Abstract
Histone deacetylase (HDAC) catalyzes the removal of the acetyl group from the lysine residues in the N-terminal tails of nucleosomal core histones. Eight human HDACs have been identified so far. Here, we report the identification of a ninth member of the HDAC family, designated HDAC9. HDAC9 is a class II HDAC and its gene resides on human chromosome 7. HDAC9 has several alternatively spliced isoforms. One of these isoforms is histone deacetylase-related protein or myocyte enhancer-binding factor 2-interacting transcriptional repressor that we and others have previously reported and which does not possess an HDAC catalytic domain. The longest of the HDAC9 isoforms contains 1,011 aa. The isoform, designated HDAC9a, is 132 aa shorter at the C terminus than HDAC9. Also, we have identified isoforms of HDAC9 that lack the nuclear localization signal. Similar to histone deacetylase-related protein, HDAC9 transcripts are expressed at high levels in brain and skeletal muscle. The ratio of HDAC9 and HDAC9a transcripts differs among the tissues examined. HDAC9 and HDAC9a contain the HDAC catalytic domain, and Flag-tagged HDAC9 and HDAC9a possess deacetylase activity. HDAC9 and HDAC9a also repress myocyte enhancer-binding factor 2-mediated transcription. In the present study, we have identified HDAC9 and a number of alternatively spliced isoforms of HDAC9 with potentially different biological activities.
The N-terminal tails of core histones are covalently modified by posttranslational modifications, including acetylation and phosphorylation (1). Evidence suggests that these covalent modifications play important roles in several biological activities involving chromatin, e.g., transcription and replication (2). Histone deacetylases (HDACs) catalyze the removal of the acetyl group from the lysine residues in the N-terminal tails of nucleosomal core histones, resulting in a more compact chromatin structure, a configuration that is generally associated with repression of transcription. Five proteins and/or ORFs in yeast [reduced potassium dependency 3 (RPD3), HDA1, HDA one similarity 1 (HOS1), HOS2, and HOS3; refs. 3 and 4) that share significant homology in the catalytic domain have been identified as HDACs based on their sequence homology to human HDAC1 (5). To date, eight HDACs have been identified in mammalian cells and classified into two classes based on their structure and similarity to yeast RPD3 or HDA1 proteins. Recently, Sir2 family proteins that are structurally unrelated to the five proteins aforementioned have been identified as nicotinamide-adenine dinucleotide (NAD)-dependent HDACs (6–8). Class I HDACs are the yeast RPD3 homologs HDAC1, -2, -3, and -8 (5, 9–15) and are composed primarily of a catalytic domain. Class II HDACs are the yeast HDA1 homologs HDAC4, -5, -6, and -7 (16–21). HDAC4, -5, and -7 contain a long noncatalytic N terminus and a C-terminal HDAC catalytic domain, whereas HDAC6 has two HDAC catalytic domains (16–21). The yeast RPD3 and HDA1 and mammalian HDAC1–8 are sensitive to inhibition by trichostatin A (TSA; refs. 3–5 and 9–22). The Sir2 family HDACs (6–8, 23), yeast HOS3 (24), and Drosophila melanogaster dHDAC6 (23) are relatively insensitive to TSA.
We and others have previously identified a protein, designated histone deacetylase 4 and 5 related protein (HDRP) (25) or MEF2-interacting transcriptional repressor (26–28) that is 50% identical to the N-terminal domain of HDAC4 and -5. Here, we report the cloning and characterization of an HDAC, designated HDAC9, of which HDRP is an alternatively spliced isoform. We found other alternatively spliced isoforms of HDAC9, including HDAC9a, which is 132 aa shorter at the C terminus, and isoforms that lack the nuclear localization signal in the N-terminal noncatalytic end. The ratio of HDAC9 and HDAC9a transcripts varies among tissues. HDAC9 and HDAC9a contain the HDAC catalytic domain, and Flag-tagged HDAC9 and HDAC9a possess deacetylase activity and are sensitive to inhibition by TSA and suberoylanilide hydroxamic acid (29). HDAC9 and HDAC9a interact with MEF2 and repress MEF2-mediated transcription.
Materials and Methods
Database Searching and Cloning of HDAC9.
