Abstract
Zinc oxide nanoparticles (ZnO-NPs) are attractive as broad-spectrum antibiotics, however, their further engineering as antimicrobial agents and clinical translation is impeded by controversial data about their mechanism of activity. It is commonly reported that ZnO-NP’s antimicrobial activity is associated with the production of reactive oxygen species (ROS). Here we disprove this concept by comparing the antibacterial potency of ZnO-NPs and their capacity to generate ROS with hydrogen peroxide (H2O2). Then, using gene transcription microarray analysis, we provide evidence for a novel toxicity mechanism. Exposure to ZnO-NPs resulted in over three-log reduction in colonies of methicillin resistant S. aureus with minimal increase in ROS or lipid peroxidation. The amount of ROS required for the same amount of killing by H2O2 was much greater than that generated by ZnO-NPs. In contrast to H2O2, ZnO-NP mediated killing was not mitigated by the antioxidant, N-acetylcysteine. ZnO-NPs caused significant up-regulation of pyrimidine biosynthesis and carbohydrate degradation. Simultaneously, amino acid synthesis in S. aureus was significantly down-regulated indicating a complex mechanism of antimicrobial action involving multiple metabolic pathways. The results of this study point to the importance of specific experimental controls in the interpretation of antimicrobial mechanistic studies and the need for targeted molecular mechanism studies. Continued investigation on the antibacterial mechanisms of biomimetic ZnO-NPs is essential for future clinical translation.
Graphical abstract
1. Introduction
The first antibiotic, penicillin, was discovered in 1928. Since then we have seen a tremendous improvement in global health and longevity. Unfortunately, emerging antimicrobial resistance is becoming one of the greatest threats to global human health. Less than two decades after his discovering of penicillin, Alexander Fleming noted that microbes become “educated” to resist penicillin1. The time from development of a new class of antibiotics to the detection of resistance has been consistently decreasing1. As more and more pathogenic bacteria develop resistance to multiple classes of antibiotics, previously treatable illnesses will become lethal.
Nanoscale engineering of surfaces and particles provides new paths for antibacterial drug design2, 3 that augment traditional tools of organic chemistry. Multiple inorganic metal oxide nanomaterials, exemplified by Ag2O, CuO, MgO, TiO2, and ZnO nanoparticles (NPs)4–9, inhibit the growth of Escherichia coli and Staphylococcus aureus6, 10–13. Among these, ZnO appears to be one of the most promising for clinical translation due to its high potency against antibiotic resistant strains14, low toxicity to mammalian cells6, 15, 16, and low cost. Besides antimicrobial activity in dispersion, surfaces coated with ZnO-NPs4, 17, for instance layer-by-layer assembled (LbL) coatings,18, 19 suppress biofilm formation of common bacterial pathogens20 as well. Staphylococcal adhesion and biofilm growth can be reduced by as much as 95% for NPs of specific shapes20. These findings motivate in-depth look into the fundamental mechanism of action for ZnO-NPs. Such knowledge is necessary for conceptual guidance of nanoscale engineering of effective and safe antimicrobials as well as the eventual translation of ZnO-NPs from laboratory phenomena to clinical reality.
A prevailing hypothesis is that ZnO-NPs spontaneously produce reactive oxygen species (ROS) and enhance intracellular oxidative stress in bacteria leading to their death10, 12, 17, 21–24. ROS include hydroxyl radicals OH•, superoxide ions O2−•, H2O2, and hydroperoxyl radicals10, 23. ROS are proposed to cause membrane damage by lipid peroxidation, triggering cell leakage, loss of energy metabolism, and cell death5, 17, 21. While being plausible and widely reported, this hypothesis requires closer evaluation. The factual support of oxidative stress as the antimicrobial mechanism in case of ZnO-NPs and other nanoscale antimicrobials is controversial. In many studies, ZnO-NPs were first excited by ultraviolet (UV) light and then exposed to bacteria for ROS detection25, 26. It is well-known that UV excitation creates oxygen vacancies and ROS on the semiconductor surfaces and therefore antibacterial activity can be associated with UV radiation25. At the same time, several studies suggested significant antibacterial effects without UV illumination27, 28 however, strong ROS generating positive controls have not been used in parallel to evaluate the respective NP ROS generating capacity. Notably, many of ZnO-NP antibacterial experiments were conducted on nonpathogenic, laboratory strains that are devoid of multiple defense/survival mechanisms (i.e., E. coli K12)21, 23 and thus elevated ROS concentrations can originate from their native metabolic state rather than from the NPs. Contrarily, pathogenic strains of S. aureus are known to have multiple mechanisms for scavenging ROS29–31; they can cope with large amounts of ROS even if NPs can inherently generate them. Finally, spontaneous production of ROS by ZnO-NPs would act indiscriminately and therefore have significant toxicity for mammalian cells. However, studies have demonstrated that ZnO-NPs have selective toxicity for prokaryotic cells over eukaryotic cells6, 16. Therefore, we sought to conclusively evaluate the oxidative stress hypothesis for ZnO-NP mediated antibacterial effect.
Despite the poor solubility of ZnO in aqueous media, many studies have attributed antibacterial activity to release of toxic Zn2+ ions32, 33. The basis of this toxicity is related to the specificity of certain essential proteins/enzymes for certain metal cofactors such as zinc34. Toxic levels of any of these metal ions lead to mis-metallation of these enzymes leading to loss of function and ultimately lysis. As with ROS, bacteria have evolved complex regulatory processes to maintain intracellular metal ion homeostasis through a wide range of environmental concentrations34. Given the low solubility of ZnO, the concentration of free Zn2+ ions is expected to be low in aqueous suspensions. However in the growth media, ZnO dissolution is complicated by Zn speciation with various components – amino acids and peptides – in the media32, 35, 36. Since most growth media is undefined soy digest products, it is difficult to predict ZnO dissolution kinetics and resulting complexes. In this work we consider dissolved Zn2+ as a potential mechanism. However, studies dedicated to evaluation of ZnO dissolution in a defined bacteria growth media are required for more focused description of the resulting complexes generated from dissolution and beyond the scope of this work. We chose S. aureus COL, a methicillin resistant strain, as our model organism because of its prevalence in human infections, virulence, ROS scavenging abilities, and clinical relevance as an antibiotic resistant organism. The ROS generating potential of two ZnO-NP preparations and the resulting effect on S. aureus were evaluated. Specifically, we examined a commercially available ZnO-NP previously reported to generate ROS25 and one synthesized in our laboratory known to inhibit planktonic and biofilm growth of methicillin resistant S. aureus (MRSA)20, 37. Oxidative stress was evaluated by multiple experimental techniques including fluorescent detection of ROS as well as resulting lipid peroxidation. Comparison was made with an equivalent degree of antibacterial activity by hydrogen peroxide (H2O2), which was chosen for its well-studied mechanism of ROS-mediated toxicity38, 39. The dissolution of Zn2+ ions or catalysis of otherwise undefined toxic but soluble species was also considered in this study. Finally, in the abscess of definitive evidence from these specific phenotypic studies, a comprehensive gene transcription microarray analysis was performed to provide new insight into the mechanism of ZnO-NP staphylococcal toxicity.
2. Materials and Methods
2.1. Bacterial strain, media, and growth conditions
Methicillin-resistant Staphylococcus aureus substrain COL was used in all experiments due to its strong ROS scavenging abilities29, 31. A glycerol stock of the strain was maintained at −80°C and was plated on tryptic soy agar. Single colony inoculates were grown in tryptic soy broth + 1% glucose w/v (TSBG) under shaking conditions at 37°C. Mid-log, optical density of 0.45–0.55 at 600nm (OD600), cultures were used in all experiments.
2.2. ZnO nanoparticles
ZnO nanopyramids with hexagonal base (ZnO-NPYs) synthesized in our laboratory and commercially available ZnO nanospheres (ZnO-NSPs) were investigated. Both of these NPs have been extensively characterized previously37, 40, which predicated our choice of the model nanoscale antimicrobials. ZnO-NPYs were synthesized without the use of surfactants or capping agents as previously described37, 40. Briefly, pyramids were synthesized by first mixing 0.2 g potassium hydroxide with 5.5 g Zn(Ac)2 · 2H2O before adding anhydrous methanol and refluxing for 48h. After synthesis, the white precipitate was purified by washing three times with methanol, and then frozen in Milli-Q® water to be lyophilized for 24 hours. ZnO-NSP were purchased from Nanostructured & Amorphous Materials, Inc (Houston, Texas)25.
