ABSTRACT
Med15 is an important subunit of Mediator Tail module and is characterized by a KIX domain present towards amino terminal. In yeast and metazoans, Med15 KIX domain has been found to interact with various transcription factors regulating several processes including carbohydrate metabolism, lipogenesis, stress response and multidrug resistance. Mechanism of Med15 functioning in Arabidopsis is largely unknown. In this study, interactome of KIX domain of Arabidopsis Med15, AtMed15a, was characterized. We found 45 proteins that interact with AtMed15a KIX domain, including 11 transcription factors, 3 single strand nucleic acid-binding proteins and 1 splicing factor. The third helix of the KIX domain was found to be involved in most of the interactions. Mapping of the regions participating in the interactions revealed that the activation domain of a transcription factor, UKTF1 interacted with AtMed15a KIX domain. Thus, our results suggest that in Arabidopsis, activation domain of transcription factors target KIX domain of AtMed15a for their transcriptional responses.
KEYWORDS: Mediator, Med15, KIX domain, transcription factors, transactivation
Introduction
Mediator (Med) is a key regulator of protein-coding genes and is responsible for development, differentiation and maintenance of homeostasis in eukaryotes from yeast to human.1-5 It is a gigantic multiprotein complex consisting of 24–34 subunits, arranged in four modules – Head module, Middle module, Tail module and CDK module.6,7 The Head module is the most conserved structure and makes contacts with RNA polymerase II. The Tail module is the least conserved, providing a platform for gene-specific regulators to interact.8-13 The Middle module is important for the transfer of regulatory signals across the Mediator.14 The separable CDK module associates with Middle module leading to transcriptional repression.15 Mediator receives signals from activators through interaction with its Tail module and ultimately relays the transcriptional signals to the pre-initiation complex (PIC) through interactions of Head and Middle modules with the RNA polymerase II transcriptional machinery.16 However, studies as of now showed that Mediator might be involved in several other aspects of transcription including initiation, elongation, termination, repression, splicing and gene looping.17-20 The Med15 subunit along with Med2, Med3, Med5, Med14 and Med16 constitute a part of the Tail module.17 Med15 was discovered as GAL11 in biochemical studies well before the discovery of Mediator and was considered as a factor required for the transcription of GAL1, GAL7 and GAL10 genes of yeast.21,22 Med15 is the node of multiple signal transduction cascades merging at the Tail module and has been found to be involved in diverse metabolism and developmental pathways.17 In yeast, mammals, C. elegans and Arabidopsis, Med15 has been found to be important for lipogenesis.23-28 We and others have deciphered the role of Med15 in multidrug resistance in fungi and animals.29-31 Med15 in plants has been found to be important for salicylic acid-mediated immune response.32 ARC105/Med15 has also been found to be required for TGFβ/Activin/Nodal/Smad2/3 signal transduction, important for certain developmental pathways in metazoans.33 Differential expression pattern of AtMed15a in response to abiotic stresses and phytohormones as well as in different tissues, suggests its role in hormonal signaling and developmental programming of the plant.34,35 In one other study, one SNP in the KIX region of OsMed15 gene was found to be significantly associated with grain size/weight of different rice cultivars.36 Thus, Med15 is an important player in transcriptional regulation of several diverse processes in eukaryotes.
Med15 contains a KIX domain at the amino terminal, which has been found to be important for interaction of Med15 with various transcription activators. KIX domain consists of combination of three α-helices arranged in a characteristic fashion to form hydrophobic pockets, which may be exploited as docking site(s) by activation domains (ADs) of transcription factors (TFs).37-42 In yeast, KIX domain of Gal11p/Med15 has been reported to interact with Pdr1p, Oaf1p, Gcn4p etc. and, in animal, Med15 has been reported to interact with SREBP and NHR-49.23,24,26,29,43 However, except for WRINKLED1, no other transcription factors interacting with Med15 KIX domain in Arabidopsis have been reported yet.28 In this study, we have characterized the proteins that interact with the KIX domain of Med15 of Arabidopsis. Our analysis revealed that in Arabidopsis, KIX domain of Med15 is targeted by diverse categories of proteins including transcription factors and suggests that the activation domain of transcription factors may target it for their transcriptional responses. Structural analysis indicated the importance of helix α3 in the protein-protein interactions. Indeed, deletion of this helix abrogated interaction of AtMed15a KIX with other proteins. Thus, KIX domain-mediated interaction of Med15 with transcription factors is conserved in all the eukaryotic kingdoms.
Results
Characterization of Arabidopsis proteins interacting with KIX domain of AtMed15a
In Arabidopsis, though five paralogs of Med15 have been reported, expression data of only three are available in public domain.34 Expression profile generated from the data of Arabidopsis eFP browser revealed higher expression of AtMed15a (At1g15780) in several tissues including root, leaf, flower and seeds suggesting it to be an important protein (Fig. S1A). AtMed15a has got a KIX domain at its amino terminal. In fungi and metazoans, several transcription factors target KIX domain of Med15 to recruit Mediator and RNA polymerase II transcriptional machinery on the target promoters.23,24,26,29,43 In Arabidopsis, transcription factors that target Med15 is largely unknown. In order to know which proteins interact with the KIX domain of Med15 in Arabidopsis, we performed yeast two-hybrid (Y2H) screening with normalized cDNA library generated from Arabidopsis tissues using KIX domain of Arabidopsis Med15a as the bait. KIX domain (6–101 amino acids) of AtMed15a was cloned in Y2H vector pGBKT7 and expressed in yeast as Myc-tagged Gal4DBD fusion protein. Expression of Myc-Gal4DBD-AtMed15a KIX in yeast was confirmed by western blot analysis (Fig. S1B) and its inability to auto-activate the reporter genes was assured by absence of growth of yeast cells on SD Trp−/His−/Ade− medium (Fig. S1C). Around 5.5 million clones were screened and around 200 putatively positive ones were sequenced. The ‘false positives’ were rid by the process mentioned in Materials and Methods (Table S1). Finally, 45 positive clones were characterized to be coding for proteins interacting with the KIX domain of AtMed15a (Fig. 1A, Table S2). Based on SUBA (http://suba.live) analysis, out of 45 interacting proteins, 21 were found to be nuclear proteins whereas localization sites of 24 proteins were found to be outside the nucleus. A couple of AtMed15a KIX-interacting proteins were found to be localized both inside and outside the nucleus. Some of these interactions were randomly selected and validated by bimolecular fluorescence complementation (BiFC) (Fig. 1B). Interactions of AtMed15a KIX domain with three proteins (MYB63, R3H and UKTF1) were found to be localized not only inside the nucleus but also outside. Interactome of AtMed15a includes proteins involved in different cellular and physiological processes including plant development, stress responses, gene expression, transport of cellular molecules, protein modification, metabolism, hormonal signaling etc. (Fig. 2A). Function of some of these proteins interacting with AtMed15a is not yet known. Since these proteins are localized at different cellular sites, we studied localization of AtMed15a-YFP and found it to be localized both inside and outside the nucleus (Fig. 2B).
