Abstract
Titanium dioxide (TiO2) nanoparticles are produced for many different purposes, including development of therapeutic and diagnostic nanoparticles for cancer detection and treatment, drug delivery, induction of DNA double-strand breaks, and imaging of specific cells and subcellular structures. Currently, the use of optical microscopy, an imaging technique most accessible to biology and medical pathology, to detect TiO2 nanoparticles in cells and tissues ex vivo is limited with low detection limits, while more sensitive imaging methods (transmission electron microscopy, X-ray fluorescence microscopy, etc.) have low throughput and technical and operational complications. Herein, we describe two in situ post-treatment labeling approaches to stain TiO2 nanoparticles taken up by the cells. The first approach utilizes fluorescent biotin and fluorescent streptavidin to label the nanoparticles before and after cellular uptake; the second approach is based on the copper-catalyzed azide-alkyne cycloaddition, the so-called Click chemistry, for labeling and detection of azide-conjugated TiO2 nanoparticles with alkyne-conjugated fluorescent dyes such as Alexa Fluor 488. To confirm that optical fluorescence signals of these nanoparticles match the distribution of the Ti element, we used synchrotron X-ray fluorescence microscopy (XFM) at the Advanced Photon Source at Argonne National Laboratory. Titanium-specific XFM showed excellent overlap with the location of optical fluorescence detected by confocal microscopy. Therefore, future experiments with TiO2 nanoparticles may safely rely on confocal microscopy after in situ nanoparticle labeling using approaches described here.
Keywords: TiO2 nanoparticles, anatase, Click reaction, biotin–streptavidin, synchrotron X-ray, fluorescence microscopy
1 Introduction
The bulk form of titanium dioxide (TiO2) is biologically inert; however, particle sizes below 20 nm result in surface instability that confers chemical reactivity to the TiO2 nanoparticles [1–4]. Specifically, bulk forms of TiO2 have their surface titanium atoms in a hexa-coordinated geometry, but in nanoparticles 20 nm or less in size, the geometry of surface titanium atoms becomes penta-coordinated and highly reactive. This makes titanium (Ti) atoms on the nanoparticle surface very reactive, especially for binding with catechols [4–6]. A high-affinity covalent bond is created between the nanoparticles and enediol ligands (e.g., alizarin and dopamine), thereby forming stable complexes. This unique feature has been exploited in coating these nanoparticles with various agents [1, 3, 4, 7–21].
TiO2 has many useful photophysical properties, especially photocatalytic properties. The ability of TiO2 to act as a photocatalyst has been known for 90 years, but this was applied to biology only recently [3, 13, 20, 22]. Due to their unique photocatalytic and physical properties, TiO2 nanoparticles and TiO2 shell nanoparticles have been investigated for use in diagnostic assays and as gene targeting agents, radiosensitizers, and cargo delivery vehicles [3, 7, 12, 14, 18, 19]. TiO2 nanoparticles have demonstrated great potential for use in cancer therapy because they can be, for example, passively or actively targeted to neoplastic cells and then actively targeted to subcellular locations such as cell nuclei; once there, TiO2 nanoparticles can be activated to release reactive oxygen species (ROS) that result in double-stranded breaks in the genomic DNA in situ [13, 20].
Despite the extensive body of research exploring the interactions between TiO2 nanoparticles and neoplastic cells, the chemical techniques to fluorescently label TiO2 nanoparticles in vitro, in vivo, and ex vivo are limited and primarily depend on use of Alizarin Red S (ARS) [8, 10, 13, 15, 19]. Although this molecule binds well to the nanoparticle surface as an enediol ligand, its fluorescent yield is small and in the range of 0.001 at best [23], while, for example, fluorescence quantum yield for Alexa Fluor 488 is 0.92 (according to Invitrogen). In addition to ARS, chemotherapy drug doxorubicin also fluorescently labels TiO2 nanoparticles; however, its binding to the surface of TiO2 nanoparticles is mediated by single OH groups, and labeled nanoparticles shed doxorubicin molecules after cellular internalization [12]. Additionally, illumination of nanoparticles by light of UV-range wavelengths used for fluorescent microscopy excites TiO2, causing ROS release, which results in rapid bleaching of the samples. To make things more complicated, TiO2 nanoparticles quench fluorescence from immediately adjacent fluorophores such as Cy3 and FAM when they are used to label single-stranded DNA oligonucleotides, and these in turn are adsorbed onto 20 nm nanoparticles by phosphate groups [24]. Several studies attached biotin to TiO2 nanoparticles [1, 9]; however, this route for indirect labeling of nanoparticles in cells is challenging because of possible artifacts associated with background streptavidin labeling of cells. This has hindered our ability to gain in-depth knowledge of the details and mechanisms of TiO2 nanoparticle cellular targeting, internalization, and subcellular localization. Techniques that do not rely on fluorescent labeling such as transmission electron microscopy (TEM) and X-ray fluorescence microscopy (XFM or SXRF) are often used to determine TiO2 internalization and intracellular stability [3, 7, 8, 10–12, 15, 25]. XFM is a quantitative technique used to detect and map nanoparticles directly in whole cells based on titanium-specific K-alpha X-ray fluorescence emitted from Ti atoms. XFM scanning registers a quantitative and qualitative map of the distribution of cellular elements such as sulfur (S), phosphorus (P), and zinc (Zn) [25]. Similarly, energy dispersive electron spectroscopy (EDS, EDX, or XEDS) can be used to identify the elemental signature of titanium, but this technique does not function in combination with whole cells. However, both EM and XFM suffer from low throughput and other limitations. TEM requires biological sample sectioning and permits analysis of only a small number of cells. Similarly, XFM is not readily available and requires synchrotron-quality X-rays. Both techniques can be expensive and technically challenging, and neither allows analysis of large numbers of cells.
In order to meet the demands of the growing body of research exploiting TiO2 nanomaterials, it has become necessary to develop an in situ post-treatment labeling approach capable of precisely detecting the subcellular localization of nanomaterials in cells as well as tissue samples; such a technique also must be quick and produce consistent results. Herein, we describe two in situ labeling techniques to label TiO2 nanoparticles in order to image them using fluorescence microscopy. In addition, XFM was used to confirm the co-registration of the Ti elemental signature and nanoparticle optical fluorescence.
2 Experimental
Nanoparticle preparation
TiO2 nanoparticles were synthesized by applying a low-temperature alkaline hydrolysis approach according to Abbas Z. et al [26], dialyzed in water at 4 °C, and stored at 4 °C (Fig. S1 in the Electronic Supplementary Material (ESM)).
Fluorescein surface coating preparation
TiO2 nanoparticles with an average diameter of 6 nm were synthesized by a low-temperature alkaline hydrolysis route as described previously and were dialyzed and stored at 4 °C in 10 mM Na2HPO4 buffer at pH 5.7. The molar ratio of fluorescein (FITC)-biotin (5(6)- (biotinamidohexanoylamido-pentylthioureidylfluorescein; B8889, Sigma-Aldrich) to TiO2 nanoparticles was such that 30% of the nanoparticle surface was covered by FITC-biotin. Binding was performed over a period of 16 h in the presence of 1 M glucose. Addition of glucose was performed in order to coat the nanoparticle surface and aid the uptake of nanoparticles. This degree of surface coverage of nanoparticles was used to ensure that all of the FITC-biotin molecules attached to the nanoparticle surface.
