ABSTRACT
Genomic analysis reveals extensive sequence variation and hot spots of recombination in surface proteins of Streptococcus pneumoniae. While this phenomenon is commonly attributed to diversifying selection by host immune responses, there is little mechanistic evidence for the hypothesis that diversification of surface protein antigens produces an immune escape benefit during infection with S. pneumoniae. Here, we investigate the biological significance of sequence variation within the S. pneumoniae cell wall-associated pneumococcal surface protein C (PspC) protein antigen. Using pspC allelic diversity observed in a large pneumococcal collection, we produced variant-specific protein constructs that span the sequence variability within the pspC locus. We show that antibodies raised against these PspC constructs are variant specific and prevent association between PspC and the complement pathway mediator, human factor H. We found that PspC variants differ in their capacity to bind factor H, suggesting that sequence variation within pspC reflects differences in biological function. Finally, in an antibody-dependent opsonophagocytic assay, S. pneumoniae expressing a PspC variant matching the antibody specificity was killed efficiently. In contrast, killing efficacy was not evident against S. pneumoniae expressing mismatched PspC variants. Our data suggest that antigenic variation within the PspC antigen promotes immune evasion and could confer a fitness benefit during infection.
KEYWORDS: PspC, Streptococcus pneumoniae, antigenic variation
IMPORTANCE
Loci encoding surface protein antigens in Streptococcus pneumoniae are highly polymorphic. It has become a truism that these polymorphisms are the outcome of selective pressure on S. pneumoniae to escape host immunity. However, there is little mechanistic evidence to support the hypothesis that diversifying protein antigens produces a benefit for the bacteria. Using the highly diverse pspC locus, we have now characterized the functional and immune implications of sequence diversity within the PspC protein. We have characterized the spectrum of biological function among diverse PspC variants and show that pspC sequence diversity reflects functional differences. Further, we show that sequence variation in PspC confers an immune escape benefit in the presence of anti-PspC variant-specific immunity. Overall, the results of our studies provide insights into the functional implications of protein sequence diversity and the role of variant-specific immunity in its maintenance.
INTRODUCTION
The bacterium Streptococcus pneumoniae (pneumococcus) is a respiratory commensal and pathogen characterized by extensive sequence variation. Diversity within the pneumococcal capsule is well characterized and central to the design of pneumococcal vaccines (1). Extensive research on capsular diversity has contributed greatly to our understanding of its biological importance and the critical role of antibody-mediated immunity in its maintenance (2, 3). Moreover, genetic diversity in S. pneumoniae extends beyond the capsule locus. Loci encoding surface protein antigens are also highly variable and subject to frequent recombination events (4, 5). Despite a growing body of evidence pointing to extensive sequence variation in pneumococcal surface protein antigens, we know little about the immune and functional implications of this diversity.
Pneumococcal surface protein C (PspC; also known as CbpA, Hic, and SpsA) is a cell wall-associated surface protein known to interact with three different human proteins via its α-helical domain (6). PspC binds the secretory component of immunoglobulin A (sIgA), complement component C3, and complement factor H (7–9). Mutants of S. pneumoniae that do not express PspC showed reduced virulence in mouse models of bacteremia, pneumonia, and nasal colonization (10–12). While the functional mechanisms by which PspC contributes to virulence are incompletely characterized, interactions between PspC and host proteins seem to be critical in this context. It has been proposed that the PspC-sIgA interaction facilitates adherence and subsequent invasion via the polymeric Ig receptor on mucosal epithelial cells (13, 14). Additionally, the association between PspC and the fH and C3 complement mediators seems to play an important role at various stages of the in vivo biology of the pneumococcus (15–22).
PspC is highly immunogenic, and anti-PspC antibodies are a major component of antibody immunity against S. pneumoniae (23–29). Several papers have demonstrated the protective capacity of anti-PspC immunity against carriage (10) or invasive challenge (6, 30) in mice.
Although the pspC gene is present in almost all pneumococci, it is highly polymorphic, and pneumococcal strains differ greatly in the particular antigenic variants that they express (6, 31). Like other surface antigens, PspC is commonly assumed to be under the control of diversifying selection to escape antibody action. Two lines of evidence support this reasoning. First, there is evidence for a concentration of nonsynonymous substitutions within the epitope regions of pneumococcal protein antigens (32). Second, there is age-specific variation in gene content, such that common variants of some core surface proteins, as well as surface proteins that are part of the accessory genome, are more common among isolates from younger (presumably immunologically naive) carriers of S. pneumoniae (5, 33). Relatedly, we recently reported that individuals with strong antibody responses to a particular variant of one protein antigen, pneumococcal surface protein C (PspC), are more likely, if carrying a pneumococcus, to carry one expressing a different PspC variant (23). Such results support the conjecture that common variants are net-beneficial in younger/more naive hosts but are subject to immune responses that select against their presence in more immunologically experienced people.
Despite the sequence evidence suggesting diversifying selection, mechanistic studies designed to assess the degree of allelic specificity of anti-PspC antibody responses have had heterogeneous results. While such variation probably partly reflects real variability in the extent of activity by antibodies raised to one allele against other alleles, it may also reflect variability due to strain background (21, 34) as well as cross-reactions with the paralogous PspA protein (6). A study of antibodies to three recombinant PspC immunogens showed a range in the extent of specificity, with antibodies to a PspC of group 8 binding by Western blotting and flow cytometry only to a strain containing that allele, anti-PspC3 antibodies showing binding to multiple PspC alleles (but narrower specificity in functional activities such as blocking of PspC interactions with host proteins), and anti-PspC4 antibodies showing binding also to PspA (35). In another study, opsonic and opsonophagocytic killing activity of anti-recombinant PspC antibodies was limited to the isolate from which the recombinant pspC gene was obtained and was not observed with two other strains (36).
Given these differing results and the potential for the strain background to affect PspC activity and the extent of antibody action against PspC, we sought to study PspC diversity using isogenic strains that differ only at the pspC locus. Within a data set of 616 asymptomatically carried S. pneumoniae isolates, we identified four PspC variants that span the sequence diversity within the pspC locus (the sequences are included in Text S1) (5). Here, we show that sequence variability within PspC, in an otherwise fixed genetic background, can create differences in the ability of strains to bind human factor H and can promote immune escape in opsonophagocytic assays in the presence of variant-specific immunity. Our results provide insight into the biological implications of PspC sequence diversity and the role of variant-specific antibodies in antipneumococcus immunity.
Sequences of full-length PspC variants and truncated protein fragments (in gray). Download TEXT S1, PDF file, 0.1 MB (67.2KB, pdf) .
Copyright © 2018 Georgieva et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RESULTS
Purification of recombinant PspC protein variant fragments and production of variant-specific antibodies.
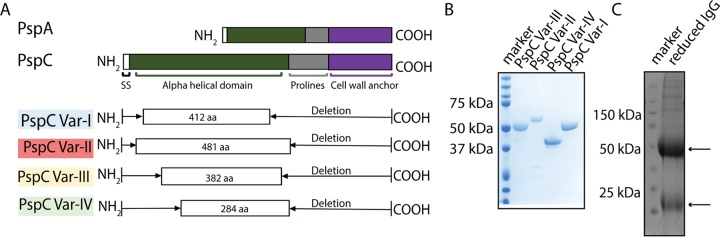
Previous analysis of whole-genome sequence data from 616 asymptomatically carried S. pneumoniae isolates showed extensive sequence variation and recombination hot spots in the S. pneumoniae PspC surface protein (5, 23). For our studies here, we selected four PspC variants that span the range of genetic diversity of the pspC locus within this pneumococcal population (5, 23, 31). We recently reported the presence of naturally acquired antibodies against these four PspC variants in individuals colonized with S. pneumoniae (23). In order to begin mechanistic studies, we first generated recombinant variant-specific PspC protein constructs (see Fig. S3 in the supplemental material). In order to capture the unique sequence characteristics of each variant, we designed truncated PspC fragments which exclude conserved epitopes common among the variants as well as any epitopes shared with PspA, another highly immunogenic pneumococcal surface protein with homology to PspC (Fig. 1A). We then cloned each of these constructs into expression vectors, thereby generating PspC expression constructs bearing a C-terminal polyhistidine tag. Each PspC variant fragment was expressed in Escherichia coli and isolated from the protein-soluble fraction using a nickel-based on-column purification strategy. The protein-rich elution fraction was further purified on a size exclusion column, and the purity of proteins was confirmed by SDS-PAGE (Fig. 1B). For production of anti-PspC variant-fragment-specific antibodies, we immunized individual rabbits with each of the recombinant PspC variant fragments. To isolate serum immunoglobulin G (IgG) with affinity for the PspC protein while also minimizing the presence of nonspecific IgG, we first preabsorbed serum samples with lysates of ΔPspC S. pneumoniae and then purified the total IgG fraction as described in Materials and Methods (Fig. 1C).