2.3. ZnO-NP characterization
ZnO-NPs were characterized by transmission electron microscopy (TEM), selected area electron diffraction (SAED), photoluminescence spectroscopy (PL), fourier transformed infrared spectroscopy (FTIR), and nanoparticle tracking analysis (NTA). A JOEL 2010 transmission electron microscope was used for NP visualization. TEM samples were prepared by adding 10μL of 167μg/mL ZnO-NPs sonicated in Milli-Q® water solution to a carbon TEM grid and drying at room temperature. The nanocrystalline structure of ZnO-NPs was confirmed by SAED from TEM samples on the same microscope. NP surface chemistry was evaluated by PL (Jobin Yvon Horiba Fluoromax-3 instrument) and FTIR (JASCO FT/IR-4100 instrument). PL samples were prepared by sonicating 167ug/mL ZnO-NPs in Milli-Q® water for 3 mins. PL spectra of the aqueous dispersions were obtained using an excitation wavelength of 320nm at room temperature. Lyophilized ZnO-NP powder was used for FTIR with attenuated total reflection crystals. Particle aggregate distributions were obtained by NTA (Malvern NanoSight NS300) on aqueous dispersions prepared similar to PL samples.
2.4. ZnO-NP dose response and bacterial killing
To prepare NP dispersions for bacterial killing in planktonic conditions, ZnO-NP powder was sonicated into Milli-Q® water for 3 minutes to ensure a stable dispersion. Concentrations of ZnO-NPs included: 8, 80, 160, 400, and 800μg/mL. H2O2 was used as a positive control of a bacterial toxin whose mechanism of action involves the production of ROS. Concentrations of H2O2 included: 0.25, 0.50, 0.75, and 1.0 mM. These concentrations were chosen as they generated a similar degree of dose-dependent killing as the ZnO-NPs. Mid-log cultures of S. aureus grown under aerobic conditions were pelleted by centrifugation and resuspended in aqueous dispersions of ZnO-NPs or solutions of H2O2. After 30 minutes, samples were serially diluted and plated on tryptic soy agar for colony enumeration (CFUs). Actual exposure times included 30 minutes in planktonic dispersion followed by multiple 10-fold dilutions and 36 hours on agar plate. Therefore, the concentration of dispersed ZnO-NPs and dissolved H2O2 were quickly diluted below toxic concentrations after just 30 minutes. It is possible that serial dilutions would not reduce the local concentration of ZnO if the NPs are complexed with the cells. However, this is not dissimilar from H2O2 which can readily diffuse through the cell membrane and is immediately reacted to form other ROS (e.g., hydroxyl radicals) which are unable to diffuse back out and remain with the cell despite dilution of the media.
2.5 ROS detection
The presence of ROS was quantified by the fluorescence of 3′-(p-aminophenyl) fluorescein (APF; ThermoFisher Scientific, Waltham, Massachusetts) and 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA; ThermoFisher Scientific, Waltham, Massachusetts). Both dyes detect the presence of a variety of ROS including but not limited to peroxyl and hydroxyl radicals, peroxynitrite anion, and hypochlorite anion. The excitation/emission peaks for APF and H2DCFDA are 490/515 nm and 517/527 nm respectively. To evaluate for potential interactions between these dyes and ZnO-NPs we first evaluated their fluorescence in the presence of ZnO-NP dispersions and H2O2 solutions at the concentrations listed above without bacterial cells. Stock solutions of the dyes were made according to manufacturer’s instructions and were added to ZnO-NP or H2O2 samples and incubated for 30minutes. Samples were then pelleted and fluorescence reading of both the supernatants and resuspended pellets were obtained using Perkin Elmer Enspire Multimode Plate Reader. Comparing the pellets versus the supernatants provided information regarding interaction between the dye and the NPs which distribute to the pellet fraction.
For bacterial experiments, mid-log cultures grown under aerobic conditions were pelleted and resuspended in water, ZnO-NPs dispersions, or H2O2 at the concentrations listed above. Cells with fluorescent ROS detection dyes were exposed to NPs or H2O2 at 37°C for 30 minutes. The pellets represent intracellular ROS while the supernatants represent extracellular ROS or ROS spilled from lysed cells. Of note, there were no significant differences in the data trends for pellets or supernatants (see Supplementary Figures 1). For clarity only the data for the pellets are reported here. All samples were then serially diluted and plated on tryptic soy agar for CFU enumeration to provide a simultaneous measurement of bacterial cell death. This allowed us to correlate the amount of ROS in a sample with the degree of bacterial killing in that same sample.
In order to confirm that ZnO-NPs dispersions can indeed generate ROS via photocatalysis, APF and H2DCFDA fluorescence was also measured on ZnO-NP dispersions without bacteria but after UV excitation. These samples were exposed to either ambient light or 2 minutes UV excitation via a UV light box (Supplemental Figure 2).
2.6 Lipid peroxidation
Lipid peroxidation was evaluated using the thiobarbituric acid reactive substances assay (TBARS): OxiSelect™ TBARS Assay Kit (Cell Biolabs, San Diego, California). Mid-log phase cultures grown under aerobic conditions were pelleted and resuspended in ZnO-NPs dispersions or H2O2 at the concentrations listed above. After 30minutes, TBARS reagents were added and incubated at 95°C for 50 minutes. Samples were centrifuged and supernatant was collected for fluorescent spectroscopy measurements – 540nm excitation and 590nm emission. Just prior to addition of TBARS reagents, an aliquot of each sample was removed for serially dilution and CFU enumeration to allow for simultaneous measurement of lipid peroxidation and bacterial cell death for each sample. A malondialdehyde (MDA) standard curve (provided by the kit) was used to convert the fluorescence measurements into MDA equivalent concentrations (Supplementary Figure 3).
2.7. Bacterial growth recovery by antioxidant
Antioxidant recovery was evaluated by adding increasing concentrations of N-acetylcysteine, NAC, to the culture media with H2O2 or ZnO-NPs. NAC concentrations between 10mM and 70mM were evaluated to determine the maximum cell growth recovery in the presence of H2O2 without toxicity. A concentration of 50mM resulted in maximal recovery with low toxicity (see Supplementary Figure 4). Mid-log culture, 10ul, was exposed to 50mM NAC (final concentration) for 15 minutes. Then 1mL of 8, 80, 160, or 400ug/mL final concentrations of ZnO-NP dispersions or 0.25mM or 0.5mM H2O2 was added to the cells. Each sample, ~100uL, were then plated in a 96 well plate, and growth curves were measured turbidometrically (OD600) each hour for 10 hours under aerobic shaking conditions. To provide a simultaneous measurement of NP and H2O2 mediated killing, a 10ul aliquot of each sample after only 30 minutes exposure were serially diluted and plated on tryptic soy agar for CFU enumeration.
2.8. ZnO-NP supernatant growth curves
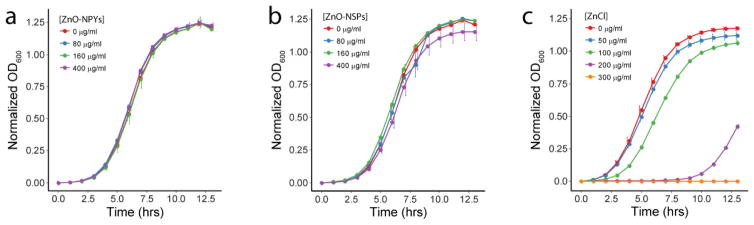
To evaluate for ZnO dissolution or catalysis of soluble toxic substances on the ZnO-NP surface, we evaluated the toxicity of media conditioned with ZnO-NPs. Briefly, ZnO-NP were sonicated into TSBG at concentrations of 0, 80, 160, and 400 μg/ml for 3 minutes and then incubated at 37°C for 30min. As a control, ZnCl2 was dissolved into TSBG at concentrations of 0, 50, 100, 200, and 300 μg/ml and treated similarly with sonication and incubation. The ZnO and ZnCl2 samples were then centrifuged at 14,000×g for 10min and the supernatants were removed. Those supernatants were then inoculated with ~ 5×107 CFU/ml in a 96-well plate, and growth curves were measured turbidometrically (OD600) each hour for 14h under aerobic shaking conditions.
2.9 Gene expression analysis
To evaluate changes in gene expression by S. aureus in response to ZnO-NPs, RNA was isolated from S. aureus after exposure to a sub-lethal but growth inhibitory concentration (160 μg/ml) of ZnO-NPYs (Supplemental Figure 5). S. aureus grown at the same time but not exposed to ZnO-NPYs were used as a control. A total of 3 ZnO-NP and 3 control samples were obtained and analyzed on independent microarray chips. Mid-log phase cells were lysed with lysostaphin followed by proteinase k digestion. Total RNA was isolated using Trizol (Invitrogen, Carlsbad, CA) followed by RNeasy column cleanup (Qiagen, Valencia, CA). RNA quantity and quality was confirmed on an Agilent 2100 Bioanalyzer before being applied to S. aureus whole genome microarray (GeneChip™, Affymetrix, Santa Clara, CA).
