), and invasion assay (F). The bar graph indicates mean ± SD, n = 3. *, p < 0.05, **, p < 0.01, ***, p < 0.001. G and H, FOXC1-overexpressing MMP7-knockdown MDA-MB-231 cells were treated with different concentrations of recombinant WNT5A protein in migration assays (G) and invasion assays (H). The bar graph indicates mean ± SD, n = 3. ***, p < 0.001. I, extravasation assays of different groups of cells injected through the tail vein, and images were taken 48h after injection. To establish FOXC1- and MMP7-overexpressing and WNT5A-knockout (FOXC1+/WNT5A-KO/MMP7+) cells, FOXC1+/WNT5A-KO cells were transfected with the pCMV6-MMP7 plasmid. pCMV6-MMP7 was constructed by cloning the MMP7 ORF into a pCMV6 vector. Representative image shows the migrated cancer cells and the lung vasculature (left panel, 40×). All the migrated cells were counted from each frozen section, and three mice were used for each group. No randomization or blinding was used. All animal experiments were performed in accordance with the approval of the Cedars-Sinai Medical Center Institutional Animal Care and Use Committee. Cells were labeled with 5μM CellTracker Green CMFDA (C2925, ThermoFisher Scientific) for 45 minutes. 1×105 cells/100μl PBS were injected into nude mice (Charles River Laboratories) through the tail vein. Forty-eight hours after injection, mice were injected with 50μg rhodamine-lectin (RL-1102, Vector Laboratories) in 100μl PBS through the tail vein and sacrificed in 30 minutes. Mouse lung tissues were collected and frozen in liquid nitrogen, then were embedded in OCT compound (Tissue-Tek). Frozen sections were cut and examined under fluorescence microscope. The bar graph indicates mean ± SD. **, p < 0.01. J, in vivo assay to assess the invasive capacity of different groups of cells injected through the tail vein. To establish FOXC1- and MMP7-overexpressing and WNT5A-knockout (FOXC1+/WNT5A-KO/MMP7+) cells, FOXC1+/WNT5A-KO cells were transfected with pEGFP-C3-MMP7. pEGFP-C3 empty vector was provided by Sandra Orsulic (Cedars-Sinai Medical Center). pEGFP-C3-MMP7 was constructed by cloning MMP7 ORF into the pEGFP-C3 vector. EGFP-labeled cells were sorted by FACS (Aria III). 5×105 cells were suspended in 100μl PBS and injected into nude mice through the tail vein. Mice were sacrificed two weeks after injection. Mouse lungs were collected and metastasis nodules were counted. Then the lungs were fixed in 4% formalin and sections were cut, followed by haematoxylin-eosin staining. Four mice were used for each group. No randomization or blinding was used. The bar graph indicates mean ± SD. ***, p < 0.001. K, schematic diagram of the involvement of FOXC1-WNT5A-NF-κB-MMP7 signaling in TNBC cell invasion.
Because MMP7 is regulated by the FOXC1-WNT5A axis, we tested whether MMP7 mediates the effect of WNT5A on migration and invasion in TNBC cells. FOXC1+/WNT5A-KO MDA-MB-231 cells were treated with different concentrations of the recombinant MMP7 protein. MMP7 treatment restored the WNT5A-KO-mediated inhibition of migration and invasion (Fig. 4D–F), to different extents. In addition, the inhibition of migration and invasion was restored by WNT5A treatment in FOXC1-knockout HCC1806 cells (Supplementary Fig. 4B and C). We also knocked down MMP7 expression in FOXC1-overexpressing MDA-MB-231 cells (Supplementary Fig. 4D). As shown in Fig. 4G and H, MMP7 reduction dramatically suppressed cell migration and invasion, which was not affected by WNT5A treatment. Similar results were also found in HCC1806 cells (Supplementary Fig. 4E–G). These data demonstrate the involvement of FOXC1-WNT5A-MMP7 signaling in regulating the invasiveness of TNBC cells.
The effect of FOXC1-WNT5A-MMP7 signaling on TNBC cell invasiveness was further confirmed in vivo. Tail vein injection of MDA-MB-231 cells in nude mice showed that knocking out WNT5A inhibited the FOXC1-induced lung extravasation and metastasis, while MMP7 overexpression reversed this inhibition (Fig. 4I and J; Supplementary Fig. 4H and I). Matrix metalloproteinase proteins have been shown to be involved in tumor progression in multiple processes, including cell migration and invasion.33 MMP7 is highly expressed in BLBC/TNBC compared with other breast cancer subtypes.34 Based on previous and our present findings, we herein propose that a FOXC1-WNT5A-MMP7 signaling axis plays an important role in the migration, invasion, and distant metastasis of TNBC cells.
