Abstract
We demonstrate that lipidomics coupled with molecular dynamics reveal unique phospholipase A2 specificity toward membrane phospholipid substrates. We discovered unexpected headgroup and acyl-chain specificity for three major human phospholipases A2. The differences between each enzyme’s specificity, coupled with molecular dynamics-based structural and binding studies, revealed unique binding sites and interfacial surface binding moieties for each enzyme that explain the observed specificity at a hitherto inaccessible structural level. Surprisingly, we discovered that a unique hydrophobic binding site for the cleaved fatty acid dominates each enzyme’s specificity rather than its catalytic residues and polar headgroup binding site. Molecular dynamics simulations revealed the optimal phospholipid binding mode leading to a detailed understanding of the preference of cytosolic phospholipase A2 for cleavage of proinflammatory arachidonic acid, calcium-independent phospholipase A2, which is involved in membrane remodeling for cleavage of linoleic acid and for antibacterial secreted phospholipase A2 favoring linoleic acid, saturated fatty acids, and phosphatidylglycerol.
1. Introduction
Phospholipase A2 (PLA2) constitutes a structurally and functionally diverse superfamily of enzymes, and distinct types can exhibit unique degradative, biosynthetic, and/or signaling functions and are implicated in a wide variety of diseases. During the last two decades, considerable effort has been devoted to cloning and purifying different types of PLA2s and defining their implications in signaling pathways as well as in various diseases.1,2 However, questions related to the specificity of these enzymes toward the huge range of naturally occurring phospholipid substrates have not been thoroughly addressed. Using a novel lipidomic-based assay described herein, we revealed significant specificity differences toward natural and synthetic phospholipids not previously feasible using traditional assays. We compared three major human PLA2s including the Group IVA cytosolic (cPLA2), Group VIA calcium-independent (iPLA2), and Group V secreted (sPLA2). The structural details and dynamics of each enzyme’s association with the membrane or mixed micelle interface, and the binding of specific phospholipids were elucidated using molecular dynamics (MD) guided by hydrogen/deuterium (H/D) exchange.3 This work constitutes the first detailed study elucidating the specificity of PLA2s toward membrane phospholipids and correlating it with the structural interactions of specific phospholipids bound to each enzyme’s active site, extending earlier studies on substrate and inhibitor binding.4−11
The PLA2 superfamily constitutes a diverse set of enzymes that have a unique structure and specific cellular and tissue localization as well as biological function.1 Each of these enzymes evolved to hydrolyze specific membrane phospholipids based on its cellular/tissue/subcellular localization and distinct biological function. Most PLA2s are water-soluble enzymes, accessing their water-insoluble substrates by first associating with the lipid–water interface and then extracting a single phospholipid molecule into the active site. Structure and dynamics of PLA2 enzymes are significant factors in defining their association with cellular membranes and their specificity toward membrane phospholipids. Consequently, each PLA2 has a unique structure that contains two very important regions: the interfacial surface through which it associates and binds to the membrane and the active site where catalysis occurs.4 Assaying the activity of PLA2 enzymes has posed significant challenges because their natural phospholipid substrates aggregate in aqueous solution to form micelles, vesicles, or liposomes.12,13 To overcome these challenges, we have employed lipidomics to develop a novel mass spectrometric-based high-throughput assay toward both natural and synthetic membrane phospholipids in mixed micelles with a nonionic surfactant. This assay was used to determine the substrate specificity of three human PLA2s toward the major phospholipid molecular species.
2. Methods
A detailed description of the experimental and computational methods used in this study is provided in the Supporting Information. Briefly, a lipidomics-based LC/MS assay was used to define the specificity of cPLA2, iPLA2, and sPLA2 toward a variety of phospholipids. Molecular dynamics-based binding computations were employed to determine the structural features of each enzyme that contribute to its specificity.
3. Results and Discussion
3.1. Lipidomics High-Throughput PLA2 Assay
A lipidomics-based HPLC/MS assay was developed using a HILIC column and multiple reaction monitoring (MRM) for targeted quantification of the assay components including the surfactant (octaethylene glycol monododecyl ether, C12E8), a free fatty acid (arachidonic acid, AA), a phospholipid (1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphocholine, PAPC), a lysophospholipid (1-palmitoyl-sn-glycero-3-phosphocholine, 16:0 LPC), and an internal standard (1-heptadecanoyl-sn-glycero-3-phosphocholine, 17:0 LPC). Details are provided in the Supporting Information. Figure S1A shows the peaks of all five analytes. The products of PLA2 activity are a free fatty acid and a lysophospholipid. The lysophospholipid was employed to measure PLA2 activity in positive ion mode because it allowed monitoring of a variety of lysophospholipids including 16:0 and 18:0 LPA, 16:0 and 18:0 LPC, 16:0 and 18:0 LPE, 16:0 and 18:0 LPG, and 16:0 and 18:0 LPS with high sensitivity (Figure S1B). An internal standard (17:0 LPC) was used for normalizing variations related to sample handling, ionization efficiency, and signal intensity fluctuations. Standard curves were generated using the ratio of each analyte peak area to the internal standard peak area as a function of the analyte concentration (Figure S2).
