ABSTRACT
Production of bacterial flagella is controlled by a multitiered regulatory system that coordinates the expression of 40 to 50 subunits and ordered assembly of these elaborate structures. Flagellar expression is environmentally controlled, presumably to optimize the benefits and liabilities of having these organelles on cell growth and survival. We recently reported a global survey of AlgU-dependent regulation and binding in Pseudomonas syringae pv. tomato DC3000 that included evidence for strong downregulation of many flagellar and chemotaxis motility genes. Here, we returned to those data to look for other AlgU-dependent influences on the flagellar regulatory network. We identified an AlgU-dependent antisense transcript expressed from within the fleQ gene, the master regulator of flagellar biosynthesis in Pseudomonas. We tested whether expression of this antisense RNA influenced bacterial behavior and found that it reduces AlgU-dependent downregulation of motility. Importantly, this antisense expression influenced motility only under conditions in which AlgU was expressed. Comparative sequence analysis of the locus containing the antisense transcript's AlgU-dependent promoter in over 300 Pseudomonas genomes revealed sequence conservation in most strains that encode AlgU. This suggests that the antisense transcript plays an important role that is conserved across most of the genus Pseudomonas.
IMPORTANCE Pseudomonas syringae is a globally distributed host-specific bacterial pathogen that causes disease in a wide-range of plants. An elaborate gene expression regulation network controls flagellum production, which is important for proper flagellum assembly and a key aspect of certain lifestyle transitions. P. syringae pv. tomato DC3000 uses flagellum-powered motility in the early stages of host colonization and adopts a sessile lifestyle after entering plant tissues, but the regulation of this transition is not understood. Our work demonstrates a link between regulation of motility and global transcriptional control that facilitates bacterial growth and disease in plants. Additionally, sequence comparisons suggest that this regulation mechanism is conserved in most members of the genus Pseudomonas.
KEYWORDS: Pseudomonas, Pseudomonas syringae, cell motility, flagella, regulation, sRNA
INTRODUCTION
Many bacteria use flagella to move toward environments beneficial to their survival and growth. Flagella are macromolecular surface appendages that actively propel cells through liquids and over surfaces. Regulation of flagellar expression is particularly important for bacterial pathogens, because they rely on motility to enter and attach to host tissues (1, 2), but flagella can also be detrimental in these contexts (3–6). Pseudomonas syringae, a species complex that cause diseases in a wide-range of agriculturally significant crops, is capable of moving over plant surfaces toward stomata or wounds in plant surfaces to access the intercellular spaces where the bacteria are adapted to grow (7). This movement requires chemotaxis sensing and flagella (8). Plant and animal immune systems respond to the presence of infecting microbes by detecting epitopes within flagellar filament proteins (3–6), and downregulation and disassembly of flagella may be an adaptive trait to facilitate infection (9–11). P. syringae and other pseudomonads secrete the AprA proteases, which degrade the remaining flagellar monomers, helping to avoid immune activation (12–14).
P. syringae encodes redundant virulence functions that help to avoid or suppress plant immune defenses activated by flagella and other molecules (15, 16). A subset of P. syringae type III secreted effectors interfere with flagellar detection and signal transduction that leads to activation of plant defenses (17–19). Culture conditions that activate expression of the P. syringae pv. maculicola type III secretion system and effector genes also downregulate flagellar gene expression, suggesting a possible regulatory link between these systems (11). This coordinated regulation requires AlgW, which is an integral component of the signal transduction pathway activating the AlgU extracytoplasmic function (ECF) sigma factor (20). AlgU is activated in response to types of stress that affect the bacterial cell envelope (21), including osmotic and water stress, and during plant infection (10, 22). Analysis of the AlgU regulon in P. syringae pv. tomato DC3000 indicates that this sigma factor acts as a master regulator in coordinating upregulation of alginate-biosynthetic genes, many effector genes, and components of the type III secretion system with downregulation of flagellar and chemotaxis genes (22).
Flagella are dynamic structures that are expressed according to environmental conditions (11, 23–25). P. syringae pv. tomato DC3000 cells and other pseudomonads produce 1 to 5 polar flagella (26, 27) using a four-tiered hierarchical transcription regulation network (28). This sophisticated control system provides for just-in-time expression of the flagellar components and for ordered assembly of the flagellar apparatus. FleQ is an NtrC-like transcription activator at the top of the transcriptional regulatory hierarchy and is necessary for transcription of more than 40 genes coding for the regulatory and structural components of the flagellar apparatus (27, 28). The fleQ gene and motility of Pseudomonas aeruginosa and Pseudomonas fluorescens are repressed by AlgU via AmrZ (29–31). AlgU also downregulates flagellar gene expression in P. syringae pv. tomato DC3000, but this is unlikely to occur via AmrZ. Expression of fleQ is not repressed by AlgU in P. syringae pv. tomato DC3000 (22), and AmrZ has a positive effect on motility and flagellar gene expression (31).
Here, we report the discovery of an AlgU-regulated antisense transcript within the P. syringae pv. tomato DC3000 fleQ gene, which we refer to as fleQ antisense (fleQas). We investigated the structure and function of this transcript and found that fleQas expression increases swimming motility but not flagellar gene expression in AlgU-expressing cells. This suggests that fleQas has a positive influence on flagellar activity, which functionally opposes AlgU-dependent downregulation of flagella. We found the predicted AlgU-dependent promoter sequences appropriately positioned to express fleQas in P. syringae pv. tomato DC3000 and in most sequenced Pseudomonas genomes that also encode an AlgU orthologue. Together, these results provide evidence of a new gene functioning within the complex regulatory cascade controlling flagellar expression and motility in Pseudomonas bacteria.
RESULTS
Functional genomic evidence for AlgU-dependent antisense transcription and AlgU binding within the fleQ gene.
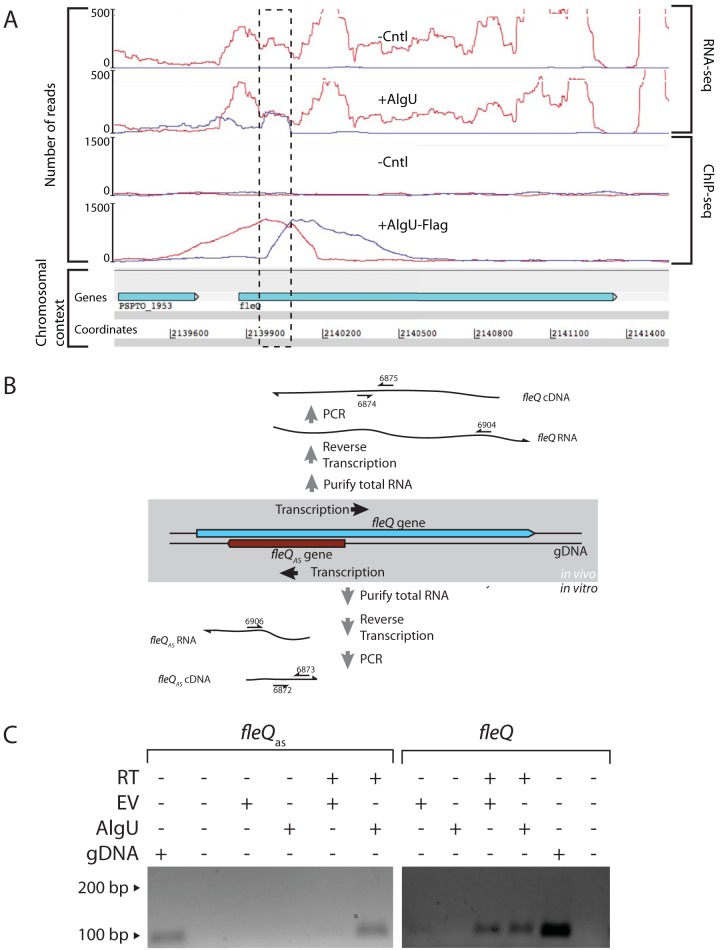
Upon further evaluation of the P. syringae pv. tomato DC3000 whole-genome AlgU-dependent expression and binding data (22), we identified a transcript that mapped to the opposite strand of the 5′ portion of fleQ (Fig. 1A). This transcript was apparent only in cells expressing AlgU, and chromatin immunoprecipitation sequencing (ChIP-seq) analysis detected AlgU binding directly upstream of the sequence coding for this transcript.
