Abstract
Objetive
Systemic vasculitides represent a heterogeneous group of rare complex diseases of the blood vessels with a poorly understood aetiology. To investigate the shared genetic component underlying their predisposition, we performed the first cross-phenotype meta-analysis of genetic data from different clinically distinct patterns of vasculitis.
Methods
Immunochip genotyping data from 2465 patients diagnosed with giant cell arteritis, Takayasu’s arteritis, antineutrophil cytoplasmic antibody-associated vasculitis or IgA vasculitis as well as 4632 unaffected controls were analysed to identify common susceptibility loci for vasculitis development. The possible functional consequences of the associated variants were interrogated using publicly available annotation data.
Results
The strongest association signal corresponded with an intergenic polymorphism located between HLA-DQB1 and HLA-DQA2 (rs6932517, P=4.16E-14, OR=0.74). This single nucleotide polymorphism is in moderate linkage disequilibrium with the disease-specific human leucocyte antigen (HLA) class II associations of each type of vasculitis and could mark them. Outside the HLA region, we identified the KDM4C gene as a common risk locus for vasculitides (highest peak rs16925200, P=6.23E-07, OR=1.75). This gene encodes a histone demethylase involved in the epigenetic control of gene expression.
Conclusions
Through a combined analysis of Immunochip data, we have identified KDM4C as a new risk gene shared between systemic vasculitides, consistent with the increasing evidences of the crucial role that the epigenetic mechanisms have in the development of complex immune-mediated conditions.
Introduction
The vasculitides constitute a heterogeneous group of diseases of complex aetiology characterised by a chronic inflammation of the blood vessels. They show a wide spectrum of clinical manifestations that depend on the affected vessels in the arterial or venous system. As a consequence, overlapping of pathophysiological mechanisms is frequent among them.1 To better define the different clinical forms of vasculitis, the Chapel Hill Consensus Conference proposed a nomenclature system in which the most common vasculitides were subdivided into three main categories, that is. large vessel vasculitis (LVV), medium vessel vasculitis and small vessel vasculitis (SVV), based on the distribution of vessel involvement. 2 In this regard, two diseases account for most cases of LVV, that is, giant cell arteritis (GCA) and Takayasu’s arteritis (TAK), which mainly involve arteries of large calibre such as the aorta and its major branches. On the other hand, IgA vasculitis (IgAV) and antineutrophil cytoplasmic antibody-associated vasculitis (AAV) are classified as SVV, as inflammation predominantly affects arterioles, small arteries, capillaries and venules.2,3
Although the aetiology of vasculitides remains unclear, cumulating data suggest that they are triggered in genetically predisposed individuals by the concurrence of certain environmental factors.4 The importance of the genetic component has been consistently supported by evidence of familial aggregation, differential prevalence depending on the ethnicity, and multiple genetic associations with disease susceptibility and progression reported during the last years.5 In this regard, the strongest association signals in most vasculitides correspond with genetic variants within the human leucocyte antigen (HLA) region, which suggests an important role of the immune system in their pathophysiology. However, each form of vasculitis has distinct HLA association markers that define them, most likely due to disease-specific differences in antigenic drivers.5
Despite the considerable increase in our understanding of the genetic basis of systemic vasculitis during the last decade, the number of identified risk loci for most types of vasculitis remains significantly lower than other immune-mediated diseases.5,6 Taking into account the low prevalence of these pathologies in the general population,1 one of the main limitations is the lack of statistical power to identify susceptibility signals with modest effects, given the large sample size required for that. Therefore, additional strategies are necessary to unravel the genetic background underlying these disorders. In this sense, the combination of the genetic data from different clinical syndromes with vasculitis as a single underpinning phenotype has been successfully applied for identifying shared genetic components.7–10 One of the most successful genetic platforms to study immune-mediated diseases is the Immunochip, which is a custom high-density array that allows the analysis of genetic variants across multiple known susceptibility loci for autoimmune and autoinflammatory disorders.11 This platform has been successful in identifying specific and common risk variants for many autoimmune and autoinflammatory diseases,12 including GCA and TAK.9,13,14
Considering the above, and with the aim to improve the knowledge of the genetic component of vasculitides, we performed a cross-phenotype meta-analysis by combining Immunochip data of individuals diagnosed with four different forms of vasculitis.
Material and methods
Study population
The study population comprised 2465 cases from four vasculitides (GCA, TAK, AAV and IgAV) and 4632 unaffected controls. A detailed description of the study population can be found in the online supplementary material.
