Abstract
Broadly neutralizing antibodies (bNAbs) that target the trimeric HIV-1 envelope glycoprotein spike (Env) are tools that can guide the design of recombinant Env proteins intended to engage the predicted human germline precursors of bNAbs (gl-bNAbs). The protein components of gl-bNAb epitopes are often masked by glycans, while mature bNAbs can evolve to accommodate and/or bypass these shielding glycans. The design of germline-targeting Env immunogens therefore includes the targeted deletion of specific glycan sites. However, the processing of glycans on Env trimers can be influenced by the density with which they are packed together, a highly relevant point given the essential contributions under-processed glycans make to multiple bNAb epitopes. We sought to determine the impact of the removal of 15 potential N-glycan sites (5 per protomer) from the germline-targeting soluble trimer, BG505 SOSIP.v4.1-GT1 using quantitative, site-specific N-glycan mass spectrometry analysis. We find that compared to SOSIP.664, there was little overall change in the glycan profile but subtle increases in the extent of processing at sites immediately adjacent to where glycans had been deleted. We conclude that multiple glycans can be deleted from BG505 SOSIP trimers without perturbing the overall integrity of the glycan shield.
Keywords: HIV-1 vaccine design, envelope, Env, immunogen design, glycosylation, mass spectrometry, virus, glycoproteomics
Graphical Abstract
INTRODUCTION
Broadly neutralizing antibodies (bNAbs) with potent activity against the majority of circulating human immunodeficiency 1 (HIV-1) strains emerge in a subset (~25%) of infected people over a period of 2–4 years (1, 2). In virus challenge studies, bNAbs can provide passive protection to macaques (3–5). The existence and characteristics of bNAbs underpin the design of immunogens intended to elicit this category of antibody responses in humans.
Like all neutralizing antibodies, bNAbs target epitopes on the trimeric HIV-1 envelope glycoprotein (Env). This trimer is composed of three heterodimers of gp120 and gp41, which together form tri-lobed spikes that are anchored onto the virion surface via the gp41 transmembrane domain (6–9). About half the mass of the trimer is accounted for by typically 80–100 N-linked glycans, most of which are located on the gp120 subunits (10). The glycans shield potentially immunogenic and antigenic regions of the underlying protein surface from the humoral immune system (11, 12), but can themselves, perhaps paradoxically, become components of multiple bNAb epitopes (8, 13–29). The high local density of glycans on the trimer can sterically constrain their processing from oligomannose- to complex-type forms by α-mannosidases in the endoplasmic reticulum (ER) and Golgi apparatus during glycoprotein egress through the secretory pathway (30–32). Consequently, there is a substantial abundance of under-processed, oligomannose-type glycans (Man5-9GlcNAc2) on the trimer surface (32–39). The oligomannose-type glycans are particularly clustered around the outer domain of gp120 as well as at the gp120-gp41 protomer interface, whereas more highly and variably processed glycans are present near the apex and base of the trimer where there are fewer steric constraints to processing enzymes (40, 41). These key details of glycan processing have emerged from analyses of recombinant, soluble mimics of the native spike, exemplified by SOSIP.664 trimers of the BG505 and other genotypes (28, 41, 42). The conserved population of oligomannose-type glycans creates a target on the trimer that is more homogeneous and conserved than the highly variable polypeptide surfaces. Hence, glycan composition is now recognized to be a critical factor in the design of Env immunogens aimed at eliciting bNAbs (43). Given the importance of glycosylation, it is now common practice to perform glycan and glycopeptide analysis on each new candidate Env immunogen (44), particularly those destined for clinical evaluation (45). The importance of this approach is underscored by the observation that presence of absence of individual glycans has been reported to influence the processing of neighboring glycans in a context-dependent manner (31, 37, 41).
Various soluble, native-like Env trimers rooted in the prototypic BG505 SOSIP.664 design are now platforms for the creation of Env immunogens aimed at inducing a broad and potent B-cell response (9, 46). They have also facilitated an ever-increasing understanding of the structure and function of the Env trimer and the multiple bNAb epitopes displayed on its surface (6, 7, 28, 47, 48). Some SOSIP.664 trimers can induce strong NAb responses against the autologous neutralization-resistant (Tier-2) primary viruses, which had been generally difficult to achieve using earlier designs of Env proteins (46). The SOSIP.664 design can be improved by introducing additional stabilizing changes to make SOSIP.v4.1 variants. These modified trimers are less immunogenic for antibodies to the immunodominant gp120 V3-region that neutralize sensitive Tier-1 viruses but not their more relevant Tier-2 counterparts (49, 50).
A central goal of various Env vaccine development programs is to improve the breadth of the elicited antibody response, a step in the path towards true bNAbs. Case studies have revealed the maturation pathways in HIV-1 infected individuals that, over several years, drove the emergence of bNAbs against epitope clusters involving the CD4-binding site (CD4bs), the trimer apex, and the mannose patch (51–53). In general, sequential rounds of sequence evolution in the virus and the responding B cells create first narrow specificity NAbs and then virus escape mutants. This process is repeated over many cycles until some antibodies mature sufficiently to be categorized as bNAbs that can target multiple virus variants. In other words, viral diversification under immune selection pressure can gradually drive substantial increases in the breadth and potency of an initially narrow and sometimes weak initial NAb response (54). This knowledge of a natural process guides vaccination strategies based on priming with an Env immunogen that is specifically designed to activate B-cell receptors on naïve cells in the germline (gl) so that they produce bNAb precursors (gl-bNAbs). These initial responses can then be boosted with one or more engineered immunogen(s) that sequentially guide the B-cells to mature and produce antibodies with a bNAb-like phenotype (55–59).
An emerging approach in the development of gl-targeting immunogens is the creation or expansion of holes in the glycan shield (60–62). We have specifically modified, and recently described, a version of the BG505 SOSIP.v4.1 trimer, termed SOSIP.v4.1-GT1 (germline-targeting trimer version 1; GT1 trimer for short) to confer the ability to bind multiple different gl-bNAbs, specifically ones directed against the trimer apex and the CD4bs; examples include the germline versions of PG9, PG16, CH01 and several members of the VRC01-class (63). Compared to the SOSIP.v4.1 trimers, each protomer in the GT1 variant contains 15 amino-acid substitutions and has a 7-residue deletion that truncates the V2 loop and potentially reduces its flexibility; together these changes facilitate gl-bNAb binding (63).
Five of the above changes involve the elimination of potential N-glycosylation sites (PNGSs), of which four are located in proximity to the CD4bs; their deletion removes impediments to gl-bNAb binding (63–67). Several other changes in V2 enhance the binding of gl-bNAbs to trimer-apex epitopes; here, deletion of 7 residues from V2 removes a PNGS near the apex. Overall, the glycan shield of the GT1 trimer lacks 15 N-glycan sites (5 per protomer). Three of the deleted sites (N197, N276 and N386) are highly conserved, as they are normally present in more than ~85% across multiple HIV-1 strains (68).
Despite these modifications, the BG505 SOSIP.v4.1-GT1 trimer retains a native-like structure and has biophysical and biochemical properties similar to the SOSIP.664 and SOSIP.v4.1 versions from which it was derived (63). However, unlike these precursor trimers, GT1 binds multiple gl-bNAbs and activates B cells expressing these in vitro. Furthermore, when tested as an immunogen in CD4bs-targeting (gl-VRC01) and N332/V3-directed (gl-PGT121) knock-in mouse models, GT1 triggers the production of epitope-specific antibodies (63). The close relationship of the glycan shield of Env and the elicitation of neutralization was recently shown supporting a strategy of selective deglycosylation for immunization priming (69).
Here, we present a quantitative, site-specific N-glycosylation analysis of BG505 SOSIP.v4.1-GT1 trimers purified from a stable CHO cell line for comparison with a similar analysis of the SOSIP.664 precursor trimer that was produced and purified in the same way. Hence, we could assess the impact of the designed removal of 15 PNGSs on the overall composition of the Env glycan shield. A key observation is that the overall processing state of the N-glycans is remarkably stable, despite the ~20% difference in the total glycan content of the two trimers. This result is in accordance with a previously published study that analyzed the consequences of removing four glycans that surround the CD4bs (70).
We did find that GT1 trimer glycans in close proximity to deleted sites were slightly more processed than on the SOSIP.664 comparator, but the effect was both modest and highly localized. Thus, the major clusters of oligomannose-type glycans that wrap around the outer domain of the trimer, and patches of complex-type glycans near the apex and base, are almost identical on the GT1 and SOSIP.664 trimers. Of particular note is that the essentially unchanged processing of the overall glycan shield leads to the preservation or increased exposure of glycan-dependent epitopes for various mature and gl-versions of bNAbs (63).
In summary, the targeted removal of individual glycan sites on this rationally designed, gl-bNAb targeting Env immunogen does not perturb the overall architecture of the glycan shield, although it does lead to small shifts in the abundance of different oligomannose-type glycans at specific sites. These findings not only support the further use of the GT1 immunogen design strategy but also highlight the relevance of assessing what happens to Env trimer glycosylation when key glycan sites are deliberately eliminated.
EXPERIMENTAL PROCEDURES
Expression and purification of SOSIP trimers
The BG505 SOSIP.v4.1-GT1 construct is described elsewhere (63). This GT1 trimer and its SOSIP.664 counterpart were expressed from stable CHO cell lines as described previously (71). Env trimers were purified from culture supernatants by 2G12-affinity chromatography followed by SEC (9, 72).
