Abstract
In mammals, GPIHBP1 is absolutely essential for transporting lipoprotein lipase (LPL) to the lumen of capillaries, where it hydrolyzes the triglycerides in triglyceride-rich lipoproteins. In all lower vertebrate species (e.g., birds, amphibians, reptiles, fish), a gene for LPL can be found easily, but a gene for GPIHBP1 has never been found. The obvious question is whether the LPL in lower vertebrates is able to reach the capillary lumen. Using purified antibodies against chicken LPL, we showed that LPL is present on capillary endothelial cells of chicken heart and adipose tissue, colocalizing with von Willebrand factor. When the antibodies against chicken LPL were injected intravenously into chickens, they bound to LPL on the luminal surface of capillaries in heart and adipose tissue. LPL was released rapidly from chicken hearts with an infusion of heparin, consistent with LPL being located inside blood vessels. Remarkably, chicken LPL bound in a specific fashion to mammalian GPIHBP1. However, we could not identify a gene for GPIHBP1 in the chicken genome, nor could we identify a transcript for GPIHBP1 in a large chicken RNA-seq data set. We conclude that LPL reaches the capillary lumen in chickens — as it does in mammals — despite an apparent absence of GPIHBP1.
Keywords: Metabolism
Keywords: Adipose tissue, endothelial cells
Lipoprotein lipase reaches the capillary lumen in chicken tissues, despite an apparent absence of the lipoprotein lipase transporter GPIHBP1.
Introduction
GPIHBP1, a glycoprotein expressed by capillary endothelial cells, is crucial for lipoprotein lipase–mediated (LPL-mediated) processing of triglyceride-rich lipoproteins (TRLs) (1–3). In mammals, GPIHBP1 can be considered as a partner protein for LPL. First, GPIHBP1 is responsible for binding LPL in the interstitial spaces and shuttling it across endothelial cells to the capillary lumen (2). In the absence of GPIHBP1, LPL is stranded within the interstitial spaces where it is useless for processing TRLs in the bloodstream (2). Second, the GPIHBP1–LPL complex on capillary endothelial cells is essential for TRL margination along capillaries (3). In the absence of GPIHBP1-bound LPL, TRLs do not stop along capillaries and simply flow on by in the bloodstream. Third, GPIHBP1 preserves the structural integrity and catalytic activity of LPL (4). In the absence of GPIHBP1, LPL’s triglyceride hydrolase domain unfolds, resulting in a rapid decline in catalytic activity (4). GPIHBP1 and LPL are equally important for intravascular lipolysis; a deficiency of either protein markedly impairs intravascular triglyceride hydrolysis and leads to severe hypertriglyceridemia (chylomicronemia) (5–19). The importance of GPIHBP1 and LPL for plasma triglyceride metabolism in mammals has been underscored by human genetics; specific missense mutations in either protein can abolish LPL–GPIHBP1 interactions, resulting in reduced delivery of LPL to the capillary lumen, impaired TRL processing, and severe hypertriglyceridemia (6, 7, 9–14).
GPIHBP1 is a GPI-anchored protein of the Ly6 superfamily (35 Ly6 family members in human; 61 in mouse) (20). All Ly6 proteins have at least one approximately 80–amino acid Ly6 domain with 8 or 10 cysteines, all arranged in a characteristic spacing pattern and all disulfide bonded, creating a 3-fingered structural domain (21). GPIHBP1 is unique among Ly6 proteins in having a disordered acidic domain at its amino terminus, with 21 of 26 consecutive residues in the human protein being aspartate or glutamate (22). Recent work by Mysling et al. (4) showed that GPIHBP1, and more specifically GPIHBP1’s acidic domain, prevents the spontaneous unfolding of LPL’s hydrolase domain (4). GPIHBP1’s Ly6 domain is responsible for high-affinity LPL binding, with the acidic domain playing only an accessory role (4). The second finger of GPIHBP1’s Ly6 domain is particularly important for LPL binding (4, 23). A variety of amino acid substitutions within the second finger abolish GPIHBP1’s capacity to bind LPL, in some cases by interfering with the proper formation of disulfide bonds (11, 23, 24). Mutations in W109 in human GPIHBP1 abolish the capacity of GPIHBP1 to bind LPL and do so without disrupting disulfide bond formation, suggesting that W109 is directly involved in LPL–GPIHBP1 interactions (24). W109 is perfectly conserved in the GPIHBP1 of all mammalian species (Supplemental Table 1).
Genes for GPIHBP1 and LPL are easily detectable in the genomes of all mammals, including marsupials and the egg-laying platypus (20, 25–27). Human and mouse LPL are 92% identical at the amino acid level; the Ly6 domains of human and mouse GPIHBP1 are 59% identical. Because LPL and GPIHBP1 are partners in plasma triglyceride metabolism, the conservation of both proteins during mammalian evolution is not surprising. However, when one examines the genomes of lower vertebrates (e.g., fish, birds, reptiles), there is a striking discrepancy between GPIHBP1 and LPL. LPL is highly conserved in every lower vertebrate species (e.g., the amino acid sequences of human and chicken LPL [cLPL] are 72% identical), but no one has yet identified a GPIHBP1 ortholog in the chicken or any other lower vertebrate species (25, 26). The function of LPL in plasma triglyceride metabolism is conserved in lower vertebrates. When goat polyclonal antibodies against cLPL were injected intravenously into chickens, they inactivated LPL, blocking TRL processing and resulting in hypertriglyceridemia (28).
Given that the structure of LPL as well as its function in TRL processing are conserved in lower vertebrates, the absence of LPL’s partner protein (i.e., GPIHBP1) is surprising. Since GPIHBP1 is essential for shuttling LPL into capillaries in mammals, the obvious question is whether the LPL in lower vertebrates is able to reach the capillary lumen. It is conceivable that TRL processing in lower vertebrates might be fundamentally different, with TRLs being transported across capillaries and undergoing processing by the LPL within the interstitial spaces. This possibility may not be farfetched, given that TRLs produced by the chicken intestine are said to move into capillaries by vesicular transport (29). Another possibility is that the LPL in lower vertebrates reaches the capillary lumen, but by a mechanism independent of GPIHBP1.
To gain further insights into TRL processing in lower vertebrate species, we used cLPL-specific antibodies to define the localization of LPL in chicken tissues. We also tested whether cLPL has the capacity to bind to human and mouse GPIHBP1.
Results
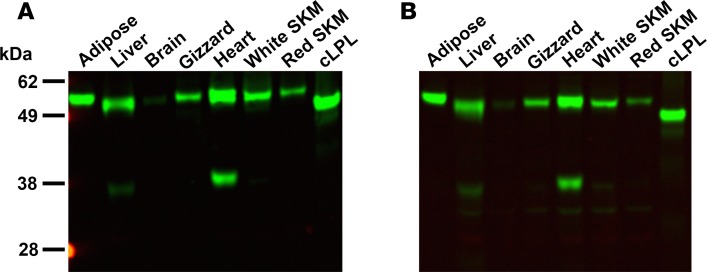
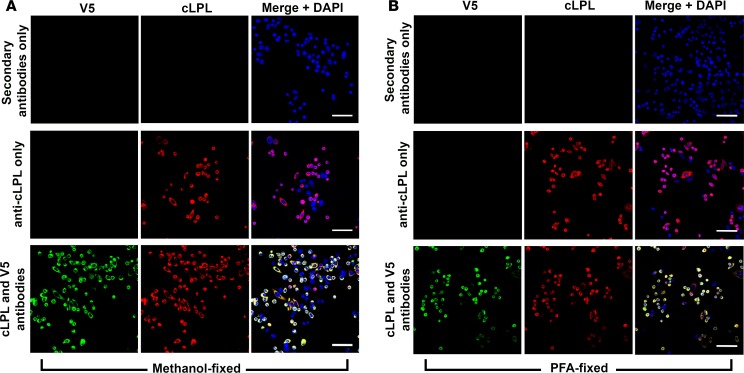
We took advantage of goat antibodies against cLPL (30, 31) to examine the localization of LPL in chicken tissues. The first step was to assess the specificity of the antibodies, which had been purified on a cLPL immunoaffinity column (generated with LPL purified from chicken adipose tissue). We examined, by Western blotting, the ability of the immunopurified antibodies to bind to LPL in crude homogenates of chicken tissues. The immunopurified IgGs against cLPL bound to a single protein of the expected size (~52 kDa), except in the heart where the IgGs also bound to a 38-kDa protein (Figure 1A). We were confident that the 38-kDa band in the heart extracts was a cLPL breakdown fragment, but to be certain about that conclusion we repeated the Western blot studies with IgGs that had been further purified on a second cLPL immunoaffinity column (generated with a cLPL polypeptide produced in E. coli). Again, the antibodies bound exclusively to LPL (along with the 38-kDa LPL breakdown fragment in the heart) (Figure 1B). To determine if the purified IgG fraction would be useful for immunohistochemistry studies, we first tested their ability to bind to CHO cells that had been transfected with an expression vector for V5-tagged cLPL. Regardless of whether the cells were fixed with methanol or paraformaldehyde, the purified IgGs against cLPL bound avidly and specifically to transfected cells, colocalizing with an antibody against the V5 tag (Figure 2).
Figure 1. Testing the specificity of an immunopurified goat IgG against chicken lipoprotein lipase (cLPL) with Western blots.
Proteins from chicken tissue extracts (30 μg/lane) were size-fractionated by SDS-PAGE and then examined by Western blotting. (A) Western blot with an immunopurified goat IgG against cLPL. The antibodies bound specifically to cLPL from E. coli and recognized a band of similar size in chicken tissues. In heart, the anti-cLPL antibodies also bound to a smaller fragment of approximately 38 kDa. (B) Western blot with the same antibodies after they had been repurified on a second cLPL immunoaffinity column generated with cLPL from E. coli. SKM: skeletal muscle.
Figure 2. Testing the ability of an immunopurified goat IgG against chicken lipoprotein lipase (cLPL) to bind to cLPL in cultured cells.
CHO pgsA-745 cells were transiently transfected with V5-tagged cLPL. Immunocytochemistry studies were performed on fixed and permeabilized cells with a goat antibody against cLPL and a mouse monoclonal antibody against the V5 tag. Binding of the primary antibodies was detected with an Alexa Fluor 555–conjugated donkey anti–goat IgG (red) and an Alexa Fluor 488–conjugated donkey anti–mouse IgG (green). As experimental controls, we included transfected cells that had been incubated with secondary antibodies alone and cells that had been incubated with the anti-cLPL antibody alone. DNA was stained with DAPI (blue). (A) Immunocytochemistry studies on cells that had been fixed with methanol. (B) Studies of cells fixed with paraformaldehyde (PFA). Scale bars: 50 μm.
