Abstract
Sleep is a biological enigma that has raised numerous questions about the inner workings of the brain. The fundamental question of why our nervous systems have evolved to require sleep remains a topic of ongoing scientific deliberation. This question is largely being addressed by research using animal models of sleep. Drosophila melanogaster, also known as the common fruit fly, exhibits a sleep state that shares common features with many other species. Drosophila sleep studies have unearthed an immense wealth of knowledge about the neuroscience of sleep. Given the breadth of findings published on Drosophila sleep, it is important to consider how all of this information might come together to generate a more holistic understanding of sleep. This review provides a comprehensive summary of the neurobiology of Drosophila sleep and explores the broader insights and implications of how sleep is regulated across species and why it is necessary for the brain.
Keywords: Sleep, Sleep regulation, Drosophila melanogaster, Neurobiology
1. Introduction
Despite being one of the most ubiquitous behaviors in the animal kingdom, sleep remains one of nature's greatest mysteries. From an evolutionary perspective, sleep is a very peculiar phenomenon. A process that prevents an organism from perceiving and interacting with their environment for extended periods of time is likely to increase the risk for predation, yet sleep has endured across the course of evolution and been systematically studied from the simple nervous systems of C. elegans (Raizen et al., 2008) all the way to the human brain (Reviewed in Hirshkowitz, 2004). In order to have persisted in such a wide range of species (Campbell and Tobler, 1984), sleep must provide some biological benefits that outweigh any maladaptive features. Following decades of scientific inquiry aimed at disentangling the biology of sleep, there is now virtually no denying the importance of sleep to animal life. The biological necessity of sleep is best highlighted by the negative consequences associated with its loss, which include impaired cognition with increased risk of accidents (Dinges et al., 1997; Belenky et al., 2003; Van Dongen et al., 2003), metabolic dysfunction (Spiegel et al., 2009), increased disease risk (van Leeuwen et al., 2009; Reviewed in Palma et al., 2013), and in extreme cases, death (Rechtschaffen et al., 1983; Montagna and Lugaresi, 2002; Shaw et al., 2002; Stephenson et al., 2007). The detrimental repercussions of sleep deprivation are evidence that there is a critical purpose for sleep, but there is still currently no scientific consensus on the core functions of sleep.
One of the most significant milestones in modern sleep research was the discovery that Drosophila melanogaster, also known as the common fruit fly, exhibits a sleep-like state (Hendricks et al., 2000; Shaw et al., 2000). Initial studies found that Drosophila sleep shares many of the same traits as mammalian sleep, such as stereotyped posture, increased arousal threshold, and a homeostatic response to sleep deprivation (Hendricks et al., 2000; Shaw et al., 2000). These common features make the fruit fly a rather useful animal model to study sleep. The characterization of Drosophila sleep has since paved the way for the widespread use of Drosophila as an animal model to study sleep at the molecular and behavioral level. Drosophila sleep is typically observed and recorded by placing flies in transparent tubes and monitoring their movements using infrared beams or video recording. Through the use of these behavioral tracking methods, it has been shown that Drosophila sleep exhibits a diurnal pattern that is most consolidated in the middle of the night and that is accompanied by high levels of wake and activity during the early and later portions of the day (Hendricks et al., 2000). During the middle of the day, Drosophila also undergo a “siesta sleep” that is sexually dimorphic, since male sleep is longer and more consolidated than sleep in female flies in the daytime (Shaw et al., 2000; Koh et al., 2006). This difference in daytime sleep largely accounts for the longer average amount of daily sleep in male flies compared to female flies. There is also variation in sleep bout duration throughout the day and night, with bouts lasting from minutes to hours (Hendricks et al., 2000). Sleep in the fruit fly is commonly defined as 5 minutes or more of rest, due to the fact that the arousal threshold – which can be measured by the response to mechanical stimuli – of resting flies is significantly increased after 5 minutes of rest (Shaw et al., 2000). Until recently, invertebrate sleep was primarily differentiated from mammalian sleep by the absence of observable sleep stages (In mammals, we can distinguish stages such as rapid eye movement (REM) from non-REM sleep). However, recent electrophysiological experiments have demonstrated that Drosophila sleep does have phases of varying intensity that have been captured by local field potential recordings in the fly brain (van Alphen, et al., 2013). While we cannot directly compare Drosophila sleep stages to the stages of mammalian sleep, it is now evident that changes in sleep depth are also a feature of invertebrate sleep.
It should be noted that our understanding of the circadian regulation of sleep and activity owes much to the fundamental studies of circadian clock proteins in the Drosophila animal model. While a comprehensive exploration of circadian rhythms is beyond the scope of this review, it is impossible to discuss sleep regulation without considering the role of the clock. The daily timing of sleep is believed to be the result of interactions between circadian rhythms and homeostatic signaling, a concept known as the two-process model of sleep (Borbély, 1982). Circadian rhythms generate daily rhythmic patterning of sleep and wake states while the sleep homeostat increases the pressure to sleep following extended periods of wake. The Drosophila circadian molecular clock still remains one of the most well understood and thoroughly characterized circadian systems (Reviewed in Nitabach and Taghert, 2008; Allada and Chung, 2010) and our understanding of the molecular machinery underlying circadian rhythms owes a great deal to the seminal studies that identified the core clock proteins in the fly. In 1971, Konopka and Benzer were the first to uncover a genetic basis underlying circadian rhythms when they conducted a forward genetic screen of Drosophila mutants that led to the seminal discovery of the circadian gene period (Konopka and Benzer 1971) (Reddy et al., 1984). Later work would identify multiple key players in Drosophila circadian regulation: timeless (Sehgal et al., 1994), dClock (Allada et al., 1998), the Drosophila homolog of mammalian Clock (King et al., 1997), and the protein DOUBLETIME, which phosphorylates PER and targets it for degradation (Price et al., 1998; Kloss et al., 1998). These genes/proteins are all required to maintain the normal diurnal rhythms of rest and activity in the fly. Most recently, Michael Young, Michael Rosbash, and Jeffrey Hall were awarded the Nobel Prize in Medicine or Physiology for their critical contributions to the understanding of circadian rhythms in Drosophila. Elucidating the biological mechanisms underlying the influence of both circadian rhythms and homeostatic signaling on sleep regulation is one of the major endeavors in sleep research today (for review, see Franken and Dijk, 2009).
This review will summarize what we have learned about sleep regulation in the fruit fly since the original description of sleep in this species and highlight the advances in our general understanding of sleep biology as a direct result of Drosophila sleep research. When relevant, we will discuss ways in which many neurobiological processes regulating sleep in the fly are conserved in higher order systems. Our goal is to provide a unified overview of the last two decades of fly sleep research and address the larger implications for understanding sleep function. We will also address the technical and scientific considerations that will be important to keep in mind as we move towards an increasingly holistic understanding of the biology of sleep.
2. Neurotransmitters Involved in Wake/Sleep Regulation
Seven neurotransmitters have hitherto been implicated in sleep and wake regulation in Drosophila (see Table 1). The wake-promoting neurotransmitters are dopamine, octopamine, and histamine, while serotonin and GABA are the major sleep promoting neurotransmitters in the fly. Acetylcholine and glutamate have been found to have both wake-promoting and sleep-promoting actions in Drosophila through the activity of separate and distinct brain circuits.
Table 1. Summary of the major neurotransmitters regulating wake and sleep in Drosophila melanogaster and identified locations of their actions.
| Neurotransmitter | Sleep/Wake Effect | Presynaptic cells | Postsynaptic Receptors/ Brain Regions |
|---|---|---|---|
| WAKE PROMOTING NEUROTRANSMITTERS | |||
| Dopamine | ↑wake | DA neurons in the PPL1and PPM3 clusters in the posterior protocerebrum (Liu et al., 2012b; Ueno et al., 2012) MB dopamine neurons (Sitaraman et al., 2015) |
DopR (dFSB) (Liu et al., 2012b; Ueno et al., 2012) D1 receptors (MBONs) (Sitaraman et al., 2015) |
| Octopamine | ↑wake | ASM cells in the medial protocerebrum (Crocker et al., 2010) | OAMB (PI) (Crocker et al., 2010) |
| Histamine | ↑wake | Unknown | HisCl1receptors (Oh et al., 2013) |
| SLEEP PROMOTING NEUROTRANSMITTERS | |||
| Serotonin | ↑sleep | DPM neurons (Haynes et al., 2015) | 5-HT1a (MB) (Yuan et al., 2006) 5-HT2b (dFSB) (Qian et al., 2017) |
| GABA | ↑sleep | DPM neurons (Haynes et al., 2015) | RdlGABAA receptor (l-LNvs) (Chung et al., 2009) GABAAB-R3 receptor (MB) (Haynes et al., 2015) |
| DUAL FUNCTION NEUROTRANSMITTERS | |||
| Acetylcholine | ↑wake | Subset of α/β core neurons in the MB (Yi et al., 2013) | nAChRs (l-LNvs) (McCarthy et al., 2011) |
| ↑sleep | Subset of α/ β core neurons in the MB (Yi et al., 2013) | Unknown | |
| Glutamate | ↑wake | Unknown | Unknown |
| ↑sleep | Unknown | NMDAR (Tomita et al., 2015) (Robinson et al., 2016) | |
2.1. Wake-promoting neurotransmitters (see Table 1)
2.1.1 Dopamine
Numerous studies have provided evidence that dopamine promotes wake in Drosophila. Dopaminergic cells occur in clusters throughout the Drosophila protocerebrum and innervate most neuropils in the central nervous system (Friggi-Grelin et al., 2003; Mao and Davis 2009). These dopaminergic cells project to the mushroom bodies and the central complex (Friggi-Grelin et al., 2003; Mao and Davis 2009), which are important sleep-regulating brain regions in the Drosophila central nervous system (Pitman et al., 2006; Joiner et al., 2006; Donlea et al., 2011 ; Liu et al., 2016).
Genetic mutations that affect dopamine signaling in Drosophila have major influences on sleep and have effectively demonstrated the arousal-promoting effects of dopamine. Kume et al., 2005 identified a short-sleeping Drosophila mutant termed fumin (fmn), the Japanese word for “sleepless”. The mutation in the fmn flies was determined to be the result of a loss-of-function mutation in the Drosophila dopamine transporter (dDAT) gene. Since dDAT is responsible for dopamine presynaptic re-uptake (Porzgen et al., 2001), it was suggested that loss-of-function of dDAT produces prolonged dopamine signal at dopaminergic synapses in fmn mutants and reduced sleep (Kume et al., 2005). Makos et al., 2009 later recorded the levels of dopamine clearance in vivo in fmn mutants using fast scan cyclic voltammetry and confirmed that the fmn mutation does in fact impair dopamine clearance.
Conversely, pharmacologically inhibiting dopamine synthesis via oral administration of the tyrosine hydroxylase inhibitor 3-iodo-tyrosine (3IY) increases sleep during the day, providing further evidence that dopaminergic signaling influences arousal (Andretic et al., 2005). It should be noted that the time-of-day specific effect of 3IY is likely due to a ceiling effect of Drosophila baseline nighttime sleep. Similarly, genetic disruption of dopamine synthesis by abolishing the expression of active tyrosine hydroxylase (TH) – an enzyme that is critical for dopamine biosynthesis – in the Drosophila CNS causes flies to have significantly increased total sleep time (Riemensperger et al., 2011). The loss of dopamine in the brain results in less time spent awake during the day and night and a higher arousal threshold, (Riemensperger et al., 2011).
Methamphetamine is a psychostimulant that acts through dopaminergic systems in mammals (Nishino et al, 1998). Methamphetamine also promotes arousal in Drosophila, as flies fed methamphetamine display longer wake bouts and extended latency to sleep after the transition from day to night (Andretic et al., 2005). The effects of methamphetamine are opposite to those caused by inhibiting the production of dopamine, and it has been suggested that the arousal-promoting effects may also act through dopaminergic signaling (Andretic et al., 2005).
Dopaminergic signaling mediates the wake-promoting effects of caffeine in Drosophila. In mammals, caffeine is a stimulant that likely promotes arousal through nonspecific antagonism of adenosine receptors, of which there are four that are known: A1, A2a, A2b, and A3 (Fredholm et al., 1999). Drosophila possesses just a single adenosine receptor, known as AdoR (Dolezelova et al., 2007). Interestingly, while the effects of caffeine in the fly are mimicked by A1 and A2 adenosine receptor antagonists (Andretic et al., 2008), studies of AdoR null mutants in Drosophila indicate that AdoR is not required for the effects of caffeine since these mutants do not exhibit any altered response to caffeine (Wu et al., 2009). Instead, the arousal-inducing effects of caffeine in Drosophila (Hendricks et al., 2000; Shaw et al., 2000) require the dopamine D1 receptor (dDA1) in the mushroom bodies (Andretic et al., 2008). The Drosophila mutant dumb1 which has deficient expression of dDA1 (Kim et al., 2007) is resistant to the arousal-promoting effects of caffeine; this resistance is rescued by the transgenic expression of wildtype dDA1 in the mushroom bodies (Andretic et al., 2008). Analysis of dDA1 mRNA from Drosophila heads further showed that caffeine treatment leads to a decrease in dDA1 mRNA expression, suggesting that caffeine promotes wakefulness in Drosophila at least in part by altering transcription of dDA1 receptors (Andretic et al., 2008).
One of the benefits of a small animal model such as Drosophila is that the task of mapping the neural circuits of behaviors is highly amenable in a nervous system comprising a smaller number of cells, cell types, and connections than higher order animal models. The dopaminergic regulation of sleep in the Drosophila brain is an excellent example of the specificity with which Drosophila genetic tools has allowed us to localize the actions of neurotransmitter circuits that generate complex behavior. Part of the wake-promoting dopamine signal has been isolated to single neurons in the PPL1 and PPM3 clusters in the posterior protocerebrum of the Drosophila brain that innervate neurons in the dorsal fan shaped body (dFSB) in each hemisphere with dDA1 receptors (Liu et al., 2012b; Ueno et al., 2012). Since the identification of these wake-promoting inputs to the dFSB, the combined approach of optogenetic stimulation of dopaminergic cells in conjunction with electrophysiological recording in dFSB neurons has confirmed that activation of TH-expressing dopaminergic cells suppresses dFSB firing to promote wake (Pimentel et al., 2016). It should also be noted that dopamine exerts wake-promoting effects outside of the dFSB, as it was found that dopaminergic activation of D1 dopamine receptors in mushroom body neurons also induces wakefulness (Sitaraman et al., 2015).
Dopamine has a conserved wake-promoting function in mammals. Similar to Drosophila fmn mutants, mice with impaired DAT activity (as a result of pharmacological administration of a DAT inhibitor) exhibit elevated amounts of wakefulness, suggesting that prolonged synaptic exposure to dopamine induces wake (Qu et al., 2010). Further evidence that dopamine stimulates wake has been observed in rats, where intracerebroventricular injections of D1 and D2 agonists increase time spent awake and decrease time spent sleeping (Isaac and Berridge, 2003). Finally, PET imaging in rhesus macaques and human subjects have determined that the wake-promoting drug modafinil increases levels of extracellular dopamine in the brain (Andersen et al., 2010; Volkow et al., 2009). It should also be noted that an interesting aspect of sleep regulation that is highlighted by the fruit fly dopaminergic system is the presence of functional and anatomical connections between wake-promoting and sleep-promoting brain regions. As previously discussed, PPL1 and PPM3 wake-promoting dopaminergic cells in the posterior protocerebrum of the Drosophila brain synapse onto DopR expressing cells in the dFSB, which is a sleep-promoting brain center (Liu et al., 2012b; Ueno et al., 2012). In mammals, arousal pathways inhibit sleep regulating centers such as the ventrolateral preoptic nucleus (VLPO) in the hypothalamus that in turn can inhibit the same ascending arousal circuits (Saper et al., 2010). This system of alternating inhibition between wake and sleep centers is the basis for the mammalian “flip-flop” model of sleep, which suggests that states transition or “switch” when a wake-promoting region or a sleep-promoting region no longer inhibits the other (Saper et al., 2010). In this case, dopaminergic synapses onto the sleep-promoting dFSB cells suggest that there may be some general conservation of structural logic that connects sleep- and wake-promoting circuits in the brain.
2.1.2. Octopamine
Octopamine is a wake-promoting neurotransmitter in Drosophila that is considered to be equivalent to the mammalian neurotransmitter norepinephrine, of which it is a structural analog. In mammals, Aston-Jones and Bloom, 1981 were the first to establish that neurons of the locus coeruleus that release norepinephrine are predominantly wake-active neurons. In the Drosophila central nervous system, there are approximately 100 octopaminergic cells that send projections to a many distinct regions in the brain including the calyx of the mushroom bodies, parts of the central complex, the protocerebrum, as well as the optic lobes (Sinakevitch and Strausfeld, 2006; Busch et al., 2009). Thus, similar to the norepinephrine cells in the locus coeruleus (Berridge and Waterhouse, 2003), octopamine cells have widespread projections in the Drosophila CNS.
Pharmacological and genetic manipulations of octopamine signaling demonstrate the arousal-promoting effects of octopamine. Drosophila that are orally administered octopamine have significantly less nighttime sleep, while flies carrying mutations that disrupt key genes in the octopamine biosynthesis pathway sleep longer during the day and have a decreased latency to sleep after the transition from day to night which is indicative of an elevated pressure to sleep (Crocker and Sehgal, 2008). It should be noted that in cases of increased sleep specific to the daytime, a ceiling effect of nighttime sleep may be a contributing factor. Furthermore, increasing the excitability of octopaminergic cells by expressing a transgene for a bacterial sodium channel in these neurons decreases total nighttime sleep, while reducing the octopaminergic cell excitability via expression of a transgenic potassium channel increases total sleep (Crocker and Sehgal, 2008).
As in the case of the dopamine arousal pathway, the neural circuitry for the wake-promoting effects of octopamine has been characterized with considerable detail. Crocker et al, 2010 utilized a mosaic analysis with a repressible cell marker (MARCM) which allows for specific temporal and spatial labeling of cells (Lee and Luo, 2001) to identify the octopaminergic cells that are responsible for the wake-promoting effect of octopamine. The wake-promoting signal of octopamine was mapped to a cluster of neurons in the medial protocerebrum; these cells activate the octopamine receptor in the mushroom body (OAMB) on insulin-like-peptide (DILP2) producing cells in the pars intercerebralis, depolarizing the DILP2 cells and increasing their cyclic-AMP (cAMP) activity (Crocker et al., 2010). The involvement of DILP2 neurons in the octopaminergic arousal pathway is supported by the reduction and increase in sleep caused by manipulating the excitability of DILP2 neurons via the transgenic expression of depolarizing or hyperpolarizing channels, respectively (Crocker et al., 2010).
In addition to being structurally similar to mammalian norepinephrine (Farooqui, 2012) octopamine also shares arousal regulating activity with norepinephrine. Norepinephrine is a critical component of the mammalian ascending arousal system and is released from the LC in the brainstem to many components of the cortex (Reviewed in Berridge et al., 2012). Early studies in rats found that noradrenergic cells in the LC produce firing bursts that precede entrance into different states of arousal (Aston-Jones and Bloom, 1981). Later work found that targeted pharmacological activation of LC cells induced arousal, which was blocked by inhibiting norepinephrine release (Berridge and Foote, 1991).
2.1.3. Histamine
Histaminergic neurons have been shown to be important wake-promoting cells in mammals (Thakkar, 2011; Parmentier et al., 2002) and evidence suggests that histamine also promotes arousal in Drosophila (Oh et al., 2013). Immunohistochemical studies have identified 18 histaminergic cell bodies in the Drosophila brain (Reviewed in Nässel, 1999). These histaminergic neurons project to regions of the ventral and lateral protocerebrum of the Drosophila brain (Pollack and Hofbauer, 1991; Reviewed in Nässel, 1999). Pharmacological administration of the histamine receptor antagonist hydroxyzine decreases the latency to sleep (Shaw et al., 2000) and genetic hypomorphic mutations that decrease activity of histidine decarboxylase (HDC) – an enzyme critical for histamine synthesis – significantly increases daytime sleep (Oh et al., 2013). There are two histamine receptors in Drosophila – the histamine-gated chloride channel subunit 1 (HisCl1) and ora transientless (Ort) – but the wake-promoting effects of histamine are likely mediated only by HisCl1 since only HisCl1 null mutants display increases in sleep (Oh et al., 2013). Future work to determine which cells are required for HisC11-mediated sleep regulation will be critical for understanding the mechanism by which histamine promotes wake in the fly.
2.2. Sleep-promoting neurotransmitters (see Table 1)
2.2.1. Serotonin
Serotonin is widely expressed in the Drosophila central nervous system (Nässel, 1988; Vallés and White, 1988; Lundell and Hirsh, 1994; Sitaraman et al., 2008) and promotes sleep in Drosophila (Yuan et al., 2006). Pharmacological elevation of serotonin levels via treatment with 5-hydroxytryptophan (5-HTP), a precursor to serotonin biosynthesis, increases sleep (Yuan et al., 2006). This effect is mimicked by transgenically overexpressing tryptophan hydroxylase (TPH, known as TRH in Drosophila), an enzyme involved in the biosynthesis of serotonin. Conversely, blocking serotonergic transmission by expressing the transgene for the tetanus neurotoxin light chain – which disrupts exocytosis of neurotransmitter – in serotonergic cells significantly decreases sleep (Yuan et al., 2006). Drosophila serotonin receptors are exclusively G protein-coupled metabotropic receptors and occur in five subtypes: 5-HT1a, 5-HT1b, 5-HT2a, 5-HT2b, and 5-HT7 (Witz et al., 1990; Saudou et al., 1992; Colas et al., 1995; Qian et al., 2017). Studies in the fly demonstrate that serotonergic regulation of sleep likely acts through multiple receptors and brain regions to promote both baseline sleep (Yuan et al., 2006; Haynes et al., 2015; Qian et al., 2017) as well as homeostatic sleep rebound after sleep deprivation (Qian et al., 2017). Yuan et al., 2006 identified 5-HT1a receptors as a regulator of sleep since loss-of function mutants display significant sleep reductions (Yuan et al., 2006). Furthermore, transgenic expression of 5-HT1a in the mushroom bodies rescues the short-sleeping phenotype of the 5-HT1a Drosophila null mutants, implicating the mushroom bodies as the site of action for 5-HT1a sleep regulation (Yuan et al., 2006). Furthermore, Haynes et al., 2015 found that expression of tryptophan hydroxylase (TRH) RNAi in the dorsal paired medial (DPM) neurons that project to the MBs is sufficient to significantly reduce sleep. Most recently Qian et al., 2017 found that in addition to 5-HT1a and TRH, genetic knockout of the 5-HT2b receptor reduces total sleep amount and sleep bout duration in Drosophila. Further genetic characterization of 5-HT2b involvement in sleep found that 5-HT2b in the dorsal fan-shaped body is required for homeostatic sleep rebound (Qian et al., 2017). Thus, serotonin mediates both sleep and sleep homeostasis via distinct sleep/wake circuits in the Drosophila brain.
