Abstract
Background
l-Arabinose occurs at economically relevant levels in lignocellulosic hydrolysates. Its low-affinity uptake via the Saccharomyces cerevisiae Gal2 galactose transporter is inhibited by d-glucose. Especially at low concentrations of l-arabinose, uptake is an important rate-controlling step in the complete conversion of these feedstocks by engineered pentose-metabolizing S. cerevisiae strains.
Results
Chemostat-based transcriptome analysis yielded 16 putative sugar transporter genes in the filamentous fungus Penicillium chrysogenum whose transcript levels were at least threefold higher in l-arabinose-limited cultures than in d-glucose-limited and ethanol-limited cultures. Of five genes, that encoded putative transport proteins and showed an over 30-fold higher transcript level in l-arabinose-grown cultures compared to d-glucose-grown cultures, only one (Pc20g01790) restored growth on l-arabinose upon expression in an engineered l-arabinose-fermenting S. cerevisiae strain in which the endogenous l-arabinose transporter, GAL2, had been deleted. Sugar transport assays indicated that this fungal transporter, designated as PcAraT, is a high-affinity (Km = 0.13 mM), high-specificity l-arabinose-proton symporter that does not transport d-xylose or d-glucose. An l-arabinose-metabolizing S. cerevisiae strain in which GAL2 was replaced by PcaraT showed 450-fold lower residual substrate concentrations in l-arabinose-limited chemostat cultures than a congenic strain in which l-arabinose import depended on Gal2 (4.2 × 10−3 and 1.8 g L−1, respectively). Inhibition of l-arabinose transport by the most abundant sugars in hydrolysates, d-glucose and d-xylose was far less pronounced than observed with Gal2. Expression of PcAraT in a hexose-phosphorylation-deficient, l-arabinose-metabolizing S. cerevisiae strain enabled growth in media supplemented with both 20 g L−1 l-arabinose and 20 g L−1 d-glucose, which completely inhibited growth of a congenic strain in the same condition that depended on l-arabinose transport via Gal2.
Conclusion
Its high affinity and specificity for l-arabinose, combined with limited sensitivity to inhibition by d-glucose and d-xylose, make PcAraT a valuable transporter for application in metabolic engineering strategies aimed at engineering S. cerevisiae strains for efficient conversion of lignocellulosic hydrolysates.
Electronic supplementary material
The online version of this article (10.1186/s13068-018-1047-6) contains supplementary material, which is available to authorized users.
Keywords: Penicillium, Transcriptome, Sugar transport, Proton symport, l-Arabinose transporter, Second-generation bioethanol, Yeast, Metabolic engineering
Background
At an annual production of 100 Mton [1], bioethanol produced by the yeast Saccharomyces cerevisiae is by volume the largest fermentation product in industrial biotechnology. Cane sugar and corn starch, which are still the predominant feedstocks for bioethanol production, almost exclusively yield sucrose and d-glucose as fermentable sugars. Alternative lignocellulosic feedstocks, derived from agricultural residues or energy crops, contain cellulose, hemicellulose, and in some cases, pectin [2]. The pentoses d-xylose and l-arabinose typically represent 10–25 and 2–3%, respectively, of the monomeric sugars in lignocellulosic hydrolysates [3]. Some industrially relevant hydrolysates, however, contain higher l-arabinose concentrations. For instance, in hydrolysates of corn fibre and sugar beet pulp, l-arabinose represents 16 and 26% of the total sugar content, respectively [4, 5].
Whilst pentose sugars are not natural substrates of S. cerevisiae, their efficient conversion to ethanol and, ultimately, other bulk products, is essential to ensure economically viable processes [6]. Extensive metabolic and evolutionary engineering has been applied to enable efficient xylose fermentation, based on expression of either a heterologous xylose reductase and xylitol dehydrogenase, or a heterologous xylose isomerase (reviewed by [7] and [8]). Construction of yeast strains capable of l-arabinose fermentation involved functional expression of bacterial genes encoding l-arabinose isomerase (AraA), l-ribulokinase (AraB), and l-ribulose-5-phosphate-4-epimerase (AraD) [9–13]. Additional overexpression of S. cerevisiae genes encoding enzymes of the non-oxidative pentose phosphate pathway (RPE1, RKI1, TAL1, and TKL1) strongly improved rates of d-xylose and l-arabinose fermentation [12, 14]. In S. cerevisiae strains whose metabolic pathways have been intensively optimized for pentose fermentation by metabolic and evolutionary engineering, uptake of l-arabinose and d-xylose is an important rate-controlling step [15–17].
Several S. cerevisiae plasma membrane hexose-transporter proteins are able to transport d-xylose and/or l-arabinose but invariably exhibit a high Km for these pentoses [18–25]. This low affinity causes sluggish pentose conversion (‘tailing’) towards the end of anaerobic batch cultures. Amongst the set of 18 S. cerevisiae hexose transporters (Hxt1-17 and Gal2), only the galactose transporter Gal2 and with, much lower activities, Hxt9 and Hxt10 support l-arabinose import [18, 19]. Gal2 has a high affinity for d-glucose and galactose but its affinity for l-arabinose is low (Km = 57–371 mM) [19, 26]. Consequently, engineered strains in which l-arabinose transport depends on Gal2 fail to grow at low l-arabinose concentrations [19]. Moreover, even when d-glucose-induced transcriptional repression of GAL2 [27–29] is prevented, kinetic competition prevents l-arabinose consumption by such strains in the presence of d-glucose.
So far, few heterologous l-arabinose transporters have been functionally expressed and characterized in S. cerevisiae [19, 26, 30]. In these previous studies, S. cerevisiae strains harbouring a functional l-arabinose fermentation pathway but no native hexose transporters proved to be excellent platforms for characterization of heterologous l-arabinose transporters. In such experiments, transporters from the yeasts Scheffersomyces stipitis (SsAraT), Pichia guilliermondii (PgAxt1) and from the plant Arabidopsis thaliana (AtStp2) were shown to support l-arabinose transport in S. cerevisiae. These transporters exhibited Km values of 0.13–4.5 mM but low transport capacities, whilst also exhibiting severe d-glucose inhibition [19, 26]. Inhibition by d-xylose was only studied for PgAxt1, where it completely blocked l-arabinose uptake [26]. Conversely, l-arabinose transporters from the fungi Neurospora crassa (Lat-1) and Myceliophthora thermophilum (MtLat-1) supported high-capacity, low-affinity (Km = 58 and 29 mM, respectively) l-arabinose uptake and were also strongly affected by d-glucose inhibition [30]. The strong inhibition of these transporters by d-glucose and/or d-xylose precludes the simultaneous utilization of d-glucose and l-arabinose in S. cerevisiae strains depending on these transporters for l-arabinose uptake.
The filamentous fungus Penicillium chrysogenum and its genome have been intensively studied in relation to its role in the production of β-lactam antibiotics [31, 32]. P. chrysogenum is able to hydrolyse arabinoxylan to l-arabinose by its Axs5 extracellular arabinofuranohydrolase, followed by uptake and metabolism of l-arabinose as a carbon and energy source [33–35]. This ability implies the presence of one or more membrane transporters capable of importing l-arabinose across the plasma membrane of this fungus.
The goal of this study was to explore the P. chrysogenum genome for l-arabinose transporters that can be functionally expressed in S. cerevisiae and support d-glucose- and d-xylose insensitive, high-affinity transport of l-arabinose. To this end, transcriptomes of l-arabinose-, ethanol- and d-glucose-limited chemostat cultures of P. chrysogenum were compared, and putative l-arabinose transporter genes were tested for their ability to support l-arabinose transport upon expression in an S. cerevisiae strain engineered for l-arabinose fermentation in which GAL2 had been deleted. A P. chrysogenum transporter identified in this screen, PcAraT, was subjected to more detailed analysis, including kinetic sugar-uptake studies with radiolabelled substrates, in vivo studies on uptake inhibition, and physiological studies with engineered S. cerevisiae strains in l-arabinose-limited chemostat cultures.
Methods
Microbial strains, growth media and maintenance
All S. cerevisiae strains constructed and used in this study (Table 1) are derived from the CEN.PK lineage [36]. Yeast strains were grown on synthetic medium (SM) [37] or on YP medium (10 g L−1 Bacto yeast extract, 20 g L−1 Bacto peptone). For shake flask cultures on synthetic medium, ammonium sulfate was replaced with urea as nitrogen source to minimize acidification. The resulting SM-urea contained 38 mmol L−1 urea and 38 mmol L−1 K2SO4 instead of (NH4)2SO4. SM and YP media were autoclaved at 121 °C for 20 min, or filter-sterilized using 0.2-µm bottle-top filters (Thermo Scientific, Waltham MA). Subsequently, synthetic media were supplemented with 1 mL L−1 of a sterile-filtered vitamin solution [37]. SM, SM-urea and YP media were further supplemented with 20 g L−1 d-glucose or l-arabinose, by adding concentrated solutions autoclaved at 110 °C for 20 min, yielding SMD or SMA, SMD-urea or SMA-urea and YPD or YPA, respectively. Yeast cultures were grown in 100 mL medium in 500-mL shake flasks at 30 °C and at 200 rpm in an Innova Incubator (New Brunswick Scientific, Edison NJ). Solid SMD, SMA, YPD and YPA contained 1.5% Bacto agar and when indicated, 200 mg L−1 G418 (Invivogen, San Diego, CA). Solid medium with ethanol and glycerol as carbon source (YPEG, SMEG, YPEG-G418) contained 2% ethanol and 3% glycerol. Selection and counter selection of the amdSYM marker cassette were performed as described previously [38]. Escherichia coli strains were grown in 5 mL Lysogeny Broth (10 g L−1 Bacto tryptone, 5 g L−1 Bacto yeast extract, 5 g L−1 NaCl) supplemented with 100 mg L−1 ampicillin in 25-mL shake flasks at 37 °C and 200 rpm in an Innova 4000 shaker (New Brunswick Scientific). Before storage at − 80 °C, yeast and E. coli cultures were mixed with glycerol (30% v/v). P. chrysogenum DS17690 was kindly provided by DSM Anti-infectives (Delft, The Netherlands) and grown in mineral medium (pH 5.5), containing 3.5 g (NH4)2SO4, 0.8 g KH2PO4, 0.5 g MgSO4·7H2O and 10 mL of trace element solution (15 g L−1 Na2EDTA·2H2O, 0.5 g L−1 Cu2SO4·5H2O, 2 g L−1 ZnSO4·7H2O, 2 g L−1 MnSO4·H2O, 4 g L−1 FeSO4·7H2O, and 0.5 g L−1 CaCl2·2H2O) per litre of demineralized water. The mineral medium was supplemented with 7.5 g L−1 d-glucose. Precultivation for chemostat cultures was carried out on mineral medium with 7.5 g L−1 d-glucose, 7.5 g L−1 l-arabinose, or 5.8 g L−1 ethanol as carbon source.