For the substrate, 100 μM phospholipid was mixed with surfactant (C12E8) in buffer. The optimum concentration of surfactant was determined by testing the activity of each enzyme as a function of surfactant concentration (Figure S3). cPLA2 and iPLA2 demonstrate optimum activity at 400 μM C12E8, whereas sPLA2 has 60% of its optimum activity at the same concentration. A surfactant concentration of 400 μM was chosen to have a uniform micelle composition for all three enzymes. Additives required for each enzyme were included (3 μM PIP2 and 90 μM calcium in 100 mM HEPES pH 7.5 buffer for cPLA2, 2 mM ATP in 100 mM HEPES pH 7.5 buffer for iPLA2, and 5 mM calcium in 50 mM Tris-HCl pH 8 buffer for sPLA2). For each enzyme, product formation was linear with the amount of enzyme (Figure S4A–C) and the reaction time (Figure S4D–F). A protein concentration in the middle of the linear range and an assay time of 30 min at 40 °C was employed with a total LC/MS analysis time of 1.6 min per sample.
3.2. Headgroup and Acyl-Chain Specificity
PLA2 enzymatic activity toward a variety of phospholipid species was determined with the goal of defining headgroup and acyl-chain specificity for all three human enzymes. Headgroup specificity was defined by keeping constant the sn-1 and sn-2 fatty acyl-chains while varying the headgroup of the phospholipid species. Positional (sn-1 or sn-2) acyl-chain specificity was defined by keeping constant the headgroup and sn-1 acyl-chain position of the phospholipid species while varying the sn-2 fatty acyl-chain.
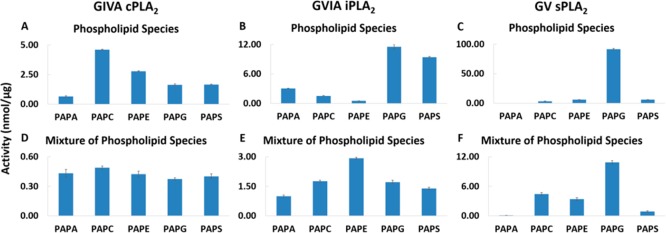
Five species of 1-palmitoyl-2-arachidonoyl-sn-glycero-3-X were employed where X was phosphatidic acid (PA), phosphocholine (PC), phosphoethanolamine (PE), phosphoglycerol (PG), or phosphoserine (PS). The sn-1 acyl chain was kept constant with the saturated palmitic acid because it is the most abundant de novo biosynthetic fatty acid in human plasma and tissue.14,15 Arachidonic acid was chosen for the sn-2 position because it has been successfully used in the past for traditional radioactive assays based on the importance of PLA2 in releasing arachidonic acid for eicosanoid production.2,13 The enzymatic activity of each enzyme toward 100 μM of each of the five phospholipid species is shown in Figure 1A–C. Based on these results, cPLA2 shows a preference for zwitterionic phospholipids such as PAPC and PAPE rather than negatively charged PAPA, PAPG, and PAPS. iPLA2 has a preference for negatively charged phospholipids and especially for PAPG and PAPS. sPLA2 is the only enzyme that exhibits remarkable specificity toward PAPG, indicating a distinct preference for that headgroup. However, to differentiate true catalytic preferences from interfacial surface association effects, the enzymatic activity of each enzyme was also determined in an equimolar mixture of all five phospholipid species (Figure 1D–F). Total lipid concentration was kept at 100 μM by employing 20 μM of each phospholipid species. cPLA2 and iPLA2 exhibit similar activity toward all five headgroups with a mild preference (factor of 2) of iPLA2 for PAPE. Interestingly, sPLA2 still shows specificity (factor of 3) toward PAPG over PAPC and PAPE and less activity toward PAPA and PAPS, indicating that this enzyme favors that headgroup in its active site.
Figure 1.
Enzymatic activity toward 100 μM of PAPA, PAPC, PAPE, PAPG, and PAPS for (A) cPLA2, (B) iPLA2, and (C) sPLA2 and toward 100 μM of a mixture of PAPA, PAPC, PAPE, PAPG, and PAPS at 20 μM each for (D) cPLA2, (E) iPLA2, and (F) sPLA2.