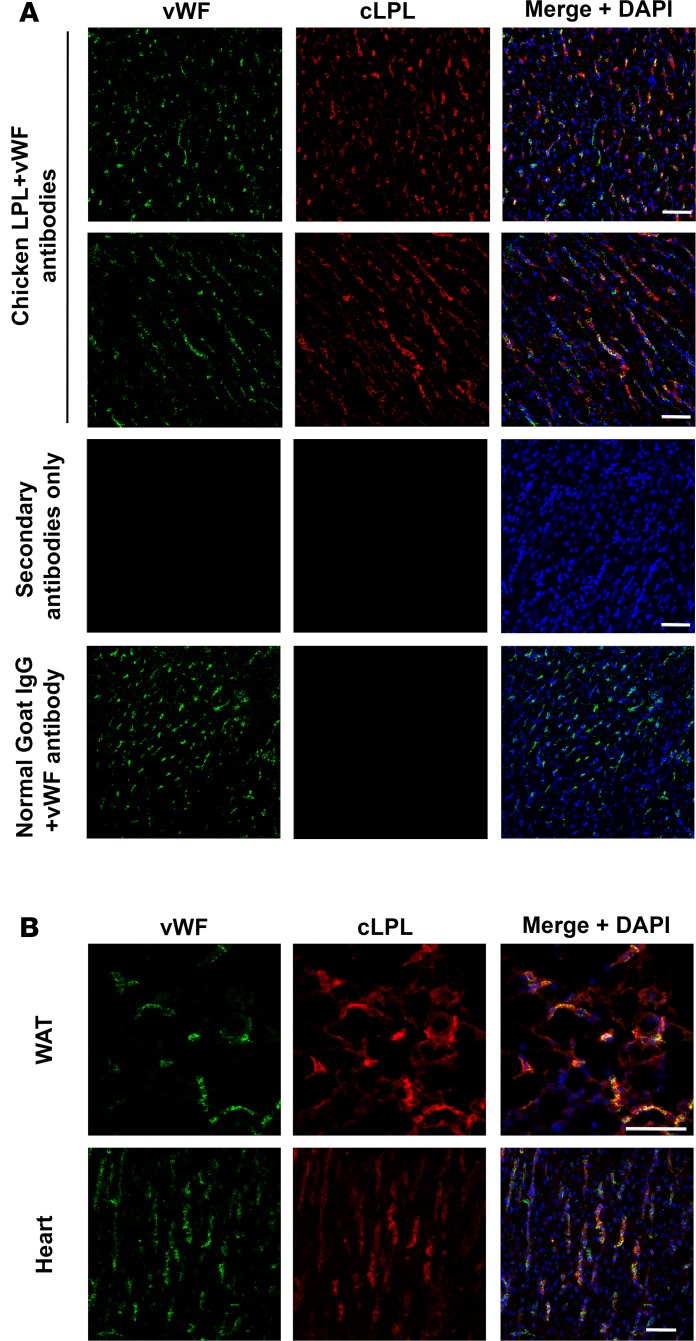
Most of the LPL in mouse tissues is bound to GPIHBP1 on capillary endothelial cells (2). In the absence of GPIHBP1, LPL remains within the interstitial spaces and never reaches the capillary lumen (2). In immunohistochemistry studies, we found that much of the LPL in the chicken heart is associated with capillary endothelial cells, colocalizing with an endothelial cell marker (von Willebrand factor; vWF) (Figure 3A). There was no binding of nonimmune goat IgGs or the secondary antibody alone to capillaries in chicken tissues (Figure 3A). The presence of cLPL on capillaries of heart and adipose tissue was confirmed in a second experiment (Figure 3B). In a third experiment, we also observed colocalization of vWF and LPL (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.96783DS1). There was occasional LPL staining that did not coincide with vWF staining, more so than in comparable experiments in mouse tissues, raising the possibility that a fraction of the cLPL in tissues remained within the interstitial spaces (Figure 3 and Supplemental Figure 1).
Figure 3. LPL in chicken tissues is associated with capillaries.
Frozen sections from chicken white adipose tissue (WAT) and heart were stained with a rabbit antibody against von Willebrand factor (vWF), followed by an Alexa Fluor 488–conjugated donkey anti–rabbit IgG (green) and an Alexa Fluor 555–conjugated goat anti–chicken lipoprotein lipase (anti-cLPL) IgG (red). DNA was stained with DAPI (blue). (A) Sections of heart stained with both cLPL and vWF antibodies, revealing colocalization of cLPL and vWF in capillaries. As controls, we examined a section incubated with secondary antibodies alone and a section stained with the vWF antibody and nonimmune goat IgG. (B) An independent chicken immunohistochemistry study in which sections of WAT and heart were stained with cLPL and vWF antibodies. Scale bars: 50 μm.
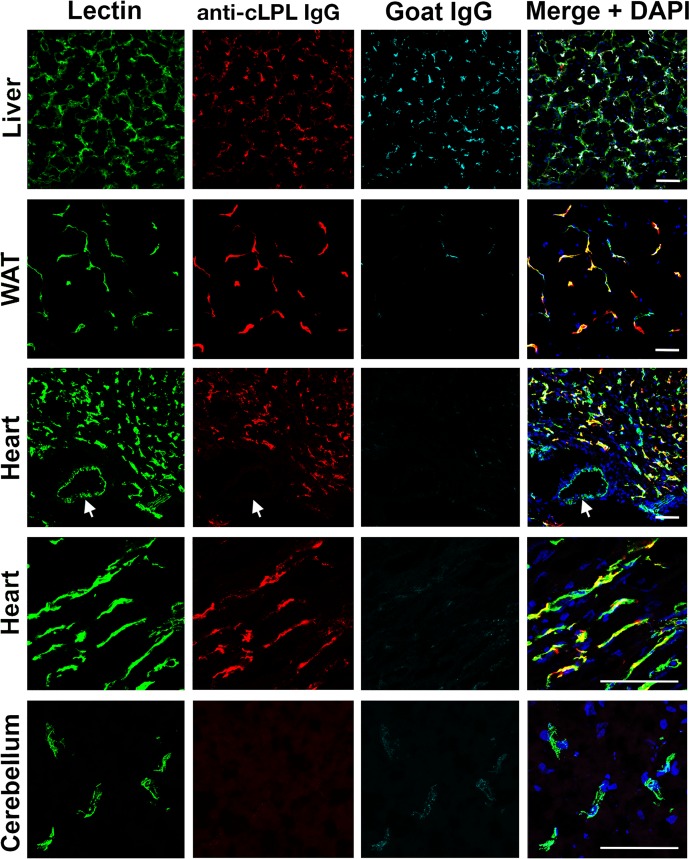
To determine if cLPL is located along the luminal surface of capillaries, 9-day-old chickens were injected intravenously with an Alexa Fluor 555–labeled goat IgG against cLPL, a fluorescein-labeled Lens culinaris agglutinin (a lectin), and an Alexa Fluor 647–labeled nonimmune goat IgG. After perfusion-fixation, white adipose tissue (WAT), heart, liver, and cerebellum were harvested for immunohistochemistry. The goat IgGs against cLPL bound to capillaries in heart and WAT, colocalizing with the lectin (Figure 4). The lectin also bound to the luminal surface of large blood vessels, whereas IgGs against cLPL did not. Also, there was no binding of cLPL antibodies to capillaries of the cerebellum (Figure 4). Only trace amounts of the nonimmune goat IgG were detectable in capillaries of chicken heart, WAT, or cerebellum, demonstrating that the perfusion of tissues was adequate (Figure 4). As expected, the nonimmune goat IgGs were bound by immunoglobulin receptors in the liver (Figure 4). Additional images showing the binding of the cLPL-specific IgGs to capillaries of chicken tissues are shown in Supplemental Figure 2. The binding of cLPL-specific IgGs to the luminal surface of heart and WAT capillaries was confirmed with further experiments in which chickens were injected with the Alexa Fluor 555–labeled goat IgG against cLPL and a fluorescein-labeled Lens culinaris agglutinin (Supplemental Figure 3).
Figure 4. Chicken lipoprotein lipase (cLPL) is located along the luminal surface of capillaries.
A 9-day-old chicken was given an intravenous injection of 0.7 mg of an Alexa Fluor 555–labeled goat IgG against cLPL (red), 0.5 mg of fluorescein-labeled Lens culinaris agglutinin (Lectin, green), and 0.7 mg of Alexa Fluor 647–labeled nonimmune goat IgG (cyan). After 4 minutes, the chicken was perfused with 50 ml of PBS followed by 30 ml of 3% paraformaldehyde (PFA) in PBS. Liver, white adipose tissue (WAT), heart, and cerebellum were harvested, fixed in 3% PFA, and 50-μm sections of WAT and 10-μm sections of heart, liver, and cerebellum were prepared. The lectin bound to endothelial cells of capillaries and larger blood vessels; the goat IgG against cLPL bound to capillaries but not larger blood vessels (arrows). The nonimmune goat IgG did not bind to blood vessels of the heart, WAT, or cerebellum (indicating an effective perfusion) but as expected did bind to macrophages in the liver. DNA was stained with DAPI (blue). Scale bars: 50 μm.