FIG 1 .
Purification of recombinant PspC protein variants and production of variant-specific antibodies. (A) Four variant-specific recombinant protein constructs were designed to capture the sequence variability within the PspC protein. Annotated are the N-terminal signal sequence (SS), the alpha helical domain, the proline-rich domain, and the choline-rich cell wall anchor region within PspC. The truncated PspC protein constructs excluded epitopes conserved among the four variants as well as regions homologous with PspA. aa, amino acids. (B) The purity of the produced proteins was checked by SDS-PAGE analysis. (C) SDS-PAGE of IgG isolated from variant-specific rabbit antisera. The arrows indicate the 50-kDa and 25-kDa molecular mass (MM) bands seen under reducing conditions, corresponding to the IgG heavy and light chains, respectively.
Anti-PspC antibodies are specific for their cognate PspC variant.
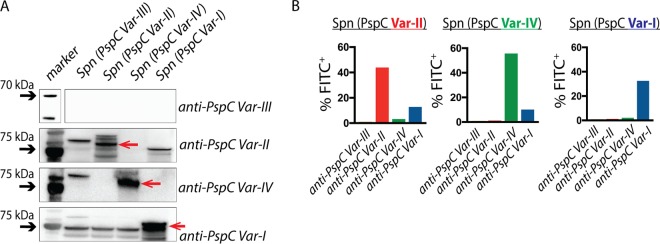
To determine the specificity of IgG for each PspC variant, we performed Western blotting of whole-cell lysates of S. pneumoniae strains expressing one of the four PspC variants (the full protein and not the fragment used for immunization). As shown in Fig. 2A, anti-PspC variant I (var.-I), anti-PspC var.-II, and anti-PspC var.-IV recognized with specificity the cognate PspC variant. In contrast, anti-PspC var.-III antibody did not recognize any PspC variant, suggesting that PspC var.-III recombinant protein might have been poorly immunogenic in rabbits or that anti-PspC var.-III IgG may have low affinity for its target. To determine whether these anti-PspC antibodies bind their cognate targets in the context of whole S. pneumoniae bacteria, we performed flow cytometry analysis of anti-PspC IgG binding to isogenic, whole S. pneumoniae bacteria that differed solely in the PspC variant that they express. Using a fluorescein isothiocyanate-positive (FITC+) conjugated anti-rabbit secondary antibody, we measured the percentages of bacterial cells with surface-deposited anti-PspC IgG (Fig. 2B). Consistent with the Western blot data, anti-PspC var.-I, anti-PspC var.-II, and anti-PspC var.-IV antibodies were specific for their cognate PspC variant in intact bacteria. Graphs show the percentages of the total bacterial population that were FITC+ for bound IgG. The total proportion of FITC+ cells was below 100% because some proportion of the cells remained unstained.
FIG 2 .
Anti-PspC antibodies are variant specific. (A) Western blot analysis of reactivity of anti-PspC var.-I, anti-PspC var.-II, anti-PspC var.-III, and anti-PspC var.-IV serum samples to whole-cell lysates of pneumococcal strains expressing different full-length PspC variants (shown at the top of the panel). Each variant-specific protein band is marked with an arrow. (B) Flow cytometry analysis for binding of variant-specific IgG to isogenic S. pneumoniae strains expressing a unique PspC variant. Using FITC+ conjugated anti-rabbit IgG secondary antibody, we evaluated binding of anti-PspC IgG to whole S. pneumoniae strains. The total percentage of FITC+ cells is shown for each S. pneumoniae strain incubated separately with anti-PspC var.-I, anti-PspC var.-II, anti-PspC var.-III, or anti-PspC var.-IV.
Anti-PspC antibodies prevent interaction between PspC and fH but not between PspC and sIgA.
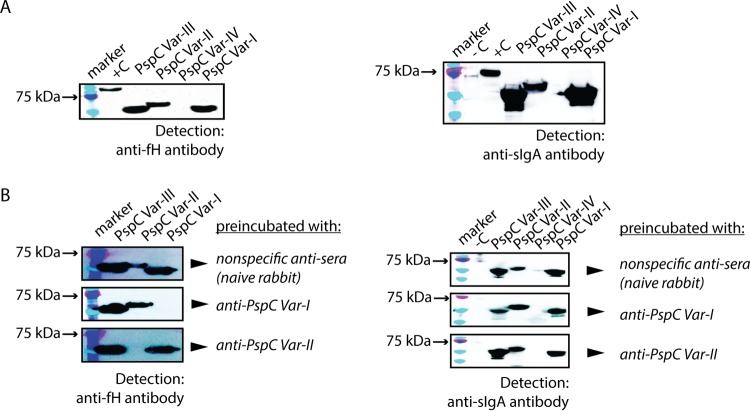
Several previous studies showed the interactions between PspC and the complement pathway mediator fH and secretory IgA (7, 8, 14, 15, 17, 19, 20, 22, 35, 37–43). Because those studies used laboratory strains of S. pneumoniae and clinical isolates bearing PspC variants different from those included here, we wanted to investigate whether each of the four PspC variants in our study interacted similarly with fH and sIgA. To look at PspC interaction with fH and sIgA, we performed Far-Western blotting. Recombinant PspC variant fragments were separated by SDS-PAGE, transferred onto a nitrocellulose membrane, and then incubated with recombinant fH or sIgA. Detection performed with fH-specific or sIgA-specific antibodies showed evidence for interactions among PspC var.-I, var.-II, var.-III, and fH/sIgA (Fig. 3A). In contrast, truncated PspC var.-IV did not bind fH or sIgA. Overall, our data on the interaction of PspC var.-I, var.-II, and var.-III with fH or sIgA are consistent with those from other studies. Our findings also suggest that, despite extensive sequence variability within the pspC locus, these biological interactions are likely important in vivo and remain preserved across a wide range of antigenic variants.
FIG 3 .
Anti-PspC IgG prevents association between PspC and the complement pathway mediator fH but not secretory IgA. (A) Western blot analysis of fH and sIgA binding to recombinant PspC proteins (truncated versions). Recombinant PspC proteins were separated by the use of 10% SDS-PAGE and transferred onto a nitrocellulose membrane, and the membrane was then incubated with recombinant human factor H protein prior to addition of anti-factor H antibody to detect fH binding. For sIgA binding, recombinant sIgA was added followed by anti-sIgA antibody. The positive control for fH detection included recombinant fH protein (+C lane). The positive control for sIgA detection included recombinant sIgA (+C lane). (B) Recombinant PspC proteins were first incubated with their respective variant-specific anti-PspC antibody followed by incubation with recombinant human factor H or sIgA. Detection was performed using anti-fH or anti-sIgA antibody. Antibody isolated from nonimmunized rabbit serum was used as a negative control. Pneumolysin pneumococcal protein (53 kDa) was included as a negative control (-C lane).