Database analyses indicate that HDRP is located on chromosome 7 (7p15–p21). The GenBank human genome database (February 2001 release) was searched using the human HDAC4 amino acid sequence. The TBLASTN program was used to identify ORFs downstream of HDRP on chromosome 7 that exhibit significant homology to the HDAC domain of HDAC4. Several fragments whose translated products exhibit over 58% identity were retrieved. Two sense primers (OL486, 5′-CCATGGAAACGGTACCCAGCAGGC-3′ and OL487, 5′-CACTCCATCGCTATGATGAAGGG-3′) and antisense primers (OL484, 5′-AGTTCCCTTCATCATAGCGATGG-3′ and OL485, 5′-AATGTACAGGATGCTGGGGT-3′) were designed based on one of these fragments whose translated products matched amino acids 842–873 of HDAC4. Reverse transcription (RT)-PCR was performed with the antisense primers and a sense primer (5′-CCCTTGTAGCTGGTGGAGTTCCCTT-3′) from the coding regions of HDRP and human brain cDNA as templates. Using the OL486 and adaptor primer 1 and marathon-ready cDNA from human brain (CLONTECH), 3′ rapid amplification of cDNA ends was performed according to the manufacturer's instruction. The products were reamplified with nested sense primer OL487 and adaptor primer 2 (CLONTECH). PCR products were cloned into pGEM-T-Easy Vector system (Promega) and sequenced with an automated DNA sequencer at the DNA Sequencing Core Facility of the Memorial Sloan–Kettering Cancer Center.
RT-PCR.
RT-PCR was performed by using Multiple Tissue cDNA panels (CLONTECH) with primers (OL516 5′-TGTGTCATCGAGCTGGCTTC-3′ and OL517 5′-ATCTTCTGCAAGTGGCTCCA-3′) that span the alternatively spliced exon 21. PCR was performed in a TGradient Thermocycler (Biometra, Tampa, FL) for 30 cycles at 95°C for 20 sec, at 60°C for 20 sec, and at 72°C for 60 sec.
Plasmid Constructs.
C-terminal FLAG-tagged HDAC9 (HDAC9-F) and HDAC9a (HDAC9a-F) expression vectors were constructed with pFLAG-CMV-5b vector (Sigma) and with PCR-amplified coding regions of HDAC9 and HDAC9a in-frame with the FLAG-tag. All constructs were confirmed by DNA sequencing.
Northern Blot Analysis.
The Human Multiple Tissue Northern Blot was obtained from CLONTECH. Hybridization was performed with ExPressHyb solution (CLONTECH) according to the manufacturer's instruction. The 32P-random priming-labeled 3′ untranslated region cDNA for human HDAC9 was used as probe.
Transfection, Immunoprecipitation, Western Blot Analysis, and Luciferase Assay.
Transfection of 293T cells, immunoprecipitation with anti-FLAG M2 agarose (Sigma), Western blot analyses, and dual luciferase assays were performed essentially as described (25).
Anti-FLAG antibody was purchased from Sigma and anti-MEF2 (C-21) was purchased from Santa Cruz Biotechnology.
HDAC Enzymatic Assay.
HDAC fluorescent activity assays using a Fluror de Lys substrate (Biomol, Plymouth Meeting, PA), which contains an acetylated lysine side chain, were performed according to manufacturer's instructions, and a SPECTRAmax GEMINI XS microplate spectrofluorometer was used with the SOFTMAX PRO V.3.1.2 system (Molecular Devices) with excitation at 355 nm and emission at 460 nm with a cut off filter of 455 nm.
HDAC enzymatic assays using [3H]histones isolated from murine erythroleukemia cells as a substrate were performed essentially as described (29). For inhibition studies, the immunoprecipitated complexes were preincubated with the different HDAC inhibitors for 30 min at 4°C. Released [3H]acetic acid was quantified by scintillation counting.
Results
Cloning of cDNA that Encodes an HDAC Designated HDAC9.
HDAC9 was cloned by PCR and 3′ rapid amplification of cDNA ends using primers designed from the sequence of human chromosome 7 whose translated product exhibited 80% identity to the HDAC domain of HDAC4. cDNA synthesized from human brain mRNA was used as the template. Two cDNAs were cloned. One encodes a protein that is 1,011 aa long. The other encodes a protein 879 aa long. Database analyses of these cDNAs against human genomic DNA sequences indicate that these two cDNAs are generated by alternative splicing (Fig. 1B).