FIG 1.
Analysis of AlgU-dependent antisense transcript expressed within the fleQ gene. (A) RNA-seq and ChIP-seq results at the fleQ locus. Strand-specific RNA-seq analysis of transcriptomes produced by P. syringae pv. tomato DC3000 ΔalgU mucAB cells containing an AlgU expression construct (+AlgU) or the empty-vector control (-Cntl) show the location of antisense transcription in the fleQ gene (dashed box). ChIP-seq analysis of P. syringae pv. tomato DC3000 ΔalgU mucAB cells containing an AlgU-Flag expression construct (+AlgU-Flag) or the empty-vector control (-Cntl) shows AlgU-Flag-dependent enrichment of sequences directly upstream of the location of fleQas transcription. Experimental details and complete data sets for the AlgU RNA-seq and ChIP-seq results are available in reference 22. The number of reads on the y axis indicates the number of sequence reads mapped per nucleotide position. The red and blue profiles indicate sequences matching the sense and antisense strands, respectively. (B) Strand-specific RT-PCR was used to test for the fleQas transcript in cells containing either the AlgU expression plasmid or the empty-vector control. PCR amplification required the product of strand-specific reverse transcription of the fleQas or the fleQ transcript. Primers complementary to either the fleQas or the fleQ transcript were used to reverse transcribe a segment of each transcript that served as the template in a PCR to detect the cDNA products. The numbered arrows refer to oligonucleotide primers (see Table S3 in the supplemental material). (C) Agarose gel showing reverse transcriptase (RT)-dependent PCR products of fleQas, which were obtained only from cells expressing AlgU. In contrast, the fleQ sense transcripts were detected in cells containing AlgU expression and the empty vector (EV). P. syringae pv. tomato DC3000 genomic DNA (gDNA) was a positive control for the gene-specific PCR.
fleQas expression is dependent on AlgU.
We used strand-specific reverse transcription and PCR (RT-PCR) to test whether the antisense transcript could be detected in cells expressing AlgU. This was done to provide independent verification of the presence of the transcript and to test whether AlgU is necessary for its production. In this experiment, PCR amplification depended on strand-specific reverse transcription of RNAs produced from within the fleQ locus (Fig. 1B). Using RNA collected from P. syringae pv. tomato DC3000 ΔalgU mucAB cells containing an AlgU expression construct or empty-vector control, we found that the fleQas transcript could be detected only in cells expressing AlgU (Fig. 1C). Note that P. syringae pv. tomato DC3000 ΔalgU mucAB cells were used to observe AlgU-dependent phenotypes in lieu of induction in response to environmental cues (22). In comparison, the fleQ sense transcript could be detected regardless of the presence of AlgU.
We used strand-specific quantitative reverse transcription (qRT)-PCR to evaluate the relative difference in antisense transcript quantity in AlgU-expressing cells versus those containing the empty vector. In this experiment, the fleQas and fleQ sense transcripts were reverse transcribed using primers complementary to the respective strands, as described above (Fig. 1B). Strand-specific primers were also used to reverse transcribe the gyrA and gap-1 housekeeping genes, which were used as controls for differential expression and normalization, respectively. We found that the fleQas transcript was more than 100-fold more abundant in AlgU-expressing cells than in those with the empty-vector control. The differences in relative transcript abundances in AlgU-expressing cells and those with the empty-vector control were as follows [averages and standard deviations (SD)]: fleQ, 1.45 (1.0); fleQas, 112.5 (64.0); and gyrA, 2.50 (2.0). (The averages and standard deviations were calculated from three biological replicates. gyrA was included as a control for AlgU-dependent changes to general transcription levels. The Tukey-Kramer honest significant difference [HSD] test results indicate that the mean expression difference for fleQas is significantly different from those of fleQ and gyrA, which are statistically equivalent.) The fleQ sense strand and gyrA housekeeping gene were unchanged by AlgU expression. Together, these results provide strong evidence that fleQas transcription is dependent on AlgU and that the transcription of the fleQ gene itself is unchanged by the AlgU-dependent transcription occurring from the opposite strand.
fleQas lengths and start sites.
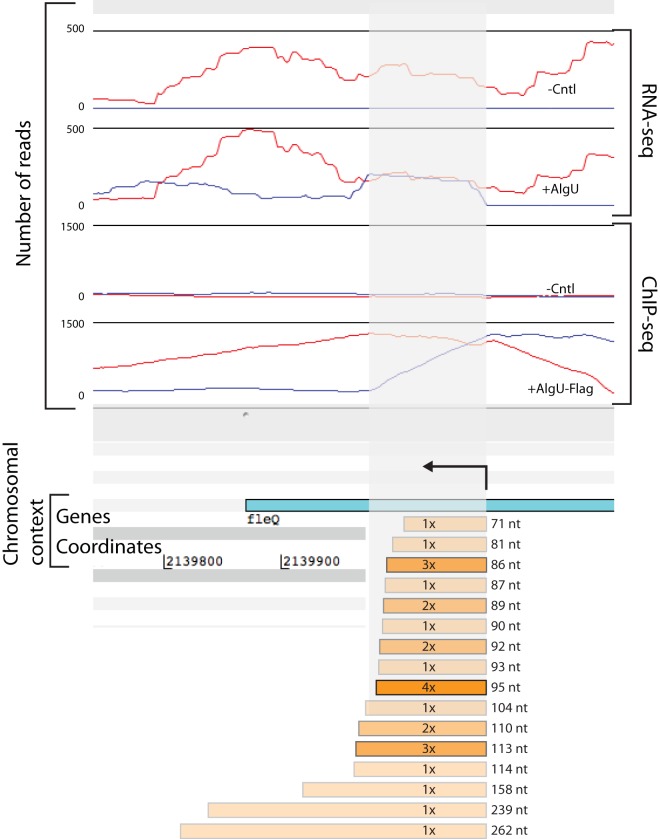
We used 5′ and 3′ rapid amplification of cDNA ends [RACE] to determine the sequences of the 5′ and 3′ ends of the fleQas transcript in AlgU-expressing cells (Fig. 2). We observed two 5′ ends that mapped over a 3-nucleotide (nt) range complementary to fleQ codons 68 and 69. We observed 16 different 3′ ends, 6 of which were observed more than once. Based on the locations of the observed 5′ and 3′ ends, we predicted transcript lengths ranging from 71 to 262 nucleotides (Fig. 2).
FIG 2.
5′ and 3′ RACE map of predicted fleQas transcription start site and lengths. Observed 5′ and 3′ ends are shown in relation to expression and AlgU binding at the fleQ locus. Two 5′ ends were observed, one at P. syringae pv. tomato DC3000 genomic coordinate 2140072 and a second that begins at either 2140073 or 2140074 (which cannot be unambiguously determined because of the addition of homopolymeric tails matching the genome sequence). The number of times each 3′ end was observed is indicated in the orange bars, with the depth of shading corresponding to the frequency of observation; the length was calculated based on the most distal 5′ end. The length of the bar is shown to scale, with the other genomic features indicated. The nucleotide coordinates are derived from the P. syringae pv. tomato DC3000 genome sequence (NC_004578.1) (55). RNA- and ChIP-seq data are available in reference 22. The arrow above the fleQ gene indicates the direction of transcription of predicted fleQas molecules.
We compared the locations of mapped fleQas transcript ends to the locations of AlgU binding and AlgU-dependent transcription observed in the functional genomic analyses (22). The locations of the mapped fleQas 5′ ends were directly downstream of the area maximally enriched in the AlgU ChIP-seq, which is consistent with those locations serving as the transcription start sites for this RNA. Additionally, there was good agreement between the most frequently observed 3′ ends (by 3′ RACE) and the ends of the majority of transcripts sequenced at this locus in the transcriptome-sequencing (RNA-seq) analysis (22).
An AlgU-dependent promoter controls fleQas expression.
