Abstract
Background
Ovarian cancer is the most lethal malignant tumor of the female reproductive system, and the metastasis is one of the major factors that contribute to the poor outcome of patients with OC. Accumulating evidence indicates that lncRNAs are expressed and play important regulatory roles in ovarian cancer.
Material/Methods
Aberrant lncRNAs in primary ovarian cancer tissues (POCTs) and paired omental metastasis tissues (OMTs) of patients with HGSOC were studied via lncRNA microarray. Real-time PCR was performed to examine CTD-2020K17.1 expression in HGSOC tissues from 38 patients, a normal ovarian surface epithelium cell line, and 4 ovarian cancer cell lines. Additionally, Transwell assays, wound healing assays, CCK-8 proliferation assays, and flow cytometry were used to explore the biological function of CTD-2020K17.1 in ovarian cancer cells. Finally, Western blot analysis was used to verify the potential target gene of CTD-2020K17.1.
Results
A novel lncRNA named CTD-2020K17.1 was identified via microarray analysis. Expression of CTD-2020K17.1 was significantly increased in OMTs and in 4 ovarian cancer cell lines compared with POCTs (P<0.05) or normal ovarian surface epithelial cell line (P<0.05). Moreover, CTD-2020K17.1 overexpression promoted migration, invasion, and proliferation of ovarian cancer cells, and CTD-2020K17.1 regulated the expression of CARD11.
Conclusions
CTD-2020K17.1 is significantly upregulated in OMTs and ovarian cancer cell lines. It can promote the migration, invasion, and proliferation of ovarian cancer cells, and CARD11 is regulated by CTD-2020K17.1.
MeSH Keywords: Neoplasm Metastasis; Ovarian Neoplasms; RNA, Long Noncoding
Background
Ovarian cancer is the most lethal malignant tumor of the female reproductive system [1,2]. More than 22 280 Americans were diagnosed with ovarian cancer in 2016, resulting in 14 240 deaths [3]. High-grade serous ovarian cancer (HGSOC) is the most common histotype of ovarian cancer [1]. The majority of HGSOC patients are diagnosed at an advanced stage. Despite the development of new surgical techniques and chemotherapeutic drugs, most patients still experience recurrent abdominal metastasis, with the 5-year survival rate below 30% [4]. Therefore, it is essential to explore the mechanism of metastasis and find novel molecular markers and targets in HGSOC for therapy to block metastasis and improve the 5-year survival rates.
Long non-coding RNA (lncRNA) is a class of non-coding RNAs with a length of more than 200 nucleotides [5]. Accumulating evidence indicates that lncRNAs are expressed and play important regulatory roles in various tumors [6–8]. For example, LncRNA UCA1 can act as a competitive endogenous RNA (ceRNA) to suppress metastasis and invasion of ovarian cancer cells through the UCA1-mir-485-5p-MMP14 axis [9]. Previous studies have reported on dysregulated lncRNAs and their biological functions in multiple histotypes of ovarian cancer, which may lead to biased results because of pathological heterogeneity. Few studies have reported the regulatory effect of lncRNAs in HGSOC, the single histotype of ovarian cancer. Thus, it is necessary to explore the differentially expressed lncRNAs in HGSOC and investigate their biological functions, which may provide more accurate and meaningful results.
In the present study, lncRNA CTD-2020K17.1 (ENST00000 585471), located on chromosome 17, was identified via microarray to be aberrantly expressed in HGSOC tissues. The full-length of CTD-2020K17.1 is 1077 nt. We studied the expression and role of CTD-2020K17.1 in HGSOC tissues and ovarian cancer cell lines, and also explored its regulatory function on CARD11 at transcription and translation levels.
Material and Methods
Clinical samples
Primary ovarian cancer tissues (POCTs) and paired omental metastasis tissues (OMTs) were collected from 38 HGSOC patients who underwent cytoreductive surgery from 2016 to 2017 at the Shengjing Hospital Affiliated to China Medical University. All samples were immediately frozen in liquid nitrogen and stored at −80°C. All patients had no prior treatment for cancer and were diagnosed by histopathological examination. This study was approved by the Ethics Committee of Shengjing Hospital Affiliated to China Medical University (2016PS36K).
LncRNA microarray analysis and coding-non-coding gene co-expression network
Three POCTs and paired OMTs from HGSOC patients were prepared for Arraystar Human LncRNA Microarray analysis. Sample labeling and microarray hybridization were performed by Kangcheng Biotech (Shanghai, China). Quantile normalization and subsequent data processing were performed using the GeneSpring GX v12.1 software package (Agilent Technologies). LncRNAs and mRNAs with significantly differential expression between the 2 groups were identified through P-value/FDR filtering.
The co-expression network of coding-non-coding genes (CNC network) was assessed by Kangcheng Biotech (Shanghai, China). The Pearson correlation coefficients (PCC) between differentially expressed lncRNAs and mRNAs were calculated for correlation analysis. The absolute value of PCC>0.99 with a P<0.05 indicated a strong relationship of each paired lncRNA and mRNA.
Cell culture
The human epithelial ovarian cancer cell lines SKOV3, OVCAR3, CAOV3, and HO8910 and normal human ovarian surface epithelial cell line HOSE were purchased from the Institute of Biochemistry and Cell Biology at the Chinese Academy of Sciences (Shanghai, China). All ovarian cancer cell lines were cultured in RPMI 1640 medium (Hyclone, USA) with 10% fetal bovine serum (FBS), while the HOSE cell line was cultured in MCDB 105: Medium 199 (1: 1, v/v) with 10% FBS. All cells were cultured at 37°C in 5% CO2.