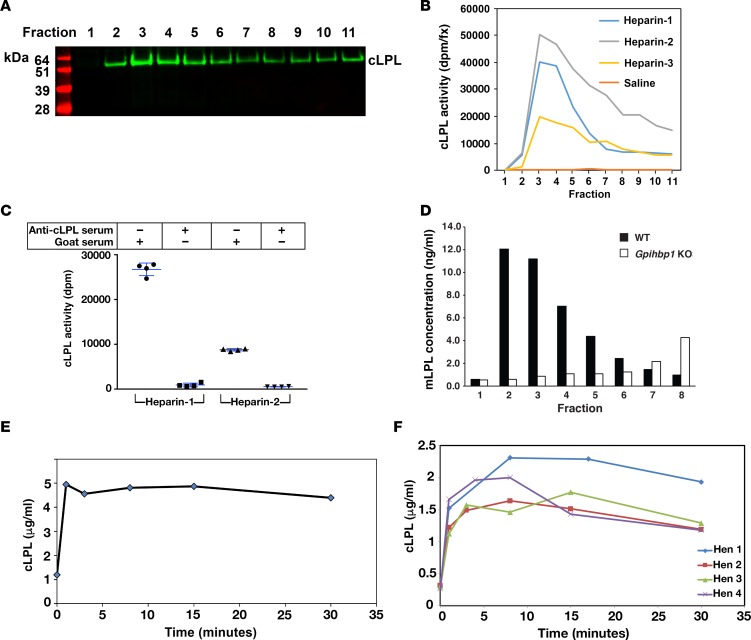
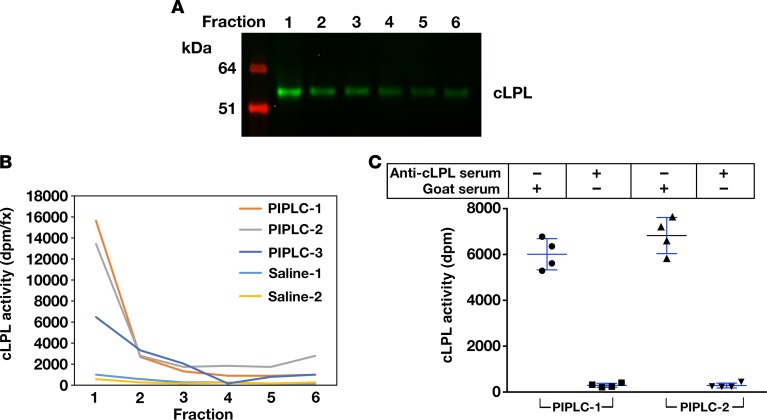
LPL was released rapidly from isolated chicken hearts by perfusing the hearts with heparin (Figure 5A). The cLPL released by heparin was catalytically active (Figure 5B), and the enzymatic activity could be abolished with the goat antiserum against cLPL (Figure 5C). The rapid release of LPL from isolated chicken hearts during the heparin perfusion was consistent with results with isolated mouse hearts. LPL was released rapidly from hearts of wild-type mice with heparin, but LPL release from hearts of Gpihbp1-deficient mice (where the LPL is mislocalized to the interstitial spaces) was reduced in amount and substantially delayed (Figure 5D). We also examined the release of LPL into the plasma compartment after giving chickens an intravenous injection of heparin. Again, the release of LPL into the plasma was rapid (Figure 5, E and F), consistent with the LPL being located inside blood vessels (2). LPL protein and LPL activity could also be released from isolated chicken hearts with a perfusion of phosphatidylinositol-specific phospholipase C (PIPLC) (Figure 6).
Figure 5. Chicken lipoprotein lipase (cLPL) can be released from tissues with heparin.
(A) Isolated chicken hearts were perfused with 20 U/ml heparin, and cLPL protein was detected in individual fractions (0.2 ml/fraction) by Western blotting with a goat antibody against cLPL. (B) cLPL activity in the fractions from 3 different chickens (Heparin-1, Heparin-2, Heparin-3) was measured with a [3H]triolein substrate. As a control, a chicken heart was perfused with saline only (Saline). (C) Inhibition of the LPL activity with a goat antiserum against cLPL. For these studies, we pooled fractions 3–5 from 2 of the chickens (Heparin-1 and Heparin-2). We aliquoted 25 μl of the pooled fractions and then added either 50 μl of the goat antiserum against cLPL or normal goat serum. We then performed LPL activity assays. The 4 data points represent duplicate lipase assays on the fractions from 2 chickens. (D) Bar graph showing rapid heparin-mediated release of mouse LPL (mLPL) from an isolated heart of a wild-type mouse (black bars). Heparin-mediated release of mLPL from the heart of a Gpihbp1-deficient mouse was delayed (white bars). Similar results were observed in 2 other pairs of wild-type and Gpihbp1-deficient mice. (E) Rapid release of cLPL (as measured by ELISA) into the plasma of a cockerel after an intravenous injection of heparin (2 U/g body weight). (F) Rapid release of cLPL into the plasma of 4 hens after an intravenous injection of heparin (2 U/g body weight).
Figure 6. Chicken lipoprotein lipase (cLPL) is released from chicken hearts by phosphatidylinositol-specific phospholipase C (PIPLC).
Isolated chicken hearts were perfused with 6 U/ml PIPLC or normal saline alone and incubated in a water bath for 10 minutes at 37°C. The hearts were then perfused with Tyrode’s buffer. Six fractions were collected, and cLPL mass and activity were measured. (A) Western blots demonstrating that PIPLC releases cLPL into the perfusate. (B) Activity assays with a [3H]triolein substrate, revealing that PIPLC releases catalytically active cLPL into the perfusate. Three independent experiments with PIPLC were performed (PIPLC-1, PIPLC-2, PIPLC-3), and 2 studies were performed with normal saline alone (Saline-1, Saline-2). (C) Inhibition of PIPLC-released cLPL activity with a goat antibody against cLPL. We pooled the fractions from the PIPLC-1 and PIPLC-2 experiments, and cLPL activity was measured in the presence of either the goat antiserum against cLPL or normal goat serum. The 4 data points represent lipase assays, performed in duplicate, on the fractions from 2 chickens.
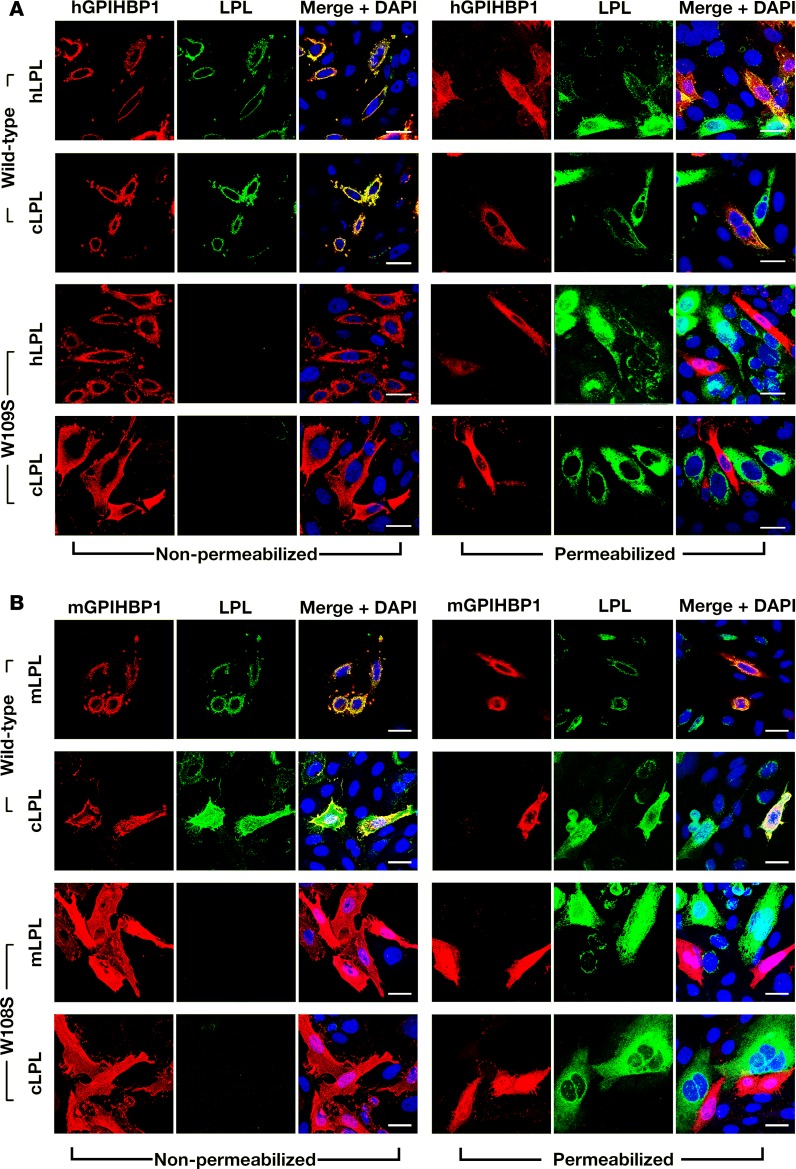
The location of cLPL within tissues (i.e., bound to capillaries but not large blood vessels of the heart; absent in brain capillaries) resembles the localization of GPIHBP1 and LPL in mouse tissues (1, 2). In mice, the presence of LPL in capillaries depends on GPIHBP1 (1, 2). To explore the possibility that the distribution of LPL in chickens might be governed by GPIHBP1, we first used a cell-based binding assay to test whether cLPL (produced in CHO cells) is capable of binding to human or mouse GPIHBP1. We also examined the ability of mouse, human, and cLPL to bind to mutant GPIHBP1 proteins that lack the ability to bind LPL (human GPIHBP1-W109S, mouse GPIHBP1-W108S) (23). As expected, human and mouse LPL bound to wild-type versions of mouse and human GPIHBP1, but not to human GPIHBP1-W109S or mouse GPIHBP1-W108S (Figure 7). Remarkably, cLPL also bound to wild-type human or mouse GPIHBP1 but not to the mutant GPIHBP1 proteins (Figure 7). We observed small amounts of cLPL binding to a few of the nontransfected cells (Figure 7B), but most of the binding of cLPL was to cells that had been transfected with GPIHBP1.
Figure 7. Chicken lipoprotein lipase (cLPL) binds to wild-type mouse GPIHBP1 (mGPIHBP1) or human GPIHBP1 (hGPIHBP1) but not to mutant GPIHBP1 proteins (mGPIHBP1-W108S, hGPIHBP1-W109S) that lack the ability to bind cLPL.
CHO pgsA-745 cells were transiently transfected with S-protein–tagged versions of wild-type or mutant GPIHBP1 and coplated with cells that had been transfected with V5-tagged versions of cLPL. Immunocytochemistry studies were performed on permeabilized and nonpermeabilized cells with a goat antibody against the S-protein tag (red) and a mouse antibody against the V5 tag (green). DNA was stained with DAPI (blue). (A) Immunocytochemistry studies showing that the secreted human (hLPL) or cLPL bound avidly to neighboring CHO cells expressing wild-type hGPIHBP1 (hence, hLPL and cLPL colocalized with hGPIHBP1) but not to CHO cells expressing hGPIHBP1-W109S (no colocalization). (B) Immunocytochemistry studies showing that the secreted mouse LPL (mLPL) or cLPL bound avidly to neighboring CHO cells expressing wild-type mGPIHBP1 (colocalization) but not to CHO cells expressing mGPIHBP1-W108S (no colocalization). Scale bars: 20 μm.