Previous studies on PspC and on fH and sIgA have also shown the role of these interactions in promoting immune evasion and epithelial adhesion during infection with S. pneumoniae (7, 14, 17, 19, 40, 44). Furthermore, a few studies have demonstrated that the fH/sIgA binding regions on PspC are targets for anti-PspC antibodies during infection and that these anti-PspC antibodies have the capacity to impede the interaction between PspC and fH/sIgA (35, 36). However, to our knowledge, in one of these studies it remained unclear whether these antibodies function in a variant-specific manner to block these interactions (36). In another study, antibodies raised against different PspC variants showed a range of specificities against pneumococcal protein lysates (35). These results showing various specificities are likely explained by the utilization of full-length PspC proteins to generate antibodies. In our study, truncated protein PspC variants (representing the unique portion of the PspC sequence) were used to generate anti-PspC IgG, allowing us to ask questions about antibody specificity while reducing the extent of cross-reactivity between PspC variants and between PspC and PspA. We separated our recombinant PspC variants by SDS-PAGE, transferred the protein sample onto a nitrocellulose membrane, and then incubated with anti-PspC antibody prior to addition of recombinant fH or sIgA. We found that anti-PspC var.-I and var.-II antibodies prevented association between recombinant PspC and fH in a variant-specific fashion (Fig. 3B). In contrast, association between PspC and sIgA remained unaffected in the presence of these antibodies, consistent with previous findings showing that fH and sIgA have distinct binding domains within PspC (38). Note that we found differences in the specificities of IgGs isolated from different rabbit antisera for PspC var.-II. A set of anti-PspC var.-II IgG isolates collected from a second rabbit was cross-reactive with PspC var.-III (Fig. S2). However, we obtained consistent results for var.-I by the use of antisera from two different rabbits.
PspC variants differ in their capacity to bind human fH.
To investigate how sequence variation within PspC may impact biological function, we compared the different capacities of the PspC protein variants to bind fH. We generated TIGR4 isogenic strains expressing a unique full-length PspC variant. We incubated each S. pneumoniae strain with IgG-depleted normal human serum as a source of factor H and measured the deposition of fH on the bacterial surface by flow cytometry as described in Materials and Methods. TIGR4 expressing var.-III bound significantly more fH (Fig. 4); in contrast, TIGR4 expressing var.-I, var.-II, or var.-IV PspC bound significantly lower levels of fH. Under these conditions, the original S. pneumoniae PspC var.-III strain similarly bound more fH than the S. pneumoniae strains bearing var.-II or var.-I. Overall, these data indicate that sequence variation within PspC encodes functional differences between antigenic variants.
FIG 4 .
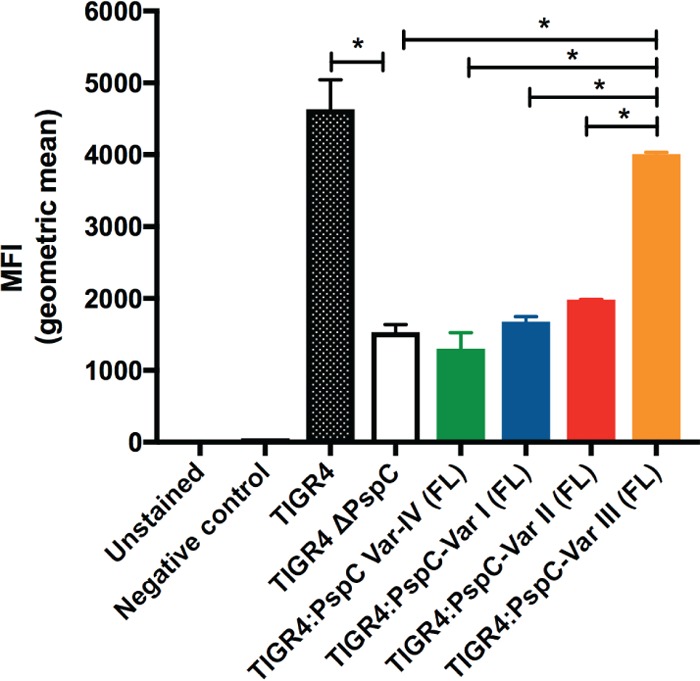
PspC variants differ in their capacity to bind fH. Flow cytometry analysis of fH binding to S. pneumoniae strains incubated with normal human serum as a source of fH was performed. Pneumococci with bound fH had increased levels of mean fluorescence intensity (MFI) compared to background fluorescence (negative control). We tested TIGR4 isogenic strains that express a unique full-length PspC variant. FL, full-length. *, P < 0.01.
Sequence diversity within pspC promotes immune escape in the presence of variant-specific immunity.
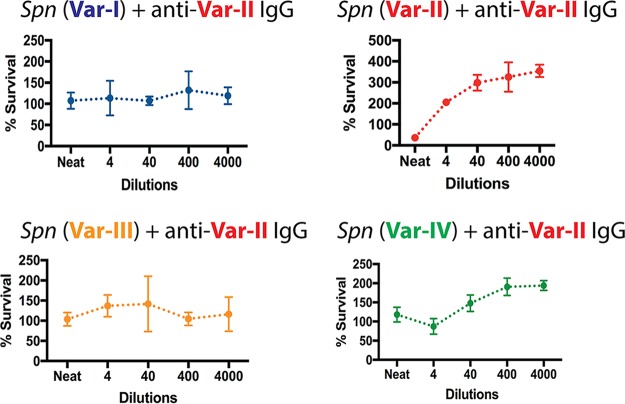
To measure the impact of variant-specific immunity against PspC on bacterial survival, we performed antibody-dependent opsonophagocytic killing assays. We found that S. pneumoniae expressing PspC var.-II was killed efficiently in the presence of anti-PspC var.-II antibody. In contrast, killing efficacy was not evident against S. pneumoniae expressing a mismatched PspC variant (Fig. 5). Within our set of anti-PspC antibodies and under the conditions tested here, only anti-PspC var.-II was opsonic whereas anti-PspC var.-I and anti-PspC var.-IV were nonopsonic (Fig. S1). While anti-PspC var.-I IgG and anti-PspC var.-IV IgG are specific for their target as shown by Western blotting and flow cytometry (Fig. 2), they seem nonfunctional in opsonophagocytic assays.
FIG 5 .
Antibody-dependent opsonophagocytic killing in the presence of anti-PspC IgG and neutrophil-like HL-60 cells. Graphs show percent survival of isogenic S. pneumoniae strains expressing one of the four PspC variants incubated with anti-PspC var.-II IgG. To determine the percentage of CFU that survived under each condition, CFU counts at the assay end time point were divided by the CFU counts at time point 0.
Anti-PspC var.-I IgG (A) and var.-IV IgG (B) are nonopsonic in a neutrophil-mediated antibody-dependent opsonophagocytic assay. Graphs show percent survival of isogenic S. pneumoniae strains incubated with anti-PspC var.-I IgG or var.-IV IgG. Download FIG S1, PDF file, 0.4 MB (374.1KB, pdf) .
Copyright © 2018 Georgieva et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Variability in anti-PspC var.-II IgG specificity depending on the source of antisera. (A) Far Western blot for fH binding in the presence of antibodies. (B) Survival data in opsonophagocytic assay using anti-PspC var.-II IgG from rabbit2 immunized with PspC var.-II. Anti-PspC var.-II IgG recognizes PspC var.-III, in addition to its cognate target, PspC var.-II. Download FIG S2, PDF file, 0.1 MB (142.5KB, pdf) .
Copyright © 2018 Georgieva et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Anti-PspC immunity is a major component of the total antiprotein antibody response during infection with S. pneumoniae. However, the impact of sequence diversity within pspC on the protein’s function or on the effectiveness of anti-PspC antibody responses has been variable in the few studies that have assessed it. In this study, we sought to address these issues and characterize the functional and immune implications of pspC diversity using antibodies to recombinant partial pspC gene products and isogenic strain sets that differ only at the pspC locus.
Despite extensive sequence variability, PspC contains several conserved regions and some regions of homology to PspA, another S. pneumoniae choline binding protein (6, 26, 31). To generate variant-specific antibodies, we produced recombinant truncated PspC protein fragments that possess only the unique sequence features that represent each PspC variant (Fig. 1; see also Text S1 and Fig. S3 in the supplemental material). In doing this, we were able to generate variant-specific instances of anti-PspC IgG that recognize their cognate target but not other PspC variants (Fig. 2). Indeed, we hypothesize that this approach may explain the greater specificity, both in recognition and in activity, observed in our study than in previous studies.
