Abstract
Human high-altitude (HA) adaptation or mal-adaptation is explored to understand the physiology, pathophysiology, and molecular mechanisms that underlie long-term exposure to hypoxia. Here, we report the results of an analysis of the largest whole-genome-sequencing of Chronic Mountain Sickness (CMS) and nonCMS individuals, identified candidate genes and functionally validated these candidates in a genetic model system (Drosophila). We used PreCIOSS algorithm that uses Haplotype Allele Frequency score to separate haplotypes carrying the favored allele from the noncarriers and accordingly, prioritize genes associated with the CMS or nonCMS phenotype. Haplotypes in eleven candidate regions, with SNPs mostly in nonexonic regions, were significantly different between CMS and nonCMS subjects. Closer examination of individual genes in these regions revealed the involvement of previously identified candidates (e.g., SENP1) and also unreported ones SGK3, COPS5, PRDM1, and IFT122 in CMS. Remarkably, in addition to genes like SENP1, SGK3, and COPS5 which are HIF-dependent, our study reveals for the first time HIF-independent gene PRDM1, indicating an involvement of wider, nonHIF pathways in HA adaptation. Finally, we observed that down-regulating orthologs of these genes in Drosophila significantly enhanced their hypoxia tolerance. Taken together, the PreCIOSS algorithm, applied on a large number of genomes, identifies the involvement of both new and previously reported genes in selection sweeps, highlighting the involvement of multiple hypoxia response systems. Since the overwhelming majority of SNPs are in nonexonic (and possibly regulatory) regions, we speculate that adaptation to HA necessitates greater genetic flexibility allowing for transcript variability in response to graded levels of hypoxia.
Keywords: adaptation, Chronic Mountain Sickness, selection sweep, high-altitude, hypoxia, Monge's disease
Introduction
Reports to date have shown that over 83 million people permanently reside at altitudes above 2,500 m (Beall 2014). Assuming an average global population growth-rate of 1.2%, the present estimate of people living above 2,500 m would be potentially >100 million, about a third of the entire US population. The real numbers could be higher if we take into account the progress made in the healthcare system of these remote areas and the job-related population influx.
The major problem that high-altitude (HA) population faces is the drop in partial pressure of oxygen with altitude, for example, at 2,500 m it is 25% less than at sea level. A constant exposure to hypoxia at HA often leads to a variety of environmentally-related clinical conditions and Chronic Mountain Sickness (CMS, also called Monge's disease; Monge 1942) mostly affecting male highlanders. An international consensus on chronic and subacute HA diseases defined CMS as multifactorial (Leon-Velarde et al. 2005) and primarily characterized by excessive erythrocytosis. Its prevalence varies with both altitude and region. For example, CMS incidence is high in Andeans (∼18%), lesser in Tibetans (1–11%), and absent from the Ethiopian population (Monge et al. 1989; Xing et al. 2008), further mystifying this disease pathogenesis. Therefore, a clear understanding of its pathophysiology would be beneficial to the large HA population at risk of developing this syndrome. It would also provide insights in understanding many disease pathophysiologies where hypoxia plays a major role, at sea level, for example, stroke, cardiac ischemia, obstructive sleep apnea, sickle cell disease.
The search for better understanding of HA adaptation has led to multiple studies primarily focusing on three major HA human populations, that is, Andean, Tibetan, and Ethiopians (Beall et al. 2010; Bigham et al. 2010; Simonson et al. 2010; Alkorta-Aranburu et al. 2012; Scheinfeldt et al. 2012; Udpa et al. 2014). The long history of their settlement at HA has led to the recognition that these populations are genetically adapted to the selection pressure, and certain regions in their genomes would display signatures of “selective sweeps”. However, CMS individuals in these populations might not have adapted to HA, and may lack alleles that are favored by selection. Various methods have been proposed for effective detection of selective sweeps, including differences in allele frequency spectrum and conservation of haplotypes carrying the favored allele (Sabeti et al. 2007). Selection pressure leaves its signature on large regions (≥200 kbp) of the genome, making detection possible even with low sample sizes, as long as the regions are densely genotyped (Zhou et al. 2013; Udpa et al. 2014). In our recent effort to understand the basis of adaptation or mal-adaptation to HA, we analyzed whole genomes of individuals from HA populations for genetic variation (Zhou et al. 2013; Udpa et al. 2014). Using robust selection detection methods, we were able to discover several regions in the genomes that were significantly associated with altitude adaptation. Instead of relying solely on statistical association, we went further and functionally validated few of these genes in a model system (Zhou et al. 2013; Udpa et al. 2014; Stobdan et al. 2015; Azad et al. 2016).
Our previous results suggested that many genes mediating the adaptation to HA remain to be discovered. In this paper, we extend the research to a significantly larger sample of 94 deeply sequenced whole genomes. We used multiple tests to detect positive selection signature in the genomes, and we identified a large number of putative candidates. To prioritize the candidates, we utilized an algorithm, PreCIOSS (Predicting Carriers of Ongoing Selective Sweeps) that we recently developed (Ronen et al. 2015). PreCIOSS utilizes the Haplotype Allele Frequency (HAF) score to separate haplotypes carrying the favored allele from the noncarriers, and allows us to prioritize genes that have a significant association between CMS/nonCMS status and the carrier/noncarrier status. Finally, in order to functionally validate the role of the prioritized genes in CMS, we used a fly model to test if the downregulation of ortholog genes would have an impact on the eclosion rate under hypoxic conditions.
Results
Genetic Ancestry of Andean HA Population
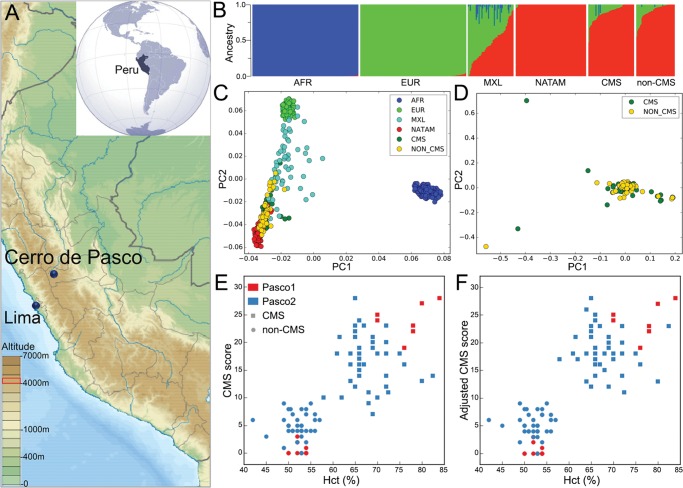
This study involves the largest whole genome sequencing investigation of CMS and nonCMS subjects from the town of Cerro de Pasco, in Peru (N = 94, Altitude >4,300 m, fig. 1A). This includes whole-genome-sequence of 20 individuals (10 CMS and 10 nonCMS) from our previous study, denoted as “Pasco1” (Zhou et al. 2013) and 74 new subjects (CMS = 40 and nonCMS = 34) denoted as “Pasco2”. To understand the genetic composition of the Andean individuals, we applied ADMIXTURE using three reference populations, AFR (150 individuals), EUR (150 individuals), and a Native American population, NATAM (100 individuals; Reich et al. 2012), as well as the MXL population (a lowland control population from 1000 Genomes project) for comparison. The analysis (fig. 1B, supplementary fig. S1, Supplementary Material online) showed the expected result that both MXL and the Andean populations have a significant NATAM ancestry. However, MXL has higher European ancestry compared with the Andeans. A statistical test for difference between European ancestry in CMS versus nonCMS could not reject the null hypothesis (P-value = 0.89). Moreover, AFR ancestry is negligible in the Andeans. Importantly, CMS and nonCMS populations have the same genetic composition. An analysis using principal components also suggested that CMS and nonCMS cluster together, and are genetically closest to NATAM (fig. 1C and D and supplementary fig. S1, Supplementary Material online).
Fig. 1.
Geographic locations, genetic admixture, and phenotypic characterization of samples collected. (A) Location of the studied population (altitude >4,300 m asl, Cerro de Pasco) in Andean Altiplano of South America. Inset: Map locations of Peru. (B) ADMIXTURE analysis (Chr. 19) using three reference populations, AFR (150 individuals), EUR (150 individuals), both from 1000 Genomes project and a Native American population, NATAM (100 individuals) from Reich et al. (2012). Although MXL (a lowland control population from 1000 Genomes project) and the Andean populations (CMS and nonCMS) have a significant NATAM ancestry; however, MXL has higher European ancestry compared with the Andeans. (C) PCA plots of the Andean (CMS and nonCMS) population projected on MXL controls and the reference populations and (D) PCA plot of CMS versus nonCMS suggested that both CMS and nonCMS cluster together, and are genetically closest to NATAM (E) Hematocrit (%) level versus the CMS score. High hematocrit is the major phenotypic characteristic for the high CMS score. (C) Adjusted CMS score (as mentioned in Results and Methods) further demarcate CMS from nonCMS subjects.