RNA extraction and real-time PCR
Total RNA was extracted from tissue samples or cell lines using TRIzol® Reagent (Ambion, USA) and reverse-transcribed to cDNA using a PrimeScriptTMRT Reagent Kit (TaKaRa Bio, China) following the manufacturer’s protocol. Relative expression levels of CTD-2020K17.1 were evaluated using SYBR® Premix Ex Taq II (TaKaRa Bio, China) on a Light Cycler 480 II Real-time PCR system (Roche Diagnostics, Switzerland). Results were normalized to the expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an endogenous reference. Primers were designed by Sangon Biotech (Shanghai, China) with the following sequences: CTD-2020K17.1: 5′-CGTAGACGGTATGACAGCCAA-3′ (forward) and 5′-CAACAGTCCAGAAATGTCGCC-3′ (reverse) and GAPDH: 5′-CGGATTTGGTCGTATTGGG-3′ (forward) and 5′-CTGGAAGATGGTGATGGGATT-3′ (reverse). H19: 5′-CTTGGAAATGAATATGCTGCAC-3′ (forward) and 5′-GTCTGGTTCCTCTAGCTTCC-3′ (reverse). The fold change of CTD-2020K17.1 was calculated by the 2-ΔΔCT method, with ΔΔCT=ΔCT OMTs lncRNA–ΔCT POCTs lncRNA(tissues), or ΔΔCT=ΔCT ovarian cancer cell line lncRNA–ΔCT HOSE lncRNA (cells). ΔCT was calculated by the following formula: ΔCT=CT target lncRNA–CT GAPDH.
Vector construction and cell transfection
CTD-2020K17.1 cDNA was synthesized and cloned into the pcDNA3.1 plasmid, and an empty plasmid, pcDNA3.1, was constructed by Gene Pharma (Suzhou, China). The CTD-2020K17.1 plasmid transfected cells and empty plasmid transfected cells were called the CTD-2020K17.1 group and the negative control (NC) group, respectively. CTD-2020K17.1 Smart Silencer and a scrambled negative control Smart Silencer were synthesized by RIOBIO (Guangzhou, China). The target sequences of CTD2020-K17.1 Smart Silence were: 5′-CTGAAGCTTGGGACTAACA-3′, 5′-CGGTATGACAGCCAAGATA-3′, 5′-CCAGGTGATTGATAAGGTT-3′, 5′-AGACGGTATGACAGCCAAGA-3′, 5′-AGGAAATCCTAGAAGGGATG-3′, and 5′-ACCTTCAGTG CAGTCACTGG-3′. Cells were then subdivided into a si-CTD-2020K17.1 group transfected with CTD-2020K17.1 Smart Silencer and a si-NC group transfected with NC Smart Silencer. Transfections were performed using Lipofectamine 2000 reagent (Invitrogen, USA) following the manufacturer’s instructions.
Transwell assays and wound healing assay
For the migration assay, 200 μL of serum-free medium containing 2×104 cells were plated in the upper chamber of a Transwell apparatus with 600 μL RPMI1640 medium containing 10% FBS placed in the lower chamber. For the invasion assay, the upper compartment of the Transwell chamber (Corning, USA) was coated with 50 μL basement membrane Matrigel (BD Biosciences, USA). We then cultured 2×104 cells in 200 μL serum-free medium upon the Matrigel and 600 μL medium supplemented with 10% serum were added to the lower chamber. After 48 h of incubation, the bottom of the chamber insert was fixed with methanol for 1 min, dyed with hematoxylin for 3 min, and then stained with eosin for 30 s. Cells migrating to the bottom of the membrane were photographed in 10 randomly selected visual fields of the filter under an inverted microscope (Leica DM3000, Germany).
To assay wound healing, 5×105 transfected cells were seeded per well in a 12-well plate and cultured to confluence. Artificial homogeneous wounds were created by scraping a sterilized 200-μL micropipette tip through the cell layer. The cells were then cultured with serum-free medium and photographed at 0, 6, 12, and 24 h with a light microscope (Leica DMI3000B, Germany). Cell-free areas were measured using Image J software.
Cell proliferation assay and cell cycle assay
Cell proliferative capacity was measured via a Cell Counting Kit 8 (CCK-8, Dojindo). Briefly, 2.5×103 cells were seeded into 96-well plates after transfection and cultured for 24, 48, 72, or 96 h. We added 10 μL of CCK-8 into each well and the cells were incubated for 1 h at 37°C. Optical density was measured using a microplate reader (Bio-Rad, USA) at a wavelength of 450 nm.
To detect cell cycle progression, flow cytometric analysis was performed. Forty-eight hours after transfection, adhered cells were trypsinized and centrifuged at 2000 rpm for 5 min and then fixed with 70% cold ethanol overnight at 4°C. A cell cycle detection kit (KeyGEN, China) was used to stain these cells according to the manufacturer’s protocol. Data were collected using the Accuri C6 Flow Cytometer (BD Bioscience, USA) and analyzed by Modfit 5.0 software.
Western blot analysis
To detect the expression of CARD11 proteins, Western blot analysis was performed. In brief, whole-cell extracts were obtained using the Protein Extraction Kit (KeyGen, China). Proteins were separated by SDS-polyacrylamide gel electrophoresis and blotted onto PVDF membranes (Millipore, USA). Membranes were immunoblotted with the specific primary antibodies CARD11 and then with peroxidase-conjugated goat anti-rabbit IgG. The bands were visualized using chemiluminescence (Thermo Fisher, China) and analyzed by Gel Capture version software (DNR Bio-Imaging Systems Jerusalem, Israel). The β-actin antibody was used as a control for total protein input. The following antibodies were used: CARD11 antibody (1: 500, ProSci) and β-actin antibody (1: 5000, Abcam).