Genotyping and quality controls
The GCA and TAK raw datasets were obtained from previously published Immunochip studies.13,14 For AAV and IgAV, new genotyping data were generated. In this case, genomic DNA was extracted from peripheral blood samples with standard methods. Genotyping was performed using the Illumina Immunochip custom genotyping array (Infinium ImmunoArray-24 V.1.0 and V.2.0 BeadChip, respectively) according to the manufacturer’s instructions. To ensure consistency among datasets, we obtained the raw data of each of them and applied the same quality control procedures in parallel (including both the previously published studies and the newly genotyped data) using PLINK V.1.07.15 In this regard, low-quality single nucleotide polymorphisms (SNPs) were filtered out if they had call rates <98%, minor allele frequencies (MAF) <0.01 and deviation from Hardy-Weinberg equilibrium (HWE; P<0.001). Samples were also removed if they showed a genotyping calling <95%, had a pair of first-degree relatives (identity by descent >0.4) or were considered outliers by plotting at >4 SD from the cluster centroids of each population using the first 10 principal components (PC; estimated using the ancestry markers included in the Immunochip). Sex chromosomes were not analysed.
Imputation
SNP genotype imputation of whole-genome data after QC was performed separately for each dataset as implemented in IMPUTE V.2.16 The 1000 Genome Project Phase 3 data were used as reference panel.17 Briefly, we updated the SNP IDs and positions to rs# and build 37 (HG19), respectively, with PLINK. Chunks of 50 000 Mbp were then generated and imputed considering the most likely call for merging genotypes (>0.9 probability threshold). Imputed data were also subjected to stringent quality filters as follows: call rate <0.98, MAF<0.01, HWE P<0.001.
Imputation of the HLA region
We performed a more comprehensive imputation of this genomic region to generate imputed data of classical HLA alleles (with two and four digits), polymorphic amino acid positions and additional SNPs. In this case, we extracted the non-imputed genotyping data from 20 000 000 to 40 000 000 base pairs at chromosome 6 (which comprises the extended HLA region) and used the SNP2HLA V.1.0.3 package18 with a reference panel of 5225 individuals of European ancestry with available genotyping data for 8961 genetic variants (common SNPs and INDEL polymorphisms) as well as types for HLA class I and II molecules at four-digit resolution, as described.13,19
Data analysis
PLINK and the R-base software under GNU Public license V.2 were used to analyse the imputed data. First, we used logistic regression on the best-guess genotypes (>0.9 probability), assuming an additive model and using the 10 first PCs and gender as covariates, to analyse each dataset separately. The results were then meta-analysed by means of the inverse variance method under a fixed effects model, except for the HLA region in which a random effects model was considered. Heterogeneity of the OR across the different datasets was estimated using both Cochran’s Q test and I2. The study-wide threshold for both statistical significance and trends of association were set at 1.28E-06 and 2.55E-05, respectively, according to the estimation by the Genetic Type I Error Calculator (CEG) software,20 which implements a Bonferroni-based validated method to control the genome-wide type I error rate at 0.05. R was used to generate the overall Manhattan plots, and the LocusZoom V.1.1 tool (http://csg.sph.umich.edu/locuszoom/)21 was used to obtain the regional association plots. Finally, the possible functional implication of the association signals was estimated using the publicly available browser resources RegulomeDB,22 HaploReg V.4.123 and Blood NESDA NTR Conditional eQTL Catalog.24
Results
A total of 2425 vasculitis cases (1008 GCA cases, 437 TAK cases, 303 IgAV cases and 677 AAV cases) and 4526 controls, as well as 191 948 genetic markers remained after QC filtering and were included in the analyses.
Cross-phenotype meta-analysis
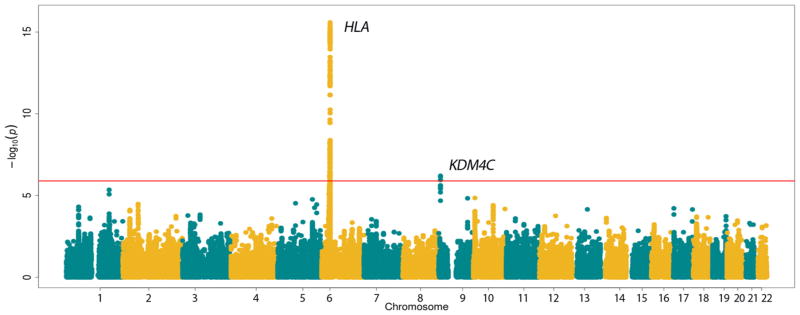
The combined analysis of the Immunochip data from all four pathologies showed evidence for common genetic associations in two loci, that is, HLA and lysine demethylase 4C (KDM4C), both surpassing the study-wide level of significance (figure 1).
Figure 1.
Manhattan plot representation of the results of the cross-disease meta-analysis of four types of vasculitis. The −log10 of the inverse variance-weighted meta-analysis P values are plotted against their physical chromosomal position. The red line represents the study-wide level of significance (P<1.28E-05). HLA, human leucocyte antigen.
