Abstract
Macrophage Receptor with COllagenous structure (MARCO) is a class A scavenger receptor that binds, phagocytoses, and modifies inflammatory responses to bacterial pathogens. Multiple candidate gene approach studies have shown that polymorphisms in MARCO are associated with susceptibility or resistance to Mycobacterium tuberculosis infection, but how these variants alter function is not known. To complement candidate gene approach studies, we previously used phylogenetic analyses to identify a residue, glutamine 452 (Q452), within the ligand-binding Scavenger Receptor Cysteine Rich domain as undergoing positive selection in humans. Herein, we show that Q452 is found in Denisovans, Neanderthals, and extant humans, but all other nonprimate, terrestrial, and aquatic mammals possess an aspartic acid (D452) residue. Further analysis of hominoid sequences of MARCO identified an additional human-specific mutation, phenylalanine 282 (F282), within the collagenous domain. We show that residue 282 is polymorphic in humans, but only 17% of individuals (rs6761637) possess the ancestral serine residue at position 282. We show that rs6761637 is in linkage disequilibrium with MARCO polymorphisms that have been previously linked to susceptibility to pulmonary tuberculosis. To assess the functional importance of sites Q452 and F282 in humans, we cloned the ancestral residues and loss-of-function mutations and investigated the role of these residues in binding and internalizing polystyrene microspheres and Escherichia coli. Herein, we show that the residues at sites 452 and 282 enhance receptor function.
Keywords: scavenger receptor, MARCO, host–pathogen interactions, positive selection, single nucleotide polymorphism
Introduction
The scavenger receptors (SRs) are an evolutionarily ancient family of pattern recognition receptors (PRRs) that are involved in homeostasis and host defense against infection. MAcrophage Receptor with COllagenous structure (MARCO) is one of the five members of the class A SRs, a family of five homotrimeric type II transmembrane glycoproteins that constitute the class A SRs. Although the class A SRs share structural homology, they differ substantially in function (Elomaa et al. 1998). The molecular structure of MARCO is composed of five domains: an intracellular cytoplasmic domain, a transmembrane domain, a spacer domain, a collagenous domain (which is critical for receptor trimerization), and a C-terminal Scavenger Receptor Cysteine Rich (SRCR) domain (Acton et al. 1993; Bowdish et al. 2009). The SRCR domain is required for binding and phagocytosing unopsonized ligands. A pocket of negatively charged arginine residues, termed the RxR or RGRAEVYY motif, located within the SRCR domain, are essential for binding and phagocytosis of ligands (Brännström et al. 2002; Ojala et al. 2007; Whelan et al. 2012; Novakowski et al. 2016).
The SRCR protein domain is evolutionarily ancient and is found in soluble and membrane-bound receptors in mammals, birds, reptiles, fish, and invertebrates, such as insects, sponges, and echinoderms (Martínez et al. 2011). Although the ancestral function of the SRCR domain is unclear, it has been hypothesized that the SRCR domain-containing proteins of the purple sea urchin may be required for processes such as cell–cell adhesion, rather than host defense (Bowdish and Gordon 2009). We have previously shown that the five modern class A SRs likely arose from ancient precursor proteins via four unique gene duplication events, followed by domain fusions, internal repeats, and deletions (Whelan et al. 2012). The SRCR domain containing transmembrane proteins involved in host defense began to arise in early fish, such as Petromyzon marinus (sea lamprey) and Callorhinchus milii (ghost shark) (Karlsson et al. 2014). This diversification of the class A SRs likely led to their specialization of function. For example, Scavenger Receptor AI (SR-AI) has been implicated as a homeostatic regulator of lipid and protein clearance, SCAvenger Receptor class A 3 (SCARA3) has been shown to be involved in cellular responses to oxidative stress, whereas MARCO has been primarily associated with host defense (Pearson 1996; Han et al. 1998). The SRCR domain of MARCO is essential for ligand binding, however, the collagenous domains of both SR-AI and SCARA3 have been shown to be the primary ligand binding site for bacteria and modified low-density lipoprotein (Rohrer et al. 1990; Ashkenas et al. 1993; Mori et al. 2014).
Several single nucleotide polymorphisms (SNPs) in MARCO have been associated with increased susceptibility to infectious disease. For example, MARCO SNPs have been associated with increased susceptibility or resistance to pulmonary tuberculosis in Chinese Han and Gambian populations (Ma et al. 2011; Bowdish et al. 2013; Thuong et al. 2016). Additional studies have linked MARCO SNPs to sepsis in patients with chronic obstructive pulmonary disease and increased disease severity in respiratory syncytial virus (RSV) infection (Thomsen et al. 2013; High et al. 2016). Unsurprisingly, the infections associated with MARCO SNPs are respiratory pathogens, since MARCO-expressing alveolar macrophages are required for clearance of unopsonized particles and bacteria in the lung (Arredouani et al. 2004, 2005). SNPs associated with increased susceptibility to infectious disease are found throughout the MARCO gene, including promoter, intron, and exon regions, and consequently, it has been challenging to attribute function to these changes.
The infections that MARCO is most strongly associated with are Mycobacterium tuberculosis, Streptococcus pneumoniae, and RSV (Ma et al. 2011; Bowdish et al. 2013; Dorrington et al. 2013; High et al. 2016; Thuong et al. 2016). Interestingly, these are human-adapted pathogens; thus, we hypothesized that there may be human-specific mutations that confer changes in MARCO function (Hertz et al. 2011; Comas et al. 2013; Chaguza et al. 2015). To test this hypothesis, we analyzed MARCO sequences from the 1000 Genomes Project, Great Apes Genome Project, and Neanderthal and Denisovan genomes to identify residues that are unique to humans.
We identified a serine-to-phenylalanine amino acid substitution at position 282 within the collagenous domain of MARCO that is unique to humans. This position is polymorphic in humans (rs6761637) and has a global minor allele frequency (MAF) of >15% and is found in higher frequencies in Asian and African populations. The rs6761637 polymorphism is in linkage disequilibrium (LD) with other MARCO polymorphisms that are associated with increased susceptibility to pulmonary tuberculosis (Ma et al. 2011; Bowdish et al. 2013; Lao et al. 2017). Next, we analyzed regions of the SRCR domain of human MARCO that we had previously identified as undergoing positive selection (Yap et al. 2015). We included recently published Neanderthal and Denisovan genomes to determine if this mutation is human-specific or hominin-specific. We identified a residue in the SRCR domain undergoing positive selection that is proximal to the RGRAEVYY motif, which is required for ligand binding (Yap et al. 2015). Residue 452 was selected for further investigation for two reasons. First, it is in close proximity to the RGRAEVYY motif, which is required for ligand binding (Brännström et al. 2002). Second, the residue at position 452 has mutated from aspartic acid, a negatively charged residue, to glutamine, a polar uncharged residue. Others have demonstrated that both acidic and basic clusters play a role in SRCR domain function (Ojala et al. 2007).
To determine the biological relevance of these two residues, we cloned constructs of Q452 and F282 variants and performed in vitro ligand binding and internalization assays. We found that both residues enhance MARCO-mediated binding and phagocytosis. Together, our findings have identified two novel residues of human MARCO that improve receptor function.
