Abstract
Understanding the strategies used by bacteria to degrade polysaccharides constitutes an invaluable tool for biotechnological applications. Bacteria are major mediators of polysaccharide degradation in nature; however, the complex mechanisms used to detect, degrade, and consume these substrates are not well-understood, especially for recalcitrant polysaccharides such as chitin. It has been previously shown that the model bacterial saprophyte Cellvibrio japonicus is able to catabolize chitin, but little is known about the enzymatic machinery underlying this capability. Previous analyses of the C. japonicus genome and proteome indicated the presence of four glycoside hydrolase family 18 (GH18) enzymes, and studies of the proteome indicated that all are involved in chitin utilization. Using a combination of in vitro and in vivo approaches, we have studied the roles of these four chitinases in chitin bioconversion. Genetic analyses showed that only the chi18D gene product is essential for the degradation of chitin substrates. Biochemical characterization of the four enzymes showed functional differences and synergistic effects during chitin degradation, indicating non-redundant roles in the cell. Transcriptomic studies revealed complex regulation of the chitin degradation machinery of C. japonicus and confirmed the importance of CjChi18D and CjLPMO10A, a previously characterized chitin-active enzyme. With this systems biology approach, we deciphered the physiological relevance of the glycoside hydrolase family 18 enzymes for chitin degradation in C. japonicus, and the combination of in vitro and in vivo approaches provided a comprehensive understanding of the initial stages of chitin degradation by this bacterium.
Keywords: chitin, chitinase, bacterial genetics, bacterial metabolism, glycoside hydrolase, polysaccharide, Cellvibrio japonicus
Introduction
Chitin, a linear polymer of β(1–4)-linked N-acetylglucosamine, is the second most abundant polysaccharide on earth after cellulose and a major source of fixed carbon and nitrogen, especially in the oceans (1). The rate of chitin degradation has been calculated to match the rate of synthesis, resulting in little net accumulation of chitin in the environment, suggesting that the microorganisms that derive nutrition from this polysaccharide are able to degrade it with high efficiency (2). Moreover, chitin is increasingly being recognized as an important feedstock for the production of renewable chemicals, and chito-oligosaccharides are being investigated for several biomedical applications (3–6). The strategies used by microbes for efficient chitin bioconversion are gaining interest, because current chemical methods used for the industrial processing of chitin are inefficient and wasteful (7, 8). Therefore, the combination of environmental importance and industrial/medical relevance has renewed interest in the bioconversion of chitin (9, 10).
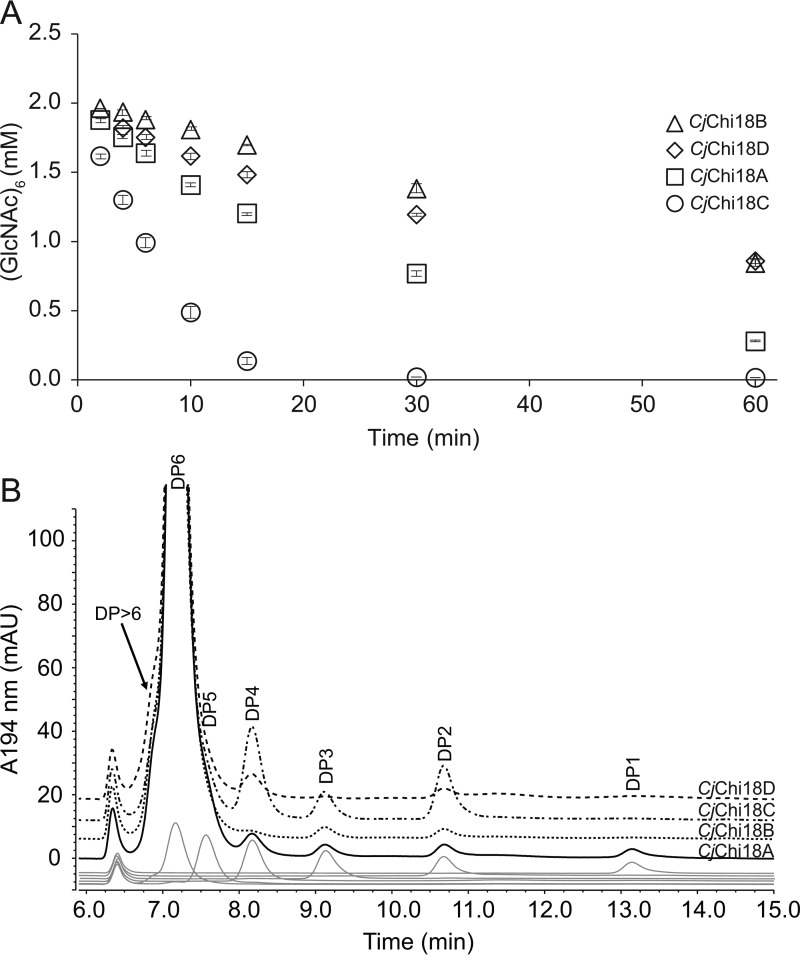
Depolymerization of chitin-rich substrates has been studied predominately from biochemical and structural perspectives, and as a consequence there is considerable understanding on certain mechanistic aspects of enzymatic degradation (11). Chitin deconstruction shares many features with cellulose degradation, which is not surprising given that chitin has a crystalline structure similar to that of cellulose and differs chemically via an N-acetyl substitution at the C2 carbon (12). Specifically, chitin depolymerization is achieved through a concerted effort of endo- and exo-acting enzymes to reduce polymer length to short oligosaccharides that are then converted to N-acetylglucosamine monomers by hexosaminidases for entry into cellular metabolism (Fig. 1) (6). Additionally, the action of lytic polysaccharide monooxygenases (LPMO)2 in aerobic microbes promotes chitin degradation by acting on crystalline regions of the substrate (13–15).
Figure 1.
Proposed model for chitin utilization in C. japonicus. CjChi18D (green) and CjLPMO10A (yellow) work together to disrupt the crystalline structure of chitin and degrade less accessible chitin fibers, whereas CjChi18B (magenta) and CjChi18C (brown) are acting on the more accessible chitin fibers to produce chito-oligosaccharides, which are taken up into the periplasm space. CjChi18A (orange) generates GlcNAc, and this lipoprotein may be acting in the periplasmic space (as shown here) or may be located on the outer side of the outer membrane and act outside the cell. IM, inner membrane; OM, outer membrane. Transporter icons (white) and phospholipids (gray) are adapted from Servier Medical Art under a Creative Commons Attribution 3.0 Unported License (https://creativecommons.org/licenses/by/3.0/).
Physiological and genetic aspects of chitin degradation, especially for terrestrial bacteria, are not well-characterized. One reason for this lack of knowledge is the multiplicity of chitinolytic enzymes in many, but not all, chitin-degrading bacteria, which makes functional analysis of the individual enzymes challenging. Although several model organisms have been used to study chitin degradation (16–18), one that is emerging as a powerful model, because of the available systems biology tools, is the saprophytic Gram-negative bacterium Cellvibrio japonicus (19, 20). This bacterium is a potent chitin degrader (13) and has a suite of nine genes encoding enzymes with predicted functions in chitin degradation (21). Specifically, the C. japonicus genome is predicted to encode four chitinases from the glycoside hydrolase family 18 (GH18), one GH19 chitinase, two hexosaminidases (GH20), one chitosanase (GH46), and one chitin-specific LPMO (auxiliary activity, AA10) (21). Secretome analysis (22) has shown that the four GH18s, and to a much lesser extent the GH19, are abundant during growth on chitin. Generally, the GH18s and LPMOs are considered key enzymes for bioconversion of crystalline chitin. The physiological role of GH19 enzymes in chitin degradation is less clear (23, 24).
The possession of large numbers of carbohydrate active enzymes (CAZymes) (25) is a hallmark feature of C. japonicus. There is increasing evidence that the CAZymes of C. japonicus that belong to the same GH family are not functionally redundant but have unique physiological functions (14, 26). In the current study, the physiological roles of the four C. japonicus GH18 chitinases were determined focusing on the initial stages of chitin degradation. Sequence analysis indicates that CjChi18B CjChi18C and CjChi18D are secreted enzymes containing at least one chitin-specific carbohydrate-binding module (CBM), whereas CjChi18A is a single domain protein that likely is membrane-anchored (Fig. 2). Through RNAseq analysis, we determined that these four chitinases were highly up-regulated while chitin was being used as the sole carbon source. Combinatorial mutational analyses determined that only the chi18D gene product is essential for chitin degradation. Biochemical characterization of the catalytic domains of these four GH18 chitinases indicated that CjChi18D is substantially more active toward insoluble chitin than the other GH18 chitinases, underpinning its importance in chitin degradation. The functional insights into the initial stages of chitin degradation by saprophytic bacteria that are provided in this study have the potential to accelerate the development of industrial chitin bioconversion strategies.