Azide surface coating preparation
The SQ Peptide Synthesis Core facility of Northwestern University prepared for us a 95% pure dopamine-PEG4-azide molecule; synthesis was performed using an NHS ester group on azido-PEG4-NHS (Click chemistry tools) and amino group of dopamine (Sigma-Aldrich). This molecule was used to coat the surface of TiO2 nanoparticles prior to cell treatment. The ratio of estimated TiO2 nanoparticle surface sites and dopamine was 1:1. Upon mixing, the clear and colorless nanoparticle solution changed to a clear and light brown solution, similar to the color change induced by dopamine binding alone, which occurs immediately upon mixing and conjugation of dopamine to nanoparticles [4]. Nanoparticles were dialyzed in 100 mM sodium phosphate buffer in dialysis tubing with 2 kDa pores. This ensured that the unbound dopamine-azide molecules (< 0.5 kDa) were removed from the nanoparticle solution prior to use in cells.
Cryo-TEM of nanoparticles
Bare TiO2 nanoparticles and azide-coated nanoparticles were diluted 1:50, plunge frozen in liquid ethane, and imaged using a TEM in the Biological Imaging Facility (BIF) at Northwestern University.
Dynamic light scattering
Bare TiO2 nanoparticles and azide-coated nanoparticles were diluted 1:100 or 1:200 in dH2O and assayed for hydrodynamic diameter size using Malvern capillary cells on the Zetasizer Nano ZSP in the Northwestern University Analytical BioNanoTechnology Equipment Core (ANTEC) facility.
Zeta potential
Bare TiO2 nanoparticles and azide-coated nanoparticles were diluted 1:100 or 1:200 in dH2O and assayed for zeta potential using Malvern capillary cells on the Zetasizer Nano ZSP in the Northwestern University ANTEC facility.
Cell culture and treatment with nanoparticles
MCF-7 cells were maintained at 37 °C with 5% CO2. Roswell Park Memorial Institute (RPMI) 1640 media was supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, 10 mM HEPES, 100 I.U.·mL−1 penicillin, 100 μg·mL−1 streptomycin, 1X non-essential amino acids, 0.25 μg·mL−1 amphotericin B, and 0.1 mg·mL−1 insulin (Sigma-Aldrich). Prior to treatment with nanoparticles, MCF-7 cells were placed in serum-free RPMI 1640 medium for 1 h. Next, the cells were treated with 0.12 μM of TiO2 nanoparticles coated with FITC-biotin (Sigma-Aldrich) overnight. The cells were washed with phosphate-buffered saline (PBS) solution and 200 mM of acidic glycine (pH 4.0) in order to remove surface-bound nanoparticles. The cells labeled by FITC were collected using fluorescence-activated cell sorting (FACS) by the DakoCytomation MoFlo flow cytometer (Dako) and seeded on formvar-coated gold electron microscopy grids (electron microscopy sciences) to allow for cell adherence. After 3 h, the cells were washed in PBS and fixed in 4% paraformaldehyde.
HeLa cells were obtained from the American Type Culture Collection (ATCC). The cell line was maintained at 37 °C with 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS and 1% penicillin and streptomycin. For visualization by confocal microscopy, ~ 50,000 HeLa cells were cultured on glass slides (BioGenex barrier slides XT134-SL) overnight. These slides were washed with 1X PBS and placed in serum-free DMEM for 1 h. About 50,000 HeLa cells were seeded per barrier slide within a surface area of 1,000 mm2. Cells were treated with dopamine-PEG4-azide-coated TiO2 nanoparticles diluted in serum-free DMEM. The final TiO2 concentration was 0.2 μg·mL−1 or, for nanoparticles averaging 5 nm in diameter, approximately 0.16 nM nanoparticles. Cells and nanoparticles were incubated at 37 °C for 1 h. As a control, we included cells without nanoparticles. After nanoparticle incubation, cells were washed once with cold acidic glycine (200 mM, pH 4.0) and then washed twice with PBS with calcium and magnesium. Cells were then fixed in 4% formaldehyde in 1X PBS.
Click chemistry labeling
The Click chemistry reaction was adapted from the manufacturer’s protocol provided for labeling of azide-conjugated proteins. Following fixation, cells were washed three times for 3 min each with 1X PBS and permeabilized with 0.5% triton-X100 in PBS for 10 min at room temperature. Then, cells were rinsed three times for 3 min each with 1X PBS and blocked with 2% bovine serum albumin (BSA) in 1X PBS for 20 min at room temperature. A cocktail of the Click reagents (Life Technologies Click chemistry kit C10269) was prepared in the following proportions: 30 μL of 100 mM CuSO4, 150 μL of 20 mg·mL−1 reactive additive buffer (component C), 1,260 μL of 1X cell reaction buffer, and 0.67 μM as the final concentration of alkyne-Alexa Fluor 488 (Life Technologies A10267). Cells were incubated with 300 μL of the Click reagent mixture per slide (1,000 mm2 surface area) for 30 min. Then, slides were washed three times for 3 min with 1X PBS, blocked for 10 min in 2% BSA in PBS, and then incubated with Hoechst 1:2,500 in PBS for 10 min.
Confocal microscopy
MCF-7 cells treated first with TiO2 nanoparticles coated with FITC-biotin were washed, fixed in 4% paraformaldehyde, and then treated with 25 μM of Cy3-labeled streptavidin (Streptavidin Cy3™ from Streptomyces avidinii, Sigma-Aldrich) for 1 h. At the same time, nuclear DNA was stained by Hoechst 33342 (Invitrogen). The samples were imaged on the LSM 510 UV Meta Microscope (Carl Zeiss Inc.). To allow visualization of cell nuclei by Hoechst staining, FITC, and Cy3, lasers of 405, 488, and 543 nm were used. The detection of the Hoechst, FITC, and Cy3 emissions was facilitated by bandpass filters of 420–480, 475–525, and 530–600 nm, respectively. After visualization of the samples with fluorescence microscopy, the position of the cells was noted, and the samples were further prepared for XFM analysis.
After Click labeling and Hoechst staining of nuclear DNA, slides with azide-dopamine nanoparticle-treated HeLa cells were washed three times for 3 min in PBS, drained of excess PBS, and covered with anti-fade mounting medium (Prolong Gold; Life Technologies); glass coverslips were applied and sealed with clear nail polish. Cells were visualized using the Nikon A1R Confocal Microscope at the Northwestern University Center for Advanced Microscopy using 405-nm and 488-nm lasers with bandpass filters of 420–480 and 505–530 nm, respectively. In addition, Z stack slices were taken every 0.22 μm. Identical microscope settings were used for all comparison images.
XFM
Imaging of cells by XFM at room temperature requires that they are prepared on suitable substrate and imaged only after they have been completely dried. Therefore, all confocal imaging had to be performed prior to XFM. We used two X-ray imaging instruments at the advanced photon source (APS) synchrotron at Argonne National Laboratory (ANL). One of them (X-ray microprobe) is located at the 2-ID-D beamline, while the other (Bionanoprobe) resides at the LS-CAT beamline. Monochromatic hard X-rays (~ 10 keV) were used at 2-ID-D; fluorescent X-ray emission was detected by a germanium detector (LEGe Detector, Canberra). The cells were raster scanned with a step size of 500 nm and dwell time of 2 s per step (Figs. 1 and 2). These data were fitted against elemental NBS standards 1832 and 1833 using MAPS software [27]. Elemental quantification and elemental 2D maps were calculated using the MAPS program [27]. At the Bionanoprobe instrument, monochromatic 10 keV hard X-rays were used as well. However, they were focused to a spot size of ~ 85 nm using Fresnel zone plates. The fluorescence spectra at each scan step were collected with a four-element silicon drift detector (Vortex ME-4, SII Nanotechnology). Data were fitted and quantified by comparison to a standard reference material (RF8-200-S2453, AXO Dresden GmbH) using the MAPS program [27].