Alignment of protein sequences corresponding to truncated PspC variants I to IV. Download FIG S3, PDF file, 0.5 MB (547.9KB, pdf) .
Copyright © 2018 Georgieva et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Biochemical data on the structural landscape of PspC-fH and PspC-sIgA interactions suggest that the fH and sIgA binding domains on PspC are surface exposed and, therefore, potential antibody targets in vivo. This motivated us to investigate whether anti-PspC antibodies recognize the fH/sIgA binding domains and whether they impede PspC-fH and PspC-sIgA interactions in a variant-specific fashion. A previous study found little evidence of allelic specificity with respect to the ability of anti-PspC antibodies to inhibit binding of these host proteins to lysates of isolates expressing homologous variants. In particular, one antiserum (to variant 3 in that study) showed limited inhibition of binding even to the homologous variant, while another (variant 5) showed inhibition of binding to the homologous and several heterologous variants (35). We found that anti-PspC var.-I antibody prevented association of PspC var.-I with fH but not with PspC var.-II or var.-III (Fig. 3A). Similarly, anti-PspC var.-II antibody prevented association of PspC var.-II with fH but not with PspC var.-I or var.-III (Fig. 3A). Together, these findings point to a variant-specific function for antibodies in blocking fH binding. Our strategy of immunizing with the most variant-specific regions of the protein (as mentioned above) may underlie the greater specificity observed here than in previous work. A different choice of variants may also contribute to this finding. Notably, we found variability in anti-PspC var.-II IgG specificity depending on the source of antisera. Anti-PspC var.-II IgG from a second rabbit immunized with PspC var.-II showed variable specificity in blocking fH binding (Fig. S2A). Similarly, in an opsonophagocytic killing assay, anti-PspC IgG from the same rabbit showed recognition of PspC var.-III in addition to its cognate target, PspC var.-II (Fig. S2B). These findings underscore the variability inherent in biological systems and highlight the importance of maintaining awareness about differences in immune responses driven by host-intrinsic factors. Interestingly, while anti-PspC antibodies prevented association between PspC and fH, they did not affect interaction between PspC and sIgA (Fig. 3B). This is consistent with other findings showing that the fH and sIgA binding domains on PspC are distinct (38).
It is important to discuss our findings in the context of other studies on anti-PspC antibodies. Data from PspC epitope mapping studies exclude the fH domain from the list of epitopes within PspC that are recognized by serum samples from healthy adult humans (45). The physical association between PspC and fH in vivo is often invoked to explain these results. It is hypothesized that because the fH binding domain is occupied, it is not seen by the naive B cells that would recognize it if it were exposed. Furthermore, it has been suggested that, due to the species-specific interaction between PspC and fH, antibodies against the fH binding domain occur only in animal models where PspC does not interact with fH (42). However, it is likely that there exists an on-off rate for PspC-fH binding such that fH is not bound to PspC at all times. Furthermore, we found evidence for interactions between PspC and rabbit fH concurrent with our finding for the presence of anti-fH domain antibodies (data not shown). At this time, it remains unclear whether this is an outcome of the particular animal model that we utilized, and dissecting the mechanistic basis for this observation is beyond the scope of the current study. Instead, we highlight the potential biological relevance of anti-fH domain antibodies. Factor H acts to dampen the alternative complement pathway, and fH recruitment to the bacterial surface is a mechanism that S. pneumoniae employs to modulate host immune activation. Therefore, antibodies with the capacity to prevent association between PspC and fH would likely have an important biological role. Our findings could inform future vaccine design and underscore the utility of antibodies with specificity against the fH binding domain.
Our data fit within the larger context of investigations on the functional implications of PspC protein diversity (43). Here, we offer additional insight into the functional range of previously uncharacterized PspC protein variants. To measure the functional consequences of sequence diversity, we set out to evaluate whether our PspC variants differ in the capacity to bind human fH (Fig. 4). Because pspC expression is controlled, it is a possibility that any differences in fH binding may be an attribute of differential gene regulation (20, 46). Additionally, it was previously demonstrated that capsule modulates the amount of fH deposited on the bacterial surface (47, 48). While these are certainly important aspects of PspC biology, our goal was to evaluate the implications of sequence diversity alone for the functional range of PspC variants, excluding any contributions of genetic background and gene regulation mechanisms. Therefore, we designed isogenic S. pneumoniae strains that differ solely in the unique full-length PspC variant that they express. Our results show that these strains bind various amounts of fH and suggest that sequence diversity within pspC reflects functional differences (Fig. 4). There was little difference in fH binding results between S. pneumoniae strains bearing PspC var.-I, var.-II, or var.-IV. Moreover, a comparison with S. pneumoniae ΔPspC suggests that, in these strains, fH binding was essentially attributable to bacterial factors other than PspC. It remains possible that in strains bearing these PspC variants, genetic background plays a crucial role in regulation of PspC gene/protein levels and thereby effectively promotes fH binding. In contrast, S. pneumoniae PspC var.-III stood out with the highest fH binding capacity. Overall, our data add to our understanding of the functional implications of sequence diversity in PspC. The range in fH binding may be a reflection of functional constraints imposed on the PspC protein. Low levels of constraint would explain how PspC accommodates such extensive sequence diversity across a wide range of protein variants. Alternatively, the range of fH binding could reflect differences in immune responses between hosts (49). This would be consistent with a model where variability in human serum fH levels introduces selection with respect to S. pneumoniae to produce PspC variants with various fH binding capacities (49, 50).
Having produced variant-specific anti-PspC antibodies, we wanted to ask questions about the benefit of sequence diversity in the context of variant-specific immunity. We performed antibody-dependent opsonophagocytic assays and found that S. pneumoniae expressing PspC var.-II matching the antibody specificity was killed efficiently. Importantly, killing efficacy was not evident against S. pneumoniae expressing a mismatched PspC variant. Our data suggest that antigenic variation within the PspC pneumococcal antigen promotes immune evasion and confers a fitness benefit in the presence of variant-specific immunity. Our observations are consistent with previously reported data showing that antibodies raised against a particular PspC variant promote neutrophil-mediated killing of a S. pneumoniae strain expressing a matched PspC variant (36). Our results extend those findings in two ways. First, we show that PspC mismatch promotes survival, while the prior report showed that it reduced phagocytic uptake but did not compare levels of killing of matched versus mismatched PspC alleles. Second, our studies were performed in otherwise-isogenic strains, thereby isolating the impact of PspC allelic variation as opposed to any other differences between the isolates employed in the earlier study. Under the conditions employed in our experiments, anti-PspC var.-I and var.-IV antibodies seemed nonfunctional in opsonophagocytic killing assays. It remains possible that these antibodies have an alternative function in vivo.
It is important to discuss our findings in the context of natural immunity, which is multifaceted and likely characterized by variant-specific as well as cross-reactive antibody responses. Dissecting the relative contributions of all of these is beyond the scope of the current study. Instead, here, we address a conceptual issue about the relevance of sequence diversity in a setting of variant-specific immunity. We highlight findings of strongly variant-specific antibody activity (here, inhibition of fH binding and opsonophagocytic activity) against a pneumococcal protein antigen. This provides evidence that sequence diversity can provide an immune escape benefit in the presence of variant-specific antibody responses. We acknowledge that, in the context of natural infection, full-length antigenic variants bearing unique as well as cross-reactive epitopes would likely generate a multifaceted antibody response marked by variable specificity. Further studies investigating how this balance between specificity and cross-reactivity is biologically meaningful are warranted and will be crucial in advancing our understanding of the interplay between sequence diversity in protein antigens and host antibody immunity.
MATERIALS AND METHODS
Cloning of recombinant PspC proteins for expression in Escherichia coli.