Results
Identification of Residues 282 and 452 as Sites of Potential Function
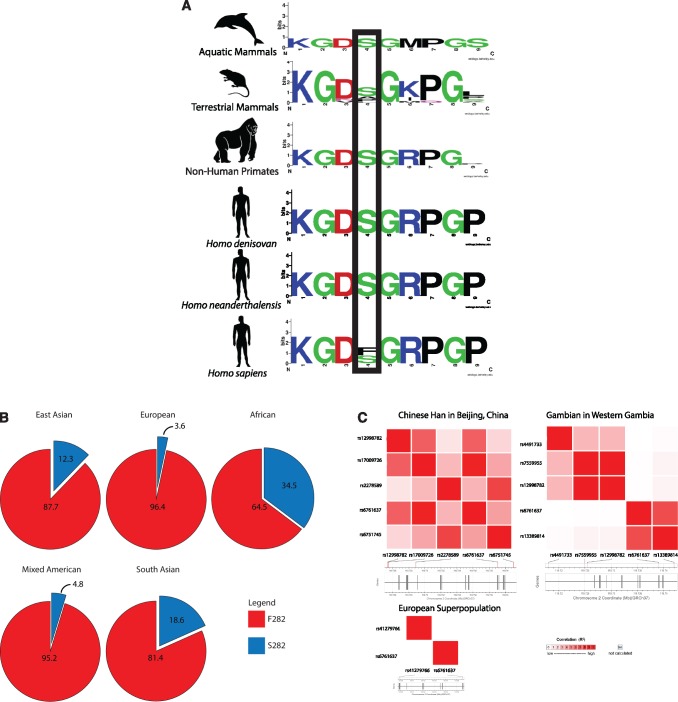
We searched for differences in the MARCO sequences between apes and hominins using sequence data from the 1000 Genomes Project, Great Apes Genome Project, and Neanderthal and Denisovan sequence data (Noonan et al. 2006; Durbin et al. 2010; Meyer et al. 2012; Prado-Martinez et al. 2013). We identified SNPs within common chimpanzee (Pan troglodytes), pygmy chimpanzee (Pan paniscus), orangutan (Pongo sp.), and gorilla (Gorilla sp.) that are mapped to the human MARCO gene. All of the ape species possessed a serine residue at position 282, whereas the majority of extant human’s sequences possess a phenylalanine residue (fig. 1A). We also identified SNPs from Neanderthals and Denisovans that are mapped to the human MARCO gene. Both Neanderthals and Denisovans also possess the serine residue. Residue 282 was found to be polymorphic in modern humans (fig. 1B) (Ma et al. 2011). Interestingly, this position is polymorphic in humans; the ancestral serine residue at the corresponding SNP (rs6761637) is observed at frequencies lower than the derived phenylalanine residue. This polymorphism was determined to have a global MAF of 16.8%. Analysis of the 1000 Genomes Project identified the rs6761637 SNP to have a frequency of 34.5%, 12.3%, and 18.6% in African, East Asian, and South Asian populations, respectively (fig. 1B). Smaller frequencies of 4.8% and 3.6% were noted for North American and European populations.
Fig. 1.
(A) Partial alignment of the collagenous domain of MARCO highlighting residue 282. Humans possess a unique phenylalanine residue at residue 282, except those with the rs6761637 polymorphism, who possess the ancestral serine residue. For a list of species included in each group, see table 2. (B) The global minor allele frequency of rs6761637 (F282S) is 16.8% and varies in frequency between 3.6% in European to 34.5% in African populations. The F282S polymorphism is found at higher frequencies in East Asian (12.3%, n = 629), South Asian (18.6%, n = 657), and African (34.5%, n = 902) populations, relative to European (3.6%, n = 535) and Mixed American (4.8%, n = 468) populations. Each population is composed of multiple subpopulations (i.e., the Mixed American population is composed of Mexican Ancestry from Los Angeles, Puerto Ricans from Puerto Rico, Colombians from Medellin, Colombia, and Peruvians from Lima, Peru). Complete descriptions of each population are available in the 1000 Genomes project. (C) The rs6761637 SNP is in linkage disequilibrium with other polymorphisms associated with infection. Haplotype mapping illustrates that rs6761637 is in LD with four SNPs associated with susceptibility to pulmonary tuberculosis in the Chinese Han population (rs2278589, rs67517405, rs12998782, and rs17009726, n = 163) and one in the Gambian population (rs13389814, n = 179). No LD was observed between rs6761637 and the rs41279766 SNP from the European population (n = 535). All analyses were considered significant if P < 0.05. P values are available in supplementary table S1, Supplementary Material online.
In order to visualize the frequency of mutation within MARCO, a site frequency spectrum for each of the primate species was created (supplementary fig. S1, Supplementary Material online). Although the samples are not randomly chosen and sometimes include different subspecies (e.g., the samples contain sequences from P. t. troglodytes and from P. t. schweinfurthiii), these data do suggest that the amount of variation within the gene is small.
The largest derived allele frequency in humans is at residue 282, with 1,915 derived alleles and 269 ancestral alleles (a breakdown of the variants within MARCO for each human population in the 1000 Genome Project are shown in supplementary table S2, Supplementary Material online) (1000 Genomes Project Consortium 2015). Using chimpanzees as the outgroup, we used the human spectra and calculated estimates of θ (θ = 4Nµ), four times the effective population size multiplied by the mutation rate, using Watterson’s method, Tajima’s method, and Fay and Wu’s method (Watterson 1975; Tajima 1983; Fay and Wu 2000). These give estimates of θ as θW = 3.15, θπ = 0.418, and θH = 1.54, respectively, based on all populations from the project. Watterson’s estimate is based on the number of segregating sites, Tajima’s estimate is based on the number of pairwise differences, while Fay and Wu’s estimate of θ is weighted by the homozygosity of derived alleles. Each estimate responds differently to population parameters. To test for the presence of selection, Tajima’s D statistic (Tajima 1989) and Fay and Wu’s H statistic were calculated. For the whole of the populations from the 1000 Genomes Project (supplementary table S2, Supplementary Material online), Tajima’s D statistic is −2.01 indicating a significant negative deviation and rejecting neutrality at this locus. Fay and Wu’s H statistic is −1.12. Although H is not significant, the strongly negative Fay and Wu’s H indicates an excess of high-frequency derived SNPs suggesting that selection may have acted to alter the protein the protein in comparison to the chimpanzee outgroup. Fay and Wu suggest that following a selective hitchhiking event both their estimate and Watterson’s estimate of θ should be larger than Tajima’s as is observed here. These data are consistent with positive selection at site 282 since the negative value of Fay and Wu’s H statistic is largely driven by the high frequency of derived nonsynonymous allele at residue 282 (supplementary fig. S1, Supplementary Material online). It should be noted that in the majority of the “populations” the high frequency of the derived allele at position 282 stands out (supplementary table S2, Supplementary Material online).
In the separate analysis of the populations, Tajima’s D and Fay and Wu’s H are also negative except that one African population (Yoruba) has a positive value of Tajima’s D (supplementary table S2, Supplementary Material online). However, the actual history of the human population is complex with population structure, size change, and gene flow, and may not meet the assumptions of the tests using Tajima’s D and Fay and Wu’s H. In fact, Tajima’s D is known to become negative when there was a recent bottleneck in the population. Therefore, we consider these tests to be only suggestive and justification to gather further experimental evidence.
Based on the above findings, we felt that further investigation was warranted to determine if the variant at 282 might be under positive selection. The allele at residue 282 has been implicated in MARCO function in candidate gene studies. rs6761637 (position 282) was previously shown to be a member of a haplotype linked to rs17009726 and associated with increased susceptibility to pulmonary tuberculosis (Ma et al. 2011). Therefore, we examined whether rs6761637 was in LD with other polymorphisms in MARCO that have been associated with human disease. We identified five SNPS, rs2278589, rs6751745, rs17009726, rs12998782, and rs13389814, all in LD with rs6761637 (fig. 1C and supplementary table S1, Supplementary Material online).
