Abstract
We evolved Thermus thermophilus to efficiently co-utilize glucose and xylose, the two most abundant sugars in lignocellulosic biomass, at high temperatures without carbon catabolite repression. To generate the strain, T. thermophilus HB8 was first evolved on glucose to improve its growth characteristics, followed by evolution on xylose. The resulting strain, T. thermophilus LC113, was characterized in growth studies, by whole genome sequencing, and 13C-metabolic flux analysis (13C-MFA) with [1,6-13C]glucose, [5-13C]xylose, and [1,6-13C]glucose + [5-13C]xylose as isotopic tracers. Compared to the starting strain, the evolved strain had an increased growth rate (~ 2-fold), increased biomass yield, increased tolerance to high temperatures up to 90 °C, and gained the ability to grow on xylose in minimal medium. At the optimal growth temperature of 81 °C, the maximum growth rate on glucose and xylose was 0.44 and 0.46 h−1, respectively. In medium containing glucose and xylose the strain efficiently co-utilized the two sugars. 13C-MFA results provided insights into the metabolism of T. thermophilus LC113 that allows efficient co-utilization of glucose and xylose. Specifically, 13C-MFA revealed that metabolic fluxes in the upper part of metabolism adjust flexibly to sugar availability, while fluxes in the lower part of metabolism remain relatively constant. Whole genome sequence analysis revealed two large structural changes that can help explain the physiology of the evolved strain: a duplication of a chromosome region that contains many sugar transporters, and a 5× multiplication of a region on the pVV8 plasmid that contains xylose isomerase and xylulokinase genes, the first two enzymes of xylose catabolism. Taken together, 13C-MFA and genome sequence analysis provided complementary insights into the physiology of the evolved strain.
Keywords: Thermophile, glucose and xylose metabolism, co-utilization, isotopic labeling, metabolic fluxes
1. INTRODUCTION
Lignocellulosic biomass is an attractive second-generation feedstock for sustainable production of biofuels and chemicals. Hydrolysis of lignocellulose results in the release of glucose and xylose as the two main sugars. However, most organisms including E. coli cannot efficiently utilize glucose and xylose at the same time due to carbon catabolite repression. Carbon catabolite repression results in inefficient fermentation processes where glucose is utilized before xylose. Thus, there is a strong interest in engineering new strains that can simultaneously utilize multiple sugars. Thermophilic organisms provide additional advantages for fermentations of lignocellulosic carbon sources (Blumer-Schuette et al., 2008; Chang and Yao, 2011; Elleuche et al., 2014; Taylor et al., 2009). Many thermophiles can naturally ferment pentoses and hexoses, and in some cases even polymeric precursors and structurally complex polycarbohydrates such as cellulose and hemicellulose (Lynd, 1989; Taylor et al., 2009). Additionally, high process temperatures allow higher rates of feedstock conversion, reduce cooling costs, reduce chances of contaminations, permit easier processing of feedstocks due to lower viscosity, and provide opportunities for enhanced product recovery (Lin and Xu, 2013; Lynd, 1989).
In this work, we have evolved a new strain of the extremely thermophilic bacterium Thermus thermophilus to efficiently co-utilize glucose and xylose at high temperatures without carbon catabolite repression. T. thermophilus is a model thermophilic organism widely used in structural biology studies (Cava et al., 2009; Yokoyama et al., 2000a; Yokoyama et al., 2000b). With its high growth rate, high cell yield, and most importantly the constitutive expression of an efficient natural competence system, T. thermophilus stands out among thermophiles as a potential host for biotechnological applications (Averhoff and Muller, 2010; Cava et al., 2009; Henne et al., 2004). In previous work, we have experimentally validated the metabolic network model of T. thermophilus HB8 using 13C metabolic flux analysis (13C-MFA) (Swarup et al., 2014), thus opening the door for more detailed studies of this organism. Here, we have applied 13C-MFA to elucidate the metabolism of the evolved strain grown on glucose, xylose, and mixture of glucose and xylose. To our knowledge, this is the first time that 13C-MFA has been applied to elucidate co-utilization of glucose and xylose in any organism. As such, this study serves as an important benchmark for future investigations of glucose and xylose co-utilization in this and other organisms.
2. MATERIALS AND METHODS
2.1. Materials and growth medium
Media and chemicals were purchased from Sigma-Aldrich (St. Louis, MO). [1,6-13C]Glucose (99.5 atom% 13C) and [5-13C]xylose (99.5% 13C) were purchased from Cambridge Isotope Laboratories (Andover, MA). [U-13C]Xylose (99% 13C) was purchased from Isotec (St. Louis, MO). Wolfe’s minerals (Cat. No. MD-TMS) and Wolfe’s vitamins (Cat. No. MD-VS) were purchased from ATCC (Manassas, VA), and Tris solution was purchased from Cellgro (Cat. No. 46-031-CM). Glucose and xylose stock solutions were prepared at 20 wt% in distilled water. The growth medium (Swarup et al., 2014) contained (per liter of medium): 0.50 g K2HPO4, 0.30 g KH2PO4, 0.50 g NH4Cl, 0.50 g NaCl, 0.20 g MgCl2.6H2O, 0.04 g CaSO4.2H2O, 40 mL of 1 M Tris, 5 mL of Wolfe’s minerals, 5 mL of Wolfe’s vitamins, and 0.05 g/L of yeast extract. Xylose and glucose were added as indicated in the text. All media and stock solutions were sterilized by filtration.
2.2. Strain, growth conditions, and adaptive evolution
The starting strain for adaptive evolution was T. thermophilus HB8 (ATCC 27634). Cells were serially propagated with daily serial transfers in aerated mini-bioreactors at 75°C with a working volume of 10 mL, as described previously (Swarup et al., 2014). T. thermophilus was first evolved on medium containing 2 g/L glucose to select for mutants with improved growth characteristics. After about one month of adaptive evolution, growth characteristics of the evolved culture were determined. While the starting strain was unable to grow on xylose, i.e. no growth was observed within 24 hr, the evolved culture was able to grow on xylose to some extent. To improve growth on xylose, the cells were further evolved on medium containing 2 g/L xylose as the main carbon source to select for mutants with enhanced xylose utilization. After about one month of adaptive evolution growth on xylose was significantly improved. The culture was then streaked on a xylose-agar plate and a single colony was isolated. A frozen stock was prepared in 15% glycerol and stored at −80°C. This frozen stock, designated T. thermophilus LC113, was used for all subsequent experiments.
To determine the optimal growth temperature of T. thermophilus LC113, cells from the frozen stock were first pre-cultured overnight at the indicated growth temperature (or at 75 °C if no detectable growth was observed overnight) in medium containing 2 g/L of glucose or xylose. Next, 1 mL of the pre-culture was used to inoculate 10 mL of fresh medium at the indicated growth temperature with either 2 g/L of glucose or 2 g/L of xylose. Maximum growth rate during the exponential growth phase was then determined as described below.