Since excessive erythrocytosis is a significant phenotype among CMS patients, there was a clear distinction in hematocrit levels between CMS and nonCMS individuals (fig. 1E, supplementary fig. S2, Supplementary Material online). In order to merge the scores from two CMS scoring systems, we normalized the newer scores to have the same mean and variance as the old scores (fig. 1F).
Genomic Regions under HA Selective Sweeps
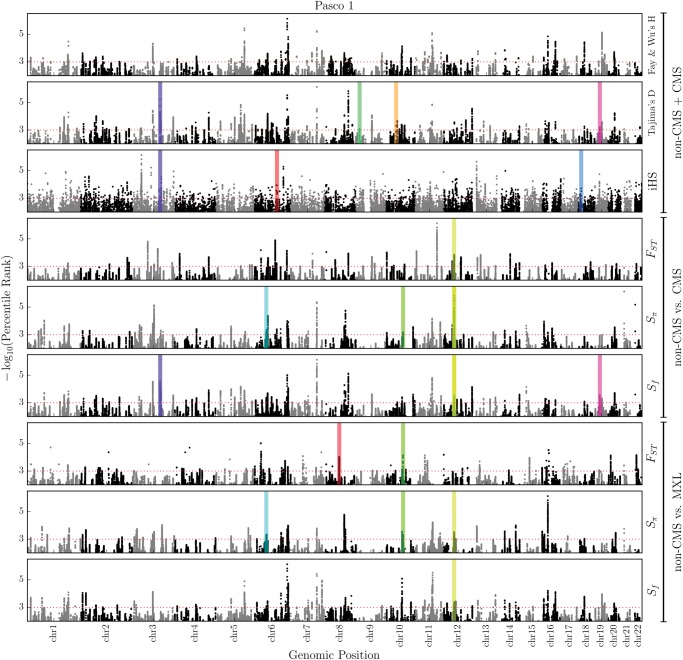
We first selected 60 (30 CMS, 30 nonCMS) of 94 individuals with the most extreme phenotypes, that is, for the CMS group a CMS-score = 20.1 ± 2.8 and for nonCMS a CMS-score = 5.5 ± 1.7. This step was aimed at getting the strongest signals of selection from comparisons between the most severe CMS patients (CMS-score = 20.1 ± 2.8) and the controls (CMS-score = 5.5 ± 1.7). Among this cohort, 20 + 20 (CMS + nonCMS) individuals were from Pasco2, and 10 + 10 (CMS + nonCMS) from Pasco1. While Pasco1 (fig. 2) and Pasco2 (supplementary fig. S3, Supplementary Material online) are samples from the same location, the individuals were sequenced at different time in different locations, using slightly different technologies. Out of an abundance of caution, we decided to separate the two in our cross-population tests. To determine which DNA region is under positive selection, we applied multiple tests of selection and a robust method to prioritize regions that we deemed important and termed “candidate regions”. A detailed account of selection criteria is provided in the Materials and Methods section. Briefly, for within population selection tests, that is, Tajima's D, Fay & Wu's H and iHS, both CMS and nonCMS individuals were combined together. The results of the ADMIXTURE and PC analysis show them largely as identical populations. Therefore, a selection in CMS subjects is anticipated to be similar to that of the nonCMS but with a lower frequency of the favored allele (fig. 2, and supplementary fig. S3, Supplementary Material online). In cross-population selection scans, we first compared nonCMS to CMS from Pasco1 and subsequently nonCMS (Pasco1) to MXL, a lowland control population from 1000 Genomes project (fig. 2). A similar test was also performed for Pasco2 (supplementary fig. S3, Supplementary Material online). We then excluded 313 gap regions, such as in short arms, centromeres, telomeres, heterochromatin, clones, and contigs, as annotated on GRCh37/hg19 (UCSC Genome Browser). These gap regions covered 196 Mbp out of a total of 2,881 Mbp in the 22 autosomes, representing only ∼6.8% of the autosomal genome (supplementary table S1, Supplementary Material online). We then applied selection scans on the autosomal chromosomes of CMS and nonCMS subjects. We retained only regions that had scores in the top 0.1% for at least one of the test statistics in both cohorts—the Pasco1 (fig. 2), and Pasco2 (supplementary fig. S3, Supplementary Material online). We also retained regions having transcripts (using RefSeq release 59 coordinates) within 50 kbp upstream or downstream of the signal resulting in 129 regions.
Fig. 2.
Genome-wide scan, for “Pasco1” (Pasco2 in supplementary material, Supplementary Material online), to detect genomic regions under selective sweep detected using individual statistical tests indicated on right Y-axis. We applied seven tests in sliding windows of 50 kbp (step size 2 kbp) along the entire genome. These tests detect either deviation from the neutral allele frequency spectrum or the frequency of carrier haplotypes. The horizontal dotted lines depict the 0.1% threshold. The prioritized candidate regions (n = 11) from PreCIOSS are highlighted with different colors at their respective genomic position.
For the final prioritization step, we used the PreCIOSS algorithm (Ronen et al. 2015), that uses the HAF score, on the entire cohort (N = 94, 20 from Pasco1 and 74 from Pasco2) to characterize and differentiate carriers of the favored mutation from noncarriers in an ongoing selective sweep, without knowledge of the favored mutation. Each haplotype in the entire cohort was assigned a carrier/noncarrier status by PreCIOSS. We hypothesized that nonCMS individuals would be enriched with “carrier haplotypes”, and the reverse would apply to CMS individuals. For each of the 129 regions previously prioritized, we applied a two-tailed Fisher Exact test with the null hypothesis of no correlation between carrier haplotypes and nonCMS status. We thus identified 11 regions containing 38 genes that were significant (P < 0.05; figs. 2 and 3, table 1, supplementary fig. S3, Supplementary Material online). Here, we describe each prioritized region based on their functional assessment.
Table 1.
Genes in the Top 11 Candidate Regions.
| Region | Chr | Carrier Haplotype Frequency |
P Valuea | Coordinates |
Gene Symbol | Gene Name | Location | ||
|---|---|---|---|---|---|---|---|---|---|
| CMS | NonCMS | Start | End | ||||||
| 1 | 8 | 0.12 | 0.28 | 5.74E-03 | 67, 620, 607 | 68, 221, 368 | PTTG3P | Pituitary tumor-transforming 3, pseudogene | 8q13.1 |
| C8orf44 | Chromosome 8 open reading frame 44 | 8q13.1 | |||||||
| SGK3 | Serum/glucocorticoid regulated kinase family member 3 | 8q13.1 | |||||||
| C8orf44-SGK3 | C8orf44-SGK3 readthrough | 8q13.1 | |||||||
| VCPIP1 | Valosin containing protein interacting protein 1 | 8q13.1 | |||||||
| PPP1R42 | Protein phosphatase 1 regulatory subunit 42 | 8q13.1 | |||||||
| CSPP1 | Centrosome and spindle pole associated protein 1 | 8q13.1-q13.2 | |||||||
| COPS5 | COP9 signalosome subunit 5 | 8q13.1 | |||||||
| TCF24 | Transcription factor 24 | 8q13.1 | |||||||
| SNHG6 | small nucleolar RNA host gene 6 | 8q13.1 | |||||||
| MCMDC2 | Minichromosome maintenance domain containing 2 | 8q13.1 | |||||||
| SNORD87 | Small nucleolar RNA, C/D box 87 | 8q13.1 | |||||||
| 2 | 10 | 0.61 | 0.78 | 1.15E-02 | 47, 618, 434 | 47, 688, 356 | ANTXRL | Anthrax toxin receptor-like | 10q11.22 |
| ANTXRLP1 | Anthrax toxin receptor-like pseudogene 1 | 10q11.22 | |||||||
| 3 | 6 | 0.2 | 0.36 | 1.45E-02 | 106, 358, 412 | 106, 503, 931 | PRDM1 | PR/SET domain 1 | 6q21 |
| 4 | 6 | 0.89 | 0.98 | 2.13E-02 | 56, 066, 358 | 56, 150, 981 | COL21A1 | Collagen type XXI alpha 1 chain | 6p12.1 |
| 5 | 3 | 0.75 | 0.89 | 2.34E-02 | 129, 094, 497 | 129, 331, 906 | MBD4 | Methyl-CpG binding domain 4, DNA glycosylase | 3q21.3 |
| IFT122 | Intraflagellar transport 122 | 3q21.3-q22.1 | |||||||
| EFCAB12 | EF-hand calcium binding domain 12 | 3q21.3 | |||||||
| RPL32P3 | Ribosomal protein L32 pseudogene 3 | 3q21.3 | |||||||
| SNORA7B | Small nucleolar RNA, H/ACA box 7B | 3q21.3 | |||||||
| 6 | 19 | 0.61 | 0.76 | 2.90E-02 | 22, 811, 834 | 23, 078, 159 | ZNF492 | Zinc finger protein 492 | 19p13.11 |
| ZNF99 | Zinc finger protein 99 | 19p12 | |||||||
| ZNF723P | Zinc finger protein 723, pseudogene | 19p12 | |||||||
| 7 | 12 | 0.73 | 0.86 | 3.03E-02 | 48, 777, 513 | 48, 886, 484 | OR8S1 | Olfactory receptor family 8 subfamily S member 1 | 12q13.2 |
| C12orf54 | Chromosome 12 open reading frame 54 | 12q13.11 | |||||||
| ANP32D | Acidic nuclear phosphoprotein 32 family member D | 12q13.11 | |||||||
| ZNF641 | Zinc finger protein 641 | 12q13.11 | |||||||
| 8 | 12 | 0.81 | 0.92 | 3.44E-02 | 48, 431, 197 | 48, 529, 882 | MIR6505 | MicroRNA 6505 | 12q13.11 |
| ASB8 | Ankyrin repeat and SOCS box containing 8 | 12q13.11 | |||||||
| SENP1 | SUMO1/sentrin specific peptidase 1 | 12q13.11 | |||||||
| PFKM | Phosphofructokinase, muscle | 12q13.11 | |||||||
| COL2A1 | Collagen type II alpha 1 chain | 12q13.11 | |||||||
| CCDC184 | Coiled-coil domain containing 184 | 12q13.11 | |||||||
| 9 | 10 | 0.8 | 0.91 | 4.12E-02 | 79, 707, 274 | 79, 893, 495 | RPS24 | Ribosomal protein S24 | 10q22.3 |
| POLR3A | RNA polymerase III subunit A | 10q22.3 | |||||||
| 10 | 9 | 0.69 | 0.82 | 4.51E-02 | 17, 053, 836 | 17, 477, 199 | CNTLN | Centlein | 9p22.2 |
| 11 | 18 | 0.15 | 0.43 | 2.08E-05 | 13, 885, 293 | 13, 918, 846 | MC2R | Melanocortin 2 receptor | 18p11.2 |
Two-Tailed Fisher Exact; the start and end position are from genome assembly version GRCh37/hg19.