As the strongest association signals were located within the HLA region, we decided to carry out a more robust analysis of this genomic region in order to fine-map and localise the putative causal variants. Therefore, we conducted another meta-analysis of imputed data of SNPs, classical alleles and polymorphic amino acid positions. Due to the complex linkage disequilibrium (LD) structure and disease-specific associations of this region, we used a random effect model. As shown in online supplementary figure 1 and table 1, several variants in high LD were significantly associated to vasculitis predisposition, with an intergenic polymorphism located between HLAD-QB1 and HLA-DQA2 representing the peak signal (rs6932517, P=4.16E-14, OR=0.74). This marker showed both evidence of nominal association with each disease separately and consistent ORs across studies (GCASpain, P=1.26E-04, OR=0.73; GCAUK, P=1.52E-03, OR=0.78; TAKTurkey, P=3.91E-02, OR=0.78; TAKUSA=4.03E-03, OR=0.60; IgAV, P=1.51E-04, OR=0.67; AAV, P=3.80E-04, OR=0.77). The remaining association peaks of the HLA lost their statistical significance after conditioning on rs6932517, thus suggesting that this SNP was sufficient to explain most of the observed HLA signals.
In order to evaluate whether rs6932517 represented a true novel susceptibility marker for the analysed vasculitides or just a marker of the established risk variants for each type of vasculitis, we decided to calculate the LD between them.13,14,25,26 As shown in online supplementary table 2, evidence of moderate LD was observed in all cases (D’>0.8).
Outside the HLA region, the most strongly associated polymorphism corresponded with the KDM4C SNP rs16925200 (P=6.23E-07, OR=1.75; table 1). No heterogeneity among the ORs and consistent OR directions were observed across studies (online supplementary table 3). This genomic region also harboured additional polymorphisms with significant P values, all of them in high LD with rs16925200, which suggests that they represent genetic markers of the same signal (supplementary figure S2). In addition, the association of this locus remained significant when the association test was performed under a random effects model (online supplementary table 3). Interestingly, one of these polymorphisms (rs12003023) showed a nominal association with Behçet’s disease (BD), a variable vessel vasculitis, in a previously published Immunochip study of this form of vasculitis (P=1.11E-02, OR=1.78, 95% CI=1.14 to 2.79).27 Consistent with this, the statistical significance of the overall meta-analysis for the KDM4C rs12003023 variant increased after including the BD data (rs10203023, P=4.08E-08, OR=1.69, P(Q)=7.70E-01, I2=0).
Table 1.
List of association and suggestive non-HLA shared signals (P<2.55E-5) between GCA, TAK, IgAV and AAV
| rs ID | Position (GRCh37) | Location | Locus | Change | Combined analysis | CI 95% | Q | I2 | |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Cases=2425 | Controls=4526 | ||||||||
|
|
|
||||||||
| P value | OR | ||||||||
| rs16925200 | 9:7045058 | Intronic | KDM4C | C<T | 6.23E-07 | 1.75 | 1.41 to 2.18 | 1.88E-01 | 33.06 |
|
| |||||||||
| rs35821841 | 9:7050249 | Intronic | KDM4C | G<T | 7.01E-07 | 1.74 | 1.39 to 2.15 | 2.06E-01 | 30.59 |
|
| |||||||||
| rs12003023 | 9:7058714 | Intronic | KDM4C | G<A | 1.10E-06 | 1.67 | 1.35 to 2.04 | 6.64E-01 | 0.00 |
|
| |||||||||
| rs10975993 | 9:7028287 | Intronic | KDM4C | G<C | 2.43E-06 | 1.68 | 1.35 to 2.07 | 1.89E-01 | 32.89 |
|
| |||||||||
| rs12343988 | 9:7026828 | Intronic | KDM4C | C<T | 2.44E-06 | 1.65 | 1.37 to 2.07 | 1.89E-01 | 32.88 |
|
| |||||||||
| rs77198751 | 9:7057185 | Intronic | KDM4C | G<C | 3.23E-06 | 1.68 | 1.35 to 2.08 | 9.31E-02 | 46.98 |
|
| |||||||||
| rs74449007 | 9:7031859 | Intronic | KDM4C | G<A | 3.85E-06 | 1.66 | 1.37 to 2.10 | 2.41E-01 | 25.72 |
|
| |||||||||
| rs6704162 | 1:192414422 | Intergenic | RGS21 | RGS1 | G<A | 4.48E-06 | 1.25 | 1.14 to 1.37 | 9.26E-01 | 0.00 |
|
| |||||||||
| rs12336927 | 9:7025932 | Intronic | KDM4C | T<C | 6.34E-06 | 1.66 | 1.35 to 2.10 | 2.63E-01 | 22.74 |
|
| |||||||||
| rs12725829 | 1:192414282 | Intergenic | RGS21 | RGS1 | C<T | 8.34E-06 | 1.24 | 1.13 to 1.36 | 9.32E-01 | 0.00 |