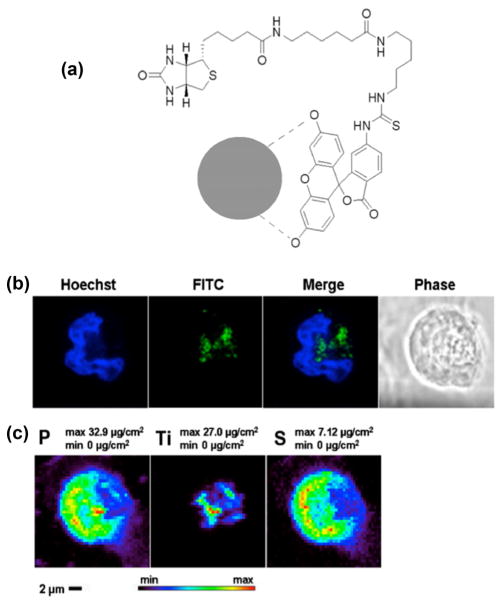
Figure 1.
Fluorescent labeling and uptake of TiO2 nanoparticles. (a) Possible interactions between nanoparticle and FITC-biotin. There are two free OH groups on the FITC molecule that are able to bind to the TiO2 surface, similar to doxorubicin molecules [12]; they are indicated by dashed lines between oxygen atoms and nanoparticle (gray circle). (b) Confocal imaging of MCF-7 cell treated with TiO2 nanoparticles coated with FITC-biotin. Separate and overlapping images of Hoechst (nuclear DNA) and FITC (nanoparticles) are shown as well as matching phase contrast image. (c) XFM maps show the distribution and concentration of phosphorus (P), titanium (Ti), and sulfur (S) in the same MCF-7 cell treated with the FITC-biotin-coated TiO2 nanoparticles. The positions of FITC signal in the confocal image and Ti signal in the XFM map are well matched.
Figure 2.
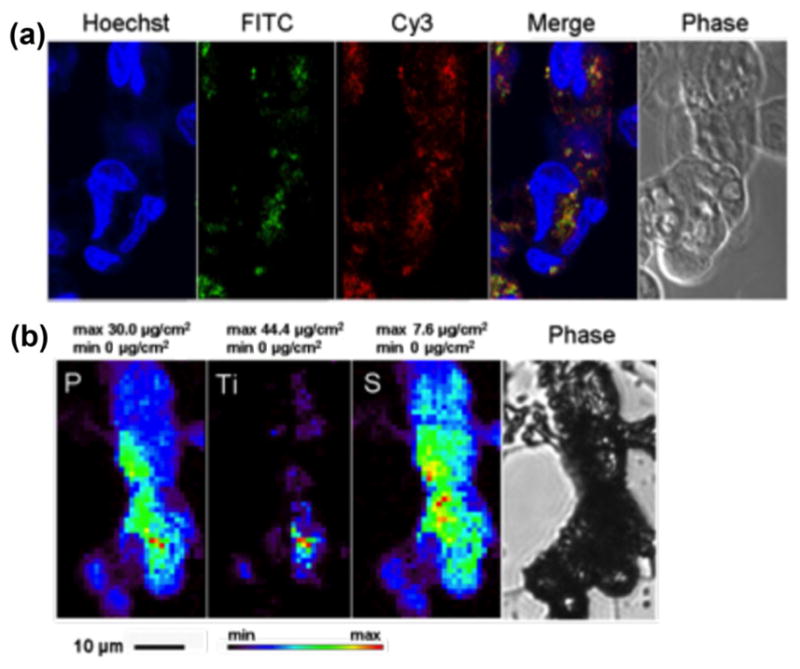
Secondary in situ fluorescent labeling of TiO2 nanoparticle aggregates. A cluster of MCF-7 cells treated with FITC- biotin TiO2 nanoparticles was chemically fixed, permeabilized, and subsequently treated with Cy3-labeled streptavidin. (a) Confocal microscopy shows matching positions of fluorescent signal of FITC-biotin-coated nanoparticles (green) and fluorescent signal of Cy3-streptavidin (red) interacting with biotin on nanoparticles after post-fixation labeling of nanoparticle-treated cells. Nuclear DNA was labeled by Hoechst. (b) XFM maps show cytoplasmic distribution of titanium (Ti) as well as 2D distribution of native cell elements P and S (last image in bottom row shows a phase contrast image of the sample prior to XFM). In this instance, combined FITC and Cy3 signals (a) and Ti fluorescent signal (b) show a similar pattern of distribution.
3 Results and discussion
We attached FITC-biotin to the TiO2 nanoparticle surface through hydroxyl groups on the FITC part of the molecule (indicated by dashed lines in Fig. 1(a)). Due to the distance between these two OH groups, it is unlikely that both of these attach to the nanoparticle at the same time. Nevertheless, a single OH group binding to the nanoparticle surface is sufficient for nanoparticle coating; we have found this to be true in the past, e.g., for binding of doxorubicin [12]. Labeled nanoparticles were incubated with MCF-7 breast cancer cells and imaged first using confocal microscopy (Fig. 1(b)) and then XFM (Fig. 1(c)) to detect the intracellular localization of the nanoparticles in a single cell. Because TiO2 nanoparticles are surface reactive and may bind to additional parts of FITC-biotin instead of to hydroxyl groups only, we were concerned about quenching of some of the fluorescent signal (similar to quenching of Cy3 and FAM [9]). To prevent this, binding between nanoparticle and FITC-biotin was performed in the presence of glucose to additionally coat the surface of TiO2.
After a 16-h treatment with nanoparticles, the cells were sorted by flow cytometry, seeded on formvar-coated gold TEM grids, and allowed to attach for 3 h prior to fixation and confocal fluorescence imaging. After optical fluorescence imaging, during which the grids were submerged in mounting media, the grids were rinsed in PBS, dehydrated in ethanol, and air dried prior to XFM.
XFM was performed using the X-ray microprobe instrument with 2ID-E beamline at the APS, ANL. The X-ray fluorescence map of sulfur showed the boundaries of the cell as expected [3, 7, 8, 10, 16–18, 20, 28, 29] and corresponded well with the cells’ outlines in optical fluorescence microscopy. Similarly, the strongest phosphorus signal area in the phosphorus XFM map overlapped with the cell nucleus; again, this matched well with the DNA (Hoechst) fluorescent signal obtained by confocal microscopy. A strong titanium signal (per-pixel signal of up to 27 μg/cm2) was detected by XFM inside the cells but outside of the nuclei. The distribution of titanium and, therefore, TiO2 nano-particles showed a good overlap with the FITC fluorescence signal in images obtained by confocal fluorescence microscopy. The slight discrepancies between the images are expected since the cells were hydrated for confocal light microscopy but dried for XFM measurements. Moreover, imaging by XFM is performed with the sample mounted vertically into a special sample holder and positioned at 45° to the incident beam direction and detector, making direct comparisons between images impossible. It should also be noted that the substrates suitable for imaging by XFM are not ideally suitable for confocal fluorescence microscopy. In many cases, when formvar-coated TEM grids are used for cell growth, cells detach from the substrate during manipulations that remove mounting media necessary for fluorescence microscopy, and often only clusters of cells remain attached to the substrate, as happened with the cells shown in Fig. 2.