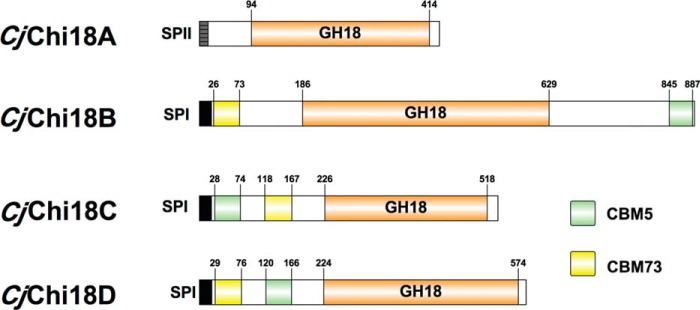
Figure 2.
Diverse architecture of the family GH18 chitinases of C. japonicus. CAZy domain representation of the family GH18 chitinases of C. japonicus is shown. The indicated domains are as follows: GH18, family GH18 catalytic domain; CBM5, chitin-binding domain; CBM73, chitin-binding domain; SPI, signal peptide, type I; SPII, signal peptide, type II. This image was generated using IBS Illustrator (65).
Results
CjChi18D is essential for the degradation of α-chitin
According to previous sequence analysis (21), C. japonicus possesses four GH18 chitinases (CjChi18A, CjChi18B, CjChi18C, and CjChi18D). A recent proteomic study (22) detected the four enzymes in the secretome of C. japonicus growing on α- and β-chitin, which suggested the importance of these enzymes for efficient chitin degradation. To elucidate the physiological relevance of these enzymes, we generated a suite of GH18 deletion mutants and assessed their fitness on insoluble chitin substrates, including an environmentally relevant substrate, crab shell.
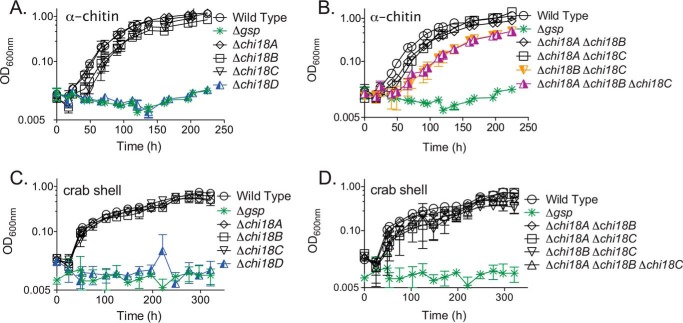
Wildtype and GH18 deletion mutant strains all grew well in defined media with either glucose (Glc) or GlcNAc as the sole source of carbon (Fig. S1). A Δgsp mutant, which lacks the entire 9.4 kb gsp operon encoding the type II secretion system that is needed for secretion of most enzymes, was also able to grow on these monosaccharides, as previously demonstrated (13, 27). When the GH18 single deletion mutants were grown using insoluble α-chitin or crab shell as the sole carbon source, distinct phenotypes emerged (Fig. 3). When α-chitin was the sole source of carbon, the Δchi18D mutant was unable to grow. The other three single deletion strains (Δchi18A, Δchi18B, and Δchi18C) all had similar growth rates and maximum optical densities (OD) as the wildtype (Table S1A). Interestingly, the Δchi18B and Δchi18C single mutant strains had more protracted lag phases than the wildtype strain when using α-chitin as carbon source (Fig. 3A). When using crab shells as the only source of carbon, the Δchi18D mutant was unable to grow, whereas the single mutants of Δchi18A, Δchi18B, and Δchi18C displayed growth similar to wildtype (Fig. 3C and Table S1B).
Figure 3.
Growth of C. japonicus mutants on chitin. Deletion mutants were grown using MOPS minimal medium supplemented with 0.25% α-chitin (A and B) or 4% crab shell (C and D). A and C show single mutants; B and D show multiple mutants. All experiments were performed in biological triplicate; error bars represent standard deviations but in many cases are too small to be observed. These growth experiments were performed simultaneously but are separated into multiple panels for clarity. As a consequence, the same control strains (wildtype and Δgsp) are repeated in each panel for a given substrate.
To test for functional redundancies between the GH18 chitinases, we generated all combinations of double deletion mutants and then assessed growth using α-chitin or crab shells as the sole carbon source. The double mutant Δchi18B Δchi18C resulted in a slower growth rate and a longer lag phase when grown on α-chitin in comparison to either of the single deletion mutants or the wildtype (Fig. 3B). The growth rate of the Δchi18B Δchi18C double mutant was reduced 37% compared with wildtype (Table S1A). The Δchi18A Δchi18B double mutant grew like the Δchi18B single mutant, whereas the Δchi18A Δchi18C double mutant grew like the Δchi18C single mutant. The Δchi18A Δchi18B Δchi18C triple mutant recapitulated the growth defect observed in the Δchi18B Δchi18C double mutant in terms of the growth rate, lag phase, and the maximum OD (Fig. 3B and Table S1A). These results suggest that the chi18B and chi18C gene products have non-redundant functions during degradation of α-chitin, whereas the chi18A gene product does not play a rate-limiting role.
Interestingly, the double mutants exhibited wildtype-like phenotypes when grown using crab shells as the sole carbon source (Fig. 3D). The growth rates of the Δchi18B and Δchi18C single mutants and the Δchi18B Δchi18C double mutant were similar to the wildtype strain when grown using crab shells. For all tested strains, the growth rates when using crab shells as the sole carbon source were substantially reduced compared with α-chitin. The results of experiments using β-chitin (Fig. S2) were very similar to the results obtained with α-chitin.
CjChi18D is a potent secreted chitinase
Sequence alignment of the catalytic domains of the four chitinase showed large sequence variation. CjChi18A and CjChi18D share higher sequence identity (32%), whereas CjChi18C and CjChi18D showed the lowest identity (19%), as determined by BLAST alignment (28). As predicted by LipoP (29) and SignalP (30) software tools, all four of the C. japonicus GH18 chitinases have a signal sequence; however, CjChi18A has a SPaseII-cleaveable sequence and is predicted to be an outer membrane associated lipoprotein. The CjChi18B, CjChi18C, and CjChi18D enzymes have a SPaseI-cleaved sequence (29) and are predicted to be secreted. The domain structure of the four GH18 chitinases is summarized in Fig. 2. The LipoP/SignalP software predictions are in agreement with a proteomics study that showed the occurrence of all four GH18 chitinases in the secretome (22).
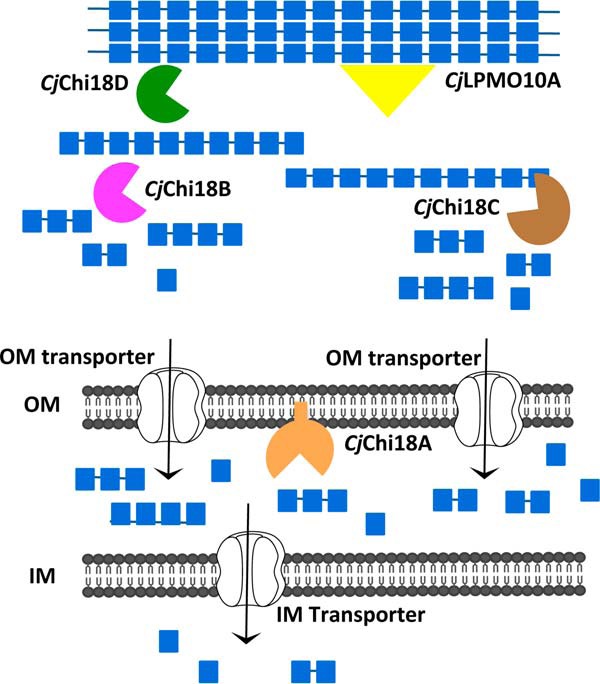
To further assess the contribution of individual GH18 enzymes as effectors of chitin degradation, we used our suite of GH18 mutants to assess secreted activity using colloidal chitin plate assays (Fig. 4 and Table S2). A wildtype strain generated a robust zone of clearing, whereas a Δgsp secretion-deficient mutant generated no zone of clearing. We observed a similarly striking phenotype with the Δchi18D single mutant strain, which also generated no zone of clearing. The remaining GH18 single mutants displayed approximately wildtype zones of clearing. Interestingly, relative to the wildtype, the Δchi18B Δchi18C double mutant showed a clear reduction in the size of the clearing zone. The Δchi18A Δchi18B Δchi18C triple mutant displayed a zone of clearing similar to the Δchi18B Δchi18C double mutant. The reduced chitin-degrading capacities of the Δchi18B Δchi18C double mutant and the Δchi18A Δchi18B Δchi18C triple mutant are in agreement with the observed growth defects of these strains when grown on α-chitin.
Figure 4.