Table 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Relevant genotype | References |
|---|---|---|
| CEN.PK 113-7D | MATa URA3 HIS3 LEU2 TRP1 MAL2-8c SUC2 | [36] |
| CEN.PK 113-5D | MATa ura3-52 HIS3 LEU2 TRP1 MAL2-8c SUC2 | [36] |
| CEN.PK102-12A | MATa ura3-52 his3-D1 leu2-3,112 TRP1 MAL2-8c SUC2 | [36] |
| IMX080 | CEN.PK102-12A glk1::SpHis5, hxk1::KlLEU2 | [75] |
| IMX581 | CEN.PK113-5D can1::cas9-natNT2 | [44] |
| IMX486 | IMX080 gal1::cas9-amdSYM | This study |
| IMX604 | IMX486 ura3-52 gre3::pTDH3-RPE1, pPGK1-TKL1, pTEF1-TAL1, pPGI1-NQM1, pTPI1-RKI1, pPYK1-TKL2 | This study |
| IMX658 | IMX604 ura3-52 gal80::(pTPI-AraA-tCYC1)*9, pPYK1-AraB-tPGI1, pPGK1-AraD-tTDH3 | This study |
| IMX660 | IMX658 hxk2::KlURA3 | This study |
| IMX728 | IMX658 hxk2::PcaraT | This study |
| IMX844 | IMX660 gal2::KanMX | This study |
| IMX869 | IMX728 gal2::KanMX | This study |
| IMX918 | IMX581 gre3::pTDH3-RPE1, pPGK1-TKL1, pTEF1-TAL1, pPGI1-NQM1, pTPI1-RKI1, pPYK1-TKL2 | This study |
| IMX928 | IMX918 gal80::(pTPI-AraA-tCYC1)*9, pPYK-AraB-tPGI1, pPGK-AraD-tTDH3 | This study |
| IMX929 | IMX918 gal80::(pTPI-AraA-tCYC1)*9, pPYK-AraB-tPGI1, pPGK-AraD-tTDH3, pUDE348 | This study |
| IMX1504 | IMX928, gal2Δ, pUDR245 | This study |
| IMX1505 | IMX928 gal2::pADH1-Pc13g04640-tPMA1 (from pPWT111), pUDR245 | This study |
| IMX1506 | IMX928 gal2::pADH1-Pc13g08230-tPMA1 (from pPWT113), pUDR245 | This study |
| IMX1507 | IMX928 gal2::pADH1-Pc16g05670-tPMA1 (from pPWT116), pUDR245 | This study |
| IMX1508 | IMX928 gal2::pADH1-Pc20g01790-tPMA1 (PcaraT) (from pPWT118), pUDR246 | This study |
| IMX1509 | IMX928 gal2::pADH1-Pc22g14520-tPMA1 (from pPWT123), pUDR245 | This study |
| DS68616 | MATa, ura3-52, leu2-112, gre3::loxP, loxP-pTPI-TAL1, loxP-pTPI-RKI1, loxP-pTPI-TKL1, loxP-pTPI-RPE1, leu2::pADH1-XKS1-tCYC1-LEU2, ura3::URA3-pTPI1-XylA-tCYC1 | DSM, The Netherlands |
| DS68625 | DS68616 his3::loxP, hxt2::loxP-kanMX-loxP, hxt367::loxP-hphMX-loxP, hxt145::loxP-natMX-loxP, gal2::loxP-zeoMX-loxP | [45] |
| DS68625-PcaraT | DS68625, pRS313-PcaraT | This study |
| DS68625-GAL2 | DS68625, pRS313-GAL2 | This study |
| DS68625-mcs | DS68625, pRS313-mcs (empty) | This study |
Molecular biology techniques
DNA fragments were amplified by PCR amplification with Phusion Hot Start II High Fidelity Polymerase (Thermo Scientific) and desalted or PAGE-purified oligonucleotide primers (Sigma-Aldrich, St. Louis, MO) performed according to the manufacturers’ instructions. Diagnostic PCRs were run with DreamTaq polymerase (Thermo Scientific). Oligonucleotide primers used in this study are listed in Additional file 1. PCR products were separated by electrophoresis on 1% (w/v) agarose gels (Thermo Scientific) in TAE buffer (Thermo Scientific) and, if required, purified with a Zymoclean Gel DNA Recovery kit (Zymo Research, Irvine, CA) or a GenElute PCR Clean-Up kit (Sigma-Aldrich). Yeast or E. coli plasmids were isolated with a Zymoprep Yeast Plasmid Miniprep II kit (Zymo Research), or a Sigma GenElute Plasmid kit (Sigma-Aldrich), respectively. A YeaStar Genomic DNA kit (Zymo Research) or an SDS/lithium acetate protocol [39] was used to isolate yeast genomic DNA. Yeast strains were transformed using the lithium acetate/polyethylene glycol method [40]. Single-colony isolates were obtained from three consecutive re-streaks on selective solid agar plates, followed by analytical PCR analysis of the relevant genotype. E. coli DH5α cultures were transformed by chemical transformation [41]. After isolation, plasmids were verified by restriction analysis and analytical PCR.
Plasmid construction
Plasmids used in this study are shown in Table 2. All synthesized gene expression cassettes were constructed by GeneArt (Regensburg, Germany). Genes encoding the five putative transporters Pc13g04640 [Genbank: CAP91533.1], Pc13g08230 [Genbank: CAP91892.1], Pc16g05670 [Genbank: CAP93237.1], Pc20g01790 (PcaraT) [Genbank: CAP85508.1] and Pc22g14520 [Genbank: CAP98740.1] were codon-pair optimized [42] for expression in S. cerevisiae and cloned into the plasmid pPWT007 [43] resulting in pPWT111, 113, 116, 118 and 123, respectively, harbouring each an expression cassette consisting of the ADH1 promoter, the codon-optimized open-reading frame of a putative transporter gene, and the PMA1 terminator. Expression cassettes for the coding regions of Lactobacillus plantarum l-arabinose isomerase araA [Genbank: ODO63149.1], l-ribulose kinase araB [Genbank: ODO63147.1] and l-ribulose-5P epimerase araD [Genbank: ODO63148.1] were codon-optimized using the most common codons present in the glycolytic genes of S. cerevisiae [10] and provided by GeneArt in pMK-RQ-based cloning vectors named, pUDE354, pUDE355 and pUDE356, respectively. The episomal plasmids used to express guide RNAs (gRNAs) were constructed from PCR amplified fragments that were ligated using the Gibson Assembly Cloning kit (New England Biolabs, Ipswich, MA). gRNA plasmids pUDR246 and pUDR245 were constructed using pROS10 as a template [44], with oligonucleotide primers listed in Additional file 1. pUDE348 was derived from pMEL10 by first PCR amplifying the plasmid backbone using primers 5792 and 5980. The gRNA sequence was introduced in the gRNA expression cassette with primers 6631 and 5979 using pMEL10 [44] as a template. Subsequently, both fragments were combined using the Gibson Assembly Cloning kit. pUD405 was obtained by integration of a Gal2-flanked KanMX cassette obtained from pUG6 with primers 944 and 945 into a pJET1.2 blunt vector according to the manufacturers’ instructions. Construction of the low-copy-number centromeric plasmid pRS313-mcs was described previously [45]. GAL2 was amplified from genomic DNA of S. cerevisiae DS68616 [45] and PcaraT was amplified from plasmid pPWT118 using primers F GAL2 Xbai and R GAL2 Cfr9i and primers F PcaraT Xbai and R PcaraT Cfr9i, respectively, and cloned into pRS313-mcs, resulting in plasmids pRS313-PcaraT and pRS313-GAL2.
Table 2.
Plasmids used in this study
| Plasmid | Characteristics | Source |
|---|---|---|
| p414-TEF1p-Cas9-CYC1t | CEN6/ARS4 ampR pTEF1-cas9-tCYC1 | [76] |
| pUG-amdSYM | Template for amdSYM marker | [38] |
| pUG-72 | Template for KlURA3 marker | [77] |
| pUG6 | Template for KanMX marker | [78] |
| pUDE327 | 2 μm, KlURA3, pSNR52-gRNA.HXK2.Y | [79] |
| pUDE335 | 2 μm, KlURA3, pSNR52-gRNA.GRE3.Y | [50] |
| pUDE348 | 2 μm, KlURA3, pSNR52-gRNA.GAL80.Y | This study |
| pUDR246 | 2 μm, KlURA3, pSNR52-gRNA.GAL2.Y pSNR52-gRNA.GAL2.Y | This study |
| pUDR245 | 2 μm, KlURA3, pSNR52-gRNA.GAL2.Y pSNR52-gRNA.GAL2.Y | This study |
| pMEL10 | pSNR52-gRNA.CAN1.Y-tSUP4 | [44] |
| pROS10 | 2 μm, KlURA3, pSNR52-gRNA.CAN1.Y pSNR52-gRNA.ADE2.Y | [44] |
| pUD344 | pJET1.2Blunt TagA-pPGI1-NQM1-TagB | [50] |
| pUD345 | pJET1.2Blunt TagB-pTPI1-RKI1-TagC | [50] |
| pUD346 | pJET1.2Blunt TagC-pPYK1-TKL2-TagF | [50] |
| pUD347 | pJET1.2BluntTagG-pTDH3-RPE1-TagH | [50] |
| pUD348 | pJET1.2Blunt TagH-pPGK1-TKL1-TagI | [50] |
| pUD349 | pJET1.2Blunt TagI-pTEF1-TAL1-TagA | [50] |
| pUD405 | pJET1.2Blunt GAL2 flanked KanMX | This study |
| pPWT111 | ampR KanMX, amdSYM, pADH1-Pc13g04640-tPMA1 | This study |
| pPWT113 | ampR KanMX, amdSYM, pADH1-Pc13g08230-tPMA1 | This study |
| pPWT116 | ampR KanMX, amdSYM, pADH1-Pc16g05670-tPMA1 | This study |
| pPWT118 | ampR KanMX, amdSYM, pADH1-Pc20g01790 (PcaraT)-tPMA1 | This study |
| pPWT123 | ampR KanMX, amdSYM, pADH1-Pc22g14520-tPMA1 | This study |
| pUD354 | pMK-RQ-pTPI1-araA-tADH3 | This study |
| pUD355 | pMK-RQ-pPYK1-araB-tPGI1 | This study |
| pUD356 | pMK-RQ-pPGK1-araD-tTDH3 | This study |
| pRS313-mcs | CEN6, ARSH4, HIS3-pHXT7, tHXT7 | [45] |
| pRS313-PcaraT | CEN6, ARSH4, HIS3, ampR, pHXT7-PcaraT-tHXT7 | This study |
| pRS313-GAL2 | CEN6, ARSH4, HIS3, ampR, pHXT7-GAL2-tHXT7 | This study |
Strain construction
Gene expression cassettes were PCR amplified with oligonucleotide primers shown in Additional file 1 and genomic DNA of CEN.PK113-7D or plasmids described in Table 2. Gene knock-outs and construct integrations were introduced with a chimeric CRISPR/Cas9 editing system [44]. To enable CRISPR/Cas9 mediated editing in strain IMX080, the SpCas9 expression cassette was amplified from p414-pTEF1-cas9-tCYC1 (Addgene plasmid # 43802) and integrated into the GAL1 locus via in vivo assembly, together with the amdSYM marker, yielding strain IMX486. For overexpression of the non-oxidative pentose phosphate pathway (PPP), IMX486 and IMX581 were co-transformed with gRNA plasmid pUDE335 and repair fragments flanked with either 60 bp homologous to GRE3 or with synthetic tags [46] assisting homologous recombination of the PPP expression cassettes (gre3flank-pTDH3-RPE1-TagH, TagH-pPGK1-TKL1-TagI, TagI-pTEF1-TAL1-TagA, TagA-pPGI1-NQM1-TagB, TagB-pTPI1-RKI1-TagC, TagC-pPYK1-TKL2-gre3flank). After counter selection of the URA3-based plasmid pUDE335, the resulting strains, IMX604 and IMX918, respectively, were co-transformed with pUDE348 and repair fragments flanked with either 60 bp homologous to GAL80 or with synthetic tags [46] (GAL80flank-pTPI1-araA-TagG, TagG-pTPI1-araA-TagA, TagA-pTPI1-araA-TagB, TagB-pTPI1-araA-TagC, TagC-pTPI1-araA-TagD, TagD-pTPI1-araA-TagM, TagM-pTPI1-araA-TagN, TagN-pTPI1-araA-TagO, TagO-pTPI1-araA-TagI, TagI-pPYK1-araB-TagK, TagK-pPGK1-araD-GAL80flank) resulting in nine copies of araA and a single copy of araB and araD integrated in the GAL80 locus. After verification of the resulting strains IMX929 and IMX658, respectively, plasmid pUDE348 was counter selected in IMX929 to yield strain IMX928. Disruption of HXK2 in IMX658 was done by PCR amplification and transformation of the KlURA3-based deletion cassette from pUG-72 [76] to obtain strain IMX660 upon transformation and plating in solid SMA. GAL2 was disrupted in IMX660 by transformation with a KanMX cassette amplified from pUD405 with primers 944 and 945 flanked with 60 bp homologous to GAL2. Transformants were incubated for 2 h in YPE before plating on YPEG-G418, yielding strain IMX844. Expression of PcaraT in IMX658 was achieved by transforming IMX658 with the gRNA plasmid pUDE327 together with an expression cassette of PcaraT (pADH1-PcaraT-tPMA1) with flanking regions homologous to the HXK2 locus amplified with the primer pair 7660 and 7676. Counter selection of the pUDE327 and subsequent transformation of a DNA fragment derived from CEN.PK113-7D using primers 2641 and 1522 repaired uracil auxotrophy and resulted in strain IMX728. GAL2 was disrupted in IMX728 by transformation with a KanMX cassette amplified from pUD405 with primers 944 and 945 flanked with 60 bp homologous to GAL2. Transformants were incubated for 2 h in YPE before plating on YPEG-G418, yielding strain IMX869. Strains IMX1505-1509 were constructed by co-transforming pUDR245 or pUDR246 and a GAL2-flanked expression cassette (pADH1-ORF-tPMA1) amplified from pPWT111, 113, 116, 118 or 123, respectively, amplified with the primer pair 10585 and 10584. IMX1504, harbouring a knockout of GAL2, was constructed by co-transforming pUDR245 and a repair fragment based on the annealed primers 9563 and 9564. Transformation of GAL2 and PcaraT plasmids, and the pRS313-mcs plasmid (as an empty plasmid/control) into the hexose-transporter deletion strain DS68625 yielded strains DS68625-GAL2, DS68625-PcaraT, and DS68625-mcs.
