Abstract
Aging is a fundamental aspect of life, yet also one of the most confounding. In individual cells, aging results in a progressive decline which affects all organelles and reduces a cell’s ability to maintain homeostasis. Because of the interconnected nature of cellular systems, the failure of even a single organelle can have cascading effects. We are just beginning to understand the dramatic physiological changes that occur during aging. Because most aging research has focused on population dynamics, or differences between wild-type and mutant populations, single-cell behavior has been largely overlooked. An open question is whether aging cells are defined by predictable sequences of physiological changes, or whether they proceed along divergent aging trajectories defined by whichever system begins to fail first. Can aging be best characterized by a cell-cycle like model with stereotyped states all cells progress through, or a Waddington landscape with divergent trajectories? Here we present work on understanding the changing physiological states of aging cells, why it will impact systems and synthetic biologists, and how the systems community can contribute significantly to the study of aging.
“Happy families are all alike; every unhappy family is unhappy in its own way.”
-Leo Tolstoy
Introduction
In a healthy, young cell, all systems operate properly, but aging has a multitude of targets to choose from, and the failure of even a single process can result in death. This abundance of opportunities to fail can be clearly seen in the widely varying lifespans of clonal organisms. Even for isogenic populations of single-celled budding yeast that are grown in identical environments, the lifespans will vary by over an order of magnitude [1]. The same is true for genetically and environmentally identical multicellular animals [2]. The fact that a population of seemingly identical individuals can have such disparate lifespans demonstrates the complexity of the aging process. Although cells have powerful networks to maintain homeostasis, the many ways cellular systems can fail means that any collection of young cells will be far more similar than any collection of middle-aged or old cells. With increasing age, the variation between cells is likely to increase significantly, suggesting that old cells will have more divergent responses to an environmental stimulus, and that the behavior of synthetic circuits will be more varied and unpredictable. Understanding how cells proceed through different physiological states will thus aid not just our understanding of the biology of aging – but how natural and synthetic circuits behave in an aging cellular chassis.
As systems begin to fail, how do homeostatic networks compensate for these failures, and do cells proceed through different physiological states based on which systems failed first? Detailed studies of individual aging cells are, for the first time, starting to grapple with the concept of divergent aging trajectories. Critically important aspects of this are the molecular identification of distinct subsets of aging cells and the penetrance within the population of different aging phenotypes. Here we will focus on the budding yeast S. cerevisiae, as it is both one of the most widely studied aging models and extremely popular within the systems biology community. The majority of the physiological changes discussed here, however, represent hallmarks of aging that are present in widely divergent organisms [3]. Thus, it seems likely that much of what we are able to decipher about modes of failure during aging in yeast will be relevant in more complex animals and even people.
Replicative aging
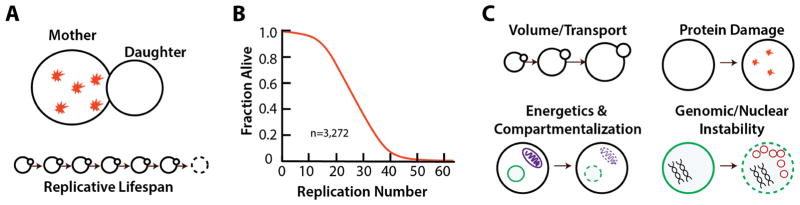
Budding yeast cells go through asymmetric divisions where the mother cell produces a newborn daughter cell [4,5]. Because this is a morphologically asymmetric division unlike fission yeast or bacteria, there is a clearly distinguished mother cell, which can be tracked as it divides (Fig 1A). During each cell cycle the mother cell retains damaged proteins, dysfunctional mitochondria and aberrant genomic material which allows daughter cells to be rejuvenated and begin life healthy [6]. As mother cells continue to divide, this accumulated damage takes a toll, and the cell cycle slows leading to senescence and death (Fig 1A). The number of daughter cells that each individual mother is able to produce before reaching senescence is defined as that cell’s replicative lifespan (Fig 1B). Although a fascinating system in its own right, studies on the replicative aging of budding yeast have uncovered many genetic modifiers of lifespan that are conserved even up through mammals [3,6–9]. Furthermore, although we know that complex systems such as stem cells undergo cell intrinsic changes during aging that affect cell behavior [10–12], there are limited in vivo models of stem cell aging. Thus, understanding the principles of how cell physiology changes and affects cellular behavior in a simple, unicellular model organism can provide valuable insight that might not otherwise be possible.