Statistical analysis
All statistical analyses were performed using SPSS 20.0 software (Chicago, USA). Data are expressed as mean ± standard deviation (SD). Each experiment was independently repeated in triplicate. The t test was performed for parametric tests. The Mann-Whitney U test was used for the correlation between CTD-2020K17.1 expression and clinicopathological parameters analysis. P<0.05 was considered statistically significant.
Results
The expression of CTD-2020K17.1 in HGSOC tissues and ovarian cancer cell lines
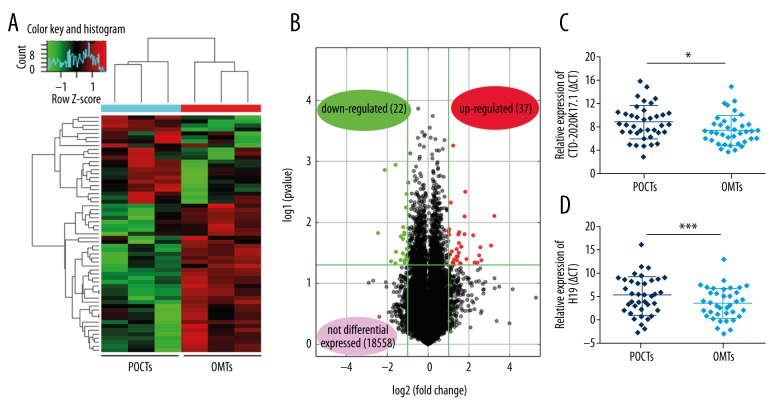
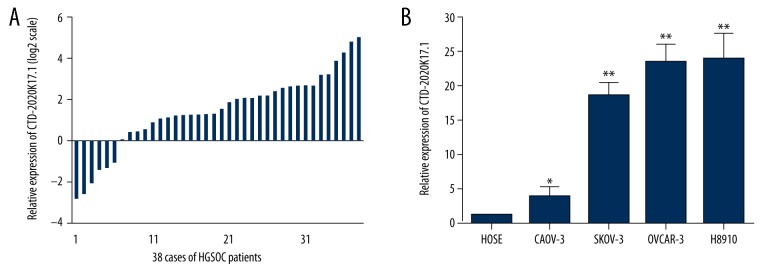
We first examined lncRNA expression profiling between 3 paired POCTs and OMTs from patients with HGSOC using the Arraystar Human LncRNAs Microarray v4.0 (Figure 1A). A total of 18 617 lncRNAs were detected, of which 37 lncRNAs were upregulated and 22 downregulated lncRNAs (fold change >2 and P<0.05) in OMTs compared to POCTs (Figure 1B). Among these upregulated lncRNAs, CTD-2020K17.1 drew our attention because it could be identified in the Gencode database and it exhibited a group-raw intensity of more than 200. We next studied the expression of CTD-2020K17.1 and a classical lncRNA H19 by real-time PCR in 38 paired HGSOC samples. CTD-2020K17.1 was significantly upregulated in OMTs compared to POCTs (Figure 1C). The lncRNA H19 was also significantly upregulated in OMTs. (Figure 1D). Thirty-two patients (84%) showed higher expression of CTD-2020K17.1 in OMTs compared with paired POCTs (Figure 2A). These data indicate that CTD-2020K17.1 has higher expression in OMTs than in primary tumor tissues (P<0.05). The relative expression of CTD-2020K17.1 (OMTs/POCTs) was correlated with the CA125 level of patients (P<0.05), but not with age, lymph node metastasis, ascites volume, or FIGO stage (Table 1).
Figure 1.
Expression levels of CTD-2020K17.1 in HGSOC tissues. (A) Hierarchical clustering analysis shows differentially expressed lncRNAs in paired POCTs and OMTs of 3 HGSOC cases (fold change >2.0, P<0.05). (B) Differentially expressed lncRNAs in OMTs compared to POCTs were determined by Volcano Plot. (C, D) CTD-2020K17.1 and H19 expression in POCTs and OMTs are presented as the mean ±SD. Being normalized to GAPDH, a lower ΔCt indicates a higher expression and the expression of CTD-2020K17.1 in OMTs is higher than POCTs. * P<0.05; *** P<0.005.
Figure 2.
Expression of CTD-2020K17.1 in ovarian cancer cell lines and tissues. (A) Relative CTD-2020K17.1 expression in OMTs compared with POCTs, presented as log base 2 of the fold change for 38 HGSOC patients. (B) Relative expression levels of CTD-2020K17.1 in 4 ovarian cancer cell lines (CAOV-3, SKOV-3, OVCAR-3, and HO8910), compared with a human ovarian surface epithelial cell line (HOSE) were calculated using the 2-ΔΔCT method. Levels of CTD-2020K17.1 in the 4 ovarian cancer cell lines were higher than in the human ovarian surface epithelial cell line. Data are shown as mean ±SD. * P<0.05, ** P<0.01.
Table 1.
Correlation between relative CTD-2020K17.1 expression and clinicopathological characteristics of HGSOC patients.
| Number | CTD-2020K17.1* | P | |
|---|---|---|---|
| Age (years) | 0.773 | ||
| <55 | 19 | 2.949 (1.464–6.409) | |
| ≥55 | 19 | 2.445 (1.357–5.897) | |
| Lymph node metastasis | 0.064 | ||
| Absent | 22 | 4.318 (2.268–6.409) | |
| Present | 16 | 2.284 (0.417–4.192) | |
| Ascites (ml) | 0.828 | ||
| <1000 | 18 | 3.870 (1.437–6.409) | |
| ≥1000 | 22 | 2.462 (1.525–6.149) | |
| FIGO stage | 0.658 | ||
| III | 31 | 2.949 (1.464–6.409) | |
| IV | 7 | 2.479 (1.357–4.563) | |
| CA125 (U/ml) | 0.044 | ||
| <1000 | 18 | 4.395 (2.268–10.528) | |
| ≥1000 | 20 | 2.395 (0.623–4.470) |
Median of relative expression, with 25th–75th percentile in parenthesis.