In order to show the selectivity and specificity of labeling TiO2 nanoparticles with FITC-biotin and mitigate the potential problem of signal quenching when nanoparticles are directly labeled with FITC-biotin, we decided to expose nanoparticle aggregates inside cells to secondary in situ labeling and use the interaction between fluorescent streptavidin and biotin attached to the nanoparticle to confirm that the two fluorescent signals overlap. Cells that contained nanoparticles modified with FITC-biotin were purified by flow cytometry sorting, seeded on formvar-coated TEM grids, fixed, permeabilized, and stained with streptavidin-Cy3. Confocal imaging of biotin-FITC, streptavidin-Cy3, and Hoechst was performed (Fig. 2(a)), and the TEM grids with cells were rinsed in PBS, dehydrated in ethanol, and air dried to be imaged by XFM. Several single cells and cell clusters were imaged by confocal microscopy, but only the cluster shown in Fig. 2 remained attached to the formvar substrate at the time of XFM imaging. The localizations of the optical fluorescence signals were compared to the 2D XFM map of titanium in the same whole cells (Fig. 2). Several cells depicted in Fig. 2 demonstrate an overlap between the position of titanium (maximal per-pixel concentration of titanium up to 44 μg/cm2) determined by XFM and sites of FITC and Cy3 dual fluorescence detected by confocal optical microscopy.
Although this approach for optical labeling of nanoparticles is currently shown to be feasible, the presence of biotin may significantly affect interactions between the cells and the nanoparticles. For example, biotin uptake occurs both in cancer cells [30] and cells of normal tissues [31]. Biotin enters cells via the ubiquitously present protein-solute carrier family 5 member 6 (SLC5M6), also known as the sodium-dependent multivitamin transporter. It is probable that the nanoparticles coated with biotin would engage in biotin receptor-mediated uptake by many different cell types in vivo. This would interfere with the intended cellular targets of nanoparticles by creating a “targeting competition” between biotin receptors and the intended cell surface epitopes. Therefore, we decided to investigate alternative approaches for nano-particle labeling in situ. Seeking nanoparticle coatings that would not target any specific cell receptors, we decided to investigate azide-PEG4-dopamine (Fig. 3) as a surface modification of TiO2 nanoparticles and use it as a basis for subsequent in situ Click reaction with commercially available Alexa Fluor 488 alkyne [26]. Azide moieties in cells are sparse, making Click labeling suitable for cell biology. In its interaction with TiO2 nanoparticles smaller than 20 nm, dopamine is known to form stable covalent bonds spontaneously; the present study and previous studies used dopamine and dopamine-linked molecules for stable surface modification of TiO2 and TiO2 shell nanoparticles [1, 3, 4, 8, 11, 13, 14, 17, 18, 20].
Figure 3.
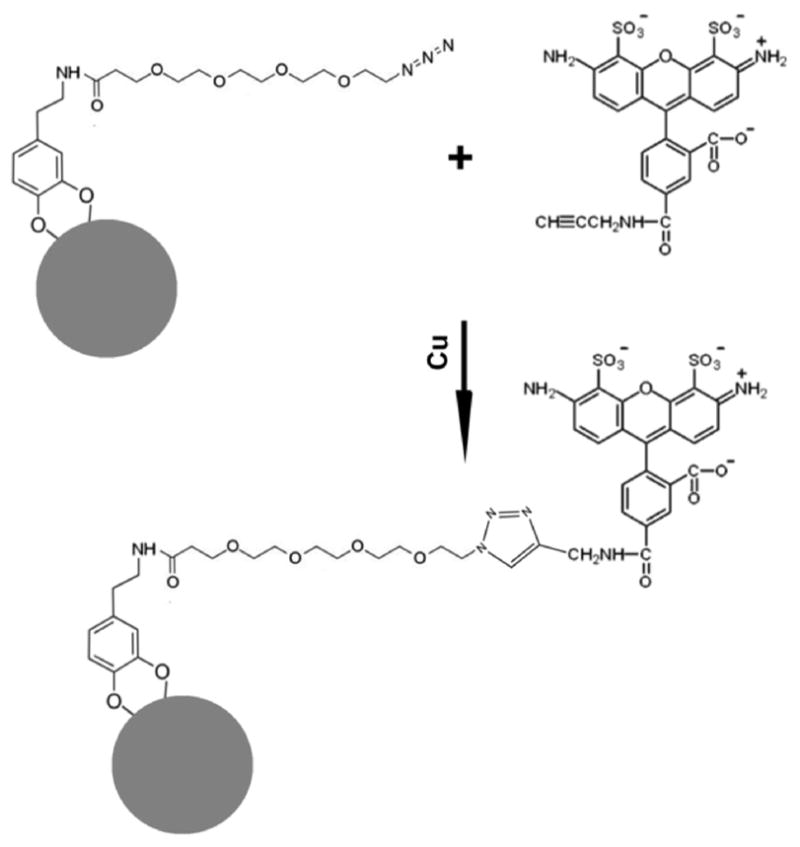
Schematic diagram of Click labeling. Steps of in situ chemical reaction between azide-PEG4-dopamine (MW = 426.47) conjugated to the surface of TiO2 nanoparticle (gray circle) and alkyne-modified Alexa Fluor 488. This Click reaction is performed in the presence of Cu in situ in permeabilized cells.
Nanoparticles conjugated to dopamine-azide showed some increase in hydrodynamic diameter compared with that of bare nanoparticles alone when assessed using dynamic light scattering (Table S1 in the ESM). In addition, we noted small differences between zeta potentials of azide-coated nanoparticles; they shifted from 34.07 ± 0.91 to 31.30 ± 0.78 mV in comparison with those of uncoated nanoparticles (Table S1 in the ESM). Finally, size and aggregation of azide-coated nanoparticles were slightly increased compared with that of bare nanoparticles by cryo-TEM (Fig. S1 in the ESM).
HeLa cells grown on microscope slides and serum-starved for 1 h were treated with azide-coated nano-particles for 1 h because we have noticed previously that non-specific uptake of nanoparticles is already substantial at this nanoparticle concentration [19]. Initially, in order to obtain a quick confirmation that new fluorescent staining matches the pattern that would be obtained by already established fluorescent dyes, we added ARS to azide-coated nanoparticles and used these doubly coated nanoparticles for cell labeling (Fig. S2 in the ESM). The overlap between ARS and Alexa Fluor 488 fluorescence we obtained was encouraging, and we proceeded to optimize the Click reaction and obtain minimal Alexa Fluor 488 background coupled with bright labeling of nanoparticle aggregates in situ (Figs. 4 and 5). We noted that oversaturation of cells with the alkyne-modified fluorophore (alkyne-Alexa Fluor 488) can produce background despite the lack of azide groups in nanoparticle-free cells; therefore, we fine-tuned the labeling reaction to prevent non-specific background fluorescence in untreated cells. Cells without nanoparticles (untreated) exhibited a very dim fluorescent signal (Fig. 5) that was easily recognized as background, non-specific fluorescence. Comparatively, cells with azide-coated TiO2 nanoparticles showed a strong fluorescent signal when excited with a 488-nm laser (Fig. 5). Using the same overall experimental conditions, we noticed even subtle differences such as a two-fold increase in concentration of alkyne-Alexa Fluor 488 or a two-fold increase in concentration of nanoparticles (Fig. 5). Thus, for example, we noticed little difference in the Alexa Fluor 488 background when two different concentrations of alkyne-Alexa Fluor 488 were used and at the same time noted much stronger signal intensity in nanoparticle-treated cells. Similarly, doubling the nanoparticle concentration increased the volume of Alexa Fluor 488-positive pixels in nanoparticle-treated cells.
