Abstract
Aminotransferases are pyridoxal 5′-phosphate–dependent enzymes that catalyze reversible transamination reactions between amino acids and α-keto acids, and are important for the cellular metabolism of nitrogen. Many bacterial and eukaryotic ω-aminotransferases that use l-ornithine (Orn), l-lysine (Lys), or γ-aminobutyrate (GABA) have been identified and characterized, but the corresponding enzymes from archaea are unknown. Here, we examined the activity and function of TK2101, a gene annotated as a GABA aminotransferase, from the hyperthermophilic archaeon Thermococcus kodakarensis. We overexpressed the TK2101 gene in T. kodakarensis and purified and characterized the recombinant protein and found that it displays only low levels of GABA aminotransferase activity. Instead, we observed a relatively high ω-aminotransferase activity with l-Orn and l-Lys as amino donors. The most preferred amino acceptor was 2-oxoglutarate. To examine the physiological role of TK2101, we created a TK2101 gene–disruption strain (ΔTK2101), which was auxotrophic for proline. Growth comparison with the parent strain KU216 and the biochemical characteristics of the protein strongly suggested that TK2101 encodes an Orn aminotransferase involved in the biosynthesis of l-Pro. Phylogenetic comparisons of the TK2101 sequence with related sequences retrieved from the databases revealed the presence of several distinct protein groups, some of which having no experimentally studied member. We conclude that TK2101 is part of a novel group of Orn aminotransferases that are widely distributed at least in the genus Thermococcus, but perhaps also throughout the Archaea.
Keywords: amino acid, archaea, enzyme, metabolism, thermophile, biosynthesis, aminotransferase, ornithine, proline, transamination, enzyme
Introduction
Aminotransferases are pyridoxal 5′-phosphate (PLP)3–dependent enzymes that catalyze reversible transamination reactions between amino acids and α-keto acids and play an important role in the cellular metabolism of nitrogen. There are two categories for aminotransferases, α-aminotransferases and ω-aminotransferases, catalyzing transamination of α- and ω-amino groups, respectively. α-Aminotransferases have been extensively studied, and several α-aminotransferases from archaea, such as branched-chain amino acid aminotransferases (1, 2), aromatic aminotransferases (2–4), aspartate aminotransferases (2, 4–6), and alanine aminotransferases (7, 8) have been identified and characterized.
The structures of ω-amino acids differ from those of α-amino acids, and the amino group involved in the transamination reaction catalyzed by ω-aminotransferase is distal to the carboxyl group (9). ω-Aminotransferases constitute the majority of the class III aminotransferases, which belong to the fold type I (aspartate aminotransferase family) of PLP-dependent enzymes (10). ω-Aminotransferases can further be divided into groups according to their natural substrates, such as putrescine, γ-aminobutyrate (GABA), N-acetylornithine, ornithine (Orn), and lysine (Lys), and they play a pivotal physiological role in their metabolism. GABA is the major inhibitory neurotransmitter in the mammalian brain. It is converted into succinic semialdehyde using 2-oxoglutarate as co-substrate by GABA aminotransferase (GABA-AT), which is considered as a viable target for drugs aimed at treating serious neurological disorders (11). N-Acetylornithine aminotransferase (Acorn-AT) catalyzes the reversible conversion of N-acetylglutamate 5-semialdehyde to N-acetylornithine in the presence of l-glutamate (Glu) and is one of the key enzymes involved in arginine (Arg) biosynthesis (12). Ornithine δ-aminotransferase (Orn-AT) has been frequently associated with proline (Pro) accumulation and Arg catabolism by catalyzing the transfer of the δ-amino group of l-Orn to 2-oxoglutarate, yielding l-glutamate 5-semialdehyde, which spontaneously cyclizes to form l-1-pyrroline-5-carboxylate (P5C) (13). Lysine ϵ-aminotransferase (Lys-AT) catalyzes the transfer of the ϵ-amino group of Lys to 2-oxoadipate to yield 2-aminoadipate 6-semialdehyde and 2-aminoadipate, a precursor of β-lactam antibiotics (14). In addition, ω-aminotransferases are of particular interest for asymmetric synthesis of chiral amines in biocatalysis (10). Although a wealth of ω-aminotransferases that act on l-Orn, l-Lys, or GABA has been identified from eukaryotes and bacteria, their presence in archaea has not been reported.
Thermococcus kodakarensis KOD1 is a hyperthermophilic archaeon isolated from Kodakara Island, Kagoshima, Japan (15, 16). It is an obligate heterotroph and grows rapidly on a variety of carbon sources. As the whole-genome sequence of T. kodakarensis has been determined (17), and gene manipulation systems established (18–22), this organism is regarded as one of the model organisms in Archaea to study their physiology and metabolism.
Here, we describe the biochemical and genetic characterization of a putative GABA-AT encoded by TK2101 in T. kodakarensis. To our surprise, the TK2101 protein is an ω-aminotransferase that exhibits high activity toward l-Orn and l-Lys. Genetic analysis of TK2101 confirmed its physiological role as an Orn-AT involved in the biosynthesis of l-Pro.
Results
Two GABA-aminotransferase homologs in T. kodakarensis
In our efforts to elucidate the mechanism of coenzyme A (CoA) biosynthesis in archaea (23–27), we identified a glutamate decarboxylase homolog encoded by TK1814 from T. kodakarensis, which catalyzed the decarboxylation of aspartate and Glu to β-alanine and GABA, respectively, in vitro (26). Gene disruption and phenotypic analyses indicated that the β-alanine formation is required for CoA biosynthesis, whereas the roles for the glutamate decarboxylase activity, if any, could not be identified. Related to this, we noticed the presence of two genes (TK1211 and TK2101) on the T. kodakarensis genome annotated as GABA-AT, and thus took interest in their functions. The TK1211 and TK2101 proteins were 37% identical to each other and, among characterized proteins, the proteins were most related to an amino acid racemase from Pyrococcus horikoshii (45 and 39% identical, respectively) (28, 29) and GABA-AT from bacteria (30–40% identical).
Overexpression of the TK2101 gene and purification of the recombinant protein
We initially expressed the TK2101 gene in Escherichia coli, but the TK2101 protein was obtained in the form of inclusion bodies. We thus examined the possibilities of expressing the gene in its native host T. kodakarensis under the control of the strong promoter Pcsg, the promoter for the cell-surface glycoprotein gene TK0895. The His-tagged TK2101 protein was purified from the cell-free extract of T. kodakarensis ETK2101 by nickel chelate affinity chromatography and gel-filtration chromatography, yielding 11.8 mg of protein from an extract prepared from 1 liter of culture. An SDS-PAGE of the purified enzyme gave a single band corresponding to a molecular mass of about 50 kDa (Fig. 1), which is consistent with the molecular mass (50,635 Da) calculated from its primary amino acid sequence containing a His6 tag. In addition, the recombinant enzyme was applied to gel-filtration chromatography to determine its quaternary structure. The purified recombinant enzyme eluted as a single peak with a retention time corresponding to ∼200 kDa, indicating that the protein assembles as a tetramer.
Figure 1.
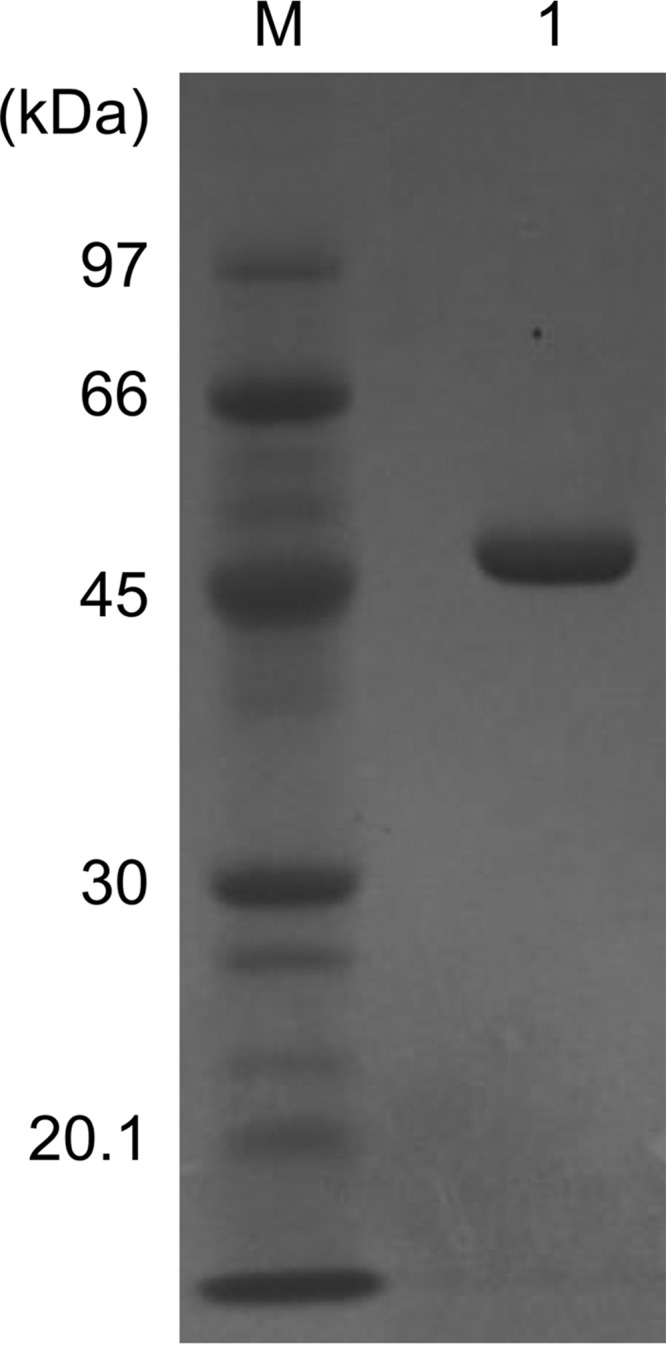
Purification of the TK2101 protein. Three micrograms of the purified TK2101 protein was applied to SDS-PAGE and stained with Coomassie Brilliant Blue (lane 1). M, molecular mass marker.