Position 452 was previously identified as one of several positions in the SRCR domain of MARCO having potentially been subject to positive selection (Yap et al. 2015). Using sequences from various groups of mammals, we generated a partial alignment of the SRCR domain of MARCO. Here, we show that position 452 encodes an aspartic acid residue in aquatic and land mammals, exclusive of primates and hominins. Primates, including Macaca mulatta (rhesus macaque), Pan troglodytes (chimpanzee), and Pongo abelii (Sumatran orangutan) all possess a histidine residue and Nomascus leucogenys (northern white-cheeked gibbon) possesses a glutamine residue. Neanderthal, Densiovan, and human sequences also shared a glutamine at position 452 (fig. 2 and table 1).
Fig. 2.
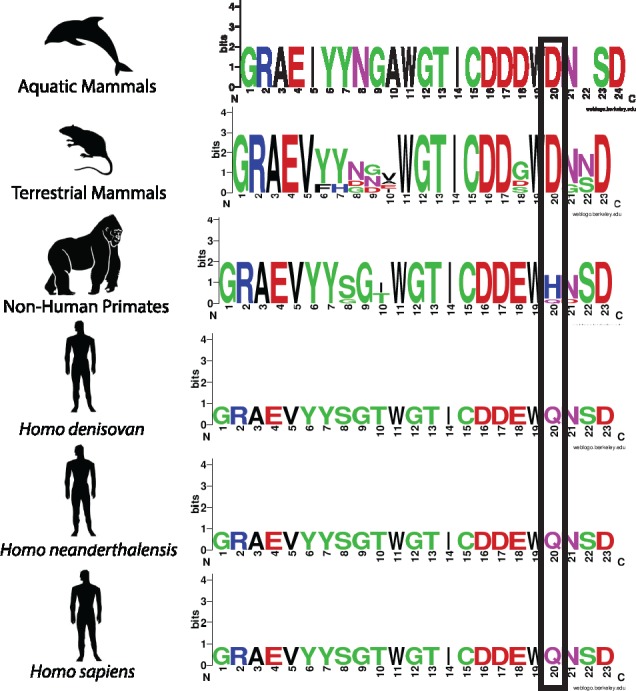
Partial alignment of the SRCR domain of MARCO highlighting a residue undergoing positive selection at position 452 (Yap et al. 2015). Aquatic (n = 2) and terrestrial (n = 15) mammals possess a conserved aspartic acid residue at position 452, nonhuman primates (n = 5) vary. Neanderthals (n = 1), Denisovans (n = 1), and humans (n = 1) possess a glutamine residue. For a list of species included in each group, see table 2.
Table 1.
SNPs Mapped from Nonhuman Primates, Neanderthals, and Denisovans to the SRCR Domain of Human MARCO.
| Chromosomal Position | Amino Acid Position | Ape Allele | Neanderthal/Denisovan Allele | Human Allele | Ape Amino Acid | Neanderthal/Denisovan Amino Acid | Human Amino Acid |
|---|---|---|---|---|---|---|---|
| 119467242 | 442 | T/C | C | C | I/T | T | T |
| 119467252 | 445 | C | A | A | T | T | T |
| 119467273 | 452 | C/A | A | A | H/Q | Q | Q |
| 119467287 | 457 | C | T | T | T | I | I |
| 119468482 | 493 | C | T | T | S | S | S |
| 119468516 | 505 | T | C | C | H | H | H |
| 119468533 | 510 | T | C | C | H | H | H |
Note.—Based on data from the Great Apes Project, common SNPs from Pan troglodytes, Gorilla gorilla, and Pan paniscus to the SRCR domain are shown. Pongo pygmaeus was incomplete and only contained SNPs mapped to the first half of the MARCO gene. For position 452, Nomascus leucogenys was also included, as it was found to have a glutamine residue. Human genome build 18 was used. The MARCO gene is located on chromosome 2 in Humans, Neanderthals, and Denisovan and on chromosome 2B in apes.
Mutation of Residue 452 Does Not Alter Expression of MARCO but May Alter Exposure of an Arginine Residue That Is Critical for Ligand Binding
In order to determine if residue 452 is required for MARCO function, we generated human MARCO constructs using site-directed mutagenesis. We generated a Q452D construct to assess if the ancestral aspartic acid residue affects receptor function relative to the human glutamine residue. We also generated a Q452A construct to replace the larger, charged residue with a smaller, neutral one to create a “functional knockout.” To determine if Q452D and Q452A substitutions caused a change in SRCR domain structure, we performed a structural and coulombic surface charge analysis. We observed minor changes in SRCR domain structure (fig. 3A and B) and surface charge (fig. 3D), but did observe changes in the surface availability of the RGRAEVYY motif (fig. 3C), suggesting that mutation of residue 452 may alter ligand binding and/or internalization by partially reducing the accessibility of the RGRAEVYY motif to ligands. We calculated Solvent Available Surface area (SAS) and found that the Q452A mutation reduced the surface availability of the first arginine residue within the RGRAEVYY motif from 114.73 to 71.90 Å2 (fig. 3E). To ensure that all MARCO constructs are equally expressed in transiently transfected HEK 293 T cells, we performed Western blotting on lysates of HEK 293 T cells at 48-h post transfection using antibodies targeting the C-terminal myc tag. We observed a similar level of total protein expression in all constructs (fig. 4A). To compare surface expression, we performed flow cytometry and observed equal surface expression of Q452A and Q452D constructs relative to MARCO (fig. 4B).
Fig. 3.
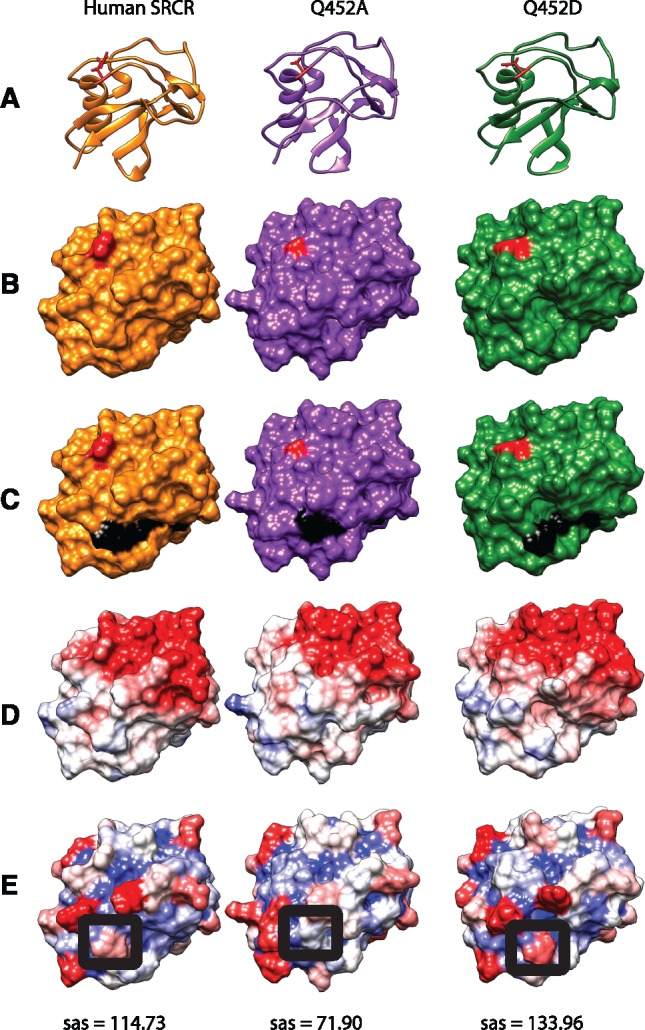
Structural comparisons of wildtype human SRCR, Q452A, and Q452D SRCR domain models. (A) Ribbon diagrams of the respective SRCR variants with residue 452 highlighted in red. (B) Molecular surface model of the respective SRCR variants with residue 452 highlighted in red. (C) Molecular surface model highlighting the RGRAEVYY motif in black and residue 452 in red. (D) Coulombic surface charge modeling applied. Red = −10, white = 0, blue = +10 in kcal/(mol·e) at 298 K. (E) Solvent Accessible Surface area (sas) modeling applied. Red = 160 Å, white = 80 Å, blue = 0 Å. Raw sas values of arginine 13, of the SRCR domain RGRAEVYY, circled in black, are shown below each model in Å2.