Figure 1.
Replicative aging and physiological failures. A) Budding yeast undergo asymmetric divisions in which mother cells retain the majority of damaged proteins and organelles. After a number of divisions, a mother cell will stop dividing and die. B) A replicative survival curve of budding yeast (BY4742) from the Kaeberlein lab showing the enormous variation in lifespan. Isogenic cells, grown in an identical environment, may only bud 3–4 times, or 50–60. C) Categories of physiological changes that cells undergo as they age. Cells grow continuously in volume, experience increased protein damage, organelles like the mitochondria become less effective (purple), and in the nucleus there is increasing instability with increased rDNA circles (red) and reduced silencing.
Changes and Physiological States During Aging
Aging drives a large number of changes of key physiological parameters within cells (Fig 1C). Fundamental cell properties that are reported to change with age in yeast include increasing cell size and cell division time [13,14], alkalization of cytoplasmic and organelle pH [15], increasing oxidation and oxidative damage [16,17], loss of mitochondrial respiratory capacity [18] and selective destruction of mitochondria [19], loss of mating competency [20], accumulation of damaged or misfolded proteins [17,21], fragmentation of the nucleolus [22], and increasing genomic instability particularly near the ribosomal DNA [23]. These changes are highly interlinked and have been suggested to act in a causal fashion driving successive failures in related systems. Within a single cell, any of these changes will be mediated by homeostatic networks, but could also cause failures in interacting components as the cell attempts to compensate [24]. For example, a reduction in mitochondrial membrane potential that leads to a reduction of available ATP, would in turn affect the ability of proton pumps to function and result in a change to cellular pH. Thus, within these physiological variables, a number of them are likely to be highly correlated [24,25]. A physiological aging state would therefore be defined by the specific changes in a subset of physiological parameters that occur together.
Until recently, technological limitations have prevented the study of the ways in which related systems fail within single cells [26]. Although many experiments have assessed phenotypic changes in aging cells, they have largely been performed in a cross-sectional fashion where aging cells are isolated from a population [15,18,19,21,27–33]. Thus, the relationship between these spontaneous physiological changes and lifespan has remained cloudy. Only with the development of microfluidic systems in recent years has it become possible to watch individual cells throughout their entire lifespans, and observe whether changes in one network are linked to failures in another [16,34–38].
The physiological changes that occur during aging can be broadly categorized into four groups: 1) volume and transport, 2) energetics and compartmentalization, 3) protein damage, and 4) genomic/nuclear stability (Fig 1C). The first category, volume, is one of the most distinctive changes that occurs, as budding yeast cells will grow continuously, in a near-linear volumetric fashion, over their whole life. Although regulation of cell size in budding and division has been extensively explored in young cells [39], how this volumetric growth of 2–3 fold affects cell physiology – and whether it is actively regulated or a passive by-product of aging – has yet to be explored. Simplistically, this continuous growth changes the surface to volume ratio, likely forcing cells to prioritize which transporters are in the plasma membrane and increasing the timescale for diffusion. The second category, energetics and compartmentalization, has been primarily studied with a focus on mitochondrial energetics [18,19,40] or on the breakdown of asymmetric inheritance between mothers and daughters [27,33,41–43]. Many organelles rely on energetic gradients across the membranes, and while the alkalization of the vacuole during aging has been well studied [15,18,19], age-related changes in other organelles such as the nucleus with the RAN-GTP/GDP gradient and nuclear pore function have been less well characterized [29,44–46]. The third category of changes, protein damage, encompasses all protein related failures including transcriptional regulation. Most work in aging yeast has focused on the accumulation of HSP104 aggregates as chaperones bind damaged proteins in aging cells [47–49]. Intriguingly, individual protein levels seem to become less regulated during aging as well [18,29], suggesting that there may be significant differences between the proteome of individual aged cells. In the fourth category, a significant portion of research has focused on reduced silencing resulting from chromatin changes in aging cells [30,31,50–52] and the accumulation of rDNA circles which are created as a result of double stranded breaks and homologous recombination during aging [23,53–55]. Although whether the rates of large-scale genome changes such as chromothripsis or kataegis increase during aging is of significant interest as a result of recent findings [56,57], so far no similar work has been done with yeast. Detailed studies of telomerase inactive mutants and the effects of on aging have been performed however [58].