Growth experiments in shake flasks
Thawed 1-mL aliquots from frozen stock cultures were used to inoculate shake flask precultures on SM-urea supplemented with either d-glucose (20 g L−1), l-arabinose (20 g L−1), or both sugars (both 20 g L−1). These precultures were used to inoculate a second culture which was subsequently used to inoculate a third culture which was inoculated at an initial OD660 of 0.1 and used to monitor growth. Optical densities at 660 nm were measured with a Libra S11 spectrophotometer (Biochrom, Cambridge, United Kingdom). Maximum specific growth rates (μmax) were derived from at least four consecutive data points derived from samples taken during the exponential growth phase of each culture.
Spot plates
l-Arabinose-metabolizing S. cerevisiae strains expressing putative P. chrysogenum l-arabinose transporter genes (IMX1504-1509) were grown on SMD medium and a total number of approximately 104, 103, 102, and 101 cells were spotted on duplicate agar plates as described previously [47, 48] containing either 20 g L−1 l-arabinose or d-glucose as carbon source (pH 6). Cell numbers were estimated from calibration curves of OD660 versus cell counts determined with an Accuri flow cytometer (Becton–Dickinson B.V., Breda, The Netherlands), derived from exponentially growing shake flask cultures of S. cerevisiae CEN.PK113-7D on SMD medium. SMA and SMD plates were incubated at 30 °C for 97 and 41 h, respectively.
Chemostat cultivation
Aerobic carbon-limited chemostat cultures of P. chrysogenum were grown at 25 °C in 3-L turbine-stirred bioreactors (Applikon, Schiedam, The Netherlands) with a working volume of 1.8 L and a dilution rate of 0.03 h−1 as described previously [49], with the exception that, in addition to cultures grown on 7.5 g L−1 d-glucose, chemostat cultures were also grown on either 7.5 g L−1 l-arabinose or 5.8 g L−1 ethanol. Aerobic, l-arabinose-limited chemostat cultures of S. cerevisiae were grown at 30 °C in 2-L Applikon bioreactors with a working volume of 1 L and at a dilution rate of 0.05 h−1. SMA (7.5 g L−1 l-arabinose) supplemented with 0.15 g L−1 Pluronic antifoam PE 6100 was used as culture medium for the initial batch phase and for chemostat cultivation, with the exception of the initial batch phase of strain IMX929 which was grown on 20 g L−1 l-arabinose. Cultures were stirred at 800 rpm, kept at pH 5.0 by automatic addition of 2 M KOH, and sparged with 0.5 L min−1 air. Upon completion of the batch phase, chemostat cultivation was initiated, ensuring a constant culture volume with an electric level sensor. When after at least five volume changes, biomass dry weight and CO2 production varied by less than 2% over two consecutive volume changes, the culture was considered to be in steady state.
Analytical methods
Penicillium chrysogenum biomass dry weight was determined in duplicate by filtration of 10 mL culture sample over pre-weighed glass fibre filters (Type A/E, Pall Life Sciences, Hoegaarden, Belgium). After filtration, filters were washed with demineralized water and dried for 10 min at 600 W in a microwave oven (Bosch, Stuttgart, Germany) prior to reweighing. Biomass dry weight in S. cerevisiae culture samples was determined with a similar procedure using nitrocellulose filters (0.45-µm pore size; Gelman Laboratory, Ann Arbor, MI) and drying for 20 min in a microwave oven at 360 W output. Optical density (OD) of the cultures was determined at 660 nm with a Libra S11 spectrophotometer (Biochrom, Cambridge, United Kingdom). Determination of CO2 and O2 concentrations in the bioreactor exhaust gas and HPLC analysis of metabolite concentrations in culture supernatant samples were performed as described previously [50].
Sampling, RNA extraction, microarrays analysis, and data analysis
Samples (60 mL) from P. chrysogenum chemostat cultures were rapidly filtered over a glass fibre filter (Type A/E, Pall Life Sciences) and further processed for total RNA extraction by phenol–chloroform extraction [49]. The cRNA sample preparation (cDNA synthesis, purification, in vitro transcription, labelling, purification, fragmentation and biotinylation) was performed according to Affymetrix recommendations [31]. Eventually cRNA samples were hybridized onto custom-made P. chrysogenum GeneChip microarrays (array code DSM_PENa520255F). Data acquisition, hybridization, quantification of processed array images, and data filtering were performed using the Affymetrix GeneChip Operating Software (GCOS version 1.2). Global array normalization was performed by scaling the global fluorescence intensity of each microarray to 100. The scaling factors of the individual arrays were highly similar and ranged from 0.21 to 0.35. Subsequently, significant variations in expression were statistically estimated by comparing replicate array experiments using the Significance Analysis of Microarray software (SAM version 2.0) [51] with the multiclass setting. A false discovery rate of 1% was applied to minimize the chance of false-positive hits. Genes with an over threefold higher transcript level in arabinose-grown cultures than in d-glucose-grown cultures and a less than threefold difference in ethanol- and d-glucose-grown cultures were deemed to show arabinose-specific expression. Transcriptome data of strain DS17690 grown on d-glucose, ethanol or arabinose are accessible at NCBI Genome Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) under Accession Numbers GSE12632, GSE24212 and GSE10449, respectively [49].
Analysis of sugar uptake kinetics
Uptake experiments with [14C] l-arabinose, [14C] d-xylose, or [14C] d-glucose, labelled at the first carbon atom (50–60 mCi/mmol) (ARC St. Louis, MO), were performed with S. cerevisiae hexose-transporter deletion strains (DS68625) harbouring a low copy plasmid with constitutively expressed PcaraT (pRS313-PcaraT) or GAL2 (pRS313-GAL2). The experimental workflow was carried out as described previously [45] with [14C] l-arabinose concentrations of 0.5–2000 mmol L−1, [14C] d-xylose concentrations of 0.5–500 mM, or [14C] d-glucose concentrations of 0.1–500 mmol L−1. Transport competition experiments were carried out in the presence of 50 mmol L−1 [14C] l-arabinose and 0–500 mmol L−1 d-glucose or d-xylose, and at [14C] l-arabinose concentration of 2 mmol L−1 together with increasing d-glucose and xylose concentrations of 0–20 mM. Maximum biomass-specific transport rates (‘Vmax’) calculated from transport assays were expressed as nmol sugar transported per milligram biomass dry weight per minute [nmol (mg biomass)−1 min−1]. As this Vmax is influenced by the expression level of the relevant transporter, it is not solely dependent on intrinsic transporter kinetics. The impact of proton-gradient uncoupling on transport activity was determined in 200 μL synthetic medium at a [14C]-l-arabinose concentration of 2 mmol L−1, by comparing transport rates upon addition of either 10 μmol L−1 CCCP (0.5 µL of a stock solution dissolved in 100% DMSO), 0.5 μL DMSO (control), or 0.5 µL water.
Phylogenetic methods
Protein sequences used for generation of a phylogenetic tree were derived from NCBI (https://www.ncbi.nlm.nih.gov/) and the Saccharomyces Genome Database (https://www.yeastgenome.org/). Mafft was used to generate a CLUSTAL format alignment of all sequences, using the L-INS-i method default settings (https://mafft.cbrc.jp/alignment/server/) [52, 53]. Alignments were further processed using neighbour-joining and a 500 times bootstrap. The resulting Newick tree file was visualized and midpoint rooted in iTOL (https://itol.embl.de/) [54]. Gene accession numbers were ScGAL2: P13181, PcaraT: CAP85508, SsaraT: XP_001382755, Atstp2: OAP13698, Kmaxt1: GZ791039, Pgaxt1: GZ791040, Amlat1: AY923868, Amlat2: AY923869, Nclat-1: EAA30346, Mtlat-1: XP_003663698.
Results
Chemostat-based transcriptome analysis of P. chrysogenum for identification of possible l-arabinose transporter genes
Filamentous fungi exhibit a much broader range of carbon source utilization than S. cerevisiae and, similar to many other ascomycetous fungi, P. chrysogenum can grow on l-arabinose as the sole carbon source [33, 55]. To identify candidate structural genes for l-arabinose transporters in P. chrysogenum, carbon-limited chemostat cultures of strain DS17690 were grown at a dilution rate of 0.03 h−1 on different carbon sources. To discriminate between alleviation of carbon repression and l-arabinose induction, duplicate d-glucose-, l-arabinose-, and ethanol-limited chemostat cultures were performed. RNA was extracted from steady-state cultures and gene expression levels were obtained using Affymetrix DNA-arrays [49]. A total of 540 genes were differentially expressed over the three conditions. Of these differentially expressed genes, 137 exhibited an over threefold higher transcript level in l-arabinose-limited cultures than in d-glucose-limited cultures, as well as a less than threefold difference in transcript level between ethanol- and d-glucose-limited cultures (Additional file 2). Genes whose transcript levels in l-arabinose- and ethanol-limited cultures were both at least twofold higher than in d-glucose-grown cultures were not considered for further analysis as their regulation could have reflected unspecific d-glucose (de)repression. An annotation screen indicated that 16 of the identified ‘arabinose-induced’ genes encoded putative transporters, whose transcript levels were 3.4- to 52-fold higher in the l-arabinose-limited cultures than in the d-glucose-limited cultures (Table 3). Five of these genes, whose transcript levels were at least 30-fold higher in l-arabinose-limited cultures than in d-glucose-limited cultures, shared similarity with the S. cerevisiae maltose transporter Mal31, the N. crassa d-glucose transporter Rco-3, the Kluyveromyces lactis high-affinity d-glucose transporter Hgt1 and the S. cerevisiae allantoate transporter Dal5. These five transporter genes (Pc13g08230, Pc16g05670, Pc20g01790, Pc22g14520, and Pc13g04640, respectively) were selected for further functional analysis.
Table 3.