Fig. 4.
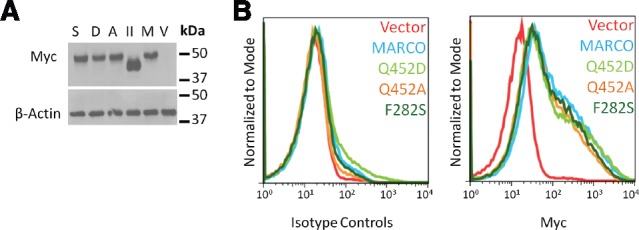
Mutation of sites Q452 and F282 does not affect expression of MARCO. HEK 293 T cells transfected with empty vector, MARCO, MARCOII, Q452A, Q452D, or F282S were examined for total protein expression by western blot (A) and surface expression by flow cytometry (B). V, vector; M, MARCO; II, MARCOII; A, Q452A; D, Q452D; S, F282S.
Mutation of Residue 282 Does Not Alter Expression of MARCO
Based on the indications that residue 282 was likely under positive selection and had been implicated in multiple candidate gene studies, we cloned the ancestral F282S substitution to assess if a serine substitution at position 282 affected receptor function. Given that a phenylalanine to serine substitution alters both the size and charge of the residue, and that the collagenous domain is critical for receptor trimerization, we hypothesized that this substitution may result in an unstable protein and subsequent lower surface expression. We observed that similar to our residue 452 constructs, substitution of residue 282 did not alter total or surface expression (fig. 4A and B). Due to the highly repetitive nature of the collagenous domain, we were unable to generate accurate structural models of the F282S SNP using currently available software.
The Residues at Positions 452 and 282 Play a Role in Receptor–Ligand Association
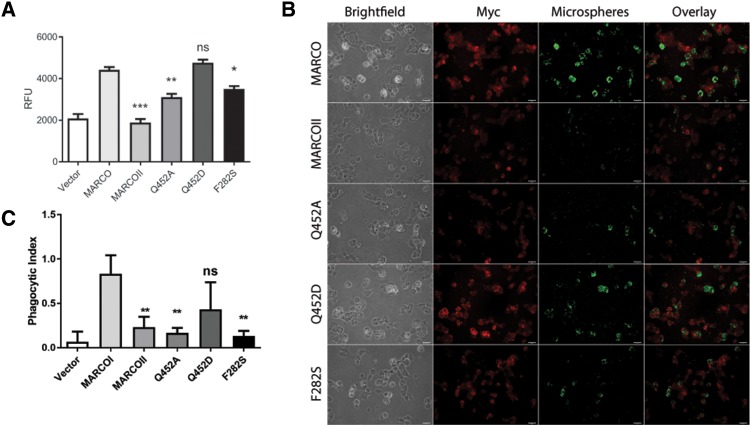
We compared the functional importance of residues Q452 and F282 in a cell-association assay using 0.5 µm maleylated bovine serum albumin (Mal-BSA)-coated polystyrene microspheres as ligands. Mal-BSA is a well-characterized MARCO ligand (Greaves and Gordon 2009; Novakowski et al. 2016). Relative to empty-vector control transfected HEK 293 T cells, we observed a 212% increase in microsphere association in MARCO-transfected cells (fig. 5A and B). Similar to our previous findings (Novakowski et al. 2016), HEK 293 T transfected with a truncated variant of MARCO lacking the SRCR domain, MARCOII, showed no significant difference in microsphere association relative to empty-vector control transfectants. Mutation of glutamine 452 to the ancestral aspartic acid residue (Q452D) resulted in no significant change in microsphere association, but mutation to alanine (Q452A) resulted in a 70% reduction in microsphere association, relative to MARCO (fig. 5A and B). Mutation of phenylalanine 282 to the ancestral serine residue resulted in a 79% reduction in microsphere association (fig. 5A and B).
Fig. 5.
Residues at positions 452 and 282 of MARCO influence ligand association and bacterial internalization. (A) Mutation of glutamine residue 452 to alanine (Q452A) reduces ligand-coated microsphere association in HEK 293 T cells, whereas mutation to the ancestral aspartic acid (Q452D) does not. Mutation of residue phenylalanine 282 to the ancestral serine (F282S) also reduces microsphere association. Statistical comparisons relative to MARCO were made using one-way ANOVA with Tukey’s post hoc test. Error bars indicate mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. Experiments were performed a minimum of three times with a minimum of three technical replicates. (B) Immunofluorescence microscopy of microsphere association in HEK 293 T transfected with empty vector, MARCO, MARCOII, or variants Q452A, Q452D, and F282S. Scale bars represent 25 µm. (C) Mutation of residue 452 to alanine (Q452A) or residue 282 to serine (F282S) greatly reduces the phagocytic index of RAW 264.7 macrophages. Phagocytic index was calculated as number of internalized bacteria divided by the number of RAW 264.7 macrophages counted. Statistical comparisons relative to MARCO were made using one-way ANOVA with Tukey’s post hoc test. Error bars indicate mean ± SEM. **P < 0.01.
The Residues at Positions 452 and 282 Greatly Enhance Phagocytosis of Bacteria
To determine if the human residues at positions 452 and 282 enhanced binding and phagocytosis of bacteria, we performed binding and phagocytosis assays using transiently transfected RAW 264.7 murine macrophages and inside–outside labeled, heat-killed Escherichia coli as a ligand. To ensure equal expression of MARCO constructs in transiently transfected RAW 264.7 macrophages, we manually traced the membrane of each cell and quantified pixel intensity. We did not observe a difference in MARCO expression between our constructs and bacterial binding did not differ between MARCO variants (data not shown). Variants expressing the A452 and S282 residues demonstrated significantly lower bacterial uptake. Relative to human MARCO-transfected cells, the Q452A had an 80% reduction in phagocytosis, and the F282S construct resulted in an 84% reduction in phagocytosis (fig. 5C).
Discussion
The SRs are a family of PRRs whose evolutionary origins predate gnathostomes. It has been hypothesized that early receptors containing SRCR domains were involved in cell–cell adhesion (Bowdish and Gordon 2009). The most ancient organism from which a class A SR (i.e., plasma membrane-expressed, with one SRCR domain) has been sequenced is Petromyzon marinus (sea lamprey), suggesting that the earliest precursors to the modern class A SRs likely arose at least 500 Ma.
We have previously shown that diversification of the class A SRs likely resulted from a single common ancestor protein that underwent gene duplication, domain fusions, and deletions (Whelan et al. 2012). This protein likely contained an SRCR domain, as three of the five modern class A SRs contain a SRCR domain (Whelan et al. 2012). This genetic diversification of the class A SRs likely would have allowed for diversification of receptor function, allowing for binding and phagocytosing of a wider variety of ligands, including pathogens and clearance of foreign or endogenous cellular debris. It is conceivable that this specialization of the SRs was the result of positive selective pressure for domains and motifs that enhanced the host defense properties and homeostatic functions of the respective receptors. Ultimately, the resulting five members of the modern class A SRs in humans are diverse in both receptor structure and function. A large number of motifs and residues of the modern class A SRs are highly conserved and/or under positive selective pressure, but which of these may play a role in the intrinsic function of the receptor remains unknown (Yap et al. 2015).