For tracer experiments, cells from the frozen stock were first pre-cultured overnight at 81 °C to early stationary phase. Next, 50 µL of this pre-culture was used to inoculate 10 mL of fresh medium containing either 1.8 g/L of [1,6-13C]glucose, 1.8 g/L of [5-13C]xylose, or a mixture of 0.9 g/L of [1,6-13C]glucose and 0.9 g/L of [5-13C]xylose. The initial optical density (OD600) of the inoculated cultures was about 0.01. Cells were then grown aerobically at 81 °C until mid-exponential phase. Biomass samples were collected for GC-MS analysis when OD600 reached 0.7.
2.3. Analytical methods
Biomass concentration was determined by measuring the optical density at 600 nm (OD600) using a spectrophotometer (Eppendorf BioPhotometer). The OD600 values were converted to cell dry weight concentrations using a pre-determined OD600-dry cell weight relationship: 1.0 OD600 = 0.34 gDW/L for T. thermophilus LC113. A molecular weight of 25.3 g/C-mol was assumed for dry biomass. Xylose concentrations were determined by GC-MS analysis using a solution containing [U-13C]xylose as internal standard as described in (Cordova and Antoniewicz, 2015), and by HPLC analysis with Agilent 1200 Series HPLC as described in (Au et al., 2014). Glucose concentrations were determined by YSI 2700 biochemistry analyzer (YSI, Yellow Springs, OH).
2.4. Determination of biomass yields and biomass specific rates
Biomass yields on glucose and xylose were determined as the slopes of least-squares regression of biomass concentration versus glucose and xylose concentration, respectively (He et al., 2014). The specific growth rate was determined as the slope of least-squares regression of ln(OD600) versus time during mid-exponential growth phase (typically for OD600 values between 0.05 and 0.7). Specific glucose and xylose uptake rates (qGluc, qXyl) were determined as the ratios of the growth rates and biomass yields.
2.5. Whole genome SMRT sequencing
High molecular weight genomic DNA was isolated from T. thermophilus LC113 strain using Genomic-tip 100/G (QIAGEN) according the manufacturer’s instructions. Single molecule real time sequencing (SMRT) sequencing was performed at the University of Delaware DNA Sequencing & Genotyping Center. SMRTbell DNA libraries were constructed according to the PacBio standard protocol. Template preparation was performed using BluePippin (Sage Science) size-selection system including DNA damage and end repair steps and ligation to hairpin adapters. Both libraries were size-selected starting at 10 kb and with an average library size of 20kb as measured by Fragment Analyzer (Advanced Analytical Technologies, Inc). Sequencing was performed on PacBio RSII (Pacific Biosciences, Menlo Park, Ca) instrument using P6-P4 chemistry, mag-bead loading and 4-hour movie time. 81,270 mapped reads with a mean read length of 11,370 bp was obtained in a single SMRT cell. To identify consensus and variant sequences, quality-filtered reads were mapped against reference sequences using the RS Resequencing protocol within the SMRT Analysis version 2.3 through the SMRT Portal, while de novo assembly was performed with the HGAP2 protocol. We obtained 337× coverage on the chromosome (accession number NC_006461), 202× coverage on plasmid pTT27 (except as noted in results, NC_006462), 45× coverage on pTT8 (NC_005792), and 484× coverage on pVV8 (except as noted in results, AB677526). The RS Modification and Motif Analysis protocol was used to identify base modifications and methyltransferase recognition motifs.
2.6. Gas chromatography mass spectrometry
GC-MS analysis was performed on an Agilent 7890B GC system equipped with a DB-5MS capillary column (30 m, 0.25 mm i.d., 0.25 µm-phase thickness; Agilent J&W Scientific), connected to an Agilent 5977A Mass Spectrometer operating under ionization by electron impact (EI) at 70 eV. The interface temperature was 280°C, MS source temperature 230°C, and MS quad temperature 150°C. Mass spectra were acquired in single ion monitoring (SIM) mode.
2.7. GC-MS analysis of biomass amino acids
Labeling of biomass amino acids was determined by GC-MS analysis of tert-butyldimethylsilyl (TBDMS) derivatives as described in (Long and Antoniewicz, 2014). For GC-MS analysis, 1 mL of culture was centrifuged, the cell pellet hydrolyzed overnight at 110 °C in 6 N HCl, and derivatized as described in (Antoniewicz et al., 2007a). 1 µL was injected at 1:40 split ratio. Helium flow was maintained at 1.0 mL/min. The injection port temperature was 250°C. The temperature of the column was started at 80 °C for 2 min, increased to 280 °C at 7 °C/min and held for 20 min. Mass isotopomer distributions were obtained by integration of ion chromatograms (Antoniewicz et al., 2007a), and corrected for natural isotope abundances (Fernandez et al., 1996).
2.8. GC-MS analysis of glucose and xylose
Labeling of glucose and xylose was determined by GC-MS analysis of aldonitrile propionate derivatives (Antoniewicz et al., 2011). For GC-MS analysis, 10 µL of culture supernatant was derivatized as described in (Antoniewicz et al., 2011). 1 µL was injected at 1:40 split ratio. Helium flow was maintained at 1.0 mL/min. The injection port temperature was 250 °C. The temperature of the column was started at 80 °C for 1 min, increased to 280 °C at 15 °C/min, and held for 6 min. Labeling of glucose was determined from the mass isotopomer distributions of fragments at m/z 370 and m/z 173, which contain carbon atoms C1-C5 and C5-C6 of glucose, respectively (Antoniewicz et al., 2011). Labeling of xylose was determined from the mass isotopomer distribution of the fragment at m/z 173, which contains carbon atoms C4-C5 of xylose (Cordova and Antoniewicz, 2015).
2.9. Biomass composition analysis
The biomass composition of T. thermophilus LC113 was determined using the methods described in (Long and Antoniewicz, 2014). Specifically, weight percentages of proteins, RNA, glycogen and lipids (wt% of dry biomass) were measured, as well as mol-fractions of amino acids and fatty acids. For these analyses, fully 13C-labeled E. coli was used as internal standard as described in (Long and Antoniewicz, 2014).