Do Distinct Aging Trajectories Exist?
Given the delicate choreography that cells engage in with each replication, and the sheer number of physiological variables that are disrupted or change during the course of aging, it is likely that individual cells follow distinct aging trajectories, at least to some extent. Whichever chance event first occurs and undermines one of the physiological variables will shift the cell into a new state and potentially determine the primary mode of failure. If true, cells could be modeled simplistically as state machine where each state predicts transition probabilities to a subsequent, more aged physiological state.
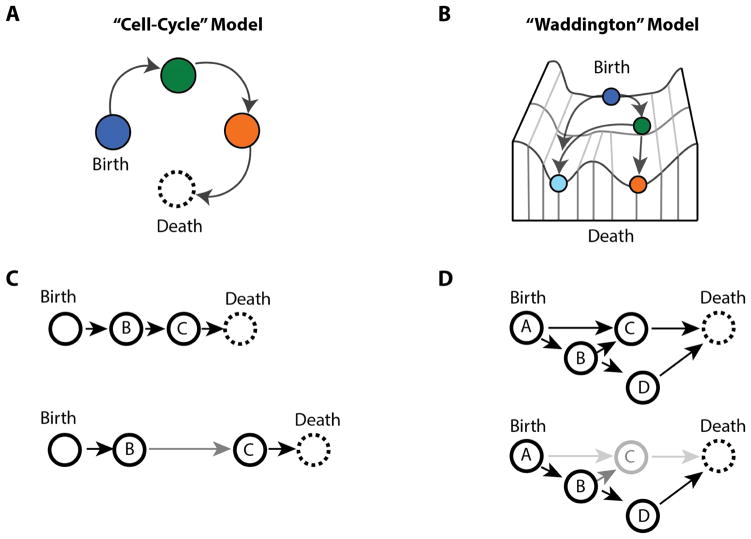
These aging states, and progression through them, could be highly stereotyped with all cells moving through the same sequence of states, but at different rates (Fig 2A). Thus, the heterogeneity we observe in lifespans and physiology in old cells would be due to cells following the same trajectory but located at different points along the trajectory. In this case, aging trajectories would be similar to the heterogeneity that arises as a result of the cell cycle. All cells follow the same path, but the cell cycle introduces a large amount of extrinsic variation in a population because cells are at different points along the stereotyped path. Alternatively, however, because entering into a specific physiological state can be caused by a chance failure event, any physiological state may only affect a fraction of cells. If a specific physiological state predisposes a cell to enter a subsequent state, then the aging progression of single cells would be more analogous to a Waddington landscape (Fig 2B). At each point in the aging trajectory, as cells enter a new physiological state, they become increasingly divergent from cells following alternative trajectories.
Figure 2.
Potential aging trajectories and systems models. A) With a “cell-cycle” model of aging, all cells move through stereotyped physiological states, but at different rates. The rate of progression through each state determines the replicative lifespan of a cell. B) In a “Waddington” model of aging, cells can move through a distinct set of physiological states that may not overlap. Both the rate of progression through states, and which physiological states an individual cell visits determines the lifespan. C) Within a “cell-cycle” model, normal cells (top) move through physiological states with certain probabilities, and interventions that increase lifespan (bottom) reduce some or all transition probabilities. D) In the “Waddington” model of aging, individual cells can proceed along different paths (top), and lifespan extending interventions might not only reduce transition probabilities, but also change which physiological states cells are likely to visit as they age (bottom).
Determining which model most accurately represents the cellular aging process will help us better understand how lifespan extending interventions function. Do genetic or environmental interventions slow aging by reducing the transition probabilities between states (Fig 2C), or do they affect both the transition probabilities and which physiological states are available to move through (Fig 2D)? If interventions remove specific physiological states from the progression, that may provide insight into how interventions can be combined to maximize lifespan. Similarly, if interventions reduce transition probabilities, do they impact all transition probabilities or only some?