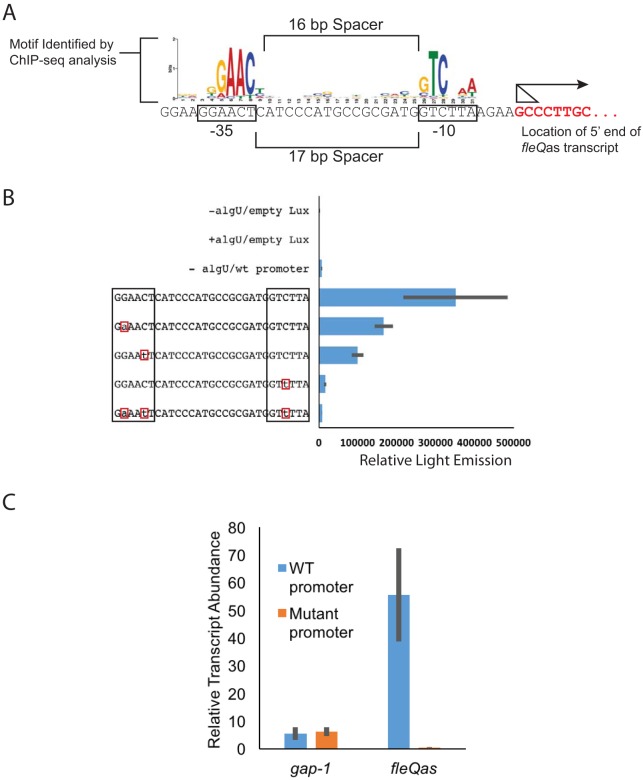
We inspected the nucleotide sequence immediately upstream of the mapped 5′ transcript ends for the presence of potential cis-acting regulatory sequences responsible for AlgU-dependent transcription of fleQas. We identified a very good match to the P. syringae pv. tomato DC3000 AlgU-dependent promoter −10 and −35 elements (22) appropriately positioned to function as a promoter for fleQas (Fig. 3A). The −10 and −35 hexamers were separated by 17 nt rather than the 16-nt spacer for the AlgU-dependent promoter consensus reported by Markel et al. (22). This is not unusual; promoters regulated by this class of sigma factors often have some variation in spacer lengths (32).
FIG 3.
An AlgU-dependent promoter controls fleQas expression. (A) Comparison of the nucleotide sequence directly upstream of the predicted fleQas transcription start site to that of the AlgU-dependent promoter reported in reference 22. The arrow shows the direction of transcription, and the triangle shows the range of potential start sites. (B) Luciferase reporter expression of wild-type (wt) and mutant fleQas promoter::lux fusions in the pBS58 vector. Empty Lux represents the reporter activity of the empty lux reporter construct in the presence and absence of an AlgU expression vector. The lux reporter vector containing the wild-type fleQas promoter is indicated by “wt promoter” or by the unchanged full promoter sequence. −algU/wt promoter, Lux expression from the wild-type promoter in the absence of AlgU. The lux reporter vectors containing the unchanged or changed promoter sequences were expressed in the presence of AlgU (indicated by the promoter sequences that show altered nucleotides [boxed in red]). Relative Light Emission indicates the light emission/OD600 unit. The sequences of the wild-type promoter and altered nucleotides are indicated. (C) qRT-PCR analysis of fleQas expression in AlgU-expressing strains (P. syringae pv. tomato DC3000 ΔalgU mucAB plus pEM53) versus that with the empty-vector control (P. syringae pv. tomato DC3000 ΔalgU mucAB plus pJN105) with the wild-type or mutant fleQas promoter. Averages and standard deviations from three biological replicates are shown for Lux and qRT-PCR expression analyses.
The AlgU-dependent function of this sequence was tested by cloning the region containing the putative promoter upstream of the lux operon and measuring reporter activity in the presence and absence of AlgU (Fig. 3B). Reporter activity from the construct was 51-fold higher in cells with the AlgU expression vector, confirming that the cloned region contains an AlgU-regulated promoter. The contributions of specific nucleotides to promoter function were further examined by substitution mutations at three conserved positions (and all three combined) of the AlgU-dependent promoter motif. The changes were chosen to avoid altering the coding sequence of the opposite-strand fleQ gene; this was done to test the suitability of these mutations for evaluating phenotypes associated with fleQas expression from the native location without interfering with fleQ expression or function (see below). The reporter activities of the fusion constructs containing these mutations were compared to that of the wild-type promoter. Each of the individual mutations reduced expression between 2- and 21-fold, with changes in the −10 element having the greatest effect, which is consistent with expectations for ECF sigma factor-regulated promoters (33, 34). Furthermore, the mutations had a slight additive effect when combined: we found that the promoter containing all three mutations reduced expression 43-fold, which is the same level of expression as in the absence of AlgU (Fig. 3B, compare −algU/wt to the triple mutant) (P = 0.53; t test).
We tested the effects of the promoter mutations on fleQas expression from the native location in the P. syringae pv. tomato DC3000 genome. In this experiment, the wild-type fleQas promoter was replaced with the mutant allele containing the three nucleotide substitutions, because that version had the lowest lux expression. We determined the relative expression of the fleQas gene in AlgU-expressing strains containing the wild-type and mutant promoters using the strand-specific qRT-PCR approach (Fig. 3C). The mutant promoter reduced fleQas expression 110-fold, which is consistent with the hypothesis that the AlgU-dependent promoter we identified is responsible for fleQas expression.
fleQas expression limits AlgU-dependent downregulation of motility.
FleQ is at the top of the flagellar gene expression cascade, serving as the master regulator of flagellar gene expression and motility in Pseudomonas (28). Colocation of the fleQ and fleQas genes suggested the hypothesis that fleQas might have a role in modulating motility. To test this idea, we compared the swimming motilities of P. syringae pv. tomato DC3000 strains that varied in their ability to produce fleQas. To do this, we used the strain containing the triple-nucleotide-substitution promoter (described above), which was designed to eliminate expression of fleQas but not to alter the FleQ protein sequence encoded by the fleQ gene on the opposite strand. By comparing the diameters of swimming colonies after a fixed incubation time, we found that AlgU-expressing cells that were able to express fleQas were 36% more motile than the AlgU-expressing cells containing the mutant promoter (Fig. 4). Specifically, we found that the motility of the bacteria was reduced in response to AlgU expression in both wild-type and mutant cells but the AlgU-expressing wild-type cells were more motile than those with the mutant fleQas promoter. The effect of fleQas expression occurs in the context of AlgU-dependent downregulation of the flagellar genes, so it was surprising to find that AlgU-regulated fleQas had a positive effect on motility. This suggests that fleQas expression interferes with, or limits, AlgU-dependent suppression of flagellar motility. Additionally, the data show that the mutant fleQas promoter does not affect motility in the absence of AlgU, which confirms that the mutations we introduced to alter fleQas promoter activity did not impact FleQ function.
FIG 4.
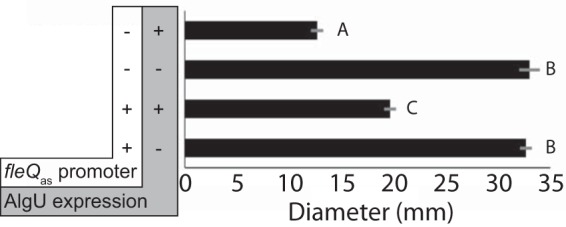
Expression of fleQas attenuates AlgU-dependent downregulation of motility. Shown are P. syringae pv. tomato DC3000 cells with a wild-type (+) or mutant (−) fleQas promoter and containing AlgU expression (+) or empty vector (−). The diameter of the colony was measured after 44 h of incubation on swimming assay medium containing 0.3% agar (59). Means and standard deviations are shown for three biological replicates. The letters indicate Tukey-Kramer HSD mean comparison results; means labeled with different letters are significantly different.
fleQas does not alter expression of flagellar genes.
We tested whether the effect of fleQas expression on swimming motility was due to a change in expression of flagellar regulatory or structural genes. To do this, we used qRT-PCR to evaluate fleQ, fliC, fleS, and fleP expression in a P. syringae pv. tomato DC3000 AlgU expression strain (P. syringae pv. tomato DC3000 ΔalgU mucAB plus pEM53) containing either the wild-type or mutant fleQas promoter (Table 1). The fleS and fleP genes are directly upstream of fleQ and were tested to ascertain whether the antisense transcript might influence their expression. We found no difference in expression for any of these genes. It is worth noting that fliC expression was unchanged, as well, suggesting that the entire flagellar expression cascade was also unchanged and that FleQ levels and activity were not affected by fleQas expression. We also tested whether other AlgU-regulated genes were affected by fleQas expression. We evaluated the expression of algU and algD but observed no difference in cells expressing fleQas.