Assessment of trimer quality and properties
The cleavage and homogeneity of purified trimers were assessed using sodium dodecyl sulfate and blue native polyacrylamide gel electrophoresis followed by Coomassie blue staining or western blotting using bNAb 2G12 (71, 73). Their thermostability was determined using DSC and their morphology by NS-EM (9, 49, 74).
Analysis of total glycan profiles by HILIC-UPLC
N-linked glycans were enzymatically released from envelope glycoproteins via in-gel digestion with Peptide-N-Glycosidase F (PNGase F), subsequently fluorescently labelled with 2-aminobenzoic acid (2-AA) and analyzed by HILIC-UPLC, as previously described (34, 41, 75). Digestion of released glycans with Endo H was used to measure the abundance of oligomannose-type glycans (34).
Creation of a glycan library by tandem ion mobility-ESI MS
The total pool of glycans was released from trimers by PNGase F digestion and analyzed using ion mobility MS, as described previously (41). Negative ion mass, collision-induced dissociation (CID) and ion mobility spectra were recorded with a Waters Synapt G2Si mass spectrometer (Waters Corp.) fitted with a nano-electrospray ion source. Waters Driftscope (version 2.8) software and MassLynxTM (version 4.1) was used for data acquisition and processing. Spectra were interpreted as described previously (76–79). The results obtained served as the basis for the creation of sample-specific glycan libraries, which were used for subsequent site-specific N-glycosylation analyses. Raw data files can be provided upon request.
Site-specific N-glycosylation analysis
Before proteolytic digestion, trimers were denatured and alkylated by incubation for 1h at room temperature (RT) in a 50 mM Tris/HCl, pH 8.0 buffer containing 6 M urea and 5 mM dithiothreitol (DTT), followed by the addition of 20 mM iodacetamide (IAA) for a further 1h at RT in the dark, and then additional DTT (20 mM) for another 1h, to eliminate any residual IAA. The alkylated trimers were buffer-exchanged into 50 mM Tris/HCl, pH 8.0 using Vivaspin columns and digested with either trypsin or chymotrypsin (Mass Spectrometry Grade, Promega) at a ratio of 1:25 (w/w) or 1:30 (w/w), respectively, according to the manufacturer’s instructions. Glycopeptides were selected from the protease-digested samples using the ProteoExtract Glycopeptide Enrichment Kit (Merck Millipore). Enriched glycopeptides were analysed by LC-ESI MS on a Q-Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific), as previously described (41), using higher energy collisional dissociation (HCD) fragmentation. Data analysis and glycopeptide identification were performed using ByonicTM (Version 2.7) and ByologicTM software (Version 2.3; Protein Metrics Inc.), as previously described (41). Searches were performed with the following Byonic settings: semi-specific (trypsin), fully specific (chymotrypsin); maximum of two (trypsin) and four (chymotrypsin) missed cleavages; 4 ppm precursor mass tolerance; 10 ppm fragment mass tolerance. Searches were performed against the GT1 and SOSIP.664 sequences in .fasta format and by using the personalized glycan library. The following modifications were included: fixed carbamidomethylation, Gln to pyro-Glu conversion (rare), Glu to pyro-Glu conversion (rare) and oxidation of methionine (rare). Manual verification of identified all glycopeptides and their relative quantification was performed by checking the correct identification of the monoisotopic peak, by assessing HCD fragmentation spectra including the presence of the correct oxonium ions and by adjusting the windows of the extracted-ion chromatograms. Where multiple charge states were identified for the same glycopeptide, hits were validated separately. The abundance of individual glycoforms was determined by adding up the intensities of the extracted-ion chromatograms over all charge states. Relative abundances = [Intensity of one glycoform] / [Intensities of all glycoforms identified on the same glycopeptide]. Raw data files can be provided upon request.
RESULTS
Generation of a CHO cell line producing BG505 SOSIP.v4.1-GT1 trimers
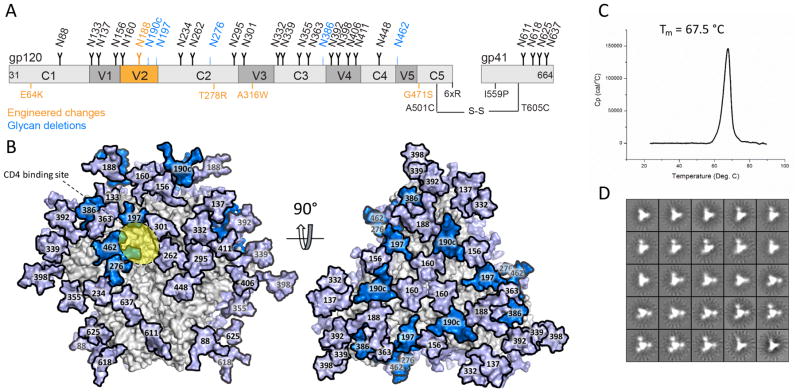
The design and structure of BG505 SOSIP.v4.1-GT1 trimers has been described elsewhere and is summarized in Figure 1A (63). We made a stable CHO cell line to express the GT1 trimers, using the same method as previously described for BG505 SOSIP.664 (71). The use of a CHO cell line is relevant as this expression system has now been used to produce a clinical grade preparation of the BG505 SOSIP.664 trimer (45). The vector system expresses both Env and furin, resulting in the production of cleaved GT1 trimers at yields of 7–10 mg/L after purification by 2G12-affinity chromatography followed by size exclusion chromatography (SEC). Visualization by negative-stain electron microscopy (NS-EM) showed that 100% of the purified GT1 trimers adopt a native, propeller-like structure (Figure 1). Their midpoint of thermal denaturation (Tm), as assessed by differential scanning calorimetry (DSC), was 67.5 °C, which is very similar to the values derived for GT1 trimers produced by transient transfection of human embryonic kidney (HEK) 293T cells, and for BG505 SOSIP.664 and SOSIP.v4.1 trimers (Figure 1 and Table S1) (9, 63).
Figure 1. Design and properties of CHO-cell produced BG505 SOSIP.v4.1-GT1 trimers.
(A) Schematic representation of the BG505 SOSIP.v4.1-GT1 construct. The constant (C1–C5) and variable (V1–V5) regions of gp120 are highlighted in light and dark grey, respectively. The stabilizing substitutions present in the SOSIP.664 are shown in black, with modifications that create the SOSIP.v4.1-GT1 construct in orange. Of note is that the E64K and A316W mutations were introduced to create the BG505 SOSIP.v4.1 trimer (49). Detailed information on the GT1 construct, including a 7 amino acid deletion in V2, are described elsewhere (63). The SOSIP.664 glycans are shown as black icons, with the ones deleted from the GT1 construct colored blue. Glycan N188 (orange) in the altered V2 region is equivalent to glycan N190 on the prototype BG505 SOSIP.664 version, and thus represents neither a glycan introduction nor a deletion. (B) The glycans deleted from the GT1 trimer are highlighted in marine blue on a model of the fully glycosylated BG505 SOSIP.664 trimer (the remaining glycans are in light blue). The approximate location of the CD4bs is highlighted in yellow. The model was constructed using PDB ID 5ACO (80) as previously described (41). (C) DSC analysis of 2G12 plus SEC-purified GT1 trimers, with the Tm value indicated (see Table S1 for additional relevant Tm values). (D) NS-EM analysis of the same purified GT1 trimers, which were 100% native-like.
Glycan-depleted GT1 trimers retain the oligomannose glycan signature
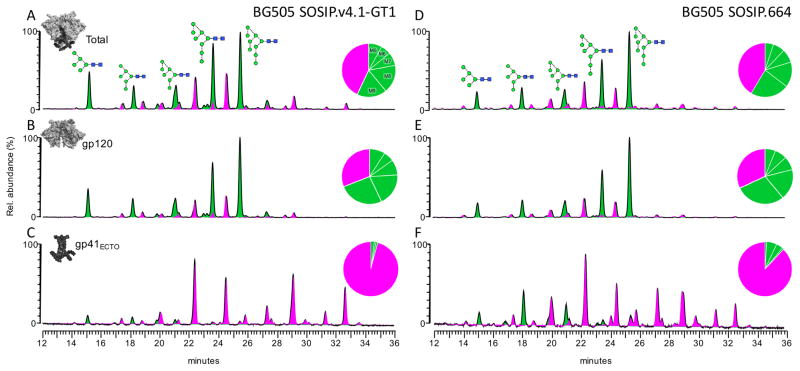
We compared the overall glycan profiles of the BG505 GT1 and SOSIP.664 trimers. The N-glycans were enzymatically released, fluorescently labeled and analyzed by hydrophilic interaction chromatography-ultra-performance liquid chromatography (HILIC-UPLC). The abundance of oligomannose-type glycans (Man5-9GlcNAc2) was determined by cleavage of the labelled glycan pool with Endoglycosidase H (Endo H) (Figure 2 and Table S2). Both trimers yielded very similar, oligomannose-dominated glycosylation profiles, as previously reported for BG505 SOSIP.664 (34, 41). The overall oligomannose contents of the GT1 and SOSIP.664 trimers were almost identical (57% and 59%, respectively). We do, however, note that the abundance of Man8GlcNAc2 in relation to Man9GlcNAc2 was elevated for GT1 (Figure 2, Table S2), similar to what was found when the same GT1 trimers were produced in HEK 293T cells (63). The abundance of Man5GlcNAc2 was also slightly elevated for GT1 compared to SOSIP.664 (8% and 5%, respectively; Figure 2, Table S2). This small but detectable producer-cell independent increase in overall α-mannosidase processing for GT1 presumably reflects the elimination of five glycans that are present on the SOSIP.664 trimer and the consequent exposure of some nearby sites (see below). The glycan profile of the GT1 trimer gp41 subunit is almost exclusively dominated by complex-type glycans, whereas oligomannose-type glycans are somewhat more abundant on gp41 from SOSIP.664 (4% on GT1 vs. 12% on SOSIP trimers) (Figure 2, Table S2). This shift may reflect subtle changes in the local environment of individual glycans neighboring the gp120 glycans (see below).