Chitinase secretion of C. japonicus GH18 mutants. Strains were grown on a plate that contained MOPS with 1.5% agar, 2% colloidal chitin, and 0.2% glucose. After incubation at 30 °C for 5 days, the plates were stained with Congo red. This experiment was conducted in biological triplicate, and quantification of the clearing zones is shown in Table S2.
The GH18 chitinases have different activities toward chitin and (GlcNAc)6
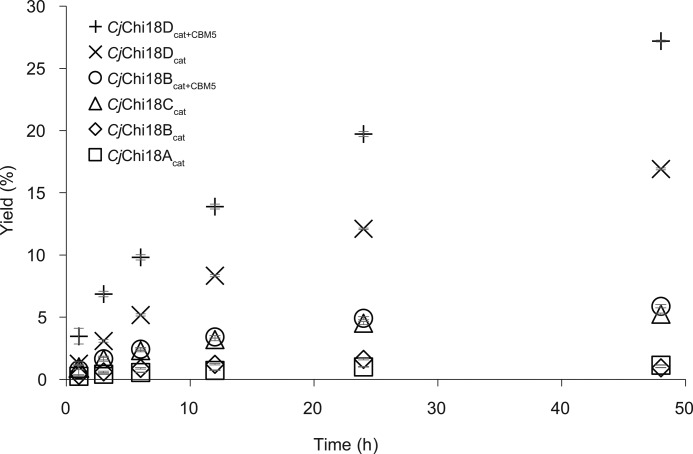
To further investigate the features of the four GH18 chitinases, they were cloned, expressed, and purified for biochemical characterization. Despite substantial efforts, soluble full-length multidomain CjChi18B, CjChi18C, and CjChi18D proteins could not be obtained. Therefore comparative biochemical analysis was conducted with overexpressed catalytic domains. All four chitinases were able to degrade α-chitin, although with greatly varying efficiency, but were clearly highest for CjChi18D (Fig. 5). As expected (31), versions of CjChi18B and CjChi18D containing their CBM5 domain gave higher yields compared with their respective solitary catalytic domains. The catalytic domain of CjChi18C gave higher yields than the catalytic domains of CjChi18A and CjChi18B, but all were poor in chitin degradation compared with CjChi18D.
Figure 5.
Degradation of α-chitin. Degradation of α-chitin (15 g/liter) at 30 °C was tested in 20 mm BisTris, pH 6.5, 0.1 mg/ml BSA. The enzyme concentration was 0.5 μm, and samples were taken at different time points. The yield refers to the degree of chitin solubilization. The α-chitin used contained 6.43% ash and 5.42% moisture, and this was taken into account when calculating yields. Each reaction was performed in triplicate; standard deviations are shown as error bars but are difficult to see because they are low and are partly covered by the symbols.
Analysis of products generated during degradation of α-chitin showed that CjChi18Acat initially produced both GlcNAc and (GlcNAc)2. The product profiles for the other variants (CjChi18Bcat, CjChi18Bcat+CBM5, CjChi18Ccat, CjChi18Dcat, and CjChi18Dcat+CBM5) showed mainly (GlcNAc)2 and smaller amounts of GlcNAc (Fig. S3), as is usual for GH18 chitinases. It is noteworthy that the product profile of CjChi18Bcat was different from CjChi18Bcat+CBM5 after 48 h, where GlcNAc is the dominating product for CjChi18Bcat, whereas (GlcNAc)2 is the dominating product for CjChi18Bcat+CBM5 (Fig. S3). For CjChi18Acat, only GlcNAc was detected after 48 h, indicating an N-acetylhexosaminidase activity for this enzyme.
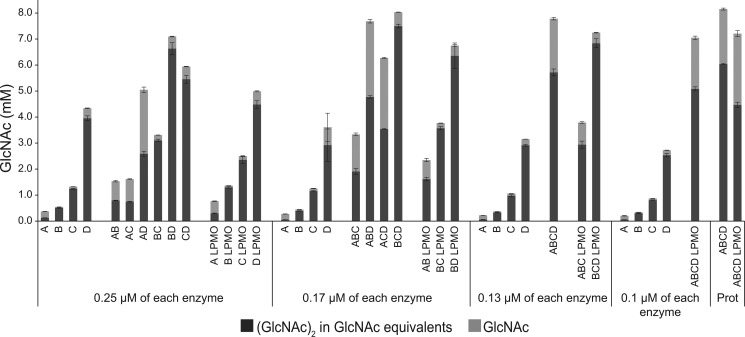
Synergy experiments (Fig. 6) confirmed the dominating role of CjChi18D in chitin bioconversion and revealed synergistic effects for various enzyme combinations, such as when mixing CjChi18Acat with CjChi18Bcat, CjChi18Bcat with CjChi18Ccat, and CjChi18Bcat with CjChi18Dcat. These synergistic effects indicate that the various chitinases must have different functionalities. As expected on the basis of the data presented above, the presence of CjChi18Acat shifted the product profile toward a higher GlcNAc/(GlcNAc)2 ratio. The effect of adding CjLPMO10A was generally small but seemed slightly larger when combined with the individual catalytic domains of CjChi18A, CjChi18B, and CjChi18C, compared with CjChi18D. Although CjChi18D alone was the most potent individual enzyme, the highest chitin solubilization yields were obtained when CjChi18D was paired with other chitinases.
Figure 6.
Synergy experiments. The catalytic domains of all chitinases and the CjLPMO10A were mixed in different ways to investigate possible synergistic effects in reactions with α-chitin (15 g/liter). The maximum total enzyme load was 0.5 μm with equal amounts of each enzyme, as indicated in the figure. In the reactions where the ratio was determined by protein quantification data from a previous proteomics study (Prot) (22), the total enzyme loading was also 0.5 μm. In the proteomics reaction without CjLPMO10A, the ratio was 21% CjChi18A, 4% CjChi18B, 22% CjChi18C, and 52% CjChi18D. In the proteomics reaction with CjLPMO10A, the ratio was 14% CjChi18A, 3% CjChi18B, 15% CjChi18C, 35% CjChi18D, and 33% CjLPMO10A. The CjLPMO10A was Cu2+-saturated before use. Reaction mixtures were incubated at 30 °C in 20 mm BisTris, pH 6.5, 0.1 mg/ml BSA, and samples were taken after 24 h. In reactions with CjLPMO10A, 0.5 mm ascorbate was added as an external electron donor. Production of GlcNAc and (GlcNAc)2 was quantified, and the amount of (GlcNAc)2 is given in GlcNAc equivalents. Three parallel reactions were done for each condition and standard deviations are shown as error bars. Reaction mixtures that contained the LPMO showed minor amounts of oxidized (GlcNAc)2, which were not quantified.
Investigation of enzyme activity toward soluble chito-oligosaccharides can give insight into preferred substrate binding modes and intrinsic catalytic rates. Thus, activity toward the soluble chito-oligosaccharide (GlcNAc)6 was determined for all four GH18 catalytic domains (Fig. 7A). The results indicated that CjChi18Ccat had the highest activity against (GlcNAc)6 with an initial rate of 300 ± 8 min−1. CjChi18Acat had an initial rate of 118 ± 1 min−1, whereas CjChi18Dcat and CjChi18Bcat had initial rates of 44.5 ± 2.6 and 27.0 ± 0.8 min−1, respectively. The product profile obtained for the enzymes shortly after initiation of the (GlcNAc)6 hydrolysis reactions (2-min reaction time) showed striking differences between the chitinases (Fig. 7B). CjChi18Acat yielded all possible product types ((GlcNAc)1–5), where GlcNAc was the dominating product (97.8 ± 7.3 μm) followed by (GlcNAc)2 (62.3 ± 2.5 μm), (GlcNAc)3 (47.7 ± 1.7 μm) and (GlcNAc)4 (33.9 ± 1.5 μm). Quantification of (GlcNAc)5 was not possible because of co-elution with (GlcNAc)6. CjChi18Bcat produced mainly (GlcNAc)2 and (GlcNAc)3, with minor amounts of (GlcNAc)4. CjChi18Ccat produced (GlcNAc)2–4 and CjChi18Dcat produced mainly (GlcNAc)2 and (GlcNAc)4. Chito-oligosaccharides longer than (GlcNAc)6 were observed in the CjChi18Dcat reaction (Fig. 7B and Fig. S4), indicating that this enzyme has transglycosylating activity. Reactions using (GlcNAc)2 as a substrate showed rapid conversion by CjChi18Acat, whereas only trace amounts of the monomer were detected for the other three chitinase (results not shown).
Figure 7.