TABLE 1.
Differences in relative transcript abundances in P. syringae pv. tomato DC3000 ΔalgU mucAB cells with the AlgU expression construct and containing either the wild-type or mutant fleQas promoter
| Gene | Relative transcript abundance [avg (SD)]a |
|---|---|
| fleQ | 1.06 (0.28) |
| fliC | 0.97 (0.08) |
| fleS | 0.68 (0.89) |
| fleP | 0.84 (1.31) |
| algU | 0.83 (0.08) |
| algD | 0.79 (0.13) |
| gyrA | 0.73 (0.21) |
Averages and standard deviations were calculated from three biological replicates. gyrA was included as a control for fleQas-dependent changes in general transcription levels.
An AlgU-dependent promoter motif upstream of fleQas is widely conserved in the genus Pseudomonas.
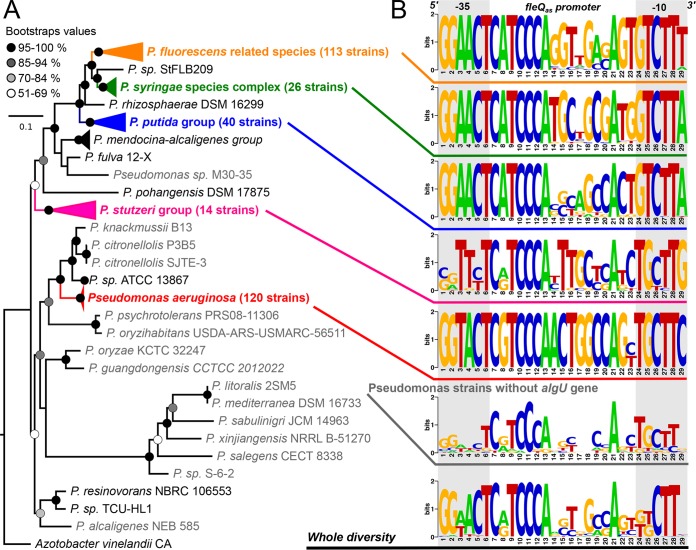
To determine the distribution of fleQas across the genus Pseudomonas, we analyzed fleQ gene sequences in all major Pseudomonas groups (35). Since we found the fleQas promoter to be AlgU dependent, we specifically tested the hypothesis that the fleQas promoter would be present only in Pseudomonas genomes that also contain the algU gene (Fig. 5).
FIG 5.
Diversity of the putative AlgU-dependent fleQas promoter in the genus Pseudomonas. (A) Rooted phylogenetic tree built from the alignment of 54 small and large ribosomal subunit protein sequences (see Table S2 in the supplemental material) of 339 complete Pseudomonas genomes (see Table S3 in the supplemental material) using the maximum-likelihood algorithm and a GTRGAMMAI model implemented in RAxML 8.2.11 (64). A total of 300 bootstrap replicates automatically determined by the MRE-based bootstrapping criterion were conducted under the rapid-bootstrapping algorithm to generate proportional support values. The tree is drawn to scale, and the branch length represents the number of substitutions per site. Genomes in which no sufficient homology with the P. syringae pv. tomato DC3000 algU gene was detected are colored light gray. (B) Sequence logos of putative AlgU-dependent promoter sequences at the fleQas locus in the major Pseudomonas groups (35) in which algU was detected in the genome. Sequence logos were also generated using the 20 Pseudomonas genomes in which no algU gene was detected (gray taxa in panel A) and another using all 339 genomes (42). Sequence logos were generated using the WebLogo application version 2.8.2 (65).
The fleQ gene was detected in all 339 Pseudomonas genomes that were analyzed (see Table S3 in the supplemental material). The 5′ region, where fleQas is located in P. syringae pv. tomato DC3000, was well conserved in most of them. When considering all strains, the Tajima D values (69) obtained both for the fleQas promoter and for algU were significantly higher than 2. This suggests that neither of the two loci evolved neutrally during the diversification of the genus Pseudomonas (Table 2). The ratio between the number of segregating sites and the fleQas promoter length, the population mutation rate (ϴ), and the nucleotide divergence average were much higher when considering all Pseudomonas genomes than when considering major Pseudomonas groups individually.
TABLE 2.
Pseudomonas fleQ, algU, and fleQas sequence polymorphism and divergencea
| Locus | Length (L) (bp)b | No. of segregating sites (S) | Ratio (S/L) | No. of alleles | Nucleotide divergencec | Population mutation rate (Θ) | Tajima's D estimated |
|---|---|---|---|---|---|---|---|
| fleQ | 1,402 | 920 | 0.66 | 239 | 0.18 | 0.21 | 1.81 (0.1 > P > 0.05 [NS]) |
| fleQas | 29 | 27 | 0.93 | 79 | 0.37 | 0.35 | 2.18 (P < 0.05) |
| algU | 983 | 720 | 0.73 | 197 | 0.28 | 0.23 | 3.05 (P < 0.01) |
Values are based on the strain list in Fig. 5.
Without missing data and gaps.
With Jukes and Cantor correction.
With statistical significance; NS, not significant.
When focusing on the fleQas promoter, the 17-nt promoter spacer region accounted for most of the sequence variability, while the −10 and −35 hexamer motifs were highly conserved within groups, with the exception of the Pseudomonas stutzeri group (Fig. 5). The −10 and −35 hexamer motifs of P. fluorescens and Pseudomonas putida were 100% and 99.2% identical to the P. syringae sequences, respectively. These groups differed from P. aeruginosa at the third position of the −35 motif and at the first, second, and sixth positions of the −10 motif.
Interestingly, the −10 and −35 hexamers showed a much lower level of conservation in a group of 20 strains (represented by 15 fleQas promoter alleles in Fig. 5) in which the algU gene was absent, supporting our hypothesis that fleQas is present and expressed only in strains that contain algU. Most of the strains missing algU and fleQas are closely related to the P. aeruginosa group (Fig. 5).
DISCUSSION
Here, we report the discovery and initial characterization of an AlgU-regulated antisense transcript expressed within the P. syringae pv. tomato DC3000 fleQ gene. The fleQas transcript was discovered through deep-sequencing analysis of the AlgU regulon and AlgU genomic binding locations (22). Our interest in the fleQas transcript developed from our desire to understand the transcriptional regulatory network responsible for AlgU-dependent downregulation of flagellar gene expression and flagellar motility. We tested the role of fleQas in motility regulation but instead found that AlgU+ cells capable of expressing the fleQas transcript were more motile than isogenic cells unable to express the fleQas transcript, indicating that fleQas has a positive effect on motility (Fig. 6). This suggests a role for fleQas in contexts where AlgU is active, such as during host interactions (10, 22). Alginate production is positively regulated by AlgU, which also downregulates flagellar gene expression (22) and motility (Fig. 4). We propose that the fleQas transcript functionally counteracts the AlgU-dependent downregulation of flagellar gene expression to provide for some movement under conditions where the presence of flagella is detrimental but low levels of movement are beneficial.
FIG 6.
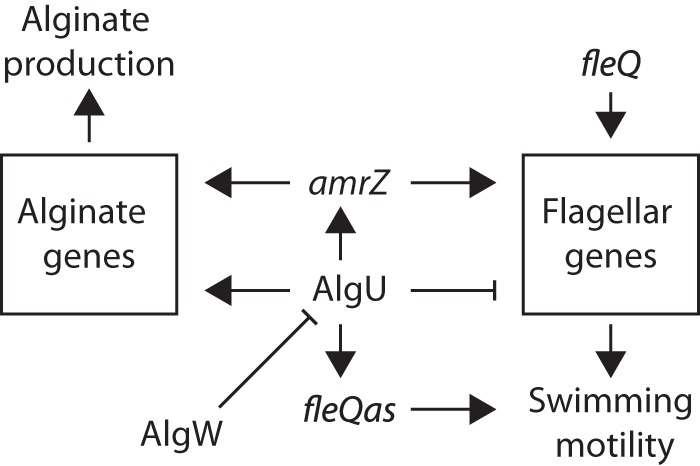
Portion of the P. syringae AlgU regulatory network controlling flagellar motility and alginate production. AlgU coordinates alginate production and suppression of motility by controlling the expression of genes necessary for these functions. AlgU also controls the expression of additional regulators, like AmrZ, which promotes alginate production and motility (67). FleQ is the master regulator of flagellar gene expression, and fleQas promotes motility when AlgU is expressed. The arrows and T-shaped bars indicate positive and negative effects on expression or function, respectively.