Our limited understanding of aging trajectories can be seen in the surprising effect of dietary restriction (low glucose) on wild type, sir2Δ, fob1Δ, and fob1Δsir2Δ cells. Fob1 acts to create a unidirectional replication fork block in the rDNA that promotes formation of rDNA circles, and Sir2 is a histone deacetylase that enhances rDNA stability and represses formation of rDNA circles. Consistent with these functions, deletion of Sir2 shortens lifespan, while deletion of Fob1 extends lifespan, and fob1Δsir2Δ double mutant cells have a lifespan that is not significantly different from wild type cells. Interestingly, these strains behave quite differently in response to calorie restriction; sir2Δ cells receive no benefit, wild type cells experience a modest ~15% increase in lifespan, fob1Δ are extended by about ~30%, and fob1Δsir2Δ cells by about ~60% [59–61]. It was speculated that these differences could arise from the relative importance of rDNA circles (genomic instability) as a determinant of lifespan in these different genotypes, with cells lacking fob1Δ less likely to experience an aging trajectory involving genomic instability [59]. In these genotypes, we are removing or worsening a specific failure mechanism, genomic instability, and this implies that death in these populations will be determined by other failure processes (Fig 2D-bottom). Thus, if caloric restriction acts primarily on one of these alternative types of molecular failure (mitochondrial function or pH homeostasis for example), this could explain the observed differential effects on lifespan from caloric restriction across these four genotypes. More generally, if caloric restriction differentially impacts the probability that cells experience specific failures during aging, this could explain the dramatic differences in the way that different genetic backgrounds respond to caloric restriction in both yeast and mammals [62,63]. Despite being proposed more than a decade ago in yeast [59], this model has still not been experimentally tested. By helping to synthesize complex physiological phenotypes into different aging trajectories, systems biology can provide an invaluable contribution to our knowledge of the biology of aging.
Effects of Physiological States on Natural & Synthetic Networks
The competition for resources is not merely something that occurs between cells or organisms. Even within a single cell, cellular subsystems must compete for resources – whether it is for ribosomes or specific tRNAs. This has been demonstrated eloquently by those studying bacterial growth laws where competition and the distribution of resources within cells changes as resource quality or ribosome effectiveness is modified [64–67]. It has also been of concern to the synthetic biology community where the presence of exogenous circuits can impose a significant burden on the cell and alter cellular function [66,68,69]. As cells age, and enter distinct physiological states, this competition within cells and between different cellular subsystems is only likely to become exacerbated.
Simplistically, the proper function of any natural or synthetic circuit depends on a large number of factors such as the availability of ATP, diffusion rates, or whether key scaffolds are preoccupied by other networks. As aging results in increasing protein damage, altered diffusion rates due to the increasing volume or molecular crowding, or different amounts of gene expression noise from reduced genomic silencing, these physiological changes will impact the function of natural and synthetic circuits in different ways. Cells in a state defined by reduced proteasome capacity may find it challenging to produce and degrade the requisite cyclins to proceed through the cell cycle; in contrast, they may be able to maintain ATP levels and thus respond properly to environmental shocks which require the removal of excess protons by an ATPase. Broadly, how a specific synthetic or natural circuit copes with age related physiological changes will depend on whether the circuit in question is vulnerable to the specific disruption [70]. We hypothesize that some circuits will be more vulnerable to specific age-associated changes in physiology than others. As synthetic biologists engineer circuits for human use, it is important to consider how designs will be impacted by the aging human host, and whether this could result in aberrant behavior.
Concluding thoughts
In spite of the tremendous progress made over the past three decades into the genetic and molecular mechanisms that underlie aging, we still have much to learn. In particular, we have an extremely limited understanding of how individual cells age and the impact of variable cellular aging trajectories on morbidity and mortality in complex animals. The changing physiological states cells move through will have important implications for how natural and synthetic circuits behave as aging results in reduced energy, or altered diffusion rates, among other effects. Grappling with this complicated interplay between emergent systems properties should be pursued using simple model organisms that the systems community is abundantly familiar with. Coupled with the technological advances in single cell sequencing and transcriptomics, and the huge strides the systems biology community has made in deciphering single-cell timelapse trajectories, yeast aging research is primed for an influx of new ideas and discoveries in this area.
Crane Highlights.