2.10. 13C-Metabolic flux analysis
13C-MFA was performed using the Metran software (Yoo et al., 2008), which is based on the elementary metabolite units (EMU) framework (Antoniewicz et al., 2007b; Young et al., 2008). The metabolic network model used for 13C-MFA is given in Supplemental Materials. The model is based on the validated model described previously for T. thermophilus HB8 (Swarup et al., 2014). The model includes all major metabolic pathways of central carbon metabolism, including glycolysis, non-oxidative pentose phosphate pathway, TCA cycle, glyoxylate shunt, anaplorotic and cataplerotic reactions, reactions for xylose metabolism, as well as lumped amino acid biosynthesis pathways, and a lumped cell growth reaction that is based on the measured biomass composition and information provided in (Lee et al., 2014). The oxidative pentose phosphate pathway was not included in the model based on the results described in (Swarup et al., 2014). For 13C-MFA, we also accounted for the exchange of intracellular and extracellular CO2 that can dilute labeling of intracellular metabolites (Leighty and Antoniewicz, 2012), and included G-value parameters to describe fractional labeling of amino acids. As described previously (Antoniewicz et al., 2007c), the G-value represents the fraction of a metabolite produced during the tracer experiment, while 1-G represents the fraction that is naturally labeled (i.e. from the inoculum). By default, one G-value parameter was included for each measured amino acid in each data set. Metabolic fluxes were estimated by minimizing the variance-weighted sum of squared residuals (SSR) between the experimentally measured and model predicted mass isotopomer distributions and substrate uptake rates using non-linear least-squares regression (Antoniewicz et al., 2006). Flux estimation was repeated at least 10 times starting with random initial values for all fluxes to find a global solution. At convergence, accurate 95% confidence intervals were computed for all estimated fluxes by evaluating the sensitivity of the minimized SSR to flux variations (Antoniewicz et al., 2006). Standard deviations of fluxes was determined as follows:
2.11. Goodness-of-fit analysis
To determine the goodness-of-fit, the 13C-MFA fitting result was subjected to a χ2-statistical test. In short, assuming the model is correct and data are without gross measurement errors, the minimized SSR is a stochastic variable with a χ2-distribution. The number of degrees of freedom is equal to the number of fitted measurements n minus the number of estimated independent parameters p. The acceptable range of SSR values is between χ2α/2(n-p) and χ21-α/2(n-p), where α is a certain chosen threshold value, for example 0.05 for 95% confidence interval (Antoniewicz et al., 2006).
3. RESULTS AND DISCUSSION
3.1. Adaptive evolution
Wild-type T. thermophilus HB8 was used as the starting strain for adaptive evolution. First, the strain was evolved on glucose through daily serial transfers to improve general growth characteristics. After about a month of adaptive evolution the growth rate on glucose was nearly doubled. Another significant improvement was that the cells no longer attached to the glass of the bioreactor, an undesired characteristic of wild-type T. thermophilus HB8; instead, the cells grew exclusively in suspension.
At this point, we tested the evolved culture for growth on other carbon sources, including xylose. While wild-type T. thermophilus HB8 is known to contain the complete machinery necessary for xylose catabolism (Ohtani et al., 2012), e.g. xylose isomerase isolated from T. thermophilus is often used to engineer other strains to improve xylose utilization (Karhumaa et al., 2005; Walfridsson et al., 1996), we and others have observed that wild-type T. thermophilus HB8 is unable to grow on xylose in minimal medium (Nunes et al., 1992). Interestingly, the evolved culture did grow to a limited extent on xylose. To improve growth on xylose, the culture was further evolved on xylose through daily serial transfers. After about a month of evolution the growth rate on xylose was dramatically improved. The evolved culture was then streaked on a xylose-agar plate and a single colony was isolated. A frozen stock was prepared that was designated T. thermophilus LC113. This frozen stock was used for all subsequent experiments.
3.2. Determination of optimal growth temperature
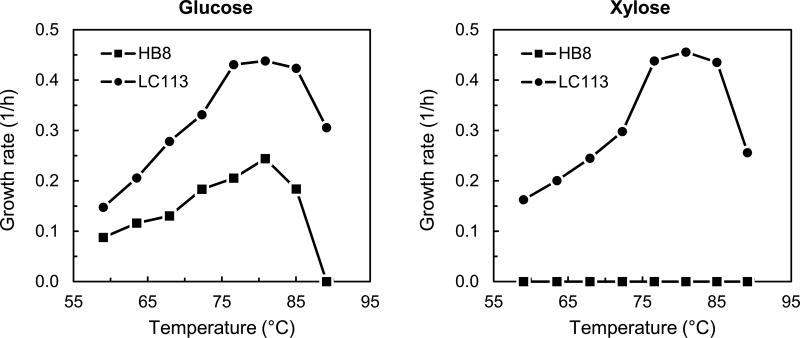
The evolved T. thermophilus strain LC113 was characterized in batch experiments to determine optimal growth conditions. First, we determined the optimal growth temperature. Figure 1 shows the measured specific growth rates of T. thermophilus LC113 grown at temperatures between 60 and 90 °C. For comparison, the specific growth rates of T. thermophilus HB8 grown on glucose, as reported in (Swarup et al., 2014), are also shown. The optimal growth temperature for T. thermophilus LC113 was 81 °C on both glucose and xylose. This is the same optimal growth temperature as for T. thermophilus HB8 (Swarup et al., 2014). The maximum growth rates of T. thermophilus LC113 on glucose and xylose were 0.44 and 0.46 h−1, respectively. The growth rate on glucose nearly doubled compared to the starting strain (0.25 h−1), and more significantly, the evolved strain was able to grow efficiently on xylose. The biomass yield was identical for glucose and xylose (0.36 ± 0.01 gDW/g, Table 1), and slightly higher than reported for the starting strain (0.33 ± 0.01 gDW/g) (Swarup et al., 2014). The maximum growth temperature of T. thermophilus LC113 was 90 °C compared to 85 °C for the starting strain (Figure 1). In summary, we determined that the evolved strain had an increased growth rate on glucose, increased biomass yield, increased tolerance to high temperatures, and gained the ability to grow on xylose. In several additional growth experiments, we also noted that T. thermophilus LC113 simultaneously co-utilized glucose and xylose without carbon catabolite repression. To elucidate this important characteristic of the evolved strain, detailed analysis by 13C-MFA was performed as is described next.
Figure 1.
Specific growth rates of wild-type T. thermophilus HB8 and the evolved strain T. thermophilus LC113 at different temperatures in medium containing 2 g/L of glucose or 2 g/L of xylose.
Table 1.
Comparison of wild-type T. thermophilus HB8 and evolved strain T. thermophilus LC113.
| T. thermophilus HB8* | T. thermophilus LC113 | |
|---|---|---|
| Optimal growth temperature | 81 °C | 81 °C |
| Maximum growth rate on glucose | 0.25 ± 0.02 h−1 | 0.44 ± 0.03 h−1 |
| Maximum growth rate on xylose | No growth** | 0.46 ± 0.03 h−1 |
| Biomass yield on glucose | 0.33 ± 0.02 gDW/g | 0.36 ± 0.01 gDW/g |
| Biomass yield on xylose | No growth** | 0.36 ± 0.01 gDW/g |
Data for T. thermophilus HB8 were taken from (Swarup et al., 2014).
No detectable growth after 24 hrs.
