Abstract
Differences in gene content are a significant source of variability within species and have an impact on phenotypic traits. However, little is known about the mechanisms responsible for the most recent gene gains and losses. We screened the genomes of 123 worldwide isolates of the major pathogen of wheat Zymoseptoria tritici for robust evidence of gene copy number variation. Based on orthology relationships in three closely related fungi, we identified 599 gene gains and 1,024 gene losses that have not yet reached fixation within the focal species. Our analyses of gene gains and losses segregating in populations showed that gene copy number variation arose preferentially in subtelomeres and in proximity to transposable elements. Recently lost genes were enriched in virulence factors and secondary metabolite gene clusters. In contrast, recently gained genes encoded mostly secreted protein lacking a conserved domain. We analyzed the frequency spectrum at loci segregating a gene presence–absence polymorphism in four worldwide populations. Recent gene losses showed a significant excess in low-frequency variants compared with genome-wide single nucleotide polymorphism, which is indicative of strong negative selection against gene losses. Recent gene gains were either under weak negative selection or neutral. We found evidence for strong divergent selection among populations at individual loci segregating a gene presence–absence polymorphism. Hence, gene gains and losses likely contributed to local adaptation. Our study shows that microbial eukaryotes harbor extensive copy number variation within populations and that functional differences among recently gained and lost genes led to distinct evolutionary trajectories.
Keywords: copy number variation, evolutionary genomics, fungi
Introduction
Differences in gene content are an extensive source of polymorphism within species that can have significant phenotypic effects (Henrichsen etal. 2009; Conrad etal. 2010). Gene gains after duplication or horizontal transfer generate significant evolutionary novelty (Ohno 1970; Lynch and Conery 2003). Large-scale gene losses following whole genome duplication events make extensive contributions to differentiation among species (Blomme etal. 2006; Wolf and Koonin 2013). Differences in gene content is also an important source of adaptive differentiation within species (Redon etal. 2006; Pezer etal. 2015; Cheeseman etal. 2016). Gene gains and losses preferentially occur in specific chromosomal locations and tend to affect genes with specific functions (Blomme etal. 2006; Albalat and Cañestro 2016). However, little is known about the mechanisms leading to gene gains and losses segregating within population and how selection acts upon gene content changes.
Gene deletions are under negative selection due to deleterious effects of loss-of-function (Conrad etal. 2006; Emerson etal. 2008; Sudmant etal. 2015). However, the degree of gene dispensability (i.e., the impact on fitness) can vary substantially among genes (Ohno 1985) and gene losses may also be adaptive (Olson 1999; Morris etal. 2012). Most gene gains originate through duplications and are assumed to be initially selectively neutral and later fixed by genetic drift, although positive selection can also lead to the fixation of a duplicated gene (Innan and Kondrashov 2010; Cardoso-Moreira etal. 2016). Adaptive gene gains through horizontal transfer can be under strong selection and spread among multiple species (Ropars etal. 2015). Adaptive gene gains and losses were well-described in pathogens of plants. In plants, the immune system triggers defense responses after detection of specific pathogen proteins, generally identified as effectors (Jones and Dangl 2006). For pathogens, the loss of genes encoding such detected proteins can be highly beneficial (Stukenbrock and McDonald 2009; Presti etal. 2015; Hartmann etal. 2017). Gene gains through expansions of specific gene families or horizontal gene transfers can contribute to host specialization and virulence on new hosts (Friesen etal. 2006; Ohm etal. 2012). The ability of pathogens to rapidly surmount host resistance and adapt to new hosts is directly linked to major economic losses in agriculture (Fisher etal. 2016). The strong selection that potentially acts on individual gene presence-absence polymorphisms makes plant pathogens excellent models to investigate the evolutionary trajectory of gene gains and losses. In addition, many plant pathogens have haploid genomes, which exposes gene gains and losses more directly to selection (Stukenbrock and Croll 2014).
Fungal plant pathogen genomes are often compartmentalized into rapidly evolving, repeat-rich and conserved, gene-rich compartments. Repeat-rich and rapidly evolving compartments often segregate presence–absence polymorphism within the species (Croll and McDonald 2012; Dong etal. 2015). Until now, analyses of within-species gene content focused on a small number of genomes or targeted specific gene categories (Yoshida etal. 2009; Syme etal. 2013; Plissonneau etal. 2016). To understand the mechanisms responsible for gene gains and losses within species, an extensive set of genomes from multiple populations needs to be analyzed. Zymoseptoria tritici is the major pathogen of wheat (Fones and Gurr 2015) with significant variation in gene content among strains. Genomic analyses showed that hundreds of genes can be lacking in individual strains compared with other strains of the species (McDonald etal. 2016; Plissonneau etal. 2016). A chromosomal rearrangement led to a major adaptation by deleting a gene that encoded for a protein recognized by the host (Hartmann etal. 2017). Hence, Z. tritici is an excellent model to investigate the evolutionary trajectories of recent gene gains and losses. In addition, the pathogen undergoes high rates of sexual reproduction (Croll etal. 2015) exposing recent gene gains and losses individually to selection. The genome of Z. tritici was assembled from telomere-to-telomere (Goodwin etal. 2011) and a comparative genomics framework of three closely related species (Grandaubert etal. 2015) enables precise characterizations of individual gene gains and losses.
In this study, we analyzed recent gene gains and losses across the genome in four worldwide Z. tritici populations. For this, we used whole genome sequences of 123 isolates to identify high-confidence gene presence–absence variations. We used orthology among three closely related species to assign each presence-absence gene polymorphism to either a recent gain or a loss event. We analyzed how recent gene gains and losses were differentially affected by chromosomal location, expression level, and gene function. Finally, we analyzed signatures of selection acting on recent gene gains and losses.
Results
Identification of Recent Gene Gains and Losses in Z. tritici Populations
We analyzed genome-wide recent gene gains and losses in four Z. tritici populations sampled from wheat fields across the geographic range of the pathogen. We used Illumina sequencing data generated for 130 haploid Z. tritici isolates (8–29× mean coverage; supplementary table S1, Supplementary Material online). First, we detected presence–absence polymorphisms of entire genes against the reference genome using mapped read depth. We excluded seven isolates from further analyses, because we found evidence of partial or complete chromosomal aneuploidy. We manually curated presence–absence polymorphisms of genes encoding small secreted proteins. Genes encoding small secreted proteins are often located in proximity to regions enriched in repeats and complex sequence rearrangements that are error-prone for deletion calls (Presti etal. 2015). We used both comparative genomics analyses and direct amplification of loci (polymerase chain reaction [PCR]) to validate gene presence–absence polymorphism calls. As the genomes of two isolates are available as telomere-to-telomere assemblies (Plissonneau etal. 2016), we used the complete genomes to validate gene presence–absence polymorphism calls. For each gene predicted to be deleted in isolate ST99CH_3D7, we performed a blast search in the complete genome sequence of the same isolate. Our analyses showed that 237 out of 251 genes predicted to be deleted were indeed absent from the ST99CH_3D7 genome. Hence, 14 (3.5%) gene deletion calls were erroneous. We performed also PCR amplification assays on 95 isolates to validate gene deletion calls in 14 different genes including six genes encoding small secreted proteins. The assay confirmed 100% of the 317 tested gene deletion calls (see supplementary text S1, Supplementary Material online, for details).
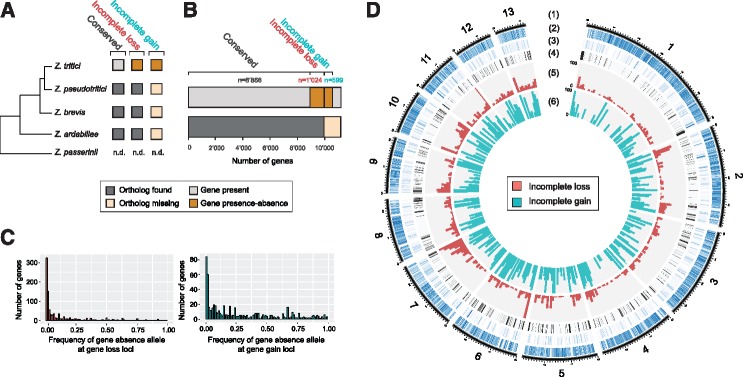
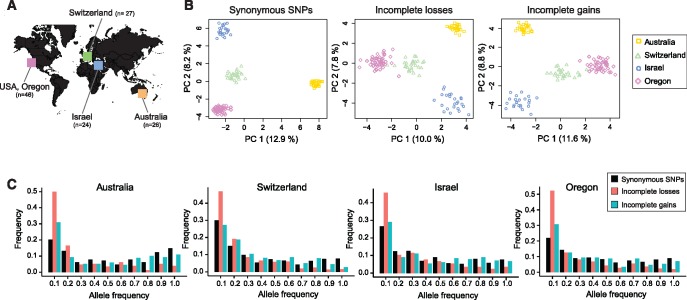
We identified a total of 37,644 presence–absence polymorphism events affecting 1,623 distinct genes in the genome (14.6% of all genes on chromosomes shared among all isolates). Then, we classified gene presence–absence polymorphism events as recent gene losses or gains in Z. tritici populations based on gene homology information in closely related species. Grandaubert etal. (2015) identified a set of core Zymoseptoria genes, that is, genes having an ortholog in at least one of the closely related species (Z. pseudotritici, Z. ardabiliae, and Z. brevis). We defined incomplete (i.e., recent) gene losses as any presence–absence polymorphism affecting a core Zymoseptoria gene. Conversely, we referred to incomplete (i.e., recent) gene gains as any presence–absence polymorphism event affecting a Z. tritici orphan gene (genes lacking an ortholog in any of the three other species). In the 123 Z. tritici isolates, we found that 1,024 genes were affected by incomplete losses (i.e., 10.4% of Zymoseptoria core genes) and 599 genes were affected by incomplete gains (i.e., 49.1% of Z. tritici orphan genes; fig. 1A). Incomplete gene gain events were segregating at higher frequency than incomplete gene loss events in the 123 isolates. The median frequency of the gene absence allele at gene loss loci was 3.3%, whereas the median frequency of the gene absence allele at gene gain loci was 16.3% among the 123 isolates (fig. 1B;supplementary tables S2 and S3, Supplementary Material online). Incomplete gene losses and gene gains were found on all 13 core chromosomes (chromosomes found in all members of the species; fig. 1C). We found that the proportions of genes affected by incomplete loss events and incomplete gain events were significantly higher in subtelomeric regions, defined here as regions within 300 kb of the telomeres, compared with the chromosome genome-wide average (Pearson’s chi-squared test; incomplete losses P < 2.2×10−16; incomplete gains P = 9.6×10−10; supplementary fig. S1, Supplementary Material online).