Microarray results were validated by performing quantitative RT-PCR on select genes identified to be significantly up or down regulated in the microarray analysis. Briefly, RNA was purified from S. aureus COL samples exposed to sub-lethal dose of ZnO-NPs, identical to microarray gene expression experiments. S. aureus grown in the same condition while not exposed to ZnO-NPs were used as controls. RNA was isolated through a lysostaphin digestion, TRizol extraction (Invitrogen, Carlsbad, CA), and then elution by RNeasy mini kit (Qiagen, Valencia, CA). RNA quality was checked using NanoDrop 2000 (ThermoFisher Scientific, Kalamazoo, MI) and RT-PCR was performed using QuantiFast SYBR Green RT-PCR Kit (Qiagen, Valencia, CA), and StepOnePlus Real-Time PCR System (Applied Biosystems, ThermoFisher Scientific, Kalamazoo, MI). Primers (Supplemental Table 1) were designed using National Center for Biotechnology Information database and ordered from Integrated DNA Technologies (Skokie, Illinois).
2.10. Statistical analyses
All data presented in this study are displayed as mean plus or minus standard error of the mean unless otherwise stated. To evaluate the role of oxidative stress on S. aureus toxicity we performed simultaneous measurements of ROS generation (by fluorescence of APF and H2DCFDA) or lipid peroxidation (by TBARS) and quantitative culture. Each measurement was performed in triplicate and at least 5 independent experiments were run on different days. We compared ROS content or lipid peroxidation associated with a greater than two-log reduction in CFUs for the two ZnO-NP preparations and H2O2. Hypothesis testing was performed via ANOVA with repeated measures. Post-hoc pairwise testing was performed using the Tukey test with p<0.05 indicating significance.
To evaluate the capacity of NAC to mitigate bacterial killing we compared the resulting CFUs after exposure to ZnO-NPs or H2O2 with and without the presence of NAC. Again ANOVA with repeated measures was used for hypothesis testing. To evaluate growth recovery by NAC, individual growth curves were summarized by calculating a growth rate constant using linear regression of the log-transformed optical density data. We then performed linear regression on the growth rate constants as a function of ZnO-NP dose with and without NAC. Hypothesis testing was performed using ANOVA of the resulting regression models.
For the microarray gene expression analysis, weighted linear models specific for microarray analysis41 were fit to the data to compare ZnO-NP treated samples to control. Specifically, we increased the power to detect differences by pooling information from all probesets to stabilize the variance estimate and samples were weighted on a gene-by-gene update algorithm42. We selected probesets with a fold change of 1.5 or greater and an adjusted p-value of 0.05 or less. The p-values were adjusted for multiple comparisons using false discovery rate of 5%43. Genes were then grouped by biological function. Finally, gene set enrichment analysis (GSEA) was used to organize genes by function, transcriptional regulation, and pathway associations (Biocyc Pathway/Genome database for S. aureus, www.biocyc.org) and identify significant biological functions or pathways altered in response to ZnO-NPs. Pathways with enrichment p-value of 0.05 or less were identified. The p-values were adjusted for multiple comparisons using a Bonferroni correction.
Fold change in gene expression between control and ZnO-NP treated bacteria was determined by quantitative RT-PCR using the delta-delta Ct method. Five separate pairs of control and test samples were performed for each gene of interest with triplicate reactions for each replicate. Student’s t-test was used to evaluate each gene for significant change in gene expression with the null hypothesis equal to zero change. Again p-values were adjusted for multiple comparisons.
3. Results
3.1. ZnO-NP characterization
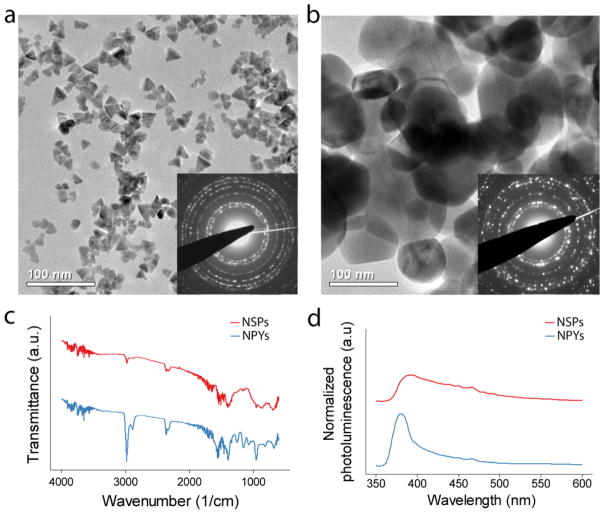
Although the two NPs are different in size and shape, their electron diffraction patterns were identical and matched the hexagonal crystalline phase of bulk ZnO (Figure 1a & b). Their average particle aggregation sizes, under the aqueous conditions used in this study, were also very similar (Supplementary Figure 6). The NPYs and NSPs also had similar FTIR patterns indicating little difference in surface chemistry (Figure 1c). The photoluminescence spectrum of both NP dispersions show only one emission peak, located at 390nm (Figure 1d). There are no other peaks observed, particularly around 520nm, which would indicate the presence of singly ionized oxygen vacancies. The NPY’s narrow luminescence peak is consistent with their small size and high crystallinity. Contrarily, the NSPs had a broader peak, corresponding to their larger size and broader spectrum of light-emitting states.
Figure 1.
Transmission electron micrographs of (a) ZnO-NPYs and (b) ZnO-NSPs. Insets show selected area electron diffraction. (c) Fourier transform infrared spectra and (d) photoluminescence spectra of corresponding ZnO-NPs. The spectra in (c & d) were shifted with respect to the ordinate for clarity.
3.2. Antimicrobial properties of ZnO-NPs
Bactericidal activities of both types of NPs and H2O2 against S. aureus demonstrate dose dependent killing in all cases (Figure 2). Exposure to ZnO-NP dispersions and H2O2 solutions resulted in greater than 1000x reduction in colony forming units (CFU) from control (5x107 CFUs of S. aureus without ZnO-NPs or H2O2 grown for 18h) for both shapes at 800μg/ml and H2O2 at 0.75mM. CFU reduction due to increasing H2O2 concentrations was similar to the potency of the ZnO-NPs. These concentrations of H2O2 were used as a positive control in the subsequent experiments.
Figure 2.
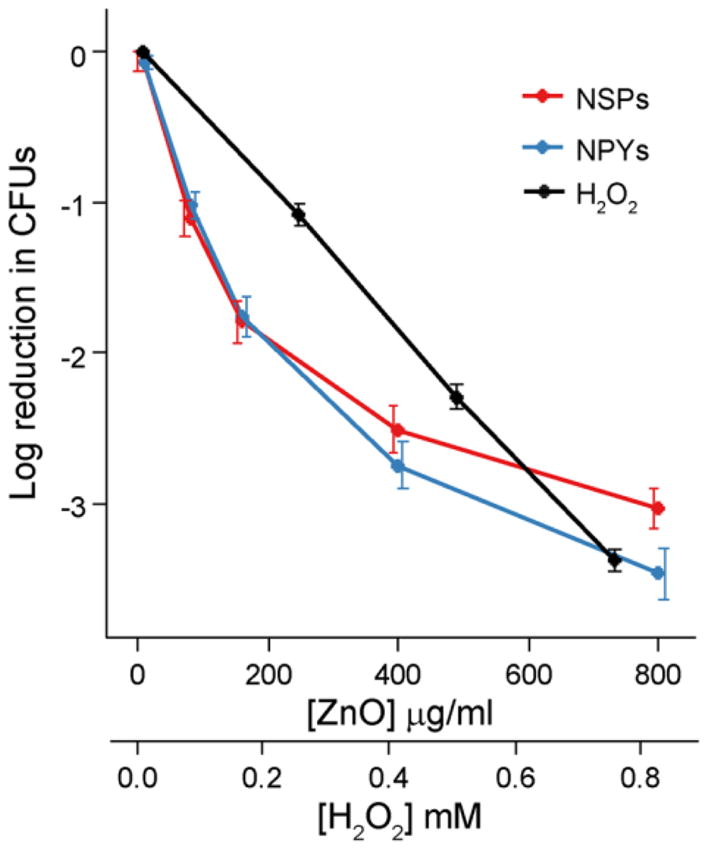
Reduction in colony counts (from 5×107) observed after exposure to increasing concentrations of ZnO-NPYs and NSPs as well as H2O2. Note the different scales of abscissa for ZnO-NPs and H2O2. Error bars represent standard error of five independent experiments run on different days.