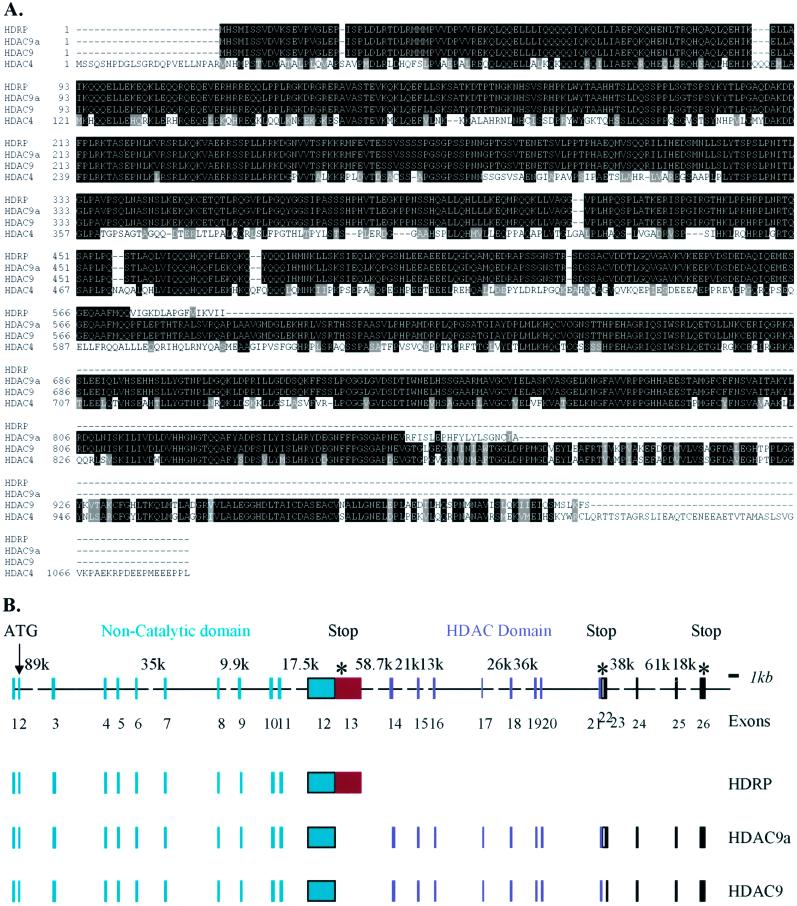
Figure 1.
Amino acid sequence alignment and HDAC9 gene structure. (A) Amino acid sequence alignment of HDAC9, HDAC9a, HDRP, and HDAC4. Deduced amino acid sequences of HDAC9 (GenBank accession no. AY032737) and HDAC9a (accession no. AY032738) are aligned with HDRP (accession no. BAA34464) and HDAC4 (accession no. NP_006028). Identical residues in all proteins are shown in reverse type on black background; similar residues are shaded in gray. (B) Human HDAC9 gene structure. The HDAC9 gene is schematically depicted. Color-coded boxes represent exons present in different isoforms; lines represent introns; broken lines represent larger introns (with size in bp at top). The 5′ untranslated region cDNA and coding region cDNA are represented here.
The HDAC9 cDNA sequences from the known 5′ end of HDRP cDNA to the 3′ untranslated region cloned in this study cover more than 511 kb of genome DNA on chromosome 7 (not shown). The coding region cDNA of HDAC9 resides in 23 exons (Fig. 1B) spanning 458 kb of genomic sequence. Exons 21, 22, and 23 are one single exon in HDAC9a, but the middle exon, number 22, which contains an in-frame stop codon, is spliced out in HDAC9. Exons 12 and 13 are a single exon used by HDRP. Exon 13 is spliced as part of an intron in HDAC9 and HDAC9a. Further, exon 7, which contains a nuclear localization signal, is alternatively spliced in an HDRP isoform (X.Z., L. Ngo, R.A.R., P.A.M., and V.M.R., unpublished data). RT-PCR analyses using primers based on sequences from exon 6 and exon 14 indicate that this alternative splicing event also occurs in HDAC9/HDAC9a (data not shown). It is currently unknown whether both HDAC9 and HDAC9a have this alternatively spliced isoform lacking exon 7. Thus, at least six proteins potentially could be generated from a single HDAC9 gene by alternative splicing of its RNA.