Figure 1.
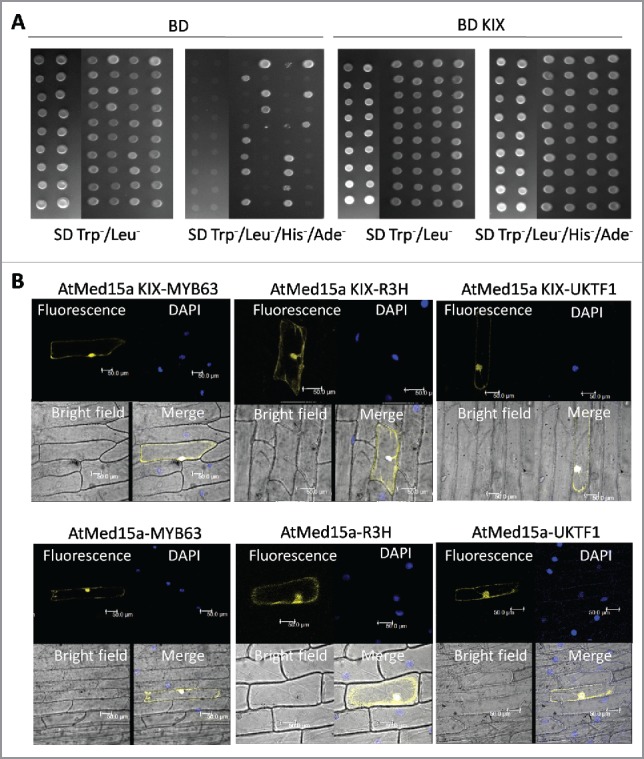
Characterization of proteins interacting with AtMed15a KIX domain. (A) Yeast two-hybrid screening was done using AtMed15a KIX as the bait. False positive clones were characterized by yeast two-hybrid analysis using vector alone (BD) as a control (left panel). Cultures of co-transformed yeast were spotted on SD Trp−/Leu− to monitor the growth and on SD Trp−/Leu−/His−/Ade− to score the interactions. Right panel shows clones of AtMed15a KIX-domain interacting protein (BD KIX). (B) Interaction of selected proteins with AtMed15a KIX domain (upper panel) and full-length AtMed15a (lower panel) was validated by BiFC analysis. AtMed15a KIX, AtMed15a and Y2H positive clones (MYB63, R3H and UKTF1) were cloned in compatible YFP BiFc vectors and bombarded on onion peel. YFP and DAPI (blue) fluorescence was observed by confocal microscopy. DAPI staining was used to identify nuclei in the cells.
Figure 2.
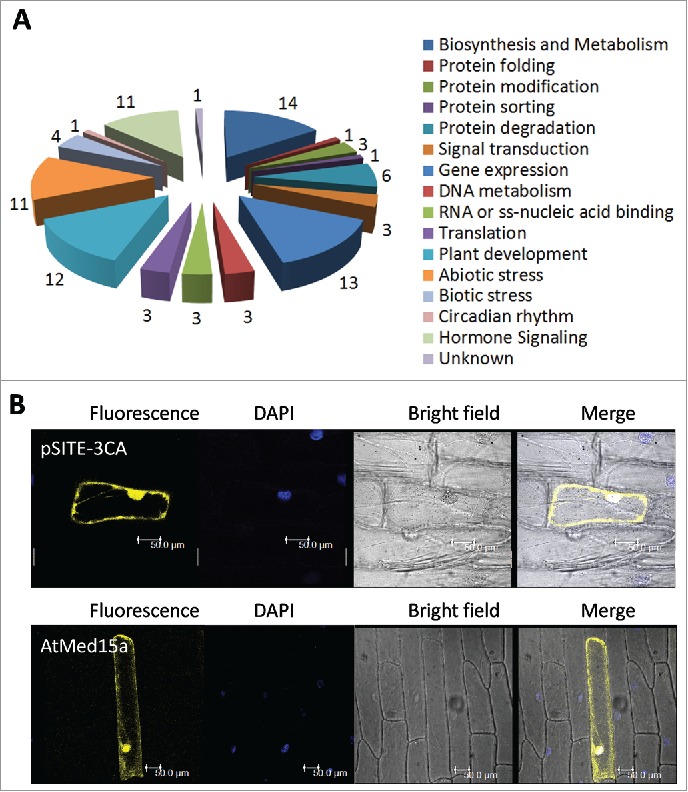
The interactome of AtMed15a KIX represents diverse proteins with different functions. (A) Graphical representation of the functional categories of proteins found to interact with AtMed15a KIX. Note that interactors with multiple functions can be present in different categories (Listed in Table S4). (B) AtMed15a was cloned in YFP vector, bombarded on onion peel and visualized using confocal microscopy.
Activation domain of transcription factors target KIX domain of AtMed15a
In the interactome of AtMed15a KIX domain, 11 transcription factors were identified including MYB63 (At1g79180) and unknown transcription factor UKTF1 (At2g20100) (Table S2). We mapped the regions in MYB63 and UKTF1 that were involved in the interaction with AtMed15a KIX. In MYB63, a minimum region of 131 to 294 amino acids was found to interact with AtMed15a KIX (Fig. 3A). Sequence analysis of this region revealed presence of eight transactivation domains (9aa TADs) in it (Fig. 3B). The ability of this region to activate the reporter genes could not be confirmed because Y187 cells (which are used for lacZ expression) transformed with this part or smaller fragments of this part did not survive. In the case of UKTF1, region spanning from 1 to 220 amino acids was found to interact with AtMed15a KIX (Fig. 4A). This region was found to harbor three 9aa TADs (Fig. 4B). We selected the interacting region of UKTF1 for further fragmentation to locate the transactivation activity (Fig. 5A). Analysis of these regions for the activation of reporter gene LacZ revealed that region from 41 to 110 amino acids of UKTF1 (U3), which harbor two 9aa TADs, possessed maximum transactivation ability (Fig. 5B). Thus, transactivation domain containing region of UKTF1 was found to interact with the KIX domain of AtMed15a.