In summary, our study reveals a novel FOXC1-mediated regulatory mechanism of non-canonical WNT5A signaling pathways in TNBC. This WNT5A-NF-κB-MMP7 signaling is essential for FOXC1-induced invasiveness of TNBC cells (Fig. 4K), suggesting new interventional strategies to inhibit and treat metastasis of TNBC.
Supplementary Material
Acknowledgments
Financial Support: This work was supported by the National Institutes of Health (CA151610), the Avon Foundation for Women (02-2014-063), and David Salomon Translational Breast Cancer Research Fund to Xiaojiang Cui, and the Fashion Footwear Charitable Foundation of New York, Inc., the Entertainment Industry Foundation, the Margie and Robert E. Petersen Foundation, and the Linda and Jim Lippman Research Fund to Armando Giuliano.
Footnotes
Disclosure of Potential Conflicts of Interest
The authors declare no conflict of interests.
Authors’ Contributions
Conception and design by BC Han and XJ Cui. Development of methodology by BC Han, B Zhou, H Tanaka, W Yang, and XJ Cui. Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.) by BC Han, B Zhou, BW Gao, YL Xu, W Yang, and XJ Cui. Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis) by BC Han, B Zhou, Y Qu, S Chung, H Tanaka, W Yang, AE Giuliano, and XJ Cui. Writing, review, and/or revision of the manuscript by BC Han, B Zhou, Y Qu, BW Gao, S Chung, H Tanaka, W Yang, AE Giuliano, and X Cui. Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases) by BC Han, B Zhou, Y Qu, BW Gao, S Chung, W Yang, and XJ Cui. Study supervision by BC Han, AE Giuliano, and XJ Cui.
References
- 1.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 2.Reis-Filho JS, Tutt AN. Triple negative tumours: a critical review. Histopathology. 2008;52:108–118. doi: 10.1111/j.1365-2559.2007.02889.x. [DOI] [PubMed] [Google Scholar]
- 3.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 4.Geyer FC, Lacroix-Triki M, Savage K, Arnedos M, Lambros MB, MacKay A, et al. β-Catenin pathway activation in breast cancer is associated with triple-negative phenotype but not with CTNNB1 mutation. Mod Pathol. 2011;24:209–231. doi: 10.1038/modpathol.2010.205. [DOI] [PubMed] [Google Scholar]
- 5.Xu J, Prosperi JR, Choudhury N, Olopade OI, Goss KH. β-catenin is required for the tumorigenic behavior of triple-negative breast cancer cells. PLoS One. 2015;10:e0117097. doi: 10.1371/journal.pone.0117097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jensen TW, Ray T, Wang J, Li X, Naritoku WY, Han B, et al. Diagnosis of Basal-Like Breast Cancer Using a FOXC1-Based Assay. J Natl Cancer Inst. 2015:107. doi: 10.1093/jnci/djv148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han B, Audeh W, Jin Y, Bagaria S, Cui X. Biology and treatment of basal-like breast cancer. In: Schatten H, editor. Molecular Biology of Breast Cancer. chapter 5. Springer-Humana Press; New York: 2013. pp. 91–109. [Google Scholar]
- 8.Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A. 2010;107:15449–15454. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han B, Qu Y, Jin Y, Yu Y, Deng N, Wawrowsky K, et al. FOXC1 Activates Smoothened-Independent Hedgehog Signaling in Basal-like Breast Cancer. Cell Rep. 2015;13:1046–1058. doi: 10.1016/j.celrep.2015.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sizemore ST, Keri RA. The forkhead box transcription factor FOXC1 promotes breast cancer invasion by inducing matrix metalloprotease 7 (MMP7) expression. J Biol Chem. 2012;287:24631–24640. doi: 10.1074/jbc.M112.375865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi J, Wang Y, Zeng L, Wu Y, Deng J, Zhang Q, et al. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell. 2014;25:210–225. doi: 10.1016/j.ccr.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoeflich KP, O’Brien C, Boyd Z, Cavet G, Guerrero S, Jung K, et al. In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clin Cancer Res. 2009;15:4649–4664. doi: 10.1158/1078-0432.CCR-09-0317. [DOI] [PubMed] [Google Scholar]
- 13.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pukrop T, Klemm F, Hagemann T, Gradl D, Schulz M, Siemes S, et al. Wnt 5a signaling is critical for macrophage-induced invasion of breast cancer cell lines. Proc Natl Acad Sci U S A. 2006;103:5454–5459. doi: 10.1073/pnas.0509703103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pierrou S, Hellqvist M, Samuelsson L, Enerback S, Carlsson P. Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. EMBO J. 1994;13:5002–5012. doi: 10.1002/j.1460-2075.1994.tb06827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujita T, Fujii H. Efficient isolation of specific genomic regions and identification of associated proteins by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) using CRISPR. Biochem Biophys Res Commun. 