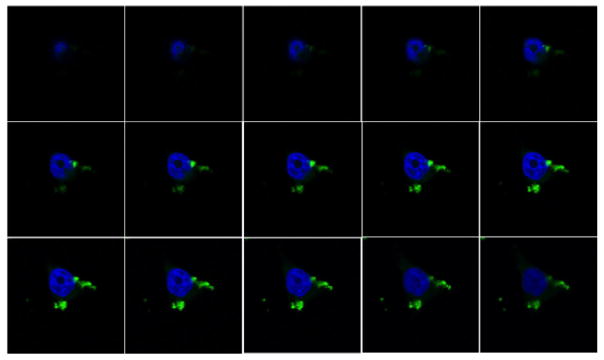
Figure 4.
Series of confocal microscopy “optical slices” of a cell treated with azide nanoparticles labeled by Click reaction with alkyne-Alexa Fluor 488 in situ. Single-cell optical slices of 0.22 μm show the cytoplasmic localization of nanoparticles Click labeled in situ (green) at different focal depths inside the cell. Cell nuclei were labeled by Hoechst staining (blue).
Figure 5.
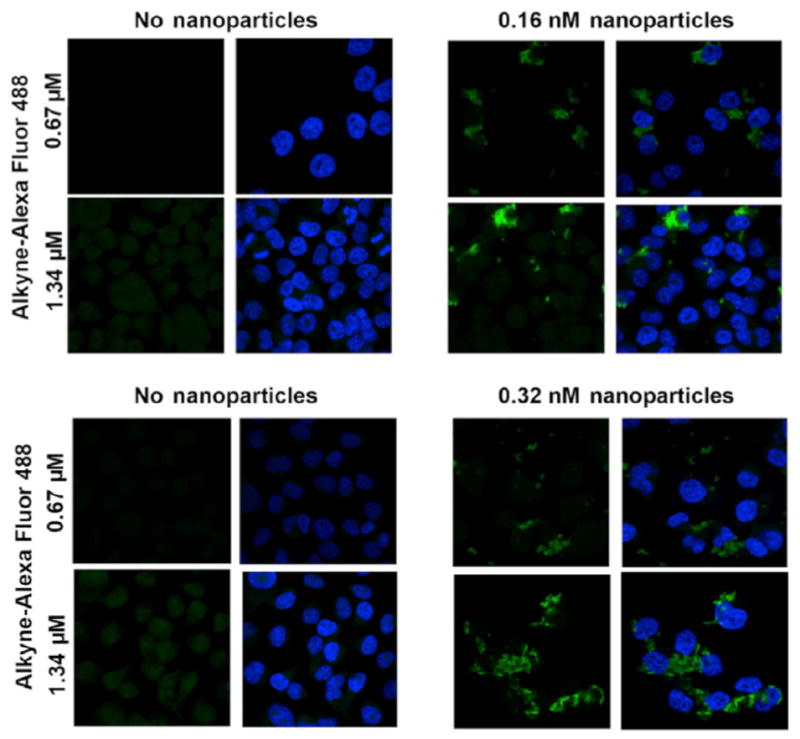
Labeling specificity of Click reaction. Increased concentration of alkyne-Alexa Fluor 488 increases fluorescent signal intensity of nanoparticle aggregates in nanoparticle-treated cells (right hand panels), while increased concentration of nanoparticles leads to greater nanoparticle uptake (bottom right). Cells not treated with nanoparticles (left panels) have low background Alexa Fluor 488 signal under the same staining and imaging conditions.
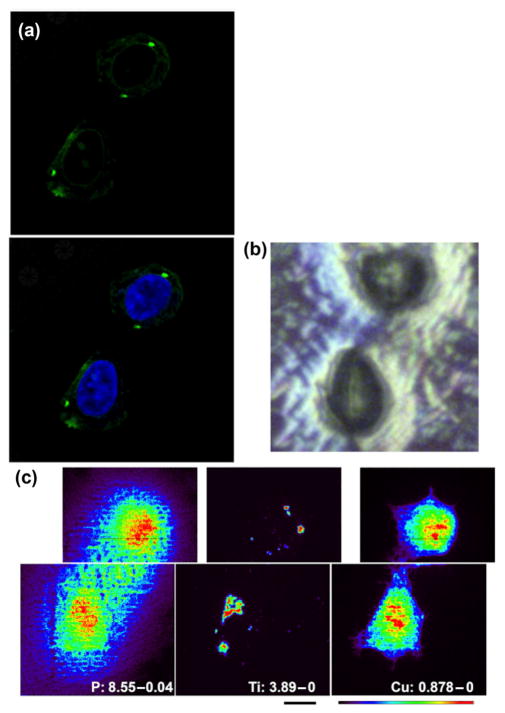
The final refined procedure for Click labeling of nanoparticles in situ yielded bright signal and low background; however, it was important to confirm that the bright green signal shown in Figs. 4 and 5 is dependent on the presence of nanoparticles. For the ultimate confirmation of the presence of nanoparticle, we used XFM. In the period between experiments shown in Figs. 1 and 2, and those in Fig. 6, APS Synchrotron obtained a new, higher resolution instrument for XFM—the Bionanoprobe [20, 28]. As detailed in the Experimental section, the X-ray beam focus of the Bionanoprobe is several fold better than that of the X-ray microprobe instrument. We decided to image the same sample by confocal microscopy and then XFM, as performed for cells in Figs. 1 and 2. In this case, cells were grown on silica nitride windows, processed, imaged by optical microscopy as if on glass slides, and finally washed, dehydrated by ethanol, and air dried as it was performed earlier. This way of sample preparation is suboptimal with regard to both preservation of cell morphology and preservation of native cellular elemental components, but the nanoparticle localization inside the cell remains unchanged. Thus, we anticipated that the only clear elemental signals from the cell would be phosphorus (indicating cell nucleus) and titanium (indicating nanoparticles); this was confirmed as anticipated, with an additional finding that copper from the Click reaction also incorporated into the cellular matter, providing (as an artifact) a clear cellular outline (Fig. 6). As anticipated, the Alexa Fluor 488 signal from confocal microscopy within the cell cytoplasm matched well with the Ti signal obtained by XFM (Fig. 6). More importantly, maximum per-pixel intensity of Ti in this image was 3.89 μg/cm2, seven times lower than the maximum signal in Fig. 1, while at the same time, signal for P differed only by three fold. Although some of these elemental signal differences could be ascribed to a different way of sample preparation, it is still likely that the cells in Fig. 6 contain less TiO2 than their counterparts in Figs. 1 and 2. However, at the same time, confocal fluorescence signal is much clearer in Alexa Fluor 488 Click-labeled samples (Figs. 4, 5 and 6) than in FITC-labeled cells (Figs. 1 and 2). Therefore, Click chemistry provides not only more specific but also more sensitive labeling for nanoparticles.