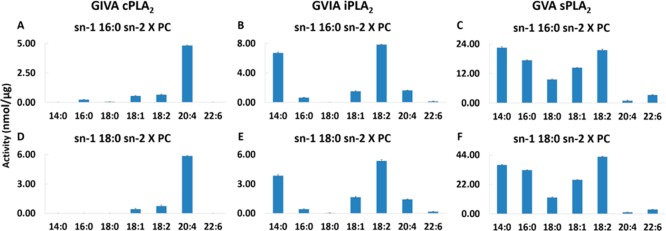
Acyl-chain specificity for each enzyme was determined by keeping the phospholipid headgroup constant as phosphocholine because it is the major headgroup in membrane phospholipids.14,16 Palmitic and stearic acid were utilized at the sn-1 position because they are synthesized de novo and constitute the major fatty acids at the sn-1 position of membrane phospholipids.14,15 For the sn-2 position, a variety of important fatty acids were compared including myristic acid (MA, 14:0), palmitic acid (PA, 16:0), stearic acid (SA, 18:0), oleic acid (OA, 18:1), linoleic acid (LA, 18:2), arachidonic acid (AA, 22:4), and docosahexaenoic acid (DHA, 22:6). cPLA2 exhibits a distinct specificity toward AA at the sn-2 position with significantly less activity toward OA and LA (Figure 2A, D). iPLA2 shows specificity toward LA and MA with slight preference for LA (Figure 2B, E). sPLA2 exhibits significant activity toward MA, PA, SA, OA, and LA with a preference for MA and LA (Figure 2C, F). Although iPLA2 and sPLA2 show measurable activity toward AA, it is not the optimal fatty acid at the sn-2 position for these two enzymes. It is also remarkable that cPLA2 does not show activity toward DHA, whereas iPLA2 and sPLA2 exhibit measurable activity with sPLA2, the most active one. Previous studies also suggested that sPLA2 shows significant activity toward DHA, whereas cPLA2 prefers AA.17,18 Even though DHA is the most abundant and significant fatty acid in mammalian brain, little is known about the regulation of its metabolism compared to AA.19 It was reported that the reduced metabolism of DHA in iPLA2 deficient mice increased their vulnerability to neuroinflammation because DHA is a precursor of anti-inflammatory neuroprotectins and resolvins.20,21
Figure 2.
Enzymatic activity toward 100 μM of PXPC for (A) cPLA2, (B) iPLA2, and (C) sPLA2 and toward 100 μM of SXPC for (D) cPLA2, (E) iPLA2, and (F) sPLA2.
For all three enzymes, no significant differences in specificity toward the sn-2 position were observed between palmitic acid (Figure 2A–C) and stearic acid (Figure 2D–F) at the sn-1 position. cPLA2 (Figure 2A, D) and sPLA2 (Figure 2C, F) activities are slightly better toward phospholipids containing steric acid (18:0) at the sn-1 position, whereas iPLA2 (Figure 2B, E) activity is somewhat better toward phospholipids containing palmitic acid (16:0) at the sn-1 position. Previously published studies suggested that, although both cPLA2 and iPLA2 contribute to LPC accumulation during stimulation of macrophages, 18:0 LPC appears to be produced primarily by cPLA2 whereas 16:0 LPC appears to be produced primarily by iPLA2.22,23
3.3. PLA2 Activity on Natural Phospholipids
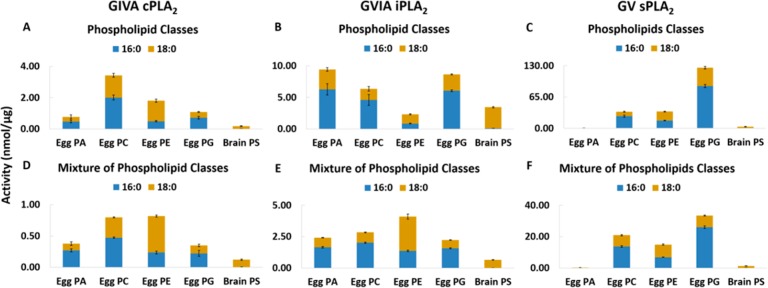
The activity of each enzyme was also measured toward 100 μM of phospholipid extracts from chicken egg or porcine brain (Figure 3A–C). Fatty acid distribution for these products is reported in Table S1 indicating that a variety of fatty acids can be esterified at the sn-1 or sn-2 position. These extracts are complex because both headgroups and fatty acyl-chains vary, and PLA2 activity is dually dependent. According to Figure 3, cPLA2 shows preference for zwitterionic phospholipids and especially for PC. iPLA2 exhibits higher activity toward negatively charged phospholipids including PA and PG. sPLA2 displays higher activity toward PG. However, in an equimolar mixture of all five extracts (Figure 3D–F), cPLA2 and iPLA2 have similar activities toward all five groups with slight variations, whereas sPLA2 has higher activity toward PG. Thus, this lipidomics-based assay can effectively be used to assay natural membrane phospholipids, and the activity will reflect the heterogeneity of the fatty acid substituents at each position and the variety of molecular species present.