We also explored the expression levels of CTD-2020K17.1 in several ovarian cancer cell lines compared with normal ovarian surface epithelium cells (HOSE). As shown in Figure 2, CTD-2020K17.1 expression was increased in CAOV-3, SKOV-3, OVCAR-3, and HO8910cell lines compared with HOSE (P<0.05 for all).
CTD-2020K17.1 upregulation facilitates ovarian cancer cell migration, invasion, and proliferation
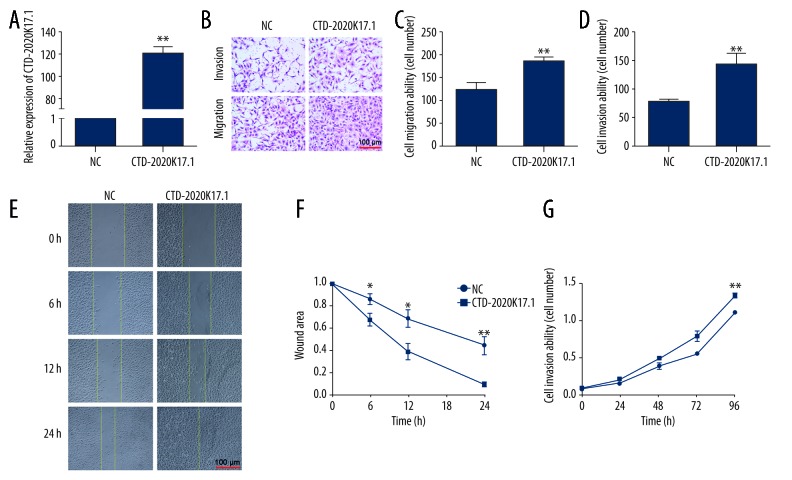
Gain-of-function studies were performed to investigate the possible biological role of CTD-2020K17.1 in the SKOV-3 cell line. Forty-eight hours after transfection, CTD-2020K17.1 levels were detected in the NC and CTD-2020K17.1 groups via real-time PCR. CTD-2020K17.1 expression was significantly increased in the CTD-2020K17.1 group (121.20±4.99-fold increase, P<0.01, Figure 3A).
Figure 3.
The effect of CTD-2020K17.1 on cell migration, invasion, and proliferation. (A) Transfection efficiency was measured by real-time RT-PCR. (B) Migration and invasion abilities were determined via Transwell assays. (C, D) Numbers of migrating or invading cells. (E, F) Migration ability was observed via wound healing assay. (G) Proliferation ability was determined by CCK-8 assays. * P<0.05, ** P<0.01.
Transwell assays were performed to evaluate the effect of CTD-2020K17.1 on cell migration and invasion capacity. The number of cells able to migrate through the basement membrane of the chamber represents the potential metastatic ability of the transfected cells (Figure 3B). The migration assay showed that significantly more CTD-2020K17.1 group cells were able to migrate through the Transwell compared to the NC group (127.25±11.77 vs. 186.25±10.46, P<0.01, Figure 3C). Likewise, for the invasion assay, there were significantly more invading cells in the CTD-2020K17.1 group than in the NC group (142.39±17.04 vs. 78.63±3.12, P<0.01, Figure 3D). The results of the Transwell assays indicate that upregulated CTD-2020K17.1 promotes cell migration and invasion. To further confirm the migration-promoting ability of CTD-2020K17.1, we performed a wound healing assay. As shown in Figure 3E, 3F, more healing was seen in the CTD-2020K17.1 group than in the NC group at 6, 12, and 24 h (P<0.05 for all).
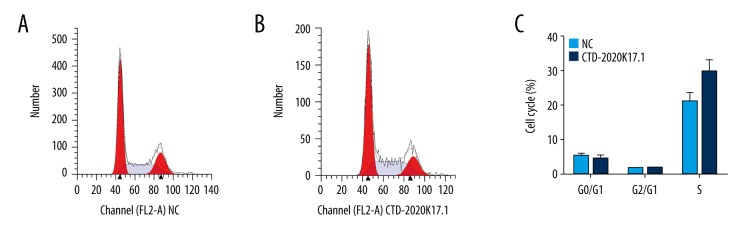
To determine whether upregulated CTD-2020K17.1 contributes to cell proliferation, we performed a CCK8 assay. The growth curve of the CTD-2020K17.1 group increased dramatically compared with the NC group (P<0.01), showing that CTD-2020K17.1 promotes proliferation (Figure 3G). However, flow cytometric analyses demonstrated that CTD2020-K17.1 had no effect on cell cycle distribution (P>0.05, Figure 4).
Figure 4.
The effect of CTD-2020K17.1 on cell cycle. (A) Flow cytometric analysis of the NC group. (B) Flow cytometric analysis of the CTD-2020K17.1 group. (C) Cell distribution between the NC and CTD-2020K17.1 groups.