Hydrolysis of (GlcNAc)6. A, hydrolysis of (GlcNAc)6 over time. The slopes of the linear parts of these curves were used to calculate the initial rates. Standard deviations are shown as error bars. B, chromatograms showing the product profile obtained 2 min after mixing chitinases with substrate. DP1–DP6 represent (GlcNAc)1–6. Chromatograms for various standards are shown as gray lines at the bottom. These experiments were done with the catalytic domains of the chitinases. The reactions contained 2 mm (GlcNAc)6, 10 mm BisTris, pH 6.5, 0.1 mg/ml BSA, and 50 nm enzyme and were done in triplicate at 30 °C. Chromatograms for samples taken after 60 min are shown in Fig. S4.
C. japonicus GH18 chitinase genes are highly expressed during chitin utilization
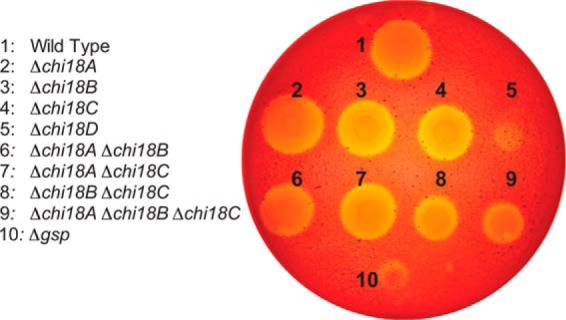
To obtain further insight into the regulation of chitinase expression and to complement the previous secretome study (22), we used RNAseq to determine changes in gene expression during growth using α-chitin relative to growth using glucose. Analysis of samples from the exponential growth phase revealed significant up-regulation of 73 CAZyme genes (Table S3A). The seven most strongly up-regulated genes were all related to chitin bioconversion, including the LPMO (6.7-fold, log2 scale), the hex20B hexosaminidase (4.1-fold), the nag9A gene (3.5-fold) that encodes a putative N-acetylhexosamine 6-phosphate deacetylase, and the four GH18 chitinases in the following order chi18D> chi18C > chi18A > chi18B, the fold change (log2 scale) being 6.7-, 5.1-, 3.9-, and 3.6-fold, respectively (Fig. 8 and Table S3A). Several other genes putatively involved in chitin bioconversion were also up-regulated, albeit to a lesser extent, including the GH19 chitinase (2.2-fold, log2 scale), the other GH20 hexosaminidase (hex20A; 2.7-fold), and the GH46 chitosanase and two polysaccharide deacetylases (2.6-, 1.6-, and 1.4-fold, respectively).
Figure 8.
Differential gene expression for C. japonicus during exponential growth. This volcano plot shows the log2(fold change) plotted against the −log10(p value) for every gene in C. japonicus during exponential growth on glucose compared with α-chitin, where each gray circle represents the expression of a single gene. The black dashed lines indicate the significance cutoff values: −log10(p value) > 2 and log2(fold change)> 1. The genes colored magenta represent CAZyme genes, and the complete list of these genes can be found in Table S3A.
The comparison of gene expression during early stationary phase yielded similar results (Fig. S5A and Table S3B). This analysis revealed the up-regulation of 47 CAZyme genes, of which seven are implicated in chitin degradation (lpmo10A, chi18D, chi18C, chi18B, nag9A, hex20A, and chi19A). Although the expression data, in particular for the exponential comparison, showed a strong up-regulation of chitin-relevant genes, there was also up-regulation of a wide variety of CAZyme genes associated with the degradation of other polysaccharides such as starch, xylan, cellulose, pectin, arabinanan, mannan, β-glucan, and xyloglucan. Therefore, the regulation of the chitin response resembles the general response previously observed for cellulose (14). When comparing the transcriptomes of the exponential and the stationary phase during growth on chitin, none of the significant changes in expression concerned CAZyme genes (Fig. S5B).
Discussion
A previous report demonstrated that C. japonicus can grow using both purified chitin and unrefined crab shells as a sole nutrient source (13). Additionally, a recent proteomic study found several secreted GH18 chitinases, suggesting a robust response to chitin (22). It has remained unknown, however, how these GH18 chitinases contribute to the degradation of chitin, and to what extent the four GH18 genes in the C. japonicus genome are equivalent. In this study, through an implementation of transcriptomic, genetic, and biochemical approaches, we assessed the physiological functions of the four GH18 enzymes. The synthesis of in vitro, in vivo, and in silico data provides a comprehensive understanding of chitin degradation by C. japonicus.
The architecture of the family GH18 chitinases indicates differential biological roles
Analysis using LipoP (29) predicts that the CjChi18A enzyme is a lipoprotein with a signal sequence cleaved by signal peptidase II (SPII). The presence of a glutamine in position +2 after the cleavage site of SPII further suggests that CjChi18A resides attached to the outer membrane (32–35). Interestingly, CjChi18A was found in high amounts in a recent secretome study (22); however, the precise location of this enzyme remains undefined. It is likely that CjChi18A functions in degrading short chito-oligosaccharides, because the absence of CBMs limits the association of this enzyme with polymeric chitin (31, 36). Such a function is supported by the biochemical data for CjChi18A showing high activity on soluble substrates and an ability to produce monomers.
Analysis of the CjChi18B, CjChi18C, and CjChi18D enzymes indicated a signal peptide that is cleaved by signal peptidase I (29). The growth defect on chitin observed in the Δgsp mutant suggests that these three enzymes are transported to the extracellular space by the type II secretion system (37). The CjChi18B, CjChi18C, and CjChi18D enzymes all possess a CBM5 and a CBM73 domain (38); however, the orientations of these domains differ between the enzymes (Fig. 2). As found with many CAZymes, the CjChi18B, CjChi18C, and CjChi18D chitinases contain serine-rich regions separating the individual domains, which likely act as flexible linkers (39).
Analysis of the domain organization of the C. japonicus chitinases, functional data, homology modeling of their 3-D structures (Fig. S6), and comparison with well-characterized GH18 chitinases from Serratia marcescens (18) all indicate that the C. japonicus chitinolytic system represents a suite of complementary activities. Available data from known processive chitinases show that there are hallmarks specific for processive enzymes, involving an α+β domain in the structure and conserved aromatic amino acids lining the catalytic cleft (40–43). Structural modeling of the C. japonicus GH18 chitinases and sequence alignment with known processive chitinases (Fig. S7) show that CjChi18B and CjChi18D have these specifications. CjChi18B has a large α+β domain (∼110 amino acids) and a “tunnel-like” substrate-binding cleft, whereas CjChi18D has a much smaller α+β domain (∼60 amino acids) and a more open active-site cleft. Compared with the chitinases from S. marcescens (Fig. S6), the structural model of CjChi18B shows a more closed active site than exo-processive SmChiA and SmChiB, whereas CjChi18D seems to have a more open active site cleft than these two Serratia enzymes. CjChi18A has the α+β domain (∼65 amino acids) but lacks most of the aromatic residues; the most similar S. marcescens enzyme seems to be SmChiD, which, like CjChi18A, primarily produces monomers (44). CjChi18C does not have features characteristic for processivity and is more similar to SmChiC, which is a non-processive endo-chitinase, displaying an open substrate binding cleft that is characteristic for non-processive glycoside hydrolases (36).
The GH18 enzymes of C. japonicus have divergent functionalities but are reliant on CjChi18D
The growth and chitinase secretion data strongly pointed to the chi18D gene product being essential for the degradation of chitinous substrates (Figs. 3 and 4 and Fig. S2). These results reinforce two features previously described for C. japonicus, specifically, that secreted enzymes are essential for recalcitrant polysaccharide degradation and that single CAZymes elicit major physiological effects (27, 45–47). Other individual enzymes did not seem essential for growth, but the Δchi18B Δchi18C double deletion mutant did show reduced growth and reduced chitin conversion efficiency (Figs. 3 and 4), indicating that these two chitinases have complementing functionalities and that they contribute to chitin conversion but are not essential. The absence of growth defects of the multiple mutants while growing using crab shells is likely a consequence of the slow overall growth rate.
Biochemical characterization of the catalytic domains revealed clear functional differences (Figs. 5 and 7, Fig. S4, and Table S4) and showed that the GH18 enzymes act synergistically during the degradation of chitin (Fig. 6). Enzyme synergy during the course of chitin degradation has been described previously for S. marcescens and is generally ascribed to collaboration between endo- and exo-acting enzymes, where endo-acting enzymes generate chain ends that are substrates for exo-acting enzymes that usually show processive action (48). The synergistic effects observed for the best combinations of C. japonicus chitinases (Fig. 6) are of similar size as the effects observed for the S. marcescens enzymes (49, 50). It is worth noting that the clearest synergy was observed when combining CjChi18B, for which there are strong indications of exo-processive action, and CjChi18C or CjChi18D, which both could be endo-acting enzymes (vide infra). Previous studies with β-chitin have shown that culture supernatants of chitin-grown S. marcescens and chitin-grown C. japonicus have similar chitinolytic power (22).