The transcription and motility data provide some insight into the mechanism of fleQas action. These data suggest that fleQas does not function by interfering with fleQ transcription or translation. This conclusion stems from the observation that transcription of fleQ and fliC is not altered by fleQas expression (Table 1). This is consistent with a model in which fleQas acts in trans by directly or indirectly affecting the expression or activity of some other component(s) of the flagellar or chemotaxis system. Escherichia coli and P. aeruginosa have mechanisms to adjust the torque generated by the flagellar motor to maintain rotation in more viscous environments (36–39). fleQas is controlled by AlgU, and the sigma factor also controls the production of alginate, which is a viscous exopolysaccharide that may increase the drag on flagellar rotation. Accordingly, there are P. aeruginosa alginate-biosynthetic mutants that are more motile than their isogenic progenitor (40). This suggests a possible role for fleQas in helping to adjust flagellar function in more viscous, alginate-rich environments, possibly by fine tuning stator composition, number, or function (36–39).
The functional tests that indicated that fleQas has a positive influence on flagellar motility relied on comparisons of wild-type and mutant strains that were no longer able to express the fleQas transcript. The mutations that we used to eliminate AlgU-dependent expression of fleQas did not alter the amino acid sequence of FleQ encoded on the opposite strand, but it is possible that expression of FleQ could have been altered because of a deviation from the naturally occurring codon sequences (41). However, in the absence of AlgU (i.e., no fleQas expression) we found no difference in the motility (Fig. 4) or expression (Table 1) of the fleQ and fliC genes with AlgU expressed when comparing cells with the wild-type mutant fleQas promoter. The most parsimonious explanation for these observations is that FleQ function was unchanged by these mutations and that wild-type and mutant FleQ proteins were expressed similarly and were equally effective at activating motility.
We considered the conservation of the fleQas promoter to ascertain whether fleQas was a genetic feature peculiar to P. syringae pv. tomato DC3000 or whether it is present in other bacteria, as well. We found evidence for widespread conservation of the fleQas promoter among the pseudomonads, and strikingly, the conservation (barring P. stutzeri) was limited to pseudomonads encoding an AlgU orthologue (Fig. 5). The divergence of the fleQas promoter in P. stutzeri might contribute to differences in motility and/or colony morphology that distinguish these bacteria from the other pseudomonads (39, 42). The finding that the fleQas promoter is mainly conserved in species that also encode an AlgU orthologue supports the hypothesis that the fleQas promoter is under purifying selection and not merely a product of selection acting on the fleQ gene present on the opposite strand. Together, these findings support the conclusion that fleQas is functional and is maintained over much of Pseudomonas diversification despite very different niche specializations (i.e., the species represent human, insect, and plant pathogens, saprophytes, and soil-dwelling bacteria).
We propose that fleQas functions as a small RNA (sRNA) and not an as mRNA coding for a small protein. Although we did not explicitly test this idea, there are not any credible open reading frames in the fleQas gene. The transcript-mapping and transcriptome data suggest that most of the fleQas molecules are on the order of 100 nt in length. The first ATG does not occur until 203 nt downstream of the observed 5′ ends (Fig. 2). This means that most of these transcripts would not include a canonical translation start site and therefore are unlikely to be translated. In general, antisense transcription is rather common in P. syringae pv. tomato DC3000: 124 antisense transcripts were detected in a whole-genome transcriptional analysis, which accounts for antisense transcription in approximately 2% of the annotated genes (43, 44). The fleQas transcript was not observed in the global transcriptome (43), but this was expected, because AlgU was not active under the conditions tested in that study.
Flagellum production in other bacteria is regulated by analogous multistage expression programs that also provide appropriate spatial and temporal expression of the flagellar subunits (45). So far, 29 trans-acting sRNAs have been found to regulate flagellum-mediated motility in E. coli (46–48). At least five of these small RNAs control flagellar expression by direct binding to the 5′ untranslated region (UTR) of the flhDC transcript (46). The flhDC genes encode the master regulator and functional analog of Pseudomonas FleQ in E. coli (45). Antisense transcription of the fliPQR genes in Salmonella enterica promotes motility (49), and in Listeria monocytogenes, a long 5′ UTR of the mogR mRNA is an antisense RNA (asRNA) for three genes involved in flagellar synthesis that may be part of a mechanism to provide temperature-dependent control of motility (50). A predicted asRNA contains sequences complementary to the fliM mRNA and possibly controls flagellar motility in Helicobacter pylori (51).
These sRNA systems may supplement or provide alternative regulatory possibilities to typical proteinaceous gene expression regulators. An emerging theme is that these RNA regulators function to optimize flagellar expression in response to environmental conditions. P. syringae fleQas likely serves in the same general capacity as the sRNA and asRNA molecules found in other organisms (46–51), that is, to provide a quick and efficient mechanism for adjusting flagellar activity to help the bacteria survive and to maximize growth in the face of changing environmental conditions.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Except where noted, P. syringae pv. tomato DC3000 and derivative strains were grown at 30°C in Kings B (KB) medium (52) or on KB medium solidified with 1.5% (wt/vol) agar, with 10 μg/ml gentamicin (Gm) and 50 μg/ml kanamycin (Km) added for selection for plasmid maintenance when necessary. E. coli DH5α and TOP10 were used as the hosts for molecular cloning and other plasmid manipulations used in this work. E. coli was grown at 37°C in LB medium or LB medium solidified with 1.5% (wt/vol) agar. All the bacterial strains used in these experiments are shown in Table 3.
TABLE 3.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference |
|---|---|---|
| P. syringae pv. tomato DC3000 | Wild type | 56 |
| PS368 | DC3000 ΔalgU mucAB | 22 |
| PS659 | DC3000 fleQas promoter mutant | This work |
| PS664 | DC3000 ΔalgU mucAB fleQas promoter mutant | This work |
| pEM53 | AlgU expression plasmid; Gmr | 22 |
| pJN105 | Empty-vector control expression plasmid; Gmr | 68 |
| pBS44 | Empty Lux promoter trap vector; Kmr | 55 |
| pHW7 | PfleQas::lux Kmr | This work |
| pEM90 | PfleQas(mut1)::lux Kmr | This work |
| pEM91 | PfleQas(mut2)::lux Kmr | This work |
| pEM92 | PfleQas(mut3)::lux Kmr | This work |
| pEM93 | PfleQas(mut1,2,3)::lux Kmr | This work |
| pK18mobsacB | Cloning vector for site-specific genomic mutations | 58 |
| pEM99 | pK18mobsacB::PfleQas(mut1,2,3) chromosomal mutagenesis vector | This work |
RNA isolation.
RNA was isolated from bacteria using the TRIzol extraction method. Briefly, bacteria were grown overnight in KB medium with 10 μg/ml gentamicin, diluted to an optical density at 600 nm (OD600) of 0.1 in fresh KB medium supplemented with 5 μg/ml gentamicin and 0.2% arabinose, and cultured to an OD600 of approximately 0.5. The cell pellets were treated with 1 ml of Life Technologies TRIzol reagent, extracted using a Zymo Research Direct-zol RNA MiniPrep kit, and treated by off-column DNase digestion using Ambion DNase I. The RNA was purified from this mixture using a Zymo Research RNA Clean and Concentrator-25 kit. The total RNA was diluted to 100 ng/μl and stored at −80°C.
RT-PCR and transcript relative quantification.
Diluted total RNA was converted to cDNA using Life Technologies Superscript III reverse transcriptase and RNase Out recombinant RNase inhibitor following the first-strand cDNA synthesis protocol with gene-specific primers (see Table S1 in the supplemental material). RNA was removed by adding RNase H and incubating at 37°C for 20 min. To test for the presence of reverse transcription products, cDNA was amplified using Life Technologies Platinum Taq DNA Polymerase High Fidelity and primers (see Table S1 in the supplemental material) to create approximately 100-bp amplicons.