Fig. 1.
Characterization of incomplete gene gains and losses among 123 completely sequenced Zymoseptoria tritici isolates. (A) Schematic tree representing the phylogenetic relationships between Z. tritici and four most closely related species. Evidence for orthologs was used to infer whether a segregating gene presence–absence polymorphism segregating in Z. tritici was due to a recent (incomplete) gain or recent (incomplete) loss. n.d., not determined. (B) Number of genes for each category of Z. tritici genes as shown in (A). (C) Frequency of the gene absence allele at incomplete gene gain and loss loci among 123 isolates. (D) Genome-wide distribution of incomplete gene gains and losses. (1) Chromosomes of the reference genome and position in Mb. (2) Percentage of genes that have an ortholog in other Zymoseptoria species shown in 10-kb nonoverlapping windows (gradient shows differences from 0% to 100%). (3) Percentage of genes with no orthologs in other Zymoseptoria species (orphans) shown in 10-kb nonoverlapping windows (gradient shows differences from 0% to 100%). (4) Content in transposable element sequences in 10-kb nonoverlapping windows (gradient shows differences from 0% to 50%). (5) Proportion of genes affected by incomplete losses in 100-kb nonoverlapping windows. (6) Proportion of genes showing an incomplete gain in 100-kb nonoverlapping windows.
Gene Functions Differentially Affected by Losses and Gains
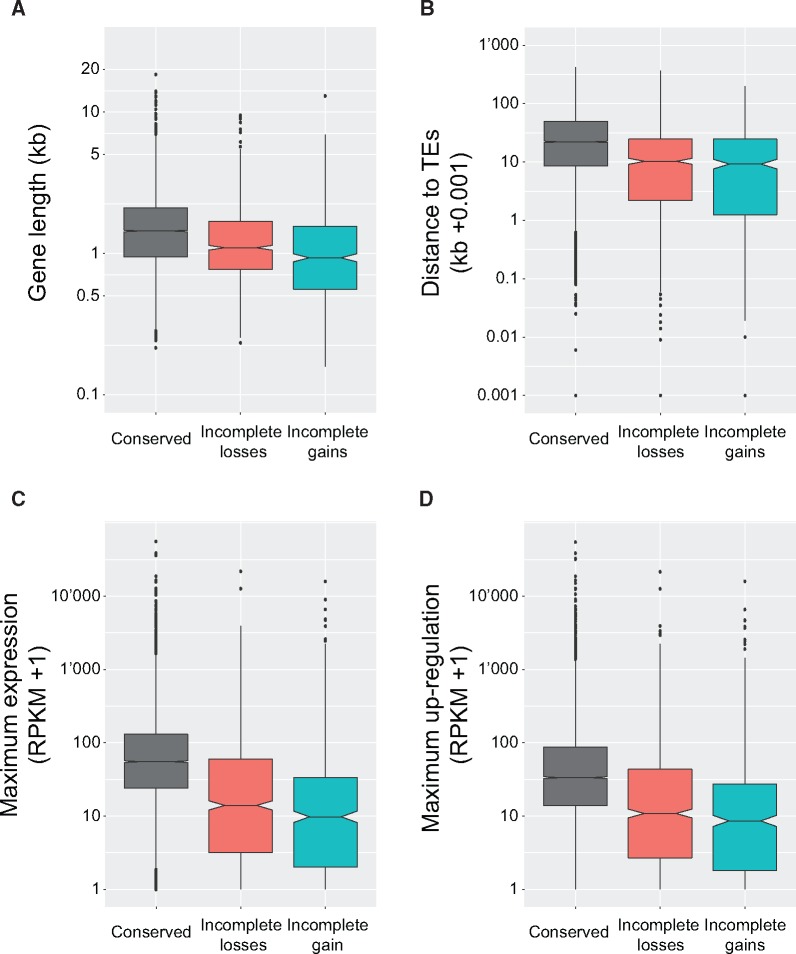
We investigated the gene functions affected by incomplete gains and losses. Genes affected by incomplete losses and gains were both significantly shorter than conserved genes (i.e., genes not affected by any loss or gain event; Kruskal–Wallis test, P < 0.05 adjusted for multiple comparisons; fig. 2A). We found that genes affected by incomplete gains were significantly shorter than genes affected by incomplete losses (P < 0.05; fig. 2A). Furthermore, we found that genes affected by incomplete gains and losses were significantly closer to transposable elements (TEs) than conserved genes (P < 0.05; fig. 2B). We found no significant difference between genes affected by gains and losses in regards to distance to TEs (P > 0.05; fig. 2B). A total of 74 out of 91 genes encoding protein domains associated with TEs, such as reverse transcriptase, transposase, or integrase domains, were affected by an incomplete gain (n = 53) or loss (n = 21).
Fig. 2.
Differences in length, genomic location, and expression level of conserved genes, genes affected by an incomplete loss and genes showing an incomplete gain. (A) Gene length (kb). (B) Distance to closest transposable element (TE). (C) Maximum gene expression level during infection of a wheat host. RPKM, reads per kilobase of exon per million mapped reads. (D) Upregulation during wheat infection. Upregulation was assessed as the maximum absolute difference in RPKM values over five time points during infection.
We analyzed transcriptomic data to identify gene expression differences. For this, we used RNA-seq data previously collected over the entire infection time course on wheat (Rudd etal. 2015). We found that genes affected by incomplete losses and gains were significantly less expressed than conserved genes (Kruskal–Wallis test, P < 0.05 multiple comparisons corrected; fig. 2C). Furthermore, genes affected by incomplete losses and gains were significantly less upregulated during infection (P < 0.05; fig. 2D). Nonetheless, we found 66 and 166 genes affected by incomplete gains and losses, respectively, that were highly upregulated during wheat infection (maximum reads per kilobase of transcript per million mapped reads, RPKM, differences >100). Genes affected by incomplete gains were significantly less expressed and less upregulated than genes affected by incomplete losses (P < 0.05).
The majority of the genes affected by incomplete losses (51.8%) or incomplete gains (79.8%) encoded proteins lacking a conserved protein family domain. For genes with a predicted function, we performed a gene ontology (GO) enrichment analysis. We tested whether some GO terms were overrepresented or underrepresented compared with the genomic background (supplementary tables S4 and S5, Supplementary Material online). GO terms for DNA integration and for proteolysis were significantly overrepresented both among genes affected by incomplete losses or gains. Among genes affected by incomplete gains, we found GO terms related to recombination processes to be overrepresented, whereas among genes affected by incomplete losses overrepresented GO terms were related to metabolism (e.g., oxidation–reduction, amide transport, pigment metabolism). The encoded proteins included peptidases, oxidoreductases, hydrolases, and methyltransferases. Genes encoding proteins with extracellular functions and association to membranes were overrepresented among genes affected by incomplete losses. GO terms for cellular processes (e.g., cellular metabolic process, gene expression, translation, RNA processing, cell communication) were significantly underrepresented among genes affected by incomplete losses and gains. Similarly, RNA and sugar binding activity as well as proteins localized in intracellular compartments were underrepresented among gene functions affected by incomplete losses and gains.