HDAC9 mRNA Is Differentially Expressed Among Human Tissues.
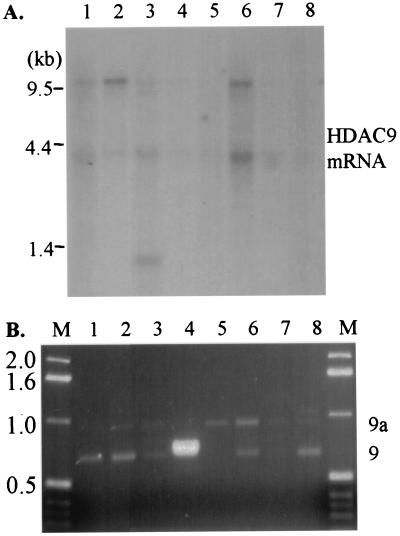
The expression of HDAC9 mRNA was determined by Northern blot analysis using Human Multiple Tissue Northern Blot (CLONTECH). The 3′ untranslated region common to HDAC9 and HDAC9a that shares no significant sequence homology with HDRP was used as a probe. Two transcripts at 9.8 and 4.1 kb were detected in all tissues examined, albeit at different levels (Fig. 2A). The 4.1-kb transcript is shorter than the 4.4-kb HDRP transcript (25). A third transcript at 1.2 kb was detected in placenta (Fig. 2A). Similarly to HDRP (25), high levels of HDAC9 transcripts are detected in brain and skeletal muscle (Fig. 2A).
Figure 2.
HDAC9 mRNA is differentially expressed among human tissues. (A) A Multiple Tissue Northern Blot (CLONTECH) was probed to determine mRNA expression of HDAC9 with a cDNA probe that recognizes both HDAC9 and HDAC9a. The tissues examined are lane 1, heart; lane 2, brain; lane 3, placenta; lane 4, lung; lane 5, liver; lane 6, skeletal muscle; lane 7, kidney; and lane 8, pancreas. Positions of the RNA size marker in kilobase (kb) are indicated on the left of the blot. (B) RT-PCR analyses of mRNA from the same tissues as examined in the Northern blot to determine the distribution of HDAC9 and HDAC9a mRNA among these tissues. PCR products were resolved by agarose gel electrophoresis and visualized by ethidium bromide under UV light. A 1-kb DNA ladder was run on both sides of the gel with the size (in kb) indicated on the left. On the right, the expected products for HDAC9 and HDAC9a are indicated as 9 and 9a, respectively.
The distribution of alternatively spliced mRNA variants among tissues was examined by RT-PCR using primers spanning the alternatively spliced exon 22 and cDNA panel from the same tissues as the Multiple Tissue Northern Blot (CLONTECH). The expected sizes of PCR products are 680 bp for HDAC9 and 993 bp for HDAC9a. The ratio of HDAC9 to HDAC9a transcripts differs among tissues (Fig. 2B). In the placenta and kidney, the levels of the two transcripts are about the same (Fig. 2B). In the brain, heart, and pancreas, there are more transcripts of HDAC9 than HDAC9a. In the other tissues examined, there are more HDAC9a transcripts than HDAC9 (Fig. 2B). Under the conditions tested, HDAC9 transcripts are undetectable in liver (Fig. 2B). The lung has a larger-than-expected size HDAC9 product that is very abundant and has very low levels of HDAC9 or HDAC9a transcripts (Fig. 2B). A larger PCR product was amplified from cDNA of the pancreas than the expected products from HDAC9 and HDAC9a (Fig. 2B). The identities of the different sized transcripts are unknown.
HDAC9 Contains HDAC Activity.