3.3. 13C-Metabolic flux analysis
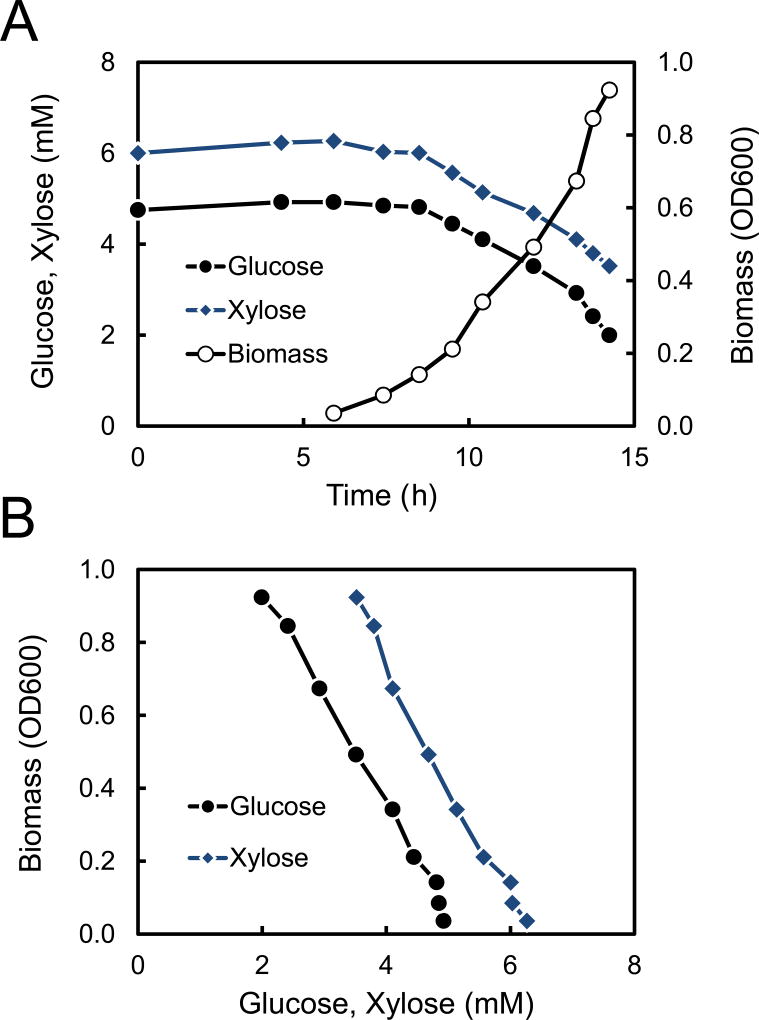
Two isotopic tracers were selected for 13C-MFA studies of T. thermophilus LC113, namely [1,6-13C]glucose and [5-13C]xylose. These two tracers were selected based on optimal tracer experiment design as described in (Antoniewicz, 2013; Antoniewicz, 2015; Crown and Antoniewicz, 2013). Parallel labeling experiments were performed to measure metabolic fluxes for three growth conditions: glucose only, xylose only, and 1:1 (wt/wt) mixture of glucose and xylose. T. thermophilus LC113 was grown aerobically at 81 °C under the three growth conditions. Figure 2 shows the growth profile for the culture with glucose + xylose. The measured glucose and xylose concentrations plotted vs. time (Figure 2A) and vs. biomass (Figure 2B) demonstrate that glucose and xylose were utilized simultaneously without any carbon catabolite repression. Table 2 shows the measured growth rates and specific glucose and xylose uptake rates for all three growth conditions. The growth rates were similar for all three conditions: 0.42 ± 0.02 for glucose only, 0.44 ± 0.02 for xylose only, and 0.45 ± 0.02 for glucose + xylose. For the culture with glucose + xylose, the specific glucose and xylose uptake rates were 4.1 ± 0.2 and 3.8 ± 0.2 mmol/gDW/h, respectively, compared to 6.6 ± 0.2 mmol/gDW/h for glucose only, and 8.2 ± 0.2 mmol/gDW/h for xylose only. No fermentation products were detected in any of the cultures, i.e. no acetate, lactate, succinate, ethanol or other common fermentation products were detected in supernatants by HPLC analysis.
Figure 2.
(A) Glucose, xylose and biomass profiles in a 13C-labeling experiment with 0.9 g/L of [1,6-13C]glucose + 0.9 g/L of [5-13C]xylose. (B) Biomass concentration plotted against glucose and xylose concentration demonstrates simultaneous utilization of glucose and xylose without carbon catabolite repression.
Table 2.
Physiological characteristics of T. thermophilus LC113 grown on 1.8 g/L of [1,6-13C]glucose, 1.8 g/L of [5-13C]xylose, and a mixture of 0.9 g/L of [1,6-13C]glucose + 0.9 g/L of [5-13C]xylose.
| [1,6-13C]glucose | [5-13C]xylose | [1,6-13C]glucose + [5-13C]xylose | |
|---|---|---|---|
| Growth rate (h−1) | 0.42 ± 0.02 | 0.44 ± 0.02 | 0.45 ± 0.02 |
| qGluc (mmol/gDW/h) | 6.6 ± 0.2 | - | 4.1 ± 0.2 |
| qXyl (mmol/gDW/h) | - | 8.2 ± 0.2 | 3.8 ± 0.2 |
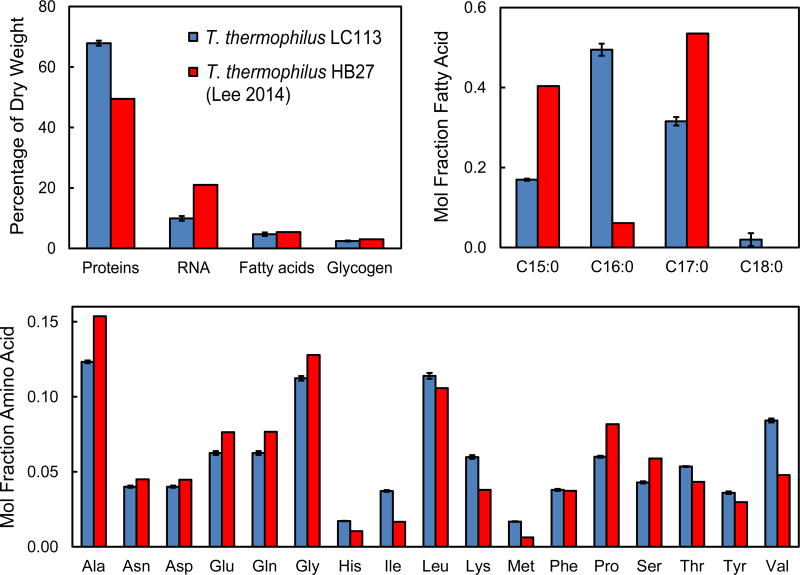
A metabolic model was constructed for 13C-MFA based on the model described in (Swarup et al., 2014). In order to construct the lumped biomass equation the composition of biomass of T. thermophilus LC113 grown at 81 °C was measured using the methods described in (Long and Antoniewicz, 2014). Figure 3 compares the measured biomass composition for T. thermophilus LC113 and the biomass composition previously reported for T. thermophilus HB27 (Lee et al., 2014). Overall, we measured a higher protein content for T. thermophilus LC113 (68% of dry weight) compared to T. thermophilus HB27 (50 wt%), and a lower RNA content (10 wt%) compared to 21 wt% for T. thermophilus HB27 (note that the value of 21 wt% was not measured directly, but was assumed the same as for E. coli). The relative distribution of fatty acids was also different between the two strains. For T. thermophilus LC113 we only detected fully saturated fatty acids, with the main fatty acids being C16:0 (50%), C17:0 (32%), and C15:0 (15%). For T. thermophilus HB27, the main fatty acids were reported to be C17:0 (54%) and C15:0 (40%). The relative distribution of amino acids was also slightly different for the two strains. For T. thermophilus LC113, we measured a higher content of valine, lysine and isoleucine and a lower content of alanine, proline and serine compared to the amino acid composition for T. thermophilus HB27. These results suggest that different T. thermophilus strains grown at different temperatures may have a significantly different biomass composition, i.e. biomass composition of T. thermophilus HB27 was characterized at 70 °C (Lee et al., 2014). It is therefore important to measure biomass composition under relevant experimental conditions to ensure that the biomass reaction used for 13C-MFA calculations is representative of the organism studied.