Aging studies of cell physiology have focused on bulk, cross-sectional analysis
Interconnected systems could result in cascading failures during aging
Limited understanding of how individual cells progress through aging states
Do cells follow a single path, or does each cell have a unique trajectory
Acknowledgments
This work was supported by NIH grants T32AG000057, R01AG056359, and P30AG013280.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1**.McCormick MA, Delaney JR, Tsuchiya M, Tsuchiyama S, Shemorry A, Sim S, Chou AC-Z, Ahmed U, Carr D, Murakami CJ, et al. A Comprehensive Analysis of Replicative Lifespan in 4,698 Single-Gene Deletion Strains Uncovers Conserved Mechanisms of Aging. Cell Metab. 2015;22:895–906. doi: 10.1016/j.cmet.2015.09.008. Authors screen the yeast deletion collection using microdissection, and compare identified longevity clusters with higher organisms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mendenhall A, Crane MM, Leiser S, Sutphin G, Tedesco PM, Kaeberlein M, Johnson TE, Brent R. Environmental Canalization of Life Span and Gene Expression in Caenorhabditis elegans. J Gerontol A Biol Sci Med Sci. 2017 doi: 10.1093/gerona/glx017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steinkraus KA, Kaeberlein M, Kennedy BK. Replicative aging in yeast: the means to the end. Annu Rev Cell Dev Biol. 2008;24:29–54. doi: 10.1146/annurev.cellbio.23.090506.123509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Longo VD, Shadel GS, Kaeberlein M, Kennedy B. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab. 2012;16:18–31. doi: 10.1016/j.cmet.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaeberlein M. Lessons on longevity from budding yeast. Nature. 2010;464:513–519. doi: 10.1038/nature08981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denoth Lippuner A, Julou T, Barral Y. Budding yeast as a model organism to study the effects of age. FEMS Microbiol Rev. 2014;38:300–325. doi: 10.1111/1574-6976.12060. [DOI] [PubMed] [Google Scholar]
- 9.Wasko BM, Kaeberlein M. Yeast replicative aging: A paradigm for defining conserved longevity interventions. FEMS Yeast Res. 2014;14:148–159. doi: 10.1111/1567-1364.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossi DJ, Bryder D, Zahn JM, Ahlenius H, Sonu R, Wagers AJ, Weissman IL. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc Natl Acad Sci U S A. 2005;102:9194–9199. doi: 10.1073/pnas.0503280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossi DJ, Jamieson CHM, Weissman IL. Stems cells and the pathways to aging and cancer. Cell. 2008;132:681–696. doi: 10.1016/j.cell.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 12.Katajisto P, Dohla J, Chaffer CL, Pentinmikko N, Marjanovic N, Iqbal S, Zoncu R, Chen W, Weinberg RA, Sabatini DM. Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science. 2015;348:340–343. doi: 10.1126/science.1260384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janssens GE, Veenhoff LM. The Natural Variation in Lifespans of Single Yeast Cells Is Related to Variation in Cell Size, Ribosomal Protein, and Division Time. PLoS One. 2016;11:e0167394. doi: 10.1371/journal.pone.0167394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frenk S, Pizza G, Walker RV, Houseley J. Aging yeast gain a competitive advantage on non-optimal carbon sources. Aging Cell. 2017;16:602–604. doi: 10.1111/acel.12582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henderson KA, Hughes AL, Gottschling DE. Mother-daughter asymmetry of pH underlies aging and rejuvenation in yeast. Elife. 2014;3:e03504. doi: 10.7554/eLife.03504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie Z, Zhang Y, Zou K, Brandman O, Luo C, Ouyang Q, Li H. Molecular phenotyping of aging in single yeast cells using a novel microfluidic device. Aging Cell. 2012;11:599–606. doi: 10.1111/j.1474-9726.2012.00821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17**.Hanzén S, Vielfort K, Yang J, Roger F, Andersson V, Zamarbide-Forés S, Andersson R, Malm L, Palais G, Biteau B, et al. Lifespan Control by Redox-Dependent Recruitment of Chaperones to Misfolded Proteins. Cell. 2016;166:140–151. doi: 10.1016/j.cell.2016.05.006. A redox regulated switch recruits chaperones to correct misfolded proteins during aging. This is Independent from pathways that chaperone recruitment during heat shock stress. Demonstrate the importance of redox signaling during aging. [DOI] [PubMed] [Google Scholar]