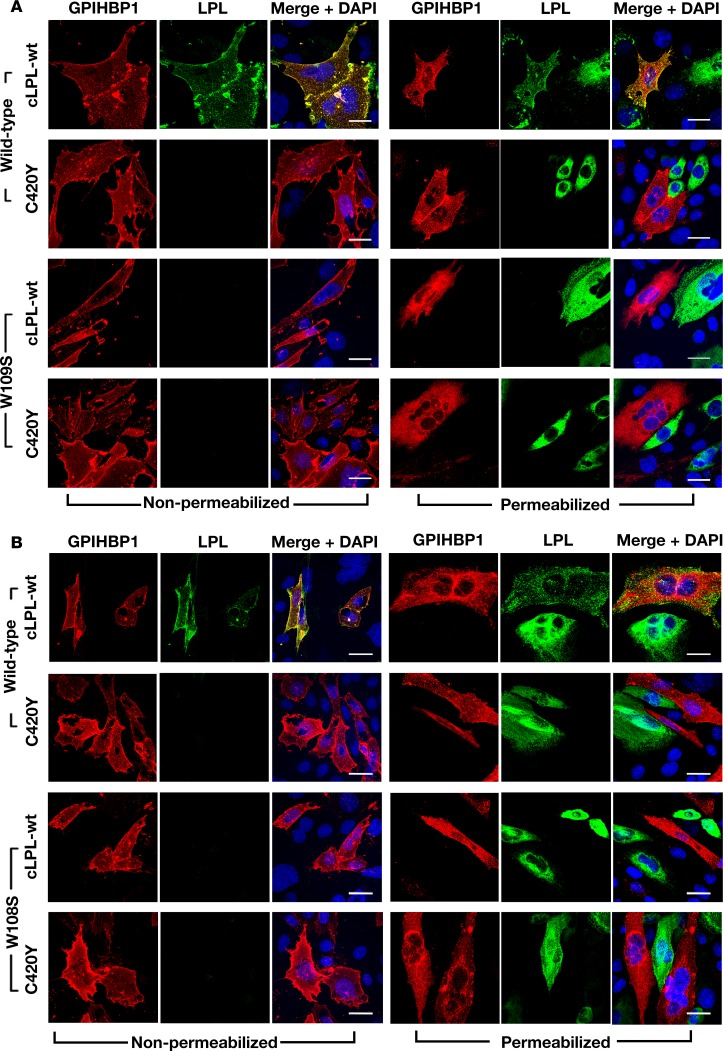
A p.C418Y substitution in human LPL abolishes LPL’s ability to bind to human or mouse GPIHBP1 (14). Introducing the corresponding mutation into cLPL (p.C420Y) abolished the ability of cLPL to bind to human or mouse GPIHBP1 (Figure 8).
Figure 8. Wild-type chicken lipoprotein lipase (cLPL-wt), but not a mutant cLPL with a p.C420Y mutation, binds to GPIHBP1.
CHO pgsA-745 cells were transiently transfected with S-protein–tagged wild-type human (h) or mouse (m) GPIHBP1 (or hGPIHBP1-W109S or mGPIHBP1-W108S) and coplated with cells that had been transfected with V5-tagged versions of cLPL (wt or C420Y). Immunocytochemistry studies were performed on permeabilized and nonpermeabilized cells with a goat antibody against the S-protein tag (red) and a mouse monoclonal antibody against the V5 tag (green). DNA was stained with DAPI (blue). (A) Immunocytochemistry studies showing that cLPL-wt bound avidly to neighboring CHO cells expressing wild-type hGPIHBP1 (hence cLPL-wt colocalized with hGPIHBP1), whereas cLPL-C420Y had no capacity to bind to cells expressing wild-type hGPIHBP1 (no colocalization). Cells expressing hGPIHBP1-W109S did not bind cLPL-wt. (B) Immunocytochemistry studies showing that cLPL-wt bound avidly to CHO cells expressing wild-type mGPIHBP1 (colocalization), while LPL-C420Y had no capacity to bind to cells expressing wild-type mGPIHBP1 (no colocalization). Cells expressing mGPIHBP1-W108S did not bind cLPL-wt. Scale bars: 20 μm.
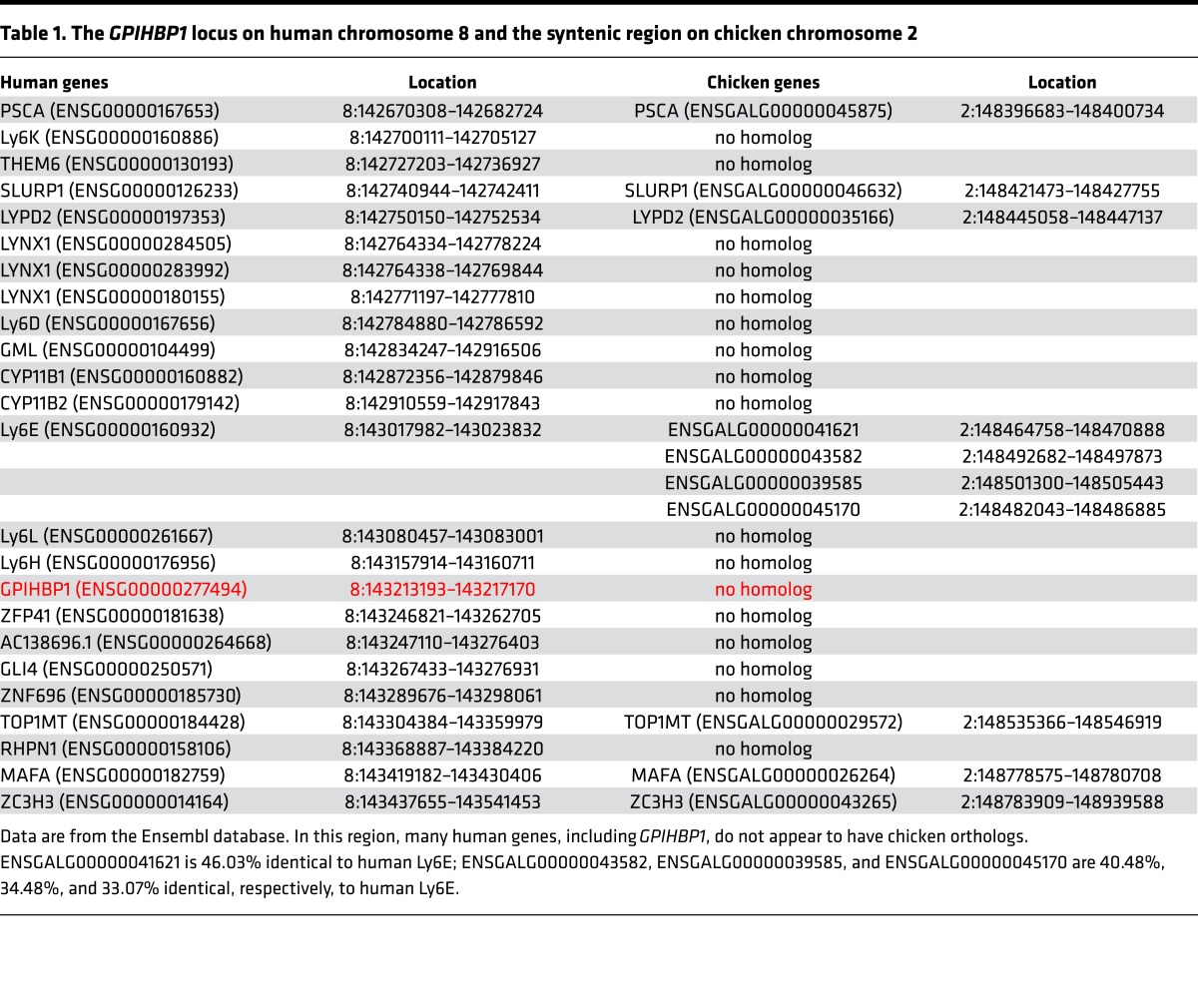
A segment of human chromosome 8 containing GPIHBP1 is syntenic to a segment of chicken chromosome 2 (Table 1). In both species, the upstream portion of the syntenic region encodes genes for several Ly6 proteins (PSCA, SLURP1, LYPD2). The Ly6 domains of chicken and human PSCA, SLURP1, and LYPD2 are 47%–65% identical. The downstream portion of the syntenic region contains genes for TOP1MT, MAFA, and ZC3H3. Fifteen genes on human chromosome 8, including GPIHBP1, are located between LYPD2 and TOP1MT, whereas only 4 genes have been identified between LYPD2 and TOP1MT on chicken chromosome 2 (Table 1); those genes encode Ly6E-like proteins (ENSGALG00000039585, ENSGALG00000041621, ENSGALG00000043582, ENSGALG00000045170). None of the 4 Ly6E-like proteins resembles human or platypus GPIHBP1, none contains an amino-terminal acidic domain, and the level of amino acid sequence identity with human or platypus GPIHBP1 is similar to the level of sequence identity to SLURP1 (a protein involved in keratinocyte differentiation) (32) (Supplemental Table 1, A and B). None of the 4 chicken Ly6E-like proteins displayed significant sequence homology to the most conserved region of mammalian GPIHBP1 (residues 106–115 in human GPIHBP1) (Supplemental Table 1C). Aside from the 4 Ly6E-like genes within the syntenic region of chicken chromosome 2, 10 other Ly6 genes exist in the chicken databases; none of those proteins resembles GPIHBP1 (Supplemental Table 2).
Table 1. The GPIHBP1 locus on human chromosome 8 and the syntenic region on chicken chromosome 2.
The expression patterns of the 4 chicken Ly6E-like proteins did not closely resemble the pattern of GPIHBP1 in mammals (high in heart, adipose tissue, lung; medium levels in skeletal muscle; low levels in liver; absent from the brain). As judged by RNA sequencing (RNA-seq), ENSGALG00000041621 was expressed at high levels in lung and gizzard; medium levels in the optic lobe, thyroid, Harderian gland, and liver; and low levels in trachea, cerebellum, and heart. ENSGALG00000039585 was expressed at high levels in the skin, medium to low levels in the Harderian gland, and very low levels in the heart. ENSGALG00000045170 was expressed at high levels in skin, Harderian gland, and thymus; medium levels in thyroid and gizzard; and low levels in the heart. ENSGALG00000043582 was expressed at high levels in the trachea and medium to low levels in heart, Harderian gland, lung, and skin.