Various studies have gathered evidence that serotonin has a role in modulating mammalian sleep, though its role is more multifaceted than in flies. Serotonergic signaling acts differentially to promote or suppress different stages of the sleep-wake cycle depending on the serotonin receptor subtype and the brain region involved (Reviewed in Ursin, 2002). Much work has yet to be done to characterize these different components of serotonergic sleep modulation in the brain. However, pharmacological manipulations of serotonergic signaling have demonstrated effects on sleep and wake in similar ways that have been seen in Drosophila. For example, injecting p-chlorophenylalanine – an inhibitor of serotonin synthesis – into the dorsal raphe nucleus acutely induces insomnia in rats (Gao et al., 2002) and early experiments in humans found that administration of the serotonin precursor 5-HTP increases REM sleep duration, though total sleep is not affected (Wyatt et al., 1971). Additionally, some antidepressant serotonin reuptake and serotonin receptor subtype inhibitors have been linked to improvements in subjective and objective sleep quality in depressed and non-depressed patients (Reviewed in Wilson and Argyropoulos, 2005).
2.2.2. GABA
γ-Aminobutyric acid (GABA) is considered to be the primary inhibitory neurotransmitter in vertebrates and invertebrates. In Drosophila, GABA-producing cells occur in small clusters that innervate large numbers of cells throughout the entire brain. (Enell et al., 2007; Okada et al., 2009). Studies in mammalian systems have found that GABAergic cells are present in the sleep-promoting region of the hypothalamus known as the ventrolateral preoptic nucleus (VLPO) (Gong et al., 2004) as well at the median preoptic nucleus (Benedetto et al., 2012) and that GABAergic transmission promotes sleep (Gallopin et al., 2000; Benedetto et al., 2012). Likewise, the net effect of GABAergic transmission in the Drosophila brain promotes sleep and advances sleep onset, since reducing the excitability of GABAergic neurons in the fly by expressing the transgene for a hyperpolarizing potassium channel significantly decreases sleep time and increases the latency to sleep at night (Agosto et al., 2008). Furthermore, administration of the GABA-A agonist 4,5,6,7-tetrahydroisoxazolo-[5,4-c]pyridine-3-ol (THIP) significantly increases sleep in wildtype flies (Dissel et al., 2015).
Work in Drosophila has demonstrated that in clock cells (Parisky et al., 2008) and also in cells critical for learning and memory (Haynes et al., 2015), GABA regulates both sleep and sleep onset. Drosophila neurons can express ionotropic GABAA and metabotropic GABAb receptors (Mezler et al., 2001). Parisky et al., 2008 found that ionotropic GABAA signaling in clock cells regulates timing of sleep onset during the night. The Resistant to dieldrin (Rdl) ionotropic GABAA receptor is expressed in the large ventral lateral neurons (l-LNvs), which are a set of neurons among the approximately 150 neurons that comprised the Drosophila circadian network (Reviewed in Peschel and Helfrich-Förster, 2011). Of the known clock cells, the LNvs are considered to be the principal circadian pacemaker cells in the Drosophila brain (Renn et al., 1999; Kaneko et al., 2000; Blanchardon et al., 2001; Sheeba et al., 2008). Rdl is one of three GABAA subunits in Drosophila and is expressed in the optic lobes, antennal lobes, mushroom bodies and the central complex (Enell et al., 2007). Knockdown of the Rdl GABAA receptor in PDF-expressing clock cells decreases sleep (Parisky et al., 2008; Chung et al., 2009) amount while overexpression increases sleep amount (Parisky et al., 2008). Electrophysiological recordings of l-LNvs further demonstrate that GABA inhibits the activity of l-LNvs, suggesting that inhibition of l-LNvs promotes sleep (Chung et al., 2009; McCarthy et al., 2011). Many of the molecular regulators of GABA signaling in the l-LNvs have further been identified. For example, the protein WIDE AWAKE (WAKE) increases Rdl expression at the cell membrane at the end of the day (Liu et al., 2014) while Drosophila Neuroligin 4 (DNlg4), a critical cell adhesion molecule, is required for proper clustering of Rdl in the l-LNvs (Li et al., 2013). Both WAKE and DNlg4 are necessary for sleep in clock cells since knockdown of WAKE and DNlg4 in PDF-expressing neurons decreases sleep amount and increases the latency to sleep in Drosophila (Liu et al., 2014; Li et al., 2013). In contrast, Fbxl4, an E3 ubiquitin ligase modulates sleep through its promotion of Rdl degradation in clock cells (Li et al., 2017). Both genetic mutation of the fbxl4 gene and knockdown of fbxl4 in PDF cells increases sleep (Li et al., 2017). Thus, molecules that regulate expression and degradation of GABA consequently affect wake behavior in the fly.
Finally, GABA release from the dorsal paired medial (DPM) neurons has been shown to promote sleep by inhibiting the MBs (Haynes et al., 2015). Inhibiting GABA release from the DPM neurons decreases sleep time. Furthermore, RNAi targeted towards Rdl GABAA receptors and GABAB-R3 receptors in the MBs recapitulates the sleep phenotype, suggesting that both ionotropic and metabotropic GABA signaling in the MBs promotes sleep. These results illustrate the role of GABA in promoting sleep and regulating sleep onset in Drosophila. In mammals, cells in the ventral lateral preoptic area (VLPO) of the hypothalamus – a critical sleep-promoting region in the brain – are GABAergic (Gallopin et al., 2000) and administration of GABA directly into the VLPO of rats increases sleep amount (Xiong et al., 2014). In this case, GABA-mediated inhibition of wake-promoting mushroom body neurons by the dorsal paired medial neurons shares similarity with the inhibitory actions of the VLPO on arousal centers in the mammalian brain.
2.3. Dual Function Neurotransmitters (see Table 1)
2.3.1. Glutamate
Glutamate is one of the primary excitatory neurotransmitters in the mammalian central nervous system. In Drosophila, glutamate is the primary excitatory neurotransmitter at the neuromuscular junction (Bogdanik et al., 2004; DiAntonio, 2006; Rival et al., 2006) similar to acetylcholine in mammals. However, glutamate is also widely expressed in the Drosophila central brain (Sinakevitch-Pean et al., 2001).
There is evidence that glutamatergic signaling has wake-promoting effects in the fly (Zimmerman et al., 2016). Transgenic activation of CNS-specific glutamatergic cells using a temperature sensitive gal80 protein to drive expression of a bacterial sodium channel in cells containing the glutamate transporter protein (VGLUT) significantly increases wake during both the day and night (Zimmerman et al., 2016). Conversely, suppressing glutamatergic signaling by expressing a heat-induced transgenic potassium channel in VGLUT cells significantly reduces wake, though the effect is only significant during the night. These results suggest that glutamate signaling has a wake-promoting effect on Drosophila sleep regulation.
In addition to evidence that glutamate promotes wake, there is also data that demonstrates that glutamate signaling has sleep-promoting effects in Drosophila. Tomita et al., 2015 found that flies with a genetic loss of the NMDA type glutamate receptor (Nmdar1) are hyperactive and demonstrate significant reductions in sleep. Additionally, flies fed the NMDAR antagonist MK-801 also displayed significant reductions in sleep (Tomita et al., 2015). Together, these results suggest that Drosophila NMDA glutamate receptors likely mediate a sleep-promoting signal in the fly under normal conditions. Interestingly, pharmacological inhibition of NMDAR activity also induces wake in hibernating arctic ground squirrels (Jinka et al., 2012). Though hibernation-induced torpor is likely regulated differently than sleep, this finding does demonstrate that some wake-inhibiting action of NMDA signaling is conserved across different species. Robinson et al., 2016 also found that enhanced Drosophila vesicular glutamate transporter (DVGLUT) expression as a result of knocking down the RNA-editing gene Adar (Adenosine deaminase acting on RNA) in neurons significantly increases total sleep time during both the day and night. These effects involve NMDAR and AMPAR signaling, since knockdown of NMDAR subunits NR1 and NR2 as well as knockdown of the AMPA receptor GluRI rescues the sleep phenotype of Adar mutants (Robinson et al., 2016). Recently, Liu et al., 2016 found that homeostatic sleep drive after sleep deprivation is regulated by glutamatergic neurons in the ellipsoid body of the Drosophila brain. Extended wakefulness induces an increase in NMDA receptor expression in a subset of neurons in the ellipsoid body is required for rebound sleep after prolonged wakefulness (Liu et al., 2016). Taken together, these data demonstrate that glutamatergic signaling has sleep-promoting effects in the fly. Since it has also been demonstrated that overall increases in glutamatergic output promote wake (Zimmerman et al., 2016), it seems likely that the location of release or the postsynaptic receptors may dictate whether glutamate signaling generates a sleep or a wake-promoting signal. Furthermore, the effects of glutamate on sleep and wake may be also be differentiated by developmental and mature brain signaling effects. These effects highlight the need to understand the role of genes and transmitters in the context of defined neural circuits.
2.3.2. Acetylcholine
Acetylcholine is a major excitatory neurotransmitter in Drosophila. In contrast to mammals, acetylcholine is most abundant not in the peripheral nervous system, but in the central nervous system. Acetylcholine is widely expressed in regions of the fly brain such as the protocerebrum, the mushroom bodies, and the central complex (Buchner et al., 1986; Yasuyama and Salvaterra, 1999; Salvaterra and Kitamoto, 2001). In mammals, acetylcholine is a wake- and REM-active neurotransmitter (Szerb, 1967; Saper et al., 2005; Celesia and Jasper, 1966; Reinoso-Suárez et al., 2001; Vazquez and Baghdoyan, 2001). Evidence in Drosophila suggests that acetylcholine acts to promote both wakefulness and sleep, depending on the neuronal groups on which it acts.
Acetylcholine provides excitatory input to the wake-promoting large ventrolateral neurons (l-LNvs) – the primary circadian pacemaker cells in the Drosophila brain – and modulates their firing (McCarthy et al., 2011). Whole-cell recording of l-LNv activity in vitro demonstrates that l-LNvs fire with a synchronized rhythm and that activation of nicotinic acetylcholine receptors (nAChRs) on the l-LNvs increases their excitation and firing (McCarthy et al., 2011). Application of acetylcholine and nicotine, both nAChR agonists, depolarized the l-LNvs and caused action potential firing while treatment with the nAChR antagonists curare and a-BuTX eliminated action potential firing of the l-LNvs (McCarthy et al., 2011). Acetylcholine also induces wakefulness via G-protein coupled signaling in a subset of wake-promoting mushroom body neurons (Yi et al., 2013). Thus, acetylcholine likely promotes wake by exciting a part of the circadian circuitry in Drosophila known to modulate wakefulness as well as by activating receptors on wake-promoting cells in the brain.
While acetylcholine provides excitatory input to wake-promoting clock cells, there is also evidence that it promotes sleep as well. Expression of RNAi against vesicular acetylcholine transporter (vAChT) – which is necessary for the release and transport of acetylcholine – in a subset of MB α/β core neurons significantly decreases in sleep, suggesting that acetylcholine promotes sleep via acetylcholine transmission from these cells (Yi et al., 2013). Notably, these sleep-promoting cholinergic neurons neighbor a layer of cholinergic wake-promoting cells as described in the previous section. Thus, neurons within the same brain region that release the same neurotransmitter can have opposing effects on behavioral state depending on their location (and likely their downstream targets as well).
These studies on individual neurotransmitter systems and their involvement in sleep regulation in Drosophila has led to an increasingly comprehensive understanding of the neuronal landscape of sleep regulation in the fly brain (See Table 1). For example, we can now identify brain regions such as the mushroom bodies or the dorsal fan-shaped bodies as sleep-regulatory centers and have also characterized different excitatory and inhibitory circuits that modulate sleep and wake in the fly (see Figure 1). In mammalian systems, there is evidence of a mutual inhibitory flip-flop mechanism between wake-active and sleep-active cells (Saper et al., 2010) and while there is currently no direct evidence of this in Drosophila, the identification of different interacting sleep and wake circuits makes this seem likely. Unlike the cell autonomous regulation of circadian clock, the interaction of neuronal circuits fundamentally controls sleep and wake in mammals, and this appears to hold true in Drosophila.
Figure 1.
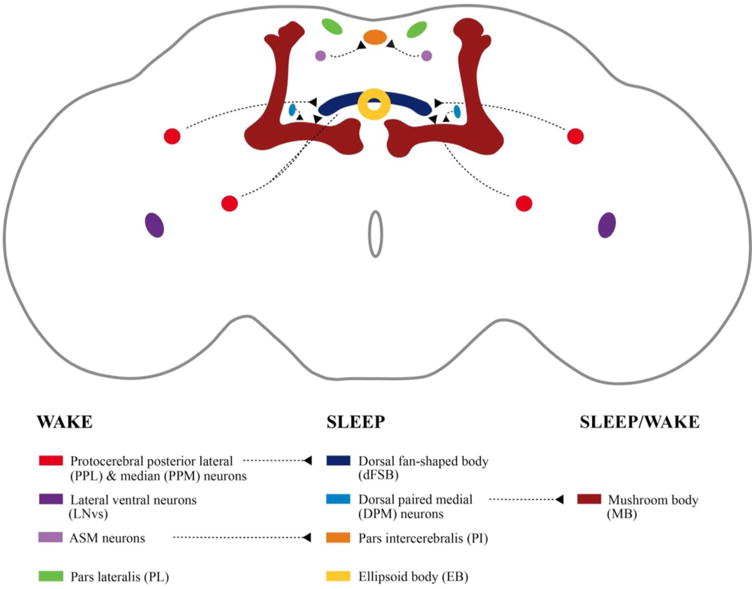
Drosophila brain regions involved in sleep and wake regulation. Dashed lines in the key indicate known functional connection between regions.
The identification of sleep- and wake-relevant neurotransmitters demonstrates that there are multiple critical brain centers that regulate Drosophila sleep (See Figure 1). Some of the primary regions that regulate sleep in the Drosophila brain are the mushroom body, the dorsal fan-shaped body, and the pars intercerebralis and pars lateralis. The lateral ventral neurons, which are a critical part of the circadian network of the fly brain, are also required for sleep and wake regulation. Some of these regions such as the dorsal fan-shaped body primarily promote sleep while regions such as the mushroom body contain both sleep and wake promoting neurons. Additionally, there are various cells and loci throughout the brain regulate sleep and wake via synaptic connections with cells is these primary regions.
3. Signaling Pathways and Molecules that Regulate Drosophila Wake/Sleep
3.1 Wake-promoting signaling pathways and molecules (see Table 2)
Table 2. Summary of Signaling Mechanisms Regulating sleep in Drosophila Melanogaster.
| WAKE-PROMOTING SIGNALS | |
|---|---|
| Pathway / Signaling Mechanism | Molecules Involved |
| Ion Channel Signaling | Ca-α 1T (Jeong et al., 2015) Dα3 (Wu et al., 2014) Sandman (Pimentel et al., 2016) TrpA1 (Lamaze et al., 2017) |
| PKA/CREB Pathway | PKA/CREB (Joiner et al., 2006) Amnesiac (Liu et al., 2008) Calcineurin (Tomita et al., 2011) (Nakai et al., 2011) |
| Monoamine Signaling | VMAT (Nall and Sehgal, 2013) |
| FMRP | FMRP (van Alphen et al., 2013) |
| Salt-inducible Kinase 3 | Salt-inducible kinase 3 (Funato et al., 2016) |
| SLEEP PROMOTING SIGNALS | |
| Pathway / Signaling Mechanism | Molecules Involved |
| Ion Channel Signaling | Shaker (Cirelli et al., 2005) Hyperkinetic (Bushey et al., 2007) Sleepless (Koh et al., 2008) dSur (Allebrandt et al., 2013) |
| EGF Pathway | Star (Foltenyi et al., 2007) Rho (Foltenyi et al., 2007) ERK (Foltenyi et al., 2007) |
| G-protein Signaling | Go protein (Guo et al., 2011) (Yi et al., 2013) SIFamide (Park et al., 2014) |
| Homer | Homer (Naidoo et al., 2012) |
| Ubiquitin-Proteosome Signaling | Cullin-3(Cul3) (Stavropoulos and Young, 2011) Fat facet (faf) (Seugnet et al., 2017) Highwire (hiw) (Seugnet et al., 2017) |
| Cellular-Stress Signaling | UPR factors (Brown et al., 2014) JNK (Takahama et al., 2012) |
| Cell Cycle Signaling | Cyclin A (Rogulja and Young, 2012) TARANIS (Afonso et al., 2015) |
| Neuroendocrine Signaling | Ecdysone (Ishimoto and Kitamoto, 2010) Dilp2 (Cong et al., 2015) sNPF (Shang et al., 2013) (Chen et al., 2013) |
| Non-Neuronal Signaling | Notch (Seugnet et al., 2011b) Relish (Williams et al., 2007) (Kuo et al., 2010) Amyloid precursor protein Like (Farca Luna et al., 2017) |
3.1.1 Wake-promoting Ion Channels
Ion channels are critical components of electrical signaling among neurons in the brain. Four different ion channels have been implicated in Drosophila wake regulation: the Ca-αlT calcium channel (Jeong et al., 2015), the Dα3 nicotinic acetylcholine receptor (Wu et al., 2014), the potassium channel Sandman (Pimentel et al., 2016), and the TrpA1 cation channel (Lamaze et al., 2017). While mammals express three different T-type calcium channels, Ca-αlT is the single T-type calcium channel that is expressed throughout the Drosophila brain and it has been demonstrated to promote wake (Jeong et al., 2015). Drosophila mutants that do not produce Ca-αlT exhibit a significant sleep increase that suggests that Ca-αlT is wake-promoting (Jeong et al., 2015). Knockdown of Ca-αlT that is limited to neurons recapitulates the effect of the Ca-αlT genetic deficiency, demonstrating a brain-specific role for Ca-αlT in sleep/wake (Jeong al., 2015). In mammals, different types of genetic and pharmacological examination of sleep regulation involving T-type calcium channels have yielded divergent effects on delta wave sleep (Anderson et al., 2005) (Kraus et al., 2010), so understanding how the phenotypes seen in the fly relate to more complex systems will require further investigation. The Drosophila nicotinic acetylcholine receptor (nAChR) Dα3 is another wake-promoting ion-channel that regulates wake (Wu et al., 2014). Wu et al., 2014 tested the hypothesis that upregulation of nicotinic acetylcholine signaling was responsible for the short-sleeping phenotype of Sleepless (SSS) and Shaker mutants (both discussed in greater detail later) by administering mutants the nAChR antagonist mecamylamine (MCA) and observing the effect on sleep (Wu et al., 2014). In support of this hypothesis, MCA rescued the short-sleep in both mutants. Furthermore, of the 10 nicotinic acetylcholine receptors in the fly, the Dα3 subunit appears to mediate changes in wakefulness since RNAi-mediated knockdown of Dα3 rescued the short sleep in the SSS mutant background (Wu et al., 2014). In the dorsal fan-shaped body, the potassium channel Sandman acts as a wake-promoting ion channel (Pimentel et al., 2016). Translocation of Sandman to the plasma membrane is responsible for suppressing activity of dFSB neurons to increase wakefulness (Pimentel et al., 2016). Finally, the cation channel TrpA1 was recently found to be responsible for Prolonged Morning Wakefulness (PMW) - a delayed onset of daytime siesta sleep - in male flies in response to elevated temperature. Lamaze et al., 2017 found that both loss-of-function mutation of TrpA1 as well as RNAi-mediated knockdown of TrpA1 in neurons suppresses PMW in male flies in response to temperature increase. Thus, TrpA1 is responsible for increasing daytime wakefulness in male flies at elevated temperatures. These results demonstrate a wake-promoting role for ion signaling in Drosophila and reveal some of the dynamics that occur at the cellular level that directly modulate the activity of sleep/wake relevant circuits to modulate sleep behavior.