Figure 3.
Enzymatic activity toward 100 μM of egg PA, egg PC, egg PE, egg PG, and brain PS for (A) cPLA2, (B) iPLA2, and (C) sPLA2 and toward 100 μM of a mixture of egg PA, egg PC, egg PE, egg PG, and brain PS at 20 μM each for (D) cPLA2, (E) iPLA2, and (F) sPLA2.
3.4. Headgroup and Acyl-Chain Binding Sites
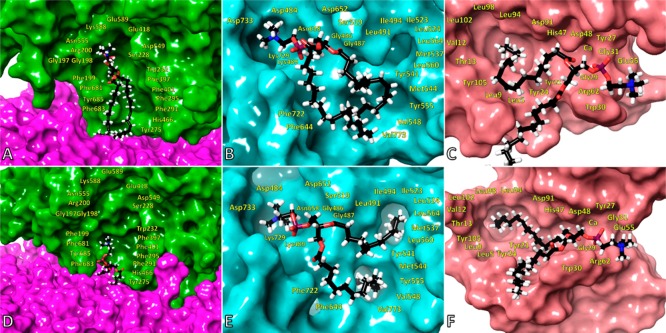
Atomistic molecular dynamics at a microsecond time scale revealed the structural characteristics contributing to the specificity of each enzyme toward each phospholipid. Simulations were carried out on four complexes of each enzyme (cPLA2, iPLA2, and sPLA2) with their optimal phospholipid substrates in the presence of a membrane patch. The sn-1 position of each phospholipid was chosen to contain an esterified palmitic acid (16:0) because the fatty acid at this position does not significantly affect the specificity of these enzymes. Arachidonic (20:4), linoleic (18:2), and myristic (14:0) acid were chosen to be esterified at the sn-2 position because cPLA2 exhibits optimum activity toward the first while iPLA2 and sPLA2 showed optimum activity toward the second and third ones. Four major headgroups were selected for the sn-3 position including phosphoethanolamine (PE), phosphoserine (PS), phosphoglycerol (PG), and phosphocholine (PC).
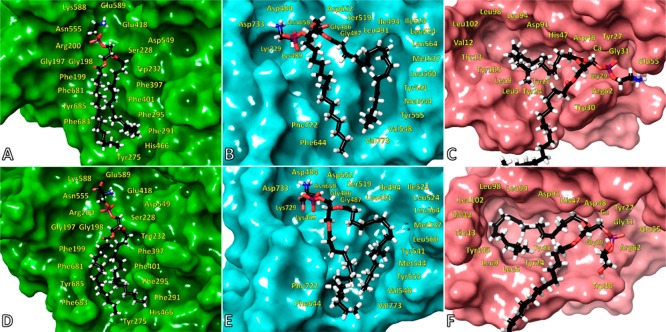
A phospholipid headgroup consists of a phosphate group that is connected to an alcohol like ethanolamine, serine, glycerol, inositol, or choline through a phosphoester bond. All three enzymes were evolved to have a positively charged residue, an arginine, or a lysine that stabilizes the phosphate group in the active site through hydrogen bonds (H-bonds) as well as electrostatic interactions (Figure 4 and Figure S5). The phosphate group in the active site of sPLA2 interacts with the calcium ion as well. The simulations showed that the binding site of the alcohol at the phospholipid headgroup in cPLA2 and iPLA2 contains residues such as aspartic or glutamic acids as well as lysines that interact through H-bonding or electrostatic interactions with PE, PS, and PC (Figure S5A, B, D, and E, Movies 1–8). At the same binding site, sPLA2 has a glutamic acid and an arginine that interact with PE (Figure S5C, Movie 9), whereas PG showed a different interaction pattern that involved an arginine and a tryptophan (Figure S5F, Movie 10). It is worth mentioning that sPLA2 is the only enzyme that showed significant preference toward PG (Figure 1C and F), and the simulations also showed a distinctive interaction pattern in comparison with PE and PC (Movies 9–12).
Figure 4.
Binding of PAPE in the active site of (A) cPLA2 (Movie 1), (B) iPLA2 (Movie 5), and (C) sPLA2 (Movie 9). Binding of PAPS in the active site of (D) cPLA2 (Movie 2), (E) iPLA2 (Movie 6), and (F) binding of PAPG in the active site of sPLA2 (Movie 10).