Candidate Region Containing SGK3 to COPS5
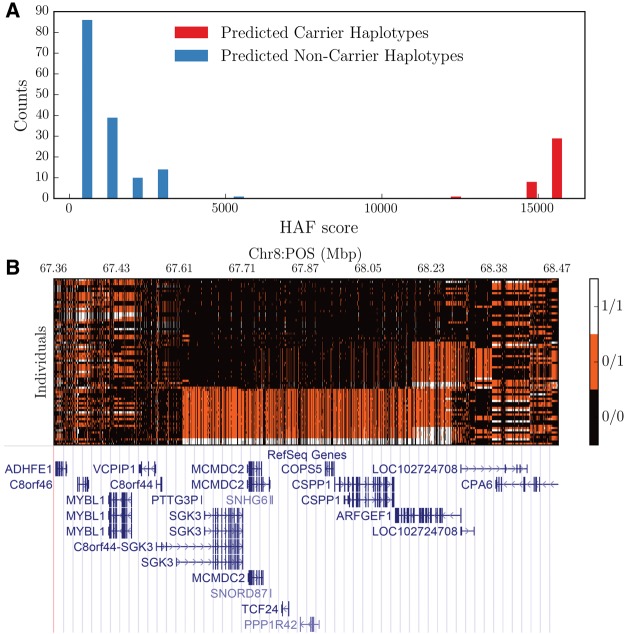
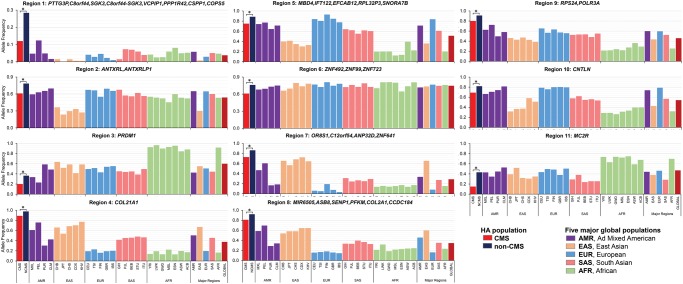
At the top of the prioritized candidate regions was a 600 kb region on chromosome 8 (chr8: 67,620,607–68,221,368). Interestingly, this region was identified in one of our previous studies (supplementary table S5, Supplementary Material online in Zhou et al. 2013). This region had a significant enrichment of the favored allele among nonCMS subjects. As depicted in figure 4A, both carrier and noncarrier haplotypes were concentrated in a few bins (HAF score of noncarriers haplotype was smaller than that in the carriers). Of the total 188 haplotypes, 37 were carriers and their frequency distribution in nonCMS was 28.4% (25/88) which was more than double that of CMS’s 12% (12/100), P = 0.006 (table 1, fig. 4B). Furthermore, the CMS group and only one population, that is, PEL (Peruvians from Lima, Peru), had a frequency of ∼12%, while the frequency in the global populations was <10% (fig. 3).
Fig. 4.
HAF score distribution in the chromosome 8 candidate region (chr8: 67, 620, 607–68, 221, 368, hg19). (A) Predicting Carriers of Ongoing Selective Sweeps (PreCIOSS) result on 94 samples (188 Haplotypes). This algorithm predicted 37 out of 188 haplotypes to be carriers of the selective sweeps. We used Two-Tailed Fisher exact test as a measure of correlation between nonCMS and carriers (table 1). (B) Genotypes of individuals at each SNV site in 94 samples. Each row is an individual, each column is a SNV, 0/0 represents homozygous of a major allele, 0/1 represents heterozygous, and 1/1 represents homozygous form of minor alleles. Haplotypes are sorted out by their HAF scores in ascending order from top to bottom.
Fig. 3.
Haplotype Allele Frequency distribution of 11 prioritized regions in CMS and nonCMS compare with the other populations from the 1000 genome project. CHB, Han Chinese in Bejing, China; JPT, Japanese in Tokyo, Japan; CHS, Southern Han Chinese; CDX, Chinese Dai in Xishuangbanna, China; KHV, Kinh in Ho Chi Minh City, Vietnam; CEU, Utah Residents (CEPH) with Northern and Western European Ancestry; TSI, Toscani in Italia; FIN, Finnish in Finland; GBR, British in England and Scotland; IBS, Iberian Population in Spain; YRI, Yoruba in Ibadan, Nigeria; LWK, Luhya in Webuye, Kenya; GWD, Gambian in Western Divisions in the Gambia; MSL, Mende in Sierra Leone; ESN, Esan in Nigeria; ASW, Americans of African Ancestry in SW USA; ACB, African Caribbeans in Barbados; MXL, Mexican Ancestry from Los Angeles USA; PUR, Puerto Ricans from Puerto Rico; CLM, Colombians from Medellin, Colombia; PEL, Peruvians from Lima, Peru; GIH, Gujarati Indian from Houston, Texas; PJL, Punjabi from Lahore, Pakistan; BEB, Bengali from Bangladesh; STU, Sri Lankan Tamil from the UK; ITU, Indian Telugu from the UK.
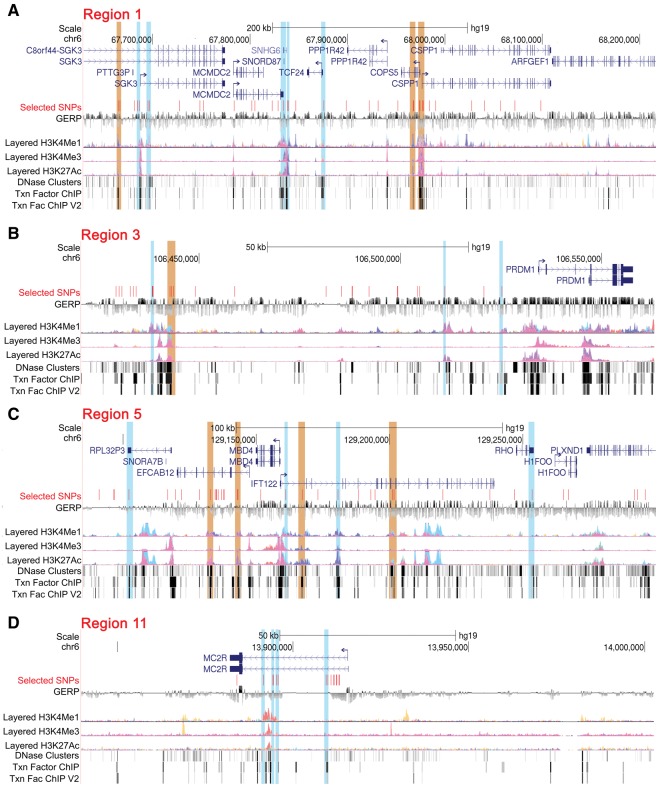
This region has a large number of genes (n = 12, see table 1) with >97% of SNVs located in nonexonic regions. Some genes are of limited relevance. For example, PTTG3P is reportedly a pseudogene, C8orf44 an open reading frame and C8orf44-SGK3 a read-through transcript that produces a protein that shares sequence identity with the downstream gene product SGK3. Both SNHG6 and SNORD87 are genes encoding small nucleolar RNA. Therefore, the plausible protein coding candidate genes includes, 1) SGK3 (serum/glucocorticoid regulated kinase family member 3), a hypoxia response gene (Hou et al. 2015; Minchenko et al. 2016), 2) TCF24 (transcription factor 24), 3) MCMDC2 (mini-chromosome maintenance domain containing 2), 4) PPP1R42 (Protein Phosphatase 1 Regulatory Subunit 42) known to be involved in meiotic recombination and centrosome activities, 5) COPS5 (also known as JAB1 or CSN5), a hypoxia-inducible factor (HIF) stabilizing gene (Bemis et al. 2004), and 6) CSPP1 (Centrosome and Spindle Pole Associated Protein 1), a candidate gene of Joubert syndrome-21 (MIM# 615636; Akizu et al. 2014). Interestingly, COPS5 was reported earlier as a candidate gene for HA adaptation in the Andean as well as Tibetan population (Bigham et al. 2009, 2010). Additionally, a large number of SNPs of the carrier haplotype overlap with known ENCODE regulatory regions that is, histone mark, transcription factor binding sites (TFBS) and DNaseI hypersensitive sites (fig. 5A). Interestingly, some of these genes such as SGK3, COPS5, and CSPP1, SNPs overlap with their promoter regions that have a large number of TFBS (>10 TFBS, orange highlighted in fig. 5A).