Putative transporter genes that showed higher relative transcript levels in aerobic, l-arabinose-limited chemostat cultures of Penicillium chrysogenum than in corresponding d-glucose- and ethanol-limited cultures
| Gene | Strong similarity to | Relative transcript levels under different nutrient limitations | ||||
|---|---|---|---|---|---|---|
| Glucose | l-Arabinose | Ethanol | Ethanol versus glucose (ratio) | l-Arabinose versus glucose (ratio) | ||
| Pc13g08230 | S. cerevisiae maltose transport protein Mal31 | 13 ± 1 | 664 ± 3 | 17 ± 1 | 1.4 | 53 |
| Pc16g05670 | Neurospora crassa glucose transporter rco-3 | 63 ± 28 | 3176 ± 40 | 69 ± 1 | 1.1 | 51 |
|
Pc20g01790
( PcaraT ) |
Kluyveromyces lactis high-affinity glucose transporter HGT1 | 32 ± 6 | 1415 ± 42 | 46 ± 3 | 1.4 | 44 |
| Pc22g14520 | S. cerevisiae allantoate permease Dal5 | 19 ± 2 | 770 ± 104 | 28 ± 1 | 1.5 | 41 |
| Pc13g04640 | K. lactis high-affinity glucose transporter HGT1 | 29 ± 5 | 971 ± 32 | 53 ± 7 | 1.8 | 34 |
| Pc21g10190 | K. lactis high-affinity glucose transporter HGT1 | 12 ± 1 | 167 ± 26 | 12 ± 1 | 1.0 | 14 |
| Pc12g00190 | Candida albicans ABC transporter CDR4 | 13 ± 2 | 164 ± 24 | 29 ± 2 | 2.2 | 12 |
| Pc14g01680 | Escherichia coli l-fucose permease fucP | 106 ± 14 | 1269 ± 172 | 68 ± 1 | 0.64 | 12.0 |
| Pc21g12210 | Aspergillus nidulans quinate transport protein qutD | 12 ± 0 | 118 ± 1 | 12 ± 1 | 1 | 9.8 |
| Pc06g01480 | S. cerevisiae maltose transport protein Mal31 | 459 ± 85 | 3551 ± 102 | 226 ± 3 | 0.5 | 7.7 |
| Pc13g10030 | S. cerevisiae high-affinity nicotinic acid permease Tna1 | 125 ± 25 | 827 ± 33 | 216 ± 3 | 1.7 | 6.6 |
| Pc21g09830 | K. lactis high-affinity glucose transporter HGT1 | 185 ± 9 | 842 ± 1 | 126 ± 3 | 0.68 | 4.6 |
| Pc16g02680 | S. cerevisiae allantoate permease Dal5 | 80 ± 29 | 360 ± 14 | 113 ± 6 | 1.4 | 4.5 |
| Pc12g05440 | S. cerevisiae maltose transport protein Mal31 | 596 ± 201 | 2633 ± 64 | 104 ± 8 | 0.17 | 4.4 |
| Pc13g15590 | S. cerevisiae glucose permease Rgt2 | 12 ± 1 | 48.0 ± 1.0 | 12 ± 1 | 1 | 4.0 |
| Pc13g06440 | S. cerevisiae high-affinity nicotinic acid permease Tna1 | 66 ± 23 | 225 ± 11 | 48 ± 5 | 0.73 | 3.4 |
P. chrysogenum DS1769 was grown in l-arabinose-, d-glucose-, or ethanol-limited chemostat cultures (dilution rate = 0.03 h−1, pH 6.5, T = 25 °C). Underlined genes were selected for further analysis based on a ≥ 30-fold higher transcript level in l-arabinose-limited cultures than in d-glucose-limited cultures. Data represent average ± mean deviation of globally scaled (target 100) Affymetrix microarrays for independent duplicate chemostat cultures
PcAraT: a P. chrysogenuml-arabinose transporter that can be functionally expressed in S. cerevisiae
Saccharomyces cerevisiae strains in which HXT transporter genes have been deleted and which express heterologous pathways for pentose metabolism have proven to be powerful platforms for screening and characterization of heterologous pentose transporter genes [19, 26, 56]. To enable screening for P. chrysogenum l-arabinose transporters, S. cerevisiae strains were first engineered for l-arabinose consumption. Using CRISPR/Cas9-mediated in vivo assembly [44], the overexpression cassettes for all structural genes involved in the non-oxidative pentose phosphate pathway (TAL1, NQM1, TKL1, TKL2, RKI1, RPE1) were stably integrated into the GRE3 locus, thereby inactivating synthesis of the Gre3 aldose reductase. Subsequently, nine copies of an expression cassette for overexpression of codon-optimized L. plantarum l-arabinose isomerase AraA and single copies of L. plantarum AraB (l-ribulokinase) and AraD (l-ribulose-5-phosphate-4-epimerase) expression cassettes were integrated into the GAL80 locus, using a strain construction strategy previously described for expression of a d-xylose pathway into S. cerevisiae [50]. This integration inactivated GAL80 and thereby alleviated transcriptional repression by d-glucose of GAL2, which encodes the major l-arabinose transporter in S. cerevisiae [57, 58]. The resulting strain IMX929 was able to grow in liquid media supplemented with l-arabinose as the sole carbon source and was used as a platform strain to test if any of the five selected putative P. chrysogenum transporter genes, placed under the control of the constitutive ADH1 promoter, could support l-arabinose transport in S. cerevisiae. To this end, single copies of codon-optimized expression cassettes were integrated into the GAL2 locus of the l-arabinose-metabolizing S. cerevisiae strain IMX928, a uracil auxotrophic daughter strain of IMX929, thereby inactivating the GAL2 gene. Consistent with previous studies [19, 26], inactivation of GAL2 in the l-arabinose metabolizing strain IMX928 yielded a strain (IMX1504) that was unable to grow on SMA plates (Fig. 1). All five strains in which GAL2 had been replaced by putative P. chrysogenum transporter genes (IMX1505-1509) showed vigorous growth on SMD plates. However, only strain IMX1508, which expressed the P. chrysogenum gene Pc20g01790, showed growth on l-arabinose (Fig. 1). Based on this observation, Pc20g01790 was designated PcaraT (P. chrysogenum Arabinose Transporter). A Blast-p search revealed strong homology of Pc20g01790 with the K. lactis gene HGT1, which encodes a high-affinity d-glucose and galactose transporter [59, 60].
Fig. 1.
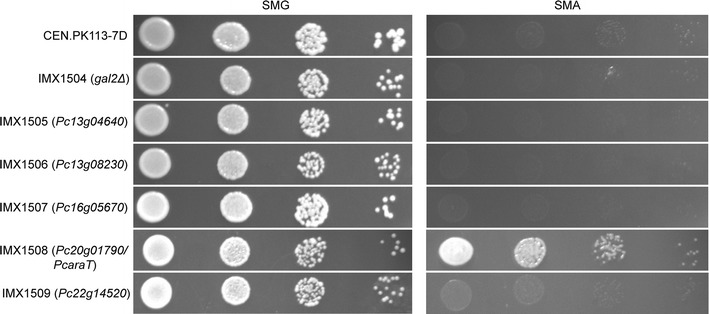
Impact of the expression of putative P. chrysogenum sugar transporter genes in an l-arabinose metabolizing S. cerevisiae strain in which GAL2 was deleted. Strains were pregrown on liquid SMD and spotted on plates containing 20 g L−1 d-glucose (SMD, left) or l-arabinose (SMA, right) as carbon source. Codes on left-hand side indicate S. cerevisiae strain names and, in brackets, the systematic name of the corresponding over-expressed P. chrysogenum gene. CEN.PK113-7D is a control strain that was not engineered for l-arabinose metabolism. SMD and SMA plates were incubated at 30 °C for 47 and 91 h, respectively. The experiment was performed in duplicate; data shown are from a single representative experiment
PcaraT encodes a high-affinity, high-specificity l-arabinose transporter
Sugar transport kinetics of PcAraT were analysed using 14C-labelled l-arabinose, d-xylose and d-glucose. To dissect transporter kinetics of PcAraT and Gal2, their structural genes were separately expressed in S. cerevisiae DS68625 [45]. Each gene was introduced on a centromeric plasmid and expressed from the HXT7 promoter. In strain DS68625, the major hexose-transporter genes (HXT1-7 and GAL2) are deleted, whilst its inability to metabolize l-arabinose enables the specific analysis of sugar uptake rather than the combination of radioactive sugar uptake and metabolism. The negative control strain DS68625-mcs (DS68625 transformed with the ‘empty’ centromeric plasmid pRS313-mcs) did not show significant [14C] l-arabinose uptake, whilst expression of either Gal2 or PcAraT (strains DS68625-GAL2 and DS68625-PcaraT, respectively) restored l-arabinose transport (Table 4). In kinetic analyses, the Km of PcAraT for l-arabinose (0.13 mmol L−1) was found to be three orders of magnitude lower than that of Gal2 (335 mmol L−1), whilst its transport capacity (Vmax) was 14-fold lower than that of Gal2 [5.3 and 75 nmol (mg biomass)−1 min−1, respectively] (Table 4). PcAraT was found to be highly l-arabinose specific, as its expression in strain DS68625 did not support transport of either [14C] d-glucose or [14C] d-xylose. Consistent with earlier reports [19, 26], expression of Gal2 in strain DS68625 enabled transport of d-glucose [Km = 1.9 mmol L−1, Vmax = 26 nmol (mg biomass)−1 min−1], whilst Gal2 has previously been shown to enable low-affinity d-xylose transport (Km = 226 mmol L−1; [20]).
Table 4.
Kinetic data for the S. cerevisiae transporter Gal2 and P. chrysogenum PcAraT derived from uptake studies with 14C-labelled l-arabinose, d-glucose and d-xylose. Sugar transport kinetics were measured by uptake of 14C-radiolabelled sugars by S. cerevisiae DS68625, an engineered strain lacking the Hxt1-7 and Gal2 transporters, expressing either GAL2 or PcaraT
| Gal2 | PcAraT | |
|---|---|---|
| Km, ARA (mmol L−1) | 335 ± 21 | 0.13 ± 0.03 |
| Vmax, ARA [nmol (mg biomass)−1 min−1] | 75 ± 5.2 | 5.3 ± 0.2 |
| Km, GLC (mmol L−1) | 1.9 | – |
| Vmax, GLC [nmol (mg biomass)−1 min−1] | 26 | – |
| l-Arabinose transport inhibition by glucose | 85% | 63% |
| Km, XYL (mmol L−1) | 226 [20] | – |
| Vmax, XYL [nmol (mg biomass)−1 min−1] | 91 [20] | – |
| l-Arabinose transport inhibition by d-xylose | 29% | 22% |
Transport inhibition was determined at 50 mmol L−1 [14C] l-arabinose and 100 mmol L−1 of either d-glucose or d-xylose and expressed relative to the transport rate observed in the absence of d-xylose or d-glucose. Values are represented as average ± mean deviation of duplicate experiments. Graphs used to calculate kinetic parameters are shown in Additional files 3–6. ARA, l-arabinose; GLC, d-glucose; XYL, d-xylose; –, no transport
The impact of the presence of d-glucose and d-xylose on l-arabinose transport by Gal2 and PcAraT was investigated in transport assays with 50 mmol L−1 [14C] l-arabinose and increasing concentrations of non-radioactive d-glucose or d-xylose. In these assays, both transporters exhibited a reduced l-arabinose transport capacity in the presence of d-glucose or d-xylose (Table 4, Additional file 3). At a concentration of 100 mmol L−1 (i.e. twice the concentration of l-arabinose), d-xylose and d-glucose inhibited l-arabinose uptake rate via Gal2 by 29 and 85%, respectively. In contrast, l-arabinose transport via PcAraT was less impaired at this concentration of d-xylose, and especially, d-glucose (22 and 63% inhibition, respectively). To study the transport mechanism of PcAraT, the impact of the protonophore uncoupler CCCP on transport kinetics was tested. Transport of l-arabinose via Gal2, which mediates facilitated diffusion of sugars [61], was not affected by CCCP, whilst this uncoupler completely abolished transport via PcAraT (Additional file 7). These results indicate that PcAraT mediates proton-coupled import of l-arabinose.