Of the five modern class A SRs, MARCO appears to be unique in that its primary function is in host defense, while other members have been shown to regulate processes such as clearance of lipids and proteins and cells undergoing oxidative stress and apoptosis (Pearson 1996; Han et al. 1998; Todt et al. 2008). We sought to identify both conserved and/or positively selected sites within the MARCO gene followed by characterizing their role in receptor function. We began by comparing the MARCO gene of nonhuman primates, Denisovans, and Neanderthals to extant humans. We identified a site within the SRCR domain, glutamine 452, that is undergoing positive selection (tables 1 and 2; fig. 2) (Yap et al. 2015). Interestingly, all currently available MARCO sequences from aquatic and terrestrial mammals, excluding nonhuman primates, possess aspartic acid, a negatively charged residue, whereas the Homo denisovan, H. neanderthalensis, and H. sapiens possess a polar, uncharged glutamine residue (fig. 2 and table 2). Furthermore, this residue is situated near the ligand-binding, RGRAEVYY motif. We identified a second site within the collagenous domain, phenylalanine 282 (F282), which we show as being unique to humans, with most ancestral organisms possessing a serine residue (fig. 1A and table 2).
Table 2.
Organisms Included in Phylogenetic Analyses.
| Nonhuman Primates | Terrestrial Mammals | Aquatic Mammals | Other |
|---|---|---|---|
| Gorilla gorilla | Dasypus novemcinctus | Tursiops truncatus | Homo sapiens |
| Macaca mulatta | Cavia porcellus | Orcinus orca | Homo neanderthalensis |
| Pan paniscus | Cricetulus griseus | Homo denisovan | |
| Pan troglodytes | Canis lupus familiaris | ||
| Pongo abelii | Loxodonta africana | ||
| Mesocricetus auratus | |||
| Mus musculus | |||
| Oryctolagus cuniculus | |||
| Sus scrofa | |||
| Rattus norvegicus | |||
| Ovis aries | |||
| Bos taurus |
In humans, MARCO is an important component of host defense against airway pathogens such as Mycobacterium tuberculosis (Mtb), Streptococcus pneumoniae, and Klebsiella pneumoniae (van der Laan et al. 1999; Bowdish et al. 2009; Dorrington et al. 2013). It is therefore unsurprising that studies of MARCO by candidate gene approach have identified SNPs that are associated with susceptibility or resistance to infection (Ma et al. 2011; Bowdish et al. 2013; Thuong et al. 2016). Interestingly, the rs6761637 polymorphism that was previously linked to increased susceptibility to pulmonary tuberculosis by Ma et al. (2011) encodes for the ancestral S282 residue. We further analyzed this polymorphism and show that ∼17% of humans possess the SNP encoding the ancestral serine residue at position 282 (fig. 1B). We discovered that the ancestral allele is found at high frequencies in the South and East Asian populations, as well as in the African population (fig. 1B). Haplotype analysis revealed that rs6761637 is in LD with other polymorphisms associated with increased susceptibility to Mtb (fig. 1C). Together, this suggested that F282 may also play a role in pathogen–receptor interactions.
To examine if mutation of residue Q452 to the ancestral aspartic acid or a loss-of-function mutation to alanine affected receptor structure, we modeled structural changes in the SRCR domain. The best reference structures for the SRCR domain are the monomeric and dimeric SRCR domains of human MARCO (Ojala et al. 2007). We observed that mutation of residue 452 to alanine partially “hid” the first arginine residue within the RGRAEVYY motif of a SRCR monomer (fig. 3). This suggested that residue 452 may not be directly involved in ligand binding, but rather may play an important role in the structure of the SRCR domain, such that the ligand-binding RGRAEVYY motif is not fully exposed to ligands. Next, we analyzed surface expression of our mutant constructs to ensure equal expression and showed that our mutations did not alter expression of our constructs relative to human MARCO (fig. 4). We then sought to determine if functional differences would be observed between ancestral and modern human residues of sites 452 and 282. We performed ligand association assays using Mal-BSA-coated polystyrene microspheres, a ligand that has previously been used to study SR function (Gough et al. 1998; Novakowski et al. 2016). We showed that mutation of residue Q452 to the ancestral aspartic acid residue had no effect on ligand association; however mutation to alanine reduced function by 70% (fig. 5A and B).
We showed that mutation of F282 to the ancestral serine residue reduced ligand association by 79%. This is surprising, given that the SRCR domain has been shown to be the primary ligand binding site for Mal-BSA microspheres (Novakowski et al. 2016).
Although the repetitive structure of collagen impedes the ability to model its structure, the hydrophobicity of the residue at position 282 may alter its location within the collagenous domain. The ancestral serine residue is slightly hydrophilic, and could reside inside or outside of the structure. In contrast, phenylalanine is hydrophobic and might localize internally within the domain. The collagenous domain of human MARCO contains two phenylalanine residues, F282 and F293. Phenylalanine residues can interact to provide additional free energy to stabilize secondary structures (Anjana et al. 2012). Given the proximity of F282 and F293, it is possible that these residues behave in a similar manner. Since the collagenous domain is important for receptor trimerization, it is possible that this mutation favors a more stable conformation of the collagenous domain that may facilitate bacterial binding or uptake (Acton et al. 1993; Brännström et al. 2002). While the SRCR domain of MARCO has been shown to be required for ligand binding, it has been shown that truncation of the collagenous domain, including residue 282, reduces but does not abolish receptor function (Brännström et al. 2002). This is in agreement with our findings (fig. 5).
Next, we demonstrated that Q452 or F282 played a role in phagocytosis of a physiologically relevant ligand, E. coli. MARCO-mediated phagocytosis of bacteria has been shown to play a central role in the clearance of infection by both phagocytosis and by enhancing inflammatory signaling through Toll-like receptors (TLRs) (van der Laan et al. 1999; Bowdish et al. 2009; Dorrington et al. 2013). MARCO responds to Mtb infection by recognizing trehalose 6,6′-dimycolate (TDM), a component of the mycobacterial cell wall (Bowdish et al. 2009). The presence of TDM enhances both MARCO-mediated phagocytosis of the bacterium and inflammatory signaling through TLR2 (Bowdish et al. 2009). Others have demonstrated that phagocytosis of Mtb results in additional activation of intracellular, proinflammatory receptors such as TLR9 and NOD2 (Bafica et al. 2005; Brooks et al. 2012). Interaction of mycobacterial ligands with TLRs 2/9 and NOD2 represents an early, but important step in activating interferon-gamma and Th-1-type protective immunity against Mtb (Hossain and Norazmi 2013). Therefore, a MARCO allele which enhances the intrinsic phagocytic function of the receptor would likely be advantageous to the host.
We did not observe a difference in binding of E. coli in RAW 264.7 cells transfected to express our MARCO constructs (data not shown), but MARCO-mediated internalization was reduced. We observed an 80% reduction in phagocytosis in the Q452A mutant and an 84% reduction in phagocytosis with our F282S mutant (fig. 5C). These findings agree with our microsphere association results (fig. 5A). Our data are complimentary to other published ligand binding studies of MARCO, whereby mutation and/or deletion of the varying regions of the either the collagenous or SRCR domain reduced, but did not totally abolish receptor function (Brännström et al. 2002).
Together, our data suggest that Q452 and F282 are important for ligand association and phagocytosis of bacteria. We show that Q452 and F282 are examples of positively selected mutations in MARCO, and that both enhance the intrinsic function of the receptor.
