Abstract
DNMT3A and 3B are the main de novo DNA methyltransferases (DNMTs) in the brain that introduce new methylation marks to non-methylated DNA in postmitotic neurons. DNA methylation is a key epigenetic mark that is known to regulate important cellular processes in neuronal development and brain plasticity. Accumulating evidence disclosed rapid and dynamic changes in DNA methylation of plasticity-relevant genes that are important for learning and memory formation. To understand how DNMTs contribute to brain function and how they are regulated by neuronal activity is a prerequisite for a deeper appreciation of activity-dependent gene expression in health and disease. This review discusses the functional role of de novo methyltransferases and in particular DNMT3A1 in the adult brain with special emphasis on synaptic plasticity, memory formation, and brain disorders.
Keywords: neuroepigenetics, cytosine methylation, DNMT, learning, memory, neuropsychiatric disorders, DNA methylation
Introduction
A rapidly increasing number of neuroepigenetic studies have been published in the past decade. This work not only contributed to a better understanding of genetic imprinting, maintenance of genomic stability, and how gene-environment interactions can influence the onset of brain diseases, but it also revealed possible mechanisms of epigenetic transmission of behavioral traits as well as transgenerational inheritance, to name only a few major advances (Blake and Watson 2016; Graff and Mansuy 2009; Sweatt 2013; Tsankova and others 2007). One driving force of this research is the question of how epigenetic modifications relate to cognition and long-term memory formation. More than 30 years ago, Francis Crick speculated that memories might be encoded in alterations of the chromosomal DNA in the brain (Crick 1984). However, only in recent years was it possible to unravel the mechanisms by which epigenetic readers and writers might induce such changes. Epigenetic modifiers place or remove epigenetic marks mainly by modification of DNA and histones. The attachment of methyl groups to the 5′-carbon of cytosine is one of the chemical labels on the backbone of DNA. DNA methyltransferases (DNMTs) are the major epigenetic building blocks of 5-methyl-cytosine methylation in DNA (Denis and others 2011) (Fig. 1), which in turn controls binding of proteins to DNA and thereby gene expression. In this review, we primarily focus on the contribution of the de novo methyltransferases DNMT3A and -B and discuss how DNA methylation is regulated by these enzymes in postmitotic neurons in the context of memory formation, behavioral plasticity, and psychiatric disorders.
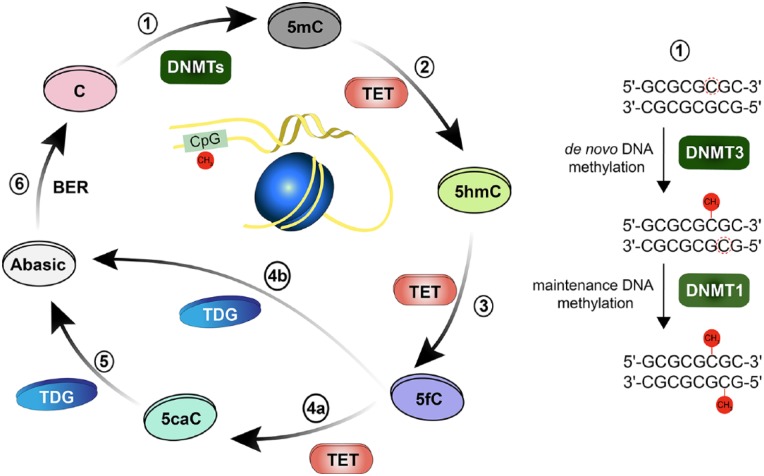
Figure 1.
Dynamic and reversible cytosine methylation. (1) De novo DNA methyltransferase enzyme DNMT3 transfer methyl group to 5′-carbon in cytosine (C), converting it to 5mC. Maintenance DNA methyltransferase DNMT1 methylates the hemimethylated DNA. (2) TET enzymes initiative successive oxidation reactions steps, initially by converting 5mC to 5-hydroxy-methylcytosine (5hmC). (3) TET enzyme converts 5hmC to 5′-formyl-cytosine (5fC). (4) TET enzyme further converts 5fC to 5′-carboxyl-cytosine (5caC). TDG either directly excises the glycosidic bond in 5fC (4b) or 5caC (5) generating an abasic site. (6) Base Excision Repair (BER) pathway includes the removal of the abasic site and replacement of the nucleotide.
Structure and Domain Organization of DNMTs
Two major DNMTs are found in mammals, DNMT1 and -3 (Jeltsch 2002). While sharing a similar domain organization and active catalytic domains in the C-terminus, the three DNMTs differ in their N-termini (Fig. 2). Of particular relevance here is the extended N-terminus of DNMT1, which is lacking in DNMT3, and which is important for the localization of the enzyme to the replication fork during DNA replication (Leonhardt and others 1992). DNMT1 largely serves as maintenance DNMT that methylates hemimethylated DNA by transferring a methyl group to the complementary DNA strand to accurately mimic the methylation pattern prior to DNA replication (Hermann and others 2004).
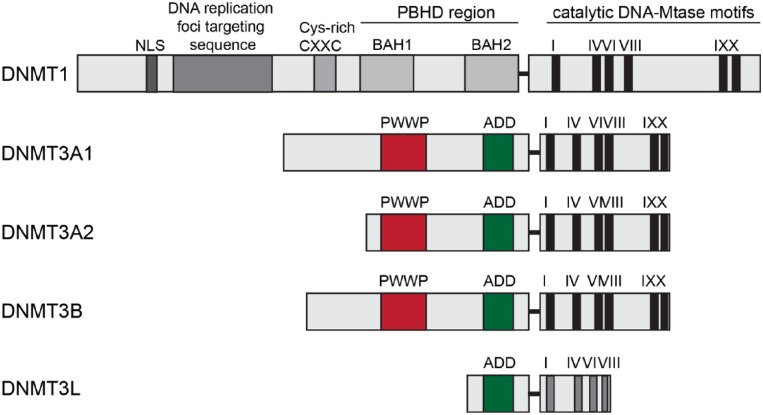
Figure 2.
Domain structure of mammalian DNMTs. The regulatory N-terminal part and conserved catalytic C-terminal part are represented. Importantly, the arrangement of different domains in DNMT1 is controlled by long linker regions, which form tight interactions with surface clefts of the domains. Both the linkers and the clefts are subject to many reported PTMs in DNMT1, including phosphorylation, acetylation, and ubiquitination. These PTMs might directly control the positioning of these domains in DNMT1. The ADD domain has been implicated in the allosteric control of DNMT3A, as it interacts with the catalytic domain of the methyltransferase and inhibits its activity, indicating that PHD-like domain-mediated interactions with other proteins could have direct regulatory impacts on the catalytic activity of the MTase. NLS = nuclear localization signal; CXXC = two cysteines separated by two other residues; BAH1/2 = tandem bromo-adjacent homology; PWWP = Pro-Trp-Trp-Pro; ADD = ATRX-DNMT3-DNMT3L.
DNMT3, in contrast, is a “de novo” DNA methyltransferase that introduces new methyl groups to previously non-methylated DNA. Two genes, Dnmt3a and Dnmt3b, give rise to different isoforms, which share high sequence and structural similarity (Fig. 2). The PWWP domain interacts with histone tails (Taverna and others 2007) and is present in all DNMT3 proteins. The PWWP domain is essential for methyltransferase (MTase) function since it specifically recognizes the repressive H3K36 trimethylation histone mark and targets DNMTs to DNA (Ge and others 2004). The structures of the PWWP domains from both DNMT3A and DNMT3B have been solved (Qiu and others 2002; Rondelet and others 2016). In addition, DNMT3A and DNMT3B directly interact with chromatin through their ADD domains that bind to H3 tails unmethylated at K4 (Ooi and others 2007; Zhang and others 2010). Two splice isoforms exist for DNMT3A, DNMT3A1, and DNMT3A2. DNMT3A2 lacks the N-terminal 219 amino acids of DNMT3A1 and is transcribed from an alternative promoter (Chen and others 2002a) (Fig. 2). Interestingly enough, DNMT3A2 is an immediate early gene whose expression in neurons can be induced by sustained synaptic activity (Oliveira and others 2012). Unfortunately, most published studies looking at mRNA expression have not distinguished both isoforms, and one can only tentatively assume that they investigated DNMT3A1. The protein transcribed from the third gene, Dnmt3l, lacks enzymatic activity since the catalytic domains are absent (Aapola and others 2000) (Fig. 2). It was postulated that although catalytically inactive, DNMT3L aids the DNA methylating function of other DNMTs. It recognizes, for instance, the unmethylated lysine 4 of histone H3 and triggers DNA methylation either by the recruitment or activation of DNMT3A2 (Ooi and others 2007; Rondelet and others 2016; Stepper and others 2016). So far no other function than DNA-methylation has been attributed to DNMT3A.