- 18.Hughes AL, Gottschling DE. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature. 2012;492:261–265. doi: 10.1038/nature11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19*.Hughes AL, Hughes CE, Henderson KA, Yazvenko N, Gottschling DE. Selective sorting and destruction of mitochondrial membrane proteins in aged yeast. Elife. 2016;5:1–25. doi: 10.7554/eLife.13943. A novel mitochondrial degradation pathway that is activated in aging yeast cells is identified. This pathway sorts and removes a subset of mitochondrial proteins from the outer membrane and degrades them through autophagy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlissel G, Krzyzanowski MK, Caudron F, Barral Y, Rine J. Aggregation of the Whi3 protein, not loss of heterochromatin, causes sterility in old yeast cells. Science. 2017;355:1184–1187. doi: 10.1126/science.aaj2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saarikangas J, Barral Y. Protein aggregates are associated with replicative aging without compromising protein quality control. Elife. 2015;4:1–24. doi: 10.7554/eLife.06197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kennedy BK, Gotta M, Sinclair DA, Mills K, McNabb DS, Murthy M, Pak SM, Laroche T, Gasser SM, Guarente L. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae Cell. 1997;89:381–391. doi: 10.1016/s0092-8674(00)80219-6. [DOI] [PubMed] [Google Scholar]
- 23.Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 24.Gottschling DE, Nyström T. The Upsides and Downsides of Organelle Interconnectivity. Cell. 2017;169:24–34. doi: 10.1016/j.cell.2017.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dillin A, Gottschling DE, Nyström T. The good and the bad of being connected: the integrons of aging. Curr Opin Cell Biol. 2014;26:107–112. doi: 10.1016/j.ceb.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen KL, Crane MM, Kaeberlein M. Microfluidic technologies for yeast replicative lifespan studies. Mech Ageing Dev. 2016 doi: 10.1016/j.mad.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thayer NH, Leverich CK, Fitzgibbon MP, Nelson ZW, Henderson KA, Gafken PR, Hsu JJ, Gottschling DE. Identification of long-lived proteins retained in cells undergoing repeated asymmetric divisions. Proc Natl Acad Sci U S A. 2014;111:14019–14026. doi: 10.1073/pnas.1416079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindstrom DL, Leverich CK, Henderson KA, Gottschling DE. Replicative age induces mitotic recombination in the ribosomal RNA gene cluster of Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1002015. doi: 10.1371/journal.pgen.1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29*.Janssens GE, Meinema AC, González J, Wolters JC, Schmidt A, Guryev V, Bischoff R, Wit EC, Veenhoff LM, Heinemann M. Protein biogenesis machinery is a driver of replicative aging in yeast. Elife. 2015;4:e08527. doi: 10.7554/eLife.08527. Authors collect multiple cohorts of aged yeast, and perform shotgun transcriptomics and proteomics. This dataset is a valuable resource, and the authors identified important changes in stoichiometry of complexes in aging cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Z, Chen K, Xia Z, Chavez M, Pal S, Seol JH, Chen CC, Li W, Tyler JK. Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes and Development. 2014;28:396–408. doi: 10.1101/gad.233221.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, Tyler JK. Elevated histone expression promotes life span extension. Mol Cell. 2010;39:724–735. doi: 10.1016/j.molcel.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hill SM, Hao X, Grönvall J, Spikings-Nordby S, Widlund PO, Amen T, Jörhov A, Josefson R, Kaganovich D, Liu B, et al. Asymmetric Inheritance of Aggregated Proteins and Age Reset in Yeast Are Regulated by Vac17-Dependent Vacuolar Functions. Cell Rep. 2016;16:826–838. doi: 10.1016/j.celrep.2016.