Basic enzymatic properties of the TK2101 protein
We first examined whether the TK2101 protein displayed amino acid racemase activity, as was reported for the PH0138 protein from P. horikoshii, which was a PLP-dependent amino acid racemase with broad substrate specificity (28, 29). Although activities with Ala, Val, Leu, and Ile were tested, we could not detect any racemase activity. The lack of activity toward Leu clearly indicated that the function of the TK2101 protein differed from that of the PH0138 protein. We then assessed the GABA-AT activity of the recombinant protein. Using GABA and 2-oxoglutarate as substrates, we measured the formation of l-Glu with HPLC after derivatizing the reaction products with fluorescamine. We observed a specific activity of 0.88 μmol·mg−1·min−1. The amino donor specificity of the TK2101 protein was investigated with various amine compounds at a fixed concentration of 5 mm 2-oxoglutarate. The enzyme preferentially catalyzed transamination of l-Orn and l-Lys with specific activity of 80.7 and 70.7 μmol·mg−1·min−1, respectively (Table 1). These values were much higher than that observed with GABA. Next, the potential amino acceptor substrates were examined in the presence of 5 mm l-Orn. As a result, 2-oxoglutarate served as the best amino acceptor among the compounds examined, although 2-oxoadipate also led to moderate activity (Table 2).
Table 1.
Amino donor specificity of the recombinant TK2101 enzyme
Activities toward different amino donors were measured at a final concentration of 5 mm with 5 mm 2-oxoglutarate as the amino acceptor. All reactions were independently carried out three times, and the results are given as mean ± S.D. ND means not detected. In addition to the compounds shown in the table, the l-forms of Ala, Cys, Asp, Glu, Phe, His, Ile, Leu, Met, Asn, Pro, Gln, Arg, Ser, Thr, Val, Trp, Tyr, along with Gly, were also examined. In the case of Glu, 2-oxoadipate was used as the amino acceptor. None of the compounds were utilized as amino donors.
| Amino donor | Specific activity | Relative activity |
|---|---|---|
| μmol·mg−1·min−1 | % | |
| γ-Aminobutyrate | 0.88 ± 0.03 | 1.1 |
| l-Ornithine | 80.7 ± 3.7 | 100 |
| d-Ornithine | 2.80 ± 0.12 | 3.5 |
| Nα-Acetylornithine | 2.60 ± 0.14 | 3.2 |
| l-Lysine | 70.7 ± 3.1 | 87.6 |
| d-Lysine | 13.5 ± 0.39 | 16.7 |
| Nα-Acetyllysine | 0.05 ± 0.002 | 0.06 |
| Putrescine | 0.01 ± 0.001 | 0.01 |
| l-2,4-Diaminobutyrate | ND | |
| Spermidine | ND | |
| Spermine | ND | |
| Agmatine | ND |
Table 2.
Amino acceptor specificity of the recombinant TK2101 enzyme
Activities toward keto acids were assayed at a final concentration of 5 mm with 5 mm l-ornithine as the amino donor. All reactions were independently carried out three times, and the results are given as mean ± S.D. ND means not detected.
| Amino donor | Specific activity | Relative activity |
|---|---|---|
| μmol·mg−1·min−1 | % | |
| 2-Oxoglutarate | 83.5 ± 3.5 | 100 |
| 2-Oxoadipate | 23.6 ± 0.90 | 28.3 |
| 2-Oxobutyrate | ND | |
| 2-Oxovalerate | ND | |
| Oxaloacetate | 1.75 ± 0.08 | 2.1 |
| Pyruvate | 2.67 ± 0.10 | 3.2 |
| Phenylpyruvate | ND |
Our activity measurements could not distinguish the amino group (2- or 5-amino group of Orn or the 2- or 6-amino group of Lys) used in the transamination reaction. If the α-amino group of Lys is used, the product after transamination is 2-oxo-6-aminohexanoic acid, and if the ω-amino group is used, this leads to the production of 2-aminoadipate 6-semialdehyde. The two compounds spontaneously react to produce 1-piperideine-2-carboxylate (P2C) and 1-piperideine-6-carboxylate (P6C), respectively. To determine which of the amino groups is utilized, we measured the absorbance of the products at different wavelengths after treatment with 2-aminobenzaldehyde (ABA). ABA has been reported to react slowly with P2C (5 h) but relatively quickly with P6C (10 min), forming dihydroquinazolinium complexes with absorption maximums at 450 and 465 nm, respectively (30). The product obtained with the TK2101 protein and l-Lys reacted quickly with ABA, and the reaction was completed within 10 min. Furthermore, the product displayed a peak in absorbance between 464 and 468 nm, indicating that the product of the transaminase reaction was 2-aminoadipate 6-semialdehyde, leading to the spontaneous formation of P6C. The TK2101 reaction with l-Lys was thus an ω-transaminase reaction utilizing the terminal amino group on the side chain. We also performed the same experiments with Orn. To the best of our knowledge, there is no report on the absorbance spectrum of P5C, corresponding to the product obtained from the ω-transaminase reaction with Orn, or 1-pyrroline-2-carboxylate, the product obtained from via the α-transaminase reaction. When the product of the reaction with l-Orn and 2-oxoglutarate was treated with ABA, we observed an absorbance spectrum gradually increasing until 460 nm with a dramatic drop between 460 and 470 nm. This is similar to the spectrum observed with P6C and ABA, suggesting that the reaction with l-Orn is also an ω-transamination. Another point that supports ω-transamination is the fact that among the substrates examined, only substrates with amino groups distant from the carboxyl groups resulted in the production of l-Glu. These included GABA, Nα-acetylornithine, and Nα-acetyl-lysine, whereas all of the other α-amino acids (l-forms of Ala, Cys, Asp, Glu, Phe, Gly, His, Ile, Leu, Met, Asn, Pro, Gln, Arg, Ser, Thr, Val, Trp, and Tyr) were not recognized as amino donors. Taken together, the results suggest that the TK2101 protein is an ω-aminotransferase with specificity toward l-Orn or l-Lys.
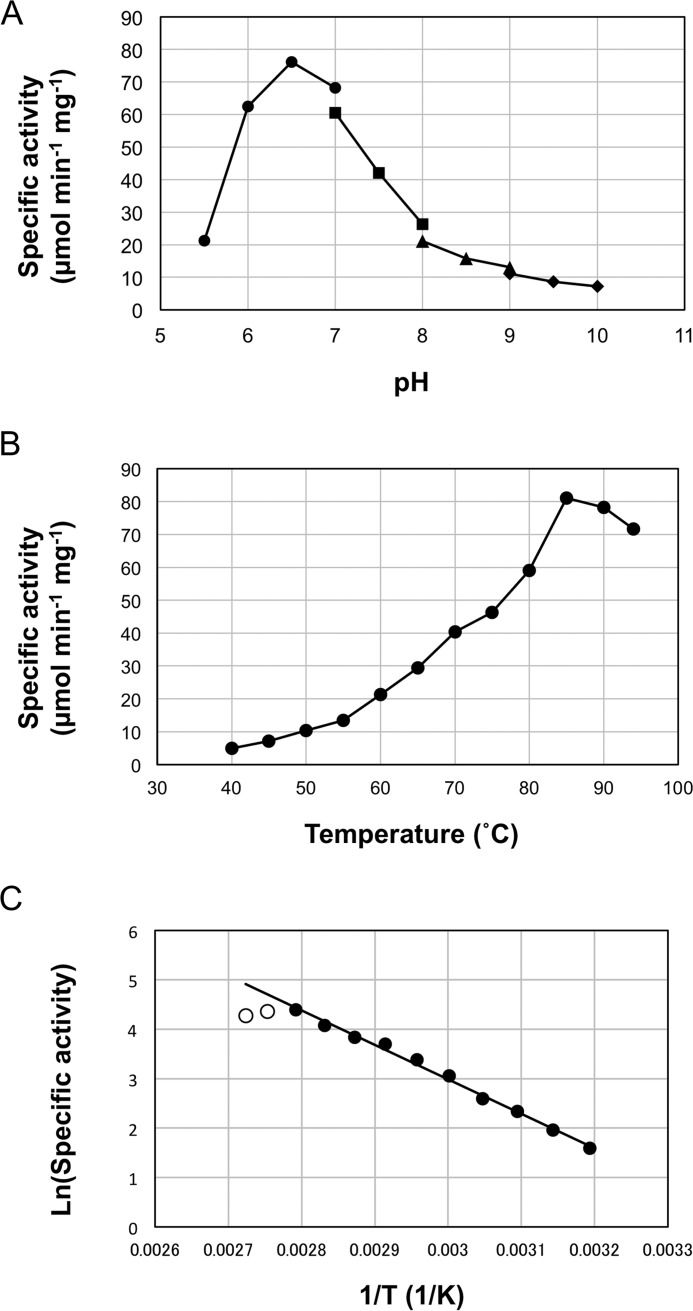
The recombinant TK2101 protein showed high activity at neutral pH with a maximum at pH 6.5 in 50 mm MES-NaOH buffer (Fig. 2A). The maximum activity of TK2101 protein was detected at 85 °C within the examined temperature range (40–94 °C), which agreed well with the optimum growth temperature of T. kodakarensis KOD1 (15). An Arrhenius plot of the data displayed linearity between 40 and 85 °C (Fig. 2B), indicating that the active site of the TK2101 protein maintains its structure within this temperature range. The activation energy for the l-Orn:2-oxoglutarate reaction was calculated to be 58.9 kJ/mol (Fig. 2C).