Posttranslational Modifications of DNMTs
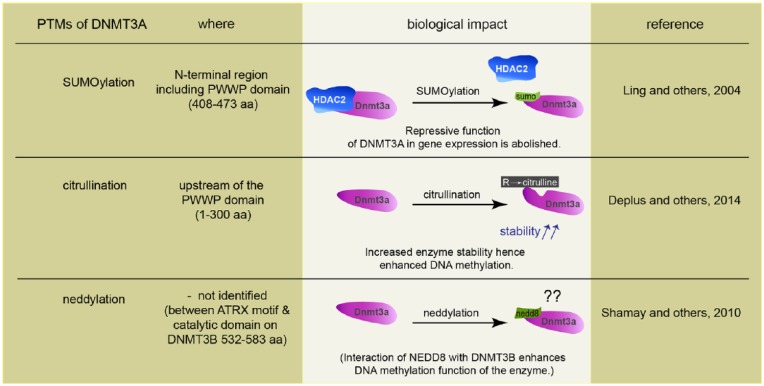
Several studies have shown how posttranslational modifications (PTMs) such as phosphorylation, acetylation, citrullination, SUMOylation, ubiquitination, neddylation, glycosylation, butyrylation, propionylation, and methylation affect the function and regulation of epigenetic modifiers and readers (Denis and others 2011). All known PTMs of DNMT1 such as acetylation, SUMOylation, methylation, and phosphorylation enhance interactions with other proteins or affect stability and catalytic properties (Denis and others 2011). DNMTs undergo complex conformational changes, capable of oligomerization and auto-inhibition, all of which might contribute to control the activity of these enzymes. Documented PTMs for de novo methyltransferases are SUMOylation, citrullination, and neddylation (see Table 1) and in particular DNMT3s are known to undergo prominent SUMOylation (Denis and others 2011). SUMOylation of DNMT3A disrupts the binding to histone deacetylase1/2 (HDAC1/2), and this abolishes the repressive function of DNMT3A in gene expression (Ling and others 2004). In addition, a direct interaction between the N-terminal PHD domain of DNMT3A and histone deacetylase (HDAC) 1 has been reported (Fuks and others 2001) and the majority of the repressive activity of the N-terminal DNMT3A can be relieved by treatment of cells with the HDAC inhibitor trichostatin A (TSA) (Fuks and others 2001). A recent study demonstrated an interaction of DNMT3A with PADI4, a Ca2+-dependent enzyme catalyzing the conversion of arginine residues to citrulline, and the authors identified a region upstream of the PWWP domain of DNMT3A as the primary site of citrullination (Deplus and others 2014). DNMT3A citrullination stabilizes the enzyme, probably by reducing its susceptibility to degradation and hence enhances DNA methylation (Deplus and others 2014).
Table 1.
|
The consequences of phosphorylation for DNMT3 activity has not been studied in any detail, although more than 24 phosphorylation sites have been identified in both enzymes in phosphoproteomic studies (Fig. 3; http://www.phosphosite.org). CK2 phosphorylates DNMT3A at two sites, S386 and S389, located next to the PWWP domain, and that CK2-mediated phosphorylation increases the heterochromatic targeting of DNMT3A and reduces its DNA methylation activity (Deplus and others 2014). These data further support the view that the combined regulation of enzymatic activity and localization is a common principle in the regulation of DNMT3A (Fig. 3).
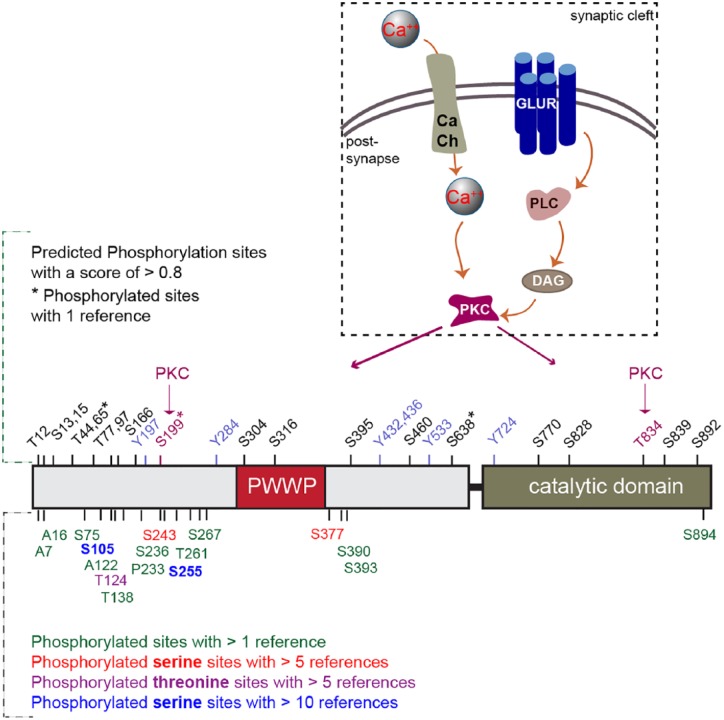
Figure 3.
Phosphorylation sites in DNMT3A (see www.phosphosite.org). In the upper panel, the predicted phosphorylation sites with a score of >0.8 are displayed. Two sites, Serine 199 and Threonine 834, are predicted to be phosphorylated by Protein Kinase C (PKC), which is activated by a series of molecular cascades upon Ca++ influx into neurons. In the lower panel, other phosphorylation sites are displayed that were identified in phospho-proteomics studies with the number of references, which reported this phosphosite.
Covalent attachment of the small ubiquitin-like neural precursor cell-expressed, developmentally downregulated 8 (NEDD8) protein leads to conformational changes that may prevent other PTMs to occur. In addition, neddylation might interfere or compete with other interaction partners or, alternatively might give rise to a new binding surface for new interaction partners (Rabut and Peter 2008). It was found that NEDD8 is directly conjugated to DNMT3A and -B and that both methylases also associate with other neddylated proteins. Moreover, it was shown that DNMT3B-dependent DNA methylation was enhanced by the interaction with NEDD8 (Shamay and others 2010). Collectively, the data point to several levels of potential regulation of DNMT function by PTMs but their relevance for neuronal gene expression has not been proven yet.
Expression of DNMT3 Family Members in the Adult Brain
Early work already showed that DNMT3 family members are crucially involved in embryonic development when these proteins are very abundant. A constitutive Dnmt3a and Dnmt3b gene knockout in mice results in embryonic or early postnatal death (Okano and others 1999). Interestingly, the two major de novo methylating enzymes, DNMT3A and -3B, show a differential age-dependent expression in brain. DNMT3B exhibits higher expression levels prenatally in the transiently amplifying neural progenitor cells, whereas DNMT3A protein levels increase postnatally and DNMT3A might replace DNMT3B (Feng and others 2005; Watanabe and others 2006). Over the lifespan the abundance of DNMT3A declines following synaptogenesis in neurons with very low levels during aging (Feng and others 2005; Goto and others 1994). Since a global DNA hypomethylation is a hallmark of neuronal aging (Jung and Pfeifer 2015), it is tempting to speculate that this decline is involved in senescence-related DNA-hypomethylation. Moreover, it is likely that this downregulation contributes to cognitive decline in aging (Oliveira and others 2012). Of note in this regard, for instance, the finding that the diminished inducible expression of DNMT3A2 in the old brain results in memory impairment, and this impairment can be rescued by viral reexpression of the protein (Oliveira and others 2012). DNMT3A2 overexpression even enhances fear memory formation and results in increased expression of synaptic plasticity-related genes (Oliveira and others 2016). Interestingly, some reports indicate that DNMT3A1 is found preferentially with heterochromatin whereas DNMT3A2 associates with euchromatin (Chen and others 2002a).