Figure 3.
Measured biomass composition for T. thermophilus LC113 (mean ± stdev, n=2) compared to T. thermophilus HB27 (Lee et al., 2014).
To determine intracellular metabolic fluxes by 13C-MFA, isotopic labeling of biomass amino acids from the 13C-labeling experiments was measured by GC-MS. The measured mass isotopomer distributions are given in Supplemental Materials. The 13C-labeling data and the measured external rates were fitted to the validated metabolic network model of T. thermophilus (Swarup et al., 2014) to quantify metabolic fluxes. Statistically acceptable fits were obtained for all three growth conditions. The minimized sum of squared residuals (SSR) were lower than the maximum statistically acceptable SSR values at 95% confidence level (Supplemental Materials), assuming a constant measurement error of 0.4 mol% for all GC-MS measurements.
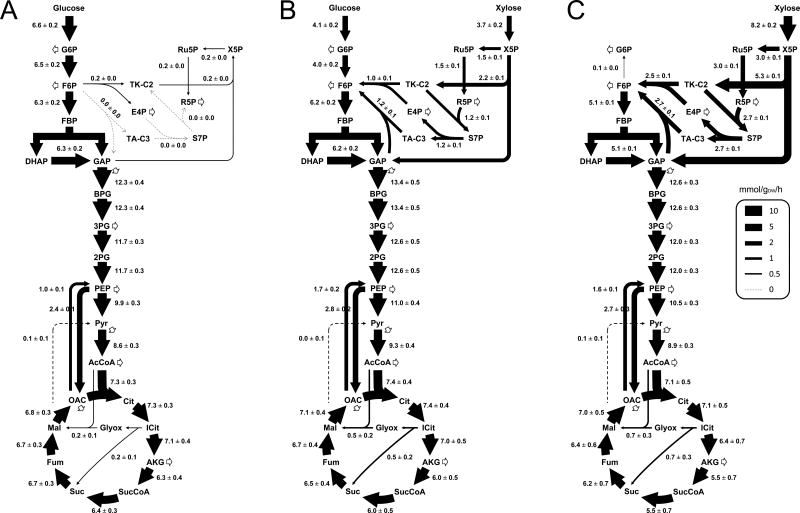
The estimated metabolic fluxes under all three growth conditions are shown in Figure 4. The complete flux results, including 95% confidence intervals, are given in Supplemental Materials. For growth on glucose only (Figure 4A), T. thermophilus LC113 catabolized glucose almost exclusively via glycolysis and the TCA cycle. The biomass precursors erythrose-4-phosphate and ribose-5-phosphate were produced via the non-oxidative pentose phosphate pathway, and largely via transketolase with little or no contribution from transaldolase. Several other reactions were also determined to carry no significant flux, including the glyoxylate shunt (0.2 ± 0.1) and malic enzyme (0.1 ± 0.1). These results are in good agreements with the results obtained for wild-type T. thermophilus HB8 grown on glucose (Swarup et al., 2014). For growth on xylose only (Figure 4C), T. thermophilus LC113 catabolized xylose mainly via the non-oxidative PPP and TCA cycle. Malic enzyme was again inactive (0.1 ± 0.1) and the glyoxylate shunt flux was very low (0.7 ± 0.3). For growth on glucose + xylose (Figure 4B), glycolysis, non-oxidative pentose phosphate pathway and the TCA cycle were all active, while malic enzyme was again inactive (0.0 ± 0.1). Interestingly, we noted that TCA cycle fluxes were very similar for all three growth conditions, e.g. the citrate synthase flux was 7.3 ± 0.3 mmol/gDW/h for glucose only, 7.1 ± 0.5 mmol/gDW/h for xylose only, and 7.4 ± 0.4 mmol/gDW/h for glucose + xylose. In contrast, metabolic fluxes in the upper part of metabolism, i.e. in upper glycolysis and non-oxidative pentose phosphate pathway, adjusted flexibly to varying sugar availability. One possible explanation for the relatively constant TCA cycle fluxes is that isocitrate dehydrogenase is the main reaction used for NADPH production by T. thermophilus LC113, as was revealed by 13C-MFA. Other reactions that could theoretically produce NADPH carried little or no flux, including malic enzyme and transhydrogenase (see Supplemental Materials). Taken together, these results suggest that NADPH production may be a limiting step in the biosynthesis of new biomass and that engineering pathways to enhance NADPH supply, e.g. by expressing an active oxidative PPP from other thermophilic organisms (Cordova et al., 2015), could be a potential strategy to further improve the growth rate of T. thermophilus LC113.
Figure 4.
Metabolic flux maps for T. thermophilus LC113 under three growth conditions: (A) glucose only, (B) glucose + xylose, (C) xylose only. Fluxes were determined by 13C-MFA (flux ± stdev; mmol/gDW/h).