Figure 2.
Enzymatic properties of the TK2101 protein. A, effect of pH on the TK2101 protein activity. Filled circles, squares, triangles, and diamonds represent the activities in MES (pH 5.5–7.0), HEPES (pH 7.0–8.0), Tricine (pH 8.0–9.0), and CHES (pH 9.0–10.0), respectively. B, effect of temperature on the TK2101 protein activity. C, Arrhenius plot of the data shown in B. Only filled circles were used to estimate the activation energy of the reaction.
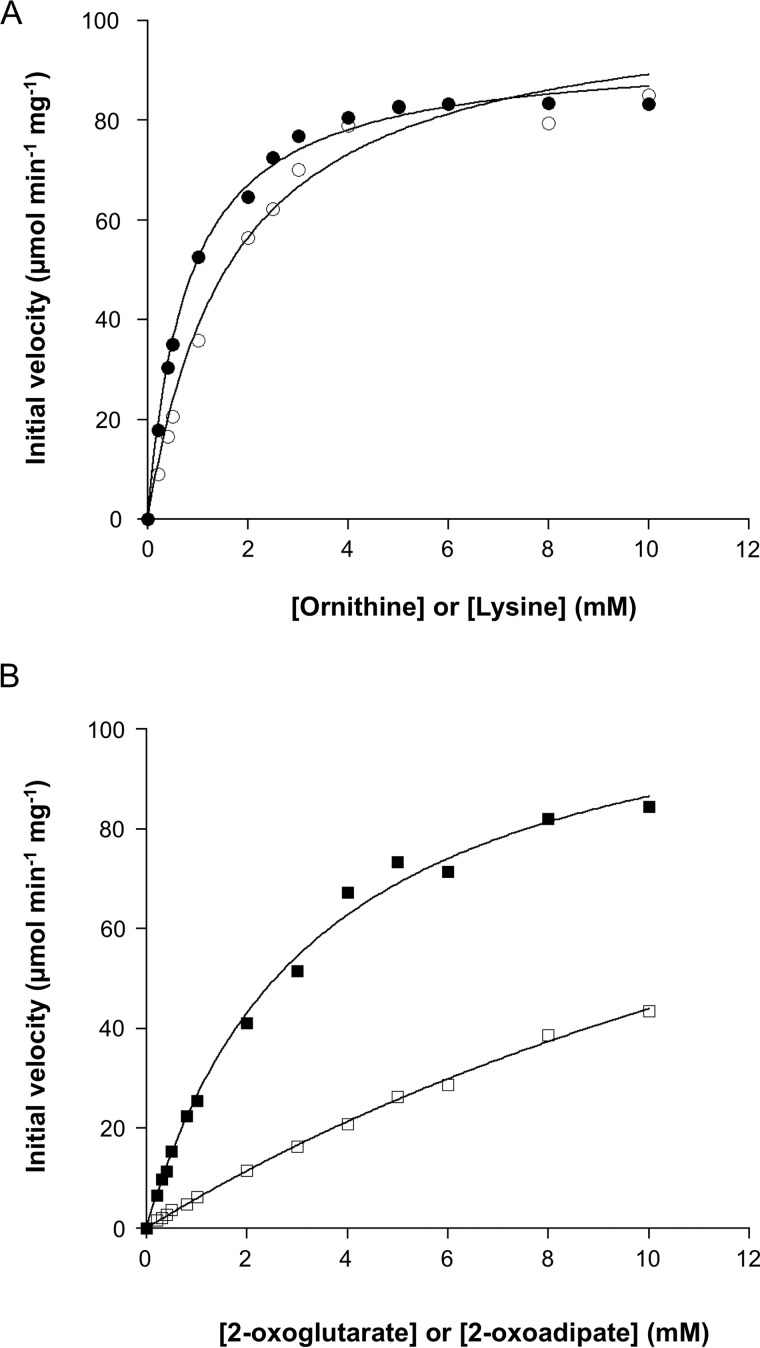
Kinetic studies
Kinetic studies on the aminotransferase activity were performed by varying the concentration of one substrate in the presence of a fixed concentration of the other (5 mm). The kinetics toward all substrates (l-Orn, l-Lys, 2-oxoglutarate, and 2-oxoadipate) followed Michaelis-Menten kinetics (Fig. 3). The obtained kinetic parameters are shown in Table 3. The kcat/Km value with l-Orn and 2-oxoglutarate was 2- and 4-fold higher than that with l-Lys and 2-oxoadipate, respectively, suggesting that l-Orn and 2-oxoglutarate are the preferred amino donor and acceptor of the TK2101 protein, respectively.
Figure 3.
Kinetic studies on the TK2101 protein. A, initial velocities of the aminotransferase reactions with varying concentrations of l-Orn and l-Lys in the presence of 5 mm 2-oxoglutarate. Filled circles and open circles represent initial velocities with l-Orn and l-Lys, respectively. B, initial velocities of the aminotransferase reactions with varying concentrations of 2-oxoglutarate and 2-oxoadipate in the presence of 5 mm l-Orn. Filled squares and open squares represent initial velocities with 2-oxoglutarate and 2-oxoadipate, respectively.
Table 3.
Kinetic parameters of the TK2101 protein reaction
| Substrate with 5 mm 2-oxoglutarate |
Substrate with 5 mm
l-ornithine |
|||
|---|---|---|---|---|
| l-Ornithine | l-Lysine | 2-Oxoglutarate | 2-Oxoadipate | |
| Vmax (μmol min−1 mg−1) | 93.8 ± 1.4 | 104 ± 4 | 116 ± 4 | 149 ± 14 |
| Km (mm) | 0.80 ± 0.05 | 1.71 ± 0.22 | 3.38 ± 0.30 | 23.9 ± 3.0 |
| kcat (s−1) | 77.6 ± 1.1 | 86.4 ± 3.5 | 95.8 ± 3.5 | 124 ± 12 |
| kcat/Km (mm−1 s−1) | 97.1 | 50.5 | 28.3 | 5.17 |
Growth of T. kodakarensis KU216 in the absence of proline
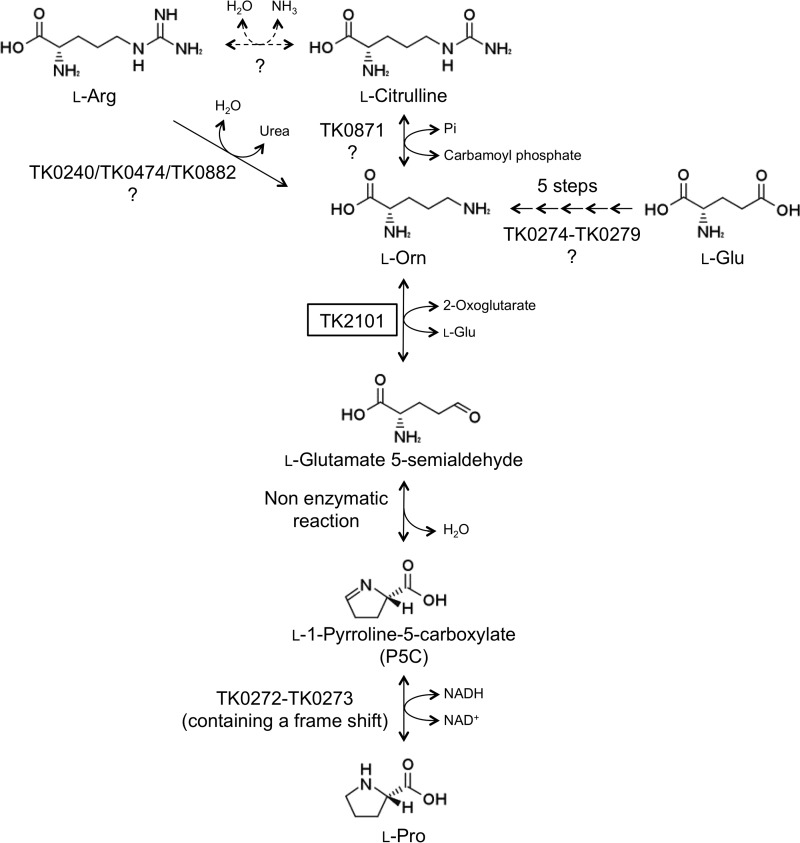
We had previously regarded the wildtype strain T. kodakarensis KOD1 as a Pro auxotroph. This was based on the fact that we could not observe growth within 16 h in the absence of l-Pro in the synthetic medium ASW-AA-S0. Another point was that during genome analysis, we observed that the gene encoding P5C reductase, the enzyme catalyzing the final reaction in l-Pro biosynthesis (Fig. 4), was split into two open reading frames (Fig. S1).