3.3. ROS production in presence of ZnO-NPs
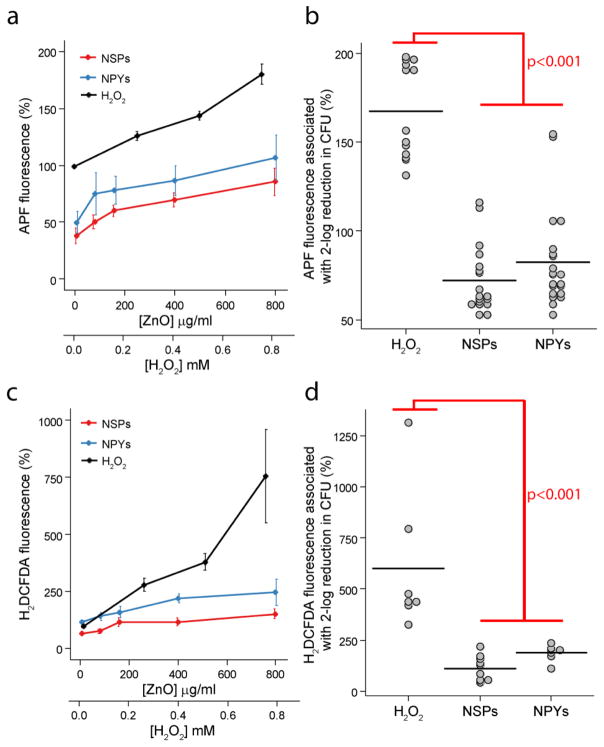
For understanding the potential for NPs to interact with a variety of fluorescent probes we first evaluated the fluorescence of APF and H2DCFDA in NP dispersions and H2O2 solutions without bacterial cells (Supplementary Figure 7). As expected, there is a dose dependent increase in fluorescence of both APF and H2DCFDA in the presence of H2O2. Likewise we observed dose dependent fluorescence of both ROS indicators in the NP dispersions. However, the absolute fluorescence of APF for low dose ZnO-NPs (80μg/ml) was slightly less than pure water. In addition, a large fraction of APF became bound to the ZnO-NPs as seen by the high fluorescence noted in the pellet fraction. Higher doses of ZnO-NPs (800μg/ml) resulted in only modest elevations in pellet fluorescence but significant increases in the supernatant. This indicates potential saturation of APF on the NPs. On the other hand, H2DCFDA appeared to have less affinity for the NPs with significantly less fluorescence in the pellet fraction. There were no significant differences between the NSPs and the NPYs. Although H2DCFDA appears to be a more reliable ROS indicator with respect to ZnO-NPs, we utilized both indicators in the subsequent studies.
To determine the baseline production of ROS by bacterial cells due to aerobic metabolism we obtained the fluorescence of APF and H2DCFDA generated by 5×107 S. aureus CFUs (the initial inoculum in all future experiments). There is a substantial increase in ROS production as indicated by the increase in APF and H2DCFDA fluorescence with the addition of bacteria (Supplemental Figure 8). It should be noted that the cells were resuspended in water (not growth media) to remove variation due to rapidly dividing cells. Given the high background ROS production by bacterial cells alone, as well as the significant sample to sample variation (note error bars), all subsequent measurements of ROS fluorescence were expressed as a percent change from a control (bacteria without H2O2 or ZnO-NPs) prepared from the same bacterial colony.
S. aureus exposure to H2O2 resulted in steady, dose-dependent increase in APF fluorescence (Figure 3a). For both ZnO-NP preparations, there was also a dose-dependent increase. However, the absolute quantity was always smaller than that of H2O2 as well as smaller than the paired controls (i.e., <100%). The difference between the two NP shapes was statistically insignificant. This decrease in APF below controls may indicate that ZnO-NPs reduce cellular metabolism and resulting metabolic ROS, although interaction with APF as seen in Supplementary Figure 8 is also possible. As such, the fluorescence of H2DCFDA was also used to detect small molecule ROS (Figure 3c). As expected, the presence of H2O2 resulted in a large dose-dependent increase in H2DCFDA fluorescence. Similarly, both ZnO-NPs had a dose-dependent increase in H2DCFDA fluorescence above control, with the effect from NPYs being modestly greater than NSPs. These findings alone might appear to confirm the initial ROS hypothesis. Indeed, ZnO-NSPs used in this study were previously shown to produce intracellular ROS25. However, these ROS can be produced in a dose-dependent manner as a side product. To be the cause of cell death, ROS from the NPs need to be produced in amounts comparable to a known ROS-based antibacterial agent, for instance H2O2, the gold standard in this field. Here, the absolute amount of ROS generated by bacteria exposed to ZnO-NPs was significantly smaller than those exposed to H2O2. To quantify the relationship between cell death and ROS production, we correlated the resulting CFU counts with the ROS fluorescence measurements. We then compared the APF and H2DCFDA fluorescence required for a 2-log or greater reduction in CFUs for H2O2 and the two ZnO-NP preparations (Figure 3b and d). H2O2 generated significantly greater amounts of ROS to kill the same amount of S. aureus as the ZnO-NPs (p<0.001). This finding leads to different conclusions about the mechanism than one might expect based on superficial evaluation of dose-dependent ROS generation.
Figure 3.
Dose and shape-dependent ROS production by (a) APF and (c) H2DCFDA fluorescence after exposure to ZnO-NPYs and -NSPs as well as H2O2. Note the different scales of abscissa for ZnO-NPs. Error bars represent standard error of five independent experiments run on different days. (b) Dot plots of APF and (d) H2DCFDA fluorescence associated with a two-log reduction in CFUs for each treatment. Black horizontal lines represent the mean value. p-values represent significant difference determine by ANOVA with repeated measures and post-hoc Tukey testing.
3.4. Lipid peroxidation in the presence of ZnO-NPs
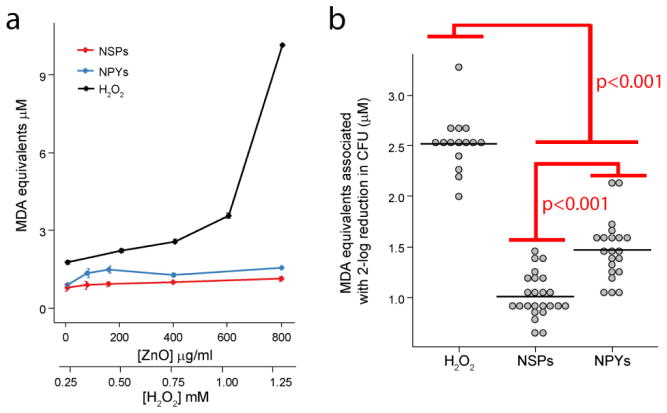
Although APF and H2DCFDA detect a wide range of small molecule ROS, many ROS are so reactive that their presence is transient and could be difficult to detect. A downstream result of intracellular oxidative stress is lipid peroxidation. To confirm ZnO-NPs were not producing a toxic but undetected ROS, we evaluated downstream lipid peroxidation. There was minimal to no increase in lipid peroxidation for all doses and shapes of ZnO-NPs tested (Figure 4a). On the other hand, the addition of H2O2 resulted in robust dose-dependent lipid peroxidation. Using a similar analysis as for small molecule ROS, we compared the degree of lipid peroxidation required for a greater than 2−log reduction in colonies. As before, we observed that H2O2 generated significantly more oxidative stress - lipid peroxidation in this case - for the same degree of bacterial eradication as ZnO-NPs (p<0.001, Figure 4b). NPYs generated higher levels of lipid peroxidation than NSPs although the absolute difference was small (p<0.001, Figure 4b).
Figure 4.
(a) Dose-dependent lipid peroxidation by ZnO-NPs and H2O2. Error bars represent standard error of five independent experiments run on different days. (b) Dot plots of lipid peroxidation associated with a two-log reduction in CFUs for each treatment. Black horizontal lines represent the mean. p-values denote significant differences determined by ANOVA with repeated measures and post-hoc Tukey testing.