3.1.2. PKA/CREB Pathway
3.1.2.1. PKA/CREB
Studies examining the role of cyclic AMP (cAMP), protein kinase A (PKA), and cAMP response element binding protein (CREB) have identified the cAMP-dependent pathway as one of the principal wake-promoting pathway in Drosophila (Hendricks et al., 2001; Joiner et al., 2006). As previously mentioned, CREB regulates circadian rhythms (Belvin et al., 1999). However, it has also been found that the activity of CREB also directly modulates sleep amount. Levels of cAMP, PKA, and CREB are inversely correlated with the amount of sleep observed in the fruit fly (Hendricks et al., 2001; Joiner et al., 2006). Joiner et al., 2006 determined that the regulation of sleep by PKA occurs in the Drosophila mushroom bodies (Joiner et al., 2006). Since CREB is a transcription factor, its involvement in regulating sleep suggests that some genes whose transcription are affected by CREB will be wake-promoting genes though these genes have not been identified. It should be noted that cAMP involvement in sleep regulation is conserved in mammals as rats that are administered the drug Rolipram – which increases cAMP levels – subsequently display increased wake (Lelkes et al., 1998). Mice who have deletion of two of these isoforms of CREB, i.e, CREB hypomorph, have reduced wakefulness, particularly in the early part of the lights off period (Graves et al., 2003). CREB is also involved in regulating the daily rhythms of rest:activity in the fly. CREB is required for normal rhythms, since flies carrying a mutation in the CREB gene leading to its disrupted expression display disrupted and shortened circadian rhythms (Belvin et al., 1999). Furthermore, a loss-of-function CREB mutation results in a concurrent decrease in the expression of the gene period, a key component of the Drosophila clock (Belvin et al., 1999). Not surprisingly, other molecules that act on the PKA pathway have been found to play a role in sleep regulation. One of these molecules is the amnesiac (AMN) neuropeptide. The amnesiac (amn) gene encodes an AMN neuropeptide that shares homology with adenylate cyclase-activating peptide (PACAP) in mammals (Liu et al., 2008). PACAPs are activators of the PKA pathway. (Figiel and Engele, 2000). The amn gene does not appear to regulate sleep amount. Instead, amn is involved in the regulation of the onset of sleep as well as the maintenance and consolidation of sleep, since flies that carry a loss-of-function amnesiac allele have fragmented sleep and a shorter latency to sleep (Liu et al., 2008). The characterization of sleep changes in amn mutants suggests that distinct genes regulate sleep duration and architecture.
3.1.2.2. Calcineurin
Calcineurin is a Ca2+/calmodulin-dependent serine/threonine protein phosphatase (Rusnak and Mertz, 2000) whose activity affects sleep amount in Drosophila (Tomita et al., 2011; Nakai et al., 2011). Calcineurin dephosphorylates target proteins to regulate their activity and studies in long-term potentiation show that calcineurin is antagonized by PKA activity (Winder et al., 1998). Tomita et al., 2011 knocked down calcineurin in neurons using RNAi and observed decreased sleep in flies. In contrast, increasing calcineurin activity by expressing a constitutively active form of calcineurin in neurons increased total sleep time. Nakai et al., 2011 similarly found that loss-of-function mutations of calcineurin subunits as well as a knockout of the calcineurin regulator Sarah (sra) decreased sleep. However, in their analysis of the effect of constitutively active calcineurin on sleep Nakai et al., 2011 also found that neuronal expression of active calcineurin decreased, rather than increased sleep. A possible explanation for the difference in results may be that the temporal differences in the calcineurin manipulations could have produced the different effects. Tomita et al., 2011 used a temperature-induced expression (McGuire et al., 2003) of the constitutively active calcineurin during adulthood while Nakai et al., 2011 expressed the active calcineurin throughout development. Increasing calcineurin activity during development could affect circuit formation and produce sleep effects that are independent of their effects in adulthood following normal development. Indeed, developmental effects of any manipulation are an important consideration in all animal model research. This may be particularly true for sleep which is inherently a circuit property.
3.1.3. Monoamine Signaling
Vesicular transporters are responsible for packaging neurotransmitters into secretory vesicles at presynaptic sites of release (Erickson and Varoqui, 2000). The Drosophila vesicular monoamine transporter (VMAT) is a vesicular transporter that packages monoamine neurotransmitters into vesicles. Monoamine neurotransmitters have been shown to promote wakefulness (see above). In Drosophila, these monoamine neurotransmitters include octopamine, dopamine, serotonin and histamine (Busch et al., 2009; Mao and Davis 2009; Sitaraman et al., 2008; Oh et al., 2013). Nall and Sehgal, 2013 conducted a small-molecule screen by treating flies with 1280 small molecules and assaying the response to sleep. From this study, it was discovered that treatment with the VMAT inhibitor reserpine – which prevents the transport of monoamines into presynaptic vesicles – significantly increased sleep in flies, a phenotype that is recapitulated in genetic null VMAT mutants (Nall and Sehgal, 2013). The effects of reserpine were not isolated to the transmission of any single monoamine, as genetic deficiency of various monoamines did not affect the response of the flies to reserpine (Nall and Sehgal, 2013). This suggests that VMAT regulates sleep through its involvement in neurotransmission for multiple neurotransmitters. These results illustrate the wake-promoting nature of VMAT activity and highlight the utility and feasibility of drug screens in Drosophila.
3.1.4. Fragile-X Mental Retardation Protein
The Fragile-X Mental Retardation Protein (FMRP) modulates the activity-dependent pruning of synapses as well as synaptic plasticity in Drosophila (Tessier and Broadie, 2008; Mercaldo et al., 2009). FMRP is the protein product of the fragile-x mental retardation gene (Fmr1), and levels of Fmr1 are inversely correlated to sleep time as evidenced by increased sleep being observed in flies with loss-of-function of Fmr1 and decreased sleep upon overexpression of the Fmr1gene (Bushey et al., 2009). FMRP been recently identified as a wake-promoting protein (van Alphen et al., 2013), as flies carrying a loss-of-function allele for FMRP display an increased sleep intensity during the day, as evidenced by a decreased responsiveness to mechanical stimuli (van Alphen et al., 2013). This finding illustrates a potential molecular connection linking synaptic plasticity and sleep regulation in the fly (see further below).
3.1.5. Salt-inducible Kinase 3
Recently, the first forward genetic analysis of sleep in mice was conducted and identified two novel regulators of sleep, Salt-inducible kinase 2 (SIK3) and NALCN (Funato et al., 2016). Random mutagenesis of SIK3 and the sodium leak channel NALCN produced dominant mutations that produced Sleepy (Slp) and Dreamless (Drl) mice that have increased non-REM and decreased REM sleep, respectively (Funato et al., 2016). While NALCN had previously been identified as a regulator of normal circadian rhythms in Drosophila (Flourakis et al., 2015), Funato et al., 2016 also demonstrated that in addition to altering sleep in mice, genetic reduction of SIK3 activation in neurons increased sleep, confirming that the wake-promoting effect of SIK3 is conserved across species. This study re-emphasizes the usefulness of unbiased forward genetic screening and also reconfirmed the conservation of pathways in sleep across species. The molecules identified through these methods have further helped identify both widespread signaling pathways and distinct physical loci in the brain that regulate the sleep and wake cycle of the fly and continue to lead to new insights that are applicable across species.
3.2. Sleep-promoting signaling pathways and molecules (see Table 2)
3.2.1. Sleep-promoting Ion Channels
One of the important discoveries that arose from unbiased forward genetic screening in Drosophila was the finding that ion channels regulate sleep. In a forward genetic screen, selected phenotypes or behavioral abnormalities are identified from a population of flies that have undergone mutagenesis in order to identify genes responsible for these behaviors. Methods for generating Drosophila genetic mutants include ethyl methanesulfonate (EMS) mutagenesis (Konopka and Benzer, 1971), P-element insertional mutagenesis (Cooley et al., 1988), piggyBac insertional mutagenesis (Horn et al., 2003), Minos insertional mutagenesis (Uchino et al., 2007) and RNA interference (RNAi) (Rogulja and Young, 2012). Cirelli et al., 2005 identified the Shaker mutant from an EMS mutagenesis screen; a point mutation in the gene encoding the Shaker fast-acting voltage-gated potassium channel that results in its loss of function induces a short-sleeping phenotype (Cirelli et al., 2005). The sleep-regulatory role of Shaker appears to be conserved in mammals, as mice that are null for the Shaker-like channel Kcna2 are also short-sleepers, though notably they also exhibit frequent seizures and have significantly shortened lifespans (Douglas et al., 2007). Other mutations in genes involved in the regulation of Shaker activity were subsequently found to also affect sleep in Drosophila. For example, loss-of-function mutants for the gene Hyperkinetic, which encodes a regulatory subunit of the Shaker channel, also exhibit a short-sleeping phenotype (Bushey et al., 2007). Additionally, the protein Sleepless (SSS), identified through an independent forward genetic screen for sleep mutants, is a regulator of Shaker that is required for normal sleep regulation. Loss of SSS results in markedly reduced sleep in Drosophila (Koh et al., 2008). SSS is predicted to be a member of the Ly-6/neurotoxin superfamily of proteins, which encompass proteins such as ion-channel-modulating snake neurotoxins (Tsetlin 1999; de Weille et al., 1991), cell-surface proteins and receptors (Davies et al., 1989; Wilhelm et al., 1999), and signaling molecules (Adermann et al., 1999; Tsuji et al., 2003; Huai et al., 2006). SSS was further shown to regulate Shaker expression and localization; genetic loss of SSS results in decreased Shaker expression and altered localization (Wu et al., 2010). In flies that are null for SSS, Shaker expression is greater in cell bodies than in the antennal nerve (Wu et al., 2010). Lastly, the Drosophila ATP-sensitive potassium channel dSur was identified as a sleep-promoting ion channel (Allebrandt et al., 2013). dSur is the fly homologue of mammalian SUR2 and knockdown of dSur in Drosophila neurons significantly reduces nighttime sleep, suggesting that dSur promotes sleep in the fly (Allebrandt et al., 2013).
3.2.2. EGF Signaling
The epidermal growth factor receptor (EGFR) is responsible for many different functions in the cell, including cell proliferation and differentiation. Epidermal growth factor receptor (EGFR) signaling is a sleep-promoting pathway in Drosophila (Foltenyi et al., 2007). This sleep-promoting role of EGFR signaling is conserved in C. elegans (Van Buskirk and Sternberg, 2007) and in mammals, EGFR signaling in the hypothalamus is required for circadian rhythms (Kramer et al., 2001) and promotes sleep (Kushikata et al., 1998). In Drosophila, there are four ligands for EGFR which must be activated by the processing proteins Star and Rho. Increasing the expression of Star and Rho increases sleep through activation of extracellular signal-regulated kinase (ERK), while decreased function of the EGFR pathway induced by RNAi of Rho decreases sleep (Foltenyi et al., 2007). Thus, activity of the EGFR pathway acts as a sleep-promoting signal in Drosophila. Interestingly, the role of EGF in sleep appears to be conserved in mammalian systems, as ICV-administered EGF promotes sleep in rabbits (Kushikata et al., 1998), the EGF receptor ligand transforming growth factor-alpha (TGF-alpha) suppresses locomotion in hamsters and mice carrying a mutation resulting in hypomorphic EGF signaling are hyperactive (Kramer et al., 2001).
3.2.3. G protein signaling
G protein-coupled receptors (GPCRs) are the most abundant family of receptors in both vertebrate and invertebrate systems. GPCRs act through G proteins which are critical for the transduction of numerous intracellular signaling cascades. In Drosophila, G protein activity promotes both sleep and wake, depending on cell types involved. Guo et al., 2011 found the first evidence that G protein signaling in the brain promotes sleep, since pharmacogenetic enhancement of both wildtype and constitutively active Go expression in neurons led to significant increases in sleep time in the fly. This wake-promoting effect of Go on sleep was found to occur in the mushroom bodies; specific activation of Go in the mushroom bodies promotes sleep while RNAi- or pertussus toxin (PTX)-mediated inhibition of Go signaling in the mushroom bodies decreases sleep time (Guo et al., 2011). This data indicates a wake-promoting role for Go signaling, but further characterization of G protein signaling in smaller subsets of cells within the mushroom body now indicate that there are cell-type specific effects of Go and that Go signaling in some mushroom body neurons promotes sleep. Yi et al., 2013 modulated Go signaling in a small subset of α/β core cells in the mushroom body and produced the opposite effects previously seen in the previous study using a different mushroom body driver. When G protein signaling is inhibited via heat shock treatment to induce temperature-sensitive PTX in this subset of cells, the result is increased sleep (Yi et al., 2013). Thus, there is evidence that location is important for determining the role that an intracellular signaling pathway may play in regulating sleep and wake. Later work found that binding to the G protein-coupled receptor SIFR in the pars intercerebralis (PI) by the neuropeptide SIFamide also promotes sleep (Park et al., 2014). Thus, G proteins act in different regions of the brain to modulate both sleep and wake.
3.2.4. Homer
Synapses are dynamic structures in the brain and evidence suggests that proteins within the synapse regulate behavioral state and are also in turn modulated by sleep (Gilestro et al., 2009) (Diering et al., 2017). Homer proteins are adaptor proteins within the synapse that bind to molecules such as group I metabotropic glutamate receptors and IP3 receptors (Tu et al., 1998) and are involved in processes such as calcium signaling (Shin et al., 2003) and receptor trafficking (Shiraishi et al., 2003). While mammalian systems carry three homer genes with 6 proteins due to alternate splicing, Drosophila contains only a single Homer gene—D-homer with one protein (Xiao et al., 1998; Kato et al., 1998). Expression of the Drosophila Homer gene was found to be upregulated during sleep deprivation and downregulated during sleep (Zimmerman et al., 2006). Experimental evidence that Homer proteins are involved in sleep regulation was uncovered through the observation that flies carrying a genetic deletion of D-homer have decreased and fragmented sleep, suggesting that D-homer promotes sleep and sleep consolidation (Naidoo et al., 2012). Furthermore, loss of Homer function affects the homeostatic response to sleep loss, as Homer null mutants do not display the increased bout durations of sleep that are characteristic of rebound sleep after sleep deprivation in wildtype flies. This indicates an inability for Homer null flies to maintain consolidated sleep despite increased sleep pressure (Naidoo et al., 2012). Recent work in rodents has demonstrated that the role of Homer in sleep may be related to the need for synaptic downscaling after wake (Diering et al., 2017). During sleep, synaptic downscaling has been observed in primary motor and somatosensory cortex (de Vivo et al., 2017) and at the postsynaptic density, the Homer protein associations with molecules such as mGluR1/5 and IP3 receptors are downregulated (Diering et al., 2017). The downregulation of these physical interactions is dependent on the dominant negative immediate early gene Homer1a (Diering et al., 2017). In Homer1a null mice, the effect of sleep on these protein interactions is lost (Diering et al., 2017). Thus, Homer signaling is a molecular mechanism of synaptic plasticity that regulates sleep and wake states and also modulates excitatory signaling at the synapse. These data strongly suggest that sleep is critically important for the regulation of synaptic homeostasis and plasticity in the brain.
3.2.5. Ubiquitin-Proteasome Signaling
A role for protein degradation pathways in sleep modulation in Drosophila was uncovered through a forward genetics approach. An EMS mutagenesis screen identified the Drosophila insomniac mutant, which displays dramatically decreased and fragmented sleep (Stavropoulos and Young 2011). The insomniacmutation was attributed to a deletion of a region of a gene encoding a putative adaptor of the Cullin-3(Cul3) ubiquitin ligase complex (Stavropoulos and Young, 2011), which is a critical component of cellular protein degradation. Insomniac physically associates with Cul3 in vitro and RNAi knockdown of Cul3 and Nedd8, an ubiquitin-like protein necessary for Cul3 activity, also decreased sleep. Two additional molecules involved in ubiquitin proteasome signaling were also recently identified as regulators of sleep. Like Cul3, the E3 ubiquitin ligase gene highwire (hiw) also promotes sleep. Knockdown of the E3 ubiquitin ligase gene highwire (hiw) in the large lateral ventral neurons (l-LNvs) significantly reduces sleep in the fly (Seugnet et al., 2017). In contrast, fat facet (faf), a deubiquitinating enzyme, is a wake-promoting molecule whose upregulation in the large lateral ventral neurons l-LNvs reduces sleep (Seugnet et al., 2017). Interestingly, hiw knockdown in the mushroom body also suppresses sleep rebound following sleep deprivation, demonstrating that hiw exerts its effects on sleep within different brain circuits (Seugnet et al., 2017). Thus, molecules required for ubiquitination such as Cul3 and hiw appear to have sleep-promoting properties. In contrast, faf which negatively regulates protein ubiquitination and protein degradation, promotes wake. These studies demonstrate that protein degradation pathways can modulate sleep in Drosophila and may also suggest that the accumulation of non-degraded proteins might somehow increase wakefulness or suppress sleep (see also further below in discussion of unfolded protein response).
3.2.6. Cellular Stress Response Signaling
3.2.6.1. Unfolded Protein Response
In the cell, the endoplasmic reticulum (ER) is responsible for the synthesis and proper processing of proteins that are either destined to be inserted in the membrane or secreted. When misfolded proteins accumulate in the cell, this leads to ER stress that triggers a cellular response known as the unfolded protein response (UPR) (Reviewed by Walter and Ron, 2011). Immunoglobulin binding protein (BiP) is a molecular chaperone protein that is upregulated during the UPR and is a key molecular component of the response to endoplasmic reticulum (ER) stress. Studies on BiP indicate that the UPR mechanism is involved in modulating the amount of recovery sleep following sleep loss in the fly. BiP mRNA and protein levels are upregulated in brain after sleep deprivation across species (Shaw et al., 2000; Naidoo et al., 2005; Naidoo et al., 2007) and BiP modulates recovery sleep in Drosophila as shown by the increased recovery sleep in BiP overexpressing flies and the decreased recovery sleep in BiP dominant negative mutants (Naidoo et al., 2007). Thus, BiP, as a component of the UPR, is involved in the homeostatic response to sleep loss in flies. It is not known whether BiP alters baseline sleep however, since BiP has not been shown to change the amounts of sleep and wake in the absence of sleep deprivation. Furthermore, Brown et al., 2014 found that inducing ER stress in young flies is sufficient to significantly fragment sleep and impair the ability to recover sleep following sleep deprivation. Pharmacological administration of UPR protein inhibitors have also been shown to reduce sleep in Drosophila (Ly et al., unpublished observations). These studies indicate a relationship between ER stress, the UPR, and sleep.
3.2.6.2. JNK Signaling
c-Jun N-terminal Kinases (JNK) are stress-activated kinases that are involved in many diverse functions such as development, cell survival, apoptosis, cellular proliferation and differentiation (Reviewed in Davis, 2000). Basket (bsk), the Drosophila homolog of mammalian JNK, is a sleep-promoting molecule in the fruit fly. RNAi knockdown of bsk in neurons using the elav-GAL4 driver produces a decrease in total sleep and the same effect is mimicked by a bsk knockdown that is specific to the mushroom bodies (Takahama et al., 2012). The results provide evidence for the involvement of bsk mushroom body activity in promoting sleep and also identify other molecular mechanisms of sleep regulation originating in the sleep-regulating mushroom body region of the Drosophila brain.
3.2.7. Cell Cycle signaling
Proteins that regulate cell cycle signaling also play a role in Drosophila sleep. Rogulja and Young, 2012 identified the cell cycle protein cyclin A as a regulator of sleep in Drosophila. Cyclins control the progression of the cell cycle by activating cyclin-dependent kinases. After conducting a screen of RNAi lines, they found that the gene Regulator of cyclin A1 (Rca1) – the Drosophila homolog of early mitotic inhibitor 1 (Emi1) in mammals – promotes sleep (Rogulja and Young, 2012). Loss of neuronal Rca1 leads to decreased sleep caused by shortened sleep bout durations (Rogulja and Young, 2012). The same effect was observed after knocking down expression of the primary target of Rca1, cyclin A (CycA). Loss of cyclin A in neurons via neuronal expression of CycA RNAi also led to an increased latency to sleep after lights off, suggesting that CycA may be involved in the transition from wakefulness to sleep. It was later found that CycA is regulated by TARANIS (TARA) – a transcriptional regulator – through a separate forward genetic screen (Afonso et al., 2015). TARA partially acts in the pars lateralis (PL), which is a neuroendocrine region of the brain that is considered analogous to the mammalian hypothalamus (de Velasco et al., 2007). Knockdown of tarain the PL significantly reduces sleep (Afonso et al., 2017). Furthermore, Cyclin-dependent kinase 1 (Cdk1), which physically interacts with CycA, acts antagonistically to CycA and TARA and promotes wake, since overexpression of Cdk1 in the PL reduces sleep (Afonso et al., 2015). Both of these studies highlight the benefits of forward genetic screening for identifying novel regulators of sleep, and are the first studies to directly demonstrate a role for cell cycle signaling pathways in sleep regulation.
3.2.8. Neuroendocrine Signaling
3.2.8.1. Ecdysone
Sleep and neuroendocrine signaling affect one another in mammalian systems (Obál et al., 2001; Meerlo et al., 2008) and flies also display evidence of neuroendocrine involvement in sleep regulation. Ecdysone is a steroid hormone that is critical for insect development and promotes sleep in Drosophila (Ishimoto and Kitamoto, 2010). Flies fed 20-hydroxyecdysone (20E), the active metabolite of ecdysone, slept longer and displayed longer consolidated sleep bouts. Furthermore, genetically reducing levels of 20E or levels of functional ecdysone receptors (EcRs) both cause decreases in sleep amount as well as decreased sleep rebound in response to sleep deprivation (Ishimoto and Kitamoto, 2010). Furthermore, overexpressing EcRs in the mushroom bodies, one of the sleep-regulating brain regions, increased sleep (Ishimoto and Kitamoto, 2010). The involvement of ecdysone in sleep promotion and regulation in Drosophila illustrates a mechanism through which neuroendocrine signaling is involved in regulation of sleep and notably provides evidence of developmental genes regulating behavior in the adult animal.