Figure 4.
Putative biosynthesis pathway for Pro in T. kodakarensis. It is expected that l-Pro can be synthesized from l-Orn through three steps containing one non-enzymatic reaction. Possible routes for l-Orn biosynthesis are also shown.
However, as described above, the TK2101 protein displayed a maximum kcat/Km value toward l-Orn/2-oxoglutarate, indicating its role as an Orn-ΑΤ. As this enzyme activity is involved in l-Pro biosynthesis from l-Orn in various organisms, we felt it necessary to reexamine the ability of T. kodakarensis to synthesize l-Pro. We thus grew T. kodakarensis KU216 in the synthetic medium ASW-AA-S0 without l-Pro. As a result, we observed a relatively slow, but highly reproducible, growth of the strain in the absence of l-Pro, indicating that T. kodakarensis does have the ability to synthesize l-Pro (data not shown).
Growth characterization of the TK2101 gene disruption strain
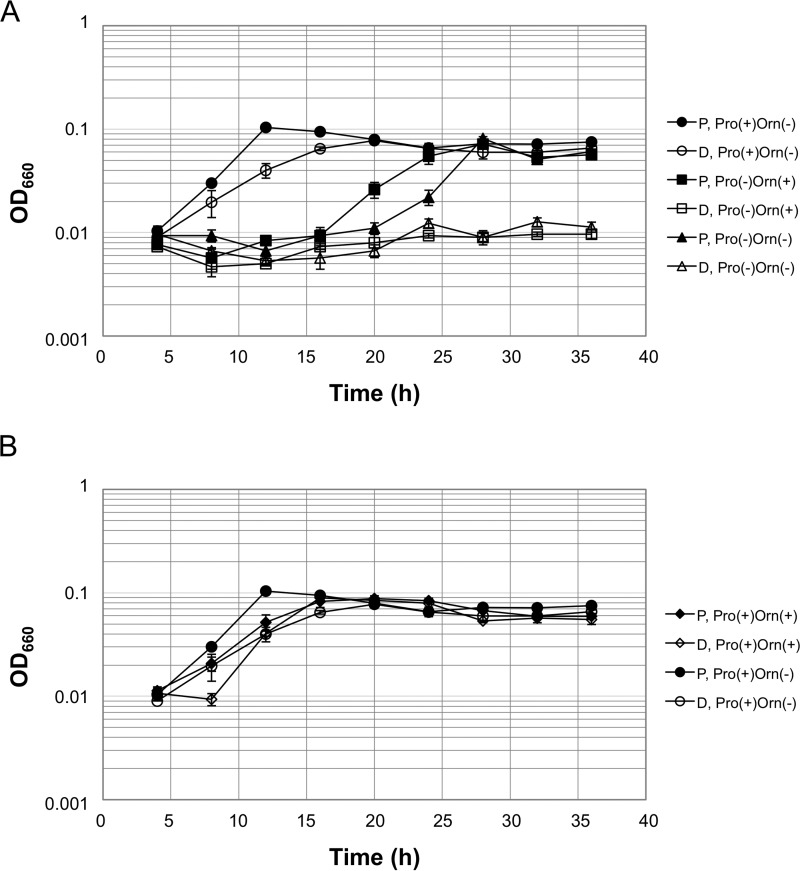
To examine the physiological role of the ω-aminotransferase activity of the TK2101 protein, a gene disruption strain was constructed, and its phenotype was compared with its parent strain T. kodakarensis KU216. The disruption plasmid was designed so that gene disruption would occur via single crossover insertion of the plasmid, followed by pop-out recombination. Cells that had undergone single crossover insertion were enriched by growing the transformants in a uracil-free medium, and cells that had further undergone pop-out recombination were selected on solid medium supplemented with 5-FOA. PCR analysis and DNA sequencing confirmed the complete deletion of TK2101 from the genome of T. kodakarensis (Fig. S2).
The TK2101 gene disruption strain (ΔTK2101) and the parent KU216 strain both exhibited growth in a synthetic medium with all 20 amino acids, including l-Pro (ASW-AA-S0-Pyr-Ura-W-Pro(+)-Orn(−)) (Fig. 5A). As described above, when l-Pro was omitted from the medium (ASW-AA-S0-Pyr-Ura-W-Pro(−)-Orn(−)), the growth of the parent strain was significantly retarded, but it eventually reached cell yields similar to those observed when the strain was grown in ASW-AA-S0-Pyr-Ura-W-Pro(+)-Orn(−). In contrast, the ΔTK2101 strain did not grow at all in the absence of l-Pro, clearly indicating the involvement of TK2101 in l-Pro biosynthesis. As expected, the addition of l-Orn did not complement the lack of l-Pro with ΔTK2101 cells (ASW-AA-S0-Pyr-Ura-W-Pro(−)-Orn(+), as l-Orn is a metabolite upstream of the TK2101 aminotransferase reaction. Interestingly, the addition of l-Orn enhanced the growth of the parent strain grown in the absence of l-Pro, but enhancement was not observed in the presence of l-Pro (Fig. 5B). Similarly, the addition of l-Orn in the presence of l-Pro did not enhance the growth of ΔTK2101 cells. This further supports the notion that l-Pro biosynthesis occurs via l-Orn and that the growth enhancement was due to the use of l-Orn to supply l-Pro.
Figure 5.
Growth properties of the parent and the TK2101 gene disruption strain in synthetic amino acid medium. A, growth comparison of the parent and ΔTK2101 strains in the absence of l-Pro and the effects of adding either l-Pro or l-Orn. B, effect of adding l-Orn in the presence of l-Pro. Each value is an average of those from three independent growth experiments. The vertical axis is represented in logarithmic scale. Filled and open symbols represent the parent strain (P) and ΔTK2101 (D), respectively. All media are based on ASW-AA-S0-Pyr-Ura-W. Strains and the presence or absence of l-Orn or l-Pro are indicated on the right side of the growth curves.
Discussion
Our biochemical analysis revealed that the TK2101 protein showed aminotransferase activity toward l-Orn, l-Lys, d-Orn, d-Lys, N-acetylornithine, GABA, N-acetyl-lysine, and putrescine (with 2-oxoglutarate) and could utilize 2-oxoglutarate, 2-oxoadipate, pyruvate, and oxaloacetate as amino acceptor (with l-Orn). The broad substrate spectrum of TK2101 is in contrast to most of the previously reported Orn-ATs (13, 14, 31–34), which were highly specific for l-Orn and 2-oxoglutarate as amino donor and acceptor, respectively. However, the specific activity of the enzyme for l-Orn and l-Lys was significantly higher than those of other candidate compounds, indicating that the TK2101 protein can be regarded as an Orn-AT or Lys-AT. The ΔTK2101 strain did not grow in the absence of l-Pro, strongly supporting that the TK2101 protein functions as an Orn-AT in T. kodakarensis. The lack of growth in this medium also suggests that there are no other proteins in T. kodakarensis that display physiologically relevant Orn-AT activity, including the TK1211 protein.
Although relatively low, the TK2101 protein displays similarity to several known eukaryotic and bacterial Orn-AT from Saccharomyces cerevisiae (23% identical), Homo sapiens (27% identical), and Bacillus subtilis (29% identical). Multiple amino acid sequence alignments of these proteins confirmed the presence of conserved domains in ω-aminotransferases (Fig. 6). Based on the structure of the protein from H. sapiens, the cofactor PLP is covalently bound through a Schiff base linkage to Lys-292, whereas Glu-235 and Asp-263 residues interact with the 3-OH and the N1 of the pyridine ring (35). The conserved nucleotide-binding sequence, GXGXXG, of ω-aminotransferase was also found in the TK2101 protein as the sequence 268GIGRTG273.
Figure 6.
Sequence alignment of the TK2101 protein and previously described Orn AT proteins. The circles indicate Lys-292, to which the PLP cofactor is bound, and Glu-235 and Asp-248, which interact with the pyridine ring, based on the structure of the protein from H. sapiens. The nucleotide-binding motif conserved in ω-aminotransferases (Gly-268–Ile–Gly–Arg–Thr–Gly-273) is indicated by a thick bar. Conserved residues are indicated with asterisks. Abbreviations: Bsu, B. subtilis; Hsa, H. sapiens; Sce, S. cerevisiae; Tko, T. kodakarensis.