Fig. 5.
The SNPs from carrier haplotype of top candidate regions that are overlapping with transcriptional regulatory elements (labelled as selected SNPs) viz, histone mark (H3K4Me1, H3K4Me3, and H3K27Ac), transcription factor binding sites (TFBS), DNaseI hypersensitive sites. SNPs overlapping with >10 TFBS are highlighted in orange and those overlapping with 5–9 TFBS or histone mark are highlighted in blue. The signals in the histone-mark tracks shows the levels of enrichment of the H3K4Me1 (found near regulatory elements), H3K4Me3 (found near promoters) and H3K27Ac (found near active regulatory elements) as determined by a ChIP-seq assay (ENCODE project). SNPs (of carrier haplotype) in the promoter region of (A) SGK3, TCF24, COPS5, and CSPP1 (candidate region 1), (B) PRDM1 (candidate region 3), and (C) IFT122 overlap with large number of regulatory elements (orange and blue highlighted). In candidate region 11, the SNPs were overlapping with the regulatory region located in the intro 1 of MC2R gene (D).
Other Candidate Regions
The frequency distribution of other candidate regions and the respective genes within each region are provided in table 1 and figure 3. Briefly, ANTXRLP1 (Anthrax Toxin Receptor-Like) is a pseudogene with little known about it. PRDM1 is a single gene on the chromosome 6 candidate region. The gene is highly conserved and, in mammals, it acts as a transcriptional suppressor. It is known to play a role as a mediator of HIF-independent hypoxia response in Caenorhabditis elegans (Padmanabha et al. 2015). IFT122 (intraflagellar transport 122) on chromosome 3 encodes a 140-kDa protein and is, coincidently, expressed highly in the pituitary gland and testis (Gross et al. 2001). Although the IFT family is reported to be essential for mammalian spermiogenesis (San Agustin et al. 2015), studies on the specific role of IFT122 are lacking. Interestingly, the 5′ region’s SNPs of both PRDM1 and IFT122 overlap with a large number of TFBS (orange and blue highlighted in fig. 5B and C). In addition to these new genes, the two genomic regions on chromosome 12 that we also reported in our previous analysis (Zhou et al. 2013) were replicated in new cohorts (Cole et al. 2014) and functionally validated (Azad et al. 2016). Like PRDM1, MC2R (melanocortin 2 receptor), also known as adrenocorticotropic hormone receptor (ACTHR), is also a single gene on chromosome 18 candidate region. This region appears interesting because of (1) the stark difference in haplotype frequency, where the SNPs are overlapping with TFBS and histone mark (fig. 5D), between CMS and nonCMS and (2) the regulatory role that MC2R may play in processing the signals of hormonal changes at HA. However, a comparable haplotype frequencies of the nonCMS group with its genetically closer population from the 1000 genome project (AMR in general and particularly MXL, P > 0.05; fig. 3) would not support the hypothesis of a strong positive selection sweep in this region. Further, the lack of a Drosophila ortholog to functionally assess its role further dampens our interest in MC2R as a priority gene.
Experimental Validation in Drosophila
From the whole genome sequencing of our subjects of HA, we identified 11 candidate regions, each consisting of a few to several genes (n = 38, table 1). One way of functionally validating the role of these candidate genes in HA adaptation is to test the function of their orthologs in regulating hypoxia tolerance in Drosophila melanogaster, a model system. We have previously developed a robust hypoxia tolerance assay in flies (Materials and Methods) and successfully used this assay to validate human ortholog candidate genes (Zhou et al. 2013; Udpa et al. 2014).
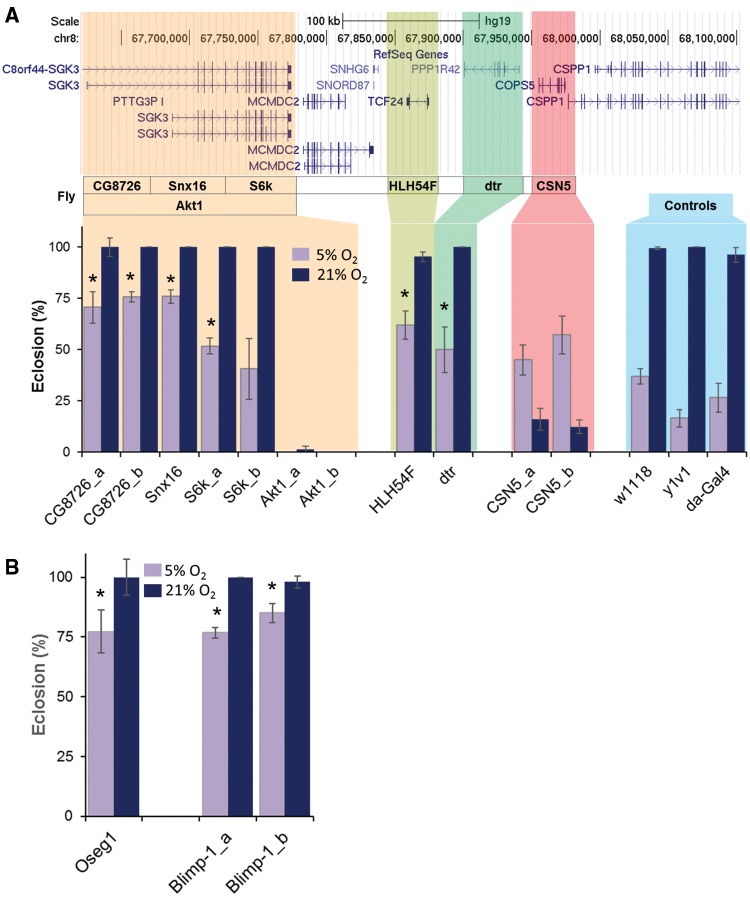
Using this approach, we first identified the fly-orthologs as mentioned in Materials and Methods (supplementary table S2, Supplementary Material online). We tested nine genes that are distributed over four candidate regions (table 2). Under normoxia, the eclosion rate was >95% for all the controls (w1118, y1v1 and da-Gal4), RNAi lines and most of the experimental crosses (UAS-RNAi × da-Gal4) except for two, Akt1 (SGK3) and CSN5 (COPS5). The Akt1-RNAi × da-Gal4 was lethal, and for CSN5-RNAi × da-Gal4 the eclosion was <25%. Both results were verified using two different RNAi lines for each gene. Under hypoxic (5% O2) environments, the eclosion rates of the background controls and all the RNAi lines were <40%. Among the experimental crosses (i.e., da-Gal4 × RNAi) targeting CG8726 and SNx16 (human ortholog C8orf44-SGK3 and SGK3) and HLH54F (human ortholog TCF24) genes, the eclosion rates were significantly higher compared with all of their corresponding controls in hypoxia (P < 0.05, fig. 6A). Similar to what we observed in normoxia, Akt1-RNAi × da-Gal4 was lethal (fig. 6A) and of the two lines tested for S6k-RNAi × da-Gal4, one was significant and the other was not. The eclosion rate for CSN5-RNAi × da-Gal4 were ∼50% in both RNAi lines (fig. 6A), which was surprisingly higher than what we observed in normoxia (fig. 6A).
Table 2.