Figure 3.
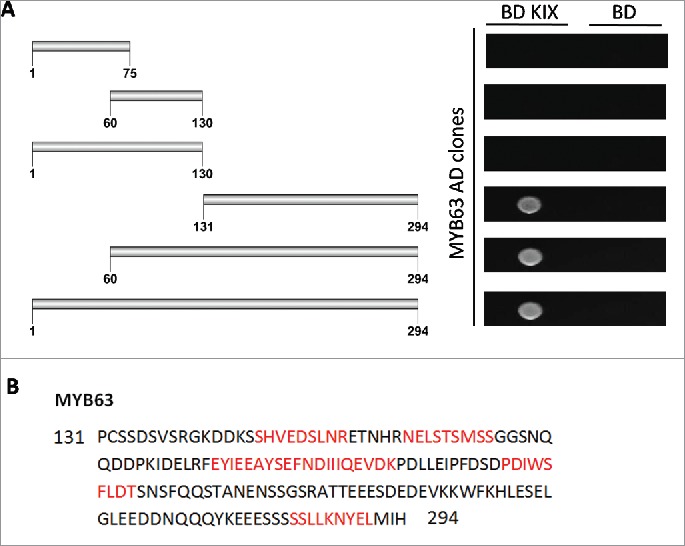
AtMed15a KIX interacting region of MYB63 harbors 9aa TADs. (A) Y2HGold co-transformed with derivatives of AD MYB63 clones and BD KIX or BD vector as control were spotted and grown on SD Trp−/Leu−/His−/Ade− with 10 mM 3-AT and 125 ng/ml Aureobasidin A agar media to score interactions. (B) 9aa TAD regions of AtMed15a KIX-interacting region of MYB63 are highlighted in red.
Figure 4.
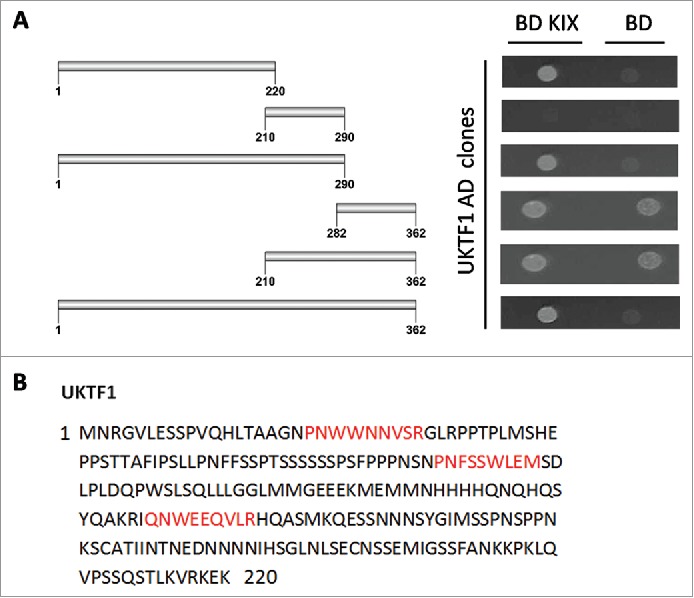
AtMed15a KIX interacting region of UKTF1 harbors 9aa TADs. (A) Y2HGold co-transformed with derivatives of AD UKTF1 clones and BD KIX or BD vector as control were spotted and grown on SD Trp−/Leu−/His−/Ade− with 10 mM 3-AT and 125 ng/ml Aureobasidin A agar media to score interactions. (B) 9aa TAD of AtMed15a KIX-interacting regions of UKTF1 are highlighted in red.
Figure 5.
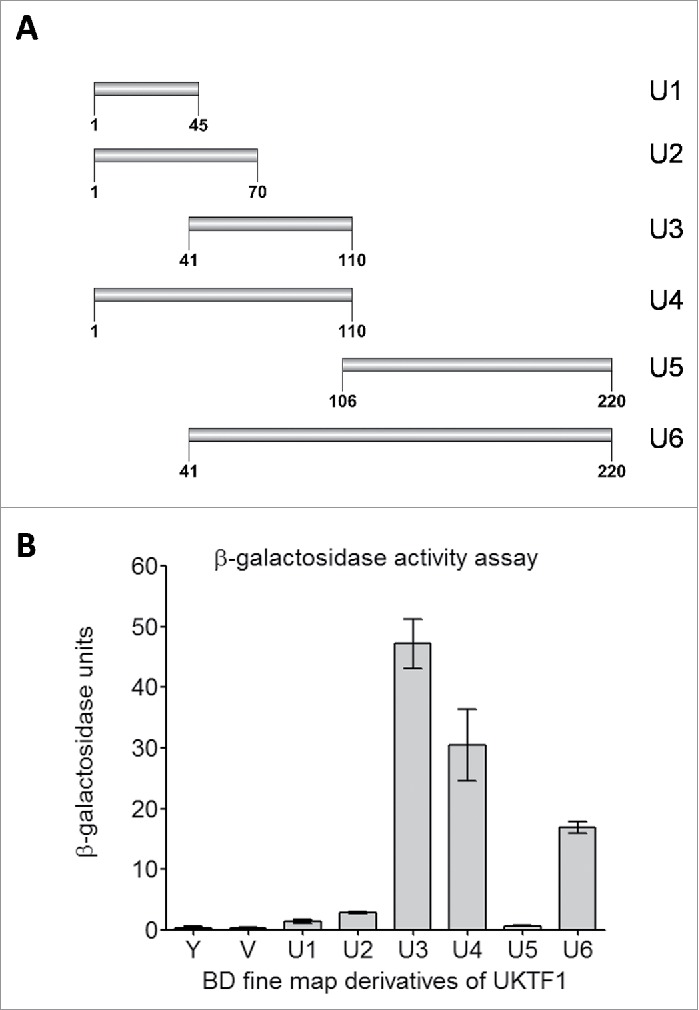
Transactivation domain of UKTF1. (A) Fragments, of AtMed15a KIX-interacting region of UKTF1 (U1 to U6), were cloned in pGBKT7 (BD) vector. (B) Y187 yeast cells were transformed with BD derivative clones (U1 to U6) of UKTF1 or BD vector control (V) and processed for β-Galactosidase assay. Graph showing β-gal units of untransformed Y187 (Y), Y187 transformed with BD vector (V) or BD clones of UKTF1 derivatives (U1-U6).