Incomplete Losses and Gains of Pathogenicity Genes and Secondary Metabolite Gene Clusters
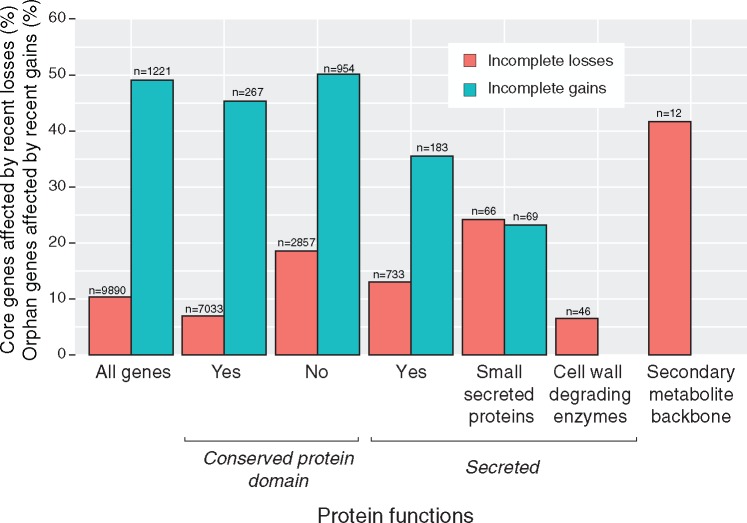
Genes gains or losses can confer significant adaptive value to colonize a host (de Jonge etal. 2011; Presti etal. 2015). We analyzed three categories of pathogenicity-related genes for evidence of incomplete gene gains and losses (fig. 3): secondary metabolite gene clusters, genes encoding cell wall degrading enzymes and small secreted proteins. Genes encoding cell wall degrading enzymes play a role in the degradation of plant cell walls and nutrient acquisition (Esquerré-Tugayé etal. 2000; de Jonge etal. 2011). Genes encoding cell wall degrading enzymes (n = 47) were affected by incomplete losses at a lower proportion (6.5%) than genes encoding proteins with a conserved domain (7.0%) and genes encoding secreted proteins (13.1%). No gene encoding cell wall degrading enzymes was recently gained.
Fig. 3.
Frequencies of incomplete gains and losses among different gene categories. The barplot shows the percentage of genes that were missing in at least one isolate. For incomplete gene losses, the percentage was calculated based on the total number of genes that have an ortholog among the closely related Zymoseptoria species, genes encoding secreted proteins or small secreted proteins, cell wall degrading enzymes or backbones of secondary metabolite gene clusters. For incomplete gene gains, the percentage was calculated based on the total number of genes that lack any ortholog (i.e., orphan gene).
Genes encoding small secreted proteins may be recognized by the host immune system (Rep 2005). Small secreted proteins were defined as secreted proteins of ≤300 amino acids and containing ≥5% cysteine residues following commonly used definitions of small secreted proteins in plant pathogenic fungi (do Amaral etal. 2012; Sperschneider etal. 2015). We identified a total of 135 genes encoding small secreted proteins and were affected by incomplete loss more frequently (24.2%) than the average for all secreted proteins (13.1%) and the genome-wide average (10.3%). Conversely, genes encoding small secreted proteins were affected by incomplete gains less frequently (23%) than the average for all secreted proteins (35.5%) and the genome-wide average (49%). Among genes encoding small secreted proteins, we found 16 genes affected by incomplete losses and 16 additional genes affected by incomplete gains. However, the proportion of genes encoding secreted proteins affected by incomplete gains was almost three times as high as the proportion affected by incomplete losses (35.5% vs. 13.1%). Presence–absence variation affected a total of 32 out of 135 genes encoding small secreted proteins (i.e., 23.7%; supplementary fig. S2, Supplementary Material online).
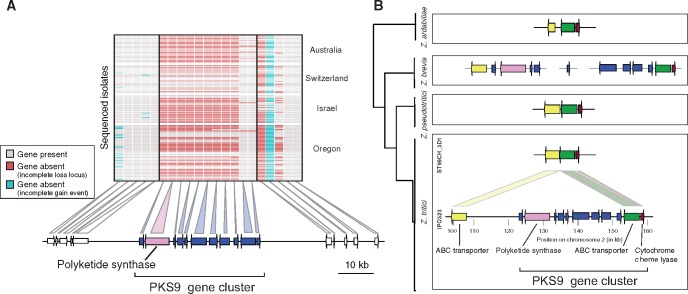
Genes encoding secondary metabolite gene clusters are involved in the biosynthesis of pigments (e.g., melanin) and toxins (e.g., antimicrobials, mycotoxins, and phytotoxins) contributing to fungal pathogenicity, nutrient acquisition, and ecological adaptation (Howlett 2006; Stergiopoulos etal. 2013). In general, each gene cluster is defined by a backbone gene (Brakhage 2013). We focused our analysis on secondary metabolite gene clusters with backbone genes encoding either a polyketide synthase (PKS) or a nonribosomal peptide synthetase (NRPS). The genome of Z. tritici contained nine PKS gene clusters, two NRPS gene clusters, and one PKS–NRPS hybrid gene cluster (Ohm etal. 2012). We considered that a secondary metabolite gene cluster was affected by a significant deletion if at least the backbone gene was deleted. Remarkably, the percentage of incomplete losses in backbone genes of secondary metabolite gene clusters was the highest of any analyzed gene category, as we found that 5 out of 12 PKS or NRPS genes (41.7%) were lost in at least one isolate (41.7%; fig. 3). Gene losses affected gene clusters either almost entirely (e.g., PKS9 gene cluster; fig. 4A) or only partially (e.g., NRPS4 or PKS10 gene cluster; supplementary fig. S3, Supplementary Material online). None of the investigated secondary metabolite gene clusters was recently gained. We analyzed the recent loss of the PKS9 gene cluster in more detail (fig. 4B). The PKS9 gene cluster is expressed in planta (Rudd etal. 2015; Palma-Guerrero etal. 2017). We analyzed the chromosomal sequence of a genome which lacked PKS9. We found that isolate ST99CH_3D1 retained the two PKS9 cluster genes encoding a transporter and a cytochrome c heme lyase. Furthermore, the isolate retained a gene encoding an additional ATP-binding cassette transporter immediately adjacent to the cluster. Next, we identified homologs of the same genes in genomes of three closely related species, Z. ardabiliae (n = 4), Z. brevis (n = 1), and Z. pseudotritici (n = 5). In all Z. ardabiliae and Z. pseudotritici genomes, the gene order within the cluster was conserved. In Z. brevis, we identified orthologs of all cluster genes, however the cluster was not assembled as a single scaffold. These results strongly suggest that the PKS9 gene cluster was gained in the ancestor of Z. brevis and Z. tritici. The absence of the cluster in Z. pseudotritici may be due to a secondary loss. Alternatively, the PKS9 gene cluster may never have reached fixation in any of the species and the lack of evidence in Z. pseudotritici may be a sampling artifact.
Fig. 4.
Evolution of the polyketide synthase (PKS) 9 gene cluster in Zymoseptoria tritici and the sister species Z. pseudotritici, Z. brevis, and Z. ardabiliae. (A) Gene presence–absence polymorphism affecting the PKS9 gene cluster among the 123 Z. tritici isolates grouped by population. The physical position of each gene is shown below. (B) Comparative genomics analyses of the PKS9 gene cluster in Z. tritici and the three closest known sister species Z. pseudotritici, Z. brevis, and Z. ardabiliae. On the left is a schematic tree representing the phylogenetic relationships between Z. tritici and three sister species. In Z. tritici, two segregating variants of the PKS9 gene cluster were found. Isolate ST99CH_3D1 is lacking nearly all genes of the PKS9 cluster compared with the IPO323 reference genome. Analyses of the homologous regions in five Z. pseudotritici and four Z. ardabiliae genomes showed that the gene cluster was missing with the exception of two genes encoding an ABC transporter and a cytochrome c heme lyase, respectively. In Z. brevis, all genes of the PKS9 cluster were present, however orthologs were split on three different genome assembly scaffolds.
Population Structure and Selection Signatures on Gene Losses and Gene Gains
Zymoseptoria tritici isolates used in this study were collected in four worldwide locations (fig. 5A). We analyzed the population structure of presence-absence polymorphism generated by incomplete gains and losses using principal component analyses (fig. 5B). The first two principal components clearly clustered the isolates by geographic origin. This clustering recapitulates the population structure identified at single nucleotide polymorphisms (SNPs).
Fig. 5.
Population structure and allele frequency spectra of incomplete gene gains and losses. (A) Sampling locations of the 123 Zymoseptoria tritici isolates. (B) Principal component analysis of the population structure found at loci segregating incomplete gene losses, incomplete gene gains and a set of 1,457 genome-wide synonymous single nucleotide polymorphisms (SNPs). The percentage of variance explained by each principal component is shown in parentheses. (C) Allele frequency spectra of incomplete gene gains and losses within populations. The allele frequency spectra were contrasted with the allele frequency spectrum of the derived allele at synonymous SNPs (n = 237,185).