Fly Ortholog Genes Tested for Hypoxia Tolerance. Eclosion Rate (%) of the F1 Progeny from the “RNAi-line X da-Gal4” at 5% and 21% O2.
| Candidate Region | Human Gene | Fly Symbol | Fly Line | Fly_Chr | Eclosion at 5% O2 | Eclosion at 21% O2 |
|---|---|---|---|---|---|---|
| 2 | SGK3 | S6k | 41,702 | 3L | High | High |
| 57,016 | 3L | NS | High | |||
| Akt1 | 31,701 | 3R | Lethal | Lethal | ||
| C8orf44-SGK3 | 33,615 | 3R | Lethal | Lethal | ||
| CG8726 | 35,281 | 2R | High | High | ||
| 57,230 | 2R | High | High | |||
| Snx16 | 38,992 | 2R | High | High | ||
| PPP1R42 | dtr | 25,812 | 2L | NS | High | |
| COPS5 | CSN5 | 28,732 | 3R | High | Low | |
| 42,781 | 3R | High | Low | |||
| TCF24 | HLH54F | 28,698 | 2R | High | High | |
| 4 | PRDM1 | Blimp-1 | 36,634 | 3L | High | High |
| 57,479 | 3L | High | High | |||
| 6 | IFT122 | Oseg1 | 51,904 | 3L | High | High |
| 7 | ZNF492 | CG15269 | 51,506 | 2L | NS | High |
| ZNF492, ZNF99 | crol | 41,669 | 2L | NS | High |
High, in 21% O2, “High” indicates higher eclosion rate of F1 progeny (from RNAi-line × da-Gal4) and in 5% O2, “High” indicates significantly higher eclosion rate when compared with all four controls, that is, RNAi-line and the three background controls (w1118, da-Gal4, y1v1) kept under similar condition and therefore indicates hypoxia tolerance; Lethal, embryonic lethal or early larval stage lethal; NS, not significant between F1 progeny and controls.
Fig. 6.
Down-regulation of Drosophila ortholog genes from the human candidate region enhances survival under hypoxic conditions. The UAS-RNAi lines for respective human genes (top) were used to ubiquitously knock-down candidate genes by crossing with da-Gal4 driver. The eclosion rates were calculated for 21% (normoxia) and 5% O2 environments. (A) The CG8726, Snx16, and S6k (C8orf44-SGK3 and SGK3), HLH54F (TCF24) RNAi flies when crossed with da-Gal4, significantly increased the eclosion rate in 5% O2 environment. The F1 from Akt1-RNAi × da-Gal4 were lethal both at 5% and 21% O2. The CSN5-RNAi × da-Gal4 eclosion rates were higher in 5% than 21% O2. (B) Eclosion rates of the fly orthologs for genes from two other human-candidate regions, Oseg1 (IFT122) and Blimp-1 (PRDM1), were also significantly high. The w1118, y1v1, and da-Gal4 were considered as background controls. Each bar represents mean ± SE of % eclosion rate for the F1 generation RNAi × da-Gal4. *, P < 0.05; human gene in parenthesis.
The other candidate gene that was validated in Drosophila was PRDM1 on chromosome 6 (fly ortholog Blimp-1). The hypoxia eclosion rate for Blimp-1 × da-Gal4 was significantly higher than in controls (<45% in controls vs. ∼75% in cross progeny, P < 0.05, fig. 6B). IFT122 (fly ortholog Oseg1) was another candidate gene from chromosome 3 region that passed hypoxia tolerance assay with ∼75% eclosion rate at 5% O2 (P < 0.05, fig. 6B). Although this region consisted of five genes, four were filtered out (two had no fly ortholog and two were pseudo/RNA-gene), leaving only IFT122. Finally, we had three genes on the fourth candidate region of which one was a pseudogene. The fly ortholog for both genes ZNF492 and ZNF99 was crol (crooked legs) and their eclosion rate was similar to controls in hypoxia (supplementary fig. S4, Supplementary Material online). From our previous experiences (Azad et al. 2012), the chances of detecting hypoxia tolerant genes would be <3%. The probability here is much high (>50%), a testament to the high likelihood that these genes have been hypoxia-selected by our methodologies.
Discussion
This study extends our previously used methodology of whole genome sequencing of adapted and mal-adapted individuals to identify genomic regions/genes under selective sweeps for HA adaptation (Zhou et al. 2013; Udpa et al. 2014). In previous studies, we were able to identify candidate genes using a limited sample but powered by robust selection methods. Those genes were replicated in other cohorts and also characterized in detail by us and others (Cole et al. 2014; Stobdan et al. 2015; Azad et al. 2016; Hsieh et al. 2016). In this study, we present the results of an expanded whole genome sequence analysis of CMS and nonCMS subjects and identify additional candidate regions that are under positive selection. Indeed, the larger sample size, the robust selection methods, and the use of a novel statistical test for prioritization all allowed us to uncover novel genes involved in HA adaptation. Additionally, using Drosophila as a model organism, we found that certain candidate genes, when downregulated in Drosophila, induced more tolerance to hypoxia than controls.
A highly prioritized region was on chromosome 8. This is an ideal example depicting a selective sweep of favored haplotypes in the nonCMS group, while in other populations, including CMS, the frequency was <50% of nonCMS (fig. 2). For example, a favored haplotype includes the alternate allele “T” of SNP rs116671744 and not “A” of rs16933182 as the former variant is significantly enriched only in the nonCMS while all the other populations have a very low frequency (supplementary fig. S5, Supplementary Material online). This region was reported in our previous study (Zhou et al. 2013) and was also the focus of several studies in which this region was reportedly influencing the switch from fetal to adult hemoglobin (Garner et al. 2002, 2004). This would correlate with HA inducing maturation of fetal red cells at levels higher than that under normal conditions (Risso et al. 2012). Among nonCMS, we anticipate a selective sweep of favored haplotype at this locus, which would negatively regulate hypoxia-induced augmentation of circulating red blood cells. We saw that some SNPs that were part of the carrier haplotype are in the promoter region of SGK3, TCF24, COPS5, and CSPP1 and overlap with a large number transcriptional regulatory elements (fig. 5A). When we tested each gene from this region by downregulating them individually in Drosophila, SGK3, TCF54, and COPS5 passed a phenotypic assay of hypoxia tolerance as the eclosion were higher. SGK3, a hypoxia response gene and its target molecules were reported to promote tumor angiogenesis (Hou et al. 2015; Minchenko et al. 2016). The N-terminal region of this gene contains phox homology (PX) domain, unique within the SGK family, and the knockdown of Drosophila orthologs aligning to this domain displays significant tolerance to hypoxia, for example, CG8726 and Snx16. Because of the PX domain, which usually acts as a downstream mediator of phosphatidylinositol 3-kinase (PI3K), SGK3 was recently proposed to have role in angiogenesis (Hou et al. 2015), an important hypoxia response phenotype. Interestingly, SGK3 also has estrogen receptor-binding regions and can be transcriptionally induced with estrogen (Wang et al. 2011), yet again indicating a hormonal role in CMS. Additionally, the catalytic domain of SGK3 is identical to that in the AKT kinases (Guo et al. 2016). Anticipated from its key role in development, the knockdown of Akt1 in Drosophila was lethal at early larvae stages irrespective of environment (fig. 6). Although very little is known about TCF24 (transcription factor 24), the eclosion rates for HLH54F (human ortholog TCF24) knockdown were significantly higher compared with their corresponding hypoxia controls. Both MCMDC2 (minichromosome maintenance domain containing 2) and PPP1R42 (Protein Phosphatase 1 Regulatory Subunit 42) were reported to have a role in meiotic recombination and centrosome activities. The eclosions in Drosophila for dtr (human ortholog PPP1R42) knockdown were low. COPS5 (also known as JAB1/CSN5) was another interesting gene in this region that was also reported in both Tibetan and Andean HA studies (Bigham et al. 2010). Because of its direct role in HIF-1alpha stabilization (Bae et al. 2002), this gene is anticipated to play an important role in controlling cellular oxygen sensing at HA. We found that downregulation of CSN5 was unusually deleterious in normoxia but its lethality was rescued by hypoxia. One plausible reason for this was the accumulation of COPS5 under hypoxia, despite its downregulation in RNAi-CSN5 × da-Gal4. A higher COPS5 would then enhance TGF-β signaling by sequestering Smad7 for degradation through the COP9-proteosome pathway (Kim et al. 2004), favoring a broad range of TGF-β related biological activities. However, in normoxia, the absence of COPS5-dependent ubiquitination of Smad7 would lead to a negative regulation of TGF-β signaling, hampering development. Adjacent to COPS5 gene lies another gene CSPP1.
We also discovered that Drosophila had a significantly increased hypoxia tolerance when the orthologs of PRDM1 (Blimp-1) and IFT122 (Oseg1) were downregulated. Both genes also had SNPs overlapping with regulatory regions (fig. 5B and C). PRDM1, a highly conserved transcriptional suppressor gene, was recently reported to be a mediator of HIF-independent response to hypoxia in C. elegans (Padmanabha et al. 2015). Interestingly, its expressions in myeloma cells are known to decrease under hypoxic environment (Kawano et al. 2013). Studies on the specific role of IFT122 are lacking but the IFT family in general is involved in spermiogenesis (San Agustin et al. 2015). The fact that this is highly expressed in the pituitary gland and testis, like ACTH-MC2R (Gross et al. 2001), is also intriguing. In addition to these new candidate genes, the two genomic regions on chromosome 12, consisting of SENP1 and ANP32D, which we reported in our previous analysis (Zhou et al. 2013), were also significant in this study. SENP1 is now recognized to be a major player leading to excessive erythrocytosis in CMS (Cole et al. 2014; Azad et al. 2016; Hsieh et al. 2016), giving credence to our approach. A region on chromosome 18 harboring MC2R had the haplotype frequency unusually low in the CMS group while it was comparable among nonCMS and other low altitude populations (fig. 3). The SNPs of the carrier haplotype in this region were although located in the intron 1 of MC2R gene, were overlapping there with the regulatory regions (fig. 5D). This gene may be interesting because of the similar phenotype in both knockout and hypoxia exposed murine models (Ou and Tenney 1979; Gosney 1984; Chida et al. 2007). In humans its role in erythroblast differentiation (Simamura et al. 2015) and the differential expression of its ligand (ACTH), when exposed to a HA environment (Bouissou et al. 1988; Ramirez et al. 1995; Kaur et al. 2002) are evidences which suggests a novel role of MC2R in CMS. However, we should not prioritize this gene, primarily because the “favored haplotype” is not enriched in the nonCMS group, when compared with other global populations.