Figure 2. Glycosylation profiles of BG505 SOSIP.v4.1-GT1 and SOSIP.664 trimers.
HILIC-UPLC profiles of PNGase F-released and fluorescently-labelled N-linked glycans from BG505 SOSIP.v4.1-GT1 (left) and SOSIP.664 (right) trimers produced in CHO cells and purified by 2G12-affinity chromatography followed by SEC. Gp140 trimer (A & D); gp120 (B & E); gp41 (C & F). Oligomannose-type and hybrid glycans (green) were identified by their sensitivities to Endo H digestion. Peaks corresponding to complex glycans are depicted in pink. The pie charts summarize the quantification of the glycan processing states, as determined by integration of the chromatogram. The corresponding percentages are listed in Table S2. Glycan sequencing data derived by exoglycosidase digestion of the GT1 trimer are shown in Figure S1.
Glycan libraries from BG505 SOSIP.v4.1-GT1 and SOSIP.664 trimers
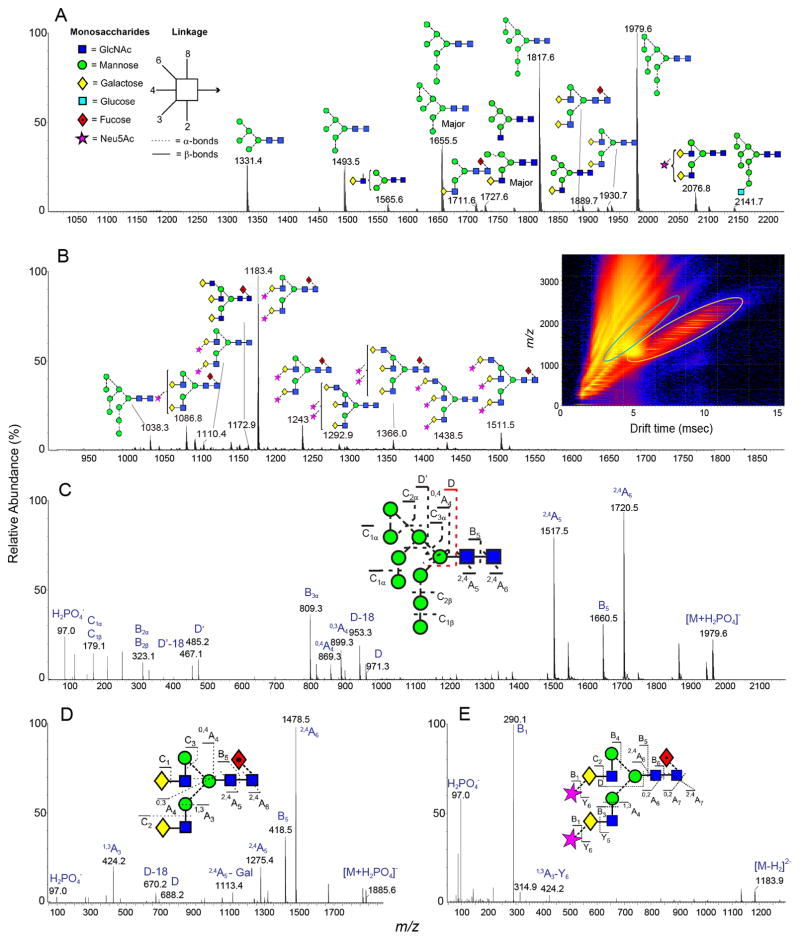
To probe the influence of glycan depletion on the glycan shield in greater detail, we performed site-specific N-glycosylation analyses on CHO cell-derived GT1 and SOSIP.664 trimers. We first generated a database of all identified glycan structures by ion mobility-electrospray ionization mass spectrometry (IM-ESI MS), both to characterize the range of different N-linked glycans and then to facilitate a subsequent site-specific N-glycan analysis (Figure 3, Table S3). Negative ion fragmentation was used to accurately assign the structure and major isomers of the glycans (81). The IM-ESI MS spectrum shows that the gp120 subunit of the GT1 trimer has a significant population of oligomannose-type glycans (Figure 3A), which is consistent with the HILIC-UPLC glycan analysis (Figure 2).
Figure 3. Ion mobility mass spectrometric analysis of BG505 SOSIP.v4.1-GT1 glycans.
Negative ion electrospray spectra of N-glycans enzymatically released from the gp120 subunit of CHO cell-produced BG505 SOSIP.v4.1-GT1 trimers. (A) Mobility-extracted singly charged negative ions found on gp120. The corresponding singly charged ions are encircled with the yellow oval in the DriftScope image (m/z against drift time) in Panel B. (B) Mobility extracted doubly charged negative ions found on gp120. The ions are encircled with a blue oval in the DriftScope image. (C–E) Collision-induced dissociation spectra for three distinct glycans derived from the transfer region of the Synapt G2Si instrument. The insets to panels C, D and E show the corresponding glycans as well as the numbering of the fragments. Panel E shows only the low mass region to emphasize the missing m/z 306 ion, the absence of which is diagnostic of α2,3-linked sialic acids (82). The numbering scheme follows that proposed elsewhere (83). The glycan symbols are shown on panel A. The complete glycan libraries of the SOSIP.664 and GT1 trimers are shown in Table S3.
Compared to BG505 SOSIP.664 trimers produced in stable HEK 293T cells (41), we found a less diverse range of complex-type glycans on the same trimers produced in CHO cells. This kind of variation reflects the different glycan-processing activities in the two cell types. For example, CHO cells do not express a functional N-acetylglucosaminyltransferase III (84), which generates bisecting glycans. No glycans with terminal sulfates, GalNAcs, or terminal α2,6-sialic acids were identified on the CHO cell-produced trimers. A higher degree of sialylation on CHO cell-derived proteins, compared to HEK 293, has been reported previously (85). Consistent with that report, we found a high abundance of sialylated glycans on the CHO cell-produced GT1 trimers, as determined by a neuraminidase digest of the glycan pool (Figure S1). The sialic acid moieties on these trimers are entirely α2,3-linked, as indicated by the absence of the characteristic m/z 306 ion from the tandem ion mobility fragmentation spectrum (Figure 3E) (82). The dominance of under-processed, oligomannose-type Man5-9GlcNAc2 glycans is, nevertheless, well conserved between the two different producer cells. Overall, the glycome analyses of CHO cell-derived BG505 SOSIP.664 and GT1 trimers and HEK 293T cell-derived BG505 SOSIP.664 trimers described here and elsewhere concur: protein-directed glycosylation events yield the predominant oligomannose glycoforms, and cell type-dependent events create sub-populations of complex-type glycans (34, 41). The conserved oligomannose patches include the epitopes for multiple bNAbs and are one of the key properties of Env immunogens that need to be preserved when trimers are further engineered.
Quantitative site-specific N-glycosylation analysis reveals a robust oligomannose-patch
To better understand the effect of eliminating selected sites on the overall glycan shield of the GT1 trimer, we performed a quantitative, site-specific N-glycosylation analysis on protease-digested glycopeptides. This method, which uses coupled, in-line liquid chromatography-electrospray mass spectrometry (LC-ESI MS), has been described and validated elsewhere (41).
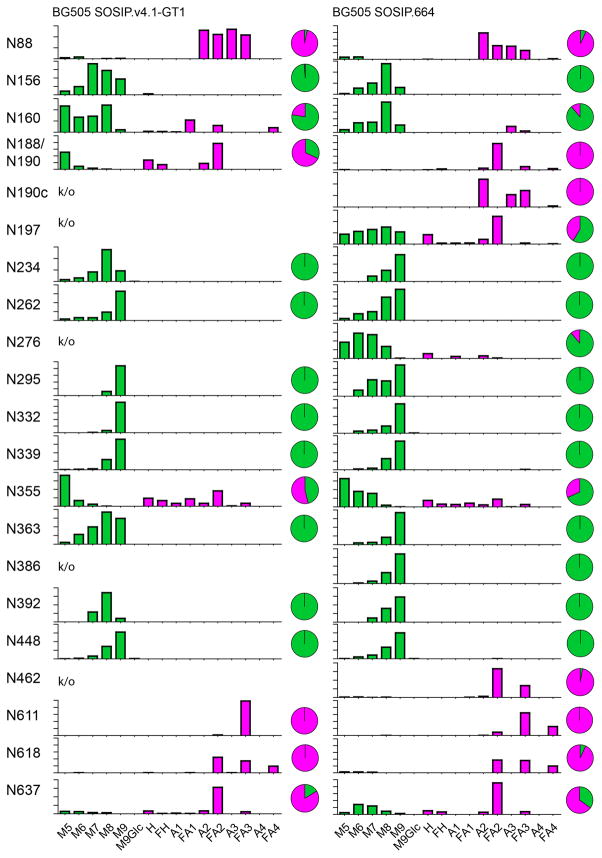
We quantified the relative abundance of specific glycan structures on individual N-glycosylation sites by summing the ion intensities of the corresponding glycopeptides. The site-analysis data show that the overall ratio between oligomannose- and complex-type glycans is highly similar for CHO cell-produced SOSIP.664 and GT1 trimers (Figure 4; inset pie charts). However, a detailed inspection of the underlying profiles reveals several examples of elevated trimming within the oligomannose glycan series on the GT1 trimer compared to SOSIP.664, exemplified by a drop in Man9GlcNAc2 and a higher abundance of Man5-8GlcNAc2. This phenomenon is particularly evident at the N234, N363 and N392 sites where the Man9GlcNAc2 to Man8GlcNAc2 ratio inverts (i.e., Man9GlcNAc2 is predominant at these sites on SOSIP.664, but Man8GlcNAc2 on GT1).