Knockdown of CTD-2020K17.1 suppresses biological function of ovarian cancer cells
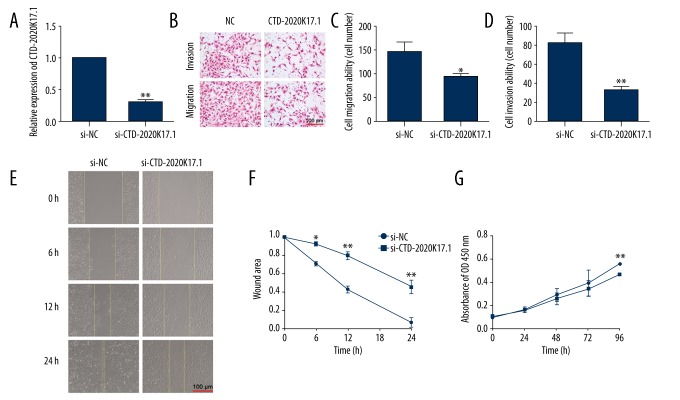
To further validate the regulatory role of CTD-2020K17.1, loss-of-function studies were performed. We first examined the knockdown efficiency of CTD-2020K17.1 Smart Silencer by real-time PCR after transfection. CTD-2020K17.1 was expressed at a lower level in the si-CTD-2020K17.1 group than in the si-NC group (0.30±0.03-fold decrease, P<0.01, Figure 5A).
Figure 5.
The effect of si-CTD-2020K17.1 on cell migration, invasion, and proliferation. (A) Transfection efficiency was measured by real-time RT-PCR. (B) Migration and invasion abilities were determined via Transwell assays. (C, D) Numbers of migrating or invading cells. (E, F) Migration ability was observed via wound healing assay. (G) Proliferation ability was determined by CCK-8 assays. * P<0.05, ** P<0.01.
Transwell assays showed that migration and invasion ability were both markedly attenuated in CTD-2020K17.1-knockdown SKOV-3 cells (Figure 5B). Compared with the si-NC group, the number of cells passing through the membrane were decreased in the si-CTD-2020K17.1 group for the migration assay (145.51±15.80 vs. 93.73±6.1, P<0.01, Figure 5C) and the invasion assay (82.59±8.48 vs. 32.73±2.51, P<0.05, Figure 5D). Moreover, wound healing was dramatically decreased at 24 h in the si-CTD-2020K17.1 group, indicating that downregulation of CTD-2020K17.1 inhibits cell migration (P<0.01, Figure 5E, 5F).
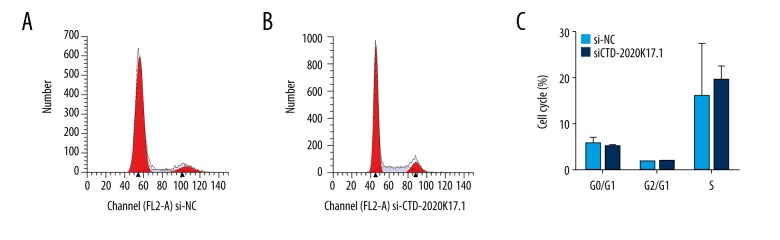
The CCK8 assay showed that CTD-2020K17.1 knockdown significantly reduced proliferation at 96 h compared with the si-NC group (P<0.01, Figure 5G). Again, no significant difference was seen in cell cycle distribution between the si-CTD-2020K17.1 and si-NC groups (P>0.05, Figure 6).
Figure 6.
The effect of si-CTD-2020K17.1 on cell cycle. (A) Flow cytometric analysis of the si-NC group. (B) Flow cytometric analysis of the si-CTD-2020K17.1 group. (C) Cell distribution between the si-NC and si-CTD-2020K17.1 groups.
CARD11 is regulated by CTD-2020K17.1 in SKOV3 cells
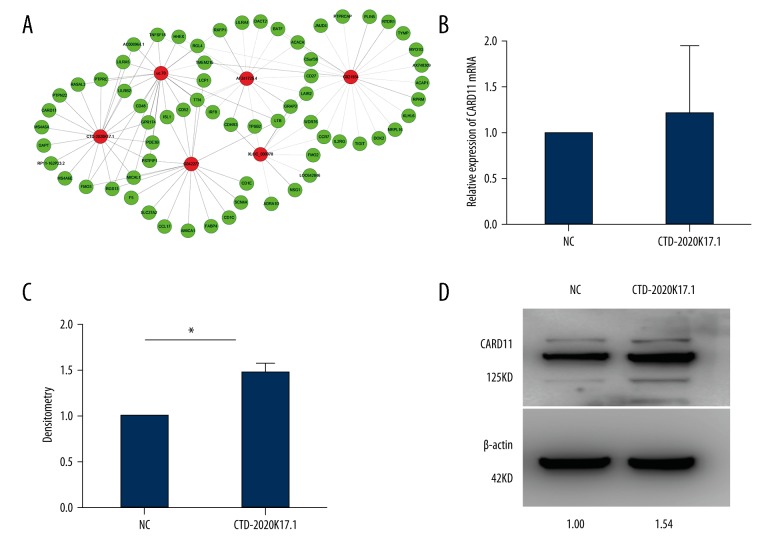
First, we performed a CNC network analysis to find the potential target mRNAs of CTD-2020K17.1, and identified CARD11, a differentially expressed gene positively correlated with CTD-2020K17.1 expression in the 3 paired POCTs and OMTs (Figure 7A).
Figure 7.
CARD11 is regulated by CTD-2020K17.1 in SKOV3 cells. (A) CNC network between the differentially expressed lncRNAs and mRNAs in POCTs and paired OMTs of HGSOC. (B) Real-time PCR revealed that the expression of CTD-2020K17.1 had no effect on CARD11 at mRNA level. (C) The densitometry (repeated 3 times) of Western blot. * P=0.019. (D) Western blot showed that CTD-2020K17.1 could upregulate CARD11 expression.
To explore the relationship between CTD-2020K17.1 and CARD11, we performed real-time PCR and Western blot assay. Forty-eight hours after transfection, there was no obvious difference at the mRNA level of CARD11 (P>0.05, Figure 7B) between the CTD-2020K17.1 group and NC group. However, we found that CARD11 protein was significantly increased in the CTD-2020K17.1 group compared with the NC group (Figure 7C).