Transcript relative quantification was done using real-time PCR performed on a My IQ5 sequence detection system (Bio-Rad) and iQ SYBR green Supermix (Bio-Rad) according to the manufacturer's protocols. Six microliters of the diluted cDNA was mixed with each primer (see Table S1 in the supplemental material) (final concentration, 0.4 μM) and 10 μl of master mix in a 20-μl final volume. The PCR was carried out with 1 cycle at 95°C for 2.5 min, followed by 40 cycles of 95°C for 15 s and 60°C for 30 s. The amount of fluorescence that resulted from incorporation of the SYBR green dye into double-stranded DNA was measured at the end of each cycle to determine the amplification kinetics. DNA contamination and formation of primer dimers were assessed using a no-reverse transcriptase control and a no-template control, respectively. Production of nonspecific products was determined by the dissociation protocol provided with the My IQ5 real-time PCR machine. The resulting threshold cycle (CT) values were calculated using the My IQ5 software and analyzed using the comparative CT method (separate tubes) described in Applied Biosystems (ABI) user bulletin no. 2 (53). The CT value of each gene tested was normalized to the CT value of housekeeping gene gap-1 as described previously 54.
5′ and 3′ RACE.
The ends of fleQas transcripts were identified using 5′ and 3′ RACE. Invitrogen 5′ RACE was performed as described by the manufacturer except where noted. The fleQas cDNAs were produced from diluted total RNA using the gene-specific primer 6906 (see Table S1 in the supplemental material) and Life Technologies SuperScript II reverse transcriptase, and the remaining RNA was degraded by incubation with RNase at 37°C for 20 min. The products were purified using Zymo Research Oligo Clean and Concentrator-25. dCTP was added to the cDNA ends with terminal deoxynucleotidyltransferase. The fleQas sequences were amplified from the deoxycytidine (dC)-tailed cDNA using gene-specific primer 6910, the provided abridged anchor primer, and Invitrogen Taq polymerase. The PCR products were amplified again with nested primer 6910, the abridged universal adapter primer, and Invitrogen Taq polymerase. The PCR products were resolved and excised from a 1% agarose gel and purified using a Zymo Research gel recovery kit. The purified products were cloned in a TOPO-TA 2.1 cloning vector using E. coli TOP10. The plasmids were recovered using a Qiagen Mini Plasmid kit and sequenced using the M13 forward and M13 reverse primers.
3′ RACE methods were adapted from those reported previously (43). Briefly, total RNA was dephosphorylated using New England BioLabs Antarctic phosphatase and purified using a Zymo Research Oligo Clean and Concentrator Kit-5. RNA oligonucleotide 6915 was ligated to the 3′ ends of the dephosphorylated total RNA using T4 single-stranded RNA (ssRNA) ligase and purified using the Zymo Research Oligo Clean and Concentrator Kit-5. cDNA was produced using Life Technologies Superscript III reverse transcriptase and RNase Out recombinant RNase inhibitor following the first-strand cDNA synthesis protocol with gene-specific primer 6914. RNA was removed by adding RNase mix and incubating at 37°C for 20 min. The cDNA was then amplified using Invitrogen Taq polymerase, gene-specific primer 6912, and anchor primer 6914. The PCR products were amplified again with a nested primer, 6913; anchor primer 6914; and Invitrogen Taq polymerase. Three distinct 3′ RACE products were excised and recovered from a 1% agarose gel using a Zymo Research gel recovery kit. These products were cloned in TOPO Vector-TA 2.1 and transformed into E. coli DH5α. Plasmids were extracted using Qiagen Mini Plasmid Prep and sequenced using M13 forward and M13 reverse primers.
Promoter::lux fusion cloning.
The region upstream of the fleQas gene (NC_004578.1; 4256861..4257095) (55) was PCR amplified from P. syringae pv. tomato DC3000 genomic DNA using primers 6916 and 6917. The correct size of the PCR product was verified by agarose gel electrophoresis, and then it was cloned in pENTR/D by directional TOPO cloning (ThermoFisher Scientific) according to the manufacturer's directions and used to transform E. coli TOP10. The four inserts containing the mutant promoter derivatives were cloned as gBlocks (IDT, Coralville, IA) in pENTR/D. The cloned inserts were sequenced using primers 2432, 2433, 6916, and 6917. These plasmids were then used to generate the lux reporter constructs by attL and attR (LR) recombination with the pBS58 destination vector using LR Clonase II enzyme mixture as described in reference 54 and are listed in Table 3. The final plasmids were sequenced to confirm the sequence and orientation of the cloned fragment.
Lux assay.
P. syringae pv. tomato DC3000 ΔalgU mucAB containing pEM53 or pJN105 and transformed with the promoter::lux fusion constructs were grown in KB medium containing Km and Gm. The cultures were diluted 1:10 in KB medium with a final concentration of 0.2% l-(+)-arabinose, 25 ng/ml kanamycin, and 5 ng/ml gentamicin. The luminescence and OD600 were measured on a BioTek Synergy 2 plate reader after shaking at room temperature for 6 h.
Chromosomal promoter mutagenesis.
The native fleQas promoter in the genome of P. syringae pv. tomato DC3000 and that of P. syringae pv. tomato DC3000 ΔalgU mucAB were replaced with the mutant version containing all three nucleotide substitutions by marker exchange mutagenesis. The plasmid directing the mutagenesis was constructed using splicing by overlap extension (SOE) PCR (56) of the ∼1-kb regions flanking the target and contained complementary sequences that included the promoter sequences with the PCR-introduced substitutions. The product of the SOE reaction was cloned in pK18MobSacB (57) to generate pEM99, which was sequenced to confirm the sequence of the cloned insert. The pEM99 construct was then used to transform P. syringae pv. tomato DC3000 and the ΔalgU mucAB mutant, and kanamycin-resistant clones were selected and then counterselected using sucrose as described in reference 58, producing the unmarked mutants. The mutant allele and flanking regions were confirmed by DNA sequence analysis.
Swimming-motility assay.
Bacteria were grown overnight in KB broth with 5 μg/ml gentamicin at 28°C with shaking and diluted to an OD600 of 0.3. Swimming medium (59) containing 0.3% agar, 5 μg/ml gentamicin, and 0.2% l-(+)-arabinose was inoculated with 2 μl of the diluted bacterial suspension and incubated for 44 h at room temperature.
Phylogenetic and computational analyses.
A total of 339 complete Pseudomonas genome sequences, representing all the major clades described to date, were downloaded from the public NCBI database in August 2017 (see Table S3 in the supplemental material). To reconstruct the evolutionary history of the genus Pseudomonas, the gene sequences coding for the 54 proteins of the large and small ribosomal subunits of P. syringae pv. tomato DC3000 (60) were aligned gene by gene against all the Pseudomonas genomes using BIGSdb open source software (61). Each gene was searched in the 339 genomes using the BLASTN algorithm and setting parameters for a locus match to a minimum of 70% sequence identity over a minimum of 50% of the query sequence length. Applying this similarity threshold, only 31 ribosomal genes could be detected in all the Pseudomonas genomes (see Table S3 in the supplemental material) and in Azotobacter vinelandii strain CA (used as the outgroup), and they were used for phylogenetic reconstruction. The sequences of the ribosomal genes were aligned individually with MAFFT 7 (62) and then concatenated into a single alignment scanned with Gblocks 0.91b (63) to remove any ambiguous position. A maximum-likelihood (ML) tree was built from this alignment with RAxML 8.2.11 (64) under a GAMMA model of rate heterogeneity using empirical base frequencies, estimating a generalized time-reversible model for nucleotide substitution and including an estimate of the proportion of invariable sites. A total of 300 bootstrap replicates automatically determined by the extended majority rule-based bootstrapping criterion (70) were conducted under the rapid-bootstrapping algorithm.
To explore the diversity of the putative AlgU-dependent promoter of fleQas, the fleQ and algU gene sequences of P. syringae pv. tomato DC3000 were aligned against all the genomes following the same approach described above for the ribosomal genes. All the fleQ alleles thus identified were then aligned, and the putative promoter of the fleQas sequence, previously identified by ChIP-seq analysis in P. syringae pv. tomato DC3000 (22), was extracted. The fleQas sequences were binned into major groups according to the ribosomal-gene-based phylogeny and NCBI taxonomy (35), and sequence variability was illustrated by a sequence logo generated by the WebLogo application version 2.8.2 (65). Several population genetic analyses were performed for each locus and group with DnaSP v5.10.1 (66) to characterize locus polymorphism and divergence, including (i) the average pairwise genetic distances (based on the Jukes-Cantor model), (ii) the population scale mutation rate (θ), and (iii) Tajima's D to test locus neutrality.