|
| |||||||||
| rs7895774 | 10:6510534 | Intronic | PRKCQ | A<G | 1.40E-05 | 0.83 | 0.77 to 0.91 | 8.52E-01 | 0.00 |
|
| |||||||||
| rs4574921 | 9:117538334 | Intergenic | TNFSF15 | C<T | 1.47E-05 | 0.83 | 0.76 to 0.90 | 9.21E-01 | 0.00 |
|
| |||||||||
| rs116760964 | 5:141510106 | Intronic | NDFIP1 | C<A | 1.72E-05 | 1.71 | 1.33 to 2.17 | 8.71E-01 | 0.00 |
|
| |||||||||
| rs12342947 | 9:7026860 | Intronic | KDM4C | G<A | 2.07E-05 | 1.54 | 1.31 to 1.95 | 1.50E-01 | 38.32 |
|
| |||||||||
| rs12517414 | 5:72558077 | Intergenic | TMEM174 | FOXD1 | A<C | 2.96E-05 | 1.27 | 1.10 to 1.37 | 2.16E-01 | 29.26 |
|
| |||||||||
| rs74995325 | 2:61351878 | Intronic | KIAA1841 | A<G | 3.29E-05 | 1.64 | 1.32 to 2.10 | 1.81E-01 | 33.98 |
|
| |||||||||
| rs6869688 | 5:158883027 | Intronic | AC008703.1 | A<G | 3.54E-05 | 0.85 | 0.79 to 0.92 | 6.72E-01 | 0.00 |
|
| |||||||||
| rs1250544 | 10:81032885 | Intronic | ZMIZ1 | A<G | 3.90E-05 | 0.85 | 0.79 to 0.92 | 2.70E-01 | 21.72 |
|
| |||||||||
| rs115069423 | 2:61349451 | 3′UTR | KIAA1841 | C<G | 4.09E-05 | 1.63 | 1.31 to 2.10 | 1.51E-01 | 38.26 |
|
| |||||||||
| rs79030953 | 2:61349916 | 3′UTR | KIAA1841 | C<T | 4.54E-05 | 1.61 | 1.30 to 2.06 | 1.92E-01 | 32.45 |
|
| |||||||||
| rs2802365 | 10:81038883 | Intronic | ZMIZ1 | A<G | 4.90E-05 | 0.85 | 0.79 to 0.93 | 2.02E-01 | 31.10 |
|
| |||||||||
| rs1890928 | 1:67929719 | 5′UTR | SERBP1 | G<A | 4.91E-05 | 0.80 | 2.28 to 2.82 | 5.35E-01 | 0.00 |
|
| |||||||||
| rs9665287 | 10:81039124 | Intronic | ZMIZ1 | C<G | 4.97E-05 | 0.85 | 0.79 to 0.93 | 1.93E-01 | 32.44 |
AAV, antineutrophil cytoplasmic antibody-associated vasculitis; GCA, giant cell arteritis; GRCh37, Genome Reference Consortium Human genome build 37; IgAV, IgA vasculitis; OR; OR for the minor allele; SNP, single nucleotide polymorphism; TAK, Takayasu’s arteritis.
Trend of associations (P<2.55E-5) were also observed for additional SNPs located in different genomic regions (table 1), including RGS21/RGS1 (rs6704162, P=4.48E-06, OR=1.25), protein kinase C theta (PRKCQ; rs7895774, P=1.40E-05, OR=0.83), LOC10012963/TNFSF15 (rs4574921; P=1.47E-05; OR=0.83), Nedd4 family interacting protein 1 (NDFIP1; rs116760964, P=1.72E-05, OR=1.71), TMEM174/FOXD1 region (rs12517414, P=2.96E-05, OR=1.27), KIAA1841 (rs74995325, P=3.29E-05, OR=1.64), AC008703.1 (rs6869688, P=3.54E-05, OR=0.85), zinc finger MIZ-type containing 1 (ZMIZ1; rs1250544, P=3.90E-05, OR=0.85) and SERPINE1 mRNA binding protein 1 (SERBP1, rs1890928, P=4.91E-05, OR=0.80).
Variants from known susceptibility loci for each individual form of vasculitis were checked in our cross-phenotype meta-analysis. Although no associations at the genome-wide level of significance were observed within those genes, three out of the five genes examined harboured at least one SNP with suggestive P values (P<0.05; online supplementary table 4).
Functional annotation of KDM4C associated polymorphisms
Given that the associated KDM4C polymorphisms were located in non-coding regions, we decided to further evaluate their possible functional implications using bioinformatics approaches. First, we identified all the SNP taggers (r2>0.8) of the lead variant (rs16925200) in the European populations of the 1000 genomes project. Fourteen SNPs met this criterion, with eight of them not being included in our merged dataset. Some of these KDM4C variants showed functional annotations of interest, such as overlapping with histone marks and DNAse hypersensitivity peaks enriched at enhancers as well as possible influence to protein binding and regulatory motifs (table 2). In addition, seven variants have been described as cis-expression quantitative trait loci (eQTLs) regulating KDM4C expression in whole blood. Specifically, rs10975993 had a score 2b in Regulome DB indicating that this SNP likely affects transcription factor binding, in particular that of small MAF proteins like MAF bZIP transcription factor K. These proteins are induced by the TGF-β pathway and seem to influence the metabolism of antioxidant and xenobiotic metabolism, being implicated in the susceptibility to various type of cancer.28 However, none of these marks were reported in cell types or tissues related to vasculitis pathology.