Relatively few studies have addressed the regional and cellular distribution of DNMTs in the brain. Collectively these studies point to a rather complex picture. DNMT3B, for instance, appears to be present though to a lesser extent compared to DNMT3A in medial prefrontal cortex (mPFC) (Elliott and others 2016), while expression levels are comparable in the subfields of the hippocampus (Mitchnick and others 2015). Surprisingly, in almost all published studies DNMT antibody staining was reported to be stronger in neuronal as compared to glial cells (Kadriu and others 2012; Nguyen and others 2007). Moreover, immunohistochemical analysis showed that DNMT expression in dorsal root ganglia (DRG) is cell-type specific. DNMT3A expression was mainly detected in the satellite cells surrounding the DRG neurons, unlike DNMT1 expression, and also in Schwann cells, whereas DNMT3B expression was mainly in DRG neurons (Pollema-Mays and others 2014). Of note, very strong immunoreactivity for DNMT1 and -3A has been reported in GAD67-positive interneurons (Kadriu and others 2012). Unfortunately, this picture is still incomplete, and this greatly hampers research on the exact role of DNMTs in the regulation of gene expression in the adult brain. Studies using well-characterized antibodies are prerequisites to provide the basis for addressing several important questions that are outlined below, and they are ultimately warranted.
The Neuronal Phenotype of Dnmt Mouse Mutants
DNMTs are broadly involved in embryo development and the targeted mutation of Dnmt1 or Dnmt3a and -3b results in early embryonic lethality (Li and others 1992; Okano and others 1999). Transgenic Dnmt mice with conditional alleles for gene deletion have provided spatial and temporal control of DNMT expression. The forebrain-specific knockout of either the Dnmt3a or Dnmt1 gene does not result in any impairment in spatial learning and memory in the Morris water maze and contextual fear conditioning (Feng and others 2010). However, the double knockout of both genes induces impaired spatial learning and memory in both tests and an attenuated slope of field excitatory postsynaptic potential in the late phase of Schaffer collateral long-term potentiation (LTP) (Feng and others 2010). This finding indirectly suggests that both enzymes might have redundant roles in learning and memory formation and that both DNMTs might be able to substitute for each other. On the contrary, a more recent study indicated that maintenance and de novo methyltransferases cannot replace for each other (Morris and Monteggia 2014). In this latter report, the authors revealed spatial and novel object recognition as well as conditioned taste aversion learning impairments in forebrain-specific conditional single Dnmt3a but not Dnmt1 knockout mice (Morris and Monteggia 2014). The Dnmt knockouts were generated using floxed Dnmt1/3a and a Cre deleter line whose expression was under the control of CamKIIα promoter (Morris and others 2016). The reasons for these discrepant reports are not clear, but it is possible that the use of different learning tasks and differences in the sex and age of subjects might have contributed to these findings. A recently published study showed that forebrain-specific deletion of DNMT1 reduces levels of anxiety, whereas the corresponding DNMT3A knockout did not have a comparable effect (Morris and others 2016). Thus, at present, it appears to be plausible that the different DNMTs contribute in a cell and circuit-specific manner to behavioral traits, learning, and memory.
The Pharmacology of DNMT Inhibitors
Prior to the generation of genetic mutant mouse models, the investigation of DNMT function in the brain depended almost exclusively on the functional block of these enzymes using pharmacological inhibitors. 5-Aza-2′-deoxycytadine (5-azadC), zebularine, and RG108 are commonly used DNMT inhibitors (Creusot and others 1982). 5-azadC and zebularine are nucleoside inhibitors that following their incorporation into DNA trap DNMTs by covalent binding of the enzyme to DNA and thereby blocking methyltransferase function irreversibly at other sites (Creusot and others 1982). It is, however, an unresolved question how cytidine analogues incorporate into the DNA of postmitotic neurons. It is rather likely that in postmitotic neurons they might have off-target effects not related to demethylation of DNA. In contrast, the Curcumin-derivative RG108 was shown to bind the active site of DNMT1 (Schirrmacher and others 2006) and inhibits the enzyme by blocking the catalytic domain without the need for incorporation into DNA. Of note, although the catalytic domains of DNMT3 and DNMT1 are highly conserved, it is not yet clear if RG108 can also block DNMT3 (Brueckner and others 2005). Nonetheless, RG108 is much less cytotoxic than nucleoside inhibitors whose cytotoxic effects are likely to be related to protein-DNA adducts induced by these compounds.
Inhibition of DNMTs and the Study of Cognitive Function
Taking into account these caveats, pharmacological inhibition of DNMT enzyme function has nonetheless contributed to a deeper appreciation of their role in neuronal function. Based on studies employing these inhibitors it is nowadays widely believed that reversible and dynamic DNA-methylation mediated by DNMTs has a role in cognition and memory (see Fig. 4). The nature of this role, however, is still essentially unclear and a matter of debate.
Figure 4.
An overview of the various external stimuli that lead to alterations in DNA-methylation in the brain to regulate plasticity-relevant gene expression. These include constraint stress, associative fear conditioning, the day and night cycle, social interaction, as well as an enriched environment.
Contextual fear conditioning induces a form of associative memory in rats that leads to the induction of Bdnf (brain-derived neurotropic factor) IV expression following the demethylation of its promoter in the CA1 region of the hippocampus (Harris and others 2016; Lubin and others 2008). Infusion of zebularine and RG108 into CA1 attenuates Bdnf IV expression, and this is correlated with impaired contextual fear memory (Lubin and others 2008). Some studies suggest that methylation regulates gene expression differentially following fear conditioning. An example is the selective differential regulation of the memory suppressor protein phosphatase 1 (PP1) and the memory enhancer reelin (Reln) (Miller and Sweatt 2007). Following contextual fear conditioning, the promoter methylation of the Reln gene is decreased, and expression of the gene is increased. On the other hand, the promoter methylation of the memory suppressor PP1 is enhanced, hence, the expression of the gene is suppressed (Miller and Sweatt 2007). Of note, both proteins will have multiple cellular functions. It is unclear how this finding relates to increased Dnmt3a and Dnmt3b transcript expression in this paradigm. Nonetheless, several studies have shown that DNA methylation is dynamic in the adult brain, and it has been speculated that regulation of methylation can be gene specific and depends on the cellular context during the process of memory formation (Day and Sweatt 2010; Miller and Sweatt 2007).
The general theoretical framework underlying most studies is based on the idea that neuronal activity is translated into altered DNA methylation, which is a prerequisite for memory maintenance. Along these lines cortical infusion of DNMT inhibitors after the establishment of short-term memory impaired memory consolidation and formation of long-term memory (Miller and others 2010). Moreover, DNMT3a and DNMT3b transcript levels are increased in the dentate gyrus (DG) whereas DNMT1 mRNA expression is unregulated in the perirhinal cortex following novel object-location learning (Mitchnick and others 2015). Thus, blocking DNMT activity impairs long-term memory. Interestingly enough, infusion of RG108 into the dentate gyrus and perirhinal cortex is effective in this regard while application of 5-azadC has only an effect following injection into the DG and perirhinal cortex. This finding might reflect the different mechanism of action and specificity of DNMT inhibitors as discussed above.