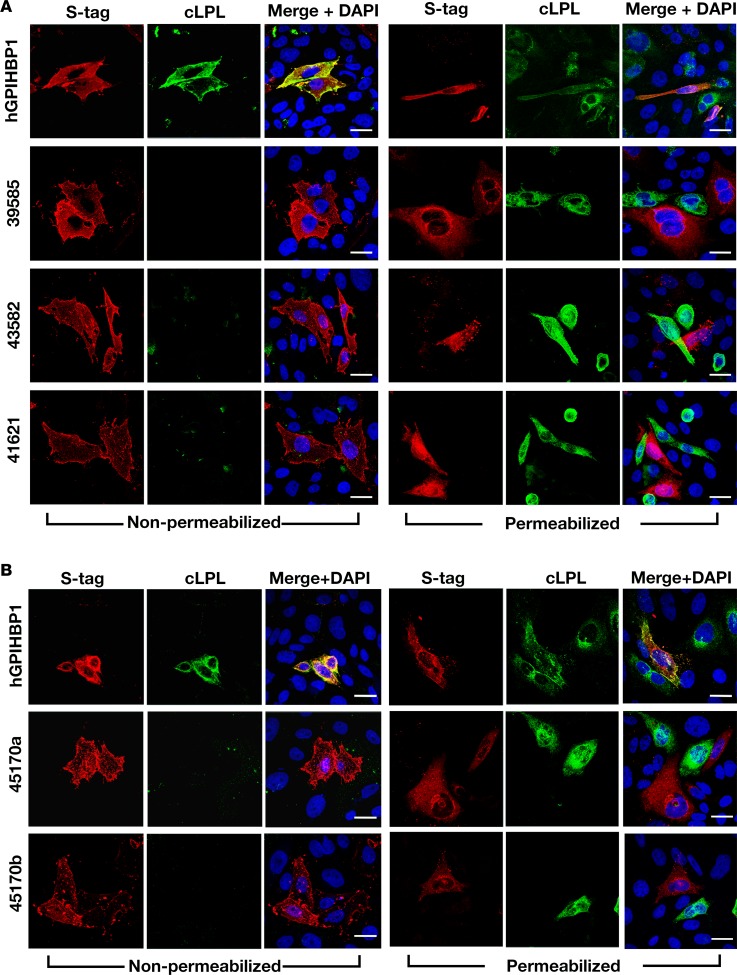
Even though the homology between GPIHBP1 and the 4 chicken Ly6E-like proteins and the pattern of expression did not seem consistent with a role in plasma triglyceride metabolism, we expressed S-protein–tagged versions of the 4 Ly6E-like proteins in CHO cells and tested their ability to bind cLPL. All of the Ly6E-like proteins could be released from the surface of cells with PIPLC (data not shown), but none bound cLPL (Figure 9). We again observed binding of cLPL to human GPIHBP1 (Figure 9).
Figure 9. Testing the ability of chicken lipoprotein lipase (cLPL) to bind to chicken Ly6E-like proteins on the surface of transfected cells.
CHO pgsA-745 cells were transfected with an expression vector for S-protein–tagged human GPIHBP1 (hGPIHBP1) or for S-protein–tagged versions of the chicken Ly6-like proteins. Immunocytochemistry studies were performed on permeabilized and nonpermeabilized cells with a goat antibody against the S-protein tag (red) and a mouse antibody against the V5 tag (green). DNA was stained with DAPI (blue). (A) Immunocytochemistry studies performed with expression vectors for ENSGALG00000039585, ENSGALG00000043582, and ENSGALG00000041621 showing than none of these chicken Ly6-like proteins bound cLPL (no colocalization). (B) Immunocytochemistry studies performed with 2 different expression vectors for ENSGALG00000045170 (45170a and 45170b). Vector 45170a contained the Ly6 domain of ENSGALG00000045170 and also included the acidic domain of hGPIHBP1; vector 45170b contained the Ly6 domain of ENSGALG00000045170. Neither vector conferred upon CHO cells the ability to bind cLPL. Scale bars: 20 μm.
It is possible that the failure to identify a chicken GPIHBP1 gene was due to gaps in the chicken genomic DNA sequences or incomplete annotation. With the assistance from the UC Santa Cruz Genomics Institute, we examined GPIHBP1 sequences from 46 mammals and examined genomes of 26 lower vertebrates for sequences similar to the sequences of the human and platypus GPIHBP1 genes, but no matches were found. To address the possibility that the chicken GPIHBP1 gene is located in an unassembled region of the genome or that the annotation was incomplete, we examined short-read RNA-seq and long-read isoform sequencing (Iso-Seq) chicken data sets for transcripts similar to mammalian GPIHBP1. The short-read RNA-seq data set was generated from 20 chicken tissues, including heart and adipose tissue (>3 billion paired-end reads). The Iso-Seq data were generated from adult chicken brain, Hamburger-Hamilton (HH) stage 26 whole embryos, and pooled embryonic heart samples (HH stages 18–20, 25, and 32).
Transcriptome annotations were generated from both the RNA-seq and Iso-Seq data, and the resulting transcript models were mapped to human, mouse, platypus, and sheep GPIHBP1 sequences and to other chicken Ly6 proteins with Blastx. All hits were manually examined for GPIHBP1 sequence motifs. We defined the GPIHBP1 sequence motifs by several conserved features in mammalian GPIHBP1, as outlined in the Methods. No chicken transcript contained these GPIHBP1 hallmarks. The short-read RNA-seq data set represented a high-depth search of the mapped exome. Thus, the failure to find candidate sequences for a chicken GPIHBP1 strongly suggests that a gene for GPIHBP1 is not present in the Gallus gallus genome assembly 4.0. The Iso-Seq data set allowed us to identify unmapped chicken transcripts (i.e., transcripts not present in the Gallus gallus genome assembly 4.0). Again, no transcripts encoding a chicken GPIHBP1 were identified (i.e., no transcripts encoding a Ly6 protein with an acidic domain; no transcripts resembling human or platypus GPIHBP1; no Ly6 proteins with homology to the most conserved region of GPIHBP1).
Discussion
In mammals, GPIHBP1 plays 3 crucial roles in intravascular triglyceride metabolism — shuttling LPL to its site of action in the capillary lumen, facilitating the margination of lipoproteins along capillaries, and stabilizing the structure and activity of LPL (2–4). However, GPIHBP1 — the protein that is so essential for intravascular lipolysis in mammals — has never been identified in the chicken or any other lower vertebrate species. In the current study, our goal was to determine if LPL in chickens, despite an apparent absence of GPIHBP1, could reach the capillary lumen. The results were clear: LPL in chickens does reach the capillary lumen. First, by immunohistochemistry, LPL is present on capillaries of chicken heart and adipose tissue, colocalizing with vWF (an endothelial cell marker). Second, a goat IgG against cLPL, when injected intravenously into chickens, binds to LPL on the luminal surface of capillary endothelial cells, whereas a nonimmune chicken IgG does not. Third, LPL protein and catalytic activity are released rapidly from isolated chicken hearts after an infusion of heparin, mirroring results obtained with isolated hearts of wild-type mice (where the LPL is unequivocally intravascular). When we performed studies on isolated hearts of Gpihbp1-deficient mice (where the LPL is extravascular), the release of LPL by heparin was substantially delayed. Finally, we found that cLPL enters the bloodstream rapidly after an intravenous injection of heparin, consistent with LPL being located within capillaries. In earlier studies of wild-type mice (2), we found that LPL enters the plasma rapidly after an intravenous injection of heparin, whereas the entry of LPL into plasma was substantially delayed in Gpihbp1-deficient mice (33).
Finding LPL within chicken capillaries implies that plasma triglyceride metabolism is similar in mammals and birds, with LPL working inside capillaries to carry out the lipolytic processing of TRLs.
While we are confident that LPL is primarily intravascular in the chicken (and presumably other avian species), we would caution against extrapolating the findings in chickens to other lower vertebrates (e.g., reptiles, amphibians, and fish). In those species, the processing of TRLs may be significantly different. Of note, the plasma triglyceride levels in Burmese pythons increase 50- to 150-fold after a meal and remain elevated for several days (34), implying that TRL processing by LPL, if it occurs intravascularly in that species, may be rather inefficient.
What explains intravascular LPL in chickens? One possibility is that chickens actually have a bona fide GPIHBP1, but this gene has not yet been uncovered, either because of gaps in genomic DNA sequences or incomplete annotation. Interestingly, the chicken gene for APOC2, an LPL cofactor, has never appeared in the genomic DNA databases, despite the fact that chicken APOC2 was identified and characterized more than 20 years ago (35). Notwithstanding the APOC2 example, we are skeptical that the apparent absence of GPIHBP1 in the chicken genomic DNA databases is due to incomplete sequences or annotation, simply because GPIHBP1 has not been identified in any lower vertebrate. Also, after examining the sequences for more than 3 billion RNA sequence tags, no transcripts for GPIHBP1 were found (i.e., no Ly6 proteins with an acidic domain; no Ly6 proteins with strong similarity to mammalian GPIHBP1). Given that GPIHBP1 should be expressed at a relatively high level in at least one of the tissue types sequenced, it seems unlikely that a GPIHBP1 transcript would evade the depth of the RNA-seq efforts. However, the results from the short-read RNA-seq data set include only genes present in the Gallus gallus genome assembly 4.0, and it is possible that the GPIHBP1 locus is simply missing from that assembly. The absence of GPIHBP1 motifs in the long-read Iso-Seq data (where unmapped transcripts were assessed) provides strong support for the notion that GPIHBP1 is absent in the chicken, particularly since these data included transcripts from embryonic chicken heart at several different HH stages. While it is still conceivable that a chicken GPIHBP1 might evade these sequence-based searches, we believe that it is rather unlikely and that our results favor the view that chickens do not have GPIHBP1 and instead express a distinct protein for transporting LPL into capillaries.
The stretch of chicken chromosome 2 syntenic to the GPIHBP1 locus on human chromosome 8 encodes 4 Ly6E-like proteins of unknown function. None of those proteins bound cLPL. The absence of LPL binding was not particularly surprising because the sequences of the 4 Ly6E-like proteins did not resemble the sequences for human or platypus GPIHBP1 (including the most highly conserved segment of GPIHBP1). Indeed, the sequences for the 4 Ly6E-like proteins were no more similar to GPIHBP1 than to SLURP1, a protein involved in keratinocyte differentiation (32). Also, the expression patterns of the 4 Ly6E-like proteins, as judged by RNA-seq, appear to be inconsistent with a role for those proteins in triglyceride metabolism. For example, none were expressed at high levels in the heart; ENSGALG00000041621 was expressed in the central nervous system; ENSGALG00000039585 and ENSGALG00000045170 were expressed at high levels in the skin; and ENSGALG00000043582 was expressed at high levels in the trachea.