Table 2.
Functional annotations of the KDM4C polymorphism rs16925200 and its proxies in the European populations of the 1000 genomes project using ENCODE data and Blood NESDA NTR Conditional eQTL Catalog
| SNP | Location (GRCh37) | Change | LD (r2) | RDM score | Regulatory chromatin marks | Protein binding and regulatory motifs | Cis eQTL | |||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| Promoter histone marks | Enhancers histone marks | DNAse hypersensitivity | Proteins bound (ChiP-seq) | Regulatory motifs altered (PWM) | ||||||
| rs12343988 | 9:7026828 | T<C | 0.80 | 5 | No | No | Yes | PPAR | No | |
|
| ||||||||||
| rs12342947 | 9:7026860 | A<G | 0.80 | 5 | No | No | Yes | NRSF | No | |
|
| ||||||||||
| rs10975993 | 9:7028287 | C<G | 0.90 | 2b | No | No | Yes | MAFK | Foxo, Nr2f2, Nrf-2 | No |
|
| ||||||||||
| rs77383525* | 9:7034271 | C<T | 0.89 | ND | No | Yes | Yes | Yes | ||
|
| ||||||||||
| rs1575452* | 9:7035554 | A<G | 0.95 | 6 | No | No | No | Homez, Nanog known1, Pouf3f2, STAT known8, STAT known9 | No | |
|
| ||||||||||
| rs74827126* | 9:7037180 | G<A | 0.94 | ND | No | Yes | No | Nanog | No | |
|
| ||||||||||
| rs78113013* | 9:7037222 | G<A | 0.94 | 6 | No | Yes | No | RREB-1 | No | |
|
| ||||||||||
| rs12686038* | 9:703739 | A<G | 0.94 | 6 | No | Yes | No | Foxc1 1, foxd3, HDAC2 disc6, NF-AT, PU.1 known2 | No | |
|
| ||||||||||
| rs35124449* | 9:7044009 | GT<G | 1.00 | 6 | No | No | Yes | Barhl1, Hmx 2, Msx2, Nkx2 10, Prrx2 1, Spz1 2 | No | |
|
| ||||||||||
| rs16925200 | 9:7045058 | T<C | – | ND | No | Yes | No | Pax-4 | Yes | |
|
| ||||||||||
| rs16925215* | 9:7047439 | T<A | 1.00 | ND | No | No | No | Yes | ||
|
| ||||||||||
| rs16925222* | 9:7048332 | G<C | 0.90 | ND | No | No | No | DMRT5 | Yes | |
|
| ||||||||||
| rs35821841 | 9:7050249 | G<T | 0.84 | 5 | No | Yes | Yes | AP-1,ATF3 | Yes | |
|
| ||||||||||
| rs77198751 | 9:7057185 | C<G | 0.84 | ND | No | Yes | No | No | ||
|
| ||||||||||
| rs12003023 | 9:7058714 | A<G | 0.84 | 5 | No | Yes | Yes | Myf,RP58 | No | |
Asterisks indicate polymorphisms not included in the combined dataset.
The most strongly associated SNP in the meta-analysis is marked in bold.
eQTL, expression quantitative trait locus; GRCh37, Genome Reference Consortium Human genome build 37; LD, linkage disequilibrium; RDM score, Regulome Data Base Score; SNP, single nucleotide polymorphism.
Discussion
The results of this study suggest the existence of a common genetic component that may influence the susceptibility to develop different forms of vasculitis. Particularly, besides the widely known association with the HLA region, we have identified KDM4C, also known as JHDM3C or JMJD2C, as a new shared locus between GCA, TAK, AAV and IgAV, which could be also shared with BD. To our knowledge, this is the first time that this gene has been implicated in the genetic predisposition to any immune-mediated disease.