PspC var.-I, var.-II, var.-III, and var.-IV were amplified from genomic DNA of the corresponding S. pneumoniae parent strain using the following primers: PspC var.-I forward primer 5’-GGAATTCAGCATATGAGTGGGGATACCCCCAAG-3’ and PspC var.-I reverse primer 5’-CCTTAAGTGGCGGCCGCTGGTTTTGGAGCTGGAGCTGG-3’; PspC var.-II forward primer 5′-GGAATTCAGCATATGAAGAATAACCTCACGGTT-3′ and PspC var.-II reverse primer 5′-CCTTAAGTGGCGGCCGCCTCTGGGTTTTCCGGCTGTGG-3′; PspC var.-III forward primer 5′-GGAATTCAGCATATGAACGAGGGAACTACCCAAGCA-3′ and PspC var.-III reverse primer 5′-CCTTAAGTGGCGGCCGCTGGATTTTCCGGCTGCGGTTT-3′; and PspC var.-IV forward primer 5′-GGAATTCAGCATATGAACGAGGGAAGTACCCAAGCA-3′ and PspC var.-IV reverse primer 5′-CCTTAAGTGGCGGCCGCTTCCGGCTCTGGTTTAACCTC-3′. Each of these constructs was then separately ligated into pET-21b (EMD Chemicals) using the restriction sites EcoRI and NotI, yielding a construct carrying an in-frame polyhistidine affinity tag (6×His tag) at the C terminus.
Expression and purification of recombinant PspC proteins in E. coli.
Plasmids pET21bPspCVar-I, pET21bPspCVar-II, pET21bPspCVar-III, and pET21bPspCVar-IV were transformed into E. coli Rosetta (DE3) (Invitrogen) for protein expression. Luria-Bertani (LB) broth (1 liter) containing 100 µg/ml ampicillin was inoculated with 5 ml of overnight culture and incubated at 37°C to an optical density at 600 (OD600) of 0.6 to 1.0. The cells were cooled to room temperature for 15 to 30 min, after which 1 mM IPTG (isopropyl β-d-thiogalactopyranoside) was added and the cells were allowed to incubate overnight at 25°C. The cells were then centrifuged at 6,500 rpm for 1 h. The pellet was resuspended in buffer (300 mM NaCl, 50 mM Tris-HCl [pH 8.0], 10 mM MgCl2, protease inhibitor cocktail, 1 µg/ml DNase), sonicated, and centrifuged at 13,000 rpm for 30 min at 4°C. The supernatant was incubated with Ni2+-charged beads and then applied to a gravity column at 4°C. The cell lysate in the gravity column was washed first with buffer A (300 mM NaCl, 50 mM Tris-HCl [pH 8.0]) and then twice with buffer A plus 20 mM imidazole. The protein was eluted with 300 mM imidazole in buffer A and dialyzed overnight in 50 mM Tris-HCl (pH 8.0). The dialyzed protein was then purified by the use of a size exclusion column, concentrated, divided into aliquots, and stored at −80°C. Proteins were subjected to SDS-PAGE and visualized by staining with 0.05% Coomassie blue R-250. The concentrations of purified proteins were determined by the use of a Qubit protein assay kit.
In silico characterization of pspC sequences.
The four pspC variant sequences in this study were previously identified within a collection of 616 asymptomatically carried S. pneumoniae isolates (5). Geneious software was used to analyze each pspC sequence. To identify regions of homology between the pspC genes as well as between PspC and PspA, we performed alignments and selected regions with low within-pair sequence identity for subsequent analysis.
Production of isogenic strains.
Four PspC isogenic variants in the TIGR4 and 603 genetic background were constructed. To generate the TIGR4:PspC var.-I, TIGR4:PspC var.-II, TIGR4:PspC var.-III, and TIGR4:PspC var.-IV strains, the PspC locus in the TIGR4 and 603 strains were replaced with a SweetJanus cassette by using a previously described transformation protocol (51). The SweetJanus cassette was then replaced by the allele corresponding to the sequence of PspC var.-I, PspC var.-II, PspC var.-III, or PspC var.-IV to generate the variant strains.
Preparation of protein extracts from S. pneumoniae strains.
Each S. pneumoniae strain was grown on solid medium (Trypticase soy agar with 5% sheep blood) for ~15 h at 37°C. Then, pneumococcal cells were harvested and resuspended in 1 ml 1× phosphate-buffered saline (PBS) buffer. Cells were then centrifuged at 4,000 × g for 10 min at room temperature, and the pellet was resuspended in 360 µl of lysis buffer with the following composition: 20 mM Tris-HCl, 2 mM EDTA, 1.2% Triton-X, and 20 µg/µl lysozyme. Samples were then incubated at 37°C for 90 min, subjected to vortex mixing for 1 min, and incubated at 55°C for 1 h. Following incubation, the lysate was centrifuged at 11,000 rpm for 30 min, and the supernatant (protein fraction) was removed for quantification using a Qubit protein assay kit. These protein fractions were subsequently used for Western blotting.
Production of rabbit antisera.
To produce custom variant-specific antisera, rabbits were inoculated with 100 μg recombinant PspC var.-I, var.-II, var.-III, or var.-IV in complete Freund’s adjuvant. At days 14, 21, 49, and 77, rabbits received a boost of 50 μg recombinant PspC in incomplete Freund’s adjuvant. Production bleeds collected at day 84 were used to isolate the anti-PspC IgG component. These immunizations were performed by Cocalico Biologicals, Inc. (Stevens, PA) per established protocols.
Western blotting.
Protein samples (10 µg per lane) were prepared in 4× SDS-PAGE loading dye, boiled for 10 min, separated on NuPAGE 10% bis-Tris gels, and transferred onto nitrocellulose membranes (Thermo Scientific). Membranes were blocked in TBST (150 mM NaCl, 25 mM Tris-HCl [pH 7.0], 0.1% Tween 20) containing 5% Blotto (Santa Cruz Biotechnology) for 1 h at room temperature and probed with antisera overnight at 4°C. Rabbit polyclonal anti-PspC var.-I, anti-PspC var.-II, anti-PspC var.-III, or anti-PspC var.-IV antisera were used at a 1:1,000 dilution in 1% Blotto. Membranes were washed in TBST and incubated for 1 h at room temperature with goat anti-rabbit IgG peroxidase-conjugated secondary antibody (Rockland). Blots were developed using a WesternSure ECL chemiluminescent substrate kit (Li-Cor) and visualized using a Bio-Rad ChemiDoc MP imaging system.
For Far-Western blots, recombinant PspC protein samples (5 µg per lane) were prepared in 4× SDS-PAGE loading dye, boiled for 10 min, separated on NuPAGE 10% bis-Tris gels, and transferred onto nitrocellulose membranes (Thermo Scientific). Membranes were blocked in TBST (150 mM NaCl, 25 mM Tris-HCl [pH 7.0], 0.1% Tween 20) containing 5% Blotto (Santa Cruz Biotechnology) for 1 h at room temperature and incubated with recombinant human fH (Quidel) or human sIgA purified from colostrum (Accurate Chemical) overnight at 4°C. The membranes were then washed and probed with murine anti-human factor H (Quidel) (clone A229) or murine anti-sIgA (Sigma-Aldrich) (clone GA-1). Antibodies were used at a 1:1,000 dilution in 1% Blotto. Finally, membranes were washed in TBST and incubated for 1 h at room temperature with goat anti-mouse IgG peroxidase-conjugated secondary antibody (Rockland). Blots were developed using a WesternSure ECL chemiluminescent substrate kit (Li-Cor) and visualized using a Bio-Rad ChemiDoc MP imaging system. To test for inhibition of fH or sIgA binding, prior to incubation with recombinant fH and sIgA, membranes were incubated with rabbit antisera against PspC var.-I, anti-PspC var.-II, anti-PspC var.-III, or anti-PspC var.-IV at a 1:1,000 dilution in 1% Blotto overnight.
Flow cytometry.