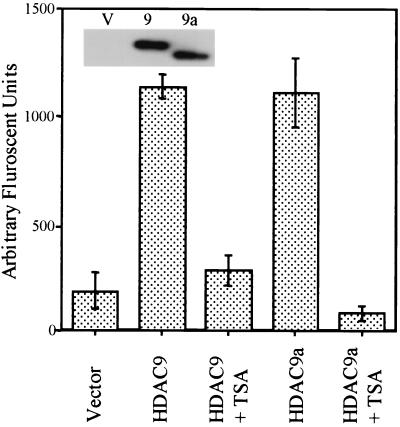
HDAC9 was named based on sequence homology to HDAC4 (Fig. 1A). To determine whether HDAC9 possesses HDAC activity, HDAC enzymatic assay was performed using anti-FLAG immunoprecipitated HDAC9-F and HDAC9a-F. As shown in Fig. 3, both HDAC9-F and HDAC9a-F deacetylate the acetylated lysine of Fluror de Lys substrate (Biomol), which contains an acetylated lysine side chain; the activity of HDAC9 and HDAC9a is comparable. TSA inhibits HDAC9 and HDAC9a deacetylase activity (Fig. 3). The Inset gel shows the amount of protein used in the assay. Suberoylanilide hydroxamic acid, a potent HDAC inhibitor (29), also can inhibit completely the HDAC activity of HDAC9-F and HDAC9a-F (data not shown). The HDAC activity of HDAC9 and HDAC9a was about 1/10 the deacetylase activity of HDAC4 when a comparable amount of protein was used under conditions tested here (data not shown). HDAC9-F and HDAC9a-F also deacetylate [3H]acetyl-histones (data not shown).
Figure 3.
HDAC9 and HDAC9a possess deacetylase activity. The HDAC enzymatic assays were performed with anti-FLAG-immunoprecipitated proteins from vector control, HDAC9-, and HDAC9a-transfected 293T cells by using Fluror de Lys substrate, which contains an acetylated lysine side chain. TSA (1 μM) was used as an HDAC inhibitor. Results are shown as the mean of two independent assays. (Inset) Anti-FLAG Western blot shows the amount of proteins used in the assay. V, vector control; 9, HDAC9-F; 9a, HDAC9a-F.
HDAC9 Represses MEF2-Mediated Transcription.
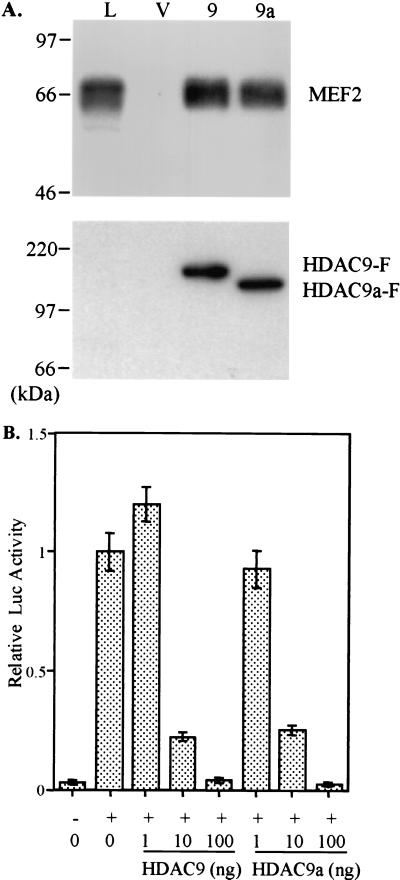
The Xenopus homolog of HDRP was identified as an MEF2-interacting transcriptional repressor (26); mouse HDRP also interacts with and represses MEF2-mediated transcription (27). We first tested whether HDAC9 and HDAC9a interact with MEF2. As shown in Fig. 4A, both HDAC9 and HDAC9a interact with MEF2 in 293T cells. We then tested whether HDAC9 and HDAC9a repress MEF2-mediated transcription. The p3XMEF2-luciferase reporter gene was transfected into 293T cells in the absence or presence of MEF2C. HDAC9-F or HDAC9a-F was included in a subset of experimental groups with MEF2C. MEF2C activates transcription over 30 times the basal level of transcription under conditions used in our experiments. HDAC9-F and HDAC9a-F repress MEF2C-mediated transcriptional activation in a dose-dependent manner and abolish the activation at the 100-ng dose for both HDAC9 and HDAC9a (Fig. 4B). The transcriptional repression effect of HDAC9 and HDAC9a on MEF2C-mediated transcription is a specific effect because a cotransfected reporter gene for transfection efficiency containing a thymidine kinase (TK) promoter was not repressed by HDAC9 or HDAC9a-F (data not shown).