Figure 6.
Confocal microscopy of nanoparticles using Click chemistry and XFM elemental mapping of titanium in cells. Cells seeded on Si3N4 windows, a substrate suitable for XFM, were treated with nanoparticles, labeled by Click, and stained by Hoechst to be imaged by confocal microscopy (a). Next, cells were rinsed with PBS, dried (b), and subjected to XFM at the Bionanoprobe, a high- resolution XFM instrument (c). (a) Confocal optical microscopy image of the cell pair that was investigated by XFM; (b) dried Si3N4 window with the same pair of cells air-dried on its surface; (c) high-resolution XFM images (beam size of ~ 85 nm; pixel size of 100 nm) of the two cells. Element and maximal elemental concentration in μg/cm2 are given in white letters. Scale bar (black) is 2 μm; color scale defines false colors map with minimal signal in black and maximal signal in red.
4 Conclusions
Two new approaches for labeling of TiO2 nanoparticles in situ were developed and confirmed by XFM. Although confocal fluorescence imaging and XFM imaging are not easily paired with each other, we managed to preserve the integrity of substrates (fragile formvar-coated TEM grids and Si3N4 windows) carrying some of the cells (many detached in the process of multiple manipulations) imaged by confocal microscopy. Thus, dual imaging was achieved, and it confirmed that the fluorescent signal and the metal oxide (elemental) signal are co-registered. XFM imaging of cells pre-screened by confocal microscopy has been accomplished only once before to our knowledge [8], showing that, as in this case, Ti signal obtained by XFM spans the entire cell volume, while confocal microscopy provides images of nanoparticles only in a given optical slice [8]. An excellent overlap was found between optical fluorescence signals (FITC and Cy3 in one case and Alexa Fluor 488 in the other) and the position of Ti in XFM-generated elemental maps. This suggests that the following can be achieved: (1) successful labeling of nanoparticles with optically competent dyes; (2) retention of fluorescent dyes on nanoparticles during cell treatments; and (3) washing out and removing sources of non-specific optical fluorescence from nanoparticle-treated cells pre- or post-stained with optically fluorescent dyes.
Labeling based on the use of FITC-biotin and streptavidin is often associated with a slightly higher background fluorescence due to the interactions between streptavidin-Cy3 and endogenous, not-nanoparticle-bound biotin. It is possible that such background could be decreased by pre-treating cells with streptavidin or neutravidin, which have been used to reduce endogenous biotin in vitro [32] and in vivo [33]; nevertheless, in this case it is possible that residual non-labeled avidin-like molecules could interact with the nanoparticles as soon as they enter the cells and reduce subsequent labeling with fluorescent avidin-like compounds.
In order to detect TiO2 nanoparticles in situ and do so with a single step without background fluorescence and without concern that biotin import into cells may be triggered, we developed a Click chemistry approach for labeling and detecting TiO2 nanoparticles in situ.
The copper-catalyzed azide–alkyne cycloaddition, also known as a Click reaction, was developed in 2008, and the following year it became a method of choice for use with biological samples [34, 35]. In nanotechnology, specifically with TiO2 nanoparticles, Click chemistry was used for the preparation of hybrid nanomaterials [36–39]. In biology, Click chemistry reactions have been used for various cell biology and biomedical applications; the virtual absence of azide-carrying moieties in cells makes this an excellent approach for labeling with low background [40]. Specifically, several Click-based techniques were developed to investigate post-translational protein modifications to identify newly synthesized proteins and to examine DNA replication in proliferating cells [41–43]. During the reaction process, a stable triazole forms in the course of a copper-catalyzed reaction between azide and alkyne moieties; this can be exploited as a simple chemical reporter to label and detect biomolecules of interest.
Commercially available kits such as those from Life Technologies can be used for labeling of nucleic acids and proteins in combination with different “standard” cell biology techniques [26]. We used components from one of such kits to label azide groups, which were attached to the nanoparticle itself, with Alexa Fluor 488 alkyne in order to label in situ azide-conjugated TiO2 nanoparticles with Alexa Fluor 488. Click chemistry staining showed the high signal intensity of Alexa Fluor 488 in confocal imaging without prominent background fluorescence, thereby permitting accurate intracellular detection of even small nanoparticle aggregates. There was an evident overlap between the strong fluorescent signal of Alexa Fluor 488 within the cytoplasm (Fig. 6), and the XFM detected localization of Ti. It is worth mentioning that the molecular weight (MW) of azide-PEG4-dopamine is about 400, while the MW of molecules used for targeted nanoparticle coating generally exceeds this by several fold. For example, the MW of B-loop peptide dopac conjugate we used previously is close to 1,500 [20]. It will remain to be evaluated whether azide-PEG4-dopamine may interfere with nanoparticle targeting or not and whether access to the azide group may be affected by the presence of targeting molecules on the nanoparticle surface. Considering the large surface area of nanoparticles and the fact that B-loop peptide covered no more than 30% of the nanoparticle surface for successful targeting [20], adapting the nanoparticles for targeting and in situ staining should be easily accomplished.
A chemical reporter labeling technique such as Click chemistry as an in situ staining approach has clear advantages over the use of nanoparticles labeled with fluorescent molecules prior to cell treatment. Moreover, the availability of alkyne-conjugated fluorescent dyes is increasing, allowing for more flexibility in the choice of detection molecules. Click chemistry itself is simple, highly selective, efficient, and reproducible. Finally, similar staining and detection techniques could be applicable to other metal oxide nanoparticles like iron oxide, which also has a high affinity for dopamine [44, 45].
Nevertheless, because Click chemistry reactions introduce a substantial amount of copper into cells (Fig. 6), the elemental content of cells treated by Click chemistry shows high Cu content as an artifact. It is possible, nevertheless, that a copper-free Click reaction can be used instead of the approach used here. Although Click chemistry may be suitable in most situations, a dual labeling approach such as the biotin-streptavidin labeling approach described in the present study has utility in pre- and post-detection studies, specifically in nanoparticle studies that require labeling through hydroxyl groups.
In conclusion, the results of this study show that it is feasible to label TiO2 nanoparticles both sequentially by biotin-FITC and then in situ by fluorescent streptavidin as well as strictly in situ by Click chemistry. Both approaches are capable of detecting TiO2 nanoparticles intracellularly through optical fluorescent microscopy. Importantly, these labeling techniques can be used as post-treatment labeling techniques to detect TiO2 nanoparticle internalization, localization, and activation. To the best of our knowledge, this is the first report on TiO2 nanoparticle detection using Click chemistry coupled with XFM. In this study, we used XFM to confirm that fluorescent signals are indeed co-localized with the nanoparticles; however, in the future, only confocal microscopy can be performed, especially with Click chemistry-labeled nanoparticles. This will increase throughput for nanoparticle imaging in situ, facilitating the evaluation of nanoparticles in a variety of cell biology scenarios.