KDM4C is a member of the Jumonji domain 2 (JMJD2) family. It is located on chromosome 9p24.1 and encodes a trimethylation-specific demethylase that controls the methylation state of histone residues, affecting gene expression and chromatin structure. 29 The main function of KDM4C is the activation of genes by removing methyl groups from the repressive histone mark trimethylated lysine 9 of the N-terminal tail of H3 (H3K9me3). Consistent with this, KDM4C has been related to cell growth in different types of cancer, and it has been reported to influence concentrations of both circulating growth factors, such as stem cell growth factor beta, and cytokines like cutaneous T-cell attracting chemokine (CCL27).30,31 Moreover, a recent GWAS performed in an Ameridian ancestry population evidenced a suggestive association of a KDM4C variant with systemic lupus erythematosus.32
Interestingly, it has been described that the H3K9me3 mark was significantly enriched in the promoters of both systemic lupus erythematosus and rheumatoid arthritis (RA)-associated genes from GWAS in the Gm12878 EBV-transformed B lymphoblastoid cell line, suggesting that epigenetics may have a relevant role in autoimmunity.33 Indeed, H3K9me3 and other histone marks (including H3K4Me3, H3K4Me2 and H3R17Me2) seem to be introduced in a transcription-coupled manner during the IFN-γ-induced activation of HLA class II genes, and it has been proposed that they may be implicated in the transcription elongation and the establishment of transcriptional memory of the HLA-DRA gene.34
On the other hand, it has been described that intronic polymorphisms located near the 3′ end of KDM4C affect the expression of this gene and, consequently, the expression levels of its target genes. Hence, genetic variation within KDM4C may have an impact on the epigenetic regulation of other genes, which evidences the close link that exists between genetic and epigenetic mechanisms.30
Genetic–epigenetic relationships like the one described above could explain many genetic associations of non-coding variants with complex diseases for which no functional implication has been established yet. In this sense, the role of epigenetics in the pathogenesis of immune-mediated diseases seems now undeniable. 35 Regarding vasculitides, insights into the high relevance of the epigenetic mechanisms for disease susceptibility have been published during the last years.36 A recent study, in which chromatin signature patterns within genetic susceptibility regions in four types of systemic vasculitis (including TAK, AAV, BD and Kawasaki disease) were analysed, identified potential specific immune cells involved in vasculitis pathogenesis through epigenetic alterations caused by genetic risk variants. Interestingly, the epigenetic changes mediating disease risk included different methylation states in different immune cells of the histone H3 such as H3K27me3 and H3K4me1 (associated with AAV and TAK, respectively).37 Additionally, it has been described that AAV patients show an increased expression of the JMJD3 protein (a histone demethylase similar to KDM4C), which lead to a decreased histone methylation within the AAV-associated genes myeloperoxidase and proteinase 3, thus resulting in a transcriptionally permissive chromatin structure at these loci.38 Furthermore, a decreased expression of histone demethylases in peripheral blood mononuclear cells, including KDM3A, was observed in IgAV patients.39 However, depletion of this gene in heterozygous mouse models does not seem to cause phenotypes that may resemble the clinical features of the vasculitides included in our study (MGI:1924054). Further studies aimed at improving the understanding of the association of KDM4C with the predisposition to vasculitis development may definitively shed light into our understanding of the molecular basis underlying these complex pathologies.
On the other hand, our analyses confirm the pivotal role of the HLA region in the genetic susceptibility to systemic vasculitis. 5 However, the fact that an HLA polymorphism represented the strongest association signal in our meta-analysis is intriguing, as specific HLA associations use to define each disease entity.5 Our results suggest the existence of a shared genetic marker for vasculitides within the HLA in an intergenic region between HLA-DQB1 and HLA-DQA2. The most significant SNP within this region, rs6932517, is in moderate LD (D’>0.8) with the different peak variants of the individual archetypal HLA class II vasculitides analysed here (GCA, AAV and IgAV; online supplementary table 2).13,25,26 Although TAK is mainly linked to HLA class I genes, previous studies suggest that some less relevant class II SNPs between HLA-DQB1 and HLA-DRB1, like rs113452171, may be also involved in its predisposition.9,14 This SNP has also a considerable LD relationship with rs6932517 (D’=1). Therefore, it is likely that the apparently shared association signal that we observed within the HLA region corresponded to a tag SNP that marks the different disease-specific associations. Despite this, rs6932517 could have a potential value as a possible diagnostic marker of vasculitides. In any case, due to the broad LD structure throughout the HLA region, more comprehensive studies interrogating this question will be necessary to establish the attribution of common susceptibility locus for vasculitides to a specific causal variant within the HLA system.
Finally, other interesting genes that have been previously related with immune-mediated diseases were suggested as shared risk factors for vasculitides in our study (table 1). Among them, it is worth mentioning the genes zinc finger, MIZ-type containing 1 (ZIMZ1), which has been described as a shared risk gene for Crohn’s disease and psoriasis,40 and protein kinase C theta (PRKCQ), a relevant locus for RA susceptibility.41 The latter, which encodes a calcium-independent and phospholipid-dependent protein kinase important for T-cell activation, has been described as a suggestive genetic marker for GCA development in a previous study.13,42 Future replication studies will be necessary to validate these suggested associations with vasculitides. In summary, we have identified KDM4C as a common risk locus for vasculitides, highlighting the relevance of the epigenetic mechanisms in the development of these complex diseases. In addition, our data suggest that genetic variation between HLA-DQB1 and HLA-DQA2 could have a high value in the understanding of the aetiological mechanisms underlying systemic vasculitides, and support the notion that the HLA class II region is also relevant in TAK.
Supplementary Material
Acknowledgments
We thank Sofía Vargas, Sonia García and Gema Robledo for their excellent technical assistance, and all the patients and healthy controls for kindly accepting their essential collaboration.