It is possible that another protein, completely unrelated to GPIHBP1, transports LPL to the capillary lumen in chickens, perhaps by interacting with sequences not present in mammalian LPL proteins. cLPL contains a 15-residue carboxyl-terminal extension enriched in positively charged residues (Supplemental Table 3). That extension is highly conserved among other avian species (Supplemental Table 3). Perhaps another transporter binds those sequences and mediates the transport of LPL across capillaries. Another possibility is that the positively charged extension augments binding to endothelial cell heparan sulfate proteoglycans (HSPGs), perhaps including GPI-anchored glypicans. The possible involvement of HSPGs in LPL transport has been proposed previously (36). In our studies, perfusing chicken hearts with PIPLC released cLPL protein and activity. That observation implies that some or all of the LPL on chicken capillaries could be bound to a GPI-anchored protein, but caution is warranted. We know that perfusing isolated mouse hearts with large amounts of PIPLC can result in lysis of endothelial cells and adjacent cardiomyocytes. In our chicken experiments, we used low amounts of PIPLC and observed no visual evidence of myocardial damage, but we cannot exclude the possibility that some or all of the LPL released by PIPLC originated from parenchymal cells.
cLPL binds avidly to mouse and human GPIHBP1 but not to mutant versions of human or mouse GPIHBP1 lacking the capacity to bind LPL (e.g., human GPIHBP1-W109S, mouse GPIHBP1-W108S) (23, 24). Also, cLPL-C420Y could not bind to human or mouse GPIHBP1. These findings suggest that the interaction between cLPL and GPIHBP1 is rather specific. One might be tempted to infer from these observations that GPIHBP1 must be present in chickens, but we doubt that any such inference is justified. We suspect that the structure of LPL was conserved during vertebrate evolution based on the protein’s ability to hydrolyze triglycerides. We further suspect that GPIHBP1 evolved to fit this highly conserved structure. According to this viewpoint, it would not be particularly surprising that cLPL would retain some ability to bind to GPIHBP1, nor would it be surprising that amino acid substitutions that disrupt the binding of human LPL to GPIHBP1 would also disrupt the binding of cLPL to GPIHBP1. The notion that GPIHBP1 in mammals evolved to fit an ancient and highly conserved enzyme is not farfetched. The estrogen receptor appeared early in animal evolution as a protein that activates gene transcription in response to a specific sterol (estrogen). During vertebrate evolution, additional sterol receptors evolved to fit the structures of sterols that had always been present as intermediates in the synthesis of estrogen (37). Also, it is noteworthy that mammalian apo-E binds avidly to a member of the LDL receptor family in chickens (the oocyte receptor for very low density lipoproteins) (38), despite the fact that chickens do not produce apo-E.
Given that LPL is capable of reaching the capillary lumen in chickens, what prompted the appearance of GPIHBP1 in mammals? We do not have a clear answer, but we speculate that intravascular lipolysis in mammals needed to be particularly efficient because mammals — unlike avian species — nurse their young. In mammals, intravascular TRL processing by LPL is crucial for the production of milk fat by the mammary gland (39, 40). We suspect that GPIHBP1 appeared in mammals to enhance the activity and stability of intravascular LPL and to increase the capacity of vital tissues to extract lipid nutrients from the plasma lipoproteins. Recent biochemical studies have proven that GPIHBP1, and in particular the acidic domain of GPIHBP1, stabilizes LPL’s hydrolase domain, limiting inactivation of the enzyme (4). Also, LPL binding to GPIHBP1 protects LPL from inactivation by ANGPTL4 (41). GPIHBP1’s ability to stabilize LPL likely increased the capacity of many tissues, including the mammary gland, to extract lipid nutrients from the plasma compartment.
Methods
Goat antibodies against cLPL.
cLPL purified from chicken adipose tissue was used to immunize a goat (30). Goat immunoglobulins against cLPL were first purified from the goat antiserum on a Sepharose–cLPL immunoaffinity column (31). The immunopurified goat IgG was further purified on a second immunoaffinity column generated with a His-tagged cLPL polypeptide from E. coli. To produce His-tagged cLPL in E. coli, the coding sequences of mature cLPL were amplified from a chicken cDNA clone with Primestar GXL polymerase (Clontech) and subcloned into the pMAPLe3 expression vector (42, 43). The plasmid was transformed into E. coli BL21 (DE3) and Rosetta (DE3). Once the culture reached an OD600 of approximately 1.0, the expression of cLPL was induced with 1 mM IPTG. After 4 hours, cells were harvested by centrifugation, and the cell pellets were resuspended in lysis buffer (50 mM Tris pH 8.0, 300 mM NaCl, 20 mM imidazole, 8 M urea). The cells were lysed using an Emulsiflex C-3 (Avestin), and the lysate was clarified by centrifugation. The supernatant fluid was loaded onto a Ni-NTA column; the column was washed with lysis buffer, and the bound protein eluted with lysis buffer containing 300 mM imidazole. The eluted proteins were concentrated and further purified by size-exclusion chromatography on a Superdex 200 column (GE Healthcare) equilibrated with 50 mM Tris (pH 8.0), 300 mM NaCl, and 8 M urea. The fractions containing cLPL were pooled, dialyzed against 50 mM Tris (pH 8.0) containing 8 M urea, loaded onto a Q-Sepharose column equilibrated with dialysis buffer, and then eluted with a 0–1 M NaCl gradient. cLPL eluted at approximately 300 mM NaCl. Fractions containing cLPL were pooled and concentrated to approximately 5 mg/ml. cLPL was then dialyzed against PBS with 4 M urea and coupled to AminoLink Coupling Resin (ThermoFisher Scientific). The cLPL–resin was used as a second immunopurification step to purify goat anti-cLPL antibodies. Briefly, the goat anti-chicken antibodies from the Sepharose–cLPL immunoaffinity column were added to the cLPL–resin affinity column. After washing with PBS, goat IgGs were eluted with 0.2 M glycine·HCl (pH 2.7). The resulting IgG fraction against cLPL was then neutralized with 1 M phosphate (pH 9).
Detecting cLPL in extracts of chicken tissues by Western blotting.
Adipose tissue, liver, brain, gizzard, heart, white skeletal muscle, and red skeletal muscle were harvested from 9-day-old chickens (approved by UCLA’s Animal Research Committee). Tissue samples (~2 mm3) were placed in 1.0 ml of ice-cold RIPA buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% NP40 [vol/vol], 0.25% sodium deoxycholate, 1 mM PMSF, and 1 mM NaF) containing a protease inhibitor cocktail (cOmplete ULTRA EDTA-free, Roche) and homogenized with a motorized homogenizer on ice. The lysate was centrifuged at 18,000 g in a microcentrifuge at 4°C, and the supernatant was collected for analysis. Tissue extracts (30 μg protein) and recombinant cLPL from E. coli were electrophoresed in 12% Bis-Tris SDS-PAGE gels in MES buffer (ThermoFisher Scientific). Size-fractioned proteins were transferred to a nitrocellulose membrane, and Western blots were performed with anti-chicken goat antibodies purified over a Sepharose–cLPL immunoaffinity column (1.75 μg/ml) or the same immunopurified antibodies that had been purified again with the immunoaffinity column generated with E. coli–expressed cLPL (1.75 μg/ml). Binding of goat IgGs was detected with an IRDye800-conjugated donkey anti–goat IgG (LI-COR; 1:2,000). Antibody binding was detected with an Odyssey infrared scanner (LI-COR).
Detecting cLPL in CHO cells that had been transfected with V5-tagged cLPL.
A modified version of CHO pgsA-745 cells (in which the small amounts of hamster LPL expression were eliminated by CRISPR/Cas9 genome editing) were electroporated with a plasmid encoding V5-tagged cLPL (2 μg DNA for 2 × 106 cells). After 24 hours, the cells were washed with PBS/Ca/Mg, fixed in methanol at –20°C for 10 minutes or with 3% paraformaldehyde (PFA) at room temperature for 15 minutes, permeabilized with 0.2% Triton X-100, and blocked with 10% donkey serum. Cells were then incubated for 1 hour at room temperature with the immunopurified goat anti-cLPL antibody (5 μg/ml) and a mouse anti–V5 tag antibody (ThermoFisher Scientific; 11.6 μg/ml), followed by a 30-minute incubation at room temperature with an Alexa Fluor 555–conjugated donkey anti-goat IgG (ThermoFisher Scientific; 1:800) and an Alexa Fluor 488–conjugated donkey anti-mouse IgG (ThermoFisher Scientific; 1:800). All antibodies were diluted in 3% donkey serum in PBS. After the staining of cells, they were fixed with 3% PFA at room temperature for 15 minutes. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI). Images were recorded with an LSM700 confocal fluorescence microscope (Zeiss) and processed with Zen 2010 software (Zeiss). Microscope exposure conditions were identical within each experiment.
Detecting cLPL in tissues by immunohistochemistry.
A 9-day-old chicken was anesthetized and perfused through the heart with 50 ml of PBS. WAT and heart were collected, rinsed in PBS, embedded in OCT compound (Tissue Tek), and frozen sections prepared (50-μm sections for WAT; 10-μm sections for heart). Sections were fixed in methanol at –20°C for 10 minutes, permeabilized with 0.2% Triton X-100 for 5 minutes, and incubated with a blocking buffer containing 10% FBS (HyClone) and 0.2% BSA in PBS/Ca/Mg. Tissues were incubated overnight at 4°C with a rabbit polyclonal antibody against human vWF (Dako; 1:200), followed by a 1-hour incubation with Alexa Fluor 555–labeled goat anti-cLPL IgG (5 μg/ml) and an Alexa Fluor 488–labeled donkey anti-rabbit IgG (ThermoFisher Scientific; 1:1,000). After washing with PBS, tissues were fixed with 3% PFA in PBS for 5 minutes at room temperature. DNA was stained with DAPI. Immunofluorescence microscopy was performed as described above. Microscope exposure conditions were identical for each tissue.
Detecting LPL in capillaries of chicken tissues.
A 9-day-old chicken was anesthetized and injected intravenously through the brachial vein with a solution containing 73 μl of Alexa Fluor 555–labeled goat anti-cLPL IgG (9.5 mg/ml), 50 μl of fluorescein-labeled Lens culinaris agglutinin (lectin, 10 mg/ml; Vector Laboratories), and 70 μl of Alexa Fluor 647–labeled nonimmune goat IgG (10 mg/ml). After 4 minutes, the chicken was perfused through the heart with 50 ml of PBS, followed by 30 ml of PBS containing 3% PFA. WAT, heart, liver, and cerebellum were harvested and fixed in 3% PFA at 4°C for 2 hours. Tissues were embedded in OCT compound, and 10-μm sections prepared. Immunofluorescence microscopy was performed as described above. Microscope exposures were identical within each experiment.