Overnight S. pneumoniae (TIGR4 isogenic strains) cultures were used to inoculate liquid THY cultures. Cultures were grown to an OD620 of 0.2 and then washed twice in 1× PBS. Approximately 10 × 106 cells were collected for staining. For fH binding to whole bacteria, S. pneumoniae cells were incubated with IgG-depleted normal human serum (Quidel) for 1 h at 37°C. Following incubation, cells were washed twice in PBS containing 2% fetal bovine serum (FBS) and incubated with goat antisera against human factor H (Quidel) for 30 min at 37°C. Cells were then incubated with Alexa Fluor 488-conjugated anti-goat IgG (Thermo Fisher) (clone A11055). For binding of anti-PspC rabbit antibodies to whole bacteria, ~10 × 106 S. pneumoniae cells were incubated with purified anti-PspC IgG for 30 min at 37°C. Afterward, cells were washed twice in PBS containing 2% FBS and stained with goat anti-rabbit IgG FITC-conjugated antibody (Abcam, Inc.) (clone ab6717). Cells were incubated at 4°C for 30 min and then washed with PBS containing 2% FBS. Finally, the cells were fixed in fixation buffer (data were immediately acquired using an LSR flow cytometer [BD Biosciences]). Data were analyzed with FlowJo software (Tree Star, San Carlos, CA).
Isolation of immunoglobulin G (IgG).
To isolate IgG from rabbit anti-PspC antisera, 1 ml of rabbit antisera was mixed with 4 ml of binding buffer (0.1 M NaH2PO4 [pH 8.0]). The sample was then applied to a prewashed protein G GraviTrap column (GE Healthcare) and incubated on column for 10 min at room temperature. The column was washed three times with 10 ml binding buffer, and IgG was eluted with 4 ml 0.1 M glycine (pH 2.5). To neutralize, 200 µl Tris (pH 8.5) was added per milliliter of eluate.
IgG depletion.
To deplete IgG from normal human serum, 1 ml of rabbit antisera was mixed with 4 ml of binding buffer (0.1 M NaH2PO4 [pH 8.0]). The sample was then applied to a prewashed protein G GraviTrap column (GE Healthcare) and incubated on the column for 10 min at room temperature. Column flowthrough containing the IgG-depleted fraction was collected.
Opsonophagocytic assay with differentiated HL-60 cells.
The HL-60 tissue culture human cell line (promyelocytic leukemia cells [American Type Culture Collection; catalog no. CCL-240]) was used as a source of effector cells. HL-60 cells were maintained in Iscove’s modified Dulbecco’s medium (IMDM) (Corning; catalog no. 10-016-CV) containing 2 mM l-glutamine and 25 mM HEPES and supplemented with 20% fetal bovine serum (American Type Culture Collection; catalog no. 30-2020). Cells were grown in suspension to ≤1 × 106 cells/ml at 37°C in a 5% CO2 atmosphere. Differentiation was carried out in supplemented IMDM (as described above) containing 0.8% N,N-dimethylformamide (Sigma; catalog no. D4551). Flasks to be differentiated were inoculated at 5 × 105 cells/ml as judged by viable counts with trypan blue exclusion. Cultures were incubated for 5 days, and the differentiation medium was not replaced during this time. For the functional assay, the bacteria (S. pneumoniae 603 isogenic strains), differentiated HL-60 cells, serum, and complement source were prepared as described below. For preparation of the serum, each serum sample was diluted in opsonophagocytosis buffer for a total of 5 dilutions. Once all serum samples were prepared, 80 µl of bacterial suspension that had been appropriately diluted (~1,000 CFU) was added to each well. Following a 30-min incubation at 37°C, 10 µl complement (baby rabbit complement [Pel-Freez; catalog no. 31064]) was added to each well. The assay plate was allowed to incubate for another 30 min at 37°C. Immediately after this, differentiated HL-60 cells were added to each well. Differentiated HL-60 cells were used in the opsonophagocytic assay at an effector/target cell ratio of 400/1. Differentiated cells were harvested by centrifugation (1,700 rpm) for 10 min at room temperature. The cell pellet was washed twice in Hanks’ buffer without Ca2+ and Mg2+ (Corning; catalog no. 21-022-CV). Finally, the cells were resuspended to 1 × 107 cells/ml in Hanks’ buffer with Ca2+ and Mg2+ (Corning; catalog no. 21-023-CV) supplemented with 0.1% gelatin. The total number of HL-60 cells to be added per well was 4 × 105 in a 40-µl volume. The assay plate was incubated at 37°C for 45 min with horizontal shaking (700 rpm) to promote the phagocytic process. All samples were run in triplicate. An aliquot from each well was plated onto solid medium (Trypticase soy agar with 5% sheep blood) to enumerate viable CFU after overnight incubation. A count of the initial number of viable bacteria added per well at time zero was included in each run. For each sample, the CFU counts at time point 45 min were divided by the CFU counts at time point 0 to determine the percentage of CFU that survived.
ACKNOWLEDGMENTS
We gratefully acknowledge Fan Zhang for help with protein purification. We thank Richard Malley and members of the Malley lab for helpful discussions and insights. We also thank Taj Azarian, Samantha Palace, and Brian Arnold as well as other members of the Lipsitch lab for helpful discussions.
Funding was provided by National Institutes of Health grant R01 AI048935 (to M. L.).
Footnotes
Citation Georgieva M, Kagedan L, Lu Y-J, Thompson CM, Lipsitch M. 2018. Antigenic variation in Streptococcus pneumoniae PspC promotes immune escape in the presence of variant-specific immunity. mBio 9:e00264-18. https://doi.org/10.1128/mBio.00264-18.
REFERENCES
- 1.Geno KA, Gilbert GL, Song JY, Skovsted IC, Klugman KP, Jones C, Konradsen HB, Nahm MH. 2015. Pneumococcal capsules and their types: past, present, and future. Clin Microbiol Rev 28:871–899. doi: 10.1128/CMR.00024-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Dam JE, Fleer A, Snippe H. 1990. Immunogenicity and immunochemistry of Streptococcus pneumoniae capsular polysaccharides. Antonie Leeuwenhoek 58:1–47. doi: 10.1007/BF02388078. [DOI] [PubMed] [Google Scholar]
- 3.AlonsoDeVelasco E, Verheul AF, Verhoef J, Snippe H. 1995. Streptococcus pneumoniae: virulence factors, pathogenesis, and vaccines. Microbiol Rev 59:591–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, Pichon B, Baker S, Parry CM, Lambertsen LM, Shahinas D, Pillai DR, Mitchell TJ, Dougan G, Tomasz A, Klugman KP, Parkhill J, Hanage WP, Bentley SD. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Croucher NJ, Finkelstein JA, Pelton SI, Mitchell PK, Lee GM, Parkhill J, Bentley SD, Hanage WP, Lipsitch M. 2013. Population genomics of post-vaccine changes in pneumococcal epidemiology. Nat Genet 45:656–663. doi: 10.1038/ng.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks-Walter AB, Briles DE, Hollingshead SK. 1999. The pspC gene of Streptococcus pneumoniae encodes a polymorphic protein, PspC, which elicits cross-reactive antibodies to PspA and provides immunity to pneumococcal bacteremia. Infect Immun 67:6533–6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammerschmidt ST, Talay SR, Brandtzaeg P, Chhatwal GS. 1997. SpsA, a novel pneumococcal surface protein with specific binding to secretory immunoglobulin A and secretory component. Mol Microbiol 25:1113–1124. doi: 10.1046/j.1365-2958.1997.5391899.x. [DOI] [PubMed] [Google Scholar]