2013;439:132–136. doi: 10.1016/j.bbrc.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Honda K. The biological role of actinin-4 (ACTN4) in malignant phenotypes of cancer. Cell Biosci. 2015;5:41. doi: 10.1186/s13578-015-0031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weng MT, Luo J. The enigmatic ERH protein: its role in cell cycle, RNA splicing and cancer. Protein Cell. 2013;4:807–812. doi: 10.1007/s13238-013-3056-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bunch H, Calderwood SK. TRIM28 as a novel transcriptional elongation factor. BMC Mol Biol. 2015;16:14. doi: 10.1186/s12867-015-0040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mazurek A, Luo W, Krasnitz A, Hicks J, Powers RS, Stillman B. DDX5 regulates DNA replication and is required for cell proliferation in a subset of breast cancer cells. Cancer Discov. 2012;2:812–825. doi: 10.1158/2159-8290.CD-12-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han SP, Tang YH, Smith R. Functional diversity of the hnRNPs: past, present and perspectives. Biochem J. 2010;430:379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- 23.Jang M, Park BC, Kang S, Chi SW, Cho S, Chung SJ, et al. Far upstream element-binding protein-1, a novel caspase substrate, acts as a cross-talker between apoptosis and the c-myc oncogene. Oncogene. 2009;28:1529–1536. doi: 10.1038/onc.2009.11. [DOI] [PubMed] [Google Scholar]
- 24.Nishita M, Enomoto M, Yamagata K, Minami Y. Cell/tissue-tropic functions of Wnt5a signaling in normal and cancer cells. Trends Cell Biol. 2010;20:346–354. doi: 10.1016/j.tcb.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Klemm F, Bleckmann A, Siam L, Chuang HN, Rietkotter E, Behme D, et al. beta-catenin-independent WNT signaling in basal-like breast cancer and brain metastasis. Carcinogenesis. 2011;32:434–442. doi: 10.1093/carcin/bgq269. [DOI] [PubMed] [Google Scholar]
- 26.Weeraratna AT, Jiang Y, Hostetter G, Rosenblatt K, Duray P, Bittner M, et al. Wnt5a signaling directly affects cell motility and invasion of metastatic melanoma. Cancer Cell. 2002;1:279–288. doi: 10.1016/s1535-6108(02)00045-4. [DOI] [PubMed] [Google Scholar]
- 27.Wang J, Ray PS, Sim MS, Zhou XZ, Lu KP, Lee AV, et al. FOXC1 regulates the functions of human basal-like breast cancer cells by activating NF-kappaB signaling. Oncogene. 2012;31:4798–4802. doi: 10.1038/onc.2011.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y, Wang CL, Li RM, Hui TQ, Su YY, Yuan Q, et al. Wnt5a promotes inflammatory responses via nuclear factor kappaB (NF-kappaB) and mitogen-activated protein kinase (MAPK) pathways in human dental pulp cells. J Biol Chem. 2014;289:21028–21039. doi: 10.1074/jbc.M113.546523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guan PP, Yu X, Guo JJ, Wang Y, Wang T, Li JY, et al. By activating matrix metalloproteinase-7, shear stress promotes chondrosarcoma cell motility, invasion and lung colonization. Oncotarget. 2015;6:9140–9159. doi: 10.18632/oncotarget.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phoomak C, Vaeteewoottacharn K, Sawanyawisuth K, Seubwai W, Wongkham C, Silsirivanit A, et al. Mechanistic insights of O-GlcNAcylation that promote progression of cholangiocarcinoma cells via nuclear translocation of NF-kappaB. Sci Rep. 2016;6:27853. doi: 10.1038/srep27853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gujral TS, Chan M, Peshkin L, Sorger PK, Kirschner MW, MacBeath G. A noncanonical Frizzled2 pathway regulates epithelial-mesenchymal transition and metastasis. Cell. 2014;159:844–856. doi: 10.1016/j.cell.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheel C, Eaton EN, Li SH, Chaffer CL, Reinhardt F, Kah KJ, et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145:926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim GE, Lee JS, Choi YD, Lee KH, Lee JH, Nam JH, et al. Expression of matrix metalloproteinases and their inhibitors in different immunohistochemical-based molecular subtypes of breast cancer. BMC Cancer. 2014;14:959. doi: 10.1186/1471-2407-14-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujita T, Fujii H. Identification of proteins associated with an IFNgamma-responsive promoter by a retroviral expression system for enChIP using CRISPR. PLoS One. 2014;9:e103084. doi: 10.1371/journal.pone.0103084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morley S, You S, Pollan S, Choi J, Zhou B, Hager MH, et al. Regulation of microtubule dynamics by DIAPH3 influences amoeboid tumor cell mechanics and sensitivity to taxanes. Sci Rep. 2015;5:12136. doi: 10.1038/srep12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 39.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.