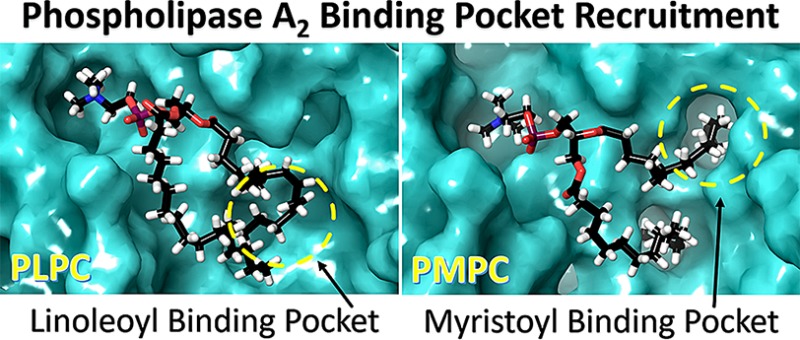
A phospholipid contains fatty acid tails at the sn-1 and sn-2 position. The sn-1 position usually contains saturated fatty acids like palmitic (16:0) or stearic (18:0), which do not significantly affect the activity of PLA2 enzymes (Figure 2). Because PLA2s catalyze the hydrolysis of the sn-2 ester bond, we hypothesized that their hydrophobic binding sites were structurally evolved to distinguish between different fatty acid tails at the sn-2 position. The hydrophobic binding site of cPLA2 is rich with aromatic residues that interact with the double bonds of arachidonic acid (20:4) through π stacking (Figure 4A and D, Movies 1 and 2) because the enzyme is specific for this fatty acid (Figure 2A and D). PLPC and PMPC are not good substrates for cPLA2, and thus, they diffuse in the membrane during the simulations (Figure 5A and D, Movies 3 and 4). As indicated in Figure 2B and E, iPLA2 exhibited optimum activity for phospholipids containing linoleic (18:2) and myristic (14:0) acid at the sn-2 position. The MD simulations showed that iPLA2 has two hydrophobic binding pockets: the first one contains aromatic and aliphatic residues and accommodates the linoleic acid tail (Figure 5B, Movie 7) and the second one contains exclusively aliphatic residues and accommodates the myristic acid tail (Figure 5E, Movie 8). The arachidonic acid tail in PAPE and PAPS (Figure 4B and E, Movies 5 and 6) was accommodated in a combined pocket and it was more unstable compared to PLPC and PMPC. In an analogous manner, sPLA2 showed optimum activity toward PLPC and PMPC (Figure 2C and F). The simulations also revealed two hydrophobic pockets: an aromatic and an aliphatic, that bind the linoleic and myristic acid tails, respectively (Figure 5C and F, Movies 11 and 12). The arachidonic acid in PAPE and PAPG (Figure 4C and F, Movies 9 and 10) was also accommodated in a combined pocket, and it was more unstable than PLPC and PMPC, analogous to iPLA2.
Figure 5.
Binding of PLPC in the active site of (A) cPLA2 (Movie 3), (B) iPLA2 (Movie 7), and (C) sPLA2 (Movie 11). Binding of PMPC in the active site of (D) cPLA2 (Movie 4), (E) iPLA2 (Movie 8), and (F) sPLA2 (Movie 12).
3.5. Active Site Properties
Each of the three enzymes contains a unique active site exhibiting specific properties. These properties define the specificity of each enzyme toward distinct types of phospholipids. The active site of each enzyme consists of a hydrophilic region where the headgroup of the phospholipid binds and a hydrophobic region where the two acyl-chains bind. The experimental and structural data showed that all three enzymes achieve substrate specificity by recruiting a specific phospholipid molecule containing the optimal fatty acid at the sn-2 position to its distinctive hydrophobic binding subsite.
cPLA2 has a deep and rigid channel-like active site that accommodates a phospholipid molecule in its entirety (Movies 1 and 2), and thus, the enzyme exhibits strict specificity for arachidonic acid at the sn-2 position. The simulations showed a well-maintained active site “pocket” volume for cPLA2 with minor changes over the time course of the simulations in the presence of PAPE, PAPS, PLPC, and PMPC (Figure S6A). The RMSD of the protein backbone atoms was stabilized below 2.0 Å for the simulations with PAPE and PAPS because these two phospholipids remained bound to the enzyme during the entire simulation, and it was stabilized above 2.0 Å for the simulations with PLPC and PMPC because they lost binding during the simulation (Figure S7A). This indicates that cPLA2 exists in two conformations on the surface of the membrane: “bound” and “unbound”. The flexibility profile of regions that are located within 4 Å of the phospholipid molecule showed that these regions are more flexible during the simulations in the presence of PLPC and PMPC because these two phospholipids diffuse into the membrane during the simulations (Figure S8 and Movies 3 and 4).