3.2.8.2. Drosophila insulin-like peptide
A component of neuroendocrine signaling in Drosophila occurs via Drosophila insulinlike peptides (DILPS). There are eight DILPS in Drosophila (Reviewed in Kannan and Fridell, 2013) whose roles in metabolism and aging reveal that neuroendocrine signaling in Drosophila is highly conserved. Cong et al., 2015 recently demonstrated that DILPs and the DILP receptor DInR regulate sleep. All mutants of the DILP1, DILP2, DILP5, DILP6 and DILP7 and a Drosophila DInR receptor mutant exhibit total sleep reductions. Conversely, increased expression of DILP2 and DInR led to increases in sleep. Cong et al., 2015 identified a subset of clock neurons that express DILP. Furthermore, dilp2 transcript levels were found to be downregulated by food restriction, providing a mechanism through which starvation suppresses sleep in the fly (Cong et al., 2015). Additionally, the genetic downregulation of Dilp2 decreases sleep while Dilp2 upregulation promotes sleep (Cong et al., 2015). This shows that starvation directly impacts a sleep-regulating pathway in the brain to inhibit sleep. Since DILPs are a critical component of Drosophila metabolic regulation (as insulin signaling is in mammals), their involvement in sleep points to some metabolic regulation of sleep.
4.2.8.3. Short Neuropeptide F
Another neuroendocrine signal that regulates sleep in Drosophila is short neuropeptide F (sNPF) (Shang et al., 2013; Chen et al., 2013; He et al., 2013). sNPF shares sequence homology to mammalian Neuropeptide Y (NPY) (Mertens et al., 2002), whose signaling regulates a wide variety of processes such as food intake (Gerald et al., 1996), response to stress (Wang et al., 2013), neuronal excitability (Colmers and Bleakman, 1994), and circadian rhythms (Wiater et al., 2011). The data on sNPF suggest different possibilities for its role in sleep. Shang et al., 2013 found that activating sNPF-expressing cells by heat-induction of the dTRPA1 cation channel significantly increased time spent asleep while inhibiting activity of sNPR-expressing cells by heat-induction of the Kir2.1 potassium channel decreased sleep during the day. Furthermore, expression of dominant-negative sNPF in peptidergic neurons results in sleep fragmentation which suggests that sNPF is involved in sustaining sleep (Shang et al., 2013). In contrast, Chen et al., 2013 found that flies deficient in sNPF or its receptor sNPFR have significantly more sleep and that flies overexpressing sNPF or sNPFR display significant reductions in sleep duration (Chen et al., 2013). sNPF increases cAMP levels in vitro as well as CREB activity in vivo (Chen et al., 2013), suggesting that the arousal promoting actions of sNPF are likely to be related to changes in the activity of the cAMP-dependent pathway. Since there are varying phenotypes observed upon different manipulations of sNPF signaling, it is important to consider some of the potential underlying causes. For example, it is possible that there are developmental effects of disrupted sNPF signaling that explain the effects of constitutive sNPF or sNPFR genetic manipulation compared to those observed upon heat-induced knockdown. This adds further support for the need to differentiate effects in adult animals from those that change development. It is also possible that the activation of sNPF-expressing cells leads to the release of some other factors that may alter the observed sleep response in a way that is different to the genetic loss of the ligand or receptor. Together, the results from these two studies suggest that sNPF signaling is critical for sleep regulation both during development as well as in the mature brain.
3.2.9. Non-Neuronal Signaling
3.2.9.1. Glial Cells
Work utilizing the Drosophila model has provided interesting insights into the involvement of non-neuronal cells in sleep modulation. Recently, studies in rodent models have led to the emergence of evidence that glial cells are involved in sleep regulation (Halassa et al., 2009; Schmitt et al., 2012; Xie et al., 2013). In Drosophila, glial cell signaling also regulates sleep, with data pointing to the Notch signaling pathway as one of the molecular mechanisms through which this occurs. Notch is a transmembrane receptor (Wharton et al., 1985) that regulates a signaling pathway critically involved in nervous system development and cell fate (Reviewed in Louvi and Artavanis-Tsakonas, 2012). In Drosophila, Notch is expressed in glial cells (Seugnet et al., 2011b) and modulates the homeostatic response to sleep loss as well as the effects of sleep loss on learning in Drosophila (Seugnet et al., 2011b). Increasing Notch activity by p-element disruption of the transcription factor bunched in the mushroom bodies – which negatively regulates Notch activity – abolishes the homeostatic response to sleep loss. Similarly, increasing Notch signaling by overexpressing its receptor ligand Delta in the mushroom bodies reduces the compensatory increase that occurs in sleep in response to sleep loss and also rescues learning impairments that occur after sleep deprivation in flies (Seugnet et al., 2011b). Since the Delta ligand is expressed in neurons, binding of Delta to glial-expressed Notch receptors is a potential mechanism through which glial cells interact with neurons to regulate Drosophila sleep. Furthermore, since glial cells are important for synaptic plasticity (Reviewed in Ben Achour and Pascual, 2010), understanding the molecular basis of glial cell-mediated sleep regulation may uncover novel mechanisms that underlie the relationship between sleep, learning, and memory. In addition to Notch signaling, glial Amyloid Precursor Protein Like (APPL), the Drosophila homologue of amyloid precursor protein (APP), was found to be a necessary promoter of wake in the fly (Farca Luna et al., 2017). Genetic knockdown of APPL in cortex glia and astrocyte-like glia – two of four different glial subtypes in the Drosophila brain – increases sleep at night while overexpression of APPL decreases sleep (Farca Luna et al., 2017). Knockdown of glial APPL leads to reduced expression of glutamine synthetase (GS) and the gap junction innexin 2 (Inx2), which are both involved in glutamate recycling at the synapse. Glial-specific double knockdown of both GS and Inx2 mimics the increased sleep phenotype seen following APPL knockdown. Furthermore, increasing the glutamate reuptake capabilities of glial cells through overexpression of the glutamate transport dEAAT1 rescues the sleep increases following APPL knockdown (Farca Luna et al., 2017). These results demonstrate a role for APP signaling in sleep regulation that occurs within glial cells and involves regulation of glutamate recycling pathways. Given that sleep dysfunction is associated with Alzheimer's disease (Ju et al., 2013; Lim et al., 2013), this may have important implications for understanding the molecular mechanisms that may regulate both sleep as well as neurodegenerative disease pathology. Both of these Drosophila studies demonstrate that signaling pathways in glial cells are relevant to sleep regulation and highlight the potential knowledge to be gained from continuing to interrogate the role of glial cells in sleep.
3.2.9.2. Fat Bodies
Sleep and immunity are processes that exert reciprocal influence on one another. Proinflammatory molecules are upregulated following insufficient sleep and sleep alterations are often associated with infection and disease (Reviewed in Imeri and Opp, 2009; Gamaldo et al., 2012). Williams et al., 2007 found that the transcription factor Relish, a critical component of the Drosophila immune response system, is upregulated during sleep deprivation and promotes sleep in the fly (Williams et al., 2007). Relish is the Drosophila homolog of NF-kappaB (NFκB) and flies heterozygous for a null mutation in the Relish gene display decreased and fragmented sleep patterns (Williams et al., 2007). Notably, altering Relish expression in the fat bodies, and not neurons, regulates sleep as shown by the sleep response to rescue or knockdown of Relish specifically in the fat bodies (Williams et al., 2007). Relish RNAi expression in the fat bodies significantly decreased sleep while expressing Relish in the fat bodies of Relish null mutants rescues their disrupted sleep. Changing expression of Relish in neurons had no effect (Williams et al., 2007). Furthermore, inducing an immune response by exposing flies to bacteria or injury increases sleep and requires Relish activity, as mutants null for Relish, do not display increases in sleep induced by immune response (Kuo et al., 2010). This is rescued by producing expression of Relish only in the fat bodies (Kuo et al., 2010). Thus, molecular signaling in the fat bodies is capable of altering sleep. Since fat bodies are equivalent to the liver/visceral fat in mammalian systems, this observation opens up a new avenue of investigation—how does fat influence sleep? Conversely, how does sleep affect metabolic function and energy balance? Given the increasing prevalence of obesity (James, 2004; Ogden 2012) this is an important question to investigate this mind-body interaction and will likely be a productive area of inquiry.
4. Conservation of Mechanisms and Insights into Sleep Function
The question of why sleep is a necessary behavior applies to nearly every animal in the animal kingdom. Sleep is observed in a broad and diverse range of organisms; elephants (Tobler, 1992), seals (Lyamin et al., 2017), giraffes (Tobler and Schwierin, 1996) cockroaches (Tobler and Neuner-Jehle, 1992), birds (Roth et al., 2006), zebrafish (Yokogawa et al., 2007), jellyfish (Nath et al., 2017) and the C. elegans roundworm (Raizen et al., 2008) are just some examples of animals that exhibit sleep states. Clearly, sleep must serve some fundamental function that makes it necessary for survival in order to have persisted in so many different animals and environments over the course of evolution. Underlying sleep and wake behavior are also many regulatory mechanisms that are conserved across multiple species, including Drosophila (Allada and Siegel, 2008; Zimmerman et al., 2008a). The cross-species examination of sleep is important to understanding this behavioral phenomenon because it highlights the core principles of sleep neurobiology. The precise function of sleep is an ongoing debate and it is still not entirely clear why such a great portion of every day is spent in an unconscious state. By understanding how the brains of different species have evolved to accomplish a similar behavioral endpoint, we can pinpoint the core biological needs that drive sleep and ultimately come to a conclusion about the biological functions of sleep.
As described in the previous sections, many of the neurotransmitters and molecules in Drosophila that regulate sleep have parallel roles in mammals. Of the neurotransmitters that are known to affect sleep and wake in Drosophila, dopamine, octopamine/norepinephrine, GABA, serotonin and glutamate are all conserved in mammalian sleep regulation (Isaac and Berridge, 2003; Aston-Jones and Bloom, 1981; Gallopin et al., 2000; Xiong et al., 2014; Ursin, 2002; Fuller et al., 2011). In addition to neurotransmitters, other molecules that share sleep-regulating function across Drosophila and other mammals include cAMP (Lelkes et al., 1998), EGFR (Kushikata et al., 1998; Kramer et al., 2001), and voltage-gated potassium channels (Douglas et al., 2007). The shared molecular components of sleep regulation in the fruit fly have further shed light on some key principles that underlie important theories about sleep function. There are currently many theories regarding the function of sleep (Kreuger et al., 2016) and emerging evidence supporting various theories has arisen from work in the Drosophila animal model. Here, we explore some of these theories about sleep function by examining evidence that is shared between Drosophila and other studied species (see Table 3).
Table 3. Some putative functions of sleep based on conserved mechanisms of sleep regulation.
Some of these references cite correlational studies
4.1. Synaptic Homeostasis and Plasticity
Plasticity is an inherent feature of all biological systems; organisms must constantly respond and adapt to changes in the environment in order to survive. Within the brain, shifting demands on different neuronal connections lead to adjustments in both synaptic transmission strength and structure (Reviewed in Holtmaat and Svoboda, 2009). There is a large amount of evidence that suggests that sleep is modulates synaptic plasticity and maintains synaptic homeostasis in the brain. Early studies found that molecular markers of synapse formation are upregulated during wakefulness and downregulated during sleep in both rats (Cirelli and Tononi, 2000a) and Drosophila (Gilestro et al., 2009). Furthermore, increased wakefulness is correlated with increases in the number of synapses and synapse size in Drosophila (Bushey et al., 2011). Using electron microscopy, de Vivo et al., 2017 recently compiled structural evidence that synaptic spine downscaling occurs during sleep in mice. Together, these data suggest that sleep is critical for modulating synaptic plasticity in the brain and that across species, sleep likely contributes to synaptic downscaling. This notion is known as the synaptic homeostasis hypothesis, which posits that sleep is required to prune and downscale synapses synaptic connections that are strengthened during wake (Reviewed in Tononi and Cirelli, 2006). This may be important for minimizing cellular stress, increasing the signal-to-noise ratio of circuits, and recalibrating the energy demands in the brain. Electrophysiological studies in mice and rats have also found evidence supporting the theory of downscaling, since spontaneous excitatory output of cortical neurons decreases in frequency and amplitude after sleep (Liu et al., 2010). During sleep, spine formation also occurs throughout the brain (Yang et al., 2014; Diering et al., 2017), though the recent evidence strongly suggests that the net effect across all synapses is a downregulation of synapse number and size following sleep (Diering et al., 2017; de Vivo et al., 2017).
Another predominating theory about sleep function that relates to its role in plasticity is that sleep is critical for the consolidation of memories and learned information and tasks (Reviewed in Walker and Stickgold, 2006; Diekelmann and Born, 2010). Numerous studies in humans, rodents, and Drosophila have shown that sleep loss is consistently followed by impairments in cognitive functioning and memory (Graves et al., 2003; Killgore et al., 2006; Palchykova et al., 2006; Li et al., 2009; Tucker et al., 2010). In Drosophila, cells that regulate sleep have been shown to mediate learning and memory (Donlea et al., 2011; Haynes et al., 2015). For example, inducing sleep through transgenic activation of sleep-promoting dFSB-projection neurons after courtship training is sufficient to generate long-term memories that last for multiple days and affect courtship behavior (Donlea et al., 2011). Additionally, sleep induced by dFSB activation suppresses the activity of MP1 and MV1 neurons that mediate memory elimination (Berry et al., 2012; Berry et al., 2015), identifying another unique learning circuit whose activity is modulated by sleep. In a different study, both transgenetic and pharmacological induction of sleep rescues learning and memory deficits in Drosophila learning mutants as well as in flies carrying a mutation in Presenilin that is relevant to Alzheimer's disease (Dissel et al., 2015). Thus, it is evident that in Drosophila, sleep and memory are functionally and anatomically linked.
4.2 Metabolism
Insufficient or disrupted sleep is associated with metabolic dysfunction in many species (Gangwisch et al., 2005; Spiegel et al., 2009; Barf et al., 2010; Wang et al., 2014), which suggests that sleep may be required for the maintenance of metabolic homeostasis. In humans, wake is associated with an upregulation of energy expenditure (Jung et al., 2011; Markwald et al., 2013) and in rodents sleep is associated with an enrichment of transcripts related to metabolic function in the periphery (Anafi et al., 2013). It has been postulated that sleep serves to regenerate energy stores that are exhausted while the brain is awake (Benington and Heller 1995) (Scharf et al., 2008). One of the ways in which sleep might mediate metabolic homeostasis is by modulating feeding behavior. In response to starvation or hunger, sleep is suppressed in rats (Borbély, 1977) and Drosophila (Keene et al., 2010); this may be a conserved adaptive response to promote foraging behaviors when energy stores are low. Indeed, sleep deprivation in humans promotes food-seeking (Chapman et al., 2013) and increases the activation of regional brain activity in response to food (St-Onge et al., 2012). Interestingly, the suppression of sleep following starvation in Drosophila requires functional taste perception (Linford et al., 2015) which validates the postulation that sleep and feeding are functionally linked. Multiple lines of evidence in Drosophila also experimentally link lipid metabolism to changes in sleep and survival. For example, Thimgan et al., 2015 found that Drosophila cycle clock mutants that are extremely sensitive to sleep deprivation (10 hours of sleep loss is enough to induce death), starvation mitigates the effects of sleep deprivation and alterations in putative lipid metabolism gene expression underlie the protective effects of starvation against sleep deprivation in these flies. Slocumb et al., 2015 also observed that flies selected for starvation-resistance express higher levels of triglyceride stores and display increased sleep compared to controls, demonstrating that individual responses to nutrient availability are correlated with changes in sleep and energy storage efficiency. Interestingly, both starvation-resistance and longer sleep can be inherited across many generations suggesting that there is genetic component to the relationship between sleep and metabolic fitness in the fruit fly (Masek et al., 2014). Recently, the RNA/DNA binding protein translin (tsrn) was identified as a critical molecular component of starvation-induced sleep. Tsrn signals in neurons expressing Leucokinin (LK) to suppress sleep (Murakami et al., 2016). RNAi-mediated knockdown of tsrn in LK cells abolishes starvation-induced sleep (Murakami et al., 2016). Genetic tsrn flies do not have altered feeding behavior after starvation, which demonstrates that tsrn regulates sleep after starvation independent of feeding (Murakami et al., 2016). To date, numerous studies have identified signaling pathways that underlie a functional relationship between sleep, feeding and metabolism (Reviewed in Shukla and Basheer, 2016). In mammals, there is a growing amount of evidence of a reciprocal interaction between sleep and energy homeostasis. Recently, work in mice has demonstrated that food consumption can directly influence sleep architecture (Perron et al., 2015). Based on work in Drosophila and other species, it seems apparent that a conserved role of sleep may be to regulate metabolic function. Given the high prevalence of metabolic disorders, there is great public health incentive to understand how sleep and metabolism are functionally linked.
4.3 Brain Development
Sleep duration is the longest during early development in humans (Iglowstein et al., 2003) and many other species (Jouvet-Mounier et al., 1970), which suggests that sleep is also vital for normal brain development. During development the growth and pruning of synapses is critical for the establishment of mature connections that will later serve the mature adult organism. Multiple lines of evidence indicate that quality of sleep during development impacts health and cognitive function later in life. Shorter sleep durations during early life correlate with decrements in cognitive performance (Touchette et al., 2007) and irregular sleep schedules during adolescence are correlated with decreased white matter integrity as measured by fractional anisotropy (Telzer et al., 2015). Similar to mammals, Drosophila melanogaster sleep for longer periods of time during early life (Shaw et al., 2000; Kayser et al., 2014) and sleep deprivation during early adult development is sufficient to induce lasting learning and memory impairments (Seugnet et al., 2011a). The longer sleep times and higher arousal thresholds observed in Drosophila during developmental sleep are mediated by decreased inhibition of dopamine onto sleep-promoting dFSB cells (Kayser et al., 2014). Artificially overexciting these cells during development disrupts the formation of a subset of olfactory glomeruli cells and impairs courtship behavior in adulthood (Kayser et al., 2014). By identifying specific loci in the brain that regulate and are affected by developmental sleep in the fly, this study also proves that sleep is necessary for proper brain development. It also shows conclusively that altering sleep during development affects behavior in adults. In rodents, one of the developmental correlates of sleep is increased synapse elimination (Yang and Gan, 2012). In the developing mouse somatosensory cortex, synaptogenesis occurs during both sleep and wake, but synapses are eliminated at higher rates during sleep (Yang and Gan, 2012). Sleep also enhances ocular dominance plasticity following monocular deprivation during the critical period of visual development (Frank et al., 2001). These data suggest that during development, sleep may be particularly important for synaptic remodeling.
4.4 Protein Homeostasis
Protein homeostasis, also known as proteostasis, is a key feature of healthy cellular function that involves tight regulation of transcription and translation, protein folding, chaperone protein activity, and protein degradation. Wakefulness likely places many cellular demands on the brain, as neuronal connections are active and proteins are turned over or generated. As previously discussed, extended wakefulness upregulates the transcription and translation of the unfolded protein response (UPR) chaperone protein BiP in Drosophila (Naidoo et al., 2007), mice (Terao et al., 2003; Naidoo et al., 2005) and rats (Cirelli and Tononi 2000b; Cirelli et al., 2004). This conserved response to extended wake suggests that sleep may be critically important for modulating cellular proteostasis. Furthermore, the study of age-related changes in sleep and overall brain health points to growing evidence that intracellular proteostatic regulation and sleep are linked. One of the behavioral consequences of aging across both flies and humans is reduced sleep time and fragmented sleep architecture, while a molecular correlate of aging is an impaired unfolded protein response to sleep deprivation (Koh et al., 2006) (Naidoo et al., 2008). The presence of both dysfunctional sleep patterns and disrupted cellular stress response during aging suggests that healthy sleep may mediate healthy protein balance in the cell. Furthermore, sleep has been implicated in the disease pathogenesis in Alzheimer's disease, in which cellular protein aggregation is a key feature. Sleep dysfunction is both a common feature and risk factor of Alzheimer's disease (Ju et al., 2013; Lim et al., 2013), which is characterized by the accumulation of intracellular tau and amyloid protein aggregation in the brain (Reviewed in Peter-Derex et al., 2015). There is mammalian evidence that sleep might promote the expulsion of unwanted proteins from the brain since injections of radiolabeled amyloid beta (Aβ) are cleared twice as fast in sleeping mice compared to those that are awake (Xie et al., 2013). Recently, it has also been demonstrated that the transgenic expression of human amyloid beta 42 (Aβ42) induces sleep fragmentation in Drosophila (Gerstner et al., 2017). Thus, there is evidence in different species that amyloidogenic alterations in proteostasis may be mediated by sleep and may conversely alter sleep patterns as well. A requirement for sleep to maintain protein homeostasis may underlie some of the correlation between poor sleep and disease risk and the fact that sleep disturbance is often a comorbidity of neurodegenerative disease pathogenesis as well as a feature of aging. As previously discussed, disrupting cellular proteostasis by inducing endoplasmic stress in Drosophila leads to impairments in normal sleep architecture (Brown et al., 2014), suggesting that the relationship between sleep and protein homeostasis is bidirectional. Similarly, in rodents, modulation of UPR pathways is sufficient to alter sleep patterns (Methippara et al., 2009). Pharmacological enhancement of the UPR PERK pathway signal via central infusion of the drug salubrinal, which prevents dephosphorylation of eukaryotic initiation factor 2α (eIF2α), increases total time spent in slow wave sleep in rats (Methippara et al., 2009). Evidence across species thus suggests that sleep regulates protein homeostasis in the brain and can be modulated by signals involved in maintaining protein homeostasis.
4.5 Oxidative Stress
Oxidative stress is another known cellular consequence of sleep deprivation (Reviewed in Villafuerte et al., 2015). Sleep loss leads to lower levels of antioxidant enzyme superoxide dismutase (SOD) in the brain of rats (Ramanathan et al., 2002), and oxidative stress markers are upregulated in the spleen and bone marrow lymphocytes of sleep-deprived mice (Lungato et al., 2016). Mackiewicz et al., 2007 conducted a microarray study of cortical and hypothalamic brain tissue in mice and found transcriptional upregulation of genes encoding antioxidant proteins during sleep. Koh et al., 2006 found that flies fed paraquat – an oxidative stress-inducing compound – on a regular basis significantly shortened their lifespans and led to reduced and fragmented sleep as well as reductions in circadian rhythmicity. These studies have not yet illustrated a causative link between sleep and oxidative stress repair but certainly emphasize a relationship between oxidative stress and disrupted sleep and point to the possibility that one of the restorative purposes of sleep is to counteract oxidative stress in the brain.