At present very little is known about upstream regulators of DNMT activity. The steroid estradiol enhances memory consolidation, and it was shown that this effect requires DNMT activity (Zhao and others 2010). In line with this finding, estradiol infusion results in elevated DNMT3B expression in the dorsal hippocampus (Zhao and others 2010). Auditory fear conditioning also leads to increased DNMT3A mRNA expression and histoneH3 acetylation in the lateral amygdala (Monsey and others 2011). Local HDAC inhibition enhances memory whereas the DNMT inhibitor 5-azadC blocks memory consolidation in this paradigm. Furthermore, intra-amygdalar infusion of 5-azadC interferes with histone acetylation and attenuates the learning-related increases in H3 acetylation (Monsey and others 2011). Memory consolidation in the basolateral amygdala depends on DNMT activity as well. Immediate local infusion of 5-azadC disrupted the ability of a conditioned stimulus to reinstate cocaine-seeking behavior in a paradigm where a conditioned stimulus was paired with cocaine self-administration (Shi and others 2015). In a similar paradigm, where rats were administered with 5-azadC into the nucleus accumbens, cue-induced cocaine seeking behavior was significantly attenuated (Massart and others 2015).
Other studies also support the notion that DNMTs regulate associative reward learning, drug addiction, and behavior (Day and others 2013; LaPlant and others 2010). LaPlant and others investigated the role of DNA methylation in chronic cocaine administration and chronic social defeat stress in the nucleus accumbens. Cocaine administration leads to a biphasic regulation of DNMT3A mRNA expression, with an increase within 4 hours and a prominent decrease 24 hours later. Cocaine only temporally regulates DNMT3A mRNA levels since its withdrawal results in upregulation of DNMT3A mRNA expression in the nucleus accumbens. Of note, DNMT activity appears to regulate spine density since infusion of the DNMT-inhibitor RG108 abolished greatly the cocaine-induced reduction in spine density (Day and others 2013; LaPlant and others 2010). The dopaminergic neurons of the ventral tegmental area are important for reward learning in animals (Hyman and others 2006). Along these lines, it was shown that DNA methylation is required to establish associative award learning in this region particularly for the expression of key plasticity genes like Fos and Egr1. Alterations in methylation, either in proximity to transcription start sites but also in intragenic regions, appear to regulate the expression of such genes and this regulation is sensitive to the RG108 infusion (Day and others 2013).
DNMT3 in Emotion and Motivated Behavior
A recent study demonstrated a role for DNMT3A1 in anxiety-related behavior in mice (Elliott and others 2016). Dnmt3a1 mRNA levels were reduced in the mPFC of adult mice that were exposed to chronic social defeat stress and after that displayed increased anxiety. The aftermath effect of the social defeat stress was overcome by virally increasing DNMT3A1 expression in the mPFC (Elliott and others 2016). In selective areas of the brain, particularly those where adult neurogenesis occurs like in the subventricular zone (SVZ), olfactory bulb, the granular zone of the DG, and in the distinct white matter tracks DNMT3A type II cells depict higher expression of DNMT3A (Hammels and others 2015). Type II cells are identified in this study by intense DNMT3A immunoreactivity while co-localizing with proliferation and immaturity markers like doublecortin in the hippocampal DG (Chouliaras and others 2011). Hammels and others found that animals resilient to chronic social defeat stress exhibited an increased number of DNMT3A type II cells in the DG, a heterogeneous population of cells with 25% doublecortin-positive immature and 60% NeuN-positive cells and that this equates with their anxiolytic, hedonic behavior (Hammels and others 2015). However, NeuN is not a reliable marker of mature neurons since immature adult-born granule cells in the DG also express NeuN prior to their maturation (Kempermann and others 2004).
In support of these observations (but see also for contradictory results Sales and others 2011), intra-hippocampus injection of DNMT inhibitors induces an anxiolytic effect in association with increased BDNF expression (Xing and others 2014). In this study, it was also found that 5-azadC injection into the ventrolateral orbital cortex induced depressive-like behavior (Xing and others 2014). These findings are not surprising. Several studies in the past decade showed that epigenetic modifiers in the hypothalamic pituitary adrenal axis regulate responses to fear and stress exposure, regulate circadian rhythm, addiction, posttraumatic stress disorder (PTSD) (Azzi and others 2014; Barbier and others 2015; Murgatroyd and others 2009; Szyf 2013; Zovkic and Sweatt 2013). Collectively these data suggest that subtle changes in the methylation status of only very few crucial cytosines in CpG sites can dictate the response of the brain to early life stress particularly if these sites modulate binding of proteins to methylated DNA (Murgatroyd and others 2009). For instance, in response to early life stress promoter hypomethylation of the arginine vasopressin hormone gene in hypothalamic nuclei was observed and this, in turn, reduced the MeCP2 occupancy on the promoter and increased the expression of the gene (Murgatroyd and others 2009). This affected later in life the behavior of the mice and led to deficits in avoidance learning (Murgatroyd and others 2009). Prenatal stress is an important factor for the development of schizophrenia (SZ), later in life (Brixey and others 1993). Bdnf IV promoter methylation increases postnatally in the hippocampus and amygdala of rats, which were exposed to prenatal stress, and hence resulted in reduced Bdnf IV expression. The correlation between reduced Bdnf IV expression and the increase in DNMT3A levels in these brain regions prompted authors to speculate about a causal relationship (Boersma and others 2014). Finally, prenatally stressed mice depict not only similar behavioral traits like psychotic patients but also similar epigenetic signatures (Dong and others 2015). DNMT1 and Ten-eleven translocation (TET) enzyme 1 level increase in prenatally stressed mice correlates with enhanced 5mCs and 5hmCs in the regulatory DNA regions and therefore decreased Bdnf gene expression (Dong and others 2015).
Regulation of DNMT Function in the Brain
The emerging picture from this vast body of literature indirectly suggests an intimate link between synaptic events and nuclear DNMT activity (Fig. 4). However, surprisingly little is known on how DNMT activity feeds back to synaptic function and conversely how synaptic activity triggers signaling that then eventually controls DNMT function. It is therefore at present still essentially unclear how neuronal activity regulates dynamic and reversible DNA methylation and how this can be influenced by DNMTs. Although there is accumulating evidence that activation of excitatory synapses increase DNMT mRNA expression (see above and Fig. 5), increased DNMT mRNA expression does not make much sense as an instantaneous regulatory event to control gene expression and also the question arises how this relates to demethylation of promoters, which is essential for the transcription of plasticity-related genes. Unfortunately, very little is known about the control of protein expression and here in particular PTMs of DNMTs. This is astonishing since it is likely that PTMs of DNMTs induced by synaptic activity affects DNA methylation. In fact, many conflicting data render it currently difficult to judge and to identify the underlying principles of synapse-to-nucleus communication in this scenario.
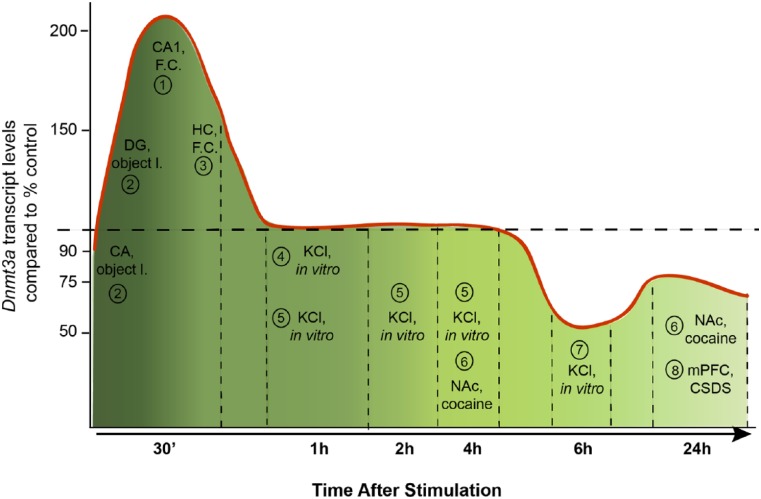
Figure 5.