The GH18 chitinases exhibit processivity during chitin hydrolysis
Although it is difficult to determine the degree of processivity from standard substrate degradation experiments, such experiments do provide indications. During degradation of crystalline chitin, processive enzymes tend to generate high (GlcNAc)2/GlcNAc ratios (43, 51). Furthermore, the degradation of (GlcNAc)6 by processive enzymes will yield a high (GlcNAc)2/GlcNAc)4 ratio because such enzymes will process the chitohexaose into three chitobiose moieties rather than releasing the chitotetraose and chitobiose into solution after a single hydrolytic event (as would be expected for a non-processive enzyme). Of the GH18 chitinases of C. japonicus, only CjChi18B showed these high ratios (Table S4), indicating that this enzyme is the most processive, as also suggested by the structural model discussed above (Fig. S6). The other three enzymes did not show clear signs of processivity. CjChi18C displays all hallmarks of a non-processive enzyme, and CjChi18A is unique because it primarily produces monomers, which is not really compatible with processivity because the repeating unit in a chitin chain is a dimer. These latter two enzymes showed the highest specific activities toward the hexamer, suggesting that they primarily act on soluble substrates. On the basis of the experimental data, it is difficult to judge whether the four enzymes are endo- or exo-acting, and it should be noted that mixed endo-/exo-modes do occur (52). CjChi18B appears to be exo-processive, whereas CjChi18C looks to be endo-non-processive. The situation for CjChi18A and CjChi18D is less clear, because the deep but still relatively open active site clefts of these enzymes are compatible with both endo- and exo-action and because the biochemical data were not conclusive regarding processivity.
The chitinolytic machinery of C. japonicus is highly responsive to the presence of chitin substrates
There was significant up-regulation of the predicted C. japonicus genes encoding chitin-active proteins (GH18, GH19, GH20, GH46, and AA10 families) during both the exponential phase and the stationary phase (Table S3). The up-regulation of a number of other CAZyme genes not involved in chitin degradation was striking but not surprising. C. japonicus has been shown to have both substrate-dependent and growth rate-dependent control of CAZyme gene expression (14). In regards to the substrate-specific response, C. japonicus seems to have two variations: one that is general (where diverse CAZyme genes are up-regulated) and one that is specific (where only substrate-specific CAZymes genes are up-regulated). The response observed for cellulose is an example of the former (14), and the response for xylan is the latter (46). The regulatory network for the chitinolytic response appears to be analogous to the cellulose response, which is not surprising given the similarities of the substrates.
The expression data also indicated that the most up-regulated chitin-relevant gene encodes the chitin-specific LPMO, CjLPMO10A. It has been shown that this enzyme will act synergistically with chitinases (13) but in the present study, the effects of adding the LPMO to the chitinases was limited (Fig. 6). This is likely an effect of the substrate used in this study, which was ball-milled into fine particles. It has previously been shown that such ball-milling reduces the crystallinity of the substrate, hence reducing the activity of the LPMO (53).
Final remarks
The proposed model (Fig. 1) for crystalline chitin degradation by C. japonicus, based on in vitro, in vivo, and in silico analyses is that the four GH18 chitinases and CjLPMO10A synergistically degrade chitin. CjLPMO10A plays an important role, as indicated by the transcriptomic data presented here and previously published biochemical data (13). CjChi18D, being the most powerful chitin degrader, likely also acts on the more recalcitrant parts of the substrate, releasing chito-oligosaccharides. CjChi18B and CjChi18C act on the more accessible chitin fibers, converting them into smaller chito-oligosaccharides, whereas CjChi18A converts chito-oligosaccharides to N-acetylglucosamine.
Experimental procedures
Growing conditions
C. japonicus strains were grown using MOPS minimal medium (54) containing 0.2% w/v glucose, 0.5% w/v N-acetylglucosamine, 0.25% (w/v) α-chitin from shrimp shells (Sigma–Aldrich), 0.2% (w/v) β-chitin from squid pen, or 4% (w/v) crab shells from Callinectes sapidus as the sole source of carbon. The following protocols were developed to prepare the chitin containing substrates. The coarse flakes of α-chitin were sieved through the top piece of a 130-mm Buchner polypropylene filter to homogenize the particle size (diameter of ∼4 mm). β-Chitin from squid pen was manually ground in a ceramic mortar and pestle and then passed through a plastic filter with 4-mm diameter holes. For the crab shells, any membranous material was discarded, and the shells were then thoroughly washed and rinsed. Clean and air-dried shells were manually ground and then sieved through a 4-mm diameter filter. To remove any generated crab shell dust, the filtrate was rinsed through a Buchner polypropylene filter. The shell pieces were subjected to autoclave sterilization. Escherichia coli strains were grown using lysogenic broth (55). For growth analysis studies, the strains were initially grown for 24 h in 5 ml of MOPS–glucose medium. Then an 18-mm test tube with 5 ml of MOPS–chitin broth was inoculated with a 1:100 dilution of the prepared overnight cultures. All cultures were incubated at 30 °C with an aeration of 225 RPM. For insoluble substrates, growth was measured as a function of the A600 value with a Spec20D+ spectrophotometer (Thermo Scientific) using biomass containment as needed (56). In growth experiments with glucose or N-acetylglucosamine, growth was monitored using a Tecan M200Pro microplate reader at 600 nm (A600) (Tecan Trading AG, Switzerland). All growth experiments were performed in biological triplicate. Plate media were solidified with 1.5% agar. When required, kanamycin was used at a concentration of 50 μg/ml.
Generation of deletion mutants
Deletion mutants of the family GH18 chitinases of C. japonicus were made and verified using previously published protocols (27, 45). A suicide vector was generated by cloning ±500 bp up- and downstream from the gene to be deleted into the pK18mobsacB vector (57) at the EcoRI and the XbaI sites. The pK18mobsacB plasmids to make the deletions of genes chi18A, chi18B, and chi18C were synthetized by GeneWiz (South Plainfield, NJ), whereas the plasmid to make the deletion of chi18D was amplified by PCR and constructed via Gibson assembly (58). All vectors were electroporated into E. coli S17 λPIR. Through tri-parental mating, the deletion plasmid was conjugated into C. japonicus using an E. coli helper strain containing the plasmid pRK2013 (59). Recombinant colonies were selected using kanamycin selection, with a counter selection carried out via sucrose toxicity. The deletion mutants were confirmed by PCR, and the primers used for the construction and verification of C. japonicus mutants are listed in Table S5.
Visualization of colloidal chitin degradation
Colloidal chitin was prepared as described in (60). Colloidal chitin plates were made using MOPS-defined medium supplemented with 2% (w/v) colloidal chitin and 0.2% (w/v) glucose. To assay for chitin degradation, 10 μl of overnight cultures of the C. japonicus strains to be analyzed were spotted onto the chitin plate and incubated for 4 days at 30 °C. Then the plates were stained with a 0.1% (w/v) Congo red solution for 10 min followed by 10 min destaining with a 1 m NaCl solution, as described previously for detection of degradation of carboxyl methyl cellulose (27, 45, 56).
Transcriptomic analysis
A transcriptomic analysis was conducted for C. japonicus grown using glucose or α-chitin. To prepare the samples, the protocol described by Gardner et al. (14) was followed. Briefly, 35 ml of cell culture were aliquoted into 50-ml conical tubes with 5 ml of a stopping solution made with ethanol and saturated phenol (19:1). The cells were pelleted by centrifugation at 8000 × g for 5 min at 4 °C. The supernatant was removed, and the cell pellets were immersed in a dry ice/ethanol bath for 5 min. The frozen cell pellets were then stored at −80 °C. For each carbon source, the samples were taken in biological triplicate and at two time points: the beginning of the exponential phase (0.1 > A600 > 0.2) and the beginning stationary phase. GeneWiz performed RNAseq using an Illumina HiSeq2500 (50-bp single-reads; >10 million reads/sample), and sequence data were analyzed using CLC Genomics Workbench (Qiagen). The log2 transformation and a Student's t test were used for the comparative analysis of C. japonicus gene expression. An adjusted p value > 0.01 and a log2 fold change > 2 were selected as significance cutoff parameters. RNAseq data for glucose grown C. japonicus cells (GSE90955) was used as done previously (26). The chitin-specific RNAseq data that was generated for this study can be found in the NCBI GEO database (GSE108935).
Bioinformatics analysis
We determined the predicted CAZy domains presented using the Database for Automated Carbohydrate-Active Enzyme Annotation (38). Using LipoP (29) and SignalP (30), we determined the putative location of the chitinolytic enzymes. Three-dimensional models of the GH18 chitinases were generated using PyMod 2.0 (61).