Conclusions
In the present study, we have identified novel genes involved in HA adaptation in the Andes. First, our use of the PreCIOSS algorithm that provides a robust method for distinguishing between carriers and noncarriers of a favored allele. Second, both HIF-dependent and independent mechanisms are involved in HA adaptation. Third, since the overwhelming majority of SNPs are in nonexonic (and possibly regulatory) regions, we suspect that this molecular adaptation allows for more genetic flexibility, that plausibly regulates transcript abundance, adjusting with the physiological responses to environmental challenges such as hypoxia.
Materials and Methods
Study Population
All individuals were volunteers from the town of Cerro de Pasco, in Peru (Altitude >4,300 m, fig. 1A), and each subject gave informed, written consent. The UCSD institutional review board approved the protocol. Individuals were assigned a composite “CMS score” based on a list of signs and symptoms (Ward et al. 2003) evaluated by expert physicians. A CMS score of ≤12 was considered as normal, while a score of >12 were considered as CMS patients. On the basis of the new international consensus CMS scoring system (Leon-Velarde et al. 2005) a score of >5 is considered as a CMS patient (fig. 1E). Both scoring methods are equally efficient in separating CMS patients from nonCMS controls (fig. 1F). When using a new scoring system we included individuals with a score of ≥7 as CMS patients and ≤4 as nonCMS controls to make sure that the phenotypes are discrete (fig. 1E and F). Genomes sequenced for this study, denoted as “Pasco2” includes 74 subjects (CMS = 40 and nonCMS = 34). In addition to this, we also included whole-genome-sequence of 20 individuals (10 CMS and 10 nonCMS) from our previous study, denoted as “Pasco1” (Zhou et al. 2013).
Library Construction and Sequencing
Blood sample (10 ml) was collected from each subject for DNA extraction. The whole genome sequencing was carried out at HLI (Human Longevity Inc., San Diego). Next Generation Sequencing (NGS) library preparation was carried out using the TruSeq Nano DNA HT kit (Illumina Inc.), essentially following manufacturer’s recommendations. Genomes were sequenced at a mean coverage of 40.3× on the Illumina HiSeqX sequencer utilizing a 150 base paired-end single index read format.
Reads were mapped to a human reference sequence (hg38 build) using ISIS Isaac Aligner (v. 1.14.02.06) in the ISIS Analysis Software (v. 2.5.26.13; Illumina; Raczy et al. 2013). The hg38 reference sequence was modified by masking the pseudoautosomal region of chrY. Single nucleotide variations and short indels were called using the ISIS Isaac Variant Caller (v. 2.0.17) with default settings. More details could be found in Telenti et al. (2016) as the same sequencing protocol and variant calling pipeline were used.
Admixture Analysis
ADMIXTURE (Alexander et al. 2009) was used to measure the genetic affinity of our Andean CMS and nonCMS individuals (Pesco1 and Pesco2 combined) with other major populations. Besides Pasco1 and Pasco2, we used 150 EUR, 150 AFR, 64 MXL, and rest of AMR (Ad Mixed American) samples from 1000 Genomes project (www.internationalgenome.org; last accessed August 30, 2017) for phasing. We also included 100 Native American (NATAM) samples with low European and African admixture as reported in Reich et al. (2012). Pasco1, Pasco2, and NATAM samples were imputed and phased using Beagle v4.1 with the 1000 Genomes AMR populations as reference panel using default parameters. Subsequently, the samples from all sources were merged together using bcftools. Markers were merged using the rsID tags. PCA was run using plink1.9 with default parameters. PCA was first run for CMS/nonCMS populations followed by all samples except 1000 Genomes AMR populations. Markers were pruned for LD using plink. Removed each SNP that has an R2 value of >0.1 with any other SNP within a 50-SNP sliding window (advanced by 10 SNPs each time). ADMIXTURE (Alexander et al. 2009) was applied to the resulting data along with three reference populations AFR, EUR and NATAM. The output was plotted using R. Since much of the genetic diversity can be modeled using any one chromosome with sufficient markers, we did all the analysis separately for chromosome 19 (fig. 1), 20, and 21 (supplementary fig. S1, Supplementary Material online).
Whole Genome Sequence Variant Call Validation Using Chip Array
Chip array was done at the IGM Genomics Center, University of California, San Diego. Briefly, 200 ng of DNA was hybridized to Illumina Human Core arrays (Illumina) and stained per Illumina’s standard protocol. Copy Number Variation (CNV) calling was carried out in Nexus CN (version 7.5) and manually inspected, visualizing the B-allele frequencies (proportion of A and B alleles at each genotype) and log R ratios (ratio of observed to expected intensities) for each sample, as described (DeBoever et al. 2017). We found that 99.6% of chip array variants were in the WGS. Chip array includes 0.6% of WGS variants and 95% of the chip array calls were the same as WGS calls.
Tests of Selection
Instead of investigating the association of individual variants with the CMS/nonCMS classification (for which the number of individuals is not sufficient), we test the genomic region for evidence of a selective sweep. The selective sweep signal was well captured by different statistical tests, which measure either deviation from the neutral allele frequency spectrum or the frequency of carrier haplotypes. We applied seven tests in sliding windows of size 50 kbp (step size 2 kbp) along the entire genome for these individuals. Among these tests, Tajima's and Fay & Wu's are based on the allele frequency spectrum (Tajima 1989; Fay and Wu 2000), iHS is haplotype based tests (Voight et al. 2006; Ferrer-Admetlla et al. 2014), the fixation index () is a cross-population test that measures the population differentiation (Hudson et al. 1992; Zhou et al. 2013; Udpa et al. 2014), while and are two cross population tests based on common estimators of the scaled mutation rate θ = 4Nμ (Zhou et al. 2013; Udpa et al. 2014). For calculating the iHS statistics we used selscan software (Szpiech and Hernandez 2014) with default parameters, and for the rest of the selection scan tests we used the software provided earlier (Zhou et al. 2013; Udpa et al. 2014). In cross-population tests, we used two sets of case–control pairs: NonCMS versus CMS, and nonCMS versus MXL. Our previous results showed that these different tests show different power depending upon the selection coefficient, time since onset of selection, and the initial frequency of the favored allele (Ronen et al. 2013). Therefore, we considered a positive result in any of these tests as an indicator of the region adapting to selection pressure. In this study, we focused only on autosomal chromosomes, as all of our study subjects are male, and the relatively lower recombination rate in the X-chromosome (and no recombination in Chr Y) makes it harder to localize the signal.
Haplotype Phasing
A few selection detection tests such as iHS require resolved haplotypes, as also the PreCIOSS algorithm, for separating carriers of the selective sweeps from noncarriers. We generated phased haplotypes by Beagle 4.1 (Browning and Browning 2007), with 1000 Genome project phase 3 as reference panel (Genomes Project et al. 2015), no imputation (impute = false), and default parameters set for the rest of parameters.
Region Prioritization
The statistical tests (see section “Tests of selection”) of selection, in sliding windows of size 50 kbp along the entire genome, identified multiple regions that are potentially under selection sweep. Our sample size does not allow enough statistical power for multiple test correction and estimation of regions with genome-wide significance. At the same time, we planned to experimentally validate selected regions. Therefore, we used extremal analysis (Kelley et al. 2006; Akey 2009) to identify regions that were the most significant in some of the tests, and further prioritized them to get a smaller list for experimental validation. In selecting prioritization methods, we were motivated by the following criteria. 1) We wanted robust signals from selection detection tests. As CMS is a polygenic disorder, the 94 individuals presented with a range of CMS scores. We expected that the extreme range of CMS scores, here we have CMS group with CMS-score = 20.1 ± 2.8 and for nonCMS group with CMS-score = 5.5 ± 1.7, would show the strongest genomic signal. Accordingly, individuals with extreme CMS scores were included in the initial selection detection tests. Since we also wanted to use all of the information available, complete cohort was later analyzed when using PreCIOSS algorithm. 2) We wanted to exclude gap regions such as short arms, centromeres, telomeres, heterochromatin, clones, and contigs, as these regions are gene poor, and have sequence with low-complexity. These make accurate mapping difficult. 3) We wanted to prioritize regions that were close to a protein coding gene, so that Drosophila orthologs could be identified for experimental validation. 4) We had the advantage of two populations (Pasco1, and Pasco2) sampled from the same geographical location to assess the reproducibility of signal in the two sets. 5) The PreCIOSS algorithm that we developed separates all carrier haplotypes from noncarriers with high significance. While we were not powered to perform associations with individual loci, we could prioritize based on strength for association of the carrier/noncarrier status of a haplotype in the selected region against the CMS/nonCMS phenotype status of the individual. For the ancestral states of all variants, needed in PreCIOSS algorithm, we used the ancestral sequences for Homo sapiens (GRCh37/hg19), release 59 from Ensemble FTP (Paten et al. 2008).