To test whether selection acted on gene gain and loss polymorphisms, we contrasted the allele frequency spectrum of incomplete gene gains and losses with derived allele frequencies at synonymous SNPs (fig. 5C). We performed comparisons of allele frequency spectra in each population separately. In all four populations, we found a strong excess of low-frequency variants (frequency ≤ 10%) and a reduction of high-frequency variants (frequency ≥ 70%) for incomplete gene losses compared with synonymous SNPs (Pearson’s chi-squared tests in Australia: χ2 = 335.99, P < 2.2×10−16; Switzerland: χ2 = 183.38, P < 2.2×10−16; Israel: χ2 = 130.79, P < 2.2×10−16; and Oregon: χ2 = 314.84, P < 2.2×10−16). This pattern suggests that gene losses were under negative selection in all populations. The allele frequency spectra of incomplete gene gains differed among populations. We found an excess of low-frequency variants (frequency ≤ 10%) for incomplete gene gains compared with synonymous SNPs in the Australian and Oregon populations, suggesting that incomplete gene gains were also under negative selection in these populations. However, the excess of rare variants of incomplete gene gains compared with synonymous SNPs was significantly lower than the one observed for incomplete gene losses (Pearson’s chi-squared tests in Australia: χ2 = 90.32, P = 1.28×10−9 and Oregon: χ2 = 82.58, P = 3.82×10−4). In contrast, in the Swiss and Israel populations, the frequency spectrum of incomplete gene gains largely matched the spectrum of synonymous SNP allele frequencies for low-frequency variants (frequency ≤ 10%). In these two populations, we also observed a slight excess of intermediate frequency variants (60% ≤ frequency ≤ 70%) for incomplete gene gains compared with synonymous SNP allele frequencies (Pearson’s chi-squared tests in Switzerland: χ2 = 79.944, P = 1.16×10−7 and Israel: χ2 = 40.988, P = 8.27×10−3). This excess could be indicative of balancing selection maintaining an allele at intermediate frequency. To investigate the differences observed between populations, we computed population size (Ne) estimates based on genome-wide SNPs (supplementary table S6, Supplementary Material online). The analyses showed that each of the four field populations had large effective population sizes.
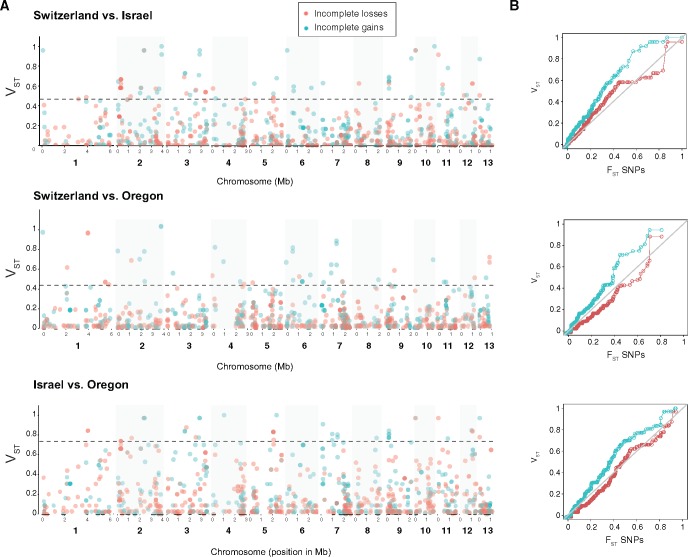
The majority of recent gene gains and losses were shared by at least two populations, although a significant proportion of events were population-specific (supplementary fig. S4, Supplementary Material online). We used VST to compute the differentiation in allele frequencies at gene gain and loss loci between pairs of the three most closely related populations (Israel, Oregon, and Switzerland). We compared the distribution of VST to the distribution of FST values of synonymous SNPs (fig. 6B). The Australian population was excluded in this analysis due to the high mean SNP FST against the other populations (mean FST = 0.28). In all three pairwise comparisons, we found that incomplete gene gains had a strong excess of high VST (>0.3) compared with synonymous SNPs and incomplete gene losses. Conversely, incomplete gene loss VST values largely followed the SNP FST values in all pairwise comparisons. There was a slight excess of extreme VST values (VST > 0.8) for incomplete gene losses compared with FST values at SNPs. Incomplete gene gains showed higher population differentiation than incomplete gene losses.
Fig. 6.
Pairwise comparisons of population differentiation of incomplete gene gain and loss frequencies. (A) Distribution of VST values of presence/absence polymorphism at gene gain and loss loci. The dashed line shows the 97.5th percentile of the distribution of corresponding pairwise FST value for genome-wide synonymous SNPs (n = 1,457). (B) Quantile–quantile plots comparing the distribution of VST for incomplete gene gains and losses and the distribution of FST for the genome-wide synonymous SNPs. Points should follow the diagonal line if distribution of FST and VST were equal.
Highly differentiated gene gain and loss allele frequencies is indicative of local adaptation. We focused on VST outliers to identify genes with excessive variation in incomplete loss and gain frequencies among populations (fig. 6A;supplementary tables S7 and S8, Supplementary Material online). We identified a total of 52 genes with highly differentiated incomplete losses frequencies. This set of genes included six genes encoding secreted proteins and genes belonging to the NRPS4 and PKS10 gene clusters (supplementary fig. S3, Supplementary Material online). Furthermore, we identified a total of 66 recently gained genes with highly differentiated allele frequencies. We found seven genes encoding secreted proteins. Three out of these were encoding small secreted proteins (genes 2_00001, 3_00158, and 8_00609). The loss of gene 8_00609 was previously found to be driven by host specialization (Hartmann etal. 2017). The gene 2_00001 (showing excess differentiation in two pairwise comparisons) is highly upregulated during infection (Rudd etal. 2015).
Discussion
We identified extensive standing genetic variation in populations of a fungal pathogen due to recent gene gains and losses. Genes affected by recent gains and losses encoded for functions in fungal virulence and ecological adaptation. Several recent gene gains and losses segregating in the species are likely linked to host specialization and under divergent selection among populations. We found generally strong selection against recent gene losses and varying strengths of selection against recent gene gains showing that these categories of gene presence-absence polymorphisms are on distinct evolutionary trajectories in populations.
High Intraspecific Gene Content Variation through Gene Gain and Loss in Compact Fungal Genomes
Variation in gene content is common within species and was found in all kingdoms of life (Schrider and Hahn 2010; Żmieńko etal. 2013). We found that gene gains and losses segregating in Z. tritici populations affected a total of 1,623 genes (14.6%). Our results were consistent with previous analyses of Z. tritici sampled in Australia (McDonald etal. 2016). Similarly, a comparison of two completely assembled Z. tritici genomes showed that each isolate was lacking several hundred of genes of the other isolate (Plissonneau etal. 2016). Intraspecific variation in gene content was also found in other fungi including the human fungal pathogen Cryptococcus gattii (Steenwyk etal. 2016) and among the compact genomes of the budding yeast Saccharomyces cerevisiae (Strope etal. 2015). Although fungal genomes are often compact and haploid, the levels of intraspecific gene content variation in fungi are comparable to the levels of gene content variation reported in plants and mammals (Żmieńko etal. 2013; Zarrei etal. 2015).
The well-studied evolutionary history of Z. tritici and multiple closely related species that diverged within <18,500 years provides a powerful comparative genomics framework (Stukenbrock etal. 2007; Stukenbrock, Quaedvlieg, etal. 2012; Grandaubert etal. 2015). A large number of well-defined orthologs allows to distinguish whether a gene presence-absence polymorphism was due to a recent gene gain or gene loss. We defined a recent gene gain as a segregating gene presence-absence polymorphism within Z. tritici for which no ortholog exists among any of the closely related species, that is, presence-absence polymorphism of a Z. tritici orphan gene. The most parsimonious explanation for such polymorphism is that the gene gain has not yet reached fixation within the species. Alternatively, the gene may have already reached fixation in the recent past and the polymorphism is evidence for a secondary gene loss. However, both scenarios imply that the gene constitutes an evolutionary novelty since the last speciation (∼10,500 years BP). A gene gain may erronously be identified if closely related species experienced independent losses of the same gene. As we analyzed multiple genomes from each of three related species of Z. tritici, the identified orphan genes were unlikely to be the product of gene losses in related species.
A species may gain an orphan gene following a gene duplication event if one gene copy diverges extensively and acquires a new function (Tautz and Domazet-Lošo 2011). Alternatively, genes may evolve de novo from noncoding DNA. This mechanism relies on the spontaneous evolution of an open reading frame (ORF) and gain of cis-regulatory elements (McLysaght and Guerzoni 2015). De novo genes are characterized by generally low expression levels and may have short lifespans in populations due to genetic drift (Carvunis etal. 2012; Palmieri etal. 2014). Some of the genes recently gained in Z. tritici shared characteristics with de novo genes including short coding sequences and low transcription levels. Alternatively, horizontal gene transfer can be a source of orphan genes in species. Orphan genes acquired from bacteria or plants led to major gains of function in plant pathogens (de Jonge etal. 2012; Ropars etal. 2015).
Both the proximity to telomeres and to repetitive elements increased the likelihood of segregating gene gain and loss polymorphisms in populations. These trends were consistent with nonallelic homologous recombination (NAHR) and retrotransposition caused by transposable elements that can cause major gene losses (Hastings etal. 2009; Carvalho and Lupski 2016). Indeed, subtelomeric regions in Z. tritici are rich in recombination hotspots and transposable elements (Goodwin etal. 2011; Croll etal. 2015). Recombination hotspots are the primary target for NAHR events. Furthermore, retrotransposition of transposable elements may also cause rearrangements as excision induces the repair of double-strand breaks (Kazazian and Goodier 2002).