iPLA2 contains a more flexible and versatile active site, and thus, this enzyme exhibits a more permissive specificity for the fatty acid at the sn-2 position. The volume of the active site changes according to the size of the bound phospholipid, indicating its high flexibility that allows binding of diverse types of phospholipids (Figure S6B). The RMSD of the protein backbone atoms for the simulations in the presence of PAPE, PAPS, and PLPC was stabilized at ∼2.0 Å because the size of these three phospholipids is comparable, whereas in the case of PMPC, it was stabilized at ∼3.0 Å because PMPC is smaller (Figure S7B). A close examination of the regions located 4.0 Å around the bound phospholipid showed that iPLA2 can recruit a diverse set of residues to achieve binding of several types of fatty acids at the sn-2 position (Figure S9). Residues like Leu491, Ile494, Ile523, Leu524, Leu564, Met537, and Leu560, which interact with myristic acid in the simulation with PMPC, showed lower RMSF values indicating lower flexibility (Figure S9, Movie 8). A similar pattern occurred during the simulations in the presence of PAPE, PAPS, and PLPC for residues like Tyr541, Tyr544, and Phe644, which interact with the arachidonic or linoleic acid (Figure S9, Movies 5, 6, 7).
The sPLA2 active site is a very shallow cavity that accommodates only the sn-2 fatty acid (Movies 9–12). The active site volume has similar values during the simulations in the presence of PAPE, PAPG, and PLPC, whereas it has a smaller value in the case of PMPC because it is significantly smaller than the other three (Figure S6C). In all four simulations, the RMSD of the protein backbone atoms was stabilized at ∼2.0 Å, indicating that the conformation of sPLA2 was not affected by the bound phospholipid (Figure S7C). sPLA2 is a small enzyme (14 kDa, 138 aa) that contains 6 disulfide bonds, making its structure very rigid.1 The hydrophobic pocket recruitment in sPLA2 was similar to iPLA2, which is consistent with the similarity of the fatty acid specificity of these two enzymes (Figure 2). Several aliphatic residues including Val12, Leu102, Leu98, and Lue94 showed lower flexibility during the simulation with PMPC because they are part of the hydrophobic pocket to which the myristic acid is bound (Figure S10). During the simulations with PAPE, PAPG, and PLPC, aromatic residues like Tyr21, Tyr24, and Tyr105 exhibited lower flexibility because they interact with the double bonds of arachidonic and linoleic acid.
3.6. Connecting Structure with Cellular Function
Molecular structure and enzymatic activity of PLA2s are tightly connected to their cellular function. cPLA2 is the main arachidonic acid provider in the eicosanoid pathway,2 and thus, it exhibits distinctive specificity for this fatty acid at the sn-2 position of a phospholipid (Figure 2A and D). The active site of cPLA2 is enriched with aromatic residues that interact with the double bonds of the arachidonic acid, making the enzyme specific for this fatty acid. iPLA2 was found to be a vital enzyme for important metabolic functions within the cells, including membrane remodeling and homeostasis,24 and thus is more permissive for the fatty acid at the sn-2 position of a phospholipid (Figure 2B and E). The active site of iPLA2 is structurally more versatile separately using aliphatic and aromatic residues to bind several types of fatty acids. sPLA2 also exhibited a more permissive specificity profile like iPLA2 for the fatty acid at the sn-2 position (Figure 2C and F) and a distinctive preference for PG over the other headgroups (Figure 1C and F). It is well-documented that some sPLA2s have antibacterial action toward Gram-positive or -negative bacteria.1 Bacterial membrane phospholipids contain high concentrations of PG, which is consistent with the preference of sPLA2s for PG.25,26 PG (Movie 10) showed a different interaction pattern with the active site of sPLA2 compared with those of PE and PC (Movies 9, 11, and 12). The sPLA2 active site contains aliphatic and aromatic residues that the enzyme recruits to bind several types of fatty acids like iPLA2 (Figure 5).