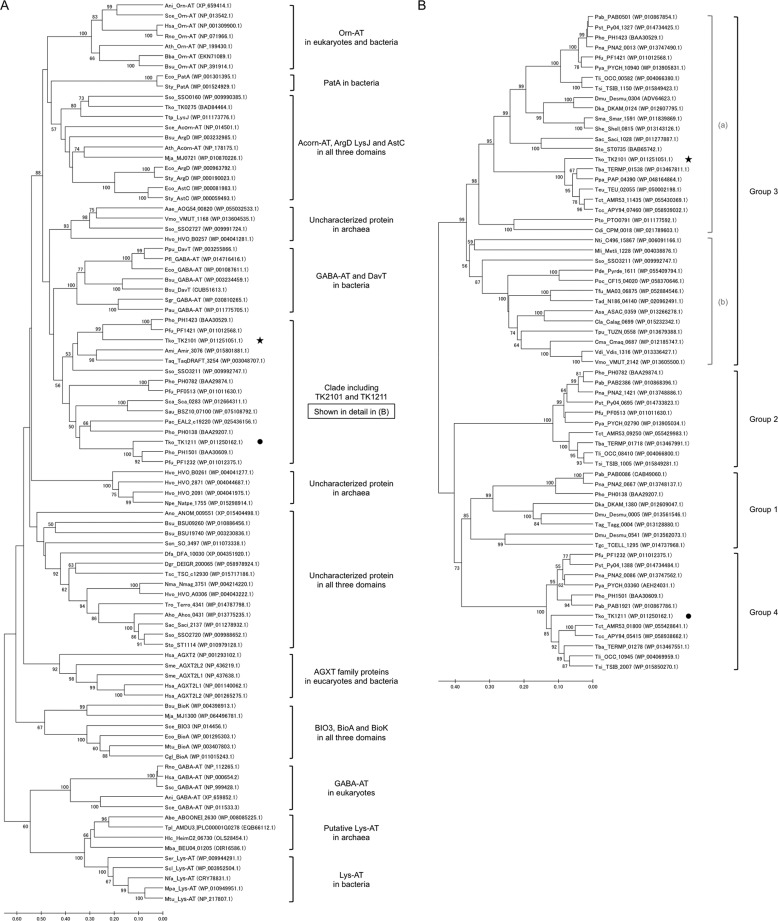
To gain insight on the structural relationships among a wide range of aminotransferases from all three domains of life, we collected sequences displaying similarity to the TK2101 protein, the GABA-AT from E. coli and S. cerevisiae, Orn-AT from B. subtilis and S. cerevisiae, and Lys-AT from Saccharopolyspora erythraea. The BLAST searches resulted in tremendous numbers of related sequences (>10,000), but we expanded the searches until members of another branch in the tree shown in Fig. 7A were identified. Thus, the tree in Fig. 7 should in principle represent all of the major sequence clades structurally related to known ω-aminotransferases, including TK2101. We observed a number of protein clades, some of which whose functions have been experimentally confirmed. These include the Orn-AT from eukaryotes and bacteria, the bacterial putrescine aminotransferase (PatA), and the acetyl-/succinylornithine aminotransferases (Acorn-AT, ArgD/AstC), and the protein-modified aminoadipate semialdehyde aminotransferase (LysJ) from all three domains. The tree clarifies that the Orn-AT identified here in T. kodakarensis resides in a clade distinct to previously characterized enzymes from eukaryotes and bacteria. The most related branch comprises bacterial GABA-AT and 5-aminovalerate aminotransferases (DavT). Among the Orn-AT sequences, the TK2101 sequence is not relatively close to the previously reported bacterial and eukaryotic Orn-ATs. The clade, including TK2101 and TK1211, harbors a large number of uncharacterized proteins from archaea and bacteria and is shown in Fig. 7B with additional sequences. The clade seems to consist of four groups, which were previously proposed in the study on the PLP-dependent amino acid racemase (PH0138 protein) from P. horikoshii (28). We can therefore presume that group 1 consists of proteins with similar function. The TK2101 sequence resides in group 3, a relatively large group that might be divided into two subgroups, 3a and 3b. We refrained from dividing this group, as we could not find an organism that harbors two proteins within this group, whereas P. horikoshii and Pyrococcus abyssi, for example, harbor one member in each of the four groups. Our results suggest that group 3 (or at least 3a), which includes TK2101, represents Orn-ATs. In contrast, the functions of enzymes belonging to groups 2 and 4 have not yet been identified. Also in the broad tree shown in Fig. 7A, there are still a number of large groups whose members have not been characterized.
Figure 7.
Phylogenetic trees of the class III aminotransferase sequences. A, tree including various class III aminotransferase sequences from all three domains, including Orn-AT from eukaryotes and bacteria. B, detailed tree of the clade containing TK1211 and TK2101, which was constructed with only aminotransferase sequences from archaea. Bootstrap values above 50 are shown. In both trees, the TK2101 (star) and TK1211 (circle) sequences are highlighted. Abbreviation of enzymes are as follows: Acorn AT, acetylornithine aminotransferase (Eukaryotes); AGXT2, alanine-glyoxylate aminotransferase; AGXT2L1, ethanolamine–phosphate phospho-lyase; AGXT2L2, 5-phosphohydroxy-l-lysine phospho-lyase; ArgD, acetylornithine aminotransferase (Bacteria); AstC, succinylornithine transaminase; BIO3, adenosylmethionine-8-amino-7-oxononanoate aminotransferase (Eukaryotes); BioA, adenosylmethionine-8-amino-7-oxononanoate aminotransferase (Bacteria); BioK, l-lysine-8-amino-7-oxononanoate transaminase; DavT, 5-aminovalerate aminotransferase; GABA AT, 4-aminobutyrate aminotransferase; Lys AT, l-lysine-ϵ-aminotransferase; LysJ, (LysW)-aminoadipate semialdehyde transaminase; Orn AT, ornithine aminotransferase; PatA, putrescine aminotransferase. Abbreviation of organisms: Aae, Acidiplasma aeolicum; Abo, Aciduliprofundum boonei; Aho, Acidianus hospitalis; Ami, Actinosynnema mirum; Ani, Aspergillus nidulans; Ano, Aspergillus nomius; Asa, Acidilobus saccharovorans; Ath, Arabidopsis thaliana; Bba, Bacillus bataviensis; Bsu, B. subtilis; Cdi, Cuniculiplasma divulgatum; Cgl, Corynebacterium glutamicum; Cla, Caldisphaera lagunensis; Cma, Caldivirga maquilingensis; Dfa, Dictyostelium fasciculatum; Dgr, Deinococcus grandis; Dka, Desulfurococcus kamchatkensis; Dmu, Desulfurococcus mucosus; Eco, E. coli; Hlc, Candidatus heimdallarchaeota archaeon LC_2; Hsa, H. sapiens; Hvo, Haloferax volcanii; Mba, Marine Group III euryarchaeote CG-Bathy1; Mja, Methanocaldococcus jannaschii; Mli, Methanofollis liminatans; Mpa, Mycobacterium paratuberculosi; Mtu, Mycobacterium tuberculosis; Nfa, Nocardia farcinica; Nma, Natrialba magadii; Npe, Natrinema pellirubrum; Nti, Natronorubrum tibetense; Pab, P. abyssi; Pac, Peptoclostridium acidaminophilum; Pau, Paenarthrobacter aurescens; Pde, Pyrodictium delaneyi; Pfl, Pseudomonas fluorescens; Pfu, P. furiosus; Pho, P. horikoshii; Pna, Pyrococcus sp. strain NA2; Poc, Pyrodictium occultum; Ppa, Palaeococcus pacificus; Ppu, Pseudomonas putida; Pst, Pyrococcus sp. ST04; Pto, Picrophilus torridus; Pya, Pyrococcus yayanosii; Rno, Rattus norvegicus; Sac, Sulfolobus acidocaldarius; Sau, Staphylococcus aureus; Sca, Staphylococcus carnosus; Sce, S. cerevisiae; Scl, Streptomyces clavuligerus; Ser, S. erythraea; Sgr, Streptomyces griseus; She, Staphylothermus hellenicus; Sma, Staphylothermus marinus; Sme, Sinorhizobium meliloti; Son, Shewanella oneidensis; Ssc, Sus scrofa; Sso, Sulfolobus solfataricus; Sto, Sulfolobus tokodaii; Sty, Salmonella typhimurium; Tad, Thermofilum adornatus; Tag, Thermosphaera aggregans; Taq, Thermus aquaticus; Tba, Thermococcus barophilus; Tcc, Thermococcus celericrescens; Tct, Thermococcus thioreducens; Teu, Thermococcus eurythermalis; Tfu, Thermofilum uzonense; Tgc, Thermogladius cellulolyticus; Tko, T. kodakarensis; Tli, Thermococcus litoralis; Tpl, Thermoplasmatales archaeon I-plasma; Tpu, Thermoproteus uzoniensis; Tro, Terriglobus roseus; Tsc, Thermus scotoductus; Tsi, Thermococcus sibiricus; Ttp, Thermus thermophiles; Vdi, Vulcanisaeta distributa; Vmo, Vulcanisaeta moutnovskia.