- 8.Dave S, Brooks-Walter A, Pangburn MK, McDaniel LS. 2001. PspC, a pneumococcal surface protein, binds human factor H. Infect Immun 69:3435–3437. doi: 10.1128/IAI.69.5.3435-3437.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith BLH, Hostetter MK. 2000. C3 as substrate for adhesion of Streptococcus pneumoniae. J Infect Dis 182:497–508. doi: 10.1086/315722. [DOI] [PubMed] [Google Scholar]
- 10.Balachandran P, Brooks-Walter A, Virolainen-Julkunen A, Hollingshead SK, Briles DE. 2002. Role of pneumococcal surface protein C in nasopharyngeal carriage and pneumonia and its ability to elicit protection against carriage of Streptococcus pneumoniae. Infect Immun 70:2526–2534. doi: 10.1128/IAI.70.5.2526-2534.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berry AM, Paton JC. 2000. Additive attenuation of virulence of Streptococcus pneumoniae by mutation of the genes encoding pneumolysin and other putative pneumococcal virulence proteins. Infect Immun 68:133–140. doi: 10.1128/IAI.68.1.133-140.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogunniyi AD, LeMessurier KS, Graham RM, Watt JM, Briles DE, Stroeher UH, Paton JC. 2007. Contributions of pneumolysin, pneumococcal surface protein A (PspA), and PspC to pathogenicity of Streptococcus pneumoniae D39 in a mouse model. Infect Immun 75:1843–1851. doi: 10.1128/IAI.01384-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang JR, Mostov KE, Lamm ME, Nanno M, Shimida S, Ohwaki M, Tuomanen E. 2000. The polymeric immunoglobulin receptor translocates pneumococci across human nasopharyngeal epithelial cells. Cell 102:827–837. doi: 10.1016/S0092-8674(00)00071-4. [DOI] [PubMed] [Google Scholar]
- 14.Asmat TM, Agarwal V, Räth S, Hildebrandt JP, Hammerschmidt S. 2011. Streptococcus pneumoniae infection of host epithelial cells via polymeric immunoglobulin receptor transiently induces calcium release from intracellular stores. J Biol Chem 286:17861–17869. doi: 10.1074/jbc.M110.212225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agarwal V, Asmat TM, Luo S, Jensch I, Zipfel PF, Hammerschmidt S. 2010. Complement regulator factor H mediates a two-step uptake of Streptococcus pneumoniae by human cells. J Biol Chem 285:23486–23495. doi: 10.1074/jbc.M110.142703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herbert AP, Makou E, Chen ZA, Kerr H, Richards A, Rappsilber J, Barlow PN. 2015. Complement evasion mediated by enhancement of captured factor H: implications for protection of self-surfaces from complement. J Immunol 195:4986–4998. doi: 10.4049/jimmunol.1501388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jarva H, Janulczyk R, Hellwage J, Zipfel PF, Björck L, Meri S. 2002. Streptococcus pneumoniae evades complement attack and opsonophagocytosis by expressing the pspC locus-encoded hic protein that binds to short consensus repeats 8–11 of factor H. J Immunol 168:1886–1894. doi: 10.4049/jimmunol.168.4.1886. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Glover DT, Szalai AJ, Hollingshead SK, Briles DE. 2007. PspA and PspC minimize immune adherence and transfer of pneumococci from erythrocytes to macrophages through their effects on complement activation. Infect Immun 75:5877–5885. doi: 10.1128/IAI.00839-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quin LR, Onwubiko C, Moore QC, Mills MF, McDaniel LS, Carmicle S. 2007. Factor H binding to PspC of Streptococcus pneumoniae increases adherence to human cell lines in vitro and enhances invasion of mouse lungs in vivo. Infect Immun 75:4082–4087. doi: 10.1128/IAI.00474-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quin LR, Carmicle S, Dave S, Pangburn MK, Evenhuis JP, McDaniel LS. 2005. In vivo binding of complement regulator factor H by Streptococcus pneumoniae. J Infect Dis 192:1996–2003. doi: 10.1086/497605. [DOI] [PubMed] [Google Scholar]
- 21.Yuste J, Khandavilli S, Ansari N, Muttardi K, Ismail L, Hyams C, Weiser J, Mitchell T, Brown JS. 2010. The effects of PspC on complement-mediated immunity to Streptococcus pneumoniae vary with strain background and capsular serotype. Infect Immun 78:283–292. doi: 10.1128/IAI.00541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janulczyk R, Iannelli F, Sjoholm AG, Pozzi G, Bjorck L. 2000. Hic, a novel surface protein of Streptococcus pneumoniae that interferes with complement function. J Biol Chem 275:37257–37263. doi: 10.1074/jbc.M004572200. [DOI] [PubMed] [Google Scholar]
- 23.Azarian T, Grant LR, Georgieva M, Hammitt LL, Reid R, Bentley SD, Goldblatt D, Santosham M, Weatherholtz R, Burbidge P, Goklish N, Thompson CM, Hanage WP, O’Brien KL, Lipsitch M. 2017. Association of pneumococcal protein antigen serology with age and antigenic profile of colonizing isolates. J Infect Dis 215:713–722. doi: 10.1093/infdis/jiw628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Croucher NJ, Campo JJ, Le TQ, Liang X, Bentley SD, Hanage WP, Lipsitch M. 2017. Diverse evolutionary patterns of pneumococcal antigens identified by pangenome-wide immunological screening. Proc Natl Acad Sci U S A 114:E357–EE366. doi: 10.1073/pnas.1613937114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCool TL, Cate TR, Tuomanen EI, Adrian P, Mitchell TJ, Weiser JN. 2003. Serum immunoglobulin G response to candidate vaccine antigens during experimental human pneumococcal colonization. Infect Immun 71:5724–5732. doi: 10.1128/IAI.71.10.5724-5732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vadesilho CF, Ferreira DM, Gordon SB, Briles DE, Moreno AT, Oliveira ML, Ho PL, Miyaji EN. 2014. Mapping of epitopes recognized by antibodies induced by immunization of mice with PspA and PspC. Clin Vaccine Immunol 21:940–948. doi: 10.1128/CVI.00239-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lebon A, Verkaik NJ, Labout JA, de Vogel CP, Hooijkaas H, Verbrugh HA, van Wamel WJ, Jaddoe VW, Hofman A, Hermans PW, Ma J, Mitchell TJ, Moll HA, van Belkum A. 2011. Natural antibodies against several pneumococcal virulence proteins in children during the pre-pneumococcal-vaccine era: the generation R study. Infect Immun 79:1680–1687. doi: 10.1128/IAI.01379-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holmlund E, Quiambao B, Ollgren J, Jaakkola T, Neyt C, Poolman J, Nohynek H, Käyhty H. 2009. Antibodies to pneumococcal proteins PhtD, CbpA, and LytC in Filipino pregnant women and their infants in relation to pneumococcal carriage. Clin Vaccine Immunol 16:916–923. doi: 10.1128/CVI.00050-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner P, Turner C, Green N, Ashton L, Lwe E, Jankhot A, Day NP, White NJ, Nosten F, Goldblatt D. 2013. Serum antibody responses to pneumococcal colonization in the first 2 years of life: results from an SE Asian longitudinal cohort study. Clin Microbiol Infect 19:E551–E558. doi: 10.1111/1469-0691.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogunniyi AD, Woodrow MC, Poolman JT, Paton JC. 2001. Protection against Streptococcus pneumoniae elicited by immunization with pneumolysin and CbpA. Infect Immun 69:5997–6003. doi: 10.1128/IAI.69.10.5997-6003.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iannelli FO, Oggioni MR, Pozzi G. 2002. Allelic variation in the highly polymorphic locus pspC of Streptococcus pneumoniae. Gene 284:63–71. doi: 10.1016/S0378-1119(01)00896-4. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Gierahn T, Thompson CM, Trzciński K, Ford CB, Croucher N, Gouveia P, Flechtner JB, Malley R, Lipsitch M. 2012. Distinct effects on diversifying selection by two mechanisms of immunity against Streptococcus pneumoniae. PLoS Pathog 8:e1002989. doi: 10.1371/journal.ppat.1002989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Regev-Yochay G, Hanage WP, Trzcinski K, Rifas-Shiman SL, Lee G, Bessolo A, Huang SS, Pelton SI, McAdam AJ, Finkelstein JA, Lipsitch M, Malley R. 2010. Re-emergence of the type 1 pilus among Streptococcus pneumoniae isolates in Massachusetts, USA. Vaccine 28:4842–4846. doi: 10.1016/j.vaccine.2010.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dieudonné-Vatran A, Krentz S, Blom AM, Meri S, Henriques-Normark B, Riesbeck K, Albiger B. 2009. Clinical isolates of Streptococcus pneumoniae bind the complement inhibitor C4b-binding protein in a PspC allele-dependent fashion. J Immunol 182:7865–7877. doi: 10.4049/jimmunol.0802376. [DOI] [PubMed] [Google Scholar]