Genome Assembly
The assembly version of the Pasco1 is GRCh37/hg19 and Pasco2 is GRCh38/hg38. We used CrossMap V0.2.5 (Zhao et al. 2014) to convert the genome assembly of Pasco2 to GRCh37/hg19. All the analysis in this paper was done in (GRCh37/hg19).
Genomic Data Used
We used the called variants of Pasco1 processed in Zhou et al. (2013) and Pasco2 as detailed above. In this paper we only focus on biallelic sites either consisting of an ancestral or a derived allele at a specific locus.
Fly Lines and Culture
We first identified the fly orthologs for the human candidate genes using DRSC Integrative Ortholog Prediction Tool (DIOPT; supplementary table S2, Supplementary Material online) and further validated these from the www.flybase.org, last accessed August 30, 2017. Out of 38 genes located in the 11 candidate regions, 20 did not have any fly orthologs. This also includes four genes encoding for noncoding RNA and one microRNA. RNAi stock lines were obtained from Bloomington Drosophila Stock Center (BDSC) at Indiana University. No RNAi lines were available at BDSC for three genes. In total, we had nine genes that were distributed over four candidate regions (supplementary table S2, Supplementary Material online). The w1118 was used as background control. To ubiquitously knock down the candidate gene in the F1 progeny the da-GAL4 driver was also obtained from BDSC. All the stock lines were raised at ∼22 °C and maintained on standard cornmeal. The UAS-RNAi × da-Gal4 crosses were considered as experimental and the RNAi line as controls. The background controls were w1118, y1v1, and da-Gal4. Both experimental and controls were first cultured in normoxia to determine whether the RNAi-mediated knockdown of each candidate gene by itself has any effect on development.
Hypoxia Tolerance
A detailed schema of the fly hypoxia tolerance assay is provided in the supplementary fig. S6, Supplementary Material online. Three to five day-old virgin females (n = 7) da-GAL4 were crossed to different UAS-RNAi lines (male, n = 6) or vice versa. Sufficient time was given (∼3 days) for the flies to mate/cross and these are referred to as “cross”. The vials were kept under ambient conditions for the flies to lay sufficient number of fertilized eggs. After 48 h, the adults were transferred to a new vial and the original vials were then transferred to a computer controlled hypoxia chamber, maintained at 5% oxygen and 12/12 h light/dark cycle (temperature ∼22 °C). The adults were discarded after 48 h from the second batch of vials and these vials were kept at ambient oxygen conditions (∼21% oxygen) to be used as “control”. The control vials were kept at ambient oxygen conditions (∼21% oxygen). After 21 days, the ratio of the empty pupae (eclosed) to the total number of pupae formed (eclosed + uneclosed) in each vial was calculated to determine the eclosion rate. Simultaneously the w1118, da-GAL4, and RNAi were “self-crossed” to be used as controls. Each set was performed in triplicates and the entire experiment was repeated twice to check for consistency. The differences in eclosion rate for the crosses at 21% and 5% oxygen were assessed using unpaired t-test and between the RNAi × daGal4 and the RNAi alone (self-crossed) were assessed using paired sample t-test. A P value of <0.05 was considered statistically significant.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Alisson Zevallos, Amilcar Tinoco, and Manuel Gasco for their help in sample collection. This work was supported by NIH grant (R01 HL127403-02) to G.G.H. T.S. was supported by the Emerald Foundation Inc. V.B. and A.A. were supported by grants from the NSF (NSF-DBI-1458557, NSF-IIS-1318386) and NIH (1R01GM114362). Collection of samples from the Cerro de Pasco (Pasco2) and Lima were supported by a Grant from Consejo Nacional de Ciencia, Tecnología e Innovación Tecnológica/Cienciactiva (CONCYTEC/Cienciactiva), Peru named “Research Circles” (Grant Number 010-2014-FONDECYT).
References
- Akey JM. 2009. Constructing genomic maps of positive selection in humans: where do we go from here? Genome Res. 195:711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akizu N, Silhavy JL, Rosti RO, Scott E, Fenstermaker AG, Schroth J, Zaki MS, Sanchez H, Gupta N, Kabra M, et al. 2014. Mutations in CSPP1 lead to classical Joubert syndrome. Am J Hum Genet. 941:80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 199:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkorta-Aranburu G, Beall CM, Witonsky DB, Gebremedhin A, Pritchard JK, Di Rienzo A.. 2012. The genetic architecture of adaptations to high altitude in Ethiopia. PLoS Genet. 812:e1003110.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad P, Zhao HW, Cabrales PJ, Ronen R, Zhou D, Poulsen O, Appenzeller O, Hsiao YH, Bafna V, Haddad GG.. 2016. Senp1 drives hypoxia-induced polycythemia via GATA1 and Bcl-xL in subjects with Monge's disease. J Exp Med. 21312:2729–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad P, Zhou D, Zarndt R, Haddad GG.. 2012. Identification of genes underlying hypoxia tolerance in Drosophila by a P-element screen. G3 (Bethesda) 210:1169–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae MK, Ahn MY, Jeong JW, Bae MH, Lee YM, Bae SK, Park JW, Kim KR, Kim KW.. 2002. Jab1 interacts directly with HIF-1alpha and regulates its stability. J Biol Chem. 2771:9–12. [DOI] [PubMed] [Google Scholar]
- Beall CM. 2014. Adaptation to high altitude: phenotypes and genotypes. Ann Rev Anthropol. 431:251. [Google Scholar]
- Beall CM, Cavalleri GL, Deng L, Elston RC, Gao Y, Knight J, Li C, Li JC, Liang Y, McCormack M, et al. 2010. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc Nat Acad Sci U S A. 10725:11459–11464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemis L, Chan DA, Finkielstein CV, Qi L, Sutphin PD, Chen X, Stenmark K, Giaccia AJ, Zundel W.. 2004. Distinct aerobic and hypoxic mechanisms of HIF-alpha regulation by CSN5. Genes Dev. 187:739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham A, Bauchet M, Pinto D, Mao X, Akey JM, Mei R, Scherer SW, Julian CG, Wilson MJ, Lopez Herraez D, et al. 2010. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 69:e1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham AW, Mao X, Mei R, Brutsaert T, Wilson MJ, Julian CG, Parra EJ, Akey JM, Moore LG, Shriver MD.. 2009. Identifying positive selection candidate loci for high-altitude adaptation in Andean populations. Hum Genomics. 42:79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouissou P, Fiet J, Guezennec CY, Pesquies PC.. 1988. Plasma adrenocorticotrophin and cortisol responses to acute hypoxia at rest and during exercise. Eur J Appl Physiol Occup Physiol. 571:110–113. [DOI] [PubMed] [Google Scholar]
- Browning SR, Browning BL.. 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 815:1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chida D, Nakagawa S, Nagai S, Sagara H, Katsumata H, Imaki T, Suzuki H, Mitani F, Ogishima T, Shimizu C, et al. 2007. Melanocortin 2 receptor is required for adrenal gland development, steroidogenesis, and neonatal gluconeogenesis. Proc Natl Acad Sci U S A. 10446:18205–18210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole AM, Petousi N, Cavalleri GL, Robbins PA.. 2014. Genetic variation in SENP1 and ANP32D as predictors of chronic mountain sickness. High Altitude Med Biol. 154:497–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBoever C, Li H, Jakubosky D, Benaglio P, Reyna J, Olson KM, Huang H, Biggs W, Sandoval E, D’Antonio M, et al. 2017. Large-scale profiling reveals the influence of genetic variation on gene expression in human induced pluripotent stem cells. Cell Stem Cell. 204:533–546 e537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu C-I.. 2000. Hitchhiking under positive Darwinian selection. Genetics 1553:1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Admetlla A, Liang M, Korneliussen T, Nielsen R.. 2014. On detecting incomplete soft or hard selective sweeps using haplotype structure. Molec Biol Evol. 315:1275–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner C, Silver N, Best S, Menzel S, Martin C, Spector TD, Thein SL.. 2004. Quantitative trait locus on chromosome 8q influences the switch from fetal to adult hemoglobin. Blood 1047:2184–2186. [DOI] [PubMed] [Google Scholar]
- Garner CP, Tatu T, Best S, Creary L, Thein SL.. 2002. Evidence of genetic interaction between the beta-globin complex and chromosome 8q in the expression of fetal hemoglobin. Am J Hum Genet. 703:793–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project C Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, et al. 2015. A global reference for human genetic variation. Nature 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosney JR. 1984. The effects of hypobaric hypoxia on the corticotroph population of the adenohypophysis of the male rat. J Pathol. 1423:163–168. [DOI] [PubMed] [Google Scholar]