The predicted pathway from l-Orn to l-Pro is shown in Fig. 4. The TK2101 protein is responsible for the conversion of l-Orn to l-glutamate 5-semialdehyde coupled with the conversion of 2-oxoglutarate to l-Glu. l-Glutamate 5-semialdehyde is spontaneously converted to P5C. The final reaction is the reduction of P5C to l-Pro catalyzed by P5C reductase (36). The T. kodakarensis genome harbors two consecutive genes (TK0272 and TK0273) that encode proteins that are 59 and 77% identical to the N- and C-terminal halves of the P5C reductase from Pyrococcus furiosus (Fig. S1A). The gene structure suggests that there has been a 2-bp insertion near the stop codon of TK0272, between nucleotides 295 and 309 (310–312 corresponds to a stop codon that can be confirmed in Fig. S1B). The two genes are separated by 14 bp. However, the translation of the intergenic region results in an amino acid sequence almost identical to that of the corresponding region in the P. furiosus P5C reductase. The two proteins in T. kodakarensis may function together to exhibit P5C reductase.
The pathways involved in the biosynthesis of l-Orn in T. kodakarensis are still not clear. Judging from the genome sequence data, there are at least three possibilities. In the first route, l-Orn is generated from l-Glu. Six genes (TK0274–TK0279) encode the proteins responsible for the biosynthesis of l-Lys from 2-aminoadipate. It has clearly been shown that the same proteins are capable of generating l-Orn from l-Glu in vitro (37). The second and third possible routes produce l-Orn from l-Arg, either directly by arginine ureohydrolase or via l-citrulline by the enzymes arginine deiminase and ornithine transcarbamoylase (38). TK0240, TK0474, and TK0882 are 28, 21, and 15% identical to the arginine ureohydrolase from B. subtilis and may be involved in the conversion of l-Arg to l-Orn. Concerning the pathway via l-citrulline, TK0871 is 89% identical to the ornithine carbamoyltransferase from P. furiosus (39). However, we could not find a gene that encodes a protein with significant similarity with arginine deiminase. Genetic studies on these three routes will be necessary to determine which routes are responsible for l-Orn generation in T. kodakarensis.
Another point that will need consideration is the ω-aminotransferase activity of the TK2101 protein toward l-Lys. The reaction, with 2-oxoglutarate as the amino acceptor, would lead to the generation of 2-aminoadipate 6-semialdehyde and l-Glu. This reaction may simply be used for the re-distribution of amino groups via l-Glu for the generation of other amino acids when l-Lys is in excess. This is regarded as the function of the multiple α-aminotransferases in T. kodakarensis. Why an ω-aminotransferase activity, and not an α-aminotransferase activity is used is not clear at present. One point that may be worthy to mention is that 2-aminoadipate 6-semialdehyde may be of more metabolic use than 2-oxo-6-aminohexanoate, the product of the α-aminotransferase activity. 2-Aminoadipate 6-semialdehyde can be oxidized to 2-aminoadipate, which can be used once again as a precursor for l-Lys biosynthesis (40). It can also be converted via P6C to pipecolate. Some bacteria use this pathway to generate pipecolate, a compatible solute that functions to relieve osmotic stress (41).
Experimental procedures
Strains, media, and culture conditions
T. kodakarensis KU216 (22) and mutant strains were cultivated under anaerobic conditions at 85 °C in nutrient-rich medium (ASW-YT) or a synthetic medium (ASW-AA). ASW-YT medium consists of 0.8× artificial seawater (ASW), 5 g liter−1 yeast extract, and 5 g liter−1 tryptone. Elemental sulfur (2 g liter−1) and sodium pyruvate (5 g liter−1) were added prior to inoculation to prepare ASW-YT-S0 and ASW-YT-Pyr medium, respectively. ASW-AA medium consisted of 0.8× ASW, a mixture of 20 amino acids, modified Wolfe's trace minerals, and a vitamin mixture (21). For all liquid media, resazurine (0.5 mg liter−1) was supplemented as an oxygen indicator, and 5.0% Na2S H2O was added until the medium became colorless. For solid medium, elemental sulfur was replaced with 2 ml of polysulfide solution (10 g of Na2S 9H2O and 3 g of sulfur flowers in 15 ml of H2O) per liter and 1% gelrite was added to solidify the medium. E. coli strain DH5α used for plasmid construction was grown at 37 °C in lysogeny broth (LB) medium containing ampicillin (100 mg liter−1). Unless otherwise stated, all chemicals were purchased from Wako Pure Chemicals (Osaka, Japan) or Nacalai Tesque (Kyoto, Japan).
Disruption of the TK2101 gene
To construct the gene disruption plasmid, the TK2101 gene along with its 5′- and 3′-flanking regions (∼1.0 kbp) was amplified from T. kodakarensis KU216 genomic DNA using the primer set 2101F1/2101R1 (Table S1). The fragment was inserted into the plasmid pUD3 at the HincII site, which contains the pyrF gene of T. kodakarensis inserted in the ApaI site of pUC118. Inverse PCR was performed with the primer set 2101F2/2101R2 (Table S1) to remove the target gene (TK2101) from the resulting plasmid. The sequences of the inserted regions were confirmed.
T. kodakarensis KU216 (ΔpyrF), exhibiting uracil auxotrophy, was used as the parent strain for TK2101 gene disruption. Cells grown in ASW-YT-S0 medium for 12 h were harvested, resuspended in 200 μl of 0.8× ASW, and kept on ice for 30 min. After addition of 3.0 μg of the constructed plasmid DNA and further incubation on ice for 1 h, cells were cultivated in ASW-AA-S0 medium without uracil for 48 h at 85 °C. Then, 200 μl of the cultures was transferred to a fresh medium and cultivated under the same conditions to enrich the desired transformants displaying uracil prototrophy. The culture was then diluted with 0.8× ASW and spread onto ASW-YT solid medium supplemented with 10 g liter−1 5-FOA and 60 mm NaOH. Only cells that have undergone a pop-out recombination can grow in the presence of 5-FOA. After cultivation at 85 °C for 2 days, colonies were selected, and their genotypes were analyzed with the primer sets 2101F1/2101R1 and 2101F3/2101R3 (Table S1). The transformants whose amplified DNA products showed the expected size were chosen and cultivated in ASW-YT-S0 medium. Relevant sequences were confirmed for the absence of unintended mutations.
Characterization of the TK2101 gene disruption strain
When we examined the growth of the TK2101 gene disruption strain (ΔTK2101), cells were grown in ASW-AA medium supplemented with 0.5 mg liter−1 resazurine, 12.5 mg liter−1 Na2S 9H2O, 5 mg liter−1 uracil, 10 μm tungsten, 2 g liter−1 elemental sulfur, and 5 g liter−1 sodium pyruvate but without l-Pro (ASW-AA-S0-Pyr-Ura-W-Pro(−)-Orn(−)). This medium supplemented either with 2.0 mm l-Pro or 2.0 mm l-Orn or both was also used and designated ASW-AA-S0-Pyr-Ura-W-Pro(+)-Orn(−), ASW-AA-S0-Pyr-Ura-W-Pro(−)-Orn(+), or ASW-AA-S0-Pyr-Ura-W-Pro(+)-Orn(+), respectively. The same number of cells, pre-cultivated until the stationary phase in ASW-AA-S0-Ura-W liquid media for 12–16 h, was inoculated to the four media and incubated at 85 °C. Growth curves were constructed by monitoring optical density at 660 nm (OD660). Growth experiments were performed in triplicate.
Construction of plasmid for overexpression of TK2101 in T. kodakarensis
The TK2101 gene was amplified from the genomic DNA of T. kodakarensis KU216 by PCR using a primer set 2101F3/2101R3 (Table S1), and a His tag sequence was incorporated in the N terminus for use in purification. Using the NdeI-SalI restriction enzyme sites incorporated during the PCR, the amplified fragment was inserted into a plasmid previously used for overexpression of TK2141 (27). The resulting TK2101 overexpression cassette (Pcsg:: TK2101::TchiA) was amplified from the plasmid with the primer set 2101F4/2101R4 (Table S1). The cassette was inserted into a T. kodakarensis–E. coli shuttle vector pRPG02.4 pRPG02 is based on pUC118 but with a replication initiator rep74 from pLC64 (19), a pyruvoyl-dependent arginine decarboxylase gene (pdaD) from P. furiosus, and a multicloning site. Proper construction of the plasmid, designated here as pRPETK2101, was confirmed by DNA sequencing.
Overexpression and purification of the recombinant TK2101 protein
T. kodakarensis KPD1 (ΔpyrF, ΔpdaD) was used as the host strain for TK2101 gene expression. pdaD corresponds to TK0149, and disruption of this gene results in agmatine auxotrophy (20, 38). For transformation, T. kodakarensis KPD1 was cultivated in ASW-YT-S0 supplemented with agmatine (0.5 mm) for 12 h at 85 °C. Cells were harvested and resuspended in 200 μl of 0.8× ASW, followed by incubation on ice for 30 min. After mixing with 3.0 μg of the expression plasmid and further incubation on ice for 1 h, cells were spread onto solid ASW-YT-S0 medium. After cultivation at 85 °C for 24 h, transformants displaying agmatine prototrophy were isolated and cultivated in ASW-YT-S0. The presence of the plasmid was confirmed by PCR.