Supplementary Material
ACKNOWLEDGMENT
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00576-17.
REFERENCES
- 1.Haiko J, Westerlund-Wikström B. 2013. The role of the bacterial flagellum in adhesion and virulence. Biology 2:1242–1267. doi: 10.3390/biology2041242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haefele DM, Lindow SE. 1987. Flagellar motility confers epiphytic fitness advantages upon Pseudomonas syringae. Appl Environ Microbiol 53:2528–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clarke CR, Chinchilla D, Hind SR, Taguchi F, Miki R, Ichinose Y, Martin GB, Leman S, Felix G, Vinatzer BA. 2013. Allelic variation in two distinct Pseudomonas syringae flagellin epitopes modulates the strength of plant immune responses but not bacterial motility. New Phytol 200:847–860. doi: 10.1111/nph.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. 2004. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 5.Dons L, Eriksson E, Jin Y, Martin E, Kristensson K, Larsen CN, Olsen JE, Rottenberg ME. 2004. Role of flagellin and the two-component CheA/CheY system of Listeria monocytogenes in host cell invasion and Virulence. Infect Immun 72:3237–3244. doi: 10.1128/IAI.72.6.3237-3244.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai MA, Quarles EK, López-Yglesias AH, Zhao X, Hajjar AM, Smith KD. 2013. Innate immune detection of flagellin positively and negatively regulates salmonella infection. PLoS One 8:e72047. doi: 10.1371/journal.pone.0072047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xin XF, He SY. 2013. Pseudomonas syringae pv. tomato DC3000: A model pathogen for probing disease susceptibility and hormone signaling in plants. Annu Rev Phytopathol 51:473–498. doi: 10.1146/annurev-phyto-082712-102321. [DOI] [PubMed] [Google Scholar]
- 8.Clarke CR, Hayes BW, Runde BJ, Markel E, Swingle BM, Vinatzer BA. 2016. Comparative genomics of Pseudomonas syringae pathovar tomato reveals novel chemotaxis pathways associated with motility and plant pathogenicity. PeerJ 4:e2570. doi: 10.7717/peerj.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu X, Lund SP, Scott RA, Greenwald JW, Records AH, Nettleton D, Lindow SE, Gross DC, Beattie GA. 2013. Transcriptional responses of Pseudomonas syringae to growth in epiphytic versus apoplastic leaf sites. Proc Natl Acad Sci U S A 110:E425–E434. doi: 10.1073/pnas.1221892110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu X, Lund SP, Greenwald JW, Records AH, Scott RA, Nettleton D, Lindow SE, Gross DC, Beattie GA. 2014. Transcriptional analysis of the global regulatory networks active in Pseudomonas syringae during leaf colonization. mBio 5:e01683-. doi: 10.1128/mBio.01683-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schreiber KJ, Desveaux D. 2011. AlgW regulates multiple Pseudomonas syringae virulence strategies. Mol Microbiol 80:364–377. doi: 10.1111/j.1365-2958.2011.07571.x. [DOI] [PubMed] [Google Scholar]
- 12.Park SH, Butcher BG, Anderson Z, Pellegrini N, Bao Z, D'Amico K, Filiatrault MJ. 2013. Analysis of the small RNA P16/RgsA in the plant pathogen Pseudomonas syringae pv. tomato strain DC3000. Microbiology 159:296–306. doi: 10.1099/mic.0.063826-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pel MJC, van Dijken AJH, Bardoel BW, Seidl MF, van der Ent S, van Strijp JAG, Pieterse CMJ. 2014. Pseudomonas syringae evades host immunity by degrading flagellin monomers with alkaline protease AprA. Mol Plant Microbe Interact 27:603–610. doi: 10.1094/MPMI-02-14-0032-R. [DOI] [PubMed] [Google Scholar]
- 14.Bardoel BW, van der Ent S, Pel MJC, Tommassen J, Pieterse CMJ, van Kessel KPM, van Strijp JAG. 2011. Pseudomonas evades immune recognition of flagellin in both mammals and plants. PLoS Pathog 7:e1002206. doi: 10.1371/journal.ppat.1002206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindeberg M, Cartinhour S, Myers CR, Schechter LM, Schneider DJ, Collmer A. 2006. Closing the circle on the discovery of genes encoding Hrp regulon members and type III secretion system effectors in the genomes of three model Pseudomonas syringae strains. Mol Plant Microbe Interact 19:1151–1158. doi: 10.1094/MPMI-19-1151. [DOI] [PubMed] [Google Scholar]
- 16.Grant SR, Fisher EJ, Chang JH, Mole BM, Dangl JL. 2006. Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu Rev Microbiol 60:425–449. doi: 10.1146/annurev.micro.60.080805.142251. [DOI] [PubMed] [Google Scholar]
- 17.Kvitko BH, Park DH, Velasquez AC, Wei CF, Russell AB, Martin GB, Schneider DJ, Collmer A. 2009. Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog 5:e1000388. doi: 10.1371/journal.ppat.1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindeberg M, Cunnac S, Collmer A. 2012. Pseudomonas syringae type III effector repertoires: last words in endless arguments. Trends Microbiol 20:199–208. doi: 10.1016/j.tim.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Cunnac S, Chakravarthy S, Kvitko BH, Russell AB, Martin GB, Collmer A. 2011. Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proc Natl Acad Sci U S A 108:2975–2980. doi: 10.1073/pnas.1013031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cezairliyan BO, Sauer RT. 2009. Control of Pseudomonas aeruginosa AlgW protease cleavage of MucA by peptide signals and MucB. Mol Microbiol 72:368–379. doi: 10.1111/j.1365-2958.2009.06654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wood LF, Leech AJ, Ohman DE. 2006. Cell wall-inhibitory antibiotics activate the alginate biosynthesis operon in Pseudomonas aeruginosa: roles of sigma (AlgT) and the AlgW and Prc proteases. Mol Microbiol 62:412–426. doi: 10.1111/j.1365-2958.2006.05390.x. [DOI] [PubMed] [Google Scholar]
- 22.Markel E, Stodghill P, Bao Z, Myers CR, Swingle B. 2016. AlgU controls expression of virulence genes in Pseudomonas syringae pv. tomato DC3000. J Bacteriol 198:2330–2344. doi: 10.1128/JB.00276-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soutourina OA, Bertin PN. 2003. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol Rev 27:505–523. doi: 10.1016/S0168-6445(03)00064-0. [DOI] [PubMed] [Google Scholar]
- 24.Vargas P, Farias GA, Nogales J, Prada H, Carvajal V, Baron M, Rivilla R, Martin M, Olmedilla A, Gallegos MT. 2013. Plant flavonoids target Pseudomonas syringae pv. tomato DC3000 flagella and type III secretion system. Environ Microbiol Rep 5:841–850. doi: 10.1111/1758-2229.12086. [DOI] [PubMed] [Google Scholar]
- 25.Quiñones B, Dulla G, Lindow SE. 2005. Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae. Mol Plant-Microbe Interact 18:682–693. doi: 10.1094/MPMI-18-0682. [DOI] [PubMed] [Google Scholar]
- 26.Roine E, Wei W, Yuan J, Nurmiaho-Lassila EL, Kalkkinen N, Romantschuk M, He SY. 1997. Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci U S A 94:3459–3464. doi: 10.1073/pnas.94.7.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nogales J, Vargas P, Farias GA, Olmedilla A, Sanjuan J, Gallegos MT. 2015. FleQ coordinates flagellum-dependent and -independent motilities in Pseudomonas syringae pv. tomato DC3000. Appl Environ Microbiol 81:7533–7545. doi: 10.1128/AEM.01798-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, Ramphal R. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol 50:809–824. doi: 10.1046/j.1365-2958.2003.03740.x. [DOI] [PubMed] [Google Scholar]