Gene Dispensability and Host–Pathogen Coevolution
We found that genes encoding functions in basic cellular processes were significantly less affected by gene gains and losses than other gene categories. This result is consistent with strong selection acting against loss of essential functions in a haploid organism (Cheeseman etal. 2016). In contrast, we found an enrichment in functions associated with membrane-bound, secretion, and nutrition-specific metabolic pathways among genes affected by incomplete gains and losses. Functions associated with secretion and nutrition are playing a major role in the interaction of pathogens with the host and environment (Monod etal. 2002; Presti etal. 2015). Recent gene losses were also frequent in gene categories with a role in fungal virulence. Functional redundancy and environment-specific dispensability (i.e., a gene function is essential only in specific environmental conditions) are two major factors thought to lead to gene dispensability (Albalat and Cañestro 2016). This is consistent with our data showing high rates of recent gain and loss events in genes involved in the interaction with the host and the environment. For plant pathogens, both hosts and the environment are highly heterogeneous over space and time.
We found strong differences in the proportions of recent gene gains and losses in three gene categories involved in host interactions. Genes encoding cell wall degrading enzymes were largely retained and constituted a clear exception to the high levels of gene dispensability for secretion and nutrition functions. This is consistent with evidence of pervasive purifying selection acting on genes encoding cell wall degrading enzymes in Z. tritici and closely related species (Brunner etal. 2013). Genes encoding small secreted proteins (a protein category frequently involved in virulence expression on the host) showed both high levels of recent gene gains and losses. This shows that this gene category is rapidly evolving and has high levels of dispensability. Recent gene losses were remarkably frequent in gene clusters responsible for secondary metabolite production. The evolution of the PKS9 cluster showed that entire secondary metabolite gene clusters can be readily gained and lost, as we found evidence that the cluster was recently acquired in an ancestor to Z. tritici and readily lost in Z. pseudotritici and Z. tritici. The highly uneven rates of recent gene gains and losses among different gene categories strongly suggests that both mechanistic factors of replication fidelity and selection for novel functions play an important role in the evolution of gene content in a species.
Evidence for Selection Driving Gene Gains and Losses in Populations
We tested for selection on gene gains and losses at two separate levels: the level of differentiation among populations at gene presence–absence frequencies among populations and deviations from neutral expectations of gene presence–absence frequencies within populations. The former analysis is designed to investigate evidence of local adaptation driven by gene gains and losses and the latter analysis tests for the overall impact of selection on gene gains and losses across the genome. We found that gene gain and loss loci showed an excess in high fixation indices compared with differentiation levels at neutral markers. Although high fixation indices at individual loci may be caused by very restricted gene flow, overall there is evidence for divergent selection acting on recent gene gains and losses.
For example, divergent selection among populations was found for the recently gained gene 8_00609 encoding a small secreted protein. The presence-absence polymorphism at this gene was previously shown to underlie host specialization on a specific wheat variety (Hartmann etal. 2017). The encoded protein is likely detected by the host immune system and, hence, isolates lacking the gene were able to evade detection during infection. Two additional, recently gained genes encoding small secreted proteins (2_00001, 3_00158) showed highly differentiated gene presence-absence frequencies among populations. Recently gene gained genes that have not yet reached fixation in the species, hence, likely play an important role in the interaction with the host and adaptation of the pathogen in different environments.
We found evidence for divergent selection on recent gene losses in PKS and NRPS secondary metabolite gene clusters. The function of secondary metabolite gene clusters in Z. tritici is largely unknown; however, secondary metabolites play important roles in pathogenicity and ecological interactions in many pathogens (Howlett 2006; Stergiopoulos etal. 2013). Gene clusters, in particular the backbone genes, are highly conserved among fungi and frequently subject to horizontal gene transfers (Wisecaver and Rokas 2015). Our study shows that recently lost gene clusters can be under divergent selection within a species. This may be due to differences in environmental conditions disfavoring the production of specific metabolites. The evidence for divergent selection for recently lost gene clusters shows how population genetic process can explain variation in gene cluster content among closely related species.
Distinct Evolutionary Trajectory of Gene Losses and Gene Gains in Populations
The distinct mechanisms by which gene gains and gene losses are generated and how recently gained genes are functionally distinct from recently lost genes strongly suggests that selection should act differently upon each category of gene presence-absence polymorphism. Selection against deleterious copy number variation (CNV) should be particularly strong in a haploid organism such as Z. tritici because deletions cannot be shielded from selection in the hemizygous state. We found indeed that incomplete gene losses showed strong signatures of negative selection, whereas the incomplete gene gains showed either weak or no signature of negative selection. Differences in the strength of selection against gene gains could be explained by differences in effective population size. However, we found no strong association between signatures of selection and population sizes. Both gene gain and loss polymorphisms recapitulated the levels of population differentiation at neutral markers. Incomplete gene gains were more stratified among populations than incomplete gene losses. This may be caused both by stronger divergent selection on gene gains and universally negative selection against gene losses among populations. Differences in selection pressure acting on gene deletions and duplications were found in populations of humans (Sudmant etal. 2015), fruit flies (Emerson etal. 2008; Cardoso-Moreira etal. 2016) and malaria parasites (Cheeseman etal. 2016). Deletions were generally more deleterious than duplications as the latter are likely to initially be selectively neutral. Our study showed that recent gene gains were on similar evolutionary trajectories within the species than gene duplications in a number of different taxa.
Positive selection on individual gene duplications or horizontally transferred genes can be an important driver to fix CNV within species (Innan and Kondrashov 2010; Ropars etal. 2015; Cardoso-Moreira etal. 2016). We showed that a subset of the genes gained de novo in Z. tritici experienced similarly strong positive selection to adapt either to the host or to the environmental conditions. The large panmictic populations maintained a large number of recent gene gains and losses at high frequency serving as a reservoir for rapid adaptive evolution.
Materials and Methods
Fungal Isolate Collection and Illumina Whole-Genome Sequencing
A total of 130 field isolates of Z. tritici were included in this study. The isolates originated from four different geographical locations including Australia, Israel, Switzerland and USA (Oregon). Each population comprised 22–50 isolates and were sampled between 1990 and 2001. Sampling years and sampling locations of these four populations were described previously (Zhan etal. 2005). Isolates were stored for long-term use in silica and saved at −80 °C after sampling. No clonal genotypes were found among these isolates in previous genetic diversity analyses (Linde etal. 2002). We used the whole-genome Illumina sequencing data of 106 isolates that were deposited on the NCBI Short Read Archive under the BioProject ID numbers PRJNA178194 and PRJNA327615 (Torriani etal. 2011; Croll etal. 2013; Hartmann etal. 2017). We generated raw sequencing data for 24 additional isolates. As for the previously available data sets, we extracted high-quality genomic DNA from liquid cultures and performed 100-bp paired-end sequencing with an insert size of ca. 500 bp on the Illumina HiSeq2000 platform. The generated raw sequencing data was deposited on the NCBI Short Read Archive under the BioProject ID PRJNA327615 (supplementary table S1, Supplementary Material online).
Read Mapping
Raw Illumina reads were screened for adapter contamination and trimmed for sequencing quality using Trimmomatic v0.32 (Bolger etal. 2014). The following settings were used: illuminaclip = TruSeq3-PE.fa:2:30:10, leading = 10, trailing = 10, slidingwindow = 5:10, minlen = 50. Then, sequence data from all isolates was aligned to the gapless, telomere-to-teleomere assembly of the reference genoms isolate IPO323 (Goodwin etal. 2011) accessed from EnsemblFungi (Flicek etal. 2014). The short read aligner Bowtie 2 version 2.2.3 (Langmead etal. 2009) was used for read alignment with the following settings: –very-sensitive-local –phred33 -X 1000. Reads were marked as PCR duplicates using the MarkDuplicates module of Picard tools version 1.118 (http://broadinstitute.github.io/picard). The genome-wide coverage of the 130 sequenced isolates was calculated as the number of mapped reads multiplied by average read length and divided by the genome size.
CNV Calling
We used CNVnator (Abyzov etal. 2011) to perform a statistical analysis of short read coverage along the 13 core chromosomes in order to predict CNV events in the 130 sequenced isolates compared with the reference genome. For each isolate, we assessed CNV events in bins of 100 bp as recommended. We retained CNV calls according to the following filtering criteria: length >500 bp, P < 0.05, and q0 < 0.5. Additionally, we filtered CNV calls for the normalized average read depth signal. We kept only deletions with normalized average read depth of <0.4 and duplications with normalized average read depth of >1.6. After CNV calling and filtration for quality, the complete data set comprised 81,550 deletions and 5,600 duplications.
Full or Partial Chromosomal Duplications
Seven isolates (a12_3B_11, a15_3B_10, AUS_1E4, ST99CH_3C4, ISY_Ar_5a, a15_2a_14, a15_2a_15) showed evidence for full or partial chromosomal duplications based on chromosome-wide analyses of predicted gene CNV events (data not shown). We calculated the per-chromosome read depth for these seven isolates using the bedtools “genomecov” command (Quinlan and Hall 2010). Suspected chromosomal duplications had a per-chromosome read depth >1.5× of the genome-wide coverage, which strongly suggested that these chromosomes were partially or entirely duplicated. As partial or full chromosomal duplications were likely caused by different mechanisms than individual gene duplications, we excluded the affected isolates from further analyses and retained 123 isolates.