Figure 4.
HDAC9 and HDAC9a interact with and repress MEF2-mediated transcription. (A) Western blot analyses of the 293T whole-cell lysate and anti-FLAG immunoprecipitates from 293T cells transfected with vector, HDAC9-F, or HDAC9a-F by using antibodies against MEF2 and FLAG. (Upper) Anti-MEF2 Western blot; (Lower) anti-FLAG Western blot. L, 293T whole-cell lysate; V, vector control immunoprecipitate (IP); 9, HDAC9-F IP; 9a, HDAC9a-F IP. (B) p3XMEF2-Luc (100 ng) and pRL-TK (5 ng) were transfected into 293T cells with pcDNA3 empty vector (−) or with pCMV-MEF2C (100 ng) (+) along with indicated amount of pFLAG-HDAC9 or pFLAG-HDAC9a. pFLAG empty vector was used to adjust the DNA to an equal amount in each transfection. The firefly luciferase activity was first normalized to the cotransfected Renilla luciferase activity, and then the value for MEF2C alone was set as 1. Results are shown as the mean of three independent transfections ± SD.
Discussion
Here, we report the identification and characterization of a ninth class II HDAC, designated HDAC9. HDAC9 has several alternatively spliced isoforms, one of which is the previously identified HDRP (25). HDAC9 and HDAC9a possess HDAC activity. However, under conditions tested here, HDAC9 and HDAC9a seem to have a lower specific enzymatic activity than HDAC4. It is possible that an essential cofactor is lost during immunoprecipitation or does not exist in 293T cells (for example, metastasis-associated protein 2 is essential for the assembly of a catalytically active HDAC1; ref. 30), the substrates used are not its natural substrate, or the FLAG tag interferes with the folding of the protein.
Alternative splicing plays an important role in expanding protein diversity and providing complexity for vertebrates (31, 32). Initial analysis of the human genome suggests that alternative splicing is a common event (33, 34). Here, in the case of HDAC9, at least six different proteins may be generated from the same gene by alternative splicing: HDAC9, HDAC9a, HDRP, and each of the three proteins lacking the exon containing a nuclear localization signal, respectively. These alternatively spliced transcripts seem to give rise to proteins with different functions—e.g., a transcriptional repressor (HDRP) without deacetylase activity or both a repressor and an active deacetylase (HDAC9/HDAC9a). Alternative splicing could also control the intracellular localization of these proteins depending upon whether the nuclear localization signal is present, and as a consequence, the function of the proteins may be different. The regulation of cellular localization of HDAC9 and its splicing isoforms by alternative splicing is very intriguing because intracellular localization of HDRP, HDAC4, and HDAC5 is regulated by posttranslational modification and protein–protein interaction. For example, HDAC4 intracellular localization is regulated by oncogenic Ras (35) and 14-3-3 proteins (36–38) and during muscle differentiation (39). Intracellular localization of HDRP, one of the alternatively spliced isoforms of HDAC9, is regulated by 14-3-3 and calmodulin-dependent kinase (ref. 28, and our unpublished observation).
Searching the human genome with the HDAC domain from either HDAC1 or HDAC9 identified a total of 10 HDACs in the presently completed human genome sequence. Based on its sequence homology to class II HDAC, HDAC10 (GenBank accession no. CAB63048) is a sequence directly deposited in the database. In addition, there are several potential pseudogenes in the human genome based on their apparent lack of intron sequences on chromosomes 1, 10, and 11. The first 164 aa of HDAC4 have a perfect match on chromosome 3, and part of the HDAC9 HDAC domain has a perfect match on chromosome 18.