Figure 4. Quantitative site-specific N-glycosylation analysis.
Relative quantification of N-glycosylation sites on the BG505 SOSIP.v4.1-GT1 and SOSIP.664 trimers. Quantitative data could be obtained for 16 and 21 glycan sites of (23 and 28) for the SOSIP.v4.1-GT1 and SOSIP.664 trimers, respectively. The trimers were digested with trypsin or chymotrypsin, enriched for glycopeptides and analysed by LC-ESI MS. For SOSIP.v4.1-GT1, the bar graphs represent the means of two analytical replicates, for SOSIP.664 one analysis was performed. The pie charts summarize the quantification of oligomannose-type (green) and complex/hybrid glycans (pink) on individual sites. The analysis was based on a glycan library generated by ion mobility ESI mass spectrometry (Table S2). Relative quantifications for individual sites are based on the peak lists shown in Table S4. The abundances of individual species in different categories have been combined according to the oligomannose series (M5 to M9; Man5GlcNAc2 to Man9GlcNAc2), hybrids (H) and fucosylated hybrids (FH), and by the number of branching antennae (A) of complex-type glycans. An = number (n) of antennae (e.g., A3 = triantennary); Gn = number (n) of galactose residues; an F before the name indicates the presence of a core fucose.
The implication of these observations is that α-mannosidase processing occurs to a greater extent at these sites on the GT1 trimer, which is understandable as these three sites are very close to positions from which PNGSs were deleted (Figure 5). For example, it is likely that mannose trimming at N392 is impeded by the presence of the nearby N386 glycan on the SOSIP.664 trimer, but can occur more readily on GT1, which lacks N386. Of note is that, despite the deletion of three surrounding glycan sites (N386, N462 and N197), the N363 glycan on the GT1 trimer was slightly more trimmed compared to SOSIP.664 but still remained oligomannose-type. The implication is that there are still significant impediments to the further processing of this glycan, such as the N392 glycan and/or ones located in V2 (Figure 1). Hence, there may be significant redundancy among the various steric factors that impede α-mannosidase trimming at sites within the intrinsic and trimer-associated mannose patches where oligomannose glycans are particularly densely clustered.
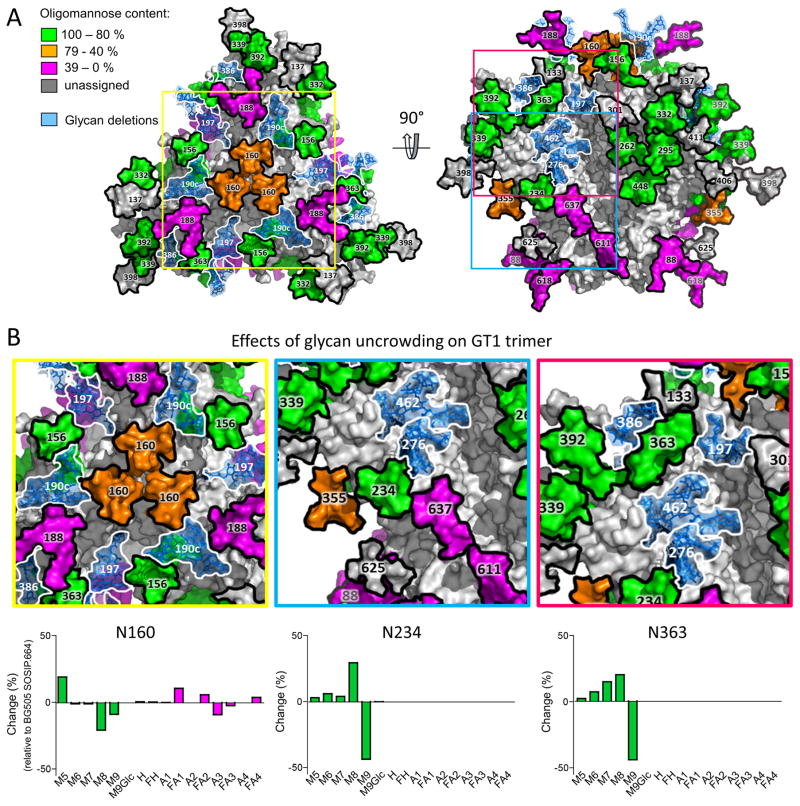
Figure 5. Processing of glycans on the glycan-depleted GT1 trimer.
(A) The model is based on the one presented in Figure 1B, but with the glycans deleted from the GT1 trimer now shown in “ghost” form (i.e., light blue with a white border). The remaining glycans are color-coded according to their processing state (see key on the figure). (B) Magnification of three sites (N160, N234 and N363) to highlight the effects of the reduction in local glycan density on the GT1 trimer. Underneath these images are difference plots of the % change in glycan abundances for GT1 compared to SOSIP.664.
Elevated processing toward smaller oligomannose-type glycans was also seen at the N156 and N160 sites in the V1-V2 region of the GT1 trimer. This may again reflect the reduction in the local glycan density caused by the removal of the nearby N190c and N197 glycans. The 7 amino-acid deletion that shortens V2 could also be contributory. Our model of the glycosylated trimer, presented in Figure 5, implies that the steric restriction to the processing of the N160 glycan may be driven by the inter-protomer clustering of the three N160 glycans where they converge at the trimer apex. Evidence for clustering is provided by cryo-EM and X-ray crystallography structures of BG505 SOSIP.664 trimers (6, 7) and has been shown to limit α-mannosidase processing by comparison of the native-like BG505 SOSIP.664 N160 glycans (68% oligomannose-type glycans) to those of monomeric (9% oligomannose-type glycans) and uncleaved gp140 pseudotrimers (34% oligomannose-type glycans) (42).
We note that the N295 glycan site (and, to a much lesser extent, N332) was more highly processed towards smaller oligomannose-type glycans on the SOSIP.664 trimer compared to GT1. Although this outcome may be rooted in differential conformational and steric constraints, we cannot fully explain the underlying mechanism using the currently available models.
The design of the GT1 trimer involved eliminating three glycans proximal to the CD4bs (N197, N276 and N462). While we have noted the moderate impact these changes have on the nearby N363 glycan, the processing of the other CD4bs-proximal glycans (N262 and N448) is little changed. This finding is consistent with there being multiple locally stabilizing factors that influence the accessibility of glycans to processing enzymes. Thus, the N262 glycan is known to form extensive interactions with the polypeptide chain (6, 86); and the N448 glycan is presumably protected by its immediate neighbors (N295 and N262). Glycan N301 is also very close to the CD4bs, but we were unable to obtain quantitative data on this site. Overall, our data confirm and extend how local glycan clustering can be sufficient to protect a glycan from processing (31).
DISCUSSION
The preservation of site-specific processing of the SOSIP.v4.1-GT1 trimer glycans has substantive implications for our understanding of the glycan shield and for Env immunogen design. That glycan processing is little changed despite the elimination of 15 PNGSs from the trimer reveals the considerable redundancy in how the local environment of each glycan acts to limit the processing activity of α-mannosidases. In other words, the loss of one glycan from a local cluster does not usually eliminate the steric constraints on the processing of its neighbors.
Our observations are consistent with and hence reinforce a recent, temporally overlapping, study of a sequential deglycosylation strategy involving glycan site deletions around the CD4bs (70). Specifically, deletion of the N197, N276, N301 and N386 sites had a minimal impact on the overall glycosylation profile of the BG505 SOSIP.664 trimer. Of those four sites, all but N301 were also eliminated from the GT1 trimer (and two more; N190c, N462), so it is encouraging that the two sets of findings were generally concordant. In the wider context of our own study with the GT1 trimer, what we have found will help in understanding the antigenicity and structural properties of this gl-bNAb targeting immunogen and the future design of improved variants based on the BG505 and other genotypes (63).
The quantitative methodology we have used in this study is clearly capable of revealing quite subtle changes in the composition of the glycan shield, particularly in respect of the abundance of different oligomannose-type glycans; this is a topic that has not been addressed previously. Our findings are also in accordance with the preservation of the mannose patch across the HIV-1 clades despite variation in its precise composition (33). They also support the hypothesis that local glycan processing is largely preserved when the glycan shield evolves over time in infected people, as part of the immune evasion process (87). An additional implication is that the presence of oligomannose-type glycans will be a fundamental feature of virion-associated Env proteins despite the extensive variation in their amino-acid sequence. By extension, variation in the location of individual glycans seems unlikely to compromise other properties that oligomannose-type glycans confer on HIV-1, such as lectin-mediated virion trafficking (88) and complement activation through the Mannose Binding Protein. Modulation of the oligomannose-triggered innate immune response, for example by the capture of cell-surface complement regulators during viral budding (89–91), may also be a fundamental property of HIV-1. This latter hypothesis is supported by the vulnerability to complement of virions artificially devoid of such regulators (92).