Funding This work was supported by the following grants: P12-BIO-1395 from Consejería de Innovación, Ciencia y Tecnología, Junta de Andalucía (Spain) and the Cooperative Research Thematic Network (RETICS) programme, RD16/0012/0004 (RIER), from Instituto de Salud Carlos III (ISCIII, Health Ministry, Madrid, Spain), and the National Institute Of Arthritis And Musculoskeletal And Skin Diseases of the National Institutes of Health under Award Number R01AR070148. FDC was recipient of a grant from the ’Ramón y Cajal’ programme of the Spanish Ministry of Economy and Competitiveness (RYC-2014–16458). RLM is supported by the Miguel Servet I programme of the Spanish Ministry of Economy and Competitiveness through the grant CP16/00033. The UKGCA Consortium was funded by Research into Ageing and The Wellcome Trust and is currently supported by the National Institute for Health Research (NIHR) Leeds Biomedical Research Centre, NIHR Leeds Diagnostic Evidence Co-operative, Medical Research Council and the Ann Wilks Memorial Fund. This article presents independent research funded in part by the NIHR
Footnotes
Disclaimer The views expressed are those of the authors and not necessarily those of the UK National Health Service, the NIHR or the Department of Health.
Competing interests None declared.
Patient consent Obtained.
Ethics approval Comité de Bioética del Consejo Superior de Investigaciones Científicas and the local ethical committees of the different participating centres.
Provenance and peer review Not commissioned; externally peer reviewed. © Article author(s) (or their employer(s) unless otherwise stated in the text of the article) 2018. All rights reserved. No commercial use is permitted unless otherwise expressly granted.
Collaborators The Collaborators details are included in online supplementary material “Supplementary_Note_R1”.
Contributors LO-F, FDC and JM were involved in the conception and design of the study as well as to the interpretation of data. LO-F performed the statistical analyses and drafted the manuscript. FDC and JM critically revised the manuscript for important intellectual content. RLM, PL, AWM, AS, KS, MAG-G and the consortium members were involved in the acquisition of data. All authors revised and approved the final version of the manuscript.
References
- 1.Katsuyama T, Sada KE, Makino H. Current concept and epidemiology of systemic vasculitides. Allergol Int. 2014;63:505–13. doi: 10.2332/allergolint.14-RAI-0778. [DOI] [PubMed] [Google Scholar]
- 2.Jennette JC, Falk RJ, Bacon PA, et al. 2012 revised International chapel hill consensus conference nomenclature of vasculitides. Arthritis Rheum. 2013;65:1–11. doi: 10.1002/art.37715. [DOI] [PubMed] [Google Scholar]
- 3.Weyand CM, Goronzy JJ. Medium- and large-vessel vasculitis. N Engl J Med. 2003;349:160–9. doi: 10.1056/NEJMra022694. [DOI] [PubMed] [Google Scholar]
- 4.Anwar S, Karim MY. Update on systemic vasculitides. J Clin Pathol. 2017;70:476–82. doi: 10.1136/jclinpath-2016-203875. [DOI] [PubMed] [Google Scholar]
- 5.Carmona FD, Martín J, González-Gay MA. Genetics of vasculitis. Curr Opin Rheumatol. 2015;27:10–17. doi: 10.1097/BOR.0000000000000124. [DOI] [PubMed] [Google Scholar]
- 6.Eyre S, Orozco G, Worthington J. The genetics revolution in rheumatology: large scale genomic arrays and genetic mapping. Nat Rev Rheumatol. 2017;13:421–32. doi: 10.1038/nrrheum.2017.80. [DOI] [PubMed] [Google Scholar]
- 7.Ellinghaus D, Jostins L, Spain SL, et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet. 2016;48:510–8. doi: 10.1038/ng.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Márquez A, Vidal-Bralo L, Rodríguez-Rodríguez L, et al. A combined large-scale meta-analysis identifies COG6 as a novel shared risk locus for rheumatoid arthritis and systemic lupus erythematosus. Ann Rheum Dis. 2017;76:286–94. doi: 10.1136/annrheumdis-2016-209436. [DOI] [PubMed] [Google Scholar]
- 9.Carmona FD, Coit P, Saruhan-Direskeneli G, et al. Analysis of the common genetic component of large-vessel vasculitides through a meta-Immunochip strategy. Sci Rep. 2017;7:43953. doi: 10.1038/srep43953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.López-Isac E, Martín JE, Assassi S, et al. Brief Report: IRF4 newly identified as a common susceptibility locus for systemic sclerosis and rheumatoid arthritis in a cross-disease meta-analysis of genome-wide association studies. Arthritis Rheumatol. 2016;68:2338–44. doi: 10.1002/art.39730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parkes M, Cortes A, van Heel DA, et al. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet. 2013;14:661–73. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 13.Carmona FD, Mackie SL, Martín JE, et al. A large-scale genetic analysis reveals a strong contribution of the HLA class II region to giant cell arteritis susceptibility. Am J Hum Genet. 2015;96:565–80. doi: 10.1016/j.ajhg.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saruhan-Direskeneli G, Hughes T, Aksu K, et al. Identification of multiple genetic susceptibility loci in takayasu arteritis. Am J Hum Genet. 2013;93:298–305. doi: 10.1016/j.ajhg.