Cloning and expression of chitinases
Synthetic genes of the four full-length chitinases, optimized for expression in E. coli, were purchased from GeneScript. These genes encoded the catalytic domain for CjChi18A and the full-length protein (without signal peptide) for CjChi18B, CjChi18C, and CjChi18D. Primers for amplification of the genes were designed so that a His6 tag followed by a TEV protease cleavage site was introduced at the N terminus of all proteins and a stop codon was introduced before the C-terminal His tag encoded in the vector. Using different primers for amplification, genes encoding different versions (Table S6) of the modular proteins were generated, which were cloned into the pNIC-CH vector (62) using ligation independent cloning (63). The DNA sequences of the genes were confirmed by Sanger sequencing. Plasmids were transformed into E. coli BL21 cells. Protein expression was tested by inoculating 50 ml of lysogenic broth + 50 μg/ml kanamycin with 500 μl of an overnight culture. The cultures were grown at 37 °C, and when the OD reached 0.6, the cells were induced by adding IPTG to a final concentration of 0.2 mm. After growth overnight at 30 °C, the cells was harvested by centrifugation (6164 × g, 12 min, 4 °C) and resuspended in 5 ml of 20 mm Tris-HCl, 150 mm NaCl, 5 mm imidazole, pH 8.0. DNaseI and PMSF were added to final concentrations of 1.4 μg/ml and 0.1 mm, respectively, before the cells were lysed by sonication (pulse 3 s on, 3 s off for 3–5 min), using a VC750 VibraCell sonicator (Sonics & Materials, Inc.). Following cell disruption, the samples were centrifuged (11,814 × g, 12 min, 4 °C), after which the supernatant was collected and filtrated (0.22 μm). Analysis by SDS-PAGE showed that only CjChi18A was soluble. Except for the full-length enzymes, which were not produced in detectable levels, all other enzyme variants were produced in large amounts but were insoluble.
Production of CjChi18A was scaled up to a 500-ml culture following the same protocol as above for production and harvesting. The filtrated supernatant containing CjChi18A was used for protein purification by nickel affinity chromatography using a HisTrap HP 5 ml column (GE Healthcare Life Sciences) connected to an Äkta pure system (GE Healthcare Life Sciences). A continuous imidazole gradient ending at 300 mm imidazole was used to elute bound protein, using a flow rate of 1 ml/min.
CjChi18Bcat, CjChi18Bcat+CBM5, CjChi18Ccat, CjChi18Dcat, and CjChi18Dcat+CBM5 were produced using a denaturing and refolding method, starting with 500-ml cultures as described above. After harvesting the cells (7025 × g, 12 min, 4 °C), the pellet was resuspended in 20 ml of 50 mm Tris, pH 8.0, 0.2 m NaCl, and 50% of the cell suspension was used in the further steps. After another centrifugation (18,459 × g, 10 min, 4 °C), the pellet was resuspended in 10 ml of 50 mm Tris, pH 8.0, 0.2 m NaCl and, after addition of lysozyme (final concentration, 200 μg/ml) and DNaseI (final concentration 0.1 mm), the samples were incubated on ice for 30 min. The samples were subsequently sonicated (pulse 3 s on, 3 s off, 2–4 min), and inclusion bodies were harvested by centrifugation (18,459 × g, 10 min), after which the protein pellet was resuspended in 25 ml of washing buffer (20 mm Tris-HCl, 100 mm NaCl, 2% Triton X-100, pH 8.0). The centrifugation and subsequent resuspension in washing buffer steps were repeated twice, and after the final centrifugation, the pellet was dissolved in 5 ml of cold denaturing solution (50 mm Tris-HCl, pH 8.5, 100 mm NaCl, 5 m guanidine HCl, 1 mm EDTA, and 20 mm DTT), vortexed for 1 min, and incubated overnight at 4 °C with slow rotation of the sample tubes. The samples were then centrifuged at 9000 × g for 10 min at 4 °C. The supernatant containing the denatured protein was then collected for refolding.
The sample containing the denatured protein (∼5 ml) was added to 250 ml of cold refolding buffer (100 mm Tris-HCl, 0.4 ml-arginine, 0.5 mm oxidized glutathione, 5 mm reduced glutathione) at a rate of 1 ml/h under intense stirring at 4 °C. After adding all protein, the solution was stirred overnight at 4 °C, before centrifugation (9000 × g, 10 min, 4 °C). The supernatant was collected and dialyzed (10-kDa molecular mass cutoff Snakeskin; Thermo Fisher Scientific) against 2.5 liters of 50 mm Tris-HCl, 0.1 m NaCl, pH 8.0, overnight. The dialysis solution was changed once, after ∼6 h. After collecting the dialyzed sample, the protein was purified with nickel affinity chromatography as described above. All refolded proteins stayed soluble during these procedures, and the yields of recovered purified soluble protein were typically 1–5 mg/liter culture.
The presence and purity of the proteins was confirmed by SDS-PAGE, and relevant fractions were pooled before the protein solution was concentrated to 26.4 mg/ml for CjChi18A and 1–3.5 mg/ml for the unfolded/refolded chitinases, with concomitant buffer exchange to 50 mm Tris-HCl, 100 mm NaCl, pH 8.0, using Amicon Ultra-15 centrifugal filters with 10,000 Nominal Molecular Weight Limit (Merck Millipore). All resulting protein solutions were stable for weeks at 4 °C. Protein concentrations were determined by measuring the absorbance at 280 nm and using the theoretical extinction coefficient (calculated using the ExPASy ProtParam tool available from https://web.expasy.org/protparam). The catalytic domain of CjLPMO10A was expressed and purified as previously described by Forsberg et al. (13).
Activity assays
Standard reactions contained 15 g/liter α-chitin (extracted from Pandalus borealis; Seagarden, Karmsund, Norway) or 2 mm chito-oligosaccharides (MegaZyme, Bray, Ireland), 0.1 mg/ml BSA and 20 mm BisTris, pH 6.5, unless stated otherwise. For chitin degradation 0.5 μm enzyme was used, whereas for chito-oligosaccharide degradation 50 nm enzyme was used, unless stated otherwise. The reaction mixtures were incubated in a thermomixer at 30 °C and 800 rpm, and enzyme activity was quenched by adding sulfuric acid to a final concentration of 25 mm. A Rezex RFQ fast acid H+ (8%) ion-exclusion column (Phenomenex) installed on a Dionex Ultimate 3000 HPLC with UV detection (194 nm) was used to analyze and quantify degradation products as described by Mekasha et al. (64). Conversion yields were calculated as percentages of the theoretical maximum after correction of the amount of chitin in the reaction for ash and water content. Reactions with (GlcNAc)6 as substrate were analyzed using a Rezex ROA-organic acid H+ (8%) ion exclusion column (Phenomenex) installed on a Dionex Ultimate 3000 HPLC, using a column temperature of 65 °C. An 8-μl sample was injected on the column, and the mono/oligosaccharides were eluted isocratically at 0.6 ml/min with 5 mm sulfuric acid as mobile phase. The chito-oligosaccharides were monitored by measuring the absorbance at 194 nm. Standards with known concentrations of GlcNAc (Sigma) and (GlcNAc)2–6 (MegaZyme) were used to determine the concentrations of (GlcNAc)6 and the degradation products.
Author contributions
E. C. M. and T. R. T. formal analysis; E. C. M. and T. R. T. investigation; E. C. M. and T. R. T. visualization; E. C. M. and T. R. T. methodology; E. C. M., T. R. T., G. V.-K., V. G. H. E., and J. G. G. writing-original draft; E. C. M., T. R. T., G. V.-K., V. G. H. E., and J. G. G. writing-review and editing; G. V.-K., V. G. H. E., and J. G. G. supervision; G. V.-K., V. G. H. E., and J. G. G. project administration; J. G. G. conceptualization; G. V.-K., V. G. H. E., and J. G. G. funding acquisition.
Supplementary Material
Acknowledgments
We thank C. E. Nelson for assistance with the collection of the RNAseq samples and A. D. Blake for technical support.
This work was supported by start-up funds (to J. G. G.); by Grant 2-R25-GM55036 from the Meyerhoff Graduate Fellows Program under the NIGMS, National Institutes of Health Initiative for Maximizing Student Development, by Chemistry and Biology Interface Program Grant T32-GM066706 (to E. C. M.); and Grant 221576 from the Research Council of Norway (to T. R. T., G. V. K., and V. G. H. E.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains references, Tables S1–S6, and Figs. S1–S7.