A diagram showing the mRNA levels of Dnmt3a isoforms from various brain regions, from various types of stimuli compared to the control levels in each study that are indicated by a dashed line. The references to these studies are as follows: 1, Miller and Sweatt 2007; 2, Morris and Monteggia 2014; 3, Mitchnick and others 2015; 4, Day and others 2013; 5, Oliveira and others 2012; 6, LaPlant and others 2010; 7, Sharma and others 2008; 8, Elliott and others 2016.
Neuronal depolarization with KCl for 1 hour, for instance, did not alter DNMT3A mRNA levels in cortical neurons (Day and others 2013); however, depolarization for 6 hours reportedly led to a reduction in DNMT3A protein levels (Sharma and others 2008). In addition, recent advances in the whole epigenome analysis have challenged the early simplistic view that DNA methylation is associated with transcriptional silencing. It is now well established that methylated CpG-rich regions of DNA are not only found in transcription initiation sites but also in gene bodies and intergenic regions (Jones 2012). Moreover, its effect on transcription may vary since, for instance, gene body CpG methylation (Yang and others 2014) and DNMT3A-dependent nonproximal methylation (Wu and others 2010) is positively associated with transcription. Additionally, non-CpG (CpH, H = A/C/T) methylation is established during synaptogenesis, in which CpH methylation accumulates in neuron but not in glial cells (Lister and others 2013) and DNMT3A is required for its active maintenance in postmitotic neurons (Guo and others 2014). In support of this, Dnmt3 knockout mice displayed largely unaltered CG methylation; however, CpH methylation was absent in the brain (Gabel and others 2015). Roles attributed to non-CpG methylation, which does occur in neurons, may also vary. Gene expression profiling from the adult mouse DG revealed the methylated CpH containing regulatory regions were associated with lower nearby gene expression independently of the CpG contexts (Guo and others 2014). Alterations in histone acetylation and DNA methylation occur in parallel following contextual fear conditioning learning and changes in DNA methylation might also occur in non-neuronal cells potentially supporting the epigenetic code for memory formation (Halder and others 2016). Unfortunately, only very few studies have looked in parallel at mRNA and protein expression as well as DNMT activity. Given this complexity and the conundrum of at first glance conflicting reports, it is at present elusive how increased DNMT transcript levels relate to protein expression and how synaptic function controls DNMT activity and selective methylation of certain genes in the context of learning and memory. This is an important question since published reports suggest an intimate link. In cultured neurons inhibition of DNMT by the antagonist RG108 results in up-scaling of excitatory synaptic strength (Meadows and others 2015) and a combined shRNA protein knockdown of DNMT1 and DNMT3A also induced intrinsic membrane excitability changes that essentially required both ongoing, AMPARs- and NMDARs-dependent synaptic transmission (Meadows and others 2016).
DNMTs in Neurodegeneration and Neuropsychiatric Disorders
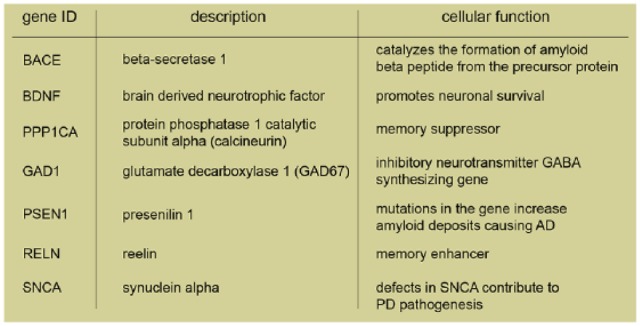
The shortcomings outlined above should also be resolved because DNMTs are apparently involved in a number of brain diseases. It is nowadays widely recognized that DNA methylation is implicated in neurodegenerative diseases as well as psychiatric disorders. Aberrant DNA methylation is, for instance, evident in SZ, bipolar disorder (BD), and major depression disorder (Mill and others 2008). In particular, SZ patients exhibit clearly altered DNA methylation patterns (Morris and Monteggia 2014) and increased DNMT1 expression (Ruzicka and others 2007). Moreover, a downregulation of genes associated with inhibitory neurons, such as glutamic acid decarboxylase67 (Gad67) and Reln, was measured in postmortem brain samples of SZ patients (Guidotti and others 2000). This downregulation was correlated with a hypermethylation of their CpG island promoter regions (Chen and others 2002b; Grayson and others 2005) and with an increase of DNMT1 (Veldic and others 2004) and DNMT3A (Zhubi and others 2009) mRNA expression in cortical GABAergic neurons of SZ patients. Interestingly, prenatal stress in mice that induces SZ-like behavior also results in high levels of DNMT1 and DNMT3A expression occurring in GABAergic neurons (Matrisciano and others 2013).
Bipolar disorder is a highly disabling and very frequent mood disorder (Gershon 2000). In SZ as well as BD patients DNMT1 expression and promoter methylation of the Reln and Gad67 gene are elevated (Grayson and Guidotti 2013). This might be relevant for disease progression since a suppression of the GABA synthesizing gene Gad1 might affect inhibitory neurotransmission (Grayson and Guidotti 2013; Murgatroyd and Spengler 2012). Additionally, DNMT1 and -3A expression were also found to be elevated in the brains of patients with epilepsy (Zhu and others 2012). The frontal cortex of patients with Alzheimer’s disease (AD) and BD exhibits enhanced DNA methylation, which is associated with hypermethylation of the BDNF gene promoter and a significant decrease in BDNF transcript levels (Kim and others 2010; Rao and others 2012). In a recent study, increased binding of DNMT1 to promoter regions of genes important for GABAergic (Reln and Gad67) or glutamatergic (BDNF IX) neurotransmission was shown in the cortex of patients with SZ and BD (Baets and others 2015). This increase in binding, however, did not correlate with enhanced promoter methylation (Baets and others 2015). Recently, a genetic study on single-nucleotide polymorphisms (SNPs) in DNMT3A revealed an association with a particular genotype in an SNP and greater cognitive decline in patients with mild cognitive impairment (Chouliaras and others 2015) (Box 1).
Box 1.
|
Hypomethylation is also observed in PD patients in the CpG-rich island of synuclein alpha (SNCA), the gene coding for α-synuclein (Jowaed and others 2010). α-Synuclein is a protein forming aberrant soluble oligomers that lead to neuronal cell death. The lower level of SNCA methylation was shown to result from a “sequestration” of DNMT1 in the cytosol in α-synuclein transgenic mice and postmortem PD brains (Desplats and others 2011). Alterations in the methylation profile of AD disease patients are discussed controversially as both hypo- and hypermethylation were reported (Coppieters and Dragunow 2011; Ellison and others 2017). Nuclear immunostaining intensity of DNMT1 and other components of the methylation machinery such as methyl-CpG binding domain protein 2/3 (MBD2/MBD3) were significantly reduced in neurons (Mastroeni and others 2010) where a loss of methylation of the amyloid precursor proteins was observed. A decrease in Bace and Psen1 (encoding for presenilin 1) methylation was also shown in AD patients and was associated with increased production of amyloid β-peptides (Mastroeni and others 2010; Scarpa and others 2003). Moreover, a pronounced global hypermethylation in the grey matter of postmortem human brain tissues was also observed in AD patients (Coppieters and others 2014). Finally, in older 5xFAD transgenic mice, which already exhibit the pathological and behavioral changes associated with AD progression, DNMT3A and -B but not DNMT1 expression, and DNA methylation was increased (Grinan-Ferre and others 2016).
Collectively the data show that the consequences of DNA methylation differ depending on the disease context. Controlling DNMT activity can in principal be proposed for the therapy of such conditions. Currently, however, the path to treatment is long since suitable small molecules to control DNMT activity specifically in disease context for a long-term treatment are not available and 5-azadC or zebularine, for instance, do not cross the blood-brain barrier.