Materials and Methods
Phylogenetic and SNP Analyses of MARCO
Amino acid sequences of MARCO were acquired using the National Center for Biotechnology Information (NCBI) and ENSEMBL databases, and aligned using multiple alignment by fast Fourier transform (MAFFT), as previously described (Yap et al. 2015). Residue 282 was identified as a site of interest by comparing human, Neanderthal, Denisovan, and ape MARCO sequences and identifying human-specific residues. To determine if residue 282 was polymorphic, we examined human SNP data for the entire chromosomal region of MARCO from the 1000 Genomes Project and the Great Apes Genome Project using PERL scripts (Durbin et al. 2010; Prado-Martinez et al. 2013). SNPs from the Neanderthal and Denisovan genomes were accessed from the Max Planck Institute for Evolutionary Anthropology (Noonan et al. 2006; Meyer et al. 2012). We then characterized the SNPs present within exons of MARCO as being nonsynonymous or synonymous substitutions using NCBI’s dbSNP database. SNP data from the Great Apes Genome Project included: Pan troglodytes, Pan paniscus, Pongo pygmaeus, Pongo abelii, Gorilla gorilla, and Gorilla beringei. We identified residue 282 as polymorphic, humans who possess the rs6761637 variant have a nonsynonymous phenylalanine to serine substitution. Estimates of θ = 4Neμ were calculated using the methods of Watterson, Tajima, and Fay and Wu (Watterson 1975; Tajima 1983; Fay and Wu 2000) and tests for selection were done using the methods of Tajima’s D statistic (Tajima 1989) and Fay and Wu’s H statistic (Fay and Wu 2000). To explore the variation within MARCO we analyzed the human genomes from Phase I of the 1000 Human Genome Project (v3.20101123). The variants from chromosome 2 were downloaded from the vcf file and the raw data were divided in individual source “populations” and “super populations.” The observed value for Fay and Wu’s estimate of θH, Fay and and Wu’s H statistic, Watterson’s estimate of θW, Tajima’s estimate of θπ and Tajima’s statistic D is shown. To test the significance of Tajima’s D and Fay and Wu’s H, simulations were done using ms (Hudson 2002), based on a simple neutral model with constant population size, no recombination, random mating and an infinite-site model. Sample sizes were chosen to match those from each “population” and each simulation was repeated 1,000 times. Watterson’s estimate of θ was used for each group (cmd: ms nsam 1000 -t θ). The results are shown in supplementary table S2, Supplementary Material online.
All LD analyses were performed using LDLink with European, Gambian, and Chinese Han populations from the 1000 Genomes project (Machiela and Chanock 2014). Alleles were determined to be in LD if R2 > 0.1 and P < 0.05. SNP frequencies were generated using the 1000 Genomes Project Version 3.1 build 144 (October 2015) (1000 Genomes Project Consortium 2015).
Several residues within the SRCR domain of MARCO were previously identified as being under positive selection using codeml, including residue 452 (Yap et al. 2015). Briefly, we identified residues undergoing positive selection using a branch-site model in PAML with free variation of ratios of nonsynonymous to synonymous substitutions (Yang 2007). Results were confirmed with a likelihood ratio test at 95% significance level (Yap et al. 2015). All sequences were visualized in WebLogo 3(Crooks et al. 2004).
Cell Lines
HEK 293 T (ATCC #CRL-3216) were propagated in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin at 37 °C and 5% CO2. RAW 264.7 were propagated under the same conditions without the presence of penicillin and streptomycin.
Plasmids
Human MARCO plasmid was kindly provided by Timo Pikkarainen (Bowdish et al. 2009). Human MARCOII plasmid was previously generated from cDNA provided by Wu-Bo Li (Invitrogen, Carlsbad). All constructs were subcloned into pcDNA3.1/Hygro(+) and C-terminal myc tags were generated by PCR, as previously described (Novakowski et al. 2016). Mutant myc-MARCO constructs Q452A, Q452D, and F282S were generated using mutagenic primers designed using NEBaseChanger and a Q5 site-directed mutagenesis kit (New England Biolabs). All plasmids were amplified and purified as previously described (Novakowski et al. 2016).
Antibodies
Primary antibodies used for Western blotting included anti-Beta actin (Sigma–Aldrich) at 0.2 µg/ml and anti-myc (9E10) at 10 µg/ml. All antibodies were diluted in 5% skim milk in Tris-buffered saline with 0.1% Tween-20 (TBST). Primary antibodies for immunofluorescence microscopy and flow cytometry included Alexa Fluor-647 conjugated anti-myc 9B11 (Cell Signaling Technology) used at 1:250 dilution. For bacterial phagocytosis assays, a Pierce anti-myc rabbit polyclonal antibody was used at 1:400 dilution (Thermofisher). Secondary antibodies for Western blotting and immunofluorescence microscopy included goat anti rabbit and goat anti mouse conjugated to horseradish peroxidase (GenWay, San Diego, CA) and donkey anti rabbit conjugated to Cy3 (Jackson ImmunoResearch, Westgrove).
Microsphere Association Assays and IF Microscopy
Microsphere binding assays were performed as previously described (Novakowski et al. 2016). Briefly, HEK 293 T cells were transfected with either pcDNA3.1/Hygro(+), MARCO, MARCOII, or one of Q452A, Q452D, and F282S mutant constructs. All MARCO constructs contained a C-terminal myc tag that we previously confirmed had no effect on microsphere binding or phagocytosis (Novakowski et al. 2016). At 48-h post transfection, cells were lifted by forceful pipetting and resuspended in Opti-Mem reduced serum growth media (Invitrogen). Cell numbers were normalized to 1×106 cells/ml by counting viable cells stained with 0.4% Trypan Blue using a hemocytometer. Cells were incubated with gentle agitation at 37 °C with 0.5 µm polystyrene yellow fluorescent microspheres (Polysciences, Warrington, PA) that were passively coated with Maleylated Bovine Serum Albumin (Mal-BSA) at a ratio of 320 microspheres: cell for 1.5 h. Cells were washed twice with phosphate-buffered saline (PBS) and then added to an opaque microtiter plate.
Fluorescence was measured on a Spectramax M3 spectrophotometer (Molecular Devices) at Ex441nm/Em486nm. Immunofluorescence microscopy experiments were performed with cells transfected as above. At 24-h post transfection, cells were lifted and seeded onto poly l-lysine-coated glass cover slips to adhere overnight. Following microsphere binding in Opti-Mem (Life Technologies), cells were washed, fixed with 2% paraformaldehyde, blocked with 5% BSA in PBS, and stained with Mouse anti-myc (9B11)-Alexa Fluor-647 (Cell Signaling Technologies, Danvers, MA). Images were obtained on a Leica DM IRE2 inverted fluorescence microscope (Leica, Wetzlar, Germany). All images were equally adjusted for brightness and contrast using ImageJ (NIH, Bethesda, MD).
Bacterial Phagocytosis Assays
Escherichia coli strain ML35 was grown to stationary phase in Lysogeny Broth at 37 °C with shaking at 200 RPM overnight. Bacteria were titered by serial dilution and plating. Bacteria were centrifuged for 1 min at 6, 000× g, resuspended in PBS, and heat killed at 70 °C for 10 min and then washed twice with PBS. Bacteria were then resuspended in 500 µl PBS (pH =8) and incubated with 0.2 mg EZ-link NHS-LC-biotin (Thermofisher) for 20 min at RT with gentle agitation. Unreacted NHS-LC-biotin was quenched with 500 µl LB broth for 10 min at RT and washed twice with PBS. Cells were subsequently resuspended once again in 500 µl PBS and fluorescently labeled with 0.5 µl CellTrace Far-Red (ThermoFisher, Waltham, MA) for 20 min at RT with gentle agitation. Unreacted CellTrace was quenched with 500 µl LB broth for 10 min at RT and washed twice with PBS.