3.4. Whole genome sequencing
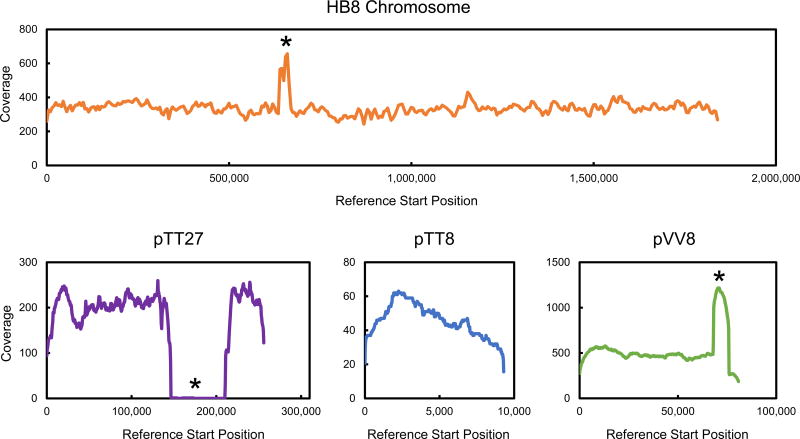
To gain more insight into the genetic basis for the improved growth of T. thermophilus LC113 on glucose and xylose, the complete genome of the evolved strain was sequenced using PacBio’s RSII third generation DNA sequencing system, known as SMRT (Single Molecule, Real-Time) sequencing. A de novo assembly using the HGAP2 protocol resulted in 8 contigs (Supplemental Materials). The largest (1.90 Mbp) contig mapped onto the chromosome of T. thermophilus HB8, the second largest (215 kbp) contig mapped onto T. thermophilus HB8 megaplasmid pTT27, the third largest (116 kbp) contig mapped onto T. thermophilus HB8 plasmid pVV8, and another (24 kbp) contig mapped onto T. thermophilus HB8 plasmid pTT8. Three other contigs mapped onto one particular region of T. thermophilus HB8 plasmid pVV8. These results confirm that T. thermophilus LC113 has retained all three plasmids that have been reported for T. thermophilus HB8 (Ohtani et al., 2012). Detailed analysis of differences between the genome sequences of T. thermophilus LC113 and T. thermophilus HB8 revealed 133 changes (14 deletions, 71 insertions, and 48 substitutions; see Supplemental Materials for details).
Several mutations occurred in genes related to glucose and xylose metabolism, for example: TTHA0685 (ABC sugar transporter), TTHA0687 (ABC sugar transporter), TTHV084 (xylulokinase), and TTHV087 (ABC sugar transporter). In addition, three large structural changes were identified that can also help to explain the physiology of the evolved strain (Figure 5, and Supplemental Materials for additional details): 1) a 28,214 bp duplication of a chromosomal region that contains many sugar transporters (HB8 chromosome coordinates 639,463–667,676); 2) a multiplication of a 7,864 bp region on the pVV8 plasmid (pVV8 coordinates 68,155–76,018) that contains, among others, xylose isomerase and xylulokinase; and 3) a 64,356 bp gap in the megaplasmid pTT27 flanked on either end with identical 32 bp sequences likely facilitating recombination (pTT27 coordinates 146,452–210,807, containing mainly “hypothetical proteins”). In both instances where a duplication/multiplication of a genetic element occurred, one end of the repeated region encodes a transposase with another transposase element nearby. The chromosomal duplication contains two transposase genes inserted within the repeated region that have homology with other regions of the published HB8 chromosome, while the repeated region in pVV8 contains one transposase gene and another located within 3 kb. This repeated pVV8 region contains xylose isomerase and xylulokinase genes. Xylose isomerase (EC 5.3.1.5) catalyzes the isomerization of xylose to xylulose (the first step of xylose metabolism), and xylulokinase (EC 2.7.1.17) catalyzes the conversion of xylulose to xylulose 5-phosphate (the second step of xylose metabolism). It is worth noting that the three smaller contigs that were found to map onto plasmid pVV8 all mapped onto the region that was multiplied. We do not believe these smaller contigs represent separate genetic elements, but rather bioinformatics-related difficulties in assembling large repeated units into a single sequence. Based on depth of coverage and the de novo assembly of contig 21 (see Supplemental Materials), we believe there are five copies of the repeated region in the pVV8 plasmid of the LC133 strain. It is interesting that these genes are present in the parent strain yet it is incapable of growth on xylose, while excess copy number facilitates growth; synthetic regulation of genes in these regions in T. thermophilus HB8 is a likely target for genome engineering.
Figure 5.
Coverage maps based on whole genome sequence analysis using T. thermophilus HB8 as a reference strain. Asterisks indicate locations where large structural changes were detected. Significant increases in coverage indicate replicated regions, and zero coverage indicates deletion of a region. Low coverage of pTT8 is in part due to plasmid size under the 10 kb size selection performed during SMRT library preparation.
3.5. Comparison with other glucose and xylose co-utilizing strains
To our best knowledge, this is the first time that co-utilization of glucose and xylose has been elucidated by 13C-MFA in any organism. Thus, a detailed comparison of flux results with other biological systems is not possible. Instead, we can compare macroscopic growth characteristics of T. thermophilus LC113 with other strains that have been engineered to co-utilize glucose and xylose. Table 3 compares the growth rates and specific glucose and xylose uptake rates of T. thermophilus LC113 and seven other strains that have been reported in literature. We only found one reference where a thermophile was shown to co-utilize glucose and xylose (Joshua et al., 2011): wild-type Sulfolobus acidocaldarius was shown to co-utilize both sugars, although the specific substrate uptake rates were much lower (4-fold lower) resulting in a much lower growth rate of 0.08 h−1. Several E. coli strains have been engineered in recent years for co-utilization of glucose and xylose. It is well known that inactivation of key components of the PTS transport system results in simultaneous utilization of glucose and xylose (Nichols et al., 2001). Liang et al (Liang et al., 2015) reported four E. coli knockout strains with varying levels of preference for glucose and xylose (Table 3), ranging from strains that strongly preferred xylose over glucose (W3110I) to strains that preferred glucose over xylose (W3110C). Gawand et al. (Gawand et al., 2013) engineered a multiple-knockout E. coli strain that stoichiometrically co-utilized glucose and xylose at high specific glucose and xylose uptake rates, although resulting in an overall reduced growth rate. Significant efforts are also on-going to engineer S. cerevisiae to simultaneously utilize glucose and xylose. Because S. cerevisiae cannot catabolize xylose natively new transporters and enzymes for xylose catabolism must be inserted, challenges that are still being investigated (Nijland et al., 2014; Reider Apel et al., 2016; Shen et al., 2015). Overall, the evolved T. thermophilus LC113 compares favorably to other strains that have been engineered to date for glucose/xylose co-utilization. It has the highest reported growth rate for growth on glucose + xylose, has a favorable ratio of glucose to xylose uptake of slightly above 1.0, and has high specific glucose and xylose utilization rates when grown on medium containing both substrates (Table 3).
Table 3.