The sensitivity of α-mannosidase processing to local steric factors is governed by the extensive substrate recognition interface that is revealed by X-ray crystallography, when the relative sizes of the enzyme and the trimer are factored in (31, 93). Previously, we used a steric-clash model based on the cryo-EM structure of a fully glycosylated trimer and the crystal structure of an enzyme inhibitor complex to reveal the extent to which the N332 glycan was sterically protected; this glycan is located near the center of the intrinsic mannose patch on the gp120 subunit of the trimer, and plays a core role in many bNAb epitopes (31). Consistent with the underlying concept, there was no processing of N332-containing glycopeptides beyond Man8GlcNAc2 among a panel of recombinant gp120 mutants that lacked various proximal glycans (31). Our site-specific observations on the GT1 trimer show that, even in regions of the trimer where the local glycan density is lower than in the outer domain, the glycan clusters are similarly protected, albeit with some trimming evident beyond Man8GlcNAc2 (Figure 4). It was notable that glycan composition changes to the glycan-depleted GT1 trimer, compared to the parental trimer, were modest by the standards of what was found in an earlier study of individual glycan site deletions from monomeric gp120 proteins (31). The different behavior of the glycan-clusters in the two Env protein contexts is likely to be attributable to the much greater conformational and steric constraints that apply to native-like trimers (28, 94, 95).
The robustness of the mannose patch on glycan-depleted trimers has implications for immunogen design. Thus, in the example examined here, localized engineering around the predicted epitopes of gl-bNAb expressing B-cells does not adversely influence glycan processing in any global way. Hence, many glycan-dependent epitopes will be present or almost fully assembled at the earliest stages of immunization. The preservation of glycan-dependent, or glycan-influenced epitopes despite engineered changes that affect the surrounding glycans may be a key element in understanding how to manipulate the B-cell response when the goal is to preserve or tolerate the trimer’s glycan shield.
Supplementary Material
Figure S1. Glycan sequencing of BG505 SOSIP.v4.1-GT1 trimers by exoglycosidase digestion
Table S1. Overview of the thermostability of BG505 SOSIP trimers
Table S2. Relative abundances of oligomannose-type glycan on BG505 SOSIP trimers
Table S3. Library of glycan structures identified on BG505 SOSIP.v4.1-GT1 and SOSIP.664 trimers
Table S4. N-linked glycopeptide compositions of trypsin- and chymotrypsin-digested BG505 SOSIP.v4.1-GT1 and SOSIP.664 trimers identified by LC-ESI MS
Acknowledgments
We thank Prof. Raymond A. Dwek FRS for insightful discussions and support. A.J.B. is the recipient of the Chris Scanlan Memorial Scholarship from Corpus Christi College, Oxford. M.C., A.B.W, and I.A.W. are supported by the Scripps CHAVI-ID (1UM1AI100663), M.C. by the International AIDS Vaccine Initiative (IAVI) Neutralizing Antibody Center CAVD grant (Glycan characterization and Outer Domain glycoform design; agreement 1981) and IAVI (VxPDC agreement 2402), and J.P.M., R.W.S., A.B.W. and I.A.W. by NIH HIVRAD grant P01 AI110657. This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 681137. M.M.-R. is a recipient of a fellowship from the Consejo Nacional de Ciencia y Tecnologıa (CONACyT) of Mexico. R.W.S. is a recipient of a Vidi grant from the Netherlands Organization for Scientific Research (NWO) and a Starting Investigator Grant from the European Research Council (ERC-StG-2011–280829-SHEV). N.Z. and A.K. were funded by Unither Virology.
Footnotes
AUTHORS CONTRIBUTIONS
A.-J.B., D.J.H., A.K., M.-M.R. A.C., K.M., V.-C.P., G.O. performed experimental work. A.J.B., D.J.H., M.C. analyzed data. A.J.B., I.A.W., N.Z., R.W.S., J.P.M, and M.C wrote the paper. I.A.W., A.B.W. J.P.M., R.W.S. and M.C. designed the study. All authors read and approved the final manuscript.
REFRENCES
- 1.Mikell I, Sather DN, Kalams SA, Altfeld M, Alter G, Stamatatos L. Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathogens. 2011;7(1):e1001251. doi: 10.1371/journal.ppat.1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landais E, Huang X, Havenar-Daughton C, Murrell B, Price MA, Wickramasinghe L, Ramos A, Bian CB, Simek M, Allen S, Karita E, Kilembe W, Lakhi S, Inambao M, Kamali A, Sanders EJ, Anzala O, Edward V, Bekker LG, Tang J, Gilmour J, Kosakovsky-Pond SL, Phung P, Wrin T, Crotty S, Godzik A, Poignard P. Broadly Neutralizing Antibody Responses in a Large Longitudinal Sub-Saharan HIV Primary Infection Cohort. PLOS Pathogens. 2016;12(1):e1005369. doi: 10.1371/journal.ppat.1005369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gautam R, Nishimura Y, Pegu A, Nason MC, Klein F, Gazumyan A, Golijanin J, Buckler-White A, Sadjadpour R, Wang K, Mankoff Z, Schmidt SD, Lifson JD, Mascola JR, Nussenzweig MC, Martin MA. A single injection of anti-HIV-1 antibodies protects against repeated SHIV challenges. Nature. 2016;533(7601):105–109. doi: 10.1038/nature17677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moldt B, Rakasz EG, Schultz N, Chan-Hui PY, Swiderek K, Weisgrau KL, Piaskowski SM, Bergman Z, Watkins DI, Poignard P, Burton DR. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proceedings of the National Academy of Sciences, USA. 2012;109(46):18921–5. doi: 10.1073/pnas.1214785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hessell AJ, Rakasz EG, Poignard P, Hangartner L, Landucci G, Forthal DN, Koff WC, Watkins DI, Burton DR. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathogens. 2009;5(5):e1000433. doi: 10.1371/journal.ppat.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342(6165):1477–83. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342(6165):1484–90. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JH, Ozorowski G, Ward AB. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science. 2016;351(6277):1043–8. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP. 664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathogens. 2013;9(9):e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lasky LA, Groopman JE, Fennie CW, Benz PM, Capon DJ, Dowbenko DJ, Nakamura GR, Nunes WM, Renz ME, Berman PW. Neutralization of the AIDS retrovirus by antibodies to a recombinant envelope glycoprotein. Science. 1986;233(4760):209–12. doi: 10.1126/science.3014647. [DOI] [PubMed] [Google Scholar]
- 11.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422(6929):307–12. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 12.Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393(6686):705–11. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 13.Blattner C, Lee JH, Sliepen K, Derking R, Falkowska E, de la Pena AT, Cupo A, Julien JP, van Gils M, Lee PS, Peng W, Paulson JC, Poignard P, Burton DR, Moore JP, Sanders RW, Wilson IA, Ward AB. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity. 2014;40(5):669–80. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, Ramirez A, Derking R, van Gils MJ, Liang CH, McBride R, von Bredow B, Shivatare SS, Wu CY, Chan-Hui PY, Liu Y, Feizi T, Zwick MB, Koff WC, Seaman MS, Swiderek K, Moore JP, Evans D, Paulson JC, Wong CH, Ward AB, Wilson IA, Sanders RW, Poignard P, Burton DR. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity. 2014;40(5):657–68. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garces F, Sok D, Kong L, McBride R, Kim HJ, Saye-Francisco KF, Julien JP, Hua Y, Cupo A, Moore JP, Paulson JC, Ward AB, Burton DR, Wilson IA. Structural evolution of glycan recognition by a family of potent HIV antibodies. Cell. 2014;159(1):69–79. doi: 10.1016/j.cell.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong L, Lee JH, Doores KJ, Murin CD, Julien JP, McBride R, Liu Y, Marozsan A, Cupo A, Klasse PJ, Hoffenberg S, Caulfield M, King CR, Hua Y, Le KM, Khayat R, Deller MC, Clayton T, Tien H, Feizi T, Sanders RW, Paulson JC, Moore JP, Stanfield RL, Burton DR, Ward AB, Wilson IA. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nature Structural & Molecular Biology. 2013;20(7):796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O’Dell S, Patel N, Shahzad-ul-Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang GY, Diwanji D, Georgiev I, Kwon YD, Lee D, Louder MK, Moquin S, Schmidt SD, Yang ZY, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang LX, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480(7377):336–43. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PNP, Spencer DIR, Seaman MS, Schuitemaker H, Feizi T, Nussenzweig MC, Bjorkman PJ. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proceedings of the National Academy of Sciences, USA. 2012;109(47):3268–77. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pancera M, Shahzad-Ul-Hussan S, Doria-Rose NA, McLellan JS, Bailer RT, Dai K, Loesgen S, Louder MK, Staupe RP, Yang Y, Zhang B, Parks R, Eudailey J, Lloyd KE, Blinn J, Alam SM, Haynes BF, Amin MN, Wang LX, Burton DR, Koff WC, Nabel GJ, Mascola JR, Bewley CA, Kwong PD. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1–V2-directed antibody PG16. Nature structural & molecular biology. 2013;20(7):804–13. doi: 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, Depetris R, Katpally U, Marozsan A, Cupo A, Maloveste S, Liu Y, McBride R, Ito Y, Sanders RW, Ogohara C, Paulson JC, Feizi T, Scanlan CN, Wong CH, Moore JP, Olson WC, Ward AB, Poignard P, Schief WR, Burton DR, Wilson IA. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334(6059):1097–103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scharf L, Scheid JF, Lee JH, West AP, Chen C, Gao H, Gnanapragasam PNP, Mares R, Seaman MS, Ward AB, Nussenzweig MC, Bjorkman PJ. Antibody 8ANC195 reveals a site of broad vulnerability on the HIV-1 envelope spike. Cell Reports. 2014;7(3):785–95. doi: 10.1016/j.celrep.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477(7365):466–70. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, Georgiev IS, Chuang GY, Druz A, Doria-Rose NA, Laub L, Sliepen K, van Gils MJ, de la Peña AT, Derking R, Klasse PJ, Migueles SA, Bailer RT, Alam M, Pugach P, Haynes BF, Wyatt RT, Sanders RW, Binley JM, Ward AB, Mascola JR, Kwong PD, Connors M. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41–gp120 interface. Nature. 2014;515(7525):138–42. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, Wilson IA, Katinger H, Dwek RA, Rudd PM, Burton DR. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1-->2 mannose residues on the outer face of gp120. Journal of Virology. 2002;76(14):7306–21. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, Kelly JW, Rudd PM, Dwek RA, Katinger H, Burton DR, Wilson IA. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300(5628):2065–71. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- 26.Freund NT, Horwitz JA, Nogueira L, Sievers SA, Scharf L, Scheid JF, Gazumyan A, Liu C, Velinzon K, Goldenthal A, Sanders RW, Moore JP, Bjorkman PJ, Seaman MS, Walker BD, Klein F, Nussenzweig MC. A new glycan-dependent CD4-binding site neutralizing antibody exerts pressure on HIV-1 In vivo. Plos Pathogens. 2015;11(10):e1005238. doi: 10.1371/journal.ppat.1005238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H, Cohen AA, Galimidi RP, Gristick HB, Jensen GJ, Bjorkman PJ. Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop. Proceedings of the Academy of Natural Sciences of Philadelphia. 2016;113(46):7151–7158. doi: 10.1073/pnas.1615939113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart-Jones GBE, Soto C, Lemmin T, Chuang GY, Druz A, Kong R, Thomas Paul V, Wagh K, Zhou T, Behrens AJ, Bylund T, Choi Chang W, Davison Jack R, Georgiev Ivelin S, Joyce MG, Kwon Young D, Pancera M, Taft J, Yang Y, Zhang B, Shivatare Sachin S, Shivatare Vidya S, Lee C, Chun D, Wu CY, Bewley Carole A, Burton Dennis R, Koff Wayne C, Connors M, Crispin M, Baxa U, Korber Bette T, Wong CH, Mascola John R, Kwong Peter D. Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B and G. Cell. 2016;165(4):1–14. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gristick HB, von Boehmer L, West AP, Jr, Schamber M, Gazumyan A, Golijanin J, Seaman MS, Fatkenheuer G, Klein F, Nussenzweig MC, Bjorkman PJ. Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nature structural & molecular biology. 2016;23(10):906–915. doi: 10.1038/nsmb.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Behrens AJ, Crispin M. Structural principles controlling HIV envelope glycosylation. Curr Opin Struct Biol. 2017;44:125–133. doi: 10.1016/j.sbi.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pritchard LK, Spencer DI, Royle L, Bonomelli C, Seabright GE, Behrens AJ, Kulp DW, Menis S, Krumm SA, Dunlop DC, Crispin DJ, Bowden TA, Scanlan CN, Ward AB, Schief WR, Doores KJ, Crispin M. Glycan clustering stabilizes the mannose patch of HIV-1 and preserves vulnerability to broadly neutralizing antibodies. Nature Communications. 2015;6:7479. doi: 10.1038/ncomms8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doores KJ, Bonomelli C, Harvey DJ, Vasiljevic S, Dwek RA, Burton DR, Crispin M, Scanlan CN. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proceedings of the National Academy of Sciences, USA. 2010;107(31):13800–5. doi: 10.1073/pnas.1006498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonomelli C, Doores KJ, Dunlop DC, Thaney V, Dwek Ra, Burton DR, Crispin M, Scanlan CN. The glycan shield of HIV is predominantly oligomannose independently of production system or viral clade. PLoS One. 2011;6(8):e23521. doi: 10.1371/journal.pone.0023521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pritchard LK, Vasiljevic S, Ozorowski G, Seabright GE, Cupo A, Ringe R, Kim HJ, Sanders RW, Doores KJ, Burton DR, Wilson IA, Ward AB, Moore JP, Crispin M. Structural constraints determine the glycosylation of HIV-1 envelope trimers. Cell Reports. 2015;11(10):1604–13. doi: 10.1016/j.celrep.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Go EP, Liao HX, Alam SM, Hua D, Haynes BF, Desaire H. Characterization of host-cell line specific glycosylation profiles of early transmitted/founder HIV-1 gp120 envelope proteins. Journal of Proteome Research. 2013;12(3):1223–34. doi: 10.1021/pr300870t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leonard CK, Spellman MW, Riddle L, Harris RJ, Thomas JN, Gregory TJ. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. Journal of Biological Chemistry. 1990;265(18):10373–82. [PubMed] [Google Scholar]
- 37.Go EP, Herschhorn A, Gu C, Castillo-Menendez L, Zhang S, Mao Y, Chen H, Ding H, Wakefield JK, Hua D, Liao HX, Kappes JC, Sodroski J, Desaire H. Comparative analysis of the glycosylation profiles of membrane-anchored HIV-1 envelope glycoprotein trimers and soluble gp140. Journal of Virology. 2015;89(16):8245–57. doi: 10.1128/JVI.00628-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panico M, Bouche L, Binet D, O’Connor MJ, Rahman D, Pang PC, Canis K, North SJ, Desrosiers RC, Chertova E, Keele BF, Bess JW, Jr, Lifson JD, Haslam SM, Dell A, Morris HR. Mapping the complete glycoproteome of virion-derived HIV-1 gp120 provides insights into broadly neutralizing antibody binding. Scientific Reports. 2016;6:32956. doi: 10.1038/srep32956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Go EP, Ding H, Zhang S, Ringe RP, Nicely N, Hua D, Steinbock RT, Golabek M, Alin J, Alam SM, Cupo A, Haynes BF, Kappes JC, Moore JP, Sodroski JG, Desaire H. A glycosylation benchmark profile for HIV-1 envelope glycoprotein production based on eleven Env trimers. Journal of Virology. 2017;91(9):e02428–16. doi: 10.1128/JVI.02428-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pritchard LK, Harvey DJ, Bonomelli C, Crispin M, Doores KJ. Cell- and protein-directed glycosylation of native cleaved HIV-1 envelope. Journal of Virology. 2015;89(17):8932–44. doi: 10.1128/JVI.01190-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Behrens AJ, Vasiljevic S, Pritchard LK, Harvey DJ, Andev RS, Krumm SA, Struwe WB, Cupo A, Kumar A, Zitzmann N, Seabright GE, Kramer HB, Spencer DI, Royle L, Lee JH, Klasse PJ, Burton DR, Wilson IA, Ward AB, Sanders RW, Moore JP, Doores KJ, Crispin M. Composition and antigenic effects of individual glycan sites of a trimeric HIV-1 envelope glycoprotein. Cell Reports. 2016;14(11):2695–2706. doi: 10.1016/j.celrep.2016.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Behrens AJ, Harvey DJ, Milne E, Cupo A, Kumar A, Zitzmann N, Struwe WB, Moore JP, Crispin M. Molecular architecture of the cleavage-dependent mannose patch on a soluble HIV-1 envelope glycoprotein trimer. Journal of Virology. 2017;91(2):e01894–16. doi: 10.1128/JVI.01894-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crispin M, Doores KJ. Targeting host-derived glycans on enveloped viruses for antibody-based vaccine design. Current Opinion in Virology. 2015;11:63–69. doi: 10.1016/j.coviro.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Behrens AJ, Struwe WB, Crispin M. Glycosylation profiling to evaluate glycoprotein immunogens against HIV-1. Expert Rev Proteomics. 2017;14(10):881–890. doi: 10.1080/14789450.2017.1376658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dey AK, Cupo A, Ozorowski G, Sharma VK, Behrens AJ, Go EP, Ketas TJ, Yasmeen A, Klasse PJ, Sayeed E, Desaire H, Crispin M, Wilson IA, Sanders RW, Hassell T, Ward AB, Moore JP. cGMP production and analysis of BG505 SOSIP.664, an extensively glycosylated, trimeric HIV-1 envelope glycoprotein vaccine candidate. Biotechnol Bioeng. 