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Auton A, Brooks LD, Durbin RM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia X, Han B, Onengut-Gumuscu S, et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One. 2013;8:e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raychaudhuri S, Sandor C, Stahl EA, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012;44:291–6. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li MX, Yeung JM, Cherny SS, et al. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum Genet. 2012;131:747–56. doi: 10.1007/s00439-011-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–7. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44:877–881. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jansen R, Hottenga JJ, Nivard MG, et al. Conditional eQTL analysis reveals allelic heterogeneity of gene expression. Hum Mol Genet. 2017;26:1444–51. doi: 10.1093/hmg/ddx043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyons PA, Rayner TF, Trivedi S, et al. Genetically distinct subsets within ANCAassociated vasculitis. N Engl J Med. 2012;367:214–23. doi: 10.1056/NEJMoa1108735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.López-Mejías R, Carmona FD, Castañeda S, et al. A genome-wide association study suggests the HLA Class II region as the major susceptibility locus for IgA vasculitis. Sci Rep. 2017;7:5088. doi: 10.1038/s41598-017-03915-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ortiz-Fernández L, Carmona FD, Montes-Cano MA, et al. Genetic analysis with the immunochip platform in behçet disease. identification of residues associated in the HLA class i region and new susceptibility loci. PLoS One. 2016;11:e0161305. doi: 10.1371/journal.pone.0161305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katsuoka F, Yamamoto M. proteins SM. MafF, MafG, MafK): History, structure and function. Gene. 2016;586:197–205. doi: 10.1016/j.gene.2016.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berry WL, Janknecht R. KDM4/JMJD2 histone demethylases: epigenetic regulators in cancer cells. Cancer Res. 2013;73:2936–42. doi: 10.1158/0008-5472.CAN-12-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gregory BL, Cheung VG. Natural variation in the histone demethylase, KDM4C, influences expression levels of specific genes including those that affect cell growth. Genome Res. 2014;24:52–63. doi: 10.1101/gr.156141.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahola-Olli AV, Würtz P, Havulinna AS, et al. Genome-wide association study identifies 27 loci influencing concentrations of circulating cytokines and growth factors. Am J Hum Genet. 2017;100:40–50. doi: 10.1016/j.ajhg.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alarcón-Riquelme ME, Ziegler JT, Molineros J, et al. Genome-wide association study in an amerindian ancestry population reveals novel systemic lupus erythematosus risk loci and the role of European admixture. Arthritis Rheumatol. 2016;68:932–43. doi: 10.1002/art.39504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dozmorov MG, Wren JD, Alarcón-Riquelme ME. Epigenomic elements enriched in the promoters of autoimmunity susceptibility genes. Epigenetics. 2014;9:276–85. doi: 10.4161/epi.27021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rybtsova N, Leimgruber E, Seguin-Estévez Q, et al. Transcription-coupled deposition of histone modifications during MHC class II gene activation. Nucleic Acids Res. 2007;35:3431–41. doi: 10.1093/nar/gkm214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farh KK, Marson A, Zhu J, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–43. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Renauer P, Coit P, Sawalha AH. Epigenetics and vasculitis: a comprehensive review. Clin Rev Allergy Immunol. 2016;50:357–66. doi: 10.1007/s12016-015-8495-6. [DOI] [PubMed] [Google Scholar]
- 37.Sawalha AH, Dozmorov MG. Epigenomic functional characterization of genetic susceptibility variants in systemic vasculitis. J Autoimmun. 2016;67:76–81. doi: 10.1016/j.jaut.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 38.Ciavatta DJ, Yang J, Preston GA, et al. Epigenetic basis for aberrant upregulation of autoantigen genes in humans with ANCA vasculitis. J Clin Invest. 2010;120:3209–19. doi: 10.1172/JCI40034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo S, Liang G, Zhang P, et al. Aberrant histone modifications in peripheral blood mononuclear cells from patients with henoch-schönlein purpura. Clin Immunol. 2013;146:165–75. doi: 10.1016/j.clim.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 40.Ellinghaus D, Ellinghaus E, Nair RP, et al. Combined analysis of genome-wide association studies for Crohn disease and psoriasis identifies seven shared susceptibility loci. Am J Hum Genet. 2012;90:636–47. doi: 10.1016/j.ajhg.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okada Y, Wu D, Trynka G, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–81. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayashi K, Altman A. Protein kinase C theta (PKCtheta): a key player in T cell life and death. Pharmacol Res. 2007;55:537–44. doi: 10.1016/j.phrs.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.