Comparison of glucose and xylose co-utilization characteristics of eight strains.
| Strain | Temp (°C) |
Growth rate (h−1) |
qGluc (mmol/gDW/h) |
qXyl (mmol/gDW/h) |
qGluc/qXyl (mol/mol) |
Reference |
|---|---|---|---|---|---|---|
| T. thermophilus LC113 | 81 | 0.45 | 4.1 | 3.8 | 1.1 | This work |
| Sulfolobus acidocaldarius DSM 639 | 75 | 0.08 | 0.8 | 1.0 | 0.8 | (Joshua et al., 2011) |
| E. coli W3110I | 37 | n/a | 1.3 | 5.9 | 0.2 | (Liang et al., 2015) |
| E. coli W3110H | 37 | n/a | 0.7 | 2.7 | 0.3 | (Liang et al., 2015) |
| E. coli W3110C | 37 | n/a | 4.0 | 2.7 | 1.5 | (Liang et al., 2015) |
| E. coli W3110G | 37 | n/a | 2.1 | 2.5 | 0.8 | (Liang et al., 2015) |
| E. coli LMSE2 | 37 | 0.38 | 3.9 | 4.9 | 0.8 | (Gawand et al., 2013) |
| S. cerevisiae HXT36-N367A | 30 | n/a | 5.2 | 1.8 | 2.9 | (Nijland et al., 2014) |
4. CONCLUSIONS
Thermus spp is one of the most widespread genera of thermophilic bacteria in nature. Isolates have been found in natural as well as in man-made thermal environments, including in many thermogenic composting sites of lignocellulosic biomass (Beffa et al., 1996; Lyon et al., 2000). T. thermophilus has several characteristics that makes it an ideal candidate for biotechnological applications (Averhoff and Muller, 2010; Cava et al., 2009; Henne et al., 2004). Although wild-type T. thermophilus HB8 is unable to grow on xylose, other T. thermophilus strains have been isolated that can catabolize xylose, and even xylan (Lyon et al., 2000). In this work, we have applied adaptive laboratory evolution to evolve T. thermophilus HB8 to efficiently co-utilize glucose and xylose. We demonstrate that the evolved strain T. thermophilus LC113 utilizes both substrates simultaneously without carbon catabolite repression, an important characteristic for the development of next generation bioprocesses. 13C-MFA results and whole genome sequence analysis provided important insights into the flexible metabolism of T. thermophilus LC113 that allows it to efficient co-utilize glucose and xylose. We also identified NADPH production as a potential bottleneck in the metabolism of this strain. Since this is the first time that 13C-MFA has been applied to elucidate co-utilization of glucose and xylose in any organism, detailed comparison with other biological systems was not possible. As such, this work serves as an important benchmark for future investigations of glucose/xylose co-utilization in other organisms.
Supplementary Material
Acknowledgments
This work was supported by NSF-MCB-1120684 grant.
References
- Antoniewicz MR. 13C metabolic flux analysis: optimal design of isotopic labeling experiments. Curr Opin Biotechnol. 2013;24:1116–21. doi: 10.1016/j.copbio.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR. Parallel labeling experiments for pathway elucidation and 13C metabolic flux analysis. Curr Opin Biotechnol. 2015;36:91–97. doi: 10.1016/j.copbio.2015.08.014. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab Eng. 2006;8:324–37. doi: 10.1016/j.ymben.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Accurate assessment of amino acid mass isotopomer distributions for metabolic flux analysis. Anal Chem. 2007a;79:7554–9. doi: 10.1021/ac0708893. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Elementary metabolite units (EMU): a novel framework for modeling isotopic distributions. Metab Eng. 2007b;9:68–86. doi: 10.1016/j.ymben.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Measuring deuterium enrichment of glucose hydrogen atoms by gas chromatography/mass spectrometry. Anal Chem. 2011;83:3211–6. doi: 10.1021/ac200012p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewicz MR, Kraynie DF, Laffend LA, Gonzalez-Lergier J, Kelleher JK, Stephanopoulos G. Metabolic flux analysis in a nonstationary system: fed-batch fermentation of a high yielding strain of E. coli producing 1,3-propanediol. Metab Eng. 2007c;9:277–92. doi: 10.1016/j.ymben.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au J, Choi J, Jones SW, Venkataramanan KP, Antoniewicz MR. Parallel labeling experiments validate Clostridium acetobutylicum metabolic network model for C metabolic flux analysis. Metab Eng. 2014;26:23–33. doi: 10.1016/j.ymben.2014.08.002. [DOI] [PubMed] [Google Scholar]
- Averhoff B, Muller V. Exploring research frontiers in microbiology: recent advances in halophilic and thermophilic extremophiles. Research in microbiology. 2010;161:506–14. doi: 10.1016/j.resmic.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Beffa T, Blanc M, Lyon PF, Vogt G, Marchiani M, Fischer JL, Aragno M. Isolation of Thermus strains from hot composts (60 to 80 degrees C) Appl Environ Microbiol. 1996;62:1723–7. doi: 10.1128/aem.62.5.1723-1727.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer-Schuette SE, Kataeva I, Westpheling J, Adams MW, Kelly RM. Extremely thermophilic microorganisms for biomass conversion: status and prospects. Curr Opin Biotechnol. 2008;19:210–7. doi: 10.1016/j.copbio.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Cava F, Hidalgo A, Berenguer J. Thermus thermophilus as biological model. Extremophiles. 2009;13:213–31. doi: 10.1007/s00792-009-0226-6. [DOI] [PubMed] [Google Scholar]
- Chang T, Yao S. Thermophilic, lignocellulolytic bacteria for ethanol production: current state and perspectives. Appl Microbiol Biotechnol. 2011;92:13–27. doi: 10.1007/s00253-011-3456-3. [DOI] [PubMed] [Google Scholar]
- Cordova LT, Antoniewicz MR. 13C Metabolic flux analysis of the extremely thermophilic, fast growing, xylose-utilizing Geobacillus strain LC300. Metab Eng. 2015 doi: 10.1016/j.ymben.2015.06.004. [DOI] [PubMed] [Google Scholar]
- Cordova LT, Long CP, Venkataramanan KP, Antoniewicz MR. Complete genome sequence, metabolic model construction and phenotypic characterization of Geobacillus LC300, an extremely thermophilic, fast growing, xylose-utilizing bacterium. Metab Eng. 2015;32:74–81. doi: 10.1016/j.ymben.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crown SB, Antoniewicz MR. Parallel labeling experiments and metabolic flux analysis: Past, present and future methodologies. Metab Eng. 2013;16:21–32. doi: 10.1016/j.ymben.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleuche S, Schroder C, Sahm K, Antranikian G. Extremozymes--biocatalysts with unique properties from extremophilic microorganisms. Curr Opin Biotechnol. 2014;29:116–23. doi: 10.1016/j.copbio.2014.04.003. [DOI] [PubMed] [Google Scholar]
- Fernandez CA, Des Rosiers C, Previs SF, David F, Brunengraber H. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J Mass Spectrom. 1996;31:255–62. doi: 10.1002/(SICI)1096-9888(199603)31:3<255::AID-JMS290>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Gawand P, Hyland P, Ekins A, Martin VJ, Mahadevan R. Novel approach to engineer strains for simultaneous sugar utilization. Metab Eng. 2013;20:63–72. doi: 10.1016/j.ymben.2013.08.003. [DOI] [PubMed] [Google Scholar]