4. Conclusions
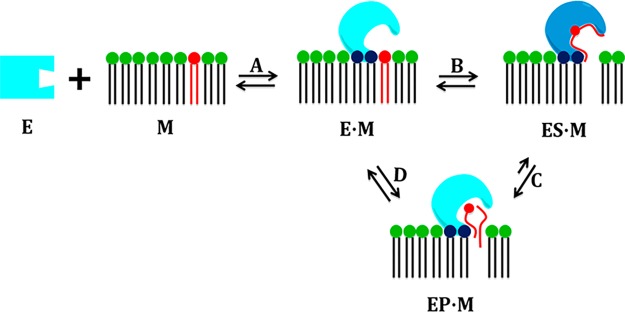
In previous studies, we introduced the novel idea that membranes serve as an allosteric ligand that enables phospholipase A2 to extract, bind, and catalyze the hydrolysis of a phospholipid molecule.3,4 We observed that these enzymes exist in a “closed” form in water and in an “open” form on the membrane surface (Figure 6). In the present study, sophisticated molecular dynamics simulations guided by experimental data indicated that, on the membrane, a PLA2 enzyme can exist in a “bound” or “unbound” form (Figure 6) with different pocket volumes. Our unique lipidomic-based enzymatic assay combined with structural studies allowed us to connect molecular structure with enzymatic activity. Our data strongly support our hypothesis that PLA2 enzymes achieve substrate specificity by recruiting the optimal fatty acid at the sn-2 position of a phospholipid molecule to its unique hydrophobic binding subsite by molecular recognition for catalysis.
Figure 6.
Phospholipase A2 exists in at least three conformations: the “closed” conformation in water (E), which associates with membranes (M) to form the “open” membrane-associated “unbound” conformation (E·M). A phospholipid molecule is then extracted into the active site, and the enzyme adopts a “bound” conformation (ES·M). Catalytic formation and release of the products (EP·M) returns the enzyme to the “open” membrane-associated “unbound” conformation (E·M) (modified from ref (3)).
Acknowledgments
This work was supported by NIH grant GM20501-41 (E.A.D.). Computational aspects were supported in part by NSF, NIH, HHMI, and NBCR (J.A.M.). We appreciate the support of Dr. Robert Konecny (UC San Diego) with operation of the computer cluster that ran the simulations in the J.A.M. laboratory. We thank Dr. Yan Wang (Shanghai Jiao Tong University) for helping us explore the potential of reverse-phase chromatography LC/MS to assay PLA2 while a visiting scientist in our laboratory (E.A.D.). We also thank Prof. Oswald Quehenberger and Aaron Armando for their support with the LC/MS system in our laboratory (E.A.D.).
Supporting Information Available
Supplementary methods, figures and tables. Supplementary movies show the binding mode of phospholipids in the active site of cPLA2, iPLA2, and sPLA2. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.7b12045.
Supplementary methods, chromatographic separations, standard curves, enzyme activities, product formation, headgroup interactions with catalytic residues, active site volumes, RMSDs, flexibilities (RMSFs), 3D models for GV sPLA2, and table of fatty acid distributions (PDF)
PAPE interactions with the catalytic residues of cPLA2 (AVI)
PAPS interactions with the catalytic residues of cPLA2 (AVI)
PLPC interactions with the catalytic residues of cPLA2 (AVI)
PMPC interactions with the catalytic residues of cPLA2 (AVI)
PAPE interactions with the catalytic residues of iPLA2 (AVI)
PAPS interactions with the catalytic residues of iPLA2 (AVI)
PLPC interactions with the catalytic residues of iPLA2 (AVI)
PMPC interactions with the catalytic residues of iPLA2 (AVI)
PAPE interactions with the catalytic residues of sPLA2 (AVI)
PAPG interactions with the catalytic residues of sPLA2 (AVI)
PLPC interactions with the catalytic residues of sPLA2 (AVI)
PMPC interactions with the catalytic residues of sPLA2 (AVI)
The authors declare no competing financial interest.