Discussion
HGSOC is a highly invasive cancer with poor prognosis. It frequently spreads within the abdominal cavity and has a predilection for omental implantation [10]. Most previous studies consider that aberrantly expressed molecules in primary tumors must determine disease fate to some extent [11,12], yet few studies have compared primary tumors and OMTs to investigate the molecular changes during tumor dissemination. Thus, it is necessary to identify molecular features in the metastatic progression of this disease. In our study, we used microarray to detect differentially expressed lncRNAs in 3 paired OMTs and POCTs of HGSOC, searching for lncRNAs with group-raw intensities over 200, meaning that they could be detected by real-time PCR, and which were listed in the Gencode database, the largest publicly available catalog of human lncRNAs, consisting of 9640 lncRNA loci [13]. We ultimately identified CTD-2020K17.1, which met the 2 above criteria and thus drew our attention.
Several researchers have recently reported on dysregulated lncRNAs in ovarian cancer. Examples include lncRNA MEG3 reducing cisplatin resistance in ovarian cancer via demethylation of curcumin [14], and lncRNA ANRIL acting as a potential biomarker in diagnosis and treatment of serous ovarian cancer [15]. However, even if CTD-2020K17.1 was differentially expressed in POCTs and OMTs according to microarray result, it may be the result of inflammation or interstitial contamination rather than the real difference between POCTs and OMTs. Therefore, larger-scale examination should be done. We examined expression of CTD-2020K17.1 in 38 HGSOC patients and found that CTD-2020K17.1 expression was highly significantly elevated in OMTs compared with their POCTs. Thus, we assumed that the difference in CTD-2020K17.1 expression may contribute to the omental localization of metastatic cells which shed from the primary tumor. Moreover, CTD-2020K17.1 was upregulated in different ovarian cancer cell lines compared with the normal ovarian epithelial cell line. Thus, it may be a metastasis-associated lncRNA of HGSOC which exerts oncogenic functions.
Metastasis is an important feature of cancers. Some lncRNA have been reported to be related with metastasis of cancers; for example, 2 classical lncRNAs– HOTAIR and H19 – can both promote cell invasion in ovarian cancer. High HOTAIR expression in epithelial ovarian cancer positively correlates with invasion [16], and lncRNA H19 may regulate ovarian cancer cell metastasis through the H19/let-7 axis [17]. Yang [18] and Zhu [19] has reported that H19 is involved in the malignant process of ovarian cancer. In our study, the Transwell assays demonstrated that upregulated CTD-2020K17.1 notably promotes migration and invasion in serous ovarian cancer cell line SKOV3, while knockdown of CTD-2020K17.1 attenuates metastasis. Furthermore, CTD-2020K17.1 could also significantly increase the wound healing rate of ovarian cancer cells. Thus, these results indicate that CTD-2020K17.1 may play oncogenic role in ovarian cancer through promoting migration and invasion.
Proliferation is another important cancer characteristic. Previous studies have reported that dysregulated lncRNAs can regulate tumor proliferation, such as lncRNA H19 silencing [20,21] suppressed the growth rate of ovarian cancer cells. Moreover, HOXA11-AS, a newly reported lncRNA, was demonstrated to act as a tumor-suppressor in epithelial ovarian cancer cells by inhibiting proliferation [22]. As demonstrated by the CCK-8 assay, overexpressing CTD-2020K17.1 had a positive impact on proliferation of SKOV3 cells, and silencing CTD-2020K17.1 significantly reduced the proliferative capacity. Subsequent flow cytometric assays showed that the alteration of CTD-2020K17.1 had no correlation with cell cycle. Thus, CTD-2020K17.1 upregulation significantly promotes ovarian cancer cell proliferation, but this effect is not caused by alteration of cell cycle. Although alteration of cell cycle could promote cell proliferation [23], there are also some other mechanism which may contribute to the cell proliferation. For example, Nrf2 can promote cell proliferation by activating autophagy and inhibiting apoptosis, while the cell cycle distribution showed no significant difference [24]. ATG5 is known to be a necessary gene for mammalian autophagy. PP2, a Src family tyrosine kinase inhibitor, can inhibit the proliferation of ATG5 knockout cell through a cell-cycle-independent mechanism [25]. Thus, the cell proliferation could also be influenced by alteration of autophagy or apoptosis without cell-cycle distribution changing. Thus, CTD-2020K17.1 can promote ovarian cancer cell proliferation through similar mechanisms, but the exact mechanism requires further exploration.