Third helix of AtMed15a KIX is responsible for most of the interactions
AtMed15a KIX comprises three alpha helices, similar to that found in the structures of different KIX domains including mCBP KIX.42 So, AtMed15a KIX structure was predicted by homology modelling using mCBP KIX (PDB id: 1KDX) as the template (Fig. 6A). mCBP KIX was used as the template because it emerged as the top hit in the search of template on the basis of similarity with AtMed15 KIX domain (Table S3). The predicted AtMed15a KIX structure showed the presence of three mutually interacting alpha helices (α1 from P28 to L42; α2 from G53 to G71 and α3 from Q75 to M89). However, there was no 310 helix in AtMed15a KIX. α1 and α2 helices of AtMed15a KIX were at an angle of ∼57 °, whereas α1 and α3 helices were found to be at an angle of ∼18 ° (Fig. 6A). On the other hand, structure of MYB63 was predicted by Phyre2 server and found to contain a helical region (181–201 amino acids) with the potential to interact with other protein (Fig. 6B, Fig. S2A, S2B). D176 to D203 of MYB63 also contains 9aa TADs (Fig. S2C). Predicted model of AtMed15a KIX and MYB63 were validated by RAMPAGE and ERRAT servers (Fig. S3). Structure of mCBP KIX-cMyb complex available in the database (PDB id: 1SB0) was used for the analysis of important features of AtMed15a KIX. Residues of mCBP KIX making contact with cMyb retrieved using PDBsum were aligned with AtMed15a KIX (Fig. 6C, Fig. S4). Replacing mCBP KIX with AtMed15a KIX in mCBP KIX-cMyb complex showed similarity of AtMed15a KIX with mCBP KIX (Fig. S5). Docking of AtMed15a KIX and MYB63 peptide (D176 to D203) showed that α1 and α3 helices of AtMed15a KIX is involved in the interactions (Fig. 6D; Fig. S6), as observed in the case of mCBP KIX-cMyb interaction. To validate these predictions, 10 amino acids (79–88) of helix α3 were deleted from the KIX domain and then its interaction with MYB63 was checked (Fig. 6E, 6F). Interestingly, deletion of these 10 amino acids abolished the interaction of not only AtMed15a KIX-MYB63 but also the interaction of AtMed15a KIX with many other proteins (Fig. 6E, 6F). Thus, a stretch of 10 amino acids (79–88) in the KIX domain of AtMed15a was found to be critical for its interaction with other proteins.
Figure 6.
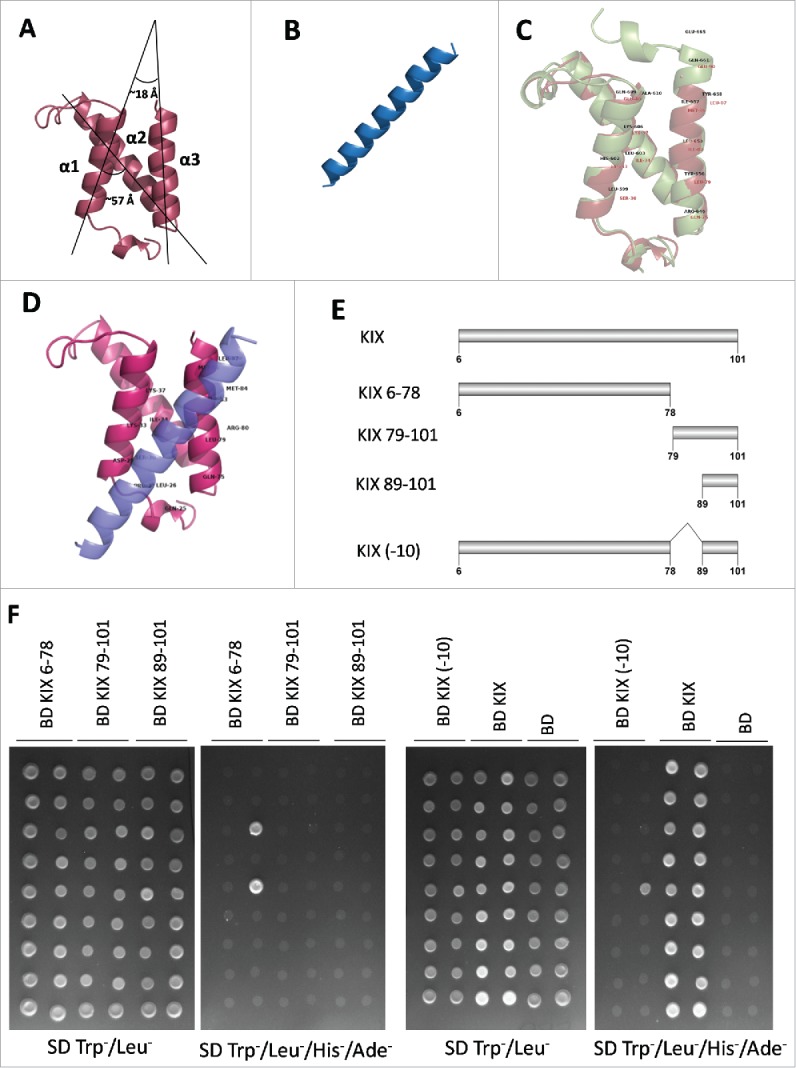
Third helix of AtMED15a KIX is required for protein-protein interactions. (A) Structure of AtMed15a KIX was predicted by homology modelling using mCBP KIX as template. (B) Based on the presence of binding region and 9aaTADs, MYB63 (D176 to D203) peptide was selected for docking studies. (C) Structures of mCBP KIX (green) and AtMed15a KIX (brown) were aligned using PyMOL, Labels show interacting residues. (D) Complex of AtMed15a KIX with MYB63 (D176 to D203), showing interacting residues of AtMed15a KIX. (E) Fragments of AtMed15a KIX were made based on the prediction of interacting residues in α3. (F) Interaction of Y2H positive clones with different fragments of AtMed15a KIX domain, as mentioned in (E), was checked. Cultures of co-transformed yeast were spotted on SD Trp−/Leu− to show proper growth, while on SD Trp−/Leu−/His−/Ade− to score the interactions.