The TK2101 overexpression strain (ETK2101) was cultivated in ASW-YT-Pyr for 12 h at 85 °C. Cells were harvested, washed with 0.8× ASW, resuspended in 20 mm sodium phosphate buffer (pH 7.4) containing 500 mm KCl, 20 mm imidazole, and 0.1 mm PLP and disrupted by sonication. After centrifugation (15,000 × g, 20 min), the supernatant was applied to His GraviTrap (GE Healthcare). The proteins with His tags were eluted with 20 mm sodium phosphate buffer (pH 7.4) containing 500 mm KCl and 500 mm imidazole. After the buffer was exchanged to 50 mm sodium phosphate buffer (pH 7.4) containing 150 mm NaCl and 0.1 mm PLP, the solution was applied to a Superdex 200 10/300 GL gel-filtration column (GE Healthcare) with a mobile phase of 50 mm sodium phosphate buffer (pH 7.4) containing 150 mm NaCl and 0.1 mm PLP at a flow rate of 0.5 ml min−1. The same chromatography conditions were used to determine the molecular mass of the recombinant TK2101 protein with the standard proteins ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), ovalbumin (44 kDa), and ribonuclease (13.7 kDa). Protein concentrations were determined with a protein assay kit (Bio-Rad) with bovine serum albumin as standard.
Enzyme assay
Aminotransferase activity of the TK2101 protein was assayed by determination of l-Glu or P5C formed. When l-Glu was used as the amino donor, 2-oxoadipate was used as the amino acceptor, and the formation of 2-aminoadipate was monitored. The standard reaction mixture (300 μl), consisting of 50 mm MES (pH 6.5 at 85 °C), 5 mm l-Orn, 5 mm 2-oxoglutarate, 0.1 mm PLP, and 1 μg ml−1 recombinant TK2101 protein, was incubated at 85 °C for 3, 5, and 10 min to confirm that product formation was linear with time. The reaction was stopped by cooling the mixture on ice, and proteins in the reaction mixture were removed by ultrafiltration using Amicon Ultra-0.5 10 K (Millipore, Billerica, MA). One unit of enzyme activity was defined as the amount of enzyme catalyzing the formation of 1.0 μmol of l-Glu or P5C. For the determination of kinetic constants, l-Orn or l-Lys was varied from 0 to 10 mm in the presence of 5 mm 2-oxoglutarate, or 2-oxoglutarate or 2-oxoadipate was varied from 0 to 10 mm in the presence of 5 mm l-Orn. The reaction was stopped after 5 min when less than 10% of the substrate was consumed. Kinetic parameters were obtained by fitting the data to the Michaelis-Menten equation using IGOR PRO, version 6.03 (Wave-Metrics, Lake Oswego, OR).
The P5C generated from l-Orn by transamination was derivatized with ABA. The reaction mixture (300 μl) was mixed with 250 μl of 0.5% (w/v) ABA in 96% ethanol for a yellow-orange color development by incubation at 37 °C for 15 min. The amount of P5C was determined at 440 nm by using the adsorption coefficient of 1.9 × 106 cm2 ml−1 reported by Voellmy and Leisinger (42).
l-Glu was detected and quantified by HPLC after derivatization with fluorescamine. The derivatization mixture (500 μl) contained 30 μl of reaction mixture, 370 μl of borate sodium hydroxide buffer (0.5 m (pH 10.0)), and 100 μl of fluorescamine (3 g liter−1). After derivatization for 3 min at room temperature in the dark, an aliquot (10 μl) of the solution was applied to a Cosmosil 5C18-PAQ 250 × 4.60 mm column (Nacalai Tesque) using an LC-VP system (Shimadzu, Kyoto, Japan). Compounds were eluted with a solution of 20 mm sodium acetate (pH 5.9) and 15% (v/v) acetonitrile at a flow rate of 1.0 ml min−1. The excitation and emission wavelengths for fluorescent detection were 390 and 460 nm, respectively.
Effects of pH and temperature on enzyme activity
For examining the pH dependence of the enzyme activity, the transamination reaction was performed at various pH values using the following buffers: MES-NaOH (50 mm (pH 5.5–7.0)), HEPES (50 mm (pH 7.0–8.0)), Tricine (50 mm (pH 8.0–9.0)), and CHES (50 mm (pH 9.0–10.0)). To investigate the effects of temperature, the aminotransferase activity was measured at various temperatures (40–94 °C) under the standard assay conditions.
Phylogenetic analysis
Sequences of aminotransferase class III proteins from various organisms were collected and aligned by using multiple sequence alignment software ClustalW of Molecular Evolutionary Genetic Analysis (MEGA) software version 7.0.26 (43). Phylogenetic trees were constructed with the method UPGMA (Unweighted Pair Group Method with Arithmetic Mean) by using the software. Boot-strap resampling was performed 1000 times.
Author contributions
R.-C. Z. and S.-i. H. formal analysis; R.-C. Z. and S.-i. H. validation; R.-C. Z., S.-i. H., and H. T. investigation; R.-C. Z., S.-i. H., and H. T. methodology; R.-C. Z., S.-i. H., and H. A. writing-original draft; T. I., Y.-G. Z., M. N., and H. A. writing-review and editing; H. A. conceptualization; H. A. supervision; H. A. funding acquisition.
Supplementary Material
This work was supported in part by the Core Research for Evolutional Science and Technology Program of the Japan Science and Technology Agency (to H. A.) within the research area “Creation of Basic Technology for Improved Bioenergy Production through Functional Analysis and Regulation of Algae and Other Aquatic Microorganisms.” The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S2 and Table S1.
K. Yoshida, T. Kanai, and H. Atomi, unpublished data.
- PLP
- pyridoxal 5′-phosphate
- CHES
- 2-(N-cyclohexylamino)ethanesulfonic acid
- Tricine
- N-[Tris(hydroxymethyl)-methyl]glycine
- -AT
- -aminotransferase
- ABA
- 2-aminobenzaldehyde
- Acorn-AT
- N-acetylornithine aminotransferase
- P5C
- l-1-pyrroline-5-carboxylate
- P2C
- 1-piperideine-2-carboxylate
- P6C
- 1-piperideine-6-carboxylate
- ASW
- artificial seawater
- 5-FOA
- 5-fluoroorotic acid.
References
- 1. Uchida Y., Hayashi H., Washio T., Yamasaki R., Kato S., and Oikawa T. (2014) Cloning and characterization of a novel fold-type I branched-chain amino acid aminotransferase from the hyperthermophilic archaeon Thermococcus sp. CKU-1. Extremophiles 18, 589–602 10.1007/s00792-014-0642-0 [DOI] [PubMed] [Google Scholar]
- 2. Xing R. Y., and Whitman W. B. (1992) Characterization of amino acid aminotransferases of Methanococcus aeolicus. J. Bacteriol. 174, 541–548 10.1128/jb.174.2.541-548.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andreotti G., Cubellis M. V., Nitti G., Sannia G., Mai X., Marino G., and Adams M. W. (1994) Characterization of aromatic aminotransferases from the hyperthermophilic archaeon Thermococcus litoralis. Eur. J. Biochem. 220, 543–549 10.1111/j.1432-1033.1994.tb18654.x [DOI] [PubMed] [Google Scholar]
- 4. Ward D. E., de Vos W. M., and van der Oost J. (2002) Molecular analysis of the role of two aromatic aminotransferases and a broad-specificity aspartate aminotransferase in the aromatic amino acid metabolism of Pyrococcus furiosus. Archaea 1, 133–141 10.1155/2002/959031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marino G., Nitti G., Arnone M. I., Sannia G., Gambacorta A., and De Rosa M. (1988) Purification and characterization of aspartate aminotransferase from the thermoacidophilic archaebacterium Sulfolobus solfataricus. J. Biol. Chem. 263, 12305–12309 [PubMed] [Google Scholar]
- 6. Tanaka T., Yamamoto S., Moriya T., Taniguchi M., Hayashi H., Kagamiyama H., and Oi S. (1994) Aspartate aminotransferase from a thermophilic formate-utilizing methanogen, Methanobacterium thermoformicicum strain SF-4: relation to serine and phosphoserine aminotransferases, but not to the aspartate aminotransferase family. J. Biochem. 115, 309–317 10.1093/oxfordjournals.jbchem.a124334 [DOI] [PubMed] [Google Scholar]
- 7. Sakuraba H., Kawakami R., Takahashi H., and Ohshima T. (2004) Novel archaeal alanine:glyoxylate aminotransferase from Thermococcus litoralis. J. Bacteriol. 186, 5513–5518 10.1128/JB.186.16.5513-5518.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ward D. E., Kengen S. W., van Der Oost J., and de Vos W. M. (2000) Purification and characterization of the alanine aminotransferase from the hyperthermophilic archaeon Pyrococcus furiosus and its role in alanine production. J. Bacteriol. 182, 2559–2566 10.1128/JB.182.9.2559-2566.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yonaha K., Nishie M., and Aibara S. (1992) The primary structure of ω-amino acid:pyruvate aminotransferase. J. Biol. Chem. 267, 12506–12510 [PubMed] [Google Scholar]
- 10. Rausch C., Lerchner A., Schiefner A., and Skerra A. (2013) Crystal structure of the ω-aminotransferase from Paracoccus denitrificans and its phylogenetic relationship with other class III aminotransferases that have biotechnological potential. Proteins 81, 774–787 10.1002/prot.24233 [DOI] [PubMed] [Google Scholar]