Possible Role of DNMTs in Demethylation
The published studies so far suggest that DNMTs are a core part of the neuronal epigenetic machinery. What is less clear is which molecular mechanisms are in control of the DNMT function. Interestingly, some studies revealed other functions of DNMT3 aside from de novo methylation that might have implications for brain development, and that might also be relevant for brain plasticity. DNMT3 enzymes were shown to be part of demethylation processes through DNA repair mechanisms (Li and others 2007). Following methylcytosine deamination, a G/T mismatch arises, and in mammals, this results in a specific base-excision repair mechanism. Thymine DNA glycosylase (TDG) is one of the two enzymes that together with methyl-CpG binding domain protein 4 (MBD4) is known to initiate this repair mechanism (Hardeland and others 2001; Krokan and others 2002). TDG might also be responsible for the removal of formylcytosine and carboxylcytosine, which are the oxidation products of methylcytosine via hydroxymethylcytosine for active demethylation initiated by Tet enzymes (He and others 2011; Maiti and Drohat, 2011). DNMT3A interacts with TDG directly via either its PWWP or even the catalytic domain (Li and others 2007). This interaction then enhances TDG activity possibly by facilitating the binding of TDG to the mismatch sites, while binding to TDG at the same time represses the DNMT3A methylating activity (Li and others 2007). Similarly, DNMT3B isoform was shown to interact with TDG and MBD4, and it was demonstrated that this interaction analogous to DNMT3A supports the G/T mismatch repair mechanism (Boland and Christman 2008). Moreover, DNMT3A and 3B enzymes can function as DNA 5-hmC dehydroxymethylases (Chen and others 2012). Evidence was provided that they could directly catalyze the conversion of 5-hmC to cytosine and control re-methylation of the cytosine (Chen and others 2012). The only supporting evidence for a demethylase function in neurons came from a very recent study, which demonstrated that a reduced supply of S-Adenosyl methionine (SAM) converts DNMT3A to a demethylase in the dentate gyrus and this switching of function impacts the gene expression and behavior (Saunderson and others 2016). The demethylase activity of DNMT3 is probably highly regulated and in concert with its de novo methyltransferase activity, a versatile means to control neuronal DNA-methylation.
Conclusions
The regulation of DNMT3A function could be an important factor by which synaptic activity regulates gene expression. The emerging picture suggests complex regulatory mechanisms at different levels. Given the suspected importance of aberrant DNA-methylation in several brain disorders, it will be imperative to learn more about the molecular underpinnings of these mechanisms.
Acknowledgments
We would like to thank Dr. Anna Karpova for the kind support on the graphics.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Supported by grants from the Bundesministerium für Bildung und Forschung “Energi” FKZ: 01GQ1421B; Deutsche Forschungsgemeinschaft (DFG Kr1879/5-1, 6-1; SFB779 TPB8); the EU Joint Programme—Neurodegenerative Disease Research (JPND) project STAD; WGL Pakt f. Forschung and People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme FP7/2007-2013/ under REA grant agreement no. 289581.
References
- Aapola U, Kawasaki K, Scott HS, Ollila J, Vihinen M, Heino M, and others. 2000. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics 65:293–8. [DOI] [PubMed] [Google Scholar]
- Azzi A, Dallmann R, Casserly A, Rehrauer H, Patrignani A, Maier B, and others. 2014. Circadian behavior is light-reprogrammed by plastic DNA methylation. Nat Neurosci 17:377–82. [DOI] [PubMed] [Google Scholar]
- Baets J, Duan X, Wu Y, Smith G, Seeley WW, Mademan I, and others. 2015. Defects of mutant DNMT1 are linked to a spectrum of neurological disorders. Brain 138:845–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier E, Tapocik JD, Juergens N, Pitcairn C, Borich A, Schank JR, and others. 2015. DNA methylation in the medial prefrontal cortex regulates alcohol-induced behavior and plasticity. J Neurosci 35:6153–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake GE, Watson ED. 2016. Unravelling the complex mechanisms of transgenerational epigenetic inheritance. Curr Opin Chem Biol 33:101–7. [DOI] [PubMed] [Google Scholar]
- Boersma GJ, Lee RS, Cordner ZA, Ewald ER, Purcell RH, Moghadam AA, and others. 2014. Prenatal stress decreases Bdnf expression and increases methylation of Bdnf exon IV in rats. Epigenetics 9:437–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland MJ, Christman JK. 2008. Characterization of Dnmt3b:thymine-DNA glycosylase interaction and stimulation of thymine glycosylase-mediated repair by DNA methyltransferase(s) and RNA. J Mol Biol 379:492–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brixey SN, Gallagher BJ, 3rd, McFalls JA, Jr, Parmelee LF. 1993. Gestational and neonatal factors in the etiology of schizophrenia. J Clin Psychol 49:447–56. [DOI] [PubMed] [Google Scholar]
- Brueckner B, Garcia Boy R, Siedlecki P, Musch T, Kliem HC, Zielenkiewicz P, and others. 2005. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res 65:6305–11. [DOI] [PubMed] [Google Scholar]
- Chen CC, Wang KY, Shen CK. 2012. The mammalian de novo DNA methyltransferases DNMT3A and DNMT3B are also DNA 5-hydroxymethylcytosine dehydroxymethylases. J Biol Chem 287:33116–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Ueda Y, Xie S, Li E. 2002. a. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J Biol Chem 277:38746–54. [DOI] [PubMed] [Google Scholar]
- Chen Y, Sharma RP, Costa RH, Costa E, Grayson DR. 2002. b. On the epigenetic regulation of the human reelin promoter. Nucleic Acids Res 30:2930–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouliaras L, Kenis G, Visser PJ, Scheltens P, Tsolaki M, Jones RW, and others. 2015. DNMT3A moderates cognitive decline in subjects with mild cognitive impairment: replicated evidence from two mild cognitive impairment cohorts. Epigenomics 7:533–7. [DOI] [PubMed] [Google Scholar]
- Chouliaras L, van den Hove DL, Kenis G, Dela Cruz J, Lemmens MA, van Os J, and others. 2011. Caloric restriction attenuates age-related changes of DNA methyltransferase 3a in mouse hippocampus. Brain Behav Immun 25:616–23. [DOI] [PubMed] [Google Scholar]
- Coppieters N, Dieriks BV, Lill C, Faull RL, Curtis MA, Dragunow M. 2014. Global changes in DNA methylation and hydroxymethylation in Alzheimer’s disease human brain. Neurobiol Aging 35:1334–44. [DOI] [PubMed] [Google Scholar]
- Coppieters N, Dragunow M. 2011. Epigenetics in Alzheimer’s disease: a focus on DNA modifications. Curr Pharm Des 17:3398–412. [DOI] [PubMed] [Google Scholar]
- Creusot F, Acs G, Christman JK. 1982. Inhibition of DNA methyltransferase and induction of Friend erythroleukemia cell differentiation by 5-azacytidine and 5-aza-2′-deoxycytidine. J Biol Chem 257:2041–8. [PubMed] [Google Scholar]
- Crick F. 1984. Memory and molecular turnover. Nature 312:101. [DOI] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. 2010. DNA methylation and memory formation. Nat Neurosci 13:1319–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Childs D, Guzman-Karlsson MC, Kibe M, Moulden J, Song E, and others. 2013. DNA methylation regulates associative reward learning. Nat Neurosci 16:1445–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis H, Ndlovu MN, Fuks F. 2011. Regulation of mammalian DNA methyltransferases: a route to new mechanisms. EMBO Rep 12:647–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplus R, Denis H, Putmans P, Calonne E, Fourrez M, Yamamoto K, and others. 2014. Citrullination of DNMT3A by PADI4 regulates its stability and controls DNA methylation. Nucleic Acids Res 42:8285–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplats P, Spencer B, Coffee E, Patel P, Michael S, Patrick C, and others. 2011. Alpha-synuclein sequesters Dnmt1 from the nucleus: a novel mechanism for epigenetic alterations in Lewy body diseases. J Biol Chem 286:9031–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E, Ruzicka WB, Grayson DR, Guidotti A. 2015. DNA-methyltransferase1 (DNMT1) binding to CpG rich GABAergic and BDNF promoters is increased in the brain of schizophrenia and bipolar disorder patients. Schizophr Res 167:35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott E, Manashirov S, Zwang R, Gil S, Tsoory M, Shemesh Y, and others. 