Supplementary Material
Acknowledgments
This research was supported by the National Institutes of Health (Nos. CA107467, EB002100, U54CA119341 and GM104530). Implementation of the Bionanoprobe is supported by NIH ARRA (No. SP0007167). Confocal optical imaging work was performed at the Northwestern University Center for Advanced Microscopy generously supported by NCI CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center. Confocal microscopy was performed on a Nikon A1R multiphoton microscope, acquired through the support of NIH 1S10OD010398-01. Work at the Advanced Photon Source at Argonne National Laboratory was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences contract No. DE-AC02-06CH11357. Metal analysis was performed at the Northwestern University Quantitative Bio-element Imaging Center generously supported by NASA Ames Research Center (No. NNA06CB93G). Use of the Simpson Querrey Institute Analytical BioNanoTechnology Equipment Core (ANTEC) facility was supported by the U.S. Army Research Office, the U.S. Army Medical Research and Materiel Command, and Northwestern University funding received from the Soft and Hybrid Nanotechnology Experimental (SHyNE) Resource (NSF NNCI-1542205). Cryo-TEM work was performed at the Northwestern University Biological Imaging Facility by Imaging Specialist Charlene Wilke. The authors thank Dr. Teng-Leong Chew for his valuable discussion and advice.
Footnotes
Electronic Supplementary Material: Supplementary material (one table and two figures) is available in the online version of this article at https://doi.org/10.1007/s12274-017-1654-8.
References
- 1.Dimitrijevic NM, Saponjic ZV, Rabatic BM, Rajh T. Assembly and charge transfer in hybrid TiO2 architectures using biotin-avidin as a connector. J Am Chem Soc. 2005;127:1344–1345. doi: 10.1021/ja0458118. [DOI] [PubMed] [Google Scholar]
- 2.Liu JQ, de la Garza L, Zhang LG, Dimitrijevic NM, Zuo XB, Tiede DM, Rajh T. Photocatalytic probing of DNA sequence by using TiO2/dopamine-DNA triads. Chem Phys. 2007;339:154–163. [Google Scholar]
- 3.Paunesku T, Rajh T, Wiederrecht G, Maser J, Vogt S, Stojićević N, Protić M, Lai B, Oryhon J, Thurnauer M, et al. Biology of TiO2-oligonucleotide nanocomposites. Nat Mater. 2003;2:343–346. doi: 10.1038/nmat875. [DOI] [PubMed] [Google Scholar]
- 4.Rajh T, Chen LX, Lukas K, Liu T, Thurnauer MC, Tiede DM. Surface restructuring of nanoparticles: An efficient route for ligand-metal oxide crosstalk. J Phys Chem B. 2002;106:10543–10552. [Google Scholar]
- 5.Urdaneta I, Keller A, Atabek O, Palma JL, Finkelstein-Shapiro D, Tarakeshwar P, Mujica V, Calatayud M. Dopamine adsorption on TiO2 anatase surfaces. J Phys Chem C. 2014;118:20688–20693. [Google Scholar]
- 6.Vega-Arroyo M, LeBreton PR, Rajh T, Zapol P, Curtiss LA. Density functional study of the TiO2-dopamine complex. Chem Phys Lett. 2005;406:306–311. [Google Scholar]
- 7.Endres PJ, Paunesku T, Vogt S, Meade TJ, Woloschak GE. DNA-TiO2 nanoconjugates labeled with magnetic resonance contrast agents. J Am Chem Soc. 2007;129:15760–15761. doi: 10.1021/ja0772389. [DOI] [PubMed] [Google Scholar]
- 8.Thurn KT, Paunesku T, Wu AG, Brown EM, Lai B, Vogt S, Maser J, Aslam M, Dravid V, Bergan R, et al. Labeling TiO2 nanoparticles with dyes for optical fluorescence microscopy and determination of TiO2-DNA nanoconjugate stability. Small. 2009;5:1318–1325. doi: 10.1002/smll.200801458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye L, Pelton R, Brook MA. Biotinylation of TiO2 nanoparticles and their conjugation with streptavidin. Langmuir. 2007;23:5630–5637. doi: 10.1021/la0626656. [DOI] [PubMed] [Google Scholar]
- 10.Thurn KT, Brown E, Wu AG, Vogt S, Lai B, Maser J, Paunesku T, Woloschak GE. Nanoparticles for applications in cellular imaging. Nanoscale Res Lett. 2007;2:430–441. doi: 10.1007/s11671-007-9081-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu AG, Paunesku T, Zhang ZL, Vogt S, Lai B, Maser J, Yaghmai V, Li DB, Omary RA, Woloschak GE. A multimodal nanocomposite for biomedical imaging. AIP Conf Proc. 2011;1365:379. doi: 10.1063/1.3625382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arora HC, Jensen MP, Yuan Y, Wu AG, Vogt S, Paunesku T, Woloschak GE. Nanocarriers enhance Doxorubicin uptake in drug-resistant ovarian cancer cells. Cancer Res. 2012;72:769–778. doi: 10.1158/0008-5472.CAN-11-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bazak R, Ressl J, Raha S, Doty C, Liu W, Wanzer B, Salam SA, Elwany S, Paunesku T, Woloschak GE. Cytotoxicity and DNA cleavage with core-shell nanocomposites functionalized by a KH domain DNA binding peptide. Nanoscale. 2013;5:11394–11399. doi: 10.1039/c3nr02203j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown EMB, Paunesku T, Wu AG, Thurn KT, Haley B, Clark J, Priester T, Woloschak GE. Methods for assessing DNA hybridization of peptide nucleic acid-titanium dioxide nanoconjugates. Anal Biochem. 2008;383:226–235. doi: 10.1016/j.ab.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurepa J, Paunesku T, Vogt S, Arora H, Rabatic BM, Lu JJ, Wanzer MB, Woloschak GE, Smalle JA. Uptake and distribution of ultrasmall anatase TiO2 Alizarin red S nanoconjugates in Arabidopsis thaliana. Nano Lett. 2010;10:2296–2302. doi: 10.1021/nl903518f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paunesku T, Ke TY, Dharmakumar R, Mascheri N, Wu AG, Lai B, Vogt S, Maser J, Thurn K, Szolc-Kowalska B, et al. Gadolinium-conjugated TiO2-DNA oligonucleotide nanoconjugates show prolonged intracellular retention period and T1-weighted contrast enhancement in magnetic resonance images. Nanomedicine. 2008;4:201–207. doi: 10.1016/j.nano.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paunesku T, Vogt S, Lai B, Maser J, Stojićević N, Thurn KT, Osipo C, Liu H, Legnini D, Wang Z, et al. Intracellular distribution of TiO2-DNA oligonucleotide nanoconjugates directed to nucleolus and mitochondria indicates sequence specificity. Nano Lett. 2007;7:596–601. doi: 10.1021/nl0624723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajh T, Dimitrijevic NM, Rozhkova EA. Titanium dioxide nanoparticles in advanced imaging and nanotherapeutics. In: Hurst SJ, editor. Methods and Protocols. Humana Press; Humana: 2011. pp. 63–75. [DOI] [PubMed] [Google Scholar]