Identification of Genes Affected by Presence–Absence Polymorphism
To identify genes affected by presence–absence polymorphism, we focused our analyses on genes affected by deletions compared with the reference genome. For this, we calculated the percentage of overlap between deletion events and genes using the bedtools “intersect” command (Quinlan and Hall 2010). We retrieved high-quality gene models predicted for the IPO323 genome using deep transcriptomics data (Grandaubert etal. 2015). A gene was considered as affected by a presence–absence polymorphism event if the deletion event was overlapping >90% of the gene. For the genes that were affected by multiple deletions, we calculated the percentage overlap based on the sum of all deletion events affecting the gene. The genes that were affected by both a deletion and a duplication event were excluded from further analyses. We validated gene presence–absence polymorphism calls using complete genome assemblies and direct amplifications of target loci (see supplementary Materials and Methods in Supplementary Material online).
Validation of Gene Presence–Absence Polymorphism Using Comparative Genomics
We compared genomic sequences of the two completely assembled genomes available for the species. The genomes included the reference genome isolate IPO323 (Goodwin etal. 2011) and a full assembly of the Swiss ST99CH_3D7 isolate (Plissonneau etal. 2016). We used blastn to screen IPO323 transcripts sequences against the ST99CH_3D7 genome assembly (Camacho etal. 2009). For each IPO323 transcript, we retained the best hit in ST99CH_3D7 based bitscores. We identified an IPO323 transcript sequence to be missing in ST99CH_3D7 if either of the following conditions were met: 1) the bitscore was inferior to 100, 2) the percent identity of the IPO323 transcript and the best ST99CH_3D7 hit was <90%, 3) the length of all blast hits in ST99CH_3D7 was <10% of the IPO323 transcript length used as a query, and 4) evidence of gene disruption such as exon loss was retrieved from blast analysis.
Manual Inspection of Gene Presence–Absence Polymorphism Events
Genes encoding small secreted proteins (SSPs) in plant pathogenic fungi are often located in proximity to regions enriched in transposable elements (TEs) and complex sequence rearrangements (de Jonge etal. 2011; Dong etal. 2015). The identification of CNV events is likely more error-prone in such regions (Teo etal. 2012). Therefore, we performed manual curation of presence–absence polymorphism events affecting genes encoding SSPs. For isolates predicted to lack particular genes encoding SSPs, we visualized the read mapping to the reference genome and presence–absence polymorphism event calls using the IGV genome browser (Robinson etal. 2011; Thorvaldsdóttir etal. 2013). We inspected the consistency of read coverage and prediction of deletion events. Furthermore, we collected independent evidence for the presence or absence of specific genes encoding SSPs using blast searches against de novo assemblies of each isolate. For this, we performed assemblies of the Illumina read data of each isolate using SPAdes 3.6.0 (Bankevich etal. 2012). We used BayesHammer read correction prior to assembly. The de novo assembly was performed with a k-mer range of “21,29,37,45,53,61,79,87” and the assembly was polished using MismatchCorrector. De novo assemblies for each of the isolates were used for BLAST searches using the blastn command of the ncbi-blast-2.2.30+ software (Camacho etal. 2009). We kept only presence–absence polymorphism events affecting genes encoding SSPs that were supported both by IGV visualization and BLAST searches. We excluded 192 out of 824 presence–absence polymorphism events of genes encoding SSPs.
Validation of Gene Presence–Absence Polymorphisms by PCR Assay
We validated a subset of gene presence–absence polymorphism calls in 95 randomly selected isolates using PCR assays. We selected 14 genes affected by CNV events belonging to the cell wall degrading enzymes, secondary metabolite gene clusters, and SSP gene categories. We aimed to have a balanced representation of gene presence–absence polymorphisms segregating at low, intermediate, and high frequencies in the populations. For each locus, we selected PCR amplicons of ca. 500 bp in a conserved region of the gene or in the immediate flanking sequences. To identify conserved regions, we used consensus sequences obtained from de novo assemblies of each isolate. Primers were designed using Primer 3.0 (Rozen and Skaletsky 2000; supplementary table S9, Supplementary Material online). For PCR reactions, we used a similar protocol as described in (Croll etal. 2013). PCR reactions were conducted in a 20-µl volume containing 5–10 ng genomic DNA, 0.5 mM each of forward and backward primers, 0.25 mM dNTP, 0.6 U Taq polymerase (DreamTaq, Thermo Fisher, Inc.), PCR buffer. PCR products were amplified for 33–35 cycles. The resulting amplicons were examined on 1% agarose gels. Each PCR reaction contained a primer pair of a microsatellite locus as a positive control to ascertain amplification (Goodwin etal. 2007).
Identification of Incomplete Gene Losses and Gene Gains
For each gene affected by presence–absence polymorphism, we defined whether the polymorphism was due to an incomplete gene loss event or an incomplete gene gain based on orthology data from three closely related species from the Zymoseptoria genus (Grandaubert etal. 2015). Grandaubert etal. identified a total of 9,890 genes that have an ortholog in at least one genome of Z. pseudotritici (n = 5), Z. brevis (n = 1), or Z. ardabiliae (n = 4). These orthologs were called Zymoseptoria core genes. A total of 1,221 that had no ortholog in any of the analyzed genomes (Grandaubert etal. 2015) were called Z. tritici orphan genes. We defined incomplete (i.e., recent) gene losses as any presence–absence polymorphism affecting a core Zymoseptoria genes and we defined an incomplete (i.e., recent) gene gain any presence–absence polymorphism affecting a Z. tritici orphan gene. Genome-wide gene losses and gains event distributions were visualized using the R package {ggplot2} (Wickham 2009) and Circos version 0.67-7 (Krzywinski etal. 2009). Pearson’s chi-squared and multiple comparison tests after Kruskal–Wallis were performed using the open source software R.
Gene Ontology Analysis and Functional Annotation
We functionally annotated the gene models for the 11,111 genes described on the core chromosomes previously (Grandaubert etal. 2015). For this, we used InterProScan v.5.16-55.0 (Jones etal. 2014) to assign protein sequence motifs to PFAM and GO terms based on hidden Markov models. Protein sequences were additionally screened for evidence of a secretion signal, transmembrane, cytoplasmic, and extracellular domains using a combination of SignalP v.4.1 (Petersen etal. 2011), Phobius v.1.01 (Käll etal. 2007), and TMHMM v.2.0 (Krogh etal. 2001). Locations of transposable elements were annotated previously (Grandaubert etal. 2015).
Identification of Orthologs of Secondary Metabolite Gene Clusters in Zymoseptoria Species
To identify homologs of genes found in secondary metabolite gene clusters, we performed BLAST analyses using blastn version 2.2.31+ (Camacho etal. 2009). The genomes of five Z. pseudotritici, four Z. ardabiliae isolates, one Z. brevis, and one Z. passerinii isolates were downloaded from NCBI under the accession numbers PRJNA63035, PRJNA277173, PRJNA63037, PRJNA63039, PRJNA46489, PRJNA63043, PRJNA277174, PRJNA63045, PRJNA63047, PRJNA63049, and PRJNA274679 (Stukenbrock etal. 2011).
SNPs Calling Procedure
SNPs calling was performed as described previously (Croll etal. 2013; Hartmann etal. 2017). In summary, the HaplotypeCaller tool of the Genome Analysis Toolkit (GATK) version 3.3-0 was used (McKenna etal. 2010). To further validate the identified SNPs, we used the independent SNP caller Freebayes v.0.9 (Garrison and Marth 2012) and filtered for quality and genotyping rate (>90%). We excluded SNPs located on accessory chromosomes, tri-allelic SNPs, and finally retained a total of 1,375,999 SNPs. To identify ancestral alleles at SNPs, we analyzed whole-genome sequencing data of the two closest known sister species of Z. tritici. We used raw Illumina reads of four Z. pseudotritici isolates (STIR04_2.2.1, STIR04_3.11.1, STIR04_5.3, STIR04_5.9.1) and four Z. ardabiliae isolates (STIR04_1.1.1, STIR04_1.1.2, STIR04_3.13.1, STIR04_3.3.2) (Stukenbrock etal. 2007; Stukenbrock, Christiansen, etal. 2012). We retained SNPs that had a genotyping rate >50% and were monomorphic within the respective sister species. Then, we assigned ancestral alleles for any Z. tritici SNP if an allele was shared and retained in both sister species. We were able to assign ancestral alleles for 584,327 SNPs. We annotated and predicted the effect of SNPs using SnpEff 4.3i (Cingolani, Platts, etal. 2012). We selected the 237,185 synonymous SNPs using SnpSift (Cingolani, Patel, etal. 2012). We generated a randomly chosen subset of synonymous SNPs at equal distance (20 kb) across the genome (1,457 bi-allelic SNPs). This subset of SNPs was assumed to be largely in linkage equilibrium given previous estimates of linkage equilibrium decay in Z. tritici populations (Croll etal. 2015).