2016. Dnmt3a in the medial prefrontal cortex regulates anxiety-like behavior in adult mice. J Neurosci 36:730–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison EM, Abner EL, Lovell MA. 2017. Multiregional analysis of global 5-methylcytosine and 5-hydroxymethylcytosine throughout the progression of Alzheimer’s disease. J Neurochem 140:383–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Chang H, Li E, Fan G. 2005. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res 79:734–46. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, and others. 2010. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci 13:423–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T. 2001. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J 20:2536–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabel HW, Kinde B, Stroud H, Gilbert CS, Harmin DA, Kastan NR, and others. 2015. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature 522:89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge YZ, Pu MT, Gowher H, Wu HP, Ding JP, Jeltsch A, and others. 2004. Chromatin targeting of de novo DNA methyltransferases by the PWWP domain. J Biol Chem 279:25447–54. [DOI] [PubMed] [Google Scholar]
- Gershon ES. 2000. Bipolar illness and schizophrenia as oligogenic diseases: implications for the future. Biol Psychiatry 47:240–4. [DOI] [PubMed] [Google Scholar]
- Goto K, Numata M, Komura JI, Ono T, Bestor TH, Kondo H. 1994. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation 56:39–44. [DOI] [PubMed] [Google Scholar]
- Graff J, Mansuy IM. 2009. Epigenetic dysregulation in cognitive disorders. Eur J Neurosci 30:1–8. [DOI] [PubMed] [Google Scholar]
- Grayson DR, Guidotti A. 2013. The dynamics of DNA methylation in schizophrenia and related psychiatric disorders. Neuropsychopharmacology 38:138–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson DR, Jia X, Chen Y, Sharma RP, Mitchell CP, Guidotti A, and others. 2005. Reelin promoter hypermethylation in schizophrenia. Proc Natl Acad Sci U S A 102:9341–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinan-Ferre C, Sarroca S, Ivanova A, Puigoriol-Illamola D, Aguado F, Camins A, and others. 2016. Epigenetic mechanisms underlying cognitive impairment and Alzheimer disease hallmarks in 5XFAD mice. Aging (Albany NY) 8:664–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Davis JM, Di-Giorgi-Gerevini V, Dwivedi Y, Grayson DR, and others. 2000. Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study. Arch Gen Psychiatry 57:1061–9. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Shin JH, Shin J, Li H, Xie B, and others. 2014. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci 17:215–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder R, Hennion M, Vidal RO, Shomroni O, Rahman RU, Rajput A, and others. 2016. DNA methylation changes in plasticity genes accompany the formation and maintenance of memory. Nat Neurosci 19:102–10. [DOI] [PubMed] [Google Scholar]
- Hammels C, Prickaerts J, Kenis G, Vanmierlo T, Fischer M, Steinbusch HW, and others. 2015. Differential susceptibility to chronic social defeat stress relates to the number of Dnmt3a-immunoreactive neurons in the hippocampal dentate gyrus. Psychoneuroendocrinology 51:547–56. [DOI] [PubMed] [Google Scholar]
- Hardeland U, Bentele M, Lettieri T, Steinacher R, Jiricny J, Schar P. 2001. Thymine DNA glycosylase. Prog Nucleic Acid Res Mol Biol 68:235–53. [DOI] [PubMed] [Google Scholar]
- Harris AP, Lennen RJ, Brydges NM, Jansen MA, Pernet CR, Whalley HC, and others. 2016. The role of brain-derived neurotrophic factor in learned fear processing: an awake rat fMRI study. Genes Brain Behav 15:221–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, and others. 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333:1303–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann A, Goyal R, Jeltsch A. 2004. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J Biol Chem 279:48350–9. [DOI] [PubMed] [Google Scholar]
- Hyman SE, Malenka RC, Nestler EJ. 2006. Neural mechanisms of addiction: the role of reward-related learning and memory. Annu Rev Neurosci 29:565–98. [DOI] [PubMed] [Google Scholar]
- Jeltsch A. 2002. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem 3:274–93. [DOI] [PubMed] [Google Scholar]
- Jones PA. 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13:484–92. [DOI] [PubMed] [Google Scholar]
- Jowaed A, Schmitt I, Kaut O, Wullner U. 2010. Methylation regulates alpha-synuclein expression and is decreased in Parkinson’s disease patients’ brains. J Neurosci 30:6355–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M, Pfeifer GP. 2015. Aging and DNA methylation. BMC Biol 13:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadriu B, Guidotti A, Chen Y, Grayson DR. 2012. DNA methyltransferases1 (DNMT1) and 3a (DNMT3a) colocalize with GAD67-positive neurons in the GAD67-GFP mouse brain. J Comp Neurol 520:1951–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G, Jessberger S, Steiner B, Kronenberg G. 2004. Milestones of neuronal development in the adult hippocampus. Trends Neurosci 27:447–52. [DOI] [PubMed] [Google Scholar]
- Kim HW, Rapoport SI, Rao JS. 2010. Altered expression of apoptotic factors and synaptic markers in postmortem brain from bipolar disorder patients. Neurobiol Dis 37:596–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokan HE, Drablos F, Slupphaug G. 2002. Uracil in DNA—occurrence, consequences and repair. Oncogene 21:8935–48. [DOI] [PubMed] [Google Scholar]
- LaPlant Q, Vialou V, Covington HE, 3rd, Dumitriu D, Feng J, Warren BL, and others. 2010. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci 13:1137–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt H, Page AW, Weier HU, Bestor TH. 1992. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71:865–73. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915–26. [DOI] [PubMed] [Google Scholar]
- Li YQ, Zhou PZ, Zheng XD, Walsh CP, Xu GL. 2007. Association of Dnmt3a and thymine DNA glycosylase links DNA methylation with base-excision repair. Nucleic Acids Res 35:390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Y, Sankpal UT, Robertson AK, McNally JG, Karpova T, Robertson KD. 2004. Modification of de novo DNA methyltransferase 3a (Dnmt3a) by SUMO-1 modulates its interaction with histone deacetylases (HDACs) and its capacity to repress transcription. Nucleic Acids Res 32:598–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, and others. 2013. Global epigenomic reconfiguration during mammalian brain development. Science 341:1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. 2008. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci 28:10576–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti A, Drohat AC. 2011. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem 286:35334–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massart R, Barnea R, Dikshtein Y, Suderman M, Meir O, Hallett M, and others. 2015. Role of DNA methylation in the nucleus accumbens in incubation of cocaine craving. J Neurosci 35:8042–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. 2010. Epigenetic changes in Alzheimer’s disease: decrements in DNA methylation. Neurobiol Aging 31:2025–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrisciano F, Tueting P, Dalal I, Kadriu B, Grayson DR, Davis JM, and others. 2013. Epigenetic modifications of GABAergic interneurons are associated with the schizophrenia-like phenotype induced by prenatal stress in mice. Neuropharmacology 68:184–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows JP, Guzman-Karlsson MC, Phillips S, Brown JA, Strange SK, Sweatt JD, and others. 2016. Dynamic DNA methylation regulates neuronal intrinsic membrane excitability. Sci Signal 9:ra83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows JP, Guzman-Karlsson MC, Phillips S, Holleman C, Posey JL, Day JJ, and others. 2015. DNA methylation regulates neuronal glutamatergic synaptic scaling. Sci Signal 8:ra61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, and others. 2008. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet 82:696–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, and others. 2010. Cortical DNA methylation maintains remote memory. Nat Neurosci 13:664–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. 2007. Covalent modification of DNA regulates memory formation. Neuron 53:857–69. [DOI] [PubMed] [Google Scholar]