Release of cLPL from isolated chicken hearts with heparin or PIPLC.
Chickens between 9 and 21 days of age were anesthetized and perfused with 20 ml of Tyrode’s buffer (136 mM NaCl, 5.4 mM KCl, 0.33 mM NaH2PO4, 1 mM MgCl2, 10 mM Hepes, pH 7.4, and 10 mM glucose) containing 1 mM CaCl2. The hearts were removed, cannulated through the aorta, and perfused with 3 ml of Tyrode’s buffer. To release LPL from blood vessels, the hearts were perfused with 20 U/ml of heparin in 4 ml of Tyrode’s buffer (or buffer alone). The LPL in the perfusate (0.2 ml/fraction) was stabilized by diluting in DOC buffer (0.1 M Tris, pH 8.5, 0.1 mM SDS, and 5 mM deoxycholate) and stored at –80°C until analysis. LPL in the perfusate was documented by Western blotting or by measuring triglyceride hydrolase activity (44). For the activity measurements, 25 μl of the sample was added to 175 μl of substrate solution composed of Intralipid containing [3H]triolein and heat-inactivated rat serum (as a source of APOC2). In some studies, LPL activity was inhibited with 50 μl of the goat antiserum against cLPL. Released fatty acids were extracted and counted in a scintillation counter (45). In other studies, isolated mouse hearts were perfused with PIPLC (6 U/ml) (1) in 1 ml Tyrode’s buffer (or with buffer alone). Hearts were incubated at 37°C for 10 minutes and then perfused with 2 ml of Tyrode’s buffer. Fractions (0.3 ml) were collected, and LPL mass and activity were measured (1).
Releasing LPL from chickens with an intravenous injection of heparin.
Young male (0.5–1.0 kg) or female chickens (0.7–0.9 kg) were anesthetized, and the brachial vein was cannulated with a 20-gauge catheter. Chickens were injected intravenously with heparin (2 U/g body weight). Blood samples (1 ml) were drawn from the contralateral brachial vein at baseline and 1, 3, 8, 15, and 30 minutes after the injection of heparin. LPL activity and mass measurements were assessed as described (46, 47).
Assessing binding of cLPL to GPIHBP1.
CHO pgsA-745 cells (2 × 106) were electroporated with 2 μg of expression vector encoding S-protein–tagged versions of mouse or human GPIHBP1 or with V5-tagged versions of human, mouse, or cLPL. The GPIHBP1-transfected cells were then coplated with the LPL-transfected cells on coverslips in 24-well plates at a ratio of 1:2 (a total of 1 × 106 cells/well). After 48 hours, cells were washed with PBS/Ca/Mg, fixed with 3% PFA, and blocked with 10% donkey serum (vol/vol). In some studies, the cells were permeabilized with 0.2% Triton X-100 (vol/vol) after PFA fixation. Cells were then incubated overnight at 4°C with a goat polyclonal antibody against the S-protein tag (Abcam; 1:800) followed by an Alexa Fluor 568–conjugated donkey anti-goat IgG (ThermoFisher Scientific; 1:800) and a mouse monoclonal antibody against the V5 tag (ThermoFisher Scientific; 11.6 μg/ml) followed by an Alexa Fluor 488–conjugated donkey anti-mouse IgG (ThermoFisher Scientific; 1:800). All antibodies were diluted in 3% donkey serum in PBS. Cells were washed 3 times with 3% donkey serum in PBS and 2 times with PBS before being fixed with 3% PFA. DNA was stained with DAPI. Images were recorded and processed as described above; microscope exposures were identical within each experiment.
Expression vectors for chicken Ly6E-like proteins.
The open reading frames for ENSGALG00000039585, ENSGALG00000041621, ENSGALG00000043582, and ENSGALG00000045170 were prepared by RT-PCR from a chicken adipose tissue cDNA library with oligo(dT), random primers, and SuperScript III (ThermoFisher Scientific). PCR fragments were initially introduced into the pGEM-T Easy Vector (Promega). The sequences for the mature proteins were amplified and subcloned into the pTriEx-4 mammalian expression vector (Novagen) as described for expression vectors for human GPIHBP1 or human CD59 (48). In addition, we also introduced the sequences for the Ly6 domain of ENSGALG00000045170 in frame with the acidic domain of human GPIHBP1 in the pTriEx-4 vector. All vectors contained an amino-terminal S-protein tag. The integrity of the expression vectors was verified by DNA sequencing.
Chicken transcriptome sequencing search.
Data were acquired from 2 chicken transcriptome sequencing projects (49, 50). The data comprise Pacific Biosciences Iso-Seq sequencing of chicken brain (PRJEB13246), whole embryo (HH stage 26, PRJEB13248), and embryonic heart (pooled HH stages 18–20, 25, and 32; SRR1177086) samples along with Illumina short-read RNA-seq for 20 tissues of adult female J-line chickens (PRJEB12891). For the Iso-Seq data, mapped and unmapped transcript models were examined with Blastx for potential GPIHBP1 transcripts, known Ly6 protein transcripts, and LPL transcripts. For the GPIHBP1 Blastx runs, we compared human, mouse, platypus, and sheep GPIHBP1 amino acid sequences to chicken transcripts. For the Ly6 protein runs, we examined chicken transcripts for the Ly6 proteins present in the Uniprot database. For the LPL runs, we used the annotated cLPL amino acid sequence. For the short-read RNA-seq data, transcript models were assembled with Cufflinks as described in the chicken transcriptome sequencing project (49) and examined with identical Blastx protocols. Mapped and unmapped chicken transcript models with hits from the GPIHBP1 and Ly6 Blastx runs were analyzed for 3 conserved features of mammalian GPIHBP1. The first is a characteristic pattern of cysteines in all mammalian GPIHBP1 proteins (CXXXXXXXXXXXXXXXCCXXXXC). We allowed a range of 7 to 19 amino acids between the first and second cysteines, and a range of 3 to 5 amino acids between the third and fourth cysteines. The second is a region with more than 15 acidic residues (D or E) within a 26–amino acid window at the amino terminus of the protein. The third feature was a WCXXXC motif (allowing a range of 3 to 5 residues between the 2 cysteines).
Study approval.
All the animals used in this study were housed in a facility accredited by Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC International). All animal protocols are approved by the Chancellor’s Animal Research Committee (ARC). The ARC-approved protocols covering the work in our laboratory include 2015-097-11, 2004-127-12B, and 2015-097-03.
Author contributions
CH, LGF, and SGY designed the research. CH, LGF, XH, RSJ, ML, YT, SDV, PK, TRP, CMA, BR, NPS, A. Bensadoun, RLW, RIK, and A.P. Beigneux performed the research. CH, LGF, RSJ, ML, HJ, and SGY analyzed the data. SGY and CH wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH National Heart, Lung and Blood Institute (HL090553, HL087228, HL125335), the National Human Genome Research Institute (4U41HG002371), and a Transatlantic Network Grant from the Leducq Foundation (12CVD04). Cuiwen He was supported by a Ruth L. Kirschstein National Research Service Award (F32HL132471). Christopher M. Allan was supported by a Ruth L. Kirschstein National Research Service Award (T32HL69766). The recombinant cLPL described in this publication was prepared by the UCLA-DOE Institute Protein Expression Technology Center, which is supported by the U.S. Department of Energy, Office of Biological and Environmental Research (BER) program under Award Number DE-FC02-02ER63421. This work was also supported in part by Texas AgriLife Research project number 8738. We thank Mark Arbing for expressing cLPL in E. coli. We thank the UCLA Brain Research Institute and Marianne Cilluffo for help in preparing chicken tissue sections.
Version 1. 10/19/2017
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2017;2(20):e96783. https://doi.org/10.1172/jci.insight.96783.
Contributor Information
Cuiwen He, Email: cuiwenhe@mednet.ucla.edu.
Xuchen Hu, Email: XuchenHu@mednet.ucla.edu.
Mikael Larsson, Email: mikael.larsson@medbio.umu.se.
Yiping Tu, Email: ytu@mednet.ucla.edu.
Sandra Duarte-Vogel, Email: SandraDuarte@mednet.ucla.edu.
Paul Kim, Email: pxkimx@ucla.edu.
Brian Raney, Email: braney@soe.ucsc.edu.
Haibo Jiang, Email: jianghaibo89@gmail.com.
Stephen G. Young, Email: sgyoung@mednet.ucla.edu.