- 19.Thurn KT, Arora H, Paunesku T, Wu AG, Brown EMB, Doty C, Kremer J, Woloschak G. Endocytosis of titanium dioxide nanoparticles in prostate cancer PC-3M cells. Nanomedicine. 2011;7:123–130. doi: 10.1016/j.nano.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan Y, Chen S, Paunesku T, Gleber SC, Liu WC, Doty CB, Mak R, Deng JJ, Jin QL, Lai B, et al. Epidermal growth factor receptor targeted nuclear delivery and high-resolution whole cell X-ray imaging of Fe3O4@TiO2 nanoparticles in cancer cells. ACS Nano. 2013;7:10502– 10517. doi: 10.1021/nn4033294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kotsokechagia T, Zaki NM, Syres K, de Leonardis P, Thomas A, Cellesi F, Tirelli N. PEGylation of nanosubstrates (titania) with multifunctional reagents: At the crossroads between nanoparticles and nanocomposites. Langmuir. 2012;28:11490–11501. doi: 10.1021/la3012958. [DOI] [PubMed] [Google Scholar]
- 22.Zhang AP, Sun YP. Photocatalytic killing effect of TiO2 nanoparticles on Ls-174-t human colon carcinoma cells. World J Gastroenterol. 2004;10:3191–3193. doi: 10.3748/wjg.v10.i21.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L, Dong S, Zhu L. Fluorescent dyes of the esculetin and alizarin families respond to zinc ions ratiometrically. Chem Commun. 2007;19:1891–1893. doi: 10.1039/b618413h. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Wang F, Liu BW, Kelly EY, Servos MR, Liu JW. Adsorption of DNA oligonucleotides by titanium dioxide nanoparticles. Langmuir. 2014;30:839–845. doi: 10.1021/la404633p. [DOI] [PubMed] [Google Scholar]
- 25.Paunesku T, Vogt S, Maser J, Lai B, Woloschak G. X-ray fluorescence microprobe imaging in biology and medicine. J Cell Biochem. 2006;99:1489–1502. doi: 10.1002/jcb.21047. [DOI] [PubMed] [Google Scholar]
- 26.Abbas Z, Holmberg JP, Hellström AK, Hagström M, Bergenholtz J, Hassellöv M, Ahlberg E. Synthesis, characterization and particle size distribution of TiO2 colloidal nanoparticles. Colloid Surf A-Physicochem Eng Aspects. 2011;384:254–261. [Google Scholar]
- 27.Vogt S. MAPS: A set of software tools for analysis and visualization of 3D X-ray fluorescence data sets. J Phys IV. 2003;104:635–638. [Google Scholar]
- 28.Chen S, Deng J, Yuan Y, Flachenecker C, Mak R, Hornberger B, Jin Q, Shu D, Lai B, Maser J, et al. The bionanoprobe: Hard X-ray fluorescence nanoprobe with cryogenic capabilities. J Synchrotron Radiat. 2014;21:66–75. doi: 10.1107/S1600577513029676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corezzi S, Urbanelli L, Cloetens P, Emiliani C, Helfen L, Bohic S, Elisei F, Fioretto D. Synchrotron-based X-ray fluorescence imaging of human cells labeled with CdSe quantum dots. Anal Biochem. 2009;388:33–39. doi: 10.1016/j.ab.2009.01.044. [DOI] [PubMed] [Google Scholar]
- 30.Russell-Jones G, McTavish K, McEwan J, Rice J, Nowotnik D. Vitamin-mediated targeting as a potential mechanism to increase drug uptake by tumours. J Inorg Biochem. 2004;98:1625–1633. doi: 10.1016/j.jinorgbio.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 31.Vadlapudi AD, Vadlapatla RK, Mitra AK. Sodium dependent multivitamin transporter (SMVT): A potential target for drug delivery. Curr Drug Targets. 2012;13:994–1003. doi: 10.2174/138945012800675650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Townsend SA, Evrony GD, Gu FX, Schulz MP, Brown RH, Langer R. Tetanus toxin C fragment-conjugated nanoparticles for targeted drug delivery to neurons. Biomaterials. 2007;28:5176–5184. doi: 10.1016/j.biomaterials.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi H, Sakahara H, Endo K, Hosono M, Yao ZS, Toyama S, Konishi J. Comparison of the chase effects of avidin, streptavidin, neutravidin, and avidin-ferritin on a radiolabeled biotinylated anti-tumor monoclonal antibody. Jpn J Cancer Res. 1995;86:310–314. doi: 10.1111/j.1349-7006.1995.tb03056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amblard F, Cho JH, Schinazi RF. Cu(I)-catalyzed huisgen azide alkyne 1,3-dipolar cycloaddition reaction in nucleoside, nucleotide, and oligonucleotide chemistry. Chem Rev. 2009;109:4207–4220. doi: 10.1021/cr9001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meldal M, Tornøe CW. Cu-catalyzed azide alkyne cycloaddition. Chem Rev. 2008;108:2952–3015. doi: 10.1021/cr0783479. [DOI] [PubMed] [Google Scholar]
- 36.Cardiel AC, Benson MC, Bishop LM, Louis KM, Yeager JC, Tan YZ, Hamers RJ. Chemically directed assembly of photoactive metal oxide nanoparticle hetero-junctions via the copper-catalyzed azide-alkyne cycloaddition “Click” reaction. ACS Nano. 2012;6:310–318. doi: 10.1021/nn203585r. [DOI] [PubMed] [Google Scholar]
- 37.Tao P, Li Y, Rungta A, Viswanath A, Gao JN, Benicewicz BC, Siegel RW, Schadler LS. TiO2 nanocomposites with high refractive index and transparency. J Mater Chem. 2011;21:18623–18629. [Google Scholar]
- 38.Upadhyay AP, Behara DK, Sharma GP, Gyanprakash M, Pala RGS, Sivakumar S. Fabricating appropriate band-edge-staggered heterosemiconductors with optically activated au nanoparticles via click chemistry for photo-electrochemical water splitting. ACS Sustainable Chem Eng. 2016;4:4511–4520. [Google Scholar]
- 39.Yadav SK, Madeshwaran SR, Cho JW. Synthesis of a hybrid assembly composed of titanium dioxide nanoparticles and thin multi-walled carbon nanotubes using “Click chemistry”. J Colloid Interface Sci. 2011;358:471–476. doi: 10.1016/j.jcis.2011.03.040. [DOI] [PubMed] [Google Scholar]
- 40.Wiederschain GY. The molecular probes handbook: A guide to fluorescent probes and labeling technologies. Biochemistry Moscow. 2011;76:1276. [Google Scholar]
- 41.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal non-canonical amino acid tagging (BONCAT) Proc Natl Acad Sci U S A. 2006;103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clark PM, Dweck JF, Mason DE, Hart CR, Buck SB, Peters EC, Agnew BJ, Hsieh-Wilson LC. Direct in-gel fluorescence detection and cellular imaging of O-GlcNAc-modified proteins. J Am Chem Soc. 2008;130:11576–11577. doi: 10.1021/ja8030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malalasekera AP, Wang HW, Samarakoon TN, Udukala DN, Yapa AS, Ortega R, Shrestha TB, Alshetaiwi H, McLaurin EJ, Troyer DL, et al. A nanobiosensor for the detection of arginase activity. Nanomedicine. 2016;13:383–390. doi: 10.1016/j.nano.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 45.Park J, Kadasala NR, Abouelmagd SA, Castanares MA, Collins DS, Wei A, Yeo Y. Polymer-iron oxide composite nanoparticles for EPR-independent drug delivery. Biomaterials. 2016;101:285–295. doi: 10.1016/j.biomaterials.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.