- Mitchnick KA, Creighton S, O’Hara M, Kalisch BE, Winters BD. 2015. Differential contributions of de novo and maintenance DNA methyltransferases to object memory processing in the rat hippocampus and perirhinal cortex—a double dissociation. Eur J Neurosci 41:773–86. [DOI] [PubMed] [Google Scholar]
- Monsey MS, Ota KT, Akingbade IF, Hong ES, Schafe GE. 2011. Epigenetic alterations are critical for fear memory consolidation and synaptic plasticity in the lateral amygdala. PLoS One 6:e19958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MJ, Monteggia LM. 2014. Role of DNA methylation and the DNA methyltransferases in learning and memory. Dialogues Clin Neurosci 16:359–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MJ, Na ES, Autry AE, Monteggia LM. 2016. Impact of DNMT1 and DNMT3a forebrain knockout on depressive- and anxiety like behavior in mice. Neurobiol Learn Mem 135:139–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmuhl Y, Fischer D, and others. 2009. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci 12:1559–66. [DOI] [PubMed] [Google Scholar]
- Murgatroyd C, Spengler D. 2012. Genetic variation in the epigenetic machinery and mental health. Curr Psychiatry Rep 14:138–49. [DOI] [PubMed] [Google Scholar]
- Nguyen S, Meletis K, Fu D, Jhaveri S, Jaenisch R. 2007. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Dev Dyn 236:1663–76. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247–57. [DOI] [PubMed] [Google Scholar]
- Oliveira AM, Hemstedt TJ, Bading H. 2012. Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities. Nat Neurosci 15:1111–3. [DOI] [PubMed] [Google Scholar]
- Oliveira AM, Hemstedt TJ, Freitag HE, Bading H. 2016. Dnmt3a2: a hub for enhancing cognitive functions. Mol Psychiatry 21:1130–6. [DOI] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, and others. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448:714–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollema-Mays SL, Centeno MV, Apkarian AV, Martina M. 2014. Expression of DNA methyltransferases in adult dorsal root ganglia is cell-type specific and up regulated in a rodent model of neuropathic pain. Front Cell Neurosci 8:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu C, Sawada K, Zhang X, Cheng X. 2002. The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nat Struct Biol 9:217–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G, Peter M. 2008. Function and regulation of protein neddylation. “Protein modifications: beyond the usual suspects” review series. EMBO Rep 9:969–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao JS, Keleshian VL, Klein S, Rapoport SI. 2012. Epigenetic modifications in frontal cortex from Alzheimer’s disease and bipolar disorder patients. Transl Psychiatry 2:e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondelet G, Dal Maso T, Willems L, Wouters J. 2016. Structural basis for recognition of histone H3K36me3 nucleosome by human de novo DNA methyltransferases 3A and 3B. J Struct Biol 194:357–67. [DOI] [PubMed] [Google Scholar]
- Ruzicka WB, Zhubi A, Veldic M, Grayson DR, Costa E, Guidotti A. 2007. Selective epigenetic alteration of layer I GABAergic neurons isolated from prefrontal cortex of schizophrenia patients using laser-assisted microdissection. Mol Psychiatry 12:385–97. [DOI] [PubMed] [Google Scholar]
- Sales AJ, Biojone C, Terceti MS, Guimaraes FS, Gomes MV, Joca SR. 2011. Antidepressant-like effect induced by systemic and intra-hippocampal administration of DNA methylation inhibitors. Br J Pharmacol 164:1711–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunderson EA, Spiers H, Mifsud KR, Gutierrez-Mecinas M, Trollope AF, Shaikh A, and others. 2016. Stress-induced gene expression and behavior are controlled by DNA methylation and methyl donor availability in the dentate gyrus. Proc Natl Acad Sci U S A 113:4830–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpa S, Fuso A, D’Anselmi F, Cavallaro RA. 2003. Presenilin 1 gene silencing by S-adenosylmethionine: a treatment for Alzheimer disease? FEBS Lett 541:145–8. [DOI] [PubMed] [Google Scholar]
- Schirrmacher E, Beck C, Brueckner B, Schmitges F, Siedlecki P, Bartenstein P, and others. 2006. Synthesis and in vitro evaluation of biotinylated RG108: a high affinity compound for studying binding interactions with human DNA methyltransferases. Bioconjug Chem 17:261–6. [DOI] [PubMed] [Google Scholar]
- Shamay M, Greenway M, Liao G, Ambinder RF, Hayward SD. 2010. De novo DNA methyltransferase DNMT3b interacts with NEDD8-modified proteins. J Biol Chem 285:36377–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma RP, Tun N, Grayson DR. 2008. Depolarization induces downregulation of DNMT1 and DNMT3a in primary cortical cultures. Epigenetics 3:74–80. [DOI] [PubMed] [Google Scholar]
- Shi HS, Luo YX, Yin X, Wu HH, Xue G, Geng XH, and others. 2015. Reconsolidation of a cocaine associated memory requires DNA methyltransferase activity in the basolateral amygdala. Sci Rep 5:13327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepper P, Kungulovski G, Jurkowska RZ, Chandra T, Krueger F, Reinhardt R, and others. 2016. Efficient targeted DNA methylation with chimeric dCas9-Dnmt3a-Dnmt3L methyltransferase. Nucleic Acids Res 45:1703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt JD. 2013. The emerging field of neuroepigenetics. Neuron 80:624–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M. 2013. DNA methylation, behavior and early life adversity. J Genet Genomics 40:331–8. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. 2007. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol 14:1025–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova N, Renthal W, Kumar A, Nestler EJ. 2007. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci 8:355–67. [DOI] [PubMed] [Google Scholar]
- Veldic M, Caruncho HJ, Liu WS, Davis J, Satta R, Grayson DR, and others. 2004. DNA-methyltransferase 1 mRNA is selectively overexpressed in telencephalic GABAergic interneurons of schizophrenia brains. Proc Natl Acad Sci U S A 101:348–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe D, Uchiyama K, Hanaoka K. 2006. Transition of mouse de novo methyltransferases expression from Dnmt3b to Dnmt3a during neural progenitor cell development. Neuroscience 142:727–37. [DOI] [PubMed] [Google Scholar]
- Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, and others. 2010. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 329:444–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing B, Liu P, Xu WJ, Xu FY, Dang YH. 2014. Effect of microinjecting of 5-aza-2-deoxycytidine into ventrolateral orbital cortex on depressive-like behavior in rats. Neurosci Lett 574:11–4. [DOI] [PubMed] [Google Scholar]
- Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. 2014. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell 26:577–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jurkowska R, Soeroes S, Rajavelu A, Dhayalan A, Bock I, and others. 2010. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res 38:4246–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Fan L, Frick KM. 2010. Epigenetic alterations regulate estradiol-induced enhancement of memory consolidation. Proc Natl Acad Sci U S A 107:5605–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Wang L, Zhang Y, Zhao FH, Luo J, Xiao Z, and others. 2012. Increased expression of DNA methyltransferase 1 and 3a in human temporal lobe epilepsy. J Mol Neurosci 46:420–6. [DOI] [PubMed] [Google Scholar]
- Zhubi A, Veldic M, Puri NV, Kadriu B, Caruncho H, Loza I, and others. 2009. An upregulation of DNA-methyltransferase 1 and 3a expressed in telencephalic GABAergic neurons of schizophrenia patients is also detected in peripheral blood lymphocytes. Schizophr Res 111:115–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovkic IB, Sweatt JD. 2013. Epigenetic mechanisms in learned fear: implications for PTSD. Neuropsychopharmacology 38:77–93. [DOI] [PMC free article] [PubMed] [Google Scholar]