4.6 Immune Function
Finally, there is a great deal of evidence that suggests that sleep plays a critical role in modulating immune function (Reviewed in Imeri and Opp, 2009). A study in humans found that short sleep durations are associated with an increased susceptibility to illness and when human subjects are exposed to rhinovirus, an increased likelihood of developing a cold is associated with short sleep durations (Prather et al., 2015). Interestingly, genetic manipulations in Drosophila that increase sleep by inhibiting the cellular activity of wake-promoting cells also increase the chance for survival after bacterial infection (Kuo and Williams, 2014b). Furthermore, acute sleep deprivation has been shown to alter immune factor levels in multiple species (Williams et al., 2007; Wilder-Smith et al., 2013). These studies highlight the importance of sleep for maintaining resilience against illness and also demonstrate that the relationship between immune function and sleep is conserved.
5. Technical Considerations and Future Directions
Here, we have provided an overview of the findings from many studies that have spanned almost twenty years of scientific research. Across the board, there are some technical considerations that are important to recognize and address, since they have broad implications for how we interpret previously published data as well as how the field of Drosophila sleep research can continue to improve as we move forward. Some of the primary issues that should be considered when looking at all of the studies in fly sleep thus far are: limitations in single-fly analysis of sleep, sex differences in sleep, and the possible role of brain development-specific effects on sleep relating to the known sleep regulators.
At present, the most notable limitation of most systems that monitor and record Drosophila sleep is that they require individual flies to be separated into tubes for data collection. This limits the analysis of sleep to flies that are socially isolated throughout the experiment and introduces the confound of social isolation effects on sleep patterns. Like rodents (Kaushal et al., 2012) and humans (Kurina et al., 2011), Drosophila sleep is affected by social experience (Lone et al., 2016; Brown et al., 2017). Social isolation reduces Drosophila sleep amount (Brown et al., 2017), while social experience increases sleep (Lone et al., 2016). The changes in sleep following social isolation also lead to cellular stress in the brain (Brown et al., 2017). For this reason, understanding the implications of findings from these traditional sleep experiments requires some consideration for the effects of social experience and studying sleep in groups (Liu et al., 2015) will provide even more information about how sleep is regulated in the brain. Additionally, there are technical differences in the way sleep is recorded from isolated flies in locomotor tubes. In the traditional set-up, fly sleep is recorded using single infrared beam breaks, where flies are placed in locomotor tubes in which a single infrared beam separates the two ends of a transparent tube. When a fly crosses this beam, activity is recorded. The benefit of this system is that it has allowed for very high throughput analysis of sleep in flies over many different genotypes and under various conditions. However, in this system, flies moving on one of side a tube may not cross the beam in the center and thus “appear” to be sleeping. Recently, work examining changes in sleep in female flies after mating found that the post-mating increases in sleep were not detectable using a single-beam system (Garbe et al., 2016). This demonstrates the necessity for higher-resolution sleep recording techniques such as multibeam systems (Garbe et al., 2016) or video systems (Zimmerman et al., 2008b) in order to capture certain sleep changes and also emphasizes the point that the absence of measurable sleep changes in a single beam system does not exclude the possibility that a sleep phenotype still exists.
Another important point to consider as we survey all of the data that has been collected on fly sleep is the sexual dimorphism of sleep in Drosophila (Shaw et al., 2000; Koh et al., 2006). New data is demonstrating that some modulation of sleep behavior is indeed specific to either females (Garbe et al., 2016) or males (Beckwith et al., 2017; Lamaze et al., 2017). Many of the seminal studies on Drosophila sleep report findings within a specific sex. It is possible that some of what we know about sleep in the fly may not apply broadly to both sexes, depending on the type of regulation that we are investigating. These current studies examining sex-specific sleep behavior will provide insight into whether this might be the case and if so, what types of sleep regulators should be examined in the context of sex-specific sleep regulation.
Finally, we must also bear in mind the possible role that development plays in producing phenotypes from specific genetic mutations in the fly. Any mutation that is present during development may exert their effect on sleep by altering the development of interacting circuits in the brain. This is especially important for understanding sleep and wake regulation. Unlike cell-autonomous clock mechanisms, sleep and wake regulation is dependent on multiple interacting circuits. Furthermore, individual proteins or molecules may have differential expression or function during development that could affect its role on sleep in the brain at various stages of life. Many more recent studies have employed acute induction or repression of genes using conditional expression systems (Nicholson et al., 2008) in adult flies to circumvent these caveats. When considering developmental changes in brain development, we should also think about the time-frame during which behavioral data is collected. Some brain circuits do not fully mature until about a week after eclosion (Sinakevitch et al., 2010; Kayser et al., 2014). Despite this, many studies are conducted in flies that are a few days post-eclosion. This raises the possibility that the same flies studied at older ages might reveal slightly different phenotypes that could differentiate the developmental role of various genes from their role in the adult brain. Overall, keeping all of these technical considerations in mind will allow researchers to better discern the implications of their own results as well as design stronger experiments to minimize technical confounds in their data.
Moving forward, we believe that it will be important to directly investigate the functional relationships between the already identified players in Drosophila sleep. Thus far, most of the results from different published studies provide support for the findings of one another. For example, the study of sleep regulation by dopamine has uncovered a functional relationship between the wake-promoting PPL and PPM neurons (Liu et al., 2012b; Ueno et al., 2012) and the dorsal fan-shaped body (Donlea et al., 2011; Pimentel et al., 2016). Even this immensely well-characterized circuit leaves room for further investigation as it has not, for example, been demonstrated that activation of only the PPL and PPM neurons directly suppresses dFSB activity. The knowledge gleaned thus far on sleep in the fly should initiate new and more focused lines of inquiry. Furthering our understanding of the known sleep and wake regulatory pathways will allow us to determine what pathways are acting in parallel to regulate sleep and which instead converge to promote either sleep or wakefulness in the fly. This will ultimately provide greater insight into how the brain operates on a large scale to orchestrate complex outputs like sleep. While this is no simple task and will require careful dissection and methodical examination, this endeavor remains well-suited to be accomplished in the fly model in ways that are similar to the methods used by the studies that have been described here.
6. Conclusion
More than a century since Thomas Hunt Morgan became one of the first scientists to use the fruit fly model to study genetic inheritance (Morgan, 1910), Drosophila melanogaster remains at the forefront of scientific inquiry. In the early days of modern Drosophila research, the fruit fly was the ideal eukaryotic model for forward genetic screening; its use led to a much deeper understanding of the genetic basis of complex behavior. Today, modern advances in genetic techniques have broadened the scope of Drosophila sleep studies. In the time since the complete sequencing of the Drosophila genome (Adams et al., 2000), a wide array of tools has been developed that present novel and elegant ways to study and manipulate neural function in the fly. For example, transgenically expressed visual markers label cells and circuits in the brain (Clyne et al., 2003; Lee and Luo, 2001; Nicolaï et al., 2010), real-time neural activity can be captured in vivo with genetically encoded calcium indicators and two-photon microscopy (Bushey et al., 2015), both light and heat can be used to activate or suppress neuronal activity (Reviewed in Owald et al., 2015), and electrode recordings of neurons can be conducted in waking flies (Maimon et al., 2010). The level of genetic tractability of the fruit fly is the current forte of the Drosophila model. These tools allow for dynamic spatiotemporal control of the fly genome with a level of precision that has in some cases led to single-cell resolution mapping of complex neural processes and behaviors (Liu et al., 2012a; Freifeld et al., 2013; Shuai et al., 2015; Yapici et al., 2016).
The study of sleep in small model organisms has important implications for our understanding of sleep in higher order systems. It is now understood that many of the major neurochemical components and signaling pathways that regulate sleep in Drosophila are conserved in mammalian sleep. New research discoveries continue to confirm that many of the general biological principles dictating how sleep and wake are controlled in the fly also exist in more complex species. For example, the identification of an interconnection between sleep and circadian rhythms has been recapitulated in many mammalian models since the early Drosophila research identifying the circadian genes and proteins that maintain the daily patterns of sleep and wake. At the circuit level, fruit fly sleep also bears resemblance to sleep in higher order organisms. For example, sleep and wake states are partially determined by the activation or inhibition of specific brain circuits. Within these sleep circuits, there is data to suggest that mutually inhibitory connections may be fundamental for state switching; this is true in both the fly and mammals. Drosophila studies have also demonstrated that there are multiple neuronal groups involved in sleep regulation that are analogous to mammalian nuclei, and a complete understanding of the Drosophila sleep circuit may further elucidate mammalian sleep circuits. Interestingly, there is also an increasing focus on sleep regulation by extraneuronal cell types that has been moved forward by some work in the fly. For example, sleep signaling from the fat bodies in Drosophila provided early experimental evidence that peripheral signals can change sleep states and work in mammals confirm not only that peripheral tissues are affected by sleep, but also that peripheral signaling can regulate behavioral state. Thus, studies in the fruit fly continue to align with the growing body of work that is demonstrating that sleep is not only controlled by the brain but also by the interaction between the brain and the rest of the body. In all species studied to date including Drosophila, it is evident that sleep affects a very wide range of biological processes. This suggests that sleep likely serves multiple functions that are not only specific to the brain but have importance for peripheral organs as well. This notion is not particularly surprising given the pervasiveness of sleep across the animal kingdom and throughout evolution.
Though sleep shares an observable similarity to death, it is critical for most life on this planet. We are hard pressed to find animals that can survive without sleep and even acute sleep perturbations carry measurable consequences across many different types of species. Our growing understanding of sleep has provided vital insight into both the biological vulnerabilities of the brain as well as its remarkable capabilities for plasticity and repair. Perhaps the most exciting aspect of our ever expanding knowledge of sleep is the deepening complexity with which we can posit new scientific questions. For example, Drosophila research has taught us that how certain intracellular signals affect behavior is just as important as where in the brain they do so. Beyond identifying new regulators of sleep, understanding what cell types or brain regions are involved in regulation by known factors will be just as critical to constructing a complete understanding of sleep's neurobiological mechanisms. The impressive resolution with which we currently understand the Drosophila nervous system certainly suggests that we are well on our way not only to fully understanding how sleep affects the brain, but also to identifying the core functions of this mysterious behavior. Though tiny, the fruit fly has revealed much about the neurobiology of sleep and the brain. Based on the current state of Drosophila sleep research, it appears likely that the Drosophila animal model will continue to expand the boundaries of our understanding of sleep and neuroscience in the years to come.
Highlights.
Drosophila sleep is regulated by multiple neurotransmitters and intracellular signaling pathways in the brain.
Many of the identified regulators of Drosophila sleep exhibit conserved role in mammalian systems.
Evidence in Drosophila and across multiple species provide evidence that sleep functions to regulate synaptic plasticity, learning and memory, metabolism, development, proteostasis, oxidative stress, and immune function
Some technical considerations related to Drosophila sleep studies are: the limitations in single-fly analysis of sleep, sex differences in sleep, and the possible role of brain development-specific effects on sleep relating to the known sleep regulators.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Sidén-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, WoodageT, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC. The genome sequence of Drosophila melanogaster. Science. 2000;287(5461):2185–95. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Adermann K, Wattler F, Wattler S, Heine G, Meyer M, Forssmann WG, Nehls M. Structural and phylogenetic characterization of human SLURP-1, the first secreted mammalian member of the Ly-6/uPAR protein superfamily. Protein Sci. 1999;8(4):810–9. doi: 10.1110/ps.8.4.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agosto J, Choi JC, Parisky KM, Stillwell G, Rosbash M, Griffith LC. Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat Neurosci. 2008;11(3):354–9. doi: 10.1038/nn2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonso DJ, Liu D, Machado DR, Pan H, Jepson JE, Rogulja D, Koh K. TARANIS Functions with Cyclin A and Cdk1 in a Novel Arousal Center to Control Sleep in Drosophila. Curr Biol. 2015;25(13):1717–26. doi: 10.1016/j.cub.2015.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93(5):791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- Allada R, Siegel JM. Unearthing the phylogenetic roots of sleep. Curr Biol. 2008;18(15):R670–79. doi: 10.1016/j.cub.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R, Chung BY. Circadian organization of behavior and physiology and Physiology in Drosophila. Annu Rev Physiol. 2010;72:605–24. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allebrandt KV, Amin N, Müller-Myhsok B, Esko T, Teder-Laving M, Azevedo RV, Hayward C, van Mill J, Vogelzangs N, Green EW, Melville SA, Lichtner P, Wichmann HE, Oostra BA, Janssens AC, Campbell H, Wilson JF, Hicks AA, Pramstaller PP, Dogas Z, Rudan I, Merrow M, Penninx B, Kyriacou CP, Metspalu A, van Duijn CM, Meitinger T, Roenneberg T. A K(ATP) channel gene effect on sleep duration: from genome-wide association studies to function in Drosophila. Mol Psychiatry. 2013;18(1):122–32. doi: 10.1038/mp.2011.142. [DOI] [PubMed] [Google Scholar]
- Anafi RC, Pellegrino R, Shockley KR, Romer M, Tufik S, Pack AI. Sleep is not just for the brain: transcriptional responses to sleep in peripheral tissues. BMC Genomics. 2013;14:362. doi: 10.1186/1471-2164-14-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen ML, Kessler E, Murnane KS, McClung JC, Tufik S, Howell LL. Dopamine transporter-related effects of modafinil in rhesus monkeys. Psychopharmacology. 2010;210(3):439–48. doi: 10.1007/s00213-010-1839-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MP, Mochizuki T, Xie J, Fischler W, Manger JP, Talley EM, Scammell TE, Tonegawa S. Thalamic Cav3.1 T-type Ca2+ channel plays a crucial role in stabilizing sleep. Proc Natl Acad Sci U S A. 2005;102(5):1743–8. doi: 10.1073/pnas.0409644102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andretic R, van Swinderen B, Greenspan RJ. Dopaminergic modulation of arousal in Drosophila. Curr Biol. 2005;15(13):1165–75. doi: 10.1016/j.cub.2005.05.025. [DOI] [PubMed] [Google Scholar]
- Andretic R, Kim YC, Jones FS, Han KA, Greenspan RJ. Drosophila D1 dopamine receptor mediates caffeine-induced arousal. Proc Natl Acad Sci U S A. 2008;105(51):20392–7. doi: 10.1073/pnas.0806776105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aston-Jones G, Bloom FE. Activity of norepinephrine-containing locus coeruleus neurons in behaving rats anticipates fluctuations in the sleep-waking cycle. J Neurosci. 1981;1(8):876–86. doi: 10.1523/JNEUROSCI.01-08-00876.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barf RP, Meerlo P, Scheurink AJ. Chronic sleep disturbance impairs glucose homeostasis in rats. Int J Endocrinol. 2010;2010:819414. doi: 10.1155/2010/819414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenky G, Wesensten NJ, Thorne DR, Thomas ML, Sing HC, Redmond DP, Russo MB, Balkin TJ. Patterns of performance degradation and restoration during sleep restriction and subsequent recovery: a sleep dose-response study. J Sleep Res. 2003;12(1):1–12. doi: 10.1046/j.1365-2869.2003.00337.x. [DOI] [PubMed] [Google Scholar]
- Ben Achour S, Pascual O. Glia: The many ways to modulate synaptic plasticity. Neurochem Int. 2010;57(4):440–5. doi: 10.1016/j.neuint.2010.02.013. [DOI] [PubMed] [Google Scholar]
- Benedetto L, Chase MH, Torterolo P. GABAergic processes within the median preoptic nucleus promote NREM sleep. Behav Brain Res. 2012;232(1):60–5. doi: 10.1016/j.bbr.2012.03.033. [DOI] [PubMed] [Google Scholar]
- Benington JH, Heller HC. Restoration of brain energy metabolism as the function of sleep. Prog Neurobiol. 1995;45(4):347–60. doi: 10.1016/0301-0082(94)00057-o. [DOI] [PubMed] [Google Scholar]
- Berridge CW, Waterhouse BD. The locus coeruleus-noradrenergic system: modulation of behavioral state and state-dependent cognitive processes. Brain Res Brain Res Rev. 2003;42(1):33–84. doi: 10.1016/s0165-0173(03)00143-7. [DOI] [PubMed] [Google Scholar]
- Berridge CW, Schmeichel BE, España RA. Noradrenergic modulation of wakefulness/arousal. Sleep Med Rev. 2012;16(2):187–97. doi: 10.1016/j.smrv.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge CW, Foote SL. Effects of locus coeruleus activation on electroencephalographic activity in neocortex and hippocampus. J Neurosci. 1991;11(10):3135–45. doi: 10.1523/JNEUROSCI.11-10-03135.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry JA, Cervantes-Sandoval I, Nicholas EP, Davis RL. Dopamine is required for learning and forgetting in Drosophila. Neuron. 2012;74(3):530–42. doi: 10.1016/j.neuron.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry JA, Cervantes-Sandoval I, Chakraborty M, Davis RL. Sleep Facilitates Memory by Blocking Dopamine Neuron-Mediated Forgetting. Cell. 2015;161(7):1656–67. doi: 10.1016/j.cell.2015.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanik L, Mohrmann R, Ramaekers A, Bockaert J, Grau Y, Broadie K, Parmentier ML. The Drosophila metabotropic glutamate receptor DmGluRA regulates activity-dependent synaptic facilitation and fine synaptic morphology. J Neurosci. 2004;24:9105–16. doi: 10.1523/JNEUROSCI.2724-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borbély AA. Sleep in the rat during food deprivation and subsequent restitution of food. Brain Res. 1977;124(3):457–71. doi: 10.1016/0006-8993(77)90947-7. [DOI] [PubMed] [Google Scholar]
- Borbély AA. A two process model of sleep regulation. Hum Neurbiol. 1982;1(3):195–204. [PubMed] [Google Scholar]