Discussion
Over two decades of research on Mediator complex has established it as a major player of transcription regulation in eukaryotes. Mediator has been found to be involved in the transcription initiation, elongation, splicing, gene looping and termination. This evolutionarily conserved protein complex has a large surface that mediates protein-protein interactions, leading to the formation of PIC, engaging TFs and RNA polymerase II transcriptional machinery.3,6,17,19 The Tail module of Mediator provides an extensive platform for interactions with gene-specific regulators. Med15 is a subunit of Tail module of Mediator. In Arabidopsis, five paralogs of Med15 has been reported.34 Chromosome 1 contains a cluster of four Med15 paralogs (At1g15780, At1g15770, At1g15772 and At1g15790) while one paralog is present on chromosome 2 (At2g10440).34 Except At1g15770, all other paralogs of Arabidopsis Med15 contain KIX domain towards amino terminal. Expression profile of three paralogs of Arabidopsis Med15 (At1g15780, At1g15790 and At2g10440) is available in public domain. At1g15780 (AtMed15a) is expressed predominantly in root, cauline leaf, senescing leaf, first node, flower, silique and dry seeds (Fig. S1A). Expression of AtMed15a can also be seen in rosette leaves. Such expression profile suggests that AtMed15a is an important protein functioning in different tissues. In contrast, At2g10440 is expressed at low level in all these tissues. Expression of At1g15790 is significantly high in senescent leaves whereas moderate expression is seen in cauline leaf suggesting its role mainly in leaf senescence.
It has been reported in fungi and animals that the KIX domain of Med15 interacts with TFs to regulate several processes.42 In this study, we have characterized the interactome of AtMed15a KIX to identify the proteins targeting it. Interestingly, in addition to 11 transcription factors, different other proteins were also found to interact with AtMed15a KIX domain (Fig. 2A). Interacting proteins of AtMed15a KIX domain belong to several categories performing different molecular functions and biological roles, which include transcription, chromatin remodelling, DNA repair, protein modification, protein degradation, signal transduction, single strand nucleic acid binding, biosynthesis, metabolism, biotic and abiotic stress response, plant development and hormone signaling (Fig. 2A). Thus, it seems that AtMed15a might have some functions beyond its involvement in transcription as a Mediator subunit. Functional studies of the novel AtMed15a KIX interactors will help to reveal the unknown functions of Med15a in Arabidopsis. Nevertheless, functional classification of AtMed15a interactome into diverse categories shows the possible involvement of AtMed15a in various physiological and biological pathways (Fig. 2A, Table S2, S4).
We found six MYB domain containing TFs (MYB10, MYB12, MYB14, MYB15, MYB63 and MYB72), three basic helix-loop-helix TFs (bHLH48, AKS1 and UKTF1), one NAC domain TF (NAC082) and one B-box domain TF (BBX8) interacting with AtMed15a KIX. MYB12 is involved in flavonoid biosynthesis, MYB15 helps in defense-induced lignifications and MYB63 is involved in lignin biosynthesis pathway during secondary cell wall formation in Arabidopsis.44-46 MYB10 and MYB72 are important for growth of plant under limited iron conditions.47 Role of MYB72 has also been implicated in induced systemic resistance.48 MYB14 is involved in cold resistance.49 NAC082 is important for growth and development of the plant.50 Thus, this study suggests that in Arabidopsis, Med15a KIX is an important target for TFs to regulate diverse pathways, just like in yeast and metazoans. There are research going on to discover classical nuclear receptors and nuclear receptor-like proteins in plants. However, till date, no nuclear receptor-like proteins have been discovered in plants. In fungi, nuclear receptor-like ligand-activated TFs Pdr1p and Oaf1p interact with KIX domain of Gal11p/Med15, while in animals, NHR-49 interacts with KIX domain of ARC105/MDT-15.23,24,26,29 In our study, we did not find any nuclear receptor-like and ligand-activated TF interacting with AtMed15a KIX domain.
In corroboration with the interaction of AtMed15a with so many different proteins including non-nuclear ones, AtMed15a was found to be localized not only inside the nucleus but outside also (Fig. 2B). Apart from nuclear functions, Mediator has also been found to be involved in cytoplasmic signaling. A study showed that cytoplasmic signaling protein Elmo1 interacts with Med31 and promotes ubiquitination and relocalization of Med31 from nucleus to cytoplasm, important for immune response during Salmonella infection of primary macrophages.51 In our study, AtMed15a KIX was found to interact with SKIP16, RPN12a and RPT5a (Table S2). SKIP1 is a part of SCF ubiquitin ligase complex, while RPN12a and RPT5a are regulatory particles of 26S proteasome. These proteins might be regulating turnover of AtMed15a in the cell. AtMed15a is a highly disordered protein.7 Disordered proteins have higher propensity to interact with other proteins. There is a possibility that being localized in the cytoplasm, AtMed15a may stabilize various cytoplasmic proteins by acting as a chaperone for them.
Structural analyses showed that AtMed15a KIX domain is highly similar to mouse CBP KIX, which is known to interact with several proteins. Based on mCBP KIX-cMyb interaction, docking of AtMed15a KIX with MYB63 (D176 to D203) revealed that a 10 amino acids-stretch (79–88) of third helix of AtMed15a KIX domain was involved in the interaction (Fig. 6D). Indeed, deletion of this stretch of 10 amino acids of helix α3 abrogated the interaction of AtMed15a KIX domain with almost all the proteins tested in the experiment (Fig. 6E, 6F).
It has been found in fungi and animals that KIX domain interacts with the activation domain of transcription factors and plays an important role in transcriptional regulation of their target genes. One of the main aims of this study was to find out the region of TFs important for targeting AtMed15a KIX domain and check if it helps in transactivation. Characterization of regions involved in its interaction with AtMed15a KIX domain revealed that the transactivation domain of UKTF1 targets AtMed15a KIX domain (Fig. 4, Fig. 5). Thus, targeting of KIX domain of Med15 by the activation domain of transcription factors is a mechanism conserved in all the eukaryotic kingdoms including plants.
Materials and methods
Cloning of AtMed15a KIX in pGBKT7 and confirmation of protein expression: CDS (16–303) of AtMed15a was cloned in pGBKT7 (BD) vector and transformed into yeast Y2HGold strain. In order to detect the expression of AtMed15a KIX in yeast Y2HGold strain, homogenized whole cell extracts were immunoprecipitated on protein A and protein G sepharose (GE Healthcare) using anti-Myc antibody (Cell Signaling Technologies). Immunoprecipitated proteins were separated on 12% SDS-PAGE gel, transferred onto Amersham Hybond-ECL membrane (GE Healthcare) using an electroblotter and detected with anti-Myc antibody. Signal was detected using the Amersham ECL-Plus Western blotting detection system, GE Healthcare.