3.5. Antioxidant recovery from oxidative stress
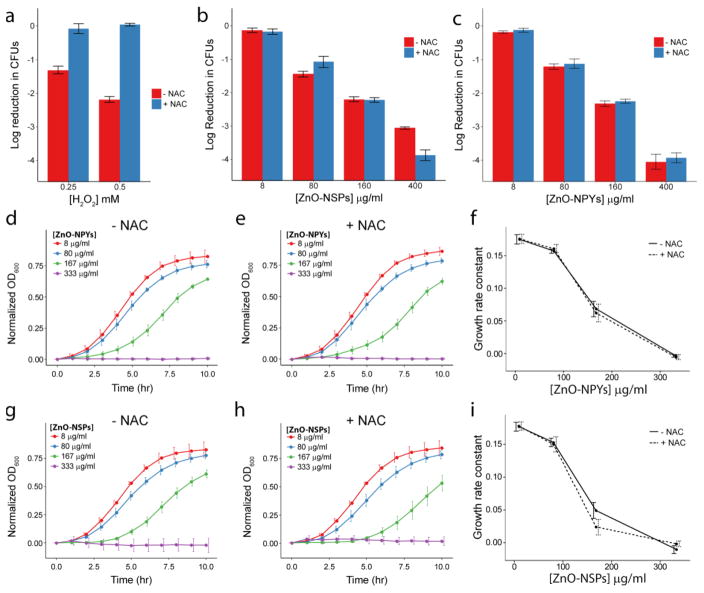
As a complementary test, we attempted to mitigate the oxidative stress to bacteria by adding the well-known antioxidant NAC to the culture media prior to the addition of ZnO-NPs or H2O2. The addition of NAC resulted in near complete recovery of bacterial colonies whose counts were dramatically reduced by H2O2 (Figure 5a). On the other hand, the addition of NAC did not mitigate the antibacterial activity of either ZnO-NP (Figure 5b and c). On the contrary, NAC improved the antibacterial effect of ZnO-NPs at 400ug/mL. This could be due to better stability of NPs at high concentrations in the presence of NAC, therefore being more effective. Alternatively, NAC can be toxic at high concentrations (See Supplemental Figure 4) and this particular concentration was synergistic with the ZnO-NPs. However, the key finding was that the presence of NAC did not recover any bacterial growth, as opposed to H2O2. The effect of NAC was similar for bacterial growth curves obtained in the presence of H2O2 and ZnO-NPs. For H2O2 almost 50% recovery was seen in the presence of NAC (Supplemental Figure 9). However, in the case of ZnO-NPs, growth curves and the resulting dose-dependent growth rates were unchanged by NAC (Figure 5d–i). The addition of NAC to NP treated cultures did not recover the S. aureus growth rate.
Figure 5.
Reduction in colony counts (from 5×107) observed after exposure to increasing concentrations of (a) H2O2, (b) ZnO-NSPs, or (c) ZnO-NPYs with and without 50 mM NAC. Growth curves of S. aureus in ZnO-NPYs dispersions of increasing concentration (d) without and (e) with NAC. Growth curves for S. aureus in ZnO-NSP dispersions (g) without and (h) with NAC. Growth rate constants as a function of (f) ZnO-NPY or (i) ZnO-NSP concentration. Error bars represent standard error of five independent experiments run on different days.
3.6. Toxicity of ZnO-NP conditioned media
The bacterial growth media (i.e., TSBG) was conditioned with ZnO-NPs as well as ZnCl2 to evaluate for any potential toxicity of dissolved Zn2+ ions or other undetermined soluble species catalyzed by the ZnO-NP surface. ZnO-NP conditioned media had minimal to no toxicity compared to ZnCl2 dissolved in media at concentrations consistent with maximum expected dissolution of ZnO (Figure 6). Although ZnCl2 has a different Zn2+ ion release profile at the selected concentrations, we demonstrate that the total amount of Zn ions incorporated into our NPs at highest concentration of 400ug/mL (which is relative to 300ug/mL ZnCl2) are less toxic to bacteria, thereby challenging the Zn ion toxicity mechanisms of ZnO-NPs against bacteria.
Figure 6.
Growth curves of S. aureus in the media condition with ZnO (a) NPY and (b) NSP dispersions as well as (c) ZnCl2. Error bars represent standard error of three independent experiments run on different days.
3.7. Gene expression changes in response to ZnO-NPs
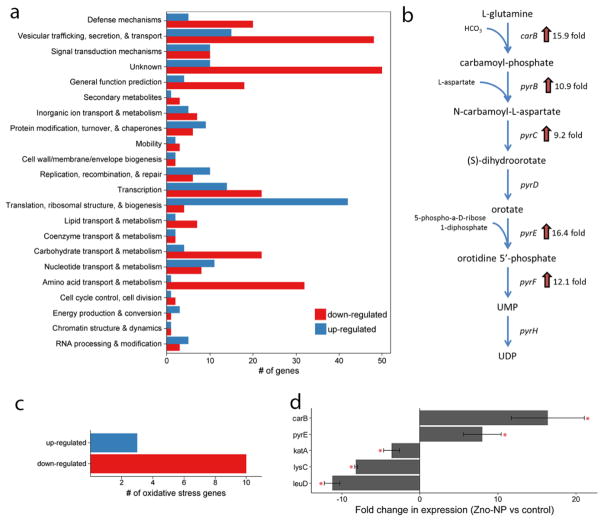
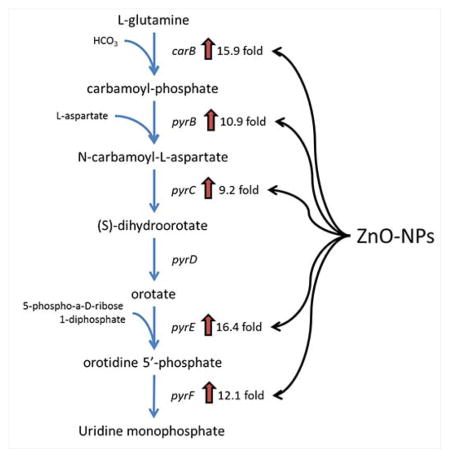
Over 800 different gene transcripts were identified as significantly changed in S. aureus exposed to ZnO-NPYs compared to controls. Of those, 556 had previously identified function. Of these, 181 genes were up-regulated and 375 were down-regulated (Supplemental Table 2). Functional classification of the genes with a significant change in expression is shown in Figure 7a. GSEA was performed to identify specific pathways or biological functions whose genes are significantly enriched in the ZnO-NP treated group (Supplemental Table 3). Specifically, ZnO-NPY treated S. aureus has up-regulation of pyrimidine biosynthesis and sugar degradation and down-regulation of amino acid synthesis. Most notably, 5 of the 10 most up-regulated genes belong to the uridine monophosphate (UMP) biosynthesis pathway (Figure 7b), a subpathway of the pyrimidine biosynthesis superpathway. Interestingly 10 of the 13 oxidative stress genes significantly altered in the microarray were down regulated upon ZnO-NPs exposure, and only 3 were upregulated (Figure 7c; Supplemental Table 4). The upregulated ROS genes experienced an average fold change of 1.91, which is not as strong as the UMP pathway, which experienced average fold changes above 10. To validate the results of the microarray experiments we selected two genes from the UMP biosynthesis pathway (carB and pyrE), two amino acid biosynthesis genes (lysC and leuD), and one oxidative stress gene (katA) for quantitative RT-PCR (Figure 7d). The results indeed confirm the upregulation of the UMP pathway and downregulation of amino acid synthesis and oxidative stress genes.
Figure 7.
(a) Functional classification of genes with significant change in transcription upon exposure to ZnO-NPs (b) UMP biosynthesis pathway with associated enzymes and their fold-change in expression after exposure to ZnO-NPs (c) Oxidative stress genes response to ZnO-NPs exposure (d) Quantitative RT-PCR of selected genes to validate microarray results. Error bars represent standard error for n=5 independent samples with triplicate reactions for each. Red star represents p<0.05.
4. Discussion
The work here not only addresses fundamental questions regarding the mechanism of action for ZnO-NP, but it also gives a new rationale for the clinical translation of ZnO-NPs. We demonstrate here that primary antibacterial mode of actions remains undiscovered between ZnO-NPs and bacteria. Literature suggests that the presence of ROS may lead to only sporadic cell death in bacterial biofilms, induce genetic variability, and possibly increase biofilm development44. Multiple studies emphasize bacteria’s ability to combat excessive ROS and how they incorporate excess ROS into many metabolic and respiratory pathways for survival24, 30, 31. Interestingly, moderate excess ROS may have unexpected consequences and increase the production of extracellular polymeric substances, accelerating the growth of biofilm. For instance, this behavior is noticed in Acinetobacter oleivorans45. Evolutionary biologists have speculated that microbial biofilm’s response to ROS may have evolved in response to increasing oxygen concentration on Earth, resulting in the integration of ROS into several different signaling pathways, including the switch between planktonic and biofilm forms30. Therefore, substances causing excessive ROS may not be the best choice to prevent biofilm growth and in particular for combating antibiotic resistant infections of indwelling medical devices. Since ZnO-NPs kill bacteria without inducing high level of oxidative stress, they can obviate ROS-inactivating pathways evolved in bacteria.