- Brown MK, Chan MT, Zimmerman JE, Pack AI, Jackson NE, Naidoo N. Aging induced endoplasmic reticulum stress alters sleep and sleep homeostasis. Neurobiol Aging. 2014;35(6):1421–41. doi: 10.1016/j.neurobiolaging.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MK, Strus E, Naidoo N. Reduced Sleep During Social Isolation Leads to Cellular Stress and Induction of the Unfolded Protein Response. Sleep. 2017;40(7) doi: 10.1093/sleep/zsx095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner E, Bucher S, Crawford G, Mason WT, Salvaterra PM, Sattelle D. Choline acetyltransferase-like immunoreactivity in the brain of Drosophila melanogaster. Cell Tissue Res. 1986;246:57–62. [Google Scholar]
- Busch S, Selcho M, Ito K, Tanimoto H. A map of octopaminergic neurons in the Drosophila brain. J Comp Neurol. 2009;513(6):643–67. doi: 10.1002/cne.21966. [DOI] [PubMed] [Google Scholar]
- Bushey D, Huber R, Tononi G, Cirelli C. Drosophila Hyperkinetic mutants have reduced sleep and impaired memory. J Neurosci. 2007;27(20):5384–93. doi: 10.1523/JNEUROSCI.0108-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey D, Tononi G, Cirelli C. The Drosophila fragile X mental retardation gene regulates sleep need. J Neurosci. 2009;29(7):1948–61. doi: 10.1523/JNEUROSCI.4830-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey D, Tononi G, Cirelli C. Sleep and synaptic homeostasis: structural evidence in Drosophila. Science. 2011;332(6037):1576–81. doi: 10.1126/science.1202839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey D, Tononi G, Cirelli C. Sleep- and wake-dependent changes in neuronal activity and reactivity demonstrated in fly neurons using in vivo calcium imaging. Proc Natl Acad Sci U S A. 2015;112(15):4785–90. doi: 10.1073/pnas.1419603112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell SS, Tobler I. Animal sleep: a review of sleep duration across phylogeny. Neurosci Biobehav Rev. 1984;8(3):269–300. doi: 10.1016/0149-7634(84)90054-x. [DOI] [PubMed] [Google Scholar]
- Celesia GG, Jasper HH. Acetycholine release from cerebral cortex in relation to state of activation. Neurology. 1966;16(11):1053–63. doi: 10.1212/wnl.16.11.1053. [DOI] [PubMed] [Google Scholar]
- Chapman CD, Nilsson EK, Nilsson VC, Cedernaes J, Rångtell FH, Vogel H, Dickson SL, Broman JE, Hogenkamp PS, Schiöth HB, Benedict C. Acute sleep deprivation increases food purchasing in men. Obesity (Silver Spring) 2013;21(12):E555–60. doi: 10.1002/oby.20579. [DOI] [PubMed] [Google Scholar]
- Chen W, Shi W, Li L, Zheng Z, Li T, Bai W, Zhao Z. Regulation of sleep by the short neuropeptide F (sNPF) in Drosophila melanogaster. Insect Biochem Mol Biol. 2013;43(9):809–19. doi: 10.1016/j.ibmb.2013.06.003. [DOI] [PubMed] [Google Scholar]
- Chung BY, Kilman VL, Keath JR, Pitman JL, Allada R. The GABAA receptor RDL acts in peptidergic PDF neurons to promote sleep in Drosophila. Curr Biol. 2009;19(5):386–90. doi: 10.1016/j.cub.2009.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C, Tononi G. Differential expression of plasticity-related genes in waking and sleep and their regulation by the noradrenergic system. J Neurosci. 2000a;20(24):9187–94. doi: 10.1523/JNEUROSCI.20-24-09187.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C, Tononi G. Gene expression in the brain across the sleep-waking cycle. Brain Res. 2000b;885(2):303–21. doi: 10.1016/s0006-8993(00)03008-0. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Gutierrez CM, Tononi G. Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron. 2004;41(1):35–43. doi: 10.1016/s0896-6273(03)00814-6. [DOI] [PubMed] [Google Scholar]
- Cirelli C, Bushey D, Hill S, Huber R, Kreber R, Ganetzky B, Tononi G. Reduced sleep in Drosophila Shaker mutants. Nature. 2005;434(7037):1087–92. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Brotman JS, Sweeney ST, Davis G. Green fluorescent protein tagging Drosophila proteins at their native genomic loci with small P elements. Genetics. 2003;165(3):1433–41. doi: 10.1093/genetics/165.3.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmers WF, Bleakman D. Effects of neuropeptide Y on the electrical properties of neurons. Trends Neurosci. 1994;17(9):373–9. doi: 10.1016/0166-2236(94)90046-9. [DOI] [PubMed] [Google Scholar]
- Cong X, Wang H, Liu Z, He C, An C, Zhao Z. Regulation of sleep by insulin-like peptide system in Drosophila melanogaster. Sleep. 2015;38(7):1075–83. doi: 10.5665/sleep.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley L, Kelley R, Spradling A. Insertional mutagenesis of the Drosophila genome with single P elements. Science. 1988;239(4844):1121–8. doi: 10.1126/science.2830671. [DOI] [PubMed] [Google Scholar]
- Crocker A, Sehgal A. Octopamine regulates sleep in Drosophila through protein kinase A-dependent mechanisms. J Neurosci. 2008;28(38):9377–85. doi: 10.1523/JNEUROSCI.3072-08a.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker A, Shahidullah M, Levitan IB, Sehgal A. Identification of a neural circuit that underlies the effects of octopamine on sleep:wake behavior. Neuron. 2010;65(5):670–81. doi: 10.1016/j.neuron.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies A, Simmons DL, Hale G, Harrison RA, Tighe H, Lachmann PJ, Waldmann H. CD59, an LY-6-like protein expressed in human lymphoid cells, regulates the action of the complement membrane attack complex on homologous cells. J Exp Med. 1989;170(3):637–54. doi: 10.1084/jem.170.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103(2):239–52. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- de Velasco B, Erclik T, Shy D, Sclafani J, Lipshitz H, McInnes R, Hartenstein V. Specification and development of the pars intercerebralis and pars lateralis, neuroendocrine command centers in the Drosophila brain. Dev Biol. 2007;302(1):309–23. doi: 10.1016/j.ydbio.2006.09.035. [DOI] [PubMed] [Google Scholar]
- de Vivo L, Bellesi M, Marshall W, Bushong EA, Ellisman MH, Tononi G, Cirelli C. Ultrastructural evidence for synaptic scaling across the wake/sleep cycle. Science. 2017;355(6324):507–10. doi: 10.1126/science.aah5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Weille JR, Schweitz H, Maes P, Tartar A, Lazdunski M. Calciseptine, a peptide isolated from black mamba venom, is a specific blocker of the L-type calcium channel. Proc Natl Acad Sci U S A. 1991;88(6):2437–40. doi: 10.1073/pnas.88.6.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiAntonio A. Glutamate receptors at the Drosophila neuromuscular junction. Int Rev Neurobiol. 2006;75:165–79. doi: 10.1016/S0074-7742(06)75008-5. [DOI] [PubMed] [Google Scholar]
- Diekelmann S, Born J. The memory function of sleep. Nat Rev Neurosci. 2010;11(2):114–26. doi: 10.1038/nrn2762. [DOI] [PubMed] [Google Scholar]
- Diering GH, Nirujogi RS, Roth RH, Worley PF, Pandey A, Huganir RL. Homer1a drives homeostatic scaling-down of excitatory synapses during sleep. Science. 2017;355(6324):511–15. doi: 10.1126/science.aai8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinges DF, Pack F, Williams K, Gillen KA, Powell JW, Ott GE, Aptowicz C, Pack AI. Cumulative sleepiness, mood disturbance, and psychomotor vigilance performance decrements during a week of sleep restricted to 4-5 hours per night. Sleep. 1997;20(4):267–77. [PubMed] [Google Scholar]
- Dissel S, Angadi V, Kirszenblat L, Suzuki Y, Donlea J, Klose M, Koch Z, English D, Winsky-Sommerer R, van Swinderen B, Shaw PJ. Sleep restores behavioral plasticity to Drosophila mutants. Curr Biol. 2015;25(10):1270–81. doi: 10.1016/j.cub.2015.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezelova E, Nothacker HP, Civelli O, Bryant PJ, Zurovec M. A Drosophila adenosine receptor activates cAMP and calcium signaling. Insect Biochem Mol Biol. 2007;37(4):318–29. doi: 10.1016/j.ibmb.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Donlea JM, Thimgan MS, Suzuki Y, Gottschalk L, Shaw PJ. Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science. 2011;332(6037):1571–6. doi: 10.1126/science.1202249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas CL, Vyazovskiy V, Southard T, Chiu SY, Messing A, Tononi G, Cirelli C. Sleep in Kcna2 knockout mice. BMC Biol. 2007;5:42. doi: 10.1186/1741-7007-5-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enell L, Hamasaka Y, Kolodziejczyk A, Nässel DR. gamma-Aminobutyric acid (GABA) signaling components in Drosophila: immunocytochemical localization of GABAb receptors in relation to the GABAA receptor subunit RDL and a vesicular GABA transporter. J Comp Neurol. 2007;505(1):18–31. doi: 10.1002/cne.21472. [DOI] [PubMed] [Google Scholar]
- Erickson JD, Varoqui H. Molecular analysis of vesicular amino transporter function and targeting to secretory organelles. FASEB J. 2000;14(15):2450–8. doi: 10.1096/fj.00-0206rev. [DOI] [PubMed] [Google Scholar]
- Everson CA. Sustained sleep deprivation impairs host defense. Am J Physiol. 1993;265(5 Pt 2):R1148–54. doi: 10.1152/ajpregu.1993.265.5.R1148. [DOI] [PubMed] [Google Scholar]
- Farca Luna AJ, Perier M, Seugnet L. Amyloid Precursor Protein in Drosophila Glia Regulates Sleep and Genes Involved in Glutamate Recycling. J Neurosci. 2017;37(16):4289–300. doi: 10.1523/JNEUROSCI.2826-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooqui T. Review of octopamine in insect nervous systems. Open Access Insect Physiol. 2012;4:1–17. [Google Scholar]
- Franken P, Dijk DJ. Circadian clock genes and sleep homeostasis. Eur J Neurosci. 2009;29(9):1820–9. doi: 10.1111/j.1460-9568.2009.06723.x. [DOI] [PubMed] [Google Scholar]
- Figiel M, Engele J. Pituitary adenylate cyclase-activating polypeptide (PACAP), a neuron-derived peptide regulating glial glutamate transport and metabolism. J Neurosci. 2000;20(10):3596–605. doi: 10.1523/JNEUROSCI.20-10-03596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flourakis M, Kula-Eversole E, Hutchison AL, Han TH, Aranda K, Moose DL, White KP, Dinner AR, Lear BC, Ren D, Diekman CO, Raman IM, Allada R. A Conserved Bicycle Model for Circadian Clock Control of Membrane Excitability. Cell. 2015;162(4):836–48. doi: 10.1016/j.cell.2015.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltenyi K, Andretic R, Newport JW, Greenspan RJ. Neurohormonal and neuromodulatory control of sleep in Drosophila. Cold Spring Harb Symp Quant Biol. 2007;72:565–71. doi: 10.1101/sqb.2007.72.045. [DOI] [PubMed] [Google Scholar]
- Frank MG, Issa NP, Stryker MP. Sleep enhances plasticity in the developing visual cortex. Neuron. 2001;30(1):275–87. doi: 10.1016/s0896-6273(01)00279-3. [DOI] [PubMed] [Google Scholar]
- Fredholm BB, Bättig K, Holmén J, Nehlig A, Zvartau EE. Actions of caffeine in the brain with special reference to factors that contribute to its widespread use. Pharmacol Rev. 1999;51(1):83–133. [PubMed] [Google Scholar]
- Freifeld L, Clark DA, Schnitzer MJ, Horowitz MA, Clandinin TR. GABAergic lateral interactions tune the early stages of visual processing in Drosophila. Neuron. 2013;78(6):1075–89. doi: 10.1016/j.neuron.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friggi-Grelin F, Coulom H, Meller M, Gomez D, Hirsh J, Birman S. Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J Neurobiol. 2003;54(4):618–27. doi: 10.1002/neu.10185. [DOI] [PubMed] [Google Scholar]
- Fuller PM, Sherman D, Pedersen NP, Saper CB, Lu J. Reassessment of the structural basis of the ascending arousal system. Journal Comp Neurol. 2011;519:933–56. doi: 10.1002/cne.22559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallopin T, Fort P, Eggermann E, Cauli B, Luppi PH, Rossier J, Audinat E, Mühlethaler M, Serafin M. Identification of sleep-promoting neurons in vitro. Nature. 2000;404(6781):992–5. doi: 10.1038/35010109. [DOI] [PubMed] [Google Scholar]
- Gamaldo CE, Shaikh AK, McArthur JC. The sleep-immunity relationship. Neurol Clin. 2012;30(4):1313–43. doi: 10.1016/j.ncl.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Gangwisch JE, Malaspina D, Boden-Albala B, Heymsfield SB. Inadequate sleep as a risk factor for obesity: analyses of the NHANES I. Sleep. 2005;28(10):1289–96. doi: 10.1093/sleep/28.10.1289. [DOI] [PubMed] [Google Scholar]
- Gao J, Zhang JX, Xu TL. Modulation of serotonergic projection from dorsal raphe nucleus to basolateral amygdala on sleep-waking cycle of rats. Brain Res. 2002;945(1):60–70. doi: 10.1016/s0006-8993(02)02625-2. [DOI] [PubMed] [Google Scholar]
- Garbe DS, Vigderman AS, Moscato E, Dove AE, Vecsey CG, Kayser MS, Sehgal A. Changes in Female Drosophila Sleep following Mating Are Mediated by SPSN-SAG Neurons. J Biol Rhythms. 2016;31(6):551–67. doi: 10.1177/0748730416668048. [DOI] [PubMed] [Google Scholar]
- Gerald C, Walker MW, Criscione L, Gustafson EL, Batzl-Hartmann C, Smith KE, Vaysse P, Durkin MM, Laz TM, Linemeyer DL, Schaffhauser AO, Whitebread S, Hofbauer KG, Taber RI, Brancheck TA, Weinshank RL. A receptor subtype involved in neuropeptide-Y-induced food intake. Nature. 1996;382(6587):168–71. doi: 10.1038/382168a0. [DOI] [PubMed] [Google Scholar]
- Gerstner JR, Lenz O, Vanderheyden WM, Chan MT, Pfeiffenberger C, Pack AI. Amyloid-β induces sleep fragmentation that is rescued by fatty acid binding proteins in Drosophila. J Neurosci Res. 2017;95(8):1548–64. doi: 10.1002/jnr.23778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilestro GF, Tononi G, Cirelli C. Widespread changes in synaptic markers as a function of sleep and wakefulness in Drosophila. Science. 2009;324(5923):109–12. doi: 10.1126/science.1166673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong H, McGinty D, Guzman-Marin R, Chew KT, Stewart D, Szymusiak R. Activation of c-fos in GABAergic neurones in the preoptic area during sleep and in response to sleep deprivation. J Physiol. 2004;556(Pt 3):935–46. doi: 10.1113/jphysiol.2003.056622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves LA, Heller EA, Pack AI, Abel T. Sleep deprivation selectively impairs memory consolidation for contextual fear conditioning. Learn Mem. 2003;10(3):168–76. doi: 10.1101/lm.48803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Yi W, Zhou M, Guo A. Go signaling in mushroom bodies regulates sleep in Drosophila. Sleep. 2011;34(3):273–81. doi: 10.1093/sleep/34.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim F, Wang Y, Carreras A, Hirotsu C, Zhang J, Peris E, Gozal D. Chronic sleep fragmentation during the sleep period induces hypothalamic endoplasmic reticulum stress and PTP1b-mediated leptin resistance in male mice. Sleep. 2015;38(1):31–40. doi: 10.5665/sleep.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halassa MM, Florian C, Fellin T, Munoz JR, Lee SY, Abel T, Haydon PG, Frank MG. Astrocytic modulation of sleep homeostasis and cognitive consequences of sleep loss. Neuron. 2009;61(2):213–9. doi: 10.1016/j.neuron.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes PR, Christmann BL, Griffith LC. A single pair of neurons links sleep to memory consolidations in Drosophila melanogaster. Elife. 2015;4:e03868. doi: 10.7554/eLife.03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Yang Y, Zhang M, Price JL, Zhao Z. Regulation of sleep by neuropeptide Y-like system in Drosophila melanogaster. PLoS One. 2013;8(9):e74237. doi: 10.1371/journal.pone.0074237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, Pack AI. Rest in Drosophila is a sleep-like state. Neuron. 2000;25(1):129–38. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Williams JA, Panckeri K, Kirk D, Tello M, Yin JC, Sehgal A. A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat Neurosci. 2001;4(11):1108–15. doi: 10.1038/nn743. [DOI] [PubMed] [Google Scholar]
- Hirshkowitz M. Normal human sleep: an overview. Med Clin North Am. 2004;88(3):551–65. vi.. doi: 10.1016/j.mcna.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Holtmaat A, Svoboda K. Experience-dependent structural synaptic plasticity in the mammalian brain. Nat Rev Neurosci. 2009;10(9):647–58. doi: 10.1038/nrn2699. [DOI] [PubMed] [Google Scholar]
- Horn C, Offen N, Nystedt S, Häcker U, Wimmer EA. piggyBac-based insertional mutagenesis and enhancer detection as a tool for functional insect genomics. Genetics. 2003;163(2):647–61. doi: 10.1093/genetics/163.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huai Q, Mazar AP, Kuo A, Parry GC, Shaw DE, Callahan J, Li Y, Yuan C, Bian C, Chen L, Furie B, Furie BC, Cines DB, Huang M. Structure of human urokinase plasminogen activator in complex with its receptor. Science. 2006;311(5761):656–9. doi: 10.1126/science.1121143. [DOI] [PubMed] [Google Scholar]
- Iglowstein I, Jenni OG, Molinari L, Largo RH. Sleep duration from infancy to adolescence: reference values and generational trends. Pediatrics. 2003;111(2):302–7. doi: 10.1542/peds.111.2.302. [DOI] [PubMed] [Google Scholar]
- Imeri L, Opp MR. How (and why) the immune system makes us sleep. Nat Rev Neurosci. 2009;10(3):199–210. doi: 10.1038/nrn2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac SO, Berridge CW. Wake-promoting actions of dopamine D1 and D2 receptor stimulation. J Pharmacol Exp Ther. 2003;307(1):386–94. doi: 10.1124/jpet.103.053918. [DOI] [PubMed] [Google Scholar]
- Ishimoto H, Kitamoto T. The steroid molting hormone Ecdysone regulates sleep in adult Drosophila melanogaster. Genetics. 2010;185(1):269–81. doi: 10.1534/genetics.110.114587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James PT. Obesity: the worldwide epidemic. Clinics in dermatology. 2004;22(4):276–280. doi: 10.1016/j.clindermatol.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Jeong K, Lee S, Seo H, Oh Y, Jang D, Choe J, Kim D, Lee JH, 1, Jonesb WD. Ca-α1T, a fly T-type Ca2+ channel, negatively modulates sleep. Scientific Reports. 2015;5:17893. doi: 10.1038/srep17893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinka TR, Rasley BT, Drew KL. Inhibition of NMDA-type glutamate receptors induces arousal from torpor in hibernating arctic ground squirrels (Urocitellus parryii) J Neurochem. 2012;122(5):934–40. doi: 10.1111/j.1471-4159.2012.07832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006;441(7094):757–60. doi: 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- Jouvet-Mounier D, Astic L, Lacote D. Ontogenesis of the states of sleep in rat, cat, and guinea pig during the first postnatal month. Dev Psychobiol. 1970;2(4):216–39. doi: 10.1002/dev.420020407. [DOI] [PubMed] [Google Scholar]
- Ju YE, McLeland JS, Toedebusch CD, Xiong C, Fagan AM, Duntley SP, Morris JC, Holtzman DM. Sleep quality and preclinical Alzheimer Disease. JAMA Neurology. 2013;70(5):587–93. doi: 10.1001/jamaneurol.2013.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CM, Melanson EL, Frydendall EJ, Perreault L, Eckel RH, Wright KP. Energy expenditure during sleep, sleep deprivation and sleep following sleep deprivation in adult humans. J Physiol. 2011;589:235–44. doi: 10.1113/jphysiol.2010.197517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JE, Lim MM, Bateman RJ, Lee JJ, Smyth LP, Cirrito JR, Fujiki N, Nishino S, Holtzman DM. Amyloid-beta dynamics are regulated by orexin and the sleep-wake cycle. Science. 2009;326(5955):1005–7. doi: 10.1126/science.1180962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan K, Fridell YW. Functional implications of Drosophila insulin-like peptides in metabolism, aging, and dietary restriction. Front Physiol. 2013;4:288. doi: 10.3389/fphys.2013.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Ozawa F, Saitoh Y, Fukazawa Y, Sugiyama H, Inokuchi K. Novel members of the Vesl/Homer family of PDZ proteins that bind metabotropic glutamate receptors. J Biol Chem. 1998;273(37):23969–75. doi: 10.1074/jbc.273.37.23969. [DOI] [PubMed] [Google Scholar]
- Kaushal N, Nair D, Gozal D, Ramesh V. Socially isolated mice exhibit a blunted homeostatic sleep response to acute sleep deprivation compared to socially paired mice. Brain Res. 2012;1454:65–79. doi: 10.1016/j.brainres.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser MS, Yue Z, Sehgal A. A critical period of sleep for development of courtship circuitry and behavior in Drosophila. Science. 2014;344(6181):289–74. doi: 10.1126/science.1250553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene AC, Duboué ER, McDonald DM, Dus M, Suh GS, Waddell S, Blau J. Clock and cycle limit starvation-induced sleep loss in Drosophila. Curr Biol. 2010;20(13):1209–15. doi: 10.1016/j.cub.2010.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killgore WD, Balkin TJ, Wesensten NJ. Impaired decision making following 49 h of sleep deprivation. J Sleep Res. 2006;15(1):7–13. doi: 10.1111/j.1365-2869.2006.00487.x. [DOI] [PubMed] [Google Scholar]
- Kim YC, Lee HG, Han KA. D1 dopamine receptor dDA1 is required in the mushroom body neurons for aversive and appetitive learning in Drosophila. J Neurosci. 2007;27(29):7640–7. doi: 10.1523/JNEUROSCI.1167-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, Turek FW, Takahashi JS. Positional cloning of the mouse circadian clock gene. Cell. 1997;89(4):641–53. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell. 1998;94(1):97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- Koh K, Evans JM, Hendricks JC, Sehgal A. A Drosophila model for age-associated changes in sleep:wake cycles. Proc Natl Acad Sci U S A. 2006;103(37):13843–7. doi: 10.1073/pnas.0605903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K, Joiner WJ, Wu MN, Yue Z, Smith CJ, Sehgal A. Identification of SLEEPLESS, a sleep-promoting factor. Science. 2008;321(5887):372–6. doi: 10.1126/science.1155942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68(9):2112–6. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Yang FC, Snodgrass P, Li X, Scammel TE, Davis FC, Weitz CJ. Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling. Science. 2001;294(5551):2511–5. doi: 10.1126/science.1067716. [DOI] [PubMed] [Google Scholar]