- Gross C, De Baere E, Lo A, Chang W, Messiaen L.. 2001. Cloning and characterization of human WDR10, a novel gene located at 3q21 encoding a WD-repeat protein that is highly expressed in pituitary and testis. DNA Cell Biol. 201:41–52. [DOI] [PubMed] [Google Scholar]
- Guo J, Chakraborty AA, Liu P, Gan W, Zheng X, Inuzuka H, Wang B, Zhang J, Zhang L, Yuan M, et al. 2016. pVHL suppresses kinase activity of Akt in a proline-hydroxylation-dependent manner. Science 3536302:929–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou M, Lai Y, He S, He W, Shen H, Ke Z.. 2015. SGK3 (CISK) may induce tumor angiogenesis (Hypothesis). Oncol Lett. 101:23–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh MM, Callacondo D, Rojas-Camayo J, Quesada-Olarte J, Wang X, Uchida N, Maric I, Remaley AT, Leon-Velarde F, Villafuerte FC, et al. 2016. SENP1, but not fetal hemoglobin, differentiates Andean highlanders with chronic mountain sickness from healthy individuals among Andean highlanders. Exp Hematol. 446:483–490 e482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Slatkin M, Maddison WP.. 1992. Estimation of levels of gene flow from DNA sequence data. Genetics 1322:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur C, Singh J, Peng CM, Ling EA.. 2002. Upregulation of adrenocorticotrophic hormone in the corticotrophs and downregulation of surface receptors and antigens on the macrophages in the adenohypophysis following an exposure to high altitude. Neurosci Lett. 3183:125–128. [DOI] [PubMed] [Google Scholar]
- Kawano Y, Kikukawa Y, Fujiwara S, Wada N, Okuno Y, Mitsuya H, Hata H.. 2013. Hypoxia reduces CD138 expression and induces an immature and stem cell-like transcriptional program in myeloma cells. Int J Oncol. 436:1809–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley JL, Madeoy J, Calhoun JC, Swanson W, Akey JM.. 2006. Genomic signatures of positive selection in humans and the limits of outlier approaches. Genome Res. 168:980–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BC, Lee HJ, Park SH, Lee SR, Karpova TS, McNally JG, Felici A, Lee DK, Kim SJ.. 2004. Jab1/CSN5, a component of the COP9 signalosome, regulates transforming growth factor beta signaling by binding to Smad7 and promoting its degradation. Molec Cell Biol. 246:2251–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Velarde F, Maggiorini M, Reeves JT, Aldashev A, Asmus I, Bernardi L, Ge RL, Hackett P, Kobayashi T, Moore LG, et al. 2005. Consensus statement on chronic and subacute high altitude diseases. High Altitude Med Biol. 62:147–157. [DOI] [PubMed] [Google Scholar]
- Minchenko DO, Riabovol OO, Tsymbal DO, Ratushna OO, Minchenko OH.. 2016. Inhibition of IRE1 signaling affects the expression of genes encoded glucocorticoid receptor and some related factors and their hypoxic regulation in U87 glioma cells. Endocr Regul. 503:127–136. [DOI] [PubMed] [Google Scholar]
- Monge C. 1942. Life in the Andes and chronic mountain sickness. Science 952456:79–84. [DOI] [PubMed] [Google Scholar]
- Monge C, Leon-Velarde F, Arregui A.. 1989. Increasing prevalence of excessive erythrocytosis with age among healthy high-altitude miners. N Engl J Med. 32118:1271.. [DOI] [PubMed] [Google Scholar]
- Ou LC, Tenney SM.. 1979. Adrenocortical function in rats chronically exposed to high altitude. J Appl Physiol Respir Environ Exerc Physiol. 476:1185–1187. [DOI] [PubMed] [Google Scholar]
- Padmanabha D, Padilla PA, You YJ, Baker KD.. 2015. A HIF-independent mediator of transcriptional responses to oxygen deprivation in Caenorhabditis elegans. Genetics 1993:739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paten B, Herrero J, Beal K, Fitzgerald S, Birney E.. 2008. Enredo and Pecan: genome-wide mammalian consistency-based multiple alignment with paralogs. Genome Res. 1811:1814–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raczy C, Petrovski R, Saunders CT, Chorny I, Kruglyak S, Margulies EH, Chuang HY, Kallberg M, Kumar SA, Liao A, et al. 2013. Isaac: ultra-fast whole-genome secondary analysis on Illumina sequencing platforms. Bioinformatics 2916:2041–2043. [DOI] [PubMed] [Google Scholar]
- Ramirez G, Herrera R, Pineda D, Bittle PA, Rabb HA, Bercu BB.. 1995. The effects of high altitude on hypothalamic–pituitary secretory dynamics in men. Clin Endocrinol (Oxf) 431:11–18. [DOI] [PubMed] [Google Scholar]
- Reich D, Patterson N, Campbell D, Tandon A, Mazieres S, Ray N, Parra MV, Rojas W, Duque C, Mesa N, et al. 2012. Reconstructing Native American population history. Nature 4887411:370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso A, Fabbro D, Damante G, Antonutto G.. 2012. Expression of fetal hemoglobin in adult humans exposed to high altitude hypoxia. Blood Cells Mol Dis. 483:147–153. [DOI] [PubMed] [Google Scholar]
- Ronen R, Tesler G, Akbari A, Zakov S, Rosenberg NA, Bafna V.. 2015. Predicting carriers of ongoing selective sweeps without knowledge of the favored allele. PLoS Genet. 119:e1005527.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronen R, Udpa N, Halperin E, Bafna V.. 2013. Learning natural selection from the site frequency spectrum. Genetics 1951:181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, et al. 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 4497164:913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Agustin JT, Pazour GJ, Witman GB.. 2015. Intraflagellar transport is essential for mammalian spermiogenesis but is absent in mature sperm. Mol Biol Cell. 2624:4358–4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinfeldt LB, Soi S, Thompson S, Ranciaro A, Woldemeskel D, Beggs W, Lambert C, Jarvis JP, Abate D, Belay G, et al. 2012. Genetic adaptation to high altitude in the Ethiopian highlands. Genome Biol. 131:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simamura E, Arikawa T, Ikeda T, Shimada H, Shoji H, Masuta H, Nakajima Y, Otani H, Yonekura H, Hatta T.. 2015. Melanocortins contribute to sequential differentiation and enucleation of human erythroblasts via melanocortin receptors 1, 2 and 5. PLoS ONE. 104:e0123232.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonson TS, Yang Y, Huff CD, Yun H, Qin G, Witherspoon DJ, Bai Z, Lorenzo FR, Xing J, Jorde LB, et al. 2010. Genetic evidence for high-altitude adaptation in Tibet. Science 3295987:72–75. [DOI] [PubMed] [Google Scholar]
- Stobdan T, Zhou D, Ao-Ieong E, Ortiz D, Ronen R, Hartley I, Gan Z, McCulloch AD, Bafna V, Cabrales P, et al. 2015. Endothelin receptor B, a candidate gene from human studies at high altitude, improves cardiac tolerance to hypoxia in genetically engineered heterozygote mice. Proc Natl Acad Sci U S A. 11233:10425–10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpiech ZA, Hernandez RD.. 2014. Selscan: an efficient multi-threaded program to perform EHH-based scans for positive selection. Molec Biol Evol 3110:2824–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1233:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telenti A, Pierce LC, Biggs WH, di Iulio J, Wong EH, Fabani MM, Kirkness EF, Moustafa A, Shah N, Xie C, et al. 2016. Deep sequencing of 10,000 human genomes. Proc Natl Acad Sci U S A. 11342:11901–11906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udpa N, Ronen R, Zhou D, Liang J, Stobdan T, Appenzeller O, Yin Y, Du Y, Guo L, Cao R, et al. 2014. Whole genome sequencing of Ethiopian highlanders reveals conserved hypoxia tolerance genes. Genome Biol. 152:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK.. 2006. A map of recent positive selection in the human genome. PLoS Biol. 43:e72.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhou D, Phung S, Masri S, Smith D, Chen S.. 2011. SGK3 is an estrogen-inducible kinase promoting estrogen-mediated survival of breast cancer cells. Mol Endocrinol. 251:72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward MP, Milledge JS, West JB.. 2003. High altitude medicine and physiology. London: Arnold. [Google Scholar]
- Xing G, Qualls C, Huicho L, Rivera-Ch M, Stobdan T, Slessarev M, Prisman E, Ito S, Wu H, Norboo A, et al. 2008. Adaptation and mal-adaptation to ambient hypoxia; Andean, Ethiopian and Himalayan patterns. PLoS ONE. 36:e2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sun Z, Wang J, Huang H, Kocher J-P, Wang L.. 2014. CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics 307:1006–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Udpa N, Ronen R, Stobdan T, Liang J, Appenzeller O, Zhao HW, Yin Y, Du Y, Guo L, et al. 2013. Whole-genome sequencing uncovers the genetic basis of chronic mountain sickness in Andean highlanders. Am J Hum Genet. 933:452–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.