Furthermore, the presence of ROS extensively damages mammalian cells, where ROS react with polyunsaturated fatty acids in the lipid bilayer and promoting lipid peroxidation, and inactivate cellular surface proteins leading to loss of cell integrity46. NPs that have an ROS mediated bactericidal effect promote mammalian cell death as well. ZnO-NPs present a distinct advantage of broad spectrum antibacterial activity4, 20 similar to H2O2 without oxidative stress to mammalian tissues or its rapid decay related to short life time of radicals in biological media.
The data presented here indicates that oxidative stress does not play the central role in antibacterial activity of ZnO-NPs with respect to MRSA. Particularly, taking into account the gene expression results of the oxidative stress genes in response to ZnO-NPs exposure, the down regulation of majority of ROS responsive genes indicates that ROS could not be the primary mode of action for an antibacterial effect. Indeed, comparison of our results with microarray experiments with S. aureus exposed to H2O2 from the literature showed less than 10% concordance in significantly up- or down- regulated genes between the two studies47, 48. Other work with ZnO-NPs has also shown down-regulation of oxidative stress genes49, 50. Some have interpreted that finding as evidence against an oxidative stress mechanism while others have interpreted it as making the cells more susceptible to ROS. These disparate interpretations have continued to fuel the ZnO-NP mechanism controversy. However, the multiple lines of evidence provided here, including gene expression, comparison with H2O2 and lack of recovery with an antioxidant, confirm that ROS mediated toxicity is not the mechanism of action for ZnO-NPs. The persistent controversy and multiple experiments required here to investigate it, highlight the complexity in evaluating the mechanism of action of any antimicrobial NP. Specifically, confirmation or rejection of any hypothesized mechanism typically requires multiple lines of evidence including both observed cellular response as well as molecular and ‘omics’ data.
A consideration of other possible mechanisms with interesting possibilities for molecular and nanoscale engineering is required to further increase and/or tune NP potency3, 51. Other mechanisms of antibacterial activity of NPs to be considered include toxic ions complexes, bacterial membrane disruption, and enzyme inhibition. In addition, the ability of NPs to suppress amyloid peptide fibrillation essential for biofilm formation may also need to be considered52.
The ROS production hypothesis for antimicrobial activity is predicated on catalysis of ROS on the ZnO surface. We acknowledge that the techniques described here are certainly not an exhaustive evaluation of toxic species dissolved from or catalyzed by ZnO-NPs. That being said, to evaluate for the production of undetected, soluble, ZnO-NP catalyzed, toxic species, we used the supernatant of ZnO-NP dispersions in growth media to generate growth curves of S. aureus. If ZnO-NPs dissolve into toxic Zn2+ ions or they catalyze the production of soluble toxic species in media then we would expect S. aureus growth to be inhibited by the supernatant of ZnO-NP dispersions, similar to ZnCl2 curves. However, almost no growth inhibition is observed with the supernatants of ZnO-NPs dispersions (Recall Figure 6). This suggests that the ZnO-NPs must remain dispersed with the cells to have their effect and the experimental conditions of the assays did not contribute to ion dissolution of the NPs. Furthermore, our gene expression data do not identify significant changes in expression of gene pathways related to metal ion transport.
With respect to membrane disruption, although lipid peroxidation results were negative in our study, it is possible that there are other membrane disruption mechanisms present between ZnO-NPs and Gram-positive cells. Previous studies show that NPs with different surface chemistry can differentially attach to various parts of the bacteria’s surface12, 17, 53, 54. Notably, lipopolysaccharides are attracted to NPs via variety of intermolecular forces including electrostatic interactions, hydrogen bonding, and van der Waals forces, that may cause lipid bilayer rupture, loss of membrane potential and cell death 55. However, it should be noted that cell membrane compromise and lysis is a common event for most antibacterial agents, including those whose mechanism does not involve the cell membrane (e.g., streptomycin). Therefore, documenting cell membrane damage in the presence of a particular NP is not sufficient. Rather the timing of loss of cell membrane integrity, proton motive force, and lysis in relation to other processes is critical. Again we highlight the need for multiple lines of evidence with well-defined controls to test any specific hypothesis. Of note, our gene expression studies do not show significant changes in expression of genes related to cell membrane synthesis/repair.
Considering enzyme inhibition, it was shown recently that ZnO-NPs can enter cells through membrane disruption10, 12 and cause enzyme inhibition 56 exemplified by strongly suppressed activity of β-galactosidase, an essential enzyme in bacterial metabolism. Enzymatic inhibition is not unique to β-galactosidase and may represent a family of biomolecular processes leading to suppression of bacterial growth or cellular death57. Ultimately, it is more than likely that several of these mechanisms together impact viability of bacteria. While the exact enzymatic targets for ZnO-NPs remain a question, our gene expression microarray data revealed a metabolic pathway, (i.e., UMP biosynthesis) not previously considered for ZnO-NP mediated bacterial toxicity. Upregulation of the UMP and other pyrimidine biosynthesis pathways have been shown to be critical for continued growth in anaerobic environment58. The combined up-regulation of this pathway with sugar catabolism suggests that ZnO-NPs may have a profound effect on carbohydrate metabolism and bioenergetics. This is a fundamentally new mechanism of action for consideration (Figure 8). Furthermore, when compared to other analyses of staphylococcal transcriptome response to various stressors, we did not find any significant concordance with heat shock, cold shock, stringent response, SOS response, anaerobic response, or oxidative stress47, 48, 59, 60. Future work on the antimicrobial effects of ZnO-NPs can now be directed at these pathways and the phenotypic manifestations of altered metabolic state.
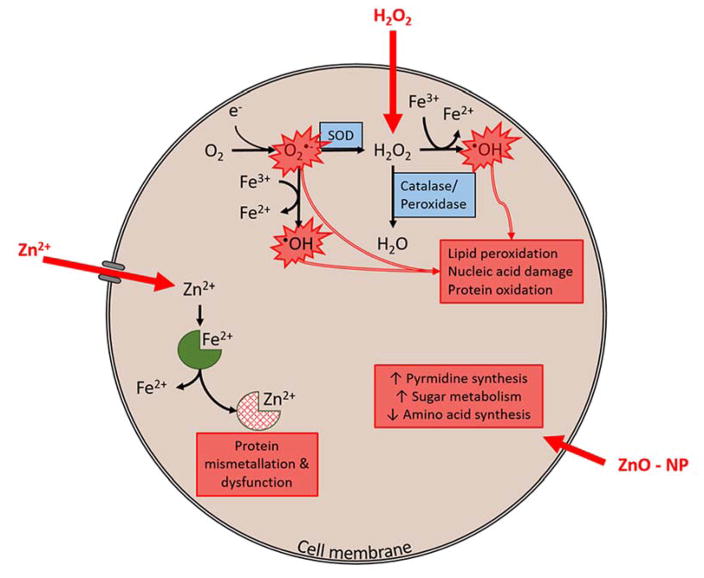
Figure 8.
H2O2 readily diffuses through the cell membrane. High concentrations overwhelm catalase and peroxidase enzymes leading to hydroxyl radical formation via Fenton reaction. In addition, high concentration of H2O2 can inhibit superoxide dismutase (SOD) driving Haber-Weiss reaction and additional production of hydroxyl radical. These ROS (hydroxyl radical and superoxide radical) cause toxicity through lipid peroxidation, nucleic acid damage and protein oxidation. Zn2+ ions require cell membrane transporters to enter the cell. In high concentrations that exceed normal homeostatic mechanism these ions displace other metal ions cofactors on proteins, causing mis-metallation and resulting protein dysfunction. Evidence here suggests an alternative mechanism for ZnO-NPs. ZnO-NPs result in dramatic increases in pyrimidine biosynthesis, sugar metabolism and decreases in amino acid synthesis. This combination or cellular processes suggests that ZnO-NPs alter energy metabolism within the cell. The precise mechanism by which ZnO-NPs enter the cell and the specific intracellular molecular targets remain unclear.