Other HDAC family members have isoforms (10). Of the eight class I and II human HDACs reported, HDAC3 has two reported isoforms—i.e., HDAC3A and HDAC3C (10). HDAC3, HDAC3A, and HDAC3C differ at the N terminus. The authors speculated that these isoforms of HDAC3 are the result of either alternative splicing or other posttranscriptional modifications. Database analyses of human genome sequences suggest that HDAC3C either is an alternatively spliced isoform of HDAC3, or it is transcribed from a different transcriptional start site. HDAC3A, which has a direct repeat of 101 bp at the 5′ end of its cDNA, could be an artifact formed during library construction, although it is possible that it is generated by other posttranscriptional modifications, as the authors suggested (10). Database analyses of other members of the HDAC family across the human genome sequence indicate that alternatively spliced isoforms exist for HDAC7. Although HDAC7 is the least homologous to HDAC9 at the N-terminal noncatalytic domain among class II HDACs, HDAC7 has a similar number of alternatively spliced isoforms as HDAC9. There appear to be two HDRP-like proteins in the database for HDAC7 and three isoforms of HDAC7. The functional significance of these splicing isoforms is unknown. The finding that the transcripts for different HDAC9 splicing isoforms are expressed in different amounts in different tissues suggests that different isoforms may have different functions in different tissues.
Acknowledgments
We thank L. Ngo and D. J. Chen for expert technical assistance and Dr. L. Butler for helpful discussion. X.Z. is a Cohen Fellow in Biomedical Research, and this work was made possible, in part, by funds granted by the Michael and Ethel Cohen Foundation (to X.Z.). These investigations were supported, in part, by Grant CA-0974823 from the National Cancer Institute (to R.A.R. and P.A.M.), and by grants from the Japan Foundation for Promotion of Cancer Research Fund, the Kleberg Foundation, and the DeWitt Wallace Fund for Memorial Sloan–Kettering Cancer Center.
Abbreviations
- HDAC
histone deacetylase
- MEF2
myocyte enhancer factor 2
- HDRP
histone deacetylase 4 and 5 related protein
- TSA
trichostatin A
- RT
reverse transcription
Footnotes
References
- 1.Davie J R, Spencer V A. J Cell Biochem. 1999;32–33,Suppl.:141–148. doi: 10.1002/(sici)1097-4644(1999)75:32+<141::aid-jcb17>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 2.Strahl B D, Allis C D. Nature (London) 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 3.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carmen A A, Rundlett S E, Grunstein M. J Biol Chem. 1996;271:15837–15844. doi: 10.1074/jbc.271.26.15837. [DOI] [PubMed] [Google Scholar]
- 5.Taunton J, Hassig C A, Schreiber S L. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 6.Smith J S, Brachmann C B, Celic I, Kenna M A, Muhammad S, Starai V J, Avalos J L, Escalante-Semerena J C, Grubmeyer C, Wolberger C, Boeke J D. Proc Natl Acad Sci USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imai S, Armstrong C M, Kaeberlein M, Guarente L. Nature (London) 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 8.Landry J, Sutton A, Tafrov S T, Heller R C, Stebbins J, Pillus L, Sternglanz R. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. . (First Published May 16, 2000; 10.1073/pnas.110148297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang W M, Inouye C, Zeng Y, Bearss D, Seto E. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang W M, Yao Y L, Sun J M, Davie J R, Seto E. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 11.Emiliani S, Fischle W, Van Lint C, Al-Abed Y, Verdin E. Proc Natl Acad Sci USA. 1998;95:2795–2800. doi: 10.1073/pnas.95.6.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dangond F, Hafler D A, Tong J K, Randall J, Kojima R, Utku N, Gullans S R. Biochem Biophys Res Commun. 1998;242:648–652. doi: 10.1006/bbrc.1997.8033. [DOI] [PubMed] [Google Scholar]
- 13.Buggy J J, Sideris M L, Mak P, Lorimer D D, McIntosh B, Clark J M. Biochem J. 2000;350:199–205. [PMC free article] [PubMed] [Google Scholar]