Population Genomics Analyses
To estimate population differentiation of allele frequencies at genes affected by incomplete loss and gain, we calculated the variant fixation index VST (Redon etal. 2006). VST is a variant of the FST index (Wright 1951; Nei 1973) and is frequently used to identify differentiated CNV between populations (Redon etal. 2006; Pezer etal. 2015; Steenwyk etal. 2016). For each pairwise population comparison, VST was calculated as VST = (VT–VS)/VT, where VT is the total variance in copy numbers between the two populations and VS is the average of the variance within each single population, weighted for its sample size. Additionally, we calculated pairwise FST values for the genome-wide 1,457 synonymous SNPs (Wright 1951; Nei 1973). We used the R package {hierfstat} (Goudet 2005) that implements Yang’s algorithm (Yang 1998). We used VST outlier detection scans to identify loci with highly differentiated loss or gain frequencies between populations. The 97.5th percentile of the distribution of FST for SNPs was used as a threshold for outlier detection in each pairwise comparison. To estimate the effective population size (Ne), we used 1,457 synonymous SNPs that were equally spaced to avoid linkage disequilibrium due to physical proximity. Ne estimates were calculated using the linkage disequilibrium approach (Waples and Do 2008) implemented in NeEstimator v2.01 (Do etal. 2014). Allele frequency spectra of incomplete gene losses and incomplete gene gains against allele frequency spectra of synonymous SNPs were compared using Pearson’s chi-squared tests on contingency tables of derived SNP allele and gene absence allele counts. Statistical tests were computed in each population separately using the open source software R.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We are grateful to Eva H. Stukenbrock and Jonathan Grandaubert for providing access to data sets. We thank Marcello Zala for assistance in the laboratory. Bruce A. McDonald provided helpful feedback on an earlier version of this manuscript. This work was supported by the ETH Zurich (Research Grant 12-03).
References
- Abyzov A, Urban AE, Snyder M, Gerstein M.. 2011. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 21:974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albalat R, Cañestro C.. 2016. Evolution by gene loss. Nat Rev Genet. 17:379–391. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomme T, Vandepoele K, De Bodt S, Simillion C, Maere S, Van de Peer Y.. 2006. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 7:R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakhage AA. 2013. Regulation of fungal secondary metabolism. Nat Rev Microbiol. 11:21–32. [DOI] [PubMed] [Google Scholar]
- Brunner PC, Torriani SFF, Croll D, Stukenbrock EH, McDonald BA.. 2013. Coevolution and life cycle specialization of plant cell wall degrading enzymes in a hemibiotrophic pathogen. Mol Biol Evol. 30:1337–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL.. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso-Moreira M, Arguello JR, Gottipati S, Harshman LG, Grenier JK, Clark AG.. 2016. Evidence for the fixation of gene duplications by positive selection in Drosophila. Genome Res. 26:787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CMB, Lupski JR.. 2016. Mechanisms underlying structural variant formation in genomic disorders. Nat Rev Genet. 17:224–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvunis AR, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, Charloteaux B, Hidalgo CA, Barbette J, Santhanam B, et al. 2012. Proto-genes and de novo gene birth. Nature 487:370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IH, Miller B, Tan JC, Tan A, Nair S, Nkhoma SC, De Donato M, Rodulfo H, Dondorp A, Branch OH, et al. 2016. Population structure shapes copy number variation in malaria parasites. Mol Biol Evol. 33:603–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Patel VM, Coon M, Nguyen T, Land SJ, Ruden DM, Lu X.. 2012. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front Genet. 3:35.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM.. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK.. 2006. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 38:75–81. [DOI] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, et al. 2010. Origins and functional impact of copy number variation in the human genome. Nature 464:704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll D, Lendenmann MH, Stewart E, McDonald BA.. 2015. The impact of recombination hotspots on genome evolution of a fungal plant pathogen. Genetics 201:1213–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll D, McDonald BA.. 2012. The accessory genome as a cradle for adaptive evolution in pathogens. PLoS Pathog. 8:e1002608.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll D, Zala M, McDonald BA.. 2013. Breakage-fusion-bridge cycles and large insertions contribute to the rapid evolution of accessory chromosomes in a fungal pathogen. PLoS Genet. 9:e1003567.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge R, Bolton MD, Thomma BP.. 2011. How filamentous pathogens co-opt plants: the ins and outs of fungal effectors. Curr Opin Plant Biol. 14:400–406. [DOI] [PubMed] [Google Scholar]
- de Jonge R, van Esse HP, Maruthachalam K, Bolton MD, Santhanam P, Saber MK, Zhang Z, Usami T, Lievens B, Subbarao KV, et al. 2012. Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc Natl Acad Sci. 109:5110–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Amaral AM, Antoniw J, Rudd JJ, Hammond-Kosack KE.. 2012. Defining the predicted protein secretome of the fungal wheat leaf pathogen Mycosphaerella graminicola. PLoS One 7:e49904.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR.. 2014. NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour. 14:209–214. [DOI] [PubMed] [Google Scholar]
- Dong S, Raffaele S, Kamoun S.. 2015. The two-speed genomes of filamentous pathogens: waltz with plants. Curr Opin Genet Dev. 35:57–65. [DOI] [PubMed] [Google Scholar]
- Emerson JJ, Cardoso-Moreira M, Borevitz JO, Long M.. 2008. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science 320:1629–1631. [DOI] [PubMed] [Google Scholar]
- Esquerré-Tugayé M-T, Boudart G, Dumas B.. 2000. Cell wall degrading enzymes, inhibitory proteins, and oligosaccharides participate in the molecular dialogue between plants and pathogens. Plant Physiol Biochem. 38:157–163. [Google Scholar]
- Fisher MC, Gow NAR, Gurr SJ.. 2016. Tackling emerging fungal threats to animal health, food security and ecosystem resilience. Phil Trans R Soc B 371:20160332.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. 2014. Ensembl 2014. Nucleic Acids Res. 42:D749–D755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fones H, Gurr S.. 2015. The impact of Septoria tritici blotch disease on wheat: an EU perspective. Fungal Genet Biol. 79:3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen TL, Stukenbrock EH, Liu Z, Meinhardt S, Ling H, Faris JD, Rasmussen JB, Solomon PS, McDonald BA, Oliver RP.. 2006. Emergence of a new disease as a result of interspecific virulence gene transfer. Nat Genet. 38:953–956. [DOI] [PubMed] [Google Scholar]
- Garrison E, Marth G.. 2012. Haplotype-based variant detection from short-read sequencing. Unpublished data [cited 2017 Apr 26]. Available from: https://arxiv.org/abs/1207.3907.