LncRNAs do not encode proteins, so they often exert their biological functions through influencing other protein coding genes. Thus, it is important to find their target genes and analyze their relative mechanisms. Based on the foundation of lncRNA microarray report and CNC network analysis, we identified CARD11, a differentially expressed gene positively correlated with CTD-2020K17.1 expression. CARD11 belongs to the caspase-associated recruitment domain (CARD) protein family and has a characteristic domain, CARD, which interacts with BCL10 and then activates the NF-κB pathway [26]. CARD11 has been demonstrated to participate in the tumorigenesis and progression of diverse tumors, such as colorectal cancer and B-cell lymphomas [27,28]. It has been reported that CARD11 is elevated in epithelial ovarian cancer [29]. Other studies reinforce that the NF-κB pathway can be activated by CARD11 [30,31] and plays a metastatic-associated role in ovarian cancer via various regulatory factors such as CD44 [32]. These studies strongly suggest that CARD11 may play an oncogenic role similar to that of CTD-2020K17.1. In our study, the CARD11 protein level was significantly elevated at 48 h after transfection of CTD-2020K17.1. However, no obvious difference in CARD11 expression was seen at the mRNA level, which is quite different from the CNC network analysis result. So, how CTD-2020K17.1 regulated CARD11 protein needs further study. Recent studies have shown that lncRNAs could regulate the production of target protein through different regulation mechanisms, including forming lncRNA-mRNA complex [33], acting as a ceRNA to bind with miRNA [34]. The protein level may also be regulated by proteolysis through various mechanisms, among which, the ubiquitin-proteasome system (UPS) [35] is the most important. For instance, the lncRNA uc.134 represses hepatocellular carcinoma progression by inhibiting ubiquitination of LATS1 [36]. Ubiquitin is a stable small-molecule protein which is widely present in eukaryotic cells and is highly conserved in structure [37]. It can covalently bind to target protein to mediates proteolysis. Deubiquitinases (DUBs) [38,39] negatively regulate ubiquitination and play critical roles in maintaining stability and homeostasis of cellular proteins. For example, UCH37, a kind of DUBs, effectively controls the rate of proteolysis, thereby promoting the progression of cancer [40]. Thus, it is also possible that CTD-2020K17.1 regulates the CARD11 protein level by affecting the production of some DUBs. However, the exact regulation relationship between CTD-2020K17.1 and CARD11 needs further investigation.
In summary, all these findings suggest that CTD-2020K17.1 exerts its function to modulate CARD11 expression during metastatic process in ovarian cancer. Our future work will focus on the exact regulatory mechanism between CTD-2020K17.1 and CARD11 in ovarian cancer.
Conclusions
In summary, CTD-2020K17.1 is a novel lncRNA which is upregulated in OMTs of HGSOC patients and in ovarian cancer cell lines. It can promote metastasis and proliferation of ovarian cancer cell line SKOV3. CARD11 expression is upregulated by CTD-2020K17.1. These findings suggest that CTD-2020K17.1 acts as an oncogene and may be a potential therapeutic target for blocking metastasis in HGSOC.
Footnotes
Source of support: This work was supported by a grant from the Key Laboratory of the Education Department of Liaoning Province (LS201602) and a grant from the Science and Technology Department of Shenyang City (F15-139-9-34)
Conflicts of interest
None.
References
- 1.Kurman RJ. Origin and molecular pathogenesis of ovarian high-grade serous carcinoma. Ann Oncol. 2013;24(Suppl 10):x16–21. doi: 10.1093/annonc/mdt463. [DOI] [PubMed] [Google Scholar]
- 2.Li X, Yu Z, Fang L, et al. Expression of adiponectin receptor-1 and prognosis of epithelial ovarian cancer patients. Med Sci Monit. 2017;23:1514–21. doi: 10.12659/MSM.899990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 4.Vig-Varga E, Benson EA, Limbil TL, et al. Alpha-lipoic acid modulates ovarian surface epithelial cell growth. Gynecol Oncol. 2006;103:45–52. doi: 10.1016/j.ygyno.2006.01.060. [DOI] [PubMed] [Google Scholar]
- 5.Maruyama R, Suzuki H. Long noncoding RNA involvement in cancer. BMB Rep. 2012;45:604–11. doi: 10.5483/BMBRep.2012.45.11.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu RT, Cao JL, Yan CQ, et al. Effects of LncRNA-HOST2 on cell proliferation, migration, invasion and apoptosis of human hepatocellular carcinoma cell line SMMC-7721. Biosci Rep. 2017;37(2) doi: 10.1042/BSR20160532. pii: BSR20160532. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Zhang S, Dong X, Ji T, et al. Long non-coding RNA UCA1 promotes cell progression by acting as a competing endogenous RNA of ATF2 in prostate cancer. Am J Transl Res. 2017;9:366–75. [PMC free article] [PubMed] [Google Scholar]
- 8.Song YX, Sun JX, Zhao JH, et al. Non-coding RNAs participate in the regulatory network of CLDN4 via ceRNA mediated miRNA evasion. Nat Commun. 2017;8:289. doi: 10.1038/s41467-017-00304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y, Jiang Y, Wan Y, et al. UCA1 functions as a competing endogenous RNA to suppress epithelial ovarian cancer metastasis. Tumour Biol. 2016;37:10633–41. doi: 10.1007/s13277-016-4917-1. [DOI] [PubMed] [Google Scholar]
- 10.Smolle E, Taucher V, Haybaeck J. Malignant ascites in ovarian cancer and the role of targeted therapeutics. Anticancer Res. 2014;34:1553–61. [PubMed] [Google Scholar]
- 11.Chai Y, Liu J, Zhang Z, Liu L. HuR-regulated lncRNA NEAT1 stability in tumorigenesis and progression of ovarian cancer. Elife. 2016;5:1588–98. doi: 10.1002/cam4.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuang D, Zhang X, Hua S, et al. Long non-coding RNA TUG1 regulates ovarian cancer proliferation and metastasis via affecting epithelial-mesenchymal transition. Exp Mol Pathol. 2016;101:267–73. doi: 10.1016/j.yexmp.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Liu J, Xu X, Li L. Curcumin suppresses cisplatin resistance development partly via modulating extracellular vesicle-mediated transfer of MEG3 and miR-214 in ovarian cancer. Cancer Chemother Pharmacol. 2017;79:479–87. doi: 10.1007/s00280-017-3238-4. [DOI] [PubMed] [Google Scholar]