It is important to note that this work is limited to S. aureus. However, the ZnO-NPs used in this study were shown to also be toxic to Staphylococcus epidermidis at these concentrations20. Others have shown broad antimicrobial spectrum for ZnO-NPs50, 61. However, the generalizability of this novel potential bioenergetics mechanism involving pyrimidine and amino acid biosynthesis as well as sugar catabolism remains a question. That being said, the UMP biosynthesis pathway is quite similar for S. aureus and E. coli58. Furthermore, enzymes in amino acid synthesis are also quite highly conserved within bacteria62. This indicates that the mechanism may be applicable to both Gram-negative and Gram-positive organisms. Although it does not appear that the UMP pathway is necessary for anaerobic metabolism in E. coli as it is in S. aureus58. This may explain why a higher dose of ZnO-NPs is typically required for E. coli. Ultimately, the surface chemistry and crystalline structure of a particular ZnO-NP preparation may have a profound effect on its interaction with the surface of a Gram-positive versus Gram-negative organism and therefore may dictate species selectivity regardless of the final mechanism63. Likewise, the three-dimensional shape may be important in accessibility of ZnO-NPs to specific molecular targets56, 64.
5. Conclusions
Here we have demonstrated that oxidative stress and toxic ion dissolution are inadequate to cause ZnO-NP mediated killing of methicillin resistant S. aureus. By means of gene expression analysis we demonstrate that ZnO-NPs may have a profound effect on anaerobic carbohydrate metabolism and energetics with upregulation of the UMP biosynthesis pathway being a key response. Altogether, the findings about the unexpectedly small role of ROS production and large role of alterations in carbohydrate metabolism and bioenergetics give strong indication of biomimetic mode of action of NPs65. Understanding the mechanism of how ZnO-NPs inhibit bacteria will help in developing new methods utilizing the extensive toolset of nanoscale and molecular scale engineering of NPs. Even subtle structural aspects of NPs, exemplified by chirality of the surface and crystal lattice, can be predictively imparted to nanocolloids66–69. A well-defined description of the antimicrobial molecular mechanism of action is necessary to guide nanoscale engineering of NPs toward a specific mechanism of antibacterial activity. Fine tuning the NP activity using biomimetic concepts65 opens the possibilities of improving both potency and selectivity of NPs as antimicrobials.
Supplementary Material
Acknowledgments
The authors would like to thank Siu On Tung for his assistance with obtaining the FTIR spectra for the ZnO-NPs. Partial support of this work was made by the National Institutes of Health (R01-GM081702, K08-AI128006), the Center for Photonic and Multiscale Nanomaterials (C-PHOM) funded by the National Science Foundation (NSF) Materials Research Science and Engineering Center program DMR 1120923 and NSF projects 1403777; 1411014. Seed funding to J.S.V and N.A.K from MCIRCC and Biointerfaces Institute is gratefully acknowledged.
Footnotes
The authors have no competing conflicts of interest related to this work.
References
- 1.Conly JM. The Canadian Journal of Infectious Diseases & Medical Microbiology. 2004;15:249–251. doi: 10.1155/2004/680349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang W, Mashayekhi H, Xing B. Environmental pollution (Barking, Essex : 1987) 2009;157:1619–1625. doi: 10.1016/j.envpol.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 3.Djurisic AB, Leung YH, Ng AM, Xu XY, Lee PK, Degger N, Wu RS. Small. 2015;11:26–44. doi: 10.1002/smll.201303947. [DOI] [PubMed] [Google Scholar]
- 4.Applerot G, Lellouche J, Perkas N, Nitzan Y, Gedanken A, Banin E. RSC Advances. 2012;2:2314–2321. [Google Scholar]
- 5.Krishnamoorthy K, Manivannan G, Kim SJ, Jeyasubramanian K, Premanathan M. Journal of Nanoparticle Research. 2012;14:1–10. [Google Scholar]
- 6.Premanathan M, Karthikeyan K, Jeyasubramanian K, Manivannan G. Nanomedicine : nanotechnology, biology, and medicine. 2011;7:184–192. doi: 10.1016/j.nano.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Romero-Urbina DG, Lara HH, Velázquez-Salazar JJ, Arellano-Jiménez MJ, Larios E, Srinivasan A, Lopez-Ribot JL, Yacamán MJ. Beilstein Journal of Nanotechnology. 2015;6:2396–2405. doi: 10.3762/bjnano.6.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Setyawati MI, Fang W, Chia SL, Leong DT. Asia-Pacific Journal of Chemical Engineering. 2013;8:205–217. [Google Scholar]
- 9.Zhang L, Ding Y, Povey M, York D. Progress in Natural Science. 2008;18:939–944. [Google Scholar]
- 10.Kumar A, Pandey AK, Singh SS, Shanker R, Dhawan A. Free radical biology & medicine. 2011;51:1872–1881. doi: 10.1016/j.freeradbiomed.2011.08.025. [DOI] [PubMed] [Google Scholar]
- 11.Liu JL, Luo Z, Bashir S. Biomaterials Science. 2013;1:194–201. doi: 10.1039/c2bm00010e. [DOI] [PubMed] [Google Scholar]
- 12.Xie Y, He Y, Irwin PL, Jin T, Shi X. Applied and environmental microbiology. 2011;77:2325–2331. doi: 10.1128/AEM.02149-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang L, Jiang Y, Ding Y, Daskalakis N, Jeuken L, Povey M, O’Neill AJ, York DW. Journal of Nanoparticle Research. 2009;12:1625–1636. [Google Scholar]
- 14.Zhang Y, Nayak TR, Hong H, Cai W. Current molecular medicine. 2013;13:1633–1645. doi: 10.2174/1566524013666131111130058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cory H, Janet L, Alex P, Reddy KM, Isaac C, Andrew C, Kevin F, Denise W. Nanotechnology. 2008;19:295103. [Google Scholar]
- 16.Reddy KM, Feris K, Bell J, Wingett DG, Hanley C, Punnoose A. Applied physics letters. 2007;90:213902-213901–213902-213903. doi: 10.1063/1.2742324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dutta RK, Nenavathu BP, Gangishetty MK, Reddy AV. Colloids and surfaces B, Biointerfaces. 2012;94:143–150. doi: 10.1016/j.colsurfb.2012.01.046. [DOI] [PubMed] [Google Scholar]
- 18.Pastoriza-Santos I, Koktysh DS, Mamedov AA, Giersig M, Kotov NA, Liz-Marzán LM. Langmuir. 2000;16:2731–2735. [Google Scholar]
- 19.Podsiadlo P, Paternel S, Rouillard JM, Zhang Z, Lee J, Lee JW, Gulari E, Kotov NA. Langmuir. 2005;21:11915–11921. doi: 10.1021/la051284+. [DOI] [PubMed] [Google Scholar]
- 20.McGuffie MJ, Hong J, Bahng JH, Glynos E, Green PF, Kotov NA, Younger JG, VanEpps JS. Nanomedicine: Nanotechnology, Biology and Medicine. 12:33–42. doi: 10.1016/j.nano.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Applerot G, Lellouche J, Lipovsky A, Nitzan Y, Lubart R, Gedanken A, Banin E. Small. 2012;8:3326–3337. doi: 10.1002/smll.201200772. [DOI] [PubMed] [Google Scholar]
- 22.Cui Y, Zhao Y, Tian Y, Zhang W, Lu X, Jiang X. Biomaterials. 2012;33:2327–2333. doi: 10.1016/j.biomaterials.2011.11.057. [DOI] [PubMed] [Google Scholar]
- 23.Horie M, Fujita K, Kato H, Endoh S, Nishio K, Komaba LK, Nakamura A, Miyauchi A, Kinugasa S, Hagihara Y, Niki E, Yoshida Y, Iwahashi H. Metallomics : integrated biometal science. 2012;4:350–360. doi: 10.1039/c2mt20016c. [DOI] [PubMed] [Google Scholar]
- 24.von Moos N, Slaveykova VI. Nanotoxicology. 2014;8:605–630. doi: 10.3109/17435390.2013.809810. [DOI] [PubMed] [Google Scholar]