2017 doi: 10.1002/bit.26498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, Arendt H, Kim HJ, Lee JH, Pugach P, Williams M, Debnath G, Moldt B, van Breemen MJ, Isik G, Medina-Ramirez M, Back JW, Koff WC, Julien JP, Rakasz EG, Seaman MS, Guttman M, Lee KK, Klasse PJ, LaBranche C, Schief WR, Wilson IA, Overbaugh J, Burton DR, Ward AB, Montefiori DC, Dean H, Moore JP. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349(6244):154–166. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garces F, Lee JH, de Val N, Torrents de la Pena A, Kong L, Puchades C, Hua Y, Stanfield RL, Burton DR, Moore JP, Sanders RW, Ward AB, Wilson IA. Affinity maturation of a potent family of HIV antibodies is primarily focused on accommodating or avoiding glycans. Immunity. 2015;43(6):1053–63. doi: 10.1016/j.immuni.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scharf L, Wang H, Gao H, Chen S, McDowall AW, Bjorkman PJ. Broadly Neutralizing Antibody 8ANC195 Recognizes Closed and Open States of HIV-1 Env. Cell. 2015;162(6):1379–90. doi: 10.1016/j.cell.2015.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Taeye SW, Ozorowski G, Torrents de la Pena A, Guttman M, Julien JP, van den Kerkhof TL, Burger JA, Pritchard LK, Pugach P, Yasmeen A, Crampton J, Hu J, Bontjer I, Torres JL, Arendt H, DeStefano J, Koff WC, Schuitemaker H, Eggink D, Berkhout B, Dean H, LaBranche C, Crotty S, Crispin M, Montefiori DC, Klasse PJ, Lee KK, Moore JP, Wilson IA, Ward AB, Sanders RW. Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell. 2015;163(7):1702–15. doi: 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanders RW, Moore JP. Native-like Env trimers as a platform for HIV-1 vaccine design. Immunological Reviews. 2017;275(1):161–182. doi: 10.1111/imr.12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, Staupe RP, Altae-Tran HR, Bailer RT, Crooks ET, Cupo A, Druz A, Garrett NJ, Hoi KH, Kong R, Louder MK, Longo NS, McKee K, Nonyane M, O’Dell S, Roark RS, Rudicell RS, Schmidt SD, Sheward DJ, Soto C, Wibmer CK, Yang Y, Zhang Z, Mullikin JC, Binley JM, Sanders RW, Wilson IA, Moore JP, Ward AB, Georgiou G, Williamson C, Abdool Karim SS, Morris L, Kwong PD, Shapiro L, Mascola JR. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509(7498):55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia SM, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BT, Kwong PD, Mascola JR, Haynes BF. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496(7446):469–76. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.MacLeod DT, Choi NM, Briney B, Garces F, Ver LS, Landais E, Murrell B, Wrin T, Kilembe W, Liang CH, Ramos A, Bian CB, Wickramasinghe L, Kong L, Eren K, Wu CY, Wong CH, Kosakovsky Pond SL, Wilson IA, Burton DR, Poignard P. Early Antibody Lineage Diversification and Independent Limb Maturation Lead to Broad HIV-1 Neutralization Targeting the Env High-Mannose Patch. Immunity. 2016;44(5):1215–26. doi: 10.1016/j.immuni.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doria-Rose NA, Mascola JR. HIV Immunology Goes Out On a Limb. Immunity. 2016;44(5):1088–90. doi: 10.1016/j.immuni.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 55.Bonsignori M, Zhou T, Sheng Z, Chen L, Gao F, Joyce MG, Ozorowski G, Chuang GY, Schramm CA, Wiehe K, Alam SM, Bradley T, Gladden MA, Hwang KK, Iyengar S, Kumar A, Lu X, Luo K, Mangiapani MC, Parks RJ, Song H, Acharya P, Bailer RT, Cao A, Druz A, Georgiev IS, Kwon YD, Louder MK, Zhang B, Zheng A, Hill BJ, Kong R, Soto C, Mullikin JC, Douek DC, Montefiori DC, Moody MA, Shaw GM, Hahn BH, Kelsoe G, Hraber PT, Korber BT, Boyd SD, Fire AZ, Kepler TB, Shapiro L, Ward AB, Mascola JR, Liao HX, Kwong PD, Haynes BF. Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody. Cell. 2016;165(2):449–63. doi: 10.1016/j.cell.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haynes BF, Kelsoe G, Harrison SC, Kepler TB. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nature Biotechnology. 2012;30(5):423–33. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao X, Chen W, Feng Y, Dimitrov DS. Maturation Pathways of Cross-Reactive HIV-1 Neutralizing Antibodies. Viruses. 2009;1(3):802–17. doi: 10.3390/v1030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malherbe DC, Doria-Rose NA, Misher L, Beckett T, Puryear WB, Schuman JT, Kraft Z, O’Malley J, Mori M, Srivastava I, Barnett S, Stamatatos L, Haigwood NL. Sequential immunization with a subtype B HIV-1 envelope quasispecies partially mimics the in vivo development of neutralizing antibodies. Journal of Virology. 2011;85(11):5262–74. doi: 10.1128/JVI.02419-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ota T, Doyle-Cooper C, Cooper AB, Huber M, Falkowska E, Doores KJ, Hangartner L, Le K, Sok D, Jardine J, Lifson J, Wu X, Mascola JR, Poignard P, Binley JM, Chakrabarti BK, Schief WR, Wyatt RT, Burton DR, Nemazee D. Anti-HIV B Cell lines as candidate vaccine biosensors. Journal of Immunology. 2012;189(10):4816–24. doi: 10.4049/jimmunol.1202165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Voss JE, Andrabi R, McCoy LE, de Val N, Fuller RP, Messmer T, Su CY, Sok D, Khan SN, Garces F, Pritchard LK, Wyatt RT, Ward AB, Crispin M, Wilson IA, Burton DR. Elicitation of Neutralizing Antibodies Targeting the V2 Apex of the HIV Envelope Trimer in a Wild-Type Animal Model. Cell Rep. 2017;21(1):222–235. doi: 10.1016/j.celrep.2017.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crooks ET, Tong T, Chakrabarti B, Narayan K, Georgiev IS, Menis S, Huang X, Kulp D, Osawa K, Muranaka J, Stewart-Jones G, Destefano J, O’Dell S, LaBranche C, Robinson JE, Montefiori DC, McKee K, Du SX, Doria-Rose N, Kwong PD, Mascola JR, Zhu P, Schief WR, Wyatt RT, Whalen RG, Binley JM. Vaccine-Elicited Tier 2 HIV-1 Neutralizing Antibodies Bind to Quaternary Epitopes Involving Glycan-Deficient Patches Proximal to the CD4 Binding Site. PLoS Pathog. 2015;11(5):e1004932. doi: 10.1371/journal.ppat.1004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCoy LE, van Gils MJ, Ozorowski G, Messmer T, Briney B, Voss JE, Kulp DW, Macauley MS, Sok D, Pauthner M, Menis S, Cottrell CA, Torres JL, Hsueh J, Schief WR, Wilson IA, Ward AB, Sanders RW, Burton DR. Holes in the Glycan Shield of the Native HIV Envelope Are a Target of Trimer-Elicited Neutralizing Antibodies. Cell Rep. 2016;16(9):2327–38. doi: 10.1016/j.celrep.2016.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Medina-Ramirez M, Garces F, Escolano A, Skog P, de Taeye SW, Moral-Sanchez I, Dosenovic P, Hua Y, McGuire AT, Gitlin Y, van der Woude P, Freund NT, Yasmeen A, Behrens AJ, Ozorowski G, Sliepen K, Blane T, Kootstra NA, van Breemen MJ, Pritchard LK, Stanfield RL, Crispin M, Ward AB, Stamatatos L, Klasse PJ, Moore JP, Nemazee D, Nussenzweig MC, Wilson IA, Sanders RW. Design and crystal structure of a native-like HIV-1 envelope trimer that engages multiple broadly neutralizing antibody precursors in vivo. Journal of Experimental Medicine. 2017;214(9):2573–90. doi: 10.1084/jem.20161160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, Nieusma T, Mathison J, Baker D, Ward AB, Burton DR, Stamatatos L, Nemazee D, Wilson IA, Schief WR. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340(6133):711–6. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Y, O’Dell S, Walker LM, Wu X, Guenaga J, Feng Y, Schmidt SD, McKee K, Louder MK, Ledgerwood JE, Graham BS, Haynes BF, Burton DR, Wyatt RT, Mascola JR. Mechanism of neutralization by the broadly neutralizing HIV-1 monoclonal antibody VRC01. Journal of Virology. 2011;85(17):8954–67. doi: 10.1128/JVI.00754-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Joyce MG, Kanekiyo M, Xu L, Biertumpfel C, Boyington JC, Moquin S, Shi W, Wu X, Yang Y, Yang ZY, Zhang B, Zheng A, Zhou T, Zhu J, Mascola JR, Kwong PD, Nabel GJ. Outer domain of HIV-1 gp120: antigenic optimization, structural malleability, and crystal structure with antibody VRC-PG04. Journal of Virology. 2013;87(4):2294–306. doi: 10.1128/JVI.02717-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McGuire AT, Hoot S, Dreyer AM, Lippy A, Stuart A, Cohen KW, Jardine J, Menis S, Scheid JF, West AP, Schief WR, Stamatatos L. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J Exp Med. 2013;210(4):655–63. doi: 10.1084/jem.20122824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Travers SA. Conservation, compensation, and evolution of N-linked glycans in the HIV-1 group M subtypes and circulating recombinant forms. ISRN AIDS. 2012;2012:823605. doi: 10.5402/2012/823605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou T, Doria-Rose NA, Cheng C, Stewart-Jones GBE, Chuang GY, Chambers M, Druz A, Geng H, McKee K, Kwon YD, O’Dell S, Sastry M, Schmidt SD, Xu K, Chen L, Chen RE, Louder MK, Pancera M, Wanninger TG, Zhang B, Zheng A, Farney SK, Foulds KE, Georgiev IS, Joyce MG, Lemmin T, Narpala S, Rawi R, Soto C, Todd JP, Shen CH, Tsybovsky Y, Yang Y, Zhao P, Haynes BF, Stamatatos L, Tiemeyer M, Wells L, Scorpio DG, Shapiro L, McDermott AB, Mascola JR, Kwong PD. Quantification of the Impact of the HIV-1-Glycan Shield on Antibody Elicitation. Cell Rep. 2017;19(4):719–732. doi: 10.1016/j.celrep.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cao L, Diedrich JK, Kulp DW, Pauthner M, He L, Park SR, Sok D, Su CY, Delahunty CM, Menis S, Andrabi R, Guenaga J, Georgeson E, Kubitz M, Adachi Y, Burton DR, Schief WR, Yates JR, III, Paulson JC. Global site-specific N-glycosylation analysis of HIV envelope glycoprotein. Nat Commun. 2017;8:14954. doi: 10.1038/ncomms14954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chung NPY, Matthews K, Kim HJ, Ketas TJ, Golabek M, de Los Reyes K, Korzun J, Yasmeen A, Sanders RW, Klasse PJ, Wilson Ia, Ward AB, Marozsan AJ, Moore JP, Cupo A. Stable 293 T and CHO cell lines expressing cleaved, stable HIV-1 envelope glycoprotein trimers for structural and vaccine studies. Retrovirology. 2014;11:33–33. doi: 10.1186/1742-4690-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ringe RP, Yasmeen A, Ozorowski G, Go EP, Pritchard LK, Guttman M, Ketas TA, Cottrell CA, Wilson IA, Sanders RW, Cupo A, Crispin M, Lee KK, Desaire H, Ward AB, Klasse PJ, Moore JP. Influences on the design and purification of soluble, recombinant native-like HIV-1 envelope glycoprotein trimers. Journal of Virology. 2015;89(23):12189–210. doi: 10.1128/JVI.01768-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, Lu M, Moore JP. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. Journal of Virology. 2002;76(17):8875–89. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pugach P, Ozorowski G, Cupo A, Ringe R, Yasmeen A, de Val N, Derking R, Kim HJ, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, Julien JP, Burton DR, Wilson IA, Sanders RW, Klasse PJ, Ward AB, Moore JP. A native-like SOSIP. 664 trimer based on an HIV-1 subtype B env gene. Journal of Virology. 2015;89(6):3380–95. doi: 10.1128/JVI.03473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Neville DC, Dwek RA, Butters TD. Development of a single column method for the separation of lipid- and protein-derived oligosaccharides. Journal of Proteome Research. 2009;8(2):681–7. doi: 10.1021/pr800704t. [DOI] [PubMed] [Google Scholar]
- 76.Harvey DJ. Fragmentation of negative ions from carbohydrates: part 2. Fragmentation of high-mannose N-linked glycans. Journal of the American Society for Mass Spectrometry. 2005;16(5):631–46. doi: 10.1016/j.jasms.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 77.Harvey DJ. Fragmentation of negative ions from carbohydrates: part 1. Use of nitrate and other anionic adducts for the production of negative ion electrospray spectra from N-linked carbohydrates. Journal of the American Society for Mass Spectrometry. 2005;16(5):622–30. doi: 10.1016/j.jasms.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 78.Harvey DJ. Fragmentation of negative ions from carbohydrates: part 3. Fragmentation of hybrid and complex N-linked glycans . J Am Soc Mass Spectrom. 2005;16(5):647–59. doi: 10.1016/j.jasms.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 79.Harvey DJ, Royle L, Radcliffe CM, Rudd PM, Dwek RA. Structural and quantitative analysis of N-linked glycans by matrix-assisted laser desorption ionization and negative ion nanospray mass spectrometry. Analytical biochemistry. 2008;376(1):44–60. doi: 10.1016/j.ab.2008.01.025. [DOI] [PubMed] [Google Scholar]
- 80.Lee JH, de Val N, Lyumkis D, Ward AB. Model building and refinement of a natively glycosylated HIV-1 Env protein by high-resolution cryoelectron microscopy. Structure. 2015;23(10):1943–51. doi: 10.1016/j.str.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harvey DJ, Sobott F, Crispin M, Wrobel A, Bonomelli C, Vasiljevic S, Scanlan CN, Scarff CA, Thalassinos K, Scrivens JH. Ion mobility mass spectrometry for extracting spectra of N-glycans directly from incubation mixtures following glycan release: application to glycans from engineered glycoforms of intact, folded HIV gp120. Journal of the American Society for Mass Spectrometry. 2011;22(3):568–81. doi: 10.1007/s13361-010-0053-0. [DOI] [PubMed] [Google Scholar]
- 82.Wheeler SF, Harvey DJ. Negative ion mass spectrometry of sialylated carbohydrates: discrimination of N-acetylneuraminic acid linkages by MALDI-TOF and ESI-TOF mass spectrometry. Analytical chemistry. 2000;72(20):5027–39. doi: 10.1021/ac000436x. [DOI] [PubMed] [Google Scholar]
- 83.Domon B, Costello CE. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconjugate Journal. 1988;5(4):397–409. [Google Scholar]
- 84.Hossler P, Khattak SF, Li ZJ. Optimal and consistent protein glycosylation in mammalian cell culture. Glycobiology. 2009;19(9):936–49. doi: 10.1093/glycob/cwp079. [DOI] [PubMed] [Google Scholar]
- 85.Croset A, Delafosse L, Gaudry JP, Arod C, Glez L, Losberger C, Begue D, Krstanovic A, Robert F, Vilbois F, Chevalet L, Antonsson B. Differences in the glycosylation of recombinant proteins expressed in HEK and CHO cells. Journal of Biotechnology. 2012;161(3):336–48. doi: 10.1016/j.jbiotec.2012.06.038. [DOI] [PubMed] [Google Scholar]
- 86.Kong L, Wilson IA, Kwong PD. Crystal structure of a fully glycosylated HIV-1 gp120 core reveals a stabilizing role for the glycan at Asn262. Proteins. 2015;83(3):590–6. doi: 10.1002/prot.24747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Coss KP, Vasiljevic S, Pritchard LK, Krumm SA, Glaze M, Madzorera S, Moore PL, Crispin M, Doores KJ. HIV-1 glycan density drives the persistence of the mannose patch within an infected individual. Journal of Virology. 2016;90(24):11132–11144. doi: 10.1128/JVI.01542-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Geijtenbeek TB, Kwon DS, Torensma R, van Vliet SJ, van Duijnhoven GC, Middel J, Cornelissen IL, Nottet HS, KewalRamani VN, Littman DR, Figdor CG, van Kooyk Y. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100(5):587–97. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 89.Yu Q, Yu R, Qin X. The good and evil of complement activation in HIV-1 infection. Cellular & Molecular Immunology. 2010;7(5):334–40. doi: 10.1038/cmi.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schmitz J, Zimmer JP, Kluxen B, Aries S, Bogel M, Gigli I, Schmitz H. Antibody-dependent complement-mediated cytotoxicity in sera from patients with HIV-1 infection is controlled by CD55 and CD59. J Clin Invest. 1995;96(3):1520–6. doi: 10.1172/JCI118190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pinter C, Siccardi AG, Lopalco L, Longhi R, Clivio A. HIV glycoprotein 41 and complement factor H interact with each other and share functional as well as antigenic homology. AIDS Res Hum Retroviruses. 1995;11(8):971–80. doi: 10.1089/aid.1995.11.971. [DOI] [PubMed] [Google Scholar]
- 92.Stoiber H, Clivio A, Dierich MP. Role of complement in HIV infection. Annual review of immunology. 1997;15:649–74. doi: 10.1146/annurev.immunol.15.1.649. [DOI] [PubMed] [Google Scholar]
- 93.Xiang Y, Karaveg K, Moremen KW. Substrate recognition and catalysis by GH47 alpha-mannosidases involved in Asn-linked glycan maturation in the mammalian secretory pathway. Proceedings of the National Academy of Sciences, USA. 2016;113(49):E7890–99. doi: 10.1073/pnas.1611213113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Go EP, Cupo A, Ringe R, Pugach P, Moore JP, Desaire H. Native Conformation and Canonical Disulfide Bond Formation Are Interlinked Properties of HIV-1 Env Glycoproteins. J Virol. 2015;90(6):2884–94. doi: 10.1128/JVI.01953-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lemmin T, Soto C, Stuckey J, Kwong PD. Microsecond Dynamics and Network Analysis of the HIV-1 SOSIP Env Trimer Reveal Collective Behavior and Conserved Microdomains of the Glycan Shield. Structure. 2017;25(10):1631–1639e2. doi: 10.1016/j.str.2017.07.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Glycan sequencing of BG505 SOSIP.v4.1-GT1 trimers by exoglycosidase digestion
Table S1. Overview of the thermostability of BG505 SOSIP trimers
Table S2. Relative abundances of oligomannose-type glycan on BG505 SOSIP trimers
Table S3. Library of glycan structures identified on BG505 SOSIP.v4.1-GT1 and SOSIP.664 trimers
Table S4. N-linked glycopeptide compositions of trypsin- and chymotrypsin-digested BG505 SOSIP.v4.1-GT1 and SOSIP.664 trimers identified by LC-ESI MS