- 15.Qiu JJ, Lin YY, Ding JX, et al. Long non-coding RNA ANRIL predicts poor prognosis and promotes invasion/metastasis in serous ovarian cancer. Int J Oncol. 2015;46:2497–505. doi: 10.3892/ijo.2015.2943. [DOI] [PubMed] [Google Scholar]
- 16.Qiu JJ, Wang Y, Ding JX, et al. The long non-coding RNA HOTAIR promotes the proliferation of serous ovarian cancer cells through the regulation of cell cycle arrest and apoptosis. Exp Cell Res. 2015;333:238–48. doi: 10.1016/j.yexcr.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Yan L, Zhou J, Gao Y, et al. Regulation of tumor cell migration and invasion by the H19/let-7 axis is antagonized by metformin-induced DNA methylation. Oncogene. 2015;34:3076–84. doi: 10.1038/onc.2014.236. [DOI] [PubMed] [Google Scholar]
- 18.Yang K, Hou Y, Li A, et al. Identification of a six-lncRNA signature associated with recurrence of ovarian cancer. Sci Rep. 2017;7:752. doi: 10.1038/s41598-017-00763-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu Z, Song L, He J, et al. Ectopic expressed long non-coding RNA H19 contributes to malignant cell behavior of ovarian cancer. Int J Clin Exp Pathol. 2015;8:10082–91. [PMC free article] [PubMed] [Google Scholar]
- 20.Medrzycki M, Zhang Y, Zhang W, et al. Histone h1.3 suppresses h19 noncoding RNA expression and cell growth of ovarian cancer cells. Cancer Res. 2014;74:6463–73. doi: 10.1158/0008-5472.CAN-13-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizrahi A, Czerniak A, Levy T, et al. Development of targeted therapy for ovarian cancer mediated by a plasmid expressing diphtheria toxin under the control of H19 regulatory sequences. J Transl Med. 2009;7:69. doi: 10.1186/1479-5876-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richards EJ, Permuth-Wey J, Li Y, et al. A functional variant in HOXA11-AS, a novel long non-coding RNA, inhibits the oncogenic phenotype of epithelial ovarian cancer. Oncotarget. 2015;6:34745–57. doi: 10.18632/oncotarget.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long F, Wang T, Jia P, et al. Anti-tumor effects of Atractylenolide-I on human ovarian cancer cells. Med Sci Monit. 2017;23:571–79. doi: 10.12659/MSM.902886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Liu Z, Hu T, et al. Nrf2 promotes progression of non-small cell lung cancer through activating autophagy. Cell Cycle. 2017;16:1053–62. doi: 10.1080/15384101.2017.1312224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang SH, Han BI, Lee M. Knockout of ATG5 leads to malignant cell transformation and resistance to Src family kinase inhibitor PP2. J Cell Physiol. 2018;233(1):506–15. doi: 10.1002/jcp.25912. [DOI] [PubMed] [Google Scholar]
- 26.Jattani RP, Tritapoe JM, Pomerantz JL. Cooperative control of caspase recruitment domain-containing Protein 11 (CARD11) signaling by an unusual array of redundant repressive elements. J Biol Chem. 2016;291(16):8324–36. doi: 10.1074/jbc.M115.683714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slattery ML, Herrick JS, Mullany LE. The co-regulatory networks of tumor suppressor genes, oncogenes, and miRNAs in colorectal cancer. Genes Chromosomes Cancer. 2017;56(11):769–87. doi: 10.1002/gcc.22481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bognar MK, Vincendeau M, Erdmann T, et al. Oncogenic CARMA1 couples NF-kappaB and beta-catenin signaling in diffuse large B-cell lymphomas. Oncogene. 2016;35:4269–81. doi: 10.1038/onc.2015.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Block MS, Charbonneau B, Vierkant RA, et al. Variation in NF-kappaB signaling pathways and survival in invasive epithelial ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2014;23:1421–27. doi: 10.1158/1055-9965.EPI-13-0962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blonska M, Lin X. NF-kappaB signaling pathways regulated by CARMA family of scaffold proteins. Cell Res. 2011;21:55–70. doi: 10.1038/cr.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Staudt LM. Oncogenic activation of NF-kappaB. Cold Spring Harb Perspect Biol. 2010;2:a000109. doi: 10.1101/cshperspect.a000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haria D, Trinh BQ, Ko SY, et al. The homeoprotein DLX4 stimulates NF-kappaB activation and CD44-mediated tumor-mesothelial cell interactions in ovarian cancer. Am J Pathol. 2015;185:2298–308. doi: 10.1016/j.ajpath.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marsh SJ, Shah JS, Cole AJ. Histones and their modifications in ovarian cancer – drivers of disease and therapeutic targets. Front Oncol. 2014;4:144. doi: 10.3389/fonc.2014.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez-Mejias A, Tay Y. Competing endogenous RNA networks: Tying the essential knots for cancer biology and therapeutics. J Hematol Oncol. 2015;8:30. doi: 10.1186/s13045-015-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collins GA, Goldberg AL. The Logic of the 26S proteasome. Cell. 2017;169:792–806. doi: 10.1016/j.cell.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ni W, Zhang Y, Zhan Z, et al. A novel lncRNA uc.134 represses hepatocellular carcinoma progression by inhibiting CUL4A-mediated ubiquitination of LATS1. J Hematol Oncol. 2017;10:91. doi: 10.1186/s13045-017-0449-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaur J, Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat Rev Mol Cell Biol. 2015;16:461–72. doi: 10.1038/nrm4024. [DOI] [PubMed] [Google Scholar]
- 38.Sun L, Chen ZJ. The novel functions of ubiquitination in signaling. Curr Opin Cell Biol. 2004;16:119–26. doi: 10.1016/j.ceb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 39.Nijman SM, Luna-Vargas MP, Velds A, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–86. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 40.Wicks SJ, Haros K, Maillard M, et al. The deubiquitinating enzyme UCH37 interacts with Smads and regulates TGF-beta signalling. Oncogene. 2005;24:8080–84. doi: 10.1038/sj.onc.1208944. [DOI] [PubMed] [Google Scholar]