- 35.Moreno AT, Oliveira ML, Ho PL, Vadesilho CF, Palma GM, Ferreira JM Jr, Ferreira DM, Santos SR, Martinez MB, Miyaji EN. 2012. Cross-reactivity of antipneumococcal surface protein C (PspC) antibodies with different strains and evaluation of inhibition of human complement factor H and secretory IgA binding via PspC. Clin Vaccine Immunol 19:499–507. doi: 10.1128/CVI.05706-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ricci S, Janulczyk R, Gerlini A, Braione V, Colomba L, Iannelli F, Chiavolini D, Oggioni MR, Björck L, Pozzi G. 2011. The factor H-binding fragment of PspC as a vaccine antigen for the induction of protective humoral immunity against experimental pneumococcal sepsis. Vaccine 29:8241–8249. doi: 10.1016/j.vaccine.2011.08.119. [DOI] [PubMed] [Google Scholar]
- 37.Achila D, Liu A, Banerjee R, Li Y, Martinez-Hackert E, Zhang JR, Yan H. 2015. Structural determinants of host specificity of complement factor H recruitment by Streptococcus pneumoniae. Biochem J 465:325–335. doi: 10.1042/BJ20141069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dave S, Carmicle S, Hammerschmidt S, Pangburn MK, McDaniel LS. 2004. Dual roles of PspC, a surface protein of Streptococcus pneumoniae, in binding human secretory IgA and factor H. J Immunol 173:471–477. doi: 10.4049/jimmunol.173.1.471. [DOI] [PubMed] [Google Scholar]
- 39.Duthy TG, Ormsby RJ, Giannakis E, Ogunniyi AD, Stroeher UH, Paton JC, Gordon DL. 2002. The human complement regulator factor H binds pneumococcal surface protein PspC via short consensus repeats 13 to 15. Infect Immun 70:5604–5611. doi: 10.1128/IAI.70.10.5604-5611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hammerschmidt S, Agarwal V, Kunert A, Haelbich S, Skerka C, Zipfel PF. 2007. The host immune regulator factor H interacts via two contact sites with the PspC protein of Streptococcus pneumoniae and mediates adhesion to host epithelial cells. J Immunol 178:5848–5858. doi: 10.4049/jimmunol.178.9.5848. [DOI] [PubMed] [Google Scholar]
- 41.Lu L, Ma Y, Zhang JR. 2006. Streptococcus pneumoniae recruits complement factor H through the amino terminus of CbpA. J Biol Chem 281:15464–15474. doi: 10.1074/jbc.M602404200. [DOI] [PubMed] [Google Scholar]
- 42.Lu L, Ma Z, Jokiranta TS, Whitney AR, DeLeo FR, Zhang JR. 2008. Species-specific interaction of Streptococcus pneumoniae with human complement factor H. J Immunol 181:7138–7146. doi: 10.4049/jimmunol.181.10.7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quin LR, Onwubiko C, Carmicle S, McDaniel LS. 2006. Interaction of clinical isolates of Streptococcus pneumoniae with human complement factor H. FEMS Microbiol Lett 264:98–103. doi: 10.1111/j.1574-6968.2006.00439.x. [DOI] [PubMed] [Google Scholar]
- 44.Asmat TM, Klingbeil K, Jensch I, Burchhardt G, Hammerschmidt S. 2012. Heterologous expression of pneumococcal virulence factor PspC on the surface of Lactococcus lactis confers adhesive properties. Microbiology 158:771–780. doi: 10.1099/mic.0.053603-0. [DOI] [PubMed] [Google Scholar]
- 45.Glennie S, Gritzfeld JF, Pennington SH, Garner-Jones M, Coombes N, Hopkins MJ, Vadesilho CF, Miyaji EN, Wang D, Wright AD, Collins AM, Gordon SB, Ferreira DM. 2016. Modulation of nasopharyngeal innate defenses by viral coinfection predisposes individuals to experimental pneumococcal carriage. Mucosal Immunol 9:56–67. doi: 10.1038/mi.2015.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Desa MN, Navaratnam P, Vadivelu J, Sekaran SD. 2008. Expression analysis of adherence-associated genes in pneumococcal clinical isolates during adherence to human respiratory epithelial cells (in vitro) by real-time PCR. FEMS Microbiol Lett 288:125–130. doi: 10.1111/j.1574-6968.2008.01345.x. [DOI] [PubMed] [Google Scholar]
- 47.Hyams C, Trzcinski K, Camberlein E, Weinberger DM, Chimalapati S, Noursadeghi M, Lipsitch M, Brown JS. 2013. Streptococcus pneumoniae capsular serotype invasiveness correlates with the degree of factor H binding and opsonization with C3b/iC3b. Infect Immun 81:354–363. doi: 10.1128/IAI.00862-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melin M, Trzciński K, Antonio M, Meri S, Adegbola R, Kaijalainen T, Käyhty H, Väkeväinen M. 2010. Serotype-related variation in susceptibility to complement deposition and opsonophagocytosis among clinical isolates of Streptococcus pneumoniae. Infect Immun 78:5252–5261. doi: 10.1128/IAI.00739-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Maten EW, Westra D, van Selm S, Langereis JD, Bootsma HJ, van Opzeeland FJH, de Groot R, Ruseva MM, Pickering MC, van den Heuvel LPWJ, van de Kar NCAJ, de Jonge MI, van der Flier M. 2016. Complement factor H serum levels determine resistance to pneumococcal invasive disease. J Infect Dis 213:1820–1827. doi: 10.1093/infdis/jiw029. [DOI] [PubMed] [Google Scholar]
- 50.Cagliani R, Forni D, Filippi G, Mozzi A, De Gioia L, Pontremoli C, Pozzoli U, Bresolin N, Clerici M, Sironi M. 2016. The mammalian complement system as an epitome of host-pathogen genetic conflicts. Mol Ecol 25:1324–1339. [DOI] [PubMed] [Google Scholar]
- 51.Trzcinski K, Thompson CM, Lipsitch M. 2003. Construction of otherwise isogenic serotype 6B, 7F, 14, and 19F capsular variants of Streptococcus pneumoniae strain TIGR4. Appl Environ Microbiol 69:7364–7370. doi: 10.1128/AEM.69.12.7364-7370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences of full-length PspC variants and truncated protein fragments (in gray). Download TEXT S1, PDF file, 0.1 MB (67.2KB, pdf) .
Copyright © 2018 Georgieva et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Anti-PspC var.-I IgG (A) and var.-IV IgG (B) are nonopsonic in a neutrophil-mediated antibody-dependent opsonophagocytic assay. Graphs show percent survival of isogenic S. pneumoniae strains incubated with anti-PspC var.-I IgG or var.-IV IgG. Download FIG S1, PDF file, 0.4 MB (374.1KB, pdf) .
Copyright © 2018 Georgieva et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Variability in anti-PspC var.-II IgG specificity depending on the source of antisera. (A) Far Western blot for fH binding in the presence of antibodies. (B) Survival data in opsonophagocytic assay using anti-PspC var.-II IgG from rabbit2 immunized with PspC var.-II. Anti-PspC var.-II IgG recognizes PspC var.-III, in addition to its cognate target, PspC var.-II. Download FIG S2, PDF file, 0.1 MB (142.5KB, pdf) .
Copyright © 2018 Georgieva et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Alignment of protein sequences corresponding to truncated PspC variants I to IV. Download FIG S3, PDF file, 0.5 MB (547.9KB, pdf) .
Copyright © 2018 Georgieva et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.