Functional expression of PcaraT in an l-arabinose-fermenting S. cerevisiae strain enables l-arabinose consumption in the presence of d-glucose
The ability to transport l-arabinose in the presence of d-glucose is a highly relevant characteristic in the construction of platform S. cerevisiae strains for conversion of lignocellulosic hydrolysates [8]. To investigate whether expression of PcaraT can confer this ability, a set of three strains was constructed that (i) could not metabolize d-glucose due to the deletion of HXK1, HXK2, GLK1 and GAL1 [20, 62]; (ii) (over)expressed non-oxidative PPP enzymes and the L. plantarum AraA, AraB and AraD genes to enable l-arabinose metabolism; and (iii) had different genotypes with respect to l-arabinose transport (GAL2, PcaraT/gal2Δ and gal2Δ in strains IMX660, IMX869 and IMX844, respectively). Since these ‘arabinose specialist strains’ cannot grow on d-glucose, the impact of the presence of d-glucose on l-arabinose metabolism can be directly measured via its effect on growth. As anticipated, strain IMX844 (gal2Δ) was unable to grow on synthetic medium supplemented with either 20 g L−1 l-arabinose or a mix of 20 g L−1 of each, l-arabinose and d-glucose. In contrast, the l-arabinose specialist strains IMX660 (GAL2) and IMX869 (PcaraT/gal2Δ) grew on synthetic medium with l-arabinose as the sole carbon source at specific growth rates of 0.240 ± 0.001 and 0.099 ± 0.001 h−1, respectively (Fig. 2a). However, when 20 g L−1 d-glucose was added to the l-arabinose medium, strain IMX660 (GAL2) did not show growth during a 120-h batch cultivation experiment (Fig. 2b), whilst strain IMX869 (PcaraT/gal2Δ) grew at 60% of the specific growth rate observed in the absence of d-glucose (µ = 0.057 ± 0.003 h−1 versus 0.099 ± 0.001 h−1, Fig. 2b). This result indicated that expression of PcAraT in strain IMX869 enabled uptake of l-arabinose in the presence of d-glucose.
Fig. 2.
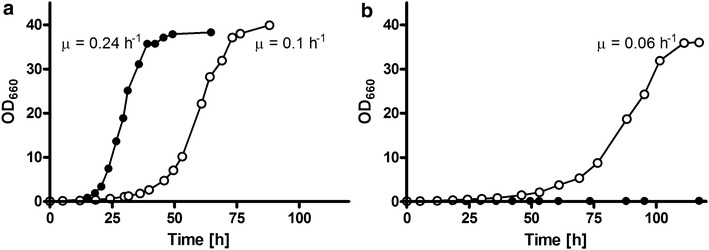
Growth curves of S. cerevisiae l-arabinose specialist strains, engineered for l-arabinose consumption and disabled for d-glucose consumption by deletion of the hexose kinase genes HXK1, HXK2, GLK1 and GAL1, and expressing either GAL2 (IMX660, filled circles) or the P. chrysogenum transporter PcAraT (IMX869, open circles) as the sole l-arabinose transporter. To assess the ability of Gal2 and PcAraT to support import of l-arabinose by growing cultures in the absence (a) and presence (b) of d-glucose, specific growth rates were estimated from shake flask cultures on synthetic media supplied with 20 g L−1 l-arabinose (a) and on synthetic media supplied with l-arabinose and d-glucose (20 g L−1 each, b)
Low residual substrate concentrations in chemostat cultures confirm high-affinity l-arabinose transport kinetics of PcAraT
To further evaluate the in vivo impact of l-arabinose transport via PcAraT, biomass-specific l-arabinose consumption rates and residual substrate concentrations were analysed in l-arabinose-limited, aerobic chemostat cultures, grown at a dilution rate of 0.05 h−1. Under these conditions, the l-arabinose-metabolizing strain IMX1508 (PcaraT/gal2Δ) exhibited a residual l-arabinose concentration of only 4.2 × 10−3 g L−1, compared to 1.8 g L−1 in cultures of strain IMX929 (GAL2) (Table 5). In these growth experiments, different promoters were used for expression of PcaraT and GAL2 (pADH1 and derepressed pGAL2, respectively). However, whilst this may moderately affect expression levels of the two transporters, this cannot explain the over 1000-fold difference in residual l-arabinose concentration. This difference was entirely consistent with the conclusion from the kinetic analyses of 14C-l-arabinose uptake, in which both transporter genes were expressed from the same promoter (pHXT7) and which also indicated that PcaraT encodes an l-arabinose transporter with a much higher affinity for l-arabinose than Gal2. In shake flask batch cultures grown on an initial l-arabinose concentration of 7.5 g L−1, these strains exhibited initial specific growth rates of 0.085 and 0.13 h−1, respectively. Based on this observation and on the high Km of Gal2 for l-arabinose ([19, 25], this study), the in vivo activity of PcAraT can be expected to exceed that of Gal2 when l-arabinose concentrations are below ca. 4 g L−1.
Table 5.
Physiological data derived from steady-state chemostat cultures of engineered l-arabinose-metabolizing S. cerevisiae strains
| IMX929 (GAL2) | IMX1508 (PcaraT) | |
|---|---|---|
| Residual l-arabinose (g L−1) | 1.77 ± 0.19 | 0.004 ± 0.002 |
| YX/S [g biomass (g l-arabinose)−1] | 0.48 ± 0.06 | 0.40 ± 0.01 |
| ql-arabinose (mmol g−1 h−1) | 0.70 ± 0.10 | 0.80 ± 0.08 |
Strains expressing either GAL2 (IMX929) or PcaraT (IMX1508) as the sole functional l-arabinose transporter were grown in aerobic, l-arabinose-limited chemostat cultures (7.5 g L−1 l-arabinose, dilution rate = 0.05 h−1, pH 5, T = 30 °C). Data are derived from independent triplicate experiments and presented as average ± mean deviation
In duplicate steady-state chemostat cultures, the biomass-specific l-arabinose consumption rate of strain IMX1508 (PcaraT) was approximately 14% higher than the one of strain IMX929 (GAL2; 0.8 ± 0.1 and 0.7 ± 0.1 mmol g−1 h−1), reflecting the slightly lower biomass yield of the former strain. This difference in biomass yield is close to the difference of 8.1% that, based on published estimates of the P/O ratio and proton stoichiometry of the plasma membrane ATPase in aerobic S. cerevisiae cultures (both close to 1.0, [63, 64]), would be expected if l-arabinose uptake via PcAraT occurred via symport with a single proton.
Discussion
Chemostat-based transcriptome analysis of P. chrysogenum proved to be an efficient method to identify candidate genes for l-arabinose transporters in this fungus. In comparison with similar studies in batch cultures, use of chemostat cultures offered several advantages. First, chemostat cultivation at a fixed dilution rate eliminated the impact of specific growth rate on transcriptional regulation [65]. Furthermore, use of l-arabinose-limited chemostat cultures of P. chrysogenum, in which residual concentrations of this pentose were very low, enabled a focus on the identification of high-affinity transporters. Finally, the use of both d-glucose- and ethanol-limited cultures as references helped to eliminate transcriptional responses of P. chrysogenum that were specific to either of these two carbon sources, e.g. as a result of CreA-mediated d-glucose repression of relevant transporter genes [66, 67]. Although this study was focused on l-arabinose transport, the P. chrysogenum transcriptome dataset from d-glucose, ethanol and arabinose grown cultures generated in this study [available via GEO, (https://www.ncbi.nlm.nih.gov/geo/) under Accession Numbers GSE12632, GSE24212, and GSE104491, respectively] may contribute to studies on other aspects on metabolism and metabolic regulation in this industrially relevant fungus.
Of five putative transporter genes that showed an over 30-fold higher transcript level in l-arabinose-limited chemostat cultures of P. chrysogenum than in d-glucose-limited cultures, only PcaraT was shown to encode an l-arabinose transporter that is functional in S. cerevisiae. Whilst the low Km of this transporter observed upon its expression in S. cerevisiae is consistent with its upregulation in l-arabinose-limited cultures of P. chrysogenum, this observation does not necessarily imply that PcAraT is the only or even the most important l-arabinose transporter active in these cultures. Problems in protein folding, plasma membrane (mis)targeting, post-translational modification and/or protein turnover [21, 68] may have affected expression of the other candidate genes. Indeed, in screening of cDNA libraries encoding putative heterologous transporters, typically only few of the candidate genes are found to enable transport of the substrate upon expression in S. cerevisiae [69, 70].
Several studies have used gal2Δ strains of S. cerevisiae to analyse transport kinetics of heterologous l-arabinose transporters (Table 6, Fig. 3). Two studies that estimated Km and Vmax of Gal2 upon its reintroduction in such a strain found different results (Table 6) [19, 26]. At l-arabinose concentrations of about 10 mmol L−1, these studies reported Gal2-mediated transport rates of 0.3 and 8.9 nmol (mg biomass)−1 min−1, respectively, as compared to a value of 2.5 nmol (mg biomass)−1 min−1 observed in the present study. One of the previous studies [26] used a strain that also expressed a functional bacterial l-arabinose pathway, thereby raising the possibility that apparent uptake rates were enhanced by subsequent metabolism of l-arabinose. Moreover, in different studies, GAL2 was expressed from different promoters (pTDH3, pADH1 and pHXT7) and either high-copy-number (2µm) [19, 26] or low-copy-number centromeric (this study) expression plasmids. d-Glucose transport kinetics via Gal2 determined in this study [Km = 1.9 mmol L−1, Vmax = 26 nmol (mg biomass)−1 min−1] were similar to previously reported values [1.5 mmol L−1 and 27 nmol (mg biomass)−1 min−1] [20].
Table 6.
Comparison of key characteristics of Gal2, PcAraT and heterologous l-arabinose transporters that were previously expressed in S. cerevisiae
| Protein | Origin | Km, ARA [mM] | Vmax, ARA [nmol (g biomass)−1 min−1] | GLC transport | XYL transport | Mechanism | References |
|---|---|---|---|---|---|---|---|
| ScGal2 | S. cerevisiae | 335 ± 21.0 57 ± 11 371 ± 19 |
75 ± 5 2.2 ± 0.3 18 ± 0.8 |
✓ | ✓ | Facilitated diffusion | This study [19] [26] |
| PcAraT | P. chrysogenum | 0.13 ± 0.03 | 5.3 ± 0.2 | ✗ | ✗ | H+ symport | This study |
| SsAraT | Scheffersomyces stipitis | 3.8 ± 1.7 | 0.4 ± 0.1 | ✓ | ✗ | nd | [19] |
| AtStp2 | Arabidopsis thaliana | 4.5 ± 2.2 | 0.6 ± 0.1 | ✗ | ✗ | H+ symport | [19] |
| KmAxt1 | Kluyveromyces marxianus | 263 ± 57 | 57 ± 6 | ✗ | ✓ | Facilitated diffusion | [26] |
| PgAxt1 | Pichia guilliermondii | 0.13 ± 0.04 | 18 ± 0.8 | ✗ | ✓ | H+ symport | [26] |
| AmLat1 | Ambrosiozyma monospora | 0.03* | 0.2 ± 0.0 | ✗ | ✗ | nd | [70, 73] |
| AmLat2 | A. monospora | nd | 4 ± 0 | ✗ | ✗ | nd | [70, 73] |
| NcLat-1 | Neurospora crassa | 58 ± 4 | 1945 ± 50 | ✓ | nd | H+ symport | [30] |
| MtLat-1 | Myceliophthora. thermophila | 29 ± 4 | 172 ± 6 | ✗ | nd | H+ symport | [30] |
nd, not determined; ARA, l-arabinose; GLC, d-glucose; XYL, d-xylose. * Km of AmLat1 was determined as a GFP-fusion protein [73]
Fig. 3.