We seeded 1×105 RAW 264.7 cells onto glass cover slips in a 12-well plate overnight. Cells were transfected with 1 µg of either pcDNA3.1/Hygro(+), MARCO, MARCOII, or one of Q452A, Q452D, and F282S mutant constructs using FuGene HD (Active Motif Inc, Carlsbad, CA) following the manufacturer’s recommended protocol. At 48-h post transfection, cells were washed once with PBS and resuspended in fresh DMEM. Labeled bacteria were added at a multiplicity of infection of 100 and samples were centrifuged for 1 min at 500×g. Samples were incubated at 37 °C for 30 min to allow phagocytosis to occur. Cells were then washed twice with PBS and stained with Alexa Fluor 488-conjugated streptavidin (BD Biosciences) at a dilution of 1:500 for 20 min at RT. Subsequently, cells were washed three times with PBS and fixed by addition of 4% paraformaldehyde (PFA) for 20 min at RT in the dark. Cells were washed three times with PBS and blocked for 30 min at RT in the dark with 5% bovine serum albumin (BSA) and 10% donkey serum in PBS. Cells were stained with a Pierce anti-c-Myc tag polyclonal antibody (ThermoFisher) at a dilution of 1:400 in 5% BSA in PBS for 30 min at RT in the dark, washed three times with PBS and then stained with donkey anti rabbit conjugated to Cy3 (Jackson ImmunoResearch). Cells were washed three times for 5 min each time, and mounted onto glass slides with PermaFluor mounting medium (ThermoFisher). Slides were imaged on a Leica DSM 6000 upright wide-field fluorescence microscope. Thirty randomly selected MARCO-expressing cells per construct were analyzed for bacterial binding and phagocytosis. To differentiate between bacterial binding versus internalization, we utilized an inside–outside staining strategy where all bacteria are stained with CellTrace Far Red, but bound, not internalized bacteria also stained with Alexa Fluor 488-conjugated streptavidin. Phagocytic index was calculated as number of internalized bacteria divided by the number of RAW 264.7 macrophages counted. To determine whether our constructs were equally expressed in transfected RAW 264.7 macrophages, we manually traced the membrane of each cell and quantified pixel intensity in ImageJ (NIH) (data not shown). Additional image processing was performed in ImageJ.
Structural Analyses
All structural and molecular analyses were performed using the iterative threading assembly refinement (I-TASSER) version 5.0 (Roy et al. 2010). Models with the highest C-score were used for analysis. Visualization was performed using UCSF Chimera 1.1 (Pettersen et al. 2004). Coulombic surface charge analyses were performed using DelPhi Web Server and visualized in UCSF Chimera 1.1 (Li et al. 2012).
Statistical Analyses
All statistical analyses were performed using Graphpad Prism 7.02 (Graphpad Software Inc., San Diego, CA).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
K.E.N., C.Y., and N.V.L.Y. designed, performed, and analyzed the experiments. G.B.G. performed the tests for selection on residue 282. K.E.N. wrote the manuscript. K.E.N., D.M.E.B., G.B.G., K.S., and B.H. planned, and edited the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
Work in the Bowdish Laboratory is supported in part by the McMaster Immunology Research Centre and the MG DeGroote Institute for Infectious Disease Research. K.E.N. is supported by an NSERC postgraduate doctoral scholarship. D.M.E.B. is supported by NSERC and a Canada Research Chair in Aging and Immunity. B.H. is supported by a Canadian Institutes for Health Research Operating Grant (MOP-123419) and Ontario Ministry of Research and Innovation Early Research Award. C.Y. is supported by a CIHR Canada Graduate Scholarship-Masters and a CIHR MD/PhD Studentship. N.V.L.Y. is supported by an Ontario Graduate Scholarship. G.B.G. was supported by NSERC (Grant # 140221-10). Work in K.Sl lab was supported by NIH Grant Number 1 R15 AI094436-01A1.
References
- 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acton S, Resnick D, Freeman M, Ekkel Y, Ashkenas J, Krieger M.. 1993. The collagenous domains of macrophage scavenger receptors and complement component C1q mediate their similar, but not identical, binding specificities for polyanionic ligands. J Biol Chem. 268:3530–3537. [PubMed] [Google Scholar]
- Anjana R, Vaishnavi MK, Sherlin D, Kumar SP, Naveen K, Kanth PS, Sekar K.. 2012. Aromatic-aromatic interactions in structures of proteins and protein-DNA complexes : a study based on orientation and distance. Bioinformatician 824:1220–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arredouani M, Palecanda A, Koziel H, Huang Y, Imrich A, Sulahian TH, Ning Y, Yang Z, Pikkarainen T, Sankala M.. 2005. MARCO is the major binding receptor for unopsonized particles and bacteria on human alveolar macrophages. J Immunol. 1759:6058–6064. [DOI] [PubMed] [Google Scholar]
- Arredouani M, Yang Z, Ning Y, Qin G, Soininen R, Tryggvason K, Kobzik L.. 2004. The scavenger receptor MARCO is required for lung defense against pneumococcal pneumonia and inhaled particles. J Exp Med. 2002:267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenas J, Penman M, Vasile E, Acton S, Freeman M, Krieger M.. 1993. Structures and high and low affinity ligand binding properties of murine type I and type II macrophage scavenger receptors. J Lipid Res. 346:983–1000. [PubMed] [Google Scholar]
- Bafica A, Scanga CA, Feng CG, Leifer C, Cheever A, Sher A.. 2005. TLR9 regulates Th1 responses and cooperates with TLR2 in mediating optimal resistance to Mycobacterium tuberculosis. J Exp Med. 20212:1715–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowdish DME, Gordon S.. 2009. Conserved domains of the class A scavenger receptors: evolution and function. Immunol Rev. 2271:19–31. [DOI] [PubMed] [Google Scholar]
- Bowdish DME, Sakamoto K, Kim M-J, Kroos M, Mukhopadhyay S, Leifer CA, Tryggvason K, Gordon S, Russell DG, Ramakrishnan L.. 2009. MARCO, TLR2, and CD14 are required for macrophage cytokine responses to mycobacterial trehalose dimycolate and Mycobacterium tuberculosis. PLoS Pathog. 56:e1000474.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowdish DME, Sakamoto K, Lack NA, Hill PC, Sirugo G, Newport MJ, Gordon S, Hill AVS, Vannberg FO.. 2013. Genetic variants of MARCO are associated with susceptibility to pulmonary tuberculosis in a Gambian population. BMC Med Genet. 141:47.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brännström A, Sankala M, Tryggvason K, Pikkarainen T.. 2002. Arginine residues in domain V have a central role for bacteria-binding activity of macrophage scavenger receptor MARCO. Biochem Biophys Res Commun. 2905:1462–1469. [DOI] [PubMed] [Google Scholar]
- Brooks MN, Rajaram MVS, Azad AK, Amer AO, Valdivia-Arenas MA, Park J, Núñez G, Schlesinger L.. 2012. NOD2 controls the nature of the inflammatory response and subsequent fate of Mycobacterium tuberculosis and M. bovis BCG in human macrophages. Cell Microbiol. 13:402–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaguza C, Cornick JE, Everett DB.. 2015. Mechanisms and impact of genetic recombination in the evolution of Streptococcus pneumoniae. Comput Struct Biotechnol J. 13:241–247.http://dx.doi.org/10.1016/j.csbj.2015.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas I, Coscolla M, Luo T, Borrell S, Holt KE, Kato-Maeda M, Parkhill J, Malla B, Berg S, Thwaites G, et al. 2013. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet. 4510:1176–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE.. 2004. WebLogo: a sequence logo generator. Genome Res. 146:1188–1190.http://dx.doi.org/10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrington MG, Roche AM, Chauvin SE, Tu Z, Mossman KL, Weiser JN, Bowdish DME.. 2013. MARCO is required for TLR2- and Nod2-mediated responses to Streptococcus pneumoniae and clearance of pneumococcal colonization in the murine nasopharynx. J Immunol. 1901:250–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin RM, Altshuler DL, Durbin RM, Abecasis GR, Bentley DR, Chakravarti A, Clark AG, Collins FS, De La Vega FM, Donnelly P.. 2010. A map of human genome variation from population-scale sequencing. Nature 4677319:1061–1073.http://dx.doi.org/10.1038/nature09534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elomaa O, Sankala M, Pikkarainen T, Bergmann U, Tuuttila A, Raatikainen-Ahokas A, Sariola H, Tryggvason K.. 1998. Structure of the human macrophage MARCO receptor and characterization of its bacteria-binding region. J Biol Chem. 2738:4530–4538. [DOI] [PubMed] [Google Scholar]