Yeast two-hybrid screening and assays: All methods for Y2H screening and assays were followed as described in the Matchmaker Gold Yeast Two-Hybrid System User Manual (Clontech). Before starting the Y2H screening, bait (pGBKT7-AtMed15a KIX) was checked for auto-activation of reporter genes. Y2H screening was done by mating Y2HGold: pGBKT7-AtMed15a KIX and Y187 harboring Arabidopsis cDNA library (Clontech). To subtract false positives, reading frames of the Y2H clones were checked and later on only in-frame Y2H clones were tested for their interaction with Gal4 DBD alone and Gal4 DBD-AtMed15a KIX on SDTrp−/Leu−/His−/Ade− agar plates. 2 µl of yeast culture at 0.2 OD was spotted on agar media.
Bimolecular fluorescent complementation (BiFC): CDS of AtMed15a, AtMed15a KIX, MYB63 (At1g79180), R3H (At3g10770) and UKTF1 (At2g20100) were first cloned in pENTR/D-TOPO vector and mobilized respectively to pSAT4-DEST-N (1–174) EYFP-C1 and pSAT5-DEST-C (175-END) EYFP-C1 vectors using Gateway cloning technology (Invitrogen).52 The primers used for cloning are given in Table S5. Recombinant plasmids were bombarded pair-wise on onion epidermal cells placed on MS plates (3% sucrose, 4.42 g/L MS, 1% agar) using PDS-1000/He Biolistic Particle Delivery System (Bio-Rad). After bombardment, plates were kept aseptically in darkness at 25°C for 18 hours. Fluorescence was observed under TCS SP2 (AOBS) laser confocal scanning microscope (Leica Microsystems).
Subcellular localization of AtMed15a: CDS of AtMed15a was cloned in pENTR/D-TOPO vector and then mobilized to destination vector pSITE-3CA using Gateway cloning technology (Invitrogen). The primers used for cloning are given in Table S5. The recombinant plasmid was used to coat gold particles and bombarded on onion epidermal cells placed on MS plates (3% sucrose, 4.42 g/L MS, 1% agar) using PDS-1000/He Biolistic Particle Delivery System (Bio-Rad). After bombardment, plates were kept aseptically in darkness at 25°C for 18 hours. Fluorescence was observed under TCS SP2 (AOBS) laser confocal scanning microscope (Leica Microsystems).
Mapping of MYB63 and UKTF1 proteins which interact with KIX domain of AtMed15a: PCR primers listed in Table S5 were used to amplify and clone CDS of genes and their derivatives in pGADT7 vector. These AD clones were tested for their interaction with Gal4 DBD alone and Gal4 DBD-AtMed15a KIX on SD Trp−/Leu−/His−/Ade− with 10 mM 3-AT and 125 ng/ml Aureobasidin A agar plates. Spotting was done with 2 µl of culture at 0.2 OD.
Transactivation assays for UKTF1: Fragments of AtMed15a KIX-interacting part of UKTF1 were cloned in pGBKT7 vector using suitable PCR primers listed in Table S5. All Gal4 DBD–UKTF1 derivative clones (U1-U6) along with BD vector as control were transformed in Y187 strain of yeast cells and selected on SD Trp− agar plates. Using these Y187 transformed colonies, β-Galactosidase liquid assays were performed in three independent replicates.
Prediction of residues important for interaction of AtMed15a KIX domain with MYB63: Protein sequences of AtMed15a and MYB63 were retrieved from NCBI protein database (https://www.ncbi.nlm.nih.gov/protein/). Homology model of AtMed15a KIX was constructed by SWISS-MODEL using mCBP KIX (PDB id: 1KDX) as a template and the predicted model was validated by RAMPAGE (http://mordred.bioc.cam.ac.uk/∼rapper/rampage.php) and ERRAT (http://services.mbi.ucla.edu/ERRAT/) servers.53 PDB structure of mCBP KIX bound to cMyb (PDB id: 1SB0) was retrieved from Protein Data Bank.54 PyMOL (https://pymol.org/2/) was used to align protein structures and replacing mCBP KIX with AtMed15a KIX in PDB structure of mCBP KIX bound to cMyb. Homology model of MYB63 was constructed by Phyre2 server and the predicted model was validated by RAMEPAGE and ERRAT servers.55 Protein binding region was predicted by ANCHOR.56 9aaTADs were analyzed by 9aaTAD Prediction Tool (http://www.med.muni.cz/9aaTAD/). PatchDock and FireDock were used for protein-protein docking studies between AtMed15a KIX and MYB63 helix (D176 to D203).57,58 Residues involved in protein-protein interaction were retrieved from PDBsum.59
Mapping of AtMed15a KIX domain: AtMed15a KIX-derived fragments derivatives were cloned in pGBKT7 vector using suitable primers listed in Table S5. The recombinant plasmids were transformed into yeast Y2HGold strain (Clontech). These BD derivative clones were tested for their interaction with 1 to 20 of the Y2H clones listed in Table S2 and AD vector on SD Trp−/Leu−/His−/Ade− agar plates. Spotting was done with 2 µl of culture at 0.2 OD.
Supplementary Material
Funding Statement
This work was supported by the Department of Science and Technology, INDIA, (EMR/2015/001336).
Abbreviations
- AD
activation domain
- AtMed
Arabidopsis thaliana Mediator
- BiFC
bimolecular fluorescence complementation
- Med
Mediator
- TF
Transcription factor
- Y2H
yeast two-hybrid
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the grant (EMR/2015/001336) funded by Science and Engineering Board, Department of Science and Technology, Government of India. We wish to thank UGC for granting Senior Research Fellowships to VK, SM and AK; CSIR for granting Senior Research Fellowship to ND, and NIPGR for granting Senior Research Fellowship to MW under short-term research fellowship program.