- LPMO
- lytic polysaccharide monooxygenase
- GH
- glycoside hydrolase family
- CAZyme
- carbohydrate active enzyme
- CBM
- carbohydrate-binding module
- Glc
- glucose
- OD
- optical density
- SPII
- signal peptidase II
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1. Gooday G. W., Humphreys A. M., and McIntosh W. H. (1986) Roles of chitinases in fungal growth. In Chitin in Nature and Technology (Muzzarelli R., Jeuniaux C., and Gooday G. W., eds.) pp. 83–91, Springer-Verlag, New York Inc., New York [Google Scholar]
- 2. Beier S., and Bertilsson S. (2013) Bacterial chitin degradation-mechanisms and ecophysiological strategies. Front. Microbiol. 4, 149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Khor E., and Lim L. Y. (2003) Implantable applications of chitin and chitosan. Biomaterials 24, 2339–2349 10.1016/S0142-9612(03)00026-7 [DOI] [PubMed] [Google Scholar]
- 4. Francesko A., and Tzanov T. (2011) Chitin, chitosan and derivatives for wound healing and tissue engineering. Adv. Biochem. Eng. Biotechnol. 125, 1–27 [DOI] [PubMed] [Google Scholar]
- 5. Matroodi S., Motallebi M., Zamani M., and Moradyar M. (2013) Designing a new chitinase with more chitin binding and antifungal activity. World J. Microbiol. Biotechnol. 29, 1517–1523 10.1007/s11274-013-1318-0 [DOI] [PubMed] [Google Scholar]
- 6. Stoykov Y. M., Pavlov A. I., and Krastanov A. I. (2015) Chitinase biotechnology: production, purification, and application. Eng. Life Sci. 15, 30–38 10.1002/elsc.201400173 [DOI] [Google Scholar]
- 7. Yan N. (2017) Sustainability: don't waste seafood waste. Nature 524, 155–157 [DOI] [PubMed] [Google Scholar]
- 8. Yan Q., and Fong S. S. (2015) Bacterial chitinase: nature and perspectives for sustainable bioproduction. Bioresources Bioprocessing 2, 31 10.1186/s40643-015-0057-5 [DOI] [Google Scholar]
- 9. Tharanathan R. N., and Kittur F. S. (2003) Chitin: the undisputed biomolecule of great potential. Crit. Rev. Food Sci. Nutr. 43, 61–87 10.1080/10408690390826455 [DOI] [PubMed] [Google Scholar]
- 10. Gortari M. C., and Hours R. A. (2013) Biotechnological processes for chitin recovery out of crustacean waste: a mini-review. Electron J. Biotechnol. 16, 10.2225/vol16-issue3-fulltext-10 10.2225/vol16-issue3-fulltext-10 [DOI] [Google Scholar]
- 11. Hemsworth G. R., Déjean G., Davies G. J., and Brumer H. (2016) Learning from microbial strategies for polysaccharide degradation. Biochem. Soc. Trans. 44, 94–108 10.1042/BST20150180 [DOI] [PubMed] [Google Scholar]
- 12. Zargar V., Asghari M., and Dashti A. (2015) A review on chitin and chitosan polymers: structure, chemistry, solubility, derivatives, and applications. ChemBioEng Rev. 2, 204–226 10.1002/cben.201400025 [DOI] [Google Scholar]
- 13. Forsberg Z., Nelson C. E., Dalhus B., Mekasha S., Loose J. S., Crouch L. I., Røhr A. K., Gardner J. G., Eijsink V. G., and Vaaje-Kolstad G. (2016) Structural and functional analysis of a lytic polysaccharide monooxygenase important for efficient utilization of chitin in Cellvibrio japonicus. J. Biol. Chem. 291, 7300–7312 10.1074/jbc.M115.700161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gardner J. G., Crouch L., Labourel A., Forsberg Z., Bukhman Y. V., Vaaje-Kolstad G., Gilbert H. J., and Keating D. H. (2014) Systems biology defines the biological significance of redox-active proteins during cellulose degradation in an aerobic bacterium. Mol. Microbiol. 95, 418–433 [DOI] [PubMed] [Google Scholar]
- 15. Vaaje-Kolstad G., Westereng B., Horn S. J., Liu Z., Zhai H., Sørlie M., and Eijsink V. G. (2010) An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330, 219–222 10.1126/science.1192231 [DOI] [PubMed] [Google Scholar]
- 16. Meibom K. L., Li X. B., Nielsen A. T., Wu C. Y., Roseman S., and Schoolnik G. K. (2004) The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. U.S.A. 101, 2524–2529 10.1073/pnas.0308707101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vaaje-Kolstad G., Bøhle L. A., Gåseidnes S., Dalhus B., Bjorås M., Mathiesen G., and Eijsink V. G. (2012) Characterization of the chitinolytic machinery of Enterococcus faecalis V583 and high-resolution structure of its oxidative CBM33 enzyme. J. Mol. Biol. 416, 239–254 10.1016/j.jmb.2011.12.033 [DOI] [PubMed] [Google Scholar]
- 18. Vaaje-Kolstad G., Horn S. J., Sørlie M., and Eijsink V. G. (2013) The chitinolytic machinery of Serratia marcescens: a model system for enzymatic degradation of recalcitrant polysaccharides. FEBS J. 280, 3028–3049 10.1111/febs.12181 [DOI] [PubMed] [Google Scholar]
- 19. Gardner J. G., and Keating D. H. (2012) Genetic and functional genomic approaches for the study of plant cell wall degradation in Cellvibrio japonicus. Methods Enzymol. 510, 331–347 10.1016/B978-0-12-415931-0.00018-5 [DOI] [PubMed] [Google Scholar]
- 20. Gardner J. G. (2016) Polysaccharide degradation systems of the saprophytic bacterium Cellvibrio japonicus. World J. Microbiol. Biotechnol. 32, 121–132 10.1007/s11274-016-2068-6 [DOI] [PubMed] [Google Scholar]
- 21. DeBoy R. T., Mongodin E. F., Fouts D. E., Tailford L. E., Khouri H., Emerson J. B., Mohamoud Y., Watkins K., Henrissat B., Gilbert H. J., and Nelson K. E. (2008) Insights into plant cell wall degradation from the genome sequence of the soil bacterium Cellvibrio japonicus. J. Bacteriol. 190, 5455–5463 10.1128/JB.01701-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tuveng T. R., Arntzen M. Ø., Bengtsson O., Gardner J. G., Vaaje-Kolstad G., and Eijsink V. G. (2016) Proteomic investigation of the secretome of Cellvibrio japonicus during growth on chitin. Proteomics 16, 1904–1914 10.1002/pmic.201500419 [DOI] [PubMed] [Google Scholar]
- 23. Itoh Y., Kawase T., Nikaidou N., Fukada H., Mitsutomi M., Watanabe T., and Itoh Y. (2002) Functional analysis of the chitin-binding domain of a family 19 chitinase from Streptomyces griseus HUT6037: substrate-binding affinity and cis-dominant increase of antifungal function. Biosci. Biotechnol. Biochem. 66, 1084–1092 10.1271/bbb.66.1084 [DOI] [PubMed] [Google Scholar]
- 24. Hoell I. A., Dalhus B., Heggset E. B., Aspmo S. I., and Eijsink V. G. (2006) Crystal structure and enzymatic properties of a bacterial family 19 chitinase reveal differences from plant enzymes. FEBS J. 273, 4889–4900 10.1111/j.1742-4658.2006.05487.x [DOI] [PubMed] [Google Scholar]
- 25. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., and Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 37, D233–D238 10.1093/nar/gkn663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nelson C. E., Rogowski A., Morland C., Wilhide J. A., Gilbert H. J., and Gardner J. G. (2017) Systems analysis in Cellvibrio japonicus resolves predicted redundancy of β-glucosidases and determines essential physiological functions. Mol. Microbiol. 104, 294–305 10.1111/mmi.13625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nelson C. E., and Gardner J. G. (2015) In-frame deletions allow functional characterization of complex cellulose degradation phenotypes in Cellvibrio japonicus. Appl. Environ. Microbiol. 81, 5968–5975 10.1128/AEM.00847-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 29. Juncker A. S., Willenbrock H., Von Heijne G., Brunak S., Nielsen H., and Krogh A. (2003) Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12, 1652–1662 10.1110/ps.0303703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Petersen T. N., Brunak S., von Heijne G., and Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods 8, 785–786 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 31. Boraston A. B., Bolam D. N., Gilbert H. J., and Davies G. J. (2004) Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 10.1042/BJ20040892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang G., Chen H., Xia Y., Cui J., Gu Z., Song Y., Chen Y. Q., Zhang H., and Chen W. (2013) How are the non-classically secreted bacterial proteins released into the extracellular milieu? Curr. Microbiol. 67, 688–695 10.1007/s00284-013-0422-6 [DOI] [PubMed] [Google Scholar]
- 33. Bendtsen J. D., Kiemer L., Fausbøll A., and Brunak S. (2005) Non-classical protein secretion in bacteria. BMC Microbiol. 5, 58 10.1186/1471-2180-5-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bendtsen J. D., Jensen L. J., Blom N., Von Heijne G., and Brunak S. (2004) Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 17, 349–356 10.1093/protein/gzh037 [DOI] [PubMed] [Google Scholar]