- Kraus RL, Li Y, Gregan Y, Gotter AL, Uebele VN, Fox SV, Doran SM, Barrow JC, Yang ZQ, Reger TS, Koblan KS, Renger JJ. In vitro characterization of T-type calcium channel antagonist TTA-A2 and in vivo effects on arousal in mice. J Pharmacol Exp Ther. 2010;335(2):409–17. doi: 10.1124/jpet.110.171058. [DOI] [PubMed] [Google Scholar]
- Kume K, Kume S, Park SK, Hirsh J, Jackson FR. Dopamine Is a Regulator of Arousal in the Fruit Fly. J Neurosci. 2005;25(32):7377–84. doi: 10.1523/JNEUROSCI.2048-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo TH, Pike DH, Beizaeipour Z, Williams JA. Sleep triggered by an immune response in Drosophila is regulated by the circadian clock and requires the NFkappaB Relish. BMC Neurosci. 2010;11:17. doi: 10.1186/1471-2202-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo TH, Williams JA. Acute sleep deprivation enhances post-infection sleep and promotes survival during bacterial infection in Drosophila. Sleep. 2014a;37(5):859–69. doi: 10.5665/sleep.3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo TH, Williams JA. Increased Sleep Promotes Survival during a Bacterial Infection in Drosophila. Sleep. 2014b;37(6):1077–86. doi: 10.5665/sleep.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushikata T, Fang J, Chen Z, Wang Y, Krueger JM. Epidermal growth factor enhances spontaneous sleep in rabbits. Am J Physiol. 1998;275(2 Pt 2):R509–14. doi: 10.1152/ajpregu.1998.275.2.R509. [DOI] [PubMed] [Google Scholar]
- Lamaze A, Öztürk-Çolak A, Fischer R, Peschel N, Koh K, Jepson JE. Regulation of sleep plasticity by a thermo-sensitive circuit in Drosophila. Sci Rep. 2017;7:40304. doi: 10.1038/srep40304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;(5):251–4. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Lelkes Z, Alföldi P, Erdos A, Benedek G. Rolipram, an antidepressant that increases the availability of cAMP, transiently enhances wakefulness in rats. Pharmacol Biochem Behav. 1998;60(4):835–9. doi: 10.1016/s0091-3057(98)00038-0. [DOI] [PubMed] [Google Scholar]
- Li Q, Li Y, Wang X, Qi J, Jin X, Tong H, Zhou Z, Zhang ZC, Han J. Fbxl4 Serves as a Clock Output Molecule that Regulates Sleep through Promotion of Rhythmic Degradation of the GABAA Receptor. Curr Biol. 2017;27(23):3616–25.e5. doi: 10.1016/j.cub.2017.10.052. [DOI] [PubMed] [Google Scholar]
- Li X, Yu F, Guo A. Sleep deprivation specifically impairs short-term olfactory memory in Drosophila. Sleep. 2009;32(11):1417–24. doi: 10.1093/sleep/32.11.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhou Z, Zhang X, Tong H, Li P, Zhang ZC, Jia Z, Xie W, Han J. Drosophila neuroligin 4 regulates sleep through modulating GABA transmission. J Neurosci. 2013;33(39):15545–54. doi: 10.1523/JNEUROSCI.0819-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AS, Kowgier M, Yu L, Buchman AS, Bennett DA. Sleep Fragmentation and the Risk of Incident Alzheimer's Disease and Cognitive Decline in Older Persons. Sleep. 2013;36(7):1027–32. doi: 10.5665/sleep.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linford NJ, Ro J, Chung BY, Pletcher SD. Gustatory and metabolic perception of nutrient stress in Drosophila. Proc Natl Acad Sci U S A. 2015;112(8):2587–92. doi: 10.1073/pnas.1401501112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Plaçais PY, Yamagata N, Pfeiffer BD, Aso Y, Friedrich AB, Siwanowicz I, Rubin GM, Preat T, Tanimoto H. A subset of dopamine neurons signals reward for odour memory in Drosophila. Nature. 2012a;488(7412):512–6. doi: 10.1038/nature11304. [DOI] [PubMed] [Google Scholar]
- Liu C, Haynes PR, Donelson NC, Aharon S, Griffith LC. Sleep in Populations of Drosophila Melanogaster. eNeuro. 2015;2(4) doi: 10.1523/ENEURO.0071-15.2015. ENEURO.0071 –15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Liu S, Kodama L, Driscoll MR, Wu MN. Two dopaminergic neurons signal to the dorsal fan-shaped body to promote wakefulness in Drosophila. Curr Biol. 2012b;22(22):2114–23. doi: 10.1016/j.cub.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Lamaze A, Liu Q, Tabuchi M, Yang Y, Fowler M, Bharadwaj R, Zhang J, Bedont J, Blackshaw S, Lloyd TE, Montell C, Sehgal A, Koh K, Wu MN. WIDE AWAKE mediates the circadian timing of sleep onset. Neuron. 2014;82(1):151–66. doi: 10.1016/j.neuron.2014.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Liu Q, Tabuchi M, Wu MN. Sleep Drive Is Encoded by Neural Plastic Changes in a Dedicated Circuit. Cell. 2016;165(6):1347–60. doi: 10.1016/j.cell.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Guo F, Lu B, Guo A. Amnesiac regulates sleep onset and maintenance in Drosophila melanogaster. Biochem Biophys Res Commun. 2008;372(4):798–803. doi: 10.1016/j.bbrc.2008.05.119. [DOI] [PubMed] [Google Scholar]
- Liu ZW, Faraquna U, Cirelli C, Tononi G, Gao XB. Direct evidence for wake-related increases and sleep-related decreases in synaptic strength in rodent cortex. J Neurosci. 2010;30(25):8671–5. doi: 10.1523/JNEUROSCI.1409-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lone SR, Potdar S, Srivastava M, Sharma VK. Social Experience Is Sufficient to Modulate Sleep Need of Drosophila without Increasing Wakefulness. PLoS One. 2016;11(3):e0150596. doi: 10.1371/journal.pone.0150596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signaling in vertebrate neural development. Nat Rev Neurosci. 2006;7(2):93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Lundell MJ, Hirsh J. Temporal and spatial development of serotonin and dopamine neurons in the Drosophila CNS. Dev Biol. 1994;165(2):385–96. doi: 10.1006/dbio.1994.1261. [DOI] [PubMed] [Google Scholar]
- Lungato L, Nogueira-Pedro A, Carvalho Dias C, Paredes-Gamero EJ, Tufik S, D'Almeida V. Effects of Sleep Deprivation on Mice Bone Marrow and Spleen B Lymphopoiesis. J Cell Physiol. 2016;231(6):1313–20. doi: 10.1002/jcp.25231. [DOI] [PubMed] [Google Scholar]
- Lyamin OI, Mukhametov LM, Siegel JM. Sleep in the northern fur seal. Curr Opin Neurobiol. 2017;44:144–51. doi: 10.1016/j.conb.2017.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackiewicz M, Shockley KR, Romer MA, Galante RJ, Zimmerman JE, Naidoo N, Baldwin DA, Jensen ST, Churchill GA, Pack AI. Macromolecule biosynthesis: a key function of sleep. Physiol Genomics. 2007;31(3):441–57. doi: 10.1152/physiolgenomics.00275.2006. [DOI] [PubMed] [Google Scholar]
- Maimon G, Straw AD, Dickinson MH. Active flight increases the gain of visual motion processing in Drosophila. Nat Neurosci. 2010;13(3):393–9. doi: 10.1038/nn.2492. [DOI] [PubMed] [Google Scholar]
- Makos MA, Kim YC, Han KA, Heien ML, Ewing AG. In vivo electrochemical measurements of exogenously applied dopamine in Drosophila melanogaster. Anal Chem. 2009;81(5):1848–54. doi: 10.1021/ac802297b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Davis RL. Eight different types of dopaminergic neurons innervate the Drosophila mushroom body neuropil: anatomical and physiological heterogeneity. Front Neural Circuits. 2009;3:5. doi: 10.3389/neuro.04.005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwald RR, Melanson EL, Smith MR, Higgins J, Perreault L, Eckel RH, Wright KP., Jr Impact of insufficient sleep on total daily energy expenditure, food intake, and weight gain. Proc Natl Acad Sci U S A. 2013;110:5695–700. doi: 10.1073/pnas.1216951110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masek P, Reynolds LA, Bollinger WL, Moody C, Mehta A, Murakami K, Yoshizawa M, Gibbs AG, Keene AC. Altered regulation of sleep and feeding contributes to starvation resistance in Drosophila melanogaster. J Exp Biol. 2014;217(Pt 17):3122–32. doi: 10.1242/jeb.103309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy EV, Wu Y, Decarvalho T, Brandt C, Cao G, Nitabach MN. Synchronized bilateral synaptic inputs to Drosophila melanogaster neuropeptidergic rest/arousal neurons. J Neurosci. 2011;31(22):8181–93. doi: 10.1523/JNEUROSCI.2017-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302(5651):1765–8. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- Meerlo P, Sgoifo A, Suchecki D. Restricted and disrupted sleep: effects on autonomic function, neuroendocrine stress systems and stress responsivity. Sleep Med Rev. 2008;12(3):197–210. doi: 10.1016/j.smrv.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Mercaldo V, Descalzi G, Zhuo M. Fragile X mental retardation protein in learning-related synaptic plasticity. Mol Cells. 2009;28(6):501–7. doi: 10.1007/s10059-009-0193-x. [DOI] [PubMed] [Google Scholar]
- Mertens I, Meeusen T, Huybrechts R, De Loof A, Schoofs L. Characterization of the short neuropeptide F receptor from Drosophila melanogaster. Biochem Biophys Res Commun. 2002;297(5):1140–8. doi: 10.1016/s0006-291x(02)02351-3. [DOI] [PubMed] [Google Scholar]
- Methippara MM, Bashir T, Kumar S, Alam N, Szymusiak R, McGinty D. Salubrinal, an inhibitor of protein synthesis, promotes deep slow wave sleep. Am J Physiol Regul Integr Comp Physiol. 2009;296(1):R178–84. doi: 10.1152/ajpregu.90765.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezler M, Müller T, Raming K. Cloning and functional expression of GABAB receptors from Drosophila. Eur J Neurosci. 2001;13(3):477–86. doi: 10.1046/j.1460-9568.2001.01410.x. [DOI] [PubMed] [Google Scholar]
- Montagna P, Lugaresi E. Agrypniaexcitata: a generalized overactivity syndrome and a useful concept in the neurophysiopathology of sleep. Clin Neurophysiol. 2002;113(4):552–60. doi: 10.1016/s1388-2457(02)00022-6. [DOI] [PubMed] [Google Scholar]
- Morgan TH. Sex Limited Inheritance in Drosophila. Science. 1910;32(812):120–2. doi: 10.1126/science.32.812.120. [DOI] [PubMed] [Google Scholar]
- Murakami K, Yurgel ME, Stahl BA, Masek P, Mehta A, Heidker R, Bollinger W, Gingras RM, Kim YJ, Ja WW, Suter B, DiAngelo JR, Keene AC. translin Is Required for Metabolic Regulation of Sleep. Curr Biol. 2016;26(7):972–80. doi: 10.1016/j.cub.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo N, Giang W, Galante RJ, Pack AI. Sleep deprivation induces the unfolded protein response in mouse cerebral cortex. J Neurochem. 2005;92(5):1150–7. doi: 10.1111/j.1471-4159.2004.02952.x. [DOI] [PubMed] [Google Scholar]
- Naidoo N, Casiano V, Cater J, Zimmerman J, Pack AI. A role for the molecular chaperone protein BiP/GRP78 in Drosophila sleep homeostasis. Sleep. 2007;30(5):557–65. doi: 10.1093/sleep/30.5.557. [DOI] [PubMed] [Google Scholar]
- Naidoo N, Ferber M, Master M, Zhu Y, Pack AI. Aging impairs the unfolded protein response to sleep deprivation and leads to proapoptotic signaling. J Neurosci. 2008;28(26):6539–48. doi: 10.1523/JNEUROSCI.5685-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo N, Ferber M, Galante RJ, McShane B, Hu JH, Zimmerman J, Maislin G, Cater J, Wyner A, Worley P, Pack AI. Role of Homer proteins in the maintenance of sleep-wake states. PLoS One. 2012;7(4):e35174. doi: 10.1371/journal.pone.0035174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo N, Davis JG, Zhu J, Yabumoto M, Singletary K, Brown M, Galante R, Agarwal B, Baur JA. Aging and sleep deprivation induce the unfolded protein response in the pancreas: implications for metabolism. Aging Cell. 2014;13(1):131–41. doi: 10.1111/acel.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nall AH, Sehgal A. Small-molecule screen in adult Drosophila identifies VMAT as a regulator of sleep. J Neurosci. 2013;33(19):8534–40. doi: 10.1523/JNEUROSCI.0253-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nässel DR. Serotonin and serotonin-immunoreactive neurons in the nervous system of insects. Prog Neurobiol. 1988;30(1):1–85. doi: 10.1016/0301-0082(88)90002-0. [DOI] [PubMed] [Google Scholar]
- Nässel DR. Histamine in the brain of insects: a review. Microsc Res Tech. 1999;44(2–3):121–36. doi: 10.1002/(SICI)1097-0029(19990115/01)44:2/3<121::AID-JEMT6>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Nath RD, Bedbrook CN, Abrams MJ, Basinger T, Bois JS, Prober DA, Sternberg PW, Gradinaru V, Goentoro L. The Jellyfish Cassiopea Exhibits a Sleep-like State. Curr Biol. 2017;27(19):2984–90.e3. doi: 10.1016/j.cub.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson L, Singh GK, Osterwalder T, Roman GW, Davis RL, Keshishian H. Spatial and temporal control of gene expression in Drosophila using the inducible GeneSwitch GAL4 system. I. Screen for larval nervous system drivers. Genetics. 2008;178(1):215–34. doi: 10.1534/genetics.107.081968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaï LJ, Ramaekers A, Raemaekers T, Drozdzecki A, Mauss AS, Yan J, Landgraf M, Annaert W, Hassan BA. Genetically encoded dendritic marker sheds light on neuronal connectivity in Drosophila. Proc Natl Acad Sci U S A. 2010;107(47):20553–8. doi: 10.1073/pnas.1010198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikonova EV, Naidoo N, Zhang L, Romer M, Cater JR, Scharf MT, Galante RJ, Pack AI. Changes in components of energy regulation in mouse cortex with increases in wakefulness. Sleep. 2010;33(7):889–900. doi: 10.1093/sleep/33.7.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino S, Mao J, Sampathkumaran R, Shelton J. Increased dopaminergic transmission mediates the wake-promoting effects of CNS stimulants. Sleep Res Online. 1998;1(1):49–61. [PubMed] [Google Scholar]
- Nitabach MN, Taghert PH. Organization of the Drosophila circadian control circuit. Curr Biol. 2008;18(2):R84–93. doi: 10.1016/j.cub.2007.11.061. [DOI] [PubMed] [Google Scholar]
- Obál F, Jr, Fang J, Taishi P, Kacsóh B, Gardi J, Krueger JM. Deficiency of growth hormone-releasing hormone signaling is associated with sleep alterations in the dwarf rat. J Neurosci. 2001;21(8):2912–8. doi: 10.1523/JNEUROSCI.21-08-02912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden CL, Carroll MD, Kit BK, Flegal KM. NCHS Data Brief. 82. 2012. Prevalence of obesity in the United States, 2009-1010; pp. 1–8. [PubMed] [Google Scholar]
- Oh Y, Jang D, Sonn JY, Choe J. Histamine-HisCl1 receptor axis regulates wake-promoting signals in Drosophila melanogaster. PLoS One. 2013;8(7):e68269. doi: 10.1371/journal.pone.0068269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada R, Awasaki T, Ito K. Gamma-aminobutyric acid (GABA)-mediated neural connections in the Drosophila antennal lobe. J Comp Neurol. 2009;514(1):74–91. doi: 10.1002/cne.21971. [DOI] [PubMed] [Google Scholar]
- Owald D, Lin S, Waddell S. Light, heat, action: neural control of fruit fly behaviour. Philos Trans R Soc Lond B Biol Sci. 2015;370(1677):20140211. doi: 10.1098/rstb.2014.0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palchykova S, Winsky-Sommerer R, Meerlo P, Dürr R, Tobler I. Sleep deprivation impairs object recognition in mice. Neurobiol Learn Mem. 2006;85(3):263–71. doi: 10.1016/j.nlm.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Palma JA, Urrestarazu E, Iriarte J. Sleep loss as a risk factor for neurologic disorders: a review. Sleep Med. 2013;14(3):229–36. doi: 10.1016/j.sleep.2012.11.019. [DOI] [PubMed] [Google Scholar]
- Park S, Sonn JY, Oh Y, Lim C, Choe J. SIFamide and SIFamide receptor defines a novel neuropeptide signaling to promote sleep in Drosophila. Mol Cells. 2014;37(4):295–301. doi: 10.14348/molcells.2014.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmentier R, Ohtsu H, Djebbara-Hannas Z, Valatx J, Watanabe T, Lin JS. Anatomical, physiological, and pharmacological characteristics of histidine decarboxylase knock-out mice: evidence for the role of brain histamine in behavioral and sleep-wake control. J Neurosci. 2002;22(17):7695–711. doi: 10.1523/JNEUROSCI.22-17-07695.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron IJ, Pack AI, Veasey S. Diet/Energy Balance Affect Sleep and Wakefulness Independent of Body Weight. Sleep. 2015;38(12):1893–903. doi: 10.5665/sleep.5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel N, Helfrich-Förster C. Setting the clock--by nature: circadian rhythm in the fruitfly Drosophila melanogaster. FEBS Lett. 2011;585(10):1435–42. doi: 10.1016/j.febslet.2011.02.028. [DOI] [PubMed] [Google Scholar]
- Peter-Derex L, Yammine P, Bastuji H, Croisile B. Sleep and Alzheimer's disease. Sleep Med Rev. 2015;19:29–38. doi: 10.1016/j.smrv.2014.03.007. [DOI] [PubMed] [Google Scholar]
- Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature. 2006;441(7094):753–6. doi: 10.1038/nature04739. [DOI] [PubMed] [Google Scholar]
- Pollack I, Hofbauer A. Histamine-like immunoreactivity in the visual system and brain of Drosophila melanogaster. Cell Tissue Res. 1991;266(2):391–8. doi: 10.1007/BF00318195. [DOI] [PubMed] [Google Scholar]
- Porzgen P, Park SK, Hirsh J, Sonders MS, Amara SG. The antidepressant-sensitive dopamine transporter in Drosophila melanogaster: a primordial carrier for catecholamines. Mol Pharmacol. 2001;59:83–95. doi: 10.1124/mol.59.1.83. [DOI] [PubMed] [Google Scholar]
- Prather AA, Janicki-Deverts D, Hall MH, Cohen S. Behaviorally Assessed Sleep and Susceptibility to the Common Cold. Sleep. 2015;38(9):1353–9. doi: 10.5665/sleep.4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. Double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94(1):83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- Qian Y, Cao Y, Deng B, Yang G, Li J, Xu R, Zhang D, Huang J, Rao Y. Sleep homeostasis regulated by 5HT2b receptor in a small subset of neurons in the dorsal fan-shaped body of drosophila. Elife. 2017;6 doi: 10.7554/eLife.26519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu WM, Xu XH, Yan MM, Wang YQ, Urade Y, Huang ZL. Essential role of dopamine D2 receptor in the maintenance of wakefulness, but not in homeostatic regulation of sleep, in mice. J Neurosci. 2010;30(12):4382–9. doi: 10.1523/JNEUROSCI.4936-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizen DM, Zimmerman JE, Maycock MH, Ta UD, You YJ, Sundaram MV, Pack AI. Lethargus is a Caenorhabditis elegans sleep-like state. Nature. 2008;451(7178):569–72. doi: 10.1038/nature06535. [DOI] [PubMed] [Google Scholar]
- Ramanathan L, Gulvani S, Nienhuis R, Siegel JM. Sleep deprivation decreases superoxide dismutase activity in rat hippocampus and brainstem. Neuroreport. 2002;13(11):1387–90. doi: 10.1097/00001756-200208070-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechtschaffen A, Gilliland MA, Bergmann BM, Winter JB. Physiological correlates of prolonged sleep deprivation in rats. Science. 1983;221(4606):182–4. doi: 10.1126/science.6857280. [DOI] [PubMed] [Google Scholar]
- Reddy P, Zehring WA, Wheeler DA, Pirrotta V, Hadfield C, Hall JC, Rosbash M. Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell. 1984;38(3):701–10. doi: 10.1016/0092-8674(84)90265-4. [DOI] [PubMed] [Google Scholar]
- Reinoso-Suárez F, de Andrés I, Rodrigo-Angulo ML, Garzón M. Brain structures and mechanisms involved in the generation of REM sleep. Sleep Med Rev. 2001;5(1):63–77. doi: 10.1053/smrv.2000.0136. [DOI] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99(7):791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Riemensperger R, Isabel G, Coulom H, Neuser K, Seugnet L, Kume K, Iché-Torres M, Cassar M, Strauss R, Preat T, Hirsh J, Birman S. Behavioral consequences of dopamine deficiency in the Drosophila central nervous system. Proc Natl Acad Sci U S A. 2011;108(2):834–9. doi: 10.1073/pnas.1010930108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rival T, Soustelle L, Cattaert D, Strambi C, Iche M, Birman S. Physiological requirement for the glutamate transporter dEAAT1 at the adult Drosophila neuromuscular junction. J Neurobiol. 2006;66:1061–74. doi: 10.1002/neu.20270. [DOI] [PubMed] [Google Scholar]
- Robinson JE, Paluch J, Dickman DK, Joiner WJ. ADAR-mediated RNA editing suppresses sleep by acting as a brake on glutamatergic synaptic plasticity. Nat Commun. 2016;7:10512. doi: 10.1038/ncomms10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogulja D, Young MW. Control of sleep by cyclin A and its regulator. Science. 2012;335(6076):1617–21. doi: 10.1126/science.1212476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth TC, 2nd, Lesku JA, Amlaner CJ, Lima SL. A phylogenetic analysis of the correlates of sleep in birds. J Sleep Res. 2006;15(4):395–402. doi: 10.1111/j.1365-2869.2006.00559.x. [DOI] [PubMed] [Google Scholar]
- Rusnak F, Mertz P. Calcineurin: form and function. Physiol Rev. 2000;80(4):1483–521. doi: 10.1152/physrev.2000.80.4.1483. [DOI] [PubMed] [Google Scholar]
- Salvaterra PM, Kitamoto T. Drosophila cholinergic neurons and processes visualized with Gal4/UAS-GFP. Brain Res Gene Expr Patterns. 2001;1(1):73–82. doi: 10.1016/s1567-133x(01)00011-4. [DOI] [PubMed] [Google Scholar]