References
- 1.Kelleher RJ III, Flanagan PM, Kornberg RD. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell. 1990;61:1209–15. doi: 10.1016/0092-8674(90)90685-8. [DOI] [PubMed] [Google Scholar]
- 2.Myers LC, Kornberg RD. Mediator of transcriptional regulation. Annu Rev Biochem. 2000;69:729–49. doi: 10.1146/annurev.biochem.69.1.729. [DOI] [PubMed] [Google Scholar]
- 3.Bourbon H-M. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional Mediator complex. Nucleic Acids Res. 2008;36:3993–4008. doi: 10.1093/nar/gkn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samanta S, Thakur JK. Importance of Mediator complex in the regulation and integration of diverse signaling pathways in plants. Front Plant Sci. 2015;6:757. doi: 10.3389/fpls.2015.00757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malik N, Agarwal P, Tyagi A. Emerging functions of multi-protein complex Mediator with special emphasis on plants. Crit Rev Biochem Mol Biol. 2017;52:475–502. doi: 10.1080/10409238.2017.1325830. [DOI] [PubMed] [Google Scholar]
- 6.Chadick JZ, Asturias FJ. Structure of eukaryotic Mediator complexes. Trends Biochem Sci. 2005;30:264–71. doi: 10.1016/j.tibs.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Nagulapalli M, Maji S, Dwivedi N, Dahiya P, Thakur JK. Evolution of disorder in Mediator complex and its functional relevance. Nucleic Acids Res. 2016;44:1591–612. doi: 10.1093/nar/gkv1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai G, Imasaki T, Yamada K, Cardelli F, Takagi Y, Asturias FJ. Mediator head module structure and functional interactions. Nat Struct Mol Biol. 2010;17:273. doi: 10.1038/nsmb.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tóth-Petróczy Á, Oldfield CJ, Simon I, Takagi Y, Dunker AK, Uversky VN, Fuxreiter M. Malleable machines in transcription regulation: The Mediator complex. PLoS Comput Biol. 2008;4:e1000243. doi: 10.1371/journal.pcbi.1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soutourina J, Wydau S, Ambroise Y, Boschiero C, Werner M. Direct interaction of RNA polymerase II and Mediator required for transcription in vivo. Science. 2011;331:1451. doi: 10.1126/science.1200188. [DOI] [PubMed] [Google Scholar]
- 11.Linder T, Gustafsson CM. The Soh1/MED31 protein is an ancient component of Schizosaccharomyces pombe and Saccharomyces cerevisiae Mediator. J Biol Chem. 2004;279:49455–9. doi: 10.1074/jbc.M409046200. [DOI] [PubMed] [Google Scholar]
- 12.Baumli S, Hoeppner S, Cramer P. A conserved Mediator hinge revealed in the structure of the MED7·MED21 (Med7·Srb7) heterodimer. J Biol Chem. 2005;280:18171–8. doi: 10.1074/jbc.M413466200. [DOI] [PubMed] [Google Scholar]
- 13.Takagi Y, Calero G, Komori H, Brown JA, Ehrensberger AH, Hudmon A, Asturias F, Kornberg RD. Head module control of Mediator interactions. Mol Cell. 2006;23:355–64. doi: 10.1016/j.molcel.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Kang JS, Kim SH, Hwang MS, Han SJ, Lee YC, Kim Y-J. The structural and functional organization of the yeast Mediator complex. J Biol Chem. 2001;276(45):42003–10. doi: 10.1074/jbc.M105961200. [DOI] [PubMed] [Google Scholar]
- 15.Elmlund H, Baraznenok V, Lindahl M, Samuelsen CO, Koeck PJB, Holmberg S, Hebert H, Gustafsson CM. The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc Natl Acad Sci USA. 2006;103:15788–93. doi: 10.1073/pnas.0607483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Myers LC, Gustafsson CM, Hayashibara KC, Brown PO, Kornberg RD. Mediator protein mutations that selectively abolish activated transcription. Proc Natl Acad Sci USA. 1999;96:67–72. doi: 10.1073/pnas.96.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11:761. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poss ZC, Ebmeier CC, Taatjes DJ. The Mediator complex and transcription regulation. Crit Rev Biochem Mol Biol. 2013;48:575–608. doi: 10.3109/10409238.2013.840259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen BL, Taatjes DJ. The Mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol. 2015;16:155–66. doi: 10.1038/nrm3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeronimo C, Robert F. The Mediator complex: At the nexus of RNA polymerase II transcription. Trends Cell Biol. 2017;27:765–83. doi: 10.1016/j.tcb.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 21.Nogi Y, Fukasawa T. A novel mutation that affects utilization of galactose in Saccharomyces cerevisiae. Curr Genet. 1980;2:115–20. doi: 10.1007/BF00420623. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki Y, Nogi Y, Abe A, Fukasawa T. GAL11 protein, an auxiliary transcription activator for genes encoding galactose-metabolizing enzymes in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4991–9. doi: 10.1128/MCB.8.11.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thakur JK, Arthanari H, Yang F, Chau KH, Wagner G, Näär AM. Mediator subunit Gal11p/MED15 is required for fatty acid-dependent gene activation by yeast transcription factor Oaf1p. J Biol Chem. 2009;284:4422–8. doi: 10.1074/jbc.M808263200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang F, Vought BW, Satterlee JS, Walker AK, Jim Sun ZY, Watts JL, DeBeaumont R, Saito RM, Hyberts SG, Yang S. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature. 2006;442:700. doi: 10.1038/nature04942. [DOI] [PubMed] [Google Scholar]
- 25.Zhao X, Xiaoli ZH, Abdulla A, Yang EST, Wang Q, Ji JY, Pessin JE, Das BC, Yang F. Inhibition of SREBP transcriptional activity by a Boron-containing compound improves lipid homeostasis in diet-induced obesity. Diabetes. 2014;63:2464–73. doi: 10.2337/db13-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taubert S, Van Gilst MR, Hansen M, Yamamoto KR. A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev. 2006;20:1137–49. doi: 10.1101/gad.1395406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hou NS, Gutschmidt A, Choi DY, Pather K, Shi X, Watts JL, Hoppe T, Taubert S. Activation of the endoplasmic reticulum unfolded protein response by lipid disequilibrium without disturbed proteostasis in vivo. Proc Natl Acad Sci USA. 2014;111:E2271–E80. doi: 10.1073/pnas.1318262111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim MJ, Jang I-C, Chua N-H. The Mediator complex MED15 subunit mediates activation of downstream lipid-related genes by the WRINKLED1 transcription factor. Plant Physiol. 2016;171:1951–64. doi: 10.1104/pp.16.00664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thakur JK, Arthanari H, Yang F, Pan S-J, Fan X, Breger J, Frueh DP, Gulshan K, Li DK, Mylonakis E. A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature. 2008;452:604. doi: 10.1038/nature06836. [DOI] [PubMed] [Google Scholar]