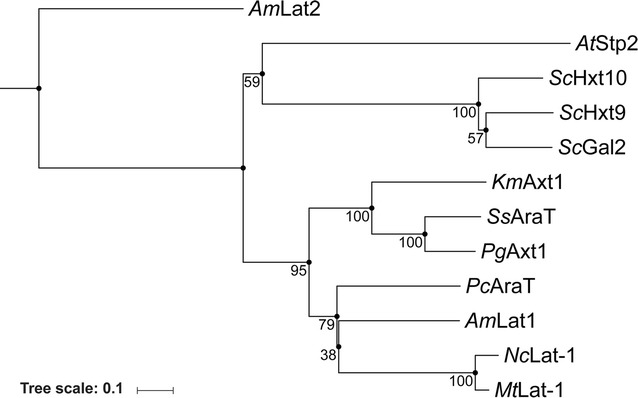
Phylogenetic tree of S. cerevisiae Gal2, PcAraT and other heterologous l-arabinose transporters that have previously been functionally expressed in S. cerevisiae. Species names are added in two-letter code in front of protein names. Numbers are derived from a 500 times bootstrap iteration. Characteristics and literature references for each transporter are provided in Table 6. Accession numbers: ScGal2: P13181, PcAraT: CAP85508, SsAraT: A3LQQ5-1, AtStp2: OAP13698, KmAxt1: GZ791039, PgAxt1: GZ791040, AmLat1: AY923868, AmLat2: AY923869, NcLat-1: EAA30346, MtLat-1: G2QFT5-1
l-Arabinose transport rates in l-arabinose-limited chemostat cultures of both Gal2- and PcAraT-dependent strains were higher than the Vmax values calculated from transporter assays with radioactively labelled l-arabinose. A similar difference between transport assays and rates of l-arabinose uptake in growing cultures was reported by Knoshaug et al. [26]. These discrepancies suggest that either the transport assays did not accurately reflect zero-trans-influx kinetics [71] or that differences in experimental conditions and/or cellular energy status between transport assays and chemostat cultures influenced l-arabinose uptake. Assuming that PcAraT mediates symport of l-arabinose with a single proton, the l-arabinose consumption rate in aerobic, l-arabinose-limited chemostat cultures of the strain IMX1508 (PcAraT gal2Δ) (0.8 mmol (g biomass)−1 h−1; Table 5) would, under anaerobic conditions, correspond to an ATP production rate of ca. 0.3 mmol ATP (g biomass)−1 h−1. This rate of ATP production is well below the reported ATP requirement of anaerobic S. cerevisiae cultures for cellular maintenance [ca. 1 mmol ATP (g biomass)−1 h−1] [72]. Consistent with this observation, no growth on l-arabinose as the sole carbon source was observed in anaerobic shake flask cultures of the l-arabinose specialist strain IMX869 strain (PcAraT gal2Δ) (data not shown).
Differences in experimental protocols for strain construction and sugar uptake studies, as well as the different kinetics observed in transport assays and growing cultures, complicate quantitative comparisons between different studies. Nevertheless, some important differences can be discerned between the heterologous l-arabinose transporters that have hitherto been expressed in S. cerevisiae (Table 6, Fig. 3). Protein sequence alignment of PcAraT and transporters that were previously shown to mediate l-arabinose import in S. cerevisiae showed that PcAraT clusters with Ambrosiozyma monospora AmLat1 (Fig. 3). In terms of its low Km, PcAraT most closely resembled AmLat1 and the P. guilliermondii PgAxt1 transporter. However, expression in S. cerevisiae of AmLat1 [70, 73] led to ~ 25-fold lower reported Vmax of l-arabinose uptake than found in the present study for PcAraT. In contrast to PcAraT, PgAxt was able to transport d-glucose, which might contribute to the strong inhibition of the latter transporter by d-glucose [26]. Although PcAraT resembled A. thaliana Stp2 [19] in being partially inhibited by d-glucose despite an inability to transport this sugar, PcAraT enabled consumption of l-arabinose in batch cultures containing 20 g L−1 d-glucose.
In common with other high-affinity sugar transporters in yeasts and fungi [26, 74], the observation that PcAraT mediates l-arabinose-proton symport should be taken into account in future strain designs, since simultaneous activity of proton symport and facilitated diffusion, e.g. via Gal2, may result in energy-consuming futile cycles [8].
In the lignocellulosic hydrolysates now used in the first industrial-scale plants for ‘second generation’ bioethanol production, l-arabinose generally represents between 2 and 3% of the total sugars [8]. At the resulting low concentrations of l-arabinose in the industrial processes, Gal2 operates far from substrate saturation and is, moreover, strongly inhibited by d-glucose. Based on its kinetic characteristics, as analysed in transport assays and growing cultures, PcAraT represents an interesting candidate transporter for evaluation of l-arabinose co-consumption under industrial conditions. If the characteristics of PcAraT determined in the present study can be reproduced in industrial strains and under industrial conditions, this transporter can contribute to a timely and efficient conversion of l-arabinose, and thereby to the overall process economics.
Conclusion
Transcriptome analyses of l-arabinose-limited P. chrysogenum chemostat cultures proved valuable for identification of the high-affinity l-arabinose transporter PcAraT. Functional expression and characterization in S. cerevisiae revealed a high affinity and specificity of this transporter for l-arabinose (Km = 0.13 mmol L−1), combined with a limited sensitivity to inhibition by d-glucose and d-xylose, which are present at high concentrations in lignocellulosic hydrolysates. These characteristics differentiate PcAraT from the endogenous S. cerevisiae transporter capable of l-arabinose transport (Gal2) and qualify it as a potentially valuable additional element in metabolic engineering strategies towards efficient and complete conversion of l-arabinose present in second-generation feedstocks for yeast-based production of fuels and chemicals.
Additional files
Additional file 1. Primers used in this study.
Additional file 2. Differentially expressed genes in P. chrysogenum chemostat cultures over three different conditions (d-glucose-, l-arabinose, and ethanol-limited). The table includes 540 genes that were differentially expressed amongst d-glucose-, l-arabinose, and ethanol-limited chemostat cultures of P. chrysogenum (7.5 g L−1, 7.5 g L−1, 5.8 g L−1, respectively, D = 0.03 h−1, T = 30 °C). 137 transcripts exhibited a fold-change higher than three on l-arabinose and lower than three on ethanol relative to the d-glucose condition (the top 137 genes presented in the table). Shown are averages from triplicate (d-glucose) or duplicate chemostat cultures (l-arabinose, ethanol), the respective standard deviation and the ratio of gene expression levels in the presence of l-arabinose over d-glucose (Ara vs. Glc), as well as ethanol over d-glucose (EtOH vs. Glc).
Additional file 3. Effect of d-glucose (a) and d-xylose (b) on the specific rate of l-arabinose uptake by PcAraT (filled triangles) and Gal2 (filled squares). Uptake experiments were performed with 50 mmol L−1 [14C-] l-arabinose in the presence of increasing concentrations of d-glucose (a) or d-xylose (b). Symbols indicate uptake rates observed with the Hxt1-7 and Gal2 deletion strain S. cerevisiae DS68625-PcaraT (filled triangles) and DS68625-GAL2 (filled squares), expressing either PcAraT or Gal2, respectively. Data are derived from duplicate experiments and shown as the average ± mean deviation.
Additional file 4. Specific rate of l-arabinose uptake by PcAraT. Uptake experiments were performed with increasing concentrations of [14C-] l-arabinose with the Hxt1-7 and Gal2 deletion strain S. cerevisiae DS68625-PcaraT expressing PcaraT on a centromeric plasmid. No [14C-] d-glucose uptake was observed for this strain. Data are derived from duplicate experiments and shown as the average ± mean deviation.
Additional file 5. Specific rate of d-glucose uptake by Gal2. Uptake experiments were performed with increasing concentrations of [14C-] d-glucose with the Hxt1-7 and Gal2 deletion strain S. cerevisiae DS68625-GAL2 expressing GAL2 on a centromeric plasmid. Data are derived from duplicate experiments and shown as the average ± mean deviation.
Additional file 6. Specific rate of l-arabinose uptake by Gal2. Uptake experiments were performed with increasing concentrations of [14C-] l-arabinose with the Hxt1-7 and Gal2 deletion strain S. cerevisiae DS68625-GAL2 expressing GAL2 on a centromeric plasmid. Data are derived from duplicate experiments and shown as the average ± mean deviation.
Additional file 7. Impact of proton-gradient uncoupling on transport activity. Transport rates of [14C]-l-arabinose of the Hxt1-7 and Gal2 deletion strains DS68625-PcaraT and DS68625-GAL2 expressing either PcAraT (DS68625-PcaraT, white bars) or Gal2 (DS68625-GAL2, grey bars) on a centromeric plasmid. Transport rates were determined in 200 μl synthetic medium at a [14C]-l-arabinose concentration of 2 mmol L−1 upon addition of either 0.5 µL water, 0.5 μL DMSO, or 10 μM CCCP (0.5 µl of a stock solution dissolved in 100% DMSO) (A). Panel (B) shows the uptake capacity in % relative to the control (H2O). Data are derived from duplicate experiments and shown as the average ± mean deviation.
Authors’ contributions
The experimental work including fermentation, strain construction and characterization was performed by JMB and MDV assisted by HWW and BC. JMB, MDV, and JGN performed transport assays, J-MGD performed transcriptome data analysis. JMB, MDV, AJMD, AJAvM, PK, J-MGD, and JTP designed the study, JMB, MDV, and JTP wrote a draft version of manuscript. All authors commented upon the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors thank Andreas K. Gombert and Isthar Snoek for help with the P. chrysogenum chemostat cultures, Aurin M. Vos for help with the generation of the phylogenetic tree (Fig. 3) and Ioannis Papapetridis and Paul de Waal for valuable input in this project.
Competing interests
Several of the authors are inventors on patent applications related to pentose fermentation by S. cerevisiae. DSM markets technology for biofuels production from lignocellulosic feedstocks, holds IP positions in this field and co-funded the research described in this publication.
Availability of data and materials
The datasets supporting the conclusions of this article are available at NCBI Genome Omnibus database (GEO) under Accession Numbers GSE12632, GSE24212, and GSE10449, respectively, in the GEO repository (https://www.ncbi.nlm.nih.gov/geo/).
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
The Ph.D. projects of J.M.B. and M.D.V. are funded by the BE-Basic R&D Program, which was granted an FES subsidy from the Dutch Ministry of Economic Affairs, Agriculture and Innovation (EL&I). The BE-Basic project ‘Omniyeast’ in which most of the research was performed receives additional financial support from DSM. The contributions of H.W.W. and B.C. were fully funded by DSM.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- SM
synthetic medium
- SMA
synthetic medium with 20 g L−1 l-arabinose
- SMAG
synthetic medium with 20 g L−1 l-arabinose and 20 g L−1 d-glucose
- SMEG
synthetic medium with 2% ethanol and 3% glycerol
- SM-urea
synthetic medium with urea
- YPD
yeast peptone medium with 20 g L−1 d-glucose
- YPA
yeast peptone medium with 20 g L−1 l-arabinose
- YPEG
yeast peptone medium with 2% ethanol and 3% glycerol
- ARA
l-arabinose
- GLC
d-glucose
- XYL
d-xylose
- OD
optical density
- ORF
open-reading frame
- nd
not determined
- PPP
pentose phosphate pathway
Footnotes
Jasmine M. Bracher and Maarten D. Verhoeven contributed equally to this work
Electronic supplementary material
The online version of this article (10.1186/s13068-018-1047-6) contains supplementary material, which is available to authorized users.
References
- 1.Renewable Fuels Association. http://ethanolrfa.org/resources/industry/statistics/. Accessed 14 June 2017.