- 14.Van den Wyngaert I, de Vries W, Kremer A, Neefs J, Verhasselt P, Luyten W H, Kass S U. FEBS Lett. 2000;478:77–83. doi: 10.1016/s0014-5793(00)01813-5. [DOI] [PubMed] [Google Scholar]
- 15.Hu E, Chen Z, Fredrickson T, Zhu Y, Kirkpatrick R, Zhang G F, Johanson K, Sung C M, Liu R, Winkler J. J Biol Chem. 2000;275:15254–15264. doi: 10.1074/jbc.M908988199. [DOI] [PubMed] [Google Scholar]
- 16.Verdel A, Khochbin S. J Biol Chem. 1999;274:2440–2445. doi: 10.1074/jbc.274.4.2440. [DOI] [PubMed] [Google Scholar]
- 17.Fischle W, Emiliani S, Hendzel M J, Nagase T, Nomura N, Voelter W, Verdin E. J Biol Chem. 1999;274:11713–11720. doi: 10.1074/jbc.274.17.11713. [DOI] [PubMed] [Google Scholar]
- 18.Grozinger C M, Hassig C A, Schreiber S L. Proc Natl Acad Sci USA. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miska E A, Karlsson C, Langley E, Nielsen S J, Pines J, Kouzarides T. EMBO J. 1999;18:5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang A H, Bertos N R, Vezmar M, Pelletier N, Crosato M, Heng H H, Th'ng J, Han J, Yang X J. Mol Cell Biol. 1999;19:7816–7827. doi: 10.1128/mcb.19.11.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kao H Y, Downes M, Ordentlich P, Evans R M. Genes Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida M, Kijima M, Akita M, Beppu T. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 23.Barlow A L, van Drunen C M, Johnson C A, Tweedie S, Bird A, Turner B M. Exp Cell Res. 2001;265:90–103. doi: 10.1006/excr.2001.5162. [DOI] [PubMed] [Google Scholar]
- 24.Carmen A A, Griffin P R, Calaycay J R, Rundlett S E, Suka Y, Grunstein M. Proc Natl Acad Sci USA. 1999;96:12356–12361. doi: 10.1073/pnas.96.22.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou X, Richon V M, Rifkind R A, Marks P A. Proc Natl Acad Sci USA. 2000;97:1056–1061. doi: 10.1073/pnas.97.3.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sparrow D B, Miska E A, Langley E, Reynaud-Deonauth S, Kotecha S, Towers N, Spohr G, Kouzarides T, Mohun T J. EMBO J. 1999;18:5085–5098. doi: 10.1093/emboj/18.18.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang C L, McKinsey T A, Lu J R, Olson E N. J Biol Chem. 2001;276:35–39. doi: 10.1074/jbc.M007364200. [DOI] [PubMed] [Google Scholar]
- 28.Zhang C L, McKinsey T A, Olson E N. Proc Natl Acad Sci USA. 2001;98:7354–7359. doi: 10.1073/pnas.131198498. . (First Published June 5, 2001; 10.1073/pnas.131198498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richon V M, Emiliani S, Verdin E, Webb Y, Breslow R, Rifkind R A, Marks P A. Proc Natl Acad Sci USA. 1998;95:3003–3007. doi: 10.1073/pnas.95.6.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Ng H H, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Black D L. Cell. 2000;103:367–370. doi: 10.1016/s0092-8674(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 32.Graveley B R. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 33.Lander E S, Linton L M, Birren B, Nusbaum C, Zody M C, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature (London) 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 34.Venter J C, Adams M D, Myers E W, Li P W, Mural R J, Sutton G G, Smith H O, Yandell M, Evans C A, Holt R A, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 35.Zhou X, Richon V M, Wang A H, Yang X J, Rifkind R A, Marks P A. Proc Natl Acad Sci USA. 2000;97:14329–14333. doi: 10.1073/pnas.250494697. . (First Published December 12, 2000; 10.1073/pnas.250494697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grozinger C M, Schreiber S L. Proc Natl Acad Sci USA. 2000;97:7835–7840. doi: 10.1073/pnas.140199597. . (First Published June 27, 2000; 10.1073/pnas.140199597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang A H, Kruhlak M J, Wu J, Bertos N R, Vezmar M, Posner B I, Bazett-Jones D P, Yang X J. Mol Cell Biol. 2000;20:6904–6912. doi: 10.1128/mcb.20.18.6904-6912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKinsey T A, Zhang C L, Olson E N. Proc Natl Acad Sci USA. 2000;97:14400–14405. doi: 10.1073/pnas.260501497. . (First Published December 12, 2000; 10.1073/pnas.260501497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKinsey T A, Zhang C L, Lu J, Olson E N. Nature (London) 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]