- 30.Monk BC, Goffeau A. Outwitting multidrug resistance to antifungals. Science. 2008;321:367. doi: 10.1126/science.1159746. [DOI] [PubMed] [Google Scholar]
- 31.Taubert S, Hansen M, Van Gilst MR, Cooper SB, Yamamoto KR. The Mediator subunit MDT-15 confers metabolic adaptation to ingested Mmaterial. PLoS Genet. 2008;4:e1000021. doi: 10.1371/journal.pgen.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Canet JV, Dobón A, Tornero P. Non-recognition-of-BTH4, an Arabidopsis Mediator subunit homolog, is necessary for development and response to salicylic acid. Plant Cell. 2012;24:4220. doi: 10.1105/tpc.112.103028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato Y, Habas R, Katsuyama Y, Näär AM, He X. A component of the ARC/Mediator complex required for TGFβ/nodal signalling. Nature. 2002;418:641. doi: 10.1038/nature00969. [DOI] [PubMed] [Google Scholar]
- 34.Pasrija R, Thakur JK. Analysis of differential expression of Mediator subunit genes in Arabidopsis. Plant Signal Behav. 2012;7:1676–86. doi: 10.4161/psb.22438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pasrija R, Thakur JK. Tissue specific expression profile of Mediator genes in Arabidopsis. Plant Signal Behav. 2013;8:e23983. doi: 10.4161/psb.23983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thakur JK, Agarwal P, Parida S, Bajaj D, Pasrija R. Sequence and expression analyses of KIX domain proteins suggest their importance in seed development and determination of seed size in rice, and genome stability in Arabidopsis. Mol Genet Genomics. 2013;288:329–46. doi: 10.1007/s00438-013-0753-9. [DOI] [PubMed] [Google Scholar]
- 37.Novatchkova M, Eisenhaber F. Linking transcriptional mediators via the GACKIX domain super family. Curr Biol. 2004;14:R54–R5. doi: 10.1016/j.cub.2003.12.042. [DOI] [PubMed] [Google Scholar]
- 38.Radhakrishnan I, Pérez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Solution structure of the KIX Domain of CBP bound to the transactivation domain of CREB: a model for Activator:Coactivator interactions. Cell. 1997;91:741–52. doi: 10.1016/S0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 39.Wei Y, Horng J-C, Vendel AC, Raleigh DP, Lumb KJ. Contribution to stability and folding of a buried polar residue at the CARM1 methylation site of the KIX domain of CBP. Biochemistry. 2003;42:7044–9. doi: 10.1021/bi0343976. [DOI] [PubMed] [Google Scholar]
- 40.Fan X, Chou DM, Struhl K. Activator-specific recruitment of Mediator in vivo. Nat Struct Mol Biol. 2006;13:117. doi: 10.1038/nsmb1049. [DOI] [PubMed] [Google Scholar]
- 41.Fan X, Struhl K. Where does Mediator bind in vivo?. PLoS One. 2009;4:e5029. doi: 10.1371/journal.pone.0005029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thakur JK, Yadav A, Yadav G. Molecular recognition by the KIX domain and its role in gene regulation. Nucleic Acids Res. 2014;42:2112–25. doi: 10.1093/nar/gkt1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jedidi I, Zhang F, Qiu H, Stahl SJ, Palmer I, Kaufman JD, Nadaud PS, Mukherjee S, Wingfield PT, Jaroniec CP. Activator Gcn4 employs multiple segments of Med15/Gal11, including the KIX Domain, to recruit mediator to target genes in vivo. J Biol Chem. 2010;285:2438–55. doi: 10.1074/jbc.M109.071589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang F, Kong W, Wong G, Fu L, Peng R, Li Z, Yao Q. AtMYB12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana. Mol Genet Genomics. 2016;291:1545–59. doi: 10.1007/s00438-016-1203-2. [DOI] [PubMed] [Google Scholar]
- 45.Chezem WR, Memon A, Li FS, Weng JK, Clay NK. SG2-Type R2R3-MYB Transcription Factor MYB15 Controls defense-induced lignification and basal immunity in Arabidopsis. Plant Cell. 2017;29:1907–26. doi: 10.1105/tpc.16.00954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou J, Lee C, Zhong R, Ye ZH. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell. 2009;21:248–66. doi: 10.1105/tpc.108.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palmer CM, Hindt MN, Schmidt H, Clemens S, Guerinot ML. MYB10 and MYB72 are required for growth under iron-limiting conditions. PLoS Genet. 2013;9:e1003953. doi: 10.1371/journal.pgen.1003953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang C, Yao X, Yu D, Liang G. Fe-deficiency-induced expression of bHLH104 enhances Fe-deficiency tolerance of Arabidopsis thaliana. Planta. 2017;246(3):421–31. doi: 10.1007/s00425-017-2703-y. [DOI] [PubMed] [Google Scholar]
- 49.Chen Y, Chen Z, Kang J, Kang D, Gu H, Qin G. AtMYB14 regulates cold tolerance in Arabidopsis. Plant Mol Biol Report. 2013;31:87–97. doi: 10.1007/s11105-012-0481-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohbayashi I, Lin CY, Shinohara N, Matsumura Y, Machida Y, Horiguchi G, Tsukaya H, Sugiyama M. Evidence for a role of ANAC082 as a Ribosomal stress response mediator leading to growth defects and developmental alterations in Arabidopsis. Plant Cell. 2017;29:2644–60. doi: 10.1105/tpc.17.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mauldin JP, Lu M, Das S, Park D, Ernst PB, Ravichandran KS. A link between the cytoplasmic engulfment protein Elmo1 and the Mediator complex subunit Med31. Curr Biol. 2013;23:162–7. doi: 10.1016/j.cub.2012.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tzfira T, Tian G-W, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V. pSAT Vectors: A modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol. 2005;57:503. doi: 10.1007/s11103-005-0340-5. [DOI] [PubMed] [Google Scholar]
- 53.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–5. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein data bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dosztányi Z, Mészáros B, Simon I. ANCHOR: Web server for predicting protein binding regions in disordered proteins. Bioinformatics. 2009;25:2745–6. doi: 10.1093/bioinformatics/btp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–W7. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andrusier N, Nussinov R, Wolfson HJ. FireDock: Fast interaction refinement in molecular docking. Proteins: Struct Funct Bioinf. 2007;69:139–59. doi: 10.1002/prot.21495. [DOI] [PubMed] [Google Scholar]
- 59.Laskowski RA. PDBsum: Summaries and analyses of PDB structures. Nucleic Acids Res. 2001;29:221–2. doi: 10.1093/nar/29.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