- Goodwin SB, van der Lee TAJ, Cavaletto JR, te Lintel Hekkert B, Crane CF, Kema GHJ.. 2007. Identification and genetic mapping of highly polymorphic microsatellite loci from an EST database of the septoria tritici blotch pathogen Mycosphaerella graminicola. Fungal Genet Biol. 44:398–414. [DOI] [PubMed] [Google Scholar]
- Goodwin SB, M’Barek SB, Dhillon B, Wittenberg AHJ, Crane CF, Hane JK, Foster AJ, Lee TAJV d, Grimwood J, Aerts A, et al. 2011. Finished genome of the fungal wheat pathogen Mycosphaerella graminicola reveals dispensome structure, chromosome plasticity, and stealth pathogenesis. PLoS Genet. 7:e1002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J. 2005. hierfstat, a package for r to compute and test hierarchical F-statistics. Mol Ecol Notes 5:184–186. [Google Scholar]
- Grandaubert J, Bhattacharyya A, Stukenbrock EH.. 2015. RNA-seq-based gene annotation and comparative genomics of four fungal grass pathogens in the genus Zymoseptoria identify novel orphan genes and species-specific invasions of transposable elements. G3 5:1323–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann FE, Sánchez-Vallet A, McDonald BA, Croll D.. 2017. A fungal wheat pathogen evolved host specialization by extensive chromosomal rearrangements. ISME J. 11:1189–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G.. 2009. Mechanisms of change in gene copy number. Nat Rev Genet. 10:551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrichsen CN, Chaignat E, Reymond A.. 2009. Copy number variants, diseases and gene expression. Hum Mol Genet. 18:R1–R8. [DOI] [PubMed] [Google Scholar]
- Howlett BJ. 2006. Secondary metabolite toxins and nutrition of plant pathogenic fungi. Curr Opin Plant Biol. 9:371–375. [DOI] [PubMed] [Google Scholar]
- Innan H, Kondrashov F.. 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 11:97–108. [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL.. 2006. The plant immune system. Nature 444:323–329. [DOI] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L, Krogh A, Sonnhammer ELL.. 2007. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res. 35:W429–W432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH Jr, Goodier JL.. 2002. LINE drive. Retrotransposition and genome instability. Cell 110:277–280. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL.. 2001. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol. 305:567–580. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol İ, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA.. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linde CC, Zhan J, McDonald BA.. 2002. Population structure of Mycosphaerella graminicola: from lesions to continents. Phytopathology 92:946–955. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS.. 2003. The origins of genome complexity. Science 302:1401–1404. [DOI] [PubMed] [Google Scholar]
- McDonald MC, McGinness L, Hane JK, Williams AH, Milgate A, Solomon PS.. 2016. Utilizing gene tree variation to identify candidate effector genes in Zymoseptoria tritici. G3 6:779–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLysaght A, Guerzoni D.. 2015. New genes from non-coding sequence: the role of de novo protein-coding genes in eukaryotic evolutionary innovation. Philos Trans R Soc B 370:20140332.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod M, Capoccia S, Léchenne B, Zaugg C, Holdom M, Jousson O.. 2002. Secreted proteases from pathogenic fungi. Int J Med Microbiol. 292:405–419. [DOI] [PubMed] [Google Scholar]
- Morris JJ, Lenski RE, Zinser ER.. 2012. The Black Queen hypothesis: evolution of dependencies through adaptive gene loss. mBio 3:e00036–e00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. 1973. Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci. 70:3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm RA, Feau N, Henrissat B, Schoch CL, Horwitz BA, Barry KW, Condon BJ, Copeland AC, Dhillon B, Glaser F, et al. 2012. Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLoS Pathog. 8:e1003037.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Berlin-Heidelberg-New York: Springer-Verlag. [Google Scholar]
- Ohno S. 1985. Dispensable genes. Trends Genet. 1:160–164. [Google Scholar]
- Olson MV. 1999. When less is more: gene loss as an engine of evolutionary change. Am J Hum Genet. 64:18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma-Guerrero J, Ma X, Torriani SFF, Zala M, Francisco CS, Hartmann FE, Croll D, McDonald BA.. 2017. Comparative transcriptome analyses in Zymoseptoria tritici reveal significant differences in gene expression among strains during plant infection. Mol Plant Microbe Interact. 303: 231–244. [DOI] [PubMed] [Google Scholar]
- Palmieri N, Kosiol C, Schlötterer C.. 2014. The life cycle of Drosophila orphan genes. eLife 3:e01311.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H.. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. [DOI] [PubMed] [Google Scholar]
- Pezer Ž, Harr B, Teschke M, Babiker H, Tautz D.. 2015. Divergence patterns of genic copy number variation in natural populations of the house mouse (Mus musculus domesticus) reveal three conserved genes with major population-specific expansions. Genome Res. 25:1114–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plissonneau C, Stürchler A, Croll D.. 2016. The evolution of orphan regions in genomes of a fungal pathogen of wheat. mBio 7:e01231-16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presti LL, Lanver D, Schweizer G, Tanaka S, Liang L, Tollot M, Zuccaro A, Reissmann S, Kahmann R.. 2015. Fungal effectors and plant susceptibility. Annu Rev Plant Biol. 66:513–545. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. 2006. Global variation in copy number in the human genome. Nature 444:444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M. 2005. Small proteins of plant-pathogenic fungi secreted during host colonization. FEMS Microbiol Lett. 253:19–27. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP.. 2011. Integrative genomics viewer. Nat Biotechnol. 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropars J, Rodríguez de la Vega RC, López-Villavicencio M, Gouzy J, Sallet E, Dumas É, Lacoste S, Debuchy R, Dupont J, Branca A, et al. 2015. Adaptive horizontal gene transfers between multiple cheese-associated fungi. Curr Biol. 2519: 2562–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H.. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 132:365–386. [DOI] [PubMed] [Google Scholar]
- Rudd JJ, Kanyuka K, Hassani-Pak K, Derbyshire M, Andongabo A, Devonshire J, Lysenko A, Saqi M, Desai NM, Powers SJ, et al. 2015. Transcriptome and metabolite profiling of the infection cycle of Zymoseptoria tritici on wheat reveals a biphasic interaction with plant immunity involving differential pathogen chromosomal contributions and a variation on the hemibiotrophic lifestyle definition. Plant Physiol. 167:1158–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Hahn MW.. 2010. Gene copy-number polymorphism in nature. Proc R Soc Lond B Biol Sci. 277:3213–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperschneider J, Dodds PN, Gardiner DM, Manners JM, Singh KB, Taylor JM.. 2015. Advances and challenges in computational prediction of effectors from plant pathogenic fungi. PLoS Pathog. 11:e1004806.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenwyk JL, Soghigian JS, Perfect JR, Gibbons JG.. 2016. Copy number variation contributes to cryptic genetic variation in outbreak lineages of Cryptococcus gattii from the North American Pacific Northwest. BMC Genomics 17:700.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergiopoulos I, Collemare J, Mehrabi R, De Wit PJGM.. 2013. Phytotoxic secondary metabolites and peptides produced by plant pathogenic Dothideomycete fungi. FEMS Microbiol Rev. 37:67–93. [DOI] [PubMed] [Google Scholar]
- Strope PK, Skelly DA, Kozmin SG, Mahadevan G, Stone EA, Magwene PM, Dietrich FS, McCusker JH.. 2015. The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res. 25:762–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock EH, Banke S, Javan-Nikkhah M, McDonald BA.. 2007. Origin and domestication of the fungal wheat pathogen Mycosphaerella graminicola via sympatric speciation. Mol Biol Evol. 24:398–411. [DOI] [PubMed] [Google Scholar]
- Stukenbrock EH, Bataillon T, Dutheil JY, Hansen TT, Li R, Zala M, McDonald BA, Wang J, Schierup MH.. 2011. The making of a new pathogen: insights from comparative population genomics of the domesticated wheat pathogen Mycosphaerella graminicola and its wild sister species. Genome Res. 21:2157–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock EH, Christiansen FB, Hansen TT, Dutheil JY, Schierup MH.. 2012. Fusion of two divergent fungal individuals led to the recent emergence of a unique widespread pathogen species. Proc Natl Acad Sci. 109:10954–10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenbrock EH, Croll D.. 2014. The evolving fungal genome. Fungal Biol Rev. 28:1–12. [Google Scholar]
- Stukenbrock EH, McDonald BA.. 2009. Population genetics of fungal and oomycete effectors involved in gene-for-gene interactions. Mol Plant Microbe Interact. 22:371–380. [DOI] [PubMed] [Google Scholar]
- Stukenbrock EH, Quaedvlieg W, Javan-Nikhah M, Zala M, Crous PW, McDonald BA.. 2012. Zymoseptoria ardabiliae and Z. pseudotritici, two progenitor species of the Septoria tritici leaf blotch fungus Z. tritici (synonym: Mycosphaerella graminicola). Mycologia 104:1397–1407. [DOI] [PubMed] [Google Scholar]
- Sudmant PH, Mallick S, Nelson BJ, Hormozdiari F, Krumm N, Huddleston J, Coe BP, Baker C, Nordenfelt S, Bamshad M, et al. 2015. Global diversity, population stratification, and selection of human copy-number variation. Science 349:aab3761.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syme RA, Hane JK, Friesen TL, Oliver RP.. 2013. Resequencing and comparative genomics of Stagonospora nodorum: sectional gene absence and effector discovery. G3 3:959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Domazet-Lošo T.. 2011. The evolutionary origin of orphan genes. Nat Rev Genet. 12:692–702. [DOI] [PubMed] [Google Scholar]
- Teo SM, Pawitan Y, Ku CS, Chia KS, Salim A.. 2012. Statistical challenges associated with detecting copy number variations with next-generation sequencing. Bioinformatics 28:2711–2718. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP.. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torriani SFF, Stukenbrock EH, Brunner PC, McDonald BA, Croll D.. 2011. Evidence for extensive recent intron transposition in closely related fungi. Curr Biol. 21:2017–2022. [DOI] [PubMed] [Google Scholar]
- Waples RS, Do C.. 2008. ldne: a program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Resour. 8:753–756. [DOI] [PubMed] [Google Scholar]
- Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer-Verlag: New York, NY. [Google Scholar]
- Wisecaver JH, Rokas A.. 2015. Fungal metabolic gene clusters—caravans traveling across genomes and environments. Microb Physiol Metab. 6:161.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Koonin EV.. 2013. Genome reduction as the dominant mode of evolution. BioEssays 35:829–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. 1951. The genetical structure of populations. Ann Eugenics 15:323–354. [DOI] [PubMed] [Google Scholar]
- Yang R-C. 1998. Estimating hierarchical F-Statistics. Evolution 52:950–956. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Saitoh H, Fujisawa S, Kanzaki H, Matsumura H, Yoshida K, Tosa Y, Chuma I, Takano Y, Win J, et al. 2009. Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell 21:1573–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrei M, MacDonald JR, Merico D, Scherer SW.. 2015. A copy number variation map of the human genome. Nat Rev Genet. 16:172–183. [DOI] [PubMed] [Google Scholar]
- Zhan J, Linde CC, Jürgens T, Merz U, Steinebrunner F, McDonald BA.. 2005. Variation for neutral markers is correlated with variation for quantitative traits in the plant pathogenic fungus Mycosphaerella graminicola. Mol Ecol. 14:2683–2693. [DOI] [PubMed] [Google Scholar]
- Żmieńko A, Samelak A, Kozłowski P, Figlerowicz M.. 2013. Copy number polymorphism in plant genomes. Theor Appl Genet. 127:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.