References
- 1.Beigneux AP, et al. Glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab. 2007;5(4):279–291. doi: 10.1016/j.cmet.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies BS, et al. GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell Metab. 2010;12(1):42–52. doi: 10.1016/j.cmet.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goulbourne CN, et al. The GPIHBP1-LPL complex is responsible for the margination of triglyceride-rich lipoproteins in capillaries. Cell Metab. 2014;19(5):849–860. doi: 10.1016/j.cmet.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mysling S, et al. The acidic domain of the endothelial membrane protein GPIHBP1 stabilizes lipoprotein lipase activity by preventing unfolding of its catalytic domain. Elife. 2016;5:e12095. doi: 10.7554/eLife.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ameis D, et al. Familial chylomicronemia (type I hyperlipoproteinemia) due to a single missense mutation in the lipoprotein lipase gene. J Clin Invest. 1991;87(4):1165–1170. doi: 10.1172/JCI115114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beigneux AP, et al. Chylomicronemia with a mutant GPIHBP1 (Q115P) that cannot bind lipoprotein lipase. Arterioscler Thromb Vasc Biol. 2009;29(6):956–962. doi: 10.1161/ATVBAHA.109.186577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charrière S, et al. GPIHBP1 C89F neomutation and hydrophobic C-terminal domain G175R mutation in two pedigrees with severe hyperchylomicronemia. J Clin Endocrinol Metab. 2011;96(10):E1675–E1679. doi: 10.1210/jc.2011-1444. [DOI] [PubMed] [Google Scholar]
- 8.Coca-Prieto I, et al. Childhood-onset chylomicronaemia with reduced plasma lipoprotein lipase activity and mass: identification of a novel GPIHBP1 mutation. J Intern Med. 2011;270(3):224–228. doi: 10.1111/j.1365-2796.2011.02361.x. [DOI] [PubMed] [Google Scholar]
- 9.Franssen R, et al. Chylomicronemia with low postheparin lipoprotein lipase levels in the setting of GPIHBP1 defects. Circ Cardiovasc Genet. 2010;3(2):169–178. doi: 10.1161/CIRCGENETICS.109.908905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olivecrona G, et al. Mutation of conserved cysteines in the Ly6 domain of GPIHBP1 in familial chylomicronemia. J Lipid Res. 2010;51(6):1535–1545. doi: 10.1194/jlr.M002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plengpanich W, et al. Multimerization of glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 (GPIHBP1) and familial chylomicronemia from a serine-to-cysteine substitution in GPIHBP1 Ly6 domain. J Biol Chem. 2014;289(28):19491–19499. doi: 10.1074/jbc.M114.558528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rios JJ, et al. Deletion of GPIHBP1 causing severe chylomicronemia. J Inherit Metab Dis. 2012;35(3):531–540. doi: 10.1007/s10545-011-9406-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Surendran RP, et al. Mutations in LPL, APOC2, APOA5, GPIHBP1 and LMF1 in patients with severe hypertriglyceridaemia. J Intern Med. 2012;272(2):185–196. doi: 10.1111/j.1365-2796.2012.02516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voss CV, et al. Mutations in lipoprotein lipase that block binding to the endothelial cell transporter GPIHBP1. Proc Natl Acad Sci USA. 2011;108(19):7980–7984. doi: 10.1073/pnas.1100992108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brunzell JD, Deeb SS. Familial lipoprotein lipase deficiency, apo C-II deficiency, and hepatic lipase deficiency. In: Scriver CR, Beaudet AL, et al., eds. The Metabolic and Molecular Bases of Inherited Disease. New York, USA:McGraw-Hill; 2001:2789–2816. [Google Scholar]
- 16.Havel RJ, Gordon RS. Idiopathic hyperlipemia: metabolic studies in an affected family. J Clin Invest. 1960;39:1777–1790. doi: 10.1172/JCI104202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayden MR, Ma Y, Brunzell J, Henderson HE. Genetic variants affecting human lipoprotein and hepatic lipases. Curr Opin Lipidol. 1991;2(2):104–109. doi: 10.1097/00041433-199104000-00008. [DOI] [Google Scholar]
- 18.Fojo SS, Brewer HB. Hypertriglyceridaemia due to genetic defects in lipoprotein lipase and apolipoprotein C-II. J Intern Med. 1992;231(6):669–677. doi: 10.1111/j.1365-2796.1992.tb01256.x. [DOI] [PubMed] [Google Scholar]
- 19.Henderson HE, Hassan F, Marais D, Hayden MR. A new mutation destroying disulphide bridging in the C-terminal domain of lipoprotein lipase. Biochem Biophys Res Commun. 1996;227(1):189–194. doi: 10.1006/bbrc.1996.1487. [DOI] [PubMed] [Google Scholar]
- 20.Loughner CL, Bruford EA, McAndrews MS, Delp EE, Swamynathan S, Swamynathan SK. Organization, evolution and functions of the human and mouse Ly6/uPAR family genes. Hum Genomics. 2016;10:10. doi: 10.1186/s40246-016-0074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fry BG, et al. Molecular evolution and phylogeny of elapid snake venom three-finger toxins. J Mol Evol. 2003;57(1):110–129. doi: 10.1007/s00239-003-2461-2. [DOI] [PubMed] [Google Scholar]
- 22.Ioka RX, et al. Expression cloning and characterization of a novel glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein, GPI-HBP1. J Biol Chem. 2003;278(9):7344–7349. doi: 10.1074/jbc.M211932200. [DOI] [PubMed] [Google Scholar]
- 23.Beigneux AP, et al. Assessing the role of the glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 (GPIHBP1) three-finger domain in binding lipoprotein lipase. J Biol Chem. 2011;286(22):19735–19743. doi: 10.1074/jbc.M111.242024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beigneux AP, et al. GPIHBP1 missense mutations often cause multimerization of GPIHBP1 and thereby prevent lipoprotein lipase binding. Circ Res. 2015;116(4):624–632. doi: 10.1161/CIRCRESAHA.116.305085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmes RS, Cox LA. Comparative studies of glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1: evidence for a eutherian mammalian origin for the GPIHBP1 gene from an LY6-like gene. 3 Biotech. 2012;2(1):37–52. doi: 10.1007/s13205-011-0026-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young SG, et al. GPIHBP1: an endothelial cell molecule important for the lipolytic processing of chylomicrons. Curr Opin Lipidol. 2007;18(4):389–396. doi: 10.1097/MOL.0b013e3281527914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holmes RS, Vandeberg JL, Cox LA. Comparative studies of vertebrate lipoprotein lipase: a key enzyme of very low density lipoprotein metabolism. Comp Biochem Physiol Part D Genomics Proteomics. 2011;6(2):224–234. doi: 10.1016/j.cbd.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kompiang IP, Bensadoun A, Yang MW. Effect of an anti-lipoprotein lipase serum on plasma triglyceride removal. J Lipid Res. 1976;17(5):498–505. [PubMed] [Google Scholar]
- 29.Fraser R, Heslop VR, Murray FE, Day WA. Ultrastructural studies of the portal transport of fat in chickens. Br J Exp Pathol. 1986;67(6):783–791. [PMC free article] [PubMed] [Google Scholar]
- 30.Bensadoun A, Hsu J, Hughes B. Large-scale lipoprotein lipase purification from adipose tissue. Methods Mol Biol. 1999;109:145–150. doi: 10.1385/1-59259-581-2:145. [DOI] [PubMed] [Google Scholar]
- 31.Bensadoun A, Kompiang IP. Role of lipoprotein lipase in plasma triglyceride removal. Fed Proc. 1979;38(12):2622–2626. [PubMed] [Google Scholar]
- 32.Favre B, et al. SLURP1 is a late marker of epidermal differentiation and is absent in Mal de Meleda. J Invest Dermatol. 2007;127(2):301–308. doi: 10.1038/sj.jid.5700551. [DOI] [PubMed] [Google Scholar]
- 33.Weinstein MM, et al. Abnormal patterns of lipoprotein lipase release into the plasma in GPIHBP1-deficient mice. J Biol Chem. 2008;283(50):34511–34518. doi: 10.1074/jbc.M806067200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riquelme CA, et al. Fatty acids identified in the Burmese python promote beneficial cardiac growth. Science. 2011;334(6055):528–531. doi: 10.1126/science.1210558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andersson Y, Nilsson S, Lindberg A, Thelander L, Olivecrona G. Apolipoprotein CII from chicken (Gallus domesticus). The amino-terminal domain is different from corresponding domains in mammals. J Biol Chem. 1996;271(51):33060–33066. doi: 10.1074/jbc.271.51.33060. [DOI] [PubMed] [Google Scholar]
- 36.Saxena U, Klein MG, Goldberg IJ. Transport of lipoprotein lipase across endothelial cells. Proc Natl Acad Sci USA. 1991;88(6):2254–2258. doi: 10.1073/pnas.88.6.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eick GN, Thornton JW. Evolution of steroid receptors from an estrogen-sensitive ancestral receptor. Mol Cell Endocrinol. 2011;334(1-2):31–38. doi: 10.1016/j.mce.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 38.Steyrer E, Barber DL, Schneider WJ. Evolution of lipoprotein receptors. The chicken oocyte receptor for very low density lipoprotein and vitellogenin binds the mammalian ligand apolipoprotein E. J Biol Chem. 1990;265(32):19575–19581. [PubMed] [Google Scholar]
- 39.Camps L, Reina M, Llobera M, Vilaró S, Olivecrona T. Lipoprotein lipase: cellular origin and functional distribution. Am J Physiol. 1990;258(4 Pt 1):C673–C681. doi: 10.1152/ajpcell.1990.258.4.C673. [DOI] [PubMed] [Google Scholar]
- 40.Goldberg IJ, Soprano DR, Wyatt ML, Vanni TM, Kirchgessner TG, Schotz MC. Localization of lipoprotein lipase mRNA in selected rat tissues. J Lipid Res. 1989;30(10):1569–1577. [PubMed] [Google Scholar]
- 41.Mysling S, et al. The angiopoietin-like protein ANGPTL4 catalyzes unfolding of the hydrolase domain in lipoprotein lipase and the endothelial membrane protein GPIHBP1 counteracts this unfolding. Elife. 2016:5. doi: 10.7554/eLife.20958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arbing MA, et al. Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions. PLoS ONE. 2013;8(11):e81753. doi: 10.1371/journal.pone.0081753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibson DG. Enzymatic assembly of overlapping DNA fragments. Meth Enzymol. 2011;498:349–361. doi: 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larsson M, Vorrsjö E, Talmud P, Lookene A, Olivecrona G. Apolipoproteins C-I and C-III inhibit lipoprotein lipase activity by displacement of the enzyme from lipid droplets. J Biol Chem. 2013;288(47):33997–34008. doi: 10.1074/jbc.M113.495366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bengtsson-Olivecrona G, Olivecrona T. Assay of lipoprotein lipase and hepatic lipase. In: Converse C, Skinner ER, eds. Lipoprotein Analysis. A Practical Approach. Oxford, UK:IRL; 1992:169–185. [Google Scholar]
- 46.Hocquette JF, Graulet B, Olivecrona T. Lipoprotein lipase activity and mRNA levels in bovine tissues. Comp Biochem Physiol B, Biochem Mol Biol. 1998;121(2):201–212. doi: 10.1016/S0305-0491(98)10090-1. [DOI] [PubMed] [Google Scholar]
- 47.Cisar LA, Hoogewerf AJ, Cupp M, Rapport CA, Bensadoun A. Secretion and degradation of lipoprotein lipase in cultured adipocytes. Binding of lipoprotein lipase to membrane heparan sulfate proteoglycans is necessary for degradation. J Biol Chem. 1989;264(3):1767–1774. [PubMed] [Google Scholar]
- 48.Gin P, et al. The acidic domain of GPIHBP1 is important for the binding of lipoprotein lipase and chylomicrons. J Biol Chem. 2008;283(43):29554–29562. doi: 10.1074/jbc.M802579200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuo RI, Tseng E, Eory L, Paton IR, Archibald AL, Burt DW. Normalized long read RNA sequencing in chicken reveals transcriptome complexity similar to human. BMC Genomics. 2017;18(1):323. doi: 10.1186/s12864-017-3691-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas S, Underwood JG, Tseng E, Holloway AK, Bench To Basinet CvDC Informatics Subcommittee Long-read sequencing of chicken transcripts and identification of new transcript isoforms. PLoS ONE. 2014;9(4):e94650. doi: 10.1371/journal.pone.0094650. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.