- 29.Tart AH, Wolfgang MC, Wozniak DJ. 2005. The alternative sigma factor AlgT represses. J Bacteriol 187:7955–7962. doi: 10.1128/JB.187.23.7955-7962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tart AH, Blanks MJ, Wozniak DJ. 2006. The AlgT-dependent transcriptional regulator AmrZ (AlgZ) inhibits flagellum biosynthesis in mucoid, nonmotile Pseudomonas aeruginosa cystic fibrosis isolates. J Bacteriol 188:6483–6489. doi: 10.1128/JB.00636-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez-Granero F, Navazo A, Barahona E, Redondo-Nieto M, Rivilla R, Martin M. 2012. The Gac-Rsm and SadB signal transduction pathways converge on AlgU to downregulate motility in Pseudomonas fluorescens. PLoS One 7:e31765. doi: 10.1371/journal.pone.0031765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guzina J, Djordjevic M. 2016. Promoter recognition by extracytoplasmic function sigma factors: analyzing DNA and protein interaction motifs. J Bacteriol 198:1927–1938. doi: 10.1128/JB.00244-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lonetto MA, Brown KL, Rudd KE, Buttner MJ. 1994. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase sigma factors involved in the regulation of extracytoplasmic functions. Proc Natl Acad Sci U S A 91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lane WJ, Darst SA. 2006. The structural basis for promoter -35 element recognition by the group IV sigma factors. PLoS Biol 4:e269. doi: 10.1371/journal.pbio.0040269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomila M, Peña A, Mulet M, Lalucat J. 2015. Phylogenomics and systematics in Pseudomonas. Front Microbiol 6:214. doi: 10.3389/fmicb.2015.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tipping MJ, Delalez NJ, Lim R, Berry RM, Armitage P. 2013. Load-dependent assembly of the bacterial flagellar motor. mBio 4:e00551-. doi: 10.1128/mBio.00551-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baker AE, O'Toole GA. 2017. Bacteria, rev your engines: stator dynamics regulate flagellar motility. J Bacteriol 199:e00088-. doi: 10.1128/JB.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luscombe NM, Laskowski RA, Thornton JM. 2001. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res 29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thormann KM, Paulick A. 2010. Tuning the flagellar motor. Microbiology 156:1275–1283. doi: 10.1099/mic.0.029595-0. [DOI] [PubMed] [Google Scholar]
- 40.Fata Moradali M, Donati I, Sims IM, Ghods S, Rehma BHA. 2015. Alginate polymerization and modification are linked in Pseudomonas aeruginosa. mBio 6:e00453-15. doi: 10.1128/mBio.00453-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Powell JR, Dion K. 2015. Effects of codon usage on gene expression: empirical studies on Drosophila. J Mol Evol 80:219–226. doi: 10.1007/s00239-015-9675-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lalucat J, Bennasar A, Bosch R, Garcı E. 2006. Biology of Pseudomonas stutzeri. Microbiol Mol Biol Rev 70:510–547. doi: 10.1128/MMBR.00047-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Filiatrault MJ, Stodghill PV, Bronstein PA, Moll S, Lindeberg M, Grills G, Schweitzer P, Wang W, Schroth GP, Luo S, Khrebtukova I, Yang Y, Thannhauser T, Butcher BG, Cartinhour S, Schneider DJ. 2010. Transcriptome analysis of Pseudomonas syringae identifies new genes, noncoding RNAs, and antisense activity. J Bacteriol 192:2359–2372. doi: 10.1128/JB.01445-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Georg J, Hess WR. 2011. cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev 75:286–300. doi: 10.1128/MMBR.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith TG, Hoover TR. 2009. Deciphering bacterial flagellar gene regulatory networks in the genomic era. Adv Appl Microbiol 67:257–295. doi: 10.1016/S0065-2164(08)01008-3. [DOI] [PubMed] [Google Scholar]
- 46.De Lay N, Gottesman S. 2012. A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol Microbiol 86:524–538. doi: 10.1111/j.1365-2958.2012.08209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomason MK, Fontaine F, De Lay N, Storz G. 2012. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol Microbiol 84:17–35. doi: 10.1111/j.1365-2958.2012.07965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bak G, Lee J, Suk S, Kim D, Young Lee J, Kim KS, Choi BS, Lee Y. 2015. Identification of novel sRNAs involved in biofilm formation, motility, and fimbriae formation in Escherichia coli. Sci Rep 5:15287. doi: 10.1038/srep15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Q, Harshey RM. 2009. Rcs signalling-activated transcription of rcsA induces strong anti-sense transcription of upstream fliPQR flagellar genes from a weak intergenic promoter: regulatory roles for the anti-sense transcript in virulence and motility. Mol Microbiol 74:71–84. doi: 10.1111/j.1365-2958.2009.06851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, Barthelemy M, Vergassola M, Nahori MA, Soubigou G, Regnault B, Coppee JY, Lecuit M, Johansson J, Cossart P. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 51.Xiao B, Li W, Guo G, Li B, Liu Z, Jia K, Guo Y, Mao X, Zou Q. 2009. Identification of small noncoding RNAs in Helicobacter pylori by a bioinformatics-based approach. Curr Microbiol 58:258–263. doi: 10.1007/s00284-008-9318-2. [DOI] [PubMed] [Google Scholar]
- 52.King EO, Ward MK, Raney DE. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 44:301–307. [PubMed] [Google Scholar]
- 53.Applied Biosystems. 2001. User bulletin number 2. ABI Prism 7700 sequence detection system. Applied Biosystems, Foster City, CA. [Google Scholar]
- 54.Swingle B, Thete D, Moll M, Myers CR, Schneider DJ, Cartinhour S. 2008. Characterization of the PvdS-regulated promoter motif in Pseudomonas syringae pv. tomato DC3000 reveals regulon members and insights regarding PvdS function in other pseudomonads. Mol Microbiol 68:871–889. doi: 10.1111/j.1365-2958.2008.06209.x. [DOI] [PubMed] [Google Scholar]
- 55.Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, Dodson RJ, Deboy RT, Durkin AS, Kolonay JF, Madupu R, Daugherty S, Brinkac L, Beanan MJ, Haft DH, Nelson WC, Davidsen T, Zafar N, Zhou LW, Liu J, Yuan QP, Khouri H, Fedorova N, Tran B, Russell D, Berry K, Utterback T, Van Aken SE, Feldblyum TV, D'Ascenzo M, Deng WL, Ramos AR, Alfano JR, Cartinhour S, Chatterjee AK, Delaney TP, Lazarowitz SG, Martin GB, Schneider DJ, Tang XY, Bender CL, White O, Fraser CM, Collmer A. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci U S A 100:10181–10186. doi: 10.1073/pnas.1731982100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 57.Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 58.Kvitko BH, Collmer A. 2011. Construction of Pseudomonas syringae pv. tomato DC3000 mutant and polymutant strains. Methods Mol Biol 712:109–128. doi: 10.1007/978-1-61737-998-7_10. [DOI] [PubMed] [Google Scholar]
- 59.Rashid MH, Rao NN, Kornberg A. 2000. Inorganic polyphosphate is required for motility of bacterial pathogens. J Bacteriol 182:225–227. doi: 10.1128/JB.182.1.225-227.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fouts DE, Abramovitch RB, Alfano JR, Baldo AM, Buell CR, Cartinhour S, Chatterjee AK, D'Ascenzo M, Gwinn ML, Lazarowitz SG, Lin NC, Martin GB, Rehm AH, Schneider DJ, van Dijk K, Tang X, Collmer A. 2002. Genomewide identification of Pseudomonas syringae pv. tomato DC3000 promoters controlled by the HrpL alternative sigma factor. Proc Natl Acad Sci U S A 99:2275–2280. doi: 10.1073/pnas.032514099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jolley KA, Maiden MCJ. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 64.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 67.Prada-Ramirez HA, Perez-Mendoza D, Felipe A, Martinez-Granero F, Rivilla R, Sanjuan J, Gallegos MT. 2016. AmrZ regulates cellulose production in Pseudomonas syringae pv. tomato DC3000. Mol Microbiol 99:960–977. doi: 10.1111/mmi.13278. [DOI] [PubMed] [Google Scholar]
- 68.Newman JR, Fuqua C. 1999. Broad-host-range expression vectors that carry the l-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197–203. doi: 10.1016/S0378-1119(98)00601-5. [DOI] [PubMed] [Google Scholar]
- 69.Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. 2009. How many bootstrap replicates are necessary? Res Comput Mol Biol 5541:184–200. doi: 10.1007/978-3-642-02008-7_13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.