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J, McCormick MA, Zheng J, Xie Z, Tsuchiya M, Tsuchiyama S, El-Samad H, Ouyang Q, Kaeberlein M, Kennedy BK, et al. Systematic analysis of asymmetric partitioning of yeast proteome between mother and daughter cells reveals “aging factors” and mechanism of lifespan asymmetry. Proc Natl Acad Sci U S A. 2015;112:1506054112. doi: 10.1073/pnas.1506054112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Luo C, Zou K, Xie Z, Brandman O, Ouyang Q, Li H. Single cell analysis of yeast replicative aging using a new generation of microfluidic device. PLoS One. 2012;7:e48275. doi: 10.1371/journal.pone.0048275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crane MM, Clark IBN, Bakker E, Smith S, Swain PS. A microfluidic system for studying ageing and dynamic single-cell responses in budding yeast. PLoS One. 2014;9:e100042. doi: 10.1371/journal.pone.0100042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu P, Young TZ, Acar M. Yeast Replicator: A High-Throughput Multiplexed Microfluidics Platform for Automated Measurements of Single-Cell Aging. Cell Rep. 2015;13:634–644. doi: 10.1016/j.celrep.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee S, Avalos I, Huberts DHEW, Lee LP, Heinemann M. Whole lifespan microscopic observation of budding yeast aging through a microfluidic dissection platform. Proc Natl Acad Sci U S A. 2012;109:4–8. doi: 10.1073/pnas.1113505109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jo MC, Liu W, Gu L, Dang W, Qin L. High-throughput analysis of yeast replicative aging using a microfluidic system. Proc Natl Acad Sci U S A. 2015;112:9364–9369. doi: 10.1073/pnas.1510328112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amodeo AA, Skotheim JM. Cell-Size Control. Cold Spring Harb Perspect Biol. 2016;8:a019083. doi: 10.1101/cshperspect.a019083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fehrmann S, Paoletti C, Goulev Y, Ungureanu A, Aguilaniu H, Charvin G. Aging yeast cells undergo a sharp entry into senescence unrelated to the loss of mitochondrial membrane potential. Cell Rep. 2013;5:1589–1599. doi: 10.1016/j.celrep.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 41.Higuchi R, Vevea JD, Swayne TC, Chojnowski R, Hill V, Boldogh IR, Pon LA. Actin dynamics affect mitochondrial quality control and aging in budding yeast. Curr Biol. 2013;23:2417–2422. doi: 10.1016/j.cub.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42*.Saarikangas J, Caudron F, Prasad R, Moreno DF, Bolognesi A, Aldea M, Barral Y. Compartmentalization of ER-Bound Chaperone Confines Protein Deposit Formation to the Aging Yeast Cell. Curr Biol. 2017 doi: 10.1016/j.cub.2017.01.069. Accumulated protein aggregates in aging yeast are sequestered to the ER. The ER is anchored in the mother cell, and this process helps ensure daughter cells have a rejuvenated lifespan. [DOI] [PubMed] [Google Scholar]
- 43.Clay L, Caudron F, Denoth-Lippuner A, Boettcher B, Frei SB, Snapp EL, Barral Y. A sphingolipid-dependent diffusion barrier confines ER stress to the yeast mother cell. Elife. 2014;2014:1–23. doi: 10.7554/eLife.01883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lord CL, Timney BL, Rout MP, Wente SR. Altering nuclear pore complex function impacts longevity and mitochondrial function in S. cerevisiae J Cell Biol. 2015;208:729–744. doi: 10.1083/jcb.201412024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lord CL, Ospovat O, Wente SR. Nup100 regulates Saccharomyces cerevisiae replicative life span by mediating the nuclear export of specific tRNAs. RNA. 2017;23:365–377. doi: 10.1261/rna.057612.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou C, Slaughter BD, Unruh JR, Eldakak A, Rubinstein B, Li R. Motility and segregation of Hsp104-associated protein aggregates in budding yeast. Cell. 2011;147:1186–1196. doi: 10.1016/j.cell.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song J, Yang Q, Yang J, Larsson L, Hao X, Zhu X, Malmgren-Hill S, Cvijovic M, Fernandez-Rodriguez J, Grantham J, et al. Essential Genetic Interactors of SIR2 Required for Spatial Sequestration and Asymmetrical Inheritance of Protein Aggregates. PLoS Genet. 2014;10:17–19. doi: 10.1371/journal.pgen.1004539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hill SM, Hao X, Liu B, Nyström T. Life-span extension by a metacaspase in the yeast Saccharomyces cerevisiae. Science. 2014;344:1389–1392. doi: 10.1126/science.1252634. [DOI] [PubMed] [Google Scholar]