- 25.Leung YH, Chan CM, Ng AM, Chan HT, Chiang MW, Djurisic AB, Ng YH, Jim WY, Guo MY, Leung FC, Chan WK, Au DT. Nanotechnology. 2012;23:475703. doi: 10.1088/0957-4484/23/47/475703. [DOI] [PubMed] [Google Scholar]
- 26.Thill A, Zeyons O, Spalla O, Chauvat F, Rose J, Auffan M, Flank AM. Environmental Science & Technology. 2006;40:6151–6156. doi: 10.1021/es060999b. [DOI] [PubMed] [Google Scholar]
- 27.Leung YH, Ng AM, Xu X, Shen Z, Gethings LA, Wong MT, Chan CM, Guo MY, Ng YH, Djurisic AB, Lee PK, Chan WK, Yu LH, Phillips DL, Ma AP, Leung FC. Small. 2014;10:1171–1183. doi: 10.1002/smll.201302434. [DOI] [PubMed] [Google Scholar]
- 28.Lyon DY, Brunet L, Hinkal GW, Wiesner MR, Alvarez PJ. Nano letters. 2008;8:1539–1543. doi: 10.1021/nl0726398. [DOI] [PubMed] [Google Scholar]
- 29.Gaupp R, Ledala N, Somerville GA. Frontiers in Cellular and Infection Microbiology. 2012;2:33. doi: 10.3389/fcimb.2012.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gambino M, Cappitelli F. Biofouling. 2016;32:167–178. doi: 10.1080/08927014.2015.1134515. [DOI] [PubMed] [Google Scholar]
- 31.Painter KL, Strange E, Parkhill J, Bamford KB, Armstrong-James D, Edwards AM. Infection and immunity. 2015;83:1830–1844. doi: 10.1128/IAI.03016-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li M, Zhu L, Lin D. Environmental Science & Technology. 2011;45:1977–1983. doi: 10.1021/es102624t. [DOI] [PubMed] [Google Scholar]
- 33.McDevitt CA, Ogunniyi AD, Valkov E, Lawrence MC, Kobe B, McEwan AG, Paton JC. PLOS Pathogens. 2011;7:e1002357. doi: 10.1371/journal.ppat.1002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemire JA, Harrison JJ, Turner RJ. Nat Rev Microbiol. 2013;11:371–384. doi: 10.1038/nrmicro3028. [DOI] [PubMed] [Google Scholar]
- 35.Wang N, Tong T, Xie M, Gaillard JF. Nanotechnology. 2016;27:324001. doi: 10.1088/0957-4484/27/32/324001. [DOI] [PubMed] [Google Scholar]
- 36.Reed RB, Ladner DA, Higgins CP, Westerhoff P, Ranville JF. Environmental Toxicology and Chemistry. 2012;31:93–99. doi: 10.1002/etc.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang M, Sun K, Kotov NA. Journal of the American Chemical Society. 2010;132:1860–1872. doi: 10.1021/ja906868h. [DOI] [PubMed] [Google Scholar]
- 38.Repine JE, Fox RB, Berger EM. The Journal of biological chemistry. 1981;256:7094–7096. [PubMed] [Google Scholar]
- 39.Thomas EL, Milligan TW, Joyner RE, Jefferson MM. Infection and Immunity. 1994;62:529–535. doi: 10.1128/iai.62.2.529-535.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan W, Xu L, Xu C, Ma W, Kuang H, Wang L, Kotov NA. Journal of the American Chemical Society. 2012;134:15114–15121. doi: 10.1021/ja3066336. [DOI] [PubMed] [Google Scholar]
- 41.Smyth GK. Statistical applications in genetics and molecular biology. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 42.Ritchie ME, Diyagama D, Neilson J, van Laar R, Dobrovic A, Holloway A, Smyth GK. BMC Bioinformatics. 2006;7:261–261. doi: 10.1186/1471-2105-7-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benjamini Y, Hochberg Y. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 44.Čáp M, Váchová L, Palková Z. Oxidative Medicine and Cellular Longevity. 2012;2012:13. doi: 10.1155/2012/601836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jang IA, Kim J, Park W. Scientific Reports. 2016;6:21121. doi: 10.1038/srep21121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stark G. The Journal of Membrane Biology. 2005;205:1–16. doi: 10.1007/s00232-005-0753-8. [DOI] [PubMed] [Google Scholar]
- 47.Chang W, Small DA, Toghrol F, Bentley WE. Journal of bacteriology. 2006;188:1648–1659. doi: 10.1128/JB.188.4.1648-1659.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nobre LS, Saraiva LM. Applied microbiology and biotechnology. 2013;97:2563–2573. doi: 10.1007/s00253-013-4730-3. [DOI] [PubMed] [Google Scholar]
- 49.Pati R, Mehta RK, Mohanty S, Padhi A, Sengupta M, Vaseeharan B, Goswami C, Sonawane A. Nanomedicine : nanotechnology, biology, and medicine. 2014;10:1195–1208. doi: 10.1016/j.nano.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 50.Raghupathi KR, Koodali RT, Manna AC. Langmuir. 2011;27:4020–4028. doi: 10.1021/la104825u. [DOI] [PubMed] [Google Scholar]
- 51.Amna S, Shahrom M, Azman S, Kaus NHM, Ling Chuo A, Siti Khadijah Mohd B, Habsah H, Dasmawati M. Nano-Micro Letters. 2015:7. [Google Scholar]
- 52.Yoo SI, Yang M, Brender JR, Subramanian V, Sun K, Joo NE, Jeong SH, Ramamoorthy A, Kotov NA. Angewandte Chemie International Edition. 2011;50:5110–5115. doi: 10.1002/anie.201007824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Planque MRR, Aghdaei S, Roose T, Morgan H. ACS nano. 2011;5:3599–3606. doi: 10.1021/nn103320j. [DOI] [PubMed] [Google Scholar]
- 54.Arakha M, Saleem M, Mallick BC, Jha S. Scientific Reports. 2015;5:9578. doi: 10.1038/srep09578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jacobson KH, Gunsolus IL, Kuech TR, Troiano JM, Melby ES, Lohse SE, Hu D, Chrisler WB, Murphy CJ, Orr G, Geiger FM, Haynes CL, Pedersen JA. Environmental Science & Technology. 2015;49:10642–10650. doi: 10.1021/acs.est.5b01841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cha SH, Hong J, McGuffie M, Yeom B, VanEpps JS, Kotov NA. ACS Nano. 2015;9:9097–9105. doi: 10.1021/acsnano.5b03247. [DOI] [PubMed] [Google Scholar]
- 57.Saptarshi SR, Duschl A, Lopata AL. Journal of Nanobiotechnology. 2013;11:26. doi: 10.1186/1477-3155-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McIllMurray MB, Lascelles J. Microbiology. 1970;64:269–277. doi: 10.1099/00221287-64-3-269. [DOI] [PubMed] [Google Scholar]
- 59.Anderson KL, Roberts C, Disz T, Vonstein V, Hwang K, Overbeek R, Olson PD, Projan SJ, Dunman PM. Journal of bacteriology. 2006;188:6739–6756. doi: 10.1128/JB.00609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fuchs S, Pané-Farré J, Kohler C, Hecker M, Engelmann S. Journal of bacteriology. 2007;189:4275–4289. doi: 10.1128/JB.00081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jones N, Ray B, Ranjit KT, Manna AC. FEMS microbiology letters. 2008;279:71–76. doi: 10.1111/j.1574-6968.2007.01012.x. [DOI] [PubMed] [Google Scholar]
- 62.Hernandez-Montes G, Diaz-Mejia JJ, Perez-Rueda E, Segovia L. Genome biology. 2008;9:R95. doi: 10.1186/gb-2008-9-6-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu F, Fang X, Jia H, Liu M, Shi X, Xue C, Chen T, Wei Z, Fang F, Zhu H, Xin H, Feng J, Wang X. Chemistry – A European Journal. 2016;22:8053–8058. doi: 10.1002/chem.201601018. [DOI] [PubMed] [Google Scholar]
- 64.Liao H, Miao X, Ye J, Wu T, Deng Z, Li C, Jia J, Cheng X, Wang X. ACS Applied Materials & Interfaces. 2017;9:13009–13015. doi: 10.1021/acsami.7b00666. [DOI] [PubMed] [Google Scholar]
- 65.Kotov NA. Science. 2010;330:188. doi: 10.1126/science.1190094. [DOI] [PubMed] [Google Scholar]
- 66.Zhou Y, Yang M, Sun K, Tang Z, Kotov NA. Journal of the American Chemical Society. 2010;132:6006–6013. doi: 10.1021/ja906894r. [DOI] [PubMed] [Google Scholar]
- 67.Govan J, Gun’ko YK. Nanoscience. Vol. 3. The Royal Society of Chemistry; 2016. pp. 1–30. [Google Scholar]
- 68.Gautier C, Bürgi T. ChemPhysChem. 2009;10:483–492. doi: 10.1002/cphc.200800709. [DOI] [PubMed] [Google Scholar]
- 69.Schaaff TG, Whetten RL. The Journal of Physical Chemistry B. 2000;104:2630–2641. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.