- Fay JC, Wu CI.. 2000. Hitchhiking under positive Darwinian selection. Genetics 1553:1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough PJ, Greaves DR, Gordon S.. 1998. A naturally occurring isoform of the human macrophage scavenger receptor (SR-A) gene generated by alternative splicing blocks modified LDL uptake. J Lipid Res. 393:531–543. [PubMed] [Google Scholar]
- Greaves DR, Gordon S.. 2009. The macrophage scavenger receptor at 30 years of age: current knowledge and future challenges. J Lipid Res. 50(Suppl):S282–S286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han HJ, Tokino T, Nakamura Y.. 1998. CSR, a scavenger receptor-like protein with a protective role against cellular damage causedby UV irradiation and oxidative stress. Hum Mol Genet. 76:1039–1046.http://dx.doi.org/10.1093/hmg/7.6.1039 [DOI] [PubMed] [Google Scholar]
- Hertz T, Nolan D, James I, John M, Gaudieri S, Phillips E, Huang JC, Riadi G, Mallal S, Jojic N.. 2011. Mapping the landscape of host-pathogen coevolution: HLA class I binding and its relationship with evolutionary conservation in human and viral proteins. J Virol. 853:1310–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- High M, Cho HY, Marzec J, Wiltshire T, Verhein KC, Caballero MT, Acosta PL, Ciencewicki J, McCaw ZR, Kobzik L.. 2016. Determinants of host susceptibility to murine respiratory syncytial virus (RSV) disease identify a role for the innate immunity scavenger receptor MARCO gene in human infants. EBioMedicine 11:73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M, Norazmi M.. 2013. Pattern recognition receptors and cytokines in Mycobacterium tuberculosis infection—the double-edged sword? Biomed Res Int. 2013:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 182:337–338.http://dx.doi.org/10.1093/bioinformatics/18.2.337 [DOI] [PubMed] [Google Scholar]
- Karlsson EK, Kwiatkowski DP, Sabeti PC.. 2014. Natural selection and infectious disease in human populations. Nat Rev Genet. 156:379–393.http://dx.doi.org/10.1038/nrg3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao W, Kang H, Jin G, Chen L, Chu Y, Sun J, Sun B.. 2017. Evaluation of the relationship between MARCO and CD36 single-nucleotide polymorphisms and susceptibility to pulmonary tuberculosis in a Chinese Han population. BMC Infect Dis. 171:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li C, Sarkar S, Zhang J, Witham S, Zhang Z, Wang L, Smith N, Petukh M, Alexov E.. 2012. DelPhi: a comprehensive suite for DelPhi software and associated resources. BMC Biophys. 5:9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M-J, Wang H-B, Li H, Yang J-H, Yan Y, Xie L-P, Qi Y-C, Li J-L, Chen M-J, Liu W, et al. 2011. Genetic variants in MARCO are associated with the susceptibility to pulmonary tuberculosis in Chinese Han population. PLoS One 68:e24069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiela MJ, Chanock SJ.. 2014. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31:3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez VG, Moestrup SK, Holmskov U, Mollenhauer J, Lozano F.. 2011. The conserved scavenger receptor cysteine-rich superfamily in therapy and diagnosis. Pharmacol Rev. 634:967–1000. [DOI] [PubMed] [Google Scholar]
- Meyer M, Kircher M, Gansauge M-T, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prüfer K, de Filippo C, et al. 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science 3386104:222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Ohtani K, Jang S, Kim Y, Hwang I, Roy N, Matsuda Y, Suzuki Y, Wakamiya N.. 2014. Scavenger receptor CL-P1 mainly utilizes a collagen-like domain to uptake microbes and modified LDL. Biochim Biophys Acta 184012:3345–3356. [DOI] [PubMed] [Google Scholar]
- Noonan JP, Coop G, Kudaravalli S, Smith D, Krause J, Alessi J, Chen F, Platt D, Pääbo S, Pritchard JK, et al. 2006. Sequencing and analysis of neanderthal genomic DNA. Science 3145802:1113–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakowski KE, Huynh A, Han S, Dorrington MG, Yin C, Tu Z, Pelka P, Whyte P, Guarné A, Sakamoto K, et al. 2016. A naturally-occurring transcript variant of MARCO reveals the SRCR domain is critical for function. Immunol Cell Biol. 947:646–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala JRM, Pikkarainen T, Tuuttila A, Sandalova T, Tryggvason K.. 2007. Crystal structure of the cysteine-rich domain of scavenger receptor MARCO reveals the presence of a basic and an acidic cluster that both contribute to ligand recognition. J Biol Chem. 28222:16654–16666. [DOI] [PubMed] [Google Scholar]
- Pearson AM. 1996. Scavenger receptors in innate immunity. Curr Opin Immunol. 81:20–28.http://dx.doi.org/10.1016/S0952-7915(96)80100-2 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE.. 2004. UCSF Chimera – a visualization system for exploratory research and analysis. J Comput Chem. 2513:1605–1612. [DOI] [PubMed] [Google Scholar]
- Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B, Veeramah KR, Woerner AE, O’Connor TD, Santpere G, et al. 2013. Great ape genetic diversity and population history. Nature 4997459:471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrer L, Freeman M, Kodama T, Penman M, Krieger M.. 1990. Coiled-coil fibrous domains mediate ligand binding by macrophage scavenger receptor type II. Nature 3436258:570–572. [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y.. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 54:725–738.http://dx.doi.org/10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. 1983. Evolutionary relationships of DNA sequences in finite populations. Genetics 1052:437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1233:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen M, Nordestgaard BG, Kobzik L, Dahl M.. 2013. Genetic variation in the scavenger receptor MARCO and its association with chronic obstructive pulmonary disease and lung infection in 10, 604 individuals. Respiration 852:1–10. [DOI] [PubMed] [Google Scholar]
- Thuong NTT, Tram TTB, Dinh TD, Thai PVK, Heemskerk D, Bang ND, Chau TTH, Russell DG, Thwaites GE, Hawn TR, et al. 2016. MARCO variants are associated with phagocytosis, pulmonary tuberculosis susceptibility and Beijing lineage. Genes Immun. 17:419–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todt JC, Hu B, Curtis JL.. 2008. The scavenger receptor SR-A I/II (CD204) signals via the receptor tyrosine kinase Mertk during apoptotic cell uptake by murine macrophages. J Leukoc Biol. 842:510–518.http://dx.doi.org/10.1189/jlb.0307135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan LJ, Döpp E. a, Haworth R, Pikkarainen T, Kangas M, Elomaa O, Dijkstra CD, Gordon S, Tryggvason K, Kraal G.. 1999. Regulation and functional involvement of macrophage scavenger receptor MARCO in clearance of bacteria in vivo. J Immunol. 1622:939–947. [PubMed] [Google Scholar]
- Watterson GA. 1975. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 72:256–276.http://dx.doi.org/10.1016/0040-5809(75)90020-9 [DOI] [PubMed] [Google Scholar]
- Whelan FJ, Meehan CJ, Golding GB, McConkey BJ, Bowdish DME.. 2012. The evolution of the class a scavenger receptors. BMC Evol Biol. 12:227.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4 : phylogenetic analysis by maximum likelihood. Mol Biol Evol. 248:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yap NVL, Whelan FJ, Bowdish DME, Golding GB.. 2015. The evolution of the scavenger receptor cysteine-rich domain of the class A scavenger receptors. Front Immunol. 6:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.