- 35. Seydel A., Gounon P., and Pugsley A. P. (1999) Testing the “+2 rule” for lipoprotein sorting in the Escherichia coli cell envelope with a new genetic selection. Mol. Microbiol. 34, 810–821 10.1046/j.1365-2958.1999.01647.x [DOI] [PubMed] [Google Scholar]
- 36. Payne C. M., Baban J., Horn S. J., Backe P. H., Arvai A. S., Dalhus B., Bjørås M., Eijsink V. G., Sørlie M., Beckham G. T., and Vaaje-Kolstad G. (2012) Hallmarks of processivity in glycoside hydrolases from crystallographic and computational studies of the Serratia marcescens chitinases. J. Biol. Chem. 287, 36322–36330 10.1074/jbc.M112.402149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nakai K., and Kanehisa M. (1991) Expert system for predicting protein localization sites in Gram-negative bacteria. Proteins 11, 95–110 10.1002/prot.340110203 [DOI] [PubMed] [Google Scholar]
- 38. Yin Y., Mao X., Yang J., Chen X., Mao F., and Xu Y. (2012) dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40, W445–W451 10.1093/nar/gks479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Black G. W., Rixon J. E., Clarke J. H., Hazlewood G. P., Theodorou M. K., Morris P., and Gilbert H. J. (1996) Evidence that linker sequences and cellulose-binding domains enhance the activity of hemicellulases against complex substrates. Biochem. J. 319, 515–520 10.1042/bj3190515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Perrakis A., Tews I., Dauter Z., Oppenheim A. B., Chet I., Wilson K. S., and Vorgias C. E. (1994) Crystal structure of a bacterial chitinase at 2.3 A resolution. Structure 2, 1169–1180 10.1016/S0969-2126(94)00119-7 [DOI] [PubMed] [Google Scholar]
- 41. van Aalten D. M., Synstad B., Brurberg M. B., Hough E., Riise B. W., Eijsink V. G., and Wierenga R. K. (2000) Structure of a two-domain chitotriosidase from Serratia marcescens at 1.9-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 97, 5842–5847 10.1073/pnas.97.11.5842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Horn S. J., Sikorski P., Cederkvist J. B., Vaaje-Kolstad G., Sørlie M., Synstad B., Vriend G., Vårum K. M., and Eijsink V. G. (2006) Costs and benefits of processivity in enzymatic degradation of recalcitrant polysaccharides. Proc. Natl. Acad. Sci. U.S.A. 103, 18089–18094 10.1073/pnas.0608909103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zakariassen H., Aam B. B., Horn S. J., Vårum K. M., Sørlie M., and Eijsink V. G. (2009) Aromatic residues in the catalytic center of chitinase A from Serratia marcescens affect processivity, enzyme activity, and biomass converting efficiency. J. Biol. Chem. 284, 10610–10617 10.1074/jbc.M900092200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tuveng T. R., Hagen L. H., Mekasha S., Frank J., Arntzen M. Ø., Vaaje-Kolstad G., and Eijsink V. G. H. (2017) Genomic, proteomic and biochemical analysis of the chitinolytic machinery of Serratia marcescens BJL200. Biochim. Biophys. Acta 1865, 414–421 10.1016/j.bbapap.2017.01.007 [DOI] [PubMed] [Google Scholar]
- 45. Gardner J. G., and Keating D. H. (2010) Requirement of the type II secretion system for utilization of cellulosic substrates by Cellvibrio japonicus. Appl. Environ. Microbiol. 76, 5079–5087 10.1128/AEM.00454-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Blake A. D., Beri N. R., Guttman H. S., Cheng R., and Gardner J. G. (2017) The complex physiology of Cellvibrio japonicus xylan degradation relies on a single cytoplasmic beta-xylosidase for xylo-oligosaccharide utilization. Mol. Microbiol. 10.1111/mmi.13903 10.1111/mmi.13903 [DOI] [PubMed] [Google Scholar]
- 47. Nelson C. E., Attia M. A., Rogowski A., Morland C., Brumer H., and Gardner J. G. (2017) Comprehensive functional characterization of the glycoside hydrolase family 3 enzymes from Cellvibrio japonicus reveals unique metabolic roles in biomass saccharification. Environ. Microbiol. 19, 5025–5039 10.1111/1462-2920.13959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jalak J., Kurasin M., Teugjas H., and Väljamäe P. (2012) Endo-exo synergism in cellulose hydrolysis revisited. J. Biol. Chem. 287, 28802–28815 10.1074/jbc.M112.381624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Suzuki K., Sugawara N., Suzuki M., Uchiyama T., Katouno F., Nikaidou N., and Watanabe T. (2002) Chitinases A, B, and C1 of Serratia marcescens 2170 produced by recombinant Escherichia coli: enzymatic properties and synergism on chitin degradation. Biosci. Biotechnol. Biochem. 66, 1075–1083 10.1271/bbb.66.1075 [DOI] [PubMed] [Google Scholar]
- 50. Vaaje-Kolstad G., Horn S. J., van Aalten D. M., Synstad B., and Eijsink V. G. (2005) The non-catalytic chitin-binding protein CBP21 from Serratia marcescens is essential for chitin degradation. J. Biol. Chem. 280, 28492–28497 10.1074/jbc.M504468200 [DOI] [PubMed] [Google Scholar]
- 51. Horn S. J., Sørbotten A., Synstad B., Sikorski P., Sørlie M., Vårum K. M., and Eijsink V. G. (2006) Endo/exo mechanism and processivity of family 18 chitinases produced by Serratia marcescens. FEBS J. 273, 491–503 10.1111/j.1742-4658.2005.05079.x [DOI] [PubMed] [Google Scholar]
- 52. Sikorski P., Sørbotten A., Horn S. J., Eijsink V. G., and Vårum K. M. (2006) Serratia marcescens chitinases with tunnel-shaped substrate-binding grooves show endo activity and different degrees of processivity during enzymatic hydrolysis of chitosan. Biochemistry 45, 9566–9574 10.1021/bi060370l [DOI] [PubMed] [Google Scholar]
- 53. Nakagawa Y. S., Eijsink V. G., Totani K., and Vaaje-Kolstad G. (2013) Conversion of α-chitin substrates with varying particle size and crystallinity reveals substrate preferences of the chitinases and lytic polysaccharide monooxygenase of Serratia marcescens. J. Agric. Food Chem. 61, 11061–11066 10.1021/jf402743e [DOI] [PubMed] [Google Scholar]
- 54. Neidhardt F. C., Bloch P. L., and Smith D. F. (1974) Culture medium for enterobacteria. J. Bacteriol. 119, 736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bertani G. (1951) Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62, 293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nelson C. E., Beri N. R., and Gardner J. G. (2016) Custom fabrication of biomass containment devices using 3-D printing enables bacterial growth analyses with complex insoluble substrates. J. Microbiol. Methods 130, 136–143 10.1016/j.mimet.2016.09.013 [DOI] [PubMed] [Google Scholar]
- 57. Schäfer A., Tauch A., Jäger W., Kalinowski J., Thierbach G., and Pühler A. (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73 10.1016/0378-1119(94)90324-7 [DOI] [PubMed] [Google Scholar]
- 58. Gibson D. G., Young L., Chuang R. Y., Venter J. C., Hutchison C. A. 3rd, and Smith H. O. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods 6, 343–345 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- 59. Figurski D. H., and Helinski D. R. (1979) Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U.S.A. 76, 1648–1652 10.1073/pnas.76.4.1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Murthy N. K. S., and Bleakley B. H. (2017) Simplified method of preparing colloidal chitin used for screening of chitinase-producing microorganisms. Internet J. Microbiol. 10, http://ispub.com/IJMB/10/2/14186 [Google Scholar]
- 61. Janson G., Zhang C., Prado M. G., and Paiardini A. (2017) PyMod 2.0: improvements in protein sequence-structure analysis and homology modeling within PyMOL. Bioinformatics 33, 444–446 [DOI] [PubMed] [Google Scholar]
- 62. Savitsky P., Bray J., Cooper C. D., Marsden B. D., Mahajan P., Burgess-Brown N. A., and Gileadi O. (2010) High-throughput production of human proteins for crystallization: the SGC experience. J. Struct. Biol. 172, 3–13 10.1016/j.jsb.2010.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Aslanidis C., and de Jong P. J. (1990) Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 18, 6069–6074 10.1093/nar/18.20.6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mekasha S., Byman I. R., Lynch C., Toupalova H., Andera L., Naes T., Vaaje-Kolstad G., and Eijsink V. G. H. (2017) Development of enzyme cocktails for complete saccharification of chitin using mono-component enzymes from Serratia marcescens. Process Biochem. 56, 132–138 10.1016/j.procbio.2017.02.021 [DOI] [Google Scholar]
- 65. Liu W., Xie Y., Ma J., Luo X., Nie P., Zuo Z., Lahrmann U., Zhao Q., Zheng Y., Zhao Y., Xue Y., and Ren J. (2015) IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 31, 3359–3361 10.1093/bioinformatics/btv362 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.