- 50.Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y, Jin M, O’Laughlin R, Bittihn P, Tsimring LS, Pillus L, Hasty J, Hao N. Multigenerational silencing dynamics control cell aging. Proceedings of the National Academy of Sciences. 2017 doi: 10.1073/pnas.1703379114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Defossez PA, Prusty R, Kaeberlein M, Lin SJ, Ferrigno P, Silver PA, Keil RL, Guarente L. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell. 1999;3:447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- 54.Denoth-Lippuner A, Krzyzanowski MK, Stober C, Barral Y. Role of SAGA in the asymmetric segregation of DNA circles during yeast ageing. Elife. 2014;2014:1–33. doi: 10.7554/eLife.03790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barral Y. A mechanism for asymmetric segregation of age during yeast budding. Nature. 2008;454:728–734. doi: 10.1038/nature07212. [DOI] [PubMed] [Google Scholar]
- 56.Maciejowski J, Li Y, Bosco N, Campbell PJ, De Lange T. Chromothripsis and Kataegis Induced by Telomere Crisis. Cell. 2015;163:1641–1654. doi: 10.1016/j.cell.2015.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang C-Z, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, Pellman D. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58**.Xie Z, Jay KA, Smith DL, Zhang Y, Liu Z, Zheng J, Tian R, Li H, Blackburn EH. Early telomerase inactivation accelerates aging independently of telomere length. Cell. 2015;160:928–939. doi: 10.1016/j.cell.2015.02.002. Accelerated aging in telomerase deficient mutants occurs through chronic replication stress and transient DNA damage response, an aging mechanism that is independent from telomere induced senescence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaeberlein M, Steffen KK, Hu D, Dang N, Kerr EO, Tsuchiya M, Fields S, Kennedy BK. Comment on “HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2006;312:1312. doi: 10.1126/science.1124608. author reply 1312. [DOI] [PubMed] [Google Scholar]
- 61.Tsuchiya M, Dang N, Kerr EO, Hu D, Steffen KK, Oakes JA, Kennedy BK, Kaeberlein M. Sirtuin-independent effects of nicotinamide on lifespan extension from calorie restriction in yeast. Aging Cell. 2006;5:505–514. doi: 10.1111/j.1474-9726.2006.00240.x. [DOI] [PubMed] [Google Scholar]
- 62.Schleit J, Johnson SC, Bennett CF, Simko M, Trongtham N, Castanza A, Hsieh EJ, Moller RM, Wasko BM, Delaney JR, et al. Molecular mechanisms underlying genotype-dependent responses to dietary restriction. Aging Cell. 2013;12:1050–1061. doi: 10.1111/acel.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liao C-Y, Rikke BA, Johnson TE, Diaz V, Nelson JF. Genetic variation in the murine lifespan response to dietary restriction: from life extension to life shortening. Aging Cell. 2010;9:92–95. doi: 10.1111/j.1474-9726.2009.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scott M, Mateescu EM, Zhang Z, Hwa T. Interdependence of Cell Growth Origins and Consequences. Science. 2010;330:1099–1102. doi: 10.1126/science.1192588. [DOI] [PubMed] [Google Scholar]
- 65.Scott M, Klumpp S, Mateescu EM, Hwa T. Emergence of robust growth laws from optimal regulation of ribosome synthesis. Mol Syst Biol. 2014;10:747. doi: 10.15252/msb.20145379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66*.Weiße AY, Oyarzún DA, Danos V, Swain PS. Mechanistic links between cellular trade-offs, gene expression, and growth. Proc Natl Acad Sci U S A. 2015;112:E1038–1047. doi: 10.1073/pnas.1416533112. A whole cell mechanistic model showing interaction, competition for resources, and how this affects synthetic circuit behavior in individual cells. Extension of these principles could be applied to aging cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Giordano N, Mairet F, Gouzé JL, Geiselmann J, de Jong H. Dynamical Allocation of Cellular Resources as an Optimal Control Problem: Novel Insights into Microbial Growth Strategies. PLoS Comput Biol. 2016;12:1–28. doi: 10.1371/journal.pcbi.1004802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ceroni F, Algar R, Stan G-B, Ellis T. Quantifying cellular capacity identifies gene expression designs with reduced burden. Nat Methods. 2015;12:415–418. doi: 10.1038/nmeth.3339. [DOI] [PubMed] [Google Scholar]
- 69.Brophy JAN, Voigt CA. Principles of genetic circuit design. Nat Methods. 2014;11:508–520. doi: 10.1038/nmeth.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khammash M. An engineering viewpoint on biological robustness. BMC Biol. 2016;14:22. doi: 10.1186/s12915-016-0241-x. [DOI] [PMC free article] [PubMed] [Google Scholar]