- 11. Lee H., Juncosa J. I., and Silverman R. B. (2015) Ornithine aminotransferase versus GABA aminotransferase: implications for the design of new anticancer drugs. Med. Res. Rev. 35, 286–305 10.1002/med.21328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rajaram V., Ratna Prasuna P., Savithri H. S., and Murthy M. R. (2008) Structure of biosynthetic N-acetylornithine aminotransferase from Salmonella typhimurium: studies on substrate specificity and inhibitor binding. Proteins 70, 429–441 [DOI] [PubMed] [Google Scholar]
- 13. Stránská J., Tylichová M., Kopecný D., Snégaroff J., and Sebela M. (2010) Biochemical characterization of pea ornithine-δ-aminotransferase: substrate specificity and inhibition by di- and polyamines. Biochimie 92, 940–948 10.1016/j.biochi.2010.03.026 [DOI] [PubMed] [Google Scholar]
- 14. Valmaseda E. M., Campoy S., Naranjo L., Casqueiro J., and Martín J. F. (2005) Lysine is catabolized to 2-aminoadipic acid in Penicillium chrysogenum by an ω-aminotransferase and to saccharopine by a lysine 2-ketoglutarate reductase. Characterization of the ω-aminotransferase. Mol. Genet. Genomics 274, 272–282 10.1007/s00438-005-0018-3 [DOI] [PubMed] [Google Scholar]
- 15. Atomi H., Fukui T., Kanai T., Morikawa M., and Imanaka T. (2004) Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1, 263–267 10.1155/2004/204953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morikawa M., Izawa Y., Rashid N., Hoaki T., and Imanaka T. (1994) Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl. Environ. Microbiol. 60, 4559–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fukui T., Atomi H., Kanai T., Matsumi R., Fujiwara S., and Imanaka T. (2005) Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 15, 352–363 10.1101/gr.3003105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matsumi R., Manabe K., Fukui T., Atomi H., and Imanaka T. (2007) Disruption of a sugar transporter gene cluster in a hyperthermophilic archaeon using a host-marker system based on antibiotic resistance. J. Bacteriol. 189, 2683–2691 10.1128/JB.01692-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Santangelo T. J., Cubonová L., and Reeve J. N. (2008) Shuttle vector expression in Thermococcus kodakaraensis: contributions of cis elements to protein synthesis in a hyperthermophilic archaeon. Appl. Environ. Microbiol. 74, 3099–3104 10.1128/AEM.00305-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Santangelo T. J., Cubonová L., and Reeve J. N. (2010) Thermococcus kodakarensis genetics: TK1827-encoded β-glycosidase, new positive-selection protocol, and targeted and repetitive deletion technology. Appl. Environ. Microbiol. 76, 1044–1052 10.1128/AEM.02497-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sato T., Fukui T., Atomi H., and Imanaka T. (2003) Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185, 210–220 10.1128/JB.185.1.210-220.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sato T., Fukui T., Atomi H., and Imanaka T. (2005) Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl. Environ. Microbiol. 71, 3889–3899 10.1128/AEM.71.7.3889-3899.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ishibashi T., Tomita H., Yokooji Y., Morikita T., Watanabe B., Hiratake J., Kishimoto A., Kita A., Miki K., Imanaka T., and Atomi H. (2012) A detailed biochemical characterization of phosphopantothenate synthetase, a novel enzyme involved in coenzyme A biosynthesis in the Archaea. Extremophiles 16, 819–828 10.1007/s00792-012-0477-5 [DOI] [PubMed] [Google Scholar]
- 24. Tomita H., Imanaka T., and Atomi H. (2013) Identification and characterization of an archaeal ketopantoate reductase and its involvement in regulation of coenzyme A biosynthesis. Mol. Microbiol. 90, 307–321 [DOI] [PubMed] [Google Scholar]
- 25. Tomita H., Yokooji Y., Ishibashi T., Imanaka T., and Atomi H. (2012) Biochemical characterization of pantoate kinase, a novel enzyme necessary for coenzyme A biosynthesis in the Archaea. J. Bacteriol. 194, 5434–5443 10.1128/JB.06624-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tomita H., Yokooji Y., Ishibashi T., Imanaka T., and Atomi H. (2014) An archaeal glutamate decarboxylase homolog functions as an aspartate decarboxylase and is involved in β-alanine and coenzyme A biosynthesis. J. Bacteriol. 196, 1222–1230 10.1128/JB.01327-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yokooji Y., Tomita H., Atomi H., and Imanaka T. (2009) Pantoate kinase and phosphopantothenate synthetase, two novel enzymes necessary for CoA biosynthesis in the Archaea. J. Biol. Chem. 284, 28137–28145 10.1074/jbc.M109.009696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kawakami R., Ohmori T., Sakuraba H., and Ohshima T. (2015) Identification of a novel amino acid racemase from a hyperthermophilic archaeon Pyrococcus horikoshii OT-3 induced by d-amino acids. Amino Acids 47, 1579–1587 10.1007/s00726-015-2001-6 [DOI] [PubMed] [Google Scholar]
- 29. Kawakami R., Sakuraba H., Ohmori T., and Ohshima T. (2017) First characterization of an archaeal amino acid racemase with broad substrate specificity from the hyperthermophile Pyrococcus horikoshii OT-3. J. Biosci. Bioeng. 124, 23–27 10.1016/j.jbiosc.2017.02.004 [DOI] [PubMed] [Google Scholar]
- 30. Soda K., Misono H., and Yamamoto T. (1968) l-Lysine-α-ketoglutarate aminotransferase. I. Identification of a product, Δ1-piperideine-6-carboxylic acid. Biochemistry 7, 4102–4109 10.1021/bi00851a045 [DOI] [PubMed] [Google Scholar]
- 31. Gafan C., Wilson J., Berger L. C., and Berger B. J. (2001) Characterization of the ornithine aminotransferase from Plasmodium falciparum. Mol. Biochem. Parasitol. 118, 1–10 10.1016/S0166-6851(01)00357-7 [DOI] [PubMed] [Google Scholar]
- 32. Jhee K. H., Yoshimura T., Esaki N., Yonaha K., and Soda K. (1995) Thermostable ornithine aminotransferase from Bacillus sp. YM-2: purification and characterization. J. Biochem. 118, 101–108 10.1093/oxfordjournals.jbchem.a124863 [DOI] [PubMed] [Google Scholar]
- 33. Takechi M., Kanda M., Hori K., Kurotsu T., and Saito Y. (1994) Purification and properties of l-ornithine δ-aminotransferase from gramicidin S-producing Bacillus brevis. J. Biochem. 116, 955–959 10.1093/oxfordjournals.jbchem.a124652 [DOI] [PubMed] [Google Scholar]
- 34. Yasuda M., Tanizawa K., Misono H., Toyama S., and Soda K. (1981) Properties of crystalline l-ornithine: α-ketoglutarate δ-aminotransferase from Bacillus sphaericus. J. Bacteriol. 148, 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shen B. W., Hennig M., Hohenester E., Jansonius J. N., and Schirmer T. (1998) Crystal structure of human recombinant ornithine aminotransferase. J. Mol. Biol. 277, 81–102 10.1006/jmbi.1997.1583 [DOI] [PubMed] [Google Scholar]
- 36. Meng Z., Liu Z., Lou Z., Gong X., Cao Y., Bartlam M., Zhang K., and Rao Z. (2009) Purification, characterization and crystallization of pyrroline-5-carboxylate reductase from the hyperthermophilic archeon Sulfolobus solfataricus. Protein Expr. Purif. 64, 125–130 10.1016/j.pep.2008.10.018 [DOI] [PubMed] [Google Scholar]
- 37. Yoshida A., Tomita T., Atomi H., Kuzuyama T., and Nishiyama M. (2016) Lysine biosynthesis of Thermococcus kodakarensis with the capacity to function as an ornithine biosynthetic system. J. Biol. Chem. 291, 21630–21643 10.1074/jbc.M116.743021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fukuda W., Morimoto N., Imanaka T., and Fujiwara S. (2008) Agmatine is essential for the cell growth of Thermococcus kodakaraensis. FEMS Microbiol. Lett. 287, 113–120 10.1111/j.1574-6968.2008.01303.x [DOI] [PubMed] [Google Scholar]
- 39. Legrain C., Villeret V., Roovers M., Tricot C., Clantin B., Van Beeumen J., Stalon V., and Glansdorff N. (2001) Ornithine carbamoyltransferase from Pyrococcus furiosus. Methods Enzymol. 331, 227–235 10.1016/S0076-6879(01)31061-3 [DOI] [PubMed] [Google Scholar]
- 40. Nishida H., Nishiyama M., Kobashi N., Kosuge T., Hoshino T., and Yamane H. (1999) A prokaryotic gene cluster involved in synthesis of lysine through the amino adipate pathway: a key to the evolution of amino acid biosynthesis. Genome Res. 9, 1175–1183 10.1101/gr.9.12.1175 [DOI] [PubMed] [Google Scholar]
- 41. He M. (2006) Pipecolic acid in microbes: biosynthetic routes and enzymes. J. Ind. Microbiol. Biotechnol. 33, 401–407 10.1007/s10295-006-0078-3 [DOI] [PubMed] [Google Scholar]
- 42. Voellmy R., and Leisinger T. (1975) Dual role for N2-acetylornithine 5-aminotransferase from Pseudomonas aeruginosa in arginine biosynthesis and arginine catabolism. J. Bacteriol. 122, 799–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tamura K., Dudley J., Nei M., and Kumar S. (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.