Supplementary Material
References
- Dennis E.; Cao J.; Hsu Y.-H.; Magrioti V.; Kokotos G. Chem. Rev. 2011, 111, 6130–6185. 10.1021/cr200085w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis E. A.; Norris P. C. Nat. Rev. Immunol. 2015, 15, 511–523. 10.1038/nri3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchlis V. D.; Bucher D.; McCammon J. A.; Dennis E. A. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, E516–E525. 10.1073/pnas.1424651112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J.; Burke J.; Dennis E. J. Biol. Chem. 2013, 288, 1806–1813. 10.1074/jbc.R112.421909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y.-H.; Bucher D.; Cao J.; Li S.; Yang S.-W.; Kokotos G.; Woods V.; McCammon J.; Dennis E. J. Am. Chem. Soc. 2013, 135, 1330–1337. 10.1021/ja306490g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher D.; Hsu Y. H.; Mouchlis V. D.; Dennis E. A.; McCammon J. A. PLoS Comput. Biol. 2013, 9, e1003156. 10.1371/journal.pcbi.1003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J.; Babakhani A.; Gorfe A.; Kokotos G.; Li S.; Woods V.; McCammon J.; Dennis E. J. Am. Chem. Soc. 2009, 131, 8083–8091. 10.1021/ja900098y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y.-H.; Burke J.; Li S.; Woods V.; Dennis E. J. Biol. Chem. 2009, 284, 23652–23661. 10.1074/jbc.M109.021857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchlis V. D.; Limnios D.; Kokotou M. G.; Barbayianni E.; Kokotos G.; McCammon J. A.; Dennis E. A. J. Med. Chem. 2016, 59, 4403–4414. 10.1021/acs.jmedchem.6b00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchlis V. D.; Morisseau C.; Hammock B. D.; Li S.; McCammon J. A.; Dennis E. A. Bioorg. Med. Chem. 2016, 24, 4801–4811. 10.1016/j.bmc.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchlis V. D.; Dennis E. A. Adv. Biol. Regul. 2016, 61, 17–24. 10.1016/j.jbior.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds L. J.; Washburn W. N.; Deems R. A.; Dennis E. A. Methods Enzymol. 1991, 197, 3–23. 10.1016/0076-6879(91)97129-M. [DOI] [PubMed] [Google Scholar]
- Yang H.; Mosior M.; Johnson C.; Chen Y.; Dennis E. Anal. Biochem. 1999, 269, 278–288. 10.1006/abio.1999.4053. [DOI] [PubMed] [Google Scholar]
- Quehenberger O.; Armando A. M.; Brown A. H.; Milne S. B.; Myers D. S.; Merrill A. H.; Bandyopadhyay S.; Jones K. N.; Kelly S.; Shaner R. L.; Sullards C. M.; Wang E.; Murphy R. C.; Barkley R. M.; Leiker T. J.; Raetz C. R. H.; Guan Z. Q.; Laird G. M.; Six D. A.; Russell D. W.; McDonald J. G.; Subramaniam S.; Fahy E.; Dennis E. A. J. Lipid Res. 2010, 51, 3299–3305. 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodson L.; Skeaff C. M.; Fielding B. A. Prog. Lipid Res. 2008, 47, 348–380. 10.1016/j.plipres.2008.03.003. [DOI] [PubMed] [Google Scholar]
- van Meer G.; Voelker D. R.; Feigenson G. W. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikano M.; Masuzawa Y.; Yazawa K.; Takayama K.; Kudo I.; Inoue K. Biochim. Biophys. Acta, Lipids Lipid Metab. 1994, 1212, 211–6. 10.1016/0005-2760(94)90255-0. [DOI] [PubMed] [Google Scholar]
- Burdge G. C.; Creaney A.; Postle A. D.; Wilton D. C. Int. J. Biochem. Cell Biol. 1995, 27, 1027–1032. 10.1016/1357-2725(95)00083-2. [DOI] [PubMed] [Google Scholar]
- Strokin M.; Sergeeva M.; Reiser G. Br. J. Pharmacol. 2003, 139, 1014–1022. 10.1038/sj.bjp.0705326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basselin M.; Rosa A. O.; Ramadan E.; Cheon Y.; Chang L.; Chen M.; Greenstein D.; Wohltmann M.; Turk J.; Rapoport S. I. J. Lipid Res. 2010, 51, 3166–73. 10.1194/jlr.M008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon Y.; Kim H. W.; Igarashi M.; Modi H. R.; Chang L.; Ma K.; Greenstein D.; Wohltmann M.; Turk J.; Rapoport S. I.; Taha A. Y. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2012, 1821, 1278–1286. 10.1016/j.bbalip.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-de-Gomez L.; Astudillo A. M.; Guijas C.; Magrioti V.; Kokotos G.; Balboa M. A.; Balsinde J. J. Immunol. 2014, 192, 752–762. 10.4049/jimmunol.1302267. [DOI] [PubMed] [Google Scholar]
- Murakami M.; Masuda S.; Ueda-Semmyo K.; Yoda E.; Kuwata H.; Takanezawa Y.; Aoki J.; Arai H.; Sumimoto H.; Ishikawa Y.; Ishii T.; Nakatani Y.; Kudo I. J. Biol. Chem. 2005, 280, 14028–14041. 10.1074/jbc.M413766200. [DOI] [PubMed] [Google Scholar]
- Balsinde J.; Bianco I. D.; Ackermann E. J.; Conde-Frieboes K.; Dennis E. A. Proc. Natl. Acad. Sci. U. S. A. 1995, 92, 8527–31. 10.1073/pnas.92.18.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. M.; Rock C. O. Nat. Rev. Microbiol. 2008, 6, 222–233. 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
- Epand R. M.; Epand R. F. J. Pept. Sci. 2011, 17, 298–305. 10.1002/psc.1319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.