- 2.Hahn-Hägerdal B, Galbe M, Gorwa-Grauslund M-F, Lidén G, Zacchi G. Bio-ethanol—the fuel of tomorrow from the residues of today. Trends Biotechnol. 2006;24:549–556. doi: 10.1016/j.tibtech.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Lynd LR. Overview and evaluation of fuel ethanol from cellulosic biomass: technology, economics, the environment, and policy. Annu Rev Energy Env. 1996;21:403–465. doi: 10.1146/annurev.energy.21.1.403. [DOI] [Google Scholar]
- 4.Micard V, Renard C, Thibault J-F. Enzymatic saccharification of sugar-beet pulp. Enzyme Microb Technol. 1996;19:162–170. doi: 10.1016/0141-0229(95)00224-3. [DOI] [Google Scholar]
- 5.Grohmann K, Bothast RJ. Saccharification of corn fibre by combined treatment with dilute sulphuric acid and enzymes. Process Biochem. 1997;32:405–415. doi: 10.1016/S0032-9592(96)00095-7. [DOI] [Google Scholar]
- 6.Lin Y, Tanaka S. Ethanol fermentation from biomass resources: current state and prospects. Appl Microbiol Biotechnol. 2006;69:627–642. doi: 10.1007/s00253-005-0229-x. [DOI] [PubMed] [Google Scholar]
- 7.Moysés DN, Reis VCB, de Almeida JRMM, Moraes LM, Torres FAG. Xylose fermentation by Saccharomyces cerevisiae: challenges and prospects. Int J Mol Sci. 2016;17:207. doi: 10.3390/ijms17030207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jansen ML, Bracher JM, Papapetridis I, Verhoeven MD, de Bruijn H, de Waal PP, Van Maris AJA, Klaassen P, Pronk JT. Saccharomyces cerevisiae strains for second-generation ethanol production: from academic exploration to industrial implementation. FEMS Yeast Res. 2017 doi: 10.1093/femsyr/fox044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becker J, Boles E. A modified Saccharomyces cerevisiae strain that consumes l-arabinose and produces ethanol. Appl Environ Microbiol. 2003;69:4144–4150. doi: 10.1128/AEM.69.7.4144-4150.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiedemann B, Boles E. Codon-optimized bacterial genes improve l-arabinose fermentation in recombinant Saccharomyces cerevisiae. Appl Environ Microbiol. 2008;74:2043–2050. doi: 10.1128/AEM.02395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bettiga M, Hahn-Hägerdal B, Gorwa-Grauslund MF. Comparing the xylose reductase/xylitol dehydrogenase and xylose isomerase pathways in arabinose and xylose fermenting Saccharomyces cerevisiae strains. Biotechnol Biofuels. 2008;1:16. doi: 10.1186/1754-6834-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wisselink HW, Toirkens MJ, del Rosario Franco Berriel M, Winkler AA, van Dijken JP, Pronk JT, van Maris AJA. Engineering of Saccharomyces cerevisiae for efficient anaerobic alcoholic fermentation of l-arabinose. Appl Environ Microbiol. 2007;73:4881–4891. doi: 10.1128/AEM.00177-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia PF, Zhang GC, Liu JJ, Kwak S, Tsai CS, Kong II, Sung BH, Sohn JH, Wang SG, Jin YS. GroE chaperonins assisted functional expression of bacterial enzymes in Saccharomyces cerevisiae. Biotechnol Bioeng. 2016 doi: 10.1002/bit.25980. [DOI] [PubMed] [Google Scholar]
- 14.Kuyper M, Hartog MM, Toirkens MJ, Almering MJ, Winkler AA, van Dijken JP, Pronk JT. Metabolic engineering of a xylose-isomerase-expressing Saccharomyces cerevisiae strain for rapid anaerobic xylose fermentation. FEMS Yeast Res. 2005;5:399–409. doi: 10.1016/j.femsyr.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 15.Richard P, Verho R, Putkonen M, Londesborough J, Penttilä M. Production of ethanol from l-arabinose by Saccharomyces cerevisiae containing a fungal l-arabinose pathway. FEMS Yeast Res. 2003;3:185–189. doi: 10.1016/S1567-1356(02)00184-8. [DOI] [PubMed] [Google Scholar]
- 16.Leandro MJ, Fonseca C, Gonçalves P. Hexose and pentose transport in ascomycetous yeasts: an overview. FEMS Yeast Res. 2009;9:511–525. doi: 10.1111/j.1567-1364.2009.00509.x. [DOI] [PubMed] [Google Scholar]
- 17.Young E, Lee S-M, Alper H. Optimizing pentose utilization in yeast: the need for novel tools and approaches. Biotechnol Biofuels. 2010;3:24. doi: 10.1186/1754-6834-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kou S, Christensen MS, Cirillo VP. Galactose transport in Saccharomyces cerevisiae II. Characteristics of galactose uptake and exchange in galactokinaseless cells. J Bacteriol. 1970;103:671–678. doi: 10.1128/jb.103.3.671-678.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subtil T, Boles E. Improving l-arabinose utilization of pentose fermenting Saccharomyces cerevisiae cells by heterologous expression of l-arabinose transporting sugar transporters. Biotechnol Biofuels. 2011;4:38. doi: 10.1186/1754-6834-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farwick A, Bruder S, Schadeweg V, Oreb M, Boles E. Engineering of yeast hexose transporters to transport d-xylose without inhibition by d-glucose. Proc Natl Acad Sci USA. 2014;111:5159–5164. doi: 10.1073/pnas.1323464111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamacher T, Becker J, Gárdonyi M, Hahn-Hägerdal B, Boles E. Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiology. 2002;148:2783–2788. doi: 10.1099/00221287-148-9-2783. [DOI] [PubMed] [Google Scholar]
- 22.Lee W, Kim M, Ryu Y, Bisson L, Seo J. Kinetic studies on glucose and xylose transport in Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2002;60:186–191. doi: 10.1007/s00253-002-1085-6. [DOI] [PubMed] [Google Scholar]
- 23.Reifenberger E, Boles E, Ciriacy M. Kinetic characterization of individual hexose transporters of Saccharomyces cerevisiae and their relation to the triggering mechanisms of glucose repression. Eur J Biochem. 1997;245:324–333. doi: 10.1111/j.1432-1033.1997.00324.x. [DOI] [PubMed] [Google Scholar]
- 24.Saloheimo A, Rauta J, Stasyk V, Sibirny AA, Penttilä M, Ruohonen L. Xylose transport studies with xylose-utilizing Saccharomyces cerevisiae strains expressing heterologous and homologous permeases. Appl Microbiol Biotechnol. 2007;74:1041–1052. doi: 10.1007/s00253-006-0747-1. [DOI] [PubMed] [Google Scholar]
- 25.Subtil T, Boles E. Competition between pentoses and glucose during uptake and catabolism in recombinant Saccharomyces cerevisiae. Biotechnol Biofuels. 2012;5:1. doi: 10.1186/1754-6834-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knoshaug EP, Vidgren V, Magalhães F, Jarvis EE, Franden MA, Zhang M, Singh A. Novel transporters from Kluyveromyces marxianus and Pichia guilliermondii expressed in Saccharomyces cerevisiae enable growth on l-arabinose and d-xylose. Yeast. 2015;32:615–628. doi: 10.1002/yea.3084. [DOI] [PubMed] [Google Scholar]
- 27.Horak J, Regelmann J, Wolf DH. Two distinct proteolytic systems responsible for glucose-induced degradation of fructose-1,6-bisphosphatase and the Gal2p transporter in the yeast Saccharomyces cerevisiae share the same protein components of the glucose signaling pathway. J Biol Chem. 2002;277:8248–8254. doi: 10.1074/jbc.M107255200. [DOI] [PubMed] [Google Scholar]
- 28.Horak J, Wolf DH. Catabolite inactivation of the galactose transporter in the yeast Saccharomyces cerevisiae: ubiquitination, endocytosis, and degradation in the vacuole. J Bacteriol. 1997;179:1541–1549. doi: 10.1128/jb.179.5.1541-1549.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Özcan S, Johnston M. Function and regulation of yeast hexose transporters. Microbiol Mol Biol Rev. 1999;63:554–569. doi: 10.1128/mmbr.63.3.554-569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Xu J, Cai P, Wang B, Ma Y, Benz JP, Tian C. Functional analysis of two l-arabinose transporters from filamentous fungi reveals promising characteristics for improved pentose utilization in Saccharomyces cerevisiae. Appl Environ Microbiol. 2015;81:4062–4070. doi: 10.1128/AEM.00165-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Den Berg MA, Albang R, Albermann K, Badger JH, Daran J-M, Driessen AJ, Garcia-Estrada C, Fedorova ND, Harris DM, Heijne WH. Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat Biotechnol. 2008;26:1161–1168. doi: 10.1038/nbt.1498. [DOI] [PubMed] [Google Scholar]
- 32.Van den Berg MA. Impact of the Penicillium chrysogenum genome on industrial production of metabolites. Appl Microbiol Biotechnol. 2011;92:45–53. doi: 10.1007/s00253-011-3476-z. [DOI] [PubMed] [Google Scholar]
- 33.Chiang C, Knight S. l-Arabinose metabolism by cell-free extracts of Penicillium chrysogenum. Biochim Biophys Acta. 1961;46:271–278. doi: 10.1016/0006-3002(61)90750-8. [DOI] [PubMed] [Google Scholar]
- 34.Chiang C, Knight S. A new pathway of pentose metabolism. Biochem Biophys Res Commun. 1960;3:554–559. doi: 10.1016/0006-291X(60)90174-1. [DOI] [PubMed] [Google Scholar]
- 35.Sakamoto T, Ogura A, Inui M, Tokuda S, Hosokawa S, Ihara H, Kasai N. Identification of a GH62 α-l-arabinofuranosidase specific for arabinoxylan produced by Penicillium chrysogenum. Appl Microbiol Biotechnol. 2011;90:137–146. doi: 10.1007/s00253-010-2988-2. [DOI] [PubMed] [Google Scholar]
- 36.Entian K-D, Kötter P. 25 Yeast genetic strain and plasmid collections. Methods Microbiol. 2007;36:629–666. doi: 10.1016/S0580-9517(06)36025-4. [DOI] [Google Scholar]
- 37.Verduyn C, Postma E, Scheffers WA, Van Dijken JP. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992;8:501–517. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- 38.Solis-Escalante D, Kuijpers NG, Bongaerts N, Bolat I, Bosman L, Pronk JT, Daran JM, Daran-Lapujade P. amdSYM, a new dominant recyclable marker cassette for Saccharomyces cerevisiae. FEMS Yeast Res. 2013;13:126–139. doi: 10.1111/1567-1364.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lõoke M, Kristjuhan K, Kristjuhan A. Extraction of genomic DNA from yeasts for PCR-based applications. Biotechniques. 2011;50:325. doi: 10.2144/000113672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/S0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 41.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-P. [DOI] [PubMed] [Google Scholar]
- 42.Roubos JA, Van Peij NNME. A method for achieving improved polypeptide expression. WO Patent App. PCT/EP2007/055,943. 2008.
- 43.Klaassen P, van der Laan JM, Gielesen BEM, van Suylekom GP. Pentose sugar fermenting cell. US Patent 8.399.215 B2. 2013.