- Saper CB, Scammell TE, Lu J. Hypothalamic regulation of sleep and circadian rhythms. Nature. 2005;437(7063):1257–63. doi: 10.1038/nature04284. [DOI] [PubMed] [Google Scholar]
- Saper CB, Fuller PM, Pedersen NP, Lu J, Scammell TE. Sleep state switching. Neuron. 2010;68(6):1023–42. doi: 10.1016/j.neuron.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudou F, Boschert U, Amlaiky N, Plassat JL, Hen R. A family of Drosophila serotonin receptors with distinct intracellular signaling properties and expression patterns. EMBO J. 1992;11(1):7–17. doi: 10.1002/j.1460-2075.1992.tb05021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf MT, Naidoo N, Zimmerman JE, Pack AI. The energy hypothesis of sleep revisited. Prog Neurobiol. 2008;86(3):264–80. doi: 10.1016/j.pneurobio.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt LI, Sims RE, Dale N, Haydon PG. Wakefulness affects synaptic and network activity by increasing extracellular astrocyte-derived adenosine. J Neurosci. 2012;32(13):4417–25. doi: 10.1523/JNEUROSCI.5689-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A, Price JL, Man B, Young MW. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science. 1994;263(5153):1603–06. doi: 10.1126/science.8128246. [DOI] [PubMed] [Google Scholar]
- Seugnet L, Suzuki Y, Donlea JM, Gottschalk L, Shaw PJ. Sleep deprivation during early-adult development results in long-lasting learning deficits in adult Drosophila. Sleep. 2011a;34(2):137–46. doi: 10.1093/sleep/34.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seugnet L, Suzuki Y, Merlin G, Gottschalk L, Duntley SP, Shaw PJ. Notch signaling modulates sleep homeostasis and learning after sleep deprivation in Drosophila. Curr Biol. 2011b;21(10):835–40. doi: 10.1016/j.cub.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seugnet L, Dissel S, Thimgan M, Cao L, Shaw PJ. Identification of Genes that Maintain Behavioral and Structural Plasticity during Sleep Loss. Front Neural Circuits. 2017;11:79. doi: 10.3389/fncir.2017.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Donelson NC, Vecsey CG, Guo F, Rosbash M, Griffith LC. Short neuropeptide F is a sleep-promoting inhibitory modulator. Neuron. 2013;80(1):171–83. doi: 10.1016/j.neuron.2013.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287(5459):1834–7. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Tononi G, Greenspan RJ, Robinson DF. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature. 2002;417(6886):287–91. doi: 10.1038/417287a. [DOI] [PubMed] [Google Scholar]
- Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou YT, Sharma VK, Holmes TC. Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr Biol. 2008;18:1537–45. doi: 10.1016/j.cub.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla C, Basheer R. Metabolic signals in sleep regulation: recent insights. Nat Sci Sleep. 2016;8:9–20. doi: 10.2147/NSS.S62365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DM, Dehoff M, Luo X, Kang SH, Tu J, Nayak SK, Ross EM, Worley PF, Muallem S. Homer 2 tunes G protein-coupled receptors stimulus intensity by regulating RGS proteins and PLCbeta GAP activities. J Cell Biol. 2003;162(2):293–303. doi: 10.1083/jcb.200210109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi Y, Mizutani A, Mikoshiba K, Furuichi T. Coincidence in dendritic clustering and synaptic targeting of homer proteins and NMDA receptor complex proteins NR2B and PSD95 during development of cultured hippocampal neurons. Mol Cell Neurosci. 2003;22(2):188–201. doi: 10.1016/s1044-7431(03)00037-x. [DOI] [PubMed] [Google Scholar]
- Shuai Y, Hirokawa A, Ai Y, Zhang M, Li W, Zhong Y. Dissecting neural pathways for forgetting in Drosophila olfactory aversive memory. Proc Natl Acad Sci U S A. 2015;112(48):E6663–72. doi: 10.1073/pnas.1512792112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinakevitch I, Strausfeld NJ. Comparison of octopamine-like immunoreactivity in the brains of the fruit fly and blow fly. J Comp Neurol. 2006;494(3):460–75. doi: 10.1002/cne.20799. [DOI] [PubMed] [Google Scholar]
- Sinakevitch I, Grau Y, Strausfeld NJ, Birman S. Dynamics of glutamatergic signaling in the mushroom body of young adult Drosophila. Neural Dev. 2010;5:10. doi: 10.1186/1749-8104-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinakevitch-Pean I, Geffard M, Plotnikova SI. Localization of glutamate in the nervous system of Drosophila melanogaster: immunocytochemical study. J Evol Biochem Physiol. 2001;37:83–88. [PubMed] [Google Scholar]
- Sitaraman D, Zar M, Laferriere H, Chen YC, Sable-Smith A, Kitamoto T, Rottinghaus GE, Zars T. Serotonin in necessary for place memory in Drosophila. Proc Natl Acad Sci U S A. 2008;105(14):5579–84. doi: 10.1073/pnas.0710168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaraman D, Aso Y, Rubin GM, Nitabach MN. Control of Sleep by Dopaminergic Inputs to the Drosophila Mushroom Body. Front Neural Circuits. 2015;9:73. doi: 10.3389/fncir.2015.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocumb ME, Regalado JM, Yoshizawa M, Neely GG, Masek P, Gibbs AG, Keene AC. Enhanced Sleep Is an Evolutionarily Adaptive Response to Starvation Stress in Drosophila. PLoS One. 2015;10(7):e0131275. doi: 10.1371/journal.pone.0131275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel K, Tasali E, Leproult R, Van Cauter E. Effects of poor and short sleep on glucose metabolism and obesity risk. Nat Rev Endocrinol. 2009;5(5):253–61. doi: 10.1038/nrendo.2009.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Onge MP, McReynolds A, Trivedi ZB, Roberts AL, Sy M, Hirsch J. Sleep restriction leads to increased activation of brain regions sensitive to food stimuli. Am J Clin Nutr. 2012;95(4):818–24. doi: 10.3945/ajcn.111.027383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson R, Chu KM, Lee J. Prolonged deprivation of sleep-like rest raises metabolic rate in the Pacific beetle cockroach, Diploptera punctata (Eschscholtz) J ExpBiol. 2007;210(Pt 14):2540–7. doi: 10.1242/jeb.005322. [DOI] [PubMed] [Google Scholar]
- Szerb JC. Cortical acetylcholine release and electroencephalographic arousal. J Physiol. 1967;192(2):329–43. doi: 10.1113/jphysiol.1967.sp008303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahama K, Tomita J, Ueno T, Yamazaki M, Kume S, Kume K. Pan-neuronal knockdown of the c-Jun N-terminal Kinase (JNK) results in a reduction in sleep and longevity in Drosophila. Biochem Biophys Res Commun. 2012;417(2):807–11. doi: 10.1016/j.bbrc.2011.12.040. [DOI] [PubMed] [Google Scholar]
- Telzer EH, Goldenberg D, Fuligni AJ, Lieberman MD, Gálvan A. Sleep variability in adolescence is associated with altered brain development. Dev Cogn Neurosci. 2015;14:16–22. doi: 10.1016/j.dcn.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terao A, Stininger TL, Hyder K, Apte-Deshpande A, Ding J, Rishipathak D, Davis RW, Heller HC, Kilduff TS. Differential increase in the expression of heat shock protein family members during sleep deprivation and during sleep. Neuroscience. 2003;116(1):187–200. doi: 10.1016/s0306-4522(02)00695-4. [DOI] [PubMed] [Google Scholar]
- Terao A, Wisor JP, Peyron C, Apte-Deshpande A, Wurts SW, Edgar DM, Kilduff TS. Gene expression in the rat brain during sleep deprivation and recovery sleep: an Affymetrix GeneChip study. Neuroscience. 2006;137(2):593–605. doi: 10.1016/j.neuroscience.2005.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier CR, Broadie K. Drosophila fragile X mental retardation protein developmentally regulates activity-dependent axon pruning. Development. 2008;135(8):1547–57. doi: 10.1242/dev.015867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakkar MM. Histamine in the regulation of wakefulness. Sleep Med Rev. 2011;15:65–74. doi: 10.1016/j.smrv.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimgan MS, Seugnet L, Turk J, Shaw PJ. Identification of genes associated with resilience/vulnerability to sleep deprivation and starvation in Drosophila. Sleep. 2015;38(5):801–14. doi: 10.5665/sleep.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobler I. Behavioral sleep in the Asian elephant in captivity. Sleep. 1992;15(1):1–12. [PubMed] [Google Scholar]
- Tobler I, Schwierin B. Behavioural sleep in the giraffe (Giraffa camelopardalis) in a zoological garden. J Sleep Res. 1996;5(1):21–32. doi: 10.1046/j.1365-2869.1996.00010.x. [DOI] [PubMed] [Google Scholar]
- Tobler I, Neuner-Jehle M. 24-h variation of vigilance in the cockroach Blaberus giganteus. J Sleep Res. 1992;1(4):231–39. doi: 10.1111/j.1365-2869.1992.tb00044.x. [DOI] [PubMed] [Google Scholar]
- Tomita J, Mitsuyoshi M, Ueno T, Aso Y, Tanimoto H, Nakai Y, Aigaki T, Kume S, Kume K. Pan-neuronal knockdown of calcineurin reduces sleep in the fruit fly, Drosophila melanogaster. J Neurosci. 2011;31(37):13137–46. doi: 10.1523/JNEUROSCI.5860-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita J, Ueno T, Mitsuyoshi M, Kume S, Kume K. The NMDA Receptor Promotes Sleep in the Fruit Fly, Drosophila melanogaster. PLoS One. 2015;10(5):e0128101. doi: 10.1371/journal.pone.0128101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tononi G, Cirelli C. Sleep function and synaptic homeostasis. Sleep Med Rev. 2006;10(1):49–62. doi: 10.1016/j.smrv.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Touchette É, Petit D, Séguin JR, Boivin M, Tremblay RE, Montplaisir JY. Associations Between Sleep Duration Patterns and Behavioral/Cognitive Functioning at School Entry. Sleep. 2007;30(9):1213–19. doi: 10.1093/sleep/30.9.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetlin V. Snake venom alpha-neurotoxins and other ‘three-finger’ proteins. Eur J Biochem. 1999;264(2):281–6. doi: 10.1046/j.1432-1327.1999.00623.x. [DOI] [PubMed] [Google Scholar]
- Tsuji H, Okamoto K, Matsuzaka Y, Iizuka H, Tamiya G, Inoko H. SLURP-2, a novel member of the human Ly-6 superfamily that is up-regulated in psoriasis vulgaris. Genomics. 2003;81(1):26–33. doi: 10.1016/s0888-7543(02)00025-3. [DOI] [PubMed] [Google Scholar]
- Tu JC, Xiao B, Yuan JP, Lanahan AA, Leoffert K, Li M, Linden DJ, Worley PF. Homer binds a novel proline-rich motif and links group 1 metabotropic glutamate receptors with IP3 receptors. Neuron. 1998;21(4):717–26. doi: 10.1016/s0896-6273(00)80589-9. [DOI] [PubMed] [Google Scholar]
- Tucker AM, Whitney P, Belenky G, Hinson JM, van Dongen HA. Effects of Sleep Deprivation on Dissociated Components of Executive Functioning. Sleep. 2010;33(1):47–57. doi: 10.1093/sleep/33.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchino K, Imamura M, Shimizu K, Kanda T, Tamura T. Germ line transformation of the silkworm, Bombyx mori, using the transposable element Minos. Mol Genet Genomics. 2007;277(3):213–20. doi: 10.1007/s00438-006-0176-y. [DOI] [PubMed] [Google Scholar]
- Ueno T, Tomita J, Tanimoto H, Endo K, Ito K, Kume S, Kume K. Identification of a dopamine pathway that regulates sleep and arousal in Drosophila. Nat Neurosci. 2012;15(11):1215–23. doi: 10.1038/nn.3238. [DOI] [PubMed] [Google Scholar]
- Ursin R. Serotonin and sleep. Sleep Med Rev. 2002;6(1):55–69. doi: 10.1053/smrv.2001.0174. [DOI] [PubMed] [Google Scholar]
- Vallés AM, White K. Serotonin-containing neurons in Drosophila melanogaster: development and distribution. J Comp Neurol. 1988;268(3):414–28. doi: 10.1002/cne.902680310. [DOI] [PubMed] [Google Scholar]
- van Alphen B, Yap MH, Kirszenblat L, Kotter B, van Swinderen B. A dynamic deep sleep stage in Drosophila. J Neurosci. 2013;33(16):6917–27. doi: 10.1523/JNEUROSCI.0061-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buskirk C, Sternberg PW. Epidermal growth factor signaling induces behavioral quiescence in Caenorhabditis elegans. Nat Neurosci. 2007;10(10):1300–7. doi: 10.1038/nn1981. [DOI] [PubMed] [Google Scholar]
- Van Dongen HP, Maislin G, Mullington JM, Dinges DF. The cumulative cost of additional wakefulness: dose-response effects on neurobehavioral functions and sleep physiology from chronic sleep restriction and total sleep deprivation. Sleep. 2003;26(2):117–26. doi: 10.1093/sleep/26.2.117. [DOI] [PubMed] [Google Scholar]
- van Leeuwen WM, Lehto M, Karisola P, Lindholm H, Luukonen R, Sallinen M, Härma M, Porkka-Heiskanen T, Alenius H. Sleep restriction increases the risk of developing cardiovascular diseases by augmenting proinflammatory responses through IL-17 and CRP. PloS One. 2009;4(2):e4589. doi: 10.1371/journal.pone.0004589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez J, Baghdoyan HA. Basal forebrain acetylcholine release during REM sleep is significantly greater than during waking. Am J Physiol Regul Integr Comp Physiol. 2001;280(2):R598–601. doi: 10.1152/ajpregu.2001.280.2.R598. [DOI] [PubMed] [Google Scholar]
- Villafuerte G, Miguel-Puga A, Rodríguez EM, Machado S, Manjarrez E, Arias-Carrión O. Sleep Deprivation and Oxidative Stress in Animal Models: A Systematic Review. Oxid Med Cell Longev. 2015;2015:234952. doi: 10.1155/2015/234952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, Fowler JS, Logan J, Alexoff D, Zhu W, Telang F, Wang GJ, Jayne M, Hooker JM, Wong C, Hubbard B, Carter P, Warner D, King P, Shea C, Xu Y, Muench L, Apelskog-Torres K. Effects of modafinil on dopamine and dopamine transporters in the male human brain: clinical implications. JAMA. 2009;301(11):1148–54. doi: 10.1001/jama.2009.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MP, Stickgold R. Sleep, memory, and plasticity. Annu Rev Psychol. 2006;57:139–66. doi: 10.1146/annurev.psych.56.091103.070307. [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334(6059):1081–6. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wang Q, Wang M, Whim MD. Neuropeptide y gates a stress-induced, long-lasting plasticity in the sympathetic nervous system. J Neurosci. 2013;33(31):12705–17. doi: 10.1523/JNEUROSCI.3132-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Carreras A, Lee S, Hakim F, Zhang SX, Nair D, Ye H, Gozal D. Chronic sleep fragmentation promotes obesity in young adult mice. Obesity (Silver Spring) 2014;22:758–62. doi: 10.1002/oby.20616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton KA, Johansen KM, Xu T, Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985;43:567–81. doi: 10.1016/0092-8674(85)90229-6. [DOI] [PubMed] [Google Scholar]
- Wiater MF, Mukherjee S, Li AJ, Dinh TT, Rooney EM, Simasko SM, Ritter S. Circadian integration of sleep-wake and feeding requires NPY receptor-expressing neurons in the mediobasal hypothalamus. Am J Physiol Regul Integr Comp Physiol. 2011;301(5):R1569–83. doi: 10.1152/ajpregu.00168.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder-Smith A, Mustafa FB, Earnest A, Gen L, Macary PA. Impact of partial sleep deprivation on immune markers. Sleep Med. 2013;14(10):1031–4. doi: 10.1016/j.sleep.2013.07.001. [DOI] [PubMed] [Google Scholar]
- Wilhelm OG, Wilhelm S, Escott GM, Lutz V, Magdolen V, Schmitt M, Rifkin DB, Wilson EL, Graeff H, Brunner G. Cellular glycosylphosphatidylinositol-specific phospholipase D regulates urokinase receptor shedding and cell surface expression. J Cell Physiol. 1999;180(2):225–35. doi: 10.1002/(SICI)1097-4652(199908)180:2<225::AID-JCP10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Williams JA, Sathyanarayanan S, Hendricks JC, Sehgal A. Interaction between sleep and the immune response in Drosophila: a role for the NFkappaB relish. Sleep. 2007;30(4):389–400. doi: 10.1093/sleep/30.4.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S, Argyropoulos S. Antidepressants and sleep: a qualitative review of the literature. Drugs. 2005;65(7):927–47. doi: 10.2165/00003495-200565070-00003. [DOI] [PubMed] [Google Scholar]
- Winder DG, Mansuy IM, Osman M, Moallem TM, Kandel ER. Genetic and pharmacological evidence for a novel, intermediate phase of long-term potentiation suppressed by calcineurin. Cell. 1998;92(1):25–37. doi: 10.1016/s0092-8674(00)80896-x. [DOI] [PubMed] [Google Scholar]
- Witz P, Amlaiky N, Plassat JL, Maroteaux L, Borrelli E, Hen R. Cloning and characterization of a Drosophila serotonin receptor that activates adenylate cyclase. Proc Natl Acad Sci U S A. 1990;87(22):8940–4. doi: 10.1073/pnas.87.22.8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MN, Ho K, Crocker A, Yue Z, Koh K, Sehgal A. The effects of caffeine on sleep in Drosophila require PKA activity, but not the adenosine receptor. J Neurosci. 2009;29(35):11029–37. doi: 10.1523/JNEUROSCI.1653-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MN, Joiner WJ, Dean T, Yue Z, Smith CJ, Chen D, Hoshi T, Sehgal A, Koh K. SLEEPLESS, a Ly-6/neurotoxin family member, regulates the levels, localization and activity of Shaker. Nat Neurosci. 2010;13(1):69–75. doi: 10.1038/nn.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Robinson JE, Joiner WJ. SLEEPLESS is a bifunctional regulator of excitability and cholinergic synaptic transmission. Curr Biol. 2014;24(6):621–9. doi: 10.1016/j.cub.2014.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt RJ, Zarcone V, Engelman K, Dement WC, Snyder F, Sjoerdsma A. Effects of 5-hydroxytryptophan on the sleep of normal human subjects. Electroencephalogr Clin Neurophysiol. 1971;30(6):505–9. doi: 10.1016/0013-4694(71)90147-7. [DOI] [PubMed] [Google Scholar]
- Xiao B, Tu JC, Petralia RS, Yuan JP, Doan A, Breder CD, Ruggiero A, Lanahan AA, Wenthold RJ, Worley PF. Homer regulates the association of group 1 metabotropic glutamate receptors with multivalent complexes of homer-related, synaptic proteins. Neuron. 1998;4:707–16. doi: 10.1016/s0896-6273(00)80588-7. [DOI] [PubMed] [Google Scholar]
- Xie L, Kang H, Xu Q, Chen MJ, Liao Y, Thiyagarajan M, O'Donnell J, Christensen DJ, Nicholson C, Iliff JJ, Takano T, Deane R, Nedergaard M. Sleep drives metabolite clearance from the adult brain. Science. 2013;342(6156):373–7. doi: 10.1126/science.1241224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong M, Li J, Wang D, Dephin E, Ye JH. Intra-ventrolateral preoptic nucleus injection of γ-aminobutyric acid induces sedation in rats. Int J Physiol Pathophysiol Pharmacol. 2012;4(2):94–8. [PMC free article] [PubMed] [Google Scholar]
- Yang G, Gan WB. Sleep contributes to dendritic spine formation and elimination in the developing mouse somatosensory cortex. Dev Neurobiol. 2012;72(11):1391–98. doi: 10.1002/dneu.20996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Lai CS, Cichon J, Ma L, Li W, Gan WB. Sleep promotes branch-specific formation of dendritic spines after learning. Science. 2014;344(6188):1173–8. doi: 10.1126/science.1249098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yapici N, Cohn R, Schusterreiter C, Ruta V, Vosshall LB. A Taste Circuit that Regulates Ingestion by Integrating Food and Hunger Signals. Cell. 2016;165(3):715–29. doi: 10.1016/j.cell.2016.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuyama K, Salvaterra PM. Localization of choline acetyltransferase-expressing neurons in Drosophila nervous system. Microsc Res Tech. 1999;45(2):65–79. doi: 10.1002/(SICI)1097-0029(19990415)45:2<65::AID-JEMT2>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Yi W, Zhang Y, Tian Y, Guo J, Li Y, Guo A. A subset of cholinergic mushroom body neurons requires Go signaling to regulate sleep in Drosophila. Sleep. 2013;36(12):1809–21. doi: 10.5665/sleep.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokogawa T, Marin W, Faraco J, Pérezon G, Appelbaum L, Zhang J, Rosa F, Mourrain P, Mignot E. Characterization of sleep in zebrafish and insomnia in hypocretin receptor mutants. PLoS Biol. 2007;5(10):e277. doi: 10.1371/journal.pbio.0050277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Joiner WJ, Sehgal A. A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr Biol. 2006;16(11):1051–62. doi: 10.1016/j.cub.2006.04.032. [DOI] [PubMed] [Google Scholar]
- Zager A, Andersen ML, Ruiz FS, Antunes IB, Tufik S. Effects of acute and chronic sleep loss on immune modulation of rats. Am J Physiol Regul Integr Comp Physiol. 2007;293(1):R504–9. doi: 10.1152/ajpregu.00105.2007. [DOI] [PubMed] [Google Scholar]
- Zimmerman JE, Rizzo W, Shockley KR, Raizen DM, Naidoo N, Mackiewicz M, Churchill GA, Pack AI. Multiple mechanisms limit the duration of wakefulness in Drosophila brain. Physiol Genomics. 2006;27(3):337–50. doi: 10.1152/physiolgenomics.00030.2006. [DOI] [PubMed] [Google Scholar]
- Zimmerman JE, Naidoo N, Raizen DM, Pack AI. Conservation of sleep: insights from non-mammalian model systems. Trends Neurosci. 2008a;31(7):371–6. doi: 10.1016/j.tins.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman JE, Raizen DM, Maycock MH, Maislin G, Pack AI. A video method to study Drosophila sleep. Sleep. 2008b;31(11):1587–98. doi: 10.1093/sleep/31.11.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman JE, Chan MT, Lenz OT, Keenan BT, Maislin G, Pack AI. Glutamate is a wake active neurotransmitter in Drosophila melanogaster. Sleep. 2016 Oct 28; doi: 10.1093/sleep/zsw046. [DOI] [PMC free article] [PubMed] [Google Scholar]