- He L, Xiao Y, Gebreselassie N, Zhang F, Antoniewicz MR, Tang YJ, Peng L. Central metabolic responses to the overproduction of fatty acids in Escherichia coli based on 13C-metabolic flux analysis. Biotechnol Bioeng. 2014;111:575–585. doi: 10.1002/bit.25124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne A, Bruggemann H, Raasch C, Wiezer A, Hartsch T, Liesegang H, Johann A, Lienard T, Gohl O, Martinez-Arias R, Jacobi C, Starkuviene V, Schlenczeck S, Dencker S, Huber R, Klenk HP, Kramer W, Merkl R, Gottschalk G, Fritz HJ. The genome sequence of the extreme thermophile Thermus thermophilus. Nat Biotechnol. 2004;22:547–53. doi: 10.1038/nbt956. [DOI] [PubMed] [Google Scholar]
- Joshua CJ, Dahl R, Benke PI, Keasling JD. Absence of diauxie during simultaneous utilization of glucose and Xylose by Sulfolobus acidocaldarius. J Bacteriol. 2011;193:1293–301. doi: 10.1128/JB.01219-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karhumaa K, Hahn-Hagerdal B, Gorwa-Grauslund MF. Investigation of limiting metabolic steps in the utilization of xylose by recombinant Saccharomyces cerevisiae using metabolic engineering. Yeast. 2005;22:359–68. doi: 10.1002/yea.1216. [DOI] [PubMed] [Google Scholar]
- Lee NR, Lakshmanan M, Aggarwal S, Song JW, Karimi IA, Lee DY, Park JB. Genome-scale metabolic network reconstruction and in silico flux analysis of the thermophilic bacterium Thermus thermophilus HB27. Microb Cell Fact. 2014;13:61. doi: 10.1186/1475-2859-13-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighty RW, Antoniewicz MR. Parallel labeling experiments with [U-(13)C]glucose validate E. coli metabolic network model for (13)C metabolic flux analysis. Metab Eng. 2012;14:533–541. doi: 10.1016/j.ymben.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Liang Q, Zhang F, Li Y, Zhang X, Li J, Yang P, Qi Q. Comparison of individual component deletions in a glucose-specific phosphotransferase system revealed their different applications. Scientific reports. 2015;5:13200. doi: 10.1038/srep13200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Xu J. Dissecting and engineering metabolic and regulatory networks of thermophilic bacteria for biofuel production. Biotechnol Adv. 2013;31:827–37. doi: 10.1016/j.biotechadv.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Long CP, Antoniewicz MR. Quantifying biomass composition by gas chromatography/mass spectrometry. Anal Chem. 2014;86:9423–7. doi: 10.1021/ac502734e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynd LR. In: Production of ethanol from lignocellulosic materials using thermophilic bacteria: Critical evaluation of potential and review. Fiechter A, editor. Springer-Verlag; Heidelberg: 1989. pp. 1–52. [Google Scholar]
- Lyon PF, Beffa T, Blanc M, Auling G, Aragno M. Isolation and characterization of highly thermophilic xylanolytic Thermus thermophilus strains from hot composts. Canadian journal of microbiology. 2000;46:1029–35. [PubMed] [Google Scholar]
- Nichols NN, Dien BS, Bothast RJ. Use of catabolite repression mutants for fermentation of sugar mixtures to ethanol. Appl Microbiol Biotechnol. 2001;56:120–5. doi: 10.1007/s002530100628. [DOI] [PubMed] [Google Scholar]
- Nijland JG, Shin HY, de Jong RM, de Waal PP, Klaassen P, Driessen AJ. Engineering of an endogenous hexose transporter into a specific D-xylose transporter facilitates glucose-xylose co-consumption in Saccharomyces cerevisiae. Biotechnology for biofuels. 2014;7:168. doi: 10.1186/s13068-014-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes OC, Donato MM, da Costa MS. Isolation and Characterization of Rhodothermus Strains from S. Miguel, Azores. System Appl Microbiol. 1992;15:92–97. [Google Scholar]
- Ohtani N, Tomita M, Itaya M. The third plasmid pVV8 from Thermus thermophilus HB8: isolation, characterization, and sequence determination. Extremophiles : life under extreme conditions. 2012;16:237–44. doi: 10.1007/s00792-011-0424-x. [DOI] [PubMed] [Google Scholar]
- Reider Apel A, Ouellet M, Szmidt-Middleton H, Keasling JD, Mukhopadhyay A. Evolved hexose transporter enhances xylose uptake and glucose/xylose co-utilization in Saccharomyces cerevisiae. Scientific reports. 2016;6:19512. doi: 10.1038/srep19512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MH, Song H, Li BZ, Yuan YJ. Deletion of D-ribulose-5-phosphate 3-epimerase (RPE1) induces simultaneous utilization of xylose and glucose in xylose-utilizing Saccharomyces cerevisiae. Biotechnol Lett. 2015;37:1031–6. doi: 10.1007/s10529-014-1759-z. [DOI] [PubMed] [Google Scholar]
- Swarup A, Lu J, DeWoody KC, Antoniewicz MR. Metabolic network reconstruction, growth characterization and 13C-metabolic flux analysis of the extremophile Thermus thermophilus HB8. Metab Eng. 2014;24:173–80. doi: 10.1016/j.ymben.2014.05.013. [DOI] [PubMed] [Google Scholar]
- Taylor MP, Eley KL, Martin S, Tuffin MI, Burton SG, Cowan DA. Thermophilic ethanologenesis: future prospects for second-generation bioethanol production. Trends Biotechnol. 2009;27:398–405. doi: 10.1016/j.tibtech.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Walfridsson M, Bao X, Anderlund M, Lilius G, Bulow L, Hahn-Hagerdal B. Ethanolic fermentation of xylose with Saccharomyces cerevisiae harboring the Thermus thermophilus xylA gene, which expresses an active xylose (glucose) isomerase. Appl Environ Microbiol. 1996;62:4648–51. doi: 10.1128/aem.62.12.4648-4651.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama K, Ohkuma S, Taguchi H, Yasunaga T, Wakabayashi T, Yoshida M. V-Type H+-ATPase/synthase from a thermophilic eubacterium, Thermus thermophilus. Subunit structure and operon. J Biol Chem. 2000a;275:13955–61. doi: 10.1074/jbc.275.18.13955. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Hirota H, Kigawa T, Yabuki T, Shirouzu M, Terada T, Ito Y, Matsuo Y, Kuroda Y, Nishimura Y, Kyogoku Y, Miki K, Masui R, Kuramitsu S. Structural genomics projects in Japan. Nat Struct Biol. 2000b;7(Suppl):943–5. doi: 10.1038/80712. [DOI] [PubMed] [Google Scholar]
- Yoo H, Antoniewicz MR, Stephanopoulos G, Kelleher JK. Quantifying reductive carboxylation flux of glutamine to lipid in a brown adipocyte cell line. J Biol Chem. 2008;283:20621–7. doi: 10.1074/jbc.M706494200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JD, Walther JL, Antoniewicz MR, Yoo H, Stephanopoulos G. An elementary metabolite unit (EMU) based method of isotopically nonstationary flux analysis. Biotechnol Bioeng. 2008;99:686–99. doi: 10.1002/bit.21632. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.