- 44.Mans R, van Rossum HM, Wijsman M, Backx A, Kuijpers NGA, van den Broek M, van Daran-Lapujade P, Pronk JT, Maris AJ, Daran J-M. CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res. 2015;15(2):fov004. doi: 10.1093/femsyr/fov004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nijland JG, Shin HY, de Jong RM, De Waal PP, Klaassen P, Driessen AJ. Engineering of an endogenous hexose transporter into a specific d-xylose transporter facilitates glucose–xylose co-consumption in Saccharomyces cerevisiae. Biotechnol Biofuels. 2014;7:168. doi: 10.1186/s13068-014-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuijpers N, Solis-Escalante D, Bosman L, van den Broek M, Pronk JT, Daran J-M, Daran-Lapujade P. A versatile, efficient strategy for assembly of multi-fragment expression vectors in Saccharomyces cerevisiae using 60 bp synthetic recombination sequences. Microb Cell Fact. 2013;12:47. doi: 10.1186/1475-2859-12-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Rossum HM, Kozak BU, Niemeijer MS, Dykstra JC, Luttik MA, Daran J-MG, van Maris AJ, Pronk JT. Requirements for carnitine shuttle-mediated translocation of mitochondrial acetyl moieties to the yeast cytosol. MBio. 2016;7:e00520-16. doi: 10.1128/mBio.00520-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bracher JM, de Hulster E, Koster CC, van den Broek M, Daran J-MG, van Maris AJ, Pronk JT. Laboratory evolution of a biotin-requiring Saccharomyces cerevisiae strain for full biotin prototrophy and identification of causal mutations. Appl Environ Microbiol. 2017 doi: 10.1128/AEM.00892-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris DM, van der Krogt ZA, Klaassen P, Raamsdonk LM, Hage S, van den Berg MA, Bovenberg RA, Pronk JT, Daran J-M. Exploring and dissecting genome-wide gene expression responses of Penicillium chrysogenum to phenylacetic acid consumption and penicillinG production. BMC Genomics. 2009;10:75. doi: 10.1186/1471-2164-10-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verhoeven MD, Lee M, Kamoen L, van den Broek M, Janssen DB, Daran J-MG, van Maris AJ, Pronk JT. Mutations in PMR1 stimulate xylose isomerase activity and anaerobic growth on xylose of engineered Saccharomyces cerevisiae by influencing manganese homeostasis. Sci Rep. 2017;7:46155. doi: 10.1038/srep46155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2017 doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katoh K, Kuma K-I, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sakamoto T, Ihara H, Kozaki S, Kawasaki H. A cold-adapted endo-arabinanase from Penicillium chrysogenum. Biochim Biophys Acta. 2003;1624:70–75. doi: 10.1016/j.bbagen.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 56.Sloothaak J, Tamayo-Ramos JA, Odoni DI, Laothanachareon T, Derntl C, Mach-Aigner AR, Martins dos Santos VAP, Schaap PJ. Identification and functional characterization of novel xylose transporters from the cell factories Aspergillus niger and Trichoderma reesei. Biotechnol Biofuels. 2016;9:1–15. doi: 10.1186/s13068-016-0564-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Torchia T, Hamilton R, Cano C, Hopper JE. Disruption of regulatory gene GAL80 in Saccharomyces cerevisiae: effects on carbon-controlled regulation of the galactose/melibiose pathway genes. Mol Cell Biol. 1984;4:1521–1527. doi: 10.1128/MCB.4.8.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nehlin JO, Carlberg M, Ronne H. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 1991;10:3373–3377. doi: 10.1002/j.1460-2075.1991.tb04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baruffini E, Goffrini P, Donnini C, Lodi T. Galactose transport in Kluyveromyces lactis: major role of the glucose permease Hgt1. FEMS Yeast Res. 2006;6:1235–1242. doi: 10.1111/j.1567-1364.2006.00107.x. [DOI] [PubMed] [Google Scholar]
- 60.Billard P, Ménart S, Blaisonneau J, Bolotin-Fukuhara M, Fukuhara H, Wésolowski-Louvel M. Glucose uptake in Kluyveromyces lactis: role of the HGT1 gene in glucose transport. J Bacteriol. 1996;178:5860–5866. doi: 10.1128/jb.178.20.5860-5866.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maier A, Völker B, Boles E, Fuhrmann GF. Characterisation of glucose transport in Saccharomyces cerevisiae with plasma membrane vesicles (countertransport) and intact cells (initial uptake) with single Hxt1, Hxt2, Hxt3, Hxt4, Hxt6, Hxt7 or Gal2 transporters. FEMS Yeast Res. 2002;2:539–550. doi: 10.1111/j.1567-1364.2002.tb00121.x. [DOI] [PubMed] [Google Scholar]
- 62.Klaassen P, de Waal PP, de Jong RM, Driessen AJM, Nijland JG, Shin HY. Polypeptides with permease activity. US Patent WO/2012/049173. 2012.
- 63.Verduyn C, Postma E, Scheffers WA, Van Dijken JP. Energetics of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. Microbiology. 1990;136:405–412. doi: 10.1099/00221287-136-3-405. [DOI] [PubMed] [Google Scholar]
- 64.Weusthuis RA, Adams H, Scheffers WA, Van Dijken JP. Energetics and kinetics of maltose transport in Saccharomyces cerevisiae: a continuous culture study. Appl Environ Microbiol. 1993;59:3102–3109. doi: 10.1128/aem.59.9.3102-3109.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Daran-Lapujade P, Daran J-M, van Maris AJ, de Winde JH, Pronk JT. Chemostat-based micro-array analysis in baker’s yeast. Adv Microb Physiol. 2008;54:257–417. doi: 10.1016/S0065-2911(08)00004-0. [DOI] [PubMed] [Google Scholar]
- 66.Cepeda-García C, Domínguez-Santos R, García-Rico RO, García-Estrada C, Cajiao A, Fierro F, Martín JF. Direct involvement of the CreA transcription factor in penicillin biosynthesis and expression of the pcbAB gene in Penicillium chrysogenum. Appl Microbiol Biotechnol. 2014;98:7113–7124. doi: 10.1007/s00253-014-5760-1. [DOI] [PubMed] [Google Scholar]
- 67.Cubero B, Scazzocchio C. Two different, adjacent and divergent zinc finger binding sites are necessary for CREA-mediated carbon catabolite repression in the proline gene cluster of Aspergillus nidulans. EMBO J. 1994;13:407. doi: 10.1002/j.1460-2075.1994.tb06275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jahn TP, Schulz A, Taipalensuu J, Palmgren MG. Post-translational modification of plant plasma membrane H+-ATPase as a requirement for functional complementation of a yeast transport mutant. J Biol Chem. 2002;277:6353–6358. doi: 10.1074/jbc.M109637200. [DOI] [PubMed] [Google Scholar]
- 69.Du J, Li S, Zhao H. Discovery and characterization of novel d-xylose-specific transporters from Neurospora crassa and Pichia stipitis. Mol BioSyst. 2010;6:2150–2156. doi: 10.1039/c0mb00007h. [DOI] [PubMed] [Google Scholar]
- 70.Verho R, Penttilä M, Richard P. Cloning of two genes (LAT1, 2) encoding specific l-arabinose transporters of the l-arabinose fermenting yeast Ambrosiozyma monospora. Appl Biochem Biotechnol. 2011;164:604–611. doi: 10.1007/s12010-011-9161-y. [DOI] [PubMed] [Google Scholar]
- 71.Teusink B, Diderich JA, Westerhoff HV, Van Dam K, Walsh MC. Intracellular glucose concentration in derepressed yeast cells consuming glucose is high enough to reduce the glucose transport rate by 50% J Bacteriol. 1998;180:556–562. doi: 10.1128/jb.180.3.556-562.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boender LG, de Hulster EA, van Maris AJ, Daran-Lapujade PA, Pronk JT. Quantitative physiology of Saccharomyces cerevisiae at near-zero specific growth rates. Appl Environ Microbiol. 2009;75:5607–5614. doi: 10.1128/AEM.00429-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Londesborough J, Richard P, Valkonen M, Viljanen K. Effect of C-terminal protein tags on pentitol and l-arabinose transport by Ambrosiozyma monospora Lat1 and Lat2 transporters in Saccharomyces cerevisiae. Appl Environ Microbiol. 2014;80:2737–2745. doi: 10.1128/AEM.04067-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Knoshaug EP, Franden MA, Stambuk BU, Zhang M, Singh A. Utilization and transport of l-arabinose by non-Saccharomyces yeasts. Cellulose. 2009;16:729–741. doi: 10.1007/s10570-009-9319-8. [DOI] [Google Scholar]
- 75.Solis-Escalante D, Kuijpers NG, Barrajon-Simancas N, van den Broek M, Pronk JT, Daran J-M, Daran-Lapujade P. A minimal set of glycolytic genes reveals strong redundancies in Saccharomyces cerevisiae central metabolism. Eukaryot Cell. 2015;14:804–816. doi: 10.1128/EC.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Güldener U, Heinisch J, Koehler G, Voss D, Hegemann J. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23-e. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Güldener U, Heck S, Fiedler T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kuijpers NG, Solis-Escalante D, Luttik MA, Bisschops MM, Boonekamp FJ, van den Broek M, Pronk JT, Daran J-M, Daran-Lapujade P. Pathway swapping: Toward modular engineering of essential cellular processes. Proc Natl Acad Sci. 2016;113:15060–15065. doi: 10.1073/pnas.1606701113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Primers used in this study.
Additional file 2. Differentially expressed genes in P. chrysogenum chemostat cultures over three different conditions (d-glucose-, l-arabinose, and ethanol-limited). The table includes 540 genes that were differentially expressed amongst d-glucose-, l-arabinose, and ethanol-limited chemostat cultures of P. chrysogenum (7.5 g L−1, 7.5 g L−1, 5.8 g L−1, respectively, D = 0.03 h−1, T = 30 °C). 137 transcripts exhibited a fold-change higher than three on l-arabinose and lower than three on ethanol relative to the d-glucose condition (the top 137 genes presented in the table). Shown are averages from triplicate (d-glucose) or duplicate chemostat cultures (l-arabinose, ethanol), the respective standard deviation and the ratio of gene expression levels in the presence of l-arabinose over d-glucose (Ara vs. Glc), as well as ethanol over d-glucose (EtOH vs. Glc).
Additional file 3. Effect of d-glucose (a) and d-xylose (b) on the specific rate of l-arabinose uptake by PcAraT (filled triangles) and Gal2 (filled squares). Uptake experiments were performed with 50 mmol L−1 [14C-] l-arabinose in the presence of increasing concentrations of d-glucose (a) or d-xylose (b). Symbols indicate uptake rates observed with the Hxt1-7 and Gal2 deletion strain S. cerevisiae DS68625-PcaraT (filled triangles) and DS68625-GAL2 (filled squares), expressing either PcAraT or Gal2, respectively. Data are derived from duplicate experiments and shown as the average ± mean deviation.
Additional file 4. Specific rate of l-arabinose uptake by PcAraT. Uptake experiments were performed with increasing concentrations of [14C-] l-arabinose with the Hxt1-7 and Gal2 deletion strain S. cerevisiae DS68625-PcaraT expressing PcaraT on a centromeric plasmid. No [14C-] d-glucose uptake was observed for this strain. Data are derived from duplicate experiments and shown as the average ± mean deviation.
Additional file 5. Specific rate of d-glucose uptake by Gal2. Uptake experiments were performed with increasing concentrations of [14C-] d-glucose with the Hxt1-7 and Gal2 deletion strain S. cerevisiae DS68625-GAL2 expressing GAL2 on a centromeric plasmid. Data are derived from duplicate experiments and shown as the average ± mean deviation.
Additional file 6. Specific rate of l-arabinose uptake by Gal2. Uptake experiments were performed with increasing concentrations of [14C-] l-arabinose with the Hxt1-7 and Gal2 deletion strain S. cerevisiae DS68625-GAL2 expressing GAL2 on a centromeric plasmid. Data are derived from duplicate experiments and shown as the average ± mean deviation.
Additional file 7. Impact of proton-gradient uncoupling on transport activity. Transport rates of [14C]-l-arabinose of the Hxt1-7 and Gal2 deletion strains DS68625-PcaraT and DS68625-GAL2 expressing either PcAraT (DS68625-PcaraT, white bars) or Gal2 (DS68625-GAL2, grey bars) on a centromeric plasmid. Transport rates were determined in 200 μl synthetic medium at a [14C]-l-arabinose concentration of 2 mmol L−1 upon addition of either 0.5 µL water, 0.5 μL DMSO, or 10 μM CCCP (0.5 µl of a stock solution dissolved in 100% DMSO) (A). Panel (B) shows the uptake capacity in % relative to the control (H2O). Data are derived from duplicate experiments and shown as the average ± mean deviation.
Data Availability Statement
The datasets supporting the conclusions of this article are available at NCBI Genome Omnibus database (GEO) under Accession Numbers GSE12632, GSE24212, and GSE10449, respectively, in the GEO repository (